-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Regulatory Circuits That Enable Proliferation of the Fungus in a Mammalian Host
article has not abstract
Published in the journal: . PLoS Pathog 9(12): e32767. doi:10.1371/journal.ppat.1003780
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003780Summary
article has not abstract
A Rich Fungal Microbiome within Us
The human body harbors trillions of microorganisms—a rich and diverse microbiota that plays key roles in human health and disease. All three domains of life—eukaryotes, archaea, and bacteria—are represented in this large assembly of microbes. Among eukaryotes, fungi are particularly prominent residents of the human body. For example, 101 species belonging to 85 fungal genera have been found in the oral cavity of healthy people [1]. Similarly, >200 fungal species (half of them novel or unnamed) representing at least 50 genera have been found in the murine gut [2]. By comparison, there are ∼150 bacterial species (or phylotypes) inhabiting the mouths of healthy individuals [3] and ∼500–1000 bacterial species in their intestines [4], [5]. Although the methodologies differ, as do the classification schemes, it seems that the taxonomical diversity of the fungal component of the microbiota may not be so different from its bacterial counterpart, even though the latter has been much more heavily catalogued. With the exception of a handful of species, however, very little is known about the biology of the members of the human “mycobiome” (i.e., fungal microbiome) and even less about the interactions that they establish with the human host.
In this review we focus on the most widely studied fungus that inhabits the human body, Candida albicans. Most, if not all, healthy adults carry this species asymptomatically in their gastrointestinal (GI) tract. Despite its status as a commensal microorganism, C. albicans is also a leading cause of mucosal disease in healthy hosts and systemic, life-threatening infections in individuals with debilitated immune systems, such as AIDS patients or people receiving chemotherapy. Not surprisingly, it is this “virulent” or “pathogenic” facet of C. albicans that has traditionally received the most attention from researchers. More recently, however, several laboratories have begun to explore the tactics that the fungus uses to proliferate in its more common niche, the mammalian gut. These studies have uncovered unexpected connections between the commensal and pathogenic lifestyles. We highlight some of these findings here.
Regulation of Nutrient Acquisition Is Pivotal for Colonizing the Mammalian Gut
The gut is a crowded environment. In the colon alone, C. albicans cohabits with ∼1011 microbial cells per milliliter of intestinal content [6]. Food resources may be plentiful but competition is obviously high (reviewed in [7]). The ability of C. albicans to colonize and proliferate in the GI tract has been studied in mice receiving antibiotics orally; while these animals are not natural hosts of C. albicans, they likely serve as reasonable proxies. Because transcription regulators are central elements within the gene network of any organism, the use of them as entry points to the dissection of this trait has proved an effective strategy. A recent screen carried with C. albicans in a mouse model of GI tract colonization [8] evaluated 77 transcription regulator mutants for their ability to endure in the intestine for several weeks after oral inoculation. This group of mutants was chosen because none of them showed overt defects under a wide variety of conditions in vitro [9], hence they would be candidates for regulating circuits needed specifically in vivo. The screen identified six transcription regulators required for C. albicans to colonize the murine gut, four of which (RTG1, RTG3, TYE7, and LYS144) controlled the expression of genes responsible for the acquisition and metabolism of nutrients, particularly carbon and nitrogen sources. A significant fraction of the target genes of these regulators (identified by full-genome chromatin immunoprecipitation experiments) are indeed upregulated in C. albicans cells growing in the mouse intestine [10]. It is noteworthy that none of the four regulators is required for the species to grow under standard laboratory conditions [9], which reinforces the notion that significant resources are devoted to simply obtain food in the GI tract. It is plausible that these stringent conditions are set, at least in part, by the presence of a competing gut microbiota; consistent with this idea, genes that enable murine gut colonization by intestinal microbes such as Citrobacter rodentium—a natural mouse pathogen—are not required to colonize the intestines of germ-free mice [11].
