-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
During animal development, cellular morphogenesis plays a fundamental role in determining the shape and function of tissues and organs. Identifying the components that regulate and drive morphogenesis is thus a major goal of developmental biology. The four-celled tip of the Caenorhabditis elegans male tail is a simple but powerful model for studying the mechanism of morphogenesis and its spatiotemporal regulation. Here, through a genome-wide post-embryonic RNAi-feeding screen, we identified 212 components that regulate or participate in male tail tip morphogenesis. We constructed a working hypothesis for a gene regulatory network of tail tip morphogenesis. We found regulatory roles for the posterior Hox genes nob-1 and php-3, the TGF-β pathway, nuclear hormone receptors (e.g. nhr-25), the heterochronic gene blmp-1, and the GATA transcription factors egl-18 and elt-6. The majority of the pathways converge at dmd-3 and mab-3. In addition, nhr-25 and dmd-3/mab-3 regulate each others' expression, thus placing these three genes at the center of a complex regulatory network. We also show that dmd-3 and mab-3 negatively regulate other signaling pathways and affect downstream cellular processes such as vesicular trafficking (e.g. arl-1, rme-8) and rearrangement of the cytoskeleton (e.g. cdc-42, nmy-1, and nmy-2). Based on these data, we suggest that male tail tip morphogenesis is governed by a gene regulatory network with a bow-tie architecture.
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1002010
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002010Summary
During animal development, cellular morphogenesis plays a fundamental role in determining the shape and function of tissues and organs. Identifying the components that regulate and drive morphogenesis is thus a major goal of developmental biology. The four-celled tip of the Caenorhabditis elegans male tail is a simple but powerful model for studying the mechanism of morphogenesis and its spatiotemporal regulation. Here, through a genome-wide post-embryonic RNAi-feeding screen, we identified 212 components that regulate or participate in male tail tip morphogenesis. We constructed a working hypothesis for a gene regulatory network of tail tip morphogenesis. We found regulatory roles for the posterior Hox genes nob-1 and php-3, the TGF-β pathway, nuclear hormone receptors (e.g. nhr-25), the heterochronic gene blmp-1, and the GATA transcription factors egl-18 and elt-6. The majority of the pathways converge at dmd-3 and mab-3. In addition, nhr-25 and dmd-3/mab-3 regulate each others' expression, thus placing these three genes at the center of a complex regulatory network. We also show that dmd-3 and mab-3 negatively regulate other signaling pathways and affect downstream cellular processes such as vesicular trafficking (e.g. arl-1, rme-8) and rearrangement of the cytoskeleton (e.g. cdc-42, nmy-1, and nmy-2). Based on these data, we suggest that male tail tip morphogenesis is governed by a gene regulatory network with a bow-tie architecture.
Introduction
Morphogenesis involves the coordinated change in the shape of cells and tissues during development, eventually giving rise to functional structures in the adult animal. Such coordinated change must occur at the correct time and in the proper position. In the case of structures that differ between the sexes, this process must also be regulated sex-specifically. While many genes and pathways are known that regulate development, the identity of genes that link regulation to the execution of morphogenesis have been more difficult to ascertain [1]. The many different cues and signals that must be integrated to control morphogenesis, combined with the complexity of the molecular machinery associated with this process, suggest that a large number of genes and gene products are involved.
To elucidate the molecular mechanisms underlying morphogenesis, the first step is thus to determine what components are involved in its regulation and execution and to determine how they interact in a network. In the pursuit of such an aim, it is advantageous to use a simple model structure that still demonstrates all the properties of cellular morphogenesis. The model we use is the male tail tip of Caenorhabditis elegans. This structure is made up of four epithelial ("hypodermal") cells, hyp8–hyp11, which are born during embryogenesis. Embryonic morphogenesis of hyp8–hyp11 leads to the formation of a pointed, whip-like tail tip. The tail tip retains this shape throughout the lifespan of the hermaphrodite. However, during the last larval stage (L4) of males, these conical cells are dramatically remodeled to form the rounded tip of the adult [2]–[4]. Male tail tip morphogenesis begins when hyp8–11 fuse to form a syncytium; fusion is followed by detachment of the cells from the overlying cuticle. Towards the middle of the L4 stage, the syncytium changes its shape from conical to round and moves anteriorly; these morphogenetic events cease at the end of the L4 stage (Figure 1A). In hermaphrodites, the tail tip cells do not fuse and do not change shape. The tail tip model thus allows the study of a sexual dimorphism at the cellular level. Another advantage of this model is that male-specific mutations in C. elegans can be propagated through the self-fertile hermaphrodites, even if the mutations affect male fertility, mating ability, or viability.
Fig. 1. Overview of tail tip morphogenesis and the experimental design for the genome-wide RNAi screen for tail tip defects in male C. elegans. 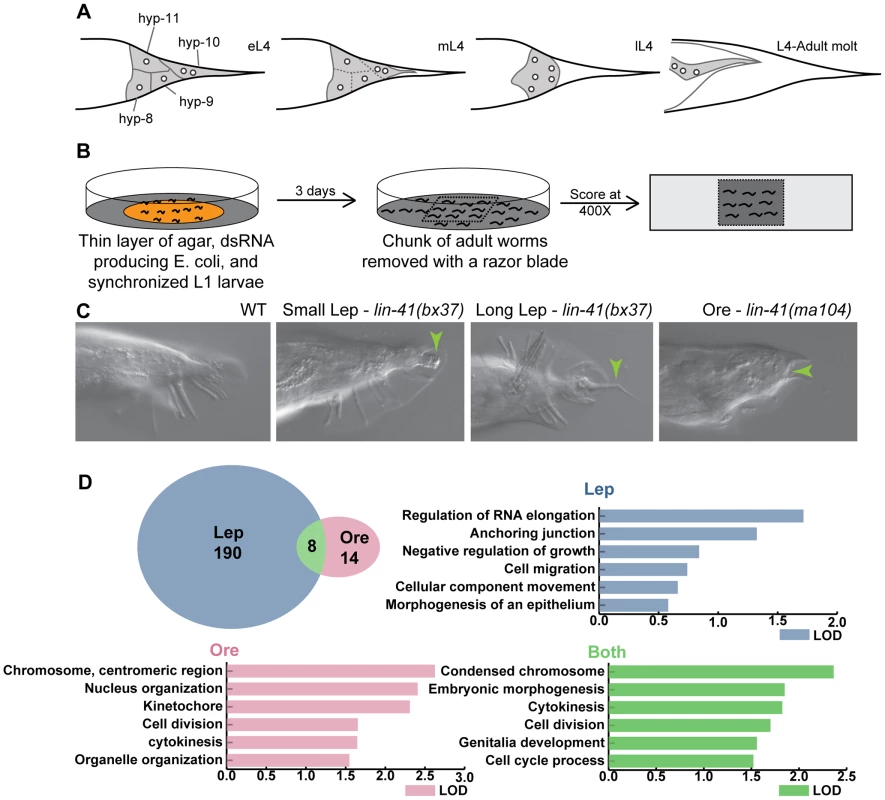
(A) Schematic of tail tip morphogenesis: the tail tip cells hyp8–hyp11 fuse and then detach from the overlying cuticle. The newly formed syncytium changes shape—becoming rounded—and migrates anteriorly. By the adult stage, the male tail has flattened out ventrally. (B) Screening methodology: L1 larvae were fed dsRNA-producing bacteria on thin films of nutrient agar and grown to adulthood. A square of agar containing worms was mounted on a slide and scored at 400× magnification. (C) The phenotypes scored included WT (wild type), Lep (leptoderan), and Ore (over-retracted). Additional phenotypes scored but not reported here were spicule morphology and general male tail defects (see http://wormtails.bio.nyu.edu). (D) Distribution of the phenotypes among the 212 positive genes; for 8 genes, Lep and Ore tails were observed in the same experiment. GO attribute enrichment via FuncAssociate [29] for genes causing Lep, Ore and Lep+Ore phenotypes upon RNAi. For each phenotype, the top six categories that were over-represented are indicated with bar graphs depicting the LOD scores, i.e. the log of the odds that the frequency of a GO category is no different than that for all genes in the genome. A few mutations in genes involved in the regulation of tail tip morphogenesis have been described previously [5]–[7]. Some of these mutations impede or completely block tail tip morphogenesis, resulting in the retention of the pointed larval tail tip in the adult male, a phenotype that we call "Lep" (Figure 1C). This phenotypic designation is derived from the term "leptoderan," which—in the taxonomic literature—describes the unretracted, pointed male tail tip in some nematodes related to C. elegans [8], [9]. Other mutations cause precocious onset of (and thus an extended total period for) tail tip retraction, which results in over-retracted ("Ore") and thus abnormally shortened adult male tails [6] (Figure 1C). Studies of these mutants have revealed a few of the important regulatory components for tail tip morphogenesis. DMD-3 was suggested to be a central regulator of tail tip morphogenesis, as it is required for tail tip retraction in males and is sufficient for inducing ectopic morphogenesis in hermaphrodite tail tips [7]. The gene encoding this DM-domain transcription factor, dmd-3, is a homolog of dmrt1 in vertebrates and doublesex in Drosophila [10], [11]. DMD-3 functions cooperatively and partially redundantly with a closely related factor, MAB-3 [7], also involved in somatic sex determination [12]. TRA-1, the most downstream global regulator in the sex-determination pathway, inhibits the expression of these genes in hermaphrodites [7], [13], [14]. The initiation of dmd-3 expression at the proper developmental stage is controlled by the "heterochronic" pathway [7]. Finally, maintenance of dmd-3 expression levels is regulated by Wnt signaling and a feedback loop involving both MAB-3 and DMD-3 [7].
Only one effector of a cellular process is known to be downstream of DMD-3 and MAB-3, namely the fusogen-encoding gene eff-1, which is important for the many fusions that occur between epithelial cells in C. elegans [15], [16]. DMD-3 and MAB-3 induce expression of eff-1 through an unknown post-transcriptional mechanism [7].
To find additional genes with roles in tail tip morphogenesis, we carried out a genome-wide, post-embryonic RNAi-feeding screen. This screen identified 212 candidates. Starting with these candidates, we used network model-building [17], transgenic reporter lines and expression epistasis analysis to construct a first draft of the gene regulatory network for tail tip morphogenesis. We found that dmd-3 expression is regulated by a nuclear hormone receptor (NHR-25), a new heterochronic gene, Hox anteroposterior patterning factors, and GATA transcription factors. NHR-25 is in turn negatively regulated by dmd-3 and mab-3. In addition, dmd-3 and mab-3 negatively regulate other signaling modules, including the TGF-β pathway. We also found that DMD-3 and MAB-3 regulate the localization or expression of genes involved in vesicular trafficking/endocytosis, cell polarity and cytoskeletal organization. Our data thus strongly support the hypothesis that DMD-3, MAB-3 and NHR-25 are central regulators of tail tip morphogenesis.
The genetic architecture for tail tip morphogenesis which emerges from this analysis closely resembles the "bow-tie" architecture, a possibly universal characteristic of robust, evolvable systems [18]. A bow-tie network has many inputs and outputs that are connected through a conserved core. Versatile weak linkages form the interface between the core and the input and output. In such a network, there is not a simple flow of information from input to output through the core; instead, there is extensive global and local feedback regulation found at every level [19].
A bow-tie architecture has been found to underlie a variety of biological networks: metabolic networks [20], [21], the Toll-like receptor signaling network [22], the epidermal growth factor receptor signaling network [23], the osmolarity glycerol signal-transduction pathway in yeast [24], stress response networks [25] and the immune system [26], [27]. In fact, it has been proposed that the bow-tie architecture of regulatory networks is ubiquitous because this structure ensures not only robustness but also evolvability [18], [19].
We found evidence for the existence of each aspect of a bow-tie architecture in the gene network governing tail tip morphogenesis. To our knowledge, this is the first time that this kind of network architecture has been explicitly identified in the context of development and morphogenesis. However, we believe that other developing systems are also governed by bow-tie genetic networks, supporting the proposition that this network architecture is universal.
