-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
The role of Wnt signaling in embryonic development and stem cell maintenance is well established and aberrations leading to the constitutive up-regulation of this pathway are frequent in several types of human cancers. Upon ligand-mediated activation, Wnt receptors promote the stabilization of β-catenin, which translocates to the nucleus and binds to the T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors to regulate the expression of Wnt target genes. When not bound to β-catenin, the TCF/LEF proteins are believed to act as transcriptional repressors. Using a specific lentiviral reporter, we identified hematopoietic tumor cells displaying constitutive TCF/LEF transcriptional activation in the absence of β-catenin stabilization. Suppression of TCF/LEF activity in these cells mediated by an inducible dominant-negative TCF4 (DN-TCF4) inhibited both cell growth and the expression of Wnt target genes. Further, expression of TCF1 and LEF1, but not TCF4, stimulated TCF/LEF reporter activity in certain human cell lines independently of β-catenin. By a complementary approach in vivo, TCF1 mutants, which lacked the ability to bind to β-catenin, induced Xenopus embryo axis duplication, a hallmark of Wnt activation, and the expression of the Wnt target gene Xnr3. Through generation of different TCF1-TCF4 fusion proteins, we identified three distinct TCF1 domains that participate in the β-catenin-independent activity of this transcription factor. TCF1 and LEF1 physically interacted and functionally synergized with members of the activating transcription factor 2 (ATF2) family of transcription factors. Moreover, knockdown of ATF2 expression in lymphoma cells phenocopied the inhibitory effects of DN-TCF4 on the expression of target genes associated with the Wnt pathway and on cell growth. Together, our findings indicate that, through interaction with ATF2 factors, TCF1/LEF1 promote the growth of hematopoietic malignancies in the absence of β-catenin stabilization, thus establishing a new mechanism for TCF1/LEF1 transcriptional activity distinct from that associated with canonical Wnt signaling.
Published in the journal: . PLoS Genet 9(8): e32767. doi:10.1371/journal.pgen.1003603
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003603Summary
The role of Wnt signaling in embryonic development and stem cell maintenance is well established and aberrations leading to the constitutive up-regulation of this pathway are frequent in several types of human cancers. Upon ligand-mediated activation, Wnt receptors promote the stabilization of β-catenin, which translocates to the nucleus and binds to the T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors to regulate the expression of Wnt target genes. When not bound to β-catenin, the TCF/LEF proteins are believed to act as transcriptional repressors. Using a specific lentiviral reporter, we identified hematopoietic tumor cells displaying constitutive TCF/LEF transcriptional activation in the absence of β-catenin stabilization. Suppression of TCF/LEF activity in these cells mediated by an inducible dominant-negative TCF4 (DN-TCF4) inhibited both cell growth and the expression of Wnt target genes. Further, expression of TCF1 and LEF1, but not TCF4, stimulated TCF/LEF reporter activity in certain human cell lines independently of β-catenin. By a complementary approach in vivo, TCF1 mutants, which lacked the ability to bind to β-catenin, induced Xenopus embryo axis duplication, a hallmark of Wnt activation, and the expression of the Wnt target gene Xnr3. Through generation of different TCF1-TCF4 fusion proteins, we identified three distinct TCF1 domains that participate in the β-catenin-independent activity of this transcription factor. TCF1 and LEF1 physically interacted and functionally synergized with members of the activating transcription factor 2 (ATF2) family of transcription factors. Moreover, knockdown of ATF2 expression in lymphoma cells phenocopied the inhibitory effects of DN-TCF4 on the expression of target genes associated with the Wnt pathway and on cell growth. Together, our findings indicate that, through interaction with ATF2 factors, TCF1/LEF1 promote the growth of hematopoietic malignancies in the absence of β-catenin stabilization, thus establishing a new mechanism for TCF1/LEF1 transcriptional activity distinct from that associated with canonical Wnt signaling.
Introduction
The Wnt/β-catenin signaling pathway plays an essential role during embryonic development and as a major regulator of stem/progenitor cell maintenance in a number of postnatal organs and tissues, including the gastrointestinal tract, the skin and the hematopoietic system [1]–[3]. Genetic alterations that lead to aberrant activation of this pathway occur very commonly in certain tumors, including colon cancer, hepatocellular carcinomas and adrenocortical adenoma [4], [5]. In other types of tumors, alternative mechanisms are more frequently responsible for Wnt/β-catenin constitutive up-regulation. Indeed, a Wnt autocrine transforming activity was initially discovered in the mouse model three decades ago [6], and we have established that this mechanism also occurs frequently in different human cancers, including breast cancer [7], non small cell lung cancer [8] and sarcoma [9].
The Wnt/β-catenin, or canonical, pathway is initiated by Wnt-mediated coupling of the seven transmembrane domain receptor Frizzled and the single-membrane-spanning low-density receptor-related protein 5/6 (LRP5/6), followed by phosphorylation of the LRP5/6 intracellular domain [10], [11]. Through a mechanism not yet fully understood, phosphorylated LRP5/6 leads to the inhibition of the so-called β-catenin destruction complex, composed of axin, glycogen synthase kinase 3, dishevelled (Dvl), casein kinase 1 and the tumor suppressor adenomatous polyposis coli, resulting in the accumulation of β-catenin in the cytoplasm and the nucleus [11], [12]. In the absence of Wnt activation, the four members of the TCF/LEF family of transcription factors form a complex with Groucho/TLE repressors and inhibit gene expression [3], [13], [14], [15], [16], [17]. Through either direct competition [18] or XIAP-mediated ubiquitylation [19], nuclear β-catenin displaces Groucho/TLE and binds to TCF/LEF factors, thus promoting a transcriptional switch that allows the expression of Wnt target genes, including Myc, cyclin D1, Axin 2 and Lef1 [4], [20].
While its role in the control of gene expression is thought to depend mainly on the interaction with TCF/LEF proteins, β-catenin can act in some contexts through binding to other transcription factors, including the homeodomain protein Prop1 [21], various nuclear receptors [17], [22], the forkhead box O factors [23] and the Krueppel-like factor 4 [24]. TCF/LEF factors can also interact with other proteins, including ALY [25], Smad [26], [27] and c-Jun [28], allowing the formation of large nuclear complexes that control the expression of genes containing a particular combination of regulatory sequences in their promoter/enhancer. While providing an additional level of regulation, such interactions do not challenge the general notion that nuclear β-catenin is required for TCF/LEF transcriptional activity, which is widely supported by biochemical and genetic data [2], [15], [29]. An exception to this paradigm may involve the hematopoietic system, where intriguing discrepancies in the phenotypes of TCF1/LEF1 and β-catenin null mice have suggested that in particular contexts canonical Wnt signals could be transduced independently of β-catenin [30]–[36].
While the β-catenin pathway has been reported to be upregulated in cancer stem cells of different leukemias [37]–[40] and in the transition from chronic to acute myelogenous leukemia [41], mutations in intracellular components that are frequently responsible for Wnt pathway activation in solid tumors are uncommon in hematopoietic tumors. Here we show that some hematopoietic tumor cells display β-catenin-independent TCF/LEF activity, whose down-regulation inhibits the expression of TCF/LEF target genes and cell growth. Using both in vitro and in vivo approaches, we further demonstrate that TCF1 retains transcriptional activity in the absence of β-catenin. Finally, we establish that ATF2 family members physically and functionally interact with TCF1/LEF1 factors to promote target gene expression and hematopoietic tumor cell growth. Together, our results uncover a new mechanism that activates TCF1/LEF1 transcriptional activity independently of β-catenin and the Wnt canonical pathway.
Results
β-catenin-independent TCF/LEF activity in hematopoietic tumor cells
Using lentiviral wild-type (Top) and mutant (Fop) TCF/LEF reporters, we investigated the status of Wnt signaling in cells derived from several types of human cancer. As a result of this screen, we identified up-regulated TCF/LEF activity in different hematopoietic tumor cells, including Ramos, K562 and Jurkat (Figure 1A). As shown in Figure S1A, TCF/LEF reporter levels in these cells were comparable to those observed in solid tumor cells exhibiting autocrine Wnt activation [8], [9]. To assess the levels of stabilized, transcriptionally active β-catenin in these same hematopoietic tumor lines, we used an approach based on the capture of uncomplexed β-catenin with GST-E-cadherin beads [7]–[9]. Surprisingly, none showed detectable levels of stabilized β-catenin (Figure 1B). It has been reported that γ-catenin can also play a role in Wnt signaling, probably through a mechanism involving stabilization of β-catenin [42], [43]. However, neither down-regulation of β-catenin nor γ-catenin had any effect on the TCF/LEF reporter in Ramos and K562 cells (Figures 1C and S1B), indicating that β-catenin and γ-catenin were dispensable for TCF/LEF transcriptional activation in these hematopoietic tumor cells.
Fig. 1. Constitutive TCF/LEF activity in hematopoietic tumor cells in the absence of β-catenin stabilization. 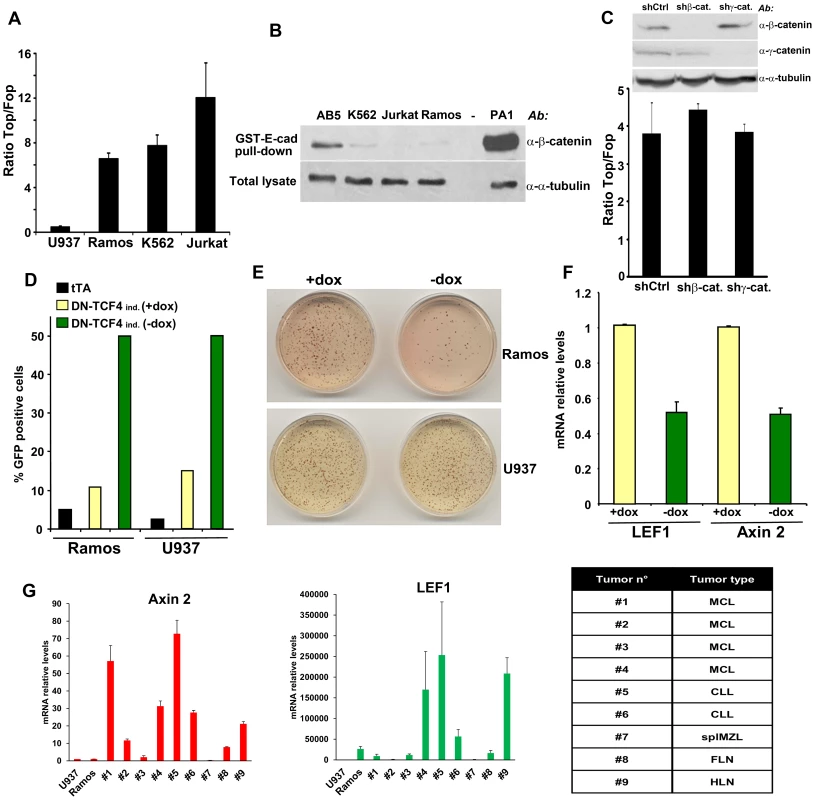
(A) The indicated hematopoietic tumor cells were transduced with wild-type (Top) or mutant (Fop) TCF/LEF firefly luciferase reporter and a renilla luciferase virus, and the TCF/LEF transcriptional activity was calculated by dividing the TOP/renilla ratio by the FOP/renilla ratio. The mean values (± s.e.m., n>5) of at least two independent experiments are shown. (B) Lack of β-catenin stabilization in K562, Jurkat and Ramos cells, as measured by GST-E-cadherin pull-down. Ovarian PA1 cancer cells and breast immortalized AB5 cells were used as positive and negative control, respectively. (C) β-catenin or γ-catenin knockdown in Ramos cells did not affect TCF/LEF activity. Ramos cells containing Top or Fop luciferase reporter were infected with β-catenin or γ-catenin shRNA and the TCF/LEF reporter activity was measured (lower panel). β-catenin and γ-catenin down-regulation was assessed by immunoblot (upper panel). (D) Ramos and U937 cells containing the tet transactivator (tTA) with or without a lentiviral inducible vector containing DN-TCF4 fused to GFP were cultured for 24 hrs in the presence or the absence of doxycycline (dox), and the proportion of GFP positive cells was assessed by FACS analysis. (E) The cells in D were used in colony formation assay in the presence or the absence of dox. (F) Effects of DN-TCF4 induction in Ramos cells on the expression of LEF1 and Axin 2 by quantitative real-time PCR. (G) mRNA levels of Axin 2 and LEF1 in human primary hematopoietic tumors. For each sample, the qPCR values were normalized with those obtained for the negative control, U937 cells. The types of malignancy corresponding to each sample are listed in the table: MCL, mantle cell lymphoma; CLL, B-cell chronic lymphocytic leukemia; splMZL, splenic marginal zone lymphoma; FLN, follicular lymphoma; HLN, Hodgkin's lymphoma. To gain insights into the biological relevance of this β-catenin-independent TCF/LEF activity, we tested the effects of a GFP-fused DN-TCF4 in Ramos lymphoma cells using a lentiviral tetracycline (Tet-off) inducible system. As a negative control for these experiments, we utilized U937 lymphoma cells, which lacked detectable constitutive TCF/LEF activity (Figure 1A). Doxycyline withdrawal triggered expression of DN-TCF4 to a similar extent in Ramos and U937 cells, as measured by FACS analysis (Figure 1D). Under these conditions, DN-TCF4 expression inhibited colony formation by Ramos cells but had no effect on U937 cells (Figure 1E). Of note, induction of DN-TCF4 expression in Ramos cells also reduced the mRNA levels of LEF1 and Axin 2 (Figure 1F), two well-established Wnt target genes. Consistent with these results, DN-TCF4 also inhibited TCF/LEF reporter activity as well as the expression of LEF1 and Axin 2 in K562 (Figures S1C and S1D). Together, these results indicated that some hematopoietic tumor lines exhibit elevated β-catenin-independent TCF/LEF activity, which positively influences the expression of Wnt target genes and tumor cell growth.
We extended our analysis to other hematopoietic tumor cell lines as well as primary tumors. Several mantle cell lymphoma lines, including Granta-519, HBL-2, JEKO-1 and JVM-2, exhibited TCF/LEF reporter levels as high or higher than Ramos (Figure S1E) and also contained undetectable levels of uncomplexed β-catenin (Figure S1F), implying their activation of β-catenin-independent TCF/LEF transcriptional activity. We also surveyed a series of primary hematopoietic tumors for expression of Axin 2 and LEF1 as compared to levels present in Ramos and U937 cells. Several exhibited high expression levels of these TCF/LEF target genes in the absence of detectable uncomplexed β-catenin (Figures 1G and S1G), arguing that activation of this novel pathway was not limited to hematopoietic tumor lines but occurred in primary human hematopoietic tumors as well.
TCF1 and LEF1, but not TCF4, are transcriptionally active independently of β-catenin
TCF1 and LEF1 were cloned in 1991 from hematopoietic cells [44]–[46] and belong to a four-member family of transcription factors containing a N-terminal β-catenin binding domain and a high mobility group DNA binding domain located closer to the C-terminus [15]. To investigate whether these factors could act as transcriptional activators in the absence of β-catenin, we expressed TCF1, LEF1 or TCF4 in 293T cells, which lack constitutive Wnt activation. Figure 2A shows that TCF1 and LEF1, but not TCF4, stimulated TCF/LEF reporter activity in these cells. The stimulatory effect of TCF1 was observed using three different reporter constructs, containing respectively 2, 4 and 8 TCF/LEF repeats generated using different backbone vectors (Figure S2A). As an additional specificity control, TCF1-induced reporter activity was strongly inhibited by an excess of DN-TCF4 (Figure S2B). Of note, deletion of the first 65 aa, containing the β-catenin binding domain, only partially reduced the TCF/LEF activation induced by TCF1 (Figure 2A), indicating that β-catenin was not strictly required for this effect. To inhibit any residual nuclear β-catenin potentially present in 293T cells, we generated a decoy construct containing the TCF1 β-catenin binding domain fused to the Gal4 DNA binding domain (BCBD). While almost completely abolishing the up-regulation of the TCF/LEF reporter induced by Wnt3a (Figure 2B), BCBD did not affect TCF1 transcriptional activity (Figure 2C), strongly arguing against the involvement of β-catenin in this activity.
Fig. 2. β-catenin-independent transcriptional activity of TCF1/LEF1 factors. 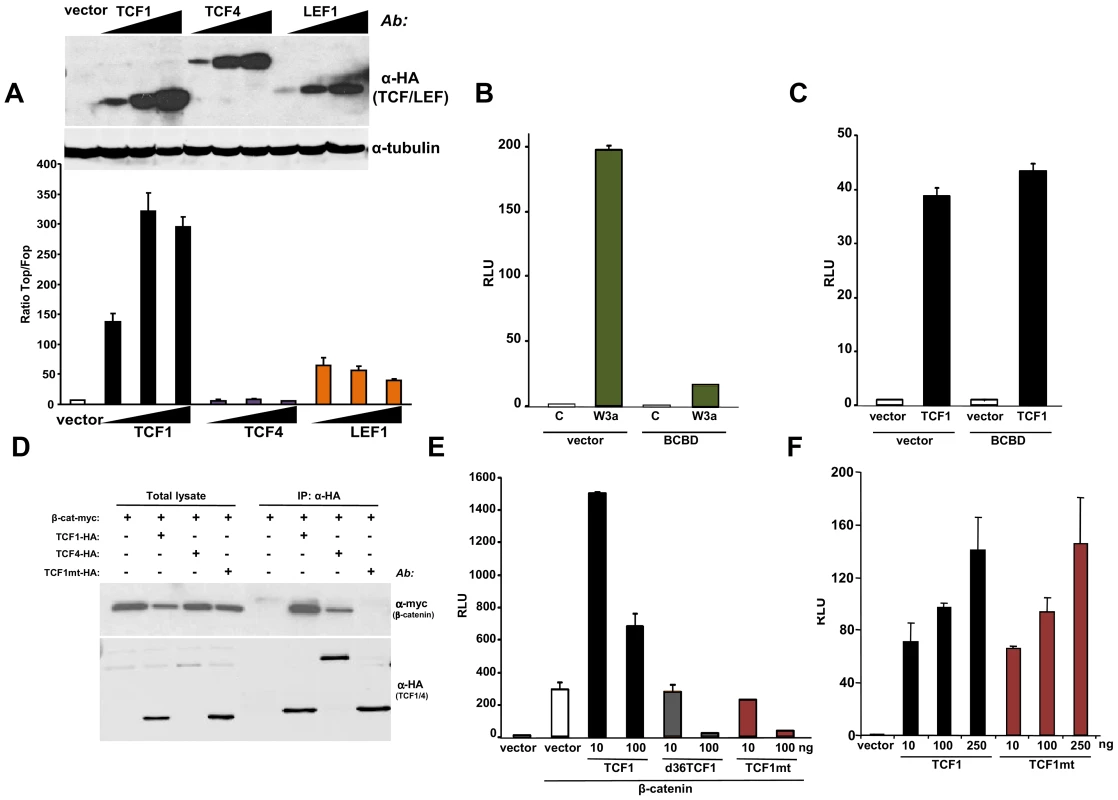
(A) Empty pcDNA3HA vector or increasing amounts of HA-tagged TCF1, LEF1 or TCF4 constructs were co-transfected with SuperTop or SuperFop and renilla luciferase plasmids in 293T cells and the Top/Fop ratio was calculated. (B–C) Effects of a decoy construct containing TCF1 β-catenin binding domain (BCBD) on Wnt3a- and TCF1-induced TCF/LEF reporter activity. 293T cells were co-transfected with SuperTop and renilla reporter plasmids, together with the Wnt3a, TCF1 and BCBD vectors as indicated. (D–E) Mutant TCF1 (mtTCF1; D21A, E29K) is unable to bind to β-catenin. (D) 293T cells were transfected with the indicated HA-tagged TCF1 or TCF4 constructs and the myc-tagged β-catenin, followed by immunoprecipitation using anti-HA antibody and immunoblot with anti-myc or anti-HA antibodies. (E) 3T3 cells were co-transfected with β-catenin, the indicated amounts of TCF1, d36TCF1 or mtTCF1, together with SuperTop and renilla luciferase plasmids. (F) Effects of TCF1 and mtTCF1 expression on TCF/LEF reporter activity. 293T cells were co-transfected with SuperTop and renilla luciferase plasmids and different amounts of TCF1 or mtTCF1, followed by reporter assay. Based on the crystal structure of the β-catenin-TCF3 complex, we mutated two residues in the TCF1 β-catenin binding domain, D21A and E29K, which should impair the ability of this transcription factor to bind to β-catenin [47]. Figures 2D and S3A show that this mutant, designated TCF1mt, was unable to physically interact with β-catenin by co-immunoprecipitation and failed to synergize with β-catenin or γ-catenin to stimulate the Wnt responsive reporter, instead exerting an antagonistic effect (Figures 2E, S3B and S3C). In contrast, both wild type TCF1 and TCF1mt expression stimulated TCF/LEF activity in 293T cells to similar extents (Figure 2F), implying that TCF1 was able to act as a transcriptional activator independently of β-catenin.
TCF1, but not TCF4, stimulates the expression of Wnt target genes in Xenopus laevis embryos independently of β-catenin
During the earliest stages of Xenopus embryonic development, locally activated Wnt signaling induces the asymmetric expression of genes of the dorsal organizer, thus allowing the establishment of dorsal-ventral polarity [48]–[50]. We used this in vivo model to assess the β-catenin-independent activity of TCF1. As previously reported [51], injection of tcf1, but not tcf4, mRNA induced ectopic expression of the Wnt direct target gene Xnr3 in animal cap explants (Figure 3A). Of note, we found that both TCF1mt and the N-terminal truncated TCF1 (del65TCF1) were able to trigger Xnr3 expression, indicating that β-catenin was not required for this effect. Under these conditions, Wnt3a, but not the TCF1 constructs, increased the expression of another organizer gene, Chordin, likely reflecting its ability to induce a higher level of Wnt pathway activation. As a specificity control, neither TCF1 nor Wnt3a had an effect on the TGF-β target gene Xbra (Figure 3A). Consistent with these data, all TCF1 constructs, but not TCF4, induced axis duplication in the embryos (Figure 3B). Together, our findings indicate that in the absence of β-catenin TCF1 does not strictly act as a transcriptional repressor, but instead can stimulate the expression of Wnt canonical target genes and functions in vivo independently of β-catenin.
Fig. 3. TCF1 activity in Xenopus laevis explants and embryos. 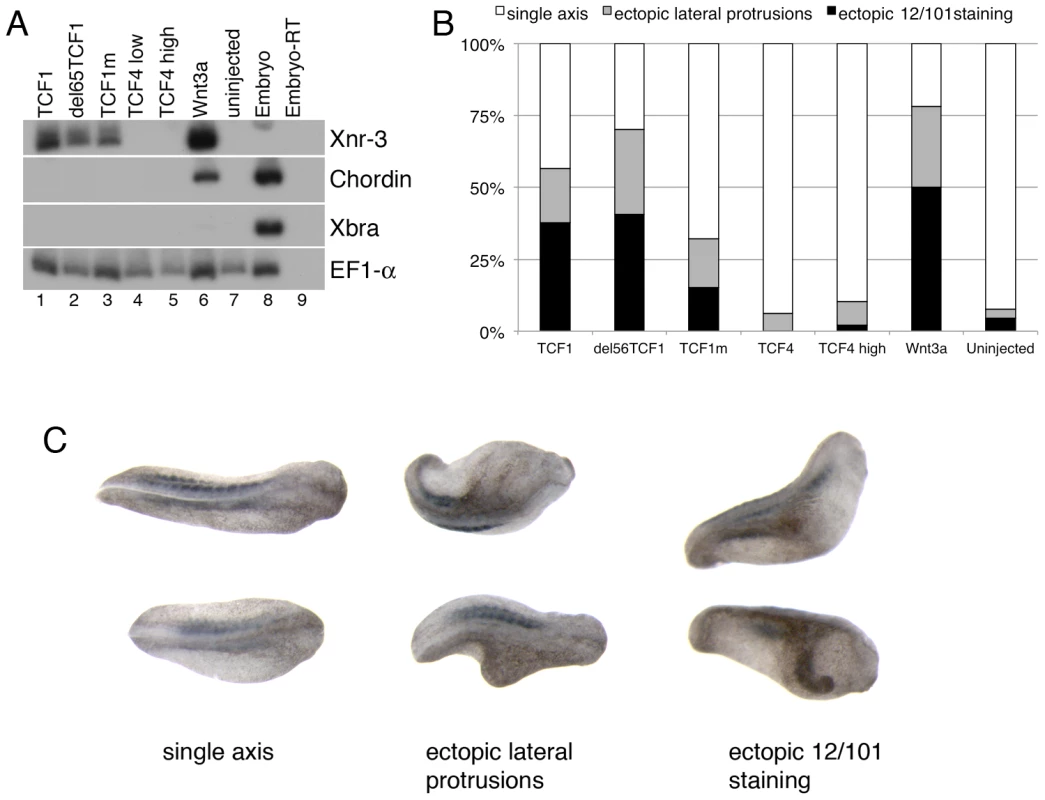
(A) Wnt3a, wild-type and β-catenin-independent mutants of TCF1, but not TCF4, induce expression of the canonical Wnt pathway-responsive gene Xnr3 in ectodermal (animal cap) explants. RT-PCR analysis of animal caps dissected at late blastula stages and cultured until stage 10.5. EF1-α is used as a loading control. The –RT lane contains all reagents except reverse transcriptase, and is used as a negative control. (B) Graph depicting embryonic perturbation by Wnt3a, TCF4, and wild-type and mutant TCF1. Embryos were injected in the ventral marginal zone at early cleavage stages and cultured until late tailbud stages. For (A) and (B), 1 ng (TCF4 high) or 500 pg (TCF1, del65TCF1, TCF1m, TCF4low, Wnt3a) of each RNA was injected, as listed. (C) Representative embryos recorded in the graph shown in (B). Dorsal views; anterior is to right. All embryos in (C) were injected with RNA encoding del65TCF1. Staining with the 12/101 antibody, which recognizes a somite specific epitope, was used to assess axis duplication. Different TCF1 domains are involved in its β-catenin-independent transcriptional activity
To gain insights into the mechanisms responsible for β-catenin-independent TCF1/LEF1 transcriptional activity, we next investigated the contribution of different TCF1 domains to its ability to stimulate TCF/LEF reporter activity. Alternative splicing gives rise to several isoforms of each TCF/LEF protein, which in some cases can affect their activity [14], [15]. While the majority of these splicing events involve the C-terminus, experiments in Xenopus have suggested that the central exon IVa might be responsible for the balance between the activating and repressive functions of these transcription factors [52], [53]. We found that the presence or the absence of exon IVa did not affect TCF/LEF1 reporter activity induced by human TCF1 in 293T cells (Figure S4), indicating that this region is not involved in β-catenin-independent TCF1 transcriptional activity.
It has been reported that certain C-terminal TCF splice variants can bind to repressors, including CtBP [54], although the biological relevance of such interactions is controversial [15], [55]. To assess whether the β-catenin independent activity of TCF1 was associated with a particular C-terminal domain, we generated a chimeric protein containing the first 299 aa of TCF1 fused to the DNA-binding domain and C-terminus of TCF4 (TCF1/4(1–299); Figure 4A). We observed that TCF1 and TCF1/4(1–299) stimulated the TCF/LEF reporter to a similar extent, while TCF4 was inactive (Figure 4B), indicating that TCF1 C-terminus and DNA binding domains were not required for this effect. Using a similar approach, we showed that swapping the N-terminal 100 aa of TCF1 with the corresponding domain of TCF4 resulted in nearly inactive fusion proteins (Figure 4B). These results implied that this region was important for TCF1 transcriptional activity but not sufficient to confer β-catenin-independent transcriptional activity to TCF4. We extended TCF1 mapping and found that deletion of the region between aa 56/101 and aa 211 partially reduced TCF1 transcriptional activity in the absence of β-catenin (Figure 4C and S5A) but had no effect on the synergy between TCF1 and β-catenin (Figure S5B). As shown in Figure 4C, a TCF1/4 fusion protein containing TCF1 aa 1–211 and the TCF4 DNA binding domain and C-terminus showed decreased TCF/LEF activity, implying that aa 212–299 are also involved in β-catenin-independent TCF1 activity.
Fig. 4. Mapping of the TCF1 domains involved in β-catenin-independent transcriptional activity. 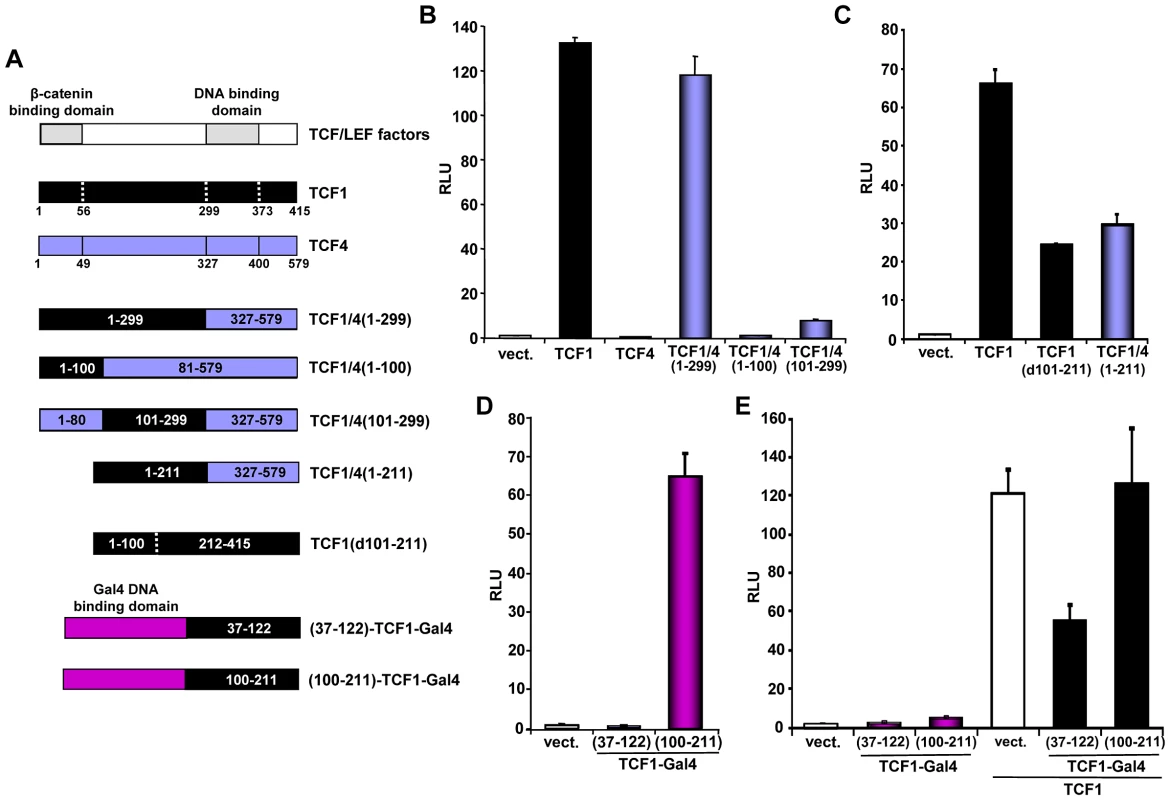
(A) Diagram of the different constructs used for the mapping. (B–D) 293T cells were co-transfected with the SuperTop reporter, the renilla plasmid and the indicated constructs, followed by reporter assay two days after transfection. (B) TCF1 N-terminal domain is required, but not sufficient, for β-catenin-independent full transcriptional activity. (C) Deletion of either aa 101–211 or 212–299 inhibits the β-catenin-independent transcriptional activity of TCF1. (D) Transcriptional activity of two different TCF1 domains fused to the Gal4 DNA binding domain. 293T cells were co-transfected with a Gal4 responsive reporter and the indicated TCF1-Gal4 fusion constructs, followed by reporter assay. (E) Expression of TCF1 aa 37–122, but not aa 100–211, inhibits TCF1-induced reporter activity. 293T cells were co-transfected with the Supertop reporter and the indicated TCF1-Gal4 fusion constructs, in the presence or the absence of TCF1, followed by reporter assay two days after transfection. As a complementary approach, we assessed the activity of different TCF1 domains fused to the Gal4 DNA binding domain. Consistent with the results shown in Figures 4B and 4C, TCF1 aa 100–211 strongly up-regulated the activity of a Gal4 reporter, while aa 37–122 had no effect (Figure 4D). Of note, expression of TCF1(37–122)-Gal4, but not TCF1(100–211)-Gal4, inhibited TCF/LEF reporter activity induced by TCF1 in 293T cells (Figure 4E), suggesting that TCF1(37–122)-Gal4 may interfere with the binding to a TCF1 molecular partner, while TCF1(100–211) may have an intrinsic transactivation activity. Together, these results indicated that these three TCF1 domains have distinct roles in its β-catenin independent activity: the region between aa 100–211 is partially active on its own, while the N-terminal 100 aa and aa 211–299 are required for full transcriptional function.
ATF2 transcription factors cooperate with TCF1/LEF1 to promote the growth of hematopoietic tumor cells
We used a candidate approach to identify partners for TCF1/LEF1 potentially involved in their β-catenin independent activity. It has been shown that c-Jun binds to TCF4 and β-catenin to promote intestinal cancer development [28]. We tested the effects of TCF1 and c-Jun co-expression in 293T cells and found that c-Jun actually decreased TCF1-induced reporter activity (Figure 5A). A similar inhibitory effect was observed upon expression of the c-Jun-related JunB (Figure 5A), indicating that the c-Jun family has opposite effects in β-catenin-dependent and β-catenin-independent TCF/LEF signaling. These results prompted us to investigate the effects of other activator protein-1 (AP-1) factors and Jun binding partners on TCF1 transcriptional activity. While c-Fos had no effect (data not shown), ATF2 strongly synergized with TCF1 in stimulating the TCF/LEF reporter (Figure 5B). Of note, expression of ATF2 alone moderately increased reporter activity (Figure 5B), presumably through endogenous TCF1/LEF1. ATF2 showed a similar synergistic effect when co-expressed with TCF1mt, as well as in the presence of either Dvl or β-catenin shRNA (Figure S6), implying that the cooperation between TCF1 and ATF2 was independent of β-catenin. The fact that ATF2 also enhanced LEF1-induced reporter activity (Figure 5C) suggested that the synergy we uncovered between TCF1 and ATF2 could be part of a more general cooperation between the TCF1/LEF1 and ATF2 families of transcription factors. Indeed, expression of the two other ATF2-like proteins, i.e. activating transcription factor 7 (ATF7) and cAMP responsive element binding protein 5 (CREB5), also increased the TCF/LEF transcriptional activity of TCF1 and LEF1 (Figures 5D, 5E and 5F). Consistent with our previous findings, TCF4 was unable to stimulate the TCF/LEF reporter even in the presence of overexpressed ATF2, CREB5 or ATF7 (Figure S7A). Of note, various combinations of TCF1/LEF1 and ATF2 factors displayed different degrees of cooperation, with TCF1-ATF2 and LEF1-CREB5 showing the strongest synergy, likely reflecting preferential interactions among distinct members of these two transcription factor families.
Fig. 5. ATF2 transcription factors cooperate with TCF1/LEF1 to stimulate TCF/LEF activity. 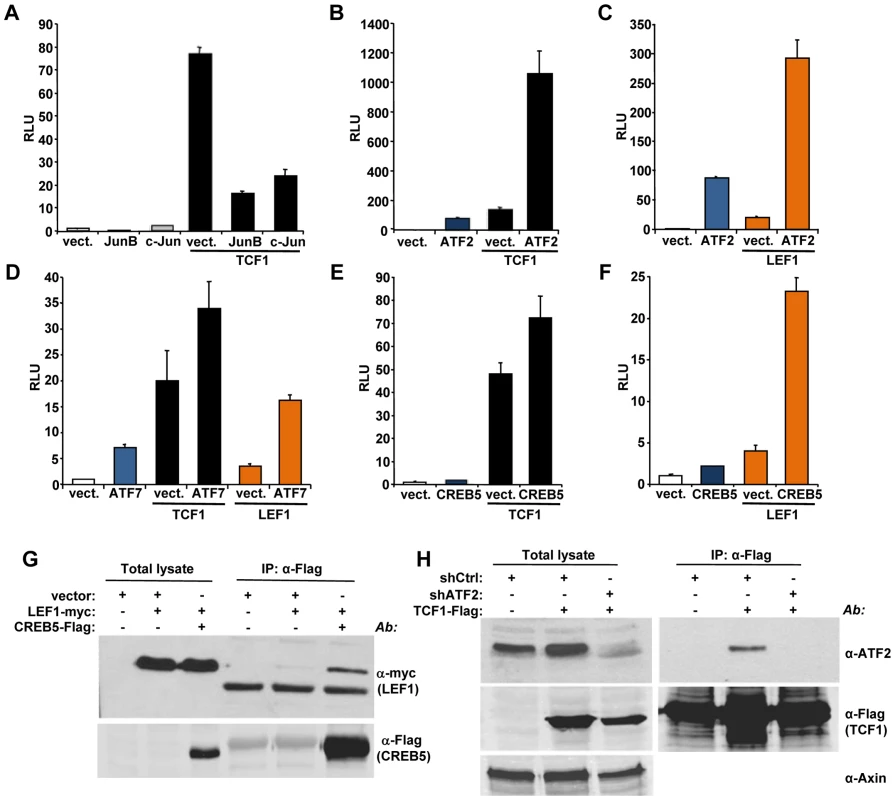
(A–F) 293T cells were co-transfected with the Supertop reporter, the renilla plasmid and the indicated TCF/LEF or AP1 transcription factors, followed by TCF/LEF reporter assay two days after transfection. (A) c-Jun and JunB inhibition of TCF1-induced reporter activity. (B–C) ATF2 synergizes with TCF1 (B) and LEF1 (C) to increase TCF/LEF activity. (D) ATF7 enhances TCF1- and LEF1-induced reporter activity. (E–F) CREB5 cooperated with TCF1 (E) and LEF1 (F). (G) myc-tagged LEF1 co-immunoprecipitates with Flag-tagged CREB5 in 293T cells. (H) Endogenous ATF2 co-immunoprecipitates with Flag-tagged TCF1 in 293T cells. Cells transduced with lentiviral shATF2 were used as a negative control and immunoblot with anti-Axin antibody was used as loading control for the total lysate. To gain insights into the mechanisms involved in the synergistic functional interactions of TCF1/LEF1 and ATF2, we investigated the ability of these proteins to form complexes. LEF1 formed a complex with CREB5 (Figure 5G), and TCF1 did so with endogenous ATF2 in 293T cells (Figure 5H), while coupling between endogenous LEF1 and ATF2/ATF7 was observed in Ramos and K562 cells (Figure S8). Of note, TCF4 showed weaker interaction with CREB5 compared to TCF1 (Figure S7B), suggesting that TCF4's lack of transcriptional activity could reflect a lower binding affinity for ATF2 factors. These findings indicated that TCF1/LEF1 proteins interact physically as well as functionally with ATF2 transcription factors.
Finally, we asked whether the association between TCF1/LEF1 and ATF2 factors plays a role in the β-catenin-independent up-regulation of TCF/LEF activity identified in some human hematopoietic tumor lines. We observed that shRNA-mediated down-regulation of ATF2 and ATF7 in Ramos cells significantly inhibited their endogenous TCF/LEF reporter activity (Figure 6A), as well as the expression of the Wnt target gene Axin 2 (Figure 6B). Consistent with our results using DN-TCF4 (Figure 1), the repression of TCF/LEF transcriptional activity induced by shATF2/7 in these cells was accompanied by inhibition of colony formation (Figure 6C). Together, our results provide compelling evidence for a new mechanism of constitutive TCF/LEF activation in tumor cells, which is independent of β-catenin and involves cooperation with ATF2 transcription factors.
Fig. 6. ATF2 and ATF7 knockdown in Ramos cells decreases TCF/LEF activity, Axin 2 expression and cell growth. 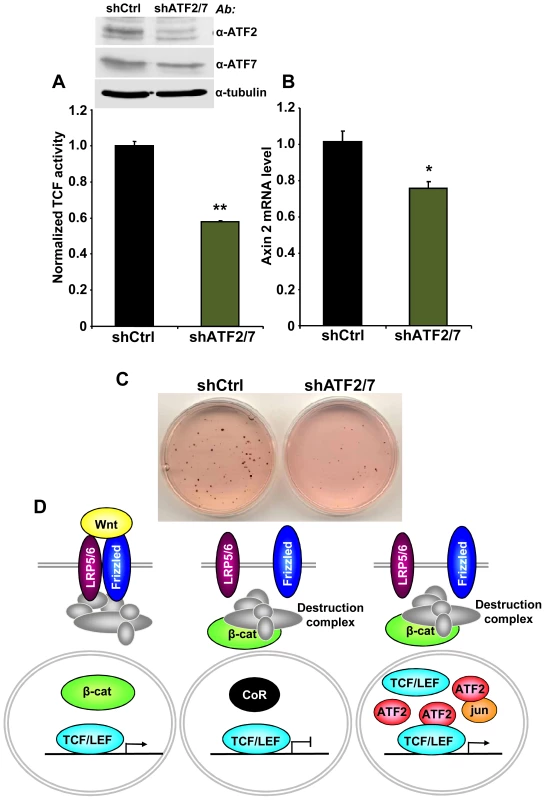
(A–C) Ramos cells containing the TCF/LEF firefly and renilla luciferase lentiviral reporters were transduced with shATF2 and shATF7 or control lentiviral vectors. ATF2 and ATF7 down-regulation assessed by immunoblot analysis. (A) shATF2/7 inhibits constitutive TCF/LEF reporter activity in Ramos cells. (B) Knockdown of ATF2 and ATF7 reduces the expression of the Wnt target gene Axin 2 in Ramos cells as measured by quantitative real-time PCR. The results normalized to the control represent the mean values ± SEM of three independent experiments. (*) P<0.05; (**) P<0.001 compared with control (two-way Anova test) (C) shATF2/7 inhibits Ramos cell colony formation. (D) Model for β-catenin-dependent and β-catenin-independent canonical Wnt signaling. See the text for details. Discussion
In the present study, we uncovered a novel mechanism of TCF/LEF dependent transcription that bypasses β-catenin and increases expression of Wnt target genes through interaction of TCF1/LEF1 and ATF2 transcription factors (Figure 6D). We utilized several different approaches, including shRNA and decoy constructs, as well as TCF1 mutants unable to bind to β-catenin, to demonstrate that TCF1/LEF1 possess transcriptional activity that is independent of β-catenin. Expression of these proteins was able to stimulate the TCF/LEF reporter in mammalian cells, and to trigger Wnt target genes in vivo, accompanied by induction of the characteristic axis-duplication phenotype in Xenopus embryos. We showed further that this mode of TCF/LEF activation in the absence of β-catenin stabilization occurs constitutively in some human hematopoietic tumors, where it plays a role in stimulating cell growth. Thus, this cooperation between TCF1/LEF1 and ATF2 represents an unexpected new strategy used by tumor cells to aberrantly activate TCF1/LEF1 signaling independently of β-catenin and the Wnt canonical pathway.
Several arguments support a role of TCF/LEF dependent Wnt signaling in the hematopoietic system [56], and it has recently been shown that moderate levels of canonical Wnt activation promote the function and/or the maintenance of various hematopoietic lineages [57]. However, the involvement of β-catenin in these cells is highly controversial [56]. While the knock-out of TCF1 and/or LEF1 demonstrated that these transcription factors are required for normal T and B cell development [30]–[32], β-catenin conditional deletion in the mature hematopoietic system using different CRE drivers only showed mild [37] or undetectable effects [33], [36]. In fact, using two different Wnt responsive gene reporters, Jeannet et al. demonstrated that thymocytes exhibited constitutive TCF/LEF activity, which was strongly inhibited upon knock-out of TCF1, but not β-catenin or γ-catenin [34]. In light of our present findings, it is tempting to speculate that this new mechanism of β-catenin-independent activation of TCF1/LEF1 identified by us here may allow a cell autonomous up-regulated level, required for the maintenance of certain hematopoietic lineages.
We identified three TCF1 domains involved in β-catenin-independent activity, all of which localized N-terminally of the high mobility group DNA binding domain. Whereas the region between aa 100 and 211 displayed some intrinsic transactivation properties when fused to the Gal4 DNA binding domain, the N-terminal 100 aa proved to be particularly important, albeit not sufficient, to confer transcriptional activity to TCF1. The N-terminal domain of TCF/LEF proteins has been previously associated with binding to β-catenin [14], [15], [47], and our demonstration that this region also participates in β-catenin-independent signaling may help in the interpretation of several previous observations. For example, it has been assumed that binding to β-catenin is required for the role of TCF1 in T cell development, since the defects in thymocyte maturation and survival observed in TCF1 null mice could be rescued by expression of the long TCF1 isoform, but not using a short TCF1 lacking the N-Terminal 116 aa [58]. The fact that the N-terminal 100 aa of TCF1 also participate in its β-catenin-independent transcriptional activity may help to reconcile these findings with the normal phenotype of β-catenin null thymocytes [33]–[35]. Human sebaceous tumors have been reported to contain LEF-1 N-terminal domain mutations with decreased ability to interact with β-catenin [59], and expression of a LEF1 construct lacking the first 32 aa driven by the keratin 14 promoter provoked the formation of sebaceous skin tumors in mice [60]. Our results indicate that N-terminally deleted LEF1 should retain β-catenin independent transcriptional activity, suggesting that the induction of such tumors might be due to this activity rather than to inhibition of Wnt canonical signaling.
We established that both TCF1 and LEF1 interact with ATF2 transcription factors to promote TCF/LEF activity, and inhibition of such activity using either DN-TCF4 or shATF2/7 decreased the expression of Wnt target genes, as well as lymphoma cell growth. The role of ATF2 proteins in cancer depends on cell context, as these factors have been associated with both oncogenic or tumor suppressive functions [61]. Contrary to what was previously reported for the β-catenin-TCF4 complex [28], we found that c-Jun and JunB inhibited TCF1-induced transcription, suggesting that Jun proteins may compete with TCF1/LEF for binding to ATF2. While c-Jun is an oncogene overexpressed or amplified in different types of cancer, including sarcomas [61], it has been demonstrated that conditional JunB inactivation [62], [63] or PU.1 related downregulation of both JunB and c-Jun [64] provoke myeloproliferative disorders and different types of leukemia in mice. It is tempting to speculate that decreased levels of JunB and/or c-Jun may perturb the balance between different AP1 factors, thus facilitating the interaction between ATF2 and TCF1/LEF1.
With the exception of few general target genes, including LEF1 and Axin 2, the transcriptional outcome of activated Wnt signaling depends on the cell/tissue context. Gene array experiments have identified hundreds of genes whose expression is modified by Wnt, with often little overlap between different cell models [15]. A number of variables likely contribute in determining which genes are regulated in a particular cell or tissue type, including the strength of the signal, cooperation with other pathways or transcription factors, expression of different LEF/TCF isoforms [65] and, possibly, even post-translational modifications of TCF/LEF factors [66], [67]. We showed that down-regulation of β-catenin-independent TCF/LEF activity inhibited the expression of LEF1 and Axin 2 genes. However, further studies will be needed to obtain a broader view of the genes regulated through this new mechanism of TCF/LEF activation and to assess similarity and differences with classical Wnt/β-catenin signaling. In a recent study, TCF1 was identified as one of the most up-regulated genes in self-renewing versus partially differentiated hematopoietic multipotential precursor cells [68]. These same authors found by ChIP-seq that TCF1 primarily binds to up-regulated genes, many of which are involved in self-renewal [68]. Yet, expression of Wnt ligands was extremely low in these cells, consistent with activation of TCF1 being independent of β-catenin and canonical Wnt signaling. The integration of this type of high-throughput dataset with those generated in other systems, in which the Wnt canonical pathway is active, may aid in dissecting the different functions of β-catenin-dependent and -independent TCF/LEF signaling.
Materials and Methods
Cell culture, transfection and lentivirus production
Ramos (Burkitt's lymphoma), U937 (histiocytic lymphoma), K562 (chronic myelogenous leukemia) and Jurkat (acute T-cell leukemia) cells were maintained in RPMI-1640 (Lonza) supplemented with 10% fetal bovine serum (FBS; Sigma). Human mantle cell lymphoma lines including Granta-519, HBL-2, JEKO-1 and JVM-2 were generously provided by Dr. Samir Parekh, Icahn School of Medicine at Mount Sinai, and were also cultured in this medium. AB5 (immortalized human breast epithelial), PA1 (ovarian teratocarcinoma), 293T (human embryonic kidney), NIH-3T3 (mouse fibroblasts) and Mel888 (melanoma) cells were maintained in DMEM (Lonza) supplemented with 10% FBS.
Transient transfection was performed using Fugene 6 (Roche) according to the manufacturer's instructions or with polyethylenimine (Polysciences). For lentivirus production, 293T cells were co-transfected with the lentiviral vector, pCMV Δ8.91 and pMD VSV-G plasmids. The conditioned medium containing the viral particles was collected two, three and four days after transfection, supplemented with 8 µg/ml polybrene and added to a pellet of hematopoietic tumor cells, followed by centrifugation for 1 h at 500 g, 4°C and overnight incubation at 37°C, 5% CO2. Two days after transduction, the cells were selected in 2 µg/ml puromycin or 10 µg/ml blasticidin.
Primary human hematopoietic tumor cells
Primary human lymphoma samples were obtained either as part of standard excisional biopsy or from peripheral blood samples from patients at the Icahn School of Medicine at Mount Sinai with informed consent reviewed and approved by the Institutional Review Board and in accordance with the Declaration of Helsinki. Specimens were processed to viable, sterile single-cell suspensions. Briefly, lymph node tissue was diced and forced through a metal sieve in a laminar flow hood into RPMI tissue culture medium. Peripheral blood mononuclear cells or disaggregated follicular lymphoma biopsy cells were pelleted by low-speed centrifugation, resuspended in media composed of 90% fetal calf serum and 10% DMSO (Sigma), frozen slowly in the vapor phase of liquid nitrogen in multiple cryotubes, and stored in liquid nitrogen. The frozen cells were thawed and maintained for 1–3 days in RPMI-1640 supplemented with 10% fetal bovine serum prior to RNA extraction for qPCR analysis or preparation of cell lysates for analysis of uncomplexed β-catenin (see below).
Constructs
Lentiviral TCF/LEF luciferase and GFP reporters, β-catenin and Dvl shRNAs, inducible and constitutive DN-TCF4 vectors and the SuperTop reporter (pTA-Luc vector) were previously described [8], [69]. The pOT (pGL3) reporter was kindly provided by B. Vogelstein. The Top-Glow reporter was purchased from Millipore. Human TCF4 was cloned by PCR from 293T cells into pcDNA3HA vector. Human TCF1 was cloned from CCRF-CEM cells into pcDNA3HA and pcDNA3flag vectors. TCF1 mutant (D21A; E29K) was generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). TCF1 N-terminal deletion and TCF1-TCF4 fusion constructs were generated by PCR in pcDNA3HA. The sequence corresponding to TCF1 β-catenin binding domain (N-terminal 60 aa), aa 37–122 and aa 100–211 were cloned downstream of Gal4 DNA binding domain into the pBIND vector from the CheckMate Mammalian Two-Hybrid System (Promega). Full-length LEF1 was cloned in pCAN-myc2 and pcDNA3HA (Figure 2A) vectors. Myc-tagged β-catenin and HA-tagged Wnt3a were cloned into pCCBS vector. Human ATF2 and CREB5 were cloned into pEF-flag vector. c-Jun and Jun-B were cloned into pcDNA3HA. The ATF7 construct was kindly provided by P.J. Hamard. The lentiviral shRNA constructs targeting β-catenin, γ-catenin, ATF2 and ATF7 were generated in VIRDH-EP or VIRHD-bla vectors, using the following targeting sequences: GTACGAGCTGCTATGTTCC (β-catenin), CACCATTCCCCTGTTTGTG (γ-catenin), AGCCCTCAGGAAGTTGATTAAA (ATF2) and CGAAGAACTCACTTCTCAGAA (ATF7). All constructs were sequence verified.
Antibodies, immunoblotting and immunoprecipitation
The following antibodies were purchased from commercial sources: mouse anti-β-catenin, mouse anti-γ-catenin (BD Biosciences), mouse anti-tubulin, mouse anti-Flag, mouse anti-HA (Sigma), rabbit anti-ATF2, rabbit anti-HA (Santa Cruz), goat anti-Axin (R&D Systems), rabbit anti-ATF7 (Abcam), mouse anti-LEF1 (Millipore). Mouse anti-myc clone 9E10 was obtained from the Mount Sinai Hybridoma Core Facility.
For immunoblot, cells were washed once with phosphate-buffered saline (PBS) and lysed on ice in lysis buffer containing 50 mM Hepes pH 7.6, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 20 mM NaF, 2 mM sodium orthovanadate, supplemented with the Complete Mini proteinase inhibitor cocktail tablets (Roche). Lysates were cleared by centrifugation at 20,000× g for 15 min at 4°C and protein concentrations were determined by using the Bio-Rad protein assay (Bio-Rad). Sodium dodecyl sulfate (SDS) loading buffer was added to equal amounts of lysate, followed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transfer to Immobilon-P membranes (Millipore). Antibodies used in immunoblot analysis were revealed by chemiluminescence using ECL Western Blotting Substrate (Thermo Fisher Scientific) or using the Odyssey Infrared Imaging System (LI-COR Biotechnology). For immunoprecipitation, equal amounts of cell lysates were incubated either with anti-Flag M2 agarose beads (Sigma) for 3 hrs at 4°C or with mouse anti-myc or mouse anti-HA antibodies for 1 hr at 4°C, followed by 3 hr incubation with Protein G Sepharose 4 Fast Flow beads (GE Healthcare). For LEF1 immunoprecipitation, equal amounts of cell lysates were incubated with aLEF1 antibody for 2 hrs at 4°C, followed by overnight incubation with Protein G Sepharose 4 Fast Flow beads. Beads were washed three times with lysis buffer and resuspended in SDS loading buffer, followed by SDS-PAGE and immunoblot.
Free β-catenin and reporter assays
Uncomplexed β-catenin was measured using glutathione S-transferase (GST)–E-cadherin or GST-TCF1 beads as described previously [70]. Briefly, bacterially expressed E-cadherin or TCF1 β-catenin binding domains fused to GST were bound to glutathione-Sepharose beads (GE Healthcare) and incubated with equal amounts of cell lysates. After pull-down, the samples were subjected to immunoblot analysis using anti-β-catenin antibody. For luciferase reporter assay, the cells were transfected or transduced with the TCF/LEF firefly luciferase reporters and the renilla luciferase control reporters or transfected with the Gal4 responsive vector pG5luc (Promega) and the renilla containing pBind plasmid. The cells were lysed and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. For GFP reporter assay, cells transduced with the Top or Fop TCF/LEF GFP reporter were transferred to polystyrene tubes (Falcon) and subjected to FACS analysis (FACScan; Becton Dickinson) using CellQuest 3.2 software (Becton Dickinson).
Colony forming assay
Growth of hematopoietic tumor cells in soft agarose was determined by seeding 2×103 cells per 60-mm dish in 0.5% sea plaque agarose (Cambrex) in RPMI supplemented with 10% FBS on a semisolid bottom layer of growth medium containing 1% agarose. Cells were fed once weekly with 0.3 mL of medium and stained after 17 days with iodonitrotetrazolium (Sigma). The cells containing Tet-off inducible DN-TCF4-GFP vector were grown overnight in the presence or the absence of 100 ng/ml doxycycline, followed by extensive washing in PBS. The cells were then counted and seeded in soft agarose dishes in the presence or the absence of 100 ng/ml doxycyline. The remaining cells were grown for one additional day with or without 100 ng/ml doxycyline, and FACS analysis was performed to assess the level of induction of DN-TCF4-GFP.
qPCR
Total RNA was extracted using the Trizol Reagent (Invitrogen), incubated with DNase I (Invitrogen) and reverse transcribed in the presence of random primers using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Quantitative PCR was performed using FastStart SYBR Green Master (Roche) on a MJ Opticon (Bio-Rad). The primers used for qPCR were: TATA Binding Protein (5′-ATCAGTGCCGTGGTTCGT and 5′-TTCGGAGAGTTCTGGGATTG), 18S (5′-GTAACCCGTTGAACCCAT and 5′-CCATCCAATCGGTAGTAG), Axin 2 (5′-ACTGCCCACACGATAAGGAG and 5′-CTGGCTATGTCTTTGGACCA), LEF1 (5′-CTTTATCCAGGCTGGTCTGC and 5′-TCGTTTTCCACCATGTTTCA).
Xenopus experiments
RNA was synthesized in vitro in the presence of cap analog using the mMessage mMachine kit (Ambion). Microinjection, explant dissection, cell culture and whole-mount antibody staining were performed as described [71]. The 12/101 antibody (Developmental Studies Hybridoma Bank, University of Iowa) was used at a 1∶1 dilution. Secondary antibody was a donkey anti-mouse IgG coupled to horseradish peroxidase (Jackson Laboratories), and was used at 1∶1000 dilution. Color reactions were performed using the Vector SG kit (Vector Laboratories). For RT-PCR, Xenopus laevis embryos were staged and harvested at appropriate stages according to morphological criteria. RNA was prepared using RNA Bee RNA isolation reagent (Tel-Test Inc.). RT-PCR was performed as described [72]. Primers used in this study are as follows: Xbrachyury (5′-GGATCGTTATCACCTCTG and 5′-GTGTAGTCTGTAGCAGCA), chordin (5′-CAGTCAGATGGAGCAGGATC and 5′-AGTCCCATTGCCCGAGTTGC), EF1-α (5′-CAGATTGGTGCTGGATATGC and 5′-ACTGCCTTGATGACTCCTAG), Xnr3 (5′-GTGAATCCACTTGTGCAGTT and 5′-ACAGAGCCAATCTCATGTGC).
Studies on Xenopus laevis embryos were performed in accordance with the guidelines of the American Veterinary Medical Association, and under the auspices of the Queens College Institutional Animal Care and Use Committee (IACUC). All experiments were undertaken with the highest regard for scientific, ethical, and humane principles.
Supporting Information
Zdroje
1. ReyaT, CleversH (2005) Wnt signalling in stem cells and cancer. Nature 434 : 843–850.
2. GrigoryanT, WendP, KlausA, BirchmeierW (2008) Deciphering the function of canonical Wnt signals in development and disease: conditional loss - and gain-of-function mutations of beta-catenin in mice. Genes & development 22 : 2308–2341.
3. CleversH, NusseR (2012) Wnt/Beta-Catenin Signaling and Disease. Cell 149 : 1192–1205.
4. CleversH (2006) Wnt/beta-catenin signaling in development and disease. Cell 127 : 469–480.
5. PolakisP (2012) Drugging Wnt signalling in cancer. The EMBO journal 31 : 2737–2746.
6. NusseR, VarmusHE (1982) Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31 : 99–109.
7. BaficoA, LiuG, GoldinL, HarrisV, AaronsonSA (2004) An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer cell 6 : 497–506.
8. AkiriG, CherianMM, VijayakumarS, LiuG, BaficoA, et al. (2009) Wnt pathway aberrations including autocrine Wnt activation occur at high frequency in human non-small-cell lung carcinoma. Oncogene 28 : 2163–2172.
9. VijayakumarS, LiuG, RusIA, YaoS, ChenY, et al. (2011) High-frequency canonical Wnt activation in multiple sarcoma subtypes drives proliferation through a TCF/β-catenin target gene, CDC25A. Cancer cell 19 : 601–612.
10. AngersS, MoonRT (2009) Proximal events in Wnt signal transduction. Nature reviews Molecular cell biology 10 : 468–477.
11. MacDonaldBT, TamaiK, HeX (2009) Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental cell 17 : 9–26.
12. LiVSW, NgSS, BoersemaPJ, LowTY, KarthausWR, et al. (2012) Wnt Signaling through Inhibition of beta-Catenin Degradation in an Intact Axin1 Complex. Cell 149 : 1245–1256.
13. CavalloRA, CoxRT, MolineMM, RooseJ, PolevoyGA, et al. (1998) Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395 : 604–608.
14. ArceL, YokoyamaNN, WatermanML (2006) Diversity of LEF/TCF action in development and disease. Oncogene 25 : 7492–7504.
15. ArchboldHC, YangYX, ChenL, CadiganKM (2012) How do they do Wnt they do?: regulation of transcription by the Wnt/β-catenin pathway. Acta physiologica (Oxford, England) 204 : 74–109.
16. ValentaT, HausmannG, BaslerK (2012) The many faces and functions of beta-catenin. The EMBO journal 31 : 2714–2736.
17. CadiganKM, WatermanML (2012) TCF/LEFs and Wnt Signaling in the Nucleus. Cold Spring Harb Perspect Biol 4: a007906.
18. DanielsDL, WeisWI (2005) Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nature structural & molecular biology 12 : 364–371.
19. HansonAJ, WallaceHA, FreemanTJ, BeauchampRD, LeeLA, et al. (2012) XIAP monoubiquitylates Groucho/TLE to promote canonical Wnt signaling. Molecular cell 45 : 619–628.
20. MosimannC, HausmannG, BaslerK (2009) Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10 : 276–286.
21. OlsonLE, TollkuhnJ, ScafoglioC, KronesA, ZhangJ, et al. (2006) Homeodomain-mediated beta-catenin-dependent switching events dictate cell-lineage determination. Cell 125 : 593–605.
22. BeildeckME, GelmannEP, ByersSW (2010) Cross-regulation of signaling pathways: an example of nuclear hormone receptors and the canonical Wnt pathway. Experimental cell research 316 : 1763–1772.
23. EssersMAG, de Vries-SmitsLMM, BarkerN, PoldermanPE, BurgeringBMT, et al. (2005) Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science (New York, NY) 308 : 1181–1184.
24. HoffmeyerK, RaggioliA, RudloffS, AntonR, HierholzerA, et al. (2012) Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science (New York, NY) 336 : 1549–1554.
25. BruhnL, MunnerlynA, GrosschedlR (1997) ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes & development 11 : 640–653.
26. LabbéE, LetamendiaA, AttisanoL (2000) Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proceedings of the National Academy of Sciences of the United States of America 97 : 8358–8363.
27. NishitaM, HashimotoMK, OgataS, LaurentMN, UenoN, et al. (2000) Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann's organizer. Nature 403 : 781–785.
28. NateriAS, Spencer-DeneB, BehrensA (2005) Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature 437 : 281–285.
29. ValentaT, GayM, SteinerS, DraganovaK, ZemkeM, et al. (2011) Probing transcription-specific outputs of beta-catenin in vivo. Genes Dev 25 : 2631–2643.
30. VerbeekS, IzonD, HofhuisF, Robanus-MaandagE, te RieleH, et al. (1995) An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 374 : 70–74.
31. OkamuraRM, SigvardssonM, GalceranJ, VerbeekS, CleversH, et al. (1998) Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity 8 : 11–20.
32. ReyaT, O'RiordanM, OkamuraR, DevaneyE, WillertK, et al. (2000) Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity 13 : 15–24.
33. CobasM, WilsonA, ErnstB, ManciniSpJC, MacDonaldHR, et al. (2004) Beta-catenin is dispensable for hematopoiesis and lymphopoiesis. The Journal of experimental medicine 199 : 221–229.
34. JeannetGg, SchellerM, ScarpellinoLo, DubouxSp, GardiolN, et al. (2008) Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood 111 : 142–149.
35. KochU, WilsonA, CobasM, KemlerR, MacdonaldHR, et al. (2008) Simultaneous loss of beta - and gamma-catenin does not perturb hematopoiesis or lymphopoiesis. Blood 111 : 160–164.
36. Ruiz-HerguidoC, GuiuJ, D'AltriT, Inglas-EsteveJ, DzierzakE, et al. (2012) Hematopoietic stem cell development requires transient Wnt/beta-catenin activity. The Journal of experimental medicine 209 : 1457–1468.
37. ZhaoC, BlumJ, ChenA, KwonHY, JungSH, et al. (2007) Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer cell 12 : 528–541.
38. WangY, KrivtsovAV, SinhaAU, NorthTE, GoesslingW, et al. (2010) The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science (New York, NY) 327 : 1650–1653.
39. YeungJ, EspositoMT, GandilletA, ZeisigBB, GriessingerE, et al. (2010) Beta-Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer cell 18 : 606–618.
40. HeidelFH, BullingerL, FengZ, WangZ, NeffTA, et al. (2012) Genetic and pharmacologic inhibition of beta-catenin targets imatinib-resistant leukemia stem cells in CML. Cell stem cell 10 : 412–424.
41. JamiesonCHM, AillesLE, DyllaSJ, MuijtjensM, JonesC, et al. (2004) Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. The New England journal of medicine 351 : 657–667.
42. ZhurinskyJ, ShtutmanM, Ben-Ze'evA (2000) Plakoglobin and beta-catenin: protein interactions, regulation and biological roles. J Cell Sci 113(Pt 18): 3127–3139.
43. MorganRG, PearnL, LiddiardK, PumfordSL, BurnettAK, et al. (2013) gamma-Catenin is overexpressed in acute myeloid leukemia and promotes the stabilization and nuclear localization of beta-catenin. Leukemia 27 : 336–343.
44. TravisA, AmsterdamA, BelangerC, GrosschedlR (1991) LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes Dev 5 : 880–894.
45. van de WeteringM, OosterwegelM, DooijesD, CleversH (1991) Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. The EMBO journal 10 : 123–132.
46. WatermanML, FischerWH, JonesKA (1991) A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes & development 5 : 656–669.
47. GrahamTA, WeaverC, MaoF, KimelmanD, XuW (2000) Crystal structure of a beta-catenin/Tcf complex. Cell 103 : 885–896.
48. McMahonAP, MoonRT (1989) Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell 58 : 1075–1084.
49. SokolS, ChristianJL, MoonRT, MeltonDA (1991) Injected Wnt RNA induces a complete body axis in Xenopus embryos. Cell 67 : 741–752.
50. WeaverC, KimelmanD (2004) Move it or lose it: axis specification in Xenopus. Development (Cambridge, England) 131 : 3491–3499.
51. StandleyHJ, DestréeO, KofronM, WylieC, HeasmanJ (2006) Maternal XTcf1 and XTcf4 have distinct roles in regulating Wnt target genes. Developmental biology 289 : 318–328.
52. GradlD, KönigA, WedlichD (2002) Functional diversity of Xenopus lymphoid enhancer factor/T-cell factor transcription factors relies on combinations of activating and repressing elements. The Journal of biological chemistry 277 : 14159–14171.
53. LiuF, van den BroekO, DestréeO, HopplerS (2005) Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/beta-catenin signalling in mesoderm development. Development (Cambridge, England) 132 : 5375–5385.
54. ChinnaduraiG (2002) CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Molecular cell 9 : 213–224.
55. HamadaF, BienzM (2004) The APC tumor suppressor binds to C-terminal binding protein to divert nuclear beta-catenin from TCF. Developmental cell 7 : 677–685.
56. StaalFJT, LuisTC, TiemessenMM (2008) WNT signalling in the immune system: WNT is spreading its wings. Nature reviews Immunology 8 : 581–593.
57. LuisTC, NaberBAE, RoozenPPC, BrugmanMH, de HaasEFE, et al. (2011) Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell stem cell 9 : 345–356.
58. IoannidisV, BeermannF, CleversH, HeldW (2001) The beta-catenin–TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nature immunology 2 : 691–697.
59. TakedaH, LyleS, LazarAJF, ZouboulisCC, SmythI, et al. (2006) Human sebaceous tumors harbor inactivating mutations in LEF1. Nature medicine 12 : 395–397.
60. NiemannC, OwensDM, HalskenJr, BirchmeierW, WattFM (2002) Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development (Cambridge, England) 129 : 95–109.
61. Lopez-BergamiP, LauE, RonaiZe (2010) Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nature Reviews Cancer 10 : 65–76.
62. PasseguéE, JochumW, Schorpp-KistnerM, Möhle-SteinleinU, WagnerEF (2001) Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell 104 : 21–32.
63. PasseguéE, WagnerEF, WeissmanIL (2004) JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 119 : 431–443.
64. SteidlU, RosenbauerF, VerhaakRGW, GuX, EbralidzeA, et al. (2006) Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nature genetics 38 : 1269–1277.
65. HoverterNP, TingJ-H, SundareshS, BaldiP, WatermanML (2012) A WNT/p21 circuit directed by the C-clamp, a sequence-specific DNA binding domain in TCFs. Molecular and cellular biology 32(18): 3648–62.
66. HikasaH, EzanJ, ItohK, LiX, KlymkowskyMW, et al. (2010) Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Developmental cell 19 : 521–532.
67. OtaS, IshitaniS, ShimizuN, MatsumotoK, ItohM, et al. (2012) NLK positively regulates Wnt/beta-catenin signalling by phosphorylating LEF1 in neural progenitor cells. The EMBO journal 31 : 1904–1915.
68. WuJQ, SeayM, SchulzVP, HariharanM, TuckD, et al. (2012) Tcf7 is an important regulator of the switch of self-renewal and differentiation in a multipotential hematopoietic cell line. PLoS genetics 8: e1002565–e1002565.
69. GrumolatoL, LiuG, MongP, MudbharyR, BiswasR, et al. (2010) Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes & development 24 : 2517–2530.
70. LiuG, BaficoA, HarrisVK, AaronsonSA (2003) A novel mechanism for Wnt activation of canonical signaling through the LRP6 receptor. Mol Cell Biol 23 : 5825–5835.
71. Hemmati-BrivanlouA, MeltonDA (1994) Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell 77 : 273–281.
72. WilsonPA, Hemmati-BrivanlouA (1995) Induction of epidermis and inhibition of neural fate by Bmp-4. Nature 376 : 331–333.
Štítky
Genetika Reprodukční medicína
Článek Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant IdentificationČlánek Bypass of 8-oxodGČlánek Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk GenesČlánek Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GGČlánek A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance inČlánek Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA OccupancyČlánek Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in StressČlánek Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Re-Ranking Sequencing Variants in the Post-GWAS Era for Accurate Causal Variant Identification
- Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations
- Past Visits Present: TCF/LEFs Partner with ATFs for β-Catenin–Independent Activity
- Functional Characterisation of Alpha-Galactosidase A Mutations as a Basis for a New Classification System in Fabry Disease
- A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits
- Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism
- Genic Intolerance to Functional Variation and the Interpretation of Personal Genomes
- Endogenous Stress Caused by Faulty Oxidation Reactions Fosters Evolution of 2,4-Dinitrotoluene-Degrading Bacteria
- Transposon Domestication versus Mutualism in Ciliate Genome Rearrangements
- Comparative Anatomy of Chromosomal Domains with Imprinted and Non-Imprinted Allele-Specific DNA Methylation
- An Essential Function for the ATR-Activation-Domain (AAD) of TopBP1 in Mouse Development and Cellular Senescence
- Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid
- Bypass of 8-oxodG
- Calpain-6 Deficiency Promotes Skeletal Muscle Development and Regeneration
- ATM Release at Resected Double-Strand Breaks Provides Heterochromatin Reconstitution to Facilitate Homologous Recombination
- Generation of Tandem Direct Duplications by Reversed-Ends Transposition of Maize Elements
- Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling
- Integrated Model of and Inherited Genetic Variants Yields Greater Power to Identify Risk Genes
- High-Throughput Genetic and Gene Expression Analysis of the RNAPII-CTD Reveals Unexpected Connections to SRB10/CDK8
- Dynamic Rewiring of the Retinal Determination Network Switches Its Function from Selector to Differentiation
- β-Catenin-Independent Activation of TCF1/LEF1 in Human Hematopoietic Tumor Cells through Interaction with ATF2 Transcription Factors
- Genetic Mapping of Specific Interactions between Mosquitoes and Dengue Viruses
- A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in
- Origin and Functional Diversification of an Amphibian Defense Peptide Arsenal
- Myc-Driven Overgrowth Requires Unfolded Protein Response-Mediated Induction of Autophagy and Antioxidant Responses in
- Integrative Modeling of eQTLs and Cis-Regulatory Elements Suggests Mechanisms Underlying Cell Type Specificity of eQTLs
- Species and Population Level Molecular Profiling Reveals Cryptic Recombination and Emergent Asymmetry in the Dimorphic Mating Locus of
- Ras-Induced Changes in H3K27me3 Occur after Those in Transcriptional Activity
- Characterization of the p53 Cistrome – DNA Binding Cooperativity Dissects p53's Tumor Suppressor Functions
- Global Analysis of Fission Yeast Mating Genes Reveals New Autophagy Factors
- Deficiency Suppresses Intestinal Tumorigenesis
- Introns Regulate Gene Expression in in a Pab2p Dependent Pathway
- Meiotic Recombination Initiation in and around Retrotransposable Elements in
- Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery
- Comparative Genomic and Functional Analysis of 100 Strains and Their Comparison with Strain GG
- A Model-Based Analysis of GC-Biased Gene Conversion in the Human and Chimpanzee Genomes
- Masculinization of the X Chromosome in the Pea Aphid
- The Architecture of a Prototypical Bacterial Signaling Circuit Enables a Single Point Mutation to Confer Novel Network Properties
- Distinct SUMO Ligases Cooperate with Esc2 and Slx5 to Suppress Duplication-Mediated Genome Rearrangements
- The Yeast Environmental Stress Response Regulates Mutagenesis Induced by Proteotoxic Stress
- Mediator Directs Co-transcriptional Heterochromatin Assembly by RNA Interference-Dependent and -Independent Pathways
- The Genome of and the Basis of Host-Microsporidian Interactions
- Regulation of Sister Chromosome Cohesion by the Replication Fork Tracking Protein SeqA
- Neuronal Reprograming of Protein Homeostasis by Calcium-Dependent Regulation of the Heat Shock Response
- A Nuclear Calcium-Sensing Pathway Is Critical for Gene Regulation and Salt Stress Tolerance in
- Cross-Species Array Comparative Genomic Hybridization Identifies Novel Oncogenic Events in Zebrafish and Human Embryonal Rhabdomyosarcoma
- : A Mouse Strain with an Ift140 Mutation That Results in a Skeletal Ciliopathy Modelling Jeune Syndrome
- The Relative Contribution of Proximal 5′ Flanking Sequence and Microsatellite Variation on Brain Vasopressin 1a Receptor () Gene Expression and Behavior
- Combining Quantitative Genetic Footprinting and Trait Enrichment Analysis to Identify Fitness Determinants of a Bacterial Pathogen
- The Innocence Project at Twenty: An Interview with Barry Scheck
- Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy
- GUESS-ing Polygenic Associations with Multiple Phenotypes Using a GPU-Based Evolutionary Stochastic Search Algorithm
- H2A.Z Acidic Patch Couples Chromatin Dynamics to Regulation of Gene Expression Programs during ESC Differentiation
- Identification of DSB-1, a Protein Required for Initiation of Meiotic Recombination in , Illuminates a Crossover Assurance Checkpoint
- Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories
- Global Analysis of the Sporulation Pathway of
- Genetic Circuits that Govern Bisexual and Unisexual Reproduction in
- Deletion of microRNA-80 Activates Dietary Restriction to Extend Healthspan and Lifespan
- Fifty Years On: GWAS Confirms the Role of a Rare Variant in Lung Disease
- The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option
- Gene Expression Regulation by Upstream Open Reading Frames and Human Disease
- Sociogenomics of Cooperation and Conflict during Colony Founding in the Fire Ant
- The Intronic Long Noncoding RNA Recruits PRC2 to the Promoter, Reducing the Expression of and Increasing Cell Proliferation
- The , p.E318G Variant Increases the Risk of Alzheimer's Disease in -ε4 Carriers
- The Wilms Tumor Gene, , Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans
- Reversible and Rapid Transfer-RNA Deactivation as a Mechanism of Translational Repression in Stress
- QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing
- Genome Wide Association Identifies Novel Loci Involved in Fungal Communication
- Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins
- A Recessive Founder Mutation in Regulator of Telomere Elongation Helicase 1, , Underlies Severe Immunodeficiency and Features of Hoyeraal Hreidarsson Syndrome
- Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci
- Causal and Synthetic Associations of Variants in the Gene Cluster with Alpha1-antitrypsin Serum Levels
- Hard Selective Sweep and Ectopic Gene Conversion in a Gene Cluster Affording Environmental Adaptation
- Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils
- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1
- Ras1 Acts through Duplicated Cdc42 and Rac Proteins to Regulate Morphogenesis and Pathogenesis in the Human Fungal Pathogen
- The DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance
- Recurrent Modification of a Conserved -Regulatory Element Underlies Fruit Fly Pigmentation Diversity
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- The Conditional Nature of Genetic Interactions: The Consequences of Wild-Type Backgrounds on Mutational Interactions in a Genome-Wide Modifier Screen
- A Critical Function of Mad2l2 in Primordial Germ Cell Development of Mice
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
- Vitellogenin Underwent Subfunctionalization to Acquire Caste and Behavioral Specific Expression in the Harvester Ant
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Chromosomal Copy Number Variation, Selection and Uneven Rates of Recombination Reveal Cryptic Genome Diversity Linked to Pathogenicity
- Genome-Wide DNA Methylation Analysis of Systemic Lupus Erythematosus Reveals Persistent Hypomethylation of Interferon Genes and Compositional Changes to CD4+ T-cell Populations
- Associations of Mitochondrial Haplogroups B4 and E with Biliary Atresia and Differential Susceptibility to Hydrophobic Bile Acid
- A Role for CF1A 3′ End Processing Complex in Promoter-Associated Transcription
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



