-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
Infectious diseases are among the major causes of mortality worldwide. Pathogens‚ invasion, colonization and persistence within their hosts depend on a tightly orchestrated cascade of events that are commonly referred to as host/pathogen interactions. These interactions are extremely diversified and every pathogen is characterized by its unique way of co-opting and manipulating host functions to its advantage. Understanding host/pathogen interactions is the key to face the threats imposed by infectious diseases and find alternative strategies to fight the emergence of multi-drug resistant pathogens. In this study, we have setup and validated a protocol for the rapid and unbiased identification of bacterial factors that regulate host/pathogen interactions. We have applied this method to the study of Coxiella burnetii, the etiological agent of the emerging zoonosis Q fever. We have isolated, sequenced and screened over 1000 bacterial mutations and identified genes important for Coxiella invasion and replication within host cells. Ultimately, we have characterized the first Coxiella invasin, which mediates bacterial internalization within non-phagocytic cells. Most importantly, our finding may lead to the development of a synthetic vaccine against Q fever.
Published in the journal: . PLoS Pathog 10(3): e32767. doi:10.1371/journal.ppat.1004013
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004013Summary
Infectious diseases are among the major causes of mortality worldwide. Pathogens‚ invasion, colonization and persistence within their hosts depend on a tightly orchestrated cascade of events that are commonly referred to as host/pathogen interactions. These interactions are extremely diversified and every pathogen is characterized by its unique way of co-opting and manipulating host functions to its advantage. Understanding host/pathogen interactions is the key to face the threats imposed by infectious diseases and find alternative strategies to fight the emergence of multi-drug resistant pathogens. In this study, we have setup and validated a protocol for the rapid and unbiased identification of bacterial factors that regulate host/pathogen interactions. We have applied this method to the study of Coxiella burnetii, the etiological agent of the emerging zoonosis Q fever. We have isolated, sequenced and screened over 1000 bacterial mutations and identified genes important for Coxiella invasion and replication within host cells. Ultimately, we have characterized the first Coxiella invasin, which mediates bacterial internalization within non-phagocytic cells. Most importantly, our finding may lead to the development of a synthetic vaccine against Q fever.
Introduction
Coxiella burnetii is an obligate intracellular Gram-negative bacterium responsible of the worldwide neglected zoonosis Q fever [1], [2]. Acute forms of the disease are characterized by a febrile illness associated with severe headache, pneumonia and hepatitis. In a small percentage (2–5%) of cases, acute Q fever develops into a chronic infection that may lead to endocarditis and chronic fatigue syndrome [1], [3]. Coxiella resists environmental stress by generating small cell variants (SCVs) that facilitate its airborne dissemination; during infections, this pathogen converts into a metabolically active large cell variant (LCV) with a unique resistance to the degradative machinery of host cells [4], [5]. These factors contribute to the extreme infectivity of this microbe, making of Coxiella a serious health concern, especially in rural areas where outbreaks are likely to occur and are accompanied by heavy economic burdens [6], [7]. Moreover, the development of Coxiella as a potential bioweapon during and since World War II, has ascribed this pathogen among class B biothreats [7]. Coxiella has two antigenic phases: phase I organisms, isolated from natural sources of infection, are extremely virulent. Phase II bacteria originate from spontaneous mutations after several in vitro passages of phase I organisms and present a truncated lipopolysaccharide (LPS) [8]. These non-reversible mutations result in a strong attenuation of virulence in vivo [9], [10]. Phase II Coxiella organisms are internalized more efficiently than phase I organisms by both professional macrophages and non-phagocytic cells [9], [11], however, once internalized, both antigenic phases replicate within host cells with similar kinetics. A phase II clone (Nine Mile phase II clone 4 or NMIIC4), which has been authorized for biosafety level 2 (BSL-2) manipulation, represents therefore an optimal model to study Coxiella infections [2], [5]. In natural infections, Coxiella has a tropism for alveolar macrophages [1], [2], however, infection of epithelial and endothelial cells has also been reported [12], [13]. Indeed, in vitro, Coxiella invades and replicates in a wide variety of phagocytic and non-phagocytic cells [5]. Coxiella internalization within host cells is a passive, endocytic process, which involves the remodeling of the host cell actin cytoskeleton [14], [15] and αVβ3 integrins have been reported as Coxiella receptors in THP-1 cells [11]. However the Coxiella factors that mediate interactions with host cell surfaces, as well as the bacterial host receptor on epithelial cells remain unknown. During the first 48 hours following internalization, bacteria reside into tight-fitting vacuoles, positive for early endosomal and autophagosomal markers [16]. As Coxiella-containing vacuoles mature along the endocytic pathway, the drop in vacuolar pH triggers the translocation of bacterial proteins by a Dot/Icm type 4b secretion system (T4SS) [17]. Effector translocation is essential for the biogenesis of a large parasitophorous vacuole (PV) that occupies the majority of the host cytoplasm [18], [19]. Such large membranous structures are highly dynamic and fusogenic and the host endocytic SNARE Vamp7 is required for optimal PV development [20]. Importantly, mature Coxiella PVs are positive for lysosomal markers and contain active degradative enzymes [5], [16]. Coxiella infections are not lytic and bacteria-filled PVs persist within infected cells, which are protected from apoptosis by a Dot/Icm-dependent mechanism [19], [21]–[25]. Importantly, due to the obligate intracellular nature of this pathogen, the microbial factors involved in host/pathogen interactions remain to date largely unknown. The homology between the T4SS of C. burnetii and L. pneumophila allowed the in silico identification of 354 candidate Coxiella effectors based on the presence of a conserved Dot/Icm regulatory motif (PmrA) [26]–[28], C-terminal translocation signals (E-block) [26]–[28], and eukaryotic-like domains [29]–[31]. Dot/Icm–dependent secretion has been validated for 108 of these using either Coxiella or Legionella as a surrogate host [18], [26]–[31]. Recent advances in Coxiella axenic culture techniques [32] rendered this pathogen genetically tractable [33], allowing for the first time to couple bioinformatics analysis to morpho-functional assays and investigate the role of candidate Coxiella virulence determinants in intracellular replication [18], [19], [27]. To date, 20 Coxiella genes encoding Dot/Icm substrates have been mutated to investigate their role in Coxiella replication within the host [27]. Here we have set up new, integrative approaches that combine transposon mutagenesis with genomics, bioinformatics and fluorescence-based functional assays aiming at the large-scale identification of intracellular bacteria virulence factors. Our approach is designed for the simultaneous investigation of multiple key steps of Coxiella infections and is based on the identification and characterization of transposon-induced phenotypes. We have generated and isolated 3000 Coxiella transposon mutants, 1082 of which have been sequenced and screened in the present study. Our analysis revealed important insights into the functionality of the Coxiella Dot/Icm apparatus and revealed a variety of bacterial factors involved in 1) internalization within host cells, 2) PV biogenesis and intracellular replication, and 3) protection of the infected cell from apoptosis. By focusing our analysis on the early events of Coxiella infections we identified the first Coxiella invasin that plays an essential role in bacterial internalization by non-phagocytic cells.
Results
Generation of a bank of Phase II Coxiella mutants
To identify the Coxiella factors involved in host-pathogen interactions, we have undertaken the generation of a library of GFP-tagged bacterial mutants by transposon mutagenesis. We have modified the Himar1-based transposon system initially developed by Heinzen and colleagues [33], [34], by inserting the enhanced green fluorescent protein (egfp) gene under the regulation of the Coxiella promoter P311, upstream of the chloramphenicol resistance cassette, thus generating pITR-CAT-ColE1-P311-GFP. To obtain stable mutants, Nine Mile Phase II clone 4 (NMIIC4) Coxiella (hereafter referred to as wt Coxiella) were electroporated using a two-plasmid system, where the transposase is encoded by a suicide plasmid that is lost during bacterial replication [34]. The eGFP-tagged Coxiella mutants thus generated were isolated on ACCM-2 agar plates in the presence of chloramphenicol and further amplified for 7 days in liquid ACCM-2 supplemented with chloramphenicol. The final concentration of each bacterial culture was calculated using the Quant-iT PicoGreen dsDNA assay. Transposon insertion sites were identified by single-primer colony PCR followed by DNA sequencing. Using the primer SP3 we amplified DNA fragments including a 278 bp region upstream of the 3′ Inverted Terminal Repeat (ITR) of the inserted transposon (Fig. 1A). The amplified fragments were then sequenced using the transposon-specific primer P3, which recognizes a sequence in the 3′ region of the Chloramphenicol Acetyltransferase (CAT) gene (Fig. 1A). The obtained sequences were then aligned on the Coxiella burnetii RSA493 annotated genome using automated sequence analysis software. The genome of Coxiella burnetii RSA493 contains 1849 coding sequences (CDS), 1814 in the bacterial chromosome and 35 in the cryptic plasmid QpH1 [35]. To date we have isolated 3000 transposon mutants, 1082 of which have been sequenced, annotated and analyzed for this study (Fig. 1B). Transposon insertions were homogeneously distributed throughout the Coxiella chromosome and plasmid, with seven “hot spots” of preferential transposon insertion (identified and annotated from 1 to 7, Fig. 1B) and a large, 52 CDS region, upstream of hot spot n. 2, which remained non-mutated. Of note, region n.7 corresponds to the locus that hosts T4SS core genes (dot/icm genes) whereas the non-mutated region between CBU_0215 and CBU_0272 is enriched in genes encoding ribosomal proteins. Overall, 926 transposon insertions were found within Coxiella annotated CDS and 156 in intergenic regions of the Coxiella genome (excluding insertions within the first 100 bp upstream of a CDS; Fig. 1C). Frequency distribution analysis revealed that mutations occurred in 483 CDS on the Coxiella chromosome and 8 CDS on the QpH1 plasmid (corresponding to 26.6% and 22.8% of the total CDS present on chromosome and plasmid respectively; Fig. 1C). The mutated CDS were then clustered according to their predicted function based on the data available on the Pathosystems Resource Integration Center (PATRIC, www.patricbrc.org; Fig. 1D).
Fig. 1. Generation of a bank of GFP-tagged Coxiella transposon mutants. 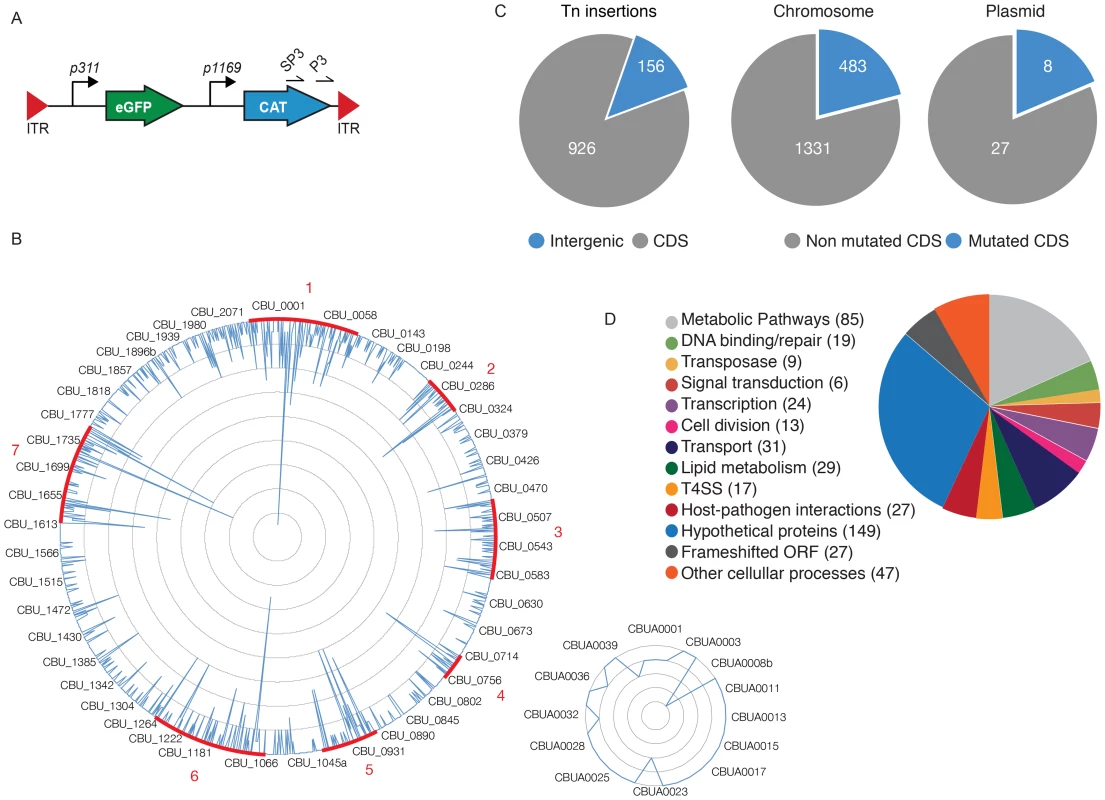
(A). The transposable element used to generate the bank of Coxiella mutants contained the eGFP gene under the regulation of the strong Coxiella promoter p311 and the chloramphenicol acetyltransferase gene (CAT) under the regulation of the mild Coxiella promoter p1169, all flanked by Inverted Terminal Repeats (ITR). Arrows labeled SP3 and P3 indicate the site of annealing of the corresponding primers. (B). Sequenced transposon insertions were annotated on the Coxiella RSA493 chromosome (large circle) and QpH1 cryptic plasmid (small circle). Peaks indicate the site of insertion of each transposon and the height of the peak corresponds to the frequency of mutants isolated presenting a transposon insertion in a given site (1 inner circle corresponds to 2 insertions). Sites of preferential transposon insertions are labeled in red. (C). Pie charts indicating the frequency of transposon insertions in Coxiella coding sequences (CDS) or intergenic regions of the genome (left chart) and the total number of mutated CDS as compared to the total number of CDS in the chromosome (middle chart) and cryptic plasmid (right chart). (D). The mutated CDS were clustered according to their predicted functions as reported by the PathoSystems Resource Integration Center (PATRIC). High-content multi-phenotypic screen of Coxiella transposon mutants
Isolated transposon mutants were used to infect Vero cells at comparable multiplicities of infection (MOI). Non-infected Vero cells were used as negative control and cells infected with GFP-NMIIC4 Coxiella (GFP-Coxiella) [33] were used as positive controls. Variations of GFP fluorescence associated with intracellular bacterial growth over 7 days of infection were monitored using a multimode micro-plate reader to obtain intracellular growth curves for all the Coxiella mutants screened (Fig. 2A). Seven days after infection, plates were imaged using an automated fluorescence microscope and images were analyzed using the automated image analysis software CellProfiler (www.cellprofiler.org). 9 morphological features were extrapolated from an average of 14000 cells for each condition (Fig. 2A). Nuclei features were used to assess the overall conditions of host cells and the potential cytotoxicity of Coxiella mutations. Coxiella features were used to score the intracellular replication of bacteria. Finally, the number of Coxiella colonies was divided by the number of host cell nuclei to estimate the efficiency of bacterial internalization within cells.
