-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
We have followed the evolution of the retroviral restriction gene, Fv1, by functional analysis. We show that Fv1 can recognize and restrict a wider range of retroviruses than previously thought including examples from the gammaretrovirus, lentivirus and foamy virus genera. Nearly every Fv1 tested showed a different pattern of restriction activity. We also identify several hypervariable regions in the coding sequence containing positively selected amino acids that we show to be directly involved in determining restriction specificity. Our results strengthen the analogy between Fv1 and another capsid-binding, retrovirus restriction factor, TRIM5α. Although they share no sequence identity they appear to share a similar design and appear likely to recognise different targets by a mechanism involving multiple weak interactions between a virus-binding domain containing several variable regions and the surface of the viral capsid. We also describe a pattern of constant genetic change, implying that different species of Mus have evolved in the face of ever-changing retroviral threats by viruses of different kinds.
Published in the journal: . PLoS Pathog 10(3): e32767. doi:10.1371/journal.ppat.1003968
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003968Summary
We have followed the evolution of the retroviral restriction gene, Fv1, by functional analysis. We show that Fv1 can recognize and restrict a wider range of retroviruses than previously thought including examples from the gammaretrovirus, lentivirus and foamy virus genera. Nearly every Fv1 tested showed a different pattern of restriction activity. We also identify several hypervariable regions in the coding sequence containing positively selected amino acids that we show to be directly involved in determining restriction specificity. Our results strengthen the analogy between Fv1 and another capsid-binding, retrovirus restriction factor, TRIM5α. Although they share no sequence identity they appear to share a similar design and appear likely to recognise different targets by a mechanism involving multiple weak interactions between a virus-binding domain containing several variable regions and the surface of the viral capsid. We also describe a pattern of constant genetic change, implying that different species of Mus have evolved in the face of ever-changing retroviral threats by viruses of different kinds.
Introduction
Viruses co-evolve with their hosts, upon which they are completely dependent for replication. As the host acquires strategies to restrict virus infection the invaders develop counter measures to evade restriction. The ensuing genetic conflict can play out over an extensive timeframe [1], [2], [3], [4]. Due to the unique replication strategy employed by retroviruses where integration of viral genetic information into the host genome occurs [5], the conflict between virus and host can take an interesting twist. When integration occurs in germ or embryonic cells, the virus can become an endogenous retrovirus (ERV) and inherited through the germ line [6], [7]. As a result, viral gene products can be conscripted to serve as defensive forces against further viral infection [8]. The murine retrovirus restriction gene, Fv1, provides perhaps the prototypic example of one such gene [9].
Fv1 restriction was first described in the early 1970s [10], [11] as an activity protecting mice against infection with murine leukemia virus (MLV). Two semi-dominant alleles were identified, Fv1n and Fv1b, that provide protection against B-tropic and N-tropic MLVs, respectively [12], [13]. The crucial difference between N-tropic and B-tropic MLV maps within the viral gag gene to a single codon encoding amino acid 110 of the mature capsid (CA) protein [14] indicating that CA represents the target for the restriction factor. MLVs insensitive to Fv1, called NB-tropic, carry further changes in CA [15], [16]. The mode of action of the Fv1 protein is not fully understood but indirect evidence suggests that it binds to CA on the cores of incoming virions shortly after virus entry into the cell without inhibiting viral reverse transcription [17] but somehow preventing entry of newly synthesized viral DNA into the nucleus [9]. Based on sequence similarity, Fv1 appears to be derived from the gag gene of an ancient ERV called MERV-L (murine endogenous retrovirus with a leucine tRNA primer binding site) though it appears only distantly related to MLV [18].
Amino acid 110 of CA also determines sensitivity of MLV to another retrovirus restriction factor, TRIM5α [19], best known for its ability to restrict HIV-1 [20]. While there is no similarity between Fv1 and TRIM5α at the primary sequence level, both molecules share a similar domain organization [9]. The N-terminal domains both contain an essential coiled coil motif involved in multimerization while the respective C-terminal domains are required for specific virus binding [21], [22]. Indeed, the C-terminal domain of Fv1 can be replaced with CypA, a molecule that binds HIV-1 CA, resulting in a factor that restricts HIV-1 [23]. TRIM5 has been isolated from a number of mammals including a variety of primates, rabbits and cows [20], [24], [25], [26], [27]. These have been shown to restrict a range of retroviruses from different genera. In particular, TRIM5 from the cotton top tamarin can restrict gammaretroviruses, lentiviruses, spumaviruses and betaretroviruses [24], [28], [29]. Comparison of the target sequences show little identity and although the gammaretroviral, betaretroviral and lentiviral CA molecules show a similar tertiary structure [30] the spumavirus target is folded very differently [31]. Residues in the C-terminal B30.2 domain of TRIM5α that determine viral recognition, and thus restriction specificity, are under strong positive selection [32], [33] and are thought to evolve under pressure imposed by retroviral infection [3], [34], [35]. However in no case have the viruses involved been identified unambiguously [36], [37], [38].
In contrast, changes in Fv1 and the acquisition of its antiviral activity are less well defined. Based on its distribution in different subgenera of Mus, it appears that the Fv1 gene was inserted around 4–7 million years ago [39], [40]. However this finding is somewhat paradoxical because the only known target for Fv1, MLV, probably arose considerably more recently as judged by the distribution of its endogenous forms [41], [42]. What then drove the spread and survival of the Fv1 open reading frame? Could it be that viruses other than MLV selected for Fv1? To address this question we have developed a panel of Fv1 genes from different mice and investigated their anti-viral activity against a variety of retroviruses. These studies reveal an extraordinary degree of plasticity in the Fv1 gene as well as two non-MLV viral targets suggesting that a number of different viruses have moulded its evolution.
Results
Isolation of Fv1 from wild mice
To study the evolution of Fv1, we set out to clone the gene from a variety of species of Mus. Consistent with previous reports [39], [40], it proved possible to clone Fv1 from multiple species of the subgenus Mus as well as single examples of the subgenera Mus nannomys and Mus pyromys (Table 1). However, we failed to amplify Fv1 from Mus coelomys, Apodemus and Rattus despite multiple attempts [43], suggesting that the insertion leading to Fv1 arose about five million years ago, at the time when the ancestors of Pyromys, Nannomys and Coelomys diverged [44]. Sequencing revealed open reading frames in all cases except Mus mus terricolor (dunni) and M. m. cookii (Figure S1). In M. m terricolor, this was due to a single base pair deletion at position 224 that causes a frameshift and premature stop, while in M. m. cookii, a base pair transition from C to T at position 650 coupled with a 5 base pair deletion causes the formation of a premature stop codon. Interestingly, in three cases, M. m. molossinus, M. m. spretus and M. m. caroli, 2 different sequences were amplified in reproducible fashion. Pairs clustered together in phylogenetic analyses, suggesting the presence of more than one segregating allele in these subspecies of mice.
Tab. 1. Sources of Fv1 clones. 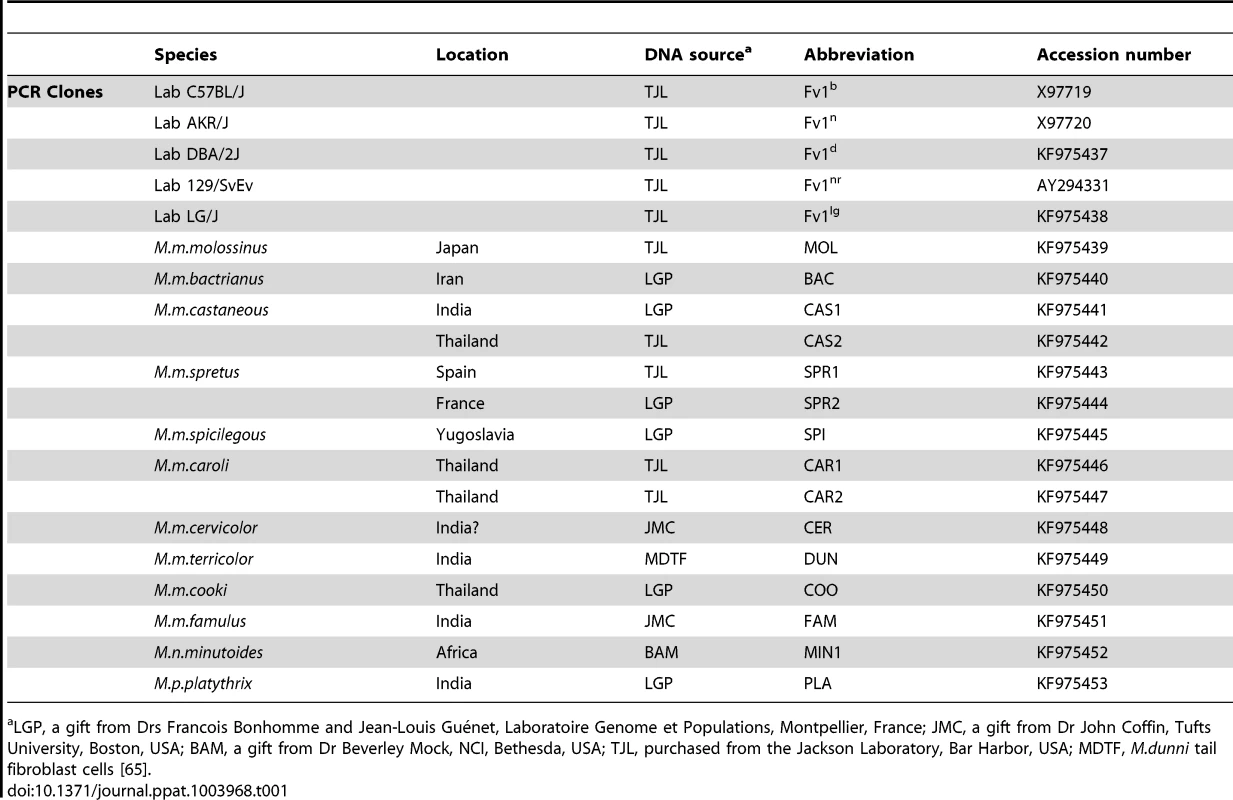
LGP, a gift from Drs Francois Bonhomme and Jean-Louis Guénet, Laboratoire Genome et Populations, Montpellier, France; JMC, a gift from Dr John Coffin, Tufts University, Boston, USA; BAM, a gift from Dr Beverley Mock, NCI, Bethesda, USA; TJL, purchased from the Jackson Laboratory, Bar Harbor, USA; MDTF, M.dunni tail fibroblast cells [65]. Sequence comparisons reveal that the N-terminal region of Fv1, which encodes an extended coiled coil region necessary for restriction activity [23], [45], is well conserved (Figure S1). Compared to M. m. caroli, M. m. famulus, Mus nannomys minutoides and Mus pyromus platythrix, all the other Fv1s contained a 3 amino acid insertion near the N-terminus (Figure S1). This change was not important for restriction activity. By contrast, the C-terminal domain shows significant variation in regions important for Fv1 function. Four variable regions, which we designate VA–D can be distinguished (Figure 1). The first variable region (residues 247–276) overlaps a sequence called the Major Homology Region (MHR) that is present in the CA protein of all retroviruses as well as Fv1 [18], [46], [47] and is essential for Fv1 function [48]. Variable regions B–D (amino acids 345–358, 375–401 and the extreme C-terminus) contain the residues we had previously shown to distinguish the predicted products of the n and b alleles of Fv1. These differences are found at amino acids 358, 399 and the very C-terminus of the Fv1 protein where an apparent deletion of 1.3 kb in genomic DNA resulted in a nineteen amino acid length difference [18]; together they appear responsible for the differences in restriction specificity [48]. The present analysis showed that the more divergent mice contained the residues that are found in Fv1n at positions 358 and 399 but they did not contain the 1.3 kb deletion. This suggested that Fv1n arose from the progenitor Fv1, which was similar in length to Fv1b, through an internal deletion, while Fv1b evolved through the substitution of the residues at positions 358 and 399.
