-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
Caspases are cysteine proteases that can drive apoptosis in metazoans and have critical functions in the elimination of cells during development, the maintenance of tissue homeostasis, and responses to cellular damage. Although a growing body of research suggests that programmed cell death can occur in the absence of caspases, mammalian studies of caspase-independent apoptosis are confounded by the existence of at least seven caspase homologs that can function redundantly to promote cell death. Caspase-independent programmed cell death is also thought to occur in the invertebrate nematode Caenorhabditis elegans. The C. elegans genome contains four caspase genes (ced-3, csp-1, csp-2, and csp-3), of which only ced-3 has been demonstrated to promote apoptosis. Here, we show that CSP-1 is a pro-apoptotic caspase that promotes programmed cell death in a subset of cells fated to die during C. elegans embryogenesis. csp-1 is expressed robustly in late pachytene nuclei of the germline and is required maternally for its role in embryonic programmed cell deaths. Unlike CED-3, CSP-1 is not regulated by the APAF-1 homolog CED-4 or the BCL-2 homolog CED-9, revealing that csp-1 functions independently of the canonical genetic pathway for apoptosis. Previously we demonstrated that embryos lacking all four caspases can eliminate cells through an extrusion mechanism and that these cells are apoptotic. Extruded cells differ from cells that normally undergo programmed cell death not only by being extruded but also by not being engulfed by neighboring cells. In this study, we identify in csp-3; csp-1; csp-2 ced-3 quadruple mutants apoptotic cell corpses that fully resemble wild-type cell corpses: these caspase-deficient cell corpses are morphologically apoptotic, are not extruded, and are internalized by engulfing cells. We conclude that both caspase-dependent and caspase-independent pathways promote apoptotic programmed cell death and the phagocytosis of cell corpses in parallel to the canonical apoptosis pathway involving CED-3 activation.
Published in the journal: . PLoS Genet 9(3): e32767. doi:10.1371/journal.pgen.1003341
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003341Summary
Caspases are cysteine proteases that can drive apoptosis in metazoans and have critical functions in the elimination of cells during development, the maintenance of tissue homeostasis, and responses to cellular damage. Although a growing body of research suggests that programmed cell death can occur in the absence of caspases, mammalian studies of caspase-independent apoptosis are confounded by the existence of at least seven caspase homologs that can function redundantly to promote cell death. Caspase-independent programmed cell death is also thought to occur in the invertebrate nematode Caenorhabditis elegans. The C. elegans genome contains four caspase genes (ced-3, csp-1, csp-2, and csp-3), of which only ced-3 has been demonstrated to promote apoptosis. Here, we show that CSP-1 is a pro-apoptotic caspase that promotes programmed cell death in a subset of cells fated to die during C. elegans embryogenesis. csp-1 is expressed robustly in late pachytene nuclei of the germline and is required maternally for its role in embryonic programmed cell deaths. Unlike CED-3, CSP-1 is not regulated by the APAF-1 homolog CED-4 or the BCL-2 homolog CED-9, revealing that csp-1 functions independently of the canonical genetic pathway for apoptosis. Previously we demonstrated that embryos lacking all four caspases can eliminate cells through an extrusion mechanism and that these cells are apoptotic. Extruded cells differ from cells that normally undergo programmed cell death not only by being extruded but also by not being engulfed by neighboring cells. In this study, we identify in csp-3; csp-1; csp-2 ced-3 quadruple mutants apoptotic cell corpses that fully resemble wild-type cell corpses: these caspase-deficient cell corpses are morphologically apoptotic, are not extruded, and are internalized by engulfing cells. We conclude that both caspase-dependent and caspase-independent pathways promote apoptotic programmed cell death and the phagocytosis of cell corpses in parallel to the canonical apoptosis pathway involving CED-3 activation.
Introduction
The elimination of unnecessary or dangerous cells is fundamental to development, tissue homeostasis and disease mitigation in multicellular organisms. The primary mechanism of cell elimination is apoptosis, a form of cell suicide that was originally defined by evolutionarily conserved morphological characteristics that include chromatin condensation, shrinkage of the cytoplasmic volume and membrane blebbing [1] and by biochemical features like phosphatidylserine exposure and DNA fragmentation [2], [3]. Apoptosis serves as a highly controlled mechanism for the removal and degradation of damaged or unnecessary cells, and blocking apoptosis can lead to catastrophic forms of cell death, such as necrosis, which can cause dangerous inflammatory responses [4]. The discovery of the CED-3 caspase as a cell-autonomous executioner of programmed cell death in the nematode Caenorhabditis elegans led to the paradigm that the caspase family of cysteine proteases drives apoptosis through the cleavage of substrate proteins at specific peptide sequences [5], [6]. Indeed, caspases have evolutionarily conserved roles in apoptosis throughout metazoa [7].
Despite the compelling causal link between caspases and apoptosis, a growing body of evidence indicates that apoptosis can occur in the absence of caspases [4]. For example, mouse cells lacking Apaf-1, an activator of the apical caspase Caspase-9, which in turn activates effector caspases, can undergo apoptosis in response to pro-apoptotic stimuli [8]. In the presence of caspase inhibitors, TNF (tumor necrosis factor) can induce a form of cell death termed necroptosis, which exhibits characteristics of both necrosis and apoptosis [4], [9]. The mitochondrial flavoprotein AIF (apoptosis-inducing factor) is thought to promote apoptotic cell death in mammals even in the presence of caspase inhibitors [10]. Furthermore, cell death with aspects of apoptotic morphology occurs in non-metazoans, including unicellular eukaryotes and prokaryotes, that lack clear caspase homologs [11], [12]. Thus, it is possible that apoptosis, as defined morphologically and biochemically, can occur in the absence of caspases.
A standard approach to assaying the caspase-dependence of apoptotic stimuli in tissue cell culture is through the pharmacological inhibition of caspases. However, it is difficult to prove that caspase activity is completely blocked in such experiments, and it is possible for caspase inhibitors to trigger non-apoptotic forms of cell death [13]. Studies of caspase-independent apoptosis in metazoans are also complicated by the existence of multiple caspases with potentially redundant functions in promoting cell death. The human genome, for example, encodes at least 10 caspase homologs, seven of which (caspases-2, -3, -6, -7, -8, -9 and -10) have demonstrated roles in apoptosis [14]. The genome of Drosophila melanogaster encodes seven caspase homologs (dcp-1, dronc, drice, dredd, decay, damm and strica) [7], several of which are essential for organismal viability. The C. elegans genome encodes three caspase homologs (csp-1, csp-2 and csp-3) in addition to ced-3 [15]. Therefore, the use of mutant animals or cell lines deleted for one or two caspases might not eliminate all caspases expressed within a specific cell. Furthermore, since caspases have different substrate specificities [16], the use of a chemical substrate-competitive caspase inhibitor might not completely block all caspase activity. Ideally, experiments that test whether apoptosis can occur in the absence of caspases should be performed using mutant animals or cells that are genetically deleted of all caspase homologs. In this regard, C. elegans is an excellent animal for studies of caspase-independent programmed cell death, because: (1) there are several examples of ced-3-independent programmed cell death in C. elegans [17]–[19]; (2) mutants of ced-3, csp-1, csp-2 and csp-3 are viable [18]–[23]; and, (3) it is relatively easy to generate multiply mutant C. elegans strains.
The ced-3 caspase gene is required for most programmed cell deaths that occur during C. elegans development [5], [20]. However, a small number of cells die in animals carrying null mutations of ced-3. The male-specific linker cell, which facilitates the connection of the vas deferens to the cloaca and then dies, undergoes a non-apoptotic cell death that bears morphological features (e.g., nuclear membrane crenellation) not seen with other C. elegans programmed cell deaths and that occurs in ced-3 mutants as well as in animals doubly mutant for ced-3 and csp-1, csp-2 or csp-3 [18], [20], [24]. We recently showed that a subset of cells fated to die in the C. elegans embryo are eliminated from ced-3 mutants via a caspase-independent shedding mechanism [19]. Interestingly, the shed cells appear apoptotic, exhibiting chromatin condensation, TUNEL-reactive DNA degradation and phosphatidylserine exposure despite the absence of all four caspases. Unlike other apoptotic programmed cell deaths of C. elegans, the shed cells do not undergo phagocytosis by engulfing cells; instead, they are extruded from the developing embryo. By contrast, a small number of apoptotic cell corpses are visible in the heads of ced-3 larvae [17]. Like other programmed cell deaths of C. elegans, these ced-3-independent cell corpses have a refractile appearance when viewed with Nomarski optics and are not extruded from the animal, suggesting that a ced-3-independent cell-killing activity contributes to these typical programmed cell deaths. The other caspase homologs, csp-1, csp-2 and csp-3, are obvious candidates for driving this ced-3-independent cell-killing activity. However, it has recently been reported that csp-2 and csp-3 inhibit apoptosis in the germline and soma, respectively [22], [23].
To date, the C. elegans caspase homolog csp-1 has no known function in vivo, including in apoptosis. An isoform of CSP-1 can cleave and possibly activate the CED-3 pro-protein in vitro [15]. We tested whether csp-1 can promote or inhibit programmed cell death and whether it is regulated by the canonical programmed cell death pathway that activates ced-3. We found that csp-1 encodes a pro-apoptotic caspase activity that promotes programmed cell death independently of the CED-3 caspase, CED-4 (the Apaf-1 homolog that activates CED-3), and CED-9 (a Bcl-2 family protein that negatively regulates CED-3 activation via inhibition of CED-4). Furthermore, we tested whether csp-1, csp-2 and csp-3 contribute to the ced-3-independent cell-killing activity that generates cell corpses in the heads of ced-3 mutant larvae and found that these apoptotic cell deaths can occur in the complete absence of caspases. Thus, during C. elegans development programmed cell death followed by cell-corpse engulfment is achieved through three redundant pathways: (1) a ced-3-dependent pathway; (2) a csp-1-dependent pathway, which is not regulated by the canonical apoptosis pathway that controls ced-3; and, (3) a caspase-independent pathway.
Results
csp-1 promotes the deaths of a subset of somatic cells fated to die
The C. elegans genes csp-1, csp-2 and csp-3 are paralogs of the pro-apoptotic ced-3 caspase gene [15], which is required for most programmed cell deaths in the worm [5], [20]. Given the conserved role of caspases in apoptosis, we tested csp-1, csp-2 and csp-3 for roles – both pro - and anti-apoptotic – in programmed cell death. We used mutations of csp-1 (n4967 and n5133) and csp-2 (n4871) that completely remove the genomic sequences encoding their respective predicted caspase active sites (SACRG in the CSP-1 protein, and VCCRG in the CSP-2 protein) and therefore eliminate any potential caspase activity encoded by these genes (ref. [19]; Figure 1A). csp-3 lacks a caspase active site (ref. [15], [22]; Figure 1A); we used the csp-3 deletion mutation n4872, which is likely a null allele [19].
