-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Initial Mutations Direct Alternative Pathways of Protein Evolution
Whether evolution is erratic due to random historical details, or is repeatedly directed along similar paths by certain constraints, remains unclear. Epistasis (i.e. non-additive interaction between mutations that affect fitness) is a mechanism that can contribute to both scenarios. Epistasis can constrain the type and order of selected mutations, but it can also make adaptive trajectories contingent upon the first random substitution. This effect is particularly strong under sign epistasis, when the sign of the fitness effects of a mutation depends on its genetic background. In the current study, we examine how epistatic interactions between mutations determine alternative evolutionary pathways, using in vitro evolution of the antibiotic resistance enzyme TEM-1 β-lactamase. First, we describe the diversity of adaptive pathways among replicate lines during evolution for resistance to a novel antibiotic (cefotaxime). Consistent with the prediction of epistatic constraints, most lines increased resistance by acquiring three mutations in a fixed order. However, a few lines deviated from this pattern. Next, to test whether negative interactions between alternative initial substitutions drive this divergence, alleles containing initial substitutions from the deviating lines were evolved under identical conditions. Indeed, these alternative initial substitutions consistently led to lower adaptive peaks, involving more and other substitutions than those observed in the common pathway. We found that a combination of decreased enzymatic activity and lower folding cooperativity underlies negative sign epistasis in the clash between key mutations in the common and deviating lines (Gly238Ser and Arg164Ser, respectively). Our results demonstrate that epistasis contributes to contingency in protein evolution by amplifying the selective consequences of random mutations.
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1001321
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001321Summary
Whether evolution is erratic due to random historical details, or is repeatedly directed along similar paths by certain constraints, remains unclear. Epistasis (i.e. non-additive interaction between mutations that affect fitness) is a mechanism that can contribute to both scenarios. Epistasis can constrain the type and order of selected mutations, but it can also make adaptive trajectories contingent upon the first random substitution. This effect is particularly strong under sign epistasis, when the sign of the fitness effects of a mutation depends on its genetic background. In the current study, we examine how epistatic interactions between mutations determine alternative evolutionary pathways, using in vitro evolution of the antibiotic resistance enzyme TEM-1 β-lactamase. First, we describe the diversity of adaptive pathways among replicate lines during evolution for resistance to a novel antibiotic (cefotaxime). Consistent with the prediction of epistatic constraints, most lines increased resistance by acquiring three mutations in a fixed order. However, a few lines deviated from this pattern. Next, to test whether negative interactions between alternative initial substitutions drive this divergence, alleles containing initial substitutions from the deviating lines were evolved under identical conditions. Indeed, these alternative initial substitutions consistently led to lower adaptive peaks, involving more and other substitutions than those observed in the common pathway. We found that a combination of decreased enzymatic activity and lower folding cooperativity underlies negative sign epistasis in the clash between key mutations in the common and deviating lines (Gly238Ser and Arg164Ser, respectively). Our results demonstrate that epistasis contributes to contingency in protein evolution by amplifying the selective consequences of random mutations.
Introduction
Understanding the factors that determine the outcome of evolution is a long-term goal of evolutionary biology. Epistasis (i.e. non-additive interactions between mutations that affect fitness) is one such determinant. Epistasis can affect the type and order of mutations that can be fixed by natural selection [1], [2]. As a result, it affects the dynamics of evolution, for instance by causing a mutation to have smaller fitness benefits in the presence than in the absence of other mutations [3], [4]. Under a specific type of epistasis called sign epistasis, interactions affect not only the size, but also the sign of the fitness effect of mutations [5]. Sign epistasis introduces particularly strong evolutionary constraints, and may create fitness landscapes with multiple adaptive peaks isolated by valleys of lower fitness [5], [6]. In such rugged fitness landscapes, epistasis can theoretically cause entire mutational pathways to be contingent on the chance occurrence of initial mutations due to a combination of two effects. First, once an initial mutation is fixed, epistasis prevents the selection of alternative initial substitutions. Next, it concurrently facilitates the selection of mutations that are beneficial in the background of this particular initial substitution, but that may be detrimental (or less beneficial) in the background of alternative initial substitutions.
The predicted evolutionary constraints stemming from epistasis also depend on the available genetic variation. For example, a population that is stuck on a suboptimal adaptive peak can escape from this peak and cross the surrounding fitness valley when mutants carrying useful combinations of mutations can be produced. This may be expected to occur in large populations with a high mutation rate [7], or possibly when there is recombination [8], [9]. For asexual populations, the evolutionary constraints determined by epistasis are the most pronounced when the supply of mutations is low and mutations arise and go to fixation one at a time. In this case, even the smallest fitness valleys form obstacles for natural selection.
To demonstrate how epistatic constraints affect evolution, one would ideally need to observe the full sequence of mutational events and subsequently test the evolutionary consequences of specific mutational interactions. While sequence divergence data may be used to study the role of epistasis [10], experimental evolution with microbes (where replicate lines evolve under identical conditions) offers a powerful experimental methodology. Such studies have shown instances of both divergent and parallel evolution, including changes in fitness, cell size and genotype [11]–[16]. A remarkable case of historical contingency was recently reported in a long-term evolution experiment with Escherichia coli, when investigators found that after more than 30,000 generations, multiple ‘potentiating’ mutations allowed one of twelve populations to evolve a key metabolic innovation [17]. Despite of such findings, identifying the exact order of mutations, and testing their individual and combined fitness effects has been challenging even with simple organisms.
Here, we used in vitro evolution of an antibiotic resistance enzyme to study the effect of epistasis on the process of mutational pathway diversification. We used TEM-1 β-lactamase, an enzyme that catalyzes the hydrolysis of ampicillin, and studied its evolution towards increased activity against the semi-synthetic antibiotic cefotaxime. Briefly, we introduced random mutations using error-prone PCR and selected mutant alleles that mediate bacterial growth in a gradient of cefotaxime concentrations. After each round of evolution, a fixed mutant allele from each line was isolated from the highest antibiotic concentration in which bacterial growth was observed. This system offers the opportunity to study entire adaptive pathways at the level of phenotype (i.e. antibiotic resistance) and genotype. Previous studies used the same system to develop protocols for directed evolution [18], to investigate evolutionary scenarios [19], [20] and to explore the potential of evolution of resistance to a range of β-lactam antibiotics [21], [22]. Collectively, these studies showed the contribution of both unique and repetitive substitutions to TEM-1's adaptation. They also identified a five-fold mutant (containing a promoter mutation and substitutions A42G, E104K, M182T and G238S) with an extremely high cefotaxime resistance [18], [23], [24]. Measurement of the resistance of constructed mutants carrying subsets of these mutations revealed substantial sign epistasis [24]–[26] and predicted the inaccessibility of most mutational pathways towards the five-fold mutant [24]. Notably, several other mutations in addition to the five mentioned above are capable of increasing cefotaxime resistance, independently or in combination with other substitutions [25], [26]. We were therefore interested in assessing whether evolution allowing all possible mutations would always lead to the fivefold mutant or whether alternative pathways to cefotaxime resistance exist, and to what extent epistasis plays a role in this.
Results
Diversity of mutational pathways
Twelve replicate lines of the TEM-1 gene were subjected to three rounds of error-prone PCR followed by selection for increased cefotaxime resistance. To broadly explore existing adaptive pathways, we varied the mutation rate (∼2 and ∼6 nucleotide substitutions per amplicon per PCR) and introduced a simplified form of recombination in half of the lines (see Materials and Methods), although neither of these two treatments appeared to affect the final level of adaptation, as measured by increases of the minimal inhibitory concentration (MIC) for cefotaxime (mutation rate: Mann-Whitney U = 12, two-tailed P = 0.394; recombination: U = 12, P = 0.394). After each round of evolution, a single clone was picked from the highest cefotaxime concentration that still permitted growth. We used this type of bottleneck selection via the fixation of a single clone [19], because it allowed us to follow the mutational pathway of the evolving genotypes. In addition, sequencing indicated that the evolved populations were dominated by a single clone (see Text S1). Plasmid from the isolated clone was purified and used as the template for the next round of evolution. Selected alleles were sequenced and assayed for increased cefotaxime resistance by measuring the MIC after cloning into the original vector and transformation into naïve bacterial cells. Figure 1A shows the changes in MIC for all lines, which levels off after the second round of evolution, suggesting the approach of one or more fitness peaks. This pattern is mirrored in the number of amino acid substitutions recorded per round for the twelve lines (30 substitutions after the first round, 29 after the second round, and 8 after the third, see Table S2). The amino acid substitutions show a remarkable pattern of parallel response. Seven of the twelve lines contained the same three substitutions, with G238S always occurring in the first round, often together with M182T, followed by E104K in the second round (see Figure 2A, mutation details and silent mutations for this experiment and subsequent in vitro evolution experiments can be found in Table S1). Since the promoter region was not allowed to evolve in our experiments, the main difference with the previously described adaptive peak (containing A42G, E104K, M182T and G238S and a promoter mutation) is the absence of A42G. There is evidence that A42G is a so-called ‘global suppressor’ substitution [27] and since many such substitutions with similar functional effects exist [28], [29], we suspect that in our experiments A42G was sometimes replaced by other global suppressor substitutions (e.g. H153L in line 1 of Figure 2A). We can not explain the absence of A42G in the lines that contain E104K, M182T and G238S after two rounds of evolution and remain unchanged after the third round (lines 5, 9 and 10 in Figure 2A). However, we would like to point out that the combination of E104K, M182T, and G238S has occasionally been reported as the evolutionary end point in other studies [26], [30]. Overall, the striking pattern of parallel evolution is consistent with adaptive constraints from epistasis. Five lines (numbers 3, 4, 7, 11 and 12) deviate from this pattern, because they lack one or more of the three parallel substitutions and evolve to lower resistance levels (Mann-Whitney, U = 1, one-tailed P = 0.003).
Fig. 1. Cefotaxime MIC of selected TEM alleles. 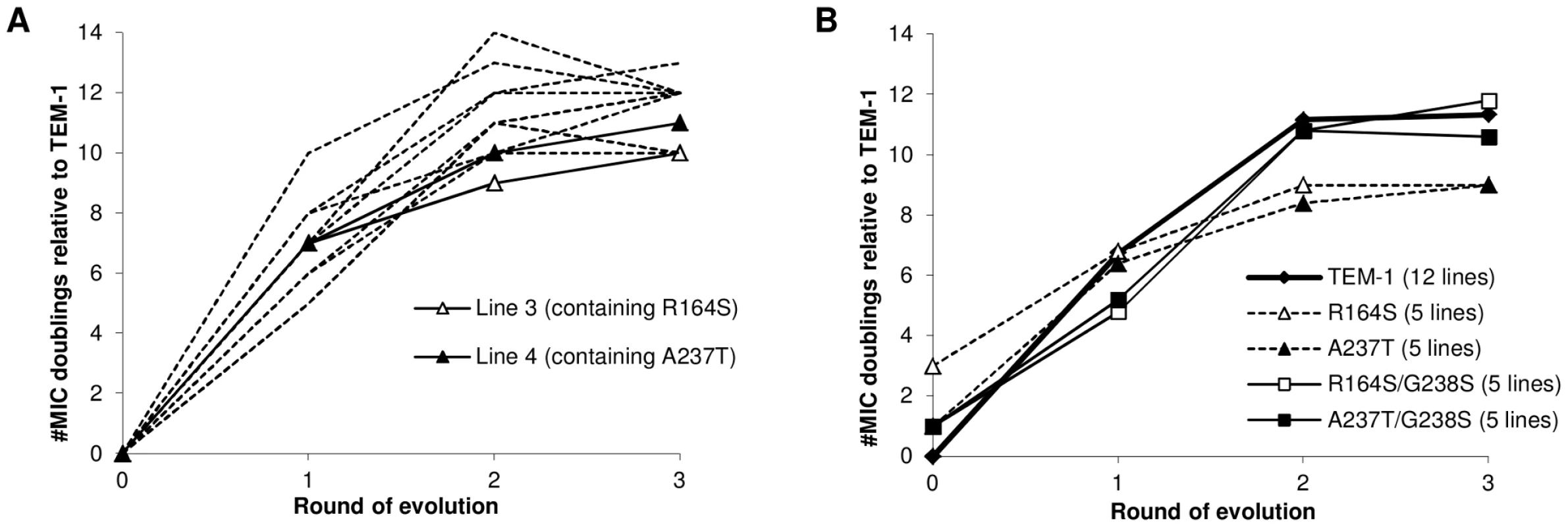
(A) Adaptation of TEM-1 β-lactamase towards improved hydrolysis of cefotaxime. Lines show changes in the average Minimal Inhibitory Concentration (MIC) of cefotaxime for the twelve lines shown in Figure 2 (see Table S3 for replicate MIC values). Aberrant lines 3 and 4 are indicated with solid lines and open and closed triangles, respectively. (B) Increase in cefotaxime MIC for TEM-1 (bold line, the average of Figure 1A) and mutants R164S (open diamonds, dashed line), A237T (diamonds, dashed line), R164S/G238S (open squares) and A237T/G238S (squares). Fig. 2. Amino acid substitutions in evolved TEM-1 alleles. 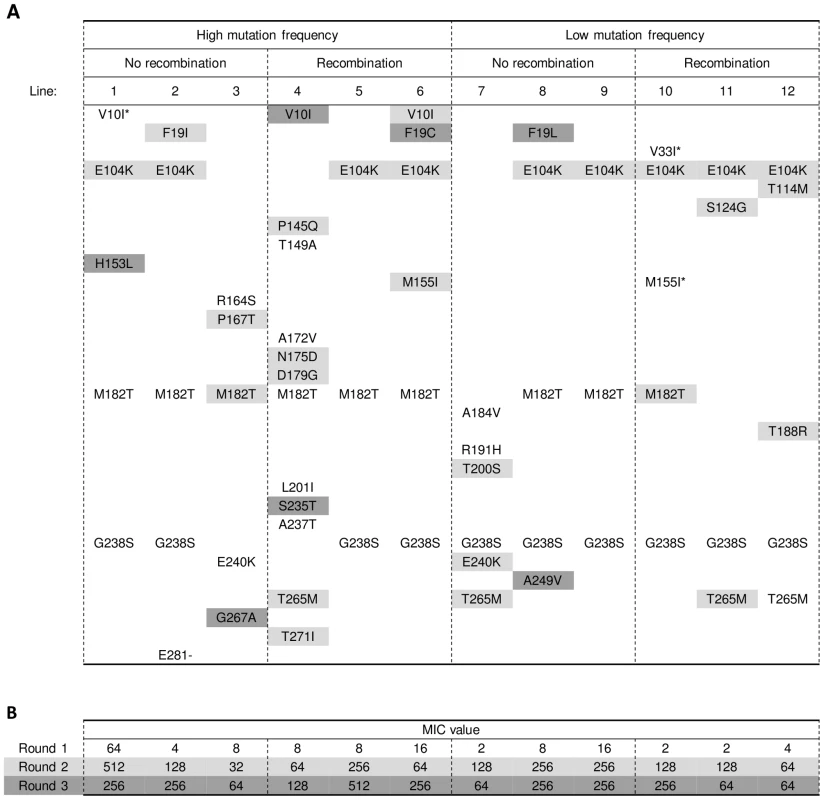
(A) Substitutions are shown as the TEM-1 amino acid according to the IUPAC single-letter code (left), the position in the protein [39] (middle), and the mutant amino acid (right). Substitutions found after the first round (and hence in all subsequent rounds) have no shade, those found after the second round have a light shade and those found after the third round have a dark shade. The three substitutions indicated with an asterisk appeared in the first round, but disappeared again after round 2 (V33I and M155I in line 10) and round 3 (V10I in line 1). Mutation details (including the official nucleotide numbering as in reference [40]) and silent mutations can be found in Table S1. (B) MIC values of the alleles isolated after the different rounds of evolution, as shown in (A). Values shown are the median value across three replicates (see Table S3 for replicate measurements). Shading as above. Epistasis between initial substitutions creates alternative adaptive pathways
We were interested in testing to what extent epistasis between different initial substitutions determined the mutational pathways taken. To test this effect of epistasis, we first had to identify the initial mutations of the common and deviating pathways. Substitution G238S was found in ten lines after the first round of evolution and is known to cause the greatest increase in cefotaxime resistance among all known single TEM-1 mutations [23]–[25]. We therefore screened the two lines that lack G238S (lines 3 and 4) for first-round adaptive substitutions with possible negative epistatic interactions with G238S, and found a likely candidate in both lines. Substitution R164S in line 3 has a positive effect on cefotaxime MIC, but shows a lower MIC when combined with G238S than both substitutions alone (see [25] and Table 1). Similarly, substitution A237T in line 4 increases activity on cefotaxime [31] and frequently occurs in clinical isolates, but never in combination with G238S (http://www.lahey.org/studies/temtable.asp). In our experiments, the MIC of the double mutant A237T/G238S was equal to that of A237T alone and only slightly higher than that of TEM-1, but considerably lower than that of G238S, thus confirming negative epistasis (see Table S3).
Tab. 1. MIC values and kinetic and stability parameters of TEM-1, single mutants R164S and G238S, and double mutant R164S/G238S. 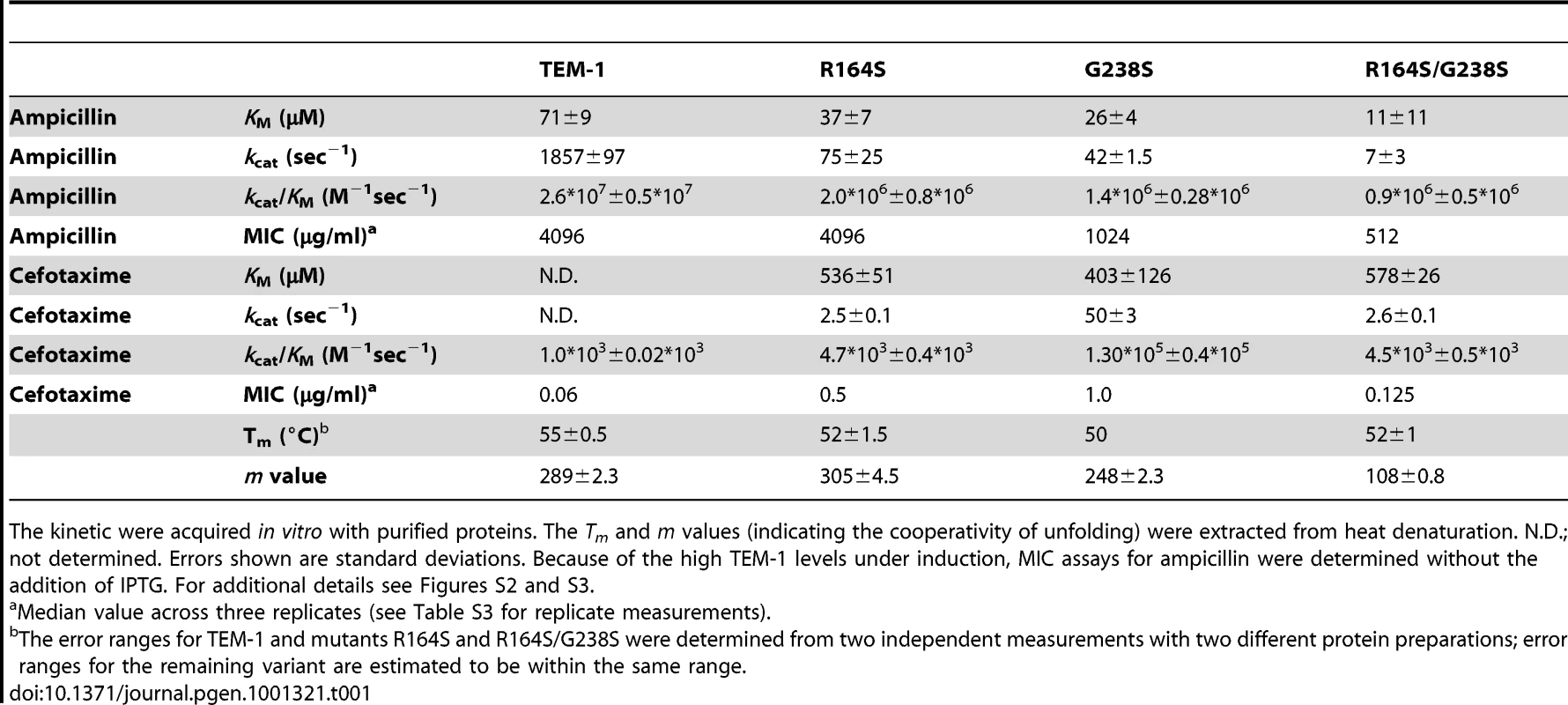
The kinetic were acquired in vitro with purified proteins. The Tm and m values (indicating the cooperativity of unfolding) were extracted from heat denaturation. N.D.; not determined. Errors shown are standard deviations. Because of the high TEM-1 levels under induction, MIC assays for ampicillin were determined without the addition of IPTG. For additional details see Figures S2 and S3. To test the predicted contingent role of these initial mutations, we ran another set of evolution experiments, this time starting with TEM-1 alleles carrying either R164S or A237T. We expected that these mutations would prevent the selection of mutations found in the common pathway, and instead facilitate the selection of other mutations. Five replicate lines of each allele were allowed to adapt to cefotaxime as before, using a mutation rate of ∼3 mutations per amplicon (see Text S2 and Table S4). Figure 1B shows the average change in resistance for all ten new lines, as well as the average for the twelve lines from the previous experiment. Again, MIC values levelled off after an initial rise, suggesting that the new lines also approach one or more adaptive peaks (see also Figure 3B). However, despite their head start due to the introduced adaptive mutation, after three rounds of evolution the resistance of these lines was significantly lower than that reached by the ten G238S-containing lines in the previous experiment (for both R164S and A237T lines: U = 3, one-tailed P<0.01), while not significantly different from each other (U = 12, two-tailed P = 1). The phenotypic assays, therefore, suggest that R164S and A237T force evolution to take different adaptive trajectories than the common trajectory followed by the TEM-1 lines.
Fig. 3. Amino acid substitutions in evolved TEM-mutants R164S and A237T. 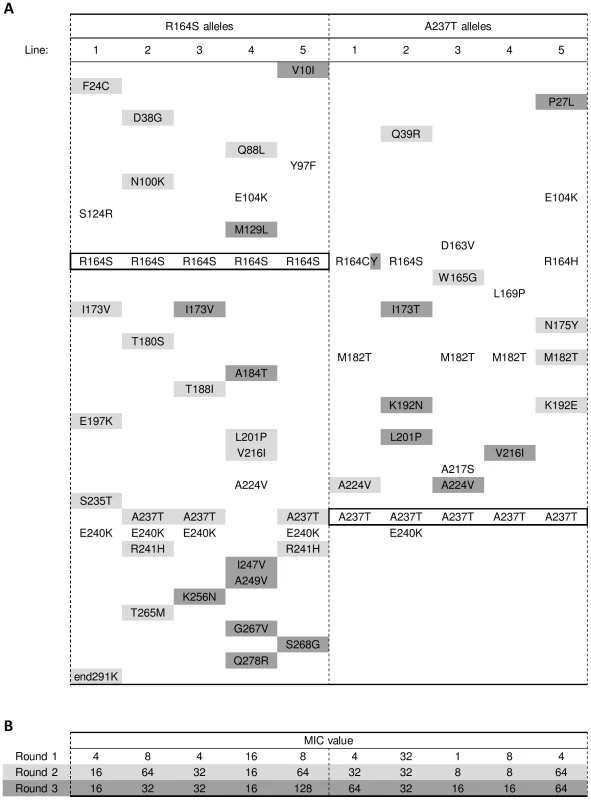
(A) See legend of Figure 2A for details. The substitutions present in the starting alleles and still present at the end of the experiment have been highlighted by a thick-lined box. Note that line 1 of the A237T alleles acquired substitution R164C after the first round of evolution, and substitution C164Y after the third round. (B) See legend of Figure 2B for details. The amino acid substitutions found in the new lines confirm this conclusion (see Figure 3A). First, despite their lower final level of resistance and lower average mutation rate, the five R164S lines accumulated more mutations than the ten G238S lines of the previous experiment (7.2 mutations, without the introduced R164S, versus 4.7 mutations including G238S, U = 4.5, one-tailed P = 0.008). Second, mutations in the background of R164S and A237T occurred at different positions, with relatively more mutations in the functionally important omega-loop (see Figure S1) compared to the ten lines carrying substitution G238S (Chi-square = 8.80, d.f. = 1, P<0.01), suggesting alternative routes towards the new enzyme functionality. Third, the presence of substitution R164S or A237T prevented the selection of G238S in all lines (Fisher's exact test, P<0.001), and often prevented the selection of other mutations repeatedly found in the G238S background, while they facilitated the selection of other mutations. For example, E104K is more often selected when G238S is present than when R164S (P = 0.004) or A237T (P = 0.001) are present, while E240K is more often found in the background of R164S than with G238S (P = 0.004). Other parallel substitutions, such as M182T, show a weaker association (i.e. with G238S compared to R164S, P = 0.036), and appear beneficial in more backgrounds. Together, the observed changes in phenotype and genotype show that the introduced substitutions are responsible for the pattern of parallel and divergent adaptation: they facilitate the selection of certain mutations, while preventing the selection of others.
Effect of epistasis between non-initial mutations
We have described the effect of epistasis between initial mutations on mutational trajectories. We were interested to see how pervasive such epistatic interactions were and whether they continue to play a role once the first mutational step in a mutational trajectory has been taken. With this in mind, our attention was drawn to line 7 of Figure 2A. Like lines 3 and 4, line 7 deviates from the other lines in Figure 2A in that it lacks two of the three main substitutions E104K, M182T, and G238S: it does contain G238S, but lacks both E104K and M182T. Because G238S is present in line 7 after the first round of evolution, we assumed that either A184V or R191H, the other two substitutions found after the first round, might be responsible for the atypical mutational pathway of this line. Because we had observed the combination of A184V and G238S in pilot experiments and because A184V is physically very close to M182T, we suspected that this substitution might be a functional equivalent of M182T. To investigate whether the double mutant A184V/G238S would prevent other substitutions of the G238S-based pathway from occurring, we constructed a TEM-1 allele carrying these two substitutions. The MIC of this double mutant was indeed higher than that of G238S by itself (see Table S3). Next, we subjected the A184V/G238S allele to three rounds of error-prone PCR and selection for increased cefotaxime resistance, again with fivefold replication, a mutation rate of on average ∼3 mutations per amplicon per error-prone PCR and without recombination. The amino acid substitutions and MIC-values recorded after each round of evolution and selection can be found in Figure 4A. Again, the MIC increases after the 1st and 2nd round, while it levels off in the 3rd round (Figure 4B). Despite of the head start of the two introduced mutations, final MIC levels were significantly lower than those of the seven lines in Figure 2A that contain mutations M182T and G238S (U = 0, one-tailed P = 0.003). The presence of A184V in the background of G238S prevented the selection of M182T (Fisher's exact test on all 13 lines of Figure 2A and Figure 4A that contain M182T or A184V in combination with G238S: P = 0.001), but E104K was selected also in this background (P = 0.07).
Fig. 4. Amino acid substitutions in evolved TEM double-mutant A184V/G238S. 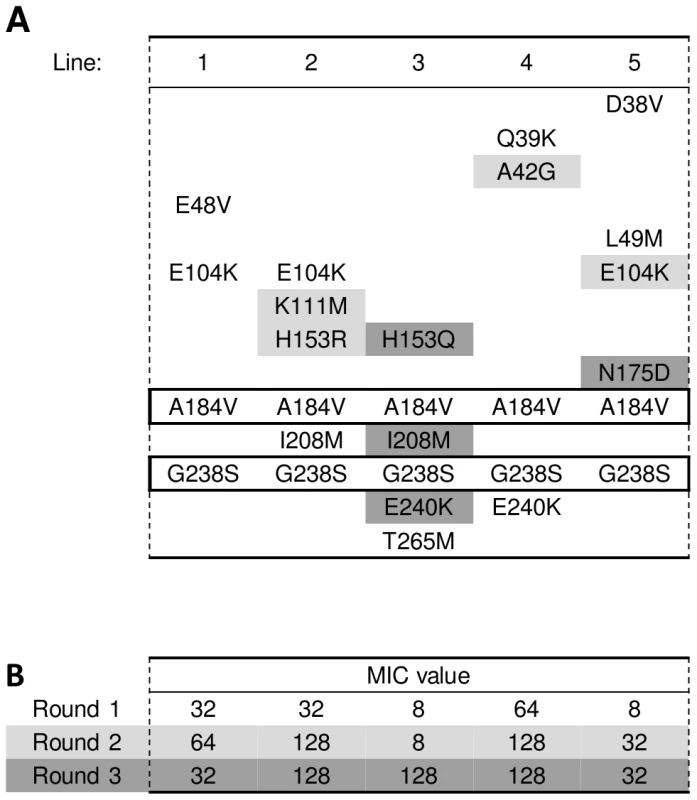
(A) See legend of Figure 2A for details. The substitutions present in the starting alleles and still present at the end of the experiment have been highlighted by a thick-lined box. (B) See legend of Figure 2B for details. More indications for sign epistatic interactions can be found in Figure 2, Figure 3, and Figure 4. For example, substitutions E104K and E240K can be found in most lines, but never co-occur in the same line. Similarly, five out of six lines that contain T265M lack M182T, and it has been speculated that both substitutions have a similar (i.e. stabilizing) function [32]. We lack statistical power to test more interactions due to their low incidence, but our examples show that sign epistasis in TEM-1 is pervasive and has adaptive consequences not only for the first, but also for later steps of the mutational pathway of this enzyme.
Adaptive constraints from pairs of epistatic mutations
Next, we were interested in testing the strength of the adaptive constraints resulting from sign epistasis directly by allowing alleles containing combinations of negatively interacting mutations to evolve. When evolution starts from an allele containing two negatively interacting mutations, can it alleviate this negative interaction by the addition of other mutations, or can it only proceed by the reversion of one of these mutations? Also, does the strength of sign epistasis affect the strength of the adaptive constraint, such that stronger sign epistasis limits the adaptive alternatives? To test this, the negatively interacting R164S/G238S double mutant, exhibiting a strong negative interaction (see MIC values for cefotaxime in Table 1), and A237T/G238S, exhibiting weaker epistasis (see Table S3), were created by site directed mutagenesis, and subjected to three rounds of directed evolution as before. The MIC trajectories of the five lines started by both double mutants show great similarity to those of the twelve TEM-1 lines in the first experiment (see Figure 1B), except that they have lower MIC-values after the first round of evolution (Figure 2B and Figure 5B; relative number of MIC doublings after round 1, U = 9, one-tailed P<0.001).
Fig. 5. Amino acid substitutions in evolved TEM double-mutants R164S/G238S and A237T/G238S. 
(A) See legend of Figure 2A for details. The substitutions present in the starting alleles and still present at the end of the experiment have been highlighted by a thick-lined box. Note that all R164S substitutions initially present in the five R164S/G238S lines eventually revert, either after the first (lines 2,3,4 and 5) or the third round of evolution (line 1). Line 1 of the A237T/G238S alleles shows a reversion of A237T after the second round of evolution, while the other four A237T/G238S lines do not show any reversion. (B) See legend of Figure 2B for details. For the A237T/G238S double mutant, only one line reverted A237T after the second round, while the other four retained the introduced mutations at residues 237 and 238 (see Figure 5A, right half). Interestingly, the line that reverted A237T is the line that ended up having the highest MIC (see Figure 5B). Three of the lines that contained both A237T and G238S after three rounds of evolution did not acquire new substitutions in the third round. Lack of reversion is unlikely to be an effect of the mutagenesis protocol, since the a(t) → g(c) transition that can revert both A237T and G238S, is relatively frequent in the mutational spectrum of the mutagenic polymerase we applied. It could be that these lines have gathered mutations in round one and two that have neutralized the negative interaction between G238S and A237T and hence made the reversions T237A and S238G either neutral or detrimental.
The combination of R164S and G238S seems to constrain evolution more severely, since all five lines eventually reverted R164S, and four lines did so directly after the first round of evolution (Figure 5A, left half). One line gathered other mutations and increased MIC during the first two rounds of evolution, and reverted R164S after the third round. In this line, in both first two rounds, at least one mutation was incorporated that is known to be beneficial in both the R164S and G238S backgrounds (E240K [33] and M182T [26], respectively). Yet, the reversion S164R in the third round, combined with other mutations, resulted in a large increase in resistance, while that of most lines with a first round reversion levelled off at this point (see Figure 5B). Thus, the level of selective constraint caused by epistasis varies for different combinations of mutations, and correlates with the strength of the negative epistatic interaction for these two cases.
Analysis of the R164S/G238S double mutant
To gain insights into the biochemical basis of the sign epistatic interaction between R164S and G238S, we purified the double mutant, and the corresponding single mutants, and examined their kinetic parameters and stability (see Table 1 and Figures S2 and S3). On ampicillin, the single mutants and double mutants exhibit very similar catalytic efficiency (kcat/KM). However, the double mutant exhibited much lower kcat than both single mutants, in particular relative to R164S, thus accounting for the lower MIC on ampicillin at concentrations above the KM. The kinetic parameters of the double mutant on cefotaxime are essentially identical to that of the single mutant R164S, but 28-fold lower in terms of kcat/KM than G238S. These differences correlate with the differences in the resistance in vivo (Table 1), but do not account for the observation that the double mutant exhibits MIC values that are lower than the R164S mutant, and that are only slightly above the MIC of TEM-1 itself. Thus, regarding the kinetic parameters, we observed epistasis between mutations R164S and G238S with respect to cefotaxime hydrolysis: the R164S substitution eliminates the much more beneficial effects of G238S. However, the kinetic parameters do not account for the sign epistasis observed for the resistance levels in vivo. The origin of the latter is likely to be lower levels of functional enzyme, which may result from the lower kinetic stability of the double mutant. The Tm-values of all three mutants are similar, and are lower than that of TEM-1. Thus the thermostability of both single mutants seems comparable to that of the double mutant. However, the double mutant exhibits a cooperativity factor (m value) that is ∼2.5-fold lower than that of the two single mutants. In fact, the fit of the double mutant to the standard two-state unfolding model [34] is poor (see Figure S2), also indicating that its folding follows a different path than that of wild-type TEM-1 and of the single mutants. Altogether, the similarity in kcat and KM values versus the lower MIC, and the perturbed folding parameters, suggest that sign epistasis relates to a combination of kinetic parameters that are much lower for the double mutant than for G238S on its own, and lower levels of properly folded and functional enzyme.
Discussion
We used laboratory evolution experiments with a model enzyme to study the topography of its fitness landscape by testing the involvement of epistasis in creating alternative mutational pathways. A previous reconstruction of the fitness landscape of TEM-1 β-lactamase for adaptation to cefotaxime based on MIC assays of all possible combinations of mutations A42G, E104K, M182T, G238S, and a promoter mutation suggested that, despite local ruggedness, it contains a single adaptive peak [24]. However, by allowing all possible mutations that may contribute to the evolution of cefotaxime resistance to be selected, we found that the fitness landscape contains more than a single adaptive peak. Our ability to monitor and manipulate the sequence of genetic changes allowed us to demonstrate that epistasis was at the basis of the landscape's ruggedness. Epistasis caused one pathway to be used preferentially, while making the particular choice of pathways historically contingent upon the first mutational step. This first step therefore resembles the crucial choice of lanes before a junction of alternative evolutionary routes. Finally, by studying how the biochemical and biophysical parameters for a specific pair of mutations correlate with the level of fitness (here, antibiotic resistance), we found that sign epistasis resulted from a combination of negative interactions at the level of enzyme activity and reduced folding efficiency (kinetic stability).
The epistatic constraints observed in our experiments depend not only on the topography of the fitness landscape, but also on the mutation supply rates of the evolving populations (i.e. population size x mutation rate). The conditions in our experiments were such that mutation supply rates were in the order of 106 (i.e. library sizes of ∼6×105 and mutation rates of ∼3 for most experiments) and multiple mutations were often selected in a single round of evolution (see Figure 2A). Assuming no mutational bias, simulations using the PEDEL program [35] indicate that all possible 2581 single mutants (i.e. 861 nucleotides×3 possible mutations) were present in our libraries, as well as up to 10% of all possible double mutants. That two of the twelve lines in our first experiment showed a deviating pattern of substitutions already after the first round of evolution, therefore, must be seen as a consequence of epistasis combined with clonal interference between genotypes carrying different sets of mutations. While epistatic constraints may be most severe during evolution by single-mutation selection (the so-called ‘weak-mutation, strong-selection’ regime), our results show that significant constraints are experienced also during evolution with much higher mutation supplies. Because of the high mutation supplies in our experiments, in some lines combinations of mutations that are individually neutral or deleterious but beneficial in combination may have been selected. The relative strength of the epistatic constraints experienced at different scales in genotype space will depend on the topography of the fitness landscape, and should be addressed in future work.
How important are our observations on a single enzyme for real organisms? In our case where mutations affect a single enzyme, sign epistasis results from the biophysical constraints underlying functionality of a single protein [36], [37]. Although possibly less severe, similar constraints are likely to exist when mutations affect different genes and functions [5], [6]. Differences observed in the molecular changes in two bacteriophage populations that evolved in parallel, indicate that mutational stochasticity and mutational order may influence the pattern of adaptation [11]. A recent analysis of epistasis between individually deleterious mutations affecting different genes of the fungus Aspergillus niger confirms the significance of sign epistasis for entire organisms [8]. Such epistatic interactions can make evolution contingent by amplifying the effects of mutational chance events in finite populations, as demonstrated here. However, our results also show that even contingent evolution may have a relatively deterministic structure once the first step has been determined.
Materials and Methods
Strains and plasmids
Escherichia coli strain DH5αE (Invitrogen) was used as a host for all plasmids. Plasmid pACSE3 [19] was used as the vector for cloning and expressing TEM alleles. This ∼5.1-kb plasmid contains a tetracycline-resistance marker and replicates at ∼10 copies per cell.
Media
LB-medium is 10g/L trypticase peptone, 5 g/L yeast extract and 10 g/L NaCl. LB-tetracycline medium is LB-medium containing 15 mg/L tetracycline and LB-kanamycin medium is LB-medium containing 50 mg/L kanamycin. Mueller Hinton (Merck) and Mueller Hinton II (BD) medium were prepared according to the manufacturers' instructions. SOC medium is 20 g/L bacto-tryptone and 5 g/L yeast extract with 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2 and 20 mM glucose. Solid media contained 16 g/L agar.
Recombinant methodology
Plasmids were prepared from overnight LB-tetracycline cultures, and purified using a GenElute Plasmid Miniprep Kit (Sigma). TEM-1 (861 bp long) was amplified from pBR322 using the high-fidelity Pfu polymerase (Stratagene), sense primer P1 (GGGGGGTCATGAGTATTCAACATTTCCGTGTCG, BspHI site underlined, this enzyme cuts 1 bp upstream of TEM-1's start codon) and antisense primer P2 (CCGAGCTCTTGGTCTGACAGTTACCAATGC, SacI site underlined, this enzyme cuts 17 bp downstream of TEM-1's stop-codon), using the following temperature cycle: 30′ at 95°C, 30′ at 61°C and 90′ at 72°C for 30 cycles, followed by 72°C for 10 min. The resulting amplicon was digested with BspHI and SacI (New England Biolabs). Plasmid pACSE3 was digested with the same restriction enzymes and dephosphorylated with Calf Intestinal Phosphatase (New England Biolabs). Digested amplicons and vectors were purified using Sigma's GenElute PCR Clean-up Kit, ligated using T4 DNA Ligase (New England Biolabs), and transformed into DH5αE by electroporation. Specific mutations were introduced into TEM-1 using the QuickChange site-directed mutagenesis kit (Stratagene). Note that in this protocol TEM-1's promoter region is not part of the amplicon.
Creation of TEM-1 libraries
Mutant TEM alleles were generated by introducing random mutations using the Genemorph I and II Mutagenesis Kits (Stratagene). Since the former kit was unexpectedly taken out of production and replaced by the latter, we carried out the initial experiment (Figure 2) using Genemorph I, but were forced to switch to Genemorph II for all other experiments. Since the error-prone polymerases in both kits differ only slightly in their mutational spectrum (see the Genemorph II manual, Stratagene), this switch has not significantly influenced the outcome of our experiments. Primers used for error-prone PCR were P3 (TCATCCGGCTCGTATAATGTGGA) and P4 (ACTCTCTTCCGGGCGCTATCAT), which flank the multiple cloning site of pACSE3. In order to make a broad exploration of the diversity of adaptive pathways, we varied the mutation rate (high rate in lines 1–6, and low rate in lines 7–12 of Figure 2A) and also introduced a simplified form of recombination in the initial experiment (in lines 4–6 and 9–12). The mutation rate was manipulated by varying the effective number of replication cycles by the application of different amounts of template DNA. Conditions were set to introduce on average ∼2 (low mutation rate) or ∼6 (high mutation rate) mutations per amplicon (0.06 and 565 ng plasmid template, respectively). The resulting amplicons were digested with BspHI and SacI, ligated into pACSE3 and electroporated into DH5αE. After recovery for 90 min in SOC medium at 37°C, the cells were diluted in 500 mL LB-tetracycline. An aliquot was taken out directly after mixing and plated onto LB-tetracycline agar to determine the library size. The remainder of the culture was incubated at 37°C overnight to amplify the library. Effective library sizes varied between ∼104 and 106 transformants with an average of ∼6×105. Aliquots of the amplified libraries were stored in 10% glycerol at −80°C.
Recombination
In the first experiment, recombination between mutant TEM-1 alleles within a library was allowed in some lines (4–6 and 10–12) by digestion of the error-prone PCR amplicons with PvuI. TEM-1 has a single PvuI restriction site roughly in the middle of the sequence, allowing recombination via a single cross-over, as confirmed by the analysis of restriction markers. The truncated amplicons were ligated into pACSE3 via a single ligation step as described above.
Expression and selection
Expression of the TEM-1 allele in pACSE3 is under control of the pTAC promoter that is tightly regulated by the lac repressor, encoded by the lacI gene on pACSE3. Expression was induced by adding 50 µM isopropyl-β-D-thiogalactopyranoside (IPTG), which was shown to mimic natural expression of TEM-1 [19]. For selection, a series of bottles containing 50 mL Mueller-Hinton medium (Merck) was inoculated with cefotaxime (Sigma, stock solution in 0.1 M NaPO4, pH 7.0) concentrations in two-fold increments, ranging from 0.0625 µg/mL, the MIC of wild-type TEM-1, to 1024 µg/mL. Each bottle was inoculated with a cell number approximately equal to 10 times the library size from the overnight amplified cultures. Cultures were incubated for 48 hours at 37°C. The culture that grew at the highest concentration of cefotaxime was plated on LB-tetracycline agar. The presumed clonal fixation of a single adaptive mutant in these 48 hour cultures [19] was confirmed by the sequence analysis of 10 randomly picked clones from a single culture (see Text S1). Therefore, the next day, a single colony was selected and grown overnight in LB-tetracycline. Plasmid from the overnight culture was isolated using GeneElute Plasmid Miniprep Kit (Sigma) and sequenced using BigDye (Perkin Elmer) or DYEnamic (AP-biotech) Terminator Cycle Sequence kits.
MIC assays
The Minimal Inhibitory Concentration (MIC) was determined from 150 µl cultures at a titre of 105 cells/mL in Mueller Hinton II medium containing 50 µm IPTG. A 150 µl solution of twofold serial dilutions of antibiotic in MH II was added to these cultures. Cultures were grown for 24 hours at 37°C, growth was determined by visual inspection, and MIC was defined as the lowest concentration of antibiotic that completely prevents visible growth. To exclude phenotypic variation in bacteria, plasmid was isolated from all selected clones and re-transformed into isogenic E. coli DH5αE cells prior to each MIC-assay. Ampicillin MICs were determined without the addition of IPTG, since this turned out to mask differences between different mutants tested.
Protein purification and thermal denaturation measurements
Measurements were performed essentially as earlier described [38]. The equation used to fit the row thermal denaturation was:Where αN is the fluorescence of the native structure at T = 25°C, p is a slope of fluorescence change of the native state, αU is a fluorescence at T = 80°C, q is a slope of fluorescence change of the unfolded state, and mN–U is the slope of transition, RT = −582.
Kinetic measurements
The kinetic parameters kcat and KM were determined by non-linear regression fit to the Michaelis-Menten equation using the KaleidaGraph program (Synergy Software). Initial velocities were recorded with increasing substrate concentrations from 1/3 KM to 3 KM. For ampicillin, hydrolysis of the β-lactam ring was monitored by loss of absorbance at 232 nm (Δε = 600 M−1 cm−1), using a quartz plate (path of 0.5 cm) in a microtitre plate reader (bioTeck). The hydrolysis of cefotaxime was monitored by loss of absorbance at 264 nm (Δε = 6360 M−1 cm−1). Because of the high initial absorbance, a quartz cell of 0.1 cm pathway was used for measurements at ≥250 µM cefotaxime. The readings in lower concentrations were performed in quartz plate in a plate reader (path of 0.5 cm).
Statistical analyses
Since MIC values are measured on a discontinuous scale (with two-fold increases in antibiotic concentration), non-parametric tests on differences in median MIC estimates were used.
Supporting Information
Zdroje
1. KondrashovFA
KondrashovAS
2001 Multidimensional epistasis and the disadvantage of sex. Proc Natl Acad Sci U S A 98 12089 12092
2. ManiGS
ClarkeBC
1990 Mutational order - a major stochastic process in evolution. Proc R Soc Biol Sci Ser B 240 29 37
3. de VisserJAGM
LenskiRE
2002 Long-term experimental evolution in Escherichia coli. XI. Rejection of non-transitive interactions as cause of declining rate of adaptation. BMC Evol Biol 2 19
4. MacLeanRC
PerronGG
GardnerA
2010 Diminishing returns from beneficial mutations and pervasive epistasis shape the fitness landscape for rifampicin resistance in Pseudomonas aeruginosa. Genetics 186 1345 1354
5. WeinreichDM
WatsonRA
ChaoL
2005 Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59 1165 1174
6. PoelwijkFJ
KivietDJ
WeinreichDM
TansSJ
2007 Empirical fitness landscapes reveal accessible evolutionary paths. Nature 445 383 386
7. WeinreichDM
ChaoL
2005 Rapid evolutionary escape by large populations from local fitness peaks is likely in nature. Evolution 59 1175 1182
8. de VisserJAGM
ParkS.-C
KrugJ
2009 Exploring the effect of sex on empirical fitness landscapes. Am Nat 174 s15 s30
9. WeissmanDB
FeldmanMW
FisherDS
2010 The rate of fitness-valley crossing in sexual populations. Genetics 186 1389 1410
10. PovolotskayaIS
KondrashovFA
2010 Sequence space and the ongoing expansion of the protein universe. Nature 465 922 926
11. WichmanHA
BadgettMR
ScottLA
BoulianneCM
BullJJ
1999 Different trajectories of parallel evolution during viral adaptation. Science 285 422 424
12. BollbackJP
HuelsenbeckJP
2009 Parallel genetic evolution within and between bacteriophage species of varying degrees of divergence. Genetics 181 225 234
13. SchoustraSE
DebetsAJM
SlakhorstM
HoekstraRF
2007 Mitotic recombination accelerates adaptation in the fungus Aspergillus nidulans. PLoS Genet 3 e68 doi:10.1371/journal.pgen.0030068
14. WoodTE
BurkeJM
RiesebergLH
2005 Parallel genotypic adaptation: when evolution repeats itself. Genetica 123 157 170
15. TravisanoM
MongoldJA
BennettAF
LenskiRE
1995 Experimental tests of the roles of adaptation, chance, and history in evolution. Science 267 87 90
16. WoodsR
SchneiderD
WinkworthCL
RileyMA
LenskiRE
2006 Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci U S A 103 9107 9112
17. BlountZD
BorlandCZ
LenskiRE
2008 Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci U S A 105 7899 7906
18. StemmerWP
1994 Rapid evolution of a protein in vitro by DNA shuffling. Nature 370 389 391
19. BarlowM
HallBG
2002 Predicting evolutionary potential: in vitro evolution accurately reproduces natural evolution of the TEM β-lactamase. Genetics 160 823 832
20. BershteinS
SegalM
BekermanR
TokurikiN
TawfikDS
2006 Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 444 929 932
21. BarlowM
HallBG
2003 Experimental prediction of the natural evolution of antibiotic resistance. Genetics 163 1237 1241
22. VakulenkoSB
GerykB
KotraLP
MobasheryS
LernerSA
1998 Selection and characterization of β-lactam-β-lactamase inactivator-resistant mutants following PCR mutagenesis of the TEM-1 β-lactamase gene. Antimicrob Agents Chemother 42 1542 1548
23. HallBG
2002 Predicting evolution by in vitro evolution requires determining evolutionary pathways. Antimicrob Agents Chemother 46 3035 3038
24. WeinreichDM
DelaneyNF
DePristoMA
HartlDL
2006 Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312 111 114
25. GiakkoupiP
TzelepiE
TassiosPT
LegakisNJ
TzouvelekisLS
2000 Detrimental effect of the combination of R164S with G238S in TEM-1 β-lactamase on the extended-spectrum activity conferred by each single mutation. J Antimicrob Chemother 45 101 104
26. ZaccoloM
GherardiE
1999 The effect of high-frequency random mutagenesis on in vitro protein evolution: a study on TEM-1 β-lactamase. J Mol Biol 285 775 783
27. HeckyJ
MullerKM
2005 Structural perturbation and compensation by directed evolution at physiological temperature leads to thermostabilization of β-lactamase. Biochemistry 44 12640 12654
28. BrownNG
PenningtonJM
HuangW
AyvazT
PalzkillT
2010 Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM β-lactamases. J Mol Biol 404 832 846
29. KatherI
JakobRP
DobbekH
SchmidFX
2008 Increased folding stability of TEM-1 β-lactamase by in vitro selection. J Mol Biol 383 238 251
30. OrenciaMC
YoonJS
NessJE
StemmerWPC
StevensRC
2001 Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol 8 238 242
31. CantuC
HuangWZ
PalzkillT
1997 Cephalosporin substrate specificity determinants of TEM-1 β-lactamase. J Biol Chem 272 29144 29150
32. SalverdaMLM
De VisserJAGM
BarlowM
2010 Natural evolution of TEM-1 β-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol Rev 34 1015 1036
33. HuangWZ
LeQQ
LaroccoM
PalzkillT
1994 Effect of Threonine-to-Methionine substitution at position 265 on structure and function of TEM-1 β-lactamase. Antimicrob Agents Chemother 38 2266 2269
34. FershtAR
1985 Enzyme Structure and Mechanism. New York Freeman 475
35. FirthAE
PatrickWM
2005 Statistics of protein library construction. Bioinformatics 21 3314 3315
36. DePristoMA
WeinreichDM
HartlDL
2005 Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet 6 678 687
37. TokurikiN
TawfikDS
2009 Stability effects of mutations and protein evolvability. Curr Opin Struct Biol 19 596 604
38. BershteinS
GoldinK
TawfikDS
2008 Intense neutral drifts yield robust and evolvable consensus proteins. J Mol Biol 379 1029 1044
39. AmblerRP
CoulsonAFW
FrereJM
GhuysenJM
JorisB
1991 A standard numbering scheme for the class-A β-lactamases. Biochem J 276 269 270
40. SutcliffeJG
1978 Nucleotide sequence of ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A 75 3737 3741
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and PhosphodiesterasesČlánek Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
- Prof. Petr Urbánek: Potřebujeme najít pacienty s nediagnostikovanou akutní intermitentní porfyrií
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



