-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Mammalian Cell Cycle Regulates Parvovirus Nuclear Capsid Assembly
Cellular and viral life cycles are connected through multiple, though poorly understood, mechanisms. Parvoviruses infect humans and a broad spectrum of animals, causing a variety of diseases, but they are also used in experimental cancer therapy and serve as vectors for gene therapy. Parvoviruses can only multiply in proliferating cells providing essential replicative and transcriptional functions. However, it is unknown whether the cell cycle regulatory machinery may also control parvovirus assembly. We found that the nuclear translocation of parvovirus MVM capsid subunits (VPs) was highly dependent on physiological cell cycle regulations in mammalian fibroblasts, including: quiescence, progression through G1/S boundary, DNA synthesis, and cell to cell contacts. VPs nuclear translocation was significantly more sensitive to cell cycle controls than viral genome replication and gene expression. The results support nuclear capsid assembly as the major driving process of parvoviruses biological hallmarks, such as pathogenesis in proliferative tissues and tropism for cancer cells. In addition, disturbing the tight coupling of parvovirus assembly with the cell cycle may determine viral persistence in quiescent and post-mitotic host tissues. These findings may contribute to understand cellular regulations on the assembly of other nuclear eukaryotic viruses, and to develop cell cycle-based avenues for antiviral therapy.
Published in the journal: . PLoS Pathog 11(6): e32767. doi:10.1371/journal.ppat.1004920
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004920Summary
Cellular and viral life cycles are connected through multiple, though poorly understood, mechanisms. Parvoviruses infect humans and a broad spectrum of animals, causing a variety of diseases, but they are also used in experimental cancer therapy and serve as vectors for gene therapy. Parvoviruses can only multiply in proliferating cells providing essential replicative and transcriptional functions. However, it is unknown whether the cell cycle regulatory machinery may also control parvovirus assembly. We found that the nuclear translocation of parvovirus MVM capsid subunits (VPs) was highly dependent on physiological cell cycle regulations in mammalian fibroblasts, including: quiescence, progression through G1/S boundary, DNA synthesis, and cell to cell contacts. VPs nuclear translocation was significantly more sensitive to cell cycle controls than viral genome replication and gene expression. The results support nuclear capsid assembly as the major driving process of parvoviruses biological hallmarks, such as pathogenesis in proliferative tissues and tropism for cancer cells. In addition, disturbing the tight coupling of parvovirus assembly with the cell cycle may determine viral persistence in quiescent and post-mitotic host tissues. These findings may contribute to understand cellular regulations on the assembly of other nuclear eukaryotic viruses, and to develop cell cycle-based avenues for antiviral therapy.
Introduction
Viruses infecting eukaryotes encounter a dynamic regulatory network of proteins expression, posttranslational modifications and signaling, ensuring fine-tuned control of the cell cycle progression and checkpoints [1–3]. These processes include the still poorly understood cell cycle-dependent vast macromolecular traffic across the central aqueous channel of the nuclear pore complex (NPC; [4,5]), which requires soluble receptors (karyopherins) for the recognition of cargos carrying nuclear transport signals [6,7]. All these cell cycle regulations affect the multiplication of many eukaryotic viruses, which must either adapt their life cycles to these factors networks, or perturb them for their benefit. This is particularly important for the many viruses that require the nuclear host cell machinery for replication. Small DNA tumor viruses encode proteins able to induce resting cells to synthesize DNA thus overcoming the G1/S restriction point [8,9]. Herpesviruses in contrast arrest cells in late G1 phase or at the G1/S interface prior to host DNA synthesis [10,11]. Other viruses provoke a cell cycle arrest commonly associated to DNA-damage responses (DDR; [12,13]), implying multiple consequences for the virus life cycles (reviewed in [14]), including the escape from innate immune sensing as e.g. in HIV infections [15]. In spite of the important body of knowledge provided by these valuable studies, little is known on how cell cycle regulations and their perturbations are connected with the assembly and maturation of viruses.
To address this fundamental issue in a mammalian system, we investigated the effects of cell cycle regulations on the nuclear assembly of the parvovirus Minute Virus of Mice (MVM) infecting mouse and human fibroblasts. The members of the Parvoviridae have a ssDNA genome, are widely spread in nature [16,17] and their productive infection largely relies on host cellular factors [18,19]. Unlike small DNA tumor viruses, parvoviruses are unable to promote entry into the S phase, although their multiplication require basic factors of proliferative cells to convert the incoming ssDNA viral genome into a double-stranded DNA, which serves as template for transcription and genome replication [20]. The reaction of ssDNA convertion requires presumably DNA polymerase δ, the proliferating cell nuclear antigen (PCNA;[21]), plus other S phase-induced factors [22]. Another cell cycle dependent process of parvovirus infection is the upregulated transcription from the early promoter at the G1/S transition [23], which may contribute to onset the viral gene expression at early S phase [24]. As the infection progresses, most parvoviruses subvert the cell cycle eliciting a DDR as strategy to support viral replication [25–28], which arrest cells at the S or G2/M phases. Viral genome amplification, as well as hijacking cellular signaling and replication factors, require activities of the multifunctional non-structural (NS, Rep) parvoviral proteins [29,30]. However, it is unknown whether the cell cycle regulatory machinery, or the S-phase environment induced by the infection, controls parvovirus assembly and maturation.
Cell cycle dependence of viral life cycles is decisive for maturation and release of progeny virions. In parvoviruses, these factors have yet another importance as these viruses are currently being used in cancer and gene therapy trials [31,32], and as their pathogenesis is restricted to proliferative tissues [33]. Consequently the knowledge of the molecular mechanisms underlying productive parvovirus infection is required for determining target cells and efficient production of vectors for therapeutical applications. To this aim, mouse and human fibroblasts subjected to several growth arrests, were analyzed along cell cycle seeking control signals exerted on the nuclear translocation of parvovirus MVM structural subunits, capsid assembly, and virus maturation.
Results
Quiescent, G1, and G1/S arrested mammalian fibroblasts retain parvovirus capsid proteins in the cytoplasm
To investigate the regulation of MVM assembly by the cell cycle we first analyzed nuclear import of VP1 and VP2 capsid proteins (summarized as VPs) in the absence of other viral components. Pooled clones of transfected mouse or human fibroblasts stably expressing VPs (respectively named MF-VPs and HF-VPs) showed capsid proteins either predominantly cytoplasmic, or nuclear, or exhibited a homogenous stain by indirect immunofluorescence (IF) using the α-VPs antibody (Fig 1A, Async. panels), which mainly reacted with disassembled capsid subunits (see Materials and Methods), suggesting that VPs localization is affected by cell physiology. For further analysis, the transfected mammalian fibroblast lines were arrested by contact inhibition at high cell density (G1), or by isoleucine/aphidicolin (a DNA pol α inhibitor) double inhibition (G1/S), showing the VPs accumulated in the cytoplasm under both arrest conditions (Fig 1A). Such nuclear VPs exclusion was also observed in serum-starved quiescent MF-VPs (G0) (Fig 1B, 0 hps), collectively indicating that the nuclear translocation of the MVM capsid proteins is sensitive to different forms of cell growth arrests.
Fig. 1. Cell cycle regulation of the nuclear translocation of MVM capsid proteins. 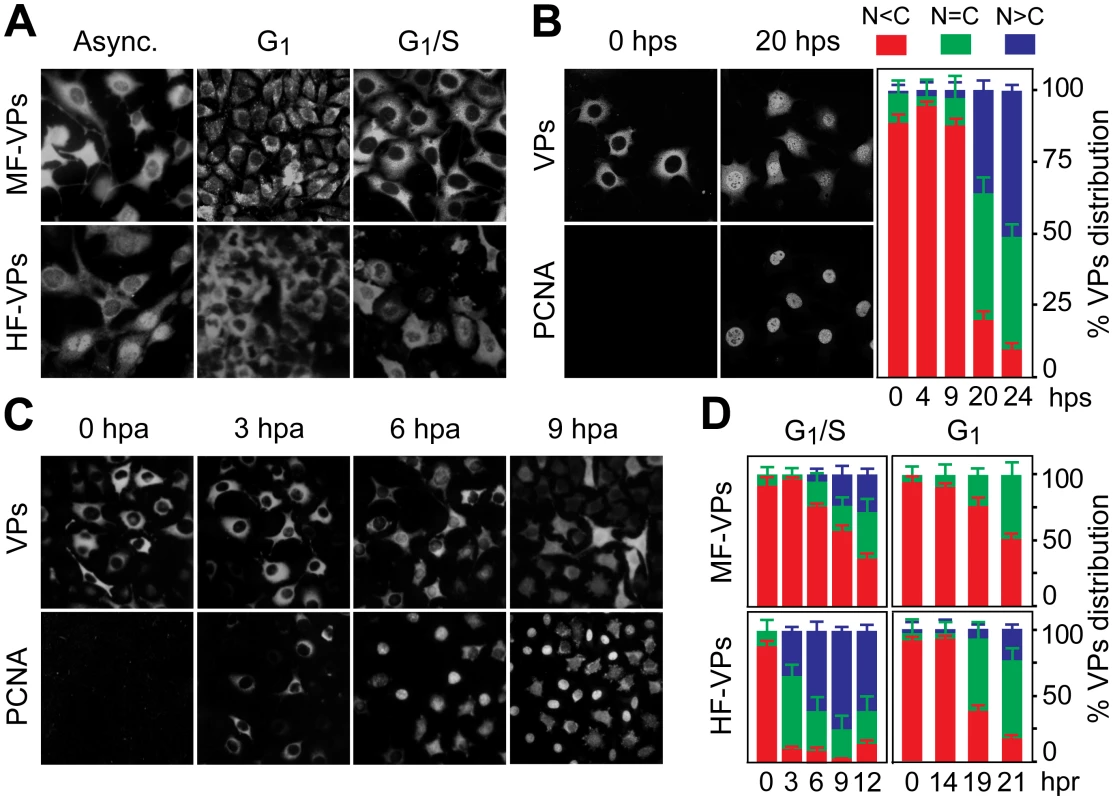
A. MVM capsid proteins (VPs) are excluded from the nucleus at G0/G1. Microscopy analysis of mouse (MF-VPs) and human (HF-VPs) fibroblasts stably expressing VPs fixed as asynchronous cultures (async.), synchronized by density arrest (G1), or by isoleucine deprivation/aphidicolin (G1/S). B. Kinetic of VPs nuclear transport in quiescent (G0) mouse fibroblast induced into cycle by serum. Left, cells stained with the α-VPs and PCNA antibodies. Right, average percentages with standard errors from three experiments of VPs subcellular distribution at the indicated hps. C. VPs nuclear transport is allowed at S phase. PCNA and VPs subcellular distribution in synchronous MF-VPs at the indicated hpa. D. VPs phenotypes scored in mouse and human fibroblasts hours post-release (hpr) of synchronization by aphidicolin (G1/S), or by density arrest (G1). Bars represent average values with standard errors from three or four experiments. (N<C (red): predominantly cytosolic, N = C (green): equal distribution between cytoplasm and nucleus, N>C (blue): predominantly nuclear). Nuclear transport of parvovirus capsid proteins proceeds at the S phase of the cell cycle
To determine the phase of the cell cycle in which VPs nuclear translocation occured, their subcellular distribution was studied in a kinetic after cells were released from the three G0/G1/S arrest methods. PCNA, a well-defined regulator of DNA replication and cell cycle control [34],[21]), was used as cell phase marker. Firstly, the G0 to G1 transition-as defined in mouse fibroblasts [35] - was applied to quiescent MF-VPs. The VPs remained in the cytoplasm all along the G1 phase, up to 9 hours post-serum addition (hps), with less than 5% of the cells exhibiting a predominantly nuclear stain (Fig 1B, quantification). As cells entered S phase a mixed cytoplasmic/nuclear VP phenotype became apparent. By 20 hps, when increased nuclear PCNA staining indicated S phase, nuclear VPs accumulation was observable in 35% of cells, progressing to >50% at 24 hps (Fig 1B, 20 hps and quantification). To study VPs nuclear transport across G1/S transition under more stringent synchronization, mouse and human fibroblasts were synchronized by the isoleucine/aphidicolin double block. All cells showed intense cytoplasmic VPs stain with nuclear exclusion even at 3 hours post-aphidicolin release (hpa), correlating to the cytoplasmic PCNA observed in most cells (Fig 1C). At the S phase onset, which was indicated by a significant nuclear PCNA accumulation (6 hpa), 20% of the cells exhibited a homogeneous VPs stain, followed by increased nuclear VPs translocation when the cell cycle progressed to S phase (Fig 1C and 1D upper left). Noteworthy, the VPs translocation into the nucleus began earlier upon aphidicolin release and was more pronounced in HF-VPs than in MF-VPs (Fig 1D, left). For final confirmation, we analyzed VPs localization in cells released from the G1 phase caused by density arrest. Upon 14 hours post-subculture at low density (hpc) favoring S phase entry, cells showed a slow though progressive increase in VPs nuclear accumulation (Fig 1D right). However, a predominant nuclear phenotype was not observed in MFs, and, as after the aphidicolin treatment, the VPs nuclear transport was more efficient in HF-VPs than in MF-VPs. Collectively these synchronization studies demonstrated that the nuclear transport of VPs proceeds at the S phase of the mammalian cell cycle, at rates lower in mouse than in human transformed fibroblasts.
S phase and the infection context govern parvovirus nuclear capsid assembly
To investigate if the cell cycle may also regulates viral capsid formation, MF-VPs and HF-VPs cultures, synchronized by the double isoleucine/aphidicolin block (G1/S) or growth to density arrest (G1). We used flow cytometry (Fig 2A and 2B) and confocal IF (Fig 2C) applying the α-VPs and α-Capsid antibodies. The specificity of this α-Capsid monoclonal antibody to recognize an epitope only conformed in the assembled MVM capsid was previously characterized by biochemical [36–38], genetic [39], and structural analyses [40]. The constitutive expression of viral structural proteins did not affect the timing of cell cycle progression from G1 to G2/M in MF-VPs and HF-VPs (cell cycle histograms in Fig 2A and 2B) as compared to untransfected controls (Fig 3A and 3B; see also Fig 4A and 4C). The VPs retained in the cytoplasm of the fibroblasts arrested at G0, G1/S or G1 phases failed to assemble into capsids (Fig 2). While VPs expression remained essentially stable throughout the cell cycle, capsid formation in cells released from the G1/S arrest started when their DNA content increased denoting entry into S phase (Fig 2A). Progression through S phase was more reliable in cells released from G1/S than from G1 arrests, and occurred faster in MFs than in HFs (Fig 2A and 2B). Capsids were exclusively formed in the nucleus of transfected fibroblasts (Fig 2C) with efficiencies strictly correlated to the rate of VPs nuclear transport shown above (Fig 1). Thereby the proportion of MF-VPs showing assembled capsid was low in G1/S (Fig 2A) and not significant in cells released from the G1 arrest (Fig 2B), as compared with the efficient capsid formation in HF-VPs (Fig 2). Cells forming capsids reached a height at late S phase (Fig 2A, 10 hpa panels), and declined as the cells underwent mitosis (14 hpa panels). We therefore concluded that MVM capsid formation is a nuclear S phase dependent process.
Fig. 2. Cell cycle dependence of MVM assembly. 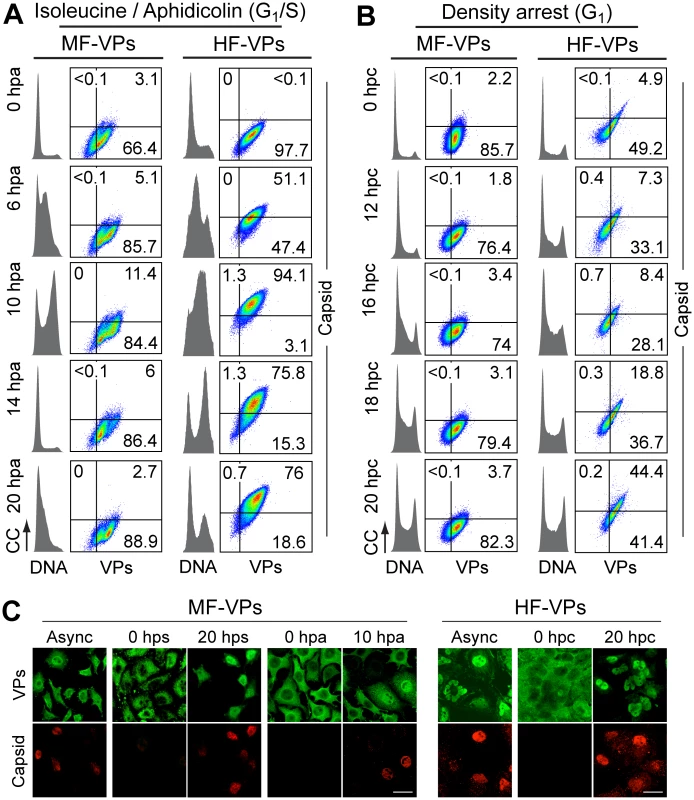
Capsid formation and cell cycle progression in VPs stably expressing mouse (MF) and human (HF) fibroblasts. A, B: analysis by flow cytometry. Cells were synchronized by (A) isoleucine deprivation+aphidicolin (G1/S), or by (B) culturing to high density (G1), and stained for DNA content with DAPI (grey histograms), VPs and Capsid (biparametric dot plots), at the indicated hours post-arrest release (hpa or hpc, respectively). CC, cell count. The percentage of cells in the corresponding gates is shown. (C) Confocal analysis of the subcellular distribution of VPs and Capsid in VPs-expressing synchronous mammalian fibroblasts at the indicated hours post-release of the arrests. Scale bar 25 μm. Representative results from more than four independent experiments of each type are illustrated. Fig. 3. Cell cycle dependence of VPs expression and capsid assembly in the synchronous MVM infection of mammalian fibroblasts. 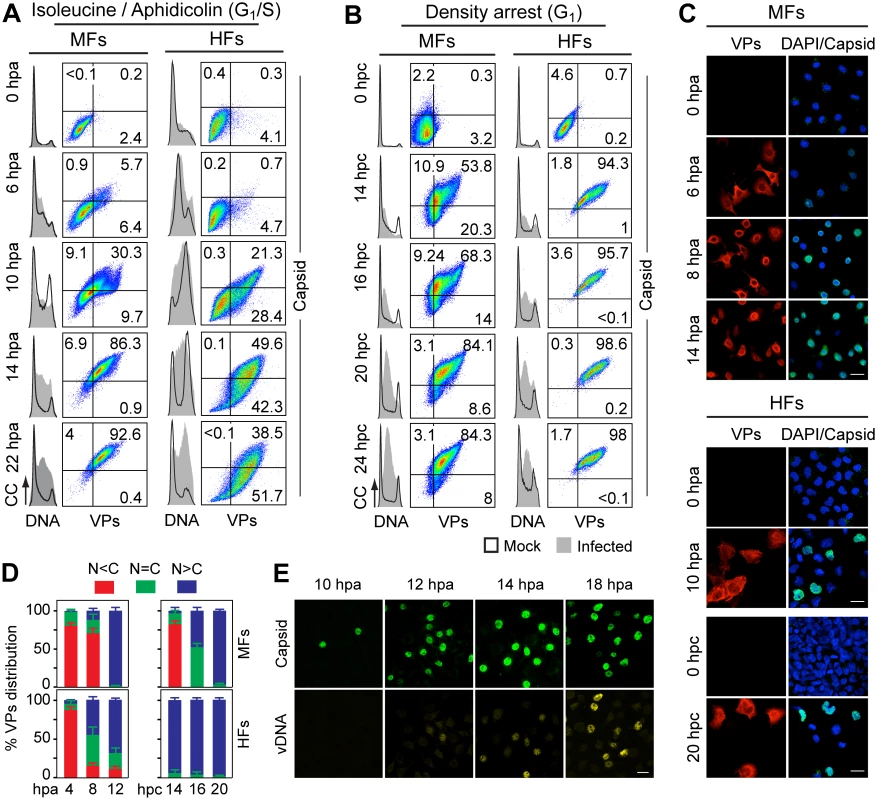
VPs expression and capsid formation in infected mouse (MFs) and human (HFs) fibroblasts analyzed by flow cytometry (A, B), or IF (C, D). Cells were synchronized by (A) isoleucine deprivation+aphidicolin (G1/S), or by (B) culturing to high density (G1), and stained for DNA content with DAPI (histograms: blank for mock, grey-filled for infected), VPs and Capsid (biparametric dot plots), at the indicated hours post-arrest releases (hpa or hpc, respectively). The percentage of cells in the corresponding gates is shown. CC, cell count. (C) Nuclear transport and assembly of MVM capsid subunits in synchronously infected mammalian fibroblasts. The figure shows representative fields of VPs and Capsid localized by double IF in MVM infected mouse and human fibroblats at the indicated hours post-release of the isoleucine/aphidicolin (G1/S) or growth to confluence (G1) cell cycle arrests. Scale bar 25 μm. (D) VPs subcellular distribution in synchronously infected mouse and human fibroblasts at the indicated hours post-release of the G1/S (hpa) or G1 (hpc) arrests. Average values with errors from four experiments are shown. (E) Synchronously infected MFs at G1/S stained for assembled capsids and viral DNA synthesis by FISH-hybridization (vDNA) along hpa. The figure shows representative confocal fields of cells from three experiments. Scale bar 10 μm. Fig. 4. Stressing cellular DNA synthesis uncouples parvovirus genome replication from nuclear capsid assembly. 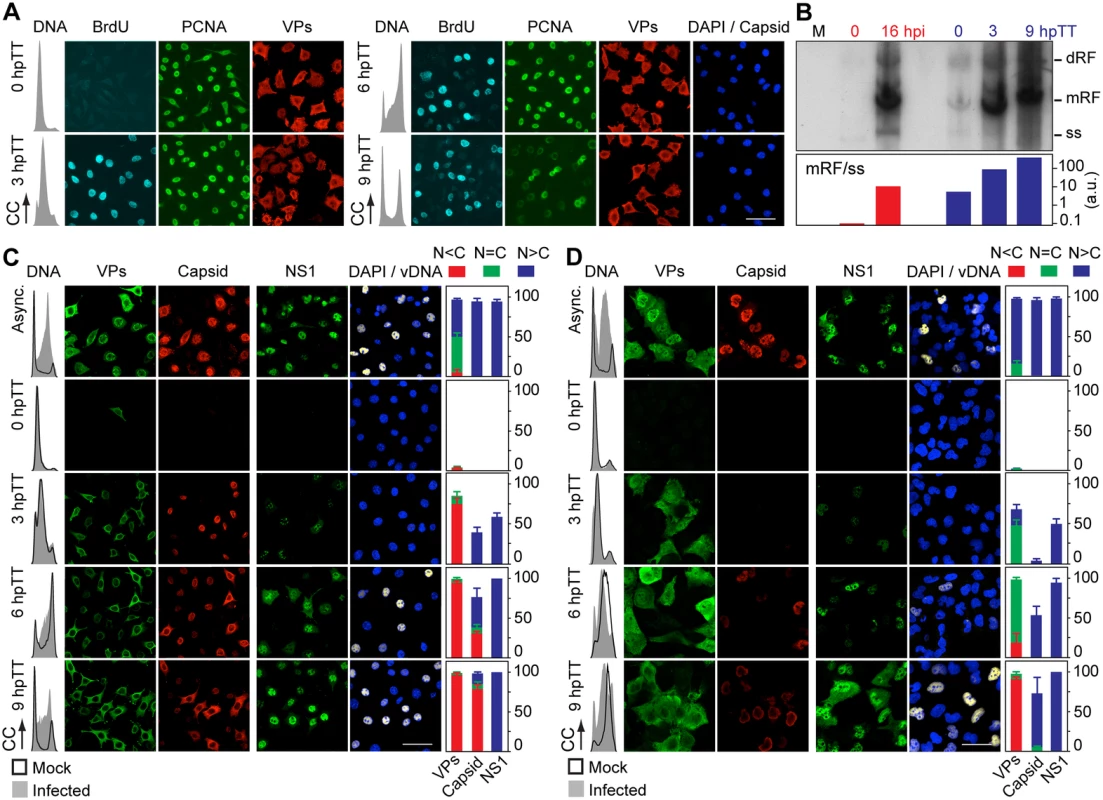
A. Cellular DNA synthesis dependence of VPs subcellular distribution and assembly in transfected MFs. The figure shows the subcellular distribution of BrdU incorporation, PCNA, VPs, and Capsid in MF-VPs analyzed by confocal microscopy at the indicated hours post-release of the double thymidine block (hpTT). Left: histograms of cell cycle analysis by flow cytometric determination of DNA content. B. Stressing cellular DNA synthesis hampers the packaging of parvovirus genome. Southern-blot analysis (performed with a full-lenght 32P-MVM probe as described [42]) of viral replication and maturation in growing (hpi) and thymidine synchronized (hpTT) infected MFs. Low molecular weight DNA isolated from 105, or from 5x105 (in 0 hpTT and 3 hpTT) cells, was loaded per lane. M, mock infection. mRF and dRF, monomeric and dimeric viral replicative intermediates, respectively; ss, single-stranded viral genomes. Below, mRF/ss ratio of synthesis measured by densitometry. a.u, arbitrary units. DNA synthesis stress hampers parvovirus capsid assembly in infected MFs (C) and HFs (D). The subcellular distribution of VPs, assembled capsid (Capsid), NS1, and viral replication analyzed by FISH-hybridization (vDNA), is shown by confocal microscopy in synchronously infected mouse and human fibroblasts at the indicated hpTT. Async: non-synchronous cultures. DNA (left columns): cell cycle phases at the respective time post-release analyzed by cytometry (histograms: blank for mock, grey-filled for infected). Right bars: percentages with standard errors of subcellular distribution of the viral antigens. Scale bar in IF is 50 μm. For verification of these findings in the context of a productive viral infection, both mammalian fibroblast cell lines synchronously infected by MVM at G1/S and G1 were studied by flow cytometry (Fig 3A and 3B), and IF as read-out (Fig 3C). Synthesis of the VPs was only detected upon release of the arrests (aphidicolin or cell density), even though the virus was inoculated several hours earlier (see Materials and Methods), showing the tight control of parvovirus gene expression in growth-arrested cells. VPs gene expression and capsid formation was first detected in MFs and HFs concomitantly with their entry in S phase as indicated by an incfreased DNA content (Fig 3A; MFs: 6 hpa, HFs: 10 hpa). In both infected fibroblast cell lines released from the two arrest methods, the number of VPs expressing cells forming capsids raised along S phase, reaching a height as the cell cycles became arrested by the infection prior G2 (see S1A and S1B Fig for quantitative cell cycle data). Importantly, an efficient VPs nuclear translocation occurred in most infected cells across S phase (Fig 3D), which was followed by an immediate assembly in nuclear capsids (Fig 3C). This high level of nuclear capsid formation in synchronously infected MFs (Fig 3) was in sharp contrast to the poor capacity observed under similar conditions in MF-VPs transfected cell (Fig 2). Of note, the VPs transport and nuclear capsid assembly were accomplished within early-mid S phase, several hours prior the bulk of viral DNA replication (Fig 3E), which occurred once infected cells have developed a pseudo-S phase or S/G2 arrest (Fig 3A and 3B, S1 Fig).
