-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genome-Wide Association Analysis in Asthma Subjects Identifies as a Novel Bronchodilator Response Gene
Bronchodilator response (BDR) is an important asthma phenotype that measures reversibility of airway obstruction by comparing lung function (i.e. FEV1) before and after the administration of a short-acting β2-agonist, the most common rescue medications used for the treatment of asthma. BDR also serves as a test of β2-agonist efficacy. BDR is a complex trait that is partly under genetic control. A genome-wide association study (GWAS) of BDR, quantified as percent change in baseline FEV1 after administration of a β2-agonist, was performed with 1,644 non-Hispanic white asthmatic subjects from six drug clinical trials: CAMP, LOCCS, LODO, a medication trial conducted by Sepracor, CARE, and ACRN. Data for 469,884 single-nucleotide polymorphisms (SNPs) were used to measure the association of SNPs with BDR using a linear regression model, while adjusting for age, sex, and height. Replication of primary P-values was attempted in 501 white subjects from SARP and 550 white subjects from DAG. Experimental evidence supporting the top gene was obtained via siRNA knockdown and Western blotting analyses. The lowest overall combined P-value was 9.7E-07 for SNP rs295137, near the SPATS2L gene. Among subjects in the primary analysis, those with rs295137 TT genotype had a median BDR of 16.0 (IQR = [6.2, 32.4]), while those with CC or TC genotypes had a median BDR of 10.9 (IQR = [5.0, 22.2]). SPATS2L mRNA knockdown resulted in increased β2-adrenergic receptor levels. Our results suggest that SPATS2L may be an important regulator of β2-adrenergic receptor down-regulation and that there is promise in gaining a better understanding of the biological mechanisms of differential response to β2-agonists through GWAS.
Published in the journal: . PLoS Genet 8(7): e32767. doi:10.1371/journal.pgen.1002824
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002824Summary
Bronchodilator response (BDR) is an important asthma phenotype that measures reversibility of airway obstruction by comparing lung function (i.e. FEV1) before and after the administration of a short-acting β2-agonist, the most common rescue medications used for the treatment of asthma. BDR also serves as a test of β2-agonist efficacy. BDR is a complex trait that is partly under genetic control. A genome-wide association study (GWAS) of BDR, quantified as percent change in baseline FEV1 after administration of a β2-agonist, was performed with 1,644 non-Hispanic white asthmatic subjects from six drug clinical trials: CAMP, LOCCS, LODO, a medication trial conducted by Sepracor, CARE, and ACRN. Data for 469,884 single-nucleotide polymorphisms (SNPs) were used to measure the association of SNPs with BDR using a linear regression model, while adjusting for age, sex, and height. Replication of primary P-values was attempted in 501 white subjects from SARP and 550 white subjects from DAG. Experimental evidence supporting the top gene was obtained via siRNA knockdown and Western blotting analyses. The lowest overall combined P-value was 9.7E-07 for SNP rs295137, near the SPATS2L gene. Among subjects in the primary analysis, those with rs295137 TT genotype had a median BDR of 16.0 (IQR = [6.2, 32.4]), while those with CC or TC genotypes had a median BDR of 10.9 (IQR = [5.0, 22.2]). SPATS2L mRNA knockdown resulted in increased β2-adrenergic receptor levels. Our results suggest that SPATS2L may be an important regulator of β2-adrenergic receptor down-regulation and that there is promise in gaining a better understanding of the biological mechanisms of differential response to β2-agonists through GWAS.
Introduction
Asthma is a chronic respiratory disease that affects over 20 million Americans and 300 million people worldwide [1], [2]. A hallmark characteristic of asthma is reversible airway obstruction, which is commonly measured via a bronchodilator response (BDR) test, in which the reduction of bronchoconstriction after administration of a short-acting reliever drug is quantified [3]. β2-agonists, the most common short-acting reliever drugs used during BDR tests and for asthma therapy, act in part by stimulating β2-adrenergic receptors (β2ARs) on airway smooth muscle cells to reduce bronchoconstriction via subsequent increases in cyclic adenosine monophosphate (cAMP) and protein kinase A (PKA) [3]. Although a comprehensive pathophysiologic understanding of BDR has not been obtained, it is a complex trait involving interactions among various tissues and cells, including inflammatory [4], airway epithelium [5], smooth muscle [6], and the autonomic nervous system [7]. In addition to being used for the diagnosis of asthma, BDR tests can be used to measure whether inhaled β2-agonists are effective in patients. Although short-acting β2-agonists are widely used clinically as asthma rescue medications, they are variably efficacious among patients [8]. Studying BDR may thus provide information regarding both the pathophysiology and pharmacogenetics of asthma.
The search for genetic variants that modify asthma susceptibility has resulted in the most recent multi-center asthma genome-wide association studies (GWAS) providing strong statistical evidence for the association of many genes, including the IKZF3-ZPBP2-GSDMB-ORMDL3 locus, HLA-DQ, IL1RL1, IL18RL1, IL33, TSLP, SLC22A5, SMAD3, and RORA, with asthma [9], [10]. Functional experiments to identify the role that these genes play in asthma pathophysiology are hindered by the complexity of the asthma phenotype. Familial aggregation [11] and genetic association studies [12] have provided suggestive evidence for a genetic contribution to interindividual differences in BDR. Candidate genes reported to be associated with BDR include β2-adrenergic receptor (ADRB2) [13], [14], adenylyl cyclase type 9 (ADCY9) [15], corticotrophin-releasing hormone receptor 2 (CRHR2) [16], and arginase 1 (ARG1) [17], [18]. While BDR is a complex phenotype, functional studies of BDR candidate genes are simpler than those for a general asthma phenotype because this pharmacogenetic phenotype can be readily simulated in vitro via stimulation of cells with β2-agonists.
In this study, we performed a GWAS of BDR in 1,644 non-Hispanic white asthmatics and found that the strongest evidence of association with BDR was at variants near the Homo sapiens spermatogenesis associated, serine-rich 2-like (SPATS2L) gene. We attempted to replicate the primary findings in two independent populations and investigated the function of SPATS2L via mRNA knockdown experiments and found evidence to support its involvement in BDR.
Results
Primary BDR GWAS
Figure 1 is an overview of our study design. Characteristics of the subjects used in the primary GWAS are provided in Table 1. We utilized 1,644 non-Hispanic white subjects from six clinical trials to measure the association of SNPs to BDR. After QC filters, 469,884 SNPs genotyped in CAMP/LOCCS/LODO/Sepracor and either genotyped or imputed in CARE and ACRN were used to test for the association of SNPs to BDR. We utilized genotyped SNPs for CAMP/LOCCS/LODO/Sepracor because these cohorts, who were all genotyped using Illumina platforms, had the largest sample size. Due to the poor overlap of Illumina and Affymetrix platform SNPs, we utilized HapMap Phase 2 imputed SNPs for CARE and ACRN, so that the maximal number of SNPs in all cohorts could be analyzed. The quantile-quantile (QQ) and Manhattan plots revealed that the distribution of association P-values was similar to that expected for a null distribution and that no P-values met genome-wide statistically significant levels (Figures S1 and S2). To expand the primary association results further, all SNPs available in the June 2010 release of the 1000 Genome Project (1000GP) data were imputed using MaCH in each of the three primary groups of genotype data and overall BDR GWAS results were re-computed. Among SNPs contained in the primary GWAS, imputed and genotyped P-values were similar, particularly for those with low P-values (Figure S3). Some imputed regions had P-values lower than those of the primary GWAS, but the results in most of these regions were not supported by primary GWAS data (Figure S2). Thus, we proceeded to attempt to validate the primary GWAS findings based on the combined genotyped CAMP/LOCCS/LODO/Sepracor SNP results and HapMap Phase 2 imputed CARE and ACRN SNP results. The top five primary GWAS SNPs with P-value<1E-05 are in Table 2. Further details on these regions and all primary GWAS SNPs with P-value<1E-04 are in Tables S1 and S2. Further details on all 1000GP imputed GWAS SNPs with P-value<1E-05 are in Tables S3
Fig. 1. Study overview. 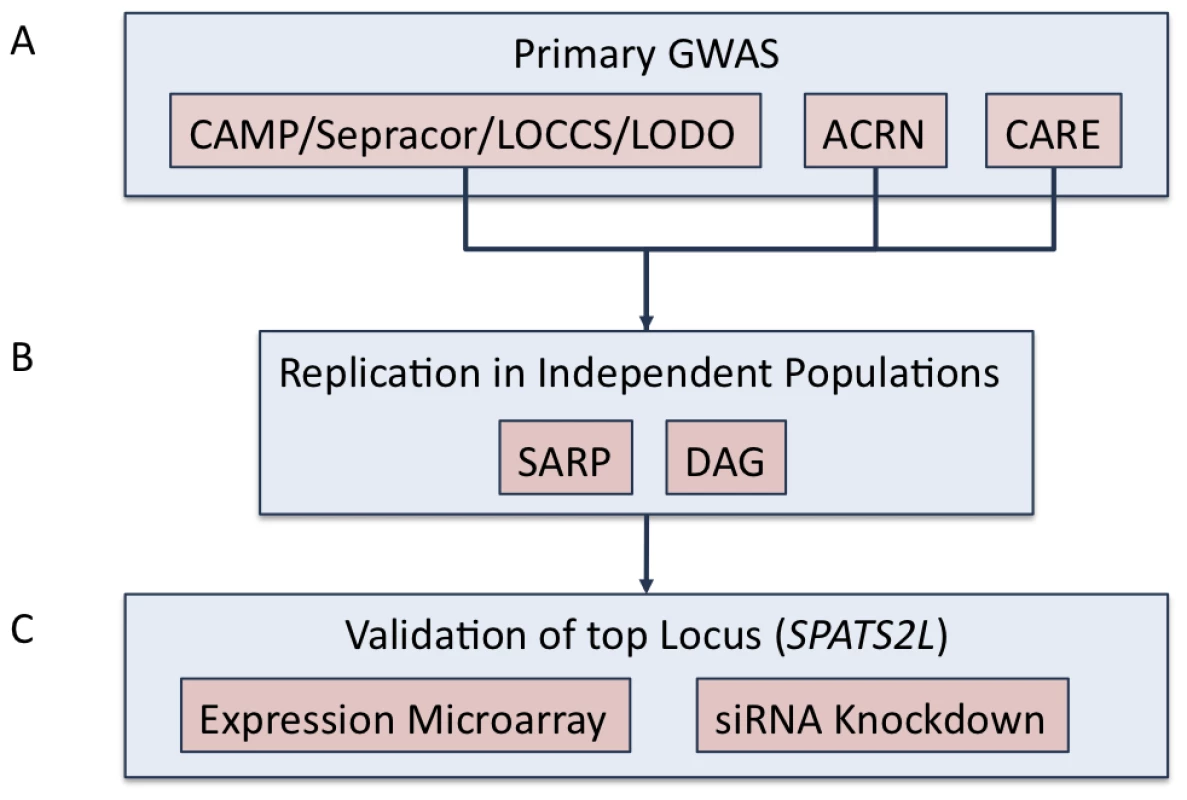
(A) Primary GWAS was conducted using subjects from CAMP, LOCCS, LODO, Sepracor, ACRN, and CARE cohort. Samples genotyped on Illumina platforms (i.e. CAMP/LOCCS/LODO/Sepracor) were pooled and analyzed first. Results from samples genotyped on Affymetrix platforms were analyzed separately and then combined to obtain the primary GWAS results. 1000GP imputed data was utilized to expand the primary GWAS association results. (B) Replication of the top (i.e. P-value<1E-04) SNPs from the primary GWAS were attempted in two independent populations: SARP and DAG. (C) The SPATS2L gene was selected for functional validation based on nominal replication of association results in SARP and analysis of publicly available resources. Tab. 1. Characteristics of GWAS subjects. 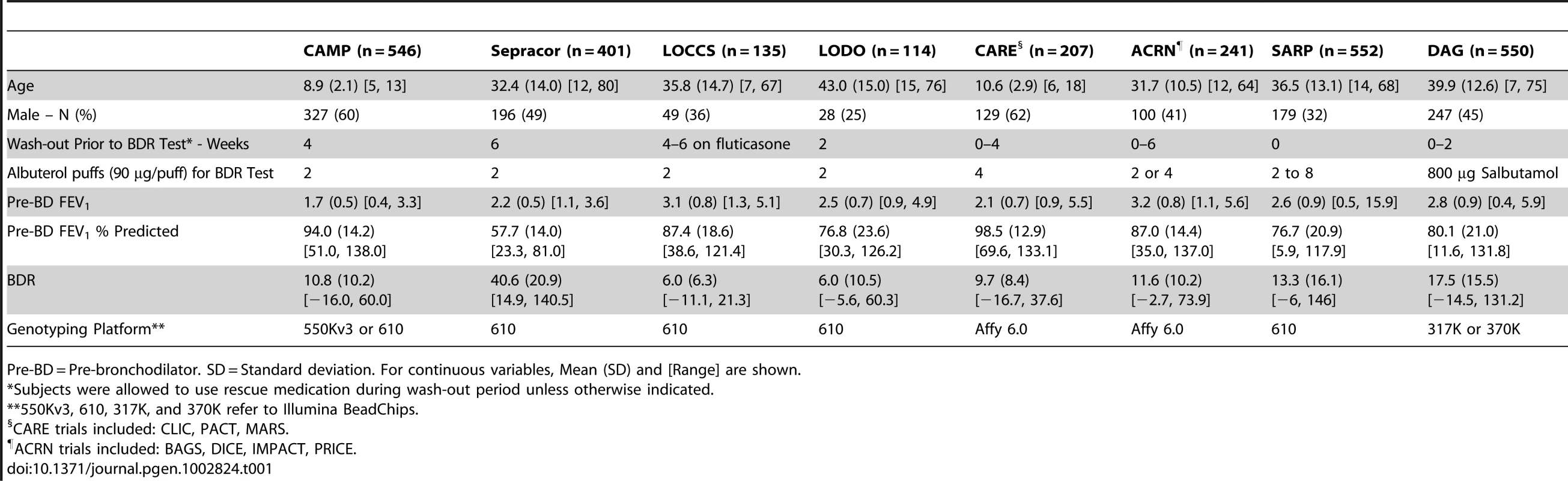
Pre-BD = Pre-bronchodilator. SD = Standard deviation. For continuous variables, Mean (SD) and [Range] are shown. Tab. 2. Primary GWAS Top Results (SNPs with Combined P-values <1e-05). 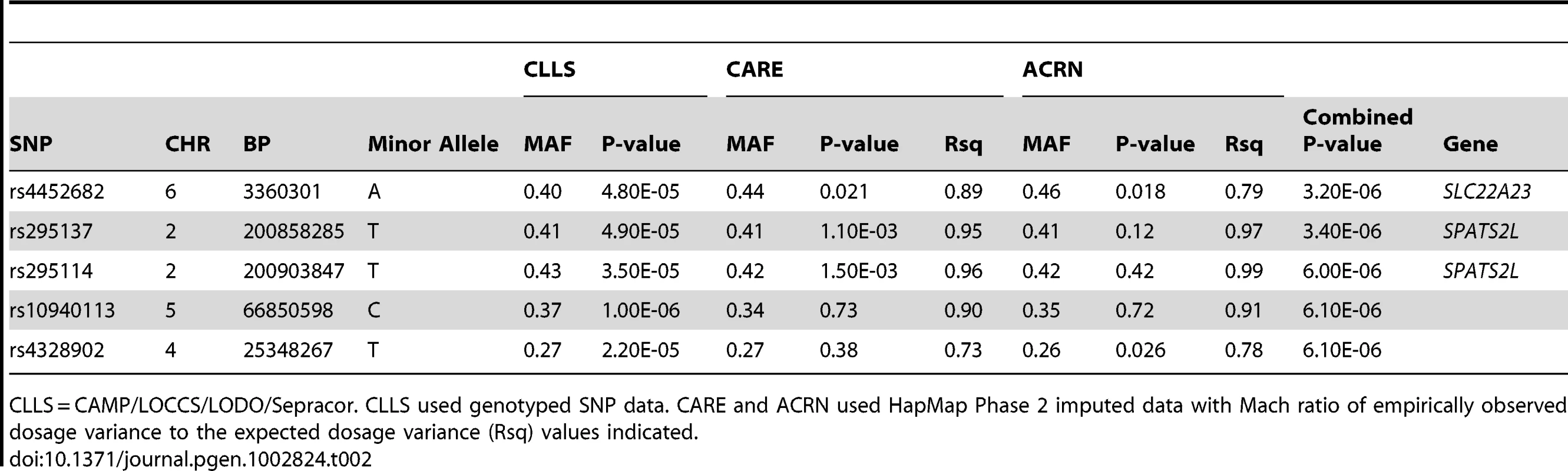
CLLS = CAMP/LOCCS/LODO/Sepracor. CLLS used genotyped SNP data. CARE and ACRN used HapMap Phase 2 imputed data with Mach ratio of empirically observed dosage variance to the expected dosage variance (Rsq) values indicated. Replication of Primary GWAS Findings
We attempted to replicate in SARP all of the SNPs with primary GWAS P-values<1E-04 (Table S5). Three had nominally significant P-values (i.e. <0.05), and two of these associations supported the top 5 primary GWAS associations (Table 3). The lowest combined P-value for all primary GWAS plus SARP data was 7.7E-07 for rs295137. The region of BDR association spanning this SNP was in the 5′UTR region of SPATS2L, a gene of unknown in vivo function and paralog of SPATS2 (Figure 2). The effect of the rs295137 genotype on BDR is shown in Figure 3, and a plot of the residuals of the linear regression fit of BDR while adjusting for age, sex, and height is shown in Figure S4. We sought further evidence of association for the two SPATS2L SNPs with lowest P-values in our primary GWAS in a second independent population: DAG. There was no evidence for association in this cohort (rs295137 P-value = 0.21; rs295114 P-value = 0.21), and combined P-values for these two SNPs across all cohorts were 9.7E-07 and 1.6E-06 (Table 3). To investigate whether our top combined association represented a biologically significant finding, we sought experimental evidence that SPATS2L was involved in bronchodilator response.
Fig. 2. Region of association near SPATS2L SNPs to BDR. 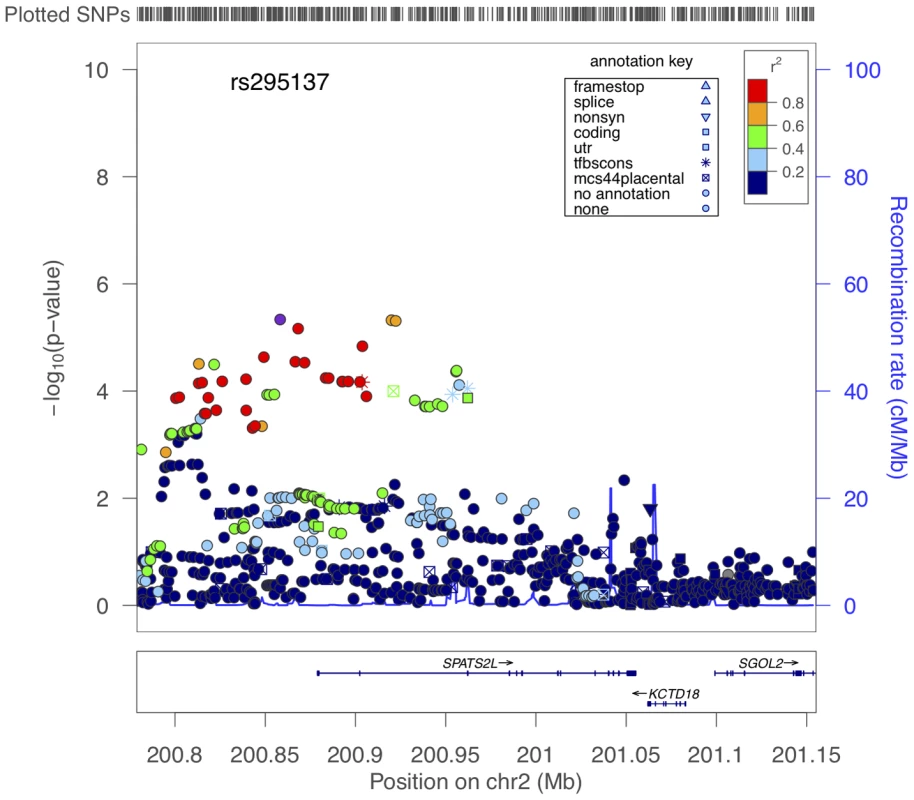
The x-axis denotes position along Chromosome 2. The y-axis denotes −Log10(P) corresponding to 1000GP imputed data P-values. LD between the SNP with the lowest P-value (rs295137) to each SNP in the plot is denoted in colors and was computed according to 1000GP June 2010 CEU data. Plot was created using LocusZoom [44]. Fig. 3. Summary of BDR by genotype of the SNP (rs295137) near SPATS2L with lowest P-value among all subjects in the primary populations. 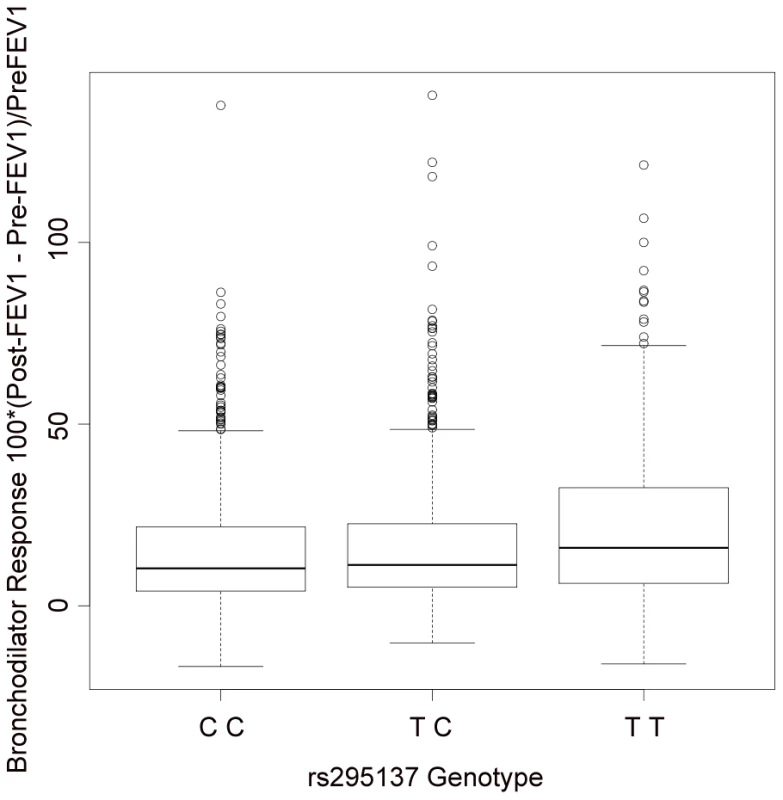
The TT genotype was associated with higher BDR (median 16.0; inter-quartile range (IQR) = [6.2, 32.4]), than the TC genotype (median 11.2; IQR = [5.2, 22.6]) or the CC genotype (median 10.3; IQR = [4.1, 21.7]). Tab. 3. Replication study of top SNPs in two independent populations (SARP and DAG). 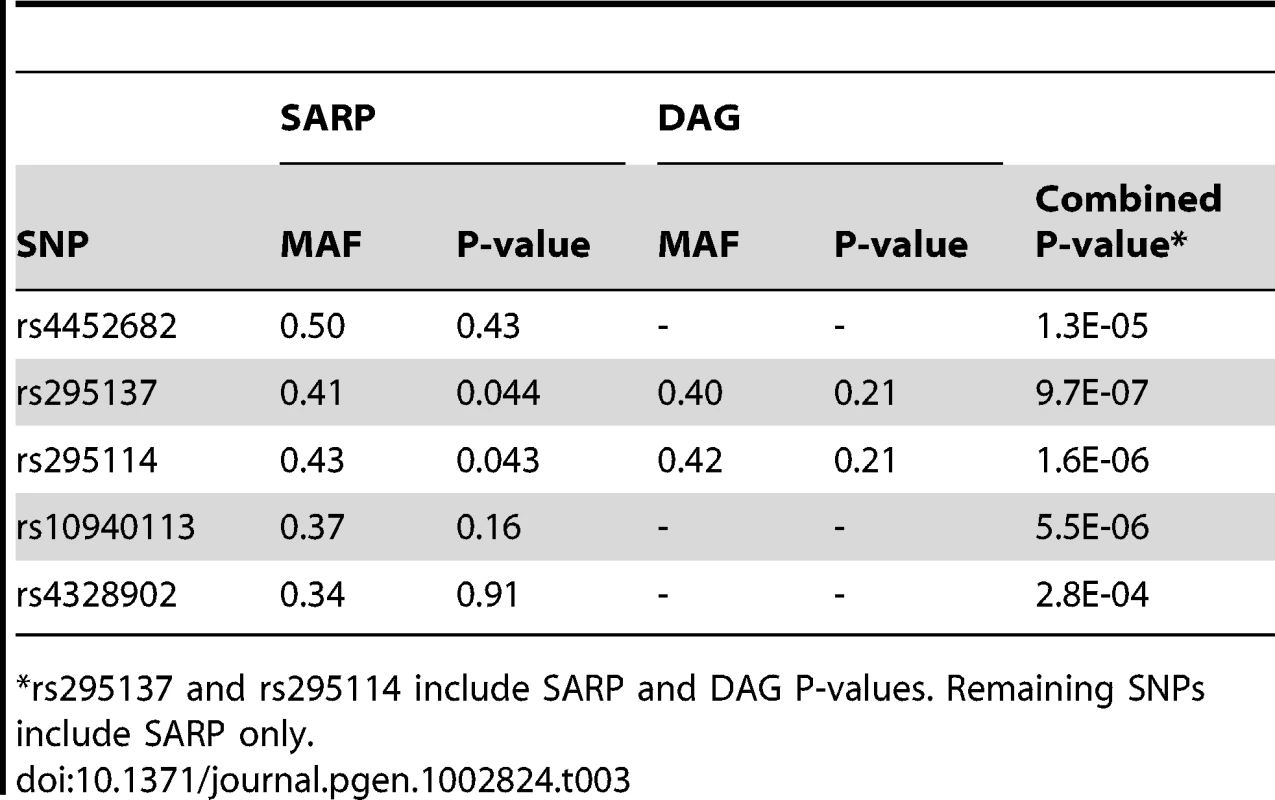
rs295137 and rs295114 include SARP and DAG P-values. Remaining SNPs include SARP only. SPATS2L mRNA Expression Changes when the β2-Agonist Pathway Is Modified in Human Airway Smooth Muscle Cells
We found one public gene expression array experiment (GSE13168) that would help to address the question of whether SPATS2L is differentially expressed in response to changes in the BDR pathway. We compared the levels of expression of two SPATS2L and one SPATS2 probes in human airway smooth muscle (HASM) cells that stably expressed a PKA inhibitor vs. a GFP control at baseline and when stimulated with the pro-asthmatic cytokines interleukin-1β (IL1β), epidermal growth factor (EGF), or both. A trend of differential expression was observed for the SPATS2 and one SPATS2L probes, but not a second SPATS2L probe (Figure S5). None of the comparisons met a Benjamini-Hochberg adjusted significance threshold, but nominally significant P-values were obtained for the SPATS2 probe under all conditions and for one of the SPATS2L probes under the condition of EGF and IL1β stimulation (Table S6). According to the Gene Enrichment Profiler, the two SPATS2L probes are highly expressed in lung, and all three probes are highly expressed in smooth muscle, especially the SPATS2 one. Overall, there are strikingly different tissue-specific expression patterns for each probe (Figure S6).
Knockdown of SPATS2L mRNA Leads to Increased β2AR Levels
We further investigated the involvement of SPATS2L in the β2-adrenergic response pathway by knocking down SPATS2L mRNA using two different small interfering RNAs (siRNA) and measuring subsequent changes in β2AR protein levels. The knockdown efficiency of the siRNAs was >80% reduction of SPATS2L mRNA as measured by qRT-PCR, and the corresponding increases in β2AR (normalized against the control β-actin protein) levels were 1.88 - (SD 0.41) and 1.86 - (SD 0.30) fold for the two SPATS2L siRNAs (Figure 4).
Fig. 4. Effect of siRNA-mediated SPATS2L knockdown on β2-adrenergic receptor levels in HASM cells. 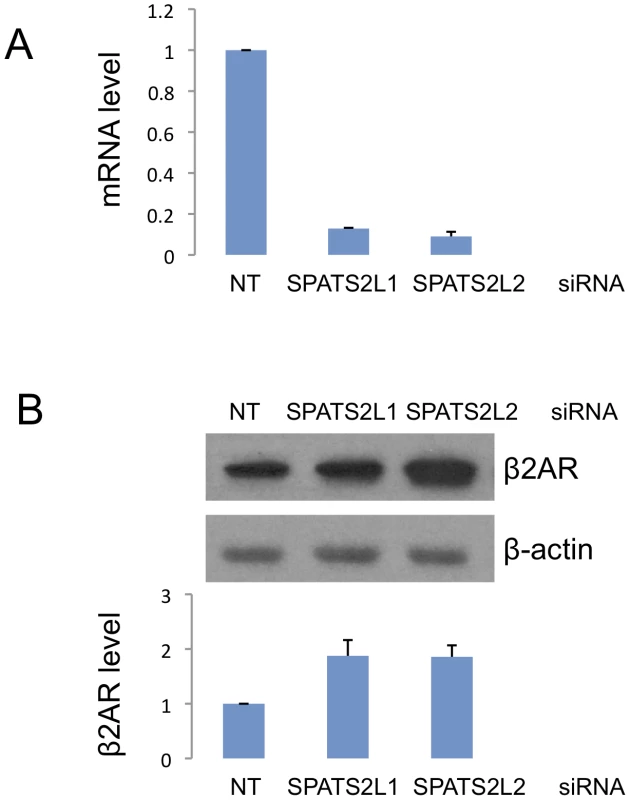
(A) Knockdown efficiency of two SPATS2L siRNAs. Quantitative real-time PCR was done on RNAs extracted from HASM cells transfected with control non-targeting (NT) or SPATS2L-specific siRNAs. (B) Western blot analysis of β2AR protein in SPATS2L knockdown HASM cells. Three independent experiments (siRNA transfection and Western blot analysis) were done in HASM cells. The upper panel is a representative of the three Western blots. Quantification of the β2AR protein amount (normalized against the control β-actin protein) from three Western blots is shown in the lower panel. Primary GWAS Results in Previously Identified BDR Candidate Genes
The association of SNPs with BDR at SNPs in/near genes (i.e. ADRB2, ADCY9, CRHR2, ARG1) previously reported as being associated with BDR was measured in our primary GWAS imputed data (Figure S7). Nominally significant (P-value<0.05) SNPs were found in ADCY9, CRHR2, and ARG1, but not in ADRB2 (Tables S7 and S8). The SNPs with lowest P-values within 50 kb of these genes were: rs2531988 for ADCY9 (3.2E-03), rs12533248 for CRHR2 (0.029), and rs6929820 for ARG1 (0.012).
Discussion
In recent years, many GWAS that have successfully identified risk-modifying loci for a wide range of complex diseases have been published, but progress toward understanding how the loci and genes identified are functionally related to diseases has been slow [19]. The relationship of genes and gene variants to pharmacogenetic traits is often easier to test functionally than that for complex diseases because pharmacogenetic traits are more amenable to in vitro testing. However, compared to GWAS of complex diseases, GWAS of pharmacogenetic traits have been challenged by the relatively small size of drug clinical trials, which has caused many studies to be underpowered for obtaining genome-wide significant associations [20]. Nonetheless, successful pharmacogenetic GWAS have led to the identification of loci involved in modulating response to inhaled corticosteroids among asthma patients [21], warfarin dose [22], and lipid-lowering response to statins [23].
One of the difficulties specific to BDR GWAS is the complexity of the BDR phenotype. Regardless of how it is quantified, BDR is highly variable among asthma patients due to the time-dependent variation in baseline FEV1 and the influence of external environmental factors [24]. BDR can be quantified in various ways, with slightly varying resulting classification of patients as responders or non-responders. For our study, we selected the definition most widely utilized in clinical and human asthma research settings: percent change in baseline FEV1 following administration of a standard dose of short-acting inhaled β2-agonist [25]. We have attempted to control for the known relationship between baseline lung function and BDR [24], [26] by (1) selecting a definition of BDR that standardizes the change in FEV1 by dividing by baseline FEV1 and (2) by using age, sex, and height, which together account for a large portion of the variability in baseline lung function, as covariates in our statistical models. Because BDR tests are routinely performed during asthma clinical trials to use as inclusion criteria and to monitor outcomes among patients, we were able to utilize subjects from several diverse asthma clinical trials that were not specifically designed to study the pharmacogenetics of BDR. Most of these trials included a wash-out period that reduces modification of BDR due to concomitant medication administration, but LOCCS and some CARE and ACRN subjects were administered BDR tests at a time when they were not necessarily off of medications (Table 1). Subjects without a placebo washout, and especially those who were on ICS (e.g. LOCCS subjects), may be expected to have less BDR than those on placebo. The relationship of the magnitude of BDR to the various gene loci could therefore be blunted and show a less significant relationship than would be expected if all studies had incorporated a placebo washout.
In addition to variable washout periods, the cohorts had other significant differences in their design. Two trials consisted of children with asthma (i.e. CAMP, CARE), while the others consisted mostly of adults. The gender composition varied from 25% to 62% male. We attempted to control for age, gender and height, all of which are known to influence BDR, by including them as covariates in the association analysis. The mean and range of BDR also varied among trials. Of most significance, because the Sepracor trial used BDR greater than 15% as a criterion for inclusion, its subjects had markedly greater BDR than those of other trials. We attempted to control for this difference and any other trial specific differences among the cohorts that were pooled in the primary analysis by including trial as a covariate in the association model. There were additional differences among trials that were not taken into account. For example, SARP and Sepracor were composed of subjects with more severe asthma than those of other cohorts. Some ACRN, CARE, and SARP subjects were administered a different amount of albuterol during their BDR tests than those of CAMP, Sepracor, LOCCS, and LODO. DAG subjects were administered a different beta-agonist (i.e. Salbutamol) at a different concentration than that used with subjects of all other trials. DAG and SARP subjects were not participants of clinical trials, so there was greater heterogeneity of subjects within those cohorts. Despite the heterogeneity among trials, we utilized as many subjects as possible in an attempt to increase our statistical power to detect associations of SNPs with BDR. We reasoned that any associations detected despite the heterogeneity of the trial populations would be those most likely to generalize to all asthma patients. Another expected consequence of the trial heterogeneity is that our association results do not replicate in all cohorts. While having the largest number of subjects provides the greatest statistical power to detect statistically significant associations that are most generalizable across the clinical trials, we may be missing associations that are specific to the individual trials. For example, the subjects within clinical trials representing different ranges of asthma severity, age, and baseline characteristics may have genetic associations that are unique to subjects with their specific trial characteristics. The small sample size of each individual clinical trial makes detection of trial-specific associations more challenging. Despite the cohort heterogeneity, our meta-analysis identified a strong association that suggests a novel gene is involved in BDR.
Our top association was at SNP rs295137, with a combined P-value across all cohorts of 9.7E-07. This P-value does not meet conventional genome-wide significance thresholds (e.g. Bonferroni corrected minimally significant P-value would be 0.05/469,884 = 1.1E-07), but performing searches through public data sources and the fact that other pharmacogenetic GWAS have discovered biologically important results without genome-wide significant associations led us to pursue our top association further. The region of association surrounding rs295137 is in the 5′UTR of SPATS2L (Figure 2). This gene maps to chromosome 2 at 2q33.1, covering 176.78 kb from 201170592 to 201347368 (NCBI 37, August 2010). According to data gathered via the AceView [27] tool, SPATS2L is a complex locus that may have at least 30 spliced variants, its in vivo function is unknown, and it is a highly expressed gene in many tissues, with the greatest number of GenBank accessions belonging to lung. In gene-trap experiments in myoblasts, SPATS2L (a.k.a. SGNP) was found to be involved in ribosomal biogenesis and translational control in response to oxidative stress [28].
The availability of one public expression array experiment that utilized HASM cells expressing a PKA inhibitor (PKI) to modify the β2-adrenergic pathway allowed us to perform a preliminary search for evidence that SPATS2L may be involved in BDR. We found that a probe for SPATS2, the paralog of SPATS2L, was significantly differentially expressed in PKI vs. control cells at baseline and when stimulated with pro-inflammatory cytokines (EGF, IL1β, or both). One SPAST2L probe followed this trend but had a nominally significant P-value only under the condition of stimulation with both EGF and IL1β, while the other SPATS2L probe did not exhibit any changes. As illustrated in Figure S6, the tissue-specific expression patterns of the three probes varied widely. While all were expressed in smooth muscle, the SPATS2 probe's relative expression in this tissue was markedly greater than that of the SPATS2L probes. Taken together, the expression patterns are consistent with tissue and isoform dependent changes in SPATS2L gene products. While the public dataset SPATS2L results were inconclusive based on the differences among probes, they suggested that SPATS2L expression may change when PKA is inhibited in HASM cells.
Knockdown of SPATS2L in HASM cells resulted in significantly increased β2AR protein levels, suggesting that SPATS2L may affect BDR by directly modulating β2AR protein expression. In HASM, β2-agonists exert their effects exclusively via the β2AR [6]. The relaxation of HASM occurs after the binding of β2-agonists to β2ARs via increased levels of cAMP followed by PKA activation. PKA activation leads to changes in gene transcription via activation of cAMP response element binding protein (CREB). Because β2ARs are the gateway to the effects of β2-agonists in HASM cells, modulations, such as SPATS2L inactivation, that increase the levels of β2ARs in HASM cells may lead to both greater relaxation in response to β2-agonists in the short term and greater differences in gene transcription in the longer term. Further study is needed to elucidate the precise mechanism by which SPATS2L regulates β2AR and consequently modifies BDR. Among our primary GWAS subjects, those whose SPATS2L SNP rs295137 has the TT genotype have greater BDR than those with CT or TT genotypes (median BDR 16.0 vs. 10.9). In one of the simplest scenarios, it is possible that the increased BDR among subjects with the TT genotype results from this genotype playing a direct role in decreasing transcription of SPATS2L, which in turn results in increased β2AR levels. Further work is required to understand how specific SNP associations in/near SPATS2L affect SPATS2L function and/or expression and how such effects impact β2AR signaling and BDR. Because the observed influence of our most strongly associated SNP genotype on BDR is relatively small (Figure 3), our current data do not support the development of any personalized therapeutics based solely on variants in/near SPATS2L.
In addition to studying top primary GWAS SNPs, we attempted to replicate findings from previous BDR candidate gene association studies. Specifically, we measured association between BDR and ADRB2 [13], [14], ADCY9 [15], CRHR2 [16], and ARG1 [17] variants. Notably, these previous findings are not entirely independent of those from the current GWAS: CAMP was a primary population utilized to identify associations in ADRB2, ADCY9, CRHR2, and ARG1 in previous reports; LODO and Sepracor were replication populations in the CRHR2, and ARG1 reports; and LOCCS was a replication population in the ARG1 report. At a nominal significance level, we replicated gene-level associations for all of the candidate genes other than ADRB2. This gene, which encodes the β2AR, is the most studied gene related to BDR and SNPs and haplotypes in this gene have been related to decreased pulmonary function [29], response to β2-agonist treatment [30], an increased frequency of asthma exacerbations [31], and BDR [13], [14]. Initial reports of ADRB2 associations were very promising and suggested that variants of this candidate gene would be reliable markers of BDR pharmacogenetics. However, a meta-analysis of 21 studies of ADRB2 polymorphisms found that most of the positive associations identified in individual studies cannot be compared to findings in other studies due to different study designs, phenotypes considered and selective reporting, making the number of variants with true replications very small and questioning the utility of ADRB2 polymorphisms for generalizable pharmacogenetic tests [32]. Our inability to find associations with ADRB2 variants is consistent with the view that BDR genetics are complex: no individual SNPs or genes are responsible for a large proportion of BDR variability observed among all asthmatics. Our results suggest that genes other than the previously identified candidate genes are more strongly associated with BDR and that functional studies of these regions may yield important insights into BDR biology despite not having strong effects or generalizing to all populations.
In summary, a BDR GWAS among asthma patients from eight cohorts found that the most strongly associated SNP, rs295137, had a combined P-value of 9.7E-07. This association led us to SPATS2L, a gene of unknown in vivo function that we showed may be involved in BDR via the down-regulation of β2AR levels. Our results support the notion that there is promise in pursuing GWAS results that do not necessarily reach genome-wide significance and are an example of the way in which results from pharmacogenetic GWAS can be studied functionally.
Materials and Methods
Ethics Statement
Each study was approved by the Institutional Review Board of the corresponding institution, which ensured that all procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation. Informed consent was obtained for all study participants.
Subjects
The primary group of subjects consisted of 1,644 non-Hispanic white asthmatics from the following drug clinical trials: Childhood Asthma Management Program (CAMP) [33], Leukotriene Modifier or Corticosteroid Salmeterol study (LOCCS) [34], Effectiveness of Low Dose Theophylline as an Add-on Treatment in Asthma trial (LODO) [35], a medication trial conducted by Sepracor, Inc. [36], [37], and subsets of clinical trials within the Childhood Asthma Research and Education (CARE) network [38], and the Asthma Clinical Research Network (ACRN) [39] participating in the NHLBI SNP Health Association Resource (SHARe) Asthma Resource project (SHARP). Some basic characteristics of these cohorts are in Table 1 and further details are provided in Text S1. BDR tests were performed according to American Thoracic Society criteria with Albuterol as the β2-agonist [25], unless otherwise noted. Baseline BDR measures were utilized, and BDR was quantified as the percent change in FEV1 in response to administration of a β2-agonist [i.e. (post-BD FEV1 – pre-BD FEV1)/pre-BD FEV1].
Genotyping and Quality Control
Genome-wide genotyping for CAMP subjects (n = 546) was performed on the HumanHap550 Genotyping BeadChip or Infinium HD Human610-Quad BeadChip by Illumina, Inc (San Diego, CA) at the Channing Laboratory. LOCCS (n = 135), LODO (n = 114), and Sepracor (n = 401) subjects were genotyped at the Riken Center for Genomic Medicine using the Infinium HD Human610-Quad BeadChip. CARE (n = 207) and ACRN (n = 241) subjects were genotyped on Affymetrix 6.0 genotyping chip by Affymetrix, Inc. (Santa Clara, CA). Data from those subjects genotyped using Illumina technologies was combined into a primary dataset with 469,884 overlapping SNPs having missingness <1%, passing HWE (P-value threshold of 1E-03), and having minor allele frequency (MAF)>0.05. EIGENSTRAT was used to identify 23 outliers (not included in counts above) based on being outside of six standard deviations of the top four principal components during five iterations [40]. The genomic inflation factor (λGC) of the remaining 1,196 subjects was 1.002, demonstrating minimal population stratification. CARE and ACRN dataset quality control (QC) also included the removal of four related subjects (i.e. CARE siblings), SNPs with MAF<0.05, missingness >5%, or not passing HWE based on a threshold of 1E-03. The λGC for CARE and ACRN genotype data were 1.02 and 0.98, demonstrating minimal population stratification among subjects within each group. Comprehensive genotyping and QC measures are provided in Text S1.
Statistical Analysis
Due to the poor overlap among SNPs genotyped on the Illumina and Affymetrix platforms, imputation of all SNPs available in HapMap Phase 2 Release 22 CEU data using the Markov Chain Haplotyping software (MaCH) [41] was performed for ACRN and CARE genotyped data. The primary GWAS consisted in the set of 469,884 SNPs that were successfully genotyped in those cohorts using Illumina arrays (i.e., CAMP/Sepracor/LOCCS/LODO) and imputed with HapMap Phase 2 data in those cohorts genotyped with Affymetrix arrays (i.e., ACRN and CARE) with a ratio of empirically observed dosage variance to the expected (binomial) dosage variance greater than 0.3, indicating good quality of imputation.
To expand the association results, imputation of all SNPs available in the June 2010 release of the 1000 Genome Project (1000GP) data using MaCH was performed for each of the three primary groups of genotype data. An overlapping set of 4,571,615 imputed SNPs had a MAF>0.05 and ratio of empirically observed dosage variance to the expected (binomial) dosage variance greater than 0.5, indicating good quality of imputation.
The association of SNPs with BDR was measured with a linear regression model as implemented in PLINK [42] in the three sets of data: 1) CAMP/Sepracor/LOCCS/LODO, 2) ACRN, 3) CARE. Association of imputed SNPs was carried out using dosage data. Covariates for the CAMP/Sepracor/LOCCS/LODO group included age, gender, height, and study. Covariates for the CARE and ACRN groups included age, gender, and height. To get the primary GWAS results, CARE and ACRN P-values were combined with those of the CAMP/Sepracor/LOCCS/LODO group by using Liptak's combined probability method [43] where hypothesis tests in CARE and ACRN had one-sided alternatives, based on the direction of the association in CAMP/Sepracor/LOCCS/LODO, so that SNPs with association tests in opposite directions would not produce inappropriately small P-values. The overall λGC was 1.002 in the primary set of GWAS results and 1.000 in the 1000GP imputed data GWAS.
Plots of association results near specific genes were created using LocusZoom with the hg18/1000 Genomes June 2010 CEU GenomeBuild/LD Population [44].
Replication Studies
1) Severe Asthma Research Program (SARP)
Replication of primary GWAS P-values was attempted in 501 European American subjects from SARP who were recruited to meet the American Thoracic Society (ATS) definition of mild to severe asthmatics, with enrichment for severe asthma [45]. Genotyping was performed on the Illumina Infinium HD Human 610-Quad Bead Chip at Wake Forest University. Tests of BDR association were performed using linear regression in R [46] with age, sex, and height as covariates.
2) Dutch Asthma GWAS (DAG)
Replication of two SNPs near SPATS2L with lowest P-values in the primary GWAS was attempted in 550 DAG subjects with asthma and BDR data that were phenotyped in a single center (i.e, Beatrixoord, Haren, the Netherlands) [18], [47]. All patients refrained from bronchodilator use for at least 8 hours, and stopped inhaled steroids for 2 weeks before pulmonary function testing if clinically possible. Reversibility was assessed measuring spirometry before and 15 minutes after inhaling 800 µg of salbutamol. Genotyping was performed using the Hapmap 317K platform or Illumina 370 Duo Chip. Tests of BDR association were performed using linear regression, with age, gender and height as covariates using Predictive Analysis Software (PASW) version 18.0.
Genome-Wide Gene Expression Data
The publicly available Gene Expression Omnibus (GEO) dataset, GSE13168, corresponding to an experiment in which human airway smooth muscle (HASM) cell cultures were generated from four donor trachea to test for the effects of glucocorticoids and PKA inhibition on the HASM transcriptome using the Affymetrix Human Genome U133A platform was used [48]. We tested for the involvement of our top primary GWAS gene in the β2-adrenergic pathway by comparing the differential expression of genes in cells stably expressing a PKA inhibitor (PKI) vs. control at baseline and in the presence of pro-inflammatory cytokines interleukin-1β (IL1β), epidermal growth factor (EGF), or both. The expression array contained two SPATS2L probes (i.e., 215617_at, 222154_s_at) and one SPATS2 probe (i.e., 218324_s_at). The probe for the paralog of SPATS2L was included to account for the possibility of non-specific binding of SPATS2L mRNA to the SPATS2 probe. Analyses were conducted in R [46]. Pre-processing of raw signal intensities was performed with RMA [49] and differential expression was quantified using the limma package [50]. Tissue-specific expression of these probes was assessed using 557 microarrays from 126 human normal primary tissues in the Gene Enrichment Profiler [51].
SPATS2L siRNA Knockdown and β2AR Western Blotting Analysis
Primary HASM cells were isolated from aborted lung transplant donors with no chronic illness. The tissue was obtained from the National Disease Resource Interchange (NDRI) and their use approved by the University of Pennsylvania IRB. HASM cell cultivation and characterization were described previously [52], [53]. Passages 4 to 7 HASM cells maintained in Ham's F12 medium supplemented with 10% FBS were used in all experiments. 2×105 HASM cells were grown overnight and then transfected with 50 nM siRNA by using DharmaFECT 1 reagent (Thermo Scientific, Lafayette, CO, USA). About 72 h post transfection, cells were washed with PBS and lysed with NP-40 lysing buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 0.5% Nonidet P-40) containing protease inhibitor cocktail (Roche Diagnostics Corporation, Indianapolis, IN, USA). Protein samples were denatured 10 min at 50°C, separated on NuPAGE 4–12% Bis-Tris gels (Invitrogen, Grand Island, NY, USA) and transferred to PVDF membranes (Bio-Rad Laboratories, Hercules, CA, USA). Immunoblot signals were developed using SuperSignal West Pico (Pierce Protein Research Products, Thermo Fisher Scientific, Rockford, IL, USA) and quantified by ImageJ software. Non-targeting control siRNA, SPATS2L-specific siRNA 1 (sense 5′ - guc agu cca uug auu guc u(dT)(dT)-3′, antisense 5′ - aga caa uca aug gac uga c(dT)(dT) -3′) and SPATS2L-specific siRNA 2 (sense 5′-caa ccu gug uug uag cag u(dT)(dT)-3′, antisense 5′ - acu gcu aca aca cag guu g(dT)(dT) -3′) were obtained from Sigma-Aldrich (Mission siRNA; St. Louis, MO, USA). Antibodies for β2AR (H20) and β-actin were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Experiments were performed in triplicate.
Supporting Information
Zdroje
1. American Lung Association 2006 Trends in Asthma Morbidity and Mortality New York, NY Epidemiology and Statistics Unit, Research and Program Services, American Lung Association
2. Global Initiative for Asthma Management and Prevention 1995 NHLBI/WHO Workshop Report National Institutes of Health, Bethesda US Department of Health and Human Services
3. NelsonHS 1995 Beta-adrenergic bronchodilators. N Engl J Med 333 499 506
4. LozaMJPennRB 2010 Regulation of T cells in airway disease by beta-agonist. Front Biosci (Schol Ed) 2 969 979
5. SalatheM 2002 Effects of beta-agonists on airway epithelial cells. J Allergy Clin Immunol 110 S275 281
6. ShoreSAMoorePE 2003 Regulation of beta-adrenergic responses in airway smooth muscle. Respir Physiol Neurobiol 137 179 195
7. JarttiT 2001 Asthma, asthma medication and autonomic nervous system dysfunction. Clin Physiol 21 260 269
8. DrazenJMSilvermanEKLeeTH 2000 Heterogeneity of therapeutic responses in asthma. Br Med Bull 56 1054 1070
9. MoffattMFGutIGDemenaisFStrachanDPBouzigonE 2010 A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 363 1211 1221
10. TorgersonDGAmplefordEJChiuGYGaudermanWJGignouxCR 2011 Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet 43 887 892
11. NiuTRogusJJChenCWangBYangJ 2000 Familial aggregation of bronchodilator response: a community-based study. Am J Respir Crit Care Med 162 1833 1837
12. HawkinsGAWeissSTBleeckerER 2008 Clinical consequences of ADRbeta2 polymorphisms. Pharmacogenomics 9 349 358
13. DrysdaleCMMcGrawDWStackCBStephensJCJudsonRS 2000 Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci U S A 97 10483 10488
14. SilvermanEKKwiatkowskiDJSylviaJSLazarusRDrazenJM 2003 Family-based association analysis of beta2-adrenergic receptor polymorphisms in the childhood asthma management program. J Allergy Clin Immunol 112 870 876
15. TantisiraKGSmallKMLitonjuaAAWeissSTLiggettSB 2005 Molecular properties and pharmacogenetics of a polymorphism of adenylyl cyclase type 9 in asthma: interaction between beta-agonist and corticosteroid pathways. Hum Mol Genet 14 1671 1677
16. PoonAHTantisiraKGLitonjuaAALazarusRXuJ 2008 Association of corticotropin-releasing hormone receptor-2 genetic variants with acute bronchodilator response in asthma. Pharmacogenet Genomics 18 373 382
17. LitonjuaAALasky-SuJSchneiterKTantisiraKGLazarusR 2008 ARG1 is a novel bronchodilator response gene: screening and replication in four asthma cohorts. Am J Respir Crit Care Med 178 688 694
18. VonkJMPostmaDSMaarsinghHBruinenbergMKoppelmanGH 2010 Arginase 1 and arginase 2 variations associate with asthma, asthma severity and beta2 agonist and steroid response. Pharmacogenet Genomics 20 179 186
19. ManolioTA 2010 Genomewide association studies and assessment of the risk of disease. N Engl J Med 363 166 176
20. DalyAK 2010 Genome-wide association studies in pharmacogenomics. Nat Rev Genet 11 241 246
21. TantisiraKGLasky-SuJHaradaMMurphyALitonjuaAA 2011 Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med 365 1173 1183
22. TakeuchiFMcGinnisRBourgeoisSBarnesCErikssonN 2009 A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet 5 e1000433 doi:10.1371/journal.pgen.1000433
23. BarberMJMangraviteLMHydeCLChasmanDISmithJD 2010 Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS ONE 5 e9763 doi:10.1371/journal.pone.0009763
24. SharmaSLitonjuaAATantisiraKGFuhlbriggeALSzeflerSJ 2008 Clinical predictors and outcomes of consistent bronchodilator response in the childhood asthma management program. J Allergy Clin Immunol 122 921 928 e924
25. American Thoracic Society 1991 Lung function testing: selection of reference values and interpretative strategies. Am Rev Respir Dis 144 1202 1218
26. WaalkensHJMerkusPJvan Essen-ZandvlietEEBrandPLGerritsenJ 1993 Assessment of bronchodilator response in children with asthma. Dutch CNSLD Study Group. Eur Respir J 6 645 651
27. Thierry-MiegDThierry-MiegJ 2006 AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol 7 Suppl 1 S12 11 14
28. ZhuCHKimJShayJWWrightWE 2008 SGNP: an essential Stress Granule/Nucleolar Protein potentially involved in 5.8s rRNA processing/transport. PLoS ONE 3 e3716 doi:10.1371/journal.pone.0003716
29. IsraelEDrazenJMLiggettSBBousheyHACherniackRM 2000 The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med 162 75 80
30. IsraelEChinchilliVMFordJGBousheyHACherniackR 2004 Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet 364 1505 1512
31. TaylorDRDrazenJMHerbisonGPYandavaCNHancoxRJ 2000 Asthma exacerbations during long term beta agonist use: influence of beta(2) adrenoceptor polymorphism. Thorax 55 762 767
32. Contopoulos-IoannidisDGAlexiouGAGouviasTCIoannidisJP 2006 An empirical evaluation of multifarious outcomes in pharmacogenetics: beta-2 adrenoceptor gene polymorphisms in asthma treatment. Pharmacogenet Genomics 16 705 711
33. Childhood Asthma Management Program Research Group 1999 The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials 20 91 120
34. PetersSPAnthonisenNCastroMHolbrookJTIrvinCG 2007 Randomized comparison of strategies for reducing treatment in mild persistent asthma. N Engl J Med 356 2027 2039
35. American Lung Association Asthma Clinical Research Centers 2007 Clinical trial of low-dose theophylline and montelukast in patients with poorly controlled asthma. Am J Respir Crit Care Med 175 235 242
36. SilvermanESPalmerLJSubramaniamVHallockAMathewS 2004 Transforming growth factor-beta1 promoter polymorphism C-509T is associated with asthma. Am J Respir Crit Care Med 169 214 219
37. SilvermanESBaronRMPalmerLJLeLHallockA 2002 Constitutive and cytokine-induced expression of the ETS transcription factor ESE-3 in the lung. Am J Respir Cell Mol Biol 27 697 704
38. GuilbertTWMorganWJKrawiecMLemanskeRFJrSorknessC 2004 The Prevention of Early Asthma in Kids study: design, rationale and methods for the Childhood Asthma Research and Education network. Control Clin Trials 25 286 310
39. DenlingerLCSorknessCAChinchilliVMLemanskeRFJr 2007 Guideline-defining asthma clinical trials of the National Heart, Lung, and Blood Institute's Asthma Clinical Research Network and Childhood Asthma Research and Education Network. J Allergy Clin Immunol 119 3 11; quiz 12–13
40. PriceALPattersonNJPlengeRMWeinblattMEShadickNA 2006 Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38 904 909
41. WillerCJSannaSJacksonAUScuteriABonnycastleLL 2008 Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40 161 169
42. PurcellSNealeBTodd-BrownKThomasLFerreiraMA 2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
43. LiptakT 1958 On the combination of independent tests. Magyar Tud Akad Mat Kutato Int Kozl 3 171 197
44. PruimRJWelchRPSannaSTeslovichTMChinesPS 2010 LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26 2336 2337
45. MooreWCBleeckerERCurran-EverettDErzurumSCAmeredesBT 2007 Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute's Severe Asthma Research Program. J Allergy Clin Immunol 119 405 413
46. R Development Core Team 2008 R: A Language and Environment for Statistical Computing Vienna, Austria R Foundation for Statistical Computing
47. KoppelmanGHMeyersDAHowardTDZhengSLHawkinsGA 2009 Identification of PCDH1 as a novel susceptibility gene for bronchial hyperresponsiveness. Am J Respir Crit Care Med 180 929 935
48. MisiorAMDeshpandeDALozaMJPascualRMHippJD 2009 Glucocorticoid - and protein kinase A-dependent transcriptome regulation in airway smooth muscle. Am J Respir Cell Mol Biol 41 24 39
49. GautierLCopeLBolstadBMIrizarryRA 2004 affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20 307 315
50. SmythGK 2005 Limma: linear models for microarray data. GentlemanRCareyVDudoitSIrizarryRHuberW Bioinformatics and Computational Biology Solutions using R and Bioconductor New York Springer 397 420 editors. pp.
51. BenitaYCaoZGiallourakisCLiCGardetA 2010 Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood 115 5376 5384
52. PanettieriRAMurrayRKDePaloLRYadvishPAKotlikoffMI 1989 A human airway smooth muscle cell line that retains physiological responsiveness. Am J Physiol 256 C329 335
53. CooperPRMesarosACZhangJChristmasPStarkCM 2010 20-HETE mediates ozone-induced, neutrophil-independent airway hyper-responsiveness in mice. PLoS ONE 5 e10235 doi:10.1371/journal.pone.0010235
Štítky
Genetika Reprodukční medicína
Článek Allelic Heterogeneity and Trade-Off Shape Natural Variation for Response to Soil MicronutrientČlánek The Chicken Frizzle Feather Is Due to an α-Keratin () Mutation That Causes a Defective RachisČlánek A Trans-Species Missense SNP in Is Associated with Sex Determination in the Tiger Pufferfish, (Fugu)
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 7- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Functional Evolution of Mammalian Odorant Receptors
- Oocyte Family Trees: Old Branches or New Stems?
- Allelic Heterogeneity and Trade-Off Shape Natural Variation for Response to Soil Micronutrient
- Guidelines for Genome-Wide Association Studies
- GWAS Identifies Novel Susceptibility Loci on 6p21.32 and 21q21.3 for Hepatocellular Carcinoma in Chronic Hepatitis B Virus Carriers
- DNA Methyltransferases Are Required to Induce Heterochromatic Re-Replication in Arabidopsis
- Genomic Data Reveal a Complex Making of Humans
- Let-7b/c Enhance the Stability of a Tissue-Specific mRNA during Mammalian Organogenesis as Part of a Feedback Loop Involving KSRP
- The Secreted Immunoglobulin Domain Proteins ZIG-5 and ZIG-8 Cooperate with L1CAM/SAX-7 to Maintain Nervous System Integrity
- RsfA (YbeB) Proteins Are Conserved Ribosomal Silencing Factors
- Gene Conversion Occurs within the Mating-Type Locus of during Sexual Reproduction
- The Chicken Frizzle Feather Is Due to an α-Keratin () Mutation That Causes a Defective Rachis
- Meta-Analysis of Genome-Wide Scans for Total Body BMD in Children and Adults Reveals Allelic Heterogeneity and Age-Specific Effects at the Locus
- Balancing Selection at the Tomato Guardee Gene Family Maintains Variation in Strength of Pathogen Defense
- Large-Scale Introgression Shapes the Evolution of the Mating-Type Chromosomes of the Filamentous Ascomycete
- OSD1 Promotes Meiotic Progression via APC/C Inhibition and Forms a Regulatory Network with TDM and CYCA1;2/TAM
- Intact p53-Dependent Responses in miR-34–Deficient Mice
- FANCJ/BACH1 Acetylation at Lysine 1249 Regulates the DNA Damage Response
- CED-10/Rac1 Regulates Endocytic Recycling through the RAB-5 GAP TBC-2
- Histone H2A Mono-Ubiquitination Is a Crucial Step to Mediate PRC1-Dependent Repression of Developmental Genes to Maintain ES Cell Identity
- F-Box Protein Specificity for G1 Cyclins Is Dictated by Subcellular Localization
- The Gene Encodes a Nuclear Protein That Affects Alternative Splicing
- A Key Role for Chd1 in Histone H3 Dynamics at the 3′ Ends of Long Genes in Yeast
- Genome-Wide Association Analysis in Asthma Subjects Identifies as a Novel Bronchodilator Response Gene
- GRHL3/GET1 and Trithorax Group Members Collaborate to Activate the Epidermal Progenitor Differentiation Program
- Brain-Specific Rescue of Reveals System-Driven Transcriptional Rhythms in Peripheral Tissue
- Recent Loss of Self-Incompatibility by Degradation of the Male Component in Allotetraploid
- Pregnancy-Induced Noncoding RNA () Associates with Polycomb Repressive Complex 2 and Regulates Mammary Epithelial Differentiation
- The HEI10 Is a New ZMM Protein Related to Zip3
- The SCF Ubiquitin E3 Ligase Ubiquitylates Sir4 and Functions in Transcriptional Silencing
- Induction of Cytoprotective Pathways Is Central to the Extension of Lifespan Conferred by Multiple Longevity Pathways
- Role of Architecture in the Function and Specificity of Two Notch-Regulated Transcriptional Enhancer Modules
- Loss of ATRX, Genome Instability, and an Altered DNA Damage Response Are Hallmarks of the Alternative Lengthening of Telomeres Pathway
- A Regulatory Loop Involving PAX6, MITF, and WNT Signaling Controls Retinal Pigment Epithelium Development
- The Three Faces of Riboviral Spontaneous Mutation: Spectrum, Mode of Genome Replication, and Mutation Rate
- Unmet Expectations: miR-34 Plays No Role in p53-Mediated Tumor Suppression In Vivo
- A Genome-Wide Association Meta-Analysis of Circulating Sex Hormone–Binding Globulin Reveals Multiple Loci Implicated in Sex Steroid Hormone Regulation
- The Role of Rice HEI10 in the Formation of Meiotic Crossovers
- A Trans-Species Missense SNP in Is Associated with Sex Determination in the Tiger Pufferfish, (Fugu)
- Influences Bone Mineral Density, Cortical Bone Thickness, Bone Strength, and Osteoporotic Fracture Risk
- Evidence of Inbreeding Depression on Human Height
- Comparative Genomics of Plant-Associated spp.: Insights into Diversity and Inheritance of Traits Involved in Multitrophic Interactions
- Detecting Individual Sites Subject to Episodic Diversifying Selection
- Regulates Rhodopsin-1 Metabolism and Is Required for Photoreceptor Neuron Survival
- Identification of Chromatin-Associated Regulators of MSL Complex Targeting in Dosage Compensation
- Three Dopamine Pathways Induce Aversive Odor Memories with Different Stability
- TDP-1/TDP-43 Regulates Stress Signaling and Age-Dependent Proteotoxicity in
- Rapid Turnover of Long Noncoding RNAs and the Evolution of Gene Expression
- The Yeast Rab GTPase Ypt1 Modulates Unfolded Protein Response Dynamics by Regulating the Stability of RNA
- Histone H2B Monoubiquitination Facilitates the Rapid Modulation of Gene Expression during Arabidopsis Photomorphogenesis
- Cellular Variability of RpoS Expression Underlies Subpopulation Activation of an Integrative and Conjugative Element
- Genetic Variants in , , and Influence Male Recombination in Cattle
- Differential Impact of the HEN1 Homolog HENN-1 on 21U and 26G RNAs in the Germline of
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Guidelines for Genome-Wide Association Studies
- The Role of Rice HEI10 in the Formation of Meiotic Crossovers
- Identification of Chromatin-Associated Regulators of MSL Complex Targeting in Dosage Compensation
- GWAS Identifies Novel Susceptibility Loci on 6p21.32 and 21q21.3 for Hepatocellular Carcinoma in Chronic Hepatitis B Virus Carriers
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



