-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3’ Tailing of Small RNAs in Arabidopsis
Small silencing RNAs are key regulators of gene expression in both plants and animals. HEN1-mediated 3’ terminal 2’-O-methylation plays a crucial role in small RNA stability control. In the absence of HEN1, several types of small RNAs become frequently uridylated (non-templated uridine addition) and trimmed, a phenomenon that is conserved across species. However, the underlying molecular mechanism is barely understood. In this study, we have discovered UTP: RNA uridylyltransferase (URT1) that acts synergistically with HESO1 in miRNA uridylation, in addition to its role in oligo-adenylated mRNA uridylation. Analyzing the miRNA profiles also reveals the existence of multiple terminal nucleotidyl transferases in the miRNA tailing process and an antagonistic action between uridylation and trimming. We believe this study will shed light on our understanding of how various terminal nucleotidyl transferases recognize their substrates and function coordinately.
Published in the journal: . PLoS Genet 11(4): e32767. doi:10.1371/journal.pgen.1005091
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005091Summary
Small silencing RNAs are key regulators of gene expression in both plants and animals. HEN1-mediated 3’ terminal 2’-O-methylation plays a crucial role in small RNA stability control. In the absence of HEN1, several types of small RNAs become frequently uridylated (non-templated uridine addition) and trimmed, a phenomenon that is conserved across species. However, the underlying molecular mechanism is barely understood. In this study, we have discovered UTP: RNA uridylyltransferase (URT1) that acts synergistically with HESO1 in miRNA uridylation, in addition to its role in oligo-adenylated mRNA uridylation. Analyzing the miRNA profiles also reveals the existence of multiple terminal nucleotidyl transferases in the miRNA tailing process and an antagonistic action between uridylation and trimming. We believe this study will shed light on our understanding of how various terminal nucleotidyl transferases recognize their substrates and function coordinately.
Introduction
MicroRNAs (miRNAs), a class of small non-coding RNAs with 20–24 nt in size, are master regulators of gene expression at post-transcriptional levels in both plants and animals [1,2]. They impact various biological processes such as development, metabolism and response to different biotic and abiotic stresses [1]. In Arabidopsis, miRNAs are typically derived from their precursor RNAs called primary miRNA transcripts (pri-miRNAs) through a sequential cleavage, from either loop proximal or loop distal, by DICER-LIKE 1 (DCL1) and its associated proteins [2–5]. After generation, the 3’ terminal riboses of miRNA/miRNA* duplexes are 2’-O-methylated by the RNA methyltransferase HUA ENHANCER1 (HEN1) [6]. One strand of the mature, methylated miRNA/miRNA* duplexes is selectively incorporated into their effector protein, mainly ARGONAUTE1 (AGO1), to form a silencing complex [7]. In the hen1 mutants, miRNAs accumulate at lower levels and become heterogeneous in size, due to different levels of trimming and non-templated nucleotides addition (tailing, also commonly referred to as uridylation because uridine is preferentially added) at their 3’ ends [6,8]. Similar process also occurs to plant small interfering RNAs (siRNAs), animal PIWI-interacting RNAs (piRNAs) and a subset of animal siRNAs [9–16]. These results reveal an evolutionary conserved role of 3’ end methylation in protecting small RNAs from 3’ end tailing, trimming and degradation.
HEN1 SUPPRESSOR1 (HESO1) is a major enzyme responsible for the small RNA uridylation in Arabidopsis [17,18]. Loss-of-function mutations in HESO1 result in reduced miRNA tailing length, increased miRNA abundance and partially restored morphological phenotypes of hen1. In contrast, over-expression of HESO1 in hen1 causes reduced miRNA accumulation and more severe morphological phenotypes. Collectively, these results demonstrate that uridylation triggers miRNA degradation [17,18]. Nevertheless, A substantial amount of miRNA uridylation is present in the null hen1 heso1 mutants, suggesting the existence of additional small RNA uridylyltransferase(s) [17,18]. Besides HESO1, the Arabidopsis genome encodes nine additional terminal nucleotidyl transferases (TNTases) [17,18]. However, their involvement, if any, in miRNA uridylation is probably masked by HESO1 because none of their single mutants has any visible effect in terms of the restoration of hen1 morphological phenotypes [18].
To identify additional miRNA uridylyltransferase (s), we screened for mutants with further recovered fertility in the hen1-2 heso1-2 background. In this study, we show that a Proline-to-Leucine substitution at amino acid 618 (P618L) in URT1 increases the silique length and enhances the miRNA function of hen1-2 heso1-2. URT1 was previously identified as an enzyme that uridylates some mRNAs with short poly(A) tails and exhibits robust U-tailing activity in vitro [19]. We find that URT1 interacts with AGO1 and is responsible for miRNA, but not heterochromatic siRNA, uridylation in hen1-2 heso1-2. miRNA uridylation is globally abolished in the hen1-2 heso1-2 urt1-3 mutant, accompanied by an extensive increase of trimming. These results demonstrate that HESO1 and URT1 act synergetically in miRNA uridylation and that trimming and tailing antagonize each other for the occupancy of miRNA 3’ ends. Moreover, a close investigation of the tailing status in hen1-2 heso1-2 urt1-3 also reveals the addition of non-uridine nucleotides by yet uncharacterized TNTase(s), suggesting the complexity of miRNA tailing. Finally, we show HESO1 and URT1 may redundantly uridylate some miRNAs in the HEN1 competent background.
Results
A point mutation in URT1 increases the fertility of hen1 heso1-2
To identify novel components in miRNA stability control, we performed another genetic screen in hen1-2 heso1-2 (note that hen1-2 is a weak allele and heso1-2 is a null allele). Fertility, as reflected by the silique length, has been proved to be an effective indicator of miRNA activity in this system [17,20]. A mutant (m37-6) with markedly longer siliques than hen1-2 heso1-2 was isolated from the M2 population of ethylmethanesulfonate (EMS) mutagenized hen1-2 heso1-2. The average silique length of m37-6 was ~1.0 cm, which was about 1.3 and 2.9 fold of those of hen1-2 heso1-2 and hen1-2, respectively (Fig 1A and 1B). Backcross analyses revealed that the increased silique length phenotype was caused by a single recessive mutation. The mutation was roughly mapped to the marker nga168 on Chr. II, where HESO1 and two other TNTases (At2g40520 and At2g45620/URT1) were nearby this marker. Sequencing of these candidate genes revealed a single C-to-T nucleotide substitution, at the position +1853 relative to the translation start site (ATG) of URT1 (the mutation was hereafter referred to as urt1-3; urt1-1 (Salk_087647C) and urt1-2 (WISCDSLOXHS208_08D) were previously characterized to be defective in the uridylation of mRNAs with shortened poly(A) tails. [19]), which resulted in a Proline-to-Leucine change at amino acid 618 (P618L) (Fig 1C and 1D). P618 is located in an uncharacterized region that links the PAP domain and the PAP-associated domain. Blast analysis suggested that P618 is rather conserved, especially among those well-characterized TNTases (Fig 1D). Introduction of a genomic fragment of URT1 fused with a GFP (pURT1-URT1-GFP) (S1 Fig) at its C-terminus into hen1-2 heso1-2 urt1-3 restored its silique length to the hen1-2 heso1-2 level, suggesting that the urt1-3 mutation is responsible for the increased fertility of hen1-2heso1-2.
Fig. 1. urt1-3 increases the silique length in hen1-2 heso1-2 and enhances miRNA function in hen1-1 heso1-2. 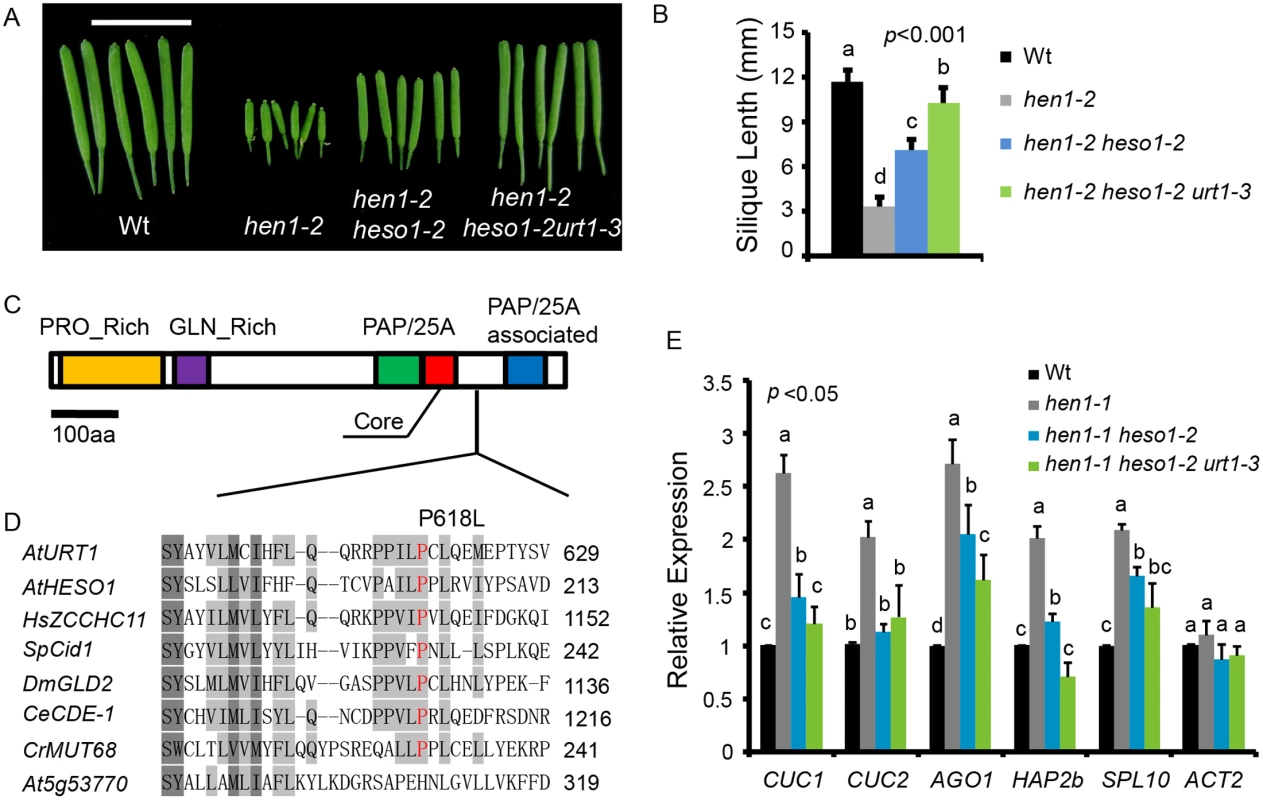
(A) Fully-expanded siliques from plants of the indicated genotypes. Scale bar, 1cm. (B) Average silique length in indicated genotypes. 40 siliques from at least 6 individual plants for each genotype were analyzed. (C) Schematic structure of URT1 protein. PRO_Rich: Proline rich domain; GLN_Rich: Glutamine rich domain; PAP/25A: Poly A polymerase domain; Core: Core regions of PAP/25A; PAP/25A associated: Poly A polymerase associated domain. (D) Conservation of URT1 P618 across species. At, A. thaliana; Hs, H. sapiens; Sp, S. pombe; Dm, D. melanogaster; Ce, C.elegans; Cr, C. reinhardtii. (E) urt1-3 enhances miRNA function in hen1-1 heso1-2. The transcript levels of five miRNA targets in indicated genotypes. Target mRNA accumulation in each genotype was quantified by qPCR using primers spanning the cleavage site and compared with those of wild type (Wt). The expression level of each gene in Wt was arbitrarily set to 1. Quantifications are normalized with GAPDH transcript. ACT2 was served as an internal control. Values are means of three biological replicates ±SD. Introgression of the urt1-3 mutation into hen1-1 heso1-2 also increased its fertility (S2 Fig). Since hen1-1 is a null allele of hen1, this observation suggested that the increase of hen1 heso1-2 fertility by the urt1-3 mutation doesn’t rely on the residual HEN1 activity. Morphological defects in hen1 are mainly due to a reduction of miRNA activities and the null heso1-2 mutation only partially restores the miRNA activities in hen1 [17,18]. To determine whether the miRNA activities in hen1-1 heso1-2 were further recovered by urt1-3, we compared the expression levels of five miRNA targets in Ler (wild type, Wt), hen1-1, hen1-1 heso1-2 and hen1-1 heso1-2 urt1-3 by quantitative real-time RT-PCR (qPCR). Confirming our previous findings [17], the expression levels of all tested targets were increased in hen1-1 as compared to those in Wt and their levels were partially restored in hen1-1 heso1-2 (Fig 1E). Notably, the expression of these transcripts in hen1-1 heso1-2 urt1-3 was further reduced, to the levels comparable to those in Wt (Fig 1E). Therefore, we conclude that the urt1-3 mutation further enhances the miRNA function of hen1-1 heso1-2.
Uridylation antagonizes 3’-to-5’ trimming of miRNAs
We next examined the expression patterns of several miRNAs in Wt, hen1-2, hen1-2 heso1-2 and hen1-2 heso1-2 urt1-3 by the small RNA northern blot assay. We found that miRNA species with sizes longer than their annotated ones were barely detected in hen1-2 heso1-2 urt1-3 for all the tested miRNAs (Fig 2). Instead, miRNAs in hen1-2 heso1-2 urt1-3 became severely shortened in size as compared with those in hen1-2 and hen1-2 heso1-2 (Fig 2). The miRNA expression patterns of the genomic complementation line (hen1-2 heso1-2 urt1-3 + pURT1-URT1-GFP) resembled those of hen1-2 heso1-2, demonstrating that the urt1-3 mutation causes the changes of miRNA profiles in hen1-2 heso1-2 (Fig 2).
Fig. 2. Small RNA northern blot analysis of miR167, miR166, miR171 and miR172 in various genotypes. 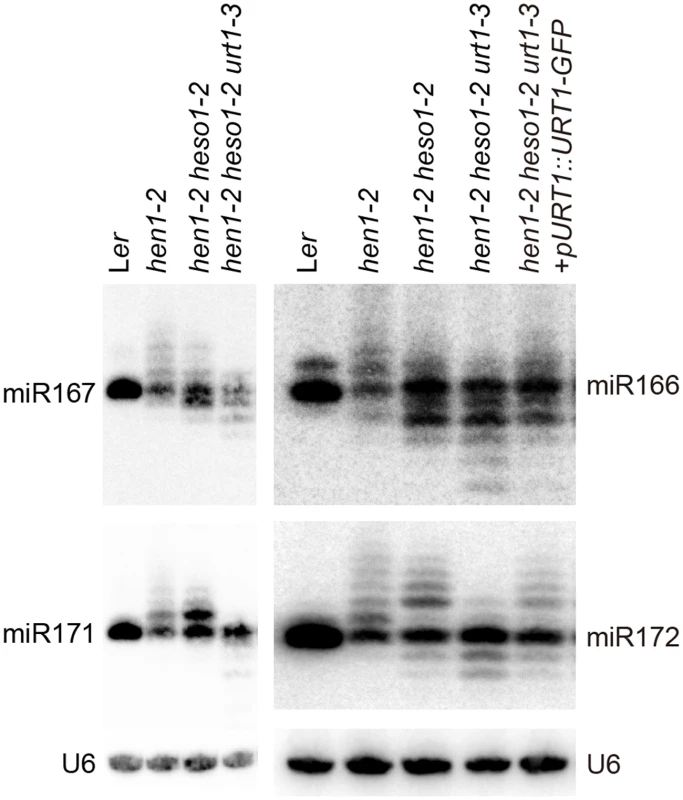
U6 was served as a loading control. Since unmethylated miRNAs can be frequently trimmed before uridylated [21], the size information given by the small RNA northern blot assay may only reflect the overall combination effect of trimming and tailing. In order to analyze the degree of uridylation reduction and trimming enhancement in detail, small RNA libraries with two biological replicates for each genotype were prepared from Wt, hen1-2, hen1-2 heso1-2 and hen1-2 heso1-2 urt1-3 and subject to the Illumina deep sequencing. Small RNA 3’ end modification analysis was performed according to a previous report with minor modifications (for details, see Materials and Methods) [18]. Based on the algorithm, small RNA reads were split into a 5’ genome matched component (5GMC) part and a 3’ tailed part, which reflects the degree of trimming and tailing, respectively. Only reads with 5GMC longer than 12nt were included in our analysis.
We first compared the overall length distribution of miRNAs among these four genotypes. As expected, about 84% of the miRNAs were 21nt in size in Wt. The size of miRNAs became heterogeneous in hen1-2, ranging from 18nt to 25nt (reads cutoff = 0.5%) with peaks at 20nt and 21nt (Fig 3A and S3A Fig). An obvious shift of this size range towards shorter ones was observed in hen1-2 heso1-2 and became more prominent in hen1-2 heso1-2 urt1-3 (Fig 3A and S3A Fig), which is consistent with our northern blot results (Fig 2). Notably, only less than 2.5% of the miRNAs were 22nt or longer in hen1-2 heso1-2 urt1-3, compared with ~9.1% of those in hen1-2 heso1-2 (p<0.0001, F-test) (Fig 3A and S3A Fig). Moreover, the 5GMC pattern of hen1-2 heso1-2 urt1-3 resembled the miRNA distribution pattern (Fig 3A and 3B and S3A and S3B Fig), indicating a drastic loss of tailing activity by the loss of functions in HESO1 and URT1. The distribution of 5GMC also revealed that a reduction of uridylation was accompanied by an increase of trimming (Fig 3B and S3B Fig). Indeed, both the levels and extents (more extensive truncation) of trimming were gradually increased in hen1-2 heso1-2 and hen1-2 heso1-2 urt1-3 when we quantified the degrees of tailing and trimming separately (Fig 3C and S3C Fig). Notably, a small peak of 5GMC at 17nt was observed in hen1-2 (Fig 3B and S3B Fig), indicating a proportion of miRNAs favor to be trimmed to 17nt before they are further tailed. However, such a peak was not observed in either hen1-2 heso1-2 or hen1-2 heso1-2 urt1-3, implicating a dynamic correlation between tailing and trimming.
Fig. 3. Global changes of miRNA profiles in hen1-1 heso1-2 urt1-3. 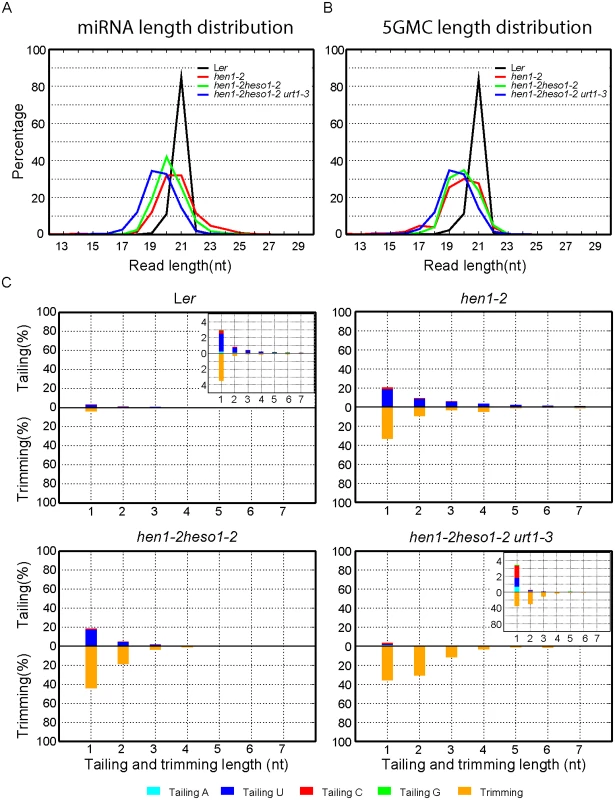
(A) Size distribution of miRNAs in indicated genotypes. (B) Size distribution of 5GMC of miRNAs in indicated genotypes. 5GMC, 5’ genome matched component, see text for details. (C) Overall 3’ end signatures (including tail length, nucleotides addition composition and trimming extent) of miRNAs in indicated genotypes. The tailing and trimming extent was calculated as % (number of reads with indicated modification divided by total analyzed reads); the nucleotides addition composition was calculated as number of a nucleotide in the tail divided by tail length. Data from biological replicate 1 were shown. See S3 Fig online for data from biological replicate 2. Analyses of individual miRNAs further support our northern blot results and global miRNA profiles (Fig 2 and Fig 3). For most of the miRNAs analyzed, a gradual loss of uridylation activity was always accompanied by an increase of trimmed species and/or normal sized ones, albeit the degree of which varied among different miRNAs (Fig 4 and S4 Fig). Notably, long-tailed species could be readily detected in hen1-2heso1-2 for some miRNAs (e.g. miR163 and miR164a) and the tails were almost abolished in hen1-2heso1-2urt1-3 (Fig 4 and S4 Fig). More intriguingly, we found that several miRNAs (e.g. miR390 and miR398b) were resistant to both tailing and trimming modifications (Fig 4 and S4 Fig). The reason for this resistance is currently unknown.
Fig. 4. urt1-3 affects miRNAs but not siRNAs uridylation in hen1-2 heso1-2. 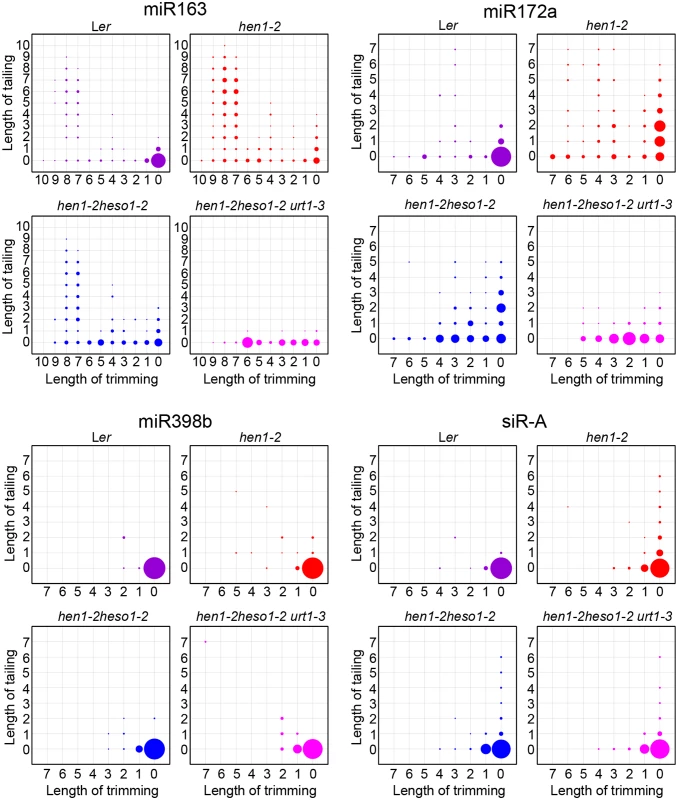
The X-axis represents the degree of trimming and the Y-axis represents the degree of tailing. The annotated miRNA sequences from miRBase v17.0 were served as standard sequences (i.e. these sequences are considered as non-tailed and non-trimmed.) For simplicity, we only analyzed reads started from the annotated 5’ ends. Thus, reads at coordinate position (0, 0) are exactly the same as annotated sequences and all reads from the same coordinate position are of same length. The relative abundance of each small RNA species is proportional to the diameter of the circles. Data from biological replicate 1 were shown. See S4 Fig online for more small RNAs and data from biological replicate 2. HESO1 and URT1 independent addition of non-uridine nucleotides by yet uncharacterized enzymes
Consistent with the overall length distribution (Fig 3A and S3A Fig), 3’ tailing was almost abolished in hen1-2 heso1-2 urt1-3 (Fig 3C, S3C Fig and S1 Table). Intriguingly, we noticed a remarkably reduced preference for uridine (39.1%) when we calculated the nucleotide composition of the residual tails in hen1-2 heso1-2 urt1-3. As controls, the percentage of uridine addition in tails of Wt, hen1-2 and hen1-2 heso1-2 was 76.7%, 88.4% and 90.3%, respectively (Fig 3C, S3C Fig and S1 Table). Further analysis revealed that the raw counts of the other three types of nucleotides additions (i.e. adenosine, cytidine and guanosine) were comparable among hen1-2, hen1-2 heso1-2 and hen1-2 heso1-2 urt1-3, although their relative proportions were increased in the triple mutant, due to an almost complete loss of uridylation (S1 Table). Based on our definition of tailing, a tiny proportion of tailed-only miRNAs might be alternatively introduced by the misprocessing, alternative processing and/or processing of pre-tailed pre-miRNAs [22,23]. We thus investigated miRNA species with 1nt trimmed and 1nt tailed, ensuring that the tails we analyzed were added after processing. We found that uridylation, but not other types of nucleotides addition were affected upon loss of function in HESO1 and URT1 (S2 Table). Taken together, these data unambiguously suggested that HESO1 and URT1 predominantly catalyze the uridylation of miRNAs and other TNTase(s) must exist to catalyze the non-uridine nucleotides addition with low activities.
Uridylation of heterochromatic siRNA is largely unaffected by URT1
We also tested whether URT1, as like HESO1, plays a role in siRNA uridylation. A reduction in 3’ uridylation and/or an increase in 3’ end trimming were observed in several ta-siRNAs, suggesting that these ta-siRNAs are also recognized by URT1 (S4 Fig). Unexpectedly, the 3’ end modification pattern was largely unaffected in all three tested heterochromatic siRNAs (hc-siRNAs) in hen1-2 heso1-2 urt1-3 as compared with those in hen1-2 heso1-2 (Fig 4 and S4 Fig), which were shown to be substrates of HESO1 [18]. The enzyme(s) responsible for siRNA uridylation in hen1heso1-2 awaits future characterization. In addition, our results may also suggest a differentiation of terminal uridyl transferases in substrate recognition.
Uridylation of miRNAs by HESO1 and URT1 in HEN1+ background
A small proportion of miRNAs in HEN1 competent background are unmethylated and these miRNAs are extensively uridylated and/or trimmed (Fig 5 and S5B Fig) [8,18,21]. We thus examined whether HESO1 and URT1 are catalyzers of miRNA uridylation in HEN1+ plants. To test this, hen1-2 heso1-2 urt1-3 was crossed to heso1-2 [17] and heso1-2 urt1-3 was obtained from the F2 progeny by genotyping. Like heso1-2, heso1-2 urt1-3 didn't show any morphological defects as compared with Wt under normal growth condition (S5A Fig) [17,24]. Small RNA deep sequencing reads ending with annotated miRNA sites were removed and the rest of miRNA variants were renormalized to 100% and subject to analysis as in Fig 4. We found the patterns for the loss of uridylation and the gain of trimming in Ler, heso1-2 and heso1-2 urt1-3 resembled those observed in hen1-2, hen1-2 heso11-2 and hen1-2 heso1-2 urt1-3 (Fig 5 and S5B Fig), suggesting that HESO1 may act redundantly with URT1 in miRNA uridylation when HEN1 is fully competent.
Fig. 5. The tailing and trimming profiles of miR158a in Ler, heso1-2, heso1-2 urt1-3 resemble those in hen1-2, hen1-2 heso1-2, hen1-2 heso1-2 urt1-3. 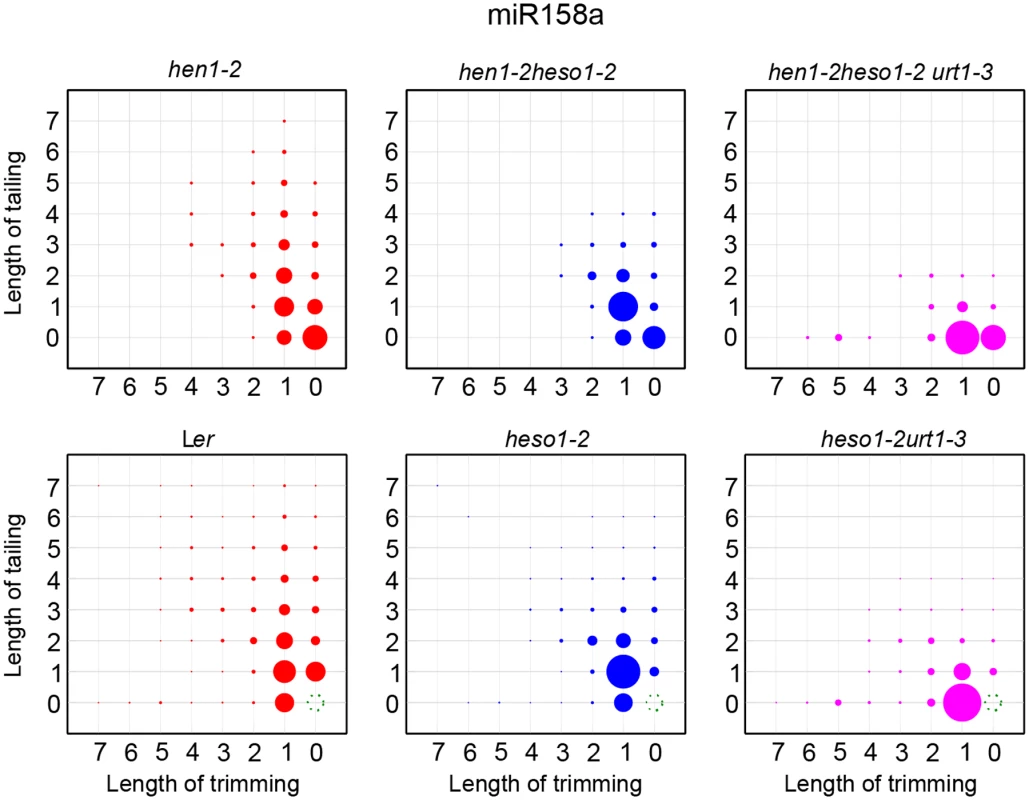
For Ler, heso1-2, heso1-2 urt1-3, reads same as annotated miR158a sequence were removed and the rest of miR158a variants were renormalized to 100%. More interpretations of the matrix are depicted in Fig 4. The P618L substitution impairs URT1 activity, affects its subcellular localization and diminishes its interaction with AGO1
Although the uridylyltransferase activity of URT1 on single stranded RNAs in vitro has been well documented [19,25], the effect of P618L substitution on its activity has not been determined and thus, how URT1P618L increases the fertility in hen1 heso1-2 and alters the miRNA profiles is unclear. To test this, we expressed both GST-URT1 and GST-URT1P618L in E. coli BL21 and purified the recombinant proteins by the Glutathione Sepharose 4B beads (Fig 6A). Consistent with previous reports [19,25], GST-URT1 but not GST alone possessed a robust activity in incorporating UTP to the 3’ end of its RNA substrate (Fig 6B). In addition, GST-URT1 was also able to add a few adenosines (A) or cytidines (C) but only one to two guanosines (G) to its RNA substrate (Fig 6B). In contrast, only residual uridylyltransferase activities were detected for GST-URT1P618L (Fig 6B), indicating the P618L single amino acid substitution severely impairs the URT1 activity without affecting its UTP preference.
Fig. 6. The P618L substitution impairs the URT1 activity, affects its subcellular localization and diminishes its interaction with AGO1. 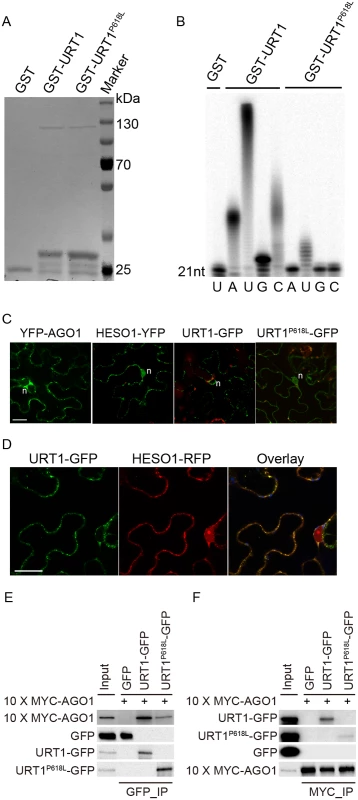
(A) Expression and purification of GST, GST-URT1 and GST-URT1P618L in E.coli BL21(DE3). After purification, proteins were subject to SDS-polyacrylamide gel analysis and monitored by the comassie brillant blue staining. (B) Terminal nucleotidyl transferase activity of URT1 and URT1P618L. 5’ end P32 labeled miR165a were incubated with GST, GST-URT1 or GST-URT1P618L in the presence of various nucleotide triphosphates for 30 minutes. After the reaction, RNAs were extracted and analyzed on a denaturing polyacrylamide gel. (C) Sub-cellular localization of YFP-AGO1, HESO1-YFP, URT1-GFP and URT1P618L-GFP. Note that only about 20% of YFP-AGO1 showed discrete cytoplasmic foci (C) while the remaining ones had a relatively even distribution[26]. For each construct, 40–50 cells were analyzed and a similar nucleus localization pattern was observed. n, nucleus. Scale bar, 20 μm. (D) URT1-GFP colocalized with HESO1-RFP. Paired constructs were coinfiltrated into N. benthamiana leaves. RFP and GFP fluorescence signals were monitored 40–48 h after infiltration by confocal microscopy. Scale bar, 20 μm. (E, F) Interactions between AGO1 and URT1 by the co-immunoprecipitation assay. 10xMYC-AGO1 and respective GFP tagged proteins were co-expressed in N. benthamiana leaves in the presence of P19 for 3 days and protein extracts were incubated with either anti-MYC-protein G-agarose beads or anti-GFP-protein G-agarose beads for 4 hrs to overnight. After reaction, proteins were resolved by the SDS-polyacrylamide gel electrophoresis and detected with respective antibodies. Input = 1%. (E) 10xMYC-AGO1 co-immunoprecipitates with URT1-GFP and URT1P618L-GFP. (F) URT1-GFP and URT1P618L-GFP co-immunoprecipitates with 10xMYC-AGO1. We next examined the localization of URT1 by transient expression in Nicotiana Benthamiana. URT1-GFP was localized to the cytoplasmic foci, a pattern similar to those observed in HESO1 and AGO1 (Fig 6C) [24,26,27]. Indeed, we found that URT1-GFP colocalized with HESO1-RFP at the cytoplasmic foci (Fig 6D), supporting a role of URT1 in miRNA uridylation. While HESO1 was also targeted to the nucleus (100%, n = 43) (Fig 6C) [17], URT1-GFP signals could only be detected around the nuclear envelope but not in the nucleoplasm (100%, n = 53) (Fig 6C). Interestingly, URT1P618L-GFP signals were readily detected in the nucleoplasm (100%, n = 50) and the number of cytoplasmic foci was greatly reduced (n = 50) (Fig 6C), suggesting a change of localization of URT1 by the P618L single amino acid substitution. Western blot analysis suggested that the change of localization of URT1P618L-GFP might be not caused by passive diffusion of truncated URT1P618L-GFP proteins (S6C Fig). Bioinformatics prediction also failed to detect a loss of nuclear export signal (NES) /a gain of nuclear localization signal (NLS) by P618L.
Our recent study suggests that HESO1 uridylates AGO1-associated miRNAs through its interaction with AGO1 [24]. We thus asked whether URT1 also interacts with AGO1 and if so, whether URT1P618L affects its interaction with AGO1. To test this, we transiently co-expressed URT1-GFP/ URT1P618L-GFP and 10xMYC-AGO1 in N. benthamiana. We were able to detect URT1-GFP in 10xMYC-AGO1 immunoprecipitates and vice versa, suggesting an interaction between URT1 and AGO1 (Fig 6E and 6F and S6 Fig). In contrast, GFP and 10xMYC-AGO1 could not co-immunoprecipitate with each other (Fig 6E and 6F and S6 Fig). Moreover, 10xMYC-DCL3 (aa1-280) failed to co-immunoprecipitate with URT1-GFP. These data suggest a direct interaction between URT1 and AGO1 (S6D Fig). The interaction between URT1-GFP and 10xMYC-AGO1 was not affected by the treatment of RNAse A (50 μg/ml), suggesting their association may be RNA-independent (S6D Fig). We also detect a positive, albeit weaker, interaction between URT1P618L-GFP and 10xMYC-AGO1 (Fig 6E and 6F and S6 Fig). However, we could not rule out the possibility that the diminution of protein-protein interaction was caused by the change of sub-cellular localization.
Discussion
We have previously shown that HESO1 is a major enzyme responsible for the uridylation of all types of small silencing RNAs in hen1 in Arabidopsis and also reveals the redundancy of small RNA uridylation [17,18]. Besides HESO1, the Arabidopsis genome encodes additional nine TNTases [17,18]. However, despite the conservation of PAP and/or PAP-associated domains, other sequences among these TNTases are highly divergent, which hinders the prediction of their exact function. Moreover, individually knock out of these nine TNTases in hen1-8 fails to rescue the morphological phenotype of hen1-8 [18]. In this study, we successfully identified URT1 as a functional homologue of HESO1 in triggering miRNA uridylation. In the accompanied paper, Tu et al also show that lack of URT1, but not other TNTases, results in an only minor reduction of uridylation of several miRNAs in hen1-8 [28]. Taken together, these observations suggest that HESO1 and URT1 can compensate for each other to some extent and function synergistically.
A single point mutation in URT1 results in the substitution of Proline by Leucine (P618L) in a yet uncharacterized region (Linker region) that bridges the poly(A) polymerase (PAP) domain and the PAP-associated domain (Fig 1C and 1D). Although P618L is rather conserved among several well-characterized TNTases (Fig 1D), sequence analysis reveals that only HESO1 and URT1 possess a complete “PAP-Linker-PAPA” structure among all ten TNTases in Arabidopsis (S3 Table), suggesting the importance of this structure for their proper function. As P618L diminishes URT1 activity without affecting its UTP preference in vitro (Fig 6B), it is likely that urt1-3 is not a null allele. In agreement with this, we found that miRNA uridylation was not completely abolished in the hen1-2 heso1-2 urt1-3 triple mutant (Fig 3C, S3C Fig and S1 Table). However, we cannot exclude the possibility that the residual miRNA uridylation is catalyzed by other uridydyl transferase(s) and urt1-3 is null in vivo.
URT1 co-localizes with HESO1 and both enzymes interact with AGO1, consistent with their roles in the uridylation of AGO1-associated miRNAs [24,28]. However, unlike HESO1, URT1 is barely detectable in the nucleoplasm and may not affect heterochromatic siRNA uridylation in the hen1heso1 background (Fig 6C, Fig 4 and S4 Fig). Moreover, URT1 appears to be less active than HESO1 in vivo since loss-of-function in URT1 alone has limited effects on the miRNA profile and growth of hen1 [17,18,28]. However, we found that HESO1 and URT1 share very similar catalytic properties in vitro in terms of the UTP preference (Fig 6B) [17]. It shall be noted that an accurate comparison of their activities will require more strict conditions such as the removal of fusion tags. Moreover, our results also suggest that URT1 may be responsible for adding long tails to the 3’ ends of at least some miRNAs (e.g. miR163 and miR164a) in vivo. Alternatively, URT1 may trigger mono - or di-uridylation, which is required for additional tailing by yet uncharacterized TNTase(s). Future studies on their expression regulation and catalytic dynamics will help to understand their differences in the functional output.
Consistent with previous notion [17], the reduced levels of miRNA uridylation are accompanied by the increased levels and extents (more extensive truncation) of trimming, demonstrating that uridylation antagonizes trimming for the occupancy of 3’ ends of miRNAs (Fig 3B and S3B Fig). More interestingly, other types of nucleotides addition (C, A, G) become evident when uridylation was drastically reduced in the hen1-2 heso1-2 urt1-3 triple mutant (Fig 3C, S3C Fig, and S1 and S2 Table), suggesting the involvement of additional TNTases in the miRNA tailing process. Indeed, these non-uridine nucleotides additions are largely unaffected by the loss of functions in HESO1 and URT1, albeit they occur at very low frequencies among different genotypes (S1 and S2 Table). Thus, the landscape of TNTases in small RNA 3’ end tailing is much more complicated than previously appreciated, although the biological relevance of non-uridine tailing modifications remains obscure.
What is the function of HESO1 and URT1 in Wt background? Although no visible morphological defects are detected in either heso1-2 or heso1-2 urt1-3 under normal growth condition, we show that uridylation of unmethylated miRNAs is greatly reduced in heso1-2 and is almost abolished in heso1-2 urt1-3 (Fig 5 and S5 Fig). Since uridylation triggers degradation of unmethylated miRNAs, it is possible that HESO1 and URT1 may play a role in the fine adjustment of miRNA abundance in Wt plants. In fact, it has been hypothesized that uridylation may act coordinately with SMALL RNA DEGRADING NUCLEASEs (SDNs), a class of 3’-to-5’ exonucleases that can remove methylated nucleotides, in the active turnover of miRNAs [29,30]. In addition, HESO1 and URT1 are likely involved in the clearance of 5’ cleavage products of miRNA guided slicing of target mRNAs. In fact, HESO1 together with one or more other TUTases can uridylates 5’ cleavage products and promotes their degradation [24]. Based on the action mode of URT1 in miRNA uridylation, it is plausible to speculate that URT1 is one of other enzyme(s) that catalyzes the uridylation of 5’ cleavage products. Moreover, URT1 has been shown to uridylate some mRNAs with short poly(A) tails [19], suggesting HESO1 and URT1 may have a broad spectrum of substrate RNAs in vivo. While the biological significance of the clearance or tentatively stabilization of their substrate RNAs through uridylation modification is unknown, one can simply envision that heso1-2 and heso1-2 urt1-3 plants are under “sub-healthy” conditions. Examining the effect of HESO1 and URT1 under various stress conditions will provide new clues for their functions.
Materials and Methods
Plant materials
All the Arabidopsis (Arabidopsis thaliana) strains used in this study are in the Landsberg erecta (Ler) background unless otherwise indicated. hen1-1, hen1-2, heso1-2, hen1-1 heso1-2 and hen1-2 heso1-2 are previously described [17,18,31]. Ethyl methanesulfonate (EMS) mutagenesis in hen1-2heso1-2 was conducted according to the Arabidopsis manual [32]. F2 population obtained from a cross between m37-6 and hen1-8 heso1-1 (in the Columbia-0 background) [18] was used for the bulk segregation analysis. To obtain the hen1-1 heso1-2 urt1-3 and heso1-2 urt1-3 mutants, hen1-2 heso1-2 urt1-3 was crossed with hen1-1 heso1-2 and heso1-2, respectively. Individuals containing respective genotypes were identified in the F2 population. hen1-1 and hen1-2 were genotyped according to [31]. urt1-3 was identified by digestion of the URT1dCAPSF/R PCR product with DdeI, which cut urt1-3 but not Wt.
Genomic complementation
For the genomic complementation assay, an approximately 4.2-kb DNA fragment containing the URT1 coding region as well as 1.4 kb promoter region was amplified from Ler genomic DNA using PrimeSTAR (Takara, R045A) with primers URT1gGWF/URT1gGWR and sub-cloned into an Entry vector pENTR-D-topo to produce pENTR-URT1g. The entry clone was transferred into the destination vector pMDC204 [33] by LR recombination to produce pURT1:URT1-GFP. The resulting plasmid was transformed into hen1-2 heso1-2 urt1-3 via floral dipping [34] and hygromycin resistance was used for the transgenic plants selection.
qPCR analysis and Small RNA northern blotting
qPCR analysis and small RNA northern blotting were performed as previously described [17,35].
Small RNA library construction and deep sequencing
Total RNA extracted from inflorescence tissue of indicated genotypes was used for small RNA library construction. Small RNA libraries were prepared as indexed (i.e. individually bar-coded) and sequenced at 51bp single-end on an Illumina Hiseq2000 following the standard protocol. The small RNA sequence data were deposited to Gene Expression Omnibus (GEO) under the accession number GSE60826. Adaptor sequence was removed using Perl scripts. Analysis of small RNA 3’ end modification was performed according to [18]. The reference miRNA sequences were obtained from miRbase v17.0 (http://www.mirbase.org/). For the determination of 5GMC, the 5’ end of miRNA was locked (i.e. miRNAs with 5’ end different from the reference were removed from analysis) and no mismatch was permitted during the alignment. Nucleotides between the annotated 3’ end and 5GMC were considered as trimmed nucleotides, while all nucleotides downstream of 5GMC, regardless of templated or not, were considered as tailed.
Terminal nucleotidyl transerases assay
URT1 CDS was amplified by PCR with primers GST-URT1F/GST-URT1R and cloned into pGEX-4T-1 to generate a Glutathione-S-transferase (GST) tagged plasmid (GST-URT1). The in vitro enzymatic assay was conducted as previously described [17].
Confocal microscopy
pH7-WGY-AGO1 (pAGO1:YFP-AGO1) [26], 35S:HESO1-YFP [17]and 35S:HESO1-RFP [24] were previously described. pURT1:URT1P618L-GFP was generated the same wasy as pURT1:URT1-GFP except that a hen1-2heso1-2urt1-3 genomic DNA was used as a template. Respective constructs were expressed in N. benthamiana leaves for 40~48hr and subjected to imaging using a Nikon A1 confocal mounted on an Eclipse 90i Nikon compound microscope.
Co-immunoprecipitation
The full-length URT1 coding sequence (CDS) was amplified from a Ler cDNA sample by PCR with primers URT1cGWF/URT1cGWR and sub-cloned into an Entry vector pENTR-D-topo to produce pENTR-URT1c. The entry clone was transferred into destination vectors pMDC83 [33] by LR recombination to produce 35S-URT1-GFP. 35S-URT1P618L-GFP was constructed the same way from a hen1-2heso1-2urt1-3 cDNA sample. URT1-GFP/URT1P618L-GFP and 10xMYC tagged AGO1 (in pGWB521 backbone) [24,36] were co-expressed in the presence of P19 in N. benthamiana leaves for 72hr and harvested. Protein extraction and Co-immunoprecipitation analysis were performed as previously described [35]. Anti-GFP (Clontech, 632459) antibody pre-coupled to the protein G-agarose beads (Santa Cruz, sc-2002) and anti-c-MYC (Sigma, A7470) agarose beads were used to immunoprecipitate the GFP tagged and 10xMYC tagged proteins, respectively. Anti-GFP (Covance, MMS-118R) and Anti-MYC (Abgent, AM1007a) were used for the western blot assay.
Bioinformatics
Online softwares cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi), NucPred (http://www.sbc.su.se/~maccallr/nucpred/cgi-bin/single.cgi) and NLStradamus (http://www.moseslab.csb.utoronto.ca/NLStradamus/) were used to predict the NLS peptide and NetNES (http://www.cbs.dtu.dk/services/NetNES/) was used to predict the NES peptide using URT1 and URT1P618L as bait sequences.
DNA and RNA Oligos
All the primers, LNA probes and other RNA/DNA oligos used in this study are listed in S4 Table.
Supporting Information
Zdroje
1. Chen XM (2009) Small RNAs and Their Roles in Plant Development. Annu Rev Cell Dev Biol 25 : 21–44. doi: 10.1146/annurev.cellbio.042308.113417 19575669
2. Ren G, Yu B (2012) Critical roles of RNA-binding proteins in miRNA biogenesis in Arabidopsis. RNA Biol 9 : 1424–1428. doi: 10.4161/rna.22740 23135480
3. Werner S, Wollmann H, Schneeberger K, Weigel D (2010) Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol 20 : 42–48. doi: 10.1016/j.cub.2009.10.073 20015654
4. Mateos JL, Bologna NG, Chorostecki U, Palatnik JF (2010) Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr Biol 20 : 49–54. doi: 10.1016/j.cub.2009.10.072 20005105
5. Bologna NG, Mateos JL, Bresso EG, Palatnik JF (2009) A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. Embo J 28 : 3646–3656. doi: 10.1038/emboj.2009.292 19816405
6. Yu B, Yang Z, Li J, Minakhina S, Yang M, et al. (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307 : 932–935. 15705854
7. Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits rnicroRNAs and short interfering RNAs. Proc Natl Acad Sci U S A 102 : 11928–11933. 16081530
8. Li J, Yang Z, Yu B, Liu J, Chen X (2005) Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis. Curr Biol 15 : 1501–1507. 16111943
9. Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, et al. (2012) PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet 8: e1002616. doi: 10.1371/journal.pgen.1002616 22536158
10. Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, et al. (2012) Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet 8: e1002702. doi: 10.1371/journal.pgen.1002702 22829772
11. Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, et al. (2012) The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet 8: e1002617. doi: 10.1371/journal.pgen.1002617 22548001
12. Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJT, et al. (2010) Hen1 is required for oocyte development and piRNA stability in zebrafish. Embo J 29 : 3688–3700. doi: 10.1038/emboj.2010.233 20859253
13. Horwich MD, Li C, Matranga C, Vagin V, Farley G, et al. (2007) The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17 : 1265–1272. 17604629
14. Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC (2007) Pimet, the Drosophila homolog of HEN1, mediates 2'-O-methylation of Piwi - interacting RNAs at their 3' ends. Genes Dev 21 : 1603–1608. 17606638
15. Kirino Y, Mourelatos Z (2007) The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13 : 1397–1401. 17652135
16. Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, et al. (2007) The 3' termini of mouse Piwi-interacting RNAs are 2'-O-methylated. Nat Struct Mol Biol 14 : 349–350. 17384646
17. Ren G, Chen X, Yu B (2012) Uridylation of miRNAs by HEN1 SUPPRESSOR1 in Arabidopsis. Curr Biol 22 : 695–700. doi: 10.1016/j.cub.2012.02.052 22464191
18. Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, et al. (2012) The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol 22 : 689–694. doi: 10.1016/j.cub.2012.02.051 22464194
19. Sement FM, Ferrier E, Zuber H, Merret R, Alioua M, et al. (2013) Uridylation prevents 3' trimming of oligoadenylated mRNAs. Nucleic Acids Res 41 : 7115–7127. doi: 10.1093/nar/gkt465 23748567
20. Yu B, Bi L, Zhai J, Agarwal M, Li S, et al. (2010) siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis. Nucleic Acids Res 38 : 5844–5850. doi: 10.1093/nar/gkq348 20448024
21. Zhai J, Zhao Y, Simon SA, Huang S, Petsch K, et al. (2013) Plant MicroRNAs Display Differential 3’ - Truncation and Tailing, Modifications Which Are ARGONAUTE1-Dependent and Conserved Across Species. Plant Cell 25 : 2417–2428. doi: 10.1105/tpc.113.114603 23839787
22. Liu C, Axtell MJ, Fedoroff NV (2012) The helicase and RNaseIIIa domains of Arabidopsis Dicer-Like1 modulate catalytic parameters during microRNA biogenesis. Plant Physiol 159 : 748–758. doi: 10.1104/pp.112.193508 22474216
23. Heo I, Ha M, Lim J, Yoon MJ, Park JE, et al. (2012) Mono-Uridylation of Pre-MicroRNA as a Key Step in the Biogenesis of Group II let-7 MicroRNAs. Cell 151 : 521–532. doi: 10.1016/j.cell.2012.09.022 23063654
24. Ren GD, Xie M, Zhang SX, Vinovskis C, Chen XM, et al. (2014) Methylation protects microRNAs from an AGO1-associated activity that uridylates 5' RNA fragments generated by AGO1 cleavage. Proc Natl Acad Sci USA 111 : 6365–6370. doi: 10.1073/pnas.1405083111 24733911
25. Kwak JE, Wickens M (2007) A family of poly(U) polymerases. Rna 13 : 860–867. 17449726
26. Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, et al. (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20 : 3255–3268. 17158744
27. Pomeranz MC, Hah C, Lin PC, Kang SG, Finer JJ, et al. (2010) The Arabidopsis Tandem Zinc Finger Protein AtTZF1 Traffics between the Nucleus and Cytoplasmic Foci and Binds Both DNA and RNA. Plant Physiol 152 : 151–165. doi: 10.1104/pp.109.145656 19897605
28. Tu B, Xu C, Liu L, Zhai JX, Li S, et al. (2014) Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet 11: e1005119.
29. Ramachandran V, Chen X (2008) Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321 : 1490–1492. doi: 10.1126/science.1163728 18787168
30. Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25 : 2383–2399. doi: 10.1105/tpc.113.113159 23881412
31. Chen X, Liu J, Cheng Y, Jia D (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129 : 1085–1094. 11874905
32. Weigel D, Glazebrook J (2002) Arabidopsis. A Laboratory Manual.
33. Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 : 462–469. 14555774
34. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743. 10069079
35. Ren GD, Xie M, Dou YC, Zhang SX, Zhang C, et al. (2012) Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci USA 109 : 12817–12821. doi: 10.1073/pnas.1204915109 22802657
36. Tanaka Y, Nakamura S, Kawamukai M, Koizumi N, Nakagawa T (2011) Development of a series of gateway binary vectors possessing a tunicamycin resistance gene as a marker for the transformation of Arabidopsis thaliana. Biosci Biotechnol Biochem 75 : 804–807. 21512216
Štítky
Genetika Reprodukční medicína
Článek Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in CrustaceansČlánek Adventures in WonderlandČlánek Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and InfectivityČlánek Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in FilamentsČlánek Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in ArabidopsisČlánek Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression ControlČlánek The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent EnhancersČlánek Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of MitochondriaČlánek The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma FormationČlánek Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 4- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Intrauterinní inseminace a její úspěšnost
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
-
Všechny články tohoto čísla
- Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- Adventures in Wonderland
- Experimental Swap of 's Assortative Mating Preferences Demonstrates Key Role of X-Chromosome Divergence Island in Incipient Sympatric Speciation
- Chromosome Replacement and Deletion Lead to Clonal Polymorphism of Berry Color in Grapevine
- The Protein Quality Control Machinery Regulates Its Misassembled Proteasome Subunits
- Genome-Wide Association Study Identifies as a Critical Gene for Susceptibility to Noise-Induced Hearing Loss
- Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and Infectivity
- Viable Neuronopathic Gaucher Disease Model in Medaka () Displays Axonal Accumulation of Alpha-Synuclein
- Multi-locus Analysis of Genomic Time Series Data from Experimental Evolution
- The Genetic Legacy of the Expansion of Turkic-Speaking Nomads across Eurasia
- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- The Pif1 Helicase, a Negative Regulator of Telomerase, Acts Preferentially at Long Telomeres
- Inhibiting K63 Polyubiquitination Abolishes No-Go Type Stalled Translation Surveillance in
- SYD-1C, UNC-40 (DCC) and SAX-3 (Robo) Function Interdependently to Promote Axon Guidance by Regulating the MIG-2 GTPase
- Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in Filaments
- Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3’ Tailing of Small RNAs in Arabidopsis
- Host Genetic Variation Influences Gene Expression Response to Rhinovirus Infection
- Contribution of Large Region Joint Associations to Complex Traits Genetics
- Volatility of Mutator Phenotypes at Single Cell Resolution
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in Arabidopsis
- A Multi-layered Protein Network Stabilizes the FtsZ-ring and Modulates Constriction Dynamics
- Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression Control
- Genome Sequencing of the Perciform Fish Provides Insights into Molecular and Genetic Mechanisms of Stress Adaptation
- Natural Variant E610G Is a Semi-dominant Suppressor of IAP-Induced RNA Processing Defects
- The Alkaline Response Pathway: Identification of a Novel Rim Pathway Activator
- Transgenerational Inheritance of Diet-Induced Genome Rearrangements in Drosophila
- A Single Nucleotide Polymorphism Uncovers a Novel Function for the Transcription Factor Ace2 during Hyphal Development
- DNA Damage Response and Spindle Assembly Checkpoint Function throughout the Cell Cycle to Ensure Genomic Integrity
- The Functional Interplay Between the t(9;22)-Associated Fusion Proteins BCR/ABL and ABL/BCR in Philadelphia Chromosome-Positive Acute Lymphatic Leukemia
- Extreme Recombination Frequencies Shape Genome Variation and Evolution in the Honeybee,
- Beyond Glycolysis: GAPDHs Are Multi-functional Enzymes Involved in Regulation of ROS, Autophagy, and Plant Immune Responses
- Comprehensive Profiling of Amino Acid Response Uncovers Unique Methionine-Deprived Response Dependent on Intact Creatine Biosynthesis
- Windpipe Controls Intestinal Homeostasis by Regulating JAK/STAT Pathway via Promoting Receptor Endocytosis and Lysosomal Degradation
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
- Cross-Population Joint Analysis of eQTLs: Fine Mapping and Functional Annotation
- The Power of Gene-Based Rare Variant Methods to Detect Disease-Associated Variation and Test Hypotheses About Complex Disease
- The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent Enhancers
- Competition between VanU Repressor and VanR Activator Leads to Rheostatic Control of Vancomycin Resistance Operon Expression
- A Missense Change in the Gene Links Aberrant Autophagy to a Neurodegenerative Vacuolar Storage Disease
- Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model
- Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of Mitochondria
- Genome-Destabilizing Effects Associated with Top1 Loss or Accumulation of Top1 Cleavage Complexes in Yeast
- Imputation-Based Population Genetics Analysis of Malaria Parasites
- Heterozygosity for a Hypomorphic Polβ Mutation Reduces the Expansion Frequency in a Mouse Model of the Fragile X-Related Disorders
- Neto-Mediated Intracellular Interactions Shape Postsynaptic Composition at the Neuromuscular Junction
- Ndd1 Turnover by SCF Is Inhibited by the DNA Damage Checkpoint in
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma Formation
- Spastin Binds to Lipid Droplets and Affects Lipid Metabolism
- Maintenance of Glia in the Optic Lamina Is Mediated by EGFR Signaling by Photoreceptors in Adult Drosophila
- Auxin Influx Carriers Control Vascular Patterning and Xylem Differentiation in
- Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
- The Lowe Syndrome Protein OCRL1 Is Required for Endocytosis in the Zebrafish Pronephric Tubule
- Postnatal Loss of Hap1 Reduces Hippocampal Neurogenesis and Causes Adult Depressive-Like Behavior in Mice
- CAPER Is Vital for Energy and Redox Homeostasis by Integrating Glucose-Induced Mitochondrial Functions via ERR-α-Gabpa and Stress-Induced Adaptive Responses via NF-κB-cMYC
- Distinct and Cooperative Activities of HESO1 and URT1 Nucleotidyl Transferases in MicroRNA Turnover in
- The Evolutionary Origination and Diversification of a Dimorphic Gene Regulatory Network through Parallel Innovations in and
- MAPK Signaling Pathway Alters Expression of Midgut ALP and ABCC Genes and Causes Resistance to Cry1Ac Toxin in Diamondback Moth
- Spatio-temporal Remodeling of Functional Membrane Microdomains Organizes the Signaling Networks of a Bacterium
- Asymmetric Transcript Discovery by RNA-seq in . Blastomeres Identifies , a Gene Important for Anterior Morphogenesis
- A Stress-Induced Small RNA Modulates Alpha-Rhizobial Cell Cycle Progression
- Systematic Profiling of Poly(A)+ Transcripts Modulated by Core 3’ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation
- The UPR Branch IRE1- in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast
- A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi
- Co-chaperone p23 Regulates . Lifespan in Response to Temperature
- Re-replication of a Centromere Induces Chromosomal Instability and Aneuploidy
- Shade Avoidance Components and Pathways in Adult Plants Revealed by Phenotypic Profiling
- Lipid-Induced Epigenomic Changes in Human Macrophages Identify a Coronary Artery Disease-Associated Variant that Regulates Expression through Altered C/EBP-Beta Binding
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



