-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
The ubiquitin-proteolytic system controls the stability of proteins in space and time. In this study, using a temperature-sensitive mutant allele of the cul-2 gene, we show that CRL2LRR-1 (CUL-2 RING E3 ubiquitin-ligase and the Leucine Rich Repeat 1 substrate recognition subunit) acts at multiple levels to control germline development. CRL2LRR-1 promotes germ cell proliferation by counteracting the DNA replication ATL-1 checkpoint pathway. CRL2LRR-1 also participates in the mitotic proliferation/meiotic entry decision, presumably controlling the stability of meiotic promoting factors in the mitotic zone of the germline. Finally, CRL2LRR-1 inhibits the first steps of meiotic prophase by targeting in mitotic germ cells degradation of the HORMA domain-containing protein HTP-3, required for loading synaptonemal complex components onto meiotic chromosomes. Given its widespread evolutionary conservation, CUL-2 may similarly regulate germline development in other organisms as well.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003375
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003375Summary
The ubiquitin-proteolytic system controls the stability of proteins in space and time. In this study, using a temperature-sensitive mutant allele of the cul-2 gene, we show that CRL2LRR-1 (CUL-2 RING E3 ubiquitin-ligase and the Leucine Rich Repeat 1 substrate recognition subunit) acts at multiple levels to control germline development. CRL2LRR-1 promotes germ cell proliferation by counteracting the DNA replication ATL-1 checkpoint pathway. CRL2LRR-1 also participates in the mitotic proliferation/meiotic entry decision, presumably controlling the stability of meiotic promoting factors in the mitotic zone of the germline. Finally, CRL2LRR-1 inhibits the first steps of meiotic prophase by targeting in mitotic germ cells degradation of the HORMA domain-containing protein HTP-3, required for loading synaptonemal complex components onto meiotic chromosomes. Given its widespread evolutionary conservation, CUL-2 may similarly regulate germline development in other organisms as well.
Introduction
The ubiquitin-proteolytic system has emerged as a central mechanism to regulate protein turnover spatially and temporally [1], [2]. In this system, ubiquitin, a small polypeptide of 76 amino acids, is covalently linked to a target protein through an enzymatic cascade, and the assembly of a poly-ubiquitin chain typically specifies that target protein for rapid degradation via the 26S proteasome [3]. The system can rapidly turn “off” regulatory proteins with high selectivity and is essential for numerous cellular processes such as transcription, signaling, DNA replication, DNA repair, cell cycle progression and differentiation. Not surprisingly, defects in the ubiquitin-proteolytic system have been implicated in a number of human diseases including cancers and neurodegenerative disorders [4]–[6].
An enzymatic cascade of three enzymes mediates the attachment of ubiquitin to substrate protein: the ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-ligase (E3) [7]. Repeated cycles of ligation to the initial ubiquitin lead to poly-ubiquitination. The assembly of poly-ubiquitin chains can occur at different lysine residues within ubiquitin, with conjugation at lysines 11 and 48 typically leading to proteasomal degradation [8]–[11]–[12].
The paramount regulatory step in the cascade is the selective recognition of substrates, which is achieved by the E3-Ligase. Many E3 enzymes are nucleated around cullin scaffold subunits, with at least five cullin family members conserved in all metazoans [13], [14]. Each cullin scaffold nucleates multiple E3-ligase complexes that all contain a similar catalytic core but use different substrate recognition modules to engage their cognate substrate(s) [15], [16]. Over the past several years, Cullin RING E3-ligases nucleated around the cullins 1, 3 and 4 have been investigated extensively. By comparison, the functions of CRL2 complexes are relatively less well understood, with the noticeable exception of the CRL2Von Hippel Lindau (VHL) complex that regulates the hypoxic response (for review see [17]). A Cul2 knockout mouse has not been reported and the roles of Cul2 during vertebrate development remain largely unknown.
In Caenorhabditis elegans, CUL-2 is highly expressed in the germline and in early embryos, where it regulates numerous developmental processes including germ cell proliferation [18]–[20], sex determination [20], progression through oocyte meiosis [21], [22], cell polarity [21], cell fate determination [23] and cell cycle progression [19]. CUL-2 accomplishes these various functions by recruiting, via the adaptor protein elongin C (ELC-1), distinct substrate-recognition subunits (SRS) that specifically engage their substrates (Figure 1A) [17]. Several SRS have been identified in C. elegans, though in most cases their relevant targets remain to be identified. For instance, the Leucine Rich Repeat protein LRR-1 is an SRS expressed throughout the germline where it is essential for germ cell proliferation [19], but the precise function of the CRL2LRR-1 E3-Ligase in the germline is still poorly understood.
Fig. 1. or209 is a conditional cul-2 temperature-sensitive allele. 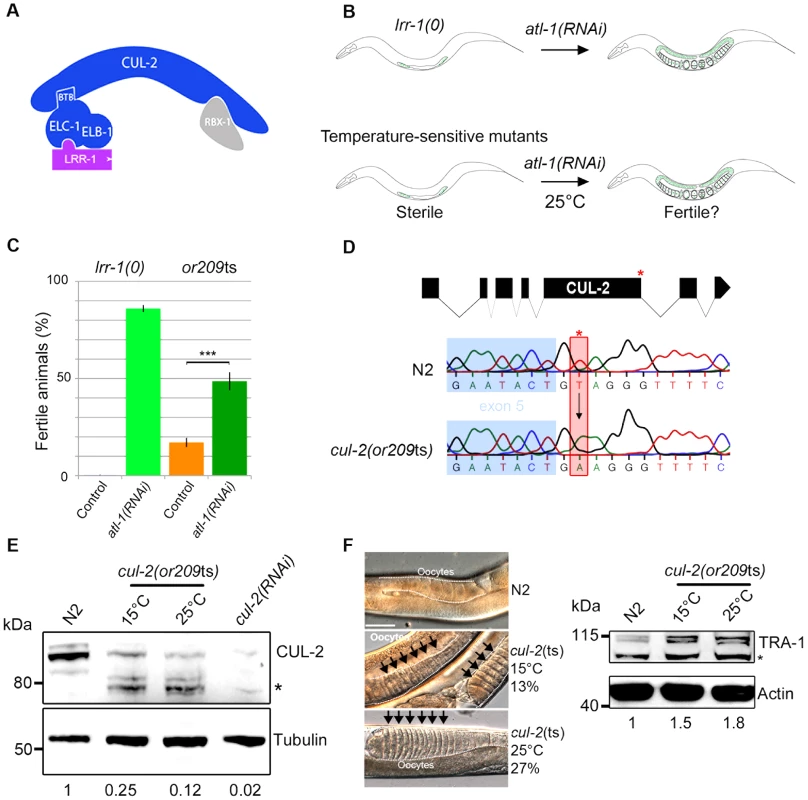
A- Schematic representation of the CRL2LRR-1 E3-ligase. This complex is composed of two modules nucleated around the CUL-2 subunit (arc-shaped in blue): the substrate recognition module and the catalytic site. The substrate recognition module comprises the adaptor protein ELC-1, ELB-1 (blue) and the LRR-1 substrate recognition subunit (purple), whereas the catalytic module contains the RING finger protein RBX-1 (grey). B- Schematic drawing of the RNAi-based screen used to search for temperature-sensitive (ts) alleles affecting the function of the CRL2LRR-1 complex. lrr-1(tm3543) mutant animals (lrr-1(0)) are sterile but recover fertility upon atl-1 depletion by RNAi (upper panel). Therefore, we searched for ts mutants that are sterile at 25°C but recover fertility upon inactivation of atl-1. The or209ts mutant fulfilled these criteria (lower panel). C- The or209ts mutation phenocopies inactivation of the CRL2LRR-1 complex. Graph showing the percentage of fertile or209 animals (25°C) after control (orange bars) and RNAi-mediated depletion of atl-1 (green bars). An average of eight different experiments is presented with 30 animals analysed in each experiment. D- Structure of the cul-2 gene (upper panel); red asterisk depicts the location of the or209 mutation. Chromatograms showing the T to A transversion found in the cul-2(or209ts) mutant (lower panels). E- Embryonic extracts of the indicated gentotype were separated by SDS-PAGE and immunoblotted with CUL-2 (upper panel) and tubulin (lower panel) antibodies (loading control). The asterisk marks the position of truncated forms of CUL-2 that probably lack the C-terminal part of the protein. The value at the bottom is the ratio between CUL-2 and tubulin signal intensities. The wild-type value was arbitrary defined as 1. F- The sex determination factor TRA-1 accumulates in cul-2(or209ts) mutant animals, presumably causing the feminisation of the germline. Micrographs of adult worms of the indicated genotypes analysed by DIC microscopy (left panel). The germline is outlined. Note the accumulation of packed oocytes (black arrows) in cul-2(or209ts) animals, which is characteristic of a feminisation (fem) phenotype. This phenotype is observed in 13% (n = 551) of animals maintained at 15°C and in 27% (n = 228) of animals shifted during 20 hours at 25°C from the L3 stage, but is never observed in wild-type (n>200). Right panel: worm extracts of synchronised cul-2(or209ts) animals cultivated at 15°C or shifted 20 hours at 25°C from the L3/L4 stage were separated by SDS-PAGE and blotted with TRA-1 (upper panel) antibodies. The asterisk marks the position of a non-specific band. The value at the bottom is the ratio between TRA-1 and actin signal intensities. The wild-type value was arbitrary defined as 1. We report here the isolation of a temperature-sensitive mutation in the C. elegans cul-2 gene. The cul-2(or209ts) mutation recapitulates all the cul-2(RNAi) and cul-2(0) phenotypes and also reveals several novel functions in the C. elegans germline. More specifically, we show that besides promoting germ cell proliferation, CUL-2 has at least two other functions in the germline: it participates in the proliferation versus meiotic entry decision and inhibits the first steps of meiotic prophase. Furthermore, we show that the CRL2LRR-1 E3-ligase inhibits meiotic prophase progression at least in part by promoting degradation of HTP-3, a HORMA domain containing protein. HTP-3 is required for progression through meiotic prophase and for the assembly of the synaptonemal complex, the zipper-like structure that tethers homologous chromosomes together.
Results
Identification of a temperature-sensitive allele in the cul-2 gene: cul-2(or209ts)
The evolutionarily conserved CRL2LRR-1 E3-ligase is essential for germline development in C. elegans: lrr-1(0) and cul-2(0) null mutants are sterile with a small germline [18], [19]. However, the precise function of this enzyme is still poorly understood as the sterility in null mutants limits analysis of gene requirements. To circumvent this problem, we screened for temperature-sensitive (ts) mutants that resemble lrr-1(0) mutants. While lrr-1(0) animals are sterile, this phenotype is suppressed by inactivation of the DNA replication ATR/ATL-1 checkpoint pathway [19]. Therefore, we screened for ts mutants that, like lrr-1(0) mutants, are sterile at the restrictive temperature of 25°C but recover fertility upon atl-1 depletion by RNAi (Figure 1B). Such mutants might be specifically defective in the function of the CRL2LRR-1 complex and allow for a more careful analysis of gene requirements in the germline at different temperatures that only partially compromise E3-ligase function.
After screening a collection of ts mutants (Figure S1, Table S1) we found one that fulfilled the above criteria: or209ts. Like lrr-1(0) mutants, most or209ts mutant animals are sterile at 25°C but recover fertility upon atl-1 inactivation by RNAi (Figure 1C). or209ts has previously been linked to chromosome III [24], and we used single nucleotide polymorphisms to further map the or209ts mutation to the end of this chromosome, which contains the cullin gene cul-2 (21.36 cM) (Figure S1). We next found that or209ts fails to complement a known cul-2 null allele (material and methods), and we then sequenced the cul-2 gene from or209ts genomic DNA and found a single nucleotide change relative to wild-type in the splicing donor site at the fifth exon-intron boundary (Figure 1D). This mutation substantially reduces CUL-2 protein levels even at the permissive temperature (15°C), but this effect is aggravated at the restrictive temperature (25°C) (Figure 1E). Consistent with the partially conditional effect on protein levels, cul-2(or209ts) animals present high levels of embryonic lethality (30%) and a severe reduction in brood size even at the permissive temperature of 15°C, compared to wild-type (Figure S1). Further analysis revealed that cul-2(or209ts) animals present typical cul-2 loss-of-function phenotypes, including the accumulation of abnormally high levels of the TRA-1 protein, presumably causing feminization of the germline (Figure 1F), and of the PIE-1 protein (Figure S2), both well-defined targets of CRL2FEM-1 and CRL2ZIF-1 E3-ligases, respectively [23], [25]. We conclude that or209ts is a conditional allele of cul-2. To our knowledge, or209ts is the first temperature-sensitive allele yet identified that affects a metazoan cullin gene.
CRL2LRR-1 E3-ligase promotes germ cell proliferation in adult germlines
Using the or209ts allele, we re-examined CUL-2 function in the germline. The C. elegans germline is spatially organized and contains - from the distal to proximal end -mitotically proliferating stem cells, meiotic germ cells, and gametes (sperm or oocyte). The mitotic zone extends 18–20 germ cell diameters (gcd) along the gonadal axis from the distal end, with the total number of germ cell nuclei exceeding 200 [26] (Figure 2A). In the transition zone (TZ), just proximal to the mitotic zone, cells enter into meiotic prophase [27], [28]. At this stage, structural components accumulate on meiotic chromosomes (e.g HIM-3 [29]) such that germ cell nuclei present a characteristic crescent shape that is readily evident by DAPI staining [30].
Fig. 2. CRL2LRR-1 E3-ligase promotes germ cell proliferation. 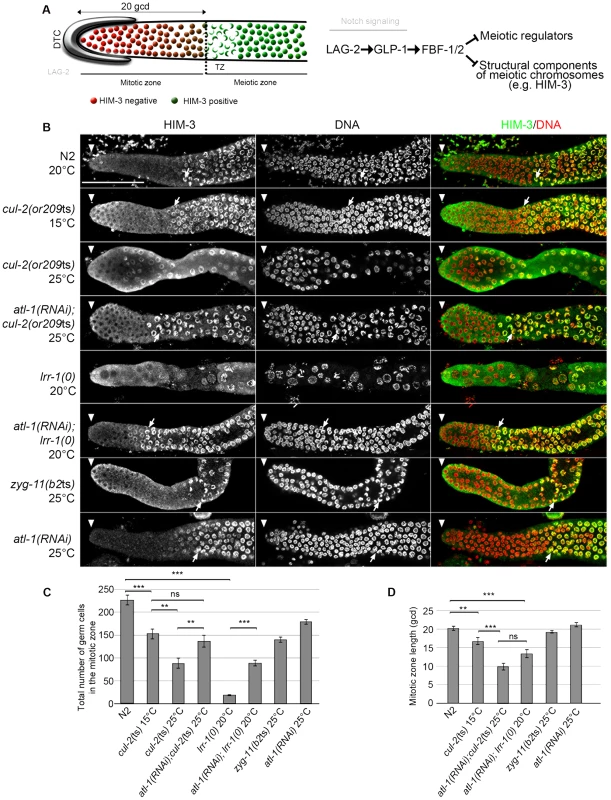
A- Schematic drawing of an adult distal germline. The distal tip cell (DTC) niche is located at the distal end. The DTC expressed the Notch ligand LAG-2, which is shown in grey. The mitotic zone contains germ cells in the mitotic cell cycle (red), including a germline stem cell (GSC) pool distally and possibly transit-amplifying germ cells proximally; at its proximal edge, some germ cells have entered pre-meiotic S-phase. The transition zone contains germ cells in meiotic S-phase and in meiotic prophase (nuclear crescents) that start to accumulate HIM-3 (green), a marker of meiotic prophase. The dashed line marks the boundary between the mitotic and transition zones (left panel). A simplified pathway with key regulators of the balance between GSC renewal and meiotic differentiation is shown in the right panel. Positive regulation (arrows) and negative regulation (bars). B- Representative images of dissected gonads of animals of the indicated genotypes taken 24 hours after the mid-L4 stage stained with HIM-3 antibodies (green) and DAPI (red). The distal end of the germline (arrowhead) is oriented toward the left, and the proximal end is oriented toward the right in this and other figures. Arrow, nuclei with crescent shape that is typical of nuclei in meiotic prophase. Germlines in each panel were treated identically and fluorescent images taken at the same settings. Scale bars: 50 µM. C- Graphs showing the mean of the total number of germ cells in the mitotic zone of the germline and D- the quantification of the size of the mitotic zone, in germ cell diameters (gcd) from the distal end in animals of the indicated genotypes. At least ten germlines of each genotype were scored. Corroborating our previous observations indicating that loss of the CRL2LRR-1 E3-ligase results in a cell cycle arrest [19], the number of germ cells in the mitotic zone was severely reduced and nuclei were enlarged in cul-2(or209ts) mutants raised from early larval stages (L1) at the restrictive temperature of 25°C. As expected, this phenotype was largely suppressed by inactivation of the ATL-1/DNA replication pathway (Figure 2). In addition, we noticed that the size of the mitotic zone was reduced in cul-2(or209ts) animals, as determined by scoring the distance, in gcd, between the distal end of the germline and the appearance of both nuclear crescents and HIM-3 positive cells (Figure 2B, arrows and 2C–D). The mitotic zone was reduced to 15 gcd in cul-2(or209ts) mutants raised to adulthood at permissive temperature (15°C), and even more reduced in mutants raised at the restrictive temperature (25°C). However, it was difficult to rigorously quantify the phenotype at 25°C because germ cell nuclei were both enlarged and reduced in number (Figure 2). We thus analyzed the size of this region in atl-1(RNAi); cul-2(or209ts) mutant hermaphrodites and also observed a smaller mitotic zone in these animals. The size of the mitotic zone was similarly reduced in atl-1(RNAi); lrr-1(0) animals but was unaffected in the control atl-1(RNAi) (Figure 2B and 2D). Finally, ZYG-11 is another substrate-recognition subunit of a CRL2 complex that appears dispensable in germline stem cells [31], and accordingly, the size of the mitotic zone was unaffected in zyg-11(b2ts) mutant germlines at 25°C (Figure 2B and 2D). Collectively, these results indicate that CUL-2, in combination with LRR-1, promotes germ cell proliferation by counteracting the ATL-1/DNA replication checkpoint but may also influence the size of the mitotic zone independently of ATL-1. Indeed, atl-1 depletion in lrr-1 and cul-2 mutants does not fully rescue the size of the mitotic zone and atl-1 depletion in WT animals does not affect the size of the mitotic zone.
cul-2 participates in the proliferation versus meiotic entry decision
We thus investigated whether cul-2, in addition to regulating mitotic proliferation, might also have a role in preventing meiotic entry, by looking for genetic interactions between cul-2 and genes regulating the proliferation versus meiotic entry decision. GLP-1/Notch signaling controls the decision between self-renewal and entry into the meiotic cell cycle [28]. Downstream of Notch, the nearly identical Puf (Pumilio and FBF)-domain RNA-binding proteins FBF-1/2 prevent meiotic entry by repressing the translation of meiotic promoting factors and structural components of meiotic chromosomes (Figure 2A) [32], [33]. We therefore constructed double mutants between cul-2(or209ts) and glp1(bn18), a temperature-sensitive glp-1 mutant in which the Notch signaling pathway is partially defective [34], and between cul-2(or209ts) and fbf-1(0) or fbf-2(0), and scored meiotic entry in the double versus single mutants at the semi-permissive temperature of 20°C. At this temperature, germ cell proliferation is only modestly affected in cul-2(or209ts) mutants (Figure 3A).
Fig. 3. 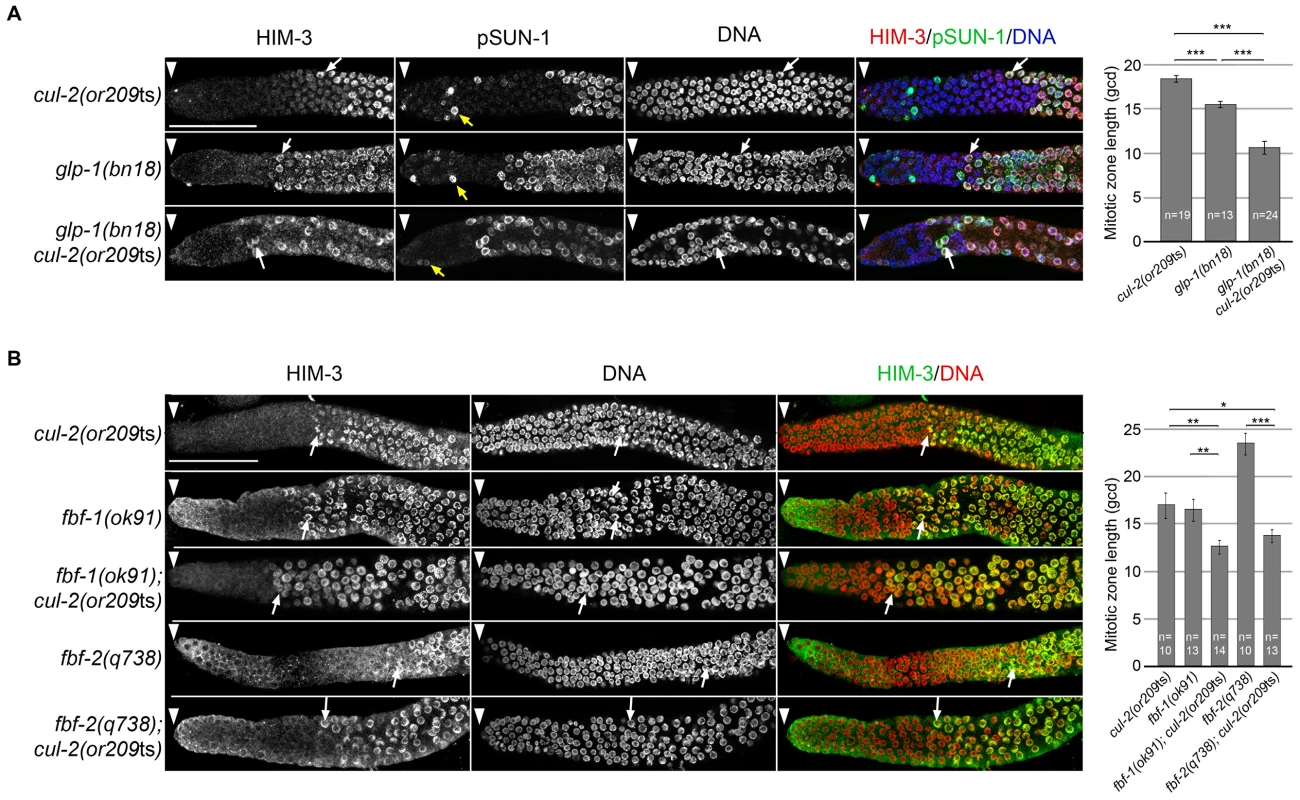
A- Representative images of dissected gonads of animals of the indicated genotypes, maintained at 20°C from L1 and taken 24 hours after L4, stained with HIM-3 (red), SUN-1 Ser8-Pi (green) antibodies and with DAPI (blue) (left panel). Arrowhead, distal end of the germline. Yellow arrow, germ cells in G2-M phase of the cell cycle and white arrow, nuclei with crescent-shape morphology typical of meiotic prophase. Quantification of the size of the mitotic zone in germ cell diameters from the distal end is shown on the right. B- Representative images of dissected gonads of animals of the indicated genotypes maintained at 20°C from L1 and taken 24 hours after the L4 stage stained with HIM-3 antibodies (green) and counterstained with DAPI (red) (left panel). Arrowhead, distal end of the germline. Arrow, nuclei with crescent-shape morphology typical of meiotic prophase. Quantification of the size of the mitotic zone in germ cell diameters from the distal end is shown on the right. Note that although <i>fbf-1</i> and <i>fbf-2</i> genes are largely redundant for promoting mitosis, they have opposite roles in regulating the size of the mitotic region. The mitotic region is smaller than normal in <i>fbf-1(0)</i> mutants but larger than normal in <i>fbf-2(0)</i> mutants. This opposite role is largely due to different spatial regulation and reciprocal 3′UTR repression; <i>fbf-2</i> expression is limited to the distal germline whereas <i>fbf-1</i> expression is much broader <em class="ref">[58]</em>. To score meiotic entry in these different mutant backgrounds, we used a combination of cellular morphology (nuclear crescents) and molecular markers. More specifically, we monitored the appearance of HIM-3 and SUN-1 Ser8-Pi (P-SUN-1) on meiotic chromosomes [35]. P-SUN-1 is first detected in the mitotic zone, at nuclear periphery, in germ cells that are in mitosis from prometaphase onward (Figure 3A, yellow arrows), and then in meiosis, in the transition zone (TZ) at foci and patches, as well as over the nuclear envelope, as reported previously [35]. As shown in Figure 3A, the cul-2ts mutant enhanced the premature meiotic entry phenotype of the glp-1(bn18) mutant, as evidenced by the premature meiotic entry in the glp-1(bn18) cul-2(or209ts) double mutants, compared to the single mutants. Likewise, when combined with cul-2(or209ts), both fbf-1 and fbf-2 mutants showed a smaller mitotic zone than single mutants (Figure 3B) indicating that cul-2 influences the size of the mitotic zone. Collectively, these results indicate that CUL-2 acts with the Notch signaling pathway and FBF-1/2 to prevent meiotic entry.
CRL2LRR-1 regulates HTP-3 stability in the mitotic zone of the germline
CRL2LRR-1 acts through HTP-3 in the C. elegans germline
Entry into meiosis requires the coordination of a number of events: upon exit from the mitotic zone, germ cells initiate the chromosome dynamics required for meiotic pairing and synapsis. In preparation for this transition, proximal cells in the mitotic zone activate the expression of both regulators of meiotic entry and chromosomal proteins required for synapsis (e.g HIM-3) that are then recruited on meiotic chromosomes. CUL-2 may thus inhibit meiotic entry by controlling the stability of regulators of meiotic entry in the mitotic zone of the germline.
Interestingly, an RNA interference (RNAi)-based screen (JM and LP, unpublished data) for suppressors of cul-2(or209ts) lethality at a semi-permissive temperature (23°C) identified, in addition to ATL-1 and its regulator MUS-101/TopBP1 [36], the axial element component HTP-3, which is required for the progression through meiotic prophase [37], [38], and SYP-1, a core component of the synaptonemal complex (SC) [39] (Figure 4A and 4B).
Fig. 4. CRL2LRR-1 E3-ligase acts through HTP-3 in the C. elegans germline. 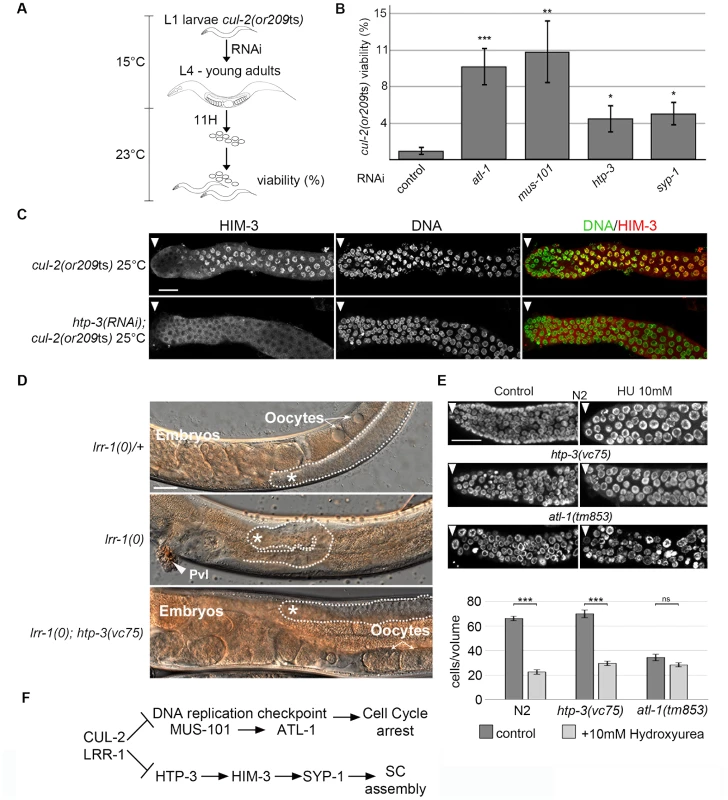
A- Flow-chart of the cul-2(ts) RNAi-based suppressor screen. At 15°C, synchronised cul-2(ts) L1 larvae were fed with bacteria expressing dsRNA, then were shifted to a semi-permissive temperature (23°C) at the L4 stage. After 11 hours, young adults were removed, and embryonic viability was determined and plotted. B- Graphs showing the percent viability of cul-2(or209ts) animals after RNAi depletion of the indicated genes. C- Representative images of dissected gonads of the indicated genotypes stained with HIM-3 antibodies (red) and DAPI (green) are shown. Scale bars: 50 µM. Arrow marks the distal end of the germline D- Reduction of htp-3 function suppresses lrr-1(0) mutant animal sterility. Micrographs of adult worms of the indicated genotypes were analysed by DIC microscopy. Thirty-four percent of lrr-1(0); htp-3(vc75) double mutants are fertile (n = 112). The germline is outlined, and the presence of oocytes and embryos is indicated. The asterisk marks the distal region of the germline. E- The DNA replication checkpoint is functional in htp-3(vc75) mutant animals. Animals of indicated genotypes were exposed to 10 mM Hydroxyurea (HU) and germlines were dissected 24 h post-L4 and stained with DAPI (upper panel). The number of germ nuclei in a given volume was determined and plotted (bottom panel). Note that HU treatment of wild-type (N2) animals leads to S-phase arrest that manifests as enlarged nuclei with an overall reduction in the number of nuclei in the mitotic zone of the germline. HU treatment similarly arrested htp-3(vc75) mutant germ cells, whereas checkpoint deficient atl-1(tm853) mutant germ cells fail to arrest cell cycle progression, indicating that the DNA replication checkpoint is functional in the htp-3(vc75) mutant. F- Pathway diagram summarizing the observed genetic interactions. CUL-2 and LRR-1 counteracts the DNA replication checkpoint pathway, which is composed of MUS-101 and ATL-1, and inhibits HTP-3, which promotes the recruitment of HIM-3, SYP-1 and the assembly of the synaptonemal complex (SC). Positive regulation (arrows) and negative regulation (bars). HTP-3 belongs to the family of HORMA (Hop1-Rev1-Mad2) domain-containing proteins [40], and is required for preparing chromosomes for meiosis. More specifically, HTP-3 is required for loading on chromosomes the meiotic cohesin REC-8 and the axial and transverse components of the synaptonemal complex, including HIM-3 and SYP-1; for the formation of double-strand breaks that initiate meiotic recombination; and for implementation of the meiotic DNA damage ATM-1/checkpoint [38], [41], [42].
Given that HTP-3 promotes HIM-3 and SYP-1 loading on chromosomes, we investigated the potential role of CRL2LRR-1 in HTP-3 regulation. As expected, the accumulation of HIM-3 on chromosomes in cul-2(or209ts) germ cells depends on HTP-3 because HIM-3 failed to accumulate on chromosomes in htp-3(RNAi); cul-2(or209ts) mutant germlines (Figure 4C). To determine whether LRR-1 acts through HTP-3 in the germline, we asked whether htp-3 reduction-of-function suppressed lrr-1(0) mutant sterility. To this end, we took advantage of the htp-3(vc75) allele, which only modestly compromises HTP-3 function [42], and constructed lrr-1(tm3543); htp-3(vc75) double mutant animals. Importantly, a significant fraction (34%, n = 115) of htp-3(vc75); lrr-1(tm3543) double mutant animals were fertile and produced embryos, in contrast to the highly penetrant sterility phenotype observed in lrr-1(tm3543) single mutants (Figure 4D). The observed suppression of the lrr-1(tm3543) sterility by the htp-3(vc75) allele is not merely resulting from an inactive ATL-1 checkpoint pathway in the htp-3(vc75) mutant because the DNA replication checkpoint is fully functional in this mutant (Figure 4E and Figure S3). Collectively, these results indicate that LRR-1 acts through HTP-3 in the germline (Figure 4F).
CRL2LRR-1 and the 26S proteasome control HTP-3 levels in the mitotic zone of the germline
To test whether CRL2LRR-1 regulates HTP-3 stability, we assessed HTP-3 protein levels by immunostaining cul-2(or209ts) and lrr-1(0) mutant germlines. In control germlines, HTP-3 was expressed at low levels in the most distally located germ cells and at high levels on chromosomes in the most proximal mitotic germ cells (Figure 5A) [38], [43]. In contrast to wild-type, HTP-3 was abundant in the most distally located germ cells in both cul-2(or209ts) and lrr-1(tm3543) mutants (Figure 5A). HTP-3 similarly accumulates in atl-1(RNAi); cul-2(or209ts) and atl-1(RNAi); lrr-1(tm3543) germlines, indicating that down-regulation of the DNA replication checkpoint does not affect HTP-3 levels. Importantly, HTP-3 also accumulates in the distal region after inactivation of PBS-5, a proteasome subunit, suggesting that CRL2LRR-1 may target HTP-3 for degradation (Figure 5A).
Fig. 5. CRL2LRR-1 E3-ligase regulates HTP-3 levels in the mitotic zone of the germline. 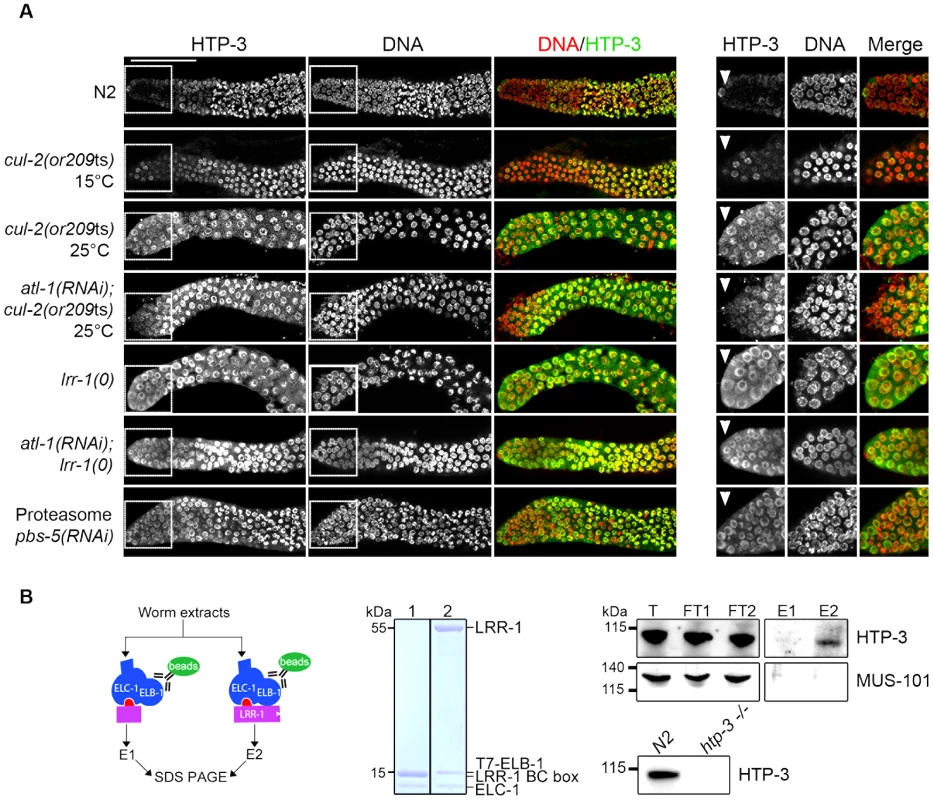
A- HTP-3 accumulates to high levels in cul-2(or209ts), lrr-1(0) and pbs-5(RNAi) germlines. Representative images of dissected gonads of the indicated genotypes stained with HTP-3 (green) antibodies and DAPI (red) (left panel). Insets show germ cells located in the most distal region of the germline (right panel) B- HTP-3 physically interacts with LRR-1. A flow chart of the experiment is presented in the left panel. Briefly, total worm extracts were incubated with the recombinant 6xHis-LRR-1/ELC-1/T7-ELB-1 trimeric complex immobilised on T7-agarose beads (green oval). As a control, we used a trimeric complex in which the substrate-binding interface LRR-1 (purple rectangle) was deleted. The middle panel shows a Coomassie-stained gel of the recombinant trimeric complexes containing either truncated or full-length LRR-1 protein (lane 1 and 2 respectively). Right panel: the trimeric complexes and associated proteins were eluted with loading buffer and analysed by western blot using MUS-101- and HTP-3-specific antibodies. T, total; FT, Flow-Through; E, elution. Lower right: total extracts of N2 and htp-3(0) worms were separated by SDS-PAGE and immunoblotted with HTP-3 antibodies. To ask whether the negative regulation of HTP-3 by CRL2LRR-1 E3 ligase involves physical interaction between the two proteins, we incubated total worm extracts with a recombinant LRR-1/ELC-1/ELB-1 trimeric complex or with a similar complex containing a truncated LRR-1 protein lacking the substrate binding interface. As shown in Figure 5B, endogenous HTP-3, but not MUS-101, was specifically retained on the trimeric complex containing full length LRR-1, but not on the complex containing the truncated form of LRR-1. Taken together, these results indicate that LRR-1 physically interacts with HTP-3 and regulates its levels in the mitotic zone of the germline, suggesting that HTP-3 is likely a direct target of the CRL2LRR-1 E3-ligase.
CRL2LRR-1 acts through HTP-3 to control HIM-3 loading on chromosomes
Our observations indicate that CRL2LRR-1 and the proteasome regulate HTP-3 stability in the mitotic zone of the germline, possibly to prevent entry into meiosis. To further investigate this possibility, we asked whether HTP-3 accumulation upon inactivation of cul-2, lrr-1 or the proteasome could force ectopic proliferative cells to enter meiosis.
GLP-1 promotes the proliferative fate, and the meiotic promoting factors GLD-1, NOS-3, GLD-2 and GLD-3 act downstream of Notch/GLP-1, in two parallel pathways (GLD-1/NOS-3 and GLD-2/GLD-3), to promote entry into meiosis. Simultaneous loss of both pathways causes a defect in meiotic entry that leads to germline overproliferation and prevention of gamete production [44]–[46]. Therefore, we asked whether loss of CUL-2 would cause ectopic proliferative germ cells to enter into meiosis in gld-3 nos-3 double mutants. As a control for this experiment, we depleted CYE-1 (Cyclin E in C. elegans) because it has been shown recently that CYE-1 depletion forces ectopic proliferative germ cells to load HIM-3 and SUN-1 Ser8-Pi (P-SUN-1) and to enter meiosis [47].
As shown in the Figure 6C, HTP-3 accumulates in tumorous gld-3 nos-3 germlines upon inactivation of cul-2, as revealed by western blot analysis. We then used immunofluorescence to test whether this accumulation is accompanied by a change in germ cell fate by monitoring the accumulation of the HIM-3 and P-SUN-1 meiotic markers. gld-3 nos-3 germlines inactivated with control dsRNA displayed ectopic proliferating germ cells with no sign of meiotic entry as revealed by the absence of HIM-3 and P-SUN-1 meiotic markers (Figure 6B and 6C). By contrast, RNAi-mediated depletion of cul-2, lrr-1 or pbs-5 forced many germ cells to express HIM-3 to high levels. However, these germ cells remain mitotic because they do not express the other meiotic marker P-SUN-1 to significant levels. Similar results were obtained using the cul-2; gld-3 nos-3 triple mutant (Figure 6B and 6C). As reported previously, CYE-1 depletion causes gld-3 nos-3 germ cells to enter into meiosis as revealed by the accumulation of HIM-3 and P-SUN-1 meiotic markers.
Fig. 6. CRL2LRR-1 E3-Ligase controls HIM-3 loading on chromosomes. 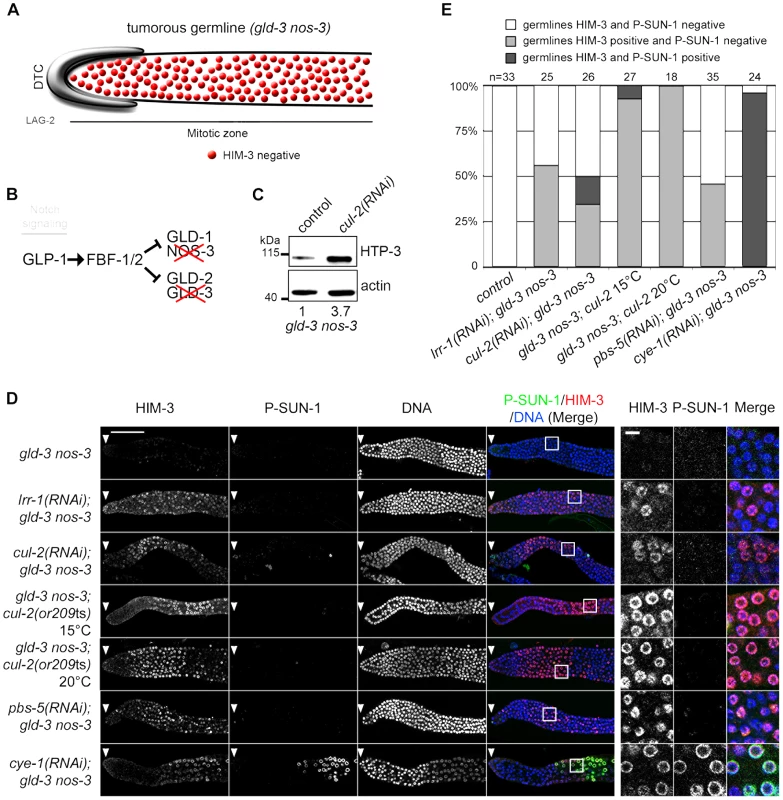
A- Schematic drawing showing that mitotic germ cells proliferate throughout the germline with no sign of meiotic entry in the gld-3 nos-3 double mutant germlines (upper panel). B- A simplified pathway with key regulators of the mitosis/meiosis decision is shown in the lower panel. C- HTP-3 accumulates in gld-3 nos-3 tumorous germlines upon cul-2(RNAi). Total gld-3 nos-3 worm extracts from control or cul-2(RNAi) were separated by SDS-PAGE and immunoblotted with HTP-3 (upper panel) and actin (lower panel) antibodies (loading control). The value at the bottom is the ratio between HTP-3 and actin signal intensities. The wild-type value was arbitrary defined as 1. D- Representative images of dissected gonads, of the indicated genotypes stained with HIM-3 (red) and SUN-1 Ser8-Pi (P-SUN-1, green) antibodies and with DAPI (blue). RNAi treatments were performed from the L1 (control, cul-2, lrr-1, cye-1) or L3 (pbs-5) stage at 20°C. Germlines in each panel were treated identically and fluorescent images taken at the same settings. Scale bars: 50 µM. The boxed regions, encompassing representative nuclei, are shown at higher magnification on the right panels. Scale bar: 5 µm. E- Graph showing the percentage of tumorous gld-3 nos-3 germlines HIM-3 and SUN-1 Ser8-Pi negative (white bars), HIM-3 positive and P-SUN-1 negative (light grey), and both, HIM-3 and P-SUN-1 positive (dark grey), as scored by counting the number of nuclei rows that contain at least 5 gcd with cells expressing HIM-3 and P-SUN-1 positive. Collectively, these results indicate that CUL-2 and LRR-1 inhibit only the first step of meiotic prophase by controlling HTP-3 stability and HIM-3 recruitment, whereas the CYE-1/CDK-2 kinase has a broader function in preventing meiotic entry.
Discussion
Through the isolation and characterization of a unique temperature-sensitive mutant allele of the cul-2 gene, we have shown that CUL-2 plays a critical role in germline development in C. elegans. In particular, CUL-2, in combination with LRR-1, i) promotes germ cell proliferation, ii) participates in the proliferation versus meiotic entry decision and iii) inhibits progression through the first step of meiotic prophase by regulating the stability of the axial element HTP-3.
cul-2(or209ts) mutant: A sensitized genetic background to analyze CRL2 functions during C. elegans development
Temperature-sensitive (ts) alleles have been instrumental for discovering the function of essential genes in C. elegans, in particular for genes like CUL-2 with roles in multiple processes. Indeed, ts alleles present numerous advantages: they often only partially reduce gene function even at the fully restrictive temperature, and they allow for modulation of activity through analysis at multiple temperatures.
CUL-2 is an essential gene in C. elegans that is highly expressed in the germline and in the early embryo where it has been implicated in numerous processes. Our phenotypic analysis revealed that the cul-2(or209ts) mutant recapitulates all the cul-2 loss of function phenotypes including feminization of the germline (Figure 1F), defects in cell fate determination with PIE-1 accumulation in somatic blastomeres (Figure S2), and defects in cell cycle progression in the early embryo [24] (data not shown). The penetrance of each phenotype varies with the duration of the shift at restrictive temperature, and thus this cul-2(or209ts) mutation provides a useful sensitized genetic background for identifying new in vivo functions of CRL2 complexes and, most importantly, for identifying its targets. Finally, other temperature-sensitive mutants affecting the ubiquitin-proteolytic system have been identified in C. elegans [48]–[50], but cul-2(or209ts) is to our knowledge the first conditional mutation reported for a metazoan cullin.
CRL2LRR-1 regulates germ cell proliferation by counteracting the DNA replication checkpoint
Using the cul-2(or209ts) allele, we confirmed our previous observations indicating that loss of the CRL2LRR-1 E3-ligase causes hyperactivation of the ATL-1/DNA replication checkpoint in germ cells, resulting in a cell cycle arrest and adult sterility [19]. Germ cell nuclei were enlarged and reduced in number in the cul-2(or209ts) mutant at 25°C, a phenotype that was partially suppressed by RNAi-mediated inactivation of atl-1 (Figure 2). Furthermore, reducing mus-101 and atl-1 function by RNAi increased the hatching rate of the cul-2(or209ts) mutant (Figure 5), further demonstrating that loss of CRL2LRR-1 function triggers activation of the ATL-1/DNA replication checkpoint pathway.
Why is the ATL-1 checkpoint pathway hyperactivated in cul-2(or209ts) and lrr-1(0) mutants? We believe that one function of the CRL2LRR-1 complex is to regulate DNA replication integrity, both in germ cells and in early embryos [19]. The ATL-1 pathway is thus activated primarily in response to DNA replication defects in the lrr-1(0) [19] and cul-2(or209ts) mutants (this study). In addition, our observations suggest that the inappropriate accumulation of structural components of meiotic chromosomes in lrr-1(0) mutants may also contribute to the activation of the ATL-1 checkpoint pathway (see below).
CUL-2 participates in the proliferation versus meiotic entry decision
Besides promoting germ cell proliferation, our results suggest that CUL-2 influences the balance between stem cell self-renewal and meiotic differentiation possibly by regulating the stability of meiotic promoting factors. In the C. elegans germline, GLP-1/Notch signaling controls the decision between self-renewal and entry into the meiotic cell cycle [28]. Downstream of Notch, the RNA-binding proteins FBF-1/2 prevent meiotic entry by repressing the translation of meiotic promoting factors, and, of structural components of meiotic chromosomes [32],[33],[51]. In addition to this regulatory network, which is largely translational in nature, there is increasing evidence that post-translational regulations play also a critical role in regulating the proliferation versus meiotic entry decision. For instance, the Cyclin E/Cdk2 kinase acts with the Notch pathway to promote the proliferative fate and to prevent meiotic entry [47], [52], and our results indicate that CUL-2 plays also a role in this process. Indeed, the cul-2ts mutant enhanced the premature meiotic entry phenotype of the glp-1(bn18) mutant and when combined with the cul-2ts mutant, both fbf-1 and fbf-2 mutants showed a smaller mitotic zone than single mutants (Figure 3). This function of CUL-2 in influencing the balance between germ cell self-renewal and meiotic entry is likely independent of the DNA replication checkpoint pathway given that atl-1 depletion does not affect the size of the mitotic zone (Figure 2).
These observations suggest the existence of a complex network of post-transcriptional and post-translational regulatory mechanisms to regulate the balance between germ cell self-renewal and meiotic entry.
What is the role of CUL-2 in this network? CUL-2 might act independently at multiple levels, for instance by regulating the activity of the CYE-1/CDK-2 kinase, and by regulating the stability both of meiotic promoting factors and of structural components of meiotic chromosomes in the mitotic zone of the germline. A gradient of high/low CYE-1/CDK-2 kinase is established along the distal to proximal end of the germline, and this gradient appears important for the self-renewal versus meiotic entry decision [47], [52]. Three complementary mechanisms establish this gradient: in the meiotic zone i) GLD-1 inhibits CYE-1 translation [53], ii) CYE-1 subunit is targeted for degradation by an E3-ligase nucleated around CUL-1 [47] and iii) the cyclin-dependent kinase inhibitor CKI-2 accumulates and inhibits CYE-1/CDK-2 activity [53]. In the mitotic zone, CKI-2 translation is inhibited by Notch and FBF-1/2. CKI-2 is not essential for germ cell proliferation but plays a role in the maintenance of the germline by influencing the proliferation versus meiotic entry decision [54]. CUL-2 has been implicated in the regulation of CKIs stability [18] and thus CUL-2 might target CKI-2 for degradation in the mitotic zone of the germline to maintain high CYE-1/CDK-2 activity in this region.
Alternatively, CUL-2 might act with CYE-1/CDK-2 to control the stability of meiotic promoting factors in the mitotic zone of the germline. Consistent with this hypothesis, substrate phosphorylation often is a pre-requisite for recognition by an E3 ligase [15], and it has been shown recently that CYE-1/CDK-2 phosphorylates GLD-1 and thereby regulates its stability in the mitotic zone of the germline [52], suggesting that CUL-2 might regulate GLD-1 degradation in germline stem cells. However, we failed to detect significant GLD-1 accumulation in germ cells located in the most distal part of the germline upon inactivation of cul-2 (data not shown), suggesting that CUL-2 might not be involved in GLD-1 degradation. Nevertheless, we cannot exclude the possibility that a small fraction of GLD-1 accumulates upon inactivation of cul-2 and thereby contributes to the cul-2 phenotype.
CRL2LRR-1 acts through HTP-3 to inhibit assembly of the synaptonemal complex
Although the mechanisms by which CUL-2 influences the proliferation versus meiotic entry decision remain to be identified, our results indicate that CRL2LRR-1 negatively regulates progression through meiotic prophase by controlling the stability of HTP-3 (Figure 7). HTP-3 accumulates in germ cells upon inactivation of cul-2, lrr-1 and the proteasome, and HTP-3 physically interacts with LRR-1 in vitro. Furthermore, reducing htp-3 function, using the htp-3(vc75) allele or with RNAi, suppressed lrr-1(tm3543) sterility and increased the hatching rate of the cul-2(or209ts) mutant, respectively. In addition, reducing syp-1 function also increased the hatching rate of the cul-2(or209ts) mutant. These results suggest that premature accumulation of HTP-3 on chromosomes allows for inappropriate HIM-3 and subsequently SYP-1 binding. This inappropriate assembly of SC components on chromosomes likely contributes to DNA replication checkpoint activation. However, although SC components appear to be expressed and loaded inappropriately on chromosomes upon inactivation of the CRL2LRR-1 E3-Ligase, germ cells do not appear to enter into meiosis because we failed to detect SUN-1 Ser8-Pi on chromosomes of ectopic proliferating germ cells upon inactivation of cul-2 or lrr-1. This observation indicates that HTP-3 accumulation is not sufficient to drive progression through meiotic prophase and that CUL-2 acts independently of HTP-3 to influence to proliferation versus meiotic entry decision, presumably by controlling the stability of meiotic regulator(s) in the mitotic zone of the germline (Figure 7).
Fig. 7. CRL2LRR-1 regulates entry into meiosis. 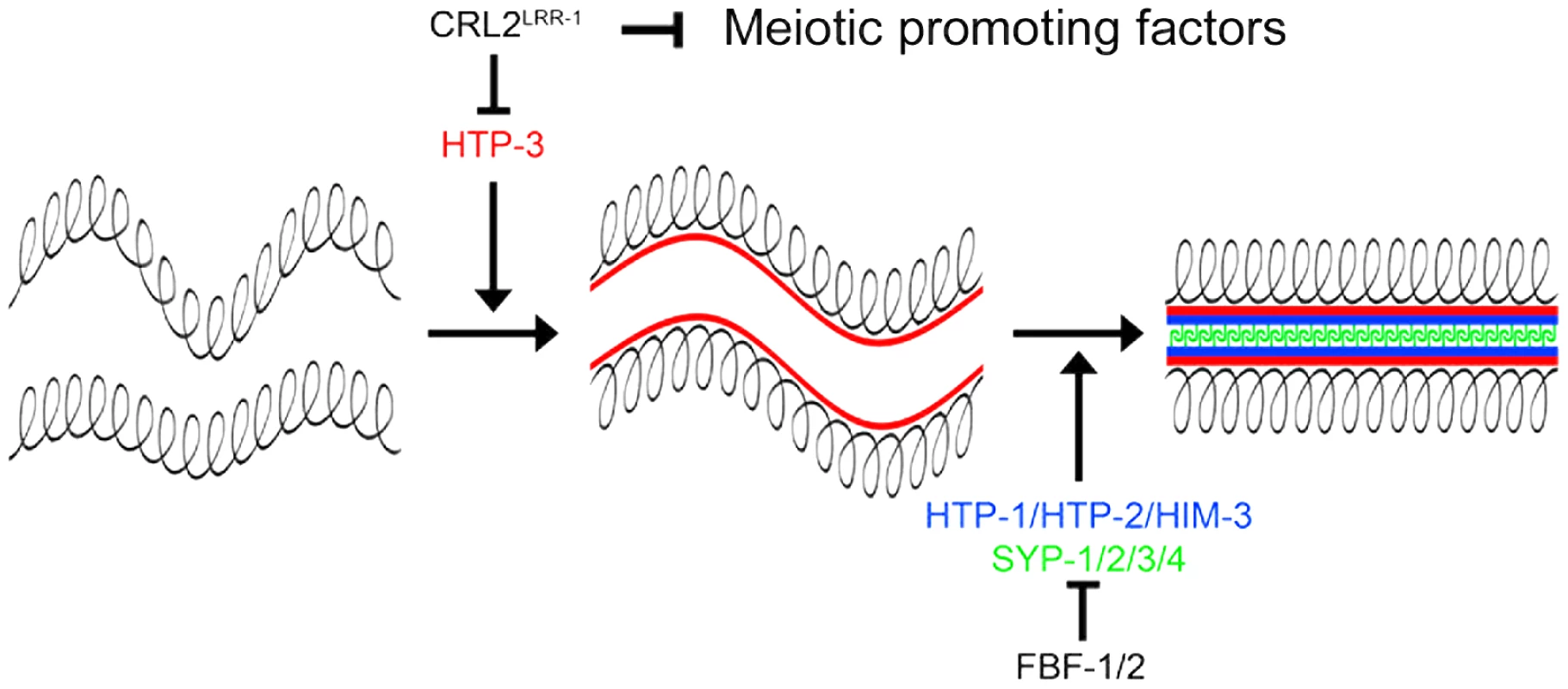
The axial element HTP-3 (red) promotes the assembly of the synaptonemal complex by triggering the recruitment of the other axial element components HIM-3, HTP-1, HTP-2 (bleu) and the transverse filaments (SYP-1/2/3/4) (green). CRL2LRR-1 inhibits the accumulation of HTP-3 whereas FBF-1/2 prevent the expression of structural components of meiotic chromosomes in the mitotic region of the germline. CRL2LRR-1 also acts independently of HTP-3 to prevent meiotic entry presumably by controlling the stability of unknown meiotic promoting factor(s). In conclusion, our results indicate that CUL-2 acts at multiple levels in the germline to coordinate germ cell proliferation and meiotic entry and thus emerged as a critical regulator of germline development in C. elegans.
Materials and Methods
Nematode strains, strain construction, and culture conditions
C. elegans strains were cultured and maintained using standard procedures [55]. Strains of the following genotypes were used: N2 Bristol (wild type); lrr-1(tm3543)II/mIn1[mIs14 dpy-10(e128)]II [19]; cul-2(or209ts)III [24]; htp-3(vc75)I [42]; htp-3(vc75)I; lrr-1(tm3543)II/mIn1[mIs14 dpy-10(e128)]II (this study); glp-1(bn18) [34]; atl-1(tm853)IV [56]; gld-3(q730) nos-3(q650)II/mIn1[mIs14 dpy-10(e128)]II [57]; gld-3(q730) nos-3(q650)II/mIn1[mIs14 dpy-10(e128)]II; cul-2(or209ts)III (this study), fbf-1(ok91)II [32], glp-1(bn18) cul-2(or209ts)III (this study); fbf-1(ok91)II; cul-2(or209ts) III (this study); fbf-2(q738)II [58], fbf-2(q738)II; cul-2(or209ts)III (this study).
Single nucleotide polymorphism (SNP) mapping and pyrosequencing
For this study, or209 worms were mated to the Hawaiian CB4856 C. elegans wild isolate, and recombinant F2 lines homozygous for the or209 mutation were isolated based on embryonic lethality at the restrictive temperature. Several SNPs located between −27.2 cM and 21.25 cM along the third chromosome were amplified by PCR from these recombinants, essentially as described [59], and genotyped using pyrosequencing technology. Briefly, PCR amplifications were performed from single worms using Taq DNA polymerase (New England Biolabs) and specific primers for each SNP. The purification of single-stranded PCR amplicons and the pyrosequencing reactions were subsequently performed according to manufacturer's instructions using a Pyromark Q96 ID instrument (Biotage). This analysis positioned the or209 mutation between +18.52 cM and the right end of chromosome III. This region contains cul-2, and we showed that all embryos from or209/cul-2(ek1) trans-heterozygotes (ek1 is a null allele of cul-2 [18]) failed to hatch.
RNA interference (RNAi)
Escherichia coli clones expressing dsRNA to deplete C. elegans genes were obtained from the MRC Geneservice (Cambridge, U) [60], [61]. RNAi feeding was performed as described using 2 mM IPTG (RNAi plates) [62].
Statistical analysis
The results are presented as means ± S.E.M. In all graphs, data were compared by a Mann-Whitney test (two-tailed p) or Student test (Figure 3 and Figure 5). All calculations were performed with InStat3 software (Graphpad). * p<0.05; ** p<0.01; *** p<0.001.
Hydroxyurea treatment
L4 animals were fed on NGM plates containing 10 mM final HU prior to analysis. Germlines were then dissected and stained with DAPI. Quantification of HU-induced cell cycle arrest was performed by counting the number of nuclei in a defined volume (20 000 µm3).
Microscopy, immunochemistry, and fluorescence microscopy
Germlines were dissected in PBS followed by freeze-crack, immersion in cold MeOH (−20°C) for 1 min, and fixation in 1× PBS, 0.08 M HEPES (pH 6.9), 1.6 mM MgSO4, 0.8 mM EGTA and 3.7% paraformaldehyde for 30 min in a humidity chamber at room temperature. Slides were washed 3×5 min, blocked for 1 h in PBT (1× PBS, 0.1% Triton X-100, and 5% BSA), and incubated overnight at 4°C with primary antibodies diluted in PBT. Working dilutions for the primary antibodies were 1∶1000 for rabbit anti-HIM-3 (M. Zetka), 1∶100 for rabbit anti-HTP-3 (M. Zetka) and 1∶1000 anti-SUN-1 Ser8-Pi (V. Jantsch). Slides were later incubated for 30 min at room temperature with secondary antibodies coupled to the Alexa 488 and 568 fluorophore (1∶600, Molecular Probes). Next, germlines were mounted in Vectashield Mounting Medium with DAPI (Vector). Fixed germlines were imaged using either a TCS SP5 confocal microscope (Leica) or a LSM 710 confocal microscope (Zeiss) with 40× objectives. Confocal images correspond to the projection of confocal Z-stacks spanning maximum 5 µm. Captured images were processed using ImageJ and Adobe Photoshop.
For whole-worm images, worms were immobilised with 20 mM levamisole and mounted on 2% agarose pads. Images were then acquired using an Axiovert 200 inverted microscope equipped with DIC optics.
Protein extracts and antibodies
Standard procedures were used for SDS–PAGE and western blotting. The following antibodies were used in this study: primary antibodies were directed against CUL-2 [19], ELC-1 [19], HTP-3 (M. Zetka), HIM-3 (M. Zetka), SUN-1 Ser8-Pi [35], Tubulin (Sigma), Actin (Sigma), TRA-1 [63] and MUS-101 [36]; secondary antibodies conjugated to peroxidase against rabbit or mouse were purchased from Sigma.
LRR-1/ELC-1/ELB-1 trimeric complexes were expressed in E. coli and purified as described [19].
Total worm extracts were prepared by cryolysis, as previously described [64], and loaded onto trimeric complexes immobilised on T7-agarose beads (Novagen) for 2 hours at 4°C. After five washes, proteins were eluted with sample buffer and separated by SDS-PAGE.
Supporting Information
Zdroje
1. HershkoA, CiechanoverA (1998) The ubiquitin system. Annu Rev Biochem 67 : 425–479.
2. ShabekN, CiechanoverA (2010) Degradation of ubiquitin: the fate of the cellular reaper. Cell Cycle 9 : 523–530.
3. HershkoA, HellerH, EliasS, CiechanoverA (1983) Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem 258 : 8206–8214.
4. WelckerM, ClurmanBE (2008) FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 8 : 83–93.
5. FrescasD, PaganoM (2008) Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer 8 : 438–449.
6. LeeJ, ZhouP (2012) Pathogenic Role of the CRL4 Ubiquitin Ligase in Human Disease. Front Oncol 2 : 21.
7. YeY, RapeM (2009) Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10 : 755–764.
8. ChauV, TobiasJW, BachmairA, MarriottD, EckerDJ, et al. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243 : 1576–1583.
9. ThrowerJS, HoffmanL, RechsteinerM, PickartCM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19 : 94–102.
10. GlotzerM, MurrayAW, KirschnerMW (1991) Cyclin is degraded by the ubiquitin pathway. Nature 349 : 132–138.
11. JinL, WilliamsonA, BanerjeeS, PhilippI, RapeM (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133 : 653–665.
12. MocciaroA, RapeM (2012) Emerging regulatory mechanisms in ubiquitin-dependent cell cycle control. J Cell Sci 125 : 255–263.
13. DeshaiesRJ, JoazeiroCA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78 : 399–434.
14. SarikasA, HartmannT, PanZQ (2011) The cullin protein family. Genome Biol 12 : 220.
15. PetroskiMD, DeshaiesRJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6 : 9–20.
16. MerletJ, BurgerJ, GomesJE, PintardL (2009) Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci
17. OkumuraF, MatsuzakiM, NakatsukasaK, KamuraT (2012) The Role of Elongin BC-Containing Ubiquitin Ligases. Front Oncol 2 : 10.
18. FengH, ZhongW, PunkosdyG, GuS, ZhouL, et al. (1999) CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat Cell Biol 1 : 486–492.
19. MerletJ, BurgerJ, TavernierN, RichaudeauB, GomesJE, et al. (2010) The CRL2LRR-1 ubiquitin ligase regulates cell cycle progression during C. elegans development. Development 137 : 3857–3866.
20. StarostinaNG, SimplicianoJM, McGuirkMA, KipreosET (2010) CRL2(LRR-1) targets a CDK inhibitor for cell cycle control in C. elegans and actin-based motility regulation in human cells. Dev Cell 19 : 753–764.
21. SonnevilleR, GonczyP (2004) Zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development 131 : 3527–3543.
22. LiuJ, VasudevanS, KipreosET (2004) CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior-posterior axis in C. elegans. Development 131 : 3513–3525.
23. DeRenzoC, ReeseKJ, SeydouxG (2003) Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424 : 685–689.
24. EncaladaSE, MartinPR, PhillipsJB, LyczakR, HamillDR, et al. (2000) DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev Biol 228 : 225–238.
25. StarostinaNG, LimJM, SchvarzsteinM, WellsL, SpenceAM, et al. (2007) A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell 13 : 127–139.
26. ByrdDT, KimbleJ (2009) Scratching the niche that controls Caenorhabditis elegans germline stem cells. Semin Cell Dev Biol 20 : 1107–1113.
27. HansenD, SchedlT (2006) The regulatory network controlling the proliferation-meiotic entry decision in the Caenorhabditis elegans germ line. Curr Top Dev Biol 76 : 185–215.
28. KimbleJ, CrittendenSL (2007) Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol 23 : 405–433.
29. ZetkaMC, KawasakiI, StromeS, MullerF (1999) Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev 13 : 2258–2270.
30. MacQueenAJ, VilleneuveAM (2001) Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev 15 : 1674–1687.
31. VasudevanS, StarostinaNG, KipreosET (2007) The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep 8 : 279–286.
32. CrittendenSL, BernsteinDS, BachorikJL, ThompsonBE, GallegosM, et al. (2002) A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417 : 660–663.
33. KershnerAM, KimbleJ (2010) Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci U S A 107 : 3936–3941.
34. QiaoL, LissemoreJL, ShuP, SmardonA, GelberMB, et al. (1995) Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics 141 : 551–569.
35. PenknerAM, FridkinA, GloggnitzerJ, BaudrimontA, MachacekT, et al. (2009) Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139 : 920–933.
36. HolwayAH, HungC, MichaelWM (2005) Systematic, RNA-interference-mediated identification of mus-101 modifier genes in Caenorhabditis elegans. Genetics 169 : 1451–1460.
37. MacQueenAJ, PhillipsCM, BhallaN, WeiserP, VilleneuveAM, et al. (2005) Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123 : 1037–1050.
38. GoodyerW, KaitnaS, CouteauF, WardJD, BoultonSJ, et al. (2008) HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell 14 : 263–274.
39. MacQueenAJ, ColaiacovoMP, McDonaldK, VilleneuveAM (2002) Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev 16 : 2428–2442.
40. AravindL, KooninEV (1998) The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci 23 : 284–286.
41. SeversonAF, LingL, van ZuylenV, MeyerBJ (2009) The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev 23 : 1763–1778.
42. CouteauF, ZetkaM (2011) DNA damage during meiosis induces chromatin remodeling and synaptonemal complex disassembly. Dev Cell 20 : 353–363.
43. HayashiM, ChinGM, VilleneuveAM (2007) C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet 3: e191 doi:10.1371/journal.pgen.0030191.
44. AustinJ, KimbleJ (1987) glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51 : 589–599.
45. KadykLC, KimbleJ (1998) Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125 : 1803–1813.
46. HansenD, HubbardEJ, SchedlT (2004) Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev Biol 268 : 342–357.
47. FoxPM, VoughtVE, HanazawaM, LeeMH, MaineEM, et al. (2011) Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 138 : 2223–2234.
48. KurzT, PintardL, WillisJH, HamillDR, GonczyP, et al. (2002) Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 295 : 1294–1298.
49. KulkarniM, SmithHE (2008) E1 ubiquitin-activating enzyme UBA-1 plays multiple roles throughout C. elegans development. PLoS Genet 4: e1000131 doi:10.1371/journal.pgen.1000131.
50. MacdonaldLD, KnoxA, HansenD (2008) Proteasomal regulation of the proliferation vs. meiotic entry decision in the Caenorhabditis elegans germ line. Genetics 180 : 905–920.
51. MerrittC, SeydouxG (2010) The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development 137 : 1787–1798.
52. JeongJ, VerheydenJM, KimbleJ (2011) Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS Genet 7: e1001348 doi:10.1371/journal.pgen.1001348.
53. BiedermannB, WrightJ, SenftenM, KalchhauserI, SarathyG, et al. (2009) Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev Cell 17 : 355–364.
54. KalchhauserI, FarleyBM, PauliS, RyderSP, CioskR (2011) FBF represses the Cip/Kip cell-cycle inhibitor CKI-2 to promote self-renewal of germline stem cells in C. elegans. EMBO J 30 : 3823–3829.
55. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
56. Garcia-MuseT, BoultonSJ (2005) Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J 24 : 4345–4355.
57. EckmannCR, CrittendenSL, SuhN, KimbleJ (2004) GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168 : 147–160.
58. LamontLB, CrittendenSL, BernsteinD, WickensM, KimbleJ (2004) FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell 7 : 697–707.
59. FayD, BenderA (2008) SNPs: introduction and two-point mapping. Worm Book 1–10.
60. FraserAG, KamathRS, ZipperlenP, Martinez-CamposM, SohrmannM, et al. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408 : 325–330.
61. KamathRS, FraserAG, DongY, PoulinG, DurbinR, et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 : 231–237.
62. KamathRS, Martinez-CamposM, ZipperlenP, FraserAG, AhringerJ (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2: RESEARCH0002 doi:10.1186/gb-2000-2-1-research0002.
63. SasagawaY, OtaniM, HigashitaniN, HigashitaniA, SatoK, et al. (2009) Caenorhabditis elegans p97 controls germline-specific sex determination by controlling the TRA-1 level in a CUL-2-dependent manner. J Cell Sci 122 : 3663–3672.
64. Luke-GlaserS, RoyM, LarsenB, Le BihanT, MetalnikovP, et al. (2007) CIF-1, a shared subunit of the COP9/signalosome and eukaryotic initiation factor 3 complexes, regulates MEL-26 levels in the Caenorhabditis elegans embryo. Mol Cell Biol 27 : 4526–4540.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