Fig. 1. Features of Fv1. 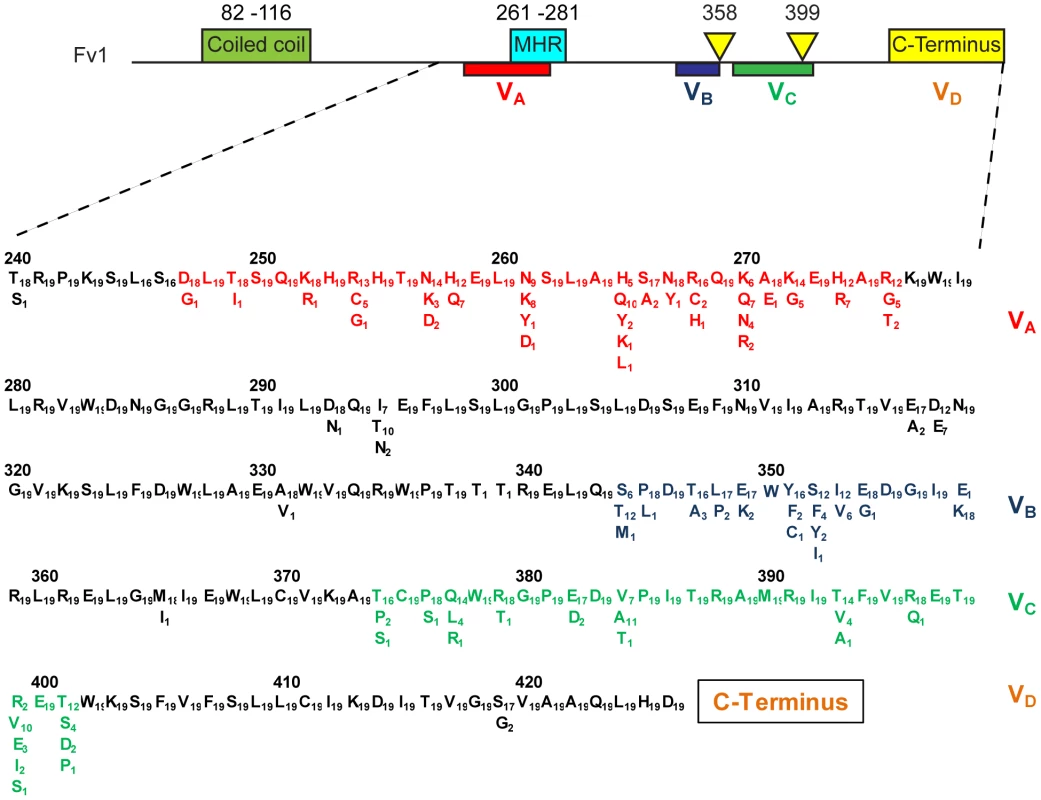
At the top of the figure is a schematic of the Fv1 protein showing the relative positions of the previously mapped functional domains including coiled coil and major homology regions as well as the host range specificity regions previously defined by a comparison of Fv1n and Fv1b [48]. Below is shown the positions of four variable regions (VA–D), the amino acid differences that define them and the number of times each amino acid occurs. Based on a comparison of 19 mice (Table 1 plus Table 2). Variable regions A–C appear to arise by point mutation but region D shows more significant changes in nucleotide sequence. The three most distantly-diverged mice, M. n. minutoides, M. m. famulus and M. p. platythrix each appear to have B1 repeat sequences inserted, apparently independently, near the deletion site that gave rise to the Fv1n allele (Figure 2). They contribute the last few amino acids of Fv1 resulting in C-termini that are rather different from either Fv1n or Fv1b. Other differences in this region arise from short insertions or deletions perhaps resulting from polymerase slippage during DNA replication. Thus the clones, SPR1 and SPR2, we amplified from M. m. spretus of French and Spanish origins differed by four amino acids; the same difference also seen between Fv1b and CAS2 (Figure 2).
Fig. 2. The C-terminal region of selected Fv1 proteins. 
Nucleic acid and predicted amino acids found at the C-terminus of selected Fv1 genes. The twelve nucleotides shown in purple for Fv1n are found 1.3 kb downstream in Fv1b [18]. Sequences shown in blue (FAM, MIN) correspond to nucleotides 1–27 of the consensus B1 repeat [69]. The sequences in red (PTX) might also come from a rearranged B1 repeat as they correspond to nucleotides 30–44 and 56–65 of the consensus sequence. The nucleotide in orange corresponds to a unique point mutation found in Mus m. spretus resulting in a novel stop codon. Different restriction activities of wild mice Fv1
By analogy with other restriction factors it seems likely that the variation seen in Fv1 arose by selection and this would be reflected in changes in restriction specificity. To test this we examined the restriction properties of Fv1 we had cloned from different mice (Table 1) as well as a number of genes synthesized on the basis of published sequences (Table 2) [40], [49]. A tree based on these sequences is shown in Figure 3; it shows good agreement with the accepted phylogeny of genus Mus [50]. The Fv1 genes were introduced into pLgatewayIYFP and tested for restriction of different MLVs using a two-colour FACS assay [19], [51]. The results are shown in Table 3.
Fig. 3. Phylogenetic tree of Fv1 sequences. 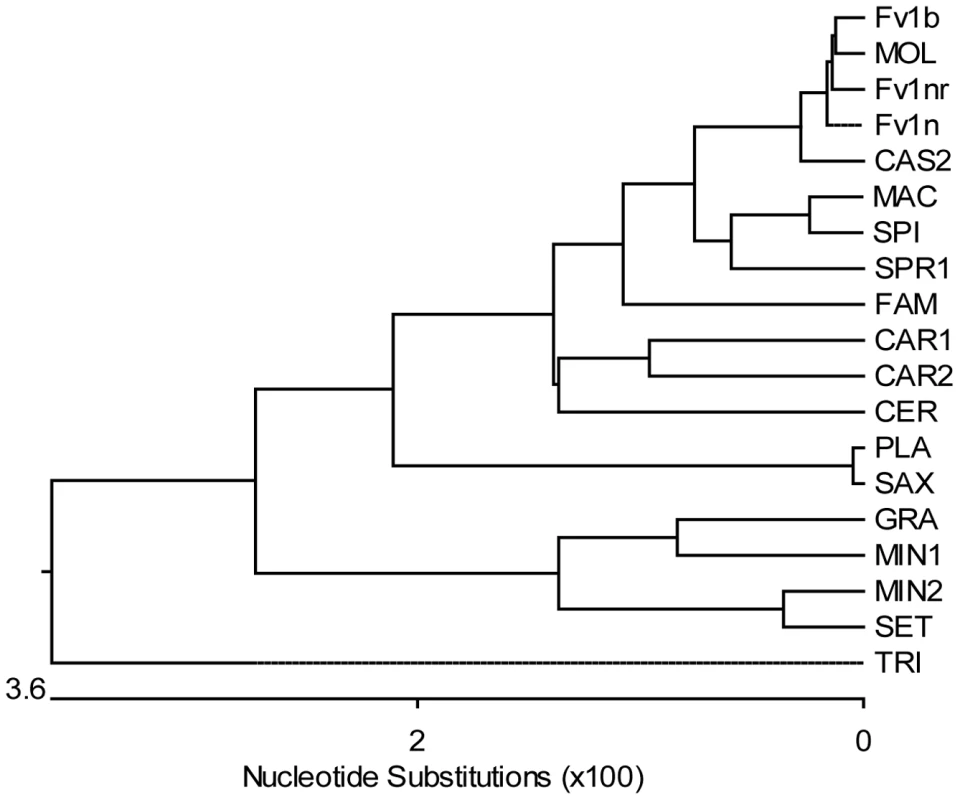
The tree was generated from the open reading frames listed in Tables 1 and 2 (bases 1 to 1278 in Fv1b) using the MegAlign programme from the DNASTAR Lasergene package. The highly divergent C-terminus was excluded from the analysis and the aspartic acid residue at position 426 (Fv1b numbering), which was the last residue conserved in all the sequences, was chosen as the cut-off point. The number of substitution events is shown at the bottom of the tree while the distances between sequence pairs is represented by the length of the branch pairs. The distance values were calculated using the Kimura distance formula that takes into account the number of non-gap mismatches and silent substitutions. Tab. 2. Sources of Fv1 synthesized clones. 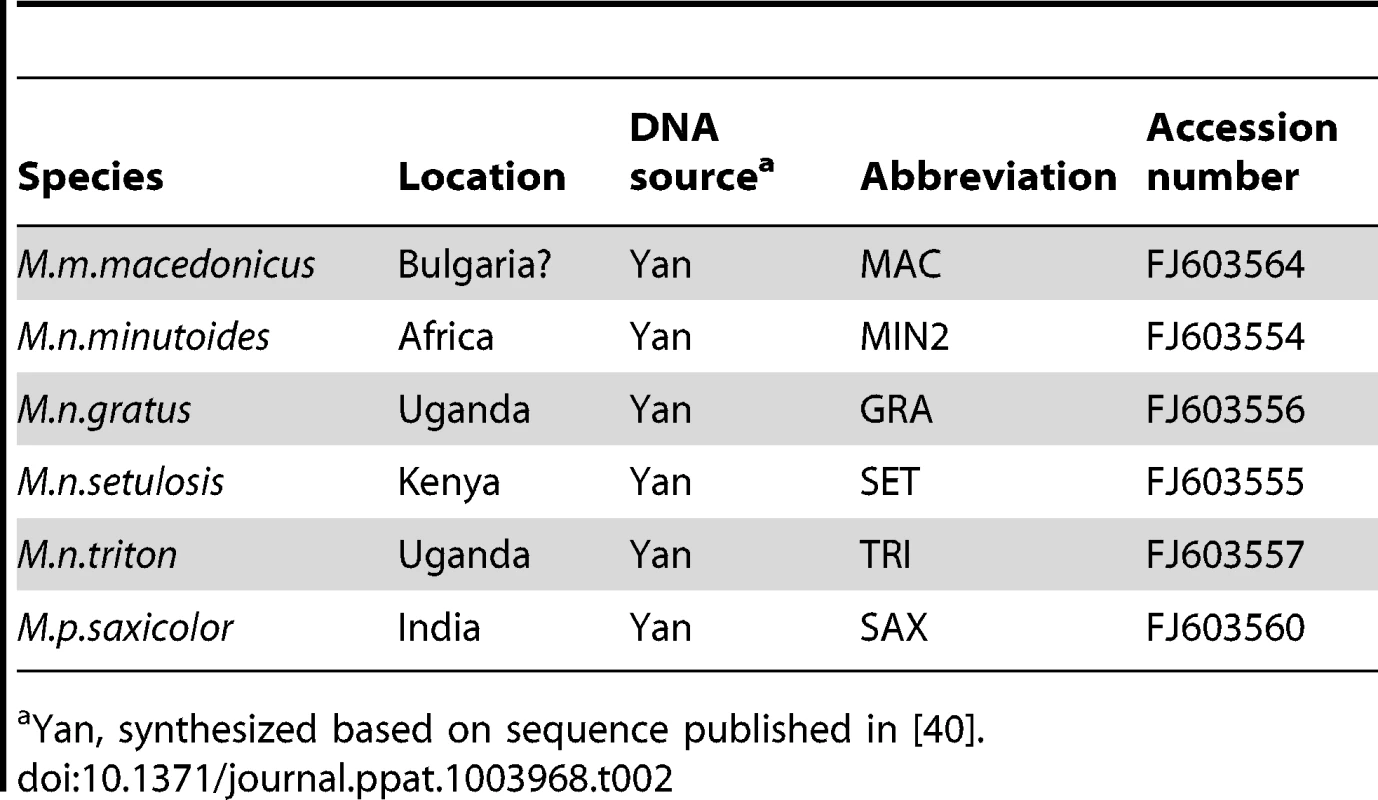
Yan, synthesized based on sequence published in [40]. Tab. 3. Restriction activity of various Fv1s against different MLVs. 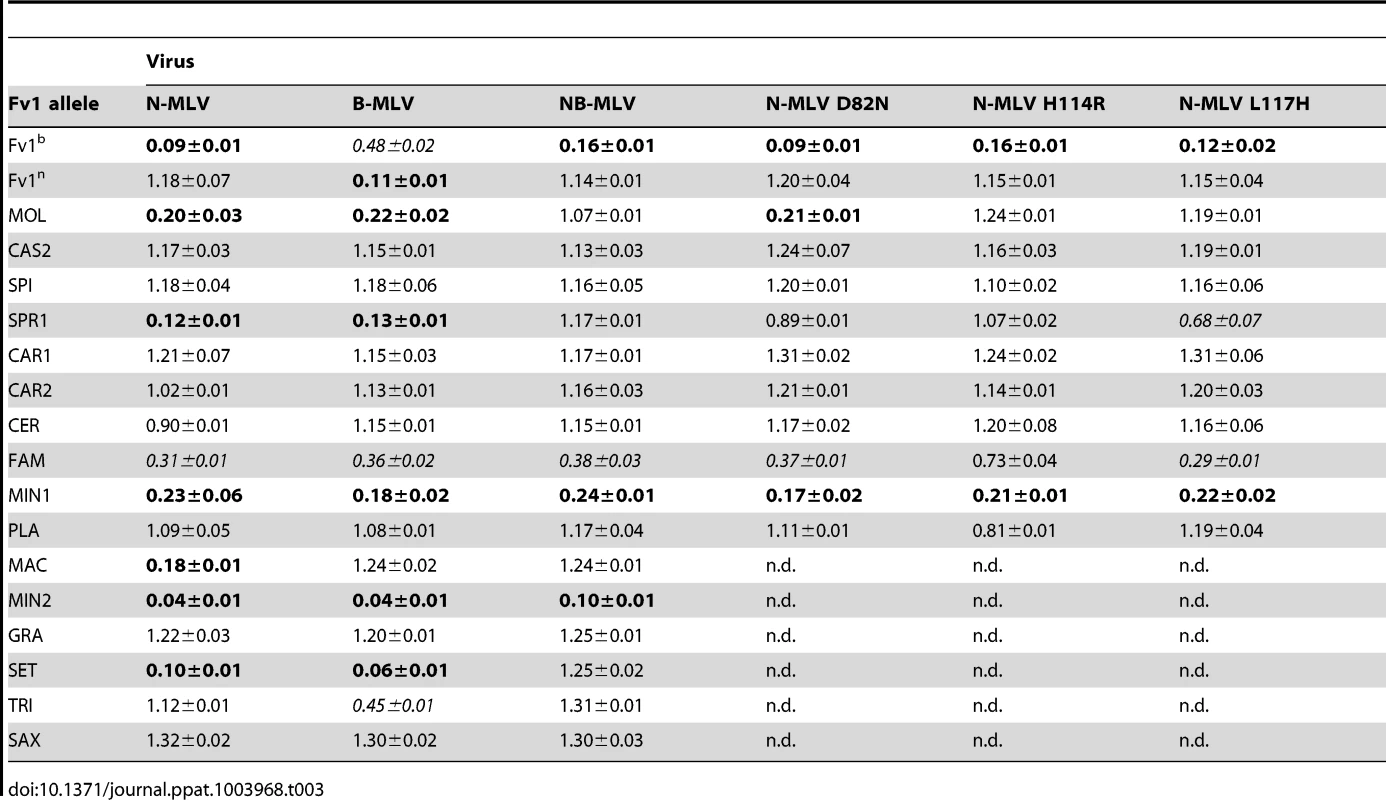
As previously shown, the Fv1n gene restricted B-MLV but not N - or NB-MLV while Fv1b, when expressed at protein levels seen in transduced cells, restricted N - and NB - MLV and also had a weak activity against B-MLV [51]. The Fv1 gene from M. m. molossinus and M. m. spretus restricted both N - and B-MLV but not NB-MLV, suggesting that they could be similar to Fv1nr [16]. Hence, we also examined three N-MLV CA variants (D82N, H114R and L117H) that confer resistance to Fv1nr [16]. Variants N-MLV H114R and N L117H, which we have found to be NR tropic, were also resistant to both the Fv1MOL1 and Fv1SPR1 proteins. In contrast, N-MLV N82D was restricted by Fv1MOL1 but not by Fv1SPR1, suggesting that Fv1MOL1 is subtly different from Fv1nr. We have cloned the Fv1nr gene from 4 different strains of mice (129SvEv, NZB/B1NJ, NZW/LacJ and RF/J); all contained a single nucleotide change compared to Fv1n, causing a serine to phenylalanine substitution at residue 352. While Fv1MOL1 also encoded phenylalanine at position 352, Fv1SPR1 possesses a serine at the corresponding position. Taken together, these results suggest that other changes could also be involved in determining the nr-specificity. Perhaps surprisingly, Fv1 from two closely related species, M. m. castaneus and M. m. spicelegus lacked perceptible Fv1 activity against MLV.
Other Fv1 genes displayed a variety of restriction phenotypes. Fv1 from two asian members of the Mus subgenus, M. m. caroli and M. m. cervicolor, lacked Fv1 activity directed against MLV as did the two members of the Pyromys subgenus that we tested, M. p. platythrix and M. p. saxicolor (Table 3). By contrast two members of the Nannomys subgenus, M. n. ninutoides and M. n setulosis were active with the M. n. minutoides clones restricting all six MLVs tested. The M. m. famulus sample, whose position in Mus phylogenetic trees is relatively poorly defined, showed weak activity against N, B, and NB-tropic MLVs. Thus more than half of the Fv1 genes with intact open reading frames did not seem to have any activity against MLV, the target that defines the Fv1 gene, even though they were expressed at similar levels to restricting genes in transduced cells (Figure S2). Further, the extent and specificity of restriction of different MLVs varies significantly. Clearly, the properties of the restriction gene have changed since the gene first became part of the mouse germ line but whether MLV alone was responsible for selecting such changes remained an open question.
Novel restriction specificities of wild mice Fv1
Prompted by the example of TRIM5α that can restrict multiple genera of retrovirus [24], [29], we decided to investigate the hypothesis that non-MLV retroviruses might play a role in shaping the evolution of Fv1, by testing a number of different retroviral vectors for restriction by Fv1 from wild mice. These included other gammaretroviruses like Gibbon Ape Leukemia Virus (GALV), Feline leukemia Virus (FeLV) and Porcine Endogenous Retrovirus-A (PERV-A), lentiviruses such as HIV-1, HIV-2, SIVmac, Equine Infectious Anemia Virus (EIAV) and Feline immunodeficiency Virus (FIV), as well as foamy viruses including Prototypic Foamy Virus (PFV), Simian Foamy Virus (SFV) and Feline Foamy Virus (FFV). Some of these results are presented in Table 4. The data show that Fv1 from M. m. caroli, that lacked activity against MLV, restricted FFV strongly and PFV weakly. Moreover, Fv1 from M. m. spretus, which restricted N - and B-MLV, and from M. m. macedonicus, which inhibited N-MLV, were also active against the lentivirus EIAV. By contrast GALV, FeLV, PERV-A, SFV, HIV-1, SIVmac and FIV were not restricted by any of the Fv1 genes in the panel (Table 4 and data not shown). Formally it remains possible that the novel specificities observed result from over expression. Unfortunately, no cell lines expressing Fv1CAR1 and Fv1SPR1 at endogenous levels are available, precluding a direct test of this idea. However we are not aware of any examples of complete restriction of novel viruses resulting from such a mechanism.
