-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
In response to insect attack and mechanical wounding, plants activate the expression of genes involved in various defense-related processes. A fascinating feature of these inducible defenses is their occurrence both locally at the wounding site and systemically in undamaged leaves throughout the plant. Wound-inducible proteinase inhibitors (PIs) in tomato (Solanum lycopersicum) provide an attractive model to understand the signal transduction events leading from localized injury to the systemic expression of defense-related genes. Among the identified intercellular molecules in regulating systemic wound response of tomato are the peptide signal systemin and the oxylipin signal jasmonic acid (JA). The systemin/JA signaling pathway provides a unique opportunity to investigate, in a single experimental system, the mechanism by which peptide and oxylipin signals interact to coordinate plant systemic immunity. Here we describe the characterization of the tomato suppressor of prosystemin-mediated responses8 (spr8) mutant, which was isolated as a suppressor of (pro)systemin-mediated signaling. spr8 plants exhibit a series of JA-dependent immune deficiencies, including the inability to express wound-responsive genes, abnormal development of glandular trichomes, and severely compromised resistance to cotton bollworm (Helicoverpa armigera) and Botrytis cinerea. Map-based cloning studies demonstrate that the spr8 mutant phenotype results from a point mutation in the catalytic domain of TomLoxD, a chloroplast-localized lipoxygenase involved in JA biosynthesis. We present evidence that overexpression of TomLoxD leads to elevated wound-induced JA biosynthesis, increased expression of wound-responsive genes and, therefore, enhanced resistance to insect herbivory attack and necrotrophic pathogen infection. These results indicate that TomLoxD is involved in wound-induced JA biosynthesis and highlight the application potential of this gene for crop protection against insects and pathogens.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1003964
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003964Summary
In response to insect attack and mechanical wounding, plants activate the expression of genes involved in various defense-related processes. A fascinating feature of these inducible defenses is their occurrence both locally at the wounding site and systemically in undamaged leaves throughout the plant. Wound-inducible proteinase inhibitors (PIs) in tomato (Solanum lycopersicum) provide an attractive model to understand the signal transduction events leading from localized injury to the systemic expression of defense-related genes. Among the identified intercellular molecules in regulating systemic wound response of tomato are the peptide signal systemin and the oxylipin signal jasmonic acid (JA). The systemin/JA signaling pathway provides a unique opportunity to investigate, in a single experimental system, the mechanism by which peptide and oxylipin signals interact to coordinate plant systemic immunity. Here we describe the characterization of the tomato suppressor of prosystemin-mediated responses8 (spr8) mutant, which was isolated as a suppressor of (pro)systemin-mediated signaling. spr8 plants exhibit a series of JA-dependent immune deficiencies, including the inability to express wound-responsive genes, abnormal development of glandular trichomes, and severely compromised resistance to cotton bollworm (Helicoverpa armigera) and Botrytis cinerea. Map-based cloning studies demonstrate that the spr8 mutant phenotype results from a point mutation in the catalytic domain of TomLoxD, a chloroplast-localized lipoxygenase involved in JA biosynthesis. We present evidence that overexpression of TomLoxD leads to elevated wound-induced JA biosynthesis, increased expression of wound-responsive genes and, therefore, enhanced resistance to insect herbivory attack and necrotrophic pathogen infection. These results indicate that TomLoxD is involved in wound-induced JA biosynthesis and highlight the application potential of this gene for crop protection against insects and pathogens.
Introduction
Higher plants respond to insect attack and wounding by activating the expression of genes involved in herbivore deterrence, wound healing, and other defense-related processes [1]–[7]. The wound response of tomato (Solanum lycopersicum) provides an attractive model to understand the signal transduction events leading from localized injury to the systemic expression of defense-related genes [7], [8]. The principle defensive markers used in these studies are genes encoding proteinase inhibitors (PIs), low molecular weight proteins that inhibit the activity of digestive enzymes in the gut of herbivores [1], [9]. In their milestone study of wound-inducible PIs in tomato, Green and Ryan proposed that specific signals generated at the wound site travel through the plant and activate the expression of PIs and other defense-related genes in remote responding leaves [10].
Systemin, an 18-amino-acid peptide signal, was purified from wounded tomato leaves on the basis of its ability to activate PI accumulation using a convenient bioassay for PI-inducing compounds [9], [11]–[13]. Systemin is derived from the cleavage of a larger precursor protein called prosystemin, which is encoded by a single copy of the Prosystemin (PS) gene [12], [14]. Transgenic tomato plants that express an antisense PS are defective in wound-induced systemic expression of PI genes and are more susceptible to insects [14]. Conversely, transgenic tomato plants (called 35S::PS) that overexpress PS constitutively express high levels of PIs without wounding and are more resistant to insects [15], [16]. In addition, genetic analysis in tomato has shown that genes required for (pro)systemin signaling are also essential for wound-induced expression of defensive genes [3], [17], [18]. Together, these genetic studies support that the peptide signal systemin acts as an upstream component of the wound-induced signaling cascades leading to defense gene expression.
It is generally believed that wounding and insect attack lead to the rapid cleavage of systemin from prosystemin. Binding of systemin to its proposed receptor on the cell surface then activates defense gene expression by increasing the endogenous levels of jasmonic acid (JA) and related pentacyclic oxylipins (collectively referred to here as JAs) that are derived from the linolenic acid via the octadecanoid pathway [1], [19]–[21]. A role for JAs in intercellular signaling is supported by the fact that application of MeJA (the methyl ester of JA) to one tomato leaf induces PI expression in distal untreated leaves [22]. JAs are now considered to be key regulators for stress-induced gene expression in virtually all plant species [1], [20], [23]–[27]. It was proposed that systemin and JA work together in the same signal transduction pathway to regulate the systemic expression of defense-related genes [1], [9], [20]. Thus, the systemin/JA signaling pathway for induced resistance in tomato provides a unique opportunity to investigate, in a single experimental system, the mechanism by which peptide and oxylipin signals interact to coordinate systemic expression of defense-related genes [7], [8].
We have been using a genetic approach to dissect the systemin/JA signaling pathway and to elucidate the role of systemin and JA in it. Genetic screen to identify mutations that suppress the constant wound signaling phenotype (i.e., constitutive expression of PIs and other defense-related genes) of 35S::PS plants has led to the identification of several important components of the systemin/JA signaling pathway 17,18,28,29. Significantly, several of the characterized spr (suppressors of prosystemin-mediated responses) mutants actually define genes that are directly involved in JA biosynthesis or signaling [17], [18], [29]. For example, Spr2 encodes a chloroplast fatty acid desaturase that catalyzes the ω3 desaturation of linoleic acid (18∶2) to linolenic acid (18∶3), the metabolic precursor for JA biosynthesis [18]. spr6, on the other hand, defines the tomato homolog of CORONATINE INSENSITIVE1 (COI1), which has been shown to be the JA receptor in Arabidopsis [29], [30]. These studies provided direct evidence that JA acts downstream of systemin in regulating wound-induced expression of defense-related genes.
Grafting experiments conducted with the JA biosynthesis mutant spr2 and the JA signaling mutant jai-1 revealed that systemic defense signaling requires both the biosynthesis of JA at the site of wounding and the ability to perceive JA in remote tissues, suggesting that JA acts as a systemic wound signal [3]. Grafting experiments also demonstrated that the graft-transmissible wound signal generated by the 35S::PS plants can be readily recognized by spr2 plants (which are insensitive to systemin), but cannot be recognized by the JA-insensitive jai-1 plants, strongly suggesting that the 35S::PS-derived wound signal is JA, rather than systemin [3]. These results challenge the previous paradigm that systemin is the long-distance mobile signal for wound-induced defense gene expression [8], [31], [32].
Genetic analyses of tomato wound response also provide insight to understand how the peptide signal systemin interacts with JA to promote systemic defense signaling. In contrast to other tomato wound response mutants that lack both local and systemic PI expression in response to wounding, spr1 plants were deficient mainly in the systemic response. Moreover, spr1 abolished JA accumulation in response to exogenous systemin, and showed reduced JA accumulation in wounded leaves [28] Analysis of reciprocal grafts between spr1 and wild-type (WT) plants showed that spr1 impedes systemic PI expression by blocking the production of the long-distance wound signal in damaged leaves, rather than inhibiting the recognition of that signal in systemic undamaged leaves. These experiments suggest that Spr1 is involved in a signaling step that couples systemin perception to the activation of the octadecanoid pathway [28]. These and other studies support that systemin acts locally at the site of wounding to amplify the production of JA, which in turn functions as a mobile signal to activate systemic defense responses [8], [28], [33]. In addition to systemin, the hydroxyproline-rich glycopeptides (HypSys peptides), which are isolated from tomato and tobacco leaves, are also powerful activators of PI expression [34]. Recent genetic data support that, similar to systemin, HypSys peptides also play a role in an amplification loop that upregulates JA production to effect strong systemic defense response [35].
Toward understanding the molecular mechanism of systemin/JA-mediated systemic defense signaling in tomato, we are conducting an enlarged genetic screen to identify more spr mutants that suppress the constitutive wound signaling phenotype of the 35S::PS plants [29]. Here we report the genetic and molecular characterization of spr8, a semidominant mutant that is defective in wound-induced expression of defense-related genes. Map-based cloning studies reveal that Spr8 encodes tomato lipoxygenase D (TomLoxD), a 13-lipoxygenase that catalyzes the hydroperoxidation of linolenic acid, a key step in JA biosynthesis [19]. We show that overexpression of TomLoxD leads to elevated wound-induced JA biosynthesis, increased expression of wound-responsive genes and, therefore, enhanced resistance to insects and necrotrophic pathogens. These results highlight the application potential of the TomLoxD gene for crop protection.
