-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
In asymmetrically dividing cells, proper positioning of the mitotic spindle relative to polarity determinants is crucial to ensure the unequal fate of daughter cells. In stem cells, derangement of the mechanisms controlling asymmetric cell division, including spindle positioning, affects the developmental fate of daughter cells and can promote tumourigenesis. The budding yeast Saccharomyces cerevisiae is an outstanding model system to study spindle positioning and its links with cell cycle progression. Indeed, budding yeast has redundant mechanisms driving spindle positioning and a “spindle position checkpoint” (SPOC) that delays cell division whenever the spindle is not properly aligned. The target of the SPOC is the small GTPase Tem1 that controls both spindle positioning and mitotic exit and whose activity can be inhibited by the GTPase-activating protein Bub2/Bfa1. Tem1, Bub2 and Bfa1 form a complex at spindle poles that becomes asymmetric and accumulates on one spindle pole when the spindle is properly aligned, while it remains symmetric in case of spindle mispositioning. Through expression of several mutant or chimeric proteins leading to symmetric distribution of the Bub2/Bfa1/Tem1 complex, we establish that asymmetry of these proteins does not drive mitotic exit but rather it contributes to spindle alignment.
Published in the journal: . PLoS Genet 11(2): e32767. doi:10.1371/journal.pgen.1004938
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004938Summary
In asymmetrically dividing cells, proper positioning of the mitotic spindle relative to polarity determinants is crucial to ensure the unequal fate of daughter cells. In stem cells, derangement of the mechanisms controlling asymmetric cell division, including spindle positioning, affects the developmental fate of daughter cells and can promote tumourigenesis. The budding yeast Saccharomyces cerevisiae is an outstanding model system to study spindle positioning and its links with cell cycle progression. Indeed, budding yeast has redundant mechanisms driving spindle positioning and a “spindle position checkpoint” (SPOC) that delays cell division whenever the spindle is not properly aligned. The target of the SPOC is the small GTPase Tem1 that controls both spindle positioning and mitotic exit and whose activity can be inhibited by the GTPase-activating protein Bub2/Bfa1. Tem1, Bub2 and Bfa1 form a complex at spindle poles that becomes asymmetric and accumulates on one spindle pole when the spindle is properly aligned, while it remains symmetric in case of spindle mispositioning. Through expression of several mutant or chimeric proteins leading to symmetric distribution of the Bub2/Bfa1/Tem1 complex, we establish that asymmetry of these proteins does not drive mitotic exit but rather it contributes to spindle alignment.
Introduction
Asymmetric cell division generates two daughter cells genetically identical but that differ in fate and/or in size and cytoplasmic material. During asymmetric cell division, polarity factors are first concentrated to specific locations to define the poles of cell division. Afterwards the spindle orients according to these polarity cues to segregate one set of chromosomes towards a given polarity determinant and the other away from it, thereby generating two unequal daughter cells (reviewed in [1–3]). Correct spindle positioning is therefore critical to preserve the right lineage of asymmetrically dividing cells. Accordingly, spindle mispositioning in asymmetrically dividing stem cells, which normally generate one daughter stem cell with self-renewal potential and one cell destined to differentiation, steers tumourigenesis by increasing the pool of undifferentiated stem cells [4, 5]. Surveillance mechanisms, or checkpoints, must therefore respond to spindle positioning errors and delay cell cycle progression until the mitotic spindle is properly oriented with respect to the cell polarity axis [6, 7].
The budding yeast Saccharomyces cerevisiae is a widely recognized model system to study asymmetric cell division. Spindle positioning in budding yeast requires either one of two redundant pathways, one that depends on the APC (Adenomatous Polyposis Coli)-related protein Kar9, and the other on dynein (reviewed in [8]). Spindle positioning errors are monitored by a surveillance mechanism, referred to as spindle position checkpoint (SPOC), that delays mitotic exit and cytokinesis to provide the time for proper spindle realignment (reviewed in [6, 9]). The target of the SPOC is a small GTPase called Tem1, which acts as molecular switch for the activation of a kinase cascade related to the Hippo pathway and named Mitotic Exit Network (MEN). In the fission yeast S. pombe a kinase cascade similar to MEN and referred to as Septation Initiation Network (SIN) triggers cytokinesis [10]. The MEN effector of Tem1 is the kinase Cdc15, which in turn promotes the activation of the downstream Mob1/Dbf2 kinase complex that ultimately leads to activation of the Cdc14 phosphatase [11]. Cdc14 is the main phosphatase that in budding yeast counteracts the activity of cyclin-dependent kinases (CDKs), and it is essential for mitotic exit and cytokinesis by dephosphorylating CDK substrates, as well as by triggering inactivation of mitotic CDKs [12]. Cdc14 is sequestered in the nucleolus in an inactive form throughout most of the cell cycle, until it is released and activated. Although the MEN is necessary for the full release of Cdc14 into the cytoplasm to promote mitotic exit [13, 14], another pathway called FEAR (Cdc Fourteen Early Anaphase Release) causes a partial release of Cdc14 from the nucleolus into the nucleus at the metaphase-to-anaphase transition [15]. The FEAR pathway involves the polo kinase Cdc5, the redundant Spo12 and Bns1 proteins, and the separase Esp1, which by inhibiting the phosphatase PP2ACdc55 allows the dissociation of Cdc14 from its nucleolar inhibitor Net1 [15, 16]. The FEAR-mediated activation of Cdc14 in anaphase is thought to regulate spindle dynamics and to contribute to timely activation of the MEN (reviewed in [17]).
Recent data have shown that MEN is not only important for triggering mitotic exit in telophase, but also has an earlier function in metaphase to promote correct spindle positioning along the polarity axis [18]. In most MEN mutants, except for cdc14, spindles are indeed misoriented relative to the cell division plane. Notably, the Mob1/Dbf2 kinase was found to phosphorylate the spindle positioning Kar9 protein, thereby favouring its concentration on astral microtubules emanating from only one of the two spindle poles [18]. Asymmetric distribution of Kar9 at spindle poles in metaphase is in turn crucial for proper spindle positioning because it targets the Kar9-decorated aster to the bud, due to Kar9 interaction with the type V myosin Myo2 [19].
The yeast centrosomes, named spindle pole bodies (SPBs), play an important role in the regulation of mitotic exit, as they act as a scaffold for MEN components, such as Tem1 and its downstream kinases (reviewed in [20]). The constitutive SPB component Nud1 recruits MEN proteins to SPBs and is essential for mitotic exit [21], suggesting that binding of one or several MEN factors to SPBs is required for mitotic exit. Consistently, Tem1 association to SPBs is critical for MEN activation [22].
Like all GTPases, Tem1 is active when bound to GTP and inactive in its GDP-bound form. The common element of the GTPase superfamily is the 160–180 residue G domain involved in nucleotide binding [23]. Within the G domain two flexible “switch regions”, referred to as Switch I and II undergo the most dramatic structural rearrangement upon GTP hydrolysis and therefore define the major conformational changes conferred by GTP versus GDP binding [24]. On the basis of sequence alignment with human Ras, Switch I and II in Tem1 correspond to residues 50–55 and 77–84, respectively.
Upon spindle misalignment the two-component GTPase-activating protein (GAP) Bub2/Bfa1 inactivates Tem1 by stimulating GTP hydrolysis [25, 26]. GTPase-activating proteins accelerate GTP hydrolysis, promoting the GDP-bound inactive form of GTPases [27, 28]. The GAP activity of the Bub2/Bfa1 complex resides on Bub2, which carries a TBC domain (Tre-2, Bub2 and Cdc16; [29]), whereas Bfa1 mediates Bub2 interaction with Tem1 and prevents Tem1 dissociation from guanine nucleotides, thereby acting as guanine dissociation inhibitor (GDI) [25, 26, 30, 31].
Often, the release of GDP from GTPases is a slow and thermodynamically unfavourable reaction. This is why GTPase activation requires in most cases the intervention of nucleotide exchange factors (GEFs) that catalyse the release of GDP, promoting its replacement by GTP [27]. The identity of the GEF(s) for Tem1, if any, remains elusive. The early proposal based on genetic data that the Lte1 protein might be the GEF for Tem1 has not been confirmed by biochemical assays [30]. Therefore, if inactivation of a GEF for Tem1 could play any role in the SPOC, besides Tem1 inhibition by the GAP Bub2/Bfa1, remains to be established.
The Kin4 protein kinase is a key component of the SPOC (reviewed in [6]). During spindle misalignment it phosphorylates Bfa1, thereby preventing the inhibitory phosphorylation of the GAP Bub2/Bfa1 by the polo kinase [32]. During the unperturbed cell cycle Kin4 is strategically restricted to the mother cell compartment, where it is thought to sense the anomalous persistence of both SPBs in anaphase. In addition, it is present on both SPBs of misaligned spindles [32–35].
Localization of MEN and SPOC proteins to the SPBs changes during the cell cycle, in that some proteins (like Cdc15, Mob1 and Dbf2) are loaded on both SPBs at the anaphase onset, whereas other proteins (like Tem1, Bfa1 and Bub2) are present on both SPBs already in metaphase and their localization becomes much more asymmetric in anaphase, when they preferentially accumulate on the old, bud-directed SPB [36–44]. Moreover, the position of the spindle seems to play a role in controlling the asymmetric localization of Tem1, Bub2 and Bfa1 in anaphase. Indeed, these proteins localize less strongly but more symmetrically on the two SPBs when a misoriented spindle elongates within the mother cell and the SPOC turns on [36, 38, 40]. Whether the asymmetric localization of Tem1 or its GAP is important for triggering MEN signalling remains to be elucidated. Remarkably, the SIN counterparts of several MEN components also localize asymmetrically on SPBs during anaphase, with the homologs of the GAP components Bub2 (Cdc16) and Bfa1 (Byr4) occupying one SPB, while the GTP-bound form of the GTPase Spg1 and its effector kinase Cdc7 occupy the other [45, 46]. Thus, in spite of the different modes of cell division in the two yeasts (asymmetric in S. cerevisiae and symmetric in S. pombe) MEN and SIN signalling is asymmetric in both. SIN asymmetry has been proposed to be crucial for timely cytokinesis, as cdc16 and byr4 mutants where Spg1 and Cdc7 are symmetric in anaphase undergo multiple rounds of septation [46, 47].
In S. cerevisiae, Kin4 was found to increase the turnover of the Bub2/Bfa1 at SPBs. However, Kin4 does not promote asymmetric localization of Bub2/Bfa1 on properly oriented spindles [36]. Chimeric proteins obtained by fusing Bub2 or Bfa1 to the structural SPB component Cnm67 cause unscheduled mitotic exit in the presence of mispositioned spindles, which led to the proposal that high turnover of Bub2/Bfa1 might be important to inhibit Tem1 in the cytoplasm during SPOC activation [36]. Conversely, a modified version of Bub2 carrying 9 myc epitopes at the C terminus localizes the GAP and Tem1 rather symmetrically at SPBs and prevents mitotic exit in some sensitized MEN mutant backgrounds [25].
In order to shed light onto the relationship between Bub2/Bfa1 symmetry and SPOC response, we report the characterization of a series of mutants altering either the Bub2/Bfa1 subunits or Tem1 and causing symmetric localization of Tem1 and its GAP during properly oriented anaphase. Remarkably, these mutant proteins as a whole tend to activate, rather than inhibit, the MEN. In addition, they lead to more symmetric distribution of Kar9 on spindle poles and to spindle positioning defects, indicating that a delicate balance between MEN activation and inactivation is required for proper spindle alignment.
Results
Bub2 GAP activity involves a ‘dual finger’ mechanism and promotes Bub2/Bfa1 disappearance from the mother SPB
The catalytic mechanism of GTPase-activating proteins (GAPs) requires an ‘arginine-finger’, where the lateral chain of a conserved arginine (R85 for Bub2, [25, 29]), interacts with the nucleotide-binding site of a G protein, thus stimulating hydrolysis of the γ−phosphate. A new catalytic mechanism, called “dual finger”, was proposed for the family of GAPs with TBC (Tre-2, Bub2 and Cdc16) domain. According to the dual finger mechanism a conserved glutamine residue contributes to stimulate GTP hydrolysis together with the canonical catalytic arginine [48]. To investigate if Bub2 acts indeed via a dual finger mechanism, we generated a mutant Bub2 variant, Bub2-Q132L, where we replaced by leucine the conserved glutamine at position 132 that identifies the glutamine finger on the basis of sequence alignment [48]. Bacterially purified His-tagged Tem1, Maltose Binding Protein (MBP)-tagged Bfa1 and glutathione-S transferase (GST)-tagged Bub2 or Bub2-Q132L proteins were used in in vitro GTPase assays, as previously described [25, 26]. The rate of GTP hydrolysis and dissociation was measured using Tem1 bound to γ[32P]-GTP, whereas the rate of GTP dissociation alone was measured using Tem1 bound to the non-hydrolysable GTP analogue γ[35S]-GTP (Fig. 1A). As shown in Fig. 1A, the kinetics of radioactivity loss from wild type Tem1 loaded with either γ[32P]-GTP or γ[35S]-GTP were very similar, suggesting that Tem1 on its own mostly dissociates GTP without hydrolysing it. The presence of Bfa1 stabilized Tem1 in the GTP-bound form (Fig. 1B), whereas Bub2 stimulated Tem1 GTPase activity in the presence of Bfa1 (Fig. 1C), but not GTP dissociation (Fig. 1B). We then compared the GAP activity of purified GST-Bub2 and GST-Bub2-Q132L. Interestingly, Bub2-Q132L did not display any GAP activity towards GTP-bound Tem1 (Fig. 1C), behaving as the GAP–dead mutant Bub2-R85A previously characterized [25]. Furthermore, it did not stimulate GTP dissociation, exactly like wild type Bub2 (Fig. 1B).
Fig. 1. Bub2 GAP activity involves a ‘dual finger’ mechanism and promotes Bub2/Bfa1 clearance from the mother SPB. 
A-C: Bacterially purified GST-Bub2 or GST-Bub2-Q132L, MBP-Bfa1 and 6xHis-Tem1 proteins were used to measure the kinetics of hydrolysis+dissociation (γ[32P]GTP) or dissociation only (γ[35S]GTP) using a filter binding assay (see Materials and Methods). Graphs show average values and standard deviations from three independent experiments. D: Exponentially growing cultures of the indicated strains were shifted to nocodazole containing medium at t = 0. Cell samples were withdrawn at the indicated time for FACS analysis of DNA contents. E: The percentage of cells with binucleate cell bodies accompanied or not by a checkpoint defect (indicated by re-budding in the absence of proper chromosome segregation) was scored in cycling cultures of the indicated strains shifted either to 14°C for 16h (left graph) or to 37°C for 3h (right graph). F-G: Exponentially growing cells with the indicated genotypes were arrested in G1 by α-factor and released into fresh medium at time 0. At 70’ after release α-factor was re-added to prevent cells from entering a second cell cycle. Cell samples were collected for FACS analysis of DNA contents (F) and for tubulin staining by indirect immunofluorescence (G). H: Cells were treated as in (F-G). TCA extracts were prepared from cell samples at the indicated time points to monitor kinetics of Bfa1-HA6 phosphorylation and Clb2 accumulation and degradation by western blot analysis. Pgk1 was used as loading control. I: Protein extracts from cells expressing the indicated tagged proteins were used for immunoprecipitation with an anti-HA affinity resin. Western blot analysis was then performed with anti-GFP and anti-HA antibodies. The input represents 1/25th of the total extract used for each IP. J-K: Localization of eGFP- tagged Bub2/Bub2-Q132L, Tem1, Bfa1 (J) and Cdc15-GFP (K) was analysed by fluorescence microscopy after formaldehyde fixation. In vivo, the Q132L substitution completely abolished the checkpoint function of Bub2. Indeed, similar to bub2Δ cells, bub2-Q132L cells escaped the mitotic arrest upon nocodazole treatment, as indicated by their ability to re-replicate their chromosomes (Fig. 1D). Checkpoint response to spindle misalignment was also impaired in bub2-Q132L cells. Indeed, when spindle mispositioning was induced by DYN1 or KAR9 deletion [49, 50] bub2-Q132L cells did not arrest in mitosis as large budded cells but re-budded, similar to bub2Δ cells (Fig. 1E). Thus, consistent with the proposed model [48], Bub2 GAP activity, and thereby its role in the SPOC, relies on a dual finger mechanism involving two catalytic residues, R85 and Q132.
The bub2-Q132L allele did not accelerate mitotic exit during the unperturbed cell cycle. Indeed, synchronized bub2-Q132L cells could divide and disassemble bipolar spindles with wild type kinetics (Fig. 1F–G). Furthermore, kinetics of degradation of the main mitotic cyclin Clb2 were very similar in wild type and bub2-Q132L cells (Fig. 1H). Interestingly, we found that cell cycle-dependent phosphorylation of Bfa1, which promotes mitotic exit [51], was abolished in bub2-Q132L cells, in agreement with the recent proposal that it requires Bub2 activity [52].
We then asked if Bub2-Q132L could still interact efficiently with Bfa1 and Tem1. Immunoprecipitations of Bub2 or Bub2-Q132L tagged with three HA epitopes showed that both proteins pulled down roughly the same amounts of GFP-tagged Bfa1. In contrast, Bub2-Q132L precipitated a higher amount of GFP-tagged Tem1 than wild type Bub2 (Fig. 1I), suggesting that abolishing the GAP catalytic activity of the Bub2/Bfa1 complex stabilizes the interaction between Tem1 and its GAP.
Because lack of Bub2 GAP activity through the bub2-R85A allele leads to increased symmetric localization of Bub2 to SPBs in anaphase [25], we analyzed the subcellular distribution of eGFP-tagged Bub2-Q132L. In contrast to wild type Bub2, which was almost exclusively present on the bud-directed SPB in 84% of anaphase cells, Bub2-Q132L-HA3 was found on both SPBs in 97% of cells in anaphase. In addition, Bfa1 and Tem1 were also more symmetrically localized on the SPBs of bub2-Q132L cells than they were in wild type cells during the same cell cycle stage (Fig. 1J). We therefore conclude that, consistent with previous data, interfering with Bub2 GAP activity affects the asymmetry of the Tem1/Bub2/Bfa1 complex on anaphase spindle poles.
Finally, we analysed the localization of the Tem1 effector kinase Cdc15 in bub2-Q132L cells. We found GFP-tagged Cdc15 on the SPBs of metaphase spindles in 55% of the cells. Deletion of BUB2 or its replacement with the bub2-Q132L allele increased both the total percentage of cells with Cdc15 at SPBs (83% and 80%, respectively, Fig. 1K) and the percentage of cells with symmetrically localized Cdc15 in metaphase (36% in bub2Δ and bub2-Q132L cells versus 11% of wild type cells). Thus, lack of Bub2 GAP activity leads to more efficient recruitment of Cdc15 at spindle poles.
Constitutive targeting of Bfa1 to SPBs facilitates mitotic exit by recruiting Tem1 to SPBs
Since spindle misalignment leads to persistent residence of Bub2/Bfa1 on both SPBs [40], we and others proposed that symmetric distribution of the GAP complex might lead to inhibition of Tem1 [25, 40]. This idea was further supported by our previous finding that a myc-tagged variant of Bub2 (Bub2-myc9) localizing mostly symmetrically on SPBs was lethal and prevented mitotic exit in sensitized backgrounds [25]. On the other hand, the Bub2/Bfa1 complex is required throughout most of the cell cycle for Tem1 association with SPB, which in turn triggers its activation [22, 40]. To assess the importance of Bub2/Bfa1 asymmetry at SPBs, we tethered Bfa1 or Bub2 to both SPBs by fusing them to the structural SPB component Spc72. We confirmed that in about 90% of the cells the Spc72-Bfa1 chimeric protein localised constitutively to the SPBs throughout the cell cycle (Fig. 2A) and was able to recruit Tem1 to both SPBs in 74% of anaphase cells, as opposed to 35% of wild type cells (Fig. 2B).
Fig. 2. Constitutive targeting of Bfa1 to SPBs facilitates mitotic exit by recruiting Tem1 to SPBs. 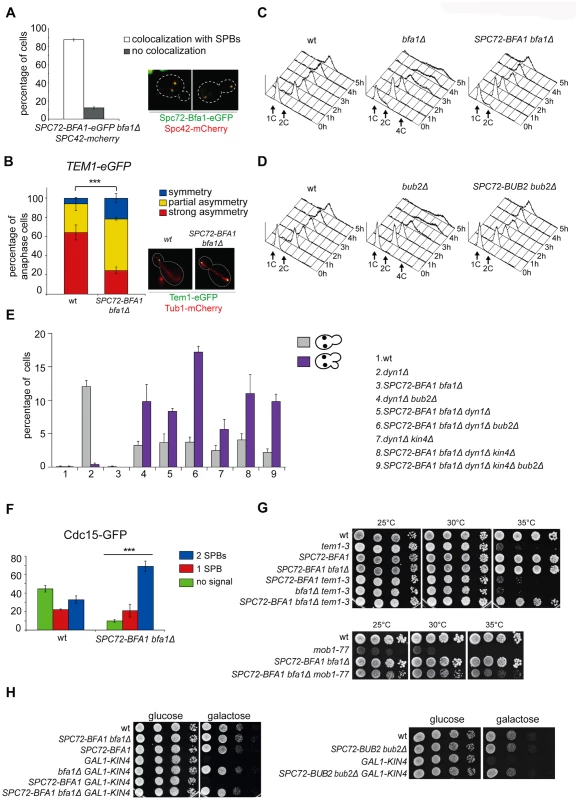
A-B: Cycling cells co-expressing Spc72-Bfa1-eGFPand Spc42-mCherry to mark the SPB (upper panel) or co-expressing Tem1-eGFP and Tub1-GFP (to mark microtubules, lower panel) were analysed to study the distribution of Spc72-Bfa1-eGFP (A) and Tem1-eGFP (B) at SPBs in SPC72-BFA1 bfa1Δ cells. C-D: Cycling cells with the indicated genotypes were shifted into nocodazole containing medium (t = 0). Cell samples were withdrawn at the indicated times for FACS analysis of DNA contents. E: The percentage of cells with binucleate cell bodies accompanied or not by a SPOC defect was scored after propidium iodide staining of cycling cultures of cells with the indicated genotypes after shift to 14°C for 16h. The histograms on the right side represent the DNA contents of the same cells as measured by FACS analysis. F: Percentage of metaphase cells with Cdc15-GFP at 0, 1 or 2 SPBs was scored in the indicated strains after formaldehyde fixation. Metaphases were identified by means of the Tub1-mCherry co-expressed marker. G: Serial dilutions of stationary phase cultures of the indicated strains were spotted on YPD and incubated at the indicated temperature. H: Serial dilutions of stationary phase cultures of the indicated strains were spotted on YP medium containing either glucose or galactose and incubated at 25°C for 48h. Both Spc72-Bfa1 and Spc72-Bub2 chimeric proteins were functional based on their ability to complement lack of endogenous BFA1 or BUB2, respectively, for what concerns the checkpoint response to microtubule depolymerisation. Indeed, in the presence of nocodazole, SPC72-BFA1 and SPC72-BUB2 cells arrested in mitosis with 2C DNA contents as well as wild type cells, whereas bub2Δ and bfa1Δ cells re-replicated their genome in the same conditions (Fig. 2C-D). Thus, the Spc72-Bfa1 and-Bub2 chimera are likely functional in that they retain their inhibitory properties towards Tem1.
Previously characterized Bub2 and Bfa1 chimeric proteins constitutively anchored to SPBs are SPOC-defective [36]. Similarly, our Spc72-Bfa1 and Spc72-Bub2 failed to activate the SPOC upon spindle mispositioning caused by DYN1 deletion (Fig. 2E). Indeed, dyn1Δ SPC72-BFA1 bfa1Δcells undergoing anaphase in the mother cell, which is symptomatic of spindle mispositioning, exited the cell cycle and re-budded, in contrast to dyn1Δ cells that arrested in mitosis as large budded cells (Fig. 2E). The SPOC failure of SPC72-BFA1 bfa1Δ cells was not worsened by deletion of BUB2 or KIN4 or both, consistent with the notion that Kin4 and Bub2/Bfa1 act in concert to inhibit Tem1. Similar results were obtained with the Spc72-Bub2chimera (S1 Fig.). Thus, constitutive targeting to both SPBs of the GAP Bub2/Bfa1, and of Tem1 as a consequence, leads to unscheduled Tem1 activation, consistent with a previous proposal [22].
We then asked if symmetric localization of Tem1 driven by the Spc72-Bfa1 chimera leads to more efficient recruitment of Cdc15 to SPBs in metaphase. This was indeed the case. Whereas GFP-tagged Cdc15 was present at the SPBs of 55% wild type metaphase cells, 90% of metaphase cells expressing the fusion SPC72-BFA1 displayed SPB-bound Cdc15 (Fig. 2F). Furthermore, Cdc15 was significantly more symmetric in SPC72-BFA1 than in wild type cells. Thus, stable tethering of Tem1 to SPBs by fusion to an SPB component [22] or by SPB recruitment via its inhibitory GAP ([36] and our data) leads in both cases to premature Tem1 activation.
We then asked if expression of the Spc72-Bfa1 chimera could have any phenotypic consequence for conditional mutants affecting the MEN. Remarkably, SPC72-BFA1 as the only source of Bfa1 in the cells suppressed the growth defects of several MEN mutants. In particular, it could partially rescue the temperature sensitivity of tem1–3 and mob1–77(Fig. 2G), as well as the cold-sensitivity of cdc15–2 and dbf2–2 mutant cells (S2A Fig.). A slight suppression, if any, was observed for the temperature-sensitivity of cdc5–2 cells, whereas the temperature-sensitivity of cdc14–3 cells was not suppressed at all (S2B-C Fig.).
Suppression of tem1–3 was recessive, as it was not observed when a wild type copy of BFA1 was present concomitant to SPC72-BFA1 in the cells (Fig. 2G). Importantly, suppression was not due to reduced GAP activity, as it could not be recapitulated by deletion of BFA1(Fig. 2G), or BUB2 or both (S2D Fig.). Since the mitotic exit defects of tem1–3 cells at high temperatures correlate with a loose interaction of the mutant Tem1 protein with SPBs [53], we conclude that Spc72-Bfa1 suppresses the temperature-sensitivity of tem1–3 cells likely by recruiting Tem1 to the SPBs. Consistent with this notion, the Spc72-Bfa1 and Spc72-Bub2 chimera suppressed the lethality caused by overexpression of the SPOC kinase Kin4 (Fig. 2H), which increases the turnover of Bub2/Bfa1, and by consequence of Tem1, at SPBs [36].
Thus, these data, together with those from a previous study [36], indicate that symmetric persistence of Bub2/Bfa1 at SPBs does not interfere with mitotic exit. Rather, in spite of being part of an inhibitory GAP complex, Bfa1 is a receptor of Tem1 at SPBs, where Tem1 promotes MEN signaling. Stable residence of Bub2/Bfa1 at SPBs causes unscheduled mitotic exit by decreasing Tem1 turnover at SPBs, as previously suggested [22].
Activation of the FEAR pathway in anaphase is required for the unscheduled mitotic exit triggered by Spc72-Bfa1 and-Bub2 chimeric proteins
The ability of Spc72-Bfa1 and-Bub2 tethers to efficiently prevent mitotic exit upon microtubule depolymerization, but not upon spindle mispositioning, was somewhat puzzling. A major difference between the two conditions lies in the activation of the spindle assembly checkpoint (SAC) after nocodazole treatment. Through inhibition of Cdc20/APC, SAC leads to securin stabilization, in turn preventing activation of separase and the FEAR pathway [15]. If inhibition of the FEAR pathway is the only reason for the failure of Spc72-Bfa1 and-Bub2 chimera to promote mitotic exit, premature FEAR activation should allow mitotic exit in cells expressing Spc72-Bfa1 and-Bub2 treated with nocodazole. Conversely, FEAR inactivation should prevent mitotic exit in the same cells undergoing spindle misalignment. To test this hypothesis, we prematurely activated the FEAR pathway and Cdc14 release from the nucleolus by either inactivation of the PP2ACdc55 phosphatase or ESP1 overexpression [16]. Remarkably, PP2ACdc55 inactivation through deletion of CDC55 had a synergistic effect with SPC72-BFA1 and SPC72-BUB2 on the kinetics of mitotic exit upon nocodazole treatment, as judged by the ability of cells to re-replicate their DNA (Fig. 3A-B). Similar data were obtained with ESP1 overexpression from the galactose-inducible GAL1 promoter (Fig. 3C-D). In contrast, reducing the levels of mitotic CDKs through CLB2 deletion did not accelerate mitotic exit in SPC72-BFA1 cells (Fig. 3E). Importantly, FEAR inactivation through deletion of both SPO12 and BNS1 reduced the unscheduled mitotic exit caused by SPC72-BFA1 in dyn1Δ cells (Fig. 3F).
Fig. 3. The activation state of the FEAR pathway influences the ability of Spc72-Bfa1 to promote unscheduled mitotic exit. 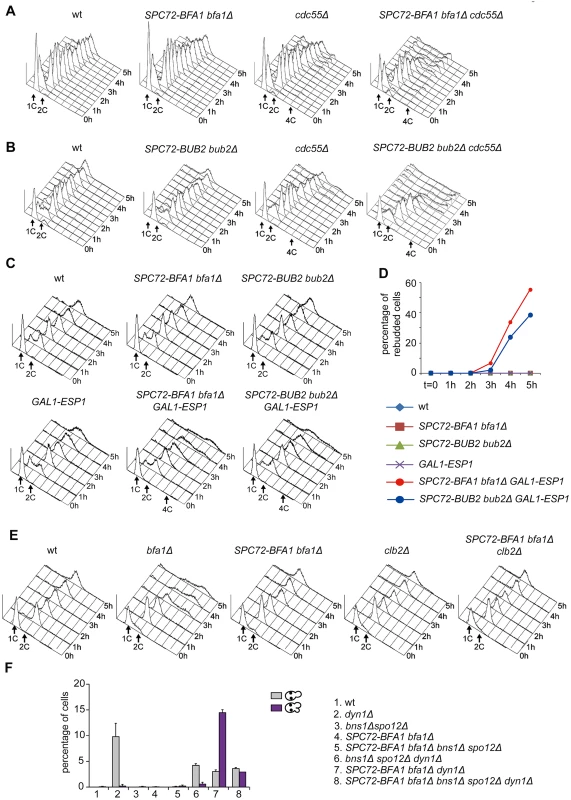
A-E: Logarithmically growing cultures of cells with the indicated genotypes were synchronized in G1 by α-factor and then released into nocodazole-containing medium (t = 0). At the indicated times cell samples were withdrawn for FACS analysis of DNA contents (A-C, E) and to score the percentage of re-budded cells (D). F: The percentage of cells with binucleate cell bodies accompanied or not by a SPOC defect was scored after DAPI staining of cycling cells with the indicated genotypes shifted to14°C for 16h. Thus, constitutive recruitment of the Bub2/Bfa1 complex to SPBs leads to precocious Tem1 activation. Whether this translates into a premature mitotic exit depends on the activation state of the FEAR pathway.
A constitutively active GTP-bound Tem1 variant is SPOC-deficient and is synthetically lethal for mutants affecting spindle positioning
To further investigate the links between Tem1 activity and the establishment of SPB asymmetry of the Tem1/Bub2/Bfa1 complex, we generated a TEM1-Q79L mutant allele, where the catalytic glutamine in the G domain (Q79, according to sequence comparison with Rab-like GTPases [27]), was replaced by leucine. We first tested the catalytic properties of Tem1-Q79L in in vitro GTPase assays in the presence of Bfa1 and Bub2. As shown in Fig. 4A, Tem1-Q79L was completely refractory to stimulation of GTP hydrolysis by the GAP Bub2/Bfa1 in vitro, suggesting that in vivo it is preferentially in its active GTP-bound form. Thus, Q79 of Tem1 likely participates directly to GTP hydrolysis along with R85 and Q132 of Bub2.
Fig. 4. The constitutively active Tem1-Q79L variant is checkpoint-deficient. 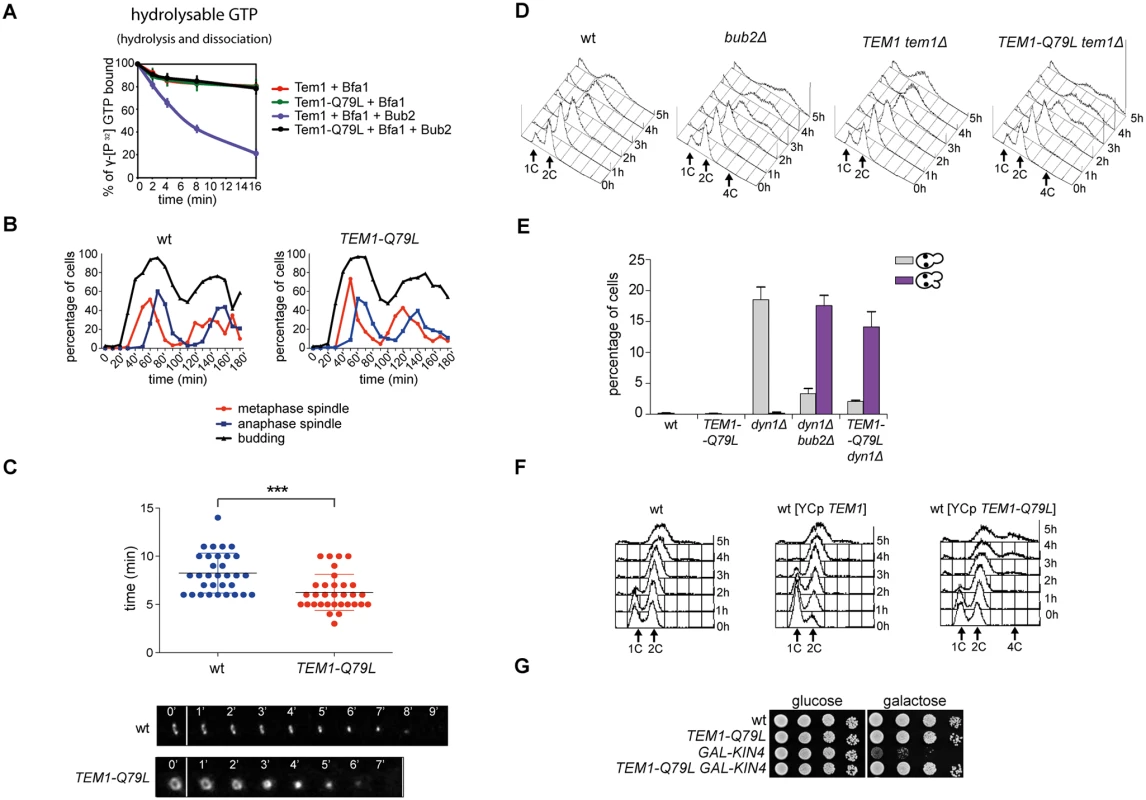
A: Bacterially purified 6XHis-Tem1 and 6XHis-Tem1-Q79L were loaded with γ[32P]GTP either in the absence or in the presence of recombinant MBP-Bfa1 and incubated at 30°C for 10 minutes. The mixture was then added to GST-Bub2 or buffer alone and kinetics of GTP hydrolysis and dissociation was followed by a filter-binding assay (see details in Material and Methods). Graphs show average values and standard deviations from three independent experiments. B: Wild type and TEM1-Q79L cells were arrested in G1 by α-factor and then released into fresh medium at 25°C (t = 0). Cell samples were withdrawn every 10’ to measure kinetics of budding and spindle formation/elongation after in situ immunostaining of tubulin. C: Actomyosin ring contraction has been visualized by live cell imaging of wild type and TEM1-Q79L expressing Myo1-GFP (n = 30). D: Logarithmically growing cultures of cells with the indicated genotypes were shifted into nocodazole containing medium (t = 0). DNA contents were analysed by flow cytometry at the indicated times. E: The percentage of cells with binucleate cell bodies accompanied or not by SPOC defect was scored after DAPI staining of cycling cells of the indicated strains shifted to 14°C for 16h. F: Logarithmically growing cultures of strains with the indicated genotypes were shifted to nocodazole containing medium (t = 0). DNA contents were analysed by flow cytometry at the indicated times. G: Serial dilutions of stationary phase cultures of the indicated strains were spotted on YPD or YP galactose plates and incubated at 30°C for 48h. When expressed in yeast cells as the sole source of Tem1, Tem1-Q79L did not cause any detectable growth defect at any temperature. Furthermore, TEM1-Q79L mutant cells showed kinetics of cell cycle progression similar to those observed in wild type cells (Fig. 4B). The absence of obvious cell cycle phenotypes in unperturbed conditions was somewhat surprising, as a similar mutation in fission yeast spg1+, encoding the SIN counterpart of the Tem1 GTPase, leads to premature cytokinesis and formation of multiple septa [54]. We therefore analysed by time-lapse video microscopy the speed of actomyosin ring contraction, as a marker of cytokinesis [55], in wild type and TEM1-Q79L cells expressing GFP-tagged myosin II (Myo1). Strikingly, contraction of the actomyosin ring took place on average 2’ faster in TEM1-Q79L relative to wild type cells (i.e. 6’ and 8’, respectively, Fig. 4C), consistently with previous data on bub2Δ cells [56]. Thus, the TEM1-Q79L allele accelerates at least some aspects of cytokinesis without affecting cell viability.
To gain further insights into the factors allowing TEM1-Q79L cells to grow at normal rates, we carried out a synthetic genetic arrays (SGA) screen to find deletions of non-essential genes that become synthetically lethal/sick with TEM1-Q79L(Table 1). This screen uncovered several genes encoding proteins involved in spindle positioning and nuclear migration, such as Kar9, the dynein light chain (DYN2) and components of the dynactin complex that cooperates with dynein for spindle positioning [57]. In addition, this screen uncovered several genes implicated in microtubule biogenesis, which might indirectly influence spindle positioning. Thus, TEM1-Q79L aggravates the sickness of cells undergoing spindle mispositioning. Interestingly, the same deletion mutants were also identified in other SGA screens as synthetically sick or lethal with BUB2 or BFA1 deletion [58–60]. A number of additional non-essential genes whose deletion displayed synthetic interactions with TEM1-Q79L was also uncovered with this screen and will be described elsewhere, since the significance of these genetic interactions has not been further explored in this context.
Tab. 1. List of non-essential genes implicated in microtubules dynamics or spindle positioning identified in the SGA screen with <i>TEM1-Q79L</i>. 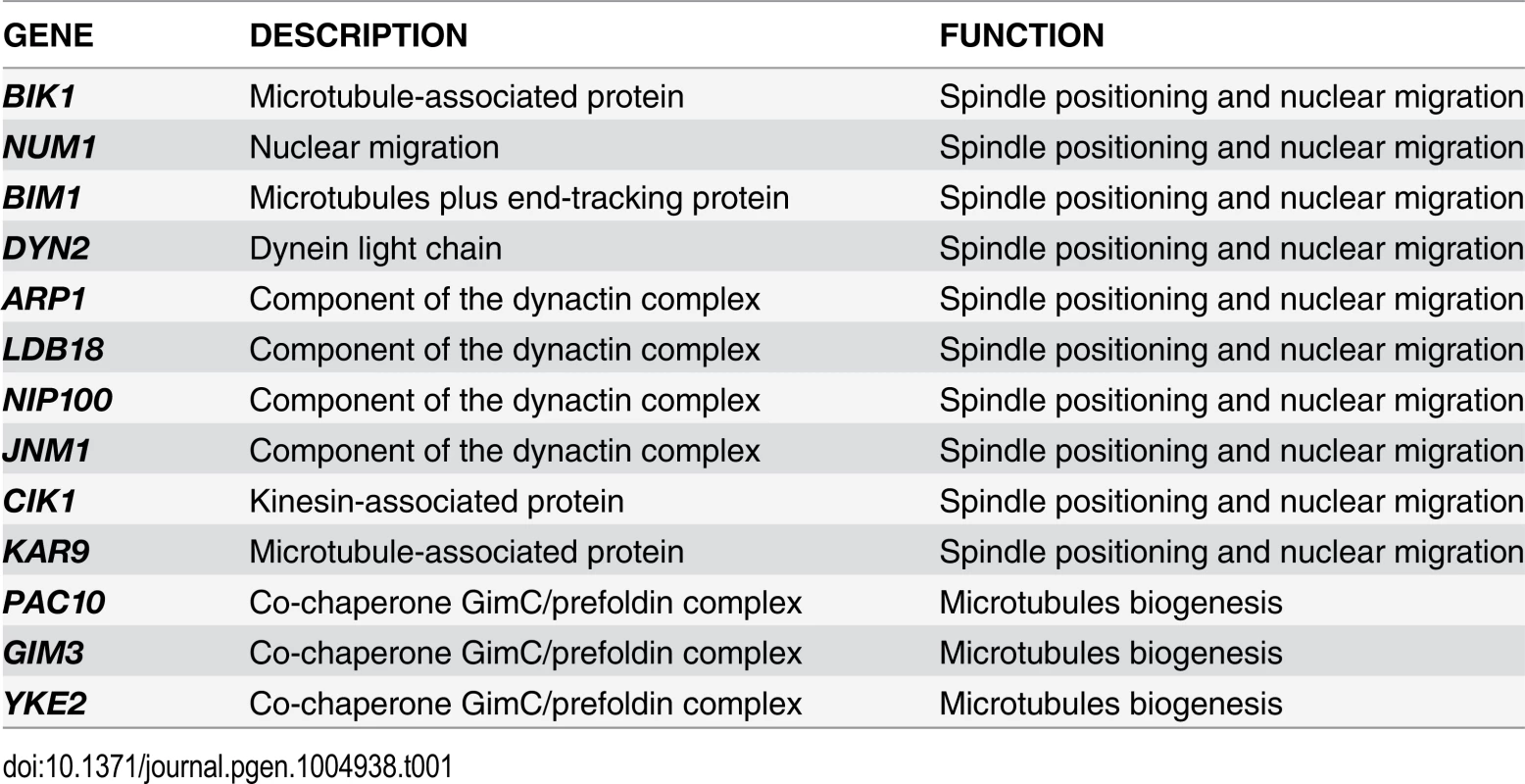
We then tested the ability of TEM1-Q79L mutant cells to respond to microtubule depolymerization and spindle mispositioning. In the presence of nocodazole, whereas wild type cells arrested in mitosis with 2C DNA contents, TEM1-Q79L cells re-replicated their genome similar to cells lacking Bub2 (Fig. 4D). In addition, TEM1-Q79L dyn1Δ cells exited mitosis and re-budded in face of spindle position defects (Fig. 4E). Thus, the TEM1-Q79L mutant allele affects SPOC response. As expected, the checkpoint defect was dominant, as the TEM1-Q79L allowed mitotic exit and re-replication in the presence of nocodazole even when expressed from an episomal plasmid in cells carrying also the endogenous TEM1 gene (Fig. 4F). Consistent with its constitutive activation, the TEM1-Q79L allele was also able to suppress the lethality associated with overexpression of KIN4 from the galactose-inducible GAL1 promoter (Fig. 4G), which delays mitotic exit by keeping the Bub2/Bfa1 GAP active [34].
The constitutively active Tem1-Q79L variant shows reduced asymmetry at spindle poles and impairs Bfa1 asymmetry
The “dual finger” model predicts that GTP hydrolysis is catalysed by the GAP and the so-called catalytic glutamine of the GTPase could stabilize the interaction between the GAP and the GTPase without directly contributing to the catalytic reaction [48]. We formally tested this idea by analysing the interaction between Tem1 or Tem1-Q79L with Bub2 and Bfa1 in co-immunoprecipitation experiments. Remarkably, HA-tagged Tem1-Q79L pulled down a higher, rather than a lower, amount of Bub2 and Bfa1 (tagged with 3PK epitopes and GFP, respectively) (Fig. 5A). Thus, constitutive binding to GTP seems to increase the affinity of Tem1 for its GAP. To further corroborate this conclusion we co-expressed Bfa1-GFP and Bub2-GFP with HA-tagged Tem1 or Tem1-Q79L. Immunoprecipitation of Tem1-Q79L-HA3 pulled down higher amounts of both Bfa1-GFP and Bub2-GFP than Tem1-HA3 (Fig. 5B), without affecting the relative proportion of Bfa1-GFP and Bub2-GFP in the immunoprecipitates. We therefore conclude that locking Tem1 in the GTP-bound form enhances its affinity for Bub2/Bfa1 without affecting the stoichiometry of the GAP complex.
Fig. 5. Constitutively active Tem1-Q79L binds more avidly Bub2/Bfa1 and shows reduced asymmetry at anaphase spindle poles. 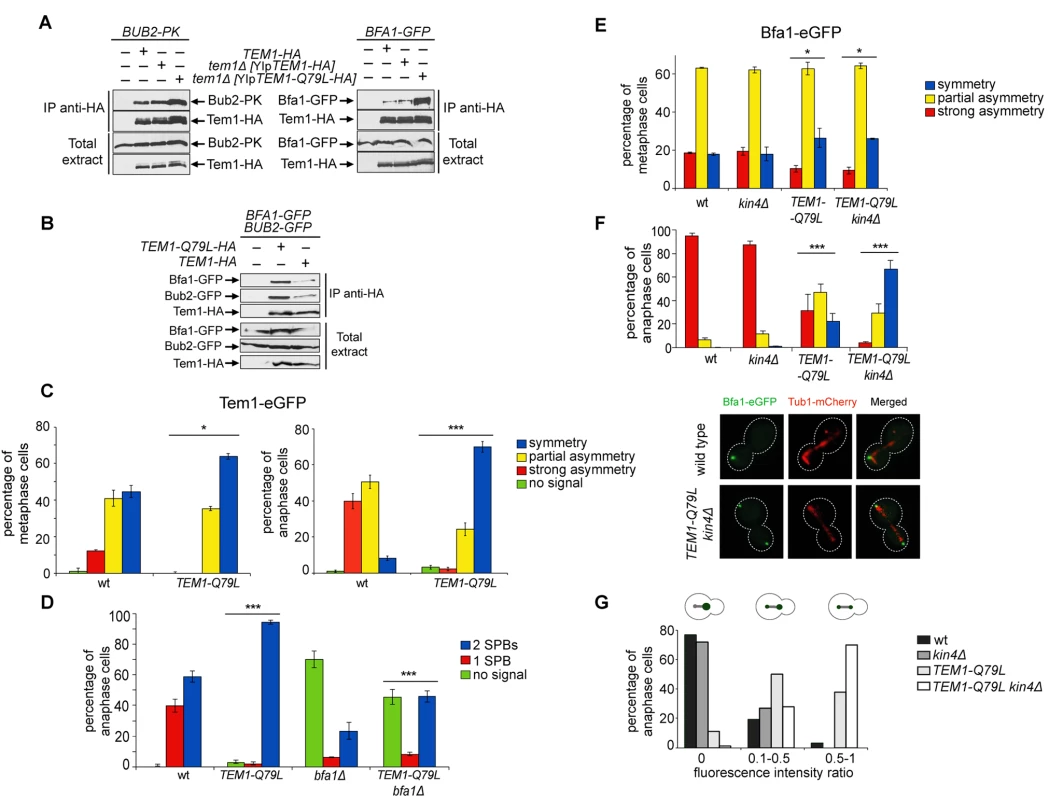
A-B: Protein extracts from cells expressing the indicated tagged proteins were used for immunoprecipitation with an anti-HA affinity resin. Western blot analysis was then performed with anti-PK, anti-GFP and anti-HA antibodies. The input represents 1/25th of the total extract used for each IP. C-F: Localization of eGFP- tagged Tem1 and Tem1-Q79L (C-D) or Bfa1-eGFP (E-F) was analysed in the indicated strains by fluorescence microscopy after formaldehyde fixation. Metaphase and anaphase cells were identified by means of the Tub1-mCherry co-expressed marker. Micrographs show representative cells of each strain in anaphase. G: Fluorescence intensity ratios were calculated between the two SPBs in anaphase cells of the indicated strains (see details in Materials and Methods). Since our data suggest that proper regulation of Tem1 GTP hydrolysis is required for asymmetry of the Tem1/Bub2/Bfa1 complex at SPBs, we analysed the subcellular distribution of Tem1-Q79L tagged with eGFP. As expected, Tem1-eGFP was already asymmetric in 53% of metaphase cells, whereas Tem1-Q79L-eGFP was asymmetric in a lower fraction of cells (35%). In anaphase, whereas Tem1-eGFP localized asymmetrically to the bud-directed SPB in 90% of cells, Tem1-Q79L-eGFP was present symmetrically at SPBs in 70% of the cells (Fig. 5C). Thus, abolishing the GAP-stimulated GTPase activity of Tem1 through different kinds of mutations invariably leads to Tem1 symmetric localization at SPBs.
Interestingly, whereas BFA1 deletion markedly affected Tem1-eGFP recruitment to SPBs as previously reported [22, 40, 61], it had a less pronounced effect on the SPB localization of Tem1-Q79L-eGFP (Fig. 5D), suggesting that loading to SPBs of constitutively active Tem1 is partially GAP-independent.
Previous data suggested that loading of Bfa1 and Bub2 on SPBs and their asymmetry in anaphase still occur in cells lacking Tem1 [30, 40]. On the other hand, experimental evidence indicates that an increased residence time of Tem1 at the SPBs or its decreased GTPase activity can influence Bub2/Bfa1 localization [22, 25, 62]. Because our results indicate that the Q79L substitution affects Tem1 activity as well as its localization in anaphase, we asked if Tem1-Q79L had any impact on localization of Bfa1. Both wild type and TEM1-Q79L mutant cells showed a similar partially asymmetrical SPB localization of Bfa1-eGFP in metaphase (Fig. 5E). In contrast, at the onset of anaphase, while Bfa1 drastically dropped to hardly detectable levels on the mother-bound SPB in 95% of wild type cells, it remained completely symmetrical on SPBs in 22% of TEM1-Q79L cells and persisted to low but clearly detectable levels on the mother-bound SPB in 46.6% of the cells (Fig. 5F). The symmetric localization of Bfa1 in TEM1-Q79L cells did not depend on premature activation of downstream MEN kinases, as it was not affected by Cdc15 inactivation through the cdc15–2 temperature-sensitive allele (S3 Fig.).
Since Kin4 promotes Bfa1 turnover at SPBs upon SPOC activation [36], we analysed Bfa1 localization in wild type and TEM1-Q79L cells lacking KIN4(Fig. 5E-F). In agreement with previous data [36], deletion of KIN4 alone did not affect Bfa1 distribution on SPBs of wild type cells in unperturbed conditions. In stark contrast, it had a synergistic impact with the TEM1-Q79L mutant allele on Bfa1 localization at SPBs specifically in anaphase, making it completely symmetrical in 65% of the cells and partially asymmetric in 30% of the cells (Fig. 5F). Consistently, the ratio in Bfa1-GFP fluorescence intensity at the mother - versus the bud-directed SPB was close to 0 for wild type and kin4Δ cells, while it significantly increased in TEM1-Q79L and TEM1-Q79L kin4Δ cells (Fig. 5G). Thus, these data reveal an unanticipated role of Kin4 in actively dislodging Bfa1 from the mother-bound SPB during anaphase of the unperturbed cell cycle. Furthermore, they indicate that Tem1 GTP hydrolysis is a primary determinant of Bfa1 asymmetry in anaphase.
The TEM1-Q79L allele enhances Cdc15 loading on SPBs, without affecting the timing of mitotic exit
To investigate further if the TEM1-Q79L allele leads to premature MEN activation, we analysed the subcellular localization of downstream MEN components, such as Cdc15 and Mob1. We observed that 99% of TEM1-Q79L mutant cells recruited Cdc15 to SPBs in metaphase, as opposed to 55% in wild type cells (Fig. 6A). Furthermore, Cdc15 was significantly more symmetric in TEM1-Q79L than in wild type cells. Deletion of KIN4, either alone or in combination with the TEM1-Q79L allele did not have any impact on Cdc15 distribution (Fig. 6A). In spite of Cdc15 enhanced loading on SPBs, recruitment of Mob1 to SPBs in metaphase was only slightly increased in TEM1-Q79L relative to wild type cells (Fig. 6B), consistent with the notion that mechanisms other than the SPOC restrain MEN activity downstream of Tem1 until late anaphase.
Fig. 6. The TEM1-Q79L allele leads to premature Cdc15, but not Mob1, loading on SPBs. 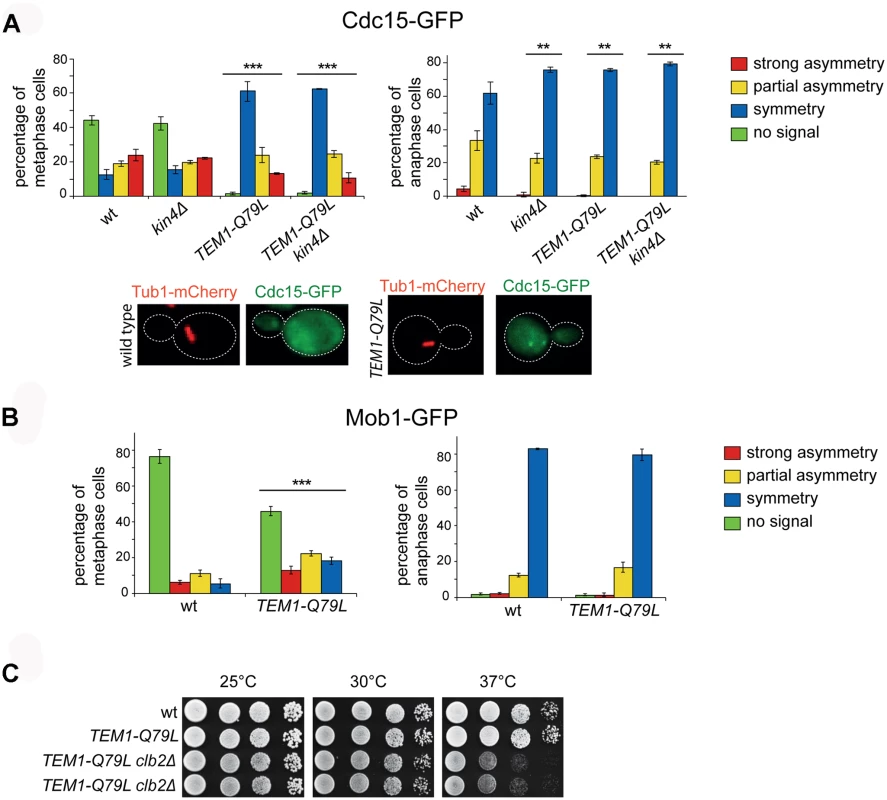
A-B: Distribution of Cdc15-GFP (A) or Mob1-GFP (B) was analysed in the indicated strains by fluorescence microscopy after formaldehyde fixation. Metaphase and anaphase cells were identified by means of the Tub1-mCherry co-expressed marker. Micrographs show representative wild type and TEM1-Q79L cells expressing Cdc15-GFP in metaphase. C: Serial dilutions of stationary phase cells with the indicated genotypes were spotted on YPD and incubated at the indicated temperatures for 48h. Phosphorylation of Cdc15 and Mob1 by cyclin B/CDKs together with Bub2/Bfa1 GAP activity provides a dual inhibition of the MEN [63]. Consistently, we found that combining the TEM1-Q79L allele with deletion of CLB2, which encodes the main mitotic cyclin B, caused synthetic growth defects at 37°C (Fig. 6C). However, no synthetic lethality or detrimental synthetic defects were induced by the additional deletion of KIN4 or BFA1, suggesting that the components of the MEN downstream of Cdc15 are likely targets of negative regulators additional to Bub2/Bfa1 and Clb2-associated CDKs.
Bub2/Bfa1 and Tem1 asymmetry is important for proper Kar9 distribution and spindle positioning
Partial asymmetry of Bub2/Bfa1 at SPBs begins already in metaphase [39]. Since during metaphase MEN induces asymmetric localization of Kar9 at spindle poles, which is in turn required for correct spindle positioning [18], we checked if symmetrical Bub2/Bfa1 and Tem1 might affect Kar9 localization and spindle positioning. To this end, we analysed the distribution of Kar9 tagged with eGFP on the metaphase spindles of wild type, SPC72-BFA1 bfa1Δ, SPC72-BUB2 bub2Δ, and TEM1-Q79L cells. Whereas 84.4% of wild type cells showed strongly asymmetric Kar9, this value dropped to 43.5%, 50.5% and 47.6% in SPC72-BFA1 bfa1Δ, SPC72-BUB2 bub2Δ and TEM1-Q79L cells, respectively, while the remaining fraction of cells displayed partial or complete symmetry (Fig. 7A).
Fig. 7. Bub2/Bfa1 and Tem1 asymmetry is important for proper Kar9 distribution and spindle positioning. 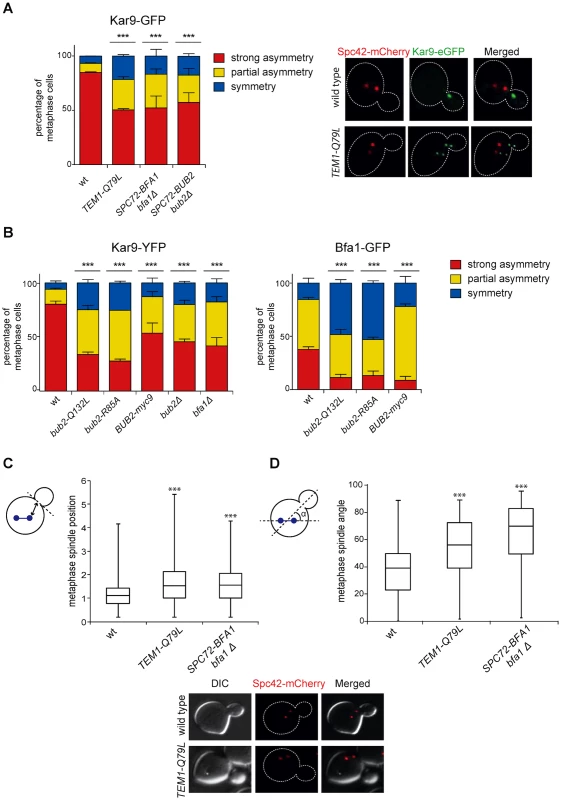
A-B: Percentage of metaphase cells carrying Kar9-eGFP (A), Kar9-YFP or Bfa1-eGFP (B) on one SPB (strong asymmetry), both SPBs unequally (partial asymmetry) or both SPBs equally (symmetry) for the indicated strains after formaldehyde fixation. Metaphase and anaphase cells were identified by means of the co-expressed SPB marker Spc42-mCherry. C-D: Spindle position (distance between the nearest SPB and the bud neck) and orientation (angle that the spindle forms with respect to the cell polarity axis) were measured in metaphase spindles of cells with the indicated genotypes after formaldehyde fixation (n≥100). Since our results indicate that the GAP activity of Bub2 and Bfa1 influences the localization of the GAP complex in the cell, we characterized Kar9 distribution in the GAP-dead Bub2 variants, bub2-Q132L and bub2-R85A cells, in cells expressing the symmetric GAP-inactive BUB2-myc9 construct [25]. All strains showed more symmetric localization of Kar9, with 33.9% of complete asymmetry for bub2-Q132L, 27.8% for bub2-R85A, 53.6% for BUB2-myc9, as opposed to 80.3% for wild type cells (Fig. 7B). Surprisingly, we found increased Kar9 symmetry also in bub2Δ and bfa1Δ mutant cells (45.7% and 41.9% of complete asymmetry, respectively), where Tem1 is present at SPBs at low levels [22], but symmetrically (Fig. 5C). Increased symmetry of Kar9 in the mutants (with the exception of bub2Δ and bfa1Δ that were not analysed) was accompanied by increased Bfa1 symmetry (Fig. 7B). Therefore, establishment of Bub2/Bfa1 and Tem1 asymmetry impacts on Kar9 asymmetry. Nonetheless, Bub2/Bfa1 symmetric distribution at SPBs is not sufficient to drive Kar9 symmetry. Indeed, when spindles were misaligned in dyn1Δ mutant cells Bub2 became increasingly more symmetric from metaphase to anaphase, whereas Kar9 symmetry increased only slightly (Fig. 8A-B), in agreement with recently published data [64, 65]. Thus, once Kar9 asymmetry has been established, it cannot be reversed by spindle misalignment, in contrast to that of Bub2/Bfa1.
Fig. 8. Bub2, but not Kar9, become symmetric on misaligned spindles. 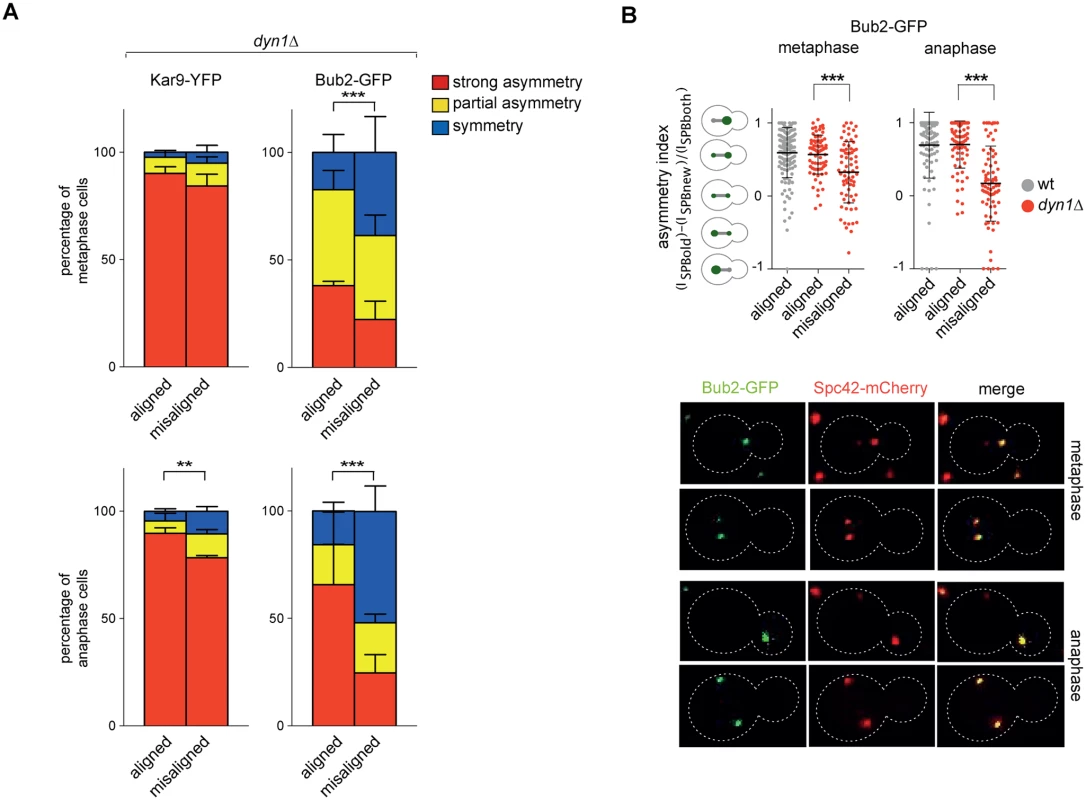
A: Distribution of Kar9-YFP and Bub2-GFP in metaphase and anaphase on aligned and misaligned (dyn1Δ) spindles. Metaphase and anaphase cells were identified by means of the co-expressed SPB marker Spc42-mCherry. Misaligned spindles in metaphase cells were identified by measuring an angle of 60°-120° between the spindle axis and a line connecting the old SPB (defined as the SPB showing brighter Spc42-mCherry fluorescence) to the bud neck. Scoring was done on 3 independent clones for each genotype with a total N ≥ 75; mean ± SD. B: Asymmetry indices were calculated for wild type and dyn1Δ cells expressing Bub2-GFP and Spc42-mCherry in (A) by dividing the difference between Bub2 fluorescence associated with the old SPB (ISPBold) and new SPB (ISPBnew) by the total Bub2 fluorescence (ISPBboth). Age of SPBs was determined by the fluorescence intensity of Spc42-mCherry (bright fluorescence: old SPB, dim fluorescence: new SPB). Micrographs show representative Bub2-GFP distribution on aligned and misaligned spindles, in either metaphase or anaphase. We then asked if Kar9 mislocalization in our mutants affects spindle positioning. SPC72-BFA1 and TEM1-Q79L cells expressing Spc42-mCherry were synchronized to collect cells in metaphase and imaged. Measurements of spindle distances from the bud neck (Fig. 7C) and spindle angles relative to the mother-bud polarity axis (Fig. 7D) indicated that both TEM1-Q79L and SPC72-BFA1 cells significantly affected the position and the orientation of metaphase spindles. Thus, hyperactive Tem1 impaired Kar9-dependent spindle positioning.
In conclusion, although constitutive symmetric localization of Tem1 and its GAP Bub2/Bfa1 does not appear to affect mitotic exit, it does compromise asymmetry of Kar9 at spindle poles, thereby causing spindle positioning and orientation defects.
Discussion
Tem1 GTP hydrolysis involves a catalytic glutamine in the switch II and the GAP Bub2/Bfa1
The mechanistic details of how GTPases switch between a GTP - and a GDP-bound state build on initial structural studies on Ras. In Ras a conserved glutamine in the switch II domain of the GTPase and a conserved arginine of the GAP both contribute to GTP hydrolysis [66, 67] and, consistently, mutations of either residue abolish GTP hydrolysis. Crystal structure of some Rab GTPases in complex with their TBC (Tre-2, Bub2 and Cdc16) GAP revealed that the conserved switch II glutamine (Q79 of Tem1) does not directly participate in GTP hydrolysis. Rather, catalysis is entirely brought about by a conserved arginine and a conserved glutamine of the TBC GAP through a mechanism referred to as “dual-finger” [48, 68]. Hence, the switch II glutamine of the GTPase was proposed to stabilize its interaction with the GAP [48]. Recently, however, Rab GTPases have been shown to be more plastic than originally anticipated in their activation/hydrolysis mechanisms. In particular, the contribution of the switch II glutamine in GTP hydrolysis is variable and for some GTPases it contributes, together with a conserved lysine in the P-loop, to activation of the GTPase by stabilizing its GEF-bound nucleotide-free form [69]. Therefore, the outcome of mutations of conserved catalytic residues varies depending on the GTPase, GEF and GAP, and is altogether unpredictable.
Here we have addressed the importance of Q79 in the switch II and the dual-finger mechanism in Tem1 GTP hydrolysis. First, we have established that the intrinsic rate of Tem1 GTP hydrolysis is negligible, and loss of GTP is mostly accounted for by nucleotide dissociation. In agreement with previous data [25, 26, 30], Bfa1 prevents nucleotide dissociation and therefore acts as guanine-nucleotide dissociation inhibitor (GDI). Second, we show for the first time that Tem1 Q79 is directly involved in the GAP-induced GTP hydrolysis without impairing its interaction with Bub2 and Bfa1. Mutation of Q79 into leucine generates a hyperactive, dominant Tem1 that is refractory to its GAP, recruits more efficiently Cdc15 to SPBs and leads to unscheduled mitotic exit in the presence of spindle positioning defects.
Finally, we show that the dual-finger mechanism applies also to GTP hydrolysis of the Tem1-Bub2-Bfa1 complex. Indeed, glutamine 132 of Bub2 is involved in GTP hydrolysis, in addition to arginine 85 that we previously showed [25]. Consistent with an important role of Q132 in Tem1 inhibition, bub2-Q132L mutant cells are SPOC-defective and undergo mitotic exit upon microtubule depolymerization.
Thus, we have defined Q79 of Tem1 together with R85 and Q132 of Bub2 as a catalytic triad for GAP-induced GTP hydrolysis.
Asymmetry of Bub2/Bfa1 and MEN activation
The Bub2/Bfa1 complex is required for efficient Tem1 binding to SPBs throughout most of the cell cycle, except in late mitosis [22, 40, 61]. In contrast, SPB recruitment of Bub2/Bfa1 does not require Tem1 [30, 40]. The amount of Tem1 at SPBs depends on the turnover of Bub2/Bfa1 at SPBs, which in turn is accelerated by spindle mispositioning through Kin4-dependent phosphorylation of Bfa1 [36, 39]. Thus, as recently proposed [61], the GAP Bub2/Bfa1 is a major Tem1 receptor at SPBs and its regulation is instrumental for establishing Tem1 asymmetry. Critical regulators of Bub2/Bfa1 asymmetry are the polo kinase Cdc5 and the phosphatase PP2ACdc55. Cdc5 phosphorylates and inactivates the Bub2/Bfa1 complex leading to Tem1 activation [51, 70], whereas PP2ACdc55 dephosphorylates Bfa1 [71]. Phosphomimetic mutations in some Cdc5-dependent Bfa1 phosphorylation sites, as well as loss of PP2ACdc55, are sufficient to induce premature asymmetry of Bub2/Bfa1 at SPBs, whereas Cdc5 inactivation or phospho-ablating mutations in Bfa1 lead to its persistent symmetry [62, 71]. Similar mechanisms might be operational in fission yeast to establish SIN asymmetry. Indeed, polo kinase has been proposed to phosphorylate the Bfa1 homolog Byr4 and promote its dissociation from SPBs [72], thereby influencing the distribution of GTP-bound Spg1 and its effector kinase Cdc7. Furthermore, PP2A regulates Byr4 asymmetry through dephosphorylation of the SIN anchor at SPBs Cdc11 [73, 74]. Indeed, Byr4 binds more efficiently to dephosphorylated Cdc11 [73], whereas the Cdc15-like kinase Cdc7 binds preferentially phosphorylated Cdc11 [75]. Thus, S. cerevisiae and S. pombe might adopt common regulatory strategies to establish MEN and SIN asymmetry.
We previously proposed that Tem1 GTP hydrolysis promotes asymmetry of both Tem1 and its GAP Bub2/Bfa1 at SPBs in anaphase [25] and results from other studies [22, 62] support this conclusion. Consistent with this idea, we now show that mutating the second catalytic finger of Bub2 (Q132) or mutating the catalytic Q79 of Tem1 leads in both cases to increased symmetry of Bub2/Bfa1 and Tem1 at SPBs. At a first glance these results appear at odds with the finding that in the complete absence of Tem1 Bub2/Bfa1 is not only recruited to SPBs with normal kinetics, but becomes asymmetric in anaphase exactly like in wild type cells [30]. However, we now show that when GTP hydrolysis is abolished Bub2 and Bfa1 bind more avidly to Tem1. Thus, the increased symmetry of Bub2/Bfa1 at SPBs in these conditions likely reflects its stronger affinity for GTP-bound Tem1. The different affinity of Bub2/Bfa1 for GTP - versus GDP-bound Tem1 has important implications for the SPOC, where active Tem1 needs to be quickly inactivated in the presence of a mispositioned spindle.
Based on our previous results using a version of Bub2 tagged with nine myc epitopes at the C-terminus (Bub2-myc9), we proposed that removal of Bub2/Bfa1 from the mother SPB is important for timely mitotic exit [25]. Indeed, Bub2-myc9 is more symmetrically localized at SPBs than wild type Bub2 and is lethal for cdc5–2 and tem1–3 mutants because it prevents mitotic exit in these sensitized backgrounds. Now we further tested this idea by expressing chimeric proteins that constitutively recruit the GAP and Tem1 to both SPBs. Contrary to our predictions, these chimeric proteins partially rescued, instead of aggravating, the temperature-sensitive growth phenotype of several MEN mutants. In particular, Spc72-Bfa1 rescued the temperature-sensitivity of tem1–3 cells likely by suppressing the SPB-binding defects of the mutant Tem1–3 protein at high temperature [53]. Importantly, suppression of MEN mutants by our chimeric proteins is not accounted for by their possible impaired GAP activity, because it could not be recapitulated by BFA1 and/or BUB2 deletion.
The chimeric proteins Spc72-Bfa1 and-Bub2 caused also unscheduled mitotic exit in the presence of mispositioned spindles, similar to Bub2 - and Bfa1-Cnm67 fusion proteins previously characterized [36]. Therefore, despite different parts of Bub2 and Bfa1 are fused to the SPB anchor (the N-terminus in our Spc72 - fusions and the C-terminus in the-Cnm67 chimera), constitutive binding of the Bub2/Bfa1 complex to SPBs invariably leads to SPOC defects. Our data are totally consistent with the notion that Tem1 recruitment to SPBs is necessary for its MEN function [22]. In this scenario, SPB-locked Bub2/Bfa1 activates Tem1 by increasing its symmetry and residence time at SPBs, thereby causing unscheduled mitotic exit in the presence of mispositioned spindles. It is worth noting that symmetry of SIN components, such as the Cdc7 kinase, at SPBs also causes unscheduled cytokinesis and repeated rounds of septation [46, 47], suggesting that asymmetry is a common strategy in S. cerevisiae and S. pombe to restrain MEN/SIN signalling.
The molecular basis for SPB-driven Tem1 activation is not known. It is possible that the GAP Bub2/Bfa1 at SPBs is constitutively kept inactive by Cdc5-mediated phosphorylation, thereby making Tem1 at SPBs refractory to GAP-mediated inhibition. Upon spindle misalignment, Kin4-mediated dislodgement of Bub2/Bfa1 from SPBs becomes essential for Tem1 inhibition in the cytoplasm and SPOC response [36, 61]. An interesting non-mutually exclusive hypothesis is that a putative GEF for Tem1 localizes at SPBs [76]. However, as mentioned above the identity of Tem1 GEF(s) remains elusive.
The finding that the chimeric proteins Spc72-Bfa1 and-Bub2, similar to Bub2 - and Bfa1-Cnm67 [36] and Cnm67-Tem1 [22], support a mitotic arrest after microtubule depolymerization, but not after spindle mispositioning, was somehow puzzling. One major difference between the SAC-mediated metaphase arrest and the SPOC-mediated anaphase arrest is that in the latter, but not in the former, the PP2ACdc55 phosphatase is inhibited by the FEAR pathway [15, 16]. We find that, indeed, CDC55 deletion or ESP1 overexpression in cells expressing Spc72-Bub2 or-Bfa1 drives unscheduled mitotic exit in the presence of nocodazole. In contrast, FEAR inhibition by deletion of SPO12 and BNS1 [15] prevents mitotic exit in cells expressing the same chimeric proteins and experiencing spindle position defects. Thus, whether the FEAR is activated or inhibited influences the impact of the chimeric proteins on mitotic exit. PP2ACdc55 has been recently shown to antagonize the Cdc5-dependent phosphorylation of Bfa1 [71], thereby providing a mechanistic explanation to our data. The FEAR-mediated partial release of Cdc14 from the nucleolus might also contribute to MEN activation by counteracting the CDK-dependent inhibitory phosphorylation of MEN components [63].
In conclusion, persistent symmetric localization of the GAP Bub2/Bfa1 does not interfere with mitotic exit. Most likely, the Bub2-myc9 protein that we described previously prevents timely MEN activation by a different mechanism. Consistently, tagging of Spc72-Bub2 at the C-terminus with nine myc epitopes causes synthetic sickness in combination with the cdc5–2 mutant allele affecting the polo kinase (S1 Table).
An unanticipated role of Bub2/Bfa1 and Tem1 asymmetry on Kar9 distribution and spindle positioning
We show that symmetric localization of the Bub2/Bfa1/Tem1 complex, independently of whether it is driven by chimeric proteins or loss of GTPase activity, interferes with Kar9 asymmetry at spindle poles in metaphase, as well as with spindle positioning and orientation relative to the cell division axis. Although Tem1 inactivation causes similar phenotypes for what concerns Kar9 localization and spindle orientation, it does not affect spindle positioning at the bud neck [18], indicating that Tem1 hyperactivation and inactivation are not equivalent in this respect. The molecular bases of this difference remain to be established. Similarly, whether Tem1 hyperactivation primarily affects Kar9 localization and, as a consequence, spindle positioning or vice-versa remains to be investigated. Although the phenotypic analyses of our mutants did not reveal any apparent alterations of astral microtubules, at the moment we cannot exclude that subtle defects in microtubule dynamics could account for spindle mispositioning and, in turn, increased Kar9 symmetry.
Previous [64] and our data indicate that not all conditions leading to symmetric distribution of the Bub2/Bfa1 complex and Tem1 cause symmetric localization of Kar9 at spindle poles. Indeed, whereas Bub2, Bfa1, Tem1 and Kar9 are all asymmetric, to different extents, on metaphase spindles, Bub2, Bfa1 and Tem1 become increasingly more symmetric upon spindle misalignment, while Kar9 remains strongly asymmetric. These data suggest that establishment of Bub2/Bfa1/Tem1 symmetry on misaligned spindles is an active process and Kar9 asymmetry is so robust that once established it cannot be reversed by bringing the Bub2/Bfa1/Tem1 complex to both spindle poles. In contrast, in our mutants Bub2, Bfa1 and Tem1 are more symmetric already in metaphase and might therefore interfere with the establishment of Kar9 asymmetry. Furthermore, we speculate that in these mutants the residence of Tem1 and the GAP Bub2/Bfa1 at SPBs is relatively stable, while these proteins turn over very fast at the SPBs of misaligned spindles [36]. An alternative explanation to our data is that Tem1 hyperactivation, rather than symmetry, is responsible for Kar9 mislocalization and spindle misalignment. Indeed, while in our mutants Tem1 is likely in the active GTP-bound form, it is inactivated and GDP-bound when the SPOC is turned on.
Factors involved in cell polarity were implicated in the asymmetry of Bub2-Bfa1 at spindle poles [25, 39]. Thus, it is tempting to speculate that in budding yeast the role of cell polarity in spindle positioning might be partly exerted through asymmetric localization of the Bub2-Bfa1-Tem1 trimeric complex at spindle poles, which in turn influences Kar9 asymmetry. Remarkably, other eukaryotic cells (i.e. nematodes, flies and mammals) employ heterotrimeric G proteins for spindle positioning during both symmetric and asymmetric cell division (reviewed in [77]. A striking parallel can be drawn between the asymmetric enrichment of their GDIs GPR-1/2, which is controlled by polarity factors and necessary for proper spindle alignment (reviewed in [78], and asymmetric localization of Bub2-Bfa1. Future work will certainly shed new light onto possible additional similarities in the mechanisms adopted by different organisms to achieve correct spindle positioning.
Materials and Methods
Strains and plasmids, media and reagents, genetic manipulations
All strains, except those used for Fig. 7B and 8 (derivatives of S288c) are derivatives of W303 (ade2–1, trp1–1, leu 2–3,112, his 3–11,15, ura3, ssd1) and listed in S2 Table. Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone and 50mg/L adenine) supplemented with 2% glucose (YEPD) or 2% galactose (YEPG). Unless otherwise stated, α-factor, nocodazole and benomyl were used at 2, 15 and 12,5 μg/ml respectively. Synchronization experiments with α-factor were performed at 25°C. Bacterial cells were grown in LD broth (1% bactotryptone, 0,5% yeast extract and 0,5% NaCl pH7,25) supplemented with 50 µg/ml ampicillin and 34 μg/ml chloramphenicol.
The SPC72-BUB2 fusion was generated by triple ligation of a HindIII/XbaI PCR fragment containing the whole ORF of 340 bp of SPC72 promoter, a XbaI/EcoRI PCR fragment containing the ORF of BUB2 spanning codons 2–172, and the LEU2-based Yiplac128 vector linearized with HindIII and EcoRI. The generated plasmid (pSP275) was linearized with BamHI for integration at the BUB2 locus, thereby generating a gene fusion under the SPC72 promoter and containing the whole ORF of SPC72 fused in frame to the entire ORF of BUB2, as well as a truncated BUB2 gene lacking the last 134 codons. Single integration of the construct at the BUB2 locus was checked by Southern blot.
The SPC72-BFA1 fusion was generated by triple ligation of a HindIII/XbaI PCR fragment containing the whole ORF of 340 bp of SPC72 promoter, a XbaI/BglII PCR fragment containing the entire ORF of BFA1 starting from the 2nd codon and 330 bp of 3’ UTR, and the LEU2-based Yiplac128 vector linearized with HindIII and BamHI. The generated plasmid (pSP371) was linearized with PstI for integration at the SPC72 locus and single integration of the construct at the SPC72 locus was checked by Southern blot.
The ORF of TEM1 and about 1000 bp of promoter region was cloned in Yiplac128 (pSP596). A variant carrying the TEM1-Q79L mutation was generated by site-directed mutagenesis (pSP597). The TEM1-bearing plasmids have been integrated at the LEU2 locus by BstXI digestion and single integrations have been checked by Southern blot.
Gene deletions were generated by one-step gene replacement [79]. One-step tagging techniques [80, 81] were used to tag Tem1, Tem1-Q79L, Bfa1, Bub2, Spc72-Bfa1 and Kar9 with multiple HA tags or eGFP.
Protein expression and purification
E. coli BL21 cells carrying pLysE plasmid (Novagen) and 6His-TEM1, 6xHis-TEM1-Q79L, MBP-BFA1, GST-BUB2 and GST-BUB2-Q132L expression plasmids were grown in LD broth containing ampicillin and chloramphenicol at 37°C for 3 h, transferred to 14°C for 1 h and induced with 0,1 mM isopropyl-1-thio-β-D-galactopyranoside for 15 h. Cells expressing MBP-Bfa1, 6His-Tem1 and GST-Bub2 fusions were resuspended, respectively, in the following cold lysis buffers: 50 mM Tris-HCl pH7.5, 200 mM NaCl and 2mM DTT supplemented with a cocktail of protease inhibitors (Complete; Boehringer); 50 mM Tris-HCl pH8, 300 mM NaCl, 2mM MgCl2 and 10mM imidazole supplemented with a cocktail of protease inhibitors; 50 mM Tris-HCl pH7.5, 200 mM NaCl supplemented with a cocktail of protease inhibitors. Cells were incubated with 1 mg/ml lysozyme in ice for 30 min, placed at 37°C for 5 min and sonicated at 4°C for 10 seconds. The extract was then clarified by centrifugation at 15,000 rpm for 30 min at 4°C. Tem1–6xHis and Tem1-Q79L-6xHis were purified by affinity chromatography with Ni-NTA columns (QUIAGEN). The MBP-Bfa1 fusion protein was purified using Amylose resin (New England Biolabs, Inc.), whereas GST-Bub2 and GST-Bub2-Q132L were purified with glutathione-Sepharose (GE Heathcare). After elution, the fusion proteins were dialyzed against 50 mM Tris-HCl pH7.5, 200 mM NaCl and stored at -80°C. For quantification, purified proteins were analyzed by Comassie staining and by Western blot with anti-GST polyclonal antibodies (Santa Cruz Biotechnology, Inc.), anti-MBP mAb (New England Biolabs, Inc.) and 6xHis mAb (CLONTECH Laboratories, Inc.).
GTPase assay and dissociation assay
GTPase assay were performed according to [25]. In brief, 240 nM of Tem1–6×His was incubated in 25 μl of loading buffer (20 mM Tris-HCl, pH 7.5, 25 mM NaCl, 5 mM MgCl2, and 0.1 mM DTT) containing 0.1 MBq of γ[32P]GTP or 0.03 MBq of γ[35S]GTP in the absence or presence of 150 nM of MBP-Bfa1 for 10 min at 30°C. The reaction was then put on ice, and 10 μl of reaction were added to 50 μl of reaction buffer (20 mM Tris-HCl, pH 7.5, 2 mM GTP, and 0.6 μg/μl BSA) containing 15 μM of GST-Bub2. The mixture was incubated at 30°C, and for each time point 10 μl of the reaction was diluted in 990 μl of cold washing buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 5 mM MgCl2). The samples were filtered through nitrocellulose filters, washed with 12 ml of cold washing buffer, and air dried, and the filter-bound radioactivity nucleotide was determined by scintillation counting.
Immunoprecipitation and western blot analysis
Immunoprecipitations were performed as described in [82]. Bub2-HA3, Bub2-Q132L-HA3, Tem1-HA3 and Tem1-Q79L-HA3 were immunoprecipitated from 1 mg of total extract by a HA-affinity resin (Roche). For Western blot analysis, protein extracts were prepared according to [83]. Proteins transferred to Protran membranes (Schleicher & Schuell) were probed with anti-PK mouse monoclonal antibodies for PK-tagged Bub2, with anti-GFP rat monoclonal antibodies for GFP-tagged Tem1, Bub2 and Bfa1 (Chromotek) and with an anti-HA monoclonal antibody (12CA5). Secondary antibodies were purchased from GE Healthcare, and proteins were detected by an enhanced chemiluminescence system according to the manufacturer.
Fluorescence microscopy
In situ immunofluorescence was performed according to [84]. Immunostaining of α-tubulin was performed with the YOL34 monoclonal antibody (Serotec) followed by indirect immunofluorescence using rhodamine-conjugated anti–rat antibody (1 : 100; Pierce Chemical Co.).
Cells expressing GFP and mCherry-tagged proteins were grown in minimum complete medium. Digital images of live cells, cells fixed with 3.7% formaldehyde or cold EtOH were taken with an oil 63X 1,4–1,6 HCX Plan-Apochromat objective (Zeiss) with a Coolsnap HQ2–1 charge device camera (Photometrics) mounted on a ZeissAxioimager Z1/Apotome fluorescence microscope controlled by the MetaMorph imaging system software. Z-stacks of 12 planes at 0.3 μm step size were acquired.
For analysis of actomyosin ring contraction (Fig. 4C) cells were mounted in SD medium on Fluorodishes and filmed at room temperature (~21°C) with a DeltaVision OMX microscope using a 63X 1.4 NA oil immersion objective and the softWoRx software (Applied Precision). Z stacks containing 31 planes were acquired every 1’ with a step size of 0.2 μm and a binning of 1. Z-stacks were deconvolved with Huygens (Scientific Volume Imaging) and max-projected.
For the analyses in Fig. 8, 120s time-lapse microscopy was performed using an Olympus BX51 microscope controlled by the TILLVision software (TILLPhotonics). For the localization of Kar9-YFP and Bub2-GFP, Z-stacks of four layers (step size 0.35 μm) and maximum intensity projections were used. Fluorescence microscopy was performed with a monochromator PolychromIV as light source and a CCD camera (Imago, TillPhotonics).
Image analysis and processing
Fluorescence intensity measurements of max intensity-projected images were performed using the ImageJ software. The index of Bfa1 symmetric distribution (σ, Fig. 5G) was measured using the following equation: σ (0<σ<1) = I1/I2, where I1 is the fluorescence intensity of the brightest of the two SPBs, and I2 is the fluorescence intensity of the dimmest.
The position and the orientation of the spindle were measured on max intensity-projected images using ImageJ. The position was determined measuring the distance between the bud neck and the nearest SPB. The orientation was determined measuring the smaller of the two angles that the spindle forms intersecting the polarity axis of the cell.
Adobe Photoshop and ImageJ were used to mount the images and to produce merged color images. No manipulations other than contrast and brightness adjustments were used.
Student’s t-test or chi-square test was used to evaluate statistical significance of differences, depending on whether one (t-test) or more parameters (chi-square test) were compared for each experimental condition.
Other techniques
Nuclear division was scored with a fluorescence microscope on cells stained with propidium iodide (Sigma Aldrich). Flow cytometric DNA quantification was determined according to [84] on a Becton-Dickinson FACScalibur.
Supporting Information
Zdroje
1. Inaba M, Yamashita YM (2012) Asymmetric stem cell division: precision for robustness. Cell Stem Cell 11 : 461–469. doi: 10.1016/j.stem.2012.09.003 23040475
2. Knoblich JA (2010) Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol 11 : 849–860. doi: 10.1038/nrm3010 21102610
3. Li R (2013) The art of choreographing asymmetric cell division. Dev Cell 25 : 439–450. doi: 10.1016/j.devcel.2013.05.003 23763946
4. Gonzalez C (2007) Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet 8 : 462–472. doi: 10.1038/nrg2103 17510666
5. Siller KH, Doe CQ (2009) Spindle orientation during asymmetric cell division. Nat Cell Biol 11 : 365–374. doi: 10.1038/ncb0409-365 19337318
6. Caydasi AK, Ibrahim B, Pereira G (2010) Monitoring spindle orientation: Spindle position checkpoint in charge. Cell Div 5 : 28. doi: 10.1186/1747-1028-5-28 21143992
7. Yamashita YM, Yuan H, Cheng J, Hunt AJ (2010) Polarity in stem cell division: asymmetric stem cell division in tissue homeostasis. Cold Spring Harb Perspect Biol 2: a001313. doi: 10.1101/cshperspect.a001313 20182603
8. Moore JK, Cooper JA (2010) Coordinating mitosis with cell polarity: Molecular motors at the cell cortex. Semin Cell Dev Biol 21 : 283–289. doi: 10.1016/j.semcdb.2010.01.020 20109571
9. Fraschini R, Venturetti M, Chiroli E, Piatti S (2008) The spindle position checkpoint: how to deal with spindle misalignment during asymmetric cell division in budding yeast. Biochem Soc Trans 36 : 416–420. doi: 10.1042/BST0360416 18481971
10. Krapp A, Gulli MP, Simanis V (2004) SIN and the art of splitting the fission yeast cell. Curr Biol 14: R722–730. doi: 10.1016/j.cub.2004.08.049 15341766
11. Stegmeier F, Amon A (2004) Closing Mitosis: The Functions of the Cdc14 Phosphatase and Its Regulation. Annu Rev Genet 38 : 203–232. doi: 10.1146/annurev.genet.38.072902.093051 15568976
12. Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, et al. (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk - dependent phosphorylation. Mol Cell 2 : 709–718. doi: 10.1016/S1097-2765(00)80286-5 9885559
13. Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, et al. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 : 233–244. doi: 10.1016/S0092-8674(00)80733-3 10219244
14. Visintin R, Hwang ES, Amon A (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus [see comments]. Nature 398 : 818–823. doi: 10.1038/19775 10235265
15. Stegmeier F, Visintin R, Amon A (2002) Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108 : 207–220. doi: 10.1016/S0092-8674(02)00618-9 11832211
16. Queralt E, Lehane C, Novak B, Uhlmann F (2006) Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125 : 719–732. doi: 10.1016/j.cell.2006.03.038 16713564
17. D’Amours D, Amon A (2004) At the interface between signaling and executing anaphase--Cdc14 and the FEAR network. Genes Dev 18 : 2581–2595. doi: 10.1101/gad.1247304 15520278
18. Hotz M, Leisner C, Chen D, Manatschal C, Wegleiter T, et al. (2012) Spindle pole bodies exploit the mitotic exit network in metaphase to drive their age-dependent segregation. Cell 148 : 958–972. doi: 10.1016/j.cell.2012.01.041 22385961
19. Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y (2003) Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112 : 561–574. doi: 10.1016/S0092-8674(03)00119-3 12600318
20. Pereira G, Schiebel E (2001) The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr Opin Cell Biol 13 : 762–769. doi: 10.1016/S0955-0674(00)00281-7 11698194
21. Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E (2000) Nud1p links astral microtubule organization and the control of exit from mitosis [In Process Citation]. Embo J 19 : 6475–6488. doi: 10.1093/emboj/19.23.6475 11101520
22. Valerio-Santiago M, Monje-Casas F (2011) Tem1 localization to the spindle pole bodies is essential for mitotic exit and impairs spindle checkpoint function. J Cell Biol 192 : 599–614. doi: 10.1083/jcb.201007044 21321099
23. Bourne HR, Sanders DA, McCormick F (1990) The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348 : 125–132. doi: 10.1038/348125a0 2122258
24. Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294 : 1299–1304. doi: 10.1126/science.1062023 11701921
25. Fraschini R, D’Ambrosio C, Venturetti M, Lucchini G, Piatti S (2006) Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J Cell Biol 172 : 335–346. doi: 10.1083/jcb.200507162 16449187
26. Geymonat M, Spanos A, Smith SJ, Wheatley E, Rittinger K, et al. (2002) Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J Biol Chem 277 : 28439–28445. doi: 10.1074/jbc.M202540200 12048186
27. Bourne HR, Sanders DA, McCormick F (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349 : 117–127. doi: 10.1038/349117a0 1898771
28. Schweins T, Wittinghofer A (1994) GTP-binding proteins. Structures, interactions and relationships. Curr Biol 4 : 547–550. doi: 10.1016/S0960-9822(00)00122-6 7922378
29. Neuwald AF (1997) A shared domain between a spindle assembly checkpoint protein and Ypt/Rab-specific GTPase-activators. Trends Biochem Sci 22 : 243–244. doi: 10.1016/S0968-0004(97)01073-6 9255064
30. Geymonat M, Spanos A, de Bettignies G, Sedgwick SG (2009) Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J Cell Biol 187 : 497–511. doi: 10.1083/jcb.200905114 19948498
31. Ro HS, Song S, Lee KS (2002) Bfa1 can regulate Tem1 function independently of Bub2 in the mitotic exit network of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 99 : 5436–5441. doi: 10.1073/pnas.062059999 11959999
32. Maekawa H, Priest C, Lechner J, Pereira G, Schiebel E (2007) The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J Cell Biol 179 : 423–436. doi: 10.1083/jcb.200705197 17967947
33. Chan LY, Amon A (2010) Spindle position is coordinated with cell-cycle progression through establishment of mitotic exit-activating and -inhibitory zones. Mol Cell 39 : 444–454. doi: 10.1016/j.molcel.2010.07.032 20705245
34. D’Aquino KE, Monje-Casas F, Paulson J, Reiser V, Charles GM, et al. (2005) The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol Cell 19 : 223–234. doi: 10.1016/j.molcel.2005.06.005 16039591
35. Pereira G, Schiebel E (2005) Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol Cell 19 : 209–221. doi: 10.1016/j.molcel.2005.05.030 16039590
36. Caydasi AK, Pereira G (2009) Spindle alignment regulates the dynamic association of checkpoint proteins with yeast spindle pole bodies. Dev Cell 16 : 146–156. doi: 10.1016/j.devcel.2008.10.013 19154725
37. Frenz LM, Lee SE, Fesquet D, Johnston LH (2000) The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J Cell Sci 113 Pt 19 : 3399–3408. 10984431
38. Molk JN, Schuyler SC, Liu JY, Evans JG, Salmon ED, et al. (2004) The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol Biol Cell 15 : 1519–1532. doi: 10.1091/mbc.E03-09-0708 14718561
39. Monje-Casas F, Amon A (2009) Cell polarity determinants establish asymmetry in MEN signaling. Dev Cell 16 : 132–145. doi: 10.1016/j.devcel.2008.11.002 19154724
40. Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E (2000) The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell 6 : 1–10. doi: 10.1016/S1097-2765(05)00017-1 10949022
41. Rock JM, Amon A (2011) Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev 25 : 1943–1954. doi: 10.1101/gad.17257711 21937712
42. Rock JM, Lim D, Stach L, Ogrodowicz RW, Keck JM, et al. (2013) Activation of the Yeast Hippo Pathway by Phosphorylation-Dependent Assembly of Signaling Complexes. Science 340 : 871–875. doi: 10.1126/science.1235822 23579499
43. Visintin R, Amon A (2001) Regulation of the mitotic exit protein kinases cdc15 and dbf2. Mol Biol Cell 12 : 2961–2974. doi: 10.1091/mbc.12.10.2961 11598184
44. Xu S, Huang HK, Kaiser P, Latterich M, Hunter T (2000) Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr Biol 10 : 329–332. doi: 10.1016/S0960-9822(00)00382-1 10744974
45. Cerutti L, Simanis V (1999) Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J Cell Sci 112 (Pt 14): 2313–2321. 10381387
46. Li C, Furge KA, Cheng QC, Albright CF (2000) Byr4 localizes to spindle-pole bodies in a cell cycle-regulated manner to control Cdc7 localization and septation in fission yeast. J Biol Chem 275 : 14381–14387. doi: 10.1074/jbc.275.19.14381 10799520
47. Sohrmann M, Schmidt S, Hagan I, Simanis V (1998) Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev 12 : 84–94. doi: 10.1101/gad.12.1.84 9420333
48. Pan X, Eathiraj S, Munson M, Lambright DG (2006) TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442 : 303–306. doi: 10.1038/nature04847 16855591
49. Miller RK, Rose MD (1998) Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol 140 : 377–390. doi: 10.1083/jcb.140.2.377 9442113
50. Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K (1995) Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol 130 : 687–700. doi: 10.1083/jcb.130.3.687 7622568
51. Hu F, Wang Y, Liu D, Li Y, Qin J, et al. (2001) Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107 : 655–665. doi: 10.1016/S0092-8674(01)00580-3 11733064
52. Valerio-Santiago M, de Los Santos-Velazquez AI, Monje-Casas F (2013) Inhibition of the mitotic exit network in response to damaged telomeres. PLoS Genet 9: e1003859. doi: 10.1371/journal.pgen.1003859 24130507
53. Asakawa K, Yoshida S, Otake F, Toh EA (2001) A Novel Functional Domain of Cdc15 Kinase Is Required for Its Interaction With Tem1 GTPase in Saccharomyces cerevisiae. Genetics 157 : 1437–1450. 11290702
54. Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V (1997) The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev 11 : 1519–1534. doi: 10.1101/gad.11.12.1519 9203579
55. Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, et al. (1998) Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol 142 : 1301–1312. doi: 10.1083/jcb.142.5.1301 9732290
56. Park SY, Cable AE, Blair J, Stockstill KE, Shannnon KB (2009) Bub2 regulation of cytokinesis and septation in budding yeast. BMC Cell Biol 10 : 43. doi: 10.1186/1471-2121-10-43 19490645
57. Kahana JA, Schlenstedt G, Evanchuk DM, Geiser JR, Hoyt MA, et al. (1998) The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol Biol Cell 9 : 1741–1756. doi: 10.1091/mbc.9.7.1741 9658168
58. Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 : 806–810. doi: 10.1038/nature05649 17314980
59. Costanzo M, Baryshnikova A, Myers CL, Andrews B, Boone C (2011) Charting the genetic interaction map of a cell. Curr Opin Biotechnol 22 : 66–74. doi: 10.1016/j.copbio.2010.11.001 21111604
60. Tong AH, Lesage G, Bader GD, Ding H, Xu H, et al. (2004) Global mapping of the yeast genetic interaction network. Science 303 : 808–813. doi: 10.1126/science.1091317 14764870
61. Caydasi AK, Lohel M, Grunert G, Dittrich P, Pereira G, et al. (2012) A dynamical model of the spindle position checkpoint. Mol Syst Biol 8 : 582. doi: 10.1038/msb.2012.15 22580890
62. Kim J, Luo G, Bahk YY, Song K (2012) Cdc5-dependent asymmetric localization of bfa1 fine-tunes timely mitotic exit. PLoS Genet 8: e1002450. doi: 10.1371/journal.pgen.1002450 22253605
63. Konig C, Maekawa H, Schiebel E (2010) Mutual regulation of cyclin-dependent kinase and the mitotic exit network. J Cell Biol 188 : 351–368. doi: 10.1083/jcb.200911128 20123997
64. Cepeda-Garcia C, Delgehyr N, Juanes Ortiz MA, ten Hoopen R, Zhiteneva A, et al. (2010) Actin-mediated delivery of astral microtubules instructs Kar9p asymmetric loading to the bud-ward spindle pole. Mol Biol Cell 21 : 2685–2695. doi: 10.1091/mbc.E10-03-0197 20534809
65. Juanes MA, Twyman H, Tunnacliffe E, Guo Z, ten Hoopen R, et al. (2013) Spindle pole body history intrinsically links pole identity with asymmetric fate in budding yeast. Curr Biol 23 : 1310–1319. doi: 10.1016/j.cub.2013.05.057 23810537
66. Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4 : 686–689. doi: 10.1038/nsb0997-686 9302992
67. Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, et al. (1997) The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277 : 333–338. doi: 10.1126/science.277.5324.333 9219684
68. Gavriljuk K, Gazdag EM, Itzen A, Kotting C, Goody RS, et al. (2012) Catalytic mechanism of a mammalian Rab.RabGAP complex in atomic detail. Proc Natl Acad Sci U S A 109 : 21348–21353. doi: 10.1073/pnas.1214431110 23236136
69. Langemeyer L, Nunes Bastos R, Cai Y, Itzen A, Reinisch KM, et al. (2014) Diversity and plasticity in Rab GTPase nucleotide release mechanism has consequences for Rab activation and inactivation. Elife 3: e01623. doi: 10.7554/eLife.01623 24520163
70. Geymonat M, Spanos A, Walker PA, Johnston LH, Sedgwick SG (2003) In Vitro Regulation of Budding Yeast Bfa1/Bub2 GAP Activity by Cdc5. J Biol Chem 278 : 14591–14594. doi: 10.1074/jbc.C300059200 12637549
71. Baro B, Rodriguez-Rodriguez JA, Calabria I, Hernaez ML, Gil C, et al. (2013) Dual Regulation of the mitotic exit network (MEN) by PP2A-Cdc55 phosphatase. PLoS Genet 9: e1003966. doi: 10.1371/journal.pgen.1003966 24339788
72. Johnson AE, Gould KL (2011) Dma1 ubiquitinates the SIN scaffold, Sid4, to impede the mitotic localization of Plo1 kinase. EMBO J 30 : 341–354. doi: 10.1038/emboj.2010.317 21131906
73. Krapp A, Cano E, Simanis V (2003) Mitotic hyperphosphorylation of the fission yeast SIN scaffold protein cdc11p is regulated by the protein kinase cdc7p. Curr Biol 13 : 168–172. doi: 10.1016/S0960-9822(02)01417-3 12546793
74. Singh NS, Shao N, McLean JR, Sevugan M, Ren L, et al. (2011) SIN-inhibitory phosphatase complex promotes Cdc11p dephosphorylation and propagates SIN asymmetry in fission yeast. Curr Biol 21 : 1968–1978. doi: 10.1016/j.cub.2011.10.051 22119525
75. Feoktistova A, Morrell-Falvey J, Chen JS, Singh NS, Balasubramanian MK, et al. (2012) The fission yeast septation initiation network (SIN) kinase, Sid2, is required for SIN asymmetry and regulates the SIN scaffold, Cdc11. Mol Biol Cell 23 : 1636–1645. doi: 10.1091/mbc.E11-09-0792 22419817
76. Hotz M, Barral Y (2014) The Mitotic Exit Network: new turns on old pathways. Trends Cell Biol 24 : 145–152. doi: 10.1016/j.tcb.2013.09.010 24594661
77. Hampoelz B, Knoblich JA (2004) Heterotrimeric G proteins: new tricks for an old dog. Cell 119 : 453–456. doi: 10.1016/j.cell.2004.10.025 15537535
78. Gonczy P (2008) Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol 9 : 355–366. doi: 10.1038/nrm2388 18431399
79. Wach A, Brachat A, Pohlmann R, Philippsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 : 1793–1808. doi: 10.1002/yea.320101310 7747518
80. Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, et al. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21 : 947–962. doi: 10.1002/yea.1142 15334558
81. Sheff MA, Thorn KS (2004) Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21 : 661–670. doi: 10.1002/yea.1130 15197731
82. Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, et al. (2001) Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. Embo J 20 : 6648–6659. doi: 10.1093/emboj/20.23.6648 11726501
83. Surana U, Amon A, Dowzer C, McGrew J, Byers B, et al. (1993) Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. Embo J 12 : 1969–1978. 8491189
84. Fraschini R, Formenti E, Lucchini G, Piatti S (1999) Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J Cell Biol 145 : 979–991. doi: 10.1083/jcb.145.5.979 10352016
Štítky
Genetika Reprodukční medicína
Článek 2014 Reviewer Thank YouČlánek Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic TranslationČlánek Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate CellsČlánek K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in ArabidopsisČlánek A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis inČlánek Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in MiceČlánek The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- 2014 Reviewer Thank You
- Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for and Myocardial Infarction
- African Glucose-6-Phosphate Dehydrogenase Alleles Associated with Protection from Severe Malaria in Heterozygous Females in Tanzania
- Genomics of Divergence along a Continuum of Parapatric Population Differentiation
- microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes
- A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in
- Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic Translation
- Partially Redundant Enhancers Cooperatively Maintain Mammalian Expression Above a Critical Functional Threshold
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Mutations in Result in Ocular Coloboma, Microcornea and Cataracts
- A Genome-Wide Hybrid Incompatibility Landscape between and
- Recurrent Evolution of Melanism in South American Felids
- Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells
- Tissue Expression Pattern of PMK-2 p38 MAPK Is Established by the miR-58 Family in
- Essential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
- Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- RR-1 Cuticular Protein TcCPR4 Is Required for Formation of Pore Canals in Rigid Cuticle
- GC-Content Evolution in Bacterial Genomes: The Biased Gene Conversion Hypothesis Expands
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
- K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
- A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in
- HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
- Mitochondrial and Cytoplasmic ROS Have Opposing Effects on Lifespan
- Structured Observations Reveal Slow HIV-1 CTL Escape
- An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging
- Combining Natural Sequence Variation with High Throughput Mutational Data to Reveal Protein Interaction Sites
- Transhydrogenase Promotes the Robustness and Evolvability of Deficient in NADPH Production
- Regulators of Autophagosome Formation in Muscles
- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Eye Selector Logic for a Coordinated Cell Cycle Exit
- Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations
- The DNA Polymerase δ Has a Role in the Deposition of Transcriptionally Active Epigenetic Marks, Development and Flowering
- Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice
- Membrane Recognition and Dynamics of the RNA Degradosome
- P-TEFb, the Super Elongation Complex and Mediator Regulate a Subset of Non-paused Genes during Early Embryo Development
- is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure
- A Pleiotropy-Informed Bayesian False Discovery Rate Adapted to a Shared Control Design Finds New Disease Associations From GWAS Summary Statistics
- Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers
- and Hyperdrive Mechanisms (in Mouse Meiosis)
- Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells
- Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma by Repressing Multiple Targets
- Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
- Abscission Is Regulated by the ESCRT-III Protein Shrub in Germline Stem Cells
- Temperature Stress Mediates Decanalization and Dominance of Gene Expression in
- Transcriptome Wide Annotation of Eukaryotic RNase III Reactivity and Degradation Signals
- The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway
- Sex-specific -regulatory Variation on the X Chromosome
- Regulation of Toll-like Receptor Signaling by the SF3a mRNA Splicing Complex
- Modeling of the Human Alveolar Rhabdomyosarcoma Chromosome Translocation in Mouse Myoblasts Using CRISPR-Cas9 Nuclease
- Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
- TIM Binds Importin α1, and Acts as an Adapter to Transport PER to the Nucleus
- Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication
- The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
- Fatty Acid Synthase Cooperates with Glyoxalase 1 to Protect against Sugar Toxicity
- Region-Specific Activation of mRNA Translation by Inhibition of Bruno-Mediated Repression
- An Essential Role of the Arginine Vasotocin System in Mate-Guarding Behaviors in Triadic Relationships of Medaka Fish ()
- Interaction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
- Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
- Prodomain Removal Enables Neto to Stabilize Glutamate Receptors at the Neuromuscular Junction
- Recent Selective Sweeps in North American Show Signatures of Soft Sweeps
- Identification and Functional Analysis of Healing Regulators in
- A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2
- Drosophila Casein Kinase I Alpha Regulates Homolog Pairing and Genome Organization by Modulating Condensin II Subunit Cap-H2 Levels
- The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Ovary
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



