-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
Polyalanine expansions in transcription factors have been associated with eight distinct congenital human diseases. It is thought that in each case the polyalanine expansion causes misfolding of the protein that abrogates protein function. Misfolded proteins form aggregates when expressed in vitro; however, it is less clear whether aggregation is of relevance to these diseases in vivo. To investigate this issue, we used targeted mutagenesis of embryonic stem (ES) cells to generate mice with a polyalanine expansion mutation in Sox3 (Sox3-26ala) that is associated with X-linked Hypopituitarism (XH) in humans. By investigating both ES cells and chimeric mice, we show that endogenous polyalanine expanded SOX3 does not form protein aggregates in vivo but rather is present at dramatically reduced levels within the nucleus of mutant cells. Importantly, the residual mutant protein of chimeric embryos is able to rescue a block in gastrulation but is not sufficient for normal development of the hypothalamus, a region that is functionally compromised in Sox3 null embryos and individuals with XH. Together, these data provide the first definitive example of a disease-relevant PA mutant protein that is both nuclear and functional, thereby manifesting as a partial loss-of-function allele.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003290
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003290Summary
Polyalanine expansions in transcription factors have been associated with eight distinct congenital human diseases. It is thought that in each case the polyalanine expansion causes misfolding of the protein that abrogates protein function. Misfolded proteins form aggregates when expressed in vitro; however, it is less clear whether aggregation is of relevance to these diseases in vivo. To investigate this issue, we used targeted mutagenesis of embryonic stem (ES) cells to generate mice with a polyalanine expansion mutation in Sox3 (Sox3-26ala) that is associated with X-linked Hypopituitarism (XH) in humans. By investigating both ES cells and chimeric mice, we show that endogenous polyalanine expanded SOX3 does not form protein aggregates in vivo but rather is present at dramatically reduced levels within the nucleus of mutant cells. Importantly, the residual mutant protein of chimeric embryos is able to rescue a block in gastrulation but is not sufficient for normal development of the hypothalamus, a region that is functionally compromised in Sox3 null embryos and individuals with XH. Together, these data provide the first definitive example of a disease-relevant PA mutant protein that is both nuclear and functional, thereby manifesting as a partial loss-of-function allele.
Introduction
Trinucleotide repeat expansions are a relatively common cause of human disease. The expanded trinucleotide can occur in an untranslated region, for example in Fragile X syndrome in which a CGG repeat adjacent to the FMRI promoter causes hypermethylation and gene silencing. Alternatively repeat expansions can occur in exonic regions and result in elongation of homopolymeric amino acid tracts. For example polyglutamine (PQ) diseases such as Huntingtons disease, are associated with long unstable PQ-encoding stretches that lead to the production of a toxic species and late onset disease characterised by the loss of a subset of neurons. In addition to PQ encoding repeats, polyalanine (PA) repeat expansion has recently emerged as a significant cause of human disease. PA expansions have been linked to nine disorders of which eight are congenital and one is late onset. Each is caused by PA expansion in a separate gene, with the eight congenital disorders linked to PA expansions in developmentally-important transcription factors. These, and the associated disorders, are SOX3 (X linked Hypopituitarism), HOXA13 (hand–foot–genital syndrome), ARX (syndromic and non-syndromic X-linked mental retardation), HOXD13 (synpolydactyly type II), PHOX2B (congenital central hypoventilation syndrome), FOXL2 (blepharophimosis, ptosis and epicanthus inversus), ZIC2 (holoprosencephaly) and RUNX2 (cleidocranial dysplasia). The ninth is the ubiquitous RNA binding protein PABPN1 which is associated with the late onset disease oculopharyngeal muscular dystrophy (OPMD).
Despite considerable functional analyses, the mechanism by which PA expansion mutations cause disease is not completely understood. Phenotype/genotype correlations in humans and mouse models indicate that many PA alleles give rise to disease phenotypes that resemble (HOXA13, FOXL2, ZIC2) or are less severe (ARX) than null alleles, consistent with complete or partial loss-of-function (LOF) [1], [2], [3], [4]. In contrast, some PA alleles cause similar but more severe disease phenotypes than null alleles consistent with toxic gain-of-function (GOF) and/or dominant negative activity (HOXD13, PHOX2B) [5], [6]. Despite these differences in mode of inheritance, all PA proteins behave very similarly in vitro, such that over-expression in cell culture results in the generation of cytoplasmic and/or nuclear aggregates, which are likely to arise through protein misfolding [7], [8], [9]. While the relevance of cellular aggregates to PA disease in general is unclear, nuclear inclusions that contain mutant PAPBN1 protein are a hallmark of OPMD [10], suggesting that aggregates may also form in patients with PA expansion mutations in developmental transcription factors. Protein aggregation also occurs in the related polyglutamine (PQ) disorders where PQ expansion confers toxic GOF [11], [12]. Together, these data suggest that aggregate formation may have a pathogenic role in PA disease alleles with GOF activity. However, the critical question of whether aggregates form in vivo and, if so, how they may be implicated in the pathogenesis of LOF PA diseases remains unresolved.
To investigate these issues, we used targeted mutagenesis of ES cells to generate a 26 alanine PA tract expansion mutation in Sox3 (Sox3-26ala). The analogous human mutation causes X linked Hypopituitarism (XH), a disease in which hemizygous males have GH deficiency (resulting in short stature) and fully penetrant intellectual disability [13]. Importantly, we find no evidence of SOX3-26ala aggregate formation in neural derivatives of targeted ES cells in vivo or in neurodifferentiated ES cell cultures. Instead, the Sox3-26ala mutation leads to a massive reduction in SOX3 protein within the nucleus. We also present developmental and biochemical evidence that residual mutant protein retains some activity, indicating that Sox3-26ala functions as a partial LOF allele.
Results/Discussion
In order to study the effects of disease-associated PA expansion at a cellular level we created R1 ES cells with a targeted mutation of Sox3. Homologous recombination was used to generate XY ES cells carrying a 36 bp expansion in the first PA tract of Sox3, extending the tract from 14 to 26 alanines (referred to hereafter as Sox3-26ala ES cells; Figure 1). Morula injection of mutant ES cells resulted in chimeras with up to 95% mutant cell contribution as assessed by coat colour (Table 1). None of these chimeras displayed any evidence of growth retardation (consistent with the absence of short stature in heterozygous female carriers of the human SOX3-26ala mutation who also contain a mixture of WT and mutant cells, due to X inactivation [13]). However, despite extensive breeding, none of these chimeras exhibited germline transmission of the mutant allele. In contrast, germline competence was demonstrated with WT parental ES cells as well as a clone carrying the PGK-Neo cassette without the PA tract expansion (referred to hereafter as Neo ES cells). These data suggest that the PA mutation in Sox3 had caused a block in male fertility. This is consistent with a recently published report demonstrating that Sox3 LOF in the germ cell lineage resulted in an early defect in spermatogenesis and the lack of any reported transmission of the human SOX3-26ala allele from hemizygous males [13], [14].
Fig. 1. Generation of Sox3-26ala ES cells. 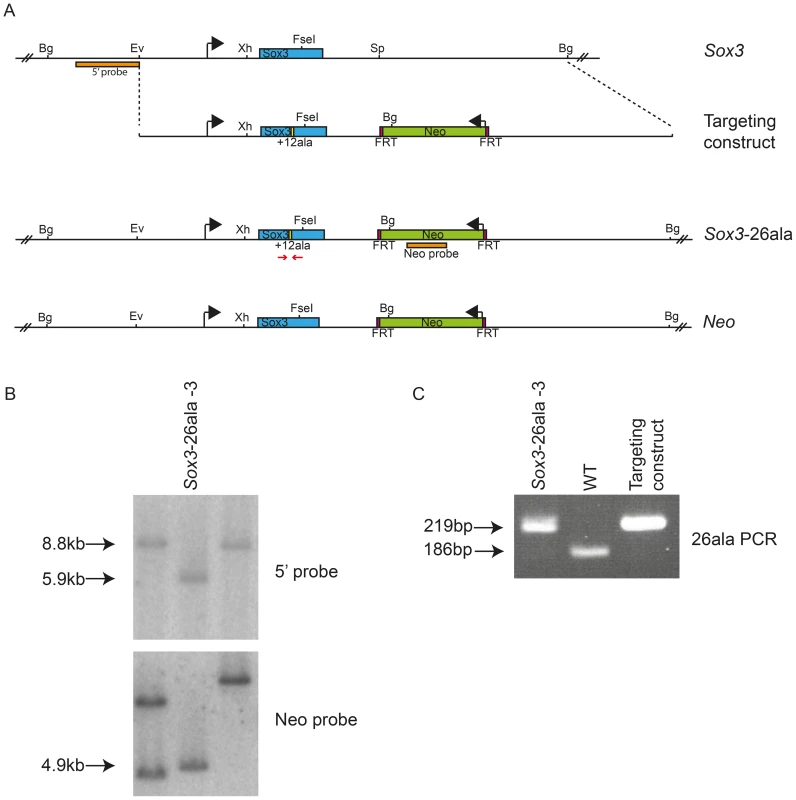
Scale representation of the Sox3 locus, targeting vector and recombinant alleles (A). Probing of BglII digested DNA from ES cell clones with the 5′ probe yielded an 8.8 kb fragment from the WT locus and a 5.9 kb fragment when the Neo cassette was recombined into the Sox3 locus (Sox3-26ala or Neo). B) Representative Southern blot of 3 clones including a targeted clone (Sox3-26ala-3) is shown. C) PCR using primers spanning the alanine expansion (red arrows in A) was used to distinguish whether targeted clones carried the expansion and gave a 219 bp product instead of 186 bp as seen in WT. Tab. 1. Failure of <i>Sox3</i>-26ala targeted ES clones to transmit through the germline. 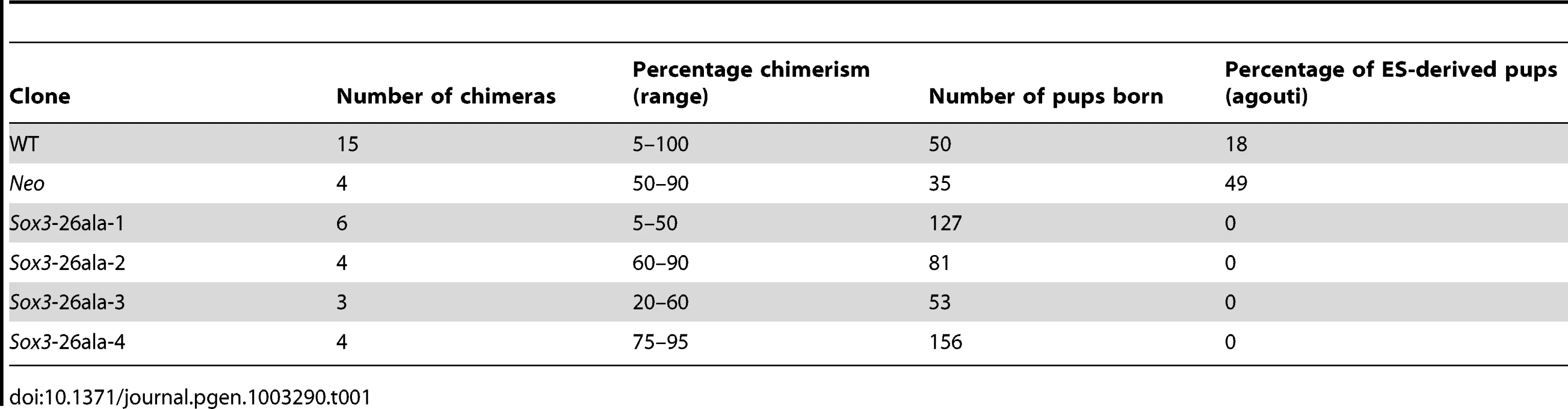
Since we were unable to transmit the Sox3-26ala mutation through the male germline, the phenotypic consequences of the mutation were initially investigated in Sox3-26ala <-> WT chimeric embryos. Immunostaining with a SOX3-specific antibody revealed a dramatic reduction in SOX3 protein in 13.5 dpc telencephalic ventricular zone cells expressing the Sox3-26ala allele (identified by NEO-immunoreactivity) in comparison to neighbouring WT cells (Figure 2A). The ability of the antibody to detect mutant protein was confirmed by staining COS-7 cells expressing exogenous mouse Sox3-26ala, in which large peri-nuclear and cytoplasmic aggregates were common (Figure S1). A striking reduction in mutant SOX3 protein was also observed in 7.5 dpc, 9.5 dpc, 10.5 dpc and 11.5 dpc chimeras (Figure 3 and Figure S2) indicating that this phenotype was not stage-dependent. High power microscopy failed to detect any cytoplasmic or nuclear aggregates of mutant protein but did reveal a very low level of protein in the nucleus (Figure 2A). Comparison of Sox3-26ala and Sox3-null embryonic CNS cells confirmed that the low level of SOX3-26ala protein that we observed was not due to background signal (Figure 2B). Mutant cells appeared morphologically normal and there was no apoptotic induction as assessed by staining for activated Caspase3 (Figure S3). To further characterise the cellular phenotype of Sox3-26ala mutant cells, we performed neurodifferentiation of Sox3-26ala ES cells (which are XY and therefore lack a WT Sox3 allele). Immunohistochemical analysis confirmed the overwhelming reduction of nuclear SOX3 protein in mutant cultures (Figure 2C) in which the expression of other neural progenitor markers were unaffected (data not shown). Rare cells with near normal levels of nuclear mutant protein were also detected but in no case was there evidence of protein aggregation (Figure 2C). Western blot analysis further supported the near complete loss of steady-state SOX3 protein levels in mutant cells (Figure 2D). As triplet repeat expansion mutations have been shown to affect mRNA transcription [12], we compared Sox3 message levels in WT and mutant cells in vitro and in vivo using qPCR and in situ hybridisation, respectively (Figure 2E–2F). No difference in Sox3 mRNA level was detected, indicating that the massive reduction in SOX3 protein levels in the mutant cells has a post-transcriptional aetiology and presumably occurs via degradation of misfolded protein [8]. This was supported by in vitro transcription/translation analysis which indicated that translation of the mutant transcript was unaffected by the PA mutation (Figure S4).
Fig. 2. Transcription is unaffected but protein is cleared from mutant cells. 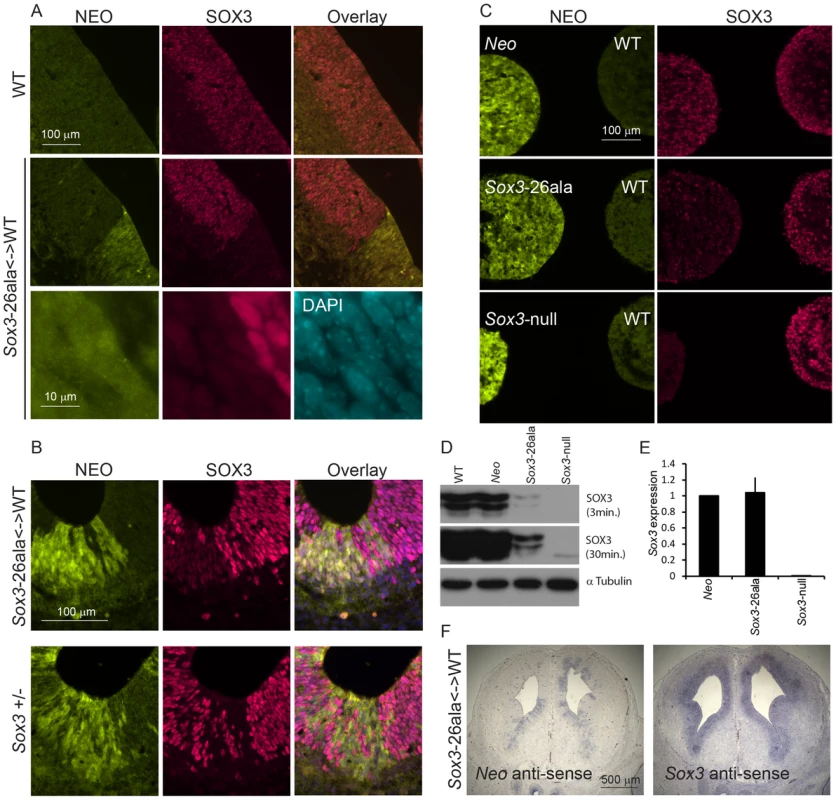
A) SOX3 protein is present in every WT cell (NEO−) of the 13.5 dpc telencephalic ventricular zone but virtually absent from equivalent tissue derived from Sox3-26ala cells (NEO+). B) Comparison of SOX3 immunostaining on Sox3-null cells (from a 14.5 dpc +/− embryo) and Sox3-26ala expressing cells (from a Sox3-26ala <-> WT chimera) confirming that the antibody is SOX3-specific and that the Sox3-26ala expressing cells exhibit a low level of residual nuclear protein. C) WT, Neo, Sox3-26ala and Sox3-null ES cells were differentiated for 5 days in CDM as multi-cellular bodies. Rare SOX3 positive cells were detected in Sox3-26ala CDMs while the majority of cells had low SOX3 protein levels in comparison to neighbouring WT CDM bodies processed on the same slide. D–E) WT, Neo, Sox3-26ala and Sox3-null ES cells were grown in N2B27 for 4 days to form neural progenitors. Western blotting for SOX3 reveals a dramatic reduction of protein in Sox3-26ala cells (D); 3 and 30 minute exposures are shown. E) Transcript levels of Sox3 are not affected in Sox3-26ala cells as determined by qPCR. Three experimental replicates are shown. Data was normalised to Sox3 levels inSox3-Neo control cells and error bars represent SEM. F) ISH confirms that Sox3 transcript is present at comparable levels in ventricular zone cells at 13.5 dpc derived from both WT (Neo−) and Sox3-26ala (Neo+) cells. ISH performed on adjacent 10 µm coronal sections of 13.5 dpc chimeric telencephalon. Fig. 3. Sox3-26ala cells cause pituitary defects indistinguishable from Sox3-null cells. 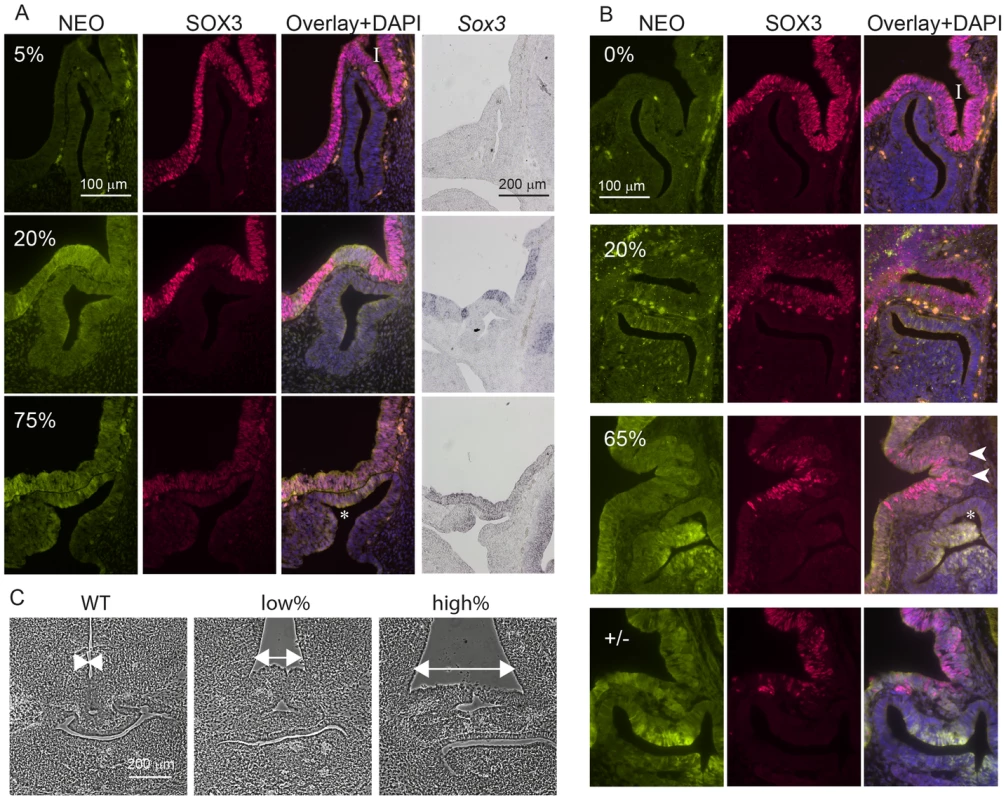
WT, Sox3+/− or Sox3-26ala<->WT chimeras were cut sagittally at 11.5 dpc (A) or 13.5 dpc (B) and immunostained for SOX3 and NEO expression. Percentage chimerism for each embryo in (A) and (B) was determined by qPCR as outlined in the methods. ISH for Neo on adjacent sections at 11.5 dpc confirmed the identification of mutant cells within the ventral diencephalon (A). Examples at 11.5 dpc show the infundibulum (I) appears unaffected in a 5% chimera, shallow in a 20% chimera and absent in a 75% chimera that also displayed a Rathke's Pouch (*) that had failed to detach from the oral ectoderm. At 13.5 dpc, heterozygous and high percentage (65%) chimeric embryos displayed a distorted infundibulum (I) with a lobular edge (arrow heads) and a branched Rathke's Pouch (*). Low percentage chimeras (20%) look similar to WT (0%). C) Phase micrographs of 13.5 dpc coronal sections through the developing pituitary show a broadening at the base of the third ventricle in chimeras (arrows). Chimerism for embryos shown in (C) was determined based on immunoreactivity for NEO in adjacent sections (data not shown). Taken together, these results indicate that Sox3-26ala is a LOF allele and argue strongly against a role for aggregation in the pathogenesis of XH. To further investigate LOF as the mechanism of disease, we compared the development of the hypothalamus, infundibulum and anterior pituitary in Sox3-26ala chimeric embryos to Sox3 heterozygous embryos that carry a complete LOF (null) allele. Sox3 +/ − embryos exhibit a defect in infundibular development that results in aberrant induction and bifurcation of the anterior pituitary primordium, Rathke's Pouch (RP) [15] accompanied by expansion of the floor of the ventral diencephalon (Figure 3). Similar defects in infundibular morphology and pituitary localisation have also been identified in XH patients [16]. Importantly, 13.5 dpc and 11.5 dpc high percentage Sox3-26ala <-> WT chimeras exhibited dysmorphology of the infundibulum, Rathke's Pouch and ventral diencephalon that was indistinguishable from Sox3+/ − embryos (Figure 3). These abnormalities were also present in low percentage chimeras, although were less severe consistent with the loss of SOX3 function being responsible for this phenotype. Notably, there was an obvious reduction in SOX3 protein in mutant (NEO+) cells thereby confirming that PA expansion has a functional impact in neural cells that are directly implicated in XH pathology.
Having established that Sox3-26ala behaves as a LOF allele, we next considered whether the low level of remnant SOX3 protein in mutant cells was functional. To investigate this, we initially performed transactivation assays in COS-7 cells using wild type and mutant human and mouse SOX3 expression constructs and a luciferase reporter containing four SOX consensus motif (SOCM) binding sites. Both mouse and human SOX3-26ala proteins showed activity in this assay that was significantly higher than background (Figure 4A). However, consistent with previous reports [7], [16], this activity was much lower than WT SOX3 protein. To investigate whether this reduction was caused by lower nuclear protein levels or an inherent defect in mutant protein transactivation activity, we measured that relative amount of WT and mutant SOX3 protein in the nucleus of transfected cells by Western Blot (Figure 4B). We observed a reduction of similar magnitude to the reduced luciferase output, suggesting that the mutant protein that is present in the nucleus has similar activity to WT. To investigate whether the residual nuclear SOX3-26ala protein is functional in vivo we compared the phenotypes of 7.5 dpc chimeras generated from either Sox3-null or Sox3-26ala ES cells, as it has been reported that Sox3-null ES cell <-> WT embryo chimeras exhibit severe gastrulation defects [15]. As previous analysis of Sox3-null ES cell chimeras was not performed with R1 ES cells (the parent line used to generate the 26ala mutant ES cells), we generated R1 Sox3-null ES cells for this experiment. Consistent with previous reports, a high proportion (52%) of Sox3-null ES cell chimeras generated abnormal gastrulae (Figure 5). In contrast, only 19% of Sox3-26ala ES cell <-> WT embryo 7.5 dpc chimeras were morphologically abnormal which was not significantly different to the proportion of abnormal chimeric embryos generated using Sox3-flox control ES cells (Figure 5). Immunostaining of Sox3-26ala chimeras revealed a reduction in SOX3 protein levels that was similar to later embryonic stages (Figure S1). These data indicate that nuclear SOX3-26ala protein is functional in vivo and is sufficiently abundant to prevent overt defects in gastrulation. We therefore conclude that Sox3-26ala is a partial LOF allele.
Fig. 4. SOX3-26ala from mouse and human retains transactivation activity. 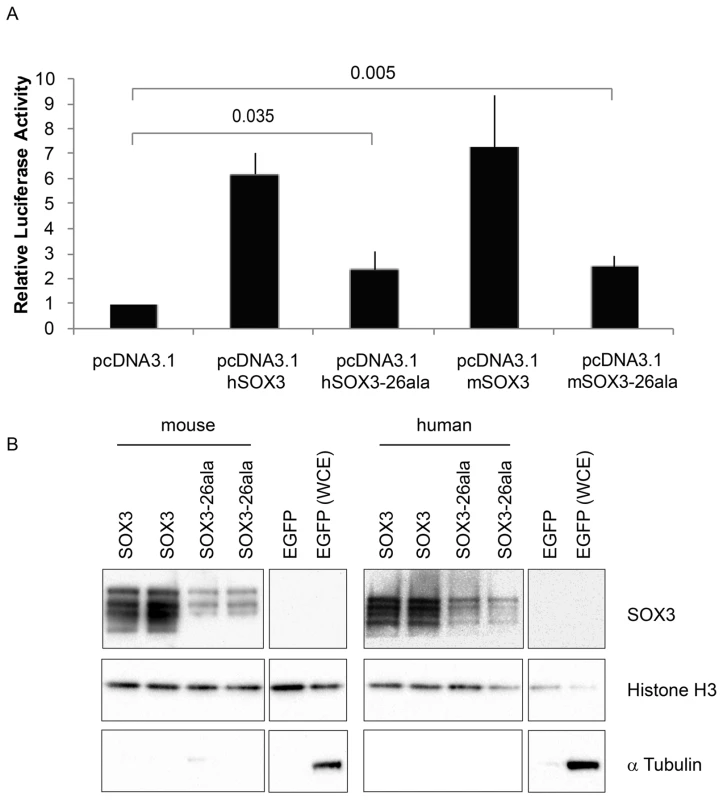
A) COS-7 cells were transfected with pcDNA3.1 expression vector containing either mouse Sox3, human SOX3, mouse Sox3-26ala, human SOX3-26ala or an empty vector control. Values represent mean normalised luciferase values plus standard deviation of four independent experiments measured 48 hours after transfection. Student's T-tests (two tailed, unequal variance) of SOX3-26ala from human or mouse compared to empty vector control show a statistically significant increase in luciferase activity. B) Nuclear protein lysates prepared from duplicate plates 48 hours after transfection show that less SOX3 is detected in the nucleus of cells expressing both mouse and human SOX3-26ala. pcDNA3.1-EGFP transfected cells were used as a control and prepared for both nuclear protein and whole cell extracts (WCE). Blotting for Histone H3, indicates equal loading and blotting for α-Tubulin shows an absence of cytoplasmic contamination in nuclear preparations. Transfection efficiency was determined by co-transfecting EGFP and counting positive cells prior to harvesting and found to be equal for all plasmids. Fig. 5. Residual nuclear SOX3-26ala protein rescues a gastrulation defect of Sox3-null <-> WT chimeric embryos. 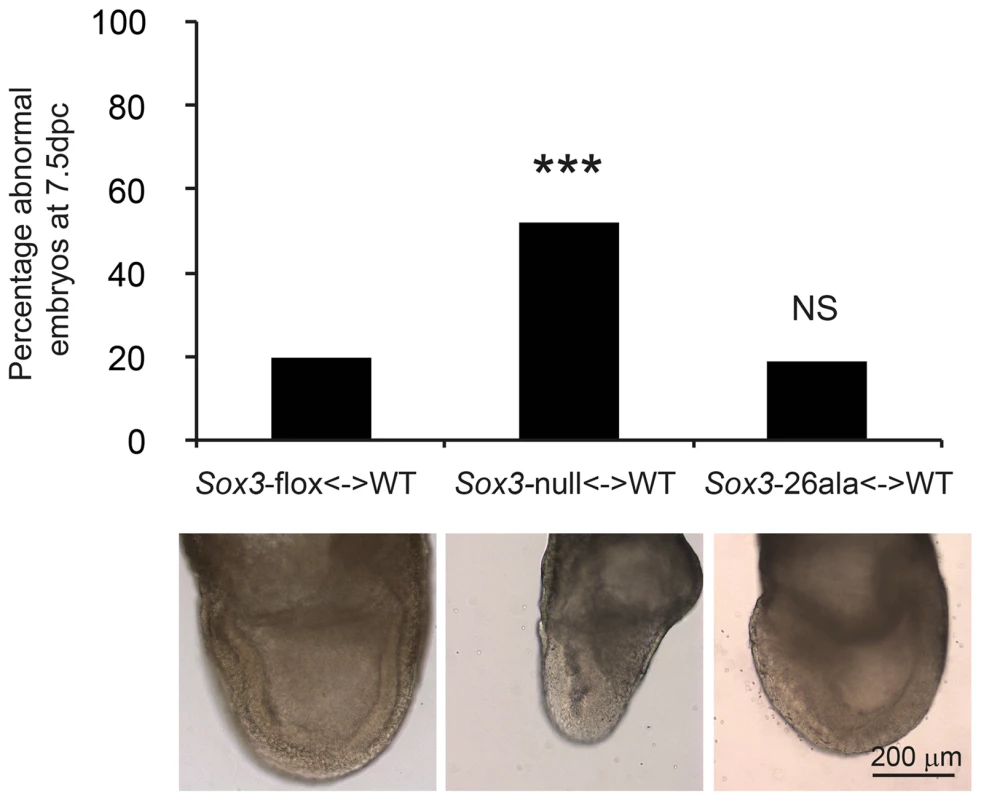
Sox3-26ala <-> WT chimeras are normal at 7.5 dpc (gastrulation) unlike Sox3-null <-> WT chimeras. A total of 15 Sox3-flox<-> WT ES chimeras, 31 Sox3-null<-> WT chimeras and 21 Sox3-26ala<-> WT chimeras were blind scored by two independent operators as morphologically normal or abnormal. The average score for each embryo was used to plot the percentage of abnormal embryos for each condition and chi squared analysis was performed with Sox3-flox<-> WT embryos used to set expected outcomes. Significantly more Sox3-null<-> WT chimeras were abnormal (p = 0.0001) while Sox3-26ala<-> WT chimeras did not deviate from expected (p = 0.95). An example of normal morphology is shown for Sox3-flox<-> WT and Sox3-26ala<-> WT chimeras and an abnormal Sox3-null<-> WT chimera is also shown that exhibits distortion of the ectodermal layer and apparent expansion of cells at the primitive streak and the adjacent extra-embryonic region. This study represents the first investigation of the disease-associated Sox3-26ala mutation under the control of the endogenous locus in a whole animal setting. The complete lack of aggregates in SOX3 positive CNS zones (including the hypothalamic-pituitary axis) provides strong evidence that aggregation is not a feature of the human disease. In contrast, our data demonstrate that PA expansion results in a dramatic reduction in SOX3 protein due to a post-translational defect. Given that PA expansion proteins co-localise with chaperones in vitro and that aggregation is promoted by pharmacological inhibition of the proteasome [8], it seems likely that mutant SOX3 protein misfolds and is cleared from the cell. Of note, the small amount of protein that remains translocates to the nucleus and is functionally active. The level of protein that remains is sufficient for some developmental contexts but not others (i.e. gastrulation but not pituitary induction). This context-specific threshold of SOX3 activity may explain why XH patients with SOX3 PA mutations, but not null mutations, have been identified.
In direct contrast to the Hoxd13 (+7ala) spdh spontaneous mouse model in which mutant protein is mislocalised to the cytoplasm [8], our data provide the first definitive example of a disease-relevant PA mutant protein that is both nuclear and functional, thereby manifesting as a partial LOF allele. Interestingly, mutant protein levels in the Hoxa13 and Arx PA mouse models, which also exhibit LOF inheritance, are diminished but not abolished by western blot and whole mount immunodetection [1], [17]. This raises the possibility that nuclear localisation of suboptimal levels of functional protein may be a feature of several PA diseases. In the case of ARX, the PA mutant mouse model has been reported to have normal nuclear localisation of mutant protein in some tissues, but to have a reduction in the total number of positive cells [17], [18]. Given our data, an alternative interpretation is that cells expressing ARX are not lost but are unable to be detected due to misfolding and clearance of the mutant protein. In situ hybridisation of Arx transcripts should provide a means of discriminating between these possibilities.
In addition to SOX3-26ala, XH is also associated with a 7 alanine expansion mutation (SOX3-22ala) in the same polyalanine tract [16], [19], [20]. Although SOX3-22ala is occasionally associated with mild learning difficulties [20], it is interesting to note that the infantile behaviour/severe MR that is fully penetrant in SOX3-26ala-carrying males is not found in affected males with the SOX3-22ala expansion mutation, suggesting that the shorter expansion is less severe. Given our data showing a massive depletion of SOX3 protein in Sox3-26ala mutant cells in vivo, we would predict that SOX3-22ala protein would also be reduced (compared to WT) but to a lesser extent than the SOX3-26ala protein. This higher level (and therefore activity) of SOX3-22ala compared to SOX3-26ala would be sufficient for “cognitive” CNS development. Consistent with this idea, the SOX3-22ala protein is less prone to aggregate formation in cell culture [7], [16], although one must be cautious in interpreting pathological mechanism from over expression studies in heterologous cell lines. While it would be useful to directly compare SOX3-26ala and SOX3-22ala levels in vivo, a mouse model of the SOX3-22ala mutation has not been generated. However, given that it is now possible to generate neural precursors directly from patients via iPS cells, it would be interesting to compare the SOX3 protein levels in neuroprogenitors derived from SOX3-22ala and SOX3-26ala carrying males. This approach might also be informative for PA disorders in general, particularly given that increased disease severity is generally associated with longer PA expansions [21].
Based on these and other published data, we propose that both GOF and LOF PA disease alleles are associated with a primary defect in protein folding but that a critical difference in the capacity of cells to clear mutant protein results in either the accumulation of mislocalised cytoplasmic protein (GOF) or a diminished level of functional nuclear protein (LOF). As indicated by earlier reports [21], the former mechanism provides scope for dominant-negative and toxic GOF through cytoplasmic sequestration of endogenous binding partners. In the latter, we propose that misfolded protein is efficiently cleared by the cell without significant perturbation of other cellular processes, manifesting as LOF or partial LOF, depending on the amount of residual functional protein. Factors that determine whether or not a cell is able to efficiently clear the misfolded protein could include the level of expression, local concentration differences within the cell, the efficiency of the degradation pathway within different affected cell types and the length of the PA expansion. We propose that the aggregates seen when SOX3-26ala and all other PA expansion proteins are expressed in vitro reflect the overloading of the cell with mutant protein such that normal degradation pathways are overwhelmed. Resolution of the factors that determine whether a cell is able to clear or tolerate mutant protein will have broader implications for proteinopathies such as Alzheimers Disease and Huntingtons Disease in which only subsets of cells display aggregation. For the future, it will be interesting to determine the behaviour of mutant protein in other PA mouse models with GOF and LOF inheritance and, where possible, in patient-derived induced pluripotent stem (iPS) cell derivatives.
Methods
ES cell targeting
Sox3-26ala allele
The targeting vector was based on a Sox3-Neo floxed vector published previously in which GFP and the loxP sites were removed and a 36 bp insertion was introduced to the existing 42 bp alanine encoding tract [15]. This was electroporated into 129 strain-derived (R1) mouse ES cells and 1000 G418 resistant clones were screened by Southern blotting. Integration at the Sox3 locus was determined by a shift in a BglII fragment from 8.8 kb to 5.9 kb when probed with a 5′Sox3 probe [15]. Clones were subsequently screened for the alanine expansion using PCR primers flanking the alanine tract; 5′-AGACGCTGCTCAAGAAGGAC-3′ and 5′-CTGCACGAGCGAGTAGGC-3′. Clones targeted with the Neo cassette but lacking the Sox3-26ala expansion were designated (Neo).
Null allele
The targeting vector for generation of the null allele was the Sox3-Neo floxed vector published previously [15]. 400 clones were screened using the above probe and homologous recombinants identified by a shift in the BglII fragment from 8.8 kb to 7.5 kb. Two correctly targeted clones were transiently transfected with a Cre recombinase expression plasmid (kind gift from Duncan Hewett), seeded at clonal density and screened by PCR for the absence of Sox3. Putative Sox3 null clones were confirmed by Southern blotting (5 kb BglII fragment using the 5′Sox3 probe, as above).
Chimera generation
129 derived ES clones were injected into c57/Bl6xB6D2F1 2.5 dpc morula, cultured overnight and transferred as blastocysts into the uterus of 2.5 dpc pseudopregnant recipients.
ES cell culture and neurodifferentiation
R1 ES cells were maintained on irradiated MEFs in standard conditions and neurodifferentiated either in aggregates in chemically defined media as described in [22] referred to as (CDM) or as monolayers as previously described [23], referred to as (N2B27).
qPCR gene expression and estimation of percentage chimerism
RNA was prepared with Trizol (Invitrogen) and reverse transcribed with a High Capacity RNA-to-cDNA kit (ABI). qPCR was performed using Fast SYBR Green Master Mix (ABI) and run on an ABI 7500 StepOnePlus system. Primer sequences and lengths of amplified products were: Sox3 (117 bp) 5′-GAACGCATCAGGTGAGAGAAG-3′ and 5′-GTCGGAGTGGTGCTCAGG-3′, β-Actin (89 bp) 5′-CTGCCTGACGGCCAGG-3′ and 5′-GATTCCATACCCAAGAAGGAAGG-3′. Sox3 expression was normalised to β-Actin and expressed as relative quantity (RQ) using ABI software. Embryonic chimerism was determined against a standard curve of Sox3 dosage generated from adult gDNA of WT and Sox3-null mixed 0∶100, 25∶75, 50∶50, 100∶0. Loading was corrected using primers against Sox1 (171 bp) 5′-GACTTGCAGGCTATGTACAACATC-3′ and 5′-CCTCTCAGACGGTGGAGTTATATT-3′ and Ngn3 (120 bp) 5′-CCCCAGAGACACAACAACCT-3′ and 5′-AGTCACCCACTTCTGCTTCG-3′. Chimerism in Figure 2C was estimated by NEO IF and ISH.
ISH and IF
Embryos were fixed in 4% PFA overnight and CDM bodies were fixed for 15 minutes. Both were then equilibrated in 30% sucrose overnight, set in OCT compound and cryosectioned on a Leica CM1900 at 10 µm. ISH on fixed frozen sections was performed using a Sox3 probe from a fragment generated with the following primers; 5′-AGCGCCTGGACACGTACAC-3′ and 5′-AGCGCCTGGACACGTACAC-3′ and a Neo probe from a fragment amplified with the following primers; 5′-GATCGATCCCCTCAGAAGAAC-3′ and 5′-GGCTATTCGGCTATGACTGG-3′. Images were captured on a Zeiss Axiophot upright microscope with AnalySIS software, using a 2.5× (Zeiss Neofluar; NA0.5) or a 10× (Zeiss Neofluar; NA0.3) objective lens. Primary antibodies and dilutions were goat anti-hSOX3 (R&D #AF2569,1∶150) and rabbit anti-NTPII (Millipore #06-747, 1∶150). Imaging of IF was performed on a Zeiss Axioplan2 upright microscope with Axiovision software, using a 20× (Zeiss Apochromat; NA0.75) or a 100× (Zeiss Apochromat; NA1.3) objective lens.
Western blotting
Day4 N2B27 neurodifferentiated ES cells were lysed in RIPA buffer supplemented with protease inhibitors. SDS loading buffer was added and the samples were resolved by 10% SDS-PAGE. Duplicate gels were subjected to immunoblot analysis using anti-hSOX3 (1∶2500) and anti-α-Tubulin (Sigma #T8203, 1∶2500) antibodies since both detect proteins at ∼40 kD. Signals were developed using ECL substrate (West Pico, Pierce). For nuclear protein lysates, cells were subjected to hypotonic lysis (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.4% NP-40, 10% Ficoll-400, 1 mM DTT, 1 mM PMSF, 1× protease inhibitors), centrifugation to pellet nuclei that were then washed in wash buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 150 mM KCl, 10% Ficoll-400, 1 mM DTT, 1 mM PMSF, 1× protease inhibitors) and lysed in nuclear extract buffer (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 0.5 mM EDTA, 20% glycerol, 0.42 M KCl, 1 mM DTT, 1 mM PMSF, 1× protease inhibitors).
Luciferase transcription assay
Transcriptional activities of mouse and human SOX3 and SOX3-26ala were determined using the Dual-Luciferase Reporter Assay System (Promega). 1.0 µg of plasmid DNA was transfected into COS-7 cells using Lipofectamine according to manufacturer's instructions. All transfections were performed in triplicate and contained luciferase reporter Sox Consensus Motif (SOCM; 4×AACAAAG) [7], Renilla luciferase plasmid pRL-CMV and one of pcDNA3.1, pcDNA3.1 hSOX3, pcDNA3.1 hSOX3-26ala, pcDNA3.1 mSOX3 or pcDNA3.1 mSOX3-26ala expression constructs (Promega). The firefly luciferase and Renilla luciferase activities were determined after 48 h on a FluorStar Optima (BMG technologies). Relative luciferase activity is the ratio of firefly to Renilla normalised to pcDNA3.1. The assay was repeated four times. Statistical analysis was performed using Student's T-test (two tailed, unequal variance).
Ethics statement
Animal experiments were approved by the University of Adelaide Animal Ethics Committee. All studies were conducted in accordance with the principles of animal replacement and reduction and experimental refinement. Animals were monitored daily for evidence of illness and, if distressed, were culled immediately by cervical dislocation by an experienced investigator/animal technician.
Supplementary methods
Cell free transcription/translation
Cell free transcription/translation was performed using the TnT Coupled Reticulocyte Lysate System (Promega, #L4610) as per manufacturer's instructions. Plasmids used were 250 ng of either pcDNA3.1 hSOX3, pcDNA3.1 hSOX3-26ala, pcDNA3.1 mSOX3 or pcDNA3.1 mSOX3-26ala. Reactions were resolved on a Bio-Rad 10% precast gel (#456-1033), transferred to PVDF membrane, exposed to phosphor-screen and scanned.
COS-7 aggregation assay
COS-7 cells were grown on glass coverslips, transfected with one of pcDNA3.1, pcDNA3.1 hSOX3, pcDNA3.1 hSOX3-26ala, pcDNA3.1 mSOX3 or pcDNA3.1 mSOX3-26ala using Fugene6 (Roche) and stained with SOX3 antibody and DAPI two days later. At least 100 cells were scored randomly from each condition. Results are shown as the mean of three independent experiments and error bars represent one standard deviation.
Supporting Information
Zdroje
1. InnisJW, MortlockD, ChenZ, LudwigM, WilliamsME, et al. (2004) Polyalanine expansion in HOXA13: three new affected families and the molecular consequences in a mouse model. Hum Mol Genet 13 : 2841–2851.
2. De BaereE, BeysenD, OleyC, LorenzB, CocquetJ, et al. (2003) FOXL2 and BPES: mutational hotspots, phenotypic variability, and revision of the genotype-phenotype correlation. Am J Hum Genet 72 : 478–487.
3. RoesslerE, LacbawanF, DubourgC, PaulussenA, HerbergsJ, et al. (2009) The full spectrum of holoprosencephaly-associated mutations within the ZIC2 gene in humans predicts loss-of-function as the predominant disease mechanism. Hum Mutat 30: E541–554.
4. ShoubridgeC, FullstonT, GeczJ (2010) ARX spectrum disorders: making inroads into the molecular pathology. Hum Mutat 31 : 889–900.
5. BrisonN, TylzanowskiP, DebeerP (2011) Limb skeletal malformations - what the HOX is going on? Eur J Med Genet 55 : 1–7.
6. DubreuilV, RamanantsoaN, TrochetD, VaubourgV, AmielJ, et al. (2008) A human mutation in Phox2b causes lack of CO2 chemosensitivity, fatal central apnea, and specific loss of parafacial neurons. Proc Natl Acad Sci U S A 105 : 1067–1072.
7. WongJ, FarlieP, HolbertS, LockhartP, ThomasPQ (2007) Polyalanine expansion mutations in the X-linked hypopituitarism gene SOX3 result in aggresome formation and impaired transactivation. Front Biosci 12 : 2085–2095.
8. AlbrechtAN, KornakU, BoddrichA, SuringK, RobinsonPN, et al. (2004) A molecular pathogenesis for transcription factor associated poly-alanine tract expansions. Hum Mol Genet 13 : 2351–2359.
9. NasrallahIM, MinarcikJC, GoldenJA (2004) A polyalanine tract expansion in Arx forms intranuclear inclusions and results in increased cell death. J Cell Biol 167 : 411–416.
10. CaladoA, TomeFM, BraisB, RouleauGA, KuhnU, et al. (2000) Nuclear inclusions in oculopharyngeal muscular dystrophy consist of poly(A) binding protein 2 aggregates which sequester poly(A) RNA. Hum Mol Genet 9 : 2321–2328.
11. DaviesSW, TurmaineM, CozensBA, DiFigliaM, SharpAH, et al. (1997) Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90 : 537–548.
12. La SpadaAR, TaylorJP (2010) Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet 11 : 247–258.
13. LaumonnierF, RonceN, HamelBC, ThomasP, LespinasseJ, et al. (2002) Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am J Hum Genet 71 : 1450–1455.
14. LarondaMM, JamesonJL (2011) Sox3 functions in a cell-autonomous manner to regulate spermatogonial differentiation in mice. Endocrinology 152 : 1606–1615.
15. RizzotiK, BrunelliS, CarmignacD, ThomasPQ, RobinsonIC, et al. (2004) SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet 36 : 247–255.
16. WoodsKS, CundallM, TurtonJ, RizottiK, MehtaA, et al. (2005) Over - and underdosage of SOX3 is associated with infundibular hypoplasia and hypopituitarism. Am J Hum Genet 76 : 833–849.
17. KitamuraK, ItouY, YanazawaM, OhsawaM, Suzuki-MigishimaR, et al. (2009) Three human ARX mutations cause the lissencephaly-like and mental retardation with epilepsy-like pleiotropic phenotypes in mice. Hum Mol Genet 18 : 3708–3724.
18. PriceMG, YooJW, BurgessDL, DengF, HrachovyRA, et al. (2009) A triplet repeat expansion genetic mouse model of infantile spasms syndrome, Arx(GCG)10+7, with interneuronopathy, spasms in infancy, persistent seizures, and adult cognitive and behavioral impairment. J Neurosci 29 : 8752–8763.
19. Burkitt WrightEM, PerveenR, ClaytonPE, HallCM, CostaT, et al. (2009) X-linked isolated growth hormone deficiency: expanding the phenotypic spectrum of SOX3 polyalanine tract expansions. Clin Dysmorphol 18 : 218–221.
20. AlatzoglouKS, KelbermanD, CowellCT, PalmerR, ArnholdIJ, et al. (2011) Increased transactivation associated with SOX3 polyalanine tract deletion in a patient with hypopituitarism. J Clin Endocrinol Metab 96: E685–690.
21. AlbrechtA, MundlosS (2005) The other trinucleotide repeat: polyalanine expansion disorders. Curr Opin Genet Dev 15 : 285–293.
22. WatayaT, AndoS, MugurumaK, IkedaH, WatanabeK, et al. (2008) Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc Natl Acad Sci U S A 105 : 11796–11801.
23. YingQL, StavridisM, GriffithsD, LiM, SmithA (2003) Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21 : 183–186.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