Fig. 1. The B and/or C isoforms of csp-1 promote programmed cell death. 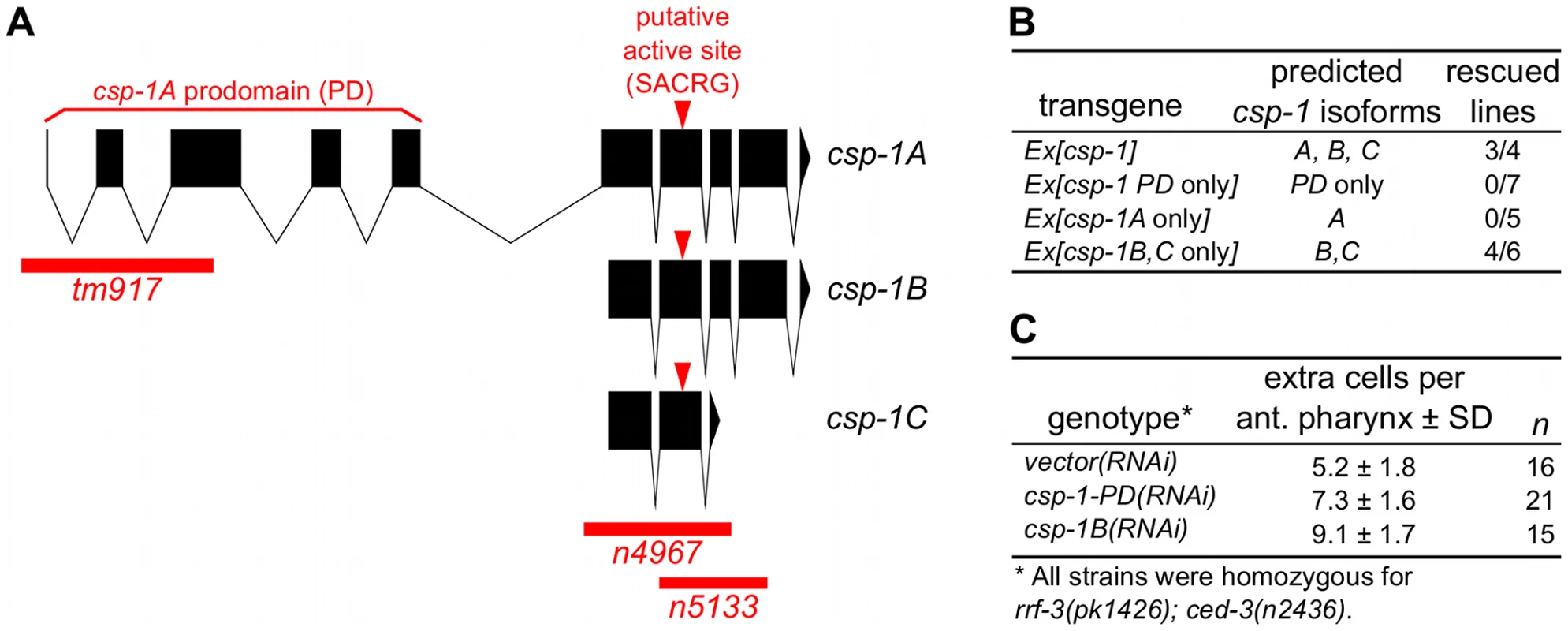
(A) Representations of the intron-exon organization of the three known csp-1 mRNA isoforms (A, B and C). Red bars indicate the csp-1 deletion alleles used in this study; arrowheads indicate the SACRG sequence that encodes the caspase active-site. The graphic was generated using the Intron-Exon Graphic Maker (N. Bhatla; www.wormweb.org). (B) Extrachromosomal arrays carrying a wild-type genomic fragment of the csp-1 locus or a mutant version that expresses only the B or C isoforms can rescue the csp-1(n4967) mutant phenotype. The csp-1 PD-only transgene contains two nonsense mutations that encode early stop codons affecting the B and C mRNA isoforms; the csp-1A-only transgene contains a mutation that changes the B/C start codon to an alanine codon; and the csp-1B/C-only transgene contains two nonsense mutations that encode early stop codons affecting the A isoform. The csp-1 transgenes were injected into csp-1(n4967); ced-3(n2436) animals, and the resulting independent lines were assayed for csp-1 rescuing activity by counting the number of extra undead cells in the anterior pharynx. The transgenes and their constructions are described in detail in Materials and Methods, and the complete data for each transgenic line are provided in Table S2. (C) RNAi knockdown of csp-1 phenocopies the csp-1 mutations. dsRNAs targeting the csp-1 pro-domain or the csp-1B isoform were in vitro transcribed and injected into the gonads of RNAi-sensitive rrf-3(pk1426); ced-3(n2436) adult hermaphrodites. Progeny of the injected adults were assayed for extra undead cells in the anterior pharynx. PD, prodomain. Recently, it was reported that mutations in csp-2 and csp-3 cause ectopic cell deaths in the germline and soma, respectively, and hence that csp-2 and csp-3 inhibit apoptosis [22], [23]. We therefore tested whether csp-1 mutants have ectopic cell deaths indicative of a loss of anti-apoptotic function. Using Nomarski optics and a Pmec-3::gfp transgene that expresses GFP in the six touch neurons (AVM, two ALM, PVM and two PLM neurons) in addition to the FLP and PVD neurons, we examined csp-1 mutants for missing cells that normally survive. We observed that csp-1(n4967) mutants contained a full complement of touch neurons and pharyngeal cells (Table S1). We also noted that csp-1(n4967) failed to cause ectopic cell deaths in sensitized animals carrying the loss-of-function mutation n2812 in the anti-apoptotic gene ced-9, a homolog of human BCL2 (Table S1; data not shown). These results indicate that csp-1 does not have an obvious anti-apoptotic function in the soma. Consistent with a previous report that csp-2 does not affect somatic cells [23], a mutation in csp-2 did not cause ectopic cell deaths in the somatic cells we examined (Table S1). However, we failed to observe the ectopic cell deaths in csp-3 mutants previously reported [22]. Ectopic somatic cell deaths have also been noted in animals with loss-of-function mutations in ced-9 [25] or tat-1 [26], [27], which encodes an aminophospholipid translocase required for the asymmetric distribution of phosphatidylserine on the inner leaflet of the plasma membrane. As expected, we found that ced-9 mutant larvae were missing pharyngeal cells and many touch neurons: more than 80% of PLM neurons were not present in ced-9(n2812) larvae (Table S1). However, we failed to detect the previously reported ectopic cell-death defect of tat-1 mutants (ref. [26], [27]; Table S1); we used the same deletion alleles for csp-3 and tat-1 and assayed the same cells that had been characterized in the previous studies.
To determine whether the C. elegans caspase homologs csp-1, csp-2 or csp-3 promote programmed cell death in the soma, we examined animals carrying csp deletion mutations for extra cells that failed to undergo programmed cell death in the anterior pharynx. As many as 16 extra cells can be counted in the anterior pharynges of mutants with strong defects in programmed cell death, e.g., ced-3(n3692) (ref. [28]; Table 1). Single mutations in csp-1, csp-2 or csp-3 failed to cause detectable defects in programmed cell death (Table 1; data not shown). However, we observed that mutations in csp-1 (but not csp-2 or csp-3) caused the survival of pharyngeal cells in sensitized strains carrying a weak mutation in the caspase gene ced-3 (Table 1). The partial loss-of-function ced-3 mutations n2427 and n2436 cause slight and intermediate defects in apoptosis, respectively (ref [17]; Table 1; data not shown). The n4967 and n5133 mutations, both of which delete the putative active site of CSP-1 (Figure 1A), enhanced the cell-death defects of ced-3(n2427) and ced-3(n2436) mutants, increasing the number of extra cells in their anterior pharynges on average by 1.4 and 2.4 cells, respectively (Table 1). These results are consistent with our RNAi experiments in which csp-1B dsRNA (which likely inactivated all csp-1 transcripts) was injected into the gonads of rrf-3(pk1426); ced-3(n2436) animals and caused an enhanced cell-death defect in their progeny (Figure 1C); we used the rrf-3 mutation to increase sensitivity to RNAi [29]. The cell-death defect conferred by the csp-1(n4967) mutation was rescued by extrachromosomal arrays carrying a 9 kb genomic csp-1 fragment that included the entire csp-1 coding region, 1.5 kb of genomic sequence 5′ of the csp-1A translational start codon and 3.5 kb of genomic sequence 3′ of the csp-1A/B translational stop codon (Figure 1B; Table S2). These results demonstrate that csp-1 encodes a detectable cell-killing activity that contributes to programmed cell death in C. elegans. Mutation of csp-2 and/or csp-3 neither enhanced nor suppressed the cell-death defects of strains mutant for csp-1 and/or ced-3 (Table 1; Table S3), suggesting that csp-1 and ced-3 are the only C. elegans caspase genes with functions in somatic programmed cell deaths.
Tab. 1. The caspase homolog csp-1 promotes programmed cell death in the C. elegans anterior pharynx. 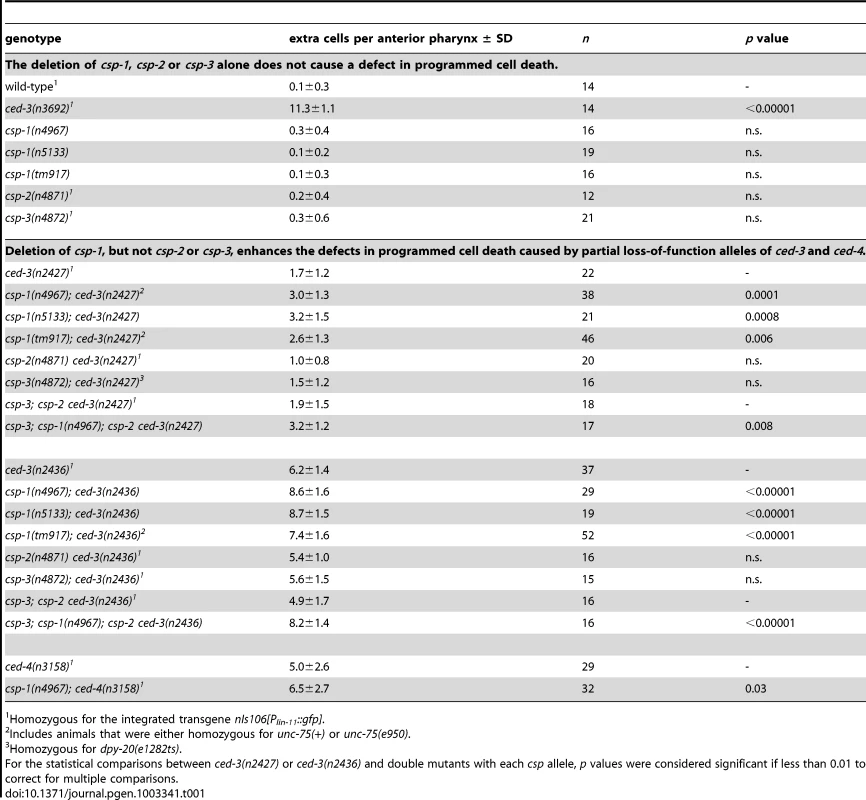
Homozygous for the integrated transgene nIs106[Plin-11::gfp]. The development of the anterior part of the C. elegans pharynx involves 16 programmed cell deaths, all of which appear to be sensitive to ced-3 [17], [28], [30]. To test whether specific pharyngeal programmed cell deaths required csp-1, we used GFP reporters to visualize the survival of cells fated to die, specifically the sister cells of the M4 and NSM neurons. csp-1 was partially required in ced-3(n2427) or ced-3(n2436) sensitized strains for the death of the M4 sister cell (Table S4); by contrast, mutation of csp-1 did not affect the cell deaths of the sister cells of the NSM neurons (data not shown). Likewise, csp-1 did not appear to function in the postembryonic programmed cell deaths of the ventral cord or postdeirid sensilla (Table S4). We conclude that csp-1 promotes cell death in a subset of cells fated to die during C. elegans development.
The csp-1B and/or C isoforms are required for the cell-killing activity of csp-1
The csp-1 locus produces three known mRNA isoforms [15], all of which include the sequence that encodes the presumptive caspase active site (Figure 1A). The csp-1A transcript contains a long prodomain not present in the other transcripts, and it uses an alternative start site that is 3 kb 5′ to the start site of the csp-1B and csp-1C isoforms. To determine which isoforms are required for the cell-killing activity of csp-1, we peformed experiments in which the csp-1 rescuing transgene was mutated to express: (1) the A isoform only, (2) the B and C isoforms only, or (3) a truncated version of csp-1A including only the prodomain (PD). Extrachromosomal arrays engineered to express only csp-1-PD or the csp-1A isoform failed to rescue the cell-death defect of csp-1(n4967) mutants (Figure 1B; Table S2). By contrast, a csp-1 transgene lacking the csp-1A translation start codon and predicted to express only the csp-1B and csp-1C transcripts rescued the csp-1(n4967) defect in programmed cell death (Figure 1B; Table S2). Consistent with these results, transgenes expressing the csp-1B cDNA, but not the csp-1A cDNA, under the control of the mec-7 promoter efficiently killed touch neurons (Figure 2A–2B; Table 2; data not shown); we also expresed the csp-1C cDNA under the control of the mec-7 promoter and failed to observe killing of the touch neurons (data not shown). Ectopic expression of csp-1B from the ser-2d and flp-15 promoters killed the OLL and I2 neurons, respectively (ref. [31]; N. Bhatla and H.R. Horvitz, unpublished results). However, we noted that tm917, a csp-1 allele that deletes coding regions of only the csp-1A transcript, enhanced significantly (albeit weakly) the cell-death defects of ced-3(n2427) and ced-3(n2436) mutants, increasing the number of extra cells in their anterior pharynges by 0.9 and 1.2 cells, respectively (Table 1). dsRNA targetting the csp-1A prodomain (csp-1-PD) caused a similar slight enhancement of the cell-death defect of ced-3(n2436) mutants (Figure 1C), suggesting that, in addition to the more robust cell-killing activity of the csp-1B transcript, csp-1A might have a weak cell-killing function.
