-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
And Baby Makes Three: Genomic Imprinting in Plant Embryos
article has not abstract
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1003981
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1003981Summary
article has not abstract
Genomic imprinting results in the preferential expression of alleles after fertilization, depending on their parent of origin. Imprinting is epigenetically controlled and has evolved independently in plants and animals [1]. This striking convergence, combined with a degree of conservation within kingdoms, strongly suggests that imprinting confers a significant fitness advantage. Current thinking holds that this advantage lies in the ability to control resource flow from mother to offspring. The drivers for selection are less clear but may include parental conflict [2], [3], adaptive integration of offspring and maternal genomes [4], or a combination of the two.
Data on imprinted gene function and expression pattern from mammals [5] support these ideas, and there is also a growing body of similar evidence from plants. For example, the parental dosage-related enhanced or repressed seed development that can follow interploidy crosses has been proposed to be mediated by disruption of allele-specific genomic imprinting [6]–[8]. There is also evidence from maize that parental dosage regulates genes controlling key stages of seed development [9], and, in Arabidopsis, imprinted Polycomb Group (PcG) proteins [10] have been shown indirectly to control seed size. Most recently, an unambiguous role for imprinting control of nutrient transfer was demonstrated using the imprinted meg1 gene in maize, which directly regulates maternal provisioning of the embryo and controls ultimate seed composition and size in a strictly gene dosage–dependent manner [11]. This clear involvement of imprinting in quantitative aspects of seed development has obvious implications for agricultural traits, and imprinted genes may represent an unrecognized pool of variation that can be exploited for crop improvement.
Plants and mammals have been reported to differ significantly in how imprinting affects the products of fertilization. While both kingdoms have evolved extra-embryonic organs to aid the process of nutrient flow from the mother to the offspring—the placenta in animals and the endosperm in plants—the placenta forms an integral part of the embryo, whereas the endosperm results from the fusion of a second sperm with a diploid accessory cell to the egg. The endosperm thus develops as a triploid structure that makes no genetic contribution to the next generation. In animals, imprinting regulates gene expression both in embryos and in other tissues during subsequent development. However, the majority of the data from plants to date has pointed to imprinting being restricted to the endosperm. Thus, while embryonic imprinting in mammals requires active reprogramming to erase and reset parent-specific imprinting marks according to the sex of the germline, it has been believed that this was not the case for plants, the endosperm being terminally differentiated (Figure 1).
Fig. 1. Imprinting, double fertilization, and the plant life history. 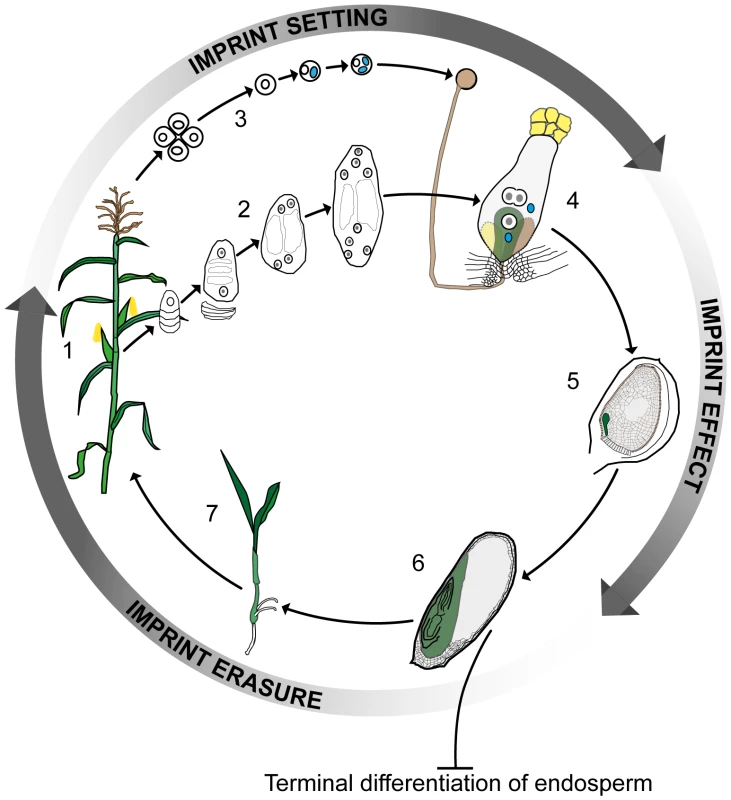
A plant at flowering (1) develops female and male reproductive organs from somatic tissue in which spore mother cells undergo meiosis and undergo a haploid phase of mega- (2) and micro- (3) gametophyte development. The megagametophyte, or embryo sac, forms two female gametes over the course of three mitotic divisions—the central cell and the egg cell—in addition to two synergid cells that assist in the fertilization process. The microgametophyte, or pollen grain, undergoes two mitotic divisions to form a vegetative cell and two male gametes, the sperm cells (blue). At fertilization (4) the pollen tube delivers the two sperm cells to the receptive synergid cell. One sperm cell fertilizes the egg cell (green) and the other sperm cell nucleus combines with the two polar nuclei of the large central cell of the embryo sac. Within the ovule (5) the fertilized central cell subsequently develops into the endosperm (grey), and the fertilized egg cell forms the embryo (green). In the seed at maturity (6), the endosperm is terminally differentiated and does not contribute genetically to the next generation, whereas the embryo germinates (7) and develops into the mature plant. Genomic imprinting in the embryo requires erasure and resetting of imprinting marks according to the sex of the gametes. Clearly, parental imprints need to be set during gametophyte development and to regulate allelic gene expression during early embryo and endosperm development. Currently available data indirectly indicate that the erasure of imprints occurs during late embryo development or germination. Color key: generative cell, sperm cell, and sperm cell nuclei: blue; central and egg cell nuclei: white with grey nucleolus; pollen grain, pollen tube, and receptive synergid: brown; egg cell, embryo, seedling, and plant: green; central cell and endosperm: grey. This view of the endosperm as the sole site of imprinting in plants was first challenged by data from maize, which showed the mee1 gene to be imprinted in the embryo [12]. More recent genome-wide approaches involving RNA sequencing (RNA-seq) of reciprocal crosses in Arabidopsis [13], rice [14], and maize [15] have also indicated the presence of several potentially imprinted genes in embryos. The study of imprinting events in plant embryos is complicated by their location deep in maternal tissue in close association with the endosperm and other tissues. This is particularly the case for Arabidopsis, where the embryo and its progenitor cells are extremely small. Despite these difficulties, in this issue of PLOS Genetics Raissig et al. [16] describe how they have risen to this challenge and, based on microarray data from gametes [17], [18] and RNA-seq results from early Arabidopsis embryos after unilateral crosses between two different ecotypes [19], identified a set of transcripts with clear hallmarks of imprinting. These transcripts are expressed de novo after fertilization and derived from only one parental allele. Using reverse transcription polymerase chain reaction (RT-PCR) product sequencing across polymorphisms, the parent-of-origin–dependent expression of several of the maternally expressed genes, and one paternally expressed gene, was validated in embryos generated from two sets of reciprocal crosses. Raissig et al. [16] also monitored allele-specific expression of promoter β-glucuronidase (GUS) fusion constructs for several of the maternally expressed genes following reciprocal crosses, obtaining data mirroring that from maize, where short imprinting control elements are located in the upstream promoter regions [20], [21]. Imprinting in plants thus differs strikingly from that in animals, where expression of clustered imprinted genes is regulated by long-range signals (generally noncoding RNAs) and chromosomal context is essential.
These new findings extend imprinting of embryo-expressed genes into the model species Arabidopsis, demonstrating that it occurs in both mono - and dicotyledonous species. Furthermore, the number of imprinted genes confirmed by Raissig et al. [16] reveals that imprinting in plant embryos is not restricted to rare events and is thus most likely to be significant for seed development. Importantly, this discovery permits the extensive genetic and “omic” resources available in Arabidopsis to be focused on not only the establishment of imprinting marks regulating monoallelic expression in the embryo but also on the resetting events that must take place as the embryonic tissue develops.
The epigenetic reprogramming events by which sex-specific marks are established in gametes and erased post-fertilization are complex and are only now starting to be understood. To date, two distinct, but in some ways interdependent, mechanisms of gene expression regulation have been shown to be involved in plant imprinting: DNA methylation and Histone 3 Lysine 27 tri-methylation (H3K27me3) by PcG complexes [22]. Rather unexpectedly, initial genetic analyses of a subset of the genes tested by Raissig et al. [16] indicate that the latter mechanism is involved in regulation of embryonic imprinting—although the actual parental imprinting marks in plant gametes have yet to be determined. Parent-specific DNA methylation patterning has long been recognized as the primary epigenetic mark involved in genomic imprinting and certainly fulfills this role in regulating imprinting in endosperm [21], [23]. However, the pattern of methylation in female gametes of maize [12], [21] also suggests that DNA methylation is unlikely to operate as a primary imprinting mark in embryos.
All the embryonically imprinted genes reported by Raissig et al. [16] showed bi-allelic expression in seedlings. This mirrors the situation in rice [14] and is consistent with the expression pattern of the imprinted mee1 gene of maize, where the active allele is transitorily demethylated in the embryo [12]. These data, and the observation that DNA methylation is highly dynamic during embryo development [24], reinforce the view that imprinting in plants acts only in the developing seed and point to erasure of imprinting marks during late embryogenesis or early seed germination. This is in striking contrast to mammals, in which imprinting is reset in the germline but, in the soma, controls development after birth by influencing resource allocation, for example, through suckling, and during early care through social interactions and kin recognition [5]. Imprinting in plants may thus be restricted to seed development simply because it is the only developmental phase where offspring are maternally dependent. These exciting discoveries do, however, serve to highlight our comparative ignorance of the nature of the epigenetic marks carried by the gametic genomes and the molecular mechanisms by which they are applied and—in the embryonic tissue—erased.
The demonstration by Raissig et al. [16] that embryonic imprinting occurs in the two major plant groupings raises a number of important questions relating to the extent of imprinting in plants, the degree of its conservation between species, and its genotype dependency. However, before we can assess the importance of imprinting in seed development and plant evolution and, of course, its applicability as a tool for plant improvement, we need to know far more about the functions of the genes affected.
Zdroje
1. FeilR, BergerF (2007) Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23 : 192–199 doi:10.1016/j.tig.2007.02.004
2. HaigD, WestobyM (1989) Parent-specific gene expression and the triploid endosperm. Am Nat 134 : 147–155.
3. TriversR, BurtA (1999) Kinship and genomic imprinting. Results Probl Cell Differ 25 : 1.
4. WolfJB, HagerR (2006) A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol 4: e380 doi:10.1371/journal.pbio.0040380
5. IslesAR, DaviesW, WilkinsonLS (2006) Genomic imprinting and the social brain. Philos Trans R Soc Lond B Biol Sci 361 : 2229–2237 doi:10.1038/nrg1062
6. LinBY (1984) Ploidy barrier to endosperm development in maize. Genetics 107 : 103–115.
7. ScottRJ, SpielmanM, BaileyJ, DickinsonHG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125 : 3329–3341.
8. KradolferD, WolffP, JiangH, SiretskiyA, KöhlerC (2013) An Imprinted Gene Underlies Postzygotic Reproductive Isolation in Arabidopsis thaliana. Dev Cell 26 : 525–535 doi:10.1016/j.devcel.2013.08.006
9. LiN, DickinsonHG (2009) Balance between maternal and paternal alleles sets the timing of resource accumulation in the maize endosperm. Proc Biol Sci 277 : 3–10 doi:10.1105/tpc.13.10.2297
10. IngouffM, HaseloffJ, BergerF (2005) Polycomb group genes control developmental timing of endosperm. Plant J 42 : 663–674 doi:10.1111/j.1365-313X.2005.02404.x
11. CostaLM, YuanJ, RousterJ, PaulW, DickinsonH, et al. (2012) Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr Biol 22 : 160–165 doi:10.1016/j.cub.2011.11.059
12. JahnkeS, ScholtenS (2009) Epigenetic resetting of a gene imprinted in plant embryos. Curr Biol 19 : 1677–1681 doi:10.1016/j.cub.2009.08.053
13. NodineMD, BartelDP (2012) Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482 : 94–97 doi:10.1038/nature10756
14. LuoM, TaylorJM, SpriggsA, ZhangH, WuX, et al. (2011) A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm. PLoS Genet 7: e1002125 doi:10.1371/journal.pgen.1002125.s014
15. WatersAJ, MakarevitchI, EichtenSR, Swanson-WagnerRA, YehCT, et al. (2012) Parent-of-Origin Effects on Gene Expression and DNA Methylation in the Maize Endosperm. Plant Cell 23 : 4221–4233 doi:10.1105/tpc.111.092668
16. RaissigMT, BemerM, BarouxC, GrossniklausU (2013) Genomic imprinting in the Arabidopsis embryo is partly regulated by PRC2. PLoS Genet 9: e1003862 doi: 10.1371/journal.pgen.1003862
17. WuestSE, VijverbergK, SchmidtA, WeissM, GheyselinckJ, et al. (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20 : 506–512 doi:10.1016/j.cub.2010.01.051
18. BorgesF, GomesG, GardnerR, MorenoN, McCormickS, et al. (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148 : 1168–1181 doi:10.1104/pp.108.125229
19. AutranD, BarouxC, RaissigMT, LenormandT, WittigM, et al. (2011) Maternal Epigenetic Pathways Control Parental Contributionsto Arabidopsis Early Embryogenesis. Cell 145 : 707–719 doi:10.1016/j.cell.2011.04.014
20. Gutiérrez-MarcosJF, CostaLM, Biderre-PetitC, KhbayaB, O'SullivanDM, et al. (2004) maternally expressed gene1 Is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16 : 1288–1301 doi:10.1105/tpc.019778
21. Gutiérrez-MarcosJF, CostaLM, Dal PràM, ScholtenS, KranzE, et al. (2006) Epigenetic asymmetry of imprinted genes in plant gametes. Nat Genet 38 : 876–878 doi:10.1038/ng1828
22. KöhlerC, WolffP, SpillaneC (2012) Epigenetic mechanisms underlying genomic imprinting in plants. Annu Rev Plant Biol 63 : 331–352 doi:10.1146/annurev-arplant-042811-105514
23. KinoshitaT, MiuraA, ChoiY, KinoshitaY, CaoX, et al. (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303 : 521–523 doi:10.1126/science.1089835
24. JullienPE, SusakiD, YelagandulaR, HigashiyamaT, BergerF (2012) DNA Methylation Dynamics during Sexual Reproduction in Arabidopsis thaliana. Curr Biol 22 : 1825–1830 doi:10.1016/j.cub.2012.07.061
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



