-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Emerging Functions for the RNome
Staphylococcus aureus is a leading pathogen for animals and humans, not only being one of the most frequently isolated bacteria in hospital-associated infections but also causing diseases in the community. To coordinate the expression of its numerous virulence genes for growth and survival, S. aureus uses various signalling pathways that include two-component regulatory systems, transcription factors, and also around 250 regulatory RNAs. Biological roles have only been determined for a handful of these sRNAs, including cis, trans, and cis-trans acting RNAs, some internally encoding small, functional peptides and others possessing dual or multiple functions. Here we put forward an inventory of these fascinating sRNAs; the proteins involved in their activities; and those involved in stress response, metabolisms, and virulence.
Published in the journal: . PLoS Pathog 9(12): e32767. doi:10.1371/journal.ppat.1003767
Category: Review
doi: https://doi.org/10.1371/journal.ppat.1003767Summary
Staphylococcus aureus is a leading pathogen for animals and humans, not only being one of the most frequently isolated bacteria in hospital-associated infections but also causing diseases in the community. To coordinate the expression of its numerous virulence genes for growth and survival, S. aureus uses various signalling pathways that include two-component regulatory systems, transcription factors, and also around 250 regulatory RNAs. Biological roles have only been determined for a handful of these sRNAs, including cis, trans, and cis-trans acting RNAs, some internally encoding small, functional peptides and others possessing dual or multiple functions. Here we put forward an inventory of these fascinating sRNAs; the proteins involved in their activities; and those involved in stress response, metabolisms, and virulence.
Introduction
Staphylococcus aureus is a commonly isolated bacterial pathogen in humans and animals and a serious threat to health. It can live as a commensal but, provided suitable opportunity, can initiate severe infections at various body sites. S. aureus is one of the most frequently isolated pathogens in hospital-associated infections but can also cause diseases in the community [1]. Nosocomial and community-acquired S. aureus infections include superficial skin lesions such as boils, abscesses, and impetigo, while invasive infections include septic arthritis, pneumonia, osteomyelitis, and endocarditis. S. aureus is an aggressive pathogen due to the combination of elevated antibiotic resistance and prominent virulence. The virulence of S. aureus is defined by a series of determinants that are often redundant in their functions. This bacterium produces an array of cell surface and secreted factors, including proteins that promote adhesion to host cells and tissues and some that bind proteins in blood toevade triggered immune responses. The organism also secretes extracellular enzymes such as proteases, a hyaluronidase, a lipase, and a nuclease that facilitate host tissue destruction and spreading. It produces membrane-damaging toxins that lyse host cells, as well as superantigens that are immunostimulatory exotoxins [2].
To face and adapt to various environmental conditions, including host colonization and spreading, S. aureus possesses many signaling pathways, some that are redundant, to coordinate the expression of its numerous virulence genes. At least 12 two-component regulatory systems and several transcription factors control these regulatory circuits, with multiple and intricate interplays to specifically reprogram the expression of target genes for continuous adaptation. Dozens of regulatory RNAs (sRNAs) are also involved in such dedicated control of gene expression, but their direct mRNA targets are, for the most part, currently unknown. Additionally, translation control and decay of selected S. aureus mRNAs, in response to specific signals during S. aureus growth and adaptation, can be achieved by specific ribonucleases [3] organized into large multi-enzyme complexes [4]. Widespread mRNA antisense transcription all over the S. aureus genome [5], as well as dedicated cis and trans sRNAs (reviewed in [6], [7]), actively participate in these gene expression controls.
More than 250 srna genes were discovered and detected as expressed transcripts in various S. aureus strains and experimental conditions [8]–[15]. The vast majority of these sRNAs are only expressed in S. aureus, a few are detected in Bacillaceae (e.g. RsaE), and several housekeeping sRNAs are detected in all eubacteria (e.g. tmRNA, RNase P RNA, 6S RNA). Most S. aureus sRNAs are located within the core genome, with a few expressed from the pathogenicity islands and from plasmids. For the most part, their functional, structural, and mechanistic details are unknown. This review will focus on the current functional understanding of cis - and trans - regulatory RNAs expressed in this organism, the unusual cases of cis sRNAs acting in trans, those expressing small peptides, and the sRNAs possessing multiple functions. We will exclude the S. aureus riboswitches that are cis-acting regulatory domains of mRNAs. The various proteins associated with S. aureus sRNA functions will be described, including the controversial roles of Hfq. The emphasis will be placed on sRNAs involved in stress response and metabolisms and on several sRNAs implicated in S. aureus pathogenesis.
A Multiplicity of sRNAs Expressed by S. aureus
Cis-encoded antisense RNAs
Cis-encoded antisense sRNAs are transcribed on the strand opposite to their target mRNAs [16], [17] and regulate gene expression by base-pairing with their complementary mRNAs (Figure 1A). Despite an extended complementarity with their primary target encoded on the opposite DNA strand, the initial interaction between the mRNA and the sRNA, “a kissing interaction,” occurs by contact between a few nucleotides usually located in accessible hairpins. This interaction is followed by additional pairings involving structural rearrangements of the two interacting RNAs [18]. In S. aureus, the first cis-encoded sRNA identified controls the rolling-circle replication of plasmid pT181 by transcriptional attenuation [13]. pT181 regulates its replication by the expression of an antisense RNA (RNAI) that blocks the expression of the plasmid-encoded replication initiation protein RepC. This mechanism involves pairings between complementary loops in the mRNA leader and the antisense RNA, which results in the formation of a transcription-termination hairpin 5′ to the Rep initiation codon. Attenuation is very efficient, aborting >90% of the Rep transcripts under standard growth conditions. Several other cis-encoded sRNAs expressed by S. aureus were detected in mobile genetic elements (PIs, plasmids, transposons) that are complementary to mRNAs expressing transposases involved in genome plasticity and integrity. RsaOX is complementary to the coding sequence of the SA0062 mRNA encoding a putative transposase [11]. Another transposase, IS1181, is probably also regulated by two additional sRNAs, RsaOW/Teg17 and Teg24as, which are complementary to its 5′ UTR, including a portion of the Shine Dalgarno (SD) sequence, and also to the 3′ UTR [10], [11]. In strain N315, the gene encoding the transposase is repeated eight times and these two sRNAs are systematically detected on the tnp locus [10]. Cis-encoded sRNAs can also interact with additional mRNA targets at distant genetic loci, in trans.
Fig. 1. A variety of mechanisms of actions for the S. aureus sRNAs. 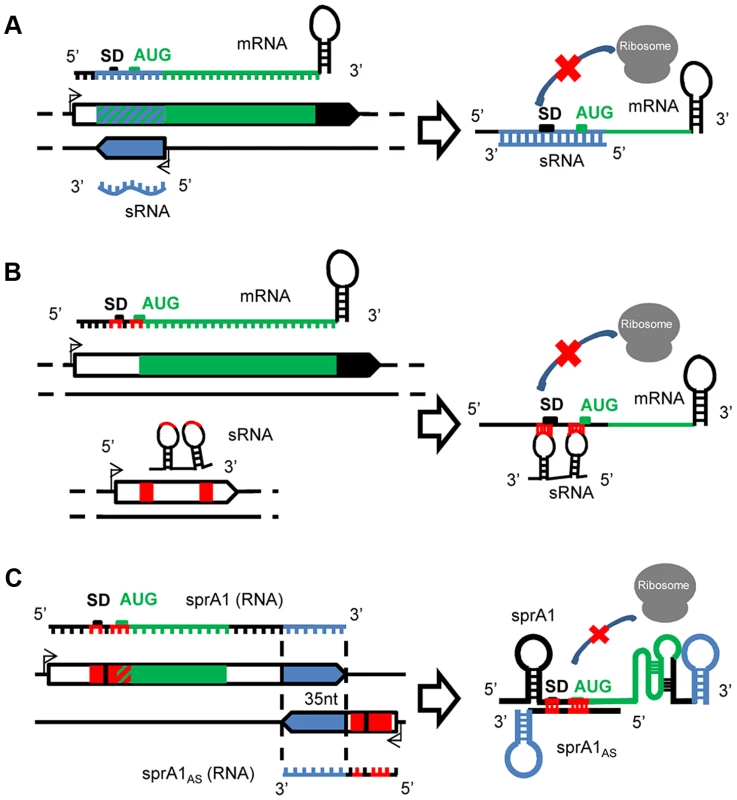
(A) Cis-encoded sRNAs bind via perfect complementarities with mRNA targets at the translation initiation sequence, preventing ribosome binding and therefore translation. (B) Trans-acting sRNAs. The trans-encoded sRNAs bind and block the ribosome binding site by interrupted pairings, using one or two hairpin(s) to repress translation initiation. (C) Cis-encoded antisense sRNAs acting in trans. In the SprA1/A1AS TA module, SprA1AS prevents SprA1 translation to prevent toxic peptide expression. On the two interacting sRNAs, the cis and trans pairing-regions are indicated in blue and red, respectively. Trans-encoded sRNAs
In contrast to the cis, the trans-encoded sRNAs are transcribed at distant genetic loci from their molecular targets and share only partial and often interrupted pairing complementarities, as for the eukaryotic microRNAs (Figure 1B). Although a seeding interaction of six to seven nucleotides is sufficient to initiate the “sRNA–mRNA” interaction in E. coli [19], pairings are usually much longer for the sRNAs expressed by S. aureus, probably due to its AT-rich genome. In most cases, the interaction involves the 5′ domains of the sRNAs that encompass the translation initiation signals (TIS) of the mRNA targets [20]. Conserved and accessible motifs were detected in several S. aureus sRNAs containing consensus sequences involved in the initial pairing with their target mRNAs. Several sRNAs from the Rsa family contain unpaired and accessible UCCC motives located in conserved hairpin regions of the sRNAs predicted to interact with the target mRNA translation initiation signals [12]. The RNAIII also harbors several UCCC motifs in the three loops H7, H13, and H14, which interact with the SD sequences of several target mRNAs. These UCCC motives were also detected in sRNAs expressed from other gram-positive bacteria such as L. monocytogenes and B. subtilis [21]–[24]. Accessible C-rich boxes could be a general signature of regulatory RNAs controlling translation initiation of various target mRNAs.
Cis-encoded antisense sRNAs acting in trans
Type I Toxin/Antitoxin (TA) pairs are present on plasmids or chromosomes, or both simultaneously [25], and consist of stable toxins and labile antitoxins encoded within small genetic modules. In S. aureus, several candidates of type I TA were detected based on sequence homology [26], among which are SprA1/A1AS and the SprG/F modules. SprA1-SprA1AS is an RNA pair transcribed from a pathogenicity island, a genetic element acquired by horizontal transfers. SprA1 is a stable and structured 208 nt-long RNA that contains an internal open reading frame (ORF) encoding a cytolytic peptide, pepA1 [27]. PepA1 inserts within the biological membranes, alters their integrity, and induces cell death [28]. A small cis-antisense RNA of SprA1, named SprA1AS (Teg152, [10]), is transcribed from the complementary DNA strand. Although 35 nt at SprA1AS 3′-end are perfectly complementary to a sequence located at SprA1 3′-end, SprA1AS acts in trans by base pairing with the 5′ domain of SprA1 to repress pepA1 translation by occluding the TIS, preventing its toxicity for the bacteria (Figure 1C). By analogy, the SprA2/A2AS pair, considered as a second copy due to elevated sequence identity with SprA1/A1AS, may also act in trans. Another unconventional case of a cis-trans RNA was detected in a plasmid from Enterococcus faecalis, another gram-positive bacterium, within the par stability determinant of a plasmid required for stable inheritance in its host [29]. In that case, overlapping RNAs I and II share a bidirectional transcription terminator but interact with one another by two non-overlapping direct repeats.
sRNAs with Multiple Functions
SsrA/tmRNA responsible for trans-translation and acting as a trans-RNA
Transfer-messenger RNA (tmRNA or SsrA) is an sRNA expressed in all bacteria that displays both tRNA and mRNA properties. tmRNA, with the help of the SmpB protein, governs trans-translation, a process that rescues the ribosomes stalled during translation of defective mRNAs, such as those lacking in-frame termination codons [30], [31]. tmRNA is recruited by the ribosomes through the smpB protein, an essential participant for ribosome rescue, and acts first as transfer RNA (tRNA) to add an alanine to the stalled polypeptide chain. Translation then switches from the problematic mRNA to a short tmRNA internal ORF that encodes a proteolytic tag [32], [33]. The stalled ribosome is released at the tmRNA termination codon, and the problematic mRNA and the tagged protein are degraded by specific RNAses and proteases, respectively. Trans-translation allows ribosome recycling and degradation of potentially toxic truncated mRNAs and proteins. A recent study [34] showed that tmRNA activity in S. aureus was not restricted to trans-translation. Inactivation of ssra-expressing tmRNA leads to an increase of pigment synthesis that is counteracted by expressing a tmRNA harboring a mutated tag. Furthermore, this phenotype is not imputable to the alteration of trans-translation since SmpB inactivation did not modify the quantity of pigments produced by S. aureus. The phenotype is due to the overexpression of the crtMN operon which encodes two enzymes involved in pigment synthesis. As the tmRNA sequence displays partial complementarity with the 5′ UTR of the crtMN mRNA, it could also act as an antisense sRNA acting in trans to regulate crtMN mRNA translation [34].
RNAIII, the Paradigm
RNAIII is the effector of the agr quorum-sensing system which coordinates gene expression in S. aureus according to the local density of bacteria [35]. The agr locus is transcribed from two divergent promoters, P2 and P3. Four genes (agrA, B, C, and D) are expressed as an operon from the P2 promoter, and RNAIII is transcribed from the P3 promoter [35]. An autoinducing peptide (AIP) is produced from agrD and secreted in the extracellular medium. The AIP binds to the agrC transmembrane protein, which, in turn, activates the agrA response regulator. AgrA, in conjunction with the global regulator SarA, activates transcription of its own operon and that of RNAIII. RNAIII controls the switch between early expression of surface proteins and late production of S. aureus exotoxins. RNAIII represents a paradigm in the field of bacterial RNAs exerting influence on pathogenesis. RNAIII controls target gene expression at multiple levels, including transcription, translation, and mRNA stability. RNAIII regulates by antisense pairings, at the post-transcriptional level, the expression of numerous targets involved in virulence and cell wall metabolism. RNAIII represses the expression of hydrolases and amidases involved in peptidoglycan turnover, an effect occurring at high cell density when RNAIII accumulates. RNAIII is a 514 nt-long RNA that possesses an intricate fold [36], is composed of 14 stem-loops (H1-H14) and three long-distance helices, and meets the definition of a multifunctional, “all-in-one” RNA. It encodes internally the δ-hemolysin peptide, which displays hemolytic and microbial activities [37]–[39]. Through various structural domains, RNAIII acts as both an activator and a repressor of dedicated mRNA targets. As for several trans-acting sRNAs, RNAIII coordinates complex regulatory networks. RNAIII activates translation of the hla mRNA encoding the α-hemolysin [40]. In the absence of RNAIII, the 5′-end of the hla mRNA forms a structure that blocks access to the ribosome on the Shine-Dalgarno (SD) sequence. RNAIII, via hairpins H2 and H3, interacts with the 5′UTR of the hla mRNA and provides accessibility to the SD site and, consequently, triggers α-hemolysin translation. RNAIII also up-regulates MAP production by interacting with the map mRNA via antisense pairings [41]. MAP, also named Eap (Extracellular adherence protein), is a surface adhesion protein involved in S. aureus immune evasion [42]. The mechanism of regulation, however, remains to be elucidated. On the other hand, RNAIII inhibits translation of various target genes by pairing at the TIS of several target mRNAs to inhibit their translation and trigger their degradations. In some cases, a single loop binding is not sufficient for down-regulation, and RNAIII binding requires additional interactions at the vicinity of the initial binding site [43], [44]. RNAIII represses translation of the SA1000 mRNA expressing a fibrinogen-binding protein involved in S. aureus adhesion to epithelial cells, of SA2353 mRNA producing a secreted antigen precursor [43], of spa mRNA encoding the immune escape protein A [14], of coa mRNA encoding a coagulase [43], [44], as well as of rot mRNA encoding a transcription factor repressing toxin production [43], [45]. Several proteins that are regulated by RNAIII are major virulence factors produced by the S. aureus clinical isolates during infection. The influence of RNAIII on membranes, surface proteins, and cell wall turnovers contributes to virulence by controlling nutrients' entries and host defences, struggle, and resistance by regulating hemolysins production and host immune evasion. We expect that RNAIII possesses additional targets involved in S. aureus virulence control, which will be progressively identified by high throughput methods using Deep RNA sequencing technologies combined with target affinity purifications.
Proteins Involved in S. aureus sRNA Functions
Hfq: A controversial factor
Since trans-encoded sRNAs display short and imperfect complementarity to their target mRNAs, the effective sRNA-mRNA annealing requires an auxiliary factor in some bacterial species. Hfq, a member of the conserved RNA-binding Sm-like protein family, is needed for the efficient annealing of some sRNAs to target mRNAs and for the intracellular stability of these sRNAs. In the case of canonical sRNA, the Hfq protein enhances the binding of sRNA on the translational start site of their mRNA targets and prevents ribosome binding. Hfq is only active in its multimeric form. The Hfq ring formed by homohexameric Hfq proteins displays a characteristic doughnut-shaped structure containing two single-stranded RNA-binding faces located on opposite sides of the ring. The proximal face binds to AU-rich sequences and sRNAs, whereas the distal face interacts with poly(A) sequences [46], [47]. The dual RNA-binding surface allows the simultaneous recruitment of an sRNA and its mRNA target on one Hfq molecule. Hfq facilitates sRNA–mRNA interaction by increasing the local concentration of the RNA species and/or by enhancing the base pairing interaction through a restructuring of these RNAs. In E. coli, the chaperon Hfq affects the turnover of some target mRNA by recruiting an RNase E in an activated state on the sRNA–mRNA duplex [48], [49]. The requirement of Hfq for riboregulations by trans-encoded sRNAs depends on the small RNAs, mRNAs, and the bacterial species. Potential links between the free energy for sRNAs–mRNA pairing, the GC-content of the bacterial genome, and the involvement of Hfq protein have been proposed [50]. A highest ΔG value and a lowest GC-content correlate with a dispensability of Hfq protein for the sRNAs–mRNAs pairing. Accordingly, Hfq is required in sRNAs regulations, which are involved in the growth, the sensitivity to various environmental stresses, and the virulence of several gram-negative strains. Moreover, Hfq or Hfq-like proteins are absent in several “low GC” gram-positive strains such as Streptococcus pneumoniae and Lactococcus lactis. In S. aureus, the function of Hfq remains unclear. Inactivation of the hfq gene in three S.aureus strains (RN6390, COL, and NEWMAN) did not affect the phenotypes of these strains [51]. In S. aureus, Hfq was not involved in more than 2,000 phenotypes tested, including sensitivity to different stress conditions, antibiotic sensibility, and virulence [51]. In agreement with the Hfq dispensability in the riboregulation, the protein has no effect on the stability/turnover of several trans-acting sRNAs [12], [27], [43], [45], [52], [53]. The S. aureus Hfq does not enhance the binding of RNAIII to spa mRNA or SA1000 mRNA, whereas an Hfq-RNAIII complex from RN6390 cells co-immunoprecipitates with an antibody against Hfq [43], [52]. However, Hfq seems to be functional in S. aureus, since the overexpression of Hfq in an agr− strain leads to the stimulation of the γ-haemolysin translation. This discrepancy between the ability of Hfq to tightly bind RNA in vitro and its inability to affect the riboregulation in vivo could be explained by the very low expression levels of Hfq in the laboratory-adapted strains RN6390 and COL. Indeed, inactivation of the hfq gene in the S. aureus 8325-4 strain expressing a detectable Hfq level alters the expression profiles of 116 genes potentially involved in the decrease of the pathogenicity of the muted strain in a murine peritonitis infection model and in the increase of expression of the surface carotenoid pigment [54]. The relationship between Hfq and carotenoid production was also revealed in another S. aureus strain. Low-fluid-shear culture of N315 cells, which promotes attachment-independent biofilm formation, leads to a decrease of carotenoid production associated with a down-regulation of the Hfq protein [55]. Hfq specifically binds to 49 of the 116 genes down-regulated in the hfq mutant of S. aureus 8325-4. In particular, some mRNA targets of sRNAs, such as sbi, sucD, and rot for respectively SprD, RsaE, and RNAIII, were copurified with Hfq, suggesting that Hfq could be implied in the translational regulation of some S. aureus genes [54]. Altogether, these studies show that the modulation of virulence and stress response could be attributed to Hfq in some strains. However, the direct involvement of sRNAs in these Hfq-dependent phenotypes and the mechanisms of actions of potential “sRNA/Hfq” complexes remain unknown. As S. aureus does not express RNase E, Hfq could recruit another endoribonuclease to affect the turnover of mRNA targets. Hfq proteins from different bacteria contain an evolutionarily conserved core of 65 amino acids and a divergent positively charged C-terminal end. The C-terminus extensions are short in gram-positive bacteria (like S. aureus, B. subtilis, and L. monocytogenes) and longer in gram-negative bacteria (102 amino acids in E. coli and Salmonella). Recently the C-terminal extension of E. coli Hfq protein was shown to be required for a non-canonical sRNA pathway, a translational regulation involving the binding of sRNA outside the canonical ribosome entry site, probably by recruiting additional RNAs or proteins on the mRNA target [56]. Thus, the short C-tail extension mainly present in S. aureus could be involved in the recruitment of specific ligands during the Hfq-dependent riboregulation by non-canonical sRNAs. In S. aureus strains that do not express the protein, the role of Hfq might be superseded by other RNA-binding molecules.
RNAse III: The major RNase involved in the sRNA-dependent mRNA turnover
Staphylococcus aureus RNase III belongs to the family of Mg2+-dependent endoribonucleases which cleave double-stranded RNA to generate short RNA duplexes with a 5′ phosphate group and two nucleotides 3′-overhang. The enzyme contains a catalytic and a dsRNA-binding domain and functions as a homodimer to recognize and cleave a variety of structures including imperfect duplexes, loop–loop interaction, and stacked helices [57]. Historically described as an endoribonuclease involved in rRNA processing and maturation in E. coli and B. subtilis, the enzyme also participates in the regulation of single and polycistronic mRNA as well as in the processing of some housekeeping RNAs in B. subtilis [58], [59]. In S. aureus, the RNase III-processing alone enhances mRNA stability and translation of the major cold-shock CspA protein through a cleavage within the 5′ leader and autoregulates its synthesis by initiating the degradation of its own mRNA [60]. In contrast with the essential involvement of the endoribonuclease RNase III in the viability of E.coli and B. subtilis, the enzyme does not influence the growth of S. aureus, but plays an important role in the pathogenicity of S. aureus in murine models [61]. RNase III is involved in the mRNA turnover of some trans-acting sRNA targets. In S. aureus the enzyme acts as an essential co-factor of the quorum-sensing regulatory RNA III for the irreversible translational repression of the mRNAs coding for the protein A (spa), the staphylocoagulase (coa) [44] and the transcriptional regulator Rot [45]. The enzyme interacts with RNAIII without promoting the repressor activity of RNAIII by improving the stability of RNAIII–mRNA duplexes. RNAse III only cleaves RNAIII when the sRNA is bound to its mRNA targets [52], [62]. RNAIII binds to the ribosome binding site of coa and rot mRNAs and recruits RNase III, which cleaves the mRNA target at an equivalent position of the loop–loop pairing. The RNase III cleavage site is independent of the sequence of the base pairs. The loop–loop interaction forms a unique hairpin motif creating a single binding site for the RNase III, which leads to a specific cleavage at single positions of the kissing interactions and irreversible repression of mRNA translation [62]. Several RNase III-binding sRNAs were identified by deep sequencing of RNA coimunoprecipitated with a wild-type RNase III and/or cleavage-defective mutants in vivo. Among the 58 sRNAs detected, many have been previously identified, such as the pathogenicity island-encoded small RNAs SprA, SprA3, SprB, SprC, SprF3/G3 [15], and RsaA, RsaE, RsaH, RsaI, RsaJ [11], [12], as well as RNAIII. Some of these sRNAs were copurified with the cleavage-defective mutant and display hairpin motifs recognizable by the enzyme, suggesting that there are substrates of RNase III [60], [62]. Thus, it appears that most of these known and unknown sRNAs are potential trans-acting factors which regulate gene expression by antisense mechanisms and recruit RNAse III to direct the mRNA decay. Also, RNase III could mediate specific cleavage of type I toxin/antitoxin pairs (SprA1/SprA1AS, SprG/SprF) to prevent toxic peptide expression. RNase III is associated with a large number of antisense transcripts, covering 44% of the annotated genes [60], [63]. These antisense (as) RNAs are issued from a genome-wide process of overlapping transcription and are perfectly complementary to the 5′ ends, 3′ ends, middle, or entire genes or operons [5], [60], [64]. RNase III-associated cis-asRNAs are usually expressed at lower levels than their complementary mRNAs, and they direct the degradation of residual mRNAs. The RNase III-mediated digestion of sense/antisense transcripts generates a large collection of short, 22 nucleotide-long, double-stranded RNAs that could also have functions [64]. Pervasive transcriptions lead to the expression of RNase III-associated mRNAs, which overlap at their 5′ or 3′ UTRs. RNase III-induced cleavages of the 5′ overlapping regions of divergent mRNAs allow the fine regulation of the expression of genes which have to be expressed in a coordinated manner as the tagG/tagH teichoic acid biosynthetic genes encoding the TagGH ABC transporter complex [60], [63]. These cleavages generate mRNAs with shorter 5′ends which could be more sensitive to degradations and/or influence their translations. In some cases, the 5′ UTRs of mRNAs extend into the coding sequence of their neighbouring genes [64]. Similar large asRNAs are encoded at particular genomic loci called the “excludons” in the gram-positive Listeria monocytogenes [65]. These long asRNAs span divergent genes or operons with related or opposing functions, and allow meticulous regulatory switches in bacteria [66], probably also occurring in S. aureus. In S. aureus, the activity of RNase III, in association with large asRNAs, could be considered a general mechanism to regulate and coordinate the expression of neighbouring genes.
Other RNases: RNase J1-J2 and RNase Y
In E. coli, sRNA-mediated mRNA decay mainly involves the recruitment of RNase E on the mRNA target. The RNase E is the central component of the degradosome that is composed of a 3′-exoribonuclease polynucleotide phosphorylase (PNPase), a RNA helicase (RhlB) and a glycolytic enzyme (enolase). RNase E catalyzes the initial endoribonucleolytic cleavage of mRNA targets which is followed by a directional, 3′ to 5′, degradation by the PNPase with the help of RhlB [67]. Most gram-positive bacteria do not contain an RNase E and use RNase E functional orthologs and other degradosome components to direct mRNA decay. Both S. aureus and B. subtilis contain a similar multicomponent ribonucleolytic degradosome complex formed around RNase Y, a functional homologue of RNase E [4], [68]. The S. aureus degradosome includes both RNases J1/J2 originally proposed to act also as functional orthologs of RNase E [69], the PNPase, the enolase, the RNA helicase CshA, and RNase P [4]. The CshA Dead-box RNA helicase plays an essential role in the regulation of quorum sensing by controlling the agrBDCA mRNA turnover [70], [71]. Both RNases Y and J1, which exhibit RNase-E–like 5′ end-dependent endonucleolytic activity, play a central role in the degradation of mRNAs in B. subtilis. The endonucleolytic cleavage by the RNase Y initiates the mRNA decay, and the resulting RNA fragments are likely to be degraded by the 5′-3′ exoribonuclease activity of RNase J1 and by the 3′–5′ activity of PNPase [72], [73]. In contrast to B. subtilis, the membrane-associated RNase Y of S. aureus is not essential for growth but is required for virulence [74], [75]. The enzyme is involved in the processing of the global virulence regulator sae and in the expression of various virulence genes by an indirect mechanism [75]. The RNase Y controls the stability of specific mRNAs and sRNAs. Interestingly, inactivation of the rnc gene encoding for RNase Y in S. aureus results in an increase of the half-life of two sRNAs, RsaA and Sau63 [8], [12], whereas the RNAIII steady-state level is unaffected. The specific activity of RNase Y represents a way to control both the expression of sRNAs and their mRNA targets in S. aureus. A similar regulation was detected in E.coli, where RNase E specifically affects the steady-state level of several sRNAs [59]. In contrast with the activity of RNase E in E. coli, an implication of RNase Y in the coupled degradation of sRNAs and their mRNA targets has not been revealed.
sRNAs Involved in Stress Response, Metabolisms, and Regulatory Networks
Sigma B-inducible small RNA encoding genes
The pathogenicity of S. aureus depends on its ability to respond quickly and specifically to a variety of environmental stresses and to control virulence genes expression. The S. aureus genome allows expression of the alternative sigma B transcriptional factor (σB) that is an essential part of the complex regulatory network controlling the expression of around 200 genes involved in virulence, cell wall metabolism, and membrane transport processes [76]–[78]. σB is involved in stress responses and contributes to pathogenesis in animal models of infections [79]. The S. aureus sigma B operon resembles that of the homologous B. subtilis operon. It contains σB, an anti-σB factor RsbW, an anti-anti-σB factor RsbV, and RsbU, a Mn2+-dependent phosphatase that positively controls σB activity by dephosphorylating RsbV [80], [81]. The sigma B regulon includes genes directly up-regulated by σB and genes indirectly regulated via σB-dependent expression of regulatory factors such as the SarA transcription factor [76], [77]. In particular, the inactivation of σB has an indirect impact on the agr quorum-sensing system by enhancing RNAIII expression [82]. By computational approaches based on the search for σB consensus binding sites (GWWT_N14–17GGGWWW) and transcriptional terminator sequence within the intergenic regions of S. aureus strain N315, three σB-regulated genes coding for new sRNAs were identified and validated [53]. Two of these sRNAs, SbrA and SbrB, are highly conserved among Staphylococci (for σB-dependent small RNAs A and B) and encode putative basic peptides of 26 and 38 amino acids, which are potential virulence factors. In contrast to sbrA, the peptide from sbrB gene is translated only in some S. aureus, whereas SbrB is expressed in all strains, which suggests a potential dual function of SbrB: a peptide-coding sRNA and activity as an sRNA regulator [53]. The third sRNA, SbrC, is a potential cis-acting antisense targeting the 3′ end of the mntABC operon encoding for an ABC transporter dedicated in the uptake of manganese. The manganese acquisition is crucial for defence systems against oxidative stress and contributes to the virulence of S. aureus [83]. In S. aureus, σB-dependent transcription is induced by the presence of MnCl2, probably via the stimulation of the Mn2+-dependent phosphatase activity of RsbU [77]. The σB-dependent induction of SbrC could be a way for σB to autoregulate its own activity and to modulate manganese uptake in function of Mn availability. The transcription of other sRNAs like RsaA, RsaD, and RsaF is induced in S. aureus strains expressing an active σB factor [12]. These sRNAs are differently transcribed in response to environmental stresses such as oxidative stress, heat stress, cold stress, osmotic stress, and acidic pH. A conserved σB promoter sequence was found upstream rsaA, suggesting its direct regulation by the σB. As RsaA is a trans-acting regulator that potentially targets three mRNAs repressed by σB [12], [76], [77], it could be an intermediate in the regulatory network controlled by σB.
Metabolisms regulations
In S. aureus, all macromolecules are synthetized from 13 biosynthetic intermediates produced by glycolytic, pentose phosphate, and tricarboxylic acid cycle (TCA cycle) pathways. These three central metabolic pathways are closely linked to the expression of several virulence factors. The alteration of pentose phosphate and glycolytic pathways affects the quorum-sensing–dependent regulation of RNAIII [84], [85], and TCA cycle inactivation induces a reduction in the production of several secreted virulence factors and cell-associated adhesion factors [86], [87], thus, slowing down central metabolism reduces bacterial virulence. Recently, RsaE, a sRNA conserved in all S. aureus strains, and also in firmicutes, has been shown to regulate several metabolic pathways [11], [12] (Figure 2). The overexpression of RsaE induces a growth defect that is partially alleviated by the addition of acetate, arguing for a role of RsaE in both catabolisms and anabolisms. Indeed, RsaE down-regulates the synthesis of enzymes from the TCA cycle (succinyl-Coa synthetase sucC and sucD, aconitase (citB), citrate synthase (citZ), and isocitrate dehydrogenase (citC)), and from the folate-dependent one-carbone metabolism (bi-functional protein fold and the formate-tetrahydrofolate ligase (Fhs)), which is involved in the purine biosynthesis pathway. RsaE also affects the amino acid pool in S. aureus. It up-regulates the expression of valine, leucine, and isoleucine operons and potentially alters aspartate biosynthesis by inducing the expression of pyruvate carboxylase. Moreover, RsaE down-regulates the opp-3 operon coding for an oligopeptide transporter involved in the uptake of specific peptides and in the regulation of extracellular protease production [88], [89]. As the transcription of genes encoding for some enzymes of the TCA cycle is regulated by the availability of amino acids [90], RsaE could indirectly modulate the TCA cycle via the regulation of the pool of free intracellular amino acids. The distinction between metabolic pathways directly and/or indirectly regulated by RsaE is difficult to apprehend because these pathways are highly interconnected. RsaE can directly regulate the TCA cycle and amino acid uptake by inhibiting the formation of ribosomal initiation complex on sucD and opp3A/opp3B mRNAs, respectively [11], [12]. The TCA cycle is involved in the energetic transition which uses the acetate accumulated in the extracellular medium during the glycolysis and amino acids as an alternative carbon source. Although the expression profile of RsaE is a subject of controversy [11], [12], in some S. aureus strains, RsaE is expressed at late exponential phase and repressed at stationary phase, and it could facilitate the transition of energy metabolisms, the purine biosynthesis, and amino acid transport in response to the nutrients' availability. Moreover, the RsaE expression seems to be dependent on the agr quorum-sensing system and σB activity, suggesting that it could modulate the metabolism profile in function of stress responses and/or virulence [12].
