-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
Although a combination of genomic and epigenetic alterations are implicated in the multistep transformation of normal squamous esophageal epithelium to Barrett esophagus, dysplasia, and adenocarcinoma, the combinatorial effect of these changes is unknown. By integrating genome-wide DNA methylation, copy number, and transcriptomic datasets obtained from endoscopic biopsies of neoplastic progression within the same individual, we are uniquely able to define the molecular events associated progression of Barrett esophagus. We find that the previously reported global hypomethylation phenomenon in cancer has its origins at the earliest stages of epithelial carcinogenesis. Promoter hypomethylation synergizes with gene amplification and leads to significant upregulation of a chr4q21 chemokine cluster and other transcripts during Barrett neoplasia. In contrast, gene-specific hypermethylation is observed at a restricted number of loci and, in combination with hemi-allelic deletions, leads to downregulatation of selected transcripts during multistep progression. We also observe that epigenetic regulation during epithelial carcinogenesis is not restricted to traditionally defined “CpG islands,” but may also occur through a mechanism of differential methylation outside of these regions. Finally, validation of novel upregulated targets (CXCL1 and 3, GATA6, and DMBT1) in a larger independent panel of samples confirms the utility of integrative analysis in cancer biomarker discovery.
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1001356
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001356Summary
Although a combination of genomic and epigenetic alterations are implicated in the multistep transformation of normal squamous esophageal epithelium to Barrett esophagus, dysplasia, and adenocarcinoma, the combinatorial effect of these changes is unknown. By integrating genome-wide DNA methylation, copy number, and transcriptomic datasets obtained from endoscopic biopsies of neoplastic progression within the same individual, we are uniquely able to define the molecular events associated progression of Barrett esophagus. We find that the previously reported global hypomethylation phenomenon in cancer has its origins at the earliest stages of epithelial carcinogenesis. Promoter hypomethylation synergizes with gene amplification and leads to significant upregulation of a chr4q21 chemokine cluster and other transcripts during Barrett neoplasia. In contrast, gene-specific hypermethylation is observed at a restricted number of loci and, in combination with hemi-allelic deletions, leads to downregulatation of selected transcripts during multistep progression. We also observe that epigenetic regulation during epithelial carcinogenesis is not restricted to traditionally defined “CpG islands,” but may also occur through a mechanism of differential methylation outside of these regions. Finally, validation of novel upregulated targets (CXCL1 and 3, GATA6, and DMBT1) in a larger independent panel of samples confirms the utility of integrative analysis in cancer biomarker discovery.
Introduction
The incidence of esophageal adenocarcinoma (EAC) is increasing at an alarming pace in the United States (>600% increase since 1975) [1]. Since most patients with EAC present at diagnosis with an advanced disease stage, the 5-year survival rate is a dismal 13% [2], underscoring the pressing need for early diagnostic biomarkers, as well as for improved therapeutic strategies, in this malignancy. Distinct pathological stages of specialized columnar epithelium (Barrett metaplasia) and low - followed by high-grade dysplasia precede adenocarcinoma [3], [4]. Barrett esophagus (BE) is defined as a change in the esophageal epithelium that can be recognized grossly by a distinct salmon pink color at endoscopy, and confirmed by the presence of specialized columnar epithelium on biopsy. The prevalence of BE is not precisely known, but it has been estimated to range between 1–10% of the general population [5]. The incidence of EAC in patients with BE is increased 100-fold above that of the general population [6]. Thus, BE, with or without associated epithelial dysplasia, provides a unique opportunity for risk stratification and secondary prevention of EAC.
Expression profiling of BE and EAC using genome-wide approaches has identified many of the transcriptomic alterations occurring during esophageal neoplastic progression [7]-[10]. In several instances, it has also been possible to identify the proximate genomic or epigenetic mechanism (for example, intragenic deletion, truncating mutation, copy number aberration, or promoter methylation, respectively) contributing to the altered expression [11]-[16]. This strategy has unequivocally yielded a rich seedbed of candidate biomarkers for diagnosis, as well as for prognostication, of BE neoplasia [17]-[20]. Nonetheless, there remains a notable lacuna in globally integrating transcriptomic abnormalities during Barrett progression with the corresponding changes occurring at the level of the genome and epigenome. We reasoned that a multi-platform integrated approach would not only enable the elucidation of novel biomarkers, but also clarify the genomic and/or epigenetic mechanisms driving transcript abnormalities during carcinogenesis. Such integration of global datasets has begun to emerge in solid tumors [21], [22], but to the best of our knowledge this has not been performed in precursor lesions, especially in the context of multistep progression occurring in a single individual. Herein, we provide an unbiased and comprehensive approach for integrating large-scale genomic, epigenetic, and transcriptomic datasets, obtained using tissue from patients undergoing endoscopic mucosal biopsy for BE. In contrast to numerous prior studies using two platforms (for example, combined copy number and expression analysis) where a “hit” on the second allele is typically inferred, the multi-platform analysis performed here can directly confirm or refute the Knudsonian paradigm for a given altered transcript. Our studies have identified striking epigenomic alterations, and in specific, widespread hypomethylation, which occurs at the earliest stages of epithelial carcinogenesis. This approach is in direct contrast to most single - or limited-locus studies that have focused on epigenetic silencing of candidate tumor suppressor genes by hypermethylation of the promoter. In addition, we have identified clustered transcripts, such as the chemokine ligands CXCL1 and 3, that are markedly upregulated via simultaneous biallelic alteration by hypomethylation and gene amplification, respectively, and whose protein products have the potential to serve as serum biomarkers of neoplastic progression in BE.
Results
Widespread hypomethylation is an early event in the progression of Barrett esophagus
Eight histologically validated endoscopic mucosal biopsies representing normal squamous mucosa and the various histopathological stages of Barrett esophagus progression were obtained from three patients undergoing repeat endoscopy for dysplasia (Figure 1A, Figure S1); two additional unmatched gastric cardia biopsies were obtained as controls for the intended profiling studies. Prospective as well as retrospective surveillance studies have convincingly established that nondysplastic BE and low-grade dysplasia (LGD) both have a significantly lower risk of progression to EAC, when compared to high-grade dysplasia (HGD); in fact, for the purposes of therapeutic decision-making between continued surveillance versus local ablation, stratification typically occurs based on the diagnosis of HGD in BE mucosa. Therefore, with the objective of pair-wise comparison, samples of non-dysplastic BE and LGD were categorized as “low”, while the two HGD samples and one EAC were categorized as “high”. Nucleic acids extracted from cryostat-embedded sections of the ten samples were utilized for three concurrent microarray-based assays: 1) gene expression profiling, 2) array comparative genomic hybridization (aCGH), and 3) genome-wide cytosine methylation, thus simultaneously querying the transcriptome, genome, and epigenome, respectively. Methylation profiling was performed using the HpaII tiny fragment Enrichment by Ligation-mediated PCR (HELP) assay, which compares HpaII (methylation-sensitive) and MspI (methylation insensitive) genomic representations to identify hypo - and hypermethylated loci in the genome.[23]-[25]. The HELP HpaII / MspI ratios were validated at over 60 independent loci by mass spectrometry-based high-throughput quantitative methylation PCR analysis (Sequenom EpiTYPER); based on this quantitative approach, a HpaII/MspI ratio of 0.3 corresponded to 50% cytosine methylation (Figure S2) and was used as a threshold for defining hypo - or hypermethylation.
