-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
Most yeast ribosomal protein genes are duplicated and their characterization has led to hypotheses regarding the existence of specialized ribosomes with different subunit composition or specifically-tailored functions. In yeast, ribosomal protein genes are generally duplicated and evidence has emerged that paralogs might have specific roles. Unlike yeast, most mammalian ribosomal proteins are thought to be encoded by a single gene copy, raising the possibility that heterogenous populations of ribosomes are unique to yeast. Here, we examine the roles of the mammalian Rpl22, finding that Rpl22−/− mice have only subtle phenotypes with no significant translation defects. We find that in the Rpl22−/− mouse there is a compensatory increase in Rpl22-like1 (Rpl22l1) expression and incorporation into ribosomes. Consistent with the hypothesis that either ribosomal protein can support translation, knockdown of Rpl22l1 impairs growth of cells lacking Rpl22. Mechanistically, Rpl22 regulates Rpl22l1 directly by binding to an internal hairpin structure and repressing its expression. We propose that ribosome specificity may exist in mammals, providing evidence that one ribosomal protein can influence composition of the ribosome by regulating its own paralog.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003708
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003708Summary
Most yeast ribosomal protein genes are duplicated and their characterization has led to hypotheses regarding the existence of specialized ribosomes with different subunit composition or specifically-tailored functions. In yeast, ribosomal protein genes are generally duplicated and evidence has emerged that paralogs might have specific roles. Unlike yeast, most mammalian ribosomal proteins are thought to be encoded by a single gene copy, raising the possibility that heterogenous populations of ribosomes are unique to yeast. Here, we examine the roles of the mammalian Rpl22, finding that Rpl22−/− mice have only subtle phenotypes with no significant translation defects. We find that in the Rpl22−/− mouse there is a compensatory increase in Rpl22-like1 (Rpl22l1) expression and incorporation into ribosomes. Consistent with the hypothesis that either ribosomal protein can support translation, knockdown of Rpl22l1 impairs growth of cells lacking Rpl22. Mechanistically, Rpl22 regulates Rpl22l1 directly by binding to an internal hairpin structure and repressing its expression. We propose that ribosome specificity may exist in mammals, providing evidence that one ribosomal protein can influence composition of the ribosome by regulating its own paralog.
Introduction
Protein synthesis is a major energy consuming process involving intricate coordination of translation machinery in response to nutrient availability and stress sensing signals, as well as hormonal and growth factor cues in multi-cellular organisms. The ribosome is comprised of two ribonucleoprotein subunits: the 40S and 60S (‘small’ and ‘large’ subunits, respectively). Together these subunits facilitate peptide bond formation, performing different roles during translation. Ribosome synthesis is a highly controlled process, whereby three distinct RNA polymerases are synchronously coordinated to produce equimolar amounts of four rRNAs and 79 mammalian ribosomal proteins (RPs) [1]–[4].
A growing number of human diseases have been linked to mutations in genes encoding factors involved in ribosome biogenesis and protein synthesis [5], [6]. These include developmental malformations, inherited bone marrow failure syndromes and cancer in a variety of organisms [5], [7]–[9]. In addition, interventions leading to reduced translation, such as dietary restriction and reduced 60S ribosomal protein expression, elicits lifespan extension in yeast, worms and files [10]–[13]. Determining the molecular pathology underlying diseases and the role of ribosomes in aging requires a better understanding of ribosome specificity and the functions of individual RPs.
RPs are generally thought to be essential components of the functional ribosome and although they do not play a direct role in catalyzing peptidyl transfer, they may be critical for both regulatory and structural functions of the ribosome [14], [15]. In addition to their role in the ribosome, many RPs, including murine Rpl22, have been shown to have extra-ribosomal functions [16]–[18]. In particular, as RNA binding proteins, RPs have been found to bind cellular and viral RNAs outside of the context of the ribosome. Some RPs also function to regulate their own expression, such as Rpl30 in yeast [19], [20] and human RPS13 [21].
RPs are often essential for viability. For example, embryonic lethality was reported in the first murine knockout of a ribosomal protein (RP) gene, Rps19 [22]. Rpl24 [23], [24], and Rps6 [25]–[27], also play essential roles. However, two reports have found that mice lacking either Rpl22 or Rpl29 survive without these RP genes [28], [29]. In yeast, approximately 85% of the RP genes are duplicated as a result of an ancient genome duplication event [30] and many of these paralogous genes are functionally redundant [31]. Generally, deletion of either paralog, but not both simultaneously, results in viability; however, yeast that lack non-duplicated RP genes or both paralogs of an individual subunit are often, but not always, inviable [32]. Cross-complementation studies in yeast, analyzing defects in growth, have shown that most RP paralogs are functionally redundant [31]; however, several recent studies suggest that some paralogs might have subtle functional differences [33]–[38]. In the case of RPL22, however, tetrad analysis indicated that the rpl22aΔ rpl22bΔ double mutant was viable, although slow growing [37], [39], while in worms disruption of rpl22 expression is lethal (http://www.shigen.nig.ac.jp/c.elegans/index.jsp).
Rpl22 is an external protein on the 60S ribosomal subunit that is incorporated into the ribosome at later stages of ribosome maturation [40]. An early study suggested that Rpl22 was not required for translation in vitro [41]; however, the protein has been identified as a component of the ribosome [40] and likely plays a role in protein translation. In addition, other activities have been attributed to Rpl22 in mammals, including association with both viral RNAs, like EBER1, and cellular RNAs, such as human telomerase RNA [42]–[44]. Mice lacking Rpl22 are viable but have a defect in T cell development attributed to p53-dependent arrest of the αβ lineage T cells [28]. Recently, RPL22 has been found to be mutated or downregulated in various cancers, including T-acute lymphoblastic leukemias [45], invasive breast carcinoma [46], and lung adenocarcinoma [47].
Here we report evidence that in mice one ribosomal protein can control composition of the ribosome by regulating expression of its own paralog. Knocking out Rpl22 results in up-regulation of Rpl22-like1 (Rpl22l1), a paralog of Rpl22 whose predicted protein sequence is highly homologous to Rpl22. Rpl22l1 was first identified (although mis-labeled Rpl22) in a screen for 14-3-3ε binding partners in mouse brain [48] and has been identified as a trace component of ribosomes in mouse liver and mammary gland tissues [49]. We find that Rpl22l1 co-sediments with actively translating ribosomes in Rpl22−/− mice and a compensatory increase in Rpl22l1 expression likely accounts for the lack of translational defects in these animals. Enhanced Rpl22l1 expression also occurs upon acute knockdown of Rpl22 expression, indicating that Rpl22 has an active role in suppressing the synthesis of its paralog. Mechanistically, we find that Rpl22 directly represses expression of Rpl22l1 mRNA by binding to an internal hairpin structure. shRNA-mediated knockdown of Rpl22l1 causes a severe growth defect in cells lacking Rpl22. Accordingly, we demonstrate that the composition of the ribosome is regulated by the novel mechanism of direct repression of one paralog by another, and offer the hypothesis that this is one mechanism by which ribosome specificity is coordinated.
Results
Rpl22−/− mice are viable
A gene-trapped mouse embryonic stem cell clone harboring a mutation in Rpl22 was obtained from Bay Genomics and used to generate Rpl22 heterozygous mice (Rpl22+/−) (see Text S1). 5' rapid amplification of cDNA ends (5′ RACE) followed by automated DNA sequencing determined that the gene-trap vector inserted between the third and fourth exons of Rpl22 (Figure S1A). Gene-trap vector disruption of Rpl22 expression was confirmed by PCR and western blot analysis (Figure S1B). Mice heterozygous for the Rpl22 mutation (Rpl22+/−) were interbred to obtain homozygous Rpl22-null (Rpl22−/−) mice. Surprisingly, Mendelian ratios of Rpl22+/+, Rpl22+/−, and Rpl22−/− were found in the resulting progeny. During the construction of our mouse line, Anderson et al. reported the generation of viable Rpl22−/− mice and observed defects in lymphocyte development [28].
Characterization of our Rpl22−/− mice indicated that they have defects in lymphocyte development (Figure S2, S3, S4) similar to that described by Anderson et al. (2007) [28]. B220+ B cells in the bone marrow were also significantly reduced in Rpl22−/− mice (Figure S4A, B). Further analysis indicated that B cell development was interrupted by Rpl22 deficiency, as evidenced by a decrease in the B220+ developing IgM-IgD - and immature IgM+IgD - B cells (Figure S4C, D).
Despite the ubiquitous expression of Rpl22 and its hypothesized role in mRNA translation, disruption of Rpl22 in mice results in a remarkably mild phenotype. Hematologic parameters are normal in these mice [50]. Also, unlike deletion of RPL22 in yeast [51], Rpl22−/− mice have no substantial difference in growth rate or size relative to Rpl22+/+ and Rpl22+/− littermates (Anderson et al. 2007 and our unpublished data). Surprisingly, no significant differences were observed in the polysome profiles of lysate from Rpl22−/− liver, lung or cultured ear fibroblasts when compared to samples collected from Rpl22+/+ mice (unpublished data), indicating that Rpl22 is not essential for translation efficiency or ribosome biogenesis in the tissues evaluated.
