-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
Plants control their flowering time in response to the temperatures of their environment, e.g. in response to the experience of winter or in response to cold and warm ambient temperatures experienced during spring. The knowledge about the evolutionary adaptation of plants to changing ambient temperatures is at present very limited. Understanding the latter is, however, becoming increasingly important due to the temperature changes associated with global warming and the anticipated changes in flowering time in ecosystems and agricultural systems. Here, we uncover an evolutionarily conserved molecular mechanism employed by Arabidopsis thaliana ecotypes for the adaptation of flowering time to cool temperatures. This structural change in the architecture of the gene FLOWERING LOCUS M can be found in multiple A. thaliana natural accessions and the knowledge gained in our study may be used to predict or modify flowering time in plants related to A. thaliana in the future.
Published in the journal: . PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005588
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005588Summary
Plants control their flowering time in response to the temperatures of their environment, e.g. in response to the experience of winter or in response to cold and warm ambient temperatures experienced during spring. The knowledge about the evolutionary adaptation of plants to changing ambient temperatures is at present very limited. Understanding the latter is, however, becoming increasingly important due to the temperature changes associated with global warming and the anticipated changes in flowering time in ecosystems and agricultural systems. Here, we uncover an evolutionarily conserved molecular mechanism employed by Arabidopsis thaliana ecotypes for the adaptation of flowering time to cool temperatures. This structural change in the architecture of the gene FLOWERING LOCUS M can be found in multiple A. thaliana natural accessions and the knowledge gained in our study may be used to predict or modify flowering time in plants related to A. thaliana in the future.
Introduction
In plants, fertilization and reproduction are directly linked to the seasonal onset of flowering. Plants enter the reproductive phase when environmental conditions are favorable for seed set and thus reproduction. Since day length and temperature as well as temperature changes throughout the year provide the crucial information about the passage of the seasons and the environment, plants sense these cues for the adjustment of their flowering time [1]. Proper flowering time and reproductive success of a given species or ecotype, on the one side, and the differences in flowering time between species or ecotypes, on the other, are the result of the differential integration of temperature and day length information.
The vernalization and the ambient temperature pathways control temperature-dependent flowering in plants. Whereas vernalization requires long periods (weeks) of cold, usually below 10°C, as experienced during the winter [2], the ambient temperature pathway modulates flowering in response to short-term (days) temperature changes in the range between 12°C and 27°C [3–5]. In A. thaliana, the central mechanism of accelerating flowering in response to prolonged cold is achieved by repression of the negative regulator FLOWERING LOCUS C (FLC), a MADS-box transcription factor [6–9]. Different mechanisms than in A. thaliana control vernalization in cereal crops such as wheat and barley, and the activity or inactivity of the vernalization pathway determines the flowering behavior of their winter and spring varieties [10, 11]. To date, the understanding of the vernalization pathway in A. thaliana is already well advanced and it is possible to make predictions on the vernalization requirement based on the plants’ genotypes [12, 13].
In contrast, the complexities of ambient temperature sensing are just beginning to be understood [5, 14, 15]. The finding that loss-of-function mutations of the gene FLM (FLOWERING LOCUS M) reduce the temperature-sensitivity of flowering in A. thaliana accessions suggested that this MADS-box transcription factor acts as a repressor in the ambient temperature pathway [16–18]. The molecular understanding of FLM is complicated by the fact that the FLM gene is alternatively spliced into at least four splice forms [18]. FLM-ß and FLM-δ, which result from the alternative use of the two exons 2 (FLM-ß) and 3 (FLM-δ), represent the two predominant splice variants in the Columbia-0 (Col-0) reference accession [19, 20]. The observation that the abundance of FLM-ß declines from 16°C to 27°C while the abundance of FLM-δ increases over the same temperature range has motivated experiments to examine the effects of the FLM-ß and FLM-δ isoforms in isolation in a flm-3 loss-of-function background. These experiments indicated that the expression of the low temperature-abundant FLM-ß and the warm temperature-abundant FLM-δ can repress and promote flowering, respectively, and consequently a model was established according to which changes in the relative abundance of FLM-ß and FLM-δ control flowering time in response to changes in ambient temperature [19].
FLM directly interacts with several other MADS-box transcription factors to control flowering through the expression of flowering time genes such as FT (FLOWERING LOCUS T) and SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO1) [19–21]. SVP (SHORT VEGETATIVE PHASE) is an important FLM interaction partner and, in this context, the flowering-repressive activity of FLM-ß and the flowering-promoting activity of FLM-δ have been explained by the differential effects of the FLM-SVP interactions [19]: It was proposed that a DNA-binding heterodimer of FLM-ß with SVP represses flowering by repressing FT and SOC1 expression. Conversely, FLM-δ could sequester SVP into an inactive complex that thereby indirectly promotes FT and SOC1 expression and consequently flowering. Although this experimentally validated model is very intriguing, it is at present not known whether the alternative splicing of FLM plays a role in flowering time adaptation in natural accessions of A. thaliana.
There is increasing evidence for global warming due to climate change [8]. Temperature changes by only a few centigrade (°C) can already lead to ecological and physiological constraints that have negative impacts on agricultural production systems [22–24]. Thus, there is a need to better understand the ambient temperature pathway and to integrate this understanding in plant breeding programs [25]. Here, we identify a structural polymorphism in the first intron of FLM as being causative for the early flowering time of the A. thaliana accession Killean-0 (Kil-0). This structural polymorphism is present in several additional accessions and directly affects FLM transcript abundance, splicing, and flowering. We further correlate the abundance of the FLM-ß and FLM-δ splice variants with flowering behavior in several A. thaliana accessions and reveal an important role of intron 1 for FLM gene expression and a predominant role of FLM in flowering time control.
Results
Killean-0 is an early flowering accession
To understand the variation in flowering time in response to temperature, we compared the flowering behavior of a collection of A. thaliana accessions at 15°C and at 21°C. In this analysis, our attention was drawn to the Scottish accession Killean-0 (Kil-0), which flowered two weeks earlier than the Columbia-0 (Col-0) reference when grown at 15°C but only one week earlier at 21°C (Figs 1A, 1B, S1A and S1B). The vernalization pathway could potentially contribute to the early flowering behavior of Kil-0 at 15°C but we detected only minor flowering time effects after a six-week vernalization treatment (S1C Fig). These flowering time effects were similar to those observed in the reference Col-0 and confirmed also the results from previous surveys that had classified Kil-0 as a summer annual [12, 26]. Since the expression of the major vernalization-responsive gene FLC was also as strongly reduced in Kil-0 as in the summer annual accession Col-0 (S1D Fig), we concluded that the temperature-dependent early flowering phenotype of Kil-0 at 15°C was vernalization-independent.
