-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
and Hyperdrive Mechanisms (in Mouse Meiosis)
article has not abstract
Published in the journal: . PLoS Genet 11(2): e32767. doi:10.1371/journal.pgen.1004950
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1004950Summary
article has not abstract
“Mendelism is a magnificent invention for fairly testing genes in many combinations, like an elegant factorial experimental design. Yet it is vulnerable at many points and is in constant danger of subversion by cheaters that seem particularly adept at finding such points.”
- James F. Crow [1]Mendelian transmission is established during meiosis, the cell division that generates haploid gametes (e.g., sperm and eggs) from diploid germ cells. Meiosis does not, however, have to be fair. Selfish genetic elements, or meiotic drivers, have evolved to cheat this process in order to be packaged into functional gametes more often than the expected 50% probability. By biasing allele transmission in their favor, meiotic drive alleles can short-circuit natural selection, causing their spread even if they are harmful to organismal fitness. Indeed, meiotic drive alleles are thought to be directly or indirectly associated with infertility in diverse eukaryotes, from fungi to flies to humans [2]. Drive occurring in male meiosis may have immediate consequences to fertility, potentially facilitating its detection. In contrast, drive in female meiosis could lead to very subtle skews in inheritance patterns and no overt signs of infertility; such driver alleles may therefore go unnoticed even if they are more pervasive [3, 4]. Surprisingly few meiotic drivers have been conclusively identified. There are three major roadblocks to the efficient identification and validation of meiotic drive alleles. First is the prevalent usage of isogenic, lab-domesticated organisms in genetics research; driver alleles can only be identified in heterozygotes. Second, the rapid evolution of suppressors to alleviate the deleterious effects of meiotic drivers can obscure drive in intraspecific crosses and also rapidly extinguish meiotic drive alleles by eliminating their evolutionary advantage [5]. Third, it can often be difficult to distinguish the actions of meiotic drive alleles from inherent viability defects associated with the underrepresented allele. Overcoming many of these limitations, a new report in this issue of PLOS Genetics by de Villena and colleagues provides compelling evidence for a massive copy number expansion that is causally linked to female meiotic drive in mice [6].
Didion et al. began their study by investigating the genetic basis of transmission ratio distortion (TRD) of a region of mouse chromosome 2. This TRD had been previously observed in a number of crosses, including, most recently, in the recombinant inbred Collaborative Cross (CC) lines designed for genetic analyses [7]. TRD was observed in favor of the WSB/EiJ allele across a ~50 Mb region in the middle of chromosome 2 (WSB/EiJ was one of the eight original inbred strains used to create recombinant inbred lines [5]). In the present report, Didion et al. first confirmed the TRD favoring the WSB/EiJ allele in the Diversity Outbred (DO) population of outbred mice derived from CC lines designed for mapping traits. Crosses of various WSB/EiJ heterozygotes to tester strains showed that TRD was specific to heterozygous females. These findings already eliminated the possibility of postmeiotic dysfunction of selective male gametes as the basis for TRD; such postmeiotic dysfunction is the basis for TRD caused by Segregation Distorter in Drosophila melanogaster [8] and the t-haplotype in mouse [9].
This finding left open two possibilities, the first being that the bias originates from a selective postfertilization defect. Indeed, Didion et al. find that the amount of TRD does negatively correlate with litter size, suggesting that lethality of oocytes or embryos inheriting the non-WSB/EiJ allele could contribute to the TRD. However, the observed decreases in litter sizes are insufficient to explain the magnitude of TRD observed. In addition, despite the decreased litter sizes, the absolute number of progeny inheriting the WSB/EiJ allele in crosses exhibiting TRD was significantly higher than the number inheriting the allele in crosses without TRD. Thus, although embryonic lethality occurs in crosses showing TRD, Didion et al. propose that “true meiotic drive” must occur to fully explain the TRD in favor of the WSB/EiJ allele. Such drive occurs because, in female meiosis, only one of four meiotic products is selected in the oocyte [10].
Based on the meiotic drive inference, the authors dubbed the causative allele R2d2 (Responder to drive on chr. 2). To map R2d2, Didion et al. again exploited the impressive genetic power of the CC and DO lines. By genotyping over 400 mice, they identified recombination events within the candidate region on chromosome 2. All eight recombinant chromosomes that showed TRD shared a 9.3 Mb region containing a 127 kb unit of DNA (R2d2) that is repeated ~36 times in WSB/EiJ (see Fig. 1). Three other mice strains previously shown to exhibit TRD also contain R2d sequences at a high copy number. In contrast, R2d is only present at 1–2X copy number in the reference mouse genome and other mouse strains that lack the TRD phenotype. In non-TRD strains, like the reference genome, the R2d sequences appear to be present only in the (presumed) ancestral location spread over a 158 kb region (R2d1) ~6 Mb distal from the R2d2 locus (Fig. 1A). These results suggested the hypothesis that whereas all mouse strains contain R2d1, a copy number expansion at R2d2 is causative for TRD (Fig. 1). A satisfying confirmation of the hypothesis emerged from the genetic instability of the R2d2 repeat cluster. Didion et al. identified a WSB/EiJ-derived female in which one of the R2d2 repeat arrays had collapsed from ~34 to ~11 copies. This collapse resulted in both the loss of TRD and increased litter sizes (Fig. 1). Together, this beautiful series of experiments, relying on both classical genetics and genomic assembly mapping, identify and confirm the R2d2 expansion as causal for female meiotic drive.
Fig. 1. Transmission ratio distortion (TRD) caused by a high copy R2d2 array. 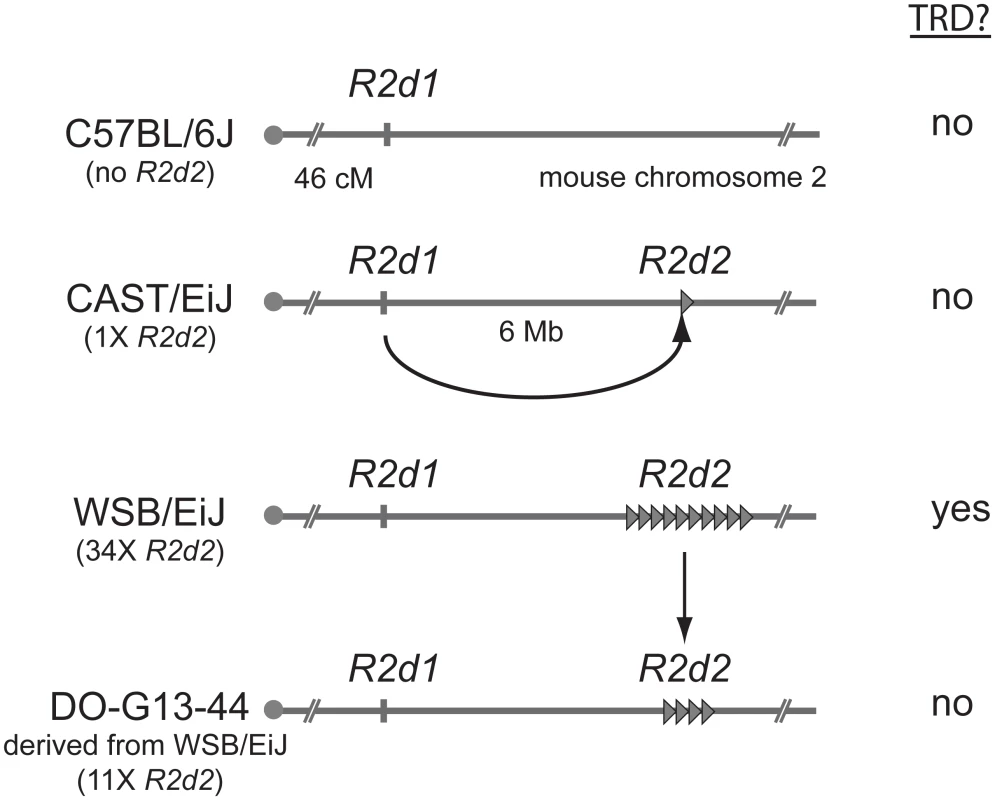
In the absence of the R2d2 sequence (e.g., C57BL/6J), or when the copy number of the sequence is low (e.g., CAST/EiJ), allele transmission through heterozygous females is Mendelian and no embryonic lethality is observed. When the copy number is high in the appropriate genetic background (e.g. WSB/EiJ), the litter sizes of heterozygous females are reduced and the R2d2 locus shows TRD. When the copy number of R2d2 repeats decreases (e.g., DO-G13–44), litter sizes increase and Mendelian segregation is restored. How does R2d2 expansion cause meiotic drive? Didion et al. favor the model that the R2d2 expansion leads to preferential inheritance in meiosis. Such non-Mendelian inheritance would occur via the formation of neocentromere-like activity on cis-acting sequences expanded in the R2d2 cluster, which favor their preferential orientation and thereby inclusion in the oocyte [11]. This situation would be highly reminiscent of the knob elements in maize that also take advantage of female meiosis asymmetries to increase their likelihood of inclusion into the oocyte [12]. Akin to knob elements, R2d2 drive might occur in Meiosis II [3, 4]. Under this model, the ensuing R2d2-dependent embryonic lethality would result from generating eggs that are aneuploid for chromosome 2.
Not all dams heterozygous for R2d2 show TRD, implicating the requirement of at least one additional modifier locus that must be present to manifest TRD. These additional loci could represent allelic variants of meiotic drive suppressors, which reduce the harmful fitness effects of drive and associated embryonic lethality. Alternatively, these could represent “trans-acting factors” (distorters) that bind R2d2 and endow it with microtubule attachment or motor function that results in neocentromere activity [12]. These findings would be at odds with predictions from the theory that distorter and responder loci ought to be tightly linked, so as to not be separated by recombination [13]. One intriguing possibility is that the only protein encoded by the R2d unit, Cwc2, might itself bind the R2d2 cluster and contribute to TRD.
In sum, the paper by Didion et al. represents a tour de force in characterizing a complex multilocus TRD system in a genetically tractable mammalian model system. Uncovering the molecular mechanisms underlying the transmission of exceptional meiotic drive alleles like R2d2, as well as their evolutionary origins, will broaden our general understanding of fertility and chromosome segregation. Such work also reinforces previous findings about the insidious and relentless nature of meiotic competition between chromosomes [1, 2, 10].
Zdroje
1. Crow JF (1988) The ultraselfish gene. Genetics 118 : 389–391. 3130288
2. Burt A, Trivers R (2006) Genes in conflict: the biology of selfish genetic elements. Cambridge, Mass.: Belknap Press of Harvard University Press. viii, 602 p., 608 p. of plates p.
3. Tao Y, Hartl DL, Laurie CC (2001) Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci U S A 98 : 13183–13188. doi: 10.1073/pnas.231478798 11687638
4. Didion JP, Morgan AP, Clayshulte AM-F, Mcmullan RC, Yadgary L, et al. (2014) A multi-megabase copy number gain causes maternal transmission ratio distortion on mouse Chromosome 2. PLOS Genet 11: e1004850.
5. Collaborative Cross C (2012) The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190 : 389–401. doi: 10.1534/genetics.111.132639
6. Larracuente AM, Presgraves DC (2012) The selfish Segregation Distorter gene complex of Drosophila melanogaster. Genetics 192 : 33–53. doi: 10.1534/genetics.112.141390 22964836
7. Ardlie KG (1998) Putting the brake on drive: meiotic drive of t haplotypes in natural populations of mice. Trends Genet 14 : 189–193. doi: 10.1016/S0168-9525(98)01455-3 9613203
8. Sandler L, Novitski E (1957) Meiotic Drive as an Evolutionary Force. The American Naturalist 91 : 105–110. doi: 10.1086/281969
9. Zwick ME, Salstrom JL, Langley CH (1999) Genetic variation in rates of nondisjunction: association of two naturally occurring polymorphisms in the chromokinesin nod with increased rates of nondisjunction in Drosophila melanogaster. Genetics 152 : 1605–1614. 10430586
10. Yu HG, Hiatt EN, Chan A, Sweeney M, Dawe RK (1997) Neocentromere-mediated chromosome movement in maize. J Cell Biol 139 : 831–840. doi: 10.1083/jcb.139.4.831 9362502
11. Buckler ESt, Phelps-Durr TL, Buckler CS, Dawe RK, Doebley JF, et al. (1999) Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153 : 415–426. 10471723
12. Malik HS (2005) Mimulus finds centromeres in the driver’s seat. Trends Ecol Evol 20 : 151–154. doi: 10.1016/j.tree.2005.01.014 16701359
13. Charlesworth B, Hartl DL (1978) Population Dynamics of the Segregation Distorter Polymorphism of DROSOPHILA MELANOGASTER. Genetics 89 : 171–192. 17248828
Štítky
Genetika Reprodukční medicína
Článek 2014 Reviewer Thank YouČlánek Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic TranslationČlánek Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate CellsČlánek K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in ArabidopsisČlánek A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis inČlánek Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in MiceČlánek The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 2- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- 2014 Reviewer Thank You
- Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for and Myocardial Infarction
- African Glucose-6-Phosphate Dehydrogenase Alleles Associated with Protection from Severe Malaria in Heterozygous Females in Tanzania
- Genomics of Divergence along a Continuum of Parapatric Population Differentiation
- microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes
- A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in
- Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic Translation
- Partially Redundant Enhancers Cooperatively Maintain Mammalian Expression Above a Critical Functional Threshold
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Mutations in Result in Ocular Coloboma, Microcornea and Cataracts
- A Genome-Wide Hybrid Incompatibility Landscape between and
- Recurrent Evolution of Melanism in South American Felids
- Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells
- Tissue Expression Pattern of PMK-2 p38 MAPK Is Established by the miR-58 Family in
- Essential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
- Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- RR-1 Cuticular Protein TcCPR4 Is Required for Formation of Pore Canals in Rigid Cuticle
- GC-Content Evolution in Bacterial Genomes: The Biased Gene Conversion Hypothesis Expands
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
- K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
- A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in
- HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
- Mitochondrial and Cytoplasmic ROS Have Opposing Effects on Lifespan
- Structured Observations Reveal Slow HIV-1 CTL Escape
- An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging
- Combining Natural Sequence Variation with High Throughput Mutational Data to Reveal Protein Interaction Sites
- Transhydrogenase Promotes the Robustness and Evolvability of Deficient in NADPH Production
- Regulators of Autophagosome Formation in Muscles
- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Eye Selector Logic for a Coordinated Cell Cycle Exit
- Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations
- The DNA Polymerase δ Has a Role in the Deposition of Transcriptionally Active Epigenetic Marks, Development and Flowering
- Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice
- Membrane Recognition and Dynamics of the RNA Degradosome
- P-TEFb, the Super Elongation Complex and Mediator Regulate a Subset of Non-paused Genes during Early Embryo Development
- is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure
- A Pleiotropy-Informed Bayesian False Discovery Rate Adapted to a Shared Control Design Finds New Disease Associations From GWAS Summary Statistics
- Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers
- and Hyperdrive Mechanisms (in Mouse Meiosis)
- Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells
- Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma by Repressing Multiple Targets
- Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
- Abscission Is Regulated by the ESCRT-III Protein Shrub in Germline Stem Cells
- Temperature Stress Mediates Decanalization and Dominance of Gene Expression in
- Transcriptome Wide Annotation of Eukaryotic RNase III Reactivity and Degradation Signals
- The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway
- Sex-specific -regulatory Variation on the X Chromosome
- Regulation of Toll-like Receptor Signaling by the SF3a mRNA Splicing Complex
- Modeling of the Human Alveolar Rhabdomyosarcoma Chromosome Translocation in Mouse Myoblasts Using CRISPR-Cas9 Nuclease
- Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
- TIM Binds Importin α1, and Acts as an Adapter to Transport PER to the Nucleus
- Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication
- The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
- Fatty Acid Synthase Cooperates with Glyoxalase 1 to Protect against Sugar Toxicity
- Region-Specific Activation of mRNA Translation by Inhibition of Bruno-Mediated Repression
- An Essential Role of the Arginine Vasotocin System in Mate-Guarding Behaviors in Triadic Relationships of Medaka Fish ()
- Interaction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
- Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
- Prodomain Removal Enables Neto to Stabilize Glutamate Receptors at the Neuromuscular Junction
- Recent Selective Sweeps in North American Show Signatures of Soft Sweeps
- Identification and Functional Analysis of Healing Regulators in
- A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2
- Drosophila Casein Kinase I Alpha Regulates Homolog Pairing and Genome Organization by Modulating Condensin II Subunit Cap-H2 Levels
- The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Ovary
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