Compensation by Rpl22l1 in Rpl22-null mice
We considered the possibility that another factor might be compensating for lack of Rpl22 in mice. A bioinformatic search identified Rpl22-like1 (Rpl22l1) as a candidate. Rpl22l1 encodes a 122 amino acid protein that is 73% identical to Rpl22 (Figure 1A). The protein sequence of Rpl22l1 is highly conserved from human to zebrafish (Figure S5). To determine if significant levels of Rpl22l1 mRNA exists in tissues from Rpl22+/+ mice and whether Rpl22l1 transcript levels increase in Rpl22−/− mice, lung, liver, spleen and kidney were harvested from Rpl22−/− mice and their littermate controls and analyzed by quantitative RT-PCR (qRT-PCR) for Rpl22 and Rpl22l1 expression with Acidic Ribosomal Protein (ARBP) mRNA levels used for normalization (Figure 1B). In Rpl22+/+ samples, high Rpl22 expression was detected, while Rpl22l1 transcripts were less abundant. In samples isolated from Rpl22−/− mice, qRT-PCR revealed a ∼3-fold induction of Rpl22l1 mRNA expression relative to littermate controls.
Fig. 1. Mouse Rpl22 has an expressed paralog, Rpl22l1. 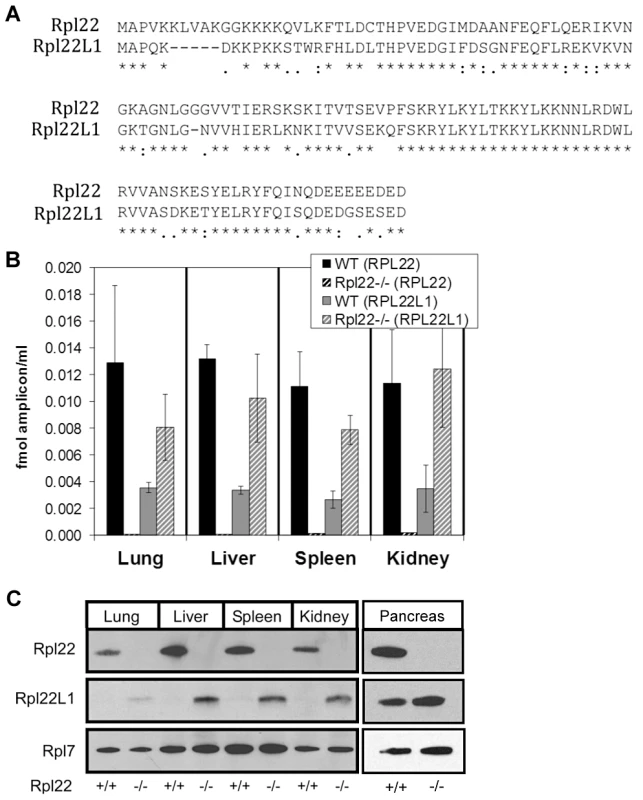
(A) Alignment of Rpl22 and Rpl22l1 protein sequences. Lung, liver, spleen, kidney and pancreas, harvested from Rpl22−/− mice and their littermate controls, were analyzed for relative Rpl22 and Rpl22l1 mRNA levels by qRT-PCR (B) or protein expression by Western blot analysis (C). Results are representative of 3 independent experiments. Rpl22l1 transcripts were found associated with actively translating ribosomes in both Rpl22+/+ and Rpl22−/− mouse ear fibroblast samples (unpublished data), suggesting that the Rpl22l1 mRNA is actively translated into protein. Consistently, equivalent increases in Rpl22l1 protein levels are observed in the absence of Rpl22 in lung, liver, spleen and kidney (Figure 1C). Similar increases in Rpl22l1 mRNA and protein expression were observed in Rpl22−/− skeletal muscle and brain (unpublished data). Additionally, Rpl22l1 was found to be abundantly expressed in both Rpl22+/+ and Rpl22−/− pancreas (Figure 1C). These results indicate that Rpl22 negatively regulates, either directly or indirectly, Rpl22l1 expression in a range of mouse tissues.
Rpl22l1 protein co-sediments with actively translating ribosomes
To determine if Rpl22l1 is incorporated into actively translating ribosomes, liver tissue was isolated from Rpl22−/− mice and their littermate controls followed by sedimentation of the lysates on sucrose gradients. Fractions collected from the gradients, were subsequently loaded onto an SDS-page gel for western blot analysis. In Rpl22−/− samples, Rpl22l1 is present in fractions containing 60S ribosome subunits and polysomes, suggesting that it is incorporated into free ribosome subunits and ribosomes actively translating mRNA in the absence of Rpl22 (Figure 2). Rpl7, a RP that is incorporated into the large subunit of the ribosome, is present in the fractions containing 60S ribosome subunits and polysomes in both Rpl22+/+ and Rpl22−/− samples. Rpl22 and Rpl22l1, but not Rpl7, were detected in fractions 1 and 2, representative of the free, non-ribosomal lysate (Figure 2C, D), consistent with the hypothesis that these RPs exist in states within the cell both associated with the ribosome and independent of the ribosome. Additionally, while Rpl22l1 levels are relatively evenly detected in 60S containing fractions of the polysome profile (Figure 2D, fractions 3–7) Rpl22 is detected at higher levels in the fractions containing free 60S subunits and messages loaded with fewer ribosomes (Figure 2C, fractions 3–5 vs 6–7).
Fig. 2. Both mouse Rpl22 and Rpl22l1 proteins can be incorporated into ribosomes. 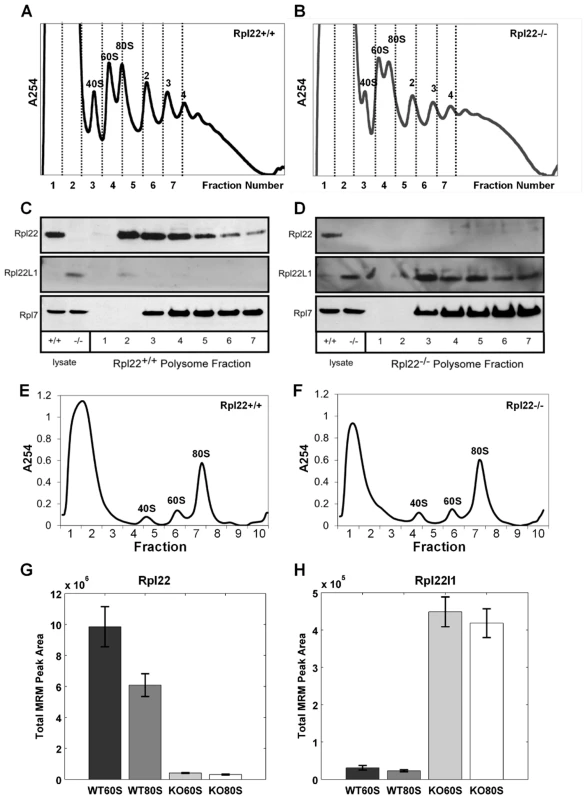
Liver tissue was isolated from Rpl22+/+ (A, C) and Rpl22−/− (B, D) mice then, after sedimentation of the lysates on sucrose gradients, fractions were collected and loaded onto an SDS-page gel for western blot analysis (C and D, respectively). Images are representative of 3 independent experiments. Multiple Reaction Monitoring Mass spectrometry (MRM-MS) analysis was performed on free 60S subunits and 80S monosomes from actively translating polysomes. Liver lysates from Rpl22+/+ and Rpl22−/− mice were subjected to a brief treatment with low amounts of RNase A to degrade mRNA between ribosomes in polysomes and release the ribosomes as 80S monomers. After inhibiting the RNase with KCl and heparin, the samples were fractionated on 10–30% sucrose gradients containing 800 mM KCl to disrupt any “nonproductive couples” of 40S and 60S subunits [79]. (E, F) Representative gradient profiles for Rpl22+/+ and Rpl22−/−samples, respectively. Fraction 6 was used to isolate 60S subunits for MRM-MS, while fraction 7 was used to isolate 80S monosomes. Summation of the integrated MRM peak areas for all transitions from all observed peptides for Rpl22 (G) and Rpl22l1 (H) proteins yielded the total MRM peak areas plotted for each of the four tested samples (WT60S, WT80S, KO60S, KO80S), indicating the relative amounts of Rpl22 and Rpl22l1 in these samples. The height of each bar represents the average of the three technical replicates performed for the given sample, and each error bar represents +/−1 standard deviation. p-values>3E-3 in both cases by paired student's t-test. Rpl22 peptides: AGNLGGGVVTIER; ITVTSEVPFSK; YFQINQDEEEEEDED. Rpl22l1 peptides: TGNLGNVVHIER; ITVVSEK. To verify that Rpl22l1 was incorporated into 60S subunits and actively translating ribosomes, free 60S subunits and 80S monosomes from actively translating polysomes were isolated from liver lysates of Rpl22+/+ (WT) and Rpl22−/− (KO) mice using sucrose density gradient fractionation (Figure 2E, F; see Text S1). Samples were then concentrated and prepared for mass spectrometry analysis using standard methods (see Text S1). In order to measure the relative amounts of Rpl22 and Rpl22l1 in the WT and KO mouse liver lysate samples, we used multiple reaction monitoring (MRM), a targeted mass spectrometry (MS) approach that is highly sensitive. MRM is a targeted type of MS and requires a list of peptide targets and their subsequent fragment targets—known as a transition list—to program the analysis on the instrument (see Text S1). The final MRM analysis consisted of 4 peptide targets for Rpl22 (5 counting uniquely modified targets) and 3 peptide targets for Rpl22l1, and was limited to the top 8 fragment ions per peptide, creating a total of 64 transitions targeted in the analysis.
