-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
Ciliated protists rearrange their genomes dramatically during nuclear development via chromosome fragmentation and DNA deletion to produce a trimmer and highly reorganized somatic genome. The deleted portion of the genome includes potentially active transposons or transposon-like sequences that reside in the germline. Three independent studies recently showed that transposase proteins of the DDE/DDD superfamily are indispensible for DNA processing in three distantly related ciliates. In the spirotrich Oxytricha trifallax, high copy-number germline-limited transposons mediate their own excision from the somatic genome but also contribute to programmed genome rearrangement through a remarkable transposon mutualism with the host. By contrast, the genomes of two oligohymenophorean ciliates, Tetrahymena thermophila and Paramecium tetraurelia, encode homologous PiggyBac-like transposases as single-copy genes in both their germline and somatic genomes. These domesticated transposases are essential for deletion of thousands of different internal sequences in these species. This review contrasts the events underlying somatic genome reduction in three different ciliates and considers their evolutionary origins and the relationships among their distinct mechanisms for genome remodeling.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003659
Category: Review
doi: https://doi.org/10.1371/journal.pgen.1003659Summary
Ciliated protists rearrange their genomes dramatically during nuclear development via chromosome fragmentation and DNA deletion to produce a trimmer and highly reorganized somatic genome. The deleted portion of the genome includes potentially active transposons or transposon-like sequences that reside in the germline. Three independent studies recently showed that transposase proteins of the DDE/DDD superfamily are indispensible for DNA processing in three distantly related ciliates. In the spirotrich Oxytricha trifallax, high copy-number germline-limited transposons mediate their own excision from the somatic genome but also contribute to programmed genome rearrangement through a remarkable transposon mutualism with the host. By contrast, the genomes of two oligohymenophorean ciliates, Tetrahymena thermophila and Paramecium tetraurelia, encode homologous PiggyBac-like transposases as single-copy genes in both their germline and somatic genomes. These domesticated transposases are essential for deletion of thousands of different internal sequences in these species. This review contrasts the events underlying somatic genome reduction in three different ciliates and considers their evolutionary origins and the relationships among their distinct mechanisms for genome remodeling.
Introduction
A transposon rearranges its host's genome when it moves from one genomic locus to another. When they invade coding or regulatory regions, transposons can alter gene expression. Furthermore, transposon-induced DNA double-strand breaks can cause chromosomal rearrangements and subsequent aneuploidy. Thus, transposons were long considered as harmful and selfish “junk DNA” [1]. However, because most eukaryotic genomes have maintained transposons and transposon-derived DNA throughout the course of evolution, it is possible that they sometimes confer an adaptive benefit to the host [2]. Because maintenance in the host genome also benefits the transposon, this would be a form of mutualism. Transposons can also accelerate genome evolution by fabricating new sequences and facilitating genome rearrangement.
Often the host manages to recruit or “domesticate” transposon-encoded genes and repurpose them for new host functions [3], [4]. A domestication event typically alters the transposon-derived sequence, curtailing its mobility. Thus it no longer meets the functional definition of a transposon. A famous example in jawed vertebrates is the evolution of the RAG1 gene from a Transib-like element. Now a key component in V(D)J recombination, it is responsible for cutting and rejoining V, D, and J segments [5],[6]. As this process is indispensable for maturation of B and T cells, the RAG1 gene domestication enabled the evolution of adaptive immunity [6]. Other examples of domesticated transposases include the yeast Klyveromyces lactis α3 MULE transposase-like protein, which enables mating-type switching [7]. In addition, C. elegans HIM-17 is a domesticated P-element–like transposase that is essential for double-strand break and chiasma formation during meiosis, as well as for the accumulation of histone H3 methylation at lysine 9 on meiotic prophase chromosomes [8]. Therefore, transposon domestication is widespread, and transposons supply toolkits for host cells to evolve new functions. However, the processes by which transposons become domesticated can vary.
Recently, three groups discovered crucial roles for transposase-related proteins in large-scale genomic rearrangements in three different ciliate species [9]–[11]. Paramecium and Tetrahymena (both Oligohymenophorea) use single-copy domesticated transposase genes for genomic rearrangements. Curiously, Oxytricha trifallax, a member of a different, deeply diverged ciliate class (Spirotrichea), requires instead the expression of thousands of active transposase genes that still reside in intact—and potentially active—transposons. Therefore, comparison of these different transposon-derived systems offers a unique opportunity to put in a broad evolutionary context two different scenarios for the recruitment, or “exaptation” [12], of either active or modified transposon functions in the emergence of new biological pathways.
Programmed Genome Remodeling in Ciliates
Ciliates are microbial eukaryotes and members of the Alveolata that include dinoflagellates and apicomplexan parasites [13]. A common feature is nuclear dimorphism, with two types of nuclei in the same cytoplasm. The larger DNA-rich somatic macronucleus [14] provides most gene expression during vegetative growth. The smaller germline micronucleus [14] is diploid and transcriptionally active mainly during conjugation. Actual numbers of macronuclei and micronuclei vary among ciliate species [14]. Oxytricha trifallax and Paramecium tetraurelia each have one macronucleus and two micronuclei in interphase vegetative cells, whereas Tetrahymena thermophila has one macronucleus and one micronucleus (Figure 1A) [14]. During asexual division, both nuclei divide; whereas during sexual conjugation, the zygotic micronucleus gives rise to both a new macronucleus and a new micronucleus, supplying the next generation with all its genetic information. However, the macronucleus and micronucleus differ substantially in their genetic content because the somatic genome undergoes an elaborate cascade of events that produces a new macronucleus from the zygotic micronucleus, after cell mating [15], [16].
