-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Recent Loss of Self-Incompatibility by Degradation of the Male Component in Allotetraploid
The evolutionary transition from outcrossing to self-fertilization (selfing) through the loss of self-incompatibility (SI) is one of the most prevalent events in flowering plants, and its genetic basis has been a major focus in evolutionary biology. In the Brassicaceae, the SI system consists of male and female specificity genes at the S-locus and of genes involved in the female downstream signaling pathway. During recent decades, much attention has been paid in particular to clarifying the genes responsible for the loss of SI. Here, we investigated the pattern of polymorphism and functionality of the female specificity gene, the S-locus receptor kinase (SRK), in allotetraploid Arabidopsis kamchatica. While its parental species, A. lyrata and A. halleri, are reported to be diploid and mainly self-incompatible, A. kamchatica is self-compatible. We identified five highly diverged SRK haplogroups, found their disomic inheritance and, for the first time in a wild allotetraploid species, surveyed the geographic distribution of SRK at the two homeologous S-loci across the species range. We found intact full-length SRK sequences in many accessions. Through interspecific crosses with the self-incompatible and diploid congener A. halleri, we found that the female components of the SI system, including SRK and the female downstream signaling pathway, are still functional in these accessions. Given the tight linkage and very rare recombination of the male and female components on the S-locus, this result suggests that the degradation of male components was responsible for the loss of SI in A. kamchatica. Recent extensive studies in multiple Brassicaceae species demonstrate that the loss of SI is often derived from mutations in the male component in wild populations, in contrast to cultivated populations. This is consistent with theoretical predictions that mutations disabling male specificity are expected to be more strongly selected than mutations disabling female specificity, or the female downstream signaling pathway.
Published in the journal: . PLoS Genet 8(7): e32767. doi:10.1371/journal.pgen.1002838
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002838Summary
The evolutionary transition from outcrossing to self-fertilization (selfing) through the loss of self-incompatibility (SI) is one of the most prevalent events in flowering plants, and its genetic basis has been a major focus in evolutionary biology. In the Brassicaceae, the SI system consists of male and female specificity genes at the S-locus and of genes involved in the female downstream signaling pathway. During recent decades, much attention has been paid in particular to clarifying the genes responsible for the loss of SI. Here, we investigated the pattern of polymorphism and functionality of the female specificity gene, the S-locus receptor kinase (SRK), in allotetraploid Arabidopsis kamchatica. While its parental species, A. lyrata and A. halleri, are reported to be diploid and mainly self-incompatible, A. kamchatica is self-compatible. We identified five highly diverged SRK haplogroups, found their disomic inheritance and, for the first time in a wild allotetraploid species, surveyed the geographic distribution of SRK at the two homeologous S-loci across the species range. We found intact full-length SRK sequences in many accessions. Through interspecific crosses with the self-incompatible and diploid congener A. halleri, we found that the female components of the SI system, including SRK and the female downstream signaling pathway, are still functional in these accessions. Given the tight linkage and very rare recombination of the male and female components on the S-locus, this result suggests that the degradation of male components was responsible for the loss of SI in A. kamchatica. Recent extensive studies in multiple Brassicaceae species demonstrate that the loss of SI is often derived from mutations in the male component in wild populations, in contrast to cultivated populations. This is consistent with theoretical predictions that mutations disabling male specificity are expected to be more strongly selected than mutations disabling female specificity, or the female downstream signaling pathway.
Introduction
The evolutionary transition from outcrossing to predominant self-fertilization (selfing) is one of the most prevalent events in flowering plants [1]–[4]. Although increased homozygosity caused by selfing often leads to reduced fitness in the offspring (inbreeding depression), the inherent transmission advantage would favor selfing [5], [6]. This is because selfers can transmit gametes in three ways—as both the ovule and pollen donor for their own selfed progeny and as the pollen donor for outcrossed progeny—whereas outcrossers cannot serve as pollen donors for their selfed progeny. Thus, an allele promoting selfing has a 3∶2 transmission advantage relative to an allele promoting outcrossing. Selfing would also be advantageous, because selfers can reproduce when pollinators or mates are scarce (“reproductive assurance” [6]–[9]). Theory suggests that selfing should evolve when these advantages outweigh the costs of inbreeding depression [6], [10], [11].
In many flowering plants, predominant selfing evolved through the loss of self-incompatibility (SI) [2], [3]. SI systems have evolved multiple times in diverse lineages of flowering plants. They generally consist of female and male specificity genes at the S-locus and other genes involved in signaling pathways [12]–[14]. In SI species, the S-locus region is subject to negative frequency-dependent selection, which is a classic example of multiallelic balancing selection [15]. The molecular basis of the SI system has been studied extensively and is well characterized in the Brassicaceae. Here, female specificity is known to be determined by the S-locus receptor kinase (SRK) and male specificity is determined by the S-locus cysteine-rich protein (SCR; also known as S-locus protein 11, SP11). SRK is a transmembrane serine/threonine receptor kinase that functions on the stigma, and SCR is a small cysteine-rich protein present in the pollen coat that acts as a ligand for the SRK receptor protein [12]–[14], [16]–[19]. SRK and SCR are tightly linked at the S-locus, where dozens of highly divergent sequence groups, called S-haplogroups (or S-haplotypes or S-alleles), are segregating. S-haplogroups confer specificity in self-recognition: direct interaction between SRK and SCR of the same S-haplogroup leads to the inhibition of pollen germination at the stigma [13], [20]. In the Brassicaceae, several genes involved in the female downstream signaling pathway of SRK have also been reported, such as M-locus protein kinase (MLPK) and ARM-repeat containing 1 (ARC1) [13], [21]–[23].
The genetic basis for the recurrent loss of SI has been a major focus from both theoretical and empirical viewpoints [4], [24]–[29]. In particular, much attention has been paid to clarifying which mutations are responsible for the loss of SI, and whether these mutations are in the female specificity gene, the male specificity gene or in downstream signaling pathways [4], [24]–[29]. With the advantage of a suite of molecular tools, the most extensively studied species in the Brassicaceae is the diploid self-compatible Arabidopsis thaliana [4], [29]–[42]. Whereas a number of mutations disabling the male and female components have been identified in wild accessions of A. thaliana [30], [33], [36], [38], [39], many accessions still retain full-length and expressed SRK sequences [38], [41]. Importantly, interspecific crossings using the self-incompatible Arabidopsis halleri revealed that some A. thaliana accessions, including Wei-1, retain a functioning female SI reaction, demonstrating that all female components including SRK and the female downstream signaling pathway are still functional [41]. In addition, Tsuchimatsu et al. [41] reported that a 213-base-pair (bp) inversion (or its derivative haplotypes) in the SCR gene is found in 95% of European accessions. When the 213-bp inversion in SCR was inverted and expressed in transgenic Wei-1 plants, the functional SCR restored the SI reaction. These results suggested that degradation of SCR (the male specificity gene) was primarily responsible for the evolutionary loss of SI of the S-haplogroup in European A. thaliana, while other mutations at genes involved in the downstream signaling pathway might have contributed to some extent [35], [39].
To understand whether such mutations in the male components of the S-locus are common in the recurrent evolution of self-compatibility in the Brassicaceae, empirical examples need to be investigated in other self-compatible species. In addition to A. thaliana, there are a few reports on the pattern of polymorphism at the S-locus in self-compatible species in Brassicaceae, such as Capsella rubella [43] and Leavenworthia alabamica [44]. However, the genetic and molecular bases responsible for the loss of SI are still unknown in these species. A major obstacle to charting the history of the S-locus in self-compatible species has been in distinguishing the primary inactivating mutation from subsequent decay of the nonfunctional S-haplogroups by further mutations. This is because all genes involved in this signaling pathway are expected to be released from selection pressure and to evolve neutrally once the SI system ceases to function [4], [28], [39], [41], although pleiotropy of these genes could play a role in maintaining the functionality of the signaling pathway [42], [45]. To study the primary mutations, a powerful approach would be to combine experiments confirming gene function with population genetic analyses finding gene-disruptive mutations.
Arabidopsis kamchatica would be a good model system to address this issue. It is a self-compatible species, and originated through allopolyploidization of two species, A. halleri and A. lyrata, which are reported to be diploid [46]–[51]. Shimizu-Inatsugi et al. [48] reported that multiple haplotypes of nuclear and chloroplast sequences of A. kamchatica are identical to those of their parental species, indicating that multiple diploid individuals of A. halleri and A. lyrata contributed to the origin of A. kamchatica. In particular, A. kamchatica and A. halleri share four chloroplast haplotypes, strongly suggesting that at least four diploid individuals of A. halleri contributed independently to the multiple origins of A. kamchatica [48]. The two parental species are predominantly self-incompatible, although some of North American populations of A. lyrata are known to be self-compatible [26], [29]. Their SI systems have been extensively studied [26], [52]–[61]. Most of these studies have focused on SRK to characterize S-haplogroups, because novel SRK haplogroups can be isolated relatively easily by using PCR primers that were designed in the conserved regions of SRK [52], [55], while much fewer SCR sequences have been isolated because of its extreme polymorphism [30], [40], [62], [63]. To date, 40 and 30 SRK haplogroups have been reported in A. lyrata and A. halleri, respectively, and studies of nucleotide polymorphisms and divergence using large sets of SRK sequences revealed various characteristics of the S-locus, such as the spatial distribution of S-haplogroups, complex dominance interactions and transspecific sharing of S-haplogroups among species [55]–[57], [59], [60], [64]. The wealth of knowledge available on these parental species enabled us to investigate nucleotide polymorphisms of the S-locus in self-compatible A. kamchatica. In addition, A. kamchatica would be a novel model to investigate the mechanism underlying the loss of SI in polyploid species. The relationship between self-compatibility and polyploidy has been debated for more than 60 years, as it is argued that polyploids have higher selfing rates than their diploid relatives [1], [65] (but see also [66] for controversy). Hypotheses have been proposed to explain this association, such as: (1) self-compatible individuals in polyploids would not suffer from limitation of mates of the same ploidy level [1], [66]–[70]; and (2) inbreeding depression would be reduced by having multiple gene copies [10], [66], [71], [72]. Despite numerous ecological and evolutionary studies, the molecular mechanisms underlying the evolution of self-compatibility of polyploid species are still poorly understood.
To understand the mechanisms underlying the loss of SI in A. kamchatica, we first isolated SRK haplogroups from A. kamchatica by examining 48 populations across its distribution range (Table S1). Based on the analyses of nucleotide divergence from parental species and the distribution of SRK haplogroups with respect to population structure, we studied how S-haplogroups in A. kamchatica have originated from the parental species. To understand the approximate timescale of this evolutionary event, we also estimated the divergence time of A. kamchatica from its parental species based on the nucleotide divergence of multiple nuclear genes other than SRK. This is because speciation time would be used as the upper boundary of the time estimate of the evolution of self-compatibility in a species, when the progenitor species was self-incompatible [43], [73]. We tested the function of SRK haplogroups through interspecific crossing with self-incompatible A. halleri, and confirmed the disomic inheritance and allelic relationships of SRK haplogroups by segregation analyses in experimental and natural populations of A. kamchatica. Most importantly, our interspecific crossing with A. halleri also indicated the retained function of the female component of SI in A. kamchatica, suggesting that the degradation of male components was responsible for the loss of SI. We suggest that the degradation of male components among the Brassicaceae might represent a general trend in the evolution of self-compatibility in wild populations in contrast to cultivated populations.
Results
Five highly diverged SRK-like sequences in A. kamchatica revealed by PCR–based screening
Through PCR-based screening, we obtained five partial SRK-like sequences from A. kamchatica, named AkSRK-A, AkSRK-B, AkSRK-C, AkSRK-D and AkSRK-E. Our five SRK sequences were aligned with S-domain sequences available for SRK from A. halleri and A. lyrata [52], [54], [55], [58], [59], [61], and a phylogenetic tree including a total of 76 SRK sequences was generated using the neighbor-joining method (Figure 1). While previous studies reported that the SRK haplogroups are transspecifically shared among A. halleri, A. lyrata and A. thaliana [59], [62], the tree clearly shows that the SRK haplogroups are also transspecifically shared between A. kamchatica, A. halleri and A. lyrata (Figure 1).
Fig. 1. Phylogeny of 76 SRK sequences of A. halleri, A. lyrata, and A. kamchatica. 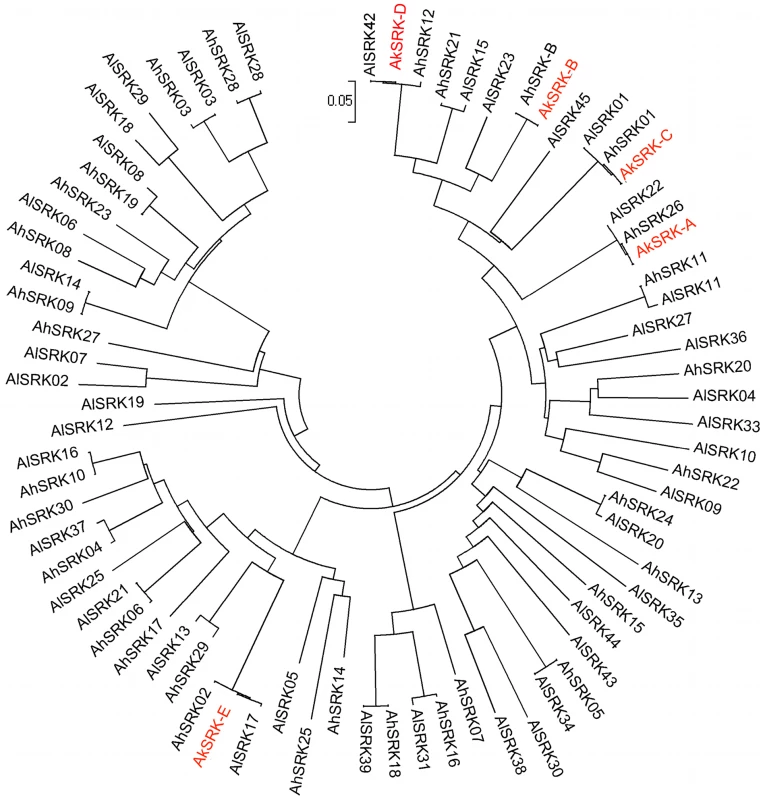
This phylogeny was obtained by the neighbor-joining method on pairwise proportions of nucleotide divergence. In total, 266 nucleotide positions were used. The evolutionary distances were computed using the Kimura two-parameter method. The tree includes 31 SRK sequences of A. halleri (AhSRK), 40 SRK sequences of A. lyrata (AlSRK) and five SRK sequences of A. kamchatica (AkSRK). See Table S10 for the accession numbers of these sequences deposited in GenBank. SRK sequences of A. halleri and A. lyrata are shown in black, and those of A. kamchatica are shown in red. In A. lyrata and A. halleri, SRK sequences that presumably share the same specificities are highly homologous, while sequences with different specificities show at most 91–92% nucleotide sequence identity [59], [74]. Here, four out of the five A. kamchatica SRK sequences (AkSRK-A, AkSRK-C, AkSRK-D and AkSRK-E) showed more than 98% sequence identity to SRK sequences previously reported in both A. halleri and A. lyrata (Figure 1), suggesting that they also share specificities with the corresponding A. halleri and A. lyrata SRK sequences. In contrast, no previously reported sequence showed any particularly high similarity with AkSRK-B. The most similar ones were AhSRK12 (81% identity over 576 bp) and AlSRK23 (87% identity over 558 bp), suggesting that they are unlikely to share specificity with AkSRK-B. Using specific primers for AkSRK-B, we successfully amplified a sequence from an A. halleri plant (lowland habitat in Western Honshu, Japan; No. 61 in Table S1) that showed 100% sequence identity to AkSRK-B of A. kamchatica and, as shown later by the interspecific crosses, they shared the functional specificity of SI. This newly discovered A. halleri ortholog was named AhSRK-B (Figure 1). Using specific primers, we also isolated orthologous sequences of AkSRK-A and AkSRK-C from A. halleri, which were nearly identical to AhSRK26 and AhSRK01, respectively (Figure S1). These sequences were named AhSRK26-Ibuki and AhSRK01-Ibuki, respectively (see the section “Retained full-length SRK sequences as well as multiple gene-disruptive mutations” for details).
Nucleotide divergence of AkSRK from orthologous genes of the parental species A. halleri and A. lyrata
Despite their transspecificity, several species-specific nucleotide substitutions have been reported within the same S-haplogroups in A. lyrata and A. halleri, respectively [61]. Thus, to obtain insight into which parental species the SRK-like sequences found in A. kamchatica were derived from, we compared the nucleotide divergence of SRK-like sequences of A. kamchatica with corresponding orthologous genes from A. halleri and A. lyrata. AkSRK-A and AkSRK-C were closer to their orthologs of A. halleri (AhSRK26 and AhSRK01, respectively) than to those of A. lyrata (AlSRK22 and AlSRK01, respectively) (Figure 2; Figure S1; Table S2). Conversely, AkSRK-D and AkSRK-E showed higher sequence identity to orthologs from A. lyrata (AlSRK42 and AlSRK17, respectively) (Figure 2; Figure S1; Table S2). These results suggest that AkSRK-A and AkSRK-C are derived from A. halleri and that AkSRK-D and AkSRK-E are derived from A. lyrata. In addition, because we found that AhSRK-B in A. halleri showed 100% identity to AkSRK-B in A. kamchatica, AkSRK-B is most likely derived from A. halleri. Based on this pattern of nucleotide divergence from the parental species, mutually exclusive distribution of SRK haplogroups and a segregation analysis in the F2 population (see below), hereafter we denote AkSRK-A, AkSRK-B and AkSRK-C as the “A. halleri-derived SRK” and AkSRK-D and AkSRK-E as the “A. lyrata-derived SRK” (see below and Discussion).
Fig. 2. Nucleotide divergence of AkSRK of five S-haplogroups in A. kamchatica from two parental species. 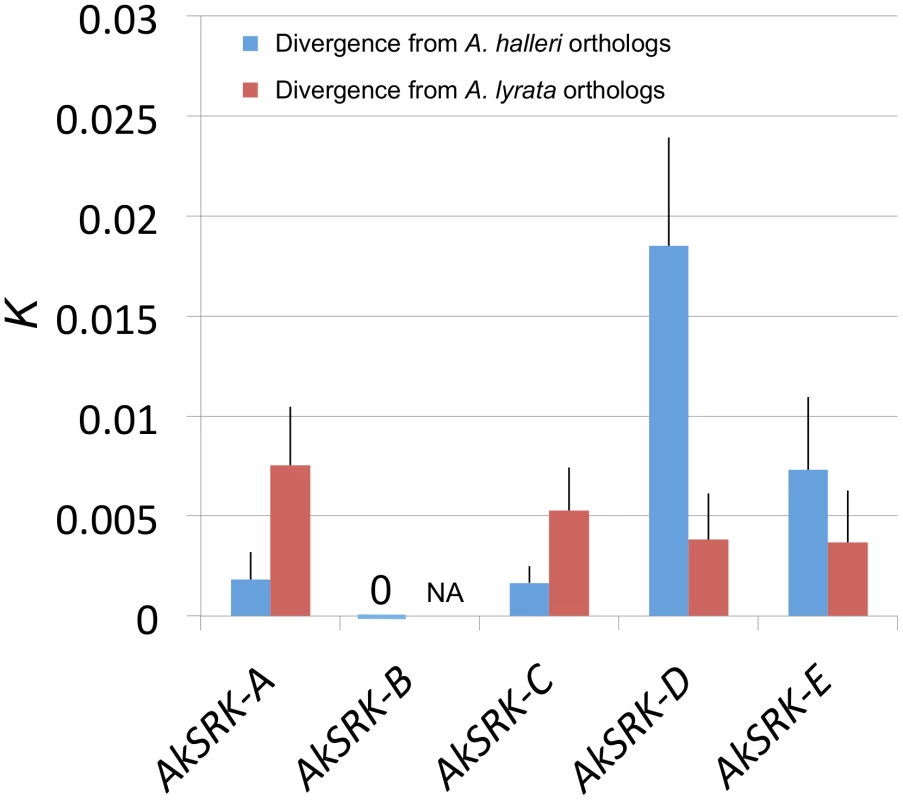
All estimates were Jukes–Cantor corrected. Bars indicate standard errors. Sequence lengths compared were: 935 bp (AkSRK-A), 3773 bp (AkSRK-B), 1067 bp (AkSRK-C), 555 bp (AkSRK-D) and 550 bp (AkSRK-E). AkSRK-B was 100% identical to AhSRK-B throughout the coding sequence (see also Figure 5). Because AkSRK-B belongs to a newly identified S-haplogroup (see Results), orthologous sequences were not yet reported from A. lyrata, while AhSRK-B from A. halleri was found in this study. Thus, nucleotide divergence from A. halleri only is shown for AkSRK-B. See also Table S2 for details. Distribution and frequency of five S-haplogroups revealed by PCR–based genotyping
Using PCR-based genotyping of SRK haplogroups, we investigated the frequencies and geographic distribution of the five S-haplogroups identified in this study. Altogether, 49 accessions from 46 populations were genotyped by primer pairs that could specifically amplify AkSRK-A, AkSRK-B, AkSRK-C, AkSRK-D or AkSRK-E (Figure 3; Figure 4; Table S1; see Table S3 for the primer pairs used). Two copies of SRK were amplified from all accessions except those from Hokkaido in Japan that had only one copy (Nos. 25, 27 and 28), and one from Kamchatka in Russia that had three copies (No. 33). This finding is consistent with reports that A. kamchatica is a self-compatible allotetraploid, which usually harbors two homeologs from the parental species, supposing rare heterozygosity because of selfing and rare duplication [46], [48].
Fig. 3. The geographic distribution of S-haplogroups and population structure in A. kamchatica. 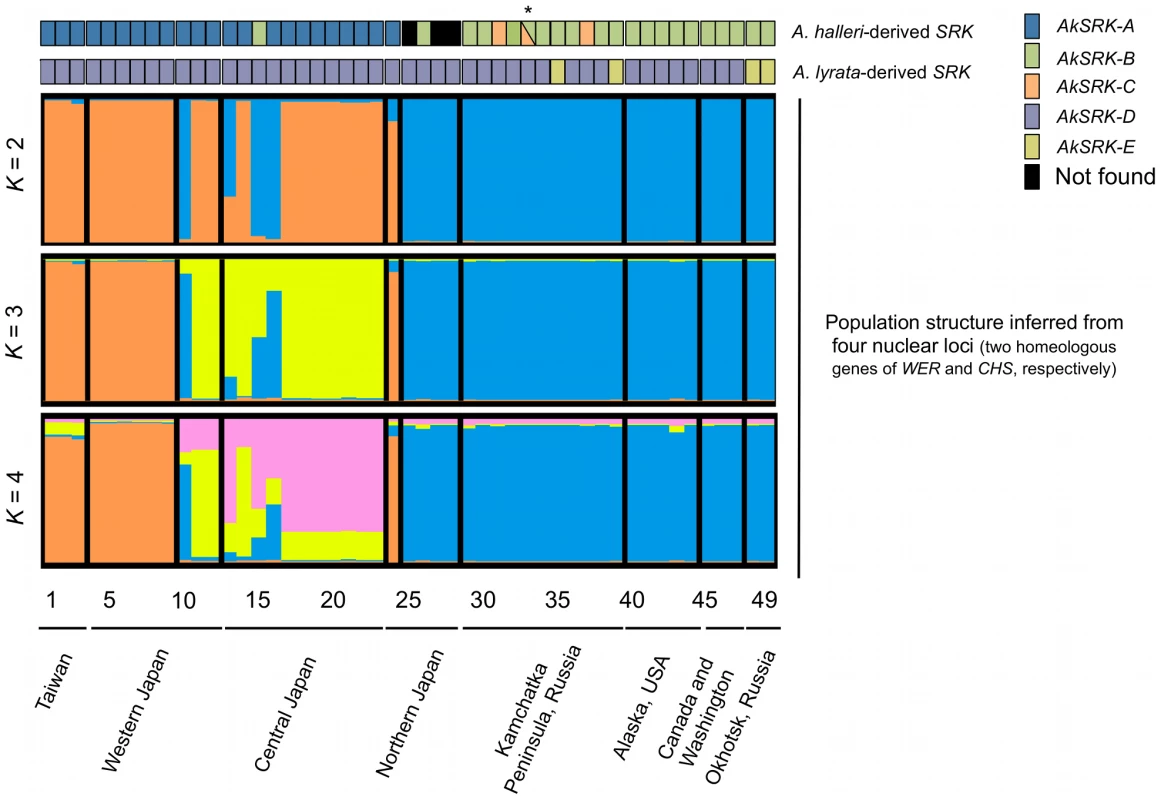
Results of PCR-based genotyping of SRK, with inference of population structure based on four loci (two homeologous genes each of nuclear WER and CHS genes) for the clustering of K = 2, 3 and 4, by the Bayesian clustering algorithm implemented in InStruct software (see Figure S2 and Table S1). Numbers below the diagrams correspond to the accessions listed in Table S1. Triangles in accession No. 33 (marked with an asterisk) indicate that AkSRK-B and AkSRK-C, as well as AkSRK-D, were found in a single individual. We confirmed these results using the software STRUCTURE instead of InStruct (see Text S1 and Figure S5 for details). Fig. 4. Geographic distribution of S-haplogroups in A. kamchatica. 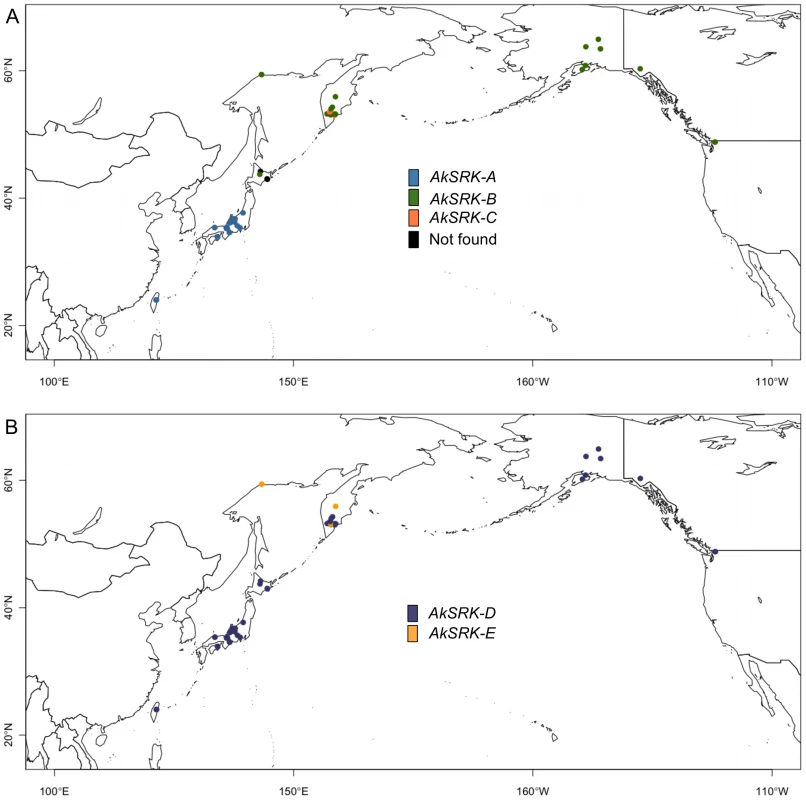
(A) Geographic distribution of AkSRK-A (blue), AkSRK-B (green) and AkSRK-C (orange). Black circles: not found. (B) Geographic distribution of AkSRK-D (purple) and AkSRK-E (dark yellow). The distributions of AkSRK-A, AkSRK-B and AkSRK-C were mutually exclusive except for one accession (No. 33, see below) and showed a strong geographic structure. AkSRK-A and AkSRK-B were found in about half of our samples (46.9% and 41.8%, respectively; Figure 3; Figure 4); all AkSRK-A were found in the southwestern part of the distribution range of the species (Taiwan and Japan), whereas AkSRK-B was mainly located in the northern and eastern part of the species range (North America and the Kamchatka Peninsula in Russia; Figure 3; Figure 4; Table S1). In contrast, the frequency of AkSRK-C was lower than those of AkSRK-A and AkSRK-B (5.1%; Figure 3; Figure 4), and was found only in three accessions from the Kamchatka Peninsula (Figure 3; Figure 4). One of them (No. 33) harbored both AkSRK-B and AkSRK-C, indicating heterozygosity or duplication of the A. halleri-derived SRK, AkSRK-B and AkSRK-C sequences. We further genotyped nine additional individuals in that population (Table S4) and found that AkSRK-B and AkSRK-C segregated at intermediate frequencies (AkSRK-B: 0.55; AkSRK-C: 0.45). This is consistent with the hypothesis that No. 33 is a rare heterozygote of AkSRK-B and AkSRK-C, because it would occasionally arise even by rare outcrossing events under predominant selfing, in particular when the allele frequencies were intermediate. Moreover, the distributions of AkSRK-D and AkSRK-E were completely mutually exclusive in our 49 samples and AkSRK-D was nearly fixed (91.8% frequency; Figure 3; Figure 4).
The geographic distribution of the A. halleri-derived S-haplogroups is concordant with the population structure inferred from polymorphisms of four other nuclear loci (two homeologous genes each of WER and CHS) (Figure 3). The high values of the mean posterior probability of data ln P(X|K), ΔK and the symmetric similarity coefficient supported the clustering of K = 2, which reflect the spatial structure of the distribution well (Figure 3; Figure S2; Table S1). The distributions of A. halleri-derived S-haplogroups—AkSRK-A, AkSRK-B and AkSRK-C—are significantly correlated with the population structure (Cramer's coefficient: 1; p = 7.62×10−12); that is, most of accessions bearing AkSRK-A belong to cluster 1 (orange in Figure 3) and most of others belong to cluster 2 (blue in Figure 3). This significant correlation indicates that the pattern of distribution of these A. halleri-derived S-haplogroups is consistent with the genome-wide pattern of polymorphism. The correlations were also significant with the clustering of K = 3 (Cramer's coefficient: 0.675; p = 6.37×10−11) and of K = 4 (Cramer's coefficient: 0.577; p = 5.39×10−12). In contrast, the distributions of A. lyrata-derived S-haplogroups—AkSRK-D and AkSRK-E—are not correlated significantly with the population structure (Cramer's coefficient: 0.302; p = 0.109). The correlations were also not significant with the clustering of K = 3 (Cramer's coefficient 0.194; p = 0.393) and of K = 4 (Cramer's coefficient 0.163; p = 0.517). In fact, the frequency of AkSRK-D is markedly higher than that of AkSRK-E and it is widespread throughout the geographically wide range, resulting in no significant correlation between these A. lyrata-derived S-haplogroups and population structure.
In A. kamchatica subsp. kawasakiana (formerly, A. lyrata subsp. kawasakiana), a previous survey of SRK identified two sequences, Aly13-1 and Aly13-22 [57]. The result of our PCR-based genotyping is consistent with those findings, as we also found AkSRK-A (orthologous to Aly13-22) in all accessions of A. kamchatica subsp. kawasakiana (Table S1; Figure 3). However, AkSRK-C (orthologous to Aly13-1) was not found in our samples of subsp. kawasakiana but only in subsp. kamchatica.
Estimation of the divergence time from parental species
Using the nucleotide divergence estimate K and the synonymous substitution rate Ks of four nuclear loci (two homeologous genes each of CHS and WER; Table S5), we estimated the divergence time of A. kamchatica from the parental species (Table S6). When we employed the mutation rate given by Koch et al. [75], which is based on the synonymous substitution rate calibrated by fossil records, the mean divergence time was 20,417 years (with a 95% confidence interval of 0–75,460 years). When we employed the mutation rate given by Ossowski et al. [76], estimated using mutation accumulation lines, the mean divergence time was 245,070 years (with a 95% confidence interval of 37,385–532,953 years). Overall, estimates based on the mutation rate given by Koch et al. [75] were smaller than those based on Ossowski et al. [76], although the 95% confidence intervals overlapped.
Two clusters were suggested in the analysis of population structure (Figure 3). Whereas here we estimated the divergence averaged over population clusters, it is possible that it varies between clusters, given that population structure is profoundly affected by the multiple origins of A. kamchatica [48]. We also calculated the clusterwise nucleotide divergence from A. halleri and A. lyrata but no significant difference from each other was found in the current dataset and clustering (Table S5).
AkSRK-A and AkSRK-B are allelic with each other and disomically inherited
Based on the pattern of nucleotide divergence from the parental species and the mutually exclusive distribution of SRK, we predicted the following allelic relationships: the A. halleri-derived SRK (AkSRK-A, AkSRK-B and AkSRK-C) should be allelic and the A. lyrata-derived SRK (AkSRK-D and AkSRK-E) should be allelic with each other, respectively. This prediction also assumes the disomic inheritance of the A. halleri - and A. lyrata-derived SRK, respectively. To confirm our predictions, we investigated the pattern of segregation of the A. halleri-derived SRK (AkSRK-A and AkSRK-B) in an F2 population that was generated by crossing individuals bearing AkSRK-A and AkSRK-D, and individuals bearing AkSRK-B and AkSRK-D (Table 1). We genotyped 95 F2 individuals with specific primers for AkSRK-A and AkSRK-B, and compared the goodness-of-fit of four inheritance models: (1) disomic and allelic, (2) disomic and nonallelic, (3) tetrasomic and allelic and (4) tetrasomic and nonallelic (Table 1). We found that the observed pattern of segregation better fitted model (1), i.e., the disomic and allelic model, rather than the other three models (p = 0.27; Table 1). We also confirmed the amplification of AkSRK-D in all 95 F2 plants and in the F1 generation, which indicates that AkSRK-D segregates neither with AkSRK-A nor with AkSRK-B in the F2 population.
Tab. 1. The pattern of segregation of AkSRK-A and AkSRK-B in 95 F2 individuals. 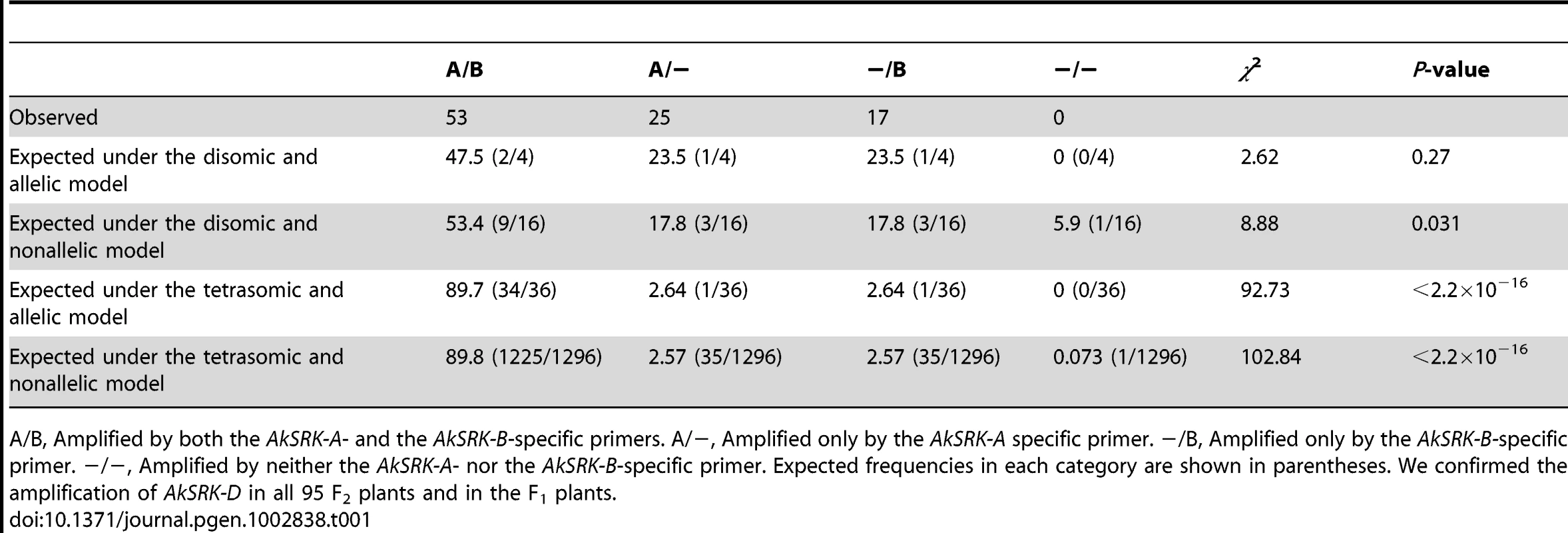
A/B, Amplified by both the AkSRK-A- and the AkSRK-B-specific primers. A/−, Amplified only by the AkSRK-A specific primer. −/B, Amplified only by the AkSRK-B-specific primer. −/−, Amplified by neither the AkSRK-A- nor the AkSRK-B-specific primer. Expected frequencies in each category are shown in parentheses. We confirmed the amplification of AkSRK-D in all 95 F2 plants and in the F1 plants. Retained full-length SRK sequences as well as multiple gene-disruptive mutations
We isolated entire coding sequences of SRK from several A. kamchatica individuals for each haplogroup (three AkSRK-A, three AkSRK-B, one AkSRK-C, five AkSRK-D and one AkSRK-E; Figure 5 and Table S1). In addition, we also isolated entire coding sequences of orthologs of AkSRK-A, AkSRK-B and AkSRK-C from A. halleri. Alignment of the coding regions of SRK from A. kamchatica and A. halleri indicates that at least one accession of four haplogroups—A, B, D and E—retains the full-length SRK, without any apparent disruptive mutations such as frameshifts or inverted repeats. Furthermore, four single nucleotide polymorphisms were identified among three sequences of AkSRK-A and 13 among five sequences of AkSRK-D (Figure 5).
Fig. 5. Entire coding sequences of SRK of five S-haplogroups from A. kamchatica and three S-haplogroups from A. halleri. 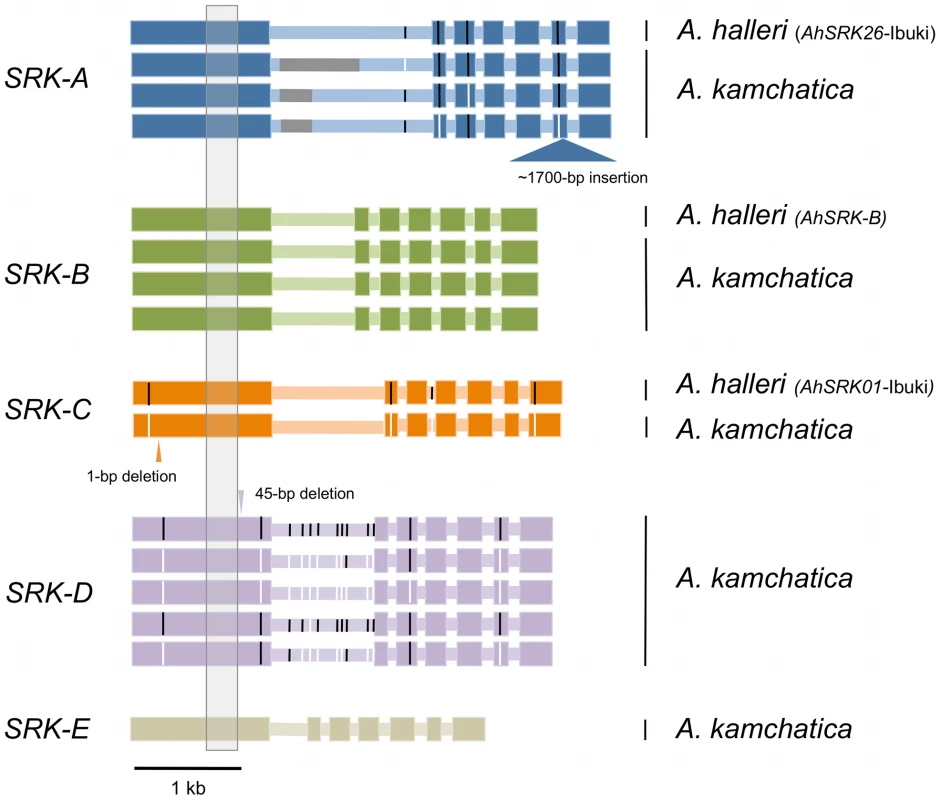
Thick and thin horizontal bars indicate exons and introns, respectively. Black and white vertical bars indicate single nucleotide polymorphisms found in each haplogroup. Nucleotide sequences were not available for part of the first intron of SRK-A because of repetitive sequences (indicated by gray bars). The gray shaded region in exon 1 was used for generating the phylogenetic tree in Figure 1. No obvious gene-disruptive mutations were found in the sequences of A. halleri, but we found three in multiple haplogroups of A. kamchatica (Figure 5; Figure S3). First, we found that AkSRK-A from an accession from Biwako, a lowland region of Japan, contained an approximately 1,700-bp insertion in exon 6. PCR-based genotyping revealed that this insertion is shared by all the accessions of A. kamchatica subsp. kawasakiana living in lowland regions in western Japan (Table S1; see Table S3 for the PCR primers used). We also found a 1-bp deletion in AkSRK-C from a Kamchatka accession that caused a frameshift. In addition, we found that AkSRK-D of an accession from mountains in central Honshu contains a 45-bp deletion in exon 1. Although this deletion does not change the reading frame, it is likely to abolish the recognition function, because it lies within the S-domain and encompasses one of the 12 conserved cysteine residues suggested to be important for protein structure [77], [78] (Figure 5; Figure S3; see also the section on interspecific crosses for the degraded female SI in the Murodo accession).
Interspecific crosses between A. halleri and A. kamchatica suggest that degradation of the male components was responsible for the loss of SI
To test whether these full-length SRK sequences and other components involved in the female signaling pathway are functional in A. kamchatica, we first conducted interspecific crosses between A. halleri (male) and A. kamchatica (female). As S-haplogroups are shared transspecifically among A. lyrata, A. halleri and A. kamchatica (Figure 1), an incompatible reaction should occur even in interspecific crosses in which pollen and stigma share the same haplogroup [41]. We used the three most frequent haplogroups in A. kamchatica—A, B and D—as all of the accessions surveyed contain at least one of them.
In eight accessions of A. kamchatica, incompatible reactions were observed when pollen of A. halleri was used to pollinate pistils of A. kamchatica sharing the same haplogroups (Figure 6). These interspecific crosses verified the shared specificities of the SI system between A. halleri and A. kamchatica. More importantly, these results indicate that the female components of the SI system are functional in these accessions of A. kamchatica. Specifically, we found incompatible reactions in the following combinations of crosses: pollen of A. halleri bearing haplogroup A with pistils of A. kamchatica accessions bearing AkSRK-A from Murodo and Tsurugi-Gozen; pollen of A. halleri bearing haplogroup D with pistils of A. kamchatica bearing AkSRK-D from Biwako and Potter; and pollen of A. halleri bearing haplogroup B with pistils of A. kamchatica bearing AkSRK-B from Darling Creek and Potter. In the Potter accession, incompatible reactions were observed when haplogroups B and D of A. halleri were pollinated, respectively, suggesting that these haplogroups are codominant on pistils.
Fig. 6. Interspecific crosses between A. halleri and A. kamchatica. 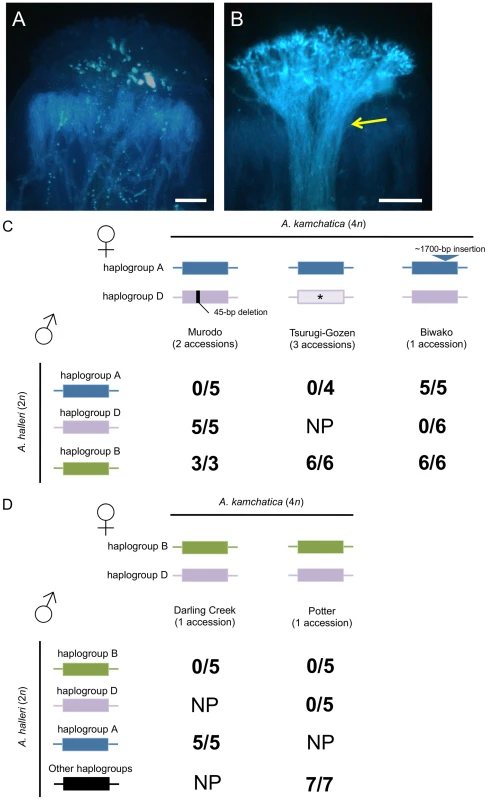
(A, B) Representative incompatible (A) and compatible (B) reactions on A. kamchatica stigmas. Crosses were carried out between A. halleri bearing haplogroup B and A. kamchatica bearing haplogroup B (A), and between A. halleri not bearing haplogroup B and A. kamchatica bearing haplogroup B (B). A bundle of pollen tubes indicates a compatible reaction (yellow arrow). Scale bars = 0.1 mm. (C) Crosses using A. kamchatica from Murodo, Tsurugi-Gozen and Biwako, bearing AkSRK-A and AkSRK-D. The AkSRK-D sequence of Murodo has a 45-bp deletion including one of the 12 conserved cysteine residues (see text). The AkSRK-A sequence of Biwako has a ∼1,700-bp insertion in exon 6 (see text and Table S1). Note that full-length sequences of AkSRK-D from Tsurugi-Gozen were not confirmed and that crosses with A. halleri bearing haplogroup D were not conducted (indicated by asterisks). (D) Crosses using A. kamchatica from Darling Creek and Potter, bearing AkSRK-B and AkSRK-D. Two A. halleri plants that do not bear SRK-A, SRK-B or SRK-D were also used as pollen donors (indicated as “Other haplogroups”; see Methods). (C, D) Numerators denote crosses where more than 20 pollen tubes penetrated the stigma, indicating compatible reactions. Denominators show the total number of crosses conducted in each combination. NP: not performed. As control experiments, we also carried out crosses of the following combinations: (1) where the S-haplogroups of pollen and stigmas differ, and (2) where the SRK of A. kamchatica possessed any gene-disruptive mutations (Figure 6). In these combinations, we consistently observed compatible reactions, indicating that the reduced pollen growth observed in this experiment was caused not by interspecific reproductive isolation between A. halleri and A. kamchatica, but by the SI system.
It is worth noting that, in the Murodo accession, incompatible reactions were observed when plants were pollinated by A. halleri containing haplogroup A, but not by those containing haplogroup D. Because all components of the signaling pathway except for SRK are thought to be shared among different specificities, the data strongly suggest that a mutation in AkSRK-D—most likely the 45-bp deletion—was responsible for this decay of the female SI reaction. Likewise, compatibility of haplogroup A of the Biwako population is also attributable to a mutation in SRK, the approximately 1,700-bp insertion, because the functionality of the downstream signaling pathway was shown by crosses with A. halleri containing haplogroup D.
Because all these accessions of A. kamchatica are self-compatible and retain functional female components of SI including downstream signaling pathways, the data indicate that the male components of S-haplogroups A, B and D are not functional in these accessions. To confirm the degradation of the male components, we conducted interspecific crosses between A. halleri (female) and A. kamchatica (male), using A. halleri bearing haplogroups A, B or D as pistil donors and A. kamchatica as a pollen donor. In these combinations, we consistently observed compatible reactions, indicating that the male components of S-haplogroups A, B and D are not functional in these accessions of A. kamchatica (Figure S4). In addition, we conducted intraspecific crosses among A. kamchatica, with the Biwako accession as a pollen donor and the Murodo accession, which retains the functional female specificity of haplogroup A, as a pistil donor (Table S7). Incompatible reactions were not observed, thus excluding the possibility that the male components of haplogroup A of the Biwako accession remain functional. Similarly, we found that the male components of haplogroup D of Murodo are also not functional. All these data confirm that the male components of haplogroups A, B and D are not functional in these accessions of A. kamchatica. These nonfunctional male components and functional female components including downstream signaling pathways suggest that degradation of the male components was primarily responsible for their loss of SI. We do not exclude the possibility that recombination events between the male and female components on the S-locus may have also been involved, although their occurrence is reported to be very rare (reviewed in [56]).
Discussion
A. halleri– and A. lyrata–derived homeologs of the S-locus in allotetraploid A. kamchatica
In this study, we identified the full-length SRK sequences of five S-haplogroups in A. kamchatica (Figure 1). Through interspecific crosses with A. halleri, we confirmed that the intact SRK sequences of the three most frequent S-haplogroups in our dataset—AkSRK-A, AkSRK-B and AkSRK-D—are indeed associated with the female specificities of SI. Although associations with SI specificities for the other less frequent haplogroups—AkSRK-C and AkSRK-E—were not confirmed experimentally, they also exhibited very high similarities with known SRK sequences from A. lyrata and A. halleri (>98%), suggesting that specificities of SI are shared between species [59], [74].
Our investigation on nucleotide polymorphism and divergence, as well as the pattern of segregation in natural and experimental populations, allows us to address how these five S-haplogroups in allotetraploid A. kamchatica originated from their parental diploid species, A. halleri and A. lyrata. First, AkSRK-A and AkSRK-C of A. kamchatica are closer to the orthologs of A. halleri than to those of A. lyrata, and AkSRK-D and AkSRK-E of A. kamchatica are closer to the orthologs of A. lyrata than to those of A. halleri (Figure 2; Figure S1; Table S2). We also found that AkSRK-B in A. kamchatica showed 100% identity to AhSRK-B in A. halleri, although its ortholog of A. lyrata was not isolated in the present study. Second, the distributions of AkSRK-A, AkSRK-B and AkSRK-C were mutually exclusive, as were those of AkSRK-D and AkSRK-E, except for a minor presumable heterozygote of AkSRK-B and AkSRK-C (Figure 3; Figure 4). Third, the pattern of segregation in the F2 population significantly supports the model in which AkSRK-A and AkSRK-B are allelic while AkSRK-D is not allelic to them (Table 1). In addition, the pattern of polymorphism within a Kamchatka population is consistent with the model in which AkSRK-B and AkSRK-C are also allelic and segregating in a local population (Table S4). These independent lines of evidence suggest that AkSRK-A, AkSRK-B and AkSRK-C are S-alleles of the A. halleri-derived S-locus and that AkSRK-D and AkSRK-E are S-alleles of the A. lyrata-derived S-locus. Although we confirmed the association between AkSRK-D and the SI specificity of A. halleri in this study, the specificity is also likely to be shared with A. lyrata (discussed in [59], [61], [62]). While there are a few reports on the evolutionary history of the S-locus in cultivated allotetraploid species, particularly Brassica napus [79], [80], to our knowledge, this study is the first to demonstrate clearly how multiple S-haplogroups in a wild allotetraploid species originated from the parental species and spread throughout a geographically wide area.
Pattern of nucleotide polymorphism and divergence of the S-locus
Shimizu-Inatsugi et al. [48] reported that multiple haplotypes of nuclear and chloroplast sequences were shared between allotetraploid A. kamchatica and its parental diploid species, suggesting independent origins of A. kamchatica. In particular, A. kamchatica and A. halleri share four identical chloroplast haplotypes, suggesting that at least four diploid individuals of A. halleri contributed independently to the multiple origins of A. kamchatica [48]. As three of the four suggested independent origins are manifested as distinct clusters of population structure, independent origins combined with range expansion out of Asia appear to affect the population structure of A. kamchatica profoundly [48]. A comparison between the geographic distribution of S-haplogroups and the population structure inferred from other loci thus illustrates how independent origins of A. kamchatica contributed to form the current pattern of polymorphism of two S-loci: the A. halleri-derived S-locus and the A. lyrata-derived S-locus. We found that the distribution of three A. halleri-derived S-haplogroups was significantly correlated with population structure. Given that the population structure of A. kamchatica is profoundly affected by its multiple independent origins, the concordance between population structure and the distribution of A. halleri-derived S-haplogroups suggests that the composition of the gene pool of the A. halleri-derived S-locus would be explained at least partially by the independent origins of this species. In contrast, there was no significant correlation found for the A. lyrata-derived S-locus, on which AkSRK-D is markedly more frequent than AkSRK-E. A similar pattern was observed in the European population of A. thaliana, in which the haplogroup A was much more frequent than the haplogroup C and thus the pattern of polymorphism at the S-locus showed deviation from the population structure [41]. A coalescent approach based on SRK sequences of species-wide samples as well as genome-wide polymorphism data would serve to unveil more precisely how the five S-haplogroups originated in A. kamchatica, especially by quantifying the effects of random genetic drift, population expansion out of Asia, and independent origins of species, respectively.
Such a population genomic approach should also reveal the demographic history during the loss of SI in A. kamchatica. The retained functionality of the female SI components including the downstream signaling pathway shown in this study implies a fairly recent loss of SI in A. kamchatica. Indeed, estimates of the average divergence time of the nuclear loci (WER and CHS) are less than 250 thousand years (kyrs), although this divergence time may not necessarily correspond to the timing of the loss of SI (Table S6). Dating of the loss of SI has also been estimated in other self-compatible lineages, A. thaliana (<314 kyrs [62]; <500 kyrs [81]), C. rubella (∼27 kyrs [43]) and L. alabamica (∼150 kyrs and 12–48 kyrs in two independent lineages [44]), assuming the substitution rate published by Koch et al. [75]. These timings of evolutionary transitions are generally concordant with the recent glacial–interglacial period that greatly influenced the distribution of many plant and animal species [82]. Indeed, recent genetic bottlenecks and/or population expansions have been suggested for A. thaliana [83]–[85], C. rubella [43], [73], L. alabamica [44] and in selfing populations of A. lyrata in North America [86]–[88]. The low nucleotide diversity of A. kamchatica in comparison with its parental species is also consistent with a bottleneck during polyploidization (0.0006–0.0012 in A. lyrata-derived homeologs of A. kamchatica and 0.0008–0.0026 in A. halleri-derived homeologs of A. kamchatica, versus 0.0150 in A. halleri, 0.0230 in A. lyrata subsp. petraea and 0.0031 in A. lyrata subsp. lyrata [48], [89]).
As range expansions could be accompanied by reduced mate availability and increased pollen limitation [7], [90]–[92], reproductive assurance might have played a role in these examples of the loss of SI. Particularly, mates with the same ploidy level would be limited after polyploidization in A. kamchatica [1], [66]–[70]. Another possibility has been suggested by recent theoretical and simulation studies, which proposed that range expansion might promote the evolution of selfing [93]. This is because inbreeding depression would be reduced in marginal populations where deleterious mutations are fixed or purged by strong genetic bottlenecks [93]–[95]. If inbreeding depression falls below a certain threshold, disrupted S-haplogroups causing self-compatibility are expected to rapidly replace all functional S-haplogroups via an inherent transmission advantage [24], [27]. Although this is not mutually exclusive to reproductive assurance [44], [96], further analyses on the demographic history and the dating of the loss of SI should contribute to our ability to address the relative importance of transmission advantage and reproductive assurance for the evolution of self-compatibility during range expansions.
Dominance interactions among S-haplogroups and the loss of SI
The molecular mechanism underlying the loss of SI in polyploid species is still not well understood, although the relationship between polyploidy and self-compatibility has been debated for more than 60 years [1], [65], [66]. Whereas polyploidization is known to disrupt SI almost invariably in gametophytic SI systems where specificities are determined by haploid genotypes of gametes [97], [98], in sporophytic SI systems where specificities are determined sporophytically by the diploid parental genotypes, polyploidization in itself does not necessarily induce the loss of SI (e.g., [57]). We speculate that, in sporophytic SI systems such as those of the Brassicaceae, dominance interactions among S-haplogroups might have played an important role in the loss of SI in polyploid species, as is also implied in a study of synthetic interspecific hybrids by Nasrallah et al. [99]. In the Brassicaceae, complex dominance interactions among S-haplogroups have been reported [52], [55], [56], [60], [100]–[103]. Given that the female components are functionally intact in A. kamchatica, as shown by our interspecific crosses, both homeologous copies of the SCR gene ought to have lost their function. However, gene-disruptive mutations at both of the homeologous SCR genes might not necessarily be required if the dominant SCR harboring a gene-disruptive mutation suppresses the expression of the recessive SCR [80], [104]. Thus, dominance interactions could facilitate the evolution of self-compatibility by a single mutation in polyploid species. This is indeed suggested for Brassica napus, which is also an allotetraploid species that originated from hybridization of B. rapa and B. oleracea [80]. In a cultivar of B. napus, neither of the homeologous SP11/SCR genes is expressed. In artificially synthesized B. napus lines, SP11/SCR alleles from B. rapa showed dominance over SP11/SCR alleles from B. oleracea, suggesting that the nonfunctional dominant SP11/SCR allele suppressed the expression of the recessive SP11/SCR allele [80].
In A. lyrata and A. halleri, such dominance relationships between S-haplogroups have been characterized. Thus, the S-haplogroup of S12 in A. halleri, which is orthologous to haplogroup D of A. kamchatica, has been suggested to belong to a relatively dominant class [60]. Given that the dominance relationship is consistent between A. halleri and A. kamchatica, haplogroup D would also be dominant in A. kamchatica and might have suppressed the expression of its homeologous counterparts, such as haplogroup C, which is orthologous to the most recessive S-haplogroup of S1 in A. lyrata and A. halleri [52], [60], [61], [103]. While we showed that haplogroups B and D are codominant in pistils in A. kamchatica (Figure 6), their dominance rank might be different in pollen, as dominance is known to occur more frequently in pollen than in pistils [60].
It is worth noting that haplogroup C, which is orthologous to the most recessive S-haplogroup of S1 in A. lyrata and A. halleri, was found at a relatively low frequency in A. kamchatica (5.1%). In contrast, S1 shows the highest frequencies in self-incompatible populations of A. lyrata (33%; [55]) and A. halleri (26.3%; [60]), because recessive haplogroups are hidden from negative frequency-dependent selection in heterozygotes, whereas the most dominant S-haplogroups are always exposed to selection [60], [105], [106]. While random genetic drift due to population bottlenecks could explain this contrasting change in frequency, we hypothesize that dominant haplogroups are more likely to be found in self-compatible populations. This is because a dominant haplogroup with gene-disruptive mutations should repress the expression of another specificity, and thus spread more rapidly than a recessive self-compatible mutation [42]. Consistent with this hypothesis, a nearly fixed haplogroup in A. thaliana has been shown experimentally to be a dominant allele in A. halleri [42], [60].
The molecular basis of the dominance relationship has recently been unveiled in Brassica, where the expression of the recessive haplogroups is specifically silenced by methylation of promoter regions induced by trans-acting small noncoding RNAs originated from dominant haplogroups [101], [107], [108]. Investigation of the dominance relationships among S-haplogroups found in A. kamchatica, the pattern of expression of the SCR genes and their methylation status in A. kamchatica will allow us to understand how dominance interactions might have contributed to the loss of SI among polyploid species.
Mutations in pollen components were responsible for the loss of SI
The retained functionality of the female SI components shown by the sequence analysis of SRK and interspecific crosses suggests that the degradation of male components, possibly the SCR gene, was responsible for the loss of SI in A. kamchatica. We also suggest that the gene-disruptive mutations found in the SRK gene of some accessions are not primary mutations causing the loss of SI but rather represent secondary and independent decay. Thus, these would represent mutations that have accumulated after the evolution of self-compatibility, although pleiotropy of SRK might have played a role in maintaining its functionality and slowed this process [42], [45].
Interestingly, evolutionary models predict that mutations disabling the male specificity of the SI system are expected to be more strongly selected than mutations disabling female specificity [25], [28]. This is because these disabled pollen grains are transmitted as outcrossing pollen to offspring with a higher probability than other wild-type (functional) pollen. In contrast, mutations disabling the female specificity are not likely to benefit from the increased fitness over other functional pistils, as long as pollinators frequently visit and supply pollen grains that belong to various specificities [109]. Our finding in A. kamchatica is thus consistent with the prediction of these models.
Mutations driving the loss of SI in the male component have also been reported in A. thaliana [41]. Similarly, the loss of SI in C. rubella might have been driven by a gene-disruptive mutation in the male component, because SRK appears to retain the full-length coding region in some accessions, indicating that mutations at SRK are unlikely to have been the primary cause for the loss of SI ([43]; reviewed in [110]). Busch et al. [44] also reported that self-compatibility in L. alabamica might have originated by the loss of function of the male component, possibly SCR. These recent extensive studies in wild Brassicaceae species demonstrate that the evolution of self-compatibility tends to be driven by mutations in the male rather than in the female components (Table 2).
Tab. 2. Numbers of examples in which the primary mutations involved in the loss of SI are attributable to male or female components of self-incompatibility. 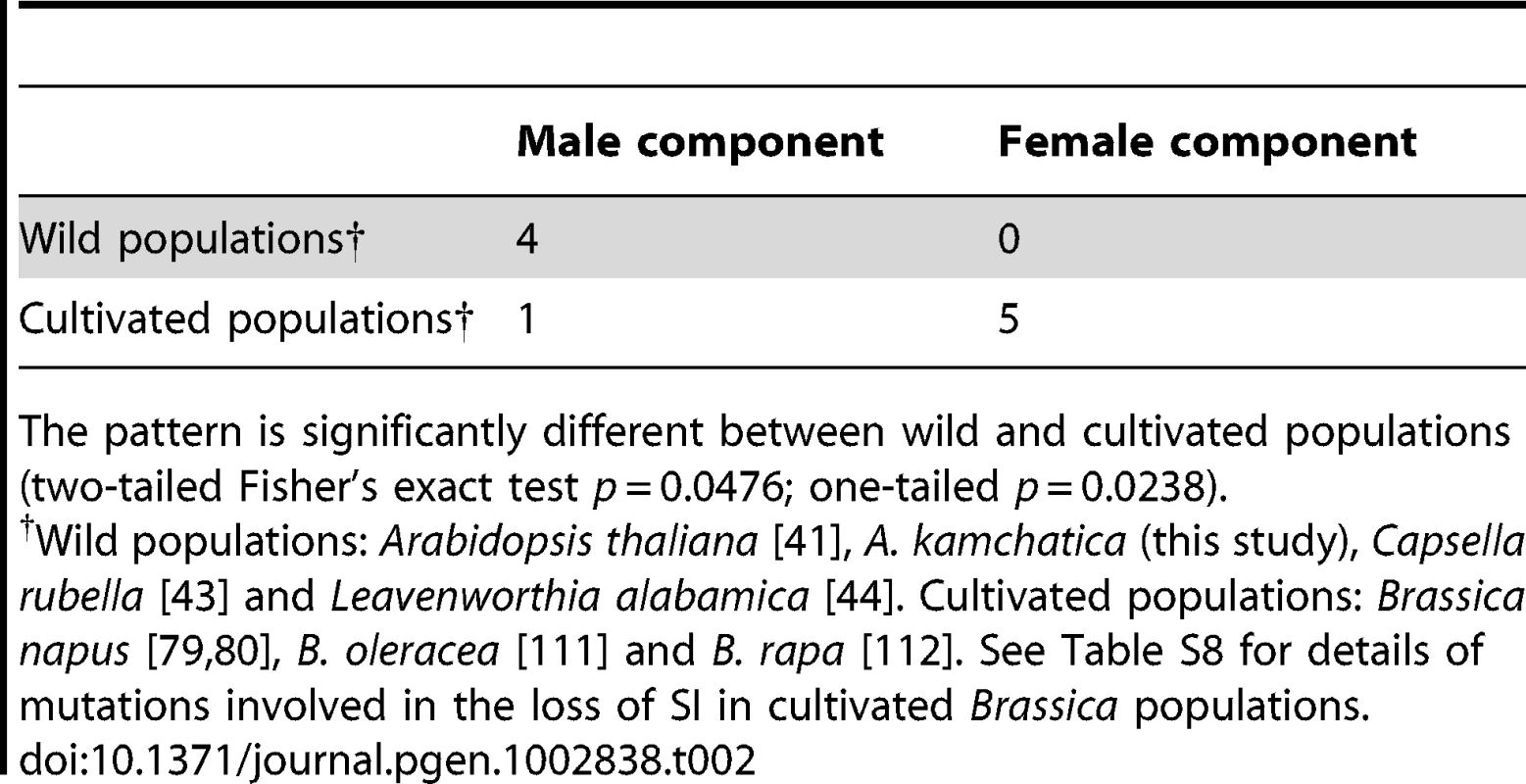
The pattern is significantly different between wild and cultivated populations (two-tailed Fisher's exact test p = 0.0476; one-tailed p = 0.0238). In contrast, in cultivated Brassica, extensive functional analyses have identified gene-disruptive mutations both in male and in female components, and the pattern is significantly different from that observed in wild populations (Table 2; Table S8; two-tailed Fisher's exact test p = 0.0476; one-tailed p = 0.0238; [79], [80], [111], [112]). This contrasting pattern suggests that the male-skewed frequency of mutations found in recent studies of wild populations could be attributed to the process of natural selection and spread in wild populations [25], [28], rather than to the mechanistic natures of mutations, such as differences in mutation rate between male and female components. If such mechanistic reasons were important, similar patterns would have been observed in both wild and cultivated populations. Indeed, the context of selective pressure would be very different between wild and cultivated populations. In a wild population, as mentioned above, mutations disabling SI would be selected in terms of the transmission advantage, i.e., number of compatible mates in a population [25], [28]. In cultivated populations, these gene-disruptive mutations would be positively selected by humans based on the self-compatible phenotype per se. Moreover, because the female SRK gene is about 10 times longer than the male SCR, it may decay even faster than the male component during domestication, as indeed shown in cultivated Brassica (Table 2; Table S8).
Conclusion and future perspectives
Our combination of population genetic analyses and crossing experiments suggests that degradation of the male components was primarily responsible for the loss of SI in A. kamchatica. As we have demonstrated in the present study, functional confirmations, such as crossings between individuals sharing the same SI specificity, will further corroborate the genetic basis for the loss of SI in other selfing species. First, sequence analysis alone cannot provide definitive evidence of gene function. For example, in previous studies of the evolution of self-compatibility in A. thaliana, a splicing mutation in the female gene SRK-B was found in the Cvi-0 accession while the male gene SCR-B did not have any obvious gene-disruptive mutations [38]. However, transgenic experiments suggested that SCR-B was also nonfunctional, possibly due to some amino acid changes, so it is not clear whether the primary mutation occurred in male or female components [40]. Second, functional analyses may reveal multiple origins of self-compatibility within species and contribute to an increase in the number of empirical examples, although in the present study, which took a conservative approach, each wild species was counted as one example (Table 2). While more empirical examples in various species should be gathered to better understand the general pattern of mutations in the evolution of self-compatibility, functional confirmations and analyses of the pattern of polymorphisms are both essential to disentangle mutational histories in the loss of SI.
Materials and Methods
Plant materials
Arabidopsis kamchatica consists of two subspecies, A. kamchatica subsp. kamchatica and A. kamchatica subsp. kawasakiana [46]. A. kamchatica subsp. kamchatica is a perennial, described originally from Kamchatka, Russia. It is also reported from East Asia (Far East Russia, China, Korea, Japan and Taiwan) and North America (Alaska, Canada and the Pacific Northwest of the USA). The second subspecies, A. kamchatica subsp. kawasakiana, is an annual found in sandy open habitats along seashores or lakeshores in lowlands in western Japan. Tetraploid chromosome number counts (2n = 32 and n = 16II) were reported from samples in Japan, Far East Russia, Alaska and Canada, representing both subspecies 46,48.
Altogether, 48 populations of A. kamchatica were sampled (43 from A. kamchatica subsp. kamchatica and five from A. kamchatica subsp. kawasakiana), including one to three individuals per population, giving a total of 60 accessions (Table S1). The sample locations included Far East Asia (Taiwan and Japan), Far East Russia (Kamchatka Peninsula and Okhotsk), Alaska, Canada and the northwest of the USA (Washington), covering the majority of the distribution range of both subspecies. In addition, five accessions of A. halleri were used for interspecific crosses with A. kamchatica and for obtaining full-length sequences of SRK (Table S1; see also the section “Interspecific crosses between A. halleri and A. kamchatica” for details).
General methods for isolation of genomic/complementary DNA, genotyping, sequencing, and construction of phylogenetic trees
Genomic DNA was isolated from young leaves using Plant DNeasy Mini kits (Qiagen, Hilden, Germany). Total RNA was extracted from floral buds and flower tissues with RNeasy kits (Qiagen). The 5′ and 3′ ends of complementary DNA (cDNA) sequences were isolated by 5′ and 3′ Rapid Amplification of cDNA Ends (RACE) with the 5′/3′ 2nd Generation RACE Kit (Roche Applied Science, Indianapolis, IN, USA). PCR was performed with ExTaq (TaKaRa Bio Inc., Shiga, Japan) or iProof High Fidelity DNA Polymerase (Bio-Rad, Hercules, CA, USA). Primers used for amplification and genotyping are shown in Table S3, with the respective annealing temperatures and elongation times. Genotyping was based on the presence or absence of PCR products. Direct DNA sequencing was conducted at the Institute of Plant Biology, University of Zurich, using a PRISM 3730 48-capillary automated sequencer (Applied Biosystems, Foster City, CA, USA). Sequence assemblies and alignments were performed in CodonCode Aligner 3.7.1 (CodonCode, Dedham, MA, USA) and BioEdit v 7.0.5 [113]. Minor adjustments to optimize the alignments were made by eye. Sequence data have been deposited in GenBank under accession numbers JX114752–JX114778. MEGA 5 was used for the construction of a phylogenetic tree [114]. Phylogenetic trees were obtained using the neighbor-joining method on pairwise proportions of nucleotide divergence. The evolutionary distances were computed using the Kimura two-parameter method.
PCR–based screening and sequencing of the entire coding region of SRK
To survey the S-haplogroups in A. kamchatica across its distribution range, we performed a PCR-based screening of the SRK gene using two kinds of primer sets: (1) “general primers”, designed in conserved regions of the S-domain of SRK and known to amplify SRK sequences that belong to a number of haplogroups (“Aly13F1” and “SLGR” [52], [55]); and (2) “haplogroup-specific primers”, designed in polymorphic regions of the S-domain of SRK [55]. Based on the obtained sequences, new haplogroup-specific primers were designed for each haplogroup (Table S3). Although we found sequences in several accessions that were highly similar to the Aly13-7 sequence, Mable et al. [55] reported that Aly13-7 is not associated with SI. Thus, these sequences were excluded from further analyses.
To investigate the functionality of SRK, the entire coding sequences of SRK were obtained by RACE PCR using the haplogroup-specific primers designed in the S-domain of SRK. The primers used for RACE PCR are listed in Table S3.
Bayesian clustering for the inference of population structures
To examine the associations between the geographic distribution of the S-haplogroups and the population structure (see the next section for details), we reexamined the population structure of A. kamchatica, which was originally inferred by Shimizu-Inatsugi et al. [48], by adding the Okhotsk population that was not included in the former study. From two accessions of this population, we obtained nucleotide sequences of the genes WER (WEREWOLF) and CHS (CHALCONE SYNTHASE). By including these data in the dataset of Shimizu-Inatsugi et al. [48], population structure was inferred by the Bayesian clustering algorithm implemented in the InStruct program [115]. The programs CLUMPP [116], DISTRUCT [117] and ΔK statistic [118] were used to summarize and interpret the outputs. The parameters used were the same as in Shimizu-Inatsugi et al. [48], except for the number of independent runs: 10 instead of 20. In addition to the software InStruct, we also used the software STRUCTURE 2.2.3, which is not able to consider inbreeding explicitly [119]. (See Text S1 and Figure S5 for details.)
Association analysis of SRK genotypes and population structures
To examine associations between the geographic distribution of the S-haplogroups and the population structure inferred from polymorphisms of other nuclear loci (WER and CHS), Cramer's coefficient V (also called φ) was calculated for two sets of SRK (A. halleri-derived AkSRK-A, AkSRK-B and AkSRK-C and A. lyrata-derived AkSRK-D and AkSRK-E). Cramer's coefficient V measures the strength of association or interdependence between two categorical variables [120], [121]. Its statistical significance was assessed using Fisher's exact test. (See Table S9 for cluster assignments based on K = 2, 3, or 4.) Heterozygotes or accessions that did not bear corresponding SRK were excluded from the analysis. All statistical analyses were conducted with R 2.10.0 [122].
Calculation of nucleotide divergence of SRK
To obtain insight into which parental species SRK of A. kamchatica were derived from, we calculated mean values of K, Ks (the proportion of synonymous substitutions per synonymous site) and Ka (the proportion of nonsynonymous substitutions per nonsynonymous site) from the outgroups A. halleri and A. lyrata for all SRK sequences obtained in A. kamchatica, using MEGA 5 [114]. We used all the sequences of A. kamchatica and A. halleri obtained in this study (see Table S1 for which accessions were used). In addition, we used the following publicly available sequences for the calculation: AlSRK22 [52], AhSRK26 [59], AlSRK01 [61], AhSRK01 [61], AlSRK42 [59], AhSRK12 [58], AlSRK17 [54] and AhSRK02 [58]. Specifically, we calculated the nucleotide divergence of the following sequence pairs: (1) AkSRK-A from AlSRK22 and from AhSRK26; (2) AkSRK-B from AhSRK-B; (3) AkSRK-C from AlSRK01 and from AhSRK01; (4) AkSRK-D from AlSRK42 and from AhSRK12; and (5) AkSRK-E from AlSRK17 and from AhSRK02.
Estimation of divergence time from two parental species
To understand the approximate timescale of the evolutionary loss of SI in A. kamchatica, the divergence time of A. kamchatica from two parental species was calculated based on four nuclear loci (CHS-lyr, CHS-hal, WER-lyr and WER-hal). We first calculated the nucleotide divergence on these loci using publicly available data [48] as well as newly obtained sequence data (see the section “Bayesian clustering for the inference of population structures”). For A. lyrata, two accessions from Far East Russia were reported to show the highest nucleotide similarities with A. lyrata-derived homeologs of A. kamchatica among those surveyed by Shimizu-Inatsugi et al. [48], suggesting that they are closest to one of the founding parents of A. kamchatica. In contrast, other individuals of A. lyrata from Europe and North America were not as close to the lyrata-derived homeologs of A. kamchatica. Therefore, we calculated the divergence of A. kamchatica from these two A. lyrata accessions from Far East Russia, as this would better represent the split from the parental species. Standard errors and 95% confidence intervals were calculated for all estimates. All estimates were corrected using the Jukes–Cantor method [123]. As two clusters were suggested in the InStruct analysis (Figure 3), we also calculated the clusterwise nucleotide divergence from A. halleri and A. lyrata (Table S10.). (See the above section, “Association analysis of SRK genotypes and population structures”, for the cluster assignment.)
Based on the calculated nucleotide divergence, we estimated the divergence time between A. kamchatica and the two parental species A. lyrata and A. halleri. The estimation was based on the expression T = K/2r, where T is the time to the most recent common ancestor, K is the nucleotide divergence and r is the substitution rate [124]–[126]. Note that our estimation of divergence time assumes a constant rate of evolution throughout the tree [127]. Here we employed two estimates of mutation rates: the synonymous mutation rate of 1.5×10−8±0.5×10−8 per site per year [75], which is commonly used in studies of the evolution of self-compatibility [44], [62], and a mutation rate of 7.1×10−9±0.7×10−9 per site per generation, which was estimated using mutation accumulation lines [76]. While we assumed a generation time of two years [128], we note that this could cause an overestimation or an underestimation, because some accessions of A. kamchatica are reported to be annual plants [47], [48] and because an effect of seed dormancy was not considered, respectively. Note that estimates based on the mutation rate given by Koch et al. (2000) are independent from the generation time, because its unit is base pair per year, not per generation. For calculating the 95% upper and lower bounds of divergence time, the 95% upper and lower bounds of nucleotide divergence and the 95% lower and upper bounds of mutation rates were used, respectively. For estimating the divergence time, we used the nucleotide divergence values of CHS-hal and WER-hal from corresponding orthologs in A. halleri, and those of CHS-lyr and WER-lyr from corresponding orthologs in A. lyrata from Far East Russia.
Interspecific crosses between A. halleri and A. kamchatica
We conducted interspecific crosses between A. halleri and A. kamchatica to test whether the full-length SRK sequences and other components involved in the female signaling pathway are functional, and also to test whether the male components are not functional in A. kamchatica. To screen A. halleri plants bearing haplogroups A, B or D, genotypes of AkSRK were surveyed in A. halleri from Mt Ibuki (35.42°N, 136.40°E), Ohara (35.16°N, 135.84°E) and Tada-Ginzan (34.90°N, 135.35°E) in Japan. We used them as both pollen and pistil donors for the crossing experiments, because individuals that bear SRK-A, SRK-B or SRK-D should bear the haplogroup A, B or D of the S-locus (encompassing SCR-A, SCR-B or SCR-D), respectively, given the tight linkage between SRK and SCR. Three A. halleri plants were used for each haplogroup. The haplogroup-specific primers listed in Table S3 were used for this screening. Two A. halleri plants from Boden and Beride in Switzerland, which bear neither SRK-A, SRK-B, nor SRK-D, were also used as pollen donors (Table S1). To confirm the SRK genotypes, eight A. kamchatica accessions were used for the crossing experiments (Table S1). Three of these eight accessions are reported to be capable of selfing [48], and the other five accessions were confirmed in this study (data not shown). Plants used in the pollination assay were grown at 22°C under a 16 h light/8 h dark cycle. We removed anthers from flower buds and carefully confirmed that stigmas were not contaminated by self-pollen using a stereomicroscope. Flowers were harvested 2–3 h after pollination when A. kamchatica was the pistil donor, or 24 h after pollination when A. halleri was the pistil donor. Harvested flowers were fixed in a 9∶1 mixture of ethanol and acetic acid, softened for 10 min in 1 M NaOH at 60°C and stained with aniline blue in a 2% K3PO4 solution. Pistils were mounted on slides to examine the pollen tubes using epifluorescence microscopy [41], [129]. In compatible crosses, more than 100 pollen tubes typically penetrate the stigma and penetration of <20 pollen tubes was considered as a criterion of incompatible crosses, following the common criteria of previous work in Arabidopsis [31], [41]. The results did not change even if a more stringent criterion of <10 pollen tubes was used. Although pollen tube growth was observed in most of the crosses to evaluate incompatible and compatible reactions, in a few combinations of crosses where A. halleri was the pistil donor, we alternatively used the lengths of siliques as a criterion of incompatible crosses (see Figure S4). A silique length of <5 mm was considered to be the criterion of an incompatible cross.
F2 generation segregation analysis
To confirm the disomic inheritance and the allelic relationship of AkSRK-A and AkSRK-B, F2 segregation analysis was conducted using A. kamchatica. F1 plants were generated using the Biwako accession from Japan as the pistil donor and the Potter accession from Alaska as the pollen donor. (See Table S1 for the detailed geographic locations of these accessions.) Ninety-five F2 plants were generated by selfing of two F1 plants and genotyped using haplogroup-specific primers for AkSRK-A, AkSRK-B and AkSRK-D (Table S3). While AkSRK-A and AkSRK-D were amplified in the Biwako accession and AkSRK-B and AkSRK-D were amplified in the Potter accessions (see Table S1 for details), all of AkSRK-A, AkSRK-B and AkSRK-D were amplified in F1 plants. Using χ2 tests with R 2.10.0 [122], the goodness-of-fit for each of the following inheritance models was calculated: (1) disomic and allelic, (2) disomic and nonallelic, (3) tetrasomic and allelic and (4) tetrasomic and nonallelic. The expected frequencies of segregants are described in Table 1.
Supporting Information
Zdroje
1. StebbinsGL 1950 Variation and evolution in plants New York Columbia University Press 643
2. StebbinsGL 1974 Flowering plants: Evolution above the species level Cambridge Belknap Press 399
3. BarrettSCH 2002 The evolution of plant sexual diversity. Nat Rev Genet 3 274 284
4. IgicBLandeRKohnJR 2008 Loss of self-incompatibility and its evolutionary consequences. Int J Plant Sci 169 93 104
5. FisherRA 1941 Average excess and average effect of a gene substitution. Ann Eugen 11 53 63
6. GoodwillieCKaliszSEckertCG 2005 The evolutionary enigma of mixed mating systems in plants: Occurrence, theoretical explanations, and empirical evidence. Annu Rev Ecol Evol Syst 36 47 79
7. DarwinC 1876 The effects of cross and self fertilisation in the vegetable kingdom London J. Murray 487
8. KaliszSVoglerDWHanleyKM 2004 Context-dependent autonomous self-fertilization yields reproductive assurance and mixed mating. Nature 430 884 887
9. EckertCGSamisKEDartS 2006 Reproductive assurance and the evolution of uniparental reproduction in flowering plants. HarderLDBarrettSCH Ecology and evolution of flowers Oxford Oxford University Press 183 203
10. LandeRSchemskeDW 1985 The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution 39 24 40
11. CharlesworthDCharlesworthB 1987 Inbreeding depression and its evolutionary consequences. Annu Rev Ecol Syst 18 237 268
12. de NettancourtD 2001 Incompatibility and incongruity in wild and cultivated plants, 2nd edn Berlin Springer 322
13. TakayamaSIsogaiA 2005 Self-incompatibility in plants. Annu Rev Plant Biol 56 467 489
14. Franklin-TongVE 2008 Self-incompatibility in flowering plants: Evolution, diversity, and mechanisms Berlin Springer 313
15. WrightS 1939 The distribution of self-sterility alleles in populations. Genetics 24 538 552
16. SchopferCRNasrallahMENasrallahJB 1999 The male determinant of self - incompatibility in Brassica. Science 286 1697 1700
17. TakasakiTHatakeyamaKSuzukiGWatanabeMIsogaiA 2000 The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403 913 916
18. TakayamaSShimosatoHShibaHFunatoMCheFS 2001 Direct ligand–receptor complex interaction controls Brassica self-incompatibility. Nature 413 534 538
19. SuzukiG 2009 Recent progress in plant reproduction research: The story of the male gametophyte through to successful fertilization. Plant Cell Physiol 50 1857 1864
20. ChapmanLAGoringDR 2010 Pollen-pistil interactions regulating successful fertilization in the Brassicaceae. J Exp Bot 61 1987 1999
21. GuTMazzurcoMSulamanWMatiasDDGoringDR 1998 Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci U S A 95 382 387
22. MazzurcoMSulamanWElinaHCockJMGoringDR 2001 Further analysis of the interactions between the Brassica S receptor kinase and three interacting proteins (ARC1, THL1 and THL2) in the yeast two-hybrid system. Plant Mol Biol 45 365 376
23. MuraseKShibaHIwanoMCheF-SWatanabeM 2004 A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303 1516 1519
24. CharlesworthDCharlesworthB 1979 The evolution and breakdown of S-allele systems. Heredity 43 41 55
25. UyenoyamaMKZhangYNewbiginE 2001 On the origin of self-incompatibility haplotypes: Transition through self-compatible intermediates. Genetics 157 1805 1817
26. MableBKRobertsonAVDartSDi BerardoCWithamL 2005 Breakdown of self-incompatibility in the perennial Arabidopsis lyrata (Brassicaceae) and its genetic consequences. Evolution 59 1437 1448
27. PorcherELandeR 2005 Loss of gametophytic self-incompatibility with evolution of inbreeding depression. Evolution 59 46 60
28. BuschJWSchoenDJ 2008 The evolution of self-incompatibility when mates are limiting. Trends Plant Sci 13 128 136
29. MableBK 2008 Genetic causes and consequences of the breakdown of self - incompatibility: Case studies in the Brassicaceae. Genet Res 90 47 60
30. KusabaMDwyerKHendershotJVrebalovJNasrallahJB 2001 Self-incompatibility in the genus Arabidopsis: Characterization of the S-locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13 627 643
31. NasrallahMELiuPNasrallahJB 2002 Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297 247 249
32. ShimizuKK 2002 Ecology meets molecular genetics in Arabidopsis. Popul Ecol 44 221 233
33. NasrallahMELiuPSherman-BroylesSBoggsNANasrallahJB 2004 Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc Natl Acad Sci U S A 101 16070 16074
34. ShimizuKKPuruggananMD 2005 Evolutionary and ecological genomics of Arabidopsis. Plant Physiol 138 578 584
35. LiuPSherman-BroylesSNasrallahMENasrallahJB 2007 A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr Biol 17 734 740
36. Sherman-BroylesSBoggsNFarkasALiuPVrebalovJ 2007 S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 19 94 106
37. TangCToomajianCSherman-BroylesSPlagnolVGuoYL 2007 The evolution of selfing in Arabidopsis thaliana. Science 317 1070 1072
38. ShimizuKKShimizu-InatsugiRTsuchimatsuTPuruggananMD 2008 Independent origins of self-compatibility in Arabidopsis thaliana. Mol Ecol 17 704 714
39. BoggsNANasrallahJBNasrallahME 2009 Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet 5 e1000426 doi:10.1371/journal.pgen.1000426
40. BoggsNADwyerKGShahPMcCullochAABechsgaardJS 2009 Expression of distinct self-incompatibility specificities in Arabidopsis thaliana. Genetics 182 1313 1321
41. TsuchimatsuTSuwabeKShimizu-InatsugiRIsokawaSPavlidisP 2010 Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464 1342 1346
42. ShimizuKKKudohHKobayashiMJ 2011 Plant sexual reproduction during climate change: gene function in natura studied by ecological and evolutionary systems biology. Ann Bot 108 777 787
43. GuoYLBechsgaardJSSlotteTNeufferBLascouxM 2009 Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci U S A 106 5246 5251
44. BuschJWJolySSchoenDJ 2011 Demographic signatures accompanying the evolution of selfing in Leavenworthia alabamica. Mol Biol Evol 28 1717 1729
45. TantikanjanaTRizviNNasrallahMENasrallahJB 2009 A dual role for the S-locus receptor kinase in self-incompatibility and pistil development revealed by an Arabidopsis rdr6 mutation. Plant Cell 21 2642 2654
46. ShimizuKKFujiiSMarholdKWatanabeKKudohH 2005 Arabidopsis kamchatica (Fisch. ex DC.) K. Shimizu & Kudoh and A. kamchatica subsp. kawasakiana (Makino) K. Shimizu & Kudoh, new combinations. Acta Phytotax Geobot 56 165 174
47. SugisakaJKudohH 2008 Breeding system of the annual Cruciferae, Arabidopsis kamchatica subsp. kawasakiana. J Plant Res 121 65 68
48. Shimizu-InatsugiRLihováJIwanagaHKudohHMarholdK 2009 The allopolyploid Arabidopsis kamchatica originated from multiple individuals of Arabidopsis lyrata and Arabidopsis halleri. Mol Ecol 18 4024 4048
49. SchmicklRJørgensenMHBrystingAKKochMA 2010 The evolutionary history of the Arabidopsis lyrata complex: A hybrid in the amphi-Beringian area closes a large distribution gap and builds up a genetic barrier. BMC Evol Biol 10 98
50. FujimotoRKinoshitaYKawabeAKinoshitaTTakashimaK 2008 Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet 4 e1000048 doi:10.1371/journal.pgen.1000048
51. HigashiHIkedaHSetoguchiH 2012 Population fragmentation causes randomly fixed genotypes in populations of Arabidopsis kamchatica in the Japanese Archipelago. J Plant Res 125 223 233
52. SchierupMHMableBKAwadallaPCharlesworthD 2001 Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158 387 399
53. CharlesworthDBartoloméCSchierupMHMableBK 2003 Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata. Mol Biol Evol 20 1741 1753
54. CharlesworthDMableBKSchierupMHBartoloméCAwadallaP 2003 Diversity and linkage of genes in the self-incompatibility gene family in Arabidopsis lyrata. Genetics 164 1519 1535
55. MableBKSchierupMHCharlesworthD 2003 Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata (Brassicaceae) with sporophytic control of self-incompatibility. Heredity 90 422 431
56. CastricVVekemansX 2004 Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Mol Ecol 13 2873 2889
57. MableBKBelandJDi BerardoC 2004 Inheritance and dominance of self-incompatibility alleles in polyploid Arabidopsis lyrata. Heredity 93 476 486
58. CastricVVekemansX 2007 Evolution under strong balancing selection: How many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae? BMC Evol Biol 7 132
59. CastricVBechsgaardJSchierupMHVekemansX 2008 Repeated adaptive introgression at a gene under multiallelic balancing selection. PLoS Genet 4 e1000168 doi:10.1371/journal.pgen.1000168
60. LlaurensVBilliardSLeducqJBCastricVKleinEK 2008 Does frequency-dependent selection with complex dominance interactions accurately predict allelic frequencies at the self-incompatibility locus in Arabidopsis halleri? Evolution 62 2545 2557
61. CastricVBechsgaardJSGrenierSNoureddineRSchierupMH 2010 Molecular evolution within and between self-incompatibility specificities. Mol Biol Evol 27 11 20
62. BechsgaardJSCastricVCharlesworthDVekemansXSchierupMH 2006 The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol Biol Evol 23 1741 1750
63. GuoYLZhaoXLanzCWeigelD 2011 Evolution of the S-locus region in Arabidopsis relatives. Plant Physiol 157 937 946
64. SchierupMHBechsgaardJSChristiansenFB 2008 Selection at work in self-incompatible Arabidopsis lyrata. II. Spatial distribution of S haplotypes in Iceland. Genetics 180 1051 1059
65. BarringerBC 2007 Polyploidy and self-fertilization in flowering plants. Amer J Bot 94 1527 1533
66. MableBK 2004 Polyploidy and self-compatibility: Is there an association? New Phytol 162 803 811
67. LevinDA 1975 Minority cytotype exclusion in local plant populations. Taxon 24 35 43
68. FelberF 1991 Establishment of a tetraploid cytotype in a diploid population: Effect of relative fitness of cytotypes. J Evol Biol 4 194 207
69. RodríguezDJ 1996 A model for the establishment of polyploidy in plants. Amer Nat 147 33 46
70. MillerJSVenableDL 2000 Polyploidy and the evolution of gender dimorphism in plants. Science 289 2335 2338
71. HedrickPW 1987 Genetic load and the mating system in homosporous ferns. Evolution 41 1282 1289
72. RonfortJ 1999 The mutation load under tetrasomic inheritance and its consequences for the evolution of the selfing rate in autotetraploid species. Genet Res 74 31 42
73. FoxeJPSlotteTStahlEANeufferBHurkaH 2009 Recent speciation associated with the evolution of selfing in Capsella. Proc Natl Acad Sci U S A 106 5241 5245
74. VekemansXSlatkinM 1994 Gene and allelic genealogies at a gametophytic self-incompatibility locus. Genetics 137 1157 1165
75. KochMAHauboldBMitchell-OldsT 2000 Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis and related genera (Brassicaceae). Mol Biol Evol 17 1483 1498
76. OssowskiSSchneebergerKLucas-LledóJIWarthmannNClarkRM 2010 The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327 92 94
77. KusabaMNishioTSattaYHinataKOckendonD 1997 Striking sequence similarity in inter - and intra-specific comparisons of class I SLG alleles from Brassica oleracea and Brassica campestris: Implications for the evolution and recognition mechanism. Proc Natl Acad Sci U S A 94 7673 7678
78. NaithaniSChookajornTRipollDRNasrallahJB 2007 Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc Natl Acad Sci U S A 104 12211 12216
79. GoringDRGlavinTLSchaferURothsteinSJ 1993 An S receptor kinase gene in self-compatible Brassica napus has a 1-bp deletion. Plant Cell 5 531 539
80. OkamotoSOdashimaMFujimotoRSatoYKitashibaH 2007 Self-compatibility in Brassica napus is caused by independent mutations in S-locus genes. Plant J 50 391 400
81. de la ChauxNTsuchimatsuTShimizuKKWagnerA 2012 The predominantly selfing plant Arabidopsis thaliana experienced a recent reduction in transposable element abundance compared to its outcrossing relative Arabidopsis lyrata. Mob DNA 3 2
82. HewittG 2000 The genetic legacy of the Quaternary ice ages. Nature 405 907 913
83. NordborgMHuTTIshinoYJhaveriJToomajianC 2005 The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3 e196 doi:10.1371/journal.pbio.0030196
84. BeckJBSchmuthsHSchaalBA 2008 Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Mol Ecol 17 902 915
85. FrançoisOBlumMGJakobssonMRosenbergNA 2008 Demographic history of European populations of Arabidopsis thaliana. PLoS Genet 4 e1000075 doi:10.1371/journal.pgen.1000075
86. FoxeJPStiftMTedderAHaudryAWrightSI 2010 Reconstructing origins of loss of self-incompatibility and selfing in North American Arabidopsis lyrata: A population genetic context. Evolution 64 3495 3510
87. HoebePNStiftMTedderAMableBK 2009 Multiple losses of self-incompatibility in North American Arabidopsis lyrata: Phylogeographic context and population genetic consequences. Mol Ecol 18 4924 4939
88. TedderAHoebePHAnsellSWMableBK 2010 Using chloroplast trnF pseudogenes for phylogeography in Arabidopsis lyrata. Diversity 2 653 678
89. Ramos-OnsinsSEStrangerBEMitchell-OldsTAguadéM 2004 Multilocus analysis of variation and speciation in the closely related species Arabidopsis halleri and A. lyrata. Genetics 166 373 388
90. BakerHG 1955 Self-compatibility and establishment after “long-distance” dispersal. Evolution 9 347 349
91. BakerHG 1967 Support for Baker's law—as a rule. Evolution 21 853 856
92. PannellJRBarrettSCH 1998 Baker's law revisited: Reproductive assurance in a metapopulation. Evolution 52 657 668
93. PujolBZhouSRSanchez VilasJPannellJR 2009 Reduced inbreeding depression after species range expansion. Proc Natl Acad Sci U S A 106 15379 15383
94. BataillonTKirkpatrickM 2000 Inbreeding depression due to mildly deleterious mutations in finite populations: Size does matter. Genet Res 75 75 81
95. GleminS 2003 How are deleterious mutations purged? Drift versus nonrandom mating. Evolution 57 2678 2687
96. BuschJWDelphLF 2011 The relative importance of reproductive assurance and automatic selection as hypotheses for the evolution of self-fertilization. Ann Bot In press
97. McClureB 2009 Darwin's foundation for investigating self-incompatibility and the progress toward a physiological model for S-RNase-based SI. J Exp Bot 60 1069 1081
98. RobertsonKGoldbergEEIgicB 2011 Comparative evidence for the correlated evolution of polyploidy and self-compatibility in Solanaceae. Evolution 65 139 155
99. NasrallahJBLiuPSherman-BroylesSSchmidtRNasrallahME 2007 Epigenetic mechanisms for breakdown of self-incompatibility in interspecific hybrids. Genetics 175 1965 1973
100. BatemanAJ 1952 Self-incompatibility systems in angiosperms. 1. Theory. Heredity 6 285 310
101. ShibaHIwanoMEntaniTIshimotoKShimosatoH 2002 The dominance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell 14 491 504
102. KusabaMTungCWNasrallahMENasrallahJB 2002 Monoallelic expression and dominance interactions in anthers of self-incompatible Arabidopsis lyrata. Plant Physiol 128 17 20
103. PrigodaNLNassuthAMableBK 2005 Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol Biol Evol 22 1609 1620
104. FujimotoRSugimuraTFukaiENishioT 2006 Suppression of gene expression of a recessive SP11/SCR allele by an untranscribed SP11/SCR allele in Brassica self-incompatibility. Plant Mol Biol 61 577 587
105. SchierupMHVekemansXChristiansenFB 1997 Evolutionary dynamics of sporophytic self-incompatibility alleles in plants. Genetics 147 835 846
106. BilliardSCastricVVekemansX 2007 A general model to explore complex dominance patterns in plant sporophytic self-incompatibility systems. Genetics 175 1351 1369
107. ShibaHKakizakiTIwanoMTarutaniYWatanabeM 2006 Dominance relationships between self-incompatibility alleles controlled by DNA methylation. Nat Genet 38 297 299
108. TarutaniYShibaHIwanoMKakizakiTSuzukiG 2010 Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466 983 986
109. VekemansXSchierupMHChristiansenFB 1998 Mate availability and fecundity selection in multi-allelic self-incompatibility systems in plants. Evolution 52 19 29
110. BuschJWUrbanL 2011 Insights gained from 50 years of studying the evolution of self-compatibility in Leavenworthia (Brassicaceae). Evol Biol 38 15 27
111. NasrallahJBRundleandSJNasrallahME 1994 Genetic evidence for the requirement of the Brassica S-locus receptor kinase gene in the self-incompatibility response. Plant J 5 373 384
112. FujimotoRSugimuraTNishioT 2006 Gene conversion from SLG to SRK resulting in self-compatibility in Brassica rapa. FEBS Lett 580 425 430
113. HallTA 1999 BIOEDIT: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41 95 98
114. TamuraKPetersonDPetersonNStecherGNeiM 2011 MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28 2731 2739
115. GaoHWilliamsonSBustamanteCD 2007 A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics 176 1635 1651
116. JakobssonMRosenbergNA 2007 CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23 1801 1806
117. RosenbergNA 2004 DISTRUCT: A program for the graphical display of population structure. Mol Ecol Notes 4 137 138
118. EvannoGRegnautSGoudetJ 2005 Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol 14 2611 2620
119. PritchardJKStephensMDonnellyP 2000 Inference of population structure using multilocus genotype data. Genetics 155 945 959
120. CramerH 1946 Mathematical method of statistics Princeton Princeton University Press 575
121. SokalRRRohlfFJ 1995 Biometry: The principles and practice of statistics in biological research, 3rd ed New York W. H. Freeman 880
122. R Development Core Team 2009 R: A language and environment for statistical computing Vienna (Austria) R Foundation for Statistical Computing Available from http://www.R-project.org
123. JukesTHCantorCR 1969 Evolution of protein molecules. MunroHN Mammalian protein metabolism New York Academic Press 32 132
124. NeiMKumarS 2000 Molecular evolution and phylogenetics Oxford Oxford University Press 336
125. CharruauPFernandesCOrozco-TerwengelPPetersJHunterL 2011 Phylogeography, genetic structure and population divergence time of cheetahs in Africa and Asia: Evidence for long-term geographic isolates. Mol Ecol 20 706 724
126. StoneACBattistuzziFUKubatkoLSPerryGHJrTrudeauE 2010 More reliable estimates of divergence times in Pan using complete mtDNA sequences and accounting for population structure. Phil Trans R Soc Lond B Biol Sci 365 3277 3288
127. DrummondAJHoSYPhillipsMJRambautA 2006 Relaxed phylogenetics and dating with confidence. PLoS Biol 4 e88 doi:10.1371/journal.pbio.0040088
128. OhwiJ 1953 Flora of Japan Washington Smithsonian Institution 1067
129. ShimizuKKOkadaK 2000 Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127 4511 4518
Štítky
Genetika Reprodukční medicína
Článek Allelic Heterogeneity and Trade-Off Shape Natural Variation for Response to Soil MicronutrientČlánek The Chicken Frizzle Feather Is Due to an α-Keratin () Mutation That Causes a Defective RachisČlánek A Trans-Species Missense SNP in Is Associated with Sex Determination in the Tiger Pufferfish, (Fugu)
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 7- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
-
Všechny články tohoto čísla
- Functional Evolution of Mammalian Odorant Receptors
- Oocyte Family Trees: Old Branches or New Stems?
- Allelic Heterogeneity and Trade-Off Shape Natural Variation for Response to Soil Micronutrient
- Guidelines for Genome-Wide Association Studies
- GWAS Identifies Novel Susceptibility Loci on 6p21.32 and 21q21.3 for Hepatocellular Carcinoma in Chronic Hepatitis B Virus Carriers
- DNA Methyltransferases Are Required to Induce Heterochromatic Re-Replication in Arabidopsis
- Genomic Data Reveal a Complex Making of Humans
- Let-7b/c Enhance the Stability of a Tissue-Specific mRNA during Mammalian Organogenesis as Part of a Feedback Loop Involving KSRP
- The Secreted Immunoglobulin Domain Proteins ZIG-5 and ZIG-8 Cooperate with L1CAM/SAX-7 to Maintain Nervous System Integrity
- RsfA (YbeB) Proteins Are Conserved Ribosomal Silencing Factors
- Gene Conversion Occurs within the Mating-Type Locus of during Sexual Reproduction
- The Chicken Frizzle Feather Is Due to an α-Keratin () Mutation That Causes a Defective Rachis
- Meta-Analysis of Genome-Wide Scans for Total Body BMD in Children and Adults Reveals Allelic Heterogeneity and Age-Specific Effects at the Locus
- Balancing Selection at the Tomato Guardee Gene Family Maintains Variation in Strength of Pathogen Defense
- Large-Scale Introgression Shapes the Evolution of the Mating-Type Chromosomes of the Filamentous Ascomycete
- OSD1 Promotes Meiotic Progression via APC/C Inhibition and Forms a Regulatory Network with TDM and CYCA1;2/TAM
- Intact p53-Dependent Responses in miR-34–Deficient Mice
- FANCJ/BACH1 Acetylation at Lysine 1249 Regulates the DNA Damage Response
- CED-10/Rac1 Regulates Endocytic Recycling through the RAB-5 GAP TBC-2
- Histone H2A Mono-Ubiquitination Is a Crucial Step to Mediate PRC1-Dependent Repression of Developmental Genes to Maintain ES Cell Identity
- F-Box Protein Specificity for G1 Cyclins Is Dictated by Subcellular Localization
- The Gene Encodes a Nuclear Protein That Affects Alternative Splicing
- A Key Role for Chd1 in Histone H3 Dynamics at the 3′ Ends of Long Genes in Yeast
- Genome-Wide Association Analysis in Asthma Subjects Identifies as a Novel Bronchodilator Response Gene
- GRHL3/GET1 and Trithorax Group Members Collaborate to Activate the Epidermal Progenitor Differentiation Program
- Brain-Specific Rescue of Reveals System-Driven Transcriptional Rhythms in Peripheral Tissue
- Recent Loss of Self-Incompatibility by Degradation of the Male Component in Allotetraploid
- Pregnancy-Induced Noncoding RNA () Associates with Polycomb Repressive Complex 2 and Regulates Mammary Epithelial Differentiation
- The HEI10 Is a New ZMM Protein Related to Zip3
- The SCF Ubiquitin E3 Ligase Ubiquitylates Sir4 and Functions in Transcriptional Silencing
- Induction of Cytoprotective Pathways Is Central to the Extension of Lifespan Conferred by Multiple Longevity Pathways
- Role of Architecture in the Function and Specificity of Two Notch-Regulated Transcriptional Enhancer Modules
- Loss of ATRX, Genome Instability, and an Altered DNA Damage Response Are Hallmarks of the Alternative Lengthening of Telomeres Pathway
- A Regulatory Loop Involving PAX6, MITF, and WNT Signaling Controls Retinal Pigment Epithelium Development
- The Three Faces of Riboviral Spontaneous Mutation: Spectrum, Mode of Genome Replication, and Mutation Rate
- Unmet Expectations: miR-34 Plays No Role in p53-Mediated Tumor Suppression In Vivo
- A Genome-Wide Association Meta-Analysis of Circulating Sex Hormone–Binding Globulin Reveals Multiple Loci Implicated in Sex Steroid Hormone Regulation
- The Role of Rice HEI10 in the Formation of Meiotic Crossovers
- A Trans-Species Missense SNP in Is Associated with Sex Determination in the Tiger Pufferfish, (Fugu)
- Influences Bone Mineral Density, Cortical Bone Thickness, Bone Strength, and Osteoporotic Fracture Risk
- Evidence of Inbreeding Depression on Human Height
- Comparative Genomics of Plant-Associated spp.: Insights into Diversity and Inheritance of Traits Involved in Multitrophic Interactions
- Detecting Individual Sites Subject to Episodic Diversifying Selection
- Regulates Rhodopsin-1 Metabolism and Is Required for Photoreceptor Neuron Survival
- Identification of Chromatin-Associated Regulators of MSL Complex Targeting in Dosage Compensation
- Three Dopamine Pathways Induce Aversive Odor Memories with Different Stability
- TDP-1/TDP-43 Regulates Stress Signaling and Age-Dependent Proteotoxicity in
- Rapid Turnover of Long Noncoding RNAs and the Evolution of Gene Expression
- The Yeast Rab GTPase Ypt1 Modulates Unfolded Protein Response Dynamics by Regulating the Stability of RNA
- Histone H2B Monoubiquitination Facilitates the Rapid Modulation of Gene Expression during Arabidopsis Photomorphogenesis
- Cellular Variability of RpoS Expression Underlies Subpopulation Activation of an Integrative and Conjugative Element
- Genetic Variants in , , and Influence Male Recombination in Cattle
- Differential Impact of the HEN1 Homolog HENN-1 on 21U and 26G RNAs in the Germline of
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Guidelines for Genome-Wide Association Studies
- The Role of Rice HEI10 in the Formation of Meiotic Crossovers
- Identification of Chromatin-Associated Regulators of MSL Complex Targeting in Dosage Compensation
- GWAS Identifies Novel Susceptibility Loci on 6p21.32 and 21q21.3 for Hepatocellular Carcinoma in Chronic Hepatitis B Virus Carriers
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



