-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Pregnancy-Induced Noncoding RNA () Associates with Polycomb Repressive Complex 2 and Regulates Mammary Epithelial Differentiation
Pregnancy-induced noncoding RNA (PINC) and retinoblastoma-associated protein 46 (RbAp46) are upregulated in alveolar cells of the mammary gland during pregnancy and persist in alveolar cells that remain in the regressed lobules following involution. The cells that survive involution are thought to function as alveolar progenitor cells that rapidly differentiate into milk-producing cells in subsequent pregnancies, but it is unknown whether PINC and RbAp46 are involved in maintaining this progenitor population. Here, we show that, in the post-pubertal mouse mammary gland, mPINC is enriched in luminal and alveolar progenitors. mPINC levels increase throughout pregnancy and then decline in early lactation, when alveolar cells undergo terminal differentiation. Accordingly, mPINC expression is significantly decreased when HC11 mammary epithelial cells are induced to differentiate and produce milk proteins. This reduction in mPINC levels may be necessary for lactation, as overexpression of mPINC in HC11 cells blocks lactogenic differentiation, while knockdown of mPINC enhances differentiation. Finally, we demonstrate that mPINC interacts with RbAp46, as well as other members of the polycomb repressive complex 2 (PRC2), and identify potential targets of mPINC that are differentially expressed following modulation of mPINC expression levels. Taken together, our data suggest that mPINC inhibits terminal differentiation of alveolar cells during pregnancy to prevent abundant milk production and secretion until parturition. Additionally, a PRC2 complex that includes mPINC and RbAp46 may confer epigenetic modifications that maintain a population of mammary epithelial cells committed to the alveolar fate in the involuted gland.
Published in the journal: . PLoS Genet 8(7): e32767. doi:10.1371/journal.pgen.1002840
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002840Summary
Pregnancy-induced noncoding RNA (PINC) and retinoblastoma-associated protein 46 (RbAp46) are upregulated in alveolar cells of the mammary gland during pregnancy and persist in alveolar cells that remain in the regressed lobules following involution. The cells that survive involution are thought to function as alveolar progenitor cells that rapidly differentiate into milk-producing cells in subsequent pregnancies, but it is unknown whether PINC and RbAp46 are involved in maintaining this progenitor population. Here, we show that, in the post-pubertal mouse mammary gland, mPINC is enriched in luminal and alveolar progenitors. mPINC levels increase throughout pregnancy and then decline in early lactation, when alveolar cells undergo terminal differentiation. Accordingly, mPINC expression is significantly decreased when HC11 mammary epithelial cells are induced to differentiate and produce milk proteins. This reduction in mPINC levels may be necessary for lactation, as overexpression of mPINC in HC11 cells blocks lactogenic differentiation, while knockdown of mPINC enhances differentiation. Finally, we demonstrate that mPINC interacts with RbAp46, as well as other members of the polycomb repressive complex 2 (PRC2), and identify potential targets of mPINC that are differentially expressed following modulation of mPINC expression levels. Taken together, our data suggest that mPINC inhibits terminal differentiation of alveolar cells during pregnancy to prevent abundant milk production and secretion until parturition. Additionally, a PRC2 complex that includes mPINC and RbAp46 may confer epigenetic modifications that maintain a population of mammary epithelial cells committed to the alveolar fate in the involuted gland.
Introduction
Noncoding RNAs (ncRNAs) are emerging as significant players in the regulatory circuitry of the cell, rivaling their protein-coding counterparts. Accumulating data have revealed the functional diversity of ncRNAs, in particular long noncoding RNAs (lncRNAs), and their relevance in regulating development and disease [1]–[6]. Previous efforts to understand the function of noncoding RNAs have predominantly focused on small/short RNAs (<200 nucleotides). However, in the past few years there has been an increased focus on lncRNAs, as large-scale analyses have revealed the abundance of these molecules in more complex organisms [7]–[11]. While initial reports of the pervasive noncoding transcription found in the eukaryotic genome were met with skepticism, experimental evidence has shown that many of these lncRNAs are expressed in tissue - and cell-specific patterns in development, suggestive of their functionality [12]–[14]. In addition, knockdown and overexpression studies have shown that an increasing number of lncRNAs play important roles in regulating a diverse spectrum of processes, including splicing [15], transcription [16], localization [17], and organization of subcellular compartments [18].
As more lncRNAs are discovered, functional categorization based on properties such as expression, localization and mechanism allow us to better predict the roles of newly discovered lncRNAs, and have vastly facilitated our understanding of those already known. One recurring feature of an emerging class of lncRNAs is an association with chromatin-modifying complexes. In fact, as many as 38% of large intergenic noncoding RNAs (lincRNAs) have been shown to interact with various chromatin-modifying complexes and 24% specifically interact with polycomb repressive complex 2 (PRC2) [19]. In addition, a recent study has identified thousands more lncRNAs that associate with PRC2, many of which do not fall into the lincRNA category [20]. PRC2 recruitment results in trimethylation of histone H3 at lysine 27 (H3K27), leading to chromatin compaction and transcriptional repression of associated genes. The role of PRC2 in the epigenetic regulation of cohorts of genes involved in the maintenance of cellular identity and differentiation during tissue development is well documented [21]. While the functional importance of the abundant association of lncRNAs with these complexes has, in most cases, not yet been validated, several lncRNAs have been shown to be necessary for targeting PRC2 to specific loci either in cis or in trans, thereby altering gene expression [13], [22]–[24].
Post-pubertal mammary gland development involves hormonally-regulated expansion and regression of the mammary epithelium as the gland undergoes the processes of pregnancy, lactation and involution [25]–[28]. During pregnancy, mammary epithelial cells (MECs) proliferate and begin to differentiate into highly specialized alveolar cells through a process known as alveologenesis. Terminal differentiation is delayed until parturition, when alveolar cells gain the capacity to produce and secrete the high levels of milk protein necessary for lactation. Following lactation, the majority of differentiated alveolar cells undergo apoptosis and the gland remodels to a pre-pregnancy state in a process called involution. However, a small population of partially committed alveolar cells remains in the involuted gland and functions as alveolar progenitor cells that rapidly differentiate into milk-producing cells in subsequent pregnancies [29]. Although several signaling pathways are known to play a role in the stepwise progression of alveologenesis [30]–[32], the molecular mechanisms that maintain alveolar progenitors in the pregnant and involuting gland are unknown. The maintenance of stem and progenitor populations has been proposed to require epigenetic modifications to stably suppress differentiation, a function well suited for a histone-modifying complex such as PRC2 [33], [34].
Previously, our laboratory identified Pregnancy-Induced NonCoding RNA (PINC) and retinoblastoma-associated protein 46 (RbAp46) as genes that are elevated in the involuted rat mammary gland compared to an age-matched virgin gland [35]. PINC and RbAp46, a member of PRC2, were shown to be highly expressed in alveolar cells of the pregnant gland and in MECs that remain in the regressed lobules following involution. PINC is a mammalian-specific, evolutionary conserved, alternatively spliced and polyadenylated lncRNA. Initial in vitro studies of mouse PINC (mPINC) splice forms, mPINC1.0 and mPINC1.6, showed potential roles in the regulation of survival and cell cycle progression of mammary epithelial cells [36]. However, the developmental regulation of PINC and RbAp46 in the post-pubertal mammary gland, as well as their cell-type specificity, suggest they may play a more specialized role in alveologenesis. Therefore, the objective of this study was to evaluate a role for mPINC in regulating alveolar development and investigate a potential interaction of mPINC and RbAp46.
In the present study, we show that mPINC expression declines in the mammary gland during the transition from late pregnancy to early lactation as well as in response to lactogenic hormone-induced differentiation of the HC11 mammary epithelial cell line. We also show that overexpression of mPINC blocks lactogenic differentiation, while knockdown of mPINC enhances lactogenic differentiation of HC11 cells. In addition, we provide evidence for an interaction between mPINC and RbAp46, as well as other members of PRC2. Finally, we identify potential mPINC targets that are differentially expressed following modulation of mPINC levels. Taken together, these data suggest that mPINC inhibits differentiation of alveolar cells and that mPINC downregulation is necessary during lactation to allow alveolar cells to undergo terminal secretory differentiation. Additionally, a PRC2 complex that includes mPINC and RbAp46 may provide the epigenetic modifications that maintain a progenitor population of MECs committed to an alveolar fate in the pregnant and involuted gland.
Results
mPINC is alternatively spliced and differentially regulated in the post-pubertal mouse mammary gland
PINC was initially isolated from the rat mammary gland. We therefore decided to investigate the localization and expression levels of PINC in the mouse. Previously, we identified two mPINC splice forms, mPINC1.0 and mPINC1.6, based on available EST data [36]. To analyze expression of mPINC during post-pubertal mouse mammary development, RT-PCR was performed using primers at the extreme ends of each splice form. We found that mPINC levels were low in the virgin gland, increased during pregnancy, declined in early lactation, and increased again in early involution (Figure 1A). Unexpectedly, mPINC1.0 and mPINC1.6 RT-PCR amplified multiple bands, indicating each splice form may be further spliced. Each band was cloned and sequenced, which verified that there are at least three mPINC1.0 splice forms and six mPINC1.6 splice forms. Of the six mPINC1.6 splice forms, three lack the highly conserved region of the unique terminal exon of mPINC1.6 and were therefore designated DCR2 (deleted conserved region 2) (Figure 1B).
Fig. 1. mPINC expression peaks in the late pregnant and early involuting gland. 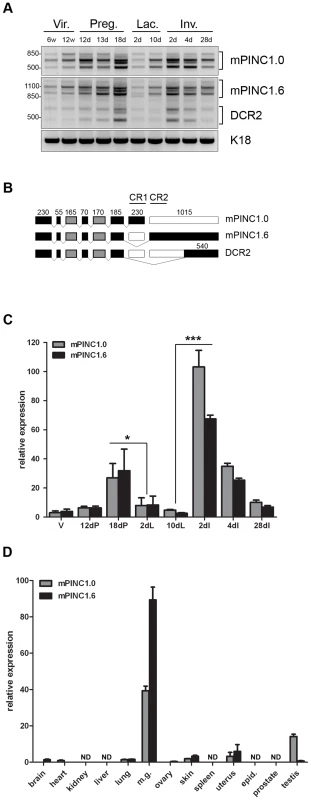
(A) RT-PCR shows multiple splice forms of mPINC1.0 and mPINC1.6 are expressed during mammary gland development. Primers designed to the extreme ends of mPINC1.0 and mPINC1.6 were used to amplify cDNA from mammary gland developmental stages. (w Vir.: weeks old virgin, d Preg.: days pregnancy, d Lac.: days lactation, d Inv.: days involution). PCR products were sequenced and found to be new splice forms of mPINC, including a new splice variant of mPINC1.6 called DCR2, for deleted conserved region 2. (B) Schematic diagram of mPINC exonic structure. Black boxes represent exons that are always included, grey boxes are sometimes included and clear boxes are never included. Nucleotide length is indicated above each exon along with black lines that overlap the most conserved regions of the PINC locus, CR1 and CR2. Exon 6 sometimes has an additional 24 nucleotides at the 3′ end in the mPINC1.0 and mPINC1.6 splice forms. This alternative splice site does not correlate with the inclusion/exclusion of any particular exon and its function is unknown. (C) qPCR shows mPINC is highest during late pregnancy and early involution. Mammary glands were harvested from 3 female Balb/c mice for each stage (V: adult virgin, dP: days pregnant, dL: days lactation, dI: days involution). Target genes were normalized to Actb and set relative to levels in the virgin mammary gland. (D) mPINC expression is most abundant in the mammary gland. Tissues were harvested from three 10 week old virgin Balb/c female mice and testis, epididymis, and prostate was harvested from three 12 week old male Balb/c mice. ND indicates tissues in which mPINC was not detected by qPCR. Target genes were normalized to Actb and set relative to levels in the lung. The RT-PCR data suggest that mPINC1.0 and mPINC1.6 levels are coordinately regulated during mammary gland development. We, therefore, performed quantitative PCR (qPCR) to more precisely quantify expression levels of mPINC splice forms. Both mPINC1.0 and mPINC1.6 increased throughout pregnancy and then declined between day 18 of pregnancy and day 2 of lactation (Figure 1C). Expression increased again at day 2 of involution, declining thereafter until 28 days post-involution, where both splice forms were still elevated compared to the virgin gland. These data are in agreement with previous observations from the rat mammary gland and verify that mPINC splice forms are, in general, coordinately regulated during mammary gland development.
PINC shows mammalian-specific conservation and previously PINC expression was found to be largely restricted to the mammary gland in the rat [36]. As mentioned above, there are only two mouse ESTs related to PINC in the GenBank database and they are both from mammary gland libraries. To obtain a more thorough analysis of PINC expression in the mouse, qPCR was performed using RNA isolated from a diverse range of tissues. The results showed that mPINC was expressed at very low levels in non-mammary tissues compared to virgin mammary gland, including the uterus and testis (Figure 1D). Interestingly, these two tissues undergo hormone-dependent cyclical expansion and regression in the adult, like the mammary gland. However, mPINC was greatly enriched in the virgin mammary gland compared to other mouse tissues, and is especially high in the pregnant and involuting mammary gland, suggesting it may have a more specialized role in mammary gland development.
mPINC is upregulated in the luminal cells during pregnancy and is enriched in luminal and alveolar progenitors
mPINC is expressed at low levels in the virgin mammary gland and is upregulated throughout pregnancy, however, the cell-type specificity of mPINC is unknown. The mammary gland contains two general populations of epithelial cells: basal and luminal [26]. Basal cells include myoepithelial cells, which are contractile and necessary for milk secretion during lactation, as well as a population of stem cells. Luminal cells are comprised of a population of mature, hormone receptor-positive cells and progenitor cells, which are thought to generate ductal and alveolar cells. To determine which epithelial cells of the mouse mammary gland express mPINC, we used fluorescence-activated cell sorting (FACS) combined with qPCR. Mammary epithelial cells from 16 day pregnant and age-matched virgin mice were sorted into basal (CD24loCD49fhi) and luminal (CD24hiCD49flo) cell populations. To confirm the purity of the sorted populations, qPCR was performed and showed elevated levels of keratin 8 (Krt8) in the luminal population and keratin 14 (Krt14) in the basal population (Figure 2A, 2B). In the virgin mouse, basal and luminal cells expressed equivalent levels of mPINC. However, mPINC specifically increased in the luminal cells during pregnancy, with expression levels increasing 5-fold compared to the basal cells (Figure 2C). These data are consistent with a role for mPINC in regulating alveolar development as the luminal cells are thought to give rise to the alveolar cells during pregnancy.
Fig. 2. mPINC rises specifically in the luminal compartment during pregnancy and is enriched in luminal and alveolar progenitors of the mammary gland. 
(A–C) RNA was isolated from FACs sorted mammary populations including, virgin luminal (VL) and basal (VB) as well as pregnant luminal (PL) and basal (PB). (A) qPCR showed Krt8 expression was enriched in the luminal populations (VL and PL), (B) Krt14 was enriched in the basal populations (VB and PB) and (C) mPINC was enriched in the pregnant luminal population, (D–F) MECs were FACs sorted into luminal and basal populations using CD24 and CD29. The luminal population was selected and further sorted into mature luminal (ML), luminal progenitors (LP), and alveolar progenitors (AP) using CD14 and ckit. (D) FACs dot plots showing CD24 and CD29 (left panel) as well as CD14 and ckit (right panel) from virgin MECs. (E) qPCR showed Ly6a and Wnt4 enriched in the ML population and Elf5 in the LP population thus verifying the purity of each population. (F) mPINC was enriched in the luminal progenitors and alveolar progenitors. Data represent mean ±SD (n = 3). Target genes were normalized to Gapdh. Next, we wanted to determine if mPINC is enriched in a particular subpopulation of luminal mammary epithelial cells. For this purpose, we used mammary epithelial cells from adult virgin mice where three luminal populations have recently been identified and characterized based on expression of CD14 and ckit [37]. These include mature luminal cells (CD14− ckit−) and two distinct luminal progenitor populations (CD14+ckit+ and CD14+ckit−/lo). The CD14+ckit+ population resembles the previously described CD24hiCD49floCD61+ luminal progenitor population [38], while the CD14+ckit−/lo population is thought to be comprised of more committed alveolar progenitor cells. Therefore, we isolated luminal mammary epithelial cells (CD29loCD24+) from virgin mice and sorted them into mature luminal cells (CD14− ckit−), luminal progenitors (CD14+ckit+), and alveolar progenitors (CD14+ckit−/lo) (Figure 2D). To validate the purity of the sorted populations, we used genes that were previously shown to be enriched in either mature luminal or luminal progenitor populations [39]–[41]. qPCR analysis showed increased expression of Wnt4 and Ly6a or Sca1 (lymphocyte antigen 6 complex locus a) in the mature luminal population as well as Elf5 (e74-like factor 5) enrichment in the luminal progenitor populations, as expected (Figure 2E). We then analyzed mPINC expression and found that it was increased more than 40-fold in the luminal progenitors and 50-fold in the alveolar progenitors compared to the mature luminal population (Figure 2F). The high level of mPINC observed in both the luminal and alveolar progenitors supports a potential role for mPINC in maintaining mammary epithelial cells in an undifferentiated state.
To verify that mPINC is enriched in alveolar cells of the luminal compartment during pregnancy, we performed in situ hybridization using probes specific to either mPINC1.0 or mPINC1.6 on mammary gland sections from mice at day 12 of pregnancy. These results showed that mPINC1.0 and mPINC1.6 were both highly expressed in alveolar cells of the mid-pregnant gland (Figure 3). The majority of alveolar cells expressed mPINC in the mid-pregnant gland, although the intensity of the in situ signal varied slightly among the alveolar clusters (Figure S1). These localization studies provide further evidence that mPINC may play a role in alveolar development.
Fig. 3. mPINC is enriched in alveolar cells of the pregnant gland. 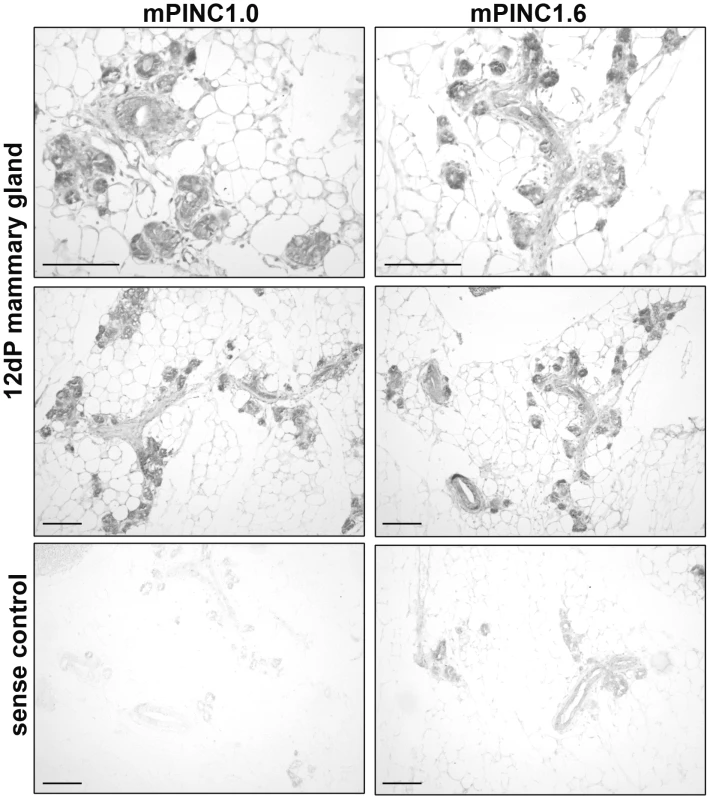
In situ hybridization using DIG-labeled probes shows mPINC1.0 and mPINC1.6 are expressed in alveolar cells of the 12 day mouse mammary gland. Sense control probes for mPINC1.0 and mPINC1.6 show very little signal in the bottom panels. Scale bars represent 100 µm. mPINC is downregulated during lactogenic hormone-induced differentiation of HC11 cells
During the transition from late pregnancy to lactation, alveolar cells undergo the final step of alveologenesis called secretory activation, which is necessary to produce and secrete high levels of milk proteins. Since a significant drop in mPINC expression levels temporally coincides with this important process, we decided to test whether mPINC plays a functional role in the terminal differentiation of alveolar cells. To do this, we used an in vitro model of lactogenic differentiation in which HC11 cells, a mouse mammary epithelial cell line derived from the Comma-D cell line originally isolated from a mid-pregnant Balb/c mouse, were treated with the lactogenic hormones hydrocortisone, insulin and prolactin to induce differentiation [42], [43]. As a read-out for differentiation, we measured the expression levels of three milk protein genes, beta-casein (Csn2), whey acidic protein (Wap) and lactotransferrin (Ltf). After one hour of hormone treatment, there was an increase in the expression of Csn2, which continued to rise at 24 and 72 hours (Figure 4B). Wap expression was unchanged after one hour, but significantly increased after 24 hours of hormone treatment and continued to rise at 72 hours (Figure 4C). Ltf expression levels only began to rise following 72 hours of hormone treatment (Figure 4D). mPINC expression was unchanged after one hour, but both mPINC1.0 and mPINC1.6 were reduced by more than 50% following 24 hours of lactogenic hormone treatment and by more than 75% after 72 hours of treatment (Figure 4A). These data are consistent with the downregulation of mPINC seen in early lactation and suggest that mPINC may play a functional role in regulating differentiation of alveolar cells. These results also indicate that HC11 cells are a suitable model to further explore the role of mPINC in regulating hormonally-induced lactogenic differentiation.
Fig. 4. mPINC expression declines during lactogenic hormone induced differentiation of HC11 cells. 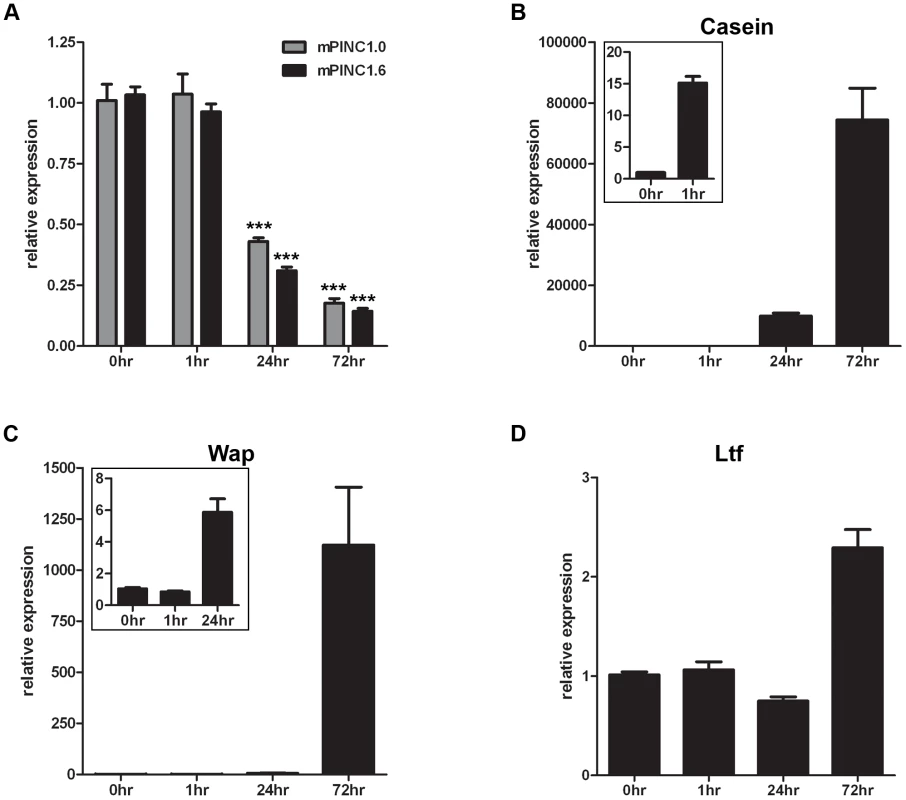
(A–C) Confluent HC11 cells were treated with lactogenic hormones for 1, 24 and 72 hrs followed by RNA isolation to detect changes in gene expression. Target genes were normalized to Gapdh and set relative to levels in untreated HC11 cells. Data are presented as mean ± SEM (n = 3). (A) qPCR shows mPINC1.0 and mPINC1.6 expression decreases following 24 and 72 hrs of hormone treatment. (B) qPCR shows Csn2 levels begin to rise after 1 hr of hormone treatment and continue to rise at 24 and 72 hrs. (C) qPCR shows Wap expression levels rise following 24 hrs of hormone treatment and continue to rise after 72 hrs. (D) Ltf expression levels rise following 72 hrs of lactogenic hormone treatment. Overexpression of mPINC inhibits differentiation
The decline in mPINC levels observed during lactogenic differentiation in vivo and in vitro suggests that mPINC downregulation may be necessary to allow alveolar cells to undergo terminal differentiation. If this is true, then maintenance of high levels of mPINC should inhibit differentiation. To test this hypothesis, we generated HC11 cells overexpressing both mPINC1.0 and mPINC1.6 (LeGO-1.0/1.6), or a vector control (LeGO-GFP) using a lentiviral vector. We then treated the overexpressing cells with lactogenic hormones to induce differentiation. Following 24 and 72 hours of treatment, qPCR analysis showed that mPINC overexpressing cells expressed significantly lower levels of Csn2, Wap and Ltf mRNA compared to control cells (Figure 5A, 5B).
Fig. 5. Overexpression of mPINC inhibits differentiation of HC11 cells. 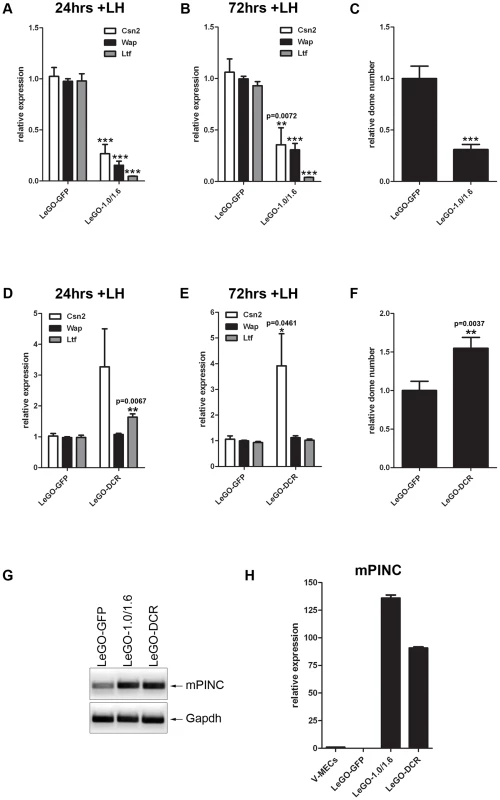
(A–C) After 24 (A) and 72 (B) hrs of hormone induction, LeGO-1.0/1.6 cells show reduced levels of Csn2, Wap, and Ltf expression by qPCR. Target genes were normalized to Gapdh and set relative to LeGO-GFP levels. Data are presented as mean ± SEM (n = 6). (C) Overexpression of mPINC also reduces formation of domes. Domes were counted at 48 hrs post-hormone treatment from nine 20× fields/experiment and data represent mean ± SEM set relative to the dome number in the LeGO-GFP control group (n = 6). (D–F) After 24 (D) and 72 (E) hrs of hormone induction, LeGO-DCR cells show increased levels of Ltf (at 24 hrs) and Csn2, but not Wap, expression compared to LeGO-GFP cells. Target genes were normalized to Gapdh and set relative to LeGO-GFP levels. Data are presented as mean ± SEM (n = 6). (F) Overexpression of the DCR mutant enhances dome formation compared to the control. Experiment was performed and analyzed as described in panel C. (G) RT-PCR shows mPINC overexpressed transcript in LeGO-1.0/1.6 and LeGO-DCR HC11 cells compared to endogenous levels in LeGO-GFP cells. (H) qPCR shows mPINC overexpressed levels are high and fairly equivalent relative to mPINC endogenous levels in MECs purified from virgin mammary gland (V-MECs). Target gene was normalized to Gapdh. Data are presented as mean ± SEM for three independent experiments. In addition to changes in gene expression, HC11 lactogenic differentiation is accompanied by morphological changes [43]. These changes include the formation of fluid-filled structures called domes that can be quantified as an indication of the extent of HC11 differentiation. Overexpression of mPINC caused a drastic reduction in the formation of dome structures compared to the control group (Figure 5C). Taken together, these results indicate that high levels of mPINC inhibit lactogenic differentiation of mammary epithelial cells, and that downregulation of mPINC may be required for terminal differentiation of alveolar cells in early lactation.
To determine whether the highly conserved regions of mPINC are necessary for its function as an inhibitor of HC11 differentiation, we generated a deletion mutant of mPINC (DCR) that lacked both terminal exons of mPINC1.0 and mPINC1.6, including CR1 and CR2. We found that overexpression of the mPINC deletion mutant (LeGO-DCR) failed to inhibit lactogenic differentiation. Instead, the DCR overexpressing cells showed slightly increased levels of Csn2 mRNA compared to control cells (LeGO-GFP) at both 24 and 72 hours after lactogenic hormone treatment, as well as increased Ltf expression at 24 hours, while Wap expression was unchanged (Figure 5D, 5E). Dome formation was also increased 1.5-fold (Figure 5F) compared to control cells. The increased markers of differentiation indicate that the DCR mutant could be interfering with endogenous mPINC function (see Discussion). However, more importantly, these results demonstrate that the conserved regions of mPINC are necessary for its function as an inhibitor of lactogenic differentiation.
To verify that the different phenotypes of the LeGO-1.0/1.6 and LeGO-DCR cells were not due to differences in overexpression levels, we performed RT-PCR and qPCR using RNA isolated from the two cell lines. RT-PCR analysis confirmed overexpression of mPINC compared to endogenous mPINC levels in control cells. In addition, qPCR analysis showed that the LeGO-1.0/1.6 and LeGO-DCR HC11 cells expressed mPINC at high (135-fold and 90-fold, respectively) and fairly equivalent levels compared to LeGO-GFP cells and MECs isolated from adult virgin mammary glands (Figure 5G, 5H). These data further suggest that the conserved regions of mPINC are necessary for its inhibitory effect on differentiation.
Knockdown of mPINC increases differentiation of HC11 cells
As mPINC overexpression inhibited differentiation of HC11 cells, we sought to determine if a reduction in mPINC levels could enhance differentiation. Because there is an endogenous mPINC isoform that lacks the conserved regions, DCR2, it was of interest to ascertain if its function is distinct from the other mPINC splice forms. Therefore, we used pairs of siRNAs to target mPINC1.0 and mPINC1.6 only (siPINC1.0/1.6) or all splice forms of mPINC, including the DCR2 isoforms (siPINC) (Figure 6A, 6B). After transfection with siRNAs, the cells were treated with lactogenic hormones to induce differentiation. At 24 and 72 hours post-treatment, knockdown of mPINC1.0 and mPINC1.6 resulted in increased expression of Wap and Ltf mRNA (Figure 6C, 6D). Knockdown of all splice forms of mPINC resulted in increased Ltf levels, but only a slight increase of Wap expression at 72 hours post-hormone treatment. Unexpectedly, knockdown of mPINC1.0 and mPINC1.6 reduced Csn2 levels, while knockdown of all mPINC isoforms had no effect on Csn2. Due to the discrepancies in these differentiation markers, we looked at the effect of knockdown on the morphological assay of differentiation. Knockdown of mPINC1.0 and mPINC1.6 increased dome formation 3.5-fold in treated cells, while knockdown of all mPINC splice forms increased dome formation 2-fold (Figure 6E, 6F). Taken together, these data provide support for the role of mPINC in inhibiting lactogenic differentiation. Interestingly, knockdown of all isoforms of mPINC, including DCR2, slightly diminished the effect on differentiation. This suggests that the endogenously expressed DCR2, that lacks the most conserved regions, may act to reduce the inhibitory effects of mPINC1.0 and mPINC1.6 on alveolar differentiation.
Fig. 6. Knockdown of mPINC enhances differentiation of HC11 cells. 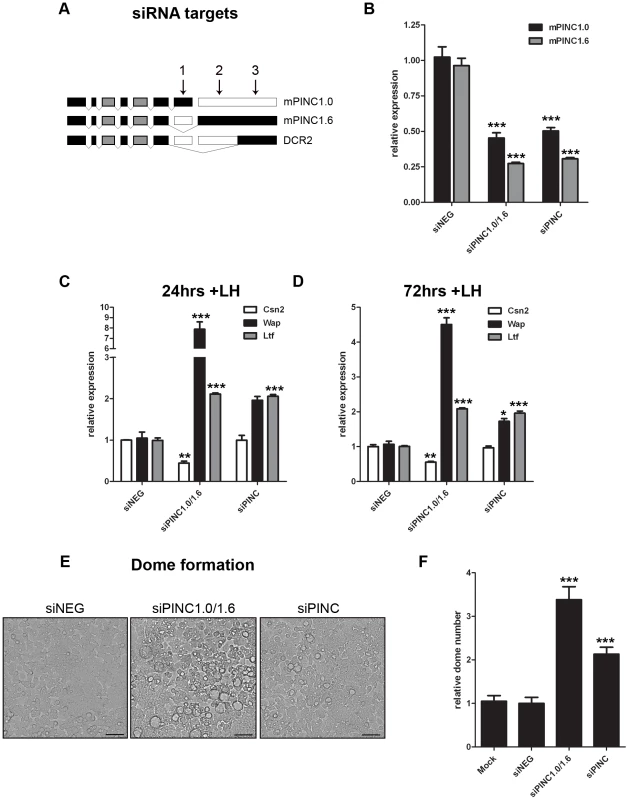
(A, B) To target mPINC1.0 and mPINC1.6 (siPINC1.0/1.6), but not DCR2, siRNAs #1 and #2 were used in combination. To target all splice variants (siPINC), siRNAs #1 and #3 were used in combination. (A) Schematic showing siRNA targets of mPINC splice forms. (B) qPCR shows knockdown of mPINC 5 days post-transfection of siRNAs. Target genes were normalized to Gapdh and set relative to levels in the siNEG control transfected HC11 cells. (C, D) Knockdown of mPNC (siPINC1.0/1.6 and siPINC) splice forms increases Wap and Ltf, but not Csn2, expression at 24 (C) and 72 (D) hrs post-hormone induction. Target genes were normalized to Gapdh and set relative to levels in siNEG treated control cells. Data are presented as mean ± SEM (n = 3). (E, F) Knockdown of mPINC also increases dome formation compared to a control. (E) Representative brightfield images show domes following 48 hrs of hormone treatment. Scale bars represent 50 µm. (F) Domes were counted at 48 hours post-hormone treatment from nine 20× fields/experiment and data represent mean ± SEM set relative to the dome number in the siNEG treated control group (n = 6). mPINC interacts with PRC2 in vitro
The previous experiments strongly suggest that mPINC plays a role in regulating alveolar differentiation. However, the mechanisms responsible are unknown. Many lncRNAs interact with chromatin-remodeling complexes such as PRC2. Interestingly, a member of PRC2, RbAp46, was identified in the same screen as mPINC, and both genes are expressed in the alveolar cells during pregnancy and in the regressed lobules of the post-involuted gland. Thus, to investigate a potential interaction with mPINC in HC11 cells, we performed RNA immunoprecipitation (RIP) assays using antibodies to members of PRC2, including enhancer of zeste homolog 2 (EZH2), suppressor of zeste 12 homolog (SUZ12) and RbAp46. RT-PCR using primers to detect mPINC showed that there was a specific interaction with all three PRC2 complex members, but not with mixed-lineage leukemia (MLL1), a member of the activating trithorax complex (Figure 7A). By qPCR, we found that both mPINC1.0 and mPINC1.6 splice forms interacted with PRC2 members (Figure 7C, 7D). Tug1, an lncRNA previously shown to interact with PRC2, was used as a positive control (Figure 7B). Together, these data indicate that mPINC interacts with PRC2 in HC11 cells, providing a potential mechanism of the inhibitory effect of mPINC on lactogenic differentiation.
Fig. 7. mPINC interacts with PRC2 in HC11 cells. 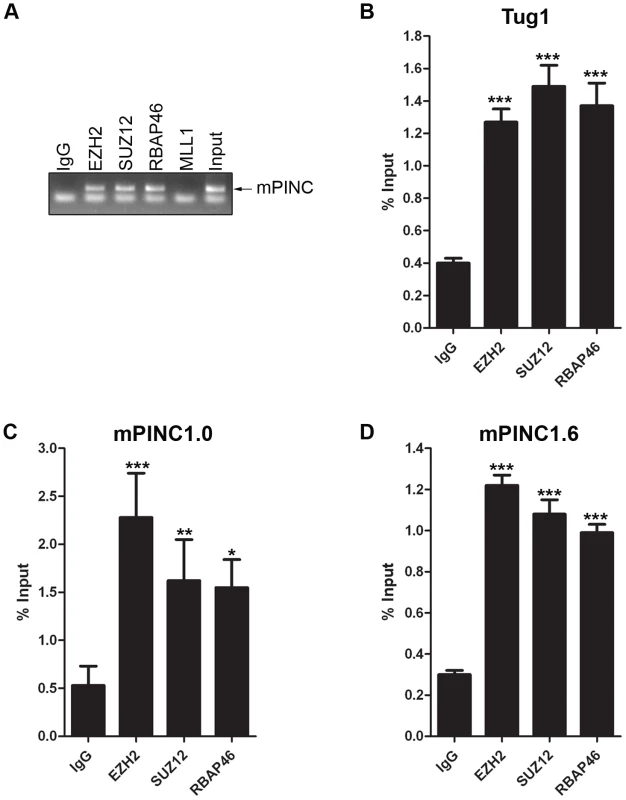
(A–D) RIP assays were performed with HC11 cells using antibodies to PRC2 members including, EZH2, SUZ12 and RbAp46. An MLL1 antibody was used as a negative control and an lncRNA known to interact with PRC2, Tug1, was used as a positive control. RNA was isolated from pull downs to detect associated RNAs. (A) RT-PCR shows mPINC is associated with PRC2 members, but not MLL1. (B–D) qPCR shows the amount of Tug1 (B), mPINC1.0 (C) and mPINC1.6 (D) transcript associated with each protein as a percentage of input RNA levels. Data represent mean ± SD (n = 3). Overexpressed mPINC and the DCR mutant interact with PRC2
To show that the overexpressed mPINC transcripts are functional, we sought to determine if they, like endogenous mPINC, interact with PRC2. Using a reverse primer to recognize a portion of the lentiviral vector that remains attached to the mPINC transcripts, we found that overexpressed mPINC1.0 and mPINC1.6 interacted with PRC2, suggesting that they are able to function like endogenous mPINC (Figure 8A). RT-PCR also showed that the DCR mutant retained the ability to interact with PRC2, providing evidence that the 5′ end of the RNA likely interacts with PRC2 and that the highly conserved regions at the 3′ end are not necessary for this interaction. qPCR analysis showed that the interaction of the overexpressed transcripts is specific to PRC2 (Figure 8B–8D). These data also suggest that the inability of the overexpressed DCR form of mPINC to inhibit lactogenic differentiation is not due to loss of binding to PRC2, but perhaps to the inability of the DCR mutant to interact with an additional unknown interacting protein or nucleic acid sequence target of mPINC.
Fig. 8. Overexpressed mPINC transcripts interact with PRC2 in HC11 cells. 
(A–D) RIP assays were performed with mPINC overexpressing HC11 cell using antibodies to PRC2 members including, EZH2, SUZ12 and RbAp46. An MLL1 antibody was used as a negative control. (A) RT-PCR shows that overexpressed mPINC1.0, mPINC1.6 and the DCR mutant interact with PRC2 members, but not IgG or MLL1. (B–D) qPCR shows the amount of mPINC1.0 (B), mPINC1.6 (C) and DCR (D) transcript associated with each protein as a percentage of input RNA levels. Data represent mean ± SD (n = 3). mPINC interacts with PRC2 in vivo
To determine whether mPINC also interacts with PRC2 in vivo, MECs were purified from mice at day 16 of pregnancy. RIP assays were performed using antibodies to PRC2 members and to the associated histone modification, H3meK27. As negative controls, antibodies to MLL1 and to the associated activating histone modification, H3meK4 were used. RT-PCR showed that mPINC is associated with PRC2 members and H3meK27, but mPINC did not interact with MLL1 or H3meK4 in MECs isolated from pregnant mice (Figure 9A). Further qPCR analysis showed that the interaction between mPINC and PRC2 members was enriched relative to the housekeeping gene, Gapdh, and that this enrichment was specific to PRC2 members and not to MLL1 (Figure 9B–9E). These results suggest that the interaction between mPINC and PRC2 is relevant in vivo, as well as in vitro, where it potentially plays a role in late pregnancy and involution to maintain alveolar progenitors by inhibiting terminal differentiation.
Fig. 9. mPINC associates with PRC2 in the 16-day pregnant mammary gland. 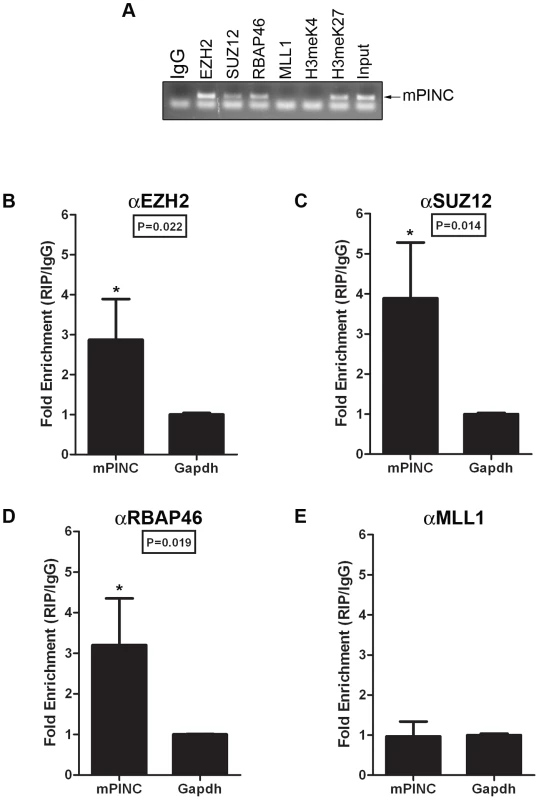
(A–D) RIP assays were performed with MECs purified from mammary glands at day 16 of pregnancy using antibodies to PRC2 members and its associated histone modification, H3meK27. Antibodies to MLL1 and its associated histone modification, H3meK4, were also used as negative controls. (A) RT-PCR shows mPINC is associated with PRC2 members and H3meK27. mPINC does not associate with MLL1 or H3meK4. (B–D) qPCR shows fold enrichment of mPINC transcript levels associated with EZH2 (B), SUZ12 (C) RpAp46 (D) and MLL1 (E) relative to Gapdh levels. Data represent mean ± SD (n = 3). Microarray analysis reveals potential targets of mPINC-PRC2
To identify potential targets of mPINC, we performed oligonucleotide microarray analysis on undifferentiated HC11 cells following mPINC knockdown (siPINC1.0/1.6 and siPINC). mPINC depletion resulted in the disrupted expression of 303 unique annotated genes (436 gene probes, shown in Table S1) (Figure 10A). Interestingly, loss of mPINC1.0 and mPINC1.6 (siPINC1.0/1.6) resulted in more gene expression changes than loss of all mPINC splice forms (siPINC). In addition, the array data identified unique genes whose expression was changed by either siPINC1.0/1.6 knockdown or siPINC knockdown alone, further indicating distinct functions for the mPINC splice forms. However, many of the genes altered by siPINC1.0/1.6 knockdown were also altered by siPINC knockdown, yet to a lesser extent. These results are in agreement with, and may help explain, the reduced effect of siPINC knockdown on lactogenic differentiation compared to siPINC1.0/1.6 knockdown. Gene ontology (GO) analysis of altered genes in the mPINC knockdown cells demonstrated GO terms were significantly enriched for developmental processes and differentiation (Figure 10B) (Table S2). GO terms more specifically related to mammary development, such as tube development and branching morphogenesis, were also enriched. These data indicate that mPINC generally regulates genes involved in development and differentiation.
Fig. 10. Microarray identifies potential targets of mPINC in HC11 cells. 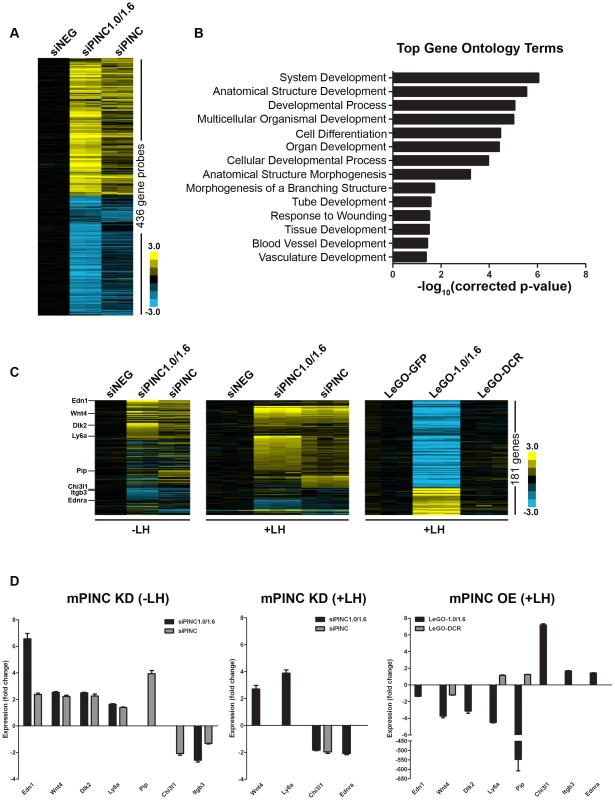
(A) Heat map showing transcript profiling of biological replicates of control siNEG cells, siPINC1.0/1.6, and siPINC 5 days post-siRNA transfection ( p<0.01 and fold change >1.8 in either siPINC1.0/1.6 or siPINC relative to siNEG). (B) Graph indicating the most significantly enriched gene ontology terms in the mPINC knockdown data set. (C) Three heat map panels depicting genes that are differentially expressed between either undifferentiated (−LH) mPINC knockdown cells (left panel) or differentiated (+LH) mPINC knockdown cells (middle panel) and differentiated (+LH) mPINC overexpression cells (right panel). 181 genes were found to be differentially regulated by mPINC (141 genes are upregulated by knockdown and downregulated by overexpression, while 40 genes are downregulated by knockdown and upregulated by overexpression at a p<0.01 and fold change >1.4, both for the overexpression and either knockdown group in the opposite direction). Genes whose expression was validated by qPCR are indicated in the heat map on the left. (D) qPCR verified differential expression of genes in mPINC knockdown (KD) compared to overexpression (OE) cells. qPCR was performed on biological triplicates, normalized using Gapdh, and shown as fold change compared to the negative control (either siNEG or LeGO-GFP). To better determine targets of mPINC that may be involved in regulating alveolar differentiation, we also performed microarray analysis on mPINC knockdown and mPINC overexpression cells treated with lactogenic hormones. As knockdown and overexpression of mPINC resulted in opposing phenotypes on differentiation, we looked for genes that were altered in opposite directions by knockdown and overexpression. For the knockdown analysis, we used the array data from undifferentiated (−LH) and differentiated (+LH) HC11 cells to reveal hormonally-regulated targets of mPINC. This analysis narrowed the list of potential mPINC targets to 181 genes (Figure 10C) (Table S3). Almost 80% of the identified genes (141 genes) were upregulated in the mPINC knockdown cells and downregulated in the overexpression cells, providing evidence that mPINC functions as part of a repressive PRC2 complex. This analysis also revealed several differentially expressed genes with known or suggested roles in mammary gland development and alveolar differentiation, including members of the Wnt and Notch signaling pathways. Genes that were upregulated by mPINC knockdown and downregulated by overexpression include endothelin 1 (Edn1), Wnt4, delta-like 2 (Dlk2), Ly6a (or Sca1) and prolactin-induced protein (Pip). Conversely, genes that were downregulated by mPINC knockdown and upregulated by overexpression include chitinase 3-like 1 (Chi3l1), integrin-beta 3 (Itgb3 or CD61), endothelin receptor a (Endra) and of course, mPINC itself. The differential expression of each of these genes was confirmed by qPCR (Figure 10D). The genes identified by microarray provide mechanistic insight into how mPINC-PRC2 may regulate alveolar differentiation as a repressive complex in the late pregnant and early involuting mammary gland.
mPINC splice forms have common, stable hairpin structures that may interact with PRC2
Although there is increasing evidence that many lncRNAs interact with PRC2 to direct their activity towards particular regions in the genome, the mechanism of this interaction remains a mystery. Recent studies identified double-stem loop structures in the RepA and Hes1-as lncRNAs that are thought to interact directly with the EZH2 subunit of PRC2 [20], but whether this will be a motif common to all PRC2-interacting lncRNAs is not yet known. To determine whether mPINC contained any structures that may interact with PRC2, we used the RNAstructure software to predict the secondary structure interactions of the mPINC isoforms mPINC1.0, mPINC1.6 and DRC2. Using SISSIz [44], we also looked for regions within the mPINC locus that showed significant evolutionary conservation of RNA secondary structure in all vertebrates (Figure 11A). As a result, we found two double hairpin regions that occurred in all three isoforms, indicated in the circular plots as structure 1 (S1) and structure 2 (S2) (Figure 11B–11D). The first is a 53 nt Y-shaped structure arising from exon 1, which showed a high level of evolutionary conservation (Figure 11E). This structure appears similar to that identified in RepA and we therefore propose that it may be a good candidate site for PRC2-interaction. The second structure is a 143 nt double hairpin that arises from exons 4 and 5 (Figure 11F). Although this region did not contain signatures of evolutionary conservation, the structure was predicted to fold with high probability. Given the high stability of the second site, we anticipate that it will also play an important role in the function of mPINC, possibly as an additional protein-interaction site. These data show that the 5′ region of mPINC, which is sufficient for interacting with PRC2, contains evolutionarily conserved stable hairpin structures that are reminiscent of double stem-loop structures of lncRNAs that have previously been shown to bind the EZH2 subunit of PRC2.
Fig. 11. Structural analysis of the PINC locus. 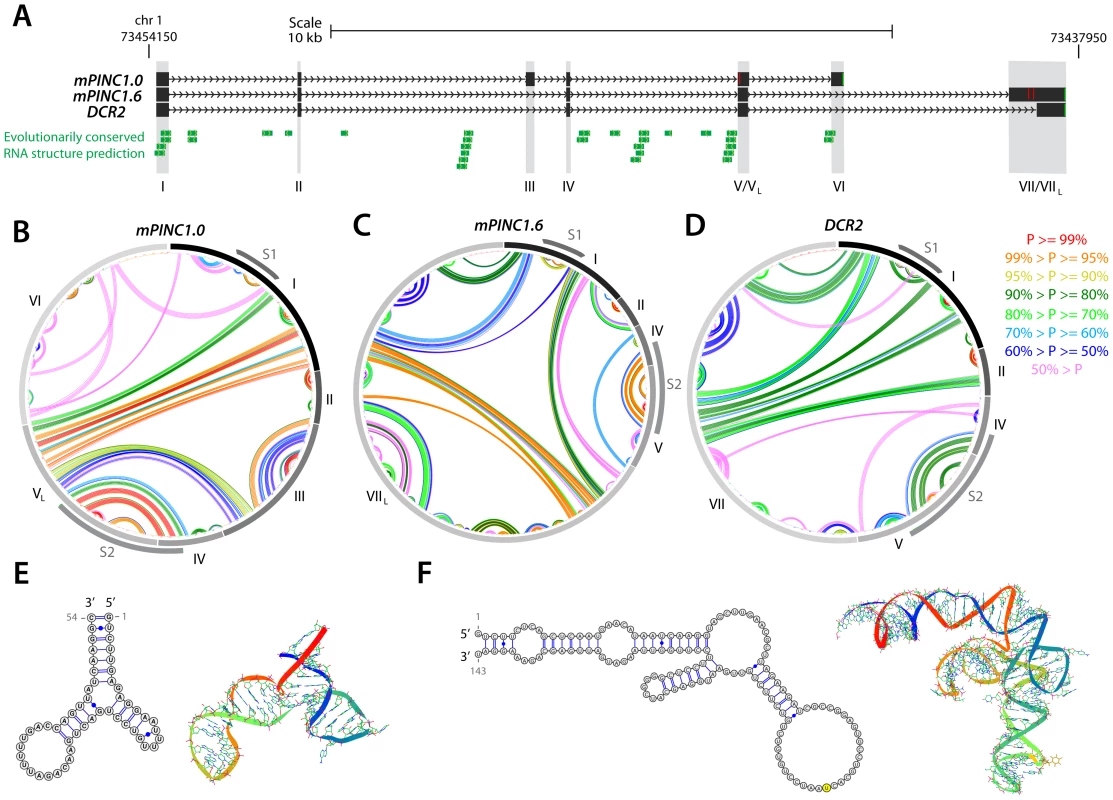
(A) Genomic representation of the PINC locus showing alternate isoforms mPINC1.0, mPINC1.6 and DCR2. Regions of evolutionarily conserved RNA structures were predicted by SISSIz (see Methods). (B–D) Circle plot secondary structure representations of interacting nucleotides in mPINC1.0 (B), mPINC1.6 (C) and DCR2 (D). The probability of the interactions is color-coded according to the legend (right). Secondary structures were computed using the RNAstructure software (see Methods). (E–F) High-confidence localized hairpin structures that represent possible protein-interaction sites and their most stable 3D representations, as modelled by MC-FOLD/MC-SYM. The corresponding regions in B–D are indicated by S1, for structure 1 (E) and by S2, for structure 2 (F) grey arcs. Discussion
Previously, we identified PINC and RbAp46, a member of the chromatin-modifying PRC2 complex, as genes that are elevated in alveolar cells of the pregnant and involuting rat mammary gland. In vitro studies of mPINC showed potential roles in regulating survival and cell-cycle progression of mammary epithelial cells, and it was further concluded that mPINC was a marker of alveolar cell fate. In this study, we extend our previous observations and demonstrate a functional role for mPINC in regulating alveolar development. First, we showed that mPINC is highly expressed in alveolar cells in the mouse mammary gland during pregnancy and downregulated in early lactation. Similarly, we found that mPINC levels decline upon lactogenic hormone-induced differentiation of HC11 cells. To investigate the functional significance of mPINC downregulation, we performed gain - and loss-of-function studies using HC11 differentiation assays. Overexpression of mPINC blocked lactogenic differentiation, while knockdown of mPINC resulted in enhanced markers of differentiation. As previous data showed that PINC and RbAp46 are co-regulated and co-localized in the rat mammary gland, we sought to determine a potential interaction in mammary epithelial cells. We provide evidence that mPINC is associated with RbAp46 and other PRC2 members in mammary epithelial cells both in vitro and in vivo. Finally, microarray analysis of mPINC knockdown and overexpression cells provided substantial insight into the putative gene networks modified by mPINC-PRC2. These results suggest that mPINC likely functions epigenetically as part of a chromatin-modifying PRC2 complex in the post-pubertal mammary gland to negatively regulate mammary alveolar differentiation.
As many lncRNAs show tissue-specific expression patterns and spatiotemporal regulation during development, it is not surprising that several lncRNAs regulate key developmental processes such as cell-fate commitment and differentiation. Large-scale analysis has revealed differentially expressed lncRNAs during mouse embryonic stem (ES) cell differentiation, thus identifying novel lncRNAs associated with either pluripotency or various stages of differentiation [45]. In addition, several lncRNAs regulate specific processes of development, such as germ layer specification [46], muscle cell differentiation [47], and terminal differentiation of erythroid cells [48] and skin cells [49]. Recently, a developmentally regulated lncRNA, Zfas1, was identified in the post-pubertal mouse mammary gland [50]. Zfas1 exhibits a 10-fold decrease between pregnancy and lactation and is downregulated in lactogenic hormone-induced differentiation of HC11 cells. Like mPINC, knockdown of Zfas1 results in enhanced lactogenic differentiation of HC11 cells. However, the molecular mechanisms that regulate the function of Zfas1 are unknown. In this study, we found that mPINC regulates mammary alveolar differentiation and we demonstrate, for the first time, the interaction of an lncRNA with PRC2 in mammary epithelial cells.
PRC2 has a well-known role in maintaining cellular identity by stably repressing different sets of genes during differentiation and development [51], [52]. Historically, PRC2 has been regarded as a critical regulator of gene expression during embryonic development [53]. However, mounting evidence suggest that PRC2 is also involved in maintaining homeostasis in adult tissues by regulating the balance between self-renewal and differentiation of specific progenitor populations [54]–[56]. For example, loss of the enzymatic subunit of PRC2, EZH2, in either cortical or epidermal progenitor cells results in accelerated differentiation, while overexpression of EZH2 in myoblasts inhibits muscle cell differentiation. Intriguingly, our data suggest an additional role for PRC2, in association with the lncRNA mPINC, in maintaining alveolar progenitors by repressing genes necessary for alveolar differentiation.
Precisely how PRC2 targets specific loci to control the fate of a particular cell is unknown. However, increasing evidence shows that lncRNAs may be, in part, responsible. The earliest and best-characterized example is the silencing of the X chromosome during dosage compensation by PRC2 recruitment, which was recently shown to be mediated in cis by a 1.6 kilobase lncRNA called RepA [22]. More recently, several more lncRNAs have been shown to be required for the targeting of PRC2 to specific loci in cis, such as Kcnq1ot1 silencing of the Kcnq1 imprinted locus and Anril repression of the p15INK4B locus [23], [24]. In this study, we showed that mPINC interacts with PRC2 in vitro and in vivo and microarray analysis of mammary epithelial cells following mPINC modulation revealed 181 potential target genes. Analysis of these target genes revealed only one gene proximal to the mPINC locus on chromosome 1, Igfbp2. Igfbp2 is located 400 kilobases downstream of mPINC and is differentially regulated during mammary development [57]. IGFBP2 levels decline throughout pregnancy and lactation with the most significant reduction occurring at day 2 of involution. Interestingly, mPINC levels are highest at day 2 of involution and may play a role in the repression of Igfbp2 at this time point. However, the function of Igfbp2 during involution, as well as during other stages of mammary development, is unknown.
LncRNAs also recruit PRC2 in trans as evidenced by the lncRNA HOTAIR, which is transcribed from the HOXC locus but is required to target PRC2 to the HOXD locus on a different chromosome [13]. mPINC may operate like HOTAIR and target genes in trans to regulate alveolar differentiation. Analysis of potential mPINC target genes showed that they are distributed throughout the genome, with the exception of Igfbp2, indicating that mPINC is most likely regulating genes in trans. The most upregulated gene in mPINC knockdown cells, Edn1, is an important cell fate determinant during embryogenesis [58], [59]. It is highly expressed in the mammary gland during lactation and is also induced in HC11 cells treated with lactogenic hormones [60]. The receptor for Edn1, Ednra, was downregulated by mPINC modulation, thus emphasizing the potential importance of this pathway. Another gene upregulated upon mPINC loss was Wnt4. Wnt4 is induced by progesterone during pregnancy and is required for proper lobuloalveolar development [61]. Wnt4 is also required for hormone-induced differentiation of HC11 cells [62]. The Notch signaling inhibitor Dlk2 [63] was also induced in the mPINC knockdown cells, while a positive regulator of Notch signaling, Mindbomb2 (Mib2) [64], was oppositely regulated. Active Notch signaling in mammary epithelial cells promotes luminal cell fate, while constitutive Notch signaling in the luminal cells during pregnancy inhibits alveolar differentiation, suggesting that repression of Notch activity is required for alveolar differentiation [65]. Chi3l1 (or Ykl40) was downregulated by mPINC knockdown and upregulated by overexpression. Treatment of mammary epithelial cells with recombinant YKL40, or ectopic expression of Ykl40 in mammary epithelial cells, inhibits lactogenic hormone-induced differentiation [66]. Chi3l1 (or Ykl40) is one of the most highly induced genes during involution and could function downstream of mPINC to maintain alveolar progenitors in the involuted gland. The identification of these potential mPINC-PRC2 target genes provides plausible mechanisms for the impact of mPINC on alveolar differentiation.
The microarray data also provided additional evidence for the differentiation status of the HC11 cells following mPINC knockdown and overexpression. There was some variability in the effect of mPINC knockdown on markers of lactogenic differentiation, as Csn2 levels were not increased similar to Ltf and Wap levels. The expression profiling, however, identified additional milk protein genes, as well as other lactation-induced genes, that were differentially expressed following mPINC knockdown and overexpression. These genes, which include Cp, Mal, Clu, Csn3, Pip and Cidea, were all induced by mPINC knockdown and decreased by overexpression [67], [68]. Additionally, overexpression of the mPINC DCR mutant not only failed to inhibit differentiation, but also showed enhanced expression of some markers of differentiation, suggesting the possibility that it acts in a dominant-negative manner. However, expression profiles of LeGO-DCR overexpressing cells clearly showed that they were very similar to the LeGO-GFP control cells, suggesting that the DCR mutant is not interfering with endogenous mPINC function. Thus, the microarray data provides further support for the hypothesis that mPINC is necessary for maintaining the undifferentiated state of mammary epithelial cells, and that the conserved regions are necessary for this function.
Lastly, the robust enrichment of mPINC in the luminal and alveolar progenitor populations further substantiates its role in maintaining cells in an undifferentiated state. Previous characterization of luminal subpopulations using CD61 and Sca-1 have shown that the luminal progenitors are CD61+ and Sca-1−, while the more differentiated, mature luminal cells are CD61− and Sca-1+ [38]. Gene expression profiling showed that depletion of mPINC results in a reduction in Itgb3 (or CD61) levels and an increase in Ly6a (or Sca1). In addition, mPINC knockdown also resulted in higher Wnt4 levels, another marker of mature luminal cells. These data suggest that by decreasing levels of mPINC, luminal progenitor cells are pushed toward a more differentiated state. Interestingly, a recent study identified a novel lncRNA called ANCR (anti-differentiation ncRNA) that is enriched in epidermal progenitors [49]. Like mPINC, loss of ANCR leads to induction of differentiation genes, suggesting ANCR is required to maintain epidermal progenitors in an undifferentiated state. Taken together, these studies provide additional evidence for the important role of lncRNAs in the maintenance of cellular homeostasis across multiple tissues.
Our current work used a well-characterized in vitro model for functional characterization of mPINC in alveologenesis. Future studies should employ loss - and gain-of-function mouse models to further investigate the role of mPINC in vivo during pregnancy and involution. We also provided evidence for an interaction between mPINC and PRC2 and identified putative stable secondary structures that may be required for this interaction. Mutations to disrupt these stable hairpin structures should be generated to determine the structure-function relationships necessary for PRC2 binding. Finally, we identified potential targets of mPINC-PRC2, several of which have known or suggested roles in regulating alveologenesis. It will be critical to verify that these loci are actually targeted by mPINC-PRC2 to provide insight into how this repressive complex regulates the stepwise differentiation of alveolar cells.
We propose a model where mPINC and RbAp46 are elevated during pregnancy and involution and interact to repress genes necessary for alveolar differentiation, thereby preserving a population of alveolar progenitors for subsequent pregnancies. PINC and RbAp46 were originally found in a screen to identify genes that are persistently altered in the parous mammary gland in an effort to determine the molecular mechanisms that govern the protective effect of an early pregnancy against tumorigenesis [35]. Future studies to better understand the epigenetic regulation of differentiation and development in the post-pubertal mammary gland will likely elucidate potential mechanisms that contribute to reduced tumorigenesis in the parous breast.
Materials and Methods
Animal care
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal research protocol was approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine (AN-504). All mice used in this study were maintained and euthanized under the strict guidelines of the Institutional Animal Care and Use Committee of Baylor College of Medicine. Balb/c mice were obtained from Harlan Laboratories. For mammary gland developmental time points, mating pairs were co-housed in the afternoon and females were checked for a vaginal plug the following morning. For involution time points, pups were force-weaned after 10 days of lactation.
Preparation of single mammary cell populations
The third, fourth, and fifth mammary fat pads were harvested from 8–10 week old virgin or 16 day pregnant Balb/c mice. The glands were subsequently weighed and minced into 1 mm×1 mm fragments using a McIlwain Tissue Chopper 800 series. Mammary organoids were derived by digesting the glands in DMEM/F12 containing 2 mg/ml Collagenase A (Roche) for 1 hr at 37°C with constant shaking at 150 rpm. Organoids were isolated from the digested preparation by differential centrifugation, consisting of a sequence of 4 to 5 brief centrifugation steps (5 sec pulses) at 1500 rpm to enrich for mammary organoids and remove contaminating stromal cells. The organoids were then resuspended in 0.25% Trypsin-EDTA for 5 min, washed, and filtered through a 0.40 µm cell strainer to obtain a single mammary epithelial cell preparation. Single cells were washed 2–3 times with HBSS prior to cell surface staining for flow cytometry or fixation for RNA immunoprecipitation.
Cell sorting
Mammary epithelial cells were resuspended at a concentration of 1×108 cells/ml in HBSS supplemented with 10 mM HEPES and 2% FBS. This cell suspension was depleted of lineage positive cells (CD45, Ter119, CD31, and BP-1) using the EasySep Mouse Epithelial Cell Enrichment Kit (Stem Cell Technologies). Mammary epithelial cells were subsequently resuspended at a density of 1×107 cells/ml and stained with anti-mouse CD24 PE (Stem Cell Technologies, 1∶100), anti-CD49f FITC (Stem Cell Technologies, 1∶100), anti-mouse CD24 APC (Biolegend, 1∶100), anti-mouse CD29 Pacific Blue (BioLegend, 1∶100), anti-mouse CD14 FITC (eBiosiences, 1∶80) and anti-mouse ckit PE (Clone ACK4, Cedarlane Laboratories, 1∶50). Cells were sorted using a BD FACS Aria II Cell Sorter.
RNA isolation
Mammary glands (#4) were excised from 10–12 week old Balb/c female mice and snap frozen. Three mice were sacrificed for each tissue type and mammary gland developmental time point. RNA was isolated using TRIzol (Invitrogen) and quantified using a NanoDrop (ThermoScientific). RNA was also isolated from MECs and cells lines using TRIzol. RNA integrity was analyzed on a 1% agarose gel prior to cDNA preparation. RNA generated from mature luminal, luminal progenitor, and alveolar progenitor fractions was amplified by the BCM Genomic and RNA Profiling Core using the NuGen WT-Ovation Kit (NuGen).
RT–PCR
For each sample, 1–3 ug of RNA was DNase treated (Invitrogen, DNase I), primed with either oligo dT or random primers and reverse transcribed using Superscript II (Invitrogen). Alternatively, RNA was DNAse treated using DNA-Free kit (Ambion) and reverse transcribed using the High Capacity RNA to cDNA kit (Applied Biosystems). To analyze splice variants of mPINC, primers designed to the first and last exons of both mPINC1.0 and mPINC1.6 were used to amplify cDNA from 12 day pregnant Balb/c mammary glands using Taq polymerase (Invitrogen). The PCR products were run on a 1% low melting point (LMP) agarose gel, excised and cloned into the TOPO TA pCR2.1 vector (Invitrogen) for sequencing. To identify the exonic structure of the new splice variants, sequences were aligned to the mPINC gene locus using UCSC Blat and Multalin.
Quantitative PCR
The ΔΔCT method was used to determine relative levels of gene expression. All primer sets were tested using a 10-fold dilution series containing 5 dilutions and primer efficiencies were measured to determine the validity of using the ΔΔCT method. For SYBR Green qPCR, mPINC primers were designed to detect each isoform specifically (mPINC1.0-SYBR-f-gctatgtgaaggaacactgcaaag, mPINC1.0-SYBR-r-tctcctcttggacagaatccactt, mPINC1.6-SYBR-f-gtgaaggaacactgcaaagagct, mPINC1.6-SYBR-r-ccccagagtgctccatgttt). Two housekeeping genes were used as normalizers, beta-actin and cyclophilin b (Actb F-gcaacgagcggttccg, Actb R-cccaagaaggaaggctgga, Ppib F-ttgccatccagccactcag, Ppib R-tgagcactggggagaaagg). Each biological replicate was run in triplicate using SYBR Green qPCR Master Mix (Applied Biosystems). The ΔΔCT method was used to obtain values of the relative levels of mPINC1.0 and mPINC1.6 by using both beta-actin and cyclophilin b as internal reference genes and setting all samples relative to the sample with the lowest amount of detectable target (Ct<35). Samples with Ct values greater than 35 were considered undetectable. For Taqman qPCR, the following probe sets were purchased from Applied Biosystems: mPINC1.0-Mm03456232_m1, mPINC1.6-Mm03456230_m1, mPINC-Mm03456228_m1, Gapdh-Mm99999915_g1, Csn2-Mm00839664_m1, Wap-Mm00839913_m1, Ltf-Mm00434787_m1, Edn1-Mm00438656_ml, Wnt4-Mm01194003_m1, Dlk2-Mm01281511_g1, Ly6a-Mm00726565_s1, Pip-Mm00476800_m1, Chi3l1-Mm00801477_m1, Itgb3-Mm00443980_m1, Ednra-Mm01243722_m1. Each biological replicate was run in triplicate using Taqman Gene Expression Master Mix (Applied Biosystems).
In situ hybridization
Mammary glands were removed from 12 day pregnant Balb/c female mice, fixed for 4 hrs in 4% PFA at 4°C then processed and embedded in paraffin. Antisense and Sense probes were PCR amplified from plasmids containing full length mPINC1.0 or mPINC1.6 and gel purified. The primers (mPINC1.0 antisense F-T7-tcctgcattaacccttcatca, mPINC1.0 antisense R-tggattctgtccaagaggaga and mPINC1.6 antisense F-T7-ttgctcacaatcatccctca, mPINC1.6 antisense R-cacttcctgctcaggtgtca) generated probes that were 167 bases for mPINC1.0 and 225 bases for mPINC1.6. Digoxigenin-labeled probes were in vitro transcribed by T7 polymerase using 500 ng of template and DIG-labeling mix (Roche). Following rehydration, sections were treated with Proteinase K (10 ug/ml), post-fixed in 4% PFA for 15 min, acetylated for 10 min and prehybridized for 1 hr in Amresco Hybe Solution (AMR-0973) at 55°C. Antisense and Sense control probes were incubated with sections overnight at 55°C diluted to 0.1 ng/ul. Excess probe was eliminated by treatment with RNase/T1 cocktail (Ambion) in 2× SSC for 30 min followed by stringent washes. Sections were then blocked in TBS with 3% sheep serum and 0.3% triton-X 100 for several hours at room temperature before adding an anti-Digoxigenin-Alkaline Phosphatase antibody (Roche, 1∶250) in block solution with 2% sheep serum overnight. The next day sections were washed and stained using BM Purple (Roche). After stopping the color reaction, sections were rinsed with water and mounted with Vectashield Mounting Medium (Vectorlabs).
HC11 cell culture
HC11 cells were grown in 6 well plates at 37°C in 5% CO2 in RPMI 1640 media (Gibco) supplemented with 10% bovine calf serum (SAFC Biosciences), 2 mM glutamine (Gibco), 5 ug/ml bovine insulin (Sigma), 10 ng/ml murine EGF (Millipore), and 50 ug/ml gentamycin (Sigma). Upon confluency, cells were grown for an additional 3 days and then incubated overnight in priming media (10% stripped donor horse serum (SAFC Biosciences), 2 mM glutamine, 5 ug/ml bovine insulin, 50 ug/ml gentamycin in RPMI 1640 media). Cells were differentiated in priming media with the addition of hormones (1 ug/ml Prolactin (National Hormone and Pituitary Program, NIDDK, National Institutes of Health, Bethesda) and 1 ug/ml Hydrocortisone (Sigma)) for either 24 hrs or 72 hrs before harvesting the cells for RNA isolation. Domes were counted following 2 days of hormone treatment from nine 20× images/experiment.
Plasmid construction
LeGO lentiviral vectors were kindly provided by Dr. Kristoffer Weber [69]. LeGO-iCer2 vector was cut with BsrGI and NotI to release the IRES-Cerulean, ends were filled with T4 polymerase and the cut vector was religated to create LeGO-IF. mPINC1.0, mPINC1.6 and the deleted conserved region (DCR) mutant were amplified from plasmids containing the full length sequences of mPINC1.0 and mPINC1.6 (GenBank #s DQ059755 and DQ059756) adding RE sites to clone into the remaining sites of the LeGO-IF MCS. All plasmids were sequence-verified prior to lentiviral production.
Lentiviral production and titering
Lentiviral vectors (LeGO-GFP, LeGO-1.0, LeGO-1.6 or LeGO-DCR) were co-transfected with packaging vectors (pCMV-VSVG, pMDLg/pRRE, pRSV-Rev) into 293-T cells using Fugene 6 (Roche). Viral supernatant was collected at 48 and 72 hrs post-transfection, pooled and filtered through 0.45 uM filters to remove cellular debris. Filtered viral supernatant was concentrated using the Beckman Coulter Optima ultracentrifuge (SW32Ti rotor) at 25,000 rpm for 1 hr and 45 min. Ultracentrifuged virus was resuspended in DMEM. LENTI-X Elisa kits (Clontech) were used to titer the virus using LeGO-GFP as a calibrator. The LeGO-GFP virus was titered first by methods previously described based on FACs analysis of GFP expression in transduced 293-T cells. HC11 cells were transduced with lentivirus at a MOI of 20. Cells were expanded following two passages, aliquoted and frozen for experiments.
Transfection of siRNAs
HC11 cells were transfected with siRNAs (10 nM/well) at 60–80% confluency in 6 well plates using Lipofectamine RNAiMAX reagent (Invitrogen). Silencer Select siRNAs (Ambion) were used to target mPINC1.0 (n273107), mPINC1.6 (n254312) and DCR2 (n254314). The controls included mock tranfection without siRNAs and a siNEG, Silencer Select Negative Control #1 (4390843).
RNA immunoprecipitation (RIP) assay
RNA immunoprecipitation (RIP) assays were carried out following published protocols with some modifications [13], [23]. Cells from 150 mm plate were harvested by trypsinization and re-suspended in 2 ml PBS, 2 ml nuclear isolation buffer (1.28 M sucrose; 40 mM Tris-HCl pH 7.5; 20 mM MgCl2; 4% Triton X-100) and 6 ml water on ice for 20 min. Nuclei were pelleted by centrifugation at 2,500 rpm for 15 min, resuspended in 1 ml of RIP buffer (150 mM KCl; 25 mM Tris pH 7.4; 5 mM EDTA; 0.5 mM DTT; 0.5% NP40; 25 µg/ml leupeptin; 1 mM benzamidine, 10 µg/ml trypsin inhibitor, 25 µg/ml aprotinin, 1 mM PMSF; 100 U/ml SUPERase-in (Ambion)) and mechanically sheared using 25 G needle with 6 strokes. Nuclear membrane and debris were pelleted by centrifugation at 14,000 rpm for 10 min. All lysates were pooled together and aliquoted at 107 cells/IP(about 500 µl of lysate). DNA was degraded with Turbo DNase (Ambion) for 20 min at 37°C followed by adding EDTA to a final concentration of 20 mM and insoluble material was removed by centrifugation for 10 min at 14,000 rpm. Supernatants were pre-cleared by adding 15 µl of rabbit IgG and 20 µl of magnetic A/G beads (Invitrogen) following incubation at 4°C for 2 hrs with gentle rotation. Antibody to EZH2, SUZ12, RbAp46 (Abcam) or MLL1 (Bethyl) were added to pre-cleared supernatants (5 µg each) together with 20 µl of A/G magnetic beads and incubated overnight at 4°C with gentle rotation. The beads were then washed 3 times with RIP buffer followed by one wash in PBS and resuspended in 1 ml of TRIzol-LS (Invitrogen). Co-precipitated RNAs were treated with DNase (Ambion) and reverse transcribed using RNA-to-cDNA kit (Applied Biosystems).
Gene expression profiling
Microarray analysis was performed on biological duplicates for undifferentiated mPINC knockdown HC11 cells and on biological triplicates for differentiated mPINC knockdown and overexpression HC11 cells. RNA was isolated using Trizol, followed by purification using RNeasey MinElute Cleanup Kit (QIAGEN). The BCM Genomic and RNA Profiling Core conducted sample quality checks using the Nanodrop ND-1000 and Agilent Bioanalyzer Nano chip. 50 ng of total RNA was amplified and Cy3-labeled using the Agilent Quick Amp Labeling Kit (for one-color) Protocol Version 6.5. Hybridization to Agilent Sure Print 3 Mouse GE 8×60K Microarrays was performed at the BCM Genomic and RNA Profiling Core. Array data were quantile normalized, after which significantly regulated genes were identified by comparing control with groups using t-test (log-transformed data, two-sided) and fold change (ratio of averages of the two groups). Java TreeView [70] represented expression patterns as color maps, where expression values were centered on the average of the control group. Microarray data are available on the Gene Expression Omnibus (GSE38052). Gene ontology (GO) analysis was carried out using DAVID [71], [72].
RNA structure prediction
The spliced, full length transcripts of interest were submitted to RNA secondary structure analysis via RNAstructure version 5.3 [73]. Folding was performed using the partition function algorithm, which enables the prediction of putative RNA pseudoknots with ProbKnot [74]. We restricted the size of helical regions in the final display to ≥4 base pairs and exported circle plots annotated with base-pair probabilities from the partition function. The 3D modeling of common, highly probable secondary structure interactions was performed with the MC-FOLD/MC-SYM pipeline [75]. MC-FOLD predictions were restricted to the topologies predicted by RNAstructure, while the resulting non-canonical secondary structure with the highest score was submitted to MC-SYM. Evolutionary conservation of RNA structure was performed using SISSIz in a sliding-window framework [44]. The 46-way Multiz alignments associated to the PINC locus were downloaded from the UCSC genome browser (hg19). Alignment blocks were then merged together with an ad-hoc perl script, realigned with Clustalw2 [76], broken into windows of 200 columns that overlap by 50, and removed identical sequence and sequences with more than 75% gaps. We qualify hits presenting a Z-score below −3 (P-value≤1.35×10E-3) as bearing evolutionary conserved structures.
Statistics
One-way ANOVA, followed by a Tukey post-test, was performed on all experiments involving 3 or more groups and the graphs in each figure show significance using asterisks to denote the p-values (***p<0.001, **p<0.01, *p<0.05). For experiments with only 2 groups, unpaired Student t-tests were performed.
Supporting Information
Zdroje
1. WapinskiOChangHY 2011 Long noncoding RNAs and human disease. Trends Cell Biol 21 354 361
2. HuarteMRinnJL 2010 Large non-coding RNAs: missing links in cancer? Hum Mol Genet 19 R152 161
3. KhalilAMRinnJL 2011 RNA-protein interactions in human health and disease. Semin Cell Dev Biol 22 359 365
4. PauliARinnJLSchierAF 2011 Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12 136 149
5. PrasanthKVSpectorDL 2007 Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev 21 11 42
6. AmaralPPMattickJS 2008 Noncoding RNA in development. Mamm Genome 19 454 492
7. KatayamaSTomaruYKasukawaTWakiKNakanishiM 2005 Antisense transcription in the mammalian transcriptome. Science 309 1564 1566
8. CarninciPKasukawaTKatayamaSGoughJFrithMC 2005 The transcriptional landscape of the mammalian genome. Science 309 1559 1563
9. BertonePStolcVRoyceTERozowskyJSUrbanAE 2004 Global identification of human transcribed sequences with genome tiling arrays. Science 306 2242 2246
10. ConsortiumEPBirneyEStamatoyannopoulosJADuttaAGuigoR 2007 Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447 799 816
11. ChengJKapranovPDrenkowJDikeSBrubakerS 2005 Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308 1149 1154
12. RavasiTSuzukiHPangKCKatayamaSFurunoM 2006 Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16 11 19
13. RinnJLKerteszMWangJKSquazzoSLXuX 2007 Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129 1311 1323
14. MercerTRDingerMESunkinSMMehlerMFMattickJS 2008 Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A 105 716 721
15. TripathiVEllisJDShenZSongDYPanQ 2010 The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39 925 938
16. FengJBiCClarkBSMadyRShahP 2006 The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20 1470 1484
17. WillinghamATOrthAPBatalovSPetersECWenBG 2005 A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309 1570 1573
18. ClemsonCMHutchinsonJNSaraSAEnsmingerAWFoxAH 2009 An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33 717 726
19. KhalilAMGuttmanMHuarteMGarberMRajA 2009 Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106 11667 11672
20. ZhaoJOhsumiTKKungJTOgawaYGrauDJ 2010 Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40 939 953
21. MargueronRReinbergD 2011 The Polycomb complex PRC2 and its mark in life. Nature 469 343 349
22. ZhaoJSunBKErwinJASongJJLeeJT 2008 Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322 750 756
23. KotakeYNakagawaTKitagawaKSuzukiSLiuN 2011 Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30 1956 1962
24. PandeyRRMondalTMohammadFEnrothSRedrupL 2008 Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32 232 246
25. VisvaderJE 2009 Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev 23 2563 2577
26. WatsonCJKhaledWT 2008 Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development 135 995 1003
27. HennighausenLRobinsonGW 2005 Information networks in the mammary gland. Nat Rev Mol Cell Biol 6 715 725
28. GjorevskiNNelsonCM 2011 Integrated morphodynamic signalling of the mammary gland. Nat Rev Mol Cell Biol 12 581 593
29. WagnerKUBoulangerCAHenryMDSgagiasMHennighausenL 2002 An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development 129 1377 1386
30. AndersonSMRudolphMCMcManamanJLNevilleMC 2007 Key stages in mammary gland development. Secretory activation in the mammary gland: it's not just about milk protein synthesis! Breast Cancer Res 9 204
31. OakesSRHiltonHNOrmandyCJ 2006 The alveolar switch: coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res 8 207
32. BriskenCRajaramRD 2006 Alveolar and lactogenic differentiation. J Mammary Gland Biol Neoplasia 11 239 248
33. MaruyamaRChoudhurySKowalczykABessarabovaMBeresford-SmithB 2011 Epigenetic regulation of cell type-specific expression patterns in the human mammary epithelium. PLoS Genet 7 e1001369 doi:10.1371/journal.pgen.1001369
34. KhavariDASenGLRinnJL 2010 DNA methylation and epigenetic control of cellular differentiation. Cell Cycle 9 3880 3883
35. GingerMRGonzalez-RimbauMFGayJPRosenJM 2001 Persistent changes in gene expression induced by estrogen and progesterone in the rat mammary gland. Mol Endocrinol 15 1993 2009
36. GingerMRShoreANContrerasARijnkelsMMillerJ 2006 A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci U S A 103 5781 5786
37. Asselin-LabatMLSutherlandKDVaillantFGyorkiDEWuD 2011 Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor caspase-14. Mol Cell Biol 31 4609 4622
38. Asselin-LabatMLSutherlandKDBarkerHThomasRShackletonM 2007 Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9 201 209
39. SleemanKEKendrickHRobertsonDIsackeCMAshworthA 2007 Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol 176 19 26
40. OakesSRNaylorMJAsselin-LabatMLBlazekKDGardiner-GardenM 2008 The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev 22 581 586
41. LimEWuDPalBBourasTAsselin-LabatML 2010 Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res 12 R21
42. BallRKFriisRRSchoenenbergerCADopplerWGronerB 1988 Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J 7 2089 2095
43. DanielsonKGObornCJDurbanEMButelJSMedinaD 1984 Epithelial mouse mammary cell line exhibiting normal morphogenesis in vivo and functional differentiation in vitro. Proc Natl Acad Sci U S A 81 3756 3760
44. GesellTWashietlS 2008 Dinucleotide controlled null models for comparative RNA gene prediction. BMC Bioinformatics 9 248
45. DingerMEAmaralPPMercerTRPangKCBruceSJ 2008 Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18 1433 1445
46. BertaniSSauerSBolotinESauerF 2011 The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell 43 1040 1046
47. CesanaMCacchiarelliDLegniniISantiniTSthandierO 2011 A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147 358 369
48. HuWYuanBFlygareJLodishHF 2011 Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 25 2573 2578
49. KretzMWebsterDEFlockhartRJLeeCSZehnderA 2012 Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 26 338 343
50. Askarian-AmiriMECrawfordJFrenchJDSmartCESmithMA 2011 SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 17 878 891
51. BoyerLAPlathKZeitlingerJBrambrinkTMedeirosLA 2006 Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441 349 353
52. BrackenAPDietrichNPasiniDHansenKHHelinK 2006 Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20 1123 1136
53. RingroseLParoR 2004 Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38 413 443
54. CarettiGDi PadovaMMicalesBLyonsGESartorelliV 2004 The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18 2627 2638
55. PereiraJDSansomSNSmithJDobeneckerMWTarakhovskyA 2010 Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A 107 15957 15962
56. EzhkovaEPasolliHAParkerJSStokesNSuIH 2009 Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136 1122 1135
57. AllarMAWoodTL 2004 Expression of the insulin-like growth factor binding proteins during postnatal development of the murine mammary gland. Endocrinology 145 2467 2477
58. AlexanderCZunigaEBlitzILWadaNLe PabicP 2011 Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development 138 5135 5146
59. GadeaAAguirreAHaydarTFGalloV 2009 Endothelin-1 regulates oligodendrocyte development. J Neurosci 29 10047 10062
60. KozakaiTSakateMMasuoYUchideTSaidaK 2002 Increased gene expression of endothelin-1 and vasoactive intestinal contractor/endothelin-2 in the mammary gland of lactating mice. Biochem Biophys Res Commun 297 1339 1343
61. BriskenCHeinemanAChavarriaTElenbaasBTanJ 2000 Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev 14 650 654
62. ZhangHSinghRRTalukderAHKumarR 2006 Metastatic tumor antigen 3 is a direct corepressor of the Wnt4 pathway. Genes Dev 20 2943 2948
63. Sanchez-SolanaBNuedaMLRuviraMDRuiz-HidalgoMJMonsalveEM 2011 The EGF-like proteins DLK1 and DLK2 function as inhibitory non-canonical ligands of NOTCH1 receptor that modulate each other's activities. Biochim Biophys Acta 1813 1153 1164
64. KooBKYoonKJYooKWLimHSSongR 2005 Mind bomb-2 is an E3 ligase for Notch ligand. J Biol Chem 280 22335 22342
65. BourasTPalBVaillantFHarburgGAsselin-LabatML 2008 Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 3 429 441
66. ScullySYanWBentleyBCaoQJShaoR 2011 Inhibitory activity of YKL-40 in mammary epithelial cell differentiation and polarization induced by lactogenic hormones: a role in mammary tissue involution. PLoS ONE 6 e25819 doi:10.1371/journal.pone.0025819
67. WangWLvNZhangSShuiGQianH 2012 Cidea is an essential transcriptional coactivator regulating mammary gland secretion of milk lipids. Nat Med 18 235 243
68. WangWJoseCKenneyNMorrisonBCutlerML 2009 Global expression profiling reveals regulation of CTGF/CCN2 during lactogenic differentiation. J Cell Commun Signal 3 43 55
69. WeberKMockUPetrowitzBBartschUFehseB 2010 Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis. Gene Ther 17 511 520
70. SaldanhaAJ 2004 Java Treeview–extensible visualization of microarray data. Bioinformatics 20 3246 3248
71. Huang daWShermanBTLempickiRA 2009 Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 44 57
72. Huang daWShermanBTLempickiRA 2009 Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37 1 13
73. ReuterJSMathewsDH 2010 RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11 129
74. BellaousovSMathewsDH 2010 ProbKnot: fast prediction of RNA secondary structure including pseudoknots. RNA 16 1870 1880
75. ParisienMMajorF 2008 The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452 51 55
76. LarkinMABlackshieldsGBrownNPChennaRMcGettiganPA 2007 Clustal W and Clustal X version 2.0. Bioinformatics 23 2947 2948
Štítky
Genetika Reprodukční medicína
Článek Allelic Heterogeneity and Trade-Off Shape Natural Variation for Response to Soil MicronutrientČlánek The Chicken Frizzle Feather Is Due to an α-Keratin () Mutation That Causes a Defective RachisČlánek A Trans-Species Missense SNP in Is Associated with Sex Determination in the Tiger Pufferfish, (Fugu)
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 7- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
-
Všechny články tohoto čísla
- Functional Evolution of Mammalian Odorant Receptors
- Oocyte Family Trees: Old Branches or New Stems?
- Allelic Heterogeneity and Trade-Off Shape Natural Variation for Response to Soil Micronutrient
- Guidelines for Genome-Wide Association Studies
- GWAS Identifies Novel Susceptibility Loci on 6p21.32 and 21q21.3 for Hepatocellular Carcinoma in Chronic Hepatitis B Virus Carriers
- DNA Methyltransferases Are Required to Induce Heterochromatic Re-Replication in Arabidopsis
- Genomic Data Reveal a Complex Making of Humans
- Let-7b/c Enhance the Stability of a Tissue-Specific mRNA during Mammalian Organogenesis as Part of a Feedback Loop Involving KSRP
- The Secreted Immunoglobulin Domain Proteins ZIG-5 and ZIG-8 Cooperate with L1CAM/SAX-7 to Maintain Nervous System Integrity
- RsfA (YbeB) Proteins Are Conserved Ribosomal Silencing Factors
- Gene Conversion Occurs within the Mating-Type Locus of during Sexual Reproduction
- The Chicken Frizzle Feather Is Due to an α-Keratin () Mutation That Causes a Defective Rachis
- Meta-Analysis of Genome-Wide Scans for Total Body BMD in Children and Adults Reveals Allelic Heterogeneity and Age-Specific Effects at the Locus
- Balancing Selection at the Tomato Guardee Gene Family Maintains Variation in Strength of Pathogen Defense
- Large-Scale Introgression Shapes the Evolution of the Mating-Type Chromosomes of the Filamentous Ascomycete
- OSD1 Promotes Meiotic Progression via APC/C Inhibition and Forms a Regulatory Network with TDM and CYCA1;2/TAM
- Intact p53-Dependent Responses in miR-34–Deficient Mice
- FANCJ/BACH1 Acetylation at Lysine 1249 Regulates the DNA Damage Response
- CED-10/Rac1 Regulates Endocytic Recycling through the RAB-5 GAP TBC-2
- Histone H2A Mono-Ubiquitination Is a Crucial Step to Mediate PRC1-Dependent Repression of Developmental Genes to Maintain ES Cell Identity
- F-Box Protein Specificity for G1 Cyclins Is Dictated by Subcellular Localization
- The Gene Encodes a Nuclear Protein That Affects Alternative Splicing
- A Key Role for Chd1 in Histone H3 Dynamics at the 3′ Ends of Long Genes in Yeast
- Genome-Wide Association Analysis in Asthma Subjects Identifies as a Novel Bronchodilator Response Gene
- GRHL3/GET1 and Trithorax Group Members Collaborate to Activate the Epidermal Progenitor Differentiation Program
- Brain-Specific Rescue of Reveals System-Driven Transcriptional Rhythms in Peripheral Tissue
- Recent Loss of Self-Incompatibility by Degradation of the Male Component in Allotetraploid
- Pregnancy-Induced Noncoding RNA () Associates with Polycomb Repressive Complex 2 and Regulates Mammary Epithelial Differentiation
- The HEI10 Is a New ZMM Protein Related to Zip3
- The SCF Ubiquitin E3 Ligase Ubiquitylates Sir4 and Functions in Transcriptional Silencing
- Induction of Cytoprotective Pathways Is Central to the Extension of Lifespan Conferred by Multiple Longevity Pathways
- Role of Architecture in the Function and Specificity of Two Notch-Regulated Transcriptional Enhancer Modules
- Loss of ATRX, Genome Instability, and an Altered DNA Damage Response Are Hallmarks of the Alternative Lengthening of Telomeres Pathway
- A Regulatory Loop Involving PAX6, MITF, and WNT Signaling Controls Retinal Pigment Epithelium Development
- The Three Faces of Riboviral Spontaneous Mutation: Spectrum, Mode of Genome Replication, and Mutation Rate
- Unmet Expectations: miR-34 Plays No Role in p53-Mediated Tumor Suppression In Vivo
- A Genome-Wide Association Meta-Analysis of Circulating Sex Hormone–Binding Globulin Reveals Multiple Loci Implicated in Sex Steroid Hormone Regulation
- The Role of Rice HEI10 in the Formation of Meiotic Crossovers
- A Trans-Species Missense SNP in Is Associated with Sex Determination in the Tiger Pufferfish, (Fugu)
- Influences Bone Mineral Density, Cortical Bone Thickness, Bone Strength, and Osteoporotic Fracture Risk
- Evidence of Inbreeding Depression on Human Height
- Comparative Genomics of Plant-Associated spp.: Insights into Diversity and Inheritance of Traits Involved in Multitrophic Interactions
- Detecting Individual Sites Subject to Episodic Diversifying Selection
- Regulates Rhodopsin-1 Metabolism and Is Required for Photoreceptor Neuron Survival
- Identification of Chromatin-Associated Regulators of MSL Complex Targeting in Dosage Compensation
- Three Dopamine Pathways Induce Aversive Odor Memories with Different Stability
- TDP-1/TDP-43 Regulates Stress Signaling and Age-Dependent Proteotoxicity in
- Rapid Turnover of Long Noncoding RNAs and the Evolution of Gene Expression
- The Yeast Rab GTPase Ypt1 Modulates Unfolded Protein Response Dynamics by Regulating the Stability of RNA
- Histone H2B Monoubiquitination Facilitates the Rapid Modulation of Gene Expression during Arabidopsis Photomorphogenesis
- Cellular Variability of RpoS Expression Underlies Subpopulation Activation of an Integrative and Conjugative Element
- Genetic Variants in , , and Influence Male Recombination in Cattle
- Differential Impact of the HEN1 Homolog HENN-1 on 21U and 26G RNAs in the Germline of
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Guidelines for Genome-Wide Association Studies
- The Role of Rice HEI10 in the Formation of Meiotic Crossovers
- Identification of Chromatin-Associated Regulators of MSL Complex Targeting in Dosage Compensation
- GWAS Identifies Novel Susceptibility Loci on 6p21.32 and 21q21.3 for Hepatocellular Carcinoma in Chronic Hepatitis B Virus Carriers
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



