-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
Genomic imprinting results in monoallelic gene expression in a parent-of-origin-dependent manner and is regulated by the differential epigenetic marking of the parental alleles. In plants, genomic imprinting has been primarily described for genes expressed in the endosperm, a tissue nourishing the developing embryo that does not contribute to the next generation. In Arabidopsis, the genes MEDEA (MEA) and PHERES1 (PHE1), which are imprinted in the endosperm, are also expressed in the embryo; whether their embryonic expression is regulated by imprinting or not, however, remains controversial. In contrast, the maternally expressed in embryo 1 (mee1) gene of maize is clearly imprinted in the embryo. We identified several imprinted candidate genes in an allele-specific transcriptome of hybrid Arabidopsis embryos and confirmed parent-of-origin-dependent, monoallelic expression for eleven maternally expressed genes (MEGs) and one paternally expressed gene (PEG) in the embryo, using allele-specific expression analyses and reporter gene assays. Genetic studies indicate that the Polycomb Repressive Complex 2 (PRC2) but not the DNA METHYLTRANSFERASE1 (MET1) is involved in regulating imprinted expression in the embryo. In the seedling, all embryonic MEGs and the PEG are expressed from both parents, suggesting that the imprint is erased during late embryogenesis or early vegetative development. Our finding that several genes are regulated by genomic imprinting in the Arabidopsis embryo clearly demonstrates that this epigenetic phenomenon is not a unique feature of the endosperm in both monocots and dicots.
Published in the journal: . PLoS Genet 9(12): e32767. doi:10.1371/journal.pgen.1003862
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003862Summary
Genomic imprinting results in monoallelic gene expression in a parent-of-origin-dependent manner and is regulated by the differential epigenetic marking of the parental alleles. In plants, genomic imprinting has been primarily described for genes expressed in the endosperm, a tissue nourishing the developing embryo that does not contribute to the next generation. In Arabidopsis, the genes MEDEA (MEA) and PHERES1 (PHE1), which are imprinted in the endosperm, are also expressed in the embryo; whether their embryonic expression is regulated by imprinting or not, however, remains controversial. In contrast, the maternally expressed in embryo 1 (mee1) gene of maize is clearly imprinted in the embryo. We identified several imprinted candidate genes in an allele-specific transcriptome of hybrid Arabidopsis embryos and confirmed parent-of-origin-dependent, monoallelic expression for eleven maternally expressed genes (MEGs) and one paternally expressed gene (PEG) in the embryo, using allele-specific expression analyses and reporter gene assays. Genetic studies indicate that the Polycomb Repressive Complex 2 (PRC2) but not the DNA METHYLTRANSFERASE1 (MET1) is involved in regulating imprinted expression in the embryo. In the seedling, all embryonic MEGs and the PEG are expressed from both parents, suggesting that the imprint is erased during late embryogenesis or early vegetative development. Our finding that several genes are regulated by genomic imprinting in the Arabidopsis embryo clearly demonstrates that this epigenetic phenomenon is not a unique feature of the endosperm in both monocots and dicots.
Introduction
Genes regulated by genomic imprinting are expressed preferentially from one allele in a parent-of-origin-dependent manner. The two alleles do not differ in their DNA sequence, but rather they are differentially marked by epigenetic modifications. In mammals, imprint establishment occurs in a sex-specific manner during gamete development [1]. The parent-of-origin-specific “imprint” is retained or interpreted after fertilization, such that the alleles can be distinguished and differentially expressed during subsequent development. The imprints are erased in the primordial germ cell lineage, which will develop into the gametes, and are reestablished according to the sex of the germ line during gametogenesis [2], [3].
Genomic imprinting evolved both in placental mammals and in flowering plants. While genes can be imprinted in both the embryo and the placenta in mammals, and even in adult tissues [2], [4], genomic imprinting in plants was primarily described for genes expressed in the endosperm, the triploid nourishing tissue that develops upon fertilization of the diploid central cell [5], [6]. The triploid endosperm does not contribute to the next generation and, therefore, there is no requirement to erase and reset parental imprints. To date, only four plant genes have been proposed to show parent-of-origin-dependent, monoallelic expression in both the embryo and the endosperm, but only for the maize gene maternally expressed in embryo1 (mee1) was this unambiguously demonstrated [7]. Although only maternal transcripts of the Os10g05750 gene were detected in rice embryos, it is not clear whether these stem from expression in the egg cell or are monoallelically expressed de novo after fertilization, as is necessary for classification as an imprinted gene [8]. In Arabidopsis thaliana, the Polycomb group gene MEDEA (MEA) and its target, the MADS-box gene PHERES1 (PHE1), are both imprinted in the endosperm and show embryonic expression [9]–[11]; but it remains controversial whether embryonic expression is imprinted or not (reviewed in [10]).
The regulation of genomic imprinting in mammals is complex and involves DNA methylation, histone modifications, and non-coding RNAs [2], [12], [13]. In Arabidopsis, DNA methylation and Polycomb Repressive Complex 2 (PRC2)-mediated trimethylation of histone 3 lysine 27 (H3K27me3) are involved in the regulation of some imprinted loci in the endosperm. The maintenance DNA-methyltransferase METHYLTRANSFERASE1 (MET1) and the DNA-glycosylase DEMETER (DME) act antagonistically to regulate imprinting at the MEA, FLOWERING WAGENINGEN (FWA), FERTILIZATION INDEPENDENT SEED2 (FIS2), and MATERNALLY EXPRESSED PAB C-TERMINAL (MPC) loci [14]–[19]. DME is preferentially expressed in the central cell and removes DNA-methylation marks on maternal alleles [14], [17], which, however, might not directly define the imprinting status at all loci [20]. In maize, most imprinted genes analyzed in detail are differentially methylated in the endosperm [7], [21]–[23], which is already established in the gametes for some loci but not for others, indicating the existence of additional, primary imprinting marks other than DNA-methylation [7], [21]. The regulation of imprinted MEA expression by DME and MET1 is also indirect and it is not the presence or absence of DNA methylation that leads to differential expression of the two parental MEA alleles [20]. Finally, the regulation of imprinted expression of MEA, PHE1, FORMIN-HOMOLOGUE 5 (AtFH5), and some other loci involves additionally or exclusively the repressive action of PRC2 [9], [17], [24]–[27]. Recent studies identified many novel candidate imprinted genes in Arabidopsis, maize, and rice using systematic, genome-wide transcriptome screens on seed tissues [8], [27]–[32]. In Arabidopsis alone, the total number of imprinted genes increased from 12 to more than 300 potentially imprinted genes [27]–[30].
In this study, we show that genomic imprinting is not restricted to the endosperm in Arabidopsis, and describe parent-of-origin-dependent, monoallelic expression in the Arabidopsis embryo. We identified 80 potentially imprinted genes from a parent-of-origin-specific embryonic transcriptome [33] and confirmed eleven MEGs and one early PEG using allele-specific expression analyses of parental transcripts and reporter gene assays. Furthermore, we found that PRC2 is involved in maintaining the imprinted expression pattern at some of these loci. In contrast to imprinting in the endosperm, imprinted expression in the embryo requires the erasure and resetting of the imprinting marks between generations. Interestingly, the MEGs and the PEG are expressed from both alleles in seedlings, suggesting that the imprints are erased during late embryogenesis or early seedling development.
Results
In-Depth Analysis of the Hybrid Embryonic Transcriptome Reveals Monoparentally Expressed Genes
To study the global parental contributions to the embryonic transcriptome and its regulation, embryonic samples were previously generated from hybrid embryos at the 2–4 cell and globular stage [33]. The hybrid embryos were derived either from a cross between Landsberg erecta (Ler) and Columbia-0 (Col-0), or from a cross between the kryptonite (kyp) mutant (Ler) and Col-0. Subsequent high-throughput sequencing of the generated cDNA libraries allowed the identification of allele-specific transcripts based on single nucleotide polymorphisms (SNPs) between the accessions [33]. For this study, we identified potentially imprinted transcripts in the embryo, for which only one parental allele was sequenced in all samples, in the 2–4 cell samples only, or in the globular samples only ([33] for details see Material and Methods). We included only genes that were not deregulated in the kyp/KYP x Col-0 sample, assuming that KYP is not a major regulator of genomic imprinting, but rather regulates the parental contributions at the genome-wide level [33]. This procedure yielded 50 potential maternally expressed genes (MEGs) and 30 potential paternally expressed genes (PEGs) in the Arabidopsis embryo (Table S1). A recent study analyzing a hybrid embryonic transcriptome in Arabidopsis [34] also detected the presence of monoallelic gene expression in the Arabidopsis embryo. The authors describe 77 maternally and 45 paternally contributed transcripts in at least one embryonic stage tested. Finally, we chose 18 MEG candidates and six PEG candidates that are highly expressed in the embryo [33] but are absent from the gametes [35], [36], suggesting de novo expression, and analyzed them in detail (i.e. allele-specific expression and reporter gene analysis; Table S1).
Monoallelic Gene Expression in the 2–4 Cell and the Globular Embryo
To confirm the identified MEGs and PEGs, we produced new embryonic cDNA libraries by crossing the Col-0 and the Ler accessions reciprocally. Embryos were isolated at the 2–4 cell embryo stage (∼2.5 days after pollination (DAP)) and at the globular embryo stage (∼4 DAP). We sampled two biological replicates of each cross and stage (8 samples in total), extracted total RNA and amplified a cDNA library (see Material and Methods). The cDNA samples from hybrid embryos were subsequently used to amplify the polymorphic, SNP-containing sequence of potentially imprinted transcripts by RT-PCR, and products were assessed for their parent-of-origin by Sanger sequencing. As a control, we performed allele-specific expression analysis of a polymorphic gene that is expressed from both parental alleles (AT1G02780, EMBRYO DEFECTIVE 2386, [33]). We readily detected both parental nucleotides at the polymorphic site in all the samples analyzed (Figure S1A), confirming that this method is suitable to detect biallelic gene expression. Importantly, we verified that all assays used in this study amplify both parental alleles with equal efficiency. To this aim we performed PCR and Sanger sequencing on genomic DNA from reciprocal F1 hybrid seedlings (Col-0 x Ler and Ler x Col-0, respectively; Figure S1B and Figure S2). We assessed also the quantitative nature of the Sanger sequencing approach by performing the different assays with various mixtures of Col-0 and Ler genomic DNA. All assays showed a good correlation between the allelic ratios and the SNP-signal, illustrating that the Sanger approach is valid to estimate allele contributions (Figure S3). In conclusion, all assays used in this study amplified both alleles with equal efficiency and, thus, do not introduce a technical bias towards one allele.
We then performed allele-specific expression analyses on the selected 18 candidate MEGs and six candidate PEGs (Table S1). For eleven of the 18 candidate MEGs we could sequence only the maternal allele in all crosses, stages, and replicates analyzed (AT1G29660, AT1G72260, AT2G47115, AT5G62210, AT3G20520, AT2G17710, AT3G21500, AT2G01520, AT1G20680, AT5G51950, AT1G29050; Figure 1A–1F and Figure S4A–S4E). Because 9 of the 11 genes showed no detectable levels in the egg cell transcriptome, our results strongly suggest that they may be regulated by genomic imprinting in the Arabidopsis embryo ([35], [36], Table S1). Two of the 18 candidate MEGs showed biallelic expression in one 2–4 cell replicate sample and were therefore excluded from further analyses (AT3G44260 and AT5G52060, Figure S4F and S4G). However, the maternal signal was much higher in these replicates, and both genes showed complete monoallelic expression at the globular stage, suggesting that they may also be regulated by genomic imprinting. From the remaining five MEGs, four genes could not be amplified at all from the embryonic cDNA libraries, and one gene was lost when a highly stringent washing procedure was applied to the embryos before RNA extraction and amplification (see below, Table S1), suggesting that they are not or only weakly expressed in the embryo.
Fig. 1. Allele-specific expression analysis of MEGs and PEGs. 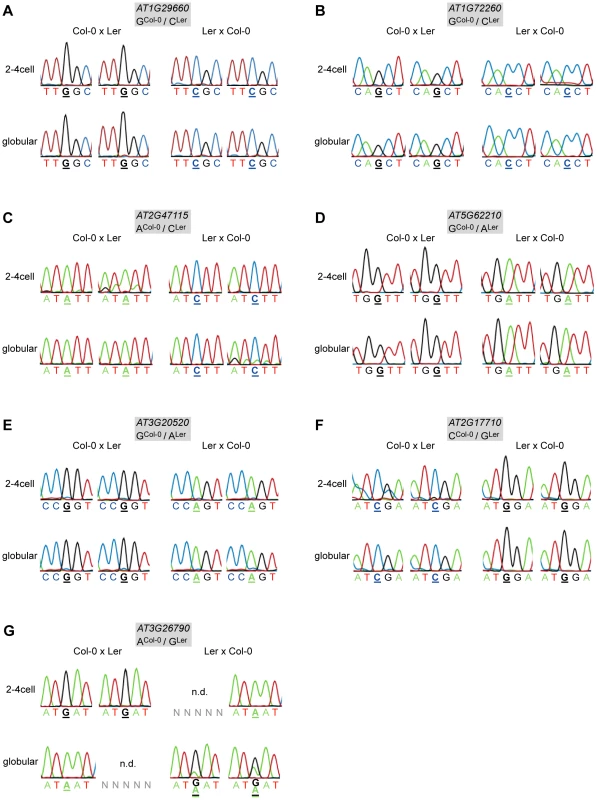
Reciprocal hybrid embryos were isolated at 2.5 DAP (2–4 cell embryos) and at 4 DAP (globular embryos) and allele-specific expression was analyzed by RT-PCR and Sanger sequencing. The direction of the cross is indicated on top of each panel, the embryonic stage on the left. Two replicates were analyzed for each stage and cross, which is represented by two individual sequencing chromatograms. The analyzed gene and the polymorphism between Col-0 and Ler are indicated in the grey box atop each panel. Furthermore, the polymorphic nucleotide is displayed in bold and underlined below each chromatogram. n.d. indicates that the transcript could not be amplified from the specific embryo replicate sample. (A) AT1G29660. (B) AT1G72260. (C) AT2G47115. (D) AT5G62210. (E) AT3G20520. (F) AT2G17710. (G) FUS3 (AT3G26790). Of the six analyzed candidate PEGs, only FUSCA3 (FUS3, AT3G26790) showed consistent monoallelic expression at the 2–4 cell stage, but not at the globular stage, at which expression was either maternal or biallelic, depending on the direction of the cross (Figure 1G). In two replicates, FUS3 could not be detected at all, which might indicate that FUS3 is expressed at a low level in the embryo; even below detection level in some samples. Two other candidate PEGs were found to be expressed from both parents (MEIDOS (AT2G20160) and AT1G63260; Figure S5), while the three remaining candidate PEGs could not be detected at all in our embryonic cDNA libraries (Table S1).
In summary, we could confirm eleven MEGs and one PEG that show parent-of-origin-dependent, monoallelic gene expression in the embryo. For nine MEGs and the PEG no transcripts were found in the gametes suggesting de novo expression and, therefore, true imprinted expression in the embryo [35], [36]. One MEG, AT1G72260, is expressed in the gametes already, and another MEG, AT2G47115, is not represented on the microarrays, such that no statement can be made about de novo transcription in the embryo [35], [36]. Nevertheless, these findings show that genomic imprinting in the embryo is more widespread than commonly thought and that imprinted expression of the maize mee1 gene is not an exceptional case.
Monoallelic Expression Patterns Are Not Due to Sporophytic or Endosperm Contamination
The embryo is surrounded by the triploid endosperm, and both embryo and endosperm are embedded in the maternal, sporophytic seed coat. Therefore, isolating embryos devoid of debris from surrounding sporophytic tissues and careful control of maternal tissue contamination is an important issue [34]. While the PEG cannot be derived from seed coat contamination, substantial contamination with maternal sporophytic tissue could explain the observed maternal expression patterns of the confirmed MEGs. Our initial samples were prepared from embryos diluted in a large volume and additionally washed one time. Although all samples were devoid of visible debris at collection, we produced two additional embryonic cDNA libraries to rule out the possibility of sporophytic contamination: following reciprocal crosses between Col-0 and Ler, we isolated 2–4 cell embryos, but washed them six times (6×) instead of one time (1×), a procedure that should result in the removal of all possible non-embryonic transcripts but may also degrade embryonic transcripts.
First, we assessed the quality of the 6× washed embryonic cDNA libraries compared to the 1× washed libraries by performing RT-PCR using primers amplifying ACTIN 11 (ACT11) and WUSCHEL-RELATED HOMEOBOX 9 (WOX9), an embryo-specific gene (Figure 2A). We amplified both control genes in the 1× and the 6× washed embryonic cDNA libraries derived from the Col-0 x Ler cross (Figure 2A). We could only weakly amplify ACT11 and WOX9 in the 6× washed Ler x Col-0 library (Figure 2A). This indicates a lower cDNA library quality of this sample, likely due to RNA degradation during the washes. Second, we confirmed the absence of seed coat and endosperm expressed genes, for which no embryo expression has been detected (based on literature, our own and available online embryo expression data) in our embryo samples by RT-PCR (Figure 2B and 2C). As seed coat markers, we tested TRANSPARENT TESTA 10 (TT10; [37]) and AT5G42530 (our own analysis based on [33]), and as endosperm markers, we tested FWA [16], AGAMOUS-LIKE 46 (AGL46; [38]), and AGAMOUS-LIKE 62 (AGL62; [39]). All genes showed distinct expression in a combined seed coat/endosperm sample, but were absent from all 1× and 6× washed embryo samples, suggesting that our previous washing regime is sufficient to remove non-embryonic transcripts.
Fig. 2. Quality, purity and allele-specific expression analyses of extensively washed embryonic control samples. 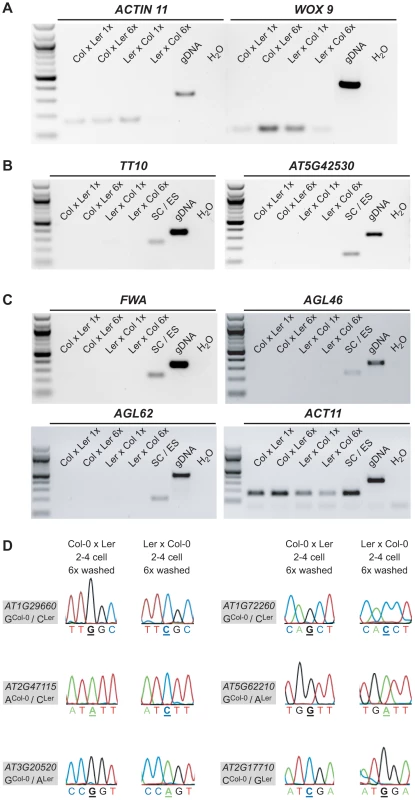
(A) RT-PCR amplifying ACT11 and WOX9, an embryo-specific gene, from 1× washed and 6× washed reciprocal, embryonic cDNA libraries (2–4 cell stage) using 32 PCR cycles. Genomic DNA was used as positive and water as negative control. (B) RT-PCR amplifying TT10 and AT5G42530, two seed coat-specific genes ([37] and our own analysis based on [33]), from 1× washed and 6× washed reciprocal, embryonic cDNA libraries (2–4 cell stage) and a combined seed coat/endosperm sample using 28 and 32 PCR cycles, respectively. Genomic DNA was used as positive and water as negative control. (C) RT-PCR amplifying FWA, AGL46 and AGL62, three endosperm-specific genes [16], [38], [39], from 1× washed and 6× washed reciprocal, embryonic cDNA libraries (2–4 cell stage) and a combined seed coat/endosperm sample using 28, 32 and 32 PCR cycles, respectively. Genomic DNA was used as positive and water as negative control. In addition, ACT11 was amplified again including the combined seed coat and endosperm sample. (D) Allele-specific expression analysis of AT1G29660, AT1G72260, AT2G47115, AT5G62210, AT3G20520, and AT2G17710 in the 6× washed embryonic cDNA libraries. The analyzed gene and the polymorphism between Col-0 and Ler are indicated in the grey box and the direction of the cross and the stage on top of the panel. The polymorphic nucleotide is displayed in bold and underlined below each chromatogram. We could readily detect the transcript of seven MEGs (AT1G29660, AT1G72260, AT2G47115, AT5G62210, AT3G20520, AT2G17710, AT3G21500) in both 6× washed 2–4 cell embryo samples and confirmed their monoallelic expression pattern by Sanger sequencing analysis (Figure 2D, Figure S6A). For the other four MEGs (AT2G01520, AT1G20680, AT5G51950, AT1G29050) we could only detect expression in the 6× washed Col-0 x Ler embryo sample, but not in the 6× washed Ler x Col-0 embryo sample, likely due to lower cDNA quality of this particular library. Nevertheless, Sanger sequencing analysis confirmed that those four MEGs show monoallelic expression in the 6× washed Col-0 x Ler sample (Figure S6A) and were thus classified as partially confirmed MEGs. Only one candidate gene that seemed to be imprinted in the 1× washed libraries could not be detected anymore in both 6× washed embryo samples, suggesting that this transcript is either lowly abundant in the embryo and/or was degraded during the extensive 6× washing procedure (AT4G11960, Figure S6C).
In conclusion, sequencing analysis of extensively washed, reciprocal 2–4 cell embryonic cDNA libraries confirmed seven MEGs and partially confirmed four MEGs as imprinted genes in the embryo, strongly suggesting that their monoallelic expression is not caused by seed coat or endosperm contamination.
MEG and PEG Reporter Lines Show Imprinted Expression in the Embryo
In order to demonstrate parent-of-origin-dependent, monoallelic expression in the embryo using an independent assay, we cloned the promoter of seven MEGs (AT1G29660, AT1G72260, AT2G47115, AT5G62210, AT3G20520, AT2G17710, AT3G21500) and the single identified PEG (AT3G26790) as transcriptional fusions with the bacterial uidA reporter gene encoding ß-glucuronidase (GUS; [40]). We screened 24 independent T1 lines for all 8 constructs for expression in the seed. Except for the PEG reporter pFUS3::GUS, all MEG reporters exhibited fairly strong staining in the seed coat, making embryo expression analyses on whole seeds impossible (Figure S7). To assess whether the gene-of-interest is indeed expressed in the embryo as indicated by our previous analyses, we isolated self-fertilized embryos at early and late stages of two to three independent lines for each MEG construct in the T1 generation and stained them for GUS activity on slides. Whereas 6 lines showed strong expression in the embryo (Figure S8), one line (pAT3G21500::GUS) showed only very weak and hardly detectable GUS staining in the embryo (Figure S9) and was, therefore, not used for further analyses.
To assess whether the promoter-GUS reporters are imprinted in the embryo, we performed reciprocal crosses between two independent reporter lines of each of the six strong MEG constructs (T2 generation) with wild-type plants (Col-0). F1 embryos were isolated at 2.5 DAP (∼2–4-cell embryos) and 4 DAP (∼globular embryos), and stained on slides for 4 days before analyzing them for GUS expression. For the MEG reporters, we expected to see the GUS signal only in the embryos that inherited the reporter gene maternally and not in the embryos that received the reporter gene from the pollen donor. We found that three of the six GUS-reporter lines are fully imprinted showing exclusive maternal expression (pAT1G72260::GUS, pAT2G47115::GUS, pAT3G20520::GUS, Figure 3B, 3C, and 3E), while the remaining three GUS-reporter lines show a very strong bias towards maternal expression (pAT1G29660::GUS, pAT5G62210::GUS, pAT2G17710::GUS, Figure 3A, 3D, and 3F). The PEG reporter line pFUS3::GUS shows very strong and embryo-specific expression starting from the 8-cell embryo stage both in whole-mount seed staining assays (Figure S7H) and after embryo isolation (Figure S10C). Since GUS expression for this gene seems to be very specific to the embryo (Figure S7H), no embryo isolation was necessary after reciprocal crosses to assess whether pFUS3::GUS shows imprinted expression. Yet, in contrast to the MEG reporter lines, we detected embryonic GUS expression no matter from which parent the reporter was inherited (Figure S10A and S10B). This suggests that the upstream regulatory region of FUS3 is not sufficient to confer imprinted paternal expression in early Arabidopsis embryos. However, pFUS3::GUS activity was first detected at 3 DAP corresponding to the (4-)8 cell stage (Figure S10A). Thus, the level of gene expression at earlier stages, where we actually detected exclusively paternal expression using allele-specific expression analysis (Figure 1G), might be below detection level in this assay.
Fig. 3. Parent-of-origin-dependent expression of MEG reporter lines in isolated embryos. 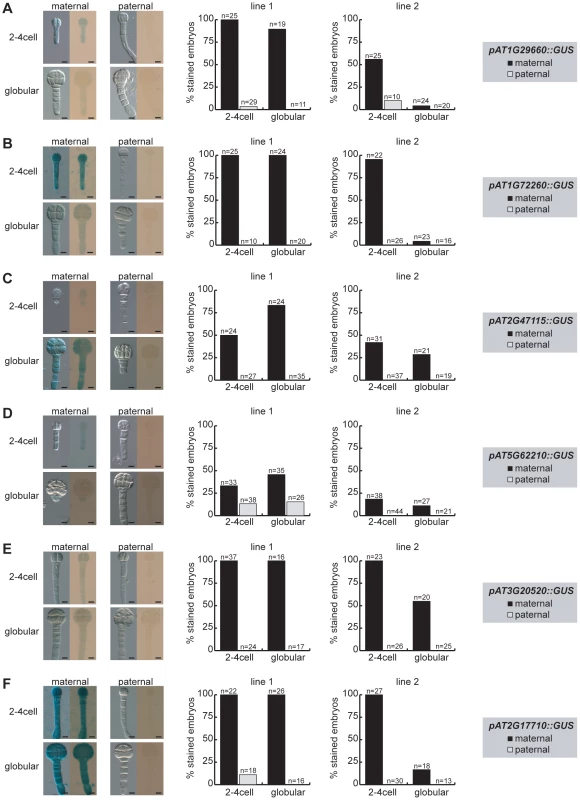
The MEG reporter lines were reciprocally crossed to wild-type Col-0 plants and embryos were isolated at 2.5 DAP (2–4 cell embryos) and at 4 DAP (globular embryos) prior to GUS staining. Embryos were stained on slides for 4 days at 37°C and then analyzed for GUS expression using bright-field microscopy. For each line, two independent transgene insertions (line 1, line 2) were analyzed and quantified for maternal (black columns, maternally inherited reporter gene) and paternal expression (grey columns, paternally inherited reporter gene) and are displayed separately (middle panel and right panel). Embryo pictures of line 1 are shown on the left, always showing a DIC picture and a bright-field picture of each stage and direction of cross. The embryonic stages, maternal or paternal GUS reporter expression and the analyzed reporter line are indicated and the numbers of the quantified embryos are shown above each column. Scale bar = 10 µm. (A) pAT1G29660::GUS. (B) pAT1G72260::GUS. (C) pAT2G47115::GUS. (D) pAT5G62210::GUS. (E) pAT3G20520::GUS. (F) pAT2G17710::GUS. In conclusion, all the MEG and PEG reporter lines cloned and analyzed are expressed in the embryo (Figure 3, Figure S8, Figure S10). Moreover, all MEG reporter lines are either fully imprinted or show a strong bias for maternal expression (Figure 3). The upstream regulatory sequences that were cloned are, thus, sufficient to confer imprinted expression during early stages of embryogenesis. On the other hand, a few loci, such as FUS3, might require additional regulatory elements for imprinting. Finally, while all MEG reporter lines are expressed in the seed coat (Figure S7) in addition to the embryo (Figure S8), they are clearly regulated by genomic imprinting in the embryo itself (Figure 3).
PRC2 but Not MET1 Is Involved in Regulating Genomic Imprinting in the Arabidopsis Embryo
In order to investigate how genomic imprinting is regulated in the embryo, we crossed mutants affecting imprinting regulators to wild-type parents of a distinct accession. DNA-methylation and histone modification, in particular H3K27me3 mediated by the PRC2, have both been shown to regulate genomic imprinting (for review see [10]). Therefore, we crossed the fertilization-independent endosperm (fie) mutant, in which PRC2-mediated repression is fully abolished, reciprocally to wild-type plants, and used the met1-3 mutant, disrupting the maintenance of DNA-methylation in the CG-context, as a male donor to pollinate wild-type plants. MET1 was thus far only implicated as a paternal repressor of imprinted loci, whereas PRC2 contributes to the regulation of MEGs (e.g. MEA) and PEGs (e.g. PHE1) (for review see [10]). Therefore, we crossed fie mutants reciprocally but the met1-3 mutant only as a pollen donor. As before, we isolated embryos from the resulting F1 hybrid seeds and proceeded with RNA extraction and library amplification, creating mutant embryonic cDNA libraries (i.e. fie/FIE x Ler, Ler x fie/FIE, and Ler x met1-3/MET1).
We tested the allele-specific expression pattern of the eleven MEGs and the PEG in the Ler x met1-3/MET1 mutant library (2–4cell stage) and found that all MEGs were still monoallelically expressed (Figure 4 and Figure S11). Thus, in contrast to some of the well-studied maternally expressed imprinted loci in the Arabidopsis endosperm (i.e. FIS2, FWA), disrupting paternal DNA-methylation maintenance does not appear to affect the imprinted expression of the embryonic MEGs at all (Figure 4A and 4B and Figure S11). Yet, the PEG could not be detected anymore in the Ler x met1-3/MET1 mutant cDNA library, indicating a potential involvement of paternal MET1 in activating the paternal FUS3 allele (Figure 4C). Since half of the embryos are expected to inherit an unaffected FUS3 allele from wild-type MET1 pollen, the remaining FUS3 transcript level may just be below our detection limit. This is in agreement with our earlier findings that, even in wild-type embryos, we are at the limit of detection for FUS3 using this assay (Figure 1).
Fig. 4. Effect of PRC2 and MET1 function on imprinted expression in the embryo. 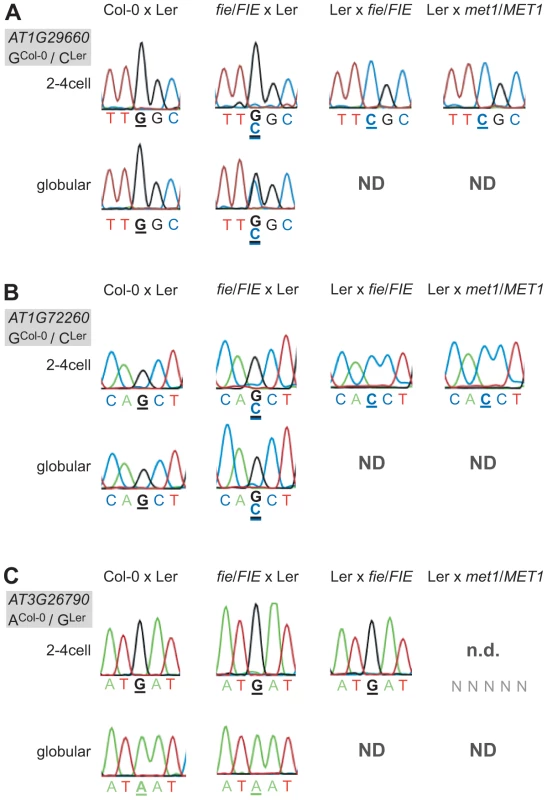
Mutant embryonic samples were generated and the confirmed MEGs and the PEG were analyzed for derepression of the silent allele. Heterozygous fie mutants (in Col-0) were crossed maternally and paternally and heterozygous met1-3 mutants (in Col-0) were crossed paternally to wild-type Ler as indicated above the chromatograms. To simplify the reading of the graph, we show again the wild-type situation of one replicate from the Col-0 x Ler cross in Figure 1. Embryos were isolated at 2.5 DAP (2–4 cell embryos) and at 4 DAP (globular embryos, only for the cross fie/FIE x Ler). The embryonic stage is indicated on the left, the analyzed gene and the polymorphism between the mutant (all in Col-0 background) and the wild-type allele (Ler) is shown in the grey box beside each panel. Furthermore, the polymorphic nucleotide is displayed in bold and underlined below each chromatogram. ND, not determined: indicates that the sample was not available; n.d., not detected: indicates that the transcript was not detected. (A) AT1G29660. (B) AT1G72260. (C) FUS3 (AT3G26790). Disrupting PRC2 function by crossing fie mutants maternally or paternally did have an effect on two MEGs and on the PEG. A maternal fie mutation was able to derepress the paternal alleles of AT1G29660 and AT1G72260 (Figure 4A and 4B). The alleles were slightly derepressed at the 2–4 cell stage in both cases, but were differently affected by the fie mutation at the globular stage. While AT1G29660 was fully derepressed and biallelically expressed (Figure 4A), the paternal allele of AT1G72260 retained a low expression level (Figure 4B). While only one replicate was tested for each cross, the data were fully consistent for the two stages we analyzed. Interestingly, a paternal fie mutation induced maternal expression of the PEG FUS3, while abolishing its paternal expression (Ler x fie/FIE, 2–4cell stage, Figure 4C). These results indicate that paternal FIE activity is required – likely indirectly – to activate the paternal FUS3 allele and that paternal FUS3 may negatively control the maternal FUS3 allele after fertilization. Interestingly, the FUS3 promoter contains two RY motifs, 484 bp and 1577 bp upstream of the start codon, respectively. FUS3, a B3-domain transcription factor, directly binds to the RY motif, which is present in many seed specific promoters [41]. This further supports the hypothesis that FUS3 autoregulates itself.
In conclusion, the PRC2 seems to be involved in regulating genomic imprinting in the embryo. Maternal PRC2 activity maintains the repression of the silent paternal allele of two MEGs after fertilization, while paternal PRC2 function together with paternal MET1 is somehow implicated in the activation of the paternal allele of the PEG FUS3. In contrast, paternal MET1 activity does not seem to play a role in regulating genomic imprinting of the eleven MEGs in the embryo. Our result indicates that there must be additional, so far unknown factors regulating genomic imprinting in the embryo.
Disrupting Embryonic MEGs or the PEG Has No Effect on Seed Viability or Early Embryogenesis
A few of the genes imprinted in the endosperm, such as MEA and FIS2, show parent-of-origin-dependent seed abortion when mutated [42], [43]. To reveal a potential role of the confirmed embryonic MEGs and the PEG during embryogenesis and seed development, we analyzed T-DNA insertions if available (Table S2). We assessed seed viability by dissecting siliques and analyzing seed set of 16 to 24 individuals of a genotyped segregating population. None of the analyzed T-DNA insertion lines showed reduced seed set (Table S2). This suggests that the MEGs and the PEG we identified play only a subtle role during embryogenesis or do not show an effect on seed development due to redundancy or an incomplete disruption of the corresponding gene. Furthermore, we tried to assess more subtle effects by dissecting and clearing seeds of heterozygous mutant individuals, followed by morphological analysis of early embryogenesis. However, we could not observe any obvious patterning defects or other developmental aberrations in the lines analyzed (Table S2). Interestingly, homozygous fus3 embryos show a phenotype late in seed development, namely a prolonged cell division phase in the embryo throughout seed maturation [44]. Yet, the late occurrence of this phenotype is unlikely to be caused by the early imprinted state of FUS3, and the zygotic recessive nature of the phenotype fits well with our observation that FUS3 has a biallelic expression late in embryogenesis.
Taken together, we did not identify any obvious fertility or embryo patterning phenotypes when analyzing the available T-DNA insertion lines disrupting the imprinted embryonic genes we identified.
Most MEGs and the PEG Are Contributed in a Parent-of-Origin-Dependent Manner in Different Ecotypes and at Later Stages
To determine whether our embryonic MEGs and the PEG are (i) indeed expressed in other embryonic samples, and (ii) monoallelically contributed in other Arabidopsis accessions and at different embryonic stages, we compared our data with the results of three recent studies. Xiang and colleagues isolated embryos from Col-0 wild-type plants manually, from zygote up to mature embryos, and performed transcriptome analysis using microarrays [45]. Eight of our MEGs and the PEG are expressed clearly above their background level at the 4-cell embryo and the globular embryo stage, whereas three MEGs are just at background level, which is considered not or lowly expressed (Table 1, Table S3). In contrast, these three MEGs are clearly expressed in young hybrid embryos from reciprocal crosses between the accession Col-0 and Cape Verde Islands (Cvi; [34]; Table 1, Table S4). This finding suggests that either the microarray technique is not sensitive enough to detect low expression levels, or that these three MEGs are stronger expressed in hybrids than in self-fertilized embryos.
Tab. 1. Embryonic expression and parent-of-origin expression of the confirmed MEGs and the PEG in other studies. 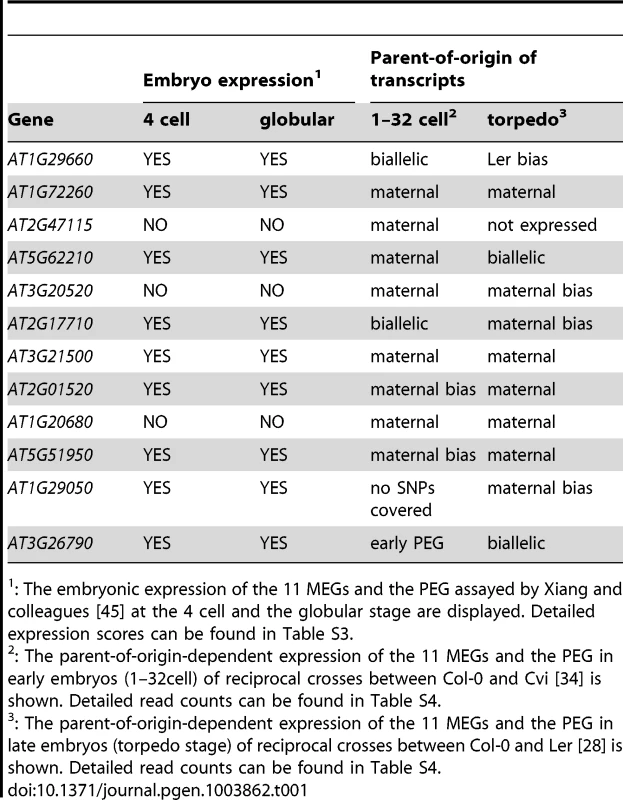
: The embryonic expression of the 11 MEGs and the PEG assayed by Xiang and colleagues [45] at the 4 cell and the globular stage are displayed. Detailed expression scores can be found in Table S3. Furthermore, we compared parent-of-origin-dependent expression of our embryonic MEGs and the PEG to the parent-of-origin-dependent expression in (i) early Col-0 x Cvi embryos (different accession, similar stage; [34]), and (ii) late torpedo-stage Col-0 x Ler embryos (same accessions, but later stage; [28]). Interestingly, eight MEGs and the PEG show a monoallelic or at least a clearly biased parent-of-origin-dependent expression in early Col-0 x Cvi embryos confirming the results of our study (Table 1, Table S4). This suggests that genomic imprinting of those loci is conserved between the Col-0, Ler, and Cvi accessions. However, analyses of additional accessions would be required to clearly demonstrate locus - versus allele-specific imprinting at these loci. Two MEGs are biallelically expressed and one is low expressed and no SNPs were covered by reads, making a parent-of-origin analysis impossible (Table 1, Table S4). Yet, when looking at the same accessions later in development (torpedo stage), we found that eight MEGs are still expressed maternally or with a maternal bias (albeit most were covered by few reads only). One MEG shows a Ler bias, one MEG and FUS3 are biallelically expressed, and one MEG is not expressed anymore (Table 1, Table S4). This indicates that at some loci imprinted expression is lost already during embryogenesis, whereas at other loci this happens later during or even after embryogenesis. This analysis shows that the majority of the analyzed loci that are imprinted in the embryo is also imprinted in different accessions [34] and maintains a parent-of-origin-dependent expression later during embryo development [28].
Embryonic Imprints Are Erased or Ineffective in the Seedling
Parent-of-origin-dependent expression seems to be maintained until late stages of embryogenesis, at least for some of the loci analyzed [28]. Eventually the imprint has to be erased, the very latest during gametogenesis. To address this question we reciprocally crossed Col-0 and Ler, grew F1 seedlings up to the 4-leaf stage (8 days after sowing), and produced hybrid F1 seedling cDNA libraries. We then performed allele-specific expression analysis using the MEG and PEG assays. 9 of 11 MEGs and FUS3 could be amplified from the seedling library and are thus expressed in the seedling. The two genes that were not amplified (AT2G47115, AT3G21500) are either not expressed in the seedling or below the detection level of our assays. Sanger sequencing revealed that all MEGs and FUS3 are expressed from both parents in F1 hybrid seedlings (Figure 5). This suggests that the imprint is erased late during embryogenesis or early during vegetative development, but long before flowering and the initiation of reproductive development. Alternatively, the imprint might persist in the seedling but is ignored by the transcriptional machinery, leading to the observed biallelic expression pattern. For instance, some loci may be transcribed from an imprinted promoter in the embryo and from an alternative, non-imprinted promoter in the seedling, or the imprinting control elements may not be accessible any more to the corresponding trans-acting factors in seedlings.
Fig. 5. Allele-specific expression analysis of confirmed MEGs and the PEG in hybrid F1 seedlings. 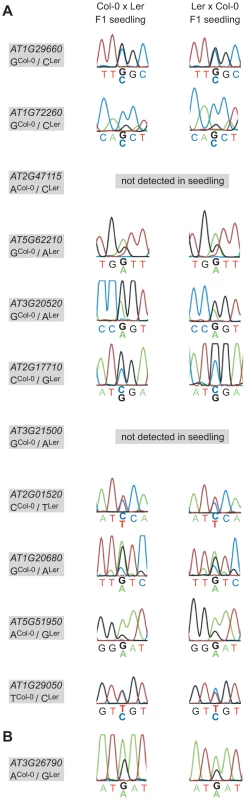
The allele-specific expression of the eleven confirmed MEGs (A) and the confirmed PEG (B) was assessed in reciprocal F1 hybrid seedling cDNA libraries (8 days after sowing). Nine MEGs and the PEG show biallelic expression in the seedling and two MEGs were not detected in the seedling samples, as indicated (AT2G47115, AT3G21500). The analyzed gene and the polymorphism between Col-0 and Ler are specified in the grey box beside each panel, whereas the direction of the cross is indicated on top. Discussion
Parent-of-Origin-Dependent, Monoallelic Gene Expression Is Not Restricted to the Endosperm in Arabidopsis
Recently, two studies independently identified monoallelically-derived transcripts in early Arabidopsis embryos when assessing the genome-wide parental contribution to plant embryogenesis [33], [34]. Analysis of reciprocal Col-0 x Cvi F1 embryos identified more than 100 potentially imprinted or maternally/paternally deposited transcripts in the Arabidopsis embryo [34], whereas we identified 50 potential MEGs and 30 potential PEGs by assessing the allele-specific transcriptome of Ler x Col-0 embryos [33]. We focused on 18 MEG and six PEG candidates that showed strong expression in the embryo but no expression in the gametes. We could confirm eleven MEGs expressed at the 2–4 cell and globular embryo stage and one PEG at the 2–4 cell stage using RT-PCR and Sanger sequencing on replicated and reciprocal hybrid, embryonic cDNA samples (Table 2, Figure 1, Figure S4). In addition, reporter gene analysis of seven MEGs and one PEG independently confirmed embryonic expression and six MEG reporter lines were fully imprinted or expressed with a strong maternal bias in the embryo (Table 2, Figure 3, Figure S8, Figure S9). Furthermore, absence of most MEG or PEG transcripts in the gametes strongly suggests de novo expression of the genes in the embryo, although the gametic trancriptome was only assessed by microarrays, which are less sensitive than RNA-Seq (Table 2, Table S1, [35], [36], [46]. However, the fact that most of those genes were found expressed in a microarray study of embryogenesis – while being absent in gametes - supports their de novo expression after fertilization ([45]; Table 1, Table S3). The MEGs were not only expressed in the embryo, but also in the surrounding seed coat (Figure S7). Therefore, examining potential maternal sporophytic contamination was essential to our study. To do so, we produced two extensively washed (6×), reciprocal 2–4 cell embryonic cDNA libraries that should be completely devoid of all potential contamination. Allele-specific expression analysis in those samples revealed pure monoallelic expression of the 11 MEGs and the PEG in the embryo, affirming their imprinted expression patterns (Figure 2, Figure S6). In addition, we isolated embryos derived from reciprocal crosses between MEG reporter lines and wild-type plants prior to GUS staining. Thus, the observed GUS signal is embryo-specific and cannot be due to diffusion from the seed coat.
Tab. 2. Overview of the embryonic MEGs and the embryonic PEG. 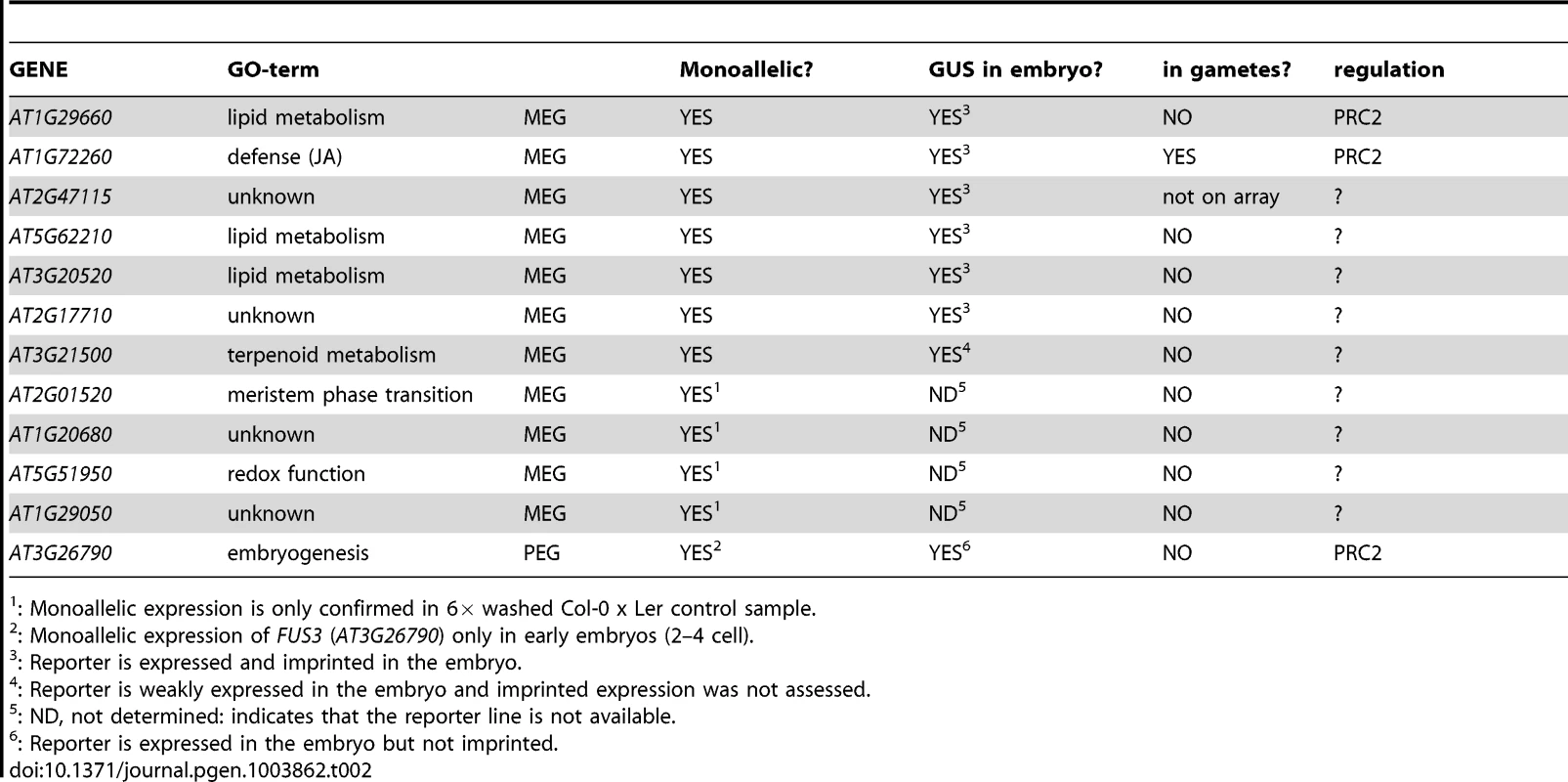
: Monoallelic expression is only confirmed in 6× washed Col-0 x Ler control sample. Apart from the controversially discussed MEA and PHE1 genes [11], the maize mee1 gene is clearly imprinted in plant embryos [7]. Other recently published studies performed genome-wide analysis of parent-of-origin allelic expression in endosperm and embryos. Hsieh and colleagues identified 37 MEGs and one PEG in the Arabidopsis embryo 7–8 DAP, but discarded them because of the possibility that these data may be due to contamination by endosperm and/or maternal tissues [27]. Gehring and coworkers found 17 MEGs and one PEG in Arabidopsis embryos 6 DAP (torpedo stage) but none was further analyzed [28]. In rice, a similar study identified seven putative MEGs in the embryo and confirmed one, Os10g05750, using RT-PCR and Sanger sequencing, but its expression status in the gametes was not assessed [8]. Furthermore, Waters and colleagues identified 29 MEGs and 9 PEGs from a dataset of maize embryos 14 DAP, but did not follow up these findings with additional experiments [31]. In all studies, many of the embryonic MEGs and PEGs are also expressed and imprinted in the endosperm, which leads the authors to attribute their pattern of expression to endosperm contamination, and do not consider it a real, intrinsic and embryo-specific feature. Similarly, the confirmed eleven MEGs of this study are not only expressed in the embryo, but also in the seed coat, the seedling and/or the endosperm (Figure S7). AT1G20680 even passed the statistical criteria used in two independent studies and is thus considered an imprinted gene in the endosperm [28], [29]. Many of our MEGs and the PEG are expressed in the endosperm and almost all MEGs are maternally expressed in the endosperm at 6 DAP (Table S5). Yet, allele-specific expression in stringently washed embryo samples and the analysis of reporter constructs strongly suggest that the embryonic parent-of-origin-dependent expression pattern is not due to contamination.
In contrast, we cannot fully exclude the possibility that mRNAs are transported from the endosperm to the embryo. To our knowledge mRNAs can only be transported symplastically via plasmodesmata [47]. However, this seems highly unlikely because the embryo is symplastically isolated from the endosperm during early seed development, due to the presence of cuticular material around the embryo ([48] and references therein). Even in the few cases where plasmodesmata between the suspensor and the endosperm have been described - only in the Fabaceae and Crassulaceae families - they are associated with electron-dense material, probably blocking the plasmodesmata [48]. Taken together, we expect that a careful reexamination of the embryonic MEG and PEG candidates found in other studies could confirm additional genes regulated by genomic imprinting in the embryo, as it is the case for the genes reported in this study.
Comparison of this study's 80 imprinted candidates with all embryonic MEG and PEG candidates identified in different studies reveals no genes to be common in all studies and only three overlapping genes between two studies (AT2G01520, AT1G49450 and AT1G57800; [27], [28], [34]). Similarly, the overlap between four recent studies [27]–[30] identifying imprinted genes in the endosperm of Arabidopsis is minor, with only 20 genes of more than 300 being shared between two studies analyzing the same ecotypes and similar developmental stage [49]. The low number of common genes may be explained, at least to some extent, by the use of different accessions and the analysis of different developmental stages. Thus, some of the potential MEGs and PEGs, in both the embryo and endosperm, seem to be imprinted allele-specifically rather than locus-specifically and/or imprinting could be specific to a given developmental stage. In addition, different statistical procedures to select potentially imprinted genes may also contribute to the discrepancies [50]. Analyzing the datasets prior to stringent filtering increases the overlap [29], as well as treating two different datasets with the same statistical pipeline [28]. Taken together, the recent rapid development of high-throughput transcriptome sequencing allows large-scale identification of imprinted genes in different organisms and tissues. Yet, the obvious discrepancy between different datasets shows that in-depth analyses and substantial validation of imprinted candidate genes using alternative methods is necessary.
In conclusion, we show that at least nine MEGs and one PEG are regulated by genomic imprinting in the embryo and, in addition, two MEGs display steady-state monoallelic expression in the embryo. Likely, the list of MEGs and PEGs in plant embryos will expand in the future by validating additional candidates or by high-throughput sequencing of additional embryonic samples.
Delayed Paternal Gene Activation in the Embryo Is Distinct from Embryonic Imprinting
In a former study, we have shown that the embryo transcriptome of intraspecific (Ler x Col-0) hybrids showed an overall maternal dominance, albeit 66% of the genes were biparentally expressed [33], [51]. Although this wide-spread maternal dominance was recently challenged by a study using different accessions and amplification techniques [34], [51], our genetic analyses further demonstrated that the underrepresentation of paternal transcripts is regulated by a maternal siRNA-based silencing mechanism [33]. However, DNA methylation and PRC2-mediated repression, two epigenetic pathways often associated with imprinting, were not involved [33]. In addition, the vast majority of genes that were maternally dominant at the 2–4 cell stage have a higher paternal contribution at the globular stage. The biallelic control gene used in this study, AT1G02780, is a good example for a gene showing biased expression in 2–4 cell embryos but an equal parental contribution at the globular stage, a pattern that we confirmed using allele-specific expression analysis (Figure S1A, [33]).
In contrast, for all potentially imprinted genes identified in this study exclusively reads from one parent only were found in all stages and replicates (Table S1). In addition, for the 11 confirmed embryonic MEGs and the PEG we never detected any paternal or maternal SNP signatures, respectively, in the chromatograms of all samples and stages analyzed. This strongly suggests that they are indeed monoallelically expressed and not only maternally biased transcripts, as it is the case for the biallelic control gene that shows a low but clearly visible paternal SNP signature already at the 2–4 cell stage (Figure S1A). Similarly, three of six MEG reporter lines were completely silent if paternally inherited, whereas another three showed paternal expression but at a very low frequency at both stages analyzed, suggesting an imprinted expression pattern in the embryo (Figure 3). In contrast, the GUS marker lines analyzed by Autran and colleagues showed a much higher frequency of paternal expression already in 2–4 cell embryos and most were fully active by the globular stage [33].
In conclusion, delayed paternal genome activation is a genome-wide, gradual process leading to a maternal bias during early embryogenesis. In contrast, the embryonically imprinted genes described in this study are completely monoallelically expressed at both the 2–4 cell and globular stage of embryogenesis. Of course, maternally expressed imprinted loci will contribute to the maternal dominance in the transcriptome of early embryos [33]. However, maternally biased expression can also be due to the deposition of maternal transcripts produced prior to fertilization, and we currently do not know what fraction of these loci are de novo transcribed in a parent-of-origin-dependent way after fertilization, i.e. are regulated by imprinting [52].
PRC2 but Not MET1 Is Involved in Maintaining Repression at Silent Alleles
The regulation of monoallelic and parent-of-origin-dependent gene expression largely depends on differential DNA methylation of the parental alleles in mammals and of some imprinted loci in the plant endosperm [10]. We analyzed the effect of a paternal met1-3 mutation, known to derepress silent paternal alleles of FWA and FIS2 [16], [18] in the endosperm, on imprinted gene expression in the embryo. We did not find any effect of paternal met1-3 on the expression of the 11 MEGs, suggesting that MET1-mediated DNA-methylation in the CG context is not important for the regulation of embryonic MEGs (Figure 4, Figure S11). By contrast, met1-3 abrogated paternal FUS3 expression, indicating a role of MET1 in activating paternal FUS3 (Figure 4C). Similarly, expression of the paternal PHE1 allele is reduced if inherited by a met1 mutant pollen [53]. However, further studies are required to confirm this observation and to rule out that FUS3 is not just below detection level in this sample, as it was the case in two wild-type samples we analyzed (Figure 1G).
The second, well-established imprinting regulator is PRC2, which mediates H3K27me3 [10]. In fact, we found that in embryos lacking maternal FIE activity, the usually silent paternal alleles of two MEGs were derepressed (Figure 4A and 4B). Similarly, the imprinted genes MEA and AtFH5 are biallelically expressed in seeds with a maternal mutation in a PRC2 subunit [17], [19], [26]. In addition, the paternal MEA allele was shown to be partially derepressed by a paternal mutation in PRC2 components, which is not the case for the identified MEGs in the embryo [26]. However, this suggests that maternal PRC2 is involved in maintaining the silent state of paternal alleles of imprinted genes in both, the endosperm and the embryo. In addition, in embryos inheriting a paternal fie mutation, the expression pattern of the PEG was inverted: Instead of being paternally expressed, FUS3 seems to be solely maternally expressed (Figure 4C). Our result suggests that paternal FIE is somehow required to activate the paternal FUS3 allele, and that paternal FUS3 is involved in the repression of the maternal FUS3 allele just after fertilization. Interestingly, the presence of RY motifs in the FUS3 promoter suggests that FUS3 can indeed bind to its own promoter. Such a negative feedback regulation of the gene product on its own imprinted expression has also been described for MEA [17], [24], [26]. However, whereas paternal PRC2 seems to be involved in the activation of the paternal FUS3 allele, imprinted expression of PHE1 requires maternal PRC2, which in this case is involved in the repression of the paternal allele of PHE1 [9]. This indicates that the requirement of PRC2 for regulation of imprinted expression of PEGs differs between the two fertilization products at least for these two loci.
We could not find any effect of mutations in MET1 and FIE on 9 MEGs. Thus, other mechanisms must be involved in regulating parent-of-origin-dependent gene expression in the embryo. As proposed for the endosperm [29], asymmetric, non-CG DNA methylation could be involved in silencing the paternal alleles. However, asymmetric DNA methylation in the CHG context involves the SUVH4 methyltransferase KYP [54] and all of the identified MEGs and PEGs are still imprinted in the mutant kyp x Ler embryonic library ([33]; Table S1). Thus, asymmetric DNA methylation in the CHG context seems unlikely to play a role in regulating the remaining 9 MEGs. Any other epigenetic mark could account for, or contribute to, the imprinted expression pattern in the embryo. Especially histone modifications might be of importance since they are more readily reversible than DNA methylation [55] and different modifications have been associated with imprinted genes in maize [56].
Erasing and Resetting the Imprint
In mammals, imprinting marks are erased and reset during germ line development. Very early during embryogenesis, as the germ line is set aside, epigenetic marks are erased and re-established according to the embryos' sex [2], [3]. In plants, no germ line is set-aside, and the gametes develop very late from differentiated sporophytic cells. In the endosperm, one-way control of imprinting is sufficient, since the it does not contribute to the next generation. In contrast, imprints on embryonic MEGs and PEGs have to be erased and reset. The only well-studied, imprinted gene in the plant embryo is the maize gene mee1 [7]. Interestingly, both alleles are fully methylated in the gametes and the maternal allele gets specifically demethylated in the zygote, indicating an additional, yet undiscovered primary imprinting mark. During embryogenesis the maternal allele continuously regains methylation and consequently becomes silent [7]. Thus, we do not know whether remethylation is cause or consequence or even involved in resetting the imprint, and we cannot speculate about the time of imprint erasure. In rice, the monoallelic expression pattern of Os10g05750 is maintained throughout development in the endosperm, but in the embryo, Os10g05750 starts to be expressed biallelically from 8 DAP. This suggests erasure of the potential imprint during late embryogenesis [8].
In Arabidopsis, we found that all MEGs and the PEG show biallelic (n = 10) or no (n = 2) expression in the early seedling; thus, the imprint must be erased – or become ineffective - either late in embryogenesis or very early during vegetative development. When analyzing the expression pattern of the 11 embryonic MEGs and the PEG in the allele-specific dataset of torpedo-staged embryos [28], we found that most genes are still expressed monoallelically or with a parental bias (n = 8), although at very low levels. Thus, complete erasure of the imprint seems to occur after the torpedo stage for most of the MEGs. Alternatively, the requirements for transcription of these genes might be different in seedlings and embryos, and the primary imprints might persist but do not control monoallelic expression any more later in development.
However, how an imprint is erased during late embryogenesis and reset during gametogenesis is unknown. In the case of the two MEGs where the repression of the maternal allele is maintained by PRC2 - likely by the seed-specific MEA-FIS2 complex [57] - it is tempting to speculate that, with decreasing expression of MEA during seed development [24], activity of the seed-specific PRC2 is decreasing and the H3K27me3 imprint might get lost by passive dilution during embryogenesis. However, a detailed analysis of epigenetic marks at embryonic MEG and PEG loci in gametes, embryos, and vegetative tissues would be required, which is extremely challenging due to the limited accessibility of gametic and embryonic tissue. In the future, advanced approaches will identify the primary imprinting mark(s) in the embryo, which will shed light on the yet unknown mechanism of erasing and resetting the imprints at embryonic MEGs and PEGs.
Biological Significance and Evolution of Genomic Imprinting in the Embryo
In placental mammals and in flowering plants, mutations in many imprinted genes cause growth defects in embryo and/or the nourishing tissue (i.e. placenta or endosperm) in a parent-of-origin-specific manner [43], [58]–[64]. Growth defects are consistent with a role of genomic imprinting in a parental conflict over resource allocation from the mother to the developing offspring [65]. Yet, when we analyzed available T-DNA lines disrupting embryonic MEGs, we could not find fertility, patterning, or obvious growth phenotypes. This might be a result of gene redundancy and/or of a subtle role these genes play during embryogenesis and seed development, which is not revealed in controlled and non-competitive laboratory conditions.
Notably, 5 of 11 MEGs have a role in metabolism, whereas 4 others are of unknown function (Table 2). In addition, all MEGs are expressed in the maternal seed coat and some MEG reporter lines and the PEG reporter line show a slightly biased expression towards the basal embryo and the suspensor (Figure 3, Figure S8, Figure S10). This suggests that embryonic MEGs might have a function at the interface between embryo and mother, possibly by linking seed coat metabolism and embryo metabolism and rendering the genes in the embryo under maternal control. This would be in line with the co-adaptation imprinting hypothesis: It predicts maternal expression of genes affecting mechanisms that are crucial at the maternal-offspring interface [66], [67]. In addition, the co-adaptation hypothesis predicts that the number of MEGs must be much higher than the number of PEGs, at least in species where the offspring develops within the mother. In fact, we find a large excess of MEGs (>90%), reminiscent of all other studies that analyzed parent-of-origin allelic expression in plant embryos. Both studies analyzing Arabidopsis embryos call 97% or 94% MEGs, respectively [27], [28]. In rice, only embryonic MEGs were called [8], and in maize embryos 76% of the imprinted candidates in the embryo show maternal expression [31]. Also in the Arabidopsis and the rice endosperm more MEGs than PEGs were identified, but the fraction of embryonic MEGs is still higher than the fraction of endosperm-specific MEGs [27]–[30]. This suggests that the co-adaptation hypothesis might be of importance for the evolution of genomic imprinting in the embryo, whereas parental conflict might drive evolution in the nourishing tissue, the endosperm. Nevertheless, the evolution of genomic imprinting is likely due to a combination of parental conflict, mother-offspring co-adaptation, and other factors, depending on the locus and the tissue of expression.
In conclusion, we describe and confirm parent-of-origin-dependent, imprinted monoallelic expression in the Arabidopsis embryo. PRC2 is involved in the regulation of parent-of-origin allelic expression at some loci analyzed, but by far not all, suggesting additional, yet undiscovered regulators of genomic imprinting in the Arabidopsis embryo. Probably, the imprint is erased late in embryogenesis or early in vegetative development since all genes are either expressed from both parental alleles or not at all in young seedlings. However, what the primary imprint is and when and how exactly it is reset, is currently unknown. Future research will likely confirm some of the embryonic MEGs and PEGs from other studies and will help elucidating the regulation, erasure and resetting of genomic imprints in the embryo.
Materials and Methods
Plant Material and Growth Conditions
Columbia-0 (Col-0) and Landsberg erecta (Ler) are the standard wild-type accession used in this study. We reciprocally crossed Col-0 and Ler to produce the hybrid embryonic samples and Col-0 was used for all Agrobacterium-mediated transformations in this study. The fie/FIE mutant (Col-0 background) used is SALK_042962 and the line has been described in detail in [68]. The met1-3/MET1 mutant (Col-0 background) used was first described in [69], and was only propagated heterozygously. It was assessed for full methylation at the 180 bp CEN-repeat by Southern blot analysis before crossing, indicating an unaltered epigenetic landscape and excluding uncontrollable, indirect effects [20]. The met1-3 genotyping assay is described in [20]. All plants were grown in a greenhouse chamber with 16 h light at ∼20°C and 8 h dark at ∼18°C with an average of 60% humidity. For crosses, plants were emasculated and pollinated 2 days later.
Calling Potentially Imprinted Genes in the Embryo
The dataset from [33] was analyzed and we called all genes that had a q-value bigger than 0.8 (strong mono-parental bias, mi>0.8 and pi>0.8 for MEGs and PEGs, respectively) in all sequenced samples (2–4 cell Ler x Col-0, 2–4 cell kyp/KYP x Col-0, globular Ler x Col-0), in the 2–4 cell samples only, or in the globular wild-type sample only. All filtered genes were then compared to the second replicate run and only kept if they still showed reads from one parent only [33]. We also accepted genes that were sequenced in the globular sample of replicate 1 only (SOLiD 2009) and were not detected in the second replicate (SOLiD 2010). This procedure yielded 50 potential MEGs and 30 potential PEGs (Table S1). Expression levels (coverage by covered base; [33]) and present/absent calls in the egg cell and sperm cell [35], [36] were used to prioritize the potential embryonic MEGs and PEGs. MEGs and PEGs being highly expressed and showing preferably absent calls in the gametes (egg cell or sperm cells) were selected for in-depth analysis (i.e. RT-PCR and Sanger sequencing).
Preparation of Hybrid Embryonic cDNA Libraries
Different wild-type accessions and/or mutant lines were reciprocally crossed as indicated in the main text, the figures, and figure legends to produce hybrid F1 seeds. For the 1×-washed wild-type embryonic samples we produced two independent biological replicates for each stage and direction of cross (i.e. 8 samples). The 2–4 cell embryos were isolated from seeds ∼2.5 days after pollination (DAP), whereas the globular embryo stage was isolated from seeds ∼4 DAP under our growth conditions. Embryo isolation was essentially performed as described in [33] with 5 additional washes after isolation for the extensively, 6× washed control samples (2–4 cell stage, reciprocally crossed). RNA was extracted using the Arcturus PicoPure RNA Isolation kit (Applied Biosystems) and the cDNA library amplified using the Ovation Pico WTA System (NuGEN) according to the manufacturer's protocol. As recommended by the Ovation Pico WTA System (NuGEN), we purified the cDNA libraries with the QIAquick PCR Purification Kit (QIAGEN) according to NuGEN's protocol. We used the Agilent 2100 Bioanalyzer (Agilent Technologies) to control cDNA library quality and measured quantity using Nanodrop. In addition, we controlled library quality and absence of genomic DNA contamination by RT-PCR amplifying ACT11 and WOX9, an embryo-specific gene [70]. Furthermore, we tested the purity of the embryonic libraries by RT-PCR amplifying TT10 and AT5G42530, two seed coat markers [37], and FWA, AGL46, and AGL62, three endosperm markers [16], [38], [39]. All primer sequences are specified in Table S6. The combined seed coat/endosperm sample consisted of apical pieces of seed coat and endosperm tissue after embryos were popped out and isolated. The cDNA library was produced like the embryonic libraries described above. In order to produce hybrid F1 seedling cDNA libraries and hybrid F1 seedling genomic DNA samples, we crossed Col-0 and Ler reciprocally, germinated the F1 hybrid seeds on plate and harvested them 8 days after sowing. Genomic DNA was extracted using the QiaQuick DNeasy kit (QIAGEN) and RNA was extracted using the NucleoSpin RNA Plant Kit (Machery-Nagel). Reverse transcription was performed as previously published [24].
RT-PCR and Sanger Sequencing
RT-PCR was performed on diluted cDNA libraries (4 ng/µl) by doing 28 to 34 cycles (94°C for 15 sec, 58°C for 20 sec, and 72°C for 30 sec) followed by 72°C for 5 min. We used Sigma Taq DNA Polymerase and PCR buffer from Sigma-Aldrich and a final concentration of 2 mM MgCl2, 0.2 mM dNTPs and 0.2–0.4 mM Primer. The resulting PCR product was analyzed on a standard DNA agarose gel and the remaining product was purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel). The purified PCR product was Sanger sequenced and the chromatograms analyzed at the site of the SNP between Ler and Col-0 to assess its parent-of-origin. All assays were tested for non-biased amplification of sequence fragments from both accessions using genomic DNA of F1 hybrid seedlings (Col-0 x Ler and Ler x Col-0). The quantitative nature of the Sanger approach was tested using the same protocol, with 30 PCR cycles and 1.0 µl of the following mixes of Ler/Col-0 genomic DNA (20 ng/µl): 1∶9, 1∶3, 1∶1, 3∶1 and 9∶1 (Ler:Col-0). All sequences of the used primer are specified in Table S6.
Reporter Lines: Cloning, Transformation, and Analysis
All GUS reporter lines were cloned using the pBGWFS7 vector (VIB, University of Gent), carrying a BASTA resistance gene (plant selection), a spectinomycin resistance gene (bacterial selection), and a Gateway-cloning cassette followed by eGFP and a uidA gene encoding ß-Glucuronidase (GUS) in frame. We amplified the upstream promoter region (from the previous gene until the start codon or a maximum of 2.5 kb of promoter sequence) of seven MEGs and one PEG containing the attB recombination sites in a two-step PCR reaction: First, we used chimeric primers comprising template-specific sequences plus the first 12 bases of the attB1 or attB2 sequence at the 5′-end. PCR was performed with the Phusion High-Fidelity DNA Polymerase (Finnzymes) and buffer, 0.2 mM dNTPs, 0.4 µM Primers using the attB adapter program 1 (98°C for 60 sec; 5 cycles of 98°C for 10 sec, 63°C for 20 sec, 72°C for 60–180 sec; 30 cycles of 98°C for 10 sec, 68°C for 20 sec, 72°C for 60–180 sec; 72°C for 300 sec). After analyzing the product on a gel, we used 1 µl of the 50× diluted first PCR product as template for the second PCR using attB adapter primers (attB1-adaptor: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′ and attB2-adaptor: 5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′) to complete the attB sites. PCR was performed as above using the attB adapter program 2 (98°C for 60 sec; 5 cycles of 98°C for 10 sec, 48°C for 20 sec, 72°C for 60–180 sec; 15 cycles of 98°C for 10 sec, 58°C for 20 sec, 72°C for 60–180 sec; 72°C for 300 sec). The resulting PCR product containing the promoter sequence and the complete attB recombination sites were precipitated with polyethylene glycol, and the BP reaction (using pDONR221) and LR reactions were performed according to the manufacturer's recommendations (Invitrogen). The resulting expression vectors were transformed into competent Agrobacterium tumefaciens (GV3101), which were used to transform Col-0 plants by floral dipping [71].
GUS Reporter Assays on Isolated Embryos
We first selected T1 lines strongly expressing the GUS reporter gene in the seed by staining young siliques overnight at 37°C after vacuum-infiltration (5–10 min) of the tissue in standard GUS staining solution (2 mM 5-bromo,4-chloro,3-indolyl-D-glucuronide (Biosynth-AG), 10 mM EDTA, 0.1% Triton X-100, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, 50 mM phosphate buffer pH 7.2). Strongly expressing lines were selected, and reciprocally crossed with wild-type Col-0 plants. We then isolated embryos 2.5 DAP (2–4 cell stage) and 3.5 to 4 DAP (globular stage) in GUS staining solution (as above but with 0.5 mM potassium ferro - and ferricyanide instead of 2.0 mM for higher GUS activity). We directly transferred the isolated embryos on a microscope slide, added fresh GUS staining solution, covered the embryos with a coverslip, and stained them without vacuum-infiltration for 4 d at 37°C in plastic boxes with high humidity to prevent drying of the samples. After 4 days we analyzed the isolated embryos for GUS reporter expression using bright-field microscopy (Leica DMR) to ensure maximum sensitivity for GUS detection.
Mutant Analysis
Available T-DNA insertion lines disrupting confirmed MEGs and PEGs (see Table S2) were ordered (2 lines/gene, if available). A mutant population (i.e. 24 individuals) was genotyped using primers flanking the insertion site (see Table S6, designed with the T-DNA primer design homepage http://signal.salk.edu/tdnaprimers.2.html) and the appropriate left border primer (for SALK lines: LBb1.3; for SAIL lines: Syg_LB1; for GABI lines: GBF_AC161_LB1; for FLAG lines: FL_LB4; for sequences see Table S6) using a standard PCR program (94°C for 15 sec, 58°C for 20 sec, and 72°C for 75 sec, 36 cycles). Then, mature siliques of each genotyped individual were opened to analyze the seed set. In addition, we harvested siliques at different developmental stages of one (usually heterozygous) mutant individual, dissected the seeds in modified Hoyer's solution (70% w/v chloralhydrate, 4% w/v glycerol, 5% w/v gum arabicum), and examined embryo patterning and development from the zygote to the torpedo stage using differential interference contrast (DIC) microscopy (Leica DMR).
Supporting Information
Zdroje
1. ReikW, WalterJ (2001) Genomic imprinting: parental influence on the genome. Nat Rev Genet 2 : 21–32.
2. BarlowDP (2011) Genomic imprinting: a mammalian epigenetic discovery model. Annu Rev Genet 45 : 379–403.
3. FengS, JacobsenSE, ReikW (2010) Epigenetic reprogramming in plant and animal development. Science 330 : 622–627.
4. FrostJM, MooreGE (2010) The importance of imprinting in the human placenta. PLoS Genet 6: e1001015.
5. FeilR, BergerF (2007) Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23 : 192–199.
6. JullienPE, BergerF (2009) Gamete-specific epigenetic mechanisms shape genomic imprinting. Curr Opin Plant Biol 12 : 637–642.
7. JahnkeS, ScholtenS (2009) Epigenetic resetting of a gene imprinted in plant embryos. Curr Biol 19 : 1677–1681.
8. LuoM, TaylorJ, SpriggsA, ZhangH, WuX, et al. (2011) A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet 7: e1002125.
9. KöhlerC, PageDR, GagliardiniV, GrossniklausU (2005) The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37 : 28–30.
10. RaissigMT, BarouxC, GrossniklausU (2011) Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23 : 16–26.
11. SpillaneC, SchmidKJ, Laoueillé-DupratS, PienS, Escobar-RestrepoJM, et al. (2007) Positive darwinian selection at the imprinted MEDEA locus in plants. Nature 448 : 349–352.
12. BartolomeiMS, Ferguson-SmithAC (2011) Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3: pii: a002592.
13. Ferguson-SmithAC (2011) Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet 12 : 565–575.
14. ChoiY, GehringM, JohnsonL, HannonM, HaradaJJ, et al. (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110 : 33–42.
15. XiaoW, GehringM, ChoiY, MargossianL, PuH, et al. (2003) Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell 5 : 891–901.
16. KinoshitaT, MiuraA, ChoiY, KinoshitaY, CaoX, et al. (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303 : 521–523.
17. GehringM, HuhJH, HsiehTF, PentermanJ, ChoiY, et al. (2006) DEMETER DNA glycosylase establishes MEDEA Polycomb gene self-imprinting by allele-specific demethylation. Cell 124 : 495–506.
18. JullienP, KinoshitaT, OhadN, BergerF (2006) Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18 : 1360–1372.
19. TiwariS, SchulzR, IkedaY, DythamL, BravoJ, et al. (2008) MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell 20 : 2387–2398.
20. WöhrmannHJ, GagliardiniV, RaissigMT, WehrleW, ArandJ, et al. (2012) Identification of a DNA methylation-independent imprinting control region at the Arabidopsis MEDEA locus. Genes Dev 26 : 1837–1850.
21. Gutiérrez-MarcosJF, CostaLM, Dal PràM, ScholtenS, KranzE, et al. (2006) Epigenetic asymmetry of imprinted genes in plant gametes. Nat Genet 38 : 876–878.
22. HaunWJ, Laoueillé-DupratS, O'connellMJ, SpillaneC, GrossniklausU, et al. (2007) Genomic imprinting, methylation and molecular evolution of maize Enhancer of zeste (Mez) homologs. Plant J 49 : 325–337.
23. HermonP, SrilunchangKO, ZouJ, DresselhausT, DanilevskayaON (2007) Activation of the imprinted Polycomb group Fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant Mol Biol 64 : 387–395.
24. BarouxC, GagliardiniV, PageDR, GrossniklausU (2006) Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev 20 : 1081–1086.
25. Fitz GeraldJN, HuiPS, BergerF, GeraldJNF, HuiPS, et al. (2009) Polycomb group-dependent imprinting of the actin regulator AtFH5 regulates morphogenesis in Arabidopsis thaliana. Development 136 : 3399–3404.
26. JullienP, KatzA, OlivaM, OhadN, BergerF (2006) Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol 16 : 486–492.
27. HsiehTF, ShinJ, UzawaR, SilvaP, CohenS, et al. (2011) Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA 108 : 1755–1762.
28. GehringM, MissirianV, HenikoffS (2011) Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 6: e23687.
29. WolffP, WeinhoferI, SeguinJ, RoszakP, BeiselC, et al. (2011) High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet 7: e1002126.
30. McKeownPC, Laouielle-DupratS, PrinsP, WolffP, SchmidMW, et al. (2011) Identification of imprinted genes subject to parent-of-origin specific expression in Arabidopsis thaliana seeds. BMC Plant Biol 11 : 113.
31. WatersAJ, MakarevitchI, EichtenSR, Swanson-WagnerRA, YehCT, et al. (2011) Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23 : 4221–4233.
32. ZhangM, ZhaoH, XieS, ChenJ, XuY, et al. (2011) Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc Natl Acad Sci USA 108 : 20042–20047.
33. AutranD, BarouxC, RaissigMT, LenormandT, WittigM, et al. (2011) Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145 : 707–719.
34. NodineMD, BartelDP (2012) Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482 : 94–97.
35. WuestSE, VijverbergK, SchmidtA, WeissM, GheyselinckJ, et al. (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20 : 506–512.
36. BorgesF, GomesG, GardnerR, MorenoN, McCormickS, et al. (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Phys 148 : 1168–1181.
37. PourcelL, RoutaboulJ-M, KerhoasL, CabocheM, LepiniecL, et al. (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17 : 2966–2980.
38. BemerM, HeijmansK, AiroldiC, DaviesB, AngenentGC (2010) An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Phys 154 : 287–300.
39. KangI-H, SteffenJG, PortereikoMF, LloydA, DrewsGN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20 : 635–647.
40. JeffersonRA, KavanaghTA, BevanMW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 : 3901–3907.
41. ReidtW, WohlfarthT, EllerströmM, CzihalA, TewesA, et al. (2000) Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J 21 : 401–408.
42. ChaudhuryAM, MingL, MillerC, CraigS, DennisES, et al. (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 94 : 4223–4228.
43. GrossniklausU, Vielle-CalzadaJP, HoeppnerMA, GaglianoWB (1998) Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280 : 446–450.
44. RazV, BergervoetJH, KoornneefM (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128 : 243–252.
45. XiangD, VenglatP, TibicheC, YangH, RisseeuwE, et al. (2011) Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Phys 156 : 346–356.
46. SchmidMW, SchmidtA, KlostermeierUC, BarannM, RosenstielP, et al. (2012) A powerful method for transcriptional profiling of specific cell types in eukaryotes: laser-assisted microdissection and RNA sequencing. PLoS ONE 7: e29685.
47. KraglerF (2013) Plasmodesmata: intercellular tunnels facilitating transport of macromolecules in plants. Cell Tiss Res 352 : 49–58.
48. Kozieradzka-KiszkurnoM, PłachnoBJ (2012) Are there symplastic connections between the endosperm and embryo in some angiosperms?–a lesson from the Crassulaceae family. Protoplasma 249 : 1081–1089.
49. PignattaD, GehringM (2012) Imprinting meets genomics: new insights and new challenges. Curr Opin Plant Biol 15 : 530–535.
50. DevealeB, Van Der KooyD, BabakT (2012) Critical evaluation of imprinted gene expression by RNA–Seq: a new perspective. PLoS Genet 8: e1002600.
51. BarouxC, AutranD, RaissigMT, GrimanelliD, GrossniklausU (2013) Parental contributions to the transcriptome of early plant embryos. Curr Opin Genet Dev 23 : 72–74.
52. Vielle-CalzadaJP, BaskarR, GrossniklausU (2000) Delayed activation of the paternal genome during seed development. Nature 404 : 91–94.
53. MakarevichG, VillarCB, ErilovaA, KohlerC (2008) Mechanism of PHERES1 imprinting in Arabidopsis. J Cell Sci 121 : 906–912.
54. JacksonJP, LindrothAM, CaoX, JacobsenSE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 : 556–560.
55. CedarH, BergmanY (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10 : 295–304.
56. HaunWJ, SpringerNM (2008) Maternal and paternal alleles exhibit differential histone methylation and acetylation at maize imprinted genes. Plant J 56 : 903–912.
57. BemerM, GrossniklausU (2012) Dynamic regulation of Polycomb group activity during plant development. Curr Opin Plant Biol 15 : 523–529.
58. LudwigT, EggenschwilerJ, FisherP, D'ErcoleAJ, DavenportML, et al. (1996) Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev Biol 177 : 517–535.
59. LauMM, StewartCE, LiuZ, BhattH, RotweinP, et al. (1994) Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev 8 : 2953–2963.
60. IngouffM, HaseloffJ, BergerF (2005) Polycomb group genes control developmental timing of endosperm. Plant J 42 : 663–674.
61. KinoshitaT, YadegariR, HaradaJJ, GoldbergRB, FischerRL (1999) Imprinting of the MEDEA Polycomb gene in the Arabidopsis endosperm. Plant Cell 11 : 1945–1952.
62. KiyosueT, OhadN, YadegariR, HannonM, DinnenyJ, et al. (1999) Control of fertilization-independent endosperm development by the MEDEA Polycomb gene in Arabidopsis. Proc Natl Acad Sci USA 96 : 4186–4191.
63. LuoM, BilodeauP, DennisES, PeacockWJ, ChaudhuryA (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97 : 10637–10642.
64. TyckoB, MorisonIM (2002) Physiological functions of imprinted genes. J Cell Physiol 192 : 245–258.
65. HaigD, WestobyM (1989) Parent-specific gene expression and the triploid endosperm. Am Nat 134 : 147–155.
66. BatesonP (1994) The dynamics of parent-offspring relationships in mammals. Trends Ecol Evol 9 : 399–403.
67. WolfJB, HagerR (2006) A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol 4: e380.
68. BouyerD, RoudierF, HeeseM, AndersenED, GeyD, et al. (2011) Polycomb Repressive Complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7: e1002014.
69. SazeH, Mittelsten ScheidO, PaszkowskiJ (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34 : 65–69.
70. WuX, ChoryJ, WeigelD (2007) Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Dev Biol 309 : 306–316.
71. CloughSJ, BentAF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743.
Štítky
Genetika Reprodukční medicína
Článek Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of SyngnathiaČlánek Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing LimbsČlánek Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory RegionsČlánek Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Stressing the Importance of CHOP in Liver Cancer
- The AmAZI1ng Roles of Centriolar Satellites during Development
- Flies Get a Head Start on Meiosis
- Recommendations from Jane Gitschier's Bookshelf
- And Baby Makes Three: Genomic Imprinting in Plant Embryos
- Bugs in Transition: The Dynamic World of in Insects
- Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids
- Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells
- Cohesinopathies of a Feather Flock Together
- Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs
- Parathyroid-Specific Deletion of Unravels a Novel Calcineurin-Dependent FGF23 Signaling Pathway That Regulates PTH Secretion
- MAN1B1 Deficiency: An Unexpected CDG-II
- Phosphate Flow between Hybrid Histidine Kinases CheA and CheS Controls Cyst Formation
- Basolateral Mg Extrusion via CNNM4 Mediates Transcellular Mg Transport across Epithelia: A Mouse Model
- Truncation of Unsilences Paternal and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model
- Autozygome Sequencing Expands the Horizon of Human Knockout Research and Provides Novel Insights into Human Phenotypic Variation
- Huntington's Disease Induced Cardiac Amyloidosis Is Reversed by Modulating Protein Folding and Oxidative Stress Pathways in the Heart
- Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data
- Targeted Ablation of and in Retinal Progenitor Cells Mimics Leber Congenital Amaurosis
- Genomic Imprinting in the Embryo Is Partly Regulated by PRC2
- Binary Cell Fate Decisions and Fate Transformation in the Larval Eye
- The Stress-Regulated Transcription Factor CHOP Promotes Hepatic Inflammatory Gene Expression, Fibrosis, and Oncogenesis
- A Global RNAi Screen Identifies a Key Role of Ceramide Phosphoethanolamine for Glial Ensheathment of Axons
- Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Species
- Cross-Modulation of Homeostatic Responses to Temperature, Oxygen and Carbon Dioxide in
- Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance
- Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont
- CRIS—A Novel cAMP-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending
- Dual Regulation of the Mitotic Exit Network (MEN) by PP2A-Cdc55 Phosphatase
- Expanding the Marine Virosphere Using Metagenomics
- Detection of Slipped-DNAs at the Trinucleotide Repeats of the Myotonic Dystrophy Type I Disease Locus in Patient Tissues
- Interaction between and during Mammalian Jaw Patterning and in the Pathogenesis of Syngnathia
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- Reactivation of Chromosomally Integrated Human Herpesvirus-6 by Telomeric Circle Formation
- Anoxia-Reoxygenation Regulates Mitochondrial Dynamics through the Hypoxia Response Pathway, SKN-1/Nrf, and Stomatin-Like Protein STL-1/SLP-2
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
- Selection on Plant Male Function Genes Identifies Candidates for Reproductive Isolation of Yellow Monkeyflowers
- Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores
- Meiotic Cohesin SMC1β Provides Prophase I Centromeric Cohesion and Is Required for Multiple Synapsis-Associated Functions
- Identification of Sphingolipid Metabolites That Induce Obesity via Misregulation of Appetite, Caloric Intake and Fat Storage in
- Genome-Wide Screen Reveals Replication Pathway for Quasi-Palindrome Fragility Dependent on Homologous Recombination
- Histone Methylation Restrains the Expression of Subtype-Specific Genes during Terminal Neuronal Differentiation in
- A Novel Intergenic ETnII-β Insertion Mutation Causes Multiple Malformations in Mice
- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- A Domesticated Transposase Interacts with Heterochromatin and Catalyzes Reproducible DNA Elimination in
- Acute Versus Chronic Loss of Mammalian Results in Distinct Ciliary Phenotypes
- MBD3 Localizes at Promoters, Gene Bodies and Enhancers of Active Genes
- Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo
- A Hereditary Spastic Paraplegia Mouse Model Supports a Role of ZFYVE26/SPASTIZIN for the Endolysosomal System
- The CCR4-NOT Complex Mediates Deadenylation and Degradation of Stem Cell mRNAs and Promotes Planarian Stem Cell Differentiation
- Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data
- Contributions of Protein-Coding and Regulatory Change to Adaptive Molecular Evolution in Murid Rodents
- Comprehensive Analysis of Transcriptome Variation Uncovers Known and Novel Driver Events in T-Cell Acute Lymphoblastic Leukemia
- A -Acting Protein Effect Causes Severe Eye Malformation in the Mouse
- Clustering of Tissue-Specific Sub-TADs Accompanies the Regulation of Genes in Developing Limbs
- Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Development
- Transcription Factor Occupancy Can Mediate Active Turnover of DNA Methylation at Regulatory Regions
- Somatic mtDNA Mutation Spectra in the Aging Human Putamen
- ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity
- Ethylene Promotes Hypocotyl Growth and HY5 Degradation by Enhancing the Movement of COP1 to the Nucleus in the Light
- The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast
- Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in
- Quantifying Missing Heritability at Known GWAS Loci
- Smc5/6-Mms21 Prevents and Eliminates Inappropriate Recombination Intermediates in Meiosis
- Smc5/6 Coordinates Formation and Resolution of Joint Molecules with Chromosome Morphology to Ensure Meiotic Divisions
- Tay Bridge Is a Negative Regulator of EGFR Signalling and Interacts with Erk and Mkp3 in the Wing
- Meiotic Crossover Control by Concerted Action of Rad51-Dmc1 in Homolog Template Bias and Robust Homeostatic Regulation
- Active Transport and Diffusion Barriers Restrict Joubert Syndrome-Associated ARL13B/ARL-13 to an Inv-like Ciliary Membrane Subdomain
- An Regulatory Circuit Modulates /Wnt Signaling and Determines the Size of the Midbrain Dopaminergic Progenitor Pool
- Variants Induce Differential Protection to Viruses in : A Phenotypic and Phylogenomic Analysis
- Base Pairing Interaction between 5′- and 3′-UTRs Controls mRNA Translation in
- Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis
- Insulin/IGF-Regulated Size Scaling of Neuroendocrine Cells Expressing the bHLH Transcription Factor in
- Sumoylated NHR-25/NR5A Regulates Cell Fate during Vulval Development
- TATN-1 Mutations Reveal a Novel Role for Tyrosine as a Metabolic Signal That Influences Developmental Decisions and Longevity in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The NuRD Chromatin-Remodeling Enzyme CHD4 Promotes Embryonic Vascular Integrity by Transcriptionally Regulating Extracellular Matrix Proteolysis
- Mutations in the UQCC1-Interacting Protein, UQCC2, Cause Human Complex III Deficiency Associated with Perturbed Cytochrome Protein Expression
- The Midline Protein Regulates Axon Guidance by Blocking the Reiteration of Neuroblast Rows within the Drosophila Ventral Nerve Cord
- Tomato Yield Heterosis Is Triggered by a Dosage Sensitivity of the Florigen Pathway That Fine-Tunes Shoot Architecture
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