Managing Iron Toxicity in the Mammalian Gut
Iron is an essential nutrient for microorganisms but at high levels it can become extremely toxic. The abundance and availability of iron vary greatly in different locales of the human body: relatively high levels (∼10−4 M) are found in the GI tract as the majority of dietary iron is not absorbed [12], whereas the concentration in the bloodstream is orders of magnitude lower (∼10−24 M free Fe3+) [13]. C. albicans, an organism that typically inhabits the gut but that can also cross into the bloodstream, harbors a regulatory circuit composed of three transcription regulators (SFU1, SEF1, and HAP43) that controls the expression of iron uptake genes and iron utilization genes [14]. SFU1 is required to persist in the GI tract [14] and for resistance to iron toxicity in vitro [9], suggesting that the primary role of this circuit in the gut is to protect C. albicans from iron's noxious effects. SEF1, on the other hand, is required not only for the fungus to endure in the intestine but also for full virulence after bloodstream infection [14]. Thus, C. albicans uses a common circuitry to balance its need to uptake iron in one niche (i.e., in the bloodstream) and protect itself from its toxic effects in other locale (i.e., the GI tract).
In addition to circuits that govern carbon, nitrogen, and iron acquisition, gene products that function as adherence molecules [15] and in the detoxification of reactive oxygen species [16] have also been shown to play roles in gut colonization. In fact, ECE1 and SOD5, which influence adhesion and protection against reactive oxygen species, respectively, are among the most highly upregulated genes when C. albicans is growing in the GI tract compared to standard laboratory conditions [15], [16]. Host cells such as macrophages and neutrophils produce reactive oxygen species, raising the possibility that SOD5's role may be to defend the fungus from these molecules [16] during colonization of the GI tract. Other transcription regulators reported to play roles in gut colonization have been shown to be rather pleiotropic (e.g., EFG1 [17], CPH2 [10], and EFH1 [15]).
A Molecular Basis for the Interplay of Commensalism and Pathogenicity
Many systemic, life-threatening infections in humans are caused by the very same bacteria or fungi that compose their microbiota. That is, these opportunistic pathogens reside in the host as harmless commensals (typically on mucosal surfaces) but can cross the host's protective barriers and colonize internal organs causing serious disease. The GI tract, for instance, appears to be the ultimate source of the majority of deep-seated C. albicans infections [18], [19]. Do entirely different gene sets account for the seemingly disparate behaviors of C. albicans as commensal versus pathogen?
Observing phenotypes on a gene-by-gene basis provides a mixed answer. On the one hand, transcription regulators such as EFG1 [16], [17], SEF1 [14], RTG1, RTG3, and HMS1 [8] are necessary for full virulence in mouse models of systemic infection (i.e., as a pathogen) as well as for GI tract colonization (i.e., as a commensal). On the other hand, mutant strains lacking the regulators TEC1 or LYS14 show reduced virulence in models of systemic infection but display no obvious defect in colonizing the murine GI tract [8], [10], [20]. The converse is true for C. albicans strains lacking TYE7 or SFU1 or ectopically expressing EFH1 [8], [14], [15]. However, genes do not function in isolation, and a clearer picture emerges by considering the existing links among some of these regulators.
Five transcription regulators that displayed a significant phenotype “exclusively” in association with the host [8] form a tightly knit circuit (Figure 1). In this circuit, the regulators are connected with one another irrespective of whether they are required for GI-tract colonization, systemic infection, or both. This observation implies that significant portions of the “pathogenic” and “commensal” lifestyles are coupled and controlled by the same core circuitry. Thus, from a gene network perspective, commensalism and pathogenicity do not appear to represent fully independent traits but rather are intertwined. The close links between these two traits may reflect the natural history of C. albicans: its association with mammals is ancient [21] and the selection pressure on the fungus has likely been as a commensal organism. Thus, it is plausible that the functions employed by C. albicans to spread from the human gut into systemic infections rely on the same regulatory circuitry that evolved to enable growth in the host as a commensal organism. Consistent with this idea, some impairment in the host immune system is typically required for C. albicans to be fully pathogenic.
Fig. 1. Gene regulatory network directing C. albicans proliferation in a mammalian host. 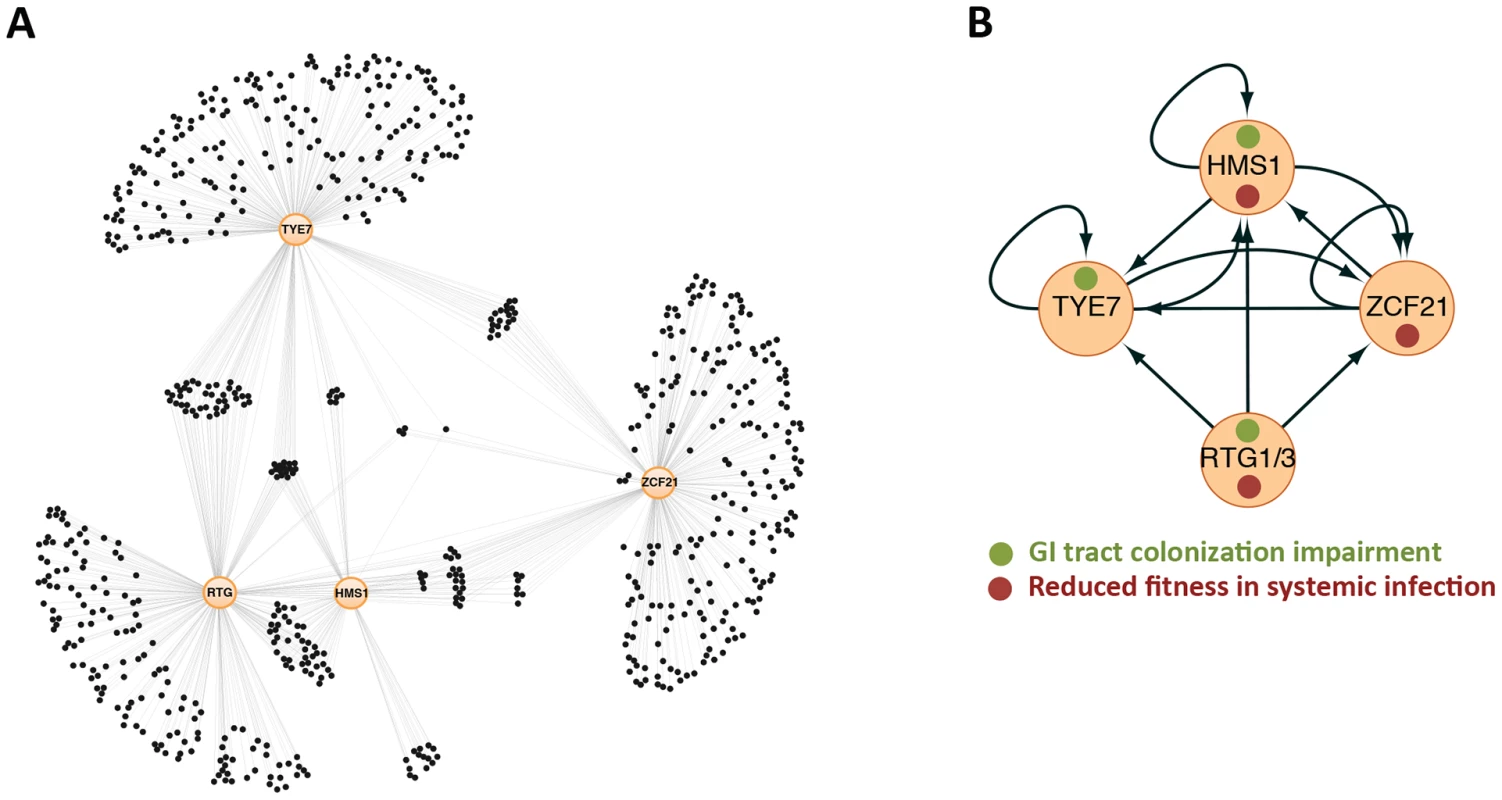
(A) Gene network composed of the transcription regulators RTG1/3, HMS1, ZCF21, and TYE7 (orange circles) and their target genes (black circles) as determined by full-genome chromatin immunoprecipitation. Thin lines indicate binding of the specified regulator to a target gene. About a quarter of the genes in the network (those in the middle) are targets shared by two or more regulators. (B) Relationships among the “master” regulators at the core of the network. Arrows represent protein-DNA interactions. Notice that the circuit displays multiple autoregulatory, feed-forward, and feedback loops. Adapted from [8]. Commensalism/Pathogenicity Transcriptional Circuit in C. albicans Has Structure in Common with Circuits Underlying Cell Differentiation
It has been shown that at the heart of the ability of C. albicans to proliferate in the mammalian host lies a highly interconnected transcriptional circuit composed of five transcription regulators (Figure 1). The distinctive feature of this circuit is the existence of multiple connections among all its components; that is the “master” regulators control one another (in addition to their target genes), and the target genes are typically bound by multiple master regulators. Thus, the circuit has many feed-forward and feedback loops. By contrast, many “textbook” genetic circuits—those that most scientists have grown used to—consist of simple, unidirectional relationships among their components, making their behavior more predictable. Because of the distinctly interwoven appearance of the C. albicans circuit, it is difficult to predict its behavior without extensive knowledge of the parameters (e.g., concentration of proteins, affinity constants, and the like). It is noteworthy that the overall topology resembles other circuits known to direct well-established cell differentiation processes such as white-opaque switching [22] and biofilm development [23] in C. albicans, filamentation in S. cerevisiae [24], and embryonic development in metazoans (e.g., see [25]). The fact that disparate functions in divergent organisms rely on a similar network architecture suggests that these transcriptional circuits may share common, underlying features. It is possible, for example, that the network structure enables the integration of multiple internal and external cues to then specify the precise pattern of target gene expression.
