-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
Neurons and other cells display a large variation in size in an organism. Thus, a fundamental question is how growth of individual cells and their organelles is regulated. Is size scaling of individual neurons regulated post-mitotically, independent of growth of the entire CNS? Although the role of insulin/IGF-signaling (IIS) in growth of tissues and whole organisms is well established, it is not known whether it regulates the size of individual neurons. We therefore studied the role of IIS in the size scaling of neurons in the Drosophila CNS. By targeted genetic manipulations of insulin receptor (dInR) expression in a variety of neuron types we demonstrate that the cell size is affected only in neuroendocrine cells specified by the bHLH transcription factor DIMMED (DIMM). Several populations of DIMM-positive neurons tested displayed enlarged cell bodies after overexpression of the dInR, as well as PI3 kinase and Akt1 (protein kinase B), whereas DIMM-negative neurons did not respond to dInR manipulations. Knockdown of these components produce the opposite phenotype. Increased growth can also be induced by targeted overexpression of nutrient-dependent TOR (target of rapamycin) signaling components, such as Rheb (small GTPase), TOR and S6K (S6 kinase). After Dimm-knockdown in neuroendocrine cells manipulations of dInR expression have significantly less effects on cell size. We also show that dInR expression in neuroendocrine cells can be altered by up or down-regulation of Dimm. This novel dInR-regulated size scaling is seen during postembryonic development, continues in the aging adult and is diet dependent. The increase in cell size includes cell body, axon terminations, nucleus and Golgi apparatus. We suggest that the dInR-mediated scaling of neuroendocrine cells is part of a plasticity that adapts the secretory capacity to changing physiological conditions and nutrient-dependent organismal growth.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1004052
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004052Summary
Neurons and other cells display a large variation in size in an organism. Thus, a fundamental question is how growth of individual cells and their organelles is regulated. Is size scaling of individual neurons regulated post-mitotically, independent of growth of the entire CNS? Although the role of insulin/IGF-signaling (IIS) in growth of tissues and whole organisms is well established, it is not known whether it regulates the size of individual neurons. We therefore studied the role of IIS in the size scaling of neurons in the Drosophila CNS. By targeted genetic manipulations of insulin receptor (dInR) expression in a variety of neuron types we demonstrate that the cell size is affected only in neuroendocrine cells specified by the bHLH transcription factor DIMMED (DIMM). Several populations of DIMM-positive neurons tested displayed enlarged cell bodies after overexpression of the dInR, as well as PI3 kinase and Akt1 (protein kinase B), whereas DIMM-negative neurons did not respond to dInR manipulations. Knockdown of these components produce the opposite phenotype. Increased growth can also be induced by targeted overexpression of nutrient-dependent TOR (target of rapamycin) signaling components, such as Rheb (small GTPase), TOR and S6K (S6 kinase). After Dimm-knockdown in neuroendocrine cells manipulations of dInR expression have significantly less effects on cell size. We also show that dInR expression in neuroendocrine cells can be altered by up or down-regulation of Dimm. This novel dInR-regulated size scaling is seen during postembryonic development, continues in the aging adult and is diet dependent. The increase in cell size includes cell body, axon terminations, nucleus and Golgi apparatus. We suggest that the dInR-mediated scaling of neuroendocrine cells is part of a plasticity that adapts the secretory capacity to changing physiological conditions and nutrient-dependent organismal growth.
Introduction
Neurons and other cells display substantial variation in size in an organism. Thus, some neuron types in the brain have large cell bodies and extensive axonal processes, whereas others are minute with restricted branches. An important question is to what extent neuron size is genetically programmed and how much growth regulation is dependent on extrinsic factors, including systemic signaling. Although there appears to be an optimal size for most cell types there is room for plasticity, for instance to accommodate for availability of nutrients during development [1]–[4]. We ask here whether there is a dynamic growth-regulation of individual neurons by systemic or paracrine factors and how allometric size scaling of cells is controlled in the CNS. These fundamental questions have not been addressed in detail previously, and available data primarily deal with growth of entire tissues or whole organisms [2]–[6].
In general growth of cells, tissues and whole organisms depends on a combination of cell autonomous nutrient sensing via the TOR (target of rapamycin) pathway and insulin/IGF signaling (IIS), both of which are evolutionarily conserved pathways [3], [7]–[15]. During organ growth both cell size and cell numbers can be regulated by components of the IIS and TOR pathways (see [3], [16], [17]). Growth of ensembles of neuroblasts and post-mitotic neurons is regulated by similar mechanisms to ensure appropriate cell proliferation and subsequent differentiation of mature properties, including size of the cell body. Neuroblasts and other stem cells remain part of their time in a quiescent state, a reversible arrest of growth and cell division [18]–[20]. Exit from this arrested state involves induction of cell growth and is nutrition dependent [21]–[23]. In feeding larvae of Drosophila the postembryonic neuroblasts (progenitors of imaginal neurons) are protected against malnutrition. Thus, whereas many tissues display reduced growth after restricted nutrition, neuroblast growth is maintained by anaplastic lymphoma kinase (ALK) signaling, and therefore the CNS as a whole grows almost normally [16], [17], [24].
What mechanisms regulate size scaling of neurons and during what part of organismal development does it occur? Neuron growth is a likely part of the post-mitotic cell differentiation process where for instance neurons destined to become peptidergic neuroendocrine cells develop specific properties, including relatively large cell bodies [25]–[27]. Thus, in snails it was shown that cell bodies of large efferent peptidergic neurons and neuroendocrine cells continue to grow in a nutrient dependent manner as the organism grows [27]. In Drosophila a class of about 300 dedicated neuroendocrine cells have been identified that are specified by the bHLH transcription factor Dimmed (DIMM) and develop a capacity for production, packaging and releasing large amounts of neuropeptide or peptide hormone [28]–[30]. Part of this specification seems to involve an enlargement of cell body size relative to many surrounding interneurons. An important question is to what extent individual neuron size is regulated cell autonomously, or by factors in its immediate niche, and how much depends on additional systemic signals. Allometric size regulation by means of systemic factors would be a suitable mechanism to regulate growth of post-mitotic neurons that need to adapt their secretory capacity to changes in body volume or physiological requirements to function optimally.
We ask here whether the size of individual post-mitotic neurons can be regulated by systemic IIS, and whether growth can be induced in specific neuron types without their neighbors growing. To address these questions, we analyzed neuron size scaling by exploring the effect of manipulating expression of the insulin receptor (dInR) and signaling components of the IIS and TOR pathways in specific sets of neurons in the CNS of Drosophila during development and in the mature organism. Initial experiments revealed a marked difference in growth of DIMM-positive and DIMM-negative neurons, with targeted IIS manipulations affecting growth in the former only. Select populations of DIMM-expressing neurons were therefore chosen for in-depth analysis. Overexpression of the dInR and some of the downstream signaling components (PI3K and Akt), as well as TOR components such as TOR, the small GTPase Rheb (Ras homolog enriched in brain) and S6 kinase (S6K) all caused increased cell body size in these neurons. Conversely, diminishing activity of these components leads to decreased neuron size. We detected no size effects of IIS manipulations in motor neurons, various interneurons or DIMM-negative neuroendocrine cells tested. Selective dInR-mediated growth control could, thus, provide plastic scaling and protection of secretory activity in neuroendocrine cells, during development or in the adult life, as a means to ensure hormone production appropriate for body volume.
Results
Manipulation of dInR expression in specific neurons affects their size
Since IIS plays an important role in growth regulation in the CNS as a whole, we decided to investigate the effect of targeted interference with the insulin receptor, dInR, in growth of individual neurons. It is likely that most, if not all, neurons and neuroblasts express the dInR, at least during developmental stages up to adult eclosion [21], [31]–[34]. To test this, we monitored receptor expression in the larval and adult CNS using three different insulin receptor antisera (described in material and methods). Two of these have been used previously in Drosophila to determine dInR localization in sensory cells of the antennae and germ line stem cells [35], [36]. All three antisera produced strong immunolabeling in the same set of neurons (Fig. 1A–C; Fig. S1, 2). In the third instar larva these were identical to a set of 20 neurons in the brain, subesophageal and abdominal ganglia expressing the neuropeptide leucokinin (LK) as seen by superposition with Lk-Gal4-driven GFP (Fig. 1A–C, F). In addition there is a weak receptor immunolabeling generally in the CNS that is insufficient to identify specific neurons, but suggests a more pan-neuronal expression. A similar set of insulin-receptor immunoreactive neurons was detected in the adult CNS and we confirmed expression of the receptor in the larval fat body (Fig. S1, 2). We could determine the effectiveness of our reagents by driving dInR-RNAi or UAS-dInR with the Lk-Gal4: this leads to reduction or increase of receptor immunolabeling in LK neurons, respectively (Fig. S1A–D). It was also clear that the size of the cell bodies of the 14 abdominal LK neurons (ABLK neurons) was altered by these dInR manipulations (Fig. S1C). Thus, we selected the ABLK neurons for a first analysis of the role of dInR in size regulation of neurons. Note that in all experiments cell body “size” was determined by measuring the largest surface area of each neuron in total z-projections of confocal images (given in µm2).
Fig. 1. Distribution of insulin receptor protein in the larval CNS of Drosophila and growth of neurosecretory cells. 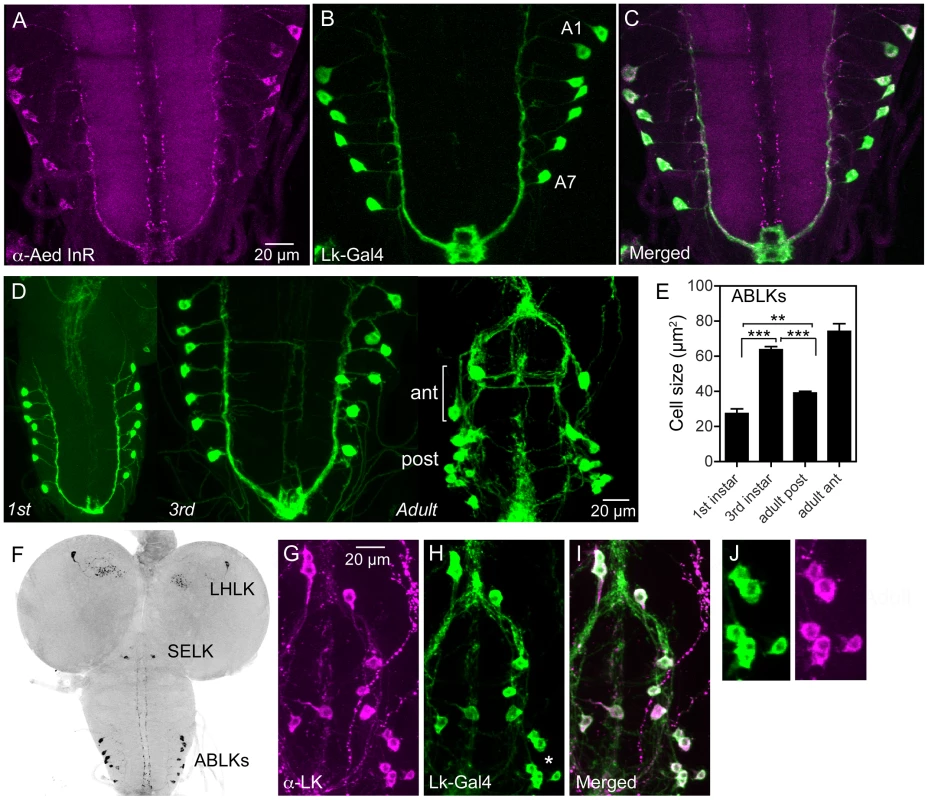
A–C An antiserum to part of the insulin receptor (InR) of the mosquito Aedes aegypti labels neurosecretory cells (designated ABLKs) in the abdominal ganglia of the third instar larva that also produce the neuropeptide leucokinin (LK), visualized here with Lk-Gal4 driven GFP. Seven pairs of LK neurons coexpress the markers in abdominal segments A1–A7. D The cell bodies of LK neurons (ABLKs) in A1–7 grow substantially from first to third instar larvae, but not during pupal development. Thus, the posterior neurons (post) in the 3 d old adult fly (corresponding to ABLKs) are even smaller than in the late larva. However a set of 8 anterior adult-specific LK neurons (ant) have larger cell bodies than the ABLKs. LK neurons are visualized by Lk-Gal4-driven GFP. E Quantification of cell body sizes in larvae and 3 d old adult flies. The ABLKs (14 posterior neurons) are significantly larger in 3rd instar larvae than in both 1st instar and 3 d old adults (unpaired Student's T-test; **p<0.01, ***p<0.001; n = 5–6 animals for each developmental stage). F Overview of the larval CNS (3rd instar) with LK-immunolabeled neurons, LHLK, SELK and ABLKs. G–J The immunolabeling of LK neurons with anti-LK (F) is a good marker for cell body size since it produces the same outline as membrane-targeted GFP (mcd8GFP) driven by the Lk-Gal4 (G). The posterior cell bodies marked by an asterisk are shown in higher magnification in J. Fig. 2. Manipulations of insulin receptor (dInR) levels alter size of the larval LK neurons in abdominal ganglia only. 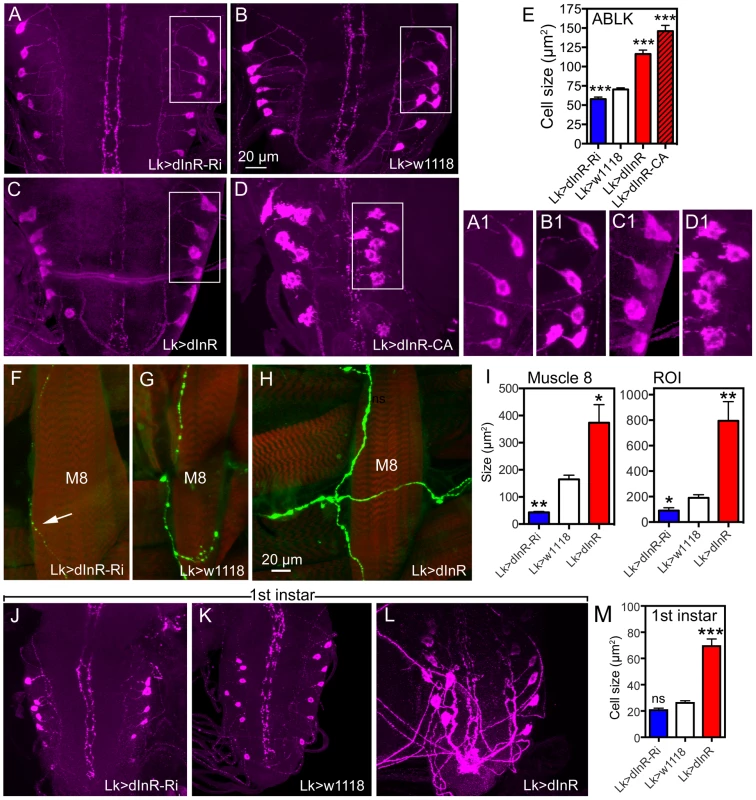
Neurons are visualized with anti-LK. In this and other figures cell body sizes are given as means (±SEM) of sizes of all specified neurons in a number (n) of specimens from at least 3 independent crosses for each genotype. A–E Knockdown or over expression of dInR using Lk-Gal4 alters the size of cell bodies (boxed areas are enlarged for details in A1–D1) monitored in late third instar larvae. The dInR-RNAi (Ri) significantly reduces size of cell bodies and, especially the constitutively active dInR (dInR-CA) drastically enlarges them. Note that the dInR-CA also induces a dislocation and irregular outlines of the cell bodies (D, D1). A quantification of the ABLK cell body size changes is shown in E (***p<0.001, n = 67–159 cell bodies from 6–13 animals of each genotype from 3 crosses; unpaired Student's T-test). F–H The peripheral axon processes of the ABLKs terminate primarily on body wall muscle 8 (M8). Muscles were stained with Rhodamine-phalloidin (red). Knockdown of dInR (in F) reduces axon diameter and bouton size of M8 branches compared to control (G), whereas overexpression of dInR leads to increased axon diameter, bouton size as well as increased branching of the axon (H). Axons were labeled with anti-LK and muscles with rhodamine-phalloidin. I Quantification of fluorescent area of axons on M8 in the three genotypes (*p<0.05, **p<0.01, n = 6–8 body walls for each genotype, 3 crosses; unpaired Student's T-test) and also in the total LK-immunolabeled axon termination in a region of interest (ROI) corresponding to the whole area shown in panel H (same for each genotype). This measurement provides combined information about branching area, as well as axon and bouton diameters (*p<0.05, **p<0.01, n = 6–8 body walls for each genotype from 3 crosses; unpaired Student's T-test). J–M The size of the ABLKs is altered already at the end of the first instar larval stage. Only the drastic increase caused by dInR overexpression is significant (***p<0.001, n = 5–10 animals for each genotype from 3 crosses; unpaired Student's T-test). The seven pairs of segmentally arranged ABLKs are neurosecretory cells with cell bodies in the abdominal ganglia and axon terminations on body wall muscle [37], [38]. It has been established that all the neurons that label with the antiserum to LK are included in the Lk-Gal4 pattern [38] (see also Fig. 1G–J). Under normal feeding conditions the cell bodies of the 14 ABLKs grow substantially (113%) from first to late third instar larvae but do not grow further during pupal stages (Fig. 1D, E). During pupal development a set of 8 additional LK neurons appear anteriorly in the abdominal ganglia [37], [39]; these have far larger cell bodies than the 14 posterior ABLKs (Fig. 1D, E). In this study we monitored size changes in the ABLKs since their large cell bodies are segmentally arranged and readily visible in every specimen.
We used the Lk-Gal4 driver to alter dInR expression in LK-producing neurons by UAS-dInR-RNAi, UAS-dInR or UAS-dInRCA, a constitutively active receptor form. To monitor the cell body size we used antiserum to LK [40]. We could show that the LK-immunolabeling provides a good measure of cell body size since it matches the size seen with a membrane targeted GFP (mcd8-GFP) driven by the Lk-Gal4 (Fig. 1G–K). The effectiveness of the different UAS-dInR-constructs used has been described in several accounts [41]–[43]. The dInR manipulations led to significant cell body phenotypes in ABLK neurons. There was about 20% decrease in cell body size after diminishing dInR, and an increase of 65% for dInR and 107% for dInRCA overexpression (Fig. 2A–E, Table 1). In addition to the prominent increase in size, the dInRCA overexpression gave rise to irregular shapes of the cell bodies and a dislocation of the cell bodies resulting in a less distinct segmental distribution (Fig. 2D). In no experimental animal we found any indication that the number of LK neurons had changed, suggesting that size, but not cell proliferation, was affected by targeted dInR activity.
Tab. 1. Manipulations of genes in Dimm positive and Dimm negative peptidergic neuroendocrine cells. 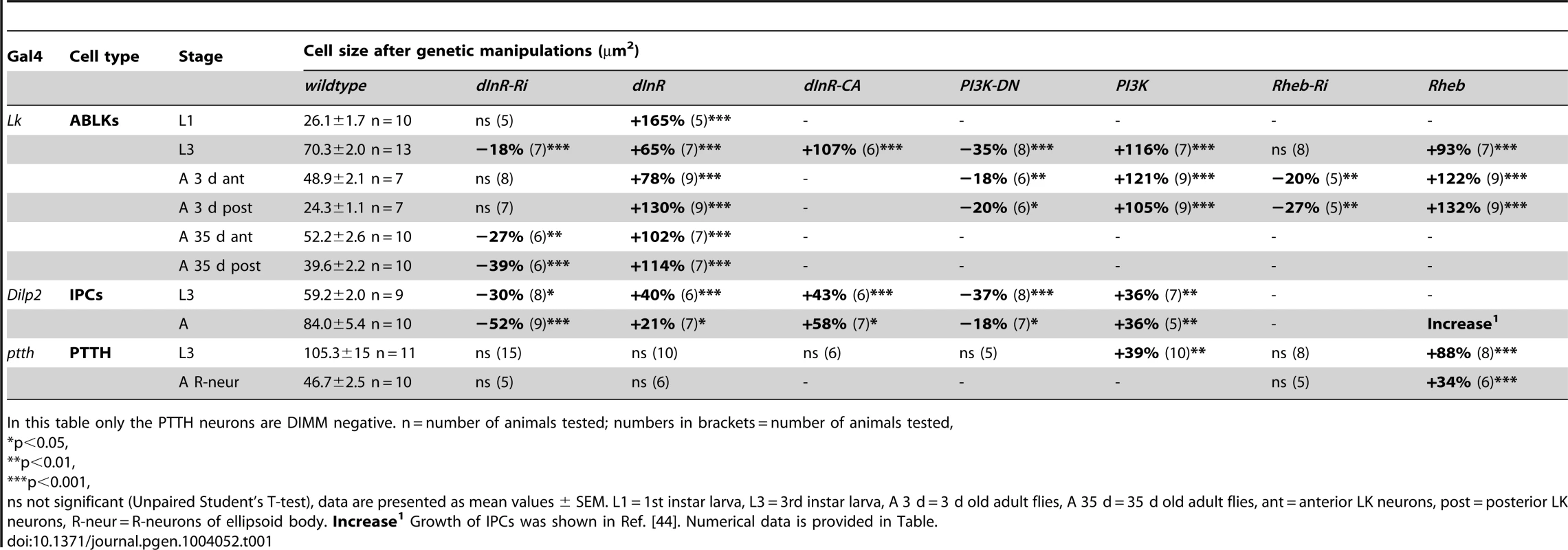
In this table only the PTTH neurons are DIMM negative. n = number of animals tested; numbers in brackets = number of animals tested, Each of the 14 larval ABLK neurons innervates muscle number 8 of the corresponding segment in the abdominal body wall [37]. To test whether the dInR-induced growth of the ABLK cell bodies is correlated with a growth of the peripheral axons we examined these axon terminations on muscle 8. Overexpression of the dInR in ABLK neurons increased the size of the axon terminations, and extra branches were seen (Fig. 2F–I). Conversely, dInR-RNAi in ABLKs led to thinner axons, smaller boutons and more restricted branching (Fig. 2F, G, I). Thus, dInR manipulations affected both the cell body size and the morphology of the peripheral axons. Importantly, we could show that the size of the ABLKs can be increased by dInR manipulations already when monitored in the first instar larva (Fig. 2J–M, Table 1). A very dramatic size increase (165%) compared to controls was noted.
Next we used a pan-neuronal driver, elav-Gal4, to manipulate the dInR widely in neurons, including the ABLK neurons. It was confirmed by anti-LK labeling that the elav-Gal4 pattern includes ABLK neurons, although the GFP expression is rather weak compared to most other neurons (not shown). Elav-driven dInR-RNAi led to smaller ABLK neurons (Fig. S3A,E), while expression of dInR or dInRCA did not result in larger ABLK neurons (Fig. S3C–E). This could be due to the Elav driver being weaker in the ABLK neurons or because the more global dInR activation induces a compensatory regulation of cell size (cell interactions preventing growth of the entire CNS). We also employed a driver that is intermediate between the Lk - and Elav-Gal4 in terms of neuron number, the c929-Gal4 [28], known to include the ABLKs [29]. Here, we found that the c929-driven dInR manipulations of neurons affected the size of the ABLK neurons at the same magnitude as the Lk-Gal4 driver (Fig. S3F–I).
The size of ABLK neurons is affected also in adult flies
We also monitored the cell body size of abdominal LK neurons after dInR manipulations in adult male flies of two different ages (3 d and 35 d old flies). Monitoring 3 d old flies we found that overexpression of the receptor by the Lk-Gal4 driver leads to significantly larger cell bodies of both the 14 posterior ABLK neurons and the anterior adult-specific ones as compared to controls (Fig. 3A–C, G, Table 1). The dInR-RNAi was, however, inefficient in producing smaller cell body size in both types of abdominal LK neurons in the younger flies (Fig. 3A–C, G). Interestingly, we found that in 35 d old adult flies both the ABLKs and the anterior adult LK cells were significantly affected by both dInR knockdown and overexpression compared to 35 d controls and 3 d old flies (Fig. 3D–G, Table 1). After 35 d the dInR over expression resulted in very large cell bodies (more than 100% growth compared to 35 d controls) with highly irregular outlines (Fig. 3F). Especially the posterior ABLKs grow substantially compared to 3 d controls. This finding suggests that the abdominal LK neurons continuously respond to insulin signaling by altered cell size also in adult flies. Similar continuous growth of adult neurons was seen for mushroom body neurons and insulin producing cells (IPCs) in Drosophila after over expression of a component of the TOR pathway, the small GTPase Rheb [44]. For these neuron types 1 d and 21 d old flies were compared. The growth of larval and adult neurons over time is summarized in Fig. 3L.
Fig. 3. Manipulations of dInR in LK neurons also affect adult cell body size. 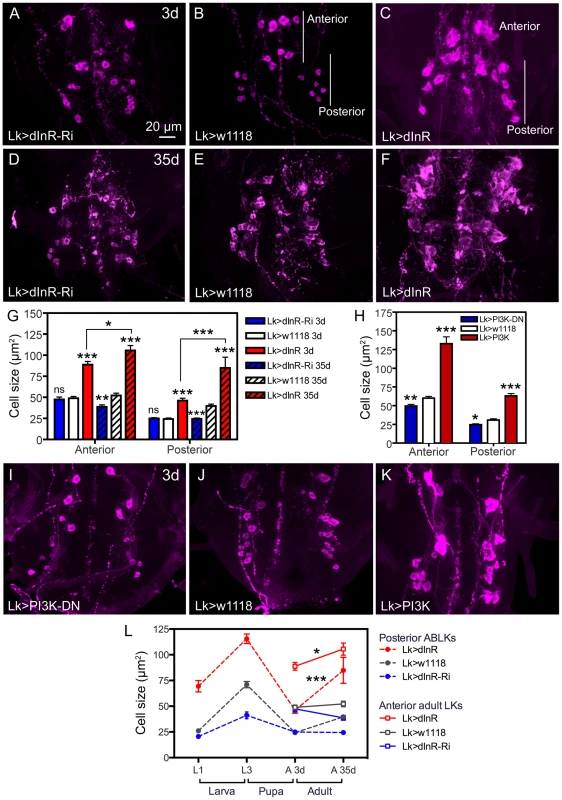
Neurons are visualized with anti-LK. A–C Monitoring adult ABLKs (Posterior) and adult-specific anterior (Anterior) LK neurons in 3 d old adult flies reveal a size increase only (no effect of dInR-RNAi) for both the anterior adult-specific neurons (that differentiate during pupal development) and the posterior ABLKs. D–F In 35 d old flies the effect of the dInR manipulations are more drastic and also the dInR-RNAi has a significant effect on cell body size. G Quantification of cell body sizes in different genotypes and developmental time (3 d and 35 d old flies) in adults (*p<0.05, **p<0.01, ***p<0.001, n = 6–10 animals from 3 crosses for each genotype, unpaired Student's T-test; comparisons were made between the same neuron type of the same age). Note that both anterior and posterior ABLKs are significantly larger in 35 d old flies than in 3 d old ones (unpaired Student's T-test). H–K manipulations of PI3K also affect cell body size of anterior and posterior LK-neurons in 3 d old adult flies. A dominant negative PI3K (PI3K-DN) expressed in Lk-Gal4 neurons diminishes cell size in both anterior neurons and posterior ones, whereas overexpression of wild type PI3K drastically increases size of both neuron types. The cell sizes are quantified in H (*p<0.05, **p<0.01, ***p<0.001, n = 6–9 animals for each genotype from 3 crosses; unpaired Student's T-test). L Summary of sizes of abdominal LK neuron cell bodies in larvae and adults over time. We plotted the sizes of 14 ABLKs and their adult counterparts the posterior ABLKs (dashed lines), as well as the 8 adult-specific anterior LK neurons solid lines). In the wild type (or control) the size of the ABLKs increase from first to third instar larva, decreases during pupal development and then increases again from 3 d adult to 35 d old adults. The adult-specific anterior LK neurons do not increase significantly in size from 3–35 d in control flies. Over expression of dInR in LK neurons increases the ABLK cell body size (compared to controls), except during pupal development. The dInR-RNAi induces a significant difference only in the third instar and the 35 d old adults. Finally, after over expression of the dInR both the anterior adult-specific (red dashed line) and the posterior LK neurons (red solid line) display a significant increase in size after 35 d compared to 3 d (unpaired Student's T-test). Manipulations of components downstream of the dInR and in the TOR pathway also produce size phenotypes in ABLKs
To further support that the cell size phenotypes in ABLKs depend on dInR-mediated signaling we manipulated the levels or activity of components that can act downstream of this receptor, such as PI3 kinase (PI3K) and the protein kinase B, Akt1 [see [7], [14], [15], [45]]. We expressed either a dominant-negative form of PI3K (PI3KDN) or a wild type PI3K in the ABLK neurons with the Lk-Gal4. LK immunolabeling in the larval CNS revealed that PI3KDN expression significantly decreased the size of ABLK cell bodies, while over expression resulted in enlarged ones with irregular outlines and displaced locations (Fig. 4A–D, Table 1). In the adult CNS overexpression of PI3K in LK neurons induces a substantial increase in both ABLK neurons and the adult-specific larger anterior LK neurons, whereas PI3KDN expression significantly reduces cell body size (Fig. 3H–L, Table 1).
Fig. 4. Manipulations of signaling components possibly downstream the dInR affect cell body size of larval ABLKs. 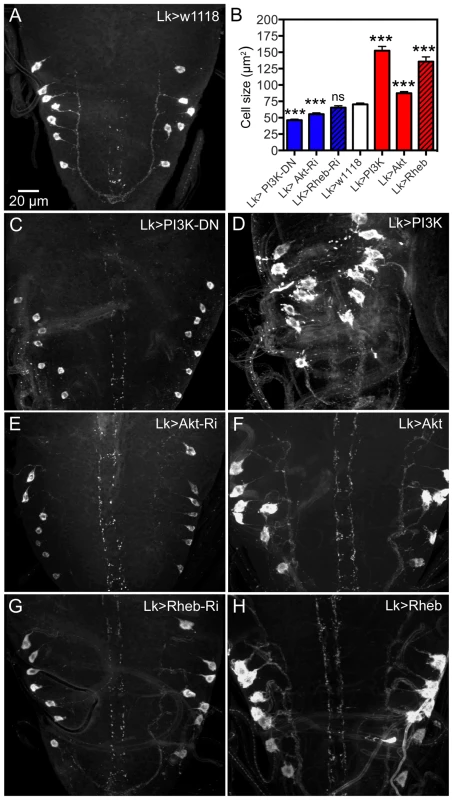
Neurons are visualized with anti-LK. A ABLKs in control flies. B Quantification of cell body sizes of LK neurons in the different experiments. C and D expression of a dominant negative form of PI3K (PI3K-DN) diminishes cell body size and overexpression of wild type PI3K increases it (and leads to dislocation and irregular shapes of cell bodies). E and F Expressing Akt-RNAi and wild type Akt gives phenotypes similar to PI3K manipulations. G and H Overexpression of Rheb drastically increases cell body size, whereas Rheb-RNAi has no significant effect (***p<0.001, n = 8–14 animals for each genotype from 3 crosses; unpaired Student's T-test). Manipulations of Akt1 expression in ABLK neurons by Akt1-RNAi and UAS-Akt1 produced larval cell body phenotypes similar to PI3K and dInR, but without the irregular outlines (Fig. 4B,E, F, Table 1). We employed an antiserum to phosphorylated Akt (pAkt) to determine whether dInR manipulations in LK neurons affected Akt phosphorylation. In control flies the antiserum to pAkt labels a set of neurons in the CNS that includes the ABLKs and LHLKs, and mushroom body Kenyon cells, as well as several other sets of unidentified neurons (Fig. S4A,B, E–G). Using the c929-Gal4 driver we manipulated dInR levels and monitored pAkt immunolabeling levels and found that dInR over expression increased pAkt immunolabeling in what appeared to be ABLKs, but dInR-RNAi had no effect (Fig. S4C,D). There seems to be a basal Akt phosphorylation in control flies (Fig. S4A–D) and possibly this is independent of dInR signaling, explaining the lack of effect of dInR-RNAi (although we cannot exclude that the RNAi knockdown is too weak to affect the immunolabeling).
Another component that can be downstream of the dInR, but also be part of the TOR signaling pathway, is the small GTPase Rheb (Ras homolog enriched in brain) [46], [47] (see also Fig. S16). Thus, Rheb can either be controlled by dInR, PI3K, Akt1 and TSC1/2 (tuberous sclerosis tumor suppressor) and activate TOR to promote cell growth via S6 kinase and 4E-BP, or activate TOR in a nutrient-dependent pathway cell autonomously in growth control [46]–[48] (see also Fig. S16). It was recently shown that overexpression of Rheb in Drosophila IPCs and mushroom body Kenyon cells resulted in enlarged cell bodies of these neurons [44]. We thus tested whether manipulations of Rheb in LK neurons affected cell body size. Overexpression of Rheb in LK neurons led to strongly enlarged and disfigured cell bodies of ABLK neurons (Fig. 4B,H, Table 1). However, there was no significant effect of Rheb-RNAi on cell body size of ABLK neurons (Fig. 4B, G). We also analyzed 3 d old adult flies after Rheb manipulations in LK-neurons. Both the anterior and the posterior abdominal LK neurons displayed enlarged cell bodies after Rheb overexpression, and smaller ones after Rheb-RNAi (S Fig. 5, Table1).
Fig. 5. Functional effects of dInR manipulations of LK neurons in adult male flies. 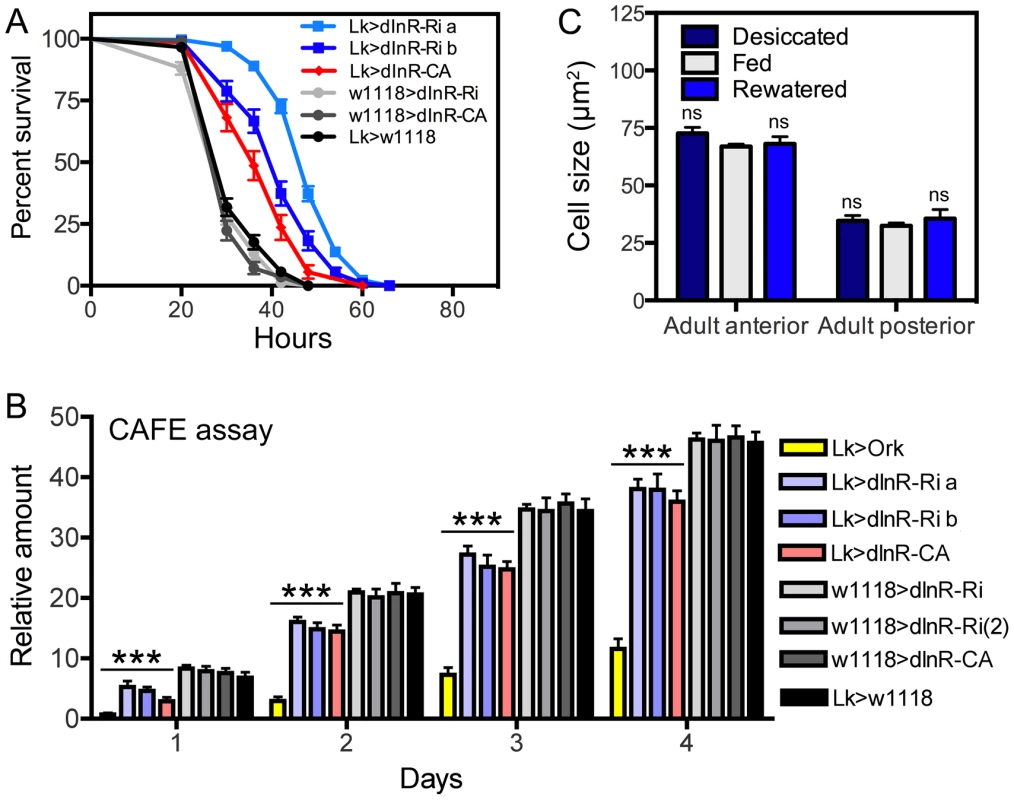
A Knockdown and overexpression of dInR in LK neurons produce the same phenotypes when flies are exposed to desiccation (dry starvation). Here the constitutively active dInR (dInR-CA) was used. Both manipulations lead to significantly increased resistance to desiccation, suggesting a malfunction in LK release that decreases diuresis in flies (***p<0.001, n = 120 flies for each genotype from 3 crosses; Log-rank (Mantel-Cox) Test). Note that two different dInR-RNAi lines were tested (a and b). B Feeding, as measured in CAFE assay, is decreased both in flies with dInR knockdown and over expression. Cumulative amount of food eaten over four days is displayed. As a control we used flies with LK neurons hyperpolarized with a constitutively active K-channel (Lk>Ork). These flies display strongly reduced feeding over four days, again suggesting that both manipulations of dInR levels produce malfunction of LK signaling; all manipulations (colored bars) produced significant decreases compared to the four controls shown in gray scale (***p<0.001, n = 10 flies for each genotype in 3 replicates, Two-way ANOVA). C To test whether cell body size of ABLK neurons is affected by activity (level of LK signaling) we exposed wild-type flies (w1118) to desiccation (dry starvation) for 18 h or 18 h desiccation followed by access to water (rewatered flies) for 2 h. As a control normally fed flies were used. No difference in cell body size was noted (ns, not significant for any comparison, n = 7–10 flies for each treatment, One-way ANOVA). Next we tried to determine whether dInR manipulations affect Rheb expression in ABLK neurons. When employing a Rheb antibody to the wild type larval CNS we found weak labeling in ABLK neurons and even less labeling elsewhere. Thus we were not able to detect any effects of dInR expression levels (not shown).
Two other components in the TOR signaling pathway were also investigated for their effect on ABLK cell body size: TOR and S6K. We expressed a dominant negative form of TOR under Lk-Gal4 control and observed decreased size of ABLKs in larvae (Fig. S6A, B, F). A dominant negative S6K also diminished the ABLK cell bodies, whereas a wild type and a constitutively active S6K increased their size (Fig. S6C–F). Thus we find that TOR signaling also affects the size of cell bodies of LK neurons.
When Drosophila larvae are exposed to conditions of restricted nutrients the growth of neuroblasts is ensured by anaplastic lymphoma kinase (Alk) signaling [16], [17], [24]. We therefore tested the effects of expressing wild type or a dominant negative form of Alk in LK neurons. As expected, we detected no effects of these manipulations under normal feeding conditions (Fig. S7A–C). Thus, we expressed a constitutively active form of Alk under Lk-Gal4 control to test its activity independent of the ligand Jelly belly. This had no effect on the size of ABLKs (Fig. S7D, E), possibly because the Lk-driver is active only post-mitotically in LK neurons [38], [39], [49].
Manipulation of dInR expression in LK neurons affects their function
To test the effect of dInR manipulations on the function of the ABLK neurons we performed two different assays in adult flies, one for feeding and the other for diuretic action. These assays were selected since it is known that in Drosophila LK is a diuretic hormone [50]–[52] and plays a role in regulation of food intake [53]. We exposed adult male flies of different genotypes to dry starvation (desiccation) and monitored survival. Both knockdown and overexpression of dInR in the LK neurons resulted in flies that were more resistant to desiccation than controls, suggesting a reduced diuretic activity in both transgenes (Fig. 5A). Also in the capillary feeding (CAFE) assay we observed that both dInR-RNAi and overexpression lead to decreased feeding (Fig. 5B). As a control we tested the effect of hyperpolarizing LK neurons by expressing the open rectifier K+-channel Ork1 [54]. The expression of this channel did not affect the size of the ABLK cell bodies, but in both assays this inactivation of LK neurons resulted in phenotypes that suggest diminished LK signaling (Fig. 5B). Thus, the phenotypes seen after dInR-RNAi in LK neurons may also be caused by decreased LK signaling; perhaps as a result of smaller neurons and a diminished pool of LK peptide. However, the finding that overexpression of the dInR also may reduce LK signaling is harder to reconcile. We speculate that constitutive overexpression of the dInR induces overproduction of LK that offsets circulating hormone levels and thus physiology, or trigger growth effects that affect neuron function adversely.
Next we asked whether the physiological state of the flies could affect the size of the ABLK cell bodies, as a consequence of altered LK production and/or release. Since the ABLKs release LK as a diuretic hormone [51] we exposed flies (4–6 d old) to 18 h desiccation or 18 h desiccation followed by 2 h of access to water (re-watering). The size of the anterior and posterior LK neurons was monitored, compared to those of normally fed flies. There was no difference in cell body size between the experimental groups (Fig. 5C), although the intensity of LK immunoreactivity differed between them (not shown). Thus, the short-term physiological conditions tested do not affect cell body size of ABLK neurons.
Manipulations of dInR in motor and interneurons do not affect cell size
To test whether the dInR-mediated growth is a feature of larger neurons with peripheral axon projections we manipulated the dInR in motor neurons. In the Drosophila ventral nerve cord of late embryos, around 80 motor neurons are stereotypically distributed in each segment [55]. They have distinct axonal projections into the periphery via the segmental nerve trunks and innervate specific target muscles of the body wall [56], [57]. We used the enhancer trap Gal4 line OK6, whose expression is predominantly in motor neurons and some interneurons in the larval CNS [58], [59], to drive dInR-RNAi, dInR and dInRCA. To monitor size of the cell bodies of motor neurons we employed an antiserum to a vesicular glutamate transporter (vGluT) known to be expressed in motor neurons [60] and to visualize their peripheral axons we used anti-HRP. Cell bodies of three types of neurons visualized by anti-vGluT antibody were measured: interneurons in the brain lobes, lateral motor neurons in segments A3–6 of the abdominal ganglia and median ones in the same segments. None of these neurons displayed any changes in cell body size after dInR manipulations (Fig. 6A–C, Table 2). Also the peripheral axonal projections of motor neurons on the body wall muscles displayed no obvious morphological effects of dInR manipulations (Fig. S8). The size of the axon terminals on muscles 12 and 13 were quantified and we found no effects of changing dInR levels (Fig. 6D, E).
Fig. 6. Manipulations of dInR expression in interneurons and motor neurons do not affect cell body size. 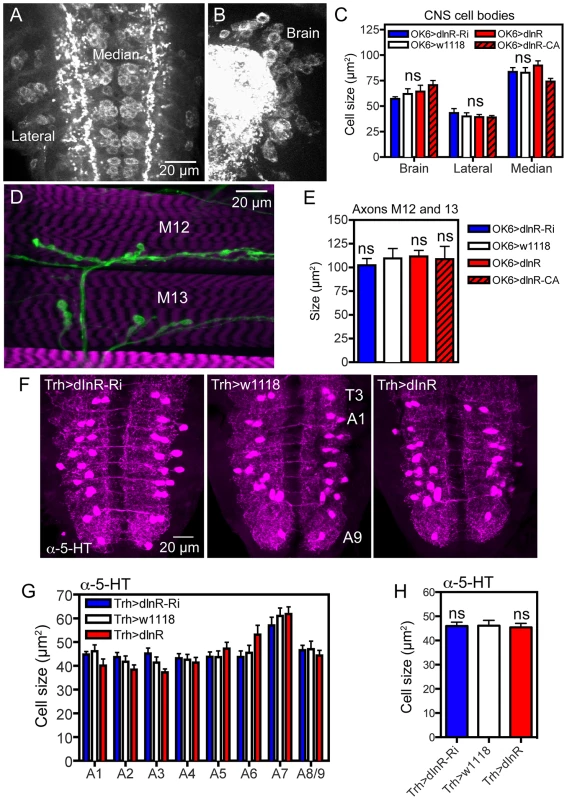
For manipulations we used the OK6-Gal4 driver, known to identify interneurons in the brain and predominantly motor neurons in the ventral nerve cord. Many of the OK6 neurons are likely to be glutamatergic. A and B Distribution of vesicular glutamate transporter immunoreactivity in abdominal motor neurons (A), and brain interneurons (B) of control flies. Two sets of cell bodies were measured in abdominal ganglia lateral and median ones. C Neither dInR-RNAi, nor over expression of dInR or dInRCA (dInR-CA) produced significant changes in cell body sizes of brain neurons or motor neurons (n = 6 animals for each genotype from 3 crosses, unpaired Student's T-test). D Analysis of the size of axon terminals of motor neurons on body wall muscles 12 and 13 (M12 and M13) was made after labeling with anti-HRP. E No effect was seen of dInR manipulations (n = 6–8 body walls for each genotype, unpaired Student's T-test). In Fig. S8 larger areas of body wall muscle innervation by motor neurons is shown for the different genotypes. F Manipulations of dInR levels in serotonergic neurons specified by a Trh-Gal4 driver did not affect cell body size of segmental interneurons in the abdominal ganglia (or elsewhere; not shown) revealed by anti-5HT immunolabeling. G Cell body size of 5-HT-immunolabeled interneurons in A1–A8/9 is not affected by dInR manipulations. H Average cell body size for all segments (ns, not significant, unpaired Student's T-test, n = 11–12 animals from 3 crosses). Tab. 2. Manipulations of genes in different neuron types (peptidergic and non-peptidergic). 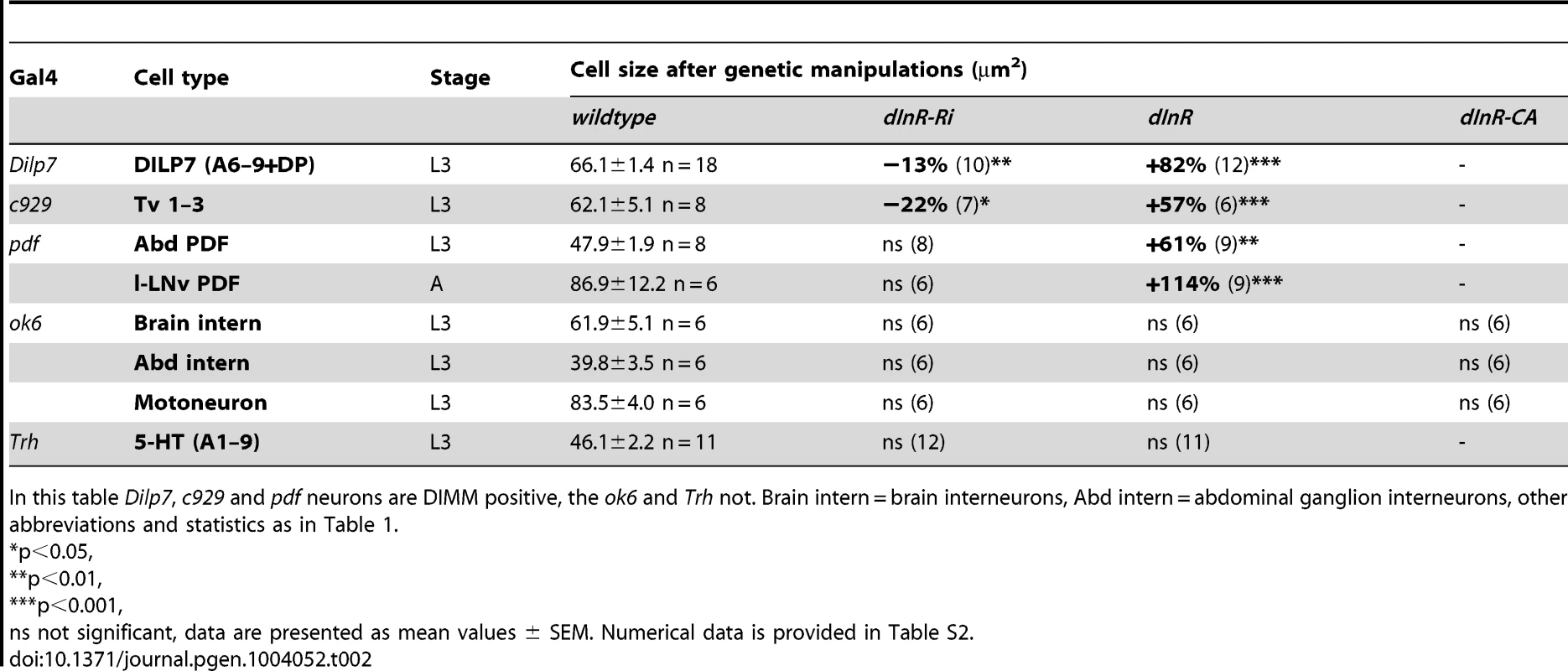
In this table Dilp7, c929 and pdf neurons are DIMM positive, the ok6 and Trh not. Brain intern = brain interneurons, Abd intern = abdominal ganglion interneurons, other abbreviations and statistics as in Table 1. To test another type of interneurons we altered dInR levels in a set of serotonin-producing neurons defined by a Trh-Gal4 line [61]. We selected cell bodies of a set of 30 segmentally distributed serotonin-expressing interneurons in abdominal neuromeres A1–A9 for analysis. No differences in cell body size could be seen after dInR manipulations (Fig. 6 F–H, Table 2). Thus, both interneurons and motor neurons of the OK6 line and 5-HT-producing interneurons seem refractory to dInR induced size changes of the type seen in ABLKs. However, both interneurons and motor neurons grow in response to targeted overexpression of PI3K and Rheb as shown in neurons of the ellipsoid and mushroom bodies of the brain and in the neuromuscular junction [44], [45], [62], [63].
Manipulation of dInR in peptidergic neurons that are DIMM positive
Since motor neurons and non-peptidergic interneurons seem not to display insulin-mediated size plasticity, whereas a set of peptidergic neurosecretory cells do, we asked whether this role of the dInR is general in neuroendocrine cells. Many of the peptidergic secretory and neuroendocrine cells express the bHLH transcription factor DIMM and it is known that DIMM expressing neurons are adapted to physiological demands for high secretory activity and often have large cell bodies [26], [29], [64]. We used the c929-Gal4, known to correspond to the DIMM expressing neurons [28], to manipulate the dInR in specific neurons. The c929 includes the ABLK neurons that also stain with DIMM antiserum from late embryo to adult stages [29], [65].
We showed already that c929-driven manipulations of dInR affected cell size in ABLK neurons (Fig. S3F–I). Next, we monitored the effect on the cell bodies of c929/DIMM neurons expressing the peptide FMRFamide. Among these are three pairs of large ventral FMRFamide-producing neurosecretory cells (Tv1–3) in the thoracic ganglia [29], [66]. Over expression of the dInR increases the size of the cell bodies of the Tv1–3 neurons and dInR-RNAi leads to a slight, but significant decrease (Fig. 7A,B, Table 2).
Fig. 7. Dimm expressing neurons respond to manipulations of the dInR and PI3K. 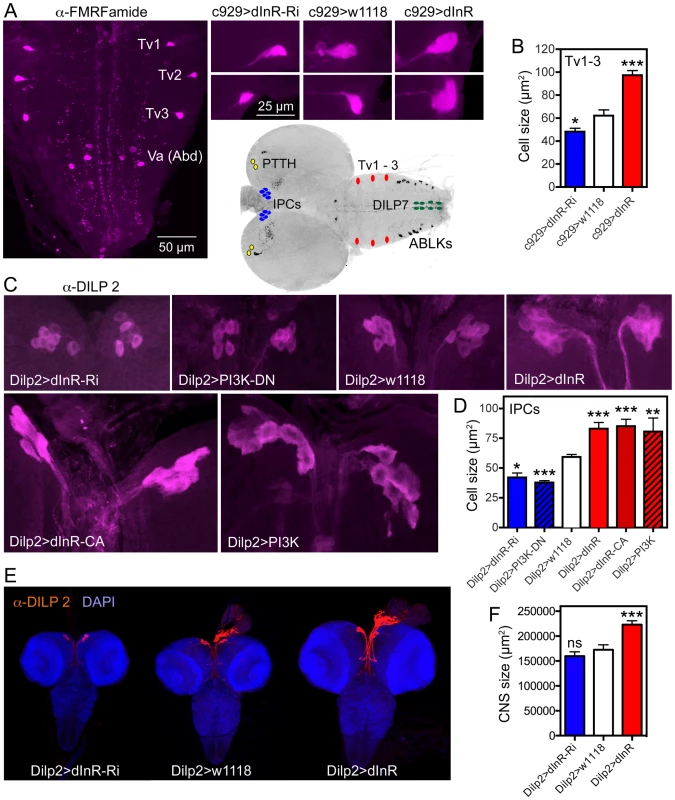
The inset at the center shows the localization in the CNS of third instar larva of neurons depicted in Fig. 6, 7 and Fig. S11. A Using the c929-Gal4 to drive dInR constructs, and an antiserum to FMRFamide, we monitored the cell body size of the thoracic Tv1 – 3 neurons, known to express Dimm. As seen in the smaller panels the cell bodies are smaller after dInR-RNAi and larger after dInR over expression. B Quantification of Tv1–3 cell body sizes (*p<0.05, ***p<0.001, n = 6–8 animals for each genotype from 3 crosses; unpaired Student's T-test). C and D Also the cell bodies of the insulin producing cells (IPCs) of the larval brain respond significantly to manipulations of the dInR as well as PI3K. A dominant negative form of PI3K (PI3K-DN) was used. D Quantification of cell body sizes of IPCs (*p<0.05, **p<0.01, ***p<0.001, n = 6–16 animals for each genotype from 3 crosses; unpaired Student's T-test). E Overexpression of dInR on IPCs also affect IIS and has systemic effects on growth of the CNS. F Quantification of the dInR mediated growth of the CNS. Only overexpression of dInR produces a significant change of CNS size (***p<0.001; unpaired Student's T-test; n = 10–12 animals for each genotype, from 3 crosses). Another group of neurosecretory cells that express DIMM are the 14 insulin producing cells (IPCs) in the brain [29]. By manipulating the dInR levels in IPCs with a Dilp2-Gal4 driver we found that over-expression of both dInR and dInRCA leads to an increase in cell body size and dInR-RNAi to a reduction of cell body size both in larval and adult brains (Fig. 7C,D, Fig. S9, Table 1). Also PI3K manipulations in the IPCs produced similar growth effects both in larvae (Fig. 7C,D, Table 1) and adults (Fig. S9). Here we also noted that Dilp2-Gal4-driven dInR in IPCs led to an increase in the size of the entire larval CNS (Fig. 7E, F), suggesting that systemic insulin signaling was affected. Feedback actions of DILPs onto the IPCs that affect IIS have been demonstrated previously [67].
About 20 neurons in the abdominal ganglia express DILP7 as well as DIMM [68]. We used a Dilp7-Gal4 to alter dInR expression in these neurons and monitored the cell body size with a DILP7 antiserum in late third instar larvae. We focused on a set of 5 pairs of median neurons in abdominal neuromeres A1 and A6–9 with large cell bodies that were easy to identify individually. The cell bodies increased in size after dInR over expression and were slightly smaller after dInR-RNAi (Fig. S10A–D, Table 2). Especially the dorsal pair (DP neurons) in A1, known to send axons to the pars intercerebralis of the brain, was strongly affected; the more posterior ones in A8–9 that send axons to the intestine, were less, but significantly, affected (Fig. S10A–D).
Finally, using a Pdf-Gal4 we found that two subsets of the pigment-dispersing factor (PDF) expressing neurons in the larval abdominal ganglion and in the adult brain known to express DIMM [29] display size phenotypes after dInR manipulations. A set of six abdominal PDF and DIMM-expressing neurosecretory cells in the larva respond to dInR alterations (Fig. S10E, G, Table 2). In the adult brain the cell bodies of the PDF and DIMM-expressing clock neurons designated large LNvs (l-LNvs) respond to dInR manipulations by size changes (Fig. S10F, G, Table 2), but the DIMM-negative small LNvs (s-LNvs), present both in larval and adult brains, do not (not shown).
In summary, our data so far, indicate that peptidergic neurons that express DIMM are affected by targeted manipulations of the dInR during development.
The size of DIMM-negative neuroendocrine cells is not influenced by dInR, but by PI3K and Rheb
To test the requirement of DIMM in regulation of cell size in large peptidergic neuroendocrine cells we analyzed a set of 4 large neurosecretory cells producing the peptide prothoracicotropic hormone (PTTH). These neurons are functional only in the larva [69] and are known to be DIMM-negative [29]. We expressed dInR, dInRCA and dInR-RNAi under the influence of a Ptth-Gal4 and monitored cell body size of the neurons in the late third instar larvae. The size was determined after anti-GFP labeling of Ptth-Gal4;UAS-gfp-driven dInR manipulations and was not affected (Fig. 8A–C, Table 1). Using the Ptth-Gal4 we then drove Rheb and Rheb-RNAi in the PTTH neurons. Here we noted a significant increase in cell body size with over expression of Rheb, but similar to the Lk-Gal4 experiments in larvae there was no effect of Rheb-RNAi (Fig. 8B, C, Table 1). Also PI3K overexpression induces enlarged cell bodies of PTTH neurons, but there was no effect of PI3KDN (Fig. 8B, C; Table 1). Growth effects of PI3K have been demonstrated in interneurons previously and it was proposed that tyrosine kinases other than the dInR could be upstream regulators [45], [63], [70]. Hence, we show that the cell size of DIMM negative neurons can be increased, but not via the dInR.
Fig. 8. Dimm-negative neurosecretory cells are not affected by dInR manipulations, but by Rheb and PI3K overexpression. 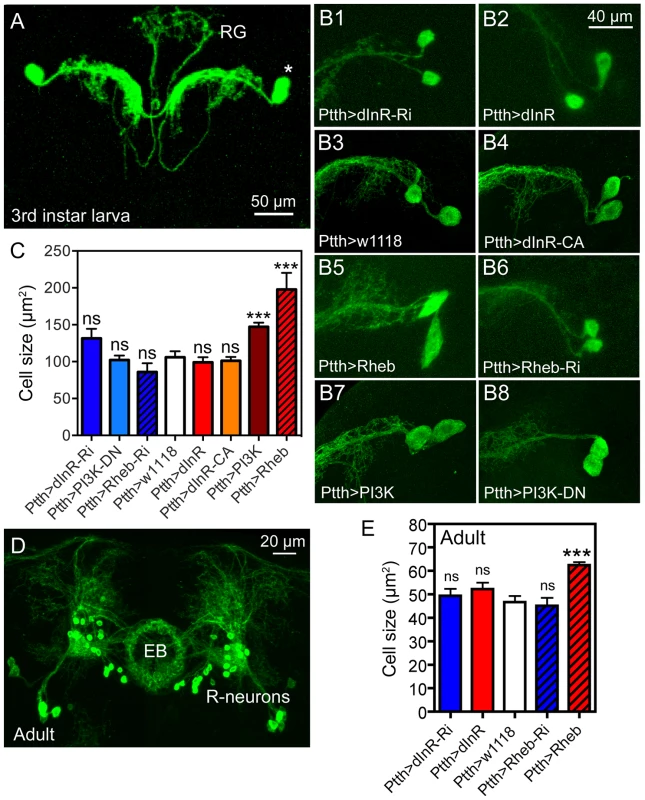
A Two pairs of larval PTTH neurons displayed with Ptth-Gal4-driven GFP. These lateral neurosecretory cells (LNCs; asterisk) supply axon terminations to the ring gland (RG). B and C Effects of manipulations of dInR, Rheb and PI3K on PTTH neurons of third instar larvae. Neurons are displayed with recombinant PtthGal4;gfp. Rheb (B5) and PI3K (B7) over expression leads to a size increase, whereas dInR and dInR-CA expression and RNAi and dominant negative (DN) manipulations have no significant effects. C Quantification of cell body size reveals that only Rheb and PI3K overexpression increases cell body size (**p<0.01, n = 5–15 animals for each genotype from 3 crosses; unpaired Student's T-test. D In the adult brain the PTTH-expressing LNCs undergo apoptosis and instead a subset of the ellipsoid body (EB) R-neurons display GFP. E Again, only the Rheb over expression leads to increased cell body size in R-neurons (***p<0.001, n = 5–10 animals for each genotype from 3 crosses; unpaired Student's T-test). CA, constitutively active; DN, dominant negative. The PTTH neurons undergo apoptosis in the early adult fly and curiously in the older fly the Ptth-Gal4 expression is seen in R-neurons that supply processes to the ellipsoid body of the central complex [69]. We analyzed the cell body size of these R-neurons in older flies and found that again only Rheb over expression increased the cell size (Fig. 8D, E, Table 1). This is consistent with the effect of Rheb of cell size in DIMM negative mushroom body neurons [44].
Origin of DILPs that regulate the size of peptidergic neurons
There are several possible sources of DILPs that could affect growth of DIMM-expressing peptidergic neurons in early larvae: DILP2, 3 and 5 released from IPCs or DILP6 from fat body could act via the circulation, DILP7 could act in a paracrine fashion from abdominal neurons with central branches (Fig. S11A), or DILP2 and 6 released from glial cells during early larval stages [7], [21], [68], [71] (see Fig. S12). Perhaps even the newly discovered DILP8 released from imaginal discs could act via the circulation [72], [73].
We tested two of these alternatives experimentally. To investigate the possible roles of brain IPCs and abdominal DILP7-producing neurons on LK neuron size we inactivated either of these insulin-producing neuron groups by expressing the hyperpolarizing potassium channel, dOrk1 [54] under control of Dilp2 or Dilp7-gal4 drivers. We measured ABLK cell bodies in third instar larvae and found that hyperpolarization of IPCs led to slightly, but significantly, decreased ABLK cell bodies, whereas the same manipulation of DILP7 neurons produced no phenotypes (Fig. S11B–F). Also ablation of the IPCs by means of Dilp2-Gal4 driven reaper reduced the size of the ABLKs (Fig. S11D). Thus DILPs from IPCs, but not DILP7, could play a role in regulation of ABLK neuron size. Not surprisingly, we found that this may be due to a general effect on somatic growth and the Dilp2>Ork larvae displayed smaller volume of the CNS and adults had smaller body and wing sizes (Fig. S11F, H–J). Furthermore, as described in a previous section we also found that the size increase in IPC cell bodies after dInR over expression was accompanied by an increase in over-all CNS size (see Fig. 7E, F). As seen in Fig. S11D and F the ablation or inactivation of IPCs does not cause a drastic decrease in ABLK size. Probably this is due to redundant sources of DILPs such as DILP6 released from the fat body or glial cells in earlier larval stages.
In 1st and 2nd instar larvae DILPs released from specific glial cells in thoracic neuromeres of the ventral ganglion terminate cell cycle arrest and growth in neuroblasts [21]. Glial cells may therefore be a source of DILPs also for neuronal growth. We confirm here that Dilp6-Gal4 driven GFP is prominent also in glial cells adjacent to ABLK neurons and elsewhere in abdominal ganglia in 1st and 2nd instar larvae (Fig. S12) and we demonstrated that dInR over expression induces strong increase in ABLK cell body size already in 1st instar larvae (Fig. 2J–M). Our findings suggest DILPs from the IPCs, but not DILP7 from abdominal neurons, are likely to be important for size regulation, and we cannot exclude a role of DILP6 from glial cells in earlier growth.
Dimm-RNAi in ABLK neurons affects cell size and dInR expression, and dInR-RNAi affects DIMM levels
DIMM is expressed in a large subpopulation of the peptidergic neurons of Drosophila, including ABLKs (Fig. 9A) and is required for activation of key genes involved in differentiation of a dynamic secretory phenotype [26], [28], [29]. Since only DIMM neurons displayed size phenotypes after dInR manipulation, we asked whether there is a functional correlation between DIMM and dInR-regulated neuronal development and growth.
Fig. 9. Possible interactions between Dimm and dInR in ABLKs and effects on nuclear size. 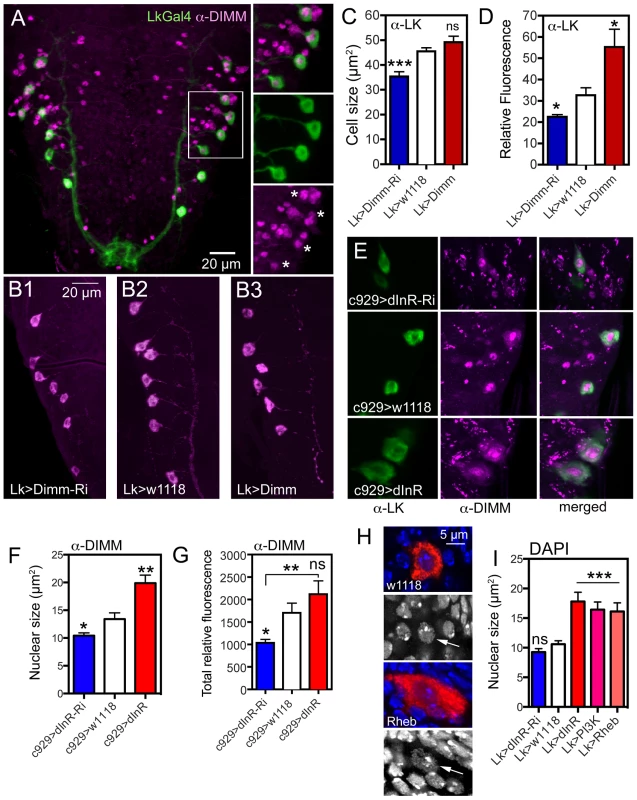
A. The ABLKs (Lk-Gal4-GFP; green) express DIMM-immunolabeling (magenta) in nuclei. Details of the framed area are shown in smaller panels (asterisks indicate nuclei of LK neurons). B–C The size of LK cell bodies diminishes with Lk-Ga4 driven Dimm-RNAi, but is not affected by Dimm overexpression (***p<0.001, n = 6–9 animals from 3 crosses for each genotype; unpaired Student's T-test). D The LK-immunofluorescence in ABLKs is affected by both Dimm-RNAi and over expression (*p<0.05, n = 6–9 flies for each genotype from 3 crosses; unpaired Student's T-test). E and F Manipulations of dInR using the c929-Gal4 affects nuclear size as determined by DIMM immunolabeling (*p<0.05, **p<0.01, n = 7–8 animals for each genotype from 3 crosses: unpaired Student's T-test). The surface area of each DIMM-labeled nucleus in LK-immunolabeled cell bodies (in A1–A4 segments) was measured using Image J and the average nuclear size (LK-neurons in A1–A4) of each fly was thereafter determined. G The total fluorescence (mean fluorescence multiplied by cell size) in abdominal DIMM immunolabeled neurons decreases with dInR-RNAi, but there is no significant increase after dInR over expression (*p<0.05, **p<0.01, n = 8 animals for each genotype from 3 crosses; unpaired Student's T-test). H and I Effects of dInR, PI3K and Rheb manipulations on size of nuclei in ABLKs. In H nuclear size is visualized by DAPI staining (blue) in ABLK neurons (red). Nuclear size (arrows) is enlarged after Rheb over expression. I Quantification of nuclear size shows that dInR-RNAi has no effect (compared to controls), whereas over expression of dInR, PI3K and Rheb produces a significant enlargement of nuclei (unpaired Student's T-test, *** p<0.001, n = 6–11 animals for each genotype from 3 crosses). To test this we knocked down or overexpressed Dimm with the Lk-Gal4 driver and monitored ABLK cell size. These cell bodies were smaller after Dimm-RNAi, but did not increase in size after over expression compared to controls (Fig. 9B, C). As shown earlier, the LK-immunoreactivity in ABLKs diminished after Dimm knockdown and increased after over expression (Fig. 9D) [65].
We next asked if there is a relation between dInR levels and DIMM expression. To test this we expressed dInR or dInR-RNAi in Lk-Gal4 or c929-Gal4 neurons and applied antiserum to DIMM to the larval CNS and monitored the immunofluorescence level in nuclei of the neurons. After dInR-RNAi there was a decrease in total DIMM immunolabeling intensity, as well as in the size of DIMM labeled nuclei (Fig. 9E–G). However after dInR overexpression the nuclear size, as determined by DIMM immunolabeling, increased (Fig. 9F), whereas total DIMM immunolabeling was not significantly affected (Fig. 9G). Thus, there is a weak, but significant effect of dInR on DIMM expression.
To test whether Dimm influences dInR expression levels in ABLK neurons we knocked down or overexpressed Dimm with the Lk-Gal4 and measured dInR immunolabeling. We found that Dimm-RNAi diminished dInR labeling in ABLKs, but not generally in neuropil, whereas overexpression resulted in significantly increased dInR (Fig. 10 A–C, E). We also measured dInR labeling in third instar larvae that had been kept for 48 h with 0% protein in the diet compared to 5% protein (Fig. 10D; Fig. S15A). There was a significant difference in dInR immunolabeling levels between these two treatments (Fig. 10D, E).
Fig. 10. Knockdown of Dimm in ABLKs affects dInR expression and cell size regulation. 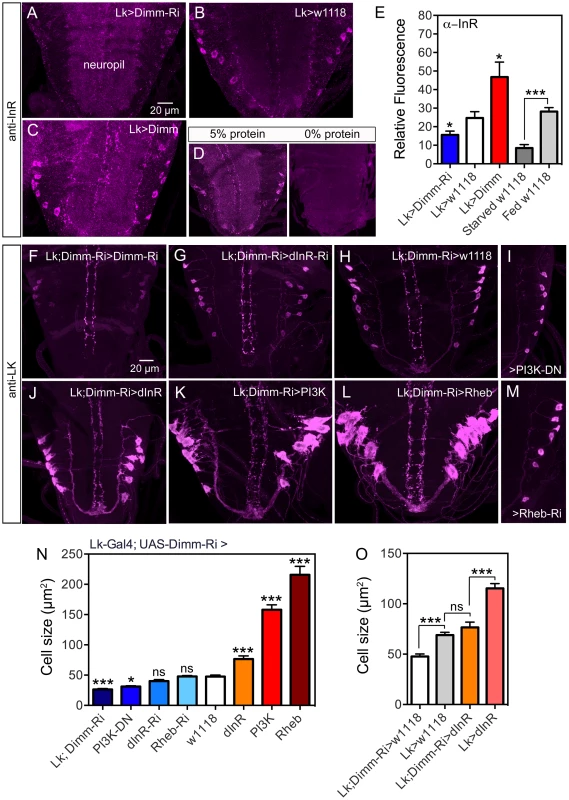
A–C InR immunolabeling (antiserum #3021) after manipulations of Dimm levels in LK neurons. Dimm knockdown drastically decreases InR immunolabeling, except in the neuropil (A) compared to controls (B). Overexpression of Dimm in LK neurons increases InR immunolabeling (C). D Insulin receptor immunoreactivity is affected by protein diet. Larvae were reared under two conditions: 20% sucrose with 5% or 0% protein (Yeast) according to the schedule in Fig. S15A. InR labeling is far stronger under 5% protein than 0% protein. Not only the ABLKs, but also general neuropil InR immunolabeling is decreased under 0% conditions. E Quantification of immunolabeling of ABLK neurons in A–D. The changes in InR immunolabeling are shown in relation to controls (Lk>w1118) (*p<0.05, ***p<0.001, n = 7–16 animals for each genotype from 3 crosses: unpaired Student's T-test). This graph also shows the effect of 48 h protein starvation on InR immunolabeling (grey bars). F–M With a double transgene (Lk-Gal4;UAS-Dimm-RNAi) we analyzed the effects of Dimm-RNAi on manipulations of dInR, PI3K and Rheb in ABLKs labeled with anti-LK in late third instar larva. In I and M only half a ganglion is depicted. N Quantification of ABLK cell body size in the different genotypes shown in F–M. Larvae with a heterozygous Dimm-RNAi genetic background displayed significantly larger ABLK cell size when either of dInR, PI3K or Rheb was overexpressed in LK neurons. Overexpression of dInR induced much weaker cell growth than PI3K or Rheb. ABLKs displayed smaller cell size when Dimm was knocked down with two copies of Dimm-RNAi (Lk-Gal4;UAS-Dimm-RNAi>Lk-Gal4; UAS-Dimm-RNAi). Knockdown of dInR in heterozygous Dimm-RNAi flies lead no decrease of cell size. Statistics: ***p<0.001; ns, not significant; n = 9–11 animals from 3 crosses for each genotype, unpaired Student's T-test; comparisons were made between each of the manipulations and the control (Lk-Gal4;UAS-Dimm-RNAi>w1118). Genotypes for each bar are as follows. Lk;DIMM-Ri, Lk-Gal4/Lk-Gal4; UAS-Dimm-RNAi/UAS-Dimm-RNAi, PI3K-DN, Lk-Gal4/UAS-PI3K-DN; UAS-Dimm-RNAi/+, Rheb-Ri, Lk-Gal4/+; UAS-Dimm-RNAi/UAS-Rheb-RNAi, dInR-Ri, Lk-Gal4/UAS-dInR-RNAi; UAS-Dimm-RNAi/+, w1118, Lk-Gal4/+; UAS-Dimm-RNAi/+, dInR, Lk-Gal4/UAS-dInR; UAS-Dimm-RNAi/+, PI3K, Lk-Gal4/UAS-PI3K; UAS-Dimm-RNAi/+, Rheb, Lk-Gal4/UAS-Rheb; UAS-Dimm-RNAi/+. O Diminished Dimm in ABLK neurons significantly restricted dInR-induced cell growth. Overexpression of dInR in Dimm-RNAi flies (Lk;Dimm-RNAi>dInR) resulted in cell size similar to controls (Lk>w1118), and much smaller than after overexpressing dInR alone. (***p<0.001; ns, not significant; n = 6–13 animals for each genotype from 3 crosses; unpaired Student's T-test). Genotypes for each bar are as follows. Lk;DIMM-Ri>w1118, Lk-Gal4/+;UAS-Dimm-RNAi/+, Lk>w1118, Lk-Gal4/+; +/+, Lk;Dimm-Ri>dInR, Lk-Gal4/UAS-dInR;UAS-Dimm-RNAi/+, Lk>dInR, Lk-Gal4/UAS-dInR; +/+. To further examine the requirement of DIMM expression for dInR-mediated size regulation we employed a double transgene (Lk-Gal4/Lk-Gal4;UAS-Dimm-RNAi/UAS-Dimm-RNAi; here abbreviated Lk-Gal4;Dimm-Ri) for interfering with dInR, PI3K, Rheb and Dimm in ABLKs. We find that flies bearing the homozygous double transgene Lk-Gal4;Dimm-Ri display very small ABLKs (Fig. 10F, N). However, driving the dInR-RNAi with Lk-Gal4;Dimm-Ri does not significantly diminish cell bodies compared to controls (Fig. 10G, H, N) and overexpression of the dInR in Dimm knockdown flies results in less increase of cell size (Fig. 10J,N). Over expression of PI3K and Rheb with the Lk-Gal4;Dimm-Ri driver on the other hand leads to a substantial size increase (Fig. 10K,L,N). Dominant negative PI3K expression decreases cell body size significantly (Fig. 10I, N), but not Rheb-RNAi (Fig. 10M, N). As discussed for PTTH neurons above, we propose that PI3K and Rheb may act independently of dInR activation. Compared to Lk-Gal4>dInR and Lk-Gal4>w1118 controls, the Lk-Gal4;Dimm-Ri>dInR flies display ABLKs that did not grow significantly (Fig. 10O), suggesting that after Dimm knockdown the dInR has little effect on cell size. In Fig. 10O it can also be seen that Dimm-RNAi (Lk;Dimm-Ri>w1118) has an effect on cell size compared to Lk>w1118 suggesting that Dimm expression alone affects cell size.
The dInR-mediated size increase affects Golgi apparatus and nuclear size
Since DIMM expression correlates with competence to produce neuropeptides in bulk we monitored the effect of dInR manipulations on cell organelles involved in protein synthesis, such as the trans-Golgi network from which secretory vesicles arise. One marker that has been associated with the trans-Golgi network in Drosophila neurons is PICK1 (the protein interacting with C kinase 1) a Bar domain protein that is known to be expressed in DIMM expressing neurons [74], [75]. We therefore employed antiserum to PICK1 to obtain an estimate of the amount of Golgi and likely secretory activity in ABLKs after dInR manipulations (using the c929-Gal4). We found that dInR over expression increases both the PICK1-immunolabeled area within the cell body and the total relative PICK1 immunofluorescence in the abdominal ganglion neurons (Fig. S13A–E). Since the punctate PICK1 immunolabeling approximately represents the extent of the trans-Golgi [74], [75] (see also Fig. S13F) we suggest that dInR over expression leads to an increased capacity for secretory vesicle production.
Using neuropeptide immunolabeling the nuclei of neurons commonly remain unlabeled and their size can be estimated. A survey of nuclei in ABLKs and IPCs (e. g. Fig. 2A–D, 4, 7C) suggest that their size increases after dInR over expression. Also DIMM immunolabeling indicated nuclear growth after IIS manipulations (Fig. 9E, F). Thus we wanted to test whether the nuclear size increase is due to DNA-endoreplication, as proposed for giant neurons in mollusks [27], [76]. Endoreplication entails DNA synthesis in absence of cell division and leads to increased cell size and either polyploidy or replication of some genomic regions, polygeny [77]. Increases in DNA ploidy can be induced by growth and IIS/TOR signaling in endoreplicating tissues [47], [78]. To obtain a measure of nuclear size, as well as estimate of DNA content, we applied DAPI staining (see [27], [44]) after dInR, PI3K and Rheb manipulations or LK neurons. We found significant increases in the size of DAPI labeled nuclei in ABLK neurons after over expression of these components, but no change in size after dInR-RNAi (Fig. 9H,I). Also in IPCs the nuclei were larger after dInR over expression (Fig. S14A–D). We quantified the DAPI fluorescence but there was no significant increase in fluorescence in nuclei of ABLKs or IPCs (Fig. 9H, Fig. S14E). Thus, there is probably no induction of DNA-endoreplication by increased IIS and TOR activity in these neuron types in the larvae.
Size scaling in Dimm positive neurons is nutrient dependent
To test whether the Dimm-positive neurons are affected by nutrient restriction or increased protein content in the diet we conducted a set of experiments. For nutrient restriction we transferred second instar larvae of different genotypes from food with 20% sucrose and 5% protein to food containing only 20% sucrose (0% protein). Another set of larvae was raised on food containing 20% sucrose and 20% protein. Controls were raised on 20% sucrose and 5% protein throughout (see Fig. S15A for diet regime). The cell body size of ABLKs was measured in late third instar larvae along with the size of abdominal serotonin-immunoreactive neurons (5-HT neurons) as a reference. We used larvae of the genotypes Lk>dInR-RNAi, Lk>w1118 and Lk>dInR.
We found that under nutrient restriction the entire CNS is significantly smaller in larvae, of all genotypes, as compared to those fed 5% or 20% protein (Fig. 11C). The size of the ABLK cell bodies was also significantly smaller for the three genotypes after 0% protein diet, compared to the other feeding conditions (Fig. 11A,B,D; Fig. S15B). In fact, the cell body sizes resembled those seen for the same genotypes in the late first instar larva (Fig. 11D). Feeding larvae protein-rich diet did not increase CNS size or ABLK cell body size compared to 5% protein for any genotype (Fig. 10C,D; Fig. S15C,D). We calculated the ratio between cell body size of 5-HT neurons and ABLKs to determine relative growth of dInR manipulated versus non-manipulated neurons. It was seen that the ratio only marginally (but significantly) increased from 0% to 5% protein after dInR overexpression in LK neurons (Fig. 11E). It can be noted that not only the CNS but also the 5-HT neurons are significantly smaller after 0% protein diet compared to 5 and 20% (Fig. 10F). In summary we show that the dInR-induced growth is protein dependent.
Fig. 11. Effects of diet on size of ABLK neurons. 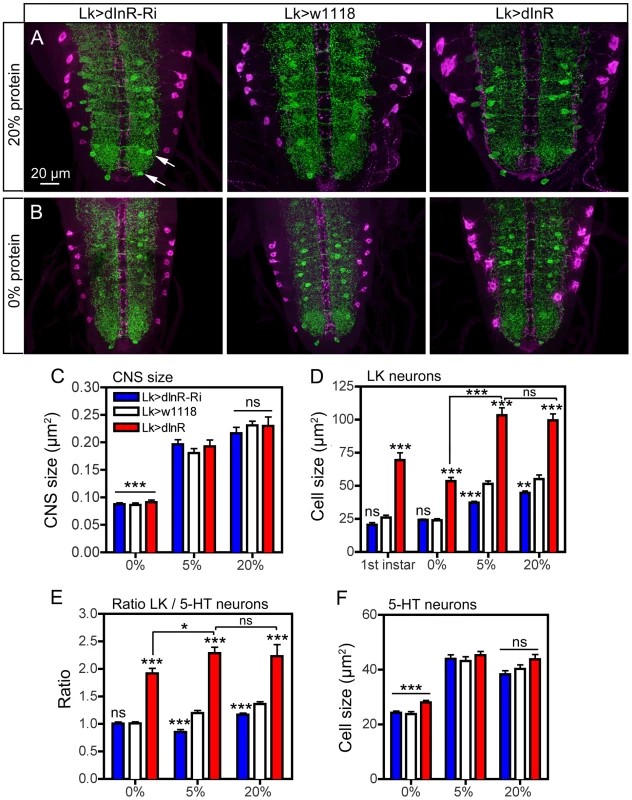
Larvae of different genotypes were reared on three different diets: 20% sucrose with either 0%, 5% or 20% protein (yeast) as detailed in the scheme in Fig. S15A. Late third instar larvae were labeled with anti-LK (magenta) and anti-5-HT (green). Statistics: *p<0.05, **p<0.01, ***p<0.001, n = 6–12 larvae for each genotype from 3 crosses; unpaired Student's T-test for comparison of genotypes; Two-way Anovas for comparisons of diet effects). A and B Manipulation of dInR expression in LK neurons affects the size of ABLK neurons in larvae fed protein-rich (20%; A) and protein-free (0%; B) and normal (5%; not shown) diet. Cell bodies of 5-HT immunolabeled neurons (arrows in Fig. 11 A) are used for reference. The size changes in CNS and different neurons are quantified in Fig. 11 C–F (cell bodies are shown in higher magnification in Fig. S15 B). C The low protein (0%) diet significantly (p<0.001) affects the size of the entire CNS compared to “standard diet” (5%), whereas 20% protein does not increase the CNS (ns; p>0.05). Note that the CNS size is not affected by the dInR manipulations. D Quantification of ABLK cell body size in third instar larvae fed the three different diets, compared to those in first instar larvae (genotypes as in C). Under 0% protein conditions all genotypes display third instar ABLK cells with sizes corresponding to those in first instar larvae. These are significantly smaller than in flies fed 5% and 20% protein (p<0.001 for all genotypes compared for 0% and 5% protein; Two-way Anova). Larvae fed protein-rich food (20%) do not display increased ABLK cell bodies compared to those fed 5% protein. The significance values indicate differences to control flies (Lk>w1118) for each protein condition (*p<0.05, **p<0.01, ***p<0.001, ns, not significant, n = 6–10 larvae for each genotype from 3 crosses; unpaired Student's T-test) and comparisons between protein diets for dInR overexpression (Two-way Anova). E The ratio between the ABLK cell size and the size of 5-HT neuron cell bodies in corresponding abdominal neuromeres (same protein conditions and genotypes as in C). These were determined to establish whether the ABLK size is affected disproportionally by protein diet (the size of the 5-HT neuron cell bodies for the three protein diets are shown in F). It can be seen that with 5-HT cells as size reference the dInR overexpression only marginally (p<0.05; Two-way Anova) increased the ABLKs after dInR overexpression when comparing 0% and 5% (and 20%) protein. For each diet dInR over expression significantly increases the ratio between ABLK and 5-HT cell body sizes compared to control flies. Only small (but significant) effects can be seen after dInR-RNAi in 5% and 20% diets. F Size of cell bodies of 5-HT immunolabeled neurons in abdominal ganglia in larvae reared in the three different diets (genotypes as in C). Note that manipulations of the dInR in LK neurons do not affect the 5-HT neurons, but 0% protein diet results in significantly smaller cell bodies compared to 5% and 20% protein. Discussion
Our study shows that insulin receptor-mediated signaling can selectively regulate the size of peptidergic neuroendocrine cells during development and in the adult organism. This may provide a mechanism for plasticity in size of secretory neurons independent on growth of other neurons, since we found that motor neurons and several types of interneurons do not display this feature. The dInR-mediated growth regulation seems to require properties mediated by, or coincident with, the expression of the transcription factor DIMM (summarized in Tables 1 and 2). Thus, several subpopulations of the approximately 300 DIMM-expressing neurons were found to respond to manipulations of dInR levels or signaling components downstream to this receptor. We also found that dInR manipulations are less effective when Dimm levels are knocked down. DIMM-expressing neurons are known to episodically release large amounts of amidated neuropeptide or peptide hormone through the regulated secretory pathway, and are thus equipped with a sizable protein synthesis machinery in the cell body [25], [26], [29], [30]. It has been proposed that secretory cells have their regulated secretory machinery scaled up during differentiation to be fit for their demanding peptide signaling and plasticity in function related to changes in environment and organismal homeostasis later in life [25]. Hence, DIMM, and MIST1 in vertebrates, are transcription factors that orchestrate the up-scaling of the secretory pathway and, thus, the size of the cell bodies [25]. A large number of genes are transcriptional targets of DIMM, but insulin-signaling components are not prominent among genes affected by over-expression of DIMM [26]. Therefore, the activation of the dInR may trigger signaling that converges on scaling events in the DIMM expressing neurons.
An extensive scaling of neuron size has been recorded in gastropod snails where many of the peptidergic neurosecretory cells are gigantic [76]. A recent study further demonstrated that the size of these peptidergic neurons increases as the adult animal grows in response to rich nutrition [27]. This growth depends on DNA-endoreplication in the specific neurons, where genomic multiplication ensures efficient protein synthesis. It was proposed that efferent neurons and neurosecretory cells, but not interneurons, adapt their sizes to the enlarged periphery and circulation as the animal grows [27]. Our results suggest that targeted increases in IIS results in larger nuclei, but not significantly increased DNA levels, as measured by DAPI labeling in nuclei. Thus up-regulated IIS probably does not induce increases in DNA ploidy in the DIMM-positive neurons. This is expected since although many cell types in Drosophila larvae are polyploid, there are no reports on endoreplication in neurons [47], [78], [79]. However, other mechanisms may regulate the size of the nucleus. For instance the gene kugelkern (kuk) affects growth of the nuclear envelope, and it is generally presumed that larger cells have larger nuclei (see [80], [81]).
In mammals brain growth depends on IGF signaling during precise time windows during development, and relies on ubiquitous neuronal expression of the IGF1 receptor [10], [82], [83]. IGF signaling, as well as insulin signaling, also partake in other developmental processes in the CNS and may do so in concert with other factors as part of a combinatorial instructive signal [5]. In Drosophila growth mediated by IIS has primarily been studied at the whole organism or tissue/organ level [3], [6], [7], [11], [16], [71], [84]. During development the IIS may act on both cell growth and proliferation. This has been shown for Drosophila neuroblasts where developmental quiescence is terminated by IIS and cell proliferation starts after neuroblast growth [21]–[23]. There seems to be no reports on IIS-mediated post-mitotic growth of neurons at the single cell level in Drosophila, although the effects of PI3K and TSC2 (gigas) on synaptogenesis and neuropil growth has been documented after targeting specific sets of neurons [45], [63].
During feeding stages post-mitotically growing neurons (as a tissue) are regulated by the cell autonomous nutrient sensing TOR signaling pathway combined with IIS [6], [15]–[17], [24], [47] (Fig. S16). In case of nutritional deficiency during development the growth of CNS neuroblasts is spared, whereas many other cell types respond to malnutrition by restricted growth [17], [85]. This sparing depends on a pathway employing Alk and its ligand jelly belly [17]. At nutritional restriction Alk signaling utilizes some of the same downstream components as the dInR mediated IIS (see Fig. S16), but is independent of TOR signaling [17].
The sets of DIMM-expressing neuroendocrine cells studied here appear to have another layer of growth control. This may enable enlargement of their cell bodies and neurites during feeding stages to adapt to somatic growth of the animal during rich nutritional conditions, or spare their growth during nutrient restriction. This regulation depends on IIS through the evolutionarily conserved pathway including dInR, the insulin receptor substrate Chico, PI3K and AKT1 [6], [7], [14], [21]. We also found that the dInR-mediate size regulation is especially prominent during nutritional restriction. Thus, we propose that post-mitotic DIMM-positive neurons are protected by the dInR in a manner resembling the action of Alk signaling in neuroblasts [17]. The size of the DIMM-positive neurons can also be altered by manipulations of TOR signaling components such as TOR, Rheb and S6K/4EBP [6], [15], [44], [46]. DIMM-negative neurons tested display increased cell body size after overexpression of Rheb but not dInR, suggesting a non-IIS dependent growth regulation. There is a previous report that targeted Rheb overexpression induces growth of IPCs and mushroom body Kenyon cells, and thus the entire mushroom body lobes [44]. Furthermore, Rheb over expression in Drosophila motor neurons increases growth of axon terminations in neuromuscular junctions, NMJs [62]. This Rheb action could occur either downstream of the dInR, or independent of the dInR as part of the TOR pathway [46], [47] (see Fig. S16). The Rheb-regulated growth of the NMJ and Kenyon cells is likely to be independent of bona fide dInR signaling since we found no dInR-mediated growth phenotypes in motor neurons and a recent paper reported that diminishment of insulin signaling to the Kenyon cells by expression of dInRDN does not induce any overt alteration of mushroom body size or morphology [86]. Thus, it is likely that the Rheb-mediated growth in DIMM negative neurons shown in the present study is independent of dInR.
Our data suggest that PI3K overexpression can increase the size of DIMM-negative neurons (PTTH neurons) and in ABLKs after Dimm knockdown. Since these neurons do not grow in response to dInR expression PI3K could possibly be activated by other upstream elements than the dInR. It has indeed been proposed that PI3K-induced growth of brain interneurons (and synapse formation) could be triggered by tyrosine kinases other than the dInR [45], [63], [70].
The source of the insulin signal that activates growth of DIMM neurons could be either globally released DILPs from the IPCs of the brain (or the fat body) or DILPs released from the local niche in a paracrine fashion. During early larval development surface glial cells in the ventral nerve cord express DILP2 and 6 [21]. These glial cells were shown to release DILPs in response to nutritional signals from the fat body and the paracrine DILPs activate IIS in dormant neuroblasts to enable reactivation of the cell cycle. This exit from dormancy involves cell growth and entry into S-phase [21], [23]. Thus, the DILP signal that regulates growth of DIMM-expressing neurons might be paracrine and glial derived in earlier larval stages, whereas systemic DILP signaling plays a role in later stages. Systemic DILP signaling is suggestive since we find that the size of DIMM-positive neurons can be affected by dInR activation or inactivation throughout larval and adult life. Also, we saw an effect on size of DIMM-positive neurons after inactivation of the DILP producing IPCs in the brain. Combined our findings suggest that there is no specific time window for the dInR mediated size regulation in DIMM-positive neurons. In this context it is important to note that DIMM-expression begins in the embryo and persists into the adult fly [65]. The level of DIMM-expression could be correlated with the expression of LK and DILP2 peptides and transient over-expression of DIMM in adults resulted in increased peptide levels [65].
The dInR-induced growth of the DIMM-positive LK neurons, that we investigated most thoroughly, includes enlarged cell bodies with increased amount of stored neuropeptide and trans-Golgi, increased nuclear size and more extensive axon terminations with larger boutons. We have no evidence that these growth phenomena are linked to DNA endoreplication. In other larval tissues, such as larval muscles of Drosophila dInR/TOR signaling is among the key regulators of endoreplication, along with FOXO and the transcription factor Myc [78]. In Drosophila DNA endoreplication has furthermore been shown in cells in epidermis, muscle, salivary glands and fat body, as well as in certain large glial cells [78], [79], [87], [88], but to our knowledge not in neurons (see also [44]).
In summary, we propose that major DIMM-positive neuroendocrine cells in Drosophila are equipped with a mechanism to maintain their hormone production at an optimum. Selective size scaling of these neurons could adjust hormone production capacity relative to somatic growth and increased demands for peptide signaling. This dInR-dependent regulation is diet-dependent.
Materials and Methods
Fly strains and husbandry
All flies (except diet experiments) were reared at 25°C and 12∶12 h light∶dark conditions on a standard yeast, corn meal, agar medium according to the standard medium recipe of Bloomington Drosophila Stock Center (BDSC), Bloomington, IN (see http://flystocks.bio.indiana.edu/). Flies with other original genetic backgrounds were backcrossed into w1118 background for four generations before experiments, and w1118 flies were used as controls in all experiments. For experiments in adult flies only males were used (unless specifically stated 3–4 d old adults were used).
The following Gal4 lines were used:
w; Lk-Gal4 (II) ([38]; gift from P. Herrero, Madrid, Spain),
w; Dilp2-Gal4 (III) ([89] from E. Rulifson, Stanford, CA),
w; Dilp7-Gal4 (III) ([90] from Y. N. Jan, San Francisco, CA),
w; elav-Gal4/TM6B (III), w; OK6-Gal4;UAS-MycRic/TM3Sb (II) ([59] from A. Ferrus, Madrid, Spain),
yw; pdf-Gal4 (II) ([91] from J. H. Park, Knoxville, TN),
w; c929-Gal4 (III) ([28] from P. H. Taghert, St Louis, MO),
w; Trh-Gal4 (II) ([61] from E. A. Kravitz, Boston, MA).
w; Dilp6-Gal4 (II) ([21] from A. Brand, Cambridge, UK)
yw, Dilp6-Gal4 (X) ([92] from the Drosophila Genetic Resource Center, Kyoto Institute of Technology, Kyoto, Japan; Stock No 103-877).
The following UAS lines were employed:
w; UAS-dInR-RNAi (III) [from Vienna Drosophila RNAi Center (VDRC), Vienna, Austria]. This line was utilized and characterized previously [41], [42].
w; UAS-InRCA and yw; UAS-InRDN ([43] from P. Shen, Athens, GA),
yw; UAS-dInR (II), w; UAS-Akt1-RNAi (III), w; UAS-Akt1 (II), w; UAS-Rheb-RNAi (III), w; UAS-Rheb (II), w; UAS-S6k.KQ (II), w; UAS-S6k.STDETE (III), w; UAS-S6k.M (II), yw;UAS-Tor.TED (II), yw;UAS-NaChBac (III), w; UAS-Ork1/TM6cSb (III), yw;UAS-mcd8-gfp (III) and w; UAS-gfp.nls (III) [all from Bloomington Drosophila Stock Center (BDSC), Bloomington, IN],
w; UAS-PI3K and yw; UAS-PI3KDN (UAS-PI3K92ED954A) [[93], [94] from A. Ferrus],
w; UAS-dInR-RNAi/CyO (II) and w; UAS-rpr-hid (II) ([95] from J. R. Martin, Gif-Sur-Yvette, France),
yw; UAS-dimm-Myc (II) and w; UAS-dimm-RNAi (II, III) ([28], [96] from P. H. Taghert, St Louis, MO).
yw; ptth-Gal4;UAS-gfp ([69] from K. F. Rewitz, Copenhagen, Denmark),
w; UAS-Alk (III), w; UAS-AlkDN (III), w; UAS-AlkCA (II) ([97], [98] from R. Palmer, Umeå, Sweden)
w; UAS-GRASP-gfp ([74] from O. Kjaerulff, Copenhagen, Denmark).
Two balancer flies were used:
w; Sco/SM1cy; Dr/TM6BTbSb and w; Sp/Cyo; Vno/TM3Sb (from C. Samakovlis, Stockholm, Sweden).
Antisera and immunocytochemistry
The central nervous system (CNS) of first, second and third instar larvae, adult CNS, as well as third instar larval body wall muscles were dissected out in 0.1 M sodium phosphate buffer (PB; pH 7.4) and fixed in ice-cold 4% paraformaldehyde (4% PFA) in 0.1 M PB for 2–4 h. All tissues were rinsed with 0.1 M PB three times over 1 hr and washed finally in 0.01 M PBS with 0.25% Triton-X (PBS-Tx) for 15 min before application of primary antisera. The primary antisera were diluted in 0.01 M PBS-Tx with 0.05% sodium azide. Incubation with primary antiserum for whole tissues was performed for 24–48 h at 4°C with gentle agitation. Tissues were rinsed thoroughly with PBS-Tx, followed by application of secondary antibody overnight and thorough wash in 0.01 M PBS and then mounted in 80% glycerol in 0.01 M PBS.
The following primary antisera were used: an antiserum to cockroach leucokinin I (LK I) (1∶2000) [40] known to recognize the conserved C-terminus of insect leucokinin peptides [37], [99] was used for identification and quantification of leucokinin. Rabbit anti-mosquito (Aedes aegypti) insulin receptor (anti-Aed InR; raised against 50 amino acids from the extracellular alpha chain), kindly provided by of M. Brown, Athens, GA) at 1∶1000, rabbit anti-phosphorylated human InR (#3021 and #3024, from Cell Signaling Technology) at 1∶2000, rabbit anti-DILP2 (1∶2000) (from J. Veenstra, Bordeaux, France; [100]), rabbit anti-DILP7 (1∶4000) (from I. Miguel-Aliaga, Cambridge, UK; [68]), mouse anti-PDF (1∶80) (C7; Developmental Studies Hybridoma Bank, University of Iowa), rabbit anti-PDH (1∶3000) ([101] from H. Dircksen, Stockholm, Sweden), guinea pig anti-DIMM (1∶2000) (from P. Taghert, St. Louis, MO; [102]), rabbit anti-FMRFamide (1∶4000) (117∶I, from C. Grimmelikhuijzen, Copenhagen, Denmark; [103]), rabbit anti-vGluT (1∶1000) (from H. Aberle, Münster, Germany [104]), rabbit anti-PICK1 (1∶500) (from O. Kjaerulff [74]), rabbit anti-serotonin (1∶1000) (s5545, Sigma), mouse anti-serotonin (1∶150) (Clone 5HT-H209; Dako, Copenhagen, Denmark), rabbit anti-horseradish peroxidase (1∶500) (anti-HRP, # 323-005-021, Jackson ImmunoResearch), rabbit anti-Rheb (1∶1000) (Abcam, Cambridge, UK), rabbit anti-phosphorylated AKT (1∶500) (pAKT; D9E) from Cell Signaling Technology (see [21]), rabbit and mouse anti-GFP (1∶1000) (Invitrogen, Carlsbad, CA), mouse anti-Repo (1∶10) (8D12; Developmental Studies Hybridoma Bank, University of Iowa).
For detection of primary antisera the following secondary antisera were used: goat anti-rabbit Alexa 546 antiserum, goat anti-rabbit Alexa 488 antiserum, goat anti-mouse Alexa 488 antiserum (all from Invitrogen), Cy3-tagged goat anti-rat antiserum and Cy3-tagged goat anti-guinea pig antiserum (Jackson ImmunoResearch, West Grove, PA) were all used at a dilution of 1∶1000.
Rhodamine-phalloidin (Invitrogen) was used at a dilution of 1∶500 to stain muscle. For insulin binding to tissue 1 mg Insulin-FITC (Sigma) was dissolved into 1 ml milli-Q water and used at a dilution of 1∶1000 in 0.01 M PBS. DNA was visualized with 4′,6-Diamidine-2-phenylindole (DAPI; Sigma) at 1∶2000.
Experiments on effect of diet
For nutrient restriction, eggs of different genotypes collected for 12 h were placed on a diet of 20% sucrose and 5% dry yeast in 2% aqueous agar and second instar larvae (48 h old) were transferred into medium containing 20% sucrose only (see Fig. S15A). The larvae were dissected at the wandering third instar stage for immunocytochemistry. For protein-rich conditions, eggs of different genotypes collected for 12 h were reared on food containing 20% sucrose and 20% yeast, and wandering third instar larvae were collected as above. As a control, flies were reared on food containing 20% sucrose and 5% dry yeast.
Image analysis
Specimens were imaged with Zeiss LSM 510 META and Zeiss LSM 780 confocal microscopes (Jena, Germany) using 20×, 40× oil or 63× oil immersion objectives. Confocal images were processed with Zeiss LSM software for either projection of z-stacks or single optical sections. Images were edited for contrast and brightness in Adobe Photoshop CS3 Extended version 10.0.
For quantification of immunofluorescence, immunostainings of tissues from different genotypes were carried out in glass vials with same amount of antibodies and all other conditions constant. Confocal images of neurons from different genotypes were obtained with identical laser intensity and scan settings. Immunofluorescence intensity in both cell bodies and tissue background was quantified in a set of regions of interest using Image J 1.40 from NIH, Bethesda, Maryland, USA (http://rsb.info.nih.gov/ij/). Mean fluorescence of projections of each cell body and image background were measured and the final immunofluorescence intensity in cell bodies was determined by subtracting the intensity of the tissue background. Total fluorescence of cell bodies (or nuclei) was calculated as mean fluorescence multiplied with cell size (or nuclear size) in some cases. These are specified in the figure legends of Fig. 9G, Fig. S1D, Fig. S4D, and Fig. S14E. For each genotype neurons in 5–15 brains were measured. The data were analyzed in Prism GraphPad 6.0, with Student's t-test. All data are presented as mean values ± SEM.
For CNS and cell size determination, the outline of CNS or cell body was extracted manually using Image J and its area determined. For each genotype CNS or neurons of 5–15 male flies from 3 independent crosses were measured. For cell size quantification in starved and re-fed flies, male w1118 flies were exposed to dry starvation for 18 h and then re-fed with 0.5% aqueous agarose gel for 2 h. For each treatment 7–10 flies were measured. For determination of axon and arborization size in larval body wall muscle, staining of a region of interest (Muscle 8 for anti-LK staining, and Muscle 12–13 for anti-HRP staining) in the 3rd segment was imaged and then quantified by outlining of each axon using Image J. For each genotype neurons in at least 6 body walls were measured.
Images of male 3 d old male flies were obtained with a Leica EZ4HD light microscope (Wetzlar, Germany) and sizes of whole body, wing and abdomen were outlined using Image J and hence quantified. 19–20 flies of each genotype from 3 crosses are measured.
Functional assays for ABLK neuron manipulations
The capillary feeding (CAFE) assay was conducted according to Ja and others [105] with slight changes. Male flies were placed into 1.5 ml Eppendorf tubes with an inserted 5 µl capillary tube with 5% sucrose, 2% yeast extract and 0.1% propionic acid. Three control food-filled capillaries were inserted in identical tubes without flies. The final consumption of food was determined as the diminished food level minus the average diminishment in control capillaries (due to evaporation). Daily consumption was measured every 24 h and calculated cumulatively over 4 consecutive days. These experiments were run in three replicates with 10 flies of each genotype for each replicate.
For desiccation (dry starvation) experiments, male flies were kept in an incubator at 25°C with 12∶12 h light∶dark (LD) conditions and controlled humidity. Flies aged 4–6 d were exposed to desiccation in bottles with neither food nor water. The number of dead flies was monitored every 12 h. These experiments were run in three replicates with at least 30 flies of each genotype per replicate.
Supporting Information
Zdroje
1. ChanYH, MarshallWF (2010) Scaling properties of cell and organelle size. Organogenesis 6 : 88–96.
2. BasergaR (2007) Is cell size important? Cell cycle 6 : 814–816.
3. SaucedoLJ, EdgarBA (2002) Why size matters: altering cell size. Curr Opinion Genetics Dev 12 : 565–571.
4. ThompsonBJ (2010) Developmental control of cell growth and division in Drosophila. Curr Opinion Cell Biol 22 : 788–794.
5. D'ErcoleAJ, YeP (2008) Expanding the mind: insulin-like growth factor I and brain development. Endocrinology 149 : 5958–5962.
6. GrewalSS (2009) Insulin/TOR signaling in growth and homeostasis: a view from the fly world. Int J Biochem Cell Biol 41 : 1006–1010.
7. BrogioloW, StockerH, IkeyaT, RintelenF, FernandezR, et al. (2001) An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol 11 : 213–221.
8. GarofaloRS, RosenOM (1989) Insulin and insulinlike growth factor 1 (IGF-1) receptors during central nervous system development: expression of two immunologically distinct IGF-1 receptor beta subunits. Mol Cell Biol 9 : 2806–2817.
9. ChenC, JackJ, GarofaloRS (1996) The Drosophila insulin receptor is required for normal growth. Endocrinology 137 : 846–856.
10. FernandezAM, Torres-AlemanI (2012) The many faces of insulin-like peptide signalling in the brain. Nat Rev Neurosci 13 : 225–239.
11. GoberdhanDC, WilsonC (2003) The functions of insulin signaling: size isn't everything, even in Drosophila. Differentiation 71 : 375–397.
12. AndersonMF, AbergMA, NilssonM, ErikssonPS (2002) Insulin-like growth factor-I and neurogenesis in the adult mammalian brain. Brain research Developmental brain research 134 : 115–122.
13. PatelPH, ThaparN, GuoL, MartinezM, MarisJ, et al. (2003) Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J Cell Sci 116 : 3601–3610.
14. BrittonJS, LockwoodWK, LiL, CohenSM, EdgarBA (2002) Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell 2 : 239–249.
15. OldhamS, HafenE (2003) Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol 13 : 79–85.
16. MirthCK, ShingletonAW (2012) Integrating body and organ size in Drosophila: recent advances and outstanding problems. Front Endocrinol 3 : 49.
17. ChengLY, BaileyAP, LeeversSJ, RaganTJ, DriscollPC, et al. (2011) Anaplastic lymphoma kinase spares organ growth during nutrient restriction in Drosophila. Cell 146 : 435–447.
18. MorrisonSJ, SpradlingAC (2008) Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132 : 598–611.
19. MooreKA, LemischkaIR (2006) Stem cells and their niches. Science 311 : 1880–1885.
20. CollerHA, SangL, RobertsJM (2006) A new description of cellular quiescence. PLoS biology 4: e83.
21. ChellJM, BrandAH (2010) Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell 143 : 1161–1173.
22. BrittonJS, EdgarBA (1998) Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125 : 2149–2158.
23. Sousa-NunesR, YeeLL, GouldAP (2011) Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature 471 : 508–512.
24. RajanA, PerrimonN (2011) Drosophila as a model for interorgan communication: lessons from studies on energy homeostasis. Dev Cell 21 : 29–31.
25. MillsJC, TaghertPH (2012) Scaling factors: transcription factors regulating subcellular domains. BioEssays : news and reviews in molecular, cellular and developmental biology 34 : 10–16.
26. ParkD, HadzicT, YinP, RuschJ, AbruzziK, et al. (2011) Molecular organization of Drosophila neuroendocrine cells by Dimmed. Curr Biol 21 : 1515–1524.
27. YamagishiM, ItoE, MatsuoR (2011) DNA endoreplication in the brain neurons during body growth of an adult slug. J Neurosci 31 : 5596–5604.
28. HewesRS, ParkD, GauthierSA, SchaeferAM, TaghertPH (2003) The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development 130 : 1771–1781.
29. ParkD, VeenstraJA, ParkJH, TaghertPH (2008) Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS ONE 3: e1896.
30. HamanakaY, ParkD, YinP, AnnangudiSP, EdwardsTN, et al. (2010) Transcriptional orchestration of the regulated secretory pathway in neurons by the bHLH protein DIMM. Curr Biol 20 : 9–18.
31. GarofaloRS, RosenOM (1988) Tissue localization of Drosophila melanogaster insulin receptor transcripts during development. Mol Cell Biol 8 : 1638–1647.
32. FernandezR, TabariniD, AzpiazuN, FraschM, SchlessingerJ (1995) The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J 14 : 3373–3384.
33. SongJ, WuL, ChenZ, KohanskiRA, PickL (2003) Axons guided by insulin receptor in Drosophila visual system. Science 300 : 502–505.
34. KimB, FeldmanEL (2012) Insulin resistance in the nervous system. Trends Endocrinol Metab 23 : 133–141.
35. RootCM, KoKI, JafariA, WangJW (2011) Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 145 : 133–144.
36. McLeodCJ, WangL, WongC, JonesDL (2010) Stem cell dynamics in response to nutrient availability. Current biology : CB 20 : 2100–2105.
37. CanteraR, NässelDR (1992) Segmental peptidergic innervation of abdominal targets in larval and adult dipteran insects revealed with an antiserum against leucokinin I. Cell Tissue Res 269 : 459–471.
38. de HaroM, Al-RamahiI, Benito-SiposJ, Lopez-AriasB, DoradoB, et al. (2010) Detailed analysis of leucokinin-expressing neurons and their candidate functions in the Drosophila nervous system. Cell Tissue Res 339 : 321–336.
39. Estacio-GomezA, Moris-SanzM, SchaferAK, PereaD, HerreroP, et al. (2013) Bithorax-complex genes sculpt the pattern of leucokinergic neurons in the Drosophila central nervous system. Development 140 : 2139–2148.
40. NässelDR, CanteraR, KarlssonA (1992) Neurons in the cockroach nervous system reacting with antisera to the neuropeptide leucokinin I. J Comp Neurol 322 : 45–67.
41. TangHY, Smith-CaldasMS, DriscollMV, SalhadarS, ShingletonAW (2011) FOXO regulates organ-specific phenotypic plasticity in Drosophila. PLoS genetics 7: e1002373.
42. ZhangW, ThompsonBJ, HietakangasV, CohenSM (2011) MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila. PLoS genetics 7: e1002429.
43. WuQ, ZhangY, XuJ, ShenP (2005) Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc Natl Acad Sci U S A 102 : 13289–13294.
44. BrownHL, KaunKR, EdgarBA (2012) The small GTPase Rheb affects central brain neuronal morphology and memory formation in Drosophila. PLoS ONE 7: e44888.
45. Martin-PenaA, AcebesA, RodriguezJR, SorribesA, de PolaviejaGG, et al. (2006) Age-independent synaptogenesis by phosphoinositide 3 kinase. J Neurosci 26 : 10199–10208.
46. DongJ, PanD (2004) Tsc2 is not a critical target of Akt during normal Drosophila development. Genes Dev 18 : 2479–2484.
47. SaucedoLJ, GaoX, ChiarelliDA, LiL, PanD, et al. (2003) Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 5 : 566–571.
48. ZhangY, GaoX, SaucedoLJ, RuB, EdgarBA, et al. (2003) Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 5 : 578–581.
49. ColombaniJ, BianchiniL, LayalleS, PondevilleE, Dauphin-VillemantC, et al. (2005) Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310 : 667–670.
50. Lopez-AriasB, DoradoB, HerreroP (2011) Blockade of the release of the neuropeptide leucokinin to determine its possible functions in fly behavior: chemoreception assays. Peptides 32 : 545–552.
51. TerhzazS, O'ConnellFC, PollockVP, KeanL, DaviesSA, et al. (1999) Isolation and characterization of a leucokinin-like peptide of Drosophila melanogaster. J Exp Biol 202 : 3667–3676.
52. RadfordJC, DaviesSA, DowJA (2002) Systematic G-protein-coupled receptor analysis in Drosophila melanogaster identifies a leucokinin receptor with novel roles. J Biol Chem 277 : 38810–38817.
53. Al-AnziB, ArmandE, NagameiP, OlszewskiM, SapinV, et al. (2010) The leucokinin pathway and its neurons regulate meal size in Drosophila. Curr Biol 20 : 969–978.
54. NitabachMN, BlauJ, HolmesTC (2002) Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109 : 485–495.
55. HongSH, LeeKS, KwakSJ, KimAK, BaiH, et al. (2012) Minibrain/Dyrk1a regulates food intake through the Sir2-FOXO-sNPF/NPY pathway in Drosophila and mammals. PLoS genetics 8: e1002857.
56. SpradlingAC, NystulT, LighthouseD, MorrisL, FoxD, et al. (2008) Stem cells and their niches: integrated units that maintain Drosophila tissues. Cold Spring Harbor symposia on quantitative biology 73 : 49–57.
57. LandgrafM, ThorS (2006) Development and structure of motoneurons. Int Rev Neurobiol 75 : 33–53.
58. AberleH, HaghighiAP, FetterRD, McCabeBD, MagalhaesTR, et al. (2002) wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron 33 : 545–558.
59. SanyalS (2009) Genomic mapping and expression patterns of C380, OK6 and D42 enhancer trap lines in the larval nervous system of Drosophila. Gene expression patterns : GEP 9 : 371–380.
60. DanielsRW, CollinsCA, GelfandMV, DantJ, BrooksES, et al. (2004) Increased expression of the Drosophila vesicular glutamate transporter leads to excess glutamate release and a compensatory decrease in quantal content. J Neurosci 24 : 10466–10474.
61. AlekseyenkoOV, LeeC, KravitzEA (2010) Targeted manipulation of serotonergic neurotransmission affects the escalation of aggression in adult male Drosophila melanogaster. PLoS ONE 5: e10806.
62. KnoxS, GeH, DimitroffBD, RenY, HoweKA, et al. (2007) Mechanisms of TSC-mediated control of synapse assembly and axon guidance. PLoS ONE 2: e375.
63. AcebesA, DevaudJM, ArnesM, FerrusA (2012) Central adaptation to odorants depends on PI3K levels in local interneurons of the antennal lobe. J Neurosci 32 : 417–422.
64. ParkD, TaghertPH (2009) Peptidergic neurosecretory cells in insects: organization and control by the bHLH protein DIMMED. Gen Comp Endocrinol 162 : 2–7.
65. HewesRS, GuT, BrewsterJA, QuC, ZhaoT (2006) Regulation of secretory protein expression in mature cells by DIMM, a basic helix-loop-helix neuroendocrine differentiation factor. J Neurosci 26 : 7860–7869.
66. WhiteK, HurteauT, PunsalP (1986) Neuropeptide-FMRFamide-like immunoreactivity in Drosophila: development and distribution. J Comp Neurol 247 : 430–438.
67. GrönkeS, ClarkeDF, BroughtonS, AndrewsTD, PartridgeL (2010) Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet 6: e1000857.
68. Miguel-AliagaI, ThorS, GouldAP (2008) Postmitotic specification of Drosophila insulinergic neurons from pioneer neurons. PLoS Biol 6: e58.
69. McBrayerZ, OnoH, ShimellM, ParvyJP, BecksteadRB, et al. (2007) Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell 13 : 857–871.
70. AcebesA, MoralesM (2012) At a PI3K crossroads: lessons from flies and rodents. Reviews in the neurosciences 23 : 29–37.
71. SlaidinaM, DelanoueR, GrönkeS, PartridgeL, LeopoldP (2009) A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev Cell 17 : 874–884.
72. ColombaniJ, AndersenDS, LeopoldP (2012) Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336 : 582–585.
73. GarelliA, GontijoAM, MiguelaV, CaparrosE, DominguezM (2012) Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 336 : 579–582.
74. JansenAM, NässelDR, MadsenKL, JungAG, GetherU, et al. (2009) PICK1 expression in the Drosophila central nervous system primarily occurs in the neuroendocrine system. J Comp Neurol 517 : 313–332.
75. HolstB, MadsenKL, JansenAM, JinC, RickhagM, et al. (2013) PICK1 Deficiency Impairs Secretory Vesicle Biogenesis and Leads to Growth Retardation and Decreased Glucose Tolerance. PLoS biology 11: e1001542.
76. GilletteR (1991) On the significance of neuronal gigantism in gastropods. Biol Bull 180 : 234–240.
77. MandrioliM, MolaL, CuoghiB, SonettiD (2010) Endoreplication: a molecular trick during animal neuron evolution. The Quarterly review of biology 85 : 159–169.
78. DemontisF, PerrimonN (2009) Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136 : 983–993.
79. UnhavaithayaY, Orr-WeaverTL (2012) Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev 26 : 31–36.
80. EdensLJ, WhiteKH, JevticP, LiX, LevyDL (2013) Nuclear size regulation: from single cells to development and disease. Trends Cell Biol 23 : 151–159.
81. BrandtA, PapagiannouliF, WagnerN, Wilsch-BrauningerM, BraunM, et al. (2006) Developmental control of nuclear size and shape by Kugelkern and Kurzkern. Curr Biol 16 : 543–552.
82. ChiuSL, ClineHT (2010) Insulin receptor signaling in the development of neuronal structure and function. Neural development 5 : 7.
83. LiuW, YeP, O'KuskyJR, D'ErcoleAJ (2009) Type 1 insulin-like growth factor receptor signaling is essential for the development of the hippocampal formation and dentate gyrus. Journal of neuroscience research 87 : 2821–2832.
84. OkamotoN, YamanakaN, YagiY, NishidaY, KataokaH, et al. (2009) A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev Cell 17 : 885–891.
85. LanetE, GouldAP, MaurangeC (2013) Protection of neuronal diversity at the expense of neuronal numbers during nutrient restriction in the Drosophila visual system. Cell reports 3 : 587–594.
86. ZhaoXL, CamposAR (2012) Insulin signalling in mushroom body neurons regulates feeding behaviour in Drosophila larvae. J Exp Biol 215 : 2696–2702.
87. PierceSB, YostC, BrittonJS, LooLW, FlynnEM, et al. (2004) dMyc is required for larval growth and endoreplication in Drosophila. Development 131 : 2317–2327.
88. SalleJ, CampbellSD, GhoM, AudibertA (2012) CycA is involved in the control of endoreplication dynamics in the Drosophila bristle lineage. Development 139 : 547–557.
89. RulifsonEJ, KimSK, NusseR (2002) Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296 : 1118–1120.
90. YangCH, BelawatP, HafenE, JanLY, JanYN (2008) Drosophila egg-laying site selection as a system to study simple decision-making processes. Science 319 : 1679–1683.
91. ParkJH, Helfrich-ForsterC, LeeG, LiuL, RosbashM, et al. (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 97 : 3608–3613.
92. BaiH, KangP, TatarM (2012) Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell 11 : 978–985.
93. LeeversSJ, WeinkoveD, MacDougallLK, HafenE, WaterfieldMD (1996) The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J 15 : 6584–6594.
94. AcebesA, Martin-PenaA, ChevalierV, FerrusA (2011) Synapse loss in olfactory local interneurons modifies perception. J Neurosci 31 : 2734–2745.
95. BelgacemYH, MartinJR (2007) Hmgcr in the corpus allatum controls sexual dimorphism of locomotor activity and body size via the insulin pathway in Drosophila. PLoS ONE 2: e187.
96. ParkD, ShaferOT, ShepherdSP, SuhH, TriggJS, et al. (2008) The Drosophila basic helix-loop-helix protein DIMMED directly activates PHM, a gene encoding a neuropeptide-amidating enzyme. Molecular and cellular biology 28 : 410–421.
97. EnglundC, LorenCE, GrabbeC, VarshneyGK, DeleuilF, et al. (2003) Jeb signals through the Alk receptor tyrosine kinase to drive visceral muscle fusion. Nature 425 : 512–516.
98. BazigouE, ApitzH, JohanssonJ, LorenCE, HirstEM, et al. (2007) Anterograde Jelly belly and Alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell 128 : 961–975.
99. HerreroP, MagarinosM, TorrojaL, CanalI (2003) Neurosecretory identity conferred by the apterous gene: lateral horn leucokinin neurons in Drosophila. J Comp Neurol 457 : 123–132.
100. VeenstraJA, AgricolaHJ, SellamiA (2008) Regulatory peptides in fruit fly midgut. Cell Tissue Res 334 : 499–516.
101. HombergU, WürdenS, DircksenH, RaoKR (1991) Comparative anatomy of pigment-dispersing hormone-immunoreactive neurons in the brain of orthopteroid insects. Cell Tissue Res 266 : 343–357.
102. AllanDW, ParkD, St PierreSE, TaghertPH, ThorS (2005) Regulators acting in combinatorial codes also act independently in single differentiating neurons. Neuron 45 : 689–700.
103. LundquistT, NässelDR (1990) Substance P-, FMRFamide-, and gastrin/cholecystokinin-like immunoreactive neurons in the thoraco-abdominal ganglia of the flies Drosophila and Calliphora. J Comp Neurol 294 : 161–178.
104. MahrA, AberleH (2006) The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene expression patterns : GEP 6 : 299–309.
105. JaWW, CarvalhoGB, MakEM, de la RosaNN, FangAY, et al. (2007) Prandiology of Drosophila and the CAFE assay. Proceedings of the National Academy of Sciences of the United States of America 104 : 8253–8256.
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