Fig. 1. Nuclear dimorphism and genome rearrangements in ciliates. 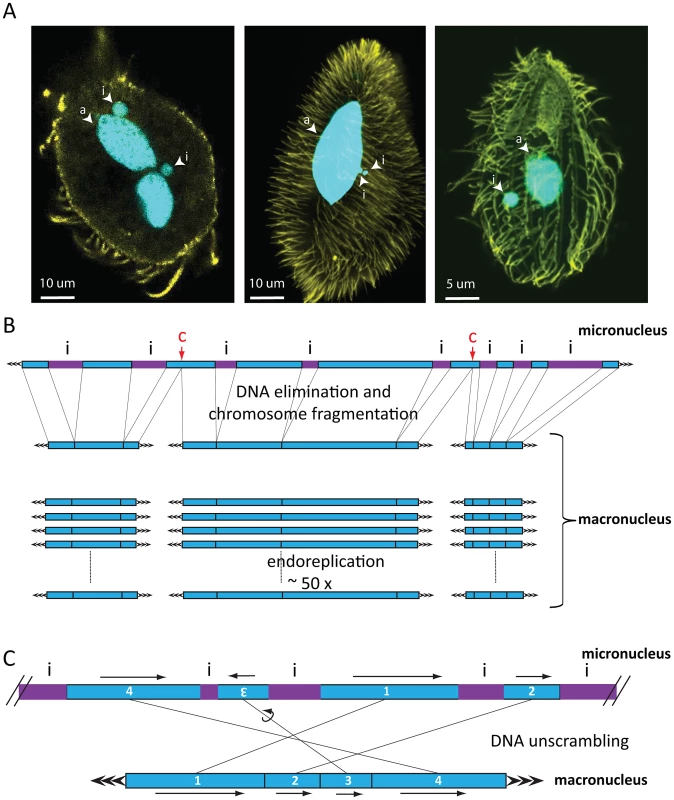
A) From left to right: Oxytricha trifallax, Paramecium tetraurelia, Tetrahymena thermophila. DNA is shown in cyan, yellow represents tubulin staining. Images were kindly provided by Wenwen Fang (Princeton University, Princeton), Kensuke Kataoka (IMBA, Vienna), and Janine Beisson (CNRS, Gif sur Yvette). Abbreviations: i = micronucleus, a = macronucleus. In Oxytricha trifallax, two lobes of a macronucleus are connected by a thin nuclear bridge (not visible in the image). B) Genome rearrangements in all ciliates shown include elimination of micronucleus (MIC)-limited sequences (i, purple IES) and chromosome breakage, which in Tetrahymena occurs at specific chromosome breakage sites (labeled c). After religation of the flanking macronuclear (MAC) sequences, Tetrahymena chromosomes undergo endoreplication to produce 50 identical copies. C) DNA unscrambling in Oxytricha involves the reshuffling and occasional inversion of precursor micronuclear (MIC) sequences (numbered blue boxes) to assemble them in the correct macronuclear order. Genome rearrangements during macronuclear development in ciliates delete large portions of germline DNA and consequently produce greater numbers of small, somatic chromosomes (on the order of 16,000 different types in Oxytricha trifallax [17]) than their longer, germline chromosomes. There is considerable variation in this process between major lineages. Macronuclear chromosomes in oligohymenophorean ciliates have an average size of 300 kbp in Tetrahymena and 800 kbp in Paramecium [18]–[20]. In contrast, spirotrichous ciliates like Oxytricha typically have gene-sized nanochromosomes in the macronucleus, which average just 3.2 kbp including short telomeres, and 90% encode just a single gene [17]. DNA elimination discards between 20% (Tetrahymena) and ∼95% (Oxytricha) of the entire germline genome during macronuclear development [16]. In some spirotrichs, as well as phyllopharyngeans [21], rearrangements in some loci also require DNA unscrambling (Figure 1C). These often complex events reorder gene pieces in the micronucleus by translocation or inversion to assemble coding information in the macronucleus [22].
Despite the genome downsizing via DNA elimination, the macronucleus contains a greater quantity of DNA than the micronucleus. This is because macronuclear chromosomes undergo endoreplication to amplification levels that typically range from 50-fold in Tetrahymena to 800-fold in Paramecium and up to 2,000-fold in Oxytricha [16]. This review focuses on DNA elimination. For a review of genome unscrambling and the role of RNA-regulated epigenetic effects in this process, as well as DNA amplification, we refer the reader to Nowacki et al. (2011) [22], and for a summary of the relationship among ciliate species with available genome information and amplification levels, we refer the reader to Figure 2 of Swart et al. (2013) [17]. Although DNA elimination is common to most ciliates, recent studies that we describe below revealed a dependence on strikingly different groups of transposase-related proteins for DNA elimination in different classes of ciliates [9]–[11].
Deletion of Germline-Limited Sequences in Oxytricha
Elimination of germline-restricted DNA sequences usually occurs at precise, nucleotide-level resolution in Oxytricha trifallax. One well-studied example of precisely removed germline-limited sequences are the Tc1/mariner transposons of the TBE (telomere-bearing element) class, which are present in thousands of copies in the micronucleus [23] and occupy roughly as much of the micronuclear genome as its estimated coding content. TBE terminal regions possess inverted repeats, with the most distal 17 bp composed of telomeric repeats ((G4T4)2G)n, and the elements are flanked by a 3 bp 5′-ANT-3′ target site duplication (Figure 2). TBE excision precisely removes one target site repeat, thereby restoring functional open reading frames (ORFs) even when TBEs interrupt protein-coding regions in the micronucleus. Mechanistically, it is likely that introduction of a double-stranded break (DSB), creating a 3 nt 5′ protruding end on one side of the transposon, initiates excision. The other target site serves as an “integration site” so that TBEs excise in a circular form, the TBE ring degrades, and macronuclear DNA religates [24] (Figure 2).
Fig. 2. TBE transposases in Oxytricha are germline-limited sequences and they participate in their own removal. 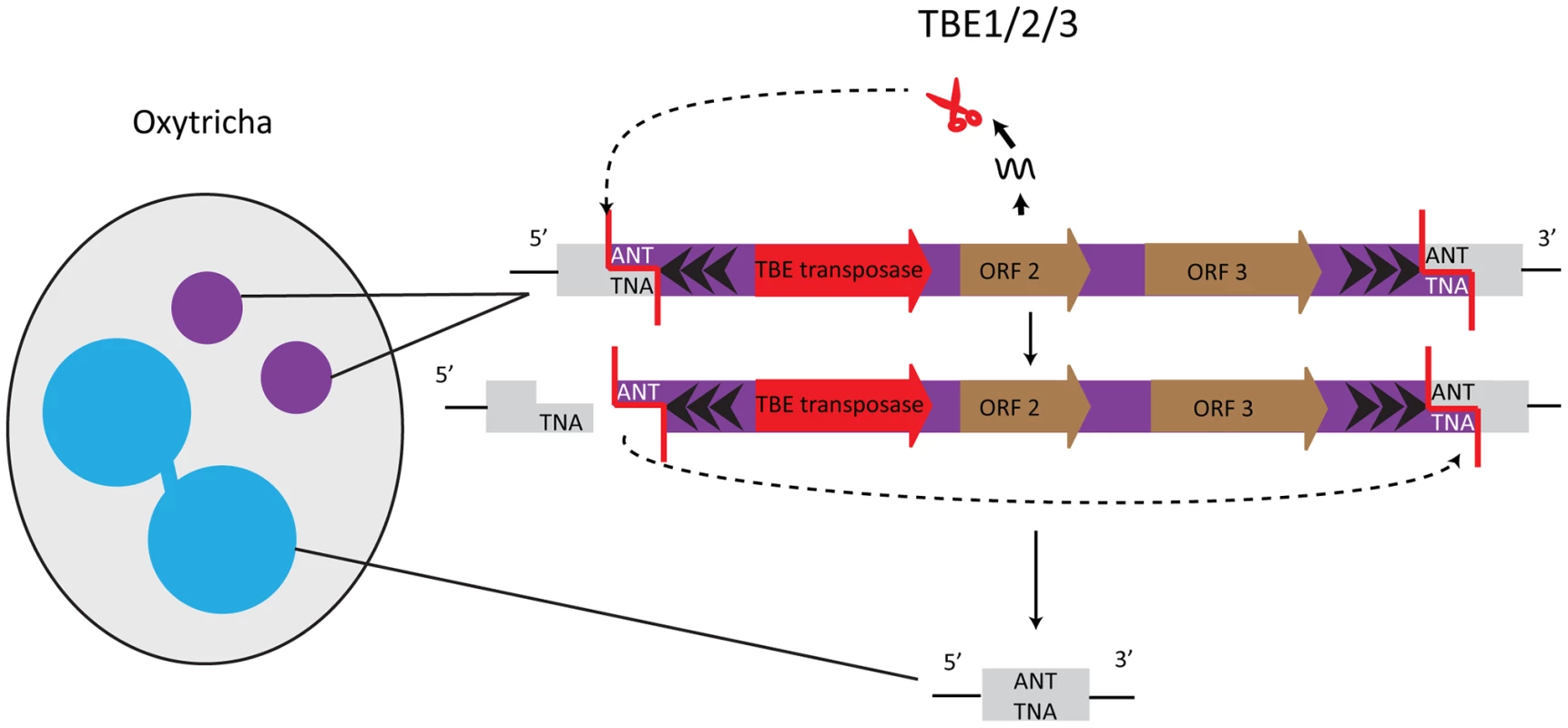
The encoded transposases of the Tc1/mariner family have a DDE catalytic motif. Cleavage of the germline-limited sequences starts with a 3 nucleotide 5′ overhang at an ANT recognition site; the second target site serves as the integration site [24]. One of the TBE-encoded genes encodes a protein belonging to the DDE transposase superfamily, suggesting involvement of this enzyme in the transposon's own removal [24], [25] (Figure 2). Furthermore, all three TBE transposon ORFs appear to be under purifying selection, which initially hinted at an important function of the transposases [26], [27]. Oxytricha trifallax has three different types of TBE transposons: TBE1, TBE2, and TBE3. The transposases encoded by these elements share ≥83% similarity at the protein level, and all three types of transposases are specifically expressed during macronuclear development when DNA rearrangements occur. RNAi against all three groups of TBE transposases in unison (but not individually) results in severe defects in elimination of both TBE transposons and non-TBE micronucleus-limited elements, as well as an accumulation of high molecular weight DNA [10]. These results lead to two non–mutually exclusive hypotheses: first, that TBE transposases act redundantly in excising both the transposons that encode them and other micronucleus-limited sequences (“internal eliminated sequences” or IESs); and second, because this experiment silenced thousands of paralogs that occupy a significant fraction of the germline genome, it suggests that a massive quantity of transposase may be required for Oxytricha genome rearrangement [10].
DNA Deletion in Paramecium and Tetrahymena
Paramecium tetraurelia has two types of eliminated sequences. Most repetitive micronucleus-limited sequences, similar to minisatellites or transposons, are eliminated imprecisely [16], [28]. In contrast, removal of approximately 45,000 different non-repetitive, single-copy IESs occurs precisely [29]. Though both types of eliminated sequences are removed reproducibly, elimination can produce microheterogeneity within a few base pairs [29]. Paramecium IESs are flanked by a 5′-TA-3′ dinucleotide, part of a weakly conserved 8 bp sequence with similarity to the recognition sequence of some Tc1/mariner transposases [30], [31]. This led Klobutcher and Herrick in an elegant model [25] to propose that some Paramecium IESs are remnants of Tc1/mariner transposons. However, IES excision starts with a double-strand break that produces a 4-base 5′-overhang [32] (Figure 3), whereas Tc1/mariner transposases yield 2-base 3′-overhangs [33], so this model would require either extensive modification of the original transposase mechanism during domestication, or recruitment of different enzymes.
Fig. 3. Transposases in Tetrahymena and Paramecium belong to the PiggyBac family. 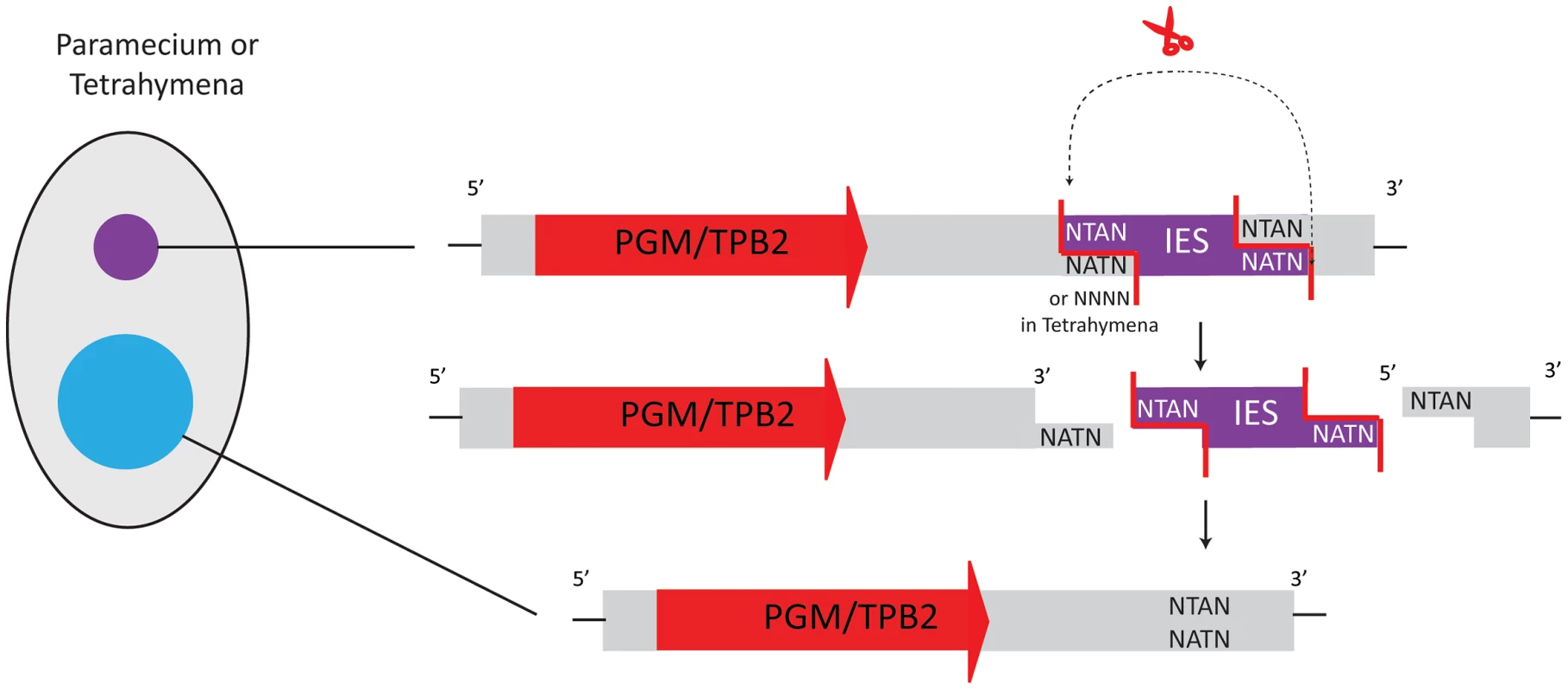
As domesticated transposases, they are present as single-copy genes in the micronuclear and macronuclear genomes. After expression from the somatic genome, they facilitate IES excision from the new macronuclear genome. IES removal occurs via a 4-base 5′ protruding end. In Paramecium, all deleted sequences have a TA dinucleotide at both boundaries, whereas Tetrahymena displays no consensus sequence. IES elimination in Tetrahymena thermophila [34] also produces double-strand breaks with 4-base 5′-overhangs. Tetrahymena removes ∼6,000–9,000 different IESs from its developing macronucleus [35], [36], an order of magnitude fewer IESs than Paramecium. Tetrahymena IESs are typically larger than in Paramecium (from ∼200 bp to >20 kbp). Most are eliminated imprecisely, leaving heterogeneity in the resulting macronuclear sequences. Hence they rarely interrupt exons, with few exceptions [37] that would be weakly conserved regions. Some IESs do bear similarity to Tc1/mariner transposons or non-LTR retrotranposons [19], [38]. Although no obvious consensus sequence exists at Tetrahymena IES boundaries, the DNA double-strand breaks (DSB) in both Paramecium and Tetrahymena produce 4-base 5′-overhangs [32], [34]. This suggested similar enzymes for DNA elimination and led researchers to search for PiggyBac transposases that could produce such ends.
Indeed, the macronuclear genomes of both species contain genes derived from PiggyBac family transposases, and Baudry et al. and Cheng et al. independently showed that a transposase of the PiggyBac family plays a crucial role in DNA elimination during maturation of the macronuclear genomes in Paramecium and Tetrahymena [9], [11]. These ciliate transposon-derived proteins are called Pgm (PiggyMac) in Paramecium and Tpb2p (Tetrahymena PiggyBac-like transposase 2) in Tetrahymena. In both Paramecium and Tetrahymena, silencing of the respective PiggyBac transposase-like genes by RNAi inhibits the processes of DNA elimination and macronuclear development [9], [11]. Both Pgm and Tpb2p have a predicted catalytic domain with conserved DDD residues, similar to PiggyBac transposases. In vitro studies with Tpb2p recombinantly expressed in E. coli revealed that Tpb2p produces a double-strand break leaving a 4-base 5′ protruding end, which correlates with the typical cleavage signature of canonical PiggyBac transposases [11], [39] and the observed form of DSB during DNA elimination in vivo [34] (Figure 3). Therefore, Pgm and Tpb2p are probably the enzymes responsible for catalyzing DNA excision during DNA elimination in Paramecium and Tetrahymena, respectively.
Both Pgm and Tpb2p localize to the newly developing macronucleus during DNA elimination. Tpb2p localizes to the subnuclear heterochromatin bodies where DNA elimination is thought to occur [9], [11]. These heterochromatin structures contain heterochromatin-specific histone modifications, tri-methylated histone H3 lysine 9 (H3K9me3) and lysine 27 (H3K27me3), and the chromodomain protein Pdd1p [40]–[42]. Localization of Tpb2p to these structures could be mediated by an interaction with some of these or other heterochromatin components. Pgm and Tpb2p share a predicted zinc finger domain and coiled-coil domain (Figure 4) that may directly interact with some of the heterochromatin components. Because heterochromatin is specifically established on Tetrahymena IESs prior to their elimination via an RNAi-related pathway [41], [42], the heterochromatin interaction of Tpb2p, and possibly other PiggyBac transposase-like proteins, may restrict their action to programmed deleted sequences.
Fig. 4. Phylogenetic analysis of representative transposases of the DDE/DDD superfamily. 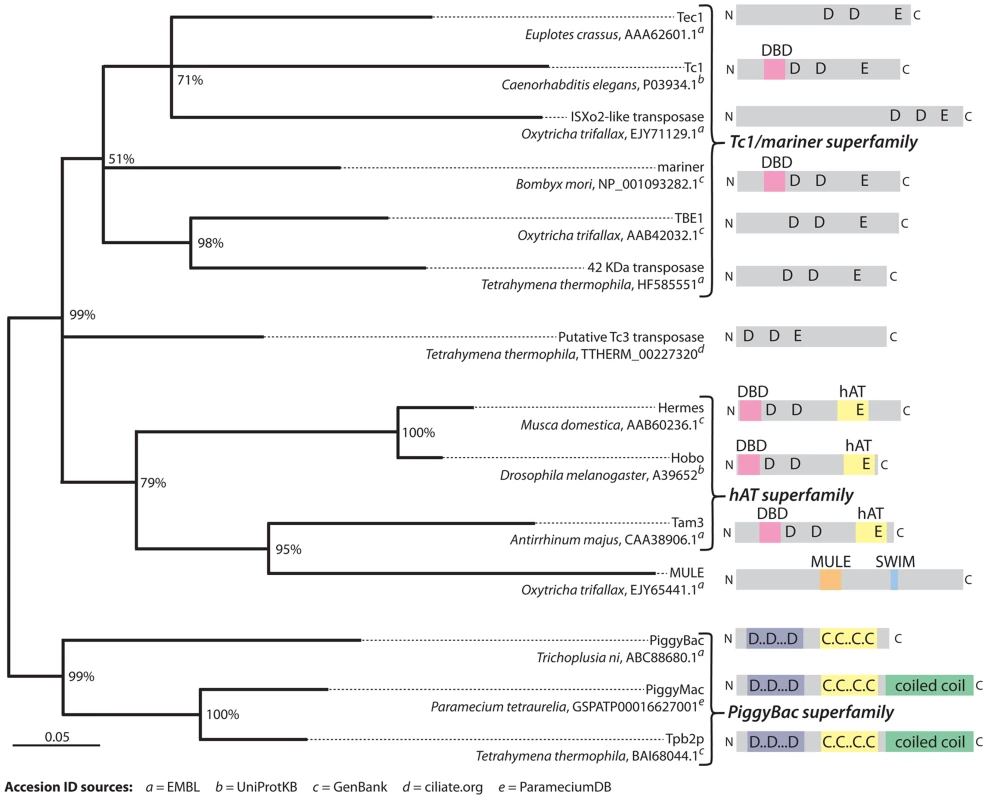
This tree supports the conclusion that TBE elements belong to the Tc1/mariner superfamily of transposons and also that there appear to be TBE-like elements present in Tetrahymena (labeled “42 kDa transposase”). Additionally, this analysis supports the conclusion that the two PiggyBac-like transposases, Pgm and Tpb2p, in Paramecium and Tetrahymena are homologous to each other. The grouping of the MULE family representative [17] within hAT transposases is unexpected [47] and possibly the result of an alignment artifact due to its disproportionately long sequence. Recently discovered Paramecium transposases Sardine, Thon, AnchoisA, and AnchoisB [29] were omitted because their inclusion in the analysis significantly lowered confidence scores for a majority of branches. The tree was created with MRBayes phylogenetic inference software [48] using the alignment shown in Dataset S1, which was edited to remove regions with gaps in the consensus sequence. The phylogeny was generated using a mixed amino acid substitution model and invariable gamma distribution rate model over 200,000 iterations with a burn-in of 25%. Branch confidence values represent conditional probabilities generated by the Bayesian inference process. The scale bar corresponds to 0.05 expected substitutions per site of the unmasked alignment positions. Domain and motif annotations were produced using the Pfam web server [49]. In addition to the association of PiggyBac transposase-like proteins with heterochromatin, their enzymatic preference for certain DNA sequences may both facilitate IES elimination and constrain it evolutionarily. For instance, the 5′-TA-3′ dinucleotide that flanks Paramecium IESs is also the smallest sequence recognized by the canonical PiggyBac transposase, whose optimal recognition sequence is 5′-TTAA-3′ [39]. Moreover, recombinant Tpb2p in solution can specifically cleave a dsDNA oligonucleotide containing 5′-TTAA-3′ before the first T [11] and cleaves the left boundary of a deleted region in Tetrahymena (5′-AGTGAT-3′) between the first A and G, when this motif is placed in the middle of an otherwise randomly designed 50 bp dsDNA oligonucleotide [11]. Therefore, although a 5′-TA-3′ is not necessary for Tpb2p cleavage, the enzyme probably recognizes limited sequence context. However, it is unlikely that primary DNA sequence is the sole determinant. Most likely, both heterochromatin interactions of Tpb2p and Pgm and their preference for certain DNA sequences determine cleavage sites for DNA elimination.
Transposon Domestication versus Mutualism: Possible Evolutionary Origins
Although Pgm and Tpb2p are similar to PiggyBac transposases, they are not present in active transposons and their genes are single-copy in the macronucleus. Thus, they are classic examples of transposon domestication by the host genome to mediate a new function—in this case, DNA elimination, consistent with [25]. Pgm and Tpb2p share 30% global identity, which either suggests a single domestication of a PiggyBac-like transposase in their oligohymenophorean ancestor or independent recruitments of related transposases. No PiggyBac-like transposase has been found in Oxytricha. After the Paramecium and Tetrahymena lineages separated, the domesticated transposases accumulated substitutions that could contribute to the differences in their DNA deletion pathways, as well as the apparent promiscuity of Tpb2p at Tetrahymena IES boundaries.
Recruitment of a single domesticated transposase in Oligohymenophorea is in sharp contrast to Oxytricha's distributed system that appears to enlist an army of thousands of TBE transposases that still reside in potentially active Tc1/mariner transposons (Figure 4) and occupy a significant fraction of the germline genome (the transposon “bloom” phase of Klobutcher and Herrick's model for IES origins from transposons [25]). Oxytricha macronuclear development requires hundreds of thousands of rearrangement events, which may explain its need for increased transposase participation. The greater complexity of genome rearrangements does not, however, explain why Oxytricha should recruit undomesticated transposases to facilitate genome rearrangement. This strategy may be easier to evolve, as active transposons would multiply in number, up to the ceiling tolerated by its host, which would ensure production of an ample quantity of transposase, in part because these enzymes also facilitate elimination of their parent transposons. This achieves a mutualistic evolutionary balance between the host and its resident germline transposons [43]. It also wonderfully displays a functional and essential role for this otherwise dispensable portion of the micronuclear genome [10]. During the divergence of spirotrichs, TBE transposases may have gained promiscuity and acquired the ability to excise off-target DNA sequences [24], [25]. Additionally, only DNA insertions with the ability to be excised by TBE transposases or other active enzymes would have been tolerated in the germline over time, and thus could accumulate. Such a mutualism [43] would have allowed not only the accumulation of germline transposons but also the production of a sufficient quantity of transposase protein to facilitate Oxytricha's elaborate process of genome remodeling and also exclude these active transposons from the soma. These requirements could have provided the selective pressure to maintain high transposon copy numbers to facilitate DNA elimination. Conversely, the DNA elimination events that domesticated PiggyBac transposases facilitate in Paramecium and Tetrahymena do not require maintenance of germline transposons. This striking difference in two evolutionary lineages separated by over a billion years may have been exaggerated over time by an evolving trend in the Oxytricha lineage to eliminate and rearrange considerably more of its micronuclear genome.
The ostensible similarities and likely homology between PiggyBac and TBE transposons (Figure 4) belie their differences. How did different ciliate lineages acquire different types of transposases and coevolve such different strategies between the transposons and their hosts to mediate different pathways of genome differentiation? Because oligohymenophorean and spirotrich ciliates are evolutionarily more distant from each other than plants and animals, a plausible explanation for the recruitment of different types of transposases for DNA elimination pathways in these distant ciliates is independent origins. However, it is also possible that the mutualistic system in Oxytricha may have predated DNA elimination by a domesticated transposase. A later transposon-domestication event or events that resulted in a high quantity of active transposase in the ancestral oligohymenophorean lineage could have lessened the dependency on feral transposons distributed throughout the genome. The modern piggyBac-like element in Paramecium and Tetrahymena might be a relic from a transposon that was initially maintained in the micronucleus by a mutualistic system more like Oxytricha's, and then later a copy of its transposase gene could have accidently lost the signals for DNA deletion and become a resident of the macronucleus as well, where it accumulated additional substitutions. Then this PiggyBac transposase, if expressed at sufficiently high levels, could have taken over the former roles of TBE or other transposases, reducing the levels of purifying selection that acted on the germline transposases until they became redundant with the function of the domesticated transposase. This relaxation of constraints on germline transposons would have permitted them to adapt or ameliorate to the background of micronuclear-limited DNA, scattering transposon remnants in the micronuclear genome, until most were eventually unrecognizable [25]. Accordingly, sequences related to TBE transposases are present in the Paramecium [29] and Tetrahymena micronuclear genomes, and some have functional open reading frames that maintain the DDE catalytic triad (Anchois, Thon, and Sardine in Paramecium [29] and the Tetrahymena sequence labeled “42 kDa transposase” in Figure 4). Therefore, these DNA sequences could be remnants from a TBE mutualistic system, and the minimal conservation suggests the possibility that TBE transposases could still contribute some role to DNA elimination in oligohymenophoreans. In this context, it would be fruitful to study the function of these newly discovered TBE transposase genes, as well as other newly discovered transposase-related genes in the Oxytricha macronucleus [17].
Conclusions
The roles of transposase proteins in programmed DNA rearrangements are just coming to light. Both structural and more functional studies are needed to understand how TBE and PiggyBac transposases interact with chromatin and induce DNA double-strand breaks. DNA elimination events, initiated by double-strand breaks, must be swiftly followed by DSB repair. Knowledge that the DNA elimination pathways in Paramecium and Tetrahymena require nonhomologous end joining (NHEJ) DSB repair machinery [44], [45] raises questions about how transposases interact with the NHEJ machinery and how they cooperatively regulate DNA elimination. In Oxytricha and other species with scrambled genes, another key set of questions is how the RNA templates [22] that provide the reordering information guide the transposases and other rearrangement machinery to form the proper religated junctions. From an evolutionary point of view, broader phylogenetic surveys are necessary to understand how two such distant groups of ciliates evolved such different DNA deletion systems, dependent on PiggyBac and TBE transposases, respectively. Because these two lineages represent just a modest fraction of ciliate biological diversity, and because some level of DNA elimination may be ancestral to ciliates [46], it would be tremendously valuable to investigate the functional and evolutionary relationships among transposases and DNA elimination events in different, deeply divergent groups of ciliates. Such comparative and functional studies are needed to achieve a better natural history of transposase recruitment and the forces of mutualism versus domestication on an evolutionary timescale.
Supporting Information
Zdroje
1. OrgelLE, CrickFH (1980) Selfish DNA: the ultimate parasite. Nature 284 : 604–607.
2. KazazianHHJr (2004) Mobile elements: drivers of genome evolution. Science 303 : 1626–1632.
3. VolffJN (2006) Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 28 : 913–922.
4. SinzelleL, IzsvakZ, IvicsZ (2009) Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell Mol Life Sci 66 : 1073–1093.
5. KapitonovVV, JurkaJ (2005) RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol 3: e181 doi:10.1371/journal.pbio.0030181
6. AgrawalA, EastmanQM, SchatzDG (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394 : 744–751.
7. BarsoumE, MartinezP, AstromSU (2010) Alpha3, a transposable element that promotes host sexual reproduction. Genes Dev 24 : 33–44.
8. ReddyKC, VilleneuveAM (2004) C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118 : 439–452.
9. BaudryC, MalinskyS, RestituitoM, KapustaA, RosaS, et al. (2009) PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev 23 : 2478–2483.
10. NowackiM, HigginsBP, MaquilanGM, SwartEC, DoakTG, et al. (2009) A functional role for transposases in a large eukaryotic genome. Science 324 : 935–938.
11. ChengCY, VogtA, MochizukiK, YaoMC (2010) A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol Biol Cell 21 : 1753–1762.
12. GouldSJ, VrbaES (1982) Exaptation - a Missing Term in the Science of Form. Paleobiology 8 : 4–15.
13. Baroin-TourancheauA, DelgadoP, PerassoR, AdoutteA (1992) A broad molecular phylogeny of ciliates: identification of major evolutionary trends and radiations within the phylum. Proc Natl Acad Sci U S A 89 : 9764–9768.
14. PrescottDM (1994) The DNA of ciliated protozoa. Microbiol Rev 58 : 233–267.
15. MochizukiK (2010) DNA rearrangements directed by non-coding RNAs in ciliates. Wiley Interdiscip Rev RNA 1 : 376–387.
16. JahnCL, KlobutcherLA (2002) Genome remodeling in ciliated protozoa. Annu Rev Microbiol 56 : 489–520.
17. SwartEC, BrachtJR, MagriniV, MinxP, ChenX, et al. (2013) The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol 11: e1001473 doi:10.1371/journal.pbio.1001473
18. AuryJM, JaillonO, DuretL, NoelB, JubinC, et al. (2006) Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444 : 171–178.
19. EisenJA, CoyneRS, WuM, WuD, ThiagarajanM, et al. (2006) Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol 4: e286 doi:10.1371/journal.pbio.0040286
20. MeyerE, CaronF, BaroinA (1985) Macronuclear structure of the G surface antigen gene of Paramecium primaurelia and direct expression of its repeated epitopes in Escherichia coli. Mol Cell Biol 5 : 2414–2422.
21. KatzLA, KovnerAM (2010) Alternative processing of scrambled genes generates protein diversity in the ciliate Chilodonella uncinata. J Exp Zool B Mol Dev Evol 314 : 480–488.
22. NowackiM, ShettyK, LandweberLF (2011) RNA-mediated epigenetic programming of genome rearrangements. Annu Rev Genomics Hum Genet 12 : 367–389.
23. HunterDJ, WilliamsK, CartinhourS, HerrickG (1989) Precise excision of telomere-bearing transposons during Oxytricha fallax macronuclear development. Genes Dev 3 : 2101–2112.
24. WilliamsK, DoakTG, HerrickG (1993) Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J 12 : 4593–4601.
25. KlobutcherLA, HerrickG (1997) Developmental genome reorganization in ciliated protozoa: the transposon link. Prog Nucleic Acid Res Mol Biol 56 : 1–62.
26. DoakTG, DoerderFP, JahnCL, HerrickG (1994) A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc Natl Acad Sci U S A 91 : 942–946.
27. WitherspoonDJ, DoakTG, WilliamsKR, SeegmillerA, SegerJ, et al. (1997) Selection on the protein-coding genes of the TBE1 family of transposable elements in the ciliates Oxytricha fallax and O. trifallax. Mol Biol Evol 14 : 696–706.
28. Le MouelA, ButlerA, CaronF, MeyerE (2003) Developmetnally regulated chromosome fragmentation is linked to imprecise elimination of repeated sequences in paramecia. Eukaryot Cell 2 : 1076–1090.
29. ArnaizO, MathyN, BaudryC, MalinskyS, AuryJ-M, et al. (2012) The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genetics 8: e1002984 doi:10.1371/journal.pgen.1002984
30. GratiasA, BetermierM (2001) Developmentally programmed excision of internal DNA sequences in Paramecium aurelia. Biochimie 83 : 1009–1022.
31. KlobutcherLA, HerrickG (1995) Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res 23 : 2006–2013.
32. GratiasA, BetermierM (2003) Processing of double-strand breaks is involved in the precise excision of Paramecium internal eliminated sequences. Mol Cell Biol 23 : 7152–7162.
33. van LuenenHG, CollomsSD, PlasterkRH (1994) The mechanism of transposition of Tc3 in C. elegans. Cell 79 : 293–301.
34. SavelievSV, CoxMM (1996) Developmentally programmed DNA deletion in Tetrahymena thermophila by a transposition-like reaction pathway. EMBO J 15 : 2858–2869.
35. SchoeberlUE, KurthHM, NotoT, MochizukiK (2012) Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev 26 : 1729–1742.
36. CoyneRS, StoverNA, MiaoW (2012) Whole genome studies of Tetrahymena. Methods Cell Biol 109 : 53–81.
37. FassJN, JoshiNA, CouvillionMT, BowenJ, GorovskyMA, et al. (2011) Genome-scale analysis of programmed DNA elimination sites in Tetrahymena thermophila. G3 (Bethesda) 1 : 515–522.
38. FillinghamJS, ThingTA, VythilingumN, KeuroghlianA, BrunoD, et al. (2004) A non-long terminal repeat retrotransposon family is restricted to the germ line micronucleus of the ciliated protozoan Tetrahymena thermophila. Eukaryot Cell 3 : 157–169.
39. MitraR, Fain-ThorntonJ, CraigNL (2008) PiggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J 27 : 1097–1109.
40. MadireddiMT, CoyneRS, SmothersJF, MickeyKM, YaoMC, et al. (1996) Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell 87 : 75–84.
41. LiuY, TavernaSD, MuratoreTL, ShabanowitzJ, HuntDF, et al. (2007) RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev 21 : 1530–1545.
42. TavernaSD, CoyneRS, AllisCD (2002) Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110 : 701–711.
43. KidwellMG, LischDR (2001) Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55 : 1–24.
44. LinIT, ChaoJL, YaoMC (2012) An essential role for the DNA breakage-repair protein Ku80 in programmed DNA rearrangements in Tetrahymena thermophila. Mol Biol Cell 23 : 2213–2225.
45. KapustaA, MatsudaA, MarmignonA, KuM, SilveA, et al. (2011) Highly precise and developmentally programmed genome assembly in Paramecium requires ligase IV–dependent end joining. PLoS Genet 7: e1002049 doi:10.1371/journal.pgen.1002049
46. RileyJL, KatzLA (2001) Widespread distribution of extensive chromosomal fragmentation in ciliates. Mol Biol Evol 18 : 1372–1377.
47. YuanaY-W, WesslerSR (2011) The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci U S A 108 : 7884–7889.
48. HuelsenbeckJP, RonquistF (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 : 754–755.
49. PuntaM, CoggillPC, EberhardtRY, MistryJ, TateJ, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–301.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



