-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
Following DNA damage or replication stress, budding yeast cells activate the Rad53 checkpoint kinase, promoting genome stability in these challenging conditions. The DNA damage and replication checkpoint pathways are partially overlapping, sharing several factors, but are also differentiated at various levels. The upstream kinase Mec1 is required to activate both signaling cascades together with the 9-1-1 PCNA-like complex and the Dpb11 (hTopBP1) protein. After DNA damage, Dpb11 is also needed to recruit the adaptor protein Rad9 (h53BP1). Here we analyzed the mechanisms leading to Mec1 activation in vivo after DNA damage and replication stress. We found that a ddc1Δdpb11-1 double mutant strain displays a synthetic defect in Rad53 and H2A phosphorylation and is extremely sensitive to hydroxyurea (HU), indicating that Dpb11 and the 9-1-1 complex independently promote Mec1 activation. A similar phenotype is observed when both the 9-1-1 complex and the Dpb4 non-essential subunit of DNA polymerase ε (Polε) are contemporarily absent, indicating that checkpoint activation in response to replication stress is achieved through two independent pathways, requiring the 9-1-1 complex and Polε.
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1002022
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002022Summary
Following DNA damage or replication stress, budding yeast cells activate the Rad53 checkpoint kinase, promoting genome stability in these challenging conditions. The DNA damage and replication checkpoint pathways are partially overlapping, sharing several factors, but are also differentiated at various levels. The upstream kinase Mec1 is required to activate both signaling cascades together with the 9-1-1 PCNA-like complex and the Dpb11 (hTopBP1) protein. After DNA damage, Dpb11 is also needed to recruit the adaptor protein Rad9 (h53BP1). Here we analyzed the mechanisms leading to Mec1 activation in vivo after DNA damage and replication stress. We found that a ddc1Δdpb11-1 double mutant strain displays a synthetic defect in Rad53 and H2A phosphorylation and is extremely sensitive to hydroxyurea (HU), indicating that Dpb11 and the 9-1-1 complex independently promote Mec1 activation. A similar phenotype is observed when both the 9-1-1 complex and the Dpb4 non-essential subunit of DNA polymerase ε (Polε) are contemporarily absent, indicating that checkpoint activation in response to replication stress is achieved through two independent pathways, requiring the 9-1-1 complex and Polε.
Introduction
The DNA replication machinery can experience various types of stress during S phase. This can happen when the replisome encounters DNA lesions that hinder its progression, while traversing slow replication zones corresponding to genomic regions difficult to replicate [1] or when encountering replication fork barriers [2]. Replication stress can also be induced by inhibiting ribonucleotide reductase (RNR) with hydroxyurea, which causes a global replication arrest by reducing the dNTPs pools [3].
Under replication stress conditions, eukaryotic cells trigger a signaling cascade, known as the replication checkpoint, which, in budding yeast, culminates with the phosphorylation of Rad53 [4]. This protein kinase is essential for the activation of the molecular mechanisms required to cope with replication arrest: it promotes stabilization of stalled replication forks and allows DNA replication re-start after removal of the blocking agent [5], [6], [7], [8]. Rad53 is also responsible for inducing the transcription of RNR genes by inhibiting the transcriptional repressor Crt1 and promoting the degradation of the RNR inhibitor Sml1 [9], [10]. Finally, Rad53 prevents the firing of late replication origins [11] and restrains spindle elongation thus preventing mitosis [12], [13], [14].
The DNA damage and replication checkpoints are genetically distinct pathways; however, they are partially overlapping since they share several of the factors involved. In fact, replication stress activates Mec1, the same apical kinase triggered by DNA damage, which is recruited to RPA-covered ssDNA by its binding partner Ddc2 [15]. After damage, Mec1 phosphorylates the Rad9 adaptor protein, which has been loaded onto DNA via chromatin-dependent and -independent pathways: the former requiring methylation of H3-K79 and the latter depending on the 9-1-1 complex and Dpb11 [16], [17], [18], [19], [20]. Phosphorylated Rad9, in turn, recruits Rad53, which becomes hyperphosphorylated in a Mec1-dependent manner. Differently, in the case of HU-induced checkpoint activation, the Rad9 adaptor protein is dispensable and its function is performed by Mrc1, a constitutive member of the replisome complex [21], [22].
It is now clear that following genotoxin treatments, primary lesions are generally recognized by specific repair factors that process them to generate ssDNA regions, which elicit the DNA damage response. On the other hand, the actual mechanism acting in the activation of the replication stress response is poorly understood. In budding yeast, it has been suggested that replication proteins may be involved in sensing blocks of the replication fork. Indeed, in addition to Dpb11, the initiation factor Sld2/Drc1 and Polε itself are required for efficient checkpoint activation in response to HU treatment, although the corresponding mutants are only mildly sensitive to the drug [23], [24], [25].
Sld2 is an essential CDK1 target required for initiation of DNA replication. Its phosphorylation and subsequent interaction with Dpb11 is essential for the loading of Polε and the firing of replication origins [26], [27]. Polε consists of four subunits: Pol2 and Dpb2 are essential for cell viability while Dpb3 and Dpb4 appear to be non-essential. These last two factors contains a histone-like fold motif and are also implicated in transcriptional regulation [28], [29]. The Polε holoenzyme is composed of two structurally distinct domains: a globular domain, made of the N-terminus of the catalytic Pol2 subunit and a tail-like domain containing the other three factors, bound to the Pol2 C-terminus [30], [31]. The catalytic subunit contains an N-terminal polymerase domain followed by a C-terminal region, where the checkpoint-defective mutations of POL2 map [24]. Surprisingly, deletion of the polymerase domain does not cause cell lethality, whereas the checkpoint domain is essential for cell viability [32].
It has been established that in response to DNA damage, the 9-1-1 clamp is loaded onto the 5′ primer-template junction adjacent to RPA-coated ssDNA [33], [34]. In higher eukaryotes, 9-1-1 then recruits TopBP1 which, through an interaction with ATRIP, stimulates the ATR kinase activity [35], [36], [37], [38]. Recent work in yeast demonstrated that Mec1 activation can proceed also through a 9-1-1-dependent, but Dpb11-independent pathway, mediated by an activation domain present in the Ddc1 subunit of the 9-1-1 complex [39]. Indeed, it has been reported that S. cerevisiae 9-1-1 can directly activate the Mec1-Ddc2 kinase in vitro [40]. The in vivo balancing between these two pathways has been recently studied, following Rad53 phosphorylation [39], which is influenced not only by Mec1 activation, but also by the Rad9 mediator [39].To determine directly the relative contributions of Ddc1 and Dpb11 to Mec1 activation in different cell cycle phases, and particularly in response to replication stress, we analyzed a direct target of Mec1 kinase, histone H2A, whose phosphorylation is not dependent upon Rad9.
In this study we found that, in G1 yeast cells, Mec1 activation induced by UV irradiation completely depends on the 9-1-1 dependent pathway, whereas Dpb11 only plays a minor role. Conversely, in response to replication stress, Mec1 activation is achieved through two independent pathways which rely on the 9-1-1 complex and Dpb11, respectively. At least one of these two pathways is necessary to efficiently activate Mec1 and to allow cell growth in the presence of HU. Finally, we provide evidence that the DNA polymerase ε complex and Sld2 are required to establish the 9-1-1 independent branch of Mec1 activation and we suggest that this could reflect strand-specificity in detecting replication stress.
Results
UV-induced Mec1 activation requires 9-1-1 and the C-terminal tail of Dpb11
We have previously shown that, in M phase, Dpb11 is required to recruit the Rad9 adaptor protein to UV-damaged DNA in a pathway that is parallel to that controlled by histone modifications [16], [20]. Dpb11 was also found to stimulate Mec1 kinase activity in vitro and this function appears to be modulated by its interaction with the 9-1-1 complex [41], [42]. To dissect the Mec1-activation role of Dpb11 in vivo and to determine the relative contribution of Dpb11 and 9-1-1 to this mechanism in different cell cycle phases, we analyzed histone H2A phosphorylation as an assay for Mec1 activity. After UV damage H2A is phosphorylated directly on serine 129 (γH2A) by Mec1 kinase; indeed mec1-1 mutant cells fail to phosphorylate H2A after DNA damage and a strain deleted in TEL1, coding for a second sensor-kinase, does not show any significant reduction in γH2A levels (Figure S1A and S1B).
We used a yeast strain carrying a C-terminal deletion of Dpb11 (Δ583―764) encoded by the dpb11-1 allele, which removes almost entirely the ATR Activation Domain (AAD) and a strain carrying the deletion of DDC1, the gene encoding the 9-1-1 subunit involved in Mec1 activation [40]. WT, dpb11-1, ddc1Δ and ddc1Δdpb11-1 cells were arrested in G1 with α-factor and in M phase with nocodazole and UV irradiated. As it is shown in Figure 1A, histone H2A is extensively phosphorylated after UV treatment in G1 and this damage-dependent modification requires the presence of a functional 9-1-1 complex, while the contribution of the AAD domain of Dpb11 is only minor. The quantification of the signal (shown in the lower panel of Figure 1A), indicates that the level of phosphorylated histone H2A (γH2A) in dpb11-1 is ∼50% of that found in WT cells.
Fig. 1. UV-induced Mec1 activation requires the 9-1-1 complex and the Dpb11 C-terminus. 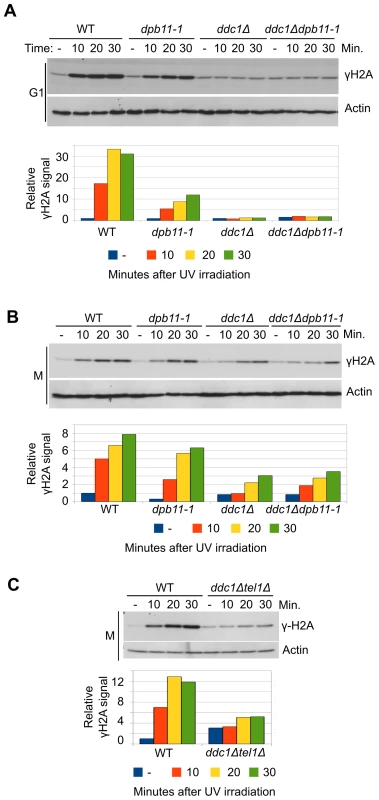
(A) K699 (WT), YFP20 (dpb11-1), YAN21/8d (ddc1Δ), YFP62/1d (ddc1Δdpb11-1) and YMIC5A3 (mec1-1) strains were grown to mid-log phase, arrested in G1 with α-factor and subjected to UV irradiation. At the indicated time-points, protein extracts were prepared and separated by SDS-PAGE. Mec1 activation was assayed by western blotting monitoring γH2A and α-actin was used as loading control. A quantification of H2A phosphorylation is shown in the lower panel. The values indicate the fold increase respect to the WT untreated sample. The mec1-1 mutation is functionally equivalent to a null mutation (B) The strains in panel A were arrested in M phase with nocodazole and subjected to the same treatment. Analysis and quantification of H2A phosphorylation was carried out as described above. (C) Strains K699 (WT) and YFP223 (ddc1Δ tel1Δ) were arrested in M phase with nocodazole and UV irradiated. At the indicated time-points Mec1 activation was assayed by western blotting monitoring γH2A. A quantification of the signal corresponding to H2A-S129 is shown in the lower panel. The values indicate the fold increase respect to the WT untreated samples. In M phase cells the basal level of phosphorylated H2A-S129 is much higher (Figure S1C), and this likely influences the magnitude of the increase measured after UV-irradiation. In these conditions, Dpb11 plays a minimal role, if any, in H2A phosphorylation and also DDC1 deletion reduces γH2A only partially (∼50%) (Figure 1B). However, the residual H2A phosphorylation observed in a ddc1Δ mutant strain is lost when TEL1 is deleted, (Figure 1C). On the other hand, deletion of TEL1in the dpb11-1 background does not significantly influence H2A phosphorylation (Figure S1D)
9-1-1 and Dpb11 act independently in signaling replication stress to the Mec1 kinase
To further elucidate the balancing between 9-1-1-dependent and Dpb11-dependent Mec1 activation in S phase, we decided to analyze this process after replication stress induced by HU. This allowed us also to minimize the side effects due to the involvement of Dpb11 in Rad9 recruitment because, during HU treatment, Rad9 does not become hyperphosphorylated and is not expected to play any role in checkpoint activation [22]. WT, dpb11-1, ddc1Δ and ddc1Δdpb11-1 cells were synchronized in G1, released into fresh medium supplemented with 200 mM HU, and checkpoint activity was monitored by measuring Rad53 phosphorylation (Figure 2A). Differently from what found in G1 and G2 cells, strains lacking either a functional 9-1-1 complex or the Dpb11 C-terminal region were fully able to phosphorylate Rad53. In these experimental conditions, ddc1Δ dpb11-1 double mutant cells showed a very severe defect in Rad53 phosphorylation, similar to that found in a Mec1-defective strain. These results suggest that a dpb11-1 ddc1Δ double mutation virtually abolishes UV-induced Mec1 activation differently from what previously reported [39], In addition, the double mutant strain showed synthetic lethality on HU plates (Figure 2B and [43]). To confirm that the dpb11-1 and ddc1Δ mutations directly affect Mec1 activity, we monitored γH2A levels in the same conditions. As shown in Figure 2C, the ddc1Δ and dpb11-1 mutations showed a synthetic defect in the ability to phosphorylate H2A-S129 (Figure 2D).
Fig. 2. Dpb11 and 9-1-1 independently activate Mec1 after replication stress. 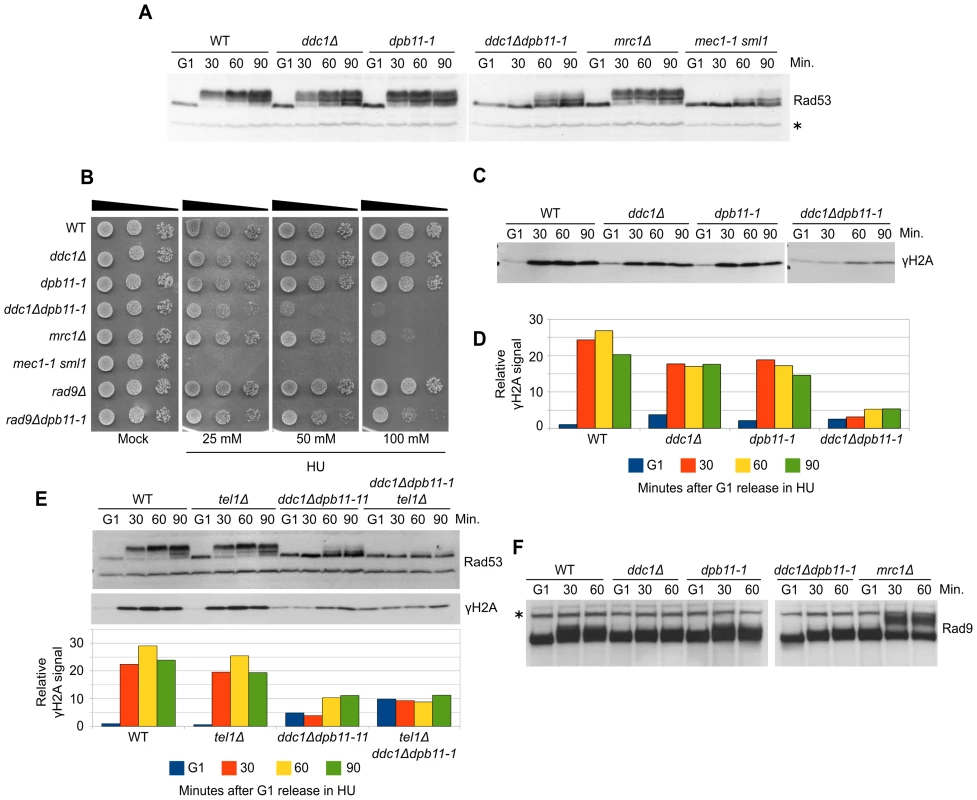
(A) K699 (WT), YFP20 (dpb11-1), YAN21/8d (ddc1Δ), YFP62/1d (ddc1Δdpb11-1), YFP125/6d (mrc1Δ) and YMIC5A3 (mec1-1) strains were grown to mid-log phase, synchronized in G1 with α-factor and released into fresh medium supplemented with 200 mM HU. At the indicated time-points, protein extracts were prepared and separated by SDS-PAGE. Rad53 activation was assayed as the phosphorylation-dependent shift of the protein. (B) 103–104 cells from overnight cultures of the strains analyzed in panel A and YFP74 (rad9Δ) and YFP161/5C (rad9Δdpb11-1) were spotted on YPD plates supplemented with HU at the indicated concentrations. Cell survival was assayed after 2–7 days. (C) The same filter in panel A was probed for Mec1 activation by analyzing the level of histone H2A phosphorylation. (D) Quantification of γH2A in the experiment shown in panel C, using α-Rad53 cross-reacting band as loading control. The values indicate the fold increase respect to the WT G1 sample. (E) Strains K699 (WT), YMIC6C3 (tel1Δ), YFP62/1d (ddc1Δdpb11-1) and YFP230 (ddc1Δdpb11-1tel1Δ) were synchronized in G1 and released into fresh medium supplemented with 200 mM HU. At the indicated time-points Rad53 and H2A phosphorylation were assayed by western blotting. A quantification of the signal is shown in the lower panel, using α-Rad53 cross-reacting band as loading control. The numbers indicate the fold increase respect to the WT G1 sample. (F) The same extracts of panel A were probed with α-Rad9 antibodies to determine the extent of Rad9 hyperphosphorylation. In all the relevant panels the loading control is indicated by an asterisk. Although displaying a severe defect in Rad53 phosphorylation, ddc1Δdpb11-1 still displays a residual low level of phosphorylated Rad53, which may be dependent upon a residual Mec1 activity. However, Figure 2E and Figure S2A show that the residual Rad53 phosphorylation in the double mutant is instead due to Tel1. Indeed, an additional mutation eliminating Tel1 function completely abolishes Rad53 phosphorylation in a dpb11-1 ddc1Δ strain and strongly sensitizes cells to HU treatment, as shown in Figure S2B. These findings further support the hypothesis that Mec1 cannot become activated in response to replication stress in the absence of both Ddc1 and Dpb11-AAD.
To verify the possibility that in dpb11-1 mutant cells an unscheduled, Ddc1-dependent, DNA damage response is triggered as a consequence of the inability to properly activate the replication stress response, similarly to what happens in an mrc1Δ strain [22], we monitored DNA damage checkpoint activation looking at Rad9 hyperphosphorylation. As shown in Figure 2F, differently from what found in the mrc1Δ control strain, no Rad9 hyperphosphorylation was detectable in ddc1Δ, dpb11-1 single or double mutant strains. Consistently, rad9Δdpb11-1 double mutant cells are far less sensitive than the ddc1Δdpb11-1 strain to HU treatment (Figure 2B and [43]).
Low levels of Rad53 activity are sufficient to prevent replication fork breakdown and premature entry into mitosis
Rad53 kinase activity is required to stabilize stalled replication forks [7]. To verify whether the increased HU sensitivity of ddc1Δdpb11-1 double mutant cells was due to their inability to fully activate Rad53 and thus to stabilize the replisomes, we performed a recovery assay. Briefly, WT, dpb11-1, ddc1Δ, ddc1Δdpb11-1 and mec1-1sml1 mutant strains were blocked in G1, released and exposed to HU for 90 minutes; cells were then washed and shifted into fresh medium lacking HU and allowed to recover. As shown in the control strain mec1-1 sml1, when Rad53 activity is impaired, cells transiently exposed to HU loose the ability to resume DNA synthesis and complete DNA replication once the drug has been removed ([6] and Figure 3A). Unexpectedly, we found that not only dpb11-1 and ddc1Δ single mutant cells, but also the double mutant strain, which has a severe Rad53 hyperphosphorylation defect, were able to recover from the HU treatment with a WT kinetics (Figure 3A). Moreover, with lower HU concentrations, ddc1Δ dpb11-1 cells were capable of completing a round of DNA replication, as demonstrated by the re-entering of the replicated chromosomes in a pulsed-field gel system (Figure 3B).
Fig. 3. Low levels of Rad53 activity are sufficient to prevent replication fork breakdown. 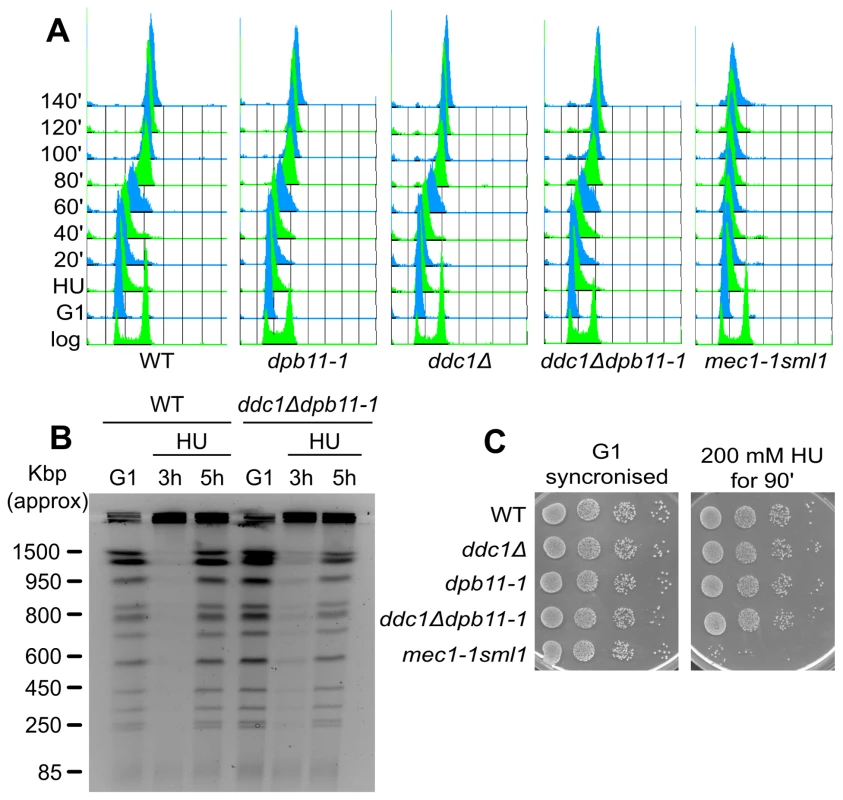
(A) HU recovery assay: K699 (WT), YFP20 (dpb11-1), YAN21/8d (ddc1Δ), YFP62/1d (ddc1Δdpb11-1) and YMIC5A3 (mec1-1) were synchronized in G1 with α-factor and released into fresh medium supplemented with 200 mM HU. 90 min later cells were transferred to fresh YPD + nocodazole and allowed to resume DNA replication. Progression into S phase was monitored by FACS analysis. (B) The indicated strains were synchronized in G1 with α-factor and released into 100 mM HU + nocodazole. 3 and 5 hours later cells were harvested and total DNA was analyzed by Pulse Field Gel Electrophoresis (PFGE). (C) The strains in panel A were synchronized in G1 and released in YPD supplemented with 200 mM HU. 90 min later 10-fold serial dilution were prepared and spotted onto YPD plates. The same was done with the G1-synchronised cultures as control. Another marker of checkpoint activation by HU is the arrest of cell cycle, preventing mitosis. When exposed to HU, checkpoint mutants fail to delay the onset of mitosis and display elongated spindles [14]. To address the hypothesis that ddc1Δ dpb11-1 cells may die as a consequence of a premature mitosis, we measured spindle length 90 minutes after HU addition. ddc1Δ dpb11-1 double mutant cells prevent spindle elongation in the presence of HU, a process which is clearly defective in a mec1-1 mutant strain (Figure S3A), suggesting that the replication checkpoint can delay mitotic entry in the double mutant [10].
In agreement with all these data, the HU sensitivity of ddc1Δ dpb11-1 double mutant cells can be observed only to chronic exposure to the drug, while it is virtually undetectable if cells are transiently exposed to HU (Figure 3C).
The inability to fully activate Rad53 causes defects in the control of RNR induction
ddc1Δ dpb11-1 mutant cells exhibit extremely low levels of Mec1 and Rad53 activation and, despite being sensitive to exposure to even low concentrations of HU (Figure 2B), they do not show some of the most common phenotypes observed in replication checkpoint defective cells. To better characterize the sensitivity to the drug, we monitored cell growth in the presence of 100 mM HU. The single and double mutant ddc1Δ dpb11-1 yeast strains were synchronized in G1, released into fresh medium supplemented with HU and cell cycle progression followed by FACS analysis. The double mutant ddc1Δ dpb11-1 showed a small delay in progressing through S-phase in the presence of HU, compared to WT and single mutant cells. Significantly, at late times (20 hours) after the release, a large fraction of double mutant cells appeared to be arrested at different stages of S-phase, while WT and single mutant cells had regained a FACS profile with 1C and 2C peaks (Figure 4A). Consistently, PFGE analysis of genomic DNA prepared from the various strains 20 hours after release from HU showed that in ddc1Δ dpb11-1 double mutant cells most of the DNA fails to enter the gel, suggesting the presence of branched intermediates (Figure 4B, 4C). It is important to note that, differently from what found in a mec1-1 strain, the ddc1Δ dpb11-1 strain did not accumulate cells with a<1C DNA content, or low molecular weight DNA fragments (Figure 4A–4C) indicating a correct segregation of chromosomes. Altogether, these findings may suggest that ddc1Δ dpb11-1 cells are unable to counteract the effect of HU by upregulating ribonucleotide reductase (RNR). Indeed, Rad53 regulates both the timely degradation of the RNR inhibitor Sml1 and the inactivation of Crt1, which represses the transcription of RNR genes [9], [10]. Consistently with this interpretation, ddc1Δ dpb11-1 cells show a modest delay in Sml1 degradation and, more significantly, CRT1 deletion suppresses, although not completely, the sensitivity of the double mutant strain to HU (Figure 4D, 4E).
Fig. 4. The inability to fully activate Rad53 causes defects in the control of RNR induction. 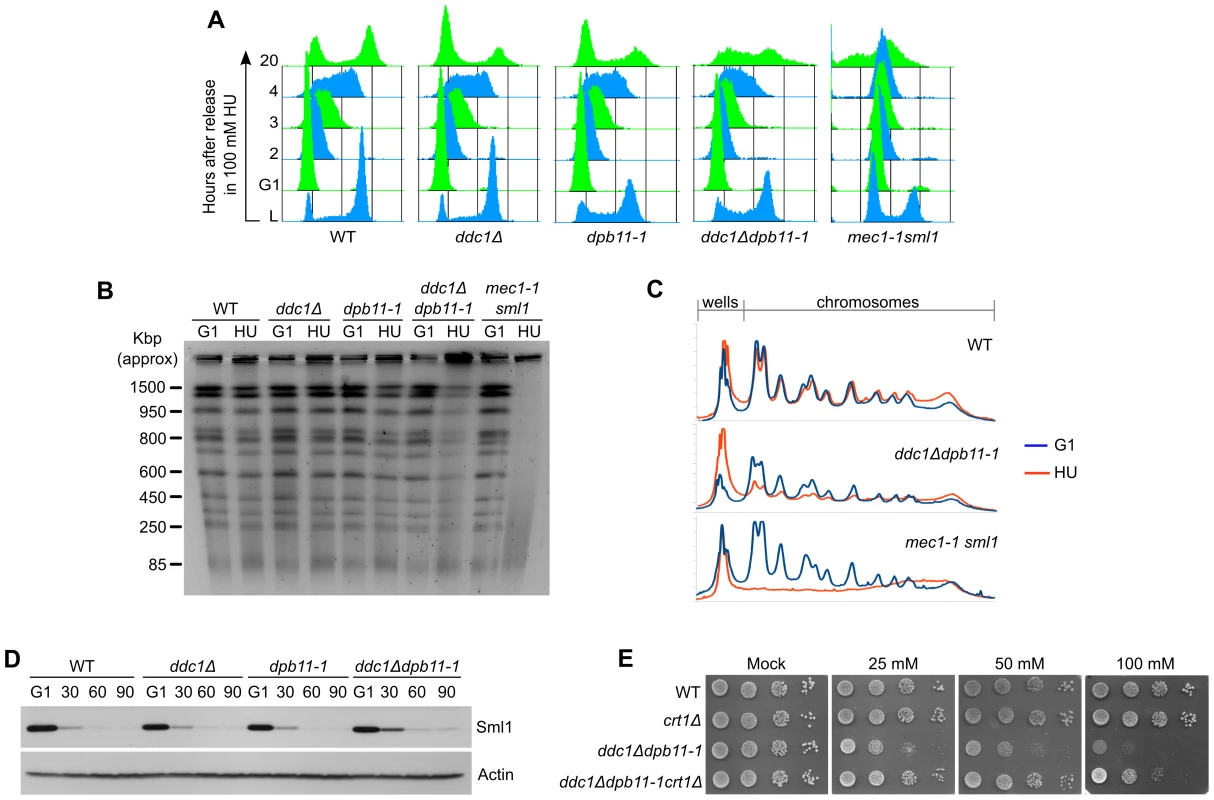
(A) K699 (WT), YFP20 (dpb11-1), YAN21/8d (ddc1Δ), YFP62/1d (ddc1Δdpb11-1) and YMIC5A3 (mec1-1) were synchronized in G1 with α-factor and released into fresh medium supplemented with 100 mM HU. At the indicated time-points after the release, progression into the cell cycle was monitored by FACS analysis. (B) DNA extracted from the G1-arrested cells and from the cells released for 20 hours in HU was separated by PFGE and stained with ethidium bromide. (C) A plot representing the intensity profile of the significant gel lanes in panel C. (D) K699 (WT), YFP20 (dpb11-1), YAN21/8d (ddc1Δ) and YFP62/1d (ddc1Δdpb11-1) strains were synchronized in G1 with α-factor and released into fresh medium supplemented with 200 mM HU. At the indicated time-points protein extracts were prepared and probed with α-Sml1 antibodies to measure the levels of Sml1 and with α-actin antibodies as a loading control. (E) Ten fold serial dilutions of overnight cultures of strains K699 (WT), YFP328 (crt1Δ), YFP62/1d (ddc1Δdpb11-1) and YFP330 (ddc1Δdpb11-1crt1Δ) were spotted on YPD plates supplemented with HU at the indicated concentration. Survival was assayed by monitoring cell growth after 6 days. Sld2 and DNA polymerase ε are required for Ddc1-independent checkpoint activation
Sld2/Drc1 and Polε participate in replication checkpoint signaling [24], [25]. Moreover, these factors were recently found to be part of the same pre-loading complex, together with Dpb11 and GINS [44]. An interesting possibility is that Sld2 and Polε exert their checkpoint function by controlling Dpb11-mediated Mec1 activation. To address this hypothesis we combined the drc1-1 allele with the DDC1 deletion. As it is shown in Figure 5A, similarly to what reported above for Dpb11, Sld2 also acts in a pathway that is parallel to that involving Ddc1; indeed, residual Rad53 phosphorylation present in ddc1Δ cells depends on Sld2. Moreover, drc1-1 cells do not show hyperphosphorylation of Rad9 in response to HU treatment, excluding the possibility of a secondary DNA damage response (Figure S4A). In agreement with these data, deletion of DDC1 displays a synergistic sensitivity to HU when combined with the drc1-1 mutation and the HU sensitivity of the double mutant strain is very similar to that observed for ddc1Δdpb11-1 cells (Figure 5B).
Fig. 5. Polε associated proteins are involved in the 9-1-1–independent checkpoint signaling branch. 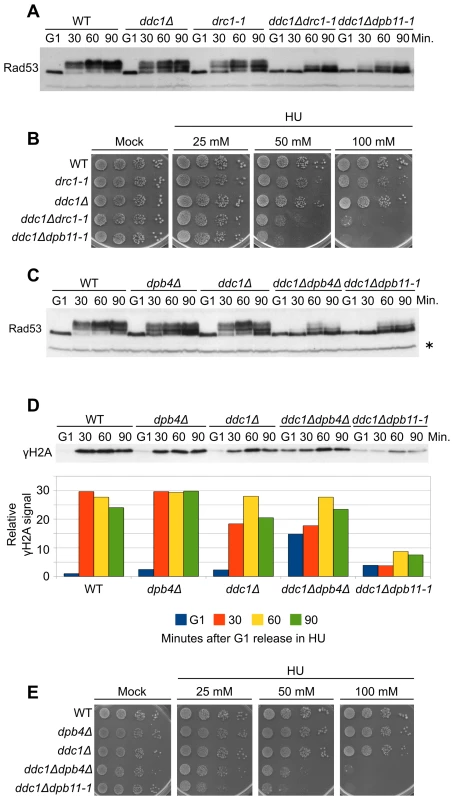
(A) Strains K699 (WT), Y799 (drc1-1), YAN21/8d (ddc1Δ), YFP218/1a (ddc1Δdrc1-1) and YFP62/1d (ddc1Δdpb11-1) were cultured to mid-log phase, synchronized in G1 with α-factor and released into fresh medium supplemented with 200 mM HU. At the indicated time-points Rad53 phosphorylation was assayed by SDS-PAGE and Western blotting. (B) Ten-fold serial dilutions of overnight cultures of the strains in panel A were spotted on YPD plates supplemented with HU at the indicated concentration. Survival was assayed by monitoring cell growth after 6 days. (C) Strains K699 (WT), YFP167/1a (dpb4Δ), YAN21/8d (ddc1Δ), YFP206/1a (ddc1Δdpb4Δ) and YFP62/1d (ddc1Δdpb11-1) were cultured to mid-log phase, synchronized in G1 with α-factor and released into fresh medium supplemented with 200 mM HU. At the indicated time-points, protein extracts were prepared and separated by SDS-PAGE. Rad53 phosphorylation was assayed by western blotting. (D) The same filter was probed for Mec1 activity by testing histone H2A phosphorylation. A quantification of γH2A, using α-Rad53 cross reacting band as loading control is shown in the lower panel. The values indicate the fold increase respect to the WT G1 sample. (E) Ten-fold serial dilutions of overnight cultures of the strains in panel C were spotted on YPD plates supplemented with HU at the indicated concentration. Cell survival was assayed monitoring cell growth after 6 days. In all the relevant panels the loading control is indicated by an asterisk. The checkpoint function of Polε appears to reside in the C-terminal domain of Pol2, which is bound, either directly or indirectly, by the three smaller subunits Dpb2, Dpb3 and Dpb4 and by Dpb11 [31], [45]. To assess if Polε participates in the Dpb11 signaling branch via its minor subunits, we deleted DPB4 in combination with the DDC1 deletion. Figure 5C shows that Rad53 phosphorylation is severely impaired in the double mutant ddc1Δdpb4Δ, closely resembling the phenotype of a ddc1Δdpb11-1 mutant. The same effect is measured by testing H2A phosphorylation in HU-treated samples (Figure 5D). The signals obtained for each time-point are quantified with respect to the signal detected in G1-arrested cells, in order to compensate for the higher basal level of γH2A observed in ddc1Δdpb4Δ double mutant cells in the absence of any treatment. Moreover, no unscheduled DNA damage checkpoint activation occurs, since no Rad9 phosphorylation is detected in dpb4Δ or dpb4Δ ddc1Δ cells treated with HU (Figure S4B). Finally, the ddc1Δdpb4Δ strain shows an HU sensitivity similar to that found in ddc1Δdpb11-1 cells (Figure 5E).
Discussion
Apical checkpoint kinases (Mec1/Tel1 in budding yeast, ATR/ATM in humans) convert a structural signal coming from damaged DNA to a phosphorylation-based signaling cascade, and a large amount of work has been devoted to clarify the underlying mechanisms. Initially, the attention was focused on the recruitment of these kinases to damaged DNA [15], based on the assumption that binding to damaged chromatin sites would lead to their activation. More recently, the finding that Dpb11/TopBP1 stimulates Mec1 activity suggests a more complex scenario [40], [41], [42].
In vitro data obtained in Xenopus egg and mammalian cell extracts demonstrate the ability of TopBP1 to increase Mec1 kinase activity [35], [38]. The significance of this TopBP1 function does not appear to be specific for multicellular eukaryotes, since an interaction between Rad4/Cut5 and the checkpoint sensor kinase Rad3-Rad26 has also been found in S. pombe [46], [47]. More recently, in S. cerevisiae cells, Dpb11 has been demonstrated to contain an ATR activation domain (AAD), which is sufficient to promote Mec1 activation in vitro [41], [42]. These findings apparently contradict a previous observation that Mec1 can normally phosphorylate Ddc2 in a dpb11-1 mutant, lacking part of the AAD, after UV damage in M phase [16], while in our hands DDC1 deletion prevents Ddc2 phosphorylation (unpublished observation). Two explanations can be envisaged: in dpb11-1 mutant cells, Mec1 activity may be sufficient to phosphorylate Ddc2, while being defective towards other substrates; alternatively, Dpb11 may play only a marginal role in response to UV irradiation in M phase. We favored the second hypothesis because dpb11-1 mutant cells are mildly sensitive to UV irradiation and are proficient in the G2/M checkpoint; moreover, the 9-1-1 complex has also been identified as an activator of Mec1 in vitro [39], [40] and may play a prominent role in M phase. If this assumption is correct, Dpb11 could play a role in Mec1 activation in response to a different kind of damage or in other cell cycle phases. Interestingly, it was demonstrated that the dpb11-1 temperature-sensitive mutant is defective in checkpoint activation after replication stress caused by HU treatment at the restrictive temperature (36°C), while it is only mildly sensitive to the drug at permissive temperature ([23], [25] and Figure 2B).
To better understand the process of Mec1 activation in vivo after DNA damage or replication stress, we analyzed the relative functions of the two putative Mec1 activators: Dpb11 and the 9-1-1 complex. We extended our previous analysis by monitoring, in different cell cycle phases, a direct target of Mec1 kinase (histone H2A) as marker of Mec1 activity. We found that, both in G1 and in M phase, the 9-1-1 complex is absolutely required for Mec1 activation in response to UV treatment, while the contribution of Dpb11 AAD is only partial (∼50%) and restricted to G1. These in vivo findings are in agreement with the current activation model inferred from in vitro biochemical data [39], indicating that 9-1-1 can stimulate Mec1 through both Dpb11-dependent and -independent pathways in G1 (Figure 6, left). Differently, in M phase, the ATR activation domain of Dpb11 is dispensable for full Mec1 activation, which relies mainly on the presence of 9-1-1 (Figure 6, right). In fact, the residual UV-induced H2A phosphorylation detectable in the ddc1Δ strain, is dependent upon the Tel1 kinase (Figure 1). Different requirements for Mec1 activation in G1 and in M phase may reflect differences in CDK-controlled processing of DNA filament ends to generate the substrate detected by checkpoint factors [48], [49].
Fig. 6. A model for 9-1-1 and Dpb11 function in Mec1 activation. 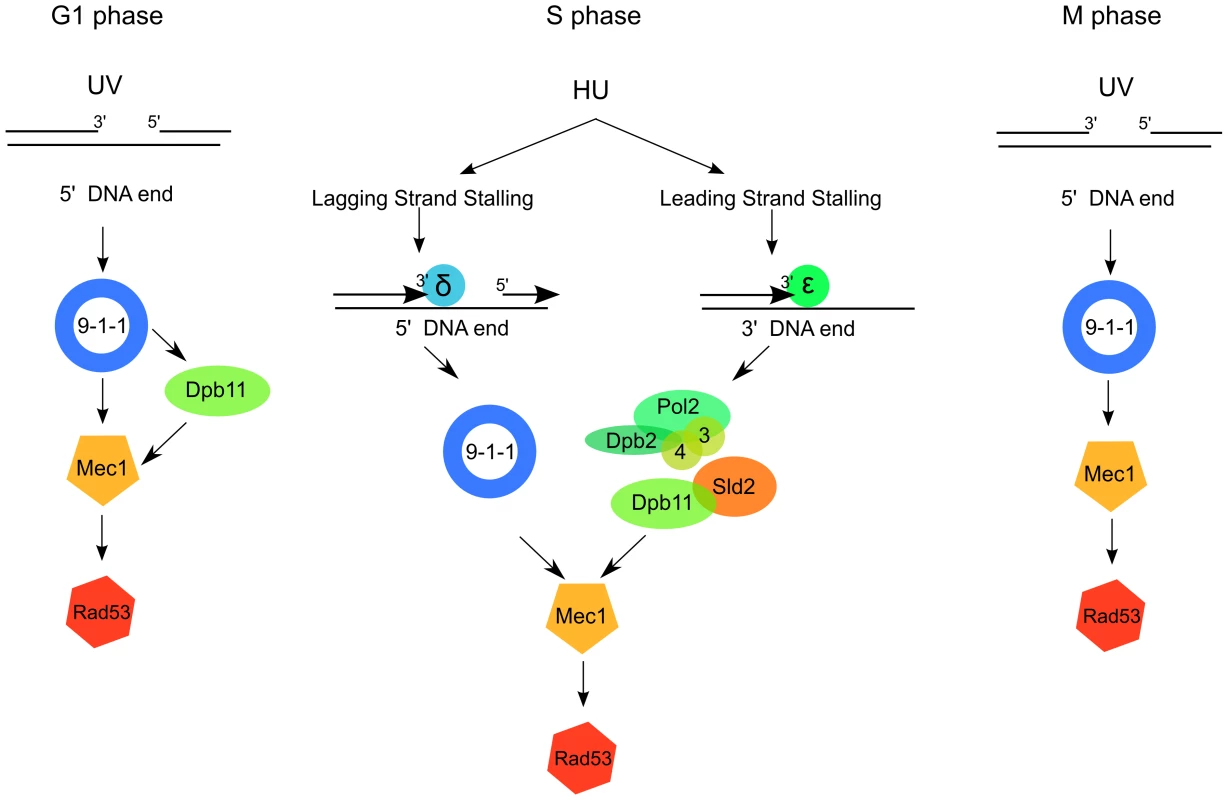
After UV irradiation in G1, Mec1 is activated by the 9-1-1 complex both directly and through the Dpb11 C-terminus (right); in M phase Mec1 activation is achieved mainly through the 9-1-1 complex, independently of Dpb11 (left). In S phase Dpb11 and the 9-1-1 complex signal replication stress to Mec1 independently from each other, likely because the detection of replication stress occurs independently on the leading and lagging strands. 9-1-1 complex could signal replication stress on the lagging strand, where the 5′ ends necessary for its loading are generated as the result of discontinuous replication. Dpb11, instead, could signal replication stress on the leading strand together with the interacting Polε. To complete studying of the pathways leading to Mec1 activation in different cell cycle stages, we analyzed the contribution of Dpb11 and Ddc1 to Mec1 activation in S phase cells challenged with replication stress. HU decreases the cellular concentration of dNTPs available for DNA synthesis and yeast cells respond by activating the replication checkpoint.
In vivo analysis of the phosphorylation state of two Mec1 substrates, H2A and Rad53, indicates that Dpb11 and 9-1-1 participate in Mec1 activation in response to HU treatment independently of each other in two parallel pathways. The possibility that dpb11-1 may cause problems to the replication process triggering a DNA damage response mediated by the 9-1-1 complex, similarly to what happens in mrc1Δ cells [22], seems unlikely. In fact, the Rad9 DNA damage-specific adaptor does not become hyperphosphorylated in both dpb11-1 and ddc1Δ single mutants. In agreement with such observation, rad9Δdpb11-1 cells are much less sensitive to HU than ddc1Δ dpb11-1 cells (Figure 2 and [43]).
We report that the HU sensitivity of ddc1Δ dpb11-1 strain is not due to replication fork collapse or premature elongation of the mitotic spindle (Figure 3 and Figure S2), two phenotypes characteristic of mutants defective in the replication checkpoint [7], [12]. Accordingly, the HU sensitivity of ddc1Δdpb11-1 double mutant cells, differently from that of a mec1-1sml1 strain, is not detectable in the case of transient HU treatment. This observation suggests that another Rad53 function activated by the replication checkpoint, and different from that responding to temporary fork arrest, is essential for sustaining growth in the constant presence of hydroxyurea. Indeed, ddc1Δ dpb11-1 double mutant cells grown in the presence of HU show defects in completing replication and accumulate replication intermediates. Moreover, ddc1Δ dpb11-1 cells are unable to counteract the effect of HU by upregulating ribonucleotide reductase. Interestingly, CRT1 deletion partially suppresses HU sensitivity of the double mutant strain (Figure 4E).
To obtain more insights on the pathways leading to Ddc1-dependent and Dpb11-dependent activation of replication checkpoint and to identify possible mechanisms specific for lagging or leading strand fork arrest, we analyzed mutants in the genes coding for proteins that are known to be involved in leading strand replication. During initiation of DNA replication, Dpb11 interacts with both Sld2 and Sld3 in a phosphorylation-dependent manner, a process that is required for origin firing [26], [27]. Moreover, temperature sensitive drc1-1 strains, mutated in Sld2, display the same checkpoint-deficient phenotype of dpb11-1 cells, when treated with HU at the non-permissive temperature, (Figure 5 and [25]). We tested whether Sld2 functions with Dpb11 in the same 9-1-1-independent pathway for Mec1 activation. Combining the drc1-1 allele with the DDC1 deletion, we found that ddc1Δ drc1-1 double mutant cells display the same Rad53 phosphorylation defect and the same HU sensitivity of a ddc1Δdpb11-1 strain, indicating that Mec1 activation by Dpb11 also requires Sld2 (Figure 5).
Mutants in the Pol2 C-terminus, the enzyme replicating the leading strand [50], are defective in the establishment of the replication checkpoint [24], [50] and this protein region of Pol2 was suggested to be involved in its interaction with other three Polε subunits: the essential Dpb2 protein and the non-essential Dpb3 and Dpb4 subunits [31], [45], [51]. Disruption of the DPB4 gene in a ddc1Δ background leads to identical phenotypes to the one observed in ddc1Δ dpb1-1 and ddc1Δ drc1-1, strongly suggesting that the 9-1-1-independent pathway involves leading strand replication factors. The observations that Dpb11 acts directly on Mec1 activity [41], [42] and that, in the dpb11-1 mutant, Polε seems to be normally loaded onto replication origins [52], strongly suggest that Dpb4, and possibly Sld2, function upstream of Dpb11 during checkpoint signaling. Unfortunately, it is impossible to perform a complete formal epistatic analysis as the dpb11-1 mutation also affects replication initiation and deletion of DPB4 or mutations in SLD2 are synthetic lethal when combined with the dpb11-1 allele [28], [53].
In conclusion our data suggest that during exposure to hydroxyurea, two independent pathways sense replication stress and signal for Mec1 activation. The first pathway depends on 9-1-1, which is known to be loaded at the 5′ of primer-template junctions, when RPA covers ssDNA ahead of the primer [34]. During unchallenged DNA replication these structures are normally formed on the lagging strand as a consequence of discontinuous DNA synthesis, and rapidly removed by refilling polymerase activity. Inhibition of DNA polymerization by HU likely stabilizes the 5′ DNA end providing the structure required for 9-1-1 loading. On the other hand, the higher processivity of leading strand synthesis makes it likely that the nearest 5′ end will be far away from the site of polymerase stalling, where ssDNA is generated and the Mec1-Ddc2 complex should be recruited. The absence of such structure could prevent the 9-1-1-dependent Mec1 activation. In this case a pathway requiring the leading strand factors Dpb4, Dpb11 and Sld2 becomes relevant to induce Mec1 activation (Figure 6, center). The hypothesis that Polε, Sld2 and Dpb11 work together in sensing replication stress is supported by the recent finding that an unstable complex containing Dpb11, Sld2, Polε and GINS is formed at the beginning of S-phase [44]. Moreover, the demonstration that under unstressed conditions Polε acts on the leading strand while Polδ works on the lagging strand [50], [54] supports the hypothesis that Polε and its interacting subunits may function in sensing replication stress on the leading strand, while the 9-1-1 complex may be more important to detect lagging strand fork arrest. Additional work will be needed to confirm this model and to identify the mechanisms leading to Dpb11 recruitment at the sites of replication fork stalling, since Dpb11 appears to co-localize with Polε during initiation of DNA replication, but not during elongation [52].
Materials and Methods
Yeast strains
All of the strains used in this work are derivatives of W303 (K699 [MATa ade2-1 trp1-1 can1-100 leu2-3,12 his3-11,15 ura3]) and are listed in Table 1. Deletion strains were generated by using the one-step PCR system [55] or by genetic crossing.
Tab. 1. Strains used in this work. 
Cell cycle synchronization and HU treatment
Cells were grown overnight at 25°C to a concentration of 5×106 cells/ml and arrested in G1 with 5 µg/ml α-factor for three hours. 60 ml of cultures were spun and resuspended in the same volume of YPD supplemented with HU (200 mM or 100 mM, depending on the experiment). 20 ml samples were taken every 30 minutes after the release. In the case of untreated samples cells were released in fresh YPD +10 µg/ml nocodazole and every 5 minutes samples were taken for SDS-PAGE and FACS analysis.
Cell cycle arrest and DNA damage treatments
Cells were grown in YPD medium at 25°C to a concentration of 5×106 cells/ml and arrested with nocodazole or α-factor (20 µg/ml). 50 ml of cultures were spun, resuspended in 500 µl of sterile water, and plated on a Petri dish (14-cm diameter). Rapidly, a 15 ml untreated sample was taken. Plates were irradiated at 75 J/m2 and cells were resuspended in 50 ml of YPD + nocodazole or α factor. Three 15 ml samples were taken every 10 minutes after irradiation.
SDS page, western blot, and quantification
Trichloroacetic acid protein extracts [56] were separated by SDS-PAGE; for the analysis of Rad9 phosphorylation, NuPAGE Tris-Acetate 3–8% gels (Invitrogen) were used following the manufacturer's instructions. Western blotting was performed with anti-Rad53, anti-H2A-S129 (Abcam #15083), anti-Actin (Sigma #A2066), anti-Sml1 and anti-Rad9 antibodies, using standard techniques. Values of phospho-H2A levels were obtained by quantifying the signal in the corresponding lanes using Quantity One software (BioRad) and normalizing it, first on the loading controls and then on the level of phospho-H2A in the untreated/G1-arrested sample of each strain.
Immunofluorescence
1 ml of a 5×106 cells/ml culture were fixed overnight at 4°C with fixation buffer (3,7% formaldehyde, 0,1 M K-phosphate pH 6,4, 0,5 mM MgCl2). Cells were then washed three times with wash buffer (0,1 M K-phosphate pH 6,4, 0,5 mM MgCl2), one time with spheroplasting solution (1,4 M sorbitol, 0,1 M K-phosphate pH 6,4, 0,5 mM MgCl2) and resuspended in 200 µl of the same solution. Spheroplasts were prepared using 5 µl of 10 mg/ml Zymolyase at 37°C. Spheroplasts were washed with the same solution and used to prepare multi-well immunofluorescence slides which were incubated overnight with α-tubulin antibody (YOL1/34, Seralab) diluted 1∶100 in PBS-5%BSA.
Sensitivity assay
HU plates were prepared by serial dilutions of the 2 M stock solution. Plates containing 25 mM, 50 mM and 100 mM HU were prepared. Overnight grown cultures were diluted to 1×106 cell/ml, then 10-fold serial dilutions were prepared and 10 µl of the suspensions were spotted on HU plates, which were incubated at 25°C. Images were taken 2 to 7 days later.
Pulsed field gel electrophoresis
Agarose plugs containing yeast chromosomes were prepared as described previously [57]. These were incubated overnight at 37°C in 0.5 ml/plug TE containing 1 mg/ml RNAseA. After extensive washes with Wash Buffer (10 mM Tris-HCl pH 7.5 50 mM EDTA), plugs were loaded on 1% agarose gel and sealed in the wells with a solution of 1% LMP agarose in TBE 0.5X. Gels were run at 4°C for 24 h at 165 V, with 60 seconds pulses for 12 h and 90 second pulses for 12 h, using an Amersham Gene Navigator system.
Supporting Information
Zdroje
1. ChaRSKlecknerN 2002 ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297 602 606
2. BrewerBJFangmanWL 1988 A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55 637 643
3. KrakoffIHBrownNCReichardP 1968 Inhibition of ribonucleoside diphosphate reductase by hydroxyurea. Cancer Res 28 1559 1565
4. BranzeiDFoianiM 2009 The checkpoint response to replication stress. DNA Repair (Amst) 8 1038 1046
5. SogoJMLopesMFoianiM 2002 Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297 599 602
6. DesanyBAAlcasabasAABachantJBElledgeSJ 1998 Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev 12 2956 2970
7. LopesMCotta-RamusinoCPellicioliALiberiGPlevaniP 2001 The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412 557 561
8. LuccaCVanoliFCotta-RamusinoCPellicioliALiberiG 2004 Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23 1206 1213
9. HuangMZhouZElledgeSJ 1998 The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94 595 605
10. ZhaoXChabesADomkinVThelanderLRothsteinR 2001 The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J 20 3544 3553
11. SantocanaleCDiffleyJF 1998 A Mec1 - and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395 615 618
12. AllenJBZhouZSiedeWFriedbergECElledgeSJ 1994 The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev 8 2401 2415
13. BachantJJessenSRKavanaughSEFieldingCS 2005 The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress. J Cell Biol 168 999 1012
14. WeinertTAKiserGLHartwellLH 1994 Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev 8 652 665
15. ZouLElledgeSJ 2003 Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300 1542 1548
16. PudduFGranataMNolaLDBalestriniAPiergiovanniG 2008 Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol 28 4782 4793
17. HuyenYZgheibODitullioRAGorgoulisVGZacharatosP 2004 Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432 406 411
18. GiannattasioMLazzaroFPlevaniPMuzi-FalconiM 2005 The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem 280 9879 9886
19. DuL-LNakamuraTMRussellP 2006 Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev 20 1583 1596
20. GranataMLazzaroFNovarinaDPanigadaDPudduF 2010 Dynamics of Rad9 Chromatin Binding and Checkpoint Function are Mediated by its Dimerization and are Cell Cycle-Regulated by CDK1 Activity. PLoS Genet 6 e1001047 doi:10.1371/journal.pgen.1001047
21. PellicioliALuccaCLiberiGMariniFLopesM 1999 Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J 18 6561 6572
22. AlcasabasAAOsbornAJBachantJHuFWerlerPJ 2001 Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3 958 965
23. ArakiHLeemSHPhongdaraASuginoA 1995 Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci U S A 92 11791 11795
24. NavasTAZhouZElledgeSJ 1995 DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell 80 29 39
25. WangHElledgeSJ 1999 DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 96 3824 3829
26. TanakaSUmemoriTHiraiKMuramatsuSKamimuraY 2007 CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445 328 332
27. ZegermanPDiffleyJFX 2007 Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445 281 285
28. OhyaTMakiSKawasakiYSuginoA 2000 Structure and function of the fourth subunit (Dpb4p) of DNA polymerase epsilon in Saccharomyces cerevisiae. Nucleic Acids Res 28 3846 3852
29. IidaTArakiH 2004 Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol 24 217 227
30. AsturiasFJCheungIKSabouriNChilkovaOWepploD 2006 Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol 13 35 43
31. DuaREdwardsSLevyDLCampbellJL 2000 Subunit interactions within the Saccharomyces cerevisiae DNA polymerase epsilon (pol epsilon) complex. Demonstration of a dimeric pol epsilon. J Biol Chem 275 28816 28825
32. KestiTFlickKKeränenSSyväojaJEWittenbergC 1999 DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell 3 679 685
33. ZouLLiuDElledgeSJ 2003 Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A 100 13827 13832
34. MajkaJBinzSKWoldMSBurgersPM 2006 RPA directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem 281 27855 27861
35. KumagaiALeeJYooHYDunphyWG 2006 TopBP1 activates the ATR-ATRIP complex. Cell 124 943 955
36. DelacroixSWagnerJMKobayashiMichi YamamotoKKarnitzLM 2007 The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 21 1472 1477
37. LeeJKumagaiADunphyWG 2007 The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem 282 28036 28044
38. MordesDAGlickGGZhaoRCortezD 2008 TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22 1478 1489
39. Navadgi-PatilVMBurgersPM 2009 The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell 36 743 753
40. MajkaJNiedziela-MajkaABurgersPMJ 2006 The Checkpoint Clamp Activates Mec1 Kinase during Initiation of the DNA Damage Checkpoint. Mol Cell 24 891 901
41. MordesDANamEACortezD 2008 Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci U S A 105 18730 18734
42. Navadgi-PatilVMBurgersPM 2008 Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem 283 35853 35859
43. WangHElledgeSJ 2002 Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics 160 1295 1304
44. MuramatsuSHiraiKTakY-SKamimuraYArakiH 2010 CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol epsilon, and GINS in budding yeast. Genes Dev 24 602 612
45. EdwardsSLiCMLevyDLBrownJSnowPM 2003 Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion. Mol Cell Biol 23 2733 2748
46. FuruyaKPoiteleaMGuoLCaspariTCarrAM 2004 Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev 18 1154 1164
47. TaricaniLWangTSF 2006 Rad4TopBP1, a scaffold protein, plays separate roles in DNA damage and replication checkpoints and DNA replication. Mol Biol Cell 17 3456 3468
48. IraGPellicioliABalijjaAWangXFioraniS 2004 DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431 1011 1017
49. AylonYLiefshitzBKupiecM 2004 The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23 4868 4875
50. McElhinnySANGordeninDAStithCMBurgersPMJKunkelTA 2008 Division of labor at the eukaryotic replication fork. Mol Cell 30 137 144
51. DuaRLevyDLCampbellJL 1998 Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase epsilon in DNA replication and the S/M checkpoint pathway. J Biol Chem 273 30046 30055
52. MasumotoHSuginoAArakiH 2000 Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol Cell Biol 20 2809 2817
53. KamimuraYMasumotoHSuginoAArakiH 1998 Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol 18 6102 6109
54. BurgersPM 2008 Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem 284 4041 4045
55. LongtineMSMcKenzieADemariniDJShahNGWachA 1998 Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953 961
56. FalconiMMPiseriAFerrariMLucchiniGPlevaniP 1993 De novo synthesis of budding yeast DNA polymerase alpha and POL1 transcription at the G1/S boundary are not required for entrance into S phase. Proc Natl Acad Sci U S A 90 10519 10523
57. LengronneAPaseroPBensimonASchwobE 2001 Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res 29 1433 1442
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and PhosphodiesterasesČlánek Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



