-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The General Transcriptional Repressor Tup1 Is Required for Dimorphism and Virulence in a Fungal Plant Pathogen
A critical step in the life cycle of many fungal pathogens is the transition between yeast-like growth and the formation of filamentous structures, a process known as dimorphism. This morphological shift, typically triggered by multiple environmental signals, is tightly controlled by complex genetic pathways to ensure successful pathogenic development. In animal pathogenic fungi, one of the best known regulators of dimorphism is the general transcriptional repressor, Tup1. However, the role of Tup1 in fungal dimorphism is completely unknown in plant pathogens. Here we show that Tup1 plays a key role in orchestrating the yeast to hypha transition in the maize pathogen Ustilago maydis. Deletion of the tup1 gene causes a drastic reduction in the mating and filamentation capacity of the fungus, in turn leading to a reduced virulence phenotype. In U. maydis, these processes are controlled by the a and b mating-type loci, whose expression depends on the Prf1 transcription factor. Interestingly, Δtup1 strains show a critical reduction in the expression of prf1 and that of Prf1 target genes at both loci. Moreover, we observed that Tup1 appears to regulate Prf1 activity by controlling the expression of the prf1 transcriptional activators, rop1 and hap2. Additionally, we describe a putative novel prf1 repressor, named Pac2, which seems to be an important target of Tup1 in the control of dimorphism and virulence. Furthermore, we show that Tup1 is required for full pathogenic development since tup1 deletion mutants are unable to complete the sexual cycle. Our findings establish Tup1 as a key factor coordinating dimorphism in the phytopathogen U. maydis and support a conserved role for Tup1 in the control of hypha-specific genes among animal and plant fungal pathogens.
Published in the journal: . PLoS Pathog 7(9): e32767. doi:10.1371/journal.ppat.1002235
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002235Summary
A critical step in the life cycle of many fungal pathogens is the transition between yeast-like growth and the formation of filamentous structures, a process known as dimorphism. This morphological shift, typically triggered by multiple environmental signals, is tightly controlled by complex genetic pathways to ensure successful pathogenic development. In animal pathogenic fungi, one of the best known regulators of dimorphism is the general transcriptional repressor, Tup1. However, the role of Tup1 in fungal dimorphism is completely unknown in plant pathogens. Here we show that Tup1 plays a key role in orchestrating the yeast to hypha transition in the maize pathogen Ustilago maydis. Deletion of the tup1 gene causes a drastic reduction in the mating and filamentation capacity of the fungus, in turn leading to a reduced virulence phenotype. In U. maydis, these processes are controlled by the a and b mating-type loci, whose expression depends on the Prf1 transcription factor. Interestingly, Δtup1 strains show a critical reduction in the expression of prf1 and that of Prf1 target genes at both loci. Moreover, we observed that Tup1 appears to regulate Prf1 activity by controlling the expression of the prf1 transcriptional activators, rop1 and hap2. Additionally, we describe a putative novel prf1 repressor, named Pac2, which seems to be an important target of Tup1 in the control of dimorphism and virulence. Furthermore, we show that Tup1 is required for full pathogenic development since tup1 deletion mutants are unable to complete the sexual cycle. Our findings establish Tup1 as a key factor coordinating dimorphism in the phytopathogen U. maydis and support a conserved role for Tup1 in the control of hypha-specific genes among animal and plant fungal pathogens.
Introduction
Dimorphism, the capacity of certain fungi to change their morphology between yeast-like growth and a filamentous state in response to environmental signals, is frequently associated with the virulence of both animal and plant pathogenic fungi [1]–[6]. This morphological conversion is controlled by several conserved signaling pathways, such as the cyclic AMP-protein kinase A pathway and a mitogen-activated protein (MAP) kinase cascade [4], [6]–[10]. Another well known transcriptional regulator controlling dimorphism is the general transcriptional repressor Tup1, which is conserved from fungi to mammals [11]–[16]. The mechanism of action for Tup1 has been best studied in the yeast Saccharomyces cerevisiae. In this fungus, Tup1p forms a transcriptional co-repressor complex with Ssn6p, a protein that contains tetratricopeptide repeat (TPR) motifs known to mediate protein-protein interactions [17]–[20]. Neither Tup1p nor Ssn6p have direct DNA binding activity and their role in transcription depends on their recruitment to promoters by specific DNA binding proteins [18], [21]. Tup1p repression mechanisms include the interaction with RNA polymerase II holoenzyme components and the alteration of chromatin structure through interaction with histones H3 and H4 and histone deacetylases [22]–[26]. Tup1p controls S. cerevisiae dimorphism in both haploid and diploid strains. Deletions of TUP1 result in reduced haploid invasive growth and reduced diploid pseudohyphal growth, which are considered the filamentous forms of this yeast [11].
Although the role of Tup1 in fungal dimorphism seems conserved, the way it controls this process frequently differs between fungi. The deletion of tup1 from the animal pathogens Candida albicans, Penicillium marneffei and Cryptococcus neoformans give clear examples of this variability. In C. albicans, the homozygous mutant for TUP1 shows a constitutive filamentation phenotype, in contrast to the situation described for S. cerevisiae, and reduced virulence [11]. In P. marneffei, however, tupA is required for the maintenance of its filamentous form, negatively regulating yeast morphogenesis instead of filament formation [12]. In the case of C. neoformans, TUP1 is required for the formation of dikaryotic hyphae due to a mating defect of TUP1 mutant strains, and for virulence [27], [28]. In addition, the molecular mechanisms and genetic pathways by which Tup1 acts in fungal dimorphism are poorly understood in most species [7], [12], [27]–[33]. This role of Tup1 in regulating the dimorphic transition is completely unknown in plant pathogenic fungi, which require different morphogenetic changes to successfully colonize their hosts and cause disease. The only data that might link Tup1 to a role in plant fungal dimorphism are a study into the role of sql1, a gene functionally homologous to S. cerevisiae SSN6, in U. maydis. Here overexpression of truncated forms of Sql1 was shown to induce morphological changes in this fungus [34].
The corn smut fungus Ustilago maydis is a well established model for studying dimorphism and virulence in plant pathogens [35]–[38]. Pathogenic development of this fungus initiates with the transition from yeast-like growth to the formation of polar filaments on the plant leaf surface. Control of this process relies on a tetrapolar mating system consisting of the biallelic a and the multiallelic b loci. Only strains differing in the allelic composition at both loci can successfully form and maintain the infectious filamentous form of the fungus [39]. Locus a encodes the pheromone-receptor system that allow cells from different mating types to detect each other, form conjugation tubes, and fuse [40], [41]. Locus b is then responsible for determining the fate of the resulting dikaryon. This locus encodes a pair of homeodomain transcription factors, bE and bW, that form a compatible heterodimer if proceeding from different alleles, triggering filamentation and pathogenicity [42], [43]. Upon dikaryon filament formation, the hypha tip differentiates to form a specialized structure for plant penetration, known as the appressorium [44], [45]. Once inside the plant, mycelium expansion takes place, leading to the formation of plant tumors. In these tumors, fungal nuclei fuse prior to the separation and rounding up of each hyphal section to form diploid spores. In favorable conditions spores germinate in a meiotic process that forms new haploid cells [46].
The highly conserved cAMP and MAP kinase pathways play a central role in the control of several of the morphological changes required during U. maydis pathogenic development [47]–[51]. Both of these pathways are activated following the recognition of pheromones by receptors of opposite mating types during the yeast to infective hyphae transition, resulting in the transcriptional and post-translational activation of the Prf1 transcription factor [47], [51]–[53]. Once activated, Prf1 promotes the expression of a and b loci genes (for review see [38]) (Figure 1). Thus, U. maydis integrates the inputs that activate both pathways through Prf1 to promote the b-dependent infectious form of the fungus. In the animal pathogen C. albicans, cAMP and MAP kinase pathways induce filamentous growth by promoting the activation of Efg1 and Cph1 transcriptional regulators, respectively, that extend down to hypha-specific target genes [2], [7], [54]–[56]. Control of filamentation in this fungus also requires the transcriptional repression of hypha-specific genes via Tup1, which acts through a third parallel pathway involving Rfg1 and Nrg1 transcriptional regulators [7], [29]–[33]. In U. maydis, as a plant pathogenic fungus, it is unknown whether or not Tup1 plays a role in dimorphism and virulence. Analyzing the function of Tup1 in this plant pathogen could help better understand how it acts within the genetic pathways controlling these processes in different biological contexts.
Fig. 1. Schematic representation of the regulation of U. maydis mating-type gene expression. 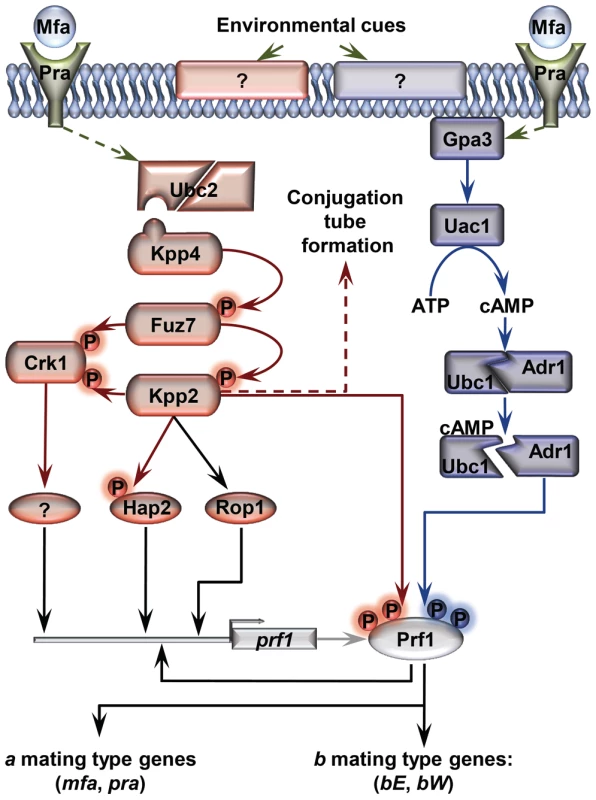
Pheromone (Mfa) recognition by the receptor (Pra) of the opposite mating type, together with environmental cues sensed by unknown receptors (represented by question marks), result in the activation of the cAMP (blue) and MAP kinase (red) pathways. The central core of the MAP kinase module is composed of Kpp4 (MAPKKK), Fuz7 (MAPKK) and Kpp2 (MAPK), and the alternative MAP kinase, Crk1. Once both pathways have been induced, the downstream transcription factor Prf1 becomes transcriptionally and post-translationally activated and the expression of a and b mating-type genes takes place. Transcriptional control of prf1 depends on Rop1, Hap2, a putative unknown factor induced by Crk1, and Prf1 itself. Activation of the MAP kinase module in compatible haploid FB1 or FB2 strains also leads to the formation of conjugation tubes through a Prf1 independent pathway (discontinuous red arrow). Transcriptional regulation is indicated by black arrows. Scheme adapted from [38]. In this work, we explore the roles of Tup1 during the life cycle of the maize pathogen U. maydis. We demonstrate that tup1 is required for normal mating and filament formation in this fungus and that it controls these processes by transcriptional activation of the Prf1 transcription factor through at least two of its direct regulators. Additionally, we show that tup1 is essential for full pathogenic development, affecting tumor formation and spore production. Our results indicate that Tup1 represents a key factor for the regulation of the pathogenic filamentous and dispersible spore forms of the corn smut fungus U. maydis.
Results
Identification of the U. maydis tup1 homologue
To identify Tup1 homologues in U. maydis we performed a blast search against the MIPS U. maydis database (MUMDB) proteome using Tup1p from the S. cerevisiae database (SGD) as the query sequence. A U. maydis protein sequence, um03280, with an e-value of 9.5e-81 and 66% similarity to S. cerevisiae Tup1p, was retrieved. This sequence, already annotated in MUMDB as Tup1, shows homology to Tup1 proteins from other fungi; including the animal pathogens C. albicans (67% similarity), C. neoformans (73%) and P. marneffei (75%) (all data in Table S1). A sequence alignment of Tup1 proteins from these organisms revealed a number of conserved domains, based on S. cerevisiae: (1) the tup_N domain, located in the N-terminal region, which is known to be required for Tup1p/Ssn6p complex formation; (2) seven WD40 domain repeats in the C-terminal region, that mediate protein-protein interactions and (3) a poorly conserved central region, which possesses histone binding activity in S. cerevisiae [24], [57], [58] (Figure 2, Table S2 and Figure S1).
Fig. 2. Comparison of conserved protein domains between different members of the Tup1 family of transcriptional repressors. 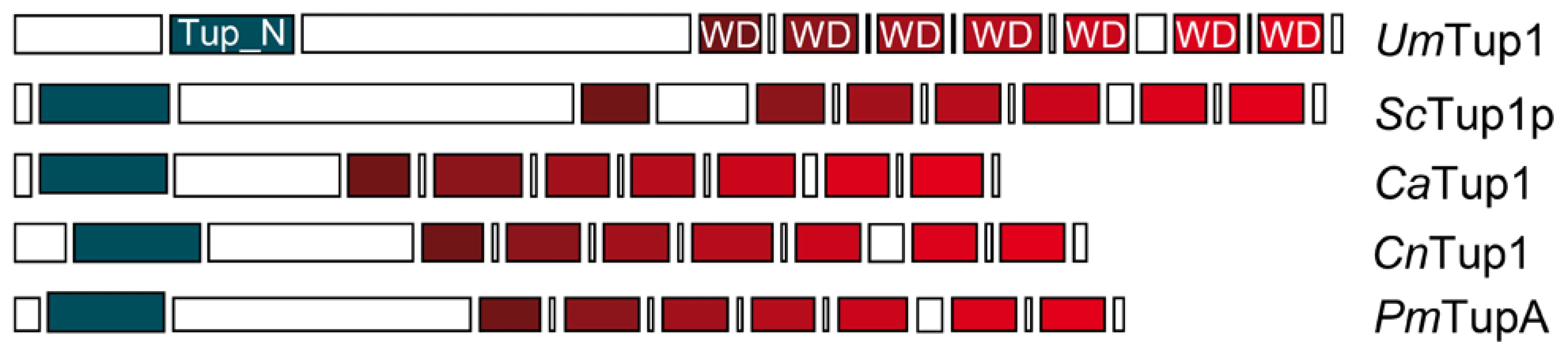
Conserved structure of Tup1 proteins in U. maydis (UmTup1), S. cerevisiae (ScTup1p), C. albicans (CaTup1), C. neoformans (CnTup1) and P. marneffei (PmTupA) (for accession numbers see Methods). Domains according to InterPro (Pfam) and functionally characterized in S. cerevisiae [19], [57] are shown. All the domains described for ScTup1 are conserved in the U. maydis Tup1 protein, including the N-terminal Tup_N domain, required for Ssn6p binding (blue square), seven WD40 domains in the C-terminal region (red tone squares), and a less conserved central region. Tup1 is required for full pathogenic development
To test if Tup1 has a role during the U. maydis life cycle, we generated deletion mutants for tup1 in both mating compatible strains, FB1 and FB2, replacing the tup1 open reading frame with the carboxin resistance cassette from pMF1-c [35]. Examination of cell growth and morphology did not reveal any statistically significant differences in either of the tup1 mutants (Figure S2).
Since the U. maydis life cycle is intrinsically linked to its host, we assayed the virulence of tup1 deletion strains. For this purpose, we infected seven day old maize seedlings with compatible mixtures of either wild-type or Δtup1 fungi, and scored tumor formation 14 and 21 days post-infection (dpi). We noticed a considerable reduction in the number of Δtup1 infected plants that developed tumors compared to wild-type infections. Moreover, the size of tumors developed by Δtup1 strains were also considerably reduced (Figure 3A, 3B, and Figure S3). In addition, we observed reduced plant mortality for tup1 mutant infections, with no dead plants observed at 14 dpi and only 11% mortality versus 57% for the wild-type strain 21 dpi. (Figure 3B and Figure S3).
Fig. 3. tup1 is required for full pathogenic development. 
(A) Representative images showing the most prevalent tumor category for wild-type and tup1 mutant infected plants. (B) Disease symptoms caused by wild-type and tup1 mutant strains are shown. Strains are indicated within the color legend. The total number of infected plants (n) is indicated below each strain combination. Symptoms were scored 14 days post-inoculation. Categories correspond to: large tumors (>5 mm), medium tumors (1–5 mm), small tumors (<1 mm). Mean values of three independent experiments and the standard deviation are shown. Asterisk (*) represents statistically significant differences in regard to the wild-type strain. (C) tup1 mutant spore development phenotypes 21 days post-infection. Left: picture of similarly sized tumors developed by the indicated strains. Strong spore formation is evident by dark coloration inside the tumor. Right: tumor sample analyzed by optical microscopy. Spores were present in tumors induced by wild-type strains. Hyphae at fragmentation or rounded cell formation stages were seen (arrowheads) in the tup1 mutant-induced tumors. Mature spores were not observed (scale bar = 20 µm). To ascertain whether tup1 mutants are able to complete the sexual cycle we assayed infected plants for the presence of spores 21 dpi. Interestingly, while we found large numbers of spores in wild-type tumors, we could not find spores in tup1 mutant infected plants. Microscopy analysis of the Δtup1 induced tumors revealed that none of the fungal hyphae observed had progressed beyond the rounded cell formation stage that occurs just before spore maturation [46] (Figure 3C).
These results indicate that tup1 is required for full pathogenic development of U. maydis and support a conserved role for tup1 in the virulence of animal and plant fungal pathogens.
Δtup1 cells are impaired in mating and infective filament formation
During U. maydis plant infection, multiple morphological changes of the fungus are required (for review see [38]). To ascertain which steps of the infectious process are responsible for the decreased amount and size of tumors generated by tup1 mutants, we first determined the extent to which they were able to successfully undergo mating and develop dikaryon filaments. To test this, we co-spotted compatible combinations of tup1 mutants and wild-type strains on PD-Charcoal plates, where the appearance of “fuzzy” white colonies indicates successful mating and the formation of dikaryon filaments. As shown in Figure 4A, crosses between tup1 mutants were unable to form white fuzzy colonies, indicating a recognition or fusion defect between compatible partners, or a post-fusion filamentation defect. Similarly, crosses between tup1 mutants and compatible wild-type strains also showed fuzzy colony formation defects. Filamentation was partially affected when FB1Δtup1 was crossed with wild-type FB2, showing an intermediate phenotype between wild-type and Δtup1 crosses. In contrast, the FB1 and FB2Δtup1 cross showed the same loss of fuzzy colony phenotype as the double mutant cross. In order to check whether the differences observed in FB1Δtup1 and FB2Δtup1 strains could lead to different rates of tumor formation, we performed a plant infection assay using FB1 vs FB2Δtup1 and FB1Δtup1 vs FB2 crosses. As shown in Figure S4A the infection rates of these two strains were similar and slightly different to the rates observed for the cross of both wild type strains.
Fig. 4. tup1 is required for mating. 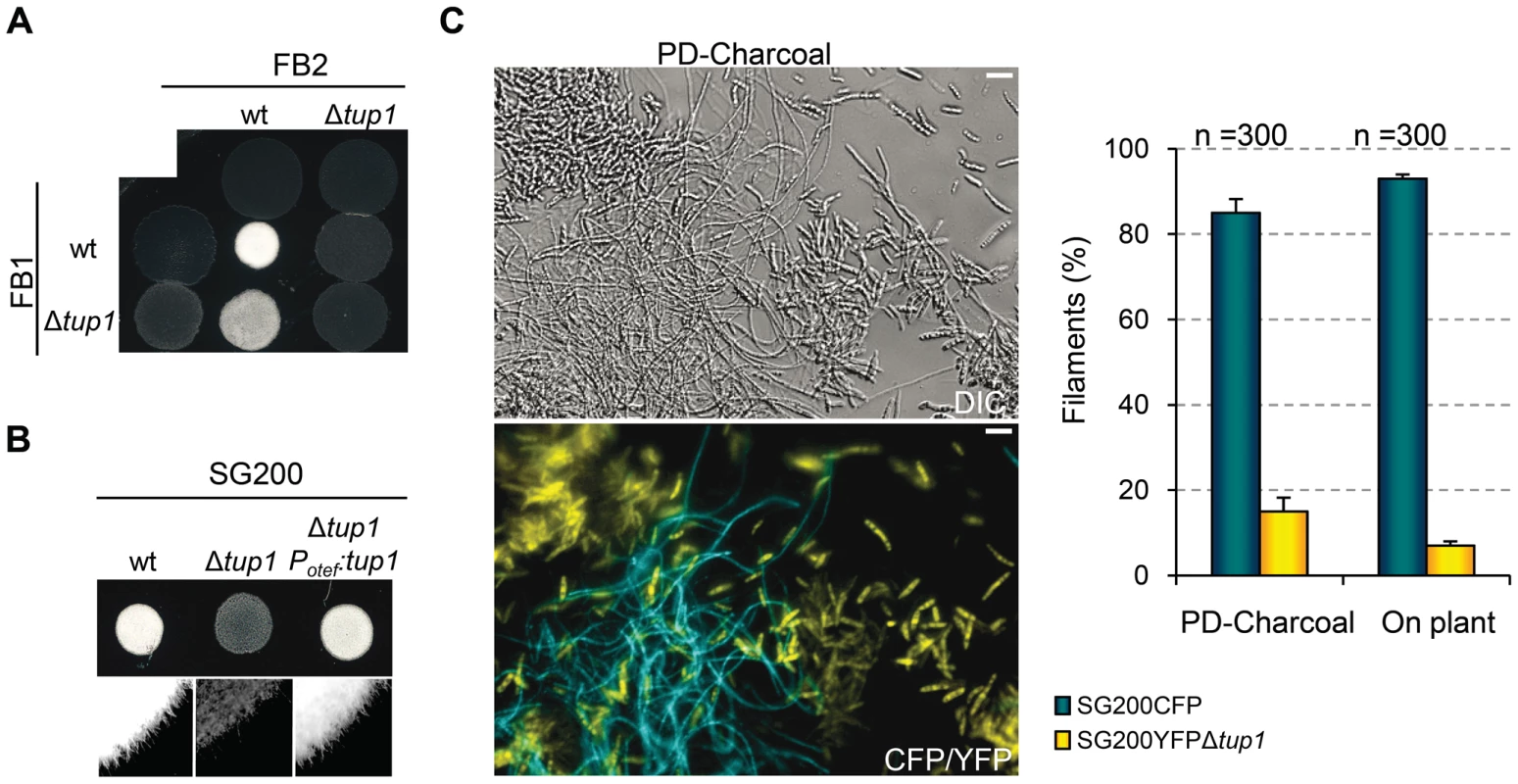
(A) Mating between compatible U. maydis strains. The strains indicated (top/left) were spotted either alone or in combination and incubated on PD-charcoal plates for 24 hours at 25°C. A white fuzzy colony appearance is indicative of successful mating and the formation of aerial dikaryotic hyphae. (B) Filament formation in SG200 and SG200Δtup1 strains. The indicated strains were spotted alone on PD-charcoal plates. The presence of white fuzzy colonies indicates the formation of filaments. (C) Quantification of filamentation defects in the tup1 deletion strain. A mixture with equal number of cells from SG200CFP and SG200YFPΔtup1 were spotted onto charcoal plates or inoculated into maize plants. Image on the left represents the filamentation capacity of both strains on charcoal containing media. Scale bar represents 20 µm. The chart on the right indicates the number of filaments that corresponded to each strain in charcoal plates or on the plant leaf surface. Strains are indicated within the color legend. The total number of filaments counted (n) is indicated above each pair of columns. Mean values of three independent experiments and the standard deviation are shown. In addition, we analyzed white fuzzy colony formation in a SG200 background, which is able to form the infective hypha without the necessity of mating with a compatible partner, because of the presence of an active bE1/bW2 heterodimer and a constitutively expressed mfa2 gene [59]. Significantly, SG200Δtup1 did not generate fuzzy colonies on charcoal plates, suggesting a post-fusion role for tup1 (Figure 4B). In order to quantify the phenotype, we performed a filamentation assay by co-spotting SG200CFP [60] and SG200YFPΔtup1 labeled strains on PD-charcoal plates. After fuzzy colony formation, colony samples were used for the quantification of filaments formed by each strain. As shown in Figure 4C, 80% of the filaments corresponded to the wild-type strain, while only 20% belonged to the mutant. Maize infection experiments with tup1 mutants in the SG200 background revealed similar virulence defects to what we had observed in FB1 and FB2 backgrounds (Figure S5A and S5B). Insertion of a single copy of tup1 under the control of the constitutive otef promoter in the ip locus [34] of SG200Δtup1, restored its filamentation and pathogenic capacity, indicating successful complementation (Figure 4B, Figure S5A and S5B). Moreover in the case of the FBD11 diploid strain, which also do not need to mate with a compatible partner to cause virulence, the heterozygous mutant FBD11Δtup1/tup1 and the homozygous FBD11Δtup1/Δtup1 were almost completely avirulent in leaf infection experiments (Figure S4B and S4C). Because of the reduced infection capacity of the FBD11 wild-type strain, we also performed flower infections (where we usually observe bigger tumors) with these strains to better reflect the differences between them. This experiment revealed big tumors in the wild-type strain, medium tumors in the heterozygous and small tumors in the homozygous mutant strains (Figure S4D and S4E).
These results point to a post-fusion filamentation defect as a plausible reason for the impaired pathogenicity of Δtup1 strains. However, it has been reported that mating or filamentation defects on PD-Charcoal plates are not always conserved on the plant leaf surface [61]. To check this, we co-infected 7 day old maize seedlings with the labeled strains, SG200CFP and SG200YFPΔtup1 and quantified filament formation on the leaf surface. As shown in Figure 4C (on plant columns), the filamentation defect seen on charcoal containing media was also apparent on the leaf surface, with only around 5% of the filaments formed corresponding to the mutant strain.
Finally, to check whether tup1 could also be implicated in other morphological changes required during the U. maydis infection process, we checked for appressoria formation and the presence of clamp-like cells during mycelium expansion in tup1 mutant strains. We observed that both of these structures were formed in the deletion mutants for tup1 (Figure 5A and 5B), although at lower frequency than the wild type, which is very likely a consequence of the filament formation defect showed by these mutants. The frequency of appressoria formation by SG200YFPΔtup1 was reduced to a similar extent as filament formation (Figure S5C), and mycelium expansion was reduced in Δtup1 infected plants at 2 dpi (Figure 5C). These results, together with the capacity, albeit reduced, of tup1 mutants to induce tumors in maize, suggest that those tup1 mutant cells that overcome the filamentation defect are then able to undergo the morphological changes required for plant penetration and expansion. Thus, the role of tup1 in the morphological changes that occur during U. maydis infection seems to be specific to the yeast-to-hypha transition.
Fig. 5. Appressorium, clamp-like cells formation and mycelium expansion of tup1 mutants. 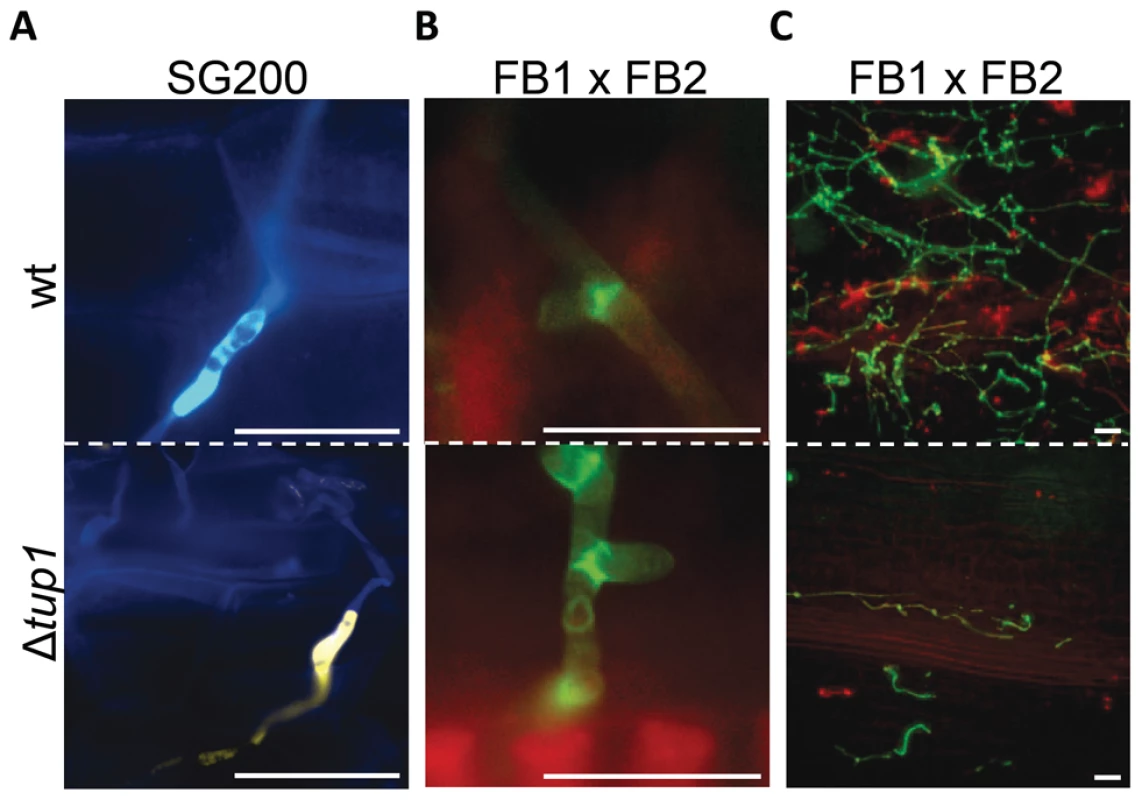
(A) Appressorium formed by wild-type SG200CFP and SG200YFPΔtup1 strains. (B) Clamp-like cells formed by wild-type and tup1 mutant cells 2 dpi. (C) Visualization of mycelium expansion inside the plant tissue of the indicated strains 2 dpi. Infected leaf samples were stained with WGA-AF and propidium iodide (see Methods). Scale bars represent 20 µm. Induction of the b locus restores the filamentation defect of tup1 mutants
As tup1 mutants are unable to form dikaryotic hyphae at wild-type levels, we wondered whether tup1 regulates genes downstream of the b locus, thus compromising the fungal dimorphic transition in tup1 mutants. To this end, we used the AB33 strain in which expression of a compatible bE1/bW2 heterodimer is under the control of the nar inducible promoter [62]. When this strain is grown in inducing conditions it forms a b-dependent filament. We found that deletion of tup1 in this background did not affect its filamentation capacity (Figure 6A; see Figure S6 for quantification). This result suggests that Tup1 is affecting processes upstream of the b locus or, alternatively, is acting on a parallel pathway regulating filamentation. To discern between these two possibilities, we extracted total RNA from SG200 and SG200Δtup1 fungi grown on charcoal-containing media for 48 hours and quantified the expression of bE and bW by Northern blot. We observed a strong decrease in both gene transcripts in SG200Δtup1 indicating that tup1 is required for the normal expression of b loci genes (Figure 6B lanes 5 and 6).
Fig. 6. Genetic interaction between tup1 and the b mating-type locus. 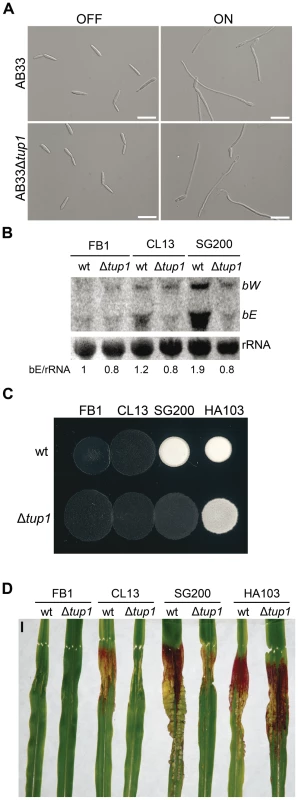
(A) Induction of b-compatible heterodimer in the AB33 background. Expression of bE and bW genes was induced by a shift from ammonium (OFF) to nitrate (ON) containing minimal media. b-dependent filament formation could be observed both in wild-type and tup1 mutant strains. Pictures were taken 5 hours post-induction. Scale bars represent 20 µm. (B) b-gene expression level in wild-type and tup1 deletion strains of CL13 (a1 bE1/bW2), SG200 (a1 mfa2 bE1/bW2) and HA103 (a1 (bE1/bW2)con). 10 µg of total RNA extracted from each strain grown on charcoal minimal media for 48 hours at 25°C was loaded per lane. Methylene blue stained rRNA was used as loading control. Numbers indicate the relative signal of bE gene in regard to rRNA. (C) Filamentation capacity of the indicated strains growing on PD-charcoal plates during 24 hours at 25°C. White fuzzy colonies indicates b-dependent filament formation. (D) Representative images showing the most prevalent disease category for wild-type and tup1 mutant infected plants. Strains are indicated below. The FB1 (a1 bE1/bW1) background, which harbors an incompatible b-heterodimer, was used as control. Scale bar represent 1 cm. To test if constitutive b expression could rescue the filamentation and virulence phenotypes of tup1 mutants, we took advantage of the HA103 strain, which harbors a compatible bE1/bW2 heterodimer under the control of constitutive promoters [52]. Deletion of tup1 in HA103 did not produce the filamentation and virulence defects described for the SG200 background (Figure 6C, 6D and Figure S7), indicating that constitutive b expression partially rescues these phenotypes. To better understand the effect of b expression on the tup1 mutant virulence phenotype, we used the HA103 parental strain, CL13 [59], which carries compatible bE1 and bW2 genes under the control of their own promoter and lacks the constitutively-expressed mfa2 gene present in SG200. Deletion of tup1 from CL13 led to a 90% reduction in maize tumor formation (Figure 6D and Figure S7), revealing an even clearer b-genes dependent rescue of tup1 mutant phenotypes. Interestingly, the expression level of the b genes correlated with the phenotype of the wild-type and Δtup1 strains (Figure 6B). Moreover, when we focused on the CL13 and SG200 backgrounds, we observed that the SG200Δtup1 strain had a b expression level, filamentation and virulence capacity comparable to the wild-type CL13 strain (Figure 6, Figure S7 and Figure S8). Thus, the effect of deleting tup1 from SG200 seems to be equivalent to removing its constitutive expression of mfa2, which would suggest a putative role for the pheromone responsive pathways in tup1 mutant phenotypes.
Tup1 is required for mfa1 gene expression, and conjugation tube formation upon pheromone stimulation
In our earlier experiment we bypassed the requirement for cell fusion by using the SG200 strain to identify a post-fusion requirement for tup1 in U.maydis filamentation. However, this experiment does not exclude a role for tup1 in mating between compatible strains as well, especially since both a and b loci genes are in the same position of the genetic pathway that controls the dimorphic transition. Moreover, as commented above, the similarity between SG200Δtup1 and CL13 strains may reflect a role for tup1 in the transduction of the pheromone signal.
To test this possibility, we extracted total RNA from a FB1Δtup1 vs FB2Δtup1 cross grown on charcoal-containing media for 24 hours and compared mfa1 and bE1 expression with a wild-type strains cross by Northern blot. In the wild-type cross, as a result of the recognition of pheromones by receptors of opposite mating types, activation of pheromone responsive pathways takes places, which is reflected in the expression of genes at both a and b loci. In the case of the tup1 mutant cross, however, we observed reduced mfa1 and bE1 expression (Figure 7A), indicating that tup1 is necessary for wild-type expression of these genes. Accordingly, FB1Δtup1 and FB2Δtup1 strains drastically reduced conjugation hyphae formation upon stimulation with synthetic pheromones of the opposite mating type (Figure 7B and 7C).
Fig. 7. Pheromone response and conjugation tube formation in tup1 mutants. 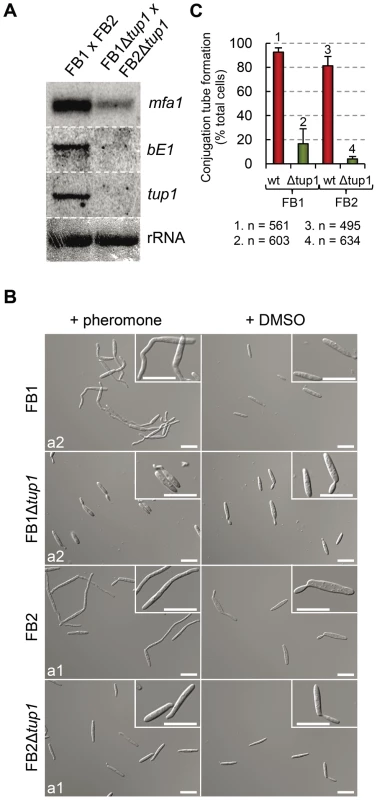
(A) Expression level of mfa1 and bE1 in FB1 vs FB2 and FB1Δtup1 vs FB2Δtup1 crosses after 24 h on charcoal containing plates. tup1 expression was used as experimental control and rRNA used as loading control. (B) DIC images of conjugation hyphae in wild-type and tup1 mutant strains. Wild-type and tup1 mutant strains were grown on CM liquid media until exponential phase and then exposed to the pheromone of the opposite mating type or DMSO (pheromone solvent) for 5 hours. Strains (left), and pheromone or DMSO treatments (top) are indicated. Type of pheromone (a1 or a2) is shown inside each picture. Scale bars indicate 20 µm. (C) Quantification of conjugation hyphae formation in wild-type and tup1 deletion strains. Total number of cells counted is indicated below the chart. Mean values of three independent experiments and the standard deviation are shown. Thus, tup1 is required for signal transduction upon stimulation with pheromone and expression of genes at both a and b loci, which is reflected in the observed pre and post-fusion defects of Δtup1 cells.
Tup1 controls a and b loci genes downstream of the MAP kinase cascade
The expression of a and b loci genes is controlled by the cAMP and MAP kinase pathways through their common effector Prf1. To situate tup1 within this genetic context, we used the FB1Pcrg1:fuz7DD strain, which harbors a constitutively active allele of fuz7 MAPKK under the control of the arabinose inducible promoter crg1 [51] (see Figure 1 for components of the MAP kinase pathway). Upon induction, this strain promotes the expression of a and b loci genes via the Prf1 transcription factor. After deleting tup1 from this strain, we checked for a and b loci gene expression under inducing conditions. As expected, increased transcription for genes at both loci was observed in the wild-type strain; however, this was not the case for the tup1 mutant, indicating that Tup1 regulates a and b gene expression downstream of Fuz7 MAPK kinase (Figure 8A). Since Tup1 is involved in regulating the expression of genes related to glucose metabolism, the expression level of Fuz7 under the control of the crg1 promoter was also examined. No difference in fuz7DD expression was observed between the wild-type and the Δtup1 strains (Figure 8A).
Fig. 8. Tup1-dependent regulation of mating-type genes and prf1 transcription factor. 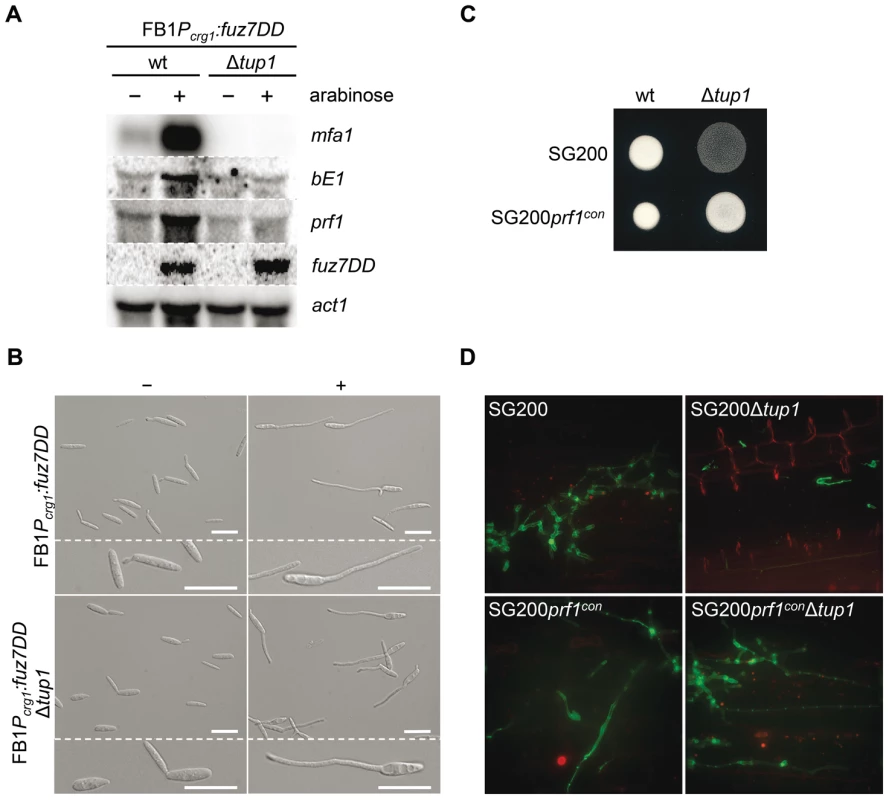
(A) mfa1, bE1, and prf1 expression levels upon fuz7DD allele induction. Expression of the fuz7DD allele was induced by a shift from a glucose to arabinose containing CM media. Total RNA was extracted 5 hours post-induction and 10 µg were loaded in each lane. U. maydis actin was used as loading control. Strains (above) and probes (right) are indicated. (B) Conjugation tube formation upon fuz7DD induction. Pictures were obtained by optical microscopy 5 hours post-induction. - (glucose) and + (arabinose) indicate non-inducing and inducing conditions, respectively. Scale bars represent 20 µm. (C) Filamentation of constitutive expressed prf1 strains on charcoal media. SG200, SG200prf1con and their derivatives were spotted alone on charcoal plates and grown at 25°C for 24 hours. White fuzzy colonies appearance indicates formation of filaments. (D) On planta filamentation of constitutive expressed prf1 strains. SG200, SG200prf1con and their derivatives were inoculated into maize plants and their filamentation capacity was determined 24 hours post-inoculation. Apart from its effect on the expression of the previously mentioned genes, induction of the fuz7DD allele, promotes conjugation tube formation through a Prf1 independent pathway that also requires the action of Kpp2 MAP kinase [51]. Thus, we wondered whether the induction of fuz7DD in the tup1 deletion strain could also induce conjugation tube formation. As shown in Figure 8B, tup1 mutants in this background were able to form conjugation hyphae at similar levels to wild-type fungi in inducing conditions (Figure S9 for quantification). This result makes it unlikely that Tup1 is regulating conjugation tube formation downstream of the MAP kinase cascade and, at the same time, strongly suggest that tup1 regulates mating-type genes downstream of Kpp2 MAP kinase.
Tup1 is required for expression of prf1 transcription factor
We have shown that tup1 seems to regulate the expression level of a and b loci genes acting downstream of the MAP kinase cascade. Since the Prf1 transcription factor is the genetic element connecting the MAP kinase cascade and the mating-type genes, we measured prf1 expression level in a FB1Pcrg1:fuz7DD background under inducing conditions. The removal of tup1 prevented the increase in prf1 expression (Figure 8A), indicating that tup1 is required for prf1 expression upon MAP kinase cascade induction. Moreover, the filamentation defects on charcoal-containing media as well as on the plant surface were rescued with the constitutive expression of prf1 (Figure 8C and 8D). These results strongly suggest that tup1 affects mating and b-dependent filament formation through control of prf1 transcription factor expression level rather than by controlling the expression of a and b loci genes directly.
Tup1 deletion affects the expression of several b and pheromone/fuz7DD regulated genes as well as the rop1 transcription factor
As constitutive bE/bW expression did not fully complement Δtup1 phenotypes, we were interested in identifying other Tup1 regulated genes, that might also have roles in the dimorphic transition and virulence in U. maydis. For this purpose we performed a microarray analysis with custom Affimetrix array (MPIUstilagoA), covering 5823 of the 6787 predicted U. maydis genes, and compared the gene expression of SG200 and SG200Δtup1 strains grown on MM-charcoal array plates for 48 hours (see Methods). We identified a total of 115 genes (around 2 % of the covered genes) with altered expression in the tup1 mutant strain. Of these, 59 were upregulated and 56 downregulated. Within this list appear the bE and bW genes together with 34 genes that have also been described as b regulated genes [63], and 17 genes described as pheromone regulated [64] (Table S3). Thus, around 36% of the genes directly or indirectly regulated by tup1 are also regulated upon bE/bW heterodimer and/or pheromone/fuz7DD induction, in agreement with our earlier results and supporting the quality of our dataset. Additionally, in order to experimentally validate our microarray data, the differential expression of some of the genes was confirmed by Northern blot analysis (Figure 9A).
Fig. 9. Microarray validation and pac2 mutants filamentation and virulence phenotypes. 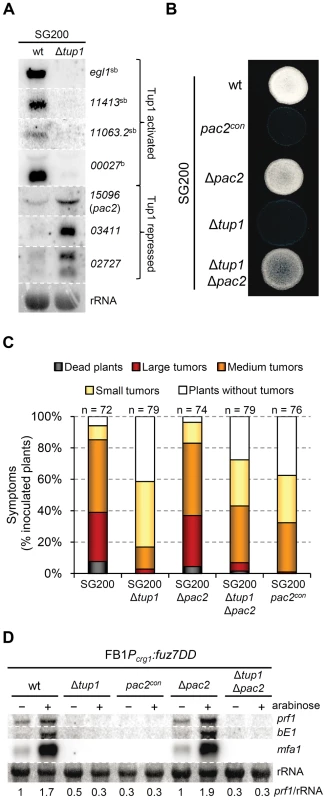
(A) Validation of microarray data by Northern blot. Probes (right) and strains (top) used are indicated. b-dependent (b) and strictly b-dependent genes (sb) according to [63], are indicated. Methylene blue stained rRNA was used as loading control. Total RNA was extracted from the indicated strains growing on minimal media charcoal-array medium for 48 hours at 25°C. A total of 10 µg of RNA was loaded per lane. (B) Filamentation capacity of the pac2 mutant strains. Strains indicated (left) were spotted alone on PD-charcoal plates and grown for 24 hours at 25°C. pac2con indicates constitutive expression of pac2 from the otef promoter. (C) Pathogenicity of pac2 mutant strains. Seven day old maize seedlings were infected with the indicated strains (color legend). Total number of infected plants (n) is indicated above each column. Symptoms were scored 14 dpi. Tumors categories correspond to: large tumors (>5 mm), medium tumors (1–5 mm) and small tumors (<1 mm). Represented are the main values of three independent experiments. (D) Pac2-dependent regulation of prf1. Prf1 expression level of the indicated strains upon fuz7DD allele induction was measured by Northern blot. Expression of the fuz7DD allele was induced by a shift from a glucose (-) to arabinose (+) containing CM media. 10 µg of total RNA were loaded per lane. rRNA was used as loading control. Numbers indicate the relative signal of prf1 gene in regard to rRNA. All the 115 Tup1-regulated genes were classified in functional categories using the Blast2Go tool [65]. Enrichment analysis of genes up-regulated by the deletion of tup1 did not reveal a significant over-representation in any of the GO categories (Table S4). Of the genes down-regulated upon tup1 deletion our analysis revealed a significant over-representation in two GO categories: “Carbohydrate metabolic process” (GO:0005975; 8 genes) and “Antioxidant activity” (GO:0016209; 3 genes) (Table S4). 4 of the 8 genes belonging to the first category were also b-regulated genes, with two of them defined as strictly b-dependent (Table S3). The second category comprises proteins involved in the inhibition of dioxygen or peroxide-induced reactions and could be related to pathogenicity since production of these compounds is a well-characterized plant defense mechanism [66], [67], and H2O2 detoxification is required for U. maydis virulence [68].
Interestingly, several tup1-regulated genes are associated with processes that could be related to the morphological switch from yeast-like to filamentous growth. Almost 10% of these genes are potentially involved in cell wall synthesis or modification, revealing that the altered yeast-to-hypha transition, promoted by deletion of tup1, results in a different cell wall composition.
Significantly, we found that rop1, that encodes a direct activator of Prf1, was down-regulated in the tup1 deletion strain (Table S3). This suggests an indirect role for tup1 in controlling prf1 expression. Rop1 has been described as being required for the mating of compatible strains on charcoal containing media, with a post-fusion role, due to the inability of SG200Δrop1 to form white fuzzy colonies on charcoal plates. It is essential for conjugation tube formation upon pheromone stimulation, and for expression of pheromone-responsive genes [61]. These phenotypes clearly resemble the situation described for tup1 mutants; however, rop1 mutants are fully pathogenic, with no mating or filamentation defects described on the plant leaf surface [61].
In addition to rop1, we identified an interesting candidate gene, um15096, that could be related to the tup1 mutant phenotypes. In Schizosaccharomyces pombe, a homologue of um15096, named pac2, has been shown to be a repressor of ste11 (the putative functional homologue of prf1) [69]. Interestingly, um15096/pac2, herein referred to as pac2, appeared over-expressed in the tup1 deletion strain. To check whether this putative prf1 repressor could also be playing a role during filamentation and pathogenic development, we over-expressed pac2 by integrating an extra copy of the gene under the control of the otef constitutive promoter in the ip locus of the SG200 strain. Filament formation of SG200pac2con was reduced on charcoal containing media (Figure 9B) and, more importantly, pathogenicity was reduced to levels comparable to tup1 mutants (Figure 9C). The fact that pac2 is over-expressed in tup1 mutants together with the observation that ectopic pac2 expression decreases filamentation and virulence in the wild-type strain, strongly suggest that pac2 expression contributes to the filament formation and pathogenic defects of Δtup1 cells. Consistent with this, the deletion of pac2 from SG200 resulted in wild-type filamentation and infection rates (Figure 9C). When prf1 expression was induced by constitutively activating the MAPK pathway at Fuz7 level, overexpression of pac2 abolished its expression, while deletion of pac2 did not apparently affect it. Similar results were observed for mfa1 and bE1 genes. The double Δtup1Δpac2 mutant showed the same level of expression as the single Δtup1 strain (Figure 9D); probably as consequence of the regulation of rop1 via Tup1. Surprisingly, pac2 deletion, weakly restored the filamentation and infection defects shown by SG200Δtup1 strain (Figure 9B and 9C), indicating that Pac2 contributes to tup1 deletion strain phenotypes.
In summary, our microarray data reveal that at least 36% of the genes whose expression is affected by deletion of tup1 seems to be a consequence of tup1-dependent regulation of a and b loci genes through prf1. Moreover, the role of Tup1 in the control of prf1 expression could be explained by the altered expression of rop1 and pac2 observed in the tup1 mutant strain.
Tup1 affects the expression of the prf1 transcriptional regulators rop1 and hap2 but not crk1
As Tup1 seems to have an indirect effect on prf1 transcription level through Rop1 and, putatively, Pac2, we wondered whether the expression of other known prf1 regulators could be affected in tup1 deletion strains. Apart from Rop1, prf1 is known to be directly regulated by Hap2 [70] and indirectly through the MAP kinase Crk1 [71]. Northern blot assays of SG200 and SG200Δtup1 grown on charcoal media showed that the expression level of crk1 was unaffected in tup1 deleted strain. In contrast, the levels of rop1 and hap2 were reduced in comparison to the wild-type strain (Figure 10). However, as Crk1 acts on prf1 indirectly, and since it has been previously reported that the effect of Crk1 on prf1 depends on the prf1 promoter UAS [71], we tested whether Tup1 could regulate prf1 via its UAS. For this purpose, we used the HA232 strain, which harbors a GFP reporter gene under the control of the prf1 promoter UAS (see [53] for details). In this strain, GFP is strongly expressed when grown on glucose-containing media, while its expression is reduced on a maltose containing media [53]. As is shown in Figure S10, the expression levels of the reporter gene were indistinguishable in Δtup1 mutants from the wild-type in all the conditions tested. This indicates that Tup1 is unlikely to act via the prf1 promoter UAS, in contrast to Crk1. Thus, the effect of Tup1 on prf1 expression seems to be mediated via Rop1 and Hap2 but not through the Crk1 pathway.
Fig. 10. tup1 is required for wild-type expression levels of the prf1 transcriptional regulators rop1 and hap2. 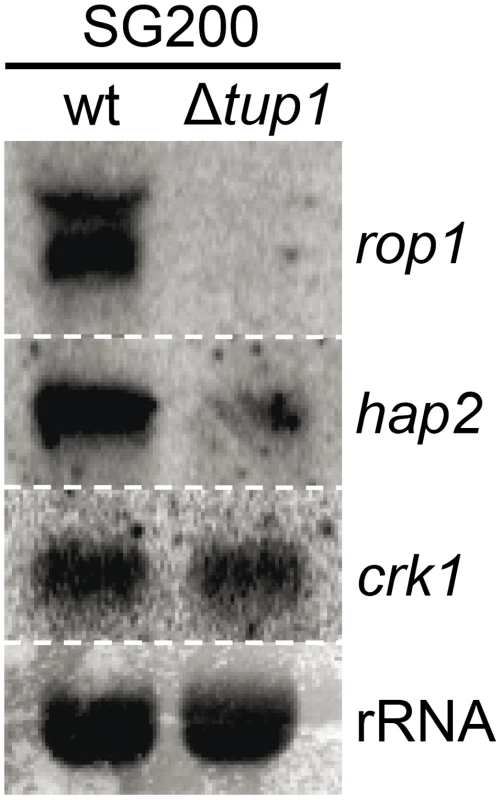
Northern blot of rop1, hap2 and crk1. 10 µg of total RNA extracted from the indicated strains growing on minimal media charcoal plates for 48 hours at 25°C was loaded per lane. rRNA was used as loading control. To sum up, although other factors may be implicated in tup1 mutant phenotypes, Tup1 seems to control the dimorphic transition and participates in the virulence program of U. maydis by indirectly regulating prf1 expression via altered rop1 and hap2 expression levels, and possibly also through pac2, which would lead to a down-regulation of prf1-dependent expression of a and b loci genes and their related phenotypes.
Discussion
In the basidiomycete phytopathogen U. maydis, the switch from non-infective yeast-like growth to an infective filament formation occurs in response to different environmental cues, and is tightly controlled by complex genetic pathways in order to ensure the coordination and timing of the different processes associated with dimorphism. In this work, we have shown that the highly conserved general transcriptional repressor Tup1 plays a central role in controlling the proper expression of the genes implicated in the genetic control of mating, filamentation, and pathogenic development of this corn smut fungus.
Tup1 has been shown to be important during growth of vegetative cells in other fungi such as S. cerevisiae, C. neoformans or P. marneffei [12], [27], [72]. In the case of Ustilago maydis, differences could be observed in the tup1 mutants, although none of these were statistically significant. Interestingly the normal growth of Δtup1 strains contrasts with the poor growth capacity described for U. maydis strains harboring a partial deletion of sql1, the functional homolog to S. cerevisiae SSN6. However because these strains were not stable, the role of Sql1 could not be completely analyzed [34]. Thus a comparison between Tup1 and Sql1 of their growth capacity on U. maydis vegetative cells cannot be properly established. In other fungi, single deletions of tup1 and ssn6 have been reported to result in different phenotypes [73]-[76]. For example, the deletion of SSN6 but not of TUP1 homologues is lethal in S. pombe [75] and Aspergillus nidulans [76]. Moreover, Tup1 and Ssn6 have been shown to regulate different set of genes [74] and to form independent complexes in C. albicans [77].
A central question in this study was whether tup1 is involved in the infectious process of plant pathogenic fungi. We have observed that infections with Δtup1 cells lead to a reduction in tumor formation, plant death, and a failure of spore formation, indicating that Tup1 is required for full pathogenic development in U. maydis, and making tup1 mutants unlikely to cause damage in natural environments. Thus, tup1 seems to play a conserved role in virulence of animal and plant fungal pathogens.
The next key question was to try to understand the mechanism by which tup1 is required for normal tumor formation. Our results suggest that the virulence phenotype of Δtup1 cells has two main causes: (i) a recognition problem between compatible partners, due to the inability of tup1 mutants to form conjugation hyphae upon pheromone stimulation, and (ii) a filamentation defect, due to the inability of SG200 to form filaments at wild-type levels both on PD-charcoal plates and on the plant leaf surface. Additionally, the fact that the differences on conjugation hypha formation between FB1Δtup1 and FB2Δtup1 strains, though not statistically significant, together with the differential filamentation showed by crosses of these strains with their respective compatible wild-type strains on charcoal plates, suggest also a role for Tup1 in cell fusion, at least in the FB2 background. These defects result in tup1 mutants being unable to properly undergo dimorphic transition. These findings suggest that the impaired pathogenicity of tup1 mutant animal and plant fungi may also depend on a conserved role in the yeast-to-hypha transition.
Consistent with the conjugation and filamentation phenotypes of tup1 mutants, the expression of a and b loci mating-type genes was reduced in tup1 deletion strains, most likely as a consequence of Tup1-dependent regulation of the prf1 transcription factor. Microarray analysis of SG200Δtup1 during filamentation on charcoal media revealed a number of mis-regulated genes whose expression was also affected upon b-compatible heterodimer and/or pheromone/fuz7DD induction, including the b locus genes themselves, supporting the proposed role for tup1 during U. maydis mating and dikaryotic filament formation. On the other hand, in our microarray analysis we did not detect tup1-dependent changes in gene expression for any of the b-dependent genes previously described as being essential for pathogenicity [60], [63], [78], which is consistent with the ability, albeit reduced, of tup1 mutants to induce tumors in maize.
Interestingly, the main effector that links tup1 to the control of dimorphism seems to be conserved between U. maydis and C. albicans. In contrast, the genetic pathways by which tup1 acts on filamentation seem to differ, depending on the genetic control of hypha-specific genes in each organism. In C. albicans, Tup1 is proposed to control filamentous growth through the repression of hypha-specific genes by forming complexes with the transcriptional repressors Rfg1 and Nrg1, rather than affecting the elements in the Cph1-mediated MAPK and Efg1-mediated cAMP pathways [7], [54]–[56]. Moreover, expression analysis of filament-specific genes in Δcph1/Δcph1, Δefg1/Δefg1 and Δtup1/Δtup1 strains revealed common and divergent target genes [7]. Thus, Tup1 integrates into the network system proposed for the control of filament-specific genes in this fungus [7], [10]. On the other hand, in U. maydis, Tup1 controls infective filament-specific gene expression via a central regulatory, the Prf1 transcription factor, which is transcriptionally and post-translationally regulated by the cAMP and MAPK pathways [47], [51]–[53]. Interestingly, U maydis Prf1 is a High Mobility Group (HMG) transcription factor, similar to C. albicans Rfg1. Thus, an analogous mechanism, implicating a Tup1-Prf1 complex, could explain the roles of Tup1 in the regulation of hypha specific genes in U. maydis. Moreover, in S. cerevisiae, a complex between Tup1p and the HMG-transcription factor Rox1p has also been proposed [19], [79]-[81]. S. cerevisiae ROX1, whose deletion can be complemented by C. albicans RFG1 [33], is known to control hypoxic gene expression in a TUP1 dependent manner [19], [79]–[81]. Additionally, the deletion of TUP1 increases the expression of ROX1 [82], [83], but Rox1p itself is also able to regulate its own expression [83]. In aerobic conditions these observations can be explained by the proposed Tup1p-Ssn6p-Rox1p complex which would regulate ROX1 expression and Rox1p-dependent hypoxic gene expression. In anaerobic conditions, however, the regulation of ROX1 expression seems to implicate an anaerobic repressor that requires Tup1p for its function [83]. Similarly, in U. maydis, the expression of prf1 is dependent on Tup1 and prf1 is also self-regulated [52]. However, when we analyzed the effect of Tup1 on prf1 expression level more deeply, we observed that at least two direct activators of Prf1 were also down-regulated upon tup1 deletion, rop1 and hap2. This finding, although not excluding a putative Tup1-Prf1 complex, points to an indirect effect of Tup1 on the expression of prf1 and its regulated genes. Rop1 is required for pheromone response and for fuzzy colony formation on charcoal-containing plates, but is dispensable for mating and filamentation on the plant leaf surface. In the case of hap2, it is known to be essential for the pheromone response and has also an effect on the filamentation capacity of SG200 that seem to be conserved on planta. Thus, we propose that the effect of Tup1 on prf1 is the sum of the effects of Tup1 in both rop1 and hap2 on artificial media, while only the effect on hap2 would be responsible for the on planta phenotypes. The drastic effect of tup1 deletion on prf1 expression levels on charcoal plates may be diminished on the plant leaf surface as rop1 is dispensable in this situation.
In this work, we have also described a new gene, pac2, which is likely to be playing a role in the tup1 mutant virulence phenotype, since its over-expression causes a decrease in the pathogenic capacity of U. maydis SG200 strain and its expression is increased in the SG200Δtup1 strain. Since the homologue of this gene in S. pombe is a repressor of ste11 [69], the putative functional homologue of prf1, we analyzed the relationship between Pac2 and Prf1 in U. maydis. We found that over-expression of pac2 in a FB1Pcrg1:fuz7DD strain abolished the prf1 expression observed in the wild type strain establishing Pac2 as a repressor of Prf1. Accordingly, the deletion of pac2 in a SG200Δtup1 strain partially restored its filamentation and virulence defects. However, the double Δtup1Δpac2 mutant in the FB1Pcrg1:fuz7DD background shows the same prf1 expression level than the single Δtup1 strain, probably because of Tup1 control of rop1 and hap2. Nevertheless since prf1 regulation on charcoal plates or during virulence integrates several imputs besides the MAPK pathway the relationship between pac2 and prf1 in the regulation of filamentation and pathogenicity cannot be fully established. Thus, the final role of tup1 in U. maydis virulence is also likely to be linked to its control of hap2 and pac2 mRNA levels (Figure 11).
Fig. 11. Proposed model for the roles of Tup1 in the control of mating-type genes. 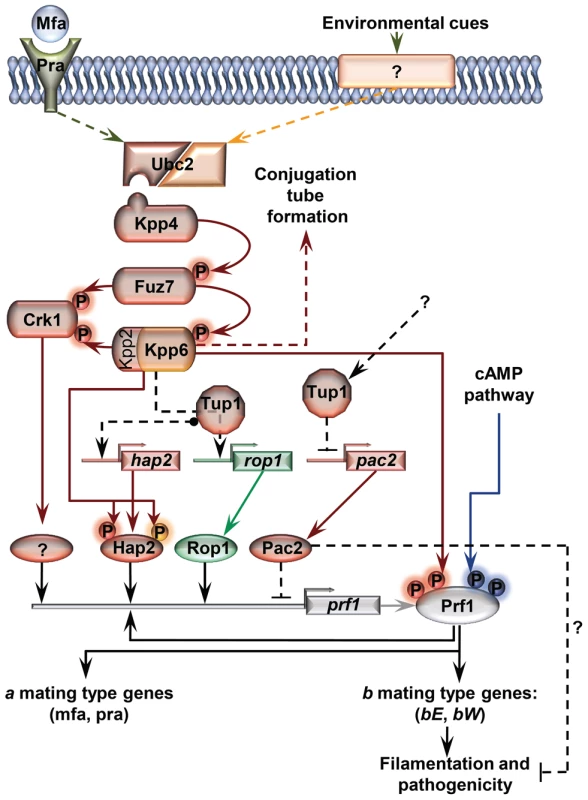
The MAP kinase pathway is shown in red. Black arrows represent transcriptional control. Components exclusively required in laboratory conditions (charcoal, pheromone stimulation, etc) are shown in green. Components specifically required during pathogenesis are shown in orange. In laboratory conditions the effect of Tup1 on prf1 expression would be mediated via its control of hap2, rop1 and pac2 expression levels. During infection, where rop1 is not required, Tup1 would control prf1 expression through hap2 and pac2. Question marks indicate putative elements or interactions. Surprisingly, although Tup1 is described as a general transcriptional repressor, the deletion of tup1 from U. maydis leads to the down-regulation of the genes that control the dimorphic transition, suggesting an activator role for tup1 in controlling them. On the other hand, determining how Pac2 controls prf1 gene expression would help to determine the role of tup1 as an activator and/or repressor during dimorphism. The way Tup1 seems to control the expression of the prf1 transcription factor, through hap2 and rop1 and, putatively, pac2, clearly reflects the complex genetic regulation that prf1-related processes require.
Similarly, the number of genes that we found to be up - or down-regulated following tup1 deletion when cultured on charcoal-containing media was equivalent. Thus, under the conditions tested, the loss of tup1 causes a similar effect on both the de-repression and repression of genes. Although this could reflect indirect changes in genes expression resulting from the repression of Tup1-gene targets, it is nevertheless an intriguing observation. Regarding an activating role for Tup1, previous studies have also shown that Tup1 can behave as an activator as well as a repressor of the same target gene in different conditions [84] or different genetic backgrounds [85] in S. cerevisiae.
Finally, we have shown that tup1 seems to be required for spore production inside maize tumors. Roles for Tup1 in sporulation have been previously reported in other fungi. In S. cerevisiae, the sporulation-specific genes DIT1 and DIT2, which are required for spore wall formation, are regulated by Tup1p [86]; in Neurospora crassa, mutants for rco-1, the homologue of TUP1, are aconidial [87]; and in C. neoformans, tup1 deletion considerably reduces spore production [27].
In summary, our work provides new insights into the complex regulatory circuits for sexual and pathogenic development of U. maydis. We have identified for the first time a requirement for tup1 at several steps of the life cycle of a pathogenic plant fungus, including in the genetic pathways controlling dimorphism and virulence. Our findings contribute to a better understanding of the role of this general transcriptional repressor in pathogenic fungi and of the precise genetic control that these pathogenesis-related processes require. We consider that the roles and mechanisms of action described for U. maydis tup1 in this work will also be extremely valuable for studying the roles of tup1 in the transcriptional regulation of morphogenetic processes in other organisms.
Methods
Strains, growth conditions and plasmids
Escherichia coli DH5α was used for cloning purposes. Growth conditions for E. coli [88] and U. maydis [42], [89] and the quantification of appressoria formation on the plant leaf surface [60] have been described previously. Quantification of filaments was performed as for the appressoria. For studies of growth rates and morphology, cells were grown on YEPSL liquid media for 12 hours, then diluted in the same media to an OD600 of 0.05 and grown until an OD600 of 0.8-1. Exponential growth cultures were examined under the microscope and transferred to solid plates for colony morphology studies. Growth rates on liquid media were determined by counting cells at different time-points. For charcoal mating and filamentation assays, cells were grown on YEPSL until exponential phase, washed twice with water, spotted onto PD-charcoal plates and grown for 24–48 hours at 25°C. For charcoal-grown cells used for RNA extractions, cells were spread out on charcoal plates at a concentration of OD600 = 0.1 per cm2. For DNA array charcoal media see below. U. maydis strains relevant to this study are listed in Table S5. Induction of nar promoter in AB33 [62] and crg promoter in FB1Pcrg1:fuz7DD [51] strains, and their derivatives, were done as previously described. Mating assays were performed as previously described in [90]. Pheromone stimulation was performed following the protocol of [51]. For pathogenicity assays, U. maydis strains were grown to exponential phase and concentrated to an OD600 of 3, washed twice in water, and injected into 7 days old maize (Zea mays) seedlings (Early Golden Bantam). Tumor formation was quantified 14 to 21 days post infection. Data are expressed as means ±SD of triplicate samples. Statistical significance was assessed using Statistical Calculators (http://www.graphpad.com/quickcalcs/index.cfm) and considered significant if p values were <0.05.
DNA and RNA procedures
Molecular biology techniques were used as described by [88]. U. maydis DNA isolation and transformation procedures were carried out following the protocol of [91]. Deletion constructs were generated according to [36]. To generate single deletion U. maydis mutants for tup1 (Um03280), pac2 (Um15096) and um04807 genes, fragments of the 5′ and 3′ flanks of their open reading frames were generated by PCR on U. maydis FB1 genomic DNA with the following primer combinations: UmTUP1KO5-1/UmTUP1KO5-2 and UmTUP1KO3-1/UmTUP1KO3-2; UmPAC2KO5-1/UmPAC2KO5-2 and UmPAC2KO3-1/UmPAC2KO3-2; Um04807KO5-1/Um04807KO5-2 and Um04807KO3-1/Um048071KO3-2; (Sequences in Table S2). These fragments were digested with SfiI and ligated with the 1.9 Kb SfiI carboxin resistance cassette, 2.7 Kb SfiI hygromycin resistance cassette, or 1.5 Kb SfiI neourseotricin resistance cassette as described previously [35]. Ligation products were then clone into pGEM-T-EASY vector (Promega). PCR generated linear DNA for each construct was used for U. maydis transformation.
For complementation of the tup1 deletion, the p123-tup1 plasmid was generated. p123-tup1 is a p123 [92] derivative in which the eGFP fragment has been substituted with the tup1 open reading frame . For this purpose, the tup1 open reading frame was amplified by PCR with the oligonucleotides Tup1-Start and Tup1-Stop, which contain NcoI and NotI restriction sequences respectively. Phusion high fidelity DNA polymerase (Invitrogen) was used. The PCR product was digested with NcoI and NotI, purified, and cloned into a p123 vector digested with the same restriction enzymes. Positive cloning was verified by restriction analysis and sequencing. To generate SG200Δtup1Potef:tup1 strain, p123-tup1 was linearized with SspI and integrated into SG200Δtup1 ip locus by homologous recombination.
For over-expression of pac2, the p123-pac2 plasmid was generated by replacing the eGFP fragment from p123 with the pac2 open reading frame. The Pac2 open reading frame was amplified using the oligonucleotides UmPac2ATGSmaXma y UmPac2StopNotI, digested with XmaI and NotI restriction enzymes and ligated into the p123 vector digested with the same enzymes. Successful cloning was verified by restriction analysis and sequencing. To generate SG200pac2con, p123-pac2 was linearized with SspI and integrated into SG200 wild-type strain ip locus.
For constitutive expression of pac2 in FB1Pcrg1:fuz7DD, we constructed the plasmid p5HOP2. This plasmid consists in 1 kb fragment of the upstream sequence of pac2 open reading frame (ORF) followed by the otef constitutive promoter, the hygromycin resistance cassette and 1 kb of the pac2 ORF integrated in a pGEM-T-EASY vector. For this purpose 1 kb fragment of the upstream sequence of pac2 was amplified with the primers Umpac2-5UTR-1 and Umpac2-5UTR-2, using FB1 genomic DNA; the otef constitutive promoter followed by 1kb of pac2 ORF was amplified with the primers Umotefpac2 and Umpac2-+1kb, using the plasmid p123-pac2 as template. Both flanks where then digested with SfiI restriction enzyme and ligated with the hygromycin resistance cassette. This construction was ligated to a pGEM-T-EASY vector. FB1Pcrg1:fuz7DDpaccon was generated by transformation of the wild-type FB1Pcrg1:fuz7DD with the mentioned construct.
Single homologous integration of the linear plasmids or PCR products transformed was verified by PCR and Southern blot.
In the expression analysis, cells grown on liquid culture were recovered by centrifugation, washed with cold water, and total RNA was isolated with QIAGEN (Valencia, CA) RNeasy mini kit. For charcoal grown cells, biomass was recovered and transferred to liquid nitrogen pre-chilled mortars. Total RNA was then extracted from the crushed powder with trizol reagent (Invitrogen) and with the QIAGEN RNeasy mini kit. Isolated RNA was separated by formaldehyde denaturing agarose gel electrophoresis, and transferred overnight by capillary action to nylon membranes. Probes were obtained by PCR with the oligonucleotides indicated in Table S6. Radioactive labelling of PCR generated probes was carried out. Radioactive bands were visualized and quantified using a Molecular Dynamics PhosphoImager.
For qRT-PCR first strand cDNA synthesis was performed using the Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacturer's protocol. As a template for the reaction 1 µg of total RNA was used. Samples were incubated at 50°C for 1 hour. Real-time PCR was performed in a ABIPRISM 7000 Sequence Detection System (Applied Biosystems) using the Power SYBR Green PCR Master Mix according to the manufacturer's protocol. Primers used for detection are shown in Table S6.
Sequence alignment and domain structure
U. maydis Tup1 sequence was obtained from MIPS U. maydis DataBase (http://mips.gsf.de/genre/proj/ustilago/). S. cerevisiae and C. albicans Tup1 sequences were obtained from SGD (http://www.yeastgenome.org/) and CGD (http://www.candidagenome.org/) databases, respectively. The rest of the Tup1 sequences were obtained from the NCBI. Multiple sequence alignments were made with ClustalW2. Domain structure analysis was performed using InterProScan Sequence Search tool from the European Bioinformatics Institute (http://www.ebi.ac.uk/). Pfam retrieved domains were used. Schematic representation of the retrieved domains was performed maintaining proportions of each domain with respect to the whole protein sequence length.
Fluorimetric measurement of GFP
Cells were grown on nitrate minimal media containing 1% glucose or 1% maltose to an OD600 of 0.6–0.8, then pelleted and resuspendend in sterile water to an OD600 of 1.0. Fluorescence from 200 µl of cell suspension transferred to a microtiter plate was measured by using a POLARstar Omega fluorescence reader (BMG LABTECH). GFP fluorescence was measured at a wavelength of 485 nm for excitation and 520 nm for emission. Fluorescence was normalized to OD600. At least three independent experiments were performed, each measured in triplicate.
Microscopy
Cell morphology of WGA-stained cells, conjugation tube and b-dependent filament formation were analyzed with a Zeiss Apotome microscope.
For on planta quantification of filament and appressoria formation in co-infection experiments with U. maydis CFP and YFP labelled strains, leaf samples were stained with calcofluor white (Sigma) to visualize fungal material and then checked for CFP or YFP fluorescence. Quantification of filament formation on charcoal plates was performed by fluorescence analysis of colony samples from co-spotted YFP and CFP strains. Post-penetration stages were visualized by WGA-AF 488 and Propidium Iodide (Sigma) staining of infected leaf samples as previously described [93]. Samples were examined using a Leica fluorescence microscope, equipped with a PlanApo x 100 lens and a Deltavision widefield microscope (Applied Precision, Issaquah, WA) equipped with 20, 40, 63 and 100 x lens. Image processing was carried out using Adobe Photoshop CS2.
DNA array
SG200 and SG200Δtup1 cells were grown on YEPSL until exponential phase, then washed twice with sterile water and cultured on minimal charcoal array plates (12.5% Holliday salts, 2% vitamins, 30 mM L-glutamine, 2% glucose, 4% agar and 2% charcoal, pH 7) during 48 hours at 25°C. 144 cm2 plates and a cell density of OD600 of0.1/cm2 was used. DNA-array analysis was performed using custom-designed Affymetrix chips (UstilagoA). Probe sets for the individual genes can be obtained from http://mips.helmholtz-muenchen.de/genre/proj/ustilago/. Target preparation, hybridization and data analysis was performed as described before [94], with the following alterations: total RNA was extracted as commented in DNA and RNA procedures for charcoal growing cells; 5 µg RNA were used for first strand cDNA synthesis at 50°C with Superscript II (Invitrogen); an adjusted P-value of ≤0.01 for the false discovery rate [95] and a change in expression of ≥2 was used for filtering. Expression values were calculated as mean of two biological replicates. Array data can be accessed at GEO/NCBI database (accession number GSE29591).
Accession numbers
U. maydis sequence data can be found in the GenBank/EMBL data libraries under accession numbers XP_759427 for Tup1, XP_762643.1 for Pac2,, XP_756724 for bE1, XP_756725 for bW1, XP_758529 for Mfa1, XP_760967 for Acf1, XP_762479 for Egl1, XP_762172 for Rop1, XP_762530 for Hap2, XP_758660 for Crk1, XP_758860 for Prf1, XP_757661 for Fuz7, XP_760954 for um04807, XP_758669 for um11413, XP_756174 for um00027, XP_759558 for um03411 and XP_758874 for um02727. Other sequences used in this study have the following accession numbers: S. cerevisiae Tup1p, NP_010007; C. albicans Tup1, AAB63195; C. neoformans Tup1, XP_570974; P. marneffei TupA, AAL99251; N. crassa Rco-1, AAB37245; A. nidulans TupA ACD46267; S. pombe Tup11, NP_592873; S. pombe Tup12, NP_592910.
Supporting Information
Zdroje
1. SchulzBBanuettFDahlMSchlesingerRSchaferW 1990 The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60 295 306
2. LoHJKohlerJRDiDomenicoBLoebenbergDCacciapuotiA 1997 Nonfilamentous C. albicans mutants are avirulent. Cell 90 939 949
3. LinXHuangJCMitchellTGHeitmanJ 2006 Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATalpha allele enhances filamentation. PLoS Genet 2 e187
4. LiuH 2002 Co-regulation of pathogenesis with dimorphism and phenotypic switching in candida albicans, a commensal and a pathogen. Int J Med Microbiol 292 299 311
5. LinX 2009 Cryptococcus neoformans: Morphogenesis, infection, and evolution. Infect Genet Evol 9 401 416
6. NadalMGarcia-PedrajasMDGoldSE 2008 Dimorphism in fungal plant pathogens. FEMS Microbiol Lett 284 127 134
7. BraunBRJohnsonAD 2000 TUP1, CPH1 and EFG1 make independent contributions to filamentation in candida albicans. Genetics 155 57 67
8. ErnstJF 2000 Transcription factors in candida albicans - environmental control of morphogenesis. Microbiology 146 1763 1774
9. LiuH 2001 Transcriptional control of dimorphism in candida albicans. Curr Opin Microbiol 4 728 735
10. Sanchez-MartinezCPerez-MartinJ 2001 Dimorphism in fungal pathogens: Candida albicans and ustilago maydis--similar inputs, different outputs. Curr Opin Microbiol 4 214 221
11. BraunBRJohnsonAD 1997 Control of filament formation in candida albicans by the transcriptional repressor TUP1. Science 277 105 109
12. ToddRBGreenhalghJRHynesMJAndrianopoulosA 2003 TupA, the penicillium marneffei Tup1p homologue, represses both yeast and spore development. Mol Microbiol 48 85 94
13. PflugradAMeirJYBarnesTMMillerDM3rd 1997 The groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development 124 1699 1709
14. FisherALCaudyM 1998 Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev 12 1931 1940
15. LevanonDGoldsteinREBernsteinYTangHGoldenbergD 1998 Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci U S A 95 11590 11595
16. GrbavecDLoRLiuYGreenfieldAStifaniS 1999 Groucho/transducin-like enhancer of split (TLE) family members interact with the yeast transcriptional co-repressor SSN6 and mammalian SSN6-related proteins: Implications for evolutionary conservation of transcription repression mechanisms. Biochem J 337 13 17
17. WilliamsFEVaranasiUTrumblyRJ 1991 The CYC8 and TUP1 proteins involved in glucose repression in saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol 11 3307 3316
18. KeleherCAReddMJSchultzJCarlsonMJohnsonAD 1992 Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68 709 719
19. TzamariasDStruhlK 1995 Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev 9 821 831
20. VaranasiUSKlisMMikesellPBTrumblyRJ 1996 The Cyc8 (Ssn6)-Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol Cell Biol 16 6707 6714
21. KomachiKReddMJJohnsonAD 1994 The WD repeats of Tup1 interact with the homeo domain protein alpha 2. Genes Dev 8 2857 2867
22. KuchinSCarlsonM 1998 Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol Cell Biol 18 1163 1171
23. CooperJPRothSYSimpsonRT 1994 The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev 8 1400 1410
24. EdmondsonDGSmithMMRothSY 1996 Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev 10 1247 1259
25. WatsonADEdmondsonDGBoneJRMukaiYYuY 2000 Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev 14 2737 2744
26. DavieJKEdmondsonDGCocoCBDentSY 2003 Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J Biol Chem 278 50158 50162
27. LeeHChangYCKwon-ChungKJ 2005 TUP1 disruption reveals biological differences between MATa and MATalpha strains of cryptococcus neoformans. Mol Microbiol 55 1222 1232
28. LeeHChangYCVarmaAKwon-ChungKJ 2009 Regulatory diversity of TUP1 in cryptococcus neoformans. Eukaryot Cell 8 1901 1908
29. BraunBRKadoshDJohnsonAD 2001 NRG1, a repressor of filamentous growth in C.albicans, is down-regulated during filament induction. EMBO J 20 4753 4761
30. MuradAMLengPStraffonMWishartJMacaskillS 2001 NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in candida albicans. EMBO J 20 4742 4752
31. KadoshDJohnsonAD 2005 Induction of the candida albicans filamentous growth program by relief of transcriptional repression: A genome-wide analysis. Mol Biol Cell 16 2903 2912
32. KhalafRAZitomerRS 2001 The DNA binding protein Rfg1 is a repressor of filamentation in candida albicans. Genetics 157 1503 1512
33. KadoshDJohnsonAD 2001 Rfg1, a protein related to the saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in candida albicans. Mol Cell Biol 21 2496 2505
34. LoubradouGBrachmannAFeldbruggeMKahmannR 2001 A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in ustilago maydis. Mol Microbiol 40 719 730
35. BrachmannAKonigJJuliusCFeldbruggeM 2004 A reverse genetic approach for generating gene replacement mutants in ustilago maydis. Mol Genet Genomics 272 216 226
36. KamperJ 2004 A PCR-based system for highly efficient generation of gene replacement mutants in ustilago maydis. Mol Genet Genomics 271 103 110
37. BolkerM 2001 Ustilago maydis--a valuable model system for the study of fungal dimorphism and virulence. Microbiology 147 1395 1401
38. BrefortTDoehlemannGMendoza-MendozaAReissmannSDjameiA 2009 Ustilago maydis as a pathogen. Annu Rev Phytopathol 47 423 445
39. BanuettFHerskowitzI 1989 Different a alleles of ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci U S A 86 5878 5882
40. SpelligTBolkerMLottspeichFFrankRWKahmannR 1994 Pheromones trigger filamentous growth in ustilago maydis. EMBO J 13 1620 1627
41. BolkerMUrbanMKahmannR 1992 The a mating type locus of U. maydis specifies cell signaling components. Cell 68 441 450
42. GillissenBBergemannJSandmannCSchroeerBBolkerM 1992 A two-component regulatory system for self/non-self recognition in ustilago maydis. Cell 68 647 657
43. KamperJReichmannMRomeisTBolkerMKahmannR 1995 Multiallelic recognition: Nonself-dependent dimerization of the bE and bW homeodomain proteins in ustilago maydis. Cell 81 73 83
44. SnetselaarKMMimsCH 1992 Sporidial fusion and infection of maize seedlings by the smut fungus ustilago maydis.. Mycologia 84 193 203
45. SnetselaarKMMimsCH 1994 Light and electron microscopy of ustilago maydis hyphae in maize. Mycol Res 98 347 355
46. BanuettFHerskowitzI 1996 Discrete developmental stages during teliospore formation in the corn smut fungus, ustilago maydis. Development 122 2965 2976
47. KaffarnikFMullerPLeibundgutMKahmannRFeldbruggeM 2003 PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in ustilago maydis. EMBO J 22 5817 5826
48. GoldSDuncanGBarrettKKronstadJ 1994 cAMP regulates morphogenesis in the fungal pathogen ustilago maydis. Genes Dev 8 2805 2816
49. GoldSEBrogdonSMMayorgaMEKronstadJW 1997 The ustilago maydis regulatory subunit of a cAMP-dependent protein kinase is required for gall formation in maize. Plant Cell 9 1585 1594
50. DurrenbergerFWongKKronstadJW 1998 Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in ustilago maydis. Proc Natl Acad Sci U S A 95 5684 5689
51. MullerPWeinzierlGBrachmannAFeldbruggeMKahmannR 2003 Mating and pathogenic development of the smut fungus ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot Cell 2 1187 1199
52. HartmannHAKahmannRBolkerM 1996 The pheromone response factor coordinates filamentous growth and pathogenicity in ustilago maydis. EMBO J 15 1632 1641
53. HartmannHAKrugerJLottspeichFKahmannR 1999 Environmental signals controlling sexual development of the corn smut fungus ustilago maydis through the transcriptional regulator Prf1. Plant Cell 11 1293 1306
54. StoldtVRSonnebornALeukerCEErnstJF 1997 Efg1p, an essential regulator of morphogenesis of the human pathogen candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J 16 1982 1991
55. LiuHKohlerJFinkGR 1994 Suppression of hyphal formation in candida albicans by mutation of a STE12 homolog. Science 266 1723 1726
56. CsankCSchroppelKLebererEHarcusDMohamedO 1998 Roles of the candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun 66 2713 2721
57. TzamariasDStruhlK 1994 Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature 369 758 761
58. ZhangZVaranasiUTrumblyRJ 2002 Functional dissection of the global repressor Tup1 in yeast: Dominant role of the C-terminal repression domain. Genetics 161 957 969
59. BolkerMGeninSLehmlerCKahmannR 1995 Genetic regulation of mating, and dimorphism in ustilago maydis. Can J Bot 73 320 325
60. Flor-ParraIVranesMKamperJPerez-MartinJ 2006 Biz1, a zinc finger protein required for plant invasion by ustilago maydis, regulates the levels of a mitotic cyclin. Plant Cell 18 2369 2387
61. BrefortTMullerPKahmannR 2005 The high-mobility-group domain transcription factor Rop1 is a direct regulator of prf1 in ustilago maydis. Eukaryot Cell 4 379 391
62. BrachmannAWeinzierlGKamperJKahmannR 2001 Identification of genes in the bW/bE regulatory cascade in ustilago maydis. Mol Microbiol 42 1047 1063
63. HeimelKSchererMVranesMWahlRPothiratanaC 2010 The transcription factor Rbf1 is the master regulator for b-mating type controlled pathogenic development in ustilago maydis. PLoS Pathog 6 e1001035
64. ZarnackKEichhornHKahmannRFeldbruggeM 2008 Pheromone-regulated target genes respond differentially to MAPK phosphorylation of transcription factor Prf1. Mol Microbiol 69 1041 1053
65. ConesaAGotzSGarcia-GomezJMTerolJTalonM 2005 Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 3674 3676
66. ApostolIHeinsteinPFLowPS 1989 Rapid stimulation of an oxidative burst during elicitation of cultured plant cells : Role in defense and signal transduction. Plant Physiol 90 109 116
67. WuGShorttBJLawrenceEBLevineEBFitzsimmonsKC 1995 Disease resistance conferred by expression of a gene encoding H2O2-generating glucose oxidase in transgenic potato plants. Plant Cell 7 1357 1368
68. MolinaLKahmannR 2007 An ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19 2293 2309
69. KunitomoHSugimotoAWilkinsonCRYamamotoM 1995 Schizosaccharomyces pombe pac2+ controls the onset of sexual development via a pathway independent of the cAMP cascade. Curr Genet 28 32 38
70. Mendoza-MendozaAEskovaAWeiseCCzajkowskiRKahmannR 2009 Hap2 regulates the pheromone response transcription factor prf1 in ustilago maydis. Mol Microbiol 72 683 698
71. GarridoEVossUMullerPCastillo-LluvaSKahmannR 2004 The induction of sexual development and virulence in the smut fungus ustilago maydis depends on Crk1, a novel MAPK protein. Genes Dev 18 3117 3130
72. WilliamsFETrumblyRJ 1990 Characterization of TUP1, a mediator of glucose repression in saccharomyces cerevisiae. Mol Cell Biol 10 6500 6511
73. HwangCSOhJHHuhWKYimHSKangSO 2003 Ssn6, an important factor of morphological conversion and virulence in candida albicans. Mol Microbiol 47 1029 1043
74. Garcia-SanchezSMavorALRussellCLArgimonSDennisonP 2005 Global roles of Ssn6 in Tup1 - and Nrg1-dependent gene regulation in the fungal pathogen, candida albicans. Mol Biol Cell 16 2913 2925
75. Fagerstrom-BillaiFDurand-DubiefMEkwallKWrightAP 2007 Individual subunits of the Ssn6-Tup11/12 corepressor are selectively required for repression of different target genes. Mol Cell Biol 27 1069 1082
76. GarciaIMathieuMNikolaevIFelenbokBScazzocchioC 2008 Roles of the aspergillus nidulans homologues of Tup1 and Ssn6 in chromatin structure and cell viability. FEMS Microbiol Lett 289 146 154
77. KanekoAUmeyamaTUtena-AbeYYamagoeSNiimiM 2006 Tcc1p, a novel protein containing the tetratricopeptide repeat motif, interacts with Tup1p to regulate morphological transition and virulence in candida albicans. Eukaryot Cell 5 1894 1905
78. SchererMHeimelKStarkeVKamperJ 2006 The Clp1 protein is required for clamp formation and pathogenic development of ustilago maydis. Plant Cell 18 2388 2401
79. BalasubramanianBLowryCVZitomerRS 1993 The Rox1 repressor of the saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol Cell Biol 13 6071 6078
80. MennellaTAKlinkenbergLGZitomerRS 2003 Recruitment of Tup1-Ssn6 by yeast hypoxic genes and chromatin-independent exclusion of TATA binding protein. Eukaryot Cell 2 1288 1303
81. KlinkenbergLGMennellaTALuetkenhausKZitomerRS 2005 Combinatorial repression of the hypoxic genes of saccharomyces cerevisiae by DNA binding proteins Rox1 and Mot3. Eukaryot Cell 4 649 660
82. ZhangMRosenblum-VosLSLowryCVBoakyeKAZitomerRS 1991 A yeast protein with homology to the beta-subunit of G proteins is involved in control of heme-regulated and catabolite-repressed genes. Gene 97 153 161
83. DeckertJPeriniRBalasubramanianBZitomerRS 1995 Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in saccharomyces cerevisiae. Genetics 139 1149 1158
84. ConlanRSGounalakiNHatzisPTzamariasD 1999 The Tup1-Cyc8 protein complex can shift from a transcriptional co-repressor to a transcriptional co-activator. J Biol Chem 274 205 210
85. BarralesRRJimenezJIbeasJI 2008 Identification of novel activation mechanisms for FLO11 regulation in saccharomyces cerevisiae. Genetics 178 145 156
86. FriesenHHepworthSRSegallJ 1997 An Ssn6-Tup1-dependent negative regulatory element controls sporulation-specific expression of DIT1 and DIT2 in saccharomyces cerevisiae. Mol Cell Biol 17 123 134
87. YamashiroCTEbboleDJLeeBUBrownREBourlandC 1996 Characterization of rco-1 of neurospora crassa, a pleiotropic gene affecting growth and development that encodes a homolog of Tup1 of saccharomyces cerevisiae. Mol Cell Biol 16 6218 6228
88. SambrookJFrischEFManiatisT 1989 Molecular cloning: A laboratory manual. Cold Spring Harbour, New York Cold Spring Harbour Laboratory Press
89. HollidayR 1974 Ustilago maydis. KingRC Handbook of Genetics New York, USA Plenum Press 575
90. GillissenBBergemannJSandmannCSchroeerBBolkerM 1992 A two-component regulatory system for self/non-self recognition in ustilago maydis. Cell 68 647 657
91. SchulzBBanuettFDahlMSchlesingerRSchaferW 1990 The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60 295 306
92. Wedlich-SoldnerRBolkerMKahmannRSteinbergG 2000 A putative endosomal t-SNARE links exo - and endocytosis in the phytopathogenic fungus ustilago maydis. EMBO J 19 1974 1986
93. DoehlemannGvan der LindeKAssmannDSchwammbachDHofA 2009 Pep1, a secreted effector protein of ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog 5 e1000290
94. EichhornHLessingFWinterbergBSchirawskiJKamperJ 2006 A ferroxidation/permeation iron uptake system is required for virulence in ustilago maydis. Plant Cell 18 3332 3345
95. BenjaminiYHochbergY 1995 Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B 57 289 300
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage EgressČlánek A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene ExpressionČlánek An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes byČlánek Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine ScrapieČlánek Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 9- Stillova choroba: vzácné a závažné systémové onemocnění
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Unconventional Repertoire Profile Is Imprinted during Acute Chikungunya Infection for Natural Killer Cells Polarization toward Cytotoxicity
- Envelope Deglycosylation Enhances Antigenicity of HIV-1 gp41 Epitopes for Both Broad Neutralizing Antibodies and Their Unmutated Ancestor Antibodies
- Co-opts GBF1 and CERT to Acquire Host Sphingomyelin for Distinct Roles during Intracellular Development
- Nrf2, a PPARγ Alternative Pathway to Promote CD36 Expression on Inflammatory Macrophages: Implication for Malaria
- Robust Antigen Specific Th17 T Cell Response to Group A Streptococcus Is Dependent on IL-6 and Intranasal Route of Infection
- Targeting of a Chlamydial Protease Impedes Intracellular Bacterial Growth
- The Protease Cruzain Mediates Immune Evasion
- High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism
- Plague and Climate: Scales Matter
- Exhausted CD8 T Cells Downregulate the IL-18 Receptor and Become Unresponsive to Inflammatory Cytokines and Bacterial Co-infections
- Maturation-Induced Cloaking of Neutralization Epitopes on HIV-1 Particles
- Murine Gamma-herpesvirus Immortalization of Fetal Liver-Derived B Cells Requires both the Viral Cyclin D Homolog and Latency-Associated Nuclear Antigen
- Rapid and Efficient Clearance of Blood-borne Virus by Liver Sinusoidal Endothelium
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene Expression
- Strain Specific Resistance to Murine Scrapie Associated with a Naturally Occurring Human Prion Protein Polymorphism at Residue 171
- Development of a Transformation System for : Restoration of Glycogen Biosynthesis by Acquisition of a Plasmid Shuttle Vector
- Monalysin, a Novel -Pore-Forming Toxin from the Pathogen Contributes to Host Intestinal Damage and Lethality
- Host Phylogeny Determines Viral Persistence and Replication in Novel Hosts
- BC2L-C Is a Super Lectin with Dual Specificity and Proinflammatory Activity
- Expression of the RAE-1 Family of Stimulatory NK-Cell Ligands Requires Activation of the PI3K Pathway during Viral Infection and Transformation
- Structure of the Vesicular Stomatitis Virus N-P Complex
- HSV Infection Induces Production of ROS, which Potentiate Signaling from Pattern Recognition Receptors: Role for S-glutathionylation of TRAF3 and 6
- The Human Papillomavirus E6 Oncogene Represses a Cell Adhesion Pathway and Disrupts Focal Adhesion through Degradation of TAp63β upon Transformation
- Analysis of Behavior and Trafficking of Dendritic Cells within the Brain during Toxoplasmic Encephalitis
- Exposure to the Viral By-Product dsRNA or Coxsackievirus B5 Triggers Pancreatic Beta Cell Apoptosis via a Bim / Mcl-1 Imbalance
- Multidrug Resistant 2009 A/H1N1 Influenza Clinical Isolate with a Neuraminidase I223R Mutation Retains Its Virulence and Transmissibility in Ferrets
- Structure of Herpes Simplex Virus Glycoprotein D Bound to the Human Receptor Nectin-1
- Step-Wise Loss of Bacterial Flagellar Torsion Confers Progressive Phagocytic Evasion
- Complex Recombination Patterns Arising during Geminivirus Coinfections Preserve and Demarcate Biologically Important Intra-Genome Interaction Networks
- An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes by
- Non-Lytic, Actin-Based Exit of Intracellular Parasites from Intestinal Cells
- The Fecal Viral Flora of Wild Rodents
- The General Transcriptional Repressor Tup1 Is Required for Dimorphism and Virulence in a Fungal Plant Pathogen
- Interferon Regulatory Factor-1 (IRF-1) Shapes Both Innate and CD8 T Cell Immune Responses against West Nile Virus Infection
- A Small Non-Coding RNA Facilitates Bacterial Invasion and Intracellular Replication by Modulating the Expression of Virulence Factors
- Evaluating the Sensitivity of to Biotin Deprivation Using Regulated Gene Expression
- The Motility of a Human Parasite, , Is Regulated by a Novel Lysine Methyltransferase
- Phosphodiesterase-4 Inhibition Alters Gene Expression and Improves Isoniazid – Mediated Clearance of in Rabbit Lungs
- Restoration of IFNγR Subunit Assembly, IFNγ Signaling and Parasite Clearance in Infected Macrophages: Role of Membrane Cholesterol
- Protease ROM1 Is Important for Proper Formation of the Parasitophorous Vacuole
- The Regulated Secretory Pathway in CD4 T cells Contributes to Human Immunodeficiency Virus Type-1 Cell-to-Cell Spread at the Virological Synapse
- Rerouting of Host Lipids by Bacteria: Are You CERTain You Need a Vesicle?
- Transmission Characteristics of the 2009 H1N1 Influenza Pandemic: Comparison of 8 Southern Hemisphere Countries
- Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine Scrapie
- Sequential Bottlenecks Drive Viral Evolution in Early Acute Hepatitis C Virus Infection
- Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen
- Genomic and Proteomic Analyses of the Fungus Provide Insights into Nematode-Trap Formation
- Influenza Virus Ribonucleoprotein Complexes Gain Preferential Access to Cellular Export Machinery through Chromatin Targeting
- Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
- Protease-Sensitive Conformers in Broad Spectrum of Distinct PrP Structures in Sporadic Creutzfeldt-Jakob Disease Are Indicator of Progression Rate
- Vaccinia Virus Protein C6 Is a Virulence Factor that Binds TBK-1 Adaptor Proteins and Inhibits Activation of IRF3 and IRF7
- c-di-AMP Is a New Second Messenger in with a Role in Controlling Cell Size and Envelope Stress
- Structural and Functional Studies on the Interaction of GspC and GspD in the Type II Secretion System
- APOBEC3A Is a Specific Inhibitor of the Early Phases of HIV-1 Infection in Myeloid Cells
- Impairment of Immunoproteasome Function by β5i/LMP7 Subunit Deficiency Results in Severe Enterovirus Myocarditis
- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Tri6 Is a Global Transcription Regulator in the Phytopathogen
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- The Next Opportunity in Anti-Malaria Drug Discovery: The Liver Stage
- Significant Effects of Antiretroviral Therapy on Global Gene Expression in Brain Tissues of Patients with HIV-1-Associated Neurocognitive Disorders
- Inhibition of Competence Development, Horizontal Gene Transfer and Virulence in by a Modified Competence Stimulating Peptide
- A Novel Metal Transporter Mediating Manganese Export (MntX) Regulates the Mn to Fe Intracellular Ratio and Virulence
- Rhoptry Kinase ROP16 Activates STAT3 and STAT6 Resulting in Cytokine Inhibition and Arginase-1-Dependent Growth Control
- Hsp90 Governs Dispersion and Drug Resistance of Fungal Biofilms
- Secretion of Genome-Free Hepatitis B Virus – Single Strand Blocking Model for Virion Morphogenesis of Para-retrovirus
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
- Membrane Remodeling by the Double-Barrel Scaffolding Protein of Poxvirus
- A Diverse Population of Molecular Type VGIII in Southern Californian HIV/AIDS Patients
- Disruption of TLR3 Signaling Due to Cleavage of TRIF by the Hepatitis A Virus Protease-Polymerase Processing Intermediate, 3CD
- Quantitative Analyses Reveal Calcium-dependent Phosphorylation Sites and Identifies a Novel Component of the Invasion Motor Complex
- Discovery of the First Insect Nidovirus, a Missing Evolutionary Link in the Emergence of the Largest RNA Virus Genomes
- Old World Arenaviruses Enter the Host Cell via the Multivesicular Body and Depend on the Endosomal Sorting Complex Required for Transport
- Exploits a Unique Repertoire of Type IV Secretion System Components for Pilus Assembly at the Bacteria-Host Cell Interface
- Recurrent Signature Patterns in HIV-1 B Clade Envelope Glycoproteins Associated with either Early or Chronic Infections
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