Conclusion
The study of opportunistic pathogens such as the fungus C. albicans, which can proliferate in disparate niches of the host either as a commensal or a pathogenic organism, allows us to genetically dissect the features of these two ways of life as well as to reveal the links between them. The results thus far suggest that there may be no clear distinction between the genetic circuitries employed during “harmless” proliferation in the gut or “disease-causing” growth after bloodstream infection. Rather, a single highly interconnected transcriptional circuit (one whose structure resembles others that orchestrate cell differentiation) may govern proliferation in all niches of the mammalian host.
Zdroje
1. GhannoumMA, JurevicRJ, MukherjeePK, CuiF, SikaroodiM, et al. (2010) Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6: e1000713 doi:10.1371/journal.ppat.1000713
2. IlievID, FunariVA, TaylorKD, NguyenQ, ReyesCN, et al. (2012) Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336 : 1314–1317.
3. AasJA, PasterBJ, StokesLN, OlsenI, DewhirstFE (2005) Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43 : 5721–5732.
4. Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486 : 207–214.
5. QinJ, LiR, RaesJ, ArumugamM, BurgdorfKS, et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464 : 59–65.
6. WalterJ, LeyR (2011) The human gut microbiome: ecology and recent evolutionary changes. Ann Rev Microbiol 65 : 411–429.
7. FischbachMA, SonnenburgJL (2011) Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10 : 336–347.
8. PérezJC, KumamotoCA, JohnsonAD (2013) Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol 11: e1001510 doi:10.1371/journal.pbio.1001510
9. HomannOR, DeaJ, NobleSM, JohnsonAD (2009) A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5: e1000783 doi:10.1371/journal.pgen.1000783
10. RosenbachA, DignardD, PierceJV, WhitewayM, KumamotoCA (2010) Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot Cell 9 : 1075–1086.
11. KamadaN, KimYG, ShamHP, VallanceBA, PuenteJL, et al. (2012) Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336 : 1325–1329.
12. MiretS, SimpsonRJ, McKieAT (2003) Physiology and molecular biology of dietary iron absorption. Annu Rev Nutr 23 : 283–301.
13. MartinRB, SavoryJ, BrownS, BertholfRL, WillsMR (1987) Transferrin binding of Al3+ and Fe3+. Clin Chem 33 : 405–407.
14. ChenC, PandeK, FrenchSD, TuchBB, NobleSM (2011) An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10 : 118–135.
15. WhiteSJ, RosenbachA, LephartP, NguyenD, BenjaminA, et al. (2007) Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog 3: e184 doi:10.1371/journal.ppat.0030184
16. PierceJV, DignardD, WhitewayM, KumamotoCA (2013) Normal adaptation of Candida albicans to the murine gastrointestinal tract requires Efg1p-dependent regulation of metabolic and host defense genes. Eukaryot Cell 12 : 37–49.
17. PierceJV, KumamotoCA (2012) Variation in Candida albicans EFG1 expression enables host-dependent changes in colonizing fungal populations. mBio 3: e00117–00112.
18. ColeGT, HalawaAA, AnaissieEJ (1996) The role of the gastrointestinal tract in hematogenous candidiasis: from the laboratory to the bedside. Clin Infect Dis 22 Suppl 2: S73–88.
19. MirandaLN, van der HeijdenIM, CostaSF, SousaAP, SienraRA, et al. (2009) Candida colonisation as a source for candidaemia. J Hosp Infect 72 : 9–16.
20. SchweizerA, RuppS, TaylorBN, RollinghoffM, SchroppelK (2000) The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol Microbiol 38 : 435–445.
21. OddsFC (1987) Candida infections: an overview. CRC Cr Rev Microbiol 15 : 1–5.
22. ZordanRE, MillerMG, GalgoczyDJ, TuchBB, JohnsonAD (2007) Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol 5: e256 doi:10.1371/journal.pbio.0050256
23. NobileCJ, FoxEP, NettJE, SorrellsTR, MitrovichQM, et al. (2012) A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148 : 126–138.
24. BornemanAR, Leigh-BellJA, YuH, BertoneP, GersteinM, et al. (2006) Target hub proteins serve as master regulators of development in yeast. Gene Dev 20 : 435–448.
25. NephS, StergachisAB, ReynoldsA, SandstromR, BorensteinE, et al. (2012) Circuitry and dynamics of human transcription factor regulatory networks. Cell 150 : 1274–1286.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete InfectionsČlánek Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?Článek Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication FidelityČlánek CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial PathogensČlánek The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite PersistenceČlánek The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 12- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Host Susceptibility Factors to Bacterial Infections in Type 2 Diabetes
- LysM Effectors: Secreted Proteins Supporting Fungal Life
- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Innate Lymphoid Cells: New Players in IL-17-Mediated Antifungal Immunity
- Cytoplasmic Viruses: Rage against the (Cellular RNA Decay) Machine
- Balancing Stability and Flexibility within the Genome of the Pathogen
- The Evolution of Transmissible Prions: The Role of Deformed Templating
- Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete Infections
- Host Defense via Symbiosis in
- Regulatory Circuits That Enable Proliferation of the Fungus in a Mammalian Host
- Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?
- Burning Down the House: Cellular Actions during Pyroptosis
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial Pathogens
- Combining Regulatory T Cell Depletion and Inhibitory Receptor Blockade Improves Reactivation of Exhausted Virus-Specific CD8 T Cells and Efficiently Reduces Chronic Retroviral Loads
- Shaping Up for Battle: Morphological Control Mechanisms in Human Fungal Pathogens
- Identification of the Virulence Landscape Essential for Invasion of the Human Colon
- Nodular Inflammatory Foci Are Sites of T Cell Priming and Control of Murine Cytomegalovirus Infection in the Neonatal Lung
- Hepatitis B Virus Disrupts Mitochondrial Dynamics: Induces Fission and Mitophagy to Attenuate Apoptosis
- Mycobacterial MazG Safeguards Genetic Stability Housecleaning of 5-OH-dCTP
- Systematic MicroRNA Analysis Identifies ATP6V0C as an Essential Host Factor for Human Cytomegalovirus Replication
- Placental Syncytium Forms a Biophysical Barrier against Pathogen Invasion
- The CD8-Derived Chemokine XCL1/Lymphotactin Is a Conformation-Dependent, Broad-Spectrum Inhibitor of HIV-1
- Cyclin A Degradation by Primate Cytomegalovirus Protein pUL21a Counters Its Innate Restriction of Virus Replication
- Genome-Wide RNAi Screen Identifies Novel Host Proteins Required for Alphavirus Entry
- Zinc Sequestration: Arming Phagocyte Defense against Fungal Attack
- The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite Persistence
- Biphasic Euchromatin-to-Heterochromatin Transition on the KSHV Genome Following Infection
- The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
- HIV-1 Vpr Accelerates Viral Replication during Acute Infection by Exploitation of Proliferating CD4 T Cells
- A Human Torque Teno Virus Encodes a MicroRNA That Inhibits Interferon Signaling
- The ArlRS Two-Component System Is a Novel Regulator of Agglutination and Pathogenesis
- An In-Depth Comparison of Latent HIV-1 Reactivation in Multiple Cell Model Systems and Resting CD4+ T Cells from Aviremic Patients
- Enterohemorrhagic Hemolysin Employs Outer Membrane Vesicles to Target Mitochondria and Cause Endothelial and Epithelial Apoptosis
- Overcoming Antigenic Diversity by Enhancing the Immunogenicity of Conserved Epitopes on the Malaria Vaccine Candidate Apical Membrane Antigen-1
- The Type-Specific Neutralizing Antibody Response Elicited by a Dengue Vaccine Candidate Is Focused on Two Amino Acids of the Envelope Protein
- Tmprss2 Is Essential for Influenza H1N1 Virus Pathogenesis in Mice
- Signatures of Pleiotropy, Economy and Convergent Evolution in a Domain-Resolved Map of Human–Virus Protein–Protein Interaction Networks
- Interference with the Host Haemostatic System by Schistosomes
- RocA Truncation Underpins Hyper-Encapsulation, Carriage Longevity and Transmissibility of Serotype M18 Group A Streptococci
- Gene Fitness Landscapes of at Important Stages of Its Life Cycle
- Phagocytosis Escape by a Protein That Connects Complement and Coagulation Proteins at the Bacterial Surface
- t Is a Structurally Novel Crohn's Disease-Associated Superantigen
- An Increasing Danger of Zoonotic Orthopoxvirus Infections
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
- Transcriptional Analysis of Murine Macrophages Infected with Different Strains Identifies Novel Regulation of Host Signaling Pathways
- Serotonergic Chemosensory Neurons Modify the Immune Response by Regulating G-Protein Signaling in Epithelial Cells
- Genome-Wide Detection of Fitness Genes in Uropathogenic during Systemic Infection
- Induces an Unfolded Protein Response via TcpB That Supports Intracellular Replication in Macrophages
- Intestinal CD103+ Dendritic Cells Are Key Players in the Innate Immune Control of Infection in Neonatal Mice
- Emerging Functions for the RNome
- KSHV MicroRNAs Mediate Cellular Transformation and Tumorigenesis by Redundantly Targeting Cell Growth and Survival Pathways
- HrpA, an RNA Helicase Involved in RNA Processing, Is Required for Mouse Infectivity and Tick Transmission of the Lyme Disease Spirochete
- A Toxin-Antitoxin Module of Promotes Virulence in Mice
- Real-Time Imaging of the Intracellular Glutathione Redox Potential in the Malaria Parasite
- Hypoxia Inducible Factor Signaling Modulates Susceptibility to Mycobacterial Infection via a Nitric Oxide Dependent Mechanism
- Novel Strategies to Enhance Vaccine Immunity against Coccidioidomycosis
- Dual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic
- —What Makes the Species a Ubiquitous Human Fungal Pathogen?
- αvβ6- and αvβ8-Integrins Serve As Interchangeable Receptors for HSV gH/gL to Promote Endocytosis and Activation of Membrane Fusion
- -Induced Activation of EGFR Prevents Autophagy Protein-Mediated Killing of the Parasite
- Semen CD4 T Cells and Macrophages Are Productively Infected at All Stages of SIV infection in Macaques
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Host Defense via Symbiosis in
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