Fig. 2. Multi-phenotypic high-content screen of Coxiella transposon mutants. 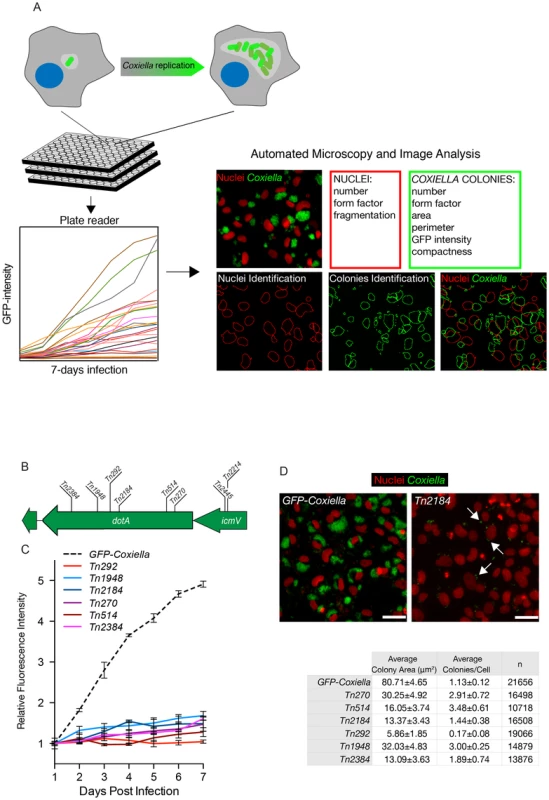
(A). Schematic representation of the screening pipeline. Cells seeded in 96-well plates in triplicate were challenged with GFP-tagged Coxiella mutants and intracellular bacterial growth was monitored over 7 days of infection using a multimode plate reader to obtain growth curves for each mutant. Plates were then fixed, and images were acquired with an automated fluorescence microscope. The automated image analysis software CellProfiler was then used to segment the acquired images and extrapolate 3 features from infected cells (red box) and 6 features from Coxiella colonies (green box) for phenotype analysis. (B). 6 independent transposon insertions in the essential dot/icm core gene dotA were used to validate the screening approach (labeled bars indicate the sites of transposon insertion within the dotA and the icmV genes). (C). Intracellular growth curves of the 6 isolated dotA transposon mutants as compared to GFP-Coxiella. From day 2 post-infection, the growth curves of all dotA mutants were significantly different from that of GFP-Coxiella (P<0.001, 2way ANOVA). (D). Representative images of colonies from GFP-Coxiella and the dotA mutant Tn2184 (green) juxtaposed to nuclei of infected host cells (red). Arrows point at intracellular Coxiella. The average area of GFP-Coxiella colonies was compared to that of the 6 dotA mutants as well as the number of colonies per cell. Data were calculated using CellProfiler; values are mean ± standard deviation of triplicate samples; the total number of analyzed cells is indicated in the right-most column of the table (n). Scale bars 20 µm. We validated our approach by comparing the growth curves and morphology of GFP-Coxiella to those of 6 mutants (Tn2384, Tn1948, Tn292, Tn2184, Tn514, Tn270) carrying independent transposon insertions in the gene CBU_1648, which encodes DotA, an essential component of the Coxiella Dot/Icm secretion system [36] (Fig. 2B). Axenic growth of the 6 dotA mutants was comparable to that of wt Coxiella (Fig. S1A). GFP signal analysis indicated that intracellular growth of GFP-Coxiella followed a typical growth curve, with bacterial replication clearly detectable from day 2 post-infection and increasing until day 7 post-infection (Fig. 2C). Morphological analysis of GFP-Coxiella-infected Vero cells reported bacterial colonies with an average area of 80.71±4.65 microns2 and an average number of Coxiella colonies/cell of 1.13±0.12 (n = 21656; Fig. 2D). As expected, the GFP signal originated by all the DotA transposon mutants failed to increase significantly during the 7 days of infection (Fig. 2C). Accordingly, morphological analysis indicated a strong reduction in the average area of intracellular Coxiella colonies, which ranged from 5.86±1.85 to 32.03±4.83 microns2 (n = 19066 and 14879 respectively, Fig. 2D). Interestingly, the majority of dotA mutations corresponded to an increased number of colonies/cell, which, in the case of mutant Tn514, reached 3.48±0.61 (Fig. 2D). This phenotype suggests that, in the absence of a functional secretion system, the Coxiella-containing vacuoles fail to coalesce to form the typical, large PV. Moreover, two independent transposon insertions in the Dot/Icm gene icmV (Tn2445 and Tn2214), which is located in the same operon as dotA, produced a comparable, strong replication phenotype (data not shown).
The data derived from the multi-phenotypic analysis of Coxiella infections of host cells were mined to identify mutations that perturbed: 1) host cell invasion, 2) intracellular replication and 3) host cell survival. To screen for invasion and replication phenotypes, we plotted the average area of Coxiella colonies against the average number of colonies/cell (Fig. 3A). Statistical analysis was used to define regions in the resulting scatter plot corresponding to mild (−4<Z-score≤−2) and severe (Z-score≤−4) phenotypes. The 1082 analyzed Coxiella mutants were found in 3 well-defined clusters: one included mutants whose phenotype did not vary significantly from that of GFP-Coxiella (Fig. 3A green dots). A second cluster was clearly shifted towards a reduction in the size of Coxiella colonies and an increase in the average number of Coxiella colonies/cell, representative of mutations that affect Coxiella intracellular replication without affecting host cell invasion (Fig. 3A light and dark red dots). A third cluster was shifted towards a reduced number of colonies/cell, indicating mutations that affect host cell invasion (Fig. 3A light and dark blue dots). In parallel, the average area of Coxiella colonies was plotted against the total number of host cells surviving the 7 days of infection, to identify Coxiella genes that are potentially involved in the protection of the host cell from apoptosis (Fig. 3B). As above, statistical analysis was used to define regions corresponding to mild (−4<Z-score≤−2) and severe (Z-score≤−4) phenotypes. The vast majority of the mutants analyzed did not affect cell survival, regardless of bacterial replication within host cells (Fig. 3B green dots). 37 mutations mildly affected host cell survival (Fig. 3B light red dots), and 7 mutations were particularly detrimental to host cell survival (Fig. 3B dark red dots). Next, the phenotypic data from mutations within CDS were integrated with the annotated transposon insertions in the Coxiella genome. We thus clustered the screened mutations according to the mutated CDS and assigned to each mutant a color-coded map based on the intensity of their replication (R), internalization (I) or cytotoxic (C) phenotype (Table S1). Intergenic transposon insertions were retained in a separate table (data not shown).Importantly, mutant Tn1832 carries an intergenic transposon insertion and phenocopies wt Coxiella and GFP-Coxiella (Fig. S2). This mutant has been used in our validation experiments as additional control.
Fig. 3. Large-scale identification of Coxiella factors involved in host/pathogen interactions. 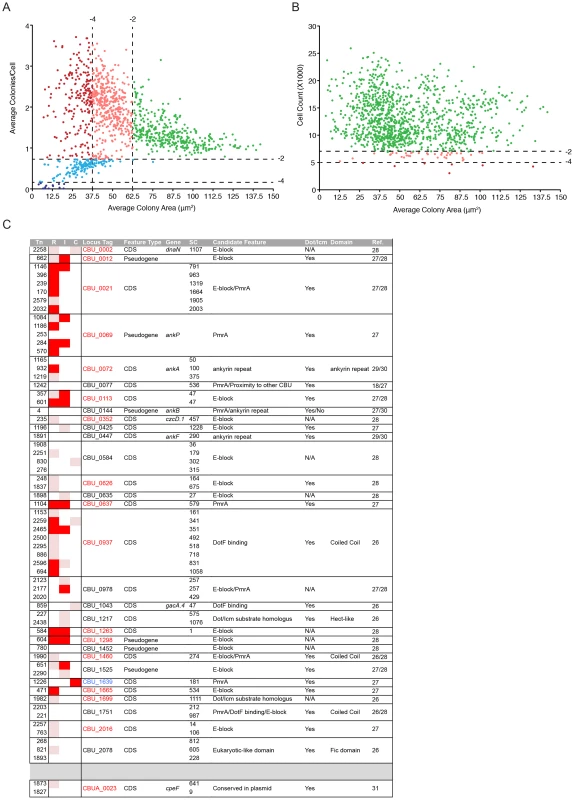
(A). For each screened mutant, the average area (in microns2) of Coxiella colonies was plotted against the relative number of colonies per cell to identify phenotypes of interest. Green dots represent phenotypes that deviate from wt Coxiella by a Z-score>−2 (not significant). Pink and light blue dots represent replication and internalization phenotypes respectively, with a Z-score between −2 and −4 (mild phenotypes). Red and dark blue dots represent phenotypes with a Z-score≤−4 (strong phenotypes). (B). For each screened mutant, the average area (in microns2) of Coxiella colonies was plotted against the total number of cells (infected and not infected) that survived 7 days of infection to estimate the cytotoxicity of transposon insertions. Green dots represent phenotypes that deviate from wt Coxiella by a Z-score>−2 (not significant). Pink dots represent cytotoxic phenotypes with a Z-score between −2 and −4 (mild phenotypes). Red dots represent cytotoxic phenotypes with a Z-score≤−4 (strong phenotypes). (C). Mutants presenting transposon insertions disrupting Coxiella genes previously identified as putative Dot/Icm substrates were clustered in rows according to the mutated gene (CDS) and their intracellular replication (R), internalization (I) and cytotoxic (C) phenotypes were illustrated. White squares represent non-significant phenotypes (Z-score>−2). Pink squares represent mild phenotypes (Z-score between −2 and −4). Red squares represent strong phenotypes (Z-score≤−4). Mutant number (Tn), information on the feature (Feature Type), annotated CDS name (Gene), distance of the transposon insertion site from the CDS start codon (SC), feature that allowed the identification of the gene as a candidate Dot/Icm substrate (Candidate Feature), Dot/Icm-mediated secretion (Dot/Icm), identified protein domains (Domain) and reference to previous publications (Ref.) were integrated in the table. CDS codes in red indicate significant intracellular replication phenotypes; CDS codes in blue indicate significant cytotoxic phenotypes. Coxiella factors required for intracellular replication
Intracellular replication of Coxiella relies on the functionality of a Dot/Icm T4SS, which is highly homologous to that of L. pneumophila [2], [37]–[39]. The Coxiella genome contains 23 homologues to the 25 known dot/icm genes of the Legionella T4SS. Accordingly, Legionella has been used as a model organism to test the secretion of candidate Coxiella effector proteins. Recently, it has been reported that the Coxiella dot/icm genes icmD, dotA, dotB and icmL.1 are essential for Coxiella replication within host cells, proving for the first time the functionality of the Coxiella T4SS [18], [19], [36]. The enrichment of transposon insertions in dot/icm genes (Fig. 1B, region n. 7), prompted us to analyze the phenotype of 38 Coxiella mutants carrying single transposon insertions in 16 Dot/Icm genes (Fig. 4A; Fig. S1). First, we followed the axenic growth of the 38 Dot/Icm transposon mutants and found it to be indistinguishable from that of wt Coxiella and of the control transposon mutant Tn1832 (Fig. S1A). Multi-phenotypic analysis confirmed the previously reported observations that icmD, dotA and icmL.1 are essential for Coxiella replication within host cells [18], [19], [36] (Fig. 4B, C; Fig. S1B). Moreover, we could observe that 12 dot/icm genes (dotA, dotB, icmV, E, D, G, J, N, C, P, K, X, L.1) are essential for bacterial replication within the host, whereas mutations in icmB and icmS showed an intermediate phenotype, which corresponded to partial intracellular replication as assessed by morphological analysis (Fig. 4B, C; Fig. S1B). Of note, transposon insertions in dot/icm genes present in operons produced consistent phenotypes. Each mutation was then trans complemented by challenging host cells in combination with wt Coxiella as previously described [19]. The intergenic mutant Tn1832 was used as positive control. As illustrated in Figure 4C, mutations resulting in severe replication phenotypes were efficiently complemented by the presence of wt Coxiella in the PV occupied by Dot/Icm mutants (black bars).
Fig. 4. Role of Dot/Icm core proteins in Coxiella infections. 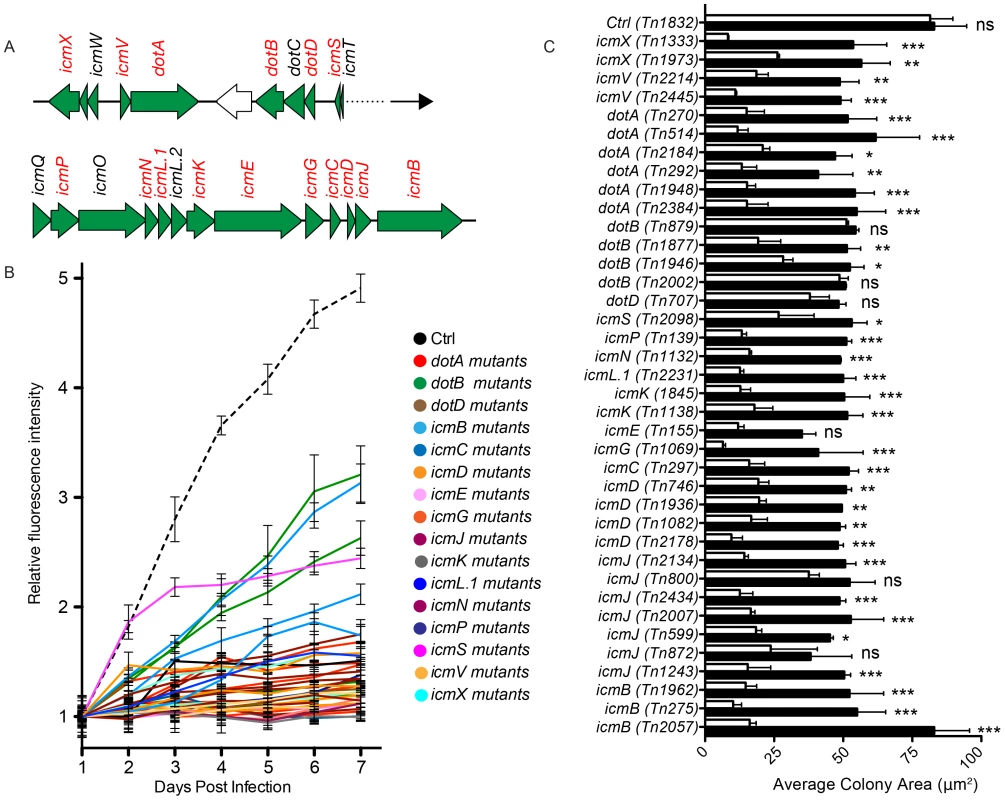
(A). Schematic representation of the region of the Coxiella genome containing Dot/Icm genes. The genes that have been mutated in this study are represented in red. (B). Intracellular growth curves of Coxiella mutants containing transposon insertions in dot/icm genes calculated as variations of GFP fluorescence over 7 days of infection and compared to GFP-tagged wt Coxiella (Ctrl, dashed black line). Mutants in the same CDS are grouped by color. With the exception of Tn2098 (icmS), the growth curves of all Dot/Icm mutants were significantly different from that of GFP-Coxiella (P<0.001, 2way ANOVA) from day 2 post-infection. The growth curve of Tn2098 (icmS) was significantly different from that of GFP-Coxiella (P<0.001, 2way ANOVA) from day 3 post-infection. (C) Vero cells were incubated either with the Dot/Icm transposon mutants alone (white bars) or in combination with wt Coxiella (black bars) to complement the replication phenotypes observed. 7 days post-infection, the average area (in microns2) was calculated using CellProfiler from an average of 6000 cells per condition. Values are mean ± standard deviation of triplicate samples (co-infection phenotypes were compared to their respective single infection condition. ns = non-significant; * = P<0.05; ** = P<0.01; *** = P<0.001, 2way ANOVA). We have also isolated and screened 63 transposon mutants in 31 CDS encoding predicted Coxiella Dot/Icm substrates (Fig. 3C). The products of 22 of these CDS have been previously reported to be positive for Dot/Icm secretion [18], [26], [27], [29], [30], [31] (Fig. 3C). Our analysis indicated that mutations in 17 of these genes produced replication phenotypes (Fig. 3C, CDS in red), further suggesting that these genes encode Coxiella effectors. Interestingly, a transposon insertion in CBU_1639 resulted in a strong cytotoxic phenotype (Fig. 3C CDS in blue), suggesting that this gene plays a role in the Coxiella-mediated inhibition of apoptosis.