Results
Genome-wide RNAi screen identifies 212 candidate genes involved in male tail tip morphogenesis
To identify tail tip morphogenesis genes, we cultured animals on dsRNA-expressing bacteria from L1 to adult and looked for evidence of defective morphogenesis. Using the Ahringer RNAi-feeding library [28], we screened 16,131 genes, approximately 83% of the genome. Genes that gave a positive Lep or Ore phenotype were screened again; only repeatable positives were kept as candidates. We identified 212 genes, of which 190 produced Lep phenotypes, 14 produced Ore phenotypes, and 8 produced both Lep and Ore phenotypes in a single experiment (Figure 1D). Positives in each category were analyzed for GO-attribute enrichment using FuncAssociate [29]. Relative to the genome, Lep positives showed enrichment of GO-attributes associated with components or processes involved in morphogenesis, such as anchoring junctions and cell migration. However, Ore and Ore/Lep positives were enriched for genes involved in cell division/cytokinesis, and nuclear/chromosome organization (Figure 1D). The 212 candidates are listed and categorized by developmental pathway or annotated cellular process (if known) in Table S1. Raw RNAi data are publicly available along with representative images via the "Male Tail Tip Database" (MTTdb, at http://wormtails.bio.nyu.edu).
We identified components of conserved and widely studied developmental regulatory pathways (e.g. Hox anteroposterior patterning factors, TGF-β signaling module, heterochronic pathway, and GATA transcription factors). Importantly, the screen also identified components of fundamental cellular structures and processes that are likely to be important for the execution of morphogenesis. These include vesicular trafficking/endocytosis, cellular polarity, cytoskeleton, cell junctions, and nuclear export/import. Genes with the most severe and/or most penetrant RNAi phenotypes were selected for further study (Table 1).
Tab. 1. Genes identified in the RNAi screen that caused the most severe and/or penetrant tail tip morphogenesis defects. 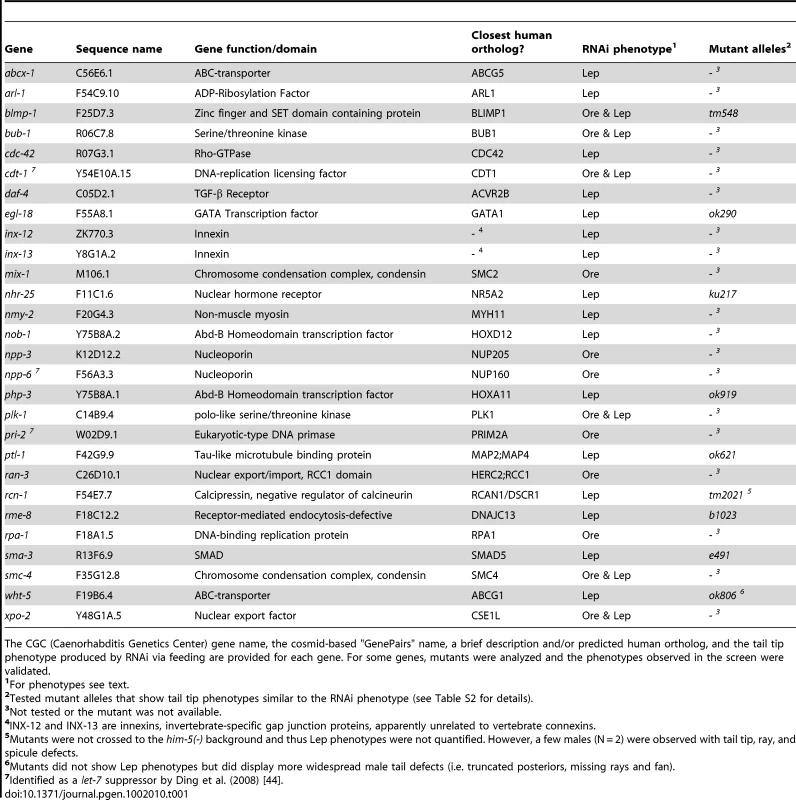
The CGC (Caenorhabditis Genetics Center) gene name, the cosmid-based "GenePairs" name, a brief description and/or predicted human ortholog, and the tail tip phenotype produced by RNAi via feeding are provided for each gene. For some genes, mutants were analyzed and the phenotypes observed in the screen were validated. Regulatory modules
Hox genes play central roles in shaping animal body plans by distinguishing different fields of cells along the anteroposterior axis [30]. Hox proteins regulate not only the expression of other transcription factors, but also of genes involved in specific processes such as morphogenesis [31]. Our screen has shown that posterior Hox patterning is crucial for tail tip morphogenesis. RNAi knockdowns of the Abd-B homologs php-3 and nob-1 postembryonically cause Lep phenotypes (Figure S1A, Tables S1 and S2). Likewise, a php-3 null allele, ok919, results in a 100%-penetrant Lep phenotype of moderate severity (Figure S1B, Table S2). The severity of the php-3(ok919) phenotype is increased with nob-1 RNAi treatment (Table S2). A nob-1::gfp translational fusion construct (containing a genomic fragment with the nob-1 gene and 9 Kb of the 5′-regulatory region) is expressed in the tail tip cells hyp8–11 throughout larval development in both sexes (Figure 2A and data not shown). A php-3::gfp translational fusion driven only by the short 500-bp intergenic region between nob-1 and php-3 shows variable expression in the nuclei of hyp8–11 during tail tip morphogenesis (data not shown). This latter transgene only partially rescues the tail tip phenotypes of php-3 mutants (Table S2), suggesting that regulatory elements upstream of both php-3 and nob-1 are required for appropriate expression of php-3. Using a tail tip-specific promoter (from the gene lin-44 [32]), we observed that expression of the PHP-3::GFP fusion protein product in hyp8–11 is sufficient to rescue the php-3(ok919) Lep phenotype (Figure S2A and Table S2). Mosaic animals that only express PHP-3::GFP in a subset of tail tip cells do not show rescue (Figure S2B). These data suggest that Hox-mediated patterning by PHP-3 and NOB-1 is carried out cell-autonomously and is required in all tail tip cells (hyp8–11) for proper morphogenesis.
Fig. 2. Expression patterns of sample regulatory genes identified in the RNAi screen. 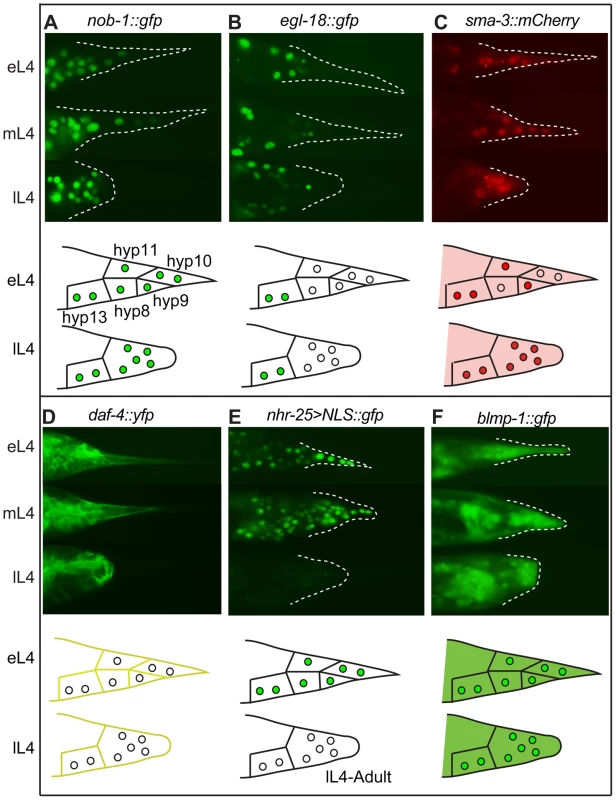
Fluorescent micrographs for early, middle and late L4 larvae and schematic drawings for early and late L4 larvae are shown in the upper and lower portions of each panel, respectively. (A) NOB-1::GFP is expressed in hyp8–11 throughout larval development. (B) EGL-18::GFP is expressed in the male tail but not in hyp8–11. (C) SMA-3::mCherry is cytoplasmic in early L4 and begins to accumulate in the nuclei of the tail tip cells. It then remains nuclear and cytoplasmic throughout tail tip morphogenesis. (D) DAF-4::YFP localizes to the membranes of hyp8–11 prior to and during morphogenesis. (E) An nhr-25 transcriptional reporter shows expression in hyp8–11 at the beginning of morphogenesis, followed by a rapid shutoff. (F) BLMP-1::GFP is expressed in the cytoplasm and nuclei of hyp8–11 throughout larval development. GATA transcription factors play important regulatory roles during the differentiation of multiple cell types in normal development [33], [34] and during tumorigenesis [35]. RNAi knockdown of the gene for the GATA factor egl-18 resulted in Lep phenotypes (Figure S1C, Tables S1 and S2). An egl-18 null allele, ok290, causes Lep phenotypes of varying severity with 45% penetrance (Figure S1D, Table S2). Another GATA transcription factor that was missed in our RNAi screen, elt-6, shares an operon with egl-18 [36]. We repeated the RNAi treatment against elt-6 and observed low-penetrance, low-severity tail tip defects (Table S2). RNAi treatment for elt-6 in the egl-18(ok290) mutant strain, however, dramatically increased the penetrance and severity of the tail tip defects, such that tail tip morphogenesis failed altogether in some individuals (Table S2). Interestingly, an egl-18::gfp translational fusion is expressed in the nuclei of the main body epidermal syncytium hyp7, but appears to be excluded from the tail tip cells hyp8–11 (Figure 2B). This observation suggests that EGL-18 and ELT-6 function in cells adjacent to the tail tip to regulate tail tip morphogenesis cell-nonautonomously. Furthermore, transforming wild-type animals with the egl-18/elt-6 operon regulated by the lin-44 promoter disrupted morphogenesis (data not shown), suggesting that morphogenesis requires exclusion of these GATA factors from hyp8–11. In other epidermal cells, it has been observed that EGL-18 and ELT-6 repress cell fusion [37]. Thus, their exclusion from the tail tip cells may be required to allow tail tip cell fusion and subsequent morphogenesis.
TGF-β signaling is a major conserved cell-signaling module which regulates multiple processes during the development of all animals and also during the progression of cancer [38]. The screen identified two components of this pathway. RNAi treatment against sma-3, which encodes a Smad protein, and daf-4, encoding the TGF-β receptor, resulted in Lep phenotypes (Figure S1E, Tables S1 and S2). A sma-3 null allele, e491, resulted in a 57%-penetrant low-severity Lep phenotype (Figure S1F, Table S2). Of the other known components of the TGF-β pathway, only sma-2 showed any RNAi phenotype: RNAi against sma-2 significantly enhanced the penetrance of the Lep phenotype of sma-3(e491) (Table S2). A transgenic strain expressing a sma-3::mCherry translational reporter shows expression at low levels in the cytoplasm of the tail tip of both sexes at the beginning of the L4 stage. In males, the SMA-3::mCherry fusion protein enters the nuclei of hyp8–11 prior to tail tip morphogenesis and remains in both the cytoplasm and nuclei during morphogenesis (Figure 2C). In hermaphrodites, SMA-3::mCherry remains cytoplasmic throughout L4 and never enters the nuclei (data not shown). The dynamic localization pattern of SMA-3::mCherry suggests that TGF-β-mediated gene expression occurs concurrently with tail tip morphogenesis. Consistent with this hypothesis, DAF-4::YFP fusion protein localizes to the plasma membranes of hyp8–11 during tail tip morphogenesis (Figure 2D). Taken together, the RNAi results, loss-of-function mutant phenotypes and expression patterns of sma-3 and daf-4 suggest that TGF-β signaling is required during tail tip morphogenesis.
Previous work has shown that Wnt signaling plays a crucial role in the regulation of tail tip morphogenesis [5]. Consistent with those findings, the RNAi screen identified additional genes that are in or interact with the Wnt pathway. RNAi treatments against sys-1 (β-catenin) and lit-1 (Nemo-like kinase) each resulted in Lep phenotypes (Table S1, Figure S3C, S3D). A transgenic strain expressing a SYS-1::GFP fusion protein shows faint cytoplasmic expression in hyp8 and hyp11 but not in hyp9 or hyp10 (Figure S3C). A strain expressing a LIT-1::GFP fusion protein shows nuclear expression in hyp9 and hyp10 but not in hyp8 or hyp11 prior to morphogenesis (Figure S3D).
