-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
Non-obligate bacterial pathogens must alter their gene expression profiles when transitioning between environments. Vibrio cholerae is a natural inhabitant of aquatic ecosystems and the etiological agent of the severe diarrheal disease, cholera. Its virulence gene regulation is controlled by a complex transcriptional cascade involving a membrane-localized regulator termed ToxR. Here we show that ToxR undergoes proteolysis under nutrient limitation at alkaline pH and this loss is associated with the entry of V. cholerae into a dormant state, similar to that found in its natural environment between epidemics. Thus, to our knowledge, we provide the first evidence of a link between the proteolysis of a virulence regulator and the entry of a bacterial pathogen into an environmentally persistent state.
Published in the journal: . PLoS Genet 11(4): e32767. doi:10.1371/journal.pgen.1005145
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005145Summary
Non-obligate bacterial pathogens must alter their gene expression profiles when transitioning between environments. Vibrio cholerae is a natural inhabitant of aquatic ecosystems and the etiological agent of the severe diarrheal disease, cholera. Its virulence gene regulation is controlled by a complex transcriptional cascade involving a membrane-localized regulator termed ToxR. Here we show that ToxR undergoes proteolysis under nutrient limitation at alkaline pH and this loss is associated with the entry of V. cholerae into a dormant state, similar to that found in its natural environment between epidemics. Thus, to our knowledge, we provide the first evidence of a link between the proteolysis of a virulence regulator and the entry of a bacterial pathogen into an environmentally persistent state.
Introduction
The ability of microorganisms to alter their gene expression profiles when transitioning between environments is fundamental for their survival. Vibrio cholerae O1 is a non-obligate pathogen that switches between the human host, where it colonizes the intestinal epithelium, and the aquatic environment where it is found as free-living or attached to biotic and abiotic surfaces [1–4]. Upon entry of V. cholerae into the human host, the expression of its two major virulence factors is induced: the toxin co-regulated pilus (TCP), an essential intestinal colonization factor [5], and cholera toxin (CT), responsible for the diarrhea associated with the disease [5,6]. The expression of these factors is coordinately regulated at the transcriptional level by a virulence cascade involving a number of regulatory proteins [7]. Central to this cascade is the cooperation between two pairs of membrane-localized transcriptional regulators, TcpPH, encoded on the Vibrio Pathogenicity Island (VPI) [8], and ToxRS, encoded in the ancestral genome and also present in non-pathogenic isolates of V. cholerae. These findings indicate that ToxR has roles outside of the human host. Both TcpPH and ToxRS are required to activate the transcription of the master virulence regulator, ToxT [9–12]. ToxT, also encoded on the VPI, directly activates the expression of TCP and CT, as well as other genes [7,13,14].
Once intestinal colonization and proliferation have taken place, V. cholerae downregulates the expression of the virulence cascade [8,15,16]. In the classical biotype of V. cholerae, termination of virulence is mediated by proteolysis of the major virulence activator ToxT [9–12,17] as well as by the regulated intramembrane proteolysis (RIP) of TcpP [18]. RIP is a widely distributed mechanism in both prokaryotes and eukaroytes for responding to extracellular signals and stresses [19–21]. The initial proteolytic event in the RIP of TcpP is catalyzed by a currently unidentified site-1 protease, resulting in a TcpP species that is further degraded by the site-2 metalloprotease RseP (YaeL) [18]. The most widely studied example of RIP activates the σE-dependent envelope stress response [22,23]. This process involves RseA, a membrane localized anti-σ factor that arrests RpoE. In response to envelope stress, RpoE is released for interaction with RNA polymerase by a RIP event in which RseA is sequentially cleaved by the serine protease DegS at a periplasmic site, and then by RseP at an intramembrane site [24–26].
The downregulation of virulence gene expression late in the infection process coincides with the upregulation of genes that promote detachment of bacteria from the mucosal surface of the intestine and that enhance the survival of V. cholerae when it returns to the environment [27–29]. Regulatory systems involved in controlling the expression of these genes include the quorum sensing regulator HapR [27], the stationary phase alternative sigma factor RpoS [28], and the VarS/VarA two component system [29]. Once in the environment, biofilm formation and entry into a dormant state known as viable but nonculturable (VBNC) or conditionally viable environmental cells (CVEC), play crucial roles in the survival of V. cholerae by facilitating its environmental persistence within aquatic habitats during periods between epidemics [4,30–33]. CVEC are clumps of dormant cells embedded in a biofilm matrix that can be recovered using enriched culturing techniques [31]. Quorum sensing has been implicated in the regulation of CVEC [31,34]. Nonetheless, the molecular mechanisms governing entry into VBNC remain to be elucidated.
Microarray analysis has revealed that ToxR influences the expression of more than 150 genes in V. cholerae [35]. Besides virulence, regulated genes include those involved in cellular transport, energy metabolism, motility, and iron uptake. In addition, ToxR reciprocally regulates the expression of two outer membrane porins, OmpU and OmpT, in response to the nutritional status of the cell [36–38]. Furthermore, unlike tcpP, toxR expression is not altered under conditions that favor the maximal expression of virulence genes in the laboratory [39,40]. In this study, we show that under nutrient limitation at alkaline pH, ToxR levels decrease by a RIP mediated event. This process is dependent upon the metalloprotease RseP, which appears to function as a site-2 protease, and the σE-mediated periplasmic stress response, which likely provides a site-1 protease that contributes to the cleavage of ToxR prior to RseP. We determined that the loss of ToxR, either by proteolysis or genetically, is associated with the entry of V. cholerae into a dormant, nonculturable state that is similar to the VBNC state observed in the natural environment.
Results
ToxR levels are reduced in response to nutrient limitation at alkaline pH
The expression of ompU is activated by ToxR in nutrient abundant environments, such as in rich medium [36] or in the host [41], whereas the expression of ompT is derepressed in nutrient limited conditions, such as in minimal medium [38] or during the late stationary phase of growth [37]. Since the levels of ToxR increase in the presence of nutrients to facilitate ompU expression [38], we hypothesized that the levels of ToxR would decrease during the growth of V. cholerae in late stationary phase to facilitate ompT expression. To test this, wild-type classical biotype strain O395 was grown in LB medium (starting pH 7.0, unbuffered) for 12 and 48 hours. As shown in Fig 1A, the levels of ToxR after 48 hours were significantly reduced compared to the 12 hour culture. The final pH of the 48 hour culture was determined to be 9.2–9.3, indicating that the medium became alkaline during growth. To determine whether alkaline pH contributed to the loss of ToxR after 48 hours, V. cholerae was grown for 12 and 48 hours in LB medium with a starting pH of 9.3 (unbuffered) and also in LB medium buffered at pH 7.0 with 100 mM HEPES. As shown in Fig 1A, ToxR was undetectable in cultures grown for 48 hours in LB at pH 9.3, whereas its levels remained high at this pH when grown for only 12 hours. This indicates that, in LB, ToxR appears to start being proteolyzed once the cultures reach both alkaline pH and nutrient limitation. When the medium was buffered at pH 7.0, ToxR levels remained high even after 48 hours (Fig 1A). Thus, in late stationary phase, ToxR levels significantly decrease after 48 hours due to an increase in the pH. As a control we examined the stability of a ToxR-unrelated protein, GbpA, and found that these conditions did not affect the stability of GbpA (S1A Fig) [42]. We then determined the dynamics of ToxR proteolysis by measuring ToxR protein levels between 12 hours and 48 hours post-inoculation in LB pH 9.3 at 6 hour intervals (Fig 1B). We found that ToxR proteolysis begins around 24 hours of growth at LB pH 9.3 (Fig 1B). ToxR becomes undetectable between 42 and 48 hours of culture (Fig 1B).
Fig. 1. Proteolysis of ToxR during late stationary phase at alkaline pH. 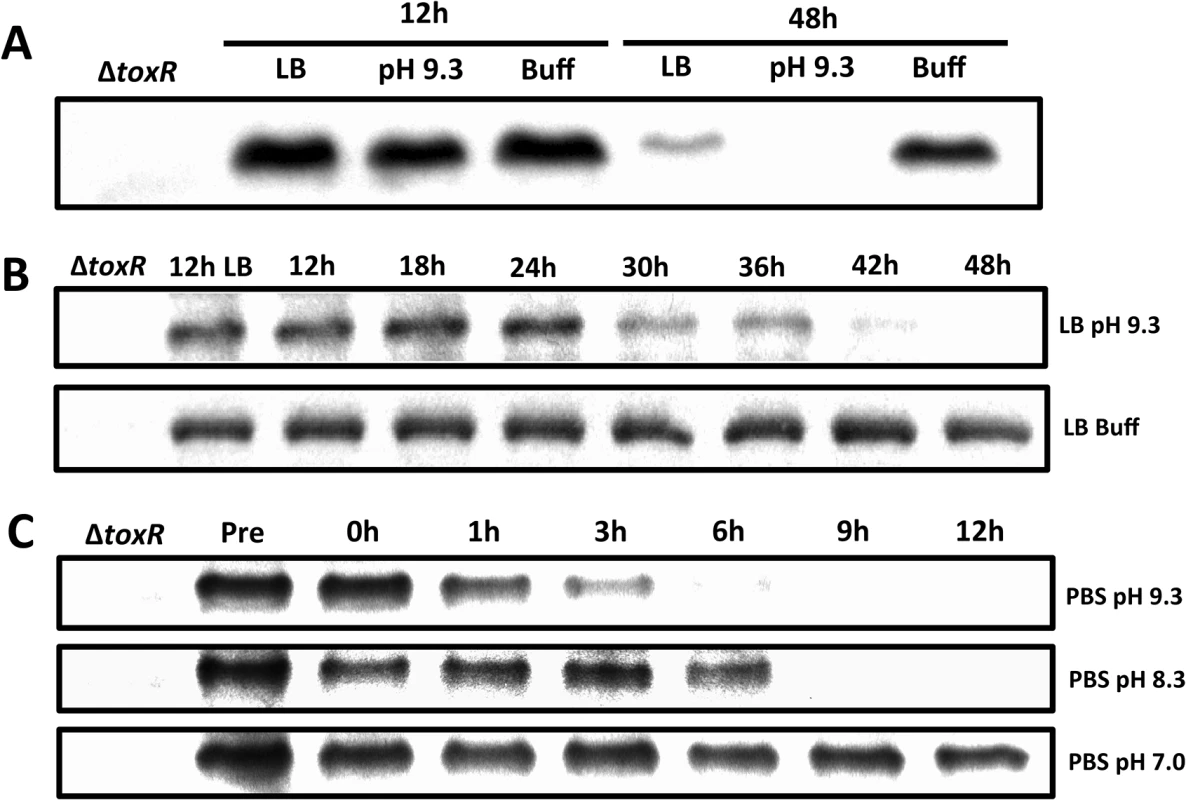
(A) ToxR immunoblot of O395 wild-type or ΔtoxR grown for either 12 or 48 hours in LB starting pH 7.0 unbuffered (LB), LB starting pH 9.3 unbuffered (pH 9.3), or LB buffered to pH 7.0 with 100 mM HEPES (Buff). (B) ToxR immunoblots of O395 wild-type grown at different time points in LB starting pH 9.3 unbuffered (pH 9.3), or LB buffered to pH 7.0 with 100 mM HEPES (Buff). (C) ToxR immunoblots of O395 wild-type grown overnight in LB starting pH 7.0 unbuffered at 37°C, pelleted, and resuspended in phosphate buffered saline (PBS) at pH 7.0, pH 8.3, or pH 9.3 for 12 hours. The observation that ToxR levels start decreasing after 24 hours at pH 9.3 (Fig 1B) suggests that nutrient limitation associated with late stationary phase might influence the levels of ToxR in stationary phase. To assess this, overnight cultures of O395 grown in LB at 37°C were pelleted and resuspended in phosphate buffered saline (PBS) at pH 7.0 or pH 9.3 for 12 hours. As shown in Fig 1C, ToxR levels decreased after only 3 hours in response to nutrient limitation at pH 9.3, whereas they did not significantly decrease in response to nutrient limitation at neutral pH. When overnight cultures were transferred to PBS at pH 8.3, the levels of ToxR started decreasing between 6 and 9 hours (Fig 1C). These results indicate that in late stationary phase, ToxR levels decrease in response to nutrient limitation at alkaline pH.
Proteolysis of ToxR during late stationary phase at alkaline pH occurs in an RseP and σE-dependent manner
The levels of TcpP have been shown to be controlled by RIP through the site-2 protease RseP [18]. We determined whether the levels of ToxR in response to nutrient limitation at alkaline pH may also be influenced by proteolysis through RseP. As shown in Fig 2A, ToxR was largely undetectable in strain O395 after growth for 48 hours in LB with a starting pH of 7.0 or 9.3, whereas the levels of ToxR in the ΔrseP mutant under these conditions were similar to wild-type grown in LB buffered at pH 7.0 with 100 mM HEPES. This finding indicates that RseP is involved in the proteolysis of ToxR during late stationary phase at alkaline pH, possibly functioning as a site-2 protease.
Fig. 2. Proteolysis of ToxR is RseP and RpoE-dependent. 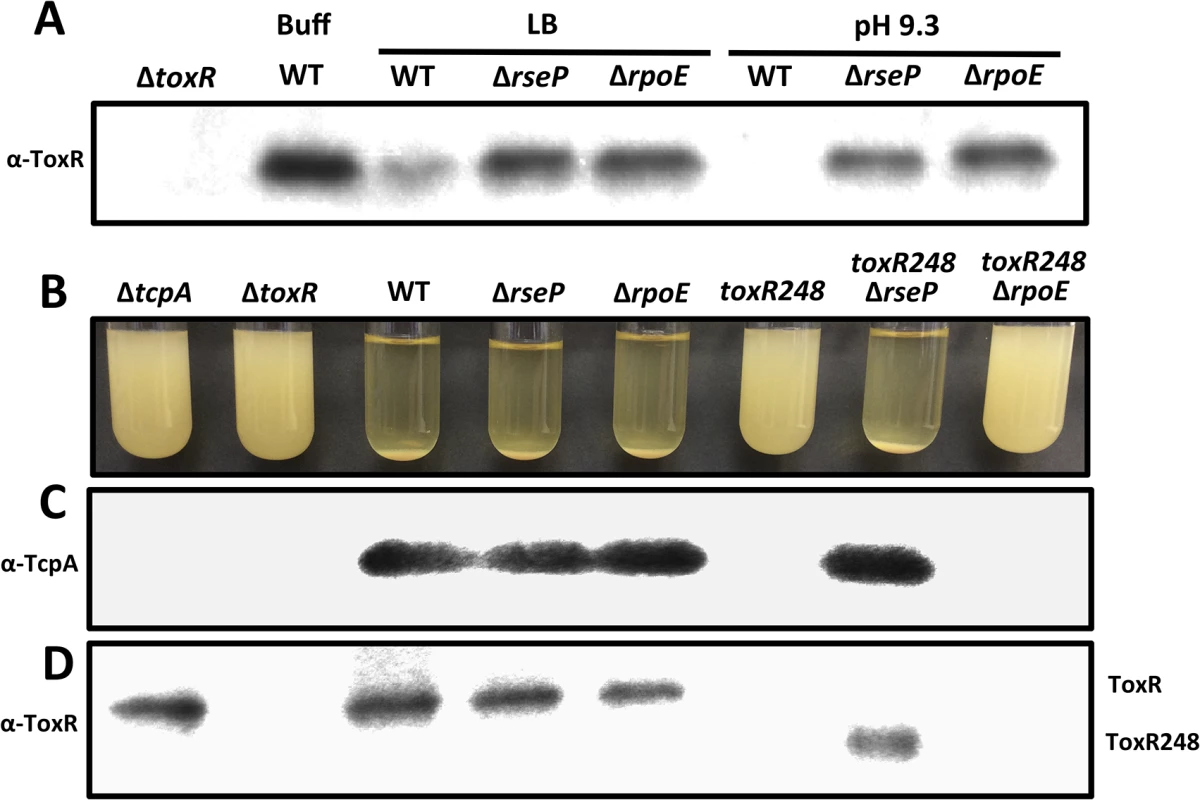
(A) ToxR immunoblot of cultures of O395 wild-type or ΔtoxR, ΔrseP or ΔrpoE grown for 48 hours in LB starting pH 7.0 unbuffered (LB), LB starting pH 9.3 unbuffered (pH 9.3), or LB buffered to pH 7.0 with 100 mM HEPES (Buff). (B) Autoagglutination of O395 ΔtcpA, ΔtoxR, wild-type (WT), ΔrseP, ΔrpoE, toxR248, toxR248ΔrseP and toxR248ΔrpoE grown under inducing conditions (LB starting pH 6.5, 30°C) for 15 hours. Autoagglutination can be visualized as a pellet at the bottom of the tube. (C) TcpA and (D) ToxR immunoblots of the cultures in (B). The ToxR activated porin OmpU functions as an outer membrane sensor responding to damage induced by antimicrobial peptides and triggers activation of the σE-dependent envelope stress response by promoting DegS-mediated cleavage of RseA [43]. We hypothesized that OmpU may also function as a sensor responding to alkaline pH during late stationary phase and induce the activation of the σE pathway, ultimately leading to the proteolysis of ToxR. To address this, we assessed the levels of ToxR in a ΔrpoE mutant after growth for 48 hours in LB with a starting pH of 7.0 or 9.3. As shown in Fig 2A, the levels of ToxR in the ΔrpoE mutant under these conditions were similar to the ΔrseP mutant. These findings indicate that both RseP and RpoE play a role in the proteolysis of ToxR during late stationary phase at alkaline pH.
Although the proteolysis of ToxR in late stationary phase at alkaline pH depends upon the σE pathway, this process is either independent of the porin OmpU, or OmpU is insufficient to trigger the proteolysis of ToxR since an ompU mutant did not restore ToxR levels under this condition (S1 Table). To further investigate the role of RpoE in the proteolysis of ToxR, we cloned rpoE into an expression vector, pBAD22, and tested whether ectopic overexpression of rpoE affected the stability of ToxR (S1B Fig). We found that ectopic expression of rpoE does not trigger the proteolysis of ToxR, as ToxR was detectable at every time point we measured (S1B Fig). In addition, we tested the effect of conditions that induce the σE pathway on the stability of ToxR (S1C Fig) [43,44]. We determined that growth in 3% ethanol or exposure to P2 antimicrobial peptide did not induce proteolysis of ToxR (S1C Fig). Thus, it appears that the σE pathway alone is not sufficient to induce the proteolytic cascade that culminates with the RIP of ToxR. It is possible that a second pathway might act in conjunction with the σE pathway to orchestrate the proteolysis of ToxR, in a similar manner as the Cpx pathway and the σE pathway work in combination in order to monitor the cell envelope in Escherichia coli [45,46]. Interestingly, the Cpx two-component system, which partially overlaps with the σE pathway in the periplasmic stress response [46,47] does not play a role in the proteolysis of ToxR. In late stationary phase at alkaline pH, deletion of cpxR, a regulator of the pathway, did not restore the levels of ToxR (S1 Table).
Periplasmic truncations of ToxR are proteolyzed in an RseP-dependent, σE-independent manner
The involvement of σE in the proteolysis of ToxR raises the possibility that the role of this pathway is to provide a site-1 protease that contributes to the cleavage of its periplasmic domain. Since RseP is known to activate the σE pathway [25,26], the function of RseP in the proteolysis of ToxR may be to activate this pathway, or to also function directly as the site-2 protease of ToxR. To assess this, we generated periplasmic truncations of ToxR that should bypass the requirement for a site-1 protease and assessed the roles of RseP and σE in the proteolysis of these truncations. If RseP functions indirectly in the proteolysis of ToxR the introduction of a ΔrseP mutation into strains carrying the truncations should have no influence on their stability. A periplasmic truncation was generated by introducing a stop codon in ToxR at position 249 (amber, toxR248). When cells are grown under inducing conditions the production of TCP allows for the formation of microcolonies, clusters of bacterial cells tethered together; this process is dependent on the production of the major pilin subunit TcpA [48]. Due to their size, microcolonies flocculate and form a pellet in a phenomenon known as autoagglutination [5,48]. As shown in Fig 2B, the strain carrying toxR248 failed to autoagglutinate, and neither TcpA nor ToxR could be detected by immunoblot (Fig 2C and2D) indicating that the truncated protein is proteolyzed. However, introduction of ΔrseP into the toxR248 strain restored autoagglutination (Fig 2B), TcpA production (Fig 2C) and produced an intermediate species of ToxR (Fig 2D) indicating that RseP influences the stability of the truncated ToxR protein. In contrast, a ΔrpoE mutation in toxR248 failed to autoagglutinate (Fig 2B) and did not stabilize the truncated ToxR protein (Fig 2D). These findings indicate that RseP plays a direct role in the cleavage of ToxR when a portion of its periplasmic domain is missing, and suggest that the effect of the σE pathway occurs prior to site-2 proteolysis of ToxR by RseP.
Culturability of V. cholerae decreases over time at alkaline pH
The proteolysis of ToxR under nutrient limitation at alkaline pH might provide an advantage to V. cholerae by preventing the expression of genes with roles in nutrient rich environments, such as OmpU, and promoting those with roles in nutrient poor conditions, such as OmpT [36–38]. Under conditions that are not conducive to active growth, such as nutrient limitation, V. cholerae is capable of entering a dormant, nonculturable state referred to as VBNC or CVEC that facilitates its survival and persistence [30,31,49–51]. We investigated whether the loss of ToxR is associated with the entry of V. cholerae into a nonculturable state. To assess this, the culturability of V. cholerae was measured by plating cultures of V. cholerae O395 between 12 and 48 hours after inoculation on LB medium with a starting pH of 9.3 and also in LB medium buffered at pH 7.0 with 100 mM HEPES at 6 hour intervals, and determining the colony forming units (CFUs) at each time point (Fig 3A). As shown in Fig 3A, the number of CFU/ml of O395 grown in LB 100 mM HEPES do not change over time whereas the number of CFU/ml of cultures grown on LB with a starting pH of 9.3 start getting reduced in some cultures around 24 hours of growth with a final CFU count nearly 5 logs lower than the cultures grown in LB buffered at pH 7.0 (Fig 3A). Interestingly, the culturability of V. cholerae is reduced more or less in parallel with the proteolysis of ToxR (Fig 1B). These results indicate that growth of V. cholerae to late stationary phase at alkaline pH, defined as ToxR proteolysis inducing (TPI) conditions, decreases the culturability of V. cholerae.
