-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
Salt stress is an important environmental factor that significantly limits crop productivity worldwide. Studies on responses of plants to salt stress in recent years have identified novel signaling pathways and have been at the forefront of plant stress biology and plant biology in general. Thus far, research on salt stress in plants has been focused on cytoplasmic signaling pathways. In this study, we discovered a nuclear calcium-sensing and signaling pathway that is critical for salt stress tolerance in the reference plant Arabidopsis. Through a forward genetic screen, we found a nuclear-localized calcium-binding protein, RSA1 (SHORT ROOT IN SALT MEDIUM 1), which is required for salt tolerance, and identified its interacting partner, RITF1, a bHLH transcription factor. We show that RSA1 and RITF1 regulate the transcription of several genes involved in the detoxification of reactive oxygen species generated by salt stress and that they also regulate the SOS1 gene that encodes a plasma membrane Na+/H+ antiporter essential for salt tolerance. Together, our results suggest the existence of a novel nuclear calcium-sensing and -signaling pathway that is important for gene regulation and salt stress tolerance.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003755
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003755Summary
Salt stress is an important environmental factor that significantly limits crop productivity worldwide. Studies on responses of plants to salt stress in recent years have identified novel signaling pathways and have been at the forefront of plant stress biology and plant biology in general. Thus far, research on salt stress in plants has been focused on cytoplasmic signaling pathways. In this study, we discovered a nuclear calcium-sensing and signaling pathway that is critical for salt stress tolerance in the reference plant Arabidopsis. Through a forward genetic screen, we found a nuclear-localized calcium-binding protein, RSA1 (SHORT ROOT IN SALT MEDIUM 1), which is required for salt tolerance, and identified its interacting partner, RITF1, a bHLH transcription factor. We show that RSA1 and RITF1 regulate the transcription of several genes involved in the detoxification of reactive oxygen species generated by salt stress and that they also regulate the SOS1 gene that encodes a plasma membrane Na+/H+ antiporter essential for salt tolerance. Together, our results suggest the existence of a novel nuclear calcium-sensing and -signaling pathway that is important for gene regulation and salt stress tolerance.
Introduction
Salt stress severely limits the quality and productivity of important agricultural crops worldwide. Therefore, a thorough understanding of the molecular basis of salt stress signal transduction pathways and salt tolerance mechanisms is of fundamental importance for the understanding of plant biology and for the generation of salt-tolerant crops through rational breeding and genetic engineering strategies.
High levels of soluble salts including chlorides of sodium, calcium, and magnesium often cause soil sodicity, alkalinity, and other soil problems. Excessive salts in soil are harmful to plants because of toxicity of Na+ and other ions [1]. To deal with salt stress, plants have evolved mechanisms in order to coordinate the activities of various ion transporters and to thereby maintain ion homeostasis in the cell cytoplasm. Salt Overly Sensitive (SOS) pathway proteins play a key role in Na+ homeostasis in plants [2]. The calcium sensor SOS3 senses salt stress-induced increases of cytosolic free calcium levels and interacts with and activates a serine/threonine protein kinase SOS2 [2]. The SOS3-SOS2 protein kinase complex then phosphorylates and thereby activates the plasma membrane-localized Na+/H+ antiporter SOS1 [2]. Overexpression of SOS1 leads to improved salt tolerance in transgenic Arabidopsis [2], [3]. SOS2 also positively controls the activities of tonoplast Na+/H+ antiporters, which sequester Na+ ions in the vacuole, and the activities of a vacuolar H+/Ca2+ exchanger CAX1 [4], [5]. In addition to its critical role in Na+ homeostasis, SOS1 is important for oxidative stress responses under salt stress [6].
Salt stress and many other abiotic and biotic stresses cause plants to produce reactive oxygen species (ROS) including superoxide (O2.−) and hydroxyl (OH.) free radicals, hydrogen peroxide (H2O2), and free singlet oxygen [7]–[9]. In many cases, the over-accumulated ROS have detrimental effects on cellular processes [10]. Below the threshold level that is not harmful to plant cells, however, H2O2 can act as a messenger molecule to initiate signal transduction cascades involving mitogen-activated protein kinase (MAPK) under various environmental cues [11], [12]. Plants have developed enzymatic (e.g., superoxide dismutase, peroxidase, and catalase) and non-enzymatic (e.g., antioxidants and some secondary metabolites) strategies to detoxify excessive ROS [7]–[9].
Here, we report the isolation and characterization of a nuclear-localized calcium-binding protein, RSA1, which was identified in a forward genetic screen for critical genes required for salt tolerance in plants. The rsa1-1 mutant plants are hypersensitive to NaCl but not to LiCl, CsCl, or general osmotic stress. The rsa1-1 plants over-accumulate ROS and are hypersensitive to exogenous H2O2 or ROS-producing agents such as methyl viologen. Protein-protein interaction studies revealed that RSA1 interacts with a bHLH transcription factor, RITF1, which in turn regulates many RSA1 downstream target genes, including SOS1, that are important for ROS detoxification and/or Na+ homeostasis. Together, our results demonstrate that RSA1 is a nucleus-localized calcium sensor that has a crucial role in gene regulation under salt stress and in salt stress tolerance.
Results
Identification of the rsa1-1 mutant
To isolate genes that play essential roles in salt stress tolerance, we used a modified root-bending assay [13], [14] and screened an ethyl methanesulfonate (EMS)-mutagenized Arabidopsis M2 population for mutants with hypersensitivity to 100 mM NaCl. These mutants were designated as short root in salt medium (rsa). One of these mutants, rsa1-1, was chosen for detailed characterization. The shoot development of rsa1-1 is normal under control conditions, but the primary root of rsa1-1 is slightly shorter than that of the wild type (Figure 1A). Both roots and shoots of rsa1-1 seedlings display a hypersensitive phenotype in response to supplemental NaCl in the growth medium (Figure 1A–1C). The roots of rsa1-1 seedlings have more root hairs than the wild type under control and salt-treated conditions (Figure S1A). The soil-grown rsa1-1 plants are more sensitive than the wild type when treated with 300 mM NaCl (Figure 1D and 1E), and germination of rsa1-1 seeds is more inhibited by NaCl than is germination of wild-type seeds (Figure 1F), suggesting that salt hypersensitivity of rsa1-1 plants does not depend on developmental stage. We then determined whether the salt hypersensitive phenotype of rsa1-1 is specific to Na+. We found that rsa1-1 mutant plants are not more sensitive to LiCl or CsCl than the wild type (Figure S1B and S1C), even though Li and Cs are in the same column of the periodic table of the elements as Na and are generally considered more toxic. The response of rsa1-1 plants to mannitol, which induces general osmotic stress, was similar to that of wild-type plants (Figure S1D). Thus, rsa1-1 is only hypersensitive to NaCl.
Fig. 1. rsa1-1 plants are hypersensitive to NaCl, and RSA1 is involved in Na+ homeostasis under salt stress. 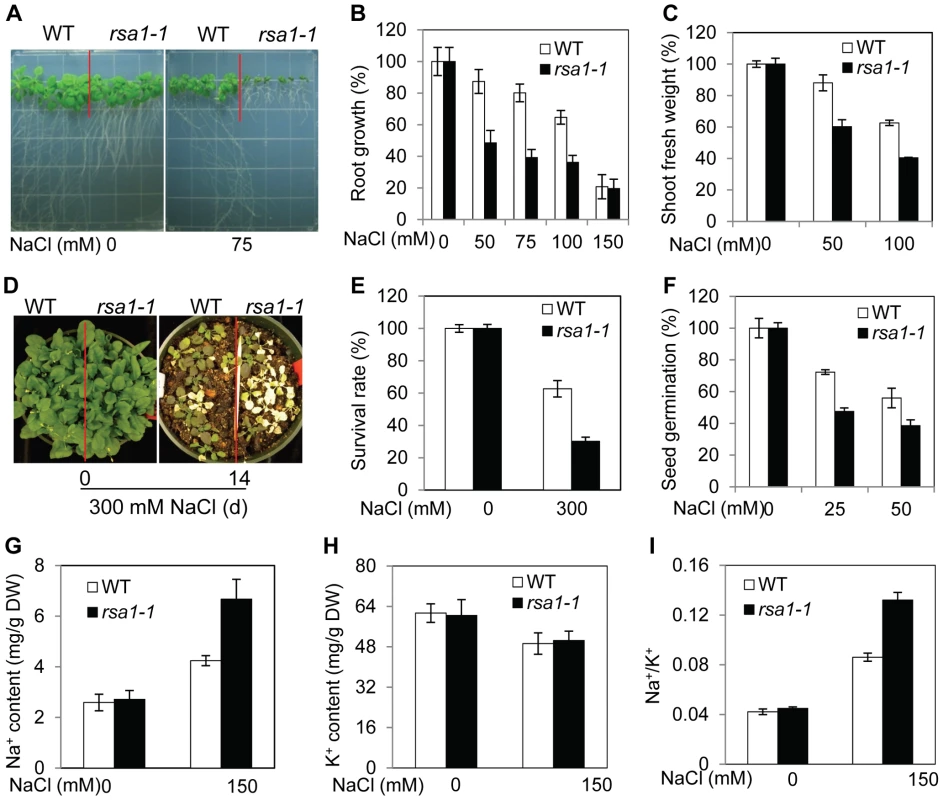
(A)–(C) Five-d-old wild-type and rsa1-1 seedlings grown on MS medium were transferred to MS medium supplemented with different levels of NaCl and allowed to grow for an additional 8 d. Root elongation or shoot fresh weight was measured and is shown as a percentage relative to growth on normal MS medium. (D) Two-week-old wild-type and rsa1-1 plants grown in soil were irrigated with 300 mM NaCl for 0 or 14 d. (E) Survival rate of wild-type and rsa1-1 plants as shown in (D). (F) Seed germination of wild type and rsa1-1 in response to various NaCl levels. There were 80–150 seeds per genotype per biological replicate. Seeds in which the radical had emerged through the seed coat were considered germinated. (G) Na+ content in soil-grown wild-type and rsa1-1 plants. DW, dry weight. (H) K+ content in soil-grown wild-type and rsa1-1 plants. (I) Ratio of Na+ to K+ accumulation in soil-grown wild-type and rsa1-1 plants. WT, wild type. Error bars indicate the standard deviation (n = 30–40). The experiments in Figure 1 were repeated at least five times with similar results, and data from one representative experiment are presented. When rsa1-1 plants were backcrossed with wild-type plants, all F1 plants displayed a wild-type phenotype, and F2 progeny from the self-pollinated F1 plants segregated at approximately 3∶1 (wild type vs. mutant; Table S1). These results suggest that rsa1-1 is a recessive mutation in a single nuclear gene.