We identified multiple nuclear hormone receptor genes in our screen: nhr-9, nhr-23, nhr-25 and nhr-165. NHR-25 is the C. elegans homolog of FTZ-F1 [39] which is a highly conserved protein with diverse functions regulating embryonic patterning [40], [41], and ecdysone-mediated molting in Drosophila [42]. Both nhr-25 RNAi and a hypomorphic allele, ku217, showed Lep phenotypes (Figure S1G and S1H, Tables S1 and S2). Furthermore, nhr-25 RNAi treatments on the ku217 strain showed a complete lack of male tail morphogenesis in 33% of the animals (N = 24, Table S2), a phenotype reminiscent of the mab-3(e1240);dmd-3(ok1327) double mutant [7]. We were not able to produce transgenic lines expressing NHR-25 fusion proteins due to lethality, as previously reported [43]. Instead, we made a transgenic strain expressing a transcriptional reporter containing the 5′ - and 3′-regulatory regions for nhr-25. This reporter shows a dynamic expression pattern in which expression in hyp8–11 is intense prior to and at the beginning of morphogenesis, rapidly shuts off in late L4, and is never on in adult animals (N = 34 adults) (Figure 2E). RNAi against another nuclear hormone receptor gene, nhr-165, showed a low-penetrant, mild, yet reproducible Lep phenotype upon RNAi treatment (Table S1, and Figure S3A). An nhr-165::gfp translational reporter shows nuclear expression in the lateral hypodermis (hyp7, but not hyp8–11) near the time of larval molts. This expression is highest and seen in the largest number of cells in the tail prior to the L3–L4 molt (Figure S3A).
New genes that temporally control morphogenesis
The heterochronic pathway ensures that tail tip morphogenesis is initiated precisely at the beginning of the L4 stage [6]. We identified genes in this pathway: the zinc-finger transcription factor gene blmp-1, and known suppressors of the miRNA let-7 [44]. BLIMP-1 is a transcriptional repressor that regulates ecdysone-mediated molts in Drosophila [45] and differentiation of multiple cell types in humans and mice, such as lymphocytes [46] and primordial germ cells [47], [48]. Intriguingly, BLIMP-1 is a target of let-7-mediated degradation in Reed-Sternberg cells, a Hodgkin-lymphoma cell line, suggesting a possibly conserved interaction with the heterochronic pathway, of which let-7 is a member [49], [50]. RNAi treatments directed against blmp-1 produced Ore phenotypes (Table S1). A deletion allele, tm548, produces an Ore phenotype with 100% penetrance (Table S2) due to precocious initiation of tail tip morphogenesis during the L3 stage (N = 26, data not shown). A transgenic line expressing a BLMP-1::GFP fusion protein shows both nuclear and cytoplasmic expression in hyp8–11 throughout development, although cytoplasmic expression is most intense during tail tip retraction (Figure 2F).
Of the 41 suppressors of let-7 lethality identified by Ding et al. [44], six were positives in our screen. RNAi knockdown of two (pri-2, npp-6) resulted in the Ore phenotype, of two others (spg-7, smo-1) in the Lep phenotype and of two further genes (cdt-1, xpo-2) in both phenotypes in a single experiment. Four of these genes—pri-2, npp-6, cdt-1, and xpo-2—are predicted by N-Browse [17] to interact in a subnetwork that includes other genes for which RNAi knockdown also produced Ore phenotypes (Figure 3). It is thus possible that these additional genes also influence the timing of tail tip morphogenesis. It is still unclear why both Ore and Lep phenotypes are observed in a single experiment. One possible explanation is that precision in timing of tail tip morphogenesis is lost when certain gene-products are removed. In this case, morphogenesis might begin too early in one animal and too late in another, leading to Ore tails and Lep tails, respectively.
Fig. 3. Genes with Ore-like phenotypes. 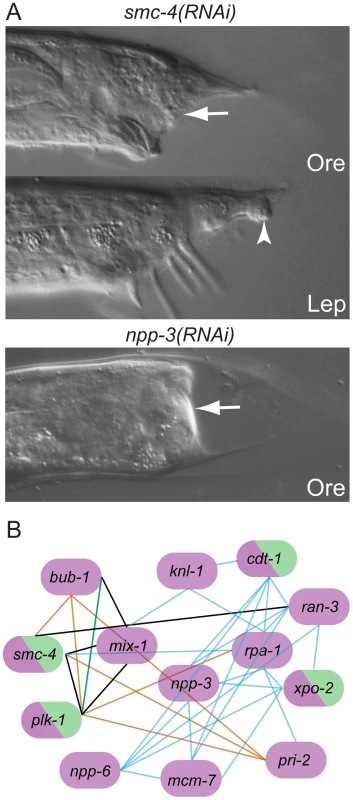
(A) DIC images of male tails: RNAi of smc-4 resulted in Ore (arrow) and Lep (arrowhead) phenotypes. npp-3 RNAi resulted in severe Ore tails (arrow). (B) Subnetwork of genes that produce the Ore phenotype as reconstructed by N-Browse [17]. Thirteen of the 22 genes with an Ore phenotype are tightly clustered: purple, Ore RNAi phenotype; green, Lep RNAi phenotype; black edges: predicted genetic interactions; orange edges: expression correlation; blue edges: phenotypic correlation in the embryo; green edges: protein-protein interaction (multiple data sources, see [17]). Post-transcriptional regulation is important for tail tip morphogenesis
Post-transcriptional regulation appears to play an important role during the coordination of tail tip morphogenesis, as our screen has identified multiple genes that encode RNA splicing factors, kinases, phosphatases and ubiquitinating enzymes. One such gene is ubc-12, which is a part of the NED-8 conjugating system and has been shown to be important in epidermal differentiation during embryogenesis in C. elegans [51]. RNAi-knockdown of ubc-12 resulted in larval lethality in the RNAi hyper-sensitive rrf-3(-) background (Table S4). However, we identified ubc-12 in a pilot screen which was carried out in the wild-type background; ubc-12 RNAi treatments produced a highly penetrant and severe tail tip defect (Figure S3B). UBC-12::GFP expresses intensely in the cytoplasm of hyp10 just prior to and during tail tip retraction (Figure S3B).
Components of junctions, organelles, and the cytoskeleton required for tail tip morphogenesis
We identified many genes known to play central roles during the execution of morphogenesis. Such genes encode proteins involved in vesicular trafficking, endocytosis, cell-cell communication, cytoskeletal rearrangement, establishment of cellular polarity and cell-cell transport.
Vesicular trafficking/endocytosis machinery
Several genes identified in our screen are predicted to function in endocytosis or vesicular trafficking, including arl-1, agef-1, rme-8, rab-6.2, dab-1 and rab-14. We focused our attention on rme-8 and arl-1 because of their strong RNAi phenotypes. RNAi-knockdown of rme-8, a gene essential for receptor-mediated endocytosis [52], produces a dramatic disruption in tail tip retraction and male tail morphogenesis in general. Adult males lack rays and a fan and the tail tip is completely unretracted; however, spicules are present (Figure 4A). The same phenotype was seen in the temperature-sensitive lethal allele, rme-8(b1023ts), at the permissive temperature (Table S2). RME-8::GFP is localized in small round structures (which we interpret as vesicles) that form at the apical surfaces of the detaching tail tip cells and are concentrated at the cell surfaces during retraction (observed in 50/52 cases) (Figure 4A). Tail tip retraction is also disrupted upon knocking down arl-1, encoding an ADP-ribosylation factor-like protein (Figure 4B). Expression of ARL-1::GFP is cytoplasmic and extremely faint in hyp8–11 during early L4, prior to tail tip retraction (28/32 animals) (Figure 4B). Occasionally, bright expression is seen in hyp10 (4/32 animals). ARL-1::GFP becomes more intense and localizes to puncta (possibly vesicles) after rounding of the cells has occurred (Figure 4B). In adult animals, hypodermal expression is absent, but neuronal expression (e.g., in PHCL and PHCR) is apparent in all animals (N = 35).
Fig. 4. Effectors of morphogenesis identified in the RNAi screen. 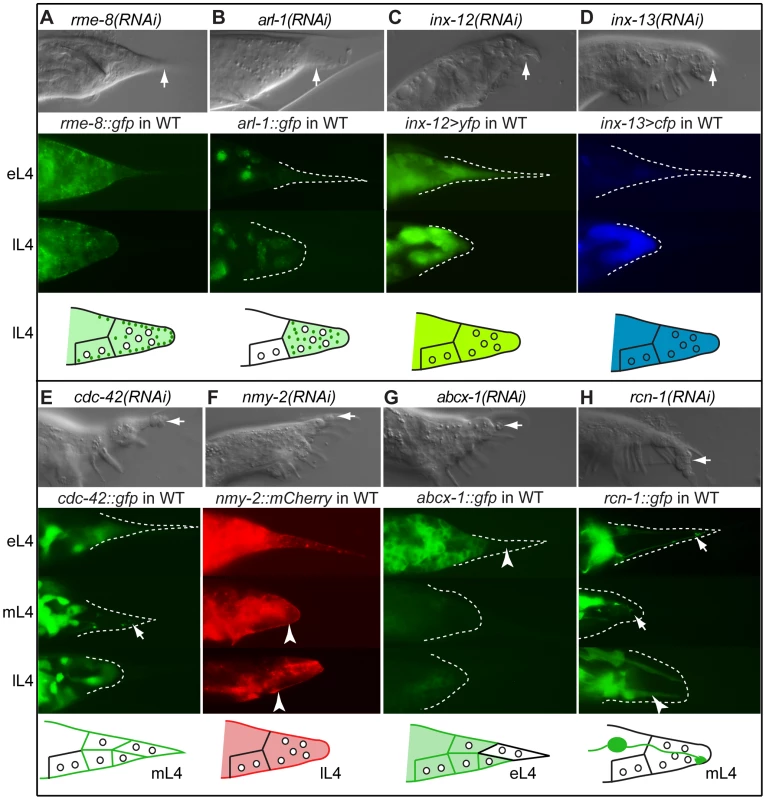
DIC images of RNAi phenotypes in adult males (top of each panel), expression patterns in the male tail of early L4 (eL4), mid L4 (mL4) and/or late L4 (lL4) (middle of each panel), and schematic of expression/localization patterns (bottom of each panel). Arrows in the DIC images indicate Lep tail tips; arrows in the fluorescent images indicate localization patterns of interest. (A) rme-8(RNAi) results in a total failure of tail tip morphogenesis. RME-8::GFP localizes to the edge of detaching and retracting tail tip cells as puncta (depicted as dark green dots in the schematic). (B) arl-1(RNAi) causes a disruption of tail tip retraction. ARL-1::GFP is expressed cytoplasmically at extremely low levels in hyp8–11 prior to morphogenesis (eL4); during morphogenesis (lL4), it localizes to distinct puncta (dark green dots in the schematic). (C, D) inx-12(RNAi) and inx-13(RNAi) result in subtle Lep phenotypes. Reporters for inx-12 and inx-13 are expressed in tail tip cells prior to and during morphogenesis. (E) cdc-42(RNAi) results in a Lep phenotype. CDC-42::GFP is cytoplasmic prior to tail tip retraction, localizes to the membranes and discrete puncta during retraction (arrow), and becomes cytoplasmic again in late L4 and adults. (F) nmy-2(RNAi) results in Lep phenotypes. NMY-2::mCherry localizes to the posterior end of retracting tail tip cells and to the ventral surface of the male tail (arrowheads). (G) abcx-1(RNAi) results in Lep phenotypes. ABCX-1::GFP is expressed in hyp8, 9 and 11 prior to but not during morphogenesis. Expression is absent from hyp10 (arrowhead). (H) rcn-1(RNAi) causes Lep phenotypes. RCN-1::GFP is expressed in PHCL and PHCR (arrows), as well as in other more anterior neurons during tail tip morphogenesis. After morphogenesis, expression is intense in the phasmid socket cells (arrowhead). Gap junctions
Transmission electron microscopic reconstruction of the tail tip revealed the presence of gap junctions between the hyp cells in the tail [3]. The gap junction protein-encoding genes, inx-12 and inx-13 show Lep phenotypes of varying severity upon RNAi treatment (Figure 4C, 4D and Table S1). Transcriptional reporters for both inx-12 (YFP) and inx-13 (CFP) are expressed at a low level in hyp8–11 prior to tail tip morphogenesis but more highly during retraction (Figure 4C, 4D). Because this increase in expression occurs after the tail tip cells have fused with each other, we predict that these innexins establish new junctions between the tail tip syncytium and other cells, although it is also possible that they form hemichannels.
