-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
Despite extensive genetic analysis, the evolutionary relationship between polar bears (Ursus maritimus) and brown bears (U. arctos) remains unclear. The two most recent comprehensive reports indicate a recent divergence with little subsequent admixture or a much more ancient divergence followed by extensive admixture. At the center of this controversy are the Alaskan ABC Islands brown bears that show evidence of shared ancestry with polar bears. We present an analysis of genome-wide sequence data for seven polar bears, one ABC Islands brown bear, one mainland Alaskan brown bear, and a black bear (U. americanus), plus recently published datasets from other bears. Surprisingly, we find clear evidence for gene flow from polar bears into ABC Islands brown bears but no evidence of gene flow from brown bears into polar bears. Importantly, while polar bears contributed <1% of the autosomal genome of the ABC Islands brown bear, they contributed 6.5% of the X chromosome. The magnitude of sex-biased polar bear ancestry and the clear direction of gene flow suggest a model wherein the enigmatic ABC Island brown bears are the descendants of a polar bear population that was gradually converted into brown bears via male-dominated brown bear admixture. We present a model that reconciles heretofore conflicting genetic observations. We posit that the enigmatic ABC Islands brown bears derive from a population of polar bears likely stranded by the receding ice at the end of the last glacial period. Since then, male brown bear migration onto the island has gradually converted these bears into an admixed population whose phenotype and genotype are principally brown bear, except at mtDNA and X-linked loci. This process of genome erosion and conversion may be a common outcome when climate change or other forces cause a population to become isolated and then overrun by species with which it can hybridize.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003345
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003345Summary
Despite extensive genetic analysis, the evolutionary relationship between polar bears (Ursus maritimus) and brown bears (U. arctos) remains unclear. The two most recent comprehensive reports indicate a recent divergence with little subsequent admixture or a much more ancient divergence followed by extensive admixture. At the center of this controversy are the Alaskan ABC Islands brown bears that show evidence of shared ancestry with polar bears. We present an analysis of genome-wide sequence data for seven polar bears, one ABC Islands brown bear, one mainland Alaskan brown bear, and a black bear (U. americanus), plus recently published datasets from other bears. Surprisingly, we find clear evidence for gene flow from polar bears into ABC Islands brown bears but no evidence of gene flow from brown bears into polar bears. Importantly, while polar bears contributed <1% of the autosomal genome of the ABC Islands brown bear, they contributed 6.5% of the X chromosome. The magnitude of sex-biased polar bear ancestry and the clear direction of gene flow suggest a model wherein the enigmatic ABC Island brown bears are the descendants of a polar bear population that was gradually converted into brown bears via male-dominated brown bear admixture. We present a model that reconciles heretofore conflicting genetic observations. We posit that the enigmatic ABC Islands brown bears derive from a population of polar bears likely stranded by the receding ice at the end of the last glacial period. Since then, male brown bear migration onto the island has gradually converted these bears into an admixed population whose phenotype and genotype are principally brown bear, except at mtDNA and X-linked loci. This process of genome erosion and conversion may be a common outcome when climate change or other forces cause a population to become isolated and then overrun by species with which it can hybridize.
Introduction
Despite polar bears' clear morphological and behavioral adaptations to their arctic environment [1], [2], their genetic relationship to brown bears remains unclear [3], [4], [5], [6]. Analysis of maternally inherited mitochondrial DNA (mtDNA) shows that polar bears fall within the range of variation of brown bears. Extant brown bears from Alaska's ABC (Admiralty, Baranof and Chichagof) Islands, some extinct brown bears from Ireland and mainland Alaska, and two ∼115,000-year-old polar bears share the mtDNA haplotype of all extant polar bears [3], [7], [8], [9], [10], [11]. The time to most recent common ancestor (TMRCA) of this mtDNA haplotype has been estimated at ∼160 thousand years ago (kya) (Figure S7) [3], [6], [9], [10]. Recent analysis of data from a panel of brown and polar bears at 14 nuclear loci showed that polar bears are generally distinct from brown bears, with genomic TMRCA averaging ∼600 kya [4]. Under a simple population split model without subsequent admixture, the population divergence should be more recent than average genomic divergence and thus polar bears became a distinct species more recently than 600 kya. A separate recent genome sequencing survey concluded that brown bear and polar bear lineages are much older. Miller and colleagues concluded that the lineage that would become polar bears diverged from that which would become brown bears more than 4 million years ago, followed by admixture that continues to the present [6]. Consistent with this, the past and present geographic ranges of both species overlap at their margins (Figure 1), and fertile hybrids are known in both captive and wild populations [2], [12].
Fig. 1. Map showing the approximate current geographic ranges of brown bears (brown) and polar bears (blue). 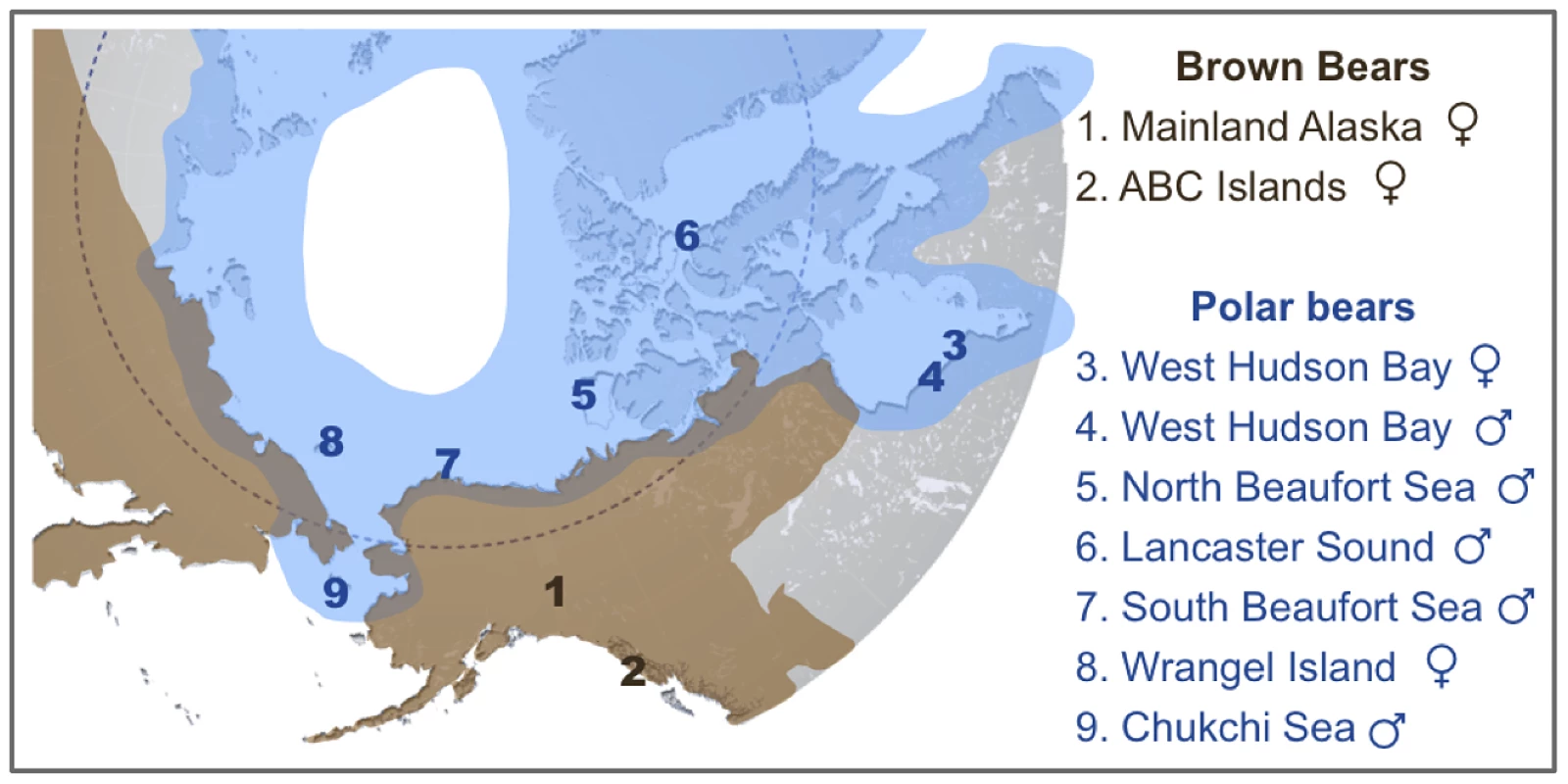
Numbers indicate the geographic location of origin of two brown bears and seven polar bears analyzed here. An American black bear from central Pennsylvania was also sequenced as part of this study. Shotgun data amounting to 4–6× coverage for polar bears and 11–12× coverage for brown and black bears (Table S2) was aligned to the current distribution of the polar bear genome [13]. The current consensus is that mtDNA and perhaps other polar bear loci are the result of past introgressions from brown bears into polar bears [3], [4], [6]. One scenario that has been proposed to reconcile the complicated discordance between the mtDNA trees and the species trees requires at least two instances of hybridization [3]. The first, which must have occurred before ∼115kya, passed the mtDNA haplotype from polar bears into brown bears, including the ancient Irish brown bears and ancestors of the ABC Island brown bears. The second passed this mtDNA haplotype back into polar bears, after which it came to fixation in all extant polar bears. This convoluted scenario is necessary if, in fact, polar bears derive their mtDNA haplotype and other loci from brown bears. Unfortunately, this prevailing consensus has gone unquestioned. Here, we present an analysis of published and newly generated genome-wide data for brown bears and polar bears. We find extensive evidence of previous admixture, from polar bears into brown bears, especially of X-linked genes.