The rsa1-1 mutation affects Na+ homeostasis
The altered responses to salt stress in rsa1-1 suggest that ion homeostasis in rsa1-1 may be disturbed. As shown in Figure 1G, soil-grown rsa1-1 plants accumulate more Na+ than the wild type when treated with 150 mM NaCl. Relative to wild-type plants, the K+ level is not altered in rsa1-1 with or without salt stress (Figure 1H). The ratio of Na+ to K+ is much higher in rsa1-1 than in the wild type under salt stress (Figure 1I). These results suggest that RSA1 is required for Na+ homeostasis under salt stress. These results also suggest that RSA1 may directly or indirectly affect the known SOS pathway for Na+ homeostasis under salt stress.
The rsa1-1 mutation causes over-accumulation of ROS
Abiotic stresses including salt stress can cause production of ROS [7], [15]. We determined the effect of the rsa1-1 mutation on ROS levels and on the response to oxidative stress. The fluorescent dye 5-(and 6)-chloromethyl-2′7′-dichlorodihydrofluorescein diacetate acetyl ester (CM-H2DCFDA) was used to visualize and quantify total ROS in the root tissues. The rsa1-1 plants accumulated slightly more ROS than the wild type under control conditions but accumulated substantially more ROS than the wild type when treated with 50 mM NaCl (Figure 2A and 2B). Similar results were obtained for the levels of hydrogen peroxide (H2O2), which was quantified with the Amplex red reagent, 10-acetyl-3,7-dihydrophenoxazine (Figure 2C). These results suggest that RSA1 is an important regulator of ROS accumulation in plants under salt stress. Furthermore, we found that rsa1-1 plants are hypersensitive to exogenous application of H2O2 or methyl viologen (MV) (Figure 2D–2F). MV can lead to an increase in the generation of toxic superoxide free radicals in chloroplasts [16].
Fig. 2. rsa1-1 mutant plants accumulate more ROS and are hypersensitive to oxidative stress. 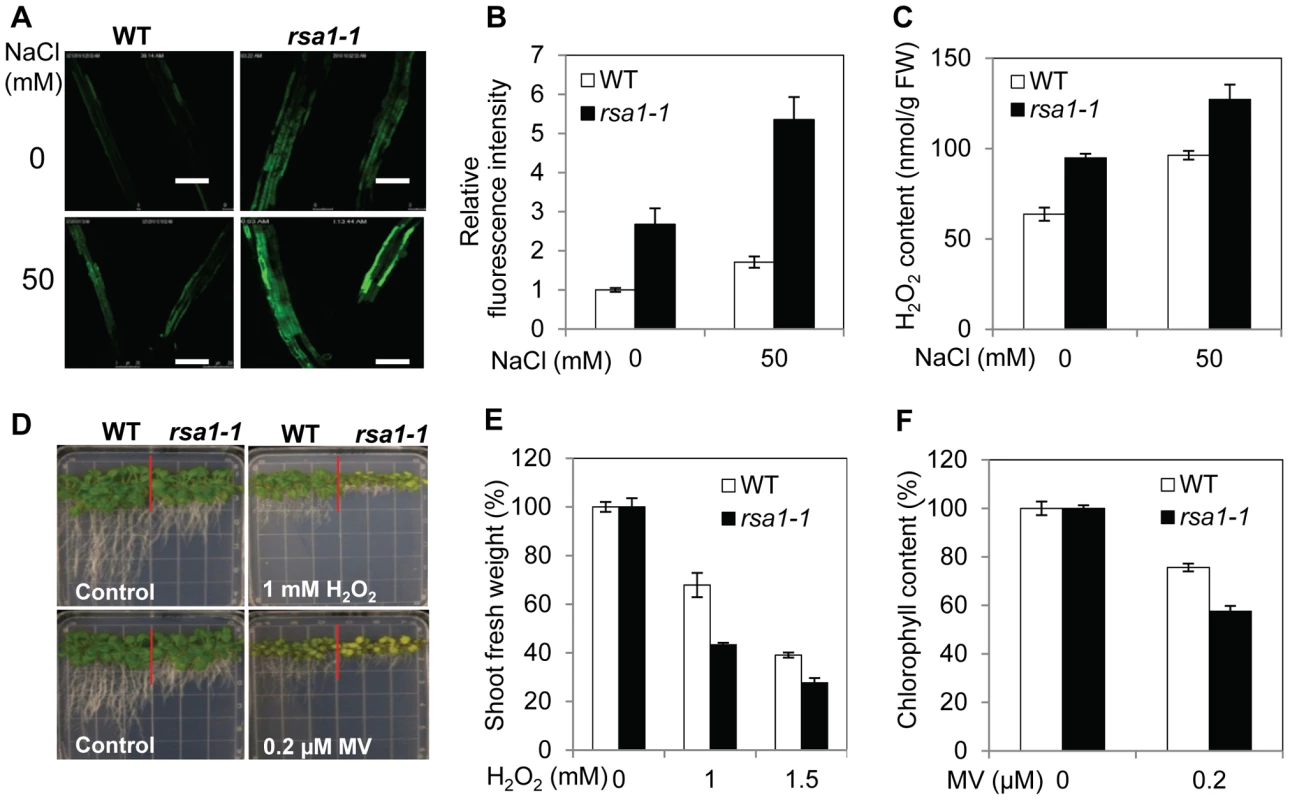
(A) Total ROS accumulation in roots of 5-d-old wild-type and rsa1-1 seedlings subjected to 0 or 50 mM NaCl for 12 h. Bars = 200 µm. (B) Fluorescence intensity in plants shown in (A). (C) H2O2 content of wild-type and rsa1-1 plants subjected to 0 or 50 mM NaCl for 12 h. (D) Growth responses of wild-type and rsa1-1 seedlings to oxidative stress-inducing reagents H2O2 and methyl viologen (MV). Seeds of the wild type and rsa1-1 were sown directly on MS medium containing 0 or 1 mM H2O2 (upper panel) and 0 or 0.2 µM MV (lower panel) and allowed to grow for an additional 14 d. (E) and (F) Shoot fresh weight and chlorophyll content in wild-type and rsa1-1 plants under treatment conditions shown in (D). Error bars indicate the standard deviation (n = 24). The experiments in Figure 2 were repeated at least four times with similar results, and data from one representative experiment are presented. RSA1 encodes a nuclear protein containing an EF-hand motif
We identified the RSA1 locus through a map-based cloning strategy. The rsa1-1 mutation is caused by a single nucleotide substitution in At2g03150, and this mutation results in the change of proline at position of 685 to leucine in the deduced RSA1 polypeptide (Figure S2A). RSA1 encodes a putative calcium-binding protein with one EF-hand motif at its C-terminal region (Figure S2A). We confirmed the identity of RSA1 by a gene complementation analysis. The wild-type RSA1 driven by its native promoter is able to fully complement the rsa1-1 mutant phenotype (Figure S2B and S2C). Two T-DNA alleles of RSA1 (rsa1-2 and rsa1-3) were obtained, and expression of RSA1 is not detectable in either rsa1-2 or rsa1-3 plants, suggesting that they are null alleles of RSA1 (Figure S2D). Like rsa1-1 plants, rsa1-2 and rsa1-3 plants are hypersensitive to NaCl stress (Figure S2B and S2E).
Strong expression of RSA1 was detected in all examined tissues of wild-type plants including guard cells (Figure S3A and S3B). RSA1 is slightly induced by salt stress (Figure 3A). Unlike most EF-hand proteins, RSA1 is predominantly localized in the nucleus of Arabidopsis root tip cells or tobacco leaf cells (Figure 3B). Upon salt stress, RSA1 continues to be localized in the nucleus (Figure 3B). The p35S:YFP-RSA1 and pRSA1:RSA1-GFP genes used in our subcellular localization studies are able to restore the rsa1-1 mutant phenotype to wild type (Figure S3C and S3D), suggesting that the p35S:YFP-RSA1 and pRSA1:RSA1-GFP genes are functional in planta.
Fig. 3. RSA1 is localized in the nucleus and has calcium-binding activity. 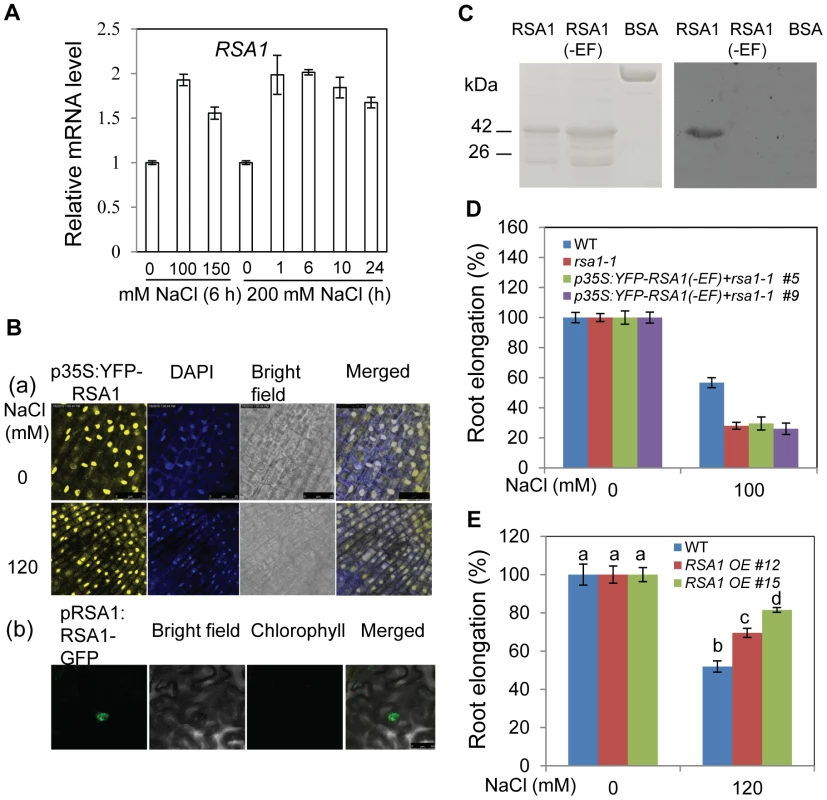
(A) RSA1 expression under salt stress. Fourteen-d-old wild-type seedlings grown on MS medium were transferred to filter paper saturated with 0, 100, 150, and 200 mM NaCl for the indicated time. (B) Localization of p35S:YFP-RSA1 in Arabidopsis root tip cells with or without salt stress ([a], DAPI staining was used to indicate the nuclei) and pRSA1:RSA1-GFP in tobacco leaves (b). Bars = 25 µm in (a) and 50 µm in (b). (C) Calcium-binding activity of RSA1 and RSA1(-EF) proteins. 6× His-tagged proteins were purified, separated on SDS-PAGE, and stained with coomassie blue (left panel) or electroblotted onto a PVDF membrane, overlayed with 45Ca2+, and autoradiographed (right panel). BSA (bovine serum albumin), negative control for 45Ca2+ binding. (D) Salt tolerance of wild-type, rsa1-1, and rsa1-1 plants expressing p35S:YFP-RSA1(-EF). (E) Salt tolerance of RSA1 overexpression plants in rsa1-1 background (Figure S3F). Five-d-old wild-type and rsa1-1 seedlings grown on MS medium were transferred to MS medium supplemented with different levels of NaCl and allowed to grow for an additional 8 d (for [D] and [E]). RSA1 (-EF), RSA1 without EF-hand motif. One-way ANOVA (Tukey-Kramer test) was performed, and statistically significant differences are indicated by different lowercase letters (p<0.01). Error bars represent the standard deviation (n = 4 in [A], 40 in [D] and [E]). The experiments in Figure 3 were performed at least three times with similar results, and data from one representative experiment are presented. RSA1 is a calcium-binding protein
Like other calcium-dependent protein kinases (CPKs) and calmodulin 2 (CAM2) in Arabidopsis, the EF-hand motif in RSA1 is highly conserved (Figure S3E) [17]. We produced a recombinant RSA1 protein with a 6× His tag at its N-terminus and purified it from E. coli to test its potential calcium-binding activity (Figure 3C). The recombinant RSA1 protein showed calcium-binding activity in the calcium overlay assays, and the calcium-binding activity was abolished when the 12-amino acid consensus sequence for calcium binding in the EF-hand motif was deleted (Figure 3C). We further showed that RSA1 protein carrying the 12-amino acid deletion in the EF-hand motif is unable to complement the rsa1-1 mutant (Figure 3D), suggesting that the calcium-binding activity of RSA1 is required for its in vivo function. We were able to identify two independent lines of Arabidopsis transgenic plants overexpressing RSA1 (p35S:YFP-RSA1) in the rsa1-1 background (Figure S3F). These RSA1 overexpression plants are substantially more tolerant to 120 mM NaCl than the wild type (Figure 3E). These results further confirm that RSA1 is a positive regulator of plant salt stress tolerance.
RSA1 interacts with a basic helix-loop-helix (bHLH) transcription factor that is important for salt stress tolerance
To determine how RSA1 functions in the salt stress tolerance pathway, we identified interaction partners of RSA1 via a yeast two-hybrid screen. Six RSA1 interacting proteins that are localized in the nucleus were identified in the yeast two-hybrid screen. One basic helix-loop-helix (bHLH) transcription factor, RITF1 (for RSA1 interacting transcription factor 1 encoded by At3g06590), appeared many times and showed the strongest interaction with RSA1 in our yeast two-hybrid screen (Figure 4A). We therefore focused on this transcription factor. We found that RITF1 is predominantly localized in the nucleus of Arabidopsis protoplasts (Figure 4B). Bimolecular fluorescence complementation (BiFC) analysis using Arabidopsis protoplasts and tobacco leaves, and split luciferase complementation (Split-LUC) analysis in tobacco leaves confirmed that RSA1 interacts with RITF1 in vivo (Figure 4C, 4D and Figure S4A). Co-immunoprecipitation (Co-IP) analysis in tobacco plants further confirmed the interaction between RSA1 and RITF1 in vivo (Figure 4E).
Fig. 4. RSA1 interacts with RITF1. 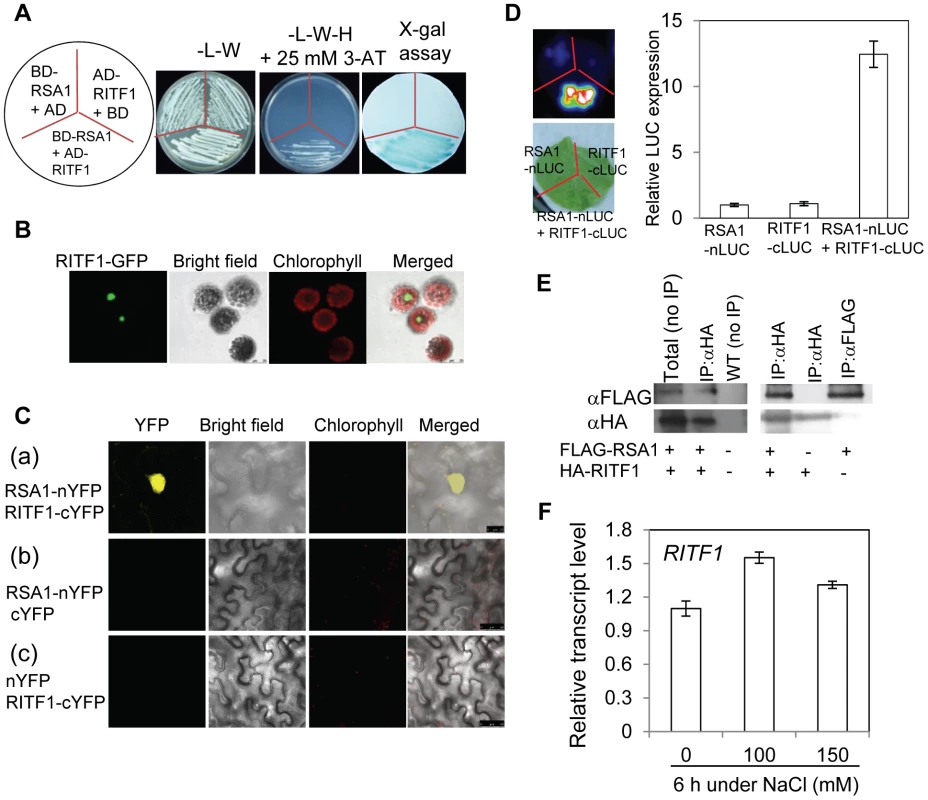
(A) RSA1 interacts with RITF1 as determined by yeast two-hybrid assays. Yeast strain AH109 co-transformed with RSA1-pDEST32 (bait) and RITF1-pDEST22 (prey) was subjected to x-gal assay. AH109 cells co-transformed with RSA1-pDEST32/pDEST22 (empty vector) or RITF1-pDEST22/pDEST32 (empty vector) were used as negative controls. Yeast cells grown on SD medium-L-W or SD medium-L-W-H+3-AT are shown. 3-AT, 3-amino-1,2,4-triazole. L, W, H, symbols for amino acids leucine, tryptophan, and histidine, respectively. SD, yeast minimal media. (B) Localization of RITF1-GFP in Arabidopsis protoplasts. Bar = 25 µm. (C) RSA1 interacts with RITF1 in vivo as determined by BiFC assays in tobacco leaf epidermal cells. Bars = 25 µm in (a), and 50 µm in (b) and (c). YFP images were detected at an approximate frequency of 4.04% (101 out of 2,501 tobacco leaf epidermal cells analyzed exhibited BiFC events). (D) RSA1 interacts with RITF1 in vivo as determined by Split-LUC assays. (E) RSA1 interacts with RITF1 in vivo as determined by Co-IP assays. (F) RITF1 expression under salt stress. The qRT-PCR analysis was carried out with 14-d-old wild-type seedlings grown for 6 h on MS medium containing 0, 100, or 150 mM NaCl. Error bars represent the standard deviation (n = 20 in [D], 4 in [F]). The experiments in Figure 4 were performed at least three times with similar results, and data from one representative experiment are presented. qRT-PCR analysis indicated that RITF1 is slightly inducible by salt stress (Figure 4F). We obtained the T-DNA knockdown plants of RITF1 (ritf1) (Figure S4B). The ritf1 plants are substantially more sensitive to salt stress than wild-type plants during seed germination and during seedling growth and development (Figure 5A and 5B). We further observed that ritf1 plants are more sensitive to oxidative stress imposed by H2O2 or MV than the wild type (Figure 5C–5E). We overexpressed RITF1 in Arabidopsis (Figure S4C), and all of the RITF1 overexpression plants displayed increased tolerance to NaCl and the oxidative stress-inducing reagents H2O2 and MV (Figure 5F–5H). These results suggest that the RSA1 interacting transcription factor, RITF1, is required for plant tolerance to salt and oxidative stresses.
Fig. 5. ritf1 mutant plants are sensitive to salt and oxidative stresses, and overexpression of RITF1 increases plant tolerance to salt and oxidative stresses. 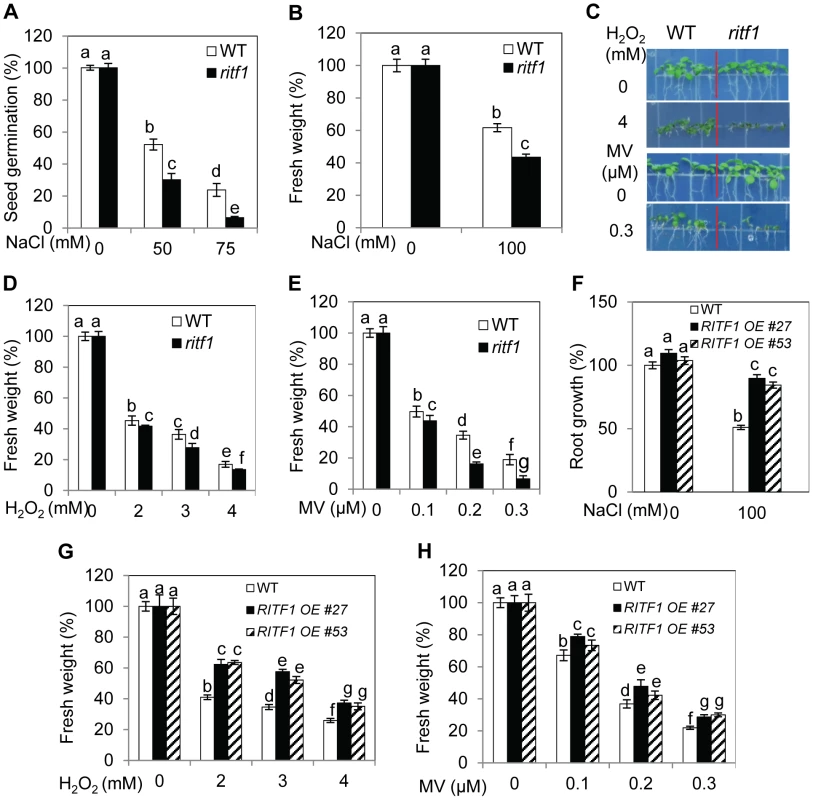
(A) Seed germination of wild type and ritf1 in response to various NaCl levels. There were 80–150 seeds per genotype per biological replicate. (B) Fresh weight of wild-type and ritf1 seedlings under salt stress. Five-d-old seedlings grown on MS medium were transferred to MS medium containing 0 or 100 mM NaCl and allowed to grow for an additional 7 d. (C) Growth responses of wild-type and ritf1 seedlings to oxidative stress-inducing reagents H2O2 and methyl viologen (MV). (D) and (E) Fresh weight of seedlings grown on MS medium containing various levels of H2O2 (D) or MV (E) as shown in (C). (F) Salt tolerance of RITF1 overexpression plants. Five-d-old seedlings grown on MS medium were transferred to MS medium containing 0 or 100 mM NaCl and allowed to grow for an additional 10 d. (G) and (H) Fresh weight of wild-type and RITF1 overexpression plants grown on MS medium containing various levels of H2O2 (G) or MV (H). In (C)–(E), (G), and (H), seeds were sown directly on MS medium supplemented with various levels of H2O2 or MV and allowed to grow for an additional 10 d. Error bars represent the standard deviation (n = 8 in [A], 40 in [B], [D]–[H]). One-way ANOVA (Tukey-Kramer test) was performed, and statistically significant differences are indicated by different lowercase letters (p<0.05). The experiments in Figure 5 were repeated at least four times with similar results, and data from one representative experiment are presented. RSA1 and RITF1 regulate the expression of genes important for oxidative and salt stress responses
Because RSA1 is localized in the nucleus, we determined whether the rsa1-1 mutation affects gene expression. We performed a whole-genome microarray analysis with Arabidopsis Affymetrix ATH1 GeneChips. Compared to genes in the wild type, 41 genes displayed higher expression levels while 54 genes showed lower expression levels in rsa1-1 by at least 2-fold (p<0.01) under control conditions (Table S2). The microarray analysis also revealed that 69 genes in rsa1-1 displayed at least a 2-fold increase in transcripts levels while 76 genes in rsa1-1 showed at least a 2-fold decrease in transcripts levels relative to the wild type (p<0.01) under salt stress conditions (Table S3). Compared to genes in the wild type, 13 genes displayed higher expression levels while 27 genes showed lower expression levels in rsa1-1 by at least 2-fold (p<0.01) under both control and salt stress conditions (Table S4). The differentially expressed genes in rsa1-1 encode proteins with diverse functions, and a large portion of these proteins have predicted functions in stress responses (Table S2, S3, and S4). In addition, as indicated by comparison with publicly available expression datasets, most of the differentially expressed genes in rsa1-1 are not responsive to salt stress treatments in the wild type (Figure S5 and S6). We validated the microarray data with qRT-PCR analysis for four genes, which encode peroxidase, zinc finger protein 5, bHLH DNA-binding superfamily protein, and root hair specific 19 with putative peroxidase activity (Figure S7A–S7D). qRT-PCR analysis also revealed that the expression levels of SOS1, At2g36690, and At5g14130 are dramatically reduced in rsa1-1 under both control and salt stress conditions (Figure 6A), suggesting that RSA1 is a positive regulator of these three genes. At2g36690 and At5g14130 encode oxidoreductase and peroxidase, respectively. Therefore, RSA1 controls genes important for ROS detoxification and signal transduction. Furthermore, we observed that expression of SOS1, At2g36690, and At5g14130 is substantially reduced in the ritf1 mutant plants under both control and salt stress conditions (Figure 6B), indicating that similar to RSA1, RITF1 is a positive regulator of these three genes. Expression of SOS1, At5g14130, and At2g36690 is substantially enhanced in transgenic plants overexpressing RSA1 or RITF1 under salt stress (Figure 6C and 6D), further confirming that RSA1 and RITF1 are positive regulators for salt-induced expression of SOS1, At5g14130, and At2g36690.
Fig. 6. RSA1 and RITF1 regulate gene expression, and RITF1 binds directly to promoters of three RSA1 target genes. 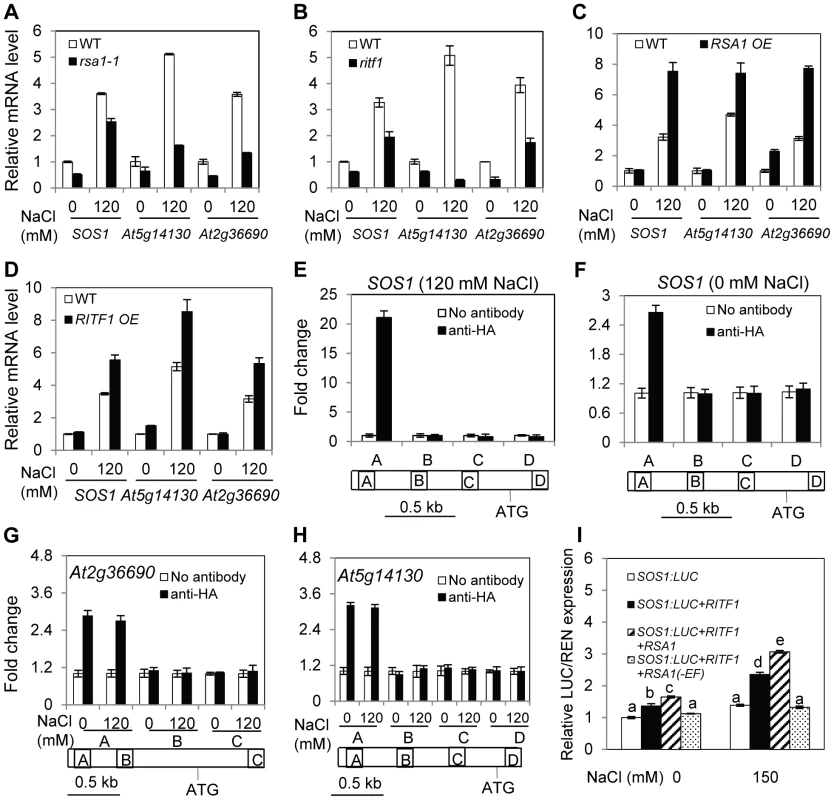
(A)–(D) Expression of SOS1, At5g14130, and At2g36690 in wild-type, rsa1-1, ritf1, RITF1 overexpression, or RSA1 overexpression plants. At5g14130 and At2g36690 encode peroxidase superfamily protein, oxidoreductase (2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein). Data in (C) and (D) are from multiple RSA1 OE or RITF1 OE plants (Figure S3F and S4C). Expression values from one representative transgenic line are presented. (E)–(H) ChIP-qPCR analysis of SOS1, At2g36690, and At5g14130 genes. Regions of amplifications in (E)–(H) are specified in Table S6A. (I) Relative luciferase activity from the dual luciferase reporter assays in tobacco leaves. RSA1(-EF), RSA1 without EF-hand motif. Error bars represent the standard deviation (n = 4 in [A]–[H], 12 in [I]). One-way ANOVA (Tukey-Kramer test) was performed, and statistically significant differences are indicated by different lowercase letters (p<0.01). These experiments were repeated at least four times with similar results, and data from one representative experiment are presented. RITF1 binds directly to cis elements in promoters of RSA1-regulated genes
bHLH transcription factors can potentially bind to a signature motif called E-box, which consists of a consensus hexanucleotide sequence of CANNTG [18], [19]. Database searches revealed that there are many such cis promoter elements in RSA1 downstream target genes as determined by the microarray analysis (Table S5). We then used a chromatin immunoprecipitation (ChIP) assay to determine whether RITF1 binds directly to these cis promoter elements in vivo. We produced transgenic Arabidopsis plants expressing the haemagglutinin (HA)-tagged RITF1 under the control of its native promoter (RITF1:RITF1-HA) (Figure S7E). RITF1 is enriched in the promoter regions of four genes (SOS1, At2g36690, At5g14130, and At2g39040) (Figure 6E–6H; Figure S7F). These results suggest that RSA1 controls expression of its downstream target genes through direct binding of RITF1 to promoters of RSA1 target genes.
We subsequently carried out a dual luciferase reporter assay to determine the effect of RSA1 and RITF1 on SOS1 promoter activity in vivo. RSA1 and RITF1 can transiently activate the SOS1 promoter that is transcriptionally fused with the firefly luciferase under both control and salt stress conditions (Figure 6I). When the EF-hand motif of RSA1 is deleted, the activation effect of RSA1 and RITF1 on the SOS1 promoter is abolished (Figure 6I), suggesting that the calcium-binding activity of RSA1 is required for the transient activation of SOS1 promoter activity. Furthermore, we observed that the rsa1-2ritf1 double mutant plants do not display any additive effect of rsa1-2 and ritf1 mutations under salt stress or oxidative stress (Figure S8). Together, these results suggest that RSA1 and RITF1 function in a common pathway for gene expression and that both proteins are required for plant tolerance to salt and oxidative stresses.
Discussion
By performing a forward genetic screen for genes crucial for salt stress tolerance, we have identified a salt hypersensitive mutant, rsa1-1. RSA1 encodes a nuclear-localized calcium-binding protein that is important for gene regulation and salt stress tolerance.
Its location in the nucleus suggested that RSA1 might play a role in gene regulation, and this was supported by three other findings. First, the microarray analysis confirmed that RSA1 controls expression of a number of genes under both control and salt stress conditions (Table S2 and S3). A large portion of the differentially expressed genes in rsa1-1 encode proteins with predicted roles in responses to abiotic or biotic stresses (Table S2 and S3). In addition, many genes in rsa1-1 were differentially expressed under control conditions (as revealed in the microarray analysis, see Table S2), and these changes may contribute to the reduced growth of the primary root. This defect does not persist, however, because later stages of rsa1-1 plants develop normally in potting soil. Second, yeast two-hybrid analysis indicated that RSA1 interacts with a bHLH transcription factor, RITF1. We confirmed this RSA1–RITF1 interaction in vivo by BiFC and Split-LUC assays and Co-IP analysis (Figure 4 and Figure S4A). Database searches indicate that RITF1 has three close homologs in Arabidopsis (bHLH transcription factors encoded by At3g17100, At3g05800, and At1g09250), and these homolog proteins do not interact with RSA1 in yeast two-hybrid assays (Figure S9A). T-DNA knockdown alleles of these three genes show normal morphology and the same responses to NaCl stress as the wild type (Figure S9B–S9E). These results suggest that RITF1 has a specialized role in salt stress responses. Interestingly, many of the RSA1 target genes revealed by the microarray analysis contain cis-promoter elements that RITF1 can potentially bind to (Table S5). Third, chromatin immunoprecipitation (ChIP) analysis indicated that RITF1 is able to bind to cis-promoter elements in four RSA1 target genes including SOS1 (Figure 6E–6H; Figure S7F). This is the first time that two positive regulators (RSA1 and RITF1) for SOS1 gene expression at the transcriptional level have been identified. Besides SOS1, the remaining three genes (At5g14130, At2g36690, and At2g39040) all encode enzymes for ROS detoxification. Thus, RSA1 is important for gene regulation through its interaction with partners such as the bHLH transcription factor RITF1. The transgenic plants overexpressing RSA1 or RITF1 also display enhanced expression of SOS1, At5g14130, and At2g36690, further confirming that RSA1 and RITF1 are important positive regulators for SOS1, At5g14130, and At2g36690 (Figure 6C and 6D). It is clear that the RSA1-RITF1 complex positively controls expression of SOS1, which encodes a plasma-membrane localized Na+/H+ antiporter (Figure 6A and 6C). Presumably, the Na+/H+ antiporter activity of SOS1 is reduced in rsa1-1 under salt stress, leading to increased accumulation of Na+ in rsa1-1 plants (Figure 1G). The SOS3-SOS2 protein kinase complex phosphorylates SOS1 and thereby activates its Na+/H+ antiporter activity [2]. Thus, the RSA1-RITF1 complex and the SOS3-SOS2 protein kinase complex regulate a common downstream target, SOS1, at two different levels in two separate cellular compartments, i.e., there is transcriptional regulation through RSA1-RITF1 in the nucleus and posttranslational modification by SOS3-SOS2 in the cytosol (Figure 7). In addition, constitutive expression of SOS1 in the rsa1-1 background can only partially rescue (∼12.5% improvement compared to the rsa1-1 plants) the hypersensitive phenotype of rsa1-1 in response to NaCl (Figure S10A and S10C). These results suggest that besides SOS1, other components in the signal transduction pathways mediated by RSA1 under salt stress are essential for salt stress tolerance.
Fig. 7. A working model for RSA1 and RITF1 function under salt stress. 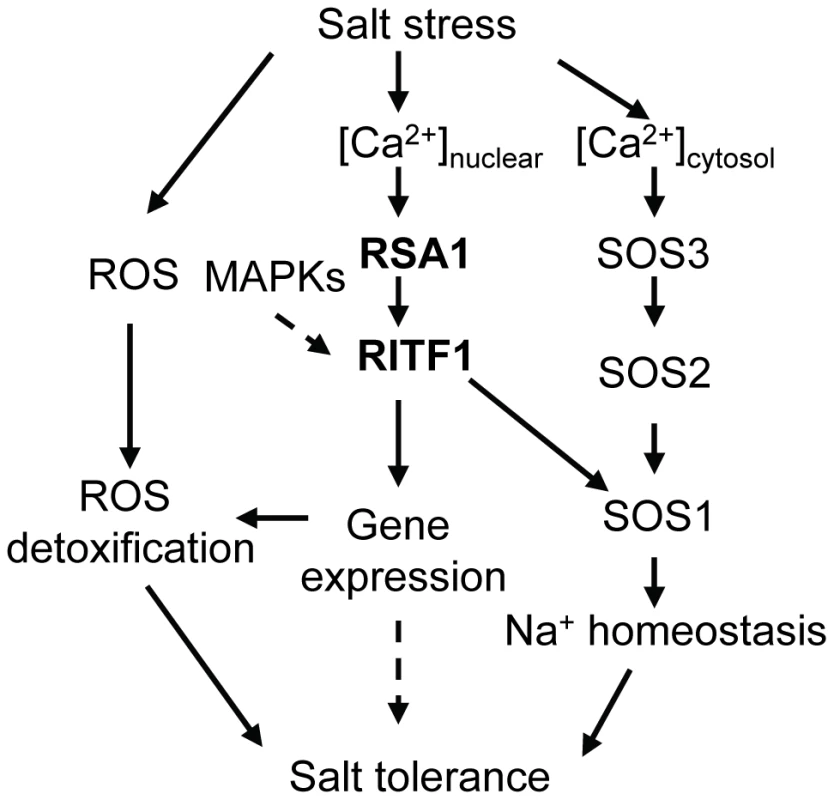
The calcium-binding protein RSA1 senses salt-induced changes in nuclear free calcium and interacts with a bHLH transcription factor, RITF1. RITF1 may be phosphorylated by nuclear-localized MAPKs. The RSA1-RITF1 complex controls expression of genes important for detoxification of salt-induced ROS and for Na+ homeostasis under salt stress. Some RITF1 target genes may play a role in salt tolerance with so far unknown mechanisms. The calcium-binding protein SOS3 senses salt-induced cytosolic calcium increases and interacts with SOS2, a protein kinase. The SOS3-SOS2 protein kinase complex then phosphorylates and thereby activates the plasma membrane-localized Na+/H+ antiporter SOS1. How does RSA1 sense changes in free calcium levels elicited by salt stress in the nucleus and then transduce signals to downstream signal transduction components and eventually activate gene expression? Calcium signals are detected in the nuclei of root nodules of legume plants [20]–[22]. Similar observations have been reported for other plant species (e.g., tobacco) in response to external stimuli [23]–[27]. Although the ultimate sources of nuclear, free calcium are debated, it is now clear that a nuclear-associated change in calcium levels acts as a secondary messenger for downstream signal transduction events as evidenced in the symbiotic signaling pathway in legumes for nodule formation [28]–[30]. Following Nod factor induction of nuclear-calcium spiking in legumes, a nuclear-localized calcium - and calmodulin-dependent protein kinase (CCaMK) and nuclear-associated transcription factors and other proteins are involved in the signaling transduction pathway for nodulation [31]–[35]. RSA1 is a nuclear-localized calcium-binding protein. Thus, RSA1 may sense changes in nuclear, free Ca2+ and then transduce this nuclear calcium signal to regulate gene expression that helps plants to cope with salt stress. We used a yeast two-hybrid screening strategy to identify RSA1 interacting partners, and a bHLH transcription factor RITF1 was isolated in this process. RSA1 physically interacts with RITF1 both in vitro and in vivo. ChIP-qPCR analysis revealed that RITF1 is able to directly bind to cis promoter elements of RSA1 downstream target genes. Determining how RSA1 or the RSA1-RITF1 complex function in the nuclear calcium-sensing pathway required for gene regulation under salt stress is very challenging because tools to manipulate calcium levels in the nucleus of intact plants are lacking. In the current study, we observed that the calcium-binding EF-hand motif in RSA1 is critical for RSA1 function in vivo (Figure 3C and 3D). Under salt stress, RSA1 and RITF1 can transiently activate the SOS1 promoter, and the calcium-binding EF-hand motif in RSA1 is crucial for the activation of the SOS1 promoter as determined in a dual luciferase reporter assay (Figure 6I). Furthermore, the rsa1-2ritf1 double mutant plants do not display any additive effect of rsa1-2 and ritf1 mutations under salt stress or oxidative stress (Figure S8) and overexpression of RITF1 in the rsa1-1 background is unable to rescue the salt hypersensitive phenotype of rsa1-1 (Figure S10B and S10D). Together, these results suggest that RSA1 senses the salt-induced changes in nuclear free calcium and interacts with a transcription factor, RITF1, for gene expression, and that both RSA1 and RITF1 are required for salt tolerance. RITF1 may require posttranslational modifications (i.e., phosphorylation) to be active, and RSA1 is likely involved in these processes. Popescu et al. [36] reported that RITF1 is phosphorylated by multiple MAPKs in the presence of different MAPKKs in a protein microarray analysis. Further investigation is required to determine which MAPK phosphorylates RITF1 and the relationship between the RSA1-mediated calcium signal and phosphorylation of RITF1 by certain MAPK(s).
The importance of RSA1 and RITF1 proteins in salt tolerance is indicated by the loss-of-function and gain-of-function studies. The rsa1-1 mutation causes a hypersensitive phenotype in response to NaCl. The increased sensitivity in rsa1-1 is specific to Na+ (Figure 1 and Figure S1). This phenotype of rsa1-1 is very different from that of previously described salt overly sensitive (sos) mutants (sos1 through sos6), which are hypersensitive to both Na+ and Li+ [37]–[40]. In addition, as indicated by comparison with publicly available expression datasets, most of the differentially expressed genes with or without salt stress in rsa1-1 (as revealed in the microarray analysis) are not responsive to salt stress treatments in the wild type (Figure S5 and S6). Therefore, mis-regulation of these genes in rsa1-1 mutant plants may contribute to the overall increased salt sensitivity. Like rsa1-1 mutant plants, ritf1 mutant plants are more sensitive to salt stress than the wild type (Figure 5A and 5B). Overexpression of RSA1 or RITF1 improves the performance of transgenic Arabidopsis plants under salt stress (Figure 3E, 5F–5H). Furthermore, the excessive production of deleterious ROS in rsa1-1 plants may contribute to the increased sensitivity of the rsa1-1 mutant to salt stress (Figure 2 and 7). Upon salt stress, ROS accumulate to higher levels in the rsa1-1 mutant than in the wild type (Figure 2), which indicates that RSA1 is required for maintenance of ROS levels. The microarray analysis showed that RSA1 controls expression of genes that encode ROS scavenging enzymes and redox-related proteins. Improper functioning of these ROS detoxifying proteins is the possible cause of over-accumulation of ROS in rsa1-1. SOS1 is involved in maintaining proper ROS levels because ROS over-accumulate in the sos1-1 mutant plants under salt stress, and the sos1-1 mutation affects expression of several ROS detoxifying genes [6]. Therefore, reduced expression of SOS1 in rsa1-1 may also contribute to the increased salt sensitivity of rsa1-1. Finally, database searches revealed that close homologs of RSA1 are present in other plant species including monocots (such as rice and maize) and dicots (such as cucumber and tomato) (Figure S11A). In all of these plant species except grape, RSA1 homologs exist as a single copy gene. In contrast, two to three RITF1 close homologs are present in Arabidopsis and other plant species with RSA1 homologs (Figure S11B). These findings suggest that the RSA1-RITF1 complex is conserved across plant species. It is also possible that mechanisms of RSA1 and RITF1 function under salt stress are conserved in these plant species. Because overexpression of RSA1 or RITF1 increases salt tolerance in Arabidopsis, manipulation of RSA1 and RITF1 (or their close homologs) levels in salt-sensitive crops such as rice and tomato may increase their salt tolerance.
Materials and Methods
Plant material and growth conditions
A firefly luciferase reporter gene driven by the stress-responsive RD29A promoter [41] was introduced into Arabidopsis plants in the Columbia glabrous1 (gl1) background. Seeds from one homozygous line (referred to as the wild type) were mutagenized with ethyl methanesulfonate (EMS), and M2 seeds were used to screen for hypersensitive mutants in the presence of 100 mM NaCl with a modified root-bending assay [13], [14], [42]. Arabidopsis seedlings on Murashige and Skoog (MS) medium agar plates (1× MS salts, 2% sucrose, 1.2% agar, pH 5.7) were routinely grown in vertical orientation under cool, white light (∼120 µmol m−2 s−1) at 21°C with a 16-h-light/8-h-dark photoperiod. Soil-grown plants were kept under cool, white light (∼100 µmol m−2 s−1) with a 16-h-light/8-h-dark photoperiod at 21°C; the potting soil was a 1∶1 mixture of Metro Mix 360 and LC1 (Sun Gro Horticulture, Bellevue, WA).
Seeds of rsa1-2 (SALK_007142), rsa1-3 (CS16026), and rift1 (CS811403) were obtained from the Arabidopsis Biological Resource Center (ABRC; Columbus, OH).
Microarray analysis and qRT-PCR analysis
Six-d-old wild type and rsa1-1 mutant seedlings grown on MS medium (1× MS salts, 2% sucrose, 1.2% agar, pH 5.7) were transferred to MS medium supplemented with 0 or 120 mM NaCl and allowed to grow for an additional 24 h. Total RNA was prepared as described [43]. Microarray analysis was carried out using Arabidopsis Affymetrix ATH1 GeneChips in the School of Medicine, University of Maryland, Baltimore as described [42], [44], [45]. The microarray data discussed in this study have been deposited in NCBI's Gene Expression Omnibus [46] and are accessible through GEO Series accession number GSE39236. All qRT-PCR experiments were performed as described [43].
Chromatin immunoprecipitation (ChIP) assay
The RITF1 gene including its native promoter (2.8 kb) was amplified by PCR and cloned into pEarlyGate301. The resulting construct (pRITF1:RITF1-HA) was then transformed into wild-type plants by a floral dip method with Agrobacterium tumefaciens (strain GV3101)-mediated transformation [47]. ChIP assays were carried out with 15-d-old seedlings expressing pRITF1:RITF1-HA that were grown on horizontally oriented MS agar plates (1× MS salts, 2% sucrose, 0.6% agarose, pH 5.7) as described [48], [49]. Briefly, seedlings were crosslinked with 1% formaldehyde, and chromatin was isolated, sonicated (Fisher Biodismembrator, model120), and pre-cleared with salmon sperm DNA/protein-A agarose beads for 1 h. Samples were then immunoprecipitated with anti-HA antibody (Sigma, H6908) at 4°C overnight. The chromatin antibody complex was precipitated with salmon sperm DNA/protein-A agarose beads, washed with four different buffers for 5 min per buffer, and reverse-crosslinked with 200 mM NaCl in elution buffer (1% SDS, 0.1 M NaHCO3) for 6 h at 65°C. Proteins in the complex were removed by proteinase K at 45°C for 1 h. DNA was precipitated in the presence of two volumes of ethanol, 1/10 volume of 3 M sodium acetate (pH 5.2), and 2 µg of glycogen. Real-time PCR analysis was performed with immunoprecipitated DNA using a Bio-Rad CFX96 Real-Time System.
Electron scanning microscopy of rsa1-1 roots
Root morphology of wild-type and rsa1-1 seedlings with or without salt stress was observed with a scanning electron microscope (SEM, AMRAY 1820D). Seedlings were first fixed on MS medium with GA fixation solution (2% glutaraldehyde, 1× MS salts, 2% sucrose, pH 5.7) for 30 min at room temperature. Roots of fixed seedlings were then excised and placed in GA fixation solution overnight at 4°C. After roots were placed in a secondary fixation solution containing 1% OsO4 in distilled H2O (pH 7.0) for 60 min, they were sequentially washed with H2O (three times, 10 min each time), and then with 70, 95, and 100% ethanol (once for 10 min for each ethanol concentration). Roots were then critical point dried using CO2. Roots were mounted on stubs and shadowed with gold and palladium (4∶1) before viewing with the SEM.
Genetic mapping of the rsa1-1 locus and generation of RSA1-related constructs
For mapping of the rsa1-1 locus, homozygous rsa1-1 plants (in Columbia) were crossed to wild-type plants of Landsberg erecta. From the segregating F2 population, 657 homozygous rsa1-1 mutants were selected, and DNA was extracted from each of these plants for mapping with simple sequence length polymorphism (SSLP) markers between Columbia and Landsberg erecta.
A 8.9-kb genomic DNA fragment of At2g03150 including its putative promoter and 3′-termination sequence was amplified by PCR with the BAC clone T18E12 as a template and was cloned into the pMDC99 vector through Gateway technology (Invitrogen). The resulting binary vector (pRSA1:RSA1) was transformed into rsa1-1 plants by a floral dip method with A. tumefaciens (strain GV3101)-mediated transformation. T2 seedlings were examined for sensitivity to 100 mM NaCl.
The DNA fragment containing the RSA1 promoter was amplified by PCR with the pMDC99-RSA1 plasmid as a template and cloned into the binary vector pMDC164. The resulting binary vector (pRSA1:GUS) was transferred to wild-type (Columbia) plants, and pRSA1:GUS activity was observed as described [40].
The RSA1 coding region was amplified by PCR and cloned into the pEarleyGate104 binary vector. The resulting construct (p35S:YFP-RSA1) was then transformed into wild-type plants through an A. tumefaciens (strain GV3101)-mediated transformation. The subcellular localization of p35S:YFP-RSA1 was determined in root tip cells of T2 seedlings subjected to 0 or 120 mM NaCl for 24 h using a Leica SP5X confocal microscope (Leica Microsystems). The p35S:YFP-RSA1 construct was also transferred to rsa1-1 mutant plants to determine whether p35S:YFP-RSA1 is functional in vivo and to identify over-expressers of RSA1 in the rsa1-1 mutant background. The RSA1 coding region without EF-hand motif (deletion of 12 consensus amino acids in the EF-hand motif) was amplified by PCR reactions and cloned into the pEarleyGate 104 binary vector. The resulting construct (p35S:YFP-RSA1[-EF]) was transformed into rsa1-1 mutant plants through an A. tumefaciens (strain GV3101)-mediated transformation to determine whether the EF-hand motif in RSA1 is required for its function in vivo.
In a separate study, the RSA1 gene including its native promoter was amplified by PCR and cloned into the pMDC107 binary vector. The resulting construct (pRSA1:RSA1-GFP) was transformed into tobacco plants, and the subcellular localization of pRSA1:RSA1-GFP protein in tobacco leaves was determined as described [43]. The same construct was also transferred to the rsa1-1 mutant plants to determine whether pRSA1:RSA1-GFP is functional in vivo.
Recombinant protein expression and calcium overlay assay
For expression of His-tagged RSA1 (corresponding to amino acids from 1171 to 1340) and RSA1ΔEF (deletion of 12 consensus amino acids in the EF-hand motif) proteins in Escherichia coli (E. coli), RSA1 or RSA1(-EF) was amplified by PCR and cloned into pDEST17 through the Gateway technology (Invitrogen). The resulting plasmids (pDEST17-RSA1 and pDEST17-RSA1ΔEF) were transformed into E. coli strain Rosetta (DE3) pLysS (EMD Millipore). A 4 mL volume of overnight culture was inoculated into 400 mL of LB media containing 100 mg/L carbenicillin and 34 mg/L chloramphenicol and was incubated at 30°C until OD600 = 0.5. Protein expression was then induced overnight by adding IPTG to a final concentration of 0.4 mM at 28°C. Recombinant proteins were purified with affinity chromatography using a His-Trap column (GE Healthcare) according to the manufacturer's instructions.
Calcium overlay assay was performed as described [50], [51] with minor modifications. A 50 µg quantity of recombinant RSA1 or RSA1(-EF) was separated on SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, 0.45-µm-pore size). After transfer, the membrane was washed three times for 20 min each time with washing buffer (60 mM KCl, 5 mM MgCl2, and 10 mM imidazole-HCl [pH 6.8]) and incubated with 2.5 µCi/mL 45CaCl2 (PerkinElmer) in washing buffer for 20 min at room temperature. The membranes were washed with distilled water for 30 min, dried, and exposed to x-ray film for 3 d at −80°C.
Yeast two-hybrid screening, subcellular localization of RITF1, BiFC assay, Split-LUC assay, and Co-IP analysis
The yeast two-hybrid system and the Arabidopsis l-ACT cDNA expression library [52] were obtained from the ABRC (stock number CD4-22). The RSA1 coding region was amplified by PCR and cloned into pAS1-CYH2 to generate RSA1-pAS1-CYH2 as bait. Yeast (Saccharomyces cerevisiae strain AH109) transformation and library screening were as described [53]. The yeast colonies expressing different combinations of bait and prey constructs were analyzed for the LacZ reporter gene with paper-lifting methods. Whatman filter paper discs (#1) were used to transfer yeast colonies from the media and were then dipped in liquid nitrogen for 10 s; the color reaction was then performed by imbibing the paper discs in a solution containing 60 mM Na2HPO4⋅7H2O, 40 mM NaH2PO4⋅H2O, 10 mM KCl, 1 mM MgSO4, 38.6 mM β-mercaptomethanol, and 0.334 mg/mL of 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-Gal) at 30°C.
For validation of the results obtained from the above yeast two-hybrid screening, RSA1 and RITF1(At3g06590) coding regions were amplified by PCR and cloned into pDEST32 (pDEST32-RSA1) as bait and pDEST22 (pDEST22-RITF1) as prey, using the ProQuest™ Two-Hybrid System with Gateway Technology (Invitrogen). For potential interactions between RSA1 and RITF1 homologs (At3g17100, At3g05800, and At1g09250), coding sequences of RITF1 homologs were amplified by PCR and cloned into pDEST22. Yeast transformation and downstream analysis were performed according to the manufacturer's instructions.
The RITF1 coding region was amplified by PCR and the PCR product was cloned into the pMDC83 vector. The resulting construct (p35S:RITF1-GFP) was then transformed into Arabidopsis protoplasts prepared from 4-week-old Arabidopsis leaves. GFP signals in transformed protoplasts were then detected with a Leica SP5X confocal microscope (Leica Microsystems). The p35S:RITF1-GFP was also transformed into Arabidopsis wild-type and rsa1-1 plants to identify transgenic plants overexpressing RITF1 in wild-type or rsa1-1 background.
Bimolecular fluorescence complementation (BiFC) assays were conducted as described [54]. Briefly, coding regions of RSA1 and RITF1 were amplified by PCR and cloned into pUC-SPYNE and pUC-SPYCE, respectively. The resulting constructs (RSA1-nYFP and RITF1-cYFP) were co-transfected into protoplasts prepared from 4-week-old Arabidopsis leaves. YFP signals in transformed protoplasts were then detected by confocal microscopy. The RSA1-nYFP and RITF1-cYFP constructs were also transformed into A. tumefaciens strain C58C1 and co-infiltrated with 35S:p19 (p19 is a RNA silencing repressor protein from tomato bushy stunt virus [55]) in A. tumefaciens strain C58C1 into the 3-week-old leaves of tobacco (Nicotiana benthamiana) plants. The infiltrated tobacco plants were grown for an additional 3 d in a growth chamber under a 16-h-light/8-h-dark photoperiod at 21°C. YFP signals in transformed tobacco leaves were then detected by confocal microscopy.
For the Split-LUC assay, coding regions of RSA1 and RITF1 were amplified by PCR and cloned into the Gateway compatible firefly luciferase complementation imaging vectors (modified from original plasmids described by [56]). The resulting constructs (RSA1-nLUC and RITF1-cLUC) were transformed into A. tumefaciens strain C58C1 and co-infiltrated with 35S:p19 in A. tumefaciens strain C58C1 into the 3-week-old leaves of tobacco plants. The infiltrated tobacco plants were grown for an additional 3 d in a growth chamber under a 16-h-light/8-h-dark photoperiod at 21°C. Luciferase expression was observed with a CCD (charge-coupled device) camera as described [41].
Co-IP analysis was performed as described [57], [58] with minor modifications. Briefly, coding regions of RSA1 and RITF1 were amplified by PCR and cloned into pEarleyGate 202 and pEarleyGate 201, respectively. The resulting constructs (FLAG-RSA1 and HA-RITF1) were transformed into A. tumefaciens strain C58C1 and co-infiltrated with 35S:p19 in A. tumefaciens strain C58C1 into the 3-week-old leaves of tobacco (Nicotiana benthamiana) plants. The infiltrated tobacco plants were grown for an additional 3 d in a growth chamber under a 16-h-light/8-h-dark photoperiod at 21°C. Proteins were extracted from leaf samples with extraction buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 2 mM DTT, 10% glycerol, 1% Triton X-100, 1 mM PMSF, 1× Halt protease inhibitor cocktail [Fisher Scientific]). The protein extracts were incubated with anti-HA antibody (Sigma; Cat. No. H6908) overnight. The immunocomplexes were collected by adding protein A agarose beads and were washed with immunoprecipitation buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 2 mM DTT, 10% glycerol, 0.15% Triton X-100, 1 mM PMSF, 1× Halt protease inhibitor cocktail [Fisher Scientific]). The pellet (immunocomplexes with beads) was resuspended in 2× SDS-PAGE loading buffer. Eluted proteins were analyzed by immunoblotting using anti-FLAG antibody (Sigma; Cat. No. F1804) or anti-HA antibody (Sigma; Cat. No. H6908). Chemiluminescence signal was detected by autoradiography.
Dual luciferase reporter assays
The putative SOS1 promoter fragment (∼1.9 kb upstream of the translation start site) containing the cis element CACTTG that is recognized by RITF1 (as determined by the ChIP-qPCR analysis) was amplified by PCR with BAC clone F14H20 as a template. The PCR product was cloned into the transient expression vector pGreenII 0800-LUC to serve as a reporter plasmid. The coding regions of RSA1 and RITF1 were amplified by PCR and cloned into the transient expression vector pGreenII 62-SK to serve as effect plasmids. The RSA1 coding region without EF-hand motif (deletion of 12 consensus amino acids in the EF-hand motif) was amplified by PCR with p35S:YFP-RSA1(-EF) as a template and cloned into the transient expression vector pGreenII 62-SK to serve as effect plasmid. The resulting plasmids (SOS1∶LUC, RSA1, RITF1, and RSA1[-EF]) were transformed into A. tumefaciens (strain GV3101). Four-week-old tobacco leaves were co-infiltrated with the A. tumefaciens (strain GV3101) harboring the above plasmids in the following combination and ratios: SOS1∶LUC (100%); SOS1∶LUC+RITF1 (1∶9); SOS1∶LUC+RITF1+RSA1 (1∶4.5∶4.5); and SOS1∶LUC+RITF1+RSA1(-EF) (1∶4.5∶4.5). The infiltrated tobacco plants were grown for an additional 3 d in a growth chamber under a 16-h-light/8-h-dark photoperiod at 21°C and were subjected to 0 or 150 mM NaCl for 24 h. Activities of firefly luciferase under the control of the SOS1 promoter (SOS1:LUC) and Renillia luciferase under the control of the 35S promoter (35S:LUC) (both the SOS1:LUC and 35S:LUC transgenes are present on the SOS1:LUC reporter plasmid, and 35S:LUC served as an internal control) were measured using the reagents contained in the Dual-Luciferase Reporter Assay System (Promega) with the Modulus Microplate Multimode Reader (Turner BioSystem) as described [59]. Normalized data (ratio of luminescent signal intensity from the SOS1:LUC reporter and luminescent signal intensity from the internal control reporter, 35S:LUC) from 12 independent biological samples are presented.
Overexpression of SOS1 in rsa1-1
The coding region of SOS1 was amplified by PCR, and the PCR product was cloned into the binary vector pEarleyGate201. The resulting construct (p35S:HA-SOS1, abbreviated as SOS1 OE) was transformed into the rsa1-1 mutant plants through an A. tumefaciens (strain GV3101)-mediated transformation.
Ion content determination
Ion content in rsa1-1 was measured as described previously [60] with minor modifications. Briefly, 1-month-old wild-type and rsa1-1 plants grown in soil side by side in the same pots were treated with 150 mM NaCl and allowed to grow for one additional week. Shoots were harvested, dried in an oven at 65°C for 72 h, and weighed. A minimum of 2 g of dried shoot tissue was ashed at 480°C for 16 h before 2 mL of concentrated HNO3 was added to the ashes. After the ashes were dried on a hot plate, 10 mL of 3 M HCl was added, and the mixture was allowed to reflux for 2 h. The digests were passed through Whatman #40 filter paper, and the filtrate volumes were brought to 25 mL with 0.1 M HCI. Levels of Na+ and K+ were then determined with an inductively coupled plasma optical emission spectrometer (ICP-OES) (PerkinElmer Optima 4300 DV).
Determination of reactive oxygen species levels
Five-d-old seedlings grown on vertically oriented MS medium (1× MS salts, 2% sucrose, 1.2% agar, pH 5.7) were transferred to MS medium supplemented with 0 or 50 mM NaCl and allowed to grow for an additional 12 h. Seedlings were incubated with 5-(and 6)-chloromethyl-2′7′-dichlorodihydrofluorescein diacetate acetyl ester (CM-H2DCFDA) for 30 min and washed with distilled H2O to remove excess CM-H2DCFDA. Fluorescence images were obtained with a Leica SPX5 confocal microscope (Leica Microsystems), and ROS levels were quantified based on the intensity of fluorescence with the ImageJ software (NIH, http://rsbweb.nih.gov/ij/).
For quantification of H2O2, 5-d-old wild-type and rsa1-1 seedlings were transferred to MS medium containing 0 or 75 mM NaCl and allowed to grow for an additional 12 h. Seedlings were then harvested, and H2O2 content was determined with an Amplex red hydrogen/peroxidase assay kit (Invitrogen) according to the manufacturer's instructions.
Determination of chlorophyll and anthocyanin accumulation
Chlorophyll was determined as described [61] with minor modifications. Chlorophyll was extracted by incubating ground seedlings in 80% acetone overnight at 4°C in darkness and with continuous shaking. The contents of chlorophyll a and b were calculated as 7.49A664.9+20.3A648.2. Anthocyanin accumulation was determined as described [62]. Briefly, anthocyanin was extracted by incubating ground seedlings in acidified methanol (1% HCl) overnight at 4°C in darkness and with continuous shaking. The amount of anthocyanin was calculated as A530 - 0.33A657.
Supporting Information
Zdroje
1. HasegawaPM, BressanRA, ZhuJ-K, BohnertHJ (2000) Plant Cellular and Molecular Responses to High Salinity. Annu Rev Plant Physiol Plant Mol Biol 51 : 463–499.
2. ZhuJ-K (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6 : 441–445.
3. YangQ, ChenZZ, ZhouXF, YinHB, LiX, et al. (2009) Overexpression of SOS (Salt Overly Sensitive) Genes Increases Salt Tolerance in Transgenic Arabidopsis. Mol Plant 2 : 22–31.
4. ChengNH, PittmanJK, ZhuJ-K, HirsschiKD (2004) The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem 279 : 2922–2926.
5. QiuQS, GuoY, QuinteroFJ, PardoJM, SchumakerKS, et al. (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J Biol Chem 279 : 207–215.
6. Katiyar-AgarwalS, ZhuJ, KimK, AgarwalM, FuX, et al. (2006) The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 103 : 18816–18821.
7. ApelK, HirtH (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55 : 373–399.
8. GillSS, TutejaN (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48 : 909–930.
9. NandaAK, AndrioE, MarinoD, PaulyN, DunandC (2010) Reactive oxygen species during plant-microorganism early interactions. J Integr Plant Biol 52 : 195–204.
10. MøllerIM, JensenPE, HanssonA (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58 : 459–481.
11. JonakC, ÖkreszL, BögreL, HirtH (2002) Complexity, cross talk and integration of plant MAP kinase signaling. Curr Opin Plant Biol 5(5): 415–424.
12. KovtunY, ChiuW-L, TenaG, SheenJ (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97 : 2940–2945.
13. WuS-J, LeiD, ZhuJ-K (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8 : 617–627.
14. ZhuJ, GongZ, ZhangC, SongCP, DamszB, et al. (2002) OSM1/SYP61: A syntaxin protein in Arabidopsis controls abscisic acid–mediated and non-abscisic acid–mediated responses to abiotic stress. Plant Cell 14 : 3009–3028.
15. BorsaniO, ValpuestaV, BotellaMA (2001) Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol 126 : 1024–1030.
16. FujibeT, SajiH, ArakawaK, YabeN, TakeuchiY, et al. (2004) A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol 134 : 275–285.
17. DayIS, ReddyVS, Shad AliG, ReddyAS (2002) Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol 3 (10): research0056.1–0056.24.
18. HeimMA, JakobyM, WerberM, MartinC, WeisshaarB, et al. (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20 : 735–747.
19. Toledo-OrtizG, HuqE, QuailPH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 : 1749–1770.
20. EhrhardtDW, WaisR, LongSR (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85 : 673–681.
21. KosutaS, HazledineS, SunJ, MiwaH, MorrisRJ, et al. (2008) Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc Natl Acad Sci USA 105 : 9823–9828.
22. Sieberer BJ ChabaudM, TimmersAC, MoninA, FournierJ, et al. (2009) A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol 151 : 1197–1206.
23. van der LuitAH, OlivariC, HaleyA, KnightM, TrewavasA (1999) Distinct calcium signaling pathways regulate calmodulin gene expression in tobacco. Plant Physiol 121 : 705–714.
24. PaulyN, KnightMR, ThuleauP, GrazianaA, MutoS, et al. (2001) The nucleus together with the cytosol generates patterns of specific cellular calcium signatures in tobacco suspension culture cells. Cell Calcium 30 : 413–421.
25. WatahikiMK, TrewavasAJ, PartonRM (2004) Fluctuations in the pollen tube tip-focused calcium gradient are not reflected in nuclear calcium level: a comparative analysis using recombinant yellow cameleon reporter. Sex Plant Reprod 17 : 125–130.
26. LecourieuxD, LamotteO, BourqueS, WendehenneD, MazarsC, et al. (2005) Proteinaceous and oligosaccharidic elicitors induce different calcium signatures in the nucleus of tobacco cells. Cell Calcium 38 : 527–538.
27. WalterA, MazarsC, MaitrejeanM, HopkeJ, RanjevaR, et al. (2007) Structural requirements of jasmonates and synthetic analogues as inducers of Ca2+ signals in the nucleus and the cytosol of plant cells. Angew Chem Int Ed Engl 46 : 4783–4785.
28. MazarsC, BourqueS, MithöferA, PuginA, RanjevaR (2009) Calcium homeostasis in plant cell nuclei. New Phytol 181 : 261–274.
29. HirschS, KimJ, MuñozA, HeckmannAB (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21 : 545–557.
30. CapoenW, SunJ, WyshamD, OteguiMS, VenkateshwaranM, et al. (2011) Nuclear membranes control symbiotic calcium signaling of legumes. Proc Natl Acad Sci USA 108 : 14348–14353.
31. KalóP, GleasonC, EdwardsA, MarshJ, MitraRM, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 : 1786–1789.
32. LévyJ, BresC, GeurtsR, ChalhoubB, KulikovaO, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 : 1361–1364.
33. SmitP, RaedtsJ, PortyankoV, DebelléF, GoughC, et al. (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 : 1789–1791.
34. HeckmannAB, LombardoF, MiwaH, PerryJA, BunnewellS, et al. (2006) Lotus japonicus nodulation requires two GRAS-domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142 : 1739–1750.
35. TirichineL, Imaizumi-AnrakuH, YoshidaS, MurakamiY, MadsenLH, et al. (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441 : 1153–1156.
36. PopescuSC, PopescuGV, BachanS, ZhangZ, GersteinM (2009) MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev 23 : 80–92.
37. ZhuJ-K, LiuJ, XiongL (1998) Genetic analysis of salt tolerance in arabidopsis. Evidence for a critical role of potassium nutrition. Plant Cell 10 : 1181–1191.
38. ShiH, XiongL, StevensonB, LuT, ZhuJ-K (2002) The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 14 : 575–588.
39. ShiH, KimYS, GuoY, StevensonB, ZhuJ-K (2003) The Arabidopsis SOS5 locus encodes a cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15 : 19–32.
40. ZhuJ, LeeBH, DellingerM, CuiX, ZhangC, et al. (2010) A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J 63 : 128–140.
41. IshitaniM, XiongL, StevensonB, ZhuJ-K (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9 : 1935–1949.
42. LiW, GuanQ, WangZY, WangY, ZhuJ (2013) A Bi-Functional Xyloglucan Galactosyltransferase Is an Indispensable Salt Stress Tolerance Determinant in Arabidopsis. Mol Plant 6 : 1344–54 DOI: 10.1093/mp/sst062
43. GuanQ, WenC, ZengH, ZhuJ (2012) A KH Domain-Containing Putative RNA-Binding Protein Is Critical for Heat Stress-Responsive Gene Regulation and Thermotolerance in Arabidopsis. Mol Plant 6 : 386–395.
44. WangZY, XiongL, LiW, ZhuJK, ZhuJ (2011) The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 23 : 1971–1984.
45. GuanQ, WuJ, ZhangY, JiangC, LiuR, et al. (2013) A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 25 : 342–356.
46. EdgarR, DomrachevM, LashAE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 : 207–210.
47. CloughSJ, BentAF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–745.
48. GendrelA-V, LippmanZ, YordanC, ColotV, MartienssenRA (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 : 1871–1873.
49. GuanQ, LuX, ZengH, ZhangY, ZhuJ (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J 74 : 840–851.
50. MaruyamaK, MikawaT, EbashiS (1984) Detection of calcium binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J Biochem 95 : 511–519.
51. IshitaniM, LiuJ, HalfterU, KimCS, ShiW, et al. (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12 : 1667–1678.
52. KimJ, HarterK, TheologisA (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94 : 11786–11791.
53. TsengT-S, SaloméPA, McClungCR, OlszewskiNE (2004) SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16 : 1550–1563.
54. WalterM, ChabanC, SchützeK, BatisticO, WeckermannK (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 : 428–438.
55. VoinnetO, RivasS, MestreP, BaulcombeD (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 : 949–956.
56. ChenH, ZouY, ShangY, LinH, WangY, et al. (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146 : 368–376.
57. LeisterRT, DahlbeckD, DayB, LiY, ChesnokovaO, et al. (2005) Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17 : 1268–1278.
58. Choi duS, HwangIS, HwangBK (2012) Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24 : 1675–1690.
59. HellensRP, AllanAC, FrielEN, BolithoK, GraftonK, et al. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Method 1 : 13.
60. CodlingEE, MulchiCL, ChaneyRL (2007) Grain yield and mineral element composition of maize grown on high phosphorus soils amended with water treatment residual. J Plant Nutr 30 : 225–240.
61. LichtenthalerHK, WellburnAR (1983) Determinations of total carotenoids and chlorophyll a and b of leaf extracts in different solvents. Biochem Soc Trans 11 : 591–592.
62. MancinelliA, WalshL (1979) Photocontrol of anthocyanin synthesis. Plant Physiol 63 : 841–846.
63. HallTA (1999) BioEdit: a user-friendly biological sequence alignment editor and analyses program for Windows 95/98/NT. Nucleic Acids Symp Ser 41 : 95–98.
64. HruzT, LauleO, SzaboG, WessendorpF, BleulerS, et al. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 420747.
65. DereeperA, GuignonV, BlancG, AudicS, BuffetS, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36 (Web Server issue) W465–469.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