The Coxiella genome encodes 4 two-component systems: PhoB-PhoR (CBU_0367-CBU_0366), GacA-GacS (CBU_0712-CBU_0760), QseB-QseC (CBU_1227-CBU_1228) and an RstB-like system (CBU_2005-CBU_2006) [2], [35]. In particular, the QseB-QseC system has been reported to be homologous to the L. pneumophila PmrA-PmrB system, a fundamental regulator of Dot/Icm secretion and its role has been indirectly confirmed [26], [40]. Consistent with these observations and with our analysis on Coxiella Dot/Icm genes, 6 independent transposon insertions in CBU_1227 (qseB) and one insertion in CBU_1228 (qseC), significantly impaired Coxiella replication within host cells (Table S1). A single transposon insertion in CBU_2006, part of the RstB-like system also produced a significant replication and entry phenotype (Table S1), however, the role of this two-component system in Coxiella remains to be defined.
The annotation of the Coxiella burnetii NMI RSA493 genome revealed the presence of 207 pseudogenes, which are not conserved among different Coxiella isolates [35]. Recent whole transcriptome analysis (RNA-Seq) has validated the previous annotation confirming that these sequences do not encode complete open reading frames (Prof. Howard Shuman personal communication). Interestingly, we have isolated 85 transposon mutants in 56 CDS annotated as pseudogenes and mutations in 32 of these resulted in a strong replication phenotype (Fig. S3). Finally, we could observe that 71 out of the 151 transposon insertions in intergenic regions of the Coxiella genome exhibited a significant replication phenotype (not shown). This may reveal small RNA-mediated regulation of Coxiella virulence. Indeed, 11 transposon insertions fall within intergenic regions where putative Coxiella sRNAs have been identified by RNA-Seq (Prof. Howard Shuman personal communication). Further investigations are currently aiming at an in-depth characterization of these new candidate factors that may regulate intracellular replication of Coxiella.
Coxiella factors involved in subversion of apoptosis in infected cells
As mentioned above, our multi-phenotypic screen identified 7 transposon mutants that exhibited a strong cytotoxic phenotype when incubated with Vero cells (Fig. 3B, Fig. S4). To further analyze the phenotype of these mutants, we investigated their intrinsic capacity of triggering apoptosis and their potential of protecting infected cells from staurosporine-induced apoptosis. HeLa cells were preferred to Vero cells for this assay as they displayed a more consistent response to staurosporine and all Coxiella strains tested displayed similar replication phenotype in these cells (data not shown). Cells were either left unchallenged or incubated with wt Coxiella, the control transposon mutant Tn1832, the DotA mutant Tn270 and the 7 cytotoxic mutants (Tn881, Tn616, Tn946, Tn926, Tn1226, Tn1232, Tn1233). Three days post-inoculation, cells were either fixed in paraformaldehyde or incubated with 1 µM staurosporine for 4 h prior to fixation. The percentage of apoptotic cells was then evaluated for each condition by the TUNEL assay. Very few TUNEL-positive cells were observed among untreated cells, these were increased to 50% of the total cell population upon staurosporine treatment. As expected, incubation of cells with wt Coxiella did not increase the number of TUNEL-positive cells as compared to untreated cells and wt Coxiella-colonized cells were efficiently protected from staurosporine-induced apoptosis. Cells challenged with the control transposon mutant Tn1832 presented the same phenotype as cells incubated with wt Coxiella and conversely, the DotA mutant Tn207 failed to protect infected cells from induced apoptosis (Fig. S4). Mutant Tn881, which carries a transposon insertion in CBU_0485, exhibited a partial protection of infected cells from induced apoptosis whereas the remaining mutants failed to effectively protect cells from the effects of staurosporine (Fig. S4). Interestingly, incubation of HeLa cells with mutants carrying transposon insertions in CBU_1639, CBU_1366 and CBU_0307a significantly increased the number of TUNEL-positive cells also in the absence of staurosporine, indicating that these mutants may possess intrinsic cytotoxic properties (Fig. S4).
Identification of a Coxiella surface protein involved in host cell invasion
As for all obligate intracellular pathogens, Coxiella invasion of host cells is a priming step of the infection. However, since the bacterial factors that mediate Coxiella invasion of host cells remain unknown, we sought to identify bacterial factors whose mutations affect Coxiella internalization. High-content screening identified 48 mutations in 37 Coxiella CDS that resulted in a significant reduction in the number of infected cells after 7 days of infection (Fig. S5). Of these, 18 CDS involved in bacterial metabolism and transcription were excluded. Among the remaining 19 candidate CDS (Fig. S5, CDS boxed in red), we have identified 5 independent transposon insertions (Tn175, Tn208, Tn27, Tn907 and Tn749) in CBU_1260, all sharing a consistent, strong internalization phenotype (Fig. S5). CBU_1260 is a 747 bp CDS on the positive strand of the Coxiella chromosome encoding a hypothetical protein of a predicted size of 23 kDa. Importantly, the gene is not part of an operon, indicating that the phenotype observed for the 5 mutants analyzed in this study was indeed due to the inactivation of CBU_1260 alone (Fig. 5A). Axenic growth of the 5 transposon mutants was comparable to that of wt Coxiella and of the control transposon mutant Tn1832 (Fig. S6A). As expected, intracellular growth curve analysis of the 5 transposon mutants in CBU_1260 indicated that GFP fluorescence failed to increase during the 7 days of infection (Fig. 5B). Indeed, all 5 mutations in CBU_1260 reduced the number of cells presenting Coxiella colonies at 7 days of infection by 60–70% compared to cells challenged with GFP-Coxiella (Fig. 5C). We next used the online analysis software i-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) and Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/) to predict the structure of the hypothetical protein encoded by CBU_1260. Bioinformatics analysis predicted 8 transmembrane beta sheets forming a beta barrel domain, an N-terminal alpha helix and 4 unstructured loops (L1 to L4), exposed at the cell surface (Fig. 5A, D). This prediction was confirmed by analyzing the sequence of the protein using TMpred (http://www.ch.embnet.org/software/TMPRED_form) and the BetAware software [41]. Sequence analysis indicated that transposon insertions occurred in the distal part of the CDS, within the 4th and 6th beta sheets, with mutants Tn27 and Tn907 presenting insertions at the same site (Fig. 5A). The predicted transmembrane domain of CBU_1260 is typical of Outer Membrane Protein A (OmpA) family of proteins, which are found in several bacteria and mediate adhesion and/or internalization within host cells [42]–[50]. We therefore named the product of CBU_1260 OmpA. Coxiella encodes 3 hypothetical proteins that contain predicted OmpA-like domains: CBU_0307, CBU_0936 and CBU_1260 (ompA). Sequence alignment of these three hypothetical proteins showed high degree of homology at the level of the transmembrane domains and the N-terminal alpha helix (Fig. S7). However, little homology was observed at the level of the 4 unstructured loops of OmpA (Fig. S7). Accordingly, a transposon insertion in CBU_0307 produced a cytotoxic phenotype whereas 4 transposon insertions in CBU_0936 produced a replication phenotype (Table S1). An OmpA-specific antibody was then raised against the predicted extracellular domain of the protein. To this aim, a 15 amino acid peptide in predicted loop 1 (KKSGTSKVNFTGVTL) was used for its immunogenic potential as compared to peptides in the other loops. When tested by Western blot on lysates from wt Coxiella and the control mutant Tn1832, the anti-OmpA antibody revealed a major band at the expected size of 23 kDa (Fig. 5E, arrow) and a faint, background band of lower molecular weight (Fig. 5E). When incubated on lysates from the 5 transposon mutants in CBU_1260, the anti-OmpA antibody only recognized the background band of lower molecular size. We next performed membrane fractionation assays on wt Coxiella and the OmpA mutant Tn208 to validate the outer membrane localization of OmpA. The protein was highly enriched in the outer membrane fraction of wt Coxiella and, as expected, absent in lysates from the OmpA mutant Tn208 (Fig. 5F). Taken together, our data indicate that CBU_1260 encodes an outer membrane protein with a predicted OmpA-like structure that plays a relevant role in host cell invasion.
Fig. 5. Characterization of CBU_1260 (OmpA), a Coxiella protein involved in host cell invasion. 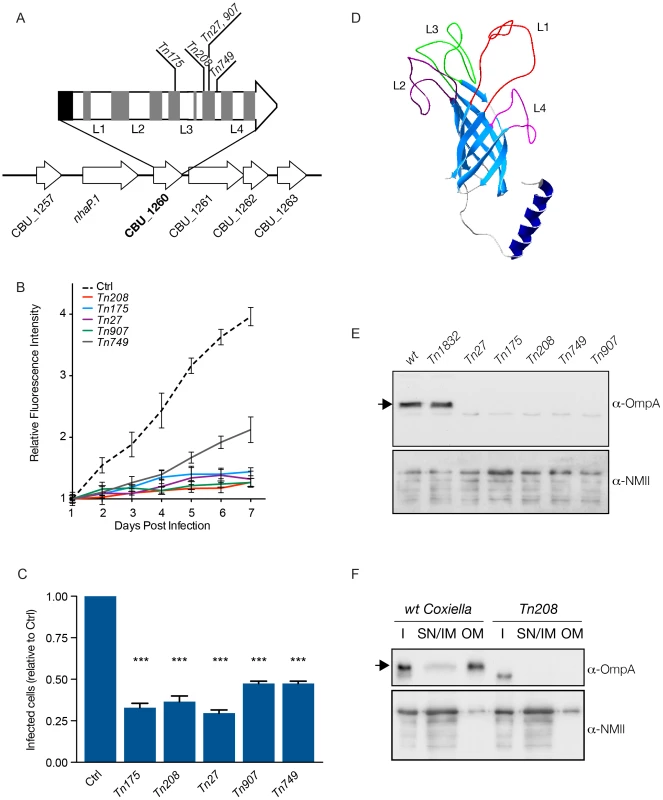
(A). Schematic representation of the genomic context of CBU_1260 and sites of transposon insertion of 5 independent isolated mutants (Tn175, Tn208, Tn27, Tn907, Tn749). Black area indicates the position of the predicted N-terminal α-helix and grey areas indicate the position of predicted β-sheets. Predicted extracellular loops are indicated from L1 to L4. (B). Intracellular growth curves of Coxiella mutants containing transposon insertions in CBU_1260 measured as variation of GFP fluorescence over 7 days of infection and compared to GFP-tagged wt Coxiella (Ctrl). The growth curves of all CBU_1260 mutants were significantly different from that of GFP-Coxiella (P<0.001, 2way ANOVA) from day 2 post-infection. (C). For each CBU_1260 mutant analyzed, the total number of Coxiella colonies identified by automated image analysis was divided by the total number of cell nuclei and expressed as relative to GFP-tagged wt Coxiella (Ctrl) to derive the efficiency of host cell invasion. Values are mean ± standard deviation of triplicate samples where an average of 12000 cells were analyzed per condition (values were compared with Ctrl condition. *** = P<0.001, 2way ANOVA). (D). iTASSER-derived prediction the CBU_1260 product. The central core of the protein is composed of an 8-β-sheet transmembrane barrel domain (light blue). The putative periplasmic domain is composed of a α-helix (dark blue), whereas the predicted extracellular domain is composed of four (L1 to 4) unstructured loops. (E). Representative Western blot of bacterial lysates from wt Coxiella, the control mutant Tn1832 and the 5 mutants presenting transposon insertions in CBU_1260. (F). Representative Western blot of wt Coxiella and mutant Tn208 subjected to membrane fractionation to assess the localization of OmpA (I = insoluble fraction; SN/IM = supernatant and inner membrane fraction, OM = outer membrane fraction). Blots were revealed with an antibody raised against the first extracellular loop (L1) of OmpA (top panels) and with a polyclonal antibody against Coxiella NMII (bottom panels) used as loading control. The arrows indicate the expected size of OmpA (23 kDa). OmpA mediates Coxiella internalization and intracellular replication within host cells
To further investigate the role of OmpA in Coxiella invasion of host cells, we validated the transposon insertion in the OmpA mutant Tn208 by PCR and used a GFP-probe to confirm by Southern blot that Tn208 contained a single transposon insertion (Fig. S6B, C). Next, non-phagocytic epithelial cells (A431), THP-1 (PMA-differentiated), J774 and RAW 264.7 macrophages were challenged either with wt Coxiella, the OmpA mutant Tn208 or the control transposon mutant Tn1832. Differential labeling of extracellular and intracellular bacteria was used to assess the efficiency of Coxiella internalization within host cells at 15, 30, 45 and 60 minutes post-infection (Fig. 6A, E, top panels). Longer time points (5 and 6 days post-infection) were analyzed to investigate the intracellular development of internalized OmpA mutants (Fig. 6A, E, bottom panels). Automated image analysis was then used to analyze approximately 8000 bacteria per condition and quantify the efficiency of bacterial internalization as well as the area occupied by intracellular Coxiella colonies. In A431 non-phagocytic cells, the internalization of Tn208 was strongly reduced as compared to that of wt Coxiella or Tn1832, which shared similar kinetics (Fig. 6B). Interestingly, when the same internalization experiment was performed using macrophages, the three bacterial strains tested (wt Coxiella, Tn208 and Tn1832) were internalized with comparable efficiency (Fig. 6F; Fig. S8A, D), with a concomitant local rearrangement of the actin cytoskeleton (Fig. 6E, top panels). Of note, despite the strong inhibition of bacterial internalization in A431 cells, OmpA mutants retained the capacity to adhere to host cells (Fig. 6A). This suggests that if OmpA plays a role in bacterial adhesion, this may be masked by the presence of alternative factors involved in Coxiella adhesion to host cells. At longer time points of infection we observed a decrease in the number of Tn208 colonies per cells in A431 cells as compared to wt Coxiella and Tn1832 colonies, which confirmed our previous observations in Vero cells (Fig. 6C). On the contrary, the number of Coxiella-colonized cells was not affected when macrophages were challenged either with wt Coxiella, the control mutant Tn1832, or the OmpA mutant Tn208 (Fig. 6G, Fig. S8B, E). Remarkably however, the average area of OmpA mutant Coxiella colonies was significantly reduced as compared to wt Coxiella and Tn1832, regardless of the cell line used for the experiment (Fig. 6D, H; Fig. S8C, F). Interestingly, this is in agreement with previously reported roles of OmpA proteins in intracellular survival of bacterial pathogens [43], [45], [46], [51]–[53].
