-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Viral OTU Deubiquitinases: A Structural and Functional Comparison
Recent studies have revealed that proteases encoded by three very diverse RNA virus groups share structural similarity with enzymes of the Ovarian Tumor (OTU) superfamily of deubiquitinases (DUBs). The publication of the latest of these reports in quick succession prevented proper recognition and discussion of the shared features of these viral enzymes. Here we provide a brief structural and functional comparison of these virus-encoded OTU DUBs. Interestingly, although their shared structural features and substrate specificity tentatively place them within the same protease superfamily, they also show interesting differences that trigger speculation as to their origins.
Published in the journal: . PLoS Pathog 10(3): e32767. doi:10.1371/journal.ppat.1003894
Category: Opinion
doi: https://doi.org/10.1371/journal.ppat.1003894Summary
Recent studies have revealed that proteases encoded by three very diverse RNA virus groups share structural similarity with enzymes of the Ovarian Tumor (OTU) superfamily of deubiquitinases (DUBs). The publication of the latest of these reports in quick succession prevented proper recognition and discussion of the shared features of these viral enzymes. Here we provide a brief structural and functional comparison of these virus-encoded OTU DUBs. Interestingly, although their shared structural features and substrate specificity tentatively place them within the same protease superfamily, they also show interesting differences that trigger speculation as to their origins.
The covalent attachment of ubiquitin (Ub) to protein substrates, i.e., ubiquitination, plays a pivotal regulatory role in numerous cellular processes [1]–[5]. Ubiquitination can be reversed by deubiquitinases (DUBs) [6] and, not surprisingly, various virus groups encode such DUBs to influence ubiquitin-mediated host cell processes [7]–[21]. Some of these viral DUBs resemble proteases belonging to the Ovarian Tumor (OTU) superfamily [22]–[28]. Makarova et al. previously identified OTU proteases as a novel superfamily of cysteine proteases from different organisms [29], and their bioinformatics-based analysis included several of the viral enzymes discussed here. Recently reported structures of these viral DUBs include the OTU domains of the nairoviruses Crimean-Congo hemorrhagic fever virus (CCHFV) [22]–[24] and Dugbe virus (DUGV) [25], the papain-like protease (PLP2) domain of the arterivirus equine arteritis virus (EAV) [26], and the protease (PRO) domain of the tymovirus turnip yellow mosaic virus (TYMV) (Figure 1A–1D) [27], [28]. These viruses are strikingly diverse, considering that nairoviruses are mammalian negative-strand RNA viruses, while the mammalian arteriviruses and plant tymoviruses belong to separate orders of positive-strand RNA viruses.
Fig. 1. Viral and eukaryotic OTU domain structures and viral protein context. 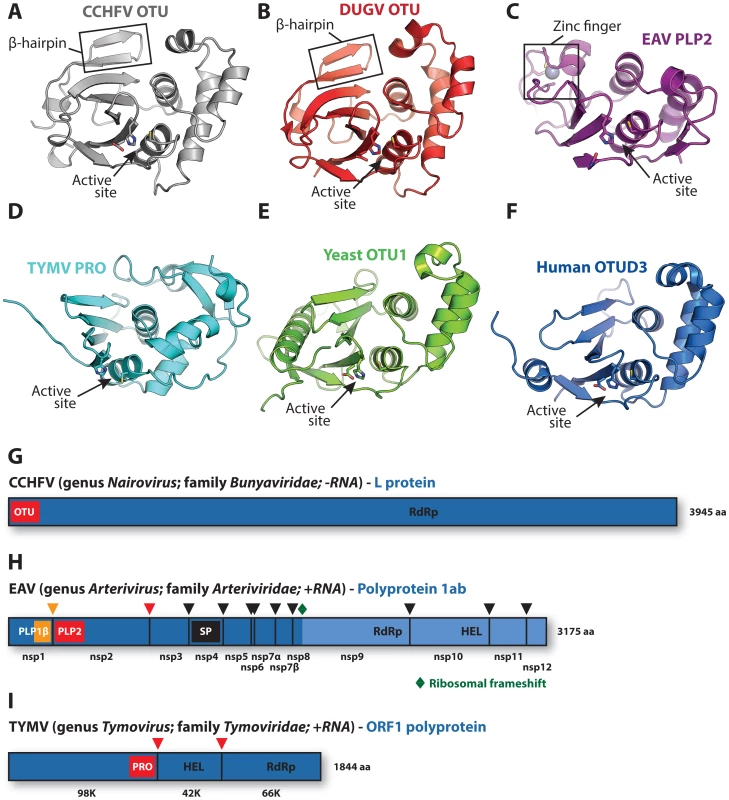