Fig. 1. Global hypomethylation is seen early during esophageal carcinogenesis. 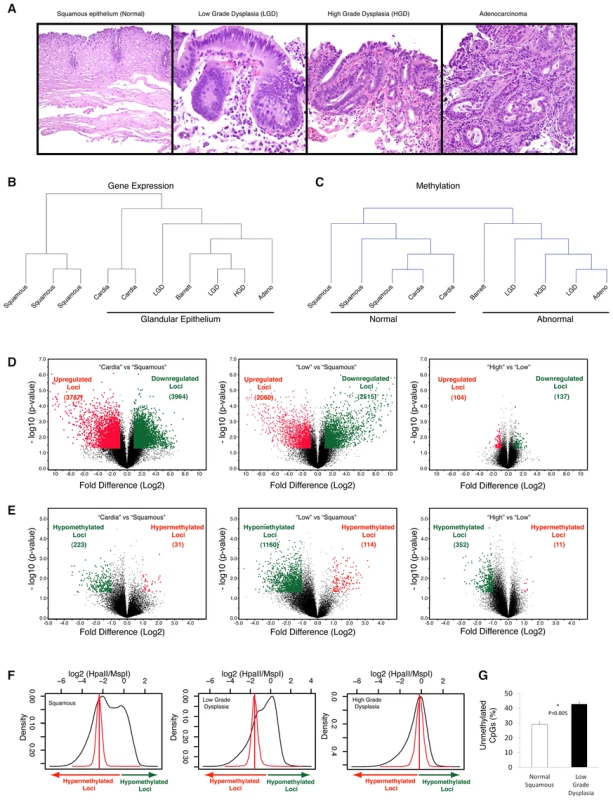
Multiple endoscopic biopsies collected at the same time from patients with clinical history of Barrett's esophagus, were classified as normal squamous epithelium, Barrett's metaplasia, low grade dysplasia (LGD), high grade dysplasia (HGD) and adenocracinoma based on histology. (Panel A). Frozen tissue samples were homogenized, used for DNA and RNA isolation and were subjected to global expression profiling, DNA methylation profiling and DNA copy number analysis. Unsupervised hierarchical clustering based on gene expression profiling shows clustering of distinct histologic tissues with glandular tissue from stomach cardia being more similar to metaplastic and dysplastic tissues (Panel B). Clustering based on methylation shows dissimilarity between normal (esophageal and stomach) and abnormal tissues (Panel C). Volcano plots demonstrating significance and magnitude of change in mean gene expression (Panel D) and DNA methylation (Panel E) are shown. Each plot represents either 36,847 genes from the microarray experiment (D) or 25,626 HpaII-amplifiable fragments (HAF) from the HELP assay (E). The log2 ratio of difference of means is shown on the x-axis and the negative log of the p-value (based on paired T-Test) is shown on the y-axis. Genes that are significantly downregulated (panel D) or hypermethylated (panel E) are shown in green and red dots indicate genes that are either upregulated (panel D) or hypomethylated (panel E). Global methylation is shown by density plots of log2 of HpaII/MspI ratios and shows bimodal distribution in normal epithelial tissues with both hypo and hypermethylated loci. The red line denotes the median HpaII/MspI ratio for random probes. A shift towards hypomethylation is observed during transformation of normal to low grade dysplasia in a representative series of samples (Panel F). Quantitative methylation for matched pairs of normal and Barrett's dysplasia as done by the LUMA assay. X axis shows mean percent hypomethylation of 3 independent paired samples. (Panel G). Unsupervised hierarchical clustering analyses demonstrated that, at the level of the transcriptome, squamous mucosa clustered discretely from “glandular” epithelium (including gastric cardiac as well as all stages of BE progression: Figure 1B); in contrast, at the level of the epigenome, “normal” mucosa (including both squamous and cardiac subtypes) clustered discretely from all “abnormal” (i.e., BE) epithelia (Figure 1C). These results suggest some degree of commonality of epigenetic profiles between otherwise normal gastrointestinal tissues, despite obvious morphological differences. A pair-wise comparison of transcriptomic profiles between normal esophageal squamous and gastric cardiac mucosal samples revealed large numbers of significantly differentially expressed transcripts, consistent with the distinct histogenesis and biologies of these normal mucosal subtypes (Figure 1D, left); in contrast, global cytosine methylation profiles between the two mucosal locations were considerably more overlapping, with significant differences in either hypo - or hypermethylation restricted to fewer loci (Figure 1E, left).
In pair-wise comparisons of gene expression during BE progression, we found large numbers of significantly differentially expressed transcripts between the early lesions of Barrett metaplasia and LGD (both classified as “Low”) versus normal squamous mucosa, confirming the previous observation [7] that even non-dysplastic Barrett epithelium may harbor profound transcriptomic aberrations, some comparable to EAC (Figure 1D, middle). Significant gene expression differences between “high and “low” BE lesions were more attenuated and restricted to only a handful of loci (Figure 1D, right). While these gene expression data were confirmatory of published results, methylation profiling revealed an unexpected dimension to epigenetic dysregulation during BE progression. Contrary to the hypermethylation reported in previous single-locus studies, we identified significant hypomethylation occurring at a large number of loci genome-wide during the transition of squamous mucosa to Barrett epithelium (1160 hypomethylated versus 114 hypermethylated loci., Figure 1E, middle,); since this epigenetic “shift” is not observed in the comparison of normal esophageal squamous versus cardiac mucosal samples, we believe these methylation alterations may be reflective of the actual BE disease process, rather than simply due to acquisition of columnar histology. A second, smaller wave of hypomethylation was observed when comparing “high” versus “low” BE categories (Figure 1E, right). This progressive hypomethylation was seen in methylation profiles of samples from the same patients (Figure 1F). Validation at the whole-genome level by the Luminometric methylation assay (LUMA) revealed a significantly large increase in unmethylated CpGs in “low” BE samples versus matched normal squamous mucosa (Figure 1G). These results demonstrate that the previously reported global hypomethylation observed in human cancers [26] can initiate at a very early stage of neoplastic transformation, such as in the non-invasive precursors of EAC.
Differential methylation within gene promoters is not restricted to CpG islands but also involves CG dinucleotides outside of these islands
In addition to this panoramic view of epigenetic shifts, we also assessed the nature of the HpaII sites showing altered methylation during BE progression. As our microarray design includes both canonical CpG islands and additional CG dinucleotide loci within gene promoters, we sought to test whether CpG islands, long considered the principal target of epigenetic dysregulation in cancer [27], were disproportionately affected. We compared the proportions of loci at which differential methylation was occurring within CpG islands versus other CG dinucleotide loci represented on the microarray. We determined that the majority of HpaII loci exhibiting differential methylation during BE progression lay, paradoxically, outside of canonical CpG islands (Figure 2A). To further validate this observation at a representative locus, we chose the example of Deleted in Malignant Brain Tumor 1 (DMBT1), whose gene promoter lacks a defined CpG island, yet demonstrates progressive hypomethylation accompanied by significant transcript upregulation during BE progression (Figure 2B). Histological examination of DMBT1 protein expression in a large archival cohort of 120 BE samples and 54 normal controls revealed significant (P<0.05) upregulation of this protein early during BE neoplasia (Figure 2C, 2D), thus validating the results obtained from our array-based analysis. These results suggest an epigenetic regulatory function for CG dinucleotide elements in the genome that do not meet the threshold for canonical CpG islands, which are strictly defined on base compositional criteria [28].
