-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
Messenger RNA (mRNA) is intrinsically prone to form higher order structures which is optimized for mRNA stability in the cell. We took advantage of recent developments in high throughput sequencing technologies and coupled them with RNA structure-probing approaches to provide a high resolution view of the mRNA secondary structure of Escherichia coli on a global, transcriptome-wide scale. Our data highlight the contribution of mRNA secondary structure as a direct effector of a variety of processes, including translation initiation and termination, mRNA abundance and degradation. This goes beyond the primary function of mRNA as an information entity in the transfer of the genetic information and places it more centrally in regulating fidelity of translation.
Published in the journal: . PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005613
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005613Summary
Messenger RNA (mRNA) is intrinsically prone to form higher order structures which is optimized for mRNA stability in the cell. We took advantage of recent developments in high throughput sequencing technologies and coupled them with RNA structure-probing approaches to provide a high resolution view of the mRNA secondary structure of Escherichia coli on a global, transcriptome-wide scale. Our data highlight the contribution of mRNA secondary structure as a direct effector of a variety of processes, including translation initiation and termination, mRNA abundance and degradation. This goes beyond the primary function of mRNA as an information entity in the transfer of the genetic information and places it more centrally in regulating fidelity of translation.
Introduction
The primary role of mRNA in cellular physiology is to act as an informational molecule for translating ribosomes. Yet, emerging evidence places mRNA more centrally in regulating biogenesis of the encoded protein, including cotranslational folding and insertion and interactions with auxiliary factors [1–3]. mRNA is intrinsically prone to form higher order structures, i.e. secondary and tertiary folded motifs. RNA structures tend to be highly dynamic and undergo conformational changes on a microsecond time scale [4]. Furthermore, one linear single-stranded RNA sequence can potentially adopt several differently complex secondary (such as hairpins and stem-loops) and tertiary folds (i.e. stabilized by interactions between distantly located sequences).
Recent developments in high throughput sequencing technologies and their coupling with RNA structure-probing approaches provided a comprehensive map of the secondary structure of the whole cellular transcriptome of yeast, plants and metazoans [5–11] and highlight the broad contribution of RNA structure to modulating gene expression. Conceptually, the mRNA structure is determined by probing its susceptibility to enzymatic cleavage (nucleases S1, P1 for single stranded and RNaseV1 for double-stranded regions), or chemical modifications (e.g. 2’-hydroxyl alkylation of exposed A, G, C or U with 1-methyl-7-nitrosatoic anhydride (1M7), methylation of exposed N1 of adenines or N3 of cytosines by dimethyl sulfate (DMS), modification of exposed N3 of uridines and to smaller extent of N1 of guanines with 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate) (reviewed in [12]). Global in vitro analysis of the total mRNA of yeast subjected to either single-strand or double-strand enzymatic digestion revealed that coding regions exhibit higher propensity to be involved in secondary structure than non-coding regions [6]. Further in vivo analysis using cell-permeable DMS to probe unpaired A and U argues that on a global level, in rapidly dividing yeast and mammalian cells, the mRNA secondary structure of the coding sequences (CDSs) does not impede translation elongation, highlighting the role of RNA-binding proteins and ATP-dependent helicases in modulating the mRNA dynamics in vivo [8]. Only few mRNA structures that are selected for regulatory purposes persist [8,13,14]. Importantly, probing all four nucleotides with 1M7 (icSHAPE) in mouse embryonic stem cells reveals that some persistent structural elements are similar in in vivo and in vitro [9,15]. Moreover, the in vitro folding landscape of an mRNA does not differ from that in vivo, but the exchange between adjacent structures in vivo is much faster than in vitro [16]. Here we combine the power of three different sequencing technologies, parallel analysis of the mRNA structure (PARS), ribosome profiling and RNA-seq to extract, to our knowledge for a first time, the structural features in mRNA selected for regulation of gene expression in Escherichia coli.
In bacteria, based on available single gene examples (summarized in [17]), it has been axiomatically assumed that secondary structure propensity correlates with mRNA stability which in turn is proportional to mRNA abundance and translatability. However, microarray-based analysis of more than 2000 genes in E. coli shows that computed secondary structure stability is not predictive of increased mRNA abundance [17]. Even highly-translated mRNAs with high abundance can be very unstable [17]. Furthermore, detailed single gene studies have shown the significant influence of tRNA abundance or mRNA secondary structure as key modulators of translation elongation rate [18–20]. Interestingly, in regions with high propensity to secondary structure, codons pairing to high-abundance tRNAs, i.e. translated faster [20],are preferentially selected; secondary structure and fast translating codons act in an opposing manner on translational speed, potentially cancelling out their individual effects and smoothing overall translational speed [21]. In physiological conditions, initiation is rather rate-limiting [22] and initiation rate is affected largely by mRNA sequence features [23]. Reduced folding of codons 3’ adjacent to the start codon enhances expression [24,25].
To address the impact of mRNA secondary structure across the E. coli transcriptome, we used parallel analysis of RNA structure (PARS) coupled to deep-sequencing [6], which reports on the intrinsic propensity of protein (ribosome)-free mRNA to partition in secondary structures. We exploited PARS to select candidate sites for regulatory RNA structures. We then complemented PARS with ribosome profiling [26] and RNA-Seq [27] to determine the impact of RNA structure on translation efficiency and mRNA abundance in the cell, respectively. With this combined approach, we uncovered structural elements that may facilitate different steps of translation. The recognition site of RNase E, a major player in mRNA decay in E. coli, was also inferred from the PARS analysis. Our global analysis corroborates early reports from single-gene studies [28–30] and features on a global level the common recognition signature of RNase E cleavage, which is composed of double-stranded and single-stranded segments. More broadly, our study provides a comprehensive foundation for understanding the impact of mRNA secondary structure in bacterial gene expression with implications in design and engineering of synthetic genes.
Results
PARS reveals globally conserved structural features among E. coli transcripts
To assess the intrinsic propensity of the E. coli transcriptome to partition in secondary structures, we isolated total mRNA (i.e. in absence of proteins and ribosomes) from exponentially growing E. coli culture and subjected it to PARS with some modifications of the original protocol [6] (Fig 1A; details are provided in the Methods section). The total mRNA was either digested with single strand-specific RNase A/T1 or with double strand-specific RNase V1 (Fig 1A) and coupled to a massively parallel sequencing to depth of ~50 million reads (~16 million uniquely mappable reads per sample). RNase T1 and RNase A cleave specifically at unpaired guanosine and pyrimidines (cytosine and uracil), respectively, while RNase V1 cleaves at all four paired bases. The results are highly reproducible across replicates (Pearson correlation coefficients R = 0.96 and R = 0.95 for V1 and A/T1 digestions, respectively, S1 Fig). The PARS score was calculated for each nucleotide, which also exhibits good reproducibility on transcriptome-wide and single-transcript levels (S1C and S1D Fig), and a positive PARS score indicates preferential involvement in double-stranded structure (Fig 1A and 1B). At a selected threshold of 1.0 [6] for reliable reads at each position (S1 Fig), we obtained structural information for ~900,000 nucleotides covering 2,536 E. coli genes. The results from PARS are in excellent agreement with known RNA structures and match four experimentally validated RNA structures (Fig 1B; S2 Fig), including also the whole 16S rRNA. Furthermore, we performed additional independent experimental validation of the ppiC transcript; the PARS values recapitulate the results from orthogonal structural probing of ppiC (S1 Fig).