Crystal structures of (A) CCHFV OTU (3PT2) [23], (B) DUGV OTU (4HXD) [25], (C) EAV PLP2 (4IUM) [26], (D) TYMV PRO (4A5U) [27], [28], (E) yeast OTU1 (3BY4) [57], and (F) human OTUD3 (4BOU) [46]. The β-hairpin motifs of CCHFV OTU and DUGV OTU are indicated in boxes in panels A and B, respectively, and the zinc-finger motif of EAV PLP2 is boxed in panel C. Active sites are indicated with arrows. The CCHFV OTU, DUGV OTU, EAV PLP2, and yeast OTU1 domains were crystallized in complex with Ub, which has been removed for clarity. Structure images were generated using PyMol [60]. (G) Schematic representation of the CCHFV large (L) protein [61], [62]. A similar organization is found in the DUGV L protein, but is not depicted. The OTU domain resides in the N-terminal region of this protein and is not involved in autoproteolytic cleavage events [48]. (H) Schematic representation of the EAV polyprotein 1ab [63]. PLP2 resides in nonstructural protein 2 (nsp2) and is responsible for the cleavage between nsp2 and nsp3 [51]. (I) Schematic representation of the TYMV ORF1 polyprotein [50]. PRO resides in the N-terminal product of this polyprotein and is responsible for two internal cleavages [49], [50]. Key replicative enzymes are indicated in G, H, and I. Colored arrowheads denote cleavage sites for the indicated protease domains. HEL, helicase; PLP, papain-like protease; RdRp, RNA-dependent RNA polymerase; SP, serine protease. Ubiquitination often involves the formation of polyubiquitin chains [1], which can target the ubiquitinated substrate to the proteasome for degradation [2] or modulate its protein–protein interactions, as in the activation of innate immune signaling pathways [3], [4]. Interestingly, several cellular OTU DUBs were found to negatively regulate innate immunity [30]–[33]. Likewise, both nairovirus OTU and arterivirus PLP2 were recently shown to inhibit innate immune responses by targeting ubiquitinated signaling factors [7]–[9], [26], [34], [35]. In contrast to eukaryotic OTU DUBs, both of these viral proteases were found to also deconjugate the Ub-like protein interferon-stimulated gene 15 (ISG15) [7], [36], which inhibits viral replication via a mechanism that is currently poorly understood [37]. Interestingly, coronaviruses (which, together with the arteriviruses, belong to the nidovirus order) also encode papain-like proteases targeting both Ub and ISG15 that were shown to inhibit innate immunity [11]–[13], [38]–[42] but belong to the ubiquitin-specific protease (USP) class of DUBs [6], [43], [44]. The presence of functionally similar, yet structurally different proteases in distantly related virus families highlights the potential benefits to the virus of harboring such enzymes.
The proteasomal degradation pathway is an important cellular route to dispose of viral proteins, as exemplified by the turnover of the TYMV polymerase [45]. Moreover, the degradation of this protein is specifically counteracted by the deubiquitinase activity of TYMV PRO, which thus promotes virus replication [10]. The functional characterization of viral OTU DUBs remains incomplete and future studies will likely reveal additional roles in replication and virus–host interplay.
Polyubiquitin chains can adopt a number of different configurations, depending on the type of covalent linkage present within the polymer [1]. A distal Ub molecule can be linked via its C-terminus to one of seven internal lysine residues present in a proximal Ub molecule via an isopeptide bond. Alternatively, in the case of linear chains, the C-terminus of the distal Ub is covalently linked to the N-terminal methionine residue of the proximal Ub via a peptide bond. While human OTU proteases often show a distinct preference for one or two isopeptide linkage types [46], nairovirus OTUs and TYMV PRO appear to be more promiscuous in their substrate preference [22], [25]. However, like most human OTU proteases, they seem unable to cleave linear polyubiquitin chains in vitro [22], [25], [46]. Arterivirus PLP2 has not been extensively studied in this respect.
