-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
Precise temporal and cell-type specific regulation of gene expression is required for development of differentiated cells even in simple organisms. Endospore development by the bacterium Bacillus subtilis involves only two types of differentiated cells, a forespore that develops into the endospore, and a mother cell that nurtures the developing endospore. During development temporal and cell-type specific regulation of gene expression is controlled by transcription factors called sigma factors (σ). An anti-sigma factor known as CsfB binds to σG to prevent its premature activity in the forespore. We found that CsfB is also expressed in the mother cell where it blocks ectopic activity of σG, and blocks the activity σE to allow σK to take over control of gene expression during the final stages of development. Our finding that CsfB directly blocks σE activity also explains how CsfB plays a role in preventing ectopic activity of σE in the forespore. Remarkably, each of the major roles of CsfB, (i.e., control of ectopic σG and σE activities, and the temporal limitation of σE activity) is also accomplished by redundant regulatory processes. This redundancy reinforces control of key regulatory steps to insure reliability and stability of the developmental process.
Published in the journal: . PLoS Genet 11(4): e32767. doi:10.1371/journal.pgen.1005104
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005104Summary
Precise temporal and cell-type specific regulation of gene expression is required for development of differentiated cells even in simple organisms. Endospore development by the bacterium Bacillus subtilis involves only two types of differentiated cells, a forespore that develops into the endospore, and a mother cell that nurtures the developing endospore. During development temporal and cell-type specific regulation of gene expression is controlled by transcription factors called sigma factors (σ). An anti-sigma factor known as CsfB binds to σG to prevent its premature activity in the forespore. We found that CsfB is also expressed in the mother cell where it blocks ectopic activity of σG, and blocks the activity σE to allow σK to take over control of gene expression during the final stages of development. Our finding that CsfB directly blocks σE activity also explains how CsfB plays a role in preventing ectopic activity of σE in the forespore. Remarkably, each of the major roles of CsfB, (i.e., control of ectopic σG and σE activities, and the temporal limitation of σE activity) is also accomplished by redundant regulatory processes. This redundancy reinforces control of key regulatory steps to insure reliability and stability of the developmental process.
Introduction
Developmental transcription networks underlie all cellular differentiation processes. These networks usually integrate a variety of environmental and cellular inputs to activate regulators such as transcription factors that control the expression of cell type-specific genes [1,2]. Superimposed onto these transcription networks, are protein-protein interaction, signal transduction and metabolic networks [3]. Overall, the combination of these different networks ensures the timely production of proteins and other components essential for the morphogenesis of newborn cells. Ultimately, a complete understanding of cellular differentiation requires detailed knowledge of how the circuitry of transcriptional regulators influences global gene expression in space and time to drive cell morphogenesis at sequential stages of development.
Spore formation in the bacterium Bacillus subtilis is an example of a prokaryotic cell differentiation process. At the onset of sporulation, triggered by severe nutrient scarcity, the rod-shaped cell divides close to one of its poles producing a small forespore, the future spore, and a larger mother cell (Fig. 1A). The mother cell nurtures development of the forespore, but undergoes autolysis to release the mature spore at the end of the process. Soon after asymmetric division, the mother cell engulfs the forespore, which becomes isolated from the external medium and separated from the mother cell cytoplasm by a double membrane and an intermembrane space. Following engulfment completion, gene expression in the mother cell drives the last stages of spore maturation by promoting the assembly of concentric protective structures. In parallel, gene expression in the forespore prepares the future spore for dormancy.
Fig. 1. The sporulation network and the action of CsfB on σG. 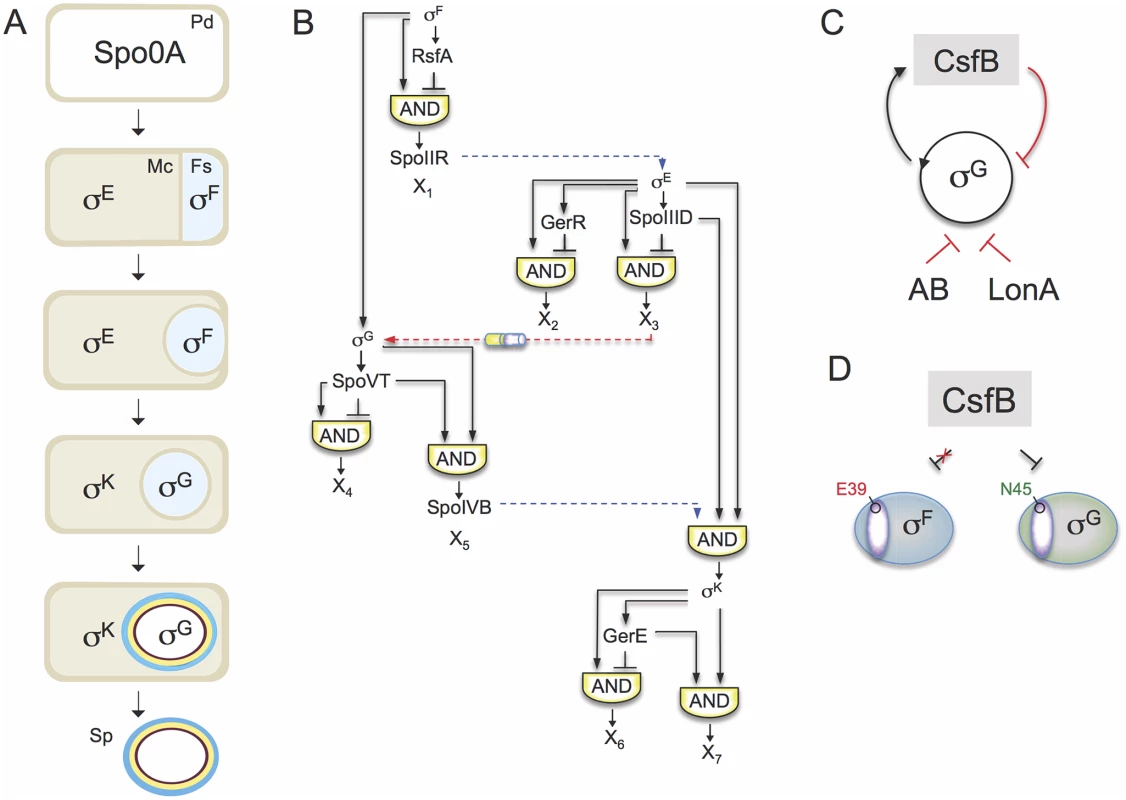
A: the main morphological stages of sporulation are represented, with the main regulatory proteins active in the indicated cells. Pd, pre-divisional cell; Mc, mother cell; Fs, forespore; Sp, mature spore. B: organization of the transcriptional network of sporulation. The broken blue lines represent cell-cell signalling pathways. First, σF drives production of a protein, SpoIIR, secreted to the intermembrane space. SpoIIR then activates a membrane-embedded protease that triggers the proteolytic activation of pro-σE [18]. σE and σF activity are required for the assembly of a cell-cell secretion system (the SpoIIQ-SpoIIIAH channel in the figure) that, following engulfment completion, allows the mother cell to nourish the isolated forespore, thus enabling continued macromoelcualr synthesis and activation of the σG auto-regulatory loop [67]. Lastly, σG controls production of a signaling protein, SpoIVB, secreted into the intermembrane space that activates the machinery responsible for the proteolytical activation of σK [18]. The negative feedback loop through which σK limits production of σE is omitted for simplicity. C: the panel represents the composite negative feedback loop that operates in pre-divisional cells, and possibly also in the forespore prior to engulfment completion, to prevent activation of the σG positive auto-regulatory loop. Transcriptional and protein-protein interactions are shown in black and red, respectively. D: a single amino acid residue in region 2.1 (purple sector) allows CsfB to discriminate between the highly similar forespore sigma factors σF and σG: N45 of B. subtilis σG allows binding by CsfB, whereas a glutamate at the same position precludes binding. Conversely, a glutamate at the homologous position of σF (E39) impedes binding by CsfB whereas an asparagine at the same position is sufficient for binding. N45 and E39 are conserved among Bacillus orthologues of σG and σF. The sporulation regulatory network includes four RNA polymerase sigma subunits that are activated in a cell type-specific manner and define a regulatory cascade that constitutes the core of the transcription network. σF and σE control early stages in development in the forespore and in the mother cell, respectively. At late stages of development, i.e., post-engulfment, σF is replaced by σG, and σE is replaced by σK (Fig. 1A). The genes for the cell type-specific sigma factors are part of a genomic signature for sporulation [4,5,6,7]. A second level of regulation results from the action of additional transcription factors, three in the mother cell (SpoIIID, GerR, and GerE), and two in the forespore (RsfA and SpoVT), which are less conserved among spore-formers. The combination of the primary regulators (i.e., the sporulation sigma factors) and the secondary regulators (ancillary transcription factors) organizes the two cell-specific lines of gene expression into a series of interlocked type-1 coherent and incoherent feed-forward loops (FFLs) [8,9](Fig. 1B). Coherent type-1 FFLs are used as persistence detectors whereas incoherent type-1 FFL generate pulses of gene expression (reviewed by [3]).
The first mother cell-specific sigma factor, σE, has a central role in controlling engulfment. Together with the ancillary factor SpoIIID, σE also turns on the genes required for the synthesis of pro-σK and the machinery that triggers proteolytic activation of σK. In parallel, SpoIIID and GerR, both acting as repressors, switch off two classes of genes initially activated by σE. Thus, the early transcription network in the mother cell includes a type-1 coherent FFL (with SpoIIID as an activator), leading to the production of σK, and two type-1 incoherent FFLs (with SpoIIID and GerR as negative regulators). Both SpoIIID and GerR are therefore critical to the early to late developmental transition characterized by the activation of σK and the inhibition of a large fraction of the σE regulon in the mother cell [9]. Once active, σK triggers a negative feedback loop that lowers production of σE and decreases the levels of SpoIIID [10,11,12].
Superimposed onto the transcriptional network are several cell-cell signaling pathways that operate at critical stages of morphogenesis, across the forespore membranes, to allow activation of the next sigma factor in the cascade, in the adjacent cell (Fig. 1B). The requirement for σG for σK activity is an example of such a pathway. σF drives the initial transcription of the gene for σG in the forespore [13], but the main period of σG activity, dependent on activation of an auto-regulatory loop, only begins after engulfment completion [14,15,16,17]. σG then controls production of a signaling protein, SpoIVB, which is secreted into the intermembrane space and activates the machinery responsible for the proteolytical activation of σK [18].
Another level of control of sigma factor activity is through the inhibitory action of anti-sigma factors. CsfB (also called Gin, for inhibitor of sigma G) is a Zn2+-containing anti-sigma factor that inhibits σG [19,20,21,22]. CsfB is part of a composite negative feedback loop that together with another anti-sigma factor, SpoIIAB, and the LonA protease, prevents activation of the auto-regulatory σG in pre-divisional cells (Fig. 1C) [20]. After the onset of sporulation CsfB is also produced in the forespore under σF control [23], and has a role in delaying the onset of σG activity until engulfment completion [14,24]. Importantly, while σF and σG are highly similar proteins, σF itself is refractory to the action of CsfB. The basis for this selectivity can be traced to a single asparagine residue in a conserved β´-interacting surface of σG [20]. Likewise, a conserved glutamate residue of σF is important for resistance (Fig. 1D). The anti-σG activity of CsfB, and the delayed transcription of the gene encoding σG relative to other σF-dependent genes are redundant mechanisms ensuring that the onset of σG activity begins at the appropriate time, possibly to coincide with engulfment completion. Accurate temporal control of σG activity is important, because it leads to the activation of σK [18] and premature activity of σK interferes with spore morphogenesis [25]. Timely activation of σK is important, as it ensures that the final stages in the assembly of the spore surface structures only initiate after the forespore is engulfed [18]. The σG-dependent production of SpoIVB, together with delayed transcription of genes required for pro-σK synthesis and processing, which require both σE and SpoIIID (Fig. 1B; above), effectively couples pro-σKactivation to engulfment completion. In total, timely activation of the cell type-specific sigma factors enforces the directionality and fidelity of the morphogenetic process [7,18].
This works focuses on an additional role of the anti-sigma factor CsfB in controlling cell type-specific gene expression during spore development. We show that in addition to its previously characterized functions in the forespore and pre-divisional cells, the anti-sigma factor CsfB also plays a role in the mother cell where its synthesis is activated under the control of σK. We show that CsfB binds to and inhibits σE in vitro and in vivo, while σK is resistant to CsfB. A single residue in conserved region 2.3 of the sigma subunit is sufficient for the discrimination by CsfB and swapping this specificity interferes with the temporal progression of the mother cell line of gene expression. Thus, CsfB is part of a negative feedback loop that acts to limit the activity of σE following engulfment completion. Furthermore, we show that CsfB is also involved in preventing ectopic activity of σG in the mother cell. Therefore, CsfB is intricately connected to both cell-specific lines of gene expression to enforce the cell-type specific action of σG and the early to late developmental transition in the two cell types undergoing sporulation.
Results
Following engulfment completion, expression of csfB is turned on in the mother cell from a σK-dependent promoter
During sporulation, expression of csfB in the forespore is controlled by σF and occurs prior to engulfment completion [20,23]. Accordingly, sequences centered at about 26 bp (GTATA) and 48 bp (GGGGAGGCTA) upstream of the csfB start codon match the consensus for σF-controlled promoters [8] (Fig. 2A). Presumably, the same σF-type promoter can also be recognized by σG in pre-divisional cells [8,20].
Fig. 2. Expression of csfB in sporulating cells. 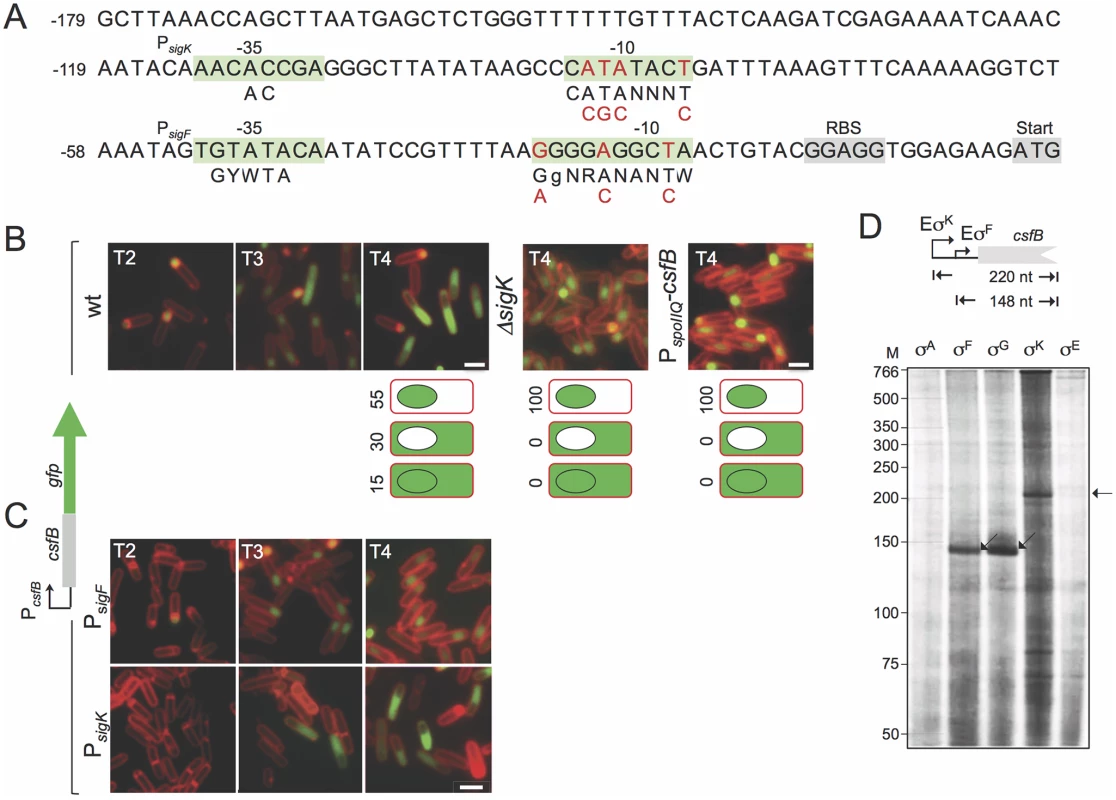
A: the panel represents the regulatory region of the csfB gene, with the -35 and -10 elements of putative σF- or σK-dependent promoters. The consensus sequences for promoter recognition by σFor σKare represented below the promoter sequence (R is A or G; W is A or T; Y is C or T; N, is any base) [8,9]. The point mutations introduced in the putative σF- or σK-dependent promoters are indicated in red. The ribosome-binding site (RBS) and start codons are also represented, for reference. B and C: the localization of a CsfB-GFP fusion protein, expressed from the amyE locus in a csfB mutant background was monitored by fluorescence microscopy at the indicated times (in hours) after the onset of sporulation in re-suspension medium, in cultures of the following strains: an otherwise wild type, a ΔsigK mutant, a strain expressing CsfB-GFP from the forespore-specific PspoIIQ promoter (in B); strains bearing the point mutations indicated in A, in the σF (PsigK) or σK promoters (PsigF) (in C). Cells were stained with the membrane dye FM4–64 prior to fluorescence microscopy. Scale bars, 1 μm. The cartoons in B represent the percentage of cells with GFP fluorescence in the coloured compartment at hour 4 of sporulation in re-suspension medium. D: in vitro transcription reactions using RNA polymerase containing the indicated B. subtilis sigma factors, and a PCR template containing the csfB promoter. Products of about 148 or 220 nucleotides were expected for transcripts initiating from the putative σF- and σK-dependent promoters, as represented. In addition, sequences matching the -10 (CATATACT) and -35 (AACACCGA) elements of the σK consensus binding sequence are present in the csfB regulatory region, upstream of the putative σF promoter [26] (Fig. 2A). This suggested to us that expression of csfB could also take place in the mother cell, at a later stage in development, after σK is activated [25]. To test this possibility, we first examined expression of a functional csfB-gfp fusion inserted at the non-essential amyE locus of a strain deleted for the native csfB gene [20]. Cells were induced to sporulate by resuspension in a nutrient poor medium and examined by phase contrast and fluorescence microscopy at different times after resuspension (which marks the beginning of sporulation under these culturing conditions). In a wild type background, expression of csfB-gfp was first detected in the forespore of sporulating cells (sporangia), two hours after the onset of sporulation, consistent with the time of activation of σF (Fig. 2B) [20]. However, by hour 3 of sporulation CsfB-GFP fluorescence was also observed in the mother cell of sporangia that had completed the process of forespore engulfment, as indicated by the loss of FM4–64 staining at the forespore membranes (Fig. 2B). At hour 4 of sporulation, 55% of sporangia exhibited a fluorescence signal restricted to the forespore, 30% fluoresced only in the mother cell, and 15% in both cell types (Fig. 2B). Moreover, the appearance of a CsfB-GFP signal in the mother cell seemed to occur concomitantly with a decrease in the forespore-specific fluorescence signal (Fig. 2B). By contrast, no CsfB-GFP fluorescence was seen in the mother cell of a sigK deletion mutant, even though these sporangia remained capable of completing engulfment (Fig. 2B). These results suggest that σK is responsible for the mother cell-specific accumulation of CsfB-GFP.
An alternative explanation, however, is that CsfB-GFP may be exported from the forespore to the mother cell upon engulfment completion, in an unindentified process that would require σK activity. To test this idea we replaced the native promoter sequences (PcsfB) driving expression of the csfB-gfp fusion by PspoIIQ, a well-characterized forespore-specific, σF-dependent promoter [22,27,28]. In this strain, accumulation of CsfB-GFP was detected in the forespore from hour 2 of sporulation onwards as expected, but was never observed in the mother cell even after σK had been activated (Fig. 2B). Thus, mother cell-specific transcription, and not transport from the forespore, is responsible for CsfB-GFP accumulation in the mother cell at late stages of development.
To more precisely delineate the respective contributions of the csfB promoters, we introduced point mutations in each of the σF - and σK-type putative -10 elements (highlighted in red in Fig. 2A) and examined production of CsfB-GFP during sporulation. In a strain bearing mutations in the putative σK-10 element, in which csfB-gfp expression is presumably only driven by the σF-dependent promoter (PsigF in Fig. 2C), CsfB-GFP is restricted to the forespore. Conversely, when point mutations were introduced in the -10 region of the putative σF-dependent promoter (PsigK in Fig. 2C), CsfB-GFP only accumulates in the mother cell.
Immunoblot analysis of whole cell extracts of sporulating cells producing CsfB-GFP under the control of the wild type promoters revealed a steady increase in the accumulation of the fusion protein, between hour 2 and 5 of sporulation (S1A Fig). By contrast, when expression of the fusion was solely dependent on PsigF, the intensity of the CsfB-GFP band strongly decreased after hour 3, whereas when expression was driven exclusively from PsigK, CsfB-GFP accumulated only from hour 3 onwards (S1A Fig). These observations are consistent with the presumed windows of activity of each of the csfB promoters (Fig. 2A), as well as the fluorescence microscopy experiments described above (Fig. 2B and C). We also carried out experiments with a lacZ transcriptional fusion and similarly detected two periods of csfB expression, the first beginning around hour 2 of sporulation and the second around hour 3–4 (S1B Fig). The first period of β-galactosidase activity was not observed when expression of the fusion relied only on PsigK, whereas the second period was absent when expression of csfB was controlled by PsigF (S1B Fig). These results are in line with the view that expression of csfB takes place at two developmental stages during sporulation, first in the forespore, under the control of σF, and later in the mother cell, under the control of σK.
Finally, we conducted in vitro transcription reactions in which core RNA polymerase, purified from B. subtilis, was reconstituted either with each of the four cell type-specific sporulation sigma factors or with the main sigma subunit, σA. All σ factors were overproduced and purified from E. coli cells (S2A Fig; see also S1 Text). While the σA or σE forms of RNA polymerase did not initiate transcription from the csfB promoter region, a run-off product of 148 nucleotides was obtained with the σF - and σG-containing holoenzymes (Fig. 2D). The size of this transcript is consistent with the location of PsigF and in line with the idea that σFand σG utilize the same csfB promoter (Fig. 2A). In addition, a product of 220 nucleotides was obtained with the σK-reconstituted holoenzyme (Fig. 2D), in agreement with the location of the predicted PsigK (Fig. 2A). In all, our findings suggest that, following engulfment completion, expression of csfB is switched off in the forespore by an unknown mechanism and activated in the mother cell, under the direct control of σK.
CsfB modulates expression of the σE and σK regulons
In pre-divisional sporangia, σG activity is inhibited by a composite negative-feedback loop involving CsfB [20]. We have argued that the σF-promoter of csfB is recognized and used by σG to achieve this mechanism of negative auto-regulation (Fig. 1C). Thus, inactivation of PsigF should mimic the previously described activity of σG in pre-divisional cells of a csfB deletion mutant [20]. In support of this hypothesis, we found that PsigK cells (carrying mutations in the σFpromoter) show activity of σG in pre-divisional cells, as measured by the expression of transcriptional fusions of lacZ and cfp to the σG-dependent sspE promoter (S3 Fig; see also S1 Text). These results confirm that the σF-type promoter of csfB is the one used by σG in pre-divisional cells.
Next, we used DNA microarrays to examine sporulation-specific gene expression in the PsigF strain, which does not express CsfB in the mother cell, in comparison to the wild type. The total RNA used for these studies was extracted and purified from cultures in resuspension medium at hour 3 of sporulation, i.e., shortly after csfB is turned on in the mother cell (above). We observed important changes in expression of several of the sporulation sigma factor regulons. Nevertheless, the expression of nearly all of the genes in the σFregulon remained unchanged in the PsigF strain versus the wild type (Fig. 3A; S3 Table); however, this does not include the group of forespore-expressed genes that are under the dual control of σFand σG (see below, σG regulon). By contrast, a large fraction of the σE-dependent genes was upregulated in the PsigF strain relative to the wild type (Fig. 3A; S3 Table). Importantly, the σE regulon can be sub-divided into 4 groups: i) genes that rely on σE alone for expression, ii) those that require SpoIIID as a positive factor, iii) those that are repressed by SpoIIID, and iv), those that are repressed by GerR [9]. A closer inspection of the transcriptional profiling data revealed that the σE-dependent genes that are upregulated in the PsigF strain are the first and second groups, i.e., those that rely exclusively on σE for expression and those that are activated by SpoIIID (Fig. 3A; S3 Table). Conversely, essentially no upregulated genes are seen among the σE-dependent genes that are repressed by GerR or SpoIIID (Fig. 3A; S3 Table). These genes correspond to pulses X2 and X3 in Fig. 1B [9]. It is likely that transcription of these genes has already been turned off by the time CsfB starts to accumulate in the mother cell, explaining the lack of effect of the mutation. In contrast, the increased expression of the σE- and σE/SpoIIID-dependent genes suggests that in the absence of CsfB, the activity of σE in the mother cell is increased or prolonged.
Fig. 3. Role of CsfB in the mother cell. 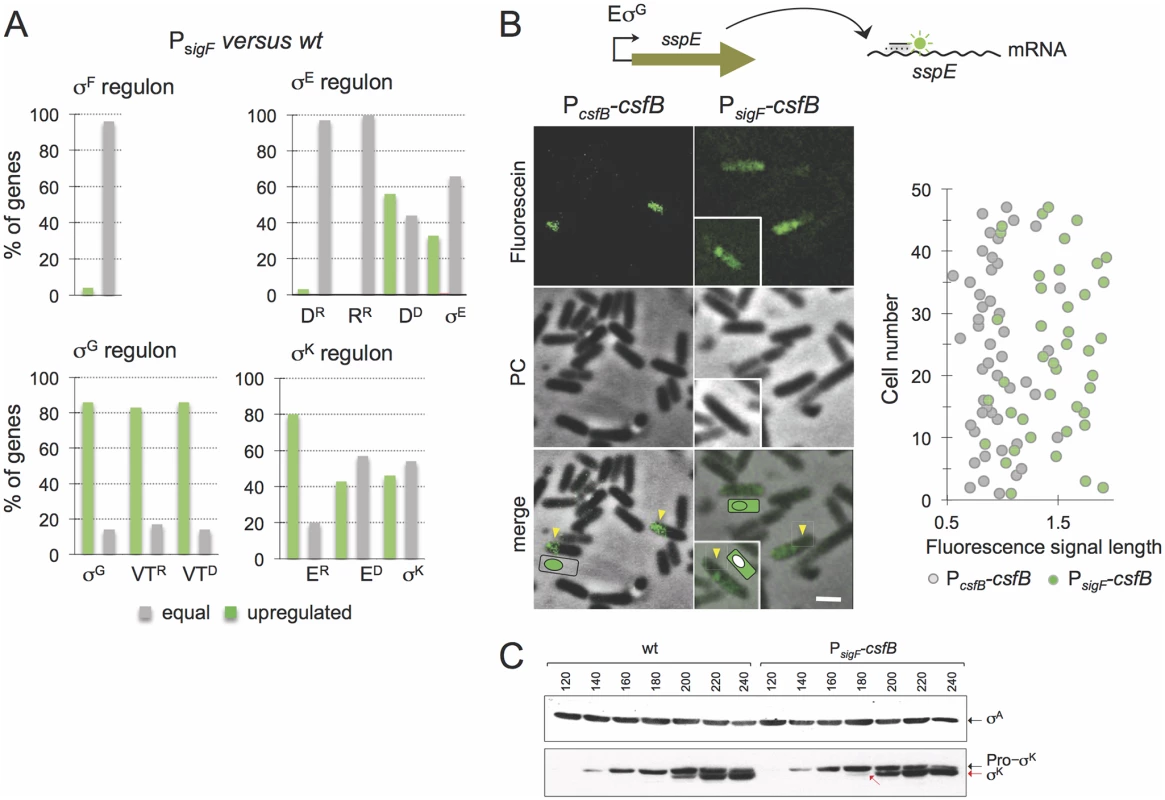
A: microarrays were used to assess the impact of mutational inactivation of the σG-dependent promoter for the csfB gene (PsigF strain) on sporulation-specific gene expression. The panel represents the percentage of genes in each of the sporulation-specific sigma regulons, whose expression was increased (green bars) or unaffected (grey bars) in the PsigF mutant relative to the wild type. Genes, repressed (“R”) by or dependent (“D”) on the following ancillary transcription factors, are shown: D, SpoIIID; R, GerR; VT, SpoVT; E, GerE. B: the panel illustrates the results of a fluorescence in situ hybridization (FISH) experiment to localize the sspE mRNA, produced under σG control, in a wild type strain and in the PsigF mutant. The mRNA was localized using a fluorescein-labelled anti-sense oligonucleotide and fluorescence microscopy. PC, phase contrast. The graph on the right shows the length (in μm) distribution of the fluorescence signal measured along the longitudinal axis of the cells. C: immunoblot analysis of pro-σK and σK. Samples from cultures of the wild type and PsigF-csfB strain were collected at the indicated times (in minutes) after the induction of sporulation by re-suspension. Whole cell extracts were prepared, proteins (15 μg) electrophoretically resolved and immunobloted with anti-σK and anti-σA (as a loading control) antibodies. Arrows indicate the position of pro-σK (in black) and σK (red). Several of the σE/SpoIIID-dependent genes are involved in production and activation of σK[9,29]. The σK regulon itself can be sub-divided into genes whose expression is activated by σK alone, those that require GerE for full expression (pulse X7 in Fig. 1B) and those repressed by GerE (pulse X6) [9]. Transcriptional profiling of the PsigF strain shows that a significant fraction of the σK-dependent genes is upregulated, regardless of GerE dependency (Fig. 3A; S3 Table). We presume that the increased expression of the σK regulon may result from augmented activity of σK (but see also below, section on the ectopic activation of the σG regulon).
In all, the genome-wide transcriptional profiling data highlight the importance of csfB for modulating the levels of σE - and σK-dependent gene expression in the mother cell, and are consistent with the interpretation that perturbations affecting the mother cell line of gene expression in a csfB mutant are mainly caused by an increased and protracted activity of σE.
CsfB contributes to the inhibition of σG and the timely activation of σK in the mother cell
A somewhat unexpected result of the global transcriptional profiling analysis was that the mutation in the σK-dependent promoter of csfB also caused a generalized increase in the expression of the σG regulon (Fig. 3A; S3 Table), including genes that are repressed (pulse X4 in Fig. 1B) or activated by SpoVT (pulse X5). We considered two possibilities. First that the increase in σE activity in the PsigF strain somehow led to an increase in the activity of σG in the forespore. Second, that eliminating expression of csfB in the mother cell alleviated some of the restrictions imposed on σG to become active in this cell type. To test these hypotheses, we first examined the localization of the fluorescence signal from a PsspE-CFP fusion in cells of the PsigF-csfB strain, in comparison with the wild type, at hour 3 of sporulation in resuspension medium. In the wild type strain, and as expected for a σG-controlled gene, the fluorescence signal was confined to engulfed forespores (S4A Fig, yellow arrows). Fluorescence from PsspE-cfp was also detected in engulfed forespores for the PsigF strain (S4A Fig, yellow arrows). However, for the PsigF mutant, a week fluorescence signal was also detected in the mother cell prior to engulfment completion (S4A Fig, white arrows). The quantitative analysis of the signal shows that the mother cell-associated fluorescence in PsigF sporangia that have not completed the engulfment sequence is consistently higher than for the wild type (S4B Fig; see also S1 Text).
In an attempt to verify this observation, we turned to fluorescence in situ hybridization (FISH) using a specific antisense DNA probe labelled with Cy3, in an attempt to localize the sspE mRNA [30]. Because at this time in sporulation the forespore is not yet recognizable by phase contrast microscopy, we relied mainly on the size of the fluorescence signal to distinguish mother cell or sporangia (larger) from forespore (smaller) localization. The FISH images suggest that the signal is confined to the forespore in the wild type strain, whereas the signal is mostly associated with whole cells (sporangia) in the PsigF strain (Fig. 3B). In cells of the mutant strain, the size of the fluorescence signal is 1.5 to 2 times longer than that observed for the wild type strain (Fig. 3B). Moreover, the size of the fluorescence signal for the PsigF strain coincides with the size of the sporangia as judged from the phase contrast images (Fig. 3B). This suggests whole cell production of the sspE mRNA. Some cells of the PsigF mutant do not show a forespore-associated signal (Fig. 3B, insert). This suggests that expression of sspE may initiate earlier in the mother cell than in the forespore, in line with the detection of PsspE-CFP in the mother cell, for the PsigF strain, prior to engulfment completion (S4 Fig; see also above). In any event, the results are consistent with the hypothesis that the sspE mRNA is also produced in the mother cell. Therefore, these data suggest that eliminating transcription of csfB from the PsigK promoter increases transcription of σG-dependent genes in the mother cell and thus that CsfB contributes to the negative regulation of σG in this cell. Presumably, CsfB is part of a composite negative feedback loop limiting σG activity in the mother cell similar to the mechanism that prevents activation of σG in pre-divisional cells. In these cells, CsfB plays a key role in inhibiting σG activity, along with the SpoIIAB anti-sigma factor and the LonA protease [20,31,32]. Previous work has shown that when the sigG gene is placed under the control of a σE-controlled promoter, SpoIIAB and LonA are also important for the inhibition of σG activity in the mother cell [15,20,32]. In an extension of this work, we now show that SpoIIAB plays the leading role in σG inhibition in the mother cell, while LonA and CsfB appear to have largely redundant supportive contributions (S5A Fig). Nevertheless, these results demonstrate the ability of CsfB to act as an inhibitor of σG activity in the mother cell, in line with the observed σG activity in the PsigF strain (Fig. 3 and S4 Fig).
Importantly, during the course of these experiments aiming at directly testing for a role of CsfB in the inhibition of σG in the mother cell (above), we noticed that the forced production of σG in the mother cell resulted in premature activity of σK (S5B Fig). Activation of σK requires pro-σK processing, which in turn depends on the activity of σG in the forespore, following engulfment completion [25,33]. Since σG shows some activity prior to engulfment completion in the PsigF strain (above), we therefore considered the possibility that the augmented transcription of the σK-controlled genes, as detected in our microarray analysis (above), could result in part from premature processing of pro-σK independently of the σG-dependent pathway that operates following engulfment completion (S5C Fig). To test this idea, we surveyed the accumulation of pro-σK and σK by immunoblotting, throughout sporulation. Pro-σK was first detected in both the wild type strain and in the PsigF strain 140 min after resuspension in sporulation medium (Fig. 3C). However, mature σK is first detected 180 min after resuspension for the PsigF strain, and only at minute 200 for the wild type (Fig. 3C). Moreover, even at minute 200, the fraction of mature σK is higher for the PsigF strain (Fig. 3C). Therefore, the lack of csfB expression in the mother cell, can lead to premature activation of pro-σK, again underscoring the need for strict inhibition of σG activity in the mother cell.
CsfB forms a complex with σE but not with σK in sporulating cells
The analysis of the global transcriptional profiling data raised the possibility that CsfB could act as an inhibitor of σEand/or σK. This prompted us to determine whether the anti-sigma factor could form complexes with either sigma factor. To isolate CsfB-GFP and interacting proteins from sporulating cultures of a strain expressing csfB-gfp, we used a GFP-binding protein (GBP) coupled to a chromatographic matrix (GFP-Trap beads). As controls, we examined a wild type strain carrying no gfp fusion and a strain producing GFP under the control of the xylose-inducible PxylA promoter. The extracts were prepared 4 hours after the onset of sporulation, when CsfB-GFP is known to accumulate in the mother cell (Fig. 2B; above). Control experiments also confirmed the accumulation of σE, σG, σK or GFP in the whole-cell extracts prepared from the various strains (Fig. 4A). The extracts from all strains were incubated with GFP-Trap beads, the bound proteins eluted and identified by immunoblot analysis with antibodies raised against σE, σG, σK or GFP. We found σE but not σK, to be pulled down efficiently from extracts of the strain producing CsfB-GFP. By contrast, σE was not recovered from extracts of the strain containing no GFP fusion or from the strain that produced unfused GFP (Fig. 4A). Thus, retention of σE by the GFP-trap beads depended on formation of a complex with CsfB-GFP. As expected, CsfB-GFP was able to pull down σG [20], a property used here as a positive control for the experiment (Fig. 4A). Interestingly, a role for CsfB in inhibiting σE activity in the forespore soon after asymmetric division was previously suggested [34]. Under the conditions of our experiments, however, σE is only expected to accumulate in the mother cell and the expression of csfB has switched to the mother cell in most sporangia in the population when the cells were harvested for the pull down assays (see above). Therefore, the result reported in Fig. 4A most likely reflects an interaction occurring between σE and CsfB in the mother cell and not in the forespore.
Fig. 4. Sigma-CsfB interactions. 
A: pull-down assays using GBP. Whole cell extracts were prepared for cultures of different strains 4 hours after the onset of sporulation in resuspension medium. The extracts were cleared and incubated with GBP bound to sepharose beads. Samples of the whole cell extracts, as well as the proteins bound to the GBP beads were visualized, following elution, by immunoblotting with anti-σG, anti-σE, anti-σK and anti-GFP antibodies. Strains in A: a wild type strain, a strain producing GFP from the xylose-inducible PxylA promoter; strains producing CsfB-GFP from its native promoter region (PcsfB), from the σF-type promoter (PsigF) or the σK-type promoter (PsigK), as indicated. The asterisk indicates a likely degradation product of σG. B: Ni2+-NTA affinity chromatography assay for CsfB-σ interactions. His6-CsfB, and untagged σE and σK, were purified from E. coli cells and analysed by SDS-PAGE (first three lanes). The three proteins were then individually applied to a column and eluted with an imidazole buffer. The eluted proteins were detected following SDS-PAG by Coomassie-staining. His6-CsfB bound to the Ni2+-NTA column whereas σE or σK did not. σE but not σK, bound to the column in the presence of His6-CsfB. The asterisk indicates a likely degradation product of σE. Molecular weight markers (M, in kDa) are shown on the left side of panels B. The asterisks in A and B indicate likely degradation products of σG or σE. C: colony lift assay (left) and assays in liquid medium (right) for the detection of β-galactosidase activity in yeast strains expressing fusions of CsfB to the GAL4 activation domain (AD) and fusions of σF, σE, σG, σK, and σA to the GAL4 binding domain (BD), as indicated. Assays in which the BD and AD were expressed from empty vectors were used as negative controls (“-“). In agreement with this interpretation, CsfB-GFP produced solely from the PsigK promoter, was still able to pull down σE (Fig. 4A). In addition, this construct could also pull down σG and conversely CsfB-GFP produced from the PsigF was able to pull down σE (Fig. 4A). Even though these observations could be explained by complex formation after the cells are disrupted in particular for the PsigF strain (provided that there was enough free CsfB-GFP), we note that more σE and less σG seemed to be pulled-down in the strain where csfB-gfp expression is restricted to the mother cell than in the strain producing CsfB-GFP in both compartments (Fig. 4A). Thus, it is possible that the amounts of σE and σG pulled-down from the PsigK-csfB-gfp carrying strains reflect the levels of both of these proteins in the mother cell. Considering that σG can accumulate and become active in the mother cell under certain conditions [15,20]; S4 Fig and S5 Fig; see also above), some σG might be present in the mother cells at hour 4 of sporulation.
CsfB binds to σE but not to σK
One of the consequences of inactivating the PsigK promoter of csfB was the protracted expression of a specific class of σE-dependent genes. A prediction that stems from these results and from the pull-down experiments described above is that CsfB binds directly to σE. To test this hypothesis, we purified His6-CsfB, σE and σK (both lacking their pro-sequences) overproduced in E. coli (Fig. 4B, first three lanes and S2A Fig) (Note that in addition to σE, which has a molecular weigth of 29 kDa, a stable proteolytic fragment of about 20 kDa accumulated in the preparations; Fig. 4B). We then asked whether His6-CsfB immobilized onto a Ni2+-NTA column could capture untagged, purified σE or σK. While His6-CsfB bound efficiently to the Ni2+-NTA column, neither σE nor σK did (Fig. 4B). However, in the presence of His6-CsfB, σE (but not σK), was efficiently retained by the column (Fig. 4B), a result fully consistent with the pull-down experiments described above (note that both σE and the 20 kDa fragment were retained; Fig. 4B, left lanes).
To confirm these results with a different technique, we also carried out a GAL4-based yeast two-hybrid assay, an approach that had been used before to dissect the interaction between σG and CsfB [20]. We constructed translational fusions of σF (as a negative control), σG (as a positive control), σE, σK, σA or CsfB to the C-terminus of the GAL4 DNA binding (BD) or activation domains (AD). The various fusion proteins were expressed in different combinations in yeast cells and checked for their ability to interact in vivo, as assessed by the expression of a lacZ gene preceded by a GAL4-responsive element. As expected, we detected an interaction of CsfB with σG, but not with σF([20]; Fig. 4C). In addition, CsfB interacted with σE (Fig. 4C), suggesting that the two proteins could indeed establish specific contacts. No interaction was detected between CsfB and σK or σA (Fig. 4C).
Thus, both the yeast two-hybrid and the affinity chromatography assays indicate that CsfB and σE directly interact.
CsfB inhibits transcription in vitro from σG - and σE-dependent promoters
While CsfB binds to both σG [20] and σE ([34]; see above), no study has shown direct inhibition of transcriptional activity by the anti-sigma factor. To test the ability of purified CsfB to inhibit σG - or σE-directed transcription, core RNA polymerase (E) was purified from B. subtilis, and reconstituted with σF, σE, σG, σK, or σA overproduced and purified from E. coli cells (S2A Fig). As templates for in vitro transcription reactions, we used PCR-generated DNA fragments corresponding to promoters of genes known to be under the control of the sigma factors tested, and whose transcriptional start site has been mapped. As such, the gcaD gene was used as the template for σA-directed transcription [35,36], spoIIQ as a template for EσF [27,37], spoIID, spoIIM, spoIIIA p1 and spoIIIA p2 as templates for EσE [38,39,40], sspB for EσG [30], and gerE for EσK [33] (Fig. 5A and S2B Fig). All forms of RNA polymerase tested directed the production of run-off transcripts of expected sizes (Fig. 5B-D and S2C Fig). No specific transcription products were seen when templates were mixed with core RNA polymerase in the absence of a σ subunit (S2C Fig). CsfB did not inhibit transcription by EσA, EσF (Fig. 5B) or EσK (Fig. 5C), but inhibited the σE-directed utilization of the spoIIM (Fig. 5B), spoIID (Fig. 5C and D), spoIIIA p1 and spoIIIA p2 promoters (S2C Fig). CsfB also inhibited the σG-directed transcription of sspB (Fig. 5B and D). Inhibition of EσA, EσFor EσK by CsfB was not observed even at molar ratios higher than those that inhibited EσE (Fig. 5C and D). Interestingly, inhibition of EσE, and to some extent of EσG, required molar ratios higher than 1 (Fig. 5B and D; S2C Fig). One possibility is that active CsfB is a dimer (or a higher order multimeric form), in agreement with the results of a previous study [21].
Fig. 5. CsfB inhibits in vitro transcription by RNA polymerase associated with σG or σE. 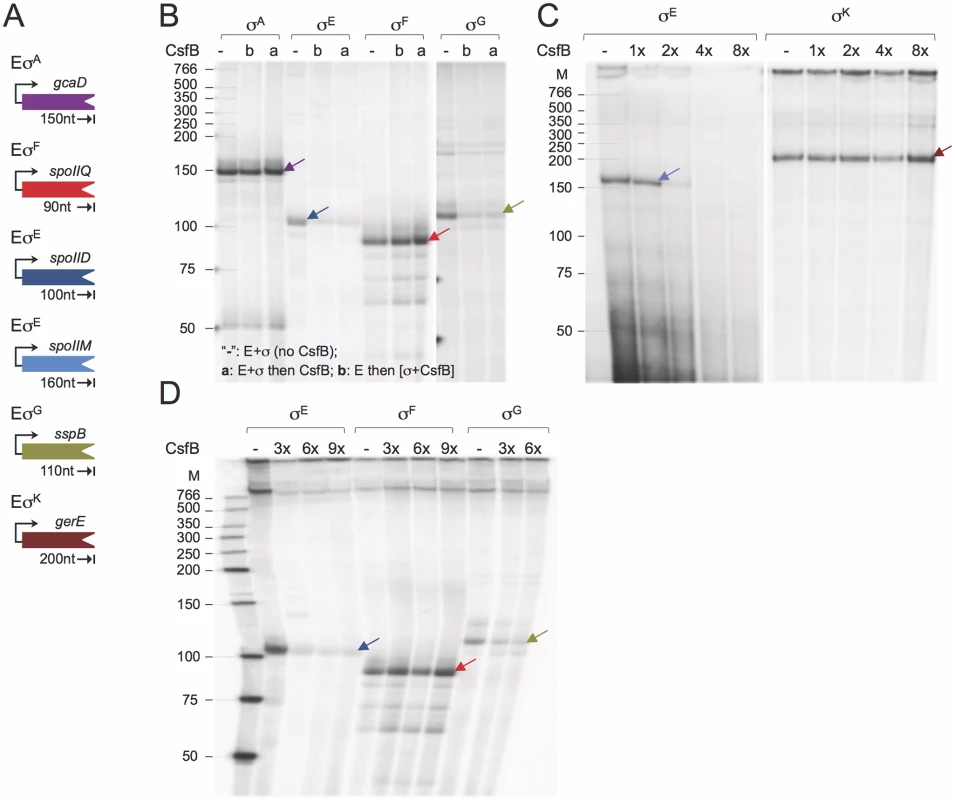
A: schematic representation of the promoter-containing PCR fragments used as templates for the in vitro transcription reactions. The expected size (in nucleotides) for each of the run-off products is indicated. B: effect of CsfB on in vitro transcription reactions with the indicated RNA polymerase holoenzymes. CsfB (130 nM) was either added to the reaction after mixing core (E; 13 nM) and the sigma subunit (“a”; 130 nM) or together with the sigma subunit to a mixture already containing core (“b”). The symbol “-”refers to a control reaction lacking CsfB. C and D: effect of the CsfB concentration, shown in molar ratio relative to RNA polymerase (13 nM), on in vitro transcription reactions with the indicated holoenzymes. CsfB was added at the same molar concentration as sigma (130 nM, 1x) or 2, 3, 4 and 8-fold excess and the mixture added to core RNA polymerase (13 nM). In B-D, arrows indicate the position of the expected run-off products, identified with arrows, which maintain the color code for the templates as represented in A. The position of molecular weight markers (in nucleotides) is shown on the left side of the panels. In total, the run-off transcription assays show that CsfB inhibits transcription by the RNA polymerases that contain the σ subunit to which it can bind in vitro, i.e., σE or σG.
Mapping the interaction between CsfB and σE
To map the region(s) involved in the CsfB-σE interaction, we performed GST-pull down and yeast two-hybrid assays, two techniques that were used before to map the interaction between CsfB and region 2.1 of σG [20]. For that purpose, full-length mature σE or different fragments of the sigma factor were fused to either GST or to GAL4 (Fig. 6A). For the GST pull-down assays, we used a CsfB-StrepII-tagged protein [20] and a GST-σE fusion protein, lacking its pro region (sigma fragment a, Fig. 6A) or GST fusions to different fragments of σE (fragments b to h; Fig. 6A), overproduced and partially purified from E. coli. We incubated CsfB-StrepII-tag with GST or the GST-σE fusion proteins immobilized on glutathione beads. After washing, the presence of CsfB in the elution samples was assessed by immunoblotting with an anti-StrepII tag antibody. The CsfB protein was retained by GST-σE (fragment a) but not by GST alone (Fig. 6B, left panel). GST fusions to σE fragments corresponding to regions 2.1 through 2.4 (fragment c, GST-σE 30–141), 2.1 through 3.1 (fragment d, GST-σE 30–172), and 2.2/2.3 (fragment e, GST-σE 79–117) pulled down CsfB, whereas fragments encompassing regions 2.1/2.2 (sigma fragment b; GST-σE 30–98), 2.3/2.4 (fragment f, GST-σE 98–141), 3.1 (fragment g, GST-σE 141–172), and 4.1/4.2 (fragment h, GST-σE 172–239), did not (Fig. 6B). With the exception of fragment e (2.2/2.3, σE 79–117), the same fragments also showed an interaction with CsfB in yeast two-hybrid assays (Fig. 6C). The lack of interaction of fragment e in the yeast two-hybrid assay may be due to the topology and/or stability of the GAL4 fusion.
Fig. 6. Mapping the region in σE involved in the interaction with CsfB. 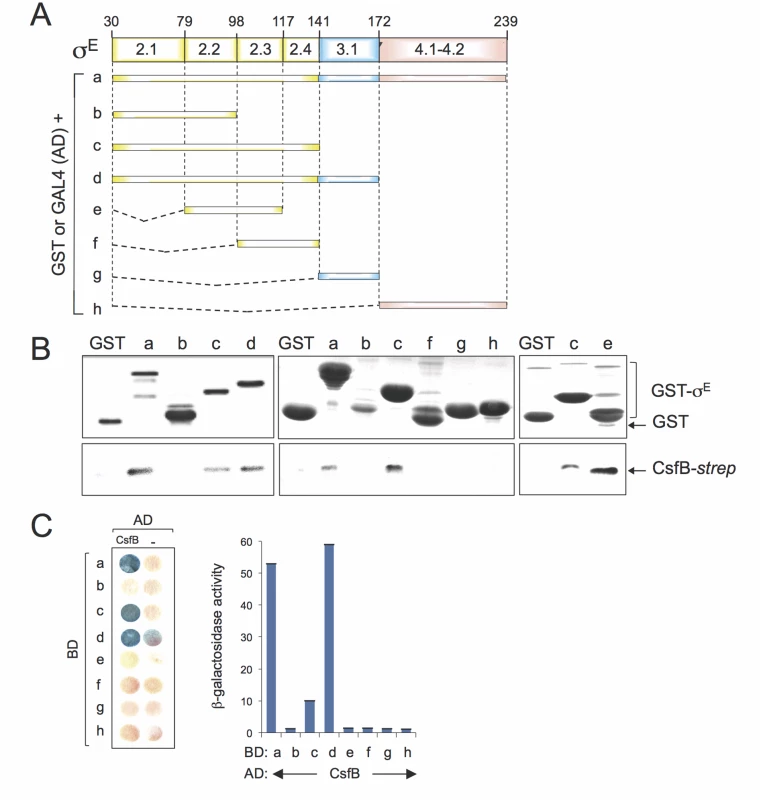
A: schematic representation of the B. subtilis σE protein, with the various regions within domains 2 and 4, represented. The full-length protein (a), or the indicated fragments (b to h) were fused to either the binding domain of GAL4 or to GST (below). The numbers refer to the residue number in the primary structure of σE. B: GST pull-down assays with purified CsfB-Strep II tag. GST and the various GST-σE fusions (as depicted in A) were bound to glutathione beads and incubated with purified CsfB-Strep II (100 nM). Bound proteins were detected, following elution, with an anti-Strep II tag antibody. The amounts of GST or the various GST fusion proteins bound to the glutathione beads were estimated by immunoblot with an anti-GST antibody. C: colony lift assay (left) and quantitative assays in liquid cultures (right) for the detection of β-galactosidase activity in yeast strains expressing fusions of CsfB to the GAL4 activation domain (AD) and fusions of different σE segments (as depicted in C) to the GAL4 binding domain (BD), as indicated. Assays in which the BD and AD were expressed from empty vectors were used as negative controls (“-“). Region 2.1 was previously implicated in the interaction of CsfB with σG [20]. However, this region does not appear to be sufficient to mediate the interaction of CsfB with σE. In contrast, because σE 79–117 (regions 2.2/2.3) interacted with CsfB, but neither σE 30–98 (regions 2.1/2.2) nor σE 98–141 (regions 2.3/2.4) did, it is likely that residues at the end of region 2.2 and/or at the beginning of region 2.3 of σE are necessary for the interaction. In any event, our results show that CsfB and σE interact directly, and that the region required for the interaction differs from that found in σG [20,22].
A single residue in region 2.3 allows CsfB to discriminate between σE and σK
The N45E substitution in σG results in loss of CsfB binding, whereas the E39N substitution in σF is sufficient for conferring CsfB binding ability [20]. However, the residue homologous to N45 in σE corresponds to a glutamate, E64 (Fig. 7A). Together with the mapping experiments described in the preceding section, this observation suggests that the interaction of CsfB with σE differs from that with σG. To more precisely delineate the determinants for CsfB binding to σE, we sought to identify residues that are necessary for the interaction. An inspection of the amino acid sequence of σE in regions 2.2 and 2.3 revealed an asparagine residue located at position 100 (i.e., at the beginning of region 2.3) and conserved among all of Bacillus σE orthologues. Strikingly, the position homologous to N100 in σK is occupied by an aspartate (E93) in B. subtilis and invariably (with the exception of the σE proteins) by an acidic residue in all other Bacillus sigma factors (Fig. 7A). N100 in σE is located in the vicinity of several residues involved in promoter melting (Fig. 7A). By contrast, E64 in σE and N45 in σG are located at the beginning of a helix within which several contacts are established with the β´ subunit of RNA polymerase (S6 Fig). Thus, depending on which sigma factor is present in the complex, CsfB appears to bind to two distinct functional surfaces of the RNA polymerase holoenzyme.
Fig. 7. A residue in σE involved in the interaction with CsfB. 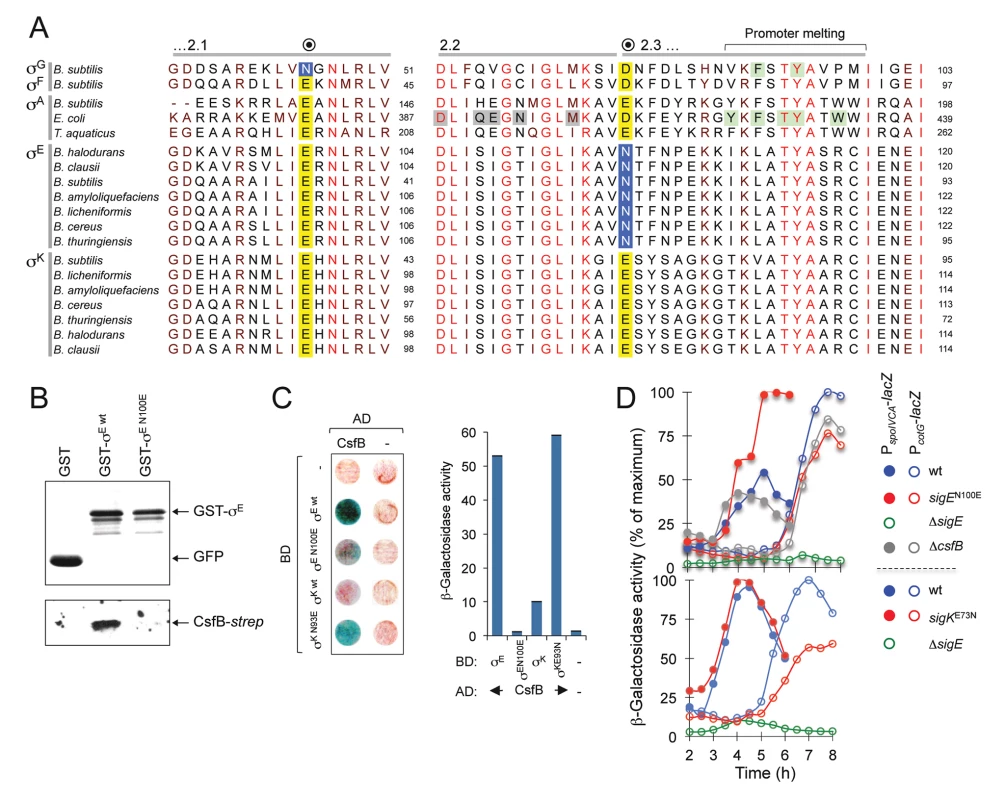
A: alignment of regions 2.1, 2.2 and the beginning of region 2.3 from the σA, σF, σG, σE, and σK proteins of the indicated species (the sequence of σA from T. thermophilus is identical to that of T. aquaticus for the segments represented). The residue in B. subtilis σG important for the interaction with CsfB is shown in a blue box. Note that the homologous position in σF and in the other sigma proteins is invariantly occupied by a glutamic acid (yellow box). The residue herein identified in region 2.3 of B. subtilis σE (N100) that is important for binding by CsfB is shown in a blue box. This residue is conserved among Bacillus orthologues of σE whereas in other sigma factors the homologous position is occupied by and acidic residue (yellow box). Residues in region 2.2. of E. coli σA implicated in core binding are highlighted in grey; residues, at the end of region 2.3, involved in promoter meting are also indicated (reviewed by [55]). The residues boxed in green were shown to affect sporulation in B. subtilis [20] and promoter melting in E. coli [68,69]. The accession numbers of the aligned sequences are given in the Material and Methods section. B: GST pull-down assays to investigate the role of E100 of σE in the interaction with CsfB. GST, GST-σE and GST-σE N100E fusions were bound to glutathione beads and incubated with purified CsfB-Strep II (100 nM). Protein complexes were captured on glutathione sepharose beads, and visualized, following elution, by immunoblotting with anti-GST, or anti-Strep tag antibodies. C: colony lift assays (left) and assays in liquid medium (right) for the detection of β-galactosidase activity in yeast strains expressing fusions of CsfB to the GAL4 activation domain (AD) and fusions of σE wt, or σE N100E, σK wt, or σK E93N, to the GAL4 binding domain (BD), as indicated. Assays in which the BD and AD were produced from empty vectors were used as negative controls (“-“). D: effect of the sigEN100E and sigKE73N alleles on σE- and σK-dependent gene expression. The figure shows the expression of PspoIVCB-lacZ (σE-dependent) and PcotG-lacZ (σK-dependent) transcriptional fusions during sporulation. Samples were withdrawn from cultures of the represented strains, at the indicated times, in hours after the onset of sporulation in re-suspension (denoted as T0), and assayed for β-galactosidase activity (shown in Miller units). We wanted to test whether the CsfB anti-sigma factor discriminated between σE and σK by differentiating an asparagine from an acidic residue at the beginning of region 2.3. We constructed GST and GAL4 fusions to forms of σE bearing the single amino acid substitution N100E and used them in pull-down and yeast two-hybrid experiments. We found that GST-σE N100E did not pull-down CsfB-strepII partially purified from E. coli cells (Fig. 7B) and showed a much-reduced interaction (when compared to the wild type sigma factor) with the anti-sigma factor in a yeast two-hybrid assay (Fig. 7C). By contrast, a GAL4 fusion to σK bearing an E93N substitution interacted strongly with the anti-sigma factor in the yeast two-hybrid assay (Fig. 7C). To determine whether the N100E and E93N substitutions in σE or σK affected their activity in vivo, the corresponding mutations were first transferred to the sigE and spoIVCB genes (spoIVCB codes for the N-terminal half of σK; see the Material and Methods section for details). We then used PspoIVCA - (σE-dependent promoter) and PcotG-lacZ (σK-dependent promoter) transcriptional fusions to monitor the activity of σE and σK respectively, during sporulation. Consistent with loss of CsfB regulation, the N100E substitution increased expression of spoIVCA-lacZ, whereas the E93N replacement reduced expression of cotG-lacZ (Fig. 7D). Expression of PspoIVCA-lacZ or PcotG-lacZ in a ΔcsfB mutant did not differ much from the wild type (Fig. 7D), presumably because premature activation of pro-σK somehow compensates for increased activity of σE (see also above). In any event, the N100E substitution is sufficient to render σE refractory to CsfB, whereas the E93N substitution suffices to make σK susceptible to CsfB.
The N100E substitution in σE affects the assembly of the spore surface layers
Assembly of a protective protein shell called the coat that forms the spore surface, involves the timely production of over 80 components under the control of σE or σK [41,42]. Since the N100E substitution increases the activity of σE and reduces that of σK we wanted to examine the coat protein composition in spores of the strain producing σE N100E. Proteins were extracted from purified spores by treatment with a buffer containing SDS and the reducing agent DTT, or by NaOH [43]. The first treatment releases about 80 proteins from wild type spores (S7A Fig, top) whereas NaoH extraction produces a much smaller collection of extractable proteins (S7A Fig, bottom). We found that several proteins were more extractbale from N100E spores than from wild type spores, among which the species labelled a-e (SDS/DTT extraction) and f-k (NaOH extraction) in S7A Fig (bottom panel). For example, the outer coat proteins CotA (in band a), CotB (in b), and Tgl (in d), as well as the inner coat proteins CotN (in bands h and i), YaaH (in c) and YybI (in band d) were more extractable from σE N100E spores (S7A Fig, see also S1 Text). Synthesis of YaaH, CotN and YybI is under σE control, whereas production of CotA, CotB, and Tgl is mainly controlled by σK [9,29,41,44,45,46]. Thus, the N100E substitution has a strong and global impact on the assembly of the spore surface layers, affecting the assembly of proteins from both the inner and outer coat layers, which are produced at different periods (controlled by σE or by σK) during spore coat assembly.
Discussion
A key finding of our study is that expression of csfB switches from the forespore to the mother cell midway into the process of sporulation. The regulatory region of csfB includes two promoters: a σF-type promoter (PsigF) utilized by σG in pre-divisonal cells and by σF in the forespore [14,17,20], and a σK-type promoter (PsigK), located further upstream, active in the mother cell following engulfment completion. Activation of csfB transcription in the mother cell coincides with a decline in forespore-specific expression (Fig. 2). It has been hypothesized that a reduction in the levels or activity of CsfB in the forespore could be a factor promoting σG activity following engulfment completion [14,17,20]. SpoIIAB also disappears from the forespore following engulfment completion [47]. It remains unclear how CsfB activity is turned off in the forespore during the late stages of sporulation, since σG can utilize PsigF in vitro and in vivo ([20]; this work). However, it has been suggested that Zn2+-depletion in the forespore, following engulfment completion, could render CsfB unstable [21]. If so, then the decrease in the CsfB-GFP signal in the engulfed forespore (Fig. 1B) could represent protein degradation.
Enforcing the transition from early to late stages of development
Genome-wide transcriptional profiling analysis showed that inactivation of PsigK, which abolishes CsfB accumulation in the mother cell, caused increased transcription of σE-controlled genes that either rely solely on σE for expression or are dependent on the type I coherent FFL formed by σE and the ancillary transcriptional activator SpoIIID [9]. Increased σE activity was also observed using a σE-responsive lacZ reporter in a strain expressing the CsfB-resistant form of σE (σE N100E). By contrast, σE-dependent genes repressed by GerR or SpoIIID (i.e., the output of the type I incoherent FFLs generating pulses X2 and X3 in Fig. 1B) did not show increased expression in the PsigF-csfB strain, most likely because their transcription has already been switched off at the time of analysis due to the action of the two repressors (Fig. 3A). In general, these observations are consistent with an increase in the activity of σE in the absence of CsfB and with the properties of the FFLs formed by σE [3]. They also support a model in which the appearance of CsfB in the mother cell promotes the transition from early to late cell-type specific gene expression (i.e., the σE to σK switch) (Fig. 8A).
Fig. 8. Model for the functions of CsfB. 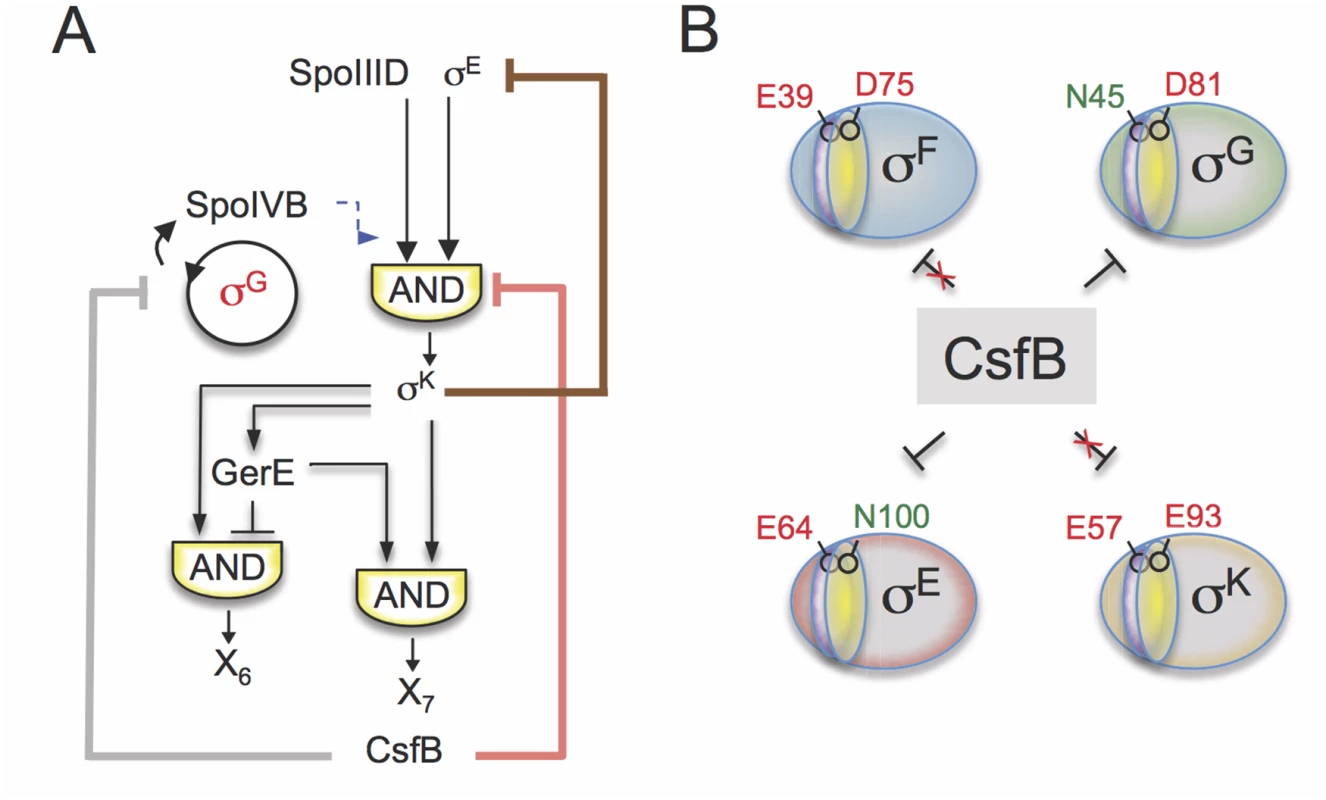
A: following engulfment completion, two negative feedback loops are established that act to limit the level (brown line) and the activity of σE (red line). The loop that acts at the level of σE activity involves the σK-dependent production of CsfB (this work), and restricts mainly the expression of the σE-and SpoIIID-dependent gene class. Both feedback loops promote proper switching from the early (pre-engulfment, σE-dependent) to late (post-engulfment completion, σK-dependent) stages in development. Together with LonA and SpoIIAB, CsfB also antagonizes σG in the mother cell (grey line). Minimizing the chances of σG becoming active in the mother cell prevents the premature, forespore-independent activation of σK. Thus, the action of CsfB superimposes both onto the transcriptional and cell-cell signaling networks. B: the panel represents the residues that allow binding of CsfB to σG (N45) and σE (N100), and that are sufficient to render σF (E39) and σK (E93) resistant to the anti-sigma factor. N45 in σG and E39 in σF are located in conserved region 2.1 (purple sector) whereas N100 in σE and E93 in σE are located in region 2.3 (yellow). While a previously described negative feedback loop driven by σK reduces the levels of σE [11,12,48], the feedback loop revealed in this work, and similarly initiated by σK, uses CsfB to limit the activity of σE in the mother cell. Thus two partially redundant feedback loops evidently function to promote proper switching from σE - to σK-dependent transcription following engulfment completion, and thus the developmental transition from early to late stages of morphogenesis (Fig. 8A).
σG and the activity of σK
Interference with csfB expression in the mother cell, as when PsigK is inactivated, also leads to increased expression of several genes under σK control (Fig. 3). Because CsfB does not bind to σK and does not inhibit σK-directed transcription in vitro, it is unlikely that the increased activity of σK is due to the loss of a direct interaction with CsfB. Instead, it is more likely to be a consequence of the ectopic activation of σG in the mother cell. Previous work has shown that two negative regulators of σG, the anti-sigma factor SpoIIAB and the LonA protease, effectively counteract σG in the mother cell, when its synthesis is artificially induced by driving expression of sigG from a σE-dependent promoter [15]. Under the same genetic conditions, we showed that CsfB contributes, along with LonA and SpoIIAB, to the inhibition of σG activity in the mother cell (S5 Fig). In the PsigF-csfB mutant, our transcriptional profiling data indicate that most of the genes in the σG regulon show increased expression. This increased expression is due in part to increased activity of σG in the mother cell of PsigF sporangia. Not only transcription from the σG-dependent PsspE promoter fused to cfp is detected in the mother cell prior to engulfment completion (S4 Fig), but FISH experiments also show accumulation of the sspE transcript in the mother cell (Fig. 3). Therefore, CsfB is one of several redundant mechanisms that act to silence σG in the mother cell.
In wild type cells, it is the activity of σG in the forespore following engulfment completion that triggers σK activation in the mother cell, via the signalling protein SpoIVB. Mutations that bypass this signalling pathway result in premature activity of σK and defects in spore morphogenesis [25]. We have shown that the production of σG in the mother cell uncouples pro-σK processing from engulfment completion and leads to premature activation of σK because SpoIVB can activate processing of pro-σK directly in the cell where it is produced (S5 Fig). Accordingly, processing of pro-σK in the PsigF mutant is detected earlier than in the wild type (Fig. 3C). By contrast, in the σE N100E strain, expression of a σK-dependent lacZ fusion is reduced and delayed, presumably because pro-σK processing remains strictly dependent on the post-engulfment, σG-dependent SpoIVB signalling from the forespore. Therefore, the silencing of σG in the mother cell by CsfB, along with the contributions of SpoIIAB and LonA, prevents premature activity of σK.
Discrimination by CsfB of the σF/ σG and σE/ σK pairs
We have previously shown that N45, an asparagine residue in region 2.2 of σG, is a critical determinant for CsfB binding [20] (Fig. 1C). Not only is this residue conserved among Bacillus orthologues of σG but a glutamic acid residue, E39, is found at the homologous position of the CsfB-resistant σF protein in B. subtilis, and an acidic residue is invariably found at the equivalent position in σF orthologues of other Bacillus species. Importantly, the N45E substitution renders σG refractory to CsfB binding and conversely, the E39N variant of σF is susceptible to the anti-sigma factor. Residue N45 of σG is most likely involved in a direct contact with the β´ subunit of RNA polymerase, suggesting that CsfB, like other anti-sigma factors interferes with the σ/β ´ interaction [20]. Our in vitro transcription assays support this model, as CsfB is sufficient to inhibit σG - and σF-directed transcription at bona fide promoters in vitro (Fig. 5).
Our results now implicate a region encompassing regions 2.2 and 2.3 of σE in CsfB binding (Fig. 6), and an asparagine residue, N100, at the beginning of region 2.3, was found to be a key determinant (Fig. 7A-C). In a striking parallel with the σF/σG pair, a homologous asparagine is found in all known orthologues of σE from related organisms (Fig. 7A). Despite the high degree of similarity between the σE and σK proteins across sporeformers [4,5,6,7], the homologous residue in the σK protein of B. subtilis is a glutamic acid, E93, and an acidic residue is invariably found among Bacillus orthologues of σK (Fig. 7A). Also reminiscent of the σF /σG pair, the N100E substitution makes σE refractory to CsfB, whereas the E73N variant of σK becomes susceptible to the anti-sigma factor (Fig. 7B to D).
Thus, binding of CsfB to any of the four sigma factors of sporulation is favored by the presence of a conserved asparagine in region 2.1 (for σF/σG) or 2.3 (σE/σK) and hindered by a glutamic acid at either position (Fig. 8B). Resistance to CsfB binding requires both positions to be occupied by acidic residues. This discrimination is essential for the control of gene expression during sporulation: it allows σG activity to be inhibited in the forespore prior to engulfment completion, while allowing σF-dependent transcription; it also enables CsfB to antagonize σE and σG in the mother cell thus enforcing the cell type-specificity of σG and facilitating the timely switching from σE to the CsfB-immune σK. Making σF or σK susceptible to CsfB or making σE or σG resistant to the anti-sigma factor interferes with proper temporal control and compartmentalization of the forespore and mother cell lines of gene expression.
The strict conservation of the discriminating residues in regions 2.1 or 2.3 of the sigma factors among Bacillus species (Fig. 7A) underscores the importance of CsfB for the activity of the cell type-specific sigma factors in sporeforming organisms closely related to B. subtilis.
Binding of CsfB to σF/σG and to σE/ σK
The crystal structure of the σ70-containing RNA polymerase holoenzyme from Thermus aquaticus shows that residue E189, the homologue of N45 in σG, is involved in a direct contact with K159 in the β´ subunit [49,50,51,52]. We have argued that an asparagine residue could also contribute to the interaction with this site of β´, consistent with the view that one mechanism by which anti-sigma factors function is by occluding sigma-core binding interfaces [53,54]. Occluding a β´-binding surface is consistent with a role for CsfB in preventing the ectopic activity of σG in pre-divisional cells [20] or the premature activation of σG in the forespore [14]. In contrast, N100 in σE is located in the beginning of region 2.3, just upstream of a motif containing several conserved residues involved in promoter melting [55] (Fig. 7A and S6 Fig). This suggests that CsfB may interfere with σE function at a step during transcription initiation subsequent to closed complex formation. In contrast to σG, whose ectopic or premature activation has to be prevented, σE is already engaged in transcription when CsfB appears in the mother cell. Therefore, the most effective mechanism for antagonizing σE activity may not be targeting a core-binding surface but rather a functional region that prevents the activity of the σE-containing RNA polymerase holoenzyme. We note, however, that N100 is also close to several residues in region 2.2 that have been implicated in core binding in E. coli σ70, and are conserved in B. subtilis σA (highlighted in grey in Fig. 7A) (reviewed by [55]). Thus, binding of CsfB in the vicinity of N100 could also possibly occlude core-binding sites in region 2.2.
Concluding remarks
The function of CsfB affects both the transcriptional and the cell-cell signalling networks that control spore differentiation (Fig. 8A). In the forespore, CsfB contributes to the inhibition of σG during early stages of development [14]. The σK-dependent expression of csfB in the mother cell limits the activity of σE and prevents the ectopic activation of σG These activities of CsfB are each one part of redundant mechanisms that work to the same end. σK activity blocks expression of sigE by an unknown mechanism [11,12], and ectopic activity of σG is limited by SpoIIAB and LonA in the mother cell. Our discovery that CsfB binds and inhibits the activity of σE also leads us to propose that in the forespore, CsfB binding to σE explains how CsfB functions as one of the several partially redundant mechanisms described by Piggot and co-workers [34] that prevent ectopic activity of σE in the forespore (see also Fig. 1B). That several of the major roles for CsfB (i.e., preventing ectopic expression of σG and σE, and facilitating the transition between σE and σK) involve partially redundant mechanisms speaks to the importance of controlling these processes, and provides an explanation for why CsfB is highly conserved among sporeformers [6,21]. Moreover, these redundancies may also explain why the phenotype of a csfB mutant is relatively mild. The original work on CsfB found no decernable phenotype [23], whereas subsequent work from Stragier’s group [22] described a small but significant germination defect, but only when a ΔcsfB mutation was combined with an allele causing premature transcription of the gene for σG [22]. The PsigF and PsigK strains formed spores with the same efficiency (77% and 88%, respectively) as the wild type strain (83%). However, we did observe a difference in the protein composition of the spore surface layers between σE N100E and wild type spores (S7A Fig). It is unknown whether this phenotype provides some of the selective pressure for maintaining CsfB during the evolution of sporeformers, but we note the important role of the spore surface layers in mediating many of the environmental interactions of spores, including with cells of host organisms [42]. On the other hand, the evolution of redundant mechanisms to control key steps in development may have promoted the stabilization or canalization of the cell type-specific patterns of gene expression that led to the establishment of the endospore differentiation pathway [56].
The role of CsfB in favouring the switch from early to late stages in spore development is likely to be conserved among Bacillus species and other sporeformers. Importantly, at least in Bacillus species, the actions of the CsfB anti-sigma rely strongly on its ability to discriminate between the highly similar σF/σG and σE/σK pairs (Fig. 8B). Interestingly, CsfB is not found in some Clostridia, a more distantly related class of sporeforming organisms, including the human intestinal pathogen C. difficile. The gene regulatory network for sporulation has been recently characterized in detail for C. difficile. It is interesting to note that in this organism, σG is active prior to engulfment completion and that σE remains active until the late stages of sporulation [57,58,59]. The looser temporal control of σ factor function in C. difficile and presumably also in other Clostridia may stem, at least in part, from the absence of a CsfB orthologue. Moreover, it may be interesting to investigate whether this looser regulation results in greater heterogeneity of morphology among spores of certain Clostridial species than among spores of Bacillus species.
Material and Methods
Strains and general techniques
Except for strain MBS3656 ([60]; see below), all other B. subtilis strains used in this work are congenic derivatives of the Spo+ strains MB24 (trpC2 metC3) or PY79 (prototrophic). Their construction is detailed in S1 Text, and they are listed, with their genotypes, in S1 Table. All plasmids used in this work are described in S1 Text. Primers used for plasmid construction, mutagenesis or sequencing are listed, with their sequences, in S2 Table. LB medium was used for routine growth or maintenance of E. coli and B. subtilis, and sporulation was induced by growth and exhaustion in Difco sporulation medium (DSM) or by the re-suspension method [43].
Purification of RNA polymerase, CsfB and the sigma factors σA, σF, σE, σG and σK
The strains for sigma factor and CsfB expression were cultured in LB at 37°C with Ampicillin (100μg/ml). Arabinose was added to final concentration of 0.5% at an OD600 of 0.6. The culture was allowed to incubate for another 1–2 hours before cells were harvested and stored at -80°C. Sigma factors and CsfB were purified using Qiagen Ni-NTA. The Ni-NTA-affinity purified protein was analyzed on a 10% SDS-PAGE. Fractions containing the sigma factor or CsfB were pooled and dialyzed against dialysis buffer (50mM Tris, 100mM NaCl, 3mM β-mercapatoenthanol). The dialyzed protein was chromatographed through a GE HiLoad 16/60 Superdex 75 column, fractions (1 ml) collected, and those containing the desired protein pooled and dialyzed against 1X in vitro transcription reaction buffer (50mM Tris, 100mM KCl, 10% Glycerol, 10mM DTT). Protein concentration was measured using a Bradford assay (Bio-Rad, Hercules, CA). Protein aliquots were stored at -20°C.
RNA polymerase (RNAP) was purified from B. subtilis strain MH5636 [60] cultured in LB with chloramphenicol (5 μg/ml) to an OD600 of 1.0. Cells was harvested by centrifugation and lysed by treatment with lysozyme (5mg/ml) for 30 minutes at 4°C followed by passage twice through a French pressure cell at a pressure of 20,000 psi. Core RNA polymerase was purified essentially as described by Burgess and colleagues [55]. Briefly the cell lysate supernatant was first purified using Qiagen Ni-NTA affinity column. Fractions containing RNAP were pooled and subjected to chromatography on a HiLoad 16/60 Superdex 200 gel filtration column. Fractions containing RNA polymerase were then purified by ion-exchange exchange chromatography on a GE Mono Q 5/50 GL column. The purity of RNAP was assayed on a 4–20% SDS-PAGE. The concentration of purified RNAP was determined using a Bradfors assay (Bio-Rad, Hercules, CA). RNAP aliquots were at -20°C.
In vitro transcription assays
Core RNAP (13 nM) was incubated in transcription reaction buffer (40 mM Tris/HCl (pH 8), 50 mM KCl, 10 mM MgCl2, 10mM DTT, 50 mg/ml acetylated BSA, 0.5 ml RNase-inhibitor [61] on ice for 30 minutes with 130 nM sigma factor and 1.0 μg of a DNA template purified through a CsCl/ethidium bromide density gradient and cleaved with a restriction enzyme (BamHI or HindIII). After 10 min at 37°C, ATP, CTP, and GTP were added to a final concentration of 1.0 mM and 5 μCi UTP was added to 50 μl transcription reactions. The mixture was incubated for 10 minutes at 377°C after which 0.2 mM unlabeled UTP was added, followed by a further incubation for 10 minutes at 37°C. Finally, the reaction mixture was extracted with phenol-chloroform and the nucleic acids precipitated and electrophoretically resolved through 10%(w/v) polyacrylamide gels containing 7M urea. In the reactions containing CsfB, the anti-sigma factor was added at an amount equal to sigma (130 nM) or 2, 3, 4 or 8-fold excess (indicated as 1x to 8x in Fig. 6C and D), and incubated with sigma for 20 min at room temperature. RNAP was added and the reactions conducted as described above, and incubated with sigma for 20 min at room temperature. RNAP was added and the reactions conducted as described above.
Transcriptional profiling of sporulating cells
These analyses were carried out as previously described in Cozy et al. [62] and Winkelman et al. [63]. Briely, the arrays obtained form Agilent include 15,744 probes covering the annotated protein-coding genes of B. subtilis [64] and the small non-coding RNAs reported in Rasmussen et al. [65] and Irnov et al [66]. The arrays were designed using the Agilent eArray application. Strains AH6825 (PcsfB-csfB) and AH6827 (PsigF-csfB) were sporulated by resuspension in Sterlini-Mandelstam medium and samples collected at hour 3 of sporulation. Cells pellets were recovered by centrifugation after mixing with an equal volume of cold methanol. Total RNA was recovered using a hot acid-phenol protocol followed by clean-up using the Qiagen RNeasy kit. cDNA was synthesized from the purified total RNA and labelled using the Agilent Fairplay III kit. After hybridization and washes using standard protocols, the arrays were scanned in an Agilent Technologies DNA microarray scanner with Surescan high-resolution technology. Processed signal from the Agilent software was subjected to standard lowess normalization using Bioconductor run in R and the geometric mean of the probes was used to give the final value for each gene and nc-RNA.
Fluorescence in situ hybridization (FISH)
For RNA-FISH, we used the following protocol: cells growing in Re-suspension medium were fixed in Histochoice solution (Ameresco) for 15 min at room temperature and 30 min on ice. The samples were centrifuged three times at 3.000xg for 2 min and washed in 1xDEPC-treated PBS. The cell pellets were resuspended in 100 μl GTE buffer (50 mM glucose, 20 mM Tris-HCl pH 7.5, 10 mM EDTA pH 8). Sixteen microlitres of a 10 mg/ml lysozyme solution (GTE, 4 mM vanadyl ribonucleoside complex (VRC), 10 mg/ml lysozyme) were added to 48 μl of cell suspension. The mixture was immediately placed onto poly-L-lysine-coated multi-well slides, and incubated for 10 min at room temperature. The excess liquid was aspirated and the slides were left 1 min to dry before putting them in -20°C methanol for 10 min. Next, the slides were dipped in -20°C acetone for 30 s. Once the slides were dry, they were incubated at 37°C for 30–60 min in a 40% formamide solution (40% formamide, 2x DEPC-treated saline-sodium citrate buffer (SSC)). LNA probe was added to the hybridization solution I (80% formamide, 1 mg/ml E. coli tRNA, 2x DEPC-treated SSC, 70 μg/ml calf-thymus DNA) at a final concentration of 250 nM, and incubated at 80°C for 5 min before mixing with the hybridization solution II (20% dextran sulphate, 4mM VRC, 40U RNase inhibitor, 0.2% RNase-free BSA, 2x DEPC-treated SSC) in a 1 : 1 ratio. The hybridization solution (25 μl) was added to each well of the slide and hybridized for 2 h. The slides were then washed twice in 50% formamide and 2x DEPC-treated SSC solution for 30 min and briefly rinsed five times in DEPC-treated PBS. DAPI was added to a final concentration of 10 μg/ml in SlowFade solution (Invitrogen) were added to each well and the slide was covered and sealed using clear nail polish. The slides were either visualized immediately or stored in the dark at -20°C. The sequence of the probes, which were labeled with Cy3, is given S2 Table.
Fluorescence microscopy
Samples (0.6 ml) of cultures were collected, resuspended in 0.2 ml of phosphate-buffered saline (PBS) and the membrane dye FM4–64 (Molecular Probes) added to a final concentration of 10 μg ml−1. Cells were observed on slides padded with 1.7% agarose. Images were acquired on Leica fluorescence microscopes DMR2A and DM6000B equipped with a Cool Snap HQ camera (Roper Scientific, Arizona, USA) and an iXonEM+ 885 camera (Andor Technology, Connecticut, USA), respectively, or on a Nikon E1000 microscope equipped with an Orca-ER camera (Hamamatsu Corporation, New Jersey, USA), using 63x or 100x lens objective plus an additional 1.6X optavar, phase-contrast optics and standard filters for visualization of GFP and FM4–64. Images were acquired using Metamorph (Molecular Devices, Berks, UK) and processed for publication using Photoshop (Adobe).
Accession numbers
The Genbank accession numbers for the sequences represented in Fig. 8A and S3C Fig, are as follows. For σG, CAB13407.1 (B. subtilis) and for σF, CAB14277.1 (B. subtilis). For σA, NP_390399.2 (B. subtilis), P00579.2 (E. coli), AAG36964.1 (T. aquaticus); WP_011172619.1 (T. thermophilus). For σE, NP_243422.1 (B. halodurans), YP_175847.1 (B. clausii), NP_389415.2 (B. subtilis), ABS73878.1 (B. amyloliquefaciens); YP_006713129.1 (B. licheniformis); YP_085245.1 (B. cereus), ABY76243.1 (B. thuringiensis). For σK, NP_242151.1 (B. halodurans), YP_175113.1 (B. clausii), WP_003237137.1 (B. subtilis), WP_015240253.1 (B. amyloliquefaciens); YP_079918.1 (B. licheniformis); YP_085663.1 (B. cereus), ABY76244.1 (B. thuringiensis). For the β´subunit of RNAP polymerase: WP_003225772.1 (B. subtilis); YP_491473.1 (E. coli); WP_003043700.1 (T. aquaticus); YP_005641350.1 (T. thermophilus).
Supporting Information
Zdroje
1. Levine M, Davidson EH (2005) Gene regulatory networks for development. Proc Natl Acad Sci U S A 102 : 4936–4942. 15788537
2. Stathopoulos A, Levine M (2005) Genomic regulatory networks and animal development. Dev Cell 9 : 449–462. 16198288
3. Alon U (2007) Network motifs: theory and experimental approaches. Nat Rev Genet 8 : 450–461. 17510665
4. Traag BA, Pugliese A, Eisen JA, Losick R (2012) Gene conservation among endospore-forming bacteria reveals additional sporulation genes in Bacillus subtilis. J Bacteriol. 195 : 253–260. doi: 10.1128/JB.01778-12 23123912
5. Abecasis A, Serrano M, Alves L, Quintais L, Pereira-Leal JB, et al. (2013) A genomic signature and the identification of new endosporulaion genes. J Bacteriol 195 : 2101–2115. doi: 10.1128/JB.02110-12 23396918
6. Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, et al. (2012) Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol 14 : 2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x 22882546
7. de Hoon MJ, Eichenberger P, Vitkup D (2010) Hierarchical evolution of the bacterial sporulation network. Curr Biol 20: R735–745. doi: 10.1016/j.cub.2010.06.031 20833318
8. Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, et al. (2006) The forespore line of gene expression in Bacillus subtilis. J Mol Biol 358 : 16–37. 16497325
9. Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, et al. (2004) The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol 2: e328. 15383836
10. Wang L, Perpich J, Driks A, Kroos L (2007) One perturbation of the mother cell gene regulatory network suppresses the effects of another during sporulation of Bacillus subtilis. J Bacteriol 189 : 8467–8473. 17890309
11. Zhang B, Struffi P, Kroos L (1999) sigmaK can negatively regulate sigE expression by two different mechanisms during sporulation of Bacillus subtilis. J Bacteriol 181 : 4081–4088. 10383978
12. Zhang B, Kroos L (1997) A feedback loop regulates the switch from one sigma factor to the next in the cascade controlling Bacillus subtilis mother cell gene expression. J Bacteriol 179 : 6138–6144. 9324264
13. Sun DX, Cabrera-Martinez RM, Setlow P (1991) Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor sigma G. J Bacteriol 173 : 2977–2984. 1902213
14. Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, et al. (2009) Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet 5: e1000566. doi: 10.1371/journal.pgen.1000566 19609349
15. Serrano M, Neves A, Soares CM, Moran CP Jr., Henriques AO (2004) Role of the anti-sigma factor SpoIIAB in regulation of sigmaG during Bacillus subtilis sporulation. J Bacteriol 186 : 4000–4013. 15175314
16. Kellner EM, Decatur A, Moran CP Jr. (1996) Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol Microbiol 21 : 913–924. 8885263
17. Camp AH, Losick R (2009) A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev 23 : 1014–1024. doi: 10.1101/gad.1781709 19390092
18. Hilbert DW, Piggot PJ (2004) Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev 68 : 234–262. 15187183
19. Camp AH, Losick R (2008) A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol 69 : 402–417. doi: 10.1111/j.1365-2958.2008.06289.x 18485064
20. Serrano M, Real G, Santos J, Carneiro J, Moran CP Jr., et al. (2011) A negative feedback loop that limits the ectopic activation of a cell type-specific sporulation sigma factor of Bacillus subtilis. PLoS Genet 7: e1002220. doi: 10.1371/journal.pgen.1002220 21935351
21. Rhayat L, Duperrier S, Carballido-Lopez R, Pellegrini O, Stragier P (2009) Genetic dissection of an inhibitor of the sporulation sigma factor sigma(G). J Mol Biol 390 : 835–844. doi: 10.1016/j.jmb.2009.05.073 19497328
22. Karmazyn-Campelli C, Rhayat L, Carballido-Lopez R, Duperrier S, Frandsen N, et al. (2008) How the early sporulation sigma factor sigmaF delays the switch to late development in Bacillus subtilis. Mol Microbiol 67 : 1169–1180. doi: 10.1111/j.1365-2958.2008.06121.x 18208527
23. Decatur A, Losick R (1996) Identification of additional genes under the control of the transcription factor sigma F of Bacillus subtilis. J Bacteriol 178 : 5039–5041. 8759874
24. Chary VK, Xenopoulos P, Piggot PJ (2007) Expression of the sigmaF-directed csfB locus prevents premature appearance of sigmaG activity during sporulation of Bacillus subtilis. J Bacteriol 189 : 8754–8757. 17921305
25. Cutting S, Oke V, Driks A, Losick R, Lu S, et al. (1990) A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell 62 : 239–250. 2115401
26. Sierro N, Makita Y, de Hoon M, Nakai K (2008) DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res 36: D93–96. 17962296
27. Londono-Vallejo JA, Frehel C, Stragier P (1997) SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol 24 : 29–39. 9140963
28. Stragier P, Losick R (1996) Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30 : 297–241. 8982457
29. Steil L, Serrano M, Henriques AO, Volker U (2005) Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151 : 399–420. 15699190
30. Nicholson WL, Sun DX, Setlow B, Setlow P (1989) Promoter specificity of sigma G-containing RNA polymerase from sporulating cells of Bacillus subtilis: identification of a group of forespore-specific promoters. J Bacteriol 171 : 2708–2718. 2468649
31. Rather PN, Coppolecchia R, DeGrazia H, Moran CP Jr. (1990) Negative regulator of sigma G-controlled gene expression in stationary-phase Bacillus subtilis. J Bacteriol 172 : 709–715. 2105300
32. Schmidt R, Decatur AL, Rather PN, Moran CP Jr., Losick R (1994) Bacillus subtilis lon protease prevents inappropriate transcription of genes under the control of the sporulation transcription factor sigma G. J Bacteriol 176 : 6528–6537. 7961403
33. Cutting S, Panzer S, Losick R (1989) Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J Mol Biol 207 : 393–404. 2474075
34. Chary VK, Xenopoulos P, Eldar A, Piggot PJ (2010) Loss of compartmentalization of sigma(E) activity need not prevent formation of spores by Bacillus subtilis. J Bacteriol 192 : 5616–5624. doi: 10.1128/JB.00572-10 20802044
35. Hove-Jensen B (1992) Identification of tms-26 as an allele of the gcaD gene, which encodes N-acetylglucosamine 1-phosphate uridyltransferase in Bacillus subtilis. J Bacteriol 174 : 6852–6856. 1328164
36. Moran CP Jr., Johnson WC, Losick R (1982) Close contacts between sigma 37-RNA polymerase and a Bacillus subtilis chromosomal promoter. J Mol Biol 162 : 709–713. 6820071
37. Amaya E, Khvorova A, Piggot PJ (2001) Analysis of promoter recognition in vivo directed by sigma(F) of Bacillus subtilis by using random-sequence oligonucleotides. J Bacteriol 183 : 3623–3630. 11371526
38. Guillot C, Moran CP Jr. (2007) Essential internal promoter in the spoIIIA locus of Bacillus subtilis. J Bacteriol 189 : 7181–7189. 17693505
39. Smith K, Youngman P (1992) Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74 : 705–711. 1391050
40. Rong S, Rosenkrantz MS, Sonenshein AL (1986) Transcriptional control of the Bacillus subtilis spoIID gene. J Bacteriol 165 : 771–779. 2419309
41. McKenney PT, Eichenberger P (2013) Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol Microbiol 83 : 245–260.
42. Henriques AO, Moran CP Jr. (2000) Structure and assembly of the bacterial endospore coat. Methods 20 : 95–110. 10610808
43. Cutting SaHPB (1990) Genetic Analysis; Harwood CRaCSM, editor: John Wiley and sons, Lta.
44. Kim H, Hahn M, Grabowski P, McPherson DC, Otte MM, et al. (2006) The Bacillus subtilis spore coat protein interaction network. Mol Microbiol 59 : 487–502. 16390444
45. Henriques AO, Moran CP Jr. (2007) Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol 61 : 555–588. 18035610
46. Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, et al. (2003) The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol 327 : 945–972. 12662922
47. Kirchman PA, DeGrazia H, Kellner EM, Moran CP Jr. (1993) Forespore-specific disappearance of the sigma-factor antagonist spoIIAB: implications for its role in determination of cell fate in Bacillus subtilis. Mol Microbiol 8 : 663–671. 8332059
48. Wang L, Perpich J, Driks A, Kroos L (2007) Maintaining the transcription factor SpoIIID level late during sporulation causes spore defects in Bacillus subtilis. J Bacteriol 189 : 7302–7309. 17693499
49. Haugen SP, Ross W, Gourse RL (2008) Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6 : 507–519. doi: 10.1038/nrmicro1912 18521075
50. Murakami KS, Darst SA (2003) Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 13 : 31–39. 12581657
51. Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002) Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296 : 1285–1290. 12016307
52. Campbell EA, Westblade LF, Darst SA (2008) Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr Opin Microbiol 11 : 121–127. doi: 10.1016/j.mib.2008.02.016 18375176
53. Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, et al. (2003) Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol Cell 11 : 1067–1078. 12718891
54. Helmann JD (1999) Anti-sigma factors. Curr Opin Microbiol 2 : 135–141. 10322161
55. Burgess RR, Anthony L (2001) How sigma docks to RNA polymerase and what sigma does. Curr Opin Microbiol 4 : 126–131. 11282466
56. Siegal ML, Bergman A (2002) Waddington's canalization revisited: developmental stability and evolution. Proc Natl Acad Sci U S A 99 : 10528–10532. 12082173
57. Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, et al. (2013) Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet 9: e1003756. doi: 10.1371/journal.pgen.1003756 24098137
58. Pereira FC, Saujet L, Tome AR, Serrano M, Monot M, et al. (2013) The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet 9: e1003782. doi: 10.1371/journal.pgen.1003782 24098139
59. Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, et al. (2013) Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet 9: e1003660. doi: 10.1371/journal.pgen.1003660 23950727
60. Qi Y, Hulett FM (1998) PhoP-P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol Microbiol 28 : 1187–1197. 9680208
61. Gebhard S, Gaballa A, Helmann JD, Cook GM (2009) Direct stimulus perception and transcription activation by a membrane-bound DNA binding protein. Mol Microbiol 73 : 482–491. doi: 10.1111/j.1365-2958.2009.06787.x 19602149
62. Cozy LM, Phillips AM, Calvo RA, Bate AR, Hsueh YH, et al. (2012) SlrA/SinR/SlrR inhibits motility gene expression upstream of a hypersensitive and hysteretic switch at the level of sigma(D) in Bacillus subtilis. Mol Microbiol 83 : 1210–1228. doi: 10.1111/j.1365-2958.2012.08003.x 22329926
63. Winkelman JT, Bree AC, Bate AR, Eichenberger P, Gourse RL, et al. (2013) RemA is a DNA-binding protein that activates biofilm matrix gene expression in Bacillus subtilis. Mol Microbiol 88 : 984–997. doi: 10.1111/mmi.12235 23646920
64. Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, et al. (2009) From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155 : 1758–1775. doi: 10.1099/mic.0.027839-0 19383706
65. Rasmussen S, Nielsen HB, Jarmer H (2009) The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol 73 : 1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x 19682248
66. Irnov I, Sharma CM, Vogel J, Winkler WC (2010) Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res 38 : 6637–6651. doi: 10.1093/nar/gkq454 20525796
67. Crawshaw AD, Serrano M, Stanley WA, Henriques AO, Salgado PS (2014) A mother cell-to-forespore channel: current understanding and future challenges. FEMS Microbiol Lett.
68. Schroeder LA, Karpen ME, deHaseth PL (2008) Threonine 429 of Escherichia coli sigma 70 is a key participant in promoter DNA melting by RNA polymerase. J Mol Biol 376 : 153–165. 18155246
69. Panaghie G, Aiyar SE, Bobb KL, Hayward RS, de Haseth PL (2000) Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex. J Mol Biol 299 : 1217–1230. 10873447
Štítky
Genetika Reprodukční medicína
Článek Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in CrustaceansČlánek Adventures in WonderlandČlánek Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and InfectivityČlánek Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in FilamentsČlánek Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in ArabidopsisČlánek Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression ControlČlánek The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent EnhancersČlánek Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of MitochondriaČlánek The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma Formation
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 4- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- Adventures in Wonderland
- Experimental Swap of 's Assortative Mating Preferences Demonstrates Key Role of X-Chromosome Divergence Island in Incipient Sympatric Speciation
- Chromosome Replacement and Deletion Lead to Clonal Polymorphism of Berry Color in Grapevine
- The Protein Quality Control Machinery Regulates Its Misassembled Proteasome Subunits
- Genome-Wide Association Study Identifies as a Critical Gene for Susceptibility to Noise-Induced Hearing Loss
- Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and Infectivity
- Viable Neuronopathic Gaucher Disease Model in Medaka () Displays Axonal Accumulation of Alpha-Synuclein
- Multi-locus Analysis of Genomic Time Series Data from Experimental Evolution
- The Genetic Legacy of the Expansion of Turkic-Speaking Nomads across Eurasia
- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- The Pif1 Helicase, a Negative Regulator of Telomerase, Acts Preferentially at Long Telomeres
- Inhibiting K63 Polyubiquitination Abolishes No-Go Type Stalled Translation Surveillance in
- SYD-1C, UNC-40 (DCC) and SAX-3 (Robo) Function Interdependently to Promote Axon Guidance by Regulating the MIG-2 GTPase
- Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in Filaments
- Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3’ Tailing of Small RNAs in Arabidopsis
- Host Genetic Variation Influences Gene Expression Response to Rhinovirus Infection
- Contribution of Large Region Joint Associations to Complex Traits Genetics
- Volatility of Mutator Phenotypes at Single Cell Resolution
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in Arabidopsis
- A Multi-layered Protein Network Stabilizes the FtsZ-ring and Modulates Constriction Dynamics
- Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression Control
- Genome Sequencing of the Perciform Fish Provides Insights into Molecular and Genetic Mechanisms of Stress Adaptation
- Natural Variant E610G Is a Semi-dominant Suppressor of IAP-Induced RNA Processing Defects
- The Alkaline Response Pathway: Identification of a Novel Rim Pathway Activator
- Transgenerational Inheritance of Diet-Induced Genome Rearrangements in Drosophila
- A Single Nucleotide Polymorphism Uncovers a Novel Function for the Transcription Factor Ace2 during Hyphal Development
- DNA Damage Response and Spindle Assembly Checkpoint Function throughout the Cell Cycle to Ensure Genomic Integrity
- The Functional Interplay Between the t(9;22)-Associated Fusion Proteins BCR/ABL and ABL/BCR in Philadelphia Chromosome-Positive Acute Lymphatic Leukemia
- Extreme Recombination Frequencies Shape Genome Variation and Evolution in the Honeybee,
- Beyond Glycolysis: GAPDHs Are Multi-functional Enzymes Involved in Regulation of ROS, Autophagy, and Plant Immune Responses
- Comprehensive Profiling of Amino Acid Response Uncovers Unique Methionine-Deprived Response Dependent on Intact Creatine Biosynthesis
- Windpipe Controls Intestinal Homeostasis by Regulating JAK/STAT Pathway via Promoting Receptor Endocytosis and Lysosomal Degradation
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
- Cross-Population Joint Analysis of eQTLs: Fine Mapping and Functional Annotation
- The Power of Gene-Based Rare Variant Methods to Detect Disease-Associated Variation and Test Hypotheses About Complex Disease
- The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent Enhancers
- Competition between VanU Repressor and VanR Activator Leads to Rheostatic Control of Vancomycin Resistance Operon Expression
- A Missense Change in the Gene Links Aberrant Autophagy to a Neurodegenerative Vacuolar Storage Disease
- Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model
- Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of Mitochondria
- Genome-Destabilizing Effects Associated with Top1 Loss or Accumulation of Top1 Cleavage Complexes in Yeast
- Imputation-Based Population Genetics Analysis of Malaria Parasites
- Heterozygosity for a Hypomorphic Polβ Mutation Reduces the Expansion Frequency in a Mouse Model of the Fragile X-Related Disorders
- Neto-Mediated Intracellular Interactions Shape Postsynaptic Composition at the Neuromuscular Junction
- Ndd1 Turnover by SCF Is Inhibited by the DNA Damage Checkpoint in
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma Formation
- Spastin Binds to Lipid Droplets and Affects Lipid Metabolism
- Maintenance of Glia in the Optic Lamina Is Mediated by EGFR Signaling by Photoreceptors in Adult Drosophila
- Auxin Influx Carriers Control Vascular Patterning and Xylem Differentiation in
- Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
- The Lowe Syndrome Protein OCRL1 Is Required for Endocytosis in the Zebrafish Pronephric Tubule
- Postnatal Loss of Hap1 Reduces Hippocampal Neurogenesis and Causes Adult Depressive-Like Behavior in Mice
- CAPER Is Vital for Energy and Redox Homeostasis by Integrating Glucose-Induced Mitochondrial Functions via ERR-α-Gabpa and Stress-Induced Adaptive Responses via NF-κB-cMYC
- Distinct and Cooperative Activities of HESO1 and URT1 Nucleotidyl Transferases in MicroRNA Turnover in
- The Evolutionary Origination and Diversification of a Dimorphic Gene Regulatory Network through Parallel Innovations in and
- MAPK Signaling Pathway Alters Expression of Midgut ALP and ABCC Genes and Causes Resistance to Cry1Ac Toxin in Diamondback Moth
- Spatio-temporal Remodeling of Functional Membrane Microdomains Organizes the Signaling Networks of a Bacterium
- Asymmetric Transcript Discovery by RNA-seq in . Blastomeres Identifies , a Gene Important for Anterior Morphogenesis
- A Stress-Induced Small RNA Modulates Alpha-Rhizobial Cell Cycle Progression
- Systematic Profiling of Poly(A)+ Transcripts Modulated by Core 3’ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation
- The UPR Branch IRE1- in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast
- A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi
- Co-chaperone p23 Regulates . Lifespan in Response to Temperature
- Re-replication of a Centromere Induces Chromosomal Instability and Aneuploidy
- Shade Avoidance Components and Pathways in Adult Plants Revealed by Phenotypic Profiling
- Lipid-Induced Epigenomic Changes in Human Macrophages Identify a Coronary Artery Disease-Associated Variant that Regulates Expression through Altered C/EBP-Beta Binding
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