Fig. 6. Coxiella OmpA is involved in host cell invasion and intracellular replication. 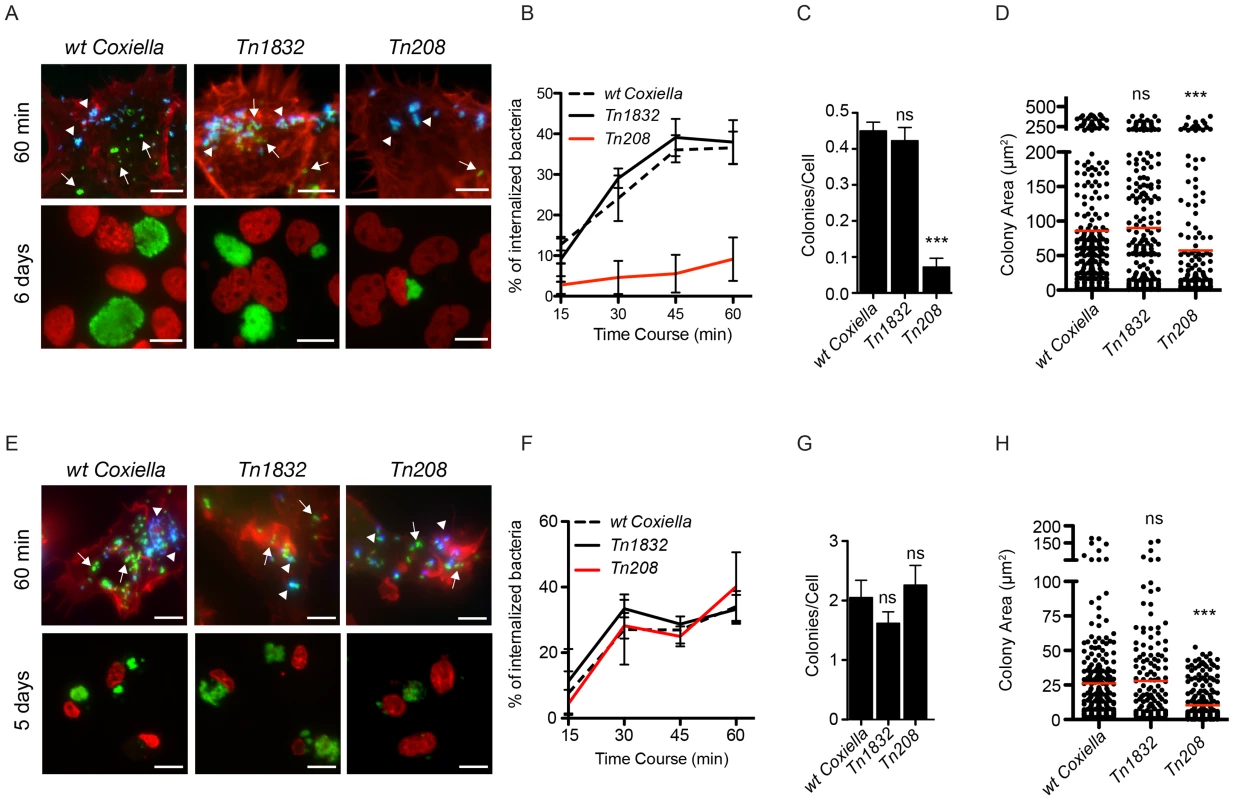
A431 (A) and THP-1 (E) cells were incubated with wt Coxiella, the control transposon mutant Tn1832 or the OmpA mutant Tn208. 60 minutes after infection (top panels) cells were fixed and labeled with an anti-Coxiella antibody coupled to Alexa Fluor 555 (blue) and with Atto-647N phalloidin (red) prior to cell permeabilization. Internalized bacteria were detected by GFP fluorescence (green) in the case of Tn208 and Tn1832 whereas for wt Coxiella infections, cells were permeabilized and bacteria were stained with the anti-Coxiella antibody as above, coupled to Alexa Fluor 488 (green). Alternatively, cells were fixed 6 (A, bottom panels) or 5 (E, bottom panels) days after infection, DNA (red) was labeled with Hoechst 33258 and wt Coxiella (green) with the specific antibody as above. The automated image analysis software CellProfiler was used to calculate the percentage of internalized bacteria (B and F), the number of colonies/cell (C and G) and the area (in microns2) of intracellular Coxiella colonies identified for each condition (D and H). Values are means ± standard deviations of triplicate experiments where an average of 8000 bacteria (B and F) or 400 vacuoles (C, D, G, H) were analyzed for each condition (values were compared to wt Coxiella infections. ns = non-significant; *** = P<0.001 2way ANOVA for B and F and t test for C, D, G, H). The difference between the percentage of internalized wt Coxiella (or the control mutant Tn1832) and the Tn208 mutant was statistically significant in B (P<0.001, 2way ANOVA) and non-significant in F. Arrows indicate internalized bacteria (green); arrowheads indicate extracellular bacteria (green and blue). Scale bars 10 µm. OmpA is necessary and sufficient to mediate internalization within non-phagocytic cells
To further dissect the role of OmpA in Coxiella interaction with host cell surfaces, we 1) purified the recombinant protein to coat inert latex beads and 2) ectopically expressed OmpA in E. coli, to assess the capacity of OmpA to confer adhesiveness and invasiveness to inert particles and extracellular bacteria, respectively. Histidine-tagged, recombinant OmpA was produced by E. coli BL21-DE3 star pLysS transformed with the pET28a vector containing ompA32-248. The first 31 amino acids of OmpA corresponding to the intracellular N-terminal alpha helix were excluded to increase the solubility of the protein. Red fluorescent latex beads were then coated with 100 µg/ml His-OmpA32-248 or GST as control and used to challenge A431 cells for 1 hour at 37°C. Unbound beads were removed by rinsing cells in PBS and cells were fixed in paraformaldehyde. Cells were then probed with an anti-histidine antibody without permeabilization to differentially label adherent and internalized beads and with fluorescent phalloidin to define the cell perimeter and volume. Alternatively, after fixation samples were further processed for scanning electron microscopy. Confocal microscopy analysis of cross sections of cells incubated with His-OmpA32-248-coated beads revealed a fraction of beads adhering to the cell surface, hence positive for the anti-histidine staining, and another fraction within the cell volume (as defined by the actin labeling) and negative to the anti-histidine staining (Fig. 7A). Three-dimensional reconstruction of confocal sections coupled to surface rendering confirmed the presence of adhering and internalized beads (Fig. 7A). GST-coated beads failed to adhere to and invade A431 cells significantly (Fig. 7B). Scanning electron microscopy analysis corroborated these observations: several His-OmpA32-248-coated beads were adhering to the surface of A431 cells (Fig. 7C, green inset) whereas others were clearly covered by the cell plasma membrane (Fig. 7C, red inset). Very few GST-coated beads were observed at the surface of cells and none seemed to be internalized (data not shown), confirming our observations by fluorescence microscopy. We next assessed the capacity of OmpA to trigger the internalization of non-invasive bacteria by non-phagocytic cells using the gentamicin protection assay. To this aim E. coli BL21-DE3 star pLysS were transformed with pET27b-OmpA, which allowed the IPTG-regulated expression and periplasmic targeting of full-length OmpA. The expression and outer membrane localization of OmpA were verified by Western blot using the OmpA-specific antibody on transformed E. coli cultures induced overnight with IPTG and processed to separate the bacterial outer membranes from the inner membranes and cytoplasmic components (Fig. 7D). Non-induced, transformed bacteria were used as control. We could observe that IPTG-induced bacteria efficiently produced OmpA, which was enriched in the outer membrane fraction of E. coli lysates (Fig. 7D). Importantly, induction of OmpA expression conferred E. coli a 20-fold increase in invasiveness as compared to the non-induced bacteria (Fig. 7E). Next, we used the E. coli ectopic expression approach to dissect the role of the 4 predicted extracellular loops of OmpA. By replacing each loop with a myc tag, we generated 4 OmpA mutants (OmpAΔL1, OmpAΔL2, OmpAΔL3, OmpAΔL4) that were used to test their capacity to confer invasiveness to E. coli in a gentamicin protection assay. Similar to wt OmpA, all mutated proteins were detected in the outer membrane fraction of induced E. coli (not shown). Interestingly, only the exchange of loop 1 with a myc tag significantly reduced E. coli internalization by non-phagocytic cells (Fig. 7E). Collectively, these data suggest that OmpA is necessary and sufficient to mediate Coxiella internalization within non-phagocytic cells and that loop 1 is primarily involved in interacting with a potential cognate receptor at the surface of host cells.
Fig. 7. OmpA is necessary and sufficient to trigger the internalization of beads and extracellular bacteria within non-phagocytic cells. 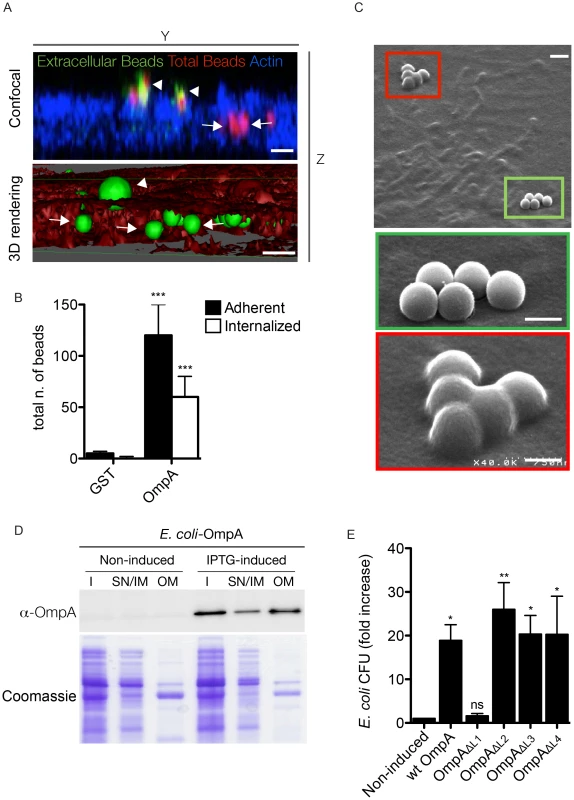
(A). A431 cells were incubated 60 minutes with Cy3 latex beads coated with His-tagged OmpA32-248 (red). To visualize extracellular beads, cells were labeled without permeabilization with an anti-histidine antibody coupled to Alexa Fluor 488 (green) and the cell cytoskeleton was labeled with Atto-647N phalloidin. Successive images were acquired along the Z-axis of the sample using a confocal microscope and the YZ view was reconstructed using ImageJ (Confocal). Alternatively, surface volume rendering (3D) was obtained using Osirix. Beads were pseudocolored in green and the cell volume in red. Arrowheads and arrows point at extracellular and intracellular beads, respectively. Scale bars 1 µm. (B). A431 cells were incubated 60 minutes with Cy3 latex beads coated with either His-tagged OmpA32-248 or GST. Extracellular beads were visualized by labeling non-permeabilized cells with an anti-histidine or an anti-GST antibody, respectively. The automated image analysis software CellProfiler was used to identify and count intracellular and extracellular (adherent) beads. Values are means ± standard deviations of triplicate experiments (values were compared to GST control conditions. *** = P<0.001, 2way ANOVA) (C). Representative image of A431 cells incubated 60 minutes with Cy3 latex beads coated with His-tagged OmpA32-248 and processed for scanning electron microscopy. Green box and inset indicate adherent beads; red box and inset indicate internalized beads. Scale bars 1 µm (top panel) and 0.5 µm (insets). (D). E. coli BL21-DE3 star pLysS transformed with pET27b-OmpA (E. coli-OmpA) were left untreated (Non-induced) or induced with IPTG (IPTG-induced) to synthesize periplasm-targeted OmpA. Insoluble bacterial fractions (I), supernatant and inner membrane fractions (SN/IM) and outer membrane fractions (OM) were probed with the anti-OmpA antibody by Western blot (upper panel). Samples stained with Coomassie blue were used as loading control (lower panel). (E). E. coli transformed with pET27b-OmpA (wt OmpA) or one of the 4 variants carrying loop substitutions (OmpAΔL1, OmpAΔL2, OmpAΔL3, OmpAΔL4) were left untreated or induced with IPTG and incubated with A431 cells. The gentamicin survival assay was used to determine the capacity of E. coli expressing wt OmpA and its mutated variants to invade non-phagocytic cells. Values are mean ± standard deviations of triplicate experiments (values were compared to non induced condition. ns = non-significant; * = P<0.05; ** = P<0.01, 2way ANOVA). Blocking OmpA interactions with the host cell inhibits Coxiella internalization
To determine whether OmpA function requires the interaction with host cell surface factors, we sought to block candidate ligand/receptor interactions, either by saturating potential OmpA receptors at the surface of host cells or by masking OmpA at the surface of bacteria, prior to infection. In the first case, A431 cells were incubated with 100 µg/ml His-OmpA32-248for 1 hour at 4°C prior to challenging with wt Coxiella and the efficiency of bacterial internalization was determined by differential bacterial labeling. A431 cells incubated in the same conditions with GST or with buffer alone were used as controls. Indeed, pretreating A431 cells with His-OmpA32-248 effectively inhibited wt Coxiella internalization as compared to buffer - or GST-treated cells (Fig. 8A, B). Alternatively, wt Coxiella were incubated with increasing concentrations (0.1, 1 and 5 µg/ml) of either anti-OmpA antibody or naïve rabbit serum prior to infection and bacterial differential labeling was used to determine Coxiella invasiveness in A431 cells. Confirming our previous observations, the pre-treatment of bacteria with the anti-OmpA antibody, but not naïve rabbit serum, inhibited Coxiella internalization in a concentration dependent manner (Fig. 8C, D). These observations suggest the presence of a receptor for OmpA at the surface of host cells, which remains to be identified, and that OmpA/receptor interactions are essential to mediate Coxiella internalization within host cells.