Fig. 2. DMBT1 upregulation by non-CpG island methylation changes. 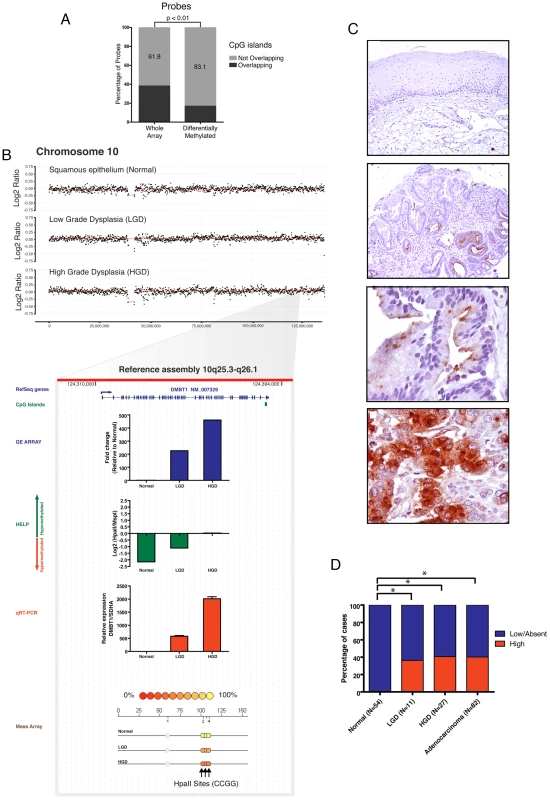
Differentially methylated loci between normal esophageal epithelium and Barretts/LGD were mapped to genomic locations of CpG islands. A large percentage of these loci were not within CpG islands. Paired T Test was used to compare to distribution of loci present on the HELP array (A). The DMBT1 gene is not amplified (panel B) and lacks a CpG island in its promoter. DMBT1 was seen to be significantly upregulated and hypomethylated during integrative analysis and marked upregulation was validated by qRT-PCR (red bar). Significant hypomethylation was seen by the HELP assay (green bar) and validated by quantitative MassArray (B). Immunohistochemical evaluation of the protein levels of the DMBT1 gene on primary tissues shows a frequent and significant protein upregulation during early stage dysplasia (panel C,D). Integrated multi-platform analysis provides experimental evidence of silencing of tumor suppressor genes by both genetic and epigenetic mechanisms in concordance with Knudson's two hit hypothesis
Subsequently, in order to generate a multi-component genetic model of BE progression, we developed an in silico algorithm for integrating data from these three high-resolution platforms (i.e., gene expression, HELP, and aCGH), using genomic coordinates for the respective probes from each of the platform arrays (Figure 3A). This algorithm, which we call Multi-dimensional Integration of Genomic data from Human Tissues (MIGHT), provides a composite three-dimensional graphical output of gene sets demonstrating significant alterations in pair-wise unbiased comparisons across the different array platforms (Figure 3B, 3C). By integrating differences in transcript expression with both methylation status and copy number at a given locus, the MIGHT algorithm not only identifies significant transcriptomic alterations during neoplastic progression, but also elucidates the relative contributions of genomic and epigenetic factors toward such deregulation. The advantage of an integrative approach in developing an accurate “patient-specific” multi-component genetic progression model is illustrated in Figure 4. A chromosome 9p21 hemizygous deletion was identified by aCGH analysis of LGD and HGD biopsies obtained from this individual, which was absent in the matched normal esophageal squamous epithelium, consistent with a somatic monoallelic loss, as confirmed by FISH analysis (Figure 4B). This region harbors two closely approximated tumor suppressor genes: CDKN2A/p16 and CDKN2B/p15. Prior copy number and other studies have implicated CDKN2A as the target of inactivation at this locus in BE [15], [17], [29] Nevertheless, in this particular example, microarray data demonstrated that the relative fold reduction in gene expression was considerably greater for CDKN2B (∼100-fold downregulation) than for CDKN2A (∼4-fold); moreover, this finding was independently validated by qRT-PCR, which confirmed the complete absence of CDKN2B transcripts in the dysplastic biopsy samples, while the expression of CDKN2A, albeit significantly reduced, was still detectable. Analysis of the third component (HELP analysis) clarified that the CDKN2B promoter in the retained allele underwent progressive hypermethylation during BE progression, while the CDKN2A promoter maintained its methylation status quo (Figure 4A, bottom). This finding suggests the importance of methylation of the remaining allele in regulating expression, and provides direct experimental evidence for genetic and epigenetic hits acting concurrently and synergistically to downregulate tumor suppressor genes during oncogenesis through bi-allelic inactivation (Figure S3).
Fig. 3. Global integrative analysis of transcriptomic, epigenomic, and DNA copy number data. 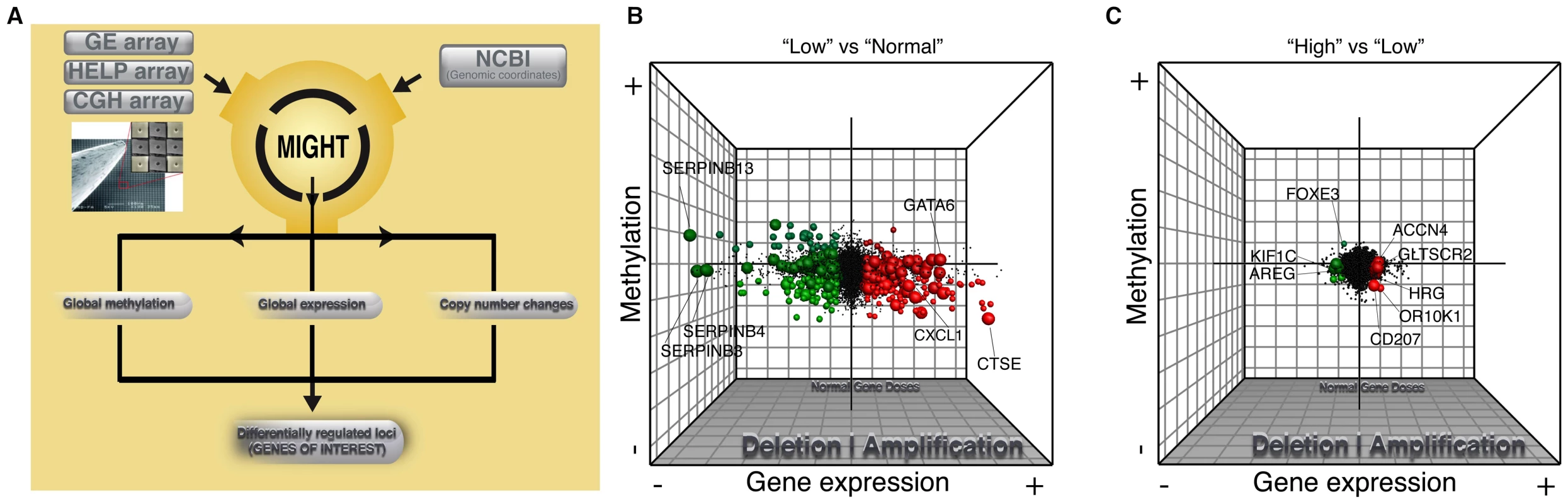
Probes from all three arrays were linked to genes on the basis of genomic coordinates isolated from NCBI build35. Pairwise analysis was performed between main histologic groups of normal squamous vs LOW (B) and LOW vs HIGH (C). The x axis represents fold-change differences (log2) in gene expression, the y axis represents fold-change differences (log2) in methylation and the z axis represents the significance of these changes (–log10 p-value) for both platforms. Upregulated genes (>2.5 fold change in GE with a p-value<0.05) are shown in green and downregulated genes are shown in red. Darker shading of these colors is used to represent genes that are significantly differentially methylated. Hypermethylated genes are at the top of the graph and hypomethylated genes on the bottom. Finally, copy number changes are defined by “volume”. Genes with copy number changes (amplified or deleted) are depicted by larger sized circles. This plot allows us to visualize differentially regulated genes and the mechanisms of their dysregulation. Fig. 4. Differential inactivation of the CDKN2A/CDKN2B locus by combination of genomic and epigenetic mechanisms. 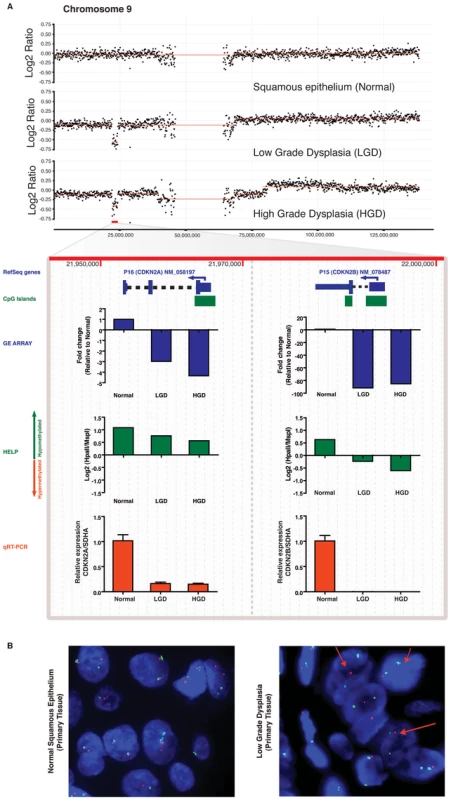
In an index patient with serial progression biopsies, there is a hemizygous deletion of the CDKN2A/CDKN2B gene locus at 9p21, beginning at the stage of LGD, and persisting into the corresponding HGD sample (Panel A), which is accompanied by expected downregulation of both transcripts in expression microarray data. Although independent qRT-PCR for CDKN2A and CDKN2B transcripts (red bars) validates the expression array data (blue bars), the magnitude of downregulation of CDKN2B is more pronounced than CDKN2A (∼100-fold versus ∼5-fold on microarray, and complete absence of CDKNA2B transcripts on qRT-PCR versus downregulated but detectable CDKN2A transcripts). MIGHT analysis confirms that this discrepancy between closely spaced hemizygously deleted loci correlates with increased promoter methylation of the residual CDKNA2B allele, as demonstrated by the HELP assay (green bars); such progressive hypomethylation is not observed on the residual CDKN2A allele and likely is responsible for the reduced but demonstrable transcripts. Uniallelic deletion of the CDKN2A/CDKN2B locus is validated by FISH (Panel B) on primary tissue as shown by loss on the LSI p16 SpectrumOrange probe (red arrows). Integrated multi-platform analysis as a tool for biomarker discovery in multistep carcinogenesis
Finally, in addition to the value of integrated datasets in understanding mechanisms of transcript disruption (as illustrated above), we explored the utility of the MIGHT algorithm as a tool for biomarker discovery in Barrett progression. In particular, we focused on genes that were significantly overexpressed in pair-wise comparisons, by hypomethylation and / or genomic amplification, and validated selected examples of patient-specific aberrations in larger sample sets.
