-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genome-Wide Association Analysis of Soluble ICAM-1 Concentration Reveals Novel Associations at the , , , and Loci
Soluble ICAM-1 (sICAM-1) is an endothelium-derived inflammatory marker that has been associated with diverse conditions such as myocardial infarction, diabetes, stroke, and malaria. Despite evidence for a heritable component to sICAM-1 levels, few genetic loci have been identified so far. To comprehensively address this issue, we performed a genome-wide association analysis of sICAM-1 concentration in 22,435 apparently healthy women from the Women's Genome Health Study. While our results confirm the previously reported associations at the ABO and ICAM1 loci, four novel associations were identified in the vicinity of NFKBIK (rs3136642, P = 5.4×10−9), PNPLA3 (rs738409, P = 5.8×10−9), RELA (rs1049728, P = 2.7×10−16), and SH2B3 (rs3184504, P = 2.9×10−17). Two loci, NFKBIB and RELA, are involved in NFKB signaling pathway; PNPLA3 is known for its association with fatty liver disease; and SH3B2 has been associated with a multitude of traits and disease including myocardial infarction. These associations provide insights into the genetic regulation of sICAM-1 levels and implicate these loci in the regulation of endothelial function.
Published in the journal: . PLoS Genet 7(4): e32767. doi:10.1371/journal.pgen.1001374
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001374Summary
Soluble ICAM-1 (sICAM-1) is an endothelium-derived inflammatory marker that has been associated with diverse conditions such as myocardial infarction, diabetes, stroke, and malaria. Despite evidence for a heritable component to sICAM-1 levels, few genetic loci have been identified so far. To comprehensively address this issue, we performed a genome-wide association analysis of sICAM-1 concentration in 22,435 apparently healthy women from the Women's Genome Health Study. While our results confirm the previously reported associations at the ABO and ICAM1 loci, four novel associations were identified in the vicinity of NFKBIK (rs3136642, P = 5.4×10−9), PNPLA3 (rs738409, P = 5.8×10−9), RELA (rs1049728, P = 2.7×10−16), and SH2B3 (rs3184504, P = 2.9×10−17). Two loci, NFKBIB and RELA, are involved in NFKB signaling pathway; PNPLA3 is known for its association with fatty liver disease; and SH3B2 has been associated with a multitude of traits and disease including myocardial infarction. These associations provide insights into the genetic regulation of sICAM-1 levels and implicate these loci in the regulation of endothelial function.
Introduction
A member of the immunoglobulin superfamily of adhesion receptors, ICAM-1 is expressed on endothelial cells where it serves as a receptor for the leukocyte integrins LFA-1 and Mac-1 [1]. A soluble form of ICAM-1 (sICAM-1) is present in plasma and is thought to arise from proteolytic cleavage of the extra-cellular domains of ICAM-1. Although the physiologic function of soluble ICAM-1 remains to be fully defined, plasma concentration of sICAM-1 have a predictive value for the risk of myocardial infarction, ischemic stroke, peripheral arterial disease and noninsulin-dependent diabetes mellitus in epidemiological studies [2]–[4].
We recently described a genome-wide association study of sICAM-1 in 6,578 apparently healthy women from the Women's Genome Health Study (WGHS), which confirmed a known association at the ICAM1 locus and identified a novel association at the ABO locus [5]. These results were subsequently replicated in large-scale genomics studies from Barbalic [6] et al. and Qi [7] et al. Nevertheless, the total variance explained by these associations remained low (8.4%) as compared to the relatively high heritability estimates (from 0.34 to 0.59) [8], [9] for sICAM-1. We therefore hypothesized that other, weaker, common genetic determinants of sICAM-1 remained to be discovered. To explore this issue, we performed a larger genome-wide association study (GWAS), evaluating 334,295 SNPs in 22,435 apparently healthy women of European ancestry from the WGHS.
Results
We found that 67 SNPs passed our pre-specified threshold of genome-wide significance of P<5×10−8 for association with sICAM-1 (Table S1 and Figure 1A). These SNPs clustered within 5 loci in the vicinity of ABO (9q34.2), RELA (11q13.1), SH2B3 (12q24.12), ICAM1 (19p13.2) and PNPLA3 (22q13.31). The ICAM1 [10], [11] and ABO [5] loci have previously been identified as contributing to sICAM-1 levels, but the SH2B3, RELA and PNPLA3 loci were not previously shown to be associated with sICAM-1. The genomic context of these three latter loci is illustrated in Figure 2A, 2B and 2C.
Fig. 1. Quantile-quantile plot of association with sICAM-1. 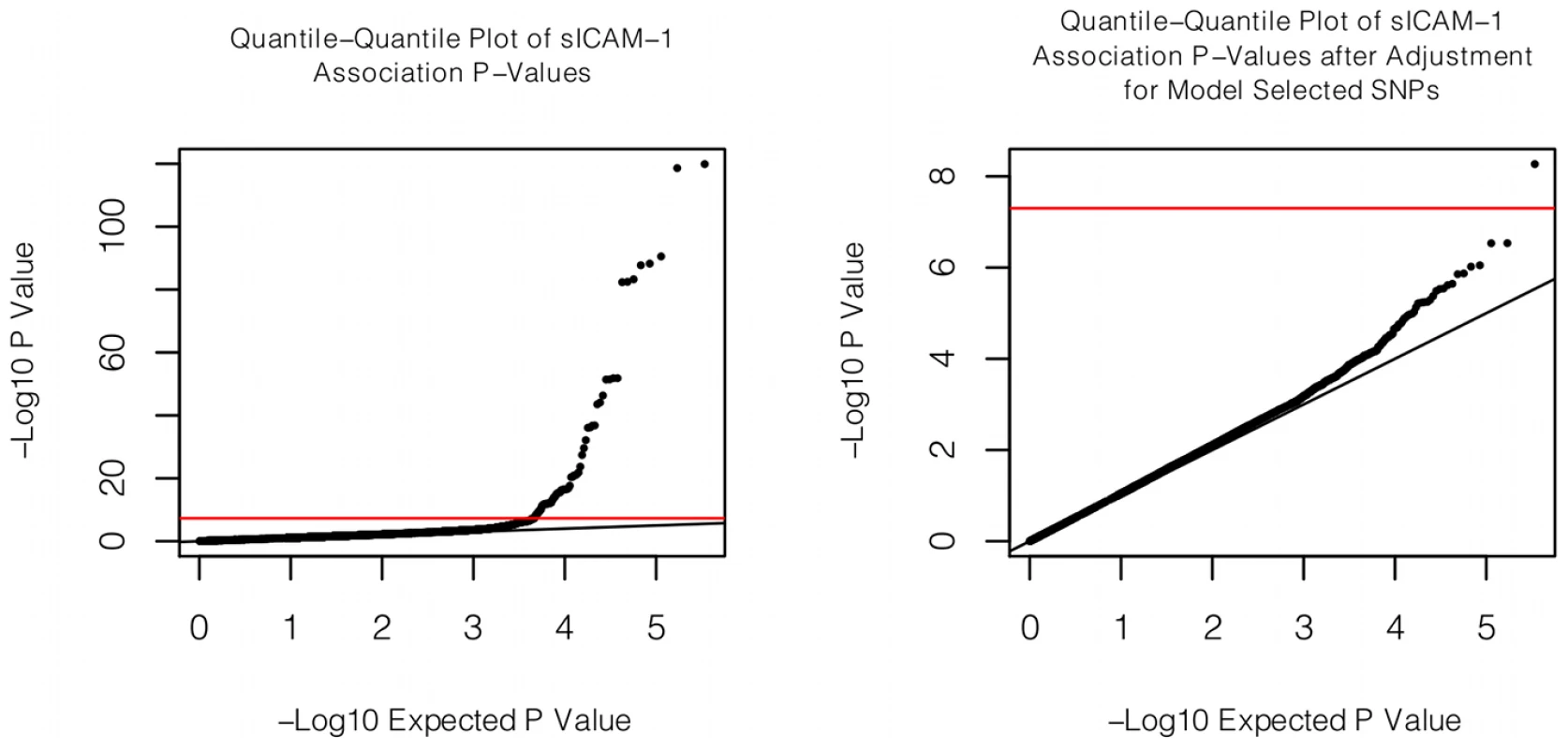
The quantile-quantile plot of sICAM-1 association P-values is shown on the left. On the right, the same quantile-quantile plot is shown, but after adjusting sICAM-1 values for the 9 SNPs retained by the model selection algorithm. Fig. 2. Genomic context of novel associations. 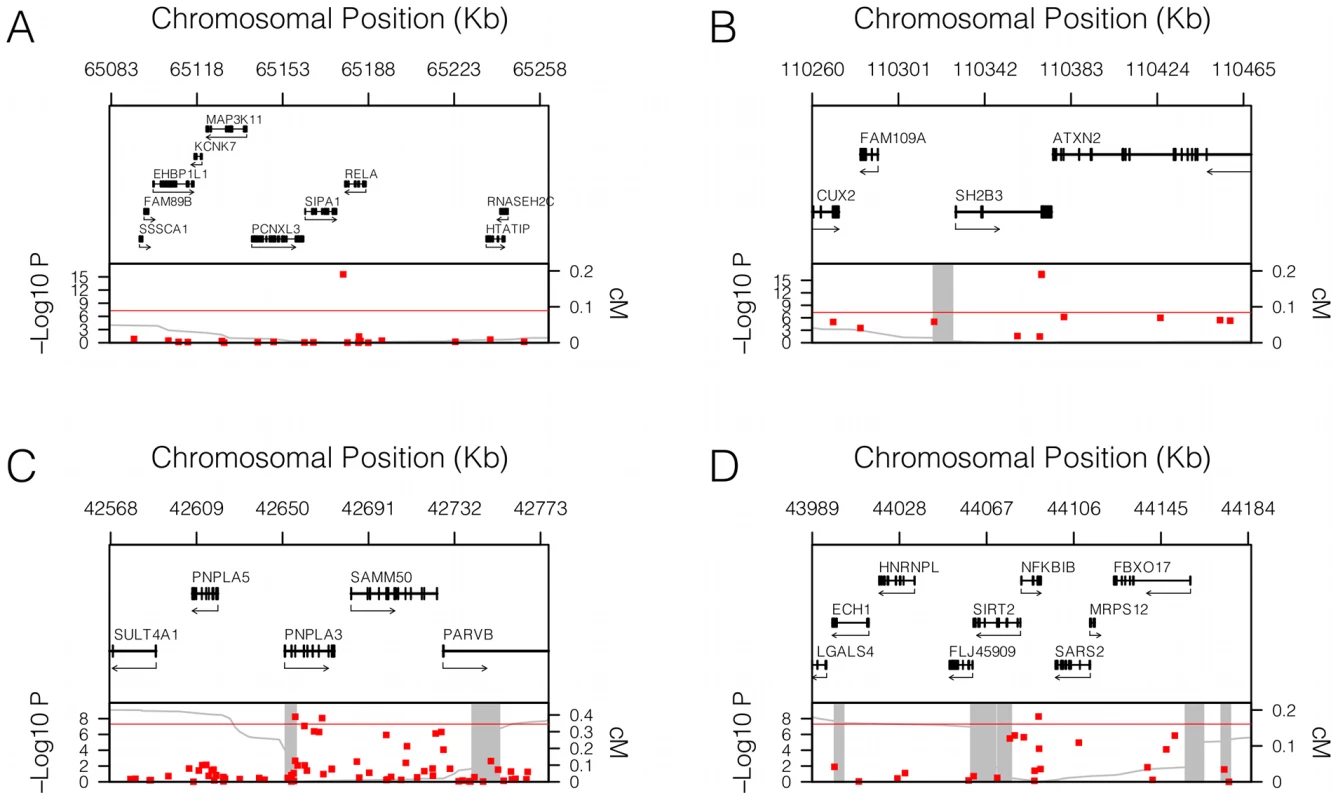
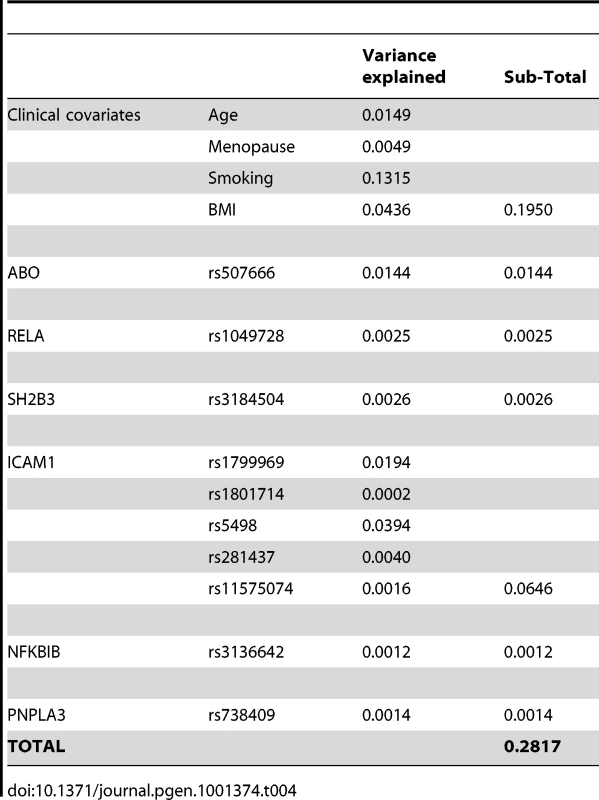
Genomic context for each of the four novel loci with significant association with sICAM-1 concentration. (A) RELA locus (11q13.1); (B) SH2B3 locus (12q24.12); (C) PNPLA3 locus (22q13.31); and (D) NFKBIB locus (19q13.2). Upper panel: Genes from RefSeq release 25. Only one isoform is shown when multiple splicing variants are known. Lower Panel: SNPs are shown according to their physical location and –log10 P-values for association with sICAM-1 (red dots). The red line represents the genome-wide significance threshold of 5×10−8. Also shown is the genetic distance in cM from the lowest P-value SNP (light grey line) along with the position of recombination hotspots (light grey vertical bars). Recombination rates and hotspots are based on HapMap data, as described by McVean et al. [43] and Winckler et al. [44]. In order to determine whether more than one non-redundant association signal could be detected at each of these five loci, we applied a model selection algorithm. The SNP with the lowest P-value for association was the only one retained at every locus with the exception of the ICAM1 locus, where 5 SNPs were selected by the model (Table 1). Interestingly, model selected SNPs at the ICAM1 locus showed lower P-value when they were all included in a single multivariate model than when considered separately. Three of the model selected SNPs at the ICAM1 locus (rs281437, rs1801714 and rs11575074) were not significant at a genome-wide level of significance in a univariate analysis. We performed two analyses to determine if the multiple SNPs selected at the ICAM1 locus were the result of an underlying association with a known but untyped variant. First, we tested all imputed SNPs (using MACH) within 1.5 Mb of rs1799969 (the lead SNP at that locus) for association with adjusted sICAM-1 levels. No imputed SNP was more significant than the directly genotyped rs1799969. Second, we tested the same set of imputed SNPs after additional adjustment of sICAM-1 levels for the effect of model selected SNPs. No additional SNP was associated at genome-wide significance. The 5 SNPs at the ICAM1 locus selected by our algorithm were also used in haplotype analysis using WHAP [12], as implemented in PLINK [13] (Table 2). The estimate of the proportion of variance attributable to haplotypes, as well as their regression coefficients, is consistent with the linear model of these same SNPs, reinforcing the adequacy of an additive model to explain the association.
Tab. 1. SNPs retained by the model selection algorithm. 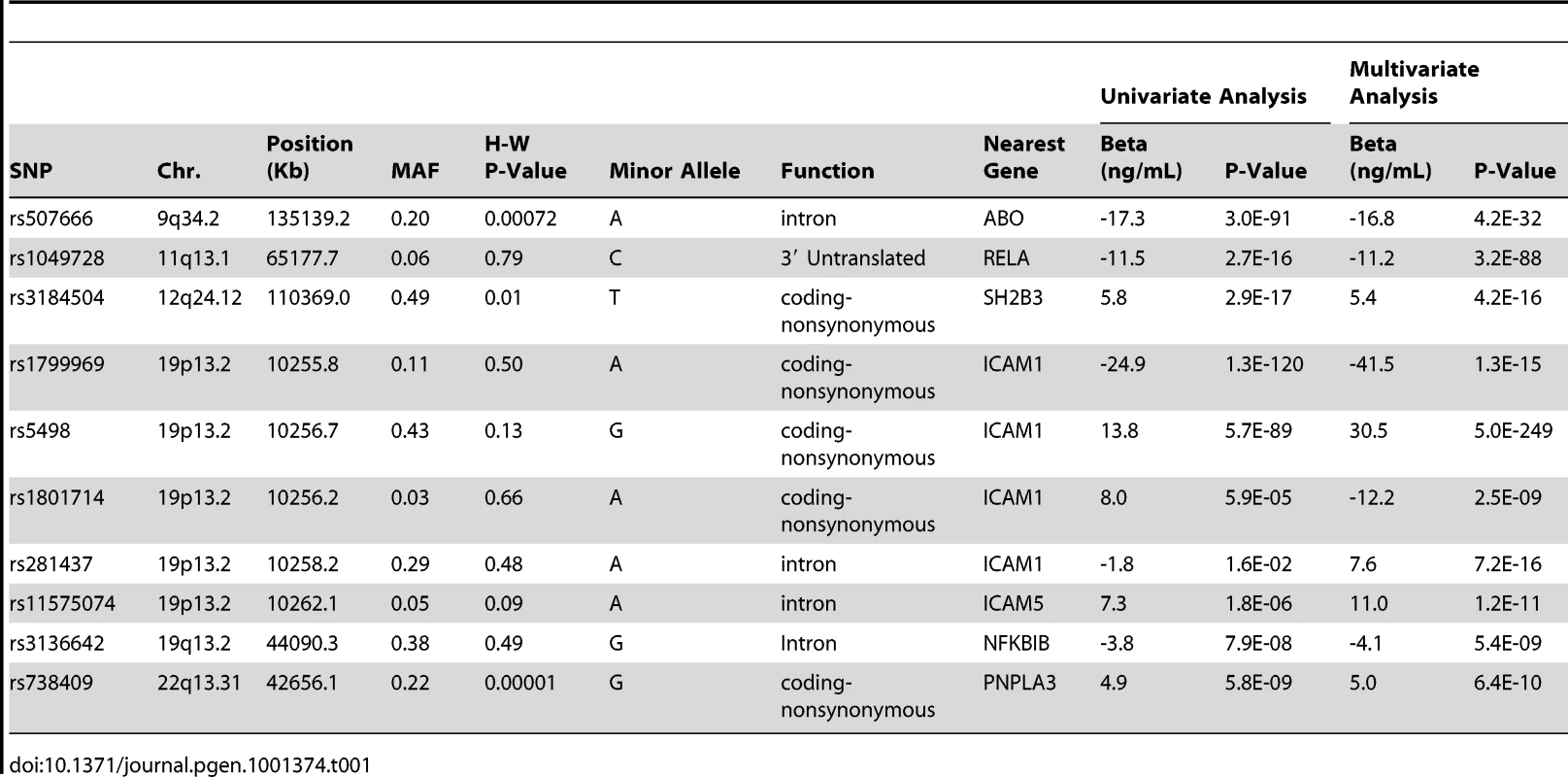
Tab. 2. Haplotype Analysis of rs1799969, rs1801714, rs5498, rs281437, and rs11575074 (19p13.2; ICAM1 locus). 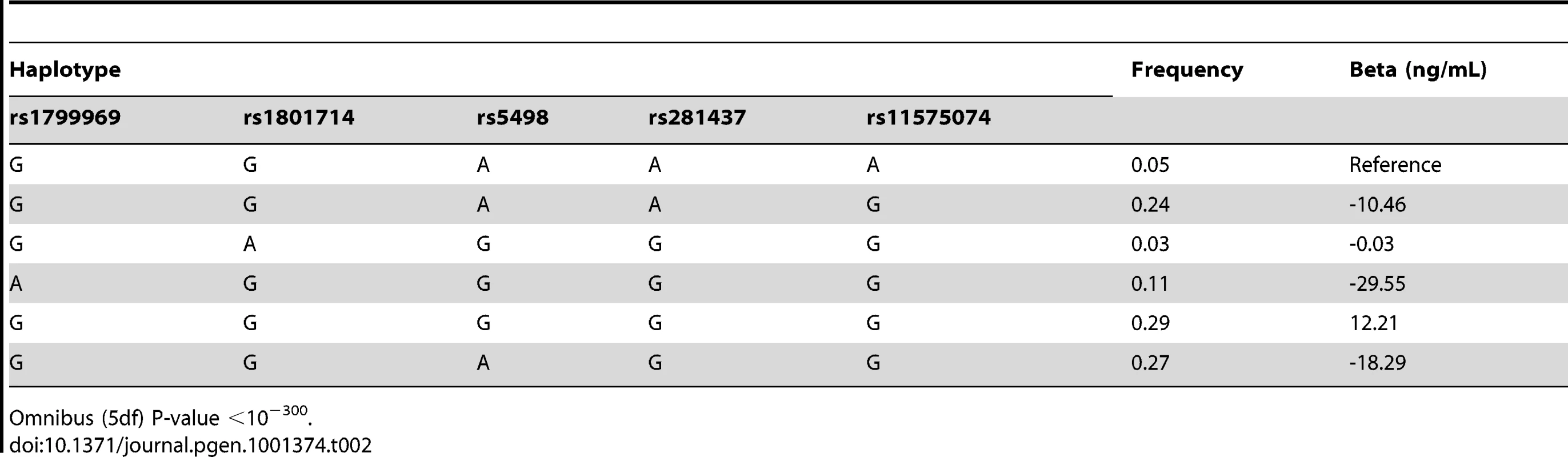
Omnibus (5df) P-value <10−300. Next we tested whether any additional SNPs are associated with sICAM-1 levels after adjustment for the model selected SNPs (see Figure 1B). A single SNP was associated with sICAM-1 at genome-wide significance (P = 5.4×10−9; −4.1 ng/mL per minor allele) in the vicinity of the NFKBIB locus at 19q13.2 (Figure 2D). This SNP, rs3136642, is intronic to NFKBIB and had a minor allele frequency of 0.38. The model selection algorithm retained no other SNP at the NFKBIB locus. Further adjustment of sICAM-1 values for rs3136642 did not identify any additional SNP with genome-wide significant association with sICAM-1. We also performed GWAS analysis using imputed genotypes (using MACH). Because no new locus reached genome-wide significance after adjustment for model selected SNPs, only results of directly genotyped SNPs are presented. These results were essentially unchanged when the first 10 components of a principal component analysis were included as covariates to account for sub-Caucasian stratification. All 4 novel loci identified in WGHS were replicated (one-sided P<0.05) in 9,813 individuals from the CHARGE consortium [14] (Table 3).
Tab. 3. Replication of novel loci in CHARGE (N = 9,813). 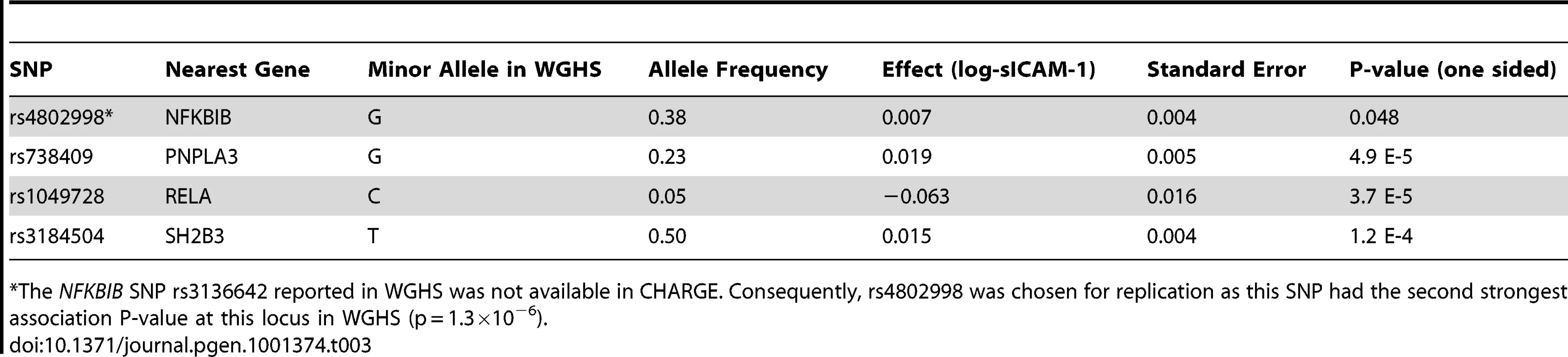
*The NFKBIB SNP rs3136642 reported in WGHS was not available in CHARGE. Consequently, rs4802998 was chosen for replication as this SNP had the second strongest association P-value at this locus in WGHS (p = 1.3×10−6). Collectively, the 5 SNPs at the ICAM1 gene locus explained 6.5% of sICAM1 total variance, whereas the other loci explained from 0.1 to 1.4% of the variance. In comparison, clinical covariates explained 19.5% of the variance (Table 4). For 4 of the loci, there was no strong evidence for non-additive effects of the minor allele as judged by lack of significance for a likelihood ratio test comparing the