-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Global Analysis of the Sporulation Pathway of
The Gram-positive, spore-forming pathogen Clostridium difficile is the leading definable cause of healthcare-associated diarrhea worldwide. C. difficile infections are difficult to treat because of their frequent recurrence, which can cause life-threatening complications such as pseudomembranous colitis. The spores of C. difficile are responsible for these high rates of recurrence, since they are the major transmissive form of the organism and resistant to antibiotics and many disinfectants. Despite the importance of spores to the pathogenesis of C. difficile, little is known about their composition or formation. Based on studies in Bacillus subtilis and other Clostridium spp., the sigma factors σF, σE, σG, and σK are predicted to control the transcription of genes required for sporulation, although their specific functions vary depending on the organism. In order to determine the roles of σF, σE, σG, and σK in regulating C. difficile sporulation, we generated loss-of-function mutations in genes encoding these sporulation sigma factors and performed RNA-Sequencing to identify specific sigma factor-dependent genes. This analysis identified 224 genes whose expression was collectively activated by sporulation sigma factors: 183 were σF-dependent, 169 were σE-dependent, 34 were σG-dependent, and 31 were σK-dependent. In contrast with B. subtilis, C. difficile σE was dispensable for σG activation, σG was dispensable for σK activation, and σF was required for post-translationally activating σG. Collectively, these results provide the first genome-wide transcriptional analysis of genes induced by specific sporulation sigma factors in the Clostridia and highlight that diverse mechanisms regulate sporulation sigma factor activity in the Firmicutes.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003660
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003660Summary
The Gram-positive, spore-forming pathogen Clostridium difficile is the leading definable cause of healthcare-associated diarrhea worldwide. C. difficile infections are difficult to treat because of their frequent recurrence, which can cause life-threatening complications such as pseudomembranous colitis. The spores of C. difficile are responsible for these high rates of recurrence, since they are the major transmissive form of the organism and resistant to antibiotics and many disinfectants. Despite the importance of spores to the pathogenesis of C. difficile, little is known about their composition or formation. Based on studies in Bacillus subtilis and other Clostridium spp., the sigma factors σF, σE, σG, and σK are predicted to control the transcription of genes required for sporulation, although their specific functions vary depending on the organism. In order to determine the roles of σF, σE, σG, and σK in regulating C. difficile sporulation, we generated loss-of-function mutations in genes encoding these sporulation sigma factors and performed RNA-Sequencing to identify specific sigma factor-dependent genes. This analysis identified 224 genes whose expression was collectively activated by sporulation sigma factors: 183 were σF-dependent, 169 were σE-dependent, 34 were σG-dependent, and 31 were σK-dependent. In contrast with B. subtilis, C. difficile σE was dispensable for σG activation, σG was dispensable for σK activation, and σF was required for post-translationally activating σG. Collectively, these results provide the first genome-wide transcriptional analysis of genes induced by specific sporulation sigma factors in the Clostridia and highlight that diverse mechanisms regulate sporulation sigma factor activity in the Firmicutes.
Introduction
Clostridium difficile is a Gram-positive, spore-forming, obligate anaerobe that causes gastrointestinal diseases including diarrhea, pseudomembranous colitis, and toxic megacolon [1]–[3]. C. difficile infections and C. difficile-related deaths have risen dramatically in the past decade, increasing the financial burden on health care systems [4]–[7]. While C. difficile is best known for causing hospital-acquired antibiotic-associated infections, recent epidemiologic studies indicate that community-acquired C. difficile infections are increasingly more common and associated with significant morbidity [6], [7]. A key element to the success of C. difficile as a pathogen is its ability to produce spores. Spores are resistant to most disinfectants and antibiotics, making them difficult to eliminate both from infected humans and the environment [1], [2], [8]. As a result, C. difficile spores disseminate readily from person to person and cause high rates of recurrent infections, which can lead to serious illness or even death [1]–[3], [9].
Although spores are critical to the pathogenesis of C. difficile, their composition and formation remain poorly characterized. Less than 25% of the spore coat proteins identified in the well-characterized spore-former Bacillus subtilis have homologs in C. difficile [10]. In contrast, the regulatory proteins that control spore coat gene expression and other sporulation events in B. subtilis are conserved in C. difficile and all other spore-forming Firmicutes [10]–[13]. These include the master sporulation transcriptional regulator, Spo0A, and the sporulation sigma factors σF, σE, σG, and σK.
In B. subtilis the sporulation sigma factors function at discrete stages during spore development to couple changes in gene expression with specific morphological changes in the cell [14]–[16]. The morphological changes begin with the formation of a polar septum, which creates two compartments, the mother cell and the forespore. The mother cell engulfs the forespore and guides the assembly of the spore until it lyses once spore maturation is complete. By coupling these developmental changes to the sequential activation of compartment-specific sporulation sigma factors, the mother cell and forespore produce divergent transcriptional profiles that coordinately lead to the formation of a dormant spore [16].
Sporulation gene transcription in B. subtilis begins with the activation of the transcription factor Spo0A, which in turn activates early sporulation gene transcription, such as the genes encoding the early sigma factors σF and σE. σF is initially held inactive by an anti-σ factor and only undergoes activation after septum formation is complete; this mode of regulation couples σF activation in the forespore to a morphological event [17], [18]. Active σF induces the transcription of genes whose products mediate cleavage of an inhibitory pro-peptide from σE in the mother cell via trans-septum signaling [19]. Active σE induces the transcription of genes whose products lead to the activation of the late sporulation sigma factor σG in the forespore, which occurs during or after engulfment [20], [21]. Activated σG in the forespore subsequently induces the expression of genes whose products proteolytically activate σK in the mother cell via trans-septum signaling [22]. Notably, the activity of each sigma factor relies on the activation of the preceding sigma factor [11], [14]–[16], [23]. As a result, the sigma factors operate in a sequential, “criss-cross” manner and collectively control the expression of hundreds of genes during sporulation [24]–[26].
The regulatory pathway controlling sporulation sigma factor activation in B. subtilis is thought to be conserved across endospore-forming bacteria, since all four sigma factors are conserved [11], [12]. However, a growing body of work in the Clostridia suggests that diverse pathways regulate sporulation sigma factor activity in the Firmicutes. In C. perfringens, a sigG− mutant still produces cleaved σK, suggesting that σG does not control the proteolytic activation of σK as it does in B. subtilis [27]. Furthermore, a C. perfringens sigK− mutant exhibits a phenotype more severe than a B. subtilis sigE− mutant in that it fails to initiate asymmetric division or produce σE [28], suggesting that in C. perfringens σK functions upstream of σE. Indeed, C. perfringens σE and σK have been suggested to be dependent on each other for full activity, in contrast with B. subtilis [28]. A similar early sporulation defect has been observed in a sigK− mutant of C. botulinum, which also exhibits reduced expression of early sporulation genes spo0A and sigF [29]. In contrast with B. subtilis and C. perfringens, however, a C. acetobutylicum sigF− mutant does not initiate asymmetric division [30], and a sigE− mutant fails to complete asymmetric division [31]. In addition, a C. acetobutylicum sigE− mutant produces wildtype levels of σG [31] in contrast with B. subtilis, and a sigG− mutant exhibits elongated forespores and pleiotropic defects in coat and cortex formation [31].
To determine how these sporulation sigma factors regulate sporulation in C. difficile, we constructed mutations in the genes encoding the sporulation transcription factor Spo0A and the sigma factors σF, σE, σG, and σK and determined the transcriptional profiles of these mutants using RNA-Sequencing (RNA-Seq). The transcriptional analyses, combined with cytological characterization of the sigma factor mutants, suggest that divergent mechanisms regulate the activity of σG and σK in C. difficile relative to B. subtilis and other Clostridium spp. In addition, these analyses have identified a set of 314 genes that are upregulated during sporulation in a Spo0A-, σF-, σE-, σG-, and/or σK-dependent manner. These sporulation-induced genes provide a framework for identifying and characterizing C. difficile spore proteins that may have diagnostic or therapeutic utility.
Results
C. difficile sporulation sigma factors are essential for mature spore formation
In order to identify genes that are regulated by the sporulation-specific sigma factors, we used a modified TargeTron gene knockout system to disrupt the genes encoding σF, σE, σG, and σK in C. difficile [32]. This system uses a group II intron to insert an erythromycin resistance cassette into the target gene (Figure S1A). JIR8094 [33], an erythromycin-sensitive derivative of the sequenced C. difficile strain 630 [34], was used as the parental strain. As a control, we also constructed a targeted disruption in spo0A, which encodes the master regulator of sporulation Spo0A [35], [36]. Colony PCR of the intron-disrupted mutants confirmed the expected size change resulting from the intron insertion into the spo0A, sigF, sigE, sigG, and sigK genes (Figure S1B).
To determine the effect of blocking sigma factor production on sporulation, the mutants were induced to sporulate on solid sporulation media and visualized by phase contrast microscopy [37]. It should be noted that sporulation is asynchronous in this assay, and the extent and timing of sporulation exhibits variability even between biological replicates (Figure S2). Nevertheless, after 18 hrs of growth, sufficient numbers of cells have initiated sporulation to detect the production of immature phase-dark forespores and mature phase-bright spores in the wildtype strain (Figure 1 and S2). In contrast, spo0A−, sigF−, sigE−, sigG−, and sigK− cultures failed to produce phase-bright spores (Figure 1). No phase-dark or phase-bright forespores were observed in the spo0A−, sigF−, or sigE− strains, suggesting a block early in sporulation.
Fig. 1. C. difficile sigF − sigE−, sigG−, and sigK− sigma factor mutants are defective in mature spore formation. 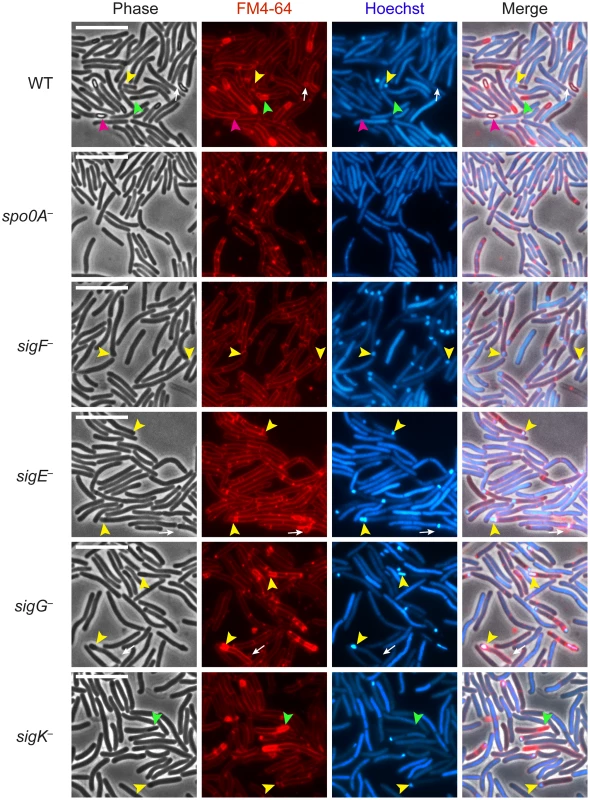
C. difficile strains wildtype (WT), spo0A−, sigF−, sigE−, sigG−, and sigK− were grown on sporulation media for 18 hrs and evaluated by live phase-contrast and fluorescence microscopy. Phase contrast, FM4-64 membrane staining (red), nucleoid staining with Hoechst (blue), and the merge of these images are shown for each strain. Yellow arrowheads indicate forespore compartments that stain with FM4-64 and Hoechst; green arrowheads indicate phase-dark, immature forespores that stain with FM4-64 but not Hoechst; and pink arrowheads indicate phase-bright mature spores that exclude both the FM4-64 and Hoechst stains. Phase-bright spores were not observed in any of the mutant strains. Circular vesicles that were labeled by FM4-64, but not visible by phase-contrast microscopy (white arrows) were frequently observed in cultures grown on sporulation media, even in the spo0A− mutant (data not shown). Scale bars represent 10 µm. Analysis of live, sporulating cultures with the lipophilic dye FM4-64 (to stain mother cell and forespore membranes) and Hoechst 33342 (to stain cell nucleoids) revealed polar septum formation in wild type and the sigma factor mutants but not in the spo0A− mutant (Figure 1). This result was consistent with the observation that Spo0A is necessary to induce the sporulation pathway in C. difficile [35], [36]. Overall, the proportion of sporulating cells detected by membrane and DNA staining in the culture was 25%, 41%, 24%, 26%, and 18% for wildtype, sigF−, sigE−, sigG−, and sigK−, respectively, as indicated by the presence of a polar septum, immature forespore compartment, or mature forespore (Table S1). Wildtype cultures contained a heterogenous population of sporulating cells at discrete stages of sporulation: 28% of sporulating cells exhibited intense DNA staining of an FM4-64-labeled forespore compartment (yellow arrows, Figure 1, Table S1); 28% showed phase-dark forespores that stained with both FM4-64 and Hoechst (Table S1), 28% exhibited phase-dark forespores that stained intensely with FM4-64 but not Hoechst (green arrows, Figure 1, Table S1), and 16% contained a phase-bright forespore that failed to be stained with either FM4-64 or Hoechst (pink arrows, Figure 1, Table S1). In contrast, sigF− and sigE− sporulating cells were arrested at the asymmetric division stage, with 95% and 92% of sporulating cells, respectively, exhibiting intense DNA staining of an FM4-64-labeled forespore compartment (yellow arrows, Figure 1, Table S1). The sigG− mutant strain was arrested at the phase-dark forespore stage, with 69% of sporulating cells exhibiting intense forespore membrane and nucleoid staining (yellow arrows, Figure 1, Table S1). While only 4% of the sigG− cells were observed to produce forespores that stained only with FM4-64, 44% of sporulating sigK− cells were captured at this stage of sporulation, a phenotype that was also observed in wildtype (green arrows, Figure 1, Table S1). Taken together, these results indicate that all four sporulation sigma factors are required to complete spore formation and suggest that σG is necessary to complete the stage of sporulation development required to exclude the Hoechst dye from staining the forespore chromosome. The results are also consistent with studies investigating B. subtilis forespore development, which indicate that nucleic acid stains are excluded earlier than membrane stains during spore development [38]–[40].
To confirm that the gene disruptions prevented sigma factor production in each of the respective sigma factor mutants, we performed Western blot analyses using antibodies raised against C. difficile sigma factors. Similar to B. subtilis, Spo0A was required for the production of all the factors, and σF was observed in the sigE−, sigG−, and sigK− strains at wildtype levels (Figure 2, [41]). σE was detected in both its pro - and cleaved form in wildtype, sigG− and sigK− strains, whereas the majority of σE was unprocessed in the sigF− strain (Figure 2). This result slightly deviates from the B. subtilis model, where pro-σE processing is completely abrogated in a B. subtilis sigF− strain [42]. In contrast, a C. perfringens sigF− mutant fails to produce pro-σE altogether [27], and σE processing has not been demonstrated in C. acetobutylicum [31]. σK was present in wildtype and sigG− mutant strains but absent in the sigF− and sigE− strains (Figure 2), analogous to observations in B. subtilis where σE is required for sigK expression. A C. perfringens sigE− strain in contrast produces low amounts of σK [28]. Consistent with the observation that C. difficile σK lacks an N-terminal pro-peptide [43], no processing of σK was observed in wildtype C. difficile (Figure 2), even though σK undergoes proteolytic activation in B. subtilis and C. perfringens [27]. σG was detected in the C. difficile sigF−, sigE− and sigK− mutants (Figure 2) in contrast with studies of other endospore-forming bacteria, where σG activity and auto-activation of sigG transcription is partially dependent on σE in B. subtilis [44]–[46], and σG production depends on σF in C. perfringens and C. acetobutylicum [27], [30].
Fig. 2. Analysis of sporulation sigma factor production in sporulation sigma factor mutants. 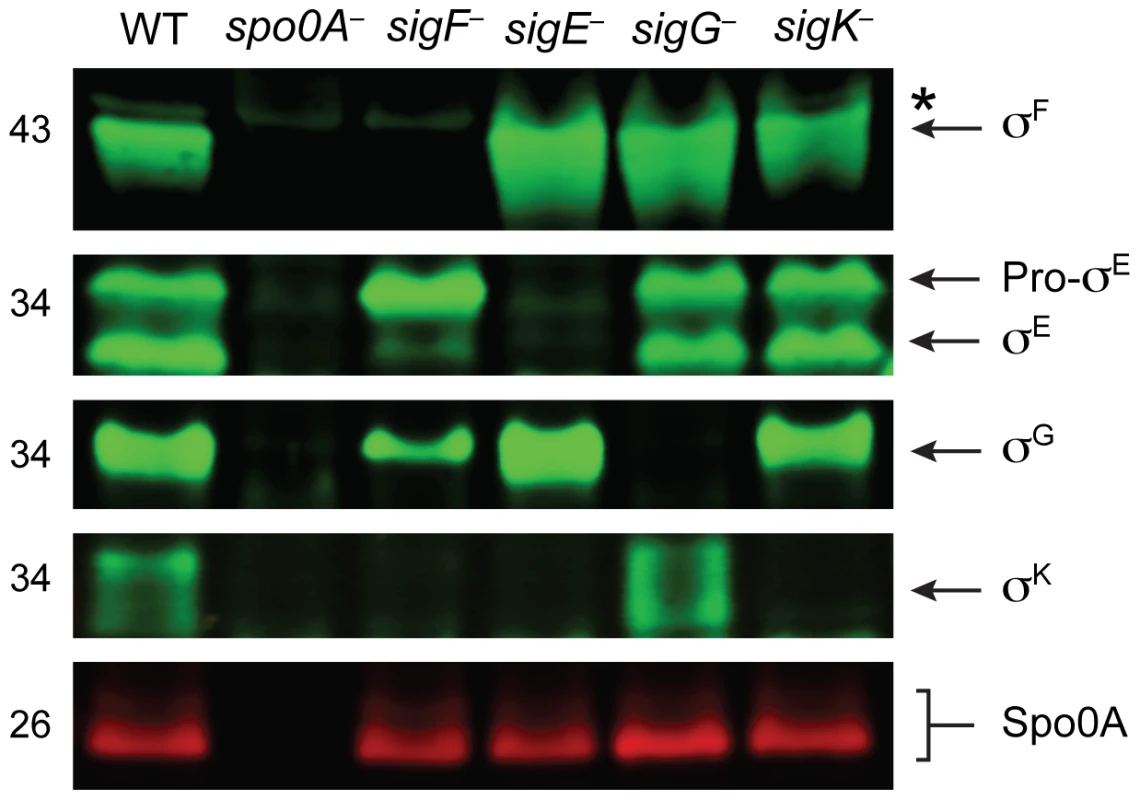
Western blot analyses of σF, σE, σG, σK, and Spo0A, respectively, in wildtype (WT), spo0A−, sigF−, sigE−, sigG−, and sigK− strains grown for 18 hr on sporulation media using antibodies raised against Spo0A and the sporulation sigma factors. The * demarcates a non-specific band observed in the sigF− and spo0A− mutants. Pro-σE indicates full-length σE prior to pro-peptide removal. We next performed transmission electron microscopy (TEM) to identify the precise developmental stage at which each sigma factor mutant was stalled. Cortex and coat layers were present on forespores in wildtype sporulating cells, while the spo0A− mutant exhibited no signs of spore formation (Figure 3). The sigF− mutant failed to progress beyond asymmetric division (Figure 3), similar to a B. subtilis sigF− mutant [47] but in contrast with a C. acetobutylicum sigF− mutant which does not initiate asymmetric division [30]. Nevertheless, unlike B. subtilis, a more electron-translucent region in the mother cell cytosol surrounded by electron dense layers was observed in some sigF− mutant cells; this region resembled mislocalized spore coat ([37], Figure 3). The C. difficile sigE− strain was arrested at the asymmetric division stage similar to the sigF− mutant, although electron-translucent regions surrounded by coat-like layers were not observed in any sigE− cell analyzed. The C. difficile sigE− mutant phenotype resembled the phenotype of sigE− mutants of B. subtilis [47] and C. perfringens [28], with frequent observations of disporic cells or cells with multiple septa at one pole (Figures 1 and 3). This observation was in contrast with a C. acetobutylicum sigE− mutant, which does not complete asymmetric division [31]. The C. difficile sigG− mutant produced forespores lacking an apparent cortex layer, similar to B. subtilis [21], [44]; however, unlike B. subtilis, the forespores were surrounded by thin layers that resembled the spore coat layers visible in wildtype cells (Figure 3). In addition, the C. difficile sigG− mutant exhibited pleiotropic defects including forespore ruffling, incomplete membrane fission during engulfment, and a septated forespore compartment (Figures 3 and S3). Quantitation of the prevalence of each phenotype revealed that forespore ruffling, incomplete engulfment, and a septated forespore compartment were observed in 98, 87 and 21% of sigG− cells, respectively (Figure S3). Lastly, the C. difficile sigK− mutant produced forespores surrounded by a layer that resembled the cortex layer of wildtype, but no coat layers were apparent (Figure 3). This phenotype was more similar to a B. subtilis sigK− mutant, which lacks both cortex and coat [22], than C. perfringens, which fails to initiate polar septum formation [28].
Fig. 3. C. difficile sigF−, sigE−, sigG−, and sigK− mutants are arrested at different stages of spore formation. 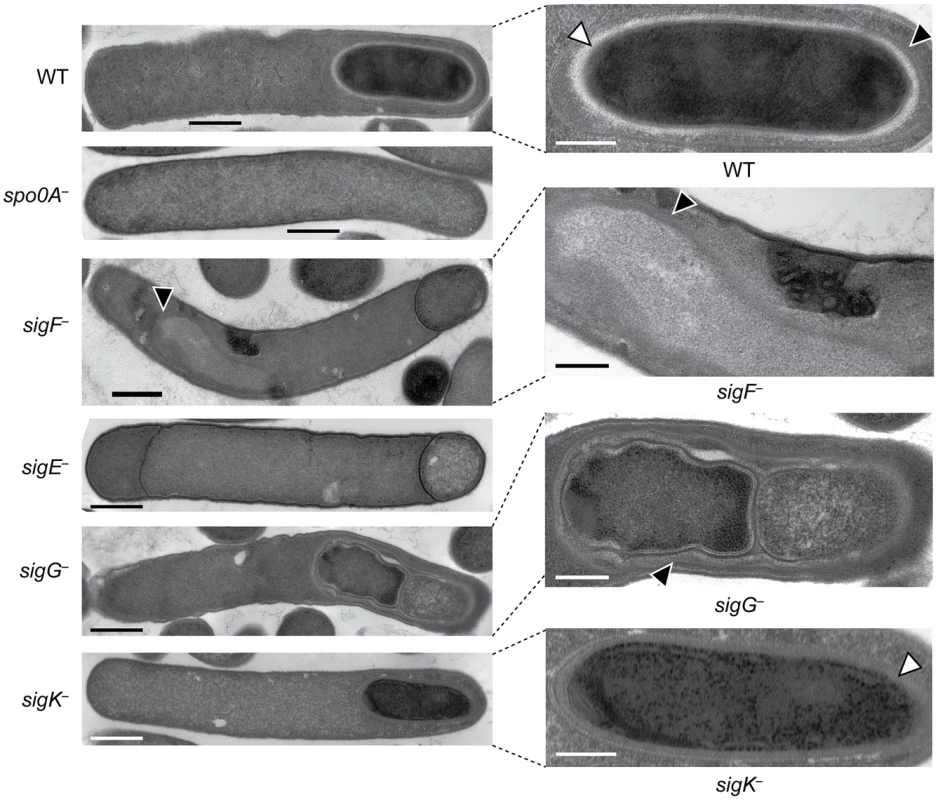
Transmission electron microscopy (TEM) of wildtype, spo0A−, sigF−, sigE−, sigG−, and sigK− strains at 18 hrs of growth on sporulation media. The forespore regions of wild type (WT), sigG−, and sigK− strains, and an electron-translucent region within the sigF− mutant mother cell cytosol, are shown on the right. Black triangles indicate regions that resemble coat layers, while white triangles indicate regions consistent with cortex. Scale bars represent 500 nm. Inset scale bars represent 250 nm. Plasmid complementation rescues the sporulation defects of sigF−, sigE− sigG−, and sigK− mutants
To validate that the observed mutant phenotypes were due to the targeted insertions, we complemented the mutant strains by expressing a wildtype copy of the gene encoding the corresponding sigma factor from a plasmid. We used either the pMTL83151 or pMTL84151 multicopy plasmids [48] to express the complementing genes or operons from their native promoters. The complementation constructs all restored production of phase-bright spores when expressed in their respective mutant backgrounds (Figure S4A), although phase-bright spore formation by the sigK complementation strain was delayed relative to wildtype. Western blot analysis further confirmed that the complementation constructs restored production of the respective sigma factor to wildtype levels (Figure S4B). TEM analysis revealed that all four complementation constructs restored coat and cortex formation to their respective mutant strains (Figure S5). Heat resistance assays to measure complementation strain sporulation efficiency revealed that the sigF− and sigE−constructs fully complemented heat resistance relative to wildtype and that the sigG− and sigK− constructs partially complemented heat resistance (70 and 23%, respectively, Figure S4C).
