-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
Neuroblastoma is a malignant neoplasm of the developing sympathetic nervous system that is notable for its phenotypic diversity. High-risk patients typically have widely disseminated disease at diagnosis and a poor survival probability, but low-risk patients frequently have localized tumors that are almost always cured with little or no chemotherapy. Our genome-wide association study (GWAS) has identified common variants within FLJ22536, BARD1, and LMO1 as significantly associated with neuroblastoma and more robustly associated with high-risk disease. Here we show that a GWAS focused on low-risk cases identified SNPs within DUSP12 at 1q23.3 (P = 2.07×10−6), DDX4 and IL31RA both at 5q11.2 (P = 2.94×10−6 and 6.54×10−7 respectively), and HSD17B12 at 11p11.2 (P = 4.20×10−7) as being associated with the less aggressive form of the disease. These data demonstrate the importance of robust phenotypic data in GWAS analyses and identify additional susceptibility variants for neuroblastoma.
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1002026
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002026Summary
Neuroblastoma is a malignant neoplasm of the developing sympathetic nervous system that is notable for its phenotypic diversity. High-risk patients typically have widely disseminated disease at diagnosis and a poor survival probability, but low-risk patients frequently have localized tumors that are almost always cured with little or no chemotherapy. Our genome-wide association study (GWAS) has identified common variants within FLJ22536, BARD1, and LMO1 as significantly associated with neuroblastoma and more robustly associated with high-risk disease. Here we show that a GWAS focused on low-risk cases identified SNPs within DUSP12 at 1q23.3 (P = 2.07×10−6), DDX4 and IL31RA both at 5q11.2 (P = 2.94×10−6 and 6.54×10−7 respectively), and HSD17B12 at 11p11.2 (P = 4.20×10−7) as being associated with the less aggressive form of the disease. These data demonstrate the importance of robust phenotypic data in GWAS analyses and identify additional susceptibility variants for neuroblastoma.
Introduction
Neuroblastoma is a pediatric cancer of the developing sympathetic nervous system and is the most common childhood solid tumor outside the central nervous system [1], [2]. Its broad spectrum of clinical behaviors is the basis for ways to categorize neuroblastoma into three risk groups: high-risk, intermediate-risk and low-risk. The approximately 50% of cases classified as high-risk show an aggressive clinical course with widespread metastases to bone and bone marrow present at diagnosis [3]. Despite intensive multimodal therapy, the long-term survival rate is less than 50% for children with high-risk neuroblastoma [1]. On the other hand, substantial portions of neuroblastoma patients show favorable clinical features including spontaneous regression of disease, and are classified as low-risk. Low-risk neuroblastoma patients have a greater than 95% survival probability with minimal, if any, chemotherapy [1]. Intermediate-risk cases are the most heterogeneous, and also the smallest subset using current definitions, comprising about 15% of all neuroblastoma patients.
We have recently performed a neuroblastoma GWAS by applying single marker analyses and identified three distinct loci significantly associated with neuroblastoma. Each of these SNP associations was within genes and particularly enriched in the high-risk group of patients: FLJ22536 at chromosome 6p22 [4], BRCA1 associated RING domain 1 (BARD1) at 2q35 [5], and LIM domain only 1 (LMO1) at 11p15 [6]. A similar approach was utilized to identify a common copy number variation (CNV) at chromosome 1q21 within the NBPF23 gene that is also robustly associated with neuroblastoma [7].
In this study, we report that by adapting statistical methods to analyze genotype data, we discovered, and successfully replicated, three distinct loci as associated with the low-risk group of neuroblastoma. Furthermore, we report several gene sets as enriched in all risk groups of neuroblastoma.
Results
Gene-centric method identifies three low-risk neuroblastoma susceptibility loci
As we are interested in studying disease causal variants that have a high likelihood of impacting protein-encoding genes, we developed a gene-centric computational method to test for association signals at the gene level. This method adapted the global test [8], developed to test association of genes groups using microarray expression data, to analyze our genotype data. Our method computes an aggregated test score based on genotype data of all SNPs on a region extending 10 kilo-bases upstream and downstream of a gene. We applied this method using a discovery set containing 1627 cases and 2575 control subjects, aimed at analyzing association to 15,885 genes annotated in the UCSC Genome Browser [9] (Materials and Methods). The replication dataset contained 398 cases and 1507 control subjects. Our methodology correctly identified the three significant genes already reported (FLJ22536, BARD1 and LMO1). In addition, our method also identified the dual-specificity phosphatase 12 gene (DUSP12) at chromosome band 1q23.3 (Table 1) as significantly associated with neuroblastoma.
Tab. 1. Summary of gene-centric analysis results for different phenotypic neuroblastomas. 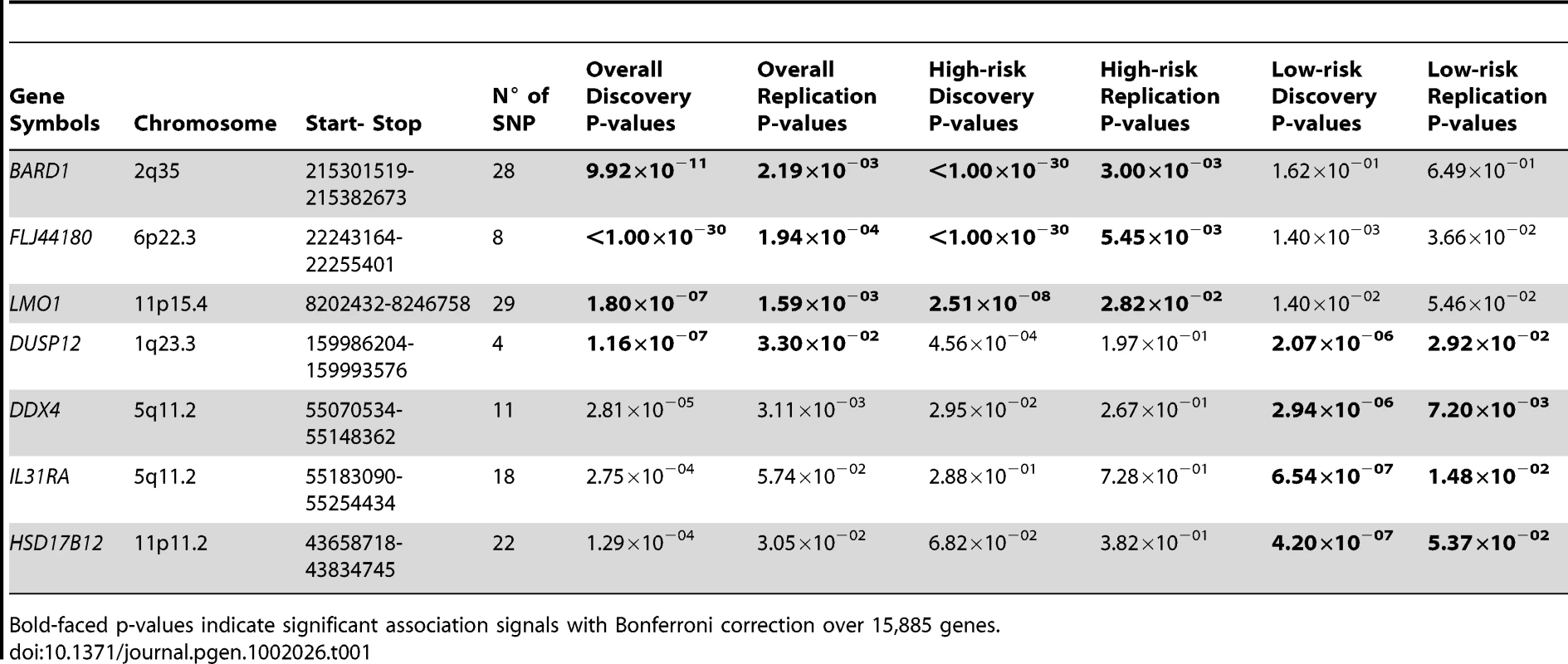
Bold-faced p-values indicate significant association signals with Bonferroni correction over 15,885 genes. We next sought to determine if association signals discovered in our unbiased scan would be further enriched, or diminished, when we restricted our analyses to the divergent phenotypes of low-risk or high-risk neuroblastoma. We first analyzed a subset of 678 high-risk neuroblastoma cases from the original discovery case series, again matched to 2575 control subjects. This analysis reconfirmed that all three previously reported signals were truly associated with high-risk neuroblastoma (Table 1), but DUSP12 did not show a strong association signal in the high-risk disease case series (P = 4.56×10−04). In parallel, we analyzed a subset of 574 low-risk cases and 1722 matched control subjects and a replication set of 124 cases and 496 matched control subjects (Materials and Methods). This analysis confirmed DUSP12 and three novel genes as associated with low-risk neuroblastoma: DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 isoform (DDX4) and interleukin-31 receptor A precursor (IL31RA) both at the same locus within chromosome band 5q11.2, and hydroxysteroid (17-beta) dehydrogenase 12 (HSD17B12) at chromosome band 11p11.2 (Table 1). All signals had significant discovery p-values using Bonferroni correction over 15,885 genes (P<3.15×10−6), and replication p-values less than 0.05.
Our gene-centric method was able to detect DUSP12 and HSD17B12, the only two genes containing at least one SNP that passed the Bonferroni correction in single marker (SNP) analysis of the low-risk neuroblastoma (Figure 1 and Figure 2) using association testing as implemented in PLINK [10]. The fact that our gene-centric results were compatible with the single marker results supported the effectiveness of our method. In addition, we were able to detect two gene-level association signals located at a single locus for DDX4 and IL31RA even though these genes did not contain any significant SNPs in the single marker analysis (Figure 1 and Table 2). These genes, however, contained several SNPs with moderate signals (Figure 2), and our gene-centric method was able to combine these effects and detected the overall significance of these two gene's signals. Being independently replicated in our study (P = 7.20×10−3 and 1.48×10−2 respectively), these two signals offered indications that our gene-centric method was more effective than single marker analysis in detecting gene-level association signals. Indeed, our power computation, adjusting for 15,885 tests, indicated that our method performed far better than the single SNP method in both our discovery and replication case series (Figures S1, S2, S3, S4).
Fig. 1. Manhattan plot of single marker analysis of the low-risk neuroblastoma data set. 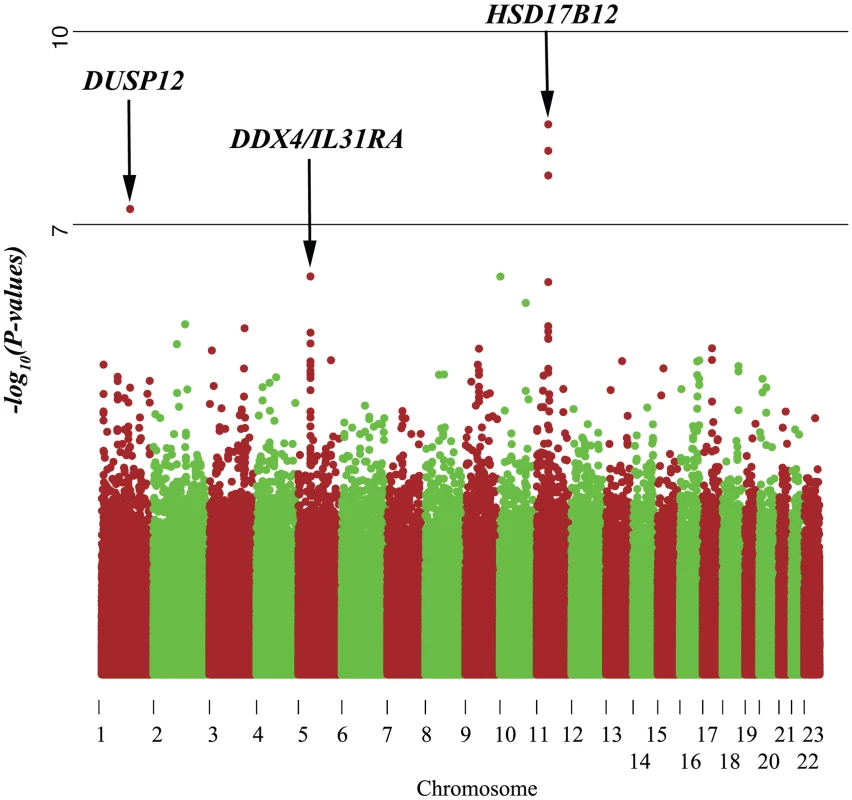
Even though the genes DDX4 and IL31RA do not contain significantly associated SNPs (P = 1.0×10−07), the combined effect of moderately associated SNPs drives these two genes to be significant in our gene-centric analysis (genome-wide gene centric threshold p-values for significance is P<3.15×10−6). Fig. 2. Haplotype view of the 4 genes significantly associated with low-risk neuroblastoma. 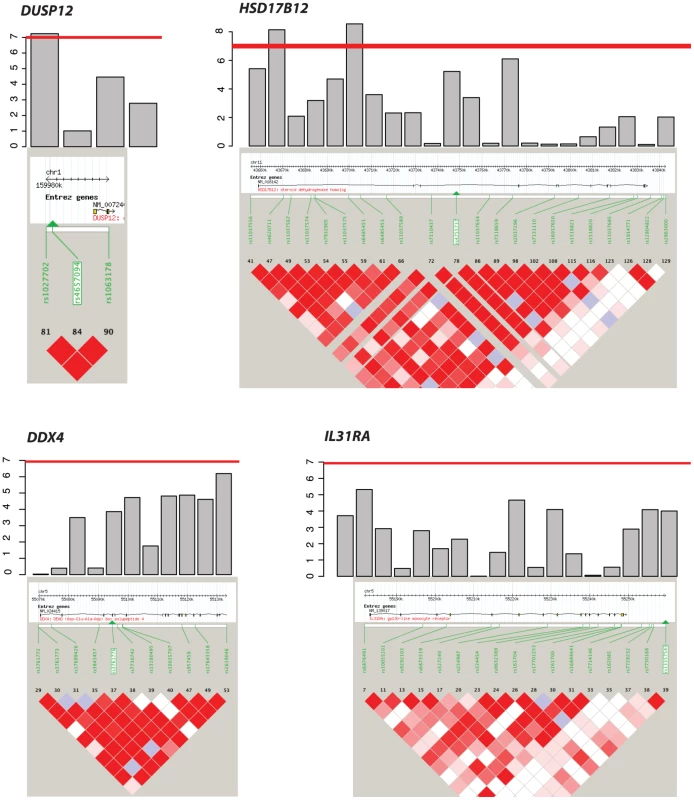
Red line indicates P<1.0×10−7. Only DUSP12 and HSD17B12 contain SNPs with significant single-marker p-values in neuroblastoma low-risk subset. While DDX4 and IL31RA do not contain significant SNPs, our gene-centric method was able to detect these genes as associated with low-risk neuroblastoma. Tab. 2. Additional summaries of gene-centric analysis results for low-risk neuroblastoma. 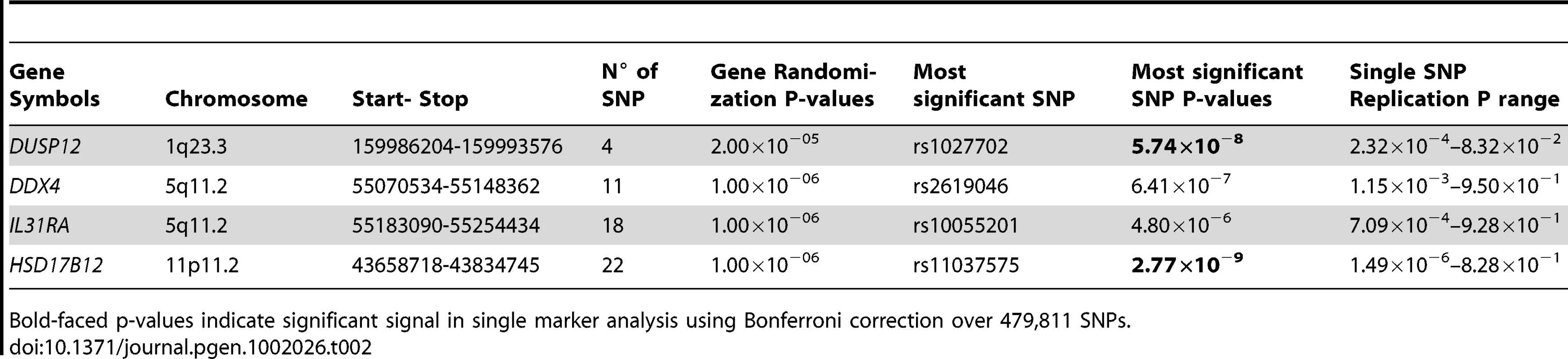
Bold-faced p-values indicate significant signal in single marker analysis using Bonferroni correction over 479,811 SNPs. To further confirm the validity of our discovery, we computed a randomization p-value for each of the four newly discovered genes. For each of these genes, we computed a separate null test statistic distribution by calculating the gene-centric test statistics of one million randomly selected pseudo-genes. These pseudo-genes were selected to contain the same number of SNPs as were contained in the referenced gene. This method of selecting pseudo-genes was based on our observation that the average gene-based test statistic was strongly correlated with the number of SNPs in these genes (Figure S5), Using these null distributions to compare against the observed test statistics of the four newly discovered genes, we arrived at the randomization p-values (Table 2). These p-values (range 2.0×10−5–1.0×10−6) were compatible with the p-values asymptotically computed by our gene-centric method, and notably strengthened the credibility of the discovery of four novel disease causal genes associated with low-risk neuroblastoma.
To assess the joint impact on disease risk of these genes, we estimated the two-locus genotype odd ratios for all pairs amongst the four most significant SNPs within these four genes (Table 3). For each SNP pair tested, the independently contributed disease risks for carriers of risk alleles at only one locus were overall slightly stronger than the disease risks of each SNP when analyzed separately. In all but one case, the odd ratios of disease risks for carriers of both risk alleles increased markedly (odd ratios range from 2.505 to 3.435). However, no significant interaction between these SNP pairs was detected (P ranges from 0.459 to 0.909). Further, we computed all SNP pair interactions amongst all four genes and again noticed no significant SNP-SNP interaction signals (the best SNP pair signals' P ranges from 0.108 to 0.523, Table S1). In the special case of DDX4 and IL31RA, the modest disease risk for carriers of both risk alleles implicated the true association signal encompassed both genes though they are 38 kilo-bases apart from each other (Figure S6).
Tab. 3. Estimates of low-risk neuroblastoma odd ratios by genotype between the most significant SNPs. 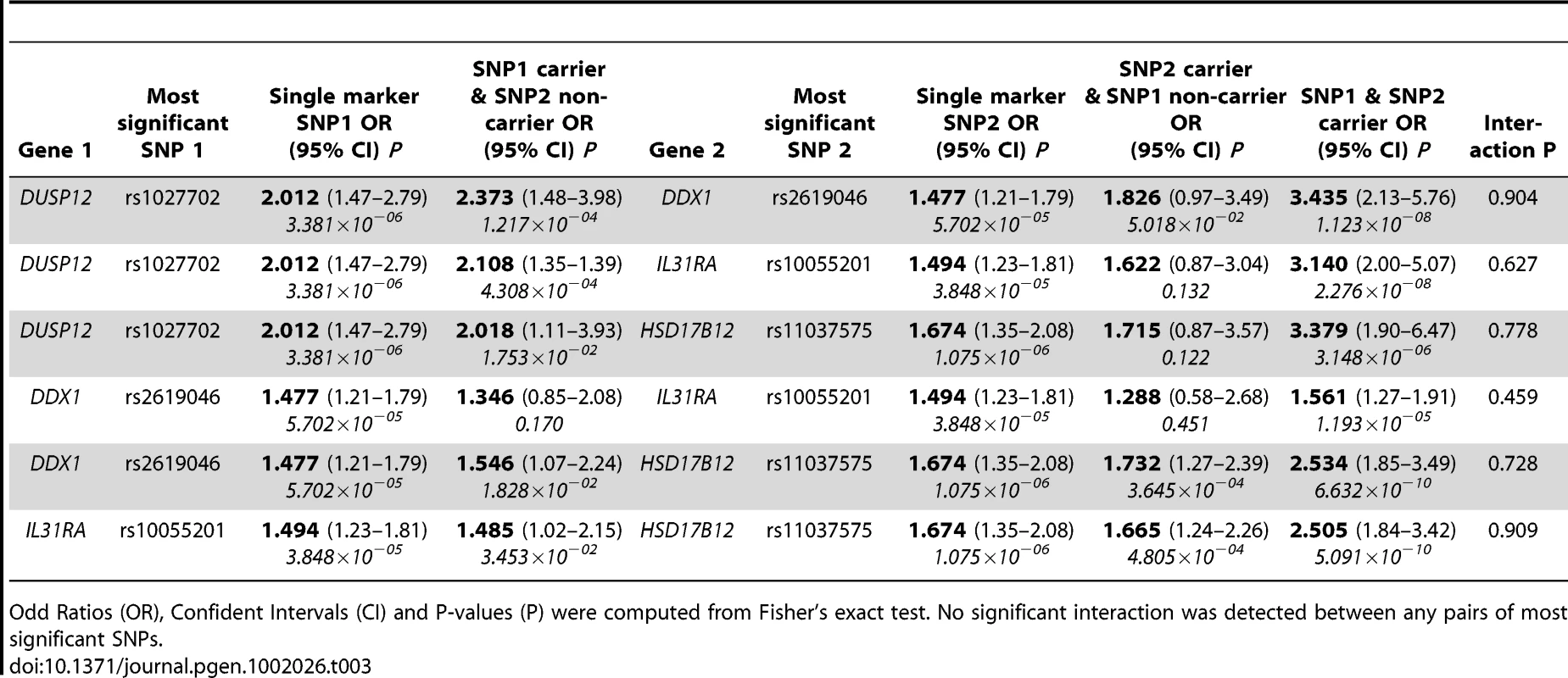
Odd Ratios (OR), Confident Intervals (CI) and P-values (P) were computed from Fisher's exact test. No significant interaction was detected between any pairs of most significant SNPs. Lastly, we sought to further replicate our results in an independent cohort in Italy of 115 low-risk cases and 680 controls. We selected to genotype the three most significant SNPs (rs1027702, rs2619046, rs11037575) in the three loci that contain DUSP12, DDX4/IL31RA, and HSD17B12 respectively. We analyzed these SNPs data sing various statistical tests listed in Table 4. Interestingly, rs1027702 showed strong replication signals for allele frequency association test as well as dominant model association test (P = 0.031 and 0.008 respectively). On the other hand, both rs2619046, rs11037575 showed strong significant signals for homozygous association test (P = 0.042 and 0.028 respectively) as well as recessive model association test (P = 0.047 and 0.037 respectively). Overall, these replication results provide unambiguous evidence to confirm these three loci as significantly associated with low-risk neuroblastoma.
Tab. 4. Single SNP replication results in Italian cohort (n = 115 low-risk neuroblastoma and 680 controls). 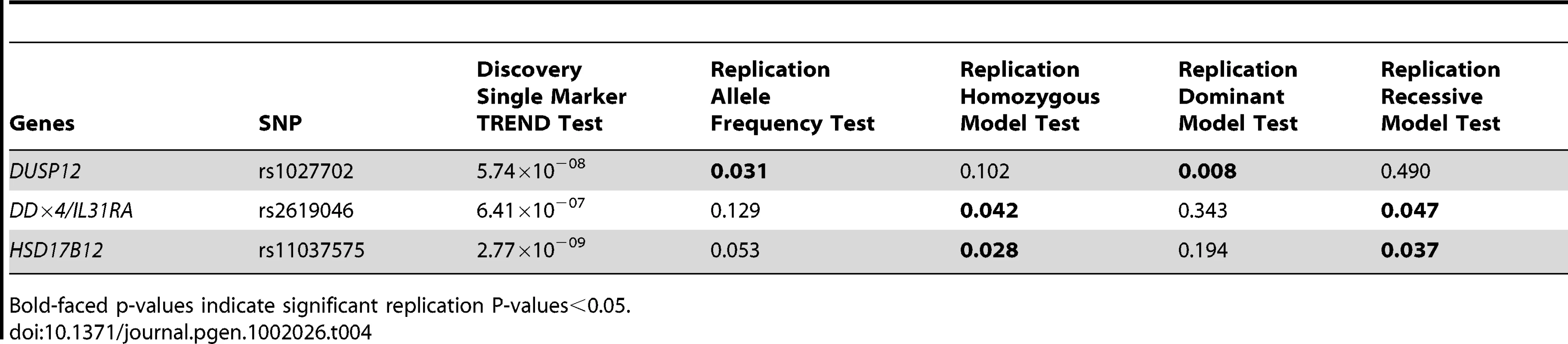
Bold-faced p-values indicate significant replication P-values<0.05. Gene-set analyses identify enriched gene sets in all phenotypes
We are also interested in gene set analyses to identify specific pathways and gene sets that are enriched in neuroblastoma. To perform this analysis, we adapted the random-set approach, which was developed to analyze gene set analysis using gene expression data. This method [11] was suitable for our purpose since it required gene-level scores, which were conveniently obtained by taking the logarithm transformation of our gene-centric p-values.
We applied this random-set procedure using the overall, high-risk and low-risk data sets described earlier over 4734 gene sets obtained from the Gene Set Enrichment Analysis site [12], and selected enriched gene sets based on Bonferroni correction criterion (P<1.05×10−5). Additionally selected based on replication p-value threshold of 0.05, three Gene Ontology [13] sets were associated with all cases of neuroblastoma: Nuclear Ubiquitin Ligase Complex, Negative Regulation of Intracellular Transport, and Regulation of Phosphorylation (Table 5). The first two gene sets were also significantly enriched in high-risk neuroblastoma (P = 1.275×10−09 and 6.332×10−07 respectively) with significant replication p-values (0.030 and 0.036 respectively). The third gene set appeared to be enriched in low-risk neuroblastoma (P = 1.678×10−06); however, we were unable to replicate this result (P = 0.96). Furthermore, we identified and successfully replicated an additional gene set that was exclusively enriched in low-risk neuroblastoma: Cytokine and Chemokine Mediate Signaling Pathway (discovery P = 8.175×10−06 and replication P = 0.040). The identification of these gene sets may elucidate biological pathways that are important in the biology of neuroblastoma.
Tab. 5. Summary of gene set analysis results for all, high-risk, and low-risk neuroblastoma. 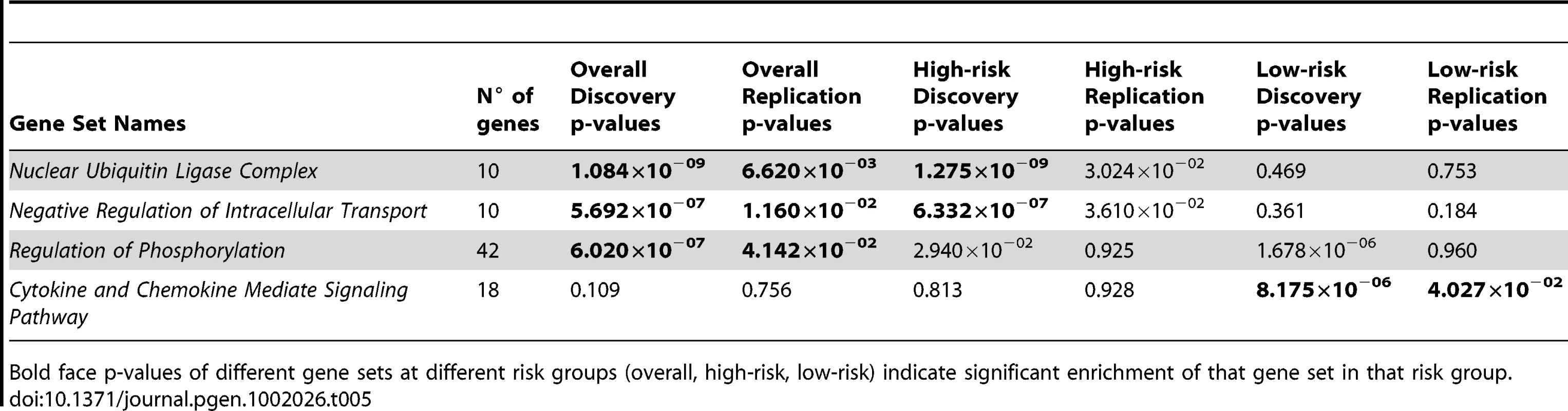
Bold face p-values of different gene sets at different risk groups (overall, high-risk, low-risk) indicate significant enrichment of that gene set in that risk group. Discussion
Taken together, this study implicates DUSP12, DDX4, IL31RA, and HSD17B12 as neuroblastoma susceptibility genes, with particular relevance for those at low-risk for malignant progression and death from disease. Methodologically, we suggest that the gene-centric method has stronger power of detection of association signals compared to the single marker method (Figures S1, S2, S3, S4). Not only was the gene-centric method able to detect the two genes harboring genome-wide significant SNPs (DUSP12 and HSD17B12), but also it was able to detect 2 genes that would have been missed by the single marker analysis (DDX4 and IL31RA). Since this method was originally developed to analyze gene expression data, its limitation is the lack of ability to take into account the haplotype effect in computing gene level test statistics. However, our efforts to replicate the discovery with two independent cohorts unequivocally verify association signals at these loci. Further studies will be required to determine if these common variations tag cis - or trans-acting disease causal variations. Interestingly, the segregation of gene-level association signals and gene set enrichment scores between high-risk and low-risk neuroblastoma (Table 1 and Table 5) supports the view that common variation in the human genome can predisposed not only to a particular disease, but also to a clinically relevant disease subsets, thus demonstrating the power of robust phenotypic data in GWAS efforts.
Materials and Methods
Subjects and quality control
Study subjects
The neuroblastoma patients in this study were children registered through the North American-based Children's Oncology Group (COG) and were diagnosed with neuroblastoma or ganglioneuroblastoma. Blood samples from the neuroblastoma cases were identified through the COG neuroblastoma repository for specimen collection at time of diagnosis. All specimens were annotated with clinical and genomic information (Table S2). Samples were assigned into three risk groups (low-risk, intermediate-risk and high-risk) based on the COG risk assignment algorithm [1], that includes patient age at diagnosis [14], International Neuroblastoma Staging System (INSS) stage [2], tumor histopathology [15], DNA index [16], and MYCN amplification status [17]. The only eligibility criterion for genotyping was availability of 1.5 µg of high quality DNA from a tumor-free source such as peripheral blood or bone marrow cells uninvolved with tumor. Since neuroblastoma in the United States is demographically a disease of Caucasian of European descendent, we limited our analyses to this ethnic group to minimize genetic heterogeneity. Summaries of clinical and genomic information of our discovery and replication cohorts are provided in Table S2.
The control group in this study included 2575 children of self-reported Caucasian ancestry who were recruited and genotyped by the Center for Applied Genomics at the Children's Hospital of Philadelphia (CHOP). Eligibility criteria for control subjects were: 1) self-reported Caucasian; 2) availability of 1.5 µg of high quality DNA from peripheral blood or mononuclear bone marrow cells; and 3) no known medical disorder, including cancer, based on self-reported intake questionnaire and/or clinician-based assessments.
The CHOP Institutional Review Board approved this study.
Genotyping and quality control for discovery cohort
SNP genotyping was performed using the Illumina Infinium II BeadChip (Illumina, San Diego, CA, USA) according to methods detailed elsewhere [4], [5]. Since a portion of the individuals in the discovery cohort was genotyped by the HumanHap550 v1 array (n = 859) while others were genotyped by the v3 array (n = 768), our analysis only concerned the markers shared by the v1 and v3 array. The HumanHap550 v1 array contains 555,175 markers, while the v3 array contains 561,288 markers, including 544,902 markers that are shared by the two arrays. We filtered out 8,749 SNP markers with call rate less than 95%. We also excluded 5,415 SNP markers whose Hardy-Weinberg Equilibrium p-values were less than 0.001. Finally, we excluded additional 50,869 SNP markers whose minor allele frequency is less than 5%.
A total of 96 cases were removed from our data set due to their low genotype call rate (<95%). Furthermore, we used Multi-Dimensional Scaling (MDS) as implemented in the PLINK [10], for inferring population structure (Figure S7). Comparing self-identified ancestry with MDS-inferred ancestry confirmed 1642 neuroblastoma patients of European ancestry. Finally, we calculated genome-wide identity-by-state (IBS) estimates for all pair-wise comparisons among all case subjects and control subjects to detect cryptic relatedness and potential duplicated genotype within our data set. This step further excluded 15 neuroblastoma patients from our analyses.
After all quality control steps, our discovery data set contained 1627 neuroblastoma case subjects of European ancestry, each of which contained 479,811 SNP markers. To correct the potential effects of population structure, 2575 matching control subjects of European ancestry were selected based on their low IBS estimates with case subjects. The genomic control inflation factor for this data set was 1.08.
Five hundred and seventy four (574) low-risk cases, selected from the above 1627 cases, were included for all low-risk neuroblastoma analyses. To keep the genomic inflation factor low, three best matching control subjects were selected for each case, based on IBS estimates, making a total of 1722 control subjects included for analyses. The genomic control inflation factor for this data set was 1.07.
Genotyping and quality control for initial replication cohort
SNP genotyping was performed using the Illumina Human610-Quad array that includes both SNP and CNV markers. The Human610-Quad array contains 620,901 SNPs. We filtered out 48,831 SNP markers with call rate less than 95%. We also excluded 13,305 SNP markers whose Hardy-Weinberg Equilibrium p-values were less than 0.001. Finally, we excluded additional 49,057 SNP markers whose minor allele frequency was less than 5%. A total of 15 cases were removed from our data set due to their low genotype call rate (<95%). After all quality control steps, our replication data set contained 398 neuroblastoma case subjects of European ancestry, each of which contained 509,708 SNP markers. To correct the potential effects of population structure, 1507 matching control subjects of European ancestry were selected based on their low IBS estimates with case subjects.
One hundred and twenty four (124) low-risk cases, selected from the above 398 cases, were included for all low-risk neuroblastoma replication analyses. For each case, four best matching control subjects were selected based on IBS estimates, making a total of 496 control subjects included for analyses.
Genotyping of second replication cohort
One hundred and fifteen (115) low-risk neuroblastoma subjects for Italy and six hundred and eighty (680) control Italian subjects were selected to be genotyped at three SNPs: rs1027702, rs2619046, and rs11037575. All samples were genotyped by Taqman SNP Genotyping Assay by Applied Biosystems.
Statistical analyses
Gene-centric analysis
Our gene-centric analysis adopted the global test method [8], developed to test association of a group of genes using microarray data. First, to mirror gene expression data, we quantified our SNP genotype data by counting the number of minor alleles for each sample at each SNP. Second, due to the analogy in relative relationship of the two concepts in global test and in our study, we substituted the concepts of “genes” and “group of genes” from global test with “SNPs” and “genes” respectively.
This method adopted the generalized linear model framework to model the relationship between Y, a vector of clinical outcomes, and X, the n×m matrix of genotypic data of n subjects and m SNPs. In this model, α is the intercept, β is a length m vector of regression coefficients, and h is a general link function such as the logit function
Testing association between genotypic data and clinical outcomes is equivalent to testing the null hypothesis H0: β1 = β2 = … = βm = 0. Since the number of SNP is much larger than the number of subjects, it is not possible to test this hypothesis in a classical way. Instead, we could test H0 if we assume β1,…,βm to be samples from a common distribution with expectation zero and variance τ2. The null hypothesis becomes simply H0: τ2 = 0. If we rewrite the model in terms ri = ∑jxijβj, with i = 1,…,n, then ri is the linear predictor, the total effect of all covariates for subject i. Let r = (r1,…,rn), then r is a random vector with expectation zero and Cov(r) = τ2XX. The original model simplifies to
This is a simple random effect model. Under the null hypothesis, the test statistic
has expectation E(Q) = trace(R) and variance:
where R = (1/m)XX' is an n×n matrix proportional to the covariance matrix of the random effects r, μ = h−1(α) is the expectation of Y under H0, and μ2 and μ4 are the second and fourth central moments of Y under H0.
The test statistic Q could be rewritten aswhere Xi is the length n vector of genotype of SNP i. The expression Qi = (1/μ2)[Xi' (Y-μ)]2 would be exactly the test statistic of SNP i if it were the only SNP on the gene of interested; or we could interpret that Qi is the “contribution” of SNP i to the overall test statistic. This means that the overall test statistic is simply the average of the statistics Q1,…, Qm of m individual SNPs. Notably, the averaging is over a squared covariance between genotype and clinical outcomes, SNPs with large variance (i.e. strong association signals) have stronger influences on the outcome of the test statistic Q than those with weaker association signals.
Using this method, we computed an aggregated effect of all SNPs that are located from 10-kilo bases upstream to 10-kilo bases downstream for the gene being tested, and computed asymptotic p-value for each gene. We performed the global test on 15,885 annotated, unannotated and predicted genes downloaded from the UCSC Genome Browser [9], and used strict Bonferroni correction criterion (P<3.15×10−6) to determine whether a gene was associated with neuroblastoma. True association signals were further selected based on replication p-value less than 0.05.
Randomization p-value computation
For each significant gene, we computed randomization p-values by comparing its test statistic and its respective null distributions. A null distribution of a gene was composed of test statistics of one million pseudo-genes having the same number of SNPs as the referenced genes. The SNPs of these pseudo-genes were randomly selected across the genome.
Odd ratios estimation
We used the Fisher's exact test to estimate the odd ratios using genotype data as well as the 95% confident interval and p-values.
SNP-SNP interaction estimation
Single marker interaction scores were computed using the general linear model to compute interaction effect between two SNPs.
Gene set analysis
To analyze the significance of gene sets, we adopted the random-set method [11] since it allows us to utilize the gene-centric results to compute enrichment score for each gene set. In this analysis, we used the logarithm transformation of our gene-centric method as gene-level scores to detect gene sets enriched in neuroblastoma.
We performed three separate gene set analyses for overall, high-risk and low-risk data sets over 4734 gene sets downloaded from the Broad Institute MsigDb [12]. These gene sets include five categories: positional gene sets, curated gene sets (chemical and genetic perturbations, and canonical pathways), motif gene sets (microRNA targets, and transcription factor targets), computational gene sets (cancer modules, and cancer gene neighborhoods), and GO gene sets (GO cellular components, GO biological process, and GO molecular function). Strict Bonferroni correction criterion was used to select gene sets that are enriched in neuroblastoma.
Data deposition
The genotypic and phenotypic information from this study is deposited in dbGAP (www.ncbi.nlm.gov/gap) under accession number phs000124.v2.p1.
Ethics statement
The Children's Hospital of Philadelphia Institutional Review Board approved this study.
Supporting Information
Zdroje
1. MarisJM 2010 Recent advances in neuroblastoma. N Engl J Med 362 2202 2211
2. BrodeurGMPritchardJBertholdFCarlsenNLCastelV 1993 Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol 11 1466 1477
3. AmbrosPFAmbrosIMBrodeurGMHaberMKhanJ 2009 International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br J Cancer 100 1471 1482
4. MarisJMMosseYPBradfieldJPHouCMonniS 2008 Chromosome 6p22 Locus Associated with Clinically Aggressive Neuroblastoma. N Engl J Med 358 2585 2593
5. CapassoMDevotoMHouCAsgharzadehSGlessnerJT 2009 Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet 41 718 723
6. WangKDiskinSJZhangHAttiyehEFWinterC 2011 Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature 469 216 220
7. DiskinSJHouCGlessnerJTAttiyehEFLaudenslagerM 2009 Copy number variation at 1q21.1 associated with neuroblastoma. Nature 459 987 991
8. GoemanJJvan de GeerSAde KortFvan HouwelingenHC 2004 A global test for groups of genes: testing association with a clinical outcome. Bioinformatics 20 93 99
9. HinrichsASKarolchikDBaertschRBarberGPBejeranoG 2006 The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34 D590 598
10. PurcellSNealeBTodd-BrownKThomasLFerreiraMA 2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
11. NewtonMAQuintanaFADen BoonJASenguptaSAhquistP 2007 Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Ann Appl Stat 1 85 106
12. SubramanianAKuehnHGouldJTamayoPMesirovJP 2007 GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23 3251 3253
13. Gene Ontology Consortium 2008 The Gene Ontology project in 2008. Nucleic Acids Res 36 D440 4
14. BreslowNMcCannB 1971 Statistical estimation of prognosis for children with neuroblastoma. Cancer Res 31 2098 2103
15. ShimadaHAmbrosIMDehnerLPHataJJoshiVV 1999 The International Neuroblastoma Pathology Classification (the Shimada system). Cancer 86 364 372
16. LookATHayesFAShusterJJDouglassECCastleberryRP 1991 Clinical relevance of tumor cell ploidy and N-myc gene amplification in childhood neuroblastoma: a Pediatric Oncology Group study. J Clin Oncol 9 581 591
17. BrodeurGMSeegerRCSchwabMVarmusHEBishopJM 1984 Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224 1121 1124
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and PhosphodiesterasesČlánek Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