Fig. 1. PARS analysis. 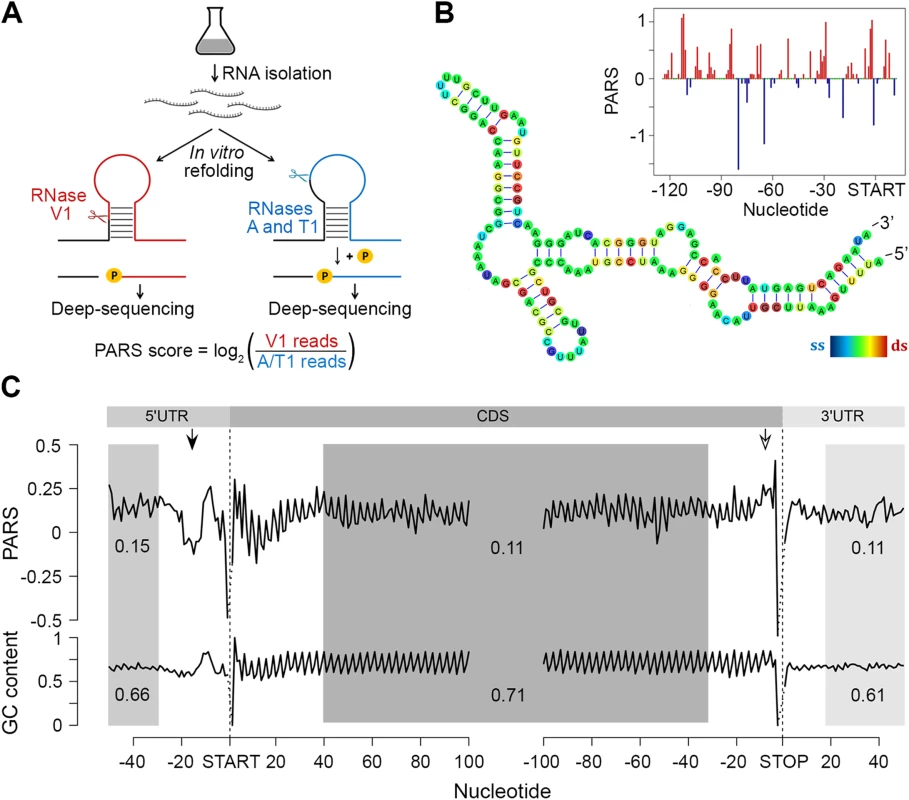
(A) Overview of modified PARS approach. RNase V1 cleaves double-stranded RNA and combination of RNases A/T1 the single stranded RNA with optimal activities at physiological pH (7.0). RNAse A/T1 usage requires an additional phosphorylation step prior to library generation. (B) The PARS score of the rpoS leader sequence (inset) was overlaid with the experimentally determined structure [64]. Double-stranded nucleotides with positive PARS score are colored red, single-stranded nucleotides with negative PARS score–blue, nucleotides with missing PARS score or equal to zero–green. The color intensity of the rpoS nucleotides reflects the PARS scores (rainbow legend). (C) Metagene analysis of protein-coding transcripts. Average PARS score for each nucleotide (top) and GC content (bottom) across the 5’UTRs, CDS and 3’UTRs of all protein-coding transcripts, aligned at the start or stop codon, respectively. For the shaded areas the average PARS scores or GC content is calculated; thus note the deviations from the total GC content of 51% in E. coli. Unstructured region upstream of the start codon and structured sequence preceding the stop codon are marked by arrows with filled and open arrow heads, respectively. Metagene analysis of the transcripts aligned at their start and stop codons shows that E. coli CDSs have a propensity to form double-stranded structure to a level that is similar to the structure propensity of the 5’ - and 3’-untranslated regions (UTRs) (Fig 1C). This global trend is different than that in eukaryotic organisms. In yeast, UTRs are less structured than CDSs [6]. Conversely, in metazoans [31] and humans [11] UTRs are, on average, more structured than coding regions. A well-defined periodic pattern is present only in the CDSs but not in the 5’ and 3’UTRs as detected by discrete Fourier transform (S3 Fig) with first nucleotide being the most structured (S3 Fig). Three nucleotide periodicity is also detected in yeast [6], A. thaliana [5], mouse [32] and human [11] and is intrinsic to the structure of the genetic code (see the periodic pattern of the GC content, Fig 1C), consistent with prior computational predictions for various genomes [33]. We noticed, however, that in some regions the mRNA structure deviates from the nucleotide content, e.g. a uniform unstructured region around 20 nt upstream of the initiation start and more structured region upstream of the termination codon (Fig 1C). These positions may provide candidate sites for functional conformation of mRNA in vivo and we address their role below.
The region 10–30 nt downstream of the initiation was also less structured than the average PARS score of the CDS (Fig 1C). Less structured regions at the 5’ start of the CDSs facilitate initiation and general gene expression [24,25], a trend which is also present in the human [11] but not in the yeast [6] transcriptome.
Intrinsic secondary structure propensity of the CDS influences elongation only locally in some genes
We next asked whether the intrinsic secondary structure propensity of the CDS influences the translation (elongation) efficiency and correlates with mRNA abundance in the cell. We complemented the PARS analysis with ribosome profiling which captures the positions of translating ribosomes with nucleotide resolution [26] which showed high reproducibility between biological replicates on a global (S4 Fig) and single gene level (S1 Fig). We hypothesized that a persisting mRNA structure would induce ribosomal pausing which would be detected by enrichment of ribosome-protected fragments (RPFs) upstream of an mRNA structured stretch. A structured stretch was defined when 6 nt within a window of 10 nt show a positive PARS score (for details see Methods section and S5 Fig). In total, within the CDSs we extracted 908 stretches with high structure propensity in vitro. For the majority of the structured stretches we did not detect an accumulation of the RPF upstream of them (Figs 2A and S5) suggesting that the majority of these structures may not persist in vivo and do not influence the elongating ribosomes that is consistent with the observation in yeast and mammalian cells [8]. Nonetheless, a sizeable fraction of structured sites in the CDS (above the 80th percentile, 87 positions) caused ribosomal pausing, i.e. L1>L2 (Eqs 2 and 3; Fig 2A). Along with the genes with previously validated structures (Fig 2B and S1 Table), our analysis revealed some promising candidates for novel functional RNA structures (Fig 2C; S1 Table). One of the genes, deaD, encodes a DEAD-box RNA helicase that functions in large ribosomal subunit assembly [34] and RNA degradation under cold shock [35]. Contrary to the prevailing views for DeaD function at only low temperature, recent evidence describes its expression over a broad temperature range but with large variation in expression level [36]. It is tempting to speculate that the newly identified persistent structure in deaD (Fig 2C) may regulate its expression level at different temperatures through a structure-induced translational pausing.
Fig. 2. Ribosomal pausing induced by secondary structure in CDS. 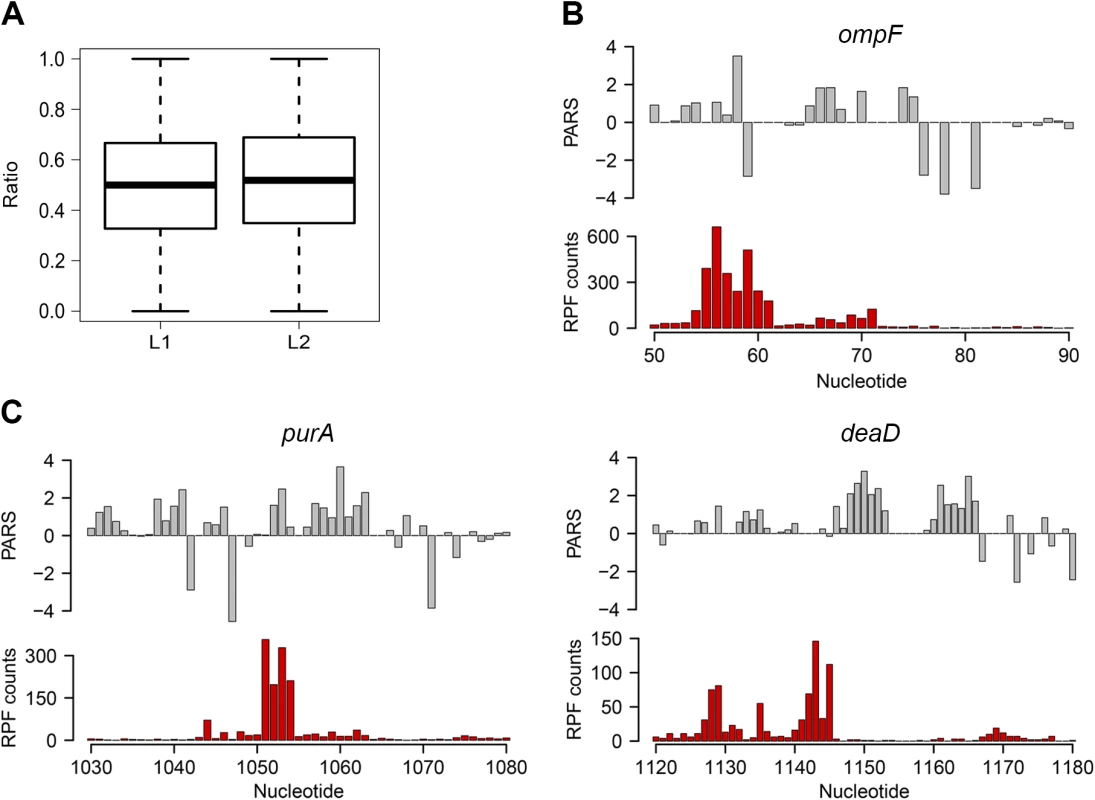
(A) Globally, ribosomal pausing is not significantly affected by the presence of secondary structure in the CDS. Box plot analysis of the ratio of RPF upstream (L1) calculated from Eq 2 and downstream (L2) calculated from Eq 3 of detected secondary structures (P = 0.1209, Kolmogorov-Smirnov test). (B,C) Ribosomal pausing is observed within coding sequences above the 80th percentile (panel A). Examples of ompF transcript with previously validated secondary structure [65] (B) as well as newly detected genes (C) for which a local secondary structure causes non-uniform ribosomal distribution. Aligned PARS score (upper panel, gray) with the RPF counts (bottom panel, red) at each nucleotide. Slow-translated regions, mostly formed by clustering of suboptimal codons, are enriched in E. coli membrane proteins at the beginning of their transmembrane domains [37]. Similarly to yeast, these regions may promote interaction with the signal recognition particle [2] and thus facilitate membrane targeting and translocation. Since a large fraction of the identified structural sites that correlated with accumulated RPF reads were in membrane proteins (S1 Table), we analyzed the distance between the pausing positions and start of the transmembrane domains. The majority of the pausing sites were within 11 to 80 amino acids downstream of the membrane domains (S5 Fig). Strikingly, this distance interval closely resembles the 30–72 amino acid span needed to exit the ribosomal tunnel [38]. Thus, secondary structure-induced ribosome stalling may play a role in membrane targeting in a manner similar to the transient pausing of translation by suboptimal codons [2,37].
mRNA abundance correlates with the mean structural propensity of the coding sequence
Clearly, under physiological conditions, the secondary structure propensity of the majority of CDSs had no impact on the elongating ribosomes. However, mRNA structure is important for a variety of processes, including maintenance of stability and half-life [39]. To quantify the transcriptome, we performed an RNA-Seq experiment [27] which exhibited high reproducibility between biological replicates (S4 Fig). Comparison of the mean PARS score over the CDS revealed a clear correlation with the mRNA abundance (Fig 3A and 3B): the 30% most abundant transcripts exhibited higher secondary structure than the 30% least abundant genes (p = 2.2*10−16, Mann-Whitney test, Fig 3C). Thus, we next asked whether low abundance transcripts are more susceptible to degradation. In E. coli, RNase E is a key enzyme in RNA metabolism and has a major influence on the mRNA life cycle [40]. Recent RNA-Seq-based analysis identified ~1,800 RNase E target sites within E. coli mRNAs [41]. Within the genes with a transcript load over the threshold of 1.0 (S1E Fig), we identified 64 RNase E cleavage positions (Fig 3D, S2 Table) which score among the first 100 cleavage sites [41]. However, those genes did not cluster within the gene group with the lowest abundance and lowest propensity to form secondary structure.
Fig. 3. mRNA structure correlates with mRNA abundance. 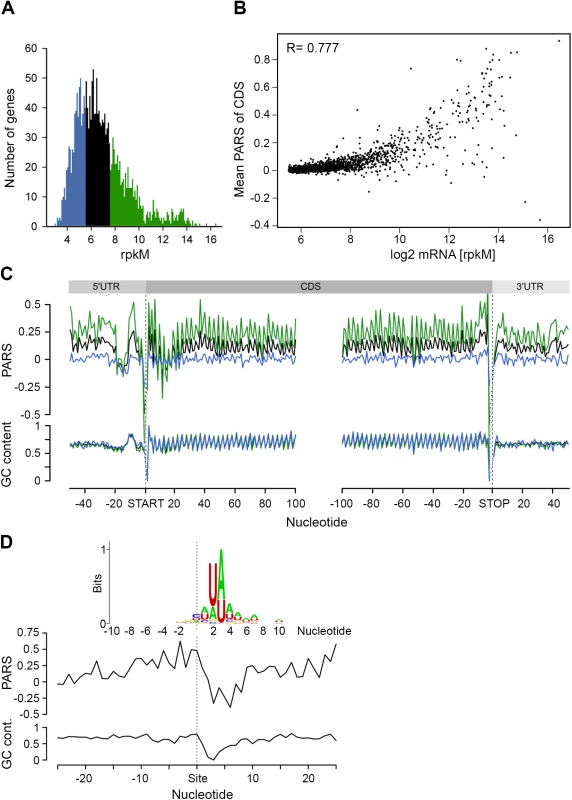
(A) Distribution of transcript abundance, expressed in gene read counts normalized by the length of CDS per kilobase and the total mapped reads per million (rpkM). The 30% least (blue) and most (green) abundant genes from the reliably detected genes (S4 Fig) are highlighted. (B) Dependence of the mean PARS score on the mRNA abundance of the middle (black) and most (green) abundant transcripts as defined in panel A. R = 0.777, Pearson correlation coefficient. (C) Average PARS score (top) and GC content (bottom) for each position of all transcripts (black curve) as well as the 30% most (green) and least (blue) abundant. (D) Average PARS score (top) and GC content (bottom) for each position around the top 64 RNase E cleavage sites (S2 Table). Inset, the sequence logo of the aligned RNase E cleavage sites, spanning from -10 to +10 nt. The cleavage site of RNase E is at an unpaired sequence [41] which lacks a specific sequence motif but is rather enriched in A and U (Fig 3D, inset). Single gene studies propose the importance of stem-loop structures 5’ adjacent to the A/U rich target sites of RNase E [28–30]. Strikingly, we observed this common signature for the 64 identified RNaseE target sites: the unpaired target region is preceded by a structured mRNA stretch (Fig 3D). Also, this structural signature is common for all additional ~1,800 RNase E target sites. Furthermore, we analyzed the structural features of additional endonucleases which have been identified under RNase E-depleted conditions [41]. The target sites of other endonucleases bears no secondary structure upstream the cleavage site and thus significantly differ than that of RNase E (S6 Fig) implying that the structural signature of the RNase E target sites is of importance for its recognition. Notably, the target sites of all endonucleases lack a specific consensus sequence motif but are rather enriched in specific nucleotides (S6 Fig). This observation is consistent with mutational study of the unpaired RNase E cleavage site, which suggests that RNase E cleavage is affected by the extent of A and U rather than their order [29].
Unstructured sequence upstream of the start codon is a general feature of E. coli genes
We detected a unique structural feature for the E. coli transcripts which is not present in yeast and human [6,11]: the region 7–12 nt upstream of the start codon is significantly more structured (mean value 0.17) than the average CDS (mean value 0.11, Fig 1C, marked with an arrow). A large fraction of genes in E. coli is initiated by Shine-Dalgarno (SD) sequence upstream of the start codon and its hybridization strength to the anti-SD of the 16S rRNA (3’-UCCUCCAC-5’) determines initiation fidelity. We computed the minimum hybridization free energy (MHE) between the anti-SD sequence and genes whose translation was initiated by SD which revealed four major groups (referred to as strong, medium, weak, and no SD groups, Fig 4A). [The complete list of all parameters plotted in Fig 4A is available on our webpage (http://www.chemie.uni-hamburg.de/bc/ignatova/tools-and-algorithms.html)]. A randomized sample of the same size displayed different MHE distribution (S7 Fig), implying the functional importance of different SD groups. Moreover, the four groups that are selected based on the strength of the SD:anti-SD pairing resemble previous definitions (which however use a threshold of MHE value of -4.4 kcal/mol to select for more stringent SD sequences) [42]. Note that we did not use any threshold and also included SDs with lower MHE (weak SD) that occur naturally, e.g., AAGG [43] with MHE of −2.9 kcal/mol.
Fig. 4. Stronger SD sequence has a higher propensity to form secondary structure which does not correlate with the translation efficiency. 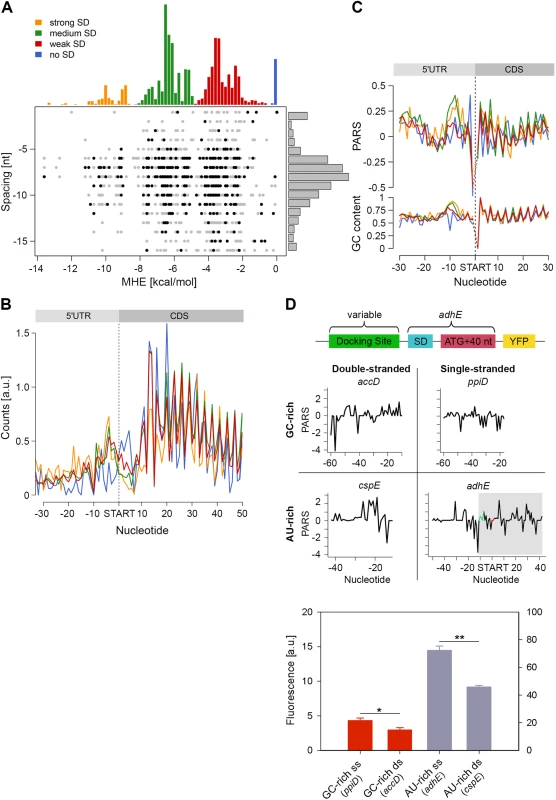
(A) SD strength does not correlate with translation efficiency (i.e. the total RPFs per coding mRNA) of a gene. SD hybridization energies fall into four major distributions: strong SD, MHE < -8.5 kcal/mol; medium SD, -8.5 < MHE < -4.4 kcal/mol; weak SD, -4.4 < MHE < -2 kcal/mol; no SD, MHE > -2.0 kcal/mol. For each gene (dot) the MHE (horizontal axis) of the SD sequence is plotted against SD spacing (vertical axis), defined as the distance between the second to last nucleotide of the SD and the start codon. Genes with the 30% highest ribosomal density are highlighted as black dots. (B) Cumulative plots of ribosomal density for all genes grouped by SD strength. Genes were aligned by the first nucleotide of the start codon. (C) Average PARS score smoothed over 3 nt (top) and GC content (bottom) for each position of the four SD strength classes, aligned by the start codon. The four different SD groups are color coded as in panel A. (D) FACS expression analysis of adhE whose original docking site was replaced by three other docking sites with clearly different sequence (AU-rich or GC-rich) and different PARS score. Only the sequence upstream of the SD (green on the PARS profiles) was replaced. The common part of adhE which is fused to YFP (schematic inset) is shadowed on the PARS profiles. The average PARS score over the docking site (12 to 30 nt upstream of the start codon, red on the PARS profiles are): adhE–-0.564, ppiD–-0.495, cspE–0.724, accD–0.665. Data are means (n = 3) ± standard error of the mean (s.e.m.). *, P <0.05; **, P <0.01. In general, the GC content of each SD group mirrored the SD strength. SD:anti-SD base pairing is crucial to align the P-site of the ribosome on the start codon, hence the optimal spacing between the SD and the start codon is 7–8 nt [44,45] which we also detected independent of the strength of the SD (Fig 4A). To our surprise, we did not observe any correlation between SD strength and translation efficiency, which was determined by the density of ribosomes (RPF) per mRNA (Pearson correlation, R = 0.03, Fig 4A). Highly translated genes did not preferably cluster in any of the SD groups (Chi-square test: p = 0.3539, black symbols, Fig 4A). Notably, even some genes lacking an SD sequence were also highly translated (Fig 4A). We also noticed that for genes with strong and medium SD more RPFs accumulated in the SD vicinity (Fig 4B); these genes were slightly more structured in the SD vicinity than genes with weak SD or those lacking an SD, which is however mirrored in the GC content in this region (Fig 4C).
By analyzing the profiles of the gene groups with different SD strength, we noticed one striking feature: the region starting at ~20 nt upstream of the start codon is the most unstructured region within each gene (mean value of -0.06 for the region -22 to -13 nt, Fig 1C). Strikingly, this feature is not recapitulated by the GC content suggesting that it is not selected through A/U-rich sequences and may play active role in regulating translation initiation. Clearly, ribosomes attach to this unstructured site since we detected reads in the ribosome profiling data set at this location (Fig 4B). The ribosome binds in a biphasic-kinetics mode to some mRNAs and both phases have clear implications for the expression of the corresponding gene [46]. While the second transition in the kinetic curves represents the positioning of the anti-SD of 16S rRNA over the SD sequence, the role of first phase is unclear [46]. Usually multiphasic transitions suggest multiple binding events, thus we hypothesized that this unpaired region might represent an additional unspecific binding site of the 30S to facilitate its positioning over the SD. To examine the physiological importance of this unpaired site in expression of the encoded protein, we compared four different sites: AU-rich sequences with low (i.e. unstructured) and high (i.e. structured) PARS score and GC-rich sequences with low and high PARS score. Each site was fused to the first 50 nt of adhE (SD and first 42 nt of the CDS) upstream of the YFP. The resulting expression was quantified by flow cytometry (schematic in Fig 4D). Notably, constructs with less structured upstream regions resulted in higher expression than their more structured counterparts with similar sequence content (compare AU-rich with single - and double-stranded docking site—adhE vs cspE, or GC-rich with single - and double-stranded docking site—ppiD vs accD; Fig 4D). The variant with unpaired AU-rich region exhibited higher expression than the one with unpaired GC-rich sequence (compare adhE and ppiD, Fig 4D). In general, AU-rich single-stranded regions are less structured than GC-rich single-stranded regions, which correlates with the mean PARS score over this region (-30 –-12 nt upstream of the start codon); the mean PARS score of unpaired AU-rich adhE is -0.564 and of the GC-rich ppiD is -0.495 (Fig 4D). The adhE gene exhibited the highest expression, which might be argued that it due to using part of adhE as an invariable backbone in our constructs (schematic Fig 4D). To exclude this argument, we replaced the invariable adhE part with a fragment of the same size originating from ppiD (SD and first 42 nt of the CDS, S7 Fig). Replacing the original ppiD region upstream of the SD with the most unstructured sequence of adhE enhanced the expression by twofold (S7 Fig).
In sum, our results feature the poorly structured region at ~20 nt upstream of the start codon as an additional binding site of the ribosome distinct from SD binding, and its secondary structure propensity correlates with the expression of the downstream CDS.
Higher secondary structure upstream of the stop codon has a likely role in termination
In the metagenome analysis we noticed that the region upstream of the stop codon is more structured than the average PARS score of the CDS and 3’-UTR, whereas a GC content of this region does not significantly differ from the average CG content of the CDS (Fig 1C). Genes terminated with the UAA codon exhibited the highest propensity to form secondary structures in the 3’-termini of the CDS (p = 2.2*10−16, Mann-Whitney test, Fig 5A). Notably, we observed an enrichment of RPF reads ~10–30 nt upstream of the UAA-termination codon (p = 6.94*10−6, Mann-Whitney test) suggesting a persistent secondary structure (Fig 5B).
Fig. 5. The stop codon of operon genes is more structured than non-operon genes. 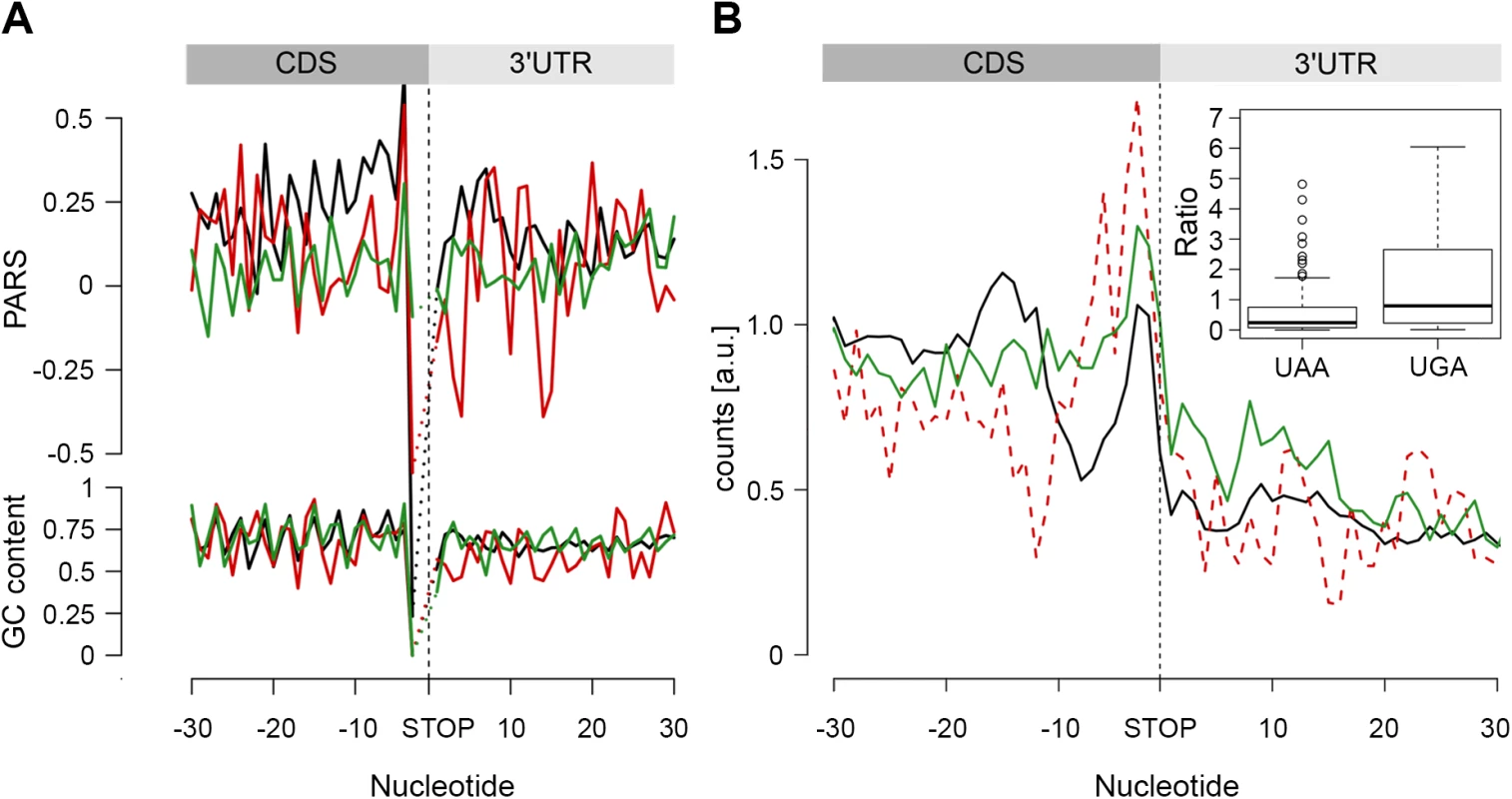
(A) Average PARS score and GC content for each position of genes terminating with UAA (black), UAG (red) and UGA (green) stop codons. (B) RPF coverage around the stop codon region for genes terminated by UAA (black), UAG (dashed red) and UGA (green) stop codons. Only genes with coverage over 60 reads (S4D Fig) were used; overlapping operon genes were excluded. Note, that UAG-terminated genes are included only for comparison; their low number prevents performing any statistical analysis. The inset shows, for both UAA- and UAG-terminated genes, the ratio between the RPFs downstream of the stop codon (3 to27 nt) and a mean of the CDS. The readthrough value for the majority of the genes was zero; only genes with a value higher than zero are plotted. In E. coli, a large fraction (53%) of protein-coding genes is organized as polycistronic mRNAs in operons to facilitate the association and physical interactions of functionally related proteins. The SD sequence of an overlapping or a closely positioned downstream gene (S8 Fig) may influence our analysis, resulting in an apparent higher structure in the 3’ vicinity of the upstream gene. Thus, we next separately analyzed the secondary structure upstream of the stop codon of protein-coding genes organized in operons from those in non-operons; the operon group is additionally divided in two groups: non-overlapping, with a distance of ≥ 30 nt from the downstream gene, and overlapping, with a downstream gene located < 30 nt to the upstream gene. Only UAA-terminated genes showed increased PARS score (p = 0.00023 for non-overlapping, p = 3.2*10−10 for overlapping, p = 4.07*10−5 for non-operon, Mann-Whitney test, S8 Fig) in the 3’ vicinity of the coding sequence and this feature is not mirrored by the GC content. Also, the frequency of the three stop codons (UAA, UAG and UGA) is similar for all gene groups and resembles stop codon usage in the genome (S8 Fig).
We hypothesized that secondary structure upstream of the stop codon may influence the termination fidelity of the UAA-terminated genes. Additional in-frame stop codons may act as safeguards against leaky termination. We reasoned that if the structure in the vicinity of the UAA stop codon influences termination, those genes would show lower frequency of ribosomes in the 3’-UTR. We analyzed the ribosome occupancy downstream of the UAA - and UGA-terminated genes (considering it in general as a readthrough). Overlapping genes were excluded from this analysis as ribosomes terminating the upstream gene cannot be unambiguously distinguished from ribosomes initiating the downstream gene. Strikingly, we observed a low but significant fraction of RPF reads downstream of the UGA stop codon while RPF reads in the 3’ UTR of the UAA-terminated genes were nearly not detectable (Fig 5B). This phenomenon occurred in the background of a similar distribution of additional in-frame stop codons downstream of all terminating codons: UAA–10.7%, UGA–8.7% and UAG–7.4%. Together, this analysis suggests that structure upstream of the stop codon may enhance the termination fidelity of the UAA-codon terminated genes.
Discussion
We provide a comprehensive analysis of the intrinsic structure propensity of the E. coli mRNAome, which combined with physiological analysis, identifies structural features implicated in the regulation of translation efficiency in E. coli. These include a universal unstructured site at ~20 nt upstream of the start codon, which we postulate to serve as a non-specific docking of the 30S ribosomal subunit; this site differs from the SD:anti-SD binding site. Within the CDSs, we identified a small set of persisting structured regions that transiently stall the ribosomes and may regulate protein integration into the membrane. On a global level, however, the secondary structure of the CDS has no effect on translation elongation in vivo, highlighting the importance of energy-dependent processes (for example ATP-dependent helicases, ribosomes) or passive elements (for example single-stranded RNA binding proteins) in regulating mRNA structures in the cell [8]. Moreover, the propensity of CDSs to form secondary structure is counterbalanced by selection of codons that pair to high-abundance tRNAs which in general smooths the overall translation speed [21]. For the majority of E. coli transcripts, translation is initiated by complementation of the anti-SD of the 30S subunit with the SD sequence upstream of the start codon. Our analysis reveals that SD sequences are often occluded in secondary structures with a highly dynamic reversible folding/unfolding kinetics on a microsecond time scale [4,47]. Thermodynamically, for an anti-SD to outcompete such a secondary structure the 30S subunit needs to be already in the close vicinity of the SD. Although in the current analysis neither ribosome profiling nor PARS analysis bear kinetic information or can reveal a sequence of binding events, we envision that the unfolded site upstream of the SD sequence may act as a primary unspecific docking site of the 30S subunit to enable interactions with the SD sequence within its unfolding window. Supportive for our model is the observed biphasic kinetics of ribosome binding to some mRNAs with an unclear first phase and a second owing to anti-SD:SD interactions [46]. Also, current approaches to predict translational rates based only on SD strength fail to accurately account for known differences in translation initiation rates [48]. Our expression analysis convincingly shows that the unstructured site at ~20 nt influences translation of the downstream CDS and the expression level correlates with the degree of its unfolding. The global genome-wide analysis features this unstructured region upstream of the start codon as the most unfolded structure in the E. coli genome but its size seems smaller than the 30S subunit (Fig 1C). The contacts with the mRNA might be established by the essential S1 protein, which is the only ribosomal protein with an mRNA-binding affinity. Furthermore, S1 protein, which is essential for unfolding of structured SD [49], attaches to mRNA 11 nt upstream of the SD [50] which is approximately the position of the unpaired region.
We also observed an enrichment of ribosomes upstream of a persistent secondary structure which is found ~4–8 nt 5’-adjacent of the UAA stop codon. Previous research on termination regulation provides appropriate context for the interpretation of these results. The efficiency of translation termination (or conversely, the rate of termination suppression) is sensitive to the 5’ and 3’ sequence in immediate proximity of the stop codon [51]. Moreover, the nature of the corresponding codon (i.e. nucleotides 4–6) upstream of the stop codon plays an important role in the efficiency of termination [52]. Systematic exchange of different codons prior the stop codon evidence the highest termination efficiency by those encoding bulky amino acids, in the absence of a broader sequence motif. Interactions of the bulky residues of the nascent peptide with the ribosomal tunnel are suggested to slow down terminating ribosome prior to termination which enhances the termination fidelity [52]. The accurate positioning of the A-site over the stop codon determines the accuracy in termination and suppresses readthrough: A-rich sequences preceding the stop codons distort the ribosomes in the P-site which alters the stop-codon decoding in the A-site [53]. In comparison, our analysis features a persistent mRNA secondary structure upstream of the UAA stop codon which is not encoded by a universal sequence motif but is similarly responsible for a ribosomal slowdown. By drawing an analogy to these studies, we suggest that the secondary structure upstream of the UAA stop codon slows down the elongating ribosome which may assist the accurate positioning of the ribosomal A-site for accurate decoding of the UAA stop codon.
Another striking aspect of our analysis is the identification of a global signature of RNase E cleavage site. Earlier single-gene studies proposed the importance of secondary structures 5’ upstream of the single-stranded cleavage site [28–30,54]. Our analysis corroborates those observations and features a structured region upstream of the A/U rich unpaired site as common signature of RNase E cleavage sites on a transcriptome-wide scale. This signature can be reconciled with the RNase E crystal structure: while a single-stranded segment only fits in the shallow channel leading to the RNase E active site [54], the internal flexibility of the quaternary structure [55] can clearly accommodate secondary mRNA structures. The latter significantly shortens the distance between the cleavage site and 5’ terminus and may explain how distant 5’ termini of the mRNA facilitate catalysis [54].
In summary, our approach of structurally probing bacterial mRNA in vitro with PARS, complemented with RNA-Seq and ribosome profiling, reveals structural features of importance for a variety of cellular processes. Although coding mRNA sequences show a frequent intrinsic propensity to form secondary structure, only a small fraction influences translation fidelity in vivo. Our combined approach features the importance of applying a variety of techniques to unambiguously evaluate structure-function relationships in physiological context.
Methods
RNA structural probing by deep sequencing
The E. coli MC4100 strain was cultured at 37°C to mid-log phase (OD600 ~ 0.4) in LB media. Total RNA was extracted using TRIzol reagent (Invitrogen) and the sample was enriched in mRNA by depleting small RNAs with GeneJET RNA Purification Kit (Fermentas) and ribosomal RNA with two cycle of MICROBExpress Bacterial mRNA Enrichment Kit (Ambion) which reduces the amount of rRNA to appr. 25% of the total sequencing reads. To probe the RNA structure, two μg of enriched mRNA were resuspended in 45 μl of DEPC water and denatured for 3 min at 95°C, refolded at 37°C, combined with 10x RNA-structure buffer with pH 7.0 (100 mM Tris, 1 M KCl, 100 mM MgCl2) and digested for 1 min at 37°C with either 0.05 U RNase V1 (Life Technologies) or a combination of 2 μg RNase A and 5 U RNase T1 (Thermo Scientific). The reaction was stopped by extracting the RNA with phenol-chlorophorm. RNases A/T1 were preferred as they exhibit a stable activity at pH 7.0 [56] compared to nuclease S1 which has a pH optimum ~5.0 and aberrant activity at pH 7.0. At pH 7.0, high concentrations of nuclease S1 are required; however at such high concentrations S1 also digests double-stranded regions [57]. The nucleolytic reaction was stopped by extracting the RNA with phenol-chlorophorm. The RNase A/T1-digested sample was phosphorylated with T4 PNK (NEB) and purified with RNA Clean & Concentrator kit (Zymo Research). Both the V1 and A/T1 digested samples were randomly fragmented in buffer with pH 9.2 (100 mM Na2CO3, 2 mM EDTA) for 12 min at 95°C. The reaction was stopped by adding 560 μl 300 mM NaOAc, pH 5.5, followed by isopropanol precipitation. RNA size selection and generation of the cDNA libraries were performed as described [26].
Ribosome profiling
To isolate mRNA-bound ribosome complexes and extract the RPFs we used a previously described approach [58] with some modifications. For the isolation of RPFs, an aliquot of 100 A260 units of ribosome-bound mRNA fraction (prior to ultracentrifugation in the sucrose gradients) was subjected to nucleolytic digestion with 10 units/μl micrococcal nuclease (Fermentas) for 10 min at room temperature in buffer with pH 9.2 (10 mM Tris pH 11 containing 50 mM NH4Cl, 10 mM MgCl2, 0.2% triton X-100, 100 μg/ml chloramphenicol and 20 mM CaCl2). The monosomal fraction was separated by sucrose density gradient (15–50% w/v). The total RNA was isolated from monosomes using the hot SDS/phenol method. Micrococcal nuclease also cleaved rRNA into fragments with a size similar to the RPFs. The sample was enriched predominantly in one rRNA fragment which was removed by subtractive hybridization at 700 C using a 5’-biotin-5’-GCCTCGTCATCACGCCTCAGCC-3’. DNA oligonucleotide along with μMACS Streptavidin Kit (Myltenyi Biotec) to remove the biotin-labeled DNA/rRNA hybrids. Both randomly fragmented mRNAs and RPFs extracted from monosomes were denatured for 2 min at 80°C, and 3’-dephosphorylated with T4 polynucleotide kinase (NEB) for 90 min at 37°C in the corresponding buffer without ATP (NEB). RNA was precipitated by standard methods. Subsequently, 20-35-nt RNA fragments were size selected on a denaturing 15% polyacrylamide gel stained with SYBR Green II (Invitrogen) using 10-100-nt leader (Affymetrix) as a standard. The gel was extracted, precipitated and resuspended in DEPC water.
Random mRNA fragmentation and cDNA libraries
To generate the RNA-Seq sample to which the ribosome profiling data are compared, 20 μl of the enriched mRNA (as described above) was mixed with equal volume of 2x alkaline fragmentation solution (2 mM EDTA and 100 mM Na2CO3 pH 9.2) and incubated for 40 min at 95°C. The reaction was stopped by adding 560 μl 300 mM NaOAc pH 5.5, followed by isopropanol precipitation. The optimal time for fragmentation of mRNA was determined using GAPDH mRNA (0.25 μg; Fermentas) and the spectra were recorded with BioAnalyzer (Agilent RNA 6000 Kit).
The cDNA libraries from RPFs and fragmented mRNAs were prepared using a modified protocol for miRNA [59] which yielded much higher resolution and allowed for calculation of the position of the ribosomes with codon precision (S4 Fig). Gel-purified RNA fragments were dissolved in 10 mM Tris pH 8.0 and used for the preparation of the cDNA library via direct adapter ligation [59] including some additional steps. As both mRNA fragments and RPFs were hydroxylated at their 5’ - and 3’-termini, after the ligation of the adapter to the 3’-end, the fragments were 5’-phosphorylated with T4 polynucleotide kinase in ATP-containing buffer (NEB) for 30 min at 37°C followed by the adaptor ligation at the 5’-termini. The fragments with adaptors at both termini were size selected on a denaturing 15% polyacrylamide gel, extracted and reverse transcribed with RevertAid H Minus Reverse Transcriptase (Fermentas) using 5’-CAAGCAGAAGACGGCATACGA-3’ primer and PCR-amplified with Pfu DNA Polymerase (Fermentas) for 10 to 20 cycles. The PCR amplified DNA library was quantified with BioAnalyzer (Agilent DNA 1000 Kit) and sequenced on the Illumina GAIIx platform.
Mapping of the sequencing reads
Sequenced reads were quality trimmed using fastx-toolkit (0.0.13.2; quality threshold: 20) and sequencing adapters were cut using cutadapt (1.2.1; minimal overlap: 1 nt) discarding reads shorter than 12 nucleotides. Processed reads were mapped to the E. coli genome (strain MG1655, version U00096.2, downloaded from NCBI) using Bowtie (0.12.9) allowing a maximum of two mismatches for the RNA-Seq and ribosome profiling data and a maximum of three mismatches for the PARS data. Strain MC4100 is a derivative of MG1655 with four major deletions [60]
The number of raw reads unambiguously aligned to ORFs in both RNA-Seq and ribosome profiling data sets, from two biological and one technical replicates were used to generate gene read counts, by counting the number of reads whose middle nucleotide (for even read length the nucleotide 5' of the mid-position) fell in the CDS. Gene read counts were normalized by the length of the unique CDS per kilobase (rpkM) and the total mapped reads per million (rpM) [27]. In this mapping round, reads aligning to rRNA and tRNA genes were excluded since a large fraction of them map non-uniquely due to the multiple copies of those genes. Mapping of 5S and 16S RNA was done separately allowing no mismatches to only one copy of the rRNA reference sequence.
Computing the PARS score
The first nucleotide of the mapped reads from V1 or A/T1 digested samples, each derived from two biological replicates, was assigned to a nucleotide position in the genome and the counts were normalized to the sequencing depth. For each position, we computed the PARS score which is defined as the log2 of the ratio between the number of reads per million (rpM) from the V1-treated and the A/T1-treated samples (to each we added a small number 1, to avoid division by zero and to reduce the potential overestimating of low-coverage bases [6]). RNase A hydrolyzes at single-stranded C and U nucleotides and RNase T1 at single-stranded G nucleotides, thus we excluded all adenines from the analyses. In addition, zero PARS score may result at positions with the same count values for A/T1 and V1 digestion, which are usually located in regions with highly flexible structure. As a minimum PARS coverage per transcript we used a threshold of 1.0 per transcript length (S1 Fig) termed transcript load [6] which is defined as the sum of combined PARS readouts of the biological replicates per transcript divided by the effective transcript length (that is the annotated transcript length minus the number of unmappable nucleotides); the same threshold was used in yeast PARS analysis named as load of a transcript [6]. For the cumulative plots, all genes were aligned either to the start or the stop codon and for each position the mean of the PARS score of the two biological replicates was calculated. The GC content was calculated considering only the non-zero PARS score entries.
Periodicity of average PARS score in the CDSs and 5’UTR and 3’UTR was analyzed by Discrete Fourier transform (S3 Fig). The following regions were analyzed: over 10 to 99 nt downstream of the start codon, 99 to10 nt upstream of the stop codon (i.e. excluding possible influences of the initiation and termination codons but keeping the translation reading frame) for the CDSs, and 50 to 11 nt upstream of the start codon or downstream of the stop codon for the 5’UTR and 3’UTR, respectively. The periodicity for each of the three nucleotides in a codon was calculated also over the same region of the CDSs (S3 Fig).
Modeling the sampling error between biological replicates
To select a reliable minimum of read counts per gene and to assess the influence of counting noise, we computed the binomial partitioning of total counts between two independent biological replicates [26] of the RNA-Seq and ribosome profiling from bacteria grown in LB. Genes were binned logarithmically based on the total number of their reads. The standard deviation of the ratio (repl#1/(repl#1 + repl#2)) across each bin was computed as a function of the mean sum of reads in each bin. In addition, a constant variance was added to the theoretical predictions accounting for other sources of error, yielding:
where p represents the probability to assign a read to replicate #1, n is the total number of sequencing reads from replicate #1 and replicate #2 and s was obtained by fitting Eq 1 to the data (S4 Fig).Detection of RPF enrichment upstream of secondary structures
To determine positions whose secondary structure may influence elongation we used two approaches: CDS were systematically screened for double-stranded stretches (1) with a window of 10 nt containing 4 to 8 structured nt (i.e. with positive PARS score), or (2) using the mean PARS score within a window with different size (10 or 20 nt) (S5 Fig). A 10-nt-window with 6 structured nt delivered the best result considering the number of the selected positions (908 positions, S5 Fig) and was chosen in the analysis.
To define RPF enrichment upstream of a selected secondary structure (L1), the RPF counts over 29 nt upstream of the double-stranded stretch (RPF1) were compared to the RPF counts over 29 nt (1st-30th nt) downstream (L2) of the detected stretch (RPF2). Read counts were normalized by the total number of reads for the whole region [61]:
Determination of codon periodicity in the RPF and RNA-Seq data sets
Reads with length of 23–25 nt which were unambiguously mapped to the 1000 most expressed genes were combined for the RNA-Seq or ribosome profiling and binned by their length. To compute the codon periodicity in the RNA-Seq and ribosome profiling data sets, we used the reads mapped to the 3’-ends of the corresponding ORFs which were positioned at one of the three stop codons (UAG, UAA and UGA).
Detection of SD sequences
For all annotated genes, the MHE was calculated between sequences 1–25 nt upstream of the start codon and anti-SD sequence (3’-UCCUCCAC-5’) using RNAsubopt (2.1.5; default parameters) from the Vienna RNA Package [62]. For each 8mer, the calculated MHE was assigned to the 8th base as described [63] and the minimum of the calculated MHE of all 8mers was taken as an identifier for the SD sequence and used to determine the corresponding spacing. To designate different SD groups based on their MHE we used a randomization control. The random sample was created in two different ways: (1) by generating all possible random 8-mer sequences (65,536 sequences) or (2) by choosing each nucleotide randomly within the 8-mer (444,000 sequences). For both randomized groups we received similar results. For comparison to the natural SD, 4,400 random sequences were selected which resemble the E. coli gene number in S7 Fig.
Footprint analysis with fluorescently-labeled mRNA
In vitro transcribed RNA of ppiC was 3’ end-labeled with 10 μM pCp-Cy3 (Jena Bioscience) using 15 U T4 Ligase 1 (NEB). 2 μg of fluorescently-labeled RNA was structure probed with 0.05 U of RNase V1 (Ambion) or with a dilution 1 : 7000 of combined RNase A/T1 (Thermo Scientific), in conditions identical to the PARS experiment. The digestion was stopped with phenol chlorophorm extraction, precipitated overnight at 4°C and resuspended in 10 μl of 2x RNA Loading Dye (Thermo Scientific). In parallel, a ddNTP-Sanger sequencing PCR reaction was performed using 20 pmol of a 3’-fluorescently(Cy3)-labeled primer, in the presence of 400 ng of DNA template, 10 μM dNTPs, 1.25 U Pfu DNA Polymerase (Thermo Scientific), Pfu Polymerase Buffer and 1 mM of each ddNTP. PCR was performed according to the manufacturer instructions in a volume of 15 μl. After addition of 2x RNA Loading Dye, all samples were boiled for 3 min at 95°C and loaded on a 6% PA, 1x TBE, 7M UREA gel (50x40 cm), already pre-run for 30 min at 50W. The gel was then run for 3 h at 50W and the fluorescence was detected using a fluorescent gel imager.
Expression analysis
In each biological replicate, cells were grown in LB medium till OD600 = 0.5 and induced with 1 mM IPTG for 90 minutes. The median expression of the YFP-fused constructs was quantified in a population of approximately 105 cells by flow cytometry on a FACSCalibur (BD Bioscience). The forward (fcs) and sideward (ssc) scatter was recorded at each measurement and the data were processed by Flowing software 2. The values were normalized to the autofluorescence background of untransformed cells transformed.
Quantitative RT-PCR
Total mRNA was extracted using the GeneJET RNA Purification Kit (Fermentas) and treated with DNase I (Fermentas). The cDNA was synthesized with RevertAid H Minus Reverse Transcriptase (Fermentas) and quantitative RT-PCR was performed on a StepOnePlus Real-Time PCR system (Applied Biosystems) using template-specific primers. The values were normalized to the amount of the total RNA.
Statistical analysis
All data analyses were performed with in-house algorithms in Pearl and R. Differences between the distributions were assessed for significance by a nonparametric Mann-Whitney test, and enrichment of RPF was assessed by a Kolmogorov-Smirnov test. Note that we used Mann-Whitney U test, also called Wilcoxon rank-sum test, which is suitable for unpaired data for which no normal distribution can be assumed. To determine codon periodicity, Kullback-Leibler divergence was used to measure the deviation of the observed distribution of the 3’-end of the mapped read from a uniform distribution. Differences in the expression (FACS experiments) were evaluated using two-tailed Student’s t-test. Differences were considered statistically significant when P< 0.05.
Data access
All sequencing data files are available from Gene Express Omnibus database, GSE63817.
Supporting Information
Zdroje
1. Kramer G, Boehringer D, Ban N, Bukau B (2009) The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16 : 589–597. doi: 10.1038/nsmb.1614 19491936
2. Pechmann S, Chartron JW, Frydman J (2014) Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol 21 : 1100–1105. doi: 10.1038/nsmb.2919 25420103
3. Zhang G, Ignatova Z (2011) Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr Opin Struct Biol 21 : 25–31. doi: 10.1016/j.sbi.2010.10.008 21111607
4. Porschke D (1973) The dynamics of nucleic-acid single-strand conformation changes. Oligo - and polyriboadenylic acids. European journal of biochemistry / FEBS 39 : 117–126. 4770785
5. Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, et al. (2014) In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505 : 696–700. doi: 10.1038/nature12756 24270811
6. Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, et al. (2010) Genome-wide measurement of RNA secondary structure in yeast. Nature 467 : 103–107. doi: 10.1038/nature09322 20811459
7. Li F, Zheng Q, Vandivier LE, Willmann MR, Chen Y, et al. (2012) Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell 24 : 4346–4359. doi: 10.1105/tpc.112.104232 23150631
8. Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS (2014) Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505 : 701–705. doi: 10.1038/nature12894 24336214
9. Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, et al. (2015) Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519 : 486–490. doi: 10.1038/nature14263 25799993
10. Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, et al. (2015) hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 519 : 491–494. doi: 10.1038/nature14280 25799984
11. Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, et al. (2014) Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505 : 706–709. doi: 10.1038/nature12946 24476892
12. Kwok CK, Tang Y, Assmann SM, Bevilacqua PC (2015) The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem Sci 40 : 221–232. doi: 10.1016/j.tibs.2015.02.005 25797096
13. Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, et al. (2011) mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol 12 : 235–245. doi: 10.1038/nrm3083 21427765
14. Takyar S, Hickerson RP, Noller HF (2005) mRNA helicase activity of the ribosome. Cell 120 : 49–58. 15652481
15. Burgess DJ (2015) RNA: Detailed probing of RNA structure in vivo. Nature reviews Genetics 16 : 255. doi: 10.1038/nrg3939 25854184
16. Mahen EM, Watson PY, Cottrell JW, Fedor MJ (2010) mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol 8: e1000307. doi: 10.1371/journal.pbio.1000307 20161716
17. Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN (2002) Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci U S A 99 : 9697–9702. 12119387
18. Chen C, Zhang H, Broitman SL, Reiche M, Farrell I, et al. (2013) Dynamics of translation by single ribosomes through mRNA secondary structures. Nat Struct Mol Biol 20 : 582–588. doi: 10.1038/nsmb.2544 23542154
19. Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, et al. (2008) Following translation by single ribosomes one codon at a time. Nature 452 : 598–603. doi: 10.1038/nature06716 18327250
20. Zhang G, Hubalewska M, Ignatova Z (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 16 : 274–280. doi: 10.1038/nsmb.1554 19198590
21. Gorochowski TE, Ignatova Z, Bovenberg RA, Roubos JA (2015) Trade-offs between tRNA abundance and mRNA secondary structure support smoothing of translation elongation rate. Nucleic Acids Res 43 : 3022–3032. doi: 10.1093/nar/gkv199 25765653
22. Bulmer M (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics 129 : 897–907. 1752426
23. Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, et al. (2014) Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol Syst Biol 10 : 770. doi: 10.15252/msb.20145524 25538139
24. Bentele K, Saffert P, Rauscher R, Ignatova Z, Bluthgen N (2013) Efficient translation initiation dictates codon usage at gene start. Mol Syst Biol 9 : 675. doi: 10.1038/msb.2013.32 23774758
25. Goodman DB, Church GM, Kosuri S (2013) Causes and effects of N-terminal codon bias in bacterial genes. Science 342 : 475–479. doi: 10.1126/science.1241934 24072823
26. Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324 : 218–223. doi: 10.1126/science.1168978 19213877
27. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5 : 621–628. doi: 10.1038/nmeth.1226 18516045
28. Ehretsmann CP, Carpousis AJ, Krisch HM (1992) Specificity of Escherichia coli endoribonuclease RNase E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev 6 : 149–159. 1730408
29. McDowall KJ, Lin-Chao S, Cohen SN (1994) A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem 269 : 10790–10796. 7511606
30. Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Blasi U (2003) Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9 : 1308–1314. 14561880
31. Li F, Zheng Q, Ryvkin P, Dragomir I, Desai Y, et al. (2012) Global analysis of RNA secondary structure in two metazoans. Cell Rep 1 : 69–82. doi: 10.1016/j.celrep.2011.10.002 22832108
32. Incarnato D, Neri F, Anselmi F, Oliviero S (2014) Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol 15 : 491. 25323333
33. Shabalina SA, Ogurtsov AY, Spiridonov NA (2006) A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res 34 : 2428–2437. 16682450
34. Iost I, Dreyfus M (2006) DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res 34 : 4189–4197. 16935881
35. Resch A, Vecerek B, Palavra K, Blasi U (2010) Requirement of the CsdA DEAD-box helicase for low temperature riboregulation of rpoS mRNA. RNA Biol 7 : 796–802. 21045550
36. Vakulskas CA, Pannuri A, Cortes-Selva D, Zere TR, Ahmer BM, et al. (2014) Global effects of the DEAD-box RNA helicase DeaD (CsdA) on gene expression over a broad range of temperatures. Mol Microbiol 92 : 945–958. doi: 10.1111/mmi.12606 24708042
37. Fluman N, Navon S, Bibi E, Pilpel Y (2014) mRNA-programmed translation pauses in the targeting of E. coli membrane proteins. Elife 3.
38. Woolhead CA, McCormick PJ, Johnson AE (2004) Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116 : 725–736. 15006354
39. Carrier TA, Keasling JD (1997) Controlling messenger RNA stability in bacteria: strategies for engineering gene expression. Biotechnol Prog 13 : 699–708. 9413129
40. Mackie GA (2013) RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol 11 : 45–57. doi: 10.1038/nrmicro2930 23241849
41. Clarke JE, Kime L, Romero AD, McDowall KJ (2015) Direct entry by RNase E is a major pathway for the degradation and processing of RNA in Escherichia coli. Nucleic Acids Res 42 : 11733–11751.
42. Ma J, Campbell A, Karlin S (2002) Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J Bacteriol 184 : 5733–5745. 12270832
43. Wood CR, Boss MA, Patel TP, Emtage JS (1984) The influence of messenger RNA secondary structure on expression of an immunoglobulin heavy chain in Escherichia coli. Nucleic Acids Res 12 : 3937–3950. 6328446
44. Osterman IA, Evfratov SA, Sergiev PV, Dontsova OA (2013) Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res 41 : 474–486. doi: 10.1093/nar/gks989 23093605
45. Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, et al. (1992) Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol Microbiol 6 : 1219–1229. 1375310
46. Studer SM, Joseph S (2006) Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol Cell 22 : 105–115. 16600874
47. de Smit MH, van Duin J (2003) Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J Mol Biol 331 : 737–743. 12909006
48. Salis HM, Mirsky EA, Voigt CA (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27 : 946–950. doi: 10.1038/nbt.1568 19801975
49. Duval M, Korepanov A, Fuchsbauer O, Fechter P, Haller A, et al. (2013) Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol 11: e1001731. doi: 10.1371/journal.pbio.1001731 24339747
50. Sengupta J, Agrawal RK, Frank J (2001) Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc Natl Acad Sci U S A 98 : 11991–11996. 11593008
51. Bonetti B, Fu L, Moon J, Bedwell DM (1995) The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol 251 : 334–345. 7650736
52. Bjornsson A, Mottagui-Tabar S, Isaksson LA (1996) Structure of the C-terminal end of the nascent peptide influences translation termination. The EMBO journal 15 : 1696–1704. 8612594
53. Tork S, Hatin I, Rousset JP, Fabret C (2004) The major 5' determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res 32 : 415–421. 14736996
54. Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, et al. (2005) Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437 : 1187–1191. 16237448
55. Koslover DJ, Callaghan AJ, Marcaida MJ, Garman EF, Martick M, et al. (2008) The crystal structure of the Escherichia coli RNase E apoprotein and a mechanism for RNA degradation. Structure 16 : 1238–1244. doi: 10.1016/j.str.2008.04.017 18682225
56. Tripathy DR, Dinda AK, Dasgupta S (2013) A simple assay for the ribonuclease activity of ribonucleases in the presence of ethidium bromide. Anal Biochem 437 : 126–129. doi: 10.1016/j.ab.2013.03.005 23499964
57. Green MR, Sambrook J (2012) Molecular Cloning: A Laboratory Manual New York: Cold Spring Haror Laboratory Press.
58. Cozzone AJ, Stent GS (1973) Movement of ribosomes over messenger RNA in polysomes of rel + and rel—Escherichia coli strains. J Mol Biol 76 : 163–179. 4578097
59. Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466 : 835–840. doi: 10.1038/nature09267 20703300
60. Peters JE, Thate TE, Craig NL (2003) Definition of the Escherichia coli MC4100 genome by use of a DNA array. J Bacteriol 185 : 2017–2021. 12618467
61. Zhang Y, Mooney RA, Grass JA, Sivaramakrishnan P, Herman C, et al. (2014) DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol Cell 53 : 766–778. doi: 10.1016/j.molcel.2014.02.005 24606919
62. Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, et al. (2011) ViennaRNA Package 2.0. Algorithms Mol Biol 6 : 26. doi: 10.1186/1748-7188-6-26 22115189
63. Li GW, Oh E, Weissman JS (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484 : 538–541. doi: 10.1038/nature10965 22456704
64. Soper TJ, Woodson SA (2008) The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14 : 1907–1917. doi: 10.1261/rna.1110608 18658123
65. Schmidt M, Zheng P, Delihas N (1995) Secondary structures of Escherichia coli antisense micF RNA, the 5'-end of the target ompF mRNA, and the RNA/RNA duplex. Biochemistry 34 : 3621–3631. 7534474
66. Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, et al. (1980) Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res 8 : 2275–2293. 6159576
67. Gutell RR, Lee JC, Cannone JJ (2002) The accuracy of ribosomal RNA comparative structure models. Curr Opin Struct Biol 12 : 301–310. 12127448
68. Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, et al. (2001) Crystal structure of the ribosome at 5.5 A resolution. Science 292 : 883–896. 11283358
Štítky
Genetika Reprodukční medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 10- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
- Prof. Petr Urbánek: Potřebujeme najít pacienty s nediagnostikovanou akutní intermitentní porfyrií
-
Všechny články tohoto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání






