-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
Recent work has identified changes in the metabolism of the aromatic amino acid tyrosine as a risk factor for diabetes and a contributor to the development of liver cancer. While these findings could suggest a role for tyrosine as a direct regulator of the behavior of cells and tissues, evidence for this model is currently lacking. Through the use of RNAi and genetic mutants, we identify tatn-1, which is the worm ortholog of tyrosine aminotransferase and catalyzes the first step of the conserved tyrosine degradation pathway, as a novel regulator of the dauer decision and modulator of the daf-2 insulin/IGF-1-like (IGFR) signaling pathway in Caenorhabditis elegans. Mutations affecting tatn-1 elevate tyrosine levels in the animal, and enhance the effects of mutations in genes that lie within the daf-2/insulin signaling pathway or are otherwise upstream of daf-16/FOXO on both dauer formation and worm longevity. These effects are mediated by elevated tyrosine levels as supplemental dietary tyrosine mimics the phenotypes produced by a tatn-1 mutation, and the effects still occur when the enzymes needed to convert tyrosine into catecholamine neurotransmitters are missing. The effects on dauer formation and lifespan require the aak-2/AMPK gene, and tatn-1 mutations increase phospho-AAK-2 levels. In contrast, the daf-16/FOXO transcription factor is only partially required for the effects on dauer formation and not required for increased longevity. We also find that the controlled metabolism of tyrosine by tatn-1 may function normally in dauer formation because the expression of the TATN-1 protein is regulated both by daf-2/IGFR signaling and also by the same dietary and environmental cues which influence dauer formation. Our findings point to a novel role for tyrosine as a developmental regulator and modulator of longevity, and support a model where elevated tyrosine levels play a causal role in the development of diabetes and cancer in people.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1004020
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004020Summary
Recent work has identified changes in the metabolism of the aromatic amino acid tyrosine as a risk factor for diabetes and a contributor to the development of liver cancer. While these findings could suggest a role for tyrosine as a direct regulator of the behavior of cells and tissues, evidence for this model is currently lacking. Through the use of RNAi and genetic mutants, we identify tatn-1, which is the worm ortholog of tyrosine aminotransferase and catalyzes the first step of the conserved tyrosine degradation pathway, as a novel regulator of the dauer decision and modulator of the daf-2 insulin/IGF-1-like (IGFR) signaling pathway in Caenorhabditis elegans. Mutations affecting tatn-1 elevate tyrosine levels in the animal, and enhance the effects of mutations in genes that lie within the daf-2/insulin signaling pathway or are otherwise upstream of daf-16/FOXO on both dauer formation and worm longevity. These effects are mediated by elevated tyrosine levels as supplemental dietary tyrosine mimics the phenotypes produced by a tatn-1 mutation, and the effects still occur when the enzymes needed to convert tyrosine into catecholamine neurotransmitters are missing. The effects on dauer formation and lifespan require the aak-2/AMPK gene, and tatn-1 mutations increase phospho-AAK-2 levels. In contrast, the daf-16/FOXO transcription factor is only partially required for the effects on dauer formation and not required for increased longevity. We also find that the controlled metabolism of tyrosine by tatn-1 may function normally in dauer formation because the expression of the TATN-1 protein is regulated both by daf-2/IGFR signaling and also by the same dietary and environmental cues which influence dauer formation. Our findings point to a novel role for tyrosine as a developmental regulator and modulator of longevity, and support a model where elevated tyrosine levels play a causal role in the development of diabetes and cancer in people.
Introduction
The aromatic amino acid tyrosine serves many metabolic roles including being a building block for protein synthesis, a source of energy, and a precursor for the synthesis of melanin and several neurotransmitters including dopamine and other catecholamines. Beyond these currently known functions for tyrosine, recent work has suggested that tyrosine could also play regulatory roles in both metabolism and the control of cell proliferation. Specifically, in people elevated serum tyrosine levels occur with obesity and represent a risk factor for the development of diabetes [1]–[6]. Additionally, the enzyme tyrosine aminotransferase (TAT), which acts to normally convert tyrosine to energy, has been identified as a tumor suppressor gene which acts to promote apoptosis and prevent the development of hepatocellular carcinoma [7]. How changes in tyrosine metabolism could contribute to these disease processes is currently unknown, but it is possible that levels of this amino acid could play a direct regulatory role for the behavior of specific cells and tissues. While consistent with the available data, direct evidence for this model is currently lacking.
The nematode Caenorhabditis elegans normally progresses through four larval stages before developing into a reproductive adult animal. However specific cues, such as crowding, low food availability, or elevated temperature, can be sensed by the developing worm and lead to developmental arrest in a diapause state called a dauer larva [8]–[11]. Entry into dauer permits worms to delay the completion of development and the initiation of reproduction in environments which are not favorable, and instead the animals can survive as a dauer for up to several months before resuming normal development when conditions become favorable for reproductive success. This developmental decision requires a complicated interplay of sensory neurons with specific cGMP, TGF-β, and insulin-like signaling cascades controlling the choice of reproductive versus dauer development [9], [10].
In the worm, the daf-2 insulin/IGF-1 receptor (IGFR) signaling pathway is involved in both dauer development and adult longevity [12]–[14]. Active signaling through the pathway during development enables animals to reach reproductive adulthood whereas reductions in daf-2/IGFR signaling due to either environmental triggers or genetic mutations lead to arrest as a dauer [9], [14]. In adult worms, daf-2/IGFR signaling is a major modulator of longevity and mutations impairing the pathway can result in 100% increases in lifespan [13].
At a molecular level, the daf-2/IGFR pathway consists of daf-28 and other insulin-like peptides, which are thought to act as ligands for the DAF-2 insulin/IGF-1 receptor [15]–[18]. Downstream of daf-2/IGFR, is the age-1 PI3 kinase and a kinase cascade consisting of the phosphoinositide-dependent kinase pdk-1 and the protein kinase B genes akt-1 and akt-2 (Figure 1A) [19]–[22]. Both akt-1 and akt-2 normally act to phosphorylate the DAF-16 FOXO transcription factor which leads to its retention in the cytoplasm [23]–[25]. Reductions in either daf-2/IGFR or combined akt-1 and akt-2 activity result in the entry of DAF-16/FOXO into the nucleus and strong activation of DAF-16/FOXO target genes [23]–[25]. In contrast, loss of only akt-1 activity leads to the translocation of DAF-16/FOXO into the nucleus but a lesser increase in the expression of DAF-16/FOXO target genes (Figure 1A and Table S1) [26], [27]. This finding suggested that additional pathways could be involved in controlling the transcriptional activity of DAF-16/FOXO (Figure 1A). One group of potential regulators is the eak (enhancer of akt-1 null) genes, which were identified in a forward genetic screen and act in a non-cell autonomous manner to control the transcriptional activity of nuclear localized daf-16/FOXO (Figure 1A) [26]–[29]. The identified eak genes lack structural homology to one another and appear to lie in one or more poorly characterized pathways that act in parallel to akt-1. The identification of the eak genes suggests that additional novel pathways either downstream or parallel to insulin signaling may await discovery.
Fig. 1. Diagram of daf-2/IGFR signaling pathway and tyrosine metabolic pathway. 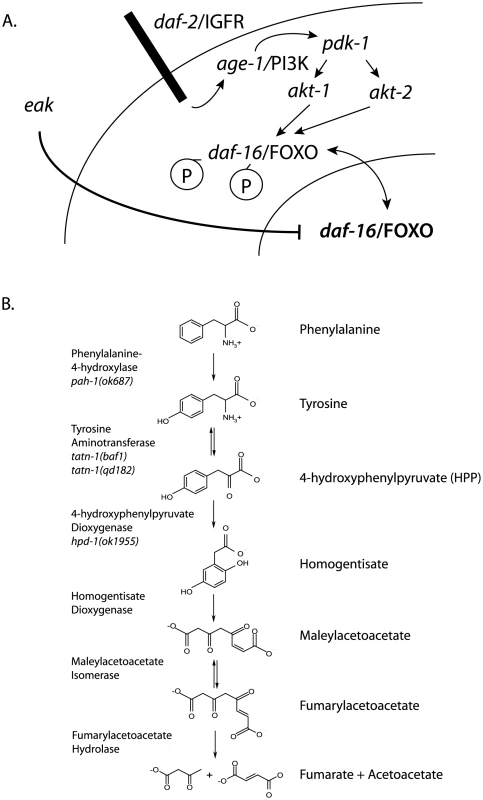
(A) For dauer formation in C. elegans the daf-2/IGFR receptor lies upstream of a PI3 kinase signaling cascade consisting of age-1/PI3 kinase, the phosphoinoside-dependent kinase pdk-1, and the protein kinase B family kinases akt-1 and akt-2. Both AKT-1 and AKT-2 act to phosphorylate DAF-16/FOXO and prevent entry of this protein into the nucleus. Inhibition of AKT-1 leads to entry of DAF-16/FOXO into the nucleus without the activation of DAF-16 target genes. This suggested that other pathways acted to control the transcriptional activity of nuclear DAF-16/FOXO. The eak genes are a group of structurally unrelated genes which act in a cell non-autonomous manner to restrain the transcriptional activity of nuclear DAF-16/FOXO through an undefined molecular mechanism. (B) Tyrosine is degraded to fumarate and acetoacetate via a five step degradation pathway. Shown are the names and structures of the intermediates as well as the names of the enzymes which catalyze each step. Also shown are mutant alleles affecting enzymes in the pathway that are studied in this work. In both vertebrates and worms, there is data suggesting a link between tyrosine metabolism and insulin signaling. TAT, which catalyzes the first step in a conserved degradation pathway that converts tyrosine to fumarate and acetoacetate (Figure 1B), has been well studied as a target of regulation by insulin signaling in vertebrates with insulin effects seen at both the transcriptional and translational level [30]–[38]. Further, in C. elegans the hpd-1 gene, which encodes the enzyme 4-hydroxyphenylpyruvate dioxygenase and lies immediately downstream of TAT in the tyrosine degradation pathway, is a target gene for the daf-16/FOXO transcription factor and positively regulated by daf-2/IGFR signaling [39]. The down-regulation of hpd-1 in daf-2/IGFR mutants could lead to a reduction in tyrosine clearance and could, at least in part, account for the increases tyrosine levels observed in these animals [40]. Furthermore, the inhibition of hpd-1 by RNAi was shown to both extend lifespan and delay dauer exit through unknown mechanisms [39]. Hence, tyrosine metabolism appears to be actively controlled by insulin signaling though the consequences of this regulation are currently unclear.
In our work, we identify tatn-1, which is the worm ortholog of TAT, to be a novel dauer formation regulator that is under the control of several dauer-inducing stimuli, including daf-2/IGFR signaling, and ultimately regulates tyrosine levels in the worm. We further find that tatn-1 mutations enhance the dauer-formation and lifespan phenotypes of both daf-2/IGFR and eak mutants suggesting that elevated tyrosine levels have inhibitory effects on insulin signaling. These effects require the aak-2/AMPK gene, and tatn-1 mutants have elevated levels of the activated phospho-AAK-2 protein consistent with activation of AAK-2 signaling in response to elevated tyrosine. The activation of AAK-2 may lead to effects on the downstream transcription factors daf-16/FOXO and crh-1/CREB. We see a partial dependence on daf-16/FOXO for some tatn-1 phenotypes and activation of daf-16/FOXO target genes, and the loss of crh-1/CREB, which is inhibited by activated aak-2, mimics some tatn-1 phenotypes. Together our findings establish a novel role for tyrosine as a metabolic signal that influences insulin signaling, development, and lifespan through effects on aak-2/AMPK signaling. While further study is necessary, our results also suggest that the recently observed associations between tyrosine metabolism and both diabetes and cancer are due to elevated tyrosine levels playing a direct causal role in disease pathogenesis.
Results
Reduced tyrosine aminotransferase activity promotes dauer arrest
The eak genes were identified in a genetic screen as enhancers of the weak dauer formation phenotype shown by akt-1 mutants, and these genes normally act to suppress the transcriptional activity of nuclear localized daf-16/FOXO [26]–[29]. To identify new genes that act in parallel to eak genes to control dauer formation, we performed a genome-wide RNAi screen for gene inactivations that enhance the weak dauer-constitutive phenotype of the eak-4(mg348) mutant. Since RNAi of dauer-constitutive genes typically yields a weaker phenotype than the corresponding mutants, we constructed an eri-3(mg408); eak-4(mg348) double mutant to enhance the sensitivity of the eak-4 mutant to RNAi [41], [42]. Control experiments demonstrated that RNAi inhibition of daf-2/IGFR, akt-1, or the 14-3-3 gene ftt-2 enhance dauer arrest by the eri-3; eak-4 mutants whereas RNAi inhibition of daf-7, daf-9, or daf-11, which encode components of TGF-β, dafachronic acid, and cGMP pathways, respectively, do not (Figure 2A). These data suggested that the screen could be enriched for genes that act in the daf-2/IGFR pathway to control dauer arrest.
Fig. 2. Tyrosine aminotransferase mutations enhance eak dauer and lifespan phenotypes. 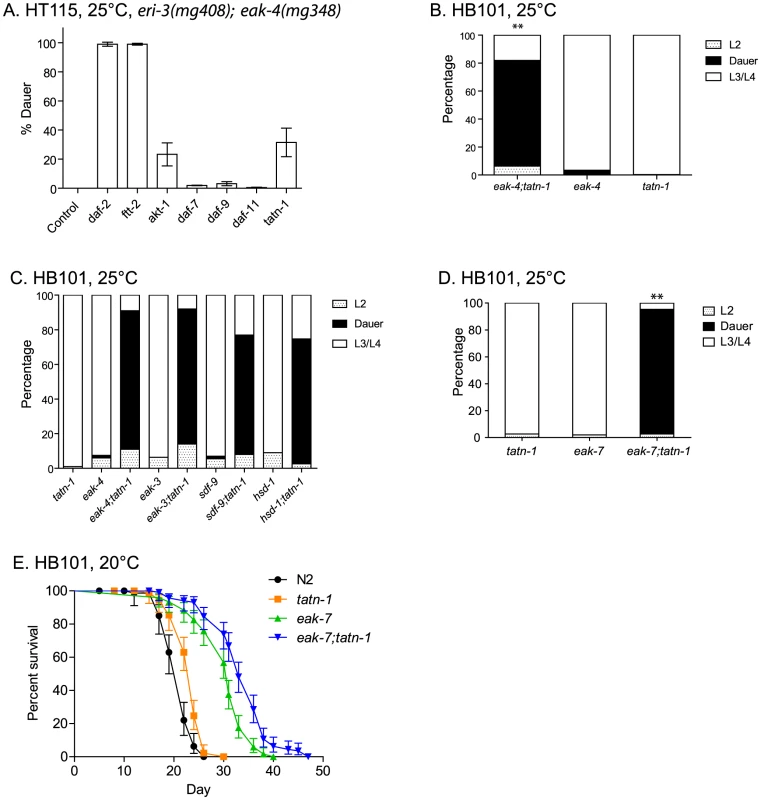
(A) Effect of treatment of eri-3(mg408); eak-4(mg348) mutants with RNAi clones for daf-2, ftt-2, akt-1, daf-7, daf-9, daf-11, and tatn-1 on dauer arrest at 25°C. Bars represent mean percentage of dauers observed in two trials, and the error bars represent standard deviation. The empty vector RNAi vector was used as a negative control. (B) Enhanced dauer formation by eak-4(mg348);tatn-1(baf1) versus the eak-4(mg348) or tatn-1(baf1) mutations alone. ** p<0.001 for pairwise Fisher's exact test. (C) tatn-1 mutations enhance dauer arrest by eak-3(mg344), sdf-9(mg337), and hsd-1(mg337) mutants. ** p<0.001 for each double mutant compared to its respective eak single mutant by Fisher's exact test. (D) tatn-1 enhances eak-7(tm3188) dauer arrest. ** p<0.001 for comparisons between eak-7;tatn-1 and either tatn-1 or eak-7 alone. (E) tatn-1(baf1) extends the lifespan of both wild type and eak-7 worms. Shown are survival curves for wild type N2, tatn-1(baf1), eak-7(3188), and eak-7(3188); tatn-1(baf1), with error bars showing the 95% confidence intervals for each point. The mean survival for N2, tatn-1, eak-7, and eak-7;tatn-1 are 21.0, 23.0, 30.0, and 33.5 days, respectively. p<0.001 for all comparisons by log-rank test. Among the RNAi clones identified from the screen was tatn-1, which encodes the worm ortholog of tyrosine aminotransferase. Inhibition of tatn-1 by RNAi enhanced dauer arrest by eri-3; eak-4 mutants to the same extent as akt-1 RNAi (Figure 2A) [43]. Tyrosine aminotransferase is the first enzyme in the conserved five step tyrosine degradation pathway present in worms and other eukaryotes, and this enzyme catalyzes the deamination of tyrosine to produce 4-hydroxyphenylpyruvate (Figure 1B). The subsequent steps in the degradation pathway convert 4-hydroxyphenylpyruvate into fumarate and acetoacetate which can be ultimately metabolized by the Krebs' cycle (Figure 1B).
Similarly to the effects of tatn-1 RNAi, we also found that the tatn-1(baf1) mutation, which has a P224S mutation in a conserved region of the protein and is likely a partial loss of function allele [43], [44], also promoted dauer arrest by the eak-4(mg348) mutants (Figure 2B and Table S2). Interestingly, the interaction between tatn-1 and eak-4 was strongly influenced by worm diet with diets consisting of the E. coli K12-derived HT115 or K12-B hybrid HB101 bacterial strains showing the strongest interaction (Figure S1). Additionally, we observed that a second tatn-1 allele, tatn-1(qd182), both enhanced dauer arrest by the eak-4 mutant and also had a weak effect on dauer formation in isolation (Figure S2). The tatn-1(qd182) allele encodes a protein with a G171E mutation affecting a highly conserved glycine residue (Figure S2). Together these findings demonstrate a novel role for tyrosine aminotransferase as a regulator of the dauer development decision.
We then tested whether tatn-1 interacted with other eak genes via the construction of tatn-1; eak mutants. We found that tatn-1(baf1) enhanced the dauer-constitutive phenotype of all eak mutants tested, including eak-4(mg348), eak-3(mg344),eak-5/sdf-9(mg337), eak-2/hsd-1(mg433), and eak-7(tm3188) (Figure 2C and 2D). eak-7 showed a particularly strong interaction with tatn-1, with 92.4% of the population forming dauers at 25°C (Figure 2D), and we observed dauers in eak-7; tatn-1 in cultures growing at lower temperatures or on other worm diets. These data suggest that tatn-1 is a general enhancer of dauer formation by eak mutants.
In addition to enhancing dauer formation, eak-7 mutations, but not other eak mutations, extend the lifespan of wild-type and akt-1 mutant worms [27]. We tested whether a tatn-1 mutation also enhanced the lifespan of eak-7 mutants by conducting survival assays using wild type N2, tatn-1(baf1), eak-7(tm3188), and eak-7(tm3188); tatn-1(baf1) worm populations. We found that tatn-1, eak-7, and eak-7; tatn-1 mutants all showed increased longevity relative to N2 (mean survivals of 21.0, 23.0, 30.0, and 33.5 days, respectively) (Figure 2E and Table S3). Specifically, tatn-1 extends mean survival 10.4%, eak-7 extends mean survival 43.1%, and eak-7; tatn-1 increases survival 59.2% over wild type (Figure 2E). These findings demonstrate a novel role for tatn-1 in modulating lifespan and also demonstrate that the effects of the eak-7 – tatn-1 genetic interaction also influence adult longevity.
Tyrosine aminotransferase interacts with insulin signaling in worms
Mutations in the eak genes enhance the dauer formation phenotype of loss-of-function mutations affecting genes in the daf-2/IGFR signaling pathway, so we tested whether tatn-1 mutations also enhanced daf-2/IGFR mutant phenotypes [28]. We used the daf-2(e1368) allele which has a strong dauer-constitutive phenotype when grown at 25°C but a weaker phenotype when grown at lower temperatures. Compared to tatn-1(baf1) and daf-2(e1368) alone, we found that daf-2(e1368); tatn-1(baf1) mutants showed increased levels of dauer formation when grown at 23°C (Figure 3A). Further, we found that the tatn-1 mutation extends the adult lifespan of worms treated with daf-2 RNAi starting at day 1 of adulthood (Figure 3B). We used RNAi treatment due to the high levels of dauer arrest that we observed with the daf-2(e1368); tatn-1(baf1) animals. In these RNAi experiments, the mean survival of worms were 22.0 days for N2 on control RNAi, 24.0 days for tatn-1 on control RNAi, 30.5 days for N2 on daf-2 RNAi, and 37.0 days for tatn-1 on daf-2 RNAi (Figure 3B and Table S3). Hence, while daf-2 RNAi treatment extends the survival of wild type N2 worms by 37.4%, the inclusion of the tatn-1 allele further extends the lifespan of daf-2(RNAi) treated worms by an additional 21%. These findings show a genetic interaction between tatn-1 and daf-2/IGFR with impaired tyrosine degradation enhancing the daf-2/IGFR dauer formation and lifespan phenotypes.
Fig. 3. The tatn-1 mutant enhances dauer formation and lifespan of worms with impaired daf-2/IGFR signaling. 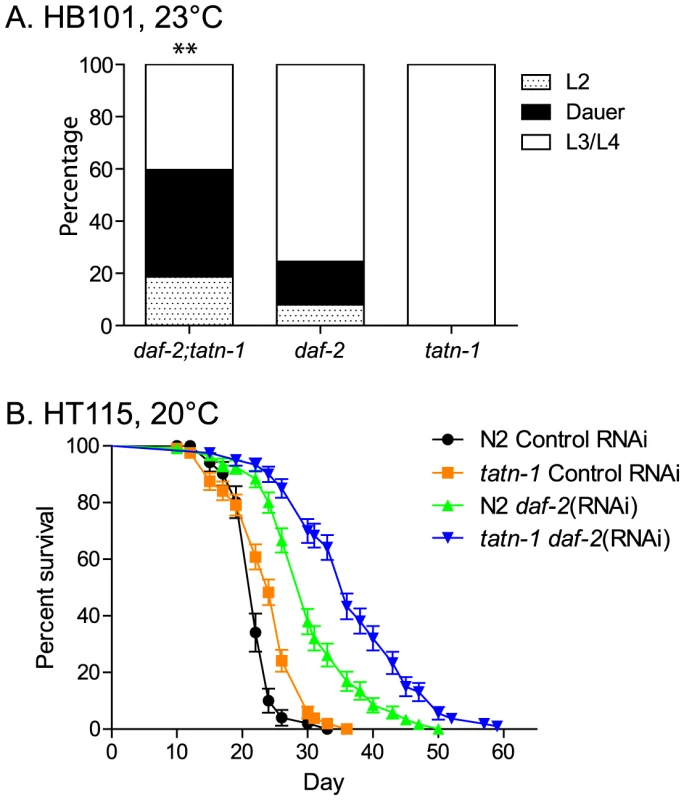
(A) Enhanced dauer formation by daf-2(e1368); tatn-1(baf1) compared to daf-2(e1368) grown at 23°C for 3 days. ** p<0.01 by Fisher's exact test. (B) tatn-1 extends the lifespan of worms treated with daf-2 RNAi from day 1 of adulthood. The mean survival for N2 control RNAi, tatn-1(baf1) control RNAi, N2 daf-2(RNAi), and tatn-1(baf1) daf-2(RNAi) are 22.0, 24.0, 30.5, and 37.0 days, respectively. p<0.001 for N2 vs. tatn-1 on control RNAi and N2 daf-2(RNAi) vs. tatn-1 daf-2(RNAi) by log-rank test. tatn-1 likely does not alter PI3 kinase or TGF-β signaling
Since the eak mutations interact with akt-1 mutations to enhance dauer formation, the effects of tatn-1 mutations could be due to inhibition of the PI3 kinase signaling pathway (Figure 1A) [20]–[22], [28], [45]. To test this possibility, we looked for genetic interactions between loss-of-function and gain-of-function mutations affecting the PI3 kinase pathway with tatn-1. First, we constructed tatn-1 mutants containing the loss-of-function mutations affecting the PKB genes akt-1 and akt-2, and the related kinase sgk-1. We found that none of these genes interacts strongly with a tatn-1 mutation to enhance dauer arrest (Figure 4A). This finding suggests that tatn-1 is not an upstream regulator of either akt-1 or akt-2. To further test whether tyrosine affected the PI3 kinase signaling cascade, we tested whether the eak-4(mg348); tatn-1(baf1) interaction was blocked by gain-of-function mutations in either pdk-1 or akt-1. These mutations were identified in genetic screens as suppressors of the dauer-constitutive phenotype of an age-1/PI3K null mutant [20], [21]. We constructed both eak-4(mg348); pdk-1(mg142), tatn-1(baf1) and eak-4(mg348); akt-1(mg144); tatn-1(baf1) mutants and examined the effects of these mutations on dauer formation. We found that pdk-1(mg142), which is a dominant gain-of-function allele of 3-phosphoinositide-dependent kinase and lies upstream of akt-1, akt-2, and sgk-1, had little effect on dauer formation by the eak-4(mg348); tatn-1(baf1) mutants (70% with pdk-1(mg142) versus 75% without) (Figure 4B). We further found that akt-1(mg144) had at most a modest effect on dauer formation by eak-4(mg348); tatn-1(baf1) mutants (51% with akt-1(mg144) versus 77.6% without) (Figure 4C). Together these results suggest that the interaction between eak-4 and tatn-1 likely does not involve changes in the PI3 kinase signaling cascade.
Fig. 4. tatn-1 effects on development do not require changes in PI3 kinase signaling. 
(A) tatn-1(baf1) does not augment dauer arrest by mutants with loss-of-function mutations in the protein kinase B genes akt-1, akt-2, or sgk-1. (B) The pdk-1(mg142) gain-of-function allele minimally affects dauer formation by eak-4(mg348); tatn-1(baf1) mutants. % dauers observed was 75.6% in eak-4; tatn-1 and 70.7% in eak-4; pdk-1(mg142); tatn-1. ** p<0.001 by Fisher's exact test. (C) The akt-1(mg144) gain-of-function allele weakly inhibited dauer formation by eak-4(mg348); tatn-1(baf1) mutants. % dauers observed was 77.6% in eak-4; tatn-1 and 51% in eak-4; akt-1(mg144); tatn-1. In addition to the daf-2/IGFR signaling pathway, dauer formation in worms is also regulated by a TGF-β signaling pathway [9]–[11]. While traditionally these two pathways are viewed as independent, more recent work has indicated that cross-talk between the pathways may occur. Specifically, genes in one pathway, such as sdf-9/eak-5 or pdp-1, have been shown to augment the dauer formation phenotypes of genes in the other pathway [46], [47]. Notably, the pdp-1 phosphatase was identified from an RNAi screen using daf-2/IGFR pathway mutants as was tatn-1. To test whether tatn-1 could act in the TGF-β signaling pathway, we blocked this pathway with either the daf-3/SMAD or daf-5/Sno mutations [41], [48], [49]. We found that neither mutant reduced dauer formation by the eak-4; tatn-1 mutants (Figure S3), which is consistent with tatn-1 acting independently of the TGF-β pathway.
AAK-2/AMPK is activated in tatn-1 mutants and required for effects on dauer formation and longevity
Since tatn-1 did not interact with akt-1, akt-2, or sgk-1, we looked for alternate signaling pathways that could be activated by a tatn-1 mutation, and then interact with the daf-2/IGFR signaling pathway. The AMP-activated protein kinase (AMPK) ortholog AAK-2 was considered as a candidate because aak-2 interacts with daf-2 signaling, plays roles in dauer development, modulates worm longevity, and acts in part through the daf-16/FOXO transcription factor which is part of the daf-2/IGFR signaling pathway [50]–[53]. To test the involvement of aak-2/AMPK, we compared dauer formation between eak-4(mg348); tatn-1(baf1) and eak-4(mg348); tatn-1(baf1), aak-2(gt33) mutants. We found that loss of aak-2 strongly reduced dauer arrest from 84.7% to 10.7% (Figure 5A and Table S2). The aak-2 mutation also reduced dauer formation by an eak-4(mg348); tatn-1(qd182) mutant though to a lesser degree than with the tatn-1(baf1) allele (Figure S4). This may be due to the tatn-1(qd182) allele being a stronger loss-of-function allele than tatn-1(baf1) as suggested by the developmental delay phenotype shown by tatn-1(qd182) and the higher tyrosine levels found in this mutant (Figure S4 and below). Together these findings support a necessary role for aak-2/AMPK for worms to respond to reductions in tatn-1 activity, especially with weaker alleles.
Fig. 5. aak-2 activity is necessary and sufficient for tatn-1 effects on development and longevity. 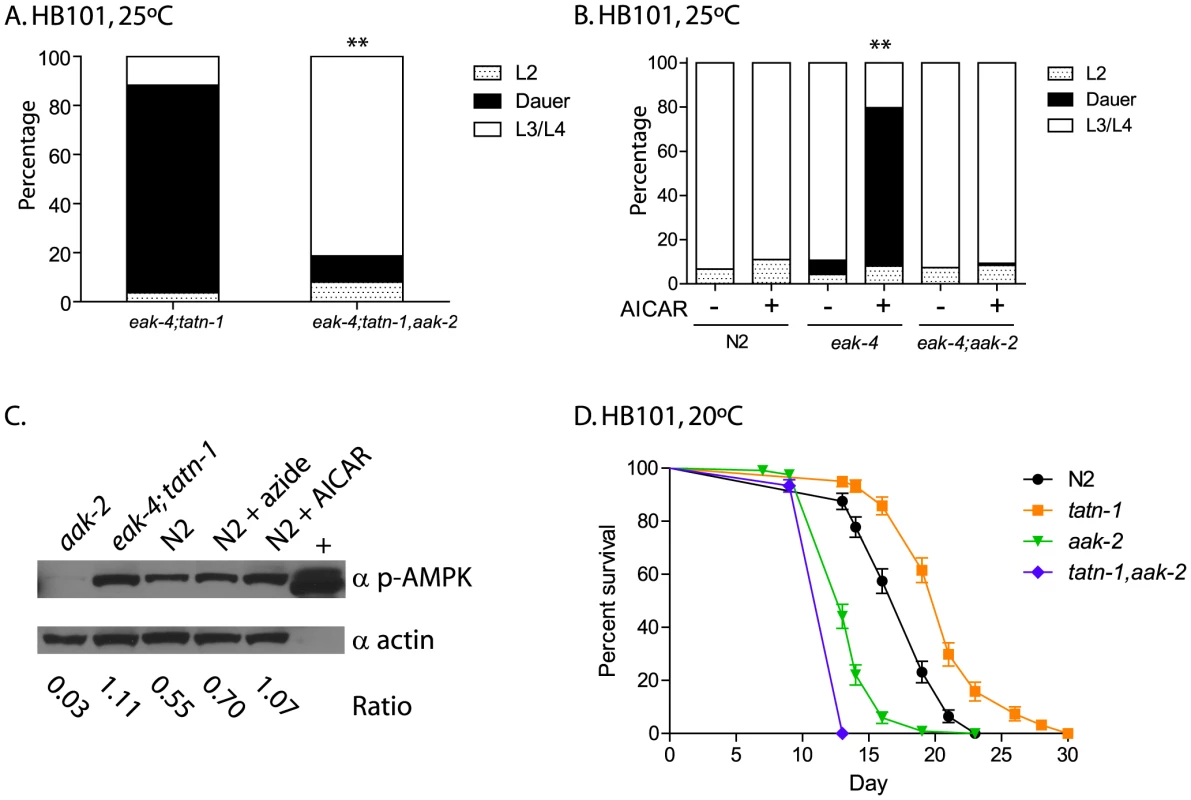
(A) The effects of tatn-1(baf1) on dauer formation require the aak-2/AMPK gene. ** p<0.001 by Fisher's exact test. (B) Activation of aak-2 with the AMPK agonist AICAR (0.125 mM) mimics the effects of tatn-1 on eak-4(mg348) mutants. The effect of AICAR depends on aak-2/AMPK because an eak-4; aak-2 mutant fails to respond to AICAR. ** p<0.001 for comparison of eak-4 with and without AICAR by Fisher's exact test. (C) Levels of phospho-AAK-2 are increased in eak-4(mg348); tatn-1(baf1) L2 larval worms or N2 L2 larval worms treated with 1 mM AICAR compared to untreated N2 L2 larval worms. Levels of phospho-AAK-2 was detected using an anti-phospho-T172 specific antibody, and actin was a loading control. The ratio represents the level of phospho-AMPK normalized for the actin level in each lane. N2 larval worms treated with 10 mM sodium azide to deplete ATP levels and the AMPK positive control extract (+) (Cell Signaling Technologies) were included as positive controls. (D) aak-2 is required for tatn-1 effects on lifespan. Mean survival for N2, tatn-1(baf1), aak-2(gt33), and tatn-1(baf1); aak-2(gt33) were 18.0, 21.0, 14.0, and 13.0 days, respectively. p<0.001 for all pairwise curve comparisons by log-rank test. To further explore the role of aak-2/AMPK in dauer formation, we treated worms with the AMPK agonist AICAR. Treatment of wild-type N2 worms with 0.125 mM AICAR had no effect on development or dauer formation, but eak-4 mutants treated with AICAR showed a significant increase in dauer formation compared to an untreated control (Figure 5B). This effect required aak-2/AMPK as an eak-4(mg348); aak-2(gt33) mutant failed to respond to AICAR (Figure 5B). These findings demonstrate that aak-2 activity is both necessary and sufficient to promote dauer formation by eak-4 mutants.
Our findings could suggest that AAK-2 is activated in the tatn-1 mutants. To test this possibility we used western blotting to measure levels of AAK-2 phosphorylated on the activating Thr243 residue, which is analogous to Thr172 in the vertebrate orthologs. We found that treatment of N2 wild-type animals with either sodium azide which inhibits mitochondrial function and depletes ATP levels by approximately 50% at this dose or with 1 mM AICAR leads to increases in phosphorylated AAK-2 levels compared with untreated L2 larval N2 worms grown in parallel (Figure 5C) [54]. Furthermore levels of phospho-AAK-2 were also increased in eak-4; tatn1 mutant L2 larvae compared to the N2 control larvae (Figure 5C). Importantly, the phopho-AAK-2 signal was lost in aak-2(gt33) mutants confirming the identity of this band as AAK-2 (Figure 5C). These findings demonstrate that both tatn-1 mutations and AICAR treatment serve to activate AMPK in C. elegans, and that this activation of AAK-2 is required for the effects of tatn-1 mutations on development.
The requirement for aak-2/AMPK for the effects of tatn-1 could reflect impaired tyrosine degradation leading to reduced production of the TCA cycle precursors fumarate and acetoacetate and a consequent decrease in energy production. To test this possibility we analyzed worm lysates for levels of AMP and ATP. We found that eak-4(mg348); tatn-1(baf1) mutants had a lower AMP/ATP ratio than N2 worms grown in parallel (Figure S5). This suggests that the role of aak-2/AMPK does not reflect reduced energy production in the tatn-1 mutants.
Finally, we tested whether the effect of tatn-1(baf1) on adult lifespan requires aak-2 activity, and we found that a tatn-1 mutation increased wild-type lifespan by 17.8%, but decreased aak-2(gt33) lifespan by 8.9% compared to aak-2(gt33) alone (N2 mean survival 18.0 days, tatn-1(baf1) 21.0 days, aak-2(gt33) 14.0 days, and tatn-1(baf1), aak-2(gt33) 13.0 days) (Figure 5C and Table S3). Hence aak-2 is both required for the tatn-1 lifespan increase and may even play a protective role in animals with reduced tatn-1 activity.
tatn-1 acts by daf-16/FOXO dependent and independent pathways
Since the effects of eak genes on dauer formation are dependent on both the FOXO transcription factor DAF-16 and the nuclear hormone receptor DAF-12, we tested whether these genes are required for dauer formation by eak-4(mg348); tatn-1(baf1) mutants through the construction of daf-16(mgDf47); eak-4(mg348); tatn-1(baf1) and eak-4(mg348); tatn-1(baf1), daf-12(rh61rh411) mutants [26]–[28]. We found that the genetic interaction is completely dependent on daf-12 (Figure 6A). However, we identified both daf-16 dependent and independent effects of tatn-1. The tatn-1 effects on dauer formation are largely blocked by the daf-16(mgDf47) mutation, but a small percentage of worms still form dauers (Figure 6A). This daf-16 independent pathway was also seen in experiments using the stronger tatn-1(qd182) allele (Figure S6). These findings suggest that daf-16/FOXO does play a vital role in the interaction between eak genes and tatn-1, but that daf-16/FOXO independent pathways are also involved.
Fig. 6. tatn-1 acts by daf-16/FOXO dependent and independent mechanisms. 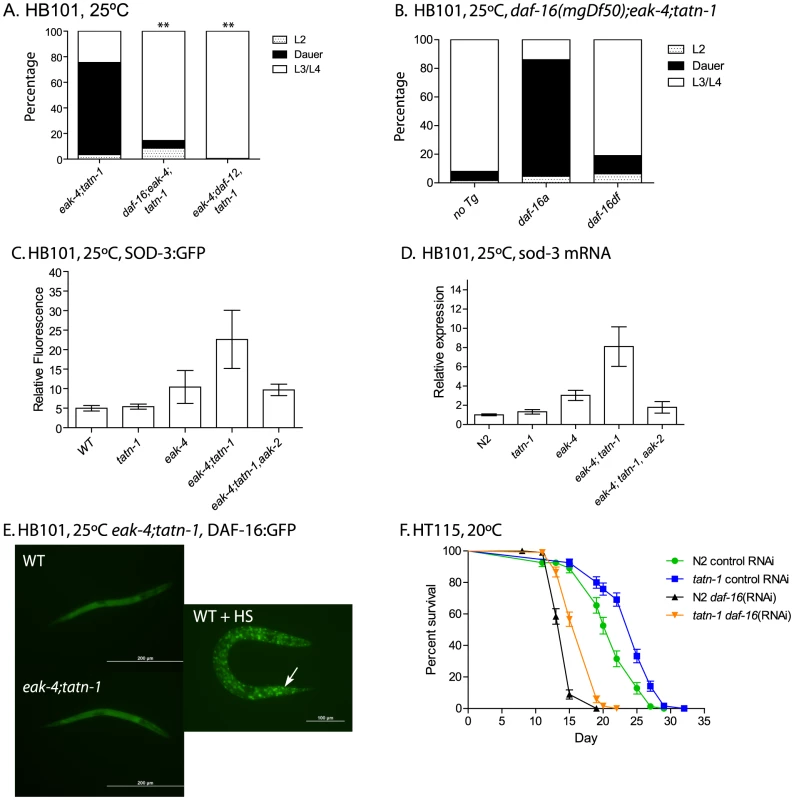
(A) Mutations in daf-16 (daf-16(mgDf47)) or daf-12 (daf-12(rh61rh411)) inhibit dauer formation by eak-4(mg348); tatn-1(baf1) mutant worms. ** p<0.001 by Fisher's exact test. (B) The daf-16(mgDf50) mutation also inhibits dauer formation by eak-4; tatn-1(baf1) mutants, and a transgene expressing the DAF-16A isoform largely rescues dauer formation by daf-16(mgDf50); eak-4; tatn-1 mutants while a transgene expressing the DAF-16DF isoform only weakly rescues. (C) eak-4; tatn-1 mutants activate expression of the daf-16 target gene sod-3 as indicated by expression of a sod-3p:GFP reporter gene in L2 larvae, and this induction requires the aak-2/AMPK gene. * p<0.05 for t-test comparisons of eak-4 vs. eak-4; tatn-1, and eak-4; tatn-1 vs. eak-4; tatn-1; aak-2. (D) Induction of the endogenous sod-3 gene in eak-4; tatn-1 mutants is seen by quantitative RT-PCR, and this induction also depends on the aak-2/AMPK gene. (E) DAF-16A:GFP is not nuclear localized in either L2 stage WT or eak-4; tatn-1 larvae, but exposure to a 1 hour 35°C heat shock produces clear nuclear localization of DAF-16A:GFP (arrow). (F) tatn-1 effects on lifespan are daf-16 independent. Treatment of N2 and tatn-1(baf1) mutants with daf-16 RNAi reduces mean survival, but tatn-1(baf1) still lives longer than N2. The mean survival of N2 control RNAi, tatn-1 control RNAi, N2 daf-16(RNAi), tatn-1 daf-16(RNAi) are 21.0, 24.0, 14.5, and 17.0 days. p<0.001 for comparison of N2 control RNAi vs. tatn-1 control RNAi, and tatn-1 daf-16 (RNAi) vs. tatn-1 control RNAi by log-rank test. The daf-16/FOXO gene encodes multiple isoforms which have been recently shown to have differential modes of regulation and have distinct effects on development and longevity [24], [55], [56]. For dauer development, three isoforms appear to be involved with DAF-16A being the predominant isoform, DAF-16DF playing a somewhat lesser role, and DAF-16B playing a modest role, at best [55]. As a result, we tested whether the developmental effects of eak-4 and tatn-1 are dependent on particular daf-16 isoforms through the use of isoform-specific transgenes to rescue the dauer-constitutive phenotypes lost in daf-16/FOXO mutants (Figure 6B). We found that a transgene encoding a DAF-16A:mRFP fusion protein was able to strongly rescue the formation of dauers by a daf-16(mgDf50); eak-4(mg348); tatn-1(baf1) mutant, whereas a transgene encoding a DAF-16DF:GFP fusion protein only weakly rescued dauer formation (Figure 6B). These data suggest that the daf-16a isoform is most involved in the developmental effects produced by tatn-1 mutants.
To further explore the role of daf-16/FOXO in tatn-1 phenotypes, we examined the effects of eak-4 and tatn-1 mutations on both daf-16/FOXO target gene expression and DAF-16 subcellular localization. We used a sod-3:GFP transgene to examine the effects of eak-4 and tatn-1 mutations on expression of the daf-16 target gene sod-3, which encodes a manganese superoxide dismutase enzyme [57], [58]. We generated combinations of eak-4(mg348) and tatn-1(baf1) with the transgene, and examined GFP expression in L2 larvae. We found that the combination of eak-4 and tatn-1 mutations resulted in the highest expression of sod-3:GFP even prior to dauer formation (Figure 6C). Furthermore, aak-2/AMPK was required for this enhanced activation of the sod-3:GFP reporter (Figure 6C). Similar effects of tatn-1 and eak-4 on sod-3 expression were also seen when mRNA levels for the endogenous gene were measured by Q-PCR in L2 larvae, and this effect on sod-3 expression also required aak-2/AMPK (Figure 6D). Together these findings demonstrate that tatn-1 and eak-4 act in an aak-2/AMPK dependent manner to promote at least some aspects of daf-16/FOXO transcriptional activity.
Since the eak genes act to inhibit DAF-16/FOXO within the nucleus, a possible explanation for the enhancement of dauer formation in eak; tatn-1 double mutants may be explained by the tatn-1 allele causing increased nuclear localization of DAF-16 as do mutations affecting akt-1 [26]. We tested for changes in DAF-16 localization via the use of transgenic animals expressing a DAF-16A:GFP fusion protein [59]. We found that eak-4; tatn-1 mutants failed to show clearly visible nuclear localization of DAF-16:GFP in synchronized L2 worms grown at 25°C (Figure 6E). In contrast, exposure of animals to a 1 hour heat-shock at 35°C led to strong nuclear localization of DAF-16A:GFP (Figure 6E). Together these findings suggest that the tatn-1 enhancement of the eak dauer formation phenotype could be due to an increase in daf-16 transcriptional activity without an accompanying significant change in DAF-16 subcellular localization.
We then used daf-16 RNAi treatment to test whether daf-16/FOXO is required for the lifespan extension of tatn-1 mutants. We found that silencing of daf-16 through RNAi results in a shortened lifespan for both tatn-1 and wild-type N2 worms compared to control RNAi treatment (Figure 6F). However in daf-16 RNAi treated worms, tatn-1 still produces lifespan extension over wild type (mean survival for N2 control RNAi 21.0 days, tatn-1(baf1) control RNAi 24.0 days, N2 daf-16(RNAi) 14.5 days, and tatn-1(baf1) daf-16(RNAi) 17.0 days). Specifically, tatn-1(baf1) produced a 13.7% increase in lifespan in control RNAi treated worms, but a 17.8% increase in lifespan in daf-16 RNAi treated worms. These data suggest that the increased lifespan resulting from tatn-1 is either independent of daf-16 or occurs in a site resistant to the effects of daf-16 RNAi.
crh-1/CREB shows overlapping gene expression and phenotypes with tatn-1
To explore the effects of impaired tyrosine metabolism in C. elegans, we performed whole transcriptome RNA sequencing (RNA-seq) to identify genes that are differentially regulated in the tatn-1 mutants. To maximize the gene expression changes seen, we used the stronger tatn-1(qd182) allele and compared its transcriptome to that of wild-type N2 animals. Using ANOVA testing with a false-discovery rate of 5%, we identified 890 up-regulated and 3732 down-regulated genes in the tatn-1(qd182) mutant relative to N2 (Table S4).
To understand how the tatn-1 mutants might affect the daf-2/IGFR pathway, we used this data set to examine whether changes in the expression of genes in pathway are seen. We found that there was no change in the expression of daf-2/IGFR, daf-16/FOXO, aak-2/AMPK, eak-4, or akt-2 (Table S4). However we did find that levels of the age-1 PI3-kinase are reduced almost 76% and levels of akt-1 are reduced almost 70% compared to N2 while levels of the daf-18/PTEN tumor suppressor, which normally inhibits signaling through the PI3-kinase signaling pathway, is reduced almost 95% compared to wild-type animals (Table S4). Despite the observed changes in the expression of genes in the PI3-kinase signaling pathway, there is likely little net effect on the regulation of downstream targets by the pathway as we failed to observe differences in DAF-16:GFP localization, which would translocate to the nucleus if the pathway was inhibited (Figure 6E).
We then used both the DAVID program and the Panther database both to identify biologic themes within the up-regulated and down-regulated genes by testing for over-represented gene classes based on structural and functional annotations and to visualize the gene classes seen in both groups of genes (Figure S7 and Table S5) [60], [61]. Within the up-regulated set, we found that genes involved in tyrosine metabolism and neuropeptide signaling were strongly over-represented (7–10 fold) (Table S5). Specifically, we found that every gene in the tyrosine degradation pathway is up-regulated in the tatn-1(qd182) mutant suggesting that the altered metabolism is detected and leads to a compensatory change in expression of the pathway (Table S4). The significance of the changes in neuropeptide signaling gene expression is currently unclear, but could suggest that impaired tyrosine metabolism or the resulting increased in AAK-2/AMPK activity produces direct changes in neuronal activity or that these changes could be the direct downstream effectors responsible for the tatn-1 phenotypes. Graphically, we saw a greater percentage of genes which were expressed in the extracellular compartment and had catalytic or receptor activity among the up-regulated genes compared to those that were down-regulated in the tatn-1 mutants (Figure S7). In contrast, within the down-regulated genes, we identified over-representation of a broad range of genes involved in germline development, cell cycle, DNA replication, and larval development (Table S5). Further, these genes were more likely to be expressed in the intracellular compartment and to have regulatory effects on translation or enzyme activity (Figure S7). The expression changes in genes involved in cell cycle regulation and development could perhaps account for the developmental delay observed in the tatn-1(qd182) mutant compared to N2 (Figure S4).
Given the requirement we found for daf-16/FOXO for aspects of the tatn-1 phenotypes, we used Gene Set Association Analysis (GSAA) to test for enrichment of genes known to be regulated by daf-16/FOXO in the context of daf-2/IGFR signaling [62], [63]. Via this approach, we found both the up-regulated and down-regulated daf-16/FOXO target genes identified by Murphy et. al. to be enriched within the tatn-1(qd182) transcriptome (Figure 7A). This suggests that the expression of a subset of daf-16/FOXO target genes is altered by changes in tyrosine metabolism.
Fig. 7. tatn-1 mutants and crh-1/CREB mutants share gene expression profiles and effects on dauer formation. 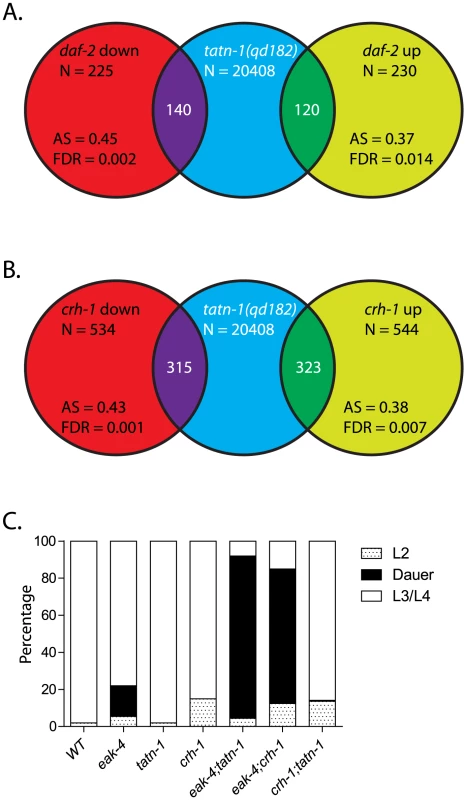
(A) Shot-gun whole transcriptome sequencing (RNA seq) was used to used to characterize and measure the transcriptome of N2 and tatn-1(qd182) mutants. From these experiments a total of 20,408 mRNA and other RNA transcripts were detected. To test for evidence of a daf-16/FOXO gene expression signature in the tatn-1(qd182) mutants Gene Set Association Analysis (GSAA) was used to determine whether the expression of daf-16/FOXO target genes regulated in the context of daf-2/IGFR signaling are associated with the tatn-1(qd182) mutant gene expression profile. GSAA calculates a differential expression score for each gene in the entire 20,408 gene RNA-seq dataset, and then uses a running weighted Kolmogorov-Smirnov test to examine association of an entire gene set with each phenotypic class. The strength of the association is measured by the association score (AS) where positive scores indicate association of the gene set with the phenotype, and statistical significance is measured by a false discovery rate (FDR) that is adjusted for multiple testing. From 225 genes down-regulated in daf-2/IGFR mutants, 140 showed association with the tatn-1(qd182) profile, and from 230 genes up-regulated in daf-2/IGFR mutants, 120 showed evidence of association by GSAA analysis. AS represents the association score with positive values indicating association, and FDR represents the false discovery rate for the association. (B) To test for evidence of a crh-1/CREB gene expression signature in the tatn-1(qd182) mutants Gene Set Association Analysis (GSAA) was used to determine whether the expression of crh-1/CREB target genes identified through microarray studies using wild-type and crh-1 mutant worms associate with the tatn-1(qd182) expression profile. From 534 genes down-regulated in crh-1/CREB mutants, 315 showed evidence of association, and from 544 genes up-regulated in crh-1/CREB mutants, 323 showed evidence of association by GSAA analysis. AS represents the association score with positive values indicating association, and FDR represents the false discovery rate for the association. (C) crh-1/CREB mutants enhance dauer formation by eak-4 mutants but not by tatn-1 mutants. Since our genetic studies suggested the involvement of a daf-16-independent pathway, we also used GSAA to test whether target genes recently identified for the CREB transcription factor crh-1 are enriched in the tatn-1(qd182) mutant [53]. We chose to focus on crh-1 because recent work has demonstrated that crh-1/CREB lies downstream of aak-2/AMPK [53]. crh-1/CREB and aak-2/AMPK are mechanistically linked because AAK-2 directly phosphorylates and inactivates the crh-1/CREB coactivator crtc-1, and as a result both aak-2/AMPK over-expressing and crh-1/CREB mutant animals are long-lived and share gene expression profiles [53]. We found that in the tatn-1(qd182) mutants, there is differential expression of both genes up-regulated and genes down-regulated in crh-1/CREB mutants (Figure 7B). This suggests that altered tyrosine metabolism could lead to changes in crh-1 target gene expression and could suggest a role for crh-1/CREB in the tatn-1 phenotypes. To test for crh-1/CREB involvement, we combined the crh-1(tz2) null allele with tatn-1 and eak-4 and examined the effects on dauer formation. We found that crh-1 showed a similar interaction as tatn-1 with eak-4, but did not promote dauer formation by the tatn-1 mutant (Figure 7C). Together these findings suggest that tatn-1 mutants share phenotypes and gene expression profiles with crh-1/CREB mutants and could be consistent with crh-1/CREB acting as an additional downstream effector of the response to impaired tyrosine metabolism.
Tyrosine aminotransferase expression is controlled by diet and environment
In vertebrates, tyrosine aminotransferase has been reported to be an insulin target gene with insulin treatment leading to reduced expression [32]–[35], [38]. As a result, we asked whether tatn-1 could also be regulated by daf-2/IGFR signaling in worms. To test the effects of daf-2 signaling on tatn-1 expression, we generated transgenic worms with an integrated transgene expressing a TATN-1:GFP fusion protein under the control of the tatn-1 promoter. This transgene rescues the tatn-1(baf1) mutation and blocks dauer formation by eak-4(mg348); tatn-1(baf-1) mutants, which demonstrates that the fusion protein is both functional and expressed in the correct anatomical locations (Figure 8A).
Fig. 8. Control of TATN-1 protein expression by daf-2/IGFR signaling, diet and temperature. 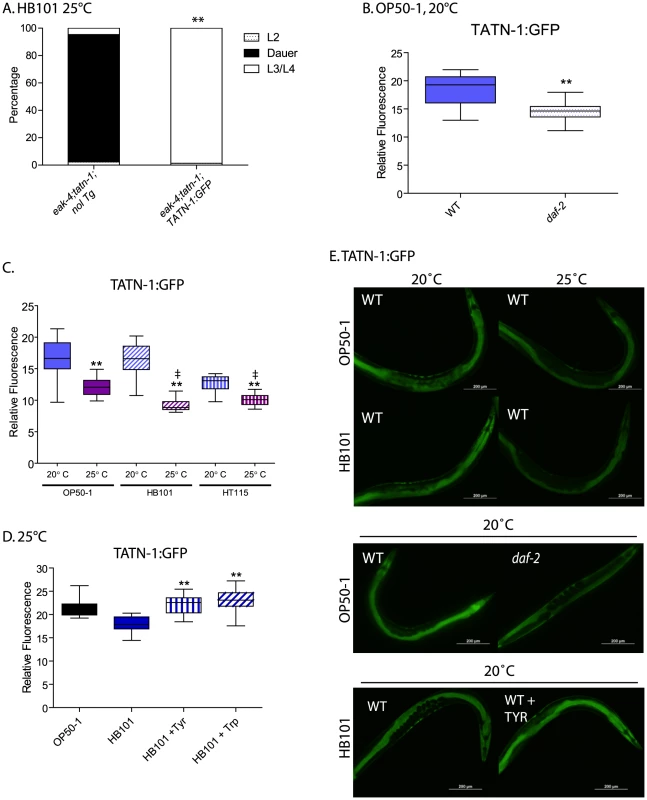
(A) Rescue of tatn-1 mediated dauer arrest by a tatn-1p:tatn-1 cDNA:GFP transgene (bafIs131). (B) daf-2 signaling positively regulates TATN-1:GFP expression. Box and whiskers plot showing comparison of GFP fluorescence between day 1 adult wild-type and daf-2(e1368) mutants expressing the TATN-1:GFP transgene and grown on OP50-1 bacteria. N≥15 worms, ** p<0.001 by t-test. (C) Bacterial food source and temperature regulate TATN-1:GFP expression. Wild-type worms expressing the TATN-1:GFP transgene were grown on the indicated bacteria until adulthood and then either kept at 20°C or shifted to 25°C overnight. N≥15 worms, ** p<0.001 for 20°C versus 25°C for each bacterial strain by t-test, and ‡ p<0.001 for 25°C treatment of OP50-1 versus HB101 or HT115. (D) Supplementation of HB101 with 1 mg/mL tyrosine or tryptophan increases TATN-1:GFP expression. Wild type worms expressing the TATN-1:GFP transgene were grown until adulthood on the indicated diet. N≥15 worms, ** p<0.001 for comparison between HB101, and HB101+Tyr or HB101+Trp by t-test. (E) Representative photos showing the effects of the treatments graphed in panels B-D. GFP fluorescence representing the TATN-1:GFP fusion protein is observed in the intestine and hypodermis of worms (Figure 8E). When we crossed the transgene into the daf-2(e1368) mutant, and we found that the presence of the daf-2/IGFR mutation led to a 20% decline in GFP expression in adult worms grown on OP50-1 at 20° (Figure 8B and Figure 8E). This is consistent with daf-2/IGFR signaling acting positively to promote TATN-1 expression in adult worms. Interestingly, the effect of daf-2/IGFR signaling on TATN-1 expression likely occurs either at the translational or protein stability level because Q-PCR experiments demonstrated almost a 50% increase in tatn-1 mRNA expression in the daf-2 mutant animals (Figure S8). As a result of the divergent regulation of tatn-1 mRNA and protein levels, we focused on TATN-1:GFP expression in our subsequent experiments.
Beyond daf-2/IGFR signaling, we found that both diet and environmental temperature affected TATN-1 levels to a similar or even greater degree. Specifically, adult worms grown on OP50-1, HB101, or HT115 show decreases in TATN-1:GFP expression when shifted from 20°C to 25°C for 24 hours with worms grown on OP50-1 showing a 26.2% decrease, on HB101 a 44.7% decrease, and on HT115 a 24.2% decrease (Figure 8C and Figure 8E). Further, we found that the OP50-1 fed worms showed greater TATN-1:GFP expression compared to HB101 and HT115 fed worms. This difference was especially apparent in worms grown at 25°C, due to the variability of GFP intensity seen at 20°C, with HB101 and HT115 fed animals showing a 25.3% and 17.3% decrease, respectively, compared to OP50-1 fed worms (Figure 8C and Figure 8E).
The effects of diet on TATN-1:GFP expression suggests that the E. coli bacterial strains vary in nutrient composition in a way that can be detected by the worms. Prior work has demonstrated that protein is the primary component of these bacteria but that the overall protein levels are not significantly different between strains [64]. However, this work also suggested that specific amino acids could vary between the strains and account for differences in fat content in worms fed each strain. Specifically, pept-1 mutants, which lack an intestinal peptide transporter, fail to show the expected differences in fat content when fed different bacterial strains [64]. In vertebrates, tyrosine aminotransferase expression is controlled by dietary amino acid intake, most notably for tryptophan [65]–[67]. To test whether dietary amino acid intake could affect TATN-1:GFP expression, we supplemented HB101 spotted NGA plates spotted with either tyrosine or tryptophan at a final concentration of 1 mg/mL, and compared TATN-1:GFP expression to worms fed HB101 alone or OP50-1. This concentration is 8 times the level found in standard NGA media (0.125 mg/mL). We found that the addition of tyrosine or tryptophan increases the GFP expression level in HB101 fed worms up to that seen in worms grown on OP50-1 (Figure 8D and Figure 8E). Together these data demonstrate that TATN-1 levels are dynamic and under the control of both daf-2/IGFR signaling as well as dietary and environmental cues. Importantly many of these signals that control TATN-1 expression also influence dauer formation suggesting that tatn-1 could be a regulated modulator of daf-2/IGFR signaling and developmental decisions.
Tyrosine mediates the effects of tatn-1 mutations
Since TATN-1 is the first enzyme in the tyrosine degradation pathway, decreased activity should both increase the levels of tyrosine and also decrease the levels of the downstream metabolites (Figure 1B). The tatn-1 mutant phenotypes could be a direct result of changes in the level of a particular metabolite. For example, elevated fumarate levels are known promote hypoxia inducible factor (HIF) activity in certain renal cancers [68]. Alternately, tatn-1 could have a novel function that is independent of its metabolic activity. For example, subunits of the phenylalanine hydroxylase enzyme are known to have an additional, non-enzymatic role as a transcriptional co-activator [69]. To explore whether either of these models accounts for the tatn-1 phenotypes, we tested for interactions between eak-4 and mutant alleles of other enzymes in the tyrosine degradation pathway (Figure 1B) by constructing pah-1; eak-4 and hpd-1; eak-4 mutants. These mutants lack pah-1, which encodes the enzyme phenylalanine-4-hydroxylase that converts phenylalanine into tyrosine, or hpd-1, which encodes 4-hydroxyphenylpyruvate dioxygenase and catalyzes the step immediately downstream of tatn-1 [39], [43], [70]. We found that pah-1 did not enhance dauer arrest by the eak-4 mutants (Figure 9A) whereas both tatn-1 and hpd-1 increased dauer formation. Lower numbers of eak-4; tatn-1 dauers were seen because the experiment was scored after 3 days due to the slower development of the hpd-1 mutant. Additionally, when we used the pah-1 mutant to block the synthesis of tyrosine in a tatn-1; eak-4 mutant, we found that this reduced dauer formation (Figure 9B). These results suggested that the effects of tatn-1 on dauer formation were directly linked to the metabolic effects of tatn-1, and that the accumulation of tyrosine instead of deficiency of a downstream metabolite could be responsible for the tatn-1 phenotype.
Fig. 9. tatn-1 acts by increasing tyrosine levels. 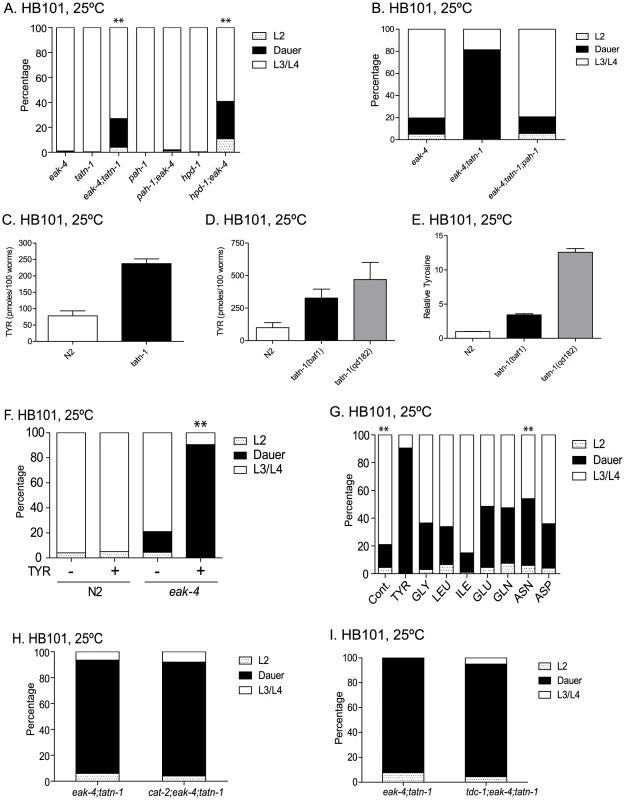
(A) Both tatn-1 and hpd-1 mutations augment dauer arrest by eak-4(mg348) mutants after three days at 25°C, whereas a pah-1 mutation does not. ** p<0.001 for comparisons between eak-4 and eak-4; tatn-1, and hpd-1; eak-4 by Fisher's exact test. (B) Inhibiting tyrosine synthesis with the pah-1 mutation reduces dauer formation by eak-4; tatn-1 mutants. (C) tatn-1(baf1) has a higher concentration of tyrosine compared to wild type N2. The bars represent the mean tyrosine concentration from four independent samples, and the error bars show the standard error of the mean. ** p<0.001 by t-test. (D) tatn-1(qd182) and tatn-1(baf1) have higher tyrosine levels compared to wild type N2. (E) tatn-1(qd182) has higher tyrosine levels than N2 and tatn-1(baf1) when the measured tyrosine levels are normalized to the levels of non-aromatic amino acids in each sample. (F) Treatment of eak-4(mg348) mutants but not N2 with tyrosine results in dauer arrest. ** p<0.001 for comparison of eak-4 + and - tyrosine using Fisher's exact test. (G) Tyrosine (1 mg/mL) is more potent than other amino acids in producing dauer arrest by eak-4(mg348) mutants. ** p<0.001 for comparison between eak-4 vs eak-4+tyrosine and eak-4+tyrosine and eak-4+asparagine using Fisher's exact test. (H) Impaired dopamine synthesis in cat-2(e1112) mutants does not block the effect of tatn-1 on dauer formation. (I) Impaired tyramine and octopamine synthesis in tdc-1(ok914) mutants does not block the effect of tatn-1 on dauer formation. As a result, we measured the levels of amino acids in wild-type N2, tatn-1(baf1), and tatn-1(qd182) larval animals grown at 25°C on HB101 plates by liquid chromatography mass spectrometry (LC-MS/MS). We found that wild-type N2 worms contained an average of 78.1 pmol of tyrosine per 100 worms whereas tatn-1(baf1) worms contained 237.4 pmol per 100 worms (Figure 9C). Hence the tatn-1(baf1) mutation produced a roughly three fold increase in tyrosine levels in the mutant animals. Further to compare the effects of tatn-1(qd182) on tyrosine levels compared to tatn-1(baf1), we measured tyrosine levels in additional samples grown and prepared in parallel. We found in these samples that wild-type N2 worms contained an average of 99.6 pmol of tyrosine per 100 worms, tatn-1(baf1) contained an average of 327.2 pmol per 100 worms, and tatn-1(qd182) contained an average of 470.2 pmol per 100 worms, which is an 43.7% increase over tatn-1(baf1) (Figure 9D). However, we noted that the tatn-1(qd182) worms were smaller than N2 or tatn-1(baf1) and the levels of many other amino acids measured in parallel were lower in tatn-1(qd182) compared to tatn-1(baf1). This suggested that the tatn-1(qd182) samples may have contained less overall biomass, and hence our normalization to worm counts alone may underestimate the effect of the tatn-1(qd182) mutation on tyrosine levels. To correct for this difference, we normalized tyrosine levels to the levels of all non-aromatic amino acids in the samples with the assumption that the net effect of these mutations on the levels of these amino acids is neutral. After normalization, we found that tatn-1(baf1) produced a 3.4 fold increase in tyrosine levels compared to N2 whereas tatn-1(qd182) produced a 12.6 fold increase compared to N2, which is also a 3.7 fold increase over tatn-1(baf1) levels (Figure 9E). These findings demonstrate that both tatn-1 alleles increase tyrosine levels compared to wild-type animals, and that the stronger phenotypes of the tatn-1(qd182) allele are likely due to the further increases in tyrosine levels observed.
To directly test whether elevated tyrosine levels are responsible for the tatn-1 phenotype, we treated worms with exogenous tyrosine cast into the NGA plates at 1 mg/mL. This treatment results in tyrosine levels in the worms that are elevated compared to untreated animals, but lower than those seen in the tatn-1 mutants (Figure S9). We found that supplementation had no effect on the development of wild-type worms but lead to dauer arrest by the eak-4 mutants (Figure 9F). These results directly demonstrate changes in tyrosine levels alter the development of eak-4 mutant worms and are responsible for the tatn-1 phenotype.
Amino acids are known to antagonize insulin actions in vertebrates, so our results could represent either the non-specific effects of any amino acid or a tyrosine-specific effect [71], [72]. To test these possibilities we directly compared the ability of a variety of amino acids to enhance dauer formation by eak-4 mutants. We grew eak-4 mutants on HB101 spotted NGA supplemented with tyrosine, glycine, leucine, isoleucine, glutamate, glutamine, asparagine, or aspartate, each at the concentration of 1 mg/mL. Since tyrosine is the largest of these amino acids, this resulted in worms being treated with higher molar equivalents of the other amino acids compared to tyrosine. We found that while other amino acids do increase the formation of dauers by eak-4 mutants, none was as potent as tyrosine (Figure 9G). This suggests that the effect on dauer formation shows selectivity for the presence of tyrosine. The effects of tyrosine and to a lesser extent the other amino acids is not due to a toxic effect of the amino acid supplementation as treated worms showed a similar lifespan to untreated worms (Figure S9).
Besides being a building block for proteins, tyrosine serves as a precursor for the synthesis of catecholamine neurotransmitters. In vertebrates, there is evidence that the levels of tyrosine as a precursor influences the synthesis of these neurotransmitters [73]. Hence, one possibility is that tatn-1 mutations raise tyrosine levels and facilitate its conversion into the neurotransmitters dopamine, octopamine, or tyramine which could produce the observed phenotypes. To test this possibility, we blocked dopamine synthesis with the cat-2 mutation, which affects the worm tyrosine hydroxylase gene, and we blocked octopamine and tyramine synthesis with the tdc-1 mutation, which removes the enzyme tyrosine decarboxylase [74], [75]. We found that both cat-2; eak-4; tatn-1 and tdc-1; eak-4; tatn-1 mutants are similar to eak-4; tatn-1 mutants with regards to the formation of dauers (Figure 9H and Figure 9I). These data demonstrate that excessive synthesis of dopamine, octopamine, or tyramine is not responsible for the tatn-1 phenotype. Instead tyrosine is directly sensed by the worms and acts as a developmental regulator.
Discussion
Tyrosine as a modulator of daf-2 insulin/IGF-1 signaling effects
Together our results identify tyrosine and tyrosine aminotransferase activity as a modifier of daf-2/IGFR effects in C. elegans (Figure 10). While the control of tyrosine aminotransferase expression and activity has been extensively studied as a target of insulin signaling in vertebrates [30]–[34], [36], [38], a connection between tyrosine aminotransferase or tyrosine metabolism and insulin action has not been demonstrated. Prior work in C. elegans has suggested that the hpd-1 gene, which encodes 4-hydroxyphenylpyruvate dioxygenase, is repressed in daf-2 mutants and that knock-down of hpd-1 by RNAi delayed dauer exit and extended lifespan [39]. However, the mechanism involved has been unclear. Our work shows that both tatn-1, and likely hpd-1, impact on daf-2/IGFR signaling through increasing tyrosine levels in the animal.
Fig. 10. Model for the regulation of TATN-1 expression, tyrosine levels, and the resulting effects of tyrosine effects on cell signaling pathways. 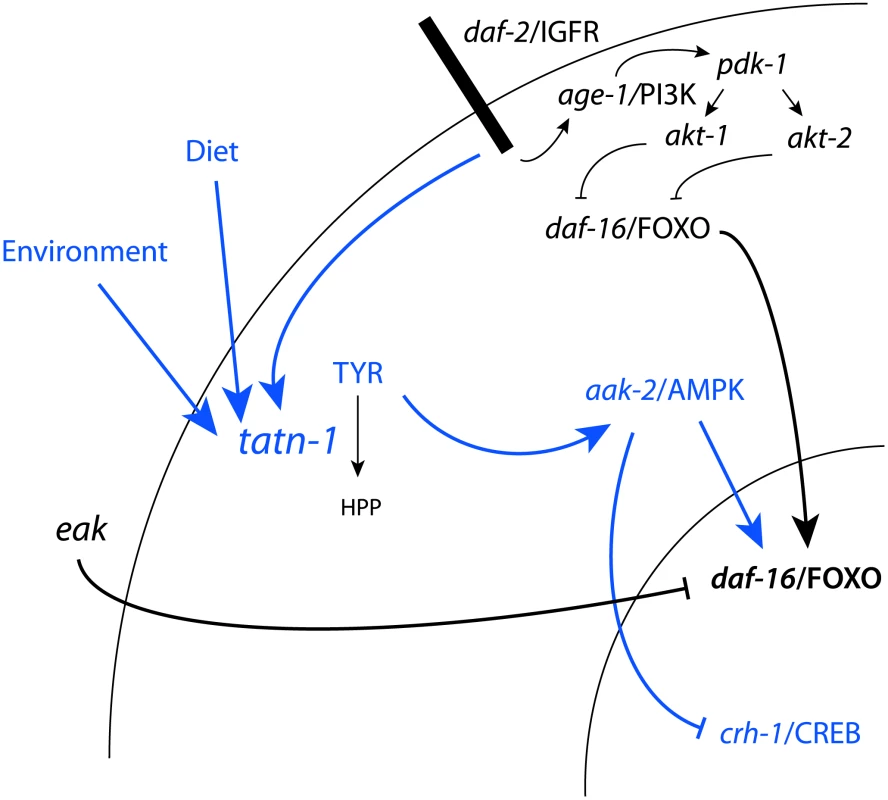
Shown is the daf-2/IGFR signaling pathway and eak genes from Figure 1A with the addition, shown in blue, of the effects of tyrosine identified in this work and the control of TATN-1 expression by daf-2/IGFR signaling and by dietary and environmental cues as demonstrated in Figure 8. In summary, worms sense the available diet and environmental temperature and these factors both contribute to the dauer decision and the regulation of TATN-1 protein levels. Conditions which promote the dauer decision also tend to reduce the expression of TATN-1. The reductions in TATN-1 expression lead to reduced removal of tyrosine through degradation and increase free tyrosine levels in the animal. These increases in tyrosine activate aak-2/AMPK and disrupt the effects of normal daf-2/IGFR signaling through positive effects on daf-16/FOXO and perhaps inhibitory effects on crh-1/CREB. The effects of tyrosine are particularly pronounced when the daf-2/IGFR pathway is compromised, such as through mutations in the daf-2/IGFR gene or the eak genes which lie in a parallel pathway. This may be due to further reductions in TATN-1 expression or reductions in inhibitory regulators of daf-16/FOXO activity. We find the effects of tyrosine on daf-2/IGFR signaling to be complex with roles for both the daf-16/FOXO transcription factor and the aak-2/AMPK seen (Figure 10). One way that high tyrosine levels could interact with daf-2/IGFR signaling would be for tyrosine to somehow activate aak-2/AMPK. AMP kinases are a known regulator of both daf-16 and the vertebrate homolog FOXO3 [51], [76]. AMPK regulates FOXO transcriptional activity through the phosphorylation of up to six sites on these proteins. In our work, aak-2/AMPK mutations suppress the dauer promoting effects of tatn-1 mutations, treatment of worms with the AMPK agonist AICAR is able to mimic the effects of tyrosine, and increases in the active phosphorylated form of AAK-2 are seen in the tatn-1 mutant. These findings demonstrate that elevated tyrosine levels activate aak-2/AMPK which could then phosphorylate daf-16/FOXO. This phosphorylation event could then interfere with the inhibitory effects of an intact daf-2/IGFR pathway on daf-16/FOXO activity. Further work would be needed to test this model, and the ability of mutants lacking aak-2/AMPK or daf-16/FOXO to still respond to elevated tyrosine levels also supports the presence of alternate, currently unknown downstream pathways.
These alternate pathways could either lie in parallel to daf-16/FOXO or could be the dominant response pathway with daf-16/FOXO only playing a permissive role, especially at lower tyrosine levels. One possible alternate pathway involves the CREB transcription factor crh-1 (Figure 10). The crtc-1 co-activator for the CREB transcription factors has been shown to be a target of regulation by aak-2/AMPK, and in vertebrates, CRTC co-activators are known to interact with insulin signaling in mediating the hepatic metabolic adaptation to the fed versus fasting state [53], [77], [78]. We find that tatn-1(qd182) mutants show evidence of crh-1/CREB-regulated gene expression, and a crh-1/CREB mutant mimics the interaction of eak-4 and tatn-1. Perhaps elevated tyrosine levels lead to the activation of aak-2/AMPK which then results in the activation of daf-16/FOXO and inhibition of crtc-1 and crh-1/CREB (Figure 10). The presence of paired downstream pathways could explain the partial requirement for daf-16/FOXO, especially at higher tyrosine levels.
Beyond effects on daf-16/FOXO and crh-1/CREB, elevated tyrosine, especially at high levels, could also have hormetic effects via changing cellular redox status, producing ER stress, or perturbing the protein folding environment [79]. These effects could account for the positive effects of increased tyrosine on longevity, and some of the genes involved in sensing hormetic stresses, such as the HSF-1 ortholog hsf-1, also interact with daf-16/FOXO and play roles in dauer formation [80]–[82]. Alternately, tyrosine could act via a novel pathway that operates independently of daf-16/FOXO or crh-1/CREB, especially at higher levels. For example, the vertebrate calcium-sensing receptor, PPARγ nuclear receptor, and aryl hydrocarbon receptor (AHR) have all been shown to respond to aromatic amino acids, though their connection to insulin action is currently unclear [83]–[85]. The recent finding that Akt and Foxo1 are largely dispensable for the control of hepatic metabolism by insulin in vivo has suggested that FOXO - independent pathways exist and play important roles in metabolic control [86].
Our work also provides insights into the eak genes which are known to act via unclear mechanisms to reduce daf-16/FOXO transcriptional activity while not significantly affecting the subcellular localization of DAF-16/FOXO [26], [27], [29]. We find that beyond enhancing the inhibitory effects of akt-1 on daf-16/FOXO activity, the eak genes also suppress the effects of amino acids and AMPK activity on daf-16/FOXO activity. Additional work will be needed to understand if the eak genes normally represent a control point where the effects of these metabolic signals on insulin signaling can be enhanced or suppressed.
The complex control of tyrosine aminotransferase
We find that the regulation of tatn-1 expression in worms is complex with daf-2 activity, diet, and environmental conditions each contributing to the expression level (Figure 10). In vertebrates, tyrosine aminotransferase has also been shown to undergo regulation at the transcriptional, translation, and degradation levels in response to hormonal and nutritional cues [66]. We find that daf-2/IGFR activity inhibits tatn-1 gene transcription but raises TATN-1 protein levels. This is consistent with work in vertebrates showing that insulin shows complex effects on tyrosine aminotransferase expression with actions at both the transcriptional and translational level [30]–[34], [36], [38]. Nutritional cues appear to also be an important regulator because we find that the E. coli strain used as food has a powerful effect on the expression of tatn-1 and these effects parallel the effects of the weaker tatn-1(baf1) allele on dauer formation. In rats, the activity of hepatic tyrosine aminotransferase varies several-fold during the day with a peak during the evening and nadir in the early morning [87]. Studies of the cyclic variation have demonstrated that dietary protein intake is a prime inducer of tyrosine aminotransferase levels [65]. Feeding animals a protein-free diet results in a constant low level of tyrosine aminotransferase, whereas feeding animals protein meals at differing times produces corresponding shifts in enzyme production. Among amino acids, some such as tryptophan are potent inducers of tyrosine aminotransferase expression [66]. The mechanisms accounting for the dietary effects of amino acids on tyrosine aminotransferase are currently unclear. This could suggest a role for additional nutrient sensitive pathways which may well be conserved as we find that both tryptophan and tyrosine act as tatn-1 inducers in worms. Finally, we find a novel role for environmental conditions on tatn-1 expression as lower temperatures promote expression and higher temperatures inhibit it. How changes in temperature translate into the observed effects is unclear but perhaps hormonal changes mediated by the cytochrome P450 daf-9 and the nuclear hormone receptor daf-12 or changes mediated by thermosensory neurons are involved [88]. Together this suggests that the control of tyrosine aminotransferase activity, which modulates tyrosine levels, could be controlled via a complex network of internal and external cues. Our finding that changes in tyrosine levels alter both signaling pathways and gene expression patterns could suggest that carefully controlling tyrosine metabolism and ultimately tyrosine levels plays an important role in overall homeostasis (Figure 10).
Aromatic amino acids in human disease
Recent work has suggested that levels of specific amino acids, particularly branched chain and aromatic amino acids, could influence insulin sensitivity in people and mice [1]–[6]. While the exact role of aromatic amino acids in metabolic disease is unknown, our results suggest that these could play a causal role in either insulin-resistance or the development of diabetes [89]. Given the complex nature of tyrosine aminotransferase regulation, subtle changes in hormone levels, diet, and perhaps other factors could lead to changes in hepatic tyrosine metabolism and contribute to changes in serum aromatic amino acid levels. There may also be significant changes during the day due to dietary intake or release from internal stores such as muscle. As tyrosine levels increase, it is possible that, as in worms, the increases modify responses to insulin signaling and augment pre-existing insulin resistance in a harmful way (Figure 10). The connection between insulin signaling and tyrosine metabolism could potentially even lead to a vicious cycle of reduced insulin signaling producing elevated tyrosine levels which then lead to a further reduction in insulin signaling
Tyrosine aminotransferase has also been found to be a tumor suppressor gene in human hepatocellular carcinoma (HCC) [7]. The human tyrosine aminotransferase gene is located on 16q, which is frequently deleted in HCC, and analysis of tumors reveals that gene deletion or silencing via hypermethylation is common [7]. Consistent with an inhibitory role in the pathogenesis of liver cancer, transfection of HCC cancer cell lines with a tyrosine aminotransferase transgene suppressed malignant behavior such as growth in soft agar and the formation of tumors in nude mice. In these cells, tyrosine aminotransferase expression also acted to inhibit tumor formation via the stimulation of apoptosis, but the exact molecular events are still unclear [7]. Our data would suggest that the activation of AMPK or the downstream effects of AMPK on FOXO transcription factors, such as FOXO3, or CREB would be attractive targets for future study. Alternately, we also saw down-regulation of genes involved in DNA repair so the elevated tyrosine levels could also promote the accumulation of additional cancer promoting mutations (Table S5). Together these findings suggest that extracellular or intracellular tyrosine levels could act as signaling molecules involved in the control of cell growth, differentiation, and physiology.
Materials and Methods
C. elegans strains and maintenance
All C. elegans strains were propagated on standard nematode growth agar (NGA) plates containing streptomycin (200 µg/mL) and spotted with OP50-1, as previously described [90]. For specific experiments, worms were fed HB101, OP50, or HT115 E. coli strains using NGA containing streptomycin (HB101) or no antibiotics (OP50 and HT115).
The following C. elegans mutants were obtained from the C. elegans Genetics Center, which is supported in part by NIH Office of Research Infrastructure Programs (P40 OD010440): daf-16(mgDf50) I, cat-2(e1112) II, pah-1(ok687) II, tdc-1(ok914) II, crh-1(tz2) III, daf-2(e1368) III, hpd-1(ok1955) III, unc-119(ed3) III, akt-1(mg144) V, akt-1(mg306) V, aak-2(gt33) X, akt-2(ok393) X, daf-12(rh61rh411) X, pdk-1(mg142) X, sgk-1(ok538) X, muIs84[pAD76(sod-3::GFP)], lpIs14 [daf-16f::GFP+unc-119(+)], and lpIs12 [daf-16a::RFP+unc-119(+)]. tatn-1(baf1) X has been described previously, and tatn-1(qd182) was identified in an unrelated mutagenesis screen and is a gift from Daniel Pagano and Dennis Kim [43], [44]. muIs109[daf-16::GFP] X has been described previously and is a gift from Malene Hansen [59]. daf-16(mgDf47), hsd-1(mg433) I, eak-3(mg344) III, eak-4(mg348) IV, eak-7(tm3188) IV, sdf-9(mg337) V have been described previously [26]–[28], [91]. Double and triple mutants were generated by standard genetic crosses, and the genotypes of strains were confirmed by PCR using oligos which detect gene deletions or RFLP's associated with the mutation (Table S6). Throughout this work tatn-1 is implied to refer to the tatn-1(baf1) allele except specifically as noted otherwise.
Dauer assays
Worm embryos were isolated by sodium hypochlorite treatment, and eggs were transferred to plates, and grown at the indicated temperature in a designated incubator. The plates were scored two or three days later under a dissecting microscope for the presence of L2, dauer, and L3/L4 and older worms. We conducted control experiments to determine the robustness and reproducibility of visual scoring by having several scorers evaluate a series of still images and corresponding movies of larvae of different developmental stages. We then compared the correlation between the scorers for the entire series via the use of a kappa statistic [92]. These experiments indicated that scoring was consistent between raters within the lab with all comparisons showing “substantial” to “almost perfect” agreement (Table S7) [92].
For each assay, approximately 100 worms were scored from each of two to three plates set up in parallel for each genotype used in an experiment. This resulted in 200 to, more typically, 300 animals being scored for each genotype within an experiment. Each experiment was repeated at least once with comparable results, which resulted in between 400–600 worms being scored per genotype in total. The percentages of each stage were graphed using Prism5 software, and the graphs show pooled data from a single trial. To perform pairwise comparisons between mutant strains, a contingency table was set up using the counts for L2, dauer, and L3/L4 categories, and p-values were calculated using Fisher's exact contingency test within SAS version 9.3. To determine SDS resistance, worms were washed from plates with 1% SDS, and then incubated for 20 minutes with gentle rocking. Worms were then pelleted and washed with water. Aliquots were scored for survival as demonstrated by movement, and each experiment was repeated at least once. The percentages of living worms were graphed with Prism5.
Lifespan assays
Lifespan assays were conducted as previously described at 20°C using either NGA or RNAi plates containing 50 µM FUDR [93]. Lifespan assays for N2, tatn-1(baf1), eak-7(tm3188), and eak-7(tm3188); tatn-1(baf1) used NGA plates spotted with HB101, and worms were grown from eggs at 16°C to minimize larval arrest. Lifespan assays for N2, tatn-1(baf1), aak-2(gt33), and tatn-1(baf1), aak-2(gt33) used NGA plates spotted with HB101. Lifespan assays for amino acid treated N2 worms used either NGA plates or NGA plates supplemented with 1 mg./mL tyrosine, glycine, or isoleucine, and then spotted with HB101. Lifespan assays using daf-2 and daf-16 RNAi treatment used NGA media supplemented with carbenicillin (50 µg/mL) and isopropyl β-d-thiogalactopyranoside (IPTG, 1 mM). RNAi treatment for daf-16 was started at egg hatching while daf-2 RNAi treatment started on day 1 of adulthood with larval development occurring on NGA plates spotted with HB101 at 20°C.
For all lifespan assays three plates containing 40 worms each, for each genotype were set up, as well as an extra plate, with worms to replace worms that had crawled off the plate, bagged, or exploded, to reduce the number of censored events. Prism5 (Graphpad Software) was used to generate graphs and perform log-rank testing for curve comparisons. SAS was used to create lifetables and calculate mean survival.
Amino acid supplementation
Amino acids (Sigma-Aldrich) were dissolved as 45 mg/mL stock solutions in water, and then added to molten NGM to obtain a 1 mg/mL final concentration. These plates were dried and spotted with HB101 before use.
Amino acid analysis
Worms were grown on HB101 spotted NGM plates for two days at 25°C before being washed from the plates and then being rinsed twice with miliQ water. For Figure S9A, NGM plates either with or without 1 mg./mL tyrosine cast into the agar were used to grow the worm culture. To normalize the samples for worm number, a 5 µL aliquot was removed and scored for worm number. Amino acids were then extracted using aqueous methanol and crushing with a mortar and pestle as previously described [94]. The methanol solution was removed by evaporation and the residue stored frozen at −80°C. Amino acid analysis was performed via liquid chromatography tandem mass spectrometry following reconstitution of the residue in 0.1 mL water as described previously for urine [95]. Amino acid content was then either normalized to total worm number in the sample (Figure 9B, Figure 9C, and Figure S9A) or normalized to the levels of individual non-aromatic amino acids and then divided by the average normalized level observed in the wild-type N2 samples (Figure 9D).
AICAR treatment
Aliquots from a 250 mM AICAR solution dissolved in water (Cell Signaling Technology) were spotted onto NGA plates spotted with HB101 to give a final concentration of 0.125 mM 1 hour before eggs were added to the plates. A comparable volume of water alone was used as a negative control.
AMP and ATP measurements
Worm embryos were isolated by sodium hypochlorite treatment from N2 and eak-4(mg348); tatn-1(baf1) adults, and the eggs were transferred to NGA plates spotted with HB101. The plates were incubated at 25°C for 24 hours so most of the population was L2 larvae. The worms were washed from plates with water and washed with water to remove bacteria. Nucleotides were then extracted from the worms as previously described [50]. The resulting extract was stored at −80°C until analysis. ATP, ADP, and AMP levels were measured by HPLC with UV detection of individual nucleotides.
Fluorescent imaging
Images of worms were obtained with a BX51 fluorescence microscope and quantified using ImageJ software as previously described [44].
Quantitative RT-PCR
For sod-3 expression, worm embryos were isolated by sodium hypochlorite treatment, and eggs were transferred to NGA plates spotted with HB101 and incubated at 25°C for 24 hours. The worms were washed from plates in water, pelleted by centrifugation, washed with water, and frozen for storage. RNA extraction, reverse transcription, and quantitative PCR were performed as previously described [27], [44]. The geometric mean level of the control genes pmp-3, cdc-42, and Y45F10D.4 were used to normalize the samples, and the relative levels of sod-3 expression were determined using the 2−ΔΔCt approach [96], [97].
For tatn-1 expression, N2 and daf-2(e1368) embryos were isolated by sodium hypochlorite treatment, and eggs were transferred to S-basal to arrest the worms at the L1 stage. L1 larvae were added to NGA plates spotted with OP50-1 and the plates were incubated at 20°C for 3 days. Adult worms were washed from plates and RNA was isolated as described above. The geometric mean level of the control genes pmp-3, cdc-42, and Y45F10D.4 were used to normalize the samples, and the relative levels of tatn-1 expression were determined using the 2−ΔΔCt approach [96], [97].
The oligos used to detect pmp-3, cdc-42, and Y45F10D.4 have been previously described [98]. The expression of sod-3 was detected using the oligos 5′-CCAACCAGCGCTGAAATTCAATGG-3′ and 5′-GGAACCGAAGTCGCGCTTAATAGT-3′ [99]. The expression of tatn-1 was detected using 5′-CTTGATCAGAGAAGAATCAGTG-3′ and 5′-GAGTGTTGATTGAAGTTGCG-3′. These oligos were designed to cross intron-exon boundaries using the PerlPrimer program [100].
Whole transcriptome RNA sequencing
N2 wild-type control and tatn-1(qd182) mutant worms were synchronized via the use of hypochlorite treatment and grown on HB101 spotted NGM plates at 25°C for 2 days. These conditions and time point correspond to the conditions used for the amino acid analysis separately performed using these strains. The worms were washed from the plates and were then washed twice with miliQ water. The worm pellet was then suspended in QIAzol lysis reagent (Qiagen, Valencia, CA) and frozen at −80°C. Total RNA was isolated using the Qiagen miRNeasy mini kit and the RNA yield was measured by spectrophotometry. Total RNA was sent to Expression Analysis (Durham, NC) for analysis including bioanalyzer electrophoresis to ensure RNA quality followed by library preparation using the Illumina TruSeq RNA sample prep kit. The resulting library was subjected to high-throughput 50 nucleotide paired end sequencing using an Illumina sequencer at a depth of 17 million reads per sample.
The resulting sequence data was clipped using internally developed software from Expression Analysis and matched to the C. elegans genome using RSEM [101]. The resulting transcript counts were then normalized using the upper quartile normalization approach [102]. Differentially expressed genes were then identified through the use of ANOVA testing and genes with a FDR score of 5% or lower were considered to be differentially expressed. This lead to the identification of 4622 genes as being differentially expressed (890 up-regulated and 3732 down-regulated) between tatn-1(qd182) and wild-type N2.
Over-represented gene classes were identified in the up-regulated and down-regulated genes through the use of DAVID [60]. Analysis of the transcriptome data for daf-16/FOXO and crh-1/CREB regulated genes was performed using the Gene Set Association Analysis for RNA-seq program (available at http://gsaa.unc.edu/login/index.html) [63]. GSAA calculates a differential expression score for each gene in the entire RNA-seq dataset, 20408 genes in all, and then uses a running weighted Kolmogorov-Smirnov test to examine association of an entire gene set with each phenotypic class. The strength of the association is measured by the association score (AS) where positive scores indicate association of the gene set with the phenotype, and statistical significance is measured by a false discovery rate (FDR) that is adjusted for multiple testing. The daf-16/FOXO regulated genes were from previously published microarray data from Murphy et. al., and the crh-1/CREB regulated genes were from recently published microarray data from Mair et. al. [53], [62].
Generation of TATN-1:GFP transgenic animals
Clones for the tatn-1 promoter and cDNA where purchased from Open Biosystems and verified by sequencing [103], [104]. A tatn-1p:tatn-1 cDNA:GFP transgene was generated using Gateway cloning and the vector pDEST-MB14 [104]. The resulting transgene or punc-119cbr, which contains the unc-119 gene from Caenorhabditis briggsae, was used to bombard HT1593 (unc-119(ed3)) as previously described [105], [106]. From bombardment we obtained bafIs130 from punc-119cbr, which carries the unc-119 gene from Caenorhabditis briggsae, and bafIs131 from the tatn-1p:tatn-1 cDNA:GFP transgene [106]. Both transgenes were outcrossed with N2 and then mated with eak-4(mg348); tatn-1(baf1) to test for rescue of the dauer formation phenotype. The bafIs131 transgene was also crossed into a daf-2(e1368) mutant to generate daf-2(e1368); bafIs131.
Anti-phospho AMPK western blotting
Worm embryos were isolated by sodium hypochlorite treatment, and eggs were transferred to NGA plates spotted with HB101 and incubated at 25°C for 24 hours. For treatment with AICAR, the plate was spotted 1 hour before adding the eggs with aliquots from a 250 mM AICAR stock that was dissolved in water. As a positive control, N2 wild-type worms were grown as above, washed off in S-basal, and then exposed to 10 mM sodium azide in S-basal. This dose of S-basal has been previously shown to result in a 50% decrease in ATP concentrations in treated worms [54]. The worms were washed from plates in water, pelleted by centrifugation, washed with water, and suspended in 1× LDS loading buffer (Invitrogen) before being heated to 70°C in a Bransonic sonicator waterbath (Branson) for 30–40 minutes [107]. After heating the samples were centrifuged and the supernatant retained for analysis. The protein levels were measured using the CB-X protein assay kit (G-Biosciences), and 30 µg of protein was run on a 10% Nupage SDS-PAGE gel (Invitrogen) and blotted to a nitrocellulose membrane. Phospho-AAK-2 was detected using a rabbit monoclonal antibody (Cell Signaling Technology #2535), and actin was detected using a rabbit anti-actin antibody (Cell Signaling Technology #4967) followed by detection by an anti-rabbit HRP secondary antibody and visualization using chemiluminescence (Bio-Rad). The resulting X-ray film was scanned and quantified using gel analysis tools in ImageJ [108].
Supporting Information
Zdroje
1. NewgardCB, AnJ, BainJR, MuehlbauerMJ, StevensRD, et al. (2009) A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9 : 311–326.
2. WurtzP, TiainenM, MakinenVP, KangasAJ, SoininenP, et al. (2012) Circulating Metabolite Predictors of Glycemia in Middle-Aged Men and Women. Diabetes Care
3. WangTJ, LarsonMG, VasanRS, ChengS, RheeEP, et al. (2011) Metabolite profiles and the risk of developing diabetes. Nat Med 17 : 448–453.
4. StancakovaA, CivelekM, SaleemNK, SoininenP, KangasAJ, et al. (2012) Hyperglycemia and a Common Variant of GCKR Are Associated With the Levels of Eight Amino Acids in 9,369 Finnish Men. Diabetes
5. ChengS, RheeEP, LarsonMG, LewisGD, McCabeEL, et al. (2012) Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation 125 : 2222–2231.
6. WurtzP, MakinenVP, SoininenP, KangasAJ, TukiainenT, et al. (2012) Metabolic signatures of insulin resistance in 7,098 young adults. Diabetes 61 : 1372–1380.
7. FuL, DongSS, XieYW, TaiLS, ChenL, et al. (2010) Down-regulation of tyrosine aminotransferase at a frequently deleted region 16q22 contributes to the pathogenesis of hepatocellular carcinoma. Hepatology 51 : 1624–1634.
8. CassadaRC, RussellRL (1975) The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol 46 : 326–342.
9. Riddle DL, Albert PS, Riddle DL, Blumenthal T, Meyer BJ, et al.. (1997) Genetic and environmental regulation of dauer larva formation. C elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. pp. 739–768.
10. HuPJ (2007) Dauer. WormBook 1–19.
11. FielenbachN, AntebiA (2008) C. elegans dauer formation and the molecular basis of plasticity. Genes Dev 22 : 2149–2165.
12. RiddleDL, SwansonMM, AlbertPS (1981) Interacting genes in nematode dauer larva formation. Nature 290 : 668–671.
13. KenyonC, ChangJ, GenschE, RudnerA, TabtiangR (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366 : 461–464.
14. KimuraKD, TissenbaumHA, LiuY, RuvkunG (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277 : 942–946.
15. PierceSB, CostaM, WisotzkeyR, DevadharS, HomburgerSA, et al. (2001) Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes and Development 15 : 672–686.
16. LiW, KennedySG, RuvkunG (2003) daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev 17 : 844–858.
17. MichaelsonD, KortaDZ, CapuaY, HubbardEJ (2010) Insulin signaling promotes germline proliferation in C. elegans. Development 137 : 671–680.
18. CornilsA, GloeckM, ChenZ, ZhangY, AlcedoJ (2011) Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development 138 : 1183–1193.
19. MaloneEA, InoueT, ThomasJH (1996) Genetic analysis of the roles of daf-28 and age-1 in regulating Caenorhabditis elegans dauer formation. Genetics 143 : 1193–1205.
20. ParadisS, RuvkunG (1998) Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes and Development 12 : 2488–2498.
21. ParadisS, AilionM, TokerA, ThomasJH, RuvkunG (1999) A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes and Development 13 : 1438–1452.
22. MorrisJZ, TissenbaumHA, RuvkunG (1996) A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382 : 536–539.
23. LinK, HsinH, LibinaN, KenyonC (2001) Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. NatGenet 28 : 139–145.
24. LeeRY, HenchJ, RuvkunG (2001) Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. CurrBiol 11 : 1950–1957.
25. HendersonST, JohnsonTE (2001) daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. CurrBiol 11 : 1975–1980.
26. ZhangY, XuJ, PuscauC, KimY, WangX, et al. (2008) Caenorhabditis elegans EAK-3 inhibits dauer arrest via nonautonomous regulation of nuclear DAF-16/FoxO activity. Dev Biol 315 : 290–302.
27. AlamH, WilliamsTW, DumasKJ, GuoC, YoshinaS, et al. (2010) EAK-7 controls development and life span by regulating nuclear DAF-16/FoxO activity. Cell Metab 12 : 30–41.
28. HuPJ, XuJ, RuvkunG (2006) Two membrane-associated tyrosine phosphatase homologs potentiate C. elegans AKT-1/PKB signaling. PLoS Genet 2: e99.
29. DumasKJ, GuoC, WangX, BurkhartKB, AdamsEJ, et al. (2010) Functional divergence of dafachronic acid pathways in the control of C. elegans development and lifespan. Dev Biol 340 : 605–612.
30. SpencerCJ, HeatonJH, GelehrterTD, RichardsonKI, GarwinJL (1978) Insulin selectively slows the degradation rate of tyrosine aminotransferase. J Biol Chem 253 : 7677–7682.
31. ReshefL, GreengardO (1969) The effect of amino acid mixtures, insulin, epinephrine and glucagon in vivo on the levels of rat liver tyrosine aminotransferase. Enzymol Biol Clin (Basel) 10 : 113–121.
32. NitschD, BoshartM, SchutzG (1993) Activation of the tyrosine aminotransferase gene is dependent on synergy between liver-specific and hormone-responsive elements. Proc Natl Acad Sci U S A 90 : 5479–5483.
33. MoorePS, KoontzJW (1989) Insulin-mediated regulation of tyrosine aminotransferase in rat hepatoma cells: inhibition of transcription and inhibition of enzyme degradation. Arch Biochem Biophys 275 : 486–495.
34. MessinaJL, ChatterjeeAK, StrapkoHT, WeinstockRS (1992) Short - and long-term effects of insulin on tyrosine aminotransferase gene expression. Arch Biochem Biophys 298 : 56–62.
35. LeeKL, IshamKR, JohnsonA, KenneyFT (1986) Insulin enhances transcription of the tyrosine aminotransferase gene in rat liver. Arch Biochem Biophys 248 : 597–603.
36. GelehrterTD, TomkinsGM (1970) Posttranscriptional control of tyrosine aminotransferase synthesis by insulin. Proc Natl Acad Sci U S A 66 : 390–397.
37. GelehrterTD, EmanuelJR, SpencerCJ (1972) Induction of tyrosine aminotransferase by dexamethasone, insulin, and serum. Characterization of the induced enzyme. J Biol Chem 247 : 6197–6203.
38. CrettazM, Muller-WielandD, KahnCR (1988) Transcriptional and posttranscriptional regulation of tyrosine aminotransferase by insulin in rat hepatoma cells. Biochemistry 27 : 495–500.
39. LeeSS, KennedyS, TolonenAC, RuvkunG (2003) DAF-16 target genes that control C. elegans life-span and metabolism. Science 300 : 644–647.
40. FuchsS, BundyJG, DaviesSK, VineyJM, SwireJS, et al. (2010) A metabolic signature of long life in Caenorhabditis elegans. BMC Biol 8 : 14.
41. TewariM, HuPJ, AhnJS, yivi-GuedehoussouN, VidalainPO, et al. (2004) Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGF-beta signaling network. MolCell 13 : 469–482.
42. DuchaineTF, WohlschlegelJA, KennedyS, BeiY, ConteDJr, et al. (2006) Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124 : 343–354.
43. FisherAL, PageKE, LithgowGJ, NashL (2008) The Caenorhabditis elegans K10C2.4 Gene Encodes a Member of the Fumarylacetoacetate Hydrolase Family: A CAENORHABDITIS ELEGANS MODEL OF TYPE I TYROSINEMIA. J BiolChem 283 : 9127–9135.
44. FergusonAA, SpringerMG, FisherAL (2010) skn-1-Dependent and -independent regulation of aip-1 expression following metabolic stress in Caenorhabditis elegans. Mol Cell Biol 30 : 2651–2667.
45. HertweckM, GobelC, BaumeisterR (2004) C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell 6 : 577–588.
46. JensenVL, AlbertPS, RiddleDL (2007) Caenorhabditis elegans SDF-9 enhances insulin/insulin-like signaling through interaction with DAF-2. Genetics 177 : 661–666.
47. NarasimhanSD, YenK, BansalA, KwonES, PadmanabhanS, et al. (2011) PDP-1 links the TGF-beta and IIS pathways to regulate longevity, development, and metabolism. PLoS Genet 7: e1001377.
48. PattersonGI, KoweekA, WongA, LiuY, RuvkunG (1997) The DAF-3 Smad protein antagonizes TGF-beta-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes Dev 11 : 2679–2690.
49. da GracaLS, ZimmermanKK, MitchellMC, Kozhan-GorodetskaM, SekiewiczK, et al. (2004) DAF-5 is a Ski oncoprotein homolog that functions in a neuronal TGF beta pathway to regulate C. elegans dauer development. Development 131 : 435–446.
50. ApfeldJ, O'ConnorG, McDonaghT, DiStefanoPS, CurtisR (2004) The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev 18 : 3004–3009.
51. GreerEL, DowlatshahiD, BankoMR, VillenJ, HoangK, et al. (2007) An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol 17 : 1646–1656.
52. NarbonneP, RoyR (2009) Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 457 : 210–214.
53. MairW, MorantteI, RodriguesAP, ManningG, MontminyM, et al. (2011) Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature 470 : 404–408.
54. LagidoC, PettittJ, FlettA, GloverLA (2008) Bridging the phenotypic gap: real-time assessment of mitochondrial function and metabolism of the nematode Caenorhabditis elegans. BMC Physiol 8 : 7.
55. KwonES, NarasimhanSD, YenK, TissenbaumHA (2010) A new DAF-16 isoform regulates longevity. Nature 466 : 498–502.
56. Robida-StubbsS, Glover-CutterK, LammingDW, MizunumaM, NarasimhanSD, et al. (2012) TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab 15 : 713–724.
57. HondaY, HondaS (1999) The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J 13 : 1385–1393.
58. LibinaN, BermanJR, KenyonC (2003) Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115 : 489–502.
59. BermanJR, KenyonC (2006) Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 124 : 1055–1068.
60. Huang daW, ShermanBT, LempickiRA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 : 44–57.
61. MiH, MuruganujanA, ThomasPD (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41: D377–386.
62. MurphyCT, McCarrollSA, BargmannCI, FraserA, KamathRS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424 : 277–283.
63. XiongQ, AnconaN, HauserER, MukherjeeS, FureyTS (2012) Integrating genetic and gene expression evidence into genome-wide association analysis of gene sets. Genome Res 22 : 386–397.
64. BrooksKK, LiangB, WattsJL (2009) The influence of bacterial diet on fat storage in C. elegans. PLoS ONE 4: e7545.
65. ZigmondMJ, ShoemakerWJ, LarinF, WurtmanRJ (1969) Hepatic tyrosine transaminase rhythm: interaction of environmental lighting, food consumption and dietary protein content. J Nutr 98 : 71–75.
66. WurtmanRJ (1974) Daily rhythms in tyrosine transaminase and other hepatic enzymes that metabolize amino acids: mechanisms and possible consequences. Life Sci 15 : 827–847.
67. RossDS, FernstromJD, WurtmanRJ (1973) The role of dietary protein in generating daily rhythms in rat liver tryptophan pyrrolase and tyrosine transaminase. Metabolism 22 : 1175–1184.
68. IsaacsJS, JungYJ, MoleDR, LeeS, Torres-CabalaC, et al. (2005) HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 8 : 143–153.
69. CitronBA, DavisMD, MilstienS, GutierrezJ, MendelDB, et al. (1992) Identity of 4a-carbinolamine dehydratase, a component of the phenylalanine hydroxylation system, and DCoH, a transregulator of homeodomain proteins. ProcNatlAcadSciUSA 89 : 11891–11894.
70. CalvoAC, PeyAL, YingM, LoerCM, MartinezA (2008) Anabolic function of phenylalanine hydroxylase in Caenorhabditis elegans. FASEB J 22 : 3046–3058.
71. TremblayF, LavigneC, JacquesH, MaretteA (2007) Role of dietary proteins and amino acids in the pathogenesis of insulin resistance. Annu Rev Nutr 27 : 293–310.
72. TremblayF, MaretteA (2001) Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem 276 : 38052–38060.
73. WurtmanRJ, LarinF, MostafapourS, FernstromJD (1974) Brain catechol synthesis: control by train tyrosine concentration. Science 185 : 183–184.
74. LintsR, EmmonsSW (1999) Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFbeta family signaling pathway and a Hox gene. Development 126 : 5819–5831.
75. AlkemaMJ, Hunter-EnsorM, RingstadN, HorvitzHR (2005) Tyramine Functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron 46 : 247–260.
76. GreerEL, OskouiPR, BankoMR, ManiarJM, GygiMP, et al. (2007) The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem 282 : 30107–30119.
77. KooSH, FlechnerL, QiL, ZhangX, ScreatonRA, et al. (2005) The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437 : 1109–1111.
78. DentinR, LiuY, KooSH, HedrickS, VargasT, et al. (2007) Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449 : 366–369.
79. CypserJR, JohnsonTE (2002) Multiple stressors in Caenorhabditis elegans induce stress hormesis and extended longevity. J Gerontol A Biol Sci Med Sci 57: B109–114.
80. WalkerGA, ThompsonFJ, BrawleyA, ScanlonT, DevaneyE (2003) Heat shock factor functions at the convergence of the stress response and developmental pathways in Caenorhabditis elegans. FASEB J 17 : 1960–1962.
81. MorleyJF, MorimotoRI (2004) Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15 : 657–664.
82. HsuAL, MurphyCT, KenyonC (2003) Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300 : 1142–1145.
83. ConigraveAD, QuinnSJ, BrownEM (2000) L-amino acid sensing by the extracellular Ca2+-sensing receptor. Proc Natl Acad Sci U S A 97 : 4814–4819.
84. BittingerMA, NguyenLP, BradfieldCA (2003) Aspartate aminotransferase generates proagonists of the aryl hydrocarbon receptor. Mol Pharmacol 64 : 550–556.
85. SchumacherU, LukacsZ, KaltschmidtC, FreudlspergerC, SchulzD, et al. (2008) High concentrations of phenylalanine stimulate peroxisome proliferator-activated receptor gamma: implications for the pathophysiology of phenylketonuria. Neurobiol Dis 32 : 385–390.
86. LuM, WanM, LeavensKF, ChuQ, MonksBR, et al. (2012) Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat Med 18 : 388–395.
87. WurtmanRJ, AxelrodJ (1967) Daily rhythmic changes in tyrosine transaminase activity of the rat liver. Proc Natl Acad Sci U S A 57 : 1594–1598.
88. LeeSJ, KenyonC (2009) Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans. Curr Biol 19 : 715–722.
89. LangenbergC, SavageDB (2011) An amino acid profile to predict diabetes? Nat Med 17 : 418–420.
90. Sulston JE, Horvitz HR (1988) Methods. In: Wood WB, editor. The nematode, Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. pp. 587–606.
91. PatelDS, FangLL, SvyDK, RuvkunG, LiW (2008) Genetic identification of HSD-1, a conserved steroidogenic enzyme that directs larval development in Caenorhabditis elegans. Development 135 : 2239–2249.
92. LandisJR, KochGG (1977) The measurement of observer agreement for categorical data. Biometrics 33 : 159–174.
93. PowolnyAA, SinghSV, MelovS, HubbardA, FisherAL (2011) The garlic constituent diallyl trisulfide increases the lifespan of C. elegans via skn-1 activation. Exp Gerontol
94. GeierFM, WantEJ, LeroiAM, BundyJG (2011) Cross-platform comparison of Caenorhabditis elegans tissue extraction strategies for comprehensive metabolome coverage. Anal Chem 83 : 3730–3736.
95. HeldPK, WhiteL, PasqualiM (2011) Quantitative urine amino acid analysis using liquid chromatography tandem mass spectrometry and aTRAQ reagents. J Chromatogr B Analyt Technol Biomed Life Sci 879 : 2695–2703.
96. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 : 402–408.
97. HoogewijsD, HouthoofdK, MatthijssensF, VandesompeleJ, VanfleterenJR (2008) Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 9 : 9.
98. HochbaumD, ZhangY, StuckenholzC, LabhartP, AlexiadisV, et al. (2011) DAF-12 regulates a connected network of genes to ensure robust developmental decisions. PLoS Genet 7: e1002179.
99. LiJ, EbataA, DongY, RizkiG, IwataT, et al. (2008) Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol 6: e233.
100. MarshallOJ (2004) PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20 : 2471–2472.
101. LiB, DeweyCN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12 : 323.
102. BullardJH, PurdomE, HansenKD, DudoitS (2010) Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11 : 94.
103. LameschP, MilsteinS, HaoT, RosenbergJ, LiN, et al. (2004) C. elegans ORFeome version 3.1: increasing the coverage of ORFeome resources with improved gene predictions. Genome Res 14 : 2064–2069.
104. DupuyD, LiQR, DeplanckeB, BoxemM, HaoT, et al. (2004) A first version of the Caenorhabditis elegans Promoterome. Genome Res 14 : 2169–2175.
105. HochbaumD, FergusonAA, FisherAL (2010) Generation of transgenic C. elegans by biolistic transformation. J Vis Exp
106. FergusonAA, CaiL, KashyapL, FisherAL (2013) Improved Vectors for Selection of Transgenic Caenorhabditis elegans. Methods Mol Biol 940 : 87–102.
107. ZaninE, DumontJ, GassmannR, CheesemanI, MaddoxP, et al. (2011) Affinity purification of protein complexes in C. elegans. Methods Cell Biol 106 : 289–322.
108. Abramoff MDMPJ (2004) RamSJ (2004) Image Processing with ImageJ. Biophotonics International 11 : 36–42.
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