It is important to note that many positive-strand RNA viruses, including arteriviruses and tymoviruses, encode polyproteins that are post-translationally cleaved by internal protease domains [47]. Thus, while CCHFV OTU is not involved in viral protein cleavage and its activity seems dispensable for replication (Figure 1G) [48], both arterivirus PLP2 and tymovirus PRO are critically required for viral replication due to their primary role in polyprotein maturation (Figure 1H, 1I) [49]–[53]. Interestingly, while both EAV PLP2 and TYMV PRO can process peptide bonds in cis and in trans [50], [51], PRO does not cleave peptide bonds in linear polyubiquitin chains in vitro [25]. To date, activity of EAV PLP2 towards linear polyubiquitin chains has not been reported.
Based on mutagenesis of putative catalytic residues, arterivirus PLP2 and tymovirus PRO were initially generally classified as papain-like cysteine proteases [51], [54], [55]. Now that crystal structures of these proteases are available, it is possible to search the DALI server [56] in order to identify structurally similar domains. Using the 3-dimensional coordinates of TYMV PRO, the most recently solved structure of a viral OTU protease, such a query identifies structural similarity with eukaryotic OTU DUBs as well as the nairovirus OTU domains and EAV PLP2 (Table 1). A superposition of these viral protease structures with yeast OTU1 [57] further highlights their similarities (Figure 2A–2C), and these comparisons together clearly position them within the OTU DUB superfamily. Sequence comparisons alone were insufficient to demonstrate this conclusively, as the similarity of viral OTU domains to each other and to eukaryotic OTU proteases is very limited and mostly restricted to the areas surrounding the active site residues [29].
Fig. 2. Superpositions of the viral OTU proteases with yeast OTU1 and one another. 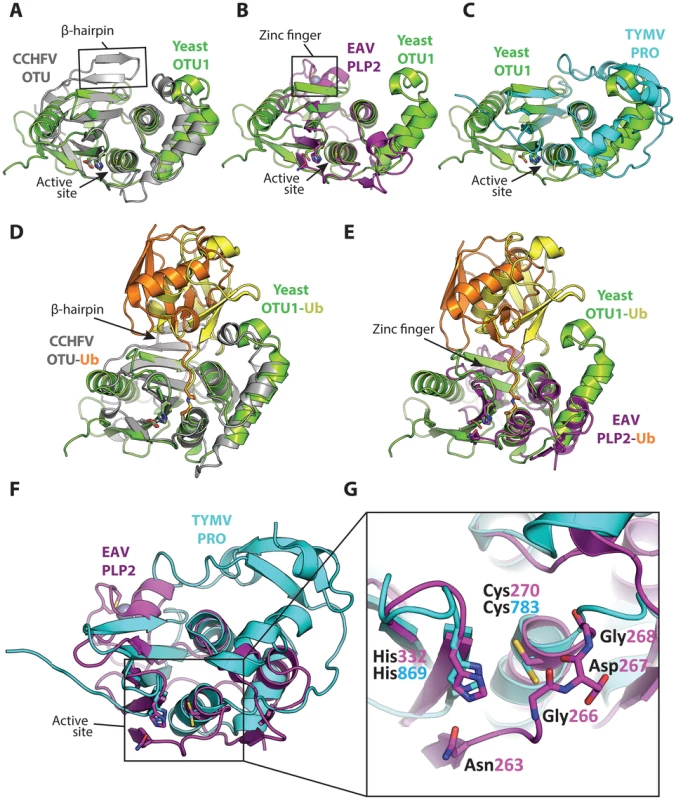
Superpositions of yeast OTU1 (3BY4) [57] with (A) CCHFV OTU (3PT2) [23], RMSD: 1.8 Å over 112 residues, (B) EAV PLP2 (4IUM) [26], RMSD: 2.8 Å over 69 residues, and (C) TYMV PRO (4A5U) [27], [28], RMSD: 1.4 Å over 76 residues. Superpositions of the yeast OTU1-Ub complex with (D) the CCHFV OTU-Ub complex and (E) the EAV PLP2-Ub complex, highlighting the difference in the orientation of Ub between the two viral OTU domains versus the eukaryotic yeast OTU1 domain. The Ub that is complexed with yeast OTU1 is depicted in yellow, while the Ub complexed with CCHFV OTU or EAV PLP2 is depicted in orange. (F) Superposition of EAV PLP2 and TYMV PRO, RMSD: 2.5 Å over 53 residues. (G) Close-up of the active site region (boxed) of the superposition depicted in F. Side chains of the catalytic cysteine (Cys270 and Cys783 for EAV PLP2 and TYMV PRO, respectively) and histidine (His332 and His869 for EAV PLP2 and TYMV PRO, respectively) residues are shown as sticks, as well as the active site Asn263 for EAV PLP2. The backbone amide group of Asp267 likely contributes to the formation of the oxyanion hole in the active site of EAV PLP2, yet a functionally equivalent residue is absent in TYMV PRO. The Gly266 and Gly268 residues flanking Asp267 in EAV PLP2 are depicted as sticks as well, for clarity. Note the alternative orientation of the active site cysteine residue of TYMV PRO which, unlike EAV PLP2, was not determined in covalent complex with an Ub suicide substrate. All alignments were generated using the PDBeFOLD server [64], and thus the reported RMSD values differ from those reported in Table 1, in which the DALI server was used. The yeast OTU1, CCHFV OTU, and EAV PLP2 domains were all crystallized in complex with Ub, which has been removed in panels A, B, C, F, and G for clarity. All images were generated using PyMol [60]. RMSD, root-mean-square deviation. Tab. 1. Three-dimensional structural alignment of viral OTU domains against selected structures in the Protein Data Bank using the DALI server [56]. ![Three-dimensional structural alignment of viral OTU domains against selected structures in the Protein Data Bank using the DALI server <em class="ref">[56]</em>.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/f1daaf751950583e9b900f5569e0218e.png)
*z-score (>2 indicates significant structural similarity [59]); % sequence identity. Structural characterization of nairovirus (CCHFV and DUGV) OTU domains and EAV PLP2 in complex with Ub revealed that while these viral proteases adopt a fold that is consistent with eukaryotic OTU DUBs, they possess additional structural motifs in their S1 binding site that rotate the distal Ub relative to the binding orientation observed in eukaryotic OTU enzymes (Figure 2D, 2E) [22]–[26]. In the case of CCHFV OTU, this alternative binding mode was shown to expand its substrate repertoire by allowing the enzyme to also accommodate ISG15. Since TYMV PRO was crystallized in its apo form [27], [28], it remains to be determined whether its S1 site binds Ub in an orientation similar to nairovirus OTU and EAV PLP2 or eukaryotic OTU DUBs.
A remarkable feature of EAV PLP2 is the incorporation within the OTU-fold of a zinc finger that is involved in the interaction with Ub (Figures 1C, 2E). The absence of similar internal zinc-finger motifs in other OTU superfamily members prompted us to propose that PLP2 prototypes a novel subclass of zinc-dependent OTU DUBs [26].
Finally, an interesting structural difference between TYMV PRO and other OTU proteases of known structure is the absence of a loop that generally covers the active site (Figure 2F, 2G). Because of this, TYMV PRO lacks a complete oxyanion hole. It also lacks a third catalytic residue that would otherwise form the catalytic triad that has been observed in other OTU proteases (Figure 2G). Lombardi et al. suggested that the resulting solvent exposure of the active site may contribute to the broad substrate specificity of TYMV PRO [28]. Interestingly, EAV PLP2 also has broad substrate specificity, cleaving Ub, ISG15, and the viral polyprotein, even though it does possess an intact oxyanion hole and an active site that is not solvent exposed. Future work may uncover additional aspects relating to the unusual architecture of the TYMV PRO active site.
The presence of structurally similar proteases, each displaying unique features, in these highly diverse virus groups suggests that their ancestors have independently acquired their respective OTU enzymes. Although their origins remain elusive, one possible scenario is the scavenging of an OTU DUB-encoding gene that directly enabled the ancestral virus to interact with the cellular ubiquitin landscape [29]. The absence of an OTU homologue in other lineages of the bunyavirus family strongly suggests that a nairoviral ancestor acquired an OTU DUB through heterologous recombination. In this scenario, the current differences between the nairoviral and eukaryotic OTU domains would reflect divergent evolution. In the case of arteriviruses, however, it is also conceivable that a preexisting papain-like protease that was initially only involved in polyprotein maturation acquired OTU-like features through a process of convergent evolution. Although rare, such a scenario would account for the limited structural similarity between eukaryotic OTU domains and EAV PLP2, which contrasts with that observed for nairovirus OTU (Figure 2A, 2B; Table 1). For tymoviruses, which encode one (OTU) protease, the existence of related viruses that do not encode a protease domain or that encode one (papain-like) or two (OTU combined with a second papain-like) protease domains complicates the development of a straightforward scenario describing PRO acquisition and evolution [58]. These and other intriguing unsolved questions should be addressed through structural and functional studies of additional OTU-like proteases, be they viral or cellular, the results of which may shed more light on the various scenarios explaining the evolution of viral OTU domains.
Zdroje
1. KomanderD, RapeM (2012) The ubiquitin code. Annu Rev Biochem 81 : 203–229.
2. ClagueMJ, UrbeS (2010) Ubiquitin: same molecule, different degradation pathways. Cell 143 : 682–685.
3. JiangX, ChenZJ (2012) The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol 12 : 35–48.
4. OudshoornD, VersteegGA, KikkertM (2012) Regulation of the innate immune system by ubiquitin and ubiquitin-like modifiers. Cytokine Growth Factor Rev 23 : 273–282.
5. HuangTT, D'AndreaAD (2006) Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol 7 : 323–334.
6. NijmanSM, Luna-VargasMP, VeldsA, BrummelkampTR, DiracAM, et al. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123 : 773–786.
7. Frias-StaheliN, GiannakopoulosNV, KikkertM, TaylorSL, BridgenA, et al. (2007) Ovarian tumor domain-containing viral proteases evade ubiquitin - and ISG15-dependent innate immune responses. Cell Host Microbe 2 : 404–416.
8. van KasterenPB, BeugelingC, NinaberDK, Frias-StaheliN, van BoheemenS, et al. (2012) Arterivirus and Nairovirus Ovarian Tumor Domain-Containing Deubiquitinases Target Activated RIG-I To Control Innate Immune Signaling. J Virol 86 : 773–785.
9. SunZ, ChenZ, LawsonSR, FangY (2010) The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J Virol 84 : 7832–7846.
10. ChenonM, CambordeL, CheminantS, JupinI (2012) A viral deubiquitylating enzyme targets viral RNA-dependent RNA polymerase and affects viral infectivity. EMBO J 31 : 741–753.
11. ChenZ, WangY, RatiaK, MesecarAD, WilkinsonKD, et al. (2007) Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J Virol 81 : 6007–6018.
12. ZhengD, ChenG, GuoB, ChengG, TangH (2008) PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res 18 : 1105–1113.
13. ClementzMA, ChenZ, BanachBS, WangY, SunL, et al. (2010) Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84 : 4619–4629.
14. WangD, FangL, LiP, SunL, FanJ, et al. (2011) The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J Virol 85 : 3758–3766.
15. KattenhornLM, KorbelGA, KesslerBM, SpoonerE, PloeghHL (2005) A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell 19 : 547–557.
16. WangJ, LovelandAN, KattenhornLM, PloeghHL, GibsonW (2006) High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J Virol 80 : 6003–6012.
17. BottcherS, MareschC, GranzowH, KluppBG, TeifkeJP, et al. (2008) Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J Virol 82 : 6009–6016.
18. SompallaeR, GastaldelloS, HildebrandS, ZininN, HassinkG, et al. (2008) Epstein-barr virus encodes three bona fide ubiquitin-specific proteases. J Virol 82 : 10477–10486.
19. GonzalezCM, WangL, DamaniaB (2009) Kaposi's sarcoma-associated herpesvirus encodes a viral deubiquitinase. J Virol 83 : 10224–10233.
20. WhitehurstCB, VaziriC, ShackelfordJ, PaganoJS (2012) Epstein-Barr virus BPLF1 deubiquitinates PCNA and attenuates polymerase eta recruitment to DNA damage sites. J Virol 86 : 8097–8106.
21. JiangJ, TangH (2010) Mechanism of inhibiting type I interferon induction by hepatitis B virus X protein. Protein Cell 1 : 1106–1117.
22. AkutsuM, YeY, VirdeeS, ChinJW, KomanderD (2011) Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc Natl Acad Sci U S A 108 : 2228–2233.
23. JamesTW, Frias-StaheliN, BacikJP, Levingston MacleodJM, KhajehpourM, et al. (2011) Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc Natl Acad Sci U S A 108 : 2222–2227.
24. CapodagliGC, McKercherMA, BakerEA, MastersEM, BrunzelleJS, et al. (2011) Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J Virol 85 : 3621–3630.
25. CapodagliGC, DeatonMK, BakerEA, LumpkinRJ, PeganSD (2013) Diversity of ubiquitin and ISG15 specificity among nairoviruses' viral ovarian tumor domain proteases. J Virol 87 : 3815–3827.
26. van KasterenPB, Bailey-ElkinBA, JamesTW, NinaberDK, BeugelingC, et al. (2013) Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc Natl Acad Sci U S A 110: E838–847.
27. RobinC, BeaurepaireL, ChenonM, JupinI, BressanelliS (2012) In praise of impurity: 30S ribosomal S15 protein-assisted crystallization of turnip yellow mosaic virus proteinase. Acta Crystallogr Sect F Struct Biol Cryst Commun 68 : 486–490.
28. LombardiC, AyachM, BeaurepaireL, ChenonM, AndreaniJ, et al. (2013) A Compact Viral Processing Proteinase/Ubiquitin Hydrolase from the OTU Family. PLOS Pathog 9: e1003560.
29. MakarovaKS, AravindL, KooninEV (2000) A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci 25 : 50–52.
30. WertzIE, O'RourkeKM, ZhouH, EbyM, AravindL, et al. (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430 : 694–699.
31. KayagakiN, PhungQ, ChanS, ChaudhariR, QuanC, et al. (2007) DUBA: a deubiquitinase that regulates type I interferon production. Science 318 : 1628–1632.
32. EnesaK, ZakkarM, ChaudhuryH, Luong leA, RawlinsonL, et al. (2008) NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem 283 : 7036–7045.
33. LiS, ZhengH, MaoAP, ZhongB, LiY, et al. (2010) Regulation of virus-triggered signaling by OTUB1 - and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem 285 : 4291–4297.
34. HolzerB, BakshiS, BridgenA, BaronMD (2011) Inhibition of interferon induction and action by the nairovirus Nairobi sheep disease virus/Ganjam virus. PLOS One 6: e28594.
35. BakshiS, HolzerB, BridgenA, McMullanG, QuinnDG, et al. (2013) Dugbe virus ovarian tumour domain interferes with ubiquitin/ISG15-regulated innate immune cell signalling. J Gen Virol 94 : 298–307.
36. SunZ, LiY, RansburghR, SnijderEJ, FangY (2012) Nonstructural protein 2 of porcine reproductive and respiratory syndrome virus inhibits the antiviral function of interferon-stimulated gene 15. J Virol 86 : 3839–3850.
37. SkaugB, ChenZJ (2010) Emerging role of ISG15 in antiviral immunity. Cell 143 : 187–190.
38. BarrettoN, JuknelieneD, RatiaK, ChenZ, MesecarAD, et al. (2005) The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 79 : 15189–15198.
39. LindnerHA, Fotouhi-ArdakaniN, LytvynV, LachanceP, SuleaT, et al. (2005) The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 79 : 15199–15208.
40. DevarajSG, WangN, ChenZ, TsengM, BarrettoN, et al. (2007) Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282 : 32208–32221.
41. WangG, ChenG, ZhengD, ChengG, TangH (2011) PLP2 of mouse hepatitis virus A59 (MHV-A59) targets TBK1 to negatively regulate cellular type I interferon signaling pathway. PLOS One 6: e17192.
42. SunL, XingY, ChenX, ZhengY, YangY, et al. (2012) Coronavirus Papain-like Proteases Negatively Regulate Antiviral Innate Immune Response through Disruption of STING-Mediated Signaling. PLOS One 7: e30802.
43. RatiaK, SaikatenduKS, SantarsieroBD, BarrettoN, BakerSC, et al. (2006) Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci U S A 103 : 5717–5722.
44. WojdylaJA, ManolaridisI, van KasterenPB, KikkertM, SnijderEJ, et al. (2010) Papain-like protease 1 from transmissible gastroenteritis virus: crystal structure and enzymatic activity toward viral and cellular substrates. J Virol 84 : 10063–10073.
45. CambordeL, PlanchaisS, TournierV, JakubiecA, DrugeonG, et al. (2010) The ubiquitin-proteasome system regulates the accumulation of Turnip yellow mosaic virus RNA-dependent RNA polymerase during viral infection. Plant Cell 22 : 3142–3152.
46. MevissenTE, HospenthalMK, GeurinkPP, ElliottPR, AkutsuM, et al. (2013) OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154 : 169–184.
47. HellenCU, KrausslichHG, WimmerE (1989) Proteolytic processing of polyproteins in the replication of RNA viruses. Biochemistry 28 : 9881–9890.
48. BergeronE, AlbarinoCG, KhristovaML, NicholST (2010) Crimean-Congo hemorrhagic fever virus-encoded ovarian tumor protease activity is dispensable for virus RNA polymerase function. J Virol 84 : 216–226.
49. BransomKL, WeilandJJ, DreherTW (1991) Proteolytic maturation of the 206-kDa nonstructural protein encoded by turnip yellow mosaic virus RNA. Virology 184 : 351–358.
50. JakubiecA, DrugeonG, CambordeL, JupinI (2007) Proteolytic processing of turnip yellow mosaic virus replication proteins and functional impact on infectivity. J Virol 81 : 11402–11412.
51. SnijderEJ, WassenaarAL, SpaanWJ, GorbalenyaAE (1995) The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J Biol Chem 270 : 16671–16676.
52. WassenaarAL, SpaanWJ, GorbalenyaAE, SnijderEJ (1997) Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J Virol 71 : 9313–9322.
53. PosthumaCC, PedersenKW, LuZ, JoostenRG, RoosN, et al. (2008) Formation of the arterivirus replication/transcription complex: a key role for nonstructural protein 3 in the remodeling of intracellular membranes. J Virol 82 : 4480–4491.
54. ZiebuhrJ, SnijderEJ, GorbalenyaAE (2000) Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 81 : 853–879.
55. BransomKL, DreherTW (1994) Identification of the essential cysteine and histidine residues of the turnip yellow mosaic virus protease. Virology 198 : 148–154.
56. HolmL, RosenströmP (2010) Dali server: conservation mapping in 3D. Nucleic Acids Research 38: W545–W549.
57. MessickTE, RussellNS, IwataAJ, SarachanKL, ShiekhattarR, et al. (2008) Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem 283 : 11038–11049.
58. MartelliGP, AdamsMJ, KreuzeJF, DoljaVV (2007) Family Flexiviridae: a case study in virion and genome plasticity. Annu Rev Phytopathol 45 : 73–100.
59. HolmL, KaariainenS, RosenstromP, SchenkelA (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24 : 2780–2781.
60. DeLano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA, USA.: DeLano Scientific.
61. KinsellaE, MartinSG, GrollaA, CzubM, FeldmannH, et al. (2004) Sequence determination of the Crimean-Congo hemorrhagic fever virus L segment. Virology 321 : 23–28.
62. HonigJE, OsborneJC, NicholST (2004) Crimean-Congo hemorrhagic fever virus genome L RNA segment and encoded protein. Virology 321 : 29–35.
63. SnijderEJ, KikkertM, FangY (2013) Arterivirus molecular biology and pathogenesis. J Gen Virol 94 : 2141–2163.
64. KrissinelE, HenrickK (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60 : 2256–2268.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule FormationČlánek Oral Bacteria and CancerČlánek A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 3- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
- Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
- Mycobacteriophages: Windows into Tuberculosis
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- Five Things to Know about Genetically Modified (GM) Insects for Vector Control
- A Missing Dimension in Measures of Vaccination Impacts
- Eosinophils Are Important for Protection, Immunoregulation and Pathology during Infection with Nematode Microfilariae
- Clonality of HTLV-2 in Natural Infection
- Production, Fate and Pathogenicity of Plasma Microparticles in Murine Cerebral Malaria
- Group B Streptococcal Infection of the Choriodecidua Induces Dysfunction of the Cytokeratin Network in Amniotic Epithelium: A Pathway to Membrane Weakening
- New Insights into How Adapts to Its Mammalian Host during Bubonic Plague
- Foodborne Transmission of Nipah Virus in Syrian Hamsters
- A Polysaccharide Virulence Factor from Elicits Anti-inflammatory Effects through Induction of Interleukin-1 Receptor Antagonist
- Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH
- Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages
- Crk Adaptors Negatively Regulate Actin Polymerization in Pedestals Formed by Enteropathogenic (EPEC) by Binding to Tir Effector
- Fatty Acid Biosynthesis Contributes Significantly to Establishment of a Bioenergetically Favorable Environment for Vaccinia Virus Infection
- A Cytosolic Chaperone Complexes with Dynamic Membrane J-Proteins and Mobilizes a Nonenveloped Virus out of the Endoplasmic Reticulum
- Intracellular Promote Invasive Cell Motility through Kinase Regulation of the Host Actin Cytoskeleton
- MAVS-MKK7-JNK2 Defines a Novel Apoptotic Signaling Pathway during Viral Infection
- RON5 Is Critical for Organization and Function of the Moving Junction Complex
- Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway
- and Exhibit Metabolic Symbioses
- The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface
- Target Cell Availability, Rather than Breast Milk Factors, Dictates Mother-to-Infant Transmission of SIV in Sooty Mangabeys and Rhesus Macaques
- Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
- Infection of Adult Thymus with Murine Retrovirus Induces Virus-Specific Central Tolerance That Prevents Functional Memory CD8 T Cell Differentiation
- Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in
- In Vivo Administration of a JAK3 Inhibitor during Acute SIV Infection Leads to Significant Increases in Viral Load during Chronic Infection
- Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
- Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
- A Compact, Multifunctional Fusion Module Directs Cholesterol-Dependent Homomultimerization and Syncytiogenic Efficiency of Reovirus p10 FAST Proteins
- The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck
- Genetic Dissection of Gut Epithelial Responses to
- Two-Component System Cross-Regulation Integrates Response to Heme and Cell Envelope Stress
- Oral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
- A Model System for Studying the Transcriptomic and Physiological Changes Associated with Mammalian Host-Adaptation by Serovar Copenhageni
- Inflammasome Sensor NLRP1 Controls Rat Macrophage Susceptibility to
- ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by
- The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses
- Caspase-1-Dependent and -Independent Cell Death Pathways in Infection of Macrophages
- The Effect of Cell Growth Phase on the Regulatory Cross-Talk between Flagellar and Spi1 Virulence Gene Expression
- Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms
- Oral Bacteria and Cancer
- Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
- Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector
- VE-Cadherin Cleavage by LasB Protease from Facilitates Type III Secretion System Toxicity in Endothelial Cells
- Dimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
- Crystal Structure of the Vaccinia Virus DNA Polymerase Holoenzyme Subunit D4 in Complex with the A20 N-Terminal Domain
- Post-Translational Regulation via Clp Protease Is Critical for Survival of
- Modulation of Phagosomal pH by Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport
- Rotavirus Activates Lymphocytes from Non-Obese Diabetic Mice by Triggering Toll-Like Receptor 7 Signaling and Interferon Production in Plasmacytoid Dendritic Cells
- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Interferon Regulatory Factor-1 Protects from Fatal Neurotropic Infection with Vesicular Stomatitis Virus by Specific Inhibition of Viral Replication in Neurons
- HMGB1-Promoted and TLR2/4-Dependent NK Cell Maturation and Activation Take Part in Rotavirus-Induced Murine Biliary Atresia
- An Immunomics Approach to Schistosome Antigen Discovery: Antibody Signatures of Naturally Resistant and Chronically Infected Individuals from Endemic Areas
- PPARγ Agonists Improve Survival and Neurocognitive Outcomes in Experimental Cerebral Malaria and Induce Neuroprotective Pathways in Human Malaria
- A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
- Viral OTU Deubiquitinases: A Structural and Functional Comparison
- Heterogeneity and Breadth of Host Antibody Response to KSHV Infection Demonstrated by Systematic Analysis of the KSHV Proteome
- Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm
- Broadly Reactive Human CD8 T Cells that Recognize an Epitope Conserved between VZV, HSV and EBV
- Oncogenic Human Papillomaviruses Activate the Tumor-Associated Lens Epithelial-Derived Growth Factor (LEDGF) Gene
- Erythrocyte Invasion: Combining Function with Immune Evasion
- IL-1α and Complement Cooperate in Triggering Local Neutrophilic Inflammation in Response to Adenovirus and Eliminating Virus-Containing Cells
- Chronic Exposure to Type-I IFN under Lymphopenic Conditions Alters CD4 T Cell Homeostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



