-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Dynamic Regulation of H3K27 Trimethylation during Differentiation
During growth of multicellular organisms, identities of stem cells and differentiated cells need to be maintained. Cell fate is epigenetically controlled by the conserved Polycomb-group (Pc-G) proteins that repress their target genes by catalyzing histone H3 lysine 27 trimethylation (H3K27me3). Although H3K27me3 is associated with mitotically stable gene repression, a large fraction of H3K27me3 target genes are tissue-specifically activated during differentiation processes. However, in plants it is currently unclear whether H3K27me3 is already present in undifferentiated cells and dynamically regulated to permit tissue-specific gene repression or activation. We used whole-genome tiling arrays to identify the H3K27me3 target genes in undifferentiated cells of the shoot apical meristem and in differentiated leaf cells. Hundreds of genes gain or lose H3K27me3 upon differentiation, demonstrating dynamic regulation of an epigenetic modification in plants. H3K27me3 is correlated with gene repression, and its release preferentially results in tissue-specific gene activation, both during differentiation and in Pc-G mutants. We further reveal meristem - and leaf-specific targeting of individual gene families including known but also likely novel regulators of differentiation and stem cell regulation. Interestingly, H3K27me3 directly represses only specific transcription factor families, but indirectly activates others through H3K27me3-mediated silencing of microRNA genes. Furthermore, H3K27me3 targeting of genes involved in biosynthesis, transport, perception, and signal transduction of the phytohormone auxin demonstrates control of an entire signaling pathway. Based on these and previous analyses, we propose that H3K27me3 is one of the major determinants of tissue-specific expression patterns in plants, which restricts expression of its direct targets and promotes gene expression indirectly by repressing miRNA genes.
Published in the journal: . PLoS Genet 7(4): e32767. doi:10.1371/journal.pgen.1002040
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002040Summary
During growth of multicellular organisms, identities of stem cells and differentiated cells need to be maintained. Cell fate is epigenetically controlled by the conserved Polycomb-group (Pc-G) proteins that repress their target genes by catalyzing histone H3 lysine 27 trimethylation (H3K27me3). Although H3K27me3 is associated with mitotically stable gene repression, a large fraction of H3K27me3 target genes are tissue-specifically activated during differentiation processes. However, in plants it is currently unclear whether H3K27me3 is already present in undifferentiated cells and dynamically regulated to permit tissue-specific gene repression or activation. We used whole-genome tiling arrays to identify the H3K27me3 target genes in undifferentiated cells of the shoot apical meristem and in differentiated leaf cells. Hundreds of genes gain or lose H3K27me3 upon differentiation, demonstrating dynamic regulation of an epigenetic modification in plants. H3K27me3 is correlated with gene repression, and its release preferentially results in tissue-specific gene activation, both during differentiation and in Pc-G mutants. We further reveal meristem - and leaf-specific targeting of individual gene families including known but also likely novel regulators of differentiation and stem cell regulation. Interestingly, H3K27me3 directly represses only specific transcription factor families, but indirectly activates others through H3K27me3-mediated silencing of microRNA genes. Furthermore, H3K27me3 targeting of genes involved in biosynthesis, transport, perception, and signal transduction of the phytohormone auxin demonstrates control of an entire signaling pathway. Based on these and previous analyses, we propose that H3K27me3 is one of the major determinants of tissue-specific expression patterns in plants, which restricts expression of its direct targets and promotes gene expression indirectly by repressing miRNA genes.
Introduction
Throughout their lifecycle, plants produce new organs through a group of undifferentiated cells which are maintained in structures called meristems. These pluripotent cells continuously divide and, in the shoot, cells in the periphery of the apical meristem differentiate to give rise to lateral organs like leaves or flowers. Stem cell maintenance is tightly controlled by numerous and interconnected pathways involving transcriptional regulation, phytohormones, microRNAs and epigenetic gene regulation (reviewed in [1]).
Epigenetic gene regulation is a key mechanism to confer stable, but reversible gene expression states. Epigenetics has been revealed as fundamental mechanism to maintain cell and tissue identity and to regulate stem cells and cancer.
Important epigenetic regulators of developmental processes are the Polycomb-group (Pc-G) and Trithorax-group (Trx-G) proteins which catalyze histone H3 lysine 27 trimethylation (H3K27me3) or H3K4me3, respectively (reviewed in [2], [3]). In Arabidopsis, around 5% of the canonical histone H3.1 is trimethylated at K27 [4]. Pc-G target genes in plants and animals are covered by H3K27me3 [5]–[8], a distribution that likely permits epigenetic inheritance of the modification [9]. Whereas Drosophila melanogaster Polycomb repressive complex 2 (PRC2) was shown to methylate H3K27 in vitro and in vivo via its histone methyltransferase subunit Enhancer of zeste [E(z)] [10]–[13], evidence for the biochemical activity of the conserved plant PRC2 is still lacking. H3K27me3 was shown to depend at least partially on plant PRC2 members as loss of the E(z) homolog CURLY LEAF (CLF) leads to reduced levels of H3K27me3 [8], [14]. In addition, immunofluorescence analyses of plants lacking the redundantly acting E(z) homologs CLF and SWINGER (SWN) revealed a reduction in euchromatic H3K27me3, but also a frequent re-distribution to chromocenters [15].
In plants, numerous developmental pathways are controlled by Pc-G proteins including seed development, flowering time, vernalization and organ identity (reviewed in [16], [17]). Loss of sporophytic Pc-G activity results in plants that show overproliferation and strong defects in organ identity [18], [19]. The severe phenotypes of Pc-G mutants indicate an essential role for Pc-G proteins in plant development which was strongly supported by whole genome analyses of H3K27me3 targets. In seedlings, more than 4000 H3K27me3 target genes were uncovered which are largely protein-coding genes and mostly exclude transposable element genes and heterochromatic regions [5], [20], [21]. A strikingly large number of developmentally important transcription factor genes showed H3K27me3 coverage [5]. However, the fact that key genes involved in the biosynthesis and inactivation of the phytohormone gibberellic acid exhibit enrichment in H3K27me3 [22] indicates an important role for Pc-G proteins in regulating developmental processes beyond the transcription factor level. Numerous genome-wide analyses of H3K27me3 and Pc-G protein binding have been performed for mammals and Drosophila [6], [23]–[29]. These studies revealed a considerably smaller number of target genes in Drosophila and mammals compared to Arabidopsis, but also key developmental transcription factor genes as H3K27me3 targets.
Presence of H3K27me3 is largely correlated with gene silencing in animals and plants, although H3K27me3 is only partially removed upon gene activation in Drosophila [30]. In Arabidopsis, H3K27me3 targets are enriched for genes with tissue-specific expression patterns or are induced by abiotic or biotic stresses suggesting that H3K27me3 is dynamically regulated in response to developmental or environmental cues [5]. Indeed, several thousand genes either lose or gain H3K27me3 when etiolated seedlings were transferred into light [22]. Furthermore, a novel approach of cell-type specific tagging and isolation of nuclei allowing cell type specific analyses of hair and non-hair cells of the Arabidopsis root epidermis revealed hundreds of genes carrying differential H3K4me3 and H3K27me3 in the two cell types and a strong correlation of gene repression with low H3K4me3 and high H3K27me3 [31].
Different cell types can harbor distinct chromatin profiles which is particularly the case for pluripotent mammalian embryonic stem (ES) cells [7], [32]. Many genes in ES cells carry bivalent marks, thus both H3K4me3 and H3K27me3, which are mostly resolved to monovalent states of either H3K4me3 or H3K27me3 during differentiation [7], [33]. In plants, however, it is currently unclear if undifferentiated cells harbor distinct chromatin profiles.
Here, we present genome-wide analyses of H3K27me3 as well as gene expression analyses from meristematic, undifferentiated cells and differentiated leaf cells using the vegetative shoot apical meristem as a model system. We confirm a large fraction of previously identified H3K27me3 targets and reveal several hundred additional, tissue-specifically methylated genes. Genes with strong expression differences in the two tissues are enriched for differential H3K27me3 suggesting that also in plants Pc-G proteins define gene ON/OFF expression states. We find that a large fraction of microRNA genes are differentially methylated in meristems and leaves and that their tissue-specific expression patterns are controlled by Pc-G proteins. In addition, a large number of genes involved in biosynthesis, transport, perception and signaling of the phytohormone auxin are among the identified target genes suggesting that entire gene regulatory networks are controlled and possibly stabilized by Pc-G mediated gene regulation. Collectively, our analyses suggest that Pc-G proteins control differentiation processes by conferring tissue-specific H3K27me3 of hundreds of genes including microRNA genes and genes involved hormonal pathways.
Results
Genome-wide analyses of gene expression and H3K27me3 in meristems and leaves
In Arabidopsis seedlings, several thousand genes are covered by H3K27me3 [5], [20].
It was previously suggested that H3K27me3 is required for stable gene repression throughout most of the plants lifecycle and is either reset to allow gene activation or acquired at later developmental stages to confer stable expression states during differentiation processes [5], [34]. In order to uncover dynamic regulation of H3K27me3 during development, undifferentiated meristematic (Me) and differentiated leaf (Le) tissues of clavata3 (clv3) mutant plants were analysed. Meristematic tissue and stem cells can be easily isolated from clv3 mutants by manual dissection as the mutant harbours larger vegetative shoot apical meristems and increased stem cell numbers [35] (Figure 1A). Importantly, leaf development is not affected by loss of CLV3. Both leaf and meristematic tissue samples were subjected to array based genome wide expression (Figure 1) and H3K27me3 (Figure 2) profiling. Therefore, this approach permits the identification of H3K27me3 targets that gain or lose H3K27me3 during differentiation of meristematic cells. In addition, the associated changes in gene expression of H3K27me3 targets can be revealed.
Fig. 1. Tissue enrichment through manual dissection of Arabidopsis clv3 mutants. 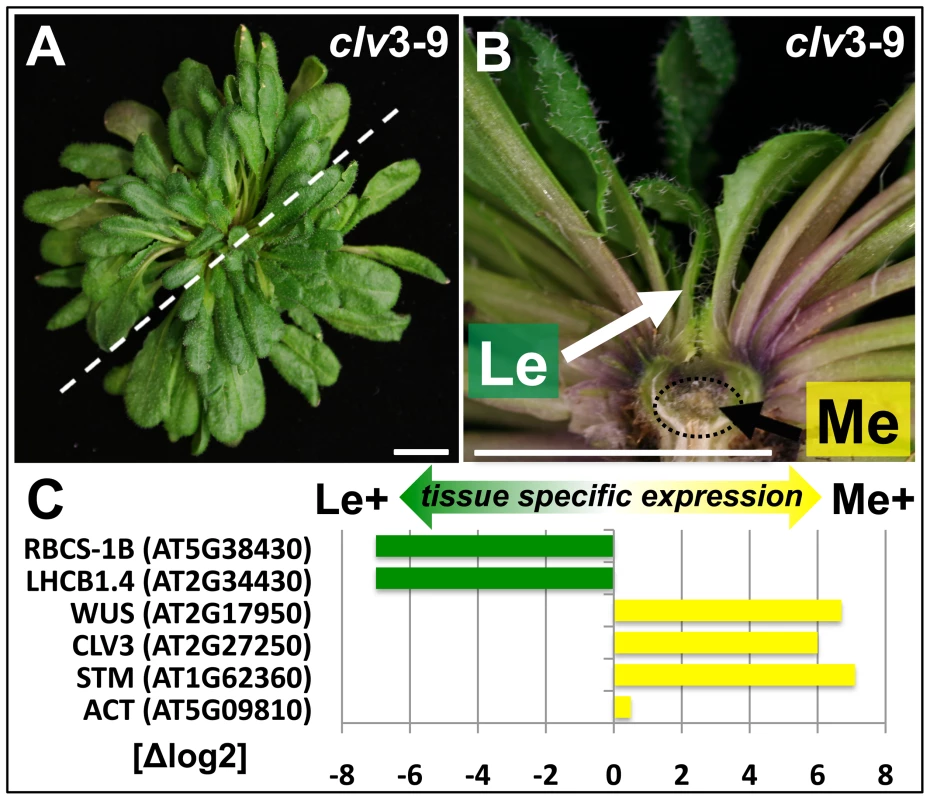
(A) Nine week old clv3–9 plant grown under short day conditions, (B) longitudinal section of plant shown in (A). The section plane is marked with a dashed line in A, scale bars indicate 1 cm. Two samples, a meristematic fraction (Me) marked with a dotted circle and young leaves (Le) were harvested. (C) Differential expression of known reference genes obtained by genome wide expression analysis in the two dissected tissues. CLV3 expression is detected as the point mutation in the clv3–9 allele does not interfere with CLV3 transcriptional regulation. Fig. 2. Tissue-specific ChIP-on-chip analysis reveals differentially methylated genes. 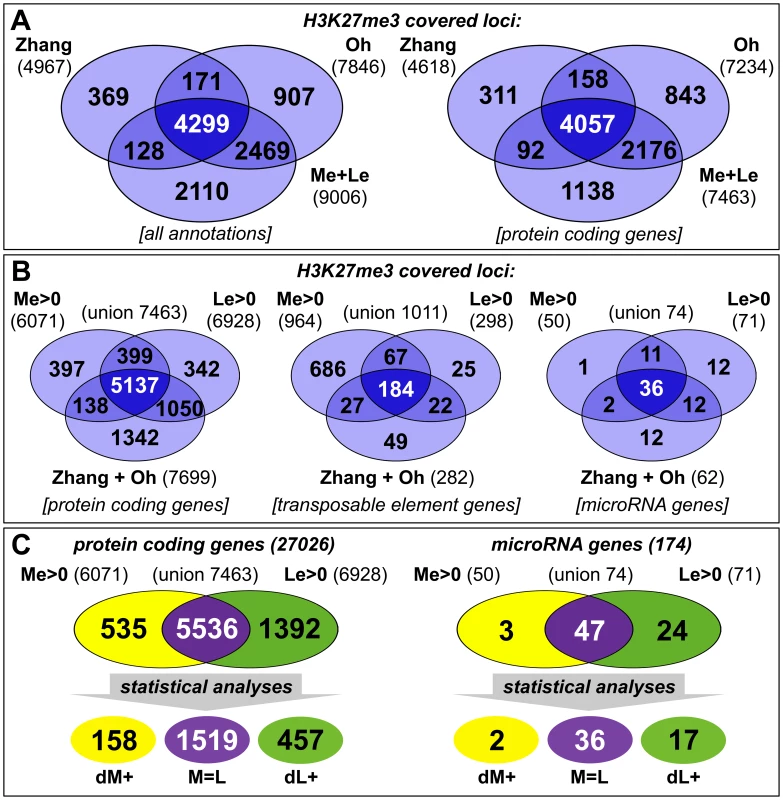
(A, B) The venn diagrams represent a comparison of H3K27me3 covered loci taken from three independent genome wide analyses of Arabidopsis plants (Zhang et al. [5]; Oh et al. [20] and this work, meristem (Me) + leaves (Le)). (A) Comparison of all annotations (left) and of protein coding genes (right) in published data and union of Me+Le. (B) Comparison of protein coding (left), transposable element (middle) and microRNA genes (right) in union of published data and Me and Le sample. (C) Comparison of Me and Le targets, protein coding (left) and microRNA genes (right). Statistical analyses (see methods) were applied to uncover significantly differentially or equally methylated genes in meristem and leaves. Annotations are based on TAIR8 genome annotation. The expression analysis verified both sample identity and specific enrichment of each dissected tissue because typical transcripts characteristic for the meristematic domain or green tissue were detected as highly and differentially expressed (Figure 1C). Meristem and stem cell identity genes like SHOOTMERISTEMLESS (STM), CLV3 and WUSCHEL (WUS) were highly expressed in the meristematic sample whereas photosynthesis-related genes encoding for subunits of RUBISCO (RBCS-1B) or light harvesting complexes (LHCB1.4) were exclusively detected in leaves.
We generated genome-wide H3K27me3 profiles for each tissue by hybridizing sheared genomic DNA that was immunoprecipitated with an antibody detecting H3K27me3 to high density tiling arrays, as was previously described [36], [37]. We identified a total of 9006 H3K27me3 target genes in the two tissues which include more than 7400 protein coding genes (27,6% of all annotated Arabidopsis protein coding genes) and interestingly also 74 miRNA genes (43% of all miRNA genes) (Figure 2). Importantly, more than 80% of protein coding H3K27me3 target genes identified in our study were also revealed in previous analyses of young seedlings [5], [20], demonstrating the validity of our approach. Both former studies were performed with 14 days old wildtype plants, whole seedlings or their aerial parts, whereas the tissue samples analyzed in our study were derived from 9 weeks old clv3 mutant plants. The large H3K27me3 overlap in the different tissues therefore suggests extensive and stable gene silencing by Pc-G mediated H3K27me3 throughout vegetative plant development. In addition to a large number of common H3K27me3 targets, the analysis identified many, presumably tissue-specific protein coding target genes (1001 genes only detected in seedlings (by Oh et al. [20]); 1230 genes only identified in meristem or young leaf tissue in our study) (Figure 2A). Interestingly, although previous studies showed that transposable elements (TE) are largely devoid of H3K27me3 [5], [20], we detected a high number of TEs as meristem-specific H3K27me3 targets (Figure 2B). This suggests an unexpected role for Pc-G proteins in the regulation of TEs specifically in the stem cell harbouring meristem.
To study the role of H3K27me3 in controlling developmentally important genes we focused our analyses on protein coding and miRNA genes and compared the methylation profiles of the individual tissues (Figure 2B, 2C). This uncovered large differences between both samples as nearly 2000 protein coding and 27 miRNA genes were exclusively methylated in one sample (Me or Le) revealing these as tissue-specific H3K27me3 targets (details for all annotations are shown in Table S1). Interestingly, in contrast to the largely meristem-specific H3K27me3 targeting of TEs, a larger set of protein coding and miRNA H3K27me3 targets was identified in the leaf compared to the meristematic sample (Figure 2B). Thus, heterochromatic and euchromatic loci may be differentially targeted by Pc-G proteins in undifferentiated and differentiated tissues.
For further analyses on the comparison of H3K27me3 presence and gene expression and on gene families we isolated genes harbouring defined methylation levels and performed conservative statistical analyses to group these into sets of equally (M = L) and differentially methylated genes [meristem - (dM+) and leaf - (dL+) specific] (see methods). These analyses uncovered 1519 protein coding genes showing equal H3K27me3 levels in both samples, 158 genes specifically methylated in the meristem and 457 genes methylated in leaves (Figure 2C).
Collectively, our analyses uncover dynamic regulation of a large number of H3K27me3 target genes during differentiation which involves acquisition and removal of H3K27me3 at protein coding, miRNA and TE genes.
Confirmation of differential methylation in wildtype meristems and leaves
We sought to confirm the genome-wide, tissue-specific H3K27me3 patterns by independent ChIP experiments on meristems and leaves of clv3 mutants (Figure S1) and wildtype (Figure 3). The transcription factors KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 (KNAT2) and KNAT6 are specifically expressed in the meristem ([38], (Figure 3)) and were identified as leaf specifically methylated (dL+) (Figure S1A representatively shows the KNAT6 locus). H3K27me3 ChIP-qPCR analysis of independently dissected clv3 tissues confirmed low meristematic and high leaf H3K27me3 levels for KNAT2 and KNAT6 and revealed an antagonistic pattern of the active mark H3K4me3 (Figure S1B, S1C). Thus, H3K4me3 and H3K27me3 distribution are consistent with meristem specific expression of both KNAT genes (Figure 3E). To further prove the significance of differential H3K27me3 identified by ChIP-chip on clv3 mutants (Figure 2) we performed ChIP-qPCR analyses of 2 months old dissected wildtype plants, grown under short day conditions. Consistent with the data on clv3, KNAT2 and KNAT6 showed leaf specific H3K27me3 (Figure 3). In addition, equal methylation for the non-expressed genes AGAMOUS and FUSCA3 and differential methylation of several transcription factor genes, PIN-FORMED (PIN) auxin efflux carriers and microRNA genes were confirmed (Figure 3). For all genes tested by ChIP, moderate to strong differential expression was detected which was highest in the tissues lacking H3K27me3 (Figure 3E). Thus, these analyses strongly suggest that the clv3 ChIP-chip results are transferable to wild type plants.
Fig. 3. Confirmation of differentially methylated genes in wildtype plants. 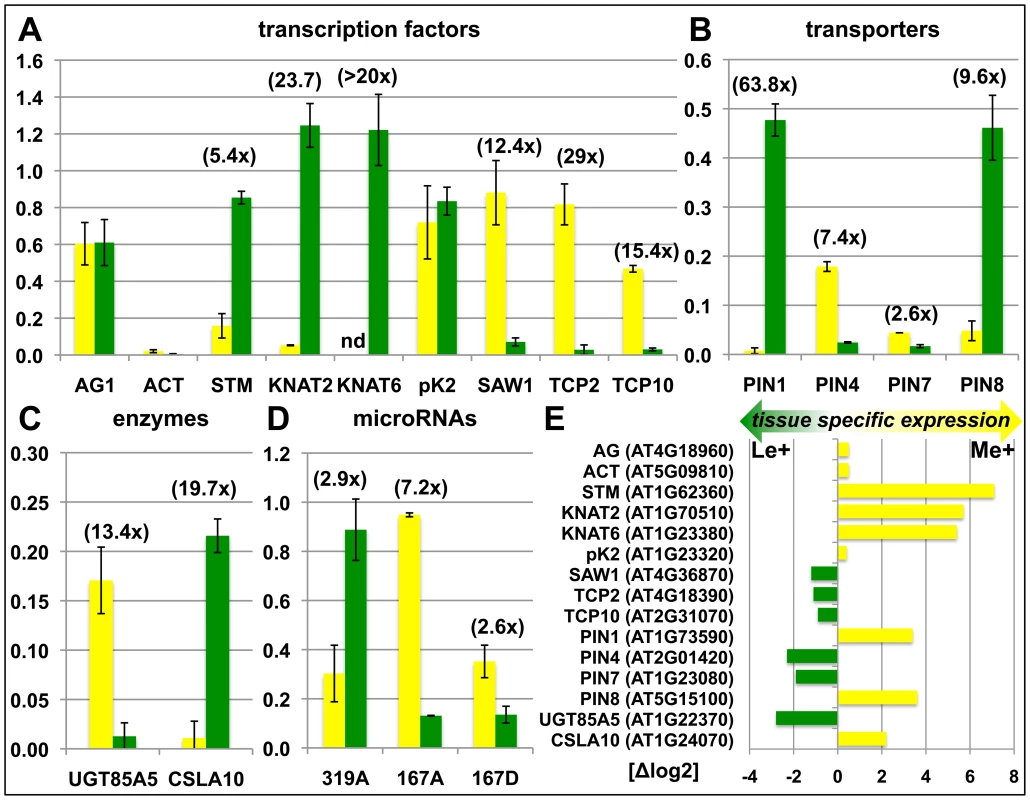
Differential H3K27me3 in meristem and leaves of wildtype was confirmed by independent H3K27me3-ChIP analyses for selected transcription factors (A), transporters (B), enzymes (C) and miRNA genes (D). Green: relative enrichment of H3K27me3 in leaves; yellow: relative enrichment of H3K27me3 in meristem. Enrichment factors between different tissues are indicated in brackets. 2 months old, short-day grown wild type plants were manually dissected. Enrichment of specific loci is normalized to the H3K27me3 target FUSCA3 harbouring equal methylation levels in both tissues. The results are shown as mean values of two independent precipitations and qPCR analyses. Methylation of KNAT6 was not detectable (nd) in the meristem sample. (E) Differential expression of genes analyzed in (A) – (D). Data was retrieved from whole transcriptome analyses of manually dissected clv3 mutant meristems and leaves. Presence of H3K27me3 is correlated with gene repression in a tissue-specific manner
Since we had identified tissue-specific H3K27me3 target genes we asked whether differential H3K27me3 is correlated with differential gene expression. In total, more than 22000 protein coding genes (85% of the genome) were expressed in at least one of the tissues. A large fraction (nearly 9500 genes) showed equal expression in both samples [X(M = L)], whereas 2.833 were at least four times higher expressed in one sample compared to the other [1.865 higher in the meristem (dXM≥2, Δlog2) and 968 higher in leaves (dXL≤−2, Δlog2)] (Figure 4A, Figure S2). We next binned genes with similar expression differences in the two tissues (Figure 4B, Table S2). These gene sets were analyzed for enrichment of H3K27me3 targets (Figure 4B) and of genes harbouring equal or differential H3K27me3 levels (M = L, dM+, dL+) (Figure 4C). In addition, we performed the reciprocal analyses (Figure 4D). Genes that were equally expressed both in the meristem and the leaves (X(M = L)) showed a significant underrepresentation of H3K27me3 targets (Figure 4B). In contrast, methylated genes were more frequent in the groups of non expressed and differentially expressed genes (Figure 4B). Interestingly, the relative abundance of target genes increased with increasing tissue specific expression in the meristem (Me+) or leaves (Le+) (Figure 4B). We next asked how differential expression relates to differential or equal H3K27me3 (Figure 4C). This detailed tissue specific analysis clearly revealed that the highly specific expression in one tissue (dXM≥2 or dXL≤−2) was correlated with a depletion of tissue specific methylation (dM+ or dL+) in the same sample and a strong enrichment of repressive histone methylation in the other – non expressing – sample. Importantly, the stronger the differences in gene expression levels between the two tissues, the more genes were identified which showed antagonistic, differential H3K27me3 (Figure 4C). Genes showing equal H3K27me3 levels in both tissues were enriched for non-expressed genes which was not the case for differentially methylated genes, revealing that most genes carrying differential methylation are expressed in at least one of the tissues (Figure 4C, 4D). We found the same correlations when the analyses were performed with the different subsets of H3K27me3 covered genes (Figure 4D). Leaf-specifically expressed genes (dXL≤−2) were more frequent in the set of meristem specifically methylated genes (dM+) and vice versa whereas genes showing equal, but detectable methylation (M = L) were depleted of expressed genes.
Fig. 4. Anti-correlation of gene expression and repressive H3K27 tri-methylation. 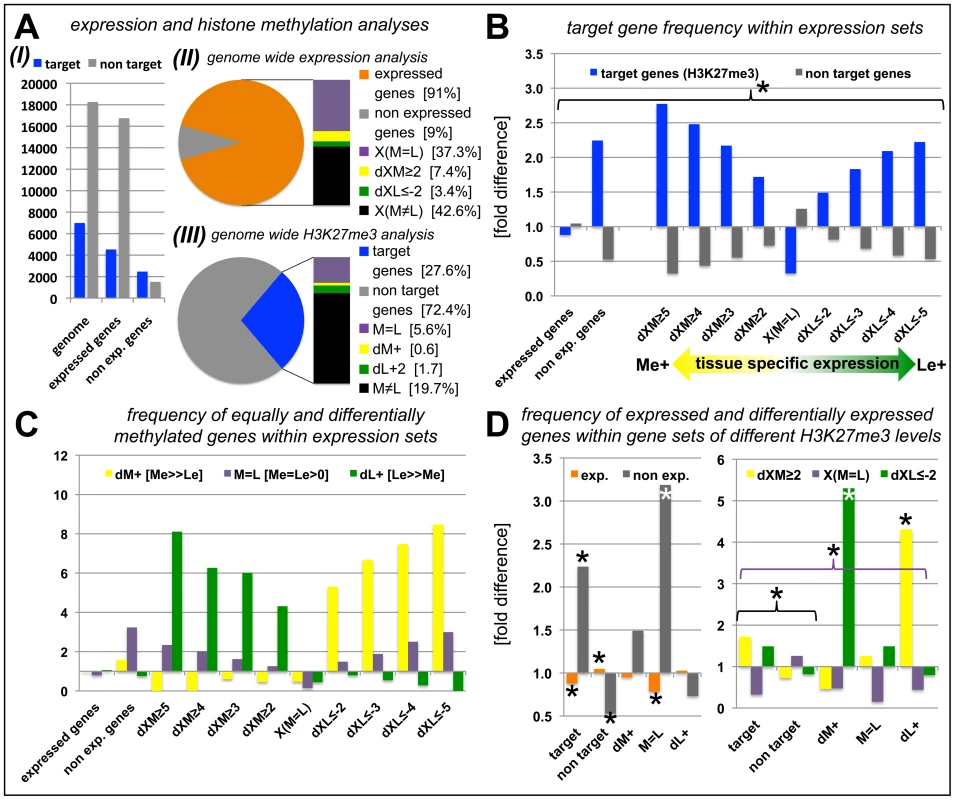
(A) Genome-wide relation of gene expression and H3K27me3 targeting (I). (II) Representation of percentage of genes for defined expression states. (III) Representation of percentage of genes for defined H3K27me3 levels. For expressed genes and H3K27me3 targets, number of genes detected in leaves or meristems of clv3 mutants was merged and compared to genes present on Agilent array (for II) or all protein coding genes (for III). (B) Relative proportion of H3K27me3 target genes within gene sets of defined expression levels, y-axis indicates fold enrichment compared to subset frequency in the genome, i.e. target genes or non target genes. (C) Relative proportion of equally and differentially methylated H3K27me3 target genes within gene sets of defined expression levels, y-axis indicates fold enrichment compared to specific subset frequency in the genome. (D) Relative proportion of differentially or equally expressed genes within gene sets of differentially, equally or non-methylated genes, y-axis indicates fold enrichment compared to specific subset frequency in the genome. Significant differences (Students tTest; p≤0.05) are marked with an asterisk (*). Coloured brackets indicate significant differences for similarly coloured bars within the bracket. Statistical analyses and gene lists are shown in Table S2. Abbreviations: exp. (expressed); dXM (preferential expression in meristem as Δlog2(Me/Le)); dXL (preferential expression in leaf as Δlog2(Me/Le)); X(M = L) (equal expression in meristem and leaf); dM+ (H3K27me3 meristem specific); dL+ (H3K27me3 leaf specific); M = L (equal level of H3K27me3 in leaf and meristem); see methods for details on subsets of differentially expressed and methylated genes. In whole genome analyses of H3K27me3 in seedlings, Oh et al. [20] identified 1001 H3K27me3 targets which were not revealed in our analyses (Figure 2A). 824 of these are expressed in at least one of the tissues we analyzed (data not shown), suggesting that these targets are seedling-specifically methylated and are subjected to dynamic changes of H3K27me3 in development.
Functional characterization of H3K27me3 target genes by Gene Ontology and gene family analyses
Previous analyses of H3K27me3 target genes in seedlings identified transcription factors as a major class of H3K27me3 target genes [5]. To reveal whether all transcription factor families and other gene families are preferentially targeted by H3K27me3 we analyzed our datasets generated from leaves and meristems.
We first exploited the resources of the Gene Ontology (GO) annotations from TAIR (http://www.arabidopsis.org/) providing functional categorizations of Arabidopsis protein coding genes (Figure S3). As expected the molecular function “transcription factor activity” was enriched in H3K27me3 target genes, whereas general cellular housekeeping functions like “DNA or RNA metabolism” and “electron transport or energy pathways” were depleted. Importantly, also tissue-specific differences were uncovered: a significant depletion of leaf-specific H3K27me3 target genes was observed in the cellular component “plastid”, consistent with the restriction of photosynthesis to leaves. In addition, genes associated with “kinase activity” and “transporter activity” were revealed as preferential H3K27me3 targets in the meristematic sample, suggesting specific repression of these sets of genes in undifferentiated cells.
We further dissected the H3K27me3 target genes by detailed analysis of gene families with a focus on transcription factor, enzyme and transporter gene families as these were enriched in the broad GO term analyses of H3K27me3 targets (Figure 5; Figure S4). Gene families involved in fundamental cellular processes like DNA replication, cell cycle regulators and protein phosphorylation/dephosphorylation were significantly depleted in H3K27me3 target genes (Figure S4; Table S3). Surprisingly, several transcription factor families were largely not regulated by H3K27me3 including the AUXIN RESPONSE FACTOR (ARF), CCAAT-HAP2 (HAP2) and SQUAMOSA-PROMOTER BINDING LIKE (SPL) gene families (Figure 5). Other transcription factor gene families showed a strong bias towards tissue-specific targeting by H3K27me3 (dM+; dL+) suggesting a specific regulation in the meristem or leaves. Leaf-specific H3K27me3 target genes comprise known regulators of meristem function like HOMEOBOX transcription factors and several other gene families (e.g. IAA and DOF transcription factors), while TCP, CONSTANS-like and GRAS-transcription factors were targeted by H3K27me3 specifically in the meristem indicating a role for theses gene families in differentiation processes or leaf development. A surprisingly large set of enzyme and transporter gene families were uncovered as H3K27me3 targets which are involved in differentiation or developmental processes (e.g. cell wall biosynthesis), hormone biosynthesis (YUCCA flavin monooxygenases, CYTOCHROME P450s) or transport of photosynthetic products (sucrose transporters) and hormones (PINFORMED (PIN)-like auxin efflux carriers) (Figure S4, Table S3). Previous analysis had identified the actin regulator formin ARABIDOPSIS FORMIN HOMOLOGUE 5 (AtFH5) as Pc-G target gene whose mis-expression is partially responsible for Pc-G mutant seed phenotypes, revealing that regulation of enzymatic genes is an important function of Pc-G proteins [39].
Fig. 5. Abundance of H3K27me3 target genes in transcription factor gene families. 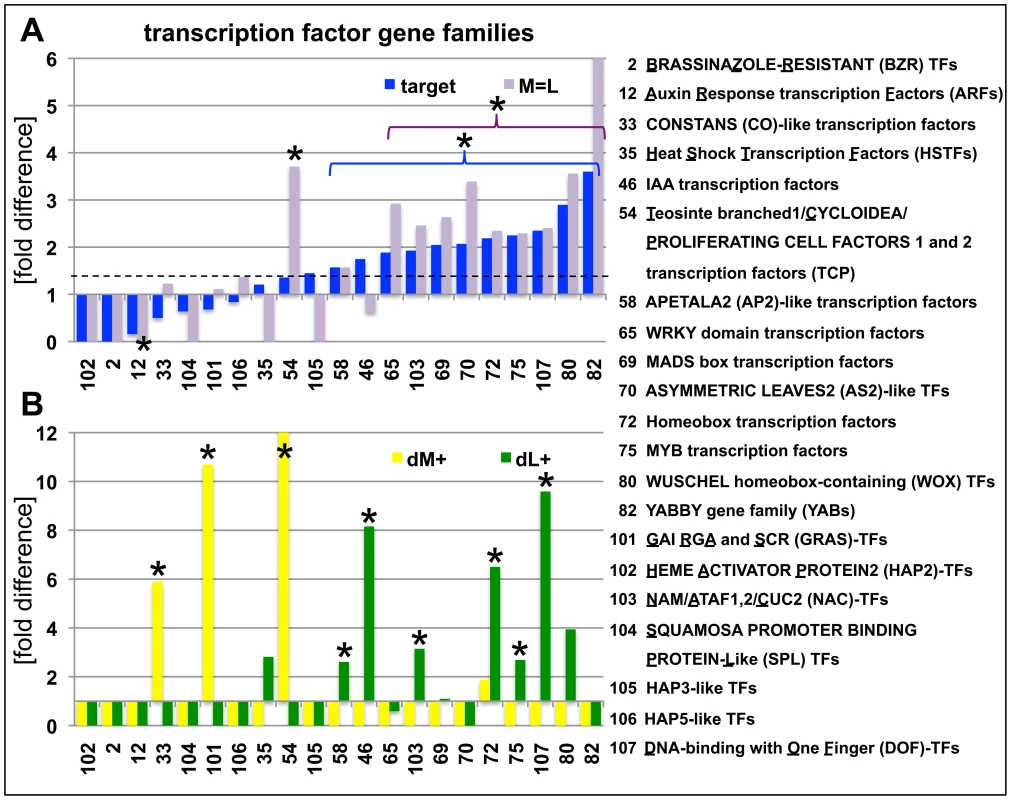
(A) Relative enrichment or depletion of H3K27me3 target genes (blue) or equally methylated genes (M = L, purple) in specific transcription factor families, y-axis indicates fold difference relative to the observed frequencies of specific subsets within the genome. The dashed line indicates relative enrichment of H3K27me3 targets in all transcription factor genes (1907 in total, taken from (http://plntfdb.bio.uni-potsdam.de/v3.0/)) compared to genome average. (B) Relative enrichment or depletion of meristem-specific (dM+, yellow) or leaf-specific (dM+, green) H3K27me3 target genes in specific transcription factor families, y-axis indicates fold difference relative to the observed frequencies of specific subsets within the genome. Asterisks indicate significant differences (p≤0.05, χ2-test) compared to total number of genes for a specific subset. Coloured brackets indicate significant differences for similarly coloured bars within the bracket. Details of statistical analyses can be found in Table S3. Analyses of additional gene families are shown in Figure S4. Lastly, we studied whether specific transposable element gene families were overrepresented in the different tissues. We generally observed a significantly larger amount of retrotransposons compared to DNA transposons as H3K27me3 target genes (Figure S4H, Table S3). Also tissue-specific differences were identified among retrotransposon gene families. Whereas gypsy-like retrotransposons were significantly enriched in the meristematic fraction, copia-like retrotransposons were overrepresented in leaves and athila-like retrotransposons almost completely non-targeted by H3K27me3 in leaves (Figure S4H).
Collectively, these analyses uncovered H3K27me3 targeting of a diverse set of gene families which are involved in biosynthesis and transport of secondary metabolites and transcriptional regulation and ultimately in cellular differentiation. These sets of genes include many known regulators of meristem or leaf development and provide an important resource for the identification of additional, novel factors involved in differentiation or stem cell regulation. In addition, also specific transposable element gene families are tissue-specifically targeted by H3K27me3 suggesting differential regulation of TE genes in the different tissues.
H3K27me3 targets specific miRNA gene families and is reduced at miRNA target genes
During the analyses of H3K27me3 targets we found that a large number of miRNA and several trans-acting siRNA (tasiRNA) genes carried H3K27me3 which was, to our knowledge, not previously revealed (Figure 2). These regulatory RNAs display differential methylation in the same way as protein coding genes, thus many show leaf-specific H3K27me3, whereas only few carry H3K27me3 exclusively in the meristem (Figure 2C). Seventy four of the annotated 174 miRNA loci were identified in our analysis and we revealed additional 12 that were detected, but not described by previous studies [5], [20]. Thus, nearly 50% of all microRNA genes are covered by H3K27me3 (Figure 2, Table 1). A high number of miRNAs (25 of 103 unique miRNAs) are encoded in multiple loci which constitute 96 of all 174 miRNA loci (Arabidopsis Small RNA Project (http://asrp.cgrb.oregonstate.edu/) [40]). Multiple and single loci miRNAs are regulated by H3K27me3 but miRNAs encoded in multiple loci are heavily targeted (72%) whereas only 22% of miRNAs present at a single locus are H3K27me3 targets.
