-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMAN1B1 Deficiency: An Unexpected CDG-II
Congenital disorders of glycosylation (CDG) are a group of rare metabolic diseases, due to impaired protein and lipid glycosylation. In the present study, exome sequencing was used to identify MAN1B1 as the culprit gene in an unsolved CDG-II patient. Subsequently, 6 additional cases with MAN1B1-CDG were found. All individuals presented slight facial dysmorphism, psychomotor retardation and truncal obesity. Generally, MAN1B1 is believed to be an ER resident alpha-1,2-mannosidase acting as a key factor in glycoprotein quality control by targeting misfolded proteins for ER-associated degradation (ERAD). However, recent studies indicated a Golgi localization of the endogenous MAN1B1, suggesting a more complex role for MAN1B1 in quality control. We were able to confirm that MAN1B1 is indeed localized to the Golgi complex instead of the ER. Furthermore, we observed an altered Golgi morphology in all patients' cells, with marked dilatation and fragmentation. We hypothesize that part of the phenotype is associated to this Golgi disruption. In conclusion, we linked mutations in MAN1B1 to a Golgi glycosylation disorder. Additionally, our results support the recent findings on MAN1B1 localization. However, more work is needed to pinpoint the exact function of MAN1B1 in glycoprotein quality control, and to understand the pathophysiology of its deficiency.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1003989
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003989Summary
Congenital disorders of glycosylation (CDG) are a group of rare metabolic diseases, due to impaired protein and lipid glycosylation. In the present study, exome sequencing was used to identify MAN1B1 as the culprit gene in an unsolved CDG-II patient. Subsequently, 6 additional cases with MAN1B1-CDG were found. All individuals presented slight facial dysmorphism, psychomotor retardation and truncal obesity. Generally, MAN1B1 is believed to be an ER resident alpha-1,2-mannosidase acting as a key factor in glycoprotein quality control by targeting misfolded proteins for ER-associated degradation (ERAD). However, recent studies indicated a Golgi localization of the endogenous MAN1B1, suggesting a more complex role for MAN1B1 in quality control. We were able to confirm that MAN1B1 is indeed localized to the Golgi complex instead of the ER. Furthermore, we observed an altered Golgi morphology in all patients' cells, with marked dilatation and fragmentation. We hypothesize that part of the phenotype is associated to this Golgi disruption. In conclusion, we linked mutations in MAN1B1 to a Golgi glycosylation disorder. Additionally, our results support the recent findings on MAN1B1 localization. However, more work is needed to pinpoint the exact function of MAN1B1 in glycoprotein quality control, and to understand the pathophysiology of its deficiency.
Introduction
Congenital Disorders of Glycosylation (CDG) are a group of genetic diseases, due to deficient protein and lipid glycosylation [1]. To date, over 60 distinct disorders have been described, comprising a very broad range of phenotypes [2]. However, the search for the culprit gene in an unsolved CDG case can be very laborious, due to its heterogeneous clinical presentation and the extensive list of candidate genes. Over the last few years, massive parallel sequencing techniques have permitted to identify the underlying genetic defect in a growing number of diseases. In case of CDG, exome sequencing led to the discovery of 7 novel disorders in less than 2 years [3].
N-glycosylation of proteins is a meticulously orchestrated process occurring in the cytosol, the endoplasmic reticulum (ER) and the Golgi apparatus. Responsible for the modification of secreted and transmembrane proteins, N-glycosylation is considered as one of the most widespread forms of glycosylation. A prerequisite for export of a protein out of the ER and its further transport through the secretory pathway is the adaptation of a native folding state [4]. However, protein folding is inherently error prone. Therefore, a quality control system has evolved to ensure that only properly folded proteins reach the plasma membrane. Glycoproteins unable to acquire a correct conformation will be recognized as terminally misfolded and marked for degradation by demannosylation of their glycan moiety.
The α(1,2)-mannosidase, MAN1B1, catalyzes the removal of the terminal mannose residue from the middle branch of Man9GlcNAc2, hence generating a degradation signal corresponding to Man8GlcNAc2 isomer B [5]. Initially, MAN1B1 was predicted to function as an ER resident protein, based on the localization of its yeast orthologue Mns1p [6]. Also the overexpressed recombinant human orthologue was found to reside within the ER [7]. However, recent studies indicate that the endogenous MAN1B1 localizes to the Golgi apparatus of mammalian cells, implying that quality control is not confined to the ER, but stretches throughout the secretory pathway [8]. In this new model, MAN1B1 functions not only as a checkpoint for misfolded proteins escaping the ER, but also as a lectin retrieving these proteins back to the ER prior to their degradation by the 26S proteasome.
Interestingly, mutations in MAN1B1 have been recently associated with non-syndromic autosomal recessive intellectual disability (NS-ARID) [9]–[10]. However, in the present study we show that MAN1B1 deficiency is associated with N-glycosylation disorders and causes a CDG-II syndrome. In addition, our findings suggest that MAN1B1 plays a role in protein quality control at the level of the Golgi apparatus.
Results
Whole exome sequencing and gene identification
To identify the causative gene in an unsolved CDG-II case, we performed whole exome sequencing by using the SeqCap EZ Human Exome Library v2.0 and the Illumina HiSeq2000. A total number of 29,518,319 reads were retrieved. 89% of the reads generated passed CASAVA quality filters, were unique, and mapped to the reference genome. The mean target coverage was 56, with 83% of the target covered at least 20-fold. Quality filtering led to a total of 29,437 variants, of those 11,026 were nonsynonymous. 702 variants had a frequency lower than 1% in the 1000 Genomes project. 8 homozygous variants and 132 compound heterozygous variants fitted with a recessive disease model. From this 140 variants, 4 were retained based on a Polyphen-2 score higher than 0.8. Out of this list, 2 were homozygous and 2 compound heterozygous (Table 1). The variant detected in MAN1B1 (MIM 604346) encoded the c.1225T>C (p.S409P) mutation in a homozygous state. The sequencing depth of this variant was 22 reads. On the basis of the described function of MAN1B1, this gene was the best candidate from the list. Using Sanger sequencing, we confirmed the presence of the homozygous missense mutation in exon 8 of MAN1B1. This serine residue is highly conserved. The software tools SIFT, PolyPhen-2 and Project HOPE predicted the mutation to be damaging (Figure S1, Figure S2). The serine 409 is located within an alpha helix of the protein. We hypothesize that the mutation would impair MAN1B1 function by either breaking or kinking this helix.
Tab. 1. Overview of candidate variants identified by exome sequencing. 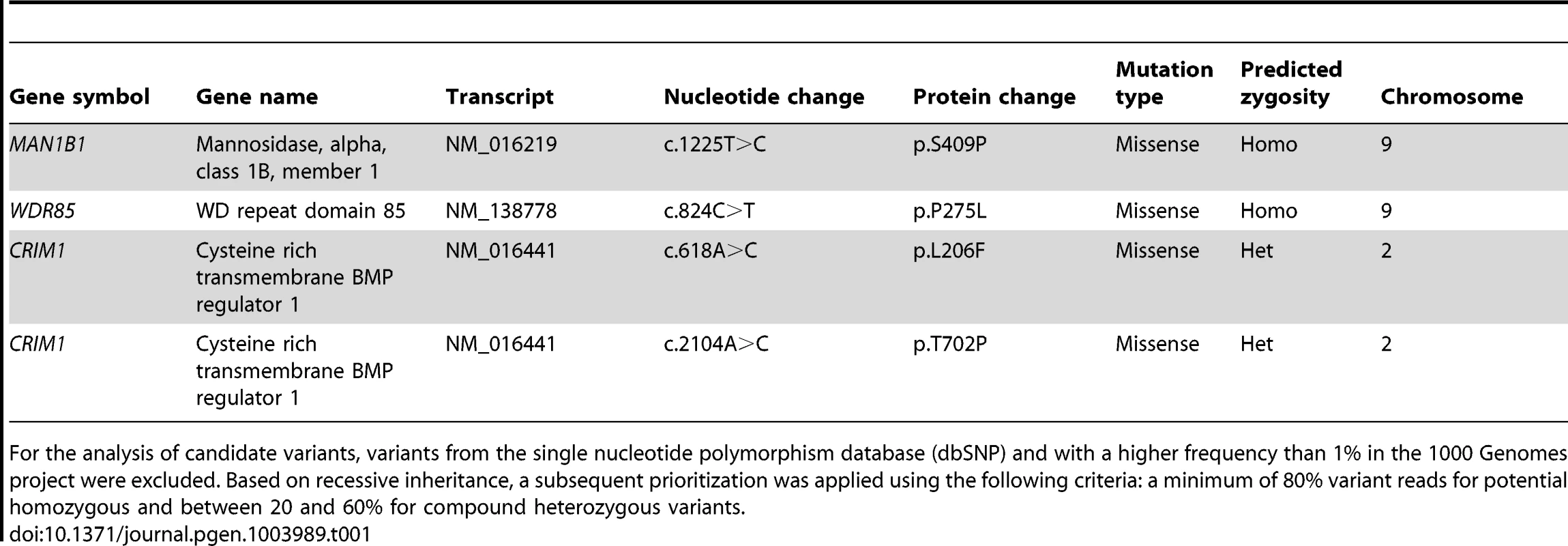
% in the 1000 Genomes project were excluded. Based on recessive inheritance, a subsequent prioritization was applied using the following criteria: a minimum of 80% variant reads for potential homozygous and between 20 and 60% for compound heterozygous variants. For the analysis of candidate variants, variants from the single nucleotide polymorphism database (dbSNP) and with a higher frequency than 1 Sequencing of MAN1B1 in a cohort of individuals with unsolved CDG-II allowed us to identify 6 other cases with MAN1B1 mutations. Case 2 is compound heterozygous for the same missense mutation c.1225T>C (p.S409P) in exon 8, and the nonsense mutation c.172G>T (pE58X) in exon 1. Case 3 was found to carry the homozygous missense mutation c.1000C>T (p.R334C) in exon 7 (Figure S1). Cases 4.1 and 4.2 are homozygous for the same missense mutation as case 3, namely p.R334C. The arginine residue at position 334 is located within an alpha helix of the protein and strictly conserved (Figure S2). Furthermore it is localized at the active site pocket of the protein, where it interacts with the substrate via hydrogen bonding. The conversion to a cysteine might then alter the electrostatic interactions necessary for binding of the substrate in the active site. Case 5 was found to carry a homozygous 2 bp deletion c.1833_1834delAG (p.T611del), causing a frameshift that results in an alternative protein, missing many of the residues required for substrate recognition. Case 6 carried a heterozygous 4 bp deletion in intron 9 (c.1445+2delTGAG). Sequencing of full MAN1B1 cDNA did not only confirm splicing of exon 9 due to the 4 bp deletion, but also revealed a heterozygous loss of exon 4. To prove that this event on cDNA was due to a deletion of exon 4, a long template PCR on genomic DNA stretching from exon 2 to exon 5 was performed. The PCR product revealed a shorter fragment of approximately 3,000 bp instead of the estimated 10,000 bp, confirming the presence of a heterozygous deletion of exon 4 (data not shown). Further sequencing of the boundaries showed a deletion of 6,453 bp (c.465+1460_620+527del). The 4 bp deletion in intron 9 was located on the maternal allele. The deletion of exon 4 occurred de novo on the paternal allele (Figure S1F).
Clinical presentation
The index case (P1, Figure 1A) is a 16 years old girl of Portuguese origin. Head circumference and length at birth were situated at percentile 97. The girl presented with psychomotor retardation and hypotonia. There was slight facial dysmorphism. A brain MRI performed at the age of 7, showed cerebellar hypoplasia with vermian atrophy. To date, she presents severe psychomotor retardation and obesity.
Fig. 1. Clinical features of the seven cases. 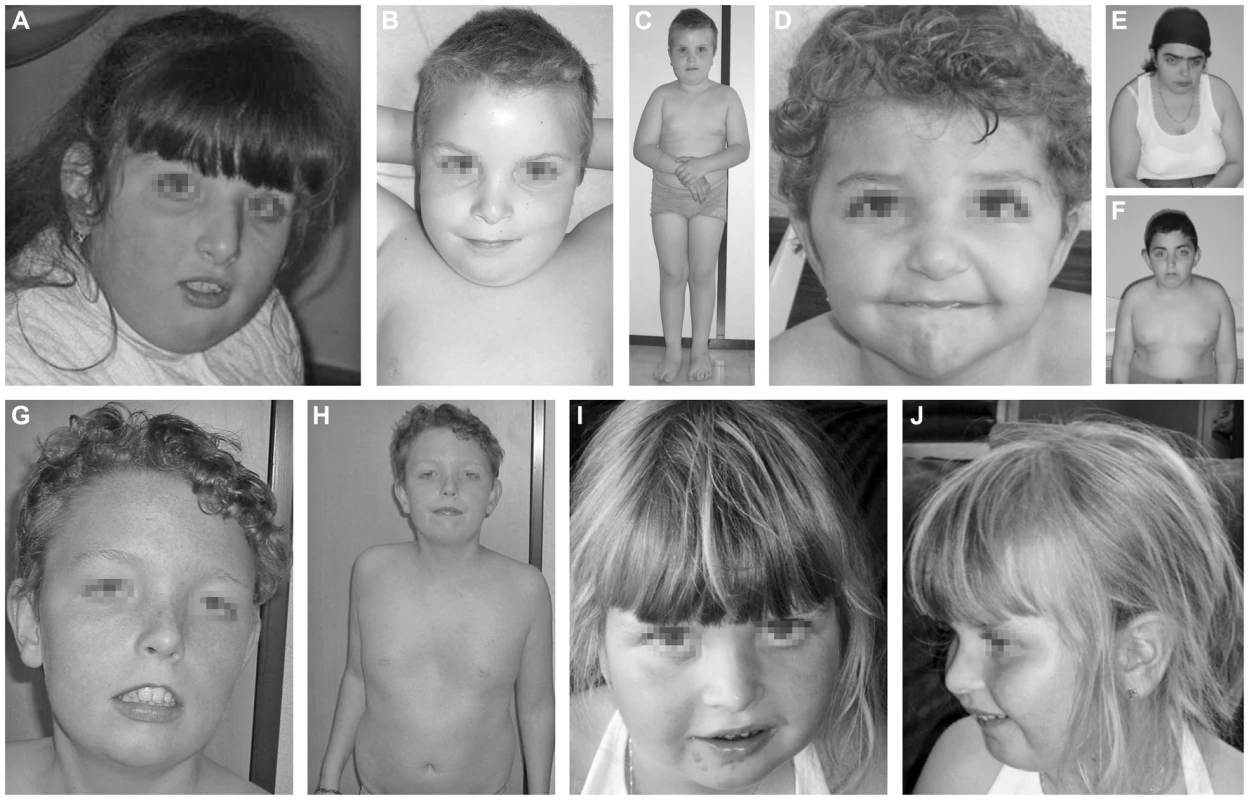
Clinical features of P1 at the age of 7 years (A), P2 at the age of 10 years (B, C), P3 at the age 3.5 years (D), P4.1 at the age of 18 years (E), P4.2 at the age of 12 years (F), P5 at the age of 13 years (G, H) and P6 at the age of 5 years (I, J). Note the facial dysmorphism, i.e. hypertelorism with downslanting palpebral fissures (A, D, E, F, G), large, low set ears (A, B, C, D, E, F, G, J), thin upper lip with hypoplastic nasolabial fold (A, B, D, E, F, G, I), tubular nose in P1, P4.1 and P4.2 (A, E, F) and a depressed nasal bridge in patients P2, P3, P5 and P6 (B, D, G, I). Note the truncular obesity (C, E, F, H) and the widely spaced, inverted nipples (B, F, H). Note the pectus excavatum in P5 (H). Case 2 (P2, Figure 1B, 1C) is a 12 years old boy. He was born as the first child of healthy, non-consanguineous parents of Portuguese origin. Developmental delay was noted at the age of 18 months. He presented with hypotonia and slight facial dysmorphism. Fingers were long and thin. Brain MRI was normal. To date, he displays moderate mental retardation (IQ 43) and obesity (BMI>>P97). Periods of auto-aggressive behaviour and repetitive movements occur.
Case 3 (P3, Figure 1D) is an 11 years old girl of Turkish origin. She presented with delayed motor development at the age of 3 years. Brain MRI was normal. She showed slight facial dysmorphism, hypermobility of the joints and skin laxity. To date she presents with mild mental retardation, short stature (length<P3), truncular obesity (weight at P50) and macrocephaly (head circumference at P97).
Case 4.1 (P4.1, Figure 1E) and case 4.2 (P4.2, Figure 1F) are siblings of Turkish origin. Parents are consanguineous (first cousins). Case 4.1 is a 24 years old woman. She came under medical attention because of mild psychomotor retardation and hypotonia. Brain MRI was normal. She showed slight facial dysmorphism, hypermobility of the joints and skin laxity. She presented an important scoliosis, which was corrected by surgery at the age of 18. To date, she presents mild mental retardation (IQ 62) and truncular obesity.
Case 4.2 is a 18 years old man. He presented with psychomotor retardation and hypotonia. Brain MRI was normal. There was only slight facial dysmorphism and skin laxity. To date, he presents mild mental retardation (IQ 69) and obesity.
Case 5 (P5, Figure 1G, 1H) is a 13 years old boy. He was born at term as the first child of healthy, non-consanguineous parents of Belgian origin. He presented with hypotonia and slight dysmorphic features. Fingers were long and thin. Hypermobility of the joints and skin laxity were noticed, as well as a pectus excavatum. Dilatation of the aortic root was seen on cardiac ultrasound. Brain MRI performed at the age of 3 months was normal. At the age of 5 years he was diagnosed with epilepsy (absences). To date, he presents mild mental retardation and is able to use simple language. Growth and weight evolution are normal, though he displays truncular fat accumulation.
Case 6 (P6, Figure 1I, 1J) is a 12 years old girl. She was born as the first child of healthy, non-consanguineous parents of Belgian origin. Reduced fetal movements were observed during pregnancy. After birth, she was diagnosed to have a muscular ventricular septum defect (VSD). Spontaneous closure occurred. Developmental delay was notified at the age of 18 months. She displayed hypotonia and a delay in motor development. There was slight facial dysmorphism, hypermobility of the joints, and skin laxity. Fingers were long and thin. At the age of 5, she was diagnosed with an autism spectrum disorder and mild mental retardation (IQ 52). Brain MRI showed multiple, small white matter lesions. Rapid bone maturation and early puberty occurred because of overweight.
Clinical features of all patients are furthermore compared in Table 2.
Tab. 2. Clinical features of patients with MAN1B1 deficiency. 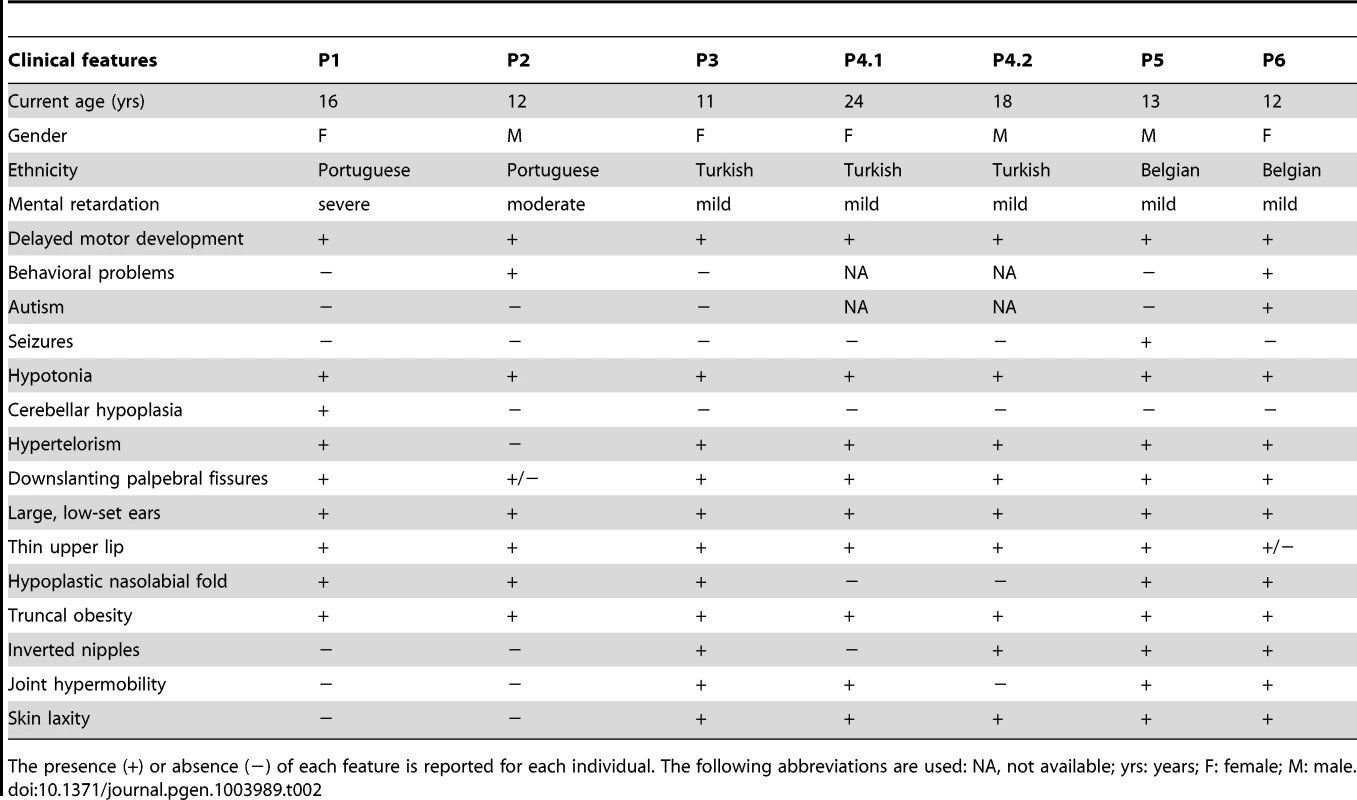
+) or absence (−) of each feature is reported for each individual. The following abbreviations are used: NA, not available; yrs: years; F: female; M: male. The presence ( Capillary zone electrophoresis (CZE) of serum showed a type 2 transferrin pattern with an increase of trisialotransferrin (representing 31 to 41% of the transferrin isoforms, normal range: 1–8%) in all affected cases (Table S1). Isoelectrofocusing of serum transferrin (sTF-IEF) confirmed these results (Figure 2). This categorized them as CDG-II.
Fig. 2. IEF profiles of serum transferrin. 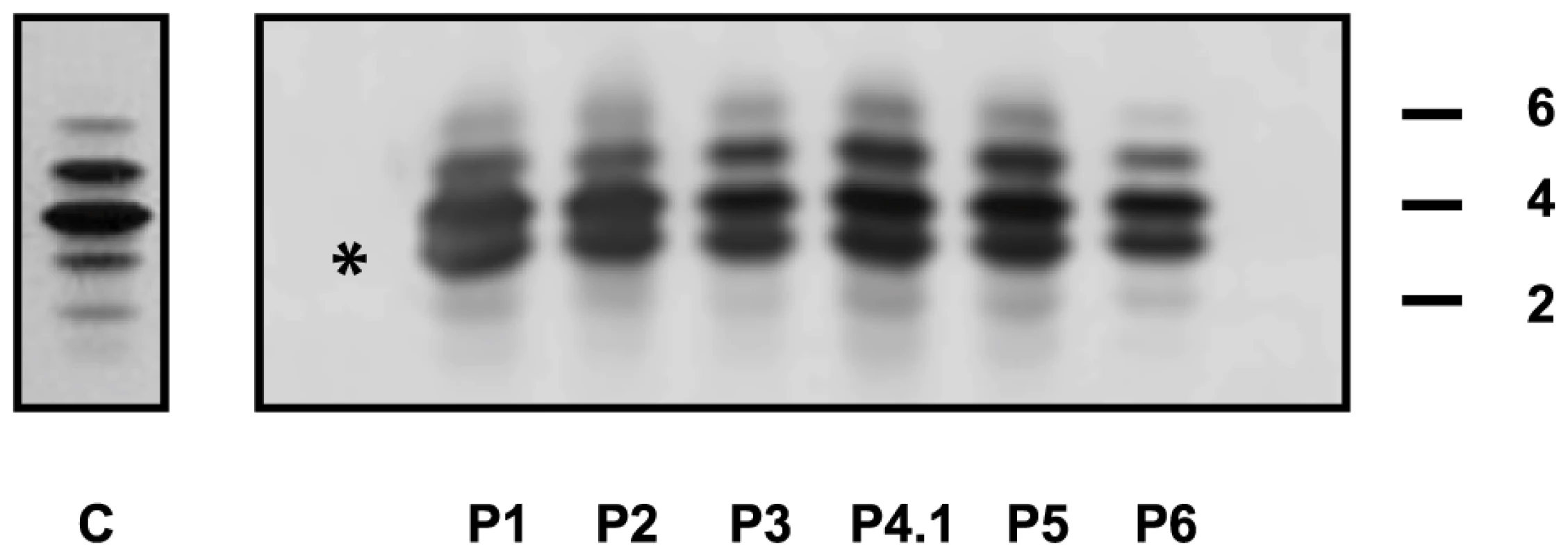
Isoelectrofocusing pattern of serum transferrin in MAN1B1-deficient individuals and a control. 2, 4 and 6 indicate disialo-, tetrasialo-, and hexasialotransferrin isoforms, respectively. The asterisk mark represents the trisialotransferrin isoform. Structural analysis of N-glycans and Golgi morphology
The type 2 sTF-IEF pattern pointed to an N-glycan remodelling defect. Therefore, we investigated the Golgi glycosylation in more detail and determined the structures of the N-linked glycans on serum glycoproteins from controls and affected cases by using mass spectrometry (Figure 3). In MAN1B1-deficient individuals (P2, P5), we observed the accumulation of the hybrid-type glycan structure NeuAc1Hex6HexNAc3 (m/z 2,390), present together with the corresponding fucosylated NeuAc1Hex6HexNAc3dHex1 (m/z 2,564). We also observed a significant increase of the sialylated NeuAc1Hex5HexNAc3 (m/z 2,186) with respect to the control. This glycan also appeared to be present together with the corresponding fucosylated one (NeuAc1Hex5HexNAc3dHex1; m/z 2,360) which was not present in the control. In MAN1B1-CDG cases, there was also a marked increase of Hex6HexNAc2 (m/z 1,783), but no significant variations of Hex5HexNAc2 (m/z 1,579).
Fig. 3. MAN1B1-deficient individuals are deficient in protein N-glycosylation. 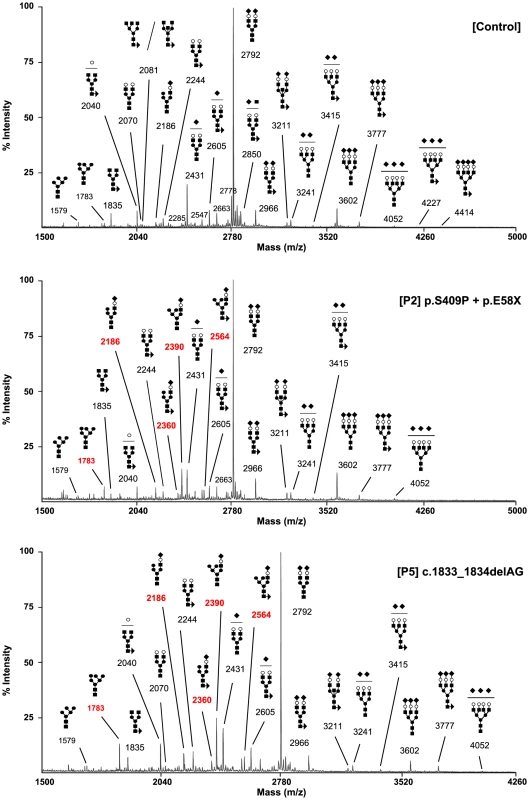
MALDI-TOF spectra of the permethylated N-glycans from sera of control and MAN1B1-deficient individuals. Respective m/z values of N-glycans structures accumulating in MAN1B1-deficient cells are displayed in red. The symbols representing sugar residues are as follows: closed square, N-acetylglucosamine; closed circle: mannose; open circle, galactose; closed diamond, sialic acid; and closed triangle, fucose. Linkages between sugar residues have been removed for simplicity. These results were confirmed in a second group of MAN1B1-CDG cases (P1, P4.1, P4.2 and P6) that were analysed independently (Figure S3). We noticed the presence of hybrid-type glycan structures NeuAc1Hex5HexNAc3 and NeuAc1Hex6HexNAc3. These glycans appeared to be present together with the corresponding fucosylated NeuAc1Hex5HexNAc3dHex1 and NeuAc1Hex6HexNAc3dHex1, respectively. These four species were not detected in controls.
Because the N-glycans abnormalities suggested a Golgi glycosylation defect, Golgi morphology was investigated. Staining with the Golgi markers TGN46 and GM130 detected significant alterations in Golgi morphology in fibroblasts of all affected cases, with marked dilatation and fragmentation (Figure 4).
Fig. 4. MAN1B1-deficient fibroblasts present alterations in Golgi structure. 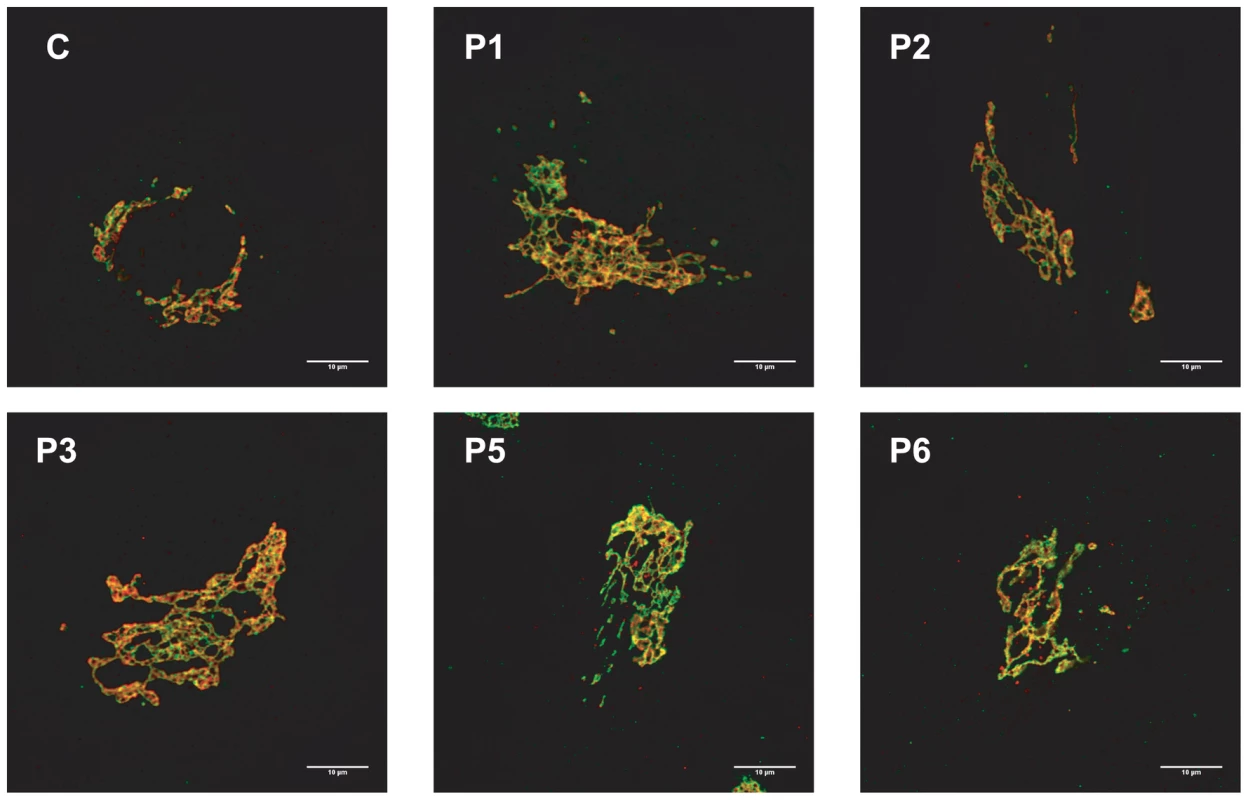
Golgi localization of GM130 (red) and TGN46 (green) in control and MAN1B1-deficient fibroblasts. Cells were fixed, double-labelled with antibodies against GM130 and TGN46, and analysed by confocal laser scanning microscopy. Scale bar represents 10 µm. Functional analysis of MAN1B1 mutations
We used quantitative real-time PCR (qPCR) to investigate the effects of the mutations on MAN1B1 expression by comparing MAN1B1 cDNA levels amplified from mRNA isolated from control and MAN1B1-deficient fibroblasts, cultured in the presence or absence of the translation inhibitor puromycin. Compared to controls, most of the MAN1B1-deficient individuals presented an expression increased 1.22 to 1.49 fold compared to the wild-type MAN1B1 transcript. Case 2, who harbours the nonsense mutation p.E58X, showed an expected reduced expression level of 40% compared to control. Case 6 showed an expression level of 2% compared to control, likely because the deletions on both alleles lead to instability of the transcript (Figure 5A, left panel). Culturing control and MAN1B1-deficient fibroblasts in presence of puromycin did not markedly alter these expression levels (Figure 5A, right panel).
Fig. 5. Functional analyses of MAN1B1 mutations. 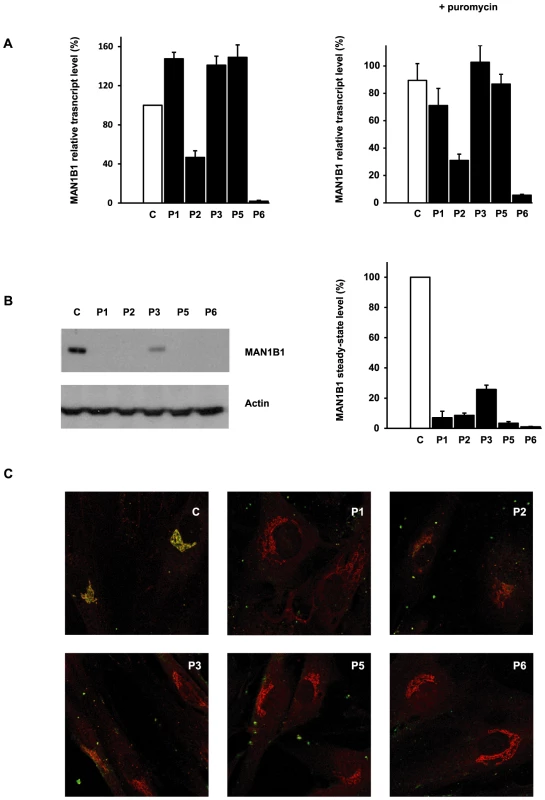
(A) Quantification of the MAN1B1 transcript in controls and affected individuals by qPCR without (left panel) and with (right panel) puromycin. MAN1B1 expression was normalized to the expression of the house-keeping gene HPRT. Values plotted with a wild-type control untreated with puromycin are set to 1. The depicted values are the mean ± SEM of at least three independent experiments. (B) Steady-state levels of the expression of MAN1B1 in control (C) and MAN1B1-deficient (P) fibroblasts. Whole-cell extracts were analysed by immunoblotting using anti-MAN1B1 antibodies. Thirty micrograms of total cell extracts were loaded into each lane and anti-β-actin antibodies were used as a loading control. Quantifications represent the mean value of three independent loadings of three independent samples. (C) Intracellular distribution of MAN1B1 in control and MAN1B1-deficient fibroblasts. Fibroblasts were double-labelled with antibodies against MAN1B1 (green) and the Golgi marker giantin (red), then fixed and processed for analysis by confocal laser scanning microscopy. Images were collected under identical settings. Depicted are the zoomed images. The independent panels are presented in Figure S4. To further investigate the effects of the mutations on MAN1B1, we performed immunoblotting using control and MAN1B1-deficient fibroblasts. As shown in Figure 5B, an extremely low level of endogenous MAN1B1 was detected in affected cases. The steady-state level of MAN1B1 in P3 was reduced to 24%, suggesting that the protein was slightly more stable than in the other affected individuals.
We then compared the subcellular localization of MAN1B1 in control and MAN1B1-deficient fibroblasts by confocal microscopy. In accordance with the data recently published by Pan and coworkers [8], the endogenous MAN1B1 showed a clear perinuclear Golgi-like distribution (Figure 5C and Figure S4, first row) rather than a typical diffuse ER localization. As expected from the western blot analysis, some of the MAN1B1-deficient individuals did not show any staining. A faint Golgi localization could be seen in some of the affected cases, despite the largely predominant Golgi staining for giantin (Figure 5C, Figure S4).
Subcellular MAN1B1 localization
As mentioned above, MAN1B1 displayed a perinuclear Golgi distribution. Double labelling of control cells with MAN1B1 and the rough ER marker PDI showed a clear separation of the two proteins (Figure 6A). A similar pattern was obtained regarding the distribution of both MAN1B1 and the ER protein GRP78, suggesting that the mannosidase was located apart from the ER compartment (data not shown). In order to gain insights into the precise MAN1B1 subcellular localization, we examined its distribution in the Golgi and the ER-Golgi intermediate compartment (ERGIC). MAN1B1 showed a high level of colocalization with both the trans-Golgi marker GPP130 and the cis-Golgi marker giantin. Only weak colocalization with the ERGIC marker ERGIC-53 was noted (Figure 6A). The intensity of the different protein markers was plotted against the distance, using the RGB profiler plugin from Image J (Figure 6B). The level of colocalization was quantified as a percentage of the area occupied by the colocalized spots from the total labelled spots, based on Pearson's coefficient. These results showed a much higher degree of colocalization for MAN1B1 versus GPP130 (80%±3) or giantin (84%±2), than for MAN1B1 versus ERGIC-53 (41%±4), which is consistent with the qualitative data. These results strongly suggest that MAN1B1 is widely distributed within the Golgi apparatus and is hardly or not detectable in the ER.
Fig. 6. Subcellular localisation of MAN1B1. 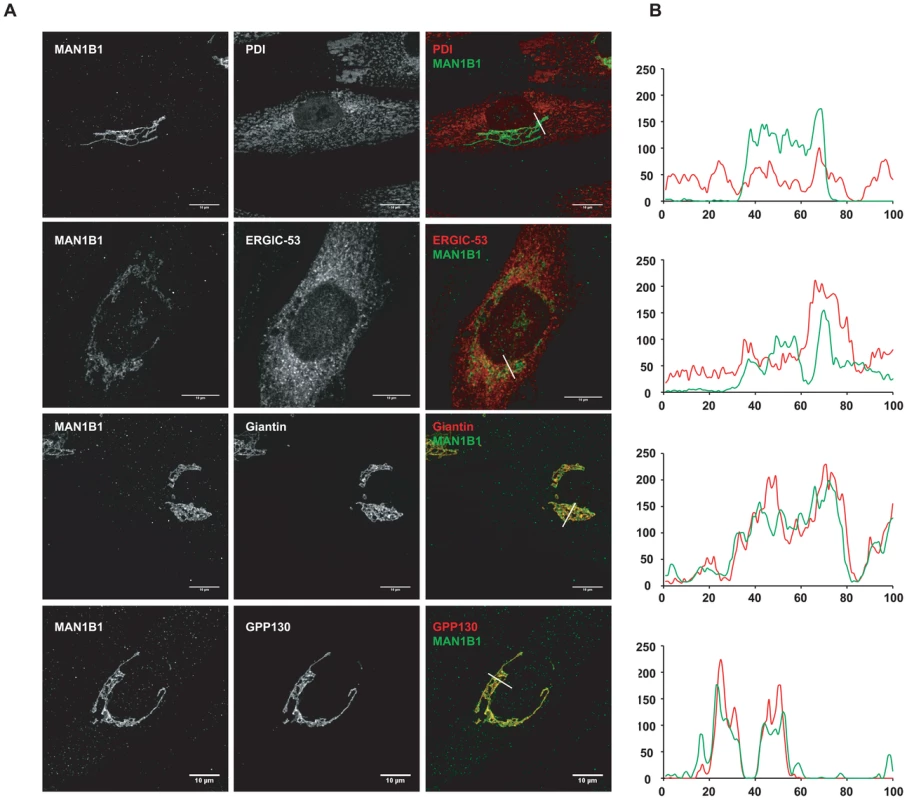
(A) Indirect double immunofluorescence staining of control and MAN1B1-deficient fibroblasts. The cells were double-labelled with antibodies against MAN1B1 and with antibodies against either the Golgi markers GPP130 and giantin, the ER-Golgi intermediate compartment (ERGIC) marker ERGIC-53, or the ER marker PDI. The cells were then examined by confocal laser scanning microscopy. Images were collected under identical settings. Scale bar represents 10 µm. (B) Confocal linescan analysis of the distribution of MAN1B1. The pixel intensity (vertical axis) of MAN1B1 (green) and PDI, ERGIC-53, giantin and GPP130 (from top to bottom, red) was hence measured along a vector drawn perpendicular across the Golgi stack and plotted versus distance (horizontal axis), using the RGB profiler plugin from Image J. MAN1B1 deficient fibroblasts present a delay of trimming from Man9GlcNAc2 to Man8GlcNAc2
The impact of MAN1B1 on Golgi glycosylation was studied in MAN1B1-deficient fibroblasts. The cells of individuals mutated in MAN1B1 were further investigated by metabolic labelling. MAN1B1 is reportedly known to be the key enzyme responsible for the trimming of Man9GlcNac2 species to Man8GlcNAc2 species [5]. To address the potential deleterious effect of the mutations on MAN1B1 function, a structural analysis of the N-linked protein glycans was performed. Therefore, cells were pulse-labelled with 2-[3H] mannose and chased for 2 h. In healthy controls as well as in MAN1B1-deficient cells, Man8GlcNAc2, Man9GlcNAc2 and Glc1Man9GlcNAc2 structures were found after the pulse period (Figure 7A, 7B and Figure S5A). As expected, when the control glycoproteins were chased for 2 hours, an increase of Man8GlcNAc2 species was observed indicating that the Man9GlcNAc2 species can serve as substrate for further trimming by ER/Golgi mannosidases. In contrast, only a slight increase of Man8GlcNAc2 species was seen in MAN1B1-deficient cells (Figure S5B). The processing efficacy from Man9 to Man8 was hence estimated for both healthy controls and MAN1B1-deficient cells. Figure 7C shows that this processing efficacy was estimated to 100%±6.9 in control cells but ranged between 40.1%±5.9 to 43.6%±2.2 for MAN1B1-deficient cells. Surprisingly, this value was much lower for P1 cells, with an average efficacy of 17.7%±3.8. These results strongly suggest that in MAN1B1-deficient cells, a delay in the trimming of Man9GlcNAc2 species occurs. For statistical relevance, we compared the mean processing efficacy of the 3 control cell lines to the mean processing efficacy of all 4 MAN1B1-deficient cell lines (Figure 7D). We conclude that the deficiency of processing from Man9GlcNAc2 species to Man8GlcNAc2 species is not complete, since this value remains of 36.1%±12.8 (p value<0.001) in cells of affected individuals (Figure 7D). This is likely due to the action of other ER/Golgi mannosidases or alpha-mannosidase like proteins such as EDEM proteins, which enable trimming of Man9 to Man8 species in mammalian cells.
Fig. 7. HPLC analysis of N-linked oligosaccharides from control and patient's fibroblasts reveal a delay of trimming from Man9GlcNAc2 to Man8GlcNAc2 in MAN1B1-deficient patients. 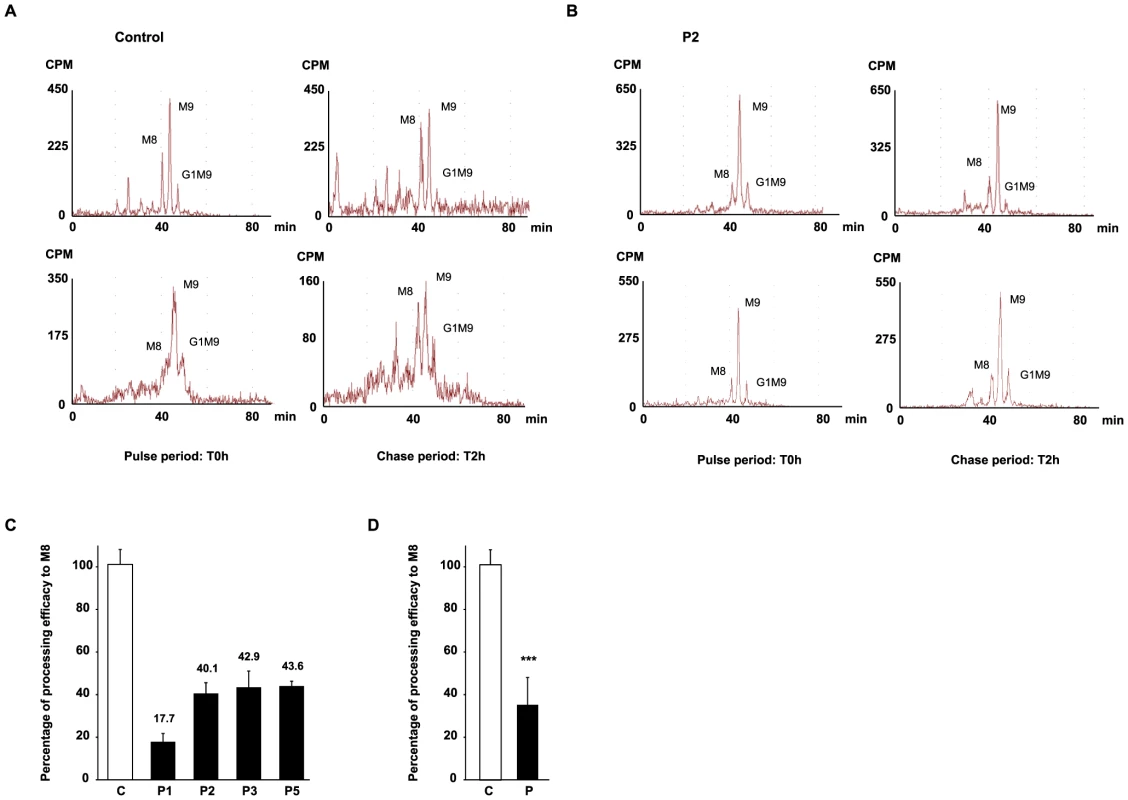
Protein N-linked oligosaccharides of two different control (A) and one MAN1B1-deficient individuals (P2, B) were separated by HPLC after the pulse period (left panels) and 2 hours of chase (right panels). Depicted for both controls and P2 are two representative chromatograms from two independent experiments, with one set of experiments presented in the upper panel, and a second one in the lower panel. Symbols: M8, M9 indicate oligosaccharide species with 8 or 9 mannose residues and possessing two GlcNAc resides at their reducing ends. G1M9 indicates oligosaccharide species with 9 mannose and 1 Glc residues and possessing two GlcNAc residues at their reducing ends. (C) Relative peak areas of M8, M9, and G1M9 were quantified and the results expressed as the percentage of processing efficiency to M8. In the calculation, the trimming efficiency from M9 to M8, which corresponds to the difference between the percentage of M8 at T0 h and T2 h, was considered as 100%. The depicted values are the mean ± SEM of independent experiments. (D) Comparison of the mean processing efficacy of the three control cell lines (C) to the mean processing efficacy of all four MAN1B1-deficient cell lines (P). Data were analysed by using Student's t test. *** indicates a p value<0.001. Discussion
MAN1B1 encodes the endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (ERManI), which cleaves the terminal mannose from the middle branch of Man9GlcNAc2, then producing Man8GlcNAc2 isomer B. It is believed to play a key role in glycoprotein quality control, by targeting terminally misfolded proteins for ERAD. The capacity to distinguish between terminally misfolded proteins and properly folded intermediates as appropriate ERAD substrates is indeed known to be one of the early steps in protein quality control. Several studies in both yeast and mammalian cells already provided evidence that the removal of terminal mannose residues from asparagine-linked Man9GlcNAc2 glycan structures generates a signal for ERAD [11]. However, the exact function of MAN1B1 is still unclear.
In the present study, we definitely link mutations in MAN1B1 to pathology that presents as a deficiency of glycosylation. The first patients with MAN1B1 deficiency were detected in a study on NS-ARID [9]. We identified MAN1B1 mutations in 7 cases with unsolved CDG-II. All patients presented a certain degree of intellectual disability, but also had facial dysmorphism and obesity. Furthermore, some of them displayed joint hypermobility and skin laxity. Dysmorphic features were similar for all cases, i.e. downslanting palpebral fissures, hypertelorism, large low set ears, a hypoplastic nasolabial fold and a thin upper lip. These clinical characteristics correspond to the phenotype of the formerly published patients, although the author assigned the dysmorphism to family traits rather than to the disease [9]. Overall, individuals with MAN1B1-CDG present a mild phenotype, which is in concordance with studies in Saccharomyces cerevisiae, since the yeast orthologue MNS1 is not essential for growth [12]. The mild disease resulting from MAN1B1 deficiency however contrasts with another defect of N-glycan trimming, which is caused by mutations in the glucosidase I (MOGS) gene. A single case of MOGS deficiency has been reported so far, characterized by dysmorphism and hypotonia, leading to the death of the affected infant [13]. We believe that MAN1B1 deficiency belongs to the relatively frequent CDG. In our cohort of solved CDG-II cases, the occurrence of MAN1B1-CDG is indeed over 25%, more frequent than TMEM165-CDG, COG5-CDG or COG7-CDG (14% respectively). From the overall number of solved CDG cases (CDG-I plus CDG-II) in our database, MAN1B1-CDG is the fifth most frequently encountered CDG type (1.9%), right behind SRD5A3-CDG (2.6%) or PMI-CDG (2.8%). PMM2-CDG and ALG6-CDG are the most frequent (69.9% and 8.5%, respectively). The genetic results were supported by a significant decrease in steady-state levels of the MAN1B1 protein. Enzymatic activity was measured indirectly by pulse-chase experiments with 2-[3H] mannose, which showed a significant delay in the trimming of Man9GlcNAc2 to Man8GlcNAc2. The trimming efficiency in MAN1B1-deficient cells was still up to 36%. This activity is likely due to the action of other ER/Golgi class I mannosidases or alpha - mannosidase-like proteins such as EDEM proteins, cleaving the terminal mannose residue at a lower rate.
Former studies localized the overexpressed recombinant MAN1B1 protein within the ER or the ERGIC. The S. cerevisiae homologue of MAN1B1, Mns1p, was originally demonstrated to function as an ER-resident protein [14]. The human MAN1B1 was then identified and cloned on the basis of its close sequence homology with MNS1 and was therefore predicted to localize and function in the ER [7] [15], despite a more than 50% sequence homology with Golgi α(1,2)-mannosidases IA, IB an IC [15]. We could clearly demonstrate that the endogenous MAN1B1 shows a Golgi distribution in primary skin fibroblasts. Further determination of its localization within the organelle revealed an equal distribution throughout the Golgi apparatus. This result confirms the initial observation of Sifers and coworkers, who used a panel of highly specific mAbs to demonstrate that human endogenous ERManI is O-glycosylated and localized in the Golgi apparatus [8]. As expected, only a faint or even no Golgi localization could be seen in MAN1B1-deficient cells.
One can wonder why the MAN1B1 protein, which is assumed to play a pivotal role in ERAD substrate generation, is physically separated from the ER. Traditionally, MAN1B1 was believed to function as a molecular clock in ER quality control. It was speculated that the slow processing activity of MAN1B1 determined the time needed for a protein to acquire its native folding state. In case of a terminally misfolded protein, futile folding cycles would provide MAN1B1 prolonged access to the glycan, resulting in its demannosylation and thereby marking the protein for proteasomal degradation [16]. However, recent findings indicate that the quality control system is not confined to the ER but stretches throughout the secretory pathway. In this model, MAN1B1 operates as a checkpoint within the Golgi apparatus, recycling misfolded proteins that escaped ERAD back to the ER, by interacting with the COP-I machinery. It is assumed that demannosylation of the glycan moiety will prevent the recycled protein from reentering the folding cycle after its retrieval to the ER. Instead, its degradation signal will be recognized by ER resident lectins, leading to proteasome mediated disposal of the misfolded protein [8].
How does MAN1B1 deficiency leads to disease? The purpose of quality control is to minimize the level and toxicity of misfolded proteins within the cell. So, failure of a checkpoint to recognize ERAD substrates might have serious repercussions on cell function. However, patients with MAN1B1-CDG present only a mild phenotype. Hence, one could assume that additional checkpoints exist in the Golgi apparatus. This hypothesis is supported by the observation that other Golgi α(1,2)-mannosidases were shown to be involved in glycoprotein quality control. In case of MAN1B1 deficiency, these Golgi mannosidases might be recruited for post-ER surveillance at the expense of their normal processing activity, leading to a delay in the trafficking of glycoproteins through the secretory pathway. If so, a relative accumulation of glycoproteins will occur within the Golgi apparatus prior to their impaired secretion. Storage of proteins will then activate a stress response increasing the capacity of the Golgi cisternae. The fact that MAN1B1 deficient cells display an altered Golgi morphology, characterized by dilatation and fragmentation of the cisternae, could support this hypothesis. However, this suggestion of a Golgi storage disease is in conflict with the model raised by Pan and coworkers, rather implying an increased secretion of misfolded glycoproteins in MAN1B1-depleted cells [8].
Furthermore, we believe that part of the phenotype might be explained by these Golgi abnormalities. First of all, disruption of Golgi morphology could explain the glycosylation deficiency observed in MAN1B1-deficient patients, by affecting the stability of key proteins responsible for the creation of the microenvironment necessary for proper Golgi glycosylation. Next, some of the observed clinical features (skin laxity, joint hypermobility) are also described in patients with COG5-, COG7 - and ATP6VOA2-CDG. Interestingly, MAN1B1-deficient cells, as well as ATP6V0A2 - and COG-deficient cells, show an altered Golgi morphology [17]–[18]. Furthermore, impaired secretion and intracellular retention of tropoelastin were already proven to result in skin laxity and joint hypermobility in patients with ATP6V0A2-CDG, implying that a similar mechanism could cause skin laxity in MAN1B1-deficient patients [19]. Finally, since the secretory pathway plays also an important role in neurotransmission and endocrine regulation, one can hypothesize that impaired protein secretion underlies the intellectual disability and obesity in our patients.
In conclusion, we have identified MAN1B1 deficiency as a relatively frequent CDG-II, since the occurrence of MAN1B1-CDG is over 25% in our cohort of unsolved CDG-II cases. Overall, the clinical picture is mild, comprising not only intellectual disability, but also truncal obesity and facial dysmorphism, hence defining MAN1B1-CDG as a syndrome. Furthermore, our results confirm that MAN1B1 is indeed localized to the Golgi apparatus. We hypothesize that part of the phenotype is linked to the disruption of Golgi morphology. Though, more work is needed to pinpoint the exact function of MAN1B1 in glycoprotein quality control and to understand the pathophysiology of its deficiency.
Materials and Methods
Ethics statement
Research on patients' cells was prospectively reviewed and approved by the Ethical Committee of the University Hospital of Leuven. PLOS consent forms were obtained for publication of clinical pictures.
Capillary zone electrophoresis and isoelectric focusing of serum transferrin
Capillary zone electrophoresis (CZE) and isoelectric focusing of serum transferrin (sTF-IEF) were performed as previously described [20].
Patients and cells
Primary fibroblasts from patients and controls were grown from a skin biopsy and cultured in Dulbecco's modified Eagle medium DMEM/F12 (Life Technologies) supplemented with 10% fetal bovine serum (Clone III, HyClones) at 37°C under 5% CO2.
Exome sequencing
For the index case, mutation analysis was achieved by whole exome sequencing. Genomic DNA was sheared by sonication, platform-specific adaptors were ligated, and the resulting fragments were size selected. The library was captured using the SeqCap EZ Human Exome Library v2.0 (Roche NimbleGen), and 2× 76 bp paired-end reads were generated on a HiSeq2000 (Illumina). Reads that did not pass Illumina's standard filters were removed prior to alignment. Remaining reads were aligned to the reference human genome (hg19), using the CASAVA pipeline. Quality filtering was applied by excluding variants found in less than 5 reads and variants detected in less than 15% variant reads. Synonymous variants were excluded. To retrieve the quality-filtered private variants, those with a higher frequency than 1% in the 1000 Genomes project were excluded. Based on recessive inheritance, a subsequent prioritization was applied using the following criteria: a minimum of 80% variant reads for potential homozygous variants and between 20 and 60% for compound heterozygous variants. Variants with a Polyphen-2 score of less than 0.8 were filtered out.
Sanger sequencing
Total RNA was isolated from the primary fibroblasts of the patient and control cells using the RNeasy Kit (Qiagen). The amount of extracted RNA was quantified using a NanoDrop spectrophotometer (Thermo Scientifics) and the purity of RNA was checked by the ratio of the absorbance at 260 and 280 nm. 2 µg of purified total RNA was then subjected to reverse transcription with the First-Strand cDNA synthesis Kit (GE Healthcare) following the manufacturer's instructions. For PCRs, cDNAs obtained after reverse transcription were diluted 1∶5.
Genomic DNA was extracted from white blood cells or fibroblasts from the patients and controls. Primers (available on demand) were designed to amplify the different exons of the MAN1B1 gene (GenBank accession number NM_016219.4), including at least 50 bp of the flanking intronic regions. Consecutive primers within intron 3 and intron 4 were designed to find deletion breakpoints. Standard cDNA - and gDNA-based PCR reactions were based on 1 µl DNA in a total volume of 25 µl, 0.2 µl Platinum Taq polymerase (Invitrogen). Standard reaction conditions were 3 min at 95°C, then 10 cycles of 30 sec at 95°C, 30 sec at 65°C (−1°C each cycle), 2 min at 72°C, followed by 25 cycles of 30 sec at 95°C, 30 sec at 55°C and 2 min at 72°C. The reaction was finished by an incubation of 5 min at 72°C.
A long template PCR stretching from exon 2 to exon 5 was performed using the Expand Long Template PCR System (Roche Applied Science). The PCR reaction was based on 4 µl DNA in a total volume of 50 µl, 0.75 µl Expand Long Template Enzyme Mix, 5 µl of Buffer 2, a final 300 nM concentration of each primer and a final 500 µM concentration of dNTPs. Reaction conditions were 2 min at 95°C, then 10 cycles of 10 sec at 95°C, 30 sec at 65°C, 8 min at 68°C, followed by 25 cycles of 15 sec at 94°C, 30 sec at 65°C and 8 min (+20 sec each cycle) at 68°C. The reaction was completed with a final elongation of 7 min at 68°C.
For the sequencing of the resulting PCR product, the BigDye Terminator Ready reaction cycle sequencing kit v.3.1 (Applied Biosystems) was used. Analysis of the results was performed on an ABI3100 Avant (Applied Biosystems).
Mass spectrometric analysis of N-linked glycans released from total plasma proteins
The glycans from total plasma N-glycoproteins were released as previously described [21]–[22]. Analysis of N-linked glycans on plasma glycoproteins was carried out by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Ciphergen) essentially as described for MALDI-TOF-MS, and performed in a positive linear ion mode [21]. The reference samples were anonymous plasma samples from patients without CDG and without a lysosomal storage disease. For cases P1, P4.1, P4.2 and P6, the lab of P. Mills and colleagues performed the analysis. For cases P2, P3 and P5, the lab of L. Sturiale and colleagues performed the analysis.
Antibodies
Anti-MAN1B1 monoclonal antibodies used for indirect immunofluorescence were kindly provided by the lab of Richard N. Sifers and colleagues (Baylor College of Medicine, Houston, USA). Monoclonal anti-MAN1B1 antibodies used for western blotting were from Novus Biologicals. Polyclonal antibodies anti-GPP130 and anti-Giantin were purchased from Covance, polyclonal antibodies anti-PDI from Cell Signalling, polyclonal antibodies anti-ERGIC53 from Sigma-Aldrich and polyclonal anti-GRP78 BiP antibodies from Abcam.
Immunoblotting
30 µg of proteins were analysed by SDS-PAGE and immunoblotted onto Hybond-ECL nitrocellulose membrane (Amersham Biosciences UK limited), with the indicated antibodies. Cells were rinsed twice with ice-cold phosphate buffer saline (PBS) and then lysed for 5 min on ice in cell lysis buffer (25 mM Tris-HCl, 150 mM NaCl pH 7.6, supplemented with 1% Triton X-100, 1% sodium deoxycholate and 0.1% SDS). Proteins were quantified by using the Micro BCA Protein BSA Assay Kit (Fischer Scientific).
Signals were detected using the Western Lightning-ECL, Enhanced Chemiluminescence Substrate (Perkin Elmer) according to the manufacturer's instructions. Signal detection was performed by autoradiography and quantified with the ImageQuant LAS 4000 (GE Healthcare), using the ImageQuant LAS 4000 Control Software for analysis.
Immunofluorescence staining
Cells were grown on glass coverslips, washed once with PBS and fixed by incubation for 30 min with 4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.2) at room temperature. The coverslips were rinsed three times with PBS for 5 min. The fixed cells were then permeabilized with PBS containing 0.5% Triton X-100 for 5 min and washed three times with PBS. Next, the fixed cells were incubated for at least 60 min with a blocking solution containing 0.1% Triton X-100 (Sigma-Aldrich), 1% BSA (Roche Applied Science), and 5% normal goat serum (Life Technologies) in PBS. After blocking, the cells were incubated overnight at 4°C with primary antibodies diluted in the previously described blocking solution. The cells were then washed again three times, followed by incubation with the Alexa 488 - or Alexa 568-conjugated secondary antibodies (1∶500, Life Technologies) for 1 h at room temperature in the dark.
Immunostaining was detected through an inverted Leica SP5 spectral microscope with a 63× oil immersion lens at room temperature. Data were therefore collected using the LAS 6000 AF software and finally processed in Adobe Photoshop 7.0 (Adobe systems). Colocalization was quantified using the colocalization plugin of ImageJ. The channel ratio was always set at 90%.
Real-time quantitative PCR
PCRs were performed for the MAN1B1 gene (GenBank NM_016219.4) and the house-keeping gene HPRT (Hypoxanthine PhosphoRibosylTransferase, GenBank NM_000194), which was used as an endogenous control for normalization. PCR primers were designed using Primer 3 software. All the primers were synthesized by Integrated DNA Technologies. PCR primers used are the followings: MAN1B1 (5′-CTGTGGATCCCCGCCCGGAA-3′ and 5′-TCAGATGCACTGGTGTGCCCTGT-3′) and HPRT (5′-GCCAGACTTTGTTGGATTTG-3′ and 5′-CTCTCATCTTAGGCTTTGTAT TTTG-3′). PCRs (20 µl) were performed using 2× LightCycler 480 SYBR Green I Master (Roche Applied Science) with 2 µl of a 1∶5 cDNA dilution and 300 nM final concentration of each primer. Data were analysed using the LightCycler 480 Software (Roche Applied Science). The comparative threshold cycle method described by Livak and Schmittgen was used to quantify the results [23]. In addition, serial dilutions were used to create standard curves for relative quantification, and the expression of the MAN1B1 transcript was normalized to HPRT expression.
Metabolic labelling
Fibroblasts (8×106 cells per labelling) were grown overnight in a 175 cm2 tissue culture flask. After 24 h, cells were preincubated for 45 min in 0.5 mM glucose and then pulse-radiolabelled for 2 h with 150 µCi of 2-[H3]-mannose (16 Ci.mmol−1, Amersham Biosciences) at the same glucose concentration. For chase experiments, the radioactive culture medium was replaced by a medium containing 5 mM glucose and 5 mM mannose. After metabolic labelling or chase period, cells were scraped with 1.1 ml MeOH/H2O (8∶3) followed by the addition of 1.2 ml CHCl3. Sequential extraction of oligosaccharide material was performed as previously described [24].
Analysis of oligosaccharide material by HPLC
Glycoproteins fraction obtained at the end of the sequential extraction were digested overnight at room temperature with trypsin (1 mg.ml−1; Sigma-Aldrich) in 0.1 M ammonium bicarbonate buffer, pH 7.9. The resulting glycopeptides were treated with 0.5 U PNGase F (Roche Applied Science) in 50 mM phosphate buffer, pH 7.2 for 4 h to release the oligosaccharides from the peptides. They were subsequently desalted on Bio-Gel P2 columns and eluted with 5% acetic acid.
The oligosaccharides were afterwards separated by HPLC on an amino derivated Asahipak NH2P-50 column (250×4.6 mm; Asahi) applying a gradient of acetonitrile/H2O ranging from 70∶30 over 50∶50 over 90 min at a flow rate of 1 ml per min. Oligosaccharides were identified on the basis of their retention time, compared to standard glycans, as previously described [24]. Elution of the labeled oligosaccharides was monitored by continuous β-counting with a flo-one β detector (Packard).
Calculation peak counts
HPLC chromatograms were analysed using the ProFSA software (Perkin Elmer). To compare the percentage of specific oligomannoside structures, a fixed amount (50,000 dpm) of radioactivity was injected into HPLC. The counts in each peak were calculated on the basis of the peak area and normalized against the total number of counts in the injected samples. Moreover, the differences in the number of mannose residues between the different oligomannoside structures were taken into account. Hence, the radioactivity associated to the Man5 species was multiplied by 9/5 to be comparable to the radioactivity associated to the Man9 species.
Supporting Information
Zdroje
1. JaekenJ, MatthijsG (2007) Congenital disorders of glycosylation: a rapidly expanding disease family. Annual review of genomics and human genetics 8 : 261–278 Available: http://www.ncbi.nlm.nih.gov/pubmed/17506657. Accessed 2 February 2013.
2. TheodoreM, MoravaE (2011) Congenital disorders of glycosylation: sweet news. Current opinion in pediatrics 23 : 581–587 Available: http://www.ncbi.nlm.nih.gov/pubmed/21970833. Accessed 7 February 2013.
3. MatthijsG, RymenD, MillónMBB, SoucheE, RaceV (2013) Approaches to homozygosity mapping and exome sequencing for the identification of novel types of CDG. Glycoconjugate journal 30 : 67–76 Available: http://www.ncbi.nlm.nih.gov/pubmed/22983704. Accessed 7 February 2013.
4. StevensFJ, ArgonY (1999) Protein folding in the ER. Seminars in cell & developmental biology 10 : 443–454 Available: http://www.ncbi.nlm.nih.gov/pubmed/10597627. Accessed 7 February 2013.
5. JakobCA, BurdaP, RothJ, AebiM (1998) Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. The Journal of cell biology 142 : 1223–1233 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2149342&tool=pmcentrez&rendertype=abstract. Accessed 7 February 2013.
6. BurkeJ, LipariF, IgdouraS, HerscovicsA (1996) The Saccharomyces cerevisiae processing alpha 1,2-mannosidase is localized in the endoplasmic reticulum, independently of known retrieval motifs. European journal of cell biology 70 : 298–305 Available: http://www.ncbi.nlm.nih.gov/pubmed/8864657. Accessed 7 February 2013.
7. GonzalezDS, KaravegK, Vandersall-NairnAS, LalA, MoremenKW (1999) Identification, expression, and characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. The Journal of biological chemistry 274 : 21375–21386 Available: http://www.ncbi.nlm.nih.gov/pubmed/10409699. Accessed 7 February 2013.
8. PanS, WangS, UtamaB, HuangL, BlokN, et al. (2011) Golgi localization of ERManI defines spatial separation of the mammalian glycoprotein quality control system. Molecular biology of the cell 22 : 2810–2822 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3154878&tool=pmcentrez&rendertype=abstract. Accessed 7 February 2013.
9. RafiqMA, KussAW, PuettmannL, NoorA, RamiahA, et al. (2011) Mutations in the alpha 1,2-mannosidase gene, MAN1B1, cause autosomal-recessive intellectual disability. American journal of human genetics 89 : 176–182 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3135808&tool=pmcentrez&rendertype=abstract. Accessed 7 February 2013.
10. NajmabadiH, HuH, GarshasbiM, ZemojtelT, AbediniSS, et al. (2011) Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478 : 57–63 Available: http://www.ncbi.nlm.nih.gov/pubmed/21937992. Accessed 5 February 2013.
11. LederkremerGZ (2009) Glycoprotein folding, quality control and ER-associated degradation. Current opinion in structural biology 19 : 515–523 Available: http://www.ncbi.nlm.nih.gov/pubmed/19616933. Accessed 7 February 2013.
12. CamirandA, HeysenA, GrondinB, HerscovicsA (1991) Glycoprotein biosynthesis in Saccharomyces cerevisiae. Isolation and characterization of the gene encoding a specific processing alpha-mannosidase. The Journal of biological chemistry 266 : 15120–15127 Available: http://www.ncbi.nlm.nih.gov/pubmed/1714453. Accessed 7 February 2013.
13. De PraeterCM, GerwigGJ, BauseE, NuytinckLK, VliegenthartJF, et al. (2000) A novel disorder caused by defective biosynthesis of N-linked oligosaccharides due to glucosidase I deficiency. American journal of human genetics 66 : 1744–1756 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1378052&tool=pmcentrez&rendertype=abstract. Accessed 7 February 2013.
14. EsmonB, EsmonPC, SchekmanR (1984) Early steps in processing of yeast glycoproteins. The Journal of biological chemistry 259 : 10322–10327 Available: http://www.ncbi.nlm.nih.gov/pubmed/6381483. Accessed 8 February 2013.
15. TremblayLO, HerscovicsA (1999) Cloning and expression of a specific human alpha 1,2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2 isomer B during N-glycan biosynthesis. Glycobiology 9 : 1073–1078 Available: http://www.ncbi.nlm.nih.gov/pubmed/10521544. Accessed 7 February 2013.
16. CabralCM, LiuY, SifersRN (2001) Dissecting glycoprotein quality control in the secretory pathway. Trends in Biochemical Sciences 26 : 619–624 Available: http://dx.doi.org/10.1016/S0968-0004(01)01942-9. Accessed 15 July 2013.
17. WuX, SteetRA, BohorovO, BakkerJ, NewellJ, et al. (2004) Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nature medicine 10 : 518–523 Available: http://www.ncbi.nlm.nih.gov/pubmed/15107842. Accessed 15 July 2013.
18. RymenD, KeldermansL, RaceV, RégalL, DeconinckN, et al. (2012) COG5-CDG: expanding the clinical spectrum. Orphanet journal of rare diseases 7 : 94 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3697985&tool=pmcentrez&rendertype=abstract. Accessed 11 June 2013.
19. KornakU, ReyndersE, DimopoulouA, van ReeuwijkJ, FischerB, et al. (2008) Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nature genetics 40 : 32–34 Available: http://www.ncbi.nlm.nih.gov/pubmed/18157129. Accessed 15 July 2013.
20. CarchonHA, ChevignéR, FalmagneJ-B, JaekenJ (2004) Diagnosis of congenital disorders of glycosylation by capillary zone electrophoresis of serum transferrin. Clinical chemistry 50 : 101–111 Available: http://www.ncbi.nlm.nih.gov/pubmed/14633925. Accessed 29 January 2013.
21. MillsPB, MillsK, MianN, WinchesterBG, ClaytonPT (2003) Mass spectrometric analysis of glycans in elucidating the pathogenesis of CDG type IIx. Journal of inherited metabolic disease 26 : 119–134 Available: http://www.ncbi.nlm.nih.gov/pubmed/12889655. Accessed 8 February 2013.
22. FaidV, ChiratF, SetaN, FoulquierF, MorelleW (2007) A rapid mass spectrometric strategy for the characterization of N - and O-glycan chains in the diagnosis of defects in glycan biosynthesis. Proteomics 7 : 1800–1813 Available: http://www.ncbi.nlm.nih.gov/pubmed/17520685. Accessed 8 February 2013.
23. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 25 : 402–408 Available: http://www.ncbi.nlm.nih.gov/pubmed/11846609. Accessed 28 January 2013.
24. FoulquierF, Harduin-LepersA, DuvetS, MarchalI, MirAM, et al. (2002) The unfolded protein response in a dolichyl phosphate mannose-deficient Chinese hamster ovary cell line points out the key role of a demannosylation step in the quality-control mechanism of N-glycoproteins. The Biochemical journal 362 : 491–498 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1222411&tool=pmcentrez&rendertype=abstract. Accessed 8 February 2013.
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



