-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
14-3-3 Proteins Regulate Exonuclease 1–Dependent Processing of Stalled Replication Forks
Replication fork integrity, which is essential for the maintenance of genome stability, is monitored by checkpoint-mediated phosphorylation events. 14-3-3 proteins are able to bind phosphorylated proteins and were shown to play an undefined role under DNA replication stress. Exonuclease 1 (Exo1) processes stalled replication forks in checkpoint-defective yeast cells. We now identify 14-3-3 proteins as in vivo interaction partners of Exo1, both in yeast and mammalian cells. Yeast 14-3-3–deficient cells fail to induce Mec1–dependent Exo1 hyperphosphorylation and accumulate Exo1–dependent ssDNA gaps at stalled forks, as revealed by electron microscopy. This leads to persistent checkpoint activation and exacerbated recovery defects. Moreover, using DNA bi-dimensional electrophoresis, we show that 14-3-3 proteins promote fork progression under limiting nucleotide concentrations. We propose that 14-3-3 proteins assist in controlling the phosphorylation status of Exo1 and additional unknown targets, promoting fork progression, stability, and restart in response to DNA replication stress.
Published in the journal: . PLoS Genet 7(4): e32767. doi:10.1371/journal.pgen.1001367
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001367Summary
Replication fork integrity, which is essential for the maintenance of genome stability, is monitored by checkpoint-mediated phosphorylation events. 14-3-3 proteins are able to bind phosphorylated proteins and were shown to play an undefined role under DNA replication stress. Exonuclease 1 (Exo1) processes stalled replication forks in checkpoint-defective yeast cells. We now identify 14-3-3 proteins as in vivo interaction partners of Exo1, both in yeast and mammalian cells. Yeast 14-3-3–deficient cells fail to induce Mec1–dependent Exo1 hyperphosphorylation and accumulate Exo1–dependent ssDNA gaps at stalled forks, as revealed by electron microscopy. This leads to persistent checkpoint activation and exacerbated recovery defects. Moreover, using DNA bi-dimensional electrophoresis, we show that 14-3-3 proteins promote fork progression under limiting nucleotide concentrations. We propose that 14-3-3 proteins assist in controlling the phosphorylation status of Exo1 and additional unknown targets, promoting fork progression, stability, and restart in response to DNA replication stress.
Introduction
DNA lesions can cause stalling and collapse of the replication fork and lead to chromosome breaks, mutations, genome rearrangements and eventually cancer [1]. To prevent this, a replication checkpoint has evolved as surveillance mechanism to control components of the replisome [2] and to allow coordinating replication with cell cycle progression and DNA repair. Maintenance of stable replication intermediates when DNA synthesis is impeded, requires replisome stability and checkpoint-dependent phosphorylation [3]. Although crucial targets for this checkpoint function await identification, nuclease activities are particularly likely to require fine-tuning, to avoid unscheduled DNA processing under DNA replication stress [4].
Exo1 is a Rad2 family DNA repair nuclease able to remove mononucleotides from the 5′ end of the DNA duplex [5] that was originally identified in the Schizosaccharomyces pombe [6] and subsequently in humans [7]. Exo1 is implicated in several DNA repair pathways including mismatch repair, post replication repair, meiotic and mitotic recombination and double strand break repair [8]–[12]. Saccharomyces cerevisiae Exo1 acts redundantly with Rad27 in processing Okazaki fragments during DNA replication [13]. More recently, Exo1 was shown to be recruited to stalled replication forks where it counteracts fork reversal [4]. Human EXO1 activity is controlled by post-translational modifications, with ATR-dependent phosphorylation targeting it to ubiquitin-mediated degradation upon replication fork stalling [14], [15], and ATM-dependent phosphorylation apparently restraining its activity during homologous recombination [16]. Analogously, Mec1-dependent phosphorylation inhibits yeast Exo1 activity at uncapped telomeres [17]. Studies in budding yeast showed that EXO1 deletion suppresses the sensitivity of rad53, but not mec1, mutant cells to agents causing reversible or irreversible stalling of replication forks [18]. Taken together, this evidence indicates that Exo1 activity is tightly controlled under DNA replication stress and DNA damage.
Eukaryotic 14-3-3 are highly conserved proteins that establish phosphorylation-dependent interactions and modulate the functions of proteins involved in processes such as metabolism, protein trafficking, signal transduction, apoptosis and cell-cycle [19]. Seven 14-3-3 isoforms exist in mammalian cells, but only two in yeast. Structural analysis showed that 14-3-3 proteins self-assemble into flexible homo - and hetero-dimers forming a central groove that is able to adapt two extended peptides [20], [21]. This feature confers them the ability to act as adaptors that integrate signals from different pathways [22], [23]. 14-3-3 proteins can also bind cruciform DNA [24] and replication initiation proteins such as Mcm2 and Orc2 [25]. Upon DNA damage and DNA replication stress, 14-3-3 proteins are required for cell cycle restart, suppression of genomic instability and viability [26]. Moreover, 14-3-3 proteins genetically and physically interact with the checkpoint protein Rad53 [27] as well as the acetyltransferases and deacetylases Esa1 and Rpd3 upon replication perturbations [28]. Although these data point to an important role of 14-3-3 during replication stress, the exact mechanism of 14-3-3 action remains unknown.
In this study, we identify 14-3-3 as novel interaction partners of Exo1 and demonstrate that they regulate phosphorylation of the nuclease. We provide evidence for an accumulation of Exo1-dependent ssDNA gaps at stalled forks in yeast 14-3-3 deficient cells and we show that this causes persistent checkpoint activation and recovery defects. We also show that 14-3-3 proteins control progression and stability of replication forks under conditions of limiting nucleotide availability. Taken together, our data demonstrate that 14-3-3 have a crucial role in regulating the function of proteins at stalled forks, among which Exo1 is a key target.
Results/Discussion
14-3-3 proteins interact with EXO1
To identify novel interaction partners for human EXO1 we designed a yeast two-hybrid screen with GAL4-bait fusion proteins that contain either full-length EXO1 or ΔN-EXO1 (EXO1366–846), which lacks the entire catalytic domain. Since the former was not expressed (data not shown), we used the latter to screen a blood peripheral cDNA library. Three 14-3-3 proteins were the highest hits (Table S1), with the β - being more represented than the ε - and ζ-isoform. The presence of an established EXO1 binding protein among the hits, MLH1 (Table S1), confirmed the reliability of this screen.
To independently verify these data, we performed co-immunoprecipitation experiments. Given the low abundance of EXO1 in the cell [14], we transiently transfected HEK-293 cells with an Omni-tagged EXO1 construct [14] and immunoprecipitated the expressed protein using a pan-14-3-3 antibody. The data showed that Omni-EXO1 and 14-3-3 proteins could be recovered as a complex (Figure 1A).
