-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPast Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
article has not abstract
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003745
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003745Summary
article has not abstract
TCF/LEF transcription factors are best known for their role as mediators of Wnt signaling, helping Wnt direct developmental transitions of stem cells in tissues or driving cell transformation and cancer when Wnt is aberrantly active. These factors possess a High Mobility Group DNA-binding domain that recognizes a motif called the Wnt Response Element (WRE: 5′-CTTTGWW-3′) and an N-terminal domain that binds β-catenin (Figure 1A). β-catenin is the cytoplasmic-nuclear mediator that communicates Wnt signals from the plasma membrane to TCF/LEFs for transcription activation (Figure 1C). The vast majority of published studies about TCF/LEFs focus on their recruitment of this mediator to Wnt target genes. This implies that “life” for TCF/LEFs began in 1996 when yeast two hybrid screens identified their mutual, strong interaction [1]–[3]. In fact, discovery of TCF/LEFs had nothing to do with Wnt and β-catenin. TCF/LEFs were first described as DNA-binding proteins that regulated transcription of lymphocyte-specific genes such as the T-Cell Receptor complex, and they did so by cooperating with transcription factors bound to juxtaposed elements in enhancers [4]–[7]. The original TCF1 and LEF1 were each characterized as a set of protein isoforms, differing by the presence or absence of N-terminal (β-catenin) and C-terminal domains, none of which were needed for enhancer activity [8]–[12]. In this issue, Grumolato et al. [13] report that TCF1 and LEF1 have constitutive, elevated activity in leukemia and lymphoma cells. They report that this activity is independent of β-catenin and instead involves direct recruitment of ATF2 and related family members (Figure 1B).
Fig. 1. β-catenin–independent and –dependent modes of Wnt signaling. 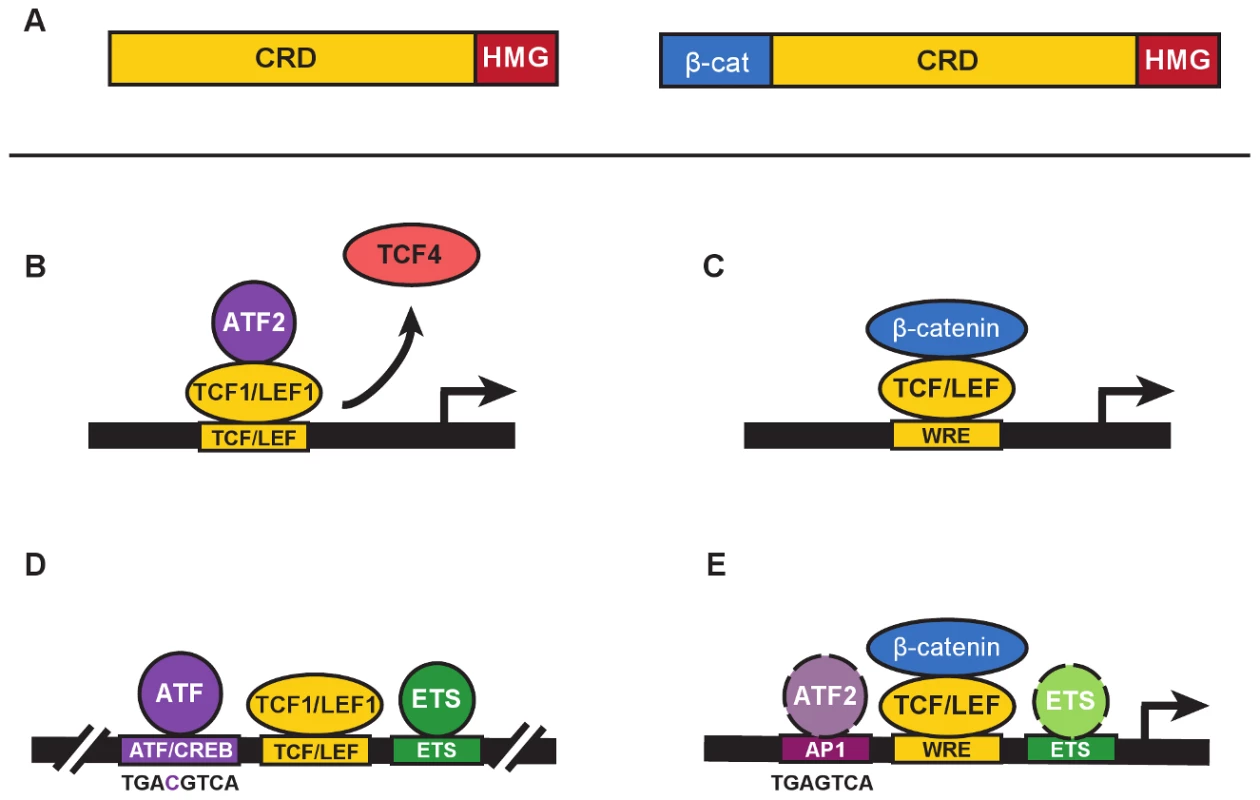
A. The general domain structure of TCF/LEF proteins includes a highly variable Context Regulatory Domain (CRD) and the well-conserved N-terminal β-catenin–binding (β-cat) and High Mobility Group (“HMG”) DNA-binding domains. N-terminal truncated forms of TCF/LEFs (left) are naturally occurring and commonly referred to as dominant negatives (e.g., dnTCF1, dnLEF1, etc.) because they block gene regulation by displacing full-length proteins from target genes (right). In the Grumolato study, dnTCF1 and dnLEF1 functioned perfectly well to activate a Wnt reporter gene. B. A simplified representation of ATF2 recruitment by TCF1 to activate transcription in a β-catenin–independent manner (referred to as a β-catenin–independent Wnt signaling pathway in Grumolato et al. [13]). In some contexts, displacement of weaker TCF activators such as TCF4 might also contribute to activation. C. β-catenin–dependent Wnt signaling requires the recruitment of β-catenin by TCF/LEFs to a Wnt Response Element (WRE) for transcriptional activation of target genes. D. A distal enhancer for the T-Cell Receptor alpha chain gene, identified as one of the very first targets for TCF/LEF binding (see references in text), contains closely juxtaposed binding sites for ATF/CREB proteins (consensus binding sequence shown below), LEF/TCFs, and ETS proteins. E. ChIP-seq studies of TCF and β-catenin genome-wide occupancy identify significant colocalization of binding motifs for AP1 and ETS transcription factors (see text for references). Consensus sequences for AP1 and ATF/CREB sites differ by a single nucleotide (see panel D for comparison), and ATF proteins are known to bind AP1 sites. Colocalized motifs suggest there is potential for interaction and cooperative crosstalk between β-catenin–bound TCF/LEFs, and ATF/CREB and ETS proteins. Grumolato et al. describe how the TOPflash reporter for Wnt signaling, a luciferase gene driven by a minimal promoter with multimers of WREs, has elevated constitutive activity in leukemia cell lines. One might assume that this activity derives from TCF/LEF recruitment of β-catenin. However, very little stabilized β-catenin could be detected and reporter activity was recapitulated using truncated forms of TCF1 missing the N-terminal β-catenin–binding domain (isoforms labelled dominant negatives, or dnTCF/dnLEF [Figure 1A]). In another twist, family member TCF4 could not substitute even though it has a β-catenin–binding domain. This meant that selective action by LEF1 and TCF1 occurred through recruitment of other transcription factors via domains distinct from the β-catenin–binding domain. Using a candidate approach, the authors tested for functional interactions with proteins that bind AP1 sites. AP1 factors are homo - and heterodimerizing leucine zipper proteins of the Jun, Fos, ATF, and JDP families [14]. Grumolato et al. report that ATF family members (especially ATF2) bind directly to TCF1 and LEF1, not TCF4, and that interactions primarily require the Context Dependent Regulatory domain (CRD; Figure 1A, B). That TCF1/LEF1-ATF2 interactions are detected in multiple types of hematopoietic tumor cell lines suggests that ATF recruitment might account for a significant portion of the “Wnt reporter activity” in these cell types. Knockdown of ATF2 reduced cell growth and lowered expression of TCF1 and LEF1 target genes, similar to effects from overexpression of a dominant negative form of TCF4. Observations such as these suggest that ATF2 is integral to the regulatory role that TCF1/LEF1 play in lymphocytes.
These discoveries highlight how TCF1/LEF1 are closely intertwined with ATF proteins. Indeed, one of the first interactions for LEF1 and, later, TCF1 was with proteins that bind an ATF/CREB element in the T-Cell Receptor alpha chain enhancer (Figure 1D; [4], [5]); interestingly, ATF4 was first discovered on the basis of its binding to this element (reviewed in [15]). Additional lymphocyte-specific enhancers were discovered as collections of ATF/CREB, TCF/LEF, and ETS elements [15]), and functional studies showed that TCF1 and LEF1 cooperated with these proteins bound to neighboring elements to create strong enhancers. Importantly, the β-catenin–binding domain was entirely dispensable, its deletion enabling even greater activity in some assays [9], [12]. Instead, it was the CRD and a strong DNA-bending function of the HMG domain that was of primary importance; DNA bending enabling a three-way, CRD-dependent interaction between TCF/LEFs and other enhancer factors [16]. The exact identities of the ATF/CREB proteins were unknown and were never fully explored. The Grumolato study brings the past back to the present by identifying specific ATF interactors for TCF1 and LEF1 for the first time, and by showing that immune system cancers possess elevated, functional interactions. The current study does not highlight the DNA binding of ATF2 because it appears to be recruited by TCF1/LEF1 to the Wnt reporters in a protein–protein interaction mode.
An emerging feature of TCF/LEFs that connects with these findings is a growing recognition of a functional split in the vertebrate family. That is, an increasing number of reports show that TCF4 and a fourth family member, TCF3, function as repressors, or at best, weak activators. More and more frequently it seems that TCF1 and LEF1 operate by opposing TCF3/TCF4 repression and providing strong activation. Since TCF1 and LEF1 are strongly active for ATF2 engagement and cooperation, and TCF4 is not, ATFs could be important players in the push-and-pull between family members. The first study to highlight a split in the family used morpholino knockdown and rescue experiments in Xenopus embryos [17]. Grumolato and colleagues use the same Xenopus system to show that overexpression of dominant negative TCF1 (dnTCF1), but not TCF4, causes axis duplication—an activity attributed to overactive Wnt signaling. It could be, as the authors posit, that dnTCF1 was recruiting ATF proteins to WREs for gene activation. But it is also possible that dnTCF1 was displacing endogenous, repressive TCFs such as TCF3 and/or TCF4 (Figure 1B). Of course, both mechanisms could be involved, but further studies are definitely warranted.
This study raises other questions about TCF/LEFs and β-catenin–independent activation of transcription. Is TOPflash the reliable indicator of Wnt signaling that its common use implies? Or can factors such as ATF2 be recruited to activate this reporter independent of β-catenin? The authors provide a “yes” to the latter question in their system, but a general answer would be best addressed with strategies that avoid overexpression of transcription factors. How much do dominant negative TCF/LEFs contribute to gene regulation? While an exact answer is not known, it is interesting to point out that these forms are expressed at significant levels in lymphocytes [8], [10]. In fact, the discovery of TCF1 came from a T lymphocyte cDNA screen in which all clones were missing the β-catenin–binding domain (the N-terminus–encoding exon discovered years later upon inspection of genomic sequences [6], [8]). Thus, even though definitive connections between β-catenin, TCF1, and LEF1 are clear in lymphocytes (reviewed in [18], [19]), the discordance between their knockout phenotypes should encourage a revisit of this issue. What about cancer? The authors point out the finding in human sebaceous tumors in which mutations have disabled the β-catenin–binding domain of LEF1. This mutation is proposed to be oncogenic because overexpression of dnLEF1 in mouse skin recapitulates sebaceous tumor development [20], [21]. Perhaps β-catenin–independent actions of TCF/LEFs are more prevalent and powerful than currently assumed. Is the ATF/CREB and TCF/LEF interaction common? The tentative answer is yes, because almost every ChIP-seq study of TCF/LEF binding in cancer genomes, and one study of β-catenin–binding to the colon cancer genome, has identified the closely related, ATF-friendly, AP1 response element as a top, cosegregating motif (Figure 1E; [22]–[25], [26]). Several studies also define cosegregating ETS elements [22], [24], [25]. Going forward, it will be important to probe how broadly ATF2 and family members crosstalk to TCF/LEFs (and perhaps β-catenin), and determine what the functional consequences of that crosstalk are in terms of gene programs and cell phenotypes.
In this Perspective, we use the protein names to refer to TCF/LEFs (i.e., LEF1, TCF1, TCF3, and TCF4). This matches the Grumolato et al. study and makes for logical reading. However, the gene names for TCF/LEFs are different: TCF1 is encoded by the TCF7 gene, and TCF3 and TCF4 by the TCF7L1 and TCF7L2 genes, respectively. LEF1 is the only respite. It is encoded by the LEF1 gene.
Zdroje
1. BehrensJ, von KriesJP, KuhlM, BruhnL, WedlichD, et al. (1996) Functional interaction of b-catenin with the transcription factor LEF-1. Nature 382 : 638–642.
2. HuberO, KornR, McLaughlinJ, OhsugiM, HerrmannB, et al. (1996) Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev 59 : 3–10.
3. MolenaarM, van de WeteringM, OosterwegelM, Peterson-MaduroJ, GodsaveS, et al. (1996) XTCF-3 transcription factor mediates b-catenin-induced axis formation in Xenopus embryos. Cell 86 : 391–399.
4. WatermanML, FischerWH, JonesKA (1991) A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev 5 : 656–669.
5. TravisA, AmsterdamA, BelangerC, GrosschedlR (1991) LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes Dev 5 : 880–894.
6. van de WeteringM, OosterwegelM, DooijesD, CleversH (1991) Identification and cloning of TCF-1, a T lymphocyte specific transcription factor containing a sequence-specific HMG box. EMBO J 10 : 123–132.
7. OosterwegelM, van de WeteringM, HolstegeFC, ProsserHM, OwenMJ, et al. (1991) TCF-1, a T cell-specific transcription factor of the HMG box family, interacts with sequence motifs in the TCR beta and TCR delta enhancers. Int Immuno 3 : 1189–1192.
8. van de WeteringM, CastropJ, KorinekV, CleversH (1996) Extensive alternative splicing and dual promoter usage generate Tcf-protein isoforms with differential transcription control properties. Mol Cell Biol 16 : 745–752.
9. CarlssonP, WatermanM, JonesK (1993) The hLEF/TCF-1a HMG protein contains a context-dependent transcriptional activation domain that induces the TCRa enhancer in T cells. Genes Dev 7 : 2418–2430.
10. HovanesK, LiTW, MunguiaJE, TruongT, MilovanovicT, et al. (2001) Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet 28 : 53–57.
11. HovanesK, LiTWH, WatermanML (2000) The human LEF-1 gene contains a promoter preferentially active in lymphocytes and encodes multiple isoforms derived from alternative splicing. Nucleic Acids Res 28 : 1994–2003.
12. GieseK, GrosschedlR (1993) LEF-1 contains an activation domain that stimulates transcription only in a specific context of factor-binding sites. EMBO J 12 : 4667–4676.
13. GrumolatoL, LiuG, HaremakiT, MungamuriSK, MongP, et al. (2013) β-catenin-independent activation of TCF1/LEF1 in human hematopoietic tumor cells through interaction with ATF2 transcription factors. PLoS Genet 9: e1003603 doi:10.1371/journal.pgen.1003603
14. Lopez-BergamiP, LauE, RonaiZ (2010) Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat Rev Cancer 10 : 65–76.
15. LeidenJM (1992) Transcriptional regulation during T-cell development: the alpha TCR gene as a molecular model. Immunol Today 13 : 22–30.
16. GieseK, KingsleyC, KirshnerJR, GrosschedlR (1995) Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev 9 : 995–1008.
17. LiuF, van den BroekO, DestreeO, HopplerS (2005) Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/{beta}-catenin signalling in mesoderm development. Development 132 : 5375–5385.
18. LentoW, CongdonK, VoermansC, KritzikM, ReyaT (2013) Wnt signaling in normal and malignant hematopoiesis. Cold Spring Harb Perspect Biol 5.
19. RoozenPP, BrugmanMH, StaalFJ (2012) Differential requirements for Wnt and Notch signaling in hematopoietic versus thymic niches. Ann N Y Acad Sci 1266 : 78–93.
20. TakedaH, LyleS, LazarAJ, ZouboulisCC, SmythI, et al. (2006) Human sebaceous tumors harbor inactivating mutations in LEF1. Nat Med 12 : 395–397.
21. NiemannC, OwensDM, HulskenJ, BirchmeierW, WattFM (2002) Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development 129 : 95–109.
22. HatzisP, van der FlierLG, van DrielMA, GuryevV, NielsenF, et al. (2008) Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol 28 : 2732–2744.
23. MokryM, HatzisP, de BruijnE, KosterJ, VersteegR, et al. (2010) Efficient double fragmentation ChIP-seq provides nucleotide resolution protein-DNA binding profiles. PLoS ONE 5: e15092 doi:10.1371/journal.pone.0015092
24. BlahnikKR, DouL, O'GeenH, McPhillipsT, XuX, et al. (2010) Sole-Search: an integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic Acids Res 38: e13.
25. FrietzeS, WangR, YaoL, TakYG, YeZ, et al. (2012) Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biol 13: R52.
26. BottomlyD, KylerSL, McWeeneySK, YochumGS (2010) Identification of b-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res 38 : 5735–5745.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