Cell polarity and cytoskeleton
Our data suggest that morphogenesis of the tail tip is carried out by an asymmetric cytoskeletal rearrangement that drives the anteriorly directed "retraction". The RNAi screen identified a role for the Rho-GTPase encoded by cdc-42, for non-muscle myosins encoded by nmy-1 and nmy-2, for a Rho-GEF encoded by ect-2, for an intermediate filament protein encoded by ifc-2, and for a microtubule binding protein encoded by ptl-1 (Table S1). RNAi-knockdown of cdc-42 results in a highly penetrant and severe Lep phenotype (Figure 4E, Table S1). A CDC-42::GFP fusion protein is diffusely distributed in the cytoplasm of the tail tip cells at the early L4 stage and then localizes to foci at the apical surfaces of the tail tip cells just before retraction begins at mid-L4 (Figure 4E). During retraction, CDC-42::GFP again becomes diffuse in the cytoplasm. RNAi-knockdown of nmy-2 also resulted in tail-tip retraction defects (Figure 4F and Table S1). Using confocal microscopy and 3-dimensional reconstruction of L4 males expressing nmy-2::gfp, we observed a cap of NMY-2::GFP that forms at the posterior end of the rounding, retracting tail tip (Video S1 and Figure S4) and along the ventral surface where the tail tip cells begin to pull away from the larval cuticle. Also, foci of NMY-2::GFP form at the apical surfaces with fibrous projections from these foci into the cells (Figure S4). Later, NMY-2::GFP becomes very intense in the region where the ray tips will form (Video S2). A strain expressing NMY-2::mCherry shows the same pattern (Figure 4F).
ABC-transporters
ATP-binding cassette (ABC) transporters play central roles in bacterial drug resistance, and mammalian metabolism and immune responses [53]. The C. elegans ABC transporter-encoding gene, abcx-1, showed a Lep phenotype upon RNAi (Figure 4G and Table S1). An ABCX-1::GFP fusion protein shows male-specific expression during the transition from L3 to L4 both on the membrane and in the cytoplasm in hyp8, 9 and 11, but not in hyp10. This expression fades and becomes extremely faint during tail tip retraction (Figure 4G).
Genes expressed in the PHC neurons
Adjacent to the tail tip cells lie the dendritic projections of the PHC neurons. Two genes that produced Lep phenotypes upon RNAi treatment are expressed in these neurons but not in the tail tip cells: the calcipressin-encoding gene rcn-1 [54] and ptl-1, a gene encoding a tau-like microtubule-associated protein [55], [56]. A null allele of ptl-1, ok621, produced Lep phenotypes with 23% penetrance (Table S2). PTL-1::GFP and RCN-1::GFP fusion proteins are expressed in the PHC neurons prior to and during hypodermal morphogenesis. In adults, PTL-1::GFP is expressed in most neurons of the tail (data not shown). RCN-1::GFP is expressed in most tail neurons and in the support cells of the phasmid neurons (socket cells, Figure 4H). This pattern is consistent with the previously described adult expression pattern of RCN-1 [54].
A genetic architecture of tail tip morphogenesis
With a list of genes required for tail tip morphogenesis, we next sought to characterize the interactions between these genes. We constructed a working hypothesis for these interactions using N-Browse, a publicly available database which integrates the information from numerous genome-wide studies to build gene networks [17]. We manually entered our candidate genes into N-Browse, excluding those that did not have predicted functions or known interactors. N-Browse produced a genetic network that included not only our candidate tail tip morphogenesis genes, but additional genes predicted to be nearest-neighbor interactors. To the resulting N-Browse network, we manually added gene interactions (edges) based on published work not represented in N-Browse [6], [7], [44], [51], [57]–[61] (Figure 5). This analysis predicts the involvement of genes not identified in our screen. We tested two of these predictions by repeating RNAi knockdown with different methods and/or by scoring larger numbers of males. We could thus validate roles in morphogenesis for elt-6 and vav-1. elt-6 has genetic interactions with egl-18 (positive in our screen) in other contexts. It showed a low-penetrance Lep phenotype when the RNAi experiment was repeated and more males were scored. In addition, elt-6(RNAi) enhances the Lep phenotype of egl-18(ok290) mutants (Table S2). Also, the network model predicts that vav-1 has interactions with php-3, cdc-42 and inx-12 (all positives in our screen, Figure 5). Although vav-1 treatment by RNAi via feeding did not result in detectable phenotypes, administering RNAi against vav-1 by soaking did cause tail tip defects (data not shown).
Fig. 5. A partial genetic network of tail tip morphogenesis as predicted by N-Browse [17] (thin lines) and other published work (thick dashed lines) [7], [44], [57]–[59], [61]. ![A partial genetic network of tail tip morphogenesis as predicted by N-Browse <em class="ref">[17]</em> (thin lines) and other published work (thick dashed lines) <em class="ref">[7]</em>, <em class="ref">[44]</em>, <em class="ref">[57]</em>–<em class="ref">[59]</em>, <em class="ref">[61]</em>.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/9c5a90af4198732077bd2a14d4220828.png)
Clusters of pathways or functional groups are surrounded by boxes to represent their proximity within the network and/or functional interactions in modules. Nodes (genes): green, Lep positives identified in the screen; purple, Ore positives identified in the screen; green/purple, Lep+Ore positives from the screen; yellow, previously identified tail tip genes; green boundary (unfilled), not identified in the screen but suggested by the network and experimentally validated; red boundary (unfilled), not positive in the screen and not further tested. Edges (lines) represent the following types of data concerning interactions: dashed black: published and experimentally validated interactions; blue: interologs (conserved interactions between pairs of proteins that have interacting homologs in other organisms); gray: predicted genetic interactions; purple: experimentally verified genetic interactions; green: protein-protein interactions; orange: expression correlations. We next asked whether the network model—developed from information in other systems—has biological relevance for tail tip morphogenesis. To this end, we tested a selection of the predicted interactions by genetic and expression epistasis analyses. The results are detailed below.
Components upstream of dmd-3 and mab-3
Previous work [7] proposed that dmd-3 and mab-3 play a central role in the regulation of tail tip morphogenesis. If so, we would predict that many regulatory pathways interact with these genes. To test this prediction, we compared dmd-3 expression in various mutant backgrounds to that in wild-type males. In wild type, the expression of a dmd-3>yfp transcriptional reporter coincides with the morphogenesis of the male tail tip, as previously reported [7] (Figure 6A). Expression of dmd-3>yfp is reduced in the php-3(ok919) mutant background (N = 32) and upon treatment with nob-1 RNAi (N = 24) (Figure 6B, 6C). Removing nob-1 by RNAi in the php-3(ok919) background results in a further reduction of dmd-3>yfp expression (N = 26) (Figure 6D). Removing egl-18 and elt-6 results in a reduction of dmd-3>yfp expression as well (N = 16) (Figure 6E). In each case, dmd-3>yfp expression is initiated at the correct time, but the levels of expression are reduced, suggesting that php-3, nob-1, egl-18, and elt-6 are positive regulators of dmd-3, but are not required for initiation of dmd-3 expression. In the nhr-25(ku217) mutant background, dmd-3>yfp expression is completely blocked in hyp8–11, while expression in the PHCL/R neurons is not affected (N = 22; Figure 6F). Thus, nhr-25 is required for the initiation of dmd-3. In the blmp-1(tm548) mutant background, dmd-3>yfp is activated precociously in hyp(8,9,11), but is not expressed in hyp10 (N = 21) (Figure 6G). We also tested if GATA expression is influenced by Hox expression; however, EGL-18::GFP expression in a php-3(ok919) background is the same as in wild type (data not shown).
Fig. 6. Expression epistasis experiments to test interactions between key regulatory genes. 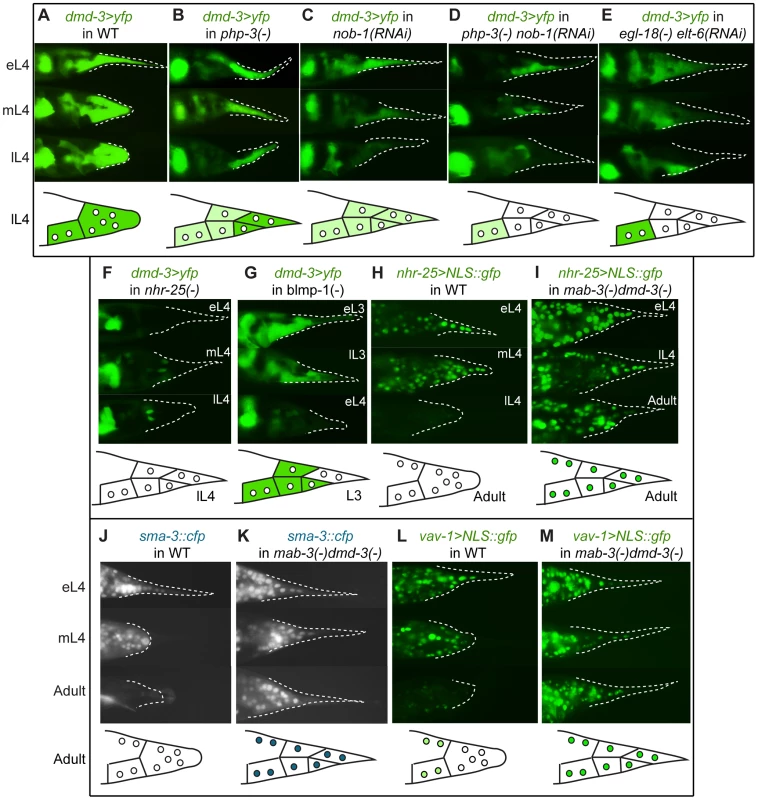
In the top of each panel are fluorescent micrographs of male tails (lateral views) at three of several stages (eL3 = early L3, lL3 = late L3, eL4 = early L4, mL4 = mid-L4, lL4 = late L4, or adult) expressing various transgenes in different genetic backgrounds. At the bottom of each panel is a schematic depicting the expression pattern at one exemplar stage. (A) Expression of a dmd-3>yfp transcriptional reporter in wild-type males. (B–G) Effects on the expression of dmd-3>yfp by RNAi-depletion or mutations of other genes (note that tail tip morphogenesis is impaired in these RNAi-treated or mutant males and rounding of the tail tip in lL4 does not occur). (B) Loss of php-3 function in ok919 mutant animals causes reduced dmd-3>yfp expression in hyp8, hyp11 and hyp13. (C) Depletion of nob-1 by RNAi causes reduced dmd-3>yfp expression in hyp8–11. (D) php-3(ok919) nob-1(RNAi) animals show no dmd-3>yfp expression in hyp8-11. (E) Although dmd-3>yfp is initially expressed in egl-18(ok290) elt-6(RNAi) animals, it shuts off prematurely in hyp8–11. (F) Expression of dmd-3>yfp is never initiated in the tail tip cells of nhr-25(ku217) reduced-function mutants. (G) blmp-1(tm548) mutants show precocious expression of dmd-3>yfp in eL3 in hyp8, hyp9 and hyp11, no expression in hyp10, and premature inactivation of dmd-3>yfp at early L4. (H) nhr-25>NLS::gfp is expressed brightly prior to and at the beginning of morphogenesis and is quickly inactivated near its end. (I) In mab-3(e1240);dmd-3(ok1327) animals, expression of the nhr-25 reporter remains bright in adult males. (J) SMA-3::CFP is expressed and nuclearly localized in hyp8–11 during morphogenesis and is not visible in adult males (the image shows autofluorescence of the spicules and the fan). (K) In mab-3(e1240);dmd-3(ok1327) adults, expression of the sma-3::cfp reporter remains very bright into adulthood. (L) vav-1>NLS::gfp is expressed brightly in hyp8–11 and in other cells of the tail prior to and during morphogenesis and at extremely low levels in adults. (M) Expression of the vav>NLS::gfp reporter remains very bright in hyp8–11 and the other cells of the male tail in adult mab-3(e1240);dmd-3(ok1327) animals. Negative regulation downstream of dmd-3 and mab-3
Because DMD-3 functions partially redundantly with MAB-3, we examined various transgenic reporters in the mab-3(e1240);dmd-3(ok1327) double mutant background. Our nhr-25 transcriptional reporter is completely shut off during late L4 in wild-type males, but maintained in mab-3(e1240);dmd-3(ok1327) double-mutant males (compare Figure 6H and 6I, respectively). Expression persists in hyp8–11 into adulthood (N = 32). Since nhr-25 is required for dmd-3 expression, this observation suggests that dmd-3 and/or mab-3 regulate nhr-25 via a negative feedback loop.