additive regression model to an alternative genotype model with an additional degree of freedom. However, the non-additive component was significant for rs507666 (P = 9.3×10−6) at the ABO locus with a tendency toward a dominant effect (mean sICAM-1 of 362.1, 342.4 and 335.4 ng/mL for 0, 1 and 2 minor alleles, respectively). The PNPLA3 SNP rs738409 also showed evidence of non-additive association (P = 4.6×10−5) with a tendency toward a recessive model (mean sICAM-1 of 352.8, 356.0 and 367.7 ng/mL for 0, 1 and 2 minor alleles, respectively). In spite of these non-additive trends, no additional locus reached genome-wide significance when a genotypic test, which does not assume an additive model of association, was conducted.
Model selected SNPs were tested for association with other available inflammation markers (C-reactive protein and fibrinogen). No significant association was noted (P>0.01) after adjusting for multiple hypothesis testing. Model selected SNPs were also tested for association with incident cardiovascular events (myocardial infarction, coronary revascularization, stroke and total cardiovascular event) over a mean follow-up period of 14 years. A Cox proportional hazard model was used adjusting for age at study entry. Only the SH2B3 SNP rs3184504 was associated with incident myocardial infarction (315 events), with each minor allele increasing the risk (P = 0.011; OR 1.23 95% CI 1.05–1.43). The association remained significant after further adjustment for sICAM-1 levels (P = 0.028; OR 1.20 95% CI 1.02–1.41). Given the known association of sICAM-1 with cardiovascular risk and the association of selected SNPs with sICAM-1, we estimated the power to detect an association between the SH2B3 SNP rs3184504 and myocardial infarction to be 6%, for alpha = 0.05. In comparison, power varied from 5% (rs281437) to 11% (rs5498) for other SNPs. The PNPLA3 SNP rs738409 was tested for association with triglyceride, LDL cholesterol, HDL cholesterol and BMI as this gene is known to be involved in lipid metabolism and association with BMI has been previously suggested [15]. No significant association was observed.
Since smoking accounts for a large fraction of the variation in sICAM-1 levels, we tested associated SNPs for interaction with smoking. A significant interaction was observed for the ICAM1 SNP rs1799969 (interaction P = 1.6×10−9) whereby current smokers had a stronger genetic association, as we previously reported [16]. A novel interaction was also observed with the ABO SNP rs507666, again with a stronger genetic association in current smokers (P = 0.0003). When restricting the GWAS analysis to current smokers, an additional association was observed with rs8034191 (P = 3.5×10−8). This latter SNP is located on chromosome 15 near the nicotinic acetylcholine receptor subunit genes CHRNA3 and CHRNA5. This locus is known to be associated with smoking behavior [17], [18] and rs8034191 has recently been associated with smoking quantity [19]. No novel association was observed when restricting the GWAS analysis to non-smokers after adjustment for the previously described loci.
We also tested whether multiple variants of individually weak effect could contribute to sICAM-1 levels. In cross-validation procedures, no increase in variance explained was observed when using P-value cut-offs less significant than 10−8 for inclusion of SNPs in gene scores (see Figure 3). In other words, selection of SNPs on the basis of P-value alone was not able to identify more of the genetic variance than could be explained by the SNPs with association P-value <10−8.
Fig. 3. Polygene analysis. 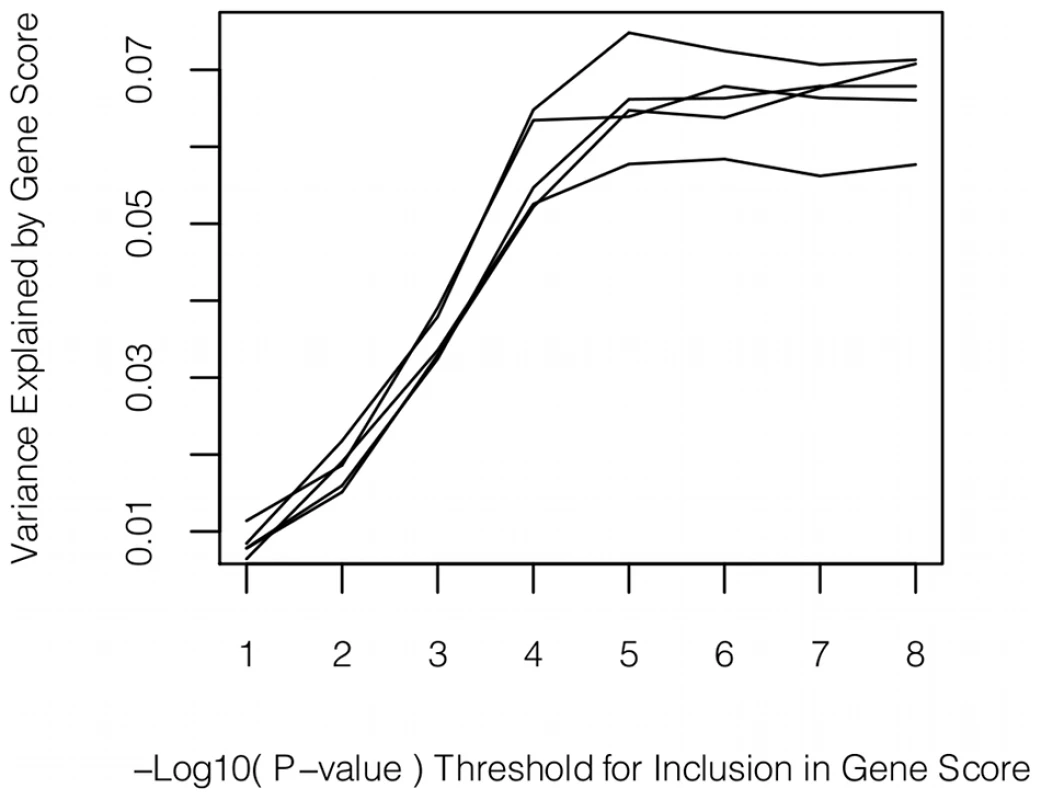
Variance explained (adjusted R2) by gene scores using varying P-value thresholds for inclusion of SNPs. Each P-value threshold was tested 5 times using a 5-fold cross-validation procedure. Discussion
Six loci – ABO, ICAM1, NFKBIK, PNPLA3, RELA and SH2B3 – have been identified in this report for association with sICAM-1. While the ABO [5] and ICAM1 [10], [11] loci had been previously reported, we extended the number of non-redundantly associated variants at the ICAM1 locus by demonstrating association of rs11575074 and rs1801714 in multivariate analysis along with the known rs1799969, rs5498 and rs281437 SNPs [5]. Neither rs1801714 nor rs11575074 are predicted eQTL (http://eqtl.uchicago.edu/Home.html), but rs1801714 is a missense variant (P352L) and rs11575074 is located in a predicted binding site for several transcription factors including PPARG [20]. The NFKBIK, PNPLA3, RELA and SH2B3 associations are novel. No strong contribution of weakly associated variants was observed in the polygene analysis whereby SNPs of varying statistical significance were included in gene scores.
Nuclear factor kB (NF-kB) proteins are a family of transcription factors involved in a number of physiological processes that include cell survival, proliferation, and activation. The NF-kB proteins (NFKB1 or NFKB2) are bound to REL, RELA, or RELB to form the NF-kB complex. These complexes are typically localized in the cytoplasm, where they are trapped by binding to IkB inhibitory proteins NFKBIA or NFKBIB. Upon inflammatory simulation, IkB kinase A and B phosphorylate IkB inhibitory proteins and mark them for degradation via the ubiquitination pathway, thereby allowing activation of the NF-kappa-B complex. Activated NF-kB complexes translocate into the nucleus and bind to NF-kB DNA binding motifs. NF-kB triggers transcription of various genes critical to inflammation, such as cytokines, chemokines and cell adhesion molecules including ICAM1 [21], [22]. Remarkably, two of the novel associations involve genes physically interacting with NF-kB. No genetic interaction, however, was noted between these two SNPs (data not shown). Taken together, these results emphasize the importance of the NFKB pathway in the regulation of sICAM-1 levels.
PNPLA3 encodes a protein of unknown function that belongs to the patatin-like phospholipase family. Members of that family are believed to complement hormone sensitive lipase for adipocyte triacylglycerol lipase activity. The methionine allele of the missense PNPLA3 SNP rs738409 (Ile148Met) has recently been associated with increased hepatic fat levels, hepatic inflammation and plasma levels of liver enzymes (traits linked to insulin resistance and obesity) [23], [24]. Nevertheless, rs738409 has been shown not to be associated with insulin resistance [25] although a previous study demonstrated an association with insulin secretion in response to oral glucose tolerance test [15]. Levels of the inflammatory marker sICAM-1 are known to be correlated with insulin resistance and obesity [4]. Consistent with rs738409 modulating the response to insulin resistance and associated phenotypes, the risk allele for fatty liver disease was associated with increased sICAM-1 levels.
SH2B3 encodes Lnk, an adaptor protein that mediates the interaction between extra-cellular receptors, such as the T-cell receptor and the thrombopoietin receptor MPL, and intracellular signaling pathways. Cells from Lnk-deficient mice show an increased sensitivity to several cytokines and altered activation of the RAS/MAPK pathway in response to IL3 and stem cell factor [26]. The same SH2B3 SNP rs3184504 identified in our study has previously been associated with multiple other traits, including blood pressure [27], [28], blood eosinophil number [29], myocardial infarction [29], celiac disease [30], type I diabetes [31], LDL-cholesterol [32], asthma [29], blood platelet number [33], hemoglobin concentration [34] and hematocrit [34]. Furthermore, rs3184504 is a non-synonymous SNP (Arg262Trp) whose derived allele (Trp) is part of a haplotype that has been suggested to have been introduced 3,400 years ago and selectively swept in European populations [33]. The derived allele is the risk allele for coronary artery disease and was the allele associated with higher sICAM-1 concentration. Association of rs3184504 with sICAM-1 further demonstrates the remarkable pleiotropy of that genetic variant by extending its effect to endothelial cell adhesion molecules. An interesting hypothesis is whether changes in sICAM-1 are mediated through increased sub-clinical atherosclerosis, but further studies will be needed to address this question.
In this report, we demonstrate genetic association of sICAM-1 with the ABO, ICAM1, NFKBIK, PNPLA3, RELA and SH2B3 loci. These findings broaden our current knowledge of the genetic architecture of sICAM-1 with identification of four novel loci. The novel association at PNPLA3 reinforces the importance of insulin resistance-related processes in the regulation of sICAM-1 levels. The observed associations also provide evidence of functional genetic variation at two genes – NFKBIK and RELA – well known for their implication in the NF-kB pathway, therefore providing a basis for the study of these polymorphisms in other conditions where this same pathway is involved. The results also extend the effect of the SH2B3 SNP rs3184504 to endothelial function.
Methods
Ethics Statement
All analyses were performed with approval of the institutional review board of the Brigham and Women's Hospital. All members of the WGHS cohort were participants in the WHS who provided an adequate baseline blood sample for plasma and DNA analysis and who gave consent for blood-based analyses and long-term follow-up.
Study Sample and sICAM-1 Measurements
All participants in this study were part of the Women's Genome Health Study (WGHS) [35]. Briefly, participants in the WGHS include North American women from the Women's Health Study (WHS) with no prior history of cardiovascular disease, diabetes, cancer, or other major chronic illness who also provided a baseline blood sample at the time of study enrollment. For all WGHS participants, EDTA anticoagulated plasma samples were collected at baseline and stored in vapor phase liquid nitrogen (−170°C). Circulating plasma sICAM-1 concentrations were determined using a commercial ELISA assay (R&D Systems, Minneapolis, Minn.); the assay used is known not to recognize the K56M (rs5491) variant of ICAM-1 [36] and the 82 Caucasian carriers of this mutation were therefore excluded from further analysis. The intra-assay coefficient of variation was 6.7% and the reported intra-individual coefficient of variation 7.6% [37]. This study has been approved by the institutional review board of the Brigham and Women's Hospital. Additional clinical characteristics of this sample are provided in Table S2.
Genotyping
Samples were genotyped with the Infinium II technology from Illumina. Either the HumanHap300 Duo-Plus chip or the combination of the HumanHap300 Duo and I-Select chips was used. In either case, the custom content was identical and consisted of candidate SNPs chosen without regard to allele frequency to increase coverage of genetic variation with impact on biological function including metabolism, inflammation or cardiovascular diseases. Genotyping at 318,237 HumanHap300 Duo SNPs and 45,571 custom content SNPs was attempted, for a total of 363,808 SNPs. Genetic context for all annotations are derived from human genome build 36.1 and dbSNP build 126.
SNPs with call rates <90% were excluded from further analysis. Likewise, all samples with percentage of missing genotypes higher than 2% were removed. Among retained samples, SNPs were further evaluated for deviation from Hardy-Weinberg equilibrium using an exact method [38] and were excluded when the P-value was lower than 10−6. Samples were further validated by comparison of genotypes at 44 SNPs that had been previously ascertained using alternative technologies. SNPs with minor allele frequency >1% in Caucasians were used for analysis. After quality control, 334,295 SNPs were left for analysis.
Population Stratification
Because population stratification can result in inflated type I error in a GWAS, a principal component analysis using 1443 ancestry informative SNPs was performed using PLINK [13] to confirm self-reported ancestry. Briefly, these SNPs were chosen based on Fst >0.4 in HapMap populations (YRB, CEU, CHB+JPT) and inter-SNP distance at least 500 kb in order to minimize linkage disequilibrium. Different ethnic groups were clearly distinguished with the two first components. 31 self-identified Caucasian women were removed from analysis because they did not cluster with other Caucasians, leaving 22,435 non-diabetic participants with non-missing sICAM-1 information for analysis. To rule out the possibility that residual stratification within Caucasians was responsible for the associations observed, a principal component analysis [39] was performed in Caucasians (only) using 64,205 SNPs chosen to have pair-wise linkage disequilibrium lower than r2 = 0.2. The first ten components were then used as covariates in the association analysis. As adjustment by these covariates did not change the conclusions, we present analysis among Caucasian participants without further correction for sub-Caucasian ancestry unless stated otherwise.
Association Analysis
Plasma concentrations of sICAM-1 were adjusted for age, smoking, menopause and body mass index using a linear regression model in R to reduce the impact of clinical covariates on sICAM-1 variance. The adjusted sICAM-1 values were then tested for association with SNP genotypes by linear regression in PLINK [13], assuming an additive contribution of each minor allele. A conservative P-value cut-off of 5×10−8 was used to correct for the roughly 1,000,000 independent statistical tests thought to correspond to all the common genetic variation of the human genome [40], [41].
Model Selection Algorithm
To investigate whether more than one SNP in each locus is independently associated with sICAM-1, a forward selection multiple linear regression model was used. For each locus with at least one genome-wide significant SNP (i.e. P<5×10−8), all genotyped SNPs within 1.5 Mb of the most significantly associated SNP and passing quality control requirements were selected for potential inclusion in our model. The forward selection algorithm then proceeded in two steps. In the first step, all SNPs not yet included in the multiple regression model were tested for association with sICAM-1. In step two, the SNP with the smallest P-value was included in the model if its multiple regression P-value was less than 5×10−8. We then repeated steps one and two, such that a single SNP was added to the multiple regression model at each iteration. The algorithm was stopped when no more SNP passed the P<5×10−8 requirement.
Polygene Analysis
To test whether multiple genetic variants of individually weak effect could explain a substantial fraction of sICAM-1 variance, we performed a “polygene” experiment as previously described [42]. Briefly, we randomly divided our dataset in 5 equal parts. We then tested SNPs for association with sICAM-1 using 4 out the 5 parts and performed linkage disequilibrium pruning as implemented in PLINK (r2>0.05 and distance <1 Mb). We then derived a gene score with non-redundant associated SNPs using varying P-value thresholds and weighting each SNP for its beta coefficient. Finally, we tested the gene score for association with sICAM-1 in the remaining one fifth of the total sample and calculated the adjusted R2. This experiment was repeated 5 times using each one of the five parts as the gene score validation group alternatively.
Replication of Novel Associations in CHARGE
We sought to replicate the 4 novel loci identified in 9,813 individuals from the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) consortium [14] for whom plasma sICAM-1 concentration and genotypes were available. The CHARGE sample consists of 4 meta-analyzed cohorts: the Framingham Heart Study, the Cardiovascular Health Study, the Atherosclerosis Risk in Communities study, and the Rotterdam Study. Complete information on each study is available as Text S1. Association analyses were performed on imputed genotypes using an additive genetic model on age and sex adjusted log-transformed sICAM-1 values.
Supporting Information
Zdroje
1. van de StolpeA
van der SaagPT
1996 Intercellular adhesion molecule-1. J Mol Med 74 13 33
2. RidkerPM
HennekensCH
Roitman-JohnsonB
StampferMJ
AllenJ
1998 Plasma concentration of soluble intercellular adhesion molecule 1 and risks of future myocardial infarction in apparently healthy men. Lancet 351 88 92
3. PradhanAD
RifaiN
RidkerPM
2002 Soluble intercellular adhesion molecule-1, soluble vascular adhesion molecule-1, and the development of symptomatic peripheral arterial disease in men. Circulation 106 820 825
4. SongY
MansonJE
TinkerL
RifaiN
CookNR
2007 Circulating levels of endothelial adhesion molecules and risk of diabetes in an ethnically diverse cohort of women. Diabetes 56 1898 1904
5. PareG
ChasmanDI
KelloggM
ZeeRY
RifaiN
2008 Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet 4 e1000118
6. BarbalicM
DupuisJ
DehghanA
BisJC
HoogeveenRC
2010 Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels. Hum Mol Genet 19 1863 1872
7. QiL
CornelisMC
KraftP
JensenM
van DamRM
2010 Genetic variants in ABO blood group region, plasma soluble E-selectin levels and risk of type 2 diabetes. Hum Mol Genet 19 1856 1862
8. BielinskiSJ
PankowJS
FosterCL
MillerMB
HopkinsPN
2007 Circulating soluble ICAM-1 levels shows linkage to ICAM gene cluster region on chromosome 19: The NHLBI Family Heart Study follow-up examination. Atherosclerosis
9. KentJWJr
MahaneyMC
ComuzzieAG
GoringHH
AlmasyL
2007 Quantitative trait locus on Chromosome 19 for circulating levels of intercellular adhesion molecule-1 in Mexican Americans. Atherosclerosis 195 367 373
10. PonthieuxA
LambertD
HerbethB
DroeschS
PfisterM
2003 Association between Gly241Arg ICAM-1 gene polymorphism and serum sICAM-1 concentration in the Stanislas cohort. Eur J Hum Genet 11 679 686
11. PuthothuB
KruegerM
BernhardtM
HeinzmannA
2006 ICAM1 amino-acid variant K469E is associated with paediatric bronchial asthma and elevated sICAM1 levels. Genes Immun 7 322 326
12. PurcellS
DalyMJ
ShamPC
2007 WHAP: haplotype-based association analysis. Bioinformatics 23 255 256
13. PurcellS
NealeB
Todd-BrownK
ThomasL
FerreiraMA
2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
14. PsatyBM
O'DonnellCJ
GudnasonV
LunettaKL
FolsomAR
2009 Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2 73 80
15. JohanssonLE
LindbladU
LarssonCA
RastamL
RidderstraleM
2008 Polymorphisms in the adiponutrin gene are associated with increased insulin secretion and obesity. Eur J Endocrinol 159 577 583
16. PareG
CookNR
RidkerPM
ChasmanDI
2010 On the use of variance per genotype as a tool to identify quantitative trait interaction effects: a report from the Women's Genome Health Study. PLoS Genet 6 e1000981
17. ThorgeirssonTE
GudbjartssonDF
SurakkaI
VinkJM
AminN
2010 Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 42 448 453
18. Tobacco and Genetics Consortium 2010 Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 42 441 447
19. LiuJZ
TozziF
WaterworthDM
PillaiSG
MugliaP
2010 Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet 42 436 440
20. XuZ
TaylorJA
2009 SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res 37 W600 605
21. CollinsT
ReadMA
NeishAS
WhitleyMZ
ThanosD
1995 Transcriptional regulation of endothelial cell adhesion molecules: NF-kappa B and cytokine-inducible enhancers. FASEB J 9 899 909
22. LedeburHC
ParksTP
1995 Transcriptional regulation of the intercellular adhesion molecule-1 gene by inflammatory cytokines in human endothelial cells. Essential roles of a variant NF-kappa B site and p65 homodimers. J Biol Chem 270 933 943
23. RomeoS
KozlitinaJ
XingC
PertsemlidisA
CoxD
2008 Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 40 1461 1465
24. YuanX
WaterworthD
PerryJR
LimN
SongK
2008 Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet 83 520 528
25. KantartzisK
PeterA
MachicaoF
MachannJ
WagnerS
2009 Dissociation between fatty liver and insulin resistance in humans carrying a variant of the patatin-like phospholipase 3 gene. Diabetes 58 2616 2623
26. VelazquezL
ChengAM
FlemingHE
FurlongerC
VeselyS
2002 Cytokine signaling and hematopoietic homeostasis are disrupted in Lnk-deficient mice. J Exp Med 195 1599 1611
27. Newton-ChehC
JohnsonT
GatevaV
TobinMD
BochudM
2009 Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet
28. LevyD
EhretGB
RiceK
VerwoertGC
LaunerLJ
2009 Genome-wide association study of blood pressure and hypertension. Nat Genet
29. GudbjartssonDF
BjornsdottirUS
HalapiE
HelgadottirA
SulemP
2009 Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet 41 342 347
30. HuntKA
ZhernakovaA
TurnerG
HeapGA
FrankeL
2008 Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet 40 395 402
31. ToddJA
WalkerNM
CooperJD
SmythDJ
DownesK
2007 Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 39 857 864
32. TalmudPJ
DrenosF
ShahS
ShahT
PalmenJ
2009 Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am J Hum Genet 85 628 642
33. SoranzoN
SpectorTD
ManginoM
KuhnelB
RendonA
2009 A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet 41 1182 1190
34. GaneshSK
ZakaiNA
van RooijFJ
SoranzoN
SmithAV
2009 Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet 41 1191 1198
35. RidkerPM
ChasmanDI
ZeeRY
ParkerA
RoseL
2008 Rationale, Design, and Methodology of the Women's Genome Health Study: A Genome-Wide Association Study of More Than 25 000 Initially Healthy American Women. Clin Chem 54 249 255
36. RegisterTC
BurdonKP
LenchikL
BowdenDW
HawkinsGA
2004 Variability of serum soluble intercellular adhesion molecule-1 measurements attributable to a common polymorphism. Clin Chem 50 2185 2187
37. EschenO
ChristensenJH
DethlefsenC
SchmidtEB
2008 Cellular Adhesion Molecules in Healthy Subjects: Short Term Variations and Relations to Flow Mediated Dilation. Biomark Insights 3 57 62
38. WiggintonJE
CutlerDJ
AbecasisGR
2005 A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet 76 887 893
39. PriceAL
PattersonNJ
PlengeRM
WeinblattME
ShadickNA
2006 Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38 904 909
40. FrazerKA
BallingerDG
CoxDR
HindsDA
StuveLL
2007 A second generation human haplotype map of over 3.1 million SNPs. Nature 449 851 861
41. Pe'erI
YelenskyR
AltshulerD
DalyMJ
2008 Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 32 381 385
42. PurcellSM
WrayNR
StoneJL
VisscherPM
O'DonovanMC
2009 Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460 748 752
43. McVeanGA
MyersSR
HuntS
DeloukasP
BentleyDR
2004 The fine-scale structure of recombination rate variation in the human genome. Science 304 581 584
44. WincklerW
MyersSR
RichterDJ
OnofrioRC
McDonaldGJ
2005 Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308 107 111
Štítky
Genetika Reprodukční medicína
Článek Incorporating Biological Pathways via a Markov Random Field Model in Genome-Wide Association StudiesČlánek Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation inČlánek Epigenetic Regulation of Cell Type–Specific Expression Patterns in the Human Mammary Epithelium
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 4- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Is Co-Expressed with Closely Adjacent Uncharacterised Genes Spanning a Breast Cancer Susceptibility Locus at 6q25.1
- The Liberation of Embryonic Stem Cells
- Genome-Wide Meta-Analysis Identifies Regions on 7p21 () and 15q24 () As Determinants of Habitual Caffeine Consumption
- A Sustained Dietary Change Increases Epigenetic Variation in Isogenic Mice
- The Exocyst Protein Sec10 Interacts with Polycystin-2 and Knockdown Causes PKD-Phenotypes
- Incorporating Biological Pathways via a Markov Random Field Model in Genome-Wide Association Studies
- Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation in
- Identification and Functional Validation of the Novel Antimalarial Resistance Locus in
- Does Positive Selection Drive Transcription Factor Binding Site Turnover? A Test with Drosophila Cis-Regulatory Modules
- Protein Phosphatase 2A Controls Ethylene Biosynthesis by Differentially Regulating the Turnover of ACC Synthase Isoforms
- Ribosomal DNA Deletions Modulate Genome-Wide Gene Expression: “–Sensitive” Genes and Natural Variation
- Reciprocal Sign Epistasis between Frequently Experimentally Evolved Adaptive Mutations Causes a Rugged Fitness Landscape
- Variable Pathogenicity Determines Individual Lifespan in
- Evolution of Vertebrate Transient Receptor Potential Vanilloid 3 Channels: Opposite Temperature Sensitivity between Mammals and Western Clawed Frogs
- Towards Establishment of a Rice Stress Response Interactome
- Mouse Genome-Wide Association and Systems Genetics Identify As a Regulator of Bone Mineral Density and Osteoclastogenesis
- Quantitative Fitness Analysis Shows That NMD Proteins and Many Other Protein Complexes Suppress or Enhance Distinct Telomere Cap Defects
- Highly Precise and Developmentally Programmed Genome Assembly in Requires Ligase IV–Dependent End Joining
- PDP-1 Links the TGF-β and IIS Pathways to Regulate Longevity, Development, and Metabolism
- Genome-Wide Association Study Using Extreme Truncate Selection Identifies Novel Genes Affecting Bone Mineral Density and Fracture Risk
- Eight Common Genetic Variants Associated with Serum DHEAS Levels Suggest a Key Role in Ageing Mechanisms
- 14-3-3 Proteins Regulate Exonuclease 1–Dependent Processing of Stalled Replication Forks
- HDA6 Regulates Locus-Directed Heterochromatin Silencing in Cooperation with MET1
- Epigenetic Regulation of Cell Type–Specific Expression Patterns in the Human Mammary Epithelium
- Enhanced Statistical Tests for GWAS in Admixed Populations: Assessment using African Americans from CARe and a Breast Cancer Consortium
- Beyond Missing Heritability: Prediction of Complex Traits
- An Evolutionary Genomic Approach to Identify Genes Involved in Human Birth Timing
- Long-Lost Relative Claims Orphan Gene: in a Wasp
- PTG Depletion Removes Lafora Bodies and Rescues the Fatal Epilepsy of Lafora Disease
- Chromatin Organization in Sperm May Be the Major Functional Consequence of Base Composition Variation in the Human Genome
- GWAS of Follicular Lymphoma Reveals Allelic Heterogeneity at 6p21.32 and Suggests Shared Genetic Susceptibility with Diffuse Large B-cell Lymphoma
- Loss-of-Function Mutations in Cause Metachondromatosis, but Not Ollier Disease or Maffucci Syndrome
- DNA Damage, Somatic Aneuploidy, and Malignant Sarcoma Susceptibility in Muscular Dystrophies
- The Phylogenetic Origin of Coincided with the Origin of Maternally Provisioned Germ Plasm and Pole Cells at the Base of the Holometabola
- Genome Analysis Reveals Interplay between 5′UTR Introns and Nuclear mRNA Export for Secretory and Mitochondrial Genes
- Genome-Wide Association Analysis of Soluble ICAM-1 Concentration Reveals Novel Associations at the , , , and Loci
- The Complete Spectrum of Yeast Chromosome Instability Genes Identifies Candidate CIN Cancer Genes and Functional Roles for ASTRA Complex Components
- Dynamic Regulation of H3K27 Trimethylation during Differentiation
- Phosphorylation-Dependent Differential Regulation of Plant Growth, Cell Death, and Innate Immunity by the Regulatory Receptor-Like Kinase BAK1
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PTG Depletion Removes Lafora Bodies and Rescues the Fatal Epilepsy of Lafora Disease
- Evolution of Vertebrate Transient Receptor Potential Vanilloid 3 Channels: Opposite Temperature Sensitivity between Mammals and Western Clawed Frogs
- Survival Motor Neuron Protein Regulates Stem Cell Division, Proliferation, and Differentiation in
- An Evolutionary Genomic Approach to Identify Genes Involved in Human Birth Timing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání