-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNdd1 Turnover by SCF Is Inhibited by the DNA Damage Checkpoint in
All cells must regulate cell division in response to extracellular and intracellular cues, and one of the most critical steps to regulate is the process of cell division, or mitosis. In response to DNA damage in budding yeast, cells activate a checkpoint that promotes DNA repair and arrests the cell cycle before division to give the cell time to repair the lesion. One of the key regulators of mitosis is an essential transcription factor called Ndd1. Ndd1 is known to be regulated by transcription and phosphorylation, both in unperturbed cells and following exposure to DNA damage. Here, we show that Ndd1 protein turnover is also regulated in both situations. Ndd1 is degraded quickly during an unperturbed cell cycle, but is strongly stabilized following exposure to DNA damage. We identify the machinery that targets Ndd1 for turnover and the signaling pathways required to stabilize Ndd1 in response to DNA damage. Maintaining high levels of Ndd1 after exposure to DNA damage may allow the cell to reactivate Ndd1 after the damage has been repaired to promote mitosis.
Published in the journal: . PLoS Genet 11(4): e32767. doi:10.1371/journal.pgen.1005162
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005162Summary
All cells must regulate cell division in response to extracellular and intracellular cues, and one of the most critical steps to regulate is the process of cell division, or mitosis. In response to DNA damage in budding yeast, cells activate a checkpoint that promotes DNA repair and arrests the cell cycle before division to give the cell time to repair the lesion. One of the key regulators of mitosis is an essential transcription factor called Ndd1. Ndd1 is known to be regulated by transcription and phosphorylation, both in unperturbed cells and following exposure to DNA damage. Here, we show that Ndd1 protein turnover is also regulated in both situations. Ndd1 is degraded quickly during an unperturbed cell cycle, but is strongly stabilized following exposure to DNA damage. We identify the machinery that targets Ndd1 for turnover and the signaling pathways required to stabilize Ndd1 in response to DNA damage. Maintaining high levels of Ndd1 after exposure to DNA damage may allow the cell to reactivate Ndd1 after the damage has been repaired to promote mitosis.
Introduction
Ndd1 is the dedicated transcriptional activator of a cluster of genes in Saccharomyces cerevisiae called the CLB2 cluster [1]. This cluster consists of thirty-three members, including the namesake cyclin CLB2, as well as other important mitotic regulators, such as the yeast polo kinase CDC5 and the activator of the anaphase promoting complex, CDC20 [2]. Transcription of this cluster is regulated during the cell cycle [1–10] and in response to DNA damage [11–14] through regulation of Ndd1.
Ndd1 is regulated both transcriptionally and post-transcriptionally during the mitotic cell cycle in order to control transcription of the CLB2 cluster. Ndd1 is itself a member of the Hcm1-regulated transcriptional cluster [15] and transcriptionally peaks in early S phase. Genome-wide studies found that the Swi4 and Swi6 transcription factors bound upstream of Ndd1, suggesting that Ndd1 may also be regulated by a G1 transcriptional regulator, the SBF, since [16]. Ndd1 is also regulated post-transcriptionally by phosphorylation. Throughout the cell cycle, Fkh2 and Mcm1 bind to CLB2 cluster promoters and coordinate both the activation and repression of the cluster by associating with Ndd1 as well as transcriptional repressors [3,4]. Ndd1 binding to Fkh2 in early mitosis requires phosphorylation by Clb2-Cdk1 and Cdc5 (the yeast Polo kinase) [5,7,9]. The phosphorylation of Ndd1 by Clb2-Cdk1 and Cdc5 also promotes the intrinsic transcriptional activator activity of Ndd1 [5,7,9]. Ndd1 activity is further regulated by Protein Kinase C (PKC), which phosphorylates Ndd1 to inhibit its activity early in S phase [6]. Recent work has also shown that Ndd1 protein turnover is regulated during the meiotic cell cycle by the Anaphase Promoting Complex and its meiotic specific substrate adaptor, Ama1 [17].
In response to DNA damage, Ndd1 is inhibited by a signal transduction cascade called the DNA damage checkpoint [11,13,14]. The checkpoint is activated by the kinases Mec1 and Tel1, homologs of human ATR and ATM, respectively. In turn, Mec1 and Tel1 activate the downstream effector kinases, Rad53, Dun1, and Chk1 [18,19]. RAD53-dependent Ndd1 phosphorylation inhibits Ndd1 by blocking its recruitment to Fkh2-bound promoters, leading to the transcriptional down-regulation of the CLB2 cluster [11,13,14]. Here, we characterize Ndd1 turnover during the mitotic cell cycle and in response to DNA damage. We find that Ndd1 turns over quickly in an unperturbed metaphase, in a Cdk1 - and SCFGrr1-dependent manner. In addition, we find that Ndd1 is stabilized by Mec1/Tel1 in response to DNA damage through inhibitory phosphorylation of Cdk1. Thus, the DNA damage checkpoint simultaneously inhibits Ndd1 function by phosphorylation, and also preserves the inhibited form. We speculate that the stabilized, inhibited Ndd1 may allow the cell to remain primed to undertake mitotic transcription once the checkpoint is inactivated.
Results
Ndd1 turns over in the cell cycle and is stable in response to DNA damage
We first asked whether Ndd1 protein turnover was regulated over a normal cell cycle. To examine protein turnover, we inhibited new protein synthesis with cycloheximide and followed the degradation of the protein over time. We found that Ndd1 was unstable in metaphase-arrested cells; however, Ndd1 was stable following treatment with the DNA damaging agent methyl methane sulfonate (MMS) (Fig 1A). Under its endogenous cell cycle-regulated promoter, Ndd1 protein was undetectable in G1-arrested cells [1,15]. In order to study Ndd1 turnover in the absence of its transcriptional regulation, we placed a Flag-tagged, second copy of NDD1 under the control of the GAL1 promoter. As we had seen with Ndd1 under its endogenous promoter, GAL1p-Ndd1 was unstable in metaphase-arrested cells and stabilized following treatment with MMS (Fig 1B, also shown in S1 Fig with FACS for cell cycle progression). Ndd1 was even more unstable in G1-arrested cells than in metaphase-arrested cells (Fig 1B), as seen by both its rate of turnover and its lower starting level (Fig 1B and quantified in Fig 1C). We next wondered whether the stabilization of Ndd1 was also induced by DNA replication checkpoint signaling. In order to test this, we arrested cells in G1 using α-factor and released them into the presence of nocodazole or hydroxyurea (HU) for 1.5 hours. Similar to what we observed with MMS treatment, cells exposed to HU treatment strongly stabilized Ndd1, while Ndd1 turned over rapidly in nocodazole treated cells (Fig 1D with FACS showing arrest; quantification shown in S2A Fig).
Fig. 1. Regulation of Ndd1 turnover. 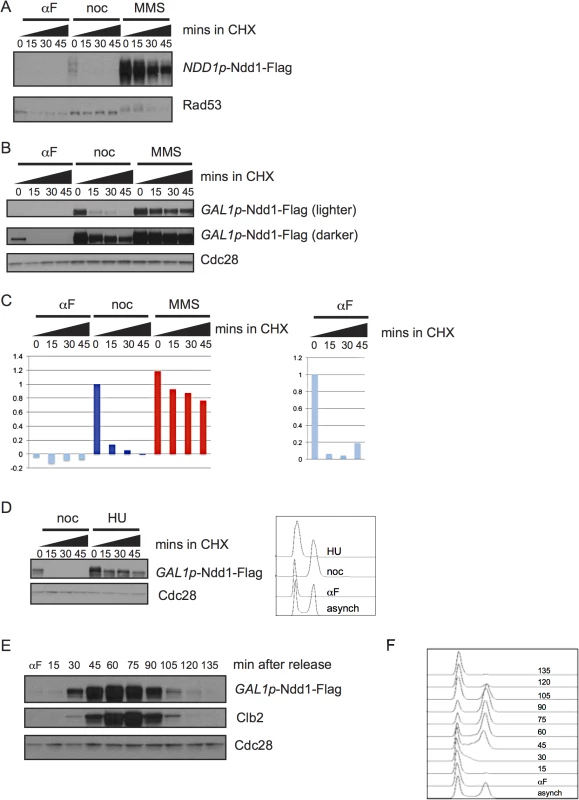
A) Cells expressing Ndd1-Flag from its endogenous promoter were arrested in G1 (with 10 μg/ml α–g/mlh (αF)), metaphase (with 10 μg/ml nocodazole (noc)), or exposed to DNA damage (with 0.05% MMS) for 2.5 hours. Cycloheximide (CHX) was added at 50 μg/ml at t = 0 and protein turnover was followed with 15 minute timepoints. Rad53 is shown as a control for damage treatment, because it hyper-shifts in response to DNA damage. B) Expression of Ndd1-Flag from the GAL1 promoter (GAL1p-Ndd1-Flag) was induced by growth in galactose for 6.5 hours until cells reached mid-log phase. Cells were treated with αF, noc, or MMS for 2.5 hours and turnover was followed as in [A]. Cdc28 is shown as a loading control. C) Quantification is shown of samples in [B]. On the left, quantification is shown from lighter exposure. Right panel shows quantification of αF samples only from the darker exposure. Y-axis shows the normalized signal above background normalized to the value at t = 0 in nocodazole arrested cells (left) or αF arrested cells (right). D) Cells expressing GAL1p-Ndd1-Flag were induced by growth in galactose for 3 hours, then arrested in G1 with 10 μg/ml αF for 2.5 hours, and released into media containing either 10 μg/ml nocodazole or 0.2 M hydroxyurea (HU). Cycloheximide was added after 1.5 hours and protein turnover was followed with 15 minute timepoints. FACS analysis shows G1 peak from the αF arrest and cell cycle progression following 1.5 hours release into HU and noc. E) GAL1p-Ndd1-Flag was induced with galactose until cells reached mid-log phase. Cells were arrested in G1 with αF for 2.5 hours, and released into the cell cycle and Ndd1 was examined. αF was re-added to the media after the 30 minute timepoint was collected to prevent the cells from entering a second cell cycle. Clb2 levels are shown to indicate cell cycle position. Cdc28 is shown as a loading control. F) FACS analysis showing cell cycle progression of timepoints shown in [F]. To determine whether the cell cycle regulated turnover we observed was sufficient to generate periodic expression of Ndd1 in the absence of transcriptional regulation, we arrested cells expressing GAL1p-Ndd1 in G1 with α-factor, released the cells into fresh media to allow them to enter the cell cycle, and followed Ndd1 levels into the next G1. Ndd1 protein levels were very low in G1, rose through the cell cycle, and dropped in the next G1 (Fig 1E, with FACS analysis of cell cycle progression shown in Fig 1F). This result confirms that protein turnover of Ndd1 is regulated during the cell cycle, as had previously been suggested [1]. Furthermore, it suggests that the changes in protein turnover rate during the cell cycle are sufficient for periodic Ndd1 expression in the absence of Ndd1 transcriptional regulation.
Cdk1 kinase promotes Ndd1 turnover
Exposure to DNA damaging agents leads to budding yeast cells arresting in metaphase; however Ndd1 turnover is strikingly different between nocodazole-arrested cells (which arrest in metaphase in the absence of DNA damage) and cells exposed to MMS. Ndd1 is phosphorylated by many kinases during the cell cycle [5–7,9]. Since phosphorylation is a common mode of regulating substrate turnover, we hypothesized that Ndd1 phosphorylation may regulate its turnover. First, we tested whether the 16 S/T-P (serine or threonine, followed by a proline) sites on Ndd1 activated its turnover in metaphase. These sites are the minimal consensus motif for CDK and MAP kinases. Clb2-Cdk1 is known to phosphorylate Ndd1 in order to promote its activity [7,9]. Because the Cdk1 phosphorylation of Ndd1 is essential for its function [7,9], we placed a Flag-tagged, second copy of NDD116A under the control of the GAL1 promoter. Ndd116A was completely stabilized in metaphase-arrested cells, consistent with phosphorylation by Cdk1 driving turnover during mitosis (Fig 2A, quantification shown in Fig 2B). However, Ndd1 was even more unstable in α-factor-arrested cells, and while Ndd116A was stabilized compared to Ndd1wt in G1, it was still quite unstable (Fig 2A), suggesting that additional kinases might phosphorylate Ndd1 in these arrested cells. In order to test whether Cdk1 itself was required for Ndd1 turnover in metaphase, we used a strain in which the yeast Cdk1 (CDC28) has been replaced with an analog sensitive allele, cdc28-as1. This allele is sensitive to inhibition by the ATP analog 1-NM-PP1, but is also hypomorphic, with a 6-fold lower kcat, even in the absence of the inhibitor [20]. We arrested cells in metaphase using nocodazole, then treated with 1-NM-PP1 (or held the cells in nocodazole without the inhibitor), and followed turnover of Ndd1 for 45 minutes following the addition of cycloheximide. Wildtype cells were unaffected by treatment with 1-NM-PP1, and Ndd1 had the expected short half-life (Fig 2C, quantification shown in S2B Fig). Consistent with the cdc28-as1 allele being hypomorphic [20], Ndd1 was significantly stabilized in cdc28-as1 cells even in the absence of the inhibitor. Moreover, this suggests that the phosphorylation of Ndd1 that promotes its turnover is very sensitive even to small changes in CDK activity. Treatment with 1-NM-PP1 in these cells led to a greater stabilization of Ndd1. Based on these data, we propose that CDK is required for Ndd1 turnover in metaphase.