RNA-Seq analysis reveals the regulatory relationships between C. difficile sporulation sigma factors
While these analyses showed that σF, σE, σG, and σK were all required for mature spore formation, they did not reveal which genes were being misregulated in the sporulation sigma factor mutants to produce their respective sporulation defects. To identify these genes and gain insight into the regulatory network controlling sporulation sigma factor activity, we used RNA-Sequencing (RNA-Seq) to transcriptionally profile our sporulation mutants and wild type during sporulation. Three biological replicates of wildtype, spo0A−, and sporulation sigma factor mutant strains were grown on sporulation media (Figure S2), and RNA was isolated. Following DNase-treatment, ribosomal RNA depletion and reverse transcription, Illumina-based RNA-Seq was used to determine the complete transcriptome of wildtype C. difficile and the sporulation mutants. Genome coverage and sequencing counts for each strain and replicate can be found in Table S2.
The DeSeq variance analysis package [49] was used to identify genes that were downregulated by ≥4-fold with an adjusted p-value of ≤0.05 in the spo0A− strain relative to wild type. This pair-wise analysis identified 276 genes as being Spo0A-dependent (Table S3). Consistent with the role of Spo0A as the master regulator of sporulation, 65 of these genes were predicted to be involved in sporulation (Table S4) [11], [50]–[52]. Six of these Spo0A-dependent genes were recently identified as encoding components of the C. difficile spore coat [50], [53], and 36 sporulation-related genes (Table S4) were shown to depend on σH, the stationary phase sigma factor that induces spo0A transcription in C. difficile [51] and B. subtilis [54]. σF-, σE-, σG-, and σK-dependent genes were identified by comparing the transcriptional profiles of the sigF−, sigE−, sigG−, and sigK− strains to wild type, respectively, using the same parameters as above. This analysis identified 183 genes as being dependent on σF for their expression (Table S5). One hundred eighteen of these σF-dependent genes were also σE-dependent (Table S6), indicating that σE has some activity in a sigF− mutant consistent with the reduced levels of cleaved σE being detected by Western blot (Figure 2); 29 of the σF-dependent genes formed a separate subset of genes that were also σG-dependent but σE-independent. Indeed, the majority of the 34 σG-dependent genes identified in this analysis were not dependent on σE for their expression (Table S7), since only four of the σG-regulated genes were also σE-regulated. Notably, none of the genes identified as being σG-dependent required σK for their expression (Table S8), suggesting that the σG produced in the sigE− and sigK− mutants is active (Figure 2). This result differs from the B. subtilis model where σE is needed to fully activate σG function [20], [21], [46], [55], [56].
Of the 169 genes that depended on σE for their expression (Table S6), 85% and 78% of these genes were dependent on Spo0A and σF, respectively (Figure 4). The expression of 29 of these genes was also σK-dependent (Table S5). Indeed the majority of the 31 σK-dependent genes were σE-dependent (Table S8; Figure 4), consistent with σE being required for σK production (Figure 2). In contrast, as described earlier, no overlap was observed between σG - and σK-dependent genes (Figure 4). Taken together, the RNA-Seq analyses suggested that (1) a small subset of σF-dependent genes are neither σE, σG, nor σK-dependent; (2) σE activity depends on Spo0A and σF but not σG or σK; (3) σK activity depends on Spo0A, σF, and σE but not σG, and (4) σG activity depends on Spo0A and σF but not σE or σK. The latter two findings differ from the B. subtilis model, where the σK-dependent genes are also σG-dependent because σK activity depends on σG [11], [15], [22], and σG-dependent genes are σE-dependent because full activation of σG requires σE [20], [21], [46], [55], [56].
Fig. 4. Venn diagram of genes identified as being dependent on either Spo0A, σF-, σE-, σG-, and/or σK-dependent as determined by RNA-Seq. 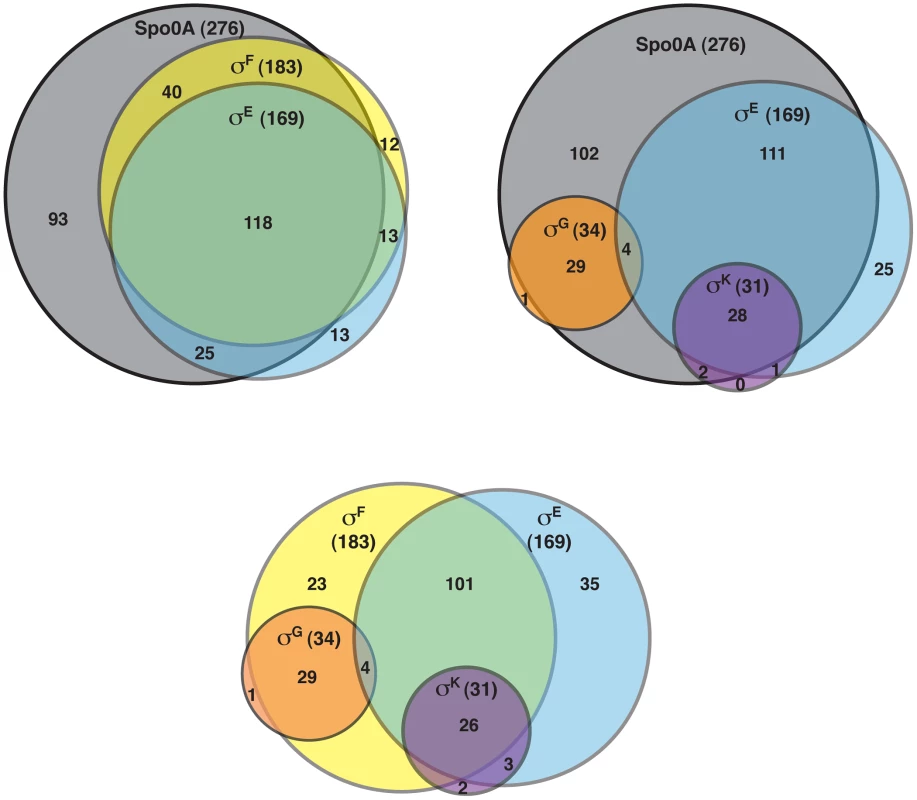
Genes were defined as being dependent on their respective sigma factor for expression if their transcript levels were decreased by ≥4-fold with an adjusted p-value of ≤0.05 in the mutant strains relative to wild type. The genes identified in these analyses are listed in Tables S3, S5, S6, S7 and S8. To visually represent the differences in gene expression profiles between the sigma factor mutants and wild type, we generated a heat map for genes downregulated by ≥4-fold with an adjusted p-value of ≤10−5 in the spo0A− strain relative to wild type. The expression levels of wild type and the sigma factor mutants relative to spo0A− strain were centered, scaled, and mapped to a red-green color scale. The heat map revealed a cluster of genes that was poorly expressed in the sigE− mutant relative to the wildtype, sigG−, and sigK− strains; these genes were also expressed at reduced levels in the sigF− mutant (Figure 5) and were primarily σE-dependent (Table S4). A separate cluster of genes was downregulated in both the sigK− and sigE− mutants relative to the wildtype and sigG− strains (Figure 5); these genes were all identified as σK-dependent genes (Table S5). Another discrete cluster of genes was downregulated in the sigG− and sigF− strains relative to the wildtype, sigE−, and sigK− strains (Figure 5); again, most of these genes were identified as σG-dependent genes, although two genes were σF-dependent but not σG-dependent (Tables S5 and S7). Thus, identification of variably expressed genes between the strains confirmed the findings of our earlier pair-wise analyses: σF-dependent genes were largely Spo0A-dependent, σE-dependent genes were largely σF-dependent, σK-dependent genes were σE-dependent, and σG-dependent genes were σF-dependent but not σE - or σK-dependent. These results support a model where (1) σF controls the activation of both σE and σG, (2) σE induces the production and activation of σK, and (3) σE and σK are dispensable for σG activation. Alternative statistical models were also employed to validate these findings (see Text S1 and Figures S6 and S7).
Fig. 5. Comparison of Spo0A-dependent gene expression in wildtype and sporulation sigma factor mutants. 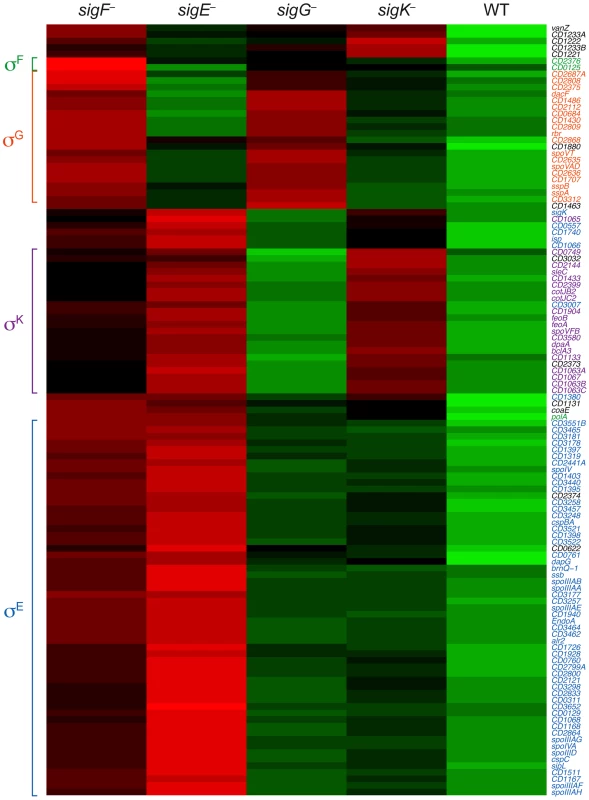
Heat map representation of the genes that were downregulated by ≥4-fold with an adjusted p-value of ≤10−5 in the spo0A− strain relative to wild type. Expression levels of these genes in wildtype (WT), sigF−, sigE−, sigG−, and sigK− strains relative to spo0A− were centered, scaled, and mapped to a red-green color scale, with green indicating that the gene was upregulated in the strain relative to the other strains, and red indicating that the gene was downregulated relative to the centered expression level. σE-regulated genes (blue), σK-regulated genes (purple), σG-regulated genes (orange), σF-dependent genes (green) are colored as indicated, and clusters of coordinately regulated genes are bracketed. Genes colored in black were identified as depending only on Spo0A for expression. Quantitative RT-PCR validates the RNA-Seq Data
To validate the RNA-Seq data, we isolated RNA from three separately prepared biological replicates of wildtype, spo0A−, sigF−, sigE−, sigG−, and sigK− strains grown on sporulation media for 18 hrs. RNA was reverse transcribed and quantitative RT-PCR (qRT-PCR) was performed using primers specific for three genes within each of the sigma factor-dependent transcriptomes. Gene expression levels in the wildtype and the sigma factor mutant strains relative to spo0A− were determined by comparative CT analysis normalized to the housekeeping gene rpoB. These analyses confirmed that the transcript levels of the σF-dependent gene gpr was reduced by >50-fold (p<0.0001) in the sigF− mutant relative to wild type, and reduced in the sigG− mutant by ∼4 fold (p<0.01); gpr expression was not affected in the sigE− and sigK− mutants. cd0125 (spoIIQ, [13]) transcription was reduced by >10-fold in the sigF− mutant relative to wild type (p<0.01), but no reduction in transcript levels was observed in sigE−, sigG−, and sigK− mutants (Figure 6A). Transcription of cd2376 was reduced by 3-fold in the sigF− relative to wild type (Figure 6A). Although this correlation was not statistically significant, it approached statistical significance (p = 0.065) (Figure 6A); this result is likely due to the low number of overall cd2376 transcripts present in the samples. Transcript levels of the σG-dependent genes spoVT, sspB, and dacF showed significant reductions in the sigG− (p<0.0004, <0.0002 and <0.0001, respectively) and sigF− mutants (p<0.0001) compared to wild type but no significant reduction in the sigE− and sigK− mutants relative to wild type (Figure 6B). This observation was consistent with the RNA-Seq data indicating that σG activity depends on σF, although it is likely that σF directly induces the transcription of some σG-dependent genes given the predicted overlap in their promoter specificities [11]. Nevertheless, given that σG is present at wildtype levels in a sigF− strain, these observations suggest that σF regulates σG activity through a post-translational mechanism.
Fig. 6. qRT-PCR validation of RNA-Seq transcriptional profiling. 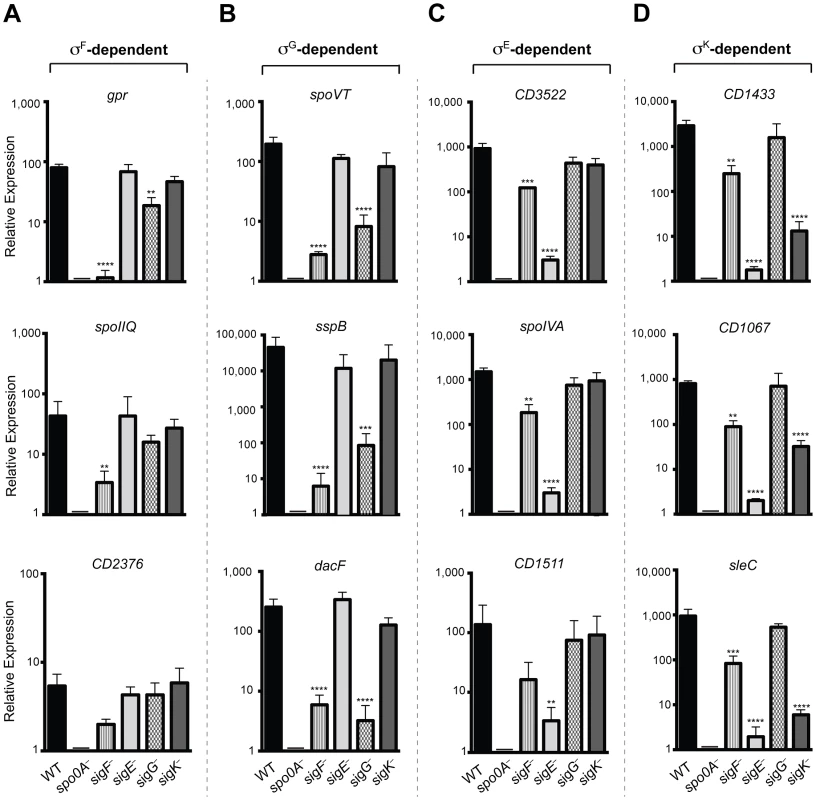
Transcript levels for three genes that were determined to be dependent on σF, σE, σG, and/or σK for expression as measured by qRT-PCR on three biological replicates. Samples were distinct from those used for RNA-Seq. (A) σF-dependent genes included gpr, CD0125 (spoIIQ [13]) and CD2376. (B) σG-dependent genes included spoVT sspB, and dacF. (C) σE-dependent genes included CD3522, spoIVA, and CD1511. (D) σK-dependent genes included CD1433, CD1067 and sleC. cDNA was produced from RNA samples harvested from wildtype (WT), spo0A−, sigF−, sigE−, sigG−, and sigK− strains grown on sporulation media for 18 hrs. Data represent the averages of three biological replicates and at least two technical replicates. Transcripts were calculated relative to spo0A− and normalized to rpoB (housekeeping gene). Error bars indicate the standard error of the mean. Statistically significant changes in transcript levels were determined relative to WT and represented by adjusted p-values determined by a Dunnett's one-way ANOVA. ****p<0.0001, ***p<0.001, **p<0.01. CD2376 transcript levels were ∼3-fold reduced in the sigF− mutant relative to wild type (p = 0.065). σE-dependent genes cd3522 and spoIVA were reduced by >100-fold, and cd1511 by >50-fold, in sigE− relative to wild type, (p<0.0001, <0.0001, and <0.006, respectively), but not in sigG− and sigK− mutants (Figure 6C). Transcript levels of these σE-dependent genes were reduced by ∼5 to 6-fold (p<0.01) in the sigF− mutant relative to wildtype, indicating that, in the absence of σF, σE activity is reduced but detectable. Transcript levels of the σK-dependent genes cd1433, cd1067 and sleC were significantly reduced by >100-fold in the sigE− (p<0.0001 for each gene) and the sigK− (p<0.0001 for each gene) strains compared to wild type (Figure 6D). σK-dependent gene expression was reduced in the sigF− mutant by 8 to 10-fold (p<0.01), suggesting that σK has reduced but detectable activity in the sigF− strain. Importantly, no statistically significant change for any of these σK-dependent genes was observed in the sigG− mutant relative to wild type, consistent with the RNA-Seq results indicating that σK activity does not depend on σG (Figures 4 and 5). Altogether, the qRT-PCR data validated the RNA-Seq data identifying σF, σE, σG, and σK-dependent genes and confirmed that (1) σE, σG, and σK activity depend on σF, (2) full σG activity requires σF but not σE, and (3) σK activity requires σE but not σG. It should be noted however that, although σF is required for full σE and σK activity, some degree of σE - and σK-dependent gene expression is observed even in the absence of σF.
Western blot analyses confirm that σK activity depends on σE but not σG
Having validated the RNA-Seq data at the transcript level, we next investigated whether changes in transcript levels correlated with changes in protein levels for σF-, σE-, σG-, and σK-regulated genes. To this end, we raised antibodies against proteins encoded by genes identified by RNA-Seq as being σF-, σE-, σG-, and σK-dependent. Western blot analyses of the germination protease Gpr confirmed that only σF is required for gpr expression, while production of the regulatory protein SpoVT and the small acid-soluble protein SspA depended on both σF and σG. These results indicate that σG can directly activate the expression of spoVT and sspA (Figure 7). Western blot analyses for CD3522, SpoIVA, and CD1511 demonstrated that their production depends on σE but not σG or σK; these proteins were detected, albeit at greatly reduced levels, in the sigF− mutant (Figure 7). These results were consistent with the observation that active, processed σE is present in both sigG− and sigK− strains, while only trace amounts of processed σE could be detected in the sigF− strain (Figure 2). Analysis of σK-dependent protein production using antibodies specific for CD1433, CD1067 and SleC confirmed that these proteins were absent in the sigE− and sigK− mutants and present in wild type and the sigG− mutant (Figure 7). Only SleC was reliably detected in the sigF− mutant, even though cd1433 and cd1067 transcripts could be detected in the sigF− strain (Figure 6D). Nevertheless, taken together these observations confirm that (1) σF does not require σE, σG, or σK for activation, (2) full σE activation requires σF, (3) full σG activation requires σF but not σE or σK, and (4) σK activation requires σF and σE but not σG.
Fig. 7. Western blot analyses of proteins encoded by genes induced by specific sigma factors during sporulation. 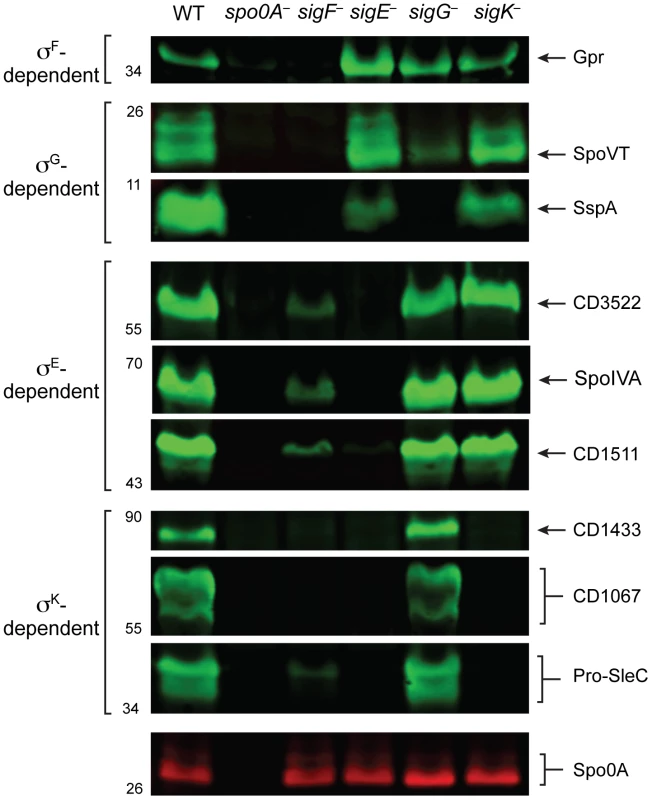
Western blot analyses of proteins encoded by genes identified as being upregulated during sporulation by specific sigma factors. Wildtype (WT), spo0A−, sigF−, sigE−, sigG−, and sigK− strains were grown on sporulation media for 18 hrs. SleC undergoes multiple processing steps [76], [85], but only the pro-SleC form is shown. Spo0A was used as a loading control. Discussion
The regulation of sporulation in the Clostridia has remained poorly characterized relative to the model spore-forming bacterium B. subtilis because the function and activity of all four sporulation sigma factors has not been simultaneously interrogated in a given Clostridium sp. to date. By constructing mutations in genes encoding for individual sporulation sigma factors in the nosocomial pathogen C. difficile and performing whole genome transcriptional profiling on these mutants, we identified 314 genes whose expression is activated during sporulation (Table S9) in a Spo0A, σF-, σE-, σG-, and/or σK-dependent manner (Tables S3, S5, S6, S7, S8). These experiments reveal that the sporulation pathway of C. difficile exhibits numerous differences relative to B. subtilis and other Clostridium spp., highlighting the diversity of mechanisms that regulate sporulation sigma factor activity in the Firmicutes.
Diverse mechanisms regulate sporulation sigma factor activity in the Firmicutes
While mutation of all four sporulation sigma factors in C. difficile abrogated functional spore formation as expected [11], the regulation and function of these sigma factors in C. difficile differed from the regulatory pathways determined for B. subtilis and other Clostridium spp. The differences between C. difficile, C. perfringens, C. acetobutylicum, and B. subtilis sporulation pathways are summarized in Figure 8, as are the similarities.
Fig. 8. Comparison of sporulation regulatory network architecture in the Firmicutes. 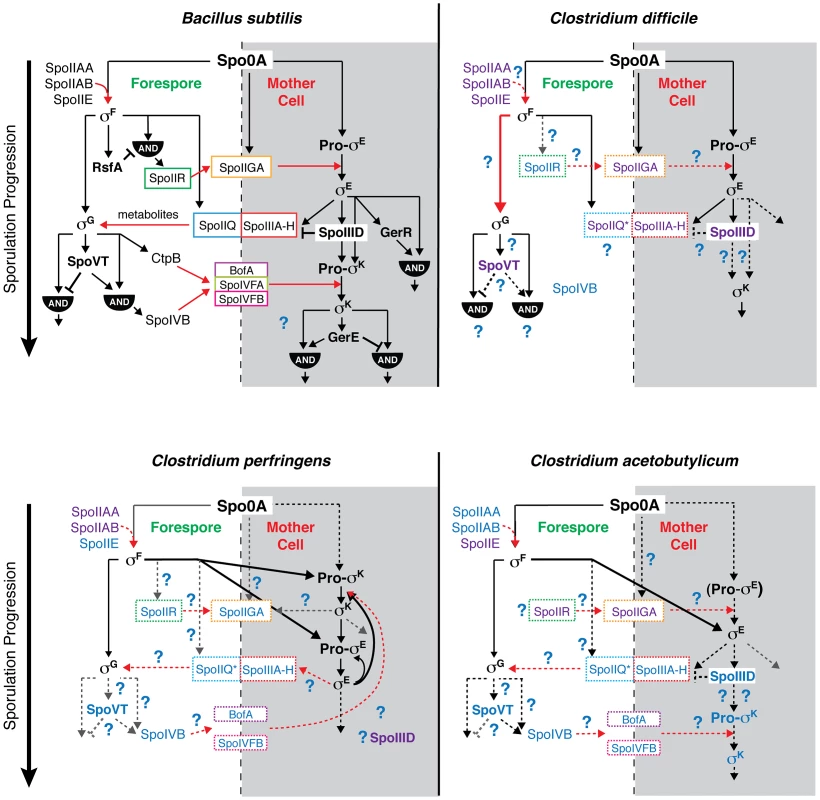
The temporal progression of sporulation is shown from top to bottom as determined in B. subtilis, C. difficile, C. perfringens, and C. acetobutylicum. Transcription factors and sigma factors are shown in bold, and proteins enclosed in boxes directly participate in trans-septum signaling. Dashed boxes indicate that the function of the proteins in trans-septum signaling has not been tested yet. Text color denotes whether the factor has been detected at both the transcript and protein level (black), at either the transcript or protein level (purple), or has not been tested yet at the transcript or protein level (blue), indicating the need for further experimentation. SpoIIQ* denotes the predicted clostridial homolog to B. subtilis SpoIIQ based on bioinformatics analyses [13]. Pro-σE in C. acetobutylicum is shown in parentheses to indicate that the pro-form has not been detected by Western blot [31]. Black arrows indicate transcriptional control of gene expression, red arrows indicate signaling pathways, dashed arrows indicate that the regulatory relationship between the factors has not been tested, and thick arrows demarcate notable points of divergence from the pathway defined in B. subtilis. AND gates are indicated. Unique features of the sporulation pathway in C. difficile include the post-translational activation of σG by σF and the absence of proteolytic activation of σK; the σE-dependent SpoIIIA-H feeding tube appears to be dispensable for σG activation. Similar to B. subtilis, our transcriptional and cytological analyses reveal that C. difficile σK functions downstream of σE to regulate late-stage sporulation events, and σG functions downstream of σF to regulate forespore maturation (Figures 2 and 6). In contrast with B. subtilis, C. difficile σG is fully active in the absence of σE, and σK is fully active in the absence of σG (Figures 6 and 7). The latter observation could have been anticipated given that C. difficile σK lacks an N-terminal pro-peptide, in contrast to all other spore formers [43]. However, the former observation was unexpected because σE-regulated gene products function to activate σG in the forespore of B. subtilis, initiating a positive feedback pathway that increases σG levels through auto-activation of the sigG promoter [44], [46], [57]. In particular, B. subtilis σG activation requires the formation of a σE-dependent “feeding tube” [20], [21], [55], [58], [59], which maintains forespore integrity by transporting small molecules from the mother cell into the forespore [20], [21], [55]. This mode of regulation couples the activation of the forespore-specific σG to σE-controlled events in the mother cell. In contrast, our results indicate that C. difficile σG is active in the absence of σE-dependent feeding tube gene expression (Figures 5 and 6, Tables S6 and S7). Nevertheless, even though σG was active at wildtype levels in the sigE− mutant (Figures 6 and 7), it remains possible that σG activity may be mislocalized in the mother cell cytosol, similar to the premature activation of σG in Lon− and anti-σG sigma factor CsfB− cells [57], [60], [61].
Even though C. difficile σG can be fully activated in the absence of σE, our results further show that σG is post-translationally activated in a σF-dependent manner (Figures 2 and 6). These results raise the intriguing question as to how σF activates σG independent of σE in C. difficile. In B. subtilis, multiple post-translational mechanisms control σG activity; however, aside from the feeding tube, these mechanisms are inhibitory rather than activating. In B. subtilis the Lon protease reduces σG activity in the mother cell [60], while the anti-σ factors SpoIIAB [57], [62] and CsfB (also known as Gin) [61], [63], [64] prevent σG activity in the forespore until engulfment is complete. Whether these factors inhibit σG activity in C. difficile is unknown, although C. difficile does not appear to encode a CsfB homolog. In future studies, it will be interesting to determine whether σF functions to activate σG directly or alleviate its inhibition, and whether C. difficile sporulation sigma factors exhibit compartment-specific activity similar to B. subtilis.
Interestingly, the morphology of the C. difficile sigG− mutant differed considerably from a B. subtilis sigG− mutant. While B. subtilis sigG− mutant forespores are normal in appearance despite lacking both a coat and cortex [44], C. difficile sigG− mutant forespores produced layers resembling spore coat around the forespore and exhibited defects in engulfment and structural integrity (Figures 3 and S3). The forespore membrane ruffling phenotype of C. difficile sigG− mutants was reminiscent of B. subtilis feeding tube mutant phenotypes [21], suggesting that σG may encode proteins required to “nurture” the C. difficile forespore. Alternatively, σG could regulate a cytoskeletal or cortex component that confers structural integrity to the forespore. Such proteins could be represented in the σG-regulated genes identified in this study (Table S7).
The phenotype of the C. difficile sigF− mutant also differed from its cognate mutant in B. subtilis, since the sigF− mutant produced low levels of σE− and σK− induced gene products (Figure 7) and regions that resembled mislocalized coat in the mother cell cytosol (Figure 3) [47]. In B. subtilis, σF is required to activate the expression of spoIIR, which encodes an intercellular signaling protein that activates SpoIIGA, the protease responsible for activating pro-σE [65], [66]. Whether the trace amounts of σE processing observed in the C. difficile sigF− mutant results from low-level expression of spoIIR or spoIIGA, or whether an unknown protease activates σE, remains to examined.
Comparison of the sporulation pathway of C. perfringens relative to C. difficile indicates that both organisms proteolytically activate σE in a σF-dependent manner (Figure 2), although it should be noted that a C. perfringens sigF− mutant does not make σE, σG, or σK [27] in contrast with C. difficile (Figure 2). Since the phenotypes of C. perfringens sigF− and sigG− mutants have not been examined by electron microscopy, the precise stage at which they are arrested remains unclear. Nevertheless, unlike C. perfringens (and C. botulinum) where σK is essential for both early and late stage sporulation events (Figure 8) [28], [29], C. difficile σK is needed only at late stages of sporulation. Furthermore, C. perfringens σK is produced at low levels in an unprocessed form in a sigE− mutant; σE is made at low levels in a C. perfringens sigK− mutant; and sigE and sigK expression appear to be auto-activated [28]. In contrast, no sigK expression was observed in the absence of σE in C. difficile.
The sporulation pathway of C. difficile appears to be most similar to the C. acetobutylicum pathway. Both C. difficile and C. acetobutylicum sigma factors σF, σE, and σG appear to function at similar stages of sporulation, although C. acetobutylicum sigF− and sigE− mutants exhibit more severe phenotypes than in C. difficile in that they fail to initiate and complete asymmetric division, respectively [30], [31], and σF is required to activate sigG transcription in C. acetobutylicum [30] unlike C. difficile. Similar to C. difficile, however, C. acetobutylicum σG does not require σE for auto-activation of sigG expression, although it is unclear whether C. acetobutylicum σG is active in the absence of σE [31]. Lastly, loss of sigG expression in C. acetobutylicum results in pleiotropic defects in coat and cortex formation and forespore integrity similar to C. difficile (Figure 3, [31]). Since a sigK− mutant in C. acetobutylicum has not been described, it will be interesting to determine whether C. acetobutylicum σK function is more similar to C. difficile than to C. perfringens and whether these differences correlate to the presence of the skin element, an ∼15 kb prophage-like element that interrupts the sigK gene in C. difficile but not other Clostridium spp. [43]. Nevertheless, our collective transcriptional and cytological analyses of sporulation sigma factor function in C. difficile suggest that novel mechanisms regulate σG and σK activation relative to other spore-forming organisms (Figure 8). Further studies are needed to determine the regulatory interplay between C. difficile sporulation sigma factors and their downstream auxiliary regulators SpoVT and SpoIIID, which modulate the expression of σG - and σK-regulated genes, respectively, in B. subtilis [24], [26], [67], [68] and are conserved in Clostridium spp.
Transcriptional profiling of sporulation in C. difficile identifies new sporulation genes
By performing whole genome transcriptional profiling on the different sporulation sigma factor mutants, we have identified distinct subsets of genes that are σF-, σE-, σG-, and σK-dependent. The number of genes determined to be σG-dependent in C. difficile was relatively small (34 genes) relative to B. subtilis, where the σG regulon comprises ∼100 genes [11], [25], [26]. Similarly, the σE and σK-dependent genes (169 and 31 genes, respectively) identified by our study were smaller than their cognate regulons in B. subtilis (270 and 150 genes, respectively, [24]). While the parameters we used to define genes as being σF-, σE-, σG-, and σK-dependent were relatively stringent, relaxing these parameters did not result in large increases in gene numbers. One explanation for the smaller size is that C. difficile activates fewer genes during sporulation than B. subtilis. A more likely explanation is that the asynchronous population of sporulating cells (Figures 1 and S2, [37]) limits the detection of genes that are transiently expressed during discrete stages of sporulation or genes that are expressed at low levels during sporulation [24]–[26], [69]. Since the RNA samples used in the RNA-Seq analysis were harvested from a sporulation timepoint in which phase-bright forespores were produced by wildtype cells (Figure S2), fewer cells in the population are likely to be at early stages sporulation. As a result, early sporulation genes may be under-represented in our data set; for example, σF-dependent early sporulation gene transcripts from spoIIR and spoIIP were almost undetectable by RNA-Seq (Table S4). In addition, genes that are regulated by more complex mechanisms beyond upregulation by a specific sigma factor are likely to be under-represented in our data set. Sporulation genes that are subject to incoherent feed forward loop regulation, in which their expression is induced by a given sigma factor and repressed by a downstream regulator such as SpoVT-mediated inhibition of sigG transcription in B. subtilis [26], may not be detected in our data set. Unraveling the complexities of sporulation gene regulation in C. difficile will require further characterization of the kinetics of sporulation and the analysis of mutants defective in auxiliary sporulation regulators.
Of the 51 genes proposed to comprise the core set of sporulation genes in spore-forming Firmicutes by bioinformatics analyses that are conserved in C. difficile [13], 34 were identified in our RNA-Seq analyses, leaving 17 that were not identified in our transcriptional analyses (Table S4). Seven of these genes do not have detectable homologs in the 630 C. difficile genome, and 6 were expressed at low levels with a base mean of expression less than 15 (Table S4).
Although some sporulation-induced genes were likely not detected in our analyses due to low levels of expression, the transcriptional profiling data presented here identify a promising set of genes that are likely to encode proteins with important roles in spore formation. Of the six spore coat proteins recently identified in a proteomic analysis of C. difficile [50], [53], all were identified in our RNA-Seq experiments. Three of these spore coat genes were determined to be σK-dependent, consistent with their predicted role as components of the outer coat (Table S4). Notably, σK-regulated genes were among the most abundantly expressed genes induced during sporulation, comprising 6 of the 10 most highly expressed sporulation genes (Table S9). The σK-regulated CD1067 gene was the most highly expressed gene induced during sporulation in C. difficile. Cysteine-rich CD1067 was also one of the most abundant spore proteins identified in proteomic analyses of purified spores and is encoded in a 7.5 kB region enriched in genes encoding spore proteins [70]. Western blot analyses of cysteine-rich CD1067 revealed that it forms higher order multimers that are highly resistant to denaturing conditions (data not shown), consistent with the proposal that CD1067 may form a rigid, disulfide-bonded structure around the spore coat upon exposure to atmospheric oxygen, for example during excretion from the host [70]. Intriguingly, CD1067 is encoded in a region enriched in highly expressed, σK-regulated genes encoding hypothetical proteins unique to C. difficile, with 8 of the 9 genes in this region being induced during sporulation and 6 of the 9 being σK-regulated. These genes may encode coat proteins that confer structural integrity and/or resistance to the C. difficile spore coat and thus may play important roles in disease transmission and/or represent good candidates for developing diagnostic reagents.
Although the number of σG-dependent genes identified by our study was small, a number of these genes encode proteins with important functions in the forespore of B. subtilis, specifically sspA, sspB, dacF, spoVT, and spoVAD [26], [67], [71]–[73]. Since B. subtilis σG induces the expression of genes encoding the germinant receptors (of which there are no homologs in C. difficile [12], [34]), it seems likely that some of the σG-dependent genes identified in our study encode proteins that transduce the germinant signal into the spore core. It will be interesting to determine whether any of the σG-regulated genes identified in our study play important roles in regulating germination and thus disease transmission.
Genes encoding hypothetical proteins were the most abundant class of genes identified in our study (82 in total, Table S10). Twenty of these hypothetical proteins were detected in proteomic analyses of C. difficile spores [70]. Indeed, two of the hypothetical proteins were previously shown to be part of the spore coat [50], and we have validated three additional proteins as localizing to the spore coat (data not shown). BLAST searches with the hypothetical proteins identified by RNA-Seq indicate that 16 have no known homologs. These C. difficile-specific proteins could comprise part of the spore coat, since coat proteins are often poorly conserved, species-specific, and categorized as hypothetical proteins [10], [11].
Taken together, by examining the regulatory interplay between sporulation sigma factors in C. difficile, our study highlights that diverse pathways regulate sporulation in the Firmicutes and that considerable work is needed to map these pathways in the Clostridia. By using whole genome transcriptional profiling to define a large set of genes that are activated by Spo0A, σF, σE, σG, and/or σK, our study also provides a framework for identifying new proteins that are necessary for sporulation and determining the role of these proteins in forming a functional, infectious spore. Studies of this nature may lead to the identification of biomarkers for C. difficile spores and candidates for vaccine development.
Materials and Methods
Bacterial strains and growth conditions
All C. difficile strains are listed in Table 1 and derive from the parent strain JIR8094 [33], an erythromycin-sensitive derivative of the sequenced clinical isolate 630 [34]. C. difficile strains were grown on solid brain heart infusion media supplemented with yeast extract (BHIS: 37 g brain heart infusion, 5 g yeast extract, 0.1% (w/v) L-cysteine, 15 g agar per liter) [74]. Taurocholate (TA; 0.1% w/v), thiamphenicol (5–10 µg/mL), kanamycin (50 µg/mL), cefoxitin (16 µg/mL), FeSO4 (50 µM), and/or erythromycin (10 µg/mL) were used to supplement the BHIS media as indicated. Cultures were grown at 37°C, under anaerobic conditions using a gas mixture containing 85% N2, 5% CO2, and 10% H2.
Tab. 1. <i>C. difficile</i> strains used in this study. 
Sporulation was induced on media containing BHIS and SMC (90 g BactoPeptone, 5 g protease peptone, 1 g NH4SO4, 1.5 g Tris base, 15 g agar per liter) [50], at 70% SMC and 30% BHIS (70∶30 media, 63 g BactoPeptone, 3.5 g Protease Peptone, 11.1 g BHI, 1.5 g yeast extract, 1.06 g Tris base, 0.7 g NH4SO4, 15 g agar per liter) [37]. 70∶30 agar (supplemented as appropriate with thiamphenicol at 5–10 µg/mL) was inoculated from a starter culture grown on solid media.
HB101/pK424 strains were used for conjugations and BL21(DE3) strains were used for protein expression. E. coli strains were routinely grown at 37°C, shaking at 225 rpm in Luria-Bertani broth (LB). Media was supplemented with chloramphenicol (20 µg/mL), ampicillin (50–100 µg/mL), or kanamycin (30 µg/mL) as indicated.
E. coli strain construction
All strains are listed in Table S11; all plasmids are listed in Table S12; and all primers used are listed in Table S13. For disruption of spo0A, sigE, sigG, sigK, and sigF, a modified plasmid containing the retargeting group II intron, pCE245 (a gift from C. Ellermeier, University of Iowa), was used as the template. Primers used to amplify the targeting sequence from the template carried flanking regions specific for each gene target and are listed as follows: spo0A (#539, 540, 541 and 532, the EBS Universal primer as specified by the manufacturer (Sigma Aldrich), sigE (#653, 654, 655 and 532), sigG (#728, 729, 730, and 532), sigK (#681, 682, 683, and 532) and sigF (#775, 776, 777, and 532). The spo0A disruption mutant was constructed using the same primers as Underwood et al. [36]. The resulting retargeting sequences were digested with BsrGI and HindIII and cloned into pJS107 (a gift from J. Sorg, University of Texas A&M), a derivative of pJIR750ai (Sigma Aldrich) [32]. The ligations were transformed into DH5α and confirmed by sequencing. The resulting plasmids were used to transform HB101/pK424.
To construct the sigE complementation construct, primers #725 and 726 were used to amplify a fragment containing 252 bp upstream and 156 bp downstream of the two gene spoIIGA-sigE operon using 630 genomic DNA as the template. To construct the sigG complementation construct, primers #835 and 836 were used to amplify 288 bp upstream and 16 bp downstream of sigG using 630 genomic DNA as the template. The sigK complementation construct was made using PCR splicing by overlap extension (SOE) [75]. Primer pair #734 and 736 was used to amplify the 5′ SOE product, while primer pair #735 and 737 was used to amplify the 3′ SOE product. The resulting fragments were mixed together, and the flanking primers #734 and #737 were used to amplify an 898 bp fragment corresponding to the sigK gene including 256 bp region of upstream sequence. This strategy was used to clone an intact sigK gene with the skin element excised [43]. To construct the sigF complementation construct, primers #954 and #956 were used to amplify 88 bp upstream and 19 bp downstream of spoIIAA-spoIIAB-sigF operon, using 630 genomic DNA as the template. All complementation constructs were digested with NotI and XhoI and ligated into pMTL83151 [48] digested with the same enzymes, with the exception of the sigF complementation construct, which was cloned into pMTL84151 digested with the same enzymes [48].
To construct strains producing recombinant CD3522, σE, σG, σF, Gpr, SpoVT, and SspA for antibody production, primer pairs #498 and 499; #596 and 597; #727 and 688; #723 and 724; #790 and 791; #883 and 884; #975 and 976; and #885 and 886 were used to amplify the cd3522, sigE, sigG, sigF, gpr, spoVT, and sspA genes lacking stop codons, respectively, using 630 genomic DNA as the template. The sigE expression construct deletes the sequence encoding the first 23 amino acids of σE, which removes its membrane-tethering domain and improves the solubility of the protein in E. coli. The resulting PCR products were digested with NdeI and XhoI, (or NheI and XhoI for gpr) ligated to pET22b (or pET21a for gpr and sspA), and used to transform DH5α. To construct a strain producing recombinant σK, PCR SOE was used to amplify the sigK gene lacking the skin element. Primer pair #689 and 736 was used to amplify the 5′ SOE product, while primer pair #735 and 737 was used to amplify the 3′ SOE product. The resulting fragments were mixed together, and the flanking #689 and #737 primers were used to amplify the sigK gene including the TAA stop codon. The resulting PCR product was digested with NcoI and XhoI, ligated to pET30a digested with the same enzymes, and used to transform DH5α. The resulting pET22b-cd3522, pET22b-sigE, pET22b-sigG, pET30a-sigK, pET22b-sigF, pET21a-gpr, pET22b-spoVT, and pET21a-sspA plasmids were used to transform BL21(DE3) for protein expression.
C. difficile strain construction
C. difficile strains were constructed using TargeTron-based gene disruption as described previously (Figure S1, [32], [37], [76]). TargeTron constructs in pJS107 were conjugated into C. difficile using an E. coli HB101/pK424 donor strain. HB101/pK424 strains containing the appropriate pJS107 construct were grown aerobically to exponential phase in 2 mL of LB supplemented with ampicillin (50 µg/mL) and chloramphenicol (10 µg/mL). Cultures were pelleted, transferred into the anaerobic chamber, and resuspended in 1.5 mL of late-exponential phase C. difficile JIR8094 cultures (grown anaerobically in BHIS broth). The resulting cell mixture was plated as seven 100 µL spots onto pre-dried, pre-reduced BHIS agar plates. After overnight incubation, all growth was harvested from the BHIS plates, resuspended in 2.5 mL pre-reduced BHIS, and twenty-one 100 µL spots per strain were plated onto BHIS agar supplemented with thiamphenicol (10 µg/mL), kanamycin (50 µg/mL), and cefoxitin (16 µg/mL) to select for C. difficile containing the pJS107 plasmid. After 24–48 hrs of anaerobic growth, single colonies were patched onto BHIS agar supplemented with thiamphenicol (10 µg/mL), kanamycin (50 µg/mL), and FeSO5 (50 µM) to induce the ferredoxin promoter of the group II intron system. After overnight growth, patches were transferred to BHIS agar plates supplemented with erythromycin (10 µg/mL) for 24–72 hrs to select for cells with activated group II intron systems. Erythromycin-resistant patches were struck out for isolation onto the same media and individual colonies were screened by colony PCR for a 2 kb increase in the size of spo0A (primer pair #556 and 557), sigE (primer pair #687 and 688), sigG (primer pair #723 and 724), sigK (primer pair #689 and 690), and sigF (primer pair #790 and 791) (Figure S1). A minimum of two independent clones from each mutant strain was phenotypically characterized.
C. difficile complementation
HB101/pK424 donor strains carrying the appropriate complementation construct were grown in LB containing ampicillin (50 µg/mL) and chloramphenicol (20 µg/mL) at 37°C, 225 rpm, under aerobic conditions, for 6 hrs. C. difficile recipient strains spo0A−, sigE−, sigG−, sigK−, and sigF−, containing group II intron disruptions, were grown anaerobically in BHIS broth at 37°C with gentle shaking for 6 hrs. HB101/pK424 cultures were pelleted at 2500 rpm for 5 min and the supernatant was removed. Pellets were transferred to the anaerobic chamber and gently resuspended in 1.5 mL of the appropriate C. difficile culture. The resulting mixture was inoculated onto pre-dried, pre-reduced BHIS agar plates, as seven 100 µL spots for 12 hrs. All spots were collected anaerobically and resuspended in 1 mL PBS. The resulting suspension was spread onto pre-dried, pre-reduced BHIS agar plates supplemented with thiamphenicol (10 µg/mL), kanamycin (50 µg/mL), and cefoxitin (10 µg/mL) at 100 µL per plate, five plates per conjugation. Plates were monitored for colony growth for 24–72 hrs. Individual colonies were struck out for isolation and analyzed for complementation by phase contrast microscopy, Western blot analysis and transmission electron microscopy. A minimum of two independent clones from each complementation strain was phenotypically characterized.
For the sigF complementation, a pMTL84151 plasmid backbone was used. The complementation protocol was followed as described except that after spots were collected from overnight growth on BHIS plates, the resulting PBS suspension was spotted onto three BHIS agar plates supplemented with thiamphenicol (10 µg/mL), kanamycin (50 µg/mL), and cefoxitin (16 µg/mL) with 7–100 µL spots per plate.
Sporulation assay
C. difficile strains were grown from glycerol stocks on BHIS plates supplemented with TA (0.1% w/v), or with both TA and thiamphenicol (5–10 µg/mL) for strains with pMTL83151-derived or pMTL84151-derived plasmids. Cultures grown on BHIS agar plates were then used to inoculate 70∶30 agar plates (with thiamphenicol at 5–10 µg/mL as appropriate) for 18–48 hrs as previously described [37]. Sporulation induced lawns were harvested in PBS, washed once, resuspended in 0.2 mL of PBS, visualized by phase contrast microscopy, and/or further processed for analysis by transmission electron microscopy or Western blotting.
Heat resistance assay
C. difficile strains grown from glycerol stocks on BHIS plates supplemented with taurocholate and thiamphenicol (described above) were inoculated on to 70∶30 media containing thiamphenicol (5–10 µg/mL). After 30 hrs of growth, cells were harvested in 1.0 mL PBS, and split into two tubes. One tube was heat shocked at 60–65°C for 25 minutes. Both heat-shocked and non-heat shocked cells were serially diluted, and cells were plated on pre-reduced BHIS-TA plates. After 20 hrs on BHIS-TA, colonies were counted, and cell counts were determined. The percent of heat-resistant spores was determined based on the ratio of heat-resistant cells to total cells, and sporulation efficiencies were determined based on the ratio of heat-resistant cells for a strain compared to wild type. Results are based on a minimum of three biological replicates. spo0A− containing empty vector was included as a control for all assays [77].
Fluorescence and light microscopy
For fluorescence microscopy studies, C. difficile strains were harvested in PBS after 18 hours of growth on 70∶30 media, pelleted, and resuspended in 1.0 mL PBS containing 1 µg/mL FM4-64 (Molecular Probes) and 15 µg/mL Hoechst 33342 (Molecular Probes). The bacterial suspension (4 µL) was added to a freshly prepared 1% agarose pad on a microscope slide, covered with a 22×22 mm #1 coverslip and sealed with VALAB (1∶1∶1 of vaseline, lanolin, and beeswax) as previously described [78]. Phase and fluorescence microscopy were performed using a Nikon PlanApo 100× Ph3 oil immersion objective (1.4 NA) on a Nikon Eclipse TE300 epifluorescence microscope. Five fields for each sample were acquired with an iXon3 885 EMCCD camera (Andor) cooled to −70°C with frame averaging set to 4 and an EM gain setting of 3, and driven by NIS-Elements software (Nikon). Images were subsequently imported into Adobe Photoshop CS6 for minimal adjustments in brightness/contrast levels and pseudocoloring.
Phase-contrast microscopy for imaging the samples used for RNA-Seq was performed as previously described [37].
Quantification of total cells undergoing sporulation was determined by analyzing multiple fields for each strain at random. Greater than 200 cells were enumerated for each strain. For cultures analyzed by fluorescence microscopy, sporulating cells were identified as either having a polar septum with or without DNA staining in the forespore, a phase-dark forespore with or without DNA staining in the forespore compartment, a phase-bright forespore without DNA staining, or a free spore (no mother cell compartment).
Electron microscopy
One hundred microliters of bacterial cell suspension samples from sporulation assays were prepared as previously described [37].
Western blot analyses
Sporulation assay C. difficile cells (50 µL of PBS suspension) were freeze-thawed three times, diluted in 100 µL EBB buffer (8 M urea, 2 M thiourea, 4% (w/v) SDS, 2% (v/v) β-mercaptoethanol), and incubated at 95°C for 20 min with vortexing every 5 min. Samples were centrifuged for 5 min at 15,000 rpm and 7 µL of 4× sample buffer (40% (v/v) glycerol, 1 M Tris pH 6.8, 20% (v/v) β-mercaptoethanol, 8% (w/v) SDS, and 0.04% (w/v) bromophenol blue), was added. Protein samples were incubated again at 95°C for 15 minutes with vortexing followed by centrifugation for 5 min at 15,000 rpm. SDS-PAGE gels (12%–15%) were loaded with 5 µL of protein prep. Gels were transferred to Bio-Rad PVDF membrane and blocked in 50% PBS:50% Odyssey Blocking Buffer with 0.1% (v/v) Tween for 30 min at RT. Polyclonal rabbit anti-σE, anti-σG, anti-σF, anti-SpoIVA [37], and anti-CD1433 [76], anti-CD1067, anti-Gpr, anti-SpoVT, and anti-SspA antibodies were used at a 1∶1,000 dilution and anti-σK, anti-CD1511, anti-SleC [76], and anti-CD3522 at a 1∶5,000 dilution. Monoclonal mouse anti-Spo0A [37] was used at a 1∶10,000 dilution. IRDye 680CW and 800CW infrared dye-conjugated secondary antibodies were used at a 1∶20,000 dilutions. The Odyssey LiCor CLx was used to detect secondary antibody fluorescent emissions for Western blots.
Antibody production
The anti-Δ230aa-σE, anti-σG, anti-σK, anti-σF, anti-CD3522, anti-Gpr, anti-SpoVT, and anti-SspA antibodies used in this study were raised in rabbits by Cocalico Biologicals (Reamstown, PA). The antigens Δ230aa-σE-His6, σG-His6, His6−σK, σF-His6, CD3522-His6, Gpr-His6, SpoVT-His6, and SspA-His6, were purified on Ni2+-affinity resin from E. coli strains #755, 743, 756, 921, 577, 853, 881, and #SspA respectively, as described above. Cultures were grown and protein expression was induced with 250 µM IPTG overnight at 19°C. E. coli cells were harvested, pelleted, and resuspended in 25 mL of low imidazole buffer (LIB; 500 mM NaCl, 50 mM Tris-HCl, pH 7.5, 15 mM imidazole, 10% (v/v) glycerol). Cells were flash frozen in liquid nitrogen, thawed, and lysed by sonication (45 sec burst followed by 5 min on ice for 3 cycles). For protein affinity purification, the lysate was centrifuged at 16,000× g for 30 min, supernatant was collected and added to pre-washed Ni2+-affinity resin for 4 hrs at 4°C. Bound beads were centrifuged at 2,000× g for 2 min at 4°C and washed once in LIB. Beads were reconstituted in 375 µL of high imidazole buffer (HIB; 500 mM NaCl, 50 mM Tris-HCl, pH 7.5, 200 mM imidazole, 10% (v/v) glycerol), incubated on a nutator for 15 min at RT, centrifuged, and eluate was collected. Beads were reconstituted with HIB for a total of five sequential elutions.
Polyclonal antibodies against CD1067 were raised in rabbits against a peptide derived from CD1067 (INSEDMRGFKKSHHC, Genscript); the polyclonal antibodies were affinity-purified using the indicated peptide (Genscript).
RNA processing
RNA for RNA-Seq was extracted from WT, spo0A−, sigE−, sigF−, sigG−, and sigK− C. difficile cell suspensions, from an 18 hr sporulation assay (described earlier), using a FastRNA Pro Blue Kit (MP Biomedical) and a FastPrep-24 automated homogenizer (MP Biomedical, setting 6.0, 45 seconds for 3 cycles). Contaminating genomic DNA was depleted using a column-bound DNase treatment with an RNeasy Kit (Qiagen) followed by two suspension DNase treatments (New England Biolabs), according to manufacturer's recommendations. Samples were tested for genomic DNA contamination using quantitative PCR for 16S rRNA and the sleC gene. DNAse-treated RNA (5 µg) was mRNA enriched using a Ribo-Zero Magnetic Kit (Epicentre).
RNA isolated for qRT-PCR was processed identically except that mRNA enrichment was done using an Ambion MICROBExpress Bacterial mRNA Enrichment Kit (Invitrogen). Reverse transcription of enriched RNA was done using the Super Script First Strand cDNA Synthesis Kit (Invitrogen) with random hexamer primers.
RNA-Seq library construction and sequencing
Enriched mRNA (100 ng) was submitted to the Advanced Technology Genome Center Core Lab at the University of Vermont for massively parallel sequencing on an Illumina HiSeq 1000. cDNA synthesis was carried out using the Ovation Prokaryotic RNA-Seq System (Nugen), according to manufacturer's instructions. Libraries were prepared using the Ovation ultralow multiplex kit (Nugen, 0304/0305-32) according to manufacturer's instructions. Briefly, samples were end-repaired, mono-adenylated, ligated to index/adaptors, and then amplified for 15 cycles (after a PCR titration was performed). Completed libraries were quantitated using a SYBR Fast Universal qPCR Kit (KAPA Biosystems). Paired end sequencing of samples was performed using a total of 10 pM of library in each flow cell lane. The samples were indexed and pooled in equal amounts to generate equal read coverage.
RNA-Seq analysis
Sequence calls and quality scores were produced in BCL format from images using Illumina RTA v1.13 with default parameters. Read pairs were mapped to libraries (demultiplexed) and converted to Fastq format using Illumina CASAVA 1.8.2 with default parameters. Adapters were clipped and reads were trimmed to remove the first 12 and last 11 cycles using Trimmomatic [79], dropping read pairs for which at least one read was less than 50 bp. The C. difficile 630 genome (NC_009089) sequence was modified by removing the sigK intervening (skin) element. The C. difficile 630 genome annotation was modified by the addition of sigK. Read pairs were aligned to the modified C. difficile 630 genome (NC_009089) using BWA 0.6.1 with default parameters with one exception (−q 20). Read pairs were mapped to NC_009089 gene annotation using the countOverlaps procedure of the R/Bioconductor IRanges package [80]–[82]. Counts associated with rRNA were removed. Counts associated with the same library were pooled. Reads were of high quality (median Phred score of 39 and a first quartile of 35) as were alignments (median mapping quality score, MAPQ of 60). Median fragment lengths were between 180 and 250.
The vast majority of unmapped sequences failed to align to sequences in the NCBI non-redundant database using a blastn and blastx search. There was no indication of highly represented reads among unmapped sequences. Since the majority of reads failed to map to known natural sequences, and since sequences can arise during library preparation particularly when the input sample is small, sequences that failed to map to the C. difficile genome likely represent spurious sequences produced during library construction.
Differential expression statistics reflecting both effect size (fold-change) and statistical significance (p-value adjusted based on the method of Benjamini and Hochberg [83]) were calculated using DESeq [49]. Duplicate reads were excluded from these analyses. Differentially expressed genes were identified based on a minimum fold-change (higher in the reference sample than the query) and maximum p-value. Tables showing genes whose expression was downregulated by ≥4-fold with an adjusted p-value of ≤0.05 during sporulation are provided in the Supplementary Information (Tables S3, S4, S5, S6, S7, S8, S9, S10). A table showing genes whose expression was upregulated by ≥4-fold with an adjusted p-value of ≤0.05 in a Spo0A-dependent manner are shown in Table S14.
The log2-transformed expression of genes that were downregulated by ≥4-fold with an adjusted p-value of ≤10−5 in the spo0A− strain relative to wild type expression were represented in a heat map using the heatmap.2 procedure of the R/Bioconductor gplots package with default options [84]. Expression levels in spo0A− were not shown because the differential expression between spo0A− and wild type was biased by the method used to select genes. Expression levels in the other four strains relative to spo0A− were centered, scaled, and mapped to a red-green color scale.
Quantitative RT-PCR
For the RNA-Seq validation, expression levels of gpr, cd0125 (spoIIQ), cd2376, cd1511, cd3522, spoIVA, cd1433, cd1067, sleC, sspB, spoVT, dacF, and rpoB (housekeeping gene) were run on WT, spo0A−, sigF− sigE−, sigG−, and sigK− cDNA templates in three replicate reactions using gene-specific primer pairs #1187 and 1188; #1213 and 1214; #1191 and 1192; #796 and 797; #989 and 990; #798 and 799; #792 and 793; #1030 and 1031; #575 and 576; #810 and 811; #995 and 996; #993 and 994; #1002 and 1003, respectively. Quantitative real-time PCR was performed using SYBR Green JumpStart Taq Ready Mix (Sigma), 50 nM of gene specific primers (Table S12), and an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Mean CT values were normalized to the spo0A− (negative control) sample and further normalized to rpoB. Relative expression values reported are representative of three biological replicates.
Supporting Information
Zdroje
1. CarrollK, BartlettJ (2011) Biology of Clostridium difficile: implications for epidemiology and diagnosis. Ann Rev Microbiol 65 : 501–521.
2. KellyC, LaMontJ (2008) Clostridium difficile–more difficult than ever. New Engl J Med 359 : 1932–1940.
3. RupnikM, WilcoxM, GerdingD (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7 : 526–536.
4. GhantojiS, SailK, LairsonD, DuPontH, GareyK (2010) Economic healthcare costs of Clostridium difficile infection: a systematic review. J Hosp Infect 74 : 309–318.
5. McGloneSM, BaileyRR, ZimmerSM, PopovichMJ, TianY, et al. (2011) The economic burden of Clostridium difficile. Clin Microbiol Infect 18 : 282–289.
6. FreemanJ, BauerM, BainesS, CorverJ, FawleyW, et al. (2010) The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23 : 529–549.
7. KhannaS, PardiD, AronsonS, KammerP, OrensteinR, et al. (2012) The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol 107 : 89–95.
8. LawleyT, ClareS, DeakinL, GouldingD, YenJ, et al. (2010) Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl Env Microbiol 76 : 6895–6900.
9. MarooS, LamontJ (2006) Recurrent Clostridium difficile. Gastroenterology 130 : 1311–1316.
10. HenriquesAO, MoranCPJr (2007) Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol 61 : 555–588.
11. de HoonM, EichenbergerP, VitkupD (2010) Hierarchical evolution of the bacterial sporulation network. Current Biol : CB 20 : 45.
12. ParedesC, AlsakerK, PapoutsakisE (2005) A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol 3 : 969–978.
13. GalperinMY, MekhedovSL, PuigboP, SmirnovS, WolfYI, et al. (2012) Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol 14 : 2870–2890.
14. ErringtonJ (2003) Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1 : 117–126.
15. HigginsD, DworkinJ (2012) Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36 : 131–148.
16. HilbertD, PiggotP (2004) Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev 68 : 234–262.
17. MargolisP, DriksA, LosickR (1991) Establishment of cell type by compartmentalized activation of a transcription factor. Science 254 : 562–565.
18. MargolisP, DriksA, LosickR (1991) Differentiation and the establishment of cell type during sporulation in Bacillus subtilis. Curr Opin Genet Dev 1 : 330–335.
19. HofmeisterAE, Londono-VallejoA, HarryE, StragierP, LosickR (1995) Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell 83 : 219–226.
20. CampAH, LosickR (2009) A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev 23 : 1014–1024.
21. DoanT, MorlotC, MeisnerJ, SerranoM, HenriquesA, et al. (2009) Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet 5: e1000566.
22. CuttingS, DriksA, SchmidtR, KunkelB, LosickR (1991) Forespore-specific transcription of a gene in the signal transduction pathway that governs Pro-σK processing in Bacillus subtilis. Genes Dev 5 : 456–466.
23. HaldenwangW (1995) The sigma factors of Bacillus subtilis. Microbiol Rev 59 : 1–30.
24. EichenbergerP, FujitaM, JensenS, ConlonE, RudnerD, et al. (2004) The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol 2: e328.
25. SteilL, SerranoM, HenriquesA, VölkerU (2005) Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiol 151 : 399–420.
26. WangS, SetlowB, ConlonE, LyonJ, ImamuraD, et al. (2006) The forespore line of gene expression in Bacillus subtilis. J Mol Biol 358 : 16–37.
27. LiJ, McClaneB (2010) Evaluating the involvement of alternative sigma factors σF and σG in Clostridium perfringens sporulation and enterotoxin synthesis. Infect Immun 78 : 4286–4293.
28. HarryK, ZhouR, KroosL, MelvilleS (2009) Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors σE and σK in Clostridium perfringens. J Bacteriol 191 : 2728–2742.
29. KirkD, DahlstenE, ZhangZ, KorkealaH, LindströmM (2012) Involvement of Clostridium botulinum ATCC 3502 σK in early-stage sporulation. Appl Environ Microbiol 78 : 4590–4596.
30. JonesS, TracyB, GaidaS, PapoutsakisE (2011) Inactivation of σF in Clostridium acetobutylicum ATCC 824 blocks sporulation prior to asymmetric division and abolishes σE and σG protein expression but does not block solvent formation. J Bacteriol 193 : 2429–2440.
31. TracyB, JonesS, PapoutsakisE (2011) Inactivation of σE and σG in Clostridium acetobutylicum illuminates their roles in clostridial-cell-form biogenesis, granulose synthesis, solventogenesis, and spore morphogenesis. J Bacteriol 193 : 1414–1426.
32. HeapJT, PenningtonOJ, CartmanST, CarterGP, MintonNP (2007) The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods 70 : 452–464.
33. O'ConnorJ, LyrasD, FarrowK, AdamsV, PowellD, et al. (2006) Construction and analysis of chromosomal Clostridium difficile mutants. Mol Microbiol 61 : 1335–1351.
34. SebaihiaM, WrenB, MullanyP, FairweatherN, MintonN, et al. (2006) The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38 : 779–786.
35. DeakinLJ, ClareS, FaganRP, DawsonLF, PickardDJ, et al. (2012) The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80 : 2704–2711.
36. UnderwoodS, GuanS, VijayasubhashV, BainesS, GrahamL, et al. (2009) Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J Bacteriol 191 : 7296–7305.
37. PutnamEE, NockAM, LawleyTD, ShenA (2013) SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J Bacteriol 195 : 1214–1225.
38. PoglianoJ, OsborneN, SharpMD, Abanes-De MelloA, PerezA, et al. (1999) A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol 31 : 1149–1159.
39. SetlowB, LoshonCA, GenestPC, CowanAE, SetlowC, et al. (2002) Mechanisms of killing spores of Bacillus subtilis by acid, alkali and ethanol. J Appl Microbiol 92 : 362–375.
40. WangKH, IsidroAL, DominguesL, EskandarianHA, McKenneyPT, et al. (2009) The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis. Mol Microbiol 74 : 634–649.
41. PiggotPJ, HilbertDW (2004) Sporulation of Bacillus subtilis. Curr Opin Microbiol 7 : 579–586.
42. JonasRM, WeaverEA, KenneyTJ, MoranCPJr, HaldenwangWG (1988) The Bacillus subtilis spoIIG operon encodes both σE and a gene necessary for σE activation. J Bacteriol 170 : 507–511.
43. HaraldsenJ, SonensheinA (2003) Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol Microbiol 48 : 811–821.
44. Karmazyn-CampelliC, BonamyC, SavelliB, StragierP (1989) Tandem genes encoding sigma-factors for consecutive steps of development in Bacillus subtilis. Genes Dev 3 : 150–7.
45. SunDX, Cabrera-MartinezRM, SetlowP (1991) Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor σG. J Bacteriol 173 : 2977–2984.
46. EvansL, FeuchtA, ErringtonJ (2004) Genetic analysis of the Bacillus subtilis sigG promoter, which controls the sporulation-specific transcription factor σG. Microbiol 150 : 2277–2287.
47. IllingN, ErringtonJ (1991) Genetic regulation of morphogenesis in Bacillus subtilis: roles of σE and σG in prespore engulfment. J Bacteriol 173 : 3159–3169.
48. HeapJ, PenningtonO, CartmanS, MintonN (2009) A modular system for Clostridium shuttle plasmids. J Microbiol Meth 78 : 79–85.
49. AndersS, HuberW (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106.
50. PermpoonpattanaP, TollsE, NademR, TanS, BrissonA, et al. (2011) Surface layers of Clostridium difficile endospores. J Bacteriol 193 : 6461–6470.
51. SaujetL, MonotM, DupuyB, SoutourinaO, Martin-VerstraeteI (2011) The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol 193 : 3186–3196.
52. RosenbuschKE, BakkerD, KuijperEJ, SmitsWK (2012) C. difficile 630Δerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLoS One 7: e48608.
53. PermpoonpattanaP, PhetcharaburaninJ, MikelsoneA, DembekM, TanS, et al. (2013) Functional characterization of Clostridium difficile spore coat proteins. J Bacteriol 195 : 1492–1503.
54. BrittonRA, EichenbergerP, Gonzalez-PastorJE, FawcettP, MonsonR, et al. (2002) Genome-wide analysis of the stationary-phase sigma factor (σH) regulon of Bacillus subtilis. J Bacteriol 184 : 4881–4890.
55. CampAH, LosickR (2008) A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol 69 : 402–417.
56. PartridgeSR, ErringtonJ (1993) The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol Microbiol 8 : 945–955.
57. CharyVK, MeloniM, HilbertDW, PiggotPJ (2005) Control of the expression and compartmentalization of (sigma)G activity during sporulation of Bacillus subtilis by regulators of σF and σE. J Bacteriol 187 : 6832–6840.
58. JiangX, RubioA, ChibaS, PoglianoK (2005) Engulfment-regulated proteolysis of SpoIIQ: evidence that dual checkpoints control sigma activity. Mol Microbiol 58 : 102–115.
59. MeisnerJ, WangX, SerranoM, HenriquesA, MoranC (2008) A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci USA 105 : 15100–15105.
60. SchmidtR, DecaturAL, RatherPN, MoranCPJr, LosickR (1994) Bacillus subtilis lon protease prevents inappropriate transcription of genes under the control of the sporulation transcription factor σG. J Bacteriol 176 : 6528–6537.
61. SerranoM, RealG, SantosJ, CarneiroJ, MoranC, et al. (2011) A negative feedback loop that limits the ectopic activation of a cell type-specific sporulation sigma factor of Bacillus subtilis. PLoS Genet 7: e1002220.
62. SerranoM, NevesA, SoaresCM, MoranCPJr, HenriquesAO (2004) Role of the anti-sigma factor SpoIIAB in regulation of σG during Bacillus subtilis sporulation. J Bacteriol 186 : 4000–4013.
63. CharyVK, XenopoulosP, EldarA, PiggotPJ (2010) Loss of compartmentalization of σE activity need not prevent formation of spores by Bacillus subtilis. J Bacteriol 192 : 5616–5624.
64. RhayatL, DuperrierS, Carballido-LopezR, PellegriniO, StragierP (2009) Genetic dissection of an inhibitor of the sporulation sigma factor σG. J Mol Biol 390 : 835–844.
65. DiezV, SchujmanGE, Gueiros-FilhoFJ, de MendozaD (2012) Vectorial signalling mechanism required for cell-cell communication during sporulation in Bacillus subtilis. Mol Microbiol 83 : 261–274.
66. RubioA, PoglianoK (2004) Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis. EMBO J 23 : 1636–1646.
67. BagyanI, HobotJ, CuttingS (1996) A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J Bacteriol 178 : 4500–4507.
68. HalbergR, KroosL (1994) Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase. J Mol Biol 243 : 425–436.
69. JonesS, ParedesC, TracyB, ChengN, SillersR, et al. (2008) The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol 9: R114.
70. LawleyT, CroucherN, YuL, ClareS, SebaihiaM, et al. (2009) Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J Bacteriol 191 : 5377–5386.
71. SchuchR, PiggotPJ (1994) The dacF-spoIIA operon of Bacillus subtilis, encoding σF, is autoregulated. J Bacteriol 176 : 4104–4110.
72. SetlowB, SetlowP (1995) Small, acid-soluble proteins bound to DNA protect Bacillus subtilis spores from killing by dry heat. Appl Environ Microbiol 61 : 2787–2790.
73. VepacheduVR, SetlowP (2007) Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme. J Bacteriol 189 : 1565–1572.
74. SorgJA, DineenSS (2009) Laboratory maintenance of Clostridium difficile. Curr Protoc Microbiol Chapter 9: Unit 9A 1.
75. HortonR, HuntH, HoS, PullenJ, PeaseL (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77 : 61–68.
76. AdamsC, EckenrothB, PutnamE, DoublieS, ShenA (2012) Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog 9: e1003165.
77. BurnsDA, MintonNP (2011) Sporulation studies in Clostridium difficile. J Microbiol Methods 87 : 133–138.
78. SmithGA, GrossSP, EnquistLW (2001) Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc Natl Acad Sci U S A 98 : 3466–3470.
79. LohseM, BolgerAM, NagelA, FernieAR, LunnJE, et al. (2012) RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40: W622–627.
80. GentlemanRC, CareyVJ, BatesDM, BolstadB, DettlingM, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
81. Pages H, Aboyoun P, Lawrence M (2012) IRanges: Infrastructure for manipulating intervals on sequences. R package version 1.14.4 ed.
82. Team RC (2012) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing.
83. HochbergY, BenjaminiY (1990) More powerful procedures for multiple significance testing. Stat Med 9 : 811–818.
84. Warnes GR (2012) gplots: Various R programming tools for plotting data. R package version 2.11.0 ed. http://cran.r-project.org/web/packages/gplots/index.html
85. OkamuraS, UrakamiK, KimataM, AoshimaT, ShimamotoS, et al. (2000) The N-terminal prepeptide is required for the production of spore cortex-lytic enzyme from its inactive precursor during germination of Clostridium perfringens S40 spores. Mol Microbiol 37 : 821–827.
86. ChastanetA, VitkupD, YuanGC, NormanTM, LiuJS, et al. (2010) Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A 107 : 8486–8491.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