Summing the integrated MRM peak areas for all transitions from all observed peptides for each protein (Rpl22 and Rpl22l1) yielded the total MRM peak areas plotted for each of the four tested samples (WT60S, WT80S, KO60S, KO80S) and indicated the relative amounts of Rpl22 and Rpl22l1 in these samples (Figure 2G, H). These data indicate that Rpl22l1 levels are significantly higher in the 60S and 80S subunits isolated from Rpl22−/− liver than in Rpl22+/+ littermate controls (Figure 2H), supporting the hypothesis that Rpl22 regulates Rpl22l1 expression and, as a result, incorporation into ribosomes.
Rpl22 directly regulates Rpl22l1 expression
Why does expression of Rpl22l1 increase in mouse tissues lacking Rpl22? We considered two potential explanations: (1) Rpl22 directly regulates Rpl22l1 expression, or (2) compensation occurs during development in Rpl22−/− mice. To distinguish between these two possibilities, Rpl22 was acutely knocked down in 3T9 fibroblasts using a lentiviral-mediated inducible knockdown system that allows doxycycline-inducible regulation of Rpl22 and changes in Rpl22l1 expression were examined. 3T9 cells were transduced with 2 different tet-on shRNA lentivirus constructs (shRNA 1 and shRNA 2) that target Rpl22 mRNA or a nonspecific control construct. Following 3 days of doxycycline treatment, Rpl22l1 mRNA expression is enhanced 1.8 fold in 3T9 cells with reduced Rpl22 expression (Figure 3A). Western blot analysis confirmed that Rpl22l1 protein levels were elevated by the knockdown of Rpl22, while expression of other RPs, such as Rpl7 remained unchanged (Figure 3B). These results confirm that Rpl22 negatively regulates the expression of Rpl22l1 acutely and raise the possibility that Rpl22-mediated regulation of Rpl22l1 is an active process with biological significance.
Fig. 3. Acute knockdown of Rpl22 enhances Rpl22l1 expression. 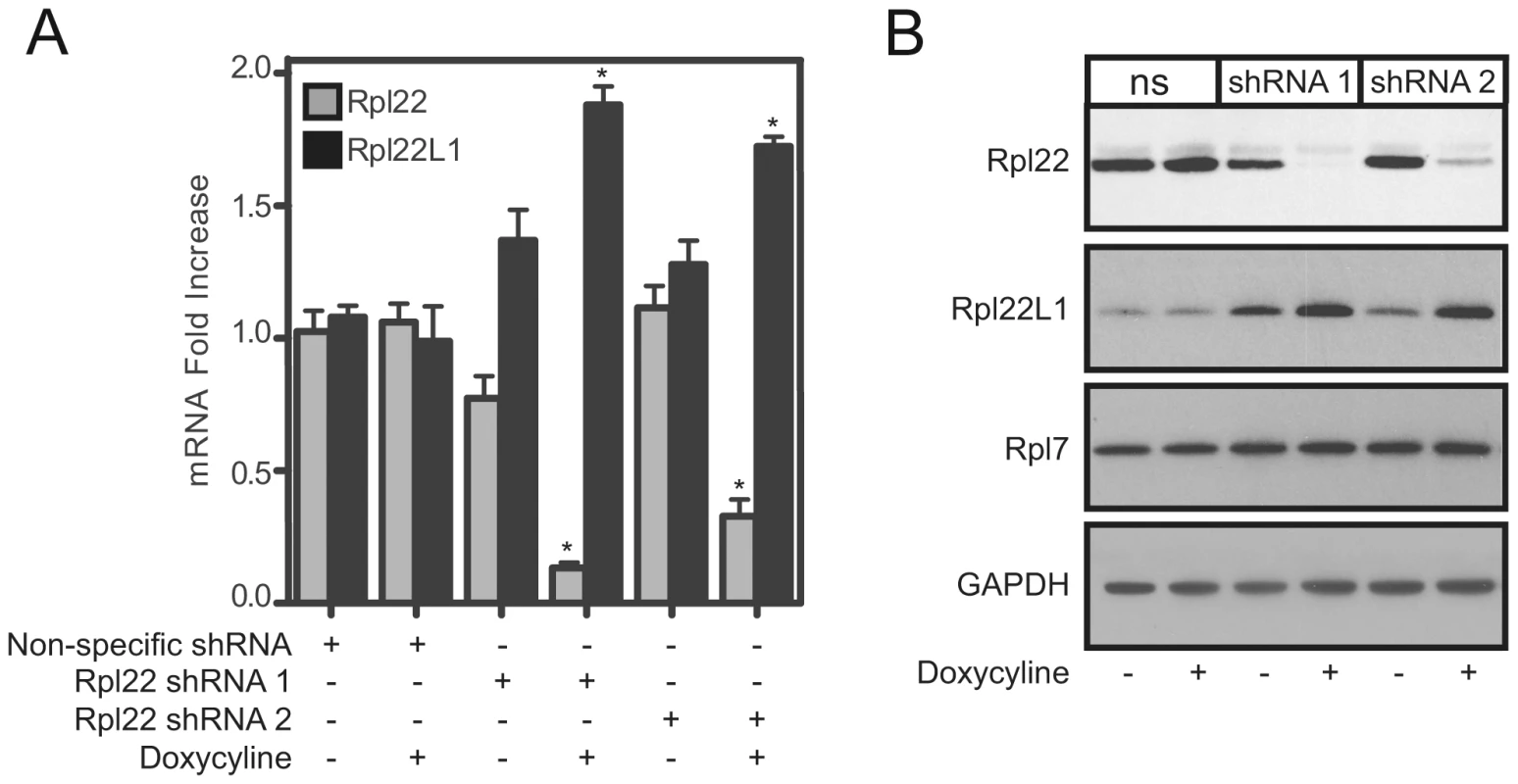
3T9 cells were transduced with doxycycline-inducible shRNA lentiviral constructs directed at Rpl22 (shRNA 1 and shRNA 2) or a non-specific (ns) shRNA construct and treated with doxycycline for 3 days to induce shRNA expression. NS, shRNA 1, or shRNA 2 expressing cells were analyzed for relative Rpl22 and Rpl22l1 mRNA levels by qRT-PCR (A) or protein expression by Western blot analysis (B). Results are the average ± SEM of 4 independent experiments (A) or representative of 3 independent experiments (B). Statistical significance is indicated (*; p<0.001 compared to untreated control). Collectively, these data suggest that Rpl22 is regulating expression of Rpl22l1 but the mechanism leading to the increased expression is unknown. To determine whether Rpl22 affects the stability of Rpl22l1 mRNA, cultures of 3T9 cells were treated with actinomycin D, which blocks transcription by all three eukaryotic polymerases [52]. After actinomycin D treatment, the levels of Rpl22l1 mRNA in Rpl22+/+ 3T9 cells decreased significantly (p<0.01) relative to the untreated control, while in Rpl22−/ − 3T9 cells Rpl22l1 levels were maintained and the rate of decay was reduced (Figure 4A). These results suggest that Rpl22 affects the stability of Rpl22l1 mRNA.
Fig. 4. Rpl22 directly binds Rpl22l1 mRNA to regulate its expression levels. 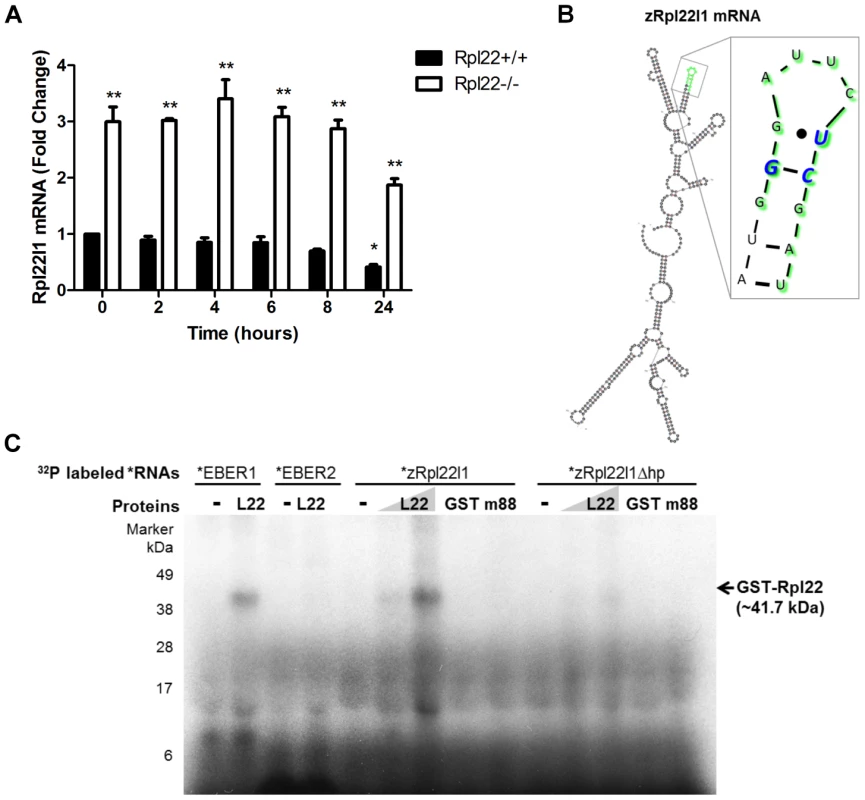
(A) In the absence of Rpl22, Rpl22l1 mRNA levels are more stable in the presence of Actinomycin D. Rpl22+/+ or Rpl22−/− 3T9 cells were treated with Actinomycin D (1 µM final concentration) and total RNA was harvested at the time points shown. Levels of Rpl22l1 mRNA were quantitated by qRT-PCR. Results are the average ± SEM of 3 independent experiments and the statistical significance indicated is (*, p<0.01, compared to Rpl22+/+ untreated; ** p<0.001, compared to Rpl22+/+ at each time point). (B) M-fold analysis [54] of zRpl22l1 mRNA reveals the presence of a consensus Rpl22 RNA-binding motif. In green are the residues deleted to remove the hairpin (zRpl22l1Δhp). In blue are the residues known to be essential for Rpl22 binding. (C) Autoradiogram of ribonuclease protection assay reveals Rpl22 protein binds to Rpl22l1 mRNA and this binding is abrogated upon removal of the hairpin. 32P labeled EBER1 (positive control), EBER 2 (negative control), zRpl22l1 or zRpl22l1Δhp RNAs were incubated in the absence or presence of GST-Rpl22 (41.7 kDa), GST (27 kDa) or m88, a GST-Rpl22 RNA binding mutant (41.6 kDa), as indicated, then UV-cross-linked, digested with RNase A, and run on a SDS protein gel. GST-Rpl22 was detected, hence, bound to EBER1 and zRpl22l1 RNAs but not Rpl22l1Δhp RNA, indicating Rpl22 binds to Rpl22l1 mRNA and this binding is abrogated upon removal of the hairpin. Numbers indicate molecular weight protein ladder in kDa. If Rpl22 is involved in mediating Rpl22l1 stability, does Rpl22 bind directly to Rpl22l1 mRNA? Previous studies determined that Rpl22 is associated with Epstein-Barr virus-expressed RNA, EBER1 [43], [53] and evaluation of the RNA binding specificity of Rpl22 suggested that Rpl22 recognizes a stem loop (hairpin) structure with a G-C at the neck followed by a U [42], [53]. To address whether regulation of Rpl22l1 expression is directly mediated by Rpl22, an algorithm termed M-fold that predicts RNA secondary structure was used to evaluate Rpl22l1 mRNA structure for potential Rpl22 RNA binding motifs [54]. Analysis revealed the presence of a consensus Rpl22 RNA-binding motif within exon 2 of zRpl22l1, suggesting that Rpl22 might interact directly with Rpl22l1 mRNA (Figure 4B).
To test whether Rpl22 can directly bind Rpl22l1 mRNA via the hairpin structure identified in Rpl22L1 mRNA by M-fold analysis an RNAse protection analysis was performed. Recombinant proteins were incubated with radiolabeled RNA, and UV-crosslinked. The RNAs were then digested with RNase A, and samples were run on a protein gel. Proteins bound to radiolabeled RNA were detected by autoradiogram at their expected molecular weight. Unbound proteins are not detected on autoradiogram. Recombinant Rpl22 was found to bind to in vitro transcribed zRpl22L1 mRNA, but not the zRpl22l1 mRNA lacking the hairpin structure (zRpl22L1Δhp) (Figure 4C), suggesting the Rpl22 directly binds to Rpl22l1 mRNA.
To determine if Rpl22 directly regulates expression of Rpl22l1, we employed a biosensor quantification assay using GFP as a fluorescent indicator of effects on expression. Zebrafish embryos were microinjected with mRNAs EGFP-zRpl22, EGFP-Rpl22l1 or a mutant form of Rpl22l1 in which the Rpl22l1 hairpin was modified (EGFP-zRpl22L1mt) in combination with constructs expressing zRpl22 or zRpl22l1. mCherry mRNA was co-injected to allow for quantification of the relative fluorescence intensity. Co-injection of zRpl22 with EGFP-Rpl22l1 led to a significant decreased fluorescence relative to those embryos co-injected with zRpl22 and EGFP-Rpl22l1mt (Figure 5A–E), suggesting that the hairpin structure within Rpl22l1 mRNA is necessary for Rpl22 to directly regulate its expression. Next, to assess if the presence of the hairpin structure within the Rpl22l1 mRNA is sufficient to regulate expression, the hairpin sequence from zRpl22l1 mRNA was fused to EGFP and evaluated in the biosensor quantification assay. The heterologous reporter mRNA, zRpl22l1-150h-EGFP, containing the minimal sequence identified by M-fold to form the hairpin structure, was co-injected with mCherry mRNA (injection control) and (Figure 5G) zRpl22 mRNA or (Figure 5I) Rpl22-Morpholino (Rpl22-MO) into zebrafish embryos. Rpl22 repressed the expression of zRpl22l1-150h-EGFP reporter, while knockdown Rpl22 can increase the expression of reporter, suggesting the hairpin structure identified in zRpl22l1 mRNA is sufficient to regulate mRNA abundance.
Fig. 5. Regulation of Rpl22l1 mRNA expression is mediated by a hairpin structure. 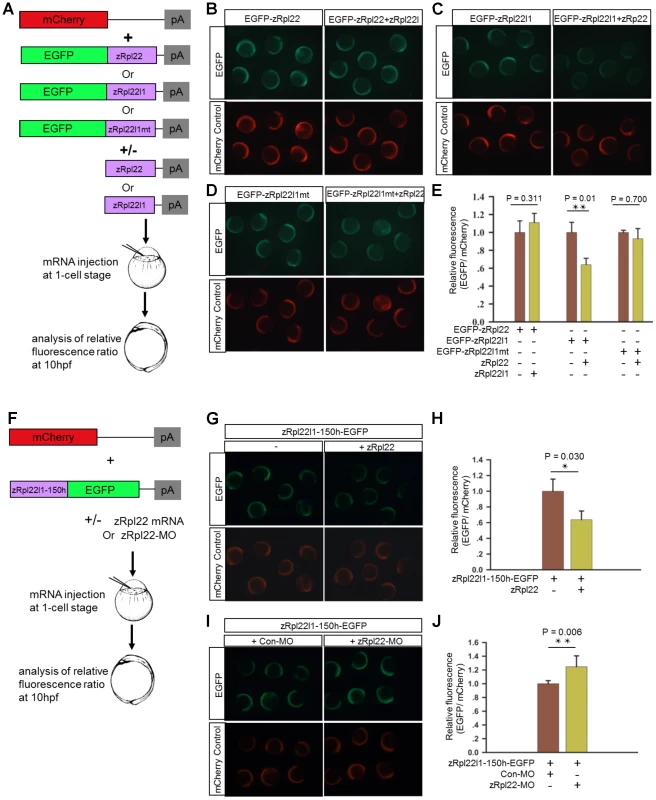
(A) Schematic representation of the biosensor quantification assay. (B–D) Stereoimages of zebrafish embryos illustrate that co-injection of zRpl22 repressed fluorescence derived from an EGFP-Rpl22l1 fusion protein upon injecting mRNA for both and assessing fluorescence at 6 hours post fertilization. Rpl22, Rpl22l1 or mutated Rpl22l1 (Rpl22l1mt) coding sequence was fused to EGFP mRNA and co-injected with mCherry mRNA (injection control) along with the corresponding inhibitor mRNAs (zRpl22 or zRpl22l1) into 1-cell stage zebrafish embryos. (F) Schematic representation of the experimental procedure. A zRpl22l1-150h-EGFP heterologous reporter mRNA, containing the minimal sequence identified by mFOLD to form the hairpin structure, was co-injected with mCherry mRNA (injection control) and (G) Rpl22 mRNA or (I) Rpl22-Morpholino (Rpl22-MO) into 1-cell stage zebrafish embryos. (E, H, J) At 10 hpf, the relative fluorescence intensity was calculated and normalized to control injections (n = 3, each group). Data are shown as mean ± standard deviation (s.d.). Rpl22l1 incorporation into ribosomes is associated with increased cell proliferation
In yeast, ribosomal protein paralogs are thought to functionally compensate for one another, each incorporating into the ribosome in the absence of the other. Deletion of both yeast RPL22 paralogs results in viable, but slow growing cells [37]. Recently Rpl22+/− or Rpl22−/− primary mouse embryonic fibroblasts (MEFs) were found to grow faster and display increased transformation potential relative to MEFs isolated from Rpl22+/+ littermates [45] To further evaluate the effect of Rpl22 and Rpl22Ll1 expression on growth rates, Rpl22+/+ or Rpl22−/− 3T9 fibroblasts were transduced with one of 2 different tet-on shRNA lentivirus constructs (shRNA#1 - and shRNA#2-Rpl22l1) that target Rpl22l1 mRNA to acutely knock-down its expression. Western blot analysis confirmed that Rpl22l1 protein levels were elevated in Rpl22−/− 3T9 fibroblasts (Figure 6A). No significant difference was observed in the growth rates of Rpl22+/+ or Rpl22−/− 3T9 fibroblasts (Figure 6B). In doxycycline-treated Rpl22+/+ or Rpl22−/− 3T9 fibroblasts expressing the shRNA constructs, Rpl22l1 protein levels were confirmed to be reduced by western blot analysis (Figure 6C, E). Knockdown of Rpl22l1 significantly reduced growth rates of Rpl22+/+ fibroblasts and greatly impaired that of Rpl22−/− 3T9 fibroblasts (Figure 6D, F and S6), indicating that cells lacking both paralogs have severe growth defects. In contrast, acute knockdown of Rpl22 in Rpl22+/+ 3T9 fibroblasts resulted in no change in the rate of proliferation (Figure S7). In summary, these cell culture studies indicate that expression of at least one paralog of Rpl22 is required for normal growth and suggests that Rpl22l1 may also affect cell proliferation by a mechanism independent of Rpl22.
Fig. 6. Acute knockdown of Rpl22l1 expression impairs cellular growth. 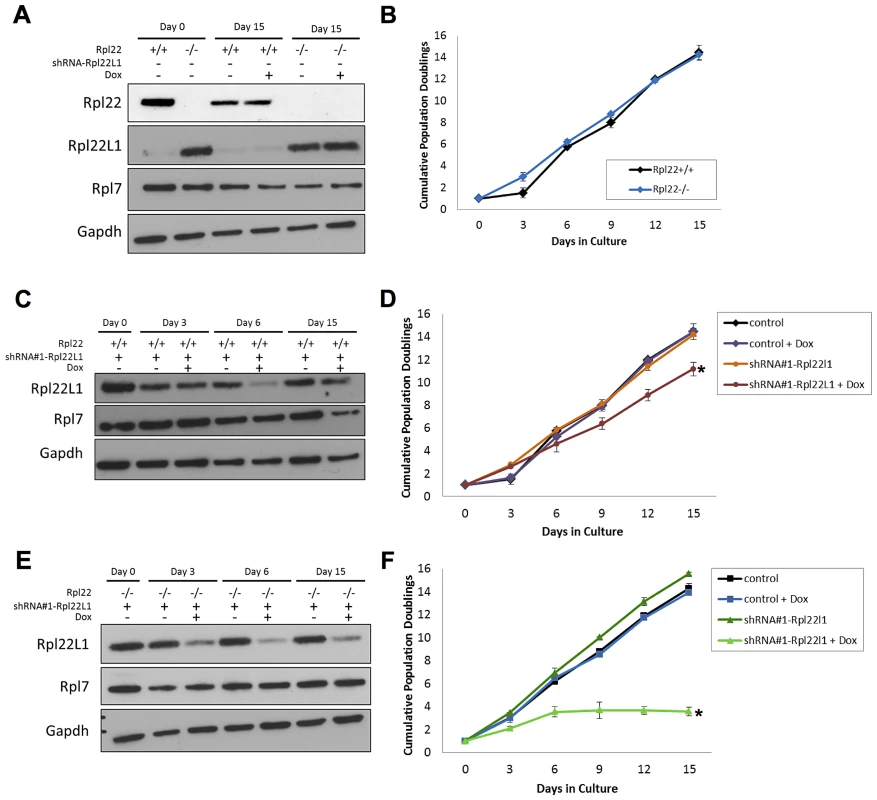
(A) Lysates isolated Rpl22+/+ and Rpl22−/− 3T9 cells treated with or without doxycycline were collected to assess levels of Rpl22 or Rpl22L1 by Western blot analysis. GAPDH was used as a loading control. (B) Growth of Rpl22+/+ and Rpl22−/− 3T9 cells was compared. Levels of Rpl22l1 were analyzed by Western blot analysis to confirm that the Rpl22l1-shRNA knocked down levels of Rpl22l1 in doxycycline-treated Rpl22+/+ (C) and Rpl22−/− (E) 3T9 cells transduced with the shRNA construct. Growth of Rpl22+/+ (D) and Rpl22−/− (F) 3T9 cells transduced with each shRNA construct was determined. Results are representative of 2 independent experiments with error bars representative of ±SD. Statistical significance is indicated (*, p<0.05 compared to untreated control). Discussion
Here we report on phenotypes of mice lacking the large subunit ribosomal protein, Rpl22. Surprisingly, Rpl22−/− mice have no developmental defects, other than previously reported defects in T and B cell development (Anderson et al. 2007 and our data). More generally, Rpl22−/− mice have no significant defects in translation, as judged by sucrose gradient sedimentation to detect ribosome occupancy on transcripts (unpublished data). This is somewhat surprising since yeast lacking Rpl22 function are slow growing with substantial defects in translation [10], [37]. One possible explanation is that the lack of a defect in translation in Rpl22−/− mice is explained by compensatory increases in expression of another gene, Rpl22l1, which shares a high degree of homology with Rpl22. While Rpl22l1 mRNA is detected at low levels in most tissues of wild-type mice, mRNA and protein levels are dramatically increased in the Rpl22 knockout. It is interesting that Rpl22l1 is expressed at relatively high levels in the pancreas even though Rpl22 is present as well. Future studies will be needed to determine why these two paralogs are jointly expressed in this tissue. When expressed, Rpl22l1 was incorporated into ribosomes and actively translating polysomes. These findings indicate that Rpl22l1 is capable of functioning within ribosomes that are actively translating mRNA. Future studies examining the function of Rpl22l1 will help decipher to what extent Rpl22 and Rpl22l1 may have redundant roles and also determine their independent functions, for which accumulating evidence is emerging [28], [45] (O'Leary et al, unpublished data).
Although poorly understood, gene compensation during development is a recurrent phenomenon in mouse knockout studies [55]; therefore, enhanced expression of Rpl22l1 might reflect developmental compensation in Rpl22−/− mice. Alternatively, Rpl22 could play an active role in the repression of Rpl22l1. To test this, we examined the consequences of acute knockdown of Rpl22 in 3T9 fibroblasts. Our findings indicate that Rpl22l1 mRNA and protein levels are rapidly increased following Rpl22 knockdown and support the conclusion that developmental compensation does not account for the increased Rpl22l1 mRNA and protein levels. Instead, we find that inhibition of transcription in cells lacking Rpl22 results a slower decay of Rpl22l1 mRNA compared to Rpl22+/+ cells, suggesting Rpl22 affects the stability of Rpl22l1 mRNA. It is possible that more than one mechanism is involved in increasing Rpl22l1 expression in the absence of Rpl22. Further studies interrogating other mechanisms are needed to full understand what regulates Rpl22l1 upon Rpl22 deficiency. We find that Rpl22 binds to a hairpin structure in the Rpl22l1 mRNA. The hairpin motif identified in Rpl22l1 mRNA is necessary and sufficient for regulation of mRNA abundance by Rpl22. These data suggest that Rpl22 might function in an extra-ribosomal capacity to bind and destabilize Rpl22l1 mRNA. Rpl22l1 is yet another RNA demonstrated to interact with Rpl22. The viral RNA EBER1 contains three Rpl22 binding sites and is thought to compete with the 28S rRNA for Rpl22 binding in Epstein-Barr virus-infected cells [42], [43], [53], [56], [57]. In yeast, recent studies have revealed regulation of the expression of one paralog in a duplicated RP gene pair by the other. Ribosomal protein S29A (Rps29a), for example, regulates its own expression along with expression of its paralog, RPS29B [58].
Interestingly, Rpl22 has also been ascribed functions independent of its role as a component of the ribosome [17], [18]; these extra-ribosomal functions include regulation of telomerase activity [44] and association with histone H1 [59]. In fact, many ribosomal proteins, including Rpl7, Rpl13a, and Rps3 [60]–[62] have extra-ribosomal functions, which will have to be considered as mechanistic links are sought to explain diseases associated with RP mutations and possibly age-related phenotypes. Given their ancient nature, it is not surprising that evolution has settled on ways of exploiting these proteins for multiple uses.
Do Rpl22 and Rpl22l1 have shared or unique functions? The observation that one ribosomal protein represses expression of its own paralog is likely to be of biological significance and determining whether mice lacking Rpl22l1 have specific phenotypes will identify tissues or conditions where Rpl22l1 is the functional RPL22 paralog that participates in translation. We propose that Rpl22 and Rpl22l1 share the ability to participate in protein translation as part of the ribosome, since both paralogs are found incorporated into ribosomes and Rpl22 has an active role in suppressing expression of Rpl22l1. However, unique roles are also well established, since knockout of mouse Rpl22 leads to specific phenotypes in T and B cell development (Anderson et al. 2007 and our data), and data from Zhang et al. indicate that both Rpl22 and Rpl22l1 are essential for T cell development in zebrafish and exhibit antagonistic functions in regulating the emergence of hematopoietic stem cells [63]. Together, these observations lead us to propose that Rpl22 and Rpl22l1 may have overlapping activities, sharing a role in enhancing large subunit ribosome function but also having distinct roles in development of the immune system [63]and, in the case of Rpl22l1, perhaps other tissues.
Based on data from S. cerevisiae, in which genes for most ribosomal proteins are duplicated, Komili et al. proposed the existence of a ribosome code, whereby ribosome subunits composed of different ribosomal proteins would have differential specificity for mRNAs [38]. This proposal was based on an accumulation of data, including differential localization of paralogs and large-scale phenotypic screens, which indicated that specific subsets of ribosomal protein genes were often identified in phenotypic screens of gene deletion strains. This is an exciting hypothesis that, if corroborated, would identify ribosome composition as a new mechanism for regulation of gene expression. Our studies in yeast raise the possibility that ribosome specificity may occur in yeast aging [10], [37]. In a long-lived, slow-growing and translation-compromised rpl22aΔ background, we find that deletion of the other paralog, RPL22B, causes no significant further reduction in translation, but blocks lifespan extension (Steffen et al., unpublished data). One possible explanation for this result is that the portion of ribosomes containing Rpl22b is increased in the rpl22aΔ background, leading to specific changes in translation conducive to enhanced replicative lifespan. Other explanations are possible and further studies will be necessary to test this important hypothesis. Structural studies do not point to an obvious mechanism by which Rpl22 paralogs would influence the pool of translated RNAs and other models involving specific non-ribosomal functions of Rpl22 and Rpl22l1 have to be identified and/or tested.
One limitation to the ribosome specificity hypothesis proposed in S. cerevisiae is that ribosomal proteins are generally not thought to be duplicated in other organisms. Therefore, if ribosome specificity indeed exists [38], it may be more prominent in yeast than mammals. Recently, Xue and Barna have suggested that specialized ribosomes might also regulate gene expression in mammals [64]. Our findings support the hypothesis, at least in the case of murine Rpl22, that one ribosomal protein may repress expression of the other, raising the possibility that ribosome specificity may extend to organisms other than yeast. In addition, a recent study of Rpl38 mutant embryos found that although global translation was unchanged, translation of Hox genes were altered [65], providing additional support for the hypothesis that differential composition of the ribosome might contribute to transcript-specific translational regulation. Cryo-EM studies of the eukaryotic 80S ribosome have demonstrated that Rpl38 is located on the surface of the ribosome and interacts with a region of the rRNA known as expansion segment 27 (ES27), which has two distinct orientations toward the L1 stalk or toward the exit tunnel [66], [67]. The location of Rpl38 in the ribosome is consistent with its proposed role in regulating transcript-specific translation.
The tissue specific defects observed in Rpl22−/− and Rpl29−/− mice, along with mice expressing mutated Rpl38 [65], suggest that temporal and spatial expression of RPs are critical for proper development and tissue patterning. In plants and Drosophila, many RPs paralogs display tissue specific variations and are differentially expressed during development [68]–[71]. In mammals, recent studies found mRNA expression patterns of RPs vary in different tissues and cell types [65], [72]. Together with the findings presented here, these studies illustrate that heterogeneous expression of RPs is tightly regulated; however additional studies will be crucial in confirming the influence of these differential expression patterns on specialized ribosome activity and message specific translation.
Further studies are also required to address the questions that have arisen from this study. Are there tissues or cell types in mice where Rpl22l1 and not Rpl22 is the predominant paralog? What is the mechanism by which Rpl22 represses expression of Rpl22l1 through interaction with its mRNA? What are the non-ribosomal functions of Rpl22 paralogs? And most importantly, do ribosomes with Rpl22l1 have different specificity for mRNAs than those with Rpl22? Findings presented in this study provide interesting leads in which to test the hypothesis that specialized ribosomes exist and, furthermore, point to a new level of regulation in ribosome biogenesis, wherein ribosomal paralogs regulate each other's synthesis to optimally maintain the organism.
Materials and Methods
Generation of Rpl22 null mice
A detailed description of the targeting vector along with the generation and genotyping of the Rpl22−/− mice can be found in the Text S1.
Materials
DMEM tissue culture medium was purchased from Mediatech (Manassas, VA), L-glutamine, penicillin/streptomycin, and trypsin were from GIBCO (Carlsbad, CA). Fetal calf serum was from Gemini (West Sacramento, CA). Rpl22 antibody was from BD Biosciences (Franklin Lakes, NJ), Rpl22l1 antibody was from Santa Cruz (Santa Cruz, CA) and the Rpl7 antibody was purchased from Novus Biologicals (Littleton, CO). GAPDH antibody was from Ambion (Austin, TX). HRP-conjugated donkey anti-rabbit and donkey anti-mouse were from Jackson Immunoresearch Laboratories, Inc (West Grove, PA). Rpl22, Rpl22L1, and ARBP primers were from Operon (Huntsville, AL). Lentiviral shRNA constructs V2MM_120192 and V2LHS_131608, directed at Rpl22 (denoted shRNA 1 and 2, respectively), along with V3LMM_473587 and V3LHS_322499, directed at Rpl22l1 (denoted shRNA 1 and 2, respectively), were purchased through Open Biosystems (Huntsville, AL) and. A non-silencing-TRIPZ lentiviral inducible shRNAmir control was also purchased through Open Biosystems.
RNA isolation and real time PCR
RNA was isolated from mouse tissue or 3T9 cells using the RNeasy kit (Qiagen) with a DNaseI treatment following the manufacturer's protocol. cDNA synthesis was performed using oligo (dT) and reverse transcriptase Superscript Preamplification System (Invitrogen). Quantitative PCR was performed using a Biorad iCycler and measuring SYBR green incorporation for product detection. The fold-increase of Rpl22 and Rpl22l1 were normalized to the housekeeping gene ARBP. The primer sequences are as follows: RPL22 (forward): 5′-GTCGCCAACAGCAAAGAGAG-3′; RPL22 (reverse): 5′-TCCTCGTCTTCCTCCTCCTC-3′; RPL22l1 (forward): 5′-TGGAGGTTTCATTTGGACCTTAC-3′; RPL22l1 (reverse): 5′-TTTCCAGTTTTTCCATTGACTTTAAC-3′. For qRT-PCR analysis of gradient fractions, values for Rpl22 and Rpl22l1 were normalized to an artificial polyadenylated RNA that was added in equal amounts to each gradient fraction.
3T9 cells were treated with 1 µM Actinomycin D for the indicated time (0, 2, 4, 6, 8 or 24 hours). Cells were washed once with PBS then total RNA was isolated and cDNA was synthesized as described above. Relative quantitation was calculated by the 2−ΔΔCT method and normalized to ARBP.
Western blot analysis
Adult mouse tissue or cultured cells were lysed in RIPA buffer (300 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0) and proteins separated and transferred to nitrocellulose membranes using the Invitrogen (Carlsbad, CA) Nu-Page system. Proteins in polysome fractions from sucrose density gradients (described above) were TCA precipitated and resuspended in 30 ul of Sample Buffer (1× Laemmli buffer with bromophenol blue and DTT), while for mouse liver polysome fractions, 32 ul of each 1 ml fraction was loaded and proteins were separated and transferred as described above. Blots were blocked with 5% milk in TBST for 1 h and incubated overnight at 4°C with the following dilutions of primary antibodies: Rpl22 (1∶250), Rpl22l1 (1∶250), Rpl7 (1∶1000), GAPDH (1∶50,000). Membranes were incubated for 2 h with HRP-conjugated donkey anti-rabbit or sheep anti-mouse (1∶10,000) secondary antibodies and antigen was detected using enhanced chemiluminescence (ECL Plus Western Blotting Detection System, Amersham).
Polysome analysis
Polysome analysis of mouse liver samples was adapted from the protocol of Oliver et al. [23]. Minced liver tissue was homogenized on ice using a Dounce homogenizer in ice-cold lysis buffer containing 1.5 mM KCl, 2.5 mM MgCl2, 5 mM Tris-HCl, pH 7.4, 1% Na deoxycholate and 1% Triton X-100. After centrifugation at 2500 g for 15 min at 4C in a microfuge, heparin and cycloheximide were added to the clarified supernatants to final concentrations of 1 mg/ml and 100 µg/ml, respectively, and centrifuged at 16,000 g for 10 min at 4°C. Samples containing 20–25 OD260 units in 1 ml were loaded onto 11 ml 7–47% (w/v) sucrose high-salt (800 mM KCl) gradients described previously [73]. Samples were centrifuged in a Beckman SW40 Ti rotor at 39,000 rpm (270,500×g) for 2 hr at 4°C and 1-ml fractions were collected from the top of the gradients with a Brandel fractionator system. The A254 gradient profiles shown in Figure 2 were digitized using a DATAQ DI-148U data recording module that converts and exports analog absorbance readings to analysis software.
Mass spectrometry
A detailed description of the sample preparation and the experimental procedure can be found in the Text S1.
MEF isolation and 3T9 cell lines
Embryos were harvested at day 13.5dpc and mouse embryonic fibroblasts (MEFs) were harvested as previously described [74]. Following isolation, MEFs were cultured in DMEM (Dulbecco's modified Eagle medium) containing 10% FBS, 1% Penicillin/Streptomycin and 1% L-Glutamine and subjected to 3T9 (Figure 6) protocol [74], [75]. Cells were removed from tissue culture plates with 0.05% trypsin and 0.02% EDTA for 3 min at 37°C, washed with DMEM, and plated overnight prior to experimentation.
Lentiviral transduction
For Rpl22 and Rpl22l1 knockdown, lentiviral pGIPZ shRNA constructs were purchased from Open Biosystems and cloned into inducible pTRIPZ constructs. Briefly, 293T cells were transfected with pTRIPZ constructs bearing shRNA against Rpl22 (shRNA 1 and 2) or a non-silencing control using calcium phosphate transfection. After 48 h, viral supernatant was harvested and used to transduce 3T9 cells. Cells were selected for 5 days in puromycin (5 µg/ml) followed by 3 days of doxycycline (1 µg/ml) treatment to induce shRNA expression before experimentation. For cell proliferation assays, Rpl22+/+ or Rpl22−/− 3T9 cells were infected with control shRNA or 1 of 2 shRNA constructs targeting Rpl22 or Rpl22l1. After 5 days of selection in puromycin, growth rates of cells transduced with each shRNA construct were determined by plating cells in triplicate at a density of 30,000 cells/well in 6-well plate in media with or without doxycycline (1 µg/ml). Every 3 days for the cells were counted and replated at 30,000 cells/well for 15 days. Protein was isolated from 10 cm plates treated in parallel to the growth assays.
Bioinformatics
Protein sequences for murine Rpl22 (NP_033105) and Rpl22l1 (NP_080793) were obtained from Genbank (http://www.ncbi.nlm.nih.gov/). Sequence alignments were performed using ClustalW2 Algorithm (http://www.ebi.ac.uk/Tools/msa/clustalw2/). mRNA sequences for mRpl22l1 (NM_026517) and zRpl22l1 (NM_001045335) were entered into mFold (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) for prediction of RNA secondary structure.
Statistical analysis
Statistical comparisons were made between groups by a 2-tail Mann-Whitney U test (Figure 6, S5, S6), by Newman-Keuls post hoc analysis, a two-tailed Student's t test (Figures 4, S2, S3, S4), or a 1-way ANOVA (Figure 3). Significant differences between groups are indicated in figure legends.
Zebrafish embryos
AB wild-type zebrafish strain was bred and maintained at 28.5°C under standard aquaculture conditions. Embryos were staged as described previously [76].
mRNAs, morpholino microinjection and biosensor quantification assay in zebrafish embryos
Full-length coding sequence for EGFP, mCherry, zebrafish Rpl22 (zRpl22) and Rpl22l1 (zRpl22l1) were cloned into pCS2+. Full - length cDNA sequences encoding EGFP-zRpl22 and EGFP-zRpl22l1 were subcloned in the pCS2+. The zRpl22l1mut, mutant form of zRpl22l1 in which hairpin GATGGGATTCTCGATT was mutated to GACGGTATCTTAGACT, was generated using GeneTailor kit (Invitrogen, Carlsbad, CA) with the modified forward primer 5′-GACTGCACTCACCCTGTGGAGGACGGTA TCTTAGACTCT GCAAACTTTG-3′. The 150 bp fragment in the zebrafish Rpl22l1 CDS containing the Rpl22-binding hairpin was amplified by PCR using the following primers: Forward: cagggatccatgcagactgttgtgagaaagaat, Reverse : tcagaattccaggttgcctgttttgccattaac. Then the PCR product was digested (BamH1+EcoR1) and ligated into pCS2+EGFP to get the in-frame chimera sequence, which was referred to as zRpl22l1-150h-EGFP. All mRNAs for microinjection were synthesized using the mMessage mMachine kit (Ambion, Austin, TX). Then embryos were injected at one-cell stage with indicated synthetic mRNAs and observed at 10 hpf stages. Injection doses: 100 pg for each mCherry, GFP-Sensor and Inhibitor mRNAs. Morpholinos were ordered from Gene Tools (Gene Tools, LLC, Philomath, OR) and dissolved in nuclease-free water. Morpholino to bind the translation start site (AUG MOs) of zebrafish Rpl22 [63] (Sequence: 5′-CCGACAGTTTTGGCAGAAAGCCAGT-3′) was injected at 6 ng in the 1-cell stage embryos. The standard control MO from Gene Tools was used as a control (Control MO, sequence: 5′-CCTCTTACCTCAGTTACAATTTATA-3′). Images were taken from the Nikon SMZ1500 stereomicroscope equipped with DS-Fi1 digital camera and Nikon Ar imaging software.
Fluorescence sensor assays was designed and performed according to the well-established method in zebrafish [77], [78]. To calculate relative fluorescence for each embryo, green or red pixel intensity was quantified using ImageJ software (NIH). Relative fluorescence was determined for each embryo (GFP/mCherry). After normalization to the average GFP/mCherry ratio in the control embryos, the sensor repression or enhancement fold was obtained in the inhibitor mRNA or Rpl22-MO injected groups.
Plasmids
The coding regions of EBER1 and EBER 2 were amplified by PCR from the cDNA of EBV-infected African Burkitt lymphoma KemIII cells (kindly provided by Jeffrey Sample, Penn State Hershey College of Medicine, Hershey, PA), adding a T7 promoter, and cloned into pCR2.1 plasmid (Invitrogen, Carlsbad, CA). The primers used for PCR were: forward 5′-TAATA CGACTCACTATAGGCAAAACCTCAGGACCTACGCTG, TAATACGACTCACTATAGGTCAAA CAGGACAGCCGTTGC and reverse 5′-GAACTGCGGGATAATGGATGC, AAGCCGAATACC CTTCTCCCAG for EBER 1 and EBER2 respectively. The zRpl22l1Δhp and EGFP-zRpl22l1mut were generated from plasmid zRpl22l1-pCS2+ and EGFP-zRpl22l1-pCS2+, respectively, with deletion of the hairpin TGGGATTCTCGA using GeneTailor mutagenesis kit according to manufacturer's instruction (Invitrogen, Carlsbad, CA). pGEX-m88 plasmid was generated from pGEX-hRpl22 (kindly provided by Joan Steitz, Yale University, New Haven, CT) using GeneTailor mutagenesis kit according to manufacturer's instruction (Invitrogen, Carlsbad, CA) creating a EEYLKE motif instead of KKYLKK.
RNAse protection assay
The plasmids used for RPA assay were EBER 1, EBER 2, zRpl22l1 and zRpl22l1Δhp. The plasmids were linearized using EcoRI for EBERs and NotI for zRpl22l1s (enzymes from New England Biolab, Ipswich, MA), and radioactive probes were prepared by in vitro transcription with T7 (EBERs) or SP6 (zRpl22like1s) RNA polymerases at 37°C (Maxiscript, Ambion, TX), in presence of 10 µCi of 3ZP-UTP (Perkin Elmer, Boston, MA), and purified by G-50 columns (Illustra Probe-Quant, GE Healthcare, Buckinghamshire, UK). The radiolabeled RNAs were renatured for 2 min at 95°C and kept on ice for 5 min. Radiolabeled RNAs and protein were incubated on ice for 12 min, in presence of 5 µg of tRNA (Roche) in RPA buffer (10 mM Tris pH 8.0, 50 mM NaCl, 0.75 mM MgCl2, 0.8 mM DTT and 2.5% glycerol), and the mixtures were cross-linked on ice pack for 30 min (using a UV Stratalinker 1800). After cross-linking, the mixture were digested by RNase A (Qiagen, Valencia, CA, 7,500 U/µl) diluted to 3,750 U/µl in RNase A buffer (20 mM Tris pH 7.0, 2 mM MgCl2, and 0.2 mM KCl) for 20 min at 37°C, then 2 µl of denaturing dye were added before denaturation for 2 min at 90°C. Nine µl of the reaction mixtures were loaded on 4–12% Bis-Tris gel, 1.5 mm (Invitrogen, Carlsbad, CA). The gels were dried and expose to BioMax MR films (Kodak, Rochester, NY) or to a phosphoimager plate read using a Fuji BAS-2500 reader.
Protein expression and purification
E. coli strain BL21DE(3)pLys (Promega, Madison, WI) was used to produce recombinant GST, GST-hRpl22 and GST-hRpl22m88 from plasmids pGEX-3X. pGEX-hRpl22 and pGEX-hRplm88. Isopropyl-β-D-thiogalactopyranoside (IPTG) at a final concentration of 100 µM was added to 2× YT media at optical density A600 = 0.4, and incubation was maintained for 4 hours at 37°C. The cells were harvested and resuspended at 0.25 mg/ml in 1× PBS (2.6 mM KCl, 1.7 mM KH2PO4, 137 mM NaCl, 11 mM Na2HPO4, pH 7.4) and lysed by 3 passages in a M-110L Pneumatic (Microfluidics, Newton, MA), in presence of Complete mini without EDTA tablets (Roche, Mannheim, Germany). The cell extracts were loaded onto glutathione sepharose beads for batch purification (GE Healthcare, Upsala, Sweden) at 2 ml/liter of culture. The beads, 0.5 ml, were washed with 1 ml of wash buffer (50 mM Tris, pH 9.5, 150 mM NaCl), then washed with 1 ml of bump buffer (50 mM Tris pH 9.5, 150 mM NaCl, 1 mM reduced GSH). The proteins were eluted with 3 fractions of 1 ml elution buffer (50 mM Tris pH 9.5, 150 mM NaCl, 10 mM reduced GSH). The protein purity was assessed by SDS gel electrophoresis, and the concentration measured using Coomassie Plus (Bradford) Protein Assay (Pierce, Rockford, IL). The proteins were stored in 10% glycerol in 20 µl aliquots at −80°C.
Ethics statement
Appropriate protocols of all work on mice have been approved by the IACUC committees at the Institution where the work was performed and is in accord with accepted national guidelines.
Supporting Information
Zdroje
1. PlantaRJ (1997) Regulation of ribosome synthesis in yeast. Yeast 13 : 1505–1518.
2. RudraD, WarnerJR (2004) What better measure than ribosome synthesis? Genes Dev 18 : 2431–2436.
3. LaferteA, FavryE, SentenacA, RivaM, CarlesC, et al. (2006) The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev 20 : 2030–2040.
4. Fromont-RacineM, SengerB, SaveanuC, FasioloF (2003) Ribosome assembly in eukaryotes. Gene 313 : 17–42.
5. LiuJM, EllisSR (2006) Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood 107 : 4583–4588.
6. NarlaA, EbertBL (2010) Ribosomopathies: human disorders of ribosome dysfunction. Blood 115 : 3196–3205.
7. ScheperGC, van der KnaapMS, ProudCG (2007) Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet 8 : 711–723.
8. EllisSR, LiptonJM (2008) Diamond Blackfan anemia: a disorder of red blood cell development. Curr Top Dev Biol 82 : 217–241.
9. CmejlaR, CmejlovaJ, HandrkovaH, PetrakJ, PospisilovaD (2007) Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat 28 : 1178–1182.
10. SteffenKK, MacKayVL, KerrEO, TsuchiyaM, HuD, et al. (2008) Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133 : 292–302.
11. ChiocchettiA, ZhouJ, ZhuH, KarlT, HaubenreisserO, et al. (2007) Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol 42 : 275–286.
12. HansenM, TaubertS, CrawfordD, LibinaN, LeeSJ, et al. (2007) Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6 : 95–110.
13. KaeberleinM, PowersRW (2005) Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310 : 1193–1196.
14. BrodersenDE, NissenP (2005) The social life of ribosomal proteins. Febs J 272 : 2098–2108.
15. NollerHF, HoffarthV, ZimniakL (1992) Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256 : 1416–1419.
16. WoolIG (1996) Extraribosomal functions of ribosomal proteins. Trends Biochem Sci 21 : 164–165.
17. WarnerJR, McIntoshKB (2009) How common are extraribosomal functions of ribosomal proteins? Molecular cell 34 : 3–11.
18. BhavsarRB, MakleyLN, TsonisPA (2010) The other lives of ribosomal proteins. Human genomics 4 : 327–344.
19. VilardellJ, WarnerJR (1994) Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes & development 8 : 211–220.
20. MaciasS, BragulatM, TardiffDF, VilardellJ (2008) L30 binds the nascent RPL30 transcript to repress U2 snRNP recruitment. Molecular cell 30 : 732–742.
21. MalyginAA, ParakhnevitchNM, IvanovAV, EperonIC, KarpovaGG (2007) Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic acids research 35 : 6414–6423.
22. MatssonH, DaveyEJ, DraptchinskaiaN, HamaguchiI, OokaA, et al. (2004) Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol 24 : 4032–4037.
23. OliverER, SaundersTL, TarleSA, GlaserT (2004) Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development 131 : 3907–3920.
24. TangQ, RiceDS, GoldowitzD (1999) Disrupted retinal development in the embryonic belly spot and tail mutant mouse. Developmental biology 207 : 239–255.
25. RuvinskyI, SharonN, LererT, CohenH, Stolovich-RainM, et al. (2005) Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes & development 19 : 2199–2211.
26. VolarevicS, StewartMJ, LedermannB, ZilbermanF, TerraccianoL, et al. (2000) Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288 : 2045–2047.
27. PanicL, TamarutS, Sticker-JantscheffM, BarkicM, SolterD, et al. (2006) Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Molecular and cellular biology 26 : 8880–8891.
28. AndersonSJ, LauritsenJP, HartmanMG, FousheeAM, LefebvreJM, et al. (2007) Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity 26 : 759–772.
29. Kirn-SafranCB, OristianDS, FochtRJ, ParkerSG, VivianJL, et al. (2007) Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev Dyn 236 : 447–460.
30. WolfeKH, ShieldsDC (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387 : 708–713.
31. RotenbergMO, MoritzM, WoolfordJLJr (1988) Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev 2 : 160–172.
32. DeanEJ, DavisJC, DavisRW, PetrovDA (2008) Pervasive and persistent redundancy among duplicated genes in yeast. PLoS genetics 4: e1000113.
33. Baudin-BaillieuA, TollerveyD, CullinC, LacrouteF (1997) Functional analysis of Rrp7p, an essential yeast protein involved in pre-rRNA processing and ribosome assembly. Mol Cell Biol 17 : 5023–5032.
34. HaarerB, ViggianoS, HibbsMA, TroyanskayaOG, AmbergDC (2007) Modeling complex genetic interactions in a simple eukaryotic genome: actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev 21 : 148–159.
35. NiL, SnyderM (2001) A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell 12 : 2147–2170.
36. EnyenihiAH, SaundersWS (2003) Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163 : 47–54.
37. SteffenKK, McCormickMA, PhamKM, MacKayVL, DelaneyJR, et al. (2012) Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191 : 107–118.
38. KomiliS, FarnyNG, RothFP, SilverPA (2007) Functional specificity among ribosomal proteins regulates gene expression. Cell 131 : 557–571.
39. CostanzoM, BaryshnikovaA, BellayJ, KimY, SpearED, et al. (2010) The genetic landscape of a cell. Science 327 : 425–431.
40. Auger-BuendiaMA, LonguetM, TavitianA (1979) Kinetic studies on ribosomal proteins assembly in preribosomal particles and ribosomal subunits of mammalian cells. Biochim Biophys Acta 563 : 113–128.
41. LavergneJP, ConquetF, ReboudJP, ReboudAM (1987) Role of acidic phosphoproteins in the partial reconstitution of the active 60 S ribosomal subunit. FEBS Lett 216 : 83–88.
42. DobbelsteinM, ShenkT (1995) In vitro selection of RNA ligands for the ribosomal L22 protein associated with Epstein-Barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J Virol 69 : 8027–8034.
43. ToczyskiDP, MateraAG, WardDC, SteitzJA (1994) The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc Natl Acad Sci U S A 91 : 3463–3467.
44. LeS, SternglanzR, GreiderCW (2000) Identification of two RNA-binding proteins associated with human telomerase RNA. Mol Biol Cell 11 : 999–1010.
45. RaoS, LeeSY, GutierrezA, PerrigoueJ, ThapaRJ, et al. (2012) Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 120 : 3764–3773.
46. FinakG, BertosN, PepinF, SadekovaS, SouleimanovaM, et al. (2008) Stromal gene expression predicts clinical outcome in breast cancer. Nature medicine 14 : 518–527.
47. BhattacharjeeA, RichardsWG, StauntonJ, LiC, MontiS, et al. (2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America 98 : 13790–13795.
48. BallifBA, CaoZ, SchwartzD, CarrawayKL (2006) Identification of 14-3-3epsilon substrates from embryonic murine brain. J Proteome Res 5 : 2372–2379.
49. SugiharaY, HondaH, IidaT, MorinagaT, HinoS, et al. (2010) Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. Journal of proteome research 9 : 1351–1366.
50. KeelSB, PhelpsS, SaboKM, O'LearyMN, Kirn-SafranCB, et al. (2012) Establishing Rps6 hemizygous mice as a model for studying how ribosomal protein haploinsufficiency impairs erythropoiesis. Experimental hematology 40 : 290–294.
51. GiaeverG, ChuAM, NiL, ConnellyC, RilesL, et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 : 387–391.
52. BensaudeO (2011) Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2 : 103–108.
53. ToczyskiDP, SteitzJA (1993) The cellular RNA-binding protein EAP recognizes a conserved stem-loop in the Epstein-Barr virus small RNA EBER 1. Molecular and cellular biology 13 : 703–710.
54. ZukerM (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research 31 : 3406–3415.
55. LiangH, LiWH (2009) Functional compensation by duplicated genes in mouse. Trends in genetics : TIG 25 : 441–442.
56. FokV, Mitton-FryRM, GrechA, SteitzJA (2006) Multiple domains of EBER 1, an Epstein-Barr virus noncoding RNA, recruit human ribosomal protein L22. RNA 12 : 872–882.
57. HoumaniJL, DavisCI, RufIK (2009) Growth-promoting properties of Epstein-Barr virus EBER-1 RNA correlate with ribosomal protein L22 binding. Journal of virology 83 : 9844–9853.
58. ParenteauJ, DurandM, MorinG, GagnonJ, LucierJF, et al. (2011) Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell 147 : 320–331.
59. NiJQ, LiuLP, HessD, RietdorfJ, SunFL (2006) Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev 20 : 1959–1973.
60. KimJ, ChubatsuLS, AdmonA, StahlJ, FellousR, et al. (1995) Implication of mammalian ribosomal protein S3 in the processing of DNA damage. The Journal of biological chemistry 270 : 13620–13629.
61. KuhnJF, TranEJ, MaxwellES (2002) Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5 kD/Snu13p snoRNP core protein. Nucleic acids research 30 : 931–941.
62. MazumderB, SampathP, SeshadriV, MaitraRK, DiCorletoPE, et al. (2003) Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115 : 187–198.
63. ZhangY, DucAC, RaoS, SunXL, BilbeeAN, et al. (2013) Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Developmental cell 24 : 411–425.
64. XueS, BarnaM (2012) Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nature reviews Molecular cell biology 13 : 355–369.
65. KondrashovN, PusicA, StumpfCR, ShimizuK, HsiehAC, et al. (2011) Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145 : 383–397.
66. ArmacheJP, JaraschA, AngerAM, VillaE, BeckerT, et al. (2010) Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proceedings of the National Academy of Sciences of the United States of America 107 : 19748–19753.
67. ArmacheJP, JaraschA, AngerAM, VillaE, BeckerT, et al. (2010) Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proceedings of the National Academy of Sciences of the United States of America 107 : 19754–19759.
68. MarygoldSJ, RooteJ, ReuterG, LambertssonA, AshburnerM, et al. (2007) The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome biology 8: R216.
69. WeijersD, Franke-van DijkM, VenckenRJ, QuintA, HooykaasP, et al. (2001) An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128 : 4289–4299.
70. DegenhardtRF, Bonham-SmithPC (2008) Transcript profiling demonstrates absence of dosage compensation in Arabidopsis following loss of a single RPL23a paralog. Planta 228 : 627–640.
71. WilliamsME, SussexIM (1995) Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. The Plant journal : for cell and molecular biology 8 : 65–76.
72. BortoluzziS, d'AlessiF, RomualdiC, DanieliGA (2001) Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics 17 : 1152–1157.
73. MacKayVL, LiX, FloryMR, TurcottE, LawGL, et al. (2004) Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics 3 : 478–489.
74. KamijoT, ZindyF, RousselMF, QuelleDE, DowningJR, et al. (1997) Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91 : 649–659.
75. TodaroGJ, GreenH (1963) Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. The Journal of cell biology 17 : 299–313.
76. KimmelCB, BallardWW, KimmelSR, UllmannB, SchillingTF (1995) Stages of embryonic development of the zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists 203 : 253–310.
77. GiraldezAJ, MishimaY, RihelJ, GrocockRJ, Van DongenS, et al. (2006) Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312 : 75–79.
78. StatonAA, KnautH, GiraldezAJ (2011) miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nature genetics 43 : 204–211.
79. MartinTE (1973) A simple general method to determine the proportion of active ribosomes in eukaryotic cells. Experimental cell research 80 : 496–498.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



