-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
Chromatin poses a barrier to the recombination process. Chromatin modification is therefore a prerequisite factor for the efficient execution of the recombination event. Chromatin remodeling and several unique histone modifications at or near DNA double strand breaks (DSBs) facilitate early recombination processes, but little is known how chromatin state impinges on post-invasion steps of recombination, such as repair synthesis through homologous template, particularly recombination subtypes such as break-induced replication (BIR) involving extensive repair synthesis. Here, we investigated the effect of deletions in chromatin modification and remodeling genes on BIR and discovered that hyper-acetylation of H3K56 selectively impairs BIR and gene conversion associated with long DNA gap synthesis. We also found that hyper-acetylation of H3K56 interferes with the recovery from replication stress in checkpoint deficient cells and induces translocation-type gross chromosomal rearrangements (GCRs). The results provide a basic understanding of how histone modification facilitates efficient fork progression in recombination, controls the types of the repair products and sustains chromosome integrity upon induction of genotoxic stress.
Published in the journal: . PLoS Genet 11(2): e32767. doi:10.1371/journal.pgen.1004990
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004990Summary
Chromatin poses a barrier to the recombination process. Chromatin modification is therefore a prerequisite factor for the efficient execution of the recombination event. Chromatin remodeling and several unique histone modifications at or near DNA double strand breaks (DSBs) facilitate early recombination processes, but little is known how chromatin state impinges on post-invasion steps of recombination, such as repair synthesis through homologous template, particularly recombination subtypes such as break-induced replication (BIR) involving extensive repair synthesis. Here, we investigated the effect of deletions in chromatin modification and remodeling genes on BIR and discovered that hyper-acetylation of H3K56 selectively impairs BIR and gene conversion associated with long DNA gap synthesis. We also found that hyper-acetylation of H3K56 interferes with the recovery from replication stress in checkpoint deficient cells and induces translocation-type gross chromosomal rearrangements (GCRs). The results provide a basic understanding of how histone modification facilitates efficient fork progression in recombination, controls the types of the repair products and sustains chromosome integrity upon induction of genotoxic stress.
Introduction
DNA damage drives mutagenesis and chromosomal rearrangements. Homologous recombination (HR) removes DNA lesions primarily at S/G2 phase of the cell cycle by pairing broken DNA ends with intact homologous template and copying across DNA breaks [1]. Break-induced replication (BIR) is the subtype of homologous recombination (HR) that eliminates one-ended DNA breaks or two-ended double strand breaks (DSBs) in the event when only one end of the DNA break is homologous to a template, such as a collapsed replication fork or eroded telomere. By searching for and copying from homologous sequences, often synthesizing hundreds of kilobases of DNA in the process, BIR has been implicated in catalyzing alternative lengthening of telomeres (ALT) and restoring replication forks [2,3]. Extensive DNA synthesis is unique to BIR or to only a subset of gene conversion events, both of which depend on Pol32, a non-essential subunit of DNA polymerase delta [4,5]. The large-scale DNA synthesis during BIR is also highly mutagenic in nature [6]. The initiation step of BIR is frequently associated with template switching, suggesting that the replication fork remains unstable and is prone to dissociation [7]. The precise mechanism behind the high propensity for mutations in BIR is not yet clear but might stem from its conservative mode of DNA synthesis and unique reliance on the helicase Pif1 for bubble migration [8,9,10].
Every DNA transaction including DNA replication and repair occurs within chromatin. Therefore, histone modification and the reconfiguring of chromatin structure are not only a prerequisite, but also dictate both the efficiency and outcome of these events. Temporally disrupted chromatin in DNA replication and repair is then restored by the deposition of new nucleosomes and the re-establishment of unperturbed chromatin architecture globally and locally [11].
Histone H3K56 is an evolutionarily conserved residue that is subjected to reversible acetylation [12,13,14,15,16,17]. Unlike many other modifications at histone tails, H3K56 is located within the globular core domain near the entry and exit sites of nucleosomes, and does not appear to affect DNA-histone interactions as well as chromatin configuration [12,13,18,19]. In Saccharomyces cerevisiae, H3K56 is acetylated by the combined action of the acetyltransferase Rtt109/Vps75 and the histone chaperone Asf1, and deacetylated by two redundant class III histone deacetylases (HDACs) Hst3 and Hst4 [16,17,20,21,22]. Functionally, acetylated H3K56 facilitates replication-coupled nucleosome assembly and replication-independent histone exchange [23,24]. H3K56 acetylation is also required for histone eviction and transcription activation at certain promoters [25]. In mammals wherein acetylation of H3K56 is less abundant than in yeast, H3K56 is also methylated by the histone lysine methyltransferase G9a and promotes PCNA docking on chromatin in G1 [26].
In addition to its role in nucleosome assembly during replication and transcription, H3K56 also contributes to DNA repair and signaling [12,27]. Dys-regulation of histone H3K56 acetylation leads to a severe sensitivity to DNA damaging agents and elevated genome instability. Both excess and reduced levels of H3K56 acetylation disrupt cell cycle progression and induce spontaneous DNA damage and hypersensitivity to DNA damaging agent treatment [16,17,20,27,28]. Both hyper - and hypo-acetylation of H3K56 result in defects in sister chromatid cohesion and recombination [29,30] and ribosomal DNA (rDNA) amplification [31], as well as increased mutation frequency and GCR rate [32]. Furthermore, H3K56 is tightly regulated by the cell cycle and DNA damage-induced checkpoint, as the amount of Hst3 fluctuates at both the transcriptional and posttranscriptional level [16,29]. Deletion of HST3 and HST4 also leads to thermo-sensitivity, defective telomere silencing and increased chromosome loss [33,34].
Considering the role of H3K56 acetylation in nucleosome assembly, some of these repair defects could be attributed to low nucleosome density and the inability to recover from cell cycle arrest upon DNA damage, particularly in the absence of other histone chaperones [35,36,37]. However, this alone cannot explain why excess acetylation of H3K56 also leads to DNA damage sensitivity and genome instability. It is likely that hyper-acetylation of H3K56 interferes with some other aspect of DNA repair and signaling beyond nucleosome assembly.
In this study, we investigated how chromatin modifiers and remodelers control BIR-mediated repair of an HO endonuclease-induced break on a disomic chromosome. Given the functional overlap of players between BIR and general DNA synthesis where chromatin modifications are tightly coupled to ongoing replication fork progression, we surmise that BIR should also depend on chromatin modification for efficient fork progression. We found that hyper-acetylation of H3K56 specifically inhibits efficient repair synthesis, therefore leading to BIR and gene conversion defects associated with long gap synthesis, but not other HR pathways. Deletion of HST3 and HST4 also impairs recovery from a collapsed replication fork in S-phase in DNA damage checkpoint deficient mrc1Δ cells and elevates translocation-type GCRs. We propose that acetylation of histone H3 impinges on the post-invasion step in recombination and thereby chromosomal integrity after DNA damage.
Results
Deletion of HST3 and HST4 leads to BIR deficiency
Extensive DNA synthesis in BIR likely relies on chromatin modification to achieve efficient fork progression. To address this question, we examined the effect of chromatin modifier gene deletions on BIR using an assay measuring BIR between disomic chromosomes [38]. Initial analysis was focused on chromatin remodeling complex factors (Swr1, INO80, RSC) and histone modifiers (Gcn5, Rtt109, Hst3, Hst4, and Dot1) implicated in DNA DSB repair. We induced a DSB at the MATa locus on a truncated copy of chromosome III, thereby triggering centromere proximal DNA ends to invade and copy from the intact MATα-inc allele on a full-length chromosome III as a DSB repair template (Fig. 1A). Since the truncated chromosome III possesses limited homology (46 bp) to the MATα-inc allele on the telomere proximal side of the DSB, BIR is the primary repair option, although a lower frequency of other repair events such as gene conversion (GC), chromosome loss or half crossover still occurs. The small percentage (>10%) of sectored Ade+/ − colony formation likely represents the cases where only one of the two sister chromatids completed repair. Different types of repair events were deduced by the presence of marker genes located on the left arm of chromosome III (ADE1 and ADE3) and the LEU2 gene of the truncated chromosome III in surviving colonies. The survival frequency is nearly 100% because cells still retain a functional chromosome III copy even if repair is aborted and the truncated chromosome is lost.