Fig. 2. csp-1B overexpression induces ectopic cell deaths. 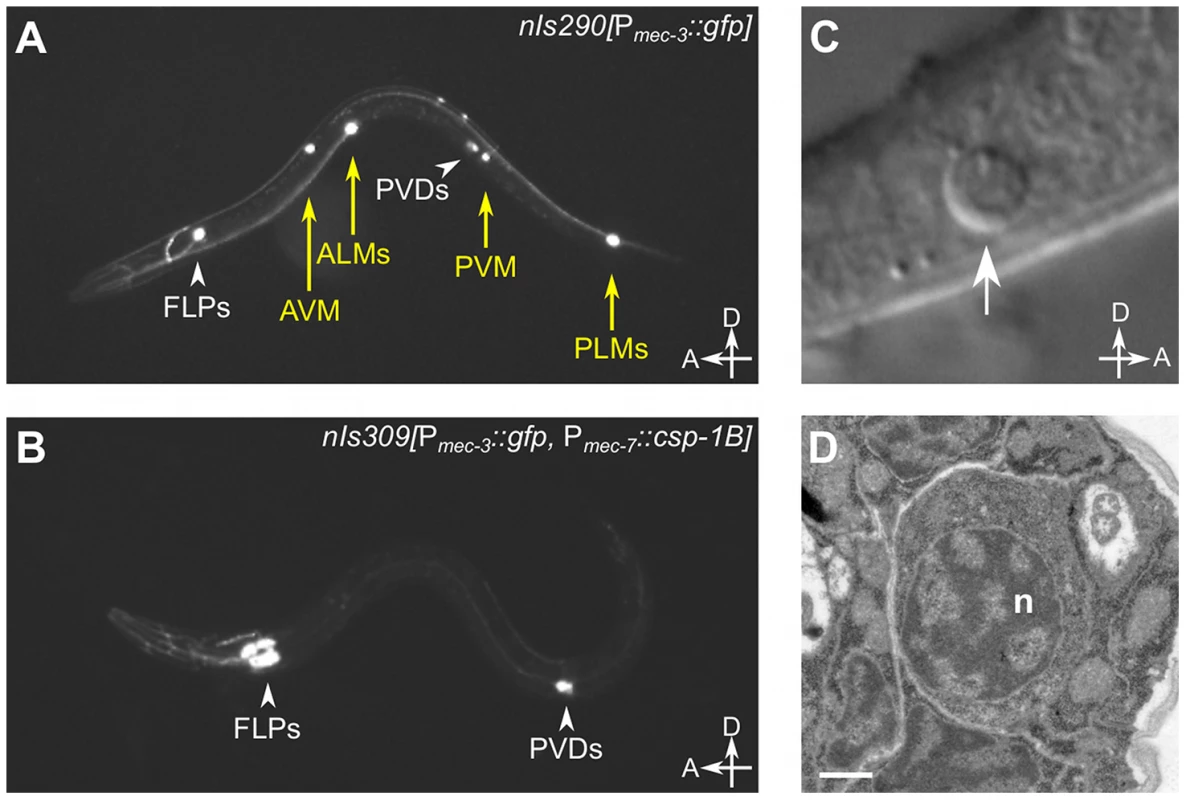
(A) Fluorescence image of a transgenic nIs290[Pmec-3::gfp] larva expressing GFP from the mec-3 promoter in the touch neurons (AVM, ALMs, PVM and PLMs, yellow arrows). mec-3 is also expressed in the FLP and PVD neurons (white arrowheads). (B) Fluorescence image of a transgenic nIs309[Pmec-7::csp-1B, Pmec-3::gfp] larva expressing CSP-1B from the mec-7 promoter (which is expressed in the AVM, ALM, PVM and PLM neurons) and GFP from the mec-3 promoter. Note the absence of touch neurons. (C) Nomarski differential interference contrast (DIC) image of a refractile PLM cell corpse (arrow) in a ced-1(e1735); ced-4(n1162); nIs309 L1 larva. (D) Transmission electron microscopic image of the cell corpse in (C). “n”, nucleus of the cell corpse; scale bar, 0.5 microns. Tab. 2. Ectopic expression of csp-1B from the mec-7 promoter can kill touch neurons, and this killing activity requires the conserved cysteine in the putative caspase active site. 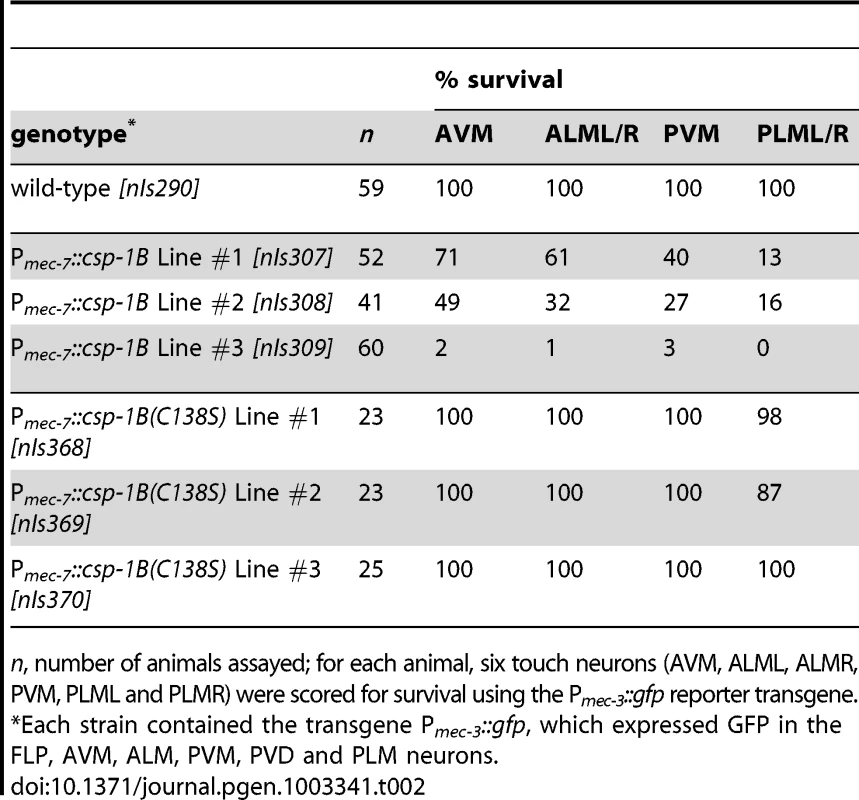
n, number of animals assayed; for each animal, six touch neurons (AVM, ALML, ALMR, PVM, PLML and PLMR) were scored for survival using the Pmec-3::gfp reporter transgene. csp-1B encodes a pro-apoptotic caspase
The proteolytic activity of caspases requires an active-site cysteine. Previously, it was shown that the CSP-1B protein can proteolytically process CED-3 in vitro and that this enzymatic activity required the active-site (SACRG) cysteine of CSP-1B, C138 [15]. We tested in vivo whether C138 was necessary by assaying the touch neuron-killing activity of mutant Pmec-7::csp-1B transgenes in which C138 was changed to a serine. We observed that the ectopic cell deaths were entirely dependent on the caspase active site (Table 2). Thus, csp-1B promotes cell death via caspase activity. The cell deaths induced by a Pmec-7::csp-1B transgene resulted in cell corpses with apoptotic characteristics (Figure 2C–2D). When observed with Nomarski optics, the csp-1B-induced cell deaths exhibited a refractile button-like appearance (Figure 2C) similar to that of developmental programmed cell deaths. Transmission electron micrographs of the cell corpses showed some contraction of the cytoplasmic volume and considerable condensation of the nuclear chromatin (Figure 2D), which are general characteristics of apoptotic cells, including those generated by ced-3 cell-killing transgenes (ref. [32]; data not shown). We conclude that csp-1B encodes a functional caspase that promotes programmed cell deaths with apoptotic morphology.
csp-1B cell-killing activity is independent of ced-9 and ced-4
CED-3, like most caspases, is expressed as an inactive zymogen with an inhibitory N-terminal prodomain. Trans-auto-proteolysis of the CED-3 pro-protein at two aspartate residues removes the pro-domain and yields two subunits that form the active caspase [33]. CED-3 auto-activation is dependent on its prodomain and is facilitated by the association of two CED-3 pro-proteins within an octameric complex formed with the Apaf-1 homolog CED-4 [34]–[36]. Under normal cellular conditions, CED-4 is sequestered by CED-9 at mitochondria through a direct protein-protein interaction [37]–[39]. In response to upstream pro-apoptotic signals and the consequent expression of the BH3-domain-only protein EGL-1, which binds to and inhibits CED-9 [40], CED-4 is released from CED-9 and translocates to the nuclear periphery [37], [41], where it facilitates CED-3 activation [38]. Thus, the activation of CED-3 is controlled by an apoptosis pathway involving a BH3-domain-only protein, a member of the Bcl-2 family of apoptosis regulators, and a homolog of the apoptosome complex protein Apaf-1. The basic elements of this apoptosis pathway are evolutionarily conserved in mammals and are responsible for the activation of caspases in response to cell-intrinsic apoptotic stimuli [7].
Consistent with the role of ced-9 in negatively regulating ced-3 activation, it was previously shown that null mutations of ced-9 enhance the touch neuron-killing activities of Pmec-7::ced-3 transgenes [32]. (These experiments were performed using a ced-3(null) background to suppress the ced-3-dependent inviability of ced-9(null) animals.) Furthermore, this enhancement is dependent on ced-4 [32], indicating that the absence of CED-9 activates endogenous CED-4 within the touch neurons and that CED-4 activation elevates CED-3 activity. Unlike the CED-3 zymogen, CSP-1B lacks a long prodomain, suggesting that it might be activated via an alternative mechanism (i.e., independently of CED-4 and CED-9). To determine whether these canonical apoptosis regulators control CSP-1B activation, we introduced the ced-9(n2812) mutation into ced-3(n3692) strains carrying Pmec-7::csp-1B transgenes and assessed the effect of this ced-9 null mutation on PLM survival. In contrast to its effects on Pmec-7::ced-3–mediated PLM killing, ced-9(n2812) failed to enhance PLM killing in Pmec-7::csp-1B strains with a ced-3(n3692) mutant background (Figure 3A). Instead, ced-9(n2812) partially suppressed csp-1B-mediated PLM death (Figure 3A). CED-9 has a poorly understood pro-apoptotic activity in addition to its anti-apoptotic role in CED-4 inhibition [42], and it is possible that this ced-9 pro-apoptotic activity contributed to the deaths of cells expressing ectopic CSP-1B. Nevertheless, our results indicate that csp-1B-mediated cell killing, unlike ced-3-mediated cell killing, is not negatively regulated by ced-9 and suggest that CSP-1B is activated independently of CED-9.
Fig. 3. csp-1B cell-killing activity is not regulated by the canonical programmed cell death pathway. 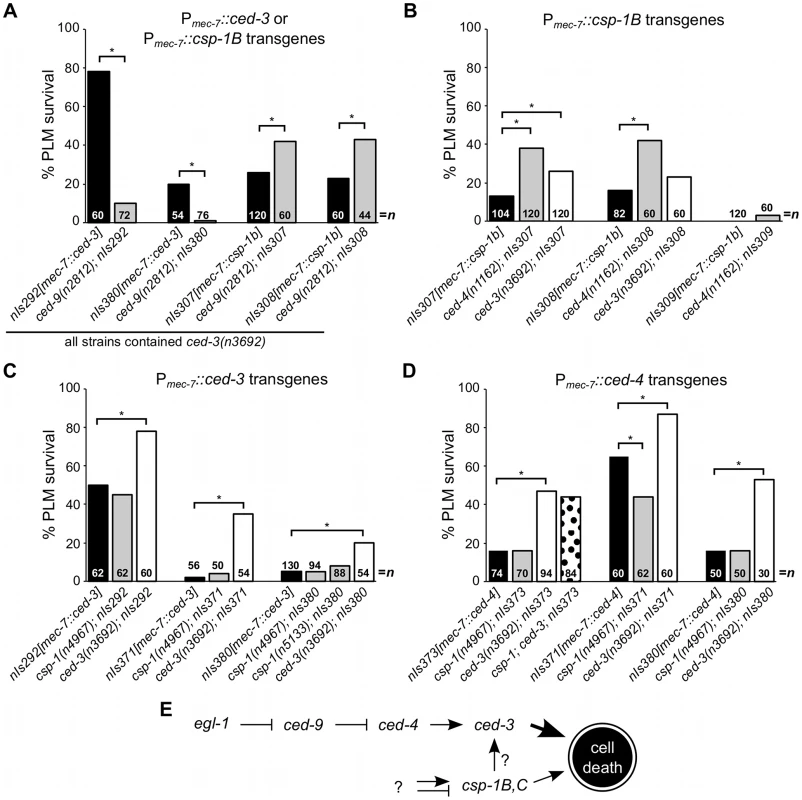
(A–D) The percentages of PLM cells that survive in strains carrying Pmec-7::ced-3, Pmec-7::csp-1B or Pmec-7::ced-4 transgenes. (A) ced-9 protects against ced-3- but not csp-1B-cell-killing transgenes. (B) The cell-killing activity of csp-1B transgenes is mostly independent of ced-3 and ced-4. The cell-killing activities of (C) ced-3 and (D) ced-4 transgenes do not require csp-1. PLM survival was scored based on the presence of GFP expressed from the mec-3 promoter. Asterisks indicate p<0.05 in a Fisher's exact test. All strains in (A) contained ced-3(n3692). (E) A model depicting the genetic pathways regulating the caspase genes csp-1 and ced-3 (see text). We also observed that the expression of a Pmec-7::csp-1A transgene in ced-3(null) mutant strains failed to cause PLM cell death, even in a ced-9(null) background (Figure S1). These results suggest that the CSP-1A isoform (which contains a long prodomain similar to that of CED-3) does not promote programmed cell death, even in the absence of the anti-apoptotic protein CED-9. A role for csp-1A in cell death cannot be excluded entirely, as it is possible that endogenous CSP-1A requires a co-factor not present in the touch neurons to mediate cell killing.