Cell cycle controls of parvovirus genome replication and capsid assembly are uncoupled
The need of S phase-related factor(s) affecting VPs nuclear transport led us to investigate if this process is supported by cellular DNA synthesis. MF-VPs were synchronized at G1/S by a double thymidine (a metabolic inhibitor) block (TT), and the VPs subcellular distribution was then analyzed during the DNA replication arrest and upon release. In arrested MF-VPs (Fig 4A, 0 hpTT panel), under cellular de novo DNA synthesis inhibition, which was shown by the flow cytometric analysis of DNA content and absence of BrdU staining, PCNA was accumulated in the nucleus as previously reported [41]. In contrast, the VPs showed mainly a cytoplasmic or mixed phenotype. When cells were released from the TT block and proceeded through S phase, which again was indicated by DNA content and synthesis activation (Fig 4A, 3 hpTT panel), nuclear PCNA stayed on at high levels, while the VPs remained mostly cytosolically. We thus conclude that although S phase is required, the activation of cellular DNA synthesis per se is not sufficient for causing VPs nuclear translocation. The cytoplasmic VPs phenotype became predominant as cells progressed through S (6 hpTT) and G2/M (9 hpTT), when PCNA staining declined and mitotic figures appeared (Fig 4A, right panels). In consistency with our findings shown in Fig 2, capsids also failed to assemble in transfected MFs under the same method of synchronization (Fig 4A, capsid panels). This experiment thus demonstrated that VPs nuclear transport operates by an S-phase dependent mechanism distinct to that of PCNA, and sensitive to the cellular DNA synthesis stress caused by thymidine.
We next investigated VPs translocation in the context of a TT-synchronous infection of mouse and human fibroblasts. Viral replication and gene expression were not observed during the DNA synthesis arrest (Fig 4C and 4D, 0 hpTT panels). Upon release of the TT block the number of VPs and NS1 expressing cells raised abruptly, in particular as cells entered and proceeded through S phase (Fig 4C and 4D, 3 hpTT panels). Noteworthy, as observed above (Fig 3E), the peak of viral DNA synthesis in both cell types occurred hours delayed with respect to VPs expression, reaching a height at 9 hpTT (Fig 4C and 4D, vDNA panels to the right). At this time the cycle of most cells showed a pseudo-S phase or drastic arrest at S/G2 caused by the MVM infection (Fig 4C and 4D, DNA content plots to the left; quantitative analyses in S1C Fig). At early time post-release most infected MFs exhibited a cytoplasmic/mixed VPs phenotype that correlated to a significant nuclear capsid staining (Fig 4C, 3 hpTT). At late S phase however the VPs proteins became increasingly retained in the cytoplasm, paralleling the phenotype observed in MF-VPs, and assembled in cytoplasmic capsids in a high proportion of cells (Fig 4C, 6 hpTT). Consistently this phenomenon gained robustness at 9 hpTT, as we observed a predominant cytoplasmic VPs phenotype and a highly efficient mislocalized capsid in most cells, under a severe S/G2 arrest (Fig 4C, lower panels, and S2 Fig). The cytoplasmic capsid assembly occurred at the time when cells were undergoing nuclear viral genome replication (Fig 4C, vDNA panels), which suggests that the virus is unable to encapsidate the genome and mature in TT synchronized cells. Accordingly, progeny single-stranded viral genomes were not resolved in MFs released from TT arrest under the accumulation of high levels of viral DNA replicative intermediates (Fig 4B)
Like in MFs, thymidine arrest of HFs irreversibly impaired VPs nuclear accumulation. At 3 hpTT we observed as a predominant mixed phenotype which was followed by increasing cytoplasmic retention when the infection proceeded to later hpTT (Fig 4D, left panels). Capsid formation in HFs started since 6 hpTT but become apparent in most cells at 9 hpTT. However, the level of assembled capsids was very low if compared to that of asynchronous infections (Fig 4D, Capsid panels). Unlike the cytoplasmic assembly found in MFs, capsid formation was confined within the nucleus of the HFs at 9hpTT (S2 Fig). We assume that the delayed capsid formation in HFs with respect to MFs may be caused by the slower progression across S phase in HFs (Fig 4C and 4D, 3–9 hpTT; and S1C Fig), whereas the distinct subcellular distribution of the capsid assembly process suggests the implication of cell-type specific factors. Noteworthy, the lasting impairment of VPs transport caused by TT in both mammalian fibroblasts did neither perturb NS1 nuclear accumulation nor viral DNA synthesis to normal levels (Fig 4C and 4D, right columns of panels). In summary these data demonstrate that nuclear capsid assembly and viral genome replication respectively require early and late S phase cellular factors. We further conclude that both steps are mediated by independent mechanisms, as only the nuclear translocation of capsid subunits was irreversibly hampered by a DNA synthesis stress.
Cell contact signals exerted on proliferating cells deregulate subcellular parvovirus assembly
We next asked whether VPs nuclear transport and capsid formation in infection is sensitive to other cell growth controls, as cellular contact inhibition, which is fundamental in the homeostasis of multicellular organisms. To mimic this physiological process in cell culture, infected MFs highly synchronized at G1/S by aphidicolin were first released from the arrest to allow cellular and viral cycles progressions (see above), and when cells had entered S phase at 6 hpa (Fig 3A, left column), an excess of growing uninfected homologous cells was added to impose general cell contacts (see scheme in S3A Fig). As shown in Fig 5A, at the end of the normal infection cycle in control cultures (22 hpa), the cells expressing VPs to higher levels were at pseudo-S phase or arrested at S/G2 (Fig 5A, right). In density arrest cultures (Fig 5A, 22 hpa/cells), while VP- cells were mostly asynchronous due to the contribution of the growing uninfected exogenous population, the high VP+ cells had also progressed to the S/G2 arrest, even though a significant proportion (close to 30%) remained at G1. Thus, contact inhibitions developed in infected cultures at S phase allowed high levels of viral gene expression, even in cells failing to progress through the cell cycle.
Fig. 5. Cell density arrest signals perturb and may misplace MVM assembly. 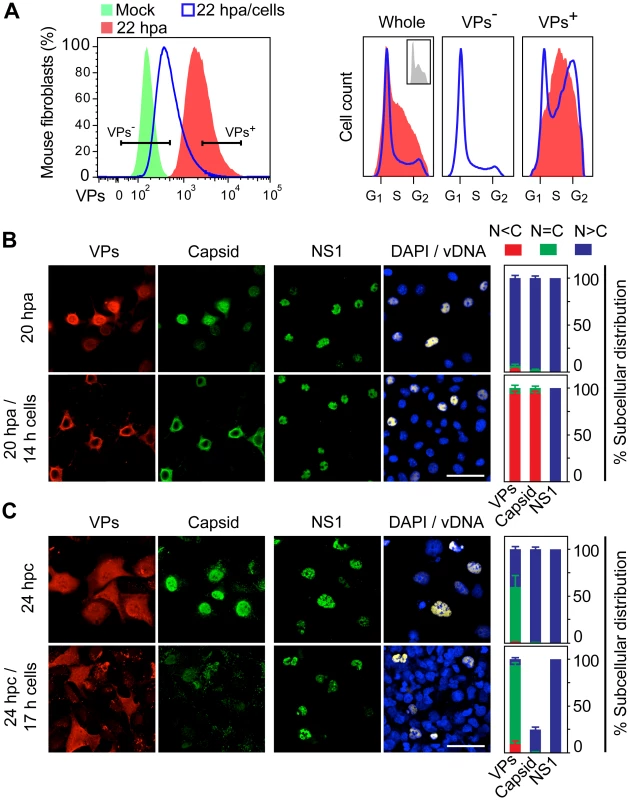
A. Flow cytometry of G1/S synchronously infected MFs sampled at 6 hpa, 22 hpa, or at 22 hpa but with density-arrest imposed since 6 hpa by uninfected exogenous cells (22 hpa/cells), analyzed for VP expression (left), and for cell cycle distribution (cell count vs. DNA content) of non-expressing (VP-) and capsid proteins expressing (VP+) cells (right). VP- cells were not detected in 22 hpa samples. Inset: cell cycle distribution at 6 hpa. B. Confocal analysis of MFs synchronously infected by MVM showing the subcellular localization of viral antigens (VPs, Capsid, NS1), and viral genome replication (vDNA), at 20 hpa without or upon the previous addition of a saturating number of cells at 6 hpa (20 hpa/14 h cells). C. Similar analysis performed in HFs synchronously infected by density arrest without (24 hpc), or upon exogenous cell addition at 7 hpc (24 hpc/17 h cells). Scale bars 50 μm. The percentage of subcellular distribution of the viral antigens within the respective infected cell populations (VP+ staining) is shown on the right. Confocal microscopy analysis under this experimental setting (Fig 5B) showed a remarkable cytoplasmic retention of the VPs in the infected MFs brought to confluence (20 hpa/14h cells). This was in contrast to their predominant nuclear accumulation in control infections maintained at low density (20 hpa). The impaired VPs nuclear transport in infected cultures brought to confluence led to an exclusive and efficient cytoplasmic capsid formation (Fig 5B), which also occurred at other time of density arrest (S3A Fig lower panels). A parallel analysis performed in HFs brought to confluence at 7 hpc (scheme in S3B Fig upper), also showed a significant cytoplasmic retention of VPs in confluent cultures, which resulted in poor nuclear capsid formation in a low proportion of the infected cells (Fig 5C, quantitative data shown to the right). Noteworthy, in both MFs and HFs, nuclear accumulation of NS1, which is the major replicative viral protein, was not impaired by the density contact signals, neither was the synthesis of viral genomes (Fig 5B and 5C, right columns; S3A and S3B Fig lower panels). We conclude that cell contact signals irreversibly deregulate MVM maturation by hampering (HFs) or misplacing (MFs) capsid assembly.
Cell cycle control of MVM assembly proceeds through oligomerization of phosphorylated VPs and is mediated by unconventional transport routes
To get insights into the molecular mechanisms involved in the cell-cycle dependent VPs nuclear translocation, we analyzed the nuclear transport of each capsid protein (VP1 and VP2) individually expressed from MVM genomic clones. VP2 nuclear translocation is driven by a so-called structured nuclear localization motif (NLM) [36], whereas the NLM of singly expressed VP1 subunits can be assisted for transport by conventional NLSs at its specific N-terminal sequence [36,42]. Transfected HFs seeded at high density to provoke growth arrest in G1 showed significant cytoplasmic retention of VP2 (Fig 6A and 6B, middle panels). In sharp contrast, VP1 was translocated into the nucleus efficiently and independently of cell growth conditions (Fig 6A and 6B, lower panels). When both proteins were co-expressed in growth arrested cells, the VPs were also significantly retained in the cytoplasm, which was not the case in growing cells (Fig 6A and 6B, upper panels). This observed difference was independent upon the MVM strain used to derive the plasmid mutants (see Materials and Methods). Since a functional NLM was required for the efficient transport of both VP subunits [42], the result shown in Fig 6A and 6B suggests that the NLM motif dominates over the NLSs of VP1, driving VP2 and VP1/VP2 complexes into the nucleus by a mechanism sensitive to density-arrest signals.