Results
To more fully delineate the genetic relationship between polar bears and brown bears, we sequenced random genomic shotgun libraries from seven polar bears, two brown bears and one black bear to learn the ancestral state for alleles (Figure 1, Text S1). We mapped these reads to the assembled genome scaffolds of polar bear (Text S1) [13]. Because the sequence coverage of each bear was uneven and too low to reliably call heterozygous sites, we down-sampled the sequence data from each bear to 1×. That is, we randomly picked a high-quality base from amongst all reads that mapped reliably at each position in the bear genome. In this way, we generated a composite haplotype for each bear and used these data for further analysis.
To gauge the level of diversity within and divergence between bear species, we made pairwise comparison between each bear, in 50 kb windows, across the bear genome (Figure 2). In agreement with previous reports [4], [14], we find that polar bears are remarkably homogeneous: polar bear alleles differ at ∼4 sites in 10,000. In contrast, brown bears have roughly four times as much genetic diversity, differing at ∼17 in 10,000 sites. We note that the level of diversity among brown bears is nearly as high as the divergence between brown and polar bears. As expected, polar bears and brown bears show similar pairwise genomic divergence from the black bear. Likewise, the polar bears, brown bears, and black bears all show similar genomic divergence from the giant panda (Ailuropoda melanoleuca) [15].
Fig. 2. Genetic diversity within and between bear species. 
(A) Pairwise differences between individuals estimated as the average number of differences per 10 thousand bases (kb) in 42,000 non-overlapping 50 kb regions. After strict quality filtering, within-sample heterozygosity was resolved by selecting a single, high-quality base at random. The Lancaster Sound polar bear showed an excess of postmortem damage, as expected for historic specimens [32], and is shown in Figure S1. Polar bears are remarkably homogenous compared to brown bears, and both polar bears and brown bears are approximately equally diverged from the American black bear. Consistent with the results of the D-statistic test, pairwise distance between the ABC Islands brown bear and all polar bears (yellow lines) is less than that between the mainland brown bear and all polar bears (red lines). (B) Schematic diagram of a representative gene tree within brown bear, polar bear, and black bear populations, with the present day at the left of the diagram. For this locus, admixture occurring more recently than the population divergence of polar bears leads to the introgression of a polar bear haplotype into brown bears. Estimate of average genomic distance for brown, black, and polar bears and for population divergence between brown bears and polar bears given different calibration points are provided in Table 1. We quantified admixture between brown and polar bears using the D-statistic [16]. In brief, D is the excess fraction of derived alleles shared between one of two conspecific individuals with a candidate admixing individual (Figure 3). Note that both incomplete lineage sorting (ILS) and admixture can lead to sharing of derived alleles, in this case between polar bears and brown bears. ILS, being a stochastic process, will result in equivalent numbers of shared, derived alleles between any two brown bears and a polar bear. Admixture, on the other hand, will result in more shared, derived alleles in the more admixed bear. Thus, under the null model of no admixture, D = 0. A significant non-zero value of D indicates more admixture with one of the two individuals.
Fig. 3. Summary of D-statistic comparisons between polar bears and brown bears. 
In each comparison, the black bear was used to define the ancestral allele. The Z-score of the D-statistic for each comparison is shown for autosomes (red) and X-chromosome (blue). Each dot represents the data from comparison of one pair of bears. In the top panel, all pairs of polar bears are compared for excess derived allele matching against the mainland brown bear. In the middle panel, all pairs of polar bears are compared against the ABC Island brown bear. The bottom panel shows the comparison of the two brown bears for excess allele matching to polar bears with each dot representing a different polar bear. Comparison of any two polar bears for admixture with brown bears found little evidence for admixture. All D-statistics comparing two polar bears to a brown bear were statistically indistinguishable from 0 (Figure 3, top and middle panels).