Since CSP-1B can proteolytically cleave CED-3 in vitro [15], we tested whether the csp-1B cell-killing activing requires the endogenous ced-3 and ced-4 genes. The ced-3(n3692) and ced-4(n1162) mutations weakly suppressed csp-1B-mediated PLM death (Figure 3B), and it is possible that the endogenous csp-1 can in part promote programmed cell death through ced-3. Nonetheless, most csp-1B cell-killing activity was independent of ced-4 and ced-3 (Figure 3B). Loss of endogenous csp-1 failed to suppress PLM death in strains carrying Pmec-7::ced-3 or Pmec-7::ced-4 transgenes (Figure 3C–3D). Together, our results are consistent with a model in which csp-1B promotes programmed cell death at least mostly independently of and in parallel to the canonical apoptosis pathway (Figure 3E).
csp-1 expression in the maternal germline contributes to embryonic programmed cell death
To determine which C. elegans cells express csp-1, we directly visualized endogenous csp-1 transcripts via fluorescence in situ hybridization (FISH) experiments using Cy5 - and ALEXA-labelled probes complementary to the csp-1B transcript (i.e., targeted to all csp-1 transcripts) or to the csp-1A prodomain (specific to the csp-1A trancript). To our surprise, csp-1 mRNA was not detectable in the somatic cells of wild-type or egl-1(n1084 n3082) mutant embryos, larvae or adult hermaphrodites (data not shown). By contrast, csp-1 transcripts were present in the germlines of L4-stage larval and adult hermaphrodites (Figure 4A–4B). This expression was restricted to the late pachytene stage of meiosis I in both L4 larval gonads (in pachytene nuclei adjacent to differentiating sperm) and adult gonads (in pachytene nuclei adjacent to the bend of the gonad arm) (Figure 4A–4B). Both csp-1A and csp-1B/C transcripts were expressed in the adult pachytene germ cells, as indicated by the presence of FISH foci recognized by the csp-1A prodomain probes and foci recognized primarily by the csp-1B probes and only weakly by the csp-1A probes (Figure 4C). Stochastic and ionizing radiation (IR)-induced germline cell deaths occur during the late pachytene stage of oocyte development in adult gonads [43], [44]. However, csp-1 (unlike ced-3) was not required for either stochastic or IR-induced germline apoptosis, even in ced-3(n2436) strains sensitized for defects in germ-cell death (Figure 4D). In these experiments, apoptotic germ cells were identified using a transgene that expresses a functional GFP::CED-1 fusion protein that envelopes dying cells engulfed by the gonadal sheath [45], [46]. We also failed to detect differences in either stochastic or IR-induced germline cell death between csp-1 mutants and wild-type animals in experiments in which apoptotic germ cells were quantified by acridine orange staining or by direct observation of their refractile morphology using Nomarski optics (data not shown). We also noted that the level of csp-1 transcript expression in the germline (as determined by FISH) was not affected by either ionizing radiation or by mutation of egl-1 or ced-3 (data not shown).
Fig. 4. csp-1 is expressed in late pachytene cells of the L4 and adult hermaphrodite germline. 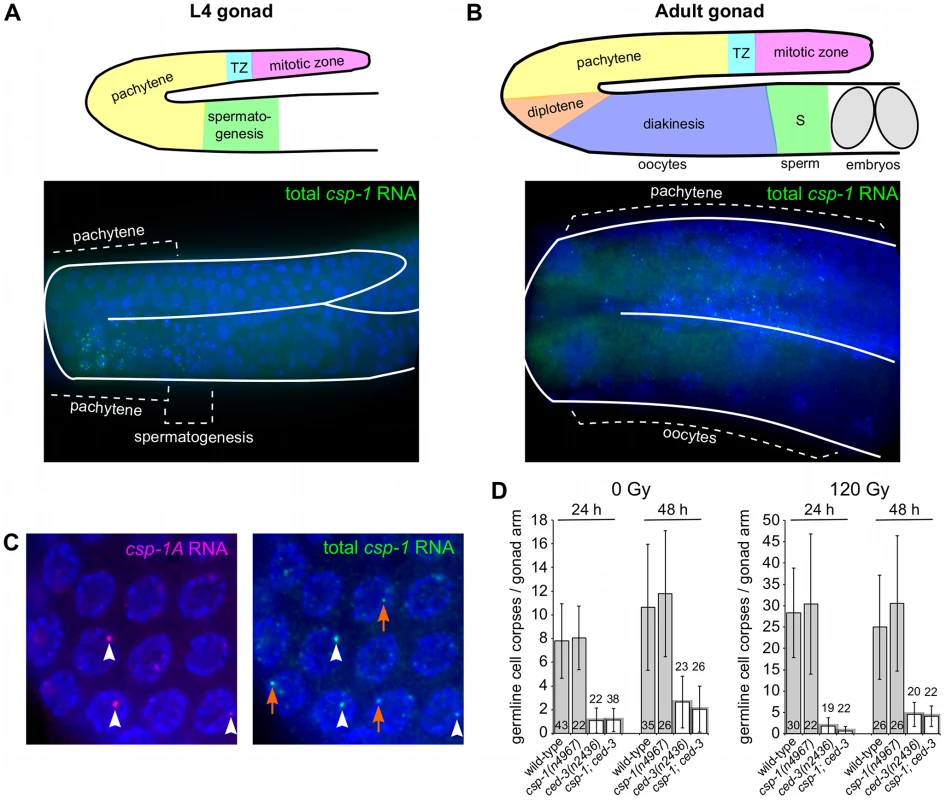
Fluorescence in situ hybridization images of gonad arms of (A) an L4 hermaphrodite and (B) an adult hermaphrodite hybridized with Cy5-labelled probes complementary to csp-1B. The Cy5-labelled probes are visible as green puncta; the gonads are outlined in white. A schematic representation is shown above each micrograph. (C) Fluorescence in situ hybridization images of an adult hermaphrodite gonad hybridized with ALEXA594-labelled probes (red puncta) complementary to the region of csp-1A that encodes the prodomain (csp-1A) and Cy5-labelled csp-1B probes (green puncta) that hybridize to all csp-1 transcript isoforms (total csp-1). White arrowheads indicate csp-1A-specific puncta; orange arrows indicate csp-1B-specific puncta, which are recognized strongly by the total csp-1 probes but only weakly by the csp-1A-specific probes. (D) The number of CED-1::GFP-positive apoptotic cells in the gonads of caspase mutants exposed to 0 Gy and 120 Gy of ionizing radiation at the L4 larval stage. The strains were scored at 24 hrs and 48 hrs post L4-stage. Error bars indicate standard deviations. Since we detected csp-1 expression in the adult germline but not in somatic cells of the embryo, we tested whether maternally supplied csp-1 transcript was necessary for the zygotic function of csp-1 in programmed cell death. Indeed, in sensitized genetic backgrounds (ced-3(n2427) and ced-3(n2436)), csp-1(+) progeny of csp-1(n4967) hermaphrodites (M−Z+ animals) had more undead pharyngeal cells than the csp-1(+) progeny of csp-1(+) hermaphrodites (M+Z+ animals) or the csp-1(n4967) progeny of csp-1(+) hermaphrodites (M+Z − animals) (Table 3). Thus, csp-1 expressed in the maternal germline is necessary for the csp-1 pro-apoptotic activity in embryonic programmed cell deaths. Given that we could not detect csp-1 expression in either embryos or larvae, it is therefore not surprising that the postembryonic programmed cell deaths of the ventral cord and postdeirid sensilla were unaffected by mutation of csp-1 (Table S4).
Tab. 3. csp-1 is maternally required for programmed cell deaths that occur embryonically in the presumptive anterior pharynx. 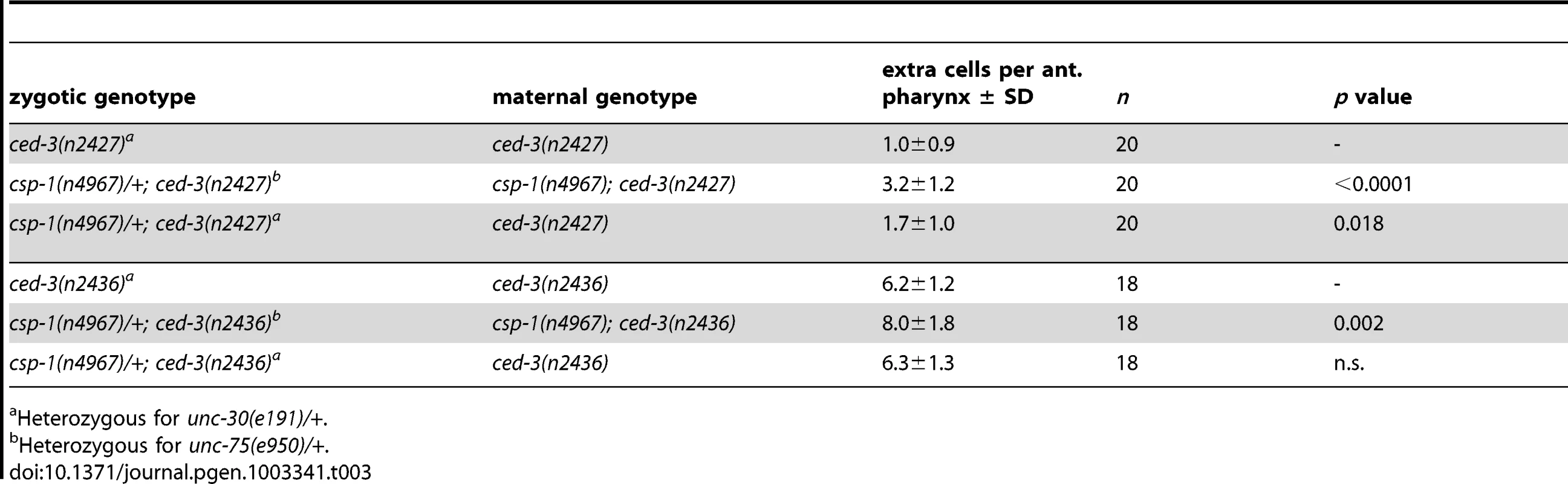
Heterozygous for unc-30(e191)/+. Programmed cell deaths occur in animals completely lacking all caspase genes
Most programmed cell deaths in C. elegans require ced-3 [20]. However, some cells die in mutants completely lacking ced-3. We previously reported that a subset of cells fated to die can be eliminated from ced-3 mutant embryos via a cell-shedding mechansm [19]. In that study, we noted that cell shedding from ced-3 mutants occurs independently of csp-1, csp-2 and csp-3: quadruple mutants lacking all four caspases also generate shed cells, indicating that cell elimination by this mechanism is completely caspase-independent [19]. Like most programmed cell deaths, the cells generated by caspase-independent extrusion are apoptotic in appearance. However, unlike caspase-dependent cell corpses, shed cells do not undergo phagocytosis by engulfing cells. The death of the male linker cell, which also occurs independently of ced-3, is non-apoptotic and requires the heterochronic protein LIN-29, its binding partner MAB-10 [47], and the polyglutamine repeat protein PQN-41 (ref. [18], [24]; Table S5). Previously it was shown that this cell death occurs in double-mutant males in which ced-3 and an additional csp gene (csp-1, csp-2 or csp-3) were inactivated [18]. We have now examined males lacking all four caspases and observed that the linker cell died in 100% of csp-3; csp-1; csp-2 ced-3 mutants (Table S5). The csp-3; csp-1; csp-2 ced-3 quadruple mutants were viable and fertile. Thus, both zygotic and maternal caspase contributions were eliminated. Our results therefore confirm that this cell death is indeed completely caspase-independent.
In addition, cell corpses are visible in the heads of larvae carrying null alleles of ced-3 (ref. [17]; Table 4). All programmed cell deaths in the developing heads of wild-type animals occur embryonically and are engulfed and degraded prior to hatching (ref. [30], [48]; Table 4). To detect ced-3-independent programmed cell deaths in larval heads, we used mutations (e.g., ced-1(e1735), ced-6(n2095) or ced-7(n1996)) that cause defects in cell-corpse engulfment and result in the persistence of many embryonic cell corpses into larval stages (ref. [49], [50]; Table 4). Like most wild-type cell corpses, the ced-3-independent cell corpses were refractile in appearance as observed with Nomarski optics and were not extruded from the animal (data not shown). We also observed that larvae mutant for ced-4 or egl-1 contained similar cell corpses, demonstrating that their generation does not require the canonical pro-apoptotic pathway that mediates most programmed cell deaths (Table 4).
Tab. 4. Cell deaths occur in the absence of all C. elegans caspase genes. 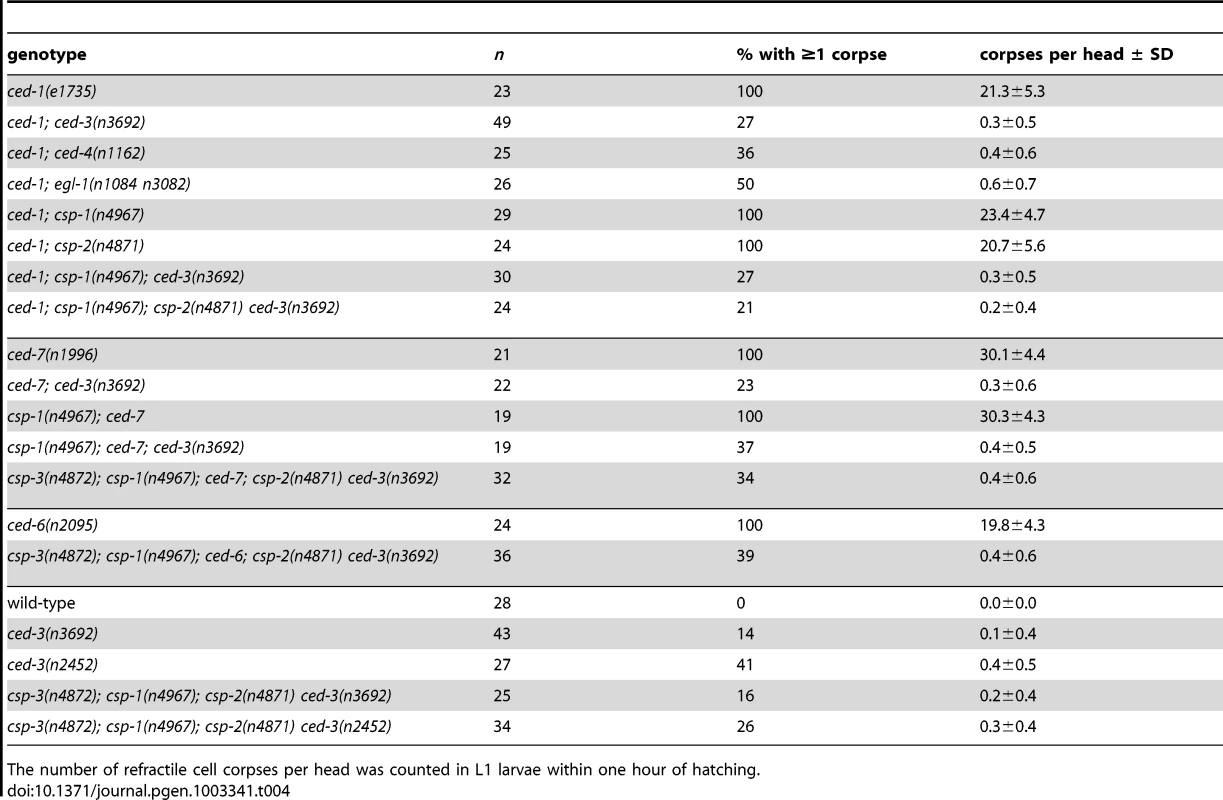
The number of refractile cell corpses per head was counted in L1 larvae within one hour of hatching. We tested whether the small number of cell corpses visible in ced-3 larval heads are generated by the other C. elegans caspase genes and found that all double, triple and quadruple caspase mutants that we examined contained a small number of refractile corpses (Table 4). For example, 39% of csp-3; csp-1; ced-6; csp-2 ced-3 mutant animals contained at least one refractile cell corpse (Table 4), indicating that these programmed cell deaths occur in animals lacking all C. elegans caspases. We observed caspase-independent cell corpses in different regions of the larval head, including positions internal and external to the pharynx, which suggests that multiple cell lineages – at low frequencies – generated caspase-independent cell corpses. Surprisingly, we discovered that engulfment-competent ced-3 and csp-3; csp-1; csp-2 ced-3 mutants also contained refractile cell corpses (Table 4). The number of cell corpses per ced-3 or csp-1; csp-2 ced-3 larva increased until 12 to 24 hours post hatching (see below; data not shown), indicating that at least some of the cell deaths occurred after embryogenesis. Given that all programmed cell deaths in the head normally occur embryonically and that cell corpses are never observed in the heads of wild-type larvae, we concluded that timing of cell deaths in these ced-3 mutants was delayed. Thus, caspase-independent cell corpses can undergo an inefficient programmed cell death with slow kinetics in the absence of CED-3 activity, indicating that these cells likely die via CED-3-mediated apoptosis in wild-type animals.
Caspase-independent cell corpses exhibit apoptotic morphology
Despite the strong causal link between caspase activation and apoptosis, recent studies have demonstrated that many morphological and biochemical changes associated with apoptosis can occur in the absence of caspases [4], [19], [21]. For example, in C. elegans the shed cells of csp-3; csp-1; csp-2 ced-3 quadruple mutants exhibit phosphatidylserine exposure, expression of the pro-apoptotic BH3-only gene egl-1, and chromatin condensation [19]. To determine whether these apoptotic attributes are evident in caspase-independent programmed cell deaths that do not involve extrusion of the dying cell from the embryo, we characterized the cell corpses visible in caspase-deleted larvae (Figure 5 and Figure 6). In most of these experiments, we used strains with the wild-type csp-3 allele, because (1) csp-3 lacks a caspase active-site [15]; (2) although previous studies reported that csp-3 has an anti-apoptotic function in somatic cells [22], we were unable to replicate those findings (Table S1); and, (3) the presence or absence of a csp-3 mutation had no effect on the frequency or appearance of caspase-independent corpses (Table 4; Figure 6B; data not shown). Like ced-3-mediated programmed cell deaths in wild-type animals, the caspase-independent corpses expressed egl-1, the upstream activator of the canonical apoptosis pathway (Figure 5A). Also, these cell corpses displayed phosphatidylserine on their cell surfaces, as indicated by the phosphatidylserine-binding reporter MFG-e8::Venus (Figure 5B), and exhibited many of the morphological hallmarks of apoptosis, including contraction of cytoplasmic volume and, in some but not all cases, condensation of nuclear chromatin (Figure 5C).
Fig. 5. The cell corpses of caspase-deleted mutants are cytologically and morphologically apoptotic. 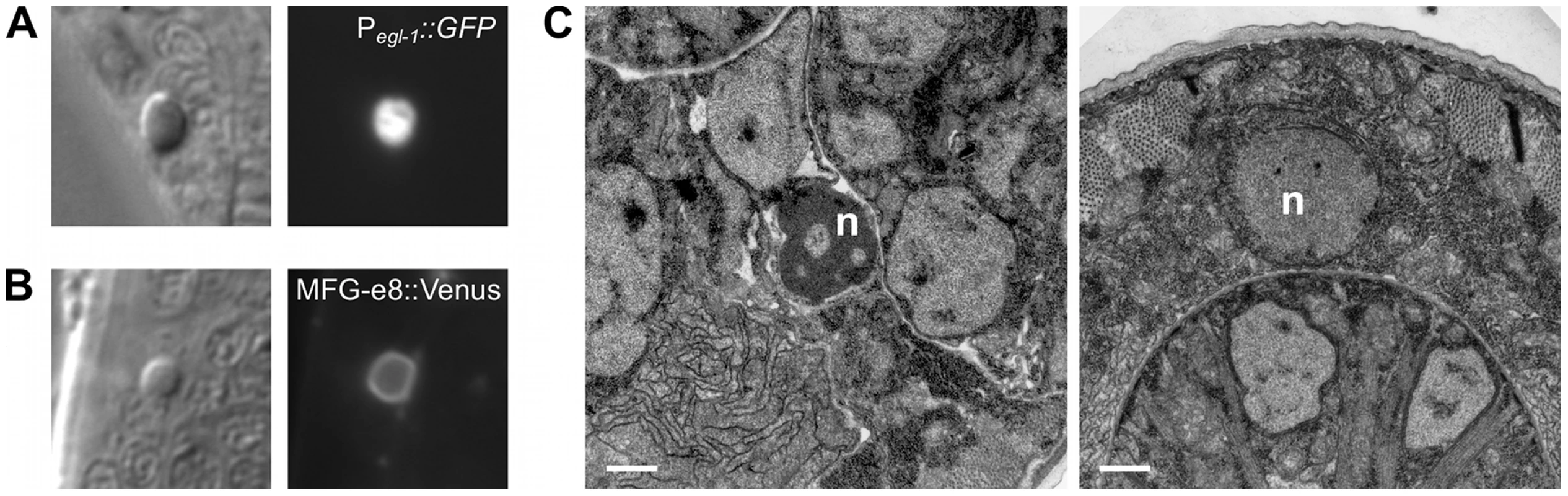
(A) Nomarski DIC and fluorescence images of a cell corpse within the head of a ced-1(e1735); csp-1(n4967); csp-2(n4871) ced-3(n3692) L1 larva carrying the integrated transgene nIs342[Pegl-1::gfp], a transcriptional reporter that expresses GFP under the control of the BH3 domain-only encoding gene egl-1. (B) Nomarski DIC and fluorescence images of a cell corpse within the head of a ced-1(e1735); csp-1(n4967); csp-2(n4871) ced-3(n3692) L1 larva carrying the extrachromosomal array nEx1646[Pdyn-1::mfg-e8::Venus], a fusion protein that binds to cell-surface exposed phosphatidylserine. (C) Representative transmission electron micrographs of cell corpses from ced-1(e1735); csp-1(n4967); csp-2(n4871) ced-3(n3692) larvae 24 hrs post hatching. “n”, nuclei of the cell corpses; scale bars, 0.5 microns. Note the difference in chromatin condensation between the two cell corpses. Fig. 6. Caspase-independent cell corpses are engulfed and degraded. 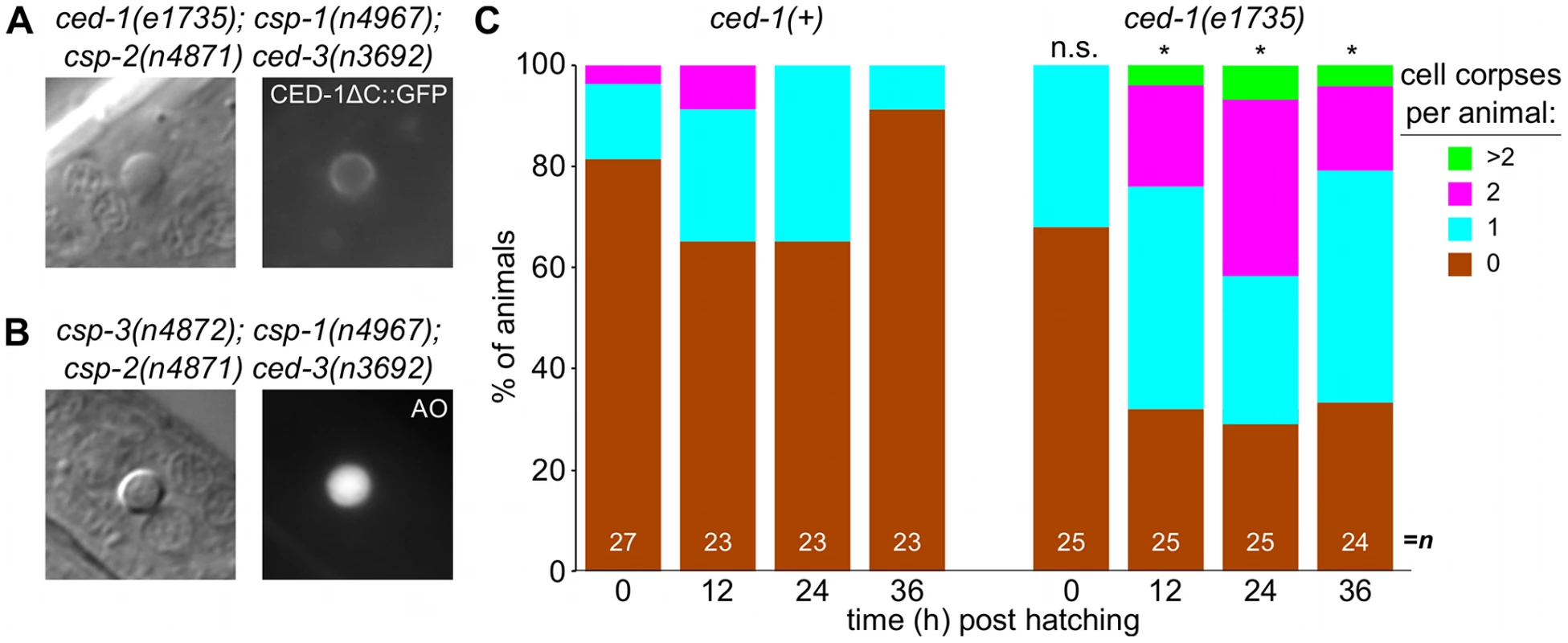
(A) Nomarski DIC and fluorescence images of a cell corpse from a ced-1(e1735); csp-1(n4967); csp-2(n4871) ced-3(n3692) L1 larva carrying the integrated transgene nIs400[Pced-1::ced-1ΔC::gfp], which expresses a non-rescuing CED-1ΔC::GFP fusion protein. CED-1 is a transmembrane receptor that is expressed on engulfing cells, binds to apoptotic cell corpses, and is required for phagocytosis [46]. (B) Nomarski DIC and fluorescence images of a cell corpse from a csp-3(n4872); csp-1(n4967); csp-2(n4871) ced-3(n3692) L1 larva stained with acridine orange (AO), which fluoresces in engulfed cell corpses undergoing degradation in endosomal compartments. (C) The fraction of csp-1(n4967); csp-2(n4871) ced-3(n3692) and ced-1(e1735); csp-1(n4967); csp-2(n4871) ced-3(n3692) with 0, 1, 2 or >2 cell corpses at different time points post hatching. Asterisks indicate p<0.05 in a Mann-Whitney test comparing the two genotypes at a given time point. Additionally, we noted that the caspase-independent cell corpses frequently stained with acridine orange (Figure 6A), suggesting that these corpses are engulfed, internalized and degraded via endosomal pathways, as are canonical programmed cell deaths [30], [48], [49], [51]. Indeed, we found that the caspase-independent corpses were recognized by CED-1 (Figure 6B), a receptor expressed on engulfing cells required for the efficient phagocytosis of cell corpses [46], [49], [50]. The recognition of caspase-independent cell corpses by CED-1 appeared to be functionally important, as ced-1; csp-1; csp-2 ced-3 larvae contained more corpses than csp-1; csp-2 ced-3 larvae (Figure 6C). Given that ced-1 and other genes that function in cell-corpse engulfment promote programmed cell death [52], [53], it is unlikely that the ced-1(e1735) loss-of-function mutation caused additional cell deaths in the caspase-deleted mutants. Instead, the extra cell corpses in ced-1 mutant larvae likely reflected an engulfment defect, consistent with the comparatively rapid degradation and disappearance of most caspase-independent corpses in ced-1(+) larvae within the 36-hour period after hatching (Figure 6C). We conclude that caspases are not required for programmed cell deaths to be recognized by the engulfment machinery, internalized and degraded. In short, many aspects of apoptosis, including phagocytosis – the ultimate fate of apoptotic cells – can occur without caspases. We conclude that a parallel, caspase-independent pathway contributes to programmed cell death in C. elegans and can execute most cellular changes associated with apoptosis.
Discussion
Our experiments revealed unexpected complexities in the execution of apoptosis in C. elegans. While the CED-3 caspase is clearly the primary effector of programmed cell death, we demonstrated the existence of additional caspase-dependent and caspase-independent contributions to developmental apoptosis. Specifically, we observed that maternally-expressed caspase gene csp-1 (but not csp-2 or csp-3) promotes the deaths of a subset of cells programmed to die during C. elegans embryogenesis (Figure 1 and Figure 4; Table 1 and Table 3). Furthermore, ectopic expression of the csp-1B isoform of csp-1 is sufficient to cell-autonomously kill cells that normally survive. These ectopic apoptotic cell deaths require the active site cysteine (C138) of CSP-1B, indicating that a caspase-like proteolytic function is responsible for its cell-killing activity (Table 2). The C. elegans genome therefore expresses at least two pro-apoptotic caspases, CED-3 and CSP-1B, to mediate programmed cell deaths. Nevertheless, the additional caspase activity conferred by csp-1 cannot account for ced-3-independent programmed cell deaths that have been observed in C. elegans. For example, the non-apoptotic death of the male linker cell and the extrusion of shed cells were already known to be caspase-independent [18], [19]. Here we demonstrate that cells in caspase-deleted animals can undergo an apoptosis-like programmed cell death followed by engulfment, indicating that the complete apoptotic program can occur in the absence of caspases. Thus, in addition to CED-3 and CSP-1B, there are caspase-independent cell-killing activities that contribute to programmed cell deaths.
CSP-1B is regulated by a mechanism distinct from that of CED-3
The caspases CED-3 and CSP-1B appear to be regulated differently. The auto-activation of CED-3 is facilitated by the Apaf-1 homolog CED-4 in a protein-protein interaction that requires the CED-3 prodomain [34]–[36]. In the absence of a pro-apoptotic signal, CED-9 sequesters CED-4 [37], thereby preventing its association with the inactive CED-3 proprotein. The CSP-1B proprotein lacks a long prodomain, suggesting that it is not activated through an association with the CED-4 octamer in cells undergoing apoptosis. Consistent with this expectation, we observed that the cell-killing activity of csp-1B transgenes, unlike that of ced-3 transgenes, was not negatively regulated by ced-9 (Figure 3). Furthermore, based on our genetic experiments (Figure 3) and the in vitro studies of Shaham [15], it does not appear that CSP-1B is activated by CED-3. We therefore propose that CSP-1B is regulated by a mechanism different from the canonical programmed cell death pathway that activates CED-3 and that CSP-1B likely promotes cell killing in parallel to CED-3 (Figure 3E).
There are no known or candidate regulators of csp-1. It is possible that csp-1 is controlled entirely at the transcriptional level and that csp-1 contributes a minor, sub-lethal pro-apoptotic activity to all cells within the C. elegans embryo. Indeed, only using sensitized backgrounds with partial defects in programmed cell death did we detect the pro-apoptotic function of csp-1. Nevertheless, we expect that it will be possible to identify regulators and effectors of csp-1 through genetic screens for mutants that modify the cell-killing activity of csp-1B transgenes.
Do the csp genes have non-apoptotic functions?
Given the minor contribution of csp-1 to programmed cell death and the lack of a detectable role of csp-2 or csp-3 in apoptosis (Table 1; Table S1; data not shown), it is tempting to speculate that the csp genes have non-apoptotic functions in C. elegans. In C. elegans, ced-3 functions in axon regeneration following laser axotomy [54]. In mammalian and Drosophila neurons, caspases have functions in dendritic pruning, axon guidance and the synaptic changes underlying long-term depression [14]. Caspase function is also required for the maturation of Drosophila sperm [55]. Interestingly, we observed robust expression of csp-1 in the germlines of L4 and adult hermaphrodites, specifically in the late pachytene nuclei (Figure 4). We also observed temporally and spatially restricted csp-2 and csp-3 mRNA expression in the late pachytene nuclei of the L4 larval germline (data not shown), suggesting that the csp genes might have functions in germ cell development. However, mutant hermaphrodites and males carrying all tested combinations of csp-1, csp-2 and csp-3, including the triple csp mutant were viable, fertile and failed to exhibit obvious brood-size defects that would suggest abnormalities in sperm or oocyte differentiation (data not shown).
csp-1B as a tool for the genetic ablation of cells
Genetically encoded cell-killing activities provide an efficient and convenient method for determining cellular function through cell ablation. Killer genes such as ced-3 have been used under the control of various promoters to ablate specific cells [32], [45], [56], [57]. However, the potent cell-killing activity of ced-3 transgenes can cause organismic inviability, particularly if the promoter expression is not exclusive to a small number of cells (see below). csp-1B overexpression using the mec-7 and flp-15 promoters efficiently killed the touch and I2 neurons, respectively (Figure 2; Table 2; N. Bhatla and H.R. Horvitz, personal communication). The mec-7 and flp-15 promoters are relatively strong, as they also robustly induced gfp expression in these cells, such that the neural processes were visible with a dissecting microscope equipped with fluorescence optics. By contrast, the odr-1 promoter did not produce detectable GFP expression in the neurites of the AWB, AWC and I1 neurons, and csp-1B under the control of the odr-1 promoter failed to kill these cells even when injected at plasmid concentrations as high as 100 ng/µl (N. Bhatla and H.R. Horvitz, unpublished results). Thus, high levels of csp-1B expression might be required to kill most cells, making the use of csp-1B as a cell-ablation tool appropriate in situations in which the promoter sequence strongly drives expression in targeted cells and/or weakly promotes expression in additional cells not intended to be targets. For example, the Pmec-7::csp-1B constructs, which were injected at a concentration of 15 ng/µl, produced csp-1B expression outside of the touch neurons that was detectable by fluorescence in situ hybridization. However, this level of csp-1B expression was sub-lethal and did not induce cell death or other cellular defects outside of the touch neurons (data not shown). By contrast, Pmec-7::ced-3 constructs were toxic to the animals when injected at concentrations above 1 ng/µl, suggesting that cells are very sensitive to ectopic ced-3 and that using ced-3 as a cell ablation tool is potentially problematic when promoter expression is not restricted to a small number of targeted cells.
What is the role of ced-3-independent cell-killing activities that have minor contributions to programmed cell death?
Although the csp-1 gene contributes a cell-killing activity to normal programmed cell deaths (Table 1), csp-1 and the other csp genes are not responsible for the ced-3-independent programmed cell deaths present in the heads of ced-3 larvae (Table 4). These deaths, like those of the male linker cell (ref. [18]; Table S5) and the embryonic shed cells [19], are caspase-independent – a surprising result in light of our observations that these cell corpses are morphologically apoptotic (Figure 5) and are engulfed (albeit with slower kinetics) like normal programmed cell deaths (Figure 6). Thus, the complete apoptotic program including cell-corpse internalization can occur in the absence of caspases in C. elegans, suggesting that the cellular changes accompanying apoptosis do not require proteolysis by the caspase family of proteases. Moreover, it is clear that apoptotic programmed cell deaths are achieved through the integration of independent cell-killing activities from CED-3, CSP-1B and an unknown caspase-independent source.
Given the minor cell-killing effects of the CSP-1B and the caspase-independent pathways, why might cell-killing activities in addition to that of CED-3 have evolved? It is possible that different cells, even within the set of C. elegans cells fated to die, are differentially sensitive to pro-apoptotic signals and that additional caspase and caspase-independent pathways ensure efficient and complete cell death under diverse environmental and developmental conditions. Interestingly, the postembryonic programmed cell deaths of the ventral cord are more sensitive to weak ced-3 mutations than are the embryonic programmed cell deaths in the presumptive anterior pharynx: ced-3 mutations that have weak effects in the anterior pharnyx typically have stronger effects in the ventral cord (ref. [17]; data not shown). We observed a complementary function for csp-1, which promotes apoptosis in the anterior pharynx (Table 1) but not in the ventral cord (Table S4).
In summary, multiple pro-apoptotic caspases function in programmed cell death in C. elegans, Drosophila and vertebrates. Furthermore, as we and others have shown, there are additional caspase-independent contributions to programmed cell deaths in C. elegans. We identified C. elegans caspase-independent cell deaths that are essentially identical to wild-type programmed cell deaths based on their apoptotic appearance and their recognition and internalization by engulfing cells. We expect that caspase-independent pro-apoptotic activities are present in other metazoans and that their identification will be of major importance to our understanding of cell death in the contexts of development and disease.
Materials and Methods
Strains
All C. elegans strains were cultured as described previously [58] and maintained at 20°C. We used Bristol N2 as the wild-type strain, and the mutations used in our experiments are listed below:
LG I. unc-75(e950), ced-1(e1735), csp-3(n4872, tm2260, tm2286), nIs177[Pceh-28::gfp] [59]
LG II. csp-1(n4967, n5133, tm917), mab-10(n5117), lin-29(n836)
LG III. ced-4(n1162, n3158), ced-6(n2095), ced-7(n1996), ced-9(n1653, n2812), tat-1(tm1034), nIs308[Pmec-7::csp-1B, Pmec-3::gfp], nIs400[Pced-1::ced-1ΔC::gfp] [19]
LG IV. csp-2(n4871), ced-5(n1812), dpy-20(e1282), unc-30(e191), ced-3(n2427, n2436, n2452, n3692), nIs309[Pmec-7::csp-1B, Pmec-3::gfp]
LG V. egl-1(n1084 n3082), bcIs39[Plim-7::ced-1::gfp] [45], nIs342[Pegl-1::4×NLS::gfp] [59], qIs56[Plag-2::gfp]
LG X. ced-8(n1891), bzIs8[Pmec-4::gfp] [22], nIs106[Plin-11::gfp] [52]
Unknown linkage. nIs290[Pmec-3::gfp]; nIs307[Pmec-7::csp-1B, Pmec-3::gfp], nIs368-370[Pmec-7::csp-1B(C138S), Pmec-3::gfp], nIs398[Pdyn-1::mfg-e8::Venus] [19], [60]
Extrachromosomal arrays. nEx1646[Pdyn-1::mfg-e8::Venus] [19], [60], nEx1465-71[csp-1(+) (pDD027)], nEx1604-9[csp-1B/C only (pDD030)], nEx1614-16[csp-1A only (pDD029)], nEx1617-19[csp-1-PD (pDD028)]
Plasmids
The Pmec-7::ced-3, Pmec-7::ced-4 [32], Pdyn-1::mfg-e8::Venus [60], Plim-7::ced-1::gfp [45], Pced-1::ced-1ΔC::gfp [46], Plin-11::gfp [52], Pegl-1::gfp and Pceh-28::gfp [59] plasmids were described previously. The csp-1 rescuing plasmid (pDD027) was constructed using PCR to amplify a 9 kb fragment of the csp-1 genomic locus with the primers 5′-gtaacgccagggttttcccagtcacgacggtgatccttcggagcttcag and 5′ - acgaggatatccgcattgag. The resulting amplicon was ligated via the TOPO-TA subcloning protocol into the pCR2.1 vector (Invitrogen). pDD028 (csp-1-PD), pDD029 (csp-1A only), and pDD030 (csp-1B/C only) were constructed using site-directed PCR mutagenesis. Two early stop codons in the csp-1B/C isoforms were generated in pDD028 using the primer 5′-ccgagaacggacgcctagtaatcgaaccataaac and its reverse complement. The csp-1B/C start codon was mutated to an alanine codon in pDD029 using the primer 5′-gactctcagagtcgagcgccgagaacggacgcc and its reverse-complement. Two early stop codons in the csp-1A isoform were generated in pDD030 using the primer 5′cctgaaaacgatagaagataattgataatcacaattcgacgatgatttgg and its reverse complement. The Pmec-7::csp-1A plasmid (pDD003) was constructed using PCR to amplify the csp-1A cDNA from pDD006 using the primers 5′-gcggctagcatggtcctgaaaacgatagaag and 5′-gcgccatggttacatcgaccttgaaaagtgcc, which incorporate the restriction sites NheI and NcoI, respectively, into the resulting amplicon. The csp-1A amplicon was digested with NheI and NcoI and then ligated into the vector pPD52.102. The Pmec-7::csp-1B plasmid (pDD002) was constructed by using PCR to amplify the csp-1B cDNA from pDD001 using the primers 5′-gcggctagcatgccgagaacggacgccaag and 5′-gcgccatggttacatcgaccttgaaaagtgcc, which incorporate the restriction sites NheI and NcoI, respectively. The csp-1B amplicon was digested with NheI and NcoI and then ligated into the vector pPD52.102, which encodes the mec-7 promoter. The Pmec-7::csp-1B(C138S) plasmid (pDD005) was constructed from pDD002 using PCR with the primers 5′-tggatgaactatacaaatagctgcgctccagcgcgttcgt and its reverse complement. The RNAi plasmid pL4440::csp-1-PD (pDD060) was constructed using PCR to amplify the prodomain encoding fragment of the csp-1A cDNA with the primers 5′-gcgagatctatggtcctgaaaacgatagaag and 5′-cgcctcgagatggcgggtttcagctgggtc, which incorporate the restriction sites BglII and XhoI, respectively. The resulting csp-1-PD amplicon was digested with BglII and XhoI and then ligated into pL4440. The RNAi plasmid pL4440::csp-1B (pDD061) was constructed using PCR to amplify the csp-1B cDNA with the primers 5′-gcgagatctatgccgagaacggacgccaag and 5′-cgcctcgagttacatcgaccttgaaaagtgcc, which incorporate the restriction sites BglII and XhoI, respectively. The resulting csp-1B amplicon was digested with BglII and XhoI and then ligated into pL4440.
RNAi experiments
The in vitro transcription, purification, preparation and microinjection of csp-1-PD (pDD060) and csp-1B (pDD061) dsRNA were performed as described previously [61].
Fluorescence in situ hybridization
The fixation of embryos and larval and adult animals, the conjugation of Cy5 or ALEXA594 fluorescent probes to in situ oligo probes, and the hybridization of oligos to fixed samples were performed as described previously [62]. All images were acquired using an inverted Nikon TE-2000 compound microscope equipped for fluorescence microscopy (Prior Scientific). Images were acquired with a PIXIS camera (Princeton Instruments) controlled by MetaMorph software (Molecular Devices) and modified for publication with ImageJ software (NIH). The “total csp-1” set of probes included 32 distinct 20-nucleotide sequences complementary to csp-1B (Biosearch Technologies, Inc). This set of oligos was conjugated to the fluorophore Cy5 (GE Healthcare) and hybridized to all three csp-1 mRNA isoforms (csp-1A, csp-1B and csp-1C). The “csp-1A” set of probes included 32 distinct 20-nucleotide sequences complementary to the region of csp-1A that encodes the prodomain. This set of oligos was conjugated to the fluorophore ALEXA594 (Invitrogen) and hybridized specifically to the csp-1A mRNA isoform. Probe sequences are listed in Table S6.
Cell-death assays and microscopy
The numbers of undead cells that failed to undergo programmed cell death in the anterior pharynges and postdeirid sensilla of L3 larvae were determined by direct observation using Nomarski optics as described previously [28]. Persistent cell corpses in larval heads also were quantified by direct observation using Nomarski optics; for this assay, larvae were staged by the time of hatching. For other cell-death assays, the ventral cord cells of young adults, the M4 neuron and its undead sister cell of L3 larvae, the touch neurons of L4 larvae, and the germ cell corpses of adult hermaphrodite gonads were identified using previously described GFP reporter transgenes [45], [52], [59]. For experiments involving ionizing radiation, L4 larvae were exposed to gamma irradiation from a Co-60 source. All strains were analyzed using a Zeiss Axioskop II compound microscope equipped for fluorescence microscopy. Images were acquired with an ORCA camera (Hammamatsu) controlled by OpenLab software (Perkin Elmer) and modified for publication using ImageJ (NIH).
Transmission electron microscopy
L1-stage larvae were fixed, stained and sectioned for transmission electron microscopy as described previously [43]. Stained sections were imaged with a JEM-1200EX II microscope (JEOL) using an AMT XR41 CCD camera.
Supporting Information
Zdroje
1. KerrJF, WyllieAH, CurrieAR (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26 : 239–257.
2. FadokVA, VoelkerDR, CampbellPA, CohenJJ, BrattonDL, et al. (1992) Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol 148 : 2207–2216.
3. WyllieAH (1980) Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284 : 555–556.
4. YuanJ, KroemerG (2010) Alternative cell death mechanisms in development and beyond. Genes Dev 24 : 2592–2602 doi:10.1101/gad.1984410.
5. YuanJ, ShahamS, LedouxS, EllisHM, HorvitzHR (1993) The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75 : 641–652.
6. YuanJY, HorvitzHR (1990) The Caenorhabditis elegans genes ced-3 and ced-4 act cell autonomously to cause programmed cell death. Dev Biol 138 : 33–41.
7. DegterevA, YuanJ (2008) Expansion and evolution of cell death programmes. Nat Rev Mol Cell Biol 9 : 378–390 doi:10.1038/nrm2393.
8. ChengEH, WeiMC, WeilerS, FlavellRA, MakTW, et al. (2001) BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX - and BAK-mediated mitochondrial apoptosis. Mol Cell 8 : 705–711.
9. CauwelsA, JanssenB, WaeytensA, CuvelierC, BrouckaertP (2003) Caspase inhibition causes hyperacute tumor necrosis factor-induced shock via oxidative stress and phospholipase A2. Nat Immunol 4 : 387–393 doi:10.1038/ni914.
10. SusinSA, LorenzoHK, ZamzamiN, MarzoI, SnowBE, et al. (1999) Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397 : 441–446 doi:10.1038/17135.
11. MadeoF, FröhlichE, FröhlichKU (1997) A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol 139 : 729–734.
12. DwyerDJ, CamachoDM, KohanskiMA, CalluraJM, CollinsJJ (2012) Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell 46 : 561–572 doi:10.1016/j.molcel.2012.04.027.
13. LockshinRA, ZakeriZ (2004) Caspase-independent cell death? Oncogene 23 : 2766–2773 doi:10.1038/sj.onc.1207514.
14. HymanBT, YuanJ (2012) Apoptotic and non-apoptotic roles of caspases in neuronal physiology and pathophysiology. Nat Rev Neurosci 13 : 395–406 doi:10.1038/nrn3228.
15. ShahamS (1998) Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades. J Biol Chem 273 : 35109–35117.
16. ThornberryNA, LazebnikY (1998) Caspases: enemies within. Science 281 : 1312–1316.
17. ShahamS, ReddienPW, DaviesB, HorvitzHR (1999) Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics 153 : 1655–1671.
18. AbrahamMC, LuY, ShahamS (2007) A morphologically conserved nonapoptotic program promotes linker cell death in Caenorhabditis elegans. Dev Cell 12 : 73–86 doi:10.1016/j.devcel.2006.11.012.
19. DenningDP, HatchV, HorvitzHR (2012) Programmed elimination of cells by caspase-independent cell extrusion in C. elegans. Nature 488 : 226–230 doi:10.1038/nature11240.
20. EllisHM, HorvitzHR (1986) Genetic control of programmed cell death in the nematode C. elegans. Cell 44 : 817–829.
21. AbrahamMC, ShahamS (2004) Death without caspases, caspases without death. Trends Cell Biol 14 : 184–193 doi:10.1016/j.tcb.2004.03.002.
22. GengX, ShiY, NakagawaA, YoshinaS, MitaniS, et al. (2008) Inhibition of CED-3 zymogen activation and apoptosis in Caenorhabditis elegans by caspase homolog CSP-3. Nat Struct Mol Biol 15 : 1094–1101 doi:10.1038/nsmb.1488.
23. GengX, ZhouQH, Kage-NakadaiE, ShiY, YanN, et al. (2009) Caenorhabditis elegans caspase homolog CSP-2 inhibits CED-3 autoactivation and apoptosis in germ cells. Cell Death Differ 16 : 1385–1394 doi:10.1038/cdd.2009.88.
24. BlumES, AbrahamMC, YoshimuraS, LuY, ShahamS (2012) Control of nonapoptotic developmental cell death in Caenorhabditis elegans by a polyglutamine-repeat protein. Science 335 : 970–973 doi:10.1126/science.1215156.
25. HengartnerMO, EllisRE, HorvitzHR (1992) Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 356 : 494–499 doi:10.1038/356494a0.
26. Darland-RansomM, WangX, SunC-L, MapesJ, Gengyo-AndoK, et al. (2008) Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science 320 : 528–531 doi:10.1126/science.1155847.
27. WangX, LiW, ZhaoD, LiuB, ShiY, et al. (2010) Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat Cell Biol 12 : 655–664 doi:10.1038/ncb2068.
28. SchwartzHT (2007) A protocol describing pharynx counts and a review of other assays of apoptotic cell death in the nematode worm Caenorhabditis elegans. Nat Protoc 2 : 705–714 doi:10.1038/nprot.2007.93.
29. SimmerF, TijstermanM, ParrishS, KoushikaSP, NonetML, et al. (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12 : 1317–1319.
30. SulstonJE, SchierenbergE, WhiteJG, ThomsonJN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100 : 64–119.
31. ChangHC, PaekJ, KimDH (2011) Natural polymorphisms in C. elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature 480 : 525–529 doi:10.1038/nature10643.
32. ShahamS, HorvitzHR (1996) Developing Caenorhabditis elegans neurons may contain both cell-death protective and killer activities. Genes Dev 10 : 578–591.
33. XueD, ShahamS, HorvitzHR (1996) The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev 10 : 1073–1083.
34. SeshagiriS, MillerLK (1997) Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis. Curr Biol 7 : 455–460.
35. YangX, ChangHY, BaltimoreD (1998) Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science 281 : 1355–1357.
36. QiS, PangY, HuQ, LiuQ, LiH, et al. (2010) Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141 : 446–457 doi:10.1016/j.cell.2010.03.017.
37. ChenF, HershBM, ConradtB, ZhouZ, RiemerD, et al. (2000) Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science 287 : 1485–1489.
38. ChinnaiyanAM, O'RourkeK, LaneBR, DixitVM (1997) Interaction of CED-4 with CED-3 and CED-9: a molecular framework for cell death. Science 275 : 1122–1126.
39. XueD, HorvitzHR (1997) Caenorhabditis elegans CED-9 protein is a bifunctional cell-death inhibitor. Nature 390 : 305–308 doi:10.1038/36889.
40. ConradtB, HorvitzHR (1998) The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93 : 519–529.
41. PourkarimiE, GreissS, GartnerA (2012) Evidence that CED-9/Bcl2 and CED-4/Apaf-1 localization is not consistent with the current model for C. elegans apoptosis induction. Cell Death Differ 19 : 406–415 doi:10.1038/cdd.2011.104.
42. HengartnerMO, HorvitzHR (1994) Activation of C. elegans cell death protein CED-9 by an amino-acid substitution in a domain conserved in Bcl-2. Nature 369 : 318–320 doi:10.1038/369318a0.
43. GumiennyTL, LambieE, HartwiegE, HorvitzHR, HengartnerMO (1999) Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126 : 1011–1022.
44. GartnerA, MilsteinS, AhmedS, HodgkinJ, HengartnerMO (2000) A Conserved Checkpoint Pathway Mediates DNA Damage–Induced Apoptosis and Cell Cycle Arrest in C. elegans. Molecular Cell 5 : 435–443 doi:10.1016/S1097-2765(00)80438-4.
45. SchumacherB, SchertelC, WittenburgN, TuckS, MitaniS, et al. (2005) C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ 12 : 153–161 doi:10.1038/sj.cdd.4401539.
46. ZhouZ, HartwiegE, HorvitzHR (2001) CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104 : 43–56.
47. HarrisDT, HorvitzHR (2011) MAB-10/NAB acts with LIN-29/EGR to regulate terminal differentiation and the transition from larva to adult in C. elegans. Development 138 : 4051–4062 doi:10.1242/dev.065417.
48. SulstonJE, HorvitzHR (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56 : 110–156.
49. HedgecockEM, SulstonJE, ThomsonJN (1983) Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220 : 1277–1279.
50. EllisRE, JacobsonDM, HorvitzHR (1991) Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129 : 79–94.
51. KinchenJM, DoukoumetzidisK, AlmendingerJ, StergiouL, Tosello-TrampontA, et al. (2008) A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol 10 : 556–566 doi:10.1038/ncb1718.
52. ReddienPW, CameronS, HorvitzHR (2001) Phagocytosis promotes programmed cell death in C. elegans. Nature 412 : 198–202 doi:10.1038/35084096.
53. HoeppnerDJ, HengartnerMO, SchnabelR (2001) Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412 : 202–206 doi:10.1038/35084103.
54. Pinan-LucarreB, GabelCV, ReinaCP, HulmeSE, ShevkoplyasSS, et al. (2012) The core apoptotic executioner proteins CED-3 and CED-4 promote initiation of neuronal regeneration in Caenorhabditis elegans. PLoS Biol 10: e1001331 doi:10.1371/journal.pbio.1001331.
55. AramaE, AgapiteJ, StellerH (2003) Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell 4 : 687–697.
56. XuK, TavernarakisN, DriscollM (2001) Necrotic cell death in C. elegans requires the function of calreticulin and regulators of Ca(2+) release from the endoplasmic reticulum. Neuron 31 : 957–971.
57. HarbinderS, TavernarakisN, HerndonLA, KinnellM, XuSQ, et al. (1997) Genetically targeted cell disruption in Caenorhabditis elegans. Proc Natl Acad Sci USA 94 : 13128–13133.
58. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
59. HiroseT, GalvinBD, HorvitzHR (2010) Six and Eya promote apoptosis through direct transcriptional activation of the proapoptotic BH3-only gene egl-1 in Caenorhabditis elegans. Proc Natl Acad Sci USA 107 : 15479–15484 doi:10.1073/pnas.1010023107.
60. VenegasV, ZhouZ (2007) Two alternative mechanisms that regulate the presentation of apoptotic cell engulfment signal in Caenorhabditis elegans. Mol Biol Cell 18 : 3180–3192 doi:10.1091/mbc.E07-02-0138.
61. AndersenEC, LuX, HorvitzHR (2006) C. elegans ISWI and NURF301 antagonize an Rb-like pathway in the determination of multiple cell fates. Development 133 : 2695–2704 doi:10.1242/dev.02444.
62. RajA, van den BogaardP, RifkinSA, van OudenaardenA, TyagiS (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5 : 877–879 doi:10.1038/nmeth.1253.
Štítky
Genetika Reprodukční medicína
Článek Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in KoreansČlánek Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal ProteomesČlánek RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria inČlánek Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein ResponseČlánek Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 3- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Power and Predictive Accuracy of Polygenic Risk Scores
- Rare Copy Number Variants Are a Common Cause of Short Stature
- Coordination of Flower Maturation by a Regulatory Circuit of Three MicroRNAs
- Ubiquitous Polygenicity of Human Complex Traits: Genome-Wide Analysis of 49 Traits in Koreans
- Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution
- Mechanistic Insight into the Pathology of Polyalanine Expansion Disorders Revealed by a Mouse Model for X Linked Hypopituitarism
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
- Problem Solved: An Interview with Sir Edwin Southern
- Long Interspersed Element–1 (LINE-1): Passenger or Driver in Human Neoplasms?
- Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis
- Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes
- A WRKY Transcription Factor Recruits the SYG1-Like Protein SHB1 to Activate Gene Expression and Seed Cavity Enlargement
- Microhomology-Mediated Mechanisms Underlie Non-Recurrent Disease-Causing Microdeletions of the Gene or Its Regulatory Domain
- Ancient Evolutionary Trade-Offs between Yeast Ploidy States
- Differential Evolutionary Fate of an Ancestral Primate Endogenous Retrovirus Envelope Gene, the EnvV , Captured for a Function in Placentation
- A Feed-Forward Loop Coupling Extracellular BMP Transport and Morphogenesis in Wing
- The Tomato Yellow Leaf Curl Virus Resistance Genes and Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases
- The U-Box E3 Ubiquitin Ligase TUD1 Functions with a Heterotrimeric G α Subunit to Regulate Brassinosteroid-Mediated Growth in Rice
- Role of the DSC1 Channel in Regulating Neuronal Excitability in : Extending Nervous System Stability under Stress
- –Independent Phenotypic Switching in and a Dual Role for Wor1 in Regulating Switching and Filamentation
- Pax6 Regulates Gene Expression in the Vertebrate Lens through miR-204
- Blood-Informative Transcripts Define Nine Common Axes of Peripheral Blood Gene Expression
- Genetic Architecture of Skin and Eye Color in an African-European Admixed Population
- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Estrogen Mediated-Activation of miR-191/425 Cluster Modulates Tumorigenicity of Breast Cancer Cells Depending on Estrogen Receptor Status
- Complex Patterns of Genomic Admixture within Southern Africa
- Yap- and Cdc42-Dependent Nephrogenesis and Morphogenesis during Mouse Kidney Development
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly
- Polycomb Group Gene Regulates Rice () Seed Development and Grain Filling via a Mechanism Distinct from
- RFX Transcription Factor DAF-19 Regulates 5-HT and Innate Immune Responses to Pathogenic Bacteria in
- Distinct Molecular Strategies for Hox-Mediated Limb Suppression in : From Cooperativity to Dispensability/Antagonism in TALE Partnership
- A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan
- TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study in Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk
- Coincident Resection at Both Ends of Random, γ–Induced Double-Strand Breaks Requires MRX (MRN), Sae2 (Ctp1), and Mre11-Nuclease
- Identification of a -Specific Modifier Locus at 6p24 Related to Breast Cancer Risk
- A Novel Function for the Hox Gene in the Male Accessory Gland Regulates the Long-Term Female Post-Mating Response in
- Tdp2: A Means to Fixing the Ends
- A Novel Role for the RNA–Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating mRNA Stability
- Association Mapping and the Genomic Consequences of Selection in Sunflower
- Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse
- A Novel Mutation in the Upstream Open Reading Frame of the Gene Causes a MEN4 Phenotype
- Ataxin1L Is a Regulator of HSC Function Highlighting the Utility of Cross-Tissue Comparisons for Gene Discovery
- Human Spermatogenic Failure Purges Deleterious Mutation Load from the Autosomes and Both Sex Chromosomes, including the Gene
- A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of piRNAs
- Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci
- Matefin/SUN-1 Phosphorylation Is Part of a Surveillance Mechanism to Coordinate Chromosome Synapsis and Recombination with Meiotic Progression and Chromosome Movement
- A Role for the Malignant Brain Tumour (MBT) Domain Protein LIN-61 in DNA Double-Strand Break Repair by Homologous Recombination
- The Population and Evolutionary Dynamics of Phage and Bacteria with CRISPR–Mediated Immunity
- Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB
- Surveillance-Activated Defenses Block the ROS–Induced Mitochondrial Unfolded Protein Response
- DNA Topoisomerase III Localizes to Centromeres and Affects Centromeric CENP-A Levels in Fission Yeast
- Genome-Wide Control of RNA Polymerase II Activity by Cohesin
- Divergent Selection Drives Genetic Differentiation in an R2R3-MYB Transcription Factor That Contributes to Incipient Speciation in
- NODULE INCEPTION Directly Targets Subunit Genes to Regulate Essential Processes of Root Nodule Development in
- Spreading of a Prion Domain from Cell-to-Cell by Vesicular Transport in
- Deficiency in Origin Licensing Proteins Impairs Cilia Formation: Implications for the Aetiology of Meier-Gorlin Syndrome
- Deficiency Reduces Adipose OXPHOS Capacity and Triggers Inflammation and Insulin Resistance in Mice
- The Conserved SKN-1/Nrf2 Stress Response Pathway Regulates Synaptic Function in
- Functional Genomic Analysis of the Regulatory Network in
- Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- CRL2 E3-Ligase Regulates Proliferation and Progression through Meiosis in the Germline
- Both the Caspase CSP-1 and a Caspase-Independent Pathway Promote Programmed Cell Death in Parallel to the Canonical Pathway for Apoptosis in
- PRMT4 Is a Novel Coactivator of c-Myb-Dependent Transcription in Haematopoietic Cell Lines
- A Copy Number Variant at the Locus Likely Confers Risk for Canine Squamous Cell Carcinoma of the Digit
- Evidence of Gene–Environment Interactions between Common Breast Cancer Susceptibility Loci and Established Environmental Risk Factors
- HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis
- Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained
- A Gene Transfer Agent and a Dynamic Repertoire of Secretion Systems Hold the Keys to the Explosive Radiation of the Emerging Pathogen
- The Role of ATM in the Deficiency in Nonhomologous End-Joining near Telomeres in a Human Cancer Cell Line
- Dynamic Circadian Protein–Protein Interaction Networks Predict Temporal Organization of Cellular Functions
- Nuclear Myosin 1c Facilitates the Chromatin Modifications Required to Activate rRNA Gene Transcription and Cell Cycle Progression
- Robust Prediction of Expression Differences among Human Individuals Using Only Genotype Information
- A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist
- The Role of the Arabidopsis Exosome in siRNA–Independent Silencing of Heterochromatic Loci
- Elevated Expression of the Integrin-Associated Protein PINCH Suppresses the Defects of Muscle Hypercontraction Mutants
- Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest
- Genome-Wide Testing of Putative Functional Exonic Variants in Relationship with Breast and Prostate Cancer Risk in a Multiethnic Population
- Heteroduplex DNA Position Defines the Roles of the Sgs1, Srs2, and Mph1 Helicases in Promoting Distinct Recombination Outcomes
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fine Characterisation of a Recombination Hotspot at the Locus and Resolution of the Paradoxical Excess of Duplications over Deletions in the General Population
- Molecular Networks of Human Muscle Adaptation to Exercise and Age
- Recurrent Rearrangement during Adaptive Evolution in an Interspecific Yeast Hybrid Suggests a Model for Rapid Introgression
- Genome-Wide Association Study and Gene Expression Analysis Identifies as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