Negative interactions are also observed between dmd-3/mab-3 and the TGF-β pathway and between dmd-3/mab-3 and the vav oncogene homolog, vav-1. In mab-3(e1240);dmd-3(ok1327) males, nuclear SMA-3::CFP remains bright in hyp8–11 into adulthood (N = 18) (Figure 6K), whereas such adult expression is never observed in wild-type males (Figure 6J, N = 31). Thus, dmd-3 and/or mab-3 negatively regulate TGF-β signaling in hyp8–11. A strain expressing a transcriptional reporter for vav-1 shows bright expression in hyp8–11 prior to and during morphogenesis (Figure 6L). Expression fades in the tail tip during adulthood and is only seen in a small subset of other cells. However, in mab-3(e1240);dmd-3(ok1327) males, expression of this reporter persists brightly in the adult tail tip and in a larger subset of cells (Figure 6M). Thus, dmd-3 and/or mab-3 negatively regulate vav-1, a new interaction not predicted by the N-Browse network model.
Cellular machinery regulated by dmd-3
We also wanted to determine how genes involved in executing morphogenesis might be affected by DMD-3 and MAB-3. In the mab-3(e1240);dmd-3(ok1327) mutant background, localization of RME-8 is disrupted. RME-8::GFP is still localized to puncta, but it is distributed more evenly in the cytoplasm (in 39/51 males) instead of concentrated near the apical surfaces of the cells (Figure 7A). Thus, dmd-3 and/or mab-3 are upstream of RME-8 localization during tail tip morphogenesis. At early L4, ARL-1::GFP is expressed intensely in the hyp10 cell in most mab-3(e1240);dmd-3(ok1327) mutant males (21/34 males), whereas this reporter was expressed intensely in only few wild-type males (4/32; Figure 7B). On the other hand, mutant adults lack the PHCL/R expression (absent in 40/41 males) that is present in all wild-type males (N = 35) (Figure 7B). Thus, arl-1 expression is controlled by dmd-3 and/or mab-3, positively in the neurons but negatively in hyp10. Finally, we examined the expression and localization of NMY-2 in mab-3(e1240);dmd-3(ok1327)males. DMD-3 and MAB-3 do not appear to affect the expression level of NMY-2::mCherry (Figure 7C). However, we did not observe the accumulation of NMY-2 at the posterior tip where rounding would occur in wild type. This lack of localization is consistent with the fact that the tail tip cells never change their shape in the mab-3(e1240);dmd-3(ok1327) mutant strain (Figure 7C). We still observe NMY-2 localization at the positions where the rays would normally form, even though rays do not form in this strain (not shown). This latter NMY-2 localization correlates with a small amount of pulling away ("anterior retraction") from the overlying cuticle in the mab-3(e1240);dmd-3(ok1327) mutant males (not shown). Thus, while transcription of nmy-2 is presumably normal, DMD-3 and MAB-3 partially affect the localization of NMY-2 in the tail tip; this effect is likely to be indirect.
Fig. 7. Cellular processes controlled by DMD-3 and MAB-3. 
Expression patterns of translational fusions are shown for wild-type males (left side of each panel) and mab-3(e1240);dmd-3(ok1327) mutant males (right side of each panel). (A) DMD-3 and MAB-3 regulate RME-8 localization but not expression. In wild-type animals, RME-8::GFP localizes as discrete puncta to the cell cortex where the tail tip cells are detaching from the cuticle (arrow), but is dispersed and cytoplasmic (arrowhead) in mab-3(e1240);dmd-3(ok1327) mutants. (B) In early L4, ARL-1::GFP shows very low levels of expression in wild-type males (arrowhead) but bright expression in hyp10 (arrow) in mab-3(e1240);dmd-3(ok1327) males. In adults, ARL-1::GFP is expressed in tail neurons in wild-type males (arrow points to phasmids), but not in mab-3(e1240);dmd-3(ok1327) males (arrowheads). (C) In wild-type males, NMY-2::mCherry localization is focused to a cap at the posterior end of the tail tip during rounding and to the region along the ventral edge where the tail tip cells are detaching from the cuticle (arrows). In mab-3(e1240);dmd-3(ok1327) animals, localization to the ventral surface still occurs (arrows), but localization to the posterior end of the tail is not observed (arrowheads). We tested 10 additional gene pairs for epistatic interactions, however, in these experiments, the expression of the reporters in the mutant background did not differ from expression in wild-type males (Table S5). Experimentally validated interactions between genes involved in male tail tip morphogenesis are shown in Figure 8. As will be discussed below, the architecture of this part of the network is consistent with a bow-tie structure, with dmd-3, mab-3 and nhr-25 at the core and a number of genes in the input and output fans.
Fig. 8. The genetic architecture of tail tip morphogenesis, showing experimentally validated connections between components. 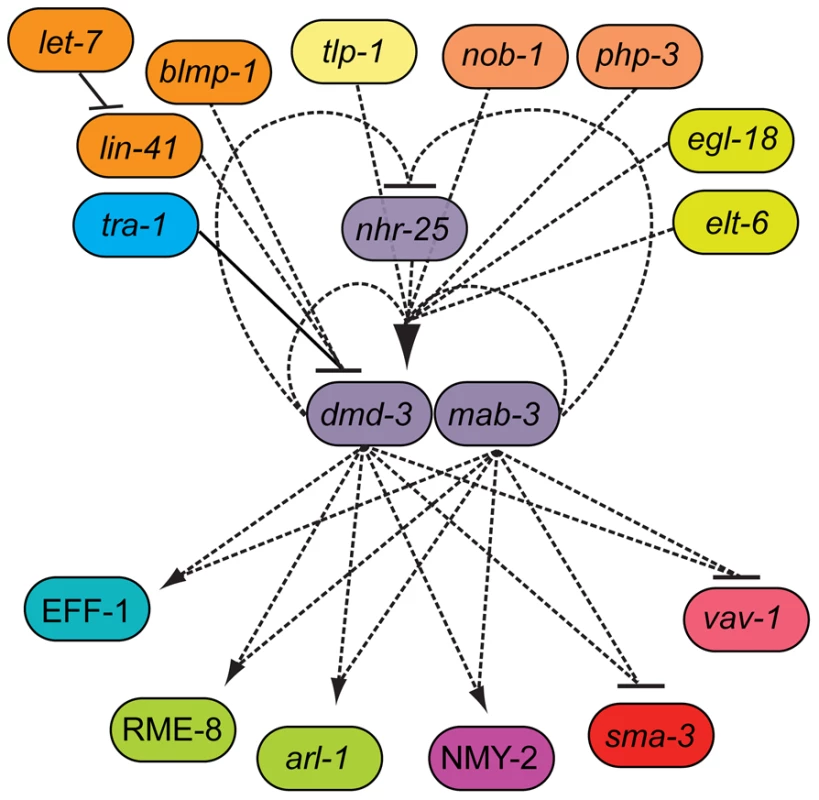
This network has a bow-tie architecture with dmd-3, mab-3 and nhr-25 in the core. The central role for dmd-3 and mab-3 was originally proposed by Mason et al. [7]. We found that nhr-25 is required for the activation of dmd-3 and forms a negative feedback loop with dmd-3 and mab-3. Multiple signaling pathways/genes feed into the core by regulating dmd-3 positively (lines with arrow at one end) or negatively (lines with bar at one end). Genes controlling morphogenesis (rme-8, nmy-2, arl-1, eff-1) and other signaling pathways (TGF-β and vav-1-mediated) are regulated by dmd-3 and mab-3. Dashed lines indicate possibly direct or indirect interactions, solid lines indicate an experimentally validated direct interactions [7]. Discussion
Genome-wide RNAi screen
Here, we used systemic RNAi to identify components that are involved in male-specific morphogenesis of the tail tip of C. elegans. RNAi via feeding in C. elegans provides a simple yet powerful means for identifying the regulatory and structural components and pathways of developmental processes [28], [62]. The methodology we employed (Materials and Methods and Figure 1B) allowed us to quickly score for subtle defects in morphogenesis at high magnification. Many of the genes we identified have roles in embryonic processes and are lethal when knocked down (e.g. nob-1, cdc-42). This justifies our approach to treat worms with RNAi postembryonically and it underscores the power of RNAi as a tool for identifying postembryonic roles of genes that have essential embryonic functions. To minimize the number of false negatives, we performed the screen on the RNAi hypersensitive strain, rrf-3 [63]. For a number of reasons, however, we believe that there are still other tail tip genes to be identified. First, our screen did not cover the entire genome (approx. 83%). Second, because of complete larval lethality, about 2% of the genes in our screen were not scored, including the known tail tip regulator lin-41 [6] (Table S4). Third, of the previously known tail tip genes, only lin-44, which encodes for the Wnt ligand, was found in the screen. Other known tail tip genes, tlp-1 and dmd-3, which have representative clones in the library, were missed. Finally, two genes (elt-6 and vav-1) not found in the screen, but predicted by the N-Browse network analysis, turned out to have RNAi-induced tail tip phenotypes when treated in a different genetic background (i.e. elt-6 RNAi in the egl-18(ok290) background) or by soaking instead of feeding (vav-1). The number of false positives is likely to be very small since only genes which were positive in the primary and secondary screen were considered. The 212 candidate genes identified in this process were significantly enriched with morphogenesis-related GO attributes relative to the genome at large, consistent with what would be predicted if our screen were successful. Some candidates were studied further to elucidate their roles in regulating or effecting tail tip morphogenesis.
Network architecture and features
Developmental decisions, such as the initiation of morphogenesis, require the input of multiple signaling pathways and result in a coordinated response by many different components of the cell. The response must be robust against perturbations from the internal and external environment. Robustness and precision of biological processes are thought to be facilitated by a bow-tie (or hourglass) architecture of the gene regulatory network [18], [19]. Characteristics of bow-tie networks include the following. (1) Many inputs and outputs are connected to a conserved core. (2) Versatile weak linkages form the interface between input and core and between core and output. (3) Systems control is facilitated by positive and negative feedback at every level. (4) Modularity and partial redundancy or degeneracy are two other properties of the bow-tie network architecture that contribute to the robustness of biological systems [19], [64].
We used the data about gene interactions described here in combination with published information to delineate a first draft of the gene regulatory network underlying tail tip morphogenesis in C. elegans males. Although the reconstruction of this network has only just begun, we already find many features that are consistent with bow-tie architecture.
Modularity
A network of interactions is called modular if it can be subdivided into relatively autonomous components (modules) that are built of highly connected parts but are more loosely connected to other modules [65]. Modularity is a major contributor to the robustness and evolvability of a system, since perturbations and mutations can occur within a module with minimal effects on the whole system [19]. It has been proposed that modularity facilitates evolutionary change by allowing new connections to be made between modules without disrupting the core function of the modules [66]. Modularity has been observed in many networks [67], [68]. The modules of metabolic networks have bow-tie structure, just like the networks themselves [21]; that is, bow-tie architecture can be nested.
In the gene regulatory network of male tail tip morphogenesis, we find evidence for many conserved regulatory and effector modules. Regulatory modules include Hox patterning, the sex-determination and heterochronic pathways and TGF-β signaling. We also identified tail tip roles for additional Wnt pathway components, i.e. the SYS-1 beta-catenin, the MIG-1 Frizzled-like receptor, and the LIT-1 Nemo-like MapK. Effector modules consist of conserved components controlling vesicular trafficking and endocytosis, establishment of cellular polarity, cytoskeletal rearrangement, and cellular fusion.
Degeneracy
Degeneracy describes the coexistence of structurally or mechanistically different components that can perform similar roles or are interchangeable under certain conditions [64]. Degeneracy confers robustness because, in a system composed of partially redundant elements, one element can compensate for the failure of another. One mechanism that generates degeneracy is gene duplication. In the male tail tip morphogenesis system, we find several examples for degeneracy due to gene paralogy. We think that MAB-3 and DMD-3 function partially redundantly because the phenotype of the mab-3(e1240);dmd-3(ok1327) double mutant is more severe than that of dmd-3(ok1327) or mab-3(e1240) alone [7]. Similarly, two other pairs of paralogs function semi-redundantly: the Hox genes php-3 and nob-1 and the GATA transcription factors egl-18 and elt-6, which form an operon. In both cases, removal of both genes results in a much more severe disruption of morphogenesis than removal of only one gene.
Both modularity and degeneracy contribute in a major way to the robustness of a system [19]. Indeed, the majority of RNAi knockdown phenotypes suggest that male tail tip morphogenesis is very robust against genetic perturbations. In most cases, the effects of RNAi (as well as some of the mutations tested) were subtle and the penetrance was low, suggesting that there is extensive buffering of the system against partial depletion of individual transcripts (or reduced functionality of genes).
Bow-tie network architecture
The conserved core
A role for dmd-3 as a central regulator of male tail tip morphogenesis has been proposed previously [7]. This gene is required for male tail tip morphogenesis and, when misexpressed in the hermaphrodite tail tip, is sufficient to cause ectopic morphogenesis [7]. Another DM-domain gene, mab-3, has a partially redundant role with dmd-3 in tail tip morphogenesis. We propose that the conserved core contains both of these genes as well as the gene for the nuclear hormone receptor NHR-25. NHR-25 is a highly conserved protein which in other contexts cooperates with pathways involved in tail tip morphogenesis. NHR-25 interacts with the Wnt pathway during asymmetric division of the T cell [57], and with the Hox protein NOB-1 during embryogenesis [59]. There is also evidence that nhr-25 is connected to the heterochronic pathway, as it is a target of the miRNA let-7 [58]. nhr-25 is the only gene we have found to be required for the initiation and not just the maintenance of dmd-3 expression. dmd-3 expression is completely eliminated in nhr-25(ku217) mutants and complete removal of nhr-25 (e.g. nhr-25 RNAi performed on the nhr-25(ku217) hypomorphic strain) results in the same phenotypes as the mab-3(e1240);dmd-3(ok1327) double mutant. dmd-3/mab-3 and nhr-25 regulate each other through a feedback loop. Thus, these three genes probably form a module in the core of the network.
Inputs to the core
Input from multiple modules regulates the sex-specific, spatial, and temporal expression of the core gene dmd-3. Sex-specificity is provided by the most downstream component of the sex-determination pathway, TRA-1, which inhibits expression of dmd-3 in hermaphrodites [7]. Previous studies [5], [7] have proposed that Wnt signaling provides spatial information. We found evidence that Hox and GATA factors also contribute to spatial patterning. Hox genes are known to govern patterning along the anteroposterior axis. As is expected, the genes involved in tail tip morphogenesis, nob-1 and php-3 are the most posterior Hox genes in C. elegans [69]. The GATA transcription factors egl-18 and elt-6 are excluded from the tail tip cells but are expressed in all the neighboring cells. They are nevertheless required for tail tip morphogenesis. We therefore postulate that GATA factors provide positional information through signaling from neighboring cells.
The heterochronic pathway provides temporal input, which ensures that morphogenesis occurs during the correct time window during the L4 stage. Overexpression of lin-41 and deletion of let-7 lead to delayed morphogenesis; removal of lin-41 leads to precocious tail tip retraction [6]. We propose here that blmp-1 is also a component of the heterochronic pathway, because removing blmp-1 results in precocious expression of dmd-3 and in precocious tail tip retraction. We are currently determining where blmp-1 acts in the heterochronic pathway.
Outputs from the core
In the tail tip system, the output downstream of the conserved core involves effectors of morphogenesis. One such output is cellular fusion driven by the fusogen protein EFF-1 [3], [7], [15], [16]. In addition, our screen has shown that genes involved with vesicular trafficking, endocytosis, establishment of cellular polarity, cytoskeletal rearrangement, cell-cell communication and transport are on the output side of the bow tie. Electron microscopy revealed an accumulation of vesicles during male tail tip morphogenesis, suggesting an important role for trafficking [3]. Consistent with this observation, two genes involved in vesicular trafficking and endocytosis that were identified in the screen display dynamic expression or localization patterns during tail tip morphogenesis: arl-1, which encodes an ADP-ribosylation factor [70], [71], and rme-8, which encodes a J-domain protein vital for receptor-mediated endocytosis [52]. Trafficking and endocytosis have been shown to play a key role in morphogenesis in other systems. For example, endocytosis of cadherins is required for the morphogenetic movement during Xenopus gastrulation [72] and the anterior migration of prechordal plate cells in zebrafish [73], [74]. The severity of the tail tip phenotype indicates that endocytosis, controlled by RME-8, is an essential process for tail tip morphogenesis. Removing rme-8 postembryonically, by either RNAi or with a temperature-sensitive allele, results in a complete failure of tail tip morphogenesis. Also, RME-8 localizes to regions of the plasma membrane which undergo dynamic change: the ventral border of the tail tip cells as they detach from the L4 cuticle, and the posterior edge of the rounding cells. This pattern is consistent with a proposed role for RME-8 during early endosome membrane trafficking and receptor recycling [61], [75], [76]. It has been observed that, in the absence of RME-8, components of signaling pathways (e.g. Wnt) that should be recycled to the membrane are instead shuttled to the lysosome and degraded [61]. It is therefore possible that RME-8 influences signaling events required for tail tip retraction which cannot occur in the absence of receptor recycling.
Morphogenesis in all systems likely involves the rearrangement of an acto-myosin network to drive cell movement and shape change. Our data suggest that, in the tail tip, these processes are controlled by CDC-42, a highly conserved Rho-GTPase [77], and NMY-2, a non-muscle myosin. Our expression analysis is consistent with NMY-2 playing a key role in reshaping the pointed tail tip cells into a rounded syncytium. NMY-2 localization to a cap at the end of the tail tip is reminiscent of what has been observed during C. elegans gastrulation where NMY-2 localizes to the apical surface of the ingressing E daughter and MS descendant cells [78]. Additionally, at the start of tail tip retraction, we observed clear foci of NMY-2 at the apical surfaces with fibrous NMY-2 densities projecting from the apical foci inside the tail tip cells. This organization is strikingly similar to the arrangements of non-muscle myosin observed for apical constriction during Drosophila gastrulation in which the apical surface area is constricted by a ratchet-like contraction of an actin network [79]. Similarly, CDC-42 localizes to foci at the apical boundaries of the tail tip cells, where it could have a role in activating the formation of actin filaments required for such "apical constriction."
Gap junctions are required for epithelial morphogenesis during embryogenesis in Drosophila [80] and play a role during the cellular changes of metastasis, such as cellular migration [81]. INX-12 and INX-13 connect the cytoplasm of neighboring cells through the formation of gap junctions [82] and are required for tail tip morphogenesis. Oddly, our reporters show their peak of expression when the tail tip cells have already fused. It is possible that these innexins are forming new gap junctions between the tail tip and other cells and that these junctions are required for efficient or continued retraction/migration.
Versatile weak linkages
Kirschner and Gerhart [83] define versatile weak linkage in biological systems as a kind of regulatory connection that can be easily broken or redirected for other functions during evolution or development. In bow-tie networks, the interfaces between input, core, and output are formed by weak linkages. Examples of components that can establish versatile weak linkages include second messengers, G-proteins, and the transcription machinery [84]. On the input side of the tail tip regulatory network, we have identified many transcription factors and signaling pathways. The control of dmd-3 transcription involves many of these inputs. It is thus possible that each input forms a weak interaction with dmd-3. One example of a weak linkage that might connect the core to the output is CDC-42, a Rho-GTPase which integrates multiple signaling pathways to mediate cellular responses [19], [85]. Additionally, processes involved in post-transcriptional gene regulation have attributes of weak linkage. We identified genes that encode for RNA splicing factors, kinases, phosphatases, proteases, ubiquitinating and neddylating proteins, and other enzymes with the capacity for post-translational modification of the cytoskeleton or other parts of the cellular machinery. One example of the latter is UBC-12, a ubiquitin-conjugating enzyme that produced a strong phenotype and was expressed throughout but not prior to tail tip morphogenesis. The fusogen-encoding gene eff-1 that is required for cell fusion may also be regulated post-transcriptionally. It has been shown that DMD-3 and MAB-3 do not influence eff-1 transcription, but they positively regulate EFF-1 protein levels [7]. Similarly, we show that DMD-3 somehow regulates the localization of RME-8 and NMY-2, but not their expression.
System controls
One feature of bow-tie networks is system control, consisting of positive and/or negative feedback which can occur between components at all levels of the network [19]. Precise system control has been emphasized as an important aspect of developmental systems since precision, robustness and versatility are stringent requirements in development and errors are particularly serious [86]. Positive autoregulation occurs when a transcription factor either directly or indirectly activates its own expression which results in the maintenance of transcription in the absence of the factors that initiated expression. Positive feedback can be used in temporal control to sustain expression over a developmental period, or in spatial control to restrict expression to a specific region [86], [87]. Patterning and duration of expression are also influenced by negative autoregulation where, for example, a transcription factor directly or indirectly represses its own expression [86], [87].
Both DMD-3 and MAB-3 are required for maintaining the expression of dmd-3, thus forming a positive feedback loop [7]. We show here that these genes are also involved in a negative feedback loop, since they inhibit NHR-25 in a pathway which regulates their own expression. We propose that these feedback loops are important for regulating the duration of morphogenesis. Specifically, the regulatory loop between NHR-25 and MAB-3/DMD-3 appears to play a key role in the temporal regulation of morphogenesis. After dmd-3 expression is initiated, its product inhibits NHR-25 production, probably in combination with translational inhibition of nhr-25 by let-7 miRNA [58]. This mechanism should prevent the kind of over-retraction (Ore) phenotype seen in lin-41(ma104lf) mutants in which dmd-3 is on for too long because its expression begins precociously in L3 and continues through L4. The specific mechanisms of this feedback loop are a topic of further study in our lab.
Another system control mechanism involves the TGF-β pathway and VAV-1. As positive regulators of morphogenesis, we would expect these pathways to either positively regulate or be positively regulated by DMD-3. However, DMD-3 and MAB-3 negatively regulate these pathways. These interactions might function to restrict morphogenesis in time and space. How this regulation is facilitated is still unknown.
In summary, we have presented evidence that the genetic network underlying C. elegans male tail tip morphogenesis is consistent with a bow-tie architecture in which dmd-3, mab-3 and nhr-25 are the central regulators in a conserved core (Figure 8). Of the 23 different interactions we tested by expression epistasis, 11 showed an epistatic effect on expression or localization; all of these involved dmd-3 and/or mab-3. Twelve interactions showed no effects; of these, only four involved dmd-3 and/or mab-3 (Table S5). We therefore believe that the position of dmd-3 and mab-3 in the core of the network is real and not merely an effect of a bias in our experimental design.
Conclusion
The architecture of the genetic network regulating male tail tip morphogenesis in C. elegans is congruent with the bow-tie model, since we find evidence for all the characteristics of bow-tie networks. We find modularity, degeneracy, a conserved core, weak linkage and positive and negative feedback loops connecting spatial, sexual and temporal inputs to the cellular responses required for morphogenesis. To our knowledge, this is the first time that a genetic network regulating a morphogenetic process has been specifically investigated for bow-tie architecture. However, it is likely that other morphogenetic events—e.g., the development of the eye in flies and possibly mammals and the formation of the pharynx in C. elegans—are also controlled by bow-tie regulatory networks. In both examples, components have been identified which are likely to be part of the conserved core. Drosophila eye development is in part controlled by the products of eight eye specification genes. Deletion of either one of these genes leads to a drastic reduction or loss of the adult eye, whereas ectopic expression of all but one results in retinal development outside of normal eye tissue [88], [89]. The fly eye specification genes are conserved with orthologs in mammals. Expression of one of them, dachshund, is regulated by at least 36 upstream factors, including the TGF-β signaling pathway, the transcription factor Zerknüllt and several other patterning genes (e.g. krüppel, snail and dorsal) [88], suggesting the existence of an extensive input fan in this system. The FoxA transcription factor PHA-4 is a central regulator of pharynx development in C. elegans. pha-4 is the only zygotic gene that deletes the entire pharynx when mutated [90]. FoxA transcription factors are conserved from Cnidaria to mammals and are always associated with the digestive tract [90]. PHA-4 has hundreds of targets, many of which are directly regulated at the transcriptional level [91], [92]. In this system, feedback loops have been identified as well [90]. Thus, there is evidence for a conserved core, an output fan and system control as elements of a bow-tie network architecture for pharynx morphogenesis. Finding bow-tie networks in multiple developmental systems supports the notion that this architecture is a universal feature of evolved gene regulatory networks and is favored by selection due to its robustness. The male tail tip thus provides a simple model for investigating not only morphogenetic mechanisms, but also the properties of a universally important genetic regulatory architecture.
The evolvable male tail
One general advantage of bow-tie architecture is the ease with which it can accommodate evolutionary changes (evolvability). Weak linkage allows the addition, deletion or switching of components without disturbing the entire system [93]–[95]. The prediction, therefore, is that a comparison of the genetic architecture for a particular process in different species will uncover differences primarily in the input or output fan of the bow-tie network, but not in the core or the conserved components of the interface. The male tail of nematodes related to C. elegans is a great model in which to study network architecture in an evo-devo context and test this prediction. One attractive aspect of the male tail tip in an evolutionary context is that it has changed shape repeatedly in multiple lineages [96], [97], thus providing a model not only to uncover the genetic architectural changes underlying the evolution of morphogenesis, but also to test if the same or different parts of the genetic architecture are altered in repeated emergences of similar morphologies. Future research will thus be focused on building a more complete picture of male tail tip regulatory architecture in C. elegans and developing genomic methods to allow comparisons with related species.
Materials and Methods
Strains and transgenic lines
Genetic manipulations and culturing of C. elegans were performed as previously described [98]. We use the following nomenclature for transgenes (similar to that used by Ziel et al. [99]). Transcriptional reporters are designated by the name of the gene associated with the promoter, followed by a “>” and the reporter gene to which it is fused (e.g., dmd-3>yfp). Translational reporters are designated by the gene, followed by “::” and the reporter to which it is fused (e.g., sma-3::mCherry). Unless otherwise stated, the endogenous promoter is used to drive expression of translational reporters. If a different promoter is used, we use both designations (e.g., lin-44>php-3::gfp represents the php-3 gene fused to the gfp gene, with expression driven by the promoter of the lin-44 gene). Unless otherwise stated, the unc-54 3′UTR is used for all constructs. The protein product of a construct is designated with capital letters (e.g., PHP-3::GFP). No construct employed cDNA; all introns were included.
Strains with transgenes generated for this paper are listed in Table S3. Other strains used for this study are listed below.
CB4088 = him-5(e1490)V. This is the otherwise wild-type, male-producing strain used as the background genotype in this study (from Caenorhabditis Genetics Center, CGC).
BW2020 = ctIs57[nob-1::gfp + rol-6] (a gift from Zhongying Zhao, University of Washington, Washington). Hermaphrodites of this strain were crossed with CB4088 males to obtain a him-59(e1490)V; ctIs57 strain.
DF125 = php-3(ok919)III; him-5(e1490)V. Made by crossing CB4088 males with RB998 hermaphrodites. RB998 = php-3(ok919)III (from CGC).
DF159 = rme-8(b1023)I; him-5(e1490)V. Made by crossing CB4088 males with DH1206 hermaphrodites. DH1206 = rme-8(b1023)I (from CGC).
DF160 = blmp-1(tm548)I; him-5(e1490)V; fsIs3[dmd-3>yfp + unc-122>gfp].
DF161 = blmp-1(tm548)I; him-5(e1490)V. Made by crossing CB4088 males with hermaphrodites carrying the blmp-1(tm548) allele (from Shohei Mitani, National BioResource Project, Tokyo Women's Medical University School of Medicine, Tokyo, Japan).
DF163 = sma-3(e491)III; him-5(e1490)V. Made by crossing CB4088 males with CB491 hermaphrodites. CB491 = sma-3(e491)III (from CGC).
DF164 = egl-18(ok290)IV; him-5(e1490)V. Made by crossing CB4088 males with JR2370 hermaphrodites. JR2370 = egl-18(ok290)IV (from CGC).
DF167 = him-5(e1490)V; nhr-25(ku217)X. Made by crossing CB4088 males with MH1955 hermaphrodites. MH1955 = nhr-25(ku217)X (from CGC).
DF171 = him-5(e1490)V; bIs34[rme-8::gfp + rol-6]. Made by crossing CB4088 males with DH1336 hermaphrodites. DH1336 = bls34[rme-8::gfp + rol-6] (from CGC).
DF177 = him-5(e1490)V; nhr-25(ku217)X; fsIs3[dmd-3>yfp + unc-122>gfp].
DF178 = mab-3(e1240)II; dmd-3(ok1327) him-5(e1490)V; bIs34[rme-8::gfp + rol-6].
DF196 = him-5(e1490)V; xnIs8[pJN343: nmy-2::mCherry + unc-119(+)]. This strain was made by crossing CB4088 males to hermaphrodites carrying the transgene xnls8 which were a generous gift from Jeremy Nance (NYU Skirball Institute, New York, New York).
DF197 = mab-3(e1240)II; dmd-3(ok1327) him-5(e1490)V; xnIs8[pJN343: nmy-2::mCherry + unc-119(+)].
DF199 = ptl-1(ok621)III; him-5(e1490)V. Made by crossing CB4088 males to RB808 hermaphrodites. RB808 = ptl-1(ok621)III (from CGC).
JJ1473 = unc-119(ed3)III; zuIs45[nmy-2::gfp + unc-119(+)] (from CGC).
KC447 = rrf-3(pk1426)II; him-5(e1490)V. A generous gift from King L. Chow (Hong Kong University of Science and Technology, Hong Kong, China).
KC529 = eri-1(mg366)IV; him-5(e1490)V. (from K. L. Chow).
UR157 = fsIs2[dmd-3>yfp + unc-122>gfp]I?; him-5(e1490)V. A generous gift from Douglas Portman (Rochester University, New York).
UR279 = mab-3(e1240)II; dmd-3(ok1327) him-5(e1490)V (from D. Portman).
WM79 = rol-6(n1270)II; neEx[lit-1::GFP + rol-6(su1006)] (from CGC). Hermaphrodites of this strain were crossed with CB4088 males to obtain a rol-6(n1270)II; him-5(e1490)V; neEx[lit-1::GFP + rol-6(su1006)] strain.
RNAi screen
The genome-wide RNAi-feeding screen was carried out in the RNAi-hypersensitive background rrf-3; him-5 (strain KC447). RNAi effects on embryogenesis were bypassed by feeding siRNA-expressing bacteria to synchronized L1 larvae. Following a recommendation by K. Chow (pers. comm.), L1 larvae were plated onto a thin film of agar (1.5 ml per 60 mm plate) containing 2 mM IPTG and 100 µg/ml ampicillin. Worms were cultured to adulthood (3 days) at 20°C at which time a square of agar with worms was removed and mounted directly onto a glass slide and covered with a coverslip (Figure 1A). All scoring was carried out at 400x with a Zeiss Axioskop equipped with Nomarski differential interference contrast. Images were recorded with a C4742-95 “Orca” Hamamatsu digital camera and Openlab software, ver. 3.0.9 (Improvision). The secondary screen was carried out in the same way but in a different RNAi hypersensitive background, eri-1 (strain KC529). RNAi clones consistently conferring a Lep or Ore tail tip phenotype were sequenced to confirm the targeted genes. All scoring data and images are available via our male tail tip database, MTTdb, at http://wormtails.bio.nyu.edu.
Generation of transgenic strains
Translational fusions and transcriptional reporters were constructed by overlap-extension PCR as previously described [100]. The 5′ upstream sequence and coding sequences for php-3 (-500 bp to the stop codon), egl-18 (-3691 to stop), rcn-1 (-4797 to stop), ptl-1 (-2105 to stop), arl-1 (-550 to stop), abcx-1 (-437 to stop), cdc-42 (-1814 to stop), sys-1 (-3411 to stop), inx-12 (-3975 to stop), blmp-1 (-4916 To stop), nhr-165 (-1559 to stop), and ubc-12 (-363 to stop), were PCR-amplified from genomic DNA and fused to gfp and the unc-54 3′-UTR amplified from pPD95.75 (Addgene). The 5′-upstream sequence and coding region of sma-3 (from -1169 bp), amplified from genomic DNA, was fused to mCherry [101] and to cfp, which were PCR-amplified from pGC326 (a gift from E. J. Hubbard) and pPD136.61 (Addgene), respectively. To make the DAF-4 reporters, the upstream sequence and coding region (from -5091) was fused to yfp that was amplified from pPD136.64 (Addgene). A transcriptional reporter of daf-4 fused the upstream sequence (-4865 to -1) to the NLS and gfp amplified from pPD122.13 (Addgene). The transcriptional reporter for nhr-25 fused the upstream sequence (-9100 to -1) to the NLS and gfp from pPD122.13 (Addgene) followed by the 3′-UTR for nhr-25 (stop to +760 bp). Transcriptional reporters for inx-12 (-3975 to -1) and inx-13 (-1557 to -1) were fused to yfp (pPD136.64 (Addgene)) and cfp (pPD136.61(Addgene)).
Transgenes were microinjected at concentrations ranging from 5–20 ng/µl along with 100 ng/µl pRF4 (rol-6(su1006)) as injection marker. Multiple lines were analyzed for each construct using epifluorescence (Axioskop with mercury lamp, 400 or 1000x). Representative images of fluorescence expression patterns are available via the MTTdb database at http://wormtails.bio.nyu.edu. Strain names and primer sequences are provided in Table S3.
Network model-building
Genes identified in our screen were manually entered into N-Browse2 (http://Aquila.bio.nyu.edu/NBrowse2/nbrowsetest.jnlp) [17]. Only a subset of these genes showed annotated interactions, and only those with an interaction one or two edges away from another candidate gene were added to our network (Figure 5). Information from other studies [6], [7], [44], [51], [57]–[60] which show genetic or direct interactions with known or newly identified tail tip genes were also included (Figure 5).
Supporting Information
Zdroje
1. HadjantonakisKSolnica-KrezelL 2010 Developmental biology 50 years-investigating the emergence of shape. Introduction. Dev Biol 341 2 4
2. SulstonJEAlbertsonDGThomsonJN 1980 The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev Biol 78 542 576
3. NguyenCQHallDHYangYFitchDH 1999 Morphogenesis of the Caenorhabditis elegans male tail tip. Dev Biol 207 86 106
4. SulstonJEHorvitzHR 1977 Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56 110 156
5. ZhaoXYangYFitchDHHermanMA 2002 TLP-1 is an asymmetric cell fate determinant that responds to Wnt signals and controls male tail tip morphogenesis in C. elegans. Development 129 1497 1508
6. Del Rio-AlbrechtsenTKiontkeKChiouSYFitchDH 2006 Novel gain-of-function alleles demonstrate a role for the heterochronic gene lin-41 in C. elegans male tail tip morphogenesis. Dev Biol 297 74 86
7. MasonDARabinowitzJSPortmanDS 2008 dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development 135 2373 2382
8. FitchDH 1997 Evolution of male tail development in rhabditid nematodes related to Caenorhabditis elegans. Syst Biol 46 145 179
9. SudhausWFitchD 2001 Comparative studies on the phylogeny and systematics of the rhabditidae (nematoda). J Nematol 33 1 70
10. HodgkinJ 2002 The remarkable ubiquity of DM domain factors as regulators of sexual phenotype: ancestry or aptitude? Genes Dev 16 2322 2326
11. RaymondCSShamuCEShenMMSeifertKJHirschB 1998 Evidence for evolutionary conservation of sex-determining genes. Nature 391 691 695
12. ShenMMHodgkinJ 1988 mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell 54 1019 1031
13. HodgkinJ 1987 A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev 1 731 745
14. ZarkowerDHodgkinJ 1992 Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell 70 237 249
15. MohlerWAShemerGdel CampoJJValansiCOpoku-SerebuohE 2002 The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell 2 355 362
16. ShemerGSuissaMKolotuevINguyenKCHallDH 2004 EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Curr Biol 14 1587 1591
17. KaoHLGunsalusKC 2008 Browsing multidimensional molecular networks with the generic network browser (N-Browse). Curr Protoc Bioinformatics Chapter 9 Unit 9 11
18. CseteMDoyleJ 2004 Bow ties, metabolism and disease. Trends Biotechnol 22 446 450
19. KitanoH 2004 Biological robustness. Nat Rev Genet 5 826 837
20. Rodriguez-CasoCCorominas-MurtraBSoleRV 2009 On the basic computational structure of gene regulatory networks. Mol Biosyst 5 1617 1629
21. ZhaoJYuHLuoJHCaoZWLiYX 2006 Hierarchical modularity of nested bow-ties in metabolic networks. BMC Bioinformatics 7 386
22. OdaKKitanoH 2006 A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol 2 2006.0015
23. OdaKMatsuokaYFunahashiAKitanoH 2005 A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol 1 2005.0010
24. KrantzMAhmadpourDOttossonLGWarringerJWaltermannC 2009 Robustness and fragility in the yeast high osmolarity glycerol (HOG) signal-transduction pathway. Mol Syst Biol 5 281
25. LiCWChenBS 2010 Identifying functional mechanisms of gene and protein regulatory networks in response to a broader range of environmental stresses. Comp Funct Genomics 408705
26. KitanoHOdaK 2006 Robustness trade-offs and host-microbial symbiosis in the immune system. Mol Syst Biol 2 2006 0022
27. TieriPGrignolioAZaikinAMishtoMRemondiniD 2010 Network, degeneracy and bow tie integrating paradigms and architectures to grasp the complexity of the immune system. Theor Biol Med Model 7 32
28. FraserAGKamathRSZipperlenPMartinez-CamposMSohrmannM 2000 Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408 325 330
29. BerrizGFKingODBryantBSanderCRothFP 2003 Characterizing gene sets with FuncAssociate. Bioinformatics 19 2502 2504
30. MalloMWellikDMDeschampsJ 2010 Hox genes and regional patterning of the vertebrate body plan. Dev Biol 344 7 15
31. PearsonJCLemonsDMcGinnisW 2005 Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6 893 904
32. HermanMAVassilievaLLHorvitzHRShawJEHermanRK 1995 The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell 83 101 110
33. KanekoHShimizuRYamamotoM 2010 GATA factor switching during erythroid differentiation. Curr Opin Hematol 17 163 168
34. HoICTaiTSPaiSY 2009 GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol 9 125 135
35. ChouJProvotSWerbZ 2010 GATA3 in development and cancer differentiation: cells GATA have it! J Cell Physiol 222 42 49
36. KohKRothmanJH 2001 ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development 128 2867 2880
37. KohKPeyrotSMWoodCGWagmaisterJAMaduroMF 2002 Cell fates and fusion in the C. elegans vulval primordium are regulated by the EGL-18 and ELT-6 GATA factors — apparent direct targets of the LIN-39 Hox protein. Development 129 5171 5180
38. IkushimaHMiyazonoK 2010 TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer 10 415 424
39. GissendannerCRSluderAE 2000 nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol 221 259 272
40. GuichetACopelandJWErdelyiMHlousekDZavorszkyP 1997 The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature 385 548 552
41. YuYLiWSuKYussaMHanW 1997 The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature 385 552 555
42. BroadusJMcCabeJREndrizziBThummelCSWoodardCT 1999 The Drosophila beta FTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol Cell 3 143 149
43. AsahinaMIshiharaTJindraMKoharaYKatsuraI 2000 The conserved nuclear receptor Ftz-F1 is required for embryogenesis, moulting and reproduction in Caenorhabditis elegans. Genes Cells 5 711 723
44. DingXCSlackFJGrosshansH 2008 The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation. Cell Cycle 7 3083 3090
45. AgawaYSarhanMKageyamaYAkagiKTakaiM 2007 Drosophila Blimp-1 is a transient transcriptional repressor that controls timing of the ecdysone-induced developmental pathway. Mol Cell Biol 27 8739 8747
46. MartinsGCalameK 2008 Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol 26 133 169
47. OhinataYPayerBO'CarrollDAncelinKOnoY 2005 Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436 207 213
48. VincentSDDunnNRSciammasRShapiro-ShalefMDavisMM 2005 The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development 132 1315 1325
49. NieKGomezMLandgrafPGarciaJFLiuY 2008 MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol 173 242 252
50. MatzukMM 2009 LIN28 lets BLIMP1 take the right course. Dev Cell 17 160 161
51. JonesDCandidoEP 2000 The NED-8 conjugating system in Caenorhabditis elegans is required for embryogenesis and terminal differentiation of the hypodermis. Dev Biol 226 152 165
52. ZhangYGrantBHirshD 2001 RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol Biol Cell 12 2011 2021
53. ParcejDTampeR 2010 ABC proteins in antigen translocation and viral inhibition. Nat Chem Biol 6 572 580
54. LeeJIDhakalBKLeeJBandyopadhyayJJeongSY 2003 The Caenorhabditis elegans homologue of Down syndrome critical region 1, RCN-1, inhibits multiple functions of the phosphatase calcineurin. J Mol Biol 328 147 156
55. GoedertMBaurCPAhringerJJakesRHasegawaM 1996 PTL-1, a microtubule-associated protein with tau-like repeats from the nematode Caenorhabditis elegans. J Cell Sci 109 Pt 11 2661 2672
56. McDermottJBAamodtSAamodtE 1996 ptl-1, a Caenorhabditis elegans gene whose products are homologous to the tau microtubule-associated proteins. Biochemistry 35 9415 9423
57. HajduskovaMJindraMHermanMAAsahinaM 2009 The nuclear receptor NHR-25 cooperates with the Wnt/beta-catenin asymmetry pathway to control differentiation of the T seam cell in C. elegans. J Cell Sci 122 3051 3060
58. HayesGDFrandARRuvkunG 2006 The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development 133 4631 4641
59. ChenZEastburnDJHanM 2004 The Caenorhabditis elegans nuclear receptor gene nhr-25 regulates epidermal cell development. Mol Cell Biol 24 7345 7358
60. SlackFJBassonMLiuZAmbrosVHorvitzHR 2000 The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5 659 669
61. ShiASunLBanerjeeRTobinMZhangY 2009 Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J 28 3290 3302
62. FireAXuSMontgomeryMKKostasSADriverSE 1998 Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806 811
63. SimmerFTijstermanMParrishSKoushikaSPNonetML 2002 Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12 1317 1319
64. WhitacreJBenderA 2010 Degeneracy: a design principle for achieving robustness and evolvability. J Theor Biol 263 143 153
65. WagnerGPPavlicevMCheverudJM 2007 The road to modularity. Nat Rev Genet 8 921 931
66. HartwellLHHopfieldJJLeiblerSMurrayAW 1999 From molecular to modular cell biology. Nature 402 C47 52
67. GunsalusKCGeHSchetterAJGoldbergDSHanJD 2005 Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature 436 861 865
68. PeterISDavidsonEH 2009 Modularity and design principles in the sea urchin embryo gene regulatory network. FEBS Lett 583 3948 3958
69. Van AukenKWeaverDCEdgarLGWoodWB 2000 Caenorhabditis elegans embryonic axial patterning requires two recently discovered posterior-group Hox genes. Proc Natl Acad Sci U S A 97 4499 4503
70. DonaldsonJGCasselDKahnRAKlausnerRD 1992 ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein beta-COP to Golgi membranes. Proc Natl Acad Sci U S A 89 6408 6412
71. LiYKellyWGLogsdonJMJrSchurkoAMHarfeBD 2004 Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J 18 1834 1850
72. WarnockDESchmidSL 1996 Dynamin GTPase, a force-generating molecular switch. Bioessays 18 885 893
73. UlrichFKriegMSchotzEMLinkVCastanonI 2005 Wnt11 functions in gastrulation by controlling cell cohesion through Rab5c and E-cadherin. Dev Cell 9 555 564
74. HammerschmidtMWedlichD 2008 Regulated adhesion as a driving force of gastrulation movements. Development 135 3625 3641
75. FujibayashiATaguchiTMisakiROhtaniMDohmaeN 2008 Human RME-8 is involved in membrane trafficking through early endosomes. Cell Struct Funct 33 35 50
76. GirardMPouponVBlondeauFMcPhersonPS 2005 The DnaJ-domain protein RME-8 functions in endosomal trafficking. J Biol Chem 280 40135 40143
77. KayAJHunterCP 2001 CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr Biol 11 474 481
78. NanceJPriessJR 2002 Cell polarity and gastrulation in C. elegans. Development 129 387 397
79. MartinAC 2010 Pulsation and stabilization: contractile forces that underlie morphogenesis. Dev Biol 341 114 125
80. BauerRLehmannCMartiniJEckardtFHochM 2004 Gap junction channel protein innexin 2 is essential for epithelial morphogenesis in the Drosophila embryo. Mol Biol Cell 15 2992 3004
81. LeitheESirnesSOmoriYRivedalE 2006 Downregulation of gap junctions in cancer cells. Crit Rev Oncog 12 225 256
82. StarichTSheehanMJadrichJShawJ 2001 Innexins in C. elegans. Cell Commun Adhes 8 311 314
83. KirschnerMGerhartJ 2005 Plausibility of life: great leaps of evolution. New Haven Yale University Press 331
84. GerhartJKirschnerM 1997 Cells, embryos and evolution: towards a cellular and developmental understanding of phenotypic variation and evolutionary adaptability. Malden, Massachusetts Blackwell Science 642
85. JordanJDIyengarR 1998 Modes of interactions between signaling pathways. Biochem Pharmacol 55 1347 1352
86. FreemanM 2000 Feedback control of intercellular signalling in development. Nature 408 313 319
87. CrewsSTPearsonJC 2009 Transcriptional autoregulation in development. Curr Biol 19 R241 246
88. AndersonJSalzerCLKumarJP 2006 Regulation of the retinal determination gene dachshund in the embryonic head and developing eye of Drosophila. Dev Biol 297 536 549
89. KumarJP 2001 Signalling pathways in Drosophila and vertebrate retinal development. Nat Rev Genet 2 846 857
90. MangoSE 2009 The molecular basis of organ formation: insights from the C. elegans foregut. Annu Rev Cell Dev Biol 25 597 628
91. GaudetJMangoSE 2002 Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295 821 825
92. ZhongMNiuWLuZJSarovMMurrayJI 2010 Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet 6 e1000848 doi:10.1371/journal.pgen.1000848
93. ConradM 1990 The geometry of evolution. Biosystems 24 61 81
94. GerhartJKirschnerM 2007 The theory of facilitated variation. Proc Natl Acad Sci U S A 104 Suppl 1 8582 8589
95. KirschnerMGerhartJ 1998 Evolvability. Proc Natl Acad Sci U S A 95 8420 8427
96. FitchDH 2000 Evolution of "rhabditidae" and the male tail. J Nematol 32 235 244
97. KiontkeKFitchDH 2005 The phylogenetic relationships of Caenorhabditis and other rhabditids. WormBook 1 11
98. BrennerS 1974 The genetics of Caenorhabditis elegans. Genetics 77 71 94
99. ZielJWHagedornEJAudhyaASherwoodDR 2009 UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nat Cell Biol 11 183 189
100. ShevchukNABryksinAVNusinovichYACabelloFCSutherlandM 2004 Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res 32 e19
101. ShanerNCCampbellRESteinbachPAGiepmansBNPalmerAE 2004 Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22 1567 1572
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and PhosphodiesterasesČlánek Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



