-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGlobal Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
Macroautophagy (autophagy) is crucial for cell survival during starvation and plays important roles in animal development and human diseases. Molecular understanding of autophagy has mainly come from the budding yeast Saccharomyces cerevisiae, and it remains unclear to what extent the mechanisms are the same in other organisms. Here, through screening the mating phenotype of a genome-wide deletion collection of the fission yeast Schizosaccharomyces pombe, we obtained a comprehensive catalog of autophagy genes in this highly tractable organism, including genes encoding three heretofore unidentified core Atg proteins, Atg10, Atg14, and Atg16, and two novel factors, Ctl1 and Fsc1. We systematically examined the subcellular localization of fission yeast autophagy factors for the first time and characterized the phenotypes of their mutants, thereby uncovering both similarities and differences between the two yeasts. Unlike budding yeast, all three Atg18/WIPI proteins in fission yeast are essential for autophagy, and we found that they play different roles, with Atg18a uniquely required for the targeting of the Atg12–Atg5·Atg16 complex. Our investigation of the two novel factors revealed unforeseen autophagy mechanisms. The choline transporter-like protein Ctl1 interacts with Atg9 and is required for autophagosome formation. The fasciclin domain protein Fsc1 localizes to the vacuole membrane and is required for autophagosome-vacuole fusion but not other vacuolar fusion events. Our study sheds new light on the evolutionary diversity of the autophagy machinery and establishes the fission yeast as a useful model for dissecting the mechanisms of autophagy.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003715
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003715Summary
Macroautophagy (autophagy) is crucial for cell survival during starvation and plays important roles in animal development and human diseases. Molecular understanding of autophagy has mainly come from the budding yeast Saccharomyces cerevisiae, and it remains unclear to what extent the mechanisms are the same in other organisms. Here, through screening the mating phenotype of a genome-wide deletion collection of the fission yeast Schizosaccharomyces pombe, we obtained a comprehensive catalog of autophagy genes in this highly tractable organism, including genes encoding three heretofore unidentified core Atg proteins, Atg10, Atg14, and Atg16, and two novel factors, Ctl1 and Fsc1. We systematically examined the subcellular localization of fission yeast autophagy factors for the first time and characterized the phenotypes of their mutants, thereby uncovering both similarities and differences between the two yeasts. Unlike budding yeast, all three Atg18/WIPI proteins in fission yeast are essential for autophagy, and we found that they play different roles, with Atg18a uniquely required for the targeting of the Atg12–Atg5·Atg16 complex. Our investigation of the two novel factors revealed unforeseen autophagy mechanisms. The choline transporter-like protein Ctl1 interacts with Atg9 and is required for autophagosome formation. The fasciclin domain protein Fsc1 localizes to the vacuole membrane and is required for autophagosome-vacuole fusion but not other vacuolar fusion events. Our study sheds new light on the evolutionary diversity of the autophagy machinery and establishes the fission yeast as a useful model for dissecting the mechanisms of autophagy.
Introduction
Macroautophagy (hereafter autophagy) is a catabolic pathway that transports cytoplasmic materials into a degradative organelle, the vacuole or lysosome. This self-digestion process is upregulated during starvation, when cells have to rely on the turnover of intracellular substances to provide the building blocks for synthesizing new macromolecules [1]. Autophagy is critically important for the survival of unicellular organisms such as yeasts, whose cells are directly exposed to a fluctuating environment [2], [3]. In recent years, diverse roles of autophagy in the development and health of multicellular organisms have also been uncovered [4], [5].
Molecular understanding of autophagy began with the identification of autophagy-related (ATG) genes in S. cerevisiae, which remains the organism where the autophagy machinery has been best characterized [6]–[8]. The Atg proteins required for all autophagy-related pathways are referred to as the core Atg proteins, and most of them are involved in the generation of a double membrane-enclosed transport vehicle called autophagosome. Two protein complexes are important for initiating the autophagosome formation process. One complex consists of Atg1 kinase and its associated proteins. The other is the phosphatidylinositol 3-kinase (PI3K) complex composed of Vps34, Vps15, Atg6, and Atg14, which generates phosphatidylinositol 3-phosphate (PI3P) at the sites where autophagosomes are assembled. PI3P is recognized by Atg18 (homolog of mammalian WIPI proteins), which together with Atg2, regulates the retrograde trafficking of Atg9, the only core Atg protein with transmembrane domains. The expansion of the autophagosome precursor, called isolation membrane or phagophore, requires the conjugation of a ubiquitin-like protein Atg8 to phosphatidylethanolamine. Factors involved in this conjugation include the Atg8 processing enzyme Atg4, the E1 enzyme Atg7, the E2 enzyme Atg3, and the E3-like complex Atg12–Atg5·Atg16. Atg12 is another ubiquitin-like protein whose conjugation to Atg5 requires the E2 enzyme Atg10.
Many ATG genes in S. cerevisiae have readily recognizable homologs in other eukaryotes, indicating that autophagy is an ancient and conserved pathway. On the other hand, differences in the autophagy machinery between S. cerevisiae and other organisms have also been documented and it has been argued that studying additional model organisms will help us better understand the evolution and mechanisms of autophagy [9], [10]. The fission yeast S. pombe is evolutionarily very distant from S. cerevisiae. Molecular clock studies estimated that these two species diverged more than 500 million years ago [11]. The existence of a homolog of mammalian Atg101 in S. pombe but not S. cerevisiae underscores the potential value of S. pombe for studying the divergences of autophagy mechanisms [12]. However, no screen for autophagy genes has been conducted in S. pombe, and published works on fission yeast autophagy have been limited to a subset of close homologs of budding yeast ATG genes [3], [13].
In this study, through unbiased genome-wide screening, we discovered new autophagy factors in S. pombe. Characterization of these and other autophagy factors in fission yeast demonstrated the utility of S. pombe in uncovering novel autophagy mechanisms.
Results
A genome-wide analysis of mating genes in fission yeast
Unlike S. cerevisiae, haploid S. pombe cells of opposite mating types (h− or h+) do not mate on the standard rich medium. Instead, mating is most efficiently triggered by nitrogen starvation, and immediately followed by meiosis and sporulation [14], [15]. From classical genetic screens, about 20 mating genes have been identified in S. pombe, which are called ste (for sterile) or ral (for ras-like) genes [16 and references therein]. According to PomBase [17], 15 ste and ral genes have been cloned thus far. The classical screens are far from saturating, as a few dozen additional genes, uncovered through other means, have been implicated in mating [15]. The mating defect of autophagy mutants, which is attributed to an inability to supply enough nitrogen intracellularly, was only discovered during a focused study on these mutants [3].
With an aim of identifying new autophagy genes, we screened the mating phenotype of a fission yeast deletion collection [18], using a barcode sequencing technology we have developed [19]. In our screening procedure (Figure 1A), we mixed a pool of haploid deletion strains, which are h+, with an equal amount of wild-type (WT) h− cells on solid mating media. After 4 days of incubation, we isolated the spores from the mating mixtures. Genomic DNA was extracted from both the input mutant pool and the spores. Barcodes associated with the deletions were amplified by PCR and sequenced. For each mutant/gene, we calculated a mating defect (MD) score, which is a normalized log2 ratio of barcode sequencing counts in input vs. spores. For mating genes, we expected MD scores higher than 0 because their barcodes should be depleted among the spores. On the other hand, genes involved in meiosis and/or sporulation but not mating should have MD scores close to 0, as their deletions are unlikely to manifest a phenotype in the heterozygous diploid state.
Fig. 1. Barcode sequencing-based screens of mating phenotype. 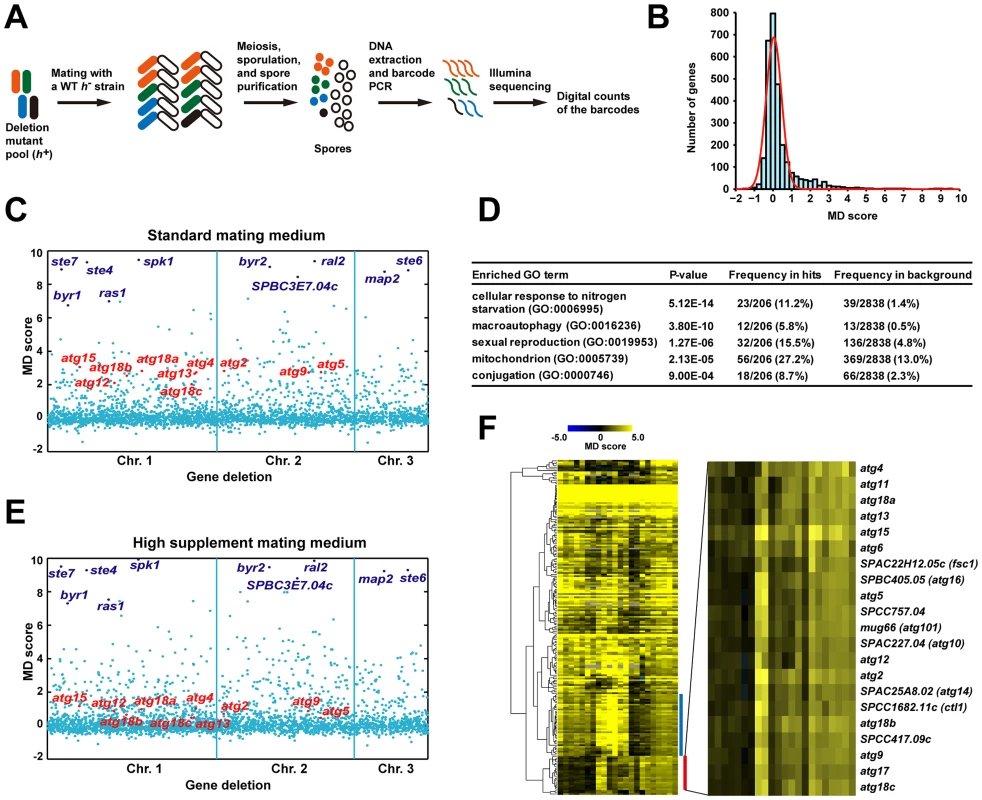
(A) Schematic of the screening procedure. (B) A histogram of the mating defect (MD) scores from the screen 0428_YES_SPA-45s conducted under standard mating conditions. The red line represents a fitted normal distribution. (C) A scatter plot of the MD scores from the screen 0428_YES_SPA-45s. The genes are ordered according to their chromosomal positions. The 10 genes with the highest average MD scores under standard conditions are highlighted in dark blue. The 10 genes known to be required for starvation-induced autophagy are highlighted in red. (D) Gene Ontology (GO) term enrichment analysis of the screen hits obtained under standard conditions. (E) A scatter plot of the MD scores from the screen 0428_YES_SPA-200s. Genes are highlighted as in C. (F) Hierarchical clustering analysis of the MD scores from the 22 screens. For a detailed view of the heat map, see Figure S2. Blue bar denotes the cluster enriched for mitochondrial protein-coding genes. Red bar denotes the autophagy gene cluster, whose close-up view is shown at right. Under the standard mating conditions (pregrowth in liquid YES medium, and mating on the SPA solid medium supplemented with 45 mg/l of leucine, uracil, and adenine) [20], we obtained MD scores for more than 2800 mutants, representing about 80% of the non-essential S. pombe genes. The distribution of MD scores largely conforms to a normal distribution centered at 0, except for a long right tail, which represents mating-defective mutants with higher than usual MD scores (Figure 1B and 1C). We repeated the screen twice, and identified the mating-defective mutants as the ones passing a false discovery rate (FDR) cutoff of 0.1 in all three screens (Figure S1). Using this stringent criterion, a total of 206 deletion mutants were found to be mating-defective, to different extents, under the standard mating conditions (Table S1).