Using the MIGHT platform, we determined that many genes not previously implicated in esophageal carcinogenesis were significantly upregulated during stepwise progression to cancer (Figure 2 and Tables S1, S2, S3, S4). These were upregulated by either loss of methylation, or gene amplification, or both occurring together. Genes previously known to be important during metaplastic transformation of squamous to columnar epithelium were also included in the list of most significantly upregulated transcripts (for example, villin and mucin genes), confirming the biological validity of our assays. Transcripts corresponding to a family of chemokine ligands were among the most significantly upregulated, and the corresponding gene cluster is present on the 4q21 chromosomal segment that was amplified in all patient samples during the process of Barrett neoplasia (Figure 5A, 5B) Integrative analysis revealed that in addition to amplification, the CXCL1 and CXCL3 gene promoters were also hypomethylated during transformation, and their relative increase in transcript expression was many fold greater (4–6 fold by array, 10–20 fold by qRT-PCR) than that of IL-8, a chemokine gene which was part of the 4q21 amplicon but whose promoter was not affected by loss of methylation (Figure 5A, Bottom). These observations were validated by qRT-PCR in larger independent set of primary samples (Figure 5C), and confirmed a greater than 30-fold mean increase in CXCL1 and CXCL3 levels when compared to IL-8 transcripts at each histological grade of BE progression, thus demonstrating the combinatorial affect of both genetic and epigenetic alterations on dysregulation of gene expression during carcinogenesis.
Fig. 5. Aberrant expression of a 4q21 chemokine cluster by gene amplification and promoter hypomethylation during esophageal carcinogenesis. 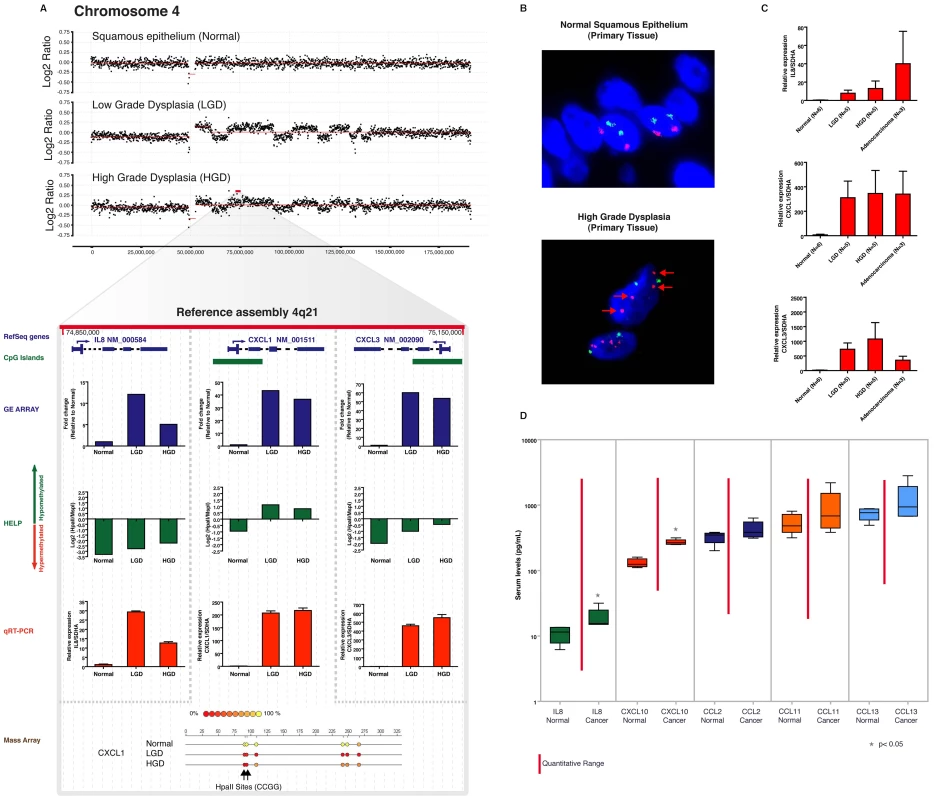
In an index patient with serial progression biopsies, amplification of chromosome 4q21, containing a cluster of genes transcribing chemokine ligands, is seen beginning at LGD, and persisting into HGD (Panel A). Expression and promoter methylation data for three representative genes in this amplicon (CXCL1, CXCL3, and IL8) are shown below the copy number window. All three transcripts are upregulated in the LGD and HGD samples by microarray data (blue bars), which is validated using independent qRT-PCR assays (red bars). Nonetheless, there is strikingly higher upregulation of CXCL1 (>200-fold on qRT-PCR compared to matched normal) and CXCL3 (500–700 fold on qRT-PCR compared to normal), versus the 10–30 fold upregulation in IL8 mRNA. Integrative analysis on the MIGHT platform confirms that this is likely arises due to progressive hypomethylation of the CXCL1 and CXCL3 promoters (as gauged by HELP assay, green bars), which is not seen at the IL8 promoter. The hypomethylation of CXCL1 observed in the dysplastic samples by the HELP assay is validated by Mass Array at the bottom. The 4q21 chemokine cluster amplification is also confirmed by FISH (Panel B) on the corresponding archival HGD sample from the same individual used in the integrative analysis. The RP11-94K4 SpectrumOrange probe representing the 4q21.1 - 4q21.2 amplicon (red arrows) was used. Moreover, the upregulation of IL8, CXCL1, and CXCL3 transcripts is also confirmed in an independent panel of endoscopic mucosal biopsy samples, including LGD (N = 5), HGD (N = 5), and adenocarcinoma (N = 3) (Panel C). Notably, the significantly greater upregulation of CXCL1 and CXCL3 transcripts observed in the index patient persists in this independent cohort of cases. Finally, chemokine detection using a multiplex chemiluminiscent assay was performed on five serum samples from patients with esophageal adenocarcinoma (designated “Cancer”) and compared to samples from controls with GERD symptoms and normal esophageal histology (designated “Normal”) (Panel D). Serum levels of only two chemokines, IL-8 and CXCL-10, both encoded by the 4q21 cluster, are significantly higher in cancer patients when compared to normals, supporting the evidence that members encoded by this amplicon are expressed at higher levels when compared to chemokines that are not part of the amplicon, such as CCL2, CCL11 and CCL13. Unfortunately, CXCL1 and CXCL3 were not included in this pre-formed multiplex assay. In light of the significant upregulation of chromosome 4q21 chemokine cluster transcripts in BE and EAC, and the likely secretion of their protein products into the circulation, we evaluated the potential of using this chemokine familyas serum biomarkers of EAC. Serum samples were collected from an independent cohort of patients with EAC and were compared to samples from patients with gastroesophageal reflux disease (GERD) symptoms without demonstrable BE on histology. Levels of chemokine ligands, IL-8 (IL8), IP-10 (CXCL10), Eotaxin (CCL11), MCP-1 (CCL2) and MCP-4 (CCL13) were determined in serum samples by a multiplexed assay on an ultra-sensitive chemiluminiscence detection platform. We observed that both chemokines that were a part of the amplified 4q21 segment (IL-8 and CXCL10) were significantly elevated in the EAC serum samples compared to the controls(P<0.05), while none of chemokines outside of this amplicon demonstrated a significant difference between cancer and control specimens (Figure 5D). These results validate the feasibility of identifying candidate biomarkers by integrative analysis in a limited number of patient samples, and extrapolating their utilization to larger, independent patient cohorts.
