-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Toxin-Antitoxin Module of Promotes Virulence in Mice
Toxin-antitoxin (TA) modules are widely prevalent in both bacteria and archaea. Originally described as stabilizing elements of plasmids, TA modules are also widespread on bacterial chromosomes. These modules promote bacterial persistence in response to specific environmental stresses. So far, the possibility that TA modules could be involved in bacterial virulence has been largely neglected, but recent comparative genomic studies have shown that the presence of TA modules is significantly associated with the pathogenicity of bacteria. Using Salmonella as a model, we investigated whether TA modules help bacteria to overcome the stress conditions encountered during colonization, thereby supporting virulence in the host. By bioinformatics analyses, we found that the genome of the pathogenic bacterium Salmonella Typhimurium encodes at least 11 type II TA modules. Several of these are conserved in other pathogenic strains but absent from non-pathogenic species indicating that certain TA modules might play a role in Salmonella pathogenicity. We show that one TA module, hereafter referred to as sehAB, plays a transient role in virulence in perorally inoculated mice. The use of a transcriptional reporter demonstrated that bacteria in which sehAB is strongly activated are predominantly localized in the mesenteric lymph nodes. In addition, sehAB was shown to be important for the survival of Salmonella in these peripheral lymphoid organs. These data indicate that the transient activation of a type II TA module can bring a selective advantage favouring virulence and demonstrate that TA modules are engaged in Salmonella pathogenesis.
Published in the journal: . PLoS Pathog 9(12): e32767. doi:10.1371/journal.ppat.1003827
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003827Summary
Toxin-antitoxin (TA) modules are widely prevalent in both bacteria and archaea. Originally described as stabilizing elements of plasmids, TA modules are also widespread on bacterial chromosomes. These modules promote bacterial persistence in response to specific environmental stresses. So far, the possibility that TA modules could be involved in bacterial virulence has been largely neglected, but recent comparative genomic studies have shown that the presence of TA modules is significantly associated with the pathogenicity of bacteria. Using Salmonella as a model, we investigated whether TA modules help bacteria to overcome the stress conditions encountered during colonization, thereby supporting virulence in the host. By bioinformatics analyses, we found that the genome of the pathogenic bacterium Salmonella Typhimurium encodes at least 11 type II TA modules. Several of these are conserved in other pathogenic strains but absent from non-pathogenic species indicating that certain TA modules might play a role in Salmonella pathogenicity. We show that one TA module, hereafter referred to as sehAB, plays a transient role in virulence in perorally inoculated mice. The use of a transcriptional reporter demonstrated that bacteria in which sehAB is strongly activated are predominantly localized in the mesenteric lymph nodes. In addition, sehAB was shown to be important for the survival of Salmonella in these peripheral lymphoid organs. These data indicate that the transient activation of a type II TA module can bring a selective advantage favouring virulence and demonstrate that TA modules are engaged in Salmonella pathogenesis.
Authors Summary
Bacteria have the capacity to rapidly adapt to and survive ever-changing environments. This aptitude is essential for a foodborne pathogen that upon ingestion by a host, and in a short period of time, will switch from a free-living state in the contaminated food to a parasitic existence in a host. During this process, the pathogen has to face various destructive surroundings such as the gastric acid of the stomach, the antimicrobiotic activities of the intestinal milieu, or the host immune defences. This raises the question of how a pathogen achieves this rapid adaptation. Bacteria are capable of regulating their biochemical activity with the help of self-toxins. These toxins are normally associated with an antitoxin that limits their toxic activity. In response to unfavourable environmental conditions, the toxin is rapidly freed from the antitoxin and slows down the biological activity of the bacterium. This makes the bacterium less sensitive to harmful environments. In the present study, we investigated whether the bacterial pathogen Salmonella possesses similar self-protecting systems and if they are necessary for virulence. We found that these systems are present in pathogenic species of Salmonella and help the bacterium establish an infection.
Introduction
Prokaryotic genomes contain toxin–antitoxin (TA) loci that induce cell dormancy in response to various stresses [1], [2]. This is mediated by the toxin components that target essential cellular processes, such as DNA replication, mRNA stability or protein synthesis (for review see [2]). Five types of TA systems have been described. In type I and III, the antitoxin is a RNA molecule that either regulates toxin gene expression (type I) or forms a complex with the toxin protein and inhibits its activity (type III) [3]. The recently described type IV and V systems refer to protein-protein modules in which the antitoxin masks the toxin activity either by interfering with binding of the toxin to its target (type IV) [4] or by cleaving specifically the toxin mRNA (type V) [5]. In type II modules, toxin and antitoxin are proteins that are co-transcribed from an operon. By binding its cognate toxin, the antitoxin blocks the toxin activity. Very often, the antitoxin binds to a palindromic stretch within the promoter region and represses the transcription of the operon. Environmental conditions that favour the degradation of the labile antitoxin raise the level of free toxin and also relieve the expression inhibition of the TA locus. This regulation loop maintains a high level of free toxin as long as the conditions supporting the antitoxin degradation are sustained.
Type II TA modules are widely prevalent in prokaryotes. In 2009, a genomic analysis in 750 complete genomes of archaea and bacteria discovered previously unnoticed protein families that are homologous to toxins and antitoxins of known type II TA and highlighted their exceptional mobility [6]. A comprehensive search on 2181 prokaryotic genomes detected more than 10000 sequences that were grouped within toxin (12) and antitoxin (20) super-families [7]. These systems have been proposed to be beneficial in hostile conditions by favouring persistence [8]. However, the circumstances under which TA modules are activated and support bacterial persistence remain poorly understood.
A study published in 2005 established that TA modules are abundant in free-living prokaryotes but rare in obligate host-associated organisms [9]. It led to the conclusion that TA modules are stress-response elements that increase the fitness of free-living prokaryotes. However, more recent data reported the presence of TA modules in pathogenic intracellular bacteria [7] and a significant association with the pathogenicity of epidemic bacteria [10]. Mycobacterium tuberculosis contains more than 60 TA systems, while the saprophytic Mycobacterium smegmatis has only two [9]. Ricketsia spp., which are obligate intracellular pathogens, possesses up to 32 TA modules [7] and are capable of inducing host cell death in a TA-dependent manner [11]. Elsewhere it has been established that TA modules can promote the colonization of the mouse bladder or kidneys by an uropathogenic strain of Escherichia coli [12]. Hence, there is increasing evidence that TA modules are related to bacterial pathogenicity and involved in host-pathogen interactions.
Salmonellae are Gram-negative bacteria responsible for severe gastroenteritis and systemic infections in humans. These bacteria are widespread and commonly found in the intestinal tract of cold - and warm-blooded animals and in the environment where their association with protozoa such as amoebae is thought to favour persistence and transmission [13], [14]. Salmonellae must therefore have evolved mechanisms to rapidly adapt to the multitude of environmental conditions encountered either as free-living organism or as a pathogen in association with a host. The present study investigated the presence of type II TA loci in strains of Salmonella and their possible role in virulence.
Results
Identification of type II TA modules in Salmonella Typhimurium
Bioinformatics analysis using the RASTA-Bacteria webtool [15] and BLASTP of already identified toxins and antitoxins confirmed the presence of eleven type II TA modules in the Salmonella enterica subsp. enterica serovar Typhimurium (S. Typhimurium) genome, nine located on the chromosome and two in the virulence plasmid (pSLT) (Table 1). They belong to five distinct super-families of toxins (see Table 1) [16]. Two of these modules (STM2954.1N-STM2955.S, STM4032.2N-STM4033) have been previously identified in S. Typhimurium LT2 [9], while the vapBC locus (STM 3033-STM3034) has been shown to be a bona fide type II TA loci [17]. Of note, we could not establish any correspondence between six other TA modules described in the afore-mentioned study [9] and the other TA modules described herein.
Tab. 1. Identification and family distribution of <i>S.</i> Typhimurium TA loci. 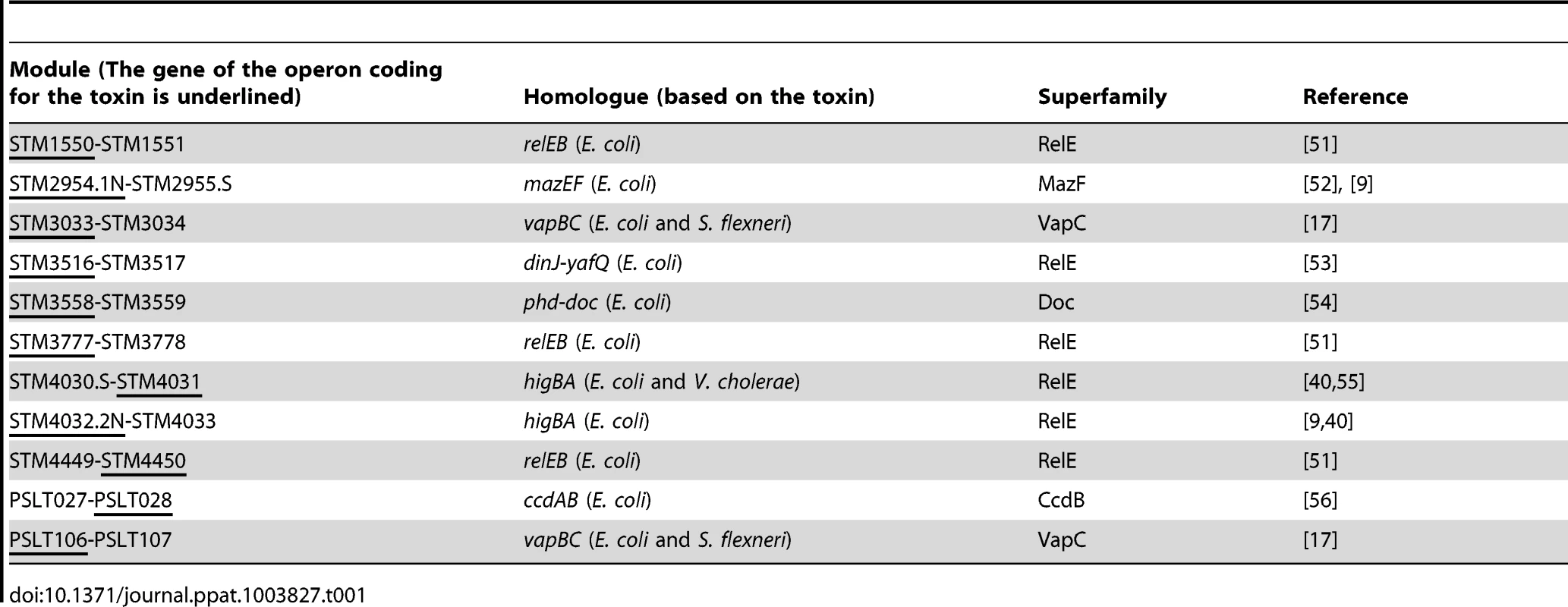
The genus Salmonella comprises two species, S. bongori and S. enterica. The majority of human infections are caused by various serovars of S. enterica while S. bongori does not cause disease in mammals [18]. We used BLASTP of S. Typhimurium chromosomal toxins and genomic context analyses to examine sequenced genomes of Salmonellae for homologs of these TA systems. Using an in-house R script [19], we performed a hierarchical clustering based on the Euclidean distance for the presence or absence of TA modules in the studied genomes as illustrated in the heatmap in Figure 1A. The clustering of bacteria in the horizontal dendrogram indicates the presence of two main clusters. The first cluster consists of serovars of S. enterica subspecies enterica, which are associated with warm-blooded host, that have between five and eight of the nine chromosomal TA modules found in S Typhimurium (Figure 1A). Interestingly, four of these TA modules are present in all serovars of S. enterica subspecies enterica. The second cluster comprises subspecies houtenae and arizonae of S. enterica, which infect cold-blooded animals and the non-pathogenic species S. bongori. The subspecies houtenae and arizonae present a low number of modules (two and four, respectively), while none are present in S. bongori. These analyses indicate that TA modules detected in S. Typhimurium are more prevalent in strains associated with warm-blooded hosts as compared to strains that infect cold-blooded animals. They also suggest a role in pathogenesis since these TA modules are absent from the non-pathogenic species S. bongori.
Fig. 1. Conservation of TA modules across the genus Salmonella. 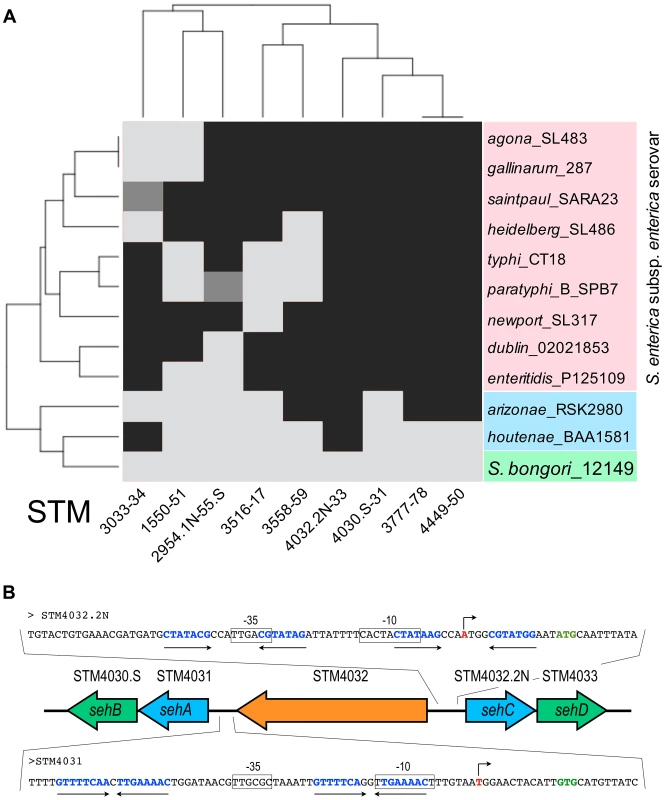
(A) The toxins of S. Typhimurium TA modules were subjected to BLASTP against various Salmonella strains. Serovars of S. enterica subsp. enterica are coloured in red. Arizonae and houtenae subspecies of S. enterica are coloured in blue. The non-pathogenic species bongori is coloured in green. All sequences were taken of NCBI and ColiBASE databases. Gene products were clustered according to their presence (black) or absence (light grey). Incomplete modules are indicated in dark grey. (B) Genetic organization of the STM4031-STM4030.S and STM4032.2N-STM4033 modules in S. Typhimurium chromosome. Toxin genes are coloured in blue and antitoxin genes in green. A single gene, STM4032 (in orange), separates the two TA modules. The promoter regions of the TA operons are shown as a blow-up. Putative transcriptional start sites are shown as broken arrows. Start sites of the first gene in the TA operons are shown in green bold letters. Putative −10 and −35 sequences are boxed. Palindromic sequences that are putative antitoxin binding sites are shown as opposing arrows. The toxin components of sehAB and sehCD are moderately toxic or non toxic for Salmonella grown in artificial media
To assess the putative functions of these modules, we expressed the predicted toxins in E. coli from an inducible promoter. The growth in Luria-Bertani (LB) broth of E. coli strains was measured over a period of 8 hours. A group of five proteins (group 1 in Figure 2A) exerted a strong bacteriostatic effect in E. coli. The expression of seven other putative toxins (group 2) only slightly attenuated bacterial growth, suggesting that these proteins are either not toxins or that their toxicity is limited in a heterologous expression system.
Fig. 2. Effect of toxins and antitoxins on bacterial growth. 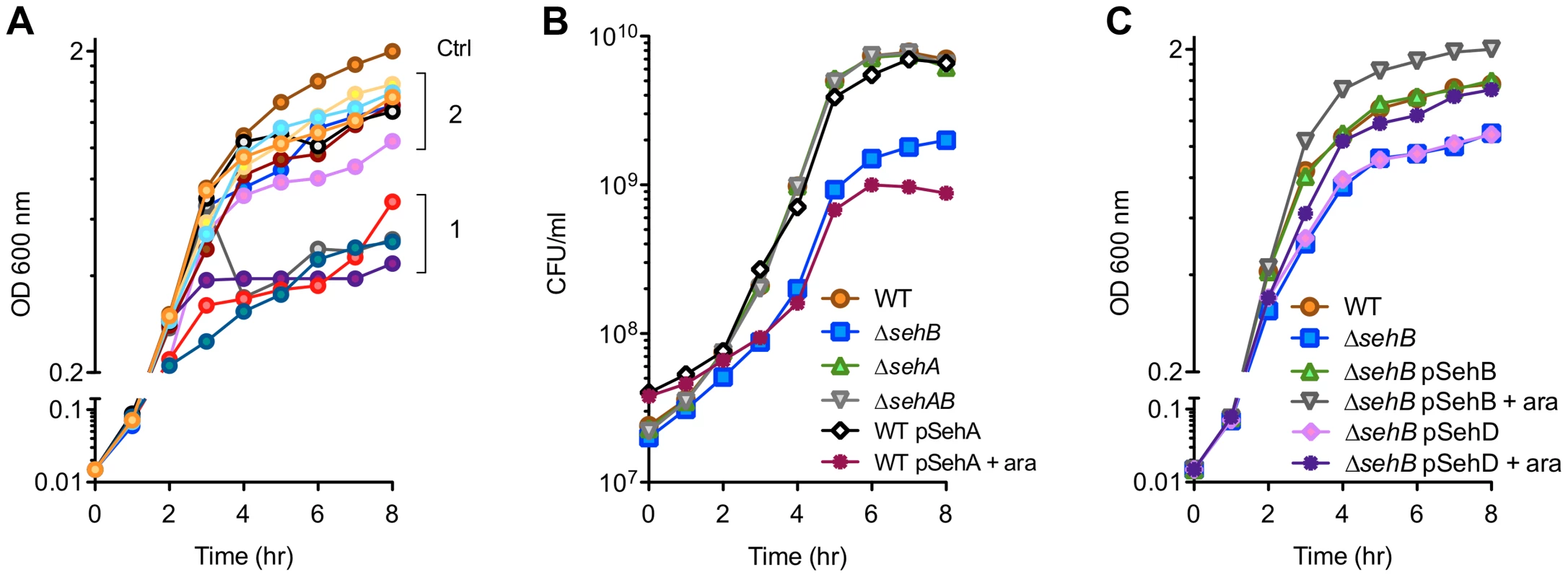
Bacteria were grown at 37°C in LB broth and the optical density was monitored (A & C) or the colony forming units (CFU) were enumerated (B). (A) Effect of Salmonella toxins over-expression on E. coli growth. BL21(DE3) cells were transformed with an empty pDEST17 (ctrl) or derived plasmids carrying the putative S. Typhimurium toxin genes. Toxin expression was induced by adding 1 mM IPTG 2 hr after inoculation. Group 1: STM2954.1n, STM3516, STM3558, PSLT028. Group 2: STM1550, STM3033, STM3777, STM4031, STM4032.2N, STM4450, PSLT106. (B & C) Growth kinetics of wild-type (WT) and sehAB mutant strains of S. Typhimurium. When indicated, strains were complemented with pDEST49-derivative vectors for the expression of SehA, SehB or SehD under the control of the arabinose-inducible promoter. Next, we focused our studies on TA modules present in all Salmonella strains capable of infecting warm-blooded animals (Figure 1A). More precisely, we targeted STM4031-STM4030.S and STM4032.2N-STM4033, which presented a close chromosomal location, being separated by a single unrelated gene, and encode transcripts on opposite chromosome strands (Figure 1B).
STM4031-4030.S encodes proteins presenting 40% and 46% identity with HigB and HigA from E. coli (Taxonomy ID 83333), respectively. The product of STM4032.2N presents 72% identity with HigB of E. coli (Taxonomy ID 561). While both STM4030.S and STM4033 products possess an HTH-XRE DNA binding domain that is found in type II antitoxins, they do not present significant shared similarity (7.7% identity). Likewise, the toxic components (STM4031 and STM4032.2N) show only 11.6% identity and none exerted strong toxicity in E. coli (Group 2 in Figure 2A). We renamed these TA gene locus seh (Salmonella enterica Hig-like) modules. Hence, STM4031-STM4030.S and STM4032.2N-STM4033 correspond to sehAB and sehCD, respectively (Figure 1).
We examined the chromosomal location and organization of sehAB and sehCD locus in several strains of Salmonella. A bioinformatics analysis revealed that sehAB and sehCD are not present in genomic islands, yet a cluster of genes similar to that found in S. Typhimurium was present in all strains of the species enterica (except arizonae in which sehAB is missing), (Figure S1). This cluster of genes has a conserved chromosomal location and is missing in the species bongori.
We generated various S. Typhimurium mutants of sehAB and sehCD and evaluated the impact of these deletions on bacterial growth. While deletion of the higA antitoxin gene in Mycobacterium tuberculosis [20], Vibrio cholerae [21] or Pseudomonas aeruginosa [22] is lethal, we were able to generate Salmonella mutants deleted of either antitoxin gene. A mutant deleted of the gene coding for the SehB antitoxin (ΔsehB) formed tiny colonies on agar plates and grew slower than the wild-type strain in LB broth, while over-expression of the SehA toxin limited the growth of the wild-type S. Typhimurium strain (Figure 2B). No growth change was observed in absence of the toxin (ΔsehA) or in absence of the module (ΔsehAB). Thus, the bacteriostatic effect was observed where there was excess of toxin or in the absence of antitoxin, suggesting that SehAB functions as a toxin-antitoxin system. The growth defect of ΔsehB was complemented by plasmid for the expression of the antitoxin (pSehB) under the control of the ara promoter (Figure 2C). Of note this complementation was also observed in absence of arabinose probably because of low level of expression. Remarkably, a ΔsehB mutant over-expressing SehB grew faster and to a higher density than the wild-type strain (Figure 2C). This suggests that SehA limits the growth of S. Typhimurium in a way that might reflect the topological organization of the sehAB operon in which the SehA toxin is transcribed first and may therefore be more abundant than its cognate antitoxin. However, wild-type and ΔsehA strains had similar growth curves (Figure 2B) and a ΔsehA strain over-grew when its cognate antitoxin was over-expressed (Figure S2A). These data indicate that the SehB antitoxin not only neutralizes its cognate toxin, but is also beneficial for Salmonella growth in a toxin-independent manner.
The ΔsehC, ΔsehD and ΔsehCD mutant strains were indistinguishable from the parental strain indicating that the toxin component is not limiting bacterial growth (Figure S2B). However, over-expression of the SehD antitoxin was able to partially complement for the absence of SehB (Figure 2C). Therefore we tested if SehD supported the growth of a ΔsehB mutant, as this mutant was unexpectedly viable. This hypothesis was tested by deleting sehD in a ΔsehB strain. We obtained a viable mutant deleted of both antitoxin genes (ΔsehB ΔsehD) indicating that SehD is not required for Salmonella growth in absence of SehB and vice versa. Altogether, these results indicate that, in LB broth, the SehC toxin does not impair S. Typhimurium growth while SehA exerts a non-lethal toxicity that is neutralized by its cognate antitoxin.
The antitoxin components of SehAB and SehCD repress their own transcription
We undertook a biochemical and biophysical characterization of SehAB and SehCD and checked whether these modules regulate their own expression by binding palindromic sequences in their promoter region as expected for type II TA systems [2]. We found two perfect or near perfect palindromic sequences in the sehAB and sehCD promoter regions, which overlap with the putative −35 and/or −10 sequences (Figure 1B). To test whether SehB and SehD repress the expression of their operons, we generated plasmid-based transcriptional reporters by fusing the sehAB and sehCD promoter regions to the gfpmut3a gene. Bacteria carrying these reporter constructs were grown in LB broth and the fluorescence level of bacteria was monitored by flow cytometry [23]. The expressions of the sehAB-gfp and sehCD-gfp transcriptional fusions were 50 - and 10-fold increased, respectively, in the absence of their relevant antitoxin or module, whereas the absence of the cognate toxin alone did not modify GFP expression (Figure 3A). By combining transcriptional reporters, mutants and plasmids we found that SehB does not repress SehCD expression and vice versa (Figure S3). This was predictable given that palindromic sequences and the nucleotide sequences between the palindromes are distinct in sehAB and sehCD promoter regions (Figure 1B).
Fig. 3. SehB and SehD antitoxins bind to their promoter region. 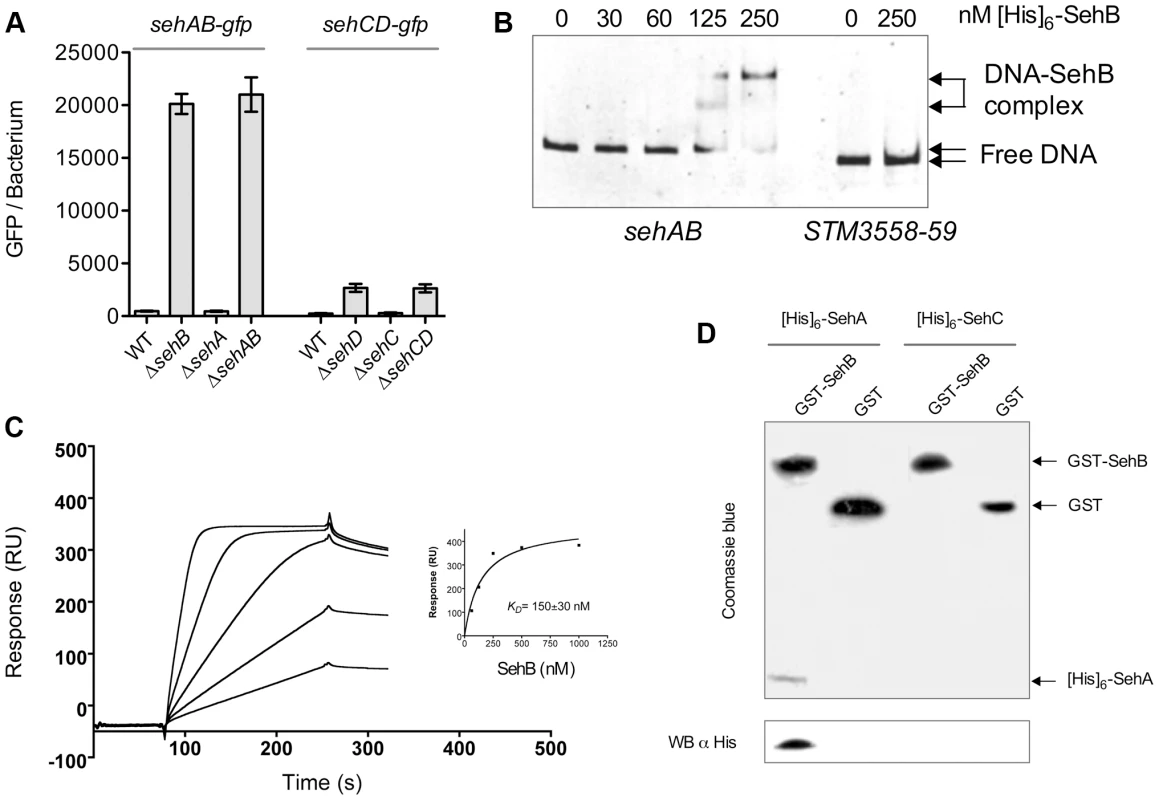
(A) Expressions of sehAB-gfp and sehCD-gfp are de-repressed in absence of their relevant antitoxin. S. Typhimurium wild-type (WT) and various mutant strains carrying a transcriptional fusion sehAB-gfp or sehCD-gfp were grown in LB broth and samples were taken at 4 hr post-inoculation. The relative fluorescence intensity of bacteria was determined by flow cytometry. Between 5000 and 10.000 bacteria were analysed to calculate the mean GFP/bacterium. (B) The SehB antitoxin binds specifically to its promoter region. An Electrophoretic mobility shift assay was carried out by incubating 100 ng PCR products covering sehAB (22.5 nM) or STM3558-59 (20 nM) promoter regions and increasing amounts [His]6-SehB as indicated. The complexes were separated on 6% polyacrylamide gels and DNA stained. (C) Surface Plasmon Resonance analysis of the binding of purified SehB to immobilized dsDNA. Sensorgrams for the interaction of various concentrations of SehB (62.5, 125, 250, 500 and 1000 nM) with immobilized DNA (300 resonance units). Inset shows fitting curve for equilibrium binding that resulted in a KD of 150±30 nM. (D) SehA is pulled-down by SehB. Purified GST or GST-SehB were incubated with an extract a E. coli cells over-expressing [His]6-SehA or [His]6-SehC. After washing, proteins bound to glutathione-beads were separated by SDS-PAGE and analysed either by Coomassie blue staining of the acrylamide gel or by electrotransfer on a PVDF membrane and Western Blotting using an anti-[His]6 antibody. Next, we investigated if the antitoxin-mediated repression was due to direct binding of antitoxins to their promoter regions [2]. We performed electrophoresis mobility shift assays (EMSA) using purified [His]6-tagged versions of antitoxins and PCR products covering the promoter regions of the operons. Both antitoxins specifically bound to their respective seh promoter region in a concentration-dependent manner (Figures 3B and S4), but not to the STM3559-58 promoter region used as a control. SehB and SehD bound with high affinity since the formation of the promoter DNA-antitoxin complexes reached a plateau below 250 and 500 nM, respectively. By comparison, the binding saturation of M. tuberculosis HigA to its promoter required one hundred times more concentrated protein [20]. We used Surface Plasmon Resonance to better characterize the interaction between SehB and its promoter region. We immobilized a biotinylated DNA oligomer corresponding to the sequence of the first palindrome of the sehAB promoter on a streptavidin chip, and assayed the binding of purified SehB. We measured a KD of 150±30 nM, thus confirming the high affinity of SehB for this palindromic sequence (Figure 3C).
Using size-exclusion chromatography coupled with online multiangle laser light scattering (ultra-violet light absorbance and refractive index detectors) we characterized the formation of complexes between the SehAB proteins and the DNA palindromic sequence. We observed that purified SehB formed a stable dimer at 32 kDa (theoretical mass 31.4 kDa) (Figure S5). In the presence of DNA, we observed a mass of 34.4 kDa for the antitoxin dimer, of 14.6 KDa (theoretical mass 14.0 kDa) for the dsDNA, and of 49 kDa for the complex. We found that purified SehA formed a tetramer and higher aggregate but we were unsuccessful at reconstituting a toxin-antitoxin or a ternary complex using these purified proteins. However, GST-SehB specifically pulled-down [His]6-SehA from a bacterial extract (Figure 3D). This indicates that proteins of the sehAB module are capable of interacting. We concluded that repression of sehAB transcription is mediated by the binding of either the SehAB toxin-antitoxin complex or the SehB homo-dimer to its promoter region.
Transcription of the sehAB locus is modulated by environmental conditions
Antitoxins of type II systems are believed to be more prone to proteolytic degradation than their cognate toxins. It results in increased availability of toxins as well as increased transcription of TA operons. In order to compare the stability of SehA and SehB, we constructed S. Typhimurium strains chromosomally expressing either SehA-3XFLAG or SehB-3XFLAG. These strains were grown for 6 hours in minimal medium after which protein synthesis was blocked with chloramphenicol. The level of pre-existing FLAG-tagged SehA and SehB was monitored during 60 min by Western Blotting. While the SehA toxin was remarkably stable, we noticed a marked decrease in SehB antitoxin signal during this period of time (Figure 4A). This result shows that SehB is labile as compared to SehA. Next, we determined if the transcriptional activity of seh loci is modulated in response to conditions encountered during interactions with animal cells. Wild-type S. Typhimurium and sehAB locus mutant strains carrying sehAB-gfp or sehCD-gfp transcriptional reporters were used to infect RAW 264.7 mouse macrophages. These strains were also grown in minimal medium, which is known to induce the expression of genes necessary for intracellular survival/replication [24]. As control, we used a transcriptional reporter for sifA, which is required for intracellular replication and is up-regulated in these growth conditions (Figure 3B). As compared to bacteria grown in LB broth, fluorescence resulting from expression of sehAB-gfp in wild-type Salmonella increased significantly both in minimal medium (4.3 times) and in macrophages (3 times) (Figure 4B). We observed a very similar profile of transcriptional activity in the absence of SehA, thus confirming that the toxin does not play a significant role in the regulation of the operon activity. As shown previously (Figure 3A), the transcriptional activity of sehAB increased dramatically in the absence of the antitoxin (ΔsehB) or the full module (ΔsehAB). Yet, in spite of the strong de-repression due to the absence of the antitoxin, sehAB-gfp expression was still approximately 1.5 times more increased in mutant strains grown in minimal medium as compared to LB broth. These results suggest that in addition of the repression mediated by the antitoxin, the transcriptional activity of the sehAB promoter is controlled by additional regulatory elements which are at least partly dependent on environmental conditions. The expression of sehCD-gfp increased about three and 1.5 times in bacteria grown in minimal medium or in macrophages, respectively (Figure 4C). The sehCD promoter was insensitive to the absence of either or both SehAB proteins. We concluded that the sehAB loci responds to conditions encountered inside host cells, therefore suggesting this module might influence the intracellular growth of S. Typhimurium.
Fig. 4. Regulation of sehAB and sehCD promoters and role of SehAB proteins in intracellular replication. 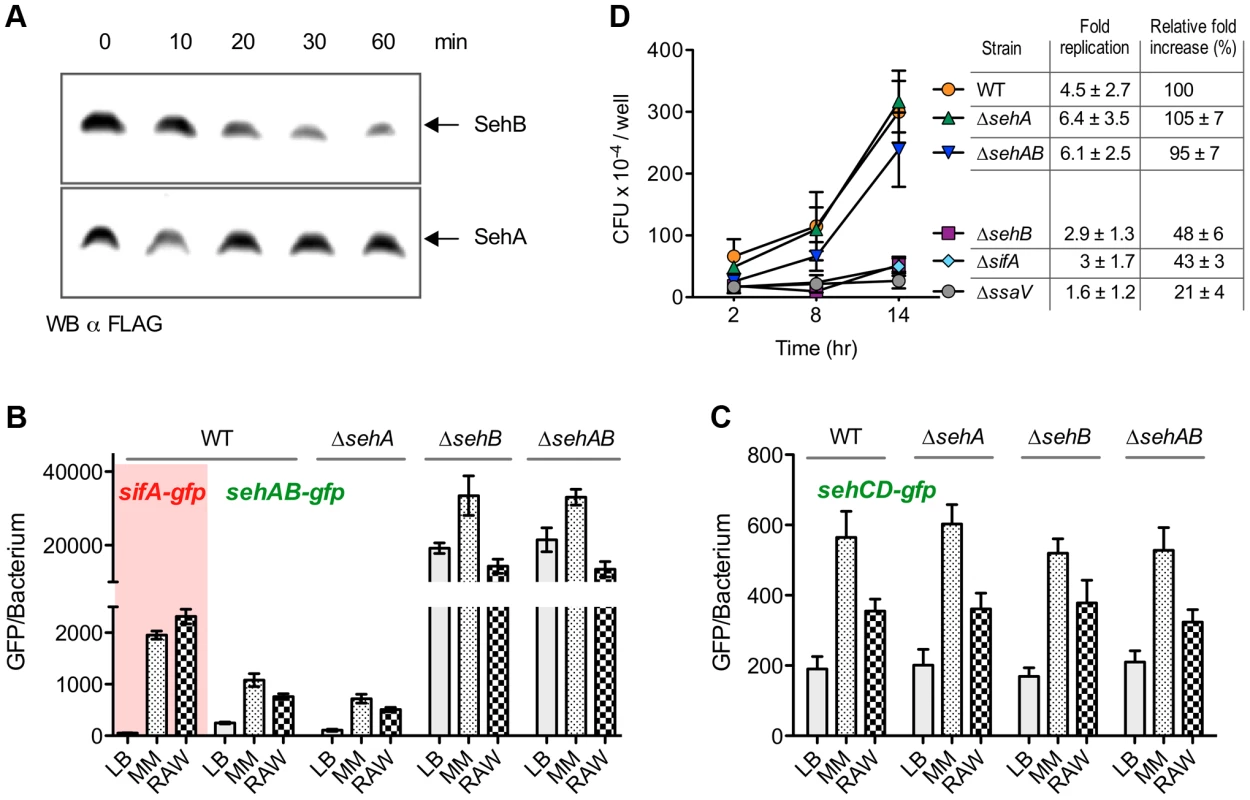
(A) SehB is prone to degradation. Strains expressing chromosomally SehB-3XFLAG or SehA-3XFLAG were grown for 6 hours in minimal medium. 100 µg/ml chloramphenicol was then added and samples were taken at the indicated times. The presence of remaining SehA and SehB was analysed by Western Blotting using an anti-FLAG antibody. (B & C) Expression of sehAB-gfp is de-repressed in minimal medium and inside mouse macrophages. Wild-type (WT) S. Typhimurium and sehAB mutant strains carrying the transcriptional fusion sifA-gfp, sehAB-gfp or sehCD-gfp were grown in LB broth, minimal medium (MM) or inside RAW 264.7 mouse macrophages. Bacteria were collected at 4 (LB) or 16 (MM) hours post-inoculation or extracted from macrophages 16 hours post-infection. The relative GFP fluorescence intensity of bacteria was determined by flow cytometry. (D) sehAB is dispensable for intracellular replication. C57BL/6 bone marrow-derived macrophages were infected with wild-type (WT) or various sehAB mutant strains. ΔsifA and ΔssaV strains were used as controls. Cells were lysed at 2, 8 and 14 hr post-infection for enumeration of intracellular bacteria. A representative experiment is shown. The graph values are the mean colony forming units (CFU) per well ± SD of triplicates. The “fold replication” for the presented experiment was calculated as a ratio of the intracellular bacteria between 14 and 2 hr. The “normalized replication” represent the fold increase calculated as a ratio of the intracellular bacteria between 16 and 2 hr and normalized to that of the wild-type strain (100%). Values are means ± SD of 3 independent experiments. sehAB and sehCD are dispensable for intracellular replication
Cell lines and primary cells were used to investigate the potential role of seh modules in intracellular replication of Salmonella. Bone marrow-derived macrophages (BMM) were infected with sehAB mutant strains and the numbers of intracellular bacteria were determined at 2, 8 and 14 hours post-infection. We also calculated the relative fold increase of intracellular bacteria between 2 and 14 h post-infection with respect to the wild-type strain (100%). As expected, we observed at 14 hours post-infection a dramatic replication defect for the control ΔssaV mutant strain [25] (Figure 4D). Bacteria deleted of sehB or sifA [26] presented a moderate intracellular replication defect (relative fold increase of 48±6% and 43±3%, respectively). Mutants deleted for sehA or the full module (ΔsehAB) replicated similar to the wild-type strain, indicating that sehAB is dispensable for replication inside BMM.
The intracellular replication of seh mutants was also tested in human HeLa cells and RAW 264.7 mouse macrophages. We also observed in these cell lines a moderate intracellular replication defect for the ΔsehB mutant while deletion of either or both sehCD genes had no effect (Figure S6A). Strains over-expressing either antitoxin replicated more than twice as much as the control, whether the absence or presence of their cognate toxin or module (Figure S7).
Next we tested whether the replication defect of a ΔsehB antitoxin mutant could be attributed, at least in part, to a direct toxic effect of the SehA toxin on eukaryotic cells. Infected macrophages were stained with fluorescent markers for death and early apoptosis and analyzed by flow cytometry. Infection by Salmonella increased the percentage of cells positive for both markers from about 1 to 10%, no matter which strain was used (Figure S8). These results indicate that the presence of high level of free SehA toxin in Salmonella does not impact the viability of infected cells.
We concluded that i) SehAB and SehCD are dispensable for intracellular replication, ii) Over-expression of either antitoxin confers to Salmonella an intracellular hyper-replicative phenotype similar to that observed in LB broth, and iii) The moderate intracellular growth defect of the ΔsehB strain is likely nonspecific and rather results from a general growth defect for this strain.
SehAB is a virulence factor in perorally inoculated mice
We compared the virulence of wild-type and seh mutant strains in mice by performing mixed infections. As controls, we again used the ΔsifA and ΔssaV strains that respectively, are moderately and highly attenuated in this model [27]–[29]. Groups of C57BL/6 mice were inoculated intraperitoneally with different two strain combinations (1∶1 mix) and bacteria were recovered from mouse spleens after 2 days to determine the competitive index (CI) [30]. We found that the ΔsehB antitoxin mutant is highly attenuated with respect to the wild-type strain (CI = 0.02±0.01). Remarkably, this attenuation is more pronounced and similar to that observed with the ΔsifA (CI = 0.16±0.05) and ΔssaV strains (CI = 0.01±0.003) (Figure 5A). Complementation with a plasmid for the expression of SehB allowed the ΔsehB mutant to recover virulence (CI = 0.75±0.2), while the ΔsehA and ΔsehAB mutant strains displayed a wild-type level of virulence. These results demonstrate that, while the sehAB module is dispensable for systemic infection, SehA exerts in vivo a very strong toxic action that is neutralized by its cognate antitoxin (Figure 5A).
Fig. 5. sehAB is transiently involved in Salmonella virulence upon peroral inoculation. 
Mice were inoculated intraperitoneally (A) or perorally (B & C) with a 1∶1 mixture of two Salmonella strains as indicated. C57BL/6 mice we used, except for panel C for which 129S2 and C57BL/6 mice were infected as indicated. Spleens were harvested two days (A), five days (B) or at various times (C) post-inoculation and bacteria were enumerated. The lowest and highest total CFU/organ in each group of CI were in (B) WT versus : ΔsehAB, 8.7×105–2.3×107; ΔsehB, 2.3×105–1.9×108; ΔsehA, 4.3×103–2.5×107; ΔsehAB pSehAB, 5.2×104–1.3×108; ΔsehAB ΔsehCD, 6×105–1.4×107; (C) WT versus ΔsehAB in 129S2: Day 5, 2.8×103-1.3×104; day 14, 3.6×104-6×104; day 18, 2.5×104-1.2×106; in C57BL/6: 8.7×105-6.2×106. Each symbol represents a mouse and horizontal bars correspond to the means ± SD. A one-sample t-test was used to determine whether a CI was significantly different of one, and unpaired t-tests to determine whether two values were significantly different. P-values: ns, not significant; *, P<0.05; **, P<0.01; ***, P<0.0005. Next, we tested if this module could play a role in mice inoculated by the natural route. We found that by peroral inoculation, a strain deleted of sehB also presents a strong virulence defect (CI = 0.16±0.06) while bacteria deleted of the toxin are unaffected (CI∼1) (Figure 5B). Interestingly, the ΔsehAB strain is significantly attenuated (CI∼0.50) (Figure 5B and 5C) but complemented by a plasmid expressing SehAB under its natural promoter (Figure 5B). We also examined the role of SehAB in 129S2 mice carrying wild-type alleles of Nramp1. 129S2 mice have been used to develop a model of persistent S. Typhimurium infection [31], [32] and we took advantage of the prolonged survival of Salmonella-infected 129S2 mice to analyze the evolution of the CI with time. At five days post-inoculation, a ΔsehAB strain presented in 129S2 mice an attenuation (CI = 0.64±0.24) similar to that observed in C57BL/6 mice (Figure 5C). Surprisingly, this value did not decrease to a great extent with time but rather tended to reach a plateau (CI = 0.41±0.05 at day eighteen post-inoculation, Figure 5C). These results suggest that after an initial deficiency in establishing an infection by the natural route, the ΔsehAB mutant is able to persist as well as the wild-type strain. It confirms that sehAB does not play a significant role in the systemic infection. Rather, this module is involved in the early phase of peroral infection.
Strains deleted of sehAB or of both sehAB and sehCD were not significantly different in their virulence attenuation when given perorally to C57BL/6 (Figure 5B). This indicates that the sehCD module, whose toxin does not seem to be active in mice (ΔsehD in Figure 5A), does not contribute significantly to Salmonella virulence in the mouse model.
sehAB is highly transcribed in mesenteric lymph nodes and is required for survival of Salmonella in these organs
The attenuation of a ΔsehAB mutant suggested that upon peroral inoculation Salmonella encounters conditions favouring the enhanced transcription of the sehAB locus. To confirm this, we inoculated mice with wild-type S. Typhimurium carrying a transcriptional reporter for sehAB (sehAB-gfp). Bacteria extracted from various organs were examined for the expression of GFP by flow cytometry and compared to reference strains grown in LB broth. Most Salmonella extracted from the mesenteric lymph nodes (MLN) of perorally inoculated mice showed GFP fluorescence levels (blue curve, Figure 6A) that were slightly higher than that of wild-type S. Typhimurium grown in LB broth (green curve, Figure 6A). This indicates that the majority of bacteria in the MLNS exhibit slightly higher expression of sehAB. Yet, we also observed in MLNs a second population with a mean GFP fluorescence similar to that observed for the ΔsehB strain grown in LB broth (red curve, Figure 6A). This population, hereafter referred to as GFPhigh bacteria, corresponded to Salmonella in which expression of sehAB is de-repressed. This GFPhigh population although not exceeding 5% of the total Salmonella population, was however significantly more prevalent in spleens and MLNs of mice inoculated via the peroral route (Figure 6B). The mean fluorescence of total bacteria was also significantly increased in populations extracted from perorally but not from intraperitoneally inoculated mice, when compared to bacteria grown in LB broth (Figure 6C). Finally, in perorally inoculated mice, the mean fluorescence of GFPhigh bacteria was significantly higher in Salmonella extracted form MLNs in comparison to their corresponding spleens (Figure 6C). Considering that having crossed the intestinal barrier the bacteria will reach the MLNs before the spleen [33], these results indicate that activation of the sehAB module is an early event in the process of infection through oral ingestion. To further emphasise this point we also analysed the consequences of the deletion of sehAB on the virulence attenuation of bacteria present in various organs. We found that the CI of wild-type versus ΔsehAB strains is much lower in MLNs (0.05±0.03) as compared to the spleen (0.51±0.19) and the liver (0.32±0.02) (Figure 6D). These results reveal that sehAB is especially important for the survival of bacteria in MLNs.
Fig. 6. sehAB is highly transcribed in a small population of bacteria upon peroral inoculation. 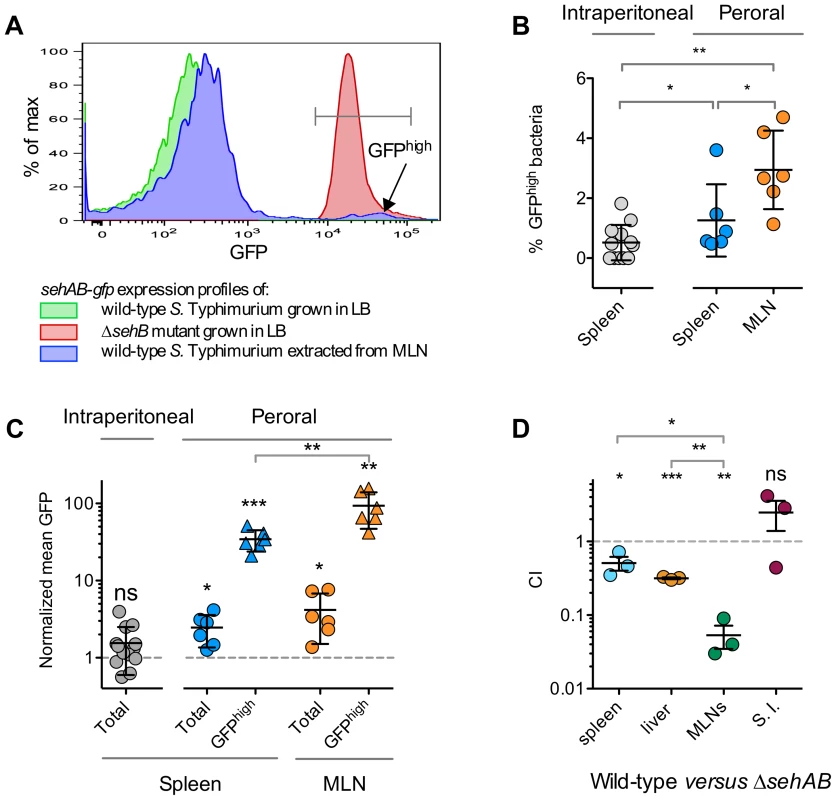
(A–C) C57BL/6 mice were inoculated intraperitoneally or perorally as indicated with wild-type S. Typhimurium carrying a transcriptional fusion sehAB-gfp. The relative GFP fluorescence intensity of bacteria extracted from the spleen or MLNs was determined by flow cytometry. Between 5000 and 10.000 bacteria were analysed for each sample. (A) A small population of Salmonella extracted from infected mice are highly fluorescent. Bacteria were extracted from MLNs of a mouse five days post-peroral inoculation. Bacteria were analyzed by flow cytometry for GFP fluorescence (blue) and compared to bacteria used for the inoculation (green) and, as control, with a ΔsehB strain (red). Bacteria extracted from MLNs contain a population expressing a high level of GFP (GFPhigh). (B) The percentage of GFPhigh bacteria is significantly higher in mice inoculated perorally versus intraperitoneally. Bacteria were extracted from spleens or MLNs of mice inoculated intraperitoneally or perorally and the percentages of GFPhigh bacteria were determined. (C) The mean GFP fluorescence of total and GFPhigh bacteria is significantly increased in bacteria extracted from perorally inoculated mice and significantly higher in MLN versus spleen. The mean GFP fluorescence was normalized to that of bacteria used for inoculation. (D) The CI of wild-type versus ΔsehAB is lower for the population of bacteria extracted from MLNs. C57BL/6 mice were inoculated perorally with a 1∶1 mixture of wild-type and ΔsehAB strains. Spleen, liver, MLNs and the small intestine (S. I.) were collected five days post-inoculation and bacteria were enumerated. Each symbol represents a mouse, and horizontal bars correspond to the means ± SD. The lowest and highest total CFU/organ in each group of CI were in : spleen, 8.8×106-7.5×107; Liver, 7×107-2.1×108; MLNs, 6.5×104-1.9×105; S. I., 7.1×105-5×106. (B–D) A one-sample t-test was used to determine whether a value was significantly different of one and an unpaired t-tests to determine whether two values were significantly different. P-values: ns, not significant; *, P<0.05; **, P<0.01; ***, P<0.0005. Discussion
Type II TA modules are inhibitors of translation that induce bacterial dormancy. Being in a dormant state helps bacteria to survive harmful environments. For example, type II TA modules favour resistance to antibiotics [1]. This study presents evidence that virulent strains of Salmonella possess multiple TA systems and shows that one of these is beneficial in the early phase of infection by the natural route.
We ascertained the presence of eleven type II TA modules in S. Typhimurium and examined the presence of homologous genes across Salmonellae. A heatmap generated using both strains and TA gene modules highlighted a set of four TA that are found across serovars of S. enterica infecting warm-blooded animals. In contrast, non-pathogenic species and strains of S. enterica, which are associated with cold-blooded animals, possess no or low numbers of TA modules found in S Typhimurium, respectively. Interestingly, previous data extracted from a transcriptional profile of intracellular S. Typhimurium [34] showed that the expression of the eleven TA modules is induced during infection of mouse macrophages, thus highlighting the possible involvement of these modules during the interaction with host cells.
It is important to note that the present study did neither perform an exhaustive review of TA modules in Salmonellae, nor investigate whether the non-pathogenic species S. bongori or strains infecting cold-blooded animals have specific TA module repertoires. It is very likely that other TA systems exist and will be discovered as illustrated by the recently published study by Slattery et al. [35]
These TA operons, which are part of the prokaryotic mobilome [36], are likely to have been acquired by horizontal transfer and positively selected because of their advantage for pathogenesis. Strikingly, the sehAB and sehCD loci delineate a gene cluster that has a conserved location on chromosomes of various virulent strains of S. enterica. This suggests that a common ancestor of the enterica species has acquired this cluster as a block, and provides further indication that these TA modules play a role in virulence.
We found that a mutant deleted of sehAB is attenuated for virulence in mice inoculated perorally but fully virulent in mice inoculated intraperitoneally. This is reminiscent to what has been observed for the SP1-encoded type three secretion system [37], which helps Salmonella to cross the intestinal barrier. Thus, sehAB is likely to play a role in an early step of the infection process that is temporally localized between the arrival of the bacterium in the stomach and the triggering of a systemic infection. Interestingly, the CI in spleens of perorally inoculated mice was the same at 5 and 18 days post-infection, thus confirming that sehAB plays a transient role and is not involved in the systemic phase of the infection. The stomach and the gut are aggressive environments in which the activation of sehAB might favour persistence. Peyer's patches are preferential sites for Salmonella to cross the intestinal barrier [38], [39]. MLNs into which Peyer's patches are drained are central sites for the induction of the mucosal immune response and possess an important antibacterial arsenal. Thus, another possibility is that sehAB helps Salmonella to survive the biochemical stresses (NO, antibacterial peptide…) and/or the innate immune cellular defences present in the subepithelial environment. Indeed, this possibility is supported by the presence of Salmonella with high sehAB promoter activity in MLNs and by the low CI of a ΔsehAB mutant in these peripheral lymphoid organs. The association of these data suggests that an up-regulation of the sehAB promoter helps Salmonella to survive the detrimental environment of the MLNs.
Five days post-inoculation we detected in MLNs a maximum of five percent of GFPhigh bacteria. We cannot exclude the possibility that the activation of sehAB occurs earlier and in a larger part of the bacterial population. However, our attempts to detect fluorescent bacteria in mice at earlier times post-inoculation were unsuccessful because the number of bacteria present in the various organs was too low.
The HigB toxin of E. coli K12 cleaves mRNAs positioned at the ribosomal A-site and stops translation [40]. The mechanism of action of the SehA toxin is unknown but, considering it shares 40% identity with HigB, these toxins are likely to have similar targets. However, our results indicate that SehA activity is dependent on an additional mechanism. Indeed, a ΔsehB strain, which expresses a high level of free toxin, presents only a partial growth defect in LB broth or in cultured cells but is severely impaired for virulence in the mouse model. Interestingly, a previous screen performed in a mouse model selected STM4030 (sehB) among the most important genes for virulence [41]. Altogether, this indicates that the toxin itself is necessary but not sufficient to limit bacterial growth and that an additional factor synergizes with SehA activity under conditions of mouse infection. This might be an activator/inhibitor factor whose association/dissociation with the toxin is controlled by environmental conditions encountered in mice. This factor is operational in intraperitoneally inoculated mice although a ΔsehAB remains fully virulent under these conditions of infection. Overall it suggests that activation of the SehA toxin and transcription of the sehAB locus are not triggered by the same environmental conditions.
In general toxins and antitoxins form hetero-oligomers that are toxin-inhibiting complexes that can bind their promoter region [42], [43]. We could not reconstitute a SehAB toxin-antitoxin complex using purified proteins. In addition, we observed an aggregation of purified SehA, which might result from non-native folding and could explain its lack of interaction with the antitoxin. Nevertheless SehA was specifically pulled-down by SehB from a bacterial extract, indicating that SehA and SehB are likely to form a canonical type II TA complex.
Stress conditions leading to the de-repression of sehAB are poorly mimicked in cultured cells. This is shown by the limited activation of the sehAB promoter and by the absence of growth defect in the ΔsehAB mutant. This suggests that this locus does not support the survival/replication of intracellular bacteria. Another possibility is that cultured cells and bone marrow-derived macrophages are not representative of cell types encountered by Salmonella during the early phase after oral inoculation. sehAB might help the bacterium to survive bactericidal activity mediated by immune cells that are recruited to the infection sites. Supporting this observation is the fact that Salmonella are not killed and can survive in dendritic cells. Of note, this survival is independent of virulence factors known to be important in macrophages [44].
We could not find any phenotype associated with sehCD. Nevertheless, this locus like sehAB, is conserved among pathogenic strains of Salmonella enterica indicating its importance for the general fitness of Salmonella. The sehCD locus might for example, be necessary for interactions with another warm-blooded Salmonella host.
The present study supports the idea that type II TA modules sustain Salmonella virulence. Likewise, M. tuberculosis contains many more TA systems than the saprophytic M. smegmatis, leading to the suggestion that TA modules play a role in virulence by supporting the long-term dormancy of the pathogen in macrophages [45]. TA systems of extra-intestinal pathogenic E. coli have recently been shown to also impact the persistence of bacteria within host tissues [12]. Thus, it is more and more apparent that TA systems are important elements in the virulence of bacterial pathogens. The next challenge will be to understand the signalling and biochemical processes supporting the full activation of the SehA toxin and when and where the conditions necessary for this activation are encountered in the course of oral inoculation. Finally, it will be essential to evaluate the importance and role of other type II TA modules in Salmonella pathogenicity.
Materials and Methods
Ethic statement
Animal experimentation was conducted in strict accordance with good animal practice as defined by the French animal welfare bodies (Law 87–848 dated 19 October 1987 modified by Decree 2001-464 and Decree 2001-131 relative to European Convention, EEC Directive 86/609). All animal work was approved by the Direction Départementale des Services Vétérinaires des Bouches du Rhône (authorization number 13.118 to S.M.).
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Table S1 in Text S1. Strains were cultured in LB broth (Difco) or minimal medium (M9, glycerol 0.2%, MgSO4 1 mM, CaCl2 200 mM, thiamine 1 mg/ml, casamino acids 1 mg/ml. Ampicillin (50 µg/ml), kanamycin (50 µg/ml), tetracycline (10 µg/ml) and chloramphenicol (50 µg/ml) were added when required. Bacterial suspensions were prepared from overnight LB broth cultures that were centrifuged and resuspended in fresh LB broth or minimal medium to an OD600 of 1. Then, 250 ml-flasks containing 50 ml of LB broth or minimal medium were inoculated with 1 ml of the bacterial suspensions and incubated at 37°C in a shaking incubator at 200 rpm.
Toxin-antitoxin databases and identification
Using RASTA webtool and standard BLASTP, we searched the S. Typhimurium genome for sequences coding for toxins and antitoxins belonging to the seven known type II TA super families: vapBC, relEB, parED, mazEF, phd-doc, ccdAB and higAB. In addition, we used others type II toxin-antitoxin sequences described for E. coli [2] (yafNO, hicAB, yefMyoeB, mqsRA, chpBIchpBK, rnlAB). For TA identification, the protein set from a whole genome was first arranged by genome coordinates. Each protein in our TA databases was then used in a BLASTP query. A hit was defined as a match to a query protein with an E-value threshold of 10−5, which should cover at least 70% of the TA with more than 30% identity. Lastly, a TA module was recognized if two contiguous proteins in the genome were possible toxin and antitoxin as suggested by the results of a BLASTP. In addition to BLASTP, we used the TBLASTN algorithm to search for the presence of toxin and antitoxin genes. Genomic locations of TA modules encoded by Salmonella sequenced strains were obtained from NCBI.
Construction of mutant strains
Non-polar gene-deletion mutants were generated by the lambda Red recombinase system [46], using gene-specific primer pairs to amplify pKD4 kanamycin or pKD3 chloramphenicol resistance genes as shown in Table S2 in Text in Text S1. Salmonella mutants were transformed with the pCP20 plasmid to excise the antibiotic cassette. This excision was performed for the ΔsehAB mutant, which was then transformed with pK3-SehAB for the expression of SehAB under its natural promoter, and for sehA::3XFLAG and sehB::3XFLAG. Gene deletions were checked by PCR.
Construction of plasmids
The sehAB-gfp and sehCD-gfp transcriptional fusions were generated by cloning PCR products corresponding to the respective promoter regions into pFPV25 [47] (see primers in Table S2 in Text S1). These PCR products were digested with EcoRI and BamHI and ligated to pFPV25 digested with the same enzymes. For E. coli expression, PCR products corresponding to Salmonella toxins were cloned in pDEST-17 using the Gateway system (Invitrogen, Ltd., Paisley, U.K.). sehB and sehD antitoxin genes were cloned both into pBAD-DEST49 and pMPM-K6. For pMPM-K6, primers contained the NcoI (5′)/HindIII (3′) restriction sites. The resulting PCR products were digested with NcoI and HindIII and ligated into pMPM-K6 [48] digested with the same restriction enzymes, generating the plasmids pK6-SehB and pK6-SehD. These plasmids contain the antitoxin genes under an arabinose inducible promoter. The sehAB locus was cloned in the pMPM-K3 plasmid for the expression of SehAB with its own promoter. The PCR product resulting from the amplification of chromosomal DNA of 12023 with the primers STA1T-EcoRI-5 and STA1T-HINDIII-3 was digested with HindIII and ligated into pMPM-K3 previously digested with SmaI-HindIII, generating the pK3-SehAB. All constructs were confirmed by DNA sequencing. Plasmids used in this study are listed in Table S1 in Text S1.
Electrophoretic mobility shift assay (EMSA)
EMSA experiments were performed as described previously [49]. PCR products corresponding to sehAB and sehCD promoter regions were the same as used to generate sehAB-gfp and sehCD-gfp. These fragments (100 ng), were mixed with increasing concentrations of [His]6-tagged SehB or SehD in PBS/50% glycerol. They were incubated 30 min at room temperature and then separated by electrophoresis in 6% polyacrylamide gels in Tris-borate-EDTA buffer. The DNA bands were visualized by staining with ethidium bromide.
Eukaryotic cells and culture conditions
RAW 264.7 and HeLa cell lines were grown in DMEM (GibcoBRL) supplemented with 10% foetal calf serum (FCS; GibcoBRL), 2 mM nonessential amino acids, and glutamine (GibcoBRL) at 37°C in 5% CO2.
Bacterial infection and replication assays
Bone marrow-derived macrophages, HeLa and RAW 264.7 macrophage were grown, infected and treated as previously described [50].
Analysis of SehAB proteins degradation and western blot analysis
Bacteria expressing SehA-3XFLAG or SehB-3XFLAG from the chromosome were grown during 6 h in minimal medium. Then, protein synthesis was stopped by addition of 100 µg/ml chloramphenicol, and samples were removed at the indicated time points. SehAB proteins were detected by Western Blotting using a monoclonal anti FLAG-tag antibody (Sigma-Aldrich) and a polyclonal goat-anti mouse IgG HRP conjugate (Sigma–Aldrich).
Competitive index
Eight - to ten-week-old C57BL/6 or 129S2 mice were inoculated intraperitoneally or perorally with equal amounts of two bacterial strains for a total of 105 bacteria per mouse. The spleens were harvested 2 or 5 days after inoculation, as indicated, and homogenized. Bacteria were recovered and enumerated after plating a dilution series onto LB agar with the appropriate antibiotics. Competitive indexes (CI) were determined for each mouse [30]. The CI is defined as the ratio between the mutant and wild-type strains within the output (bacteria recovered from the mouse after infection) divided by their ratios within the input (initial inoculum). Statistical analyses were performed using Prism (GraphPad, San Diego, CA, USA).
Flow cytometry analysis of bacteria extracted from synthetic medium, macrophages or mouse organs
Bacteria grown in synthetic medium (LB broth or MM) or extracted from infected cells or mouse organs were treated and analyzed by flow cytometry as previously described [23].
Supporting Information
Zdroje
1. GerdesK, MaisonneuveE (2012) Bacterial persistence and toxin-antitoxin Loci. Annu Rev Microbiol 66 : 103–123 doi:10.1146/annurev-micro-092611-150159
2. YamaguchiY, ParkJ-H, InouyeM (2011) Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet 45 : 61–79 doi:10.1146/annurev-genet-110410-132412
3. FineranPC, BlowerTR, FouldsIJ, HumphreysDP, LilleyKS, et al. (2009) The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci USA 106 : 894–899 doi:10.1073/pnas.0808832106
4. MasudaH, TanQ, AwanoN, WuK-P, InouyeM (2012) YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol Microbiol 84 : 979–989 doi:10.1111/j.1365-2958.2012.08068.x
5. WangX, LordDM, ChengH-Y, OsbourneDO, HongSH, et al. (2012) A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol 8 : 855–861 doi:10.1038/nchembio.1062
6. MakarovaKS, WolfYI, KooninEV (2009) Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4 : 19 doi:10.1186/1745-6150-4-19
7. LeplaeR, GeeraertsD, HallezR, GuglielminiJ, DrèzeP, et al. (2011) Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Research 39 : 5513–5525 doi:10.1093/nar/gkr131
8. LewisK (2007) Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5 : 48–56 doi:10.1038/nrmicro1557
9. PandeyDP, GerdesK (2005) Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Research 33 : 966–976 doi:10.1093/nar/gki201
10. GeorgiadesK, RaoultD (2011) Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PLoS ONE 6: e17962 doi:10.1371/journal.pone.0017962
11. AudolyG, VincentelliR, EdouardS, GeorgiadesK, MediannikovO, et al. (2011) Effect of rickettsial toxin VapC on its eukaryotic host. PLoS ONE 6: e26528 Available: http://www.ncbi.nlm.nih.gov.gate1.inist.fr/pubmed?term=Effect%20of%20Rickettsial%20Toxin%20VapC%20on%20Its%20Eukaryotic%20Host.
12. NortonJP, MulveyMA (2012) Toxin-Antitoxin Systems Are Important for Niche-Specific Colonization and Stress Resistance of Uropathogenic Escherichia coli. PLoS Pathog 8: e1002954 doi:10.1371/journal.ppat.1002954
13. Douesnard-MaloF, DaigleF (2011) Increased persistence of Salmonella enterica serovar Typhi in the presence of Acanthamoeba castellanii. Appl Environ Microbiol 77 : 7640–7646 doi:10.1128/AEM.00699-11
14. BleasdaleB, LottPJ, JagannathanA, StevensMP, BirtlesRJ, et al. (2009) The Salmonella pathogenicity island 2-encoded type III secretion system is essential for the survival of Salmonella enterica serovar Typhimurium in free-living amoebae. Appl Environ Microbiol 75 : 1793–1795 doi:10.1128/AEM.02033-08
15. SevinEW, Barloy-HublerF (2007) RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol 8: R155 doi:10.1186/gb-2007-8-8-r155
16. JørgensenMG, PandeyDP, JaskolskaM, GerdesK (2009) HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J Bacteriol 191 : 1191–1199 doi:10.1128/JB.01013-08
17. WintherKS, GerdesK (2009) Ectopic production of VapCs from Enterobacteria inhibits translation and trans-activates YoeB mRNA interferase. Mol Microbiol 72 : 918–930 doi:10.1111/j.1365-2958.2009.06694.x
18. BäumlerAJ (1997) The record of horizontal gene transfer in Salmonella. Trends Microbiol 5 : 318–322 doi:10.1016/S0966-842X(97)01082-2
19. RDC T (2010) R: A Language and Environment for Statistical Computing.
20. Fivian-HughesAS, DavisEO (2010) Analyzing the regulatory role of the HigA antitoxin within Mycobacterium tuberculosis. J Bacteriol 192 : 4348–4356 doi:10.1128/JB.00454-10
21. BuddePP, DavisBM, YuanJ, WaldorMK (2007) Characterization of a higBA toxin-antitoxin locus in Vibrio cholerae. J Bacteriol 189 : 491–500 doi:10.1128/JB.00909-06
22. HoodRD, SinghP, HsuF, GüvenerT, CarlMA, et al. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7 : 25–37 doi:10.1016/j.chom.2009.12.007
23. AusselL, ZhaoW, HébrardM, GuilhonA-A, VialaJPM, et al. (2011) Salmonella detoxifying enzymes are sufficient to cope with the host oxidative burst. Mol Microbiol 80 : 628–640 doi:10.1111/j.1365-2958.2011.07611.x
24. LinehanSA, RytkönenA, YuX-J, LiuM, HoldenDW (2005) SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect Immun 73 : 4354–4362 doi:10.1128/IAI.73.7.4354-4362.2005
25. Hensel, SheaJE, WatermanSR, MundyR, NikolausT, et al. (1998) Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol 30 : 163–174.
26. SteinMA, LeungKY, ZwickM, PortilloFG-D, FinlayBB (1996) Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol Microbiol 20 : 151–164 doi:10.1111/j.1365-2958.1996.tb02497.x
27. SheaJE, BeuzonCR, GleesonC, MundyR, HoldenDW (1999) Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun 67 : 213–219.
28. HenryT, CouillaultC, RockenfellerP, BoucrotE, DumontA, et al. (2006) The Salmonella effector protein PipB2 is a linker for kinesin-1. Proc Natl Acad Sci USA 103 : 13497–13502 doi:10.1073/pnas.0605443103
29. BeuzónCR, UnsworthKE, HoldenDW (2001) In vivo genetic analysis indicates that PhoP-PhoQ and the Salmonella pathogenicity island 2 type III secretion system contribute independently to Salmonella enterica serovar Typhimurium virulence. Infect Immun 69 : 7254–7261 doi:10.1128/IAI.69.12.7254-7261.2001
30. BeuzónCR, HoldenDW (2001) Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect 3 : 1345–1352.
31. RubyT, McLaughlinL, GopinathS, MonackD (2012) Salmonella's long-term relationship with its host. FEMS Microbiol Rev 36 : 600–615 doi:10.1111/j.1574-6976.2012.00332.x
32. MonackDM, BouleyDM, FalkowS (2004) Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med 199 : 231–241 doi:10.1084/jem.20031319
33. CarterPB, CollinsFM (1974) The route of enteric infection in normal mice. J Exp Med 139 : 1189–1203.
34. ErikssonS, LucchiniS, ThompsonA, RhenM, HintonJCD (2003) Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47 : 103–118.
35. SlatteryA, VictorsenAH, BrownA, HillmanK, PhillipsGJ (2013) Isolation of highly persistent mutants of Salmonella enterica serovar typhimurium reveals a new toxin-antitoxin module. J Bacteriol 195 : 647–657 doi:10.1128/JB.01397-12
36. KooninEV, WolfYI (2008) Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Research 36 : 6688–6719 doi:10.1093/nar/gkn668
37. GalánJE, CurtissR (1989) Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA 86 : 6383–6387.
38. JonesBD, GhoriN, FalkowS (1994) Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med 180 : 15–23.
39. LelouardH, HenriS, de BovisB, MugnierB, Chollat-NamyA, et al. (2010) Pathogenic bacteria and dead cells are internalized by a unique subset of Peyer's patch dendritic cells that express lysozyme. Gastroenterology 138 : 173–84.e1–3 doi:10.1053/j.gastro.2009.09.051
40. Christensen-DalsgaardM, JørgensenMG, GerdesK (2010) Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol Microbiol 75 : 333–348 doi:10.1111/j.1365-2958.2009.06969.x
41. ChaudhuriRR, PetersSE, PleasanceSJ, NorthenH, WillersC, et al. (2009) Comprehensive identification of Salmonella enterica serovar typhimurium genes required for infection of BALB/c mice. PLoS Pathog 5: e1000529 doi:10.1371/journal.ppat.1000529
42. MatéMJ, VincentelliR, FoosN, RaoultD, CambillauC, et al. (2011) Crystal structure of the DNA-bound VapBC2 antitoxin/toxin pair from Rickettsia felis. Nucleic Acids Research doi:10.1093/nar/gkr1167
43. BrownBL, GrigoriuS, KimY, ArrudaJM, DavenportA, et al. (2009) Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog 5: e1000706 doi:10.1371/journal.ppat.1000706
44. NiedergangF, SirardJC, BlancCT, KraehenbuhlJP (2000) Entry and survival of Salmonella typhimurium in dendritic cells and presentation of recombinant antigens do not require macrophage-specific virulence factors. Proc Natl Acad Sci USA 97 : 14650–14655 doi:10.1073/pnas.97.26.14650
45. RamageHR, ConnollyLE, CoxJS (2009) Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5: e1000767 doi:10.1371/journal.pgen.1000767
46. DatsenkoKA, WannerBL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97 : 6640–6645 doi:10.1073/pnas.120163297
47. ValdiviaRH, FalkowS (1996) Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol 22 : 367–378.
48. MayerMP (1995) A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163 : 41–46.
49. De la CruzMA, Fernández-MoraM, GuadarramaC, Flores-ValdezMA, BustamanteVH, et al. (2007) LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol Microbiol 66 : 727–743 doi:10.1111/j.1365-2958.2007.05958.x
50. SchroederN, HenryT, de ChastellierC, ZhaoW, GuilhonA-A, et al. (2010) The virulence protein SopD2 regulates membrane dynamics of Salmonella-containing vacuoles. PLoS Pathog 6: e1001002 doi:10.1371/journal.ppat.1001002
51. GotfredsenM, GerdesK (1998) The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol 29 : 1065–1076.
52. AizenmanE, Engelberg-KulkaH, GlaserG (1996) An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3″,5-″bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA 93 : 6059–6063.
53. MotiejūnaitėR, ArmalytėJ, MarkuckasA, SužiedėlienėE (2007) Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module. FEMS Microbiol Lett 268 : 112–119 doi:10.1111/j.1574-6968.2006.00563.x
54. HazanR, SatB, RechesM, Engelberg-KulkaH (2001) Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J Bacteriol 183 : 2046–2050 doi:10.1128/JB.183.6.2046-2050.2001
55. Christensen-DalsgaardM, GerdesK (2006) Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol Microbiol 62 : 397–411 doi:10.1111/j.1365-2958.2006.05385.x
56. AfifH, AllaliN, CouturierM, Van MelderenL (2001) The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison-antidote system. Mol Microbiol 41 : 73–82.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete InfectionsČlánek Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?Článek Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication FidelityČlánek CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial PathogensČlánek The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite PersistenceČlánek The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 12- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Host Susceptibility Factors to Bacterial Infections in Type 2 Diabetes
- LysM Effectors: Secreted Proteins Supporting Fungal Life
- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Innate Lymphoid Cells: New Players in IL-17-Mediated Antifungal Immunity
- Cytoplasmic Viruses: Rage against the (Cellular RNA Decay) Machine
- Balancing Stability and Flexibility within the Genome of the Pathogen
- The Evolution of Transmissible Prions: The Role of Deformed Templating
- Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete Infections
- Host Defense via Symbiosis in
- Regulatory Circuits That Enable Proliferation of the Fungus in a Mammalian Host
- Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?
- Burning Down the House: Cellular Actions during Pyroptosis
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial Pathogens
- Combining Regulatory T Cell Depletion and Inhibitory Receptor Blockade Improves Reactivation of Exhausted Virus-Specific CD8 T Cells and Efficiently Reduces Chronic Retroviral Loads
- Shaping Up for Battle: Morphological Control Mechanisms in Human Fungal Pathogens
- Identification of the Virulence Landscape Essential for Invasion of the Human Colon
- Nodular Inflammatory Foci Are Sites of T Cell Priming and Control of Murine Cytomegalovirus Infection in the Neonatal Lung
- Hepatitis B Virus Disrupts Mitochondrial Dynamics: Induces Fission and Mitophagy to Attenuate Apoptosis
- Mycobacterial MazG Safeguards Genetic Stability Housecleaning of 5-OH-dCTP
- Systematic MicroRNA Analysis Identifies ATP6V0C as an Essential Host Factor for Human Cytomegalovirus Replication
- Placental Syncytium Forms a Biophysical Barrier against Pathogen Invasion
- The CD8-Derived Chemokine XCL1/Lymphotactin Is a Conformation-Dependent, Broad-Spectrum Inhibitor of HIV-1
- Cyclin A Degradation by Primate Cytomegalovirus Protein pUL21a Counters Its Innate Restriction of Virus Replication
- Genome-Wide RNAi Screen Identifies Novel Host Proteins Required for Alphavirus Entry
- Zinc Sequestration: Arming Phagocyte Defense against Fungal Attack
- The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite Persistence
- Biphasic Euchromatin-to-Heterochromatin Transition on the KSHV Genome Following Infection
- The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
- HIV-1 Vpr Accelerates Viral Replication during Acute Infection by Exploitation of Proliferating CD4 T Cells
- A Human Torque Teno Virus Encodes a MicroRNA That Inhibits Interferon Signaling
- The ArlRS Two-Component System Is a Novel Regulator of Agglutination and Pathogenesis
- An In-Depth Comparison of Latent HIV-1 Reactivation in Multiple Cell Model Systems and Resting CD4+ T Cells from Aviremic Patients
- Enterohemorrhagic Hemolysin Employs Outer Membrane Vesicles to Target Mitochondria and Cause Endothelial and Epithelial Apoptosis
- Overcoming Antigenic Diversity by Enhancing the Immunogenicity of Conserved Epitopes on the Malaria Vaccine Candidate Apical Membrane Antigen-1
- The Type-Specific Neutralizing Antibody Response Elicited by a Dengue Vaccine Candidate Is Focused on Two Amino Acids of the Envelope Protein
- Tmprss2 Is Essential for Influenza H1N1 Virus Pathogenesis in Mice
- Signatures of Pleiotropy, Economy and Convergent Evolution in a Domain-Resolved Map of Human–Virus Protein–Protein Interaction Networks
- Interference with the Host Haemostatic System by Schistosomes
- RocA Truncation Underpins Hyper-Encapsulation, Carriage Longevity and Transmissibility of Serotype M18 Group A Streptococci
- Gene Fitness Landscapes of at Important Stages of Its Life Cycle
- Phagocytosis Escape by a Protein That Connects Complement and Coagulation Proteins at the Bacterial Surface
- t Is a Structurally Novel Crohn's Disease-Associated Superantigen
- An Increasing Danger of Zoonotic Orthopoxvirus Infections
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
- Transcriptional Analysis of Murine Macrophages Infected with Different Strains Identifies Novel Regulation of Host Signaling Pathways
- Serotonergic Chemosensory Neurons Modify the Immune Response by Regulating G-Protein Signaling in Epithelial Cells
- Genome-Wide Detection of Fitness Genes in Uropathogenic during Systemic Infection
- Induces an Unfolded Protein Response via TcpB That Supports Intracellular Replication in Macrophages
- Intestinal CD103+ Dendritic Cells Are Key Players in the Innate Immune Control of Infection in Neonatal Mice
- Emerging Functions for the RNome
- KSHV MicroRNAs Mediate Cellular Transformation and Tumorigenesis by Redundantly Targeting Cell Growth and Survival Pathways
- HrpA, an RNA Helicase Involved in RNA Processing, Is Required for Mouse Infectivity and Tick Transmission of the Lyme Disease Spirochete
- A Toxin-Antitoxin Module of Promotes Virulence in Mice
- Real-Time Imaging of the Intracellular Glutathione Redox Potential in the Malaria Parasite
- Hypoxia Inducible Factor Signaling Modulates Susceptibility to Mycobacterial Infection via a Nitric Oxide Dependent Mechanism
- Novel Strategies to Enhance Vaccine Immunity against Coccidioidomycosis
- Dual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic
- —What Makes the Species a Ubiquitous Human Fungal Pathogen?
- αvβ6- and αvβ8-Integrins Serve As Interchangeable Receptors for HSV gH/gL to Promote Endocytosis and Activation of Membrane Fusion
- -Induced Activation of EGFR Prevents Autophagy Protein-Mediated Killing of the Parasite
- Semen CD4 T Cells and Macrophages Are Productively Infected at All Stages of SIV infection in Macaques
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Host Defense via Symbiosis in
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