Tab. 1. Regulation of miRNA and tasiRNA genes and their targets by H3K27me3. 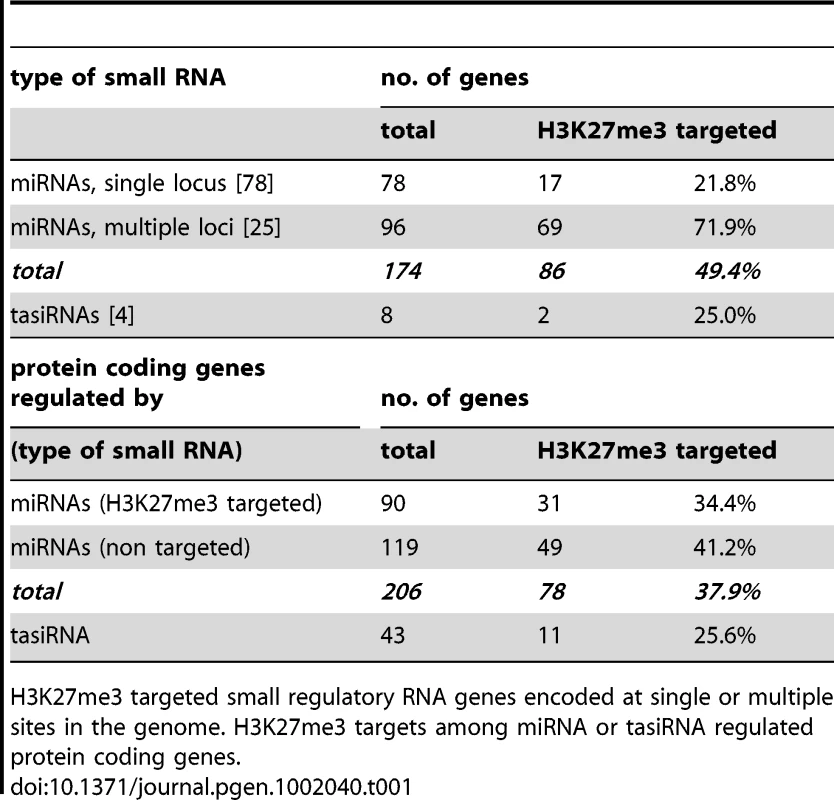
H3K27me3 targeted small regulatory RNA genes encoded at single or multiple sites in the genome. H3K27me3 targets among miRNA or tasiRNA regulated protein coding genes. Currently 206 genes including many developmentally important transcription factors are known which are negatively regulated by miRNAs at a post-transcriptional level. Interestingly, genes that are regulated by a H3K27me3 covered miRNA genes less frequently carry H3K27me3 compared to genes whose expression is controlled by a miRNA gene devoid of H3K27me3 (Table 1). This bias is even more apparent when specific miRNA and tasiRNA gene families and their targets are analyzed (Table 2): whereas most miRNA169, miRNA156/157, miRNA167 and tasiRNA3 loci carry H3K27me3, their target genes (HAP2, SPL or ARF genes) are largely devoid of H3K27me3. The lack of H3K27me3 targeting of these transcription factor genes is in contrast to the generally high enrichment of transcription factors as H3K27me3 targets (Figure 5; compare HAP2, SPL and ARF families to average enrichment in H3K27me3 targets of all transcription factor genes, p<0.05 (χ2-test) for HAP2 and ARF genes (Table S3)). However, H3K27me3-mediated repression of the miRNA genes likely permits an indirect positive regulation of these specific transcription factor families by Pc-G proteins. Interestingly, for other gene families (e.g. TCP and HD-ZIP transcription factors) both miRNA genes and their targets carry H3K27me3, suggesting that expression of these transcription factors is regulated transcriptionally by Pc-G proteins and post-transcriptionally by miRNAs.
Tab. 2. H3K27me3 targeted miRNAs or tasiRNAs encoded by multiple loci and H3K27me3 regulation of their target genes. 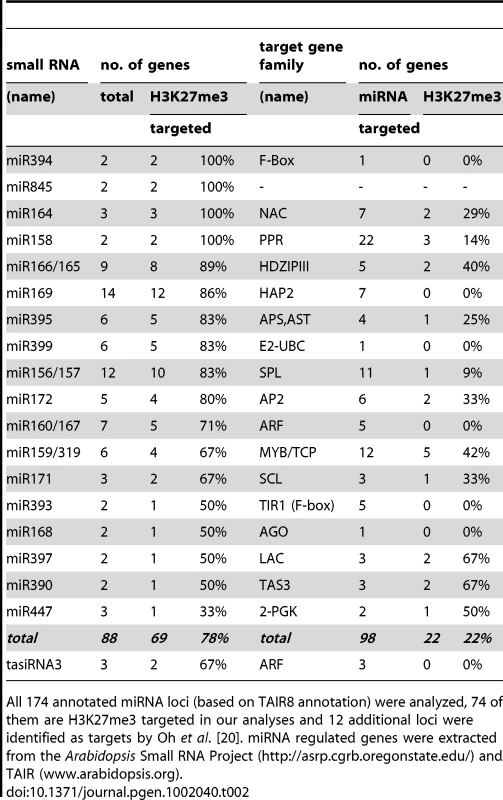
All 174 annotated miRNA loci (based on TAIR8 annotation) were analyzed, 74 of them are H3K27me3 targeted in our analyses and 12 additional loci were identified as targets by Oh et al. [20]. miRNA regulated genes were extracted from the Arabidopsis Small RNA Project (http://asrp.cgrb.oregonstate.edu/) and TAIR (www.arabidopsis.org). Biosynthesis, transport, perception, and signaling genes of the phytohormone auxin are regulated by Pc-G/H3K27me3
Our in-depth gene family analyses revealed enrichment of specific biosynthetic enzymes, transporters and transcription factors (Figure 5). Therefore, we inquired whether H3K27me3 targets entire developmental pathways which involve local biosynthesis of a signaling molecule, its transport, perception and signal transduction. We focused on the phytohormone auxin as we initially realized that the PIN auxin efflux carriers are largely H3K27me3 targets (Figure S4). Several pathways and gene families (YUCCA monooxygenases, CYTOCHROME P450s and TRYPTOPHANE AMINO TRANSFERASES) participate in the biosynthesis of the phytohormone auxin from tryptophan (reviewed in [41], [42]). Auxin is transported by several different carrier proteins in the plant. The AUXIN RESISTANT1 (AUX1)/LIKE AUX1 (LAX) gene family enables auxin influx and PIN - and several ATP-BINDING CASSETTE (ABC) transporters mediate auxin efflux (reviewed in [43]). The analysis of gene families uncovered that most of these gene families are enriched for H3K27me3 targets (Figure 5, Figure S4), including the genes for which a role in auxin regulation was shown (Table 3, Table 4). Interestingly, although the auxin receptor gene TRANSPORT INHIBITOR RESPONSE 1 (TIR1) and its homologues are largely devoid of H3K27me3, miRNA393a which negatively regulates these is targeted by H3K27me3 in leaves (Table 4). Similarly, the ARF transcription factor genes which are involved in repressing auxin inducible genes (reviewed in [44]) are depleted in H3K27me3, however, miRNAs and tasiRNAs controlling ARF expression levels are largely targeted by H3K27me3 (Figure 5, Table 2, Table 4). On the other hand, IAA class of transcription factors which positively regulate auxin responsive genes are highly enriched for H3K27me3 targets, similar to other transcription factor families. Thus, auxin biosynthesis, transport, perception and transcriptional responses are controlled by H3K27me3, both by direct targeting of the regulatory genes or by indirect regulation through miRNAs. The analyses of the auxin pathway therefore reveal that not only specific gene families but also entire pathways involving diverse gene families can be controlled by H3K27me3.
Tab. 3. Genes involved in auxin biosynthesis and transport are regulated by PcG/H3K27me3. 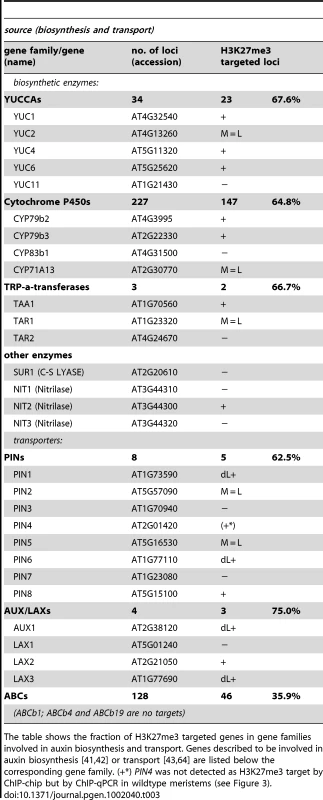
The table shows the fraction of H3K27me3 targeted genes in gene families involved in auxin biosynthesis and transport. Genes described to be involved in auxin biosynthesis [41], [42] or transport [43], [64] are listed below the corresponding gene family. (+*) PIN4 was not detected as H3K27me3 target by ChIP-chip but by ChIP-qPCR in wildtype meristems (see Figure 3). Tab. 4. Genes involved in auxin perception and signaling are regulated by PcG/H3K27me3. 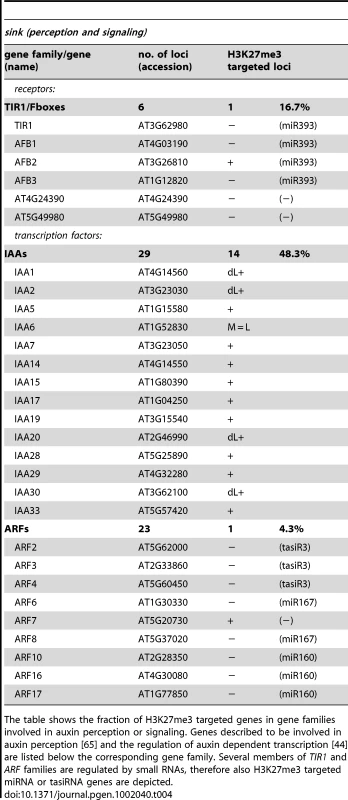
The table shows the fraction of H3K27me3 targeted genes in gene families involved in auxin perception or signaling. Genes described to be involved in auxin perception [65] and the regulation of auxin dependent transcription [44] are listed below the corresponding gene family. Several members of TIR1 and ARF families are regulated by small RNAs, therefore also H3K27me3 targeted miRNA or tasiRNA genes are depicted. H3K27me3 is entirely dependent on Pc-G proteins
Only selected plant genes expressed in vegetative tissues were previously analyzed for an overlap of H3K27me3 and Pc-G protein binding and dependence of H3K27me3 on Pc-G proteins [8], [14], [45], [46]. Although immunofluorescence analyses on the severe clf/swn mutants revealed a reduction in euchromatic H3K27me3 [15], it is currently unclear whether the H3K27me3 is completely dependent on Pc-G proteins. To reveal whether presence of H3K27me3 is likely corresponding to Pc-G binding and regulation we used immunoblot analyses to study histone methylation levels in various PRC2 mutants. We used mutants that are partially or completely deficient for Arabidopsis homologs of Drosophila E(z) [clf-28 (null allele), swn-7 (null allele)] or Suppressor of zeste 12 [vernalization2-1 (vrn2-1) (null allele), embryonic flower2–10 (emf2–10) (weak allele of emf2)] [18], [47]. We studied single and double mutants of these as they show different degrees of reduction in somatic Pc-G activity: moderate reduction in clf-28 and emf2–10 more severe reduction in vrn2/emf2–10 double mutants and complete loss of somatic Pc-G activity in the clf/swn double mutant (Figure 6) [18]. H3K27me3 was completely lost in the clf/swn mutants, strongly reduced in vrn2/emf2-10 mutants and only mildly affected in clf or emf2–10 mutants (Figure 6). Loss of H3K27me3 in clf/swn was correlated with a strong increase in H3K4me3. H3K27me1, a modification preferentially present in heterochromatin, was not affected in any of the mutants whereas H3K27me2 showed a clear reduction in vrn2/emf2–10, but increase in clf/swn mutants (Figure S5). Thus, our study of histone methylation in Pc-G mutants strongly suggests that all H3K27me3 is catalyzed by PRC2 proteins and that a possible re-distribution to heterochromatic regions [15] involves only marginal amounts of H3K27me3.
Fig. 6. Immunoblot analyses of histone modifications in Pc-G mutants. 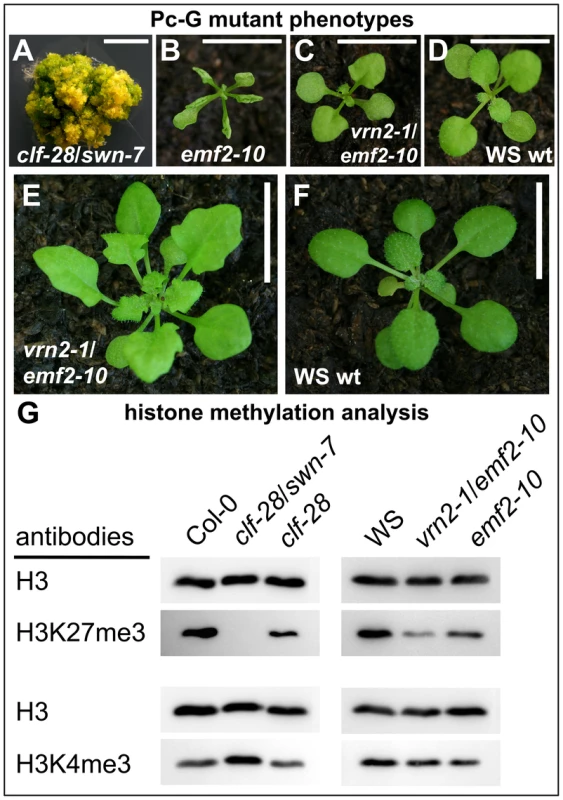
(A) clf-28/swn-7 double mutants, 60 d old and grown in axenic conditions in long days, (B) emf2–10, (C) vrn2–1/emf2–10 double mutants, (D) Wassilewskija (WS) wildtype. Plants in (B)–(D) are 17 old and grown in short day conditions. (E), (F) 45 d old plants grown under short day conditions, (E) vrn2-1/emf2-10 double mutant, (F) wildtype. Scale bars in pictures (A) to (D) indicate 0.5 cm and in (E) and (F) 1 cm. (G) Immunoblot analyses of H3K4me3 and H3K27me3 levels in Pc-G mutants. Similar amounts of histone H3 were loaded as confirmed with a H3 specific antibody detecting the unmodified C-terminal part of H3. Immunoblot analyses of additional histone modifications are shown in Figure S5. Since the global analysis of H3K27me3 abundance revealed a strong reduction even in the vrn2/emf2–10 double mutant showing relatively mild morphological alterations (Figure 6A) we were interested if a H3K27me3 reduction occurs at most genes or gene-specifically. We therefore studied H3K27me3 of selected genes in leaves from wildtype and vrn2/emf2–10 plants by ChIP-qPCR (Figure S6). This analysis identified three different classes: STM and FUS3 displayed no difference in H3K27me3 in the mutants, several genes including KNAT2 or PIN8 had intermediate H3K27me3 levels and most analyzed genes (e.g. PIN1 and PIN6) showed a strong reduction or a complete loss in the mutants. Thus, the observed global reduction in H3K27me3 is rather caused by the loss of H3K27me3 at some loci than by an equal reduction of H3K27me3 at all target loci (Figure S6).
Pc-G proteins control tissue-specific expression of their target genes
To study possible changes in gene expression resulting from a reduction in H3K27me3, we performed qRT-PCR based expression analyses of clf/swn and vrn2/emf2–10 double mutants (Figure 7, Figure 8). Clf/swn mutants show complete loss of H3K27me3, do not maintain organ identity post-embryonically and are therefore only comparable to wildtype shortly after germination. However, vrn2/emf2–10 mutants show strong leaf serration especially in older leaves, but produce leaves and flowers (Figure 6), thus tissue-specific changes in gene expression can also be analyzed at later developmental stages. In 9 d old clf/swn seedlings, we observed mis-expression of several previously analysed H3K27me3 targets [8], [48], [49], ranging from mild (KNAT2) to strong (KNAT6, STM and AG) mis-expression (Figure 7A). Surprisingly, for TCP2, TCP4, TCP10 and SPL3 we observed a strong down-regulation in clf/swn mutants although they likely lost H3K27me3 (Figure 7B). However, TCP2, TCP4 TCP10 are negatively regulated by miRNA319 [50] which is targeted by H3K27me3 (Figure 3). Consistently, we observed mis-expression of the miRNA319a precursor in clf/swn mutants which is likely responsible for post-transcriptional down-regulation of TCP2, 4 and 10 in the Pc-G mutant. Expression of TCP5 which is not targeted by miRNA319 is not affected in the mutant. Similarly, SPL3 whose abundance is regulated by the H3K27me3 targeted miRNA156 showed reduced expression in clf/swn mutants (Figure 7B).
Fig. 7. Pc-G proteins control tissue-specific expression of H3K27me3 target genes. 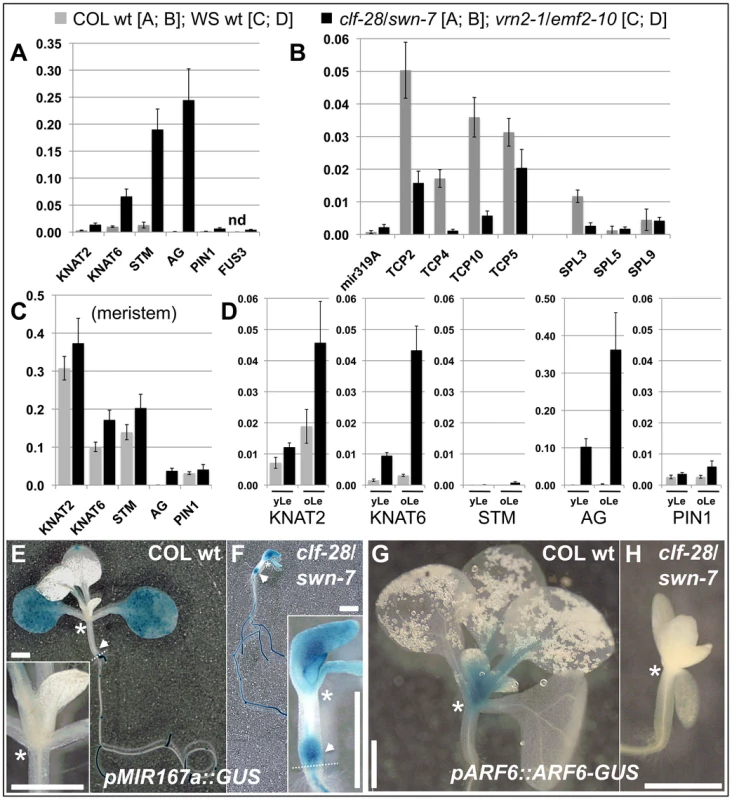
(A) – (C) qRT-PCR based expression analyses of (A, B) 9 d old wildtype and clf-28/swn-7 plants or (C, D) 45 days old, dissected wildtype and vrn2-1/emf2–10 plants. Samples were enriched for meristem (Me) (C), young leaves (yLe, up to 3 mm blade length) or older leaves (oLe, 3 – 10 mm) (D). Expression values were normalized to At5g60390 (EF-1α). (E, F) Colorimetric analyses of pMir167a::GUS expression in 9 d old wildtype (E) and clf-28/swn-7 double mutants (F). Insets are close-ups. (G, H) Expression of pARF6::ARF6-GUS 9 d old wildtype (G) and clf-28/swn-7 double mutants (H). pARF6::ARF6-GUS carries the mir167 binding site. Asterisks mark meristematic region, white arrow base of hypocotyl. The border between the hypocotyl and the root is indicated with a dashed white line. Scale bars indicate 1 cm. All plants were grown in short day conditions. Fig. 8. Expression analysis of transposable element genes. 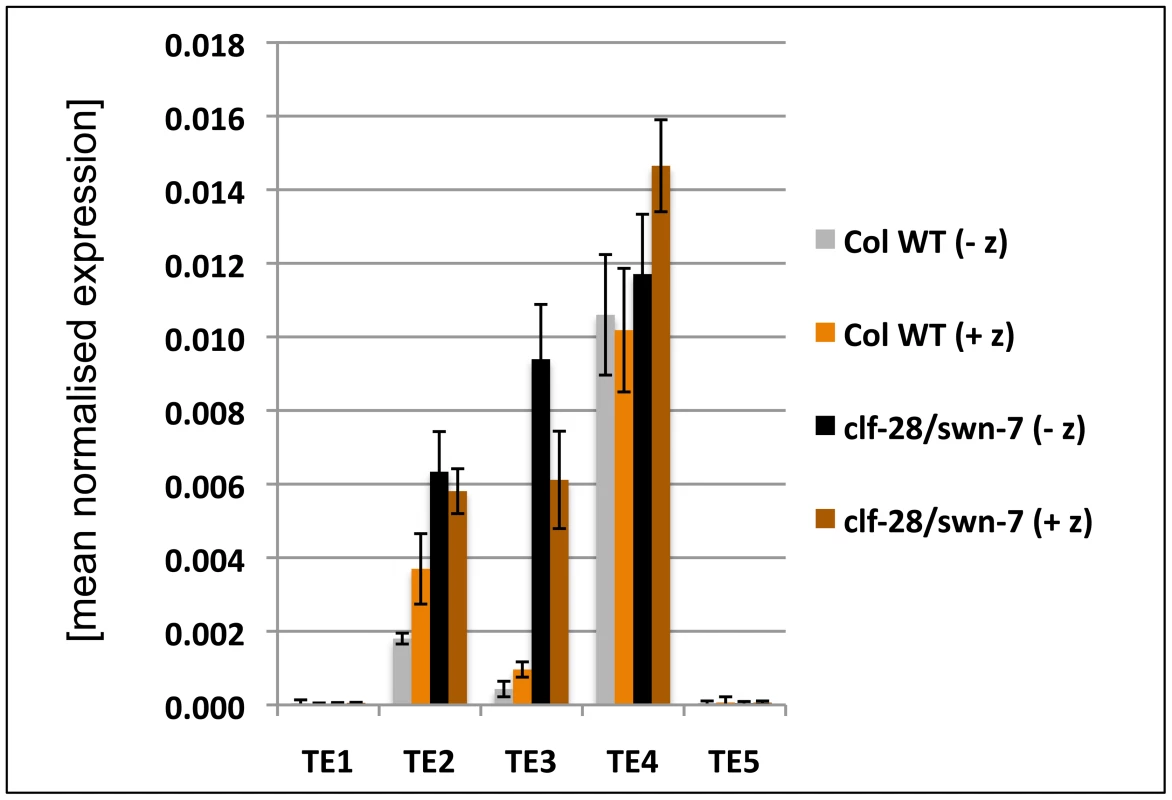
The expression of several transposable element genes was analysed in 9 d old wildtype and clf-28/swn-7 seedlings by qRT-PCR. Plants were grown on media with (+ z) and without the DNA methyltransferase inhibitor zebularine (− z). Expression values were normalized to At5g60390 (EF-1α). [TE1 (AT3G28400; hAT transposase), TE2 (AT3G21020; copia element), TE3 (AT3G21040; copia element), TE4 (AT3G43690; copia element), TE5 AT4G02960; copia element)]. H3K27me3 targets 6 of the 8 PIN gene members (Table 3), we therefore also analysed their expression in clf/swn and vrn2/emf2–10 mutants. Except for PIN1, none of the PIN genes showed upregulation in the mutants (Figure 7A, Figure S6). PIN3, PIN4 and PIN7 which are all not targeted by H3K27me3 in leaves even showed a strong down-regulation of expression in clf/swn mutants.
As we also revealed H3K27me3 targeting of several TE genes, we studied expression of several H3K27me3 targeted TE genes. We observed upregulation of two TE genes in the clf/swn mutant (Figure 8), suggesting that Pc-G proteins and H3K27me3 can prevent expression of TE genes, which is consistent with a previous study revealing Pc-G mediated silencing of a TE gene in the endosperm [51]. To study whether interference with DNA methylation can further enhance activation of TE genes, we grew wildtype and clf/swn plants on media containing the DNA methyltransferase inhibitor zebularine [52] and analysed TE activation (Figure 8). The tested TE genes were neither activated by zebularine in wildtype nor further transcriptionally activated in clf/swn suggesting DNA methylation independent regulation of the TE genes which are activated in the Pc-G mutants.
Lastly, we investigated whether Pc-G mutants control tissue-specific expression of differentially methylated H3K27me3 targets. Mir167a and 167d genes show higher levels of H3K27me3 in the meristem compared to leaves (Figure 3). Reporter transgenes in which β-GLUCURONIDASE (GUS) was fused to the promoters of mir167a and mir167d [53] revealed preferential expression of the miRNA genes in cotyledons, but exclusion from the meristem, consistent with the H3K27me3 patterns (Figure 7, Figure S8). In clf/swn mutants, the expression domains of mir167a and mir167d reporters were expanded to the shoot apical meristem, the petioles, the main root and the base of the hypocotyl (Figure 7F, Figure S8). The genes ARF6 and ARF8 genes are regulated by mir167 which is in agreement with their mutually exclusive expression domains [53] (Figure 7E, 7G). Consistent with mis-expression of mir167 in clf/swn mutants we observed loss of shoot meristematic expression of a pARF6::ARF6-GUS reporter gene construct that harbours the miRNA binding site (Figure 7H). This further demonstrates the expansion of the miRNA expression domain to the meristem.
To uncover tissue-specific mis-expression of differentially methylated H3K27me3 targets in the vrn2/emf2–10 mutant we dissected plants and analyzed gene expression in meristematic tissue, young leaves (up to 3 mm blade length) and older leaves (3–10 mm). In wildtype, KNAT2, KNAT6, STM and PIN1 are strongly expressed in the meristem and carry H3K27me3 preferentially in leaves (Figure 3, Figure 7). Expression of these genes was not altered in meristems of vrn2/emf2–10 double mutants as expected (Figure 7C). In older leaves of the mutant, however, the KNAT genes were activated which correlates with a reduction in H3K27me3 (Figure 7D, Figure S6). STM displayed only moderate mis-expression in leaves, consistent with similar levels of H3K27me3 in wildtype and vrn2/emf2–10 mutants. Although H3K27me3 of PIN1 is depleted in vrn2/emf2-10 mutant leaves the gene is only slightly activated in the mutant (Figure 7, Figure S6). For several H3K27me3 target genes exhibiting differential methylation we revealed tissue-specific mis-expression in Pc-G mutants indicating that Pc-G proteins restrict expression of their targets by depositing tissue-specific H3K27me3. We also showed that Pc-G proteins transcriptionally regulate miRNA consistent with an up-regulation of miRNA genes and concomitant down-regulation of miRNA target genes in Pc-G mutants. In addition, although no global effect on DNA methylation and TE activation was revealed in clf/swn mutants [15], a subset of TE genes may be regulated by Pc-G proteins (Figure 8 and [51]).
Discussion
In this study, we analyzed the dynamic regulation of the PRC2-mediated modification H3K27me3 in undifferentiated meristematic and differentiated leaf tissue. Our analyses revealed differential methylation of hundreds of H3K27me3 target genes including protein coding, transposable element and microRNA genes and expand previous observations that plant Pc-G proteins have important roles in controlling tissue-specific expression patterns of gene families and regulatory networks.
Enrichment of meristematic and leaf tissue for H3K27me3 and expression analyses
To study the dynamics of H3K27me3 in Arabidopsis development and its function in tissue differentiation we focused on the vegetative shoot apical meristem which harbors continuously dividing stem cells and gives rise to terminally differentiated organs (leaves) and also differentiates into the flower producing inflorescence meristem. Manual dissection of meristems and young leaves was performed on clv3 mutants as these have strongly enlarged meristems and increased stem cell numbers [35], but show no obvious defects in leaf development. Independent analyses on dissected wildtype meristems and leaves confirmed differential H3K27me3 of all tested genes (Figure 3). Furthermore, a large overlap of H3K27me3 targets with previously published data on seedlings [5], [20] revealed a similar set of H3K27me3 targets both in wildtype seedlings and clv3 mutant leaves and meristems. Thus, overall the experimental system appeared highly suitable to study the dynamics of H3K27me3 on a whole genome level during differentiation.
H3K27me3 is dynamically regulated during differentiation and correlates with gene repression in a tissue-specific manner
Our analyses identified several hundred protein coding genes, transposable elements and microRNA genes exhibiting differential H3K27me3 patterns between meristematic and leaf tissue. Genes that acquired H3K27me3 during differentiation included several meristematic regulators like STM or KNAT2 and 6, but also many previously non-characterized genes which may present novel regulators of meristem regulation and maintenance. Leaf tissue is developmentally derived from continuously dividing cells in the shoot tip which differentiate in the periphery of the meristem. Thus, H3K27me3 at the subset of genes showing meristem-specific H3K27me3 must be actively or passively removed during differentiation. H3K27me3 demethylases have not been identified in plants yet, however, our analyses strongly suggest the contribution of these enzymatic activities to confer dynamic regulation of H3K27me3. Charron et al [22] observed differential H3K27me3 patterns of protein coding and transposable element genes when dark grown seedlings were compared with dark grown seedlings shifted to light. In addition, dynamic changes in H3K27me3 and/or Pc-G protein binding were also unveiled both during Drosophila development and differentiation of mammalian embryonic stem cells suggesting conserved mechanisms which confer developmental dynamics of H3K27me3 [23], [28], [32], [54].
We confirmed that H3K27me3 is largely excluded from heterochomatic regions and transposable element (TE) genes, especially in leaf tissue (only 4% of all targets) [5], [20]. Interestingly, more than 15% of H3K27me3 targets detected in the meristem were TE genes (Figure 2). H3K27me3 detection at TEs was likely not due to cross-reaction of the antibody to H3K27me1 which is found in heterochromatin, because the same anti-H3K27me3 antibody was used for both analyzed tissues. In addition, the antibody was not detecting histones in clf/swn mutants which lacked all H3K27me3 but showed no alterations in H3K27me1 (Figure 6, Figure S5). Transposable elements are usually repressed throughout sporophytic development, thus loss of H3K27me3 in leaves likely does not result in transcriptional activation. The stem cells are required for plant germline formation and therefore must be particularly protected from transposable element activation. Thus, H3K27me3 may represent an additional, meristem-specific silencing mechanism besides the canonical heterochromatic marks H3K9me2 and H3K27me1 [55], [56]. Also in mammalian embryonic stem cells, transposable elements are marked by a specific set of repressive histone modifications, suggesting that pluripotent cells are protected by a large array of modifications [32]. For a few TE genes we observed up-regulation in the clf/swn Pc-G mutant, consistent with a previous report showing up-regulation of a TE gene in Pc-G mutant endosperm [51], suggesting that Pc-G proteins contribute to the regulation of TEs.
Our data revealed that non-expressed genes are preferential H3K27me3 target genes (Figure 4) as previously reported [5], [20], [21], [31]. Interestingly, protein coding genes which showed strong expression differences between meristem and leaf are enriched for H3K27me3 targets. In addition, differential H3K27me3 was anti-correlated with tissue-specific gene expression patterns (Figure 4). Thus, similar to Drosophila, Pc-G proteins likely control ON/OFF expression states of their targets in Arabidopsis [30]. However, tissue-specific H3K27me3 is not the only determinant which generates tissue-specific expression patterns as many genes showing differential expression are not covered by H3K27me3.
Tissue-specific targeting of particular gene families by H3K27me3 in Arabidopsis
Previous analyses of H3K27me3 identified transcription factors as one major class of H3K27me3 targets, but preference for certain families was not unveiled [5]. We confirmed this finding as most transcription factor families including MADS-, WOX-, HOMEOBOX - and YABBY-transcription factors were preferentially targeted by H3K27me3. However, others including SPL-, ARF - and HAP2-transcription factor genes were largely devoid of H3K27me3, indicating that transcription factor genes are not per se enriched for H3K27me3 (see also below). In addition, certain transporter gene families (e.g. sucrose transporters, auxin carriers) and gene families involved in biosynthetic pathways (peroxidases, cytochrome P450 genes) were among the H3K27me3 targets. In Drosophila and mammals, transporter and biosynthetic genes (hydrolases, cytochrome P450 genes) were also revealed as preferential H3K27me3 targets in addition to transcription factor genes [6], [23], [26], [29]. This strongly suggests an unexpected level of conservation not only of Pc-G proteins but also of their target gene families between animals and plants.
We also identified gene families and functional categories showing preferential H3K27me3 targeting in the meristem (e.g. TCP transcription factor genes, Cytochrome P450 genes; functional categories “plastid” and “kinase activity”) or in leaves (e.g. MYB and HOMEOBOX transcription factor genes). In particular the differential methylation of HOMEOBOX transcription factor genes confirmed the validity of our approach as these are repressed in leaves by Pc-G proteins ([48] and this study).
Thus, our in-depth analyses of H3K27me3 targets confirmed specific gene families as preferential H3K27me3 targets, but also identified novel pathways and gene families that are likely controlled by Pc-G proteins and epigenetic gene regulation.
Dynamic regulation of miRNA genes by H3K27me3 controls expression of miRNA target genes
Our study discovered a large fraction of microRNA genes as H3K27me3 targets many of which exhibited tissue-specific differences in H3K27me3 (Figure 2, Figure 3, Table 1, Table 2). In addition, we uncovered mis-expression of several miRNA genes and down-regulation of their target genes in Pc-G mutants (Figure 7). Although miRNAs largely post-transcriptionally regulate their target genes, some miRNAs recruit the DNA methylation machinery to their target genes resulting in transcriptional repression [57]–[59]. Similarly to H3K27me3, miRNA genes target a multitude of transcription factor genes [60], [61], raising the possibility that miRNAs mediate recruitment of Pc-G proteins to some of their targets. Our analyses suggest that this is likely a rare phenomenon as transcription factor genes which are miRNA targets, are largely not targeted by H3K27me3 (Table 1, Table 2), despite the general enrichment of transcription factors as H3K27me3 targets ([5] and this study). Nonetheless, we also identified gene pairs, for which both the miRNA gene (e.g. mir319a) and their targets (e.g. TCP2, TCP10) displayed differential, but antagonistic H3K27me3 patterns. Thus, for a subset of genes miRNA-mediated targeting of H3K27me3 is an attractive possibility.
In summary, our studies provide evidence that Pc-G proteins and H3K27me3 do not only negatively regulate expression of transcription factor genes by direct association, but also positively control their expression by restricting expression of miRNA genes (Figure 9). This regulation appears to be particularly important in meristematic and leaf tissues as a large fraction of miRNA genes is differentially methylated in these two tissues (Figure 2). Regulation of miRNA genes by Pc-G proteins is likely a conserved mechanism. Recent analyses of miRNA gene regulation during mouse and human lymphopoiesis and genome-wide profiling of Pc-G protein binding sites in Drosophila uncovered H3K27me3-mediated control of miRNA gene expression [62], [63]. In addition, more than 30 miRNA loci were discovered as H3K27me3 targets in human embryonic stem cells [26].
Fig. 9. Models of the H3K27me3-mediated regulation of protein coding genes and auxin signaling. 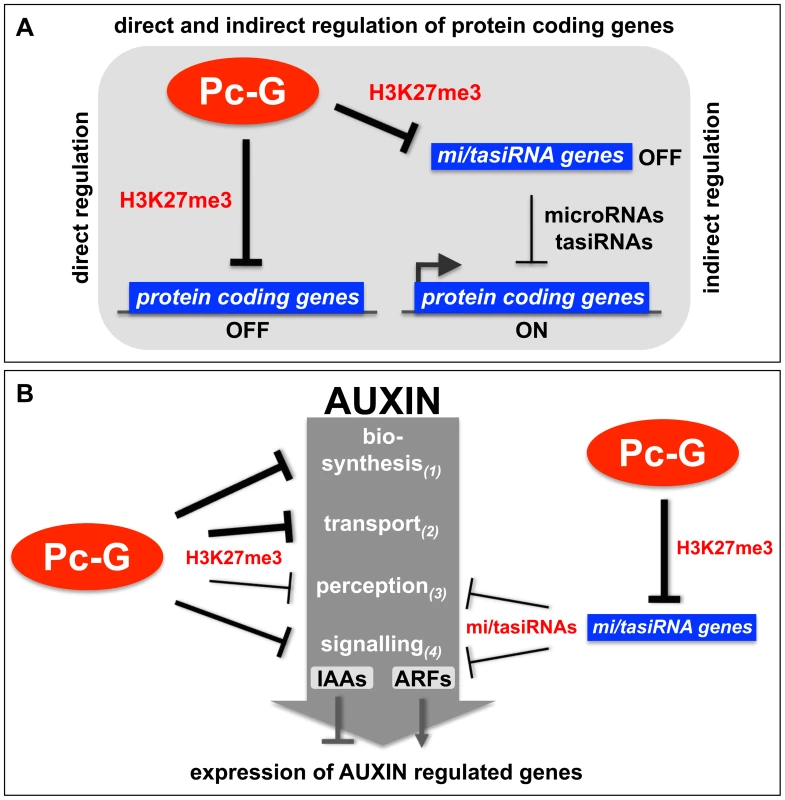
(A) Protein coding genes can be directly repressed by Pc-G proteins through the deposition of H3K27me3 or indirectly positively regulated by Pc-G proteins through the H3K27me3-mediated repression of small RNA genes (tasiRNAs and miRNAs) (see Table 1, Table 2 for details). (B) Pc-G proteins directly repress auxin biosynthesis, transport, perception and signaling (IAA transcription factors). Thickness of arrows is relative to number of genes targeted by H3K27me3. Pc-G proteins indirectly activate genes involved in auxin perception and signaling (ARF transcription factors) through the repression of miRNA (mir160, 167, 393) and tasiRNA (TAS3) genes. Positive regulation of the ARF transcriptional activator and auxin perception genes and negative regulation of IAA transcriptional repressor genes likely promotes expression of auxin inducible genes (see Table 3, Table 4 for details). H3K27me3 regulates diverse pathways in auxin biosynthesis, transport, perception, and signaling
Organ-specific auxin maxima are generated by both local auxin biosynthesis and auxin transport, which is largely mediated by the PIN efflux carrier proteins (reviewed in [43], [64], [65]). To date, three different pathways for the synthesis of auxin from tryptophan have been identified or proposed for plants (reviewed in [42]). Our analyses identified most of the key regulators of auxin transport and biosynthesis to be H3K27me3 targets several of which are leaf-specifically methylated (Figure 3, Figure 5, Table 3, Table 4). Additionally, our results suggest that auxin perception is controlled by Pc-G proteins as H3K27me3 targets the mir393a gene which in turn regulates the auxin receptor genes TIR1/AFB (Table 4) [66], [67]. In addition, H3K27me3 regulates the transcriptional responses to auxin as half of the AUX/IAA repressor genes are covered by H3K27me3. Although only one of the 23 ARF transcriptional activator genes carries H3K27me3, all known miRNAs or tasiRNAs that regulate a total of 8 ARF genes are H3K27me3 targets (Table 2). Thus, we propose that auxin responsive genes are largely suppressed in Pc-G mutants as H3K27me3 restricts expression of the AUX/IAA repressors and promotes expression of the ARF activators by controlling the expression of ARF-regulating miRNAs (Figure 7, Figure 9, Table 2). Consistent with this hypothesis, we observed down-regulation of several auxin-inducible PIN genes (PIN3, PIN4, PIN7) [68] in clf/swn mutants (Figure S7). In addition, the Pc-G mutant terminal flower2 was recently shown to have lower expression of auxin responsive genes including the synthetic reporter DR5::GUS [69] and genome-wide expression profiles of embryonic flower 1 (emf1) and emf2 Pc-G mutants identified more down - than upregulated auxin regulatory genes in the mutants [70]. Loss of vegetative Pc-G activity in clf/swn or vrn2/emf2 mutants is accompanied by loss of organ identity, generation of somatic embryos and callus-like appearance [18], [19] and all these phenotypes are associated with deregulated auxin responses. Although we cannot rule out that these phenotypes are auxin-independent, we showed mis-regulation of PIN, ARF and miRNA167 genes in Pc-G mutants revealing that loss of H3K27me3 is indeed associated with mis-regulation of genes involved in auxin responses (Figure 7). Other studies have also linked Pc-G proteins to hormonal control of gene expression: Charron and colleagues [22] uncovered strong H3K27me3 targeting of genes involved in gibberellic acid biosynthesis and inactivation. Global gene expression in emf1 and emf2 mutants revealed mis-regulation of many genes involved in diverse hormonal pathways [70]. However, many of these effects are likely indirect, thus a rigorous comparison of Pc-G protein binding, H3K27me3 targeting and gene expression profiles in Pc-G mutants in defined tissues will be essential to uncover the mis-regulated target genes that determine the Pc-G mutant phenotypes.
Role of H3K27me3 in Pc-G silencing and epigenetic inheritance
This and previous studies uncovered at least 25% of all Arabidopsis protein coding genes as H3K27me3 targets, some of which are tissue-specifically methylated [5], [20]–[22], [31]. Although H3K27me3 is entirely dependent on Pc-G proteins, H3K4me3 is enriched in clf/swn mutants (Figure 6) and H3K27me3 is strongly correlated with gene repression (Figure 4), only a limited number of H3K27me3 targets are mis-expressed in Pc-G mutants or in tissues in which H3K27me3 is lost (this study and [70]). Also in animals lack of Pc-G proteins results in mis-expression of only a small number of target genes [24]. Thus, loss of H3K27me3 may poise genes for activation but gene expression may require additional environmental or developmental cues. Thus, Pc-G proteins and H3K27me3 may confer stability of developmental processes and gene regulatory networks.
Pc-G proteins are considered as epigenetic regulators conferring mitotically stable gene expression states. The large number of H3K27me3 targets implies that either more than 25% of all protein coding genes are epigenetically regulated or that H3K27me3 has a more general gene regulatory role. Certainly, epigenetic phenomena like the vernalization response are controlled by Pc-G proteins and H3K27me3 [8], [71], [72]. In addition, Pc-G target genes are covered with H3K27me3 which may ensure perpetuation of the modification through replication [9]. Interestingly, for small genes like miRNA genes the number of modified nucleosomes may not be sufficient to guarantee high fidelity of inheritance.
This and previous studies unveiled tissue-specific dynamics of H3K27me3 distribution [22], [31], [51], suggesting that if H3K27me3 is generally mitotically heritable it may persist for only limited cell divisions until it is reset again to allow gene activation. The mechanisms of H3K27me3 inheritance and resetting can now be studied with the identified Pc-G target genes which are dynamically regulated during differentiation.
Materials and Methods
Plant material and growth conditions
All plants were grown under either long day (16 h light/8 h darkness) or short day conditions (8 h light/16 h darkness). The clavata3–9 mutant in Columbia background carries a point mutation in the CLV3 coding region which disrupts CLV3 function. clf-28 (SALK_139371), swn-7 (SALK_109121), clf-28 swn-7, vrn2-1 [47], emf2-10 [18] and vrn2-1/emf2–10 mutants were used in this study. Pmir167a::GUS, Pmir167d::GUS and pARF6::ARF6-GUS were previously described [53]. For expression and ChIP-chip analyses, clv3 mutant plants were grown for 9 weeks under short day conditions and enriched for meristematic and leaf tissue by manual dissection. Tissues for expression and ChIP-chip analyses were simultaneously harvested. For zebularine treatment, plants were germinated on plates containing 40 µM zebularine (Sigma, Munich) and harvested 9 days after germination.
Array-based expression analysis of different tissue samples
Array hybridization
Total RNA was extracted with QIAGEN plant RNeasy kit (Qiagen, Hilden), Superscript II (Invitrogen, Carlsbad) was used for cDNA synthesis. Probe labelling and array hybridisation on Agilent 44 k arrays [G2519F, V4 (Agilent, Santa Clara)] were performed by the Imagenes Corp. (Berlin, Germany).
Gene expression data analysis
The quality of the array hybridization was tested with scatter plots of the raw intensities (Figure S9). Pearson correlation coefficients were determined showing high similarity of biological replicates. The normalized and background subtracted data received from Imagenes was processed with a spreadsheet program (Excel, Microsoft) resulting in 22988 expressed genes (total), 22237 in the meristem (Me) and 22017 in the leaf (Le) sample (Figure S2). For the comparison of different tissues the log2 values [log2(Me); log2(Le)] of the normalized intensities and their difference between the tissues [Δlog2(Me-Le)] were calculated. Gene sets of different expression levels were binned to compare gene expression and histone methylation (Figure 4). Genes with a minimal difference in expression of Δlog2|Me-Le|≥2 which were statistically significantly different [determined by Students tTest (p≤0.05)] were considered as tissue specifically expressed. Genes with Δlog2|Me-Le| <1 and a standard deviation of the samples ≤0.05% were considered as equally expressed (Figure S2). Genes expressed in the different tissues area are provided in Table S4.
Tissue-specific H3K27me3 ChIP and ChIP-chip analyses
ChIP and array hybridization
For each tissue, 200 mg plant material was harvested on ice, crosslinked with 1% formaldehyde for 15 minutes under vacuum, frozen in liquid nitrogen and stored at −70° until chromatin extraction. A previously described protocol [8] was used for ChIP. Antibodies specific for H3K27me3 (#07-449) and H3K4me3 (#07-473) were retrieved from Millipore (Billerica, USA). Chromatin was sheared with a Bioruptor sonicator (10 pulses, 30 seconds each) (Diagenode, Liege, Belgium). High density tiling arrays with probes tiled approximately every 220 bp over the entire sequenced Arabidopsis genome were used as described [36], [37]. Input and precipitated DNA was amplified with a whole genome amplification kit (WGA2, Sigma, St. Louis) as described in “PROT30” of The Epigenome NoE web-page (http://www.epigenome-noe.net/). Labelling and array hybridisation were performed by the Imagenes NimbleGen service (Berlin, Germany).
H3K27me3 ChIP-chip data analysis
The array hybridization data was received from Imagenes (Berlin, Germany) as intensity ratios of precipitate and input samples in the GFF-file format. In a first processing step M-Values [M = log2(IP/I) where IP is signal intensity from hybridized immunoprecipitated DNA and I is signal intensity from hybridized input DNA] were determined. The quality of the array hybridization was analyzed with scatter plots of the M values for biological replicates (Figure S10). The correlation of the two biological replicates for each tissue was estimated with the Spearman's rank correlation coefficient (SCC) (Figure S10). The data was analyzed with SignalMap1.9 software (Nimblegen) and the Bioconductor R packages [73] in an R programming environment. M values were used as input for the Bioconductor package Ringo. The Ringo clustering algorithm has been developed specifically for widely occurring enrichments including those found for histone modifications [74]. Prior to cluster detection Ringo applies a running median to smooth the datasets. A window size of 1400 bp was used for running median. The cluster algorithm was executed with a distance cut off of 700 bp and an intensity threshold defined by the 80th percentile. The identified methylation clusters were used to calculate gene specific methylation scores for each annotation [based on TAIR8 genome annotation (www.arabidopsis.org)] and tissue (Me or Le) and scaled so that the maximum equals 100. Further analyses and comparisons of loci-specific methylation scores were calculated with a regular spreadsheet program (Excel, Microsoft). A scale normalisation on the level of methylation scores was performed for the different tissues to account for differences in precipitation efficiency. Since the array hybridisations were performed as replicates we only considered genes for which a positive methylation score was detected in both samples. This resulted in 6071 protein coding genes methylated in the meristematic sample and 6928 in leaf sample (Figure S11). Other annotations are depicted in Table S1. We grouped methylated genes in specific classes for further analyses after retrieving methylation scores (ms) for each annotation: methylated [ms (Me)>0 (Me>0 in the text) or ms(Le)>0 (Le>0) (target)], non methylated [ms(Me) = 0 and ms(Le) = 0 (Me = Le = 0) (non target)], equally [ms(Me) = ms(Le)>0 (M = L)] or differentially methylated in the meristem (ms(Me)≫ms(Le) [dM+]) or leaf sample (ms(Le)≫ms(Me) [dL+]). Two criteria were used to detect differentially methylated loci: ms(Me)/ms(Le)≥2 or ms(Le) = 0 (p≤0.05, Students tTest) and |ms(Me)-ms(Le)|≥10 for meristem specifically methylated loci (dM+) and ms(Le)/ms(Me)≥2 or ms(Me) = 0 (p≤0.05, Student’s tTest) and |ms(Me)-ms(Le)|≥10 for leaf specifically methylated loci (dL+). Equally methylated loci (M = L) were isolated with the criteria 0.5<ms(Me)/ms(Le)<2; |ms(Me)-ms(Le)|<10 and a standard deviation ≤0.05%. H3K27me3 target genes identified in the different tissues are listed in Table S5.
Gene Ontology (GO) term and gene family analyses of target genes
Specific gene ontologies (GO) were extracted for specific gene sets with the GO analysis tool on TAIR (www.arabidopsis.org/tools/bulk//go/index.jsp) and calculated as enrichment or depletion over the background of all protein coding genes in the genome (based on TAIR8). Significance was determined by a hypergeometric test. Gene families were extracted from TAIR and the plant transcription factor database (http://plntfdb.bio.uni-potsdam.de/v3.0/) [75] and compared to specific subsets of methylated genes. Significance of over - or under-representation of target genes within a gene family was tested with a χ2-test (in comparison with the observed frequency of targets or subsets within the genome).
qPCR based analysis of gene expression (qRT-PCR) and histone modifications (ChIP-qPCR)
All qPCR analyses were performed with a CHROMO4 cycler (BioRad, Munich) and a MesaGreen reaction mix (Eurogentec, Liege) with a two step program. The genes At5g60390 (EF-1α), At4g34270 and At1g13320 were used as reference genes in qRT-PCR based expression analyses [76]. For ChIP-qPCR analyses the PCRs were performed on input and immunoprecipitated samples, % of input was calculated and normalized to the reference gene FUSCA3 (FUS3, AT3G26790) which carries H3K27me3 and is not expressed in both meristematic and leaf sample. For H3K4me3 analyses, ACTIN2 (AT5G09810) was used as reference as it carries H3K4me3 and is equally expressed in both samples. Sequences of oligonucleotides used for gene expression and ChIP analyses are listed in Table S6.
Analyses of global histone modification in Pc-G mutants by immunoblot analyses
Histone enriched plant extracts were prepared from ten day old seedlings as described [77]. Various histone modifications were detected by standard immunoblot analyses using antibodies specific for H3 (Abcam; ab1791), H3K27me1 (Millipore; 07-448), H3K27me2 (Millipore; 07-452), H3K27me2 (Active Motif; 39245), H3K27me3 (Millipore; 07-449), H3K4me3 (Diagenode; pAb-003-050).
Histochemical analysis of Arabidopsis plants
Seven days old plants were histochemically assayed as previously described [78]. Photos were taken with a stereomicroscope equipped with AxioCam ICC1 camera and AxioVision4.8 software (Zeiss).
Accession numbers
Data files for transcriptome and H3K27me3 analyses can be accessed via GEO accession number GSE24507.
Supporting Information
Zdroje
1. BartonMK 2010 Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol 341 95 113
2. SchuettengruberBChourroutDVervoortMLeblancBCavalliG 2007 Genome regulation by polycomb and trithorax proteins. Cell 128 735 745
3. MullerJVerrijzerP 2009 Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev 19 150 158
4. JohnsonLMollahSGarciaBAMuratoreTLShabanowitzJ 2004 Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res 32 6511 6518
5. ZhangXClarenzOCokusSBernatavichuteYVPellegriniM 2007 Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5 e129 doi:10.1371/journal.pbio.0050129
6. SchwartzYBKahnTGNixDALiXYBourgonR 2006 Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38 700 705
7. BernsteinBEMikkelsenTSXieXKamalMHuebertDJ 2006 A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125 315 326
8. SchubertDPrimavesiLBishoppARobertsGDoonanJ 2006 Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25 4638 4649
9. HansenKHBrackenAPPasiniDDietrichNGehaniSS 2008 A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10 1291 1300
10. MullerJHartCMFrancisNJVargasMLSenguptaA 2002 Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111 197 208
11. CaoRWangLWangHXiaLErdjument-BromageH 2002 Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298 1039 1043
12. CzerminBMelfiRMcCabeDSeitzVImhofA 2002 Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111 185 196
13. KuzmichevANishiokaKErdjument-BromageHTempstPReinbergD 2002 Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16 2893 2905
14. JiangDWangYWangYHeY 2008 Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3 e3404 doi:10.1371/journal.pone.0003404
15. LindrothAMShultisDJasencakovaZFuchsJJohnsonL 2004 Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J 23 4286 4296
16. SchatlowskiNCreaseyKGoodrichJSchubertD 2008 Keeping plants in shape: Polycomb-group genes and histone methylation. Semin Cell Dev Biol 19 2291 2305
17. KohlerCVillarCB 2008 Programming of gene expression by Polycomb group proteins. Trends Cell Biol 18 236 243
18. ChanvivattanaYBishoppASchubertDStockCMoonYH 2004 Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131 5263 5276
19. SchubertDClarenzOGoodrichJ 2005 Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol 8 553 561
20. OhSParkSvanNS 2008 Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet 4 e1000077 doi:10.1371/journal.pgen.1000077
21. TurckFRoudierFFarronaSMartin-MagnietteMLGuillaumeE 2007 Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3 e86 doi:10.1371/journal.pgen.0030086
22. CharronJBHeHEllingAADengXW 2009 Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21 3732 3748
23. SchuettengruberBGanapathiMLeblancBPortosoMJaschekR 2009 Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7 e13 doi:10.1371/journal.pbio.1000013
24. BrackenAPDietrichNPasiniDHansenKHHelinK 2006 Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20 1123 1136
25. BoyerLAPlathKZeitlingerJBrambrinkTMedeirosLA 2006 Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441 349 353
26. LeeTIJennerRGBoyerLAGuentherMGLevineSS 2006 Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125 301 313
27. SquazzoSLO'GeenHKomashkoVMKrigSRJinVX 2006 Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res 16 890 900
28. OktabaKGutierrezLGagneurJGirardotCSenguptaAK 2008 Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell 15 877 889
29. TolhuisBde WitEMuijrersITeunissenHTalhoutW 2006 Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38 694 699
30. PappBMullerJ 2006 Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev 20 2041 2054
31. DealRBHenikoffS 2010 A Simple Method for Gene Expression and Chromatin Profiling of Individual Cell Types within a Tissue. Developmental Cell 18 1030 1040
32. MikkelsenTSKuMJaffeDBIssacBLiebermanE 2007 Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448 553 560
33. AzuaraVPerryPSauerSSpivakovMJorgensenHF 2006 Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8 532 538
34. LafosMSchubertD 2009 Balance of power—dynamic regulation of chromatin in plant development. Biol Chem 390 1113 1123
35. FletcherJCBrandURunningMPSimonRMeyerowitzEM 1999 Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911 1914
36. ZilbermanDGehringMTranRKBallingerTHenikoffS 2007 Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39 61 69
37. Thibaud-NissenFWuHRichmondTRedmanJCJohnsonC 2006 Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J 47 152 162
38. SemiartiEUenoYTsukayaHIwakawaHMachidaC 2001 The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771 1783
39. FitzgeraldJNHuiPSBergerF 2009 Polycomb group-dependent imprinting of the actin regulator AtFH5 regulates morphogenesis in Arabidopsis thaliana. Development 136 3399 3404
40. GustafsonAMAllenEGivanSSmithDCarringtonJC 2005 ASRP: the Arabidopsis Small RNA Project Database. Nucleic Acids Res 33 D637 D640
41. ChandlerJW 2009 Local auxin production: a small contribution to a big field. Bioessays 31 60 70
42. ZhaoY 2010 Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61 49 64
43. VannesteSFrimlJ 2009 Auxin: a trigger for change in plant development. Cell 136 1005 1016
44. LiscumEReedJW 2002 Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49 387 400
45. SungSSchmitzRJAmasinoRM 2006 A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev 20 3244 3248
46. CalonjeMSanchezRChenLSungZR 2008 EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20 277 291
47. GendallARLevyYYWilsonADeanC 2001 The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107 525 535
48. KatzAOlivaMMosqunaAHakimOOhadN 2004 FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J 37 707 719
49. XuLShenWH 2008 Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18 1966 1971
50. PalatnikJFAllenEWuXSchommerCSchwabR 2003 Control of leaf morphogenesis by microRNAs. Nature 425 257 263
51. WeinhoferIHehenbergerERoszakPHennigLKohlerC 2010 H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet 6 e1001152 doi:10.1371/journal.pgen.1001152
52. BaubecTPecinkaARozhonWMittelstenSO 2009 Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J 57 542 554
53. WuMFTianQReedJW 2006 Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133 4211 4218
54. NegreNHennetinJSunLVLavrovSBellisM 2006 Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol 4 e170 doi:10.1371/journal.pbio.0040170
55. JacksonJPJohnsonLJasencakovaZZhangXPerezBurgosL 2004 Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112 308 315
56. JacobYFengSLeBlancCABernatavichuteYVStroudH 2009 ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16 763 768
57. GriggSPCanalesCHayATsiantisM 2005 SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437 1022 1026
58. ChellappanPXiaJZhouXGaoSZhangX 2010 siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res doi 10.1093/nar/gkq590
59. BaoNLyeKWBartonMK 2004 MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7 653 662
60. Jones-RhoadesMWBartelDP 2004 Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14 787 799
61. RhoadesMWReinhartBJLimLPBurgeCBBartelB 2002 Prediction of plant microRNA targets. Cell 110 513 520
62. KuchenSReschWYamaneAKuoNLiZ 2010 Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32 828 839
63. EnderleDBeiselCStadlerMBGerstungMAthriP 2011 Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res doi:10.1101/gr.114348.110
64. ZhaoY 2008 The role of local biosynthesis of auxin and cytokinin in plant development. Curr Opin Plant Biol 11 16 22
65. ZazimalovaEMurphyASYangHHoyerovaKHosekP 2010 Auxin transporters—why so many? Cold Spring Harb Perspect Biol 2 a001552
66. ParryGCalderon-VillalobosLIPriggeMPeretBDharmasiriS 2009 Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci U S A 106 22540 22545
67. NavarroLDunoyerPJayFArnoldBDharmasiriN 2006 A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 436 439
68. GodaHSasakiEAkiyamaKMaruyama-NakashitaANakabayashiK 2008 The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55 526 542
69. RizzardiKLandbergKNilssonLLjungKSundås-LarssonA 2010 TFL2/LHP1 is involved in auxin biosynthesis through positive regulation of YUCCA genes. Plant J DOI:10.1111/j.1365-313X.2010.04470.x
70. KimSYZhuTSungZR 2010 Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis. Plant Physiol 152 516 528
71. SungSAmasinoRM 2004 Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427 159 164
72. BastowRMylneJSListerCLippmanZMartienssenRA 2004 Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164 167
73. GentlemanRCCareyVJBatesDMBolstadBDettlingM 2004 Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80
74. ToedlingJSkylarOKruegerTFischerJJSperlingS 2007 Ringo—an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 8 221
75. Perez-RodriguezPRiano-PachonDMCorreaLGRensingSAKerstenB 2010 PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38 D822 D827
76. CzechowskiTStittMAltmannTUdvardiMKScheibleWR 2005 Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5 17
77. YanDZhangYNiuLYuanYCaoX 2007 Identification and characterization of two closely related histone H4 arginine 3 methyltransferases in Arabidopsis thaliana. Biochem J 408 113 121
78. Colon-CarmonaAYouRHaimovitch-GalTDoernerP 1999 Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20 503 508
Štítky
Genetika Reprodukční medicína
Článek Incorporating Biological Pathways via a Markov Random Field Model in Genome-Wide Association StudiesČlánek Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation inČlánek Epigenetic Regulation of Cell Type–Specific Expression Patterns in the Human Mammary Epithelium
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 4- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Is Co-Expressed with Closely Adjacent Uncharacterised Genes Spanning a Breast Cancer Susceptibility Locus at 6q25.1
- The Liberation of Embryonic Stem Cells
- Genome-Wide Meta-Analysis Identifies Regions on 7p21 () and 15q24 () As Determinants of Habitual Caffeine Consumption
- A Sustained Dietary Change Increases Epigenetic Variation in Isogenic Mice
- The Exocyst Protein Sec10 Interacts with Polycystin-2 and Knockdown Causes PKD-Phenotypes
- Incorporating Biological Pathways via a Markov Random Field Model in Genome-Wide Association Studies
- Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation in
- Identification and Functional Validation of the Novel Antimalarial Resistance Locus in
- Does Positive Selection Drive Transcription Factor Binding Site Turnover? A Test with Drosophila Cis-Regulatory Modules
- Protein Phosphatase 2A Controls Ethylene Biosynthesis by Differentially Regulating the Turnover of ACC Synthase Isoforms
- Ribosomal DNA Deletions Modulate Genome-Wide Gene Expression: “–Sensitive” Genes and Natural Variation
- Reciprocal Sign Epistasis between Frequently Experimentally Evolved Adaptive Mutations Causes a Rugged Fitness Landscape
- Variable Pathogenicity Determines Individual Lifespan in
- Evolution of Vertebrate Transient Receptor Potential Vanilloid 3 Channels: Opposite Temperature Sensitivity between Mammals and Western Clawed Frogs
- Towards Establishment of a Rice Stress Response Interactome
- Mouse Genome-Wide Association and Systems Genetics Identify As a Regulator of Bone Mineral Density and Osteoclastogenesis
- Quantitative Fitness Analysis Shows That NMD Proteins and Many Other Protein Complexes Suppress or Enhance Distinct Telomere Cap Defects
- Highly Precise and Developmentally Programmed Genome Assembly in Requires Ligase IV–Dependent End Joining
- PDP-1 Links the TGF-β and IIS Pathways to Regulate Longevity, Development, and Metabolism
- Genome-Wide Association Study Using Extreme Truncate Selection Identifies Novel Genes Affecting Bone Mineral Density and Fracture Risk
- Eight Common Genetic Variants Associated with Serum DHEAS Levels Suggest a Key Role in Ageing Mechanisms
- 14-3-3 Proteins Regulate Exonuclease 1–Dependent Processing of Stalled Replication Forks
- HDA6 Regulates Locus-Directed Heterochromatin Silencing in Cooperation with MET1
- Epigenetic Regulation of Cell Type–Specific Expression Patterns in the Human Mammary Epithelium
- Enhanced Statistical Tests for GWAS in Admixed Populations: Assessment using African Americans from CARe and a Breast Cancer Consortium
- Beyond Missing Heritability: Prediction of Complex Traits
- An Evolutionary Genomic Approach to Identify Genes Involved in Human Birth Timing
- Long-Lost Relative Claims Orphan Gene: in a Wasp
- PTG Depletion Removes Lafora Bodies and Rescues the Fatal Epilepsy of Lafora Disease
- Chromatin Organization in Sperm May Be the Major Functional Consequence of Base Composition Variation in the Human Genome
- GWAS of Follicular Lymphoma Reveals Allelic Heterogeneity at 6p21.32 and Suggests Shared Genetic Susceptibility with Diffuse Large B-cell Lymphoma
- Loss-of-Function Mutations in Cause Metachondromatosis, but Not Ollier Disease or Maffucci Syndrome
- DNA Damage, Somatic Aneuploidy, and Malignant Sarcoma Susceptibility in Muscular Dystrophies
- The Phylogenetic Origin of Coincided with the Origin of Maternally Provisioned Germ Plasm and Pole Cells at the Base of the Holometabola
- Genome Analysis Reveals Interplay between 5′UTR Introns and Nuclear mRNA Export for Secretory and Mitochondrial Genes
- Genome-Wide Association Analysis of Soluble ICAM-1 Concentration Reveals Novel Associations at the , , , and Loci
- The Complete Spectrum of Yeast Chromosome Instability Genes Identifies Candidate CIN Cancer Genes and Functional Roles for ASTRA Complex Components
- Dynamic Regulation of H3K27 Trimethylation during Differentiation
- Phosphorylation-Dependent Differential Regulation of Plant Growth, Cell Death, and Innate Immunity by the Regulatory Receptor-Like Kinase BAK1
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PTG Depletion Removes Lafora Bodies and Rescues the Fatal Epilepsy of Lafora Disease
- Evolution of Vertebrate Transient Receptor Potential Vanilloid 3 Channels: Opposite Temperature Sensitivity between Mammals and Western Clawed Frogs
- Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation in
- An Evolutionary Genomic Approach to Identify Genes Involved in Human Birth Timing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