Fig. 8. Recombinant OmpA and anti-OmpA antibody perturb Coxiella interactions with the host cell surface. 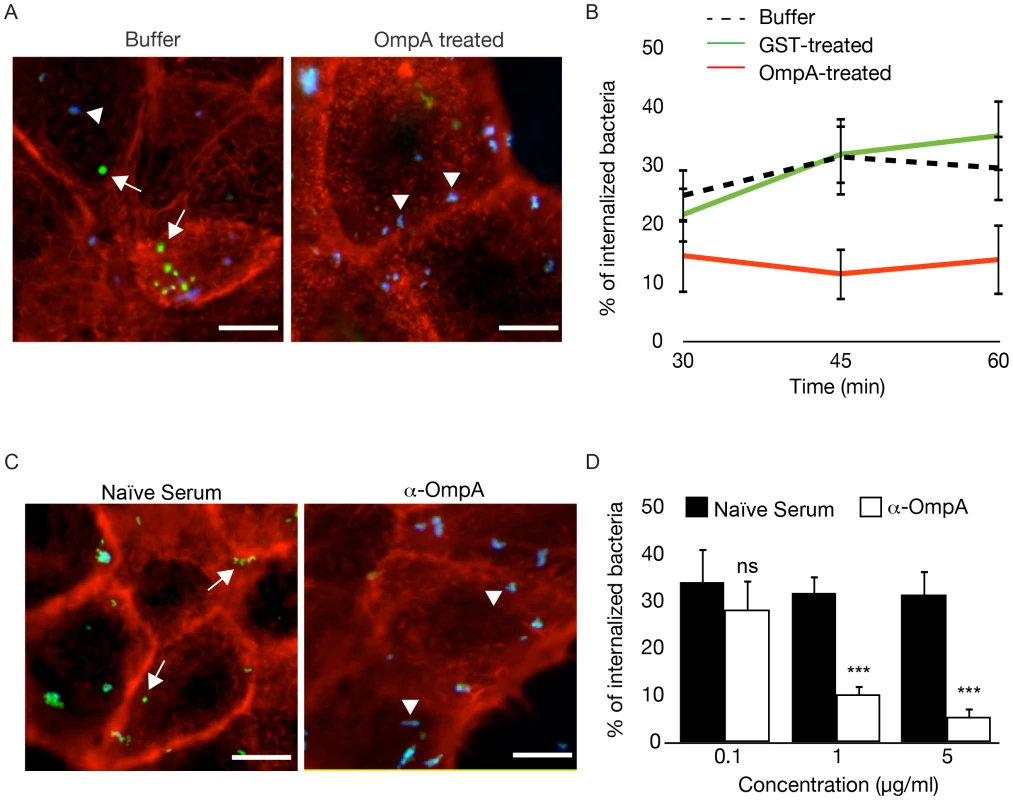
(A). Representative images of A431 cells pretreated with OmpA32-248 buffer or with 100 µg/ml His-tagged OmpA32-248 and incubated with wt Coxiella for 60 minutes. Cells were fixed and labeled with an anti-Coxiella antibody coupled to Alexa Fluor 555 (blue) and with Atto-647N phalloidin (red) prior to cell permeabilization. Internalized bacteria were detected by using the same anti-Coxiella antibody as above coupled to Alexa Fluor 488, on permeabilized cells (green). Arrowheads and arrows point at extracellular and intracellular bacteria, respectively. (B). The automated image analysis software CellProfiler was used to analyze cells pretreated with OmpA32-248 buffer, 100 µg/ml GST or 100 µg/ml His-tagged OmpA32-248 and incubated with wt Coxiella for 30, 45 and 60 minutes. For each condition, intracellular and extracellular bacteria were automatically identified, counted and the percentage of internalized bacteria was calculated. (C). Representative images of A431 cells incubated for 60 minutes with wt Coxiella pretreated with 1 µg/ml rabbit naïve serum or anti-OmpA antibody. After fixation, cells were treated as in A. Arrowheads and arrows point at extracellular and intracellular bacteria, respectively. (D). CellProfiler was used to analyze cells incubated for 60 minutes with wt Coxiella pretreated with naïve serum or anti-OmpA antibody at increasing concentrations as indicated. For each condition, intracellular and extracellular bacteria were automatically identified, counted and the percentage of internalized bacteria was calculated. In all cases, values are means ± standard deviations of triplicate experiments where an average of 6000 bacteria were analyzed for each condition. In B, the difference between the percentage of internalized wt Coxiella in cells treated with buffer alone (or with GST) and cells treated with recombinant OmpA was statistically significant from 45 min of infection (P<0.001, 2way ANOVA). In D, values were compared to naïve serum condition. ns = non-significant; * = P<0.05; *** = P<0.001, 2way ANOVA. Scale bars 10 µm. OmpA mutation attenuates Coxiella virulence in the in vivo model system Galleria mellonella
Larvae of the wax moth Galleria mellonella are an emerging, efficient model for the study of host/pathogen interactions in vivo. Like other insects, Galleria larvae present essential aspects of the innate immune response to microbial infections, which are conserved in mammals. In particular, insects possess humoral and cellular defense responses, the first including antimicrobial peptides (galiomycin, gallerimycin and lysozyme in the case of Galleria) and the latter consisting of specialized phagocytic cells, known as hemocytes or granulocytes [54]–[57]. Importantly, in the case of several bacterial pathogens, typical phenotypes observed in mammalian infection models were efficiently reproduced using Galleria [58]–[61]. It has been recently demonstrated that the Coxiella closely-related pathogen Legionella pneumophila invades Galleria hemocytes and replicates within large membranous compartments that present the same morphology and characteristics of Legionella-containing vacuoles generated during infection of macrophages and amoeba [62]. Infections by L. pneumophila result in severe damage to insect organs, which is accompanied by an immune response, including larvae melanization and nodule formation [62]. Moreover, the role of bacterial virulence factors previously characterized in higher mammalian models is conserved during infections of Galleria mellonella [61], [62]. Importantly, Galleria mellonella larvae are also susceptible to phase II Coxiella infections (Norville et al. Unpublished data). We thus investigated the phenotype associated with the OmpA mutation carried by Tn208 in the context of Galleria infections. Larvae were exposed to Coxiella by injecting 106 bacteria (either wt Coxiella, Tn1832 or Tn208) in the upper right proleg and larvae were incubated at 37°C up to 300 h post-infection to determine survival rates. Alternatively, larvae were incubated up to 24 and 96 h prior to fixation in paraformaldehyde and processing for immunofluorescence. In all cases, larvae injected with PBS were used as a negative control. Larvae injected with PBS alone did not show any survival defect throughout the incubation time, whereas larvae infected with wt Coxiella or the control mutant Tn1832 died significantly faster compared to those infected with the OmpA mutant Tn208 (Fig. 9A). Immunofluorescence analysis revealed that at 96 h post-inoculation, wt Coxiella as well as the control mutant Tn1832 organisms triggered the formation of large, highly infected nodules of hemocytes (Fig. 9B). These were often juxtaposed to larval organs that also appeared severely infected and damaged (Fig. 9C, top panels). In contrast, when Galleria were challenged with the OmpA mutant Tn208 fewer nodules of smaller size were observed throughout the larvae and only a small fraction of these presented signs of infection (Fig. 9B). When these nodules were observed at higher magnification, we could detect small Tn208 colonies (Fig. 9C bottom panels) that were reminiscent of what we had previously observed in cultured macrophages. Our observations indicate that the OmpA-associated phenotypes observed in cultured cells can be reproduced during in vivo infections.
Fig. 9. OmpA mutation perturbs Coxiella infections in vivo. 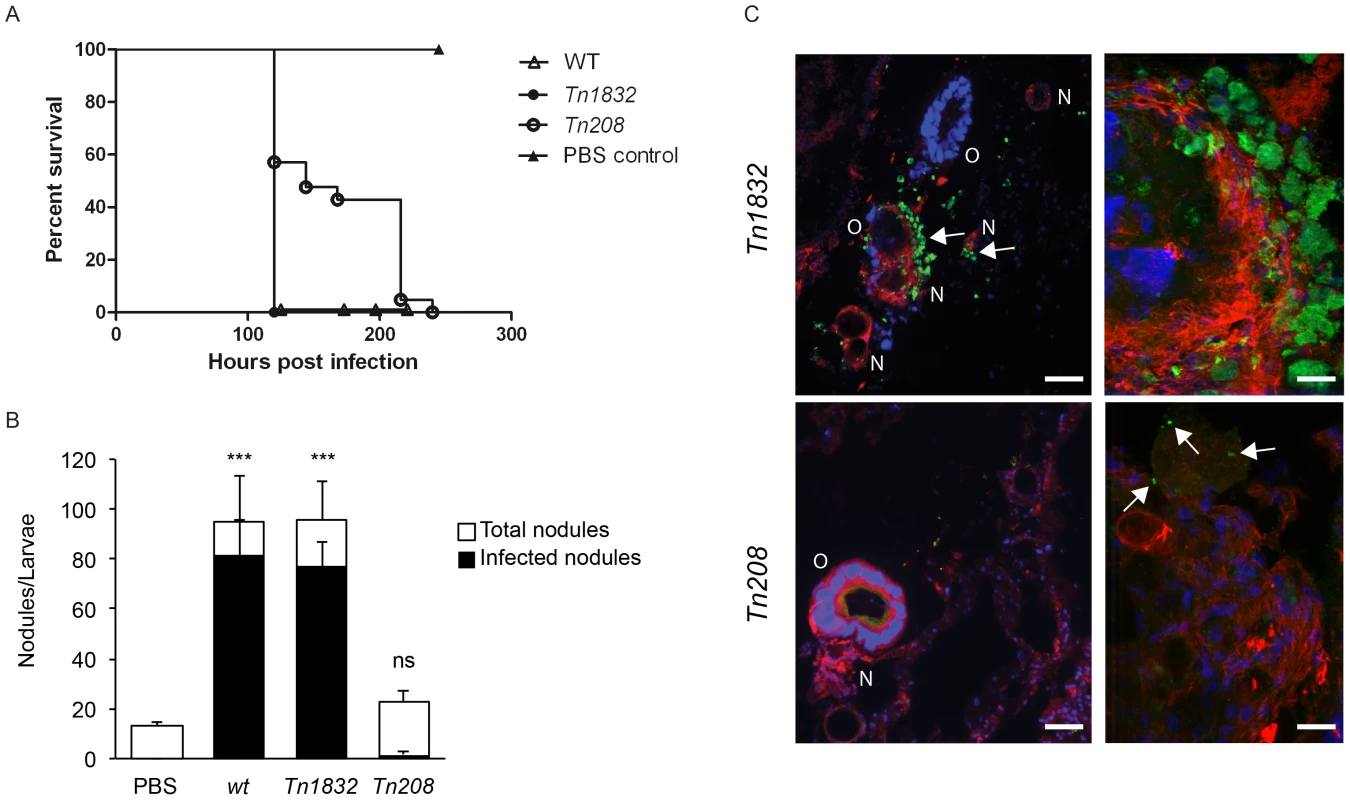
(A). Survival chart of Galleria mellonella larvae injected with either wt Coxiella (empty triangles), the control transposon mutant Tn1832 (full circles) or with the OmpA mutant Tn208 (empty circles). PBS-injected larvae were used as control (full triangles). Values are means of three replicates, each with 10 injected larvae per condition. P<0.05. (B). The total number of nodules (white bars) and the fraction of infected nodules (black bars) from Galleria mellonella larvae treated as in A were quantified. Values are means ± standard deviations from 3 larvae analyzed per condition (values were compared to PBS condition. ns = non-significant; *** = P<0.001, 2way ANOVA). (C). Representative images of the larval nodules (N) and organs (O) from Galleria mellonella larvae treated as in A, fixed and processed for immuno-histochemistry. Samples were stained with Hoechst 33258 (blue), Atto-647N phalloidin (red) and an anti-GFP antibody (green). Arrows indicate the presence of Coxiella in nodules and organs. Scale bars 50 µm (left panels) and 20 µm (right panels). Discussion
Infection by bacterial pathogens depends on the subversion of host functions, which is tightly orchestrated by an array of bacterial proteins referred to as virulence factors. In the last decade, cellular microbiology has stressed the importance of studying pathogens in relation to their host, however, the effective, global identification of bacterial virulence determinants and the characterization of their diverse mechanisms of action requires the development of new high-throughput and high-content screens (HTS and HCS respectively) [63]. Here, we have set up protocols for the multi-phenotypic screen of bacterial factors that are involved in host cell invasion and colonization. Our approach integrates transposon mutagenesis, genomics, bioinformatics and fluorescence-based functional assays that have been adapted for the large-scale identification of virulence factors from virtually any intracellular bacterium. The advantage of our screening technique lies in the possibility of analyzing every bacterial mutations for multiple phenotypes, such as 1) internalization within the host/cell, 2) intracellular replication and 3) cytotoxicity, simultaneously. This analysis, integrated with the map of genome mutations, allows a global overview of bacterial genes involved in host/pathogen interactions. The emerging bacterial pathogen Coxiella burnetii is an excellent model system to apply our strategy. To date, very little is known about the bacterial factors that regulate Coxiella interactions with the host; however, the recent development of axenic culture techniques now allows genetic manipulation of this microbe and in-depth analysis of its virulence factors [32], [64]. Moreover, previous in silico identification of putative Coxiella virulence factors [18], [26]–[31], provides an excellent database to cross-reference bioinformatics analysis to our functional assays. Importantly, our assay allows the identification of Coxiella virulence factors on a whole-genome scale, based on the phenotypes associated with the random mutagenesis of Coxiella CDS.
Our aim being the generation of the first bank of C. burnetii transposon mutants, we chose to sequence all isolated mutants independently of their phenotype during infections. This has provided a global survey of the distribution of transposon insertions and allowed us to pinpoint also those genes, suspected to encode virulence factors, which failed to produce a phenotype during Coxiella colonization of host cells. Mapping of transposon insertions revealed an overall homogeneous distribution of mutations, with regions of preferential transposon insertion as well as other regions that remained non-mutated. Of note, the lack of transposon insertions in the region between CBU_0678 and CBU_0698 is expected, having used the annotated genome of Coxiella NMI (RSA493) to map transposon insertions in Coxiella NMII (RSA439) in which this region is missing. Interestingly however, we failed to isolate transposon mutants from the large region between CBU_0215 and CBU_0272, enriched in essential genes encoding ribosomal proteins. This suggests that non-mutated regions may have been targeted by the transposon but gene disruption was lethal for the bacterium. Non-mutated regions may be thus exploited to map essential genes in the Coxiella chromosome and plasmid. When analyzing transposon mutants exhibiting a strong replication phenotype, we have occasionally observed internalization phenotypes that were not consistently reproduced in all transposon mutants isolated for a given gene. We believe that this is due to a technical limitation of our screening technique, imposed by the extremely different sizes and associated fluorescence of Coxiella colonies. In some cases the signal to noise ratio was very close to the threshold imposed to the image analysis software and resulted in colonies and/or bacteria that were not detected when images were segmented. Ongoing implementation of our automated analysis pipeline will allow the identification of these outlier phenotypes.
Independent studies have formally proven the essential role of the Coxiella Dot/Icm secretory apparatus by isolating mutants in 4 of the 23 dot/icm Coxiella genes (dotA, dotB, icmD and icmL.1) [18], [19], [36]. In all cases dot/icm mutants retained the capacity of invading host cells but failed to generate large PVs and replicate therein [18], [19], [36]. The enrichment of transposon insertions in the region of the Coxiella genome hosting dot/icm core genes allowed us to validate our assays exploiting existing data and, more importantly, provided a comprehensive overview of the role of the different components of the Coxiella T4SS. Interestingly, we have identified mutations resulting in intermediate phenotypes that allowed partial bacterial replication. Of particular interest is a mutation in icmS, which confers a multivacuolar phenotype to mutants. The icmS gene encodes a chaperone protein that mediates the secretion of a subclass of bacterial effectors [40]. The observation of a multivacuolar phenotype suggests that in Coxiella, IcmS may be involved in the secretion of effectors that mediate membrane fusion events required for the biogenesis of the PV. To facilitate the identification of putative IcmS substrates, a machine learning approach is currently being used to identify other transposon mutants that share the same multivacuolar phenotype.
To date, candidate Coxiella effectors have been identified by bioinformatics analysis based on conserved Dot/Icm regulatory motif (PmrA) [26], [27], C-terminal translocation signals (E-block) [27], [28], and eukaryotic-like domains. So far, 354 candidate effectors have been thus identified, however Dot/Icm-dependent translocation assays using either Coxiella or Legionella as a surrogate model, indicated that the majority of these might be false positives [18], [26]–[28]. In addition, the lack of efficient methods for the genetic manipulation of Coxiella severely hampered the functional study of these putative effectors. In a recent study, transposon mutants were obtained from 20 Coxiella candidate effectors with 10 exhibiting a significant replication phenotype [27]. Here we report the entry, replication and cytotoxic phenotype of 63 transposon mutants in 31 previously identified Coxiella candidate Dot/Icm substrates. Indeed, some of these candidates play a role during infection whereas some others fail to produce a phenotype, stressing the importance of coupling high-content screens to in silico analysis to identify bacterial effectors. Moreover, further studies of other Coxiella genes sharing none of the features of Dot/Icm substrates, can be exploited to enrich existing databases for the bioinformatics-based identification of Coxiella effectors, thus creating a feedback loop that would significantly improve and accelerate the study of Coxiella pathogenesis.
A considerable number of transposon insertions were mapped outside Coxiella CDS. By excluding mutations that affected the first 100 bp upstream of annotated genes (to exclude mutations that might affect promoter regions of genes), we obtained a list of 151 intergenic transposon insertions. Interestingly, 71 of these resulted in a significant replication phenotype, suggesting that these non-coding regions of the Coxiella genome may play a role in host/pathogen interactions. sRNAs are emerging as regulators that enable pathogens to adapt their metabolic needs during infection and timely express virulence factors [65], [66]. However, recent studies in other organisms revealed the existence of a number of putative sRNAs higher than initially expected, suggesting the presence of many non-functional sRNAs and complicating the identification of relevant sRNAs. The functional data obtained by our screening approach are being cross referenced with a list of putative Coxiella sRNAs identified by RNA-seq to facilitate the identification of Coxiella sRNAs that may coordinate host/pathogen interactions. Similarly, the interesting observation that a number of mutations in Coxiella CDS annotated as pseudogenes have an effect in host cell infection suggests that these genomic regions may have an important regulatory role.
Bacterial adhesion and invasion of host cells is a fundamental step of the infection by intracellular bacterial pathogens [67]. These processes can be active or passive depending on the nature of the pathogen. “Triggering” bacteria commonly use a type 3 secretion system (T3SS) to inject effectors across the host cell plasma membrane to trigger actin rearrangements and pathogen internalization by phagocytosis, whereas “zippering” bacteria use surface proteins that interact with cognate receptors at the surface of host cells [67]. This activates a ligand/receptor signaling cascade that leads to the internalization of large particles by an endocytosis-like mechanism [68]–[71]. The lack of a T3SS in Coxiella suggests that these organisms adhere to and invade host cells by a zippering mechanism. Indeed, it has been reported that Coxiella is passively internalized by host cells by a yet undefined mechanism, which is accompanied by the local rearrangement of the actin cytoskeleton [11], [14], [15]. αVβ3 integrins have been shown to mediate Coxiella adhesion to THP-1 cells [11], however, the lack of these integrins at the surface of epithelial cells, which are effectively colonized by Coxiella, suggest the presence of additional/alternative receptors. Similarly, the Coxiella surface determinants for host cell adhesion and invasion remain to be defined. Here we have identified the product of CBU_1260 as the first Coxiella invasin. Predictive analysis on the primary sequence of CBU_1260 revealed the presence of 8 beta sheets forming an OmpA-like domain highly homologous to that identified and characterized in several other bacterial pathogens [49], [50]. Examples are the OmpA proteins encoded by E. coli K1 [44], [47], Yersinia pestis [45], Francisella tularensis [46], Klebsiella pneumoniae [48] and Shigella flexneri [72]. These outer membrane proteins are involved in bacterial adhesion and/or internalization within host cells, as well as in the NF-κB-mediated modulation of the immune response to infection, which is required for intracellular bacterial development. Importantly, OmpA-like proteins with similar functions in bacterial adhesion and internalization have been reported in other bacterial pathogens such as Rickettsia conorii, Anaplasma phagocytophilum and Ehrlichia chaffeensis [73]–[75], however these proteins share no structural homology with the OmpA proteins described above. In agreement with in silico predictions, membrane fractionation experiments performed in Coxiella as well as in E. coli ectopically expressing OmpA, showed that the protein is indeed enriched in the outer membrane fraction of bacterial lysates. Our multi-phenotypic analysis revealed that five independent transposon insertions that disrupted CBU_1260 sequence severely affected Coxiella internalization and replication within host cells. The internalization phenotype was specific of non-phagocytic cells, whereas OmpA mutants were still internalized by phagocytic cells. This observation indicated that, in the absence of an active phagocytic process, OmpA is able to actively trigger Coxiella internalization by means of ligand/receptor interactions. Importantly however, the intracellular replication of OmpA mutants was severely affected in both epithelial and macrophage cell lines. This phenotype is in line with a reported role of OmpA proteins in facilitating bacterial survival within host cells [46], [48], [51], [53]. Importantly, OmpA-like proteins share conserved transmembrane domains but are characterized by extremely variable extracellular domains, which are unique to each pathogen, and confer specific functions [50]. OmpA was predicted to have 4 unstructured loops exposed at the bacterial surface. By replacing each loop with a myc tag, we have generated 4 OmpA mutants (OmpA ΔL1, ΔL2, ΔL3 and ΔL4) and showed that loop 1 is essential to confer invasiveness to E. coli ectopically expressing the OmpA mutants. Accordingly, a specific antibody against loop 1 effectively blocks OmpA function. Bioinformatics analysis indicated the presence of 2 additional OmpA-like proteins in the Coxiella genome, CBU_0307 and CBU_0936, sharing with OmpA a good degree of homology at the level of the transmembrane OmpA-like domain but no significant homology in the 4 extracellular loops. In line with these observations, CBU_0307 and CBU_0936 failed to produce internalization phenotypes when mutated by transposon insertions. Finally, experiments aiming at blocking potential OmpA interactions with a cognate receptor, effectively blocked Coxiella internalization, indicating the presence of an interacting partner at the surface of host cells, which remains to be identified. Using Galleria mellonella larvae as a surrogate in vivo model system we could reproduce the OmpA mutant phenotypes observed in cultured cells. Indeed, only wt Coxiella and the control mutant Tn1832 were able to induce the formation of nodules that were abundantly colonized by Coxiella and disrupt the organization of Galleria peripheral organs. The OmpA mutant Tn208 induced a milder formation of nodules that presented few, isolated bacteria. Accordingly, larvae infected with the OmpA mutant survived Coxiella infections longer than those infected with the control mutants.
In summary, multi-phenotypic screening of host/pathogen interactions is an efficient method for the study of infectious diseases. Here we have applied this method to Coxiella infections and identified a bacterial protein that is essential for Coxiella internalization within non-phagocytic cells. Understanding how intracellular bacteria adhere to and invade their host is essential to 1) understand the cell biology of infection and identify the candidate targets of anti-infectious molecules and 2) to develop targeted vaccines. Of note, bacterial OmpA proteins are considered as new pathogen-associated molecular patterns (PAMPs) and are among the most immuno-dominant antigens in the outer membrane of Gram-negative bacteria [50], [76]. Our laboratory currently investigates the possibility of using OmpA to develop a synthetic vaccine against Q fever.
Materials and Methods
Bacterial strains, cell lines and growth conditions
Strains used in this study are listed in Fig. S9. Escherichia coli strains were grown in Luria-Bertani (LB) medium supplemented with ampicillin (100 µg/ml), kanamycin (50 µg/ml) or chloramphenicol (30 µg/ml) as appropriate. Coxiella burnetii NMII and transposon mutants were grown in ACCM-2 [77] supplemented with kanamycin (340 µg/ml) or chloramphenicol (3 µg/ml) as appropriate in a humidified atmosphere of 5% CO2 and 2.5% O2 at 37°C. Cells were routinely maintained in RPMI (Vero, THP-1, J774 and RAW 264.7) or DMEM (A431 and HeLa), containing 10% fetal calf serum (FCS) in a humidified atmosphere of 5% CO2 at 37°C. For experiments, THP-1 were allowed to differentiate into macrophages for 2 days in the presence of 200 nM phorbol myristate acetate (PMA, Sigma).
Antibodies and reagents
Hoechst 33258, rabbit anti poly-His, anti mouse and anti-rabbit HRP-conjugated antibodies and Atto-647N phalloidin were purchased from Sigma. Rabbit anti Coxiella NMII antibodies were kindly provided by Robert Heinzen. Synthesis and production of the peptide KKSGTSKVNFTGVTL, as well as the generation of the cognate antibody in rabbit (named anti-OmpA in this study) were performed by Eurogentec, Belgium. Mouse and rabbit IgG conjugated to Alexa Fluor 488 and 555 as well as Prolong Gold antifade mounting reagent were purchased from Invitrogen. Paraformaldehyde was provided by Electron Microscopy Sciences, PA.
Plasmids
Plasmids and primers used in this study are listed in Fig. S9. DNA sequences were amplified by PCR using Phusion polymerase (New England Biolabs) and gene-specific primers (Sigma). To create the plasmid pITR-CAT-ColE1-P311-GFP, the promoter of CBU_0311 (P311) was amplified from Coxiella RSA439 NMII genomic DNA using primers P311-XhoI-Fw and P311-Rv, GFP was amplified from pEGFP-N1 (Clontech) using primers EGFP-Fw and GFP-PITR-Rv, and P1169-CAT-ColE1 was amplified from plasmid pITR-CAT-ColE1 using primers GFP-PITR-Fw and XhoI-PITR-Rv. PCR fragments P311, GFP and P1169-CAT-ColE1 were fused by overlapping PCR. The resulting PCR product was digested with XhoI and ligated to obtain circular pITR-CAT-ColE1-P311-GFP. OmpA32-248 was amplified from Coxiella RSA439 NMII genomic DNA using primers OmpA32-248-BamHI-Fw and OmpA-EcoRI-Rv and cloned into pET28a to obtain pET28a-OmpA32-248. OmpA was amplified from Coxiella RSA439 NMII genomic DNA using primers OmpA-BamHI-shift-Fw and OmpA-EcoRI-Rv and cloned into pET27b to obtain pET27b-OmpA. Plasmids pET27b-OmpAΔL1, pET27b-OmpAΔL2, pET27b-OmpAΔL3 and pET27b-OmpAΔL4 were generated by PCR using pET27b-OmpA as template and primer pairs loop1-myc-HindIII-Fw/loop1-myc-HindIII-Rv, loop2-myc-HindIII-Fw/loop2-myc-HindIII-Rv, loop3-myc-HindIII-Fw/loop3-myc-HindIII-Rv, loop4-myc-HindIII-Fw/loop4-myc-HindIII-Rv. The PCR products were digested with HindIII and ligated to obtain the corresponding plasmids.
Generation of a bank of Coxiella transposon mutants
C. burnetii RSA439 NMII organisms were electroporated with the pITR-CAT-ColE1-P311-GFP and pUC19::Himar1C9 plasmids [34] using the following setup: 18 kV, 400 Ω, 25 µF. Bacteria were then grown overnight in ACCM-2 supplemented with 1% FBS and the following day 3 µg/ml chloramphenicol were added to bacterial cultures. Bacteria were then amplified for 4 days and plated on solid ACCM-2 for clone isolation. Seven days post-inoculation, colonies were isolated and amplified for 6 days in liquid ACCM-2 supplemented with 3 µg/ml chloramphenicol. The concentration of each isolated mutant was quantified using the PicoGreen (Invitrogen) assay according to manufacturer's instructions. To map transposon insertions, single primer colony PCR was performed on 1 µl of C. burnetii transposon mutant in stationary phase in ACCM-2. The PCR mix contained 1× HF buffer (New England Biolabs), 200 µM dNTPs, 1 µM primer SP3 and 1 U of Phusion polymerase (New England Biolabs). The PCR cycle consisted in initial denaturation (98°C, 1 min), 20 high stringency cycles (98°C, 10 sec; 50°C, 30 sec; 72°C, 90 sec), 30 low stringency cycles (98°C, 10 sec; 30°C, 30 sec; 72°C, 90 sec) and 30 high stringency cycles (98°C, 10 sec; 50°C, 30 sec; 72°C, 90 sec) followed by a final extension at 72°C for 7 min. PCR products were then sequenced at Beckman Coulter Genomics (Stansted, UK) using primer P3. Insertion sites were mapped on the annotated C. burnetii RSA493 NMI genome using MacVector (MacVector Inc.) and recorded in a relational database (FileMaker).
Axenic growth of Coxiella
106 GE/ml of bacteria were inoculated in 4 ml ACCM-2 and allowed to grow for 8 days in a humidified atmosphere of 5% CO2 and 2.5% O2 at 37°C. Where needed, 3 µg/ml chloramphenicol were added to bacterial cultures. At the indicated time points bacterial concentrations were evaluated from 100 µl of cultures, using the PicoGreen (Invitrogen) assay according to manufacturer's instructions.
Mutant library screening
Vero cells were seeded into triplicate 96-wells plates (Greiner Bio one) 2 days prior to infection. Cells were then challenged with C. burnetii RSA439 NMII transposon mutants at an MOI of 100. For each plate, cells in well A1 were left uninfected and cells in wells A2 and A3 were incubated with GFP-C. burnetii RSA439 NMII at multiplicities of infection of 100 and 200. Bacterial contact with cells was promoted by centrifugation (10 min, 400 g, RT) and cells were incubated in a humidified atmosphere of 5% CO2 at 37°C. Unbound bacteria were removed after 1 h of incubation and cells were further incubated in fresh culture medium for 7 days. Plates were analyzed at a 24-hours interval using a TECAN Infinite 200 Pro operated by the Magellan software (TECAN) to monitor the variations of GFP fluorescence associated with the intracellular growth of Coxiella. Raw data were analyzed for background subtraction, normalization and quality control among triplicates using in-house developed methods. Seven days after infection, plates were fixed in 3% paraformaldehyde in PBS at room temperature for 30 minutes, rinsed in PBS and incubated in blocking solution (0.5% BSA, 50 mM NH4Cl in PBS, pH 7.4). Cells were then incubated in Hoechst 33258 diluted 1∶200 in blocking solution for 30 minutes at room temperature, rinsed and incubated in PBS. Images were acquired with an Arrayscan VTI Live epifluorescence automated microscope (Cellomics) equipped with an ORCA ER CCD camera. 6 fields/well of triplicate 96-wells plates were imaged with a 20× objective in the GFP, DAPI and Bright-field channels. Images were then processed and analyzed using CellProfiler. Briefly, the GFP channel was subtracted from the corresponding DAPI channel to avoid false identification of large Coxiella colonies as host cell nuclei, images were thresholded using the Otsu global method and host cell nuclei as well as Coxiella colonies were identified and segmented. The number, form factor and fragmentation of host cell nuclei and the number, form factor, area, perimeter, GFP intensity and compactness of Coxiella colonies were then calculated per object and per image. An average of 14000 cells per condition (infection with a given Coxiella mutant) were thus analyzed. Raw data were processed for background subtraction, normalization and quality control among the 6 fields per well and plate triplicates using in-house developed methods. Data were recorded in a relational database (FileMaker) that allowed clustering of phenotypes according to the annotated transposon insertions.
Immunofluorescence staining and microscopy
Cells were fixed in 3% paraformaldehyde in PBS at room temperature for 30 minutes, rinsed in PBS and incubated in blocking solution (0.5% BSA, 50 mM NH4Cl in PBS, pH 7.4). When appropriate, 0.05% saponin was added to the blocking solution for cell permeabilization. Cells were then incubated with the primary antibodies diluted in blocking solution for 1 h at room temperature, rinsed five times in PBS and further incubated for 45 min with the secondary antibodies diluted in the blocking solution. Fluorescent phalloidin was added to the secondary antibodies to label actin, where needed. After labeling, coverslips were mounted using Prolong Gold antifade mounting medium (Invitrogen) supplemented with Hoechst 33258 for DNA staining. For differential labeling, extracellular bacteria or beads were stained using specific antibodies without permeabilizing the cells. Intracellular bacteria or beads were visualized by green and red fluorescence, respectively. Alternatively, a second staining was performed after cellular permeabilization. Secondary antibody labeling using two different fluorochromes (before and after permeabilization) allowed discrimination between adherent extracellular bacteria/beads and those that have been internalized. Samples were analyzed with a Zeiss Axioimager Z1 epifluorescence microscope (Carl Zeiss) connected to a Coolsnap HQ2 CCD camera. Images were acquired alternatively with 63× or 40× oil immersion objectives and processed with MetaMorph (Universal Imaging Corp.). Image J and CellProfiler software were used for image analysis and quantifications. 3D reconstruction and surface rendering were performed using the Osirix software.
Complementation assay
Transposon insertions in Dot/Icm core genes were complemented in trans as previously described [19]. Briefly, Vero cells grown on 96-wells plates were either challenged with the transposon mutants alone or in combination with wt Coxiella at a 1∶1 ratio for a total MOI of 100. Bacterial contact with cells was promoted by centrifugation (10 min, 400 g, RT) and cells were incubated in a humidified atmosphere of 5% CO2 at 37°C. Unbound bacteria were removed after 1 h of incubation and cells were further incubated in fresh culture medium for 7 days. Plates were then fixed in 3% paraformaldehyde in PBS at room temperature for 30 minutes, rinsed in PBS and incubated in blocking solution (0.5% BSA, 50 mM NH4Cl in PBS, pH 7.4). Cells were then labeled with the anti-NMII antibody to detect wt Coxiella and with Hoechst 33258 as described above for DNA labeling. 96-well plates were imaged and analyzed essentially as described above for the mutant library screening protocol with the additional acquisition of the TRITC channel to detect and segment wt Coxiella colonies. Object masking was applied using CellProfiler to specifically calculate the area of mutant Coxiella colonies growing within wt Coxiella-occupied PVs.
TUNEL assay
The terminal deoxyribonucleotidyl transferase-mediated triphosphate (dUTP)-biotin nick end labeling (TUNEL) method was used for detection of DNA fragmentation of nuclei using the In Situ Cell Death Detection Kit TMR (Roche) according to the manufacturer's instructions. Briefly, HeLa cells grown on 96-well plates in triplicate were either left untreated or challenged with the indicated Coxiella strains at an MOI of 100 and incubated at 37°C for 3 days. Cells were then either fixed and permeabilized as described above or incubated with 1 µM staurosporine for 4 hours prior to fixation. Samples were then incubated 1 h at 37°C in the dark with TUNEL reaction mixture. Samples were then washed three times with PBS, and incubated with Hoechst 33258 for DNA staining and with an anti-NMII antibody to detect bacteria in the samples infected with wt Coxiella. 96-wells plates were analyzed with an Arrayscan VTI Live epifluorescence automated microscope (Cellomics) equipped with an ORCA ER CCD camera. 10 fields/well were imaged with a 20× objective in the GFP (bacteria), TRITC (TUNEL), DAPI (nuclei) and Bright-field (cells) channels. Images were then processed and analyzed using CellProfiler. Briefly, the GFP channel was subtracted from the corresponding DAPI channel to avoid false identification of large Coxiella colonies as host cell nuclei, images were thresholded using the Otsu global method and host cell nuclei, Coxiella colonies and fragmented nuclei were identified and segmented. The percentage of fragmented nuclei over the total number of nuclei was then calculated on an average of 6000 cells per condition.
Scanning electron microscopy
Cells were washed in PBS and fixed with 2.5% glutaraldehyde in Sorensen buffer, pH 7.2 for an hour at room temperature, followed by washing in Sorensen buffer. Fixed samples were dehydrated using a graded ethanol series (30–100%), followed by 10 minutes in graded Ethanol-Hexamethyldisilazane and finally Hexamethyldisilazane alone. Subsequently, the samples were sputter coated with an approximative 10 nm thick gold film and then examined under a scanning electron microscope (Hitachi S4000, at CRIC, Montpellier France) using a lens detector with an acceleration voltage of 20 kV at calibrated magnifications.
Immuno-histology
Galleria mellonella larvae were fixed overnight in paraformaldehyde 4% in PBS (pH 7.4). Larvae were then rinsed 3 times in PBS and cryo protected by successive incubations in PBS containing increasing concentrations of sucrose (10%, 20%, 30%). Samples were then frozen in isopentane at −80°C using a SnapFrost machine (Excilone). Consecutive 20 µm sections were then obtained from each sample using a Leica CM3050S cryostat. For indirect immuno-fluorescence, sections were permeabilized and blocked in PBS, 10% goat serum, 0.3% Triton X-100 for 1 h at room temperature. Samples were then incubated 48 hours at 4°C with the anti GFP antibody, then rinsed in PBS. Samples were then incubated with the appropriate secondary antibodies, Atto-647N phalloidin and Hoechst 33258 for 24 hours at 4°C. Samples were then washed in PBS and mounted on glass slides for microscopy analysis. Samples were analyzed either with an EVOS microscope (AMG) or with an ApoTome-equipped Zeiss Axioimager Z1 epifluorescence microscope (Carl Zeiss) connected to a Coolsnap HQ2 CCD camera. Images were acquired alternatively with a 10× objective (EVOS) or with a 63× oil immersion objective (Axioimager Z1) and processed with Image J and AxioVision (Carl Zeiss).
Protein expression and purification
Genes cloned into pET27b or pET28a vectors were expressed in E. coli BL21-DE3 star pLysS (Invitrogen). Bacterial cultures were grown at 25°C to mid-exponential phase (OD600 nm = 0.5) and were induced overnight with 400 µM isopropyl-β-D - thiogalactopyranoside (IPTG). For GST expression, E. coli XL1-blue were transformed with pGEX-4T1, grown at 37°C to mid-exponential phase (OD600 nm = 0.5) and induced for 4 h with 1 mM IPTG. Bacteria were harvested by centrifugation, resuspended in lysis buffer (20 mM Tris pH 8, 300 mM NaCl, 5% glycerol, complete anti-protease (Roche)) and lysed with BugBuster (Novagen) following the manufacturer's recommendations. Lysates were then cleared by centrifugation (11 000 g, 20 min, 4°C). Proteins were purified by gravity flow using Ni2+ agarose His-select resin column (Sigma) for His-tagged proteins or glutathione-sepharose (Sigma) for GST. His-tagged and GST proteins were eluted with lysis buffer supplemented with 250 mM imidazole or 25 mM reduced glutathione, respectively.
Membrane fractionation
For Coxiella membrane fractionation, 100 ml of wt Coxiella or Tn208 mutant grown in ACCM-2 for 7 days were pelleted and resuspended in 200 µl 20 mM Tris pH 8 containing 1× Complete protease inhibitor (Roche). For E. coli membrane fractionation, 30 ml of IPTG-induced or non-induced E. coli BL21-DE3 star pLysS pET27b-OmpA, pET27b-OmpAΔL1, pET27b-OmpAΔL2, pET27b-OmpAΔL3 or pET27b-OmpAΔL4 were pelleted and resuspended in 5 ml 20 mM Tris pH 8 containing 1× Complete protease inhibitor (Roche). Bacteria were sonicated using a Branson Sonifier S-450 (6 pulses of 20 s at 40% intensity) and cleared by centrifugation at 10000 g for 5 min at 4°C. Inner membrane proteins were extracted by incubation with sarkosyl (0.5% final concentration) at RT for 15 min. Outer membrane proteins were pelleted by ultracentrifugation (TLA-100, 32000 r.p.m., 30 min, 4°C) and resuspended in 2× Laemmli sample buffer. Insoluble, soluble/sarkosyl-solubilized and outer membrane fractions were resolved by SDS-PAGE and analyzed by Coomassie staining (Sigma) or immunoblotting with anti-OmpA and anti NMII antibodies.
Preparation of protein-coated beads
0.5 µm fluorescent red sulfate-modified polystyrene beads (Sigma) were washed three times with 25 mM MES pH 6.1 (MES buffer). The sulfate-modified beads (7.2×109) were then mixed with either 100 µg/ml purified GST or His-OmpA32-248 and incubated at room temperature (RT) for 4 h. The GST - or His-OmpA32-248-coated beads were then washed three times with MES buffer and resuspended in MES buffer containing 1% BSA.
Internalization assays
For fluorescent beads internalization assay, 7×107 GST - or His-OmpA32-248-coated beads in DMEM were applied to 1×105 A431 cells seeded onto glass coverslips in 24-well plates and contact was promoted by centrifugation (10 min, 400 g, RT). Cells were incubated in a humidified atmosphere of 5% CO2 at 37°C. Cells were washed three times with PBS and fixed in 4% paraformaldehyde before being processed for immunofluorescence staining. Bacteria internalization assays were performed as follow: 6×106 C. burnetii NMII GE were applied to cells (MOI 100) and contact was promoted by centrifugation (10 min, 400 g, RT). Cells were then fixed in 4% paraformaldehyde before being processed for immunofluorescence staining. For protein blocking experiments, A431 cells were pre-incubated for 1 h at 4°C with either 100 µg/ml of GST or 100 µg/ml of His-OmpA32-248 prior to the internalization assay described above. For antibody inhibition experiments, 6×106 C. burnetii RSA439 NMII GE were incubated at 4°C with increasing concentrations of either naïve rabbit serum or anti-OmpA antibodies (0.1 to 5 µg/ml) prior to the internalization assay described above. Gentamicin protection assays were performed as follow: IPTG-induced and non-induced BL21-DE3 star pLysS pET27b-OmpA, pET27b-OmpAΔL1, pET27b-OmpAΔL2, pET27b-OmpAΔL3 or pET27b-OmpAΔL4 were diluted to OD600 nm = 0.5 in DMEM and applied to cells at an MOI of 10 and contact was promoted by centrifugation (10 min, 400 g, RT). Following 1 h incubation at 37°C/5% CO2, cells were washed 5 times with PBS and incubated for 2 h with DMEM supplemented with 100 µg/ml gentamicin. Cells were then washed 5 times with PBS, lysed with 1 ml 0.1% Triton X-100 in ddH2O and serial dilutions were plated onto LB agar plates supplemented with the appropriate antibiotic for colony-forming units (CFU) assessment. Internalization frequency was determined as the number of CFU surviving the gentamicin challenge out of the total bacterial input. The results are representative of at least three independent experiments.
Supporting Information
Zdroje
1. MaurinM, RaoultD (1999) Q fever. Clin Microbiol Rev 12 : 518–553.
2. Van SchaikEJ, ChenC, MertensK, WeberMM, SamuelJE (2013) Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat Rev Microbiol 11 : 561–573 doi:10.1038/nrmicro3049
3. KazarJ (2005) Coxiella burnetii infection. Ann N Y Acad Sci 1063 : 105–114 doi:10.1196/annals.1355.018
4. McCaulTF, WilliamsJC (1981) Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J Bacteriol 147 : 1063–1076.
5. VothDE, HeinzenRA (2007) Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol 9 : 829–840 doi:10.1111/j.1462-5822.2007.00901.x
6. Arricau-BouveryN, RodolakisA (2005) Is Q Fever an emerging or re-emerging zoonosis? Vet Res 36 : 327–349 doi:10.1051/vetres
7. MadariagaMG, RezaiK, TrenholmeGM, WeinsteinRA (2003) Review Q fever: a biological weapon in your backyard. The Lancet infectious diseases 3 : 709–721.
8. HackstadtT, PeacockMG, HitchcockPJ, ColeRL (1985) Lipopolysaccharide Variation in Coxiella burnetii: Intrastrain Heterogeneity in Structure and Antigenicity. Infect Immun 48 : 359–365.
9. Moosa, HackstadtT (1987) Comparative virulence of intra - and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect Immun 55 : 1144–1150.
10. AndohM, Russell-LodrigueKE, ZhangG, SamuelJE (2005) Comparative virulence of phase I and II Coxiella burnetii in immunodeficient mice. Ann N Y Acad Sci 1063 : 167–170 doi:10.1196/annals.1355.026
11. CapoC, LindbergFP, MeconiS, ZaffranY, TardeiG, et al. (1999) Subversion of monocyte functions by Coxiella burnetii: impairment of the cross-talk between alphavbeta3 integrin and CR3. J Immunol 163 : 6078–6085.
12. Russell-LodrigueKE, ZhangGQ, McMurrayDN, SamuelJE (2006) Clinical and pathologic changes in a guinea pig aerosol challenge model of acute Q fever. Infect Immun 74 : 6085–6091 doi:10.1128/IAI.00763-06
13. JensenTK, MontgomeryDL, JaegerPT, LindhardtT, AgerholmJS, et al. (2007) Application of fluorescent in situ hybridisation for demonstration of Coxiella burnetii in placentas from ruminant abortions. APMIS 115 : 347–353.
14. MeconiS, JacomoV, BoquetP (1998) Coxiella burnetii Induces Reorganization of the Actin Cytoskeleton in Human Monocytes. Infect Immun 66(11): 5527.
15. RosalesEM, AguileraMO, SalinasRP, CarminatiSa, ColomboMI, et al. (2012) Cortactin is involved in the entry of Coxiella burnetii into non-phagocytic cells. PLoS One 7: e39348 doi:10.1371/journal.pone.0039348
16. RomanoPS, GutierrezMG, BerónW, RabinovitchM, ColomboMI (2007) The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol 9 : 891–909 doi:10.1111/j.1462-5822.2006.00838.x
17. NewtonHJ, McDonoughJa, RoyCR (2013) Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One 8: e54566 doi:10.1371/journal.pone.0054566
18. CareyKL, NewtonHJ, LührmannA, RoyCR (2011) The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 7: e1002056 doi:10.1371/journal.ppat.1002056
19. BeareP, GilkS, LarsonC, HillJ (2011) Dot/Icm Type IVB Secretion System Requirements for Coxiella burnetii Growth in Human Macrophages. MBio 2: e00175–11 doi:10.1128/mBio.00175-11
20. CampoyEM, MansillaME, ColomboMI (2013) Endocytic SNAREs are involved in optimal Coxiella burnetii vacuole development. Cell Microbiol 15 : 922–941 doi:10.1111/cmi.12087
21. VothDE, HoweD, HeinzenRA (2007) Coxiella burnetii inhibits apoptosis in human THP-1 cells and monkey primary alveolar macrophages. Infect Immun 75 : 4263–4271 doi:10.1128/IAI.00594-07
22. VothDE, HeinzenRA (2009) Sustained activation of Akt and Erk1/2 is required for Coxiella burnetii antiapoptotic activity. Infect Immun 77 : 205–213 doi:10.1128/IAI.01124-08
23. LührmannA, RoyCR (2007) Coxiella burnetii inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect Immun 75 : 5282–5289 doi:10.1128/IAI.00863-07
24. LührmannA, NogueiraCV, CareyKL, RoyCR (2010) Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci U S A 107 : 18997–19001 doi:10.1073/pnas.1004380107
25. KlingenbeckL, EckartRA, BerensC, LührmannA (2012) The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell Microbiol [epub ahead of print]. doi:10.1111/cmi.12066
26. ChenC, BangaS, MertensK, WeberMM, GorbaslievaI, et al. (2010) Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A 107 : 21755–21760 doi:10.1073/pnas.1010485107
27. WeberMM, ChenC, RowinK, MertensK, GalvanG, et al. (2013) Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol 195 : 3914–3924 doi:10.1128/JB.00071-13
28. LifshitzZ, BursteinD, PeeriM, ZusmanT, SchwartzK, et al. (2013) Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A 110: E707–15 doi:10.1073/pnas.1215278110
29. PanX, LührmannA, SatohA, Laskowski-ArceMA, RoyCR (2008) Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320 : 1651–1654 doi:10.1126/science.1158160
30. VothDE, HoweD, BearePA, VogelJP, UnsworthN, et al. (2009) The Coxiella burnetii Ankyrin Repeat Domain-Containing Protein Family Is Heterogeneous, with C-Terminal Truncations That Influence Dot/Icm-Mediated Secretion. J Bacteriol 191 : 4232–4242 doi:10.1128/JB.01656-08
31. VothDE, BearePa, HoweD, SharmaUM, SamoilisG, et al. (2011) The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol 193 : 1493–1503 doi:10.1128/JB.01359-10
32. OmslandA, CockrellDC, HoweD, FischerER, VirtanevaK, et al. (2009) Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci U S A 106 : 4430–4434 doi:10.1073/pnas.0812074106
33. BearePA, SandozKM, OmslandA, RockeyDD, HeinzenRA (2011) Advances in genetic manipulation of obligate intracellular bacterial pathogens. Front Microbiol 2 : 97 doi:10.3389/fmicb.2011.00097
34. BeareP, HoweD, CockrellD (2009) Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J Bacteriol 191 : 1369–1381 doi:10.1128/JB.01580-08
35. BearePA, UnsworthN, AndohM, VothDE, OmslandA, et al. (2009) Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun 77 : 642–656 doi:10.1128/IAI.01141-08
36. BearePA, LarsonCL, GilkSD, HeinzenRA (2012) Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol 78 : 4580–4589 doi:10.1128/AEM.00881-12
37. VogelJP (2004) Turning a tiger into a house cat: using Legionella pneumophila to study Coxiella burnetii. Trends Microbiol 12 : 103–105.
38. ZamboniDS, McGrathS, RabinovitchM, RoyCR (2003) Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol 49 : 965–976 doi:10.1046/j.1365-2958.2003.03626.x
39. ZusmanT, YerushalmiG, SegalG (2003) Functional Similarities between the icm/dot Pathogenesis Systems of Coxiella burnetii and Legionella pneumophila. Infect Immun 71 : 3714–3723 doi:10.1128/IAI.71.7.3714
40. ZusmanT, AloniG, HalperinE, KotzerH, DegtyarE, et al. (2007) The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol 63 : 1508–1523 doi:10.1111/j.1365-2958.2007.05604.x
41. SavojardoC, FariselliP, CasadioR (2013) BETAWARE: a machine-learning tool to detect and predict transmembrane beta-barrel proteins in prokaryotes. Bioinformatics 29 : 504–505 doi:10.1093/bioinformatics/bts728
42. NambaA, ManoN, TakanoH, BeppuT, UedaK, et al. (2008) OmpA is an adhesion factor of Aeromonas veronii, an optimistic pathogen that habituates in carp intestinal tract. J Appl Microbiol 105 : 1441–1451 doi:10.1111/j.1365-2672.2008.03883.x
43. SerinoL, NestaB, LeuzziR, FontanaMR, MonaciE, et al. (2007) Identification of a new OmpA-like protein in Neisseria gonorrhoeae involved in the binding to human epithelial cells and in vivo colonization. Mol Microbiol 64 : 1391–1403 doi:10.1111/j.1365-2958.2007.05745.x
44. FaganRP, LambertMa, SmithSGJ (2008) The hek outer membrane protein of Escherichia coli strain RS218 binds to proteoglycan and utilizes a single extracellular loop for adherence, invasion, and autoaggregation. Infect Immun 76 : 1135–1142 doi:10.1128/IAI.01327-07
45. BartraSS, GongX, LoricaCD, JainC, NairMKM, et al. (2012) The outer membrane protein A (OmpA) of Yersinia pestis promotes intracellular survival and virulence in mice. Microb Pathog 52 : 41–46 doi:10.1016/j.micpath.2011.09.009
46. MahawarM, AtianandMK, DotsonRJ, MoraV, RabadiSM, et al. (2012) Identification of a novel Francisella tularensis factor required for intramacrophage survival and subversion of innate immune response. J Biol Chem 287 : 25216–25229 doi:10.1074/jbc.M112.367672
47. DattaD, VaidehiN, FlorianoWB, KimKS, PrasadaraoNV, et al. (2003) Interaction of E. coli outer-membrane protein A with sugars on the receptors of the brain microvascular endothelial cells. Proteins 50 : 213–221 doi:10.1002/prot.10257
48. MarchC, MorantaD, RegueiroV, LlobetE, TomásA, et al. (2011) Klebsiella pneumoniae outer membrane protein A is required to prevent the activation of airway epithelial cells. J Biol Chem 286 : 9956–9967 doi:10.1074/jbc.M110.181008
49. SmithSGJ, MahonV, LambertMA, FaganRP (2007) A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett 273 : 1–11 doi:10.1111/j.1574-6968.2007.00778.x
50. ConferAW, AyalewS (2013) The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol 163 : 207–222 doi:10.1016/j.vetmic.2012.08.019
51. SelvarajSK, PrasadaraoNV (2005) Escherichia coli K1 inhibits proinflammatory cytokine induction in monocytes by preventing NF-kB activation. Microbes and Infection 78 : 544–54 doi:10.1189/jlb.0904516.1
52. SoulasC, BaussantT (2000) Outer membrane protein A (OmpA) binds to and activates human macrophages. J Immunol 165 : 2335–2340.
53. KrüllM, BockstallerP, WuppermannFN, KluckenAC, MühlingJ, et al. (2006) Mechanisms of Chlamydophila pneumoniae-mediated GM-CSF release in human bronchial epithelial cells. Am J Respir Cell Mol Biol 34 : 375–382 doi:10.1165/rcmb.2004-0157OC
54. LavineMD, StrandMR (2002) Insect hemocytes and their role in immunity. Insect Biochem Mol Biol 32 : 1295–1309.
55. O'CallaghanD, VergunstA (2010) Non-mammalian animal models to study infectious disease: worms or fly fishing? Curr Opin Microbiol 79–85 doi:10.1016/j.mib.2009.12.005
56. FerrandonD, ImlerJ-L, HetruC, HoffmannJA (2007) The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol 7 : 862–874 doi:10.1038/nri2194
57. VogelH, AltincicekB, GlöcknerG, VilcinskasA (2011) A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics 12 : 308 doi:10.1186/1471-2164-12-308
58. OlsenRJ, WatkinsME, CantuCC, BeresSB, MusserJM (2011) Virulence of serotype M3 Group A Streptococcus strains in wax worms (Galleria mellonella larvae). Virulence 2 : 111–119 doi:10.4161/viru.2.2.14338
59. ChampionOL, KarlyshevAV, SeniorNJ, WoodwardM, La RagioneR, et al. (2010) Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J Infect Dis 201 : 776–782 doi:10.1086/650494
60. BerginD, ReevesE (2005) Superoxide Production in Galleria mellonella Hemocytes: Identification of Proteins Homologous to the NADPH Oxidase Complex of Human Neutrophils. Infect Immun 73 : 4161–70 doi:10.1128/IAI.73.7.4161
61. MukherjeeK, AltincicekB, HainT, DomannE, VilcinskasA, et al. (2010) Galleria mellonella as a model system for studying Listeria pathogenesis. Appl Environ Microbiol 76 : 310–317 doi:10.1128/AEM.01301-09
62. HardingCR, SchroederGN, ReynoldsS, KostaA, CollinsJW, et al. (2012) Legionella pneumophila pathogenesis in the Galleria mellonella infection model. Infect Immun 80 : 2780–2790 doi:10.1128/IAI.00510-12
63. BrodinP, ChristopheT (2011) High-content screening in infectious diseases. Curr Opin Chem Biol 15 : 534–539 doi:10.1016/j.cbpa.2011.05.023
64. OmslandA, HeinzenRA (2011) Life on the outside: the rescue of Coxiella burnetii from its host cell. Annu Rev Microbiol 65 : 111–128 doi:10.1146/annurev-micro-090110-102927
65. PapenfortK, VogelJ (2010) Regulatory RNA in bacterial pathogens. Cell Host Microbe 8 : 116–127 doi:10.1016/j.chom.2010.06.008
66. Toledo-AranaA, RepoilaF, CossartP (2007) Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol 10 : 182–188 doi:10.1016/j.mib.2007.03.004
67. Pizarro-CerdáJ, CossartP (2006) Bacterial adhesion and entry into host cells. Cell 124 : 715–727 doi:10.1016/j.cell.2006.02.012
68. VeigaE, GuttmanJa, BonazziM, BoucrotE, Toledo-AranaA, et al. (2007) Invasive and adherent bacterial pathogens co-Opt host clathrin for infection. Cell Host Microbe 2 : 340–351 doi:10.1016/j.chom.2007.10.001
69. BonazziM, VasudevanL, MalletA, SachseM, SartoriA, et al. (2011) Clathrin phosphorylation is required for actin recruitment at sites of bacterial adhesion and internalization. 195 : 525–536 doi:10.1083/jcb.201105152
70. BonazziM, KühbacherA, Toledo-AranaA, MalletA, VasudevanL, et al. (2012) A common clathrin-mediated machinery co-ordinates cell-cell adhesion and bacterial internalization. Traffic 13 : 1653–1666 doi:10.1111/tra.12009
71. Pizarro-CerdáJ, BonazziM, CossartP (2010) Clathrin-mediated endocytosis: what works for small, also works for big. Bioessays 32 : 496–504 doi:10.1002/bies.200900172
72. PoreD, ChakrabartiMK (2013) Outer membrane protein A (OmpA) from Shigella flexneri 2a: a promising subunit vaccine candidate. Vaccine 31 : 3644–3650 doi:10.1016/j.vaccine.2013.05.100
73. HillmanRD, BaktashYM, MartinezJJ (2013) OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with α2β1 integrin. Cell Microbiol 15 : 727–741 doi:10.1111/cmi.12068
74. OjogunN, KahlonA, RaglandSA, TroeseMJ, MastronunzioJE, et al. (2012) Anaplasma phagocytophilum outer membrane protein A interacts with sialylated glycoproteins to promote infection of mammalian host cells. Infect Immun 80 : 3748–3760 doi:10.1128/IAI.00654-12
75. PopovVL, YuXJ, WalkerDH (2000) The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb Pathog 28 : 71–80 doi:10.1006/mpat.1999.0327
76. JeanninP, MagistrelliG, GoetschL, HaeuwJ-F, ThieblemontN, et al. (2002) Outer membrane protein A (OmpA): a new pathogen-associated molecular pattern that interacts with antigen presenting cells-impact on vaccine strategies. Vaccine 20 Suppl 4: A23–7.
77. OmslandA, BearePa, HillJ, CockrellDC, HoweD, et al. (2011) Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol 77 : 3720–3725 doi:10.1128/AEM.02826-10
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule FormationČlánek Oral Bacteria and CancerČlánek A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 3- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
- Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
- Mycobacteriophages: Windows into Tuberculosis
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- Five Things to Know about Genetically Modified (GM) Insects for Vector Control
- A Missing Dimension in Measures of Vaccination Impacts
- Eosinophils Are Important for Protection, Immunoregulation and Pathology during Infection with Nematode Microfilariae
- Clonality of HTLV-2 in Natural Infection
- Production, Fate and Pathogenicity of Plasma Microparticles in Murine Cerebral Malaria
- Group B Streptococcal Infection of the Choriodecidua Induces Dysfunction of the Cytokeratin Network in Amniotic Epithelium: A Pathway to Membrane Weakening
- New Insights into How Adapts to Its Mammalian Host during Bubonic Plague
- Foodborne Transmission of Nipah Virus in Syrian Hamsters
- A Polysaccharide Virulence Factor from Elicits Anti-inflammatory Effects through Induction of Interleukin-1 Receptor Antagonist
- Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH
- Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages
- Crk Adaptors Negatively Regulate Actin Polymerization in Pedestals Formed by Enteropathogenic (EPEC) by Binding to Tir Effector
- Fatty Acid Biosynthesis Contributes Significantly to Establishment of a Bioenergetically Favorable Environment for Vaccinia Virus Infection
- A Cytosolic Chaperone Complexes with Dynamic Membrane J-Proteins and Mobilizes a Nonenveloped Virus out of the Endoplasmic Reticulum
- Intracellular Promote Invasive Cell Motility through Kinase Regulation of the Host Actin Cytoskeleton
- MAVS-MKK7-JNK2 Defines a Novel Apoptotic Signaling Pathway during Viral Infection
- RON5 Is Critical for Organization and Function of the Moving Junction Complex
- Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway
- and Exhibit Metabolic Symbioses
- The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface
- Target Cell Availability, Rather than Breast Milk Factors, Dictates Mother-to-Infant Transmission of SIV in Sooty Mangabeys and Rhesus Macaques
- Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
- Infection of Adult Thymus with Murine Retrovirus Induces Virus-Specific Central Tolerance That Prevents Functional Memory CD8 T Cell Differentiation
- Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in
- In Vivo Administration of a JAK3 Inhibitor during Acute SIV Infection Leads to Significant Increases in Viral Load during Chronic Infection
- Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
- Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
- A Compact, Multifunctional Fusion Module Directs Cholesterol-Dependent Homomultimerization and Syncytiogenic Efficiency of Reovirus p10 FAST Proteins
- The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck
- Genetic Dissection of Gut Epithelial Responses to
- Two-Component System Cross-Regulation Integrates Response to Heme and Cell Envelope Stress
- Oral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
- A Model System for Studying the Transcriptomic and Physiological Changes Associated with Mammalian Host-Adaptation by Serovar Copenhageni
- Inflammasome Sensor NLRP1 Controls Rat Macrophage Susceptibility to
- ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by
- The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses
- Caspase-1-Dependent and -Independent Cell Death Pathways in Infection of Macrophages
- The Effect of Cell Growth Phase on the Regulatory Cross-Talk between Flagellar and Spi1 Virulence Gene Expression
- Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms
- Oral Bacteria and Cancer
- Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
- Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector
- VE-Cadherin Cleavage by LasB Protease from Facilitates Type III Secretion System Toxicity in Endothelial Cells
- Dimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
- Crystal Structure of the Vaccinia Virus DNA Polymerase Holoenzyme Subunit D4 in Complex with the A20 N-Terminal Domain
- Post-Translational Regulation via Clp Protease Is Critical for Survival of
- Modulation of Phagosomal pH by Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport
- Rotavirus Activates Lymphocytes from Non-Obese Diabetic Mice by Triggering Toll-Like Receptor 7 Signaling and Interferon Production in Plasmacytoid Dendritic Cells
- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Interferon Regulatory Factor-1 Protects from Fatal Neurotropic Infection with Vesicular Stomatitis Virus by Specific Inhibition of Viral Replication in Neurons
- HMGB1-Promoted and TLR2/4-Dependent NK Cell Maturation and Activation Take Part in Rotavirus-Induced Murine Biliary Atresia
- An Immunomics Approach to Schistosome Antigen Discovery: Antibody Signatures of Naturally Resistant and Chronically Infected Individuals from Endemic Areas
- PPARγ Agonists Improve Survival and Neurocognitive Outcomes in Experimental Cerebral Malaria and Induce Neuroprotective Pathways in Human Malaria
- A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
- Viral OTU Deubiquitinases: A Structural and Functional Comparison
- Heterogeneity and Breadth of Host Antibody Response to KSHV Infection Demonstrated by Systematic Analysis of the KSHV Proteome
- Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm
- Broadly Reactive Human CD8 T Cells that Recognize an Epitope Conserved between VZV, HSV and EBV
- Oncogenic Human Papillomaviruses Activate the Tumor-Associated Lens Epithelial-Derived Growth Factor (LEDGF) Gene
- Erythrocyte Invasion: Combining Function with Immune Evasion
- IL-1α and Complement Cooperate in Triggering Local Neutrophilic Inflammation in Response to Adenovirus and Eliminating Virus-Containing Cells
- Chronic Exposure to Type-I IFN under Lymphopenic Conditions Alters CD4 T Cell Homeostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