Fig. 2. Ndd1 turnover requires Cdk1, Hog1, and three of the GSK3 family kinases. 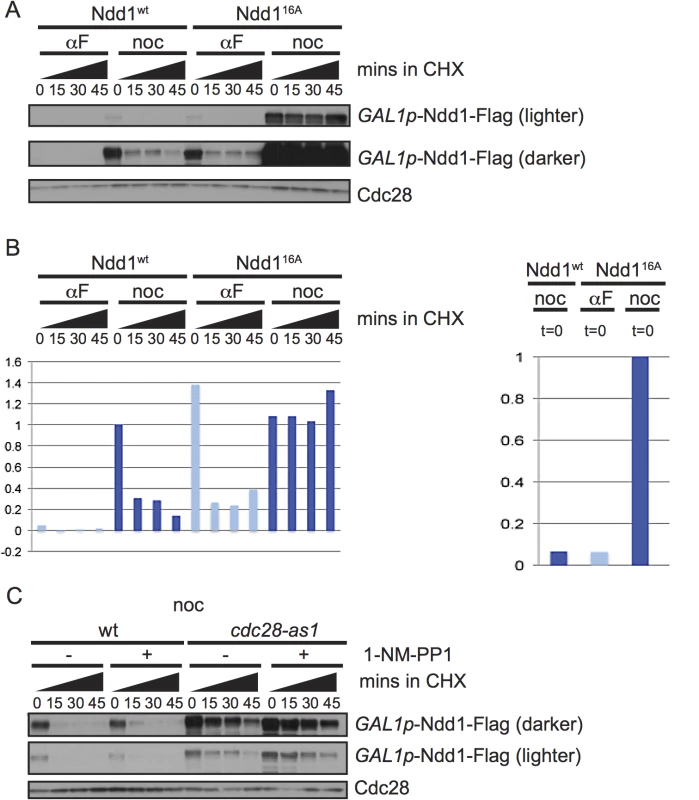
A) Cells expressing wildtype Ndd1-Flag (GAL1p-Ndd1wt-Flag) or an allele with 16 S/T-P consensus sites mutated (GAL1p-Ndd116A-Flag) were induced with galactose, arrested with αF or noc for 2.5 hours and examined after cycloheximide (CHX) addition as in Fig 1B. Cdc28 is shown as a loading control. B) Quantification of the experiment in [A] is shown. On the left is the quantification from the darker exposure, while right side shows relative abundance quantified from the lighter exposure. Y-axis shows the normalized signal above background normalized to the value at t = 0 in nocodazole arrested Ndd1wt cells (left) or Ndd116A cells (right). C) GAL1p-Ndd1wt-Flag was induced with galactose in a wildtype or cdc28-as1 strain. Cells were then arrested in metaphase with nocodazole for 3.5 hours, with treatment of 1 μM 1-NM-PP1 for the last hour, as noted. Cycloheximide was then added at t = 0 and turnover was followed as in [A]. Cdc28 is shown as a loading control. Ndd1 is turned over by SCFGrr1
We next wanted to know which E3 ligase targeted Ndd1 for degradation. Each member of the SCF family of E3 ligases contains the cullin Cdc53, Skp1, and the Ring finger subunit Rbx1, in addition to any one of several substrate adaptors called F-box proteins [21]. Budding yeast have 20 putative F-box proteins, most of which are thought to target a specific suite of substrates [22]. We found that Ndd1 was strongly stabilized at the non-permissive temperature in the temperature-sensitive mutant cdc53-1 (Fig 3A, quantification shown in S2C Fig), suggesting that Ndd1 is a substrate of an SCF ligase.
Fig. 3. SCFGrr1 turns over Ndd1. 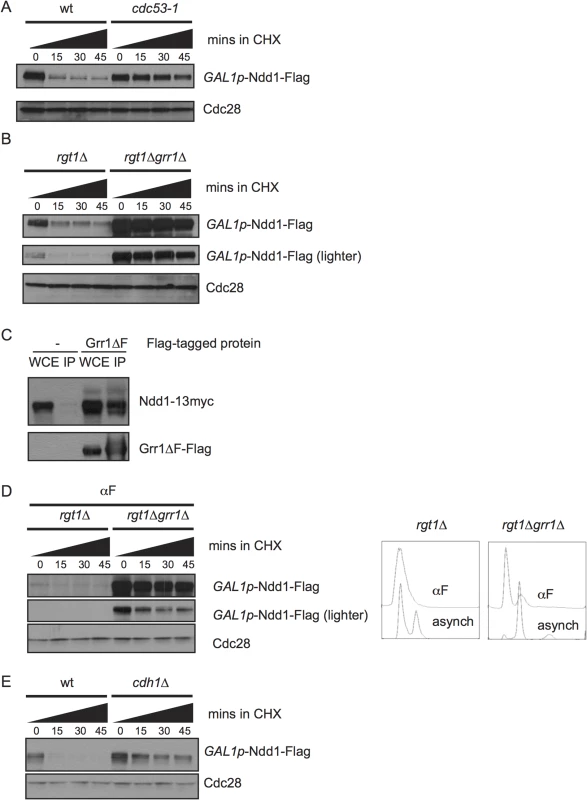
A) GAL1p-Ndd1-Flag was induced with galactose in wildtype or cdc53-1 strains at 23°C for 5 hours. All strains were then shifted to 37°C for 2.5 hours to inactivate the temperature-sensitive allele before addition of cycloheximide (CHX). Turnover was then examined as in Fig 1B, except at 37°C. Cdc28 is shown as a loading control. B) GAL1p-Ndd1-Flag was induced with galactose in rgt1Δ or rgt1Δ grr1Δ strains for 2.5 hours. Protein turnover was followed as in [A]. C) Strains expressing NDD1p-Ndd1-13Myc with either an empty vector (pRS426) or pYES-Grr1ΔF-Flag (containing a galactose-inducible copy of Grr1-Flag lacking its F-box domain) were grown in galactose media overnight to double twice. Whole cell extracts (WCE) were prepared and immunoprecipitated with anti-Flag antibody, Western blotted and probed for Flag and Myc. D) Experiment was performed as in [B], expect that cells were pre-arrested in G1 using 10 μg/ml α–/ml im (αF) for 6 hours. grr1Δ cells do not arrest well, but FACS analysis shows strong enrichment of G1 peak. E) Experiment was performed as in [B] in wt or cdh1Δ strains. The F-box protein Grr1 targets several cell cycle and metabolic proteins for turnover, including the regulator of glycolysis Pfk27 and the G1 cyclins [22–25]. grr1Δ strains grow very slowly [26,27], and simultaneous deletion of RGT1, a glucose-responsive transcription factor, partially rescues this growth defect [28]. Therefore, to examine the effect of GRR1 deletion on Ndd1 stability, we compared Ndd1 turnover in rgt1Δ cells and grr1Δ rgt1Δ cells. We found that Ndd1 was strongly stabilized by deletion of GRR1 (Fig 3B, quantification shown in S2D Fig), which suggests that Grr1 is the primary F-box protein that targets Ndd1 for turnover. In order to test whether Ndd1 was a direct substrate of Grr1, we tested whether Ndd1 physically bound Grr1. We used an allele of Grr1 lacking its F-box (Grr1Δ F), such that it could not interact with the rest of the SCF, but was still able to bind substrates. This allele allowed us to overexpress Grr1 without promoting substrate turnover [22,23]. Grr1Δ F-Flag was immunoprecipitated from cells, and extracts were probed for Ndd1-myc. As shown in Fig 3C, Ndd1-myc is specifically pulled down with Grr1Δ F-Flag, suggesting that Ndd1 is a direct substrate of Grr1. Grr1 interacts with its known substrates by binding to negatively charged phosphorylation on the substrates through Grr1’s positively charged leucine rich repeats, but there is no known consensus site for the Grr1 phosphodegron [29]. We found that Grr1 also contributed to the very fast turnover of Ndd1 in G1-arrested cells (Fig 3D, quantification shown in S2E Fig), suggesting that Cdk1-dependent phosphorylation of Ndd1 cannot be the only mechanism by which Grr1 recognizes Ndd1 as its substrate. In addition, we tested whether the Anaphase Promoting Complex (APC) was involved in Ndd1 turnover. Cdh1 is the APC adaptor protein active during G1. In cdh1Δ cells, Ndd1 is also stabilized (Fig 3E, quantification shown in S2F Fig), suggesting that APCCdh1 may also contribute to the very quick turnover of Ndd1 observed in G1.
Ndd1 stability in response to DNA damage requires Swe1
Our finding that Ndd1 was strongly stabilized in cells treated with the DNA damaging agent MMS (Fig 1A and Fig 1B) provided an explanation for previous work that had found Ndd1 was expressed at high levels in damaged-arrested cells [14,30,31]. Since Ndd1 turnover is mediated by Cdk1 activity, we examined whether Ndd1 stabilization in response to DNA damage required the kinase Swe1 (Fig 4A, quantified in Fig 4B). Swe1 is the S. cerevisiae homolog of Wee1, which inhibits Cdk1 by phosphorylation on a conserved tyrosine residue [32]. Wee1 homologs in other organisms are important for cell cycle arrest in response to DNA damage, but Swe1 is dispensable for this arrest in budding yeast [33,34]. Nonetheless, both Swe1 and its downstream inhibitory phosphorylation on Cdk1 accumulate in response to DNA damage [33,34], although there have been conflicting reports in the literature of the involvement of the damage checkpoint signaling pathway on the accumulation of Swe1 [30,35,36]. We find that both Swe1 and its downstream inhibitory phosphorylation on Cdk1 are present at high levels in cells arrested with MMS compared to cells arrested in metaphase with nocodazole (Fig 4C). Swe1 phosphorylation specifically inhibits Clb2-Cdk1 kinase function [37]. While Swe1 and the inhibitory phosphorylation of Cdk1 are dispensable for cell cycle arrest in response to DNA damage, Clb2 activity is partially inhibited [33,34]. Consistently, Cdk phosphorylation of Ndd1 has been shown to be mediated by Clb2-Cdk1 [7,38], suggesting that the checkpoint-mediated block to Ndd1 turnover is through Clb2/Cdk inhibition by Swe1 (see Discussion for more detail).
Fig. 4. Ndd1 stabilization in response to DNA damage requires Swe1 and Mec1/Tel1. 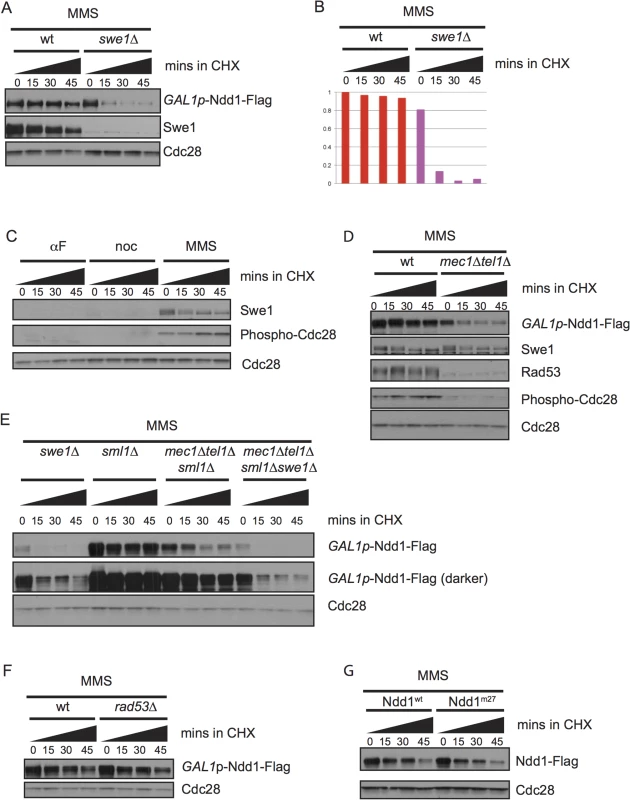
A) GAL1p-Ndd1-Flag was induced with galactose for 2.5 hours in wildtype or swe1Δ strains. Cells were then exposed to MMS for 2.5 hours in galactose. Cycloheximide (CHX) was added at t = 0, samples were taken every 15 minutes, blotted and probed for Swe1, Flag, and total Cdc28, as a loading control. B) Quantification of experiment shown in [A]. Y-axis shows the normalized signal above background normalized to the value at t = 0 in wildtype cells. C) Samples from Fig 1B were re-run and probed for Swe1 and Phospho-Y19 Cdc28 to follow Swe1 turnover in αF, noc, or MMS. Cdc28 loading control is reproduced from Fig 1B. D) GAL1p-Ndd1-Flag was induced with the addition of galactose in wildtype or mec1Δ tel1Δ strains. Cells were then exposed to 0.05% MMS for 4.5 hours. CHX was added at t = 0 and turnover was followed as in [A]. Rad53 is shown as a control for mec1Δ tel1Δ. Swe1 and Phospho-Y19 Cdc28 shown are from the same samples but run on a separate gel. Cdc28 is shown as a loading control. E) Experiment was performed as in [D] using swe1Δ, wt, mec1Δtel1Δ, swe1Δmec1Δtel1Δ strains. All mec1Δtel1Δ strains were rescued by simultaneous deletion of sml1Δ. F) Experiment was performed as in [D] with sml1Δ and rad53Δsml1Δ strains. G) Ndd1wt-Flag and Ndd1m27-Flag under the endogenous Ndd1 promoter were grown to mid-log phase (3–4 hours) and then treated with MMS for 2.5 hours. Cycloheximide was added at t = 0 and turnover was followed for 45 minutes with 15 minute timepoints taken as in [D]. We also found that stabilization of Ndd1 required the DNA damage checkpoint, as mec1Δtel1Δ mutants failed to stabilize Ndd1 after exposure to MMS (Fig 4D, quantification shown in S2G Fig). We found that Swe1 was stable independent of Mec1 and Tel1, consistent with an earlier report that Swe1 stability in response to DNA damage and replication stress was independent of Mec1 and its downstream kinase Rad53 [36]. We do, however, observe less Cdc28 inhibitory phosphorylation in the mec1Δtel1Δ mutants. A previous mass spectrometry study identifying proteome-wide phosphorylations in budding yeast treated with MMS did identify a phosphorylated residue on the phosphatase Mih1 that removes the Cdk1 inhibitory phosphorylation [39]. Though neither the signaling pathways upstream of the phosphorylation of Mih1 nor the downstream effects have been characterized, this result suggests that Mec1 and Tel1 could modulate Cdk1 signaling by affected Mih1. However, since Swe1 is required for the Cdk1-inhibitory phosphorylation, we found that the destabilization of Ndd1 observed in swe1Δ mutants was epistatic to that observed in mec1Δtel1Δ mutants (Fig 4E, quantification shown in S2H Fig).
Previously, our lab and others have identified Rad53-dependent phosphorylation of Ndd1 that inhibits its function in response to DNA damage signaling [11,13,14]. We therefore tested whether the stabilization of Ndd1 in response to DNA damage was controlled by the same pathway. Surprisingly, we found that Ndd1 stabilization is independent of both Rad53 (Fig 4F, quantification shown in S2I Fig) and the 27 Rad53-dependent phosphorylation sites that inhibit its function in response to DNA damage (Fig 4G, quantification shown in S2J Fig), suggesting that two opposing methods of regulation (inhibition and stabilization) independently converge on Ndd1 in response to DNA damage.
Discussion
In this work, we show that Ndd1 protein turnover is regulated during the cell cycle and in response to DNA damage. During G1, Ndd1 turnover reinforces the transcriptional regulation of Ndd1 to keep Ndd1 levels low. During metaphase, Ndd1 turnover requires Cdk1 activity (Fig 2C). We find that Ndd1 is stabilized in response to DNA damage (Fig 1A–1C), a result that explains several older observations that Ndd1 protein accumulated in response to DNA damage [14,30,31]. In response to DNA damage, Ndd1 is inhibited by phosphorylation [11,13,14], however the stabilization of Ndd1 we observe is independent of the known signaling and phosphorylation sites that we previously identified (Fig 4F and 4G). We propose a model in which Ndd1 is stabilized in damage, while its activity is independently inhibited (see Fig 5). Our speculation is that accumulated Ndd1 can be quickly de-phosphorylated during recovery following the repair of DNA damage to allow for rapid activation of its transcriptional targets to promote mitosis. Unfortunately, it is difficult to experimentally test this model, due to the fact that CDK phosphorylations promote both activation of Ndd1 and its destruction.
Fig. 5. Model for mitotic turnover of Ndd1. 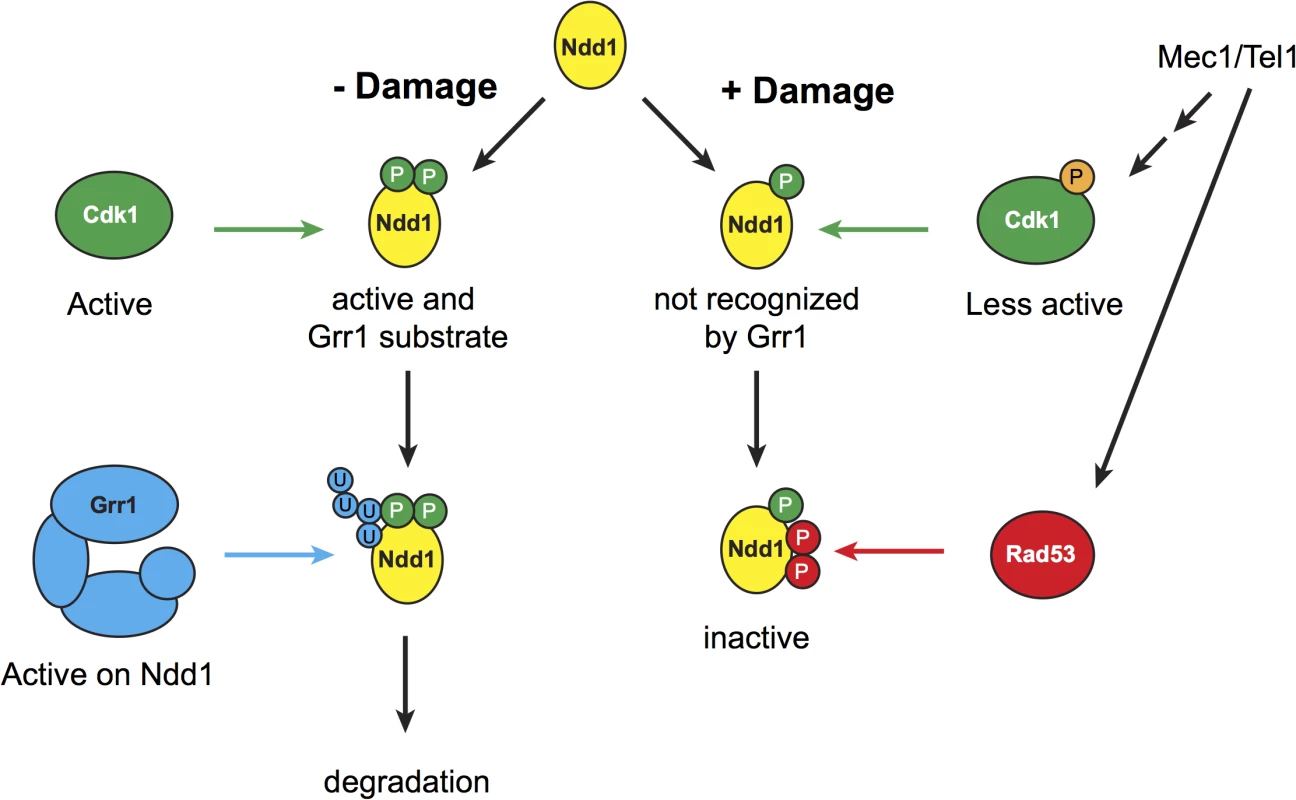
In the absence of DNA damage, Ndd1 is phosphorylated by Cdk1, which promotes Ndd1 transcriptional activity but also enables SCFGrr1 to recognize Ndd1 as a substrate. Ndd1 is therefore ubiquitinated and subsequently degraded. In response to DNA damage, Swe1 levels are increased, and Cdk1 is inhibited by phosphorylation which lowers its activity several fold. Ndd1 is no longer recognized by Grr1 and is therefore stabilized. Mec1 and Tel1 are important for maintaining high levels of phospho-Cdk1, and therefore help to stabilize Ndd1, either by promoting Swe1 activity or by inhibiting the phosphatase Mih1 (homolog of Cdc25). While Ndd1 is stabilized, its activity is independently inhibited by Rad53-dependent phosphorylation, leading to the accumulation of inhibited Ndd1. We speculate that maintaining this inhibited pool of Ndd1 may allow the cell to rapidly re-enter the cell cycle once the damage is repaired and the checkpoint is turned off. During metaphase, Cdk1 is required for Ndd1 activity. Phosphorylation of Ndd1 by Cdk1 is required both for its recruitment to target gene promoters, by generating a binding site for Fkh2, and for Ndd1’s intrinsic transactivator activity [7,9]. Here, we find that Cdk1 activity is also required to turn over Ndd1 in metaphase (Fig 2C). Recent work on a different cell cycle transcription factor, Hcm1, found that Cdk1 similarly promoted both the activity and the turnover of Hcm1 [40]. This suggests that dual, opposing regulation by Cdk1 may be a common theme in cell cycle regulation. Ndd1 has 16 potential Cdk1 sites that fit its minimal consensus sequence. A subset of these sites has been shown to be essential [9], and therefore presumably required for Ndd1 activation; however, it is still unknown whether other Cdk1 sites on Ndd1 are also essential or important for activation, and whether turnover of Ndd1 and activation of Ndd1 involve the same Cdk1 phosphorylation sites.
In other organisms, Cdk1 inhibitory phosphorylation by Wee1 is an important component of the cellular response to DNA damage and replication stress, with the checkpoint ultimately leading to high levels of Cdk1 phosphorylation to inhibit its function and block the G2/M transition [41–45]. In budding yeast, Cdk1 inhibitory phosphorylation is not required for the checkpoint to arrest cells in response to DNA damage [33,34]. Instead, Swe1 responds to cell wall stress through the morphogenesis checkpoint [46]. In addition, DNA damage has been shown to affect Swe1, although there have been conflicting reports on the effect of DNA damage or replication stress on Swe1 levels. One group found that Swe1 was stabilized in a MEC1 - and RAD53-independent manner [36]. Others have shown that Swe1 levels drop in response to prolonged HU treatment (which leads to replication stress by depleting nucleotides), in a checkpoint-dependent manner [35]. In addition, Swe1 has been shown to transcriptionally accumulate in response to DNA damage, in a Rad53-dependent manner [30]. Here, we find that our 2.5 hour treatment with MMS leads to high levels of Swe1, which is stable in our cycloheximide chase assays (Fig 4C). In addition, we find levels of Swe1 are high independent of DNA damage checkpoint signaling (Fig 4D). We find that DNA damage checkpoint signaling through Mec1 and Tel1 is required for the accumulation of high levels of phosphorylated Cdk1. Mec1 and Tel1 may modify Swe1 activity or may affect Cdk1 inhibition through modulation of the Mih1 phosphatase. In support of this idea, a phosphorylated residue on Mih1 was identified in a proteome-wide study from yeast treated with MMS, but the signaling pathways and the functional consequences have not been studied [39]. Different treatments (either with different drugs to induce damage or replication stress) or different timeframes could explain the observed differences in checkpoint signaling requirements for Swe1 and Cdk1 inhibitory phosphorylation.
Though Swe1 and the inhibitory phosphorylation of Cdk1 are not required for cell cycle arrest in response to DNA damage, our data (Fig 4C) and others [33,34] do find that Cdk1 inhibitory phosphorylation accumulates in response to DNA damage. While many groups have found that Cdk1 kinase is still active in response to DNA damage, Clb2-Cdk1 activity towards H1 is lowered 2 - to 4-fold relative to mitotic cells [33,34,47]. Phosphorylated Cdk1 specifically inhibits Clb2-dependent kinase activity [37], and, consistently, Ndd1 is a Clb2-Cdk1 substrate [7,38]. Furthermore, our results using the hypomorphic cdc28-as1 allele, which has 6-fold lower catalytic activity and significantly stabilizes Ndd1 even in the absence of the 1-NM-PP1 inhibitor, suggests that even modest decreases in Cdk1 activity strongly affect Ndd1 turnover. Even with the 6-fold kcat change in cdc28-as1, the doubling time of these cells is not particularly slow (140 minutes v 120 minutes), suggesting that for many of its functions, this level of activity must be sufficient [20]. Therefore, we propose that the two - to four-fold change in kinase activity in response to DNA damage may not affect the phosphorylation of many substrates; perhaps there are other residues in unfavorable positions that are very sensitive to even small changes in Cdk1 activity. Regardless, our results illustrate one of the first significant functions of Cdk1 inhibition in response to DNA damage.
Because Ndd1 is such an important regulator of mitosis through its regulation of transcription of the CLB2 cluster, many different kinases regulate Ndd1 activity during the cell cycle and in response to stresses such as DNA damage [5–7,9,11,13,14,38]. Here, we find that Ndd1 protein turnover is also controlled during the cell cycle and in response to DNA damage. Moreover, the regulation of Ndd1 protein turnover is independent of the Rad53-dependent inhibition of Ndd1 in response to DNA damage (Fig 4F and 4G). Ndd1 may function as an integration point for many different signals, allowing the cell to ultimately control mitosis.
Materials and Methods
Yeast methods
Yeast strains were grown in YM-1 media with 2% dextrose at 30°C unless otherwise noted. Strains were made using standard techniques. Expression from the GAL1 promoter was induced with 2% galactose. For experiments with temperature-sensitive strains, cells were maintained at 23°C until the experiment.
Cell cycle and damage treatments
For cell cycle experiments, cells were arrested with 10 μg/ml αg/ml ll μg/ml nocodazole, or 0.05% MMS for 2.5 hours or as noted. For 1-NM-PP1 inhibition of cdc28as-1, cells were arrested in nocodazole for 2.5 hours, then held in nocodazole or additionally treated with 1 μM 1-NM-PP1 for 1 more hour.
For FACS analysis of cell cycle progression, cells were fixed in 70% ethanol and stored at 4°C until analysis. Cells were sonicated and treated with 0.25 mg/ml RNase A for 1 hour at 50°C and then digested with 0.125 mg/ml Proteinase K for 1 hour at 50°C. Cells were then labeled with 1 μM Sytox green. Data were collected on a FACSCalibur machine and analyzed with FlowJo software.
Western blots
From cultures in mid-log phase, cell pellets of equivalent optical densities were collected, washed with 1 ml 4°C H2O, and frozen on dry ice. Pellets were thawed in boiling sample buffer (50 mM Tris pH 7.5, 5% SDS, 5 mM EDTA, 10% glycerol, 0.5% β-mercaptoethanol, bromophenol blue, 1 μg/ml leupeptin, 1 μg/ml bestatin, 0.1 mM benzamidine, 1 μg/ml pepstatin A, 5 mM NaF, 1 mM Na3VO4, 80 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride). Cells were boiled for 3 minutes, bead-beaten with glass beads for 3 minutes, and clarified by centrifugation. Extracts were analyzed by SDS-PAGE and Western blotting. Western blots were performed with low-salt phosphate buffered saline with Tween-20 (PBS-T) (15 mM NaCl, 1.3 mM NaH2PO4, 5.4 mM Na2HPO4, 0.05% Tween-20). Primary antibody incubations were performed in 5% nonfat dry milk and low salt PBS-T. Antibodies were used as follows: α-ilk and low salt PBS-T. Antibodies were used as follows:α-ilk and low salt PBS-T. Antibodies were used asαilk and low salt PBS-T. Antibodies were used as for α-Myc (9E10), α-Myc (9E10), saltD. Kellogg), and α-phospho-Cdc2 (Y15) (antibody was raised against human Cdc2 but recognizes S. cerevisiaie Cdc28 inhibitory phosphorylation as well, from Cell Signaling Technology #9111).
Half-life assays
Cells were grown as for Western blotting to mid-log. Cycloheximide was added to cultures for a final concentration of 50 μg/ml after collection of the t = 0 timepoint. Equivalent ODs were collected for each time point, and processed for Western blots as described above. For all experiments with Ndd1 under the control of the GAL1 promoter, cells were grown in galactose until the cells reached mid-log phase, from 2.5 to 8 hours, before the addition of cycloheximide or treatment for cell cycle arrest or with DNA damaging agent. For half-life experiments in arrested cells, cells were treated with αours, benocodazole, or MMS (still in the presence of galactose) for 2.5 hours before addition of cycloheximide. For cdc53-1 experiment, cells (including wild-type control) were grown in galactose for 5 hours and then shifted to 37°C for 2.5 hours.
To quantify gels, blots were scanned and quantified using Fiji from most informative exposure. Background levels from the same exposure gel were subtracted from each value. Quantified values were then normalized to corresponding Cdc28 loading control.
Grr1 binding experiment
Experiment was performed as in [22]. Briefly, strains carrying either pRS426 or pYES2-GRR1ΔF-FLAG-URA3 plasmids were grown in synthetic media lacking uracil with 2% raffinose. Grr1ΔF-Flag was induced with 2% galactose and cultures were allowed to double twice. Cells were lysed in a buffer containing 100 mM Tris-HCl (pH 7.5), 300 mM NaCl, 2 mM EDTA, 0.2% NP-40 with a Roche Complete protease inhibitor tablet without EDTA (one tablet/25 ml), 1 mM PMSF, and four Roche PhosSTOP Phosphatase Inhibitor Tablets (four tablets/25 ml). Lysis was carried out by bead beating, and cleared by centrifugation at 4°C. Lysates were incubated with a 25 μl slurry of anti-Flag M2 Magnetic Beads (Sigma-Aldrich) overnight while rotating at 4°C. Beads were washed three times with PBS buffer containing 0.1% NP-40. Proteins were eluted by mild vortexing in PBS buffer containing 0.1% NP-40 and 500 ng/ml 3X FLAG peptide (Sigma-Aldrich) for 45 min at room temperature. Samples shown are 0.2% of the total input and 20% of the Flag elution.
Supporting Information
Zdroje
1. Loy CJ, Lydall D, Surana U (1999) NDD1, a high-dosage suppressor of cdc28-1N, is essential for expression of a subset of late-S-phase-specific genes in Saccharomyces cerevisiae. Mol Cell Biol 19 : 3312–3327. 10207056
2. Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, et al. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9 : 3273–3297. 9843569
3. Koranda M, Schleiffer A, Endler L, Ammerer G (2000) Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406 : 94–98. 10894549
4. Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, et al. (2000) Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406 : 90–94. 10894548
5. Darieva Z, Bulmer R, Pic-Taylor A, Doris KS, Geymonat M, Sedgwick SG, et al. (2006) Polo kinase controls cell-cycle-dependent transcription by targeting a coactivator protein. Nature 444 : 494–498. 17122856
6. Darieva Z, Han N, Warwood S, Doris KS, Morgan BA, Sharrocks AD (2012) Protein kinase C regulates late cell cycle-dependent gene expression. Mol Cell Biol 32 : 4651–4661. doi: 10.1128/MCB.06000-11 22966207
7. Darieva Z, Pic-Taylor A, Boros J, Spanos A, Geymonat M, Reece RJ, et al. (2003) Cell cycle-regulated transcription through the FHA domain of Fkh2p and the coactivator Ndd1p. Current Biology 13 : 1740–1745. 14521842
8. Pic-Taylor A, Darieva Z, Morgan BA, Sharrocks AD (2004) Regulation of cell cycle-specific gene expression through cyclin-dependent kinase-mediated phosphorylation of the forkhead transcription factor Fkh2p. Mol Cell Biol 24 : 10036–10046. 15509804
9. Reynolds D, Shi BJ, McLean C, Katsis F, Kemp B, Dalton S (2003) Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev 17 : 1789–1802. 12865300
10. Veis J, Klug H, Koranda M, Ammerer G (2007) Activation of the G2/M-specific gene CLB2 requires multiple cell cycle signals. Mol Cell Biol 27 : 8364–8373. 17908798
11. Edenberg ER, Vashisht A, Benanti JA, Wohlschlegel J, Toczyski DP (2014) Rad53 downregulates mitotic gene transcription by inhibiting the transcriptional activator Ndd1. Mol Cell Biol 34 : 725–738. doi: 10.1128/MCB.01056-13 24324011
12. Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO (2001) Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell 12 : 2987–3003. 11598186
13. Jaehnig EJ, Kuo D, Hombauer H, Ideker TG, Kolodner RD (2013) Checkpoint kinases regulate a global network of transcription factors in response to DNA damage. Cell Rep 4 : 174–188. doi: 10.1016/j.celrep.2013.05.041 23810556
14. Yelamanchi SK, Veis J, Anrather D, Klug H, Ammerer G (2014) Genotoxic Stress Prevents Ndd1-Dependent Transcriptional Activation of G2/M-Specific Genes in Saccharomyces cerevisiae. Mol Cell Biol 34 : 711–724. doi: 10.1128/MCB.01090-13 24324010
15. Pramila T, Wu W, Miles S, Noble WS, Breeden LL (2006) The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev 20 : 2266–2278. 16912276
16. MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E (2006) An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7 : 113. 16522208
17. Okaz E, Arguello-Miranda O, Bogdanova A, Vinod PK, Lipp JJ, Markova Z, et al. (2012) Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell 151 : 603–618. doi: 10.1016/j.cell.2012.08.044 23101628
18. Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9 : 616–627. doi: 10.1038/nrm2450 18594563
19. Edenberg ER, Downey M, Toczyski D (2014) Polymerase stalling during replication, transcription and translation. Curr Biol 24: R445–452. doi: 10.1016/j.cub.2014.03.060 24845677
20. Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, et al. (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407 : 395–401. 11014197
21. Petroski MD, Deshaies RJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6 : 9–20. 15688063
22. Mark KG, Simonetta M, Maiolica A, Seller CA, Toczyski DP (2014) Ubiquitin ligase trapping identifies an SCF(Saf1) pathway targeting unprocessed vacuolar/lysosomal proteins. Mol Cell 53 : 148–161. doi: 10.1016/j.molcel.2013.12.003 24389104
23. Benanti JA, Cheung SK, Brady MC, Toczyski DP (2007) A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat Cell Biol 9 : 1184–1191. 17828247
24. Landry BD, Doyle JP, Toczyski DP, Benanti JA (2012) F-box protein specificity for g1 cyclins is dictated by subcellular localization. PLoS Genet 8: e1002851. doi: 10.1371/journal.pgen.1002851 22844257
25. Barral Y, Jentsch S, Mann C (1995) G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev 9 : 399–409. 7883165
26. Spielewoy N, Flick K, Kalashnikova TI, Walker JR, Wittenberg C (2004) Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol Cell Biol 24 : 8994–9005. 15456873
27. Flick KM, Spielewoy N, Kalashnikova TI, Guaderrama M, Zhu Q, Chang HC, et al. (2003) Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol Biol Cell 14 : 3230–3241. 12925759
28. Vallier LG, Coons D, Bisson LF, Carlson M (1994) Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics 136 : 1279–1285. 8013905
29. Hsiung YG, Chang HC, Pellequer JL, La Valle R, Lanker S, Wittenberg C (2001) F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol Cell Biol 21 : 2506–2520. 11259599
30. Bastos de Oliveira FM, Harris MR, Brazauskas P, de Bruin RA, Smolka MB (2012) Linking DNA replication checkpoint to MBF cell-cycle transcription reveals a distinct class of G1/S genes. EMBO J 31 : 1798–1810. doi: 10.1038/emboj.2012.27 22333912
31. Mazumder A, Pesudo LQ, McRee S, Bathe M, Samson LD (2013) Genome-wide single-cell-level screen for protein abundance and localization changes in response to DNA damage in S. cerevisiae. Nucleic Acids Res 41 : 9310–9324. doi: 10.1093/nar/gkt715 23935119
32. Booher RN, Deshaies RJ, Kirschner MW (1993) Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J 12 : 3417–3426. 8253069
33. Amon A, Surana U, Muroff I, Nasmyth K (1992) Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature 355 : 368–371. 1731251
34. Sorger PK, Murray AW (1992) S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature 355 : 365–368. 1731250
35. Enserink JM, Smolka MB, Zhou H, Kolodner RD (2006) Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae. J Cell Biol 175 : 729–741. 17130284
36. Liu H, Wang Y (2006) The function and regulation of budding yeast Swe1 in response to interrupted DNA synthesis. Mol Biol Cell 17 : 2746–2756. 16571676
37. Keaton MA, Bardes ES, Marquitz AR, Freel CD, Zyla TR, Rudolph J, et al. (2007) Differential susceptibility of yeast S and M phase CDK complexes to inhibitory tyrosine phosphorylation. Curr Biol 17 : 1181–1189. 17614281
38. Koivomagi M, Valk E, Venta R, Iofik A, Lepiku M, Morgan DO, et al. (2011) Dynamics of Cdk1 substrate specificity during the cell cycle. Mol Cell 42 : 610–623. doi: 10.1016/j.molcel.2011.05.016 21658602
39. Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7 : 1389–1396. doi: 10.1074/mcp.M700468-MCP200 18407956
40. Landry BD, Mapa CE, Arsenault HE, Poti KE, Benanti JA (2014) Regulation of a transcription factor network by Cdk1 coordinates late cell cycle gene expression. EMBO J 33 : 1044–1060. doi: 10.1002/embj.201386877 24714560
41. Jin P, Gu Y, Morgan DO (1996) Role of inhibitory CDC2 phosphorylation in radiation-induced G2 arrest in human cells. J Cell Biol 134 : 963–970. 8769420
42. O'Connell MJ, Raleigh JM, Verkade HM, Nurse P (1997) Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J 16 : 545–554. 9034337
43. Rhind N, Furnari B, Russell P (1997) Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev 11 : 504–511. 9042863
44. Rhind N, Russell P (1998) Tyrosine phosphorylation of cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol Cell Biol 18 : 3782–3787. 9632761
45. Rowley R, Hudson J, Young PG (1992) The wee1 protein kinase is required for radiation-induced mitotic delay. Nature 356 : 353–355. 1549179
46. Lew DJ (2003) The morphogenesis checkpoint: how yeast cells watch their figures. Curr Opin Cell Biol 15 : 648–653. 14644188
47. Stueland CS, Lew DJ, Cismowski MJ, Reed SI (1993) Full activation of p34CDC28 histone H1 kinase activity is unable to promote entry into mitosis in checkpoint-arrested cells of the yeast Saccharomyces cerevisiae. Mol Cell Biol 13 : 3744–3755. 8388545
Štítky
Genetika Reprodukční medicína
Článek Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in CrustaceansČlánek Adventures in WonderlandČlánek Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and InfectivityČlánek Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in FilamentsČlánek Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in ArabidopsisČlánek Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression ControlČlánek The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent EnhancersČlánek Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of MitochondriaČlánek The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma FormationČlánek Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 4- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- Adventures in Wonderland
- Experimental Swap of 's Assortative Mating Preferences Demonstrates Key Role of X-Chromosome Divergence Island in Incipient Sympatric Speciation
- Chromosome Replacement and Deletion Lead to Clonal Polymorphism of Berry Color in Grapevine
- The Protein Quality Control Machinery Regulates Its Misassembled Proteasome Subunits
- Genome-Wide Association Study Identifies as a Critical Gene for Susceptibility to Noise-Induced Hearing Loss
- Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and Infectivity
- Viable Neuronopathic Gaucher Disease Model in Medaka () Displays Axonal Accumulation of Alpha-Synuclein
- Multi-locus Analysis of Genomic Time Series Data from Experimental Evolution
- The Genetic Legacy of the Expansion of Turkic-Speaking Nomads across Eurasia
- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- The Pif1 Helicase, a Negative Regulator of Telomerase, Acts Preferentially at Long Telomeres
- Inhibiting K63 Polyubiquitination Abolishes No-Go Type Stalled Translation Surveillance in
- SYD-1C, UNC-40 (DCC) and SAX-3 (Robo) Function Interdependently to Promote Axon Guidance by Regulating the MIG-2 GTPase
- Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in Filaments
- Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3’ Tailing of Small RNAs in Arabidopsis
- Host Genetic Variation Influences Gene Expression Response to Rhinovirus Infection
- Contribution of Large Region Joint Associations to Complex Traits Genetics
- Volatility of Mutator Phenotypes at Single Cell Resolution
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in Arabidopsis
- A Multi-layered Protein Network Stabilizes the FtsZ-ring and Modulates Constriction Dynamics
- Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression Control
- Genome Sequencing of the Perciform Fish Provides Insights into Molecular and Genetic Mechanisms of Stress Adaptation
- Natural Variant E610G Is a Semi-dominant Suppressor of IAP-Induced RNA Processing Defects
- The Alkaline Response Pathway: Identification of a Novel Rim Pathway Activator
- Transgenerational Inheritance of Diet-Induced Genome Rearrangements in Drosophila
- A Single Nucleotide Polymorphism Uncovers a Novel Function for the Transcription Factor Ace2 during Hyphal Development
- DNA Damage Response and Spindle Assembly Checkpoint Function throughout the Cell Cycle to Ensure Genomic Integrity
- The Functional Interplay Between the t(9;22)-Associated Fusion Proteins BCR/ABL and ABL/BCR in Philadelphia Chromosome-Positive Acute Lymphatic Leukemia
- Extreme Recombination Frequencies Shape Genome Variation and Evolution in the Honeybee,
- Beyond Glycolysis: GAPDHs Are Multi-functional Enzymes Involved in Regulation of ROS, Autophagy, and Plant Immune Responses
- Comprehensive Profiling of Amino Acid Response Uncovers Unique Methionine-Deprived Response Dependent on Intact Creatine Biosynthesis
- Windpipe Controls Intestinal Homeostasis by Regulating JAK/STAT Pathway via Promoting Receptor Endocytosis and Lysosomal Degradation
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
- Cross-Population Joint Analysis of eQTLs: Fine Mapping and Functional Annotation
- The Power of Gene-Based Rare Variant Methods to Detect Disease-Associated Variation and Test Hypotheses About Complex Disease
- The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent Enhancers
- Competition between VanU Repressor and VanR Activator Leads to Rheostatic Control of Vancomycin Resistance Operon Expression
- A Missense Change in the Gene Links Aberrant Autophagy to a Neurodegenerative Vacuolar Storage Disease
- Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model
- Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of Mitochondria
- Genome-Destabilizing Effects Associated with Top1 Loss or Accumulation of Top1 Cleavage Complexes in Yeast
- Imputation-Based Population Genetics Analysis of Malaria Parasites
- Heterozygosity for a Hypomorphic Polβ Mutation Reduces the Expansion Frequency in a Mouse Model of the Fragile X-Related Disorders
- Neto-Mediated Intracellular Interactions Shape Postsynaptic Composition at the Neuromuscular Junction
- Ndd1 Turnover by SCF Is Inhibited by the DNA Damage Checkpoint in
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma Formation
- Spastin Binds to Lipid Droplets and Affects Lipid Metabolism
- Maintenance of Glia in the Optic Lamina Is Mediated by EGFR Signaling by Photoreceptors in Adult Drosophila
- Auxin Influx Carriers Control Vascular Patterning and Xylem Differentiation in
- Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
- The Lowe Syndrome Protein OCRL1 Is Required for Endocytosis in the Zebrafish Pronephric Tubule
- Postnatal Loss of Hap1 Reduces Hippocampal Neurogenesis and Causes Adult Depressive-Like Behavior in Mice
- CAPER Is Vital for Energy and Redox Homeostasis by Integrating Glucose-Induced Mitochondrial Functions via ERR-α-Gabpa and Stress-Induced Adaptive Responses via NF-κB-cMYC
- Distinct and Cooperative Activities of HESO1 and URT1 Nucleotidyl Transferases in MicroRNA Turnover in
- The Evolutionary Origination and Diversification of a Dimorphic Gene Regulatory Network through Parallel Innovations in and
- MAPK Signaling Pathway Alters Expression of Midgut ALP and ABCC Genes and Causes Resistance to Cry1Ac Toxin in Diamondback Moth
- Spatio-temporal Remodeling of Functional Membrane Microdomains Organizes the Signaling Networks of a Bacterium
- Asymmetric Transcript Discovery by RNA-seq in . Blastomeres Identifies , a Gene Important for Anterior Morphogenesis
- A Stress-Induced Small RNA Modulates Alpha-Rhizobial Cell Cycle Progression
- Systematic Profiling of Poly(A)+ Transcripts Modulated by Core 3’ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation
- The UPR Branch IRE1- in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast
- A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi
- Co-chaperone p23 Regulates . Lifespan in Response to Temperature
- Re-replication of a Centromere Induces Chromosomal Instability and Aneuploidy
- Shade Avoidance Components and Pathways in Adult Plants Revealed by Phenotypic Profiling
- Lipid-Induced Epigenomic Changes in Human Macrophages Identify a Coronary Artery Disease-Associated Variant that Regulates Expression through Altered C/EBP-Beta Binding
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