Fig. 1. Early flowering is controlled by one major recessive locus in Kil-0. 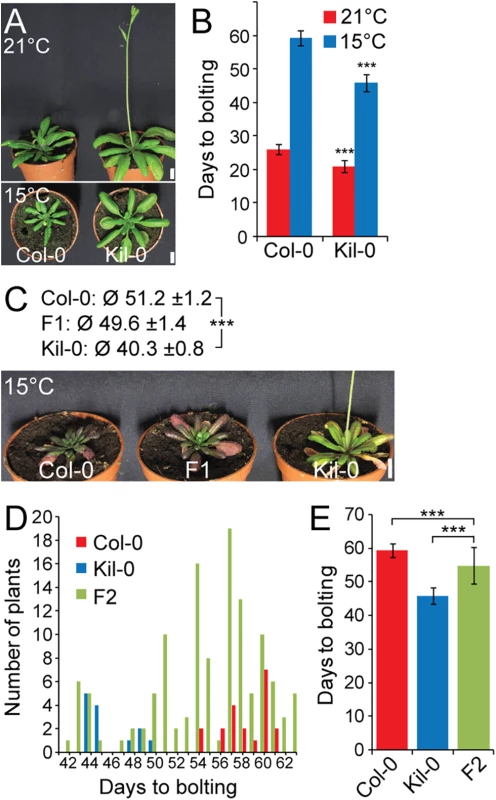
(A) Representative photographs and (B) quantitative flowering time analysis (days to bolting ± SD) of Col-0 and Kil-0 grown under continuous light at 21°C and 15°C. (C) Representative photographs of 50 day-old plants of Col-0, Kil-0, and their F1 hybrid grown under continuous light at 15°C. The average days to bolting ± SD are indicated above the photograph. (D) Distribution of flowering time and (E) average days to bolting ± SD in a population of 124 F2 segregants from a Col-0 x Kil-0 hybrid as well as among Col-0 (n = 20) and Kil-0 (n = 15) plants. Student’s t-tests were performed in comparison to the wild type unless indicated otherwise in the figure: *** = p ≤ 0.001. Scale bar = 1 cm. FLM is the causative locus for early flowering in Kil-0
The prominent early flowering of Kil-0 at 15°C (hitherto FT15) reliably allowed distinguishing between Kil-0 and Col-0. Analyses of F1 and F2 Kil-0 x Col-0 plants indicated that the Kil-0 flowering phenotype was determined by a major-effect recessive locus (Fig 1C, 1D and 1E). We subsequently mapped the FT15 locus to a 968 kb genomic region (S2A and S2B Fig). After selfing F2 plants with a recombination event in this interval, we identified 49 F3 plants with an early (Kil-0) and 41 F3 plants with a late (Col-0) flowering behavior (S2C and S2D Fig). We further narrowed down the interval of interest to a 151 kb region between 28.9 and 29.1 Mb on chromosome 1 (S2E Fig) and sequenced pools of 15 early and 9 late flowering F3 recombinants (S1 Table). Additionally, we sequenced the Kil-0 genome and identified 309 high confidence SNPs (single nucleotide polymorphisms) in the 151 kb mapping interval (S2E Fig). We smoothed the allele frequencies of the 309 SNPs of the two pools of early and late flowering F3 plants using LOESS (locally weighted scatterplot smoothing) and calculated the difference (Δf) between them. A fraction of Δf > 25% defined a final mapping interval of 31.3 kb that comprised eleven annotated genes (S2E Fig). Since the flowering phenotype of Kil-0 segregated in a recessive manner (Fig 1C), we assumed that a potential candidate gene might show reduced transcript abundance in comparison to Col-0. When we investigated RNA-seq data from 10 day-old Kil-0 and Col-0 plants grown at 21°C, we identified FLOWERING LOCUS M (FLM) as the only gene within the 31.3 kb region that was expressed at a significantly lower level in Kil-0 than in Col-0 (Fig 2A and S2 Table). FLM was also the most strongly downregulated gene in Kil-0 when we specifically analyzed 267 genes with a role in flowering time regulation (Fig 2A and S3 Table). These data suggested that FLM (FLMKil-0) may be causative for early flowering in Kil-0.
Fig. 2. Identification of the Kil-0 early flowering locus. 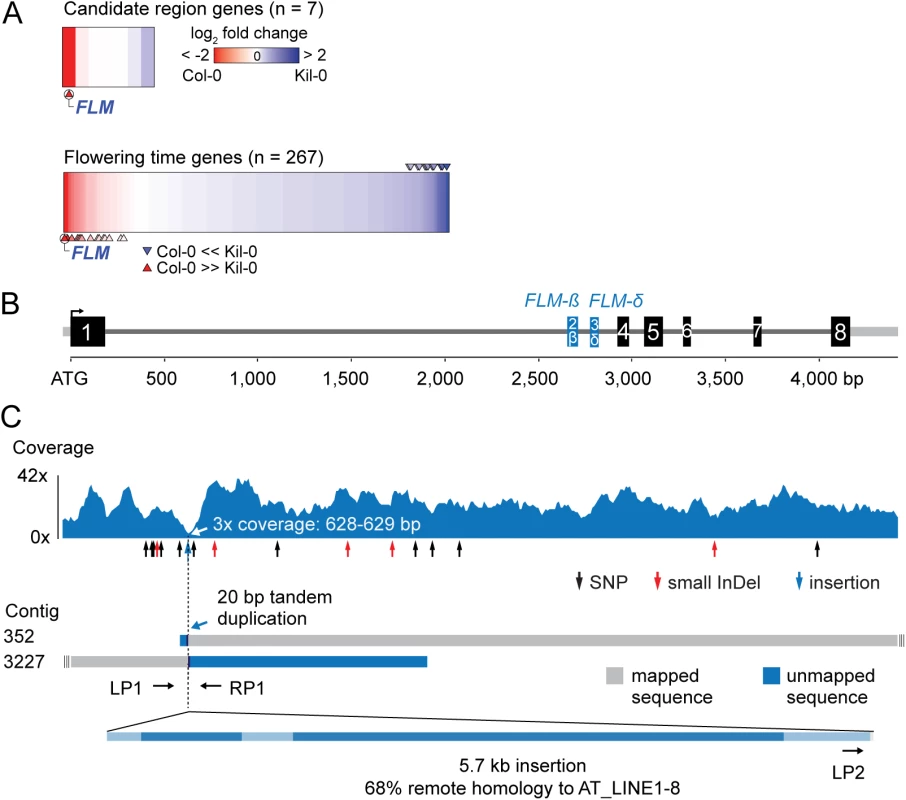
(A) Heatmap of differentially expressed genes in Kil-0 versus Col-0 in 21°C as analyzed by RNA-seq. The upper panel shows the fold change expression between Kil-0 and Col-0 of the seven expressed genes in the 31.3 kb mapping interval depicted in S2E Fig. The lower panel shows expression values of 267 flowering time genes. Fold changes are log2 transformed, blue represents upregulation and red downregulation in Kil-0 compared to Col-0. Significantly differentially (p < 0.01) expressed genes are marked with arrows. (B) Gene model of the FLMKil-0 locus of the Col-0 reference gene with the alternatively used exons 2 and 3 that give rise to FLM-ß and FLM-δ are shown in blue. Black boxes indicate exons, grey lines are 5’- or 3’-untranslated regions or introns. (C) Read coverage of the Kil-0 genomic data on the Col-0 reference gene model. Small sequence polymorphisms are indicated below the graph. Distribution of two independently assembled contigs from Kil-0 genomic sequence reads reveal the presence of a 5.7 kb insertion in FLM intron 1 with homology to the A. thaliana transposon At_LINE1-8. Sequence elements with homology to the LINE element are shown in dark blue in the lowermost panel. The respective positions of the primers used for the screening of further A. thaliana accessions are depicted. Kil-0 FLM harbors a LINE retrotransposon
Since the gene expression analyses had indicated that FLM may be the causative locus for early flowering im Kil-0, we compared the FLM genomic loci from Kil-0 and Col-0 at the molecular level. We found, however, no SNPs in the FLM coding sequence and only a few SNPs in the FLM promoter or introns. Interestingly, read coverage was greatly reduced at the beginning of the first FLM intron in Kil-0 (Fig 2B and 2C). Since a structural polymorphism could cause such a read coverage reduction, we de novo assembled the Kil-0 genomic sequencing reads and identified an insertion in FLMKil-0 that was flanked by two contigs corresponding to two halves of the FLMCol-0 locus at the predicted insertion site (Fig 2C). We PCR-amplified this region and confirmed the presence of a 5.7 kb insertion in the first intron of FLMKil-0 (Fig 2C). The inserted sequence showed similarity (68% identity, 86% coverage) to A. thaliana ATLINE1_8 (hereafter called LINE insert), a non-LTR (NON-LONG TERMINAL REPEAT LONG INTERSPERSED NUCLEAR ELEMENTS) retrotransposon with six typical retrotransposon domains as well as a perfect copy of the second exon of a RIBONUCLEASE H-LIKE (AT1G04625) gene from chromosome 1 of the Col-0 genome (S4 Table and Figs 2C and S3). We considered this FLM insertion polymorphism as the most likely cause for early flowering in Kil-0 and subsequently conducted genetic experiments to confirm this hypothesis.