The mutants of 9 ste and ral genes (byr1/ste1, ste4, ras1/ste5, ste6, ste7, byr2/ste8, ste20, ral2, scd2/ral3) were among the deletion strains screened. It was satisfying to see all of them scored as mating-defective by our analysis. Moreover, seven of them were among the top 10 hits ranked by the average MD scores (Table S1). It is probably no coincidence that all of these ste and ral genes are involved in the nutrient sensing or pheromone response pathways, as mutants blocking these signaling pathways are known to have the most severe mating defect [14], [15].
As expected, Gene Ontology (GO) term analysis showed that among the 206 screen hits, genes involved in starvation response, sexual reproduction, and macroautophagy are significantly enriched (Figure 1D). Surprisingly, genes encoding mitochondrial proteins are also heavily enriched, suggesting that mitochondria may play a previously under-appreciated role in mating.
Using barcode sequencing-based analysis, we could recapitulate the finding that the severity of the mating defect of autophagy mutants is influenced by the mating conditions [3]. Raising the concentrations of supplements in the mating medium from 45 mg/l to 200 mg/l led to a significant reduction of the MD scores of autophagy mutants, but did not change those of the top-ranked signaling mutants (compare Figure 1C and 1E). This observation suggests that extending our analysis to alternative mating conditions may help classifying mating genes into different functional categories. Thus, in addition to the three screens performed under the standard mating conditions, we conducted 19 screens under 18 non-standard conditions (Table S2), resulting in a total of 63146 MD score measurements for 2915 mutants (Table S3).
As predicted, hierarchical clustering of the data from the 22 screens revealed conspicuous patterns of phenotypic variations among the mating genes, with many falling into tight clusters (Figure 1F and Figure S2). Two of the most distinct clusters are enriched for mitochondrial protein-coding genes and autophagy genes, respectively. In this study, we focused on the genes in the autophagy cluster, but our extensive phenotyping data should be a useful resource for future investigation on other genes and cellular processes.
The autophagy cluster contains previously unknown fission yeast autophagy genes
There are 21 genes in the autophagy cluster (Figure 1F), including 10 genes known to be required for starvation-induced autophagy (atg2, atg4, atg5, atg9, atg12, atg13, atg15, atg18a, atg18b, atg18c) [3], [13]. These 10 genes represent all known fission yeast autophagy factors detectable by our analysis, as the other 4 characterized autophagy genes did not have MD scores, two due to lack of deletion strains (atg3 and atg8), one due to lack of decoded barcodes (atg1), and one due to low barcode read counts (atg7). Among the remaining 11 genes in the autophagy cluster, atg6, atg11, atg17, and atg101 have reported homology to autophagy genes in other organisms [21], but no experimental data on these 4 genes have been published. We hypothesized that these 4 genes, as well as the other 7 genes, which are unnamed and have no reported connections to autophagy, may also function in starvation-induced autophagy.
To test this hypothesis, we monitored nitrogen starvation-induced autophagy using the Atg8 fusion protein processing assay [13], [22], [23] (Figure 2A). We constructed a strain expressing from the endogenous promoter an Atg8 protein N-terminally tagged with cyan fluorescent protein (CFP), and then introduced the deletions of the autophagy cluster genes individually into this strain by PCR-based gene targeting. When wild-type (WT) cells expressing CFP-Atg8 were shifted from a growth medium (EMM) to a nitrogen-free medium (EMM-N), immunoblotting analysis showed the appearance of a free CFP band, due to the autophagic delivery of CFP-Atg8 into vacuoles and the subsequent proteolysis that releases the protease-resistant CFP. The processing of CFP-Atg8 was not observed in the 14 mutants known to be defective in autophagy or vacuolar proteolysis (atg1, atg2, atg3, atg4, atg5, atg7, atg9, atg12, atg13, atg15, atg18a, atg18b, atg18c, and isp6). In addition, we found that atg6, atg11, atg17, and atg101 are also required for CFP-Atg8 processing, thus providing for the first time evidence that they are required for autophagy. Among the 7 unnamed genes, five are also required for CFP-Atg8 processing. For reasons that will be explained below, we give these five genes the names of atg10, atg14, atg16, ctl1, and fsc1, respectively. The other two genes (SPCC757.04 and SPCC417.09c), when deleted, had no effect on CFP-Atg8 processing. We suspect that the deletion library strains for these two genes may harbor background mutations that interfere with autophagy. These two genes were not pursued further.
Fig. 2. CFP-Atg8 processing defect of autophagy mutants and the identification of Atg10, Atg16, and Atg14. 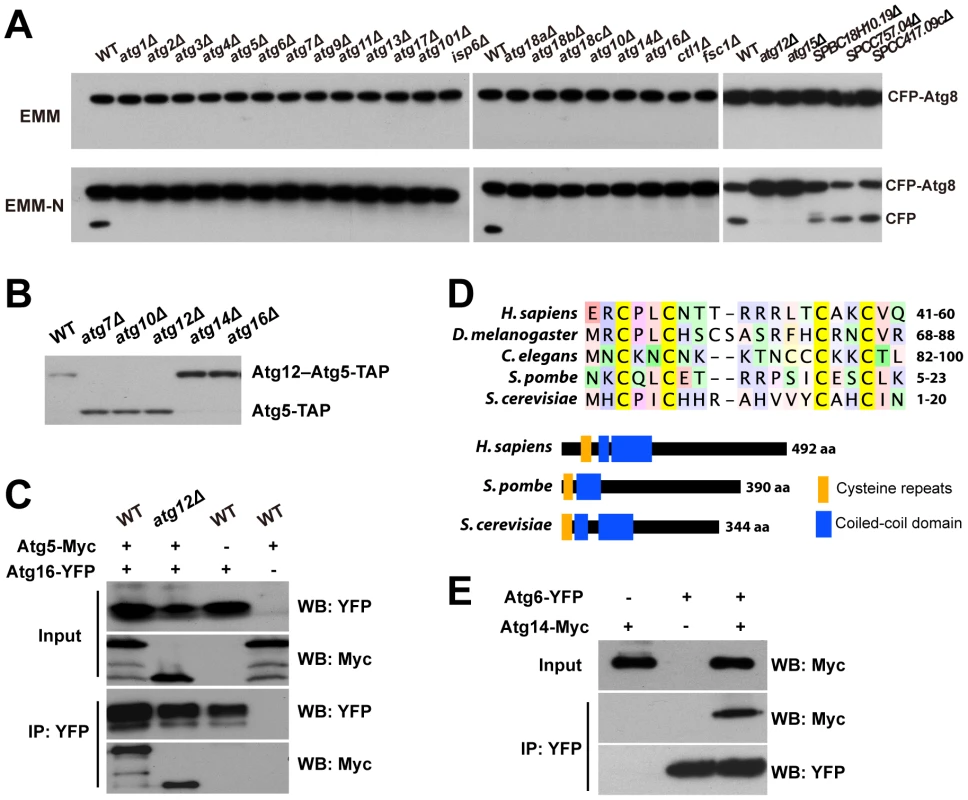
(A) CFP-Atg8 processing assay. Cells were collected before and 8 h after shifting to a nitrogen-free medium (EMM-N). (B) The conjugation of Atg12 to Atg5 requires the atg10 gene. (C) Atg5-Myc was co-immunoprecipitated with Atg16-YFP in both wild-type and atg12Δ cells. Input, 1%; IP, 20%. (D) The “cysteine repeats” region and the domain organization of Atg14 proteins. Coiled-coil domains are predicted as in Figure S4. (E) Atg14-Myc was co-immunoprecipitated with Atg6-YFP. Input, 1%; IP, 20%. Identification of S. pombe Atg10, Atg16, and Atg14
Three core components of the autophagy machinery, Atg10, Atg16, and Atg14, have not been identified in S. pombe [21], [24]. We found that three new autophagy genes uncovered in our screens share distant homology with genes encoding these three Atg proteins in other species.
SPAC227.04 contains a Pfam domain (PF07238) associated with Atg3 and Atg10 proteins. Our sequence homology analysis suggested that SPAC227.04 is more closely related to Atg10 proteins in metazoa and plants than to any Atg3 proteins (Figure S3). If SPAC227.04 is indeed Atg10 in S. pombe, removing it should abolish the conjugation of Atg12 to Atg5. In crude extracts made from wild-type cells expressing TAP-tagged Atg5 (Atg5-TAP), we detected by immunoblotting one major band of Atg5-TAP, presumably in the form of Atg12–Atg5 conjugate, as this band disappeared in atg12Δ extracts, as well as in atg7Δ extracts, which is defective in the E1 enzyme (Figure 2B). A faster-migrating band, likely representing the free form of Atg5, appeared in atg12Δ and atg7Δ extracts. When SPAC227.04/atg10 was deleted, only the free form of Atg5 was detected, thus confirming our prediction.
SPBC405.05 is currently annotated as a sequence orphan. We found that it shares homology with S. cerevisiae Atg16 in both the N-terminal Atg5-binding domain and the C-terminal coiled-coil domain (Figure S4). Similar to what has been reported for S. cerevisiae [25], we found in a co-immunoprecipitation (co-IP) experiment that, S. pombe SPBC405.05/Atg16 protein interacts with Atg5 both in the presence and in the absence of Atg12 (Figure 2C).
SPAC25A8.02, also annotated as a sequence orphan, was shown by our PSI-BLAST analysis to be related to metazoan Atg14 proteins. The most conserved sequence feature in Atg14 proteins is a pair of CXXC motifs termed “cysteine repeats” [26]. SPAC25A8.02 contains such a sequence feature, as well as a coiled-coil domain following the cysteine repeats, thus sharing the same domain arrangement with Atg14 proteins in other organisms (Figure 2D). Consistent with our homology analysis, we could co-immunoprecipitate SPAC25A8.02/Atg14 with Atg6, the expected binding partner of Atg14 in a PI3K complex (Figure 2E).