Tab. 4. Restriction activity of various Fv1s against different viruses. 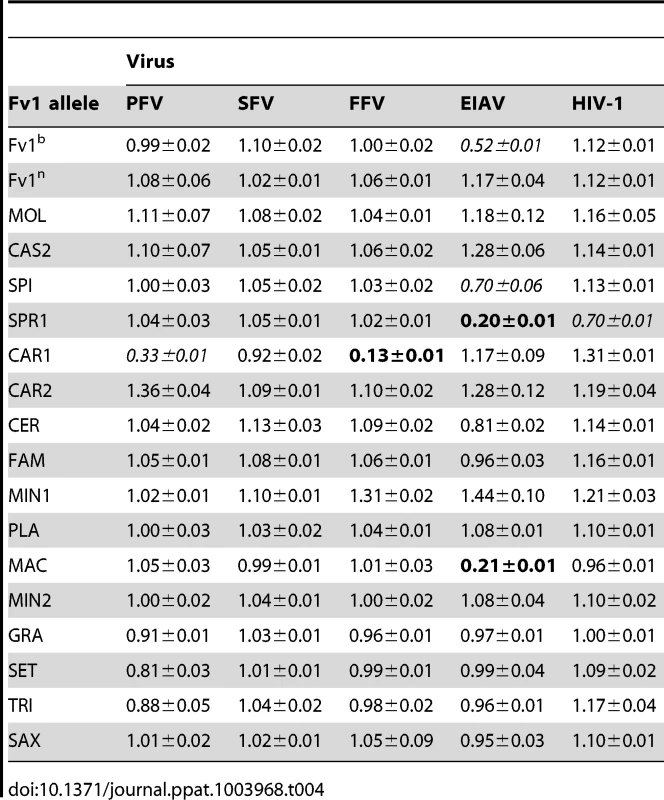
To further characterize restriction mediated by Fv1CAR1 and Fv1SPR1 stable cell lines were derived by transducing MDTF cells with retroviral vectors carrying these genes and selecting for G418-resistant single cell clones. These cell lines were used in virus titrations by measuring the percentage of transduced cells by FACS with different amounts of virus (Figure 4A,B). As expected, the titre of EIAV was dramatically reduced in the cell line expressing Fv1SPR1 compared to the untransduced MDTF control (Figure 4A). Similarly titres of FFV and PFV were greatly reduced in MDTF cells expressing Fv1CAR1 compared to untransduced while titres of SFV were unaffected by the presence of the Fv1 gene (Figure 4B). These results confirm the observations made with the 2 colour FACS assay that Fv1 from some wild mice can restrict non-MLV retroviruses.
Fig. 4. Staging restriction blocks in novel Fv1s. 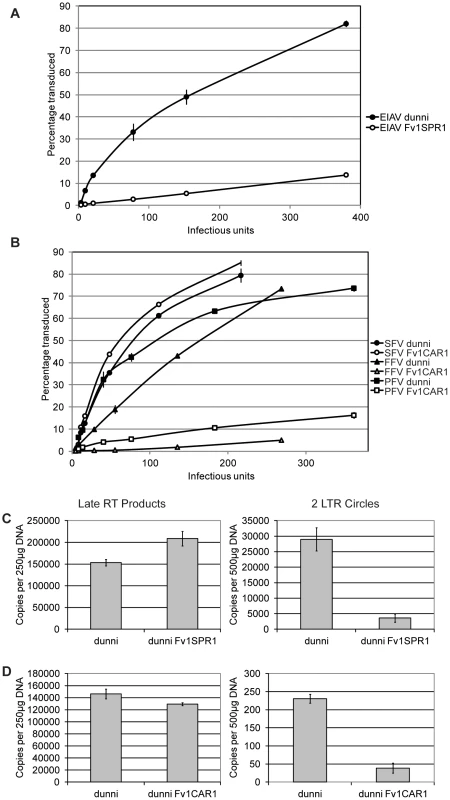
MDTF cells were transduced with Fv1CAR1 and Fv1SPR1, then stable Fv1-expressing cell lines selected and tested for restriction of virus replication by FACS (A, B) or by PCR to measure viral DNA synthesis or formation of circular viral DNA containing two LTRs (C, D). Fv1 is thought to interfere with MLV replication by preventing nuclear import of newly synthesized viral DNA [9]. To test whether this was also true for EIAV and FFV, we examined the fate of viral DNA in restricting cell lines. Testing EIAV replication in Fv1SPR1 cells shows no inhibition of reverse transcription as measured by levels of newly synthesized late DNA products (Figure 4C). However levels of 2-LTR circles, which are thought to form only after nuclear entry [52], are substantially reduced suggesting a block in nuclear uptake. In Fv1CAR1 cells a reduction in FFV 2-LTR circles, with no change in late RT products, was also observed (Figure 4D), again consistent with a block in nuclear import. However, interpretation of these data is complicated by the fact that the majority of FFV DNA synthesis is thought to occur in the producer cells [53]. Nevertheless, it appears likely that Fv1 is acting to block lentivirus and foamy virus replication at the same stage in the viral life cycle as seen with MLV.
Mapping the specificity determinants of the novel restriction activities
To identify the specificity determinants of these novel restriction activities, chimeric Fv1 genes were constructed and tested for restriction. To look at FFV restriction, we made chimeras between Fv1CAR1, which restricted only FFV, and Fv1n, which restricted B-MLV. Schematic views of the constructs made and the corresponding restriction data are shown in Figure 5A. Replacement of a C-terminal fragment of Fv1n (from residue 318) with the corresponding fragment from Fv1CAR1 generated a chimera (Fv1nC4) capable of restricting FFV. Replacement with a shorter fragment starting from residue 353 (Fv1nC5) was insufficient to confer restriction, suggesting that the determinants of FFV restriction were found between residues 316 and 352 of Fv1 from M. m. caroli. In the reciprocal chimeras, replacing the small C-terminal segment of Fv1CAR1 beginning from residue 352 with that from Fv1n did not result in any loss of activity against FFV. However, when a larger fragment beginning at residue 316 was replaced, activity was lost, confirming the presence of the determinants of FFV restriction within the region of Fv1CAR1 between residues 316 and 352. Within this region, there are 5 residues that differ between Fv1n and Fv1CAR1. These were systematically changed to identify the residues involved in specificity determination (Figure 5B). No single change could endow Fv1n with the ability to restrict FFV (Figure 5B). However two single changes at positions 349 and 352 of Fv1CAR1 resulted in loss of FFV restriction. We therefore mutated both these positions in Fv1n to the corresponding amino acids found in Fv1CAR1. This generated a construct (Fv1nE349KS352Y) capable of restricting FFV. Taken together, these results indicate that both lysine 349 and tyrosine 352 in Fv1 from M. m. caroli are crucial for FFV restriction. We had previously shown that MLV recognition maps downstream of this region [48]; it was therefore interesting to see that Fv1nE349KS352Y (and chimera Fv1Cn5) could recognize both B-MLV and FFV in an additive fashion.
Fig. 5. Mapping the determinants of FFV restriction by Fv1 from M. m. caroli. 
(A) Analysis of restriction by chimeric Fv1 constructs. (B) Analysis of restriction by site directed mutant forms of Fv1. To examine EIAV restriction by Fv1 from M. m. spretus, a second set of chimeras was made between Fv1SPR1, which restricts N-MLV, B-MLV and EIAV, and Fv1n, which only restricts B-MLV. Restriction of EIAV was seen with chimeras only when amino acids from positions 191 and 271 were derived from Fv1SPR1 (Fig. 6A) suggesting that the determinants of EIAV restriction lay between these residues. Interestingly, the determinants for MLV restriction were slightly different from those of EIAV. Replacing a short segment of C-terminus of Fv1n (from residue 366) with that from Fv1SPR1 in Fv1nS3 was sufficient to confer restriction of N-MLV, suggesting that this region contained determinants of N-MLV restriction. However, a reciprocal change in Fv1SPR1 (Fv1Sn3) did not abolish N-MLV restriction. It was only when a C-terminal segment beginning with residue 191 was replaced from Fv1SPR1 (Fv1Sn1) that the restriction of N-MLV was lost. This suggested that additional requirements for N-MLV restriction were found between residues 191 and 271 of Fv1 from M. m. spretus, perhaps overlapping with those that determined EIAV restriction.
Fig. 6. Mapping the determinants of EIAV restriction by Fv1 from M. m. spretus. 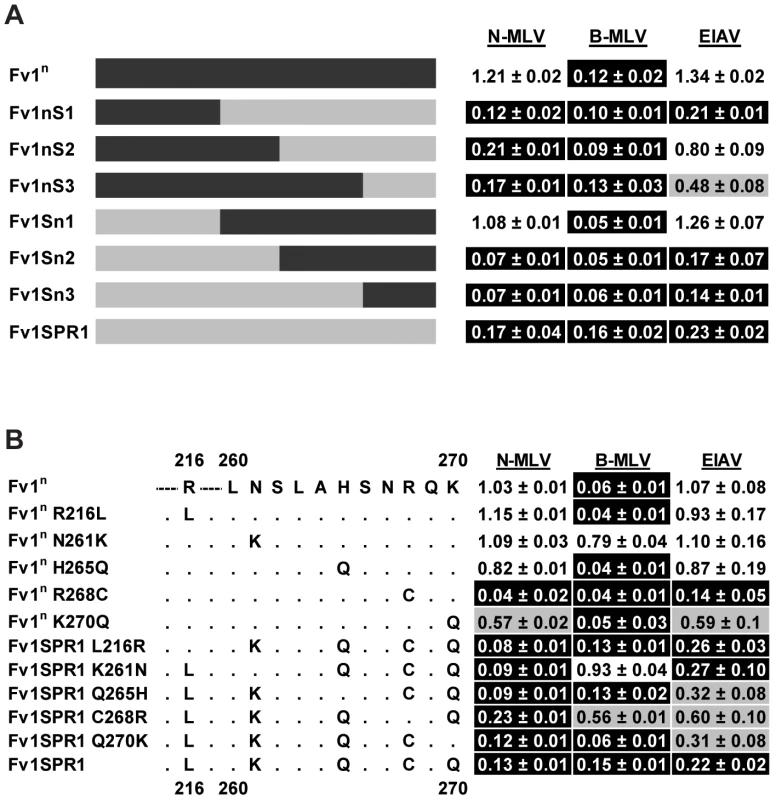
(A) Analysis of restriction by chimeric Fv1 constructs. (B) Analysis of restriction by site directed mutant forms of Fv1. There are 5 differences between Fv1n and Fv1SPR1 in the segment between residues 191 and 271 (Figure 6B). To identify the residues involved in restriction, site-directed mutagenesis was employed to change the residues in Fv1n to those present in Fv1SPR1. Reciprocal mutations were also made in Fv1SPR1. These mutants were tested for restriction of EIAV, N - and B-MLV. The substitution from arginine to cysteine at position 268 in Fv1n was sufficient to confer the ability to restrict both N-MLV and EIAV. The reciprocal change in Fv1SPR1 resulted in a partial reduction in restriction of all three viruses. These results indicated that residue 268 was the major determinant of EIAV restriction by Fv1SPR1 and had an influence on MLV restriction but that other neighbouring residues were also important. A lysine to glutamine change at residue 270 in Fv1n resulted in low but reproducible restriction of EIAV and N-MLV though the reciprocal change in Fv1SPR1 had little effect. Interestingly, substitution of the residue at position 261 in both Fv1n and Fv1SPR1 seemed to abolish the restriction of B-MLV, indicating that this residue was involved in the interaction with B-MLV. We conclude that residues 261, 268 and 270 in Fv1 from M. m. spretus are all involved in virus recognition. However, it would appear that recognition of EIAV by Fv1 from M. m. macedonicus has arisen in a different manner as it contains arginine rather than cysteine at position 268.
Discussion
In this study of Fv1 evolution we have demonstrated that Fv1 shows substantial sequence variation in its C-terminal half, the region of the protein thought to contain determinants of restriction specificity. In addition we have shown that Fv1 is capable of restricting viruses other than its previously defined targets and identified the sequence variation responsible for these novel targets. We note that some Fv1 alleles do not appear to possess an associated restriction activity; it would be of considerable interest to determine whether they recognize other targets.
A previous study had identified six codons, specifying Fv1 amino acids 261, 265, 270, 362, 299 and 401, that show evidence for positive selection during the course of Mus evolution [40]. These represent potential sites of interaction between Fv1 and its target viruses. Combining these data with our previous studies of Fv1 specificity [16], [48], it seems reasonable to conclude that the four variable regions defined in Figure 1 constitute four domains collectively or individually involved in target selection and binding (Figure 7). Thus VRA (amino acids 247–276) includes the positively selected residues 261, 265 and 270 as well as three residues, 261, 268 and 270, shown to be important for EIAV restriction by Fv1SPR1 while VRB (amino acids 345–358) has positively selected amino acid 352, amino acids 349 and 352 important for FFV recognition by Fv1CAR1 as well as residues 352 and 358 important for NR - and N - versus B-tropism, respectively [16], [48]. Variable region C (amino acids 375–401) contains positively selected amino acids 399 and 401 while residue 399 was also implicated in determining N - versus B-tropism [48]. The nature of the length variation at the C-terminus precludes computational analysis for positive selection; nevertheless functional studies [48] provide compelling evidence that this region can also alter restriction specificity.
Fig. 7. Properties of the variable regions of Fv1. 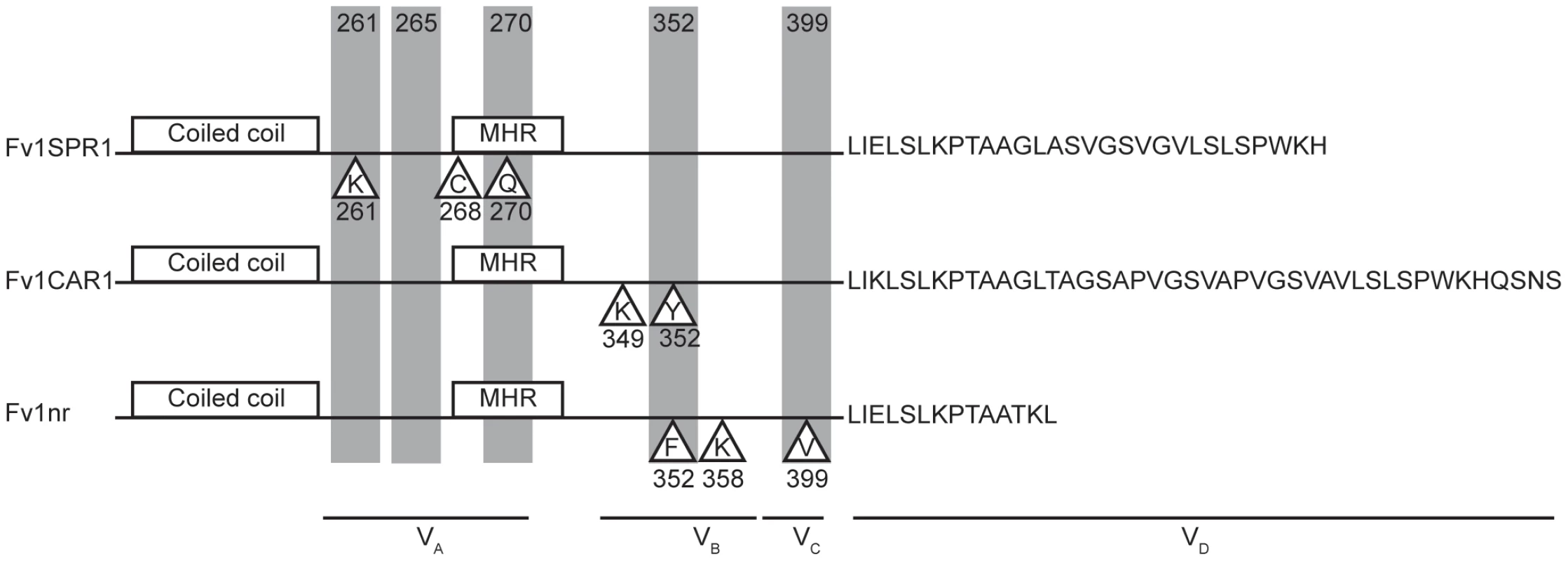
Proteins corresponding to the Fv1SPR1, Fv1CAR1 and Fv1nr alleles are illustrated. Shaded positions indicate amino acids showing positive selection [40], residues in triangles have been implicated in restriction specificity (this paper, [16], [48]). We have previously noted that CA binding restriction factors Fv1 and TRIM5α share certain design features despite lack of sequence similarity [9]; the present study strengthens this analogy. Both factors possess an N-terminal coiled-coil region allowing dimer formation. They also contain other sequences facilitating the formation of higher order multimers. Both contain a C-terminal domain responsible for virus binding that can be substituted with the cellular CA binding cyclophilin A protein to give a fusion protein capable of restricting HIV-1 and other lentiviruses [23], [54]. We now provide evidence that the CA binding domain of Fv1, like TRIM5α [24], [32], [55], [56], appears to comprise multiple variable regions, showing attributes of positive selection, implying virus driven evolution [3]. Further, Fv1 is capable of recognizing multiple genera of retrovirus. It seems possible that the ability to recognize multiple viruses by low affinity binding with avid binding provided by multimerisation [57] represents a common theme in restriction factor design. Further insights into the interaction between virus and restriction factor requires detailed structural information; unfortunately both Fv1 and TRIM5α are relatively recalcitrant to such studies.
The origin of Fv1 remains unclear. It is only present in Mus and appears related to the gag gene of the endogenous retrovirus family ERV-L [18], [47]. This suggests that Fv1 might be derived from an endogenous retrovirus following the loss of both LTRs and pol coding sequences [58]. Interestingly a significant increase in MERV-L copy number took place at around the time of the separation of Mus subgenera [59], the time when Fv1 became part of the Mus germline. However sequence alignments indicate that Fv1 and MERV-L share only 43% amino acid identity whereas the different genomic MERV-L elements are much more closely related to one another (<5% nucleic acid divergence). BLAST searches of the NCBI non-redundant genome databases reveal no sequences intermediate between Fv1 and MERV-L. This suggests that Fv1 might be derived from an exogenous virus related to ERV-L that has not made its home as an intact ERV, at least not in any species so far sequenced, and may no longer exist in infectious form. As such Fv1 might be the last remnant of an ancient extinct virus, or paleovirus [2]. Unfortunately this inability to identify the proximal precursor for Fv1 prevents us from determining whether or not the original transgene showed restriction activity and, if so, against which virus.
The selection and continuing existence of the Fv1 open reading frame implies that it provides an evolutionary advantage, presumably by providing protection against retroviral infection. The observation of multiple restriction specificities suggests that a variety of unknown viruses have contributed to this process. Taken together with frequent genetic changes to inactivate [60] or block MLV receptors [61], these data imply that multiple virus epidemics have occurred in the course of mouse evolution [62]. One might postulate that at least four significant virus exposures have occurred during Mus evolution (Figure 8). One took place after the divergence of Nannomys; a second occurred in M. m. caroli; a third in mice in countries surrounding the Mediterranean Sea and a fourth in the Mus musculus subfamily. In turn this prompts the question of how the current properties of a restriction factor reflect the properties of the viruses involved in selection, a question that is as relevant for TRIM5α as for Fv1. Specifically one might ask whether the ability to restrict one genus of retrovirus reflects prior exposure to that kind of virus. An affirmative answer might resolve the vexed question of whether foamy viruses have deleterious effects on their hosts [63], possibly as co-pathogens [64] since both Fv1 (this paper) and TRIM5α [29] have evolved to see one or more such virus. Alternatively, changes selected by, say, a gammaretrovirus like MLV, might fortuitously result in recognition of a lentivirus like EIAV or a foamy virus like FFV. In light of the shorter generation time of mice compared to primates Fv1 could provide a more useful system for studying evolution of restriction specificity than does TRIM5α. The observation of multiple alleles of Fv1 might also suggest that selection is an ongoing process offering opportunities for experimental analysis. In particular, the evolution of restriction activity against the lentivirus EIAV, which appears to have happened in two different ways in M. m. spretus and M. m. macedonicus as well as the kind and source(s) of the virus(es) involved would appear worthy of more detailed investigation.
Fig. 8. Events in the evolution of the Fv1 gene. 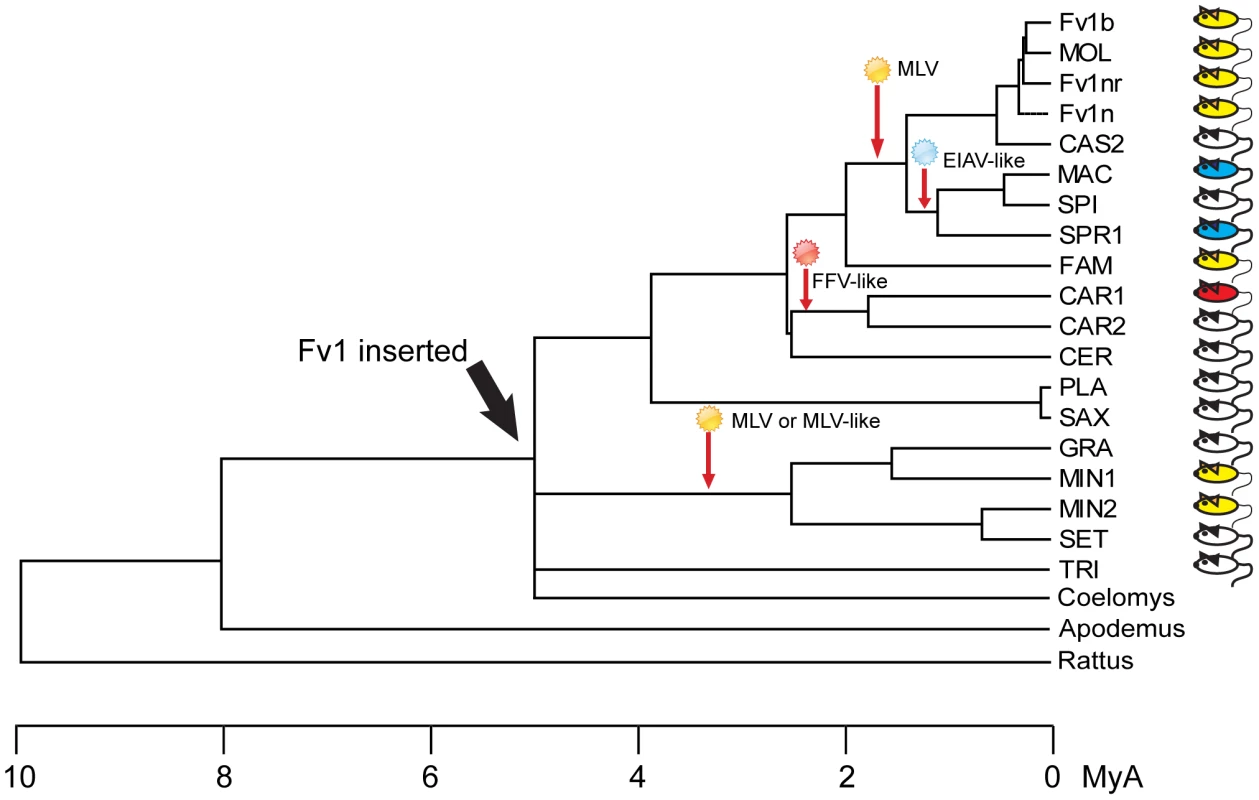
A phylogenetic tree showing the approximate times of Fv1 acquisition and hypothetical virus infections leading to selection of new retroviral restriction activities. Colored mice indicate Fv1 activity against at least one virus. Materials and Methods
Fv1 cloning
Genomic DNA samples for Mus musculus laboratory mouse strains C57BL/6J, AKR/J, DBA/2J, 129/SvEv, and LG/J, M. m. spretus (M. spretus), M. m. caroli (Mus caroli), M. m. molissinus (MOLD/Rk) and M. m. castaneous were purchased from the Jackson Laboratory. Genomic DNA from M. p. platythrix, M. m. cookii, M. m. spicilegus, M. m. spretus, M. m. castaneous and M. m. bactrianus were gifts from Dr. F. Bonhomme (Laboratoire Genome et Populations, Universite de Montpellier II, CNRS). M. n. minutoides genomic DNA was a gift from Dr. B. Mock (National Cancer Institute, NIH), while M. m. famulus and M. m. cervicolor genomic DNA were gifts from Dr. John Coffin (Tufts University School of Medicine, Boston). M. m. terricolor (dunni) genomic DNA was prepared from a Mus dunni tail fibroblast (MDTF) line [65] using the DNeasy blood and tissue kit (Qiagen). The Fv1 ORF was PCR amplified from mouse genomic DNA using primers PL80 and GT17 (see Table S1 for primer sequences) that permit the amplification of a sequence starting from 3056 bp upstream of the start codon of Fv1 to 2684 bp downstream of the start codon. Sequence analysis of this region from in-bred mice identified 2 SacI sites downstream of the PL80 primer-binding site, while GT17 contained a SalI site. The PCR products were hence cloned initially as SacI/SalI fragments into M13 phage and sequenced. Subsequent subcloning of Fv1 ORFs was carried out following amplification with the primers GatewayFv1F and Gateway Fv1rev. The PCR product was used in a second amplification reaction with primers UniversalF and UniversalRev to attach the attB sites to the ends of the fragment. This was then inserted into pDNR221, which is an entry vector to the Gateway Cloning system, using BP clonase (Invitrogen). Finally, the entry clone was used in a LR reaction with LR clonase to insert the Fv1 ORF into either pLgatewayIRESEYFP or pLgatewaySN to generate retroviral delivery vectors carrying either the EYFP or G418 resistance marker. Details of these different clones as well as the abbreviations used for their designation are summarized in Table 1.
Fv1 open reading frames from M. m. macedonicus, M. n. minutoides, M. n. gratus, M. n. setulosis, M. n. triton and M. p. saxicolor were synthesized chemically (GENEART, Life Technologies) based on their published sequences [40] with added attB sites and introduced into pLgatewayIRESEYFP via pDNR221. These clones are also summarized in Table 2.
Construction of chimeric Fv1s
Fv1 chimeras were generated by overlapping PCR. Briefly, a 5′ fragment was amplified from one parental sequence while a 3′ fragment was amplified from the other. The two fragments were then combined in a third amplification reaction using forward and reverse primers that annealed to the 5′ and 3′ ends of Fv1 respectively. Internal primer pairs were designed to target regions of identity between the two parental sequences. The sequences of the primers are shown in Table S1.
To generate the Fv1nC series, the 5′ fragments were amplified from Fv1n using TopoFv1F and either C1Rev, C3Rev, C4Rev or C5Rev, while the 3′ fragments were amplified from Fv1caroli (CAR1) using either C1F, C3F, C4F or C5F and Fv1caroliRev. The 2 fragments were joined in a reaction using TopoFv1F and Fv1caroliRev to yield Fv1nC1, Fv1nC3, Fv1nC4 and Fv1nC5. Similarly, the 5′ fragments for the reciprocal series Fv1Cn were amplified from Fv1caroli (CAR1), using the same primer pairs as the Fv1nC series, while the 3′ fragments were amplified from Fv1n using either C1F, C3F, C4F or C5F and Fv1nRev. These fragments were joined using primer pair TopoFv1F and Fv1nRev, yielding Fv1Cn1, Fv1Cn3, Fv1Cn4 and Fv1Cn5.
The 5′ fragments for the Fv1nS series were amplified from Fv1n using TopoFv1F and either S1Rev, S2Rev or S3Rev, while the 3′ fragments were amplified from Fv1spretus (SPR1) using either S1F, S2F or S3F and Fv1spretusRev. The 2 fragments were joined together in a reaction using TopoFv1F and Fv1spretusRev to yield Fv1nS 1, Fv1nS2 and Fv1nS3. Similarly, the 5′ fragments for the reciprocal series Fv1Sn were amplified from Fv1spretus (SPR1), using the same primer pairs as the Fv1nS series, while the 3′ fragments were amplified from Fv1n using either S1F, S2F or S3F and Fv1nRev. These fragments were joined using primer pair TopoFv1F and Fv1nRev, yielding Fv1Sn1, Fv1Sn2 and Fv1Sn3.
The chimeric fragments were cloned into pENTR/D-TOPO (Invitrogen) and verified by sequencing before transferring into the retroviral vector pLgatewayIRESEYFP.
Site directed mutagenesis
The point mutants were generated by site directed mutagenesis using the primer pairs listed in Table S1. Mutagenesis was carried out in 50 microlitre reactions containing 2.5 units of Pfu ultra, 10 ng of template, 0.2 mM dNTP and 125 ng each of the forward and reverse primer. The reaction was performed in a thermal cycler at 95°C for 2 minutes followed by 18 cycles of 95°C for 30 seconds, 55°C for 1 minute and 68°C for 9 minutes 30 seconds. The PCR product was then digested with10 units of DpnI (Roche) for 1 hour before transforming XL10Gold cells (Agilent technologies). Colonies were screened by restriction digest and the mutations were verified by sequencing.
Cells and virus production
MDTF and 293T cells were maintained in DMEM containing 10% foetal calf serum and 1% penicillin and streptomycin. Viruses were made by the transient transfection of 293T cells as previously described [19], [51]. Delivery viruses were produced by co-transfecting pcz-VSVG, pHIT60 and a retroviral vector containing Fv1 and either the EYFP or G418 resistance gene. N-, B - and NB-tropic MLV tester viruses were generated by co-transfection of pczVSVG, pczCFG2fEFPf and either pCIGN, pCIGB or pHIT60 respectively, while the NR-tropic viruses were made using a mutagenized form of pCIGN as previously described [16]. EIAV tester viruses were made using pczVSVG, pONY3.1 and pONY8.4ZCG or pONY4.1Z [66], while PFV, SFV and FFV were produced with pciSFV-1envwt and either pczDWP001, pcDWS001 or pcDWF003 respectively [29]. HIV-1 tester viruses were generated by co-transfecting pczVSVG with p8.91 and pCSGW. MLV and HIV-1 were frozen in aliquots at −80°C while EIAV and foamy viruses were freshly prepared for each experiment.
Restriction assays
Restriction activity was routinely assayed using transient two colour FACS analyses as described previously [19], [51]. Briefly, Fv1 was introduced into MDTF cells together with an EYFP marker in a retroviral delivery vector. Three days post-transduction, the cells were challenged with tester viruses carrying the EGFP markers. The cells were then subjected to FACS analyses three days later and the percentages of tester virus positive cells in EYFP (i.e. Fv1) - positive and - negative cells determined and compared. Ratios of less than 0.3 were taken as restriction while those that were greater than 0.7 were taken to represent no restriction. Numbers between 0.3 and 0.7 were taken to represent partial restriction.
Alternatively, single cell clones stably expressing restricting Fv1s were derived by transducing MDTF cells in 12 well plates with limiting dilutions of retroviral vectors carrying Fv1 and a G418 resistance marker. The cells from each well were transferred to a 10 cm dish and G418 was added to a concentration of 1 mg/ml. Well-separated colonies were picked from the dishes when they appeared 7 to 10 days after antibiotic selection was started. Typically, 6 to 8 colonies were picked for each Fv1 cell line, expanded and tested for restriction before being used for virus titration. To titrate tester viruses, MDTF cells and their derivatives were seeded in 12 well plates at a density of 5×104 cells per well 24 hours prior to infection. Increasing amounts of viruses carrying the EGFP marker were then added to the wells and the percentage of infected cells was determined by FACS 3 days post infection.
Quantitative PCR
MDTF cells and their derivatives stably expressing Fv1 were seeded in 6 well plates at a density of 5×105 cells per well 24 hours prior to infection. The cells were transduced at an m.o.i. of 1 with equal amounts of viral vectors that had been pre-treated with 10 units/ml of DNase (Promega) for 1 hour at room temperature. The cells were harvested 7 or 18 hours post-infection for quantification of late RT products and 2 LTR circles respectively. Total genomic DNA was extracted using the DNeasy blood and tissue kit (Qiagen) and 250 mg or 500 mg was used for quantitative PCR to detect late RT products and 2 LTR circles respectively. Primers and probes directed against EGFP [67] were used for quantifying late RT products from MLV and FFV while those directed against LacZ were used for EIAV. The retroviral vectors fEGFPf and pHIT111 were used as standards for EGFP and LacZ quantification respectively. Primers and probes for the detection of MLV 2 LTR circles have been described previously [68]. In order to detect EIAV and FFV 2 LTR circles, primers and probes that amplified and bound to a fragment spanning the 2LTRs were designed. For EIAV 2 LTR circle detection, EIAV2LTRCF (5′ACTCAGATTCTGCGGTCTGAG3′), EIAV2LTRCRev (5′ACCCCTCATAAAAACCCCAC3′) and EIAV2LTRCprobe (5′FAM-CTCAGTCCCTGTCTCTAGTTTGTCTGTTCG-Tamra3′) were used while FFV2LTRCF (5′CCAGAACTCACATGAGTGGTG3′), FFV2LTRCRev (5′CTCATCGTCACTAGATGGCAG3′) and FFV2LTRCprobe (5′FAM-GAAGGACTAACCTATCCCAGGTATAGGCCG3-Tamra') were used for the quantification of FFV 2LTR circles. The primer pairs were used to amplify fragments spanning the 2 LTRs from genomic DNA of EIAV or FFV infected cells. The fragments were cloned into pCR-BluntII-TOPO (Invitrogen) to be used as standards. Quantitative PCR was performed in 25 ml reactions using the ABsolute QPCR Rox mix from Abgene with 300 nM of each primer and 200 nM of probe. A programme of 50°C for 2 minutes, 95°C for 15 minutes followed by 40 cycles of 95°C for 15 s and 60°C for 1 minute was employed in the Applied Biosystems 7500 real time PCR system.
Phylogenetic analysis
Trees were generated using the MegAlign programme from the DNASTAR Lasergene package. The distance values were calculated using the Kimura distance formula that takes into account the number of non-gap mismatches and silent substitutions.
Supporting Information
Zdroje
1. ComptonAA, HirschVM, EmermanM (2012) The host restriction factor APOBEC3G and retroviral Vif protein coeveolve due to ongoing genetic conflict. Cell Host and Microbe 11 : 91–98.
2. EmermanM, MalikHS (2010) Paleovirology–modern consequences of ancient viruses. PLoS Biol 8: e1000301.
3. MeyersonNR, SawyerSL (2011) Two-stepping through time: mammals and viruses. Trends Microbiol 19 : 286–294.
4. SauterD, UnterwegerD, VoglM, UsmaniSM, HeigeleA, et al. (2012) Human tetherin exerts strong selection pressure on the HIV-1 group N Vpu protein. PLoS Pathog 8: e1003093.
5. Goff SP (2007) Retroviridae: The retroviruses and their replication. In: Knipe DM, Griffin DE, Lamb RA, Strauss SE, Howley, Marting MA, Roizman B, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins. Chapter 55: : pp1999–2069.
6. FeschotteC, GilbertC (2012) Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13 : 283–296.
7. StoyeJP (2012) Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10 : 395–406.
8. AswadA, KatzourakisA (2012) Paleovirlogy and virally derived immunity. Trends in Ecology and Evolution 27 : 627–36.
9. Sanz-RamosM, StoyeJP (2013) Capsid-binding retrovirus restriction factors: discovery, restriction specificity and implications for the development of novel therapeutics. J Gen Virol 94 : 2587–98.
10. LillyF (1970) Fv-2: identification and location of a second gene governing the spleen focus response to Friend leukemia virus in mice. J Natl Cancer Inst 45 : 163–169.
11. PincusT, HartleyJW, RoweWP (1971) A major genetic locus affecting resistance to infection with murine leukemia viruses. I. Tissue culture studies of naturally occurring viruses. J Exp Med 133 : 1219–1233.
12. RoweWP (1972) Studies of genetic transmission of murine leukemia virus by AKR mice. I. Crosses with Fv-1 n strains of mice. J Exp Med 136 : 1272–1285.
13. RoweWP, HartleyJW (1972) Studies of genetic transmission of murine leukemia virus by AKR mice. II. Crosses with Fv-1 b strains of mice. J Exp Med 136 : 1286–1301.
14. KozakCA, ChakrabortiA (1996) Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225 : 300–305.
15. HopkinsN, SchindlerJ, HynesR (1977) Six NB-tropic leukemia viruses derived from a B-tropic virus of BALB/c have altered p30. J Virol 21 : 309–318.
16. StevensA, BockM, EllisS, LeTissierP, BishopKN, et al. (2004) Retroviral capsid determinants of Fv1 NB - and NR-tropism. J Virol 78 : 9592–9598.
17. JolicoeurP, RassartE (1980) Effect of Fv-1 gene product on synthesis of linear and supercoiled viral DNA in cells infected with murine leukemia virus. J Virol 33 : 183–195.
18. BestS, Le TissierP, TowersG, StoyeJP (1996) Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382 : 826–829.
19. YapMW, NisoleS, LynchC, StoyeJP (2004) Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci U S A 101 : 10786–10791.
20. StremlauM, OwensCM, PerronMJ, KiesslingM, AutisslerP, et al. (2004) The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427 : 848–853.
21. StremlauM, PerronM, LeeM, LiY, SongB, et al. (2006) Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5a restriction factor. Proc Natl Acad Sci U S A 103 : 5514–5519.
22. HilditchL, MatadeenR, GoldstoneDC, RosenthalPB, TaylorIA, et al. (2011) Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc Natl Acad Sci U S A 108 : 5771–5776.
23. YapMW, MortuzaGB, TaylorIA, StoyeJP (2007) The design of artificial retroviral restriction factors. Virology 365 : 302–314.
24. OhkuraS, YapMW, SheldonT, StoyeJP (2006) All three variable regions of the TRIM5a B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol 80 : 8554–8565.
25. SchallerT, HuéS, TowersGJ (2007) An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J Virol 81 : 11713–11721.
26. YlinenLMJ, KeckesovaZ, WebbBLJ, GiffordRJM, SmithTPL, et al. (2006) Isolation of an active Lv1 gene from cattle indicates that tripartite motif protein-mediated innate immunity to retroviral infection is widespread among mammals. J Virol 80 : 7332–7338.
27. SiZ, VandegraaffN, O'hUiginC, SongB, YuanW, et al. (2006) Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc Natl Acad Sci U S A 103 : 7454–7459.
28. DiehlWE, StansellE, KaiserSM, EmermanM, HunterE (2008) Identification of Post-entry Restrictions to Mason-Pfizer Monkey Virus Infection in New World Monkey Cells. J Virol 82 : 11140–11151.
29. YapMW, LindemannD, StankeN, RehJ, WestphalD, et al. (2008) Restriction of foamy viruses by primate Trim5alpha. J Virol 82 : 5429–5439.
30. MortuzaGB, GoldstoneDC, PashleyC, HaireLF, PalmariniM, et al. (2009) Structure of the capsid amino-terminal domain from the betaretrovirus, Jaagsiekte sheep retrovirus. J Mol Biol 386 : 1179–1192.
31. GoldstoneDC, FlowerTG, BallNJ, Sanz-RamosM, WYM, et al. (2013) A unique spumavirus Gag N-terminal domain with functional properties of orthoretroviral matrix and capsid. PLoS Pathog 5: e1003376 doi:1003310.1001371/journal.ppat.1003376
32. SawyerSL, WuLI, EmermanM, MalikHS (2005) Positive selection of primate TRIM5a identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A 102 : 102.
33. HanK, LouDI, SawyerSL (2011) Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet 7: e1002388.
34. GoldschmidtV, CiuffiA, OrtizM, BrawandD, MunozM, et al. (2008) Antiretroviral activity of ancestral TRIM5alpha. J Virol 82 : 2089–2096.
35. JohnsonWE, SawyerSL (2009) Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics 61 : 163–178.
36. KaiserSM, MalikHS, EmermanM (2007) Restriction of an extinct retrovirus by the human TRIM5a antiviral protein. Science 316 : 1756–1758.
37. Perez-CaballeroD, SollSJ, BieniaszPD (2008) Evidence for restriction of ancient primate gammaretroviruses by APOBEC3 but not TRIM5a proteins. PLoS Pathogens 4: e1000181.
38. YapMW, StoyeJP (2013) Apparent effect of rabbit endogenous lentivirus type K acquisition on retrovirus restriction by lagomorph Trim5αs. Philos Trans R Soc Lond B Biol Sci 368 doi: 10.1098/rstb.2012.0498
39. QiCF, BonhommeF, Buckler-WhiteA, BucklerC, OrthA, et al. (1998) Molecular phylogeny of Fv1. Mamm Genome 9 : 1049–1055.
40. YanY, Buckler-WhiteA, WollenbergK, KozakCA (2009) Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc Natl Acad Sci U S A 106 : 3259–3263.
41. KozakCA, O'NeillRR (1987) Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J Virol 61 : 3082–3088.
42. TomonagaK, CoffinJM (1999) Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs. J Virol 73 : 4327–4340.
43. Ellis SA (2000) Evolutionary and functional studies of the mouse retrovirus restriction gene, Fv1. PhD thesis, University of London.
44. GuénetJ-L, BonhommeF (2003) Wild mice: an ever-increasing contribution to a popular mammalian model. Trends in Genetics 19 : 24–31.
45. BishopKN, MortuzaGB, HowellS, YapMW, StoyeJP, et al. (2006) Characterization of an amino-terminal dimerization domain from retroviral restriction factor Fv1. J Virol 80 : 8225–8235.
46. CravenRC, Leure-duPreeAE, R. A. WeldonJ, WillsJW (1995) Genetic analysis of the major homology region of the Rous Sarcoma Virus Gag protein. J Virol 69 : 4213–4227.
47. BénitL, de ParsevalN, CasellaJ-F, CallebautI, CordonnierA, et al. (1997) Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and a gag coding sequence closely related to the Fv1 restriction gene. J Virol 71 : 5652–5657.
48. BishopKN, BockM, TowersG, StoyeJP (2001) Identification of the regions of Fv1 necessary for MLV restriction. J Virol 75 : 5182–5188.
49. LundriganBL, JansaSA, TuckerPK (2002) Phylogenetic relationships in the genus Mus, based on paterrnally, maternally and biparentally inherited characters. Syst Biol 51 : 410–431.
50. ChevretP, JenkinsP, CatzeflisF (2003) Evolutionary systematics of the Indian mouse Mus famulus Bonhote, 1898: molecular (DNA/DNA hybridization and 12S rRNA sequences) and morphological evidence. Zoological Journal of the Linnean Society 137 : 385–401.
51. BockM, BishopKN, TowersG, StoyeJP (2000) Use of a transient assay for studying the genetic determinants of Fv1 restriction. J Virol 74 : 7422–7430.
52. SloanRD, WainbergMD (2011) The role of unintegrated DNA in HIV infection. Retrovirology 8 : 52.
53. YuSF, BaldwinDN, GwynnSR, YendapalliS, LinialML (1996) Human Foamy Virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271 : 1579–1582.
54. SayahDM, SokolskajaE, BerthouxL, LubanJ (2004) Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430 : 569–573.
55. SongB, GoldB, O'hUiginC, JavanbakhtM, LiX, et al. (2005) The B30.2(SPRY) domain of retroviral restriction factor TRIM5a exhibits lineage-specific length and sequence variation in primates. J Virol 79 : 6111–6121.
56. Perez-CaballeroD, HatziioannouT, YangA, CowanS, BieniaszPD (2005) Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J Virol 79 : 8969–8978.
57. LiX, SodroskiJ (2008) The TRIM5{alpha} B-box 2 Domain Promotes Cooperative Binding to the Retroviral Capsid by Mediating Higher-order Self-association. J Virol 82 : 11495–11502.
58. BestS, Le TissierPR, StoyeJP (1997) Endogenous retroviruses and the evolution of resistance to retroviral infection. Trends Microbiol 4 : 313–318.
59. BénitL, LallemandJ-B, CasellaJ-F, PhilippeH, HeidmannT (1999) ERV-L elements: a family of endogenous retrovirus-like elements active through the evolution of mammals. J Virol 73 : 3301–3308.
60. BamunusingheD, LiuQ, LuX, OlerA, KozakCA (2013) Endogenous gammaretrovirus acquisition in Mus musculus subspecies carrying functional variants of the XPR1 virus receptor. J Virol 87 : 9845–9855.
61. OdakaT, IkedaH, YoshikuraH, MoriwakiK, SuzukiS (1981) Fv-4: gene controlling resistance to NB-tropic Friend murine leukemia virus. Distribution in wild mice, introduction into genetic background of BALB/c mice, and mapping of chromosomes. J Natl Cancer Inst 67 : 1123–1127.
62. KozakCA (2013) Evolution of different antiviral strategies in wild mice exposed to different gammaretroviruses. Curr Opin Virol 3 : 657–63.
63. LinialML (2000) Why aren't foamy viruses pathogenic? Trends Micribiol 8 : 284–289.
64. ChoudharyA, GalvinTA, WilliamsDK, BerenJ, BryantMA, et al. (2013) Influence of naturally occurring simian foamy viruses (SFVs) on SIV disease progression in the rhesus macaque (Macaca mulatta) model. Viruses 5 : 1414–1430.
65. LanderMR, ChattopadhyaySK (1984) A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic and mink cell focus-forming viruses. J Virol 52 : 695–698.
66. GoldstoneDC, YapMW, RobertsonLE, HaireLF, TaylorWR, et al. (2010) Structural and functional analysis of prehistoric lentiviruses uncovers an ancient molecular interface. Cell Host Microbe 8 : 248–259.
67. IkedaY, CollinsMK, RadcliffePA, MitrophanousKA, TakeuchiY (2002) Gene transduction efficiency in cells of different species by HIV and EIAV vectors. Gene Therapy 9 : 932–938.
68. SerhanF, PenaudM, PetitC, Leste-LasserreT, TrajcevskiS, et al. (2004) Early detection of a two-long-terminal-repeat junction molecule in the cytoplasm of recombinant murine leukemia virus-infected cells. J Virol 78 : 6190–6199.
69. LabudaD, SinnettD, RicherC, DeragonJ-M, StrikerG (1991) Evolution of mouse B1 repeats: 7SL RNA folding pattern conserved. J Mol Evol 32 : 405–414.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule FormationČlánek Oral Bacteria and CancerČlánek A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 3- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
- Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
- Mycobacteriophages: Windows into Tuberculosis
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- Five Things to Know about Genetically Modified (GM) Insects for Vector Control
- A Missing Dimension in Measures of Vaccination Impacts
- Eosinophils Are Important for Protection, Immunoregulation and Pathology during Infection with Nematode Microfilariae
- Clonality of HTLV-2 in Natural Infection
- Production, Fate and Pathogenicity of Plasma Microparticles in Murine Cerebral Malaria
- Group B Streptococcal Infection of the Choriodecidua Induces Dysfunction of the Cytokeratin Network in Amniotic Epithelium: A Pathway to Membrane Weakening
- New Insights into How Adapts to Its Mammalian Host during Bubonic Plague
- Foodborne Transmission of Nipah Virus in Syrian Hamsters
- A Polysaccharide Virulence Factor from Elicits Anti-inflammatory Effects through Induction of Interleukin-1 Receptor Antagonist
- Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH
- Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages
- Crk Adaptors Negatively Regulate Actin Polymerization in Pedestals Formed by Enteropathogenic (EPEC) by Binding to Tir Effector
- Fatty Acid Biosynthesis Contributes Significantly to Establishment of a Bioenergetically Favorable Environment for Vaccinia Virus Infection
- A Cytosolic Chaperone Complexes with Dynamic Membrane J-Proteins and Mobilizes a Nonenveloped Virus out of the Endoplasmic Reticulum
- Intracellular Promote Invasive Cell Motility through Kinase Regulation of the Host Actin Cytoskeleton
- MAVS-MKK7-JNK2 Defines a Novel Apoptotic Signaling Pathway during Viral Infection
- RON5 Is Critical for Organization and Function of the Moving Junction Complex
- Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway
- and Exhibit Metabolic Symbioses
- The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface
- Target Cell Availability, Rather than Breast Milk Factors, Dictates Mother-to-Infant Transmission of SIV in Sooty Mangabeys and Rhesus Macaques
- Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
- Infection of Adult Thymus with Murine Retrovirus Induces Virus-Specific Central Tolerance That Prevents Functional Memory CD8 T Cell Differentiation
- Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in
- In Vivo Administration of a JAK3 Inhibitor during Acute SIV Infection Leads to Significant Increases in Viral Load during Chronic Infection
- Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
- Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
- A Compact, Multifunctional Fusion Module Directs Cholesterol-Dependent Homomultimerization and Syncytiogenic Efficiency of Reovirus p10 FAST Proteins
- The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck
- Genetic Dissection of Gut Epithelial Responses to
- Two-Component System Cross-Regulation Integrates Response to Heme and Cell Envelope Stress
- Oral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
- A Model System for Studying the Transcriptomic and Physiological Changes Associated with Mammalian Host-Adaptation by Serovar Copenhageni
- Inflammasome Sensor NLRP1 Controls Rat Macrophage Susceptibility to
- ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by
- The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses
- Caspase-1-Dependent and -Independent Cell Death Pathways in Infection of Macrophages
- The Effect of Cell Growth Phase on the Regulatory Cross-Talk between Flagellar and Spi1 Virulence Gene Expression
- Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms
- Oral Bacteria and Cancer
- Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
- Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector
- VE-Cadherin Cleavage by LasB Protease from Facilitates Type III Secretion System Toxicity in Endothelial Cells
- Dimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
- Crystal Structure of the Vaccinia Virus DNA Polymerase Holoenzyme Subunit D4 in Complex with the A20 N-Terminal Domain
- Post-Translational Regulation via Clp Protease Is Critical for Survival of
- Modulation of Phagosomal pH by Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport
- Rotavirus Activates Lymphocytes from Non-Obese Diabetic Mice by Triggering Toll-Like Receptor 7 Signaling and Interferon Production in Plasmacytoid Dendritic Cells
- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Interferon Regulatory Factor-1 Protects from Fatal Neurotropic Infection with Vesicular Stomatitis Virus by Specific Inhibition of Viral Replication in Neurons
- HMGB1-Promoted and TLR2/4-Dependent NK Cell Maturation and Activation Take Part in Rotavirus-Induced Murine Biliary Atresia
- An Immunomics Approach to Schistosome Antigen Discovery: Antibody Signatures of Naturally Resistant and Chronically Infected Individuals from Endemic Areas
- PPARγ Agonists Improve Survival and Neurocognitive Outcomes in Experimental Cerebral Malaria and Induce Neuroprotective Pathways in Human Malaria
- A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
- Viral OTU Deubiquitinases: A Structural and Functional Comparison
- Heterogeneity and Breadth of Host Antibody Response to KSHV Infection Demonstrated by Systematic Analysis of the KSHV Proteome
- Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm
- Broadly Reactive Human CD8 T Cells that Recognize an Epitope Conserved between VZV, HSV and EBV
- Oncogenic Human Papillomaviruses Activate the Tumor-Associated Lens Epithelial-Derived Growth Factor (LEDGF) Gene
- Erythrocyte Invasion: Combining Function with Immune Evasion
- IL-1α and Complement Cooperate in Triggering Local Neutrophilic Inflammation in Response to Adenovirus and Eliminating Virus-Containing Cells
- Chronic Exposure to Type-I IFN under Lymphopenic Conditions Alters CD4 T Cell Homeostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