Fig. 2. RsaE controls central metabolic pathways. 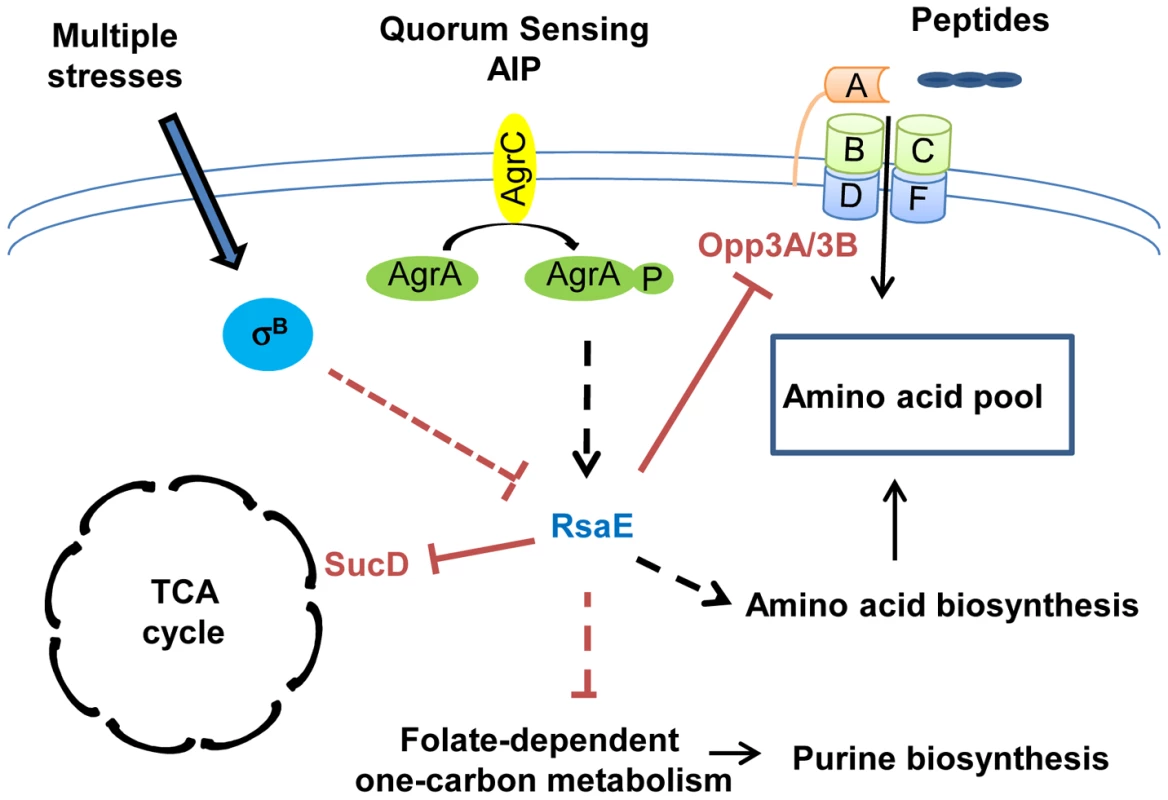
RsaE regulates, directly or indirectly, the expression of several genes involved in amino acid synthesis, peptides transport, carbohydrate metabolism, and the TCA cycle. RsaE directly regulates the TCA cycle by inhibiting sucD mRNA translation coding for one of the subunits of the succinyl-Coa synthase. It alters the purine biosynthetic pathway via the down-regulation of some enzymes involved in the folate-dependent, one-carbon metabolism. RsaE uses multiple binding sites for the regulation of the opp3BCDFA mRNA expressing an oligopeptide transporter involved in nutrient transport. RsaE pairs directly with sites overlapping the ribosome binding site of the upstream (opp3B) and distal (opp3A) genes from the operon to inhibit their translations. RsaE modulates the intracellular pool of amino acid by down-regulating the expression of an oligopeptide transporter and by up-regulating genes that produce amino acid synthesis enzymes. In some S. aureus strains, RsaE expression is controlled by the agr quorum-sensing system in response to autoinducing peptide (AIP), and it depends on the σB regulon. The plain and dashed lines indicate the direct and indirect gene regulations, respectively (red bars: inhibitions, black arrows: stimulations). sRNAs involved in global regulatory networks
S. aureus expresses a large array of extracellular and cell-wall–associated virulence factors at different stages of the infectious process. The exoproteins and cell-wall–associated adhesins, respectively involved in host immune evasion and host cell adhesion, are expressed early during the initial colonization, while the production of toxins that facilitate S. aureus growth and spread in the host tissues occurs late during infection. Their temporal expressions are controlled by two component regulatory systems (e.g. saeRS, arlRS, lytSR, srrAB) and global transcriptional regulatory factors (e.g. the sarA protein family, spX). The overlapping regulation between two component systems and global transcriptional factors constitutes a fine-tuning system for an efficient transcriptional control of virulence genes expression [91]. These factors affect the expression of virulence genes by directly binding to the promoters of target genes and/or indirectly through the regulation of the expression of global regulatory elements targeting the same set of virulence genes. RNAIII has characteristics similar to global regulatory factors that regulate directly and indirectly the expression of virulence genes, such as spa and hla genes [14]. The expression of spa encoding an adhesin acting as an host immune evasion protein is directly controlled, at the translational level, by an “RNAIII-mRNA” direct pairing mechanism [14], as well as at the transcriptional level by three members (SarA, SarS, and Rot) of the SarA family of transcriptional regulators (Figure 3) [92]. spa is positively regulated by the transcriptional factors Rot (Repressor of toxins) and SarS and negatively regulated by SarA. RNAIII affects the mRNA level of spa by inhibiting rot translation by a base pairing mechanism [45]. As Rot activates SarS transcription, RNAIII-mediated inhibition of Rot expression down-regulates the two transcriptional activators of spa. These transcriptional and translational controls avoid putative leakages in spa mRNA expression. RNAIII uses similar double controls to up-regulate the expression of hla encoding the α-hemolysin [93]. RNAIII enhances α-hemolysin translation by a pairing interaction at hla mRNA 5′UTR [40] and up-regulates hla mRNA expression by down-regulating Rot, which acts as a repressor of hla transcription in SaeRS - and SarS-dependent ways [93], [94]. In accordance with the antagonism between Rot and RNAIII, cellular amounts of Rot are inversely correlated to the RNAIII level in most S. aureus strains [95]. However, the transcriptomes of S. aureus strains deleted in RNAIII or in Rot only partially overlap, suggesting that RNAIII affects the expression levels of additional transcription factors [96]. One potential target of RNAIII could be the transcriptional factor SarT, which is down-regulated by agr at the post-exponential phase of growth [97] and contains a putative pairing interaction with RNAIII at its 5′UTR [43]. RNAIII-mediated down-regulation of SarT, which acts as a positive and negative transcriptional regulator of sarS and hla, respectively, could be another way to control spa and hla expression. The involvement of S. aureus sRNAs in the global gene regulatory network is not restricted to RNAIII. Recently, a new sRNA named ArtR (AgrA-repressed, toxin-regulating sRNA) was reported to activate α-hemolysin expression by binding to the sarT mRNA, promoting its degradation [98]. Although RNAIII and ArtR both similarly regulate hla expression, they display different expression patterns. In contrast to RNAIII, ArtR transcription is repressed by agrA, suggesting that ArtR-mediated hla up-regulation could be enhanced in agr-deficient strains. Multiple sRNAs controlling the expression of a similar component from a regulatory network allows the sharp regulation of virulence genes. Given the importance of multiple components from regulatory networks to express virulence genes and the elevated variability of their expression levels among the S. aureus strains, it is most likely that S. aureus expresses many other sRNAs that deeply interact with this network to influence bacterial virulence.
Fig. 3. Schematic overview of the multiple interactions between sRNAs and transcriptional regulators involved in spa (protein A) and hla (α-hemolysin) expression in S. aureus strain 8325-4. 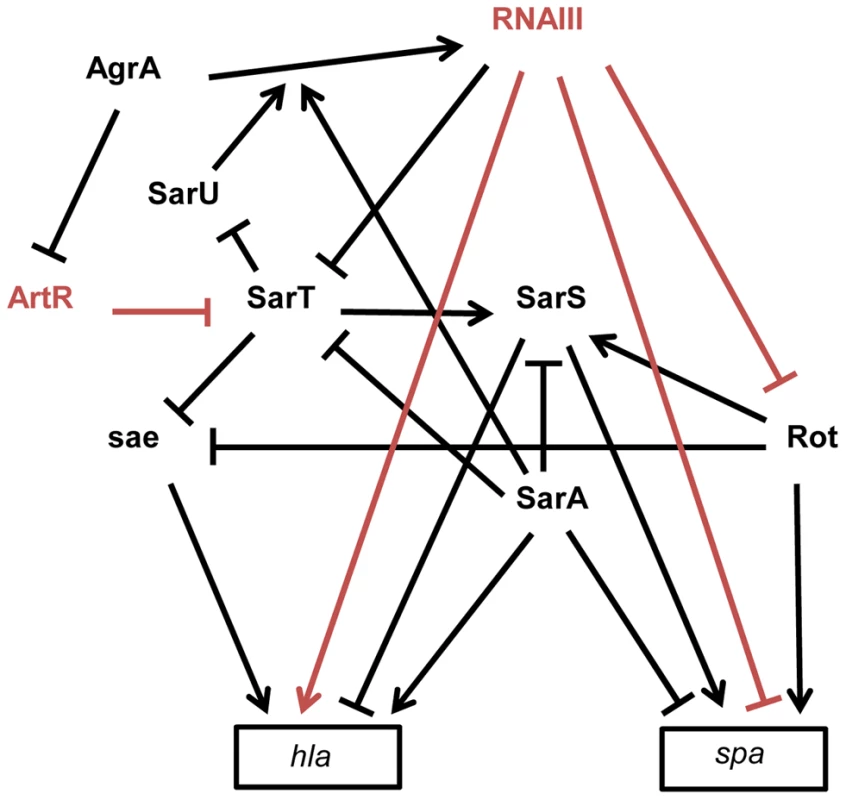
The arrows indicate the stimulations and the bars, the repressions. The direct effects of two sRNAs on gene expression are indicated in red. RNAIII represses rot and spa translation by direct pairing interactions [14], [45]. Rot requires SarT to stimulate SarS in the presence of SarA [92], [120]. In contrast to SarA, Rot and SarS are direct activators of spa expression [92], [121]. In the exponential phase of growth, spa transcription is stimulated by Rot and by SarS. In the post-exponential phase, spa transcription and translation are repressed by SarA and RNAIII, respectively, and the direct inactivation of Rot by RNAIII leads to the repression of the Rot and the SarS-dependent transcription activations of spa. hla is up-regulated by SarA and down-regulated by SarS [93]. Rot and SarT repress hla transcription by a sae-dependent way [94]. In the post-exponential phase of growth, RNAIII enhances hla translation by direct pairings at the hla mRNA 5′UTR and stimulates hla transcription by down-regulating the expression of SarT and Rot. AgrA, the master transcriptional regulator of quorum sensing, stimulates RNAIII expression [122] but also represses ArtR expression. ArtR indirectly activates hla transcription by repressing sarT translation [98]. SarA stimulates the AgrA-dependent expression of RNAIII [122]. SarT directly represses SarU which activates agr (RNAIII) transcription [123]. sRNAs and Virulence Gene Regulations
The dual-function SCCmec-encoded psm-mec RNA suppresses agrA translation and attenuates MRSA virulence
SCCmec is a mobile genetic element that confers methicillin resistance to the methicillin-resistant S. aureus (MRSA) strains. SCCmec contains several genes, including the cytolysin psm-mec gene, whose transcription product suppresses colony spreading and the expression of phenol-soluble modulin α, a cytolytic toxin [99]. The psm-mec RNA binds the agrA mRNA, encoding a virulence regulatory factor and inhibits its translation [100]. Deletion of psm-mec in MRSA clinical isolates increases virulence on mice skin infection models (Figure 4). The psm-mec RNA suppresses MRSA virulence by agrA translation inhibition, and the absence of psm-mec in community-acquired (CA) MRSA strains is responsible for their elevated virulence.
Fig. 4. sRNAs from the S. aureus RNome implicated in bacterial virulence. 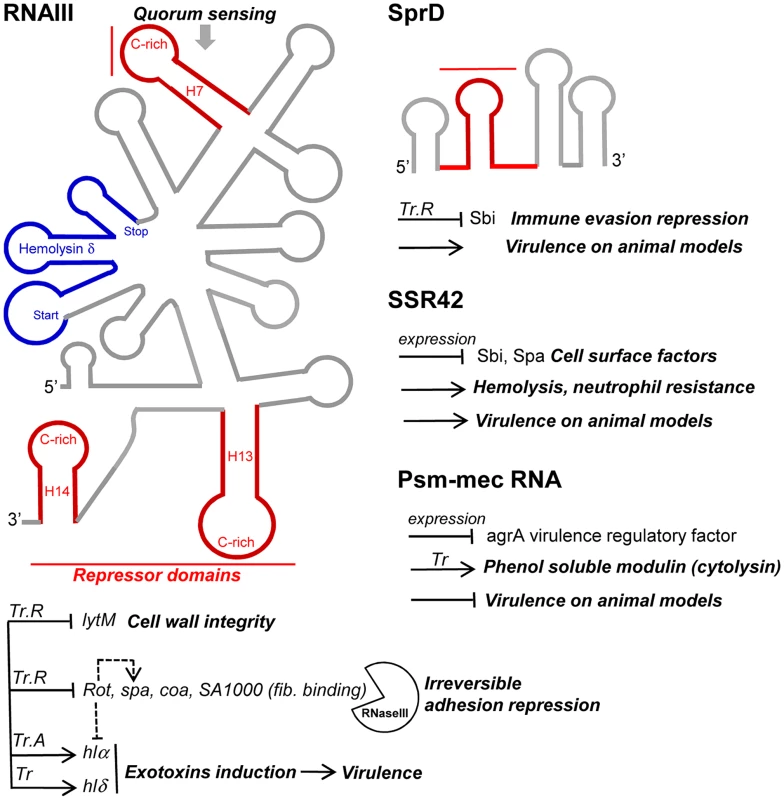
Multitasking RNAIII is the effector of quorum sensing to perceive bacterial population density and regulates multiple targets involved in peptidoglycan metabolism, adhesion, exotoxins production, and virulence. RNAIII internally encodes hemolysin δ (blue). RNAIII contains at least three repressor domains (red) containing accessible UCCC motifs that interact, by antisense pairings, with the ribosome binding sites of numerous target mRNAs for translational repression (Tr.R), some triggering endoribonuclease III (RNase III) cleavages to induce target mRNA degradations and irreversible gene expression decay. Translation of at least two exotoxins is activated by RNAIII, one encoded (hlδ), and another (hlα) by translation activation (Tr.A). SprD is expressed from the genome of a converting phage and interacts, by antisense pairings, with the 5′ part of the sbi mRNA encoding an immune evasion molecule. SprD possesses an important role in S. aureus virulence, but the mechanism of its control is yet to be elucidated, with Sbi being only one player among others. The 891-nucleotide long SSR42 affects extracellular virulence expression, hemolysis, neutrophil virulence, and pathogenesis and contains a putative internal ORF. The mechanisms of target regulation remain to be elucidated. The SCCmec-encoded psm-mec RNA suppresses agrA translation and attenuates MRSA virulence, acting as a dual-function RNA regulator. SprD, a pathogenicity island-encoded RNA regulating an immune-evasion molecule from the core genome
Small pathogenicity island rNA D′ (SprD) is among the few S. aureus RNAs with an identified function. SprD is expressed from the genome of a converting phage [15], a horizontally-acquired pathogenicity island (PI) being the repository of toxins, adherence, invasion factors, superantigens, and secretion systems [101]. SprD down-regulates, at the translational level, the expression of the Sbi immune evasion molecule located on the core genome [102]. One of its four hairpins binds the 5′ UTR of the sbi mRNA by an antisense pairing mechanism. The initial binding involves the hairpin loop, and the interaction extends farther upstream and downstream from that initial binding site. The “SprD-sbi mRNA” interaction sequesters the sbi mRNA TIS and, consequently, prevents translation initiation of the Sbi protein. Sbi is an immunoglobulin-binding protein expressed by S. aureus [103] that impairs the host immune response. Sbi acts as a complement inhibitor and forms a tripartite complex with host complement factors H and C3b [104]. SprD contains four hairpins, one of which interacts with the ribosome binding site of sbi mRNA to form a long imperfect duplex that prevents translation initiation in vivo. SprD contributes to causing disease in a mouse model of infection, although this effect is not only linked to the deregulation of Sbi production. It suggests that SprD regulates the expression of other targets playing important roles during host infection.
The implication of the 891 nucleotides-long small stable RNA42 in S. aureus virulence
Small stable RNAs (SSRs) are RNAs specifically produced and/or stabilized in response to various environmental conditions [104]. Among the SSRs, SSR42 is involved in host erythrocyte lysis, resistance to human polymorphonuclear leukocyte killing, and pathogenesis in a murine model of bacterial infection [3]. SSR42 is primarily expressed during the stationary phase of S. aureus growth, is a stable RNA with a ∼30 minute half-life, and appears to control the expression of a large set of target genes (∼80) including virulence factors, which is the rationale of its involvement in S. aureus pathogenesis and virulence.
sRNAs Expressions during Infections
Staphylococcus aureus is a common resident of human skin and nasopharynx. It is also a cause of life-threatening illness, producing virulence factors that enable survival and spreading in various hosts. Its switch from commensalism to an infectious pathogen is poorly understood, whereas nasal carriage and clinical isolates belong to the same genetic clusters [105]. The few S. aureus sRNAs with known targets regulate major biochemical pathways, some ultimately implicated in virulence [6]. The S. aureus sRNAs were detected and studied in various strains and their specific expression profiles during infection in humans are, for the vast majority of the ∼250 sRNAs expressed by this bacterium, unknown. However, RNAIII expression in clinical samples, such as nasal secretions or cystic fibrosis sputa, has been monitored [106]–[108]. The majority of clinical isolates isolated from acute infections has functional agr and produces RNAIII in vivo [109]. These data suggests that RNAIII influences the virulence phenotype. Agr-defective mutants, however, were detected in infected patients, and a mixture of agr positive and defective strains were detected in healthy humans [110]. Thus, agr is involved during acute infection, while agr mutants can be selected during chronic infections and dormant states. A recent study reported the expression profiles of the five sRNAs (RNAIII, RsaA, RsaE, RsaG, and RsaH) in strains isolated from patients with acute cutaneous infection, chronic cystic fibrosis, or nasal colonization [111]. The expression profiles of these five sRNAs are strain-specific and do not correlate to the type of infections or colonization, but the authors noticed that sRNA expression was more uniform among the strains from colonization compared to those responsible for infections. This observation might reflect the fact that S. aureus was primarily a commensal and then became an opportunistic pathogen [112], [113]. Deep RNA sequencing technologies now allow global analyses of the S. aureus RNome in various clinical isolates to detect putative differences of expression of all sRNAs, with possible applications in the early diagnostic of strains that are susceptible to cause life-threatening infections.
Phenotypes Associated with sRNAs Expressions
sRNAs can be differentially expressed in “normal” versus “small-colony variant” (SCV) phenotypes, as identified in a S. aureus clinical strain recovered from a patient with osteomyelitis [8]. SCVs grow slowly, lose their pigmentations, have reduced hemolytic activity, have decreased susceptibility to aminoglycosides, have lower toxins production, and have improved intracellular persistence [112], [114]–[117]. The “normal” phenotype corresponds to the “virulent” strain, whereas the SCVs are persister cells. In SCVs, the expression of RNAIII is phenotype-specific, detected in the normal phenotype but switched off in the SCVs [8]. The absence of RNAIII in the SCVs may account for their decreased output of toxins and their lower virulence [115]. Moreover, the expression of several pathogenicity islands (PIs)-encoded sRNAs, sprA-G, is turned off in the SCVs at late growth phases with at least one of these RNAs, SprD, that possesses a major role in virulence [102]. The lower expression levels of the Sprs in the SCVs could also account for their reduced pathogenicity compared to the normal phenotype. Expression of another sRNA, Sau-13, is up in the normal phenotype but down in the SCVs [8]. SCVs have electron transport deficiencies [115], [118]. Sau-13 could regulate ions' transport and metabolism by its antisense action against the alkaline phosphatase precursor phoB. An sRNA, Sau-66, is only expressed in the SCVs but absent in the normal phenotype [8]. Sau-66 has antisense potential on a nearby gene coding for a protein involved in folate biosyntheses. Since folate is a carbon donor during purine biosynthesis, Sau-66 may influence the formation of thymidin-auxotrophs SCVs [119]. Altogether, these data show that the expression of a subset of S. aureus sRNAs is associated with preferred phenotypes. The identification of their molecular targets, however, will be required to assess their roles in phenotypes-associated lifestyles and their putative implications in virulence.
Conclusion
Recent advances in the characterization of the plethora of regulatory RNAs expressed by S. aureus have provided novel insights about how they monitor various cellular activities. Most of the few sRNAs whose physiological roles have been determined control the expression of genes involved in central metabolisms, in response to quorum sensing, and on virulence by pairing to target mRNAs to modulate their translational activities and stabilities. Several sRNAs encode and express small peptides that may play important roles in virulence or in bacterial growth control. As is the case for some sRNAs expressed in other bacteria, it is likely that other mechanisms of action are used by S. aureus sRNA such as molecular mimicry (e.g. the 6S RNA) or binding to regulatory proteins. The number of sRNAs identified in S. aureus has considerably increased in the past decade, up to 250 members, but the biological functions of most of them remain unknown. Some sRNAs that are expressed from mobile genetic elements can regulate target genes located on the core genome, as for SprD with Sbi, implying an efficient functional integration of the accessory genetic elements into the overall regulatory networks from the S. aureus genome required for virulence. The sRNAs expressed from the core genome probably are involved in wider biological functions. Most of the well characterized sRNAs act as fine-tuning regulators by repressing the translational level of only one gene (Table 1), but it is likely that they target other genes and that one gene is regulated by different sRNAs. The identification of their molecular targets becomes a critical step to further understand their roles in bacterial homeostasis and pathogenesis. Bioinformatics approaches based on the prediction of sRNA base pairing within the TIS of mRNAs allowed identifying antisense targets of some sRNAs. However, these approaches often lead to false positive predictions and do not highlight the interactions outside the TIS that are not uncommon in S. aureus [44] and also in other bacterial species. In rare cases, quantitative proteomics and microarray analyses of sRNA mutant strains have allowed the identification of target genes, but these genetic approaches are not well suited to detect the primary targets of the sRNAs involved in broad regulatory networks, such as RNAIII, which regulates the expression of the Rot transcription factor. Until now, few global regulatory sRNAs were identified. The identification of new sRNAs that have an impact on the regulatory network control and the characterization of mechanisms that allow them to connect environmental responses to other cellular processes are future challenges. In S. aureus, the characterization of sRNA functions is complicated by the elevated genetic variability between the strains. Such a high variance in the expression of virulence and transcription factors among the S. aureus strains makes it difficult to generalize the functional impacts of a given sRNA to all the S. aureus strains. The characterization of the input of these sRNAs in global gene expression will provide a better understanding of the processes allowing the extraordinary adaptation of S. aureus in its various environments and its elevated pathogenicity in humans and animals. It should provide fundamental insights for potential therapeutic applications in using some of these sRNAs as early diagnostic markers and putative drug targets. Other future challenges will be to comprehend the contribution of S. aureus sRNAs during the various steps of the infectious process, host–pathogen interactions, colonization, spread, and antibiotic resistance. To tackle these ambitious goals, it will require developing elegant technologies in living animals to analyze the implications of the S. aureus RNome during infection.
Tab. 1. Cis- and trans-acting sRNAs, their corresponding mRNA targets, and physiological consequences. 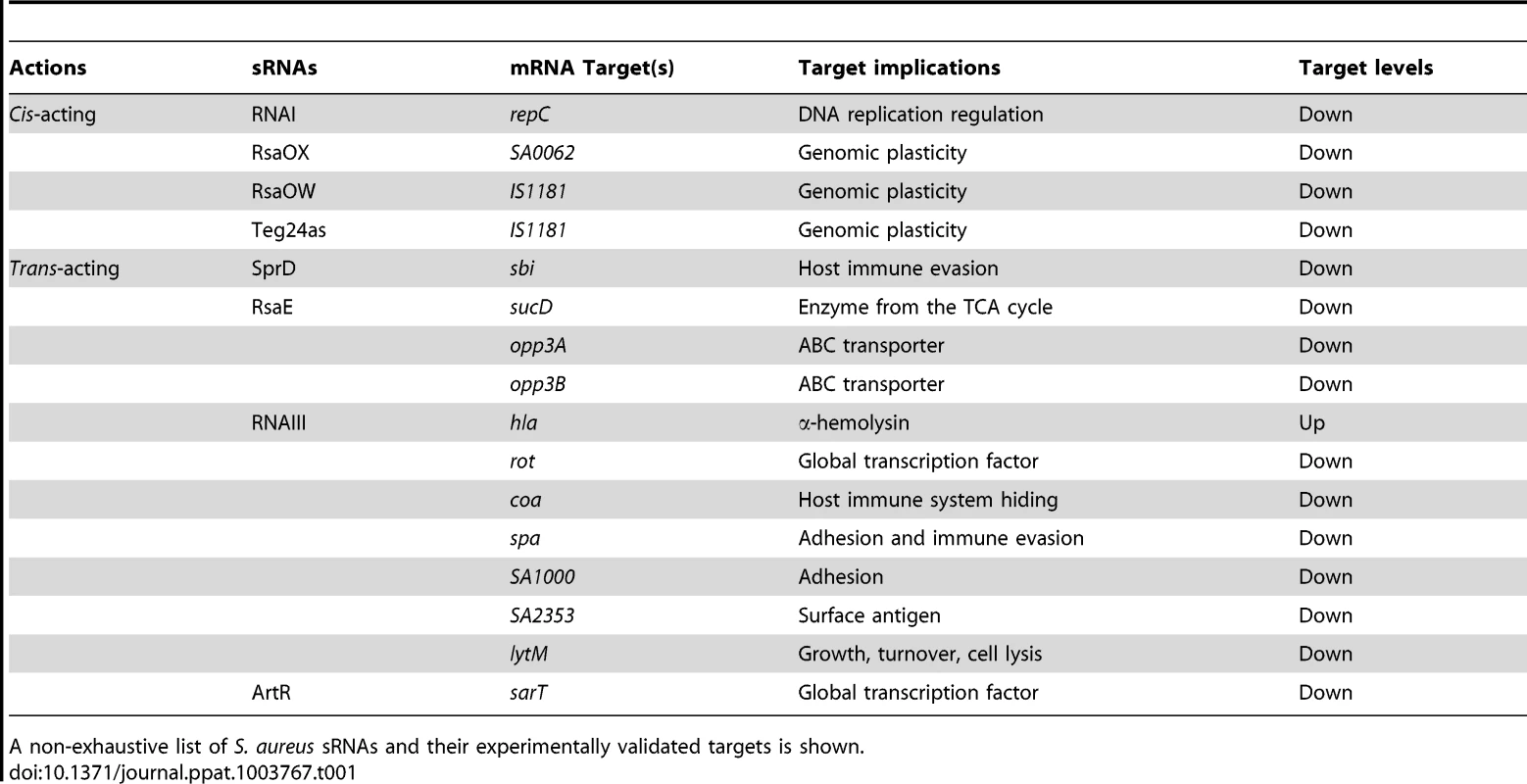
A non-exhaustive list of S. aureus sRNAs and their experimentally validated targets is shown.
Zdroje
1. DavidMZ, DaumRS (2010) Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23 : 616–687.
2. XuSX, McCormickJK (2012) Staphylococcal superantigens in colonization and disease. Front Cell Infect Microbiol 2 : 52.
3. MorrisonJM, MillerEW, BensonMA, AlonzoF3rd, YoongP, et al. (2012) Characterization of SSR42, a novel virulence factor regulatory RNA that contributes to the pathogenesis of a Staphylococcus aureus USA300 representative. J Bacteriol 194 : 2924–2938.
4. RouxCM, DeMuthJP, DunmanPM (2011) Characterization of components of the Staphylococcus aureus mRNA degradosome holoenzyme-like complex. J Bacteriol 193 : 5520–5526.
5. LasaI, Toledo-AranaA, DobinA, VillanuevaM, de los MozosIR, et al. (2011) Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci U S A 108 : 20172–20177.
6. FeldenB, VandeneschF, BoulocP, RombyP (2011) The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog 7: e1002006 doi:10.1371/journal.ppat.1002006
7. RomillyC, CaldelariI, ParmentierD, LioliouE, RombyP, et al. (2012) Current knowledge on regulatory RNAs and their machineries in Staphylococcus aureus. RNA Biol 9 : 402–413.
8. Abu-QatousehLF, ChinniSV, SeggewissJ, ProctorRA, BrosiusJ, et al. (2010) Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J Mol Med (Berl) 88 : 565–575.
9. AndersonKL, RobertsC, DiszT, VonsteinV, HwangK, et al. (2006) Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol 188 : 6739–6756.
10. BeaumeM, HernandezD, FarinelliL, DeluenC, LinderP, et al. (2010) Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One 5: e10725 doi:10.1371/journal.pone.0010725
11. BohnC, RigoulayC, ChabelskayaS, SharmaCM, MarchaisA, et al. (2010) Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res 38 : 6620–6636.
12. GeissmannT, ChevalierC, CrosMJ, BoissetS, FechterP, et al. (2009) A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res 37 : 7239–7257.
13. NovickRP, IordanescuS, ProjanSJ, KornblumJ, EdelmanI (1989) pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell 59 : 395–404.
14. NovickRP, RossHF, ProjanSJ, KornblumJ, KreiswirthB, et al. (1993) Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. Embo J 12 : 3967–3975.
15. PichonC, FeldenB (2005) Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc Natl Acad Sci U S A 102 : 14249–14254.
16. GottesmanS, StorzG (2011) Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3: a003798.
17. WatersLS, StorzG (2009) Regulatory RNAs in bacteria. Cell 136 : 615–628.
18. StorzG, VogelJ, WassarmanKM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43 : 880–891.
19. PapenfortK, VogelJ (2010) Regulatory RNA in bacterial pathogens. Cell Host Microbe 8 : 116–127.
20. RombyP, VandeneschF, WagnerEG (2006) The role of RNAs in the regulation of virulence-gene expression. Curr Opin Microbiol 9 : 229–236.
21. GaballaA, AntelmannH, AguilarC, KhakhSK, SongKB, et al. (2008) The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A 105 : 11927–11932.
22. MandinP, RepoilaF, VergassolaM, GeissmannT, CossartP (2007) Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res 35 : 962–974.
23. NielsenJS, OlsenAS, BondeM, Valentin-HansenP, KallipolitisBH (2008) Identification of a sigma B-dependent small noncoding RNA in Listeria monocytogenes. J Bacteriol 190 : 6264–6270.
24. Toledo-AranaA, DussurgetO, NikitasG, SestoN, Guet-RevilletH, et al. (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459 : 950–956.
25. FozoEM, HemmMR, StorzG (2008) Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev 72 : 579–589.
26. FozoEM, MakarovaKS, ShabalinaSA, YutinN, KooninEV, et al. (2010) Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res 38 : 3743–3759.
27. SayedN, JousselinA, FeldenB (2011) A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat Struct Mol Biol 19 : 105–112.
28. SayedN, Nonin-LecomteS, RetyS, FeldenB (2012) Functional and structural insights of a Staphylococcus aureus apoptotic-like membrane peptide from a toxin-antitoxin module. J Biol Chem 287 : 43454–43463.
29. GreenfieldTJ, EhliE, KirshenmannT, FranchT, GerdesK, et al. (2000) The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol Microbiol 37 : 652–660.
30. DulebohnD, ChoyJ, SundermeierT, OkanN, KarzaiAW (2007) Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry 46 : 4681–4693.
31. GilletR, FeldenB (2001) Emerging views on tmRNA-mediated protein tagging and ribosome rescue. Mol Microbiol 42 : 879–885.
32. HaebelPW, GutmannS, BanN (2004) Dial tm for rescue: tmRNA engages ribosomes stalled on defective mRNAs. Curr Opin Struct Biol 14 : 58–65.
33. KeilerKC (2008) Biology of trans-translation. Annu Rev Microbiol 62 : 133–151.
34. LiuY, WuN, DongJ, GaoY, ZhangX, et al. (2010) SsrA (tmRNA) acts as an antisense RNA to regulate Staphylococcus aureus pigment synthesis by base pairing with crtMN mRNA. FEBS Lett 584 : 4325–4329.
35. NovickRP, GeisingerE (2008) Quorum sensing in staphylococci. Annu Rev Genet 42 : 541–564.
36. BenitoY, KolbFA, RombyP, LinaG, EtienneJ, et al. (2000) Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 6 : 668–679.
37. KregerAS, KimKS, ZaboretzkyF, BernheimerAW (1971) Purification and properties of staphylococcal delta hemolysin. Infect Immun 3 : 449–465.
38. MellorIR, ThomasDH, SansomMS (1988) Properties of ion channels formed by Staphylococcus aureus delta-toxin. Biochim Biophys Acta 942 : 280–294.
39. VerdonJ, BerjeaudJM, LacombeC, HechardY (2008) Characterization of anti-Legionella activity of warnericin RK and delta-lysin I from Staphylococcus warneri. Peptides 29 : 978–984.
40. MorfeldtE, TaylorD, von GabainA, ArvidsonS (1995) Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. Embo J 14 : 4569–4577.
41. LiuY, MuC, YingX, LiW, WuN, et al. (2011) RNAIII activates map expression by forming an RNA-RNA complex in Staphylococcus aureus. FEBS Lett 585 : 899–905.
42. ChavakisT, HussainM, KanseSM, PetersG, BretzelRG, et al. (2002) Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat Med 8 : 687–693.
43. BoissetS, GeissmannT, HuntzingerE, FechterP, BendridiN, et al. (2007) Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21 : 1353–1366.
44. ChevalierC, BoissetS, RomillyC, MasquidaB, FechterP, et al. (2010) Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog 6: e1000809 doi:10.1371/journal.ppat.1000809
45. GeisingerE, AdhikariRP, JinR, RossHF, NovickRP (2006) Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 61 : 1038–1048.
46. LinkTM, Valentin-HansenP, BrennanRG (2009) Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A 106 : 19292–19297.
47. SchumacherMA, PearsonRF, MollerT, Valentin-HansenP, BrennanRG (2002) Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. Embo J 21 : 3546–3556.
48. BandyraKJ, SaidN, PfeifferV, GornaMW, VogelJ, et al. (2012) The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell 47 : 943–953.
49. UrbanJH, VogelJ (2007) Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res 35 : 1018–1037.
50. JousselinA, MetzingerL, FeldenB (2009) On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol 17 : 399–405.
51. BohnC, RigoulayC, BoulocP (2007) No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol 7 : 10.
52. HuntzingerE, BoissetS, SaveanuC, BenitoY, GeissmannT, et al. (2005) Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. Embo J 24 : 824–835.
53. NielsenJS, ChristiansenMH, BondeM, GottschalkS, FreesD, et al. (2011) Searching for small sigmaB-regulated genes in Staphylococcus aureus. Arch Microbiol 193 : 23–34.
54. LiuY, WuN, DongJ, GaoY, ZhangX, et al. (2010) Hfq is a global regulator that controls the pathogenicity of Staphylococcus aureus. PLoS One 5: e13069 doi:10.1371/journal.pone.0013069
55. CastroSL, Nelman-GonzalezM, NickersonCA, OttCM (2011) Induction of attachment-independent biofilm formation and repression of Hfq expression by low-fluid-shear culture of Staphylococcus aureus. Appl Environ Microbiol 77 : 6368–6378.
56. SalimNN, FanerMA, PhilipJA, FeigAL (2012) Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res 40 : 8021–8032.
57. ChevalierC, HuntzingerE, FechterP, BoissetS, VandeneschF, et al. (2008) Staphylococcus aureus endoribonuclease III purification and properties. Methods Enzymol 447 : 309–327.
58. HerskovitzMA, BechhoferDH (2000) Endoribonuclease RNase III is essential in Bacillus subtilis. Mol Microbiol 38 : 1027–1033.
59. SteadMB, MarshburnS, MohantyBK, MitraJ, Pena CastilloL, et al. (2011) Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res 39 : 3188–3203.
60. LioliouE, SharmaCM, CaldelariI, HelferAC, FechterP, et al. (2012) Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet 8: e1002782 doi:10.1371/journal.pgen.1002782
61. LiuY, DongJ, WuN, GaoY, ZhangX, et al. (2011) The production of extracellular proteins is regulated by ribonuclease III via two different pathways in Staphylococcus aureus. PLoS One 6: e20554 doi:10.1371/journal.pone.0020554
62. RomillyC, ChevalierC, MarziS, MasquidaB, GeissmannT, et al. (2012) Loop-loop interactions involved in antisense regulation are processed by the endoribonuclease III in Staphylococcus aureus. RNA Biol 9 : 1461–1472.
63. LioliouE, SharmaCM, AltuviaY, CaldelariI, RomillyC, et al. (2013) In vivo mapping of RNA-RNA interactions in Staphylococcus aureus using the endoribonuclease III. Methods 63 : 135–143.
64. LasaI, Toledo-AranaA, GingerasTR (2012) An effort to make sense of antisense transcription in bacteria. RNA Biol 9 : 1039–1044.
65. WurtzelO, SestoN, MellinJR, KarunkerI, EdelheitS, et al. (2012) Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol 8 : 583.
66. SestoN, WurtzelO, ArchambaudC, SorekR, CossartP (2013) The excludon: a new concept in bacterial antisense RNA-mediated gene regulation. Nat Rev Microbiol 11 : 75–82.
67. CarpousisAJ (2007) The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol 61 : 71–87.
68. Lehnik-HabrinkM, NewmanJ, RotheFM, SolovyovaAS, RodriguesC, et al. (2011) RNase Y in Bacillus subtilis: a Natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J Bacteriol 193 : 5431–5441.
69. EvenS, PellegriniO, ZigL, LabasV, VinhJ, et al. (2005) Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res 33 : 2141–2152.
70. OunS, RedderP, DidierJP, FrancoisP, CorvagliaAR, et al. (2013) The CshA DEAD-box RNA helicase is important for quorum sensing control in Staphylococcus aureus. RNA Biol 10 : 157–165.
71. RedderP, LinderP (2012) DEAD-box RNA helicases in gram-positive RNA decay. Methods Enzymol 511 : 369–383.
72. DurandS, GiletL, BessieresP, NicolasP, CondonC (2012) Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet 8: e1002520 doi:10.1371/journal.pgen.1002520
73. Lehnik-HabrinkM, LewisRJ, MaderU, StulkeJ (2012) RNA degradation in Bacillus subtilis: an interplay of essential endo - and exoribonucleases. Mol Microbiol 84 : 1005–1017.
74. KaitoC, KurokawaK, MatsumotoY, TeraoY, KawabataS, et al. (2005) Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol Microbiol 56 : 934–944.
75. MarincolaG, SchaferT, BehlerJ, BernhardtJ, OhlsenK, et al. (2012) RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol Microbiol 85 : 817–832.
76. BischoffM, DunmanP, KormanecJ, MacapagalD, MurphyE, et al. (2004) Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J Bacteriol 186 : 4085–4099.
77. Pane-FarreJ, JonasB, ForstnerK, EngelmannS, HeckerM (2006) The sigmaB regulon in Staphylococcus aureus and its regulation. Int J Med Microbiol 296 : 237–258.
78. ShawLN, AishJ, DavenportJE, BrownMC, LithgowJK, et al. (2006) Investigations into sigmaB-modulated regulatory pathways governing extracellular virulence determinant production in Staphylococcus aureus. J Bacteriol 188 : 6070–6080.
79. JonssonIM, ArvidsonS, FosterS, TarkowskiA (2004) Sigma factor B and RsbU are required for virulence in Staphylococcus aureus-induced arthritis and sepsis. Infect Immun 72 : 6106–6111.
80. Pane-FarreJ, JonasB, HardwickSW, GronauK, LewisRJ, et al. (2009) Role of RsbU in controlling SigB activity in Staphylococcus aureus following alkaline stress. J Bacteriol 191 : 2561–2573.
81. SennMM, GiachinoP, HomerovaD, SteinhuberA, StrassnerJ, et al. (2005) Molecular analysis and organization of the sigmaB operon in Staphylococcus aureus. J Bacteriol 187 : 8006–8019.
82. LauderdaleKJ, BolesBR, CheungAL, HorswillAR (2009) Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun 77 : 1623–1635.
83. HorsburghMJ, WhartonSJ, CoxAG, InghamE, PeacockS, et al. (2002) MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol Microbiol 44 : 1269–1286.
84. SeidlK, StuckiM, RueggM, GoerkeC, WolzC, et al. (2006) Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother 50 : 1183–1194.
85. ZhuY, NandakumarR, SadykovMR, MadayiputhiyaN, LuongTT, et al. (2011) RpiR homologues may link Staphylococcus aureus RNAIII synthesis and pentose phosphate pathway regulation. J Bacteriol 193 : 6187–6196.
86. SomervilleGA, ChausseeMS, MorganCI, FitzgeraldJR, DorwardDW, et al. (2002) Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun 70 : 6373–6382.
87. SomervilleGA, Said-SalimB, WickmanJM, RaffelSJ, KreiswirthBN, et al. (2003) Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect Immun 71 : 4724–4732.
88. Borezee-DurantE, HironA, PiardJC, JuillardV (2009) Dual role of the oligopeptide permease Opp3 during growth of Staphylococcus aureus in milk. Appl Environ Microbiol 75 : 3355–3357.
89. HironA, Borezee-DurantE, PiardJC, JuillardV (2007) Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J Bacteriol 189 : 5119–5129.
90. ZhuY, XiongYQ, SadykovMR, FeyPD, LeiMG, et al. (2009) Tricarboxylic acid cycle-dependent attenuation of Staphylococcus aureus in vivo virulence by selective inhibition of amino acid transport. Infect Immun 77 : 4256–4264.
91. PriestNK, RudkinJK, FeilEJ, van den ElsenJM, CheungA, et al. (2012) From genotype to phenotype: can systems biology be used to predict Staphylococcus aureus virulence? Nat Rev Microbiol 10 : 791–797.
92. OscarssonJ, HarlosC, ArvidsonS (2005) Regulatory role of proteins binding to the spa (protein A) and sarS (staphylococcal accessory regulator) promoter regions in Staphylococcus aureus NTCC 8325-4. Int J Med Microbiol 295 : 253–266.
93. OscarssonJ, KanthA, Tegmark-WisellK, ArvidsonS (2006) SarA is a repressor of hla (alpha-hemolysin) transcription in Staphylococcus aureus: its apparent role as an activator of hla in the prototype strain NCTC 8325 depends on reduced expression of sarS. J Bacteriol 188 : 8526–8533.
94. LiD, CheungA (2008) Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect Immun 76 : 1068–1075.
95. JelsbakL, HemmingsenL, DonatS, OhlsenK, BoyeK, et al. (2010) Growth phase-dependent regulation of the global virulence regulator Rot in clinical isolates of Staphylococcus aureus. Int J Med Microbiol 300 : 229–236.
96. Said-SalimB, DunmanPM, McAleeseFM, MacapagalD, MurphyE, et al. (2003) Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 185 : 610–619.
97. SchmidtKA, MannaAC, GillS, CheungAL (2001) SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect Immun 69 : 4749–4758.
98. XueT, ZhangX, SunH, SunB (2013) ArtR, a novel sRNA of Staphylococcus aureus, regulates alpha-toxin expression by targeting the 5′ UTR of sarT mRNA. Med Microbiol Immunol E-pub ahead of print. doi: 10.1007/s00430-013-0307-0
99. KaitoC, SaitoY, NaganoG, IkuoM, OmaeY, et al. (2011) Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog 7: e1001267 doi:10.1371/journal.ppat.1001267
100. KaitoC, SaitoY, IkuoM, OmaeY, MaoH, et al. (2013) Mobile Genetic Element SCCmec-encoded psm-mec RNA Suppresses Translation of agrA and Attenuates MRSA Virulence. PLoS Pathog 9: e1003269 doi:10.1371/journal.ppat.1003269
101. NovickRP, ChristieGE, PenadesJR (2010) The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol 8 : 541–551.
102. ChabelskayaS, GaillotO, FeldenB (2010) A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog 6: e1000927 doi:10.1371/journal.ppat.1000927
103. ZhangL, JacobssonK, VasiJ, LindbergM, FrykbergL (1998) A second IgG-binding protein in Staphylococcus aureus. Microbiology 144 : 985–991.
104. HauptK, ReuterM, van den ElsenJ, BurmanJ, HalbichS, et al. (2008) The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with host complement Factor H and C3b. PLoS Pathog 4: e1000250 doi:10.1371/journal.ppat.1000250
105. LamersRP, StinnettJW, MuthukrishnanG, ParkinsonCL, ColeAM (2011) Evolutionary analyses of Staphylococcus aureus identify genetic relationships between nasal carriage and clinical isolates. PLoS One 6: e16426 doi:10.1371/journal.pone.0016426
106. BurianM, WolzC, GoerkeC (2010) Regulatory adaptation of Staphylococcus aureus during nasal colonization of humans. PLoS One 5: e10040 doi:10.1371/journal.pone.0010040
107. CheungAL, BayerAS, ZhangG, GreshamH, XiongYQ (2004) Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol 40 : 1–9.
108. GoerkeC, CampanaS, BayerMG, DoringG, BotzenhartK, et al. (2000) Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect Immun 68 : 1304–1311.
109. TraberKE, LeeE, BensonS, CorriganR, CanteraM, et al. (2008) agr function in clinical Staphylococcus aureus isolates. Microbiology 154 : 2265–2274.
110. ShopsinB, Drlica-WagnerA, MathemaB, AdhikariRP, KreiswirthBN, et al. (2008) Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J Infect Dis 198 : 1171–1174.
111. SongJ, LaysC, VandeneschF, BenitoY, BesM, et al. (2012) The expression of small regulatory RNAs in clinical samples reflects the different life styles of Staphylococcus aureus in colonization vs. infection. PLoS One 7: e37294 doi:10.1371/journal.pone.0037294
112. von EiffC, BeckerK, MachkaK, StammerH, PetersG (2001) Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med 344 : 11–16.
113. WertheimHF, VosMC, OttA, van BelkumA, VossA, et al. (2004) Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364 : 703–705.
114. von EiffC, HeilmannC, ProctorRA, WoltzC, PetersG, et al. (1997) A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J Bacteriol 179 : 4706–4712.
115. ProctorRA, von EiffC, KahlBC, BeckerK, McNamaraP, et al. (2006) Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4 : 295–305.
116. VaudauxP, KelleyWL, LewDP (2006) Staphylococcus aureus small colony variants: difficult to diagnose and difficult to treat. Clin Infect Dis 43 : 968–970.
117. von EiffC, BeckerK, MetzeD, LubritzG, HockmannJ, et al. (2001) Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with darier's disease. Clin Infect Dis 32 : 1643–1647.
118. LannergardJ, von EiffC, SanderG, CordesT, SeggewissJ, et al. (2008) Identification of the genetic basis for clinical menadione-auxotrophic small-colony variant isolates of Staphylococcus aureus. Antimicrob Agents Chemother 52 : 4017–4022.
119. ChatterjeeI, KriegeskorteA, FischerA, DeiwickS, TheimannN, et al. (2008) In vivo mutations of thymidylate synthase (encoded by thyA) are responsible for thymidine dependency in clinical small-colony variants of Staphylococcus aureus. J Bacteriol 190 : 834–842.
120. SchmidtKA, MannaAC, CheungAL (2003) SarT influences sarS expression in Staphylococcus aureus. Infect Immun 71 : 5139–5148.
121. GaoJ, StewartGC (2004) Regulatory elements of the Staphylococcus aureus protein A (Spa) promoter. J Bacteriol 186 : 3738–3748.
122. ReyesD, AndreyDO, MonodA, KelleyWL, ZhangG, et al. (2011) Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J Bacteriol 193 : 6020–6031.
123. MannaAC, CheungAL (2003) sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect Immun 71 : 343–353.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete InfectionsČlánek Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?Článek Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication FidelityČlánek CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial PathogensČlánek The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite PersistenceČlánek The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 12- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Host Susceptibility Factors to Bacterial Infections in Type 2 Diabetes
- LysM Effectors: Secreted Proteins Supporting Fungal Life
- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Innate Lymphoid Cells: New Players in IL-17-Mediated Antifungal Immunity
- Cytoplasmic Viruses: Rage against the (Cellular RNA Decay) Machine
- Balancing Stability and Flexibility within the Genome of the Pathogen
- The Evolution of Transmissible Prions: The Role of Deformed Templating
- Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete Infections
- Host Defense via Symbiosis in
- Regulatory Circuits That Enable Proliferation of the Fungus in a Mammalian Host
- Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?
- Burning Down the House: Cellular Actions during Pyroptosis
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial Pathogens
- Combining Regulatory T Cell Depletion and Inhibitory Receptor Blockade Improves Reactivation of Exhausted Virus-Specific CD8 T Cells and Efficiently Reduces Chronic Retroviral Loads
- Shaping Up for Battle: Morphological Control Mechanisms in Human Fungal Pathogens
- Identification of the Virulence Landscape Essential for Invasion of the Human Colon
- Nodular Inflammatory Foci Are Sites of T Cell Priming and Control of Murine Cytomegalovirus Infection in the Neonatal Lung
- Hepatitis B Virus Disrupts Mitochondrial Dynamics: Induces Fission and Mitophagy to Attenuate Apoptosis
- Mycobacterial MazG Safeguards Genetic Stability Housecleaning of 5-OH-dCTP
- Systematic MicroRNA Analysis Identifies ATP6V0C as an Essential Host Factor for Human Cytomegalovirus Replication
- Placental Syncytium Forms a Biophysical Barrier against Pathogen Invasion
- The CD8-Derived Chemokine XCL1/Lymphotactin Is a Conformation-Dependent, Broad-Spectrum Inhibitor of HIV-1
- Cyclin A Degradation by Primate Cytomegalovirus Protein pUL21a Counters Its Innate Restriction of Virus Replication
- Genome-Wide RNAi Screen Identifies Novel Host Proteins Required for Alphavirus Entry
- Zinc Sequestration: Arming Phagocyte Defense against Fungal Attack
- The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite Persistence
- Biphasic Euchromatin-to-Heterochromatin Transition on the KSHV Genome Following Infection
- The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
- HIV-1 Vpr Accelerates Viral Replication during Acute Infection by Exploitation of Proliferating CD4 T Cells
- A Human Torque Teno Virus Encodes a MicroRNA That Inhibits Interferon Signaling
- The ArlRS Two-Component System Is a Novel Regulator of Agglutination and Pathogenesis
- An In-Depth Comparison of Latent HIV-1 Reactivation in Multiple Cell Model Systems and Resting CD4+ T Cells from Aviremic Patients
- Enterohemorrhagic Hemolysin Employs Outer Membrane Vesicles to Target Mitochondria and Cause Endothelial and Epithelial Apoptosis
- Overcoming Antigenic Diversity by Enhancing the Immunogenicity of Conserved Epitopes on the Malaria Vaccine Candidate Apical Membrane Antigen-1
- The Type-Specific Neutralizing Antibody Response Elicited by a Dengue Vaccine Candidate Is Focused on Two Amino Acids of the Envelope Protein
- Tmprss2 Is Essential for Influenza H1N1 Virus Pathogenesis in Mice
- Signatures of Pleiotropy, Economy and Convergent Evolution in a Domain-Resolved Map of Human–Virus Protein–Protein Interaction Networks
- Interference with the Host Haemostatic System by Schistosomes
- RocA Truncation Underpins Hyper-Encapsulation, Carriage Longevity and Transmissibility of Serotype M18 Group A Streptococci
- Gene Fitness Landscapes of at Important Stages of Its Life Cycle
- Phagocytosis Escape by a Protein That Connects Complement and Coagulation Proteins at the Bacterial Surface
- t Is a Structurally Novel Crohn's Disease-Associated Superantigen
- An Increasing Danger of Zoonotic Orthopoxvirus Infections
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
- Transcriptional Analysis of Murine Macrophages Infected with Different Strains Identifies Novel Regulation of Host Signaling Pathways
- Serotonergic Chemosensory Neurons Modify the Immune Response by Regulating G-Protein Signaling in Epithelial Cells
- Genome-Wide Detection of Fitness Genes in Uropathogenic during Systemic Infection
- Induces an Unfolded Protein Response via TcpB That Supports Intracellular Replication in Macrophages
- Intestinal CD103+ Dendritic Cells Are Key Players in the Innate Immune Control of Infection in Neonatal Mice
- Emerging Functions for the RNome
- KSHV MicroRNAs Mediate Cellular Transformation and Tumorigenesis by Redundantly Targeting Cell Growth and Survival Pathways
- HrpA, an RNA Helicase Involved in RNA Processing, Is Required for Mouse Infectivity and Tick Transmission of the Lyme Disease Spirochete
- A Toxin-Antitoxin Module of Promotes Virulence in Mice
- Real-Time Imaging of the Intracellular Glutathione Redox Potential in the Malaria Parasite
- Hypoxia Inducible Factor Signaling Modulates Susceptibility to Mycobacterial Infection via a Nitric Oxide Dependent Mechanism
- Novel Strategies to Enhance Vaccine Immunity against Coccidioidomycosis
- Dual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic
- —What Makes the Species a Ubiquitous Human Fungal Pathogen?
- αvβ6- and αvβ8-Integrins Serve As Interchangeable Receptors for HSV gH/gL to Promote Endocytosis and Activation of Membrane Fusion
- -Induced Activation of EGFR Prevents Autophagy Protein-Mediated Killing of the Parasite
- Semen CD4 T Cells and Macrophages Are Productively Infected at All Stages of SIV infection in Macaques
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Host Defense via Symbiosis in
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