In PomBase, another gene, SPBC18H10.19, is currently annotated as atg14 because of its match to a Pfam domain (PF10186) associated with budding yeast Atg14. This domain is also found in metazoan UVRAG and Atg14 proteins, which are mutually exclusive subunits of Beclin 1-containing PI3K complexes [27]. In budding yeast, the likely counterpart of UVRAG, Vps38, resides in a PI3K complex distinct from the Atg14-containing complex and is dispensable for autophagy [28]. SPBC18H10.19 lacks the N-terminal cysteine repeats typical for the Atg14 proteins. Furthermore, the deletion library strain of SPBC18H10.19 showed no mating defect in our screens and an independent deletion made in the CFP-Atg8 strain did not block starvation-induced CFP-Atg8 processing (Figure 2A). Thus, we conclude that SPAC25A8.02 is the S. pombe Atg14, and SPBC18H10.19 may be the fission yeast equivalent of metazoan UVRAG and budding yeast Vps38.
Subcellular localization of S. pombe Atg proteins
Our analysis of the autophagy cluster genes increased the number of experimentally defined fission yeast autophagy factors from 14 to 23, and the identification of Atg10, Atg16, and Atg14 completed the roster of expected core autophagy components. We were, therefore, afforded an opportunity to comprehensively characterize the autophagy machinery in this organism for the first time. To survey the properties of fission yeast autophagy factors, we expressed them as fluorescent protein-tagged forms under the control of their endogenous promoters, and examined their subcellular localization by live cell imaging.
Atg8 is the only fission yeast Atg protein whose localization has been investigated [3], [13]. As reported by previous studies, we found that nitrogen starvation triggered the formation of bright CFP-Atg8 puncta in the cytoplasm. Co-expressing other autophagy proteins tagged with YFP in the CFP-Atg8 strain showed that 14 Atg proteins and Ctl1 colocalized with Atg8 on the punctate structure (Figure 3A). In S. cerevisiae, the same set of Atg proteins also colocalize at a punctuate structure, which has been termed the pre-autophagosomal structure or phagophore assembly site (PAS) [29]–[31]. Because of the similarity in the way Atg proteins assemble together, we propose that the structure where fission yeast Atg proteins colocalize during starvation is the counterpart of PAS in budding yeast, and will refer to it as PAS hereafter.
Fig. 3. Subcellular localization of fission yeast autophagy factors. 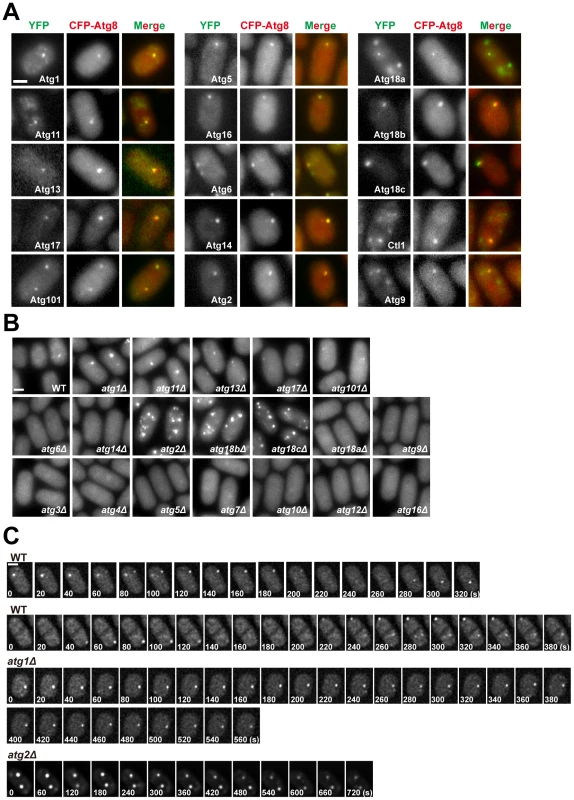
(A) Fifteen Atg proteins colocalized with CFP-Atg8 at cytoplasmic puncta induced by starvation. Images were acquired 2 h after starvation. (B) The distribution of CFP-Atg8 in atg mutants. Images were acquired 3 h after starvation. (C) Time-lapse analysis of CFP-Atg8 puncta in wild type, atg1Δ, and atg2Δ cells. Bars, 3 µm. The majority of the PAS-localizing fission yeast Atg proteins do not accumulate on distinct subcellular structures under non-starvation conditions. The exceptions are Atg1, Atg11, Atg6, Atg18a, Atg9, and Ctl1. In vegetatively growing cells, Atg1, Atg11 and Atg18a were observed on the vacuole membrane (Figure S5A). Atg18a also formed puncta co-localizing with an endosomal marker (Figure S5B). Atg6 was observed on punctate structures labeled by an endosomal marker as well, presumably reflecting its role in the vacuolar protein sorting pathway [28] (Figure S5C). The localization patterns of Atg9 and Ctl1 will be described below.
In S. cerevisiae, the localization of Atg8 at PAS is influenced by many other autophagy factors [29], [31]. To assess how fission yeast autophagy factors act, we analyzed the localization of CFP-Atg8 in atg mutants during starvation (Figure 3B). Mutants of the Atg8 conjugation system, atg3Δ, atg4Δ, atg5Δ, atg7Δ, atg10Δ, atg12Δ, and atg16Δ, completely abolished Atg8 puncta formation, so did the PI3K mutants atg6Δ and atg14Δ. In contrast, Atg8 puncta were readily detected in atg1Δ, atg11Δ, atg13Δ, atg17Δ, atg101Δ, atg2Δ, atg18bΔ, and atg18cΔ. These eight mutants can be classified into three groups based on the number, intensity, and emergence timing of the Atg8 puncta. Group 1 consists of atg1Δ and atg11Δ, in which Atg8 puncta appeared relatively normal in the first hour after starvation, but their numbers did not decline afterwards as happened in the wild type. Group 2 consists of atg13Δ, atg17Δ, and atg101Δ, which lacked obvious Atg8 puncta during the first hour after starvation, and the puncta emerged later appeared dimmer than those found in the wild type. Group 3 consists of atg2Δ, atg18bΔ, and atg18cΔ, in which the Atg8 puncta were much more numerous than in the wild type at all time points, and some of the puncta were notably brighter than those in the wild type.
Despite the superficial resemblance of the snapshot images of the Atg8 puncta in atg1Δ cells and wild type cells, time-lapse imaging analysis showed that unlike wild type cells, in which Atg8 puncta were dynamic structures with durations mostly in the range of 100 to 200 seconds, Atg8 puncta persisted much longer in atg1Δ cells (Figure 3C). This is similar to the observations in S. cerevisiae [32], [33]. In addition, we found that Atg8 puncta in atg2Δ cells were also long-lasting structures (Figure 3C).
Atg18a is required for the recruitment of the Atg12–Atg5·Atg16 complex to PAS
One particularly intriguing observation was the lack of Atg8 puncta in atg18aΔ cells (Figure 3B), suggesting that Atg18a plays a role different from that of the other two Atg18/WIPI paralogs, Atg18b and Atg18c. To assess how atg18aΔ may affect the PAS organization, we examined the localization of several representative Atg proteins in this mutant (Figure 4A). Atg1, Atg13, Atg14, and Atg2 still formed puncta in starved atg18aΔ cells. In contrast, neither Atg5 nor Atg16 formed detectable puncta. Thus, atg18aΔ blocked the recruitment of Atg5 and Atg16 to PAS, and probably as a consequence, indirectly abolished the PAS localization of Atg8.
Fig. 4. Atg18a is required for the PAS targeting of the Atg12–Atg5·Atg16 complex. 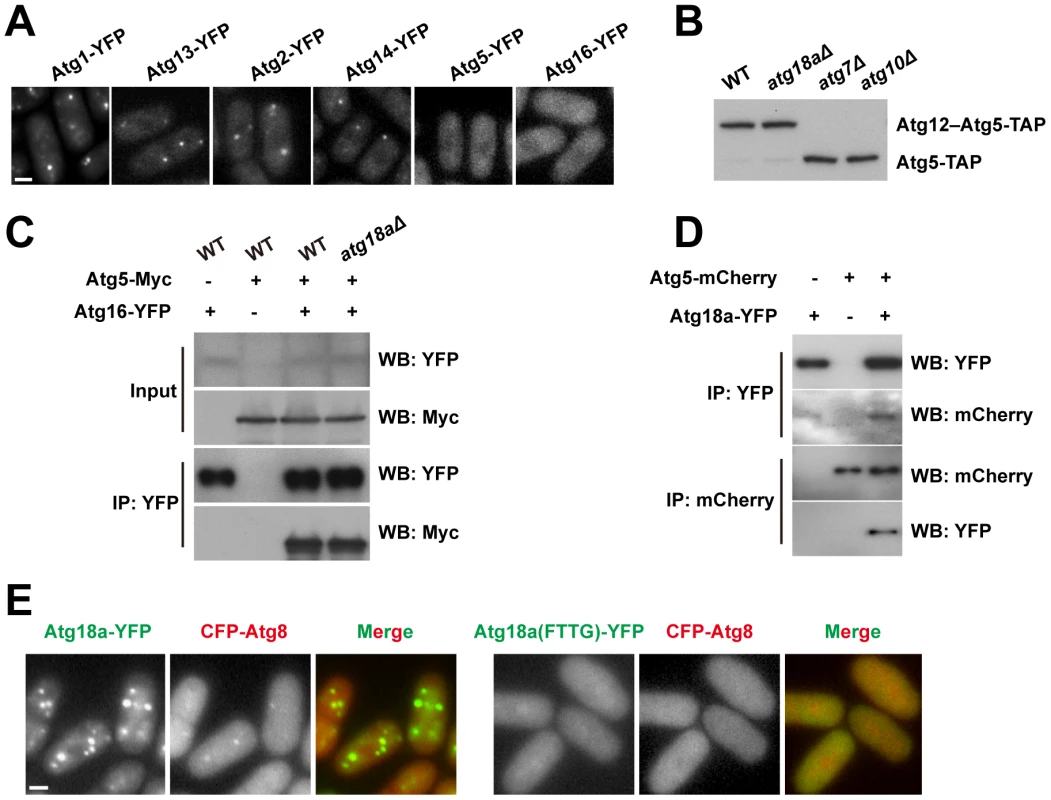
(A) atg18aΔ abolished the starvation-induced puncta formation by Atg5 and Atg16. (B) Atg12–Atg5 conjugation is normal in atg18aΔ cells. (C) The interaction between Atg5 and Atg16 is intact in atg18aΔ cells. Input, 1%; IP, 20%. (D) Atg5 and Atg18a co-immunoprecipitated with each other. (E) Mutating the FRRG motif in Atg18a abolished its own puncta and the Atg8 puncta in starved cells. Bars, 3 µm. We have shown that in S. pombe, Atg5 mainly exists in the form of Atg12–Atg5 conjugate, and physically interacts with Atg16 (Figure 2C). Neither Atg12–Atg5 conjugate formation (Figure 4B), nor the interaction between Atg5 and Atg16 (Figure 4C), was affected by atg18aΔ. Thus, the Atg12–Atg5·Atg16 complex remains intact in atg18aΔ cells, and the localization defect is probably due to a failure to recruit this complex as a whole to PAS.
We hypothesized that Atg18a may physically interact with the Atg12–Atg5·Atg16 complex. To test this idea, we performed co-IP experiments and found that, indeed, Atg5 was co-precipitated with Atg18a, and in a reciprocal IP, Atg18a was co-precipitated with Atg5 (Figure 4D). Thus, Atg18a may serve as a binding platform for the recruitment of the Atg12–Atg5·Atg16 complex to PAS.
The Atg18 family proteins bind PI3P in a manner dependent on a conserved FRRG motif [34], [35]. When the FRRG motif in Atg18a was mutated to FTTG, the protein became diffusely distributed and could no longer support the puncta formation by Atg8 (Figure 4E). Together, our data support a sequential recruitment model in which Atg18a is targeted to PAS by PI3P binding, and then in turn recruits the Atg12–Atg5·Atg16 complex.
Ctl1 is required for starvation-induced autophagy
We gave the previously uncharacterized gene SPCC1682.11c the name ctl1 because it encodes the sole member of the choline transporter-like (CTL) protein family (Pfam PF04515) in S. pombe. This protein family is ubiquitous in eukaryotes, with one member in S. cerevisiae (Pns1), one member in C. elegans (CHTL-1), two members in D. melanogaster, and five members in humans [36] (Figure S6). One vertebrate CTL protein, CTL1/SLC44A1, was shown to be a choline transporter on the plasma membrane and in mitochondria [37], [38]. However, S. cerevisiae Pns1 and C. elegans CHTL-1 do not act as choline transporters [39], [40]. Thus, choline transport does not appear to be a universal function of CTL proteins. Like other members of the CTL family, S. pombe Ctl1 is predicted to be a multi-transmembrane protein, with several methods agreeing on the same prediction that Ctl1 contains 10 transmembrane helices with both its N terminus and C terminus facing the cytoplasm (Figure 5A).
Fig. 5. Ctl1 is required for autophagic transport and normal PAS organization. 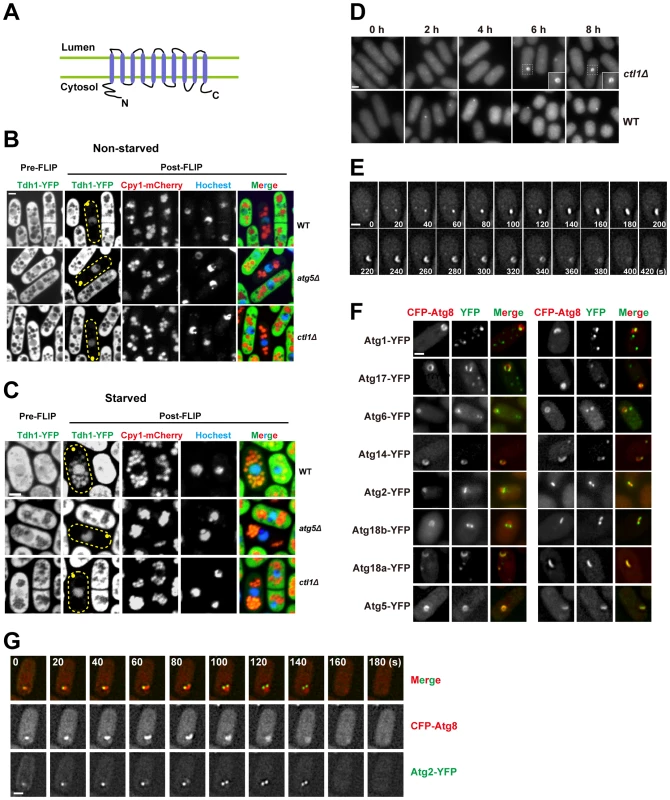
(A) The predicted membrane topology of Ctl1. (B,C) A fluorescence loss in photobleaching (FLIP) assay revealed the immobilized and non-diffusible pools of Tdh1-YFP. Yellow dots mark the sites of photobleaching. (B) In non-starved cells, only nuclear YFP signal remained in post-FLIP images. (C) In starved cells, vacuolar YFP signal was observed in post-FLIP images of wild-type, but not atg5Δ or ctl1Δ cells. (D) Ring-shaped and C-shaped structures labeled by CFP-Atg8 appeared in ctl1Δ cells after prolonged starvation. (E) Time-lapse images of a ctl1Δ cell. (F) The localization patterns of other Atg proteins in ctl1Δ cells containing CFP-Atg8-labeled structures. (G) Time-lapse images of a ctl1Δ cell expressing both CFP-Atg8 and YFP-Atg2. Bars, 3 µm. To corroborate the result of our CFP-Atg8 processing assay, we used a fluorescence loss in photobleaching (FLIP) assay to monitor the non-specific autophagy of an abundant cytosolic protein Tdh1, which is the major form of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in fission yeast [41]. In this assay, the fluorescence signal of the diffusible pool of Tdh1-YFP is depleted by repetitive photobleaching of a small region near one tip of the cell. If Tdh1-YFP is trapped or immobilized in certain cellular compartments and cannot freely diffuse to the site of photobleaching, such cellular compartments should stand out in post-FLIP images due to the remaining YFP signal. In non-starved cells, the only compartment with visible YFP signal in post-FLIP images is the nucleus (Figure 5B), perhaps due to the reported association of Tdh1 with RNA polymerase II [42]. Upon starvation, in post-FLIP images of wild-type cells (Figure 5C), YFP signal became detectable in vacuoles, which were labeled by mCherry-tagged Cpy1 (carboxypeptidase Y, or CPY), a vacuolar lumenal protein [43]. The starvation-induced vacuolar YFP signal is due to the autophagic delivery of Tdh1-YFP, because deletion of atg5 abolished such signal (Figure 5C). ctl1Δ also blocked the starvation-induced vacuolar targeting of Tdh1-YFP (Figure 5C). This result confirmed that Ctl1 is required for non-specific autophagy.
Ctl1 is required for the normal organization of PAS
To understand how Ctl1 contributes to autophagy, we examined the localization of Atg8 in ctl1Δ cells. In wild type cells, the level of starvation-induced CFP-Atg8 puncta peaked at around 1 h after starvation. However, in ctl1Δ cells, no Atg8 puncta were observed in the first few hours after starvation (Figure 5D). After prolonged starvation (>4 h), Atg8 became concentrated on cytoplasmic structures in ctl1Δ cells. Remarkably, some of these CFP-Atg8 labeled structures are not dot-like as seen in wild type, but rather C-shaped or ring-shaped (Figure 5D). Time-lapse analysis showed that these distinctly shaped structures represent different stages of a dynamic process, in which a CFP-Atg8 labeled structure first emerges as a dot, then elongates and bends into a C-shape, and subsequently grows to form a ring before its eventual disappearance, in a total time frame of several minutes (Figure 5E). The C shape may correspond to a cup-like structure in three-dimensional space, and the ring shape may correspond to a hollow sphere-like structure.
Interestingly, YFP-tagged Atg proteins localized to different regions of the Atg8-labeled structure in ctl1Δ cells: Atg5 perfectly co-localized with Atg8, whereas Atg1, Atg17, Atg6, Atg14, Atg2, and Atg18b localized to sub-regions of the Atg8-labeled structures, with Atg2 and Atg18b concentrating at the tips of the expanding structures (Figure 5F and G). Thus, even though this structure is a pathological outcome of the loss of Ctl1, it resembles the PAS in that many Atg proteins localize on it. We suspect that the distinct localization patterns of the Atg proteins on this structure may reflect their intrinsic properties. Consistent with this idea, we found that unlike Atg2 and Atg18b, Atg18a perfectly colocalized with Atg8 on this structure (Figure 5F), echoing its role in promoting the PAS localization of Atg5 and Atg8.
Ctl1 and Atg9 interact with each other and influence each other's localization
To determine whether Ctl1 is associated with other autophagy factors, we performed affinity purification coupled with mass spectrometry analysis and found that Atg9 co-purified with Ctl1 (unpublished data). Reciprocal co-IP experiments demonstrated that Ctl1 and Atg9 indeed interact with each other (Figure 6A).
Fig. 6. Ctl1 and Atg9 interact with each other and influence each other's localization. 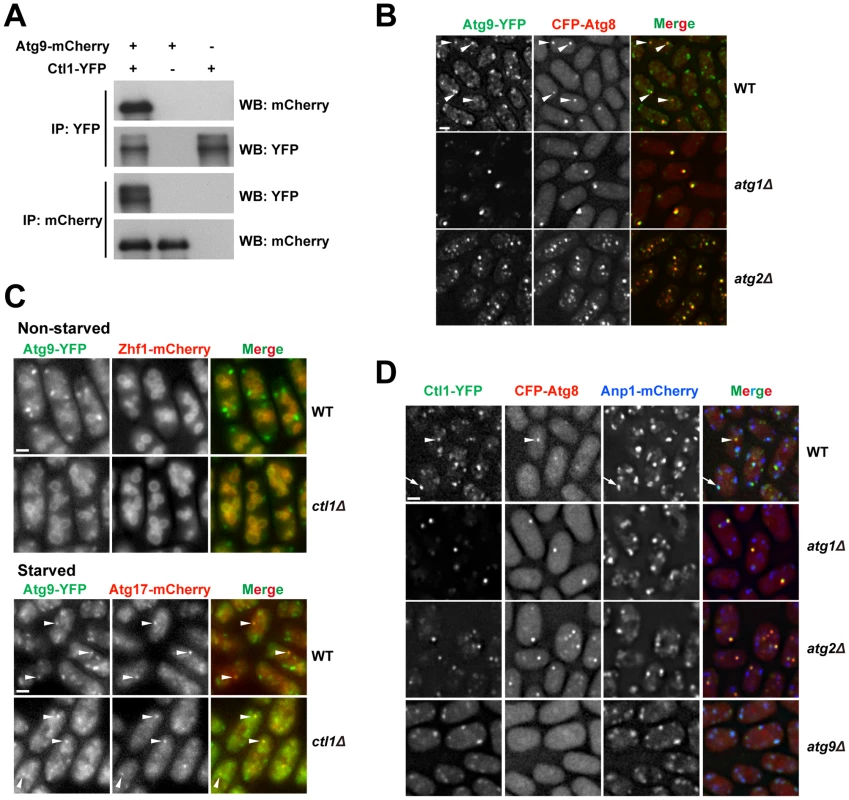
(A) Atg9 and Ctl1 co-immunoprecipitated with each other. (B) Localization patterns of Atg9 and Atg8 in starved cells. Arrowheads point to the puncta where Atg9 and Atg8 colocalized in the wild-type cells. (C) ctl1Δ altered the localization pattern of Atg9 in non-starved cells. Zhf1 is a vacuole membrane marker [77], and Atg17 is a PAS marker. Arrowheads point to puncta where Atg9 and Atg17 colocalized. (D) Localization patterns of Ctl1 in starved cells. Atg8 and Anp1 are PAS and Golgi markers, respectively. The arrowhead points to a punctum where Ctl1 and Atg8 colocalized. The arrow points to a punctum where Ctl1 and Anp1 colocalized. Bars, 3 µm. In S. cerevisiae, Atg9 cycles between PAS and non-PAS compartments, and the retrograde trafficking of Atg9 from PAS requires Atg1 and Atg2 [44]–[46]. Similarly, we found that in S. pombe, Atg9-YFP localized to punctate cytoplasmic structures, and partially co-localized with Atg8 puncta during starvation (Figure 6B). In starved atg1Δ or atg2Δ cells, Atg9 puncta almost completely overlapped with Atg8 puncta (Figure 6B), suggesting that recycling of Atg9 from PAS requires Atg1 and Atg2, like in S. cerevisiae.
The Ctl1-Atg9 interaction prompted us to examine whether Ctl1 influences the distribution of Atg9. In non-starved wild-type cells, besides the punctate structures, we also found weak Atg9-YFP signal on the vacuole membrane (Figure 6C). In non-starved ctl1Δ cells, Atg9 puncta no longer stood out, whereas the vacuole membrane signal was more noticeable, suggesting that Atg9 may be partially mislocalized (Figure 6C). In starved ctl1Δ cells, Atg9 puncta became more prominent, and some of them co-localized with mCherry-tagged Atg17 (Figure 6C), suggesting that Atg9 can still traffic to the PAS in the absence of Ctl1.
Similar to Atg9, Ctl1 tagged with YFP localized to cytoplasmic punctate structures, and upon starvation, a fraction of Ctl1-YFP puncta colocalized with CFP-Atg8 puncta (Figure 6D). In addition, Ctl1 became largely restricted to PAS in starved atg1Δ or atg2Δ cells (Figure 6D), suggesting that like Atg9, Ctl1 is recycled from PAS in an Atg1 and Atg2-dependent manner.
In S. cerevisiae, it has been shown that Atg9 traffics from Golgi apparatus to PAS via small vesicular carriers [46]. As Ctl1 and Atg9 may share similar cycling routes, we monitored the spatial relationship between Ctl1 and a Golgi marker, Anp1 [47]. In starved wild-type cells, Ctl1 puncta partially colocalized with Anp1, whereas in starved atg9Δ cells, nearly 100% of Ctl1 puncta colocalized with Anp1 (Figure 6D), indicating that Ctl1 travels from Golgi to PAS in an Atg9-dependent manner.
Fsc1 localizes to the vacuole membrane
We gave the previously uncharacterized gene SPAC22H12.05c the name fsc1 because it encodes a protein containing five fasciclin domains (Pfam PF02469) (Figure 7A and Figure S7). Fasciclin domain-containing proteins exist in animals, fungi, plants, bacteria, and cyanobacteria [48]. In animals and plants, this type of protein is usually found at the cell surface and mediate cell adhesion [49], [50]. However, one mammalian fasciclin domain protein, stabilin-1, was reported to have intracellular trafficking roles [51].
Fig. 7. Fsc1 localizes to the vacuole membrane and forms starvation-induced puncta. 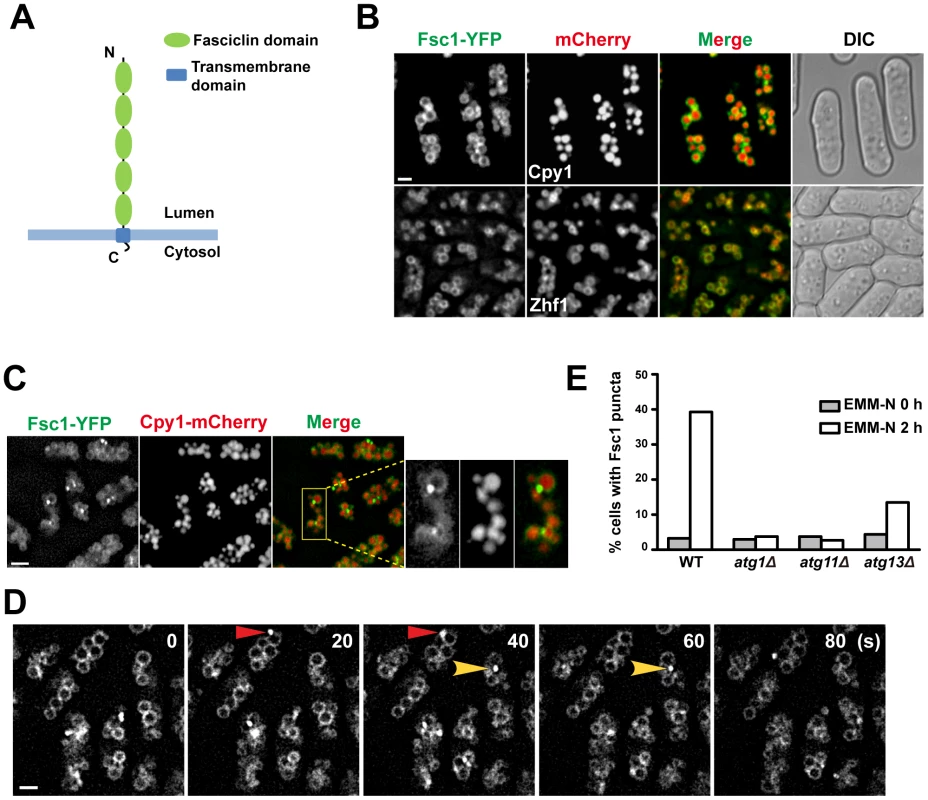
(A) The predicted membrane topology and domain organization of Fsc1. (B) Localization of Fsc1 in non-starved cells. Cpy1 and Zhf1 are vacuole lumen and vacuole membrane markers, respectively. Bar, 3 µm. (C) Localization of Fsc1 in starved cells. Bar, 6 µm. (D) Time-lapse images of Fsc1 puncta induced by starvation. Bar, 3 µm. (E) Fsc1 puncta induced by starvation are dependent on Atg1, Atg11, and partially dependent on Atg13. Two hundred cells were examined for each data point. Fsc1 is predicted to be a type I transmembrane protein, with the bulk of its amino acids exposed in the lumenal/extracellular space (Figure 7A). In vegetatively growing cells, Fsc1 tagged at its C-terminus with YFP localized to the vacuole membrane (Figure 7B). Interestingly, upon starvation, bright puncta of Fsc1-YFP were observed on the vacuolar rim of a small number of vacuoles (Figure 7C), indicating a dramatic concentration of Fsc1 at special sites on the vacuole membrane. The starvation-induced Fsc1 puncta were dynamic structures with durations of less than a minute (Figure 7D), and they were abolished in atg1Δ and atg11Δ cells, and diminished in atg13Δ cells (Figure 7E). No overlap between Fsc1 puncta and Atg8 puncta was observed (Figure S8), indicating that these two types of starvation-induced structures are spatially distinct entities.
Fsc1 is required for the fusion of autophagosomes with vacuoles
The fact that Fsc1 localizes to the destination compartment of autophagic trafficking led us to hypothesize that Fsc1 may be required for a late step of autophagy, the fusion between autophagosomes and vacuoles. To test this idea, we applied the Tdh1-YFP FLIP assay to fsc1Δ cells. Upon starvation, YFP signal in round cytoplasmic structures became visible in post-FLIP images of fsc1Δ cells (Figure 8A). However, unlike wild-type cells (Figure 5C), the YFP signal in fsc1Δ cells did not overlap with Cpy1-mCherry. Thus, Tdh1 entered a closed membrane compartment but did not reach the vacuole. Such YFP-labeled structures were absent in the post-FLIP images of fsc1Δ atg5Δ cells, suggesting that the Tdh1-containing membrane structures accumulated in fsc1Δ cells are autophagosomes.
Fig. 8. Fsc1 is required for autophagosome-vacuole fusion. 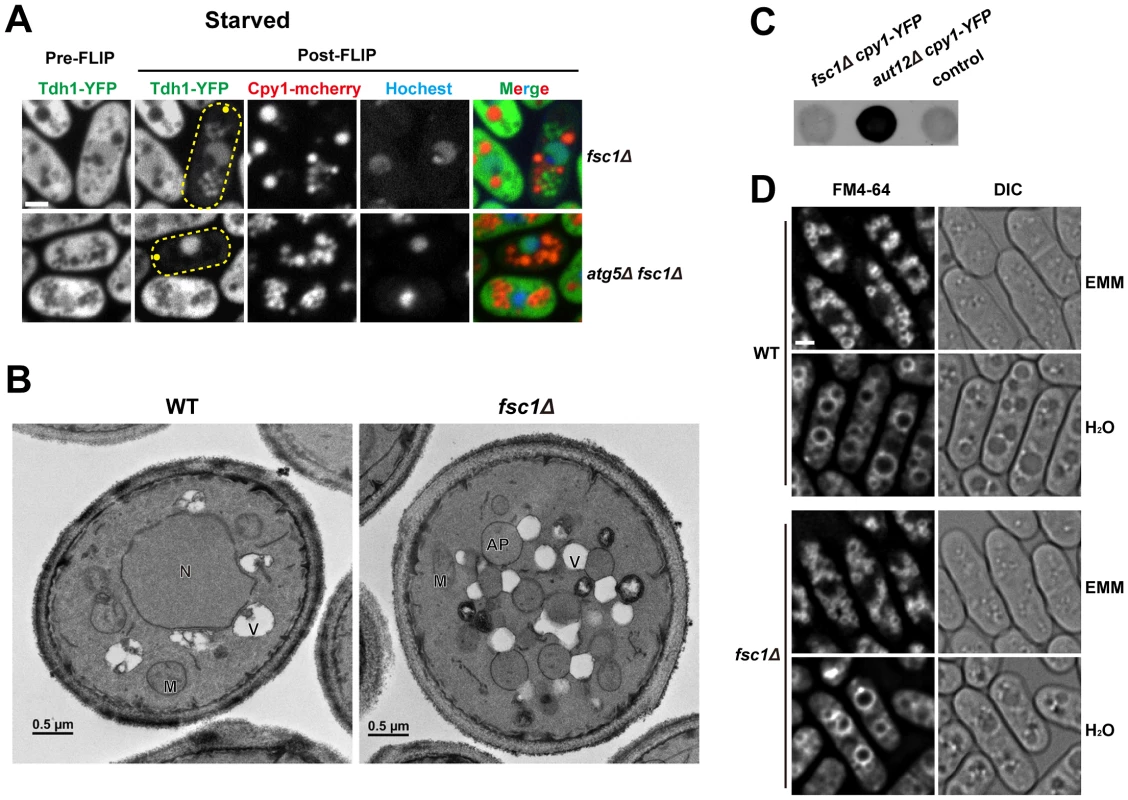
(A) In starved fsc1Δ cells, Tdh1-YFP entered closed cytoplasmic membrane structures, which are not vacuoles. These structures are dependent on Atg5, thus are likely autophagosomes. Bar, 3 µm. (B) TEM analysis of starved wild-type and fsc1Δ cells. N, nucleus; M, mitochondrion; V, vacuole; AP, autophagosome. (C) Unlike the mutant lacking a general vacuolar fusion factor Aut12, fsc1Δ cells did not secret Cpy1-YFP, which was detected by a colony blot assay with an antibody recognizing YFP. The control is a strain not expressing Cpy1-YFP. (D) fsc1Δ did not affect homotypic vacuole fusion occurring when cells were shifted from EMM medium to water. Vacuoles were stained with the vital dye FM4-64. Bar, 3 µm. To verify the results obtained by the FLIP assay, we performed transmission electron microscopy (TEM) analysis. In starved fsc1Δ cells, we observed the accumulation of spherical membrane structures whose lumenal contents had the same electron opacity as the cytosol, suggesting that they are autophagosomes (Figure 8B). Moreover, TEM images showed that autophagosomes in these cells were often in extensive contact with vacuoles, suggesting that docking of autophagosomes onto vacuoles had occurred but membrane fusion was blocked.
To our knowledge, all known autophagosome-vacuole fusion factors in S. cerevisiae, such as Mon1/Aut12 and Vam3, are required not only for autophagosome-vacuole fusion, but also for other vacuolar fusion events, such as those occurring in the CPY trafficking [52], [53], and homotypic vacuole fusion [54]. In contrast, Fsc1 appears to be dispensable for these processes, as unlike aut12Δ cells, Cpy1 was not missorted to cell surface in fsc1Δ cells (Figure 8C), and homotypic vacuole fusion induced by hypo-osmotic stress occurred normally in fsc1Δ cells (Figure 8D) [55]. Thus, Fsc1 is specifically required for autophagosome-vacuole fusion.
Discussion
In this study, we quantitatively assessed the mating efficiencies of the fission yeast deletion library strains under 19 different mating conditions, and generated an extensive phenotyping dataset that allows hundreds of genes to be clustered in a way that reflects their functional relationships. The autophagy gene cluster represents a comprehensive inventory of fission yeast autophagy factors. Through systematic analyses of these autophagy factors, we uncovered novel autophagy mechanisms, gained insights into how autophagy pathway has evolved, and established the fission yeast as a model for deciphering the inner workings of the autophagy machinery.
Organization of PAS in fission yeast versus budding yeast
In S. cerevisiae, PAS is the site where the Atg proteins involved in autophagosome formation assemble together, as observed by fluorescence microscopy. Here, we identified a similar entity in S. pombe by live cell imaging of fluorescent protein-tagged Atg proteins, and named it PAS due to its resemblance to the PAS in S. cerevisiae. The similarities include: (1) In both species, PAS is a dot-like structure whose finer details cannot be resolved by conventional light microscopy; (2) At any given time during starvation, in the majority of PAS-containing cells, only one PAS punctum can be observed; (3) The same set of Atg proteins can be colocalized at PAS, with the exception of Atg29 and Atg31, which are absent in S. pombe, and Atg101, which is absent in S. cerevisiae; (4) PAS is a dynamic structure with a duration in the range of minutes, as revealed by the time-lapse analysis of Atg8 puncta; (5) The assembly of Atg proteins at PAS is controlled in a hierarchical manner, with Atg8 being one of the most downstream factors, whose recruitment to PAS or dynamics at PAS is altered in the mutants defective in any other PAS-localizing Atg proteins.
There are also notable differences between these two organisms in terms of PAS organization and the roles of PAS-localizing Atg proteins: (1) In S. cerevisiae, a constitutive biosynthetic route termed cytoplasm-to-vacuole targeting (Cvt) pathway utilizes the Atg proteins to transport cytosolic hydrolases into the vacuole [56], and thus the assembly of Atg proteins at PAS occurs under nutrient-rich conditions; in contrast, PAS cannot be detected in S. pombe under nutrient-rich conditions, presumably due to the lack of the Cvt pathway, whose key factors Ape1 and Atg19 do not have apparent homologs in S. pombe; (2) Atg11 in S. cerevisiae is dispensable for starvation-induced autophagy, whereas Atg11 in S. pombe is essential for starvation-induced autophagy and appears to have a closer relationship with Atg1 than the other putative Atg1 regulators, consistent with a proposition that S. pombe Atg11 may be more similar to mammalian FIP200 than to budding yeast Atg11 [12]; (3) There are three Atg18/WIPI proteins in each species, but only one of the three paralogs in S. cerevisiae (Atg18/Svp1) is essential for starvation-induced autophagy, whereas all three paralogs in S. pombe are needed for starvation-induced autophagy.
Functions of the Atg18/WIPI proteins
We found that the mutants of fission yeast Atg18 paralogs exhibited different phenotypes, with atg18aΔ abolishing the Atg8 puncta, and atg18bΔ or atg18cΔ elevating the levels of Atg8 puncta. This is analogous to the situation in mammalian cells, where LC3 (a mammalian homolog of Atg8) puncta increased upon the depletion of either WIPI1 or WIPI4 but decreased upon the depletion of WIPI2 [57]–[59]. Such functional distinctions cannot be readily explained by the phylogenetic relationships among Atg18/WIPI proteins (Figure S9 and S10). In mammals, WIPI1 and WIPI2 are much more similar to each other than to WIPI4, and in S. pombe, Atg18b and Atg18c do not show significantly higher sequence homology to each other than to Atg18a.
Our analysis suggests that the lack of Atg8 puncta in atg18aΔ cells is due to a defect in the PAS targeting of the Atg12–Atg5·Atg16 complex, which physically interacts with Atg18a. To our knowledge, this is the first time a physical interaction between a WIPI/Atg18 protein and the Atg12–Atg5·Atg16 complex has been observed. Similar interactions may underlie the roles of S. cerevisiae Atg18 and its paralog Atg21 in promoting the PAS localization of Atg5 and Atg16 [35], [60], and the role of mammalian WIPI2 in the recruitment of LC3 to the omegasome, which may be the equivalent of PAS in mammalian cells [57].
As Atg18a accumulates on subcellular structures other than PAS, it probably cooperates with additional factors for the specific targeting of Atg12–Atg5·Atg16 to PAS. Atg2 is unlikely to be such a factor, as its mutant behaved like atg18bΔ and atg18cΔ.
The role of Ctl1 in autophagy
Ctl1 is a novel autophagy factor uncovered by our screens. Our phylogenetic analysis showed that fungal CTL proteins fall into two clades, with S. pombe Ctl1 in one clade, and S. cerevisiae Pns1 in the other (Figure S6). Pns1, whose function is unknown, has been localized at the plasma membrane [61], and it does not appear to be required for starvation-induced autophagy (unpublished data). Thus, among the fungal CTL proteins, perhaps only the ones falling into the same clade as Ctl1 are autophagy factors. As fungal species in many lineages have both Ctl1-like and Pns1-like proteins (Figure S6), these two types of proteins might have co-existed in the common ancestor of fungi, but one of them was lost in the lineage leading to S. cerevisiae, while the other was lost in the lineage leading to S. pombe.
In ctl1Δ cells, autophagosome formation appears to be defective, as we did not observe any cytoplasmic signal of Tdh1-YFP in post-FLIP images. The late emerging Atg8-labeled structures in ctl1Δ cells may be aberrant isolation membranes that cannot mature into completely sealed autophagosomes. Ctl1 may regulate the distribution of Atg proteins on the expanding isolation membrane, and in its absence, Atg proteins occupy different regions of the isolation membrane, instead of concentrating at one subregion.
It is interesting to note that the distinct localization patterns of Atg proteins we observed in ctl1Δ cells bear remarkable resemblance to the three types of Atg protein distribution patterns observed in S. cerevisiae when Ape1 is overexpressed [62]. In both ctl1Δ S. pombe cells and Ape1-overexpressing S. cerevisiae cells, Atg8 and Atg5 are distributed all over a cup-shaped structure, whereas Atg2 and an Atg18 family protein (Atg18 in S. cerevisiae and Atg18b in S. pombe) concentrate at the edge of this structure. In Ape1-overexpressing S. cerevisiae cells, Atg17, Atg6, and Atg14 localize to a subregion of the cup-shaped structure, termed vacuole-isolation membrane contact site (VICS); in ctl1Δ cells, these three proteins also localize to subregions of the cup-shaped structure. Thus, the spatial separation of Atg proteins under these two circumstances probably reflects evolutionarily conserved functional distinctions among the Atg proteins.
Fsc1 and autophagosome-vacuole fusion
In S. cerevisiae, all known mutants blocking autophagosome-vacuole fusion are also defective for vacuolar fusion in the CPY and ALP pathways, as well as vacuole–vacuole homotypic fusion [63]. Thus, it is unclear whether there are mechanisms specifically regulating autophagosome-vacuole fusion in budding yeast. Here, we showed that, in S. pombe, a vacuole membrane protein Fsc1 is uniquely required for autophagosome-vacuole fusion, thus revealing a specific control of autophagic traffic at the vacuolar fusion step, and providing a molecular entry point for dissecting the mechanism of such a control. Fsc1 formed puncta on the vacuole membrane during starvation. We speculate that these structures may be in some way connected to autophagosome-vacuole fusion. For example, they may correspond to fusion-ready zones on the vacuole membrane, or the actual fusion sites, or special post-fusion structures.
Many fungal species have at least one protein sharing the exact same domain organization as Fsc1. The S. cerevisiae homolog of Fsc1 is Ylr001c (Figure S7), which like Fsc1, also localizes to the vacuole membrane [61]. However, Ylr001c seems to be dispensable for starvation-induced autophagy (unpublished data), perhaps due to functional redundancy in S. cerevisiae, or differences in vacuole physiology between the two organisms. One obvious difference is that an S. cerevisiae cell has one or a few large vacuoles, whereas an S. pombe cell has about 80 small vacuoles [55]. Thus, there may be a need for more elaborate fusion target selection in S. pombe to avoid overwhelming the degradative capacities of some vacuoles while leaving other vacuoles idle. Mammalian cells, where a large number of lysosomes are present in each cell, may share this need. Several lines of recent evidence suggest that autophagosome-lysosome fusion in mammalian cells utilizes mechanisms distinct from other lysosomal fusion events [64]–[66]. We expect that further analysis of Fsc1 may provide mechanistic insights relevant to autophagosome-lysosome fusion in mammalian cells.