Fig. 3. V. cholerae shows reduced culturability over time at alkaline pH. 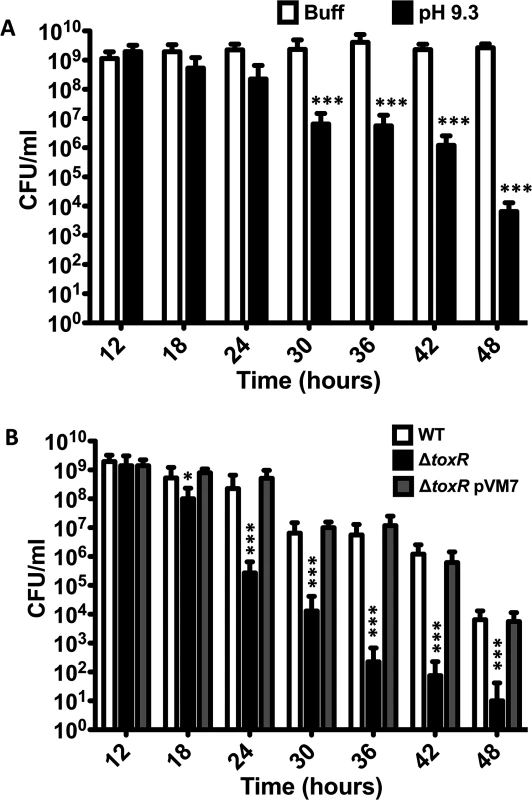
(A) CFU/ml of O395 wild-type strain grown at different time points in LB pH 7.0 with 100 mM HEPES (Buff), or LB starting pH 9.3 unbuffered (pH 9.3). The bars represent the mean of four independent experiments and the error bars indicate the standard deviation. Statistical comparisons were made using the student’s t-test and compare samples relative to 12h Buff. ***P < 0.0005. (B) CFU/ml of O395 wild-type (WT), ΔtoxR, or ΔtoxR pVM7 strains grown at different time points in LB starting pH 9.3 unbuffered. The bars represent the mean of four independent experiments and the error bars indicate the standard deviation. Statistical comparisons were made using the student’s t-test and compare samples relative to wild-type on LB pH 9.3 at that specific time point. *P < 0.05, ***P < 0.0005. There appears to be a correlation between the proteolysis of ToxR and the loss of culturability of V. cholerae as the levels of the protein decrease at a similar time point as V. cholerae begins to lose culturability (Fig 1A and3A). We assessed whether a ΔtoxR mutant strain would become nonculturable at a faster rate than the wild-type as, given that it will not produce ToxR from the beginning of the incubation period, it might lose culturability as soon as the suitable conditions are met. We found that, after 18 hours of growth on LB pH 9.3, when the nutrients are becoming scarce as the bacterium reaches mid-stationary phase, ΔtoxR starts losing culturability (Fig 3B and S2B Fig). After 48 hours of growth at LB pH 9.3 the number of CFUs recovered ranged between 0 and 102 (Fig 3B), indicating that ΔtoxR becomes nonculturable at a faster rate than the wild-type strain when cultured on LB pH 9.3. On the other hand, the number of CFUs remained relatively constant when ΔtoxR was cultured on LB buffered at pH 7.0 with 100mM HEPES, confirming that both alkaline pH and nutrient limitation are required for loss of culturability (S2 Fig). We also determined the culturability of a ΔtoxR strain harboring a plasmid that constitutively expresses toxR (pVM7) [9]. The strain ΔtoxR pVM7 shows a similar number of CFU/ml as the wild-type strain indicating that ectopic expression of ToxR recovers wild-type phenotype in the ΔtoxR strain (Fig 3B).
Growth curves for both wild-type and ΔtoxR on LB buffered and LB pH 9.3 show a similar pattern, indicating that there is no growth difference between the strains in these conditions (S2B Fig). Furthermore, a similar pattern, ΔtoxR losing culturability faster than wild-type at pH 9.3, was found when the wild-type strain and ΔtoxR were cultured on PBS pH 7 and PBS pH 9.3 (S3 Fig). The ΔtoxR strain starts losing culturability as soon as the cultures are transferred to nutrient limiting conditions (PBS) at alkaline pH (S3B Fig).
The loss of ToxR is required for loss of culturability of V. cholerae after 48 hours at alkaline pH
The same conditions that trigger the proteolysis of ToxR induce the loss of culturability of V. cholerae (Fig 1A and 3A). Additionally, a ΔtoxR mutant becomes nonculturable at a significantly faster rate than the wild-type strain and its phenotype can be complemented by ectopic expression of ToxR (Fig 3B). To further investigate the relationship between ToxR and the culturability of V. cholerae, the CFUs under TPI conditions were determined for wild-type O395, ΔtoxR, ΔrseP and ΔrpoE mutants. As shown in Fig 4, the number of CFU/ml for wild-type, which is able to proteolyze ToxR, was around 104 and the ΔtoxR mutant ranged between 0 and 102. In contrast, the number of CFU/ml for ΔrseP and ΔrpoE mutants that are unable to proteolyze ToxR was approximately 109 under this condition, similar to the number of CFUs recovered when the wild-type strain was grown on LB buffered at pH 7.0 with 100 mM HEPES (Fig 4). Introduction of the ΔtoxR mutation into the ΔrseP and ΔrpoE backgrounds decreased the CFUs under TPI conditions similar to that of the ΔtoxR mutant alone.
Fig. 4. Reduction in culturability of V. cholerae depends on the loss of ToxR. 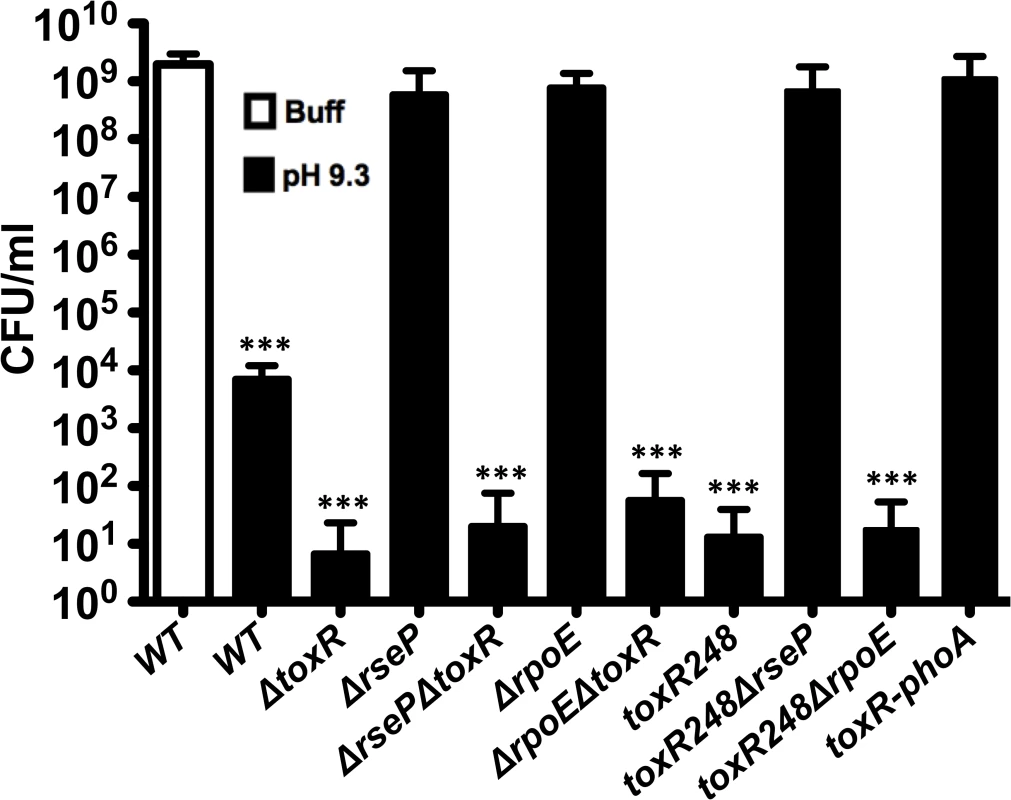
CFU/ml of O395 wild-type (WT) and mutant strains in LB pH 7.0 with 100 mM HEPES (Buff), or LB starting pH 9.3 unbuffered (pH 9.3) for 48 hours. The bars represent the mean of at least four independent experiments and the error bars indicate the standard deviation. Statistical comparisons were made using the student’s t-test and compare samples relative to wild-type 48h Buff. ***P < 0.0005. ToxR undergoes proteolysis in strains toxR248 and toxR248ΔrpoE (Fig 2). We found that both strains show a highly reduced number of CFU/ml under TPI conditions, similar to the ΔtoxR strain (Fig 4). The strain toxR248ΔrseP does not proteolyze ToxR, and an intermediate species of the regulator can be detected for this strain (Fig 2D). Consistently, the number of CFU/ml under TPI conditions for toxR248ΔrseP is similar to that of strains that cannot proteolyze ToxR such as wild-type under buffered conditions (Fig 4).
To further study the association between the loss of ToxR and loss of culturability of V. cholerae, we constructed a fusion strain, toxR-phoA, in which ToxR does not undergo proteolysis under TPI conditions (S3C Fig) [52]. The toxR-phoA fusion strain does not lose culturability under TPI conditions and show a similar number of CFUs as the strains that do not proteolyze ToxR (wild-type on LB buffered, ΔrseP, ΔrpoE, or toxR248ΔrseP) (Fig 4). Thus, the absence of ToxR, either by proteolysis or genetically, appears to be required for the loss of culturability of the strains.
V. cholerae remains viable and displays an altered cellular morphology under TPI conditions
To determine whether the decreased culturability of the wild-type and some of the mutants under TPI conditions is due to their entry into a dormant state and not cell death, the viability of the cells in the cultures in Fig 4 were examined using the LIVE/DEAD BacLight Bacterial Viability and Counting Kit. This kit allows for the discernment between viable cells (green) and dead cells (red) using fluorescent microscopy. The entry of V. cholerae into a dormant state is associated with loss of culturability (Fig 4), maintenance of viability (green cells), and a change in its morphology from elongated rods to round, coccoid-shaped cells [53,54]. Dead cells (O/N HK) can be seen as red under fluorescence (F), whereas cells from an overnight culture (O/N) are green and elongated (F and DIC) (Fig 5). We found that when grown in LB pH 7.0 buffered with 100 mM HEPES (48h Buff) the wild-type shows a morphology and viability similar to O/N (Fig 5), indicative of a culturable state. Under TPI conditions (48h pH 9.3), the cells remain viable and alive (green) and their morphology changes to a coccoid form, indicative of entry into a dormant state (Fig 5).
Fig. 5. Viability and morphology of V. cholerae after 48 hours at alkaline pH. 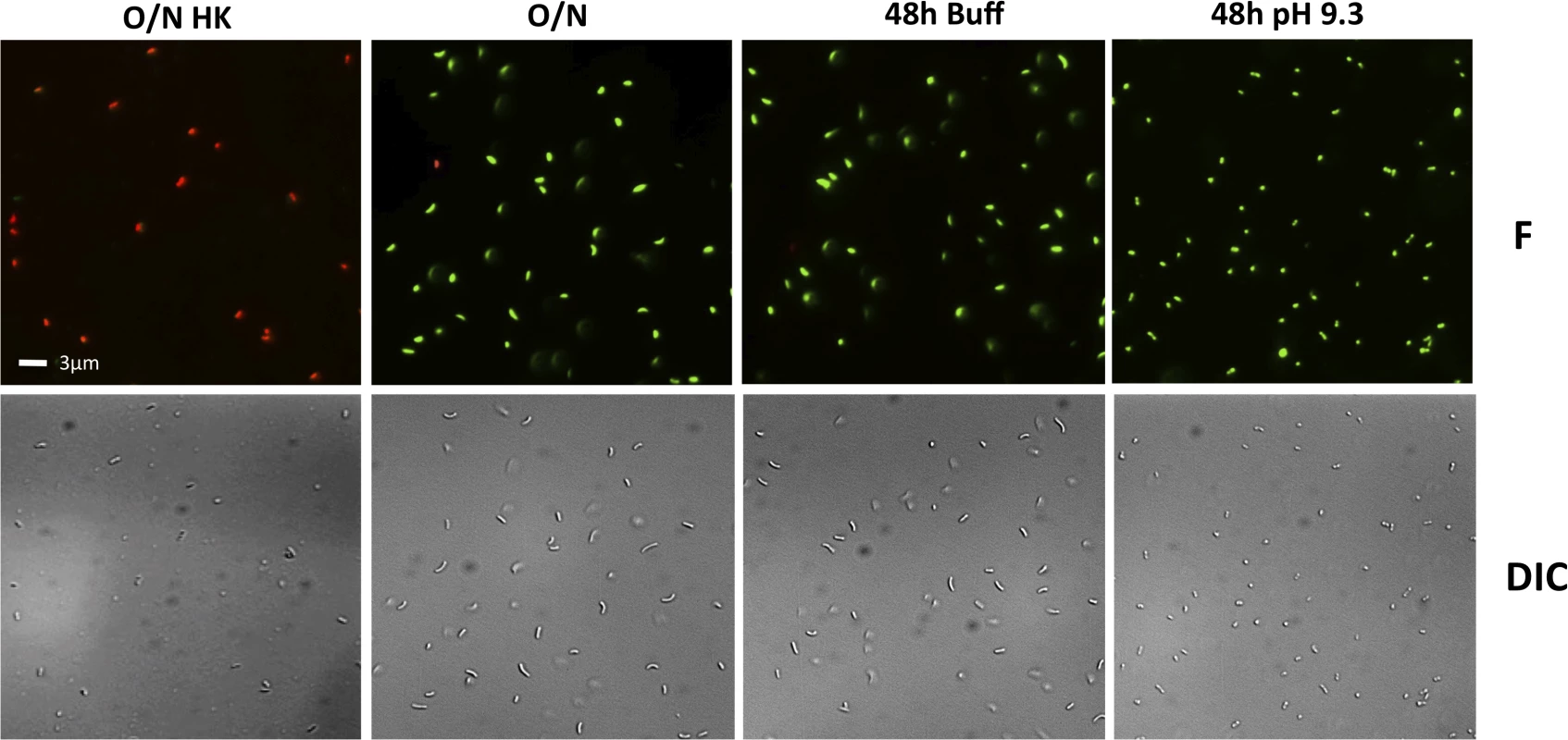
Fluorescent (F) and differential interference contrast (DIC) images of O395 wild-type grown in LB starting pH 7.0 unbuffered overnight and heat killed (O/N HK), LB starting pH 7.0 unbuffered overnight (O/N) as controls, 48 hours in LB buffered to pH 7.0 with 100 mM HEPES (48h Buff), and 48 hours in LB pH 9.3 unbuffered (48h pH 9.3). The viability and morphology of the mutant strains tested in Fig 4 is consistent with their culturability (Fig 6). The strains that cannot proteolyze ToxR (ΔrseP, ΔrpoE, toxR248ΔrseP, and toxR-phoA) show a similar morphology and viability to cells in O/N cultures (Figs 5 and 6), whereas the mutants that do not encode ToxR (ΔtoxR, ΔrsePΔtoxR, and ΔrpoEΔtoxR) or proteolyze it (toxR248 and toxR248ΔrpoE) are viable and round (Fig 6). These findings indicate that in V. cholerae the loss of ToxR is associated with entry into a dormant state.
Fig. 6. Viability and morphology of V. cholerae mutants after 48 hours at alkaline pH. 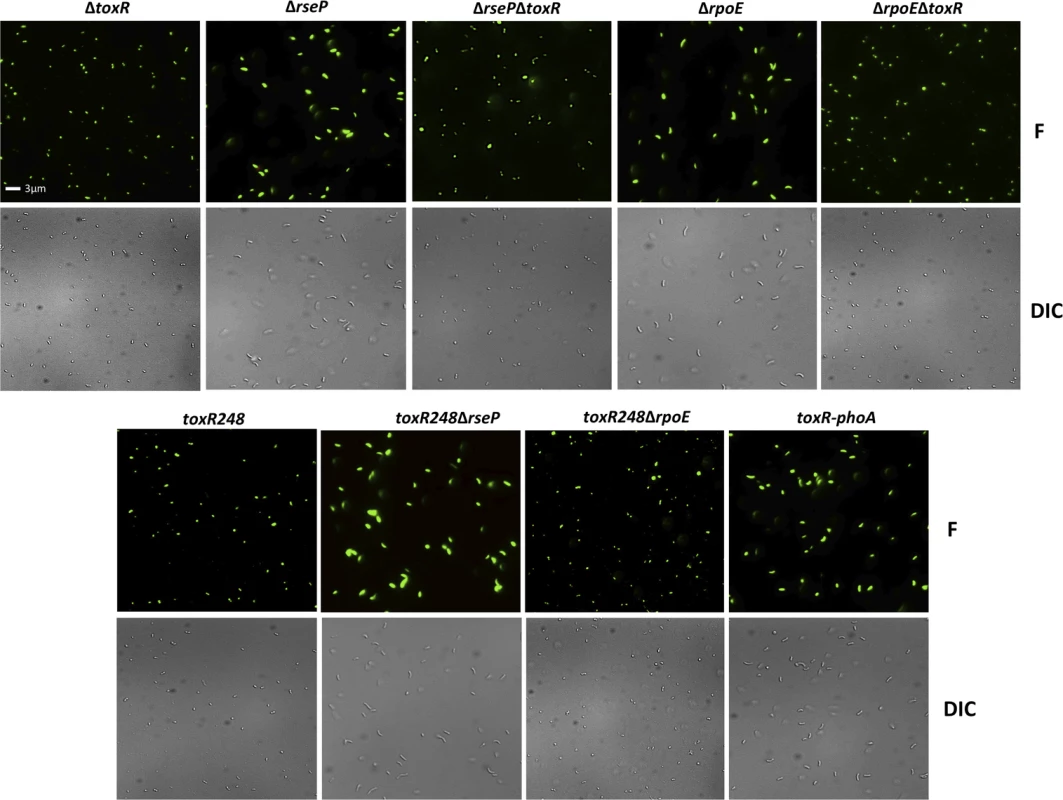
Fluorescent (F) and differential interference contrast (DIC) images of O395 ΔtoxR, ΔrseP, ΔrsePΔtoxR, ΔrpoE or ΔrpoEΔtoxR, toxR248, toxR248ΔrseP, toxR248ΔrpoE, and toxR-phoA grown for 48 hours in LB starting pH 9.3 (unbuffered). The cells were observed after treatment with the LIVE/DEAD BacLight Bacterial Viability and Counting Kit. Viable and culturable cells appear green and elongated; viable but dormant cells appear green and round; dead cells appear red and round. The loss of ToxR is also associated with entry of V. cholerae El Tor biotype and Vibrio parahaemolyticus into a dormant state
V. cholerae is classified into more than 200 serogroups, however, only the O1 serogroup causes epidemic cholera. V. cholerae O1 is further classified into two biotypes, classical and El Tor. Strain O395, where previous studies regarding termination of virulence had been made, belongs to the classical biotype [17,18]. Strains of the El Tor biotype are the source of the current pandemic of cholera and were recently shown to enter VBNC differentially when compared with classical [55]. We determined the effect of TPI conditions in the El Tor biotype strain N16961. We found that severely reduced levels of ToxR in late stationary phase were also observed with the El Tor biotype strain N16961 (S4A Fig). In this strain, it was necessary to incubate for 72 hours at pH 9.3 to visualize complete loss of ToxR by immunoblot. For N16961, the number of CFU/ml of cultures grown for 72 hours in LB with a starting pH of 9.3 was over 4 logs lower than the number of CFU/ml of cultures grown in LB buffered at pH 7.0 with 100mM HEPES (Fig 4B). We also found that when grown in LB pH 7.0 buffered with 100 mM HEPES, N16961 remains viable (green) and elongated (S4C Fig), indicative of a culturable state whereas under TPI conditions, the cells are viable and round (S4C Fig), indicative of entry into a dormant state.
V. parahaemolyticus is an intestinal pathogen that causes bloody diarrhea and is generally transmitted through the consumption of raw or uncooked fish. Recently, Whitaker et al showed that ToxR is required for intestinal colonization of V. parahaemolyticus in the adult murine model [56]. V. parahaemolyticus is known to enter VBNC under starvation conditions [57]. We determined whether the entry of V. parahaemolyticus into dormancy was also associated with loss of ToxR. Unexpectedly, V. parahaemolyticus cannot grow on LB with a starting pH of 9.3, however, like V. cholerae, it also alkalinizes the pH of the media during growth. Nonetheless, we found that ToxR undergoes proteolysis in V. parahaemolyticus RIMD2210633 when the bacterium is cultured on LB pH 7.0 unbuffered for 48 hours (Fig 7A). Furthermore, the bacterium loses culturability and adopts a viable coccoid form under unbuffered conditions and nutrient limitation (Fig 7B and 7C). Our data suggests that entry into dormancy might be mediated by ToxR proteolysis among other species of the family Vibrionaceae encoding ToxR.
Fig. 7. Effect of nutrient limitation in the stability of ToxR, culturability and morphology of Vibrio parahaemolyticus. 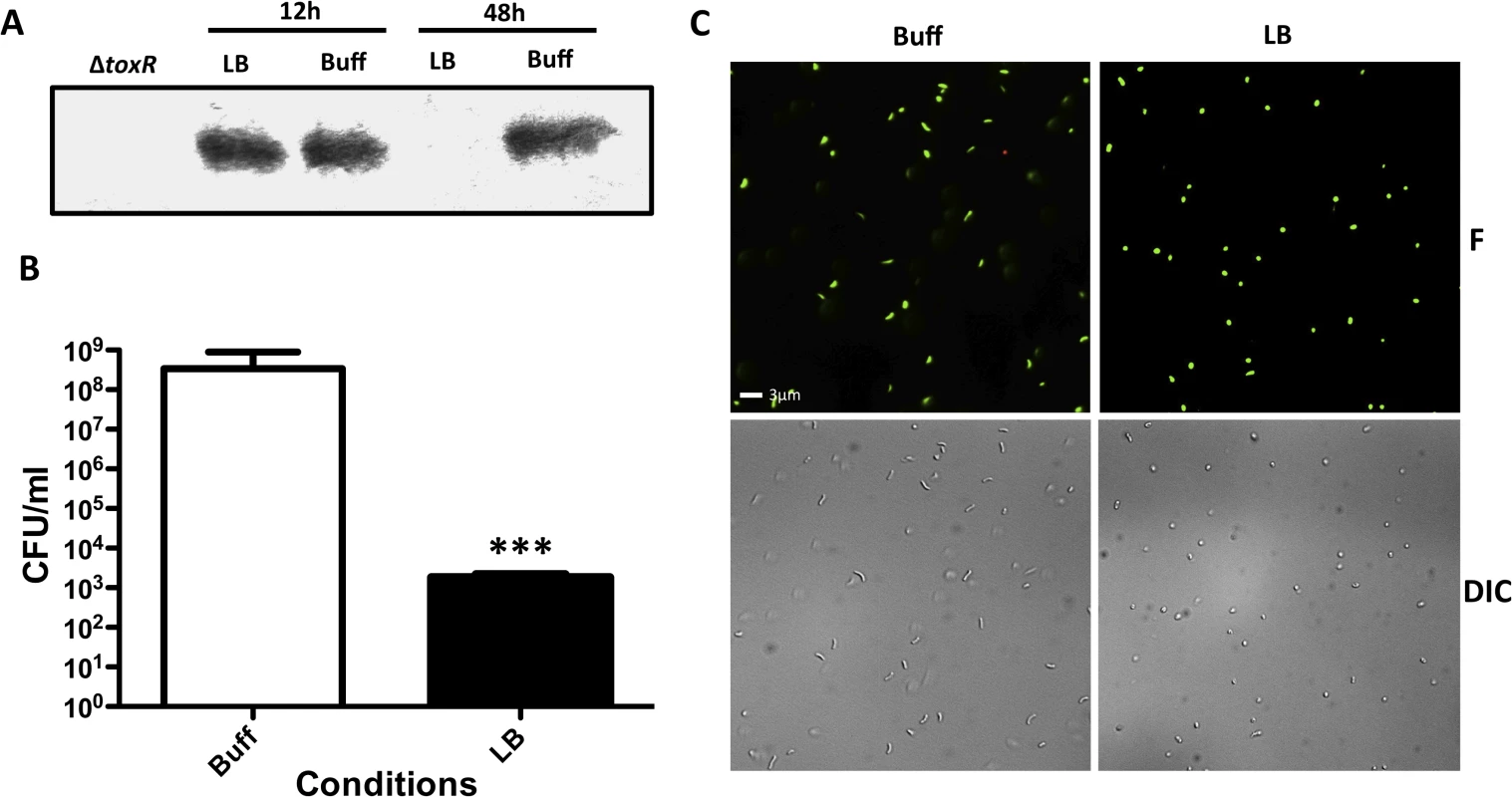
(A) ToxR immunoblot of V. parahaemolyticus RIMD2210633 wild-type or ΔtoxR grown for either 12 or 48 hours in LB starting pH 7.0 unbuffered (LB), or LB buffered to pH 7.0 with 100 mM HEPES (Buff) (B) Culturability of V. parahaemolyticus RIMD2210633 grown for 48 hours in LB starting pH 7.0 unbuffered (LB), or LB buffered to pH 7.0 with 100 mM HEPES (Buff). The bars represent the mean of four independent experiments and the error bars indicate the standard deviation. Statistical comparisons were made using the student’s t-test and compare samples relative to 48h Buff. ***P < 0.0005. (C) Fluorescent (F) and differential interference contrast (DIC) images of V. parahaemolyticus RIMD2210633 grown for 48 hours under conditions as in (B). The cells were observed after treatment with the LIVE/DEAD BacLight Bacterial Viability and Counting Kit. Viable and culturable cells appear green and elongated; viable but dormant cells appear green and round; dead cells appear red and round. Discussion
The mechanisms by which V. cholerae terminates virulence and prepares for entry into the aquatic environment have recently begun to be elucidated. In this study, we found a link between the loss of virulence regulator ToxR and entry of V. cholerae into a nonculturable, dormant state. Unlike TcpP, which functions primarily in virulence gene regulation, ToxR is present in non-pathogenic strains of V. cholerae and other members of the family Vibrionaceae, indicating it has roles in addition to virulence. The association between the proteolysis of ToxR and entry into a dormant state sheds new light on the function of ToxR outside of the human host and the molecular mechanisms for entry into an environmentally persistent state.
We show here that ToxR in V. cholerae O395 undergoes proteolysis in late stationary phase (by 48 hours) as the medium is depleted for nutrients and becomes alkaline. That alkaline pH decreases the stability of ToxR is interesting in light of the fact that ToxR mediated activation of ompU expression plays a protective role for V. cholerae during organic acid stress [58]. This suggests that upregulation of ToxR repressed genes, such as ompT, once ToxR has been proteolyzed may protect V. cholerae under alkaline stress. The conditions identified here that induce the proteolysis of ToxR appear to be specific since we examined a wide variety of other potential stimuli and none were found to influence the stability of ToxR in V. cholerae.
The finding that RseP, the site-2 protease that influences the levels of TcpP through RIP [18], also influences the levels of ToxR in response to nutrient limitation at alkaline pH raised the possibility that ToxR might be subject to RIP. RIP is a mechanism commonly used by bacteria to rapidly adapt to changes in their environment that are important for their survival and establishment [19,20]. One of the best-studied examples of RIP activates the expression of the stress response genes associated with the σE pathway [22,23]. Other important processes regulated by RIP include sporulation [59], cell division [60], pheromone production [61], quorum sensing [62] and biofilm formation [63,64].
We also found a link between the σE dependent stress response pathway and the proteolysis of ToxR. The σE pathway has been shown to play important roles in the virulence of V. cholerae and other Vibrio species and is induced under alkaline pH in Yersinia pseudotuberculosis [44,65–68]. The role of RpoE in the proteolysis of ToxR raised the possibility that RseP might not function directly as the site-2 protease of ToxR but instead might act indirectly through its ability to activate the σE pathway. To assess this, a periplasmic truncation of ToxR that would bypass the requirement for a site-1 protease, and which has been previously shown to decrease ToxR stability, was analyzed [52,69]. This truncation was found to be proteolyzed in an RseP-dependent, RpoE-independent manner, indicating that RseP plays a direct role in the proteolysis of ToxR.
We have thus far been unable to identify any other factors that play a role in the proteolysis of ToxR. Deletion of ompU, encoding a protein that acts as an outer membrane sensor that responds to damage induced by antimicrobial peptides and triggers activation of the σE pathway [43] did not restore the levels of ToxR in late stationary phase at alkaline pH. This led us to determine whether the σE pathway was sufficient to trigger the proteolysis of ToxR. Given that ectopic expression of rpoE and induction of the σE pathway are not sufficient to trigger the proteolysis of ToxR, it appears that a second pathway might work synergistically with the σE pathway in order to initiate the RIP of ToxR. Deletion of Cpx, which partially overlaps with the σE pathway did not restore wild-type levels of ToxR under TPI conditions [46,47]. RpoS, a stationary phase sigma factor, which has been shown to influence the culturability of classical biotype V. cholerae [55], did not influence the levels of ToxR in late stationary phase at alkaline pH. DegS, the site-1 protease involved in activation of the σE pathway in E. coli, also did not influence the proteolysis of ToxR, nor did the σE-regulated proteases DegP or VC0554 (S1 Table) [70].
It seemed that the proteolysis of ToxR under nutrient limitation at alkaline pH might provide an advantage to V. cholerae by increasing its ability to survive in nutrient depleted environments outside of the host. Bacteria have evolved a variety of adaptive responses that allow them to survive when conditions are not conducive to active growth. One such response is their ability to enter a dormant state where they remain viable, but are no longer culturable [71,72]. We found that under nutrient limitation at alkaline pH V. cholerae becomes nonculturable, and this occurs as the levels of ToxR are reduced. The cells remained viable as they became nonculturable, consistent with their entry into a dormant state [53]. Nutrient limitation, along with other factors such as temperature or salinity, have been previously shown to influence entry of V. cholerae into VBNC [51,72–74]. To our knowledge this is the first time that alkaline pH has been found to affect the entry of V. cholerae or other member of the Vibrionaceae into a dormant state.
From our data it can be gleaned that V. cholerae has evolved a tightly regulated response to avoid premature proteolysis of ToxR. As our results show, the presence of both nutrient limitation and alkaline pH are required to initiate the proteolysis of ToxR and the changes associated with it. Furthermore, short-term exposure to these conditions does not immediately trigger the proteolytic cascade of ToxR, indicating that the cells possess mechanisms that prevent untimely proteolysis of the virulence regulator. Given the drastic metabolic changes associated with entry into dormancy, it is possible that cells avoid proteolysis of ToxR when nutrient limitation and alkaline conditions are only transient.
We found a time difference in the proteolysis of ToxR between V. cholerae N16961 and O395. While ToxR cannot be detected in O395 cultures grown in LB for 48 hours at pH 9.3, ToxR can still be detected, even though at significantly reduced levels, in N16961 cultures under the same conditions. Nonetheless, ToxR becomes undetectable when the cultures of N16961 are incubated for a longer time. The loss of ToxR was also associated with entry into a dormant state of N16961. Our results are consistent with previous findings showing differential entry into dormancy between classical and El Tor biotypes [55]. In the aquatic environment, a significant proportion of toxigenic V. cholerae can be found as VBNC [30–32]. Thus, in the wild-type, the proteolysis of ToxR is associated with the formation of a dormant state that appears similar to the VBNC/CVEC state that is observed in the natural environment. Further work is needed to determine how V. cholerae can be recovered from this dormant state and whether this recovery is associated with the presence of ToxR.
V. parahaemolyticus, an intestinal pathogen that causes a bloody diarrhea, is known to enter VBNC under starvation conditions [57]. Interestingly, ToxR is also required for intestinal colonization of this pathogen [56]. We found that ToxR undergoes proteolysis in V. parahaemolyticus under alkaline conditions and nutrient limitation. Furthermore, similar to V. cholerae, the bacterium also loses culturability and adopts a viable coccoid form under these conditions. ToxR is encoded among other members of the family Vibrionaceae [75–78]. Our data suggests that the association between the loss of ToxR and entry into VBNC might be a widespread phenomenon among those species. In Photobacterium profundum ToxR becomes undetectable by immunoblot when cultured at high pressure (272 atm) relative to atmospheric pressure (1 atm) [79]. It has been suggested that ToxR is proteolyzed under high pressure and its loss may serve to increase the production of the porin OmpH (analogous to that of OmpT), which has a larger channel than OmpL (analogous to OmpU) [79,80]. This trait could be important in the deep-sea where nutrients are particularly scarce [79,80]. Furthermore, strains of P. profundum with mutations in an rpoE-like locus are pressure sensitive, suggesting that RpoE might also play a role in the proteolysis of ToxR in this species [81].
It remains to be determined at which stage of the V. cholerae life cycle ToxR undergoes proteolysis. It is tempting to speculate that the proteolysis of ToxR occurs during the late stages of infection, as a consequence of the resulting nutrient limitation and alkaline pH in the intestine [27,29,82]. The proteolysis of ToxR at this stage in the life cycle of V. cholerae would contribute to the termination of virulence and upregulate genes repressed by ToxR, such as ompT, that play a role in environmental survival. Consistent with this hypothesis is the finding that several genes involved in glycerol metabolism, which are downregulated by ToxR [35], have been found to play a role in survival in pond water [27]. Unlike ToxR, proteolysis of TcpP does not appear to be involved in the entry into VBNC since cultures transferred from inducing to non-inducing conditions, which triggers proteolysis of TcpP, do not lose culturability and keep growing in the media they are transferred to (S5 Fig).
A sequential model for the RIP of ToxR during late stationary phase is shown in Fig 8. In the early stages of colonization, when nutrients are abundant, ToxR upregulates the expression of genes such as ompU and those involved in virulence and downregulates the expression of genes such as ompT with roles in environmental survival. During the late stages of colonization, as nutrients become depleted and the environment becomes alkalinized, ToxR is proteolyzed. This occurs due to activation of the σE pathway via sequential degradation of RseA by either DegS or another site-1 protease and RseP, the site-2 protease, which releases RpoE to activate its regulated genes. It is possible that one of these genes may encode a site-1 protease that cleaves a periplasmic portion of ToxR, however, at least a second protease/system appears to be necessary for the site-1 proteolytic event to occur. Next, RseP cleaves at an intramembrane site within ToxR leading to full proteolysis of the regulator. This process induces and prevents, respectively, expression of ToxR repressed and activated genes, providing an advantage in the environment. The loss of ToxR ultimately leads to entry into dormancy.
Fig. 8. Sequential model for the RIP of ToxR during late stationary phase. 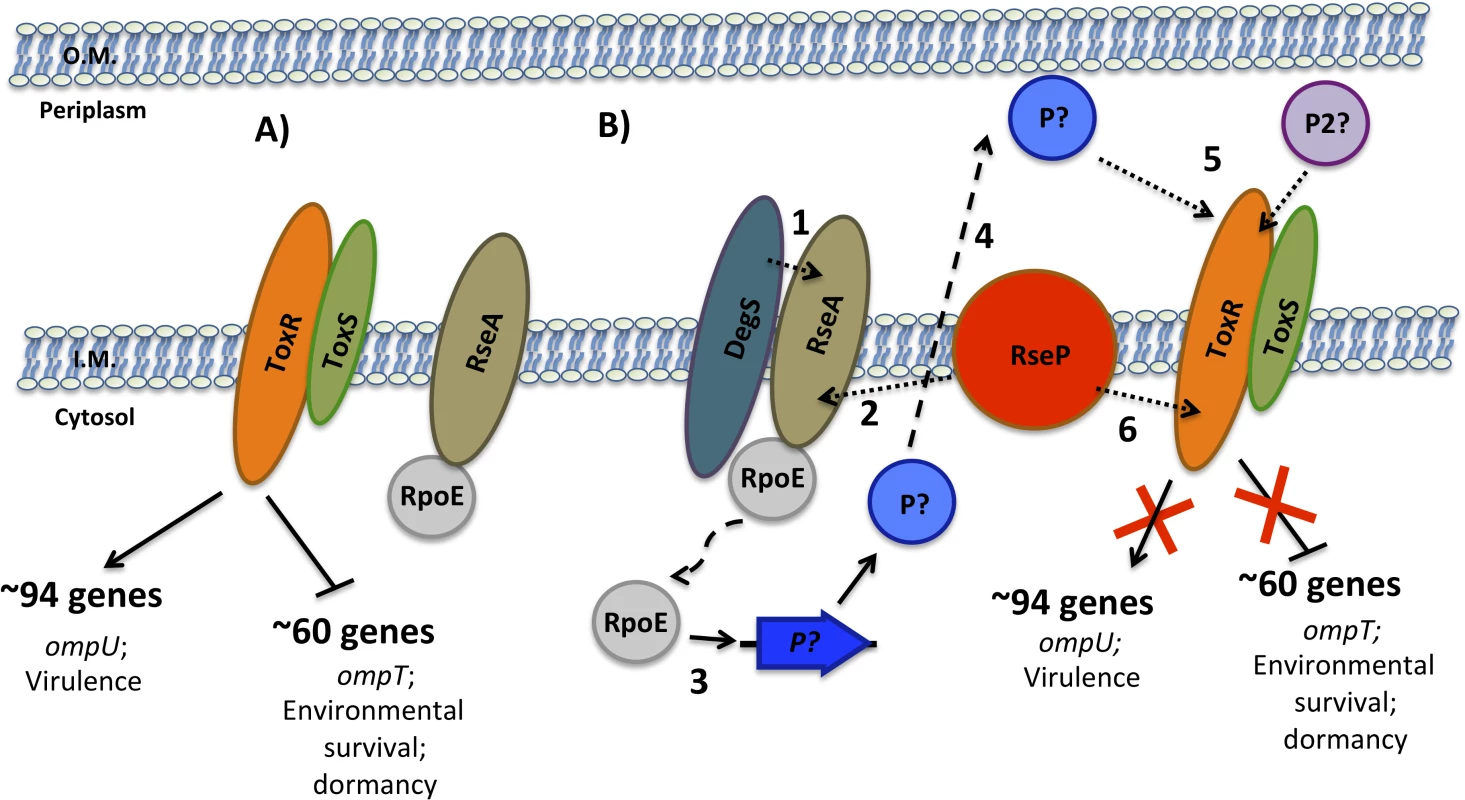
(A) In the early stages of colonization, when nutrients are abundant, ToxRS upregulate the expression of genes such as ompU important for growth under these conditions and downregulate the expression of genes such as ompT with roles in environmental survival. (B) During the late stages of colonization, as nutrients become depleted and the environment becomes alkalinized, ToxR is proteolyzed. This occurs due to activation of the σE pathway via sequential degradation of RseA by either DegS or another site-1 protease (1) and RseP (2), which releases RpoE to activate its regulated genes (3). One of these genes may encode a site-1 protease (P) that enters the periplasm (4) and cleaves a periplasmic portion of ToxR, however, at least a second protease/system (P2) appears to be necessary for the site-1 proteolytic event to occur (5). This event is followed by proteolysis of an inner-membrane site of ToxR by RseP (6), which then induces and prevents, respectively, expression of ToxR repressed and activated genes. This study has identified a RIP cascade involving RseP and RpoE that is responsible for the proteolysis of ToxR under nutrient limitation at alkaline pH. Further work is necessary in order to fully understand the pathway. This includes the identification of the site-1 protease/s that cleave ToxR as well as other genes regulated by ToxR that provide an advantage under this condition.
Materials and Methods
Bacterial strains, plasmids and culture conditions
V. cholerae O395 (O1 classical), V. cholerae N16961 (O1 El Tor), and V. parahaemolyticus RIMD2210633 were the wild-type strains for this study. E. coli S17-1λpir [83] was used for both cloning purposes and conjugation with V. cholerae. Unless otherwise indicated, cultures were grown O/N in LB at 37°C on a rotary shaker. To induce expression of V. cholerae virulence genes, cultures were grown in LB pH 6.5 at 30°C. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/ml; gentamicin (Gm), 30 μg/ml; streptomycin (Sm), 1 mg/ml.
Mutant construction
Deletion and substitution mutations were constructed by PCR amplifying two approximately 500 bp fragments of DNA upstream and downstream of the region of interest. For the substitutions, nucleotide changes were introduced into the primers. After amplification, the inserts were digested with the appropriate restriction enzymes, ligated into pKAS32 [84] and electroporated into E. coli S17-1λpir [83]. The various mutations were then transferred into V. cholerae by allelic exchange [84]. V. cholerae classical biotype contained plasmid pMIN1 [85] conferring Gmr as a counter-selection for conjugation. DNA sequencing was used to confirm the correct deletion or mutant sequence in the V. cholerae genome. The sequences of the primers used in this study are available upon request.
Protein electrophoresis and western blotting
Whole cell protein extracts were prepared from cultures grown in LB pH 6.5 at 30°C. Protein concentration was quantitated using Pierce BSA Protein Assay quantitation kit from Thermo Scientific. Protein samples were normalized and an equal amount of protein was loaded per well. The extracts were subjected to SDS-PAGE on 16% Tris Glycine gels (Invitrogen) and transferred to nitrocellulose using iBlot (Invitrogen). The membranes were blocked O/N in Tris-Buffered Saline, 3% BSA. Primary antibodies were diluted 1 : 10,000 in TBST (Tris-Buffered Saline, 0.5% Tween-20). Membranes were incubated with primary antibody for 2 hours at room temperature. After incubation, the membranes were washed with TBST four times. Goat anti-rabbit secondary antibodies (BioRad) were diluted 1 : 10,000 in TBST and incubated for 30 minutes at room temperature. The membranes were washed 4 times with TBS (Tris-Buffered Saline). Reactive protein bands were detected via ECL (Amersham).
Fluorescent microscopy
From each bacterial suspension, a 1 ml aliquot was centrifuged at 10,000 rpm for 1 min and the pellet was resuspended in phosphate buffered saline (PBS) twice. 1 ml of the mixture was then transferred to a 50 ml centrifuge tube with 24 ml PBS, and was centrifuged at 7,840 rpm for 10 minutes. The pellet was resuspended in 10 ml PBS, and a 1 ml aliquot was stained with a 3 μl mixture (1 : 1) of SYTO9 and propidium iodide (PI) nucleic acid stain (Molecular Probes, OR). After incubation in the dark for 15 min at 25°C, the stained cells were mounted on a glass slide and low-fluorescence immersion oil was added on the cover slide. The cells were then examined with a Zeiss Ax-iovert inverted microscope, and pictures were taken with AxioVision microscopy software (Zeiss).
CFU counts
Samples were serially diluted and 5 μl of each dilution was plated on LB. Plates were incubated overnight at 37 °C, and colony forming units were counted. 1 ml of initial culture was plated for those strains where no colonies were recovered after plating 5 μl. Values were plotted using Prism software. The bars represent the mean of at least four independent experiments and the error bars indicate the standard deviation.
Supporting Information
Zdroje
1. Huq A, Small EB, West PA, Huq MI, Rahman R, et al. (1983) Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol 45 : 275–283. 6337551
2. Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR (1990) Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol 56 : 1977–1980. 2383016
3. Almagro-Moreno S, Taylor RK (2013) Cholera: Environmental Reservoirs and Impact on Disease Transmission. Microbiol Spectrum 1(2):OH-0003-2012.
4. Lutz C, Erken M, Noorian P, Sun S, McDougald D (2013) Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front Microbiol 4 : 375. doi: 10.3389/fmicb.2013.00375 24379807
5. Taylor RK, Miller VL, Furlong DB, Mekalanos JJ (1987) Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA 84 : 2833–2837. 2883655
6. Sanchez J, Holmgren J (2008) Cholera toxin structure, gene regulation and pathophysiological and immunological aspects. Cell Mol Life Sci 65 : 1347–1360. doi: 10.1007/s00018-008-7496-5 18278577
7. Matson JS, Withey JH, DiRita VJ (2007) Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun 75 : 5542–5549. 17875629
8. Karaolis DK, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, et al. (1998) A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA 95 : 3134–3139. 9501228
9. Miller VL, Taylor RK, Mekalanos JJ (1987) Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell 48 : 271–279. 3802195
10. Carroll PA, Tashima KT, Rogers MB, DiRita VJ, Calderwood SB (1997) Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol Microbiol 25 : 1099–1111. 9350866
11. Häse CC, Mekalanos JJ (1998) TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA 95 : 730–734. 9435261
12. Krukonis ES, Yu RR, DiRita VJ (2000) The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol Microbiol 38 : 67–84. 11029691
13. DiRita VJ, Mekalanos JJ (1991) Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell 64 : 29–37. 1898871
14. Champion GA, Neely MN, Brennan MA, DiRita VJ (1997) A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol 23 : 323–331. 9044266
15. Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, et al. (2002) Host-induced epidemic spread of the cholera bacterium. Nature 417 : 642–645. 12050664
16. LaRocque RC, Harris JB, Dziejman M, Li X, Khan AI, et al. (2005) Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect Immun 73 : 4488–4493. 16040959
17. Abuaita BH, Withey JH (2011) Termination of Vibrio cholerae virulence gene expression is mediated by proteolysis of the major virulence activator, ToxT. Mol Microbiol 81 : 1640–1653. doi: 10.1111/j.1365-2958.2011.07798.x 21883522
18. Matson JS, DiRita VJ (2005) Degradation of the membrane-localized virulence activator TcpP by the YaeL protease in Vibrio cholerae. Proc Natl Acad Sci USA 102 : 16403–16408. 16254052
19. Urban S (2009) Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat Rev Microbiol 7 : 411–423. doi: 10.1038/nrmicro2130 19421188
20. Heinrich J, Wiegert T (2009) Regulated intramembrane proteolysis in the control of extracytoplasmic function sigma factors. Res Microbiol 160 : 696–703. doi: 10.1016/j.resmic.2009.08.019 19778605
21. Makinoshima H, Glickman MS (2005) Regulation of Mycobacterium tuberculosis cell envelope composition and virulence by intramembrane proteolysis. Nature 436 : 406–409. 16034419
22. Alba BM, Gross CA (2004) Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol 52 : 613–619. 15101969
23. Ades SE (2008) Regulation by destruction: design of the sigmaE envelope stress response. Curr Opin Microbiol 11 : 535–540. doi: 10.1016/j.mib.2008.10.004 18983936
24. Ades SE, Connolly LE, Alba BM, Gross CA (1999) The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev 13 : 2449–2461. 10500101
25. Kanehara K, Ito K, Akiyama Y (2002) YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev 16 : 2147–2155. 12183368
26. Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA (2002) DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev 16 : 2156–2168. 12183369
27. Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, et al. (2007) Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2 : 264–277. 18005744
28. Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, et al. (2006) RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog 2: e109. 17054394
29. Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A (2013) Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog 9: e1003800. doi: 10.1371/journal.ppat.1003800 24385900
30. Alam M, Sultana M, Nair GB, Siddique AK, Hasan NA, et al. (2007) Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc Natl Acad Sci USA 104 : 17801–17806. 17968017
31. Kamruzzaman M, Udden SMN, Cameron DE, Calderwood SB, Nair GB, et al. (2010) Quorum-regulated biofilms enhance the development of conditionally viable, environmental Vibrio cholerae. Proc Natl Acad Sci USA 107 : 1588–1593. doi: 10.1073/pnas.0913404107 20080633
32. Faruque SM, Islam MJ, Ahmad QS, Biswas K, Faruque ASG, et al. (2006) An improved technique for isolation of environmental Vibrio cholerae with epidemic potential: monitoring the emergence of a multiple-antibiotic-resistant epidemic strain in Bangladesh. J Infect Dis 193 : 1029–1036. 16518766
33. Xu HS, Roberts N, Singleton FL, Attwell RW, Grimes DJ, et al. (1982) Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol 8 : 313–323. doi: 10.1007/BF02010671 24226049
34. Bari SMN, Roky MK, Mohiuddin M, Kamruzzaman M, Mekalanos JJ, et al. (2013) Quorum-sensing autoinducers resuscitate dormant Vibrio cholerae in environmental water samples. Proc Natl Acad Sci USA 110 : 9926–9931. doi: 10.1073/pnas.1307697110 23716683
35. Bina J, Zhu J, Dziejman M, Faruque S, Calderwood S, et al. (2003) ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc Natl Acad Sci USA 100 : 2801–2806. 12601157
36. Miller VL, Mekalanos JJ (1988) A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170 : 2575–2583. 2836362
37. Li CC, Merrell DS, Camilli A, Kaper JB (2002) ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol Microbiol 43 : 1577–1589. 11952906
38. Mey AR, Craig SA, Payne SM (2011) The effects of amino acid supplementation on porin expression and ToxR levels in Vibrio cholerae. Infect Immun. 80 : 518–528. doi: 10.1128/IAI.05851-11 22144480
39. Medrano AI, DiRita VJ, Castillo G, Sanchez J (1999) Transient transcriptional activation of the Vibrio cholerae El Tor virulence regulator toxT in response to culture conditions. Infect Immun 67 : 2178–2183. 10225872
40. DiRita VJ, Neely M, Taylor RK, Bruss PM (1996) Differential expression of the ToxR regulon in classical and E1 Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc Natl Acad Sci USA 93 : 7991–7995. 8755590
41. LaRocque RC, Krastins B, Harris JB, Lebrun LM, Parker KC, et al. (2008) Proteomic analysis of Vibrio cholerae in human stool. Infect Immun 76 : 4145–4151. doi: 10.1128/IAI.00585-08 18591230
42. Kirn TJ, Jude BA, Taylor RK (2005) A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438 : 863–866. 16341015
43. Mathur J, Davis BM, Waldor MK (2007) Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol Microbiol 63 : 848–858. 17181782
44. Kovacikova G, Skorupski K (2002) The alternative sigma factor sigma(E) plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect Immun 70 : 5355–5362. 12228259
45. Raivio TL, Silhavy TJ (1999) The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr Opin Microbiol 2 : 159–165. 10322173
46. Ruiz N, Silhavy TJ (2005) Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol 8 : 122–126. 15802241
47. Slamti L, Waldor MK (2009) Genetic analysis of activation of the Vibrio cholerae Cpx pathway. J Bacteriol 191 : 5044–5056. doi: 10.1128/JB.00406-09 19542291
48. Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK (2000) Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol 35 : 896–910. 10692166
49. Wai SN, Mizunoe Y, Yoshida S (1999) How Vibrio cholerae survive during starvation. FEMS Microbiology Letters 180 : 123–131. 10556702
50. Pruzzo C, Tarsi R, Lleò MM, Signoretto C, Zampini M, et al. (2003) Persistence of adhesive properties in Vibrio cholerae after long-term exposure to sea water. Environ Microbiol 5 : 850–858. 14510838
51. Gonzalez-Escalona N, Fey A, Höfle MG, Espejo RT, A Guzmán C (2006) Quantitative reverse transcription polymerase chain reaction analysis of Vibrio cholerae cells entering the viable but non-culturable state and starvation in response to cold shock. Environ Microbiol 8 : 658–666. 16584477
52. Crawford JA, Krukonis ES, DiRita VJ (2003) Membrane localization of the ToxR winged-helix domain is required for TcpP-mediated virulence gene activation in Vibrio cholerae. Mol Microbiol 47 : 1459–1473. 12603748
53. Chaiyanan S, Chaiyanan S, Grim C, Maugel T, Huq A, et al. (2007) Ultrastructure of coccoid viable but non-culturable Vibrio cholerae. Environ Microbiol 9 : 393–402. 17222137
54. Krebs SJ, Taylor RK (2011) Nutrient-dependent, rapid transition of Vibrio cholerae to coccoid morphology and expression of the toxin co-regulated pilus in this form. Microbiol 157 : 2942–2953. doi: 10.1099/mic.0.048561-0 21778208
55. Pradhan S, Mallick SK, Chowdhury R (2013) Vibrio cholerae classical biotype is converted to the viable non-culturable state when cultured with the El Tor biotype. PLoS ONE 8: e53504. doi: 10.1371/journal.pone.0053504 23326443
56. Whitaker WB, Parent MA, Boyd A, Richards GP, Boyd EF (2012) The Vibrio parahaemolyticus ToxRS Regulator Is Required for Stress Tolerance and Colonization in a Novel Orogastric Streptomycin-Induced Adult Murine Model. Infect Immun 80 : 1834–1845. doi: 10.1128/IAI.06284-11 22392925
57. Wong HC, Wang P (2004) Induction of viable but nonculturable state in Vibrio parahaemolyticus and its susceptibility to environmental stresses. J Appl Microbiol 96 : 359–366. 14723697
58. Merrell DS, Bailey C, Kaper JB, Camilli A (2001) The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU. J Bacteriol 183 : 2746–2754. 11292792
59. Rudner DZ, Fawcett P, Losick R (1999) A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA 96 : 14765–14770. 10611287
60. Bramkamp M, Weston L, Daniel RA, Errington J (2006) Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol Microbiol 62 : 580–591. 17020588
61. An FY, Sulavik MC, Clewell DB (1999) Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J Bacteriol 181 : 5915–5921. 10498702
62. Stevenson LG, Strisovsky K, Clemmer KM, Bhatt S, Freeman M, et al. (2007) Rhomboid protease AarA mediates quorum-sensing in Providencia stuartii by activating TatA of the twin-arginine translocase. Proc Natl Acad Sci USA 104 : 1003–1008. 17215357
63. Heinrich J, Lundén T, Kontinen VP, Wiegert T (2008) The Bacillus subtilis ABC transporter EcsAB influences intramembrane proteolysis through RasP. Microbiol 154 : 1989–1997.
64. Qiu D, Eisinger VM, Rowen DW, Yu HD (2007) Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 104 : 8107–8112. 17470813
65. Rattanama P, Thompson JR, Kongkerd N, Srinitiwarawong K, Vuddhakul V, et al. (2012) Sigma E Regulators Control Hemolytic Activity and Virulence in a Shrimp Pathogenic Vibrio harveyi. PLoS ONE 7: e32523. doi: 10.1371/journal.pone.0032523 22384269
66. Chatterjee E, Chowdhury R (2013) Reduced virulence of the Vibrio cholerae fadD mutant is due to induction of the extracytoplasmic stress response. Infect Immun 81 : 3935–3941. doi: 10.1128/IAI.00722-13 23918781
67. Haines-Menges B, Whitaker WB, Boyd EF (2014) The alternative sigma factor RpoE is important for Vibrio parahaemolyticus cell envelope stress response and intestinal colonization. Infect Immun 89 : 3667–3677.
68. Palonen E, Lindström M, Somervuo P, Korkeala H (2013) Alternative sigma factor σE has an important role in stress tolerance of Yersinia pseudotuberculosis IP32953. Appl Environ Microbiol 79 : 5970–5977. doi: 10.1128/AEM.01891-13 23872565
69. Pfau JD, Taylor RK (1998) Mutations in toxR and toxS that separate transcriptional activation from DNA binding at the cholera toxin gene promoter. J Bacteriol 180 : 4724. 9721317
70. Ding Y, Davis BM, Waldor MK (2004) Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol Microbiol 53 : 345–354. 15225327
71. Colwell R (2009) Viable but Not Cultivable Bacteria. In: Epstein SS, editor. Microbiology Monographs. Springer Berlin Heidelberg, 10 : 121–129.
72. Oliver JD (2010) Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev 34 : 415–425. doi: 10.1111/j.1574-6976.2009.00200.x 20059548
73. Ravel J, Knight IT, Monahan CE, Hill RT, Colwell RR (1995) Temperature-induced recovery of Vibrio cholerae from the viable but nonculturable state: growth or resuscitation?. Microbiol 141 : 377–383.
74. Thomas KU, Joseph N, Raveendran O, Nair S (2006) Salinity-induced survival strategy of Vibrio cholerae associated with copepods in Cochin backwaters. Mar Pollut Bull 52 : 1425–1430. 16764894
75. Reich KA, Schoolnik GK (1994) The light organ symbiont Vibrio fischeri possesses a homolog of the Vibrio cholerae transmembrane transcriptional activator ToxR. J Bacteriol 176 : 3085–3088. 8188612
76. Lee SE, Shin SH, Kim SY, Kim YR, Shin DH, et al. (2000) Vibrio vulnificus has the transmembrane transcription activator ToxRS stimulating the expression of the hemolysin gene vvhA. J Bacteriol 182 : 3405–3415. 10852871
77. Montieri S, Suffredini E, Ciccozzi M, Croci L (2010) Phylogenetic and evolutionary analysis of Vibrio parahaemolyticus and Vibrio alginolyticus isolates based on toxR gene sequence. New Microbiol 33 : 359–372. 21213595
78. Ruwandeepika HAD, Defoirdt T, Bhowmick PP, Shekar M, Bossier P, et al. (2010) Presence of typical and atypical virulence genes in Vibrio isolates belonging to the Harveyi clade. J Appl Microbiol 109 : 888–899. doi: 10.1111/j.1365-2672.2010.04715.x 20345385
79. Welch TJ, Bartlett DH (1998) Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol Microbiol 27 : 977–985. 9535087
80. Bartlett D, Chi E (1994) Genetic characterization of ompH mutants in the deep-sea bacterium Photobacterium sp. strain SS9. Arch Microbiol 162 : 323–328. 7857197
81. Chi E, Bartlett DH (1995) An rpoE-like locus controls outer membrane protein synthesis and growth at cold temperatures and high pressures in the deep-sea bacterium Photobacterium sp. strain SS9. Mol Microbiol 17 : 713–726. 8801425
82. Patnaik BK, Ghosh HK (1966) Histopathological studies on experimental cholera. Br J Exp Pathol 47 : 210–214.
83. de Lorenzo V, Timmis KN (1994) Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5 - and Tn10-derived minitransposons. Meth Enzymol 235 : 386–405. 8057911
84. Skorupski K, Taylor RK (1996) Positive selection vectors for allelic exchange. Gene 169 : 47–52. 8635748
85. Nye MB, Pfau JD, Skorupski K, Taylor RK (2000) Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J Bacteriol 182 : 4295–4303. 10894740
Štítky
Genetika Reprodukční medicína
Článek Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in CrustaceansČlánek Adventures in WonderlandČlánek Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and InfectivityČlánek Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in FilamentsČlánek Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in ArabidopsisČlánek Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression ControlČlánek The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent EnhancersČlánek Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of MitochondriaČlánek The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma FormationČlánek Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 4- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- Adventures in Wonderland
- Experimental Swap of 's Assortative Mating Preferences Demonstrates Key Role of X-Chromosome Divergence Island in Incipient Sympatric Speciation
- Chromosome Replacement and Deletion Lead to Clonal Polymorphism of Berry Color in Grapevine
- The Protein Quality Control Machinery Regulates Its Misassembled Proteasome Subunits
- Genome-Wide Association Study Identifies as a Critical Gene for Susceptibility to Noise-Induced Hearing Loss
- Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and Infectivity
- Viable Neuronopathic Gaucher Disease Model in Medaka () Displays Axonal Accumulation of Alpha-Synuclein
- Multi-locus Analysis of Genomic Time Series Data from Experimental Evolution
- The Genetic Legacy of the Expansion of Turkic-Speaking Nomads across Eurasia
- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- The Pif1 Helicase, a Negative Regulator of Telomerase, Acts Preferentially at Long Telomeres
- Inhibiting K63 Polyubiquitination Abolishes No-Go Type Stalled Translation Surveillance in
- SYD-1C, UNC-40 (DCC) and SAX-3 (Robo) Function Interdependently to Promote Axon Guidance by Regulating the MIG-2 GTPase
- Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in Filaments
- Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3’ Tailing of Small RNAs in Arabidopsis
- Host Genetic Variation Influences Gene Expression Response to Rhinovirus Infection
- Contribution of Large Region Joint Associations to Complex Traits Genetics
- Volatility of Mutator Phenotypes at Single Cell Resolution
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in Arabidopsis
- A Multi-layered Protein Network Stabilizes the FtsZ-ring and Modulates Constriction Dynamics
- Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression Control
- Genome Sequencing of the Perciform Fish Provides Insights into Molecular and Genetic Mechanisms of Stress Adaptation
- Natural Variant E610G Is a Semi-dominant Suppressor of IAP-Induced RNA Processing Defects
- The Alkaline Response Pathway: Identification of a Novel Rim Pathway Activator
- Transgenerational Inheritance of Diet-Induced Genome Rearrangements in Drosophila
- A Single Nucleotide Polymorphism Uncovers a Novel Function for the Transcription Factor Ace2 during Hyphal Development
- DNA Damage Response and Spindle Assembly Checkpoint Function throughout the Cell Cycle to Ensure Genomic Integrity
- The Functional Interplay Between the t(9;22)-Associated Fusion Proteins BCR/ABL and ABL/BCR in Philadelphia Chromosome-Positive Acute Lymphatic Leukemia
- Extreme Recombination Frequencies Shape Genome Variation and Evolution in the Honeybee,
- Beyond Glycolysis: GAPDHs Are Multi-functional Enzymes Involved in Regulation of ROS, Autophagy, and Plant Immune Responses
- Comprehensive Profiling of Amino Acid Response Uncovers Unique Methionine-Deprived Response Dependent on Intact Creatine Biosynthesis
- Windpipe Controls Intestinal Homeostasis by Regulating JAK/STAT Pathway via Promoting Receptor Endocytosis and Lysosomal Degradation
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
- Cross-Population Joint Analysis of eQTLs: Fine Mapping and Functional Annotation
- The Power of Gene-Based Rare Variant Methods to Detect Disease-Associated Variation and Test Hypotheses About Complex Disease
- The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent Enhancers
- Competition between VanU Repressor and VanR Activator Leads to Rheostatic Control of Vancomycin Resistance Operon Expression
- A Missense Change in the Gene Links Aberrant Autophagy to a Neurodegenerative Vacuolar Storage Disease
- Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model
- Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of Mitochondria
- Genome-Destabilizing Effects Associated with Top1 Loss or Accumulation of Top1 Cleavage Complexes in Yeast
- Imputation-Based Population Genetics Analysis of Malaria Parasites
- Heterozygosity for a Hypomorphic Polβ Mutation Reduces the Expansion Frequency in a Mouse Model of the Fragile X-Related Disorders
- Neto-Mediated Intracellular Interactions Shape Postsynaptic Composition at the Neuromuscular Junction
- Ndd1 Turnover by SCF Is Inhibited by the DNA Damage Checkpoint in
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma Formation
- Spastin Binds to Lipid Droplets and Affects Lipid Metabolism
- Maintenance of Glia in the Optic Lamina Is Mediated by EGFR Signaling by Photoreceptors in Adult Drosophila
- Auxin Influx Carriers Control Vascular Patterning and Xylem Differentiation in
- Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
- The Lowe Syndrome Protein OCRL1 Is Required for Endocytosis in the Zebrafish Pronephric Tubule
- Postnatal Loss of Hap1 Reduces Hippocampal Neurogenesis and Causes Adult Depressive-Like Behavior in Mice
- CAPER Is Vital for Energy and Redox Homeostasis by Integrating Glucose-Induced Mitochondrial Functions via ERR-α-Gabpa and Stress-Induced Adaptive Responses via NF-κB-cMYC
- Distinct and Cooperative Activities of HESO1 and URT1 Nucleotidyl Transferases in MicroRNA Turnover in
- The Evolutionary Origination and Diversification of a Dimorphic Gene Regulatory Network through Parallel Innovations in and
- MAPK Signaling Pathway Alters Expression of Midgut ALP and ABCC Genes and Causes Resistance to Cry1Ac Toxin in Diamondback Moth
- Spatio-temporal Remodeling of Functional Membrane Microdomains Organizes the Signaling Networks of a Bacterium
- Asymmetric Transcript Discovery by RNA-seq in . Blastomeres Identifies , a Gene Important for Anterior Morphogenesis
- A Stress-Induced Small RNA Modulates Alpha-Rhizobial Cell Cycle Progression
- Systematic Profiling of Poly(A)+ Transcripts Modulated by Core 3’ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation
- The UPR Branch IRE1- in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast
- A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi
- Co-chaperone p23 Regulates . Lifespan in Response to Temperature
- Re-replication of a Centromere Induces Chromosomal Instability and Aneuploidy
- Shade Avoidance Components and Pathways in Adult Plants Revealed by Phenotypic Profiling
- Lipid-Induced Epigenomic Changes in Human Macrophages Identify a Coronary Artery Disease-Associated Variant that Regulates Expression through Altered C/EBP-Beta Binding
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



