-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Systematic Detection of Polygenic Regulatory
Evolution
The idea that most morphological adaptations can be attributed to changes in the
cis-regulation of gene expression levels has been gaining
increasing acceptance, despite the fact that only a handful of such cases have
so far been demonstrated. Moreover, because each of these cases involves only
one gene, we lack any understanding of how natural selection may act on
cis-regulation across entire pathways or networks. Here we
apply a genome-wide test for selection on cis-regulation to two
subspecies of the mouse Mus musculus. We find evidence for
lineage-specific selection at over 100 genes involved in diverse processes such
as growth, locomotion, and memory. These gene sets implicate candidate genes
that are supported by both quantitative trait loci and a validated
causality-testing framework, and they predict a number of phenotypic
differences, which we confirm in all four cases tested. Our results suggest that
gene expression adaptation is widespread and that these adaptations can be
highly polygenic, involving cis-regulatory changes at numerous
functionally related genes. These coordinated adaptations may contribute to
divergence in a wide range of morphological, physiological, and behavioral
phenotypes.
Published in the journal: . PLoS Genet 7(3): e32767. doi:10.1371/journal.pgen.1002023
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002023Summary
The idea that most morphological adaptations can be attributed to changes in the
cis-regulation of gene expression levels has been gaining
increasing acceptance, despite the fact that only a handful of such cases have
so far been demonstrated. Moreover, because each of these cases involves only
one gene, we lack any understanding of how natural selection may act on
cis-regulation across entire pathways or networks. Here we
apply a genome-wide test for selection on cis-regulation to two
subspecies of the mouse Mus musculus. We find evidence for
lineage-specific selection at over 100 genes involved in diverse processes such
as growth, locomotion, and memory. These gene sets implicate candidate genes
that are supported by both quantitative trait loci and a validated
causality-testing framework, and they predict a number of phenotypic
differences, which we confirm in all four cases tested. Our results suggest that
gene expression adaptation is widespread and that these adaptations can be
highly polygenic, involving cis-regulatory changes at numerous
functionally related genes. These coordinated adaptations may contribute to
divergence in a wide range of morphological, physiological, and behavioral
phenotypes.Introduction
To what extent the evolution of gene expression cis-regulation drives the evolutionary innovations of life is an important unresolved question. While some contend that changes in cis-regulation are responsible for the majority of morphological adaptations [1], others point out that only a few such cases have been demonstrated [2], [3] (we distinguish here between cis-regulatory changes that have been shown to affect phenotypes, of which there are a moderate number [4], [5], and those that have further been shown to be adaptive, of which there have been far fewer [2], [3]; adaptive changes are those that are subject to positive selection as a result of increasing fitness).
This long-standing paucity of examples of adaptive cis-regulatory divergence was due in large part to the fact that historically it has not been possible to formally demonstrate the presence of cis-regulatory adaptation from genome-wide data [3]. Sequence-based approaches have often been used to scan the genome for accelerated divergence in non-coding regions [6]–[9], but what fraction of these represent positive selection on cis-regulation remains unknown; other possible explanations include changes in local mutation rate or biased gene conversion rate [10], or selection on non-coding RNAs, recombination control elements, DNA replication origins, or any other non-coding feature of genomes (e.g. [6]). Moreover, even when accelerated evolution does reflect cis-regulatory adaptation, the target genes often cannot be identified, since transcriptional enhancers can act on distant genes in many species.
Alternatively, many studies have attempted to detect genes under positive selection from genome-wide gene expression data, but have been unable to demonstrate the presence of positive selection due to the lack of a null model of neutrality [3], [11]. For example, the finding that gene expression divergence among three populations of Fundulus fish species correlates better with the species' environment than with their phylogeny [12] is consistent either with widespread adaptation to the environment, or with a neutral mutation affecting many gene expression levels being shared between two populations by chance; these cannot be distinguished without a null model of neutral change. Similarly, studies that rank genes by their ratio of gene expression divergence between species to diversity within species [13]–[14] can identify promising candidates for follow-up studies, but cannot distinguish between neutral and adaptive evolution without knowing how the expression of a “neutral gene” would evolve [3].
Several studies have succeeded in developing accurate neutral models of gene expression change by quantifying expression divergence when selection is artificially weakened in the lab [15]–[17]. In these studies positive selection on a gene's expression would be indicated by a greater divergence between species than expected from the neutral model; less divergence than expected would reflect negative selection. Although these studies have had the potential to discover positive selection, they have only uncovered negative selection—i.e. all genes have shown less divergence between species than expected under neutrality. However since these studies can only measure “average” selection pressures (much like the dN/dS metric for coding regions), genes even with fairly frequent episodes of positive selection on expression would go undetected if they are most often subject to negative selection [3]. Therefore the lack of any positive selection on gene expression identified in these studies is not evidence against the existence of such positive selection.
This landscape has changed with the recent publication of two studies of selection on genome-scale gene expression data in Saccharomyces yeast [3], . In one of these [18], we used the directionality of gene expression quantitative trait loci (eQTL; reviewed in [20]) to demonstrate that at least 242 gene expression levels (and likely many more) have been subject to lineage-specific selection (i.e. different selective regimes between two lineages) since the divergence of two strains of S. cerevisiae, and then employed population-genetic analyses to show that most of these represent positive selection, as opposed to relaxed negative selection. Although this work expanded the number of known cases of gene expression adaptation (across all species) by over 10-fold, it revealed little insight into the higher-level traits being selected. In another important recent study, Bullard et al. [19] examined the allele-specific expression (ASE) levels of gene sets (e.g. pathways, co-expressed gene clusters, etc.) in a hybrid between S. cerevisiae and another yeast, S. bayanus. ASE implies the presence of a cis-acting polymorphism affecting expression, and consistent directionality of ASE within a gene set implies lineage-specific selection (see below for further explanation). This method has great promise for identifying the biological processes affected by gene expression adaptation, though it remains unknown if the gene sets implicated in this work have been subject to positive (as opposed to relaxed negative) selection [19]. Interestingly, parallel analysis of the genomic sequences of these same gene sets revealed no cases of either promoters or protein-coding regions under positive selection [19].
Here we apply a gene set-based test of selection on gene expression to M. musculus. Although mouse is a heavily studied model organism, both in the lab and in the wild, no cases of gene expression adaptation have been demonstrated in this species (one example, the Agouti gene, has been found in Peromyscus deer mice [21]). Our results show that both “traditional” eQTL mapping in an F2 population as well ASE analysis in an F1 hybrid can be used to detect lineage-specific selection on gene sets, and that data from additional strains can be used to polarize the changes and infer the probable action of positive selection. Moreover, we expand the known extent of gene expression adaptation in M. musculus from zero genes to over 100, and find that a great deal of such adaptation may occur in parallel on many genes of small effect, in contrast to all previously known cases of gene expression adaptation [1], [2] aside from our work in yeast [18]. Finally, our results suggest that gene expression adaptations can affect behavioral and physiological phenotypes, in addition to their more well-established role in morphological evolution [1].
Results
A test of selection on cis-regulation
The test of lineage-specific selection we use is based upon an idea first formalized by Orr [22] in an elegant test of selection on quantitative traits: under neutrality, QTLs for any given trait are expected to be unbiased with respect to their directionality. In other words, given two parents (A and B) of a genetic cross, A alleles at any QTL would be expected to be equally likely as B alleles to increase the trait value. If a significant bias is seen—e.g., among 20 QTLs for a trait, the A allele increases the trait value at all of them—neutrality may be rejected in favor of lineage-specific selection (in the absence of ascertainment bias [see Text S1]). At present, no gene expression levels have been mapped to a sufficient number of eQTLs to reject neutrality for any single gene. However, if the expression levels of an entire group of genes is treated as a single trait, and each eQTL used in the test is independent (i.e. caused by a distinct polymorphism), then lineage-specific selection can be detected as a bias in the directionality of eQTLs for the gene set being tested [3], [19] (This approach will have the greatest power for gene sets containing genes that predominantly have the same direction of effect on a trait under selection; for gene sets with a significant fraction of genes that act in opposition, selection in one direction could result in upregulation of some, and downregulation of others.).
The independence of eQTLs for different genes is critical for this test, since a single eQTL that affected many genes could lead to a strong bias in the directionality of effect even in the absence of lineage-specific selection (Figure 1, strain A versus B). To ensure that each eQTL is independent, we considered only local eQTLs—that is, eQTLs located at genetic markers that are close in the genome to the gene whose expression they control. These local eQTLs have been shown to be primarily cis-acting [23] (so we refer to these as cis-eQTL for brevity), though we note that our test of selection is equally valid for local trans-acting eQTLs. Since a single cis-eQTL could conceivably control multiple nearby genes, and thus violate the requirement for independence, we also discard genes that are located close to others in the same gene set (see Methods).
Fig. 1. Illustration of our test for lineage-specific selection. 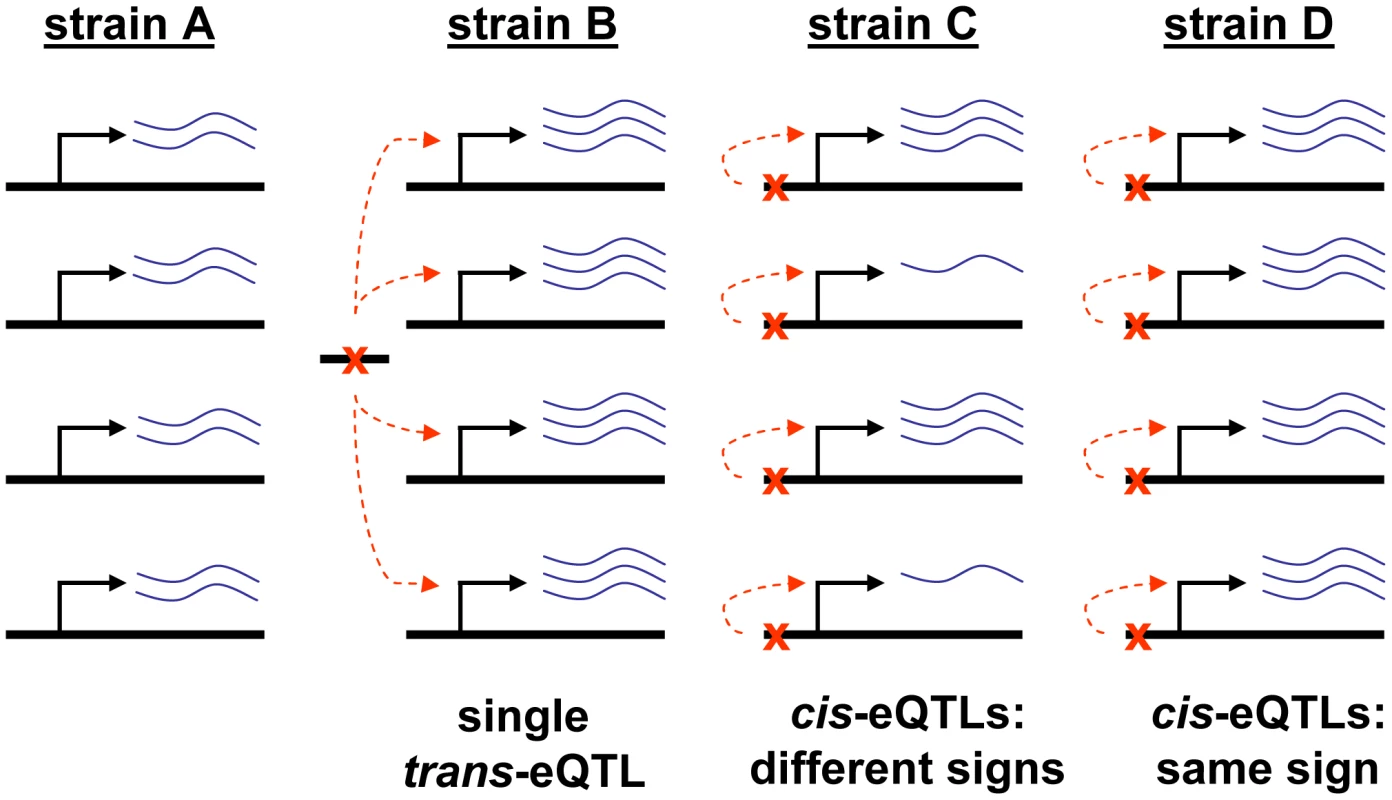
At any eQTL, either the allele from parent A up-regulates expression (and thus parent B's down-regulates), or the allele from parent A down-regulates expression (and thus parent B's up-regulates). In our test we include an equal number of each type (arbitrarily termed “+” and “–”), so that any gene set that is not under lineage-specific selection should have close to the same number of genes in each eQTL direction (Figure 1, strain A versus C). This null expectation requires no assumptions about gene sets or eQTLs or the complex biological networks involved, but follows simply from the fact that we constrain the total number of + and – eQTLs to be equal (relaxing this constraint to allow different numbers of + and – eQTLs is straightforward, and requires only adjusting the null expectation; e.g. if we adjust our cutoffs so that 60% of all eQTLs are +, then any random or non-lineage-specific-selected gene set is expected to have ∼60% + eQTLs). A hypergeometric p-value, testing whether the observed data deviate from this expectation by having an excess of either + or – eQTLs (Figure 1, strain A versus D), constitutes the test. Although this method will have greater power for gene sets with many cis-eQTLs, any variation in the total number of cis-eQTLs per gene set (whether due to real biological differences, or experimental design, e.g. gene sets not well-represented on the expression array) will not lead to false-positive results, since these will affect + and – eQTLs equally. Further, the test is sensitive to both positive selection and relaxed negative selection acting on a gene set, as long as that selection is present in only one of the two lineages; thus it is a test of lineage-specific selection, although positive selection can be inferred with additional data (see below). In this sense, it is similar to the McDonald-Kreitman test [24], which also cannot distinguish between positive and relaxed negative selection [25]. However unlike the McDonald-Kreitman test, as well as nearly all other previous tests of selection (on both gene expression levels and DNA/protein sequences), this is not dependent on any assumptions about either demographic histories or a subset of neutral sites (see Text S1).
Inferring selection in mouse
We applied our test of selection to eQTL data from a cross between two diverged inbred mouse strains, C57BL/6J (B6) and CAST/EiJ (CAST). B6, like most commonly used lab strains, is a mosaic of several lineages [26], but primarily Mus musculus domesticus. CAST represents Mus musculus castaneus, a subspecies thought to have diverged from the primary B6 progenitor strains ∼500,000 years ago [27]. This divergence, as well as recent selection during inbreeding in the lab, provides ample opportunity for adaptive changes to have accumulated in each lineage.
To map cis-eQTLs, we produced 442 F2 animals, either with B6 as the F0 paternal strain (referred to here as CxB F2 animals) or maternal strain (referred to as BxC F2 animals). Each mouse was genotyped at 1,438 informative genetic markers, and genome-wide gene expression was measured in adult brain, liver, and skeletal muscle (see Methods). Cis-eQTLs were found by linear regression of gene expression levels on genotypes separately in each of four cohorts: CxB females, CxB males, BxC females, and BxC males. A total of 5,000 cis-eQTLs in each cohort—the strongest 2,500 in each direction (corresponding to a false discovery rate [FDR] <10% in each cohort)—were retained for analysis. Using the same number of + and – eQTLs allows us to apply our simple yet robust null expectation of neutrality to any gene set: regardless of what complex biological networks and population histories underlie the eQTLs, any gene set not subject to lineage-specific selection (including random gene sets) will show an approximately equal number of + and – eQTLs, following the binomial distribution. Throughout this work we report gene sets significant at either a high-confidence (<2% FDR) or medium-confidence (<15% FDR) cutoff, with FDRs estimated by testing randomly generated gene sets matched in size to the real ones (see Methods).
We began by testing gene sets from the Gene Ontology (GO) Consortium [28], since these have been shown to be useful in a wide range of applications (while any particular gene's GO classification and expression data may be imperfect, the sheer number of genes and expression measurements being used make this a potentially powerful test; any inaccuracies in the gene set assignments may lead to false negatives, but are unlikely to result in false positives). Applying the hypergeometric test to 531 GO gene sets (each with at least 50 members) separately in each tissue, we found one high-confidence set (FDR = 1.5%, meaning that there is approximately a 1.5% probability that this enrichment is due to chance, given the number of gene sets tested, and the overlap in content between gene sets): genes in the “mitochondria” set were biased towards increased expression from B6 cis-eQTL alleles (“B6-upregulation”) in liver (Table 1; see Table S1 for gene lists). These results were consistent across all four cohorts (Figure 2a), not only at the gene-set level, but also for specific genes within those sets (see Text S1), underscoring their robustness. SNPs that could disrupt microarray probe hybridization are unlikely to explain the results, since these did not show any enrichment in the B6-upregulated mitochondria-related genes (see Text S1). The number of genes affected by selection can be estimated as the difference between the numbers of cis-eQTLs in each direction (see Text S1); in mitochondria, this is estimated separately in each cohort as 32-35 genes in females and 47-48 genes in males (Figure 2a, green numbers). We note this will be conservative if any of the CAST-upregulated cis-eQTLs were fixed by positive selection as well. No additional gene sets were observed with medium confidence.
Fig. 2. Results of the selection test for two gene sets. 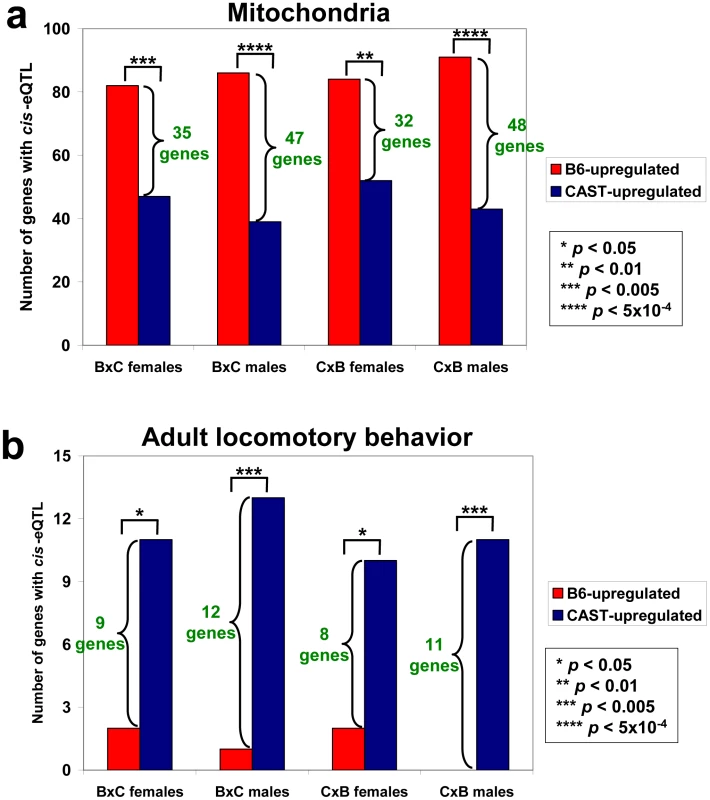
To increase our statistical power, we combined results across tissues, since many cis-eQTLs in our data were not tissue-specific. Seven additional gene sets were found: one at high-confidence and six at medium-confidence (Table 1; see Table S2 for results from all 531 gene sets). Two of the seven sets were related to mitochondria at different levels of the GO hierarchy (“mitochondrial inner membrane” and “intracellular organelle”), while the other five represented a diverse collection of functions. As an example, locomotory genes—which are biased towards CAST-upregulation in all three tissues—are shown in Figure 2b. Similar to the mitochondria gene set, the specific genes implicated in each cohort overlapped extensively (see Text S1). In sum, these results suggest that lineage-specific selection involving these subspecies can be inferred for several functional categories.
We also applied our method to other types of gene sets. Testing 41 modules of genes co-expressed in each F2 population (see Methods), we did not find any significant enrichments for biased directionality of cis-eQTLs. However testing 75 pathways from the KEGG database [29], we found one at medium confidence (FDR = 4.5%): the JAK/STAT pathway was biased towards cis-upregulation in CAST brain (Table 1).
Inferring selection via mRNA sequencing
To complement the microarray-based approach described above, we turned to sequencing RNA isolated from F1 mice to directly identify allele-specific expression (ASE). While this approach does not offer the richness in terms of understanding genetically regulated networks and their interactions that can be achieved in a large F2 cross, it does address two drawbacks of the microarray approach described above: 1) our microarrays cannot provide direct evidence of cis-regulation (since local eQTLs can occasionally be trans-acting [23]), so we cannot be confident that our results truly reflect selection solely on cis-acting elements; and 2) there is considerable time and expense associated with rearing, genotyping, and expression profiling of hundreds of F2 mice.
We and others have shown that high-throughput mRNA sequencing (RNA-seq) in F1 hybrid mice is an effective approach to studying ASE [30]–[32]. mRNA levels can be accurately estimated by simply counting the density of reads from each transcript. Since heterozygous SNPs are present at a 1∶1 ratio in the genome, any significant deviation from this ratio in the number of sequence reads that can be mapped to each individual allele (as a result of containing a heterozygous SNP) indicates ASE. When the allele-specificity associates in reciprocal crosses with SNP genotype—as opposed to parent-of-origin, as seen for imprinted loci [30]–[31]—this implies the presence of a cis-acting eQTL. These cis-eQTL target genes can then be used as input for our selection test, in exactly the same fashion as those found using microarrays in an F2 population.
We searched for ASE in a set of ∼78 million sequence reads from F1 hybrid BxC and CxB embryos we generated previously [30]. Because this is not only a different technology, but also a different developmental stage (embryonic day 9.5) and tissue (whole embryos), we were encouraged to see several of our strongest hits replicate. For example, mitochondrial genes show a bias towards higher expression of B6 alleles, whereas locomotory-related genes show the opposite (Figure 3a). Gene sets that were biased in adults but not in F1 embryos might be tissue and/or stage-specific, or may be missing due to lower power of our RNA-seq data for weakly expressed genes (this is not an inherent limitation of RNA-seq, since power is limited only by the number of reads). In addition, genes lacking any B6/CAST sequence polymorphisms are not assayable by allele-specific RNA-seq.
Fig. 3. Results of the selection test in RNA-seq data. 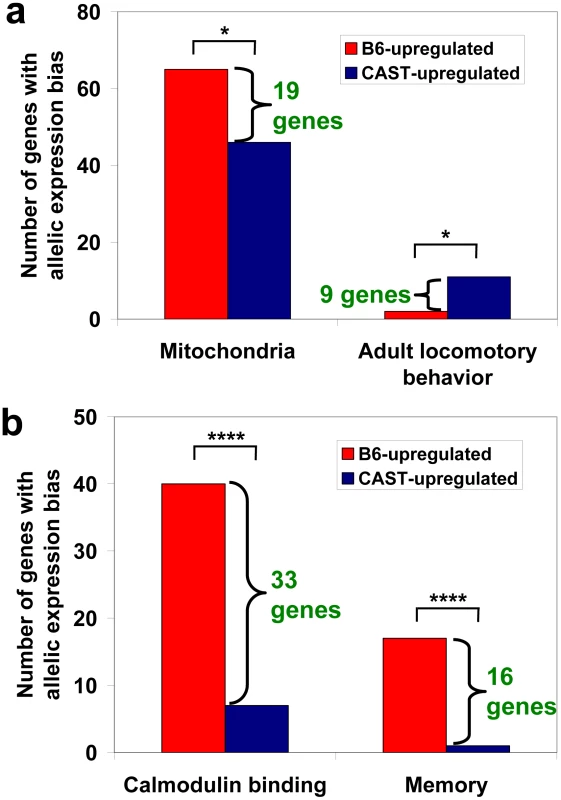
In addition to replicating some hits from adult mice, the F1 embryo data revealed new significant gene sets as well. Two gene sets reached high-confidence: “calmodulin binding” and “memory” (Table 1 and Figure 3b), both showing a bias towards B6-upregulation. Although unannotated SNPs overlapping RNA-seq reads can cause a marginal alignment bias resulting in an apparent up-regulation of the B6 reference genome alleles, our analysis indicates this is unlikely to underlie the significance of these gene sets (see Text S1). Consistent with previous work in yeast [19], we conclude that RNA-seq is a cost-effective alternative for measuring selection on cis-regulation, particularly between lineages with a high density of exonic sequence differences.
Connecting cis-regulatory selection to phenotypes
An important question is whether the lineage-specific selection we detected has had any detectable impact on organismal phenotypes. Examination of the gene sets in Table 1 reveals that specific predictions can be made for the gene sets belonging to the GO “biological process” and “cellular component” ontologies (Table 2). For example, cis-eQTLs leading to higher expression of growth or locomotory genes may be (at least naively) expected to increase growth or locomotion, since these gene annotations were typically identified by observing a reduction of growth or locomotion in a gene knockout/knockdown model; genes leading to increased growth or locomotion when inactivated are far less common (for example, among genes annotated as growth regulators [28], 40 have a mutant phenotype of decreased body size, whereas only two are associated with increased body size [33]). These effects could either be strong, like all previous examples of adaptive cis-regulatory adaptation in metazoans [1], [2]; or subtle, considering that many loci are being selected in parallel and thus may only exert major phenotypic effects in aggregate. We were unable to make any phenotypic predictions for the GO molecular function terms (“calmodulin binding”, “G-protein coupled receptor activity”, “receptor activity”, or “enzyme inhibitor activity”), or the JAK-STAT pathway.
Tab. 2. Phenotype predictions and evidence. 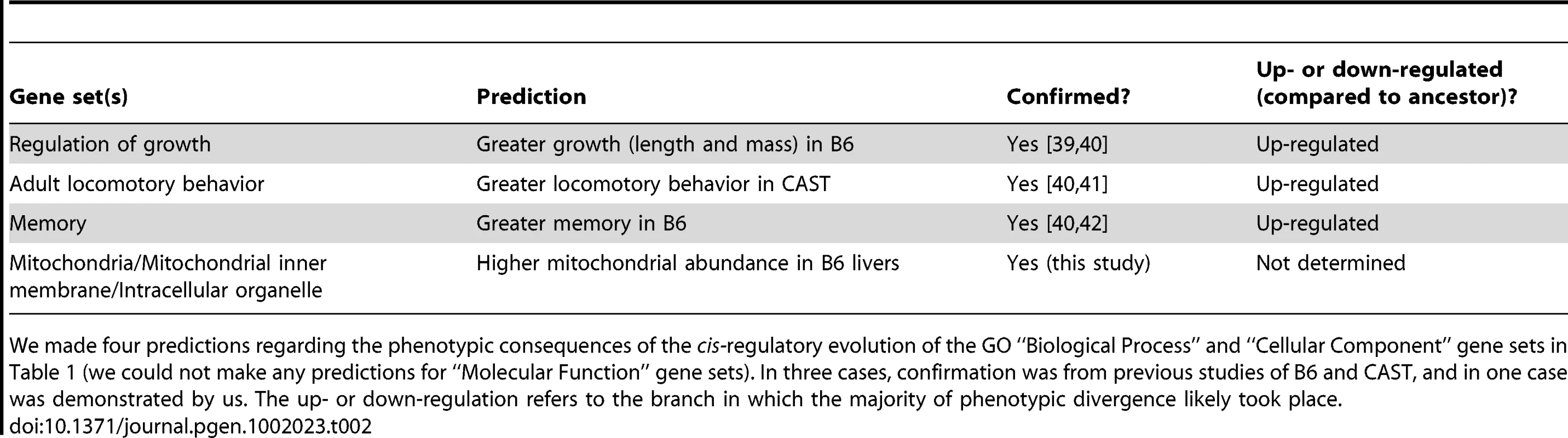
If the loci we identified have major phenotypic effects, they should be detectable by QTL mapping in our F2 mice. One phenotype we predicted to be affected was measured for every F2 individual in our cross: naso-anal length, which approximately reflects the sum of growth over the lifetime of the mice. In females, we found two significant QTLs for length, on chromosomes 2 and 15, while in males the strongest QTL was on chromosome 5 (Figure 4, red lines). In all three cases, the B6 alleles were associated with greater length, as expected since B6 alleles tend to increase expression of growth-related genes (whose knockout/knockdown phenotype is typically a reduction in growth). Strikingly, the strongest QTL from each gender overlapped almost perfectly with two of the strongest (genotype versus expression level r2>0.5) cis-eQTLs in the growth-related gene set (Figure 4, blue lines), and the weaker female length QTL coincided with a weaker (r2>0.2) but still highly significant growth-related gene cis-eQTL (Figure 4a, green line). This overlap is unlikely to occur by chance, considering that only ∼0.5% of cis-eQTLs are as close to the length QTLs as each of these are (probability of overlap by chance, p<0.001; see Methods). The three genes are Dcaf13 (also known as WDSOF1), an rRNA processing factor; Ept1, a CDP-alcohol phosphatidyltransferase (orthologous to human SELI); and Sp3, a transcription factor. All three are well-conserved, and have been implicated in positive regulation of growth either by mouse knockout [34], or RNAi experiments involving their orthologs in Caenorhabditis elegans [35]. This highly significant overlap suggests that these genes may be responsible for the length QTLs.
Fig. 4. Overlap between morphological and expression QTLs. 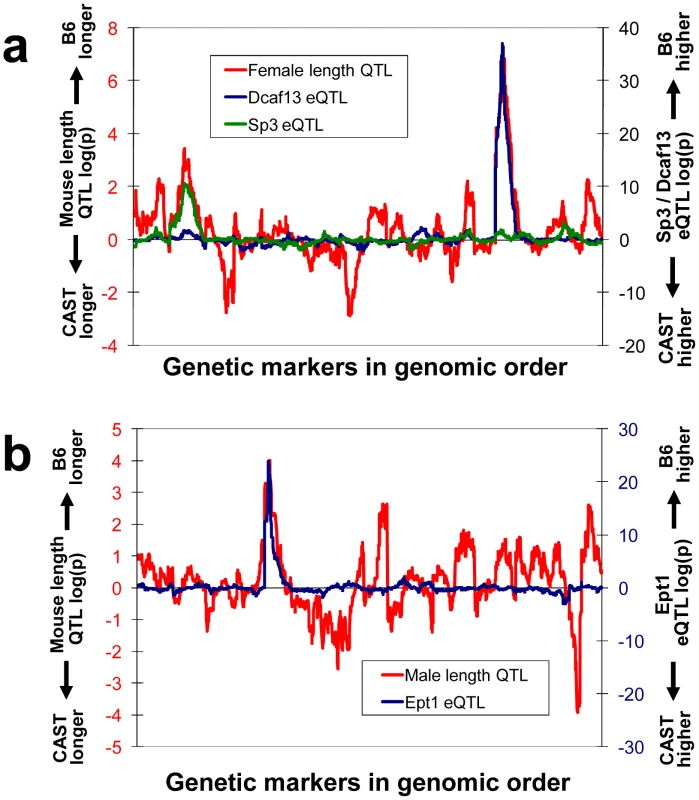
To further test the hypothesis that the cis-eQTLs for these three genes affect mouse length, we applied a statistical approach for inferring causality of eQTLs for other traits [36]–[37] that has been extensively tested and validated using transgenic mice [38]. For all three genes, causality for length was strongly (p<0.001) supported in at least one tissue. This provides further support for a role of these eQTLs in the length phenotype.
An alternative method to assess the phenotypic importance of these gene sets is to compare the predictions to phenotypes of B6 and CAST mice. Although QTL mapping cannot be performed with only two strains (typical mapping populations consist of hundreds of F2 individuals or recombinant inbred lines)—and thus causal loci cannot be implicated—concordance of predictions with observed phenotypes can at least serve as evidence that the selection on cis-regulation of these gene sets is phenotypically relevant. To this end, we searched the literature for studies where phenotypes we predicted to be affected by selection (Table 2) were measured in B6 and CAST. For three of our four predictions, we found multiple studies testing the relevant phenotypes. From the growth regulator gene set, we predicted larger size of B6 mice (measured by length, as above, or by total body mass), and indeed they are known to have nearly twice the mass of CAST mice, from an early age through adulthood [39], [40]. From the adult locomotory-related gene set showing CAST-upregulation (found in both the microarray and RNA-seq data, Figure 1 strain B, and Figure 2a), we predicted higher locomotor activity in CAST, which has indeed been observed [40], [41]. In fact, one study [41] found that daytime activity of CAST was over six times higher than that of B6. The B6-upregulation of the memory-related gene set (Figure 2b) predicted increased memory in B6 (since knockout of most memory-related genes results in reduced, not increased, memory). In two studies employing the Morris Water Maze (MWM) to measure learning and memory, B6 significantly outperformed CAST [40], [42]. In fact, CAST showed no capacity at all for memory in this context (see Text S1). In sum, all three of our predictions that have been addressed in previous publications were confirmed by multiple independent studies. We did not find any studies contradicting these predictions.
Our fourth prediction—that mitochondria would be more abundant in B6, as a result of the B6-upregulation of many mitochondrial genes (most notably genes related to the inner membrane, but also mitochondrial small ribosomal subunits [combined-tissue p = 4.5×10−8], among others) observed in both the microarray and RNA-seq data—has not, to our knowledge, been tested by previous studies. Therefore we isolated nuclear and mitochondrial genomic DNA from livers (the tissue with the strongest B6-upregulation of mitochondrial genes) of B6 and CAST adult mice, and measured the ratio of their mitochondrial to nuclear genome copy number by qPCR (see Methods). Consistent with our prediction, we found a small but highly significant (p<0.001) difference between B6 and CAST, with B6 showing a 12.9% increase in abundance. Therefore, all four of our predictions have been confirmed—three retrospectively and one prospectively—underscoring the ability of our selection test to predict phenotypic differences, and suggesting that these differences may have been shaped by lineage-specific selection on cis-regulation (though we note that other traits could also have been affected by, or been the primary targets of, the lineage-specific selection in these gene sets).
Inferring positive selection by polarizing the changes
To better understand the selection that has acted on these phenotypes, we sought to determine on which lineage the majority of changes in each trait had occurred. This can be achieved by including an outgroup species in the analysis: for example, if a trait value in B6 is much further from the outgroup than is the CAST trait value, then the most parsimonious explanation is that the majority of divergence occurred on the B6 lineage. As with all parsimony-based methods, this indicates the most likely evolutionary scenario (i.e. that requiring the fewest changes), but cannot formally rule out any less parsimonious explanation.
To perform this analysis we searched for measurements of the four traits in Table 2 from additional mouse strains. Mus spretus (SPRET) is an ideal outgroup, being the species most closely related to Mus musculus. We found published measurements from SPRET for two of the traits, growth and memory. For growth, the adult mass of SPRET was found to be statistically indistinguishable from CAST [39]—and about half of that of B6—indicating that the change in growth likely took place along the B6 lineage. Similarly for memory, SPRET showed no evidence of recall in the MWM [42], similar to CAST but in stark contrast to B6—again implicating the B6 lineage as the probable source of divergence. In fact, B6 showed significantly greater recall than all of the 12 other strains tested [42].
Although locomotory behavior has not been measured in an outgroup (to our knowledge), it was measured in nine strains in addition to B6 and CAST [41], including seven wild-derived strains that are more closely related to CAST than is B6 or other lab strains [43]. Since CAST had over twice the daytime locomotory activity of any other strain tested [41]—including the closely related wild strains—the majority of divergence can be inferred to have likely taken place on the CAST lineage, after its divergence from the other wild strains (in this case, B6 is the outgroup). The much lower daytime activity level of B6 was similar to most of the wild strains, as well as another lab strain [43].
In sum, the phenotypic changes can be polarized for three of the traits. These results rest on the logic of parsimony: that a phenotypic change in one lineage is more likely than independent changes in the same trait—of the same direction and magnitude—in two lineages. Under the assumption that the phenotypic divergence was driven by (and thus occurred on the same branch as) the expression divergence, all three cases can be inferred to have likely been caused by cis-upregulation of the relevant gene sets.
As mentioned above, our test of lineage-specific selection cannot by itself distinguish between positive selection and relaxed negative selection (analogous to the McDonald-Kreitman test [24], [25]). However recent evidence from saturation mutagenesis studies showing that the vast majority of random cis-regulatory mutations cause downregulation (see Text S1) suggests that relaxed negative selection would likewise be biased towards downregulation. If this is indeed the case for the gene sets we have implicated, then relaxed negative selection is unlikely to explain the upregulation of these three traits/gene sets, leading to the conclusion that their divergence was most likely due to the action of positive selection for upregulation. However given the qualitative nature of this argument, we cannot yet quantify the precise probability that positive selection has been acting upon the cis-regulation of these gene sets.
Discussion
Using a systematic genome-scale approach to inferring lineage-specific selection acting on cis-regulation, we found that over 100 genes belonging to several gene sets have undergone lineage-specific selection in mouse, which may have impacted diverse morphological and behavioral phenotypes. This work reports the first cases of adaptive cis-regulatory evolution in M. musculus, and expands the classes of traits (in any species) known to be affected by gene expression adaptation, which previously did not include any behavioral phenotypes. Methodologically, we augment previous work [19] by showing that adding information from an outgroup can suggest the likely action of positive selection (as opposed to relaxed negative selection) when that selection was for cis-acting upregulation. Two interesting questions for future work are how much of this selection occurred since the introduction of these strains to the lab, and for selection that occurred on the wild B6 ancestors, how much occurred in Mus musculus domesticus (the primary ancestor of B6 [26]) as opposed to Mus musculus musculus. Interestingly, wild M. m. domesticus tend to be larger than wild M. m. castaneus when reared in a common laboratory environment (C. Pfeifle, personal communication), suggesting that this adaptation was likely to have occurred in the wild. Another question raised by these findings is what are the relevant “units of selection” [44] for these polygenic adaptations; though regardless of the answer, our conclusions regarding the extent of selection on cis-regulation will not be affected.
Because the RNA-seq version of this approach can be applied rapidly and inexpensively to hybrids between any two diverged lineages (including outbred lineages), we expect it will find use in a wide range of taxa. In fact, it can be applied to any ASE data from a hybrid between diverged lineages. Published ASE data sets from a variety of species (e.g. [45], [46]) can now be similarly re-analyzed for cis-regulatory selection. This approach can also be applied to any of the numerous published eQTL data sets involving crosses between diverged parental lines.
Our approach is quite different from all previous studies of metazoan cis-regulatory adaptation [1]–[4], which have identified single genes with extremely strong effects on phenotypes such as pigmentation (e.g. [21], [47], [48]) or skeletal structure (e.g. [49]). Our results reveal several important insights that could not have been found at this single-gene level. For example, the only previously known case of pathway-level gene expression adaptation was from our work on the ergosterol biosynthesis pathway in S. cerevisiae, where six genes clustered in the pathway have undergone selection for down-regulation [18]. Our present results extend this considerably, demonstrating that polygenic cis-regulatory adaptation can operate in parallel on dozens of genes within a single functional group or pathway, and that this has occurred in multiple gene sets during recent mouse evolution. Although each gene under such coordinate selection may be expected to have a less extreme phenotypic effect than those previously reported [1], [2], [21], [47]–[49], the sum of their effects could be quite strong. One important question that can now start to be addressed is how often cis-regulatory adaptation proceeds via dramatic changes in single genes, as opposed to more subtle changes distributed across an entire gene set [3]. Much of the answer may ultimately depend on factors such as the strength/duration of selection (with intense/short-term selection pressure likely favoring extreme single-locus changes) and the genetic architecture of the trait in question.
A second open question is how often cis-regulatory adaptation occurs by upregulation versus downregulation of genes; our results suggest that the majority of the adaptation we discovered was due to upregulation, in contrast to most previous (single-locus) studies, which have predominantly identified cases of trait loss via downregulation [2]. Interestingly, we previously observed a preponderance of upregulation in a genome-wide study of gene expression adaptation in S. cerevisiae [18], suggesting that this pattern may be widespread. Again, which of these is more common in a particular species may depend on the nature of the selective pressure and the underlying genetic architecture.
Third, it has been proposed that gene expression adaptation may be responsible for most morphological adaptations in part because it offers a solution to the issue of pleiotropy. For a gene expressed in many tissues or stages of development, an amino acid change (in a constitutive exon) will affect the protein produced in all of these different contexts. Even if this change is adaptive in one or two of them, it has been argued that it would be highly unlikely to be advantageous in all of them [1]. In contrast, the modular nature of cis-regulation allows for a change in expression in just one tissue or stage, without affecting any other; thus pleiotropic constraints should not be as severe, and adaptation should be able to proceed [1]. Predictions from this are that genes expressed more broadly will be more likely to adapt via cis-regulation, and that these adaptations will only affect a small part of the genes' expression patterns. Two recent studies attempted to test this idea. In one of these [50], genes near noncoding elements with accelerated evolution in the human lineage were proposed to have undergone human-specific selection on cis-regulation (though the authors acknowledged that such acceleration need not indicate positive selection); however no enrichment was found for these genes to be expressed in more tissues than average. In the other [51], genes were classified as either “morphogenes” or “physiogenes” based on their mouse knockout phenotypes; morphogenes (which tend to be expressed in fewer tissues) had higher dN/dS (an indicator of selection on protein-coding regions), while physiogenes had a higher magnitude of expression change between human and mouse, consistent with the prediction of greater adaptive expression change in broadly expressed genes. However this study did not distinguish between adaptive versus non-adaptive change, or cis versus trans regulation, or tissue-specific versus non-specific expression changes, so the relevance to theories of tissue-specific adaptive cis-regulatory evolution is not clear. Our results suggest that although most of the genes in our most significant gene sets are broadly expressed (not shown), their expression in all three tissues was affected by the recent selection on cis-regulation we detected (Table S2; all gene sets from Table 1 were significant in all three tissues, except for the JAK/STAT pathway); thus these adaptations were not tissue-specific, so do not support pleiotropy-based arguments for the expected prevalence of tissue-specific gene expression adaptation (we note that while the adaptations did not result in tissue-specific expression changes, the selection may have acted to change expression in just one tissue, with the rest changing as a side-effect). Of course, since we have only examined three tissues in two mouse strains, much more work is required to determine how general this conclusion is.
Finally, because of its genome-scale perspective, our approach may eventually help to address many other fundamental questions that cannot be addressed by single-locus studies [3], such as what fraction of gene expression divergence is adaptive, and what fraction of evolutionary adaptation occurs at the level of cis-regulation.
Materials and Methods
Data production
Ethics statement: All mouse work was conducted according to Institutional Animal Care and Use Committee regulations.
C57BL/6J (B6) mice were intercrossed with M. m. castaneus (CAST/EiJ) mice to generate 442 F2 progeny (276 females, 166 males). All mice were maintained on a 12 h light–12 h dark cycle and fed ad libitum. Mice were fed Purina Chow until 10 wk of age, and then fed western diet (Teklad 88137, Harlan Teklad) for the subsequent 8 wk. Mice were fasted overnight before they were killed. Their tissues were collected, flash frozen in liquid nitrogen, and stored in −80°C prior to RNA isolation.
RNA preparation and array hybridizations were performed at Rosetta Inpharmatics. The custom ink-jet microarrays used were manufactured by Agilent Technologies. The array used consisted of 2,186 control probes and 23,574 non-control oligonucleotides extracted from mouse Unigene clusters and combined with RefSeq sequences and RIKEN full-length cDNA clones.
Mouse tissues were homogenized, and total RNA extracted using Trizol reagent (Invitrogen) according to manufacturer's protocol. Three micrograms of total RNA was reverse transcribed and labeled with either Cy3 or Cy5 fluorochrome. Labeled complementary RNA (cRNA) from each F2 animal was hybridized against a cross-specific pool of labeled cRNAs constructed from equal aliquots of RNA from 150 F2 animals and parental mouse strains for each of the three tissues. The hybridizations were performed to single arrays (individuals F2 samples labeled with Cy5 and reference pools labeled with Cy3 fluorochromes) for 24 h in a hybridization chamber, washed, and scanned using a confocal laser scanner. Arrays were quantified on the basis of spot intensity relative to background, adjusted for experimental variation between arrays using average intensity over multiple channels, and fitted to a previously described error model to determine significance (type I error) [52]. All microarray data are available at NCBI GEO (GSE16227).
Genomic DNA was isolated from tail sections using standard methods and genotyping was performed by Affymetrix (Santa Clara, CA) using the Affymetrix GeneChip Mouse Mapping 5K Panel.
The RNA-seq data were described previously [30]. All data are available at the NCBI SRA (accession SRA008621.10).
Data analysis
eQTL scans were performed by linear regression of expression log ratios against genotypes (coded as 0, 1, and 2), separately in each tissue for each of the four cohorts (CxB females, CxB males, BxC females, and BxC males). eQTL were designated as “local” (and likely cis-acting) if the regression between the expression level of a gene and a genetic marker within 1 megabase of the transcription start site was significant (where significance was defined as the cutoff resulting in 2,500 eQTLs in each direction; see below). Testing for dominance (comparing the average heterozygote value to the average of the two average homozygote values) revealed evidence for non-additivity at only a small fraction of local eQTLs (as expected for cis-eQTLs, which typically act additively), so dominance effects were not included in our eQTL mapping.
We implemented the following strategy to isolate local eQTL effects in the presence of unlinked marker correlations. First the strongest local eQTL was identified, and expression of the target gene was then corrected for its effects by taking the residuals of expression when regressed against the eQTL genotype. The corrected expression level was then subjected to a whole-genome eQTL scan to identify the strongest trans-eQTL. Once this trans-eQTL was identified, its effects were regressed out of the original expression levels for the gene. These trans-corrected expression levels were then regressed against all local genetic markers once again, to identify the strength and direction of effect for the cis-eQTL. This process allows us to achieve a more accurate estimate of local eQTL effect sizes, even in the presence of unlinked trans-eQTLs or correlations between unlinked genetic markers (we note that removing trans effects is not necessary for our test, though we have found it to improve our ability to estimate cis effects). More generally, our focus on local eQTLs allows us to isolate the effect of the local polymorphism(s) on gene expression, regardless of other effects (e.g. environmental effects, trans-eQTL not captured in our regression approach, epistatic interactions, feedback, etc.); of course such effects are widespread, but they will only weaken the correlation between a genetic marker's genotype and a nearby gene expression level, potentially causing us to miss some local eQTLs, but not resulting in false-positive results.
A total of 5,000 genes with the strongest cis-eQTLs (2,500 in each direction) in each tissue/cohort combination were analyzed. The decision to use an equal number of eQTLs in each direction does not reflect any biological aspects or assumptions, but instead is merely an arbitrary choice. Whether the total “true” numbers of cis-eQTLs in each direction are actually equal is not addressed here (nor is it directly relevant for interpreting our test's results). Altering the proportion of eQTLs in each direction by up to 10% (a 60/40 ratio) in either direction did not have any impact on our results (i.e. the gene sets in Table 1 were not affected, although FDRs were changed slightly).
FDRs for each tissue/cohort combination were estimated by randomization. We first shuffled genotype labels so that one individual's entire set of genotypes was paired with another individual's expression levels. Then the entire eQTL detection procedure was carried out, and the number of cis-eQTLs above the cutoffs associated with the top 5,000 eQTLs in the real data were counted. Randomizations were repeated at least 1,000 times. The estimated FDR equals the average number of significant eQTLs in the randomized data divided by 5,000 (the number in the real data). This procedure yielded a maximum FDR of 9.7% in the smaller cohorts (BxC), and an FDR of <2% in the larger (CxB) ones. An equal number of eQTLs were used in each cohort so that results between cohorts would be directly comparable. We note that 5,000 eQTLs represents an average of ∼3.5 eQTLs per genetic marker, which is not surprising given that linkage disequilibrium extends for many megabases in a mouse F2 cross, so a single marker captures many polymorphisms.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) classifications were tabulated for each gene on the microarray. Only the 531 GO gene sets (from all levels of the GO hierarchy and all three GO branches: Biological Process, Molecular Function, and Cellular Component) and 75 KEGG gene sets containing at least 50 genes on our microarray were tested, since small gene sets have little statistical power in our test. If multiple genes from a gene set had cis-eQTLs and were located within 2 mb of each other in the genome, all but one in the cluster were discarded from the analysis, to ensure that the eQTLs being tested are all independent (the 2 mb cutoff was chosen since the most distant known cis-regulation is an enhancer ∼1 mb from its target gene; so allowing 1 mb from a cis-regulatory mutation in each direction yields 2 mb). All of the cases of clustered eQTLs within a gene set showed the same direction of effect (either up-regulated by the B6 allele, or by the CAST allele, but not a mixture of both), so the choice of which gene(s) to exclude had no effect on the test's results. Relaxing our distance cutoff results in a small increase in the sample size and gene set enrichment significance. Likewise, increasing the distance cutoff excludes a small fraction of genes, marginally decreasing the enrichment significance.
The effect directions for the cis-eQTLs of a gene set were then tested for departure from the expected 1∶1 ratio of +/ – alleles by comparing to the hypergeometric expectation. The results are similar to testing using the binomial expectation, but the hypergeometric takes into account the fact that if many + alleles have already been observed in a gene set, further genes in that set are actually slightly less likely to have + alleles by chance (since the total number of + and – alleles included in our list is equal). Coexpression modules were constructed for each tissue as previously described [53]. A total of 41 modules containing at least 50 genes were tested (10 in brain, 14 in liver, and 17 in muscle).
Hypergeometric p-values for each gene set in each tissue/cohort were then combined across cohorts by Fisher's method, to yield the single-tissue p-values for each gene set. The FDR was estimated in two ways. In the first approach, genotype labels were permuted as described above, and the entire eQTL detection and directionality test procedure was carried out. This yielded zero false positives even over many thousands of randomizations. However this randomization strategy does not account for the fact that a gene with a B6-upregulating cis-eQTL in one cohort is likely to have B6-upregulating alleles in other cohorts as well. In order to capture this effect in our permutations, we carried out a second randomization procedure. We used the cis-eQTL results from the real data, but randomly shuffled the gene set assignments for each gene. In this test, the consistency of eQTL directions across tissues and cohorts is perfectly preserved, and only the effect of the gene set assignments is randomized. With this procedure, false positives were found at all cutoffs tested; FDRs were estimated at several cutoffs, and are shown in Table 1. We note that although the data from different tissues are not entirely independent, since they come from the same mice, this does not present a problem for estimating FDRs because we combined the p-values in the same way for both real and permuted data. In addition, the non-independence of gene sets is not a problem, since this overlap is perfectly captured by our randomization procedure.
For the multi-tissue analysis, the three single-tissue p-values for each gene set were combined by Fisher's method, both for real and randomized data. This was expected to increase power because it decreases false-positive eQTLs, though it is also possible that the non-tissue-specific eQTLs this procedure enriches are more likely to be the result of recent selection. FDRs were estimated as described above for the single-tissue analysis. We also tested combining only results from mice of each gender, but did not find any sex-specific gene set enrichments.
The RNA-seq data were analyzed as follows. Sequence reads overlapping heterozygous SNPs were assigned to alleles as described [30]. All reads from each allele of each RefSeq gene were then summed to generate the total number of reads from each allele. Distinct transcripts from the same gene cannot be discerned with this approach (as with the vast majority of microarrays), so each gene was treated as if it produced a single transcript (we note that since GO annotations are typically for genes, and not individual transcripts, having transcript-specific data would not substantially affect our results). SNPs with no reads from one allele were discarded, since these are likely to reflect SNP annotation errors. Binomial p-values were calculated for each gene, using the expected 1∶1 ratio of reads from each allele. The most extreme 25% of genes with allele-specific information (2,037 genes) in each direction were retained for GO analysis. The GO analysis was carried out with the hypergeometric test as described above, except that no p-values were combined because only a single tissue/cohort was used. Randomizations were performed by replacing the cis-eQTL target genes with randomly chosen genes, and repeating the hypergeometric test.
The probability of QTLs for naso-anal length overlapping with eQTLs for the growth regulator gene set was calculated as follows. The peaks for all three eQTLs shown in Figure 4 were within the 0.5 LOD support interval of the top three length QTLs (one in males, two in females). Across all 3,834 eQTLs at this strength (r2>0.2), only 0.6% were within this interval of the male length QTL and 0.3% for each female length QTL. Since these are independent, and 27 eQTLs from the growth gene set reached this cutoff, the chance of all three overlaps is 27×0.006×26×0.003×25×0.003 = 0.00095. Interestingly, the eQTL overlapping the strongest length QTL in each gender were both in the top 12 strongest growth eQTL (r 2>0.5), so even just the overlap of those two is significant at p = 0.002. Testing the overlap with the three length QTLs in random groups of 27 eQTLs supported these calculations. In males there is one length QTL where the CAST allele is associated with greater length, but this was not included in our overlap analysis because we only posit that the alleles increasing B6 growth have been under positive selection and are present in the list of growth genes with B6-upregulating cis-eQTL. eQTL scans shown in Figure 4 were performed using CxB brain; brain was chosen because it is the tissue with the strongest growth gene eQTL direction bias, and CxB was chosen because it is the larger of the two cohorts. Expression levels were from CxB female brains in Figure 4a, and CxB male brains in Figure 4b, to match genders with the length QTL shown.
qPCR
We performed quantitative PCR with SYBR green, amplifying both nuclear and mitochondrial DNA from B6 and CAST liver tissue. The ratio of mitochondrial/nuclear DNA gives an estimate of the mitochondrial abundance in each strain, and the ratio of these ratios indicates their relative levels. The following primer sequences were used: nuclear, CCTTGGACATTAGCACATGG and AACTGTCTCCCCTGACCAAC; mitochondrial, ACAATGTTAGGGCCTTTTCG and GTTCCCAGAGGTTCAAATCC. No off-target effects were observed for either primer pair. Each reaction was repeated 48 times to ensure consistency. The 99% confidence interval for the B6:CAST ratio of mitochondrial/genomic DNA (a ratio of ratios) was 1.06 – 1.20, and the 99.9% confidence interval was 1.04 – 1.23.
Supporting Information
Zdroje
1. Prud'hommeBGompelNCarrollSB
2007
Emerging principles of regulatory evolution.
Proc Natl Acad Sci U S A
104
Suppl 1
8605
8612
2. HoekstraHECoyneJA
2007
The locus of evolution: Evo devo and the genetics of
adaptation.
Evolution
61
995
1016
3. FraserHB
Gene expression adaptation: from single genes to genomes.
Bioessays, accepted for publication
4. WrayGA
2007
The evolutionary significance of cis-regulatory
mutations.
Nat Rev Genet
8
206
5. SternDLOrgogozoV
2008
The loci of evolution: how predictable is genetic
evolution?
Evolution
62
2155
6. PollardKSSalamaSRLambertNLambotMACoppensS
2006
An RNA gene expressed during cortical development evolved rapidly
in humans.
Nature
443
167
172
7. HaygoodRFedrigoOHansonBYokoyamaKDWrayGA
2007
Promoter regions of many neural - and nutrition-related genes have
experienced positive selection during human evolution.
Nat Genet
39
1140
1144
8. PrabhakarSNoonanJPPääboSRubinEM
2006
Accelerated evolution of conserved noncoding sequences in
humans.
Science
314
786
9. AndolfattoP
2005
Adaptive evolution of non-coding DNA in
Drosophila.
Nature
437
1149
1152
10. GaltierNDuretL
2007
Adaptation or biased gene conversion? Extending the null
hypothesis of molecular evolution.
Trends Genet
23
273
277
11. FayJCWittkoppPJ
2008
Evaluating the role of natural selection in the evolution of gene
regulation.
Heredity
100
191
199
12. OleksiakMFChurchillGACrawfordDL
2002
Variation in gene expression within and among natural
populations.
Nat Genet
32
261
13. BlekhmanROshlackAChabotAESmythGKGiladY
2008
Gene regulation in primates evolves under tissue-specific
selection pressures.
PLoS Genet
4
e1000271
doi:10.1371/journal.pgen.1000271
14. GiladYOshlackASmythGKSpeedTPWhiteKP
2006
Expression profiling in primates reveals a rapid evolution of
human transcription factors.
Nature
440
242
15. DenverDRMorrisKStreelmanJTKimSKLynchM
2005
The transcriptional consequences of mutation and natural
selection in Caenorhabditis elegans.
Nat Genet
37
544
16. RifkinSAHouleDKimJWhiteKP
2005
A mutation accumulation assay reveals a broad capacity for rapid
evolution of gene expression.
Nature
438
220
17. LandryCRLemosBRifkinSADickinsonWJHartlDL
2007
Genetic properties influencing the evolvability of gene
expression.
Science
317
118
18. FraserHBMosesASchadtEE
2010
Evidence for widespread adaptive evolution of gene expression in
budding yeast.
Proc Natl Acad Sci U S A
107
2977
2982
19. BullardJHMostovoyYDudoitSBremRB
2010
Polygenic and directional regulatory evolution across pathways in
Saccharomyces.
Proc Natl Acad Sci U S A
107
5058
5063
20. GibsonGWeirB
2005
The quantitative genetics of transcription.
Trends Genet
21
616
623
21. SteinerCCWeberJNHoekstraHE
2007
Adaptive variation in beach mice produced by two interacting
pigmentation genes.
PLoS Biol
5
e219
doi:10.1371/journal.pbio.0050219
22. OrrHA
1998
Testing natural selection versus genetic drift in phenotypic
evolution using quantitative trait locus data.
Genetics
149
2099
2104
23. DossSSchadtEEDrakeTALusisAJ
2005
Cis-acting expression quantitative trait loci in
mice.
Genome Res
15
681
691
24. McDonaldJHKreitmanM
1991
Adaptive protein evolution at the Adh locus in
Drosophila.
Nature
351
652
25. HughesAL
2007
Looking for Darwin in all the wrong places: the misguided quest
for positive selection at the nucleotide sequence level.
Heredity
99
364
373
26. FrazerKAEskinEKangHMBogueMAHindsDA
2007
A sequence-based variation map of 8.27 million SNPs in inbred
mouse strains.
Nature
448
1050
1053
27. GeraldesABassetPGibsonBSmithKLHarrB
2008
Inferring the history of speciation in house mice from autosomal,
X-linked, Y-linked and mitochondrial genes.
Mol Ecol
17
5349
5363
28. The Gene Ontology Consortium
2000
Gene Ontology: tool for the unification of
biology.
Nat Genet
25
25
29
29. KanehisaMGotoSHattoriMAoki-KinoshitaKFItohM
2006
From genomics to chemical genomics: new developments in
KEGG.
Nucleic Acids Res
34
D354
D357
30. BabakTDevealeBArmourCRaymondCClearyMA
2008
Global survey of genomic imprinting by transcriptome
sequencing.
Curr Biol
18
1735
1741
31. WangXSunQMcGrathSDMardisERSolowayPD
2008
Transcriptome-wide identification of novel imprinted genes in
neonatal mouse brain.
PLoS ONE
3
e3839
doi:10.1371/journal.pone.0003839
32. BabakT
2010
Genetic validation of whole-transcriptome sequencing for mapping
expression affected by cis-regulatory
variation.
BMC Genomics
11
473
33. BabakTGarrett-EngelePArmourCDRaymondCKKellerMP
2009
The Mouse Genome Database genotypes::phenotypes.
Nucl Acids Res
37
D712
D719
34. BouwmanPGöllnerHElsässerHPEckhoffGKarisA
2000
Transcription factor Sp3 is essential for post-natal survival and
late tooth development.
EMBO J
19
655
661
35. SimmerFMoormanCvan der LindenAMKuijkEvan den BerghePV
2003
Genome-wide RNAi of C. elegans using the
hypersensitive rrf-3 strain reveals novel gene
functions.
PLoS Biol
1
e12
doi:10.1371/journal.pbio.0000012
36. SchadtEELambJYangXZhuJEdwardsS
2005
An integrative genomics approach to infer causal associations
between gene expression and disease.
Nat Genet
37
710
717
37. MillsteinJZhangBZhuJSchadtEE
2009
Disentangling molecular relationships with a causal inference
test.
BMC Genet
10
23
38. YangXDeignanJLQiHZhuJQianS
2009
Validation of candidate causal genes for obesity that affect
shared metabolic pathways and networks.
Nat Genet
41
415
423
39. BachmanovAAReedDRBeauchampGKTordoffMG
2002
Food intake, water intake, and drinking spout side preference of
28 mouse strains.
Behav Genet
32
435
443
40. Le RoyIRoubertouxPLJamotLMaaroufFTordjmanS
1998
Neuronal and behavioral differences between Mus musculus
domesticus (C57BL/6JBy) and Mus musculus
castaneus (CAST/Ei).
Behav Brain Res
95
135
142
41. KoideTMoriwakiKIkedaKNikiHShiroishiT
2000
Multi-phenotype behavioral characterization of inbred strains
derived from wild stocks of Mus musculus.
Mamm Genome
11
664
670
42. BrownREWongAA
2007
The influence of visual ability on learning and memory
performance in 13 strains of mice.
Learn Mem
14
134
144
43. SakaiTKikkawaYMiuraIInoueTMoriwakiK
2005
Origins of mouse inbred strains deduced from whole-genome
scanning by polymorphic microsatellite loci.
Mamm Genome
16
11
19
44. SchlosserG
2002
Modularity and the units of evolution.
Theory in Biosciences
121
1
80
45. GrazeRMMcIntyreLMMainBJWayneMLNuzhdinSV
2009
Regulatory divergence in Drosophila melanogaster
and D. simulans, a genomewide analysis of allele-specific
expression.
Genetics
183
547
561
46. ZhangXBorevitzJO
2009
Global analysis of allele-specific expression in
Arabidopsis thaliana.
Genetics
182
943
954
47. GompelNPrud'hommeBWittkoppPJKassnerVACarrollSB
2005
Chance caught on the wing: cis-regulatory evolution and the
origin of pigment patterns in Drosophila.
Nature
433
481
487
48. MillerCTBelezaSPollenAASchluterDKittlesRA
2007
cis-Regulatory changes in Kit ligand expression
and parallel evolution of pigmentation in sticklebacks and
humans.
Cell
131
1179
1189
49. ShapiroMDMarksMEPeichelCLBlackmanBKNerengKS
2004
Genetic and developmental basis of evolutionary pelvic reduction
in threespine sticklebacks.
Nature
428
717
723
50. HaygoodRBabbittCCFedrigoOWrayGA
2010
Contrasts between adaptive coding and noncoding changes during
human evolution.
Proc Natl Acad Sci U S A
107
7853
7857
51. LiaoBYWengMPZhangJ
2010
Contrasting genetic paths to morphological and physiological
evolution.
Proc Natl Acad Sci U S A
107
7353
7358
52. HeYDDaiHSchadtEECavetGEdwardsSW
2003
Microarray standard data set and figures of merit for comparing
data processing methods and experiment designs.
Bioinformatics
19
956
965
53. ZhuJLumPYLambJGuhaThakurtaDEdwardsSW
2004
An integrative genomics approach to the reconstruction of gene
networks in segregating populations. Cytogenet.
Genome Res
105
363
374
Štítky
Genetika Reprodukční medicína
Článek Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis inČlánek c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and PhosphodiesterasesČlánek Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 3- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
- Prof. Petr Urbánek: Potřebujeme najít pacienty s nediagnostikovanou akutní intermitentní porfyrií
-
Všechny články tohoto čísla
- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- Testing for an Unusual Distribution of Rare Variants
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Repeat Associated Non-ATG Translation Initiation: One DNA, Two Transcripts, Seven Reading Frames, Potentially Nine Toxic Entities!
- Initial Mutations Direct Alternative Pathways of Protein Evolution
- Dopamine Signalling in Mushroom Bodies Regulates Temperature-Preference Behaviour in
- Sensing of Replication Stress and Mec1 Activation Act through Two Independent Pathways Involving the 9-1-1 Complex and DNA Polymerase ε
- Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in
- Identification of a Novel Type of Spacer Element Required for Imprinting in Fission Yeast
- Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I
- Global Analysis of the Relationship between JIL-1 Kinase and Transcription
- H3K9me2/3 Binding of the MBT Domain Protein LIN-61 Is Essential for Vulva Development
- REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock
- A Novel Unstable Duplication Upstream of Predisposes to a Breed-Defining Skin Phenotype and a Periodic Fever Syndrome in Chinese Shar-Pei Dogs
- Polycomb Repressive Complex 2 Controls the Embryo-to-Seedling Phase Transition
- A Role for Set1/MLL-Related Components in Epigenetic Regulation of the Germ Line
- Genome-Wide Association Analysis Identifies Variants Associated with Nonalcoholic Fatty Liver Disease That Have Distinct Effects on Metabolic Traits
- A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium
- Ancestral Mutation in Telomerase Causes Defects in Repeat Addition Processivity and Manifests As Familial Pulmonary Fibrosis
- Ultra-Deep Sequencing of Mouse Mitochondrial DNA: Mutational Patterns and Their Origins
- Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci
- The Toll-Like Receptor Gene Family Is Integrated into Human DNA Damage and p53 Networks
- Polycomb Targets Seek Closest Neighbours
- Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis
- c-di-GMP Turn-Over in Is Controlled by a Plethora of Diguanylate Cyclases and Phosphodiesterases
- Estimating Divergence Time and Ancestral Effective Population Size of Bornean and Sumatran Orangutan Subspecies Using a Coalescent Hidden Markov Model
- Rif1 Supports the Function of the CST Complex in Yeast Telomere Capping
- A Tradeoff Drives the Evolution of Reduced Metal Resistance in Natural Populations of Yeast
- Quantifying the Underestimation of Relative Risks from Genome-Wide Association Studies
- Population-Based Resequencing of Experimentally Evolved Populations Reveals the Genetic Basis of Body Size Variation in
- Triplet Repeat–Derived siRNAs Enhance RNA–Mediated Toxicity in a Drosophila Model for Myotonic Dystrophy
- The FUN30 Chromatin Remodeler, Fft3, Protects Centromeric and Subtelomeric Domains from Euchromatin Formation
- Viral Genome Segmentation Can Result from a Trade-Off between Genetic Content and Particle Stability
- Environmental Sex Determination in the Branchiopod Crustacean : Deep Conservation of a Gene in the Sex-Determining Pathway
- Systematic Detection of Polygenic Regulatory Evolution
- The SUMO Isopeptidase Ulp2p Is Required to Prevent Recombination-Induced Chromosome Segregation Lethality following DNA Replication Stress
- Uncoupling Antisense-Mediated Silencing and DNA Methylation in the Imprinted Cluster
- Role of the Drosophila Non-Visual ß-Arrestin Kurtz in Hedgehog Signalling
- Differential Genetic Associations for Systemic Lupus Erythematosus Based on Anti–dsDNA Autoantibody Production
- COMPASS-Like Complexes Mediate Histone H3 Lysine-4 Trimethylation to Control Floral Transition and Plant Development
- H3 Lysine 4 Is Acetylated at Active Gene Promoters and Is Regulated by H3 Lysine 4 Methylation
- Diverse Roles and Interactions of the SWI/SNF Chromatin Remodeling Complex Revealed Using Global Approaches
- A Bow-Tie Genetic Architecture for Morphogenesis Suggested by a Genome-Wide RNAi Screen in
- Roles of () in Oocyte Nuclear Architecture, Gametogenesis, Gonad Tumors, and Genome Stability in Zebrafish
- A Molecular Phylogeny of Living Primates
- Roles of the Espin Actin-Bundling Proteins in the Morphogenesis and Stabilization of Hair Cell Stereocilia Revealed in CBA/CaJ Congenic Jerker Mice
- A Cholinergic-Regulated Circuit Coordinates the Maintenance and Bi-Stable States of a Sensory-Motor Behavior during Male Copulation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Whole-Exome Re-Sequencing in a Family Quartet Identifies Mutations As the Cause of a Novel Skeletal Dysplasia
- Origin-Dependent Inverted-Repeat Amplification: A Replication-Based Model for Generating Palindromic Amplicons
- FUS Transgenic Rats Develop the Phenotypes of Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration
- Limited dCTP Availability Accounts for Mitochondrial DNA Depletion in Mitochondrial Neurogastrointestinal Encephalomyopathy (MNGIE)
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání