-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Envelope Deglycosylation Enhances Antigenicity of HIV-1 gp41 Epitopes for Both Broad Neutralizing Antibodies and Their Unmutated Ancestor Antibodies
The HIV-1 gp41 envelope (Env) membrane proximal external region (MPER) is an important vaccine target that in rare subjects can elicit neutralizing antibodies. One mechanism proposed for rarity of MPER neutralizing antibody generation is lack of reverted unmutated ancestor (putative naive B cell receptor) antibody reactivity with HIV-1 envelope. We have studied the effect of partial deglycosylation under non-denaturing (native) conditions on gp140 Env antigenicity for MPER neutralizing antibodies and their reverted unmutated ancestor antibodies. We found that native deglycosylation of clade B JRFL gp140 as well as group M consensus gp140 Env CON-S selectively increased the reactivity of Env with the broad neutralizing human mAbs, 2F5 and 4E10. Whereas fully glycosylated gp140 Env either did not bind (JRFL), or weakly bound (CON-S), 2F5 and 4E10 reverted unmutated ancestors, natively deglycosylated JRFL and CON-S gp140 Envs did bind well to these putative mimics of naive B cell receptors. These data predict that partially deglycoslated Env would bind better than fully glycosylated Env to gp41-specific naïve B cells with improved immunogenicity. In this regard, immunization of rhesus macaques demonstrated enhanced immunogenicity of the 2F5 MPER epitope on deglyosylated JRFL gp140 compared to glycosylated JRFL gp140. Thus, the lack of 2F5 and 4E10 reverted unmutated ancestor binding to gp140 Env may not always be due to lack of unmutated ancestor antibody reactivity with gp41 peptide epitopes, but rather, may be due to glycan interference of binding of unmutated ancestor antibodies of broad neutralizing mAb to Env gp41.
Published in the journal: . PLoS Pathog 7(9): e32767. doi:10.1371/journal.ppat.1002200
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002200Summary
The HIV-1 gp41 envelope (Env) membrane proximal external region (MPER) is an important vaccine target that in rare subjects can elicit neutralizing antibodies. One mechanism proposed for rarity of MPER neutralizing antibody generation is lack of reverted unmutated ancestor (putative naive B cell receptor) antibody reactivity with HIV-1 envelope. We have studied the effect of partial deglycosylation under non-denaturing (native) conditions on gp140 Env antigenicity for MPER neutralizing antibodies and their reverted unmutated ancestor antibodies. We found that native deglycosylation of clade B JRFL gp140 as well as group M consensus gp140 Env CON-S selectively increased the reactivity of Env with the broad neutralizing human mAbs, 2F5 and 4E10. Whereas fully glycosylated gp140 Env either did not bind (JRFL), or weakly bound (CON-S), 2F5 and 4E10 reverted unmutated ancestors, natively deglycosylated JRFL and CON-S gp140 Envs did bind well to these putative mimics of naive B cell receptors. These data predict that partially deglycoslated Env would bind better than fully glycosylated Env to gp41-specific naïve B cells with improved immunogenicity. In this regard, immunization of rhesus macaques demonstrated enhanced immunogenicity of the 2F5 MPER epitope on deglyosylated JRFL gp140 compared to glycosylated JRFL gp140. Thus, the lack of 2F5 and 4E10 reverted unmutated ancestor binding to gp140 Env may not always be due to lack of unmutated ancestor antibody reactivity with gp41 peptide epitopes, but rather, may be due to glycan interference of binding of unmutated ancestor antibodies of broad neutralizing mAb to Env gp41.
Introduction
Two rare human monoclonal antibodes (mAbs), 2F5 and 4E10, bind to linear epitopes in the gp160 membrane proximal external region (MPER) [1], [2]. The core sequence of 2F5 epitope is aa 662–668 (ELDKWAS), and that of 4E10 is aa 671–676 NWFDIT [3]. The crystal structures of the 2F5 and 4E10 Fabs in complex with peptides containing their gp41 core epitopes demonstrated that only a relatively small portion of the CDRH3 antibody regions bound the MPER [4], [5]. Both mAbs 2F5 and 4E10 are polyreactive for lipids and have long hydrophobic heavy chain complementarity determining regions (HCDR3s) [6]. Hydrophobic HCDR3 loops of the variable region of the heavy chain (VH) of both mAbs 2F5 and 4E10 bind viron lipids in a two-step conformational change model that is required for antibody neutralization [7]–[11]. Thus, mAbs 2F5 and 4E10 use their autoreactive specificities to mediate anti-HIV-1 activity [8], [12]. The autoreactivity of mAbs 2F5 and 4E10 has raised the hypothesis that their rarity is due to tolerance control due to their polyreactivity with host antigens [6]. Indeed, the creation of knock-in mice with 2F5 [13] and 4E10 [14] VHs have demonstrated this to be the case in these mice. MAbs 2F5 and 4E10 rarely bind well to gp140 oligomers, possibly in part due to lack of formation of the gp41 intermediate form that binds to mAbs 2F5 and 4E10 [15]. Xiao et al. [16] have suggested that an additional reason for the rarity of broadly neutralizing antibodies in general, and 2F5-like antibodies in particular, is the lack of reactivity of HIV-1 Env with the unmutated ancestors of broad neutralizing antibodies. That is, this hypothesis states that there are “holes” in the naïve B cell repertoire resulting in lack of unmutated antibodies that can bind and respond to conserved Env broadly neutralizing epitopes.
Glycosylation can modulate the binding of antibodies to gp120 with gain or loss of a glycosylation site in a virus mutant affecting both Env antigenicity and virus neutralization sensitivity [17]–[23]. Analysis of HIV-1 Env mutated by site-directed mutagenesis has indicated that Env glycans in the first and second variable (V1/V2) loops [24], the third (V3) [25] and fourth variable (V4) loops of gp120 [26] modulate the binding of antibodies to Env. Yuste et al. demonstrated that glycosylation site deletion mutants of SIVmac239 induced neutralizing antibodies to a novel conserved region C-terminal to the gp41 immunodominant region [23]. In addition, two recent studies have demonstrated that the N-linked glycosylation site at Env aa 413 is associated with induction of broad neutralizing antibodies [27], [28].
In this study, we have partially deglycosylated JRFL gp140 and group M consensus gp140 (CON-S) [29] Env proteins under native, non-denaturing conditions, and demonstrated enhanced binding of mAbs 4E10 and 2F5 to deglycosylated Env. While the reverted unmutated ancestor antibodies of 2F5 and 4E10 were either non-reactive (with glycosylated JRFL Env), or poorly reactive (with glycosylated CON-S Env), they reacted well with partially deglycosylated gp140 HIV-1 envelope, indicating for some HIV-1 envelopes, an intact naïve B cell repertoire for 2F5 and 4E10 gp41 neutralizing epitopes.
Results
Analysis of Deglycosylated HIV-1 gp140 Env Proteins in SDS-PAGE, Western Blot Analysis and ELISA
To deglycosylate maximally HIV-1 Env under non-denaturing conditions, while in order to maintaining the native conformations, HIV-1 JRFL recombinant gp140 Env was treated with a wide dose range of PNGase F. Figure 1 shows JRFL gp140 Env protein treated with progressive deglycosylation (PNGase F 5, 10, 20, 50, 100 and 500 U per µg of Env protein) analyzed in SDS-PAGE and Western blots. After deglycosylation, the molecular mass of HIV-1 JRFL gp140 Env protein was progressively reduced with increasing doses of PNGase F, with maximal deglycosylation under non-denaturing conditions using 500 U PNGase F per 1 µg of Env. At this dose, JRFL Env gp140 molecular mass was reduced to near that of Env treated with PNGase F under denaturing conditions (approximately 80 kDa Env shown in lanes with an asterisk in Figure 1). In blue BN-PAGE gel, analysis, JRFL gp140 Env deglycosylated under native conditions using 500U/ug Env PNGase F migrated as monomers, dimers and trimers in the same manner as did wild-type (WT) glycosylated JRFL Env gp140 (Figure S1A).
Fig. 1. Analysis of deglycosylated JRFL protein in reducing SDS-PAGE and Western-blotting with a panel of HIV-1 MAbs. 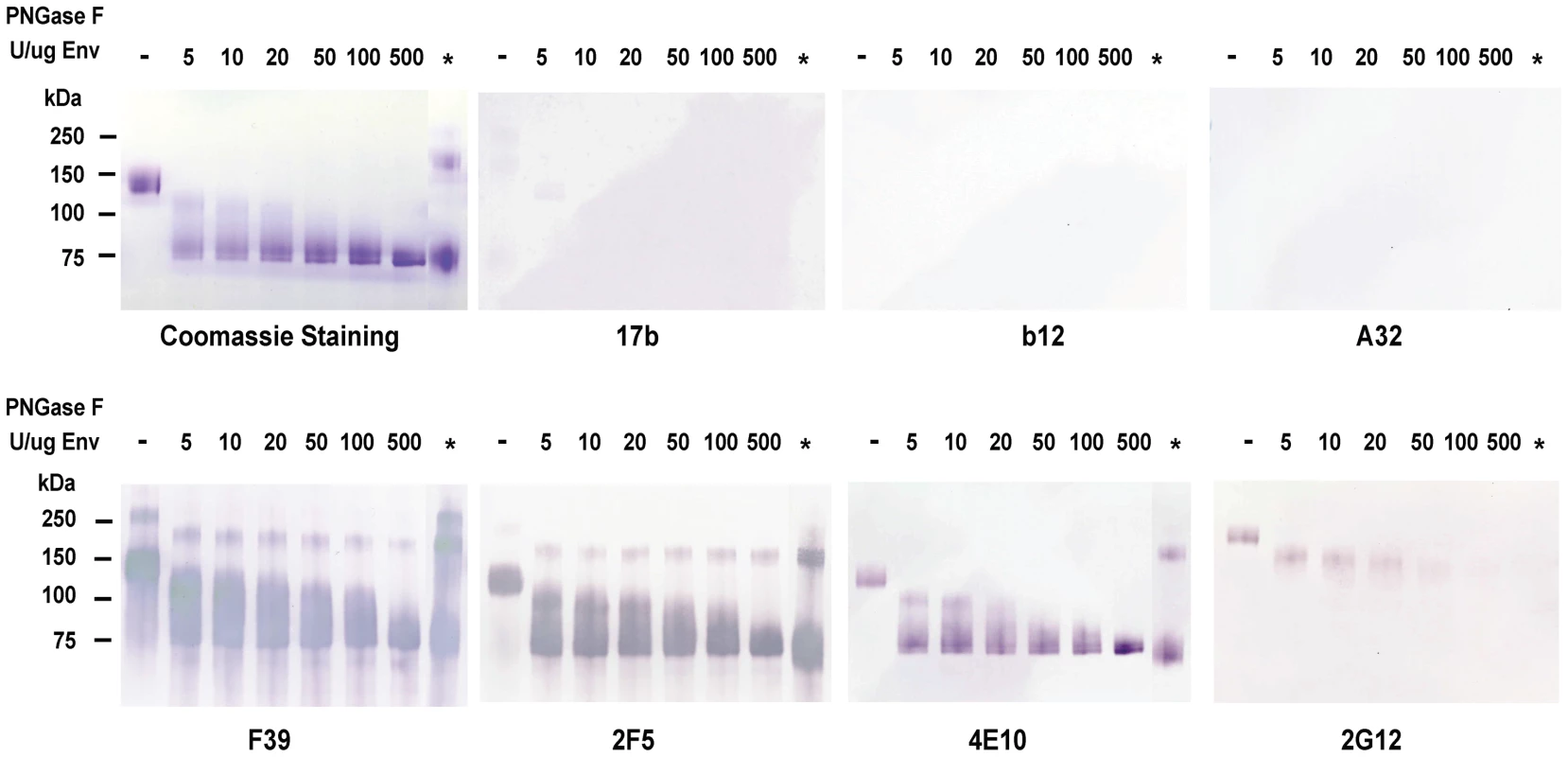
WT glycosylated and deglycosylated JRFL gp140 Env proteins were fractionated in 4–12% SDS-PAGE under reducing conditions, stained with coommassie blue or were transferred to nitrocelose membranes and blotted with a panel of human anti-HIV-1 mAbs as indicated. WT glycosylated JRFL gp140 Env protein is labeleled as [-] on the top of the lane. The amount of PNGase F (in U) used in the reaction per µg of protein is indicated on the top of lanes with progressively deglycosylation. Lane * refers to treatment of denatured JRFL gp140 protein with PNGaseF (500 U) at 37 degrees for 2 hours. Progressive reductions in molecular weight of JRFL gp140 with increasing amounts of PNGase used to deglycosylate JRFL gp140 Env are shown in Coomassie staining gels and Western blots using mAbs F39, 2F5 and 4E10. The higher molecular weight band(s) shown in lane * in the coomassie stained gel and in Western blots by F39, 2F5 and 2G12 were due to the incomplete reduction of JRFL gp140 Env protein (see coomassie stained gel in Figure S1). In SDS-PAGE and Western blot analysis under reducing conditions, there was weak or no binding of deglycosylated JRFL Env gp140 to the co-receptor binding site mAb 17b [30], to the CD4 binding site mAb 1b12 [20], or to the gp120 conformational-dependent mAb, A32 [31] (Figure 1). However, the binding of anti-V3 loop mAb F39F [31] to JRFL gp140 was not affected by deglycosylation when assayed under these conditions (Figure 1). In contrast, the binding of anti-carbohydrate mAb 2G12 to deglycosylated JRFL gp140 was decreased with progressive native deglycosylation. The binding of gp41 MPER human mAb 2F5 was not inhibited by deglycosylation or reduction, while progressive deglycosylation appeared to increase binding of the gp41 MPER antibody 4E10 to HIV-1 JRFL gp140 protein. Maximal binding of mAb 4E10 to JRFL Env protein was seen by using the deglycosylated Env treated with the highest level of PNGase F (500 U per µg of Env). This enhanced level of 4E10 binding was similar to the level of 4E10 binding to fully PNGase F-deglycosylated Env under denaturing conditions (asterisk, Figure 1).
To compare antigenic and functional epitopes, glycosylated and natively deglycosylated JRFL Envs were assayed for their ability to bind sCD4, and for their ability to undergo CD4 and mAb A32 induction of the CCR5 binding site as defined by binding of mAb 17b to Env. Thus, sCD4, mAb A32, or gp120 C1 control mAb T8 was immobilized on a surface plasmon reasonance (SPR) sensor chip and then either WT glycosylated or deglycosylated JRFL gp140 were flowed over the chip, followed by flowing over mAb 17b (Figures 2A, 2B). First, we found that WT glycosylated JRFL gp140 Env bound to sCD4 with Kd of 5.7 nM, while deglycosylated JRFL gp140 protein bound sCD4 with Kd of 35.3 nM (Figure 2A). The WT glycosylated JRFL gp140 Env was induced to bind mAb17b following binding to either sCD4 or mAb A32 with Kd of 9.5 nM and 6.0 nM, respectively, while the deglycosylated JRFL gp140 Env was induced to bind mAb17b following binding to either sCD4 or MAb A32 with slightly higher Kd of 25.0 nM and 7.5 nM, respectively. No binding of mAb 17b binding was observed following the binding of either WT glycosylated or native deglycosylated Env to gp120 C1 mAb T8 (Figure 2B). Thus, native deglycosylation of JRFL gp140 Env did not perturb the ability of the Env to undergo sCD4 and mAb A32-induced exposure of the CCR5 Env binding site.
Fig. 2. Analysis of antigenic epitopes expressed on WT glycosylated and deglycosylated JRFL gp140 by surface plasmon resonance (SRP). 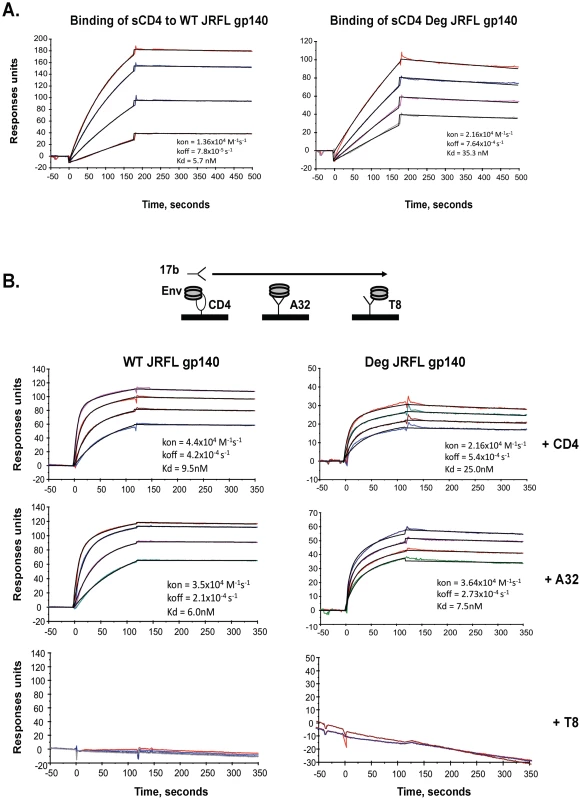
SPR assays were performed as described in Materials and Methods. Shown are the ability of WT glycosylated and deglycosylated JRFL gp140 to bind to sCD4 (Panel A) and to mAb 17B (Panel B). sCD4 or HIV-1 mAbs T8 and A32 were covalently immobilized to a CM5 sensor chip (BIAcore), and WT glycosylated (10 to 75 µg/ml) and deglycosylated JRFL gp140 were injected over each surface (100 and 300 ug/ml, respectively). To determine induction of 17b mAb binding to WT glycosylated and deglycosylated JRFL gp140, Env proteins were captured on individual flow cells immobilized with sCD4 (80–120 response units or RU) or mAb A32 (∼130–300RU) or T8 (∼340–450 RU), with about the same RU of Env gp140 captured on each surface prior to injection of varying concentrations of 17 mAb. Following stabilization of each of the surfaces, mAb 17b (10–100 µg/ml) was injected and flowed over each of the Env captured surfaces as illustrated in the diagram above the SPR profile in Panel B. Each analysis was performed at least twice. Increased Affinity of 4E10 and 2F5 mAbs for Deglycosylated Env
To ask if indeed, the deglycosylation of JRFL oligomer induced exposure of the MPER as suggested by reduced SDS-PAGE Western blot analysis, we performed indirect ELISAs using serially diluted human mAbs and a fixed amount of Env proteins captured on ELISA plates by anti-gp120 C1 mouse mAb 3B3 [32]. In this assay, both mAbs 2F5 and 4E10 bound glycosylated JRFL Env protein with the apparent Kd of approximately 67 nM and 33 nM, respectively, while both mAbs 2F5 and 4E10 bound significantly better to the natively deglycosylated JRFL Env with apparent Kds of 4.9 nM and 6.1 nM, respectively (Figure 3). The apparent Kds of mAbs 2F5 and 4E10 binding to natively deglycosylated JRFL Env proteins treated with either 500 U, 20 U or 5 U per µg Env were all similar with apparent Kds of 3.7 nM, 4.9 nM and 4.5 nM for mAb 2F5, and 4.4 nM, 5.1 nM and 6.1 nM for mAb 4E10 at these PNGase F concentrations, respectively (Figure 3).
Fig. 3. Binding kinetics of mAbs 2F5 and 4E10 to WT glycosylated and deglycosylated JRFL gp140. 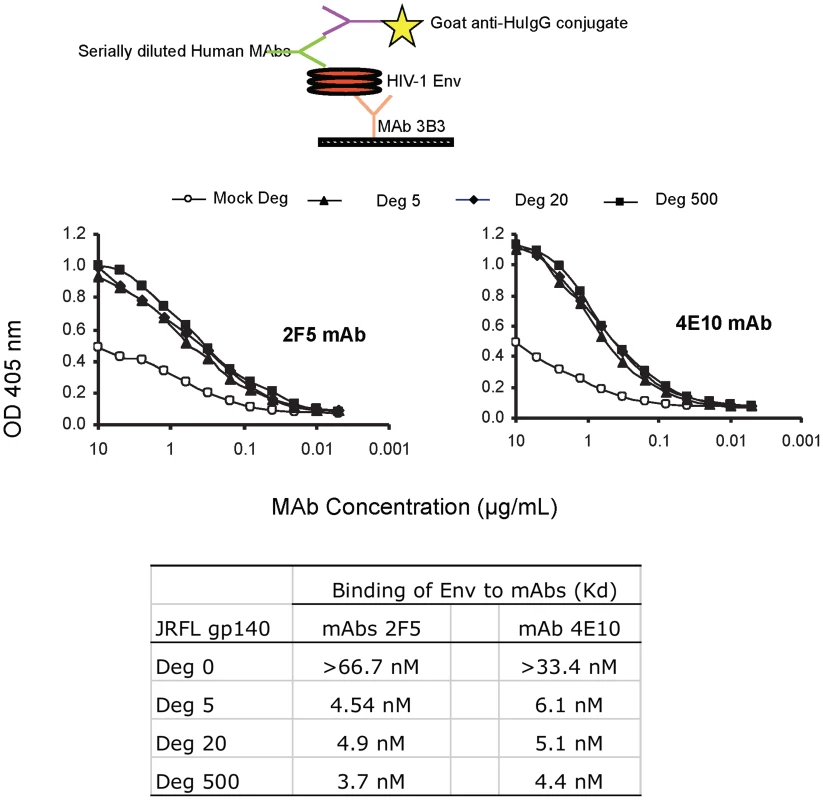
Binding of mAbs 2F5 and 4E10 to WT glycosylated and progressively deglycosylated JRFL gp140 proteins was determined in direct ELISA. An anti-HIV-1 Env C1 region antibody 3B3 [31] was coated on the 96-well plates to capture glycosylated and progressively deglycosylated JRFL gp140 proteins following by incubation with mAbs 2F5 and 4E10. The mAbs 2F5 and 4E10 were assayed at the concentrations as indicated on the x-axis. Data presented are representative of 3 independent experiments. We next performed SPR analysis of binding of mAbs 2F5 and 4E10 to WT glycosylated and deglycosylated JRFL Env proteins by flowing either glycosylated or deglycosylated JRFL Env proteins over mAbs 4E10 or 2F5 captured on an SPR sensor chip (Figure 4). Marked enhancement of mAb 4E10 binding to deglycosylated JRFL Env (Figure 4A) was observed compared to the binding of mAb 4E10 to the WT glycosylated JRFL (Figure 4B). SPR analysis demonstrated that the enhancement of 4E10 binding to deglycosylated JRFL proteins was due to both a faster on - rate (Kon) and a slower off rate (Koff) that resulted in a net 15-fold decrease in the overall dissociation constant (Kd) of 4E10 binding from 154 nM for glycosylated JRFL Env to 10 nM for native deglycosylated JRFL Env (Figures 4B and 4A). Similar but weaker enhancement (6-fold) of 2F5 mAb binding to deglycosylated JRFL Env was seen in SPR analysis (Kd = 38 nM for 2F5 binding to deglycosylated Env vs. Kd = 212 nM of 2F5 binding to WT glycosylated Env) (Figures 4C and 4D).
Fig. 4. SPR assays for native and deglycosylated JRFL gp140 binding to mAbs 4E10 and 2F5. 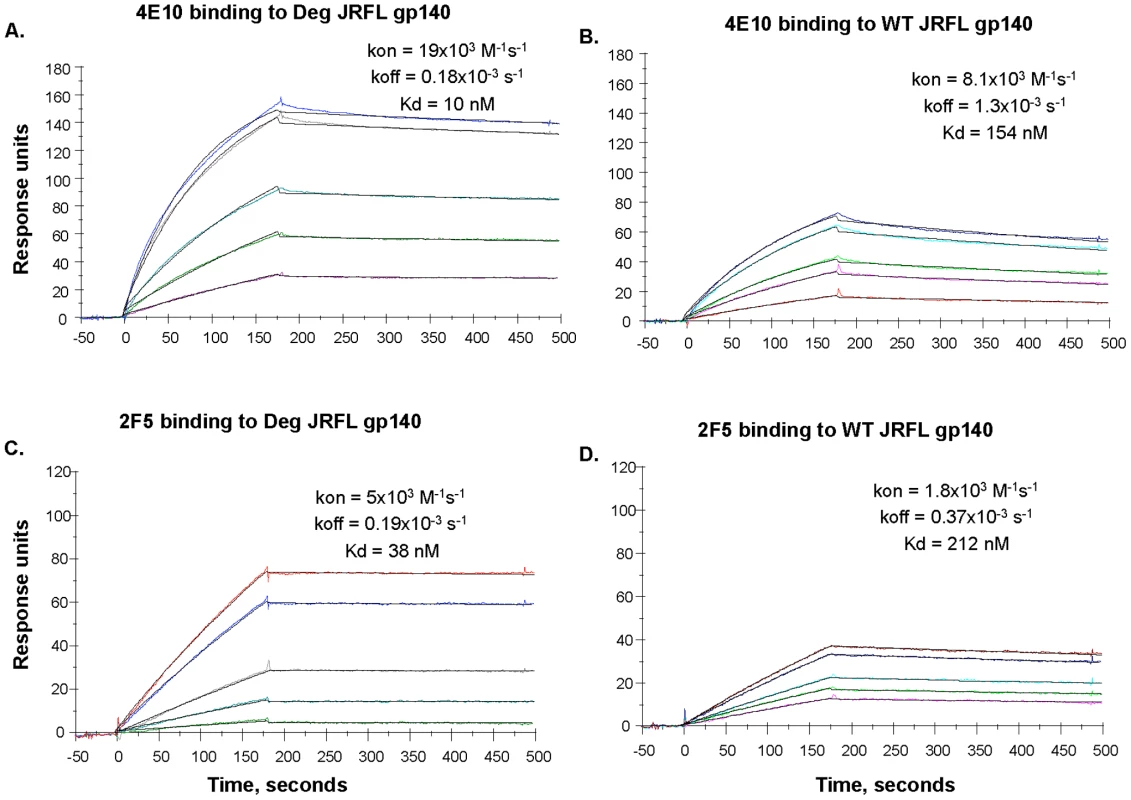
MAbs 4E10 (Panel A, B) and 2F5 (Panel C, D) were bound to anti-human Ig Fc immobilized on the CM5 sensor chip. Varying concentrations (ranging from 5 to 100 ug/mL) of native (WT) and deglycosylated (Deg) JRFL gp140 Envs were injected over the mAbs on the chip and binding kinetics were recorded. Rate constants and Kd measurements were made using the 1∶1 Langmuir model and data recorded are indicated in the individual panels. Deglycosylation of JRFL gp140 enhanced the Kd of 4E10 and 2F5 binding by ∼15-fold and ∼6-fold respectively. Data shown are representative of two experiments. Preserved DC-SIGN Binding By Natively Deglycosylated JRFL Env
We next evaluated the binding of natively partially deglycosylated Env to DC-SIGN using AF647-labeled JRFL Env. We found that neither partial deglycosylation nor labeling of AF647 had any effect on the binding of JRFL gp140 Env protein to CD4 expressing TZM-b1 cells (not shown). Natively deglycosylated JRFL gp140 Env (treated with 500 U PNGase F per µg of Env) retained the ability to bind to DC-SIGN-transfected 3T3 cells but not control 3T3 cells (Figure 5) indicating preserved high mannose residues on the PNGase-treated natively partially deglycosylated Env.