To further functionally validate the utility of our integrative discovery platform, we focused on a transcription factor, GATA6, which was significantly overexpressed early in Barrett metaplasia, and was predicted by MIGHT to be amplified without concomitant alterations in methylation (Figure 3). Assessment of the aCGH data and FISH on primary tissues readily validated the copy number alterations at the GATA6 locus during esophageal carcinogenesis (Figure 6A, 6B). Thereafter, using independent cohorts of snap-frozen and archival BE samples, respectively, we observed significant upregulation of GATA6 transcript expression (>500 fold mean increase in LGD, HGD and EAC samples, Figure 6C, 6D) and of the Gata6 protein levels (No Gata6 staining observed in 54 normal controls when compared to mean 60% positivity in a total of 201 LGD, HGD and EAC samples; Figure 6F–6I), confirming the results from the genomic analysis. To validate an oncogenic role in esophageal carcinogenesis, we used an esophageal adenocarcinoma-derived cell line, OE33 [30] and observed significant overexpression of Gata6 protein in these cells (Figure 6J). GATA6 was successfully knocked down using lentiviral short hairpin RNAs in OE33 cells (Figure 6K). Loss of Gata6 function did not affect proliferation, (Figure 6L) but resulted in significantly decreased anchorage independent growth of OE33 cells and also led to decrease in invasion and migration (Figure 6M–6P), thus providing a putative functional association between GATA6 amplification and disease progression in BE.
Fig. 6. GATA6 is amplified and functions as an oncogene in esophageal adenocarcinoma. 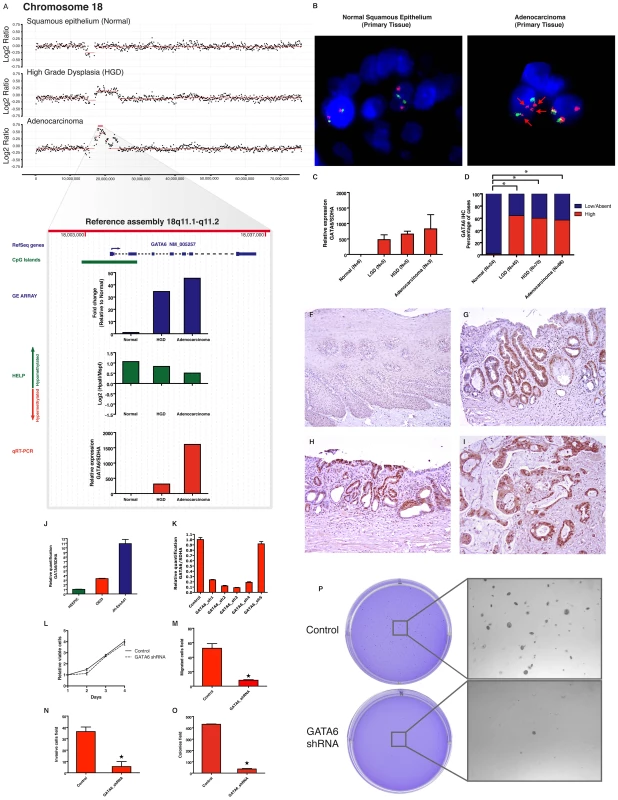
In an index patient with serial progression biopsies, aCGH shows a progressive copy number gain on the region 18q11.1-q11.2 from normal to dysplasia to cancer. (Panel A). Integrative approach demonstrates GATA6 and LAMA3 as two genes contained on this cluster (from 17–23,000,000 bp windows) with the marked upregulation during transformation of normal to dysplasia as well as from dysplasia to cancer. qRT-PCR (red bar) validates the findings on the GE array (blue bar) and also correlates with the increasing copy number doses seen in the aCGH. No relevant change is detected by the HELP assay (green bar), showing persistence of unmethylated HpaII sites on the promoter region. GATA6 amplification was corroborated by FISH in the primary adenocarcinoma tissue from this patient (panel B), RP11-18K7 spectrumOrange represent the 18q11.1-q11.2 region containing the GATA6 gene (red arrows). qRT-PCR on independent frozen primary tissues demonstrates high detectable mRNA levels in dysplasia and cancer (Mean +/− SEM). Immunohistochemical analysis on large number of primary tissues demonstrates elevated protein levels arising as early as in the LGD group. Percentage frequency of combined results from TMA and EMR are shown (Panel D) and examples of high IHC scores are shown (Panel F-I). To test the functional role of GATA6 upregulation, its expression was characterized in Barrett's associated adenocarcinoma cell lines (Panel J). Stable knock down of GATA6 from five shRNAs were screened by qRT-PCR in OE33 cell line (Panel K). shRNA-3 was selected for further experiments. Although there is no effect on cell viability (panel L), GATA6 loss significantly decreases cell migration (Panel M), Invasion (Panel N) and anchorage independent growth (panel O-P). Discussion
Esophageal cancer is the cancer with the fastest-growing prevalence in the United States [1] and arises from the metaplastic transformation of normal squamous mucosa, through the intermediate stages of dysplasia, culminating in cancer. Newer insights into the pathogenesis of this process are critically needed for prevention and early diagnosis of these lesions. Although alterations in DNA methylation have been described during esophageal carcinogenesis, studies performed thus far have focused on the aberrant hypermethylation of CpG islands located within promoters of selected tumor suppressors such as CDKN2A/p16, HPP, RUNX3, REPRIMO, amongst others. In contrast, we have determined that hypomethylation, rather than hypermethylation, is the more pervasive epigenetic alteration that occurs during Barrett progression. Additionally, we determined that global cytosine hypomethylation occurs very early during multistep carcinogenesis, observed within the first discernible metaplastic lesions within the native squamous esophagus. Even though global hypomethylation was reported in the pioneering epigenetic studies in cancer [31], most investigators have subsequently focused on hypermethylation in CpG islands within selected gene promoters. Hypomethylation has been hypothesized to lead to carcinogenesis by encouraging genomic instability [32] as well as by aberrant activation of oncogenes [33]. Additionally, as illustrated in the example of DMBT1, hypomethylation alone can lead to transcriptional upregulation during multistep progression to high-grade dysplasia and cancer. Furthermore, our data identify CG dinucelotide loci that can be targeted by differential methylation during neoplastic progression and are located outside of canonical CpG islands. Importantly, these CG alterations are not merely stochastic in nature, but appear to have bona fide regulatory influence on transcript expression. Recent work has similarly shown that cytosines present outside of CpG islands can be aberrantly methylated/ hypomethylated in cancer, and assays that cover these loci are critical to discovering the full landscape of altered methylome of malignancies [34], [35].
Since carcinogenesis is multifactorial and gene inactivation and activation can be influenced by either genetic or epigenetic mechanisms, we performed an integrative analysis to dissect the relative contributions of these alterations during this process. Our study provides direct experimental evidence of deletions and promoter methylation acting in concert to silence tumor suppressors in a bi-allelic manner, as first postulated by Knudson's two-hit paradigm. We expand on this paradigm by demonstrating that gene amplifications and hypomethylation can function in concert to upregulate gene expression of various genes. Of note, the use of an integrated approach facilitated by the MIGHT algorithm provides accurate in silico insights into mechanisms of deregulation, particularly when closely spaced gene clusters harbor discrepant alterations in transcript level, as exemplified by CDKN2A and CDKN2B, or the chemokine family in the 4q21 amplicon. In each of these instances, a subsequent validation step (such as FISH analysis for deletion, or MassArray for promoter methylation, respectively) confirmed the suggested mechanism of transcript deregulation implicated by MIGHT, underscoring the robustness of the analysis platform. Finally, we demonstrate that multiplatform high resolution integrative analysis of limited number of well annotated samples (N = three patients) can lead to findings that can be extrapolated to larger independent sample cohorts. We have illustrated this paradigm using multiple examples throughout the text, such as with the 4q21 chemokine cluster, DMBT1, and GATA6. In each of these examples, we validated the findings elucidated in the “index” patients in cohorts of either snap frozen or paraffin embedded BE and EAC tissues, In the case of the 4q21 chemokine cluster, we extended the validation one-step further, using serum samples to confirm significantly elevated circulating levels of two of the cytokines in EAC patients compared to controls. Notably, other chemokine ligands not included within the 4q21 amplicon failed to demonstrate any significant differences between cancer and control specimens, reiterating the biological relevance of the in silico MIGHT data. Chemokine ligands have recently been shown to be secreted by malignant cells and have been shown to participate in neoplastic progression of melanoma, breast, cervical and colorectal cancers [36]-[38]. CXCL1, CXCL3 and IL-8 bind to the CXCR2 receptor that has important roles in oncogene-induced senescence. It has been suggested that these chemokines potentiate tumor progression especially with cells with p53 inactivation, an event seen commonly in esophageal neoplasms [39]. These chemokines have also been implicated in tumor associated angiogenesis [37] and are a part of growing evidence of inflammatory mediators implicated in tumor growth and progression [40]. Our data reveals mechanisms associated with their upregulation in EAC and also demonstrates that this upregulation may occur early during neoplastic transformation, potentially allowing the development of a serum-based assay to screen subjects with BE for neoplastic progression.