Fig. 1. Deletion of HST3 and HST4 inhibits BIR. 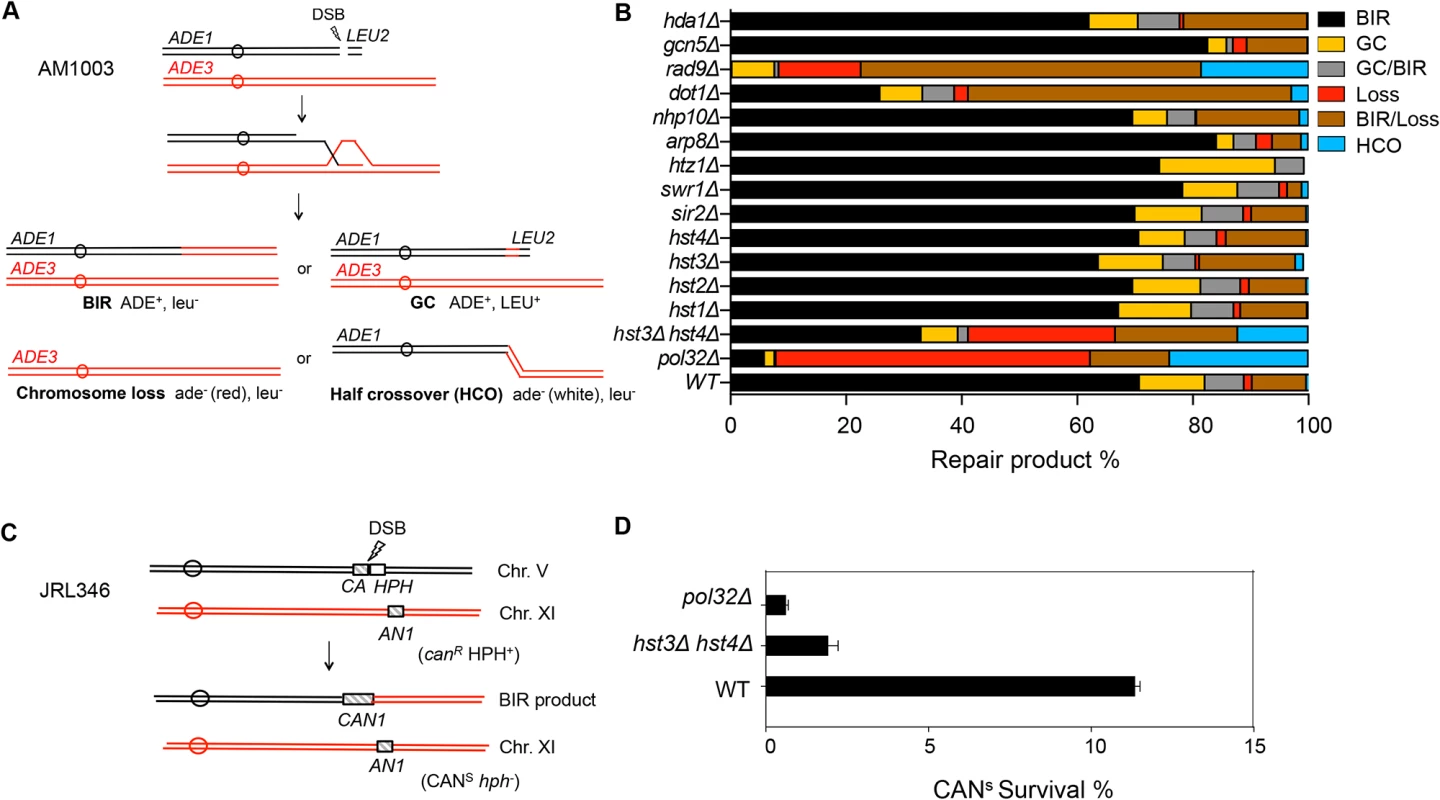
A, Schematic view of disomic chromosome III system (AM1003) for analyzing break-induced replication (BIR) in Saccharomyces cerevisiae. Four major types of pathways and the corresponding repair products are shown and distinguished by markers (ADE1, ADE3 and LEU2) and the sizes on PFGE gels. BIR, break-induced replication; GC, gene conversion; HCO, half crossover. B, Deletion of chromatin related factors and their effect on repair outcome in disomic BIR assay. C, Schematic view of an ectopic BIR assay. HO recognition site is positioned adjacent to the C-terminally truncated CAN1 gene (CA) on chromosome V and the N-terminally truncated CAN1 (AN1) on Chromosome XI as the homologous template. HPH, hygromycin B. D, Efficiency of BIR in pol32Δ, and hst3Δ hst4Δ cells, as measured by canavanine sensitive colony formation following DSB induction. Error bars represent s.d. Inactivation of most chromatin modification factors altered the repair profile and BIR frequency but to a moderate extent (Fig. 1B). Increased BIR frequency in several of these mutants (htz1, arp8, and swr1) could be explained by their reduced resection function because extensive resection is known to suppress BIR [39,40]. However, the effect of SWR1 or ARP8 deletion on resection is variable among studies and modest at best [41,42,43]. We also found that deletion of DOT1 led to a dramatic decrease in BIR but increase in BIR/chromosome loss events. Since Dot1 recruits the checkpoint adaptor protein Rad9 at DNA break sites and plays an important role in DNA damage checkpoint activation [44], BIR deficiency in dot1Δ cells can be attributed to the lack of a functional cell cycle checkpoint. Indeed, dot1Δ and rad9Δ mutants showed very similar repair profiles with elevated BIR/chromosome loss (sectoring) events. Alternatively, BIR defects in dot1Δ or rad9Δ might be due to the increased resection in these mutants reported earlier [45].
Interestingly, deletion of two class III HDACs, HST3 and HST4 reduced BIR frequency but increased chromosome loss and half crossover events. The result somewhat mirrored that of pol32Δ mutants, albeit with less severity. Deletion of HST3 or HST4 alone or other sirtuin genes including SIR2 did not reduce BIR frequency. We also monitored BIR product formation by pulsed-field gel electrophoresis (PFGE) of genomic DNA isolated at various time points post-HO expression and Southern blot hybridization with a probe that anneals to the ADE1 gene on chromosome III. We found that deletion of HST3 and HST4 led to a 2-fold reduction in BIR product formation by Southern blot assay (S1 Fig.). To further confirm whether hst3Δ hst4Δ mutant cells are defective in BIR, we performed another ectopic BIR assay, wherein a galactose-inducible HO endonuclease creates a DSB on the left end of chromosome V and BIR restores an intact CAN1 gene that renders the cells canavanine sensitive, but leads to the loss of the HPH gene distal to the DSB (Fig. 1C). Using this ectopic assay we found that deletion of HST3 and HST4 led to a 5-fold reduction in BIR (Fig. 1D). These results indicate that hst3Δ hst4Δ mutants are significantly deficient in BIR.
The strong resemblance in repair profiles between hst3Δ hst4Δ and pol32Δ mutants prompted us to examine if hst3Δ hst4Δ cells exhibit reduced expression of Pol32. We assessed the amount of triple HA-tagged Pol32 in hst3Δ hst4Δ cells via immunoblot assay with anti-HA antibody. We found that deletion of HST3 and HST4 did not reduce the expression level of Pol32 (S2 Fig.).
Hyper-acetylation of H3K56 inhibits break-induced replication
Histone H3K56 is the best-known target of Hst3/Hst4 and represents the marker of newly deposited nucleosomes during replication or repair [12,17]. We therefore tested whether H3K56R or H3K56Q mutations that either block or partially mimic H3K56 acetylation will alter BIR frequency. We found that the H3K56R mutation alone did not exert any effect on BIR efficiency, whereas H3K56Q led to a slight but reproducible BIR defect (Fig. 2A). The modest BIR deficiency in H3K56Q mutants compared to hst3Δ hst4Δ cells supports the notion that H3K56Q does not fully mimic acetylated H3K56 [12]. We then analyzed the effect of the H3K56R mutation on BIR frequency in hst3Δ hst4Δ. If the observed BIR deficiency in hst3Δ hst4Δ is due to hyper-acetylation of H3K56, then the K56R substitution should offset this BIR defect. Indeed, H3K56R almost fully rescued the BIR defect in hst3Δ hst4Δ cells (Fig. 2A). These results suggest that hyper-acetylated H3K56 inhibits BIR.
Fig. 2. Hyper-acetylation of H3K56 causes BIR defects. 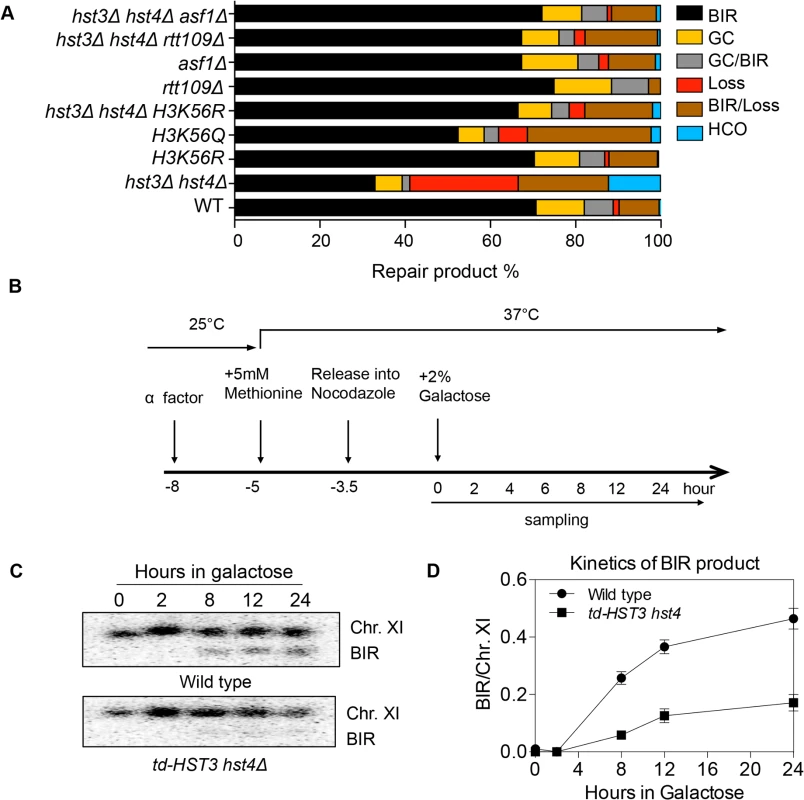
A, Various combination of mutants as shown were tested in the disomic BIR system. Shown are the averages of at least two independent experiments. BIR, break-induced replication; GC, gene conversion; HCO, half crossover. An average of 500–1000 events were scored for each experiment. B, Flowchart showing experimental procedure. C, Southern blot analysis of BIR product formation in cells depleted of Hst3. Chromosomes were separated by PFGE and a DNA probe specific for MCH2 was used to detect BIR products. D, Quantification of Southern blot results. Rtt109 is an acetyltransferase responsible for acetylation of H3K56 [20,21]. Asf1 is a histone chaperone specific for H3K56 acetylation [22]. We thus examined the effect of RTT109 or ASF1 deletion on BIR frequency in wild-type and hst3Δ hst4Δ cells using the disomic BIR assay. As predicted, rtt109Δ or asf1Δ alone did not influence BIR frequency (Fig. 2A). However, deletion of ASF1 or RTT109 in hst3Δ hst4Δ mutants improved BIR frequency to a level almost identical to that in wild-type cells. The level of BIR product formation observed in this genetic assay was also confirmed by Southern blot analysis (S1 Fig.). These results further demonstrate that hyper-acetylation of H3K56 inhibits BIR.
Hyper-acetylation of H3K56 does not inhibit other recombination subtypes
BIR deficiency in hst3Δ hst4Δ cells prompted us to test whether deletion of HST3 and HST4 generally affects other HR processes. We thus tested the effect of HST3 and HST4 deletion on mating-type switch recombination, ectopic gene conversion, and single strand annealing using genetic assays wherein the repair of an HO break occurs by different recombination subtypes (S3 Fig.). The recombination frequency was determined by survival frequency after HO expression and physical monitoring of the repair products by Southern blot-based assay. We found that none of the recombination subtypes tested were impaired in hst3Δ hst4Δ cells (S3 Fig.). We also determined the frequency of non-homologous end joining (NHEJ) in donorless hst3Δ hst4Δ cells by inducing an HO break at the MAT locus. Deletion of HST3 and HST4 did not lead to any changes in survival frequency following HO expression (S3 Fig.). This suggests that hst3Δ hst4Δ mutants are deficient in BIR but not other recombination or NHEJ processes.
Since H3K56 acetylation marks newly assembled nucleosomes, half of the nucleosomes in each cell should bear this mark after DNA synthesis. Based on this premise, we set out to test if the physiologically relevant level of H3K56 acetylation is sufficient to inhibit BIR. To this end, we engineered a yeast strain expressing a degron-fused and the MYC epitope-tagged Hst3 as the sole H3K56 deacetylase, and then examined BIR efficiency in cells arrested at G2 after one round of DNA synthesis upon depletion of Hst3 using the ectopic BIR assay (Fig. 2B). Depletion of Hst3 at non-permissive temperature (37°C) was confirmed by thermosensitivity and immunoblot assay with anti-MYC antibody (S4A and S4C Fig.). Cell cycle progression was monitored by fluorescence activated cell sorting (FACS) after propidium iodide (PI) staining of cells (S4B Fig.). We found that depletion of Hst3 decreased BIR frequency 3-fold in cells arrested at G2 as compared to those expressing Hst3 (Fig. 2C, 2D). The relatively small reduction (3-fold) in BIR in Hst3-depleted cells compared to the five-fold reduction observed in hst3 hst4 deleted cells suggests that the level of BIR is proportional to the amount of intracellular H3K56 acetylation (S4D Fig.). Inactivation of Hst3 and Hst4 by the pan–sirtuin inhibitor nicotinamide (NAM) also reduced BIR frequency 2-fold (S5 Fig.). As shown previously, deletion of SIR2, HST1 and HST2 did not alter BIR frequency (Fig. 2A). The results suggest that physiological levels of H3K56 acetylation are sufficient to suppress BIR in cells.
Hyper-acetylation of H3K56 suppresses recovery from replication stress in checkpoint deficient mrc1 cells
Replication forks stall when deoxyribonucleotide triphosphate (dNTP) pools are depleted and even collapse without a functional S-phase checkpoint [46]. Mrc1 is an S-phase checkpoint protein that stabilizes replication forks when hydroxyurea (HU) is used to induce dNTP pool depletion [47,48]. In the absence of Mrc1, stalled replication forks harbor extensive single strand DNA (ssDNA) and become collapsed [49]. Thus cell survival relies on pathways that repair collapsed replication forks [50]. We examined HU sensitivity of mrc1Δ cells deleted for HST3, HST4 or POL32 genes upon acute HU treatment. Unlike typical plate based assays where cells are exposed chronic DNA damage and dys-regulation of H3K56 acetylation leads to HU sensitivity [28], the acute HU exposure (1 to 4 h) employed here likely addresses the repair of collapsed fork [51].
We found that hst3Δ hst4Δ, pol32Δ or mrc1Δ cells are only mildly sensitive to HU treatment, but hst3Δ hst4Δ mrc1Δ or pol32Δ mrc1Δ cells showed far greater sensitivity than each single gene deletion mutant (Figs. 3A and S6A Fig.). Deletion of POL32 also sensitizes tof1 - or csm3-deleted cells to HU treatment (S6B–S6C Fig.). Furthermore, the HU sensitivity of hst3Δ hst4Δ mrc1Δ mutants is fully rescued by an asf1Δ mutation (Fig. 3B). Deletion of ASF1 or RTT109 did not sensitize mrc1Δ to HU treatment (Figs. 3B and S6D). These results suggest that the observed HU-induced stall or collapse of replication forks in mrc1Δ cells relies upon Pol32 - and H3K56 deacetylation dependent process for recovery.
Fig. 3. Synergistic HU sensitivity between hst3Δ hst4Δ and mrc1Δ but not rad53Δ. 
Cells with indicated genotypes grown in log phase were alpha factor arrested for 3 hours and released into YEPD or SC-Ura with 150 mM HU up to 5 hours. Percent survival of hst3Δ hst4Δ, mrc1Δ, hst3Δ hst4Δ mrc1Δ (A), asf1Δ, mrc1Δ, asf1Δ mrc Δ, asf1Δ hst3Δ hst4Δ mrc1Δ (B), hst3Δ hst4Δ mrc1Δ supplemented with the plasmids expressing Mrc1 or mrc1AQ (C), hst3Δ hst4Δ, rad53Δ and hst3Δ hst4Δ rad53Δ (D) are shown. Percent survival was calculated by the number of colonies treated with HU divided by the number of colonies before HU treatment and determined at every hour after release from α factor arrest. Plotted are the mean values of three independent experiments ± s.d. Rad53 is another S-phase checkpoint protein that sustains replication fork integrity in response to HU treatment [46]. Surprisingly, in contrast to mrc1Δ cells, deletion of HST3 and HST4 or POL32 did not further sensitize rad53Δ mutants to HU treatment Fig. 3D). These results suggest that not all collapsed forks depend on Hst3/Hst4 and Pol32-dependent process for repair.
Mrc1 contributes to both damage-induced checkpoint induction and replication fork stabilization [47]. These two functions can be uncoupled by the mrc1AQ allele that abolishes Mrc1’s checkpoint function but retains its role in replication fork stabilization [47]. To determine which function of Mrc1 contributes to HU resistance in hst3Δ hst4Δ or pol32Δ mutants, we examined HU sensitivity in a strain bearing the mrc1AQ mutation and hst3 hst4 or pol32 gene deletions. We found that the mrc1AQ mutation did not confer synergistic HU sensitivity to hst3Δ hst4Δ or pol32Δ cells, suggesting that Mrc1’s checkpoint function is dispensable for HU sensitivity in hst3Δ hst4Δ or pol32Δ mutants (Figs. 3C and S6E). Additionally, deletion of RAD9, another DNA damage checkpoint adaptor but not a fork stabilizer, did not sensitize hst3Δ hst4Δ cells to HU treatment (S6F Fig.). The results suggest that stalled/collapsed replication forks but not checkpoint dysfunction lead to HU sensitivity in hst3Δ hst4Δ or pol32Δ mutants.
The lethality upon acute HU treatment in hst3Δ hst4Δ mrc1∆ cells could be due to an inability to repair a collapsed fork or alternatively to a more extensive fork collapse. It has been shown that HU treatment induces Rad52-green fluorescent protein (GFP) focus formation in mrc1Δ but not in wild-type nuclei, likely marking the site of stressed and collapsed replication forks and their repair [51]. We thus monitored the kinetics of Rad52-GFP foci as a surrogate for the repair of collapsed replication forks in order to dissect HU-induced fork collapse in mrc1∆ cells. We found that Rad52-GFP foci appeared in mrc1Δ cells upon HU treatment, reached a maximum level after releasing cells into fresh YEPD medium, and gradually disappeared most likely due to repair (Fig. 4). Importantly, in hst3Δ hst4Δ mrc1Δ cells and pol32Δ mrc1Δ cells, the Rad52-GFP foci persisted for up to 8 h after HU treatment, suggesting that hst3Δ hst4Δ or pol32Δ cells are deficient in repairing collapsed replication forks (Fig. 4). To test if the repair deficiency in hst3Δ hst4Δ mrc1Δ mutants could be attributed to hyper-acetylation of H3K56, we deleted ASF1 and monitored Rad52-GFP kinetics following HU treatment. Deletion of ASF1 led to a high level of Rad52-GFP foci at 6–8 h after release from HU, consistent with the role of Asf1 in fork stability [52], yet significantly offset the level of Rad52-GFP in hst3Δ hst4Δ mrc1Δ cells (Fig. 4A and B). Deletion of HST3 and HST4 also led to an increase in spontaneous Rad52-GFP focus formation and sensitivity to acute HU treatment in the BY4741 strain, but not in another genetic background (JKM139, see Fig. 4A and C, and S6 Fig.). We surmise that Hst3 and Hst4 contribute to the integrity of replication forks in certain genetic backgrounds. The results support the role of Hst3, Hst4 and Pol32 in the repair of collapsed replication forks.
Fig. 4. Rad52-GFP foci in hst3Δ hst4Δ mrc1Δ and pol32Δ mrc1Δ cells persist after HU treatment. 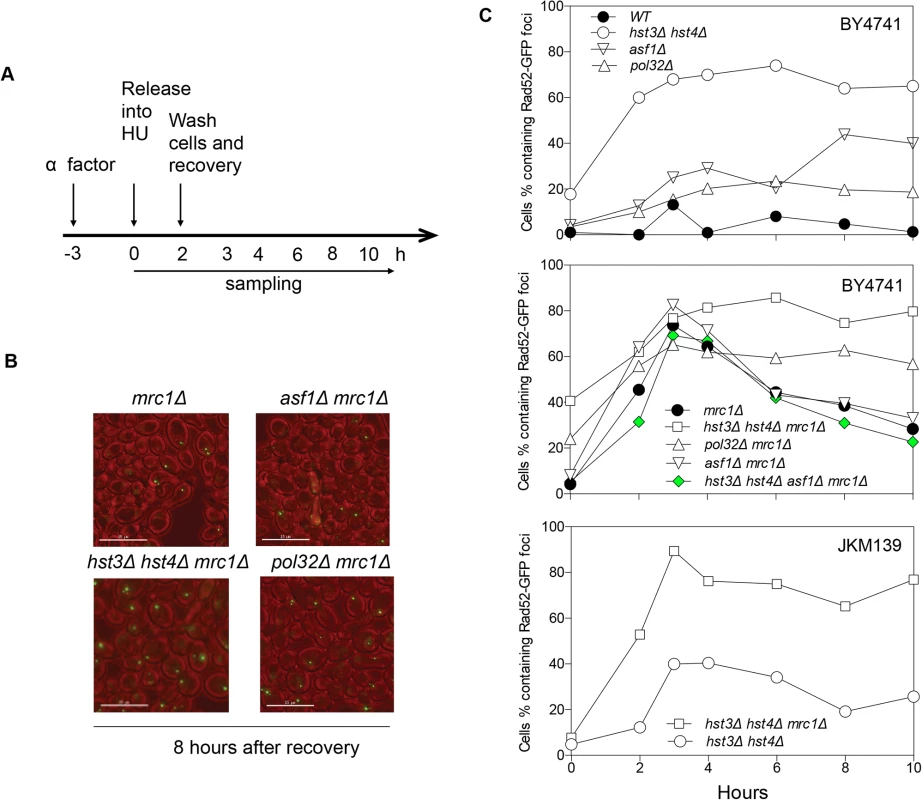
A, Flowchart showing experimental procedure. B, Representative images for different cells showing persistent Rad52-GFP foci at 8 hours after recovery from HU treatment. Size bar, 15 µm. C, Quantification of percentage of Rad52-GFP foci in the cell population. At least 300 individual cells were counted for each genotype and time point. Hyper-acetylation of H3K56 increases gross chromosomal rearrangements
Improper repair of replication stress is a likely source of gross chromosomal rearrangements (GCRs) [53]. Furthermore, deletion of ASF1 or RTT109 increases GCR rate [20,54]. We thus analyzed GCR frequency in hst3Δ hst4Δ or pol32Δ cells using the assay that measures the loss of GCR reporter genes on the left arm of chromosome V (Table 1). We found that deletion of HST3 and HST4 increased GCRs 457-fold (Table 1). Deletion of ASF1 partially reduced GCR rate to the level indistinguishable from that observed in asf1Δ cells, suggesting that hyper-acetylation of H3K56 is the likely cause of elevated GCRs in hst3Δ hst4Δ mutants. Interestingly, deletion of POL32 leads to only a mild increase in GCR. Analysis of the breakpoints of GCRs showed that most of the GCR events in hst3Δ hst4Δ cells are DNA translocations with 3–12 bp of microhomology at the junction, whereas all GCRs in pol32Δ cells are the result of de novo telomere formation (Tables 1 and S1).
Tab. 1. Gross Chromosomal Rearrangement (GCR). 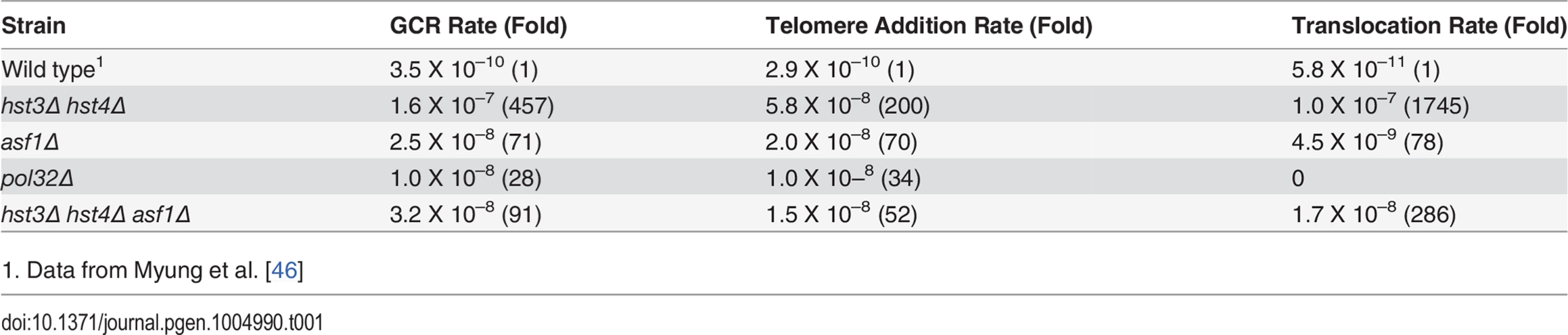
1. Data from Myung et al. [46] BIR defect in hst3Δ hst4Δ is distinct from thermosensitivity and DNA damage sensitivity
Cells bearing the hst3Δ hst4Δ mutations do not grow at 37°C but the deletion of genes involved in DNA damage checkpoint induction (RAD17) or sister chromatid establishment (CTF4) partially offset their thermosensitivity and DNA damage sensitivity phenotypes [28]. To test if the observed thermosensitivity and BIR defect share a common genetic cause, we deleted RAD17 or CTF4 and examined whether the gene deletions improve BIR in hst3Δ hst4Δ cells. We found that deletion of RAD17 or CTF4 partially offsets the growth deficiency at 37°C and resistance to MMS treatment as predicted, but does not affect defects in BIR in hst3Δ hst4Δ mutants (Fig. 5A and B). The results suggest that thermosensitivity, DNA damage sensitivity, and BIR deficiency are not genetically linked to each other in hst3Δ hst4Δ mutants and likely represent somewhat distinct molecular defects.
Fig. 5. Deletion of genes improving thermo- and MMS resistance of hst3Δ hst4Δ cells does not rescue BIR defects. 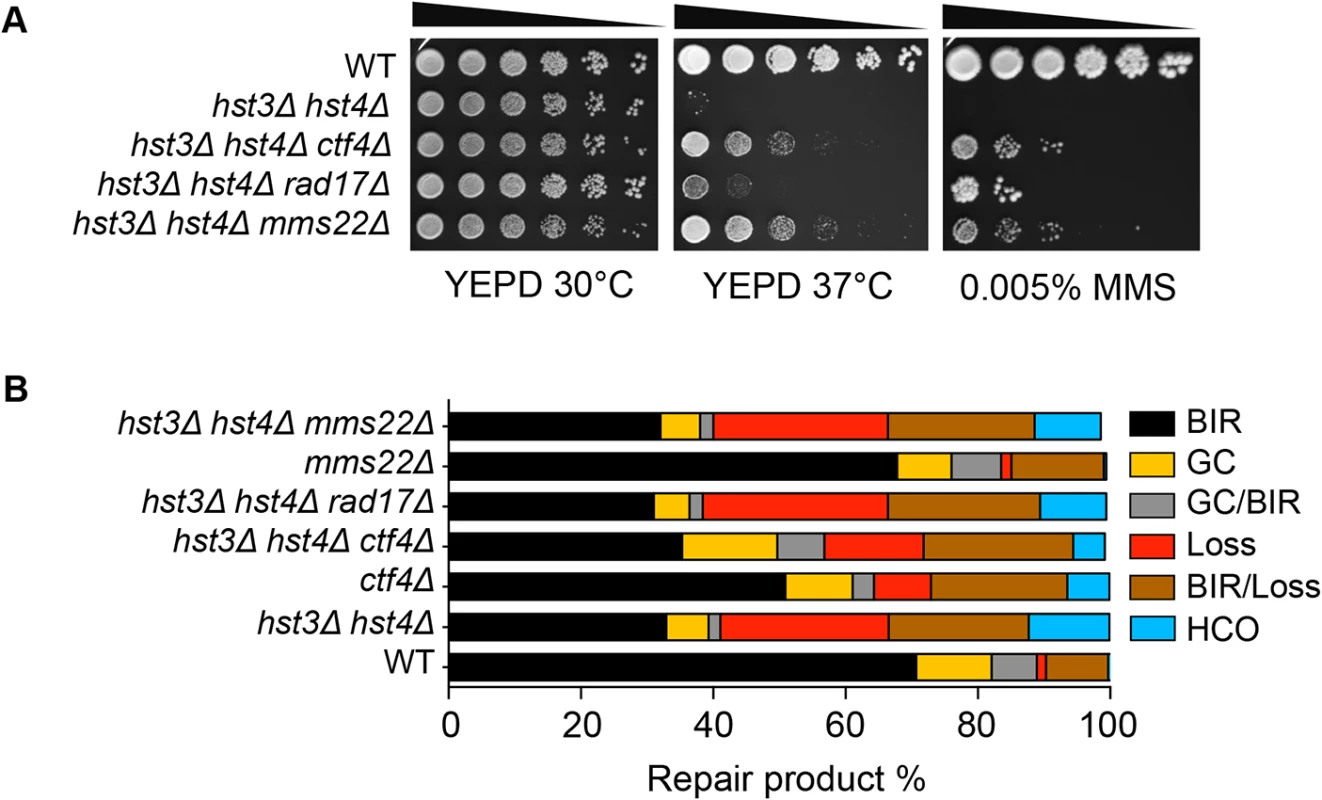
A, Serial dilutions of cells with indicated genotypes were spotted and cultured at 30°C or 37°C with or without 0.005% MMS. Pictures were taken after 4 days. B, Repair outcomes of wild-type (WT) and indicated mutant cells. Shown are the averages of at least two independent experiments. BIR, break-induced replication; GC, gene conversion; HCO, half crossover. An average of 500–1000 events were scored for each experiment. Most recently, a ubiquitin ligase complex containing Rtt101, Mms1, and Mms22 has been implicated in H3K56 acetylation-dependent mutation avoidance and damage tolerance pathways [32]. Deletion of MMS22 also offsets the thermosensitivity of hst3Δ hst4Δ cells (Fig. 5A and [55]). We thus tested the effect of MMS22 gene deletion on BIR using the disomic BIR assay. We found that deletion of MMS22 did not alter the repair profile in wild-type or hst3Δ hst4Δ cells (Fig. 5B). The results further support the model that the BIR defect in hst3Δ hst4Δ mutants is genetically distinct from underlying molecular defects causing thermosensitivity, elevated mutagenesis and sensitivity to DNA damaging agents.
Evidence suggests that acetylated H3K56 promotes SWR-C dependent exchange of H2AZ to H2A at promoter proximal regions and thereby regulates transcription [56]. If the observed BIR defect in hst3Δ hst4Δ mutants is due to excessive exchange of H2AZ to H2A, deletion of HTZ1 should lead to BIR defects as in hst3Δ hst4Δ cells. However, we found that deletion of HTZ1 or SWR1 does not result in BIR deficiency in the disomic BIR assay (Fig. 1B). Furthermore, deletion of HTZ1 or SWR1 did not rescue the BIR defect in hst3Δ hst4Δ cells (S7 Fig.). The results suggest that BIR deficiency in hst3Δ hst4Δ mutants is not due to excessive removal of H2AZ or dynamic histone exchange at promoters.
Hyper-acetylation of H3K56 inhibits repair synthesis
We then asked how hyper-acetylation of H3K56 inhibits BIR, which involves a few common and unique recombination steps [2]. Following DSB induction, the broken DNA ends are processed by 5’ to 3’ resection to yield single stranded DNA (ssDNA) that serves as the binding site for Rad51 recombinase. The ssDNA-Rad51 complex (a.k.a. pre-synaptic filament) then invades into a homologous template and initiates strand pairing. The 3’ non-homologous tail should be removed from the annealed DNA so that repair synthesis will begin copying from the template until the end of the chromosome. We analyzed the formation of ssDNA, binding of Rad51 at donor and recipient DNA molecules, 3’ flap removal, and initiation of DNA synthesis in hst3Δ hst4Δ mutant cells using PCR-based assays and chromatin immunoprecipitation (ChIP) using anti-Rad51 antibody (S8, S9, S10 Figs.).
We found that deletion of HST3 HST4 did not impair resection or the kinetics of Rad51 enrichment at either the break or homologous donor sites (see S8, S9 Figs. for experimental details). We discovered a slight reduction in flap removal efficiency in both hst3Δ hst4Δ and pol32Δ cells, but such a mild deficiency does not likely account for the BIR defects observed in these mutants (S10 Fig.). Furthermore, hst3Δ hst4Δ and pol32Δ mutants are not defective in single strand annealing wherein 3’ flap removal becomes the rate-limiting step, dictating repair frequency (S3 Fig.). We next examined the kinetics of initial repair synthesis by PCR amplification of repair products using a pair of primers in which one anneals to the donor site and the other anneals to the recipient DNA molecule. We discovered that both hst3Δ hst4Δ and pol32Δ mutants were severely defective in repair synthesis (Fig. 6). A single round of DNA replication after depletion of Hst3 in hst4 Δ cells also inhibits repair synthesis during BIR (S11 Fig.). In contrast, deletion of HST3 and HST4, or POL32 did not significantly impair or alter the kinetics of repair synthesis in mating type switching consistent with the finding that mating type switching is proficient in these mutants (S3 Fig.). The results suggest that the BIR defect in hst3Δ hst4Δ cells might be due to inefficient DNA synthesis. The results also reinforce that two-end break repair with no or limited gap is fundamentally different from two-end break repair with large gap or one-end break repair [57]. To further test if hst3Δ hst4Δ mutants are deficient in long repair DNA synthesis, we determined the frequency of gene conversion that requires 3.4-kb gap synthesis in wild-type, hst3Δ hst4Δ, and pol32Δ cells (Fig. 7A). As evident in Fig. 7B, deletion of HST3 and HST4 significantly reduces gene conversion requiring large gap synthesis (tGI354-Gap), although pol32Δ cells exhibited a more severe defect (Fig. 7B). This is consistent with the relatively greater deficiency of pol32Δ mutants in BIR and repair synthesis (see Fig. 1B). Neither hst3Δ hst4Δ nor pol32Δ exhibited reduced gene conversion frequency without a gap (tGI354). Cumulatively, our data support the model that hyper-acetylation of H3K56 inhibits extensive repair synthesis.
Fig. 6. hst3Δ hst4Δ and pol32Δ mutants are defective in the DNA synthesis step of BIR. 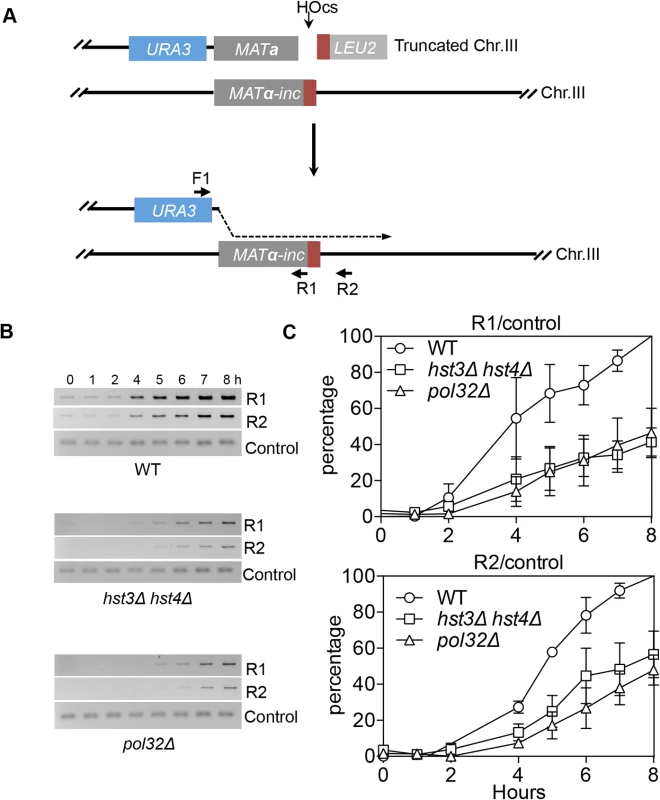
A, BIR synthesis was determined by semi-quantitative PCR using upstream PCR primer (F1) annealing to the URA3 gene inserted upstream of the MATa gene and downstream primers annealing either to a MATα-inc specific sequence (R1) or downstream (R2). B, Representative DNA agarose gel images of semi-quantitative PCR. C, Quantification of the gel images. Plotted are the means of percent repair products at indicated time points post-HO expression from three independent experiments ± s.d. Fig. 7. hst3Δ hst4Δ and pol32Δ mutants are defective in DNA synthesis during gene conversion with a 3.4-kb gap. 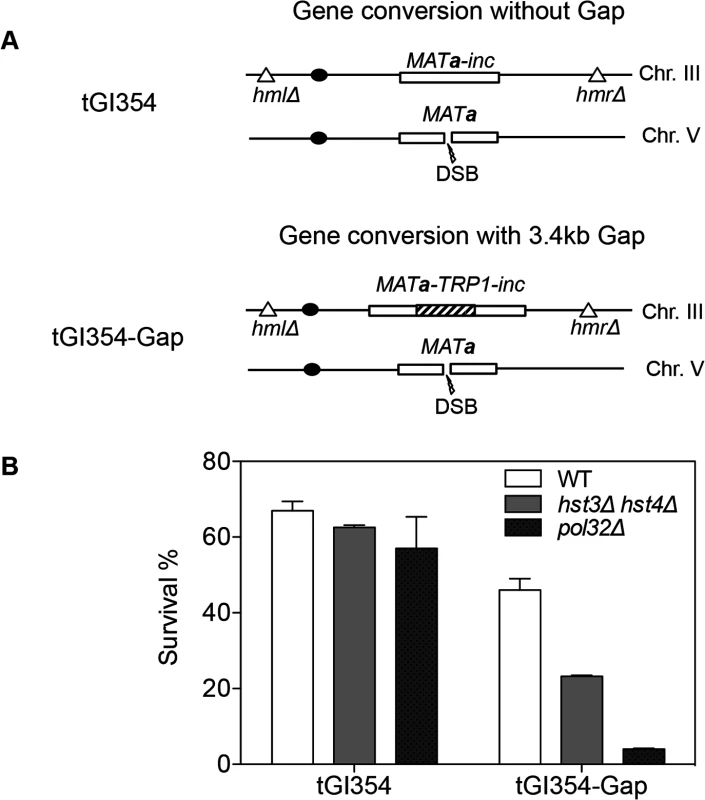
A, The TRP1 gene was inserted into MATa-inc on chromosome III in tGI354 to generate tGI354-gap strain. B, Percent survival in isogenic strains with indicated genotypes and/or a 3.4-kb gap. Plotted are the mean values of three independent experiments ± s.d. Discussion
Chromatin modification is an integral part of DNA replication and repair. By searching for chromatin modifiers involved in BIR, we discovered that deletion of two redundant sirtuins HST3 and HST4 impairs extensive repair synthesis to suppress BIR and gene conversion with long gap synthesis. We also demonstrated that Hst3/Hst4 and Pol32 facilitate recovery from collapsed replication forks when either of the S-phase checkpoint proteins MRC1 or TOF1 is deleted. Deletion of hst3 hst4 also led to an increase in translocation-type GCRs. These results suggest that histone modification modulates BIR and chromosomal integrity.
Hyper-acetylation of histone H3K56 inhibits BIR
BIR entails extensive DNA synthesis even to the end of the chromosome. Since chromatin may impede processive DNA synthesis [11], we surmise that BIR likely depends on chromatin modification, which then enables extensive DNA synthesis to the chromosome end. By surveying several candidate genes that have already been implicated in DNA synthesis and recombination, we discovered multiple chromatin modifiers that impinge on BIR events, supporting our original premise that chromatin modification represents an integral part of BIR. Interestingly, the effect of deleting several chromatin modifier genes on BIR can be explained by their role in resection, highlighting the impact of resection regulation on the frequency of BIR events [39,40,58]. The results also indicate that chromatin regulation comprises a major component in early repair steps, often contributing to the commitment stage of DNA repair. Consistent with this, chromatin configuration has been implicated in repair pathway channeling in several model systems [59,60].
Our results also led to the discovery that deletion of other histone modifiers, specifically HST3 and HST4, renders cells severely deficient in BIR using two different BIR assays, with a repair profile closely resembling that of pol32Δ mutants. Several observations indicated that hyper-acetylation of H3K56 causes BIR deficiency in hst3Δ hst4Δ cells. First, the H3K56Q mutation that mimics persistent acetylation reduced the frequency of BIR, albeit more mildly than hst3Δ hst4Δ. Second, deletion of ASF1 or RTT109, or replacement of lysine 56 with arginine in a histone H3 mutation that blocks H3K56 acetylation rescued the BIR defect in hst3Δ hst4Δ mutants. Third, hst3Δ hst4Δ cells are sensitive to HU in the presence of a collapsed replication fork, but become resistant to HU when ASF1 is deleted. We also showed that the intracellular level of H3K56 is proportional to BIR frequency and that a physiologically relevant level of H3K56 acetylation is sufficient to repress BIR. Lastly, deletion of HST3 HST4 elevated the frequency of translocation-type GCRs, but this increase was partially suppressed by the concomitant deletion of ASF1. Together these results suggest that H3K56 status, but no other HST3 and HST4 targets, is responsible for BIR deficiency and the associated chromosomal instability.
To define how H3K56 acetylation inhibits BIR, we assessed the integrity of the four major recombination steps in BIR: resection, Rad51 binding at donor and recipient DNA molecules, 3’ flap removal and repair synthesis. Our findings suggest that deletion of HST3 and HST4 specifically impairs repair synthesis. This helps explain why only BIR and gene conversion via long gap repair synthesis, but no other recombination pathways, are deficient in hst3Δ hst4Δ cells. The results also support our premise that epigenetic regulation represents a key component of BIR.
How does hyper-acetylation of H3K56 impede repair synthesis? It should be possible that acetylation of H3K56 might compromise the establishment of a stable replication fork at a BIR template. Interestingly, results from chromatin immunoprecipitation (ChIP) assays demonstrated that enrichment of Pol delta is reduced in hst3Δ hst4Δ cells, whereas Pol alpha and Pif1 still associate at the replication fork with almost equal efficiency (S12 Fig.). However, reduced association of Pol delta at a BIR template could simply be the effect but not the cause for BIR deficiency in hst3Δ hst4Δ mutants. Alternatively, acetylation might block replication fork movement by conferring nucleosomes inherently resistant to sliding or evicting from the template strand. Indeed, deletion of HST3 and HST4 in one genetic background (BY4741) led to an increase in spontaneous Rad52-GFP focus formation and elevated sensitivity to HU treatment. However, structural and biochemical analysis of histone octamers carrying H3K56Q did not reveal any major change in nucleosome architecture [18]. Furthermore, hyper-acetylation of H3K56 might impair access to or the activities of topoisomerase I or helicases, each of which relieves helical torsions or catalyzes strand unwinding ahead of a replication fork. In either case, we surmise that frequent fork stalling may ensue in hst3Δ hst4Δ mutants due to non-processive DNA synthesis, which may underlie the increased spontaneous DNA lesions and mutagenesis exhibited by these cells. The observations that many phenotypes of hst3Δ hst4Δ cells can be suppressed at least partially by improving replication processivity support this model [28]. Further studies are needed to determine precisely how H3K56 acetylation inhibits repair synthesis.
In mitotic cells, limiting extensive repair synthesis in recombination should be beneficial as it could reduce mutagenesis inherent to repair synthesis [61]. The size of gene conversion track length also positively correlates with the frequency of crossovers and the rate of loss of heterozygosity [62]. Limiting the amount of repair synthesis should also helps avoid unequal sister chromatid exchange event and sustain repeat stability as seen in rDNA or triplet repeats. Interestingly, DNA damage induces proteolytic degradation of Hst3 [16,29], preserving intracellular level of H3K56 acetylation, which could then help limit excessive repair synthesis during recombinational repair of DNA damage.
Hyper-acetylation of H3K56 interferes with recovery from collapsed forks
Since collapsed replication forks produce one-ended DSB, BIR has been implicated in the restart of replication upon fork collapse [2,3]. Nevertheless, multiple means are still available in cells to complete full genome duplication without re-establishing forks by BIR: a) use of a replication fork approaching from a neighboring origin; b) template switching or re-priming. Accordingly, the precise role of BIR in recovery from a collapsed fork has not yet been fully defined.
Interestingly, we discovered that deletion of POL32 or HST3 and HST4 caused a severe sensitivity to HU when MRC1 is also deleted. Synergism between MRC1 and HST3/HST4 in resistance to HU treatment can be attributed to deficient maintenance of fork integrity in mrc1 mutants but not to an S-phase checkpoint defect, since the checkpoint-deficient mrc1AQ allele fully rescued HU sensitivity. Puzzlingly however, in rad53Δ, where a majority of replication forks collapse, deletion of POL32 or HST3 and HST4 did not sensitize cells to HU treatment. Evidence suggests that the structure of a collapsed fork in mrc1 mutants upon HU treatment associates with extensive ssDNA formation (2–3 kb long) due to uncoupling of the replicative helicase from stalled fork [49]. Extensive ssDNA at the fork then leads to DNA breakage and the repair requires extensive repair synthesis through newly deposited, H3K56 acetylated nucleosomes on the donor DNA molecule (S13 Fig.). The unique architecture associated with stalled forks in mrc1 cells might also be ill-suited for re-priming or bypass-mediated recovery, and thereby relies on the role of BIR like process as a bona fide replication restart mechanism. Alternatively, hyper-HU sensitivity in hst3Δ hst4Δ mrc1Δ mutant could be attributed to the excessive fork collapse due to a deficiency in stalled fork protection under HU treatment. However, we did not detect massive fork collapses in hst3Δ hst4Δ mrc1Δ or mrc1Δ cells as compared to rad53Δ using the assays that monitor the frequency of Rad52-GFP foci or DNA fragmentation by PFGE (Figs. 4 and S14).
The typical collapsed replication forks in cells do not likely feature long ssDNA and such forks in principle contain acetylated H3K56 only at or behind the fork. This raises a possibility that the effect of persistent H3K56 acetylation shown in this study is not applicable to general collapsed fork recovery, but only to those with forks collapsed with long ssDNA as seen in mrc1 - or tof1-deleted cells. Furthermore, H3K56 acetylation is increased in multiple types of cancers [14] and the results shown here could be more relevant to those pathological conditions or at regions of chromatin with more persistent H3K56 acetylation.
Deletion of HST3 and HST4 leads to GCR
Previous studies suggest that both hypo - and hyper-acetylation of histone H3K56 elevates GCR events [20,32,54]. Consistent with this, we also detected dramatically increased GCRs in hst3Δ hst4Δ mutants. However, unlike in an asf1 deletion [54], most GCR events in hst3Δ hst4Δ cells are chromosomal translocations. The results indicate that the molecular defects and pathways leading to GCR events in these strains are distinctly different. Accordingly, deletion of ASF1 partially decreased the GCR rate in hst3Δ hst4Δ cells to a level comparable to that in asf1Δ cells.
We have evidence to suggest that it is unlikely that increased GCR observed in hst3Δ hst4Δ mutants is derived from a BIR defect. If BIR deficiency alone is sufficient for elevated GCR, deletion of POL32 that results in a more severe deficiency in BIR should increase GCR events at a greater extent than in hst3Δ hst4Δ cells. Instead, we found that deletion of POL32 only slightly increases GCRs. Analysis of the types of GCR and the breakpoint junctions in these two mutants further demonstrated that the BIR defect in hst3Δ hst4Δ cells cannot account for its elevated GCR because pol32Δ produces de novo telomere addition but not chromosomal translocations as seen in hst3Δ hst4Δ cells. Why then is GCR increased in hst3Δ hst4Δ mutants? One possibility is that DNA replication forks are unstable in hst3Δ hst4Δ cells and lead to frequent stall and collapse during S-phase. Spontaneous Rad52-GFP, DDC2-GFP, and Mre11-GFP focus formation, as well as Rad53 phosphorylation supports the increase in replication-associated DNA damage in hst3Δ hst4Δ cells, which then probably fuels the formation of GCRs. In fact, many cellular phenotypes of hst3Δ hst4Δ mutants can be explained by fork instability that might lead to such pleiotropic phenotypes. The idea is further supported by the observation that multiple phenotypes associated with hst3Δ hst4Δ mutants including thermosensitivity, drug sensitivity and spontaneous mutagenesis can be partially offset by RAD17, ELG1, or MMS22 deletion that catalyzes the S-phase checkpoint, PCNA unloading or replication fork stability, respectively. However, these mutations did not offset BIR deficiency, indicating that the basis of the observed BIR defect is distinctly different from that of other phenotypes.
Analysis of GCR breakpoint junctions shows that most GCR events in hst3Δ hst4Δ cells are chromosomal translocations with microhomology (MH) at the breakpoint junctions. The presence of MH suggests that GCR events in hst3Δ hst4Δ mutants proceed by microhomology-mediated (MM)-BIR or microhomology-mediated end joining (MMEJ) mechanisms. Notably, deletion of POL32 not only impairs BIR but also the MMEJ pathway [5,63]. Therefore, lack of GCRs in pol32-deleted cells may be due to their inability to carry out MMEJ. Indeed, all the GCR events in pol32 are de novo telomere formation. We also found that hst3Δ hst4Δ mutants are not deficient in MMEJ. Alternatively, it is possible that the GCR events derived from residual BIR mechanisms in hst3Δ hst4Δ cells and the severe BIR deficiency in pol32Δ cells disable the formation of translocation-type GCRs.
In all, our study has revealed the role of H3K56 acetylation in inhibiting extensive repair synthesis and restarting of stressed replication forks in checkpoint-deficient cells. It will be interesting to test if acetylation of H3K56 also inhibits collapsed fork recovery and BIR in human cells. It is noteworthy that HDAC or sirtuin inhibitors have been widely regarded as promising cancer therapeutic agents. Our study has implications for how these inhibitors target checkpoint-defective tumors cells and highlight their utility to treat cancer cells in combination with replication stress agents.
Materials and Methods
Strains
Disomic BIR strains are derivatives of AM1003 with genotype MATa-LEU2-tel/MATα-inc ade1 met13 ura3 leu2-3,112/leu2 thr4 lys5 hmlΔ::ADE1/hmlΔ::ADE3 hmrΔ::HYG ade3::Gal-HO FS2Δ::NAT/FS2 [38]. Ectopic BIR strains are derivatives of JRL346 [40]. HU sensitivity was tested on derivatives of JKM139 with genotype hoΔ MATa hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG’ ura3-52 ade3::GAL::HO [64]. JKM161, YMV80, tGI354 were used for mating-type switch, SSA, and ectopic recombination assays, respectively [65,66]. Gene conversion with a 3.4-kb gap strain was constructed by insertion of a TRP1 containing DNA fragment originating from the pFA6a-TRP1 plasmid into the MATa-inc template of tGI354. The yeast strain expressing MYC epitope tags and degron fusion was constructed by PCR amplification of degron cassette as described in [17,67]. The strain list is shown in S2 Table.
BIR assay
The assay was performed as previously described with minor changes in cell culture[38]. BIR derivative strains were cultured in SC-Ade medium overnight and then transferred to YEP-glycerol medium and cultured for more than 6 h before plating on YEP-galactose plates. Cells cultured in this way are selected against spontaneous chromosome loss. An aliquot of cells was also plated on YEPD plates. After colonies grew up on the plates for 3–5 days, they were replica-plated onto SC-Leu and SC-Ade plates to determine their genotypes. At least 500 individual colonies were scored and at least 2 different isolates with the same genotype were analyzed. Colonies replica-plated from YEPD plates were also examined to determine the background chromosome loss. BIR frequency of ectopic BIR assay was determined as described in [5].
To physically monitor BIR products, 3×107 cells were harvested to make DNA plugs for each time point. Pulsed-field gel electrophoresis (PFGE) was run in a 1.2% agarose gel with switch time 10–35s, voltage 6v/cm, angle 120°, 14°C for 32–40 h (disomic BIR assay) or with switch time 50–70 s, voltage 6v/cm, angle 120°, 14°C for 21–24 h (ectopic BIR assay). PCR products from ADE1 gene or MCH2 fragment were radiolabeled and used to detect BIR bands by Southern blot assay.
To test BIR in Hst3-depleted cells or those treated with NAM, HST4-deleted cells were arrested in G1 by treating them with 10 µg/ml alpha-factor, subjected to incubation at 37°C (non-permissive conditions) for an additional 1.5 h, releasing cells into nocodazole containing (15 µg/ml) media for 2.5 h and inducing HO endonuclease by addition of 2% (w/v) galactose. Aliquots of cells were harvested at each indicated time point and subjected to Western blot to investigate the level of Hst3, H3K56 acetylation using anti-MYC (Sigma) and H3K56 acetylation antibodies (Upstate) and FACS analysis for cell cycle progression. Cells were harvested to prepare DNA plugs for PFGE to analyze BIR products and PCR analysis of repair synthesis. PCR products from ADE1 gene or MCH2 fragment were radiolabeled and used to detect BIR bands or initial repair products by Southern blot.
HU sensitivity test
Logarithmically growing cells were alpha-factor arrested for 3 h and released into YEPD medium containing 150 mM HU. Cells were harvested at each time point, washed twice with water and plated onto YEPD plates. The survival rate was calculated by the number of colonies treated with HU divided by the number of colonies before HU treatment. For the mrc1AQ experiment, Uracil dropout medium instead of YEPD medium was used for cell culture.
Microscopy
Images were taken with a DeltaVision microscopy system (Applied Precision). Each original image contained 12–15 Z-stacks. After deconvolution, images were subjected to quick-projection. The projected pictures were then counted for Rad52-GFP foci using SoftwoRx.
Measurement of 3’ flap cleavage efficiency during gene conversion
3’ flap cleavage efficiency during gene conversion was determined by measuring the retention frequency of yeast centromeric plasmids pFP120 and pFP122 after HO induced DSB as described in [68]. The percentage of plasmid retention is calculated as the fraction of colonies retaining the repaired plasmid on SC–Ura divided by the total number of colonies on YEPD.
ChIP assay
Chromatin immunoprecipitation (ChIP) assays were performed as described previously [69]. Briefly, JKM161 cell cultures grown to a density between 1 × 107 and 2 × 107 cells/ml in pre-induction medium (YEP-glycerol) were induced for HO endonuclease by adding 2% galactose. For immunoprecipitation, the sonicated extracts were incubated with Rad51 antibody (kindly provided by Dr. Patrick Sung P). Quantitative PCR was performed using MATa (break) specific primers and HML (donor) specific primers.
Quantitative PCR-based resection assay
JKM139 derivative cells grown to a density between 1 × 107 and 2 × 107 cells/ml in pre-induction medium (YEP-glycerol) were induced for HO endonuclease by adding 2% galactose. Five ml of cells at different time points were harvested and processed for genomic DNA isolation using Masterpure yeast genomic DNA purification kit (Epicentre). DNA equal to 0.4 ml of cells was digested by 20 units of BsaJ1 enzyme or mock digested without BsaJ1 for 2 h in a 50 μl reaction volume. Samples were then diluted 10 times and subject to quantitative PCR to determine the percentage of resection. The principle of this assay is shown in S8 Fig. The rate of single stranded DNA formation (r) is calculated by r = (RSBsaJ1 digested/ RSMock digested)/(InputBsaJ1 digested/ InputMock digested), where RS indicates PCR value at the BsaJ1 restriction site flanking the HO break and Input indicates PCR value at a site from a different chromosome and does not contain the BsaJ1 restriction site. Since r = X/(X+2(1-X)) where X is the rate of resection, the percentage of resection was calculated by: 100X% = 200r/(r+1). Primer sequences for determining resection at 3.3 kb, 12.6 kb or input are 5’TCCGAACCAATGTCGTCATCCATAGTATC and 5’GGCGCCTTTAATTCATGGTCTTTACCTTT; 5’TTCTACGGCGACTGGTGGAATTGC and 5’ATGTAGCTTGGCTCTTGCTCAAATGC; 5’ACTTAGTTCTGGAGATTATCGCCTTATCG and 5’CTGTCTTTCGCTTAGTTCACCTCTACC, respectively.
Analysis of initial DNA synthesis in BIR
Semi-quantitative PCR was performed with Iproof polymerase (Biorad) with a 2 minute extension time and 20 cycles (allelic BIR assay) or 28 cycles (ectopic BIR assay) using Forward primer F (5’ TACTGGAGTTAGTTGAAGCATTAGG3’) upstream of the HO break inside the URA3 gene and Reverse primer R1 (5’ TGACTTCCAGACGCTATCCTGTGAA 3’) inside the MAT locus at an alpha-specific sequence or R2 (5’GATATTAAGTCCTCCGTCCAATCTG3’) downstream of MATalpha-inc, inside the TAF2 gene (allelic BIR assay) or Forward primer F2 (5’AATATTGTGTGTATGGGCACAAACCCTTG3’) inside the NPR2 gene and Reverse primer R3 (5’ CCTCAATGTCTCTTCTATCGGAAT3’) that anneals to the carboxy terminus of CAN1 gene (ectopic BIR assay). Control PCR was performed with 1 minute extension time for 20 cycles using Forward primer F3 (5’TCCATGCTAGATTAGCACACAGTAA3’) and reverse primer R4 (5’CTTTTGTAGGTGTCCTTAATTTCCA3’). PCR products were resolved in a 1% agarose gel and stained with ethidium bromide. The gel pictures were captured by GEL-LOGIC system and PCR bands were quantified by Carestream software. Inversed images were showed in the Fig. 6 and S11 Fig.
GCR assay
A fluctuation-based test was performed to determine gross chromosomal rearrangements [70]. In brief, 3–5 ml of 11 confluent cultures in YEPD from single colonies were plated on SC medium lacking arginine and supplemented with 60 mg/L L-canavanine and 1 g/L 5-FOA for scoring GCRs. Cells were also diluted and plated on YEPD plates for counting total number of cells. GCR plates were cultured for 5–10 days at 30°C and colony numbers counted. Calculation of GCR rate and determination of junction type (de novo telomere formation or translocation) were as previously described [70].
Supporting Information
Zdroje
1. Pâques F, Haber JE (1999) Multiple Pathways of Recombination Induced by Double-Strand Breaks in Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews 63 : 349–404. 10357855
2. McEachern MJ, Haber JE (2006) Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75 : 111–135. 16756487
3. Llorente B, Smith CE, Symington LS (2008) Break-induced replication: what is it and what is it for? Cell Cycle 7 : 859–864. 18414031
4. Lydeard JR, Lipkin-Moore Z, Sheu, Stillman B, Burgers PM, et al. (2010) Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes & Development 24 : 1133–1144. doi: 10.1155/2015/201379 25632406
5. Lydeard JR, Jain S, Yamaguchi M, Haber JE (2007) Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448 : 820–823. 17671506
6. Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, et al. (2011) Break-induced replication is highly inaccurate. PLoS Biol 9: e1000594. doi: 10.1371/journal.pbio.1000594 21347245
7. Smith CE, Llorente B, Symington LS (2007) Template switching during break-induced replication. Nature 447 : 102–105. 17410126
8. Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, et al. (2013) Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502 : 389–392. doi: 10.1038/nature12584 24025772
9. Donnianni RA, Symington LS (2013) Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci U S A 110 : 13475–13480. doi: 10.1073/pnas.1309800110 23898170
10. Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, et al. (2013) Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 502 : 393–396. doi: 10.1038/nature12585 24025768
11. Groth A, Rocha W, Verreault A, Almouzni G (2007) Chromatin challenges during DNA replication and repair. Cell 128 : 721–733. 17320509
12. Masumoto H, Hawke D, Kobayashi R, Verreault A (2005) A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436 : 294–298. 16015338
13. Xu F, Zhang K, Grunstein M (2005) Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121 : 375–385. 15882620
14. Das C, Lucia MS, Hansen KC, Tyler JK (2009) CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459 : 113–117. doi: 10.1038/nature07861 19270680
15. Xie W, Song C, Young NL, Sperling AS, Xu F, et al. (2009) Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell 33 : 417–427. doi: 10.1016/j.molcel.2009.02.004 19250903
16. Maas NL, Miller KM, DeFazio LG, Toczyski DP (2006) Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell 23 : 109–119. 16818235
17. Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, et al. (2006) The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol 16 : 1280–1289. 16815704
18. Watanabe S, Resch M, Lilyestrom W, Clark N, Hansen JC, et al. (2010) Structural characterization of H3K56Q nucleosomes and nucleosomal arrays. Biochim Biophys Acta 1799 : 480–486. doi: 10.1016/j.bbagrm.2010.01.009 20100606
19. Neumann H, Hancock SM, Buning R, Routh A, Chapman L, et al. (2009) A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 36 : 153–163. doi: 10.1016/j.molcel.2009.07.027 19818718
20. Driscoll R, Hudson A, Jackson SP (2007) Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315 : 649–652. 17272722
21. Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, et al. (2007) Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315 : 653–655. 17272723
22. Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, et al. (2006) Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci U S A 103 : 6988–6993. 16627621
23. Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, et al. (2008) Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134 : 244–255. doi: 10.1016/j.cell.2008.06.018 18662540
24. Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, et al. (2008) Cell cycle - and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet 4: e1000270. doi: 10.1371/journal.pgen.1000270 19023413
25. Williams SK, Truong D, Tyler JK (2008) Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci U S A 105 : 9000–9005. doi: 10.1073/pnas.0800057105 18577595
26. Yu Y, Song C, Zhang Q, DiMaggio PA, Garcia BA, et al. (2012) Histone H3 lysine 56 methylation regulates DNA replication through its interaction with PCNA. Mol Cell 46 : 7–17. doi: 10.1016/j.molcel.2012.01.019 22387026
27. Wurtele H, Kaiser GS, Bacal J, St-Hilaire E, Lee EH, et al. (2012) Histone H3 lysine 56 acetylation and the response to DNA replication fork damage. Mol Cell Biol 32 : 154–172. doi: 10.1128/MCB.05415-11 22025679
28. Celic I, Verreault A, Boeke JD (2008) Histone H3 K56 hyperacetylation perturbs replisomes and causes DNA damage. Genetics 179 : 1769–1784. doi: 10.1534/genetics.108.088914 18579506
29. Thaminy S, Newcomb B, Kim J, Gatbonton T, Foss E, et al. (2007) Hst3 is regulated by Mec1-dependent proteolysis and controls the S phase checkpoint and sister chromatid cohesion by deacetylating histone H3 at lysine 56. J Biol Chem 282 : 37805–37814. 17977840
30. Munoz-Galvan S, Jimeno S, Rothstein R, Aguilera A (2013) Histone H3K56 acetylation, Rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet 9: e1003237. doi: 10.1371/journal.pgen.1003237 23357952
31. Ide S, Saka K, Kobayashi T (2013) Rtt109 prevents hyper-amplification of ribosomal RNA genes through histone modification in budding yeast. PLoS Genet 9: e1003410. doi: 10.1371/journal.pgen.1003410 23593017
32. Kadyrova LY, Mertz TM, Zhang Y, Northam MR, Sheng Z, et al. (2013) A reversible histone H3 acetylation cooperates with mismatch repair and replicative polymerases in maintaining genome stability. PLoS Genet 9: e1003899. doi: 10.1371/journal.pgen.1003899 24204308
33. Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, et al. (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev 9 : 2888–2902. 7498786
34. Yang B, Miller A, Kirchmaier AL (2008) HST3/HST4-dependent deacetylation of lysine 56 of histone H3 in silent chromatin. Mol Biol Cell 19 : 4993–5005. doi: 10.1091/mbc.E08-05-0524 18799617
35. Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, et al. (2008) Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134 : 231–243. doi: 10.1016/j.cell.2008.06.035 18662539
36. Kim JA, Haber JE (2009) Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc Natl Acad Sci U S A 106 : 1151–1156. doi: 10.1073/pnas.0812578106 19164567
37. Feser J, Truong D, Das C, Carson JJ, Kieft J, et al. (2010) Elevated histone expression promotes life span extension. Mol Cell 39 : 724–735. doi: 10.1016/j.molcel.2010.08.015 20832724
38. Deem A, Barker K, VanHulle K, Downing B, Vayl A, et al. (2008) Defective Break-Induced Replication Leads to Half-Crossovers in Saccharomyces cerevisiae. Genetics 179 : 1845–1860. doi: 10.1534/genetics.108.087940 18689895
39. Marrero VA, Symington LS (2010) Extensive DNA end processing by exo1 and sgs1 inhibits break-induced replication. PLoS Genet 6: e1001007. doi: 10.1371/journal.pgen.1001007 20628570
40. Lydeard JR, Lipkin-Moore Z, Jain S, Eapen VV, Haber JE (2010) Sgs1 and exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS Genet 6: e1000973. doi: 10.1371/journal.pgen.1000973 20523895
41. Adkins NL, Niu H, Sung P, Peterson CL (2013) Nucleosome dynamics regulates DNA processing. Nat Struct Mol Biol 20 : 836–842. doi: 10.1038/nsmb.2585 23728291
42. van Attikum H, Fritsch O, Gasser SM (2007) Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J 26 : 4113–4125. 17762868
43. Chen X, Cui D, Papusha A, Zhang X, Chu CD, et al. (2012) The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489 : 576–580. doi: 10.1038/nature11355 22960743
44. Wysocki R, Javaheri A, Allard S, Sha F, Cote J, et al. (2005) Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol 25 : 8430–8443. 16166626
45. Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, et al. (2008) Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. Embo j 27 : 1502–1512. doi: 10.1038/emboj.2008.81 18418382
46. Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, et al. (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412 : 557–561. 11484058
47. Osborn AJ, Elledge SJ (2003) Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev 17 : 1755–1767. 12865299
48. Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, et al. (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3 : 958–965. 11715016
49. Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, et al. (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424 : 1078–1083. 12944972
50. Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P (2005) Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19 : 699–706. 16137625
51. Alabert C, Bianco JN, Pasero P (2009) Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J 28 : 1131–1141. doi: 10.1038/emboj.2009.75 19322196
52. Han J, Zhou H, Li Z, Xu RM, Zhang Z (2007) Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem 282 : 28587–28596. 17690098
53. Myung K, Datta A, Kolodner RD (2001) Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104 : 397–408. 11239397
54. Myung K, Pennaneach V, Kats ES, Kolodner RD (2003) Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci U S A 100 : 6640–6645. 12750463
55. Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 : 806–810. 17314980
56. Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL (2013) A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science 340 : 195–199. doi: 10.1126/science.1229758 23580526
57. Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, et al. (2009) A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev 23 : 291–303. doi: 10.1101/gad.1751209 19204116
58. Chung WH, Zhu Z, Papusha A, Malkova A, Ira G (2010) Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet 6: e1000948. doi: 10.1371/journal.pgen.1000948 20485519
59. Pai CC, Deegan RS, Subramanian L, Gal C, Sarkar S, et al. (2014) A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat Commun 5 : 4091. doi: 10.1038/ncomms5091 24909977
60. Scully R, Xie A (2013) Double strand break repair functions of histone H2AX. Mutat Res 750 : 5–14. doi: 10.1016/j.mrfmmm.2013.07.007 23916969
61. Hicks WM, Kim M, Haber JE (2010) Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329 : 82–85. doi: 10.1126/science.1191125 20595613
62. Lo YC, Paffett KS, Amit O, Clikeman JA, Sterk R, et al. (2006) Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol Cell Biol 26 : 4086–4094. 16705162
63. Villarreal DD, Lee K, Deem A, Shim EY, Malkova A, et al. (2012) Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet 8: e1003026. doi: 10.1371/journal.pgen.1003026 23144625
64. Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, et al. (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94 : 399–409. 9708741
65. Prakash R, Satory D, Dray E, Papusha A, Scheller J, et al. (2009) Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev 23 : 67–79. doi: 10.1101/gad.1737809 19136626
66. Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, et al. (2002) Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell 10 : 373–385. 12191482
67. Sanchez-Diaz A, Kanemaki M, Marchesi V, Labib K (2004) Rapid depletion of budding yeast proteins by fusion to a heat-inducible degron. Sci STKE 2004: PL8. 15010550
68. Li F, Dong J, Pan X, Oum JH, Boeke JD, et al. (2008) Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol Cell 30 : 325–335. doi: 10.1016/j.molcel.2008.02.028 18471978
69. Shim EY, Ma J-L, Oum J-H, Yanez Y, Lee SE (2005) The Yeast Chromatin Remodeler RSC Complex Facilitates End Joining Repair of DNA Double-Strand Breaks. Molecular and Cellular Biology 25 : 3934–3944. 15870268
70. Motegi A, Myung K (2007) Measuring the rate of gross chromosomal rearrangements in Saccharomyces cerevisiae: A practical approach to study genomic rearrangements observed in cancer. Methods 41 : 168–176. 17189859
Štítky
Genetika Reprodukční medicína
Článek 2014 Reviewer Thank YouČlánek Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic TranslationČlánek Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate CellsČlánek K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in ArabidopsisČlánek A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis inČlánek Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in MiceČlánek The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- 2014 Reviewer Thank You
- Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for and Myocardial Infarction
- African Glucose-6-Phosphate Dehydrogenase Alleles Associated with Protection from Severe Malaria in Heterozygous Females in Tanzania
- Genomics of Divergence along a Continuum of Parapatric Population Differentiation
- microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes
- A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in
- Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic Translation
- Partially Redundant Enhancers Cooperatively Maintain Mammalian Expression Above a Critical Functional Threshold
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Mutations in Result in Ocular Coloboma, Microcornea and Cataracts
- A Genome-Wide Hybrid Incompatibility Landscape between and
- Recurrent Evolution of Melanism in South American Felids
- Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells
- Tissue Expression Pattern of PMK-2 p38 MAPK Is Established by the miR-58 Family in
- Essential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
- Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- RR-1 Cuticular Protein TcCPR4 Is Required for Formation of Pore Canals in Rigid Cuticle
- GC-Content Evolution in Bacterial Genomes: The Biased Gene Conversion Hypothesis Expands
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
- K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
- A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in
- HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
- Mitochondrial and Cytoplasmic ROS Have Opposing Effects on Lifespan
- Structured Observations Reveal Slow HIV-1 CTL Escape
- An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging
- Combining Natural Sequence Variation with High Throughput Mutational Data to Reveal Protein Interaction Sites
- Transhydrogenase Promotes the Robustness and Evolvability of Deficient in NADPH Production
- Regulators of Autophagosome Formation in Muscles
- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Eye Selector Logic for a Coordinated Cell Cycle Exit
- Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations
- The DNA Polymerase δ Has a Role in the Deposition of Transcriptionally Active Epigenetic Marks, Development and Flowering
- Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice
- Membrane Recognition and Dynamics of the RNA Degradosome
- P-TEFb, the Super Elongation Complex and Mediator Regulate a Subset of Non-paused Genes during Early Embryo Development
- is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure
- A Pleiotropy-Informed Bayesian False Discovery Rate Adapted to a Shared Control Design Finds New Disease Associations From GWAS Summary Statistics
- Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers
- and Hyperdrive Mechanisms (in Mouse Meiosis)
- Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells
- Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma by Repressing Multiple Targets
- Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
- Abscission Is Regulated by the ESCRT-III Protein Shrub in Germline Stem Cells
- Temperature Stress Mediates Decanalization and Dominance of Gene Expression in
- Transcriptome Wide Annotation of Eukaryotic RNase III Reactivity and Degradation Signals
- The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway
- Sex-specific -regulatory Variation on the X Chromosome
- Regulation of Toll-like Receptor Signaling by the SF3a mRNA Splicing Complex
- Modeling of the Human Alveolar Rhabdomyosarcoma Chromosome Translocation in Mouse Myoblasts Using CRISPR-Cas9 Nuclease
- Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
- TIM Binds Importin α1, and Acts as an Adapter to Transport PER to the Nucleus
- Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication
- The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
- Fatty Acid Synthase Cooperates with Glyoxalase 1 to Protect against Sugar Toxicity
- Region-Specific Activation of mRNA Translation by Inhibition of Bruno-Mediated Repression
- An Essential Role of the Arginine Vasotocin System in Mate-Guarding Behaviors in Triadic Relationships of Medaka Fish ()
- Interaction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
- Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
- Prodomain Removal Enables Neto to Stabilize Glutamate Receptors at the Neuromuscular Junction
- Recent Selective Sweeps in North American Show Signatures of Soft Sweeps
- Identification and Functional Analysis of Healing Regulators in
- A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2
- Drosophila Casein Kinase I Alpha Regulates Homolog Pairing and Genome Organization by Modulating Condensin II Subunit Cap-H2 Levels
- The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Ovary
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