Results
spr8 Impairs Wound-Induced Expression of Defensive Genes
spr8 is one of the newly identified mutants that can block the constitutively high activity of polyphenol oxidase (PPO) in the 35S::PS plants [29]. Further characterization of spr8 was carried out using a spr8/spr8 homozygous line in which the 35S::PS transgene was removed by five successive backcrosses to the WT cv. Castlemart (CM). The overall plant morphology, flower development and pollen viability of spr8 plants were indistinguishable from those of WT plants (Figure S1). The wound response of spr8 was compared with that of WT using the classical radial immunodiffusion assay for the measurement of wound-induced accumulation of proteinase inhibitor II (PI-II) [11], [29], [36]. For these experiments, 16-day-old seedlings containing two fully expanded leaves were wounded and the accumulation of PI-II protein was quantified. Wounding the lower leaves of WT caused the well-known accumulation of PI-II both in the wounded leaves (local response) and in the upper unwounded leaves (systemic response) (Figure 1A). In contrast, spr8 seedlings accumulated no detectable PI-II protein in the wounded leaves and the upper unwounded leaves (Figure 1A). Consistent with the PI-II protein data, quantitative real-time PCR (qRT-PCR) assays indicated that the PI-II transcripts were expressed very weakly in wounded spr8 leaves as compared to those in WT leaves (Figure 1B). It has been shown that, similar to the PI genes [37], protein products of the tomato wound-responsive genes threonine deaminase (TD) [16] and leucine amino peptidase A (LapA) [38] have a direct role in deterring insect performance. Our parallel experiments indicated that the wound-induced expression levels of TD (Figure S2A) and LapA (Figure S2B) were also largely reduced in spr8 plants compared to those in WT plants. These results demonstrate that the spr8 mutation impairs wound-induced expression of defensive genes.
Fig. 1. spr8 impairs the wound-induced expression of PI-II. 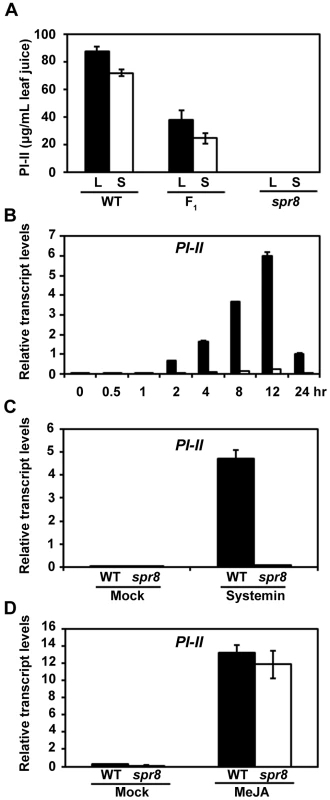
(A) PI-II protein accumulation in tomato leaves in response to mechanical wounding. Sixteen-day-old wild-type (WT), (WT×spr8) F1 (F1) and spr8 seedlings were wounded using a hemostat as described in Materials and Methods. Twenty-four hours after wounding, PI-II levels were measured in the wounded leaf (black bar; L, local response) and the upper unwounded leaf (white bar; S, systemic response). Values represent the mean ± SD of six plants. (B) Time-course of wound-induced expression of PI-II in WT and spr8 plants. Sixteen-day-old seedlings of WT (black bar) and spr8 (white bar) plants containing two fully expanded leaves were mechanically wounded with a hemostat on both leaves for indicated times before total RNAs were extracted for qRT-PCR assays. Data presented are mean values of three biological repeats with SD. (C) Expression of PI-II in WT (black bar) and spr8 (white bar) plants in response to exogenous systemin. Sixteen-day-old seedlings WT and spr8 seedlings were excised at the base of the stem and supplied with 15 mM sodium phosphate buffer (white bar), or buffer solution with 2.5 pmol systemin. PI-II transcription levels were measured 12 h after treatment. Data presented are mean values of three biological repeats with SD. (D) MeJA-induced PI-II expression in WT and spr8 plants. Sixteen-day-old seedlings of WT (black bar) and spr8 (white bar) plants were treated with MeJA for 12 hours before PI-II expression were quantified with qRT-PCR. Data presented are mean values of three biological repeats with SD. To gain additional insight into the wound response phenotype of spr8, we examined the capacity of the mutant to respond to various PI-inducing compounds. As previously reported [28], exogenous application of systemin led to strong expression of PI-II transcripts in WT plants (Figure 1C). But spr8 plants failed to express significant levels of PI-II transcripts in response to the same concentrations of systemin (Figure 1C), indicating that spr8 plants are insensitive to systemin. These results are consistent with the fact that spr8 was identified as a suppressor of prosystemin-mediated responses. We then examined the response of spr8 to the methyl ester of JA, MeJA, which is a potent elicitor of PI-II expression in WT plants (Figure 1D). As shown in Figure 1D, exogenous application of MeJA readily restored the PI-II expression of spr8 mutants to levels comparable to those of WT plants. These results led us to classify spr8 into the group of wounding/systemin-insensitive, but JA-sensitive mutants. It is most likely that the spr8 mutant defines a signaling step that couples the perception of systemin to activation of the JA pathway.
spr8 Affects Glandular Trichome Development
Trichome density and volatile emissions of glandular trichomes provide a formidable protective barrier to invasion by herbivores and pathogens [39]–[41]. Cultivated tomato contains two morphologically distinct types of glandular trichomes. Type I trichomes have an elongated multicellular stalk with a small unicellular vesicle at the tip (Figure 2A and 2B). Type VI trichomes have a unicellular stalk with a four-celled glandular head (Figure 2A and 2B) [42], [43]. In order to determine whether spr8 affects trichome development, we used scanning electron microscopy to observe the adaxial leaf surface to compare trichome morphology and density between WT and spr8 plants. A striking feature of spr8 leaves is the significant reduction of trichome number of both types (Figure 2A and 2B). Quantification of trichomes of five-week-old WT plants (containing at least five leaves) showed that the density of type VI trichome was ∼10 trichomes/mm2 on the base region of the third leaflet. Analysis of comparable spr8 leaflets showed that, type VI trichome density of the mutant was about 70% of that of WT leaflets (Figure 2C).
Fig. 2. spr8 impairs trichome development and exhibits defect in type VI glandular trichome exudates. 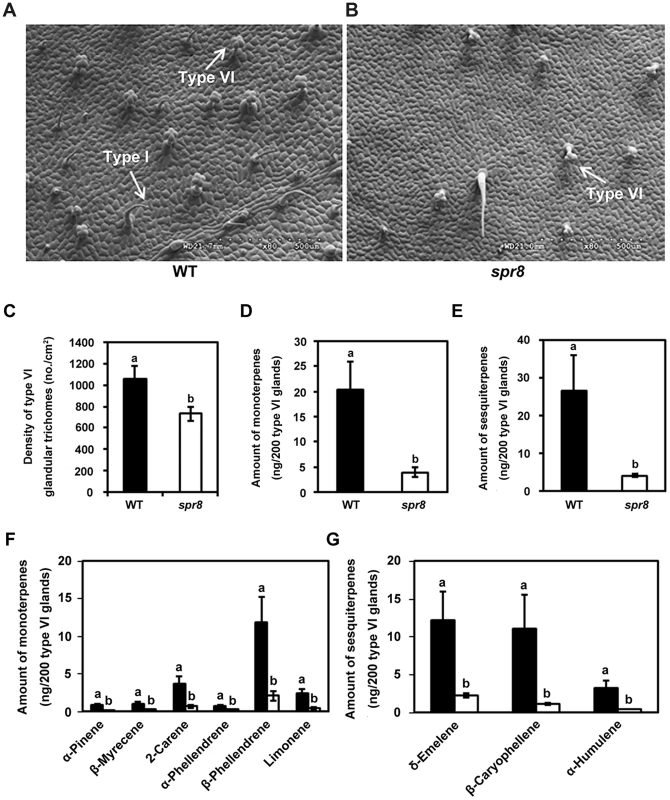
(A) and (B) Scanning electron micrographs of the adaxial surface of a leaflet from WT (A) and spr8 (B) plants. Five-week-old plants were used for all images. The type I and type VI glandular trichomes were indicated using white arrows, respectively. (C) Mean density (no. per cm2 ± SD) of type VI glandular trichomes on the leaflets adaxial surface of WT (black bar) and spr8 (white bar) plants (n = 10). Samples with the different letters are significantly different at P<0.01 between WT and spr8. (D–G) Monoterpene and sesquiterpene content of the type VI glandular trichomes from the adaxial surface of WT (black bar) and spr8 (white bar) plants leaves. Data presented are mean values of six biological repeats with SD. Samples with the different letters are significantly different at P<0.01 between WT and spr8. (D) and (E) Total contents of monoterpene (D) and sesquiterpene (E) of the type VI glandular trichome exudates from WT (black bar) and spr8 (white bar) leaves. (F) and (G) Comparison of monoterpene (F) and sesquiterpene (G) levels extracted from WT (black bar) and spr8 (white bar) leaves. Next, we used gas chromatography analysis to determine whether spr8 affects the production of compounds that are synthesized in trichome glands. For these experiments, type VI glandular trichomes were selectively collected by using a stretched-glass pipette and were extracted with methyl tert-butyl ether (MTBE) (see method). Trichome exudates were then analyzed by gas chromatography to measure the terpene composition. From 1, 000 type VI glands collected from the adaxial surface of WT leaves, six monoterpenes (α-pinene, β-myrecene, 2-carene, α-phellandrene, β-phellandrene and limonene; Figure 2F) and three sesquiterpenes (δ-elemene, β-caryophyllene, and α-humulene; Figure 2G) were identified. Comparison of terpene profiles revealed that, all of these compounds were also detected in exudates from the same number of type VI glandular trichomes of spr8 leaflets, but their accumulation levels were significantly decreased in the mutant (Figure 2F and 2G). In spr8 glandular trichomes, the accumulation levels of total monoterpenes and sesquiterpenes were reduced to 19.5% and 15.2%, respectively, of those of their WT counterparts (Figure 2D and 2E). These results support the hypothesis that the spr8 mutation affects the terpene metabolic pathway that mainly operates in type VI trichome glands.
spr8 Plants Are Compromised in Resistance against Chewing Insects
The inability of spr8 plants to express significant levels of defensive genes in response to mechanical wounding and systemin (Figure 1 and Figure S2) suggests that this mutant is compromised in resistance to herbivorous insects. To test this hypothesis, newly hatched cotton bollworm (Helicoverpa armigera) larvae were placed on leaves of 5-week-old plants to initiate a feeding trial. Sustaining long-term feeding by insects, spr8 plants were severely damaged (Figure 3A, right), while WT plants showed relatively few signs of macroscopic damage during the period of the feeding trial (Figure 3A, left). After termination of the feeding trial, PI-II protein accumulation in the remaining leaf tissues was measured, as was the weight gain of larvae reared on both of the host genotypes. In contrast with high levels of PI-II accumulation in herbivore-damaged WT leaves, very little or no PI-II protein accumulation was detected in hornworm-challenged spr8 plants (Figure 3B). These results indicate that WT plants have relatively high levels of natural resistance to the cotton bollworm larvae and that this resistance is severely compromised by the spr8 mutation. Consistently, the average weight of larvae reared on the mutant was 2.0-fold greater than that of larvae reared on WT plants (Figure 3C and 3D). These results demonstrate that Spr8 is required for the resistance of tomato plants to attacking hornworm larvae.
Fig. 3. spr8 plants show reduced resistance to cotton bollworm larvae (Helicoverpa armigera). 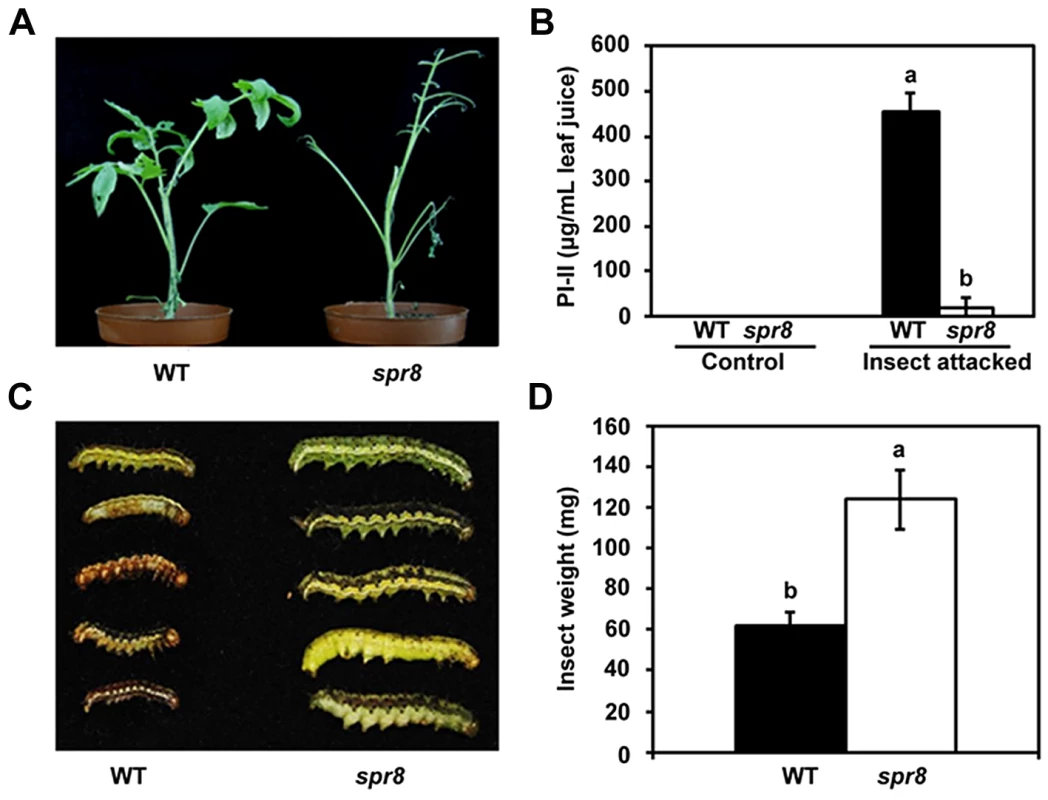
(A) Representative WT (left) and spr8 (right) plants at the end of cotton bollworm larvae feeding trial. (B) PI-II proteins accumulation in WT (black bar) and spr8 (white bar) leaves in response to cotton bollworm larvae feeding (n = 5). (C) Size of larvae recovered at the end of cotton bollworm feeding trial. (D) Larval weight recovered at the end of 7 d of feeding trial on whole plants of WT (black bar) and spr8 (white bar) (n = 15). In (B) and (D), data shown are the mean with SD. Bars with different letters are significantly different from each other (P = 0.05). The feeding trails on whole plants were repeated three times with similar results. In each experiment, 10 newly hatched larvae were placed on at least six five-week-old plants of each genotype. Larvae were allowed to feed on the same plant for the duration of the trial. The Wound-Response Phenotype of spr8 Results from a Defect in the TomLoxD Gene
Genetic analysis revealed that spr8 is a semi-dominant mutant, given that the wound-response phenotype of the heterozygous (Spr8/spr8) plants was intermediate between those of the homozygous spr8 plants and their WT counterparts (Figure 1A and Figure S3). The deficiency in wound-induced PI-II protein accumulation of spr8 provides a facile assay for map-based cloning studies to determine the genetic basis of this defect. A combination of cleaved amplified polymorphic sequence (CAPS) and simple sequence repeat (SSR) markers was used to localize Spr8 to a region on the long arm of chromosome 3 between SSR markers TES0023 and TES1203 (Figure 4A). Fine mapping using 354 backcrossed (BC1) individuals showing a WT wound response delimited the Spr8 locus to a region between the markers SSR601 and M140 in the scaffold SL2.40sc03701 of the sequenced tomato genome [44], [45]. Among the genes predicted by the International Tomato Annotation Group (ITAG2.3 release, http://solgenomics.net) in this interval, Solyc03g122340, which encodes TomLoxD (tomato lipoxygenase D), a wound-inducible lipoxygenase [46], is considered to be a strong candidate of Spr8. DNA sequencing revealed that spr8-derived TomLoxD complementary DNA (cDNA) contains a single C-to-T mutation (Figure 4B). This C-to-T mutation, which was confirmed by sequencing of PCR-amplified genomic DNA from spr8 plants, destroys a BamHI restriction site, and a CAPS marker was developed to detect the spr8 mutant allele (Figure 4C). The single base pair change in the TomLoxD gene is predicted to replace a highly conserved (i.e., invariant among plant and animal lipoxygenases) Pro residue at position 598 with an Leu (Figure 4D and Figure S4).
Fig. 4. Map-based cloning of Spr8. 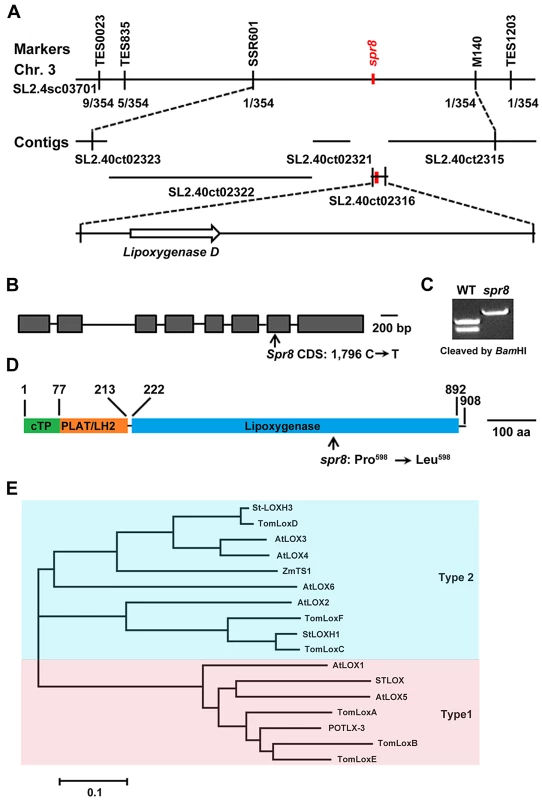
(A) Fine genetic and physical mapping of Spr8. Numbers below the line indicate the number of recombination events identified between markers. Placement of Spr8 on SL2.40ct02316 was determined by the phenotypic data. (B) Gene structure of Spr8/TomLoxD. Introns and exons are indicated by horizontal lines and closed boxes, respectively, and are drawn to scale. Arrow indicates the point mutation site of spr8, which is the BamHI recognition site GGATCC. Bar = 100 bp. (C) Development of a CAPS marker to distinguish spr8 mutants from WT plants. Parts of the TomLoxD gene were amplified from genomic DNAs of both WT and spr8 alleles using the PCR primer pair P1 (5′-TTTCCAATGTCAGTATATAACTC-3′) and P2 (5′-CCATTTCTCGATCGGATCAATG -3′). BamHI cleaved the 680 bp DNA fragment at the recognition site GGATCC in 229 bp and 451 bp, but not from the spr8 mutant, in which the recognition site was altered to GGATCT by the mutation. (D) The TomLoxD protein contains a predicted chloroplast transit peptide (cTP, green), the PLAT/LH2 beta-barrel (orange), and the lipoxygenase domain (blue). Arrow indicates the point mutation site of Spr8 protein, at which Pro598 changes to Leu598. Bar = 100 amino acid (aa). (E) Phylogenetic tree of various lipoxygenases from tomato, potato, Arabidopsis and maize. Shown is the sequence relatedness between the deduced amino acid sequence of TomLoxD and other lipoxygenases. Considering that spr8 is a semi-dominant mutation, we performed the following experiments to show that the missense mutation in TomLoxD accounts for the wound response phenotype of spr8. First, transgenic plants overexpressing a WT allele of TomLoxD (TomLoxD-OE) showed increased wound response in term of wound-induced defense gene expression (See below). Second, similar to spr8 plants, transgenic plants expressing a TomLoxD RNA interference (RNAi) construct (TomLoxD-RNAi) were defective in wound-induced expression of PI-II (Figure S5A and S5B). Third, the wound response phenotype of transgenic plants overexpressing a mutant allele of TomLoxD (TomLoxDP598L-OE) was intermediate between that of the homozygous spr8 plants and their WT counterparts (Figure S5A and S5B). Finally, overexpression of a WT allele of TomLoxD in the spr8 background failed to fully rescue the wound response defects of the mutant (Figure S5C and S5D). Collectively, these results support that the identified C-to-T mutation in the TomLoxD gene is responsible for the wound response phenotype of spr8 plants and that the spr8 allele of TomLoxD (i.e., TomLoxDP598L) acts as a dominant negative regulator of the tomato wound response pathway.
Lipoxygenases are nonheme iron-containing fatty acid dioxygenases that catalyze the peroxidation of polyunsaturated fatty acids such as linoleic acid, α-linolenic acid, and arachidonic acid [47]. Based on the positional specificity of linoleic acid oxygenation, they are classified as 9-lipoxygenases (oxygenation occurs at carbon 9 of the hydrocarbon backbone) and 13-lipoxygenases (oxygenation occurs at carbon 13 of the hydrocarbon backbone). 13-lipoxygenases can be further divided as types 1 and 2 based on the presence of a putative chloroplast transit peptide (cTP) [47]. ChloroP (http://www.cbs.dtu.dk/services/TargetP/)-based analysis predicted that the deduced amino acid sequence of TomLoxD contains a putative cTP (TomLoxD1–77), a small N-terminal PLAT/LH2 domain (TomLoxD78–213) that forms a β-barrel, and a C-terminal domain (TomLoxD222–892) that forms α-helices (Figure 4D). It is generally believed that the N-terminal β-barrel domain is involved in membrane or substrate binding, whereas the C-terminal domain harbors the catalytic site of the enzyme [48]. This primary protein structure suggests that TomLoxD is a member of the type 2 plastid-localized 13-lipoxygenases [47]. This prediction is supported by our phylogenetic analysis of plant lipoxygenases, which places TomLoxD in a clade including functionally characterized and predicted type 2 13-lipoxygenases (Figure 4E). To confirm the chloroplast localization of the TomLoxD protein, full-length of the TomLoxD cDNA was fused to the green fluorescent protein (GFP) reporter gene and subsequently transformed into Arabidopsis leaf protoplast cells. As shown in Figure S6, the GFP fluorescence was co-localized with the red chlorophyll autofluorescence, suggesting that TomLoxD is a chloroplast-localized protein. Notably, in our phylogenetic analysis, TomLoxD was most similar to the Arabidopsis LOX3 and LOX4 (71.7% and 71.3% amino acid identity, respectively) (Figure 4E), which has recently been shown to be type 2 chloroplast-localized 13-lipoxygenases that are involved in JA biosynthesis [49]. It is noteworthy that the TomLoxDP598L mutation in spr8 occurs in the C-terminal α-helices domain, presumably impairs the catalytic activity of the enzyme (Figure 4D).
The spr8 Mutation Impairs Wound-Induced JA Biosynthesis
The above-described results point to a possibility that TomLoxD is a functional 13-lipoxygenase involved in wound-induced JA biosynthesis and that the spr8 allele of TomLoxD (hereafter referred to as TomLoxDP598L) impairs wound-induced JA biosynthesis. As the first step to prove this, we examined the expression of TomLoxD or TomLoxDP598L in response to wounding. Consistent with a previous investigation [46], the levels of TomLoxD transcripts were induced by wounding within 30 min and peaked at 1 h after wounding, TomLoxD transcripts then showed a tendency of decline and returned to control levels within 8 h (Figure 5A), indicating that TomLoxD is an early wound-inducible gene. Interestingly, the wound-induced expression kinetics of TomLoxDP598L was essentially similar to that of TomLoxD, albeit its expression levels were somehow reduced as compared to that of the latter (Figure 5A). These results indicate that TomLoxDP598L is still responsive to wounding.
Fig. 5. spr8 impairs wound-induced JA biosynthesis. 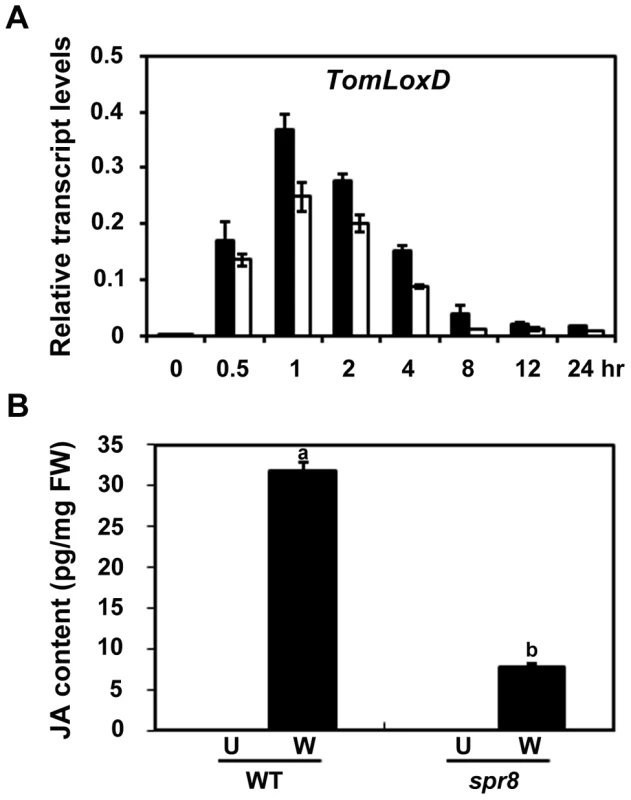
(A) Time-course transcript levels of TomLoxD in response to mechanical wounding. Sixteen-day-old seedlings of WT (black bar) and spr8 (white bar) plants were mechanically wounded for indicated times before total RNAs were extracted for qRT-PCR assays. Data presented are mean values of three biological repeats with SD. (B) JA levels in response to wounding. WT and spr8 plants (16-day-old) were mechanically wounded as described above, and JA levels were measured 1 h after wounding (W, Wounded; black bar). JA was also extracted from leaves of unwounded plants (U, Unwounded; white bar). Data show the mean ± SD of three independent samples and are indicative of three independent experiments. Bars with different letters are significantly different compared spr8 mutant plants with WT plants (P = 0.01). FW, fresh weight. To determine the contribution of TomLoxD and TomLoxDP598L in wound-induced JA biosynthesis, we used liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) to measure endogenous JA levels in WT and spr8 plants in response to wounding. We consistently observed that the JA levels in unwounded WT and mutant leaves were below the detection limit (Figure 5B). One hour after wounding, the average JA level was increased to 31.7±1.1 pg per milligram of fresh weight (pg/mg FW) in WT leaves, whereas the average JA level in mutant leaves was only 7.9±0.3 pg/mg FW (P<0.0001, Student's t test) (Figure 5B), confirming that spr8 plants are defective in wound-induced JA biosynthesis. These results indicate that TomLoxD is required for wound-induced JA biosynthesis and that the TomLoxDP598L mutant allele largely impairs this capability.
Taken together, our data support that, even though the expression of TomLoxDP598L is still responsive to mechanical wounding (Figure 5A), this mutant version of TomLoxD impairs wound-induced JA biosynthesis (Figure 5B).
The Wound-Induced Expression of TomLoxD Is Directly Regulated by the MYC Transcription Factor SlMYC2
In the model plant of Arabidopsis, much of our understanding of the JA signaling has come from the recent elucidation of the molecular details of JA-regulated gene transcription through the basic helix-loop-helix (bHLH)-type transcription factor MYC2, a master regulator of JA responses [50]–[54]. Considering that in Arabidopsis MYC2 directly regulates the expression of several JA biosynthetic genes including LOX2 [55], it is reasonable to speculate that SlMYC2, the tomato homolog of MYC2, may directly regulate the expression of TomLoxD. Indeed, several lines of evidence lends support to this hypothesis. First, wound-induced expression levels of TomLoxD were substantially reduced in SlMYC2-RNAi plants as compared to those in WT plants (Figure 6A, 6B and Figure S7), indicating that SlMYC2 positively regulates the wound-induced expression of TomLoxD; Second, chromatin immunoprecipitation (ChIP) assays using 35Spro:SlMYC2-4myc plants indicated that SlMYC2 associates with a G-box-like motif (CCATGTG) in the promoter region of TomLoxD (Figure 6C and 6D); Third, DNA electrophoretic mobility shift assays (EMSA) indicated that a maltose binding protein (MBP)-SlMYC2 fusion protein binds the promoter of TomLoxD in a G-box-like motif-dependent manner (Figure 6E). Finally, using the transient expression assay of Nicotiana benthamiana leaves, we verified the activation effect of SlMYC2 on the expression of a reporter containing the TomLoxD promoter fused with the firefly luciferase gene (LUC) (Figure 6F and 6G). Together, these data demonstrate that the wound-induced expression of TomLoxD is under the direct regulation of SlMYC2.
Fig. 6. SlMYC2 regulates TomLoxD expression through a direct association with its promoter. 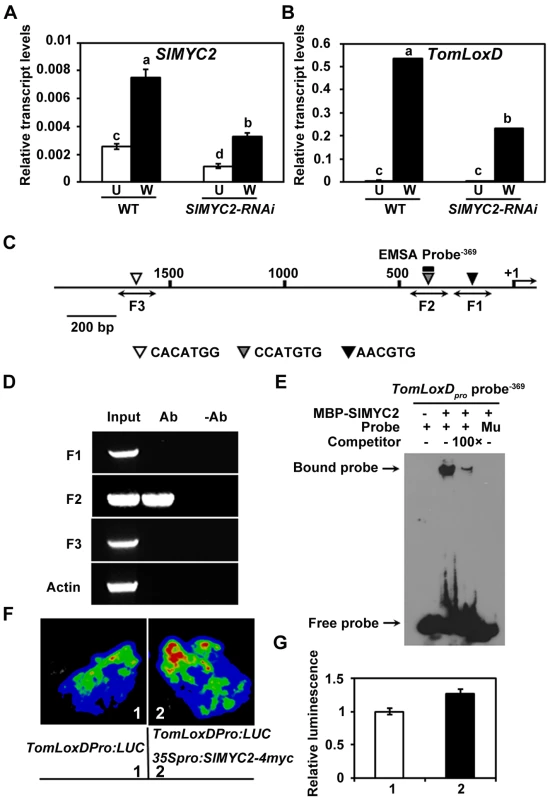
(A) and (B) Expression of SlMYC2 (A) and TomLoxD (B) in response to wounding. Sixteen-day-old seedlings of WT and SlMYC2-RNAi plants were mechanically wounded. Total RNAs were extracted 1 hour after wounding (W, Wounded; black bar) for qRT-PCR. RNAs were also extracted from leaves of unwounded plants (U, Unwounded; white bar) as control. Data presented are mean values of three biological repeats with SD. Data sets marked with different letters are significantly different from each other as assessed by Student's t test at P<0.001. (C) Schematic diagram of the promoter region of TomLoxD. Black lines represent the TomLoxD promoter region, including potential SlMYC2 binding G-box-like motif (black, gray and white triangles), DNA fragments (F1, F2 and F3) used for ChIP-PCR, and probe used for EMSA. The translational start sites (ATG) is shown as +1. Bar = 200 bp. (D) Enrichment of the DNA fragment F2 following ChIP using anti-myc antibody. 35Spro:SlMYC2-4myc transgenic seedlings and anti-myc antibody (Millipore) were used in ChIP assays. 16-day-old 35Spro:SlMYC2-4myc plants were mechanically wounded on both leaves, one hour after wounding, leaf tissues were harvested for crosslinking. The “no antibody” (-Ab) immunoprecipitates serve as negative controls. Three biological replicates were performed with similar results. (E) EMSA showing that the MBP-SlMYC2 fusion protein binds to the TomLoxDpro probe−369 of TomLoxD in vitro. Biotin-labeled probes were incubated with MBP-SlMYC2 purified proteins, and the free and bound probes were separated in an acrylamide gel. As indicated, unlabeled probes were used as competitors. Similar results were obtained in three independent experiments. Mu, mutated labeled probe in which the G-box motif was deleted. (F–G) Transient expression assays showing that SlMYC2 activates the expression of TomLoxD. Representative images of N. benthamiana leaves 72 h after infiltration are shown. The bottom panel indicates the infiltrated constructs. (G) Quantitative analysis of luminescence intensity in (F). Values are mean ± SD of five independent determinations. Overexpression of TomLoxD Leads to Increased Plant Immunity to Insects and Necrotrophic Pathogens
Our findings that TomLoxD is required for wound-induced JA biosynthesis and defense gene expression raised the possibility that overexpression of this gene could enhance wound-induced JA biosynthesis, which, in turn, leads to increased plant resistance. To test this hypothesis, we generated transgenic tomato plants overexpressing the TomLoxD cDNA driven by the cauliflower mosaic virus 35S promoter (OE plants). Increased expression of TomLoxD in transgenic lines including OE-1, OE-3 and OE-5 was confirmed by qRT-PCR analysis (Figure 7A). Under normal growth conditions, the overall growth and morphology of these OE plants was essentially similar to those of WT plants (Figure S1). We then compared the expression levels of defensive genes between these OE plants and WT plants. Similar steady-state levels of PI-II, TD and LapA transcripts were detected between the noninduced OE plants and WT plants (Figure 7B–7D). A marked increase in the accumulation levels of these transcripts was, however, observed in the TomLoxD overexpression plants in response to mechanical wounding (Figure 7B–7D). These results demonstrate that overexpression of TomLoxD leads to enhanced wound-induced activation of PI-II and other defense-related genes.
Fig. 7. Resistance of TomLoxD overexpression plants to cotton bollworm larvae and Botrytis cinerea. 
(A–D) Expression of TomLoxD (A), PI-II (B), TD (C) and LapA (D) in TomLoxD overexpression plants in response to wounding. Two-leaf-stage plants of WT and TomLoxD overexpression lines (OE-1, OE-3 and OE-5) were mechanically wounded (black bar). After 1 hour and 12 hours, leaf tissues were harvested for RNA extraction and gene expression analysis. Unwounded leaves (white bar) of each genotype were used as control. Data show the mean ± SD of three independent sample. (E) JA levels in unwounded (U, Unwounded; white bar) and wounded 1 hour (W, Wounded; black bar) leaves of two-leaf-stage WT and OE-5 plants in response to wounding. Data show the mean ± SD of three independent sample preparations. Bars with different letters are significantly different compared OE-5 plants with WT plants (P = 0.01). FW, fresh weight. (F–H) OE-5 plants show increased resistance to insects attack. (F) Representative WT (left) and OE-5 (right) plants at the end of cotton bollworm larvae feeding trial. (G) Larval weight recovered at the end of the 14-day-feeding trial on whole plants of WT (black bar) and OE-5 (white bar) (n = 15). Data represent the mean with SD. Bars with different letters are significantly different from each other (P = 0.05). (H) Size of larvae recovered at the end of cotton bollworm feeding trial. The feeding trails on whole plants were performed as described above and were repeated three times with similar results. (I–K) OE-5 plants exhibit increased resistance to B. cinerea. (I) and (J) Detached leaves from five-week-old WT (left), spr8 (middle) and OE-5 (right) plants were inoculated with B. cinerea. Photograph was taken (I) and the disease lesion diameter analyzed in Botrytis-inoculated leaves of WT (black bar), spr8 (gray bar) and OE-5 (white bar) at 3 DAI (J). Error bars represent the SD from three independent experiments (n = 30). Data sets marked with different letters are significantly different from each other as assessed by Student's t test at P<0.001. (K) Expression of PR1b1 in response to B. cinerea infection. Sixteen-day-old seedlings of WT (black bar), spr8 (gray bar) and OE-5 (white bar) were inoculated as described in Material and Method. At different times as indicated, samples were harvested for RNA extraction and qRT-PCR analysis. Data presented are mean values of three biological repeats with SD. To test that the increased wound-induced defense gene expression in these OE lines may be resulted from enhanced wound-induced accumulation levels of JA, we examined wound-induced JA accumulation between OE-5 and WT plants. Similar steady-state levels of JA were detected between OE-5 and WT plants (Figure 7E), indicating that overexpression of TomLoxD does not lead to constant accumulation of high levels of JA. In response to mechanical wounding, however, a substantial increase in the accumulation of JA was observed in OE-5 plants (Figure 7E), indicating that overexpression of TomLoxD leads to enhanced wound-induced accumulation of the defense hormone JA.
The ability of TomLoxD overexpresser lines to accumulate higher levels of JA and to express increased levels of defensive genes in response to mechanical wounding suggested that these transgenic plants may be more resistant to herbivorous insects. To test this possibility, five-week-old OE-5 and WT plants were challenged with Helicoverpa armigera larvae. After termination of the feeding trial, we examined the weight of the larvae to assess the resistance of plants. The average weight of larvae reared on OE-5 plants was only 32.5% of that of larvae reared on WT plants (Figure 6F–6H), demonstrating that overexpression of TomLoxD leads to enhanced plant resistance to herbivorous insects.
Considering that the JA-signaled plant resistance is also effective to the necrotrophic pathogen Botrytis cinerea [50], [51], [56]–[58], we examined the performance of OE-5 plants to the Hy2-1 strain of B. cinerea. For these experiments, detached leaves from five-week-old tomato plants were inoculated with 5 µL 5×105 per mL spore suspension and disease development was analyzed 3 days after inoculation (DAI). As measured by the size of necrotic lesions, whereas spr8 plants were more susceptible than WT plants to B. cinerea infection, OE-5 plants were more resistant than WT plants to this pathogen (Figure 7I and 7J). In another pathogen infection assay, 16-day-old seedlings were inoculated in planta with spore suspensions of B. cinerea and the expression levels of the pathogenesis-related (PR) gene PR1b1 [59] was examined with qRT-PCR. As shown in Figure 7K, whereas B.cinerea-induced expression levels of PR1b1 were reduced in spr8 plants than those in WT plants, expression levels of PR1b1 were much higher in OE-5 plants than those in WT plants, suggesting that the resistance of plants to pathogen is correlated with the expression levels of defense-related genes.
Discussion
TomLoxD Is Required for Wound-Induced JA Biosynthesis
Here, we provide several lines of evidence demonstrating that the wound response defect of the tomato spr8 mutant results from a mutation in TomLoxD that is required for wound-induced JA biosynthesis. First, positional cloning studies reveal that spr8 plants harbor a dominant negative mutation in TomLoxD, a 13-lipoxygenase that catalyzes the oxygenation of the polyunsaturated fatty acid linolenic acid, which is the metabolic precursor of JA. Second, spr8 leaves accumulate very little JA in response to wounding. The deficiency in wound-induced JA biosynthesis accounts for the defective wound-induced PIs expression in spr8 plants and is consistent with the fact that the wound response phenotype of the mutant can be rescued by exogenous JA. These results lead us to conclude that TomLoxD is responsible for the majority of wound-induced JA production in tomato leaves.
It is worth to note that the spr8 mutation affects a highly conserved Pro residue (Pro598) in the lipoxygenase domain of TomLoxD (Figure S4). As an α-amino acid, Pro contains a distinct cyclic structure and therefore this amino acid exhibits an exceptional conformational rigidity compared to other amino acids [48]. In this context, it is reasonable to speculate that the spr8 mutation affects the formation of the secondary structure of the TomLoxD protein and hence impairs its activity. Indeed, our data support that, even though the expression of TomLoxDP598L is still responsive to mechanical wounding (Figure 5A), this mutant version of TomLoxD impairs wound-induced JA biosynthesis (Figure 5B). Considering that the spr8 mutation occurs in the C-terminal α-helices domain of TomLoxD (Figure 4D), it is most likely that, in spr8 plants, the TomLoxDP598L protein still can bind the substrate (i.e., linoleic acid) as the WT TomLoxD does, but this mutant protein loses its catalytic activity. Competition between TomLoxD and TomLoxDP598L for substrate binding could underlie that spr8 acts genetically as a semi-dominant mutant.
As in other higher plants, in tomato lipoxygenases are encoded by a gene family consisting of 6 members (Figure 4E). It has been shown that TomLoxA, TomLoxB, TomLoxC and TomLoxE are mainly expressed in fruits during development and ripening [60]. Among them, TomLoxC is specifically involved in the generation of C6 aldehydes and alcohols, which are important constituents of volatile flavor of tomato fruits [61]–[63]. The expression of TomLoxD and TomLoxF is stimulated by the non-pathogenic rhizobacteria Pseudomonas putida BTP1 and these two genes are likely to be involved in rhizobacteria mediated-induced systemic resistance [64]. The deduced amino acid sequence of TomLoxD show high similarity to several chloroplast-localized lipoxygenases in Arabidopsis that have been shown to be involved in JA biosynthesis. Among them, LOX3 and LOX4 are involved in male fertility [49], [65] whereas LOX2 is specifically involved wound response [66], [67]. TomLoxD also shows high sequence similarity to the maize TASSELSEED1 (TS1) protein, which also encodes a plastid-localized lipoxygenase and plays a critical role in flower development and sex determination [68]. Here, we show that the tomato TomLoxD gene is specifically involved in the wound response, but shows minor, if any, effect on general plant growth and flower development (Figure S1). Taken together, these studies indicate that individual lipoxygenase isoforms are differentially regulated and have distinct physiological functions.
Transgenic Manipulation of TomLoxD Leads to Enhanced Resistance of Tomato to Insect and Pathogen Attack
Over two decades ago, Ryan and colleagues discovered the role of JAs in regulating defense gene expression in tomato [1], [20], [22]. Since then an ever growing body of evidence supports the view that the intracellular levels of JA plays a major role in controlling the strength of JA responses. Genetic engineering of plant cells for elevated endogenous JA levels therefore provides a strategy for increasing JA-dependent defenses. Indeed, the Ryan group showed that 35S::PS plants contain elevated JA levels and constantly express a spectrum of defense-related proteins that provide protection against insect attack [15], [69], [70]. Other examples of genetic alterations that cause constitutive JA accumulation include overexpression of a mitogen-activated protein kinase in tobacco [71] and mutation of the cellulose synthase CeSA3 in Arabidopsis [72], [73]. It is noteworthy that even though genetic engineering of the tomato PS gene or the Arabidopsis CeSA3 gene leads to increased JA-dependent resistance against insects or pathogens, the resulting transgenic plants show growth retardation and other physiological defects in normal growth conditions [15], [72], [73], which may limit the application potential of these genes in crop protection.
Attempts to increase endogenous JA levels and thus JA-dependent resistance by overexpression of individual JA biosynthetic genes in tomato and other plants have met with limited success [18], [33], [74], a plausible explanation is that the JA levels are mainly controlled by substrate availability [47], [75], [76]. In contrast to these unsuccessful examples, we show here that TomLoxD-OE plants exhibited increased expression levels of wound-induced defense-related genes and are more resistant to H. armigera. TomLoxD-OE plants also displayed enhanced resistance to the necrotrophic pathogen B. cinerea. These results indicated that genetic manipulation of TomLoxD leads to enhanced resistance of tomato plants to arthropod herbivores and microbial pathogens.
It is important to note that in the absence of insect attack or pathogen infection, the overall growth and fertility of TomLoxD-OE plants were essentially comparable with those of WT plants (Figure S1), indicating there was no fitness cost associated with overexpressing TomLoxD in our growth conditions. This is important because the maintenance of constitutive proteins or the continuous mounting of defenses often has severe impacts on plant growth or fertility [77]. Because the overexpression of TomLoxD does not impose a significant fitness cost to the plant, the TomLoxD-OE plants are viable candidates for field trials to improve insect and pathogen resistance in crop tomato.
Enhanced expression of defense-related genes in TomLoxD-OE plants is only observed after mechanical wounding, insect attack or pathogen infection suggests that the activation of the TomLoxD activity is regulated by the JA signaling. Indeed, we found that the wound-induced expression of TomLoxD is under the direct regulation of SlMYC2, the functional homolog of the Arabidopsis MYC2, a master regulator of JA-responsive gene expression. These findings are consistent with the long-standing observations that JA-signaling and synthesis form an apparent positive feedback regulatory loop [25], [26], [78]. It is also possible that the activity of TomLoxD for wound-induced JA biosynthesis is under posttranscriptional modification and that this modification is regulated by environmental stimuli including wounding, insect attack or pathogen infection. Alternatively, these environmental stimuli could lead to the accumulation of more substrates available for TomLoxD. Given the application potential of TomLoxD for crop protection, it is of significant in future studies to further explore the functional mechanisms of TomLoxD in wound-induced JA biosynthesis.
Materials and Methods
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum L.) cv Castlemart (CM) was used as the wild-type (WT) for all experiments. The plant material 35S::PS used in this study was previously described [15], [17], [29]. Tomato seedlings were grown in growth chambers and maintained under 16 h of light (200 µE m−2 s−1) at 28°C and 8 h of dark at 18°C and 60% relative humidity.
Mutant Isolation and Genetic Analysis
Mutagenesis of 35S::PS plants with ethyl methanesulfonate (EMS) and the isolation of suppressor of prosystemin-mediated responses (spr) mutants were performed as previously described [17], [29]. spr8 is one of the identified mutant lines and is deficient in both PPO activity and PI-II protein accumulation.
The original spr8 mutant in the 35S::PS genetic background was backcrossed to tomato cv CM as previously described [18]. The identified homozygous spr8/spr8 mutant plants were crossed to the WT and F1 plants were allowed to self-pollinate. The wound response phenotype of F1 and F2 plants was assessed by measuring PI-II accumulation following wounding treatment.
Map-Based Clone of Spr8
Map-based cloning procedures similar to those described [18], [79] were used to identify the Spr8 locus. A homozygous spr8 plant (S. lycopersicum) was crossed to the wild tomato species S. pennellii (LA716), and the resulting F1 plant was backcrossed to the spr8 parental line to generate a BC1 mapping population. The wound-response phenotype of individual BC1 plants was scored by measuring PI-II protein levels in response to mechanical wounding, as described above.
Using the BC1 population described above, bulked segregant analysis was used in combination with simple sequence repeats (SSR) analysis to identify molecular markers linked to Spr8. Equal amounts of genomic DNA from10 randomly selected wound-responsive (i.e., wild-type) and 10 nonresponsive (i.e., mutant) BC1 plants were pooled to construct a wild-type DNA bulk (B+) and a mutant DNA bulk (B−), respectively. Rough mapping using the 20 BC1 plants indicated that the target gene is linked to the marker TES0023 on the long arm of chromosome 3. Analysis of linkage between Spr8 and known SSR markers in this region demonstrated that Spr8 is located between TES0023 and TES1203. A high-resolution genetic map of the Spr8 region was constructed by scoring 354 BC1 plants for recombination events within the SSR601-Spr8-M140 interval in the scaffold SL2.40sc03701 of the sequenced tomato genome. Sequence analyses of genes in this interval revealed a C-to-T mutation in the TomLoxD gene. DNA primers for molecular markers used in map-based cloning were listed in Table S1.
For complementation analysis, the 35Spro:TomLoxD-GFP construct was introduced into the spr8 plants using Agrobacterium tumefaciens-mediated transformation for the complementation analysis. The TomLoxD-RNAi and 35Spro:TomLoxDP598L-GFP constructs were introduced into WT plants using Agrobacterium tumefaciens-mediated transformation.
DNA Constructs and Plant Transformation
DNA constructs for plant transformation were generated following standard molecular biology protocols and Gateway (Invitrogen) technology. Full-length coding sequence of TomLoxD was amplified with Gateway-compatible primers. The PCR product was cloned by pENTR Directional TOPO cloning kits (Invitrogen) and then recombined with the binary vector pGWB5 (35S promoter, C-GFP) to generate the 35Spro:TomLoxD-GFP construct. Similarly, we generated 35Spro:TomLoxDP598L-GFP construct, which was amplified from spr8 cDNAs, using the same primers as 35Spro:TomLoxD-GFP construct. Full-length coding sequence of SlMYC2 was also cloned into the pGWB17 vector (35S promoter, C-4myc) to generate the 35Spro:SlMYC2-4myc constructs.
To generate a TomLoxD-RNAi construct, fragments of the TomLoxD open read frame (106–570 bp), which were amplified from the cDNAs, were digested by XhoI and SpeI, and then inserted into XhoI-SpeI sites and SalI-XbaI sites in PUCCRNAi vector by steps. So this second ligation inserts the PCR product was in inverted orientation with respect to first cloned fragment, yielding an inverted repeat separated by the first intron fragment of GA20 oxidase from potato. The two reversed repeated DNAs were transferred to pCAMBIA-1301 (CAMBIA) from PUCCRNAi by PstI digestion. The plasmid pCAMBIA-1301 had been modified by adding a CaMV 35S promoter. Similarly, the SlMYC2-RNAi construct was performed. All primers used for DNA construct generation are listed in Table S3 online.
The above constructs were then transformed into Agrobacterium tumefaciens strain AGLO and used to transform tomato cotyledon explants as described previously [18]. Transformants were selected based on their resistance to hygromycin. Homozygous T3 or T4 transgenic seedlings were used for phenotype and molecular characterization.
PI-II Protein Accumulation Assays
The wound response of tomato plants was determined using a radial immunodiffusion assay for the detection of PI-II accumulation in leaf tissue as previously described [11], [36]. Two-leaf-stage (16-day-old) seedlings were used for the wounding treatment as described [29] and then the wounded leaf (local response) and the unwounded leaf (systemic response) were harvested separately to assay PI-II protein level.
Wounding, Systemin and MeJA Treatment of Tomato Plants
For wounding treatment, 16-day-old seedlings were wounded with a hemostat across the midrib of all leaflets on the lower leaf and the upper leaf. Then, the same leaflets were wounded again, proximal to the petiole. Wounded plants were incubated under continuous illumination conditions. For each time point of sampling, five whole plants leaves were harvested for the extraction of RNAs.
Systemin feeding experiments were performed using 16-day-old tomato seedlings as previously described with minor modifications [18], [28], [29]. Briefly, 2.5 pmol systemin was diluted from stock solutions into 300 µL 15 mM sodium phosphate, pH 6.5, prior to use. Tomato seedlings were excised at the base of the stem and placed in 0.5 mL microfuge tubes containing 300 µL dilutions. When >90% of the elicitor solution had been imbibed (approximately 2 hours), plants were transferred to glass vials containing 20 mL of water, and incubated in a Lucite Box under continuous light. Twelve hours later, leaf tissues of five plants were pooled for RNA extraction and gene expression assays. Control plants were fed with sodium phosphate buffer. Systemin was commercially synthesized by Shanghai Sangon Biological Engineering & Technology and Service Co. Ltd (Shanghai, PR China).
Sixteen-day-old tomato seedlings were treated with MeJA as described previously [80]. Control plants were incubated in a separate container in which ethanol was applied to cotton wicks. Twelve hours later, leaf tissues of five plants were pooled for RNA extraction. MeJA was purchased from Sigma-Aldrich.
Gene Expression Analysis
For qRT-PCR analysis, leaf tissues were harvested and frozen in liquid nitrogen for RNA extraction. RNA extraction and qRT-PCR analysis were performed as previously described [50]. Expression levels of target genes were normalized to those of the tomato Actin2 gene. Primers used to quantify gene expression levels are listed in Table S2.
Analysis of Trichomes
To examine the general pattern of trichome distribution on the adaxial surface of leaves, small pieces of tissue (5×5 mm), on the same base region of the third leaves from bottom to upper, were fixed, dehydrated, critical point dried in CO2, and coated with a film of gold as described [81]. Observations were performed with a HITACHI S-3000N scanning electron microscope (Japan) at an accelerating voltage of 15 kV. The density of type VI trichomes on the adaxial surface of leaves was determined by counting trichomes with a dissecting microscope equipped with a stage micrometer. All measurements were performed on WT and spr8 plants grown side by side under the same growth conditions.
Five-week-old plants were used to isolated type VI trichomes of leaves to obtain trichome exudates as previously described with minor modified [43]. Briefly, 1, 000 heads of Type VI glandular trichomes were selectively collected with a stretched-glass pipette and dissolved into 200 µL methyl tert-butyl ether (MTBE, Sigma) to analysis the chemical structures of compounds by GC-MS as described [43]. Different concentrations of external standards were run under the same GC conditions to develop standard curves to quantify volatiles (2-carene for monoterpenes, β-caryophyllene for sesquiterpenes).
Insect Feeding Trials
General procedures for rearing and handling cotton bollworm (Helicoverpa armigera) were described previously [18], [79]. The average larval weight at the beginning of the feeding trial was ∼5 mg. After termination of the feeding trial, PI-II protein accumulation in the remaining leaf tissues was measured [11], [36], as was the weight gain of larvae reared on both of the host genotypes.
Plant Infection with Botrytis cinerea
Detached leaves of five-week-old plants were inoculated as previously described [51]. For qRT-PCR experiments, the inoculation tests were performed in planta as described [58]. The same experiment was done with mock-pretreated plants as control. After inoculated for different times, the samples were then harvested for RNA extraction.
Sequence Analysis
The BLAST search program [82] was used for sequence analysis. The software ClusterX and T-coffee (http://www.ebi.ac.uk/Tools/t-coffee/) were used for sequence alignment. The phylogenetic relationship of TomLoxD in plants is inferred from protein sequences using a Bayesian approach in MrBayes [83]. The node labels are measures of support, which indicate the proportion of trees in the posterior distribution to containing the node.
JA Quantification
For JA content measurement, 16 - to 18-day-old plant leaves were wounded as described above. Approximately 200 mg leaf tissue (fresh weight) from five different plants was pooled for JA quantification as described previously [84]. Leaf tissues were also harvested from unwounded plants as controls.
ChIP-PCR Assays
ChIP assays were performed following a published protocol [50], [51], [54], [85] with minor modifications. Briefly, 1 hour after wounding treatment, 2.0 gram of 16-day-old 35Spro:SlMYC2-4myc plant leaves were harvested and cross-linked in 1% formaldehyde for ChIP experiment. myc antibody (Millipore) was used to immunoprecipitate the protein-DNA complex, and the precipitated DNA was purified using a PCR purification kit (Qiagen) for PCR analysis. Chromatin precipitated without antibody was used as negative control, while the isolated chromatin before precipitation was used as input control. Primers used for ChIP-PCR are listed in Table S4 online.
Electrophoretic Mobility Shift Assay
For plasmid construction of maltose binding protein (MBP) fusions with SlMYC2, the cDNA was amplified and cloned into the pMAL-c2 vector (New England Biolabs, Beverly, MA) via BamHI and PstI restriction sites. The MBP-SlMYC2 recombinant protein was expressed in the BL21 Escheichia coli (E. coli) strain and purified by binding onto an amylose resin (New England Biolabs) column, according to the instructions provided by the manufacturer. The 50-bp TomLoxD promoter probes containing G-box-like motif at the -369 site were synthesized and labeled with biotin at the 3′ end (Invitrogen), which containing the same sequences as that of the competitor probes without biotin-labled, while the mutated labeled probes were deleted the G-box-like motif. EMSA assays were performed using a LightShift Chemiluminescent EMSA kit (Thermo Scientific) as described [54]. Probe sequences are shown in Table S4 online.
Transient Expression Assay in N. benthamiana Leaves
The transient expression assays were performed in N. benthamiana leaves as previously described [51], [54]. The TomLoxD promoter was amplified and cloned into the pCAMBIA1381-Z (CAMBIA) via EcoRI and PstI restriction sites to generate the reporter construct TomLoxDpro:LUC. The SlMYC2 effector construct was the above-described 35Spro:SlMYC2-4myc. We used a low-light cooled CCD imaging apparatus (NightOWL II LB983 with indigo software) to capture the LUC image and to count luminescence intensity. The leaves were sprayed with 100 mM luciferin and were placed in darkness for 3 min before luminescence detection.
Transient Expression Assay in Arabidopsis Protoplast Cells
For plasmid construction of 35Spro:TomLoxD-GFP, the full length cDNA was amplified and cloned into the pGFP-2 vector [86] via XhoI and KpnI restriction sites to generate 35Spro:TomLoxD-GFP. Protoplast isolation and analysis of the subcellular location of transiently expressed GFP fusions by confocal fluorescence microscopy were performed as described [87].
Pollen Viability Assays
Alexander's triple staining was employed to measure viability of pollens, which were freshly harvested, as described previously [88].
Accession Numbers
The accession number of the sequenced tomato genome for the scaffold containing the Spr8/TomLoxD gene is SL2.40sc03701. The accession number from SGN database as following: TomLoxD (Solyc03g122340); SlMYC2(Solyc08g076930). Sequence data from this article can be found in the in the Arabidopsis Genome or GenBank databases under accession number as following: Arabidopsis thaliana AtLOX1 (AT1G55020), AtLOX2 (AT3G45140), AtLOX3 (AT1G17420), AtLOX4 (AT1G72520), AtLOX5 (AT3G22400), AtLOX6 (AT1G67560); Solanum lycopersicum TomLoxA (P38415), TomLoxB (P38416), TomLoxC (AAB65766), TomLoxD (AAB65767), TomLoxE (AAG21691), TomLoxF (NP_001234259); Zea mays ZmTS1 (ACL81190); Solanum tuberosum LOXH3 (CAA65269), StLOXH1 (CAA65268), STLOX (AAD09202), POTLX-3 (AAB67865), St13s-LOX2-1 (O24370), St13s-LOX3-1 (O24371); Nicotiana tabacum NtLOX (CAA58859); Glycine max Gm13-LOX3-1 (XP_003528556); Oryza sativa Japonica Group OsLOX6 (NP_001049158); Rattus norvegicus RnLOX3 (NP_001099263); Mus musculus Mm5-LOX (NP_033792); Homo sapiens HsLOX3 (CAC12843).
Supporting Information
Zdroje
1. RyanCA (2000) The systemin signaling pathway: differential activation of plant defensive genes. Biochimica et biophysica acta 1477 : 112–121.
2. WallingLL (2000) The myriad plant responses to herbivores. J Plant Growth Regulation 19 : 195–216.
3. LiL, LiC, LeeGI, HoweGA (2002) Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc Natl Acad Sci USA 99 : 6416–6421.
4. HoweGA, JanderG (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59 : 41–66.
5. WuJ, BaldwinIT (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44 : 1–24.
6. DickeM, BaldwinIT (2010) The evolutionary context for herbivore-induced plant volatiles: beyond the “cry for help”. Trends Plant Sci 15 : 167–175.
7. SunJ, JiangH, LiC (2011) Systemin/jasmonate-mediated systemic defense signaling in tomato. Mol Plant 4 : 607–615.
8. SchilmillerAL, HoweGA (2005) Systemic signaling in the wound response. Curr Opin Plant Biol 8 : 369–377.
9. RyanCA, PearceG (1998) Systemin: a polypeptide signal for plant defensive genes. Annu Rev Cell Dev Bio 14 : 1–17.
10. GreenTR, RyanCA (1972) Wound-induced proteinase inhibitors in plant leaves: a possible defense mechanism against insets. Science 175 : 776–777.
11. RyanCA (1967) Quantitative determination of soluble cellular proteins by radial diffusion in agar gels containing antibodies. Anal Biochem 19 : 434–440.
12. PearceG, StrydomD, JohnsonS, RyanCA (1991) A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253 : 895–897.
13. RyanCA (1992) The search for the proteinase inhibitor-inducing factor, PIIF. Plant Mol Biol 19 : 123–133.
14. McGurlB, RyanCA (1992) The organization of the prosystemin gene. Plant Mol Biol 20 : 405–409.
15. McGurlB, Orozco-CardenasM, PearceG, RyanCA (1994) Overexpression of the prosystemin gene in transgenic tomato plants generates a systemic signal that constitutively induces proteinase inhibitor synthesis. Proc Natl Acad Sci USA 91 : 9799–9802.
16. ChenH, WilkersonCG, KucharJA, PhinneyBS, HoweGA (2005) Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc Natl Acad Sci USA 102 : 19237–19242.
17. HoweGA, RyanCA (1999) Suppressors of systemin signaling identify genes in the tomato wound response pathway. Genetics 153 : 1411–1421.
18. LiC, LiuG, XuC, LeeI, BauerP, et al. (2003) The tomato suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15 : 1646–1661.
19. VickBA, ZimmermanDC (1984) Biosynthesis of jasmonic acid by several plant species. Plant physiol 75 : 458–461.
20. FarmerEE, RyanCA (1992) Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 4 : 129–134.
21. SchallerF (2001) Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot 52 : 11–23.
22. FarmerEE, RyanCA (1990) Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Natl Acad Sci USA 87 : 7713–7716.
23. DevotoA, TurnerJG (2003) Regulation of jasmonate-mediated plant responses in Arabidopsis. Ann Bot 92 : 329–337.
24. BrowseJ (2005) Jasmonate: an oxylipin signal with many roles in plants. Vitam Horm 72 : 431–456.
25. WasternackC (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 100 : 681–697.
26. WasternackC, HauseB (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111 : 1021–1058.
27. BalbiV, DevotoA (2008) Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol 177 : 301–318.
28. LeeGI, HoweGA (2003) The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J 33 : 567–576.
29. LiC, ZhaoJ, JiangH, WuX, SunJ, et al. (2006) The wound response mutant suppressor of prosystemin-mediated responses6 (spr6) is a weak allele of the tomato homolog of CORONATINE-INSENSITIVE1 (COI1). Plant Cell Physiol 47 : 653–663.
30. YanJ, ZhangC, GuM, BaiZ, ZhangW, et al. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21 : 2220–2236.
31. RyanCA, MouraDS (2002) Systemic wound signaling in plants: a new perception. Proc Natl Acad Sci USA 99 : 6519–6520.
32. StratmannJW (2003) Long distance run in the wound response - jasmonic acid is pulling ahead. Trends Plant Sci 8 : 247–250.
33. StenzelI, HauseB, MaucherH, PitzschkeA, MierschO, et al. (2003) Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato - amplification in wound signalling. Plant J 33 : 577–589.
34. PearceG, RyanCA (2003) Systemic signaling in tomato plants for defense against herbivores: Isolation and characterization of three novel defense-signaling glycopeptide hormones coded in a single precursor gene. J Biol Chem 278 : 30044–30050.
35. Narváez-VásquezJ, Orozco-CárdenasML, RyanCA (2007) Systemic wound signaling in tomato leaves is cooperatively regulated by systemin and hydroxyproline-rich glycopeptide signals. Plant Mol Biol 65 : 711–718.
36. TrautmanR, CowanKM, WagnerCG (1971) Data processing for radial immunodiffusion. Immunochemistry 8 : 901–916.
37. JohnsonR, NarvaezJ, AnG, RyanCA (1989) Expression of proteinase inhibitors I and II in transgenic tobacco plants: Effects on natural defense against Manduca sexta larvae. Proc Natl Acad Sci USA 86 : 9871–9875.
38. FowlerJH, Narváez-VásquezJ, AromdeeDN, PautotV, HolzerFM, et al. (2009) Leucine aminopeptidase regulates defense and wound signaling in tomato downstream of jasmonic acid. Plant Cell 21 : 1239–1251.
39. WagnerGJ (1991) Secreting glandular trichomes: more than just hairs. Plant physiol 96 : 675–679.
40. WilkensRT, SheaGO, HalbreichS, StampNE (1996) Resource availability and the trichome defenses of tomato plants. Oecologia 106 : 181–191.
41. HareJD (2005) Biological activity of acyl glucose esters from Datura wrightii glandular trichomes against three native insect herbivores. J Chem Ecol 31 : 1475–1491.
42. Luckwill LC (1943) The genus Lycopersicon: historical, biological, and taxonomic survey of the wild and cultivated tomatoes. (Aberdeen, Scotland: Aberdeen University Press).
43. SchilmillerAL, SchauvinholdI, LarsonM, XuR, CharbonneauAL, et al. (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA 106 : 10865–10870.
44. The Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485 : 635–641.
45. RenJ, LiC, LiC (2012) Tomato genome gets fully decoded – paves way to tastier and healthier fruits. J Genet Genomics 39 : 303–305.
46. HeitzT, BergeyDR, RyanCA (1997) A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant physiol 114 : 1085–1093.
47. FeussnerI, WasternackC (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53 : 275–297.
48. AndreouA, FeussnerI (2009) Lipoxygenases - structure and reaction mechanism. Phytochemistry 70 : 1504–1510.
49. CaldelariD, WangG, FarmerEE, DongX (2011) Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol Biol 75 : 25–33.
50. ChenR, JiangH, LiL, ZhaiQ, QiL, et al. (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24 : 2898–2916.
51. ZhaiQ, YanL, TanD, ChenR, SunJ, et al. (2013) Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet 9: e1003422.
52. BoterM, Ruíz-RiveroO, AbdeenA, PratS (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18 : 1577–1591.
53. LorenzoO, ChicoJM, Sánchez-SerranoJJ, SolanoR (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 : 1938–1950.
54. ChenQ, SunJ, ZhaiQ, ZhouW, QiL, et al. (2011) The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23 : 3335–3352.
55. HouX, LeeLY, XiaK, YanY, YuH (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19 : 884–894.
56. Glazebrook (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 : 205–227.
57. El OirdiM, BouarabK (2007) Plant signalling components EDS1 and SGT1 enhance disease caused by the necrotrophic pathogen Botrytis cinerea. New Phytol 175 : 131–139.
58. El OirdiM, El RahmanTA, RiganoL, El HadramiA, RodriguezMC, et al. (2011) Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23 : 2405–2421.
59. AhnIP, LeeSW, KimMG, ParkSR, HwangDJ, et al. (2011) Priming by rhizobacterium protects tomato plants from biotrophic and necrotrophic pathogen infections through multiple defense mechanisms. Mol Cells 32 : 7–14.
60. FerrieBJ, BeaudoinN, BurkhartW, BowsherCG, RothsteinSJ (1994) The cloning of two tomato lipoxygenase genes and their differential expression during fruit ripening. Plant physiol 106 : 109–118.
61. GriffithsA, BarryC, Alpuche-SolisAG, GriersonD (1999) Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. J Exp Bot 50 : 793–798.
62. GriffithsA, PrestageS, LinforthR, ZhangJ, TaylorA, et al. (1999) Fruit-specific lipoxygenase suppression in antisense-transgenic tomatoes. Postharvest Biol Technol 17 : 163–173.
63. ChenG, HackettR, WalkerD, TaylorA, LinZ, et al. (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol 136 : 2641–2651.
64. MariuttoM, DubyF, AdamA, BureauC, FauconnierM-L, et al. (2011) The elicitation of a systemic resistance by Pseudomonas putida BTP1 in tomato involves the stimulation of two lipoxygenase isoforms. BMC Plant Bio 11 : 29.
65. BannenbergG, MartínezM, HambergM, CastresanaC (2009) Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids 44 : 85–95.
66. BellE, CreelmanRA, MulletJE (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92 : 8675–8679.
67. GlauserG, DubugnonL, MousaviSA, RudazS, WolfenderJ-L, et al. (2009) Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J Biol Chem 284 : 34506–34513.
68. AcostaIF, LaparraH, RomeroSP, SchmelzE, HambergM, et al. (2009) tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323 : 262–265.
69. BergeyDR, HoweGA, RyanCA (1996) Polypeptide signaling for plant defensive genes exhibits analogies to defense signaling in animals. Proc Natl Acad Sci USA 93 : 12053–12058.
70. ChenH, JonesAD, HoweGA (2006) Constitutive activation of the jasmonate signaling pathway enhances the production of secondary metabolites in tomato. FEBS Lett 580 : 2540–2546.
71. SeoS, SanoH, OhashiY (1999) Jasmonate-based wound signal transduction requires activation of WIPK, a tobacco mitogen-activated protein kinase. Plant Cell 11 : 289–298.
72. EllisC, TurnerJG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13 : 1025–1033.
73. EllisC, KarafyllidisI, WasternackC, TurnerJG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 : 1557–1566.
74. LaudertD, SchallerF, WeilerEW (2000) Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta 211 : 163–165.
75. SchallerF, SchallerA, StintziA (2005) Biosynthesis and metabolism of jasmonates. J Plant Growth Regul 23 : 179–199.
76. SchallerA, StintziA (2009) Enzymes in jasmonate biosynthesis - structure, function, regulation. Phytochemistry 70 : 1532–1538.
77. BostockRM (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 43 : 545–580.
78. CreelmanRA, MulletJE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48 : 355–381.
79. LiC, SchilmillerAL, LiuG, LeeGI, JayantyS, et al. (2005) Role of β-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 17 : 971–986.
80. LiL, HoweGA (2001) Alternative splicing of prosystemin pre-mRNA produces two isoforms that are active as signals in the wound response pathway. Plant Mol Biol 46 : 409–419.
81. LiL, ZhaoY, MccaigBC, WingerdBA, WangJ, et al. (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16 : 126–143.
82. AltschulSF, MaddenTL, SchäfferAA, ZhangJ, ZhangZ, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 : 3389–3402.
83. RonquistF, HuelsenbeckJP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 : 1572–1574.
84. FuJ, ChuJ, SunX, WangJ, YanC (2012) Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal Sci 28 : 1081–1087.
85. GendrelA-V, LippmanZ, MartienssenR, ColotV (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nature methods 2 : 213–218.
86. KostB, SpielhoferP, ChuaNH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16 : 393–401.
87. Abdel-GhanySE, Müller-MouléP, NiyogiKK, PilonM, ShikanaiT (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17 : 1233–1251.
88. AlexanderMP (1980) A versatile stain for pollen, fungi, yeast and bacteria. Stain Technol 55 : 13–18.
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