FLMKil-0 is a functional temperature-sensitive FLM allele
Loss of FLM results in early flowering in the flm-3 allele in 23°C-grown Col-0 [17, 18]. To be able to compare the effect of the FLMKil-0 locus with that of FLMCol-0 and the flm-3 loss-of-function allele, we backcrossed FLMKil-0 six times into Col-0 to establish ColFLM-Kil. ColFLM-Kil flowered much earlier than Col-0 but not as early as the flm-3 knock-out mutant (Fig 3). We also introgressed flm-3 with four backcrosses into Kil-0 to obtain Kilflm-3. This introgression line flowered earlier than Kil-0. Although we performed marker assisted backcrossing, we cannot exclude the possibility that a background effect has an additional influence on flowering time regulation in the two described backcross lines. However, since the results between the two backcross lines are consistent with their effects in the original backgrounds, we considered it highly likely that the genetic data reflect FLM activity and concluded that the activity of FLMKil-0 was intermediate between that of a functional FLMCol-0 allele and the flm-3 loss-of-function allele (Fig 3).
Fig. 3. FLMKil-0 accelerates flowering. 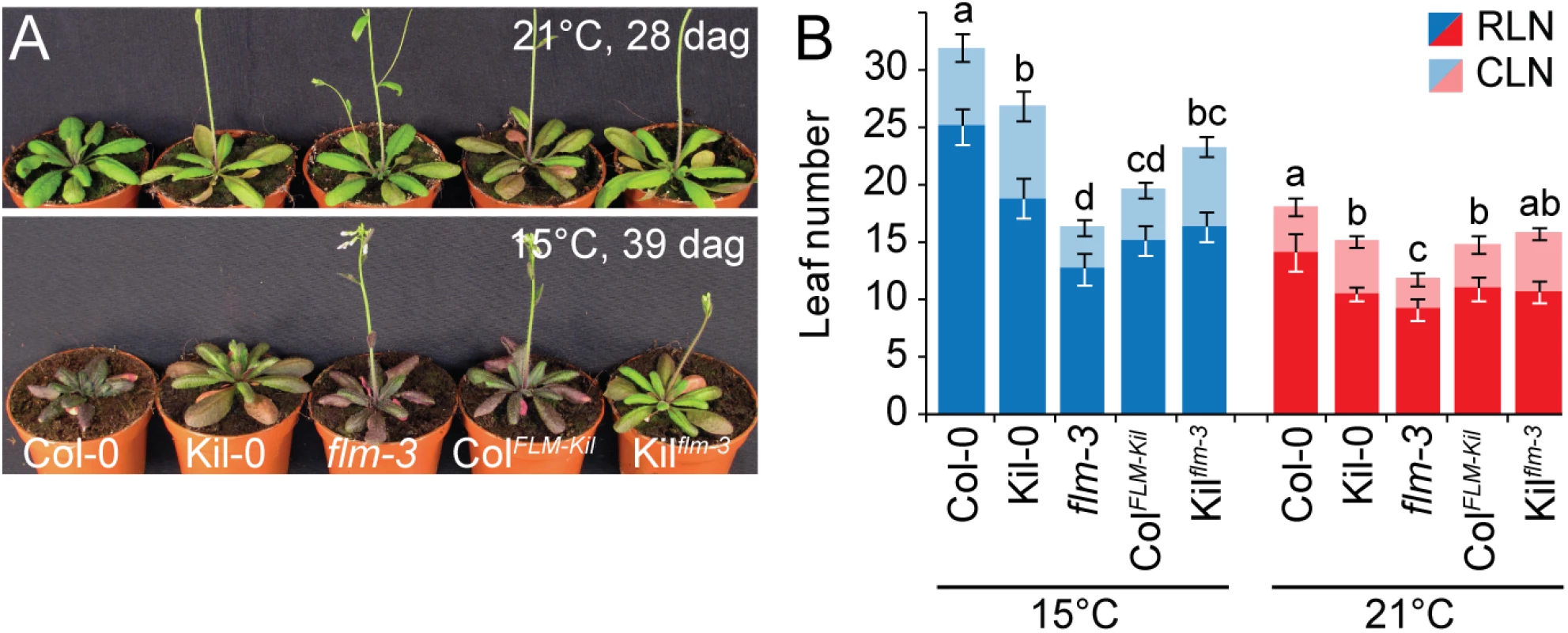
(A) Photographs of representative 28 and 39 day-old plants of the ColFLM-Kil and Kilflm-3 backcross population in comparison to Col-0, Kil-0, and flm-3 grown at 15°C and 21°C in long day photoperiod. (B) Averages ± SD of the quantitative flowering time analysis (RLN, CLN; rosette and cauline leaf number) of the genotypes shown in (A). Similar letters indicate no significant difference of total leaf number (Tukey HSD, p < 0.05). The phenotypic differences between Col-0 and ColFLM-Kil as well as those between Col-0 and flm-3 were more pronounced at 15°C than at 21°C (Fig 3). This was in agreement with our analysis of flm-3 in a range of growth temperatures, which had revealed that FLM makes a particularly prominent contribution to flowering time control at 15°C (S4 Fig). Since 15°C is closer to the average temperature in the native range of the species than the commonly used 21°C, the strong effect of FLM on flowering at 15°C should be considered a physiologically and ecologically relevant phenotype [11, 27].
Gene expression and alternative splicing of FLM are differentially regulated in Kil-0
FLM produces two major splice isoforms, FLM-ß and FLM-δ, in the Col-0 accession [19, 20]. FLM-ß is the predominant splice form in cooler and FLM-δ is the predominant splice form in warmer temperatures [19]. Since the FLMKil-0 allele had weaker effects on flowering time than the flm-3 loss-of-function allele and since FLMKil-0 did not have polymorphisms in the FLM coding region, we hypothesized that the temperature-sensitive flowering of Kil-0 may be caused by changes in FLM expression or FLM alternative splicing. To examine this, we transferred seven day-old 21°C-grown Col-0 and Kil-0 plants for three days to 9°C, 15°C, 21°C or 27°C and examined the effects on FLM-ß and FLM-δ transcript abundance. In agreement with published data, we observed a respective decrease of FLM-ß and an increase of FLM-δ in response to warmer temperatures (Fig 4A) [19, 20]. Importantly, temperature-dependent changes in the abundance of the FLM isoforms were maintained in Kil-0 as well as in ColFLM-Kil but the overall FLM transcript abundance was strongly reduced compared to Col-0. For example, when comparing the values at 21°C, Kil-0 and ColFLM-Kil had six-times less FLM-ß and 27-times less FLM-δ than Col-0 (Fig 4A).
Fig. 4. The large insertion in the first intron affects splicing efficiency and gene expression of FLM. 