Fig. 5. Binding of WT glycosylated and deglycosylated JRFL gp140 Env on DC-SIGN-expressing NIH 3T3 cells. 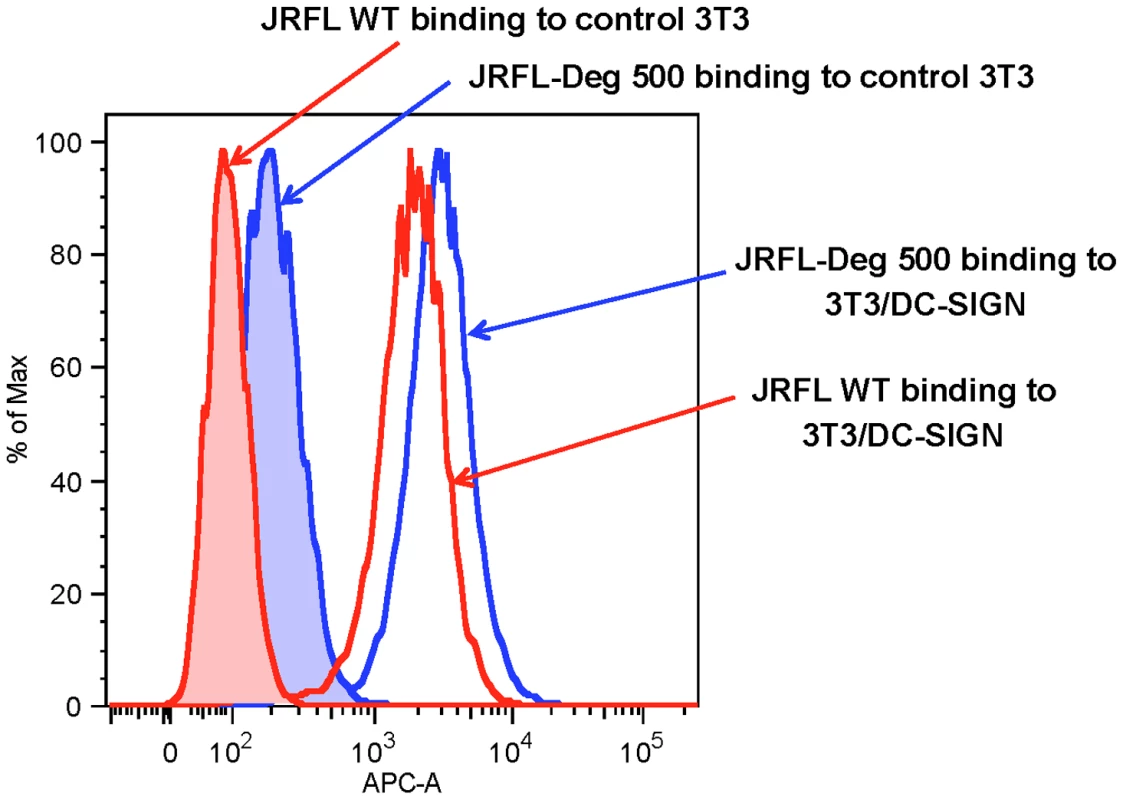
The binding of AF647-labeled WT glycosylated (histogram in red lines and deglycosylated JRFL Env gp140 (histograms in blue lines) to parent NIH 3T3 (histograms shaded) and DC-SIGN-expressing NIH 3T3 cells (histograms not shaded) were analyzed by flow cytometry. Data shown are representative of three experiments. The 2G12 Epitope was Maintained on JRFL Env After Deglycosylation Under Native Conditions
It has previously been reported that binding of mAb 2G12 requires gp120 Env N-linked glycan at positions N286, N323, and N330 [33]. Since native deglycosylation with PNGase F is not complete, and the maximally deglycosylated JRFL gp140 Env (treated with 500 U PNGase F per µg of Env) bound to DC-SIGN transfected 293T cells, we asked if 500 U PNGase F-deglycosylated gp140 could also bind the anti-gp120 carbohydrate-dependent mAb 2G12.
Although the binding of 2G12 to deglycosylated JRFL protein was weak in reducing SDS-PAGE Western blot analysis (Figure 1), in SPR analysis in which Env is flowed over captured mAb, binding of 2G12 to JRFL gp140 protein was maintained when Env was treated with PNGase F (Figure 6). While 5U and 20U of PNGase F did not decrease mAb 2G12 binding, 500U of PNGase F treatment did decrease, but did not completely abrogate, mAb 2G12 binding (Figure 6).
Fig. 6. SPR assays for binding of native and deglycosylated JRFL gp140 to mAb 2G12. 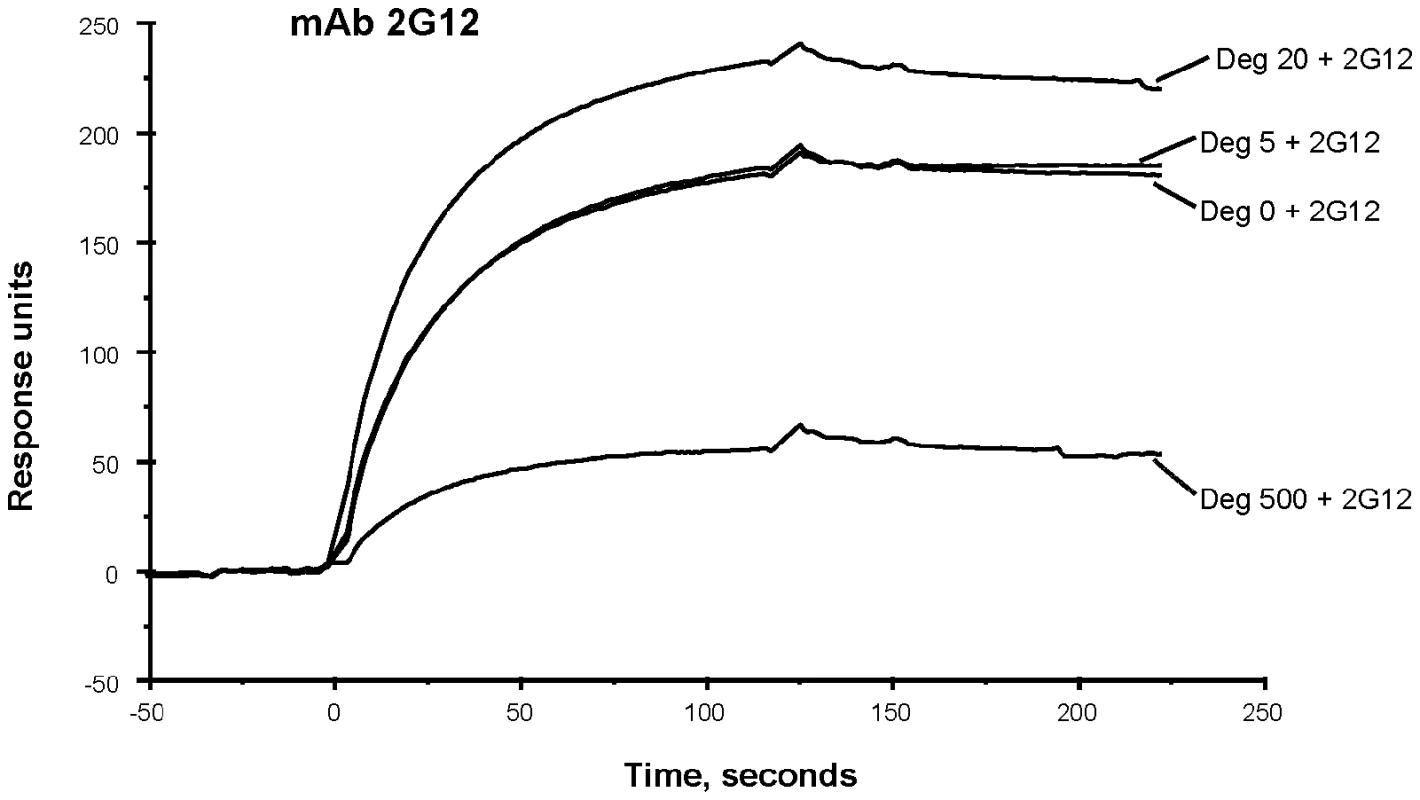
MAb 2G12 was immobilized on the CM5 chip. The saturated amounts of native and representative deglycosylated proteins were then flown over the mAbs. The binding kinetics were recorded at 25°C. Deglycosylation of JRFLgp140 using lower dose of PGNase (20 U: µg) enhanced the binding of 2G12 but substantially decreased binding of 2G12. Data show binding curves of each deglycosylated and WT glycosylated protein injected at 50 µg/mL and 200 µg/mL respectively, and are representative of two experiments. Characterization of Env Carbohydrates Remaining on JRFL gp140 After Deglycosylation Under Non-denaturing Conditions
PNGase F 500 U-treated JRFL gp140 was subjected to enzymatic digestion and LC-MS/MS analysis to define sites and forms of glycan remaining on optimally deglycosylated gp140 (500 U PNGase F) [34], [35]. JRFL gp140 has 27 potential N-linked glycosylation sites, and the occupied sites are composed of high mannose, hybrid, and complex glycans [34], [35]. The remaining glycosylation on these sites was identified using a combination of high resolution MS and MS/MS data of the endoglycosidase-treated glycopeptides, as described in the section od Materials and Methods. The resulting MS data is summarized in Supplemental Table 1. Each peptide containing a potential glycosylation site was detected as either a glycopeptide, a deglycosylated peptide, or as a natively nonglycosylated speices.
Based on the amino acid sequence of JRFL gp140 (including the signal peptide), all but four of the sites in the untreated protein were fully or partially occupied with glycans (Figure 7 and Table S1) (34). The unoccupied sites were: N141, N188, N611and N637. Upon extensive deglycosylation (500 U PNGase F per 1 µg of protein), the glycans at 12 out of 23 sites were removed: N88 (C1), N135 (V1/V2), N138 (V1/V2), N156 (V1/V2), N160 (V1/V2), N187 (V1/V2), N262 (C2), N276 (C2), N356 (C3) N462 (C4), N616, and N625 (Figure 7). The 10 sites on which glycosylation still remained were fractionally populated with glycans. Thus, PNGase F cleaved a fraction of the glycans at these sites, but the reaction was incomplete. Most of the conserved glycans were of the high mannose type and included those glycans involved in the 2G12 binding (N295, N332, N386) [33] (Figure 7). Figure 8 shows a model of JRFL gp140 Env before PNGase-F treatment (Figure 8A) and after PNGase-F treatment (Figure 8B) based on the cryo-EM structure (white mesh) of Liu et al. [36] and specific-site glycan analysis before PNGase-F treatment [34] and after treatment (Figure 7). Red molecules indicate gp120 amino acids, blue indicates gp120 glycans, white indicates 2G12 glycans [37], and yellow indicates gp41 glycans. Figure 8 depicts the loss of many complex glycans on gp120 and the loss of two gp41 glycans after PGNase-F treatment.
Fig. 7. Glycan analysis of native partially deglycosylated JRFL gp140 protein. 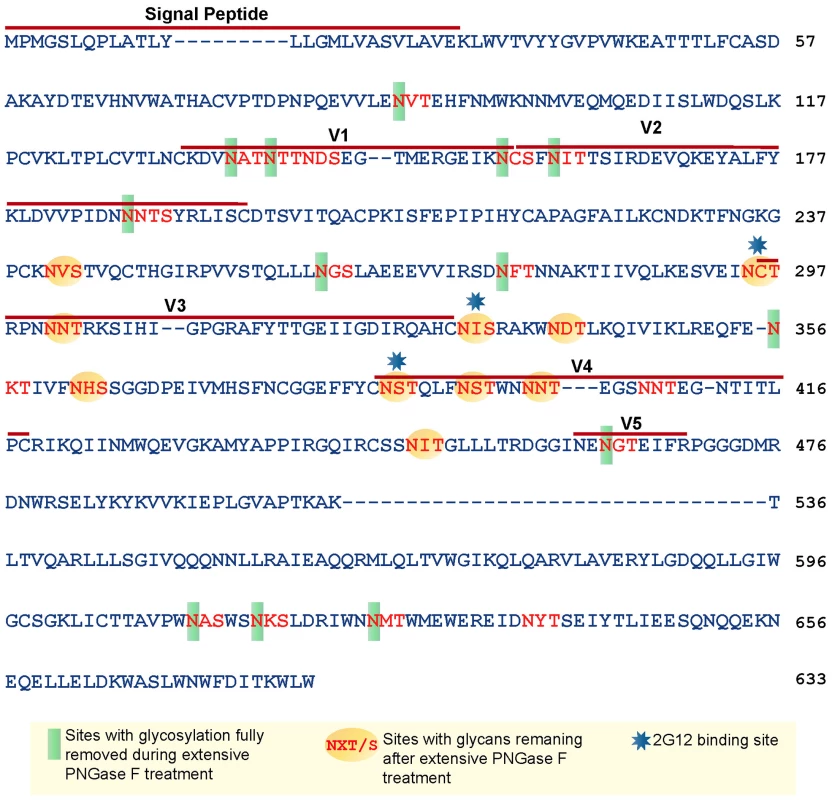
Native deglycosylation of JRFL gp140 protein was performed using PNGase F (500 unit/µg protein) as described in the materials and methods. Amino acid sequence numbers were based on the HXB2 sequence. The signal peptide and variable regions of the sequence were identified above the sequence. See Table S1 for mass spectrometry analysis data summaries. Fig. 8. Model of glycosylation patterns of recombinant WT JRFL gp140 and partially deglycosylated gp140 under non-denaturing conditions. 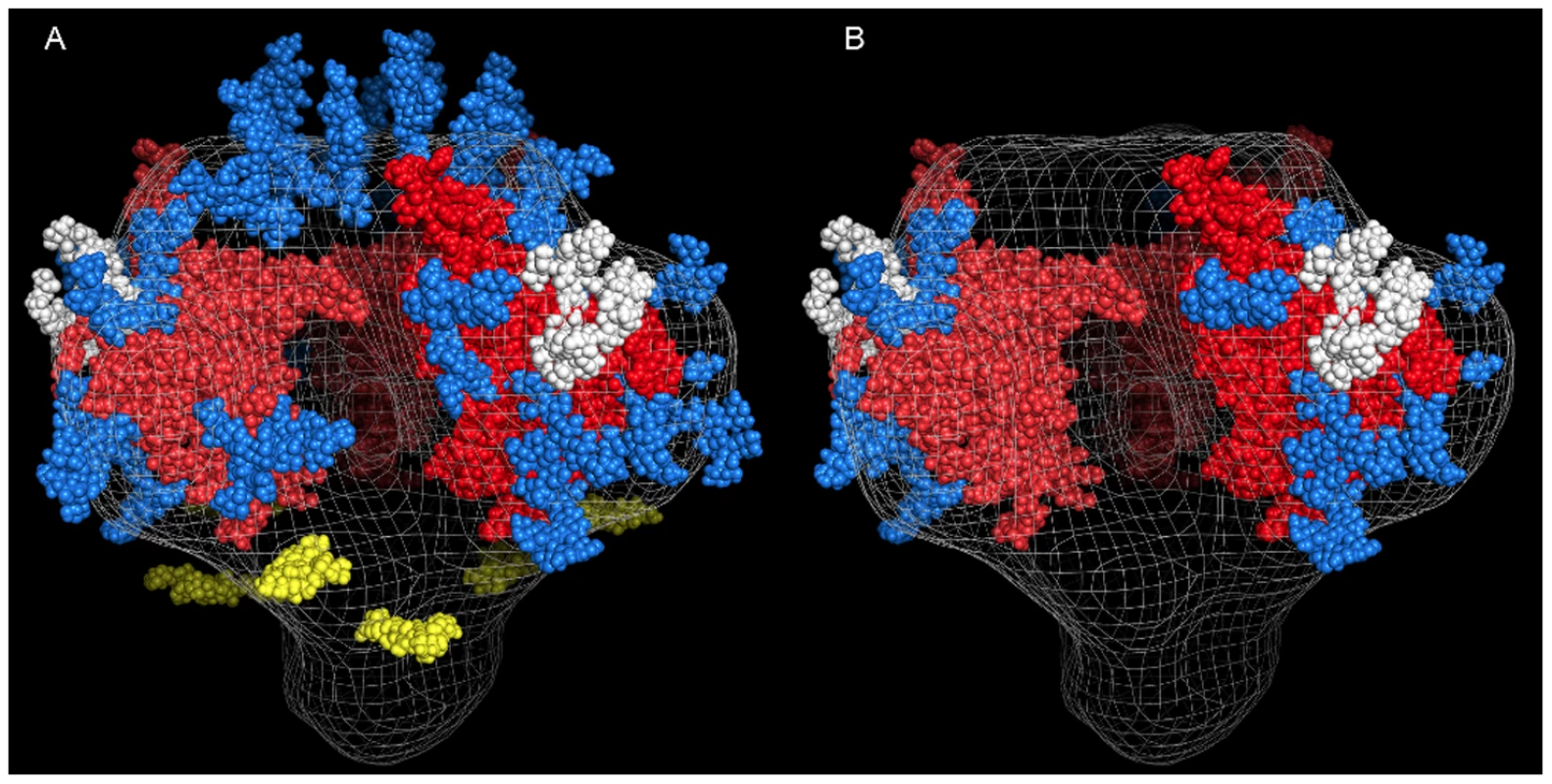
Shown are the images that were modeled based on the cryo-electron tomography as described by Schief [60]. Panel A: HIV-1 recombinant JRFL Env exhibits a number of high mannose glycans as determined by mass spectrometry (Figure 7 and Table S1). In this figure, the gp120 bearing the V3 loop [61] (PDB 2B4C)] trimer (red) is modeled over the cryo-EM structure (white mesh) (36), and its representative glycans are indicated in blue with those that are bound by the antibody 2G12 [37] in white. The fold of the prefusion gp41 trimer is not known, so gp41 glycans (yellow) have been positioned approximately equidistant around the stalk. Glycans bound to gp120 residues in the V1 and V2 loops have similarly been placed in reasonable locations across the top of the trimer. Depicted in Panel B are the glycans remaining after treatment with PNGase-F. In general, PNGase is able to remove the glycans that are attached to flexible parts of the gp120 structure, presumably allowing the enzyme to gain access to recognition elements and cleavage points. Glycans attached to Asn residues occuring in relatively stable structural elements of gp120 persist after PNGase-F treatment. Binding of mAbs 4E10 and 2F5 to gp140 Is to gp41 and Not to gp120
To determine if there were ancillary binding sites located on gp120, as previously suggested [38], binding of mAbs 4E10 and 2F5 to deglycosylated HIV-1 Env 89.6 gp120 was determined using the same non-denaturing conditions as used for binding of mAbs 2F5 or 4E10 mAb binding to deglycosylated JRFL gp140. We found that while sCD4 bound well to HIV-1 89.6 Env gp120 deglycosylated with up to 20 U PNGase F treatment, no binding of either mAbs 2F5 or 4E10 to gp120 was seen (Figure S2). Thus, the enhanced binding of mAbs 4E10 and 2F5 to the deglycosylated JRFL gp140 protein was indeed due to the binding to the gp41 component of gp140.
Next, we asked if absorption of 50 µg/ml of 4E10 by increasing amounts of either deglycosylated JRFL gp140 or deglycosylated HIV-1 Env 89.6 gp120 could absorb the 4E10 binding activity for the P-4E10 nominal peptide epitope.
As shown in Figure S3, addition of dilutions of absorbing JRFL Env gp140 protein to 50 µg/ml mAb 4E10 absorbed 4E10 binding activity to nominal 4E10 gp41 peptide, while addition of Env 89.6 gp120 (both WT glycosylated and PNGase F 20 U-treated) did not (Figure S3). Thus, the expressed epitope on JRFL gp140 is located on gp41 and PNGase F-treated gp140 absorbed 4E10 binding activity to the normal gp41 4E10 epitope peptide.
MAb 4E10 binding was the most enhanced of MPER antibodies to PNGase F-treated JRFL Env. To determine if the 4E10 neutralizing epitope was the target for 4E10 binding, we next asked if the nominal 4E10 binding epitope peptide (P-4E10: SLWNWFNITNWLWYIK) could absorb out the mAb 4E10 binding activity to deglycosylated JRFL gp140 (Figure S4). As shown in Figure S4, increasing amounts of the 4E10 epitope peptide absorbed the binding activity of 5 µg/ml mAbs 4E10 to the deglycosylated JRFL gp140.
Analysis of Deglycosylated JRFL Env Binding to 2F5 and 4E10 Reverted Unmutated Ancestors (Putative B Cell Receptors of Naïve B Cells)
While the somatically mutated 4E10 and 2F5 mAbs bound better to natively deglycosylated Env than to glycosylated Env, an immunogen needs to trigger the unmutated germline B cell receptors on the surface of naïve B cells [39], [40]. It has been previously demonstrated that a 2F5 RUA did not bind to fully glycosylated Env [16]. To assess Env binding to reverted unmutated antibody ancestors of the mutated mAbs 4E10 and 2F5, we used the program SoDA-2 [41] and Phylip's NAML [42] to infer RUAs of mAbs 2F5 and 4E10 resulting in two inferred RUAs of mAb 2F5 and one RUA of mAb 4E10 (Table S2 in SOM). These RUAs were produced in 293T cells as whole IgG1 antibodies by transfection and tested for binding to HIV-1 JFRL gp140 Env proteins. We found that indeed, the inferred RUAs of mAbs 4E10 and 2F5 did not bind to glycosylated JRFL gp140 Env, as previously shown for the 2F5 RUA by Xiao et al. [16]. However, both 2F5 and 4E10 RUAs did bind to natively deglycosylated JRFL gp140 Env oligomer (Figure 9). While the binding of the 2F5 RUAs to fully glycosylated JRFL gp140 Env was unmeasurable (Figures 9A and 19C), the binding Kds for partially deglycosylated JRFL gp140 Env were 258 nM and 270 nM, respectively (Figures 9B and 9D). Similarly, the binding of the 4E10 RUA for glycosylated Env was also unmeasurable (Figure 9E), while the binding Kd of the 4E10 RUA for the natively deglycosylated Env was 318 nM (Figure 9F). Thus, gp140 glycans mask or modulate the availability of MPER neutralizing epitopes for 2F5 and 4E10 RUA binding to gp41 linear neutralizing epitopes on the JRFL Env gp140 oligomers.
Fig. 9. SPR measurements of binding of inferred RUAs of 2F5 and 4E10 to WT glycosylated and natively deglycosylated JRFL gp140 Env proteins. 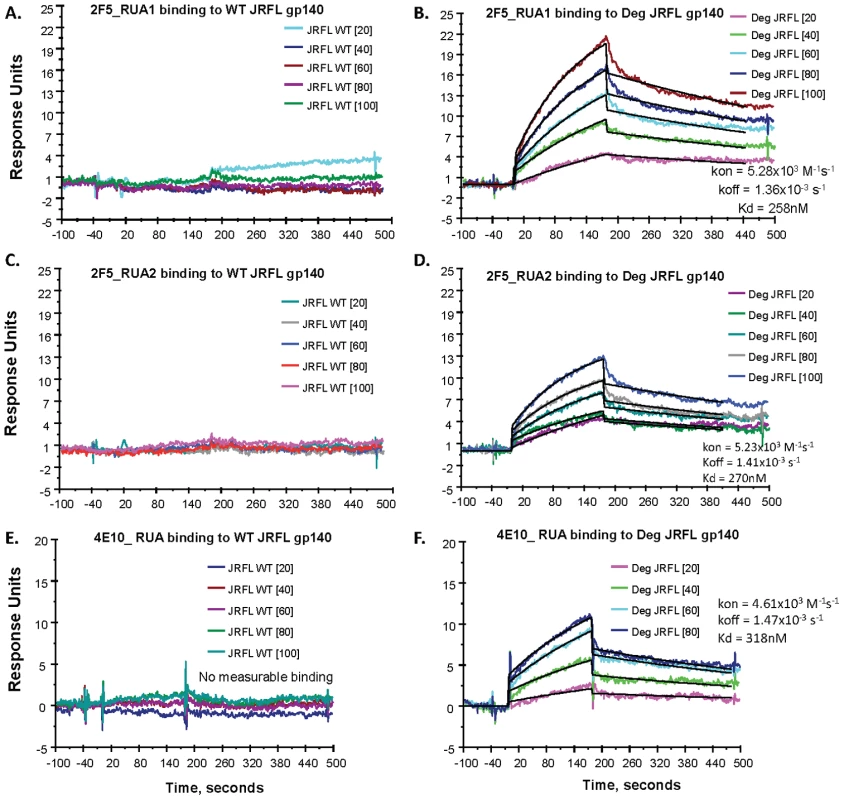
A dose range (from 20–100 µg/mL as indicated) of RUA1 (A–B) and RUA2 (C–D) inferred from the mutated 2F5 antibody sequence and one RUA (4E10 RUA, 20–80 µg/mL) (E–F) inferred from the 4E10 mutated antibody sequence were tested by SPR for binding to WT glycosylated (WT) and maximally deglycosylated (Deg) JRFL gp140 Env proteins immobilized on adjacent flow cells of the same sensor chip. Each antibody was captured on an anti-Ig Fc immobilized chip as described [7]. Neither of the RUAs of 2F5 (Panels 9A–9C) nor 4E10 (Panel 9E) bound to WT glycosylated JRFL gp140 Env protein, while reactivity of the RUAs of 2F5 (Panels 9B–9D) and 4E10 (Panel 9F) to deglycosylated JRFL gp140 Env proteins was present with the Kds and rate constants shown in each panel. Rate constants were derived by curve fitting analysis and using a 1∶1 Langmuir model. Demonstration that Native Partial Deglycosylation of Another Env, the Group M Consensus Env, CON-S gp140, also Exposes the MPER Region and Enhances Binding of Deglycosylated Env to 2F5 and 4E10 RUAs
To determine if the effect of PNGase F on JRFL gp140 is generalizable to other Envs, we studied the effect of partial native deglycosylation on the group M consensus gp140 Env CON-S [29]. This Env has previously been reported to induce a degree of breadth among clade A, B and C Tier 1 virus strains [29]. We found that, like JRFL gp140 oligomers, PGNgase F 500 U treatment under native conditions of CONS gp140 resulted in reduced MW of monomers, dimers and trimers, and did not lead to Env degradation (Figure S1). Second, native deglycosylation of CON-S gp140 resulted in maintainance of sCD4 binding (Kd = 7.4 nM for CON-S glycosylated Env and Kd = 9.4 nM for deglycosylated Env) (Figure S5). Third, like deglycosylated JRFL gp140, deglycosylated CON-S gp140 Env could undergo sCD4 and A32 mAb-induced 17b binding to the CCR5 co-receptor gp120 binding site (Figure S6), and both mAbs 2F5 (Figures S7A and S7B) and 4E10 (Figures S7C and S7D) bound with lower Kds to deglycosylated CON-S gp140 when compared to WT glycosylated CON-S gp140.
Interestingly, the WT glycosylated CON-S gp140 constitutively bound (before deglycosylation) to both 2F5 RUAs 1 and 2 (Figures S8A and S8C), demonstrating that some glycosylated recombinant Envs do have the ability to bind to broadly neutralizing antibody germline antibodies. 2F5 RUA1 bound better to glycosylated CON-S gp140 than did 2F5 RUA2 (Kd = 118 nM for RUA1 vs Kd = 4.0 µM for RUA2). The enhancement of binding to the deglycosylated CON-S was only two-fold for RUA1 (Kd = 67 nM) while the enhancement of binding of 2F5 RUA2 for deglycosylated CON-S gp140 Env was an order of magnitude (Kd = 4.0 µM for WT glycosylated CON-S gp140 and Kd = 0.36 µM for deglycosylated CON-S Env).
For CON-S gp140, the pattern of binding to the 4E10 RUA was similar to that of JRFL gp140, with no measurable binding of glycosylated CON-S gp140 to the 4E10 RUA, while the deglycosylated CON-S gp140 bound to the 4E10 RUA with a Kd of 0.23 µM (Figures S8E and S8F). That WT glycosylated CON-S gp140 binds to the RUAs of 2F5 in this setting is not surprising since we have previously published that CON-S gp140 produced in 293T cells (as the CON-S gp140 was produced in this study) is not glycosylated in gp41 [34]. That the expression of the epitopes for both mAbs 2F5 and 4E10 are further enhanced on deglycosylated CON-S therefore suggests that removing glycans on gp120 can also affect exposure of MPER neutralizing epitopes. Regardless, the effect of PNGase F native deglycosylation is not unique to JRFL gp140 Env.
Immunogenicity of HIV-1 JRFL gp140 and Deglycosylated JRFL gp140 Env Proteins in Rhesus Macaques
To determine if the enhanced binding of mAbs 4E10 and 2F5 and their RUAs to deglycosylated HIV-1 JRFL gp140 Env might have resulted in enhanced immunogenicity for induction of MPER antibodies, 4 rhesus macaques per group were immunized with either deglycosylated JRFL Env gp140 or WT JRFL gp140. Plasma samples were obtained 2 weeks after the first priming and second boosting immunizations and evaluated for binding antibody to the immunizing Env and to 2F5 and 4E10 nominal membrane proximal external region (MPER) peptides. Both the deglycosylated HIV-1 JRFL gp140 Env protein and WT JRFL gp140 Env were immunogenic and induced similar levels of antibodies to the immunizing Envs after 2 immunizations (Figure 10A). Whereas the WT glycosylated JRFL gp140 Env induced minimal levels of antibodies after the first and second immunizations to a 2F5 epitope peptide (QQEKNEQELLELDKWASLWN), in contrast, the deglycosylated JRFL gp140 Env induced enhanced levels of 2F5 epitope antibodies after the second immunization in 3 of 4 animals (Figure 10B) (p = 0.035, Student's t test). However, neither the deglycosylated HIV-1 JRFL Env proteins nor WT JRFL gp140 Env induced antibodies to HIV-1 MPER 4E10 epitope peptide, SLWNWFNITNWLWYIK (data not shown). Finally, while antibodies were present in both groups that neutralized the SF162.B pseudovirus, neither group neutralized the 2F5-sensitive HIV-1 Env BG1168.B pseudovirus (data not shown), indicating that the induced rhesus MPER antibodies either were non-neutralizing or were of insufficient titers to neutralize. Nonetheless, these data demonstrate proof-of-concept of enhanced 2F5 epitope immunogenicity of native deglycosylated JRFL gp140 Env.
Fig. 10. Enhanced immunogenicity of deglycosylated JRFL gp140 Env as determined by plasma levels of 2F5 epitope binding antibodies in immunized rhesus macaques. 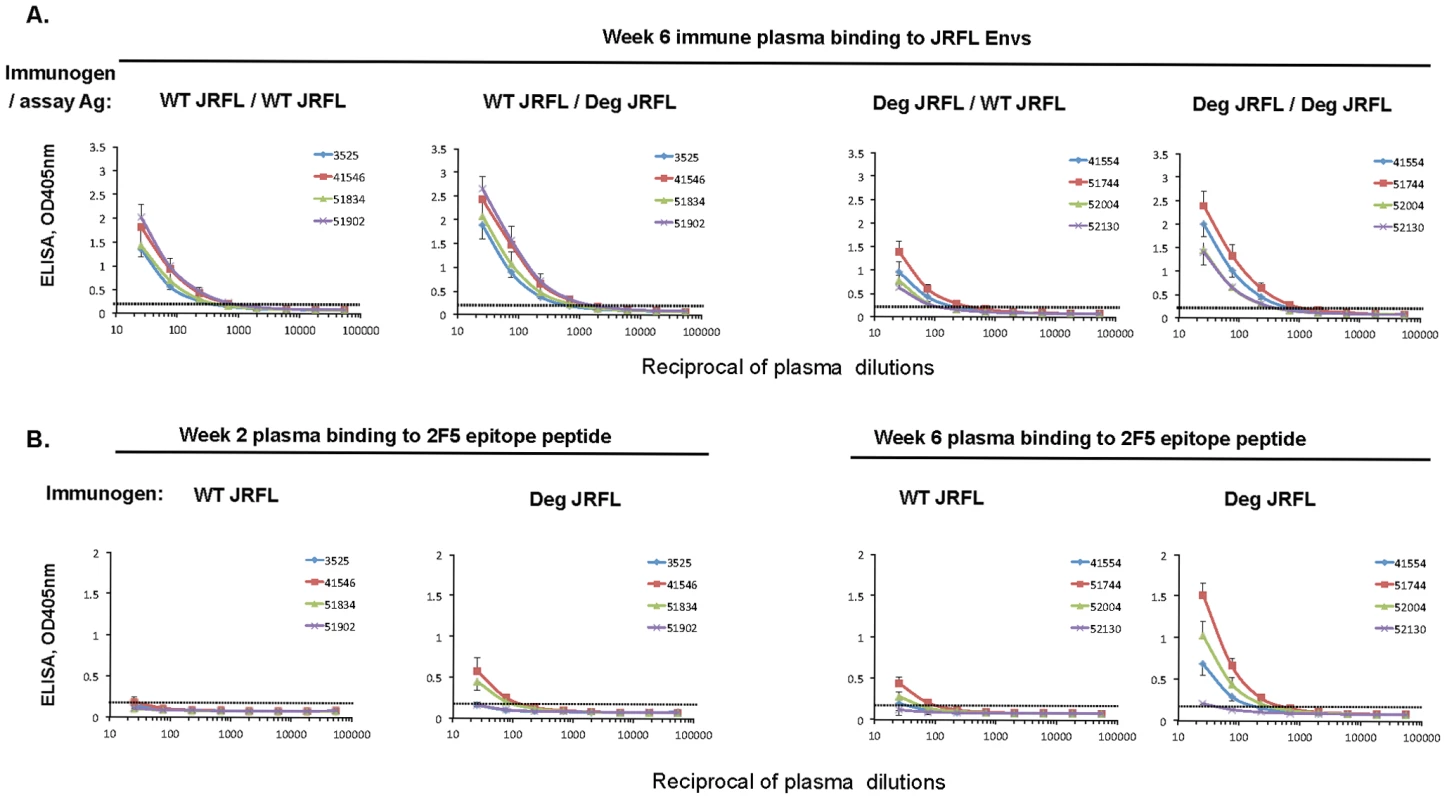
Rhesus macaques (4 per group) were immunized at weeks 0 (prime) and 4 (boost) with 100 ug of either glycosylated (WT) or natively deglycosylated (Deg) JRFL gp140 Env. Plasma samples were collected at weeks 2 and 6 from each group (as indicated on the top of the panels) and were assessed for binding to WT JRFL gp140 Env and Deg JRFL gp140 Env (Panel 10A). The antibody responses to wild-type JRFL Env or to Deg JRFL Env were undetectable after the prime and are not shown. The responses to WT JRFL Env and Deg JRFL Env after the week 4 boost (the week 6 blood draws) are shown. The antibody response to 2F5 nominal MPER peptide (QQEKNEQELLELDKWASLWN) after the prime (week 2) and boost (week 6) is shown (Panel 10B). Shown on the x axis are the reciprocal dilutions of plasma of the immunized moneys used in ELISA. The dotted lines indicate the negative cut-off values. The level of 2F5 epitope antibody response at the initial dilution (1∶25) in week 6 plasma were higher in animals immunized with deglycosylated JRFL Env than in animals immunized with WT JRFL Env (p = 0.035, Student's t test). Data shown are mean values +/− standard deviation derived from 3 separate experiments performed. Discussion
We have demonstrated that non-denaturing (native) deglycosylation of HIV-1 JRFL and the group M consensus gp140 envelope oligomers with PNGase F resulted in enhanced Env binding to gp41 broadly neutralizing mAbs 4E10 and 2F5, and as well, induced or enhanced binding of gp140s to 2F5 and 4E10 reverted unmutated ancestor antibodies. Moreover, immunization of the JRFL deglycosylated gp140 Env resulted in enhanced antibody induction to the gp41 2F5 peptide epitope.
Treatment of Env with glycosylation inhibitors and neuraminidase has been reported to enhance HIV-1 binding and neutralization by mannose-binding lectin, and to facilitate immunocapture of virions by both mAbs 2G12 and 2F5 [43]. Thus, the selective removal of complex glycans by non-denaturing PNGase F treatment may similarly expose the 2F5 and 4E10 MPER binding site for enhanced mAbs 2F5 and 4E10 binding. MAb 2G12 has been reported to bind to gp120 N-linked high mannose residues at N295, N332, and N392, as well as with peripheral glycans from 386 and 448 [3], [37]. Interestingly, our mass spectrometry analysis of deglycosylated JRFL gp140 demonstrated selective loss of N-linked glycans particularly around V1 and V2 with high mannose residues at N295, N332, and N392 remaining intact [34], [35]. The non-denatured deglycosylated JRFL gp140 Env bound well to mAb 2G12 and also bound to DC-SIGN. In this regard, the gp120 high mannose residues involved in 2G12-gp120 binding have been reported to be involved in gp120-DC-SIGN binding [44]. Thus, selective removal of gp140 glycans may induce conformational changes or unmasking of the gp41 MPER 4E10 epitope that results in enhanced on-rate (ka) of mAbs 4E10 and 2F5 binding.
Walker et al. have recently reported that 25% chronically infected HIV-1 subjects with broad neutralizing antibodies had neutralizing plasma antibody activity targeted at Env glycans at N332, and at N295 in some subjects [45]. Although N332 and N295 glycans are involved in mAb 2G12 binding, their N332 and/or N295 activity was found to bind a glycan epitope distinct from that of 2G12 [45]. That PNGase treatment of HIV-1 Env under non-denaturing condition retained all N-linked glycans involved in 2G12 and in the novel N295/N332 neutralizing epitopes, suggests natively deglycosylated HIV-1 gp120 or gp140 Env may be of use in vaccine formulations targeted at elicitation of anti-glycan gp120 neutralizing antibodies.
Yuste et al., has demonstrated that mutations in gp41 N-linked glycosylation sites exposed a neutralization epitope N-terminal to the MPER [23]. From Yuste et al., we would predict that cleavage of N-linked glycans at 611 616, 625 and/or 637 might also be responsible for directly unmasking the 2F5 and 4E10 gp41 epitopes, or inducing conformational changes of the Env that indirectly expose the gp41 membrane proximal external region, or both. Indeed, three out of four glycosylation sites in gp41 were occupied in recombinant untreated JRFL gp140, and were removed in natively deglycosylated JRFL gp140 (Figure 7). Additionally, we and others have previously shown that enhanced exposure of the MPER epitope by single amino acid changes can increase the binding and neutralizing activity by broadly neutralizing MPER antibodies [46]–[48].
Since the 4E10 antibody is markedly polyreactive [6], it was important to consider that the 4E10 epitope exposed on PNGase F-treated Env gp140 might be distant from the 672WTDIT-NWLWY681 gp41 4E10 nominal epitope. Hages-Braun et al. have suggested that two ancillary 4E10 binding sites are located on gp120 at 34 LWVTVYYGVPVWK46 and on gp41 at 512AVGIGAVFLGFLGAAGSTMGAASMTLTVQAR542 [38]. However, our studies demonstrated no 4E10 binding to PNGase F-treated gp120, and PNGase F-treated gp140, but not gp120, absorbed the binding of mAb 4E10 to the gp41 peptide (P-4E10, SLWNWFNITNWLWYIK). These data suggested that the reason that mAbs 2F5 and 4E10 RUAs can bind PNGase-treated JRFL gp140 is most likely due to enhanced exposure via glycan unmasking of the gp41 MPER, or an induced gp41 MPER conformational change that allowed for a faster on-rate for mAbs 2F5 and 4E10 to bind to gp41. That most of the deglycosylated forms of JRFL and CON-S gp140 are oligomeric and only a minor species is monomeric for both Envs (Figure S1), demonstrates that the exposure of the MPER is on oligomeric Env forms.
A number of studies have suggested that deglycosylation can affect HIV-1 envelope antigenicity and immunogenicity [17], [19], [21]–[23], [49]–[54]. A non-glycosylated outer domain of gp120 has been reported to be immunogenic and induce weak neutralizing antibodies in rabbits [50]. For example, loss of sialic acids on gp120 has been reported to improve Env immunogenicity [49], [55]. Strategies for native deglycosylation of Env have been described that can improve both immunogenicity and sensitivity of resulting virions to neutralization [51], [53].
Frey et al. have reported weak or no binding of recombinant HIV-1 Envs to mAbs 2F5 and 4E10, but strong binding of these antibodies to a stabilized gp41 intermediate molecule [15]. Whether the epitope on natively deglycosylated recombinant JRFL gp140 is in the intermediate conformation of gp41 seen by mAbs 2F5 and 4E10 during the infection process is not known. However, we have recently shown that glycosylated JRFL gp140 Env can prime for an MPER peptide-liposome boost and induce antibodies focused on the DKW core 2F5 epitope that bind to the gp41 intermediate epitope (Sekaran, M. Alam, SM, Haynes, BF, unpublished). That deglycosylated JRFL gp140 is more immunogenic in rhesus macaques than glycosylated JRFL for 2F5 epitope MPER antibodies suggests that deglycosylated JRFL may have enhanced immunogenicity as a prime in this prime-boost setting.
Other factors in addition to the conformation of the MPER neutralizing epitopes appear to contribute to regulation of anti-MPER broad neutralizing antibodies. For example, we now have experimental data with mAbs 2F5 and 4E10 knock-in mice that B cells expressing these VHs are controlled by both central and peripheral tolerance mechanisms in these animals [13], [14]. Thus, immunogen design for optimal neutralizing epitope exposure is only one-component of the problem for inducing anti-MPER antibodies. Nonetheless, the increased affinity of the non-denaturing PNGase F-treated gp140 Env may render it a preferred Env immunogen in the setting of formulation of Envs with potent adjuvants that are designed to circumvent peripheral tolerance [56], [57].
While the RUAs in this paper are close approximations of the unmutated ancestor B cell receptors on naïve B cells, it should be noted that they are only inferred and may not precisely mimic the binding of membrane bound IgM B cell receptors on naïve B cells. Our data demonstrate that RUAs of both mAbs 2F5 and 4E10 can recognize the MPER well in the context of natively deglycosylated JRFL gp140, but not in fully glycosylated JRFL gp140. Interestingly, the glycosylated group M consensus Env CON-S gp140 constitutively bound to both 2F5 RUAs. This finding is consistent with our previously published site-specific glycan analysis of the CON-S gp140 in which we found, unlike JRFL gp140, that CON-S gp140 when expressed in 293T cells, has no occupied gp41 glycan sites [34]. That native deglycosylation of CON-S gp140 still resulted in enhanced binding to unmutated ancestors of 2F5 and 4E10 suggests that removal of gp120 glycans modulates access to gp41 as well. Thus, the inability of mAbs 2F5 and 4E10 reverted unmutated ancestors to react with recombinant HIV-1 gp140 Envs in some instances may be due to glycan masking or glycan modulation of conformations of MPER neutralizing epitopes, or may be due to “holes” in the native B cell repertoire for glycosylated HIV-1 Env. That the MPER 2F5 and 4E10 gp41 epitopes can be exposed with native deglycosylation, to us suggests the importance of glycan masking on glycosylated gp140 to 2F5 and 4E10 unmutated ancestor antibody non-reactivity. In light of the host immunoregulatory controls that also appear to play roles in control of MPER broad neutralizing antibody induction, both structural considerations of immunogen design targeted to optimized binding to reverted unmutated ancestor antibodies, in concert with methods of immunogen formulation to access and drive MPER-specific naïve and memory B cell proliferation likely will be required for the ultimate safe induction of MPER broad neutralizing antibodies.
Materials and Methods
Cloning and Expression of HIV-1 Env Proteins
A codon-optimized gene encoding the gp140CF (C = gp120-gp41 cleavage site deleted, F = fusion domain-deleted) of HIV-1 WT JRF Env in plasmid pR-JRFL140 (kindly provided by Feng Gao, M.D., Duke, Durham, North Carolina) was subcloned to pcDNA3.1(-)/Hygro eukaryotic expression vector (Invitrogen, Carlsbad, CA) via Xba I site at the 5′ end and a BamHI site at the 3′ end under the regulation of the CMV immediate early promoter. The resulting recombinant plasmid, pcDNA/H-JRFL 140, was linearized by restriction endonuclease Ssp I (Invitrogen, Carlsbad, CA) digestion. Two µg of purified plasmid was transfected into HEK293T cells (ATCC, Bethesda, MD) at approximately 80% confluence with LipoFectamine 2000 (Invitrogen, Carlsbad, CA) per the manufacturer's protocol. Two days post transfection, cells were digested with trypsin-EDTA, resuspended with DMEM HG media (Sigma, St. Louis, MO) containing 20% fetal bovine serum (DMEM-20) and 200 µg/mL of hygromycin B (Sigma) (selection media), and dispersed to 10–20 100 mm polystyrene Petri dishes (Becton-Dickinson, Franklin Lakes, NJ). Hygromycin B-resistant colonies were picked, digested and re-suspended in selection media. Stably transfected cells were cloned by limiting dilution and adapted in serum-free media Pro293A (BioWhittaker, Switzerland), and used for production of purified recombinant WT JRFL gp140 protein using lectin agarose beads (Galanthus Nivalis, Vector Laboratories, Burlingame, CA) and stored at −80°C until use [29]. The group M consensus gp140 Env, CON-S gp140 CFI (C = gp120-gp41 cleavage site deleted, F = fusion domain-deleted, I = immunodominant gp41 region deleted) protein was expressed and produced as previously described [29].
Cells and Viruses
DC-SIGN-expressing NIH 3T3 cells, parent NIH 3T3 cells, TZM-BL cells that express CD4/CCR5/CXCR4 were obtained from NAID/NIH AIDS Reagent Repository. Cell lines were grown in Dulbecco's modified Eagle's medium (DMEM, Life Technologies/Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS).
Monoclonal Antibodies (mAbs) and Soluble CD4 (sCD4) Human
MAbs known to bind epitopes on gp120 (A32) [31] the gp120 V3 loop (F39F), and the CCR5 binding site (17b) [30], were kindly provided by James Robinson (Tulane Medical School, New Orleans, LA). Anti-gp41 membrane proximal external region (MPER) mAbs 2F5, and 4E10, and 2G12 [33] were purchased from Polymun Scientific, Vienna, Austria. T8 is a murine mAb that binds to the gp120 C1 region (a gift from P. Earl, NIH). Mouse mAb 3B3 binds near the N-terminus of gp120 and has broadly cross reactivity with HIV-1 gp120 proteins of different subtypes [32]. Mouse anti-DC-SIGN monoclonal antibodies [58] and recombinant sCD4 were obtained from the NIAID.NIH AIDS Reagent Repository. MAb 1b12 against the CD4BS was the gift of Dennis Burton, Scripps, La Jolla, CA [20].
Deglycosylation of Proteins
Peptide-N-glycosidase F (PNGase F) that removes N-linked carbohydrates was purchased from New England Biolabs (Beverly, MA). Progressive native deglycosylation was performed using 100 µg of HIV-1 JRFL Env protein with varied amounts of PNGase F ranging from 5, 10, 20, 50, 100 and 500 units (U) per µg of HIV-1 JRFL gp140 Env protein. For each experiment, 100 µg of Env protein were digested with indicated amount of PNGase F in 200 µL of volume (resulting in the protein concentration of 0.5 µg/µL). The reaction was carried out at 37°C for 24 h and then stopped by freezing the reaction mixtures at −80°C. Deglycosylation in denaturing conditions was performed by digesting every 100 µg of pre-denatured protein in denaturing buffer (0.5% SDS and 1.0% β-ME) at 100°C for 10 min with 1 µL (500 U) of PNGase F at 37°C for 2 h using the protocol as recommended by the manufacturer. Deglycosylation of the JRFL Env protein under the native condition was optimized by either prolonging the incubation time or increasing the amount of enzyme used in order to achieve the maximal deglycosylation. In the former case, the incubation time extended up to 7 days only minimally improved the extent of native deglycosylation of gp140 (data not shown). On the other hand, the prolonged incubation increased the risks for protein to be degraded. In contrast, with the fixed reaction time (approximately 24 h), the increased amounts of the enzyme dramatically increased the completeness of the native deglycosylation of Env. Deglycosylation under non-denaturing conditions with PNGase F (500 U per µg of JRFL Env) reduced the size of the deglycosylated JRFL gp140 Env protein close to the theoretical polypeptide size (approximately 80 kDa) as analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), which indicated that approximately 80% of carbohydrates have been removed. Deglycosylation of CON-S gp140CFI was performed under non-denaturing conditions using PNGase F at 500 U per µg of Env protein.
SDS-PAGE and Western Blot Analysis of the Env Proteins
Untreated and deglycosylated HIV-1 JRFL gp140CF and CON-S gp140CFI Env proteins were fractionated on 4–12% Bis-Tris SDA-PAGE gels (Invitrogen, Carlsbad, CA) under reducing (4% Beta-mercaptoethanol, β-ME, Fisher Scientific, Fair Lawn, NJ) or non-reducing conditions, stained with coomassie blue dye, or transferred onto nitrocellulose filters and probed with anti-Env human mAbs 17b, 1b12, 2F5, 2G12, 4E10, A32, F39F. Goat anti-human IgG alkaline phosphatase (AP) conjugates (Sigma Aldrich, Inc, St. Louis, MO; 1∶3,000 dilution) was used as secondary antibodies in Western-blots.
Surface Plasmon Resonance (SPR)
SPR assays using a BIAcore 3000 instrument were carried out to characterize the binding of untreated and deglycosylated HIV-1 Env proteins to anti-HIV-1 MPER mAbs 2F5 and 4E10 as well as to their inferred reverted unmutated ancestors. Data analysis was performed with BIAevaluation 3.0 software (BIAcore Inc, Upsaala, Sweden). To measure the affinity (Kd) and kinetics of the interaction of the Env proteins with mAbs, the Env glycoproteins at varying concentrations were flowed over mAb 2F5 or 4E10 that were captured on anti-Fc IgG immobilized on the sensor chip CM5 with amine coupling kit (Biacore AB) and as described earlier (7, 8, 59). The binding data were recorded in real time at 25°C with a continuous flow of phosphate-buffered saline (150 mM NaCl, 0.005% surfactant P20 [pH 7.4]) at 30 µL/min following subtraction of non-specific signal over the control mAb surface [7], [8], [59].
ELISA
EIA plates (Costar 3590, Corning, NY) were coated with JRFL gp140 protein (0.2 µg/well) in 100 µL of carbonate-bicarbonate (CBC) buffer (pH 9.6) over night at 4°C. Plates were then blocked with CBC containing 3% BSA (Sigma, St. Louis, MO) for 2 h at room temperature (RT). To evaluate the antibody binding kinetics, serial dilutions of each mAb (17B, 1b12, 2F5, 2G12, 4E10, and F39F) starting from 10 µg/ml in 50 µL in dilution buffer (PBS, 0.05% Tween-20, 2% goat serum and 3% BSA) were added to the plate followed by the incubation at RT for 1 h. After washing with dilution buffer, goat anti-human IgG-AP (Sigma, St. Louis, MO) was added and incubated in the same way to detect the binding of human antibodies to Env proteins. Alternatively, equal volume of native deglycosylation and mock deglycosylation reaction solutions containing equal amount of initial Env protein were serially diluted in CBC buffer (pH 9.6) starting from 10 µg/mL (2-fold serial dilution for 12 point titrations). One hundred (100) µl of the diluted solution were coated on the 96-well EIA plate (Costar 3590, Corning, NY) each well and incubated at 4°C overnight and then blocked with CBC containing 3% BSA (Sigma, St. Louis, MO) for 2 h at RT.
Each human mAb was diluted in buffer (PBS, 0.05% Tween-20, 2% goat serum and 3% BSA) to 1.0 µg/mL and 100 µl per well of each diluted antibody was incubated at RT for 1 h. For sCD4, 100 µl of 0.2 µg/mL was used. Goat anti-human IgG-AP (Sigma) was used to detect the binding of human antibodies to Env proteins. For detection of binding of sCD4, biotin-labeled anti-CD4 mAb (0.1 µg/mL in 100 µl/well) and the Strepavidin (Promega, 1∶1000, 100 µl/well) were used. In selected experiments, Env oligomers were captured on plates pre-coated with mAb 3B3 and then tested for mAb reactivity as above. ELISA results were read at 405 nm using ELISA radar (Bio-Rad, Berkeley, California).
Rhesus macaque binding antibodies to HIV-1 WT glycosylated JRFL gp140 Env and deglycosylated JRFL gp140 Env were assayed by indirect ELISA. HIV-1 WT glycosylated JRFL gp140 Env and deglycosylated (Deg) JRFL gp140 Env, 2F5 nominal MPER epitope peptide SF62 (QQEKNEQELLELDKWASLWN) and 4E10 eiptope peptide P-4E10 (SLWNWFNITNWLWYIK) were coated at 200 ng/well in 96-well ELISA plates (4 degrees C) followed by incubation with dilutions of immune plasma. AP conjugated goat anti-monkey IgG heavy - and light chain specific antibody was used as seconday antibody.
DC-SIGN Binding Assay
To assess the reactivity of untreated and deglycosylated proteins to bind DC-SIGN, WT glycosylated and deglycosylated JRFL gp140 proteins were chemically labeled with the Alexa Fluor 647 dye (AF647) using a protocol as recommended by the manufacturer (Invitrogen, Carlsbad, CA). NIH 3T3/DC-SIGN or the parent NIH 3T3 cells (5×105 cells in 100 µL of PBS containing 1% BSA in the presence of 1 mM Ca++ and Mg++ ions) were incubated with 1.0 µg/mL of the AF647-labeled undeglycosylated or deglycosylated JRFL gp140 proteins at 4°C for 30 min. The cells were washed for 4 times with the PBS buffer. The cells were resuspended in 400 100 µL of PBS buffer and analyzed with an LSRII flow cytometer (Becton Dickinson) [58]. CD4 expressing TZM-b1 cells were used as a positive control for binding of the AF647-labeled undeglycosylated and deglycosylated JRFL gp140 proteins.
Analysis of Carbohydrates by Mass Spectrometry
We have previously analyzed and compared JRFL gp140CF and CON-S gp140CFI Envs for glycan site-occupation and content [34]. To determine the glycosylation profile of both native and the deglycosylated JRFL gp140 Env proteins, the latter were treated with proteases and the digestion products were analyzed by LC/MS, as described previously and below [34], [35].
Deglycosylated JRFL gp140 Env envelope proteins were either treated with Endo H or (separately) Endo F3, to reduce the glycan heterogeneity to a single N-acetlyhexosamine, attached to the glycosylation site. Core fucosylated residues also remained fucosylated in this procedure. Typical conditions included incubating the Env (approximately 7 fmol) with enzyme, at a ratio of at least 30 U/mg of Env protein. For Endo H deglycosylation experiment, the sample was denatured with 2 M urea in 100 mM tris buffer (pH 5.5) followed by the addition of 2 µL of Endo H. After thorough mixing, the reaction was incubated for 48 hours at 37°C. For Endo F3 deglycosylation experiment, samples were incubated with 6 µL of 250 µM NH4C2H2O2 (pH 4.5) and 2 µL of Endo F3 for two weeks at 37°C.
After deglycosylation, proteins were split into two fractions and subjected to enzymatic digestion using trypsin and Glu-C. Prior to enzyme addition, proteins were unfolded using 6 M urea, and reduced and alkylated at room temperature using 10 mM DTT and 15 mM IAA at RT, respectively. Digestion conditions were carried out in tris buffer with 3 mM EDTA overnight at 37°C. For tryptic digest, pH was 8.5 and protein to enzyme ratio was 30∶1; for Glu-C digest, pH was 7.8 and the protein to enzyme ratio was 20∶1. Both reactions were quenched with the addition of 1 µL glacial acetic acid. The digestion protocols were performed three times on different days, with the same initial batch of protein, to ensure reproducibility and reliability. As an additional control experiment, WT glycosylated protein was subjected to the same protocols to verify that the method would effectively detect all sites on the protein that remained glycosylated.
For carbohydrate analysis, peptides were detected by LC-MS, using a Dionex UltiMate capillary LC system (Sunnyvale, CA) equipped with a FAMOS well plate autosampler; adjoined to a Fourier transform ion cyclotron resonance mass spectrometer (LTQ-FT, ThermoScientific, San Jose, CA). Separation of the sample (5 µL) was achieved using a C18 PepMap 300 column (300 µm i.d. ×15 cm, 300 Å, LC Packings, Sunnyvale, CA) at a flow rate of 5 µL/minute. Separation was achieved using a linear gradient over 65 minutes, and the eluted peptides were ionized using a 2.8 kV source voltage for ESI. High resolution MS and low resolution MS/MS data were collected using a data-dependent scanning protocol, where the five most intense ions were selected for MS/MS, after each high resolution MS run, and those ions were subsequently dynamically added to an exclusion list for 3 minutes. Peptides (both deglycosylated, and those with remaining glycosylation) were detected using the MS and MS/MS data with the assistance of a Mascot search (Matrix Science, London, UK, version 2.2.04), after inputting a custom HIV Env database. All automated assignments were further verified by manually assigning the relevant MS/MS spectra. All glycosylation sites were detected as glycosylated or deglycosylated, except for N406, which was not detected by MS analysis.
Ethics Statement
This study involved in the use of rhesus macaques was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health in BIOQUAL (Rockville, MD.) BIOQUAL is fully accredited by AAALAC and through OLAW, assurance number A-3086. The protocol was approved by the BIOQUAL IACUC. All physical procedures associated with this work were done under anesthesia to minimize pain and distress. Four rhesus macaques per group were immunized intramuscularly with 100 ug/dose of HIV-1 JRFLgp140 Env wild-type (WT) or JRFL gp140 CF Env deglycosylated using PNGase F at 500 U/µg Env using AS01B adjuvant (generously provided by Gerald Voss, GlaxoSmithKline Biological, Rixensart, Belgium) at week 0 and 4. Blood samples were collected at week 2 and week 6 for isolation of immune plasma.
Supporting Information
Zdroje
1. MusterTGuineaRTrkolaAPurtscherMKlimaA 1994 Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol 68 4031 4034
2. ZwickMBLabrijnAFWangMSpenlehauerCSaphireEO 2001 Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol 75 10892 10905
3. ZwickMBJensenRChurchSWangMStieglerG 2005 Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J Virol 79 1252 1261
4. OfekGTangMSamborAKatingerHMascolaJR 2004 Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol 78 10724 10737
5. CardosoRMZwickMBStanfieldRLKunertRBinleyJM 2005 Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22 163 173
6. HaynesBFFlemingJSt ClairEWKatingerHStieglerG 2005 Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308 1906 1908
7. AlamSMMcAdamsMBorenDRakMScearceRM 2007 The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol 178 4424 4435
8. AlamSMMorelliMDennisonSMLiaoHXZhangR 2009 Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci U S A 106 20234 20239
9. DennisonSMStewartSMStempelKCLiaoHXHaynesBF 2009 Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. J Virol 83 10211 10223
10. OfekGMcKeeKYangYYangZYSkinnerJ 2010 Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol 84 2955 2962
11. SchererEMLeamanDPZwickMBMcMichaelAJBurtonDR 2010 Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc Natl Acad Sci U S A 107 1529 1534
12. HaynesBFAlamSM 2008 HIV-1 hides an Achilles' heel in virion lipids. Immunity 28 10 12
13. VerkoczyLDiazMHollTMOuyangYBBouton-VervilleH 2010 Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci U S A 107 181 186
14. VerkoczyLBouton-VervilleHHutchinsonJScearceRMLiaoHX 2010 Role of immunoglobulin light chain usage and MPER specificity in counterselecting B cells expressing the broadly neutralizing antibody 2F5. AIDS Res Hum Retroviruses 26 Abstract No. P04.55LB A-155
15. FreyGPengHRits-VollochSMorelliMChengY 2008 A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A 105 3739 3744
16. XiaoXChenWFengYZhuZPrabakaranP 2009 Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun 390 404 409
17. DooresKJBurtonDR 2010 Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J Virol 84 10510 10521
18. ReitterJNMeansREDesrosiersRC 1998 A role for carbohydrates in immune evasion in AIDS. Nat Med 4 679 684
19. RichmanDDWrinTLittleSJPetropoulosCJ 2003 Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A 100 4144 4149
20. SaphireEOParrenPWPantophletRZwickMBMorrisGM 2001 Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293 1155 1159
21. WalkerLMPhogatSKChan-HuiPYWagnerDPhungP 2009 Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326 285 289
22. WeiXDeckerJMWangSHuiHKappesJC 2003 Antibody neutralization and escape by HIV-1. Nature 422 307 312
23. YusteEBixbyJLifsonJSatoSJohnsonW 2008 Glycosylation of gp41 of simian immunodeficiency virus shields epitopes that can be targets for neutralizing antibodies. J Virol 82 12472 12486
24. ColeKSSteckbeckJDRowlesJLDesrosiersRCMontelaroRC 2004 Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J Virol 78 1525 1539
25. McCaffreyRASaundersCHenselMStamatatosL 2004 N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J Virol 78 3279 3295
26. ZhangCWanYShiJZhouMMengZ 2010 Deglycosylation of HIV-1 AE Gp140 enhances the capacity to elicit neutralizing antibodies against the heterologous HIV-1 clade. AIDS Res Hum Retroviruses 26 569 575
27. GnanakaranSDanielsMGBhattacharyaTLapedesASSethiA 2010 Genetic signatures in the envelope glycoproteins of HIV-1 that associate with broadly neutralizing antibodies. PLoS Comput Biol 6 e1000955
28. KirchherrJLHamiltonJWLuXGnanakaranSMuldoonM 2010 Identification of amino acid substitutions associated with neutralization phenotype in the human immunodeficiency virus type-1 subtype C gp120. Virology 1 12
29. LiaoHXSutherlandLLXiaSMBrockMEScearceRM 2006 Group M consensus Env oligomers induce antibodies that neutralize subtype C HIV - primary isolates. Virology 353 268 282
30. ZhangWGodillotAPWyattRSodroskiJChaikenI 2001 Antibody 17b binding at the coreceptor site weakens the kinetics of the interaction of envelope glycoprotein gp120 with CD4. Biochemistry 40 1662 1670
31. WyattRMooreJAccolaMDesjardinERobinsonJ 1995 Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol 69 5723 5733
32. GaoFScearceRMAlamSMHoraBXiaS 2009 Cross-reactive monoclonal antibodies to multiple HIV-1 subtype and SIVcpz envelope glycoproteins. Virology 394 91 98
33. CalareseDALeeHKHuangCYBestMDAstronomoRD 2005 Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc Natl Acad Sci U S A 102 13372 13377
34. GoEPIrunguJZhangYDalpathadoDSLiaoHX 2008 Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J Proteome Res 7 1660 1674
35. IrunguJGoEPZhangYDalpathadoDSLiaoHX 2008 Comparison of HPLC/ESI-FTICR MS versus MALDI-TOF/TOF MS for glycopeptide analysis of a highly glycosylated HIV envelope glycoprotein. J Am Soc Mass Spectrom 19 1209 1220
36. LiuJBartesaghiABorgniaMJSapiroGSubramaniamS 2008 Molecular architecture of native HIV-1 gp120 trimers. Nature 455 109 113
37. SandersRWVenturiMSchiffnerLKalyanaramanRKatingerH 2002 The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol 76 7293 7305
38. Hager-BraunCKatingerHTomerKB 2006 The HIV-neutralizing monoclonal antibody 4E10 recognizes N-terminal sequences on the native antigen. J Immunol 176 7471 7481
39. ShihTAMeffreERoedererMNussenzweigMC 2002 Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol 3 570 575
40. Dal PortoJHabermanAKelsoeGShlomchikM 2002 Very low affinity B cells form germinal centers, become memory B cells, and participate in secondary immune responses when higher affinity competition is reduced. J Exp Med 195 1215 1221
41. MunshawSKeplerTB 2010 SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements. Bioinformatics 26 867 872
42. FelsensteinJ 1981 Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17 368 376
43. HartMLSaifuddinMSpearGT 2003 Glycosylation inhibitors and neuraminidase enhance human immunodeficiency virus type 1 binding and neutralization by mannose-binding lectin. J Gen Virol 84 353 360
44. WangSKLiangPHAstronomoRDHsuTLHsiehSL 2008 Targeting the carbohydrates on HIV-1: Interaction of oligomannose dendrons with human monoclonal antibody 2G12 and DC-SIGN. Proc Natl Acad Sci U S A 105 3690 3695
45. WalkerLMSimekMDPriddyFGachJSWagnerD 2010 Limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6 8 e1001028
46. ShenXParksRJMontefioriDCKirchherrJLKeeleBF 2009 In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J Virol 83 3617 3625
47. ShenXDennisonSMLiuPGaoFJaegerF 2010 Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc Natl Acad Sci U S A 107 5972 5977
48. BlishCANguyenMAOverbaughJ 2008 Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med 5 1 e9
49. BenjouadAGluckmanJCRochatHMontagnierLBahraouiE 1992 Influence of carbohydrate moieties on the immunogenicity of human immunodeficiency virus type 1 recombinant gp160. J Virol 66 2473 2483
50. BhattacharyyaSRajanRESwarupaYRathoreUVermaA 2010 Design of a non-glycosylated outer domain-derived HIV-1 gp120 immunogen that binds to CD4 and induces neutralizing antibodies. J Biol Chem 285 27100 27110
51. BinleyJMBanYECrooksETEgginkDOsawaK 2010 Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J Virol 84 5637 5655
52. BinleyJMWyattRDesjardinsEKwongPDHendricksonW 1998 Analysis of the interaction of antibodies with a conserved enzymatically deglycosylated core of the HIV type 1 envelope glycoprotein 120. AIDS Res Hum Retroviruses 14 191 198
53. KochMPanceraMKwongPDKolchinskyPGrundnerC 2003 Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313 387 400
54. SagarMWuXLeeSOverbaughJ 2006 Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol 80 9586 9598
55. KongLSheppardNCStewart-JonesGBRobsonCLChenH 2010 Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J Mol Biol 403 131 147
56. BusconiLLauCMTaborASUccelliniMBRuheZ 2006 DNA and RNA autoantigens as autoadjuvants. J Endotoxin Res 12 379 384
57. ShlomchikMJ 2009 Activating systemic autoimmunity: B's, T's, and tolls. Curr Opin Immunol 21 626 633
58. WuLMartinTDVazeuxRUnutmazDKewalRamaniVN 2002 Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. J Virol 76 5905 5914
59. AlamSMPaleosCALiaoHXScearceRRobinsonJ 2004 An inducible HIV type 1 gp41 HR-2 peptide-binding site on HIV type 1 envelope gp120. AIDS Res Hum Retroviruses 20 836 845
60. SchiefWRBanYEStamatatosL 2009 Challenges for structure-based HIV vaccine design. Curr Opin HIV AIDS 5 431 40
61. HuangCCTangMZhangMYMajeedSMontabanaE 2005 Structure of a V3-containing HIV-1 gp120 core. Science 310 1025 8
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage EgressČlánek A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene ExpressionČlánek An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes byČlánek Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine ScrapieČlánek Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 9- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Unconventional Repertoire Profile Is Imprinted during Acute Chikungunya Infection for Natural Killer Cells Polarization toward Cytotoxicity
- Envelope Deglycosylation Enhances Antigenicity of HIV-1 gp41 Epitopes for Both Broad Neutralizing Antibodies and Their Unmutated Ancestor Antibodies
- Co-opts GBF1 and CERT to Acquire Host Sphingomyelin for Distinct Roles during Intracellular Development
- Nrf2, a PPARγ Alternative Pathway to Promote CD36 Expression on Inflammatory Macrophages: Implication for Malaria
- Robust Antigen Specific Th17 T Cell Response to Group A Streptococcus Is Dependent on IL-6 and Intranasal Route of Infection
- Targeting of a Chlamydial Protease Impedes Intracellular Bacterial Growth
- The Protease Cruzain Mediates Immune Evasion
- High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism
- Plague and Climate: Scales Matter
- Exhausted CD8 T Cells Downregulate the IL-18 Receptor and Become Unresponsive to Inflammatory Cytokines and Bacterial Co-infections
- Maturation-Induced Cloaking of Neutralization Epitopes on HIV-1 Particles
- Murine Gamma-herpesvirus Immortalization of Fetal Liver-Derived B Cells Requires both the Viral Cyclin D Homolog and Latency-Associated Nuclear Antigen
- Rapid and Efficient Clearance of Blood-borne Virus by Liver Sinusoidal Endothelium
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene Expression
- Strain Specific Resistance to Murine Scrapie Associated with a Naturally Occurring Human Prion Protein Polymorphism at Residue 171
- Development of a Transformation System for : Restoration of Glycogen Biosynthesis by Acquisition of a Plasmid Shuttle Vector
- Monalysin, a Novel -Pore-Forming Toxin from the Pathogen Contributes to Host Intestinal Damage and Lethality
- Host Phylogeny Determines Viral Persistence and Replication in Novel Hosts
- BC2L-C Is a Super Lectin with Dual Specificity and Proinflammatory Activity
- Expression of the RAE-1 Family of Stimulatory NK-Cell Ligands Requires Activation of the PI3K Pathway during Viral Infection and Transformation
- Structure of the Vesicular Stomatitis Virus N-P Complex
- HSV Infection Induces Production of ROS, which Potentiate Signaling from Pattern Recognition Receptors: Role for S-glutathionylation of TRAF3 and 6
- The Human Papillomavirus E6 Oncogene Represses a Cell Adhesion Pathway and Disrupts Focal Adhesion through Degradation of TAp63β upon Transformation
- Analysis of Behavior and Trafficking of Dendritic Cells within the Brain during Toxoplasmic Encephalitis
- Exposure to the Viral By-Product dsRNA or Coxsackievirus B5 Triggers Pancreatic Beta Cell Apoptosis via a Bim / Mcl-1 Imbalance
- Multidrug Resistant 2009 A/H1N1 Influenza Clinical Isolate with a Neuraminidase I223R Mutation Retains Its Virulence and Transmissibility in Ferrets
- Structure of Herpes Simplex Virus Glycoprotein D Bound to the Human Receptor Nectin-1
- Step-Wise Loss of Bacterial Flagellar Torsion Confers Progressive Phagocytic Evasion
- Complex Recombination Patterns Arising during Geminivirus Coinfections Preserve and Demarcate Biologically Important Intra-Genome Interaction Networks
- An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes by
- Non-Lytic, Actin-Based Exit of Intracellular Parasites from Intestinal Cells
- The Fecal Viral Flora of Wild Rodents
- The General Transcriptional Repressor Tup1 Is Required for Dimorphism and Virulence in a Fungal Plant Pathogen
- Interferon Regulatory Factor-1 (IRF-1) Shapes Both Innate and CD8 T Cell Immune Responses against West Nile Virus Infection
- A Small Non-Coding RNA Facilitates Bacterial Invasion and Intracellular Replication by Modulating the Expression of Virulence Factors
- Evaluating the Sensitivity of to Biotin Deprivation Using Regulated Gene Expression
- The Motility of a Human Parasite, , Is Regulated by a Novel Lysine Methyltransferase
- Phosphodiesterase-4 Inhibition Alters Gene Expression and Improves Isoniazid – Mediated Clearance of in Rabbit Lungs
- Restoration of IFNγR Subunit Assembly, IFNγ Signaling and Parasite Clearance in Infected Macrophages: Role of Membrane Cholesterol
- Protease ROM1 Is Important for Proper Formation of the Parasitophorous Vacuole
- The Regulated Secretory Pathway in CD4 T cells Contributes to Human Immunodeficiency Virus Type-1 Cell-to-Cell Spread at the Virological Synapse
- Rerouting of Host Lipids by Bacteria: Are You CERTain You Need a Vesicle?
- Transmission Characteristics of the 2009 H1N1 Influenza Pandemic: Comparison of 8 Southern Hemisphere Countries
- Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine Scrapie
- Sequential Bottlenecks Drive Viral Evolution in Early Acute Hepatitis C Virus Infection
- Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen
- Genomic and Proteomic Analyses of the Fungus Provide Insights into Nematode-Trap Formation
- Influenza Virus Ribonucleoprotein Complexes Gain Preferential Access to Cellular Export Machinery through Chromatin Targeting
- Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
- Protease-Sensitive Conformers in Broad Spectrum of Distinct PrP Structures in Sporadic Creutzfeldt-Jakob Disease Are Indicator of Progression Rate
- Vaccinia Virus Protein C6 Is a Virulence Factor that Binds TBK-1 Adaptor Proteins and Inhibits Activation of IRF3 and IRF7
- c-di-AMP Is a New Second Messenger in with a Role in Controlling Cell Size and Envelope Stress
- Structural and Functional Studies on the Interaction of GspC and GspD in the Type II Secretion System
- APOBEC3A Is a Specific Inhibitor of the Early Phases of HIV-1 Infection in Myeloid Cells
- Impairment of Immunoproteasome Function by β5i/LMP7 Subunit Deficiency Results in Severe Enterovirus Myocarditis
- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Tri6 Is a Global Transcription Regulator in the Phytopathogen
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- The Next Opportunity in Anti-Malaria Drug Discovery: The Liver Stage
- Significant Effects of Antiretroviral Therapy on Global Gene Expression in Brain Tissues of Patients with HIV-1-Associated Neurocognitive Disorders
- Inhibition of Competence Development, Horizontal Gene Transfer and Virulence in by a Modified Competence Stimulating Peptide
- A Novel Metal Transporter Mediating Manganese Export (MntX) Regulates the Mn to Fe Intracellular Ratio and Virulence
- Rhoptry Kinase ROP16 Activates STAT3 and STAT6 Resulting in Cytokine Inhibition and Arginase-1-Dependent Growth Control
- Hsp90 Governs Dispersion and Drug Resistance of Fungal Biofilms
- Secretion of Genome-Free Hepatitis B Virus – Single Strand Blocking Model for Virion Morphogenesis of Para-retrovirus
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
- Membrane Remodeling by the Double-Barrel Scaffolding Protein of Poxvirus
- A Diverse Population of Molecular Type VGIII in Southern Californian HIV/AIDS Patients
- Disruption of TLR3 Signaling Due to Cleavage of TRIF by the Hepatitis A Virus Protease-Polymerase Processing Intermediate, 3CD
- Quantitative Analyses Reveal Calcium-dependent Phosphorylation Sites and Identifies a Novel Component of the Invasion Motor Complex
- Discovery of the First Insect Nidovirus, a Missing Evolutionary Link in the Emergence of the Largest RNA Virus Genomes
- Old World Arenaviruses Enter the Host Cell via the Multivesicular Body and Depend on the Endosomal Sorting Complex Required for Transport
- Exploits a Unique Repertoire of Type IV Secretion System Components for Pilus Assembly at the Bacteria-Host Cell Interface
- Recurrent Signature Patterns in HIV-1 B Clade Envelope Glycoproteins Associated with either Early or Chronic Infections
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



