-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
RNA Polymerase II (RNAPII) is the enzyme responsible for the transcription of all protein-coding genes, many non-coding genes and retrotransposons. RNAPII has a unique C-terminal domain (CTD) that is composed of repeats of a seven amino acid sequence and is conserved across species. The CTD functions as a recruiting platform for regulatory and RNA processing factors, making the CTD a master orchestrator of transcription. Mutants containing reduced CTD length show defects in steady state transcription during growth in normal conditions, as well as defects in induced gene expression. However, how CTD length affects retrotransposon gene expression remains to be investigated. Here, we uncovered a direct role for CTD length in limiting retrotransposon gene expression and mobility, revealing a new role for the RNAPII-CTD in maintaining genome integrity.
Published in the journal: . PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005608
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005608Summary
RNA Polymerase II (RNAPII) is the enzyme responsible for the transcription of all protein-coding genes, many non-coding genes and retrotransposons. RNAPII has a unique C-terminal domain (CTD) that is composed of repeats of a seven amino acid sequence and is conserved across species. The CTD functions as a recruiting platform for regulatory and RNA processing factors, making the CTD a master orchestrator of transcription. Mutants containing reduced CTD length show defects in steady state transcription during growth in normal conditions, as well as defects in induced gene expression. However, how CTD length affects retrotransposon gene expression remains to be investigated. Here, we uncovered a direct role for CTD length in limiting retrotransposon gene expression and mobility, revealing a new role for the RNAPII-CTD in maintaining genome integrity.
Introduction
RNA polymerase II (RNAPII) is the enzyme responsible for the transcription of a diverse set of genomic loci, including most protein coding genes, many non-coding genes, and retrotransposons. Rpb1, the largest subunit of RNAPII, contains a unique C-terminal domain (CTD) that is composed of heptapeptide repeats (Y1 S2 P3 T4 S5 P6 S7), the number of which increases with genomic complexity [1,2]. The CTD plays key roles in the regulation and coordination of co-transcriptional processes in vivo, a function linked to its ability to be differentially phosphorylated during the transcription cycle [3–5]. Generally, the RNAPII-CTD is phosphorylated at S5 and S7 residues at the 5’ end of genes, where RNAPII-CTD S5 phosphorylation functions in the release of RNAPII from promoter elements [6–8]. Conversely, the RNAPII-CTD is phosphorylated at S2 residues towards the 3’ end of genes, and this modification plays important roles in coordinating the sequential recruitment of elongation and termination factors [9,10]. In the budding yeast, Saccharomyces cerevisiae, deletion of the entire CTD is lethal, while strains carrying shortened versions are viable but display a range of conditional phenotypes, including reduced growth when exposed to high or low temperatures, inositol-deplete conditions, or to the genotoxic agents formamide and hydroxyurea. [11–15]. CTD truncation mutants also lead to alterations in steady state transcription when grown under normal conditions, as evidenced by decreased or increased mRNA and RNAPII levels at a subset of genes, the later primarily regulated by the transcription factor Rpn4 [12]. In addition, CTD truncation mutants have induction defects at the INO1 and GAL4 genes [14]. Interestingly, loss of the gene encoding for the Cdk8 kinase subunit of the Mediator complex restores many CTD length-dependent growth and gene expression alterations, establishing it as an important contributor to the RNAPII-CTD regulatory circuitry [11,12].
Retrotransposons constitute a major group of genetic elements transcribed by RNAPII, comprising over 3% of the genome and accounting for 5–10% of the total mRNA in haploid yeast [16,17]. In S. cerevisiae, retrotransposons are flanked by long terminal repeats (LTR), which contain promoter and termination sequences required for their transcription [18]. Retrotransposons contain a gag gene, which encodes a structural coat protein, and a pol gene, which encodes a polypeptide that is processed into the enzymes reverse transcriptase, protease and integrase. A crucial step in the replication cycle of retrotransposons is the production of a RNA intermediate by RNAPII [19]. Retrotransposon RNA is required for the synthesis of its proteins and as a template for the synthesis of cDNA, the material that becomes integrated into a new genomic location. Newly integrated copies of retrotransposon cDNA can be transcribed by RNAPII thus giving rise to a new replication cycle.
Transposition events can have grave consequences for genome structure and function, making retrotransposons important sources of genome instability [20]. Specifically, integration within host genes, although rare, can result in disruption of genetic information, while insertion within a transcription regulatory region can alter the expression of adjacent genes [21,22]. To restrict genome instability caused by transposition, all stages in the retrotransposon’s multiplication cycle are kept under tight control by the host cell. For example, in diploid yeast Ty1 gene expression is limited by the a1-alpha2 mating repressor pair [23]. Nonetheless, cellular stress resulting from genetic alterations, exposure to DNA damage conditions or adenine starvation, can result in transcriptional activation of retrotransposons leading to subsequent challenges to genomic integrity through increased Ty1 mobility [24–26]. To drive their expression, retrotransposons exploit several host transcriptional activators including Ste12 and Tec1, both of which drive basal Ty1 transcription in haploid yeast. Supporting a functional role, loss of TEC1 significantly reduces both steady state Ty1 mRNA levels and transposition rates, while its overexpression results in increased rates of Ty1 transposition [27–34].
The S. cerevisiae genome contains multiple retrotransposons, belonging to five families called Ty1 to Ty5 [17,35]. These families are closely related but differ in the order and sequence composition of their encoded genes, with Ty1 and Ty2 elements being further divided into subfamilies. Furthermore, only members of Ty1, Ty2 and Ty3 families are capable of transposition, whereas Ty4 and Ty5 elements are likely inactive due to the accumulation of deleterious mutations [36]. In addition, the yeast genome also contains LTR fragments and lone LTRs, known as delta, sigma, tau, and omega elements [17]. These are LTR sequences remaining in the genome following homologous recombination between the almost identical LTRs flanking retrotransposon, and as such their sequence and location provide a record of previous retrotransposon integration events.
Building on previous work from our laboratory, which investigated the role of the RNAPII-CTD in the expression of most protein coding genes, we focused here on examining its contribution to retrotransposon biology [12]. We found that the RNAPII-CTD had an important role in regulating RNAPII and mRNA levels at Ty1 retrotransposons. Shortening RNAPII-CTD length increased Ty1 mRNA levels and transposition rates, suggesting that the structural integrity of the RNAPII-CTD was important for limiting genome instability caused by transposition events. Several lines of evidence suggested that early events in transcription were important in mediating the enhanced expression of retrotransposons in cells with a shortened RNAPII-CTD. We recapitulated the effect using promoter-based reporter assays, and showed that the transcription factors Ste12 and Tec1, and the mediator subunit Cdk8 were required for the increased Ty1 mRNA levels in the RNAPII-CTD truncation mutant. Cdk8 contributed to retrotransposon gene expression by altering the levels of RNAPII-CTD S5 phosphorylation, Tec1 and Ste12 at Ty1 promoters. Lastly, suggesting a broader role for these factors in RNAPII CTD function, we found that loss of STE12 or TEC1 suppressed RNAPII-CTD truncation mutant growth phenotypes, the latter likely in conjunction with CDK8.
Results
Truncation of the RNAPII-CTD increased RNAPII occupancy at a subset of retrotransposons
Our previous characterization of genes whose expression is dependent on CTD length focused on protein coding genes [12]. To test whether the RNAPII-CTD had a role in the regulation of retrotransposons, we determined whether truncation of the RNAPII-CTD led to alterations in RNAPII (Rpb3 subunit) occupancy at retrotransposons using chromatin immunoprecipitation followed by hybridization to high-density microarrays (ChIP-on-chip). Grouping all 50 retrotransposon elements present in the S. cerevisiae genome revealed significantly increased RNAPII occupancy levels in the rpb1-CTD11 mutant, which contained only 11 heptapeptide repeats, compared to wild type (Fig 1A) (Table 1). We then focused on retrotransposon families rather than individual retrotransposons, as the high degree of sequence similarity amongst retrotransposon family members limited our ability to uniquely identify single elements in the ChIP-on-chip platform. Overall, the Ty1 (31 elements) and Ty2 (13 elements) family of retrotransposons had significant CTD length-dependent increases in RNAPII levels, although the effect at Ty1 elements was more pronounced than at Ty2 elements. For Ty1 and Ty2 elements, representative examples of individual retrotransposons further illustrated the differences apparent from the average occupancy profiles (Fig 1B). In addition, average gene profiles revealed that at Ty1 elements truncation of the RNAPII-CTD resulted in elevated RNAPII levels along the length of the entire element, while at Ty2 elements the increased RNAPII levels occurred primarily at their 3’ ends, suggesting different regulatory roles for the RNAPII-CTD in Ty1 and Ty2 biology (Fig 1C and 1D). The Ty3, Ty4 and Ty5 family of retrotransposons (having 2, 3 and 1 element respectively) were not investigated further because the limited number of members in each family prevented meaningful statistical analysis. However, relative RNAPII occupancy profiles at individual retrotransposons revealed that truncation of the RNAPII-CTD resulted in elevated RNAPII levels at Ty3 elements, while no effects were observed at Ty4 and Ty5 elements (S1 Fig).
Fig. 1. Genome-wide occupancy profiles of RNAPII suggested a role for the RNAPII-CTD in retrotransposon regulation. 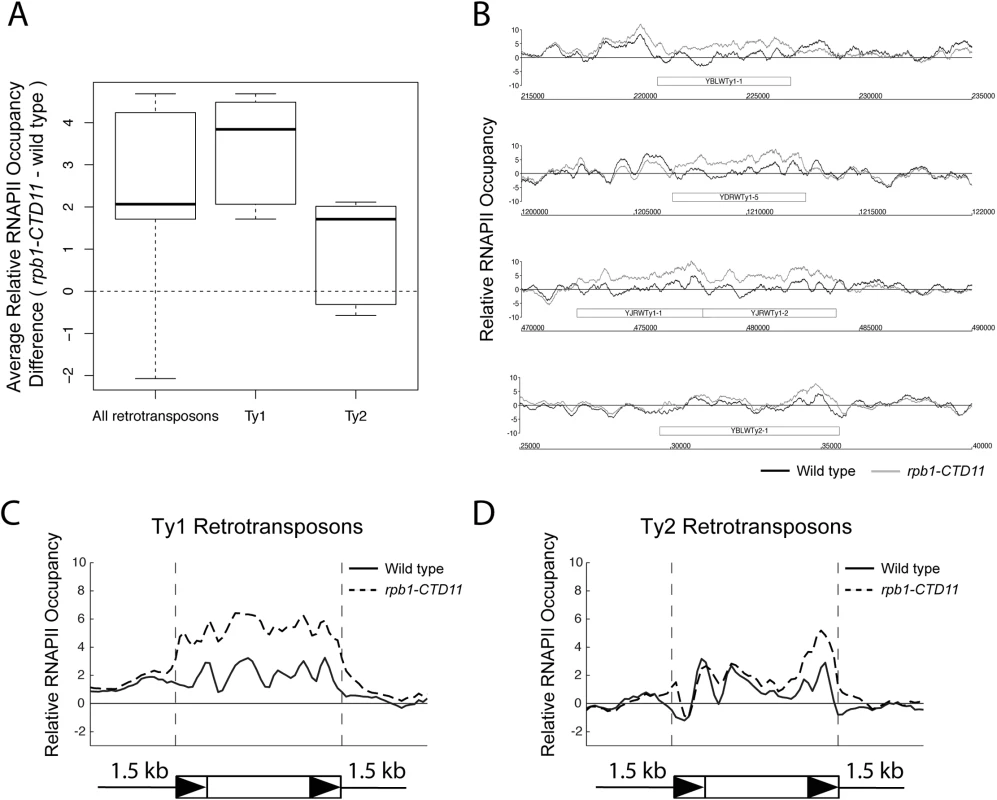
(A) Box plot showing differences in average MAT RNAPII (Rpb3) occupancy scores between the wild type and the rpb1-CTD11 mutant at all, Ty1, or Ty2 retrotransposons. (B) Chromosome plots of relative RNAPII occupancy at representative retrotransposons. Increased RNAPII levels were observed in the rpb1-CTD11 mutant compared to wild type. Labeled boxes indicate the retrotransposon. (C) Average gene profile of RNAPII occupancy at Ty1 retrotransposons showed increased levels along the length of the feature. Below, schematic of an average retrotransposon. Black triangles indicate the LTRs. (D) Average gene profile of RNAPII occupancy at Ty2 retrotransposons revealed increased levels towards the 3’ end of the feature. Tab. 1. Paired t-test p values comparing RNAPII levels in wild type vs <i>rpb1-CTD11</i> at Ty1 and Ty1 retrotransposons and derived-LTRs. 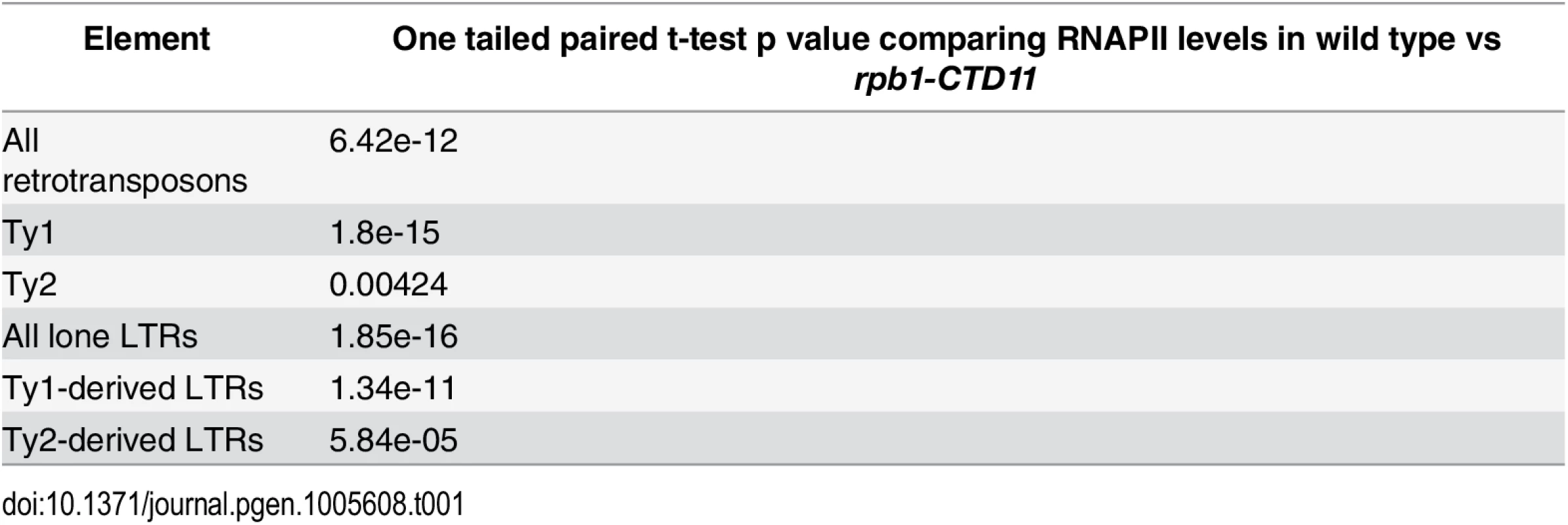
To test whether the effect of truncating the RNAPII-CTD was specific to intact retrotransposons, we determined RNAPII occupancy at lone LTRs and found significantly increased levels in the rpb1-CTD11 mutant compared to the wild type (S2 Fig) (Table 1). Focusing exclusively on delta elements, which are derived from Ty1 and Ty2 elements, revealed that these had significantly increased RNAPII levels in the rpb1-CTD11 mutant when compared to wild type, consistent with our findings at intact retrotransposons (S2 Fig) (Table 1).
Truncation of the RNAPII-CTD resulted in altered occupancy of transcription-associated factors at Ty1 and Ty2 retrotransposons
To mechanistically understand the effect of truncating the RNAPII-CTD at retrotransposons, we next determined if the increased binding coincided with changes in the occupancy of transcription - or chromatin-related factors at these loci. As such, we took advantage of our previously generated genome-wide occupancy maps of the general transcription factor TFIIB, the Mediator subunit Cdk8, the mRNA capping enzyme Cet1, the elongation factor Elf1, and the transcription elongation-associated chromatin mark H3K36me3 in wild type and the rpb1-CTD11 mutant [12]. Truncation of the RNAPII-CTD resulted in significantly increased Cdk8, Cet1 and Elf1 occupancy at Ty1 retrotransposons, albeit with clearly different magnitudes (Table 2) (Fig 2A). In contrast, truncating the RNAPII-CTD had no significant effect on the occupancy of TFIIB and H3K36me3 at Ty1 retrotransposons. At Ty2 elements Cdk8 and Cet1 occupancy also showed significantly increased levels in the rbp1-CTD11 mutant, and no changes were observed for TFIIB and H3K36me3 occupancy (Fig 2B). In contrast to the effect of truncating the RNAPII-CTD at Ty1 elements, Ty2 elements did not show any significant changes in Elf1 occupancy.
Fig. 2. Truncation of the RNAPII-CTD resulted in altered association of a subset of transcription related factors at retrotransposons. 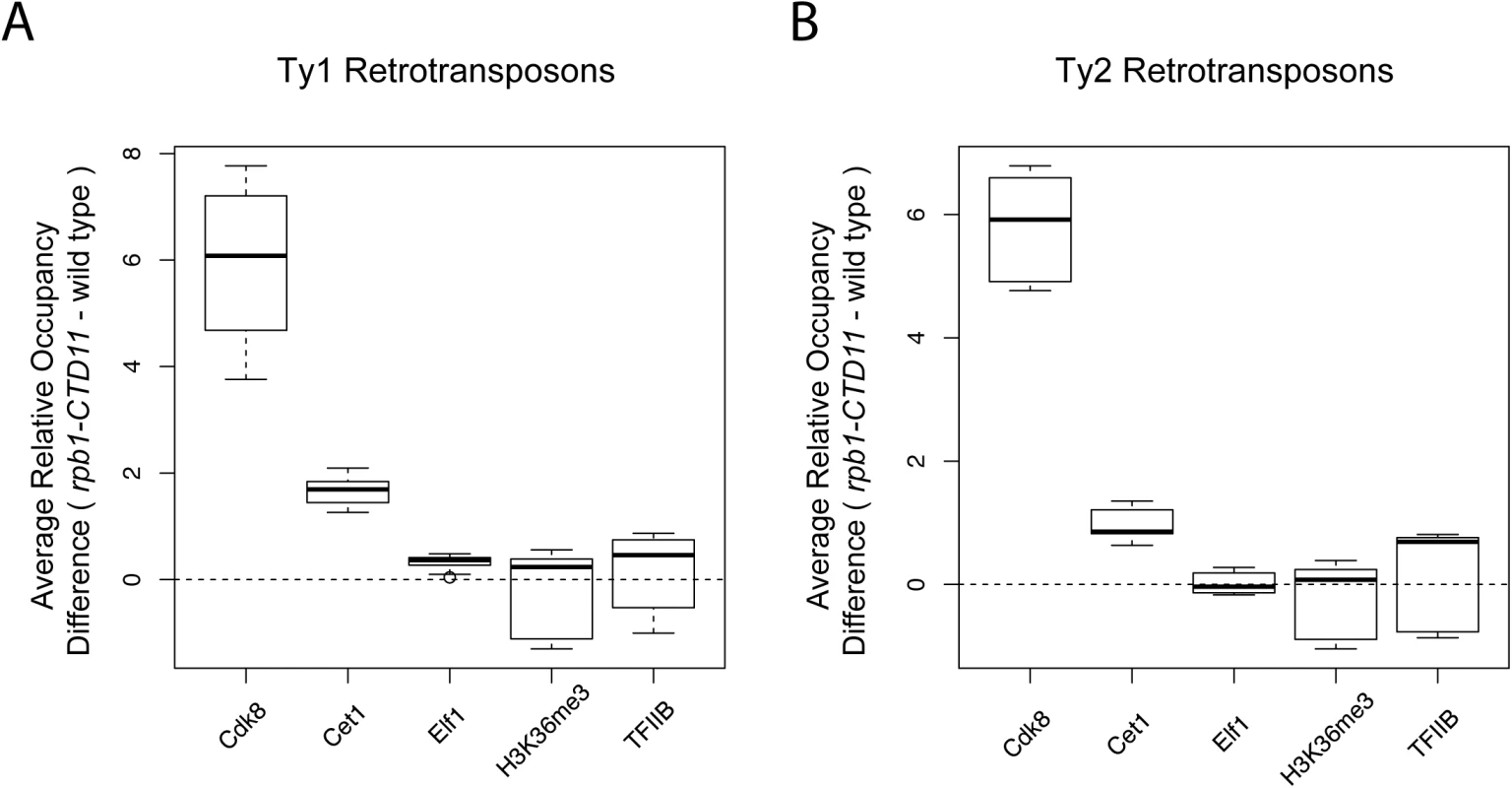
Comparison of MAT average occupancy scores at Ty1 (A) or Ty2 (B) retrotransposons for the Mediator subunit, Cdk8, the mRNA capping enzyme, Cet1, the elongation factor, Elf1, the transcription elongation-associated chromatin mark, H3K36me3, and the general transcription factor, TFIIB, under wild type and rpb1-CTD11 conditions. Tab. 2. Paired t-test p values comparing the levels of transcription or chromatin-related factors in wild type vs <i>rpb1-CTD11</i> at all retrotransposons. 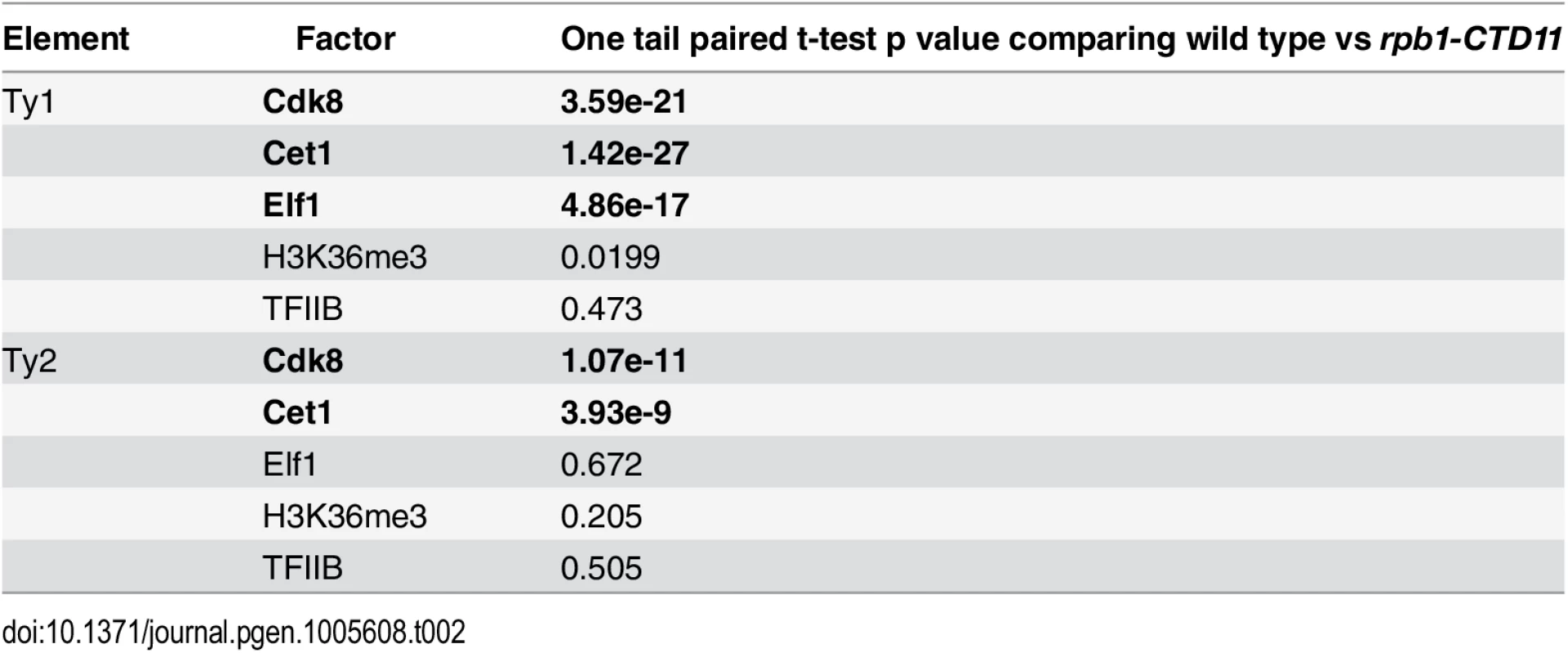
Structural integrity of the RNAPII-CTD was important for maintaining normal Ty1 mRNA levels and transposition rates
Given the increased RNAPII levels at Ty1 and Ty2 retrotransposons in the rpb1-CTD11 mutant, we hypothesized that concurrent changes in the mRNA levels of these elements would occur. We designed an RT-qPCR based assay to quantitatively measure Ty mRNA levels, focusing on regions that were unique to all members of a single retrotransposon family, and compared the levels to those of a control protein coding gene, TUB1 whose mRNA levels are not altered upon truncation of the RNAPII-CTD [12]. Mirroring the RNAPII occupancy data, mRNA levels of Ty1 retrotransposons were significantly increased in the rpb1-CTD11 mutant compared to wild type (Fig 3A). Ty2 retrotransposons had a tendency for modestly increased mRNA levels although this did not reach statistical significance (Fig 3B). The latter was consistent with the weaker effect of truncating the RNAPII-CTD on RNAPII occupancy levels at Ty2 elements. Taking advantage of a set of available mutants lacking S2 (rpb1-S2A) and S7 (rpb1-S7A) phosphorylation sites on the RNAPII-CTD [37], we also investigated the importance of CTD phosphorylation on retrotransposon gene expression. Loss of S7 phosphorylation had no effect on Ty1 or Ty2 mRNA levels (Fig 3C and 3D). In contrast, the rpb1-S2A mutant resulted in significantly increased Ty1 and Ty2 mRNA levels, revealing a broader role for the RNAPII-CTD in retrotransposon gene expression regulation.
Fig. 3. Structural integrity of the RNAPII-CTD was important for normal retrotransposon mRNA levels and transposition rates. 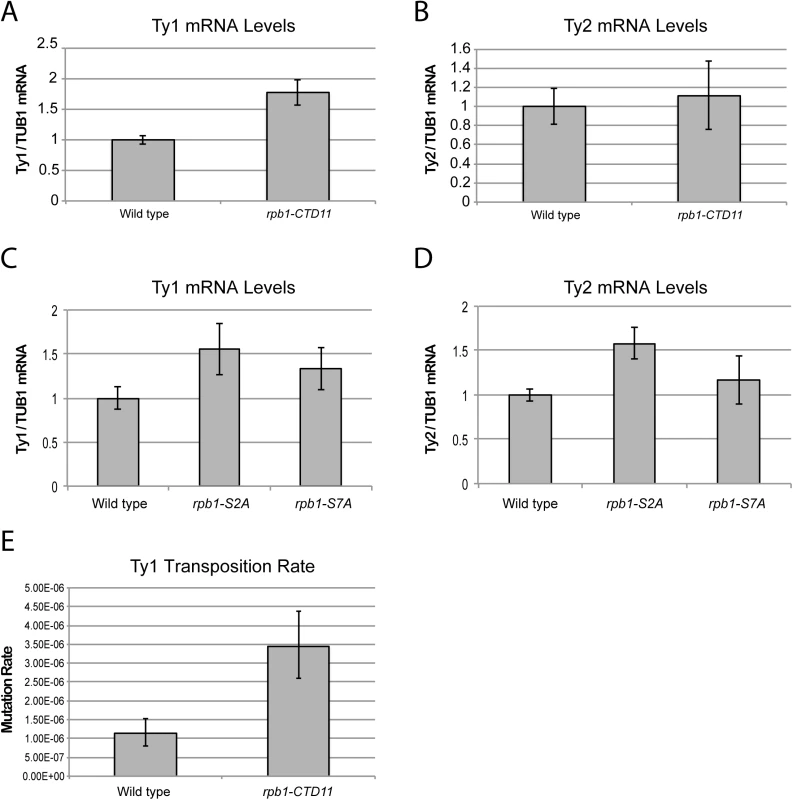
(A) Ty1 mRNA levels were significantly increased in the rpb1-CTD11 mutant compared to wild type. (B) RT-qPCR analysis of Ty2 mRNA levels in the rpb1-CTD11 mutant compared to wild type. RNAPII-CTD S2 phosphorylation was important for maintaining normal Ty1 (C) and Ty2 mRNA levels (D). (E) Transposition rates for Ty1 were increased in the rpb1-CTD11 mutant compared to wild type. Error bars represent 95% confidence intervals. Transposition rates and confidence intervals were calculated using the Fluctuation AnaLysis CalculatOR (FALCOR) web tool. Having established that upon truncation of the RNAPII-CTD Ty1 mRNA levels significantly increased, we sought to determine if this had functional consequences on genome stability manifested by increased transposition rates. Using an established Ty1 cDNA-mediated mobility assay in living yeast cells [38,39], we measured transposition rates in wild type and rpb1-CTD11 mutants. Demonstrating that genomic integrity was indeed compromised upon loss of the RNAPII-CTD repeats, we found that truncation of the RNAPII-CTD resulted in a 3-fold increase in Ty1 transposition rates compared to wild type (Fig 3E).
Loss of CDK8 normalized the increased RNAPII and mRNA levels at Ty1 retrotransposons
Given that loss of CDK8 can suppress a number of RNAPII-CTD truncation mutant phenotypes [11,12], and Cdk8 occupancy was increased at Ty1 and Ty2 retrotransposons in the rpb1-CTD11 mutant, we hypothesized that Cdk8 might contribute to the increased RNAPII occupancy and mRNA levels at Ty elements upon truncation of the CTD. Focusing on Ty1 and Ty2 elements, we did find that RNAPII levels were restored to wild type levels in the rpb1-CTD11 cdk8Δ double mutant, as evidenced by average occupancy scores and average gene profiles (Fig 4A and 4B) (Table 3). Specifically, average RNAPII binding scores at Ty1 and Ty2 elements in the rpb1-CTD11 cdk8Δ double mutant were significantly lower than the scores of the rpb1-CTD11 mutant and were not statistically different from the scores of wild type cells. Furthermore, RNAPII occupancy patterns at representative individual retrotransposons also showed restoration to wild type levels caused by loss of CDK8 in the rpb1-CTD11 mutant (Fig 4C). A similar effect was observed at Ty1 - and Ty2-derived LTRs (Fig 4D). Importantly, changes in RNAPII occupancy were mirrored by changes in mRNA levels at Ty1 retrotransposons, as loss of CDK8 also restored the elevated mRNA levels in the rpb1-CTD11 mutant to wild type levels (Fig 4E).
Fig. 4. Loss of CDK8 normalized the elevated RNAPII and mRNA levels at Ty1 and Ty2 retrotransposons. 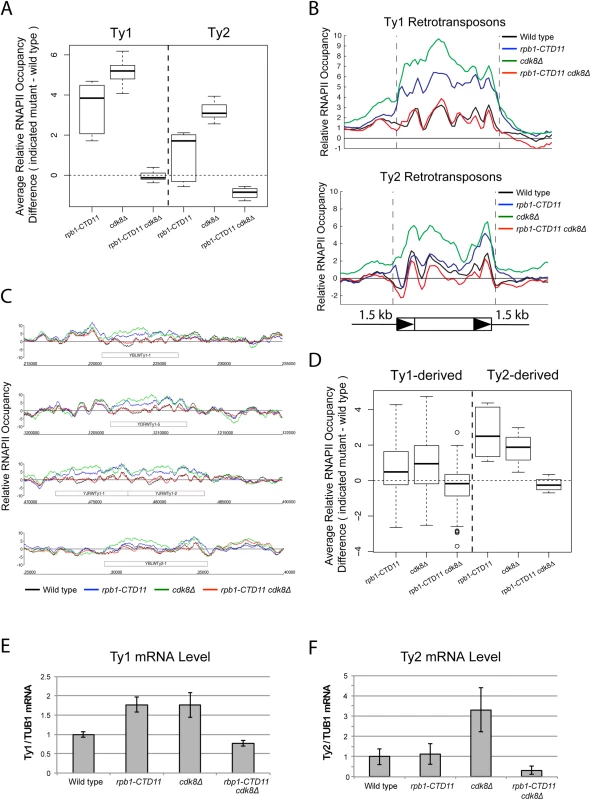
(A) MAT average RNAPII (Rpb3) occupancy scores at retrotransposons revealed elevated levels at Ty1 and Ty2 elements in the single cdk8Δ mutant. In the rpb1-CTD11 background, loss of CDK8 resulted in normalized RNAPII levels at Ty1 and Ty2 retrotransposons. (B) Average gene profiles of RNAPII occupancy at Ty1 (top) or Ty2 (bottom) retrotransposons showed normalized RNAPII levels upon loss of CDK8 in the rpb1-CTD11 mutant. (C) Chromosome plots of RNAPII levels at representative retrotransposons. (D) Loss of CDK8 normalized the elevated RNAPII levels at Ty1- and Ty2-derived LTRs observed in the rpb1-CTD11 mutant. (E) RT-qPCR analysis of wild type, rpb1-CTD11, cdk8Δ and rpb1-CTD11 cdk8Δ revealed that loss of CDK8 significantly normalized the elevated mRNA levels of Ty1 elements in the rpb1-CTD11 background. (F) Ty2 mRNA levels were significantly elevated in the cdk8Δ mutant, an effect that was normalized when combined with an RNAPII-CTD truncation. Tab. 3. Paired t-test p values comparing the levels of RNAPII at Ty1 and Ty2 retrotransposons and Ty-derived LTRs. 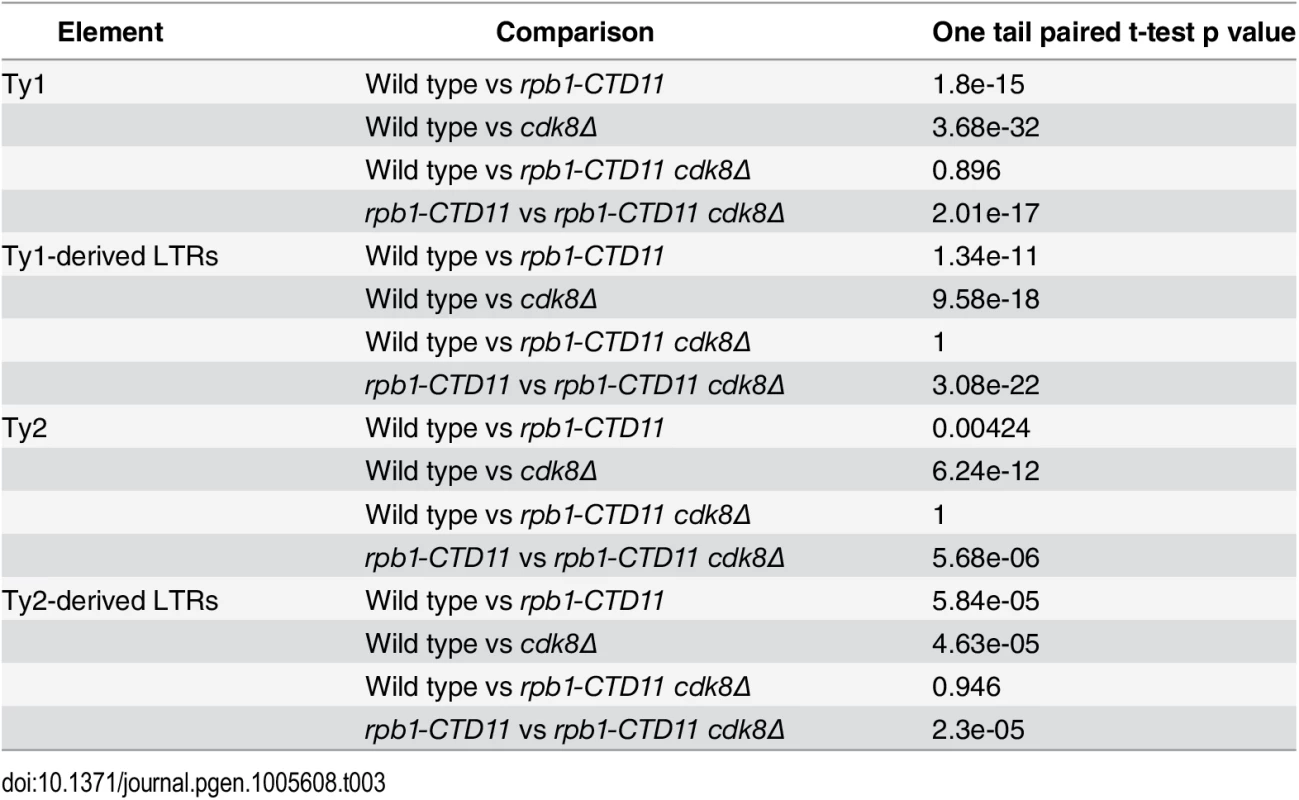
Additional inspection of the RNAPII occupancy profiles revealed that loss of CDK8 alone resulted in significantly elevated average RNAPII levels at Ty1 and Ty2 retrotransposons when compared to wild type (Fig 4A, 4B and 4C) (Table 3). Most interestingly, the increased RNAPII levels in the cdk8Δ mutant were significantly reduced in the rpb1-CTD11 cdk8Δ double mutant, demonstrating that the suppression of the elevated RNAPII levels at Ty1 and Ty2 elements between the RNAPII-CTD mutant and CDK8 deletion was reciprocal. The changes in RNAPII levels coincided with changes in Ty1 and Ty2 mRNA levels, as loss of CDK8 resulted in significant increased Ty1 and Ty2 mRNA levels, which were normalized upon truncation of the RNAPII-CTD (Fig 4F). Collectively, these data suggested that the specific CDK8-dependent phenotypes could be normalized by functional alteration of the RNAPII-CTD, consistent with a reciprocal repressive relationship at Ty1 and Ty2 elements.
The rpb1-CTD11 mutant had elevated RNAPII-CTD S5 phosphorylation levels at Ty1 retrotransposons which were normalized by loss of CDK8
Cdk8 is involved in directly and indirectly regulating RNAPII-CTD phosphorylation levels [7,40,41], thus we determined if CTD phosphorylation levels were associated with the observed changes in retrotransposon gene expression using ChIP-on-chip. Importantly, our profiles of RNAPII-CTD phosphorylation on all genes in the genome were consistent with previously published profiles (S3 Fig)[42–45]. At Ty1 and Ty2 retrotransposons, RNAPII-CTD S2 phosphorylation levels increased in the rpb1-CTD11 mutant compared to wild type and levels remained high in the rpb1-CTD11 cdk8Δ double mutant (Fig 5A, 5B and 5C). RNAPII-CTD S5 phosphorylation levels were also increased at Ty1 and Ty2 elements in the rpb1-CTD11 mutant compared to wild type (Fig 5D, 5E and 5F). Most interestingly, RNAPII-CTD S5 phosphorylation levels were reduced at Ty1 elements in the rpb1-CTD11 cdk8Δ double, an effect that was less prominent at Ty2 elements. Given that the wild type and rpb1-CTD11, cdk8Δ and rpb1-CTD11 cdk8Δ mutants differed substantially in the levels of RNAPII at Ty1 and Ty2 element, we also normalized each RNAPII-CTD S2 and S5 phosphorylation profile to its corresponding RNAPII profile. Although it is likely that this approach strongly penalizes the signal from strains carrying the rpb1-CTD11 alleles, given that their potential for acquiring phosphorylation marks is significantly reduced compared to strains with full length CTDs, we observed similar normalizing effects for S5 phosphorylation at Ty1 retrotransposons. Specifically, the rpb1-CTD11 mutant had significantly increased scores compared to wild type and these were normalized upon loss of CDK8 (S4 Fig).
Fig. 5. Loss of CDK8 decreased the elevated RNAPII-CTD S5 phosphorylation levels at Ty1 retrotransposons observed in the rpb1-CTD11 mutant. 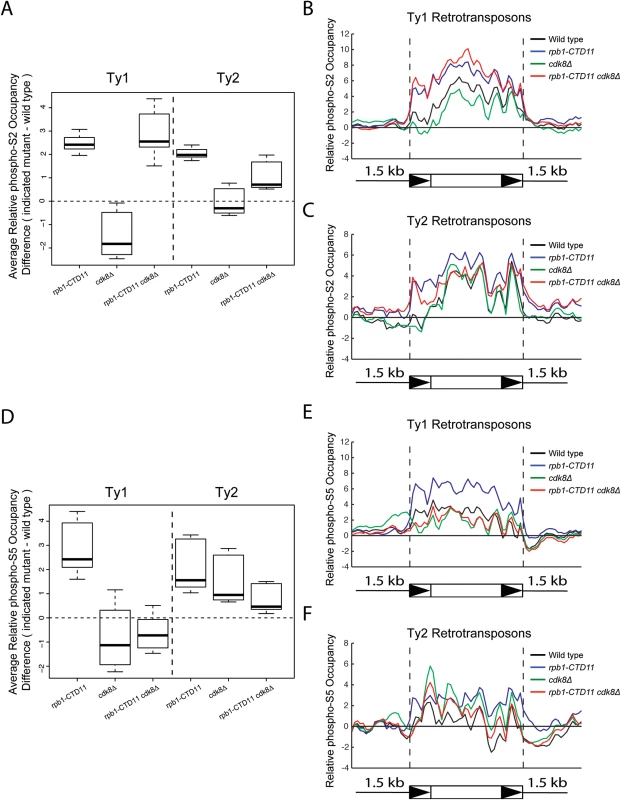
(A) Box plot showing differences in average RNAPII-CTD S2 phosphorylation occupancy scores between the wild type and the rpb1-CTD11, cdk8Δ and rpb1-CTD11 cdk8Δ mutants at Ty1 or Ty2 retrotransposons. Average gene profiles of phospho-S2 occupancy at Ty1 (B) or Ty2 (C) retrotransposons. (D) The elevated average RNAPII-CTD S5 phosphorylation scores at Ty1 and Ty2 elements in the rpb1-CTD11 mutant were reduced upon loss of CDK8. Average gene profiles of phospho-S5 occupancy at Ty1 (E) or Ty2 (F) retrotransposons. Increased Ty1 mRNA alterations were in part due to changes in promoter activity mediated by Ste12 and Tec1
In S. cerevisiae the mediator subunit Cdk8 plays major roles in transcription initiation via phosphorylation of transcription factors and the CTD [7,40,46,47]. Thus, finding that loss of CDK8 normalized the elevated Ty1 mRNA and RNAPII-CTD S5 phosphorylation levels of the rpb1-CTD11 mutant suggested that the regulation likely occurred at the level of transcription initiation. To formally test this possibility, we employed a reporter strategy wherein we inserted more than 1 kb of promoter sequences from representative Ty1 and Ty2 elements into a LacZ reporter plasmid. The representative Ty1 elements selected contained features found in most Ty1 promoters, including putative binding sites for the transcription factors Ste12 and Tec1, which often bind as a heterodimer (Fig 6A) [48,49]. For all representative promoters tested, the reporter assays showed significantly increased β-galactosidase activity in the rpb1-CTD11 and cdk8Δ mutant compared to wild type, suggesting that these promoter sequences alone were sufficient to recapitulate the expression changes of the endogenous retrotransposons observed in these mutants (Fig 6B–6E). However, β-galactosidase activity generally remained high compared to wild type in the rpb1-CTD11 cdk8Δ double mutant, suggesting that events beyond those controlled by promoter sequences were involved in normalizing the elevated mRNA and RNAPII levels at retrotransposons.
Fig. 6. The increased retrotransposon mRNA levels in the rpb1-CTD11 and cdk8Δ mutant were in part due to alterations to promoter activity. 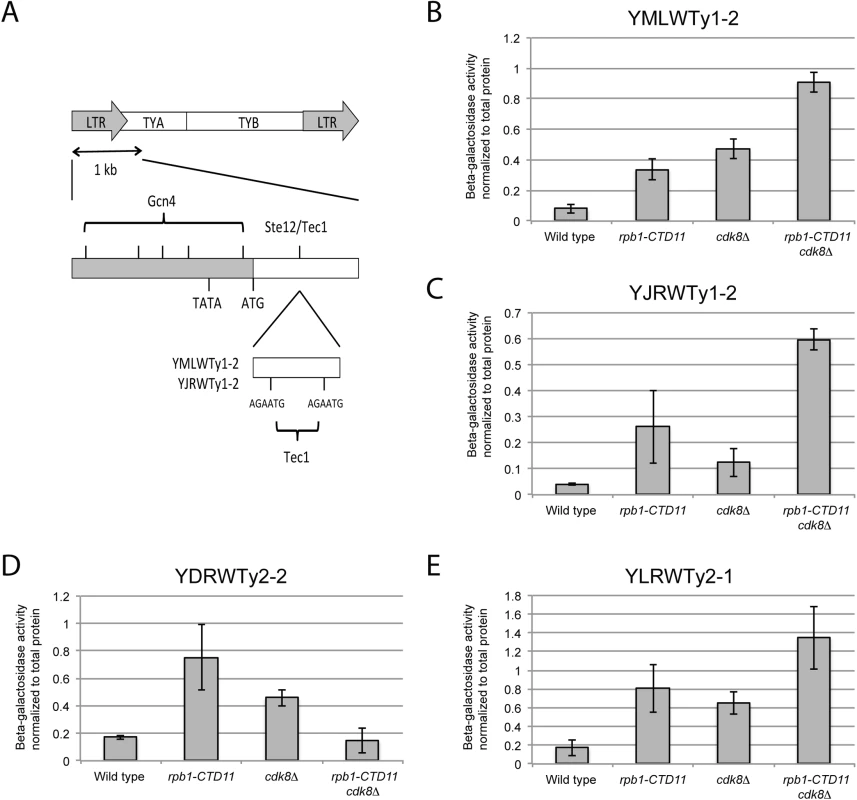
(A) Schematic of an average Ty1 promoter with binding sites for Gcn4 and Ste12/Tec1 labeled [48]. TYA and TYB are retrotransposon genes that encode for the coat protein, and reverse transcriptase, protease, integrase, and RNase H respectively. YMLWTy1-2 and YJRWTy1-2 contain two Tec1 binding sites downstream of the ATG start codon, a feature observed on some Ty1 elements. Reporter assays for YMLWTy1-2 (B), YJRWTy1-2 (C), YDRWTy2-2 (D), and YLRWTy2-1 (E) in wild type, rpb1-CTD11, cdk8Δ, and rpb1-CTD11 cdk8Δ. Further expanding the mechanistic details of the RNAPII-CTD-dependent regulation of Ty1 elements, we found that removal of the binding sites corresponding to the Tec1 consensus sequence affected expression of our reporter constructs. While, the baseline expression from the YMLWTy1-2 and YJRWTy1-2 reporter constructs was not dependent on intact Tec1 binding sites, their removal was sufficient to abolish the increased transcription of the reporter caused by truncation of the RNAPII-CTD (Fig 7A and 7B). The effect of removing the Tec1 binding sites in the cdk8Δ and the rpb1-CTD11 cdk8Δ mutant background was more nuanced. For the YMLWTy1-2 reporter construct, removal of the Tec1 binding sites reduced the increased reporter activity. In contrast, for the YJRWTy1-2 reporter construct removal of the Tec1 binding sites did not reduced β-galactosidase activity. In fact, in the cdk8Δ mutant removing the Tec1 binding sites exacerbated the transcription defect. In conclusion, although Tec1-dependent regulation was required for the increased Ty1 expression levels observed in the rpb1-CTD11 mutant, individual Ty1 elements differed in their requirement for Tec1, as revealed when CDK8 was mutated.
Fig. 7. The increased Ty1 gene expression levels observed in the rpb1-CTD11 mutant were dependent on TEC1 or STE12. 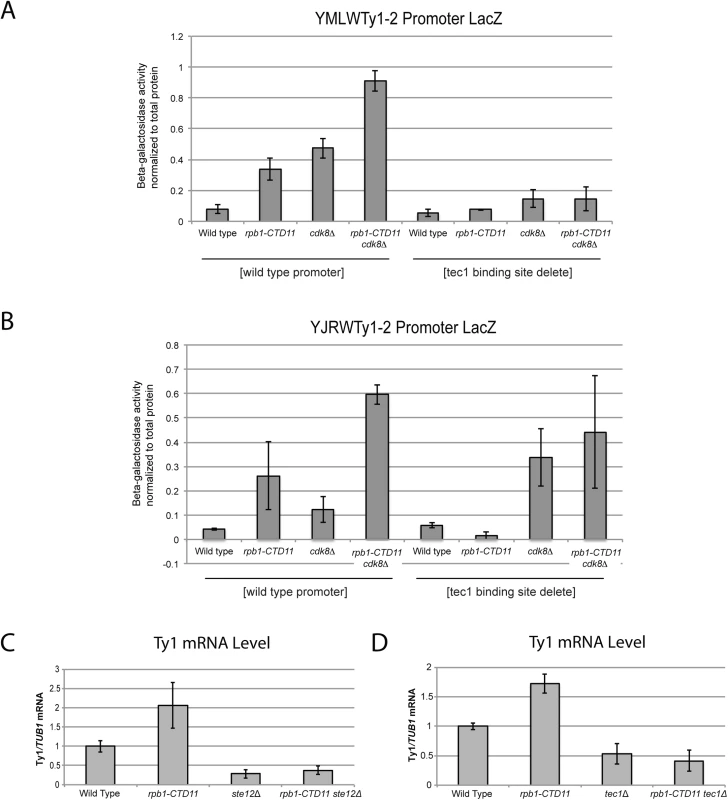
(A and B) Reporter assay for YMLWTy1-2 or YJRWTy1-2, with or without deletion of Tec1 binding sites. Tec1 binding sites were required for the increased promoter activity of Ty1 reporter constructs upon truncation of the RNAPII-CTD. (C and D) RT-qPCR analysis of wild type, rpb1-CTD11, ste12Δ and rpb1-CTD11 ste12Δ or wild type, rpb1-CTD11, tec1Δ and rpb1-CTD11 tec1Δ revealed that loss of STE12 or TEC1 in the rpb1-CTD11 background led to Ty1 mRNA levels similar to those observed in the wild type. Given that the increased expression of Ty1 elements in the RNAPII-CTD truncation mutant resulted in part from Tec1 binding site-dependent alterations in transcription initiation, we next focused on the effect of loss of TEC1 and its regulatory partner STE12 on endogenous Ty1 mRNA levels. The connection to both Ste12 and Tec1 as regulatorss of Ty1 expression was particularly intriguing given that both are also directly (Ste12) and indirectly (Tec1) regulated by Cdk8 [50,51]. Consistent with their known roles in Ty1 gene expression [28,33], ste12Δ and tec1Δ single mutants both had reduced Ty1 mRNA levels compared to wild type (Fig 7C and 7D). More importantly, loss of TEC1 or STE12 reduced the elevated Ty1 mRNA levels in the rpb1-CTD11 mutant. One explanation for the increased Ty1 mRNA levels in the rpb1-CTD11 mutant could be that the protein levels of Ste12 or Tec1 were increased in the rpb1-CTD11 mutant, similar to what we observed for Rpn4 [12]. To this end, we observed no significant differences in total mRNA or bulk protein levels for Ste12 and Tec1 in the rpb1-CTD11 mutant compared to wild type (S5A–S5D Fig). To determine if the relative occupancy of Ste12 or Tec1 was altered at individual genes, their occupancy profiles were determined using ChIP-on-chip. Overall, the ChIP-on-chip profiles of Ste12 and Tec1 were consistent with previously reported profiles and their known relationship to Cdk8 [50,52] (S6 Fig). Although we observed alterations to Ste12 and Tec1 occupancy in the rpb1-CTD11 mutant, these did not easily explain the increased Ty1 mRNA levels in this mutant (Fig 8A–8D). However, in the cdk8Δ mutant the levels of Ste12 correlated with the increased Ty1 and Ty2 mRNA levels observed, and their normalization upon truncation of the RNAPII-CTD (Fig 8A, 8C and 8E). Consistent with Cdk8’s role in targeting Ste12 for degradation [50], the cdk8Δ mutant had significantly increased Ste12 occupancy and increased mRNA levels at Ty1 and Ty2 elements. Upon truncation of the RNAPII-CTD, Ty1 and Ty2 mRNA levels were decreased, as were Ste12 levels at Ty1 and Ty2 promoters. Thus, in the cdk8Δ mutant, CTD length-dependent increases in Ste12 occupancy likely contributed to increased Ty1 and Ty2 mRNA levels.
Fig. 8. Increased levels of Ste12 at Ty1 and Ty2 promoters in the cdk8Δ mutant were normalized by truncation of the RNAPII-CTD. 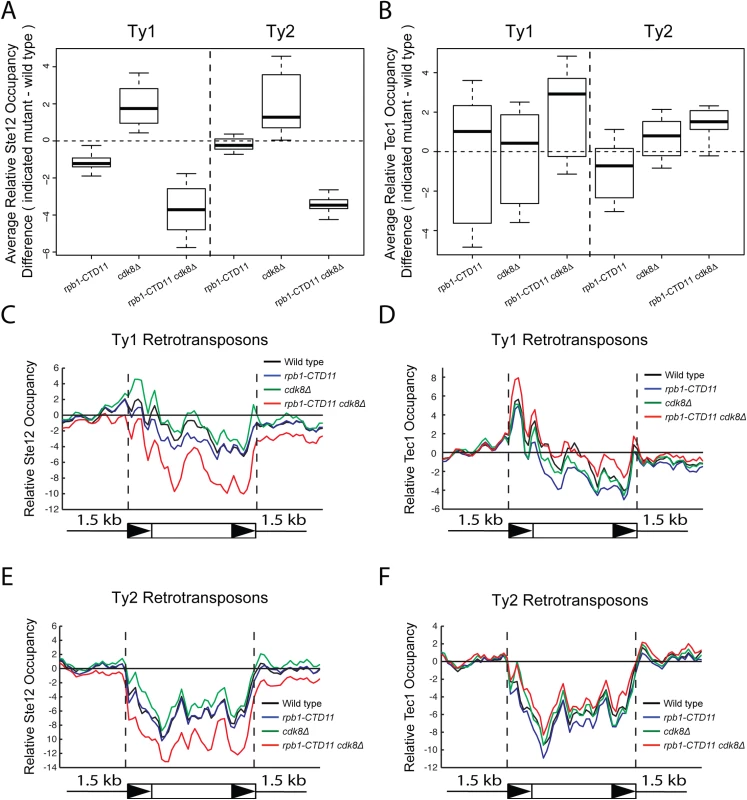
Box plot showing differences in average Ste12 (A) or Tec1 (B) occupancy scores between the wild type and the rpb1-CTD11, cdk8Δ and rpb1-CTD11 cdk8Δ mutant at Ty1 or Ty2 retrotransposons. Average gene profiles of Ste12 (C) or Tec1 (D) occupancy at Ty1 retrotransposons. Average gene profiles of Ste12 (E) or Tec1 (F) occupancy at Ty2 retrotransposons. Finally, given that Ty1 gene expression regulation is mediated by a number of different pathways, we focused on the a1-alpha2-mediated Ty1 repression and determined whether truncation of the RNAPII-CTD also resulted in increased retrotransposons expression in this biologically distinct situation [23]. Since a1-alpha2 repression is exclusive to diploid cells, we generated diploid strains homozygous for the rpb1-CTD11 allele and observed unaltered Ty1 mRNA levels when compared to a wild type diploid strain (S7 Fig). Therefore, the repressive effect of the a1-alpha2 repressor pair was not overcome by truncation of the RNAPII-CTD, suggesting that the increased Ty1 mRNA levels observed in haploid yeast resulted from specific alterations to Ste12/Tec1-mediated regulatory pathway.
A broader role for TEC1 and STE12 in the regulatory circuitry of the RNAPII-CTD
Truncation of the RNAPII-CTD results in both increases and decreases in gene expression under normal growth conditions [12,14]. Having established that loss of TEC1 and STE12 normalized the elevated expression levels of Ty1 elements caused by RNAPII-CTD truncation, we tested whether this relationship extended more broadly to other CTD length-dependent genes. Focusing on four representative protein-coding genes whose expression level is elevated in the rpb1-CTD11 mutant, we found that further loss of TEC1 showed a trend towards reduced mRNA levels, although the effects were small and not statistically significant (S8A-S8D Fig) [12]. As shown by RT-qPCR and sequencing of genomic DNA, loss of TEC1 also did not normalize the reduced RPB1 levels observed in the rpb1-CTD11 mutant, nor did it affect the truncation status of the rpb1-CTD11 allele (S8E Fig). Despite a small effect at representative genes, loss of STE12 or TEC1 robustly suppressed growth phenotypes associated with CTD truncations, suggesting a broader involvement in the cellular manifestation of truncating the RNAPII-CTD. Specifically, deletion of STE12 or TEC1 in the rpb1-CTD11 background robustly normalized the slow growth phenotype of the rpb1-CTD11 mutants when grown at 30 and 16°C (Fig 9A and 9B). In contrast to STE12, loss of TEC1 also suppressed the growth defects of rpb1-CTD11 mutants when grown at 37°C and when exposed to genotoxic agents such as hydroxyurea or formamide, indicating that the tec1Δ mutation was a more robust suppressor of rpb1-CTD11 growth phenotypes than STE12. Overall, the suppression pattern observed for loss of TEC1 was similar to that previously reported by loss of CDK8 [12]. Therefore, we used genetic analysis to test whether this was achieved via independent or overlapping pathways. The strength and condition spectrum of suppression in the rpb1-CTD11 cdk8Δ tec1Δ triple mutant was similar to that of the rpb1-CTD11 cdk8Δ and rpb1-CTD11 tec1Δ double mutants respectively (Fig 9C). Thus, CDK8 and TEC1 were epistatic when suppressing RNAPII-CTD truncation phenotypes, suggesting that they functioned in the same pathway.
Fig. 9. Loss of STE12 or TEC1 suppressed growth defects associated with rpb1-CTD11 and the latter functioned in the same pathway as CDK8. 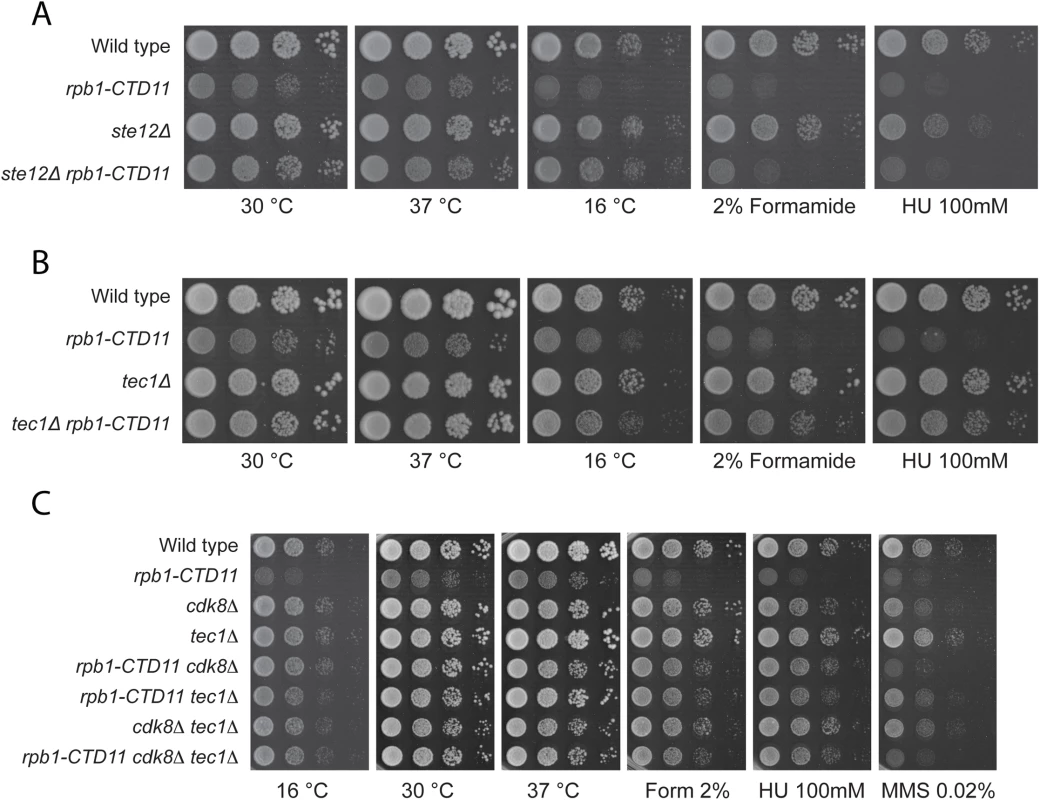
(A-B) Sensitivity of the rpb1-CTD11 mutant to growth under normal and low temperature conditions was suppressed by deletion of STE12 or TEC1. Loss of TEC1 also suppressed the growth defects of the rpb1-CTD11 mutant upon exposure to high temperatures, formamide and hydroxyurea. Ten-fold serial dilutions of the indicated mutants were plated on YPD media at 16, 30 and 37°C or media containing the indicated concentrations of hydroxyurea or formamide. (C) Loss of TEC1 and CDK8 suppressed the sensitivity of the rpb1-CTD11 mutant to growth under low and high temperatures and upon exposure to formamide and hydroxyurea. Ten-fold serial dilutions of the indicated mutants were plated and incubated on YPD media at 16, 30 and 37°C and media containing the indicated concentrations of hydroxyurea or formamide. Discussion
The work presented here highlights an unexpected role for the RNAPII-CTD in the regulation of retrotransposons, leading us to propose that by limiting retrotransposon gene expression, the RNAPII-CTD plays an important role in the maintenance of genomic integrity. Several lines of evidence pointed to a direct role for the RNAPII-CTD in restricting retrotransposon mobility and gene expression. First and foremost, truncation of the RNAPII-CTD unmasked this inhibitory role as it caused a significant increase in the rate of transposition of Ty1 elements. Second, higher mRNA and RNAPII occupancy levels underpinned this effect across different families of retrotransposons. Third, Cdk8 regulated the high RNAPII occupancy and mRNA expression caused by shortening the RNAPII-CTD, at least in part through promoter-mediated events. Furthermore, the close regulatory circuitry between the RNAPII-CTD, Tec1 and Cdk8 was not limited to retrotransposon expression, as loss of TEC1 suppressed additional CTD truncation phenotypes in a manner similar to loss of CDK8.
Our key finding, that various aspects of RNAPII-CTD integrity were important for inhibiting retrotransposition, is consistent with an increasing appreciation of a broader involvement of RNAPII and its C-terminus in diverse aspects related to the maintenance of genome integrity. For example, yeast strains with shortened RNAPII-CTDs are sensitive to several DNA damaging drugs, including the DNA replication inhibitor hydroxyurea [15]. Interestingly, we found that these sensitivities were suppressed by loss of TEC1. Furthermore, strains with critically short CTDs spontaneously revert to RNAPIIs with increased CTD lengths, suggesting enhanced facility for genomic rearrangements [11]. A role for the RNAPII-CTD is also evident in the critical process of transcription coupled repair, a process which preferentially monitors the integrity of biologically relevant loci that if damaged result in RNAPII stalling, a signal for DNA repair [53]. Repair is attempted first by the nucleotide excision repair pathway, and if unsuccessful, by other repair mechanisms which first require poly-ubiquitination - and proteasome-dependent removal of RNAPII from the template [53]. The latter is dependent on the phosphorylation status of the RNAPII-CTD, which regulates the recruitment and activity of key factors involved in RNAPII ubiquitination such as the E3 ubiquitin ligase, Rsp5 [54].
We observed a 3-fold increase in Ty1 mobility in strains with truncated RNAPII-CTDs, an effect within the range but at the lower end of retrotransposon mobility spectrum [38]. The effect of the rbp1-CTD11 mutant on Ty1 mobility was comparable to that of deleting other classical Ty1 regulators like RTT106. The increase in transposition, cause by altering CTD-length, was most likely a result of increased Ty1 mRNA levels due to higher levels of transcription. Consistent with this, the increased RNAPII levels at Ty elements and concomitant mRNA increases in the rpb1-CTD11 mutant were restored to wild type levels upon loss of the transcription factors Tec1 or Ste12, or the mediator subunit Cdk8. Recapitulation of the increased expression and its dependency on Tec1 in a Ty1 promoter reporter assay provided further support for this mechanism. Furthermore, the increased levels of RNAPII at lone LTRs supported our reporter assays by revealing that the core promoter sequences were sufficient for the initial recruitment of RNAPII with shortened CTDs. However, our analysis of these sites also suggested a nuanced mechanism of Ty1 transcription activation. Specifically, lone LTR genomic loci lack functional Ste12/Tec1 binding sites which tend to be located downstream of the ATG translation start codon. Reconciling this with a clear requirement for Ste12 and Tec1 in mediating the increased expression levels at Ty1 retrotransposons caused by shortening the RNAPII-CTD, suggested that Ste12 and Tec1 functioned to enhance transcription complex assembly on the core promoter sequence. In support of this model, cdk8Δ mutants, which had increased Ste12 occupancy at the promoter also showed increased RNAPII levels at the 5’ end of Ty1 and Ty2 elements, indicative of higher rates of initiation. Thus, these data point to a multi layer approach to transcriptional control at Ty1 promoters, where sequences upstream of the ATG start codon were sufficient for RNAPII recruitment, but additional regulatory layers down-stream functioned to increase rates of transcription initiation. These observations are consistent with previous reports that indicated that the full integrity of the Ty1 promoter was important for full activation [24]. Finally, our results suggest that this model also extends to Ty2 elements, even though differences in their regulation have been reporter. Primarily Ty1 elements depend on TEC1 for their expression while Ty2 elements do not [28].
The effect of the RNAPII-CTD on Ty1 gene expression was reminiscent of the role of the RNAPII-CTD on a subset of Rpn4-regulated genes. Specifically, under normal growth conditions Rpn4-regulated genes [12] and retrotransposons had increased RNAPII and mRNA levels in the rpb1-CTD11 mutant which were dependent on CDK8, and were mediated by alterations to transcription initiation. However, despite the similarities, distinct roles of Cdk8 suggested different transcriptional regulatory processes. Specifically, while Cdk8 was normally present at Rpn4-regulated genes, its loss did not change their expression level. In contrast, at Ty1 elements, Cdk8 levels increased upon truncation of the RNAPII-CTD and Cdk8 alone played a role in their regulation as evidenced by increased RNAPII and mRNA levels in the CDK8 deletion mutant. The increased Ty1 and Ty2 mRNA levels observed in the cdk8Δ mutant compared to the wild type, likely resulted from its function as a negative regulator of Ste12 protein levels [50]. As such, Cdk8 normally functioned to repress Ty1 mRNA levels and its loss led to increased Ste12 levels and transcription initiation (Fig 10). Importantly, the effect of Cdk8 on Ste12 was modulated by RNAPII-CTD length, resulting in decreased Ste12 and Ty1 and Ty2 mRNA levels in the rpb1-CTD11 cdk8Δ double. A different mechanism of Ty1 gene expression regulation was elicited when the RNAII-CTD was truncated. Although truncation of the RNAPII-CTD also led to increased Ty1 mRNA levels, these were not a result of increased Ste12 occupancy. In fact, we observed lower than normal Ste12 levels at Ty1 retrotransposons in the rpb1-CTD11 mutant, an effect likely mediated by the observed increase in Cdk8 recruitment to Ty1 and Ty2 elements. Instead, the increased Ty1 mRNA levels in the rpb1-CTD11 mutant correlated with increased S5 phosphorylation levels, a post initiation event associated with promoter clearance. Thus, it is likely that truncating the RNAPII-CTD elevated the rates of RNAPII promoter clearance, resulting in increased transcription, an effect that was in turn dependent on CDK8. Highlighting two distinct roles for Cdk8 in the regulation of Ty1 gene expression, it also functioned as an activator by stimulating RNAPII-CTD S5 phosphorylation and promoter release. The latter is consistent with its reported role for some protein-coding genes in mammals [55,56]. This model would also be consistent with the reciprocal suppression between Cdk8 and the RNAPII-CTD we observed at Ty1 and Ty2 elements.
Fig. 10. A role for the RNAPII-CTD and Cdk8 in Ty1 gene expression regulation. 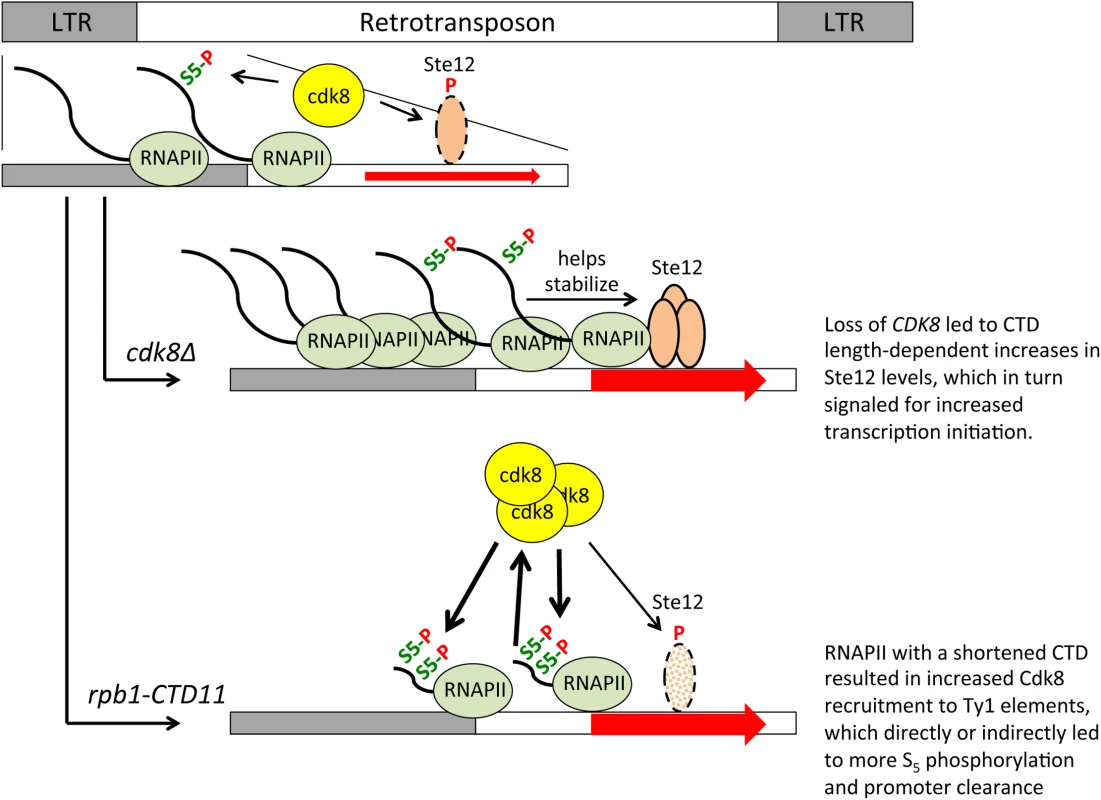
Cdk8 normally functions to target Ste12 for degradation, thus loss of CDK8 stabilized Ste12 at Ty1 promoters resulting in increased transcription initiation. Stability of Ste12 at Ty1 promoters was dependent on full length RNAPII-CTD, thus in the rpb1-CTD11 cdk8Δ double mutant Ste12 levels were normalized leading to wild type levels of transcription initiation. Truncation of the RNAPII-CTD alone resulted in increased Cdk8 recruitment to Ty1 elements and increased S5 phosphorylation levels, a key mark for promoter clearance. The increased S5 phosphorylation levels in the rpb1-CTD11 mutant were direct or indirectly dependent on Cdk8, thus in the rpb1-CTD11 cdk8Δ double mutant S5 phosphorylation levels were normalized resulting in decreased levels of promoter clearance. It is unclear to what extent the RNAPII-CTD length-dependent transcriptional regulatory pathways are linked. Integration of retrotransposons at transcription regulatory sites can alter the transcription of adjacent genes and thus alterations to Ty1 gene expression regulation and transpositions could underlie some of the observed transcriptional defects observed at other genes [12,20]. However, this effect is limited to genes near retrotransposons and we observed no correlation between being in the vicinity of a retrotransposon and having altered mRNA levels in the rbp1-CTD11 mutant. Suggestive of a different type of connection, we noted that truncation of the RNAPII-CTD resulted in STE12-dependent increases in Ty1 mRNA levels and a concomitant decreased expression of protein-coding genes primarily regulated by Ste12 [12]. Given that bulk Ste12 levels were unaltered in the rbp1-CTD11 mutant, one possibility is that the increased transcriptional output at Ty1 elements reduced the cellular pool of Ste12 protein necessary to drive the expression of other genes. Careful examination of the Ste12 ChIP-on-chip profiles did not support this hypothesis indicating that more work beyond the scope of this investigation will be necessary to illuminate the degree of network connectivity between the distinct transcriptional programs found in the rpb1-CTD11 mutant strains, and their detailed mechanistic underpinning. Nonetheless, finding that TEC1 acted as a gene with classical SRB-like phenotypes in that it could suppress the temperature, HU, and formamide sensitivity of the rpb1-CTD11 mutant, suggested that it played fundamental roles in RNAPII-CTD biology. In addition, our results indicate that CDK8, the RNAPII-CTD, TEC1, and to a lesser extent STE12, worked together in a broad network aimed at maintaining genome integrity, which in part involved limiting genome instability caused by Ty1 mobility.
Materials and Methods
Yeast strains
Strains are listed in Table 4. Complete or partial gene deletions were achieved via the one-step gene replacement method [57]. CTD truncations were generated previously by addition of a TAG stop codon followed by a NAT, kanamycin or hygromycin resistance marker at the endogenous RPB1 locus [12]. All strains containing the rpb1-CTD11 allele were confirmed by sequencing. All double mutant strains were generated via mating and tetrad dissection. For STE12 deletion mutants, strains were complemented with pRS316 [STE12] prior to mating. The pRS316 [STE12] plasmid was a gift from Dr. Ivan Sadowski and the pJC573—Ty1his3AI[Δ1] plasmid was a gift from Dr. Joan Curcio. The rpb1-S2A and rpb1-S7A mutants were obtained from Dr. Damien Herman. Plasmids are listed in Table 5.
Tab. 4. Strains used in this study. 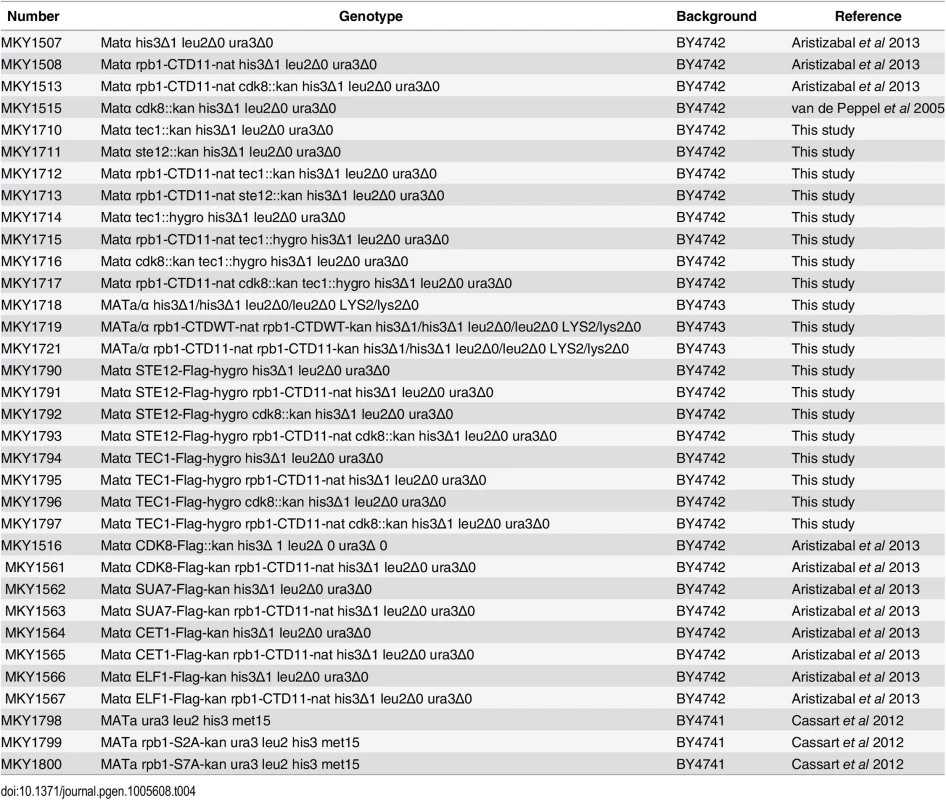
Tab. 5. Plasmids used in this study. 
Genome-wide ChIP-on-chip
Rpb3 and transcription associated factor ChIP-on-chip data used were generated previously [12]. Complete datasets can be found in array-express, code E-MTAB-1341, E-MTAB-1379, and E-MTAB-3906. Briefly, overnight cultures were diluted to 0.15 OD600 and grown to 0.5–0.6 OD600 units. Cross-linking was done with 1% formaldehyde for 20 min. Chromatin was prepared as described previously [58]. Five μl of anti-Rpb3 (Neoclone), or 4.2 μl of anti-FLAG (Sigma) were coupled to 60 μl of protein A Dynabeads (Invitrogen). DNA was amplified using a double T7 RNA polymerase method, biotin labeled, and hybridized to Affymetrix 1.0R S. cerevisiae microarrays. Rpb3 samples were normalized to input and flag tagged samples were normalized to a mock control using the rMAT software [59]. Relative occupancy scores were calculated for all probes using a 300 bp sliding window. Experiments were carried out in duplicate; quantile normalized and averaged data were used for calculating average enrichment scores. To obtain average RNAPII scores at retrotransposons and LTRs, we averaged probes whose start sites fell within the feature start and end positions. For the box plots, the middle line represents the median and the hinges represent the first and third quartile. To obtain average Ste12 and Tec1 occupancy scores, all probes whose start sites fell within the first 1500 bp of the retrotransposon were averaged, as such they included the 5’ LTR and about 1000 bp downstream of the start codon.
Genome-wide ChIP-on-chip of CTD Phospho-isoforms
Cells were grown and cross-linked as described above. All buffers contained protease (roche) and phosphatase inhibitors (10 mM NaPPi, 5 mM EGTA, 5mM EDTA, 0.1 mM Sodium orthovanadate, 5 mM NaF). Chromatin was prepared as described previously [58] and incubated overnight with 50 μl of anti-phospho-serine 2 (3E10) or anti-phospho-serine 5 (3E8) antibodies [8]. Twenty ul of protein G Sepharose (GE Healthcare) was added and incubated for 1.5 hours at 4°C. Beads were washed 3 times with FA lysis buffer with protease and phosphatase inhibitors (50 mM HEPES-KOH (pH 7.5), 1 mM EDTA, 1% Triton X-100, 0.1% SDS), 3 times with FA lysis buffer plus NaCl (500mM), one times with ChIP wash buffer (10 mM Tris-HCL (pH 8.0), 0.25 M LiCl, 1mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate) followed by TE. Immunoprecipitated and purified DNA was amplified, labeled, and hybridized as described above. Phospho-Serine 2 and 5 profiles were normalized to an input or corresponding Rpb3 profiles, and processed as described above [59].
Reporter assays
Reporter plasmids were generated by cloning ~1300 bp of the desired promoter region into the Sal1 and BamH1 sites of pLG669-Z [60]. Specifically, for YJRWTy1-2 and YMLWTy1-2 1321bp were cloned, starting at 517bp upstream of the ORF start and ending 804 bp downstream. For YLRWTy2-1 and YDRWTy2-2 1304 bp were cloned, starting 500 bp upstream of the ORF start and ending 804 bp downstream. For the Ty1 genes, the cloned sequences were selected such that they included previously reported Tec1 and Ste12 binding regions [52]. Tec1 binding sequences were deleted using nested PCR-based methods and cloned into pLG669-Z using the Sal1 and BamH1 sites. A complete list of plasmids can be found in Table 5. Reporter plasmids were transformed into the indicated mutants. Whole cell extracts were generated and clarified as described previously [61]. β-galactosidase activity was normalized to total protein levels determined using the Bradford assay. Measurements were obtained from three independent cultures and error bars represent standard deviations.
Growth assays
Overnight cultures grown on YPD were diluted to 0.5 OD600, 10-fold serially diluted and spotted onto YPD plates with or without the indicated amounts of hydroxyurea (Sigma) or formamide (Sigma). Plates were incubated at the indicated temperatures for 2–4 days.
Reverse Transcriptase PCR (RT-PCR)
RNA was extracted and purified using the Qiagen RNeasy Mini Kit. cDNA was generated using the Qiagen QuantiTect Reverse Transcription Kit. cDNA was analyzed using a Rotor-Gene 600 (Corbett Research) and PerfeCTa SYBR Green FastMix (Quanta Biosciences). Samples were analyzed in triplicate from three independent RNA preparations and the protein-coding gene TUB1 was used as a control given that its expression is not altered upon truncation of the RNAPII-CTD [62]. For measuring Ty1 mRNA levels 6 pg/μl of cDNA were used in a 15 μl PCR reaction. For measuring Ty2 mRNA levels 60 pg/μl of cDNA were used. Retrotransposon specific primers were designed, such that the targeted region was unique to all members of a single retrotransposon family. Primer specificity was evaluated by melt curve analysis of the PCR products. A complete list of primers used in this study can be found in Table 6. Error bars represent standard deviations.
Tab. 6. Primers used in this study. 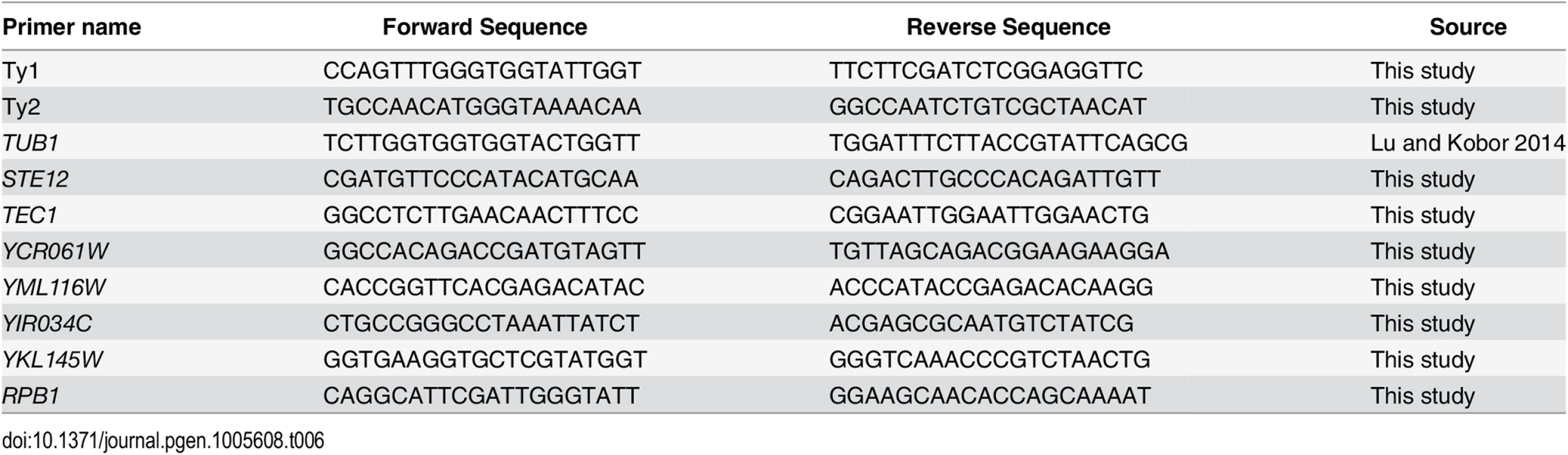
Ty1 cDNA-mediated mobility assay
This assay tracks the mobility of a genome encoded Ty1 element [38,39]. The element has a HIS3 coding sequence inserted in the opposite orientation compared to the Ty1 element. The HIS3 gene is rendered nonfunctional by the insertion of an artificial intron in the same orientation as the Ty1 element, such that it is only spliced when transcribed from the Ty1 element. During the Ty1 transposition cycle, the Ty1 element is transcribed, the intron is spliced and the mature RNA is used for the synthesis of cDNA, which is then integrated into a new genomic location. Newly integrated Ty1 elements contain an undisrupted HIS3 open reading frame and can confer a HIS+ phenotype. Briefly, wild type and rpb1-CTD11 mutants were transformed with Pac1 digested pJR573 DNA. Transformants were selected in SC-URA media and all subsequent growth procedures were done in this media. Overnight cultures for 12 independent colonies for each strain were started. The next morning, cultures were diluted to OD600 0.3 and incubated at 20°C for 24 hours. Following, an aliquot was plated onto SC-URA and SC-HIS-URA to count the total number of cells, and cells with retrotransposition events, respectively. Plates were grown for 2–3 days until colonies were visible and counted. Results were analyzed using the Fluctuation AnaLysis CalculatOR (FALCOR) web tool using the MSS Maximum Likelihood Method to calculate mutation rates. [63]. Error bars represent 95% confidence intervals as calculated by the FALCOR web tool. http://www.mitochondria.org/protocols/FALCOR.html
Supporting Information
Zdroje
1. Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell 1985 09/01;42(2):599–610. 3896517
2. Corden JL, Cadena DL, Ahearn JM Jr, Dahmus ME. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci U S A 1985 Dec;82(23):7934–7938. 2999785
3. Heidemann M, Hintermair C, Voss K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta 2012 Sep 7.
4. Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 2012 Oct 1;26(19):2119–2137. doi: 10.1101/gad.200303.112 23028141
5. Zhang DW, Rodriguez-Molina JB, Tietjen JR, Nemec CM, Ansari AZ. Emerging Views on the CTD Code. Genet Res Int 2012;2012 : 347214. doi: 10.1155/2012/347214 22567385
6. Max T, Sogaard M, Svejstrup JQ. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem 2007 May 11;282(19):14113–14120. 17376774
7. Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell 1998 Jul;2(1):43–53. 9702190
8. Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 2007 12/14;318(5857):1780–2. 18079404
9. Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 2012 Jun 29;336(6089):1723–1725. doi: 10.1126/science.1219651 22745433
10. Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 2000 Oct 1;14(19):2452–2460. 11018013
11. Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 1989 12/01;123(4):715–24. 2693207
12. Aristizabal MJ, Negri GL, Benschop JJ, Holstege FC, Krogan NJ, Kobor MS. High-throughput genetic and gene expression analysis of the RNAPII-CTD reveals unexpected connections to SRB10/CDK8. PLoS Genet 2013 Aug;9(8):e1003758. doi: 10.1371/journal.pgen.1003758 24009531
13. Nonet M, Sweetser D, Young RA. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 1987 Sep 11;50(6):909–915. 3304659
14. Scafe C, Chao D, Lopes J, Hirsch JP, Henry S, Young RA. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature 1990 10/04;347(6292):491–4. 2215664
15. Wong JMS, Ingles CJ. A compromised yeast RNA Polymerase II enhances UV sensitivity in the absence of global genome nucleotide excision repair. Molecular and General Genetics MGG 2001 02/22;264(6):842–851. 11254132
16. Elder RT, St John TP, Stinchcomb DT, Davis RW, Scherer S, Davis RW. Studies on the transposable element Ty1 of yeast. I. RNA homologous to Ty1. II. Recombination and expression of Ty1 and adjacent sequences. Cold Spring Harb.Symp.Quant.Biol. 1981;45(2):581.
17. Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 1998 May;8(5):464–478. 9582191
18. Havecker ER, Gao X, Voytas DF. The diversity of LTR retrotransposons. Genome Biol 2004;5(6):225. 15186483
19. Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell 1985 Mar;40(3):491–500. 2982495
20. Lesage P, Todeschini AL. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet Genome Res 2005;110(1–4):70–90. 16093660
21. Williamson VM, Young ET, Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell 1981 Feb;23(2):605–614. 6258806
22. Roeder GS, Fink GR. DNA rearrangements associated with a transposable element in yeast. Cell 1980 Aug;21(1):239–249. 6250713
23. Errede B, Cardillo TS, Sherman F, Dubois E, Deschamps J, Wiame JM. Mating signals control expression of mutations resulting from insertion of a transposable repetitive element adjacent to diverse yeast genes. Cell 1980 Nov;22(2 Pt 2):427–436. 6256080
24. Todeschini AL, Morillon A, Springer M, Lesage P. Severe adenine starvation activates Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol Cell Biol 2005 Sep;25(17):7459–7472. 16107695
25. Bradshaw VA, McEntee K. DNA damage activates transcription and transposition of yeast Ty retrotransposons. Mol Gen Genet 1989 Sep;218(3):465–474. 2555668
26. Sacerdot C, Mercier G, Todeschini AL, Dutreix M, Springer M, Lesage P. Impact of ionizing radiation on the life cycle of Saccharomyces cerevisiae Ty1 retrotransposon. Yeast 2005 Apr 30;22(6):441–455. 15849797
27. Morillon A, Benard L, Springer M, Lesage P. Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons. Mol Cell Biol 2002 Apr;22(7):2078–2088. 11884596
28. Laloux I, Dubois E, Dewerchin M, Jacobs E. TEC1, a gene involved in the activation of Ty1 and Ty1-mediated gene expression in Saccharomyces cerevisiae: cloning and molecular analysis. Mol Cell Biol 1990 Jul;10(7):3541–3550. 2192259
29. Errede B. MCM1 binds to a transcriptional control element in Ty1. Mol Cell Biol 1993 Jan;13(1):57–62. 8380228
30. Gray WM, Fassler JS. Isolation and analysis of the yeast TEA1 gene, which encodes a zinc cluster Ty enhancer-binding protein. Mol Cell Biol 1996 Jan;16(1):347–358. 8524314
31. Gray WM, Fassler JS. Role of Saccharomyces cerevisiae Rap1 protein in Ty1 and Ty1-mediated transcription. Gene Expr 1993;3(3):237–251. 8019126
32. Madison JM, Dudley AM, Winston F. Identification and analysis of Mot3, a zinc finger protein that binds to the retrotransposon Ty long terminal repeat (delta) in Saccharomyces cerevisiae. Mol Cell Biol 1998 Apr;18(4):1879–1890. 9528759
33. Morillon A, Springer M, Lesage P. Activation of the Kss1 invasive-filamentous growth pathway induces Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol Cell Biol 2000 Aug;20(15):5766–5776. 10891512
34. Conte D Jr, Curcio MJ. Fus3 controls Ty1 transpositional dormancy through the invasive growth MAPK pathway. Mol Microbiol 2000 Jan;35(2):415–427. 10652102
35. Bleykasten-Grosshans C, Friedrich A, Schacherer J. Genome-wide analysis of intraspecific transposon diversity in yeast. BMC Genomics 2013 Jun 14;14 : 399-2164-14-399.
36. Voytas DF, Boeke JD. Yeast retrotransposon revealed. Nature 1992 Aug 27;358(6389):717.
37. Cassart C, Drogat J, Migeot V, Hermand D. Distinct requirement of RNA polymerase II CTD phosphorylations in budding and fission yeast. Transcription 2012 Sep-Oct;3(5):231–234. doi: 10.4161/trns.21066 22771993
38. Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 2001 Dec;159(4):1449–1465. 11779788
39. Bryk M, Banerjee M, Conte D Jr, Curcio MJ. The Sgs1 helicase of Saccharomyces cerevisiae inhibits retrotransposition of Ty1 multimeric arrays. Mol Cell Biol 2001 Aug;21(16):5374–5388. 11463820
40. Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol Cell Biol 2004 02/01;24(4):1721–35. 14749387
41. Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 2000 Sep 7;407(6800):102–106. 10993082
42. Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 2010 Oct;17(10):1272–1278. doi: 10.1038/nsmb.1903 20818391
43. Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, et al. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 2010 Sep;17(9):1154–1161. doi: 10.1038/nsmb.1900 20802488
44. Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 2010 Oct;17(10):1279–1286. doi: 10.1038/nsmb.1913 20835241
45. Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, et al. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 2012 Jan 27;45(2):158–170. doi: 10.1016/j.molcel.2011.11.024 22284676
46. Galbraith MD, Donner AJ, Espinosa JM. CDK8: a positive regulator of transcription. Transcription 2010 Jul-Aug;1(1):4–12. doi: 10.4161/trns.1.1.12373 21327159
47. Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol Cell 1999 05/01;3(5):673–8. 10360183
48. Servant G, Pennetier C, Lesage P. Remodeling yeast gene transcription by activating the Ty1 long terminal repeat retrotransposon under severe adenine deficiency. Mol Cell Biol 2008 Sep;28(17):5543–5554. doi: 10.1128/MCB.00416-08 18591253
49. Wong Sak Hoi J, Dumas B. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot Cell 2010 Apr;9(4):480–485. doi: 10.1128/EC.00333-09 20139240
50. Nelson C, Goto S, Lund K, Hung W, Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 2003 Jan 9;421(6919):187–190. 12520306
51. Raithatha S, Su TC, Lourenco P, Goto S, Sadowski I. Cdk8 regulates stability of the transcription factor Phd1 to control pseudohyphal differentiation of Saccharomyces cerevisiae. Mol Cell Biol 2011 Nov 28.
52. Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, et al. Divergence of transcription factor binding sites across related yeast species. Science 2007 Aug 10;317(5839):815–819. 17690298
53. Wilson MD, Harreman M, Svejstrup JQ. Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim Biophys Acta 2013 Jan;1829(1):151–157. doi: 10.1016/j.bbagrm.2012.08.002 22960598
54. Somesh BP, Sigurdsson S, Saeki H, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Communication between distant sites in RNA polymerase II through ubiquitylation factors and the polymerase CTD. Cell 2007 Apr 6;129(1):57–68. 17418786
55. Gold MO, Rice AP. Targeting of CDK8 to a promoter-proximal RNA element demonstrates catalysis-dependent activation of gene expression. Nucleic Acids Res 1998 Aug 15;26(16):3784–3788. 9685496
56. Belakavadi M, Fondell JD. Cyclin-dependent kinase 8 positively cooperates with Mediator to promote thyroid hormone receptor-dependent transcriptional activation. Mol Cell Biol 2010 May;30(10):2437–2448. doi: 10.1128/MCB.01541-09 20231357
57. Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998 Jul;14(10):953–961. 9717241
58. Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, et al. Linking Cell Cycle to Histone Modifications: SBF and H2B Monoubiquitination Machinery and Cell-Cycle Regulation of H3K79 Dimethylation. Mol Cell 2009 11/09;35(5):626–641. doi: 10.1016/j.molcel.2009.07.017 19682934
59. Droit A, Cheung C, Gottardo R. rMAT—an R/Bioconductor package for analyzing ChIP-chip experiments. Bioinformatics 2010 Mar 1;26(5):678–679. doi: 10.1093/bioinformatics/btq023 20089513
60. Guarente L, Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 1981 Apr;78(4):2199–2203. 6264467
61. Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol 1983;101 : 181–191. 6310321
62. Lu PY, Kobor MS. Maintenance of Heterochromatin Boundary and Nucleosome Composition at Promoters by the Asf1 Histone Chaperone and SWR1-C Chromatin Remodeler in Saccharomyces cerevisiae. Genetics 2014 Feb 27.
63. Hall BM, Ma CX, Liang P, Singh KK. Fluctuation AnaLysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbr?ck fluctuation analysis. Bioinformatics 2009 Jun 15;25(12):1564–1565. doi: 10.1093/bioinformatics/btp253 19369502
64. Hentrich T, Schulze JM, Emberly E, Kobor MS. CHROMATRA: a Galaxy tool for visualizing genome-wide chromatin signatures. Bioinformatics 2012 Mar 1;28(5):717–718. doi: 10.1093/bioinformatics/bts007 22238257
65. Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 1998 Nov 25;95(5):717–728. 9845373
Štítky
Genetika Reprodukční medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 10- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