Fig. 1. EXO1 interacts with 14-3-3 proteins. 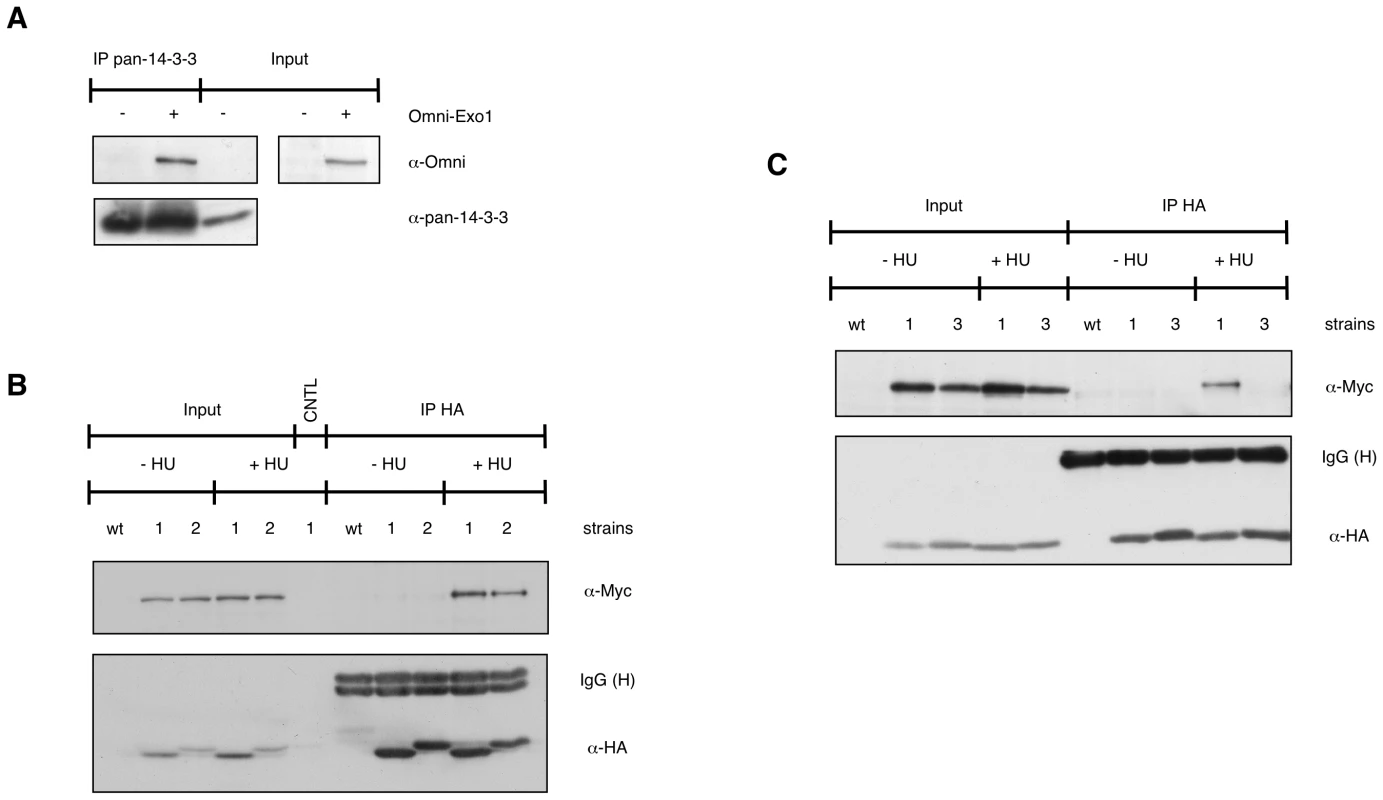
(A) HEK-293 cells were transiently transfected with empty vector (−) or pcDNA3.1-Omni-EXO1 (+). Whole cell extracts (WCE, 2.5 mg) were immunoprecipitated with a pan-14-3-3 antibody and proteins were detected as indicated. Input = 50 µg WCE. (B) Control yeast culture (wt) or cultures expressing Bmh1-HA Exo1-Myc (1) or Bmh2-HA Exo1-Myc (2) were treated for 90 min with 150 mM HU. WCE (10 mg) were immunoprecipitated with the monoclonal antibody to HA, proteins were resolved on an 8% SDS-polyacrylamide gel and detected as indicated. CNTL = immunoprecipitation performed in the absence of the antibody. Input = 100 µg WCE. (C) WCE from control yeast culture (wt) or cultures expressing Bmh1-HA Exo1-Myc (1) or Bmh1-280-HA Exo1-Myc (3) were immunoprecipitated as described in (B). To assess the physiological significance of the EXO1/14-3-3 interaction we selected Sacc. cerevisiae, a system where only two 14-3-3 proteins are present, namely Bmh1 and Bmh2. In preliminary experiments we examined whether yeast Exo1 and 14-3-3 proteins interact. A C-terminal Myc - or a HA-tag was added to the endogenous EXO1 or BMH1/BMH2 genes, respectively. Immunoprecipitation experiments showed that Exo1 formed complexes with Bmh1 or Bmh2 in a HU-dependent manner (Figure 1B). We next explored a possible direct Bmh/Exo1 interaction by Far Western blot analysis. Exo1-Myc immunoprecipitated from control or HU-treated cells was resolved by SDS-PAGE and denatured/renatured on PVDF. Probing the membrane with purified GST-Bmh1 revealed that a direct interaction with Exo1 occurred both in the case of untreated and HU-treated cells (Figure S1). These data possibly indicate that an Exo1 domain normally hidden in non-treated cells may become available for interaction with 14-3-3 proteins upon HU-treatment. Such domain may be artificially exposed during Far Western analysis due to incomplete renaturation of Exo1.
Taken together, these data suggest that the EXO1/14-3-3 interaction is conserved from yeast to mammalian cells. While the interaction is HU-independent in mammalian cells, it requires HU in yeast. This may reflect the different modes of EXO1 regulation in the two systems [15], [17].
14-3-3–deficient cells cannot restart stalled replication forks, but their recovery defect is partially suppressed by EXO1 deletion
Genetic and flow cytometric analysis evidenced the sensitivity of 14-3-3-deficient cells to DNA replication stress, with distinct bmh1 (bmh2Δ) alleles showing different defects upon nucleotide depletion (HU) or treatment with DNA damaging agents (UV or methylmethansulfonate, MMS) [26]. However, despite the evidence that 14-3-3 proteins bind origins of replication and cruciform DNA [29], suggesting a regulatory role in DNA replication [25], the issue of possible direct involvement of 14-3-3 in fork stability or processing under genotoxic stress conditions remained to be clarified. Given the comprehensive molecular characterization of yeast Exo1 as component of the replisome and of its role, in checkpoint defective cells, in the processing of forks stalled by nucleotide depletion [4], we focused our investigations on the bmh1-280 bmh2Δ double mutant (bmh1bmh2 hereafter). This mutant shows normal cell cycle progression in unperturbed conditions, but selective sensitivity and cell cycle recovery defects in response to HU [26]. The mutant Bmh1-280 protein carries a single point mutation (E136>G) in helix αE at a residue neighboring amino acids that form salt bridges and hydrogen bonds with the ligand [21]. Interestingly, immunoprecipitation experiments showed that the mutant Bmh1-280 protein did not interact with Exo1 in untreated nor HU-treated cells (Figure 1C). Thus, we asked whether the cell cycle recovery defects of this mutant reflect a direct role of 14-3-3 proteins at replication forks and whether Exo1 is also implicated in these processes. We performed neutral-neutral bidimensional gel electrophoresis (2D gel) on the early origin of replication ARS305, which is known to be activated in HU-treated cells [3]. Although the 2D gel pattern looked normal in HU-treated bmh1bmh2 cells, we observed that replication intermediates (RIs) in 14-3-3 defective cells were still present close to the origin 60 min after HU removal and were only restarted at 90–120 min (Figure 2A and data not shown). This suggests that misregulation of the replisome, without dramatic physical processing of the forks, might be sufficient to impair fork restart. This effect was not detectably suppressed by EXO1 deletion (Figure 2A).