Conversely, D-statistic comparisons between the ABC Islands and mainland brown bears for polar bear admixture were consistently and equivalently non-zero (Figure 3, bottom), regardless of the polar bear used in the comparison (D = 0.016, which translates to roughly 0.75% of the genome; Z-score = 1.24). Remarkably, when the analysis is restricted to the 12 scaffolds (∼74 Mb of sequence) identified as X-chromosome (Text S1), D = 0.22, or ∼6.5% of the X-chromosome (Z-score = 4.52) (Figure S2; Tables S3, S4, S5). We find this same enrichment of the X chromosome, compared to the autosome, for admixture with polar bears when analyzing genome sequence data from two additional, recently published ABC Islands brown bears (Figure S3, Table S6) [6]. The ABC Islands bears therefore share not only their mtDNA but also a significant portion of their X-chromosomes with polar bears. A parsimonious explanation for these observations is that the same admixture event that resulted in sharing of the polar bear mtDNA haplotype with ABC Island brown bears also results in sharing of much of the X-chromosome.
To test the direction of X-chromosome gene flow between polar bears and the ABC Islands bear we simulated the effect of having 6.5% ancestry (roughly the amount estimated above) in either polar bear or mainland brown bear X chromosome from the reciprocal species (Figure 4, Figure S5). The simulation was carried out by randomly selecting 6.5% of the X-chromosome of the candidate recipient species to be replaced by sequence from the candidate donor species (Figure S4, Text S1). We then measured the distribution of pairwise divergences that would result following this simulated admixture.
Fig. 4. Simulated admixture reveals the direction of gene flow on the X chromosome. 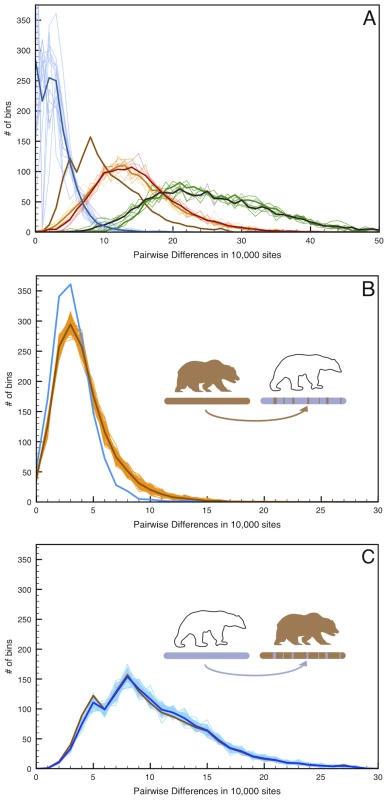
(A) Pairwise distance as in Figure 2 but limited to the 12 scaffolds identified as X-chromosome. (B) 100 replicate simulations in which 6.5% of the female West Hudson Bay polar bear X-chromosome is replaced with that of the mainland Alaska brown bear in randomly inserted 20 kb fragments, simulating admixture from the brown bear genome into polar bear ∼50kya. Pairwise differences are calculated between the simulated genome (light brown lines; mean highlighted in dark brown) and the plot comparing the two female polar bears (blue line), to maximize the number of informative sites in the test. The addition of brown bear DNA to the polar bear genome markedly increases the number of high-diversity bins (>10 differences/10 kb), indicating that any introgression of brown bear DNA into polar bears should be easily detectable. (C). As in (B), but with 6.5% of the mainland Alaska brown bear X-chromosome is replaced with that of the female West Hudson Bay polar bear. In this instance, we find no difference between the simulated (blue lines) and real (brown line) data. Given the low genetic diversity within polar bears, this amount of brown bear ancestry would be clearly identifiable as an excess of deeply diverging regions between polar bears, even in unphased data from which a random allele is chosen at each site. Conversely, simulating 6.5% polar bear ancestry in the mainland brown bear X-chromosome is more consistent with the observed level of genomic regional divergence between brown bear X-chromosomes. Thus, we deduce that the direction of gene flow was from polar bear into the ABC Islands brown bear X-chromosome.
Recently published genome sequence data from a ∼115ky polar bear [6] allow us to further probe when and in which direction admixture might have happened. Using this ancient polar bear in the D-statistic test gives nearly identical results to the extant polar bears (autosome D = 0.015; X-chromosome D = 0.212). That is, ABC island brown bears are equally enriched for polar bear matching derived alleles, even when this ∼115ky polar bear is used in the comparison. Therefore, if the admixture was from the ABC Island brown bears (or a closely related population) into polar bears, it must have occurred prior to ∼115ky. Furthermore, no significant subsequent admixture could have occurred, since the modern polar bears are nearly homogeneous for the ABC Islands brown bear D-statistic signal. Finally, if gene flow was from brown bears into polar bears, it would had to have been from a population of brown bears that lived more than ∼115kya that today finds itself restricted to a group of islands that only became habitable for brown bears since the end of the last glacial maximum, about ∼16kya. Given the unlikeliness of this scenario and the incompatibility of polar bear X-chromosomes genetic divergence with brown bear ancestry, we conclude that the direction of gene flow was from polar bears into the ABC Island brown bears.
Discussion
The genome-wide analysis presented here indicates that (1) polar bears are a remarkably homogeneous species and show no evidence of brown bear ancestry, (2) the ABC Islands brown bears show clear evidence of polar bear ancestry, and (3) this polar bear ancestry of ABC Islands brown bears is conspicuously enriched in the X-chromosome. ABC Islands brown bears show a simple positive correlation between how maternally biased a genetic locus is (mtDNA>X chromosome>autosomes) and how much polar bear ancestry is present (100%, 6.5%, 1%). Given this observation, and our knowledge about the natural history of these islands through the Pleistocene and Holocene, we present the following model.
During the peak of the last ice age, brown bears were likely absent from the region that now comprises the ABC Islands. Although fossil remains dating to this period are abundant on the more southerly islands of the Alexander Archipelago, brown bears are not among the species present during the period spanning 26-12kya, when glacial conditions were at their peak [17], [18], [19], [20]. Geological and climatological data suggest that if any habitat suitable for brown bears persisted on the ABC Islands during the LGM it would have been limited to the western part of Baranof Island, the most distant of the ABC Islands from the Alaskan mainland [17]. By itself, however, this potential refugium would have been too small to support viable populations of brown bears [21].
Polar bears, alternately, would likely have colonized the sea ice adjacent to the ABC Islands as the ice advanced southward. Notably, marine mammals dominate the fossil remains dating to this interval [19], including ringed seals, an ideal food source for polar bears [2]. As the climate warmed and ice retreated, polar bears may have been stranded on or near the ABC Islands. As the habitat became increasingly hospitable to brown bears [17], the early colonizers from the mainland would have been predominantly the more peripatetic sub-adult males [14]. Admixture involving an influx of mostly or exclusively male brown bears with the stranded polar bears would have resulted in a gradual erosion of the polar bear genome within the isolated population. The sex bias of admixing brown bears would have made genomic erosion more rapid in the autosomes, confining the vestiges of polar bear ancestry in extant ABC Islands bears primarily to matrilineal-biased genetic loci (Figure S12).
Our simplified model - little or no brown bear ancestry in polar bears and matrilineal-biased polar bear ancestry in the ABC Islands brown bears - is consistent with several important comparative genomic observations. First, mtDNA and nuclear genome diversity within both extant and a ∼115kya polar bear is extremely low. This low level of polar bear diversity is consistent with no admixture from brown bears. Brown bears, in contrast, show much higher levels of diversity including many deep genetic lineages that have not completely sorted since their population divergence from polar bears. The ABC Islands brown bears show genome-wide evidence of admixture with polar bears concentrated on the X chromosome. Importantly, the level of admixture inferred from D-statistic analyses is only compatible with polar bear admixture into the ABC Islands brown bear X chromosomes and not the other way around. Conveniently, this model explains the presence of the polar bear mtDNA haplotype in all ABC Islands brown bears: the mtDNA haplotype of the male brown bear immigrants is lost, regardless of how many male brown bear immigrants arrived.
The model for historic admixture proposed here is distinct from the traditional framework for admixture, including the scenario involving early humans and Neandertals for which the D-statistic analysis was originally developed [16], [22]. Usually, the goal is to find the signal of a potentially small amount of admixture from a single or few admixture episodes that took place many generations ago (Figure S8). While such a model is consistent with the ABC Islands brown bear autosomal D-statistic results, it is insufficient to explain the large difference in the X-chromosome or the fixation of the polar bear mtDNA haplotype in the ABC Islands brown bears (Text S1). In fact, reasonable parameter values for a model that assumes a single episode of admixture from polar bears into brown bears do not result in a ratio of D for the X and autosomes that exceeds 2.7; our observed ratio is ∼14. Alternately, a long process of sex-biased immigration of brown bears into what was initially a polar bear population can result in much higher ratios of polar bear ancestry for the X and autosomes (Table S8; Figures S9, S10, S11), consistent with the empirical observations presented here.
Spatially explicit modeling has been used to probe the dynamics of gene flow from introgression during species expansions [23]. These simulations have yielded insight into the often non-intuitive patterns seen in various loci such as the apparent asymmetry in gene flow from the native species into the invading species. An extension of this approach to incorporate a migration barrier to female, but not male, gene flow and a dwindling native population of polar bears, may more fully reveal the demographic details of the brown bear invasion. Of particular note, there is evidence that brown bear migration between the mainland and ABC Islands may be ongoing. Analysis of variation at 17 rapidly evolving microsatellite loci indicated that brown bears from Admiralty Island, the closest of the ABC Islands to the mainland, are more similar to mainland Alaskan brown bears than were bears from Baranof and Chichagof Islands [14]. Assuming no disruption of the salient features of this migration, its final state, which has not yet been realized, would be complete conversion of the population, i.e., the fixation of brown bear alleles in all genomic loci in the ABC Island bears except the strictly maternal mtDNA.
We note that our data cannot resolve the timing of the origin of polar bears as a distinct lineage. Such an estimate has been hindered mainly by the paucity of preserved ancient polar bear remains [5], [24], and consequent lack of fossil calibrations. However, our data do provide insight into the relative timing of divergence between the three bear lineages sampled here. To generate a hypothetical scenario for the timing of the origin of polar bears, we apply several previously suggested calibration strategies to our data (Table 1; Figure 2B). Regardless of the calibration strategy applied, our data support a long interval between the initial divergence between black bears and the brown bear/polar bear lineage, and the later divergence between brown bears and polar bears. This is similar to that observed by Hailer et al [4], and in contrast to the scenario predicted by the model of Miller et al [6].
Tab. 1. Estimates of genomic TMRCA. 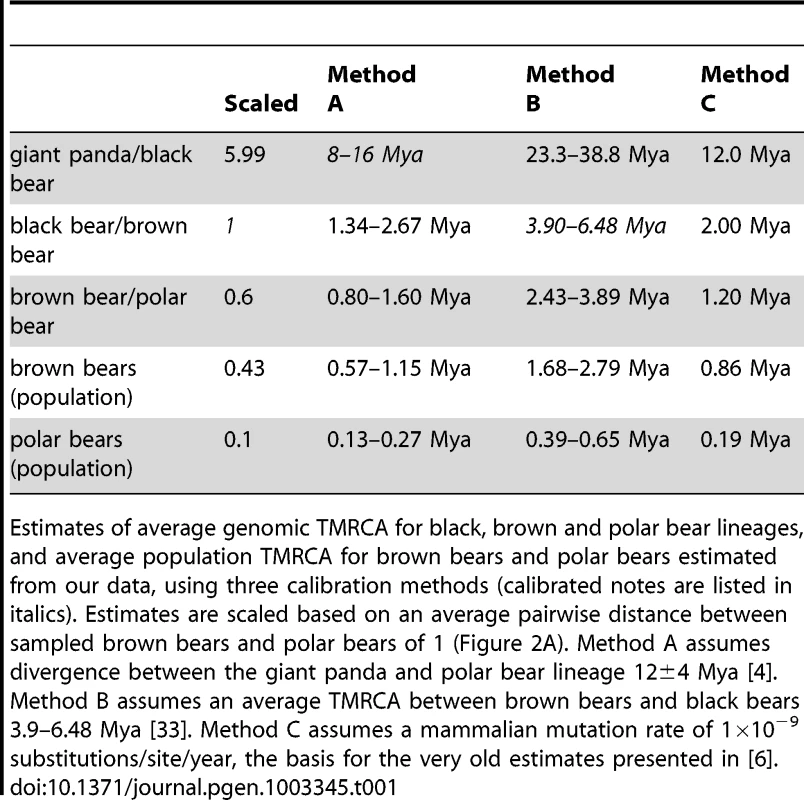
Estimates of average genomic TMRCA for black, brown and polar bear lineages, and average population TMRCA for brown bears and polar bears estimated from our data, using three calibration methods (calibrated notes are listed in italics). Estimates are scaled based on an average pairwise distance between sampled brown bears and polar bears of 1 (Figure 2A). Method A assumes divergence between the giant panda and polar bear lineage 12±4 Mya [4]. Method B assumes an average TMRCA between brown bears and black bears 3.9–6.48 Mya [33]. Method C assumes a mammalian mutation rate of 1×10−9 substitutions/site/year, the basis for the very old estimates presented in [6]. From analysis of the data presented here, we infer that polar bears most likely became a distinct lineage sometime during the Pleistocene. This timing is consistent with previous molecular (Table 1) and morphological [5] estimates. Polar bears and brown bears were clearly established as a morphologically distinct species by at least ∼115kya – the age of the oldest known polar bear fossil [10], [24]. Regardless of this timing, our data suggest that polar bears have remained a small, distinct lineage since their origin (Figure S6), with lineage-specific adaptations reinforced by the ecological constraints of their extreme environment (Text S1, Table S7) [6]. Brown bears, in contrast, have had a larger effective population size (Figure S6), with segregating polymorphism that often predates their split with polar bears (Figure 2B).
The process of genomic erosion we propose here may not be unique to the stranded ABC Islands polar bears. Past changes in the distribution of polar ice, for example, may have also stranded polar bears or hybrids on present-day Ireland, explaining the appearance of polar bear mtDNA in the remains of extinct Irish brown bears [3]. Long-term climate change may often strand populations on islands or island-like habitats, such as lakes or mountain plateaus. If these stranded populations then hybridize with closely related immigrants, we predict substantial variability in the apparent level of admixture indicated by D-statistics. Furthermore, in the case of sex-biased immigration, the ratio of D-statistics for the X and autosomes will be highly dependent on the rate and duration of immigration.
Materials and Methods
We extracted DNA from nine of the ten bears in a modern DNA laboratory using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer's specifications. The historic Lancaster Sound polar bear (Smithsonian Natural History Museum ID 512133; Table S1) was extracted in a dedicated ancient DNA laboratory at Penn State University that is geographically isolated from modern molecular biology research, using a column-based extraction protocol for ancient DNA [25].
We physically sheared the DNA of the modern bears using a Diagenode Bioruptor UCD-200 instrument. Fifty µl of each of the six modern polar bear extracts were transferred into 1.5 ml tubes and exposed to four rounds of sonication for 7 min, using the energy setting “HIGH” and an “ON/OFF interval” of 30 seconds. To attain a longer insert size, we slightly modified the procedure to include two 7-min rounds and one 5-min round of sonication for the brown bears, black bear, and second round of sequencing for two polar bears (West Hudson Bay X3249106A; and Chukchi Sea UP08.010; Table S1). We then purified and concentrated the extracts using the Agencourt AMPure XP PCR purification kit, according to manufacturer's instructions, and eluted in 20 µl of 1×TE, with 0.05% Tween20. The historic bear sample was already fragmented due to degradation, and was not sonicated.
We prepared indexed Illumina libraries using 15 µl of each extract following the protocol described in [26], with reaction volumes scaled to total volume of 40 µl. To verify final DNA concentration and the distribution of insert sizes, we ran each library on an Agilent 2100 Bioanalyzer. We then sequenced each polar bear on a separate lane of an Illumina HiSeq 2000 instrument using 100 base-pair (bp) paired-end chemistry at the UC Santa Cruz Core Genomics Facility. We sequenced one lane each of the two brown bears, the black bear, and an additional lane for two polar bears (Table S2) using an Illumina HiSeq 2000 instrument with 150-bp paired-end chemistry at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley.
From the Illumina sequence data, we removed the index and adapter sequence and merged paired reads using a script provided by M. Kircher [27]. We then trimmed each read to remove low quality bases by trimming inward from the 3′-end of the read until detecting a base with quality score ≥13 (∼95% confidence). We mapped the resulting data to the draft polar bear genome [13] using BWA [28]. We removed duplicated reads created by PCR amplification using rmdup program from samtools [29]. We then applied GATK's [30] base quality score recalibration and indel realignment, and performed SNP genotyping across all samples simultaneously using default settings in GATK [31]. Total coverage is shown in Table S2.
Supporting Information
Zdroje
1. SlaterGJ, FigueiridoB, LouisL, YangP, Van ValkenburghB (2010) Biomechanical consequences of rapid evolution in the polar bear lineage. PLoS ONE 5: e13870 doi:10.1371/journal.pone.0013870.
2. Stirling I (2011) Polar Bears: The Natural History of a Threatened Species. Brighton, Mass: Fitzhenry and Whiteside.
3. EdwardsCJE, SuchardMA, LemeyP, WelchJJ, BarnesI, et al. (2011) Ancient hybridization and a recent Irish origin for the modern polar bear matriline. Current Biology 21 : 1251–1258.
4. HailerF, KutscheraVE, HallstromBM, KlassertD, FainSR, et al. (2012) Nuclear Genomic Sequences Reveal that Polar Bears Are an Old and Distinct Bear Lineage. Science 336 : 344–347.
5. KurténB (1964) The evolution of the polar bear, Ursus maritimus (Phipps). Acta Zool Fenn 108 : 1–26.
6. Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, et al.. (2012) Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proceedings of the National Academy of Sciences of the United States of America.
7. BarnesI, MatheusP, ShapiroB, JensenD, CooperA (2002) Dynamics of Pleistocene population extinctions in Beringian brown bears. Science 295 : 2267–2270.
8. CroninMA, AmstrupSC, GarnerGW, VyseER (1991) Interspecific and Intraspecific Mitochondrial-DNA Variation in North-American Bears (Ursus). Canadian Journal of Zoology-Revue Canadienne De Zoologie 69 : 2985–2992.
9. DavisonJ, HoSYW, BraySC, KorstenM, TammelehtE, et al. (2011) Late-Quaternary biogeographic scenarios for the brown bear (Ursus arctos), a wild mammal model species. Quaternary Science Reviews 30 : 418–430.
10. LindqvistC, SchusterSC, SunY, TalbotSL, QiJ, et al. (2010) Complete mitochondrial genome of a Pleistocene jawbone unveils the origin of polar bear. Proc Natl Acad Sci U S A 107 : 5053–5057.
11. TalbotSL, ShieldsGF (1996) Phylogeography of brown bears (Ursus arctos) of Alaska and paraphyly within the Ursidae. Molecular phylogenetics and evolution 5 : 477–494.
12. PreußA, GansloßerU, PurschkeG, MagieraU (2009) Bear-hybrids: behaviour and phenotype. Zool Garten N F 78 : 204–220.
13. Li B, Zhang G, Willerslev E, Wang J, Wang J (2011) Genomic data from the Polar Bear (Ursus maritimus). Available from: http://dx.doi.org/10.5524/100008. GigaScience.
14. PaetkauD, ShieldsGF, StrobeckC (1998) Gene flow between insular, coastal and interior populations of brown bears in Alaska. Molecular Ecology 7 : 1283–1292.
15. LiR, FanW, TianG, ZhuH, HeL, et al. (2010) The sequence and de novo assembly of the giant panda genome. Nature 463 : 311–317.
16. GreenRE, KrauseJ, BriggsAW, MaricicT, StenzelU, et al. (2010) A Draft Sequence of the Neandertal Genome. Science 328 : 710–722.
17. CarraraPE, AgerTA, BaichtalJF (2007) Possible refugia in the Alexander Archipelago of southeastern Alaska during the late Wisconsin glaciation. Canadian Journal of Earth Sciences 44 : 229–244.
18. ChiverrellRC, ThomasGSP (2010) Extent and timing of the Last Glacial Maximum (LGM) in Britain and Ireland: a review. Journal of Quaternary Science 25 : 535–549.
19. Heaton TH, Grady F (2003) The Late Wisconsin vertebrate history of Prince of Wales Island, southeast Alaska. In: Schubert BW, Mead JI, Graham RW, editors. Ice Age Cave Fauna of North America: Indiana University Press. pp. 17–53.
20. HeatonTH, TalbotSL, ShieldsGF (1996) An Ice Age refugium for large mammals in the Alexander Archipelago, southeastern Alaska. Quaternary Research 46 : 186–192.
21. Alaska Department of Fish and Game (2012) Brown Bear (Ursus arctos), Available: http://www.adfg.alaska.gov/index.cfm?adfg=brownbear.main, Accessed Septemeber 28, 2012.
22. ReichD, PattersonN, KircherM, DelfinF, NandineniMR, et al. (2011) Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. American Journal of Human Genetics 89 : 516–528.
23. CurratM, RuediM, PetitRJ, ExcoffierL (2008) The hidden side of invasions: Massive introgression by local genes. Evolution 62 : 1908–1920.
24. IngólfssonÓ, WiigØ (2009) Late Pleistocene fossil find in Svalbard: the oldest remains of a polar bear (Ursus maritimus Phipps, 1744) ever discovered. Polar Research 28 : 455–462.
25. RohlandN, SiedelH, HofreiterM (2010) A rapid column-based ancient DNA extraction method for increased sample throughput. Molecular Ecology Resources 10 : 677–683.
26. Meyer M, Kircher M (2010) Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor protocols 2010: pdb prot5448.
27. KircherM (2012) Analysis of high-throughput ancient DNA sequencing data. Methods in molecular biology 840 : 197–228.
28. COSEWIC (2009) COSEWIC assessment and update status report on the Northern Leopard Frog Lithobates pipiens, Rocky Mountain population, Wester Boreal/Prairie populations and Eastern populations, in Canada. Ottawa. 69 p.
29. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079.
30. McKennaA, HannaM, BanksE, SivachenkoA, CibulskisK, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20 : 1297–1303.
31. DePristoMA, BanksE, PoplinR, GarimellaKV, MaguireJR, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics 43 : 491–+.
32. GreenRE, MalaspinasAS, KrauseJ, BriggsAW, JohnsonPLF, et al. (2008) A complete neandertal mitochondrial genome sequence determined by high-throughput Sequencing. Cell 134 : 416–426.
33. KrauseJ, UngerT, NoconA, MalaspinasAS, KolokotronisSO, et al. (2008) Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. Bmc Evolutionary Biology 8 : 220.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