Overall, our studies suggest that widespread changes in DNA methylation, especially hypomethylation, as well as genomic copy number alterations, can occur early during the multistep process of esophageal carcinogenesis, and may act in concert to deregulate the expression of important potential cancer-related pathogenic genes.
Methods
Patient samples
Specimens were obtained from patients who underwent endoscopic surveillance. All patients were diagnosed with Barrett's esophagus. After signed informed consent approved by the Johns Hopkins University IRB, endoscopic biopsies were collected and snap frozen in liquid Nitrogen, de-linked from direct patient identifiers and stored at −80 °C. DNA and RNA was extracted from the same biopsy samples.
Tissue microarray
All specimens were obtained from the surgical pathology files of the Johns Hopkins Hospital, Memorial Sloan-Kettering Cancer Center and Karmanos Cancer Center. Tissue microarrays (TMA) were generated from formalin-fixed paraffin-embedded archival tissues from 92 patients with Barrett's Esophagus and included esophageal squamous epithelium (60 cases), low-grade and high-grade dysplasia (19 and 38 cases), and adenocarcinoma (80 cases). Four 1.8 mm tissue cores represented each case and included two cores from the neoplastic compartment in order to account for potential tumor heterogeneity, and two cores from adjacent normal esophageal parenchyma as an internal control.
Additionally for GATA6 IHC, 120 endoscopic mucosal resection (EMR) specimens from 67 patients with BE were analyzed, including 31 cases of low grade dysplasia, 40 cases of high grade dysplasia and 10 cases of adenocarcinoma. All specimens were obtained from the surgical pathology files of the Johns Hopkins Hospital.
Immunohistochemistry
Rabbit polyclonal GATA6 (H-92) antibody (Santa Cruz Inc, sc-9055) was used at 1 : 500 dilution and visualized using the PowerVision+ Poly-HRP IHC kit (Immunovision Technologies) following the standard protocol for immunohistochemistry (IHC) described previously [41]. Immunohistochemical labeling was assessed in an outcome-blinded fashion by two of the authors (J.C.R and A.M) on a compound microscope. Intensity of labeling was evaluated as previously published [42], [43].
FISH analysis
The UCSC genome browser was used to select for the BAC clones spanning the 18q11.2 region: RP11-18K7 (GATA6, SpectrumOrange); and RP11-49H23 (18q22.2, Control SpectrumGreen). For the CXCL1-3 gene cluster, BAC clones covering the 4q21.1 - 4q21.2 region were RP11-94K4 (SpectrumOrange) and RP11-259E13 (4q11-q12, Control SpectrumGreen). The BAC clones were obtained from the Children's Hospital Oakland Research Institute in Oakland, USA. We also used commercial LSI p16/CEP probes (Vysis) for spanning the genetic loci for p16 and p15 (SpectrumOrange) and the alpha satellite sequences specific to chromosome 9 (SpectrumGreen).
Cell lines and culture conditions
Barrett's associated adenocarcinoma cell lines OE33 (European Collection of Cell Cultures, Wiltshire, UK) and JH-EsoAd1, recently described by our group [44], were maintained in RPMI-1640 and supplemented with 10% or 20% FBS respectively and 100 U/mL penicillin, 100 mg/mL streptomycin.
Lentiviral shRNA knockdown of GATA6 in OE33 cells
GATA6-expressing OE33 cells were seeded into 24-well plates at 9×104 cells per well concentration, and infected with either scrambled pLKO.1 (18 bp stuffer) lentiviral vector or with lentivirus expressing GATA6 shRNA (Open Biosystems, Huntsville, AL). Stable clones were selected by adding 5 µg/ml of puromycin to the cell culture media. Quantitative reverse transcription PCR (qRT-PCR) analysis was used to select the best short hairpin constructs for GATA6 mRNA knockdown. Downstream experiments were performed with GATA6_sh3 and compared with scrambled control.
Quantitative reverse transcription PCR (qRT–PCR)
One microgram of total RNA, isolated with the RNAgents kit (Promega, Madison, WI), were reversed transcribed by using the Superscript II First Strand kit (Invitrogen) as per manufacturer's protocol. 1 µL of cDNA was amplified in a 25 µl volume containing 12.5 µL of 2× SYBRGreen PCR Master Mix (Applied Biosystems) and 0.5 µM of each primer. Reactions were performed in triplicate using a 7300 Real Time PCR machine (Applied Biosystems, CA, USA) using PCR conditions and data analysis as described earlier[41]. The melting curve was constructed for each primer to ensure reaction specificity. Following PCR, the threshold cycle (CT) was obtained and relative quantities were determined by normalization with the housekeeping gene SDHA.
Statistical analysis
Data are presented as mean and S.E.M. and were compared using a Student's t-test (or Mann-Whitney U-test, as appropriate). A five-parameter logistic equation was used to calculate the curve fit in the non-linear asymmetric regression. Calculations were done with Graphpad Prims 4.0.
In vitro cell viability assays
Cell viability assays using the The CellTiter 96AQueousOne Solution Cell Proliferation Assay (Promega, Madison, WI) were performed on control-transfected and shRNA-expressing OE33 cells, as described previously [42]. At each time point evaluation, 20 µl/well of the Cell Titer 96 solution was added and incubated for 1 hour. Plates were read on a Wallac-1420 Plate reader at OD of 490 nm (PerkinElmer, Boston, MA). All experiments were set up in triplicate to determine means and standard deviations.
Boyden chamber migration - and invasion assay
The Boyden chamber migration - and invasion assays were carried out on OE33 cells. For the invasion assay, 5×104 cells were suspended in medium containing 0.2% FBS and plated in the inner chamber of a matrigel-coated 8-µm polypropylene filter inserts (BD Matrigel Matrix, BD Falcon). The bottom chamber contained normal growth media. After 24 h, the cells remaining in the insert were removed with a cotton swab, and the cells on the bottom of the filter were fixed and migrated cells were counted under the microscope. All experiments were set up in triplicate. Boyden chamber migration assay was carried out following the procedure described for the invasion assay except that the cells were plated on uncoated 8-µm pore polypropylene filter inserts in the Boyden chambers.
Anchorage-independent growth in soft agar
Anchorage-independent growth was assessed by colony formation assays in soft-agar, as previously described [41]. Briefly, the soft agar assays were set up in 6-well plates, each well containing a bottom layer of 1% agarose (Invitrogen), a middle layer of 0.7% agarose including 5×103 cells and a top layer comprising of medium only. Subsequently, the plates were kept in a tissue culture incubator maintained at 37°C and 5% CO for 14 days to allow for colony growth, with top medium being changed on a weekly basis. The assay was terminated at day 14, when plates were stained with 0.005% crystal violet (Sigma-Aldrich) solution. Colony counting was performed for each triplicate condition using an automated ChemiDoc XRS instru-ment (Bio-Rad, Hercules, CA).
Global Gene expression profiling (GE)
Total RNA, was isolated using the RNAgents kit (Promega, Madison, WI) as per the manufacturer's instructions and RNA integrity was corroborated with the Bioanalizer 2100. One microgram of RNA was reverse transcribed as published previously [9] and cDNA was submitted to Roche NimbleGen Systems, Inc. (Madison, WI) for labeling and hybridization onto the NimbleGen array (2006-10-26_Human_60mer_1in2) containing at least 10 (60mer) probes designed for 37,364 genes from GenBank build 35. Arrays were scanned using a GenePix 4000B scanner (Axon Instruments) and microarray quality controls were done as previously described [9]. Gene expression microarray data will be submitted to the GEO database for public access. Raw data reports (.pair files) were combined and analyzed with the RMA 7 (Robust Multi-array Analysis) package from NimbleScan 2.3 software, including the algorithm for background correction (data based background correction) and normalization (quantile normalization). After analysis, results of 36,846 unique nimblegene probes are reported as log2 values. Accession numbers from which NimbleGen probes were designed were linked to GenBank accessions from where Entrez IDs and additional annotations were isolated.
Global DNA methylation profiling: the HELP assay
The HELP assay was performed as published previously[23], [24], in summary, genomic DNA was isolated from cell lysates (0.1 M Tris-HCL pH 8.0; 10 mM EDTA pH 8.0 and 1% SDS), digested overnight at 50C on Proteinase K, follow by several organic extractions (TE-saturated phenol and Phenol-Chloroform-Isoamyl alcohol), DNA was purified and concentrated by ethanol precipitation and resuspended in LOTE pH 7.5 (3 mM Tris-Hcl and 0.2 mM EDTA). Intact DNA of high molecular weight was corroborated, by electrophoresis on 1% agarose gel, in all cases. DNA was digested with the restriction enzyme HpaII (methylation-sensitive) and MspI (methylation insensitive). Adapters are ligated to the fragments created by digestion that are subsequently used for ligation-mediated PCR amplification. The two digestion products are differentially labeled with two fluorophores and submitted to Roche NimbleGen Systems, Inc. (Madison, WI) for hybridization onto a human HG17 custom-designed oligonucleotide array (2006-10-26_HG17_HELP_Promoter). The focused array design stand for 25,626 HpaII-amplifiable fragments (HAF) defined by those fragments where two HpaII sites are located 200–2000 bp apart with at least some unique sequence between them and selected those located at gene promoters and imprinted regions. Each HAF was represented on the microarray by at least 14 oligonucleotides, each 50 nucleotides in length and randomly distributed across the microarray slide. HELP microarray data will be submitted to the GEO database for public access. Raw data reports (.pair files) went through an analytical pipeline of array performance and quality assessment previously described [45]. 10 and included removal of failed probes, summarization of the remaining probes belonging to the 20%-trimmed mean for each HAF. The functions in this pipeline also allows for quantile normalization that solves the fragment size-dependency of the MspI, HpaII and HpaII/MspI ratio distributions seen in this assay. The R statistical package (http://www.r-project.org/) was used to calculate the normalized log2 HpaII/MspI ratios allowing us to semi-quantitatively categorize each loci as methylated or hypomethylated based quantitative methylation detected using MassArray Epityping as a validation approach described below. In all instances, HpaII-amplifiable fragments (HAF) sequence were linked to gene promoters (defined as 2000 bp upstream from transcription start site) by using genomic coordinates from National Center for Biotechnology Information Build 35.
Array Comparative genomic hybridization (aCGH)
DNA was hybridized to a oligonucleotide array CGH (NimbleGen Systems), consisting of 388,560 isothermal probes (length 45–75 bp) covering unique genomic regions (median probe spacing of 6 kb). Hybridizations were performed at NimbleGen Facility and a reference sample of Human Male Genomic DNA was used as a normal control. Intensity data underwent through a qspline fit normalization algorithm 11 that was used to compensate for inherent differences in signal between the two dyes. To avoid false positive calls due to local variation in signal intensity, a second pass filter was employed. This filter discards change points if their means are not at least 1.5 SD from each other. Log2-ratio values of the probe signal intensities (Cy3/Cy5) were calculated and 120,000 bp windows-average were generated to visualize copy number changes using the circular binary segmentation algorithm 12 contained on the Roche NimbleScan software. Oligonucleotide array data will be submitted to the GEO database for public access. In all instances, DNA sequence coordinates are from National Center for Biotechnology Information Build 35.
Data integration and 3D visualization
An analytical pipeline was developed to integrate all platforms. In summary, Gene expression (GE), HELP and aCGH probes were linked to genes base on the basis of genomic coordinates isolated from NCBI build35. Probe to gene links containing data from all three platforms, after proper normalization described above, were considered for 3D integration. TTEST pairwise comparisons of GE and HELP were performed between main histological groups (squamous vs dysplasia and dysplasia vs adenocarcinoma) and fold change differences with their p-values were calculated. The last two variables for each pairwise were plotted simultaneously for each gene on a k-space where the x axis represents fold-change differences (log2) in GE and the y axis represents fold-change differences (log2) in methylation. The “z” axis is used to represent the significance level in –log10 p-value for both platforms. Color-coding for significant cut-offs was defined for those genes over 2.5 fold change in GE with a p-value < 0.05, with green representing downregulated and red upregulated genes. The same criterion was used to detect significant changes in methylation. A darker shade of color was added to the red and green labels to define the hypermethylated genes (on top of the graph) and lighter shade of red and green was used to define significantly hypomethylated genes.
Copy number alterations were defined by two fold criteria: the gene center approach and the genomic segmentation approach. In the gene center approach, the aCGH probes contained in the intragenic region or promoter region (when intergenic area was not available), were averaged. Computational based approach to find accumulative changes on 50% of DNA content in all three histological stages for each patient was considered gain or loss depending if the progressive trend was positive or negative respectively. A similar approach was also taken for the Circular binary segmentation (CBS) analysis, this time visual inspection at genomic windows of 120,000 bp was applied and genomic segments isolated and classified as gain or losses. Genomic windows were linked to genes and gene copy number changes for each patient was defined as those that agreed on last two criteria. Finally, on the integrative figure, copy number changes were depicted by larger circles. Genes classified as amplified and deleted had a 3 fold more larger size compared to their normal counterparts.
Quantitative DNA methylation analysis by MassArray Epityping
Quantitative high-throughput DNA methylation analysis to validate HELP results was carried out by MALDI-TOF mass spectrometry using Sequenom's EpiTYPER platform (Sequenom. San Diego, CA). The same DNA aliquots used for HELP were bisulfite-converted; PCR amplified and follow by an in vitro RNA transcription and a base specific cleavage. MALDI TOF MS analysis of the cleavage product was performed as described originally [24], [25]. MassArray primers were designed and are available upon request.
Chemokine and cytokine detection
Five serum samples from patients with gastroesophageal reflux disease (GERD) symptoms and normal esophageal histology, and five serum samples from esophageal adenocarcinoma patients were collected for analysis. Frozen endoscopic tissues from the same patients were also collected and used for tissue lysates. To determine cytokine levels in the serum, Meso Scale Discovery System multiplex assay were used for human Eotaxin (CCL11), IL-8 (IL8), IP-10 (CXCL10), MCP-1 (CCL2) and MCP-4 (CCL13) according to the manufacturer's instructions.
Luminometric methylation assay (LUMA)
Genomic DNA (200–500 ng) was cleaved with HpaII + EcoRI or MspI + EcoRI in two separate 20 µl reactions containing 33 mM Tris-acetate, 10 mM Mg-acetate, 66 mM K-acetate pH 7.9, 0.1 mg/ml BSA and 5 units of each restriction enzymes. The reactions were set up in a 96-well format and incubated at 37°C for 4 h. Then 20 µl annealing buffer (20 mM Tris-acetate, 2 mM Mg-acetate pH 7.6) was added to the cleavage reactions, and samples were placed in a PSQ96™MA system (Biotage AB, Uppsala, Sweden). The instrument was programmed to add dNTPs in four consecutive steps including Step 1: dATP (the derivative dATPαS is used since it will not react directly with Luciferase and prevents non-specific signals); Step 2: mixture of dGTP + dCTP; Step 3: dTTP; and Step 4: mixture of dGTP + dCTP. Peak heights were calculated using the PSQ96™MA software. The HpaII/EcoRI and MspI/EcoRI ratios were calculated as (dGTP + dCTP)/dATP for the respective reactions. The HpaII/MspI ratio was defined as (HpaII/EcoRI)/(MspI/EcoRI) [46].
Supporting Information
Zdroje
1. ZhangHY
SpechlerSJ
SouzaRF
2009 Esophageal adenocarcinoma arising in Barrett esophagus. Cancer Lett 275 170 177
2. EloubeidiMA
MasonAC
DesmondRA
El-SeragHB
2003 Temporal trends (1973-1997) in survival of patients with esophageal adenocarcinoma in the United States: a glimmer of hope? Am J Gastroenterol 98 1627 1633
3. MontgomeryE
BronnerMP
GoldblumJR
GreensonJK
HaberMM
2001 Reproducibility of the diagnosis of dysplasia in Barrett esophagus: a reaffirmation. Hum Pathol 32 368 378
4. MontgomeryE
GoldblumJR
GreensonJK
HaberMM
LampsLW
2001 Dysplasia as a predictive marker for invasive carcinoma in Barrett esophagus: a follow-up study based on 138 cases from a diagnostic variability study. Hum Pathol 32 379 388
5. SpechlerSJ
2002 Clinical practice. Barrett's Esophagus. N Engl J Med 346 836 842
6. ShaheenNJ
SharmaP
OverholtBF
WolfsenHC
SamplinerRE
2009 Radiofrequency ablation in Barrett's esophagus with dysplasia. N Engl J Med 360 2277 2288
7. WangS
ZhanM
YinJ
AbrahamJM
MoriY
2006 Transcriptional profiling suggests that Barrett's metaplasia is an early intermediate stage in esophageal adenocarcinogenesis. Oncogene 25 3346 3356
8. RazviMH
PengD
DarAA
PowellSM
FriersonHFJr
2007 Transcriptional oncogenomic hot spots in Barrett's adenocarcinomas: serial analysis of gene expression. Genes Chromosomes Cancer 46 914 928
9. SelaruFM
ZouT
XuY
ShustovaV
YinJ
2002 Global gene expression profiling in Barrett's esophagus and esophageal cancer: a comparative analysis using cDNA microarrays. Oncogene 21 475 478
10. XuY
SelaruFM
YinJ
ZouTT
ShustovaV
2002 Artificial neural networks and gene filtering distinguish between global gene expression profiles of Barrett's esophagus and esophageal cancer. Cancer Res 62 3493 3497
11. PowellSM
PapadopoulosN
KinzlerKW
SmolinskiKN
MeltzerSJ
1994 APC gene mutations in the mutation cluster region are rare in esophageal cancers. Gastroenterology 107 1759 1763
12. MeltzerSJ
YinJ
HuangY
McDanielTK
NewkirkC
1991 Reduction to homozygosity involving p53 in esophageal cancers demonstrated by the polymerase chain reaction. Proc Natl Acad Sci U S A 88 4976 4980
13. HuangY
BoyntonRF
BlountPL
SilversteinRJ
YinJ
1992 Loss of heterozygosity involves multiple tumor suppressor genes in human esophageal cancers. Cancer Res 52 6525 6530
14. JinZ
MoriY
YangJ
SatoF
ItoT
2007 Hypermethylation of the nel-like 1 gene is a common and early event and is associated with poor prognosis in early-stage esophageal adenocarcinoma. Oncogene 26 6332 6340
15. LaiLA
PaulsonTG
LiX
SanchezCA
MaleyC
2007 Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes Chromosomes Cancer 46 532 542
16. PowellEL
LeoniLM
CantoMI
ForastiereAA
Iocobuzio-DonahueCA
2005 Concordant loss of MTAP and p16/CDKN2A expression in gastroesophageal carcinogenesis: evidence of homozygous deletion in esophageal noninvasive precursor lesions and therapeutic implications. Am J Surg Pathol 29 1497 1504
17. SchulmannK
SterianA
BerkiA
YinJ
SatoF
2005 Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett's-associated neoplastic progression and predicts progression risk. Oncogene 24 4138 4148
18. JinZ
ChengY
GuW
ZhengY
SatoF
2009 A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett's esophagus. Cancer Res 69 4112 4115
19. SatoF
JinZ
SchulmannK
WangJ
GreenwaldBD
2008 Three-tiered risk stratification model to predict progression in Barrett's esophagus using epigenetic and clinical features. PLoS ONE 3 e1890 doi:10.1371/journal.pone.0001890
20. PrasadGA
WangKK
HallingKC
ButtarNS
WongkeesongLM
2008 Utility of biomarkers in prediction of response to ablative therapy in Barrett's esophagus. Gastroenterology 135 370 379
21. FigueroaME
ReimersM
ThompsonRF
YeK
LiY
2008 An integrative genomic and epigenomic approach for the study of transcriptional regulation. PLoS ONE 3 e1882 doi:10.1371/journal.pone.0001882
22. LearyRJ
LinJC
CumminsJ
BocaS
WoodLD
2008 Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A 105 16224 16229
23. KhulanB
ThompsonRF
YeK
FazzariMJ
SuzukiM
2006 Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res 16 1046 1055
24. OdaM
GlassJL
ThompsonRF
MoY
OlivierEN
2009 High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res 37 3829 3839
25. FigueroaME
LugthartS
LiY
Erpelinck-VerschuerenC
DengX
DNA Methylation Signatures Identify Biologically Distinct Subtypes in Acute Myeloid Leukemia. Cancer Cell 17 13 27
26. BaylinSB
HermanJG
GraffJR
VertinoPM
IssaJP
1998 Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 72 141 196
27. ToyotaM
IssaJP
1999 CpG island methylator phenotypes in aging and cancer. Semin Cancer Biol 9 349 357
28. FazzariMJ
GreallyJM
2004 Epigenomics: beyond CpG islands. Nat Rev Genet 5 446 455
29. AlbrechtB
HausmannM
ZitzelsbergerH
SteinH
SiewertJR
2004 Array-based comparative genomic hybridization for the detection of DNA sequence copy number changes in Barrett's adenocarcinoma. J Pathol 203 780 788
30. AbdallaSI
SandersonIR
FitzgeraldRC
2005 Effect of inflammation on cyclooxygenase (COX)-2 expression in benign and malignant oesophageal cells. Carcinogenesis 26 1627 1633
31. FeinbergAP
VogelsteinB
1983 Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301 89 92
32. RodriguezJ
FrigolaJ
VendrellE
RisquesRA
FragaMF
2006 Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res 66 8462 9468
33. EhrlichM
2002 DNA methylation in cancer: too much, but also too little. Oncogene 21 5400 5413
34. DoiA
ParkIH
WenB
MurakamiP
AryeeMJ
2009 Differential methylation of tissue - and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41 1350 1353
35. FigueroaME
SkrabanekL
LiY
JiemjitA
FandyTE
2009 MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood 114 3448 3458
36. SparmannA
Bar-SagiD
2004 Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell 6 447 458
37. WangD
WangH
BrownJ
DaikokuT
NingW
2006 CXCL1 induced by prostaglandin E2 promotes angiogenesis in colorectal cancer. J Exp Med 203 941 951
38. DhawanP
RichmondA
2002 Role of CXCL1 in tumorigenesis of melanoma. J Leukoc Biol 72 9 18
39. AcostaJC
O'LoghlenA
BanitoA
GuijarroMV
AugertA
2008 Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133 1006 1018
40. CoussensLM
WerbZ
2002 Inflammation and cancer. Nature 420 860 867
41. KoorstraJB
KarikariCA
FeldmannG
BishtS
RojasPL
2009 The Axl receptor tyrosine kinase confers an adverse prognostic influence in pancreatic cancer and represents a new therapeutic target. Cancer Biol Ther 8 618 626
42. HanselDE
DharaS
HuangRC
AshfaqR
DeaselM
2005 CDC2/CDK1 Expression in Esophageal Adenocarcinoma and Precursor Lesions Serves as a Diagnostic and Cancer Progression Marker and Potential Novel Drug Target. Am J Surg Pathol 29 390 399
43. MontgomeryE
MamelakAJ
GibsonM
MaitraA
SheikhS
2006 Overexpression of claudin proteins in esophageal adenocarcinoma and its precursor lesions. Appl Immunohistochem Mol Morphol 14 24 30
44. AlvarezH
KoorstraJB
HongSM
BoonstraJJ
DinjensWN
2008 Establishment and characterization of a bona fide barrett esophagus-associated adenocarcinoma cell line. Cancer Biol Ther 7
45. ThompsonRF
SuzukiM
LauKW
GreallyJM
2009 A pipeline for the quantitative analysis of CG dinucleotide methylation using mass spectrometry. Bioinformatics 25 2164 2170
46. KarimiM
JohanssonS
StachD
CorcoranM
GranderD
2006 LUMA (LUminometric Methylation Assay)--a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res 312 1989 1995
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and PhosphodiesterasesČlánek Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