Fig. 2. Pattern of HU recovery in wild-type, exo1Δ, bmh1-280 bmh2Δ, and bmh1-280 bmh2Δ exo1Δ strains. 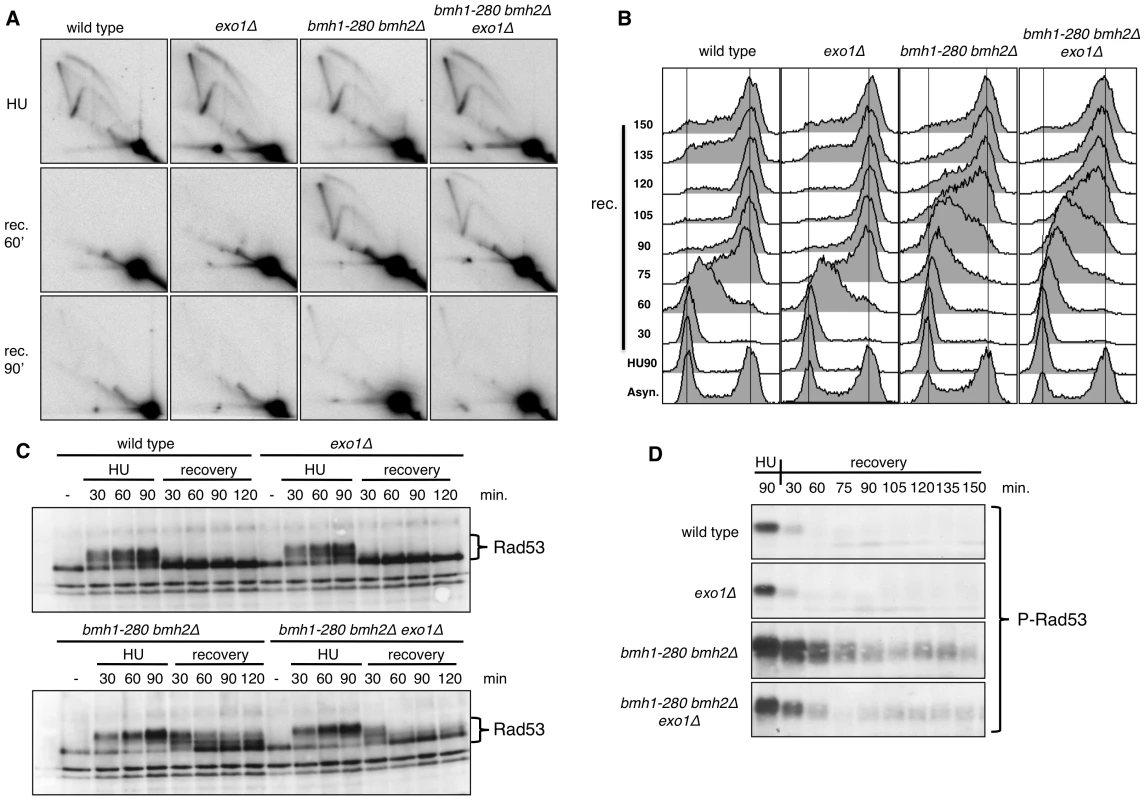
(A) Representative 2D gels of replication intermediates (RIs) at ARS305 analyzed after 90 min HU-treatment and upon HU removal (recovery, 60 and 90 min). (B) Time-course flow cytometric analysis of the DNA content in the indicated strains upon recovery from a HU-arrest. (C) Western blot analysis showing in the indicated strains the phosphorylation-dependent mobility shift of Rad53 during the HU-arrest and the recovery phase. (D) Western blot analysis performed with monoclonal antibody F9 to phosphorylated Rad53 showing in the indicated strains the status of Rad53 activation in HU-arrested cells and during the recovery phase. Flow cytometric analysis of HU-released cells confirmed the slow recovery of the bmh1bmh2 strain and showed that lack of Exo1 per se did not alter the pattern of cell cycle progression (Figure 2B). On the other hand, EXO1 deletion in a bmh1bmh2 background led to a partial rescue of the recovery defect, particularly evident at late time points (≥120 min) after release from HU (Figure 2B). This evidence prompted us to ask whether EXO1 deletion in this background may affect Rad53 activity. Western blot analysis with total and phosphospecific Rad53 antibodies [30] showed that, compared to wild type cells, Rad53 was hyperphosphorylated in HU-treated bmh1bmh2 cells and that its dephosphorylation was retarded during the HU-recovery phase (Figure 2C and 2D), thus correlating with the described replication restart defect. Importantly, deletion of EXO1 in 14-3-3-deficient cells re-established to a great extent the pattern of rapid Rad53 dephosphorylation in the recovery phase (Figure 2C and 2D), substantiating the flow cytometry data (Figure 2B).
Overall these data suggest that 14-3-3 proteins are directly implicated in the effective restart of stalled DNA replication forks. Alternatively, they may assist rapid Rad53 dephosphorylation, which is in turn required for fork restart upon HU removal [31]. The latter interpretation is however unlikely as EXO1 deletion, which markedly restores Rad53 dephosphorylation upon HU removal, does not detectably improve the defective fork restart observed in 14-3-3 deficient cells on the ARS305 replicon. Thus, in the 14-3-3 defective background, Exo1 activity does not directly impact the rate of fork restart, but slows down checkpoint inactivation and delays cell cycle resumption.
Reversible Exo1 phosphorylation in response to HU is dependent on 14-3-3 proteins
Exo1 is controlled in a phosphorylation-dependent manner upon replication fork stalling in mammalian cells [14] and upon a variety of genotoxic insults in yeast [17]. We obtained evidence that yeast Exo1 is phosphorylated in a Mec1-dependent manner also in response to HU (Figure 3A). Notably, the improved resolution of Exo1 phospho-forms by Phos-tag SDS-PAGE [32] allowed us visualizing the complete pattern of Exo1 phosphorylation in response to replicative stress (Figure 3A and 3B).
Fig. 3. Exo1 phosphorylation pattern in response to HU in wild-type, mec1Δ, and bmh1-280 bmh2Δ strains. 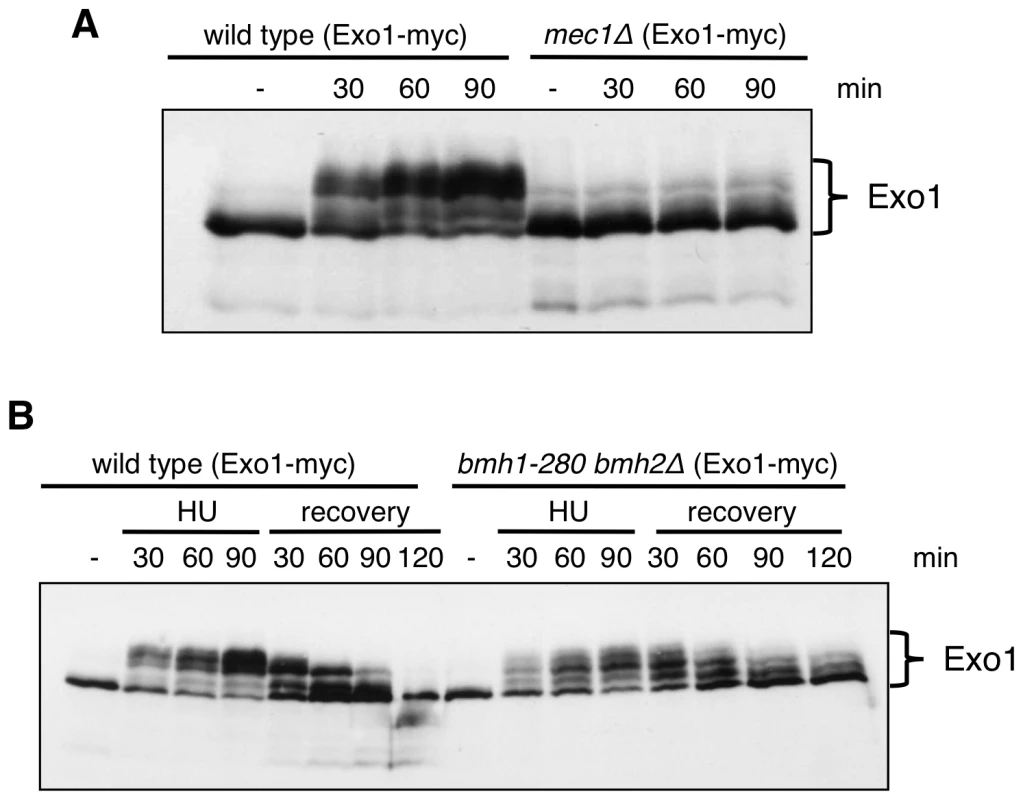
(A) Western blot analysis of Exo1 phosphorylation in HU-arrested cells of wild type and mec1Δ sml1Δ strains. (B) Western blot analysis of Exo1 phosphorylation in HU-arrested cells and during the recovery phase of the indicated strains. Both in (A) and (B) proteins were resolved on an 8% Phos-tag SDS-polyacrylamide gel. Next, we asked whether 14-3-3 proteins might be involved in the regulation of Exo1 phosphorylation and stability. Western blot analysis showed that in 14-3-3-deficient cells total Exo1 levels were unchanged (Figure S2), but Exo1 was not phosphorylated to the same stoichiometry observed in wild type cells (Figure 3B, 90 min). Moreover, the rate of Exo1 dephosphorylation upon recovery from HU was considerably reduced in mutant cells, with Exo1 being completely dephosphorylated in wild type but not in 14-3-3-deficient cells (Figure 3B, 120 min). Defective Exo1 phosphorylation in HU-treated 14-3-3-deficient cells is not an indirect consequence of defective checkpoint activation, as under these conditions Rad53, another Mec1-dependent checkpoint target, is promptly phosphorylated (Figure 2C and 2D). Since phosphorylation restrains yeast Exo1 activity [17], we propose that 14-3-3 proteins play an important role in the dynamic control of Exo1 activity upon DNA replication stress and may act as platform for the control of Exo1 phosphorylation. In this respect, attempts to assess the phosphorylation status of Bmh-bound Exo1 were unfortunately inconclusive, as - differently from TCA extracts (Figure 3) - the extracts used for immunoprecipitation fail to be resolved in discrete bands by Phos-tag SDS-PAGE (data not shown). Given that 14-3-3 proteins bind ligands in phospho-dependent and -independent manner [21], it will be important to overcome these technical limitations to address the role of 14-3-3 proteins in controlling the phosphorylation of Exo1 and, possibly, additional targets in the DNA damage response (see below).
Exo1 is responsible for the accumulation of ssDNA gaps behind the fork in bmh1bmh2 cells
As Exo1 activity and Rad53 phosphorylation have been linked to the processing of stalled DNA replication forks, we decided to assess whether defective Rad53 and Exo1 phosphorylation in 14-3-3-deficient cells could reflect changes in the fine architecture of stalled forks. To answer this question, we synchronized the cells in G1, released them for 1 h in YPD medium containing 0.2 M HU and examined RIs by electron microscopy (EM) under non-denaturing conditions [33]. For each strain, about 100 RIs were analyzed in duplicate. 14-3-3-deficient cells showed a dramatic accumulation of ssDNA gaps behind the replication fork (Figure 4A). Statistical analysis indicated that approximately 50% of all RIs analyzed contained one or more ssDNA gaps (Figure 4B). Interestingly, deletion of EXO1 in the bmh1bmh2 background completely suppressed this phenotype, leading to a reduction of the ssDNA gaps behind the fork to a level similar to wild type or exo1Δ cells (Figure 4B). The comparison of ssDNA gaps length scored by EM evidenced a striking difference: whereas bmh1bmh2 cells displayed a significant number of large size gaps (>0.5 Kb), the latter were absent in bmh1bmh2exo1Δ cells (Figure 4C). The resolution limit of 50–70 nucleotides may have impaired detection of nicks/small gaps in this as well as in previous EM studies with HU [34]. Such structures, however, become visible in 14-3-3-deficient cells, where the unleashed Exo1 activity would enlarge gaps above the detection limit.
Fig. 4. Exo1–dependent generation of ssDNA gaps in bmh1-280 bmh2Δ cells. 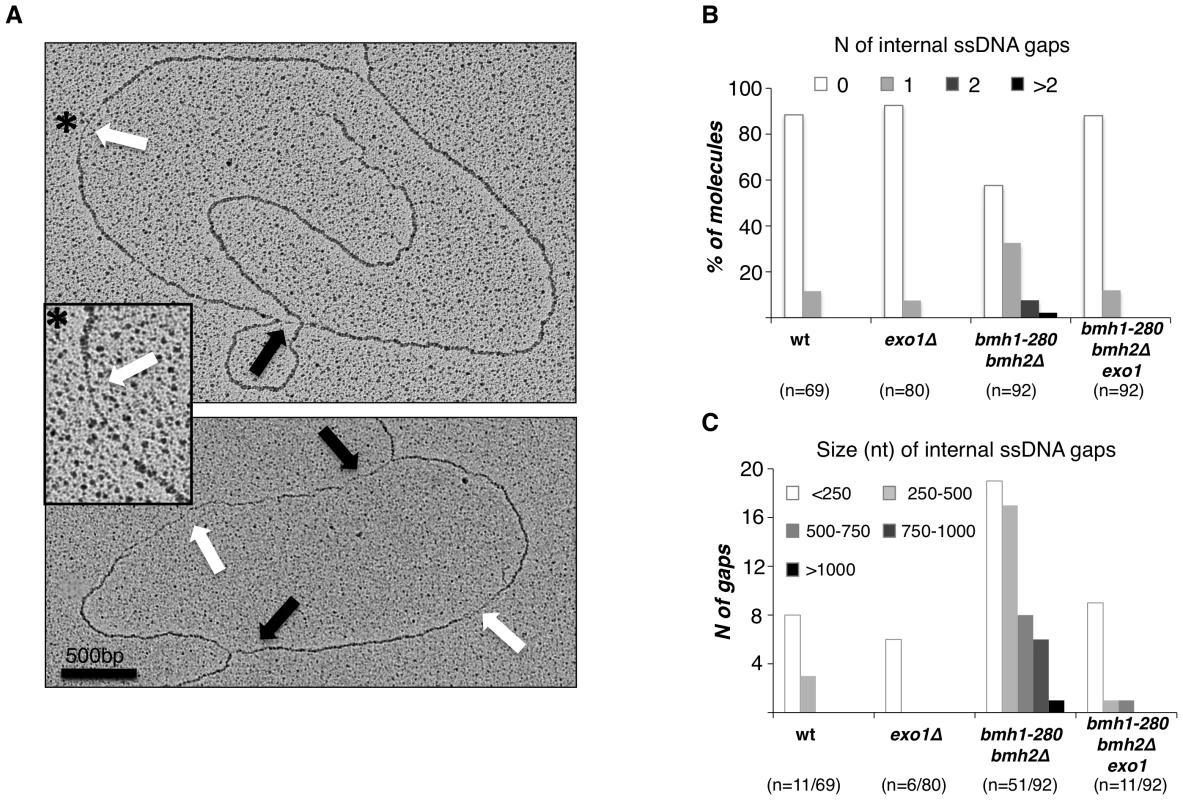
(A) Representative RIs visualized by EM in bmh1-280 bmh2Δ cells released synchronously from G1 phase in 0.2 M HU for 1 h: the magnified inset (asterisk) shows a representative ssDNA gap located behind the replication fork. Black arrows: ssDNA gap at the fork; White arrows: internal ssDNA gap located behind the fork. (B) Statistical analysis of ssDNA gap number. The number of analyzed molecules is indicated in brackets. (C) Statistical analysis of ssDNA gap length. The number of analyzed gaps/molecules is indicated in brackets. These data suggest that 14-3-3 proteins are required to prevent unscheduled Exo1 activity behind stalled replication forks in a checkpoint-proficient background. The implications of these observations are of great significance. A loose control of Exo1 activity may render DNA synthesis more discontinuous in conditions of replicative stress, either promoting additional uncoupling/repriming events or enlarging existing ssDNA gaps via 5′-3′ exonucleolytic processing. Although additional work is required to directly address this point, it is conceivable that continuous polymerase stall due to insufficient deoxynucleotide levels might per se lead to increased repriming events, thus raising the number of 5′-ends available for processing by Exo1. In this setting, a strict control of Exo1 activity would be needed to limit damage. We observed no bias for the presence of gaps in leading vs. lagging strands - whenever these could be identified [35] - and we could occasionally detect gaps on opposite strands within the same molecule (Figure S3), suggesting that unscheduled Exo1 activity in 14-3-3 defective cells is not restricted to leading or lagging strand.
Defective fork progression in 14-3-3–defective cells is independent on EXO1
Replication recovery defects have been previously described and usually reflect replication fork collapse detectable by 2D gel analysis [3]. On the contrary, stalled replication forks in 14-3-3 deficient cells, albeit unable to restart DNA synthesis and abnormally processed by Exo1 activity, upon prolonged HU treatment show a 2D gel pattern indistinguishable from that of wild type cells. We thus decided to investigate in more detail the structure and progression of these forks, performing 2D gel analysis at different time points after HU addition. To this end, cells synchronized in G1 by α-factor were released into medium containing HU and RIs were examined by 2D gels. Figure 5B shows the probes designed to visualize replication fork progression in a region of Chromosome III that contains, besides the early active origin ARS305 [36], a contiguous passively replicated region (Part A) and a region including the dormant origin ARS301 (Part D) [3]. As compared to wild type, bmh1bmh2 cells showed the same kinetics of origin firing, albeit with slightly lower efficiency as revealed by the intensity of the bubble arc at 30 min (Figure 5C). Progression of the forks in HU from ARS305 across the region of Part A (∼5 Kb to the left of ARS305) was completed after 2–3 h in wild type cells, with the peak of intermediates detectable after ∼1 h. In bmh1bmh2 cells the first intermediates appeared on this region with 30 min delay, whereas the peak of intermediates was delayed of ∼2 h as compared to wild type cells (Figure 5C), indicative of a significant decrease in the rate of the replication fork progression in HU. Slow RIs disappearance from the origin and delayed invasion of adjacent chromosomal regions may in principle result also from asynchronous firing of ARS305 during the HU arrest. However we consider this alternative interpretation unlikely for the following reasons: a) by budding experiments, 14-3-3 defective cells do not display asynchronous entrance in S-phase (data not shown); b) the comparable intensity of the Y arc on fragment A in the wt (60 min) and in the 14-3-3 mutant (180 min) suggests that forks progress synchronously but slower from the early origin ARS305; c) accordingly, the progressive accumulation of the Y signal on fragment A in 14-3-3 defective cells strictly correlates with the disappearance of the bubble ark on the ARS305 fragment, further suggesting slow but synchronous progression of replication forks on the ARS305 replicon.
Fig. 5. 2D gel analysis of RIs from wild-type, bmh1-280 bmh2Δ, and rad53-K227A strains. 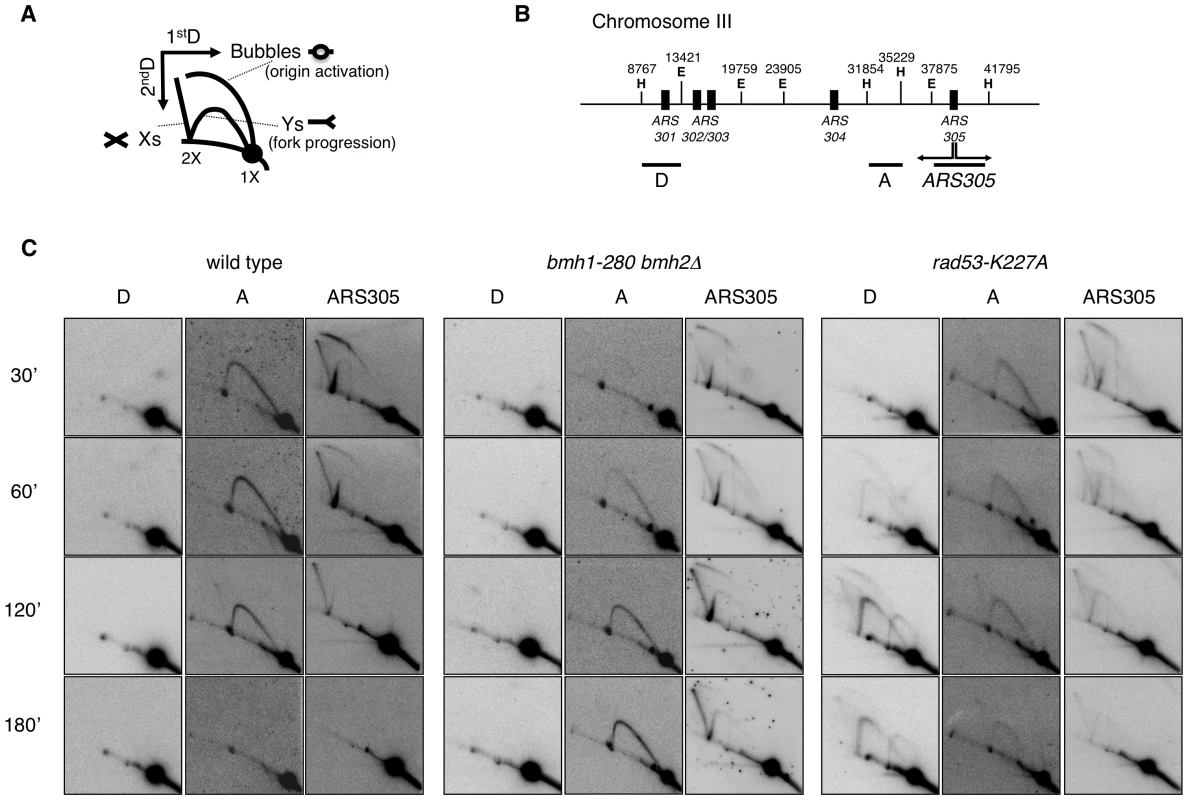
(A) Schematic representation of RIs visualized by 2D gel electrophoresis. (B) Chromosome III region adjacent to ARS305 with indication of restriction sites and probes used in 2D gel analysis (E = EcoRV; H = HindIII). (C) Time-course resolution of RIs obtained from the indicated strains arrested in G1 (α-factor) and released in S phase in the presence of 0.2 M HU. Genomic DNA was extracted at the indicated time points and digested with EcoRV and HindIII prior to standard 2D gel analysis. It was previously shown that yeast 14-3-3 proteins bind to the checkpoint kinase Rad53 and directly influence its DNA damage-dependent functions [27]. Therefore, we asked whether the slow fork progression in bmh1bmh2 cells might be solely due to checkpoint defects. To address this issue, we used checkpoint defective Rad53-mutant cells (rad53-K227A). The latter displayed striking differences when compared to bmh1bmh2 cells. Both the destabilization of RIs (ARS305 and Part A) and the uncontrolled firing of dormant origins displayed by rad53-K227A cells (Part D) [3], were absent in bmh1bmh2 cells (Figure 5C). Furthermore, EM did not display any fork reversal or accumulation of ssDNA at replication forks, typical of HU-treated rad53 cells [34] (data not shown). Finally, 2D gel analysis (Figure S4C, S4F and S4H) and drop assays (Figure S5) revealed synergistic effects of 14-3-3 and Rad53 on both fork stability and survival. Overall, these data indicate that the phenotype observed in 14-3-3 deficient cells reflects a genuine role of 14-3-3 proteins at replication forks and that 14-3-3 and Rad53 have crucial but distinct roles at HU-challenged forks.
Deletion of EXO1 partially rescued the HU-sensitivity of rad53-K227A cells, but not of a bmh1bmh2 strain (Figure S5). Furthermore, in contrast to checkpoint defective cells, where stability of RIs could be rescued by EXO1 deletion [4], fork progression defects of bmh1bmh2 cells were not rescued by loss of EXO1 (Figure S4G). Thus, while the processing defect that leads to accumulation of ssDNA gaps in 14-3-3-deficient cells was completely suppressed by EXO1 deletion, this did not reflect in suppression of HU sensitivity nor of defective fork progression in HU-treated 14-3-3 deficient cells. Altogether this evidence suggests that 14-3-3 proteins might regulate additional targets during replication stress, possibly through modulation of their phosphorylation. This is not unexpected, given the role of 14-3-3 as integrators of signaling pathways [19] and considering the multiplicity of 14-3-3 targets [37], [38]. Our data implicate 14-3-3 proteins as possible central regulator of the checkpoint response. In analogy with previously reported cases [22] and according to structural data on the dynamic nature of 14-3-3 dimers [39], one may envisage a role for 14-3-3 proteins as docking clamp tethering Exo1 - and other unknown targets - with the kinase controlling its/their activity. Notably, 14-3-3 proteins were reported to bind Rad53 [27], one of the candidate checkpoint kinases required for Exo1 phosphorylation [17].
In conclusion, this work sheds further light on processes occurring at stalled replication forks, proposing 14-3-3 proteins as central integrators of signals that regulate fork stability and processing. Challenges lying ahead consist in the identification of components of the replisome, or proteins controlling them, that may be 14-3-3 targets, as well as in the elucidation of the exact mechanism by which 14-3-3 modulate Exo1 phosphorylation and activity.
Materials and Methods
Materials
The antibodies used in this study were: goat polyclonal anti-Omni-probe (M21, sc-499, Santa Cruz Biotechnology); rabbit polyclonal anti-pan 14-3-3 (SA-483, Biomol); mouse monoclonal anti-HA (12CA5, Sigma) and anti-Myc (9E10, Santa Cruz Biotechnology); rabbit polyclonal anti-Rad53 (a kind gift from C. Santocanale, Galway, Ireland); mouse monoclonal F9 to phosphorylated Rad53 [30] (a kind gift of M. Foiani, Milano, Italy).
The chemicals and peptides used in this study were: Hydroxyurea (Sigma); α1-Mating Factor (Sigma).
Saccharomyces cerevisiae strains
The yeast strains used in this study are isogenic to W303-1A (wild type) [40] and are listed in Table 1. All strains have been obtained by one step replacement using the indicated markers and tags that have been generated by PCR. The isolated clones have been verified by colony PCR and Southern Blot or Western Blot.
Tab. 1. List of <i>Saccharomyces cerevisiae</i> strains used in this study. 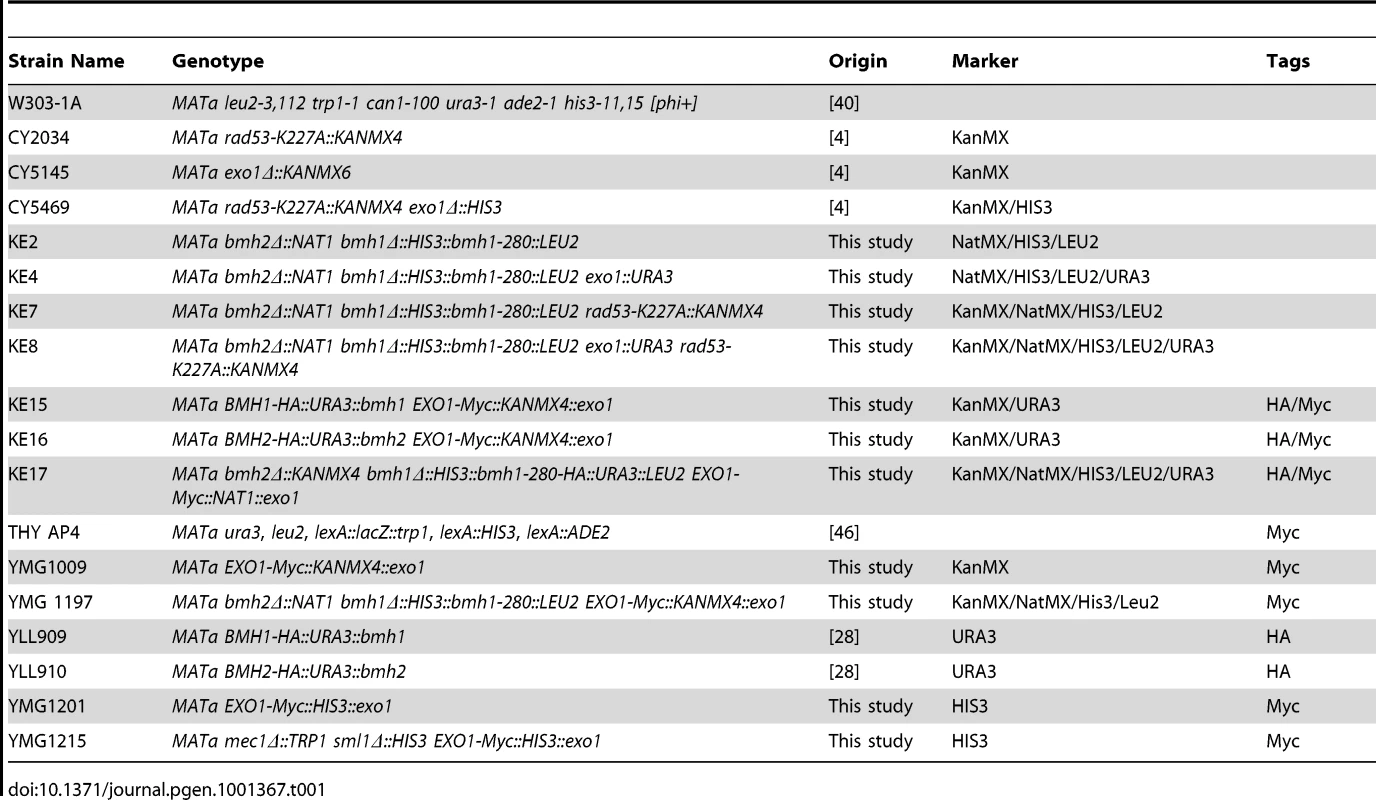
All deletion (Δ) strains lack the entire coding sequence. All strains containing the bmh1-280 mutation have been generated from strain YLL1090 [26]. The strain KE17 has been generated from DMP4644/4A (M.P. Longhese, unpublished) and is a derivative of YLL1090 [26].
Yeast two-hybrid screen
The yeast two-hybrid screening was performed with ΔN-EXO1 (EXO1366–846) as bait on a cDNA library generated from human peripheral blood mRNA (a kind gift of I. Stagljar, Toronto, Canada) as described previously [41] and using THY AP4 as reporter strain.
Protein extraction, Western and Far Western blotting, immunoprecipitation
Western blot analysis of yeast proteins was carried out upon TCA extraction [42]. To visualize Exo1, an optimized Phos-tag system (50 µM Phos-tag reagent) was employed according to [32]. Proteins were transferred to nitrocellulose (porablot NCP, 0.45 µm pore size, Machery-Nagel) overnight at room temperature applying constant amperage (200 mA). Far Western blotting [43] was performed using purified recombinant GST-Bmh1 [44] to probe Exo1 that was immunoprecipitated from control or HU-treated yeast cells.
HEK293T cells were grown and lysed as described [14] and protein concentration was determined using the Bio-Rad Protein Assay Reagent (Bio-Rad). Immunoprecipitation of Omni-EXO1 or 14-3-3 proteins from 2.5 mg total cell extracts with specific antibodies was performed as previously described [14].
For immunoprecipitation of yeast proteins, cells were lysed using ice-cold lysis buffer (25 mM Tris-HCl pH 7.4, 15 mM NaCl, 15 mM EGTA, 1 mM NaF, 1 mM Na orthovanadate, 4 mM p-Nitro-Phenyl-Phosphate (pNPP), 0.1% Triton X-100, 1 mM PMSF, complete protease inhibitors cocktail (Roche)). 14-3-3-HA was immunoprecipitated from 10 mg total yeast cell extracts using the monoclonal HA antibody.
2D gel electrophoresis and electron microscopy
DNA extraction with the CTAB method and neutral-neutral two-dimensional gel electrophoresis were performed as described [45]. EM analysis was performed as described [33].
Supporting Information
Zdroje
1. BranzeiD
FoianiM
2010 Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11 208 219
2. Muzi-FalconiM
LiberiG
LuccaC
FoianiM
2003 Mechanisms controlling the integrity of replicating chromosomes in budding yeast. Cell Cycle 2 564 567
3. LopesM
Cotta-RamusinoC
PellicioliA
LiberiG
PlevaniP
2001 The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412 557 561
4. Cotta-RamusinoC
FachinettiD
LuccaC
DoksaniY
LopesM
2005 Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17 153 159
5. LeeBI
WilsonDM3rd
1999 The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem 274 37763 37769
6. SzankasiP
SmithGR
1992 A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem 267 3014 3023
7. TishkoffDX
AminNS
ViarsCS
ArdenKC
KolodnerRD
1998 Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res 58 5027 5031
8. SzankasiP
SmithGR
1995 A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 267 1166 1169
9. FiorentiniP
HuangKN
TishkoffDX
KolodnerRD
SymingtonLS
1997 Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol 17 2764 2773
10. KirkpatrickDT
FergusonJR
PetesTD
SymingtonLS
2000 Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics 156 1549 1557
11. TsubouchiH
OgawaH
2000 Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell 11 2221 2233
12. MimitouEP
SymingtonLS
2009 DNA end resection: Many nucleases make light work. DNA Repair (Amst)
13. QiuJ
QianY
ChenV
GuanMX
ShenB
1999 Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem 274 17893 17900
14. El-ShemerlyM
JanscakP
HessD
JiricnyJ
FerrariS
2005 Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res 65 3604 3609
15. El-ShemerlyM
HessD
PyakurelAK
MoselhyS
FerrariS
2008 ATR-dependent pathways control hEXO1 stability in response to stalled forks. Nucleic Acids Res 36 511 519
16. BoldersonE
TomimatsuN
RichardDJ
BoucherD
KumarR
2010 Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res 38 1821 1831
17. MorinI
NgoHP
GreenallA
ZubkoMK
MorriceN
2008 Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J 27 2400 2410
18. SeguradoM
DiffleyJF
2008 Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev 22 1816 1827
19. MorrisonDK
2009 The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol 19 16 23
20. XiaoB
SmerdonSJ
JonesDH
DodsonGG
SonejiY
1995 Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376 188 191
21. GardinoAK
SmerdonSJ
YaffeMB
2006 Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: a comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin Cancer Biol 16 173 182
22. BraselmannS
McCormickF
1995 Bcr and Raf form a complex in vivo via 14-3-3 proteins. EMBO J 14 4839 4848
23. BridgesD
MoorheadGB
2005 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE 2005 re10
24. YahyaouiW
CallejoM
PriceGB
Zannis-HadjopoulosM
2007 Deletion of the cruciform binding domain in CBP/14-3-3 displays reduced origin binding and initiation of DNA replication in budding yeast. BMC Mol Biol 8 27
25. YahyaouiW
Zannis-HadjopoulosM
2009 14-3-3 proteins function in the initiation and elongation steps of DNA replication in Saccharomyces cerevisiae. J Cell Sci 122 4419 4426
26. LottersbergerF
RubertF
BaldoV
LucchiniG
LongheseMP
2003 Functions of Saccharomyces cerevisiae 14-3-3 proteins in response to DNA damage and to DNA replication stress. Genetics 165 1717 1732
27. UsuiT
PetriniJH
2007 The Saccharomyces cerevisiae 14-3-3 proteins Bmh1 and Bmh2 directly influence the DNA damage-dependent functions of Rad53. Proc Natl Acad Sci U S A 104 2797 2802
28. LottersbergerF
PanzaA
LucchiniG
LongheseMP
2007 Functional and physical interactions between yeast 14-3-3 proteins, acetyltransferases, and deacetylases in response to DNA replication perturbations. Mol Cell Biol 27 3266 3281
29. AlvarezD
NovacO
CallejoM
RuizMT
PriceGB
2002 14-3-3sigma is a cruciform DNA binding protein and associates in vivo with origins of DNA replication. J Cell Biochem 87 194 207
30. FioraniS
MimunG
CalecaL
PicciniD
PellicioliA
2008 Characterization of the activation domain of the Rad53 checkpoint kinase. Cell Cycle 7 493 499
31. SzyjkaSJ
AparicioJG
ViggianiCJ
KnottS
XuW
2008 Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev 22 1906 1920
32. KinoshitaE
Kinoshita-KikutaE
MatsubaraM
YamadaS
NakamuraH
2008 Separation of phosphoprotein isotypes having the same number of phosphate groups using phosphate-affinity SDS-PAGE. Proteomics 8 2994 3003
33. LopesM
2009 Electron microscopy methods for studying in vivo DNA replication intermediates. Methods Mol Biol 521 605 631
34. SogoJM
LopesM
FoianiM
2002 Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297 599 602
35. LopesM
FoianiM
SogoJM
2006 Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21 15 27
36. NewlonCS
TheisJF
1993 The structure and function of yeast ARS elements. Curr Opin Genet Dev 3 752 758
37. Pozuelo RubioM
GeraghtyKM
WongBH
WoodNT
CampbellDG
2004 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J 379 395 408
38. JinJ
SmithFD
StarkC
WellsCD
FawcettJP
2004 Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol 14 1436 1450
39. YangX
LeeWH
SobottF
PapagrigoriouE
RobinsonCV
2006 Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc Natl Acad Sci U S A 103 17237 17242
40. ThomasBJ
RothsteinR
1989 Elevated recombination rates in transcriptionally active DNA. Cell 56 619 630
41. JiaoR
BachratiCZ
PedrazziG
KusterP
PetkovicM
2004 Physical and functional interaction between the Bloom's syndrome gene product and the largest subunit of chromatin assembly factor 1. Mol Cell Biol 24 4710 4719
42. Muzi FalconiM
PiseriA
FerrariM
LucchiniG
PlevaniP
1993 De novo synthesis of budding yeast DNA polymerase alpha and POL1 transcription at the G1/S boundary are not required for entrance into S phase. Proc Natl Acad Sci U S A 90 10519 10523
43. WuY
LiQ
ChenXZ
2007 Detecting protein-protein interactions by Far western blotting. Nat Protoc 2 3278 3284
44. DemmelL
BeckM
KloseC
SchlaitzAL
GloorY
2008 Nucleocytoplasmic shuttling of the Golgi phosphatidylinositol 4-kinase Pik1 is regulated by 14-3-3 proteins and coordinates Golgi function with cell growth. Mol Biol Cell 19 1046 1061
45. LopesM
Cotta-RamusinoC
LiberiG
FoianiM
2003 Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol Cell 12 1499 1510
46. ObrdlikP
El-BakkouryM
HamacherT
CappellaroC
VilarinoC
2004 K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci U S A 101 12242 12247
Štítky
Genetika Reprodukční medicína
Článek Incorporating Biological Pathways via a Markov Random Field Model in Genome-Wide Association StudiesČlánek Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation inČlánek Epigenetic Regulation of Cell Type–Specific Expression Patterns in the Human Mammary Epithelium
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 4- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Is Co-Expressed with Closely Adjacent Uncharacterised Genes Spanning a Breast Cancer Susceptibility Locus at 6q25.1
- The Liberation of Embryonic Stem Cells
- Genome-Wide Meta-Analysis Identifies Regions on 7p21 () and 15q24 () As Determinants of Habitual Caffeine Consumption
- A Sustained Dietary Change Increases Epigenetic Variation in Isogenic Mice
- The Exocyst Protein Sec10 Interacts with Polycystin-2 and Knockdown Causes PKD-Phenotypes
- Incorporating Biological Pathways via a Markov Random Field Model in Genome-Wide Association Studies
- Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation in
- Identification and Functional Validation of the Novel Antimalarial Resistance Locus in
- Does Positive Selection Drive Transcription Factor Binding Site Turnover? A Test with Drosophila Cis-Regulatory Modules
- Protein Phosphatase 2A Controls Ethylene Biosynthesis by Differentially Regulating the Turnover of ACC Synthase Isoforms
- Ribosomal DNA Deletions Modulate Genome-Wide Gene Expression: “–Sensitive” Genes and Natural Variation
- Reciprocal Sign Epistasis between Frequently Experimentally Evolved Adaptive Mutations Causes a Rugged Fitness Landscape
- Variable Pathogenicity Determines Individual Lifespan in
- Evolution of Vertebrate Transient Receptor Potential Vanilloid 3 Channels: Opposite Temperature Sensitivity between Mammals and Western Clawed Frogs
- Towards Establishment of a Rice Stress Response Interactome
- Mouse Genome-Wide Association and Systems Genetics Identify As a Regulator of Bone Mineral Density and Osteoclastogenesis
- Quantitative Fitness Analysis Shows That NMD Proteins and Many Other Protein Complexes Suppress or Enhance Distinct Telomere Cap Defects
- Highly Precise and Developmentally Programmed Genome Assembly in Requires Ligase IV–Dependent End Joining
- PDP-1 Links the TGF-β and IIS Pathways to Regulate Longevity, Development, and Metabolism
- Genome-Wide Association Study Using Extreme Truncate Selection Identifies Novel Genes Affecting Bone Mineral Density and Fracture Risk
- Eight Common Genetic Variants Associated with Serum DHEAS Levels Suggest a Key Role in Ageing Mechanisms
- 14-3-3 Proteins Regulate Exonuclease 1–Dependent Processing of Stalled Replication Forks
- HDA6 Regulates Locus-Directed Heterochromatin Silencing in Cooperation with MET1
- Epigenetic Regulation of Cell Type–Specific Expression Patterns in the Human Mammary Epithelium
- Enhanced Statistical Tests for GWAS in Admixed Populations: Assessment using African Americans from CARe and a Breast Cancer Consortium
- Beyond Missing Heritability: Prediction of Complex Traits
- An Evolutionary Genomic Approach to Identify Genes Involved in Human Birth Timing
- Long-Lost Relative Claims Orphan Gene: in a Wasp
- PTG Depletion Removes Lafora Bodies and Rescues the Fatal Epilepsy of Lafora Disease
- Chromatin Organization in Sperm May Be the Major Functional Consequence of Base Composition Variation in the Human Genome
- GWAS of Follicular Lymphoma Reveals Allelic Heterogeneity at 6p21.32 and Suggests Shared Genetic Susceptibility with Diffuse Large B-cell Lymphoma
- Loss-of-Function Mutations in Cause Metachondromatosis, but Not Ollier Disease or Maffucci Syndrome
- DNA Damage, Somatic Aneuploidy, and Malignant Sarcoma Susceptibility in Muscular Dystrophies
- The Phylogenetic Origin of Coincided with the Origin of Maternally Provisioned Germ Plasm and Pole Cells at the Base of the Holometabola
- Genome Analysis Reveals Interplay between 5′UTR Introns and Nuclear mRNA Export for Secretory and Mitochondrial Genes
- Genome-Wide Association Analysis of Soluble ICAM-1 Concentration Reveals Novel Associations at the , , , and Loci
- The Complete Spectrum of Yeast Chromosome Instability Genes Identifies Candidate CIN Cancer Genes and Functional Roles for ASTRA Complex Components
- Dynamic Regulation of H3K27 Trimethylation during Differentiation
- Phosphorylation-Dependent Differential Regulation of Plant Growth, Cell Death, and Innate Immunity by the Regulatory Receptor-Like Kinase BAK1
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PTG Depletion Removes Lafora Bodies and Rescues the Fatal Epilepsy of Lafora Disease
- Evolution of Vertebrate Transient Receptor Potential Vanilloid 3 Channels: Opposite Temperature Sensitivity between Mammals and Western Clawed Frogs
- Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation in
- An Evolutionary Genomic Approach to Identify Genes Involved in Human Birth Timing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