Materials and Methods
Fission yeast strains and media
The fission yeast strains used in this study are listed in Table S4. Genetic methods for strain construction and composition of media are as described [20]. To construct a strain expressing CFP-Atg8 under the control of the endogenous promoter, we amplified by overlap-extension PCR the atg8 promoter and the N-terminal region of the atg8 ORF using primers 5′-GATCTAGAGAAGCGCTTATTTGTTTAC-3′, 5′-CGagatctTTGAGAACGCATGAGAACTCTCAAACTTCTTGC-3′, 5′-CTCATGCGTTCTCAAagatctCGTTCTCAATTCAAGG-3′, and 5′-GCGTCGACACCAACTGTAAGGTCAGATGG-3′. The final PCR product contained a BglII site (lowercase letters in the primer sequences) inserted near the start codon. The PCR product was digested with XbaI and SalI, and inserted into an integrating vector pJK148 [67]. DNA encoding the CFP tag was inserted into the BglII site to obtain the pJK148-CFP-Atg8 plasmid. The plasmid was linearized with SpeI, which cuts in the middle of the N-terminal region of the atg8 ORF, and transformed into fission yeast. Most of the deletion strains used in this study were constructed by PCR amplifying the deletion cassettes in the Bioneer deletion strains and transforming the PCR product into strains from our lab strain collection. The exceptions are atg1, atg3, atg6, and atg12, whose deletion strains were made without the aid of Bioneer strains, by standard PCR-based gene targeting [68]. Strains expressing Atg proteins fused with the YFP-FLAG-His6 (YFH) tag under native promoters were constructed by an overlap-extension PCR approach [69], using the ORFeome plasmids as templates [70]. Strains expressing proteins fused with other tags were made by PCR-based tagging [68]. Tdh1-YFH was expressed from an ORFeome plasmid under the control of the nmt1 promoter.
Mating phenotype screens
Deletion strain pools of Bioneer version 1.0 haploid library (catalog number M-1030H) and Bioneer version 1.0 upgrade package (catalog number M-1030H-U) were constructed as described [19]. Frozen aliquots of the two mutant pools were thawed at room temperature, mixed together, washed once with YES medium, and pre-grown in YES or EMM medium for 3 generations at 30°C. An equal amount of log-phase wild-type h− strain (DY3984) grown in YES medium was mixed with the deletion mutants, and washed twice with water. The cell suspension was diluted to 100 OD600 units/ml in water, spotted on the solid mating medium, and incubated for 4 days. The mating mixtures were treated with 0.5% (v/v) glusulase overnight at room temperature, and the spores were purified with a Percoll step gradient [71]. The spore preparations were more than 99% pure as judged by microscopy. Genomic DNA extraction, barcode PCR, and Illumina sequencing were performed as described [19]. The sequencing data are publicly available at NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under the accession number SRA068523. The data are split into 26 runs, which correspond to 4 input samples and 22 spore samples. Descriptions of the 26 runs are in Table S5.
Barcode sequencing data analysis
Mating phenotype screen data were processed as described [19], with a few modifications. For read count normalization, we used the upper-quartile normalization method [72]. To avoid noises associated with very small read counts, for a MD score to be computed for a gene, we required the read count of at least one of its barcodes to be no smaller than 1/40 of the upper-quartile read count, and also no smaller than 12, in either the input sample or the spore sample. For a gene with a single barcode decoded, its MD score is the normalized log2 fold change (input versus spore) of that barcode. For a gene with both uptag and dntag decoded, its MD score is a weighted average of the normalized log2 fold change of the two barcodes, where the weight for a barcode is the ratio of the sum of the read counts of that barcode in input and spore samples to the sum of the read counts of both barcodes. To select mating defective mutants, we calculated for each gene a robust Z-score, which is the deviation of its MD score from the median MD score expressed in the number of the normalized interquartile range (NIQR). Tail area-based FDR values were calculated from the robust Z-scores using the software fdrtool version 1.2.8 [73]. Genes with FDR values <0.1 in all three screens performed under standard conditions were deemed the screen hits. GO term enrichment analysis was conducted with AmiGO version 1.8 using GO database release 2013-02-02 [74]. Hierarchical clustering analysis was performed using the correlation (uncentered) similarity metric and the complete linkage clustering method.
CFP-Atg8 processing assay
Cell lysates were prepared using a post-alkaline extraction method [75]. Samples were separated by 12% SDS-PAGE and immunoblotted with an anti-GFP antibody (Roche).
Atg12–Atg5 conjugate analysis and co-immunoprecipitation
The Peroxidase Anti-Peroxidase (PAP) soluble complex (Sigma) was used in immunoblotting to recognize the TAP tag fused to Atg5. For immunoprecipitation, cell lysates were made by glass bead beating. GFP-trap and RFP-trap agarose beads (ChromoTek) were used for immunoprecipitating YFP - and mCherry-tagged proteins, respectively.
Light microscopy
Except for the FLIP assay, light microscopy was performed using a DeltaVision PersonalDV system (Applied Precision) equipped with a CFP/YFP/mCherry filter set (Chroma 89006 set) and a Photometrics CoolSNAP HQ2 camera. Images were acquired with a 100×, 1.4-NA objective, and analyzed with the SoftWoRx software.
FLIP assay
Photobleaching of the Tdh1-YFP signal and image acquisition were carried out with a PerkinElmer Ultraview VoX spinning disk system, using a 100× objective. Image analysis was performed with the Volocity software.
Electron microscopy
Cells were prepared for electron microscopy by fixation with glutaraldehyde and KMnO4 [76]. Samples were dehydrated with graded ethanol series, and embedded in Spurr's resin. Thin sections were stained with uranyl acetate and Sato's lead, and visualized on a transmission electron microscope.
Supporting Information
Zdroje
1. TakeshigeK, BabaM, TsuboiS, NodaT, OhsumiY (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 119 : 301–311.
2. TsukadaM, OhsumiY (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333 : 169–174.
3. KohdaTA, TanakaK, KonomiM, SatoM, OsumiM, et al. (2007) Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells 12 : 155–170 doi:10.1111/j.1365-2443.2007.01041.x
4. LevineB, KroemerG (2008) Autophagy in the pathogenesis of disease. Cell 132 : 27–42 doi:10.1016/j.cell.2007.12.018
5. MizushimaN, KomatsuM (2011) Autophagy: renovation of cells and tissues. Cell 147 : 728–741 doi:10.1016/j.cell.2011.10.026
6. XieZ, KlionskyDJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9 : 1102–1109 doi:10.1038/ncb1007-1102
7. NakatogawaH, SuzukiK, KamadaY, OhsumiY (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10 : 458–467 doi:10.1038/nrm2708
8. MizushimaN, YoshimoriT, OhsumiY (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27 : 107–132 doi:10.1146/annurev-cellbio-092910-154005
9. KlionskyDJ (2007) The importance of diversity. Autophagy 3 : 83–84.
10. KingJS (2012) Autophagy across the eukaryotes: Is S. cerevisiae the odd one out? Autophagy 8 : 1159–1162.
11. BerbeeML, TaylorJW (2010) Dating the molecular clock in fungi – how close are we? Fungal Biology Reviews 24 : 1–16 doi:10.1016/j.fbr.2010.03.001
12. MizushimaN (2010) The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 22 : 132–139 doi:10.1016/j.ceb.2009.12.004
13. MukaiyamaH, KajiwaraS, HosomiA, Giga-HamaY, TanakaN, et al. (2009) Autophagy-deficient Schizosaccharomyces pombe mutants undergo partial sporulation during nitrogen starvation. Microbiology 155 : 3816–3826 doi:10.1099/mic.0.034389-0
14. Yamamoto M, Imai Y, Watanabe Y (1997) Mating and Sporulation in Schizosaccharomyces pombe. The molecular and cellular biology of the yeast Saccharomyces: Cell cycle and cell biology. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. pp. 1037–1106.
15. Nielsen O (2004) Mating-Type Control and Differentiation. The molecular biology of Schizosaccharomyces pombe: genetics, genomics and beyond. Berlin: Springer Verlag. pp. 281–296.
16. LeupoldU, SipiczkiM (1991) Sterile UGA nonsense mutants of fission yeast. Curr Genet 20 : 67–73.
17. WoodV, HarrisMA, McDowallMD, RutherfordK, VaughanBW, et al. (2012) PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 40: D695–699 doi:10.1093/nar/gkr853
18. KimD-U, HaylesJ, KimD, WoodV, ParkH-O, et al. (2010) Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotech 28 : 617–623 doi:10.1038/nbt.1628
19. HanTX, XuX-Y, ZhangM-J, PengX, DuL-L (2010) Global fitness profiling of fission yeast deletion strains by barcode sequencing. Genome Biol 11: R60 doi:10.1186/gb-2010-11-6-r60
20. ForsburgSL, RhindN (2006) Basic methods for fission yeast. Yeast 23 : 173–183 doi:10.1002/yea.1347
21. MukaiyamaH, NakaseM, NakamuraT, KakinumaY, TakegawaK (2010) Autophagy in the fission yeast Schizosaccharomyces pombe. FEBS Lett 584 : 1327–1334 doi:10.1016/j.febslet.2009.12.037
22. ShintaniT, KlionskyDJ (2004) Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem 279 : 29889–29894 doi:10.1074/jbc.M404399200
23. CheongH, KlionskyDJ (2008) Biochemical methods to monitor autophagy-related processes in yeast. Meth Enzymol 451 : 1–26 doi:10.1016/S0076-6879(08)03201-1
24. MeijerWH, van der KleiIJ, VeenhuisM, KielJAKW (2007) ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3 : 106–116.
25. MizushimaN, NodaT, OhsumiY (1999) Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J 18 : 3888–3896 doi:10.1093/emboj/18.14.3888
26. MatsunagaK, MoritaE, SaitohT, AkiraS, KtistakisNT, et al. (2010) Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol 190 : 511–521 doi:10.1083/jcb.200911141
27. ItakuraE, KishiC, InoueK, MizushimaN (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19 : 5360–5372 doi:10.1091/mbc.E08-01-0080
28. KiharaA, NodaT, IshiharaN, OhsumiY (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 152 : 519–530.
29. SuzukiK, KirisakoT, KamadaY, MizushimaN, NodaT, et al. (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20 : 5971–5981 doi:10.1093/emboj/20.21.5971
30. KimJ, HuangW-P, StromhaugPE, KlionskyDJ (2002) Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem 277 : 763–773 doi:10.1074/jbc.M109134200
31. SuzukiK, KubotaY, SekitoT, OhsumiY (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12 : 209–218 doi:10.1111/j.1365-2443.2007.01050.x
32. CheongH, NairU, GengJ, KlionskyDJ (2008) The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell 19 : 668–681 doi:10.1091/mbc.E07-08-0826
33. XieZ, NairU, KlionskyDJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19 : 3290–3298 doi:10.1091/mbc.E07-12-1292
34. ObaraK, SekitoT, NiimiK, OhsumiY (2008) The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem 283 : 23972–23980 doi:10.1074/jbc.M803180200
35. NairU, CaoY, XieZ, KlionskyDJ (2010) Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J Biol Chem 285 : 11476–11488 doi:10.1074/jbc.M109.080374
36. TraiffortE, RuatM, O'ReganS, MeunierFM (2005) Molecular characterization of the family of choline transporter-like proteins and their splice variants. J Neurochem 92 : 1116–1125 doi:10.1111/j.1471-4159.2004.02962.x
37. O'ReganS, TraiffortE, RuatM, ChaN, CompaoreD, et al. (2000) An electric lobe suppressor for a yeast choline transport mutation belongs to a new family of transporter-like proteins. Proc Natl Acad Sci USA 97 : 1835–1840 doi:10.1073/pnas.030339697
38. MichelV, BakovicM (2009) The solute carrier 44A1 is a mitochondrial protein and mediates choline transport. FASEB J 23 : 2749–2758 doi:10.1096/fj.08-121491
39. ZuffereyR, SantiagoTC, BrachetV, Ben MamounC (2004) Reexamining the role of choline transporter-like (Ctlp) proteins in choline transport. Neurochem Res 29 : 461–467.
40. MullenGP, MathewsEA, VuMH, HunterJW, FrisbyDL, et al. (2007) Choline transport and de novo choline synthesis support acetylcholine biosynthesis in Caenorhabditis elegans cholinergic neurons. Genetics 177 : 195–204 doi:10.1534/genetics.107.074120
41. MorigasakiS, ShimadaK, IknerA, YanagidaM, ShiozakiK (2008) Glycolytic enzyme GAPDH promotes peroxide stress signaling through multistep phosphorelay to a MAPK cascade. Mol Cell 30 : 108–113 doi:10.1016/j.molcel.2008.01.017
42. MitsuzawaH, KimuraM, KandaE, IshihamaA (2005) Glyceraldehyde-3-phosphate dehydrogenase and actin associate with RNA polymerase II and interact with its Rpb7 subunit. FEBS Lett 579 : 48–52 doi:10.1016/j.febslet.2004.11.045
43. TabuchiM, IwaiharaO, OhtaniY, OhuchiN, SakuraiJ, et al. (1997) Vacuolar protein sorting in fission yeast: cloning, biosynthesis, transport, and processing of carboxypeptidase Y from Schizosaccharomyces pombe. J Bacteriol 179 : 4179–4189.
44. NodaT, KimJ, HuangWP, BabaM, TokunagaC, et al. (2000) Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 148 : 465–480.
45. ReggioriF, TuckerKA, StromhaugPE, KlionskyDJ (2004) The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 6 : 79–90.
46. YamamotoH, KakutaS, WatanabeTM, KitamuraA, SekitoT, et al. (2012) Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 198 : 219–233 doi:10.1083/jcb.201202061
47. VjesticaA, TangX-Z, OliferenkoS (2008) The actomyosin ring recruits early secretory compartments to the division site in fission yeast. Mol Biol Cell 19 : 1125–1138 doi:10.1091/mbc.E07-07-0663
48. KawamotoT, NoshiroM, ShenM, NakamasuK, HashimotoK, et al. (1998) Structural and phylogenetic analyses of RGD-CAP/beta ig-h3, a fasciclin-like adhesion protein expressed in chick chondrocytes. Biochim Biophys Acta 1395 : 288–292.
49. ElkinsT, HortschM, BieberAJ, SnowPM, GoodmanCS (1990) Drosophila fasciclin I is a novel homophilic adhesion molecule that along with fasciclin III can mediate cell sorting. J Cell Biol 110 : 1825–1832.
50. HuberO, SumperM (1994) Algal-CAMs: isoforms of a cell adhesion molecule in embryos of the alga Volvox with homology to Drosophila fasciclin I. EMBO J 13 : 4212–4222.
51. ZhangJ, GratchevA, RiabovV, MamidiS, SchmuttermaierC, et al. (2009) A novel GGA-binding site is required for intracellular sorting mediated by stabilin-1. Mol Cell Biol 29 : 6097–6105 doi:10.1128/MCB.00505-09
52. DarsowT, RiederSE, EmrSD (1997) A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol 138 : 517–529.
53. WangC-W, StromhaugPE, ShimaJ, KlionskyDJ (2002) The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J Biol Chem 277 : 47917–47927 doi:10.1074/jbc.M208191200
54. WangC-W, StromhaugPE, KauffmanEJ, WeismanLS, KlionskyDJ (2003) Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J Cell Biol 163 : 973–985 doi:10.1083/jcb.200308071
55. BoneN, MillarJB, TodaT, ArmstrongJ (1998) Regulated vacuole fusion and fission in Schizosaccharomyces pombe: an osmotic response dependent on MAP kinases. Curr Biol 8 : 135–144.
56. UmekawaM, KlionskyDJ (2012) The Cytoplasm-to-Vacuole Targeting Pathway: A Historical Perspective. Int J Cell Biol 2012 : 142634 doi:10.1155/2012/142634
57. PolsonHEJ, de LartigueJ, RigdenDJ, ReedijkM, UrbéS, et al. (2010) Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6 : 506–522.
58. BehrendsC, SowaME, GygiSP, HarperJW (2010) Network organization of the human autophagy system. Nature 466 : 68–76 doi:10.1038/nature09204
59. LuQ, YangP, HuangX, HuW, GuoB, et al. (2011) The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell 21 : 343–357 doi:10.1016/j.devcel.2011.06.024
60. StrømhaugPE, ReggioriF, GuanJ, WangC-W, KlionskyDJ (2004) Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell 15 : 3553–3566 doi:10.1091/mbc.E04-02-0147
61. HuhW-K, FalvoJV, GerkeLC, CarrollAS, HowsonRW, et al. (2003) Global analysis of protein localization in budding yeast. Nature 425 : 686–691 doi:10.1038/nature02026
62. SuzukiK, AkiokaM, Kondo-KakutaC, YamamotoH, OhsumiY (2013) Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci 126 : 2534–2544 doi:10.1242/jcs.122960
63. KlionskyDJ (2005) The molecular machinery of autophagy: unanswered questions. J Cell Sci 118 : 7–18 doi:10.1242/jcs.01620
64. GanleyIG, WongP-M, GammohN, JiangX (2011) Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 42 : 731–743 doi:10.1016/j.molcel.2011.04.024
65. ChenD, FanW, LuY, DingX, ChenS, et al. (2012) A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell 45 : 629–641 doi:10.1016/j.molcel.2011.12.036
66. ItakuraE, Kishi-ItakuraC, MizushimaN (2012) The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151 : 1256–1269 doi:10.1016/j.cell.2012.11.001
67. KeeneyJB, BoekeJD (1994) Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136 : 849–856.
68. Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, et al.. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: : 943–951. doi:10.1002/(SICI)1097-0061(199807)14 : 10<943::AID-YEA292>3.0.CO;2-Y.
69. YuY, RenJ-Y, ZhangJ-M, SuoF, FangX-F, et al. (2013) A proteome-wide visual screen identifies fission yeast proteins localizing to DNA double-strand breaks. DNA Repair (Amst) 12 : 433–443 doi:10.1016/j.dnarep.2013.04.001
70. MatsuyamaA, AraiR, YashirodaY, ShiraiA, KamataA, et al. (2006) ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 24 : 841–847 doi:10.1038/nbt1222
71. EspositoRE, DresserM, BreitenbachM (1991) Identifying sporulation genes, visualizing synaptonemal complexes, and large-scale spore and spore wall purification. Meth Enzymol 194 : 110–131.
72. BullardJH, PurdomE, HansenKD, DudoitS (2010) Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11 : 94 doi:10.1186/1471-2105-11-94
73. StrimmerK (2008) fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24 : 1461–1462 doi:10.1093/bioinformatics/btn209
74. CarbonS, IrelandA, MungallCJ, ShuS, MarshallB, et al. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25 : 288–289 doi:10.1093/bioinformatics/btn615
75. MatsuoY, AsakawaK, TodaT, KatayamaS (2006) A rapid method for protein extraction from fission yeast. Biosci Biotechnol Biochem 70 : 1992–1994.
76. KaiserCA, SchekmanR (1990) Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61 : 723–733.
77. RyukoS, MaY, MaN, SakaueM, KunoT (2012) Genome-wide screen reveals novel mechanisms for regulating cobalt uptake and detoxification in fission yeast. Mol Genet Genomics 287 : 651–662 doi:10.1007/s00438-012-0705-9
78. YamaguchiM, NodaNN, YamamotoH, ShimaT, KumetaH, et al. (2012) Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate. Structure 20 : 1244–1254 doi:10.1016/j.str.2012.04.018
79. MatsushitaM, SuzukiNN, ObaraK, FujiokaY, OhsumiY, et al. (2007) Structure of Atg5.Atg16, a complex essential for autophagy. J Biol Chem 282 : 6763–6772 doi:10.1074/jbc.M609876200
80. FujiokaY, NodaNN, NakatogawaH, OhsumiY, InagakiF (2010) Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem 285 : 1508–1515 doi:10.1074/jbc.M109.053520
81. DelorenziM, SpeedT (2002) An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18 : 617–625.
82. BiegertA, MayerC, RemmertM, SödingJ, LupasAN (2006) The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res 34: W335–339 doi:10.1093/nar/gkl217
83. KatohK, MisawaK, KumaK, MiyataT (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30 : 3059–3066.
84. PriceMN, DehalPS, ArkinAP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490 doi:10.1371/journal.pone.0009490
85. WaterhouseAM, ProcterJB, MartinDMA, ClampM, BartonGJ (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 : 1189–1191 doi:10.1093/bioinformatics/btp033
86. GouetP, CourcelleE, StuartDI, MétozF (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15 : 305–308.
87. WatanabeY, KobayashiT, YamamotoH, HoshidaH, AkadaR, et al. (2012) Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem 287 : 31681–31690 doi:10.1074/jbc.M112.397570
88. RieterE, VinkeF, BakulaD, CebolleroE, UngermannC, et al. (2013) Atg18 function in autophagy is regulated by specific sites within its β-propeller. J Cell Sci 126 : 593–604 doi:10.1242/jcs.115725
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8- Délka menstruačního cyklu jako marker ženské plodnosti
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Vztah užívání alkoholu a mužské fertility
- Intrauterinní inseminace a její úspěšnost
- Růst a vývoj dětí narozených pomocí IVF
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