(A) qRT-PCR analysis of FLM-ß, FLM-δ, and SVP expression in ten day-old Col-0, Kil-0, and ColFLM-Kil plants. Seven day-old seedlings grown at 21°C were transferred to the indicated temperature and grown for further three days. The fold change differences between Col-0 and Kil-0 at 21°C are indicated in the graph. Error bars represent SE of three biological replicates. (B) Quantitative flowering time analysis of four—five independent T2 lines in the Kil-0 background that are either hemi- or homozygous for the respective transgene construct. 14–20 transgenic plants grown at 21°C under long day photoperiod were analyzed for each genotype. Transgenic T2 lines expressing the empty pGREEN0229 vector control construct (BAR+) in the Col-0, Kil-0, and flm-3 lines were analyzed for comparison. Outliers were determined based on 1.5 x IQR (interquartile range). (C) qRT-PCR analysis of FLM-ß and FLM-δ expression of four independent homozygous T3 lines of the indicated construct in the flm-3 background. Plants were grown for ten days under 21°C long day photoperiod. Error bars represent SE of three biological replicates. (D) Graph displaying normalized read counts for the seven FLM exons in Col-0 and Kil-0, including the differentially spliced exon 2 that gives rise to the FLM-ß and FLM-δ isoforms. (E) Quantification of the normalized read counts shown in (D). The average ratios and SD from three biological replicates of the read counts of exon 1 compared to the read counts of exon 2 to exon 7 are shown. (F) qRT-PCR-based verification of the RNA-seq result shown in (E) using fragments corresponding to FLM exon 1 and exon 4. The ratios ± SE of the respective expression values are shown from three biological replicates. (G) qRT-PCR quantification of unspliced pre-mRNA from isolated nuclei. Primers are located in FLM introns 1, 2, and 6 and ACT8 introns 2 and 3, respectively. Primer sequences are provided in S7 Table. The position of the respective reverse primer relative to the ATG start codon is indicated. Fold changes are averages ± SE of two biological replicates are indicated. Student’s t-tests: ** = p ≤ 0.01; n.s., not significant. (H) Normalized read counts for FLM as detected by RNA-seq in Col-0 and Kil-0 mapped to the genomic FLM locus from exon 1 through exon 5. For both accessions, the number of mapped RNA-seq reads was normalized to range from 0 (no expression) to 1 (maximum expression). Lines indicate the mean expression level and light blue and light red areas indicate the 5% and 95% confidence intervals that were determined across the biological replicates of one genotype. Exons are represented as boxes, untranslated regions and introns as lines. Note the relative increase in intron 1 reads in Kil-0 as indicated with a dotted line. The position of the LINE insertion in FLMKil-0 is indicated by an arrow. Student’s t-tests were performed as indicated: * = p ≤ 0.05; ** ≤ 0.01; *** = p ≤ 0.001; n.s., not significant. (I) Schematic representation of the primers used for the qRT-PCR quantification of FLM intron 1 sequence-containing transcripts and the 3’-RACE PCR as indicated by arrows below the gene model; PTC, premature termination codon. A schematic representation of the intron 1 sequence-containing polyadenylated transcripts is indicated in the lower part of the panel. Numbers on the left indicate the frequency of each transcript among the 32 individual sequenced clones. The black line indicates the length of each transcript respective to the ATG start codon. It was previously shown that FLM-ß represses and that FLM-δ promotes flowering at 16°C when introduced as transgenes in the flm-3 background [19–21]. It was further proposed that FLM-ß forms heterodimers with SVP (SHORT VEGETATIVE PHASE) to prevent flowering by direct DNA-binding to repress the transcription of FT and SOC1. Conversely, FLM-δ would form inactive heterodimers with SVP and would thereby indirectly induce flowering by relieving the repression from FT and SOC1. To understand the effects of this differential regulation, we measured SVP, FT, and SOC1 expression levels. Whereas SVP expression was similar between the different genotypes, FT and SOC1 were expressed more strongly in the early flowering Kil-0 or ColFLM-Kil than in Col-0 (Figs 4A and S5). We thus concluded that the reduced expression of FLM was likely the cause for the early flowering of Kil-0 and ColFLM-Kil and that the temperature-dependent differential accumulation of FLM-ß and FLM-δ could be the basis of the temperature-sensitive flowering time in Kil-0. Since Kil-0 flowered earlier than Col-0, we assumed that the effect of the downregulation of the repressive FLM-ß isoform was dominant over the downregulation of the flowering activating isoform FLM-δ.
The LINE insertion affects FLM splice isoform abundance
To test whether the large insertion in FLMKil-0 was the causative polymorphism for low FLM expression and the specific reduction in the FLM-ß isoform, we transformed Kil-0 with a genomic fragment of FLMCol-0 (including 2 kb promoter plus 5'-UTR [untranslated region] and 0.5 kb 3'-UTR sequence) as well as an FLMKil-0 genomic variant with an engineered deletion of the LINE-insertion (FLMKil-0ΔLINE). We found that FLMCol-0 as well as FLMKil-0ΔLINE delayed flowering in Kil-0 (Fig 4B). We also tested whether the 5.7 kb insertion had a comparable effect on FLM transcript abundance when engineered into the FLMCol-0 reference and introduced a FLMCol-0 transgene with an engineered 5.7 kb LINE insertion into the flm-3 loss-of-function mutant. Indeed, the LINE insertion reduced FLM expression and changed FLM splicing, similar to the FLMKil-0 allele (Fig 4C). We thus considered it very likely that the LINE insertion in the first intron of FLM was the causative polymorphism for reduced FLM transcript abundance, differential FLM splice isoform accumulation, and early flowering in Kil-0.
To gain an understanding of the molecular effect of the LINE insertion on FLM transcription and splicing, we compared the exon usage of Col-0 and Kil-0 using RNA-seq data. In Col-0, we detected a strong differential use of alternative exons 2 (FLM-ß) and 3 (FLM-δ) that define the two dominant FLM isoforms (Fig 4D), with the FLM-ß-specific exon 2 being more abundant than the FLM-δ-specific exon 3. In Kil-0, we detected generally fewer reads for all exons including exon 1, indicating that the LINE insertion between exon 1 and exon 2 controls the overall FLM transcript abundance (Fig 4D). Furthermore, we noted that the read coverage of the exons following intron 1, including the alternatively used exons 2 (ß) and 3 (δ) was more strongly reduced in Kil-0 than in Col-0, suggesting that the inserted LINE element may also negatively control FLM splicing efficiency (Fig 4D, 4E and 4F). To approximate the rate of FLM transcription, we determined the levels of unspliced FLM pre-mRNA using primers located in intron 1 (upstream of the LINE insertion), and intron 2 and 6. No significant differences in pre-mRNA levels were detected when we amplified intron 1 indicating a comparable transcription rate between the two accessions. However, the relative abundance of intron 2 and intron 6, which are located downstream from the insertion was strongly reduced in Kil-0, indicative for a partial premature termination of FLM transcription (Figs 4G, S6A and S6B). We next mapped the RNA-seq reads from Col-0 and Kil-0 to the FLMCol-0 genomic locus paying particular attention to the exon-intron junctions (Fig 4H). Whereas the read coverage dropped sharply at the exon 1-intron 1 junction in Col-0, reads covering the beginning of intron 1 could be readily retrieved in Kil-0 (Fig 4H). This finding suggested that the LINE insertion resulted in aberrant splicing within Kil-0 intron 1. This change in the splicing pattern had an impact on the abundance of the FLM-ß and FLM-δ full-length transcripts but, importantly, the respective full-length mRNAs were still generated as confirmed by semi-quantitative RT-PCR (S6C Fig). We then used qRT-PCR with primers spanning only exon 1 or exon 1 and parts of intron 1 to validate the occurrence of transcripts containing intron 1 sequences (Fig 4I). In Col-0 as well as in Kil-0, we found transcripts including intron 1 sequences until 350 bp from the start codon (Figs 4I and S6D). The abundance of intron 1 sequence-containing reads was much higher in Kil-0 than in Col-0 indicating that the insertion may indeed promote premature transcription termination possibly in combination with aberrant splicing. Since transposon insertions were reported to induce alternative polyadenylation [28–30], we examined whether the corresponding intron 1 sequence-containing transcripts were polyadenylated and performed 3’-RACE (rapid amplification of cDNA ends) experiments. The RACE PCR yielded two abundant fragments, one corresponding in size to the full length transcript and one smaller fragment that was much more abundant in Kil-0 than in Col-0 (S6E Fig). We cloned and sequenced products and determined six different polyadenylated transcripts containing intron 1 sequences. One of these fragments represented the most abundant species (75%) among the 32 independent sequences (Fig 4I). Premature translation termination codons (PTC) located distantly from splice sites frequently trigger the degradation of aberrant transcripts through the NMD - (non-sense mediated decay-) pathway [31, 32]. We identified a PTC in intron 1, just two bases downstream from the exon1-intron1 border and hypothesized that the aberrant FLMKil-0 transcripts may be NMD targets (Fig 4I). When we used the translation inhibitor CHX (cycloheximide) to mimic the molecular phenotype of NMD-defective mutants [32, 33], we detected indeed a significant increase in the abundance of two aberrant transcripts containing intron 1 sequences (S6F and S6G Fig). In summary, we concluded that the early flowering of Kil-0 correlated with the presence of a LINE insertion in FLM intron 1. Further, the LINE insertion did not affect de novo FLM transcription initiation but partially impaired the formation of full length transcripts, possibly as a result of premature transcription termination and the formation of aberrantly spliced polyadenylated transcripts that are targeted for NMD. Alternative molecular mechanisms that were not examined here may of course also be suitable to explain the overall reduction in FLM transcript abundance in Kil-0.
The geographic distribution of the FLMKil-0 allele is consistent with a recent adaptive selective sweep
To gain information about the distribution of the FLMKil-0 structural polymorphism across the native range of the species, we screened a genetically highly diverse set of accessions from a previously published HapMap population, which we supplemented with selected laboratory accessions to obtain a final population with 419 accessions (S6 Table and Figs 2E and S7) [34]. Through PCR-based screening, we identified nine additional accessions with a LINE insertion in the same position as in Kil-0 from Scotland (Kil-0) and Sweden (Ull2-3) to Germany (8 accessions) (S5 Table and Fig 5A and 5B). We subsequently also analyzed genome sequences from 1128 A. thaliana accessions (www.1001genomes.org) and confirmed by this approach seven FLMLINE accessions and identified El-0 as an additional FLMLINE accession from Germany (S7 Fig).
Fig. 5. Geographical distribution and molecular analysis of FLMLINE accessions. 
(A) Geographical distribution of the FLMLINE accessions (blue pentagons) with the centroid (green star). Please note that one the accession with an ambiguous name was excluded due to the unavailability of geographic information. (B) Neighbour-Joining tree showing the genetic relationship among FLM sequences. The FLM sequences of the 10 FLMLINE accessions were analyzed together with Col-0 and further 88 randomly selected sequences. The clade harboring all FLMLINE alleles is marked in blue; see S8 Fig for a detailed representation of the entire tree. (C) qRT-PCR analyses of FLM-ß and FLM-δ transcript abundance of ten day-old seedlings of all FLMLINE accessions when grown in 21°C long day photoperiod (upper panel). Averages ± SE of three measurements after normalization to Col-0 as a control are shown. The inserted graph shows the summarized log2-transformed fold changes of FLMLINE accessions only (Col-0 excluded). The lower panel displays the ratios of FLM-δ over FLM-ß when normalized to Col-0. (D) Quantitative flowering time analysis of FLMLINE accessions grown at 21°C and 15°C under long day photoperiod. Dashed lines mark the respective values for Col-0. Please note that Sp-0 and Ste-0 are vernalization-sensitive accessions. (E) The right graph shows the summarized rosette leaf number of the eight vernalization-insensitive FLMLINE accessions. Student’s t-tests were performed in comparison to Col-0 unless indicated otherwise: * = p ≤ 0.05; ** ≤ 0.01; *** = p ≤ 0.001; n.s., not significant. El-0 was identified relatively late in this study and not included in this detailed analysis. Sequence analyses of the LINE insertions revealed a high sequence similarity between the ten FLMLINE-accessions (S9 Fig). Taking into account a spontaneous mutation rate of 6 to 7×10−9 per site per generation, approximately one to three seed generations per year, and the absence of any selective pressure on the insertion, we calculated that the common ancestor probably originated only 8.000 to 30.000 years ago [35, 36]. We found that the FLMLINE accessions belonged to genetically differentiated clades and were thus truly independent [37]. Additionally we performed a phylogenetic analysis using the genomic sequence of the FLM locus of these ten FLMLINE accessions together with Col-0 and 88 randomly selected accessions, which revealed that the FLMLINE lines clustered into one clade when the FLM locus was analyzed in isolation (Figs 5B and S8).
Similarly to Kil-0, all FLMLINE-accessions had low expression of the FLM-ß and FLM-δ isoforms (Fig 5C). Apart from two late-flowering vernalization-dependent accessions (Sp-0 and Ste-0), seven of the remaining eight FLMLINE accessions flowered earlier than Col-0 at 15°C (Figs 5D, S9A and S9B) [12, 26]. Consistent with the fact that these accessions come from genetically highly diverse groups, these lines showed a substantial variation in flowering time between them. However, when flowering time data of the vernalization-independent FLMLINE accessions was averaged, we measured a significant reduction in flowering time at 21°C and 15°C in comparison to Col-0 (Fig 5E). Thus, the LINE insertion correlated with early flowering in summer annual accessions. Our data thus suggests that the FLMKil-0 allele arose comparatively recently and subsequently spread geographically to contribute to flowering time regulation in a background - and temperature-dependent manner.
When we analyzed the ten FLMLINE-accessions for FLM expression by qRT-PCR, we noted a prominent variation in the abundance of the FLM-δ isoform but relatively stable expression levels of FLM-ß (Fig 5C). We therefore asked whether FLM polymorphisms other than the LINE insertion could explain these differences. However, all ten FLMLINE accessions were highly similar in this region and we identified only four different additional polymorphic sites (S9C Fig). Since a Mann-Whitney test (p > 0.05) indicated that none of these polymorphisms was significantly associated with FLM-ß and FLM-δ abundance or the ratio between these two isoforms, we concluded that the regulation of the FLM-ß and FLM-δ transcript abundance may be regulated in trans.
The first intron carries important regions for isoform-specific FLM regulation
Specifically within the family of MADS-box transcription factor genes, various cases are known where structural polymorphisms within the first intron enhance or repress gene expression [10, 38–41]. To test if FLM intronic sequences contributed to FLM expression, we transformed flm-3 mutants with constructs for the expression of the FLM-ß or FLM-δ coding sequences under control of a 2.1 kb FLM promoter fragment (S10 Fig). Importantly, none of the resulting FLM-ß or FLM-δ T1 transformants expressed significant levels of the respective transgene or showed a suppression of the flm-3 early flowering phenotype (S10 Fig). Since FLM was expressed from corresponding genomic constructs containing all introns and since it was previously shown that FLM is expressed from the above-described construct when only intron 1 is included [19], we concluded that intron 1 was essential for FLM expression.
To address whether sequence identity, sequence length or the specific insertion site within intron 1 conferred the effect of the LINE element on FLM expression and splicing, we examined the effects of T-DNA or DS (Dissociator) transposon element insertions in FLM intron 1 [42–44]. These intron 1 insertions were of similar size (4.5 kb to 5.3 kb) to the 5.7 kb LINE insertion and present in the Col-0 and No-0 (Nossen-0) accessions: Salk_068360 (Salk, Col-0), RATM13-4593-1 (RIKEN, No-0) and GK_487H01 (GABI, Col-0) (Fig 6A). Additionally, we included Co-1 that carries a ~1.5 kb insertion in FLM intron 1 (Fig 6A). When we determined the effects of these structural variants on FLM transcript accumulation and alternative splicing in plants grown at 15°C and 21°C, we found that the insertion correlated in each case with reduced FLM transcript abundance when compared to the controls (Fig 6B). Furthermore, FLM-δ was in each case more strongly reduced than FLM-ß, inviting the conclusion that increases in intron length, regardless of the molecular identities of the insertions, resulted in decreased FLM-ß and FLM-δ expression and changes in the ratio between the isoforms. This was further confirmed by the molecular analysis of pre-mRNA transcript abundance, the formation of aberrant polyadenylated transcripts and transcript targeting to the NMD pathway, which we performed in parallel for the Salk insertion line, Col-0, and Kil-0 (Figs 4G and S6). In this analysis, we identified in each case the same molecular defects in the Salk insertion line as in Kil-0. At the same time, we noted that the temperature-sensitive regulation of the FLM isoforms was maintained in all lines. Interestingly, insertions in the second half of the intron as present in the GABI and RIKEN lines caused a particularly strong reduction in the expression of FLM-δ expression (Fig 6B). We thus concluded that insertions in the second half of the intron may have additional effects on FLM-δ splicing.
Fig. 6. The first intron carries important regions for isoform-specific FLM abundance. 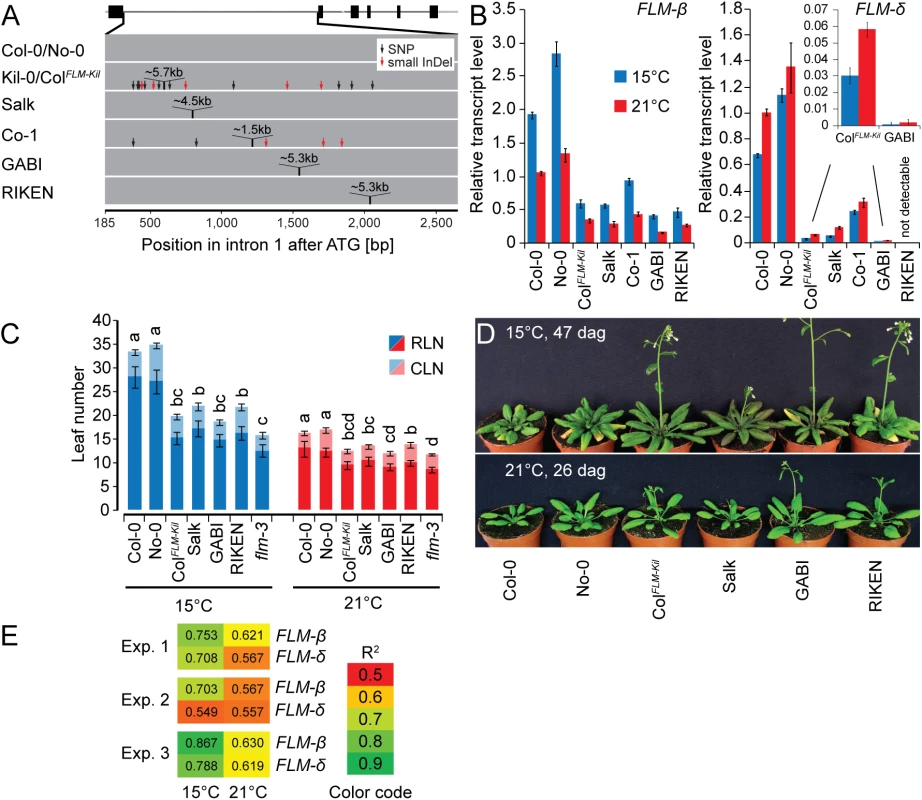
(A) Schematic representation of the insertion lines used in this study. The FLM-ß gene model is depicted as shown in Fig 3C, intron 1 of the respective lines is depicted as a grey bar. (B) FLM-ß and FLM-δ expression of the lines shown in (A). Seven day-old seedlings were grown at 21°C and then transferred to 21°C or 15°C for further three days. Shown are averages and SE of three biological replicates. (C) Quantitative flowering time analysis of the lines shown in (A) with Col-0 and No-0 as wild type controls. Similar letters indicate no significant difference of total leaf number (Tukey HSD, p < 0.05). (D) Representative photographs of the lines shown in (A) at 26 and 47 days after germination (dag) when grown at 21°C and 15°C temperature and under long day photoperiod. (E) Correlation analysis of the flowering time experiment shown in Fig 6C and further two independent experiments (15°C and 21°C) with the expression values shown in Fig 6B. Note that flowering time values of the loss-of-function allele flm-3 were also included in this analysis but that its FLM expression was set to 0 (no expression). R2 values from linear regression are shown. To examine the phenotypic consequences of the observed transcriptional changes of the FLM insertion lines, we evaluated their flowering at 15°C and 21°C. We thereby focused on the Salk and ColFLM-Kil as well as the GABI and RIKEN lines, which had contrasting phenotypes with regard to the abundance of the FLM-δ isoform. Regardless of the differences in expression of the FLM-δ isoform, all lines flowered earlier than the respective wild type, and this effect was particularly prominent at 15°C (Fig 6C and 6D). Importantly, we did not notice any pleiotropic effects on plant growth or plant height for the tested alleles suggesting that FLM acts specifically on flowering time regulation (Figs 6D and S11). We next evaluated to what extent changes in FLM-ß or FLM-δ abundance could explain the observed difference in flowering time. To this end, we correlated datasets on flowering time and FLM expression from three independent flowering time experiments (Fig 6E). In each experiment, expression of FLM-ß correlated much better with flowering time than FLM-δ and this effect was particularly pronounced at 15°C (Fig 6E). Thus, FLM-ß expression levels alone rather than the ratio between FLM-ß and FLM-δ have a prominent effect on flowering time regulation in A. thaliana.
Discussion
We have identified a recently evolved FLM allele from the accession Kil-0. The insertion of a LINE element in intron 1 of FLMKil-0 resulted in reduced FLM transcript abundance and correlated with an overall acceleration of flowering time that was particularly prominent at 15°C (Fig 7). We identified additional nine FLMLINE accessions that mainly represented lines collected from Germany. Although these FLMLINE accessions were highly homologous over the FLM locus, they represented accessions from genetically different clades indicating that FLMLINE was involved in recent adaptation to early flowering and that its rather narrow geographical distribution is likely due to the young demographic history of this allele.
Fig. 7. Model of the proposed mode of FLM action. 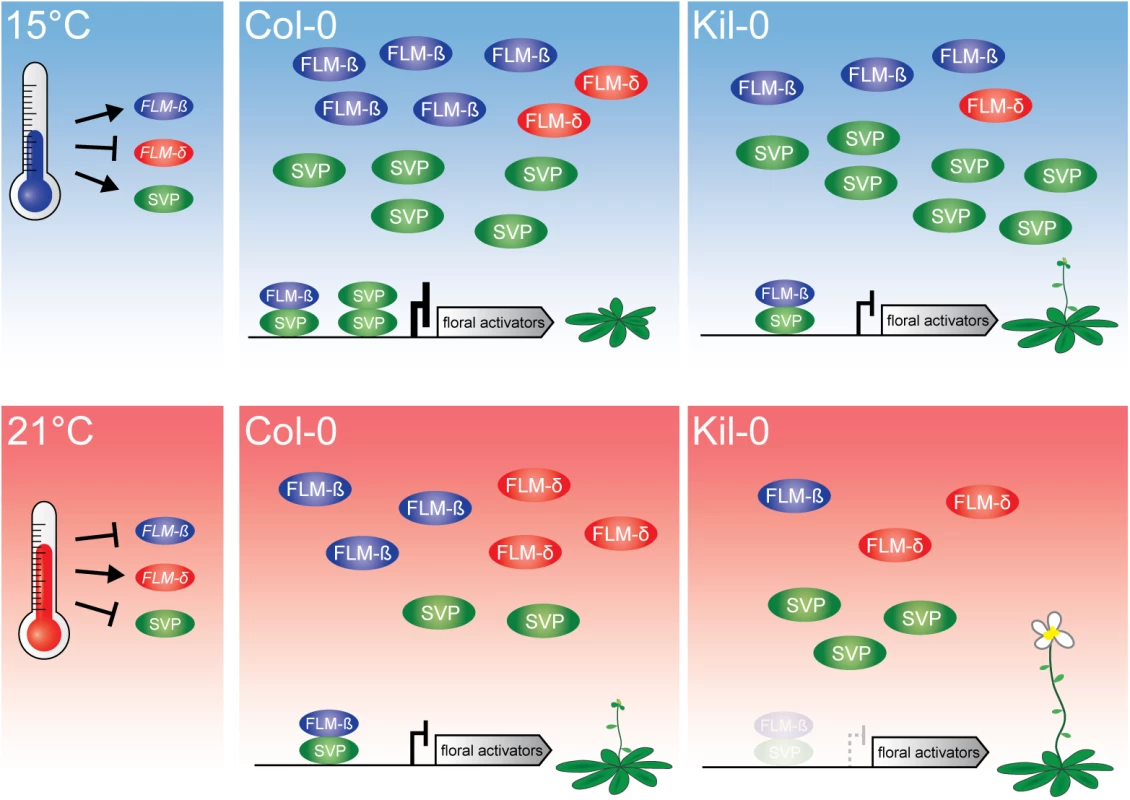
At 15°C, flowering is delayed due to an active repression of floral activators by FLM-ß-SVP heterodimers. An increased ambient temperature (21°C) results in decreased levels of the transcriptional repressive FLM-ß-SVP protein complexes and floral activator genes are expressed. Flowering in Kil-0 (right) might be accelerated in comparison to Col-0 due to a decreased abundance of FLM-ß at 15°C as well as at 21°C while SVP is unaltered in the different temperatures and between Kil-0 and Col-0. Note that our study suggests a minor role of FLM-δ in the control of flowering time in the lines and conditions examined. The present model assumes that protein abundance follows transcript abundance except for SVP, which is degraded in response to increasing temperature [20]. The LINE element insertion of FLMLINE shares 68% homology with LINE class I retrotransposons from A. thaliana. Transposable elements are typically suppressed by epigenetic mechanisms and this suppression can also negatively interfere with the expression of neighboring genes [45–48]. Epigenetic regulation is also known to control the expression of MADS-box transcription factor genes. For example, the chromatin of FLC is modified during vernalization by lysine 27 methylation of histone 3 (H3K27me), a repressive mark, and several natural variants interfere with regulatory regions in the FLC intron 1 [41, 49–53]. Thus, the LINE insertion could interfere with the direct transcriptional regulation but, alternatively, also with the epigenetic control of the FLMKil-0 locus. However, previous genome-wide studies failed to identify epigenetic marks such as H3K27me on intron 1 of FLM [54] and we detected no differences in transcription rate between the insertion lines and the Col-0 reference. Furthermore, transposon and T-DNA insertions were reported to mediate alternative polyadenylation through the utilization of alternative polyadenylation sites [28–30]. Although we detected a higher abundance of short aberrant polyadenylated transcripts in the insertion lines, we consider it unlikely that the insertion itself provides cis elements that result in their synthesis. Aberrrant transcripts did not extend into the insertion and no differences in quantity and composition of these transcripts were detected between Kil-0 and the Salk line, which have molecularly distinct insertions. We concluded that the reduction of FLM full-length transcript abundance in Kil-0 is caused by a combination of partial premature termination of transcription and aberrant splicing due to the enlargement of the first intron.
Conversely, through experiments with FLM transgenes where intronic sequences were deleted, we could conclude that intron 1 was strictly required for FLM expression and activity. A contribution of intronic sequences in gene expression regulation, generally referred to as IME (intron-mediated enhancement), was previously reported for many genes, and in plants specifically for members of the MADS-box transcription factor family [55, 56]. Several studies have already identified corresponding intronic cis-regulatory elements, e.g. in intronic regions of the floral homeotic genes AG (AGAMOUS) and members of the AGL6 (AGAMOUS-LIKE6)-subfamily [38–40, 57]. Several independent structural intron polymorphisms were also reported for the MADS-box factor and flowering time regulator VRN1 (VERNALIZATION1) from wheat and barley. There, these structural differences in intron 1 composition can promote high VRN1 expression and these differences are the main molecular cause for vernalization-independent flowering in many spring barley and wheat cultivars [10]. Thus structural intron polymorphisms, e.g. through transposon insertions, are a recurrent theme in the expression control of MADS-box transcription factors and the adaptation to the environment through these factors.
Interestingly, our expression analysis showed that the expression of the two FLM isoforms, FLM-ß and FLM-δ were differentially affected by the intron 1 insertions. Since this behavior was found in several accessions and was recapitulated by inserting a LINE-bearing FLM transgene in the Col-0 FLM allele, we judge that this regulation again is not related to the nature of the insertion in FLM but rather to the position of the insertion or the corresponding increase in intron length. It remains to be investigated, however, what the underlying molecular basis of the differential effect of the insertions on FLM-ß and FLM-δ abundance is.
Through investigations of insertion lines from different sources and ecotypes, we found that the position of the inserted sequence affected the relative abundance of FLM-ß and FLM-δ. In combination, the availability of these lines allowed the examination of flowering time and its correlation with the abundance of the two FLM splice variants.
Importantly, our study did not support a role for FLM-δ in flowering time control but rather suggests that FLM-ß abundance alone is the predominant determinant of flowering time in natural variants under the ecologically relevant temperature of 15°C. Thus, previous experiments exploring the contribution of FLM-ß and FLM-δ to flowering time control using transgenic approaches may have overestimated the contribution of FLM-δ, at least in the diverse genetic material and at the temperatures used in our study. At the same time, it cannot be ruled out that FLM-δ levels reached a subcritical level in the insertion lines analyzed in our study so that the contribution of FLM-δ could not be accurately determined. Along these lines, a recent study showed no contribution of the FLM locus to flowering time regulation in a large set of wild A. thaliana ectoypes in warm (27°C) temperature where FLM-δ is upregulated [58] indicating that even under temperature conditions that promote FLM-δ abundance no important regulatory role could be ascribed to it. Once a detailed understanding of factors controlling FLM gene expression and splicing will be obtained, it will be important to reexamine this aspect in more detail.
The role of FLM in flowering time variation was previously established in the temperature-insensitive accessions Nd-1 and Ei-6 where FLM is deleted [16, 17]. In our study, we report the first gene expression variation-allele for FLM and describe a molecular mechanism for the control of flowering time in ambient temperature through structural changes in FLM intron 1. It is interesting to note that loss-of-function alleles of flowering time genes are generally very rare and are typically distributed within a small geographic region. This may indicate that adaptation through gene loss may be disadvantageous outside of the specific ecological niche simply because the loss of the gene will prevent its future reactivation [59, 60]. In the case of FLM, this is exemplified by the Nd-1 and Ei-6 accessions, but also among the many known FLC alleles, only a few null alleles have been reported whereas gene expression-modulatory FLC alleles are more common [6, 12, 49, 50, 61]. In this regard, we perceive at least a trend towards a similar distribution among the A. thaliana FLM alleles. A deeper analysis about the FLM coding sequence polymorphisms will be required to make conclusive statements about the importance of strong and weak FLM alleles during adaptation of flowering time.
We conclude that structural variations of FLM intron 1 as described here represent an adaptive mechanism for the control of flowering time in A. thaliana and possibly also in other closely related Brassicaceae. FLM might have a very specific role in flowering time regulation because modulations of its expression seemingly affect only flowering and not other plant growth traits. We, therefore, think that FLM is an excellent candidate gene to precisely and steadily modulate flowering time in a dynamic manner over a broad range of temperature conditions to overcome the impacts of climate change on flowering in plants.
Material and Methods
Biological material
The following A. thaliana accessions and genotypes were used in this study and, unless stated otherwise, provided by the Nottingham Arabidopsis Stock Centre (NASC; Nottingham, UK): The Arabidopsis accessions Killean-0 (Kil-0), Columbia-0 (Col-0), and Nossen (No-0); the insertion mutants flm-3 (Salk_141971; Col-0), GABI-KAT GK487H01 (GABI; Col-0), Salk_068360 (Salk; Col-0) as well as RIKEN-13-4593-1 (RIKEN; No-0) from the RIKEN Stock Center. In each case, the positions of the insertions were verified by DNA sequencing. The transgenic line gFLMCol-0 (flm-3) was previously described [19], flc-3 and the line FRISF-2 FLC [6] were a gift from Franziska Turck and George Coupland (Max-Planck Institute of Plant Breeding Research, Cologne, Germany). A list of the A. thaliana accessions screened by PCR with the primers LP1, LP2, and RP1 for the FLMLINE structural polymorphism is provided as S6 Table. Primer sequences are provided as part of S7 Table.
Physiological experiments
For flowering time analyses, plants were randomly arranged in trays and grown under constant white light (70–90 μmol m-2 s-1) or in long day-conditions with 16 hrs white light (110–130 μmol m-2 s-1)/8 hrs dark in MobyLux GroBanks (CLF Plant Climatics, Wertingen, Germany) or MLR-351 SANYO growth chambers (Ewald, Bad Nenndorf, Germany). Trays were rearranged every two days and water was supplied by subirrigation. Analysis of large plant sets was performed in a walk-in chamber with constant white light as described above. Flowering time was quantified by determining the time until the macroscopic appearance of the first flower bud (days to bolting, DTB) or by counting rosette and cauline leaf numbers (RLN, CLN). Student’s t-tests, ANOVA, and Tukey HSD tests were calculated with Excel (Microsoft) and R (http://www.r-project.org/), respectively.
Kil-0 genome sequencing
For the resequencing of the Kil-0 genome or the late and early flowering F3 recombinant pools, libraries were prepared from 600 ng genomic DNA following the standard protocol of the TruSeq DNA Sample Preparation Kit v2 (Illumina, San Diego, CA). Paired-end sequencing with a read length of 100 bp was performed on a HiSeq 2500 (Illumina, San Diego, CA). Post-sequencing quality trimming was performed with the CLC Genomics Workbench (v. 7.0) and the following parameters: low quality limit = 0.05; ambiguous nucleotide = maximum 1; length minimum = 15. Post-trimming, 15 x 106 and 20 x 106 reads were obtained for the early and late flowering samples, respectively. Read mapping was performed using the TAIR10 release of the A. thaliana reference (The Arabidopsis Genome Initiative, 2000) genome reference sequence with the stringent settings: mismatch cost = 2; insertion cost = 2; deletion cost = 2; length fraction = 0.9; similarity = 0.9. An average 57 - or 79-fold coverage was obtained from the early and late flowering DNA pools. Variant calling was performed using the probabilistic variant calling tool of the CLC Genomics Workbench (v. 7.0) and default settings. SNPs with a 30–120-fold coverage and frequency f > 20% as well as a presence call in both pooled samples were selected for allele frequency mapping. From those, SNPs that showed a frequency of < 80% in the resequencing analysis of the homozygous Kil-0 parental line were discarded. Smoothing using locally weighted scatterplot smoothing (LOESS) of SNP frequency values was achieved with R (http://www.r-project.org/). 95% confidence intervals, Δf > 25%, and Δfmax were calculated from the LOESS values. The de novo assembly of Kil-0 resequencing reads was performed using the CLC Genomics Workbench v. 7.0 with default settings. Contigs were identified by a simple search and were reassembled to the Col-0 genomic FLM sequence. The Kil-0 genomic sequence is available as LN866842 at www.ebi.ac.uk/ena.
Mapping and backcrossing
To identify the causative locus for early flowering in Kil-0, the FT15 locus was mapped with polymorphic markers selected from a previously described marker collection [62]. Additional SSLP (single sequence length polymorphisms) markers were generated by searching the publicly available Kil-0 genomic sequence (www.1001genomes.org) or the genome sequence that was determined as part of this project for InDel (insertion/deletion) polymorphisms. PCR primers spanning these sites were designed with Primer3 [63] and tested on Col-0 and Kil-0 genomic DNA. The PCR fragments were generally between 200 and 700 bp long and separated on 2–3.5% agarose gels or using the QIAxcel Advanced Capillary Electrophoresis high resolution kit (Qiagen, Hilden, Germany). For rough mapping of FT15, ten early and ten late flowering F2 plants were selected from the extreme phenotypic borders of an F2 population (n = 124). The genetic marker distances were calculated from the genotype data of all screened F2 plants with JoinMap v.4.1 (Kyazma B.V.). A list of markers and the respective primers is provided as S8 Table.
ColFLM-Kil and Kilflm-3 were generated by marker-assisted backcrosses. In brief, heterozygous F1 plants were genotyped with the primers LP1, LP2, and RP1 to examine the lines for the presence of the Col-0 or Kil-0 FLM allele and with Salk_141971 forward and reverse primers to test for the flm-3 T-DNA insertion, respectively. Primer sequences are provided as part of S7 Table.
Quantitative real-time PCR
For qRT-PCR analyses, total RNA was isolated from three biological replicates using the NucleoSpin RNA Plant kit (Machery-Nagel, Düren, Germany). DNA was removed by an on-column treatment with rDNase (Machery-Nagel, Düren, Germany). 2–3 μg total RNA were reverse transcribed with an oligo(dT) primer and M-MuLV Reverse Transcriptase (Fermentas, St. Leon-Rot, Germany) and the cDNA equivalent of 30–50 ng total RNA was used in a 10 μl PCR reaction with SsoAdvanced™ Universal SYBR Green Supermix (BioRad, München, Germany) in a CFX96 Real-Time System Cycler (BioRad, München, Germany). The relative quantification was calculated with the ΔΔCt method with ACT8 as a control [64]. See S7 Table for a list of qRT-PCR primers.
RNA-sequencing
To investigate significant gene expression differences between Col-0 and Kil-0, RNA was prepared from three biological replicate samples from ten day-old seedlings (21°C, long days) as described above. Sequencing libraries were prepared following the standard protocol of the TruSeq RNA Sample prep v2 Kit (Illumina, San Diego, CA). Paired-end sequencing with a read length of 100 bp was performed on a HiSeq 1000 (Illumina, San Diego, CA). RNA-seq reads from each biological replicate were then aligned against the Col-0 TAIR10 release genome using Bowtie (version 2.1.0) and Tophat2 (version 2.0.8) with default parameters [65, 66]. On the basis of the structural gene annotation for A. thaliana TAIR10, exon - and gene-level transcription was quantified by using HTSeq, differential expression tests were performed by using DESeq2 [67, 68]. Significantly differentially expressed genes were defined as genes with Benjamini-Hochberg-adjusted p-values < 0.01. Tests for differential exon usage were conducted on basis of RNA-seq exon counts with DEXseq [69]. The RNA-seq data are publically available as PRJEB9470 at www.ebi.ac.uk/ena.
A list of genes with a role in flowering time regulation was generated by searching the TAIR database (www.arabidopsis.org) for the term “flowering time”. The resulting list was reviewed manually and is presented in S3 Table. To subsequently analyze gene expression of the FLM locus at a nucleotide resolution, the corresponding Col-0 gene sequences were extracted from the Col-0 TAIR10 reference genome and the Kil-0 genome sequence as determined as part of this study. Subsequently, RNA-seq reads were aligned against the Kil-0 and Col-0 genomic sequences using Bowtie and Tophat2 as described above. The number of mapped RNA-seq reads from Col-0 and Kil-0 per nucleotide were counted with the toolset Bedtools and subsequently normalized to range between 0 (no expression) and 1 (maximum expression). For visualization, the mean expression level and 5% and 95% confidence intervals were determined across the biological replicates of one sample.
Cloning procedures
To obtain a genomic FLM fragment from Kil-0 with a deletion of the 5.7 kb LINE insertion, a 6,981 bp gFLM(Kil-0)ΔLINE deletion fragment was amplified by overlap extension PCR from Kil-0 genomic DNA. Fragment 1 (bp—2367 to bp + 631) was amplified with ULC-1 and ULC-2 and fragment 2 (bp + 632 to bp + 4156 and 251 bp 3’-UTR plus 207 bp downstream sequence) with ULC-3 and ULC-4. After purification of the two subfragments, the full fragment was generated by overlap-extension PCR reaction with ULC-1 and ULC-4. The insert of the full-length fragment in pCR2.1-TOPO (Life Technologies, Carlsbad, CA) was sequenced and subcloned as a BamHI and XhoI fragment into pGreen0229 [70].
To insert the LINE insertion into pDP34, a previously described construct with the Col-0 genomