-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Global Analysis of the Relationship between JIL-1 Kinase and Transcription
The ubiquitous tandem kinase JIL-1 is essential for Drosophila development. Its role in defining decondensed domains of larval polytene chromosomes is well established, but its involvement in transcription regulation has remained controversial. For a first comprehensive molecular characterisation of JIL-1, we generated a high-resolution, chromosome-wide interaction profile of the kinase in Drosophila cells and determined its role in transcription. JIL-1 binds active genes along their entire length. The presence of the kinase is not proportional to average transcription levels or polymerase density. Comparison of JIL-1 association with elongating RNA polymerase and a variety of histone modifications suggests two distinct targeting principles. A basal level of JIL-1 binding can be defined that correlates best with the methylation of histone H3 at lysine 36, a mark that is placed co-transcriptionally. The additional acetylation of H4K16 defines a second state characterised by approximately twofold elevated JIL-1 levels, which is particularly prominent on the dosage-compensated male X chromosome. Phosphorylation of the histone H3 N-terminus by JIL-1 in vitro is compatible with other tail modifications. In vivo, phosphorylation of H3 at serine 10, together with acetylation at lysine 14, creates a composite histone mark that is enriched at JIL-1 binding regions. Its depletion by RNA interference leads to a modest, but significant, decrease of transcription from the male X chromosome. Collectively, the results suggest that JIL-1 participates in a complex histone modification network that characterises active, decondensed chromatin. We hypothesise that one specific role of JIL-1 may be to reinforce, rather than to establish, the status of active chromatin through the phosphorylation of histone H3 at serine 10.
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1001327
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001327Summary
The ubiquitous tandem kinase JIL-1 is essential for Drosophila development. Its role in defining decondensed domains of larval polytene chromosomes is well established, but its involvement in transcription regulation has remained controversial. For a first comprehensive molecular characterisation of JIL-1, we generated a high-resolution, chromosome-wide interaction profile of the kinase in Drosophila cells and determined its role in transcription. JIL-1 binds active genes along their entire length. The presence of the kinase is not proportional to average transcription levels or polymerase density. Comparison of JIL-1 association with elongating RNA polymerase and a variety of histone modifications suggests two distinct targeting principles. A basal level of JIL-1 binding can be defined that correlates best with the methylation of histone H3 at lysine 36, a mark that is placed co-transcriptionally. The additional acetylation of H4K16 defines a second state characterised by approximately twofold elevated JIL-1 levels, which is particularly prominent on the dosage-compensated male X chromosome. Phosphorylation of the histone H3 N-terminus by JIL-1 in vitro is compatible with other tail modifications. In vivo, phosphorylation of H3 at serine 10, together with acetylation at lysine 14, creates a composite histone mark that is enriched at JIL-1 binding regions. Its depletion by RNA interference leads to a modest, but significant, decrease of transcription from the male X chromosome. Collectively, the results suggest that JIL-1 participates in a complex histone modification network that characterises active, decondensed chromatin. We hypothesise that one specific role of JIL-1 may be to reinforce, rather than to establish, the status of active chromatin through the phosphorylation of histone H3 at serine 10.
Authors Summary
Active chromatin is characterised by a wealth of histone modifications that collectively define a decondensed form of chromatin that is accessible to regulators and more easily transcribed. The kinase JIL-1 associates with active chromatin and contributes to chromatin decondensation by phosphorylation of a key serine residue (S10) on the N-terminus of histone H3. We mapped the binding sites for JIL-1 on chromosomes in cells and found that the enzyme associates all along transcribed genes. A basal level of association can be found at most genes and correlates best with the presence of active chromatin exemplified by methylation of H3K36. A double dose of JIL-1 can be observed at active genes of the hyperactive male X chromosome that also bear an acetylation of histone H4 at lysine 16. Phosphorylation of H3 by JIL-1 creates a mark that characterises active chromatin and could play a role in preventing the formation of repressive chromatin. Surprisingly, depletion of the kinase has clear, but relatively modest, effects on transcription. We hypothesise that JIL-1 may help to reinforce the active state of chromatin that is primarily established by other factors.
Introduction
JIL-1 is a ubiquitously expressed, nuclear tandem kinase of Drosophila melanogaster that associates with chromatin at all stages of development. JIL-1 phosphorylates histone H3 in chromatin and the kinase is thought to be responsible for the majority of phosphorylation of histone H3 at serine 10 (H3S10ph) in interphase [1]. At the low level of resolution afforded by staining larval polytene chromosomes JIL-1 is seen to associate with active chromatin such as the decondensed interbands of chromosomes [2]. Ectopic recruitment of JIL-1 to a series of Lac operator repeats by fusion to a lacI DNA binding domain leads to local decondensation of polytene chromatin. This reorganisation of chromatin depends on JIL-1's kinase activity [3]. The essential function of JIL-1 can be explained, at least in part, by the involvement of the kinase in chromosome organisation [4], [5], where it plays a role in maintaining the balance between euchromatin and heterochromatin [6]–[8]. This is supported by the fact that Su(var)3-1 alleles of the JIL-1 gene are strong suppressors of position effect variegation, which provides a sensitive assay for heterochromatin propagation [9]. In the absence of JIL-1 the H3K9me2 mark and HP1 are redistributed genome-wide and tend to be enriched on the X chromosome in both males and females [6]. In addition, the lethal loss-of-function mutation of JIL-1 is rescued by reduced levels of the H3K9 methyltransferase Su(var)3-9 but not by reduced levels of the major HP1 isoform HP1a [10] suggesting that the spreading of H3K9me2 might be the cause of the lethality of the JIL-1 mutants. However, reducing the dose of Su(var)3-7, another essential component of heterochromatin, also rescues the lethality of the JIL-1 mutant. In this case the redistribution of H3K9me2 was still observed [11]. Su(var)3-7 may thus be an essential effector in the pathway of heterochromatin spreading, which is counteracted by JIL-1.
In mammalian cells the H3S10ph mark has been implicated in transcriptional activation, notably in immediate early response to mitogen, stress or steroid hormones, which trigger the transient phosphorylation of H3S10 and H3S28 by the kinases Msk1/2 or PIM-1 (for review see [12]. Whereas some of the inducing effects involve the dissociation of euchromatic HP1 isoforms [13], [14], the H3S10ph mark also serves as docking site for 14-3-3 proteins, which in turn recruit additional activating enzymes [13], [15], [16]. In D. melanogaster, the role of JIL-1 and the H3S10ph mark in transcription regulation is less clear. Corces and colleagues reported that upon heat shock gene activation, H3S10 phosphorylation by JIL-1 leads to recruitment of 14-3-3 and of the elongator protein 3 (Elp3) [17] that in turn would trigger the transition of an initiated RNA polymerase II (Pol II) into the elongation mode [18]. Johansen and co-workers by contrast argued that JIL-1 and JIL-1-dependent Ser10 phosphorylation did not have a direct function in heat shock gene activation [19].
We are interested in the potential involvement of JIL-1 in the process of X chromosome dosage compensation in D. melanogaster, which is hypothesized in light of a number of intriguing links between JIL-1 and the compensated male X chromosome. The process of dosage compensation serves to increase the transcription of genes on the single male X chromosome approximately twofold to match the combined transcription of the two alleles on the female X chromosomes [20], [21]. JIL-1 is enriched on the male X chromosome and this enrichment depends on a functional DCC [2], [22]. Severe reduction of JIL-1 levels leads to a global change in chromatin structure that is best appreciated on polytene chromosomes [1]. The euchromatic interbands appear much reduced in the absence of JIL-1 and replaced by atypical chromatin, in agreement with the above-mentioned role of JIL-1 in restricting the spread of heterochromatin. Interestingly, the dosage-compensated X chromosome shows an enhanced sensitivity to reduced JIL-1 levels, and appears diffuse and lacking polytene banding pattern [1], [4]. The Dosage Compensation Complex (DCC, also known as Male-Specific-Lethal [MSL] complex) associates along the transcribed regions of target genes and may therefore boost transcription elongation [23], [24]. The activation of genes on the male X chromosome involves the DCC subunit MOF on the X chromosome, an acetyltransferase, which selectively acetylates histone H4 at K16 (H4K16ac) [25], [26]. H4K16ac opposes the folding of the nucleosomal fibre in vitro and should render chromatin more accessible [27]. Accordingly, chromatin from the male X chromosome is released more efficiently from nuclei after shearing [28]. However, the precise twofold enhancement of transcription cannot be explained by H4K16ac alone [26].
We now mapped for the first time at high resolution the chromosomal distribution of JIL-1 kinase in SL2 cells using chromatin immunoprecipitation (ChIP) and hybridisation of associated DNA to oligonucleotide tiling microarrays. JIL-1 binding was compared to the distribution of elongating RNA polymerase II and related to steady-state mRNA levels. We also compared them to profiles of a number of histone modifications, and of DCC subunits. The vast majority of active genes is marked by a basal level of JIL-1 on gene bodies, independently of the actual transcription rates. The presence of H4K16ac on X-linked genes correlates with roughly twofold elevated levels of JIL-1. JIL-1 thus marks transcribed genes, but it also senses dosage-compensated chromatin. The phosphorylation of H3S10 is enriched on active genes in the same manner as JIL-1 itself. Perhaps surprisingly, the depletion of JIL-1 by RNA interference (RNAi) has only mild effects of transcription, which suggests that JIL-1 is not absolutely required for transcription in general or for dosage compensation in particular. In the light of the previous genetic analysis our data suggest that JIL contributes to a composite histone mark that may serve to reinforce the active state of chromatin by preventing the inappropriate association of silencing factors.
Results
JIL-1 binds most transcribed genes and is enriched on dosage-compensated genes
We raised two polyclonal antisera (R69 and R70) against a fragment (amino acids 79–571) of JIL-1 and established their specificity. In agreement with previous results [22], we confirmed that JIL-1 is a nuclear protein that associates with active, decondensed chromatin represented by interbands on larval polytenized chromosomes (Figure S1). We also visualized the clear enrichment of JIL-1 on the dosage-compensated, hyperactive male X chromosome. This enrichment on the X chromosomal territory can also be seen in nuclei of the Drosophila SL2 cells, which have a male genotype. By contrast, nuclei of female KC cells display a homogenous staining (Figure S1).
The enrichment of JIL-1 on the male X chromosome and the conflicting reports about the role of JIL-1 in the control of early transcription elongation of heat shock genes [17]–[19] prompted us to generate a high-resolution in vivo chromosomal interaction profile of the kinase and to compare it to elongating RNA polymerase II (ePol). We combined chromatin immunoprecipitation (ChIP) with probing oligonucleotide tiling arrays as before [24], [26], [29], using the JIL-1 antibodies and a well-established monoclonal antibody specific for ePol (H5). This antibody recognises phosphorylated serine 2 in the heptad repeat of the C-terminal domain (CTD) of the largest RNA polymerase II subunit. Data from four biological replicates starting with independent chromatin preparations were combined for each profile. The two JIL-1 antibodies gave very coherent results (Figure S2). We also generated a profile for histone H4 acetylated at K16 (H4K16ac). Figure 1 displays the profiles of JIL-1, ePol and H4K16ac along representative 250 kb portions of the chromosomes X and 3R (Figure 1A, 1B, respectively). The profiles resemble the distribution of histone H3 methylated at lysine 36 (H3K36me3), a modification that is placed co-transcriptionally by the CTD-associated methylase Set2 and therefore serves as a hallmark of transcribed chromatin [30], [31].
Fig. 1. JIL-1 is a mark for gene activity and for dosage compensation in SL2 cells. 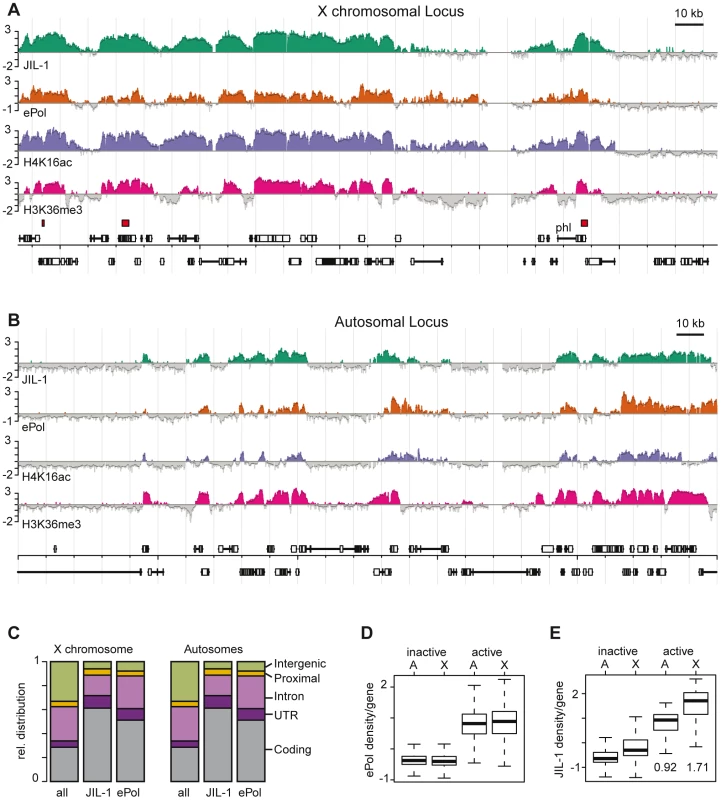
High resolution ChIP on chip profiles of JIL-1, ePol, H4K16ac and H3K36me3 [30]. The data is represented as average log2 signal ratio of IP over input. Bars are coloured when the signal is above zero. The dark grey line toping each profile represents the mean signal within a 500 bp window centred on the probe. High affinity sites (HAS) for the DCC according to [29] are depicted as red boxes above the gene annotations. A representative 250 kb portion has been selected (A) for the X chromosome and (B) for chromosome 3R. (C) Fractional distribution of oligonucleotides according to genomic features is shown for all probes on the array (n = 384680) and compared to the ones within JIL-1 (n = 112954) and ePol (n = 125303) binding regions as determined by hidden markov modelling. (D) The density per gene was defined as the average ratio (IP/input) of probes within a gene. The densities of ePol on autosomal genes (labelled A) and X-linked genes (labelled X) in a box plot representation. Activity status was determined by microarray expression profiling. (E) JIL-1 densities on the same sets of genes as in D, the median values for active autosomal and X-linked genes are given. The boxplot representation in all figures define 25th to 75th percentiles (boxes), 50th percentile (lines in boxes), and ranges (whiskers, 1.5 times the interquartile range extended from both ends of the box). Outliers were removed from the analysis. Qualitatively, the distribution of JIL-1 and ePol were similar on the X chromosome and on autosomes with a preference for coding sequences. In general, binding of JIL-1 to introns is lower than binding of ePol (Figure 1C). As expected, ePol is mainly found at active genes as assessed by Affymetrix expression profiling (Figure 1D) and there is a good correlation between the density of ePol along genes and the corresponding steady-state mRNA level (Figure S3). The distribution of JIL-1 correlates well with several subunits of the DCC on the X chromosome: at the probe level, the best correlations are observed with the targeting subunit MSL1 [24] and with the spreading subunit MOF, which is the acetyltransferase responsible for H4K16 acetylation (Figure S4). On the X chromosome 1413 genes are detectably transcribed according to ePol occupancy. Among those, the vast majority (93%) have both JIL-1 and MSL1 bound, a minority of 55 genes (4%) show no detectable JIL-1, 86 genes (6%) have no MSL1 bound, and 46 genes (3%) have neither JIL-1 nor MSL1 bound. Remarkably, the densities of ePol on active genes are comparable on autosomes and on the X chromosome (Figure 1D), although the latter ones benefit from dosage compensation (see discussion). By contrast, the mean density of JIL-1 on active genes is 0.92 for autosomal and 1.71 for X-linked genes, thus the calculated enrichment of JIL-1 on X chromosomal genes relative to autosomal ones is about 1.86 (Figure 1E). These results quantitatively reproduces the picture obtained from polytene chromosome staining (Figure S1), where a twofold enrichment had been estimated [22].
The enrichment of JIL-1 at various loci was confirmed by quantitative PCR (qPCR) on the non-amplified immunoprecipitate. Figure 2A shows as example the X chromosomal gene pole hole (phl) where JIL-1 accumulates towards the 3′ end of the gene, together with H4K16ac and H3K36me3. We also confirmed the male-specific enrichment of JIL-1 on several X-linked genes by qPCR of immunoprecipitates of chromatin prepared from sorted male and female flies (Figure 2B). The X-linked genes phl, CG14804, sta, Pgd and RpII215 show elevated levels of JIL-1 in males as compared to females. As an example for autosomal genes, we chose the Cortactin gene, which is similarly bound by JIL-1 in males and females. The H3K36me3 levels at the same genes are rather similar in both sexes, whereas H4K16ac is clearly found there only in males.
Fig. 2. JIL-1 is enriched at the 3′ end of phl and is twofold enriched at X-linked loci in male flies. 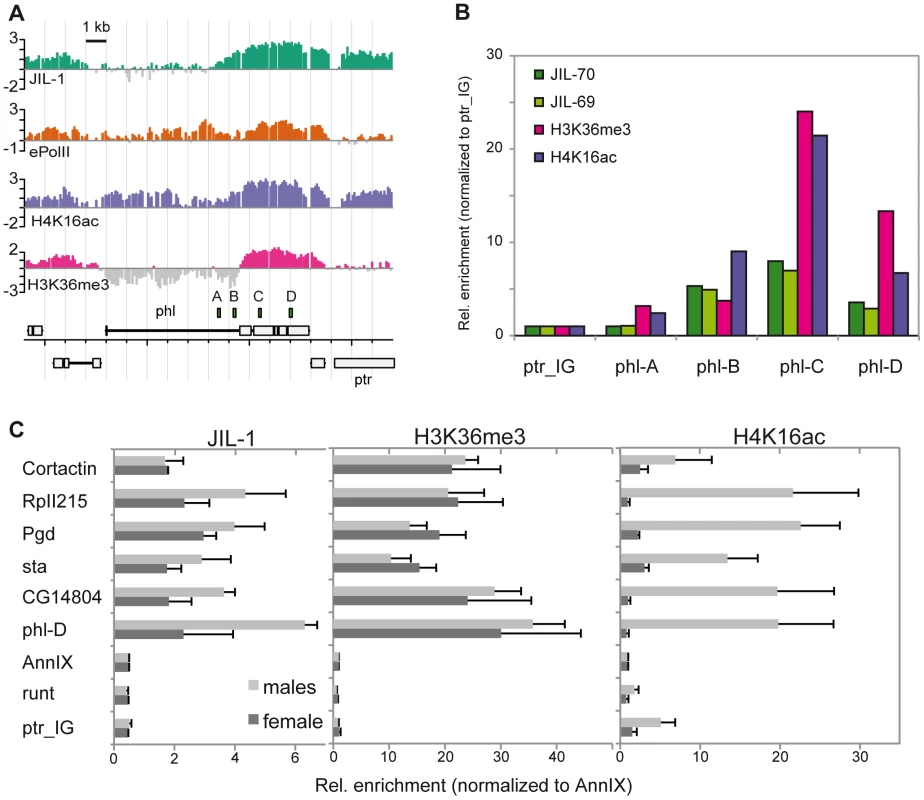
Comparison of the distribution of JIL-1, ePol, H4K16ac and H3K36me3 on the X-linked gene phl in SL2 cells. (A) Signals as revealed by ChIP-chip profiling were compared to (B) the qPCR-based quantification of unamplified material. The position of the amplicons used for qPCR is indicated above the gene annotations. The relative enrichments from amplicons A to D were calculated using the intergenic region located ∼2 kb left of the gene ptr (ptr_IG) as a reference. (C) Comparison of the enrichment of JIL-1, H3K36me3 and H4K16ac at the 3′ end of the active X-linked genes phl, CG14804, sta, Pgd, RpII215 and of the active autosomal gene Cortactin. The inactive X-linked gene runt, the promoter region of the annexin IX gene flanking the Cortactin gene and the intergenic region close to ptr served as negative control regions. Signals were normalized to the annexin IX locus and error bars represent the standard error of the mean for 3 independent biological replicates. We conclude that JIL-1 binds active genes in both sexes and exhibits a twofold enrichment on X chromosomal genes in males.
H3 phosphorylation by JIL-1 contributes to a composite phospho-acetyl mark associated with active chromatin
In order to characterise the potential of JIL-1 to phosphorylate histones we carried out in vitro kinase assays using baculovirus-expressed FLAG-tagged JIL-1. Recombinant histone H3 was phosphorylated, but not histones H2A, H2B, H4 (Figure S5A). Histone H1 prepared from Drosophila embryos, which can be phosphorylated to some extent at serine 10 [32] was not a substrate for JIL-1 in vitro (not shown). We did not detect phosphorylation of H3 in nucleosomes even by increasing the salt concentration in the assay (Figure S5A). A JIL-1 derivative, in which the catalytic domain was impaired by mutating an aspartate to alanine, had no H3 kinase activity (JIL-1D392A; [33]). We used H3-derived peptides to characterise the substrate specificity further and found that JIL-1 was unable to phosphorylate S28 in the context of the H321-34 peptide but efficiently phosphorylated the peptide H31–21. The phosphorylation occurred at serine 10 (S10) since it was abolished by incorporation of phospho-serine during peptide synthesis. At saturating amounts of peptide, neighbouring modifications of the H3 tail do not impair S10 phosphorylation (Figure S5).
H3S10 phosphorylation, particularly in the context of K14 acetylation (H3S10phK14ac) had previously been shown to be enriched on the male X chromosome [1]. By ChIP the enrichment of the straight H3S10ph epitope is difficult to document (a 1.09-fold enrichment in the data of Zhang and Oliver [28]), presumably due to the considerable amount of H3S10ph contributed by aurora kinase in mitotic cells (for review, see [34], [35]. Consistent with this, JIL-1 depletion by RNAi in SL2 cells did not lead to reproducible decrease of global H3S10ph levels (Figure S6). In order to estimate the contribution of mitotic H3S10ph in asynchronously growing SL2 cells, we arrested cells in G1/S by combined aphidicholine and hydroxyurea treatment and found the level of H3S10ph 8-fold reduced as compared to asynchronously growing cells. In the presence of the potent aurora kinase inhibitor ZM447439 [36] the global H3S10ph signal was 20 times reduced (Figure S6C). Since the proportion of mitotic SL2 cells in a culture has been estimated to be 4% [37], we conclude that the level of interphase H3S10ph is two orders of magnitude lower than that of mitotic H3S10ph. This may be an overestimate since an H3S10ph profile from ZM44743-treated cells showed very little signal over background (Figure S6E).
The detection of interphase H3S10ph in salivary glands has been shown to greatly depend on the antibody itself as well as on the procedure [19]. In particular, some antibodies may be occluded by neighbouring modifications in the H3 tail and not recognize S10ph as part of a composite epitope [38]. The two antisera used in this study, anti-H3S10ph [39] and anti-H3S10phK14ac [40] were therefore tested for their ability to recognise the S10ph mark in the presence of neighbouring modifications on the H3 tail using a custom-made peptide microarray (Figure S7). Interaction of the anti-S10ph serum, but not of the anti-S10phK14ac serum was adversely affected by the K14 acetylation. However, the latter also detected S10ph in the absence of K14 acetylation. This may explain why upon JIL-1 depletion by RNAi in SL2 cells, no decrease in H3S10phK14ac was detected by Western blotting (Figure S6D). Because the H3S10phK14ac epitope had been found enriched on the X [1] and the anti-H3S10phK14ac had been successfully used in ChIP experiments [40] we generated a global ChIP profile with the latter antibody (Figure 3A). A view of this profile along 140 kb of the X chromosome showed that H3S10phK14ac coincides with the presence of JIL-1. In addition, the H3S10phK14ac signal was enhanced on X chromosomal probes (Figure 3B) and preferentially mapped to the CDS of genes. Average binding profiles revealed that an enrichment of H3S10phK14ac throughout the transcription units can be observed in particular on X chromosomal JIL-1 target genes (Figure 3C). The global depletion of H3S10phK14ac could not be assessed by Western blot (Figure S6D), given the cross-reactivity of the antibody with (mitotic) H3S10ph but, the enrichment at the 3′ end of tested target genes is reduced upon JIL-1 RNAi (Figure 3D). Taken together, the data suggest that JIL-1 contributes to generating the composite S10phK14ac modification on histone H3 at active genes.
Fig. 3. JIL-1–dependent H3S10phK14ac is enriched on the compensated X chromosome together with JIL-1. 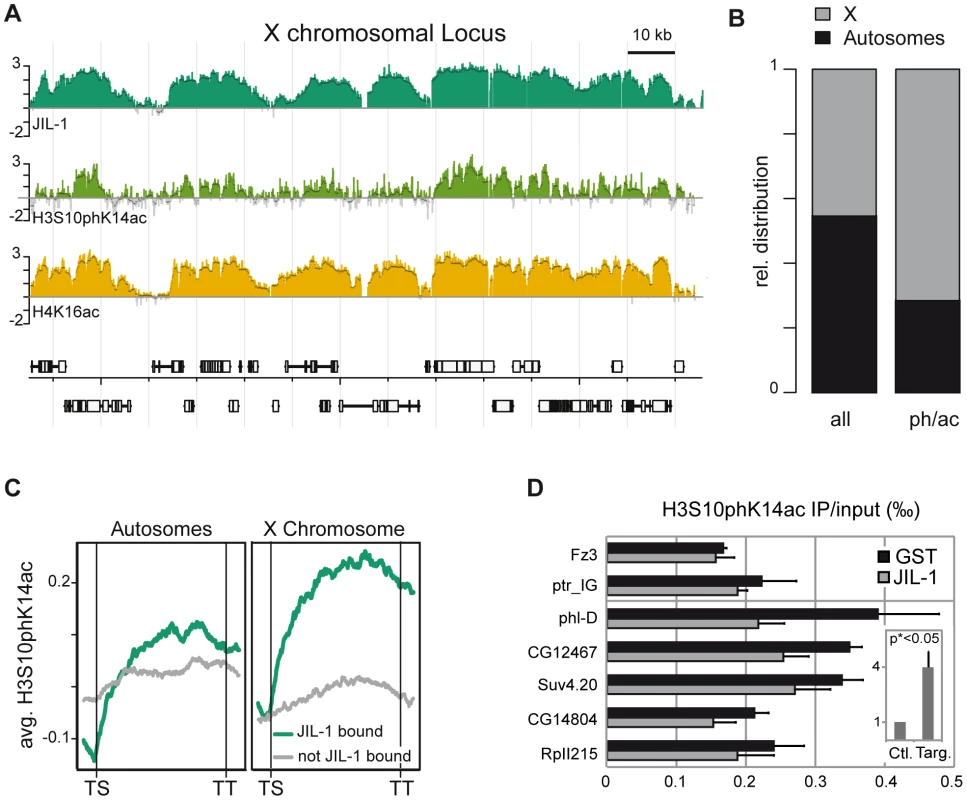
(A) Genome-wide distribution of H3S10phK14ac in comparison to JIL-1 and H4K16ac on a 140 kb X-chromosomal locus close to the gene phl. (B) Fractional distribution of probes significantly enriched for H3S10phK14ac (n = 4478) with respect to chromosomal location. As a comparison, the distribution of all probes (n = 384680) represented on the array is shown. (C) Cumulative binding profiles of H3S10phK14ac along genes that are either bound (n = 3362) or not bound (n = 2050) by JIL-1. (D) Decrease of H3S10phK14ac ChIP signal at the 3′ end of the genes phl, CG12467, Suv4.20, CG14804 and RpII215 after JIL-1 RNAi as compared to a control GST RNAi. The intergenic region close to ptr and the inactive gene Fz3 are shown as negative control regions. Results are the mean of 3 biological replicates and error bars represent the standard error of the mean. In the insert, we showed that the difference of the H3S10phK14ac ChIP signal after JIL-1 depletion is 4 times higher for target loci compared to control loci after normalization. This difference is statistically significant since the 2-sided unpaired t-test applied to the data gave a p value <0.05. JIL-1 increases the transcription efficiency of genes on the male X chromosome
The H3S10ph and H3S10phK14ac marks were correlated with active transcription in various model systems. In order to assess the contribution of JIL-1 to transcriptional output we reduced the levels of the kinase by RNAi in SL2 cells. Efficient depletion (up to 95%) was reproducibly achieved while ePol remained unaffected (Figure 4). This observation is in support of the results from the Johansen laboratory on salivary glands of the JIL-1 null mutant [19]. Removal of JIL-1 did not visibly affect the association of the DCC with the X chromosomal territory, which in Figure 4C is visualised by the diagnostic H4K16ac mark. Although flies with decreasing JIL-1 levels show increasing lethality [1] the proliferation of SL2 and KC cells appears only slightly affected (proliferation was monitored for up to 19 days in RNAi conditions, not shown).
Fig. 4. Effects of JIL-1 RNAi on gene expression and H4K16ac localization in SL2 cells. 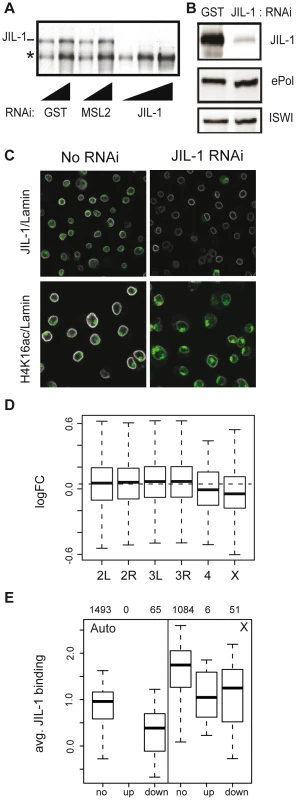
(A) The knock-down of JIL-1 was assessed by western blotting on whole cell extracts after 7 days of RNA interference (RNAi). RNAi against GST and MSL2 were used as controls. The affinity-purified serum of R69 was used to detect JIL-1. A cross-reacting protein of smaller size (*) was not affected. Two and three serial dilutions of the extract were loaded for controls and JIL-1 RNAi samples, respectively. (B) Levels of ePol detected with the H5 antibody were unaffected upon JIL-1 RNAi. The ATPase ISWI served as a loading control. (C) SL2 cells were immunostained for JIL-1, H4K16ac (in green) and lamin (in white) and analyzed by confocal imaging. (D) Log2 fold changes in gene expression after JIL-1 RNAi as compared to GST or GFP RNAi (ctr RNAi) summarized by chromosomes in a box plot. The dashed horizontal line indicates the median change of all active genes. (E) The average binding of JIL-1 per gene was plotted for autosomal genes on the left (Auto) and X-linked genes on the right (X) distinguishing genes with no expression change over JIL-1 depletion (‘no’), up-regulated genes (‘up’) and down-regulated genes (‘down’). The numbers on top indicate the number of genes in each category. Comparison of steady-state mRNA levels in JIL-1-depleted versus control cells were assessed by Affymetrix profiling. Two sets of experiments each consisting of three biological replicates were conducted with different sets of dsRNA. The statistical analysis of effects incorporates the results of all experiments. Among several thousand active genes bound by JIL-1 in SL2 cells, we found that after JIL-1 depletion the expression level was significantly reduced for 276 genes and increased for 25 genes. Globally, transcription of X-linked genes was significantly reduced as compared to the autosomal genes (p-value 1.2e-06) suggesting that the enrichment of JIL-1 on X-linked genes is functionally relevant for dosage compensation (Figure 4D). Interestingly, the few active genes on the fourth chromosome also appear to be particularly affected. We observed that total amount (Figure 4B) and distribution of ePol (not shown) were not changed after RNAi against JIL-1 and suggest that in SL2 cells, JIL-1 is probably not essential for the release of RNA polymerase II in productive elongation. Alternatively, the few percent of JIL-1 remaining after RNAi may be sufficient to fulfil this function. It is also noteworthy that, contrary to many proteins having a function in transcriptional activation, those genes whose transcription is sensitive to JIL-1 depletion tend to have lower JIL-1 levels to start with (Figure 4E).
JIL-1 association to genes does not follow ePol
To compare the distribution of ePol and JIL-1 along genes, we scaled all genes to equal length and aligned them from transcriptional start sites (TS) to transcriptional termination sites (TT). The genes were furthermore divided into six equally sized groups based on their average ePol enrichment and average distribution profiles were computed for each group (Figure 5A upper panels). The distribution of ePol along the transcribed regions is similar for autosomal and X-linked genes with a modest enrichment towards the 3′ end. Monitoring the binding of JIL-1 along the gene bodies as a function of ePol density, we found that while JIL-1 clearly associated with active genes, its binding was not proportional to ePol levels (Figure 5A centre panels). Rather, three out of the four groups with significant association of ePol showed the same average binding of JIL-1, except that there was roughly twice as much JIL-1 on X-chromosomal genes compared to autosomal genes, as observed before. This property was also illustrated when the density per gene was plotted against the corresponding steady-state transcript levels. Whereas the ePol density and transcript levels correlated well, JIL-1 binding was characterized by two clouds representing autosomal and X chromosomal genes (Figure S3B). These two representations show that the density of JIL-1 on active genes is not proportional to their transcription level. This was also observed for H3K36me3 and MSL1 distributions using the same representations (Figure S3C, S3D; Figure S8B, S8C) whereas the density of H3K4me2 and ePol were roughly proportional (Figure S8D). The discordance between ePol and JIL-1 distributions along gene bodies was even more pronounced when monitored as a function of gene length (Figure 5B). On autosomes, ePol was found enriched towards the 3′ end of short genes, but for genes longer than 1.8 kb the maximal density of ePol gradually shifted towards their 5′ ends as a function of length. This is also illustrated by visual inspection of very long genes both on the X and on autosomes (Figure S9). It is conceivable that the risk of premature transcription termination increases with gene length and thus leads to relative depletion of ePol at 3′ ends. Interestingly, this trend was less pronounced on the X chromosome, in keeping with the attractive hypothesis that dosage compensation facilitates transcriptional elongation through chromatin. By contrast, the relative distribution of JIL-1 along genes of various lengths is comparable on autosomes and the X chromosome, except for the twofold enrichment on the latter (Figure 5B).
Fig. 5. JIL-1 distributes differently than elongating polymerase on active chromatin. 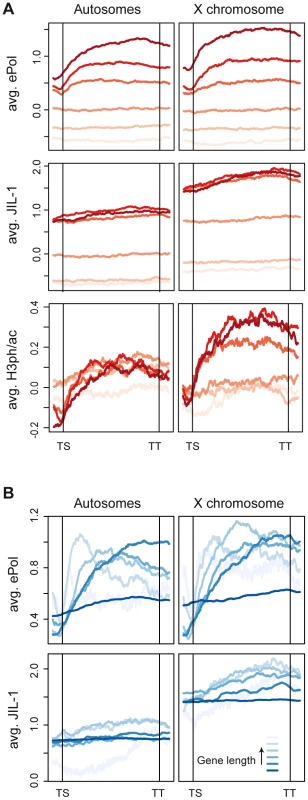
Average distribution profiles of chromatin features along genes scaled for length from transcription start (TS) to termination sites (TT). (A) All genes covered by the tiling array were grouped into 6 equally-sized bins based on increasing densities of ePol. Average profiles of ePol (top), JIL-1 (centre) and H3S10phK14ac (bottom) were calculated for each group. The increasing darkness of the red lines indicate an increasing density of ePol on the genes between the groups. (B) Genes were binned based on their length. The length distribution within the bins is the following: 287-1204, 1204-1780, 1780-2477, 2477-3778, 3778-7400 and 7400-162904 bp. Average profiles on the grouped genes are displayed for ePol (top) and JIL-1 (bottom). JIL-1 recruitment upon gene activation
A recent suggestion that JIL-1-dependent H3S10 phosphorylation was crucial to regulate the release of RNA polymerase into productive elongation in Drosophila in particular for the transcription of heat shock genes has remained controversial [17]–[19]. In our hands, heat shock treatment for 2, 5 and 10 min in SL2 cells triggered a clear enrichment of ePol at the Hsp70 locus as monitored by ChIP without strikingly affecting the distribution of JIL-1. In particular, we found that there was already some ePol and some JIL-1 at that locus in control cells as evidenced by the comparison with the active X-linked locus sta (Figure S10). Western blotting showed that the overall level of ePol drops upon heat shock, whereas the levels of JIL-1 varied only slightly. Because heat shock had been shown to trigger a massive loss of nucleosomes over a large domain [41], and because the genome-wide localization of JIL-1 appeared to correlate best with histone modifications (see below), we speculate that the lack of recruitment might be a consequence of nucleosome depletion.
We next made use of an inducible reporter system in flies ([26], [42]; Figure 6A), which harbours an insertion of a reporter gene cassette consisting of the lacZ gene downstream of five binding sites for the yeast transcription factor Gal4 (‘upstream activating sequences’; UASGal). The reporter was activated by crossing the reporter fly line with a line expressing either Gal4 alone or MOF fused to the Gal4 DNA binding domain (Gal4DBD). The basal level of β-galactosidase expression in the absence of activation was very low and could be induced about 1000-fold in male and female flies by expression of Gal4 under the control of a tubulin promoter. We monitored the levels of JIL-1, H3K36me3 and H4K16ac at the 5′ and 3′ ends of the lacZ gene by ChIP. As reference for the normalisation of the ChIP signals from different chromatin preparations we used the intergenic locus close to ptr (ptr_IG). In accordance with the results presented in Figure 2C, the X-chromosomal gene phl showed robust enrichment of JIL-1 in female flies and approximately twofold elevated levels in males. Very little recruitment of JIL-1 was observed at the reporter gene upon activation via Gal4 (Figure 6B). However, we did also not detect robust levels of H3K36me3 as compared to the endogenous loci. Since the tubulin-Gal4 driver leads to strong activation, which can be visualized by clear puffing at the level of polytene chromosomes, it is possible that these histone marks have been depleted along with nucleosomes due to vigorous transcription, a situation akin to the heat-induced transcription mentioned earlier. In order to trigger a more modest induction of the reporter gene we expressed the acetyltransferase MOF fused to the Gal4DBD (Gal4-MOF) at a low level from its endogenous promoter [26]. We limited this analysis to females in order to avoid the complications associated with all other members of the DCC. We had shown earlier that recruitment of Gal4-MOF to the reporter locus in females leads to H4K16ac and to a substantial transcriptional activation [26]. In the current experiment the activation was determined to be about 20-fold in the β-galactosidase assay. This activation was accompanied by an enrichment of JIL-1 at the 3′ end of the lacZ reporter (13-fold over the intergenic locus ptr_IG), which was about twofold higher than the enrichment at the endogenous X-linked locus phl (Figure 6C). In this case of a mild activation, we found an enrichment of H3K36me3 at the 3′ of the LacZ reporter, which was comparable to the endogenous loci phl and Cortactin. Upon induction with MOF, a 7-fold enrichment of H4K16ac is detected at the 5′ end of the LacZ reporter, which decreased towards the 3′ end. The Cortactin gene, localised about 2-3 kb downstream of the UASGAL as mapped by PCR-rescue of the transgene, also showed an increased level of H4K16ac upon Gal4-MOF targeting. Interestingly, the presence of H4K16ac in addition to H3K36me3 on the reporter gene LacZ correlated with an enrichment of JIL-1 comparable to that of X-linked genes in males. On the neighbouring Cortactin gene the enrichment was also increased. We found that the enrichment of both H4K16ac and JIL-1 at the Cortactin gene only increased the expression of the gene by a factor 1.2 in females. In accordance with previous data [26], in males H4K16ac spreads out more broadly from the recruitment sites due to the presence of the DCC and the relative expression of the Cortactin gene was increased by a factor 1.6 (Figure S11). Taken together the recruitment experiments suggest that the association of JIL-1 with active genes rather correlates with the presence of H3K36me3 and H4K16ac than with transcription as such. This conclusion can be substantiated by visual inspection of the genome-wide ChIP-chip profiles from SL2 cells, where numerous instances can be found where JIL-1 binding closely parallels the presence of H3K36me3, whereas the presence of high levels of ePol alone and sometimes also significant enrichment of H4K16ac alone do not coincide with JIL-1 binding (see Figure S9 for a selection of views).
Fig. 6. JIL-1 distribution upon activation of a reporter construct in flies. 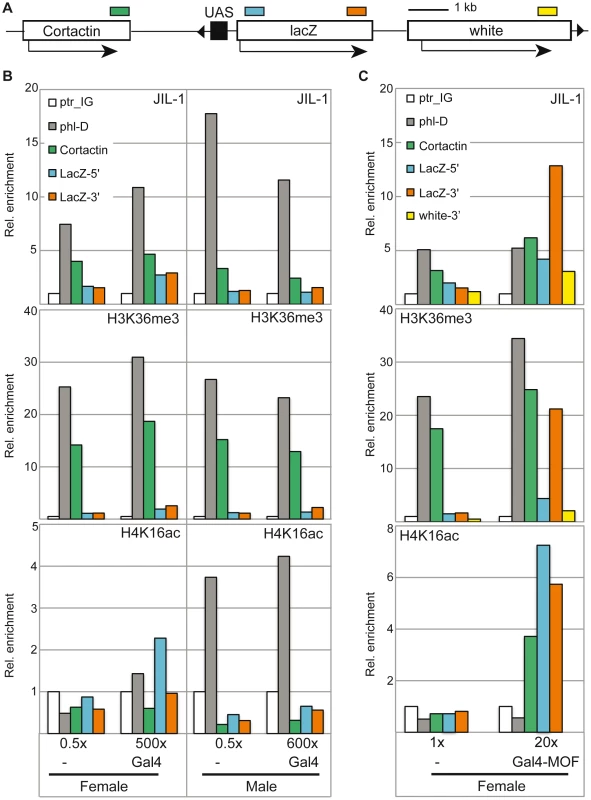
(A) The reporter construct encompassing five UAS in front of a LacZ reporter carries a miniwhite gene [42]. It is inserted on 3R at position 93B8 with the UAS close to the 3′ end of the Cortactin gene. The cartoon is drawn to scale, except for the negligible size of the UAS in order to visualize the distances between the amplicons (coloured boxes) used for qPCR analysis in panels B and C. (B) Activation of the LacZ reporter was achieved by crossing the homozygote reporter line to the Tubulin-Gal4/TM3 driver line expressing Gal4fl under control of the tubulin promoter. Relative β-galactosidase activity as measured in the corresponding flies is provided on the bottom of the panels. The reference was set to 0.5× for the heterozygote reporter line in the absence of activation. The ChIP signals for JIL-1 (upper panel), H3K36me3 (centre panel) and H4K16a (lower panel) at the intergenic locus (ptr_IG), at the 3′ of active X-linked gene (phl-D), at the 3′ of the Cortactin gene and at the 5′ and 3′ of the LacZ reporter gene are displayed. The data is provided as relative enrichments over the intergenic locus (ptr_IG). (C) For activation of the reporter by Gal4-MOF, the homozygote reporter line was used as a reference and compared to the stable line carrying both the homozygote reporter construct and the homozygote transgene leading to Gal4-MOF expression [26]. Only females were analyzed. The activity measurements for the two different types of females are mentioned below the graphs and can be compared to the values in B. In addition to the amplicons used in (B) we also analyzed binding to the 3′ end of the miniwhite gene. Because of the inherent variability of chromatin preparation one representative biological replicate is presented here. JIL-1 recruitment and dosage compensation
In order to further explore the relationship between JIL-1, transcription, H3K36me3 and H4K16ac, we looked in more detail at the relative twofold enrichment of JIL-1 on the male X chromosome. Johansen and colleagues [22] had documented earlier that the enrichment of JIL-1 on the male X chromosome depends on the DCC. Their key experiment was the ectopic expression of MSL2 in female larvae, which leads to inappropriate assembly of the DCC. The association of the DCC with the female X chromosome led to an enrichment of JIL-1, just like in males [22]. In agreement with these data, in similar experiments we found that if high levels of MSL2 are expressed from two NOPU alleles [43], [44] the X chromosome is decorated with DCC and JIL-1 is enriched concomitantly at sites of DCC binding. However, when limiting levels of MSL2 are expressed from the SXB1-2 allele [43], [44] the DCC only associates with about 50 high affinity sites (HAS) on polytene chromosomes. In this case, the enrichment of JIL-1 can only be seen at sites of DCC binding (Figure 7A). This enrichment requires the assembly of a complete DCC, including MOF, as it is not observed in the absence of MLE or MSL3 ([22] and data not shown).
Fig. 7. Distribution of JIL-1 on the X-chromosome. 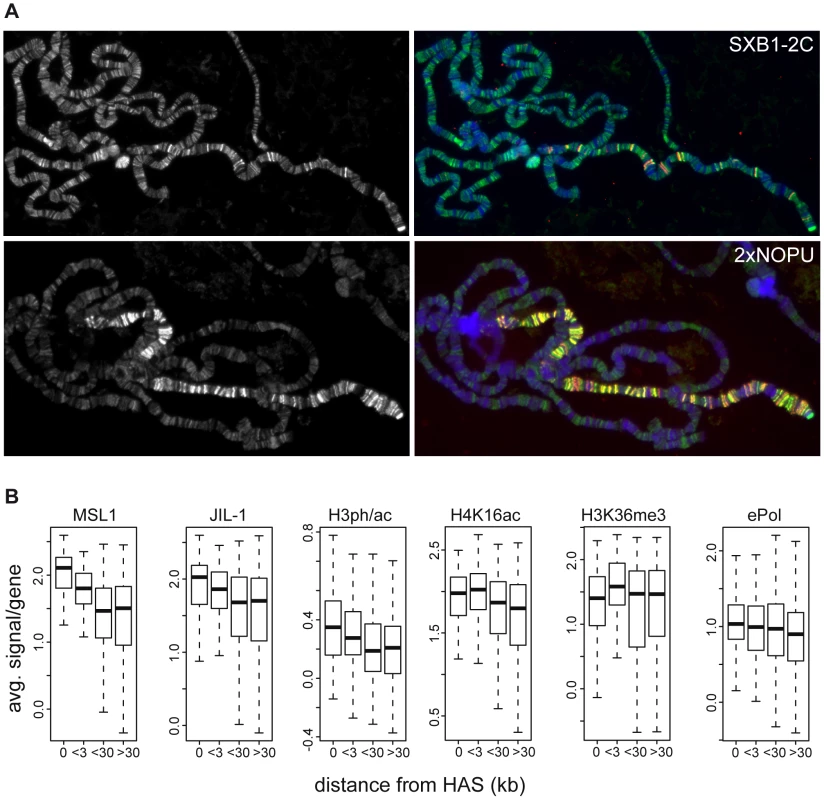
(A) Localization of JIL-1 (left panels, green in the merged pictures) in SXB1-2C and 2xNOPU females expressing increasing amounts of MSL2 (coloured red in the merged pictures on the right). (B) Densities of various features on active X-chromosomal genes grouped by increasing distance from high affinity sites (HAS) for the DCC. There are 115, 121, 405 and 465 genes in the group of genes located at increasing distances from HAS, respectively. The close relationship between DCC association and JIL-1 was also apparent at the high resolution achieved by ChIP-chip mapping. We had earlier identified 131 HAS that are preferentially bound by limiting amounts of DCC [29]. Plotting the binding of MSL1 to genes as a function of their distance to the closest HAS shows that, in general, the closer a gene resides to a HAS, the more MSL1 and H4K16ac can be found (Figure 7B). The same trend can be observed for JIL-1 as well as for the linked H3S10phK14ac mark: their densities are decreasing with increasing distance to a HAS. This is, however, not true for H3K36me3 or ePol association (Figure 7B).
In summary, diverse experiments establish two hallmarks of active chromatin that correlate with the presence of JIL-1. First, there is an excellent correlation of JIL-1 binding to H3K36me3-marked chromatin. On top of this basal level of binding, the presence of the DCC leads to an approximately twofold enrichment of JIL-1. Indeed, a global comparison of ChIP-chip probe signals (Figure 8) reveals that the correlations of JIL-1 signals with the ones of H3K36me3 (R = 0.758) or H4K16ac (R = 0.76) are very good. Remarkably, the combination of both marks (sum of the intensities of the H3K36me3 and H4K16ac signals) correlated even better with JIL-1 signals (R = 0.899) and displayed an almost linear relationship between signal intensities. This is true if all probes are considered, but also if autosomal or X chromosomal probes are plotted separately (Figure 8). This correlation can be recapitulated to some extent when the distribution of those marks is considered along genes (Figure S12). In contrast, correlation of JIL-1 and ePol (R = 0.69) is less pronounced (Figure 8). This result is consistent with the earlier observation that ePol and JIL-1 are both marks for active genes but that their relative distributions along genes differ (Figure 5).
Fig. 8. Correlations of H3K36me3, H4K16ac, and JIL-1 binding. 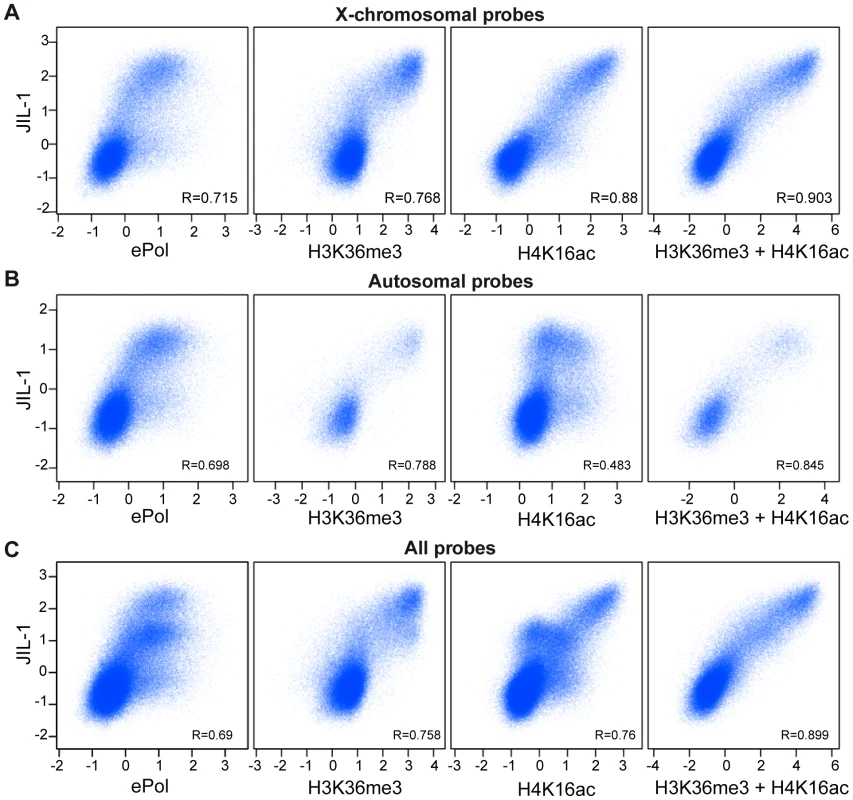
Pair-wise correlations of JIL-1 versus ePol, H3K36me3, H4K16ac and the sum of H3K36me3 and H4K16ac signals in unified 200 bp sampling windows. (A) Results for the X chromosome (B) for autosomes (C) and for the entire genome. Pearson correlation coefficients are provided for each panel. We conclude that two distinct targeting principles that are represented by H3K36me3 and H4K16ac marks determine the level of JIL-1 recruitment to active genes.
Discussion
Targeting of JIL-1
The genome-wide mapping of JIL-1 and ePol in male SL2 cells together with the reporter assay in flies suggest that JIL-1 is part of a network of factors that collectively define the state of active chromatin. Taken together with previous genetic analyses, our data lead us to consider to a role for JIL-1 not in the establishment of active chromatin, but in the reinforcement of the active state, independently of the extend of transcription.
Although JIL-1 binds the bodies of most active gene and is recruited to a reporter gene upon activation, the degree of binding does not correlate well with ePol occupancy. In this respect JIL-1 belongs to a class of several other active chromatin components whose presence in chromatin does not scale with ePol, such as H3K36me3, the DCC subunit MSL1, or H4K16ac mark. By contrast, H3K4me2 levels are proportional to ePol levels. Conceivably, JIL-1 marks active chromatin independent of whether it is currently transcribed or not. A basal level of JIL-1 binding correlates strongly with H3K36me3, an elongation marker that is placed co-transcriptionally by ePol-associated dSet2 [30]. The turnover time of this modification is not known and hence it is possible that H3K36me3 remains on chromatin between pulses of transcription.
Most recently, high-resolution mapping of 56 chromatin components using Dam-ID revealed five types of chromatin in Drosophila KC cells [45]. A colour code was used to illustrate those types. The two types of active chromatin, ‘red’ and ‘yellow’, differ in that active genes in red chromatin tend to be depleted in H3K36me3 and MRG15, which are abundant constituents of yellow chromatin. Monitoring the relative distributions of ePol and JIL-1 in those types of chromatin we found that whereas ePol is equally distributed in red and yellow chromatin, JIL-1 is clearly mostly found in yellow chromatin (Figure S13). This correlation again highlights the relationship between JIL-1 and H3K36me3. The precise link between H3K36me3 and JIL-1 recruitment remains to be explored (see below).
A second targeting principle is evident from the fact that JIL-1 levels are approximately twofold increased on the male X chromosome, where due to the action of the DCC H4K16ac levels are high. The situation is complex since the recruitment of the DCC to transcribed genes on the X chromosome is promoted by the potential interaction of the DCC subunit MSL3 with H3K36me3 [30]. Because JIL-1 binding is insensitive to different transcription rates on autosomes, it is not plausible that a presumed twofold increase in transcription on the X chromosome is directly responsible for this elevated association. Likewise, we do not think that direct interactions between JIL-1 and the DCC can explain the targeting. Although some interactions have been observed in vitro between JIL-1, MSL1 and MSL3 [22], a quantitative co-purification of endogenous JIL-1 with the DCC was never documented. Indeed, activation of a reporter gene in female flies through recruitment of Gal4-MOF is sufficient to recruit JIL-1 to male levels on the reporter gene and on the adjacent Cortactin gene. Altogether, we favour the hypothesis that the enrichment of JIL-1 on X-linked genes is a consequence of a modulating feature of dosage-compensated chromatin. The genome-wide distribution of JIL-1 indeed correlates best with the combination of the two chromatin modifications, H3K36me3 and H4K16ac, on the X-chromosome suggesting that JIL-1 recruitment occurs downstream of those two histone marks. Interestingly, the correlation still holds for autosomal probes, although autosomal H4K16ac is at least in part placed at the 5′ end of genes by alternative MOF complexes [26], [46], [47]. JIL-1 recruitment has also been linked to the H4K12ac mark, which is placed by the GCN5-containing ATAC complex [48]. However, Workman and colleagues recently found that ATAC also contains an acetyltransferase with specificity for H4K16 (atac2), which may contribute to JIL-1 recruitment [49].
In the cases of very strong transcription activation by high levels of Gal4 activator, we did not see an accumulation of JIL-1 to endogenous levels. Conversely, JIL-1 is also not enriched at developmental puffs on polytene chromosomes where robust transcription takes place according to the strong ePol staining (Figure S1). The massive heat-induced activation leads to extensive chromatin decondensation (puffing) on polytene chromosomes, which is probably accompanied by nucleosome loss [41].
In summary, we favour the idea that the recruitment of JIL-1 relies on several, partly redundant features of active chromatin. JIL-1 harbours an H3-tail binding domain in its C-terminus and another determinant of chromatin targeting in its N-terminal domain [3], but it does not contain any domain known to directly bind modified histone tails. Its recruitment to active chromatin is, therefore, most probably indirect. We know of no histone modification or any other chromatin-associated feature to be enriched on the male X chromosome except for JIL-1 and the JIL-1-dependent phospho-acetyl mark. An appealing model for JIL-1 targeting is that its general recruitment mode (correlating with H3K36me3) would be the same on all chromosomes. The additional presence of H4K16ac might enhance the access of JIL-1 to the first targeting principle, since it has been shown that H4K16ac prevents the folding of the nucleosomal fibre into more compact structures [27].
Effects of H3S10 phosphorylation by JIL-1
It has been observed that histone H3S10ph can enhance acetylation of histone H3K14 [50], [51] and inhibit methylation of histone H3K9 [52]. The biochemical analysis of JIL-1 kinase showed that a number of other modifications of the H3 N-terminus are compatible with the phosphorylation by JIL-1. Therefore, JIL-1 phosphorylation is compatible with the prior existence of other modifications, consistent with a downstream function of JIL-1.
Our data suggest that in Drosophila the function of H3S10ph may at least in part differ from that in other organisms, where it is implicated in the fast and transient induction of promoters in response to various inducers [13], [16], [53], [54]. In Drosophila, JIL-1 is not particularly enriched at promoters, but associates with genes along their entire length. This distribution is more compatible with a role in fine-tuning transcriptional elongation. The effects of JIL-1 may – at least in part – be mediated by 14-3-3 proteins, which are able to recognize the interphase H3S10ph mark [17], [54].
The depletion of JIL-1 has a clear but small effect on overall transcription. The fact that JIL-1 binds gene bodies rather that promoters and the the binding is independent of actual transcription rates leads us to speculate that the kinase may influence transcription efficiency only very indirectly by reinforcing the active chromatin state – once established by co-activators – by protecting it from neighbouring repressive chromatin. This could become important when moderately active genes are not constantly transcribed. Highly transcribed genes experience nucleosome depletion and therefore do not provide anchoring sites for silencing factors. Moderately transcribed genes, however, may experience periods without active elongation [55]-[57]. These ‘gaps’ bear the risk of placement of silencing marks, most notable H3K9me2. In this context JIL-1 may safeguard active genes (marked by H3K36me3 as a sign of recent transcriptional activity). This function may be achieved by phosphorylation of H3S10, which prevents the recognition of HP1 by occlusion of H3K9me2 [58], [59] but possibly also by phosphorylation of other substrates, like Su(var)3,9 [33]. In addition JIL-1 might have a scaffolding function for other factors, like lamins [60]. The observation that genes sensitive to the depletion of JIL-1 tend to have less JIL-1 bound both on the X chromosome and on autosomes also support a model where JIL-1 is associated to active chromatin domains to prevent the spreading of more repressive structures. We speculate that the role of JIL-1 in maintaining the balance between heterochromatin and euchromatin is not only true at the hetero/euchromatin boundary, but also within euchromatin at the level of active chromatin domains.
Materials and Methods
JIL-1 antibodies and kinase assay
Polyclonal antisera were raised in two rabbits against a fragment of JIL-1 corresponding to amino acids 79–571, which was expressed in fusion with an N-terminal MBP-tag in E. coli (Eurogentec). The resulting sera R69 and R70 were either used crude or after affinity purification on a column with covalently coupled antigen according to a standard protocol. For Western blot, the sera were diluted 1/2000 and affinity-purified antibodies were used at 0.5 µg/ml. Recombinant Flag-tagged JIL-1 and kinase assay conditions were as previously described [33].
Reporter lines and β-galactosidase activity measurements
The transgenic line used for the reporter assay in flies was originally published as U/l5 [42]. The P-element insertion site was mapped by PCR-rescue to the 3′end of the Cortactin gene (Figure 6A). To activate the reporter by the Gal4 activator homozygote flies of the reporter line were crossed with males of the activator line, which carry a transgene driving the expression of the Gal4 activator under the control of the tubulin promoter (Bloomington Stock Center #5138). Gal4 is expressed from the cellular blastoderm onwards. The F1 flies were hand-sorted according to their genotype and sex in order to obtain 190–250 mg of flies for chromatin preparation. β-galactosidase activity in extracts prepared from 2–10 flies were measured as previously described [26]. The flies carrying the balancer were used as negative controls for the flies carrying the Gal4 activator. To activate the reporter by Gal4-MOF recruitment, we used a stable line homozygote for the reporter construct and prepared chromatin for ChIP from sorted 200–500 female flies [26].
Chromatin immunoprecipitation
Chromatin from SL2 cells was prepared after crosslinking in 1% formaldehyde as previously described [29]. To preserve the phospho-epitopes and the acetylation status of the chromatin, we used 5 mM NaF, 20 mM β-glycerophosphate, 0.1 mM NaVanadate, 10 mM NaButyrate and a mixture of protease inhibitors (1mM PMSF, 1 µg/ml of each aprotinin, pepstatin and leupeptin) or the complete protease inhibitors (Roche, Cat No. 04693132011). In the particular case of the chromatin used for mapping H3S10phK14ac, 1 µM microcystin was also added. The chromatin from sorted male and/or female flies was done according to following references [26], [61]. Chromatin shearing to 300–1000 bp fragments was assessed on a Bioanalyzer (Agilent) and DNA concentration was measured using NanoDrop (Thermo Scientific). 7.5 to 10 µg of DNA per chromatin was used for ChIP. For JIL-1 ChIPs we used either crude sera R69 and R70 (5 µl/IP) or the affinity-purified antibodies (5 µg/IP). A LiCl wash was done for ChIP-chip analysis. For ePol ChIPs, 5 µl of the anti-CTD S2ph monoclonal antibody Cone H5 (ab24758, Abcam) was first coupled to a 50/50 mix of protein A - and protein G-agarose using a bridging antibody (5 µg of Goat anti-mouse IgM, Jackson ImmunoResearch) and the LiCl wash was omitted. The following volumina of antibodies were used for ChIP: H3S10phK14ac: 3 µl; H4K16ac: 5 µl of antibody #39167 (Active Motif); H3K36me3 : 5 µl of ab9050 (Abcam). MSL2 antibodies were used as control as previously published [62].
Quantitative PCR
Quantitative PCR (qPCR) was carried out in an ABI PRISM 7000 Sequence detection system in a 25 µl reaction with Power SYBR Green Master Mix (Applied Biosystems) or in LightCycler (Roche) in 10 µl reactions with Fast SYBR Green Master mix (Applied Biosystems). Input DNA were diluted to 10 ng/µl and ChIPs were diluted 1/30, 5 µl and 2 µl template were used for ABI PRISM 7000 and LightCycler, respectively. The results were comparable on the two machines. Primer sequences are provided in Table S1.
ChIP-chip and data analysis
ChIP-chip data analysis was essentially performed as in [29]. Briefly, input and IP DNA were amplified using the WGA kit (Sigma) according to the online protocol (http://www.epigenome-noe.net/WWW/researchtools/protocol.php?protid=30). Labeling and hybridization to NimbleGen dual-color arrays was carried out at ImaGenes (Berlin, Germany). The layout of the array (approx. 1 probe/100 bases, isothermal probe design) was customized [24]. Data analysis was performed using R/Bioconductor (www.bioconductor.org). Experimental replication was as follows: JIL-1 : 4 biological replicates; H3S10phK14ac: 2 biological replicates including dye swap; elongating RNA Polymerase II: 3 biological replicates including one dye swap; H4K16ac: 2 biological replicates including dye swap. Raw signals of corresponding biological samples were log2 transformed and quantile-normalized. Enrichment statistics (IP versus input signals) were computed using the ‘sam’ algorithm within Bioconductor [63]. Fdr values of the sam statistic were determined using ‘locfdr’ [64] region summarization was performed using tileHMM [65] with the following parameters: fragment size of 700, maximal gap of 400. Genes were considered ‘bound’ if covered by at least 4 probes with a posterior probability above 0.5. All data correspond to Drosophila genome version dm3 and annotation version gadfly 5.2.2.
RNAi in SL2 cells and purification of total RNA
SL2 cells were cultured according to standard methods. RNA interference (RNAi) was carried out as published [66] except that 2×106 cells were treated with 10 µg dsRNA and splitted 1/3 after 3 days of treatment. After 7 days cells were collected and counted prior to total RNA purification. For the 2 sets of expression analysis we used different sets of dsRNA for RNAi. In experiment 1, the 5′ coding sequence of JIL-1 was targeted by a corresponding dsRNA (T7_JIL-15′_for: TTAATACGACTCACTATAGGGAGATGAGTCGCTTG, T7_JIL-15′_rev: TTAATACGACTCACTATAGGGAGACATCCTCCTCCA), and the controls involved treating cells with dsRNA targeting GST sequences (T7_GST_for: TTAATACGACTCACTATAGGGAGAATGTCCCCTATACTAGGTTA, T7_GST_rev: TTAATACGACTCACTATAGGGAGAACGCATCCAGGCATTG). In the second set JIL-1 was knock - down by targeting its 3′ coding sequences (T7_JIL-13′_for: TTAATACGACTCACTATAGGGAGACAGCAGCGTCG, T7_JIL-13′_rev:TTAATACGACTCACTATAGGGAGATTGGAACTGAT) and the control dsRNAs represented GFP sequences (T7_GFP_for: TTAATACGACTCACTATAGGGTGCTCAGGTAGTGGTTGTCG, T7_GFP_rev:TTAATACGACTCACTATAGGGCCTGAAGTTCATCTGCACCA). Off-target effects were minimized using http://dscheck.rnai.jp/. Efficiency of the depletion was assessed on whole cell extracts and quantified with the Odyssey imaging system (LI-COR, Biosciences) using anti-tubulin (T9026, clone DM1A, Sigma) or anti-lamin (Clone T40, [67]) as reference. Total RNA was purified from 20-80×106 cells using either the RNA purification kit (RNeasy mini kit, Qiagen) or the lysis reagent (Qiazol, Qiagen) according to manufacturers instructions. The quality of the RNA was determined on the Bioanalyzer (Agilent).
Expression analysis after JIL-1 RNAi
Total RNA was processed for hybridization at ImaGenes (Berlin, Germany). Five biological replicates for each control and JIL-1 RNAi were analysed. Microarray data were processed using R/Bioconductor (www.bioconductor.org). Unless indicated otherwise we used standard parameters in all function calls. Expression values were calculated using ‘gcrma’. Probe sets were kept for differential expression analysis if there were more ‘present’ calls (calculated using ‘mas5calls’) in one of the two treatment groups than non-‘present’ calls and if their expression level variance was higher than 0 across all arrays. “One gene to many probe set“ relationships were resolved by retaining only the probe set with the highest variance across all arrays. Differential expression statistics were obtained using a linear model (library 'limma) including a batch effect coefficient as two sets of arrays were processed at different times. A significant response was defined if the local false discovery (‘locfdr’ package) rate calculated on the moderated t statistic was smaller than 0.2.
Microscopy
Polytene chromosome staining was according to [44]. The affinity-purified antibodies R70 or R69 were used diluted 1/150 and the H5 monoclonal antibody at 1/50. Staining of SL2 cells was according to a standard protocol. In brief, after fixation with 3.7% (w/v) formaldehyde in PBS buffer, the cells were permeabilised with 0.3% triton X-100 (v/v) in PBS in the presence of 1% formaldehyde. The affinity-purified antibodies of R70 or R69 were diluted at 0.4 µg/ml, the anti-H4K16ac antibody #39167 (ACTIVE MOTIF) was diluted 1/1000 for counterstaining of the X-chromosome territory, and the anti-lamin clone T40 [67] was diluted at 1/100 to delimitate the nuclear territory. Epifluorescence microscopy was done on the Axiovert 200 inverted microscope (Zeiss) and confocal images were taken using LSM510 Meta system (Zeiss).
Accession numbers
Microarray data has been deposited at GEO (accession GSE22620). The Nimblegen tiling data have been deposited at GEO (accession GSE22618).
Supporting Information
Zdroje
1. WangY
ZhangW
JinY
JohansenJ
JohansenKM
2001 The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 105 433 443
2. JinY
WangY
WalkerDL
DongH
ConleyC
1999 JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol Cell 4 129 135
3. BaoX
CaiW
DengH
ZhangW
KrencikR
2008 The COOH-terminal domain of the JIL-1 histone H3S10 kinase interacts with histone H3 and is required for correct targeting to chromatin. J Biol Chem 283 32741 32750
4. DengH
ZhangW
BaoX
MartinJN
GirtonJ
2005 The JIL-1 kinase regulates the structure of Drosophila polytene chromosomes. Chromosoma 114 173 182
5. DengH
BaoX
CaiW
BlacketerMJ
BelmontAS
2008 Ectopic histone H3S10 phosphorylation causes chromatin structure remodeling in Drosophila. Development 135 699 705
6. ZhangW
DengH
BaoX
LerachS
GirtonJ
2006 The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development 133 229 235
7. LerachS
ZhangW
BaoX
DengH
GirtonJ
2006 Loss-of-function alleles of the JIL-1 kinase are strong suppressors of position effect variegation of the wm4 allele in Drosophila. Genetics 173 2403 2406
8. BaoX
DengH
JohansenJ
GirtonJ
JohansenKM
2007 Loss-of-function alleles of the JIL-1 histone H3S10 kinase enhance position-effect variegation at pericentric sites in Drosophila heterochromatin. Genetics 176 1355 1358
9. EbertA
SchottaG
LeinS
KubicekS
KraussV
2004 Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev 18 2973 2983
10. DengH
BaoX
ZhangW
GirtonJ
JohansenJ
2007 Reduced levels of Su(var)3-9 but not Su(var)2-5 (HP1) counteract the effects on chromatin structure and viability in loss-of-function mutants of the JIL-1 histone H3S10 kinase. Genetics 177 79 87
11. DengH
CaiW
WangC
LerachS
DelattreM
2010 JIL-1 AND SU(VAR)3-7 Interact Genetically and Counteract Each Other's Effect on Position Effect Variegation in Drosophila. Genetics
12. VermeulenL
BergheWV
BeckIM
De BosscherK
HaegemanG
2009 The versatile role of MSKs in transcriptional regulation. Trends Biochem Sci 34 311 318
13. WinterS
SimboeckE
FischleW
ZupkovitzG
DohnalI
2008 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J 27 88 99
14. VicentGP
BallareC
NachtAS
ClausellJ
Subtil-RodriguezA
2006 Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell 24 367 381
15. MacdonaldN
WelburnJP
NobleME
NguyenA
YaffeMB
2005 Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol Cell 20 199 211
16. ZippoA
SerafiniR
RocchigianiM
PennacchiniS
KrepelovaA
2009 Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138 1122 1136
17. KaramCS
KellnerWA
TakenakaN
ClemmonsAW
CorcesVG
2010 14-3-3 Mediates Histone Cross-Talk during Transcription Elongation in Drosophila. PLoS Genet 6 e1000975 doi:10.1371/journal.pgen.1000975
18. IvaldiMS
KaramCS
CorcesVG
2007 Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev 21 2818 2831
19. CaiW
BaoX
DengH
JinY
GirtonJ
2008 RNA polymerase II-mediated transcription at active loci does not require histone H3S10 phosphorylation in Drosophila. Development 135 2917 2925
20. StraubT
BeckerPB
2007 Dosage compensation: the beginning and end of generalization. Nat Rev Genet 8 47 57
21. GelbartME
KurodaMI
2009 Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136 1399 1410
22. JinY
WangY
JohansenJ
JohansenKM
2000 JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the male specific lethal (MSL) dosage compensation complex. J Cell Biol 149 1005 1010
23. AlekseyenkoAA
LarschanE
LaiWR
ParkPJ
KurodaMI
2006 High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 20 848 857
24. GilfillanGD
StraubT
de WitE
GreilF
LammR
2006 Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev 20 858 870
25. GelbartME
LarschanE
PengS
ParkPJ
KurodaMI
2009 Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol 16 825 832
26. PrestelM
FellerC
StraubT
MitlöhnerH
BeckerPB
2010 The activation potential of MOF is constrained for doasege compensation. Molecular Cell 38 815 826
27. Shogren-KnaakM
IshiiH
SunJM
PazinMJ
DavieJR
2006 Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311 844 847
28. ZhangY
OliverB
2010 An evolutionary consequence of dosage compensation on Drosophila melanogaster female X-chromatin structure? BMC Genomics 11 6
29. StraubT
GrimaudC
GilfillanGD
MitterwegerA
BeckerPB
2008 The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet 4 e1000302 doi:10.1371/journal.pgen.1000302
30. LarschanE
AlekseyenkoAA
GortchakovAA
PengS
LiB
2007 MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28 121 133
31. LeeJS
ShilatifardA
2007 A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat Res 618 130 134
32. Villar-GareaA
ImhofA
2008 Fine mapping of posttranslational modifications of the linker histone H1 from Drosophila melanogaster. PLoS ONE 3 e1553 doi:10.1371/journal.pone.0001553
33. BoekeJ
RegnardC
CaiW
JohansenJ
JohansenKM
2010 Phosphorylation of SU(VAR)3-9 by the chromosomal kinase JIL-1. PLoS ONE 5 e10042 doi:10.1371/journal.pone.0010042
34. GietR
GloverDM
2001 Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol 152 669 682
35. NowakSJ
CorcesVG
2004 Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet 20 214 220
36. GadeaBB
RudermanJV
2005 Aurora kinase inhibitor ZM447439 blocks chromosome-induced spindle assembly, the completion of chromosome condensation, and the establishment of the spindle integrity checkpoint in Xenopus egg extracts. Mol Biol Cell 16 1305 1318
37. Morales-MuliaS
ScholeyJM
2005 Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol Biol Cell 16 3176 3186
38. ClaytonAL
HazzalinCA
MahadevanLC
2006 Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell 23 289 296
39. ClaytonAL
RoseS
BarrattMJ
MahadevanLC
2000 Phosphoacetylation of histone H3 on c-fos - and c-jun-associated nucleosomes upon gene activation. EMBO J 19 3714 3726
40. BrunmeirR
LaggerS
SimboeckE
SawickaA
EggerG
2010 Epigenetic regulation of a murine retrotransposon by a dual histone modification mark. PLoS Genet 6 e1000927 doi:10.1371/journal.pgen.1000927
41. PeteschSJ
LisJT
2008 Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134 74 84
42. ZinkD
ParoR
1995 Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J 14 5660 5671
43. KelleyRL
WangJ
BellL
KurodaMI
1997 Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387 195 199
44. DahlsveenIK
GilfillanGD
ShelestVI
LammR
BeckerPB
2006 Targeting determinants of dosage compensation in Drosophila. PLoS Genet 2 e5 doi:10.1371/journal.pgen.0020005
45. FilionGJ
van BemmelJG
BraunschweigU
TalhoutW
KindJ
2010 Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143 212 224
46. KindJ
VaquerizasJM
GebhardtP
GentzelM
LuscombeNM
2008 Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell 133 813 828
47. RajaSJ
CharapitsaI
ConradT
VaquerizasJM
GebhardtP
2010 The nonspecific lethal complex is a transcriptional regulator in Drosophila. Molecular Cell 38 827 841
48. CiurciuA
KomonyiO
BorosIM
2008 Loss of ATAC-specific acetylation of histone H4 at Lys12 reduces binding of JIL-1 to chromatin and phosphorylation of histone H3 at Ser10. J Cell Sci 121 3366 3372
49. SuganumaT
GutierrezJL
LiB
FlorensL
SwansonSK
2008 ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol 15 364 372
50. LoWS
TrievelRC
RojasJR
DugganL
HsuJY
2000 Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell 5 917 926
51. CheungP
TannerKG
CheungWL
Sassone-CorsiP
DenuJM
2000 Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 5 905 915
52. ReaS
EisenhaberF
O'CarrollD
StrahlBD
SunZW
2000 Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406 593 599
53. ZippoA
De RobertisA
SerafiniR
OlivieroS
2007 PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol 9 932 944
54. WinterS
FischleW
SeiserC
2008 Modulation of 14-3-3 interaction with phosphorylated histone H3 by combinatorial modification patterns. Cell Cycle 7 1336 1342
55. FudaNJ
ArdehaliMB
LisJT
2009 Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461 186 192
56. HagerGL
McNallyJG
MisteliT
2009 Transcription dynamics. Mol Cell 35 741 753
57. WijgerdeM
GrosveldF
FraserP
1995 Transcription complex stability and chromatin dynamics in vivo. Nature 377 209 213
58. FischleW
TsengBS
DormannHL
UeberheideBM
GarciaBA
2005 Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438 1116 1122
59. HirotaT
LippJJ
TohBH
PetersJM
2005 Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438 1176 1180
60. BaoX
ZhangW
KrencikR
DengH
WangY
2005 The JIL-1 kinase interacts with lamin Dm0 and regulates nuclear lamina morphology of Drosophila nurse cells. J Cell Sci 118 5079 5087
61. NegreN
HennetinJ
SunLV
LavrovS
BellisM
2006 Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol 4 e170 doi:10.1371/journal.pbio.0040170
62. GilfillanGD
KonigC
DahlsveenIK
PrakouraN
StraubT
2007 Cumulative contributions of weak DNA determinants to targeting the Drosophila dosage compensation complex. Nucleic Acids Res 35 3561 3572
63. TusherVG
TibshiraniR
ChuG
2001 Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98 5116 5121
64. EfronB
2007 Correlation and large scale simultaneous significance testing. Jour Amer Stat Assoc 102 99 103
65. HumburgP
BulgerD
StoneG
2008 Parameter estimation for robust HMM analysis of ChIP-chip data. BMC Bioinformatics 9 343
66. StraubT
NeumannMF
PrestelM
KremmerE
KaetherC
2005 Stable chromosomal association of MSL2 defines a dosage-compensated nuclear compartment. Chromosoma 114 352 364
67. RisauW
SaumweberH
SymmonsP
1981 Monoclonal antibodies against a nuclear membrane protein of Drosophila. Localization by indirect immunofluorescence and detection of antigen using a new protein blotting procedure. Exp Cell Res 133 47 54
68. BoehmAK
SaundersA
WernerJ
LisJT
2003 Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol 23 7628 7637
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and PhosphodiesterasesČlánek Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