Fig. 6. Signals and phosphorylations regulating the nuclear transport of MVM trimeric capsid assembly intermediates. 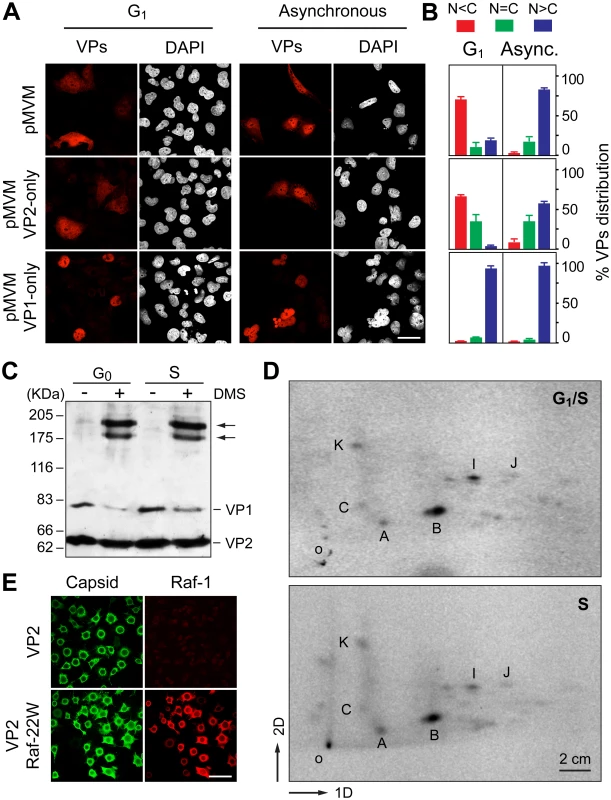
A. Cell cycle dependence of VP1 and VP2 nuclear accumulation. Confocal images of HFs seeded at high (G1 arrest) or low (asynchronous growth) density, stained with the α-VPs serum upon 48 h postransfection with the indicated genomic MVM plasmids. Scale bar, 25 μm. B. Phenotypes of the VPs subcellular distribution for the indicated plasmids 48 h postransfection in HFs seeded at high (G1 arrest) or low (asynchronous growth) density, stained with the α-VPs antibody based on four independent experiments. C. The VPs assemble into trimers in the cytoplasm of growth arrested mouse cells. Homogenates of MF-VPs made quiescent by serum starvation (G0), or upon 8 h post-stimulation into cycle by serum addition (S), were cross-linked with DMS and analyzed by 5% SDS-PAGE and Western-blot (α-VPs antibody). Arrows indicate the two types of VP trimers [37]. D. Pattern of VP2 phosphorylation in the G1 to S transition. 2D-tryptic phosphopetides analysis of the VP2 protein purified from HF-VPs, 32P-labeled either during isoleucine/aphidicolin arrest (G1/S), or 10 h upon arrest release (S). Phosphopeptides were monitored by phosphoimaging and are named as previously reported [43]. 1D, 2D: first and second dimensions; o, origin. E. Raf-1 phosphorylated VP2 fails to translocate into the nucleus of insect cells. H5 monolayers infected by the Bac-VP2wt alone (upper panels), or co-infecting with the Bac-Raf1-22W (lower panels) baculovirus, stained at 24 hpi with the α-capsid and α-Raf1 antibodies. Scale bar 50 μm. The role of VPs oligomerization and post-translational modifications during their S-phase dependent transport was next evaluated. In quiescent and S-phase induced MF-VPs, the VPs could be cross-linked with similar efficiency into oligomers (Fig 6C). They correspond in their size to the two types of trimers (2VP2/1VP1, and 3VP2) which were found as the major capsid assembly intermediates in the infection [37]. Both types of trimers also assembled in HF-VPs (S4 Fig) at times post-release of G1 during which VPs predominantly localized in the cytoplasm (Fig 1D). These results indicate that the cell cycle arrest did not inhibit the cytoplasmic formation of VPs trimers, which is required for their nuclear transport [37]. We next studied the phosphorylation pattern of the VP2 subunits, which is the major component of the capsid assembly intermediates, at distinct cell cycle phases. Phosphorylation at Ser residues by the Raf-1 kinase of the MAPK pathway was shown to be required for VP2 nuclear import [38]. VP2 proteins isolated from the cytoplasm of HF-VPs arrested by aphidicolin at G1/S, or from the nucleus at 10 h post-release (S phase), showed an identical 2D tryptic phosphopeptides map (Fig 6D). This pattern was similar to that found in the MVM infection [43] and VP2 phosphorylation by Raf-1 in vitro [38]. These data suggested that cytoplasmic phosphorylation of VP2 by Raf-1 is required, but may not be sufficient, to explain the S-phase dependent nuclear import of the VPs trimers.
We finally addressed the import pathway used by the VP2 trimer (t). In a heterologous insect system, VP2 assembled into capsid but failed to enter the nucleus alone or upon co-expressed with a constitutively active form of Raf-1 (Fig 6E), even though the VP2(t) isolated from insect cells under these experimental conditions was phosphorylated by Raf-1, and this post-translational modification sufficed VP2(t) for nuclear transport in human cells [38]. This experiment thus suggested that, unlike multiple nucleocytoplasmic transport routes [44], [45], the cellular machinery transporting VP2(t) into the nucleus is not evolutionary conserved. To study the transport pathway relevant for the MVM infection of mammalian cells, phosphorylated transport competent VP2(t) purified from HFs transfected at large scale, was used as substrate for an in vitro transport assay in digitonin-permeabilized HeLa cells. We analyzed the need of nuclear import receptors of the karyopherin (importin) α/β system binding classical NLSs [46,47], and of the transportin (Kap β2) translocating M9 domain-exposing proteins (as representative of the PY-NLS sequences) [48,49]. VP2(t) were efficiently translocated into the nucleus when a rabbit reticulocyte lysate (RRL) was used as source of transport factors (Fig 7, left column). The import of VP2(t) into the nucleus though could not be blocked by tenfold molar excess of either BSA-NLS or BSA-M9 marker conjugates (Fig 7, second and third columns). Consistently, a fifty fold molar excess of a NLS peptide failed to compete VP2(t) import, while the import of BSA-NLS was inhibited (Fig 7, fourth and fifth columns). In agreement with these observations, a replacement of the cytosolic RRL extract by Ran-GTP and purified α2 and β1 importins failed to translocate the VP2(t), despite of an active nuclear transport of the BSA-NLS cargo using the same conditions (Fig 7, right column). These results thus indicate that the phosphorylated VP2(t) access a route(s) distinct from both the importins α2/β1 and the transportin pathways for its cell cycle dependent nuclear import.
Fig. 7. Importins α/2β1 and transportin are not involved in VP2 nuclear import. 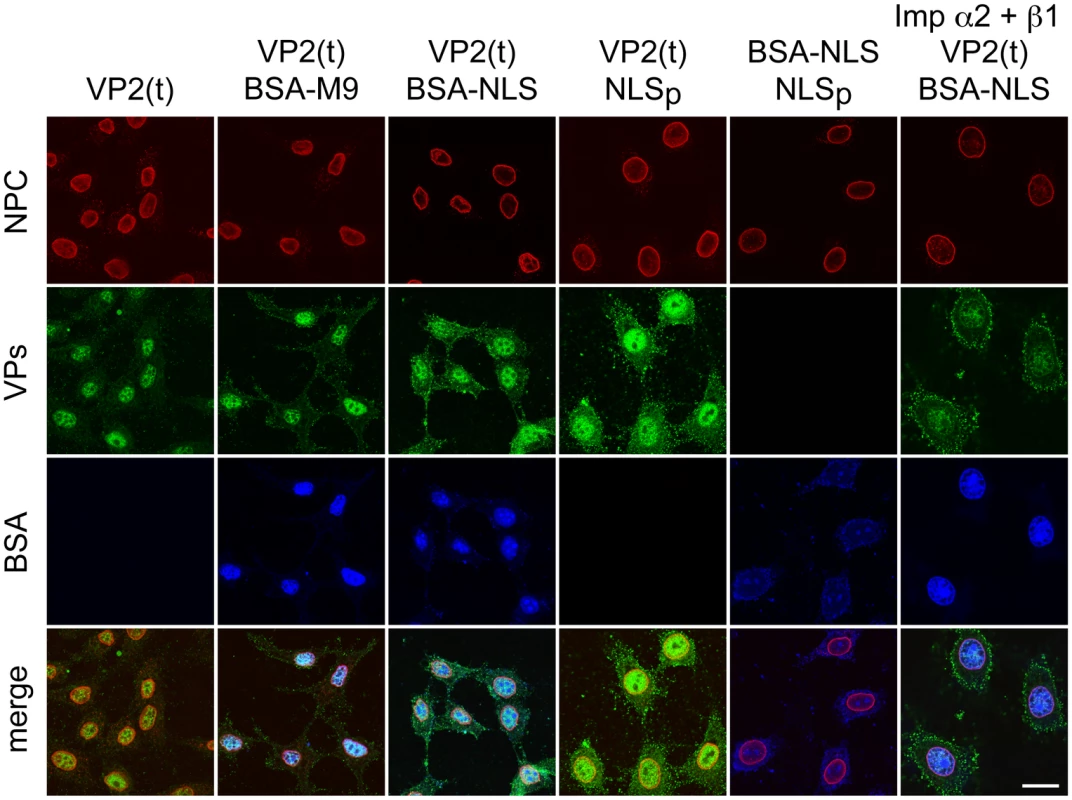
Transport of VP2-trimer (t) analyzed by confocal microscopy in Digitonine-permeabilized HeLa cells, in combination with competitor markers. The transport phenotype of 200 ng of gradient-purified VP2(t) in the presence of 10 μg of BSA conjugated to peptides of characterized import routes (BSA-NLS and BSA-M9), or of 3.5 μM of NLS peptide alone (NLSp), are shown. Transport factors were provided by rabbit reticulocyte lysates (first five columns) or by purified importins (right columns). Representative results from three experiments are shown. Scale bar 10 μm. Discussion
Cell cycle regulated nuclear transport and assembly of parvovirus capsid subunits
Our study shows an unprecedented cell cycle regulation of viral nuclear assembly. The capsid proteins of the parvovirus MVM were excluded from the nucleus of mammalian fibroblasts as quiescent (G0), arrested at G1, or at G1/S boundary, but they were translocated into the nucleus upon release of the arrests, at efficiency depended upon the inhibitor and cell type (Fig 1). VPs nuclear import was not directly stimulated by the numerous serum responsive genes immediately induced by growth factors at the G1/S restriction point [35,50], since it only proceeded as transfected and infected fibroblasts traversed through S phase (Figs 1–3). Putative cellular factors induced at S phase were sufficient to provoke high VPs nuclear transport in HFs (Figs 1 and 2), but failed to do so in MFs, the latter requiring MVM infection (Figs 1–3). A major factor contributing to the different efficiency of transport between both transfected fibroblast cells may be the level of VPs phosphorylation, a modification essential for efficient VP2 nuclear targeting catalyzed by the Raf-1 kinase, which is part of the MAPK signaling cascade ([38]; see also below). In agreement with this assumption, the endogenous Raf-1 kinase activity was found to be much lower in MFs than in HFs [38].
The VPs translocated into the nucleus of the mammalian fibroblasts immediately assembled into capsids at S phase. Accordingly, capsid formation efficiently occurred in HF-VPs traversing S phase, whereas MF-VPs showed a limited capacity (Fig 2). The efficient capsid forming capacity of infected MFs (Fig 3) suggests that expressed viral factor(s) contribute to VPs nuclear transport. This activity may well be mediated by the NS2 protein, which was dispensable for the MVM infection of human transformed fibroblasts but required for MVM capsid assembly in MFs ([51] and references therein). In the synchronous infection therefore, gene expression, VPs nuclear translocation, and capsid formation, occurred at early-mid S phase, several hours prior the accumulation of viral DNA replicative intermediates at the late pseudo-S phase arrest (Fig 3). As illustrated in Fig 8A, this timely orchestrated virus life cycle across S phase implies that gene expression results from the input ssDNA genomes converted to dsDNA templates or from basal levels of replicated viral genomes. A timely dissociated use of templates for transcription and for DNA amplification is commonly found in virus life cycles, and may have evolved also to maximize genome encapsidation into preformed capsids.
Fig. 8. Temporal course of functional and disrupted cell cycle control on parvovirus assembly. 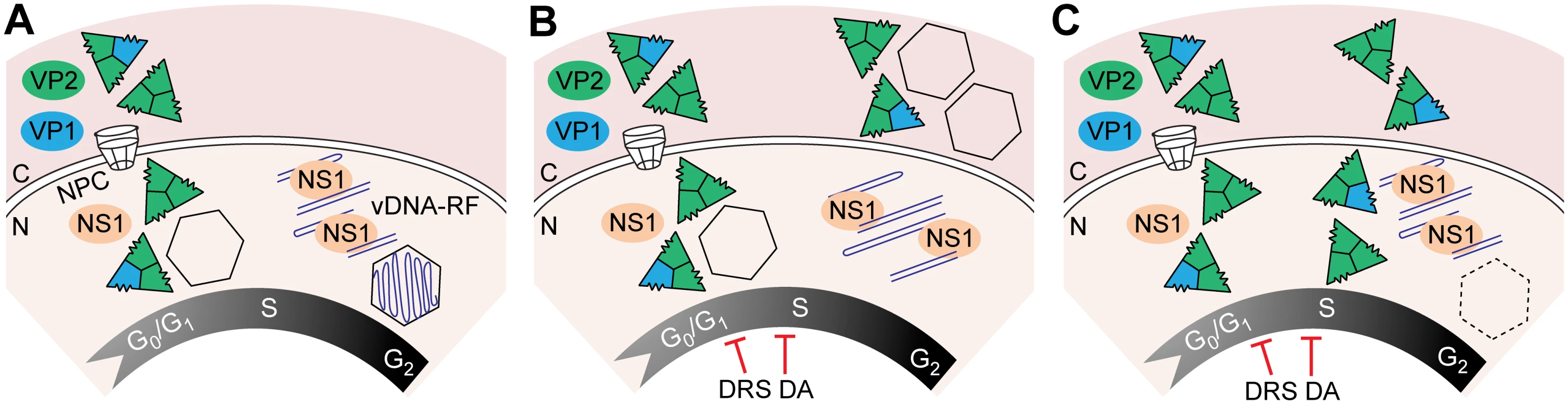
A. Parvovirus assembly and maturation is timely coupled to cell cycle progression from G1 to G2. Phosphorylated VPs trimers translocate into the nucleus at early S phase driven by the NLM, and assemble into empty capsids. Viral genome replication (vDNA-RF) and virions maturation occur as cells become arrested at the late S/G2 phases by the infection. Infected cells subjected to thymidine DNA replication stress (DRS), or density arrest by cell contacts (DA), interrupt drastically VPs nuclear transport, resulting into cytoplasmic empty capsids formation in MFs (B), or inefficient nuclear assembly in HFs represented by dotted capsid (C). Acronyms: C, cytoplasm; N, nucleus; NPC, nuclear pore complex. VPs nuclear transport and cellular DNA synthesis
The cellular DNA synthesis was identified as one major S-phase process contributing to the control of VPs nuclear transport. The VPs retained in the cytoplasm by the G1/S aphidicolin arrest translocated into the nucleus of transfected fibroblasts at S phase concomitantly with PCNA translocation (Fig 1). However, inhibition of DNA synthesis by thymidine hampered VPs transport although allowing efficient PCNA nuclear accumulation (Fig 4A), as did other cellular DNA synthesis stresses [41]. The differential transport requirements of VPs and PCNA may reflect their specificity of interacting partner proteins. PCNA nuclear accumulation was regulated by export signals [52] and a non-classical NLS segment directly binding importin-β [53], whereas classical NLSs and importin-β were not used by the VPs (Figs 6 and 7). Moreover, restored cellular DNA synthesis upon thymidine release inhibited VPs nuclear transport across S-G2 phases of infected mammalian fibroblasts (Fig 4C and 4D). This was in sharp contrast with the unrestricted transport at early-mid S phase of infected cells released from the aphidicolin arrest (Fig 3). The basis of the distinct sensitivity of VPs transport to the DNA pol α inhibition by aphidicolin or the metabolic inhibition by thymidine remains unclear. Putative mechanisms could involve a decay of transient transport factors provoked by the altered S phase progression that follows thymidine inhibition [54]. The availability of transient factors for viral VPs transport would also be more restricted upon thymidine than aphidicolin arrests because the substantial shortening of the S phase length in TT-treated cells (compare Figs 4C and 4D vs 3A, and also S1C vs S1A Fig). Even though the involved mechanisms remain unknown, the subcellular distribution of viral antigens and the levels of viral DNA amplification shown in Fig 4B–4D demonstrate that VPs nuclear transport is most sensitive to cellular DNA synthesis stresses. This implies that this step is the bottleneck of parvovirus life cycle, which may largely influence the biology of these viruses in nature.
Perturbing cell cycle progression inhibits or misplaces parvovirus capsid assembly
The cell cycle regulations also determined the subcellular compartment of parvovirus capsid assembly. While transfected and infected cells normally form MVM capsids in the nucleus (Figs 2 and 3), infected fibroblasts subjected to cell cycle perturbations—here exemplified by a DNA replication stress caused by thymidine (Fig 4), and by exogenous density-contact signals (Fig 5)—abruptly impaired capsid assembly. In HFs these cycle perturbations inhibited assembly, although allowed low levels of nuclear capsid formation both at 6–9 hpTT (Fig 4D and S2 Fig) and in density-arrested cells (Fig 5C and S3B Fig). In MFs some nuclear capsid formed only at 3 hpTT (Fig 4C). The different hpTT at which capsids preferently assembled in HFs and MFs may relate to their characteristic timings of G1–G2 cell cycle progression (Fig 4C and 4D, S1C Fig), which would affect the availability of early-mid S phase transient transport factors (suggested above). However both cell cycle perturbations eventually led in infected MFs to a remarkable misplaced cytoplasmic capsid assembly (Figs 4C and 5B; S2 and S3A Figs), reflecting the capacity of these mouse cells to form MVM capsids regardless the subcellular compartment of VPs accumulation. Moreover, none of the assayed conditions inhibiting or misplacing capsid assembly altered other key cell cycle dependent viral functions, like high levels of gene expression, genome replication, or NS1 nuclear accumulation (Figs 4B–4D and 5, S3 Fig). The Fig 8 illustrates the two types of virus assembly defects observed in fibroblasts subjected to cell cycle perturbations (DNA synthesis stress and density-arrest): either accumulation of empty capsids in the cytoplasm of MFs (Fig 8B), or poor nuclear capsid formation in HFs (Fig 8C). In natural infections, an inhibited or misplaced capsid assembly may critically hamper parvovirus maturation and spreading in cells with stressed DNA replication conditions or dense cell-to-cell contacts, thereby restricting their pathogenesis, therapeutic efficacy, or favoring persistence in host tissues.
Signals and mechanisms controlling the cell cyle regulated VPs nuclear transport
The nuclear transport of VP1 and VP2 capsid subunits showed different sensitivity to cell cycle controls. VP1 transport was not hampered by cell-density arrest signals (Fig 6A and 6B, bottom panels), suggesting that the NLS and other entry functions on its N-terminal domain [42,55,56] still allow the delivery of the incoming viral genome into the nucleus of quiescent and growth arrested cells. This is consistent with classical reports on the capacity of parvoviruses to infect resting and proliferating cells with similar efficiencies [57–59]. However, VP2 and VP2/VP1 complexes transport responded to cellular growth arrest signals (Fig 6A and 6B), which is based on cytoplasmic trimeric assembly intermediates (Fig 6C and S4 Fig). This supports that the 3VP2 and 2VP2/1VP1 trimeric assembly intermediates of MVM [37] follow a transport pathway sensitive to signals regulated by the cell cycle, and which are distinct from that of VP1. A nonconventional cell cycle-dependent nuclear import route accessed by the two types of VPs trimers is in agreement with previous findings on a structured NLM motif driving the process [36], substantially distinct from the disordered conventional NLSs [46,47,49], which interact with evolutionary conserved transport receptors of the importin/karyopherin family [60]. Consistently, permeabilized cells showed nuclear import of VP2 trimers independent of importin (NLS) and transportin (M9) (Fig 7), and the transport route accessed by the Raf-1 phosphorylated VP2 trimer in growing human cells [38] was non-conserved in insects (Fig 6E), unlike the common evolutionary conservation of multiple conventional nucleocytoplasmic transport mechanisms and karyopherins functions across the Eukaryota [44,45]. The NLM configuration at the internal capsid surface [36] would explain that capsids remain in the cytoplasm of infected mouse fibroblasts when assembly is misregulated (Figs 4C, 5B and 8B, S2 Fig). Whether this restriction on VPs nuclear transport and capsid assembly plays a role in parvovirus (and other nuclear viruses) host range deserves further investigations.
A plausible molecular explanation of the cell cycle-dependent parvovirus assembly may rely on the finding that VP2 nuclear translocation required specific phosphorylation by the Raf-1 kinase [38] of the MAPK cell cycle regulatory pathway [61]. Signaling in density arrested cells also involves permanent or pulses of ERK activation [62], which is phosphorylated by MEK in the cytoplasm, a main target of Raf-1 [61]. Thus, VP2 phosphorylation and oligomerization into nuclear transport competent trimers would enable the MVM assembly process to be coupled to subtle signals promoting cell cycle progression and proliferation. However VPs trimers containing VP2 subunits specifically phosphorylated by Raf-1 accumulated in the cytoplasm of growth arrested human fibroblasts (Fig 6D), suggesting that other modifications affecting the configuration of the trimers, or the concourse of an S phase-dependent transport machinery, may be required for VPs nuclear import. Interestingly, other Raf-1 interacting partners harbouring regulatory properties on the cell cycle also fail to immediately translocate into the nucleus ensuing phosphorylation. For example, the physical interaction of Raf-1 to the retinoblastoma tumor suppressor protein (Rb) at early G1 regulates its function and initiates a phosphorylation cascade required for cell cycle progression through the G1/S boundary [63,64], suggesting that further research on factors mediating parvovirus assembly may unravel major regulators of the mammalian cell cycle.
Materials and Methods
Mammalian cells
The A9 ouabr11 mouse fibroblasts, a variant of the mouse L cells (kindly provided by Jean Rommelaere, DKFZ, Heidelberg), and the NB324K simian virus 40–transformed human newborn kidney fibroblasts (kindly provided by Bernard Hirt, Epalinges, Switzerland) cell lines described as hosts for the MVM strains [43,65,66], as their respective derived transfected pools of clones (HF-VPs and MF-VPs, see below), were maintained under minimal number of passages in Dulbecco modified Eagle medium (DMEM) supplemented with 5% heat-inactivated fetal calf serum (FCS; Gibco). The Hela human cell line (ATCC: CCL2) was used in transport assays.
Cellular synchronizations
The following synchronization methods were used for mouse and/or human fibroblasts as indicated. In all cases synchronization and cycle progression were controlled by DNA content in flow cytometry. G0, quiescence: mouse fibroblasts (A9 cell line and MF-VPs) were seeded (1.5x105 cells/P60 mm dish) in DMEM-5% FCS for 16 h, washed several times with PBS, and then incubated in DMEM-0.1% FCS for 60–72 h. Cells were induced into cycle replacing the medium for fresh DMEM supplemented with 20% FCS [35]. G1, density arrest: mouse and human fibroblasts were seeded to a density of 5x105 cells/P60 dish in DMEM-5% FCS. The medium was replaced daily to avoid acidification and after 72 h the cultures had reached confluence and arrested in G1 [67]. Cells were then set into cycle by sub-culturing seeding at low density (usually 2x105 cells/P60 mm dish) in DMEM-5%FCS. G1-S, aphidicolin: mouse fibroblasts were para-synchronized either culturing in low serum (as above), or in isoleucine-free DMEM [68] supplemented with 5% dialyzed FCS. After 48 h, the medium was replaced with complete DMEM-5% FCS containing 5 μg/ml of the DNA polymerase-α antagonist aphidicolin ([69]; Sigma), and incubated 16 h prior release. The isoleucine/aphidicolin double block procedure was also used to highly synchronize NB324K human fibroblasts. G1/S, thymidine: a double thymidine block [70] was used to synchronize mouse fibroblasts. Cells seeded (2.5x105 cells/P60 mm dish) in DMEM-5%FCS for 24h were incubated for 16h in fresh medium supplemented with 2 mM thymidine, a nucleoside that competes for thymine incorporation into DNA. The medium was removed and replaced by normal medium without thymidine for 9h, followed by a second 16h treatment in fresh medium with 2 mM thymidine. Cells were released from the G1/S blocks by removing the medium with the inhibitor, washing with PBS several times and adding fresh pre-warmed DMEM-5% FCS.
Flow cytometric analysis
For viral antigens determination, cells were fixed for 24h in 70% ethanol at 4°C, permeabilized (PBS, 0.1% Triton X-100) for 20 min, and blocked (PBS, 0.1% Triton X-100, 1% FCS) for 10 min. Primary and secondary antibodies (mentioned below) were sequentially incubated for 1 h at 37°C in blocking solution. For DNA content determination, fixed and permeabilized cells were stained for 1 h in PBS, 0.1% Nonidet P-40, and 3 μM DAPI. All samples were analyzed in a BD Biosciences FACSCanto II Flow Cytometer equipped with 405, 488 and 633 nm laser lines. The BD FACSDiva (v6.1.2, BD Biosciences) and FlowJo (v9.3 for mac and v7.5 for PC, TreeStar) softwares were respectively used for sample acquisition and data analysis. The Cell Cycle Platform of FlowJo (v7.5) with Watson Pragmatic algorithm were used for quantitative analyses.
Virus and synchronous infections
The prototype strain of the parvovirus Minute Virus of Mice (MVMp; [22]), referred simply as MVM for this study, was grown in NB324K cells and titrated as plaque forming units (pfu) as previously described [71]. Virus stocks were prepared from infections at low multiplicity, purification by density gradients free of empty capsids, and stored at −70°C [43,72]. For synchronous infections, the purified virus was added at a multiplicity of infection (moi) of 10 pfu/cell, in the media and timings accordingly to the synchronization methods, as follows. G1 arrest at confluence: the virus was inoculated in DMEM-0.1% FCS and, upon 1h adsorption, the arrest was maintained in DMEM-5% FCS at least 14 h prior sub-culturing. G1/S, aphidicolin: in cultures para-synchronized by low serum (mouse fibroblasts only), or isoleucine deprivation (mouse and human fibroblasts, the virus was inoculated in normal or isoleucine-deprived DMEM, respectively. After 1h adsorption, the inoculum was removed and cells were further highly synchronized at G1/S by culturing for 16 h in 5 μg/ml aphidicolin in DMEM-5%FCS. G1/S, thymidine: when synchronizing by the double thymidine block (TT), the 1 h virus adsorption was performed in DMEM-0.1% FCS supplemented with 2mM thymidine in the middle of the second block (8 h post-second thymidine addition), and afterwards the thymidine treatment was maintained 7 h further in fresh medium. Time of virus cycle in the figures is referred to either hours post-inoculation (hpi), post-serum addition (hps), post-subculturing (hpc), post-aphidicolin release (hpa), post-thymidine blocks (hpTT), or general post-release of the inhibitor (hpr).
Infectious plasmids
The respective infectious molecular clones of the prototype (MVMp; [73]) and the immunosuppressive (MVMi, [74]) viral strains have been previously described [65,75]. Our MVM-derived genomic clones allowing single expression of each of the structural proteins (VP1/ΔVP2 (or VP1-only), and ΔVP1/VP2 (or VP2-only) plasmids), were constructed for MVMp by cDNA cloning [72], and for MVMi by mutating the minor splicing donor site and the VP1 specific open reading frame [42]. Genomic plasmids were transformed in the Escherichia coli JC8111 strain that permits deletion-resistant propagation of plasmid clones bearing terminal parvovirus palindromes [76].
Stable expressions of MVM capsid proteins
Fibroblasts permanently expressing the capsid proteins (VPs) of MVMp were derived from the A9 and NB324K cell lines. Clones were obtained co-transfecting (at molar ratio 1 : 5) the selection plasmid pSV2neo (carrying the bacterial aminoglycoside phosphotransferase gene that confers resistance to neomycin analogous) with the pSVtk-VPs plasmid, which contains the coding regions of the VP1 and VP2 proteins of MVMp cloned under the control of the SV40-thymidine kinase enhancer-promoter [77]. Mouse cells were transfected by a conventional calcium phosphate precipitate method, whereas the human cells were electroporated as described [36,72]. Cells were maintained at low density in medium containing the selective antimetabolite geneticin (G418, Gibco) at 400 μg/ml active product for 2–4 weeks to allow the growth of resistant clones. Twenty pick-isolated clones were grown from each transfected cell line and screened for the expression of both VP1 and VP2 structural proteins by indirect IF and western-blot with the α-VPs serum (see below). Homogenates from several VPs-expressing clones allowed the purification of MVM empty capsids, as described for other parvovirus VP-expressing systems (e.g. [78–82], which showed hemagglutinating activity with mouse erythrocytes [71]. Eight transfected clones were selected and pooled from each cell type on the basis of their normal growth and morphology, viability in colony forming units assay [71], and VPs expression to levels fairly detectable by blot at the VP1:VP2 ratio of 1 : 5 found in the natural MVM infection [79,83]. The mouse and human pools of clones (respectively named MF-VPs and HF-VPs) showed stable VPs expression over the passages.
Analysis of VPs expression
To determine VPs synthesis by immunoprecipitation (IP), cells were 35S-Met/Cys pulse-labeled for 20 min with 100 μCi/ml of (35S) methionine-cysteine (Pro-mixTM, Amersham) in methionine-free DMEM-10% dialyzed FCS supplemented with 5% normal medium. Cells were washed twice with cold PBS, scraped in 150 mM NaCl, 50 mM Tris pH 8.0, 1% NP40, 0.3% SDS, 0.5% β-mercaptoethanol, and incubated overnight at 4 oC with a 1/100 dilution of the α-VPs serum. Immune complexes were precipitated with protein A-Sepharose (10% wt/vol), washed with cold PBS, 0.05% NP40, and 1% BSA, and bound proteins subjected to 10% SDS-PAGE. Gels were fixed, dried and exposed for autoradiography to Kodak X - films. To determine VPs accumulation by western-blot, proteins resolved in polyacrilamide gels (10%SDS-PAGE) were transferred onto nitrocellulose membranes (Scheider & Schull) by a wet transfer (Trans blot Electrophoretic-transfer Cell, Biorad) for 1 h at 100 V, and developed with the α-VPs serum by a chemiluminescence method (ECL, Amersham).
2D phosphopeptides resolution
Cells were labeled with 2 mCi/ml of 32P-orthophosphate in phosphate-free medium as described [43], and the VPs proteins were isolated from cellular extracts by IP with the α-VPs serum, resolved by 8%SDS-PAGE, and blotted to membranes. The purified 32P-labelled VP2 protein was cut-off from the membranes and subjected to 2-D tryptic phosphopeptide analysis by digestion with N-tosyl-L-phenylalanine chloromethyl ketone (TPCK)-trypsin (sequencing grade, Boehringer) and resolution in 20x20 cm TLC plates (Merck, Darmstadt, Germany) following our reported methods [38,43]. Plates were exposed to a radioanalytic imaging system (Fujix BAS 1000, Fuji) for ten days.
Cross-linking of capsid proteins
Infected or transfected cells washed in PBS were centrifuged and resuspended in 50 mM Hepes pH 8.0, 150 mM NaCl, 2 mM MgCl2, 1 mM EDTA (HNEM buffer), and proteases (0.1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml leupeptin, 10 μg/ml pepstatin, 10 μg/ml aprotinin, 100 μg/ml TPCK) and phosphatases (20 mM glicerol phosphate, 5 mM NaF) inhibitors. Cells were lysed by ultrasounds at 4°C and the homogenates were clarified by centrifugation at 10000 g, 30 min at 4°C. Wherever indicated purified capsids were used as controls. The dimethyl suberimidate imidoester (DMS; Sigma) cross-linker, freshly dissolved in 50 mM in Hepes-NaOH pH 8.0, was added at 10 mM to the prewarmed samples, and cross-linking allowed to proceed for 10 min at 20°C as reported earlier [37]. The reactions were finished by adding ethanolamine to a final concentration of 0.36 M.
Antibodies
To cellular proteins we used: a rabbit polyclonal (α-Raf, Santa Cruz Biotechnology) recognizing the conserved C-terminus of human Raf-1; a mouse monoclonal antibody (α-NPC; mAb414, BAbCO, Richmond, CA) as marker of the nuclear envelope; and the α-PCNA and α-BrdU antibodies mentioned below. Two anti-MVM antibodies were used in this study: for the general recognition of both capsid proteins independently of the configuration of structured epitopes, an antiserum (α-VPs) was raised in rabbit against denatured VP2, which coding region is common to VP1 [42,79,83]. For this purpose, purified virus like particles (VLP) self-assembled by VP2 expression in a baculovirus system [81] were subjected to preparative 8%SDS-PAGE. A small vertical strip was stained, the region of the gel corresponding to the VP2 protein was excised, and the protein was electro-eluted into chambers running at 24 V overnight in 100 mM glycine-20 mM Tris base-0.01% SDS as previously described [84]. The purified denatured VP2 was directly used to raise an antibody in rabbit by injection (100 μg per dose) a first dose emulsified in Freund´s complete adjuvant followed by three boost injections in incomplete adjuvant. Bleeding was done ten days after the last injection. This rabbit α-VPs antibody was probed to recognized efficiently VP1 and VP2 when denatured in western-blots [72], whereas failed to react with purified MVM capsids by immune-precipitation or electron immune-gold staining. Intact capsids and virions were recognized by a mouse anti-MVM monoclonal antibody (α-Mab, B7), which failed to react with isolated VP subunits or trimers [37,38], in agreement with the available genetic [39] and 3-D structural data on its specific recognition of a capsid epitope [40]. The NS1 non-structural protein was recognized by a polyclonal antibody raised in rabbit against a recombinant β-gal-NS1 fusion protein expressed in bacteria [85].
Immunofluorescence
For subcellular localization of proteins by double-label indirect immunofluorescence (IF), cells seeded onto glass coverslips in dishes (Nunc) were washed twice with PBS and fixed for 8 min in 4% paraformaldehyde (PFA) at room temperature, or in Methanol:Acetone 1 : 1 at -20°C. Cells were permeabilized with 0.1% Triton X-100 in PBS and, after blocking in PBS, 0.1% Triton X-100, 1% FCS, the primary antibodies were applied diluted 1 : 50 to 1 : 500 in blocking buffer for 1 h at 37 oC. The bound IgG were visualized with a secondary goat anti-rabbit antibody conjugated to Texas red (TXRD) and with a goat anti-mouse antibody conjugated to fluorescein isothiocyanate (FITC) (Jackson Immuno Research Laboratories, Inc.; used at 1/500). The simultaneous antibody staining and FISH - hybridization for the analysis of MVM replication and gene expression was performed as reported elsewhere (Gil-Ranedo et al., in preparation). For in situ monitoring cellular DNA replication, cells cultured on coverslips were incubated with 20 μg/ml bromodeoxiuridine (BrdU, Sigma) for a 15 min - pulse, washed in PBS, and immediately fixed in methanol for 20 min at 4°C. The DNA was partially denaturized in 2N HCl for 1h at 37°C, and then neutralized washing three times in borate buffer (pH 8.5) and PBS. Samples were analyzed by IF with an anti-BrdU specific antibody (Boehringer). The Proliferating Cell Nuclear Antigen (PCNA), a cofactor of the DNA polymerase δ [86,87], was used as marker of S phase (PC10 antibody, Santa Cruz biotechnology). Samples were mounted in DAPI Fluoromont-G (Southern Biotech) and left to dry overnight. Phenotypes were scored by epifluorescence using a Zeiss Axiovert2000 inverted microscope coupled to a SPOT RT Slider digital camera and MetaVue 5.07 software, and images were taken in a Zeiss LSM 710 Laser Scanning confocal microscope and ZEN 2008 software. The images captured by 1.5 μm optical sections were processed using Fiji package. For each antigen, the scored cells were classified in three categories: mainly nuclear (N>C), comparable nuclear and cytoplasmic (N = C), and mainly cytoplasmic (N<C). The percentages of N/C subcellular distribution shown in the figures were obtained from at least 400 IF-stained cells accumulated from three to four consistent and independent experiments.
VP2 and Raf-1 expression by baculoviruses
The recombinant baculoviruses of the Autographa californica (AcMNPV) species, expressing the wt VP2 of MVMp [81], or the Raf-22W constitutively active mutant form of the Raf-1 kinase [88], were grown in Sf9 cells and titrated as fluorescence focus (IFU). For expression of the recombinant proteins, H5 insect cells cultured at 27°C in TC100 medium supplemented with 5% FCS were infected at a multiplicity of 10 IFU/cell for each baculovirus and fixed in 4% PFA at 48–72 hpi following previously described methods [38].
Nuclear transport of purified VP2 trimer in permeabilized cells
A previously described methodology [38] was followed with minor modifications. In brief, trimers of the VP2 protein (VP2t) were purified by continuous sucrose gradients from NB324K cells transfected with the pVP2-K153A plasmid at large scale as recently optimized [72]. A total amount of 200 ng of VP2t dialyzed against transport buffer (2 mM Mg-acetate, 20 mM Hepes pH 7.3, 110 mM K-acetate, 1 mM EDTA, 5 mM Na-acetate, 2 mM DTT) was used as substrate per reaction. HeLa cells permeabilized with 40 μg/ml Digitonin (Sigma) in DMEM for 5 min at 37°C were subjected to the transport reaction for 15 min at 37°C including 12 mg/ml rabbit reticulocyte lysate (RRL; Promega), protease inhibitors and energy sources (1 mM ATP, 5 mM creatine phosphate (Sigma), and 20 U/ml of creatine phosphokinase (Calbiochem). In some experiments the RRL was substituted by the purified α2 (0.15 μM) and β1 (0.15 μM) importins (Calbiochem), and Ran-GDP (2 μM). Transport controls were BSA conjugated with either the SV-40TAg NLS (PKKKRKVED) of the α2/β1 importin route, or the M9 domain (YNNQSSNFGPMK) of the transportin route. Peptides were conjugated to BSA coupled with fluor agents (Alexa Fluor 488, 594, and 647; Molecular Probes) by the chemical cross-linker SMCC (Calbiochem), cleaned by chromatography in PD10 columns (Amersham), and concentrated by centrifugation through filters (3 kDa Microcon, Millipore). For control inhibition of the nuclear import, an excess of free NLS peptide (3.5 μM), or 100 μg/ml of wheat germ agglutinin (WGA, Calbiochem), were added. The transport reaction was terminated by fixation in 4% PFA.
Ethics statement
Animal experimental protocols were approved by the Ethical Committees of the Centro de Biología Molecular Severo Ochoa (CSIC-UAM) and Universidad Autónoma de Madrid for animal health research (permit number CEI 27–652) in strict accordance with Spanish national Royal Decree (RD 1201/2005) and international EU guidelines 2010/63/UE about protection of animals used for experimentation and other scientific purposes, and Spanish national law 32/2007 about animal welfare in their exploitation, transport and sacrifice.
Supporting Information
Zdroje
1. Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D (2013) The translational landscape of the mammalian cell cycle. Mol Cell 52 : 574–582. doi: 10.1016/j.molcel.2013.09.018 24120665
2. Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, et al. (2001) Transcriptional regulation and function during the human cell cycle. Nat Genet 27 : 48–54. 11137997
3. Mukherji M, Bell R, Supekova L, Wang Y, Orth AP, et al. (2006) Genome-wide functional analysis of human cell-cycle regulators. Proc Natl Acad Sci U S A 103 : 14819–14824. 17001007
4. Weis K (2003) Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112 : 441–451. 12600309
5. Makhnevych T, Lusk CP, Anderson AM, Aitchison JD, Wozniak RW (2003) Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell 115 : 813–823. 14697200
6. Gorlich D, Prehn S, Laskey RA, Hartmann E (1994) Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79 : 767–778. 8001116
7. Stade K, Ford CS, Guthrie C, Weis K (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90 : 1041–1050. 9323132
8. Imperiale MJ, Major EO (2007) Polyomaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Fifth Edition ed. Philadelphia, PA: Lippincott Williams & Wilkins. pp. 2263–2298.
9. Hebner CM, Laimins LA (2006) Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev Med Virol 16 : 83–97. 16287204
10. Flemington EK (2001) Herpesvirus lytic replication and the cell cycle: arresting new developments. J Virol 75 : 4475–4481. 11312317
11. Paladino P, Marcon E, Greenblatt J, Frappier L (2014) Identification of herpesvirus proteins that contribute to G1/S arrest. J Virol 88 : 4480–4492. doi: 10.1128/JVI.00059-14 24501404
12. Chaurushiya MS, Weitzman MD (2009) Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst) 8 : 1166–1176. doi: 10.1016/j.dnarep.2009.04.016 19473887
13. Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461 : 1071–1078. doi: 10.1038/nature08467 19847258
14. Davy C, Doorbar J (2007) G2/M cell cycle arrest in the life cycle of viruses. Virology 368 : 219–226. 17675127
15. Laguette N, Bregnard C, Hue P, Basbous J, Yatim A, et al. (2014) Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156 : 134–145. doi: 10.1016/j.cell.2013.12.011 24412650
16. Tijssen P, Agbandje-McKenna M, Almendral JM, Bergoin M, Flegel TW, et al. (2011) Family Parvoviridae. In: King AMQ, Adams JM, Carsterns EB, Lefkowitz EJ, editors. ICTV Report 2011. Oxford, U.K.: Elsevier. pp. 405–425.
17. Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, et al. (2013) The family Parvoviridae. Arch Virol 159 : 1239–1247. doi: 10.1007/s00705-013-1914-1 24212889
18. Berns N, Parrish CR (2007) Parvoviridae. Parvoviridae. 5th ed. Philadelphia, PA: Lippincott Willians and Wilkins.
19. Weitzman MD (2006) The parvovirus life cycle: an introduction to molecular interactions important for infection. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. London, U.K.: Hodder Arnold. pp. 143–156.
20. Cotmore SF, Tattersall P (1987) The autonomously replicating parvoviruses of vertebrates. Adv Virus Res 33 : 91–174. 3296697
21. Moldovan GL, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129 : 665–679. 17512402
22. Bashir T, Horlein R, Rommelaere J, Willwand K (2000) Cyclin A activates the DNA polymerase delta-dependent elongation machinery in vitro: A parvovirus DNA replication model. Proc Natl Acad Sci U S A 97 : 5522–5527. 10792046
23. Deleu L, Fuks F, Spitkovsky D, Horlein R, Faisst S, et al. (1998) Opposite transcriptional effects of cyclic AMP-responsive elements in confluent or p27KIP-overexpressing cells versus serum-starved or growing cells. Mol Cell Biol 18 : 409–419. 9418888
24. Deleu L, Pujol A, Faisst S, Rommelaere J (1999) Activation of promoter P4 of the autonomous parvovirus minute virus of mice at early S phase is required for productive infection. J Virol 73 : 3877–3885. 10196282
25. Raj K, Ogston P, Beard P (2001) Virus-mediated killing of cells that lack p53 activity. Nature 412 : 914–917. 11528480
26. Luo Y, Kleiboeker S, Deng X, Qiu J (2013) Human parvovirus B19 infection causes cell cycle arrest of human erythroid progenitors at late S phase that favors viral DNA replication. J Virol 87 : 12766–12775. doi: 10.1128/JVI.02333-13 24049177
27. Adeyemi RO, Landry S, Davis ME, Weitzman MD, Pintel DJ (2010) Parvovirus minute virus of mice induces a DNA damage response that facilitates viral replication. PLoS Pathog 6: e1001141. doi: 10.1371/journal.ppat.1001141 20949077
28. Adeyemi RO, Fuller MS, Pintel DJ (2014) Efficient parvovirus replication requires CRL4Cdt2-targeted depletion of p21 to prevent its inhibitory interaction with PCNA. PLoS Pathog 10: e1004055. doi: 10.1371/journal.ppat.1004055 24699724
29. Cotmore SF, Tattersall P (1992) In vivo resolution of circular plasmids containing concatemer junction fragments from minute virus of mice DNA and their subsequent replication as linear molecules. J Virol 66 : 420–431. 1530771
30. Berthet C, Raj K, Saudan P, Beard P (2005) How adeno-associated virus Rep78 protein arrests cells completely in S phase. Proc Natl Acad Sci U S A 102 : 13634–13639. 16157891
31. Geletneky K, Huesing J, Rommelaere J, Schlehofer JR, Leuchs B, et al. (2012) Phase I/IIa study of intratumoral/intracerebral or intravenous/intracerebral administration of Parvovirus H-1 (ParvOryx) in patients with progressive primary or recurrent glioblastoma multiforme: ParvOryx01 protocol. BMC Cancer 12 : 99. doi: 10.1186/1471-2407-12-99 22436661
32. Dismuke DJ, Tenenbaum L, Samulski RJ (2013) Biosafety of recombinant adeno-associated virus vectors. Curr Gene Ther 13 : 434–452. 24195602
33. Siegel G (1988) Patterns of parvovirus disease in animals. In: Pattison JR, editor. Parvoviruses and Human Disease. Boca Raton, FL: CRC Press.
34. Maga G, Hubscher U (2003) Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci 116 : 3051–3060. 12829735
35. Almendral JM, Sommer D, Macdonald-Bravo H, Burckhardt J, Perera J, et al. (1988) Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol 8 : 2140–2148. 2898731
36. Lombardo E, Ramirez JC, Agbandje-McKenna M, Almendral JM (2000) A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J Virol 74 : 3804–3814. 10729155
37. Riolobos L, Reguera J, Mateu MG, Almendral JM (2006) Nuclear transport of trimeric assembly intermediates exerts a morphogenetic control on the icosahedral parvovirus capsid. J Mol Biol 357 : 1026–1038. 16469332
38. Riolobos L, Valle N, Hernando E, Maroto B, Kann M, et al. (2010) Viral oncolysis that targets Raf-1 signaling control of nuclear transport. J Virol 84 : 2090–2099. doi: 10.1128/JVI.01550-09 19939915
39. Lopez-Bueno A, Mateu MG, Almendral JM (2003) High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host. J Virol 77 : 2701–2708. 12552010
40. Kaufmann B, Lopez-Bueno A, Mateu MG, Chipman PR, Nelson CD, et al. (2007) Minute virus of mice, a parvovirus, in complex with the Fab fragment of a neutralizing monoclonal antibody. J Virol 81 : 9851–9858. 17626084
41. Bravo R, Macdonald-Bravo H (1985) Changes in the nuclear distribution of cyclin (PCNA) but not its synthesis depend on DNA replication. EMBO J 4 : 655–661. 2861088
42. Lombardo E, Ramirez JC, Garcia J, Almendral JM (2002) Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J Virol 76 : 7049–7059. 12072505
43. Maroto B, Ramirez JC, Almendral JM (2000) Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J Virol 74 : 10892–10902. 11069983
44. O'Reilly AJ, Dacks JB, Field MC (2011) Evolution of the karyopherin-beta family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PLoS One 6: e19308. doi: 10.1371/journal.pone.0019308 21556326
45. Enenkel C, Blobel G, Rexach M (1995) Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem 270 : 16499–16502. 7622450
46. Kalderon D, Roberts BL, Richardson WD, Smith AE (1984) A short amino acid sequence able to specify nuclear location. Cell 39 : 499–509. 6096007
47. Robbins J, Dilworth SM, Laskey RA, Dingwall C (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64 : 615–623. 1991323
48. Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, et al. (1996) A novel receptor-mediated nuclear protein import pathway. Cell 86 : 985–994. 8808633
49. Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, et al. (2006) Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126 : 543–558. 16901787
50. Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, et al. (1999) The transcriptional program in the response of human fibroblasts to serum. Science 283 : 83–87. 9872747
51. Cotmore SF, D'Abramo AM Jr., Carbonell LF, Bratton J, Tattersall P (1997) The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology 231 : 267–280. 9168889
52. Bouayad D, Pederzoli-Ribeil M, Mocek J, Candalh C, Arlet JB, et al. (2012) Nuclear-to-cytoplasmic relocalization of the proliferating cell nuclear antigen (PCNA) during differentiation involves a chromosome region maintenance 1 (CRM1)-dependent export and is a prerequisite for PCNA antiapoptotic activity in mature neutrophils. J Biol Chem 287 : 33812–33825. 22846997
53. Kim BJ, Lee H (2008) Lys-110 is essential for targeting PCNA to replication and repair foci, and the K110A mutant activates apoptosis. Biol Cell 100 : 675–686. doi: 10.1042/BC20070158 18498247
54. Churchill JR, Studzinski GP (1970) Thymidine as synchronizing agent. 3. Persistence of cell cycle patterns of phosphatase activities and elevation of nuclease activity during inhibition of DNA synthesis. J Cell Physiol 75 : 297–303. 5449696
55. Zadori Z, Szelei J, Lacoste MC, Li Y, Gariepy S, et al. (2001) A viral phospholipase A2 is required for parvovirus infectivity. Dev Cell 1 : 291–302. 11702787
56. Porwal M, Cohen S, Snoussi K, Popa-Wagner R, Anderson F, et al. (2013) Parvoviruses cause nuclear envelope breakdown by activating key enzymes of mitosis. PLoS Pathog 9: e1003671. doi: 10.1371/journal.ppat.1003671 24204256
57. Tattersall P (1972) Replication of the parvovirus MVM. I. Dependence of virus multiplication and plaque formation on cell growth. J Virol 10 : 586–590. 4673484
58. Rhode SL 3rd (1973) Replication process of the parvovirus H-1. I. Kinetics in a parasynchronous cell system. J Virol 11 : 856–861. 4736535
59. Siegl G, Gautschi M (1973) The multiplication of parvovirus Lu3 in a synchronized culture system. II. Biochemical characteristics of virus replication. Arch Gesamte Virusforsch 40 : 119–127. 4266337
60. Mattaj IW, Englmeier L (1998) Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem 67 : 265–306. 9759490
61. Qi M, Elion EA (2005) MAP kinase pathways. J Cell Sci 118 : 3569–3572. 16105880
62. Aoki K, Kumagai Y, Sakurai A, Komatsu N, Fujita Y, et al. (2013) Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol Cell 52 : 529–540. doi: 10.1016/j.molcel.2013.09.015 24140422
63. Wang S, Ghosh RN, Chellappan SP (1998) Raf-1 physically interacts with Rb and regulates its function: a link between mitogenic signaling and cell cycle regulation. Mol Cell Biol 18 : 7487–7498. 9819434
64. Dasgupta P, Sun J, Wang S, Fusaro G, Betts V, et al. (2004) Disruption of the Rb—Raf-1 interaction inhibits tumor growth and angiogenesis. Mol Cell Biol 24 : 9527–9541. 15485920
65. Gardiner EM, Tattersall P (1988) Mapping of the fibrotropic and lymphotropic host range determinants of the parvovirus minute virus of mice. J Virol 62 : 2605–2613. 3392768
66. Tattersall P, Bratton J (1983) Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J Virol 46 : 944–955. 6602222
67. Baserga R (1978) Resting cells and the G1 phase of the cell cycle. J Cell Physiol 95 : 377–382. 649675
68. Tobey RA (1973) Production and characterization of mammalian cells reversibly arrested in G1 by growth in isoleucine-deficient medium. Methods Cell Biol 6 : 67–112. 4585084
69. Huberman JA (1981) New views of the biochemistry of eucaryotic DNA replication revealed by aphidicolin, an unusual inhibitor of DNA polymerase alpha. Cell 23 : 647–648. 6784928
70. Bootsma D, Budke L, Vos O (1964) Studies on Synchronous Division of Tissue Culture Cells Initiated by Excess Thymidine. Exp Cell Res 33 : 301–309. 14109144
71. Rubio MP, Lopez-Bueno A, Almendral JM (2005) Virulent variants emerging in mice infected with the apathogenic prototype strain of the parvovirus minute virus of mice exhibit a capsid with low avidity for a primary receptor. J Virol 79 : 11280–11290. 16103180
72. Sanchez-Martinez C, Grueso E, Carroll M, Rommelaere J, Almendral JM (2012) Essential role of the unordered VP2 n-terminal domain of the parvovirus MVM capsid in nuclear assembly and endosomal enlargement of the virion fivefold channel for cell entry. Virology 432 : 45–56. doi: 10.1016/j.virol.2012.05.025 22727830
73. Crawford LV (1966) A minute virus of mice. Virology 29 : 605–612. 5945715
74. Bonnard GD, Manders EK, Campbell DA Jr., Herberman RB, Collins MJ Jr. (1976) Immunosuppressive activity of a subline of the mouse EL-4 lymphoma. Evidence for minute virus of mice causing the inhibition. J Exp Med 143 : 187–205. 1244418
75. Merchlinsky MJ, Tattersall PJ, Leary JJ, Cotmore SF, Gardiner EM, et al. (1983) Construction of an infectious molecular clone of the autonomous parvovirus minute virus of mice. J Virol 47 : 227–232. 6345805
76. Boissy R, Astell CR (1985) An Escherichia coli recBCsbcBrecF host permits the deletion-resistant propagation of plasmid clones containing the 5'-terminal palindrome of minute virus of mice. Gene 35 : 179–185. 3896934
77. Ramirez JC, Santaren JF, Almendral JM (1995) Transcriptional inhibition of the parvovirus minute virus of mice by constitutive expression of an antisense RNA targeted against the NS-1 transactivator protein. Virology 206 : 57–68. 7831812
78. Clemens DL, Wolfinbarger JB, Mori S, Berry BD, Hayes SF, et al. (1992) Expression of Aleutian mink disease parvovirus capsid proteins by a recombinant vaccinia virus: self-assembly of capsid proteins into particles. J Virol 66 : 3077–3085. 1313919
79. Clemens KE, Cerutis DR, Burger LR, Yang CQ, Pintel DJ (1990) Cloning of minute virus of mice cDNAs and preliminary analysis of individual viral proteins expressed in murine cells. J Virol 64 : 3967–3973. 2164605
80. Christensen J, Storgaard T, Bloch B, Alexandersen S, Aasted B (1993) Expression of Aleutian mink disease parvovirus proteins in a baculovirus vector system. J Virol 67 : 229–238. 8380073
81. Hernando E, Llamas-Saiz AL, Foces-Foces C, McKenna R, Portman I, et al. (2000) Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles. Virology 267 : 299–309. 10662625
82. Kajigaya S, Fujii H, Field A, Anderson S, Rosenfeld S, et al. (1991) Self-assembled B19 parvovirus capsids, produced in a baculovirus system, are antigenically and immunogenically similar to native virions. Proc Natl Acad Sci U S A 88 : 4646–4650. 1711206
83. Tattersall P, Cawte PJ, Shatkin AJ, Ward DC (1976) Three structural polypeptides coded for by minite virus of mice, a parvovirus. J Virol 20 : 273–289. 988192
84. Knowles WJ, Bologna ML (1983) Isolation of the chemical domains of human erythrocyte spectrin. Methods Enzymol 96 : 305–313. 6656634
85. Rubio MP, Guerra S, Almendral JM (2001) Genome replication and postencapsidation functions mapping to the nonstructural gene restrict the host range of a murine parvovirus in human cells. J Virol 75 : 11573–11582. 11689639
86. Almendral JM, Huebsch D, Blundell PA, Macdonald-Bravo H, Bravo R (1987) Cloning and sequence of the human nuclear protein cyclin: homology with DNA-binding proteins. Proc Natl Acad Sci U S A 84 : 1575–1579. 2882507
87. Bravo R, Frank R, Blundell PA, Macdonald-Bravo H (1987) Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature 326 : 515–517. 2882423
88. Williams NG, Paradis H, Agarwal S, Charest DL, Pelech SL, et al. (1993) Raf-1 and p21v-ras cooperate in the activation of mitogen-activated protein kinase. Proc Natl Acad Sci U S A 90 : 5772–5776. 8390681
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage TraffickingČlánek An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived NeuronsČlánek Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPsČlánek Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African TrypanosomiasisČlánek Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin ReleaseČlánek HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T CellsČlánek Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 6- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Introducing “Research Matters”
- Exploring Host–Pathogen Interactions through Biological Control
- Analysis of Bottlenecks in Experimental Models of Infection
- Expected and Unexpected Features of the Newly Discovered Bat Influenza A-like Viruses
- Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage Trafficking
- Recombinant Murine Gamma Herpesvirus 68 Carrying KSHV G Protein-Coupled Receptor Induces Angiogenic Lesions in Mice
- TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING
- Targeting Human Transmission Biology for Malaria Elimination
- Two Cdc2 Kinase Genes with Distinct Functions in Vegetative and Infectious Hyphae in
- An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived Neurons
- Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPs
- Virulence Factors of Induce Both the Unfolded Protein and Integrated Stress Responses in Airway Epithelial Cells
- Peptide-MHC-I from Endogenous Antigen Outnumber Those from Exogenous Antigen, Irrespective of APC Phenotype or Activation
- Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African Trypanosomiasis
- An Ultrasensitive Mechanism Regulates Influenza Virus-Induced Inflammation
- The Role of Human Transportation Networks in Mediating the Genetic Structure of Seasonal Influenza in the United States
- Host Delivery of Favorite Meals for Intracellular Pathogens
- Complement-Opsonized HIV-1 Overcomes Restriction in Dendritic Cells
- Inter-Seasonal Influenza is Characterized by Extended Virus Transmission and Persistence
- A Critical Role for CLSP2 in the Modulation of Antifungal Immune Response in Mosquitoes
- Twilight, a Novel Circadian-Regulated Gene, Integrates Phototropism with Nutrient and Redox Homeostasis during Fungal Development
- Surface-Associated Lipoproteins Link Virulence to Colitogenic Activity in IL-10-Deficient Mice Independent of Their Expression Levels
- Latent Membrane Protein LMP2A Impairs Recognition of EBV-Infected Cells by CD8+ T Cells
- Bank Vole Prion Protein As an Apparently Universal Substrate for RT-QuIC-Based Detection and Discrimination of Prion Strains
- Neuronal Subtype and Satellite Cell Tropism Are Determinants of Varicella-Zoster Virus Virulence in Human Dorsal Root Ganglia Xenografts
- Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2'-Fluoro-4'-Chloromethyl-Cytidine Triphosphate
- Structure of the Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome
- Activated Brain Endothelial Cells Cross-Present Malaria Antigen
- Fungal Morphology, Iron Homeostasis, and Lipid Metabolism Regulated by a GATA Transcription Factor in
- Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin Release
- A Macrophage Subversion Factor Is Shared by Intracellular and Extracellular Pathogens
- A Novel AT-Rich DNA Recognition Mechanism for Bacterial Xenogeneic Silencer MvaT
- Reovirus FAST Proteins Drive Pore Formation and Syncytiogenesis Using a Novel Helix-Loop-Helix Fusion-Inducing Lipid Packing Sensor
- The Role of ExoS in Dissemination of during Pneumonia
- IRF-5-Mediated Inflammation Limits CD8 T Cell Expansion by Inducing HIF-1α and Impairing Dendritic Cell Functions during Infection
- Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection
- Crystal Structure of USP7 Ubiquitin-like Domains with an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target USP7
- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- HPV16 Down-Regulates the Insulin-Like Growth Factor Binding Protein 2 to Promote Epithelial Invasion in Organotypic Cultures
- The νSaα Specific Lipoprotein Like Cluster () of . USA300 Contributes to Immune Stimulation and Invasion in Human Cells
- RSV-Induced H3K4 Demethylase KDM5B Leads to Regulation of Dendritic Cell-Derived Innate Cytokines and Exacerbates Pathogenesis
- Leukocidin A/B (LukAB) Kills Human Monocytes via Host NLRP3 and ASC when Extracellular, but Not Intracellular
- Border Patrol Gone Awry: Lung NKT Cell Activation by Exacerbates Tularemia-Like Disease
- The Curious Road from Basic Pathogen Research to Clinical Translation
- From Cell and Organismal Biology to Drugs
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
- A 21st Century Perspective of Poliovirus Replication
- Is Development of a Vaccine against Feasible?
- Waterborne Viruses: A Barrier to Safe Drinking Water
- Battling Phages: How Bacteria Defend against Viral Attack
- Archaea in and on the Human Body: Health Implications and Future Directions
- Degradation of Human PDZ-Proteins by Human Alphapapillomaviruses Represents an Evolutionary Adaptation to a Novel Cellular Niche
- Natural Variants of the KPC-2 Carbapenemase have Evolved Increased Catalytic Efficiency for Ceftazidime Hydrolysis at the Cost of Enzyme Stability
- Potent Cell-Intrinsic Immune Responses in Dendritic Cells Facilitate HIV-1-Specific T Cell Immunity in HIV-1 Elite Controllers
- The Mammalian Cell Cycle Regulates Parvovirus Nuclear Capsid Assembly
- Host Reticulocytes Provide Metabolic Reservoirs That Can Be Exploited by Malaria Parasites
- The Proteome of the Isolated Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components
- NK-, NKT- and CD8-Derived IFNγ Drives Myeloid Cell Activation and Erythrophagocytosis, Resulting in Trypanosomosis-Associated Acute Anemia
- Successes and Challenges on the Road to Cure Hepatitis C
- BRCA1 Regulates IFI16 Mediated Nuclear Innate Sensing of Herpes Viral DNA and Subsequent Induction of the Innate Inflammasome and Interferon-β Responses
- A Structural and Functional Comparison Between Infectious and Non-Infectious Autocatalytic Recombinant PrP Conformers
- Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria
- Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration
- Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
- EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival
- Single-Cell and Single-Cycle Analysis of HIV-1 Replication
- TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase
- The Herpes Simplex Virus Protein pUL31 Escorts Nucleocapsids to Sites of Nuclear Egress, a Process Coordinated by Its N-Terminal Domain
- Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections – A Prospective Cohort Study
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- Battling Phages: How Bacteria Defend against Viral Attack
- A 21st Century Perspective of Poliovirus Replication
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



