-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
Since it was first discovered in the early 1980s, Human Immunodeficiency Virus 1 (HIV-1), the causative agent of Acquired Immunodeficiency Syndrome (AIDS), has been the focus of intense research. In untreated individuals, the number of CD4+ T-cells in the blood slowly drops over time and the patient eventually succumbs to an opportunistic infection. Although current therapies are capable of managing the virus; they do not represent a true cure. As a retrovirus, HIV-1 incorporates itself into the host genome and survives in the long-lived population of memory T-cells found in the human host. In this study, we examine the roll of a T-cell specific transcription factor (RUNX1) in the control of HIV-1 replication. Through various molecular studies, we show that RUNX1 represses HIV-1 replication in T-cells. By examining samples from patients with HIV-1, we are able to show a negative correlation between viral replication and RUNX1 expression. Finally, we show that an inhibitor of RUNX1 synergizes with Vorinostat, a current lead compound in the quest to re-active HIV-1 and purge the latent pool.
Published in the journal: . PLoS Pathog 10(3): e32767. doi:10.1371/journal.ppat.1003997
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003997Summary
Since it was first discovered in the early 1980s, Human Immunodeficiency Virus 1 (HIV-1), the causative agent of Acquired Immunodeficiency Syndrome (AIDS), has been the focus of intense research. In untreated individuals, the number of CD4+ T-cells in the blood slowly drops over time and the patient eventually succumbs to an opportunistic infection. Although current therapies are capable of managing the virus; they do not represent a true cure. As a retrovirus, HIV-1 incorporates itself into the host genome and survives in the long-lived population of memory T-cells found in the human host. In this study, we examine the roll of a T-cell specific transcription factor (RUNX1) in the control of HIV-1 replication. Through various molecular studies, we show that RUNX1 represses HIV-1 replication in T-cells. By examining samples from patients with HIV-1, we are able to show a negative correlation between viral replication and RUNX1 expression. Finally, we show that an inhibitor of RUNX1 synergizes with Vorinostat, a current lead compound in the quest to re-active HIV-1 and purge the latent pool.
Introduction
Human Immunodeficiency Virus type I (HIV-1) is the etiologic agent of Acquired Immunodeficiency Syndrome (AIDS). HIV-1 has a complex life cycle that in part involves a unique transcriptional interaction between the viral Tat protein and its target RNA element (TAR) found in the R sequence of the LTR [1]–[3]. In the absence of treatment, most HIV-1 infected individuals will experience a steady decline in the number of CD4+ T-cells, progress to AIDS and eventually die as the result of acquiring opportunistic infections.
Transcriptional control of HIV-1 occurs in two phases. Basal transcription of the integrated provirus first occurs at a low level in a Tat independent manner [4]. Once the Tat protein is synthesized, viral transcription transits to a Tat-dependent route. Tat binds TAR RNA and recruits a complex of cyclin T1 and CDK9 to the start site of transcription [1] leading to the phosphorylation of the c-terminal domain (CTD) of the RNA Pol II to induce more processive transcription. Tat has also been shown to help initiate transcription through interaction with the TATA Binding Protein as well as various histone modifying enzymes such as CBP/p300 and the PBAF complex [5].
The HIV-1 LTR contains a myriad of transcription factor-binding sites, such as those for SP1 and NF-κB. It is believed that interactions of cellular factors with the HIV-1 LTR determine active transcription versus the establishment of transcriptional latency. HIV-1 latency, a state in which the infected cell produces little to no viral RNA, represents a major barrier to viral eradication in an infected individual [6]–[8].
In mammals, there are three RUNX proteins that can interact with a cofactor, core-binding factor β (CBF-β), to form an active transcription factor complex [9], [10]. RUNX protein binding to CBF-β allows transport of the complex into the nucleus via a localization signal in the RUNX protein [11]. In turn, CBF-β increases the affinity of RUNX proteins for DNA. This complex is essential for proper differentiation of cells of the hematopoietic lineage. Of particular interest is the involvement of RUNX1 in the differentiation and fate selection of CD4+ T-cells [12]–[15]. Specifically, RUNX1 is drastically down regulated when thymocytes progress from double-negative to double-positive during development, and it is also down-regulated when naïve CD4+ T-cells are stimulated through the T-cell receptor (TCR) to become effector cells. In the latter case, RUNX1 down-regulation is associated with increased IL-2 production which is likely critical for CD4+ T-cell function [14].
Mechanistically, RUNX serves a role in the initiation of transcription that is likely achieved through p300 histone acetyltransferase recruitment. Intriguingly, RUNX family members may also recruit repressive factors such as mSin3A, Suv39H1 and histone deacetlyases and serve transcriptional repressor functions. Indeed, RUNX1 is important in repressing CD4 expression in double-negative thymocytes and mature CD8+ T-cells. The choice to act as an activator or repressor is influenced by post-translational modifications of the RUNX protein in the core binding factor [16], [17] and through the recruitment of selective factors to a specific promoter [18], [19].
Three recent publications have highlighted a role for CBF-β in Vif function [20]–[22]. Specifically, CBF-β is capable of binding to Vif and this interaction increases Vif mediated APOBEC3G degradation. A recent study has presented evidence that CBF-β binding by Vif influences RUNX responsive genes [23]. These studies suggest that the Vif protein has evolved to bind CBF-β. The presumption made in the first two studies is that this mechanism is specific to protection of the virus from APOBEC3G. However, the follow-up study suggests the intriguing possibility that Vif may be binding to CBF-β in order to influence RUNX mediated gene expression.
Our current work suggests that RUNX1 may be an important HIV-1 LTR-binding factor that serves a role in latency. RUNX1, as a T-cell specific transcription factor capable of suppression or activation of target promoters, may alter the transcription of the viral LTR during cellular infection. We find that inhibition of RUNX1 by a small molecule inhibitor synergizes with the HDAC inhibitor SAHA to activate HIV-1 latency. Our findings indicate that RUNX1 with its binding partner CBF-β may act to repress LTR transcription and show that this repression might be countered by the viral Vif protein.
Results
RUNX1 and CBF-β over-expression reduce expression of viral proteins and viral replication
To assess the involvement of RUNX1 in HIV-1 transcription, RUNX1 and CBF-β expression plasmids were co-transfected with an HIV-1 molecular clone (pNL4-3) into HeLa cells (Fig 1A). Twenty-four hours post transfection, cells were harvested and protein extracts were prepared and subjected to Western blot analysis of HIV-1 protein expression. Interestingly, the transfection of either RUNX1 or CBF-β alone reduced HIV Gag expression in a dose dependent manner (Fig. 1A, compare lanes 2+3, 4+5 and 7+8 to lane 1). Transfection of RUNX1 and CBF-β together reduced Gag expression to nearly undetectable levels (Fig. 1A, lanes 6, 9, 10 and 11).
Fig. 1. RUNX1 and CBF-β are capable of repressing HIV-1 replication. 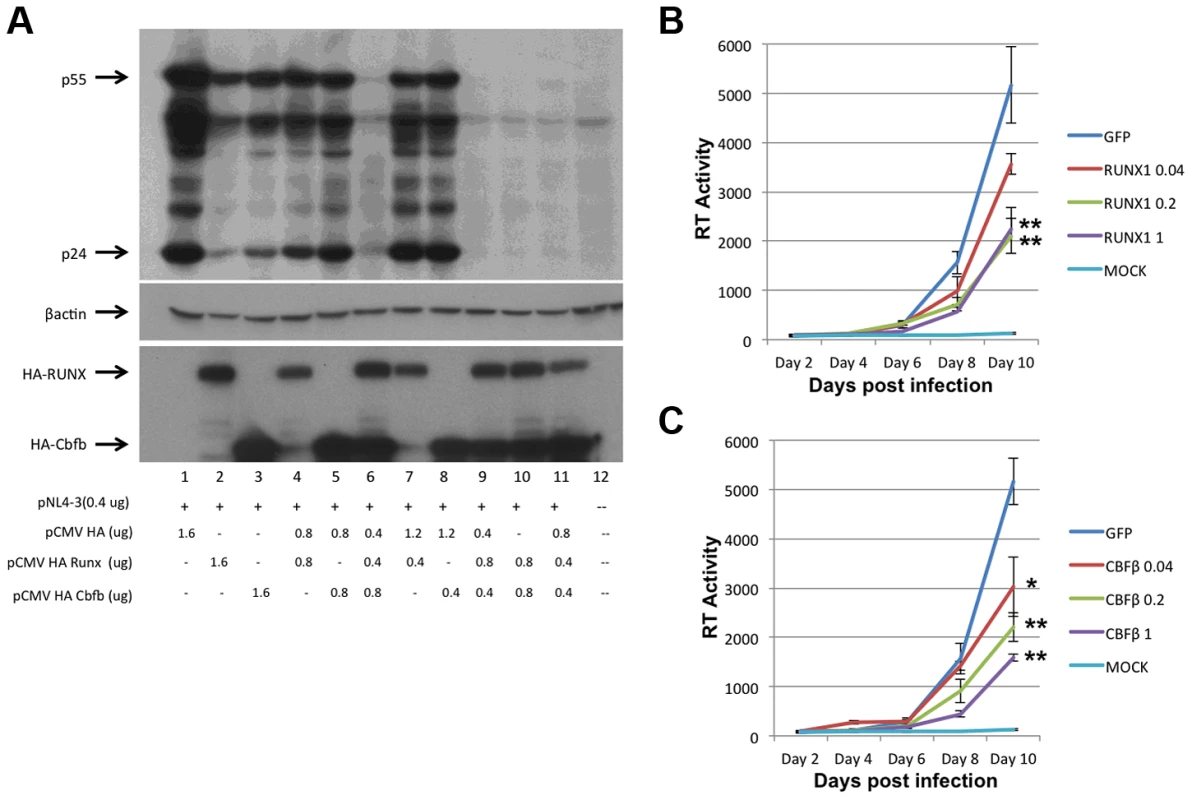
A) HeLa cells were transfected with the pNL4-3 proviral plasmid and the indicated amounts of HA-tagged RUNX1 or CBF-β expression vectors. Forty-eight hours post transfection cells were harvested, protein extracts were prepared and extracts were Western blotted for p24 and p55 Gag using human immune serum (upper panel), β-actin (middle panel) or HA (lower panel). Jurkat T-cells were transfected with1 ug of pMAXGFP or 0.04, 0.2 or 1 ug of B) pRUNX1 or C) pCBF-β and infected with NL4-3 twenty-four hours post transfection. Viral replication was followed by RT at days 2, 4, 6, 8 and 10 post infection. We next tested the effect of RUNX1 and CBF-β on spreading HIV-1 infection of Jurkat T-cells. In brief, Jurkat cells were transfected with 0.04, 0.2 or 1 ug of pMaxGFP control, pRUNX1 or pCBF-β expression vector using Amaxa nucleofection. Observation of GFP expression in transfected control cells showed that the transfection efficiency was >75%. Western blot analysis was performed to confirm over-expression of CBF-β and RUNX1 (Supplementary Figure S1). Twenty-four hours after transfection, cells were infected with the NL4-3 virus and cell culture supernatants were sampled at 2, 4, 6, 8 and 10 days post infection. Quantification of reverse transcriptase activity (RT) in the culture supernatants showed typical HIV-1 spreading infection in the cells transfected with the control vector (Figure 1B, C). However, increasing the transfected amounts of either RUNX1 (Fig. 1B) or CBF-β (Fig. 1C) produced reduced HIV-1 replication; with RT levels repressed to 43% and 30% of the control on day 10. Conversely, knock down of RUNX1 or CBF-β expression by ∼60% in a cell line model of latency induced a two-fold increase in RT production (Supplementary Figure S2). Taken together, these data are consistent with an important role served by RUNX1 and CBF - β in HIV-1 replication.
RUNX1 and CBF-β repress HIV-1 transcription
To further characterize the effects of RUNX1 and CBF-β on the transcription of an integrated HIV-1 LTR, we used the TZMbl reporter cell line. TZMbl cells are derived from HeLa cells to express CD4 and HIV-1 entry co-receptors, CXCR4 and CCR5; they contain an integrated HIV-1 LTR driving expression of firefly luciferase and a second integrated HIV-1 LTR driving the beta-galactosidase reporter gene. TZMbl were transfected with a Tat expression vector, together with RUNX1 or CBF-β expression vectors or a combination of the two. Forty-eight hours after transfection, the cells were harvested and equal protein amounts were used to determine β-galactosidase activity (Fig. 2A). We observed that the transfection of one microgram of either RUNX1 or CBF-β expression vector suppressed LTR-driven β-galactosidase expression by 88% and 90% respectively. Transfecting both vectors together repressed LTR-driven β-galactosidase expression by 94%.
Fig. 2. RUNX1 and CBF-β suppress LTR-driven promoter expression of the integrated and unintegrated LTR. 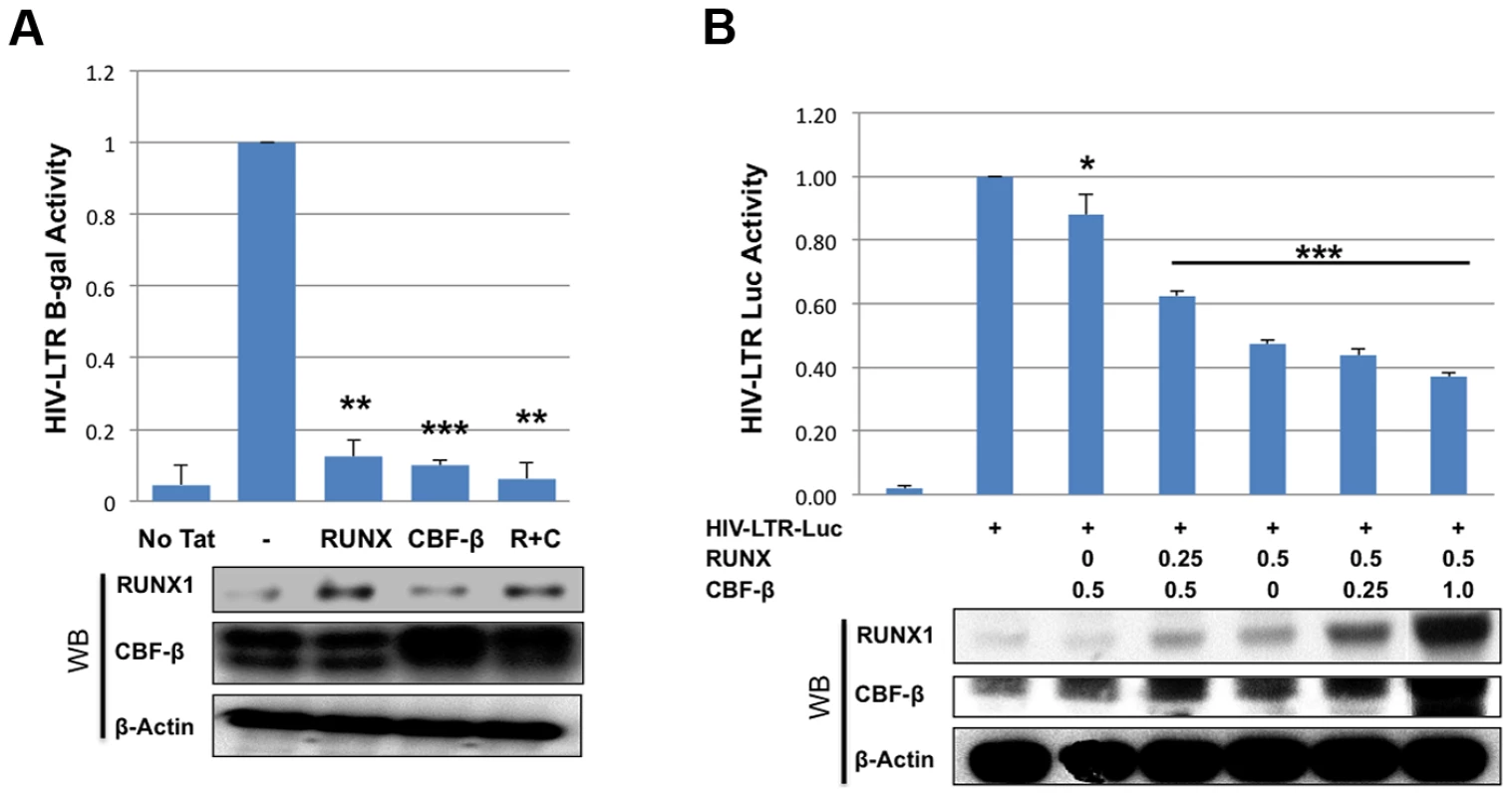
A) TZMbl indicator cells were seeded in a six well plate and transfected with 0.01 μg Tat expression vector and 1 μg of RUNX1 or CBF-β expression vectors or a combination of the two. Forty-eight hours after transfection cells were lysed and β-galactosidase activity was measured by a luminescence assay. β-galactosidase is represented as relative to transfection of Tat alone. B) 293T cells were seeded in a 6-well plate and transfected with pLTR-GL3 and RUNX1 or CBF-β vectors as indicated. Forty-eight hours after transfection the cells were harvested and cell lysates prepared. Cell extracts were then used to determine the production of luciferase. Results are displayed as luciferase signal relative to transfection of reporter construct alone. Western blotting was performed on transfected cells to assess the levels of RUNX1 and CBF-β expression. * p≤0.05, ** p≤0.01 and *** p≤0.001 We also tested the effect of RUNX1 and CBF-β on the basal activity of the HIV-LTR in 293T cells. 293T cells were transfected with 1 ug pLTR-GL3 (an HIV LTR driven luciferase reporter), 0.5 ug of RUNX1 or CBF-β expressing plasmid and increasing amounts of CBFβ or RUNX1 respectively (Fig. 2B). Transfection of CBF-β alone only modestly repressed LTR-driven transcription (Fig. 2B, 88% of control, 3rd column). Cotransfecting 0.25 ug of RUNX1 plasmid further repressed reporter expression to 62% of baseline (Fig. 2B, 4th column). Transfection with RUNX1 alone reduced reporter activity to 47% of control (Fig. 2B, 5th column). Increasing the levels of transfected CBF-β further reduced activity to a modest degree (Fig. 2B, compare columns 5 to 6 and 7). RUNX1 and CBF-β over-expression had no effect on either CMV or EF1 control promoter constructs and were capable of activating a murine leukemia virus promoter known to be responsive to RUNX1 (BXH2 LTR) (Supplementary Figure S3). Taken together these data demonstrate that RUNX1 and CBF-β are capable of suppressing the promoter activity of integrated or un-integrated HIV-1 LTRs, which is consistent with our earlier findings [24].
The HIV-1 LTR contains potential RUNX binding sites
The DNA binding site, consensus sequence TGYGGT [25], for the RUNX proteins has been described for several promoters including MHC I, KIR, the BXH2 LTR and RUNX1 itself [25]–[28]. In order to identify potential binding sites in the HIV-1 LTR the TransFac matrix of RUNX1 binding sites was used to search the DNA sequence of the NL4-3 provirus from U3 through Gag using a search algorithm available from the University of Pennsylvania [29]. This analysis revealed 10 potential binding sites in the LTR and none in the 5′ portion of the Gag coding region. Of these 10 sites, six exist in the positive orientation, while four reside on the complementary strand (Fig. 3A and 3B). The conservation of these potential sites was analyzed using fifty-eight clade B sequences that contain a complete 5′LTR from the Los Alamos HIV Sequence Database (Fig. 3B) [30]. This analysis revealed that sites 3, 4 and 5 are not well conserved amongst clade B viruses. However, the remaining seven sites are well conserved. Indeed, these sequences appear to be better conserved than other regions of the LTR, suggesting that they may be selected for function.
Fig. 3. RUNX1 binds to the HIV-1 LTR. 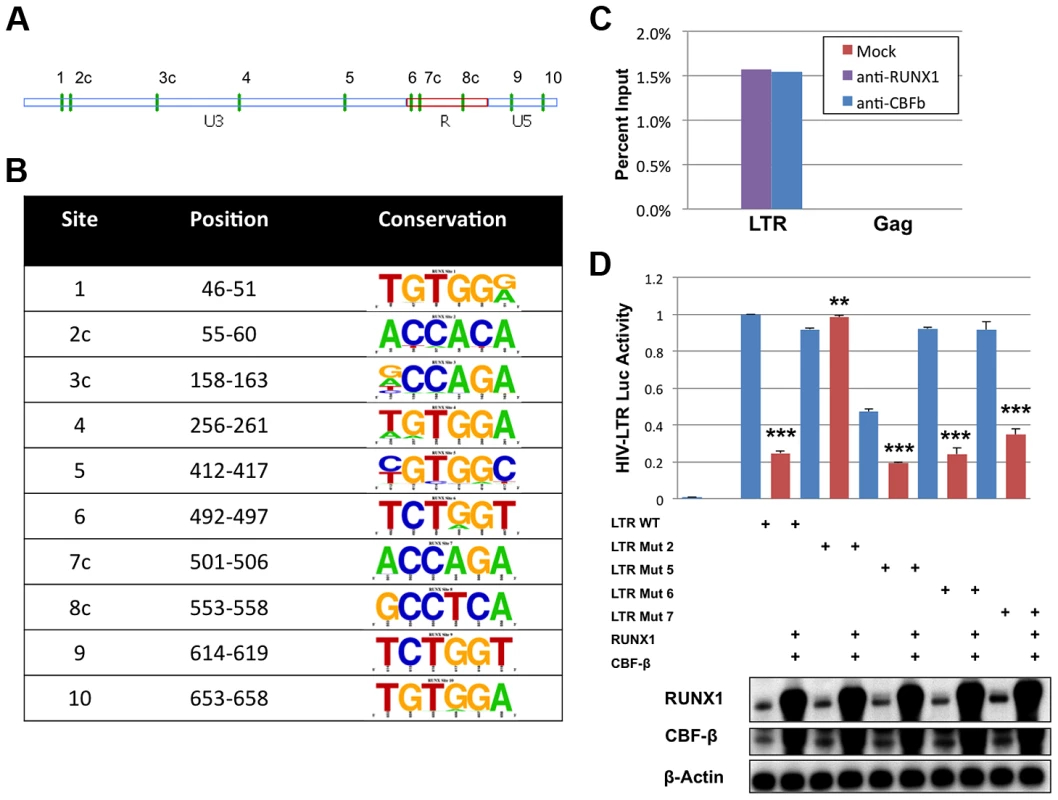
A) The position of the ten predicted binding sites (green vertical dashes) represented graphically on the LTR. The letter ‘c’ following the number indicates orientation on the reverse strand. B) The sequences corresponding to the potential binding sites were imported into UC-Berkley's WebIcon program and used to generate graphical representations of conservation. The height of each letter in the vertical column represents the percentage of sequences that contain that base at that position. Positions where only one letter is visible represent 100% conservation within the 58 sequences examined. C) HIV-1 infected ACH2 T-cell line was treated with 1% formaldehyde, lysed and then sonicated to shear chromatin to ∼1000 bp. Sheared chromatin was incubated with antibodies against RUNX1, CBF-β or no antibody control. Antibody bound chromatin complexes were immuno-precipitated with protein A/G agarose beads and DNA was detected by qPCR against the LTR or Gag sequence. D) 293T cells were seeded in a 6-well plate and transfected with pLTR-GL3 WT, mutant 2, mutant 5, mutant 6 or mutant 7 and RUNX1 or CBF-β vectors as indicated. Forty-eight hours after transfection the cells were harvested and cell lysates prepared. Cell extract were then used to determine the production of luciferase. Results are displayed as luciferase signal relative to transfection of the wild-type promoter construct. Western blotting was performed on transfected cells to assess the levels of RUNX1 and CBF-β expression. ** p≤0.01 and *** p≤0.001 Chromatin immuno-precipitations (ChIP) were performed to evaluate binding of RUNX1 to the potential LTR sites. It is known that CBF-β binds to RUNX1 and increases its affinity for DNA; thus, our ChIP analysis was performed in the context of both proteins. Chromatin was isolated from the HIV-1 latently infected ACH2 T-cell line, sheared by sonication, and immunoprecipitated with antibodies for RUNX1, CBF-β or no antibody control. The association of RUNX1 or CBF-β with HIV-1 LTR DNA or Gag DNA as a negative control was measured by qPCR (Fig 3C). ChIP analysis revealed the binding of RUNX1 and CBF-β to the LTR but not Gag DNA.
To specifically identify RUNX1 binding sites within the LTR, mutant reporter constructs were generated. Site-directed mutagenesis was used to change the final two nucleotides (consensus GT will be changed to TG) in potential RUNX binding sites 2, 5, 6 and 7 (Fig. 3A, B). This alters the important fifth residue in the binding site that has previously been shown to abrogate RUNX binding [31]. The four mutant reporter constructs were then used to evaluate RUNX1 responsiveness in 293T cells, which were transfected with 1 μg reporter (pGL3-LTR WT, pGL3-LTR mut2, pGL3-LTR mut5, pGL3-LTR mut6 and pGL3 mut7), 0.5 μg of CBF-β expression plasmid and 0.5 μg of RUNX1 expression vector. Forty-eight hours after transfection, the cells were harvested, lysed and 20 μg of protein extract used for luciferase assay (Figure 3D). RUNX1 and CBF-β expression repressed WT LTR to 25% of control (compare columns 1 and 2). Mutants 5, 6 and 7 were also repressed by RUNX1 and CBF-β. However, mutant 2 was not repressed by RUNX1 and CBF-β expression (compare columns 3 and 4). This suggests that the predicted binding site 2 is a physiological RUNX1 binding site in the LTR.
Vif expression relieves RUNX1 and CBF-β repression of HIV-1 transcription
Recently it was shown that HIV-1 Vif can bind to CBF-β and that this interaction increases APOBEC3G degradation [20]–[22]. The ability of Vif, a cytoplasmic protein, to bind CBF-β may alter the ability of CBF-β to function as a cofactor for the RUNX proteins. CBF-β itself has no nuclear localization signal. Instead, it relies on binding to a RUNX protein partner to be ferried into the nucleus [11]. We wondered if binding of CBF-β by Vif may sequester CBF-β in the cytoplasm. To test this hypothesis, Vif and CBF-β expression vectors were transfected alone or in combination into HeLa cells (Fig 4A). 24 hours after transfection, the cells were stained for Vif, CBF-β and the nuclear marker Lamin B. Vif alone localized to the cytoplasm; and CBF-β alone was seen throughout the cell (Panel A, 2nd column), which is consistent with prior studies [11]. However, in cells that over expressed both Vif and CBF-β (Panel A, 3rd and 4th column), CBF-β was localized entirely in the cytoplasm consistent with the notion that Vif sequesters CBF-β in the cytoplasm.
Fig. 4. HIV-1 Vif sequesters CBF-β in the cytoplasm. 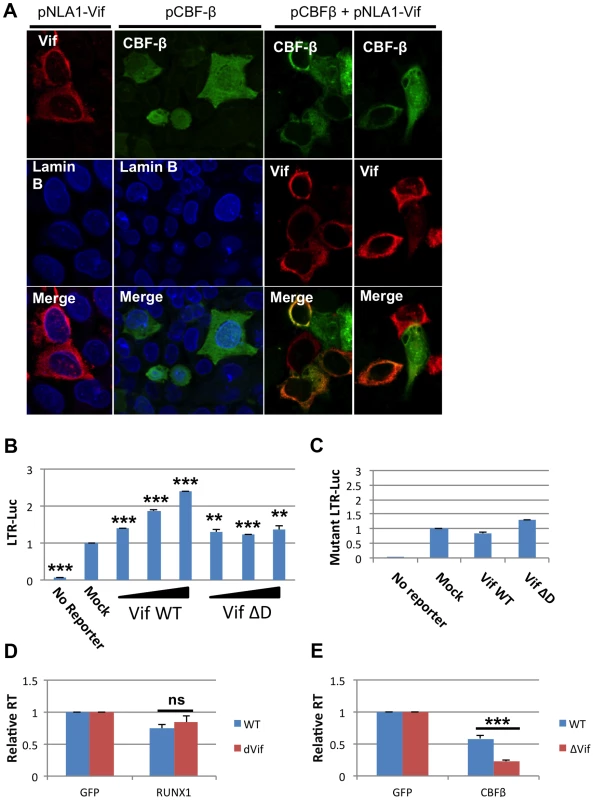
A) HeLa CBF-β knockdown (KD) cells (5×106) were cotransfected with the Vif expression vector pNL-A1 (2.5 μg) and/or the CBF-β vector pCBF-β (1 μg). Total amounts of transfected DNA were adjusted to 5 μg using empty vector DNA as appropriate. Three hours after transfection, cells were trypsinized and seeded onto cover slips. Cells were fixed 24 hr later in methanol (10 min, -20°C) and then stained with rabbit antibodies to Vif (Vif93; 1:100) or CBF-β (Thermo Fisher; 1:100). Bound antibodies were visualized by Texas-Red or Cy2-conjugated secondary antibodies (Jackson Labs; 1:100). Images were collected on a Zeiss LSM410 confocal microscope using a Plan-Apochromat 63x/1.4 oil immersion objective (Zeiss). B) 293T cells were seeded in a 6-well plate and transfected with 0.1 ug pLTR-GL3 and 0.04, 0.2 and 1 ug pVif or pVif ΔD. Forty eight hours post transfection cells were collected and used to prepare protein extracts. Cell extracts were then used to determine the production of luciferase. Results are displayed as luciferase signal relative to transfection of reporter construct alone. C) 293T cells were seeded in a 6-well plate and transfected with 0.1 ug pLTR-GL3 mutant 2 and 1 ug of pVif or pVif ΔD. Luciferase activity was determined as above. Jurkat cells were transfected with GFP plasmid and D) RUNX1 or E) CBF-β plasmid and infected twenty-four post transfection with equivalent doses of NL4-3 WT or NL4-3 ΔVif. RT values were determined at days 2, 4, 6, 8 and 10 post infection. Data is presented as RT activity relative to control (GFP transfected) cells. ** p≤0.01 and *** p≤0.001 Based on the published association of Vif with CBF-β and the above data, we explored Vif expression as a model for CBF-β depletion. 293T cells were transfected with 0.1 μg LTR-luciferase reporter and Vif expression vector (Fig 4B). Luciferase activity at forty-eight hours post-transfection was determined. Transfection of increasing amounts of Vif led to increased luciferase expression. Vif binds CBF-β through its n-terminal region, and mutation of residues 21 and 38 in Vif disrupts this binding [20], [22]. To test that Vif binding to CBF-β is required to mediate the observed increase in LTR expression, we utilized a vector that expresses a Vif protein lacking residues 23–43 (Vif ΔD). Transfection with Vif ΔD showed a small, but not dose responsive, increase in LTR driven luciferase expression.
To further confirm the involvement of Vif in rescuing RUNX1 mediated repression of the LTR, we employed the LTR mutant 2 reporter construct that was demonstrated above (Fig 3D) to be un-responsive to RUNX1 or CBF-β overexpression (Fig 4C). Transfection of neither Vif WT nor Vif ΔD was able to substantially increase Luciferase expression from this vector, supporting that the Vif effect on the LTR involves RUNX1 and CBF-β.
The ability of Vif to counteract repression mediated by RUNX1 and CBF-β in a spreading infection was monitored using a ΔVif virus. Jurkat cells were again used as they have been classified as ‘permissive’ due to their lack of APOBEC3G expression and ability to support the replication of viruses lacking Vif [32], [33]. Jurkat cells were transfected with 0.04 μg pRUNX1 or pCBF-β (Fig 4D and E). Twenty-four hours post-infection cells were infected with NL4-3 or NL4-3 ΔVif. Reverse transcriptase activity at day 10 demonstrated no difference in the susceptibility of the viruses to RUNX1 overexpression (Panel D). However, NL4-3 ΔVif was significantly more sensitive to CBF-β overexpression (Panel E). CBF-β repressed WT virus to 58% of the control, while NL4-3 ΔVif was repressed to 23% of the control. This data confirms that Vif is able to counteract CBF-β repression of HIV infection.
RUNX1 expression in CD4+ memory T-cells of viremic HIV-1 patients correlates negatively with viral load and positively with CD4+ T-cell count
In order to determine the relevance of RUNX1 expression in HIV-1 infection in human patients we examined the expression of RUNX1, CBF-β and RUNX3 in primary T-cells. Analysis of CD4+ T-cell populations (Supplementary Figure S4) shows that upon activation of naïve CD4+ T-cells (a population relatively refractory to infection) the expression levels of RUNX1, RUNX3 and CBF-β drop (correlating with greater susceptibility to infection). Interestingly, the expression of these proteins in naïve cells is fairly uniform, but expression in the memory pool (the primary target cells during infection) is much more variable. This person-to-person variability leaves open the possibility that RUNX1 expression levels may correlate with clinical outcomes.
To determine the relevance of RUNX1 expression in HIV-1 infection in human hosts we compared RUNX1 expression to viral load and CD4+ T-cell counts in viremic HIV-1 patients who were not on therapy. Previous studies have shown that two promoters drive RUNX1 expression: a proximal and distal promoter. Each promoter codes a transcript that varies in the 5′ coding region and studies suggest that one isoform may be more important than the other in terms of function [34], [35]. In keeping with this, no significant correlation was seen between total RUNX1 levels and either viral load or CD4+ T-cell counts (Data not shown). However, when examining the specific levels of the promoter proximal RUNX1 transcript in memory CD4+ T-cells we noted a significant negative correlation with viral load and a significant positive correlation with CD4+ T-cell count (Fig 5A and 5B). This was in contrast to promoter distal RUNX1 transcripts that showed no correlation with viral load or CD4+ T-cell count (Fig 5C and 5D). Memory CD4+ T-cells are the primary target of infection and account for the majority of virus seen in the blood of patients. Therefore, these findings suggest the intriguing possibility that RUNX1 expression may have a role to play in the clinical progression of HIV-1 in patients.
Fig. 5. RUNX1 expression in memory CD4+ T-cells correlates with clinical metrics in patients. 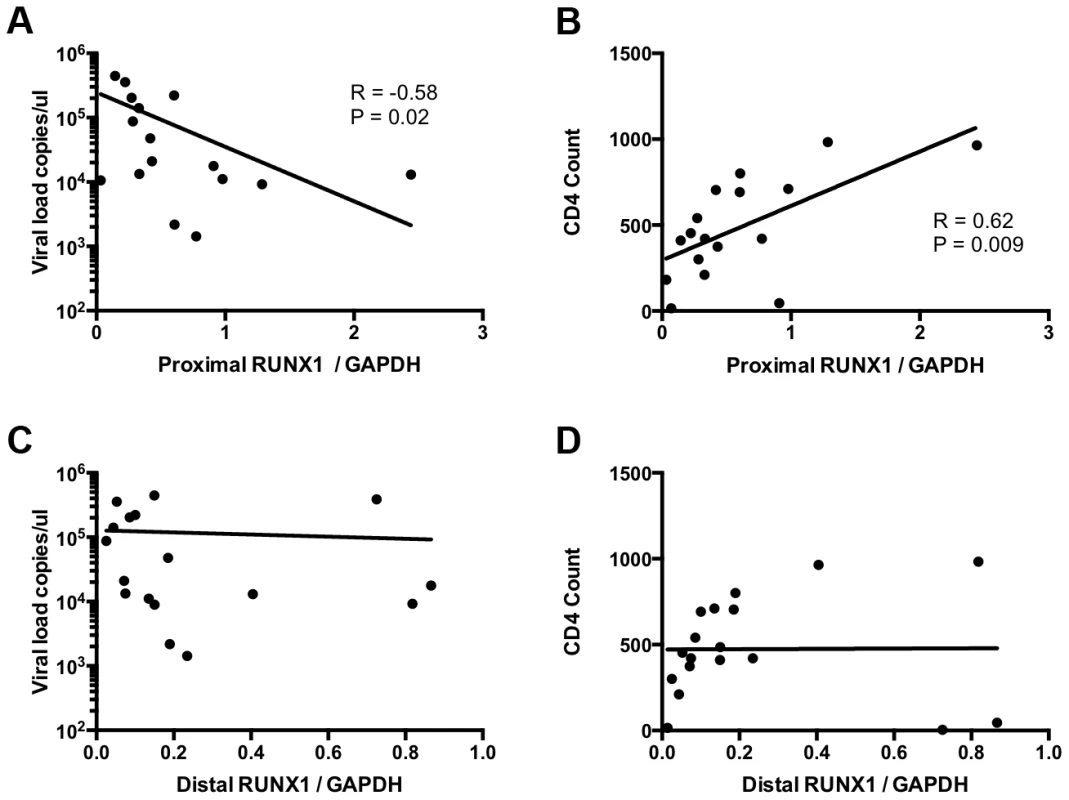
CD4+ memory T-cells were sorted from total PBMC of viremic HIV-1 patients in the absence of therapy. RNA extract from these cells was used to measure the expression of promoter proximal and distal RUNX1 as normalized to GAPDH. Expression level were then graphed as follows: A) promoter proximal RUNX1 vs viral load, B) promoter proximal RUNX1 versus CD4+ T-cell count, C) promoter distal RUNX1 versus viral load and D) promoter distal RUNX1 versus CD4+ T-cell count. R and P values were computed using Spearman test. An inhibitor of RUNX1:CBF-β interaction synergizes with SAHA to reactivate latent HIV
The benzodiazepine compound Ro5-3335 was recently identified as an inhibitor of RUNX1:CBF-β interaction and their functions in transcriptional regulation and hematopoiesis [24]. Suppression of RUNX1 or CBF-β by siRNA was capable of reactivating latent cells, suggesting that a pharmacologic inhibitor of RUNX1/CBF-β function may have a similar effect (Supplementary Figure S2). To test this hypothesis the Jlat model of HIV-1 latency [36] was used wherein viral reactivation can be followed by GFP expression. Jlat cells were treated with DMSO, 0.5, 5 or 50 μM Ro5-3335 for 72 hours and re-activation was assayed by flow cytometry (Fig 6A). Treatment with 50 uM Ro5-3335 induced a 2.2-fold increase in the number of GFP positive cells (1.9 to 4.2%). This two-fold activation is similar in magnitude to the effect seen with siRNA (Supplementary Figure S2). We next tested if the effect of Ro5-3335 could be increased by treatment with a second drug. For this purpose, we chose the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA), also known as Vorinostat, which has previously been used to reactivate latently infected T-cells in HIV-1 patients. Treatment of Jlat cells with 1 μM SAHA increased the number of GFP positive cells by 5.1-fold (1.9 to 9.6%) – similar to the induction of RT activity in ACH2 (Supplementary Figure S5). Treatment with increasing concentrations of Ro5-3335 increased the percentage of GFP positive cells in a multiplicative fashion. Treatment with 50 uM Ro5-3335 was capable of activating another 2.9-fold above SAHA alone, for a total activation of 28%. The combination of Ro5-3335 and SAHA yielded similar results in ACH2 (Fig 6B) and TZMbl (Fig 6C) cell lines. Of potential concern to the development of any therapeutic is the issue of toxicity. To address this we treated JLat, ACH2, TZMbl and J-LTR-G (a Jurkat cell line that carries an integrated LTR-GFP reporter) with SAHA and Ro5-3335 (Supplementary Figure S6). Ro5-3335 alone had minimal effects on cell viability. Treatment with SAHA induced significant cell death in all four cell types. Treatment of JLat, ACH2, TZMbl and J-LTR-G with SAHA and increasing concentrations of Ro5-3335 induced further cell death beyond SAHA alone (Fig S5, compare final four data sets for panels A–D). Interestingly, JLat and ACH2 had the greatest cell death (15% and 12% live when treated with SAHA and 50 uM Ro5-3335) as compared to TZMbl and J-LTR-G (50% and 44%). JLat and ACH2 also experienced greater cell death with SAHA treatment alone (55% and 51% live vs 81% and 63% live). ACH2 and JLat both carry complete copies of the virus, whereas TZMbl and J-LTR-G are reporter only cell lines. It is tempting to posit that the increased cell death seen in ACH2 and JLat is due to reactivation of the virus. The multiplicative effect on HIV reactivation seen with Ro5-3335 and SAHA in the three cell lines tested is indicative of possible synergy.
Fig. 6. Ro5-3335 RUNX inhibitor synergizes with SAHA HDAC inhibitor. 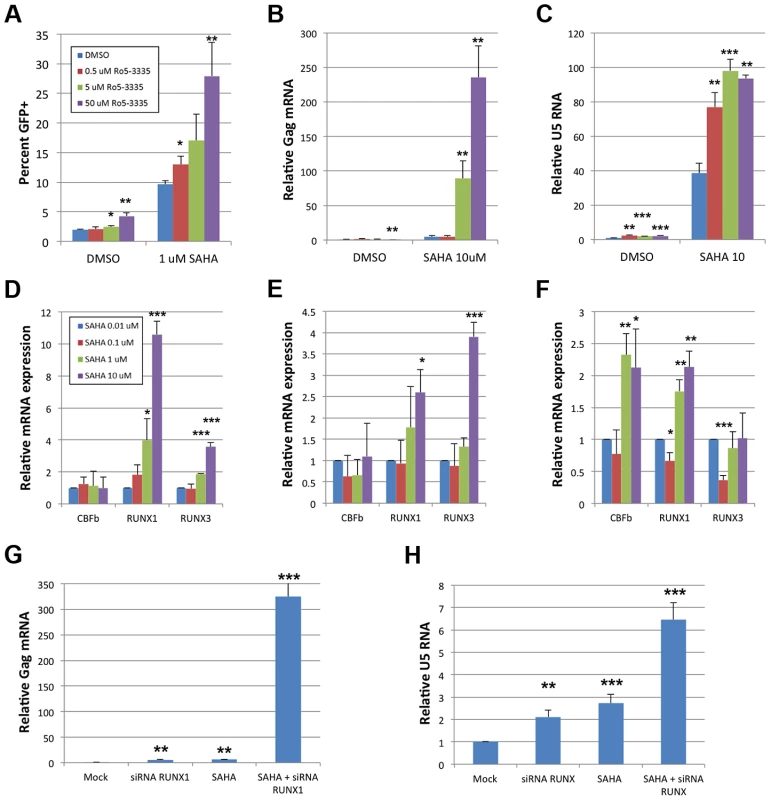
A) JLat cells were cultured in the presence of increasing concentrations of Ro5-3335 in the presence or absence of 1 μM SAHA. Seventy-two hours after treatment the percentage of GFP positive cells was determined by flow cytometry. B) ACH2 cells were cultured with Ro5-3335 and 10 μM SAHA as in panel A. Forty-eight hours post treatment RNA was extracted from the cells and used to determine the relative increase in HIV-1 Gag mRNA as normalized to GAPDH. C) TZMbl cells were cultured with Ro5-3335 and 10 μM SAHA as above. Forty-eight hours post treatment RNA was extracted from the cells and used to determine the relative increase in HIV-1 U5 mRNA as normalized to GAPDH. D) Jlat, E) ACH2 and F) TZMbl were cultured in the presence of the indicated concentrations of SAHA. Forty-eight hours post treatment RNA was extracted from the cells and used to determine the relative changes in RUNX1, CBF-β or RUNX3. G) ACH2 or H) TZMbl cells were transfected with 50 pMol siRNA against RUNX1 and/or treated with 10 μM SAHA at twenty-four hours post transfection. Forty-eight hours post transfection RNA was extracted and expression measured as above. * p≤0.05, ** p≤0.01 and *** p≤0.001 True synergy implies that two compounds are working on the same pathway or mechanism. There are two likely points of interaction that would cause synergy between Ro5-3335 and SAHA. RUNX proteins are capable of recruiting HDACs [19] and this may be happening at the HIV-1 promoter. Alternatively, the inhibition of HDACs by SAHA will induce broad changes in gene expression and this may include changes in RUNX1 and CBF-β expression. To test the later point, we treated cells with SAHA and measured the expression of CBF-β, RUNX1 and RUNX3 by quantitative RT-PCR (Fig 6D–F). Treatment of Jlat with SAHA triggered increased expression of RUNX1 and RUNX3 (Panel D) and a similar pattern was observed for ACH2 (Panel E). Treatment of TZMbl with SAHA also induced RUNX1 expression and induced CBF-β as well (Panel F).
To further verify that the observed synergy of SAHA and Ro5-3335 is due to the involvement of RUNX1 we asked if siRNA knockdown of RUNX1 would synergize with SAHA in the same way as the drug. ACH2 (Fig 6G) and TZMbl (Fig 6H) cells were again transfected with 50 pMol of siRNA against RUNX1 and then treated with either DMSO or 10 μM SAHA at 24 hours post transfection. Twenty-four hours post drug-treatment RNA was extracted from the cells and activation was measured by quantitative RT-PCR against Gag (ACH2) or U5 (TZMbl). Treatment of ACH2 with siRNA alone induced a 4.8-fold increase in expression. siRNA treatment reduced expression of RUNX1 and CBF-β by 52% and 63% respectively. Treatment with SAHA induced a 6.2-fold increase. Treatment with both siRNA against RUNX1 and SAHA led to a greater than 300 fold activation. In TZMbl cells, a similar, but smaller effect was seen. Treatment with siRNA or SAHA alone induced a 2.1 and 2.7-fold increase respectively. Treatment of TZMbl with both siRNA against RUNX1 and SAHA yielded a 6.5-fold increase in transcription. These results are consistent with that seen with Ro5-3335 and SAHA and strongly suggest that RUNX1/CBF-β inhibition may improve the ability of SAHA to activate HIV-1.
The elucidation of RUNX1 involvement in HIV-1 latency described above is heavily dependent upon cell line models of latency. Although these minimalistic systems are useful in determining the possible mechanism of action of a given protein or pathway, they do not represent an ideal model system for evaluating potential therapeutic intervention. The gold standard for evaluating latency is the ability to reactivate latent virus from the PBMCs of HIV-1 patients on suppressive therapy. In order to evaluate the effect of Ro5-3335 in primary cells we obtained PBMCs from five HIV-1 patients on suppressive therapy who had undetectable viral loads for at least 6 months. PBMCs were then treated with SAHA and Ro5-3335 to induce reactivation. In brief, 10×106 PBMCs were divided as needed for multiple conditions and placed in 10 ml RPMI +10%FBS. Cells from one patient were treated with 250 nM SAHA, 250 nM SAHA plus 5 uM Ro5-3335 or DMSO control. A SAHA dose of 250 nM was chosen as it has been shown to be equivalent to the concentration of available drug in the sera of patients [37]. Cells from two patients were treated with the above combinations plus 5 uM Ro5-3335 alone. Finally, cells from the final two patients were incubated with the three conditions plus 1 uM phorbol myristate acetate (PMA). Twenty-four hours after treatment the cells were collected, a small portion (∼300 k cells) was used for flow cytometry to detect activation of T-cells by Ki67 and cell death by vital stain. RNA was extracted from the remaining cells and used for RT-qPCR to detect HIV Gag mRNA (Fig 7A). The background level of Gag mRNA varied from 0.005 to 0.0633 copies/106 GAPDH. In three of the five patient samples treatment with SAHA induced a noticeable increase in Gag mRNA ranging from 1.4 to 6.7-fold. In all five patient samples treatment with SAHA and Ro5-3335 increased the levels of Gag mRNA beyond the levels seen in SAHA treatment (from 3.2 to 75-fold). A Wilcoxon matched pairs test showed a p-value that was approaching significance (p = 0.0625). Control cultures treated with 5 uM Ro5-3335 alone showed no increase in activation as compared to control. Flow cytometric analysis of the cells revealed no increase in T-cell activation (Fig 7B) or cell death (Supplementary Figure S5E).
Fig. 7. Ro5-3335 improves re-activation of HIV-1 in patient samples. 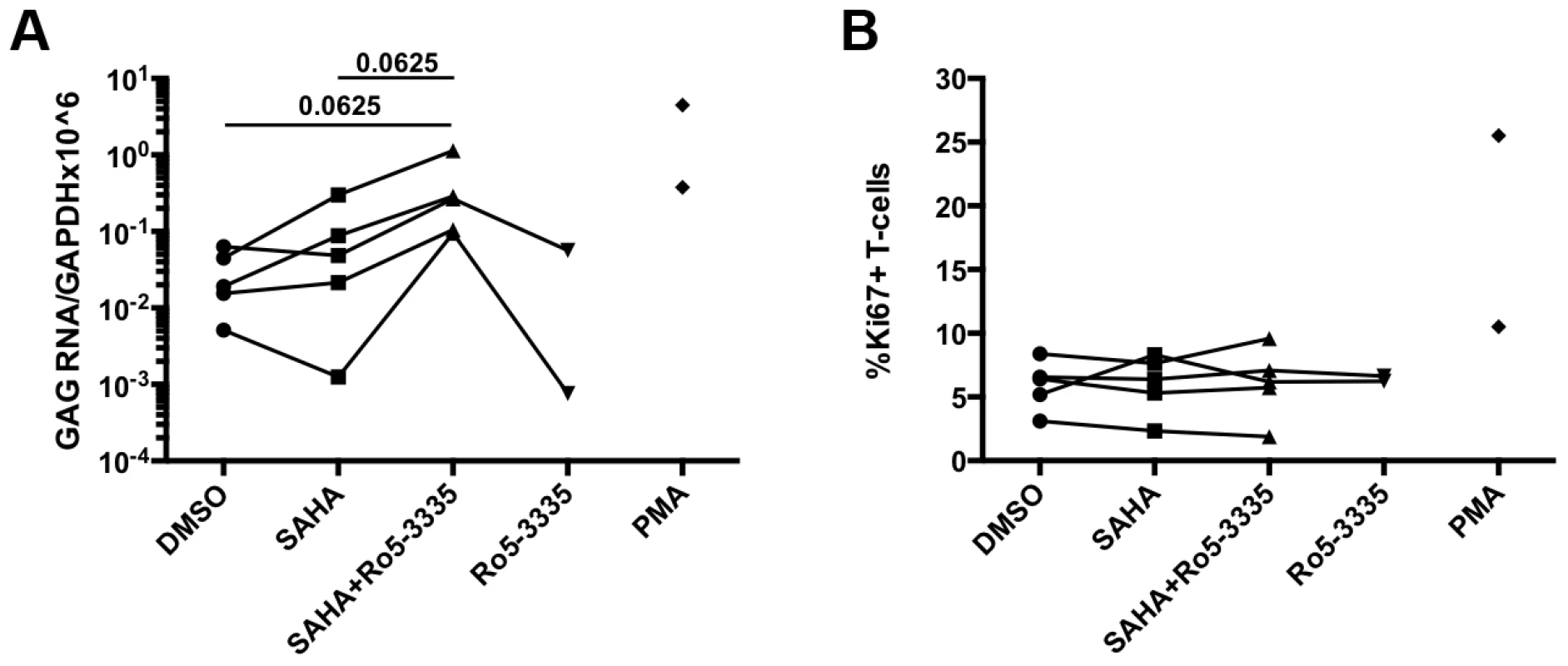
PBMCs from HIV-1 patients suppressed on therapy were treated with DMSO, 250 nM SAHA, 5 uM Ro5-3335 or both drugs in combination. PMA treatment was used as a positive control. A) Twenty-four hours after treatment RNA was isolated from cells and used to detect Gag mRNA by RT-qPCR. B) Flow cytometry was used to determine the percentage of activated T-cells by staining for intracellular Ki67. Discussion
In this study, we report a role for the RUNX family of transcription factors in repressing HIV-1 transcription driven by the viral LTR. The RUNX1 protein is involved in fate determination of T-cells and control of CD4 expression [12], [14], [38], [39] making its potential involvement in HIV replication of physiological interest. We have identified interaction of RUNX1 and the co-factor CBF-β with the viral LTR through a potential binding site (Fig 3) and that alteration of RUNX1 and CBF-β expression alters viral replication (Fig 1).
Work in in the field of HIV Vif biology has identified a role for CBF-β in Vif mediated degradation of APOBEC3G [21], [22]. We have uncovered new mechanisms for the involvement of CBF-β in HIV infection in the context of Vif expression, demonstrating that another functional consequence of Vif:CBF-β interaction is rescue of the virus from transcriptional repression by RUNX1 (Fig 4). A Vif mutant lacking the region necessary for CBF-β binding [20], [22] is incapable of counteracting RUNX1 repression of transcription, thus codifying the role of Vif in protecting the viral promoter.
Perhaps most interestingly we show that repression of RUNX1 activity by a pharmacologic inhibitor (Ro5-3335) is capable of synergizing with the HDAC inhibitor SAHA (Fig 6 and 7). True synergy implies that two compounds are working on the same pathway or mechanism. There are two likely points of interaction that would cause synergy between Ro5-3335 and SAHA. RUNX proteins are capable of recruiting HDACs [19] and this may be happening at the HIV-1 promoter. Alternatively, the inhibition of HDACs by SAHA will induce broad changes in gene expression and this may include changes in RUNX1 and CBF-β expression. Blocking RUNX1 function during SAHA treatment (via either siRNA or drug) significantly increases the activity of SAHA on the LTR. SAHA has been the recent drug of choice employed in studies attempting to reactivate latently infected memory T-cells – a necessary step in clearing the latent reservoir [37], [40]–[43]. Although the drug potently activates cells in culture and has a measurable effect on viral transcription in the T-cells of patients, it has not been successful in reducing the percentage of infected memory cells [44]. Combination therapy in which SAHA is delivered alongside a RUNX inhibitor may provide greater activation of virus, which in turn could lead to greater cell death or greater response by anti-HIV CD8+ T-cells.
It is of note that Ro5-3335 and a related drug have crossed the HIV literature before. A 1991 screen by Hsu et al. identified Ro5-3335 as a potential inhibitor of HIV-1 transcription [45]. However, the exact mechanism of this inhibition is unknown and several follow up studies paint a very complex picture. Another analysis of potential Tat inhibitors determined that Ro5-3335 did not inhibit Tat-TAR interaction [46]. We found recently that Tat binds RUNX1 with high affinity and inhibits Tat-mediated transcription together with CBF-β [24]. Combined with data presented here, our findings suggest that RUNX1 potentially influences Tat transactivation, and is the true target of Ro5-3335.
An analysis of Ro5-3335 and a related drug (Ro 24-7429) determined that they had no suppressive effect in chronically infected cells [47]. A later analysis by Cupelli and Hsu concluded that the drug might act at the level of initiation [48]. A clinical trial using Ro 24-7429 failed to reduce viral load and the presence of infectious virus in patient plasma, or to increase CD4 T-cell count [49]. Indeed, in our hands the activation of HIV-1 by Ro5-3335 happens in a timeframe beyond what was examined in the initial studies. This may explain why no inhibition was seen in chronically infected cells and suggests that the drug acts in a positive fashion on a fully integrated and chromatinized promoter.
What remains unanswered is how RUNX proteins might shape the clinical outcomes during HIV infection. Analysis of CD4+ T-cell populations (Supplementary Figure S4) shows that upon activation of naïve CD4+ T-cells (a population relatively refractory to infection) the expression levels of RUNX1, RUNX3 and CBF-β drop (correlating with greater susceptibility to infection). These findings are similar to what has been demonstrated in mice [13], [50], [51], although the detection of RUNX3 in CD4+ T-cells suggests a possible difference between humans and rodents. Interestingly, the expression of these proteins in naïve cells is fairly uniform, but expression in the memory pool (the primary target cells during infection) is much more variable. Although microarray databases show general expression of RUNX1 and CBF-β across tissues [52], no definitive studies have yet been performed in human cells which examine expression in specific T-cell subsets. Data presented in figure 5 shows a significant negative correlation between RUNX1 expression and viral load, as well as a significant positive correlation between RUNX1 expression and CD4+ T-cell counts in the absence of therapy. However, further research is needed to identify the causal relationship that drives this correlation.
Materials and Methods
Ethics statement
The human sample collection protocol was approved by the NIH Clinical Center Institutional Review Board as part of a separate ongoing study. Written informed consent was obtained in all cases and all applicable protections of patient rights and privacy applied. For this study specific samples were requested from the sample bank based on given criteria.
Cell culture and transfections
Adherent cell lines (293T and TZMbl) were maintained in DMEM supplemented with 10% fetal bovine serum, L-glutamine and penicillin/streptomycin. 293T and TZMbl cells were transfected using Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions.
Suspension cell lines (Jurkat, Jlat, J-LTR-G and ACH2) were maintained in RPMI supplemented with 10% fetal bovine serum, L-glutamine and penicillin/streptomycin. Jurkat and ACH2 cell lines were transfected using the Amaxa Nucleofector (Lonza) with reagent kit V and programs X-005 or T-014 respectively.
Chromatin immunoprecipitation
Cells for chromatin immunoprecipitation (ChIP) were cross-linked using 1% formaldehyde for 10 minutes at 37°C. Following crosslinking, cells were lysed in 1% SDS, 10 mM EDTA and 50 mM Tris-HCl pH 8.1 and sheared by sonication to less than 1000 bp. Lysate was clarified by centrifugation and diluted 1∶10 with 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl and 167 mM NaCl for antibody incubation. Following overnight incubation with antibody, complexes were precipitated using protein A/G agarose beads and washed with low salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl and 150 mM NaCl), high salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl and 500 mM NaCl) and lithium chloride (0.25 M LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA and 10 mM Tris-Hcl) buffers. Complexes were eluted in 1% SDS, 0.1 M NaHCO3 and reverse crosslinked by overnight incubation at 65°C. Proteinase K was used to digest remaining protein before DNA was cleaned using phenol:chloroform and precipitated prior to qPCR with primers for the LTR (174-464) or Gag (bp 1852-2153). Data is represented as signal for each primer pair relative to total input DNA (before IP).
Although positive signal in this LTR primer set might suggest that the relevant RUNX binding site is located between base pairs 174-464, the chromatin used in this analysis was sheared to less than 1000 bases meaning that the binding could be anywhere within the LTR.
Site-directed mutagenesis
RUNX binding site mutants were created in pLTR-Luc using QuikChange XL site directed mutagenesis kit (Stratagene) and primer pairs RUNXMut2, RUNXMut5, RUNXMut6 and RUNXMut7 (see below). Mutants were verified by sequencing.
Confocal microscopy
HeLa CBF-β knockdown (KD) cells (5×106) were transfected with the Vif expression vector pNL-A1 (2.5 μg), the CBF-β vector pCBF-β (1 μg) or were cotransfected with both vectors (2.5∶1 plasmid ratio). Total amounts of transfected DNA were adjusted to 5 μg using empty vector DNA as appropriate. Three hours after transfection, cells were trypsinized and seeded onto cover slips. Cells were fixed 24 hr later in methanol (10 min, −20°C) and then stained with rabbit antibodies to Vif (Vif93; 1∶100) or CBF-β (Thermo Fisher; 1∶100). Nuclear membranes were stained with a mouse monoclonal antibody to lamin B (RDI; 1∶100). Bound antibodies were visualized by Texas-Red or Cy2-conjugated secondary antibodies (Jackson Labs; 1∶100). Images were collected on a Zeiss LSM410 confocal microscope using a Plan-Apochromat 63x/1.4 oil immersion objective (Zeiss).
Flow cytometry and primary cell analysis
To analyze the expression levels of RUNX1 and CBF-β in primary human T-cells, I sorted resting and activated naïve and memory CD4+ and CD8+ T-cells from PBMCs. In brief, PBMC were cultured overnight in the presence of absence of SEB to broadly activate T-cells. Flow cytometry was then used to sort CD3+ T-cells into different populations. Resting cells in the unstimulated population were identified as CD69 - and activated cells from the SEB treatment as CD69+. Cells were then further classed as naïve (CD27+, CD45RO−) or memory (CD45RO+, CD27−). Finally, T-cell populations were sorted by the presence of CD4 or CD8 T-cell co-receptor. For the reactivation work, cells were analyzed for cell death using Live/Dead fixable Aqua (Life Technologies) and stained for CD3 and Ki67. Populations of T-cells were used to prepare RNA using Trizol reagent and RNA was submitted for RT-qPCR for RUNX1.
Statistical analysis
Data represented in graphical form is always the average of at least three replicates with standard deviation. Statistical significance was determined using an unpaired Student's t-test (cell culture) Mann-Whitney (Primary cells) or a Spearman's exact test with a cutoff of p<0.05.
Primers. HIV U5 F CTGCATGGGATGGAGGA
HIV U5 R GTTAGCCAGAGAGCTCCCAG
HIV Gag2 F GGTGCGAGAGCGTCAGTATTAAG
HIV Gag2 R AGCTCCCTGCTTGCCCATA
RUNXMut2 F atccttgatctgtggatctcacacacacaaggctacttcc
RUNXMut2 R ggaagtagccttgtgtgtgtgagatccacagatcaaggat
RUNXMut5 F CCAGGGAGGTGTGTGCTGGGCGGGACTG
RUNXMut5 R CAGTCCCGCCCAGCACACACCTCCCTGG
RUNXMut6 F CTGTACTGGGTCTCTCTGTGTAGACCAGATCTGAGCCT
RUNXMut6 R AGGCTCAGATCTGGTCTACACAGAGAGACCCAGTACAG
RUNXMut7 F GGGTCTCTCTGGTTAGCACAGATCTGAGCCTGGG
RUNXMut7 R CCCAGGCTCAGATCTGTGCTAACCAGAGAGACCC
GAPDH F GCTCACTGGCATGGCCTTCCGTGT
GAPDH R TGGAGGAGTGGGTGTCGCTGTTGA
RUNX1 F GATGGCACTCTGGTCACTGTGA
RUNX1 R CTTCATGGCTGCGGTAGCAT
RUNX3 F TTCCTAACTGTTGGCTTTCC
RUNX3 R TAGGTGCTTTCCTGGGTTTA
CBF-β F ACAGCGACAAACACCTAGCC
CBF-β R CAGCCCATACCATCCAGTCT
RUNX1 Proximal F TGCATGATAAAAGTGGCCTTGT
RUNX1 Proximal R CGAAGAGTAAAACGATCAGCAAAC
RUNX1 Distal F TGGTTTTCGCTCCGAAGGT
RUNX1 Distal R CATGAAGCACTGTGGGTACGA
Supporting Information
Zdroje
1. GreeneWC, PeterlinBM (2002) Charting HIV's remarkable voyage through the cell: Basic science as a passport to future therapy. Nat Med 8 : 673–680.
2. BerkhoutB, JeangKT (1989) trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J Virol 63 : 5501–5504.
3. BerkhoutB, SilvermanRH, JeangKT (1989) Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59 : 273–282.
4. YedavalliVS, BenkiraneM, JeangKT (2003) Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J Biol Chem 278 : 6404–6410.
5. BradyJ, KashanchiF (2005) Tat gets the "green" light on transcription initiation. Retrovirology 2 : 69.
6. HakreS, ChavezL, ShirakawaK, VerdinE (2014) Epigenetic regulation of HIV latency. Curr Opin HIV AIDS 6 : 19–24.
7. KarnJ (2011) The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS 6 : 4–11.
8. MargolisDM (2010) Mechanisms of HIV latency: an emerging picture of complexity. Curr HIV/AIDS Rep 7 : 37–43.
9. BlythK, CameronER, NeilJC (2005) The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer 5 : 376–387.
10. LiX, DeckerM, WestendorfJJ (2010) TEThered to Runx: novel binding partners for runx factors. Blood Cells Mol Dis 45 : 82–85.
11. AdyaN, StacyT, SpeckNA, LiuPP (1998) The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol 18 : 7432–7443.
12. TaniuchiI, OsatoM, EgawaT, SunshineMJ, BaeSC, et al. (2002) Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111 : 621–633.
13. WongWF, KohuK, ChibaT, SatoT, SatakeM (2011) Interplay of transcription factors in T-cell differentiation and function: the role of Runx. Immunology 132 : 157–164.
14. WongWF, KurokawaM, SatakeM, KohuK (2011) Down-regulation of Runx1 expression by TCR signal involves an autoregulatory mechanism and contributes to IL-2 production. J Biol Chem 286 : 11110–11118.
15. DjureticIM, Cruz-GuillotyF, RaoA (2009) Regulation of gene expression in peripheral T cells by Runx transcription factors. Adv Immunol 104 : 1–23.
16. BaeSC, LeeYH (2006) Phosphorylation, acetylation and ubiquitination: the molecular basis of RUNX regulation. Gene 366 : 58–66.
17. WeeHJ, VoonDC, BaeSC, ItoY (2008) PEBP2-beta/CBF-beta-dependent phosphorylation of RUNX1 and p300 by HIPK2: implications for leukemogenesis. Blood 112 : 3777–3787.
18. TaniuchiI, LittmanDR (2004) Epigenetic gene silencing by Runx proteins. Oncogene 23 : 4341–4345.
19. DurstKL, HiebertSW (2004) Role of RUNX family members in transcriptional repression and gene silencing. Oncogene 23 : 4220–4224.
20. Hultquist JF, McDougle RM, Anderson BD, Harris RS (2012) HIV Type 1 Viral Infectivity Factor and the RUNX Transcription Factors Interact with Core Binding Factor beta on Genetically Distinct Surfaces. AIDS Res Hum Retroviruses.
21. JagerS, KimDY, HultquistJF, ShindoK, LaRueRS, et al. (2012) Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 481 : 371–375.
22. ZhangW, DuJ, EvansSL, YuY, YuXF (2012) T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481 : 376–379.
23. Kim DY, Kwon E, Hartley PD, Crosby DC, Mann S, et al.. (2013) CBFbeta Stabilizes HIV Vif to Counteract APOBEC3 at the Expense of RUNX1 Target Gene Expression. Mol Cell.
24. CunninghamL, FinckbeinerS, HydeRK, SouthallN, MaruganJ, et al. (2012) Identification of benzodiazepine Ro5-3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFbeta interaction. Proc Natl Acad Sci U S A 109 : 14592–14597.
25. LevanonD, GronerY (2004) Structure and regulated expression of mammalian RUNX genes. Oncogene 23 : 4211–4219.
26. HuangG, ShigesadaK, WeeHJ, LiuPP, OsatoM, et al. (2004) Molecular basis for a dominant inactivation of RUNX1/AML1 by the leukemogenic inversion 16 chimera. Blood 103 : 3200–3207.
27. YamashitaN, OsatoM, HuangL, YanagidaM, KoganSC, et al. (2005) Haploinsufficiency of Runx1/AML1 promotes myeloid features and leukaemogenesis in BXH2 mice. Br J Haematol 131 : 495–507.
28. PresnellSR, ZhangL, RamiloCA, ChanHW, LutzCT (2006) Functional redundancy of transcription factor-binding sites in the killer cell Ig-like receptor (KIR) gene promoter. Int Immunol 18 : 1221–1232.
29. Schug J (2008) Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics Chapter 2: Unit 2 6.
30. CrooksGE, HonG, ChandoniaJM, BrennerSE (2004) WebLogo: a sequence logo generator. Genome Res 14 : 1188–1190.
31. SorensenKD, Quintanilla-MartinezL, KunderS, SchmidtJ, PedersenFS (2004) Mutation of all Runx (AML1/core) sites in the enhancer of T-lymphomagenic SL3-3 murine leukemia virus unmasks a significant potential for myeloid leukemia induction and favors enhancer evolution toward induction of other disease patterns. J Virol 78 : 13216–13231.
32. SheehyAM, GaddisNC, ChoiJD, MalimMH (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418 : 646–650.
33. WangJ, ReuschelEL, ShackelfordJM, JeangL, ShiversDK, et al. (2011) HIV-1 Vif promotes the G(1) - to S-phase cell-cycle transition. Blood 117 : 1260–1269.
34. ChallenGA, GoodellMA (2010) Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp Hematol 38 : 403–416.
35. WongWF, NakazatoM, WatanabeT, KohuK, OgataT, et al. (2010) Over-expression of Runx1 transcription factor impairs the development of thymocytes from the double-negative to double-positive stages. Immunology 130 : 243–253.
36. JordanA, BisgroveD, VerdinE (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22 : 1868–1877.
37. ArchinNM, EspesethA, ParkerD, CheemaM, HazudaD, et al. (2009) Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses 25 : 207–212.
38. CollinsA, LittmanDR, TaniuchiI (2009) RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol 9 : 106–115.
39. EgawaT (2009) Runx and ThPOK: a balancing act to regulate thymocyte lineage commitment. J Cell Biochem 107 : 1037–1045.
40. ContrerasX, SchwenekerM, ChenCS, McCuneJM, DeeksSG, et al. (2009) Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem 284 : 6782–6789.
41. EdelsteinLC, Micheva-VitevaS, PhelanBD, DoughertyJP (2009) Short communication: activation of latent HIV type 1 gene expression by suberoylanilide hydroxamic acid (SAHA), an HDAC inhibitor approved for use to treat cutaneous T cell lymphoma. AIDS Res Hum Retroviruses 25 : 883–887.
42. WightmanF, EllenbergP, ChurchillM, LewinSR (2012) HDAC inhibitors in HIV. Immunol Cell Biol 90 : 47–54.
43. ArchinNM, LibertyAL, KashubaAD, ChoudharySK, KurucJD, et al. (2012) Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487 : 482–485.
44. ShanL, DengK, ShroffNS, DurandCM, RabiSA, et al. (2012) Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36 : 491–501.
45. HsuMC, SchuttAD, HollyM, SliceLW, ShermanMI, et al. (1991) Inhibition of HIV replication in acute and chronic infections in vitro by a Tat antagonist. Science 254 : 1799–1802.
46. MeiHY, MackDP, GalanAA, HalimNS, HeldsingerA, et al. (1997) Discovery of selective, small-molecule inhibitors of RNA complexes—I. The Tat protein/TAR RNA complexes required for HIV-1 transcription. Bioorg Med Chem 5 : 1173–1184.
47. DunneAL, SiregarH, MillsJ, CroweSM (1994) HIV replication in chronically infected macrophages is not inhibited by the Tat inhibitors Ro-5-3335 and Ro-24-7429. J Leukoc Biol 56 : 369–373.
48. CupelliLA, HsuMC (1995) The human immunodeficiency virus type 1 Tat antagonist, Ro 5-3335, predominantly inhibits transcription initiation from the viral promoter. J Virol 69 : 2640–2643.
49. HaubrichRH, FlexnerC, LedermanMM, HirschM, PettinelliCP, et al. (1995) A randomized trial of the activity and safety of Ro 24-7429 (Tat antagonist) versus nucleoside for human immunodeficiency virus infection. The AIDS Clinical Trials Group 213 Team. J Infect Dis 172 : 1246–1252.
50. CohenMMJr (2009) Perspectives on RUNX genes: an update. Am J Med Genet A 149A: 2629–2646.
51. Collins A, Hewitt SL, Chaumeil J, Sellars M, Micsinai M, et al. RUNX Transcription Factor-Mediated Association of Cd4 and Cd8 Enables Coordinate Gene Regulation. Immunity 34 : 303–314.
52. WuC, OrozcoC, BoyerJ, LegliseM, GoodaleJ, et al. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10: R130.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule FormationČlánek Oral Bacteria and CancerČlánek A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 3- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
- Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
- Mycobacteriophages: Windows into Tuberculosis
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- Five Things to Know about Genetically Modified (GM) Insects for Vector Control
- A Missing Dimension in Measures of Vaccination Impacts
- Eosinophils Are Important for Protection, Immunoregulation and Pathology during Infection with Nematode Microfilariae
- Clonality of HTLV-2 in Natural Infection
- Production, Fate and Pathogenicity of Plasma Microparticles in Murine Cerebral Malaria
- Group B Streptococcal Infection of the Choriodecidua Induces Dysfunction of the Cytokeratin Network in Amniotic Epithelium: A Pathway to Membrane Weakening
- New Insights into How Adapts to Its Mammalian Host during Bubonic Plague
- Foodborne Transmission of Nipah Virus in Syrian Hamsters
- A Polysaccharide Virulence Factor from Elicits Anti-inflammatory Effects through Induction of Interleukin-1 Receptor Antagonist
- Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH
- Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages
- Crk Adaptors Negatively Regulate Actin Polymerization in Pedestals Formed by Enteropathogenic (EPEC) by Binding to Tir Effector
- Fatty Acid Biosynthesis Contributes Significantly to Establishment of a Bioenergetically Favorable Environment for Vaccinia Virus Infection
- A Cytosolic Chaperone Complexes with Dynamic Membrane J-Proteins and Mobilizes a Nonenveloped Virus out of the Endoplasmic Reticulum
- Intracellular Promote Invasive Cell Motility through Kinase Regulation of the Host Actin Cytoskeleton
- MAVS-MKK7-JNK2 Defines a Novel Apoptotic Signaling Pathway during Viral Infection
- RON5 Is Critical for Organization and Function of the Moving Junction Complex
- Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway
- and Exhibit Metabolic Symbioses
- The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface
- Target Cell Availability, Rather than Breast Milk Factors, Dictates Mother-to-Infant Transmission of SIV in Sooty Mangabeys and Rhesus Macaques
- Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
- Infection of Adult Thymus with Murine Retrovirus Induces Virus-Specific Central Tolerance That Prevents Functional Memory CD8 T Cell Differentiation
- Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in
- In Vivo Administration of a JAK3 Inhibitor during Acute SIV Infection Leads to Significant Increases in Viral Load during Chronic Infection
- Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
- Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
- A Compact, Multifunctional Fusion Module Directs Cholesterol-Dependent Homomultimerization and Syncytiogenic Efficiency of Reovirus p10 FAST Proteins
- The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck
- Genetic Dissection of Gut Epithelial Responses to
- Two-Component System Cross-Regulation Integrates Response to Heme and Cell Envelope Stress
- Oral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
- A Model System for Studying the Transcriptomic and Physiological Changes Associated with Mammalian Host-Adaptation by Serovar Copenhageni
- Inflammasome Sensor NLRP1 Controls Rat Macrophage Susceptibility to
- ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by
- The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses
- Caspase-1-Dependent and -Independent Cell Death Pathways in Infection of Macrophages
- The Effect of Cell Growth Phase on the Regulatory Cross-Talk between Flagellar and Spi1 Virulence Gene Expression
- Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms
- Oral Bacteria and Cancer
- Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
- Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector
- VE-Cadherin Cleavage by LasB Protease from Facilitates Type III Secretion System Toxicity in Endothelial Cells
- Dimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
- Crystal Structure of the Vaccinia Virus DNA Polymerase Holoenzyme Subunit D4 in Complex with the A20 N-Terminal Domain
- Post-Translational Regulation via Clp Protease Is Critical for Survival of
- Modulation of Phagosomal pH by Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport
- Rotavirus Activates Lymphocytes from Non-Obese Diabetic Mice by Triggering Toll-Like Receptor 7 Signaling and Interferon Production in Plasmacytoid Dendritic Cells
- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Interferon Regulatory Factor-1 Protects from Fatal Neurotropic Infection with Vesicular Stomatitis Virus by Specific Inhibition of Viral Replication in Neurons
- HMGB1-Promoted and TLR2/4-Dependent NK Cell Maturation and Activation Take Part in Rotavirus-Induced Murine Biliary Atresia
- An Immunomics Approach to Schistosome Antigen Discovery: Antibody Signatures of Naturally Resistant and Chronically Infected Individuals from Endemic Areas
- PPARγ Agonists Improve Survival and Neurocognitive Outcomes in Experimental Cerebral Malaria and Induce Neuroprotective Pathways in Human Malaria
- A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
- Viral OTU Deubiquitinases: A Structural and Functional Comparison
- Heterogeneity and Breadth of Host Antibody Response to KSHV Infection Demonstrated by Systematic Analysis of the KSHV Proteome
- Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm
- Broadly Reactive Human CD8 T Cells that Recognize an Epitope Conserved between VZV, HSV and EBV
- Oncogenic Human Papillomaviruses Activate the Tumor-Associated Lens Epithelial-Derived Growth Factor (LEDGF) Gene
- Erythrocyte Invasion: Combining Function with Immune Evasion
- IL-1α and Complement Cooperate in Triggering Local Neutrophilic Inflammation in Response to Adenovirus and Eliminating Virus-Containing Cells
- Chronic Exposure to Type-I IFN under Lymphopenic Conditions Alters CD4 T Cell Homeostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



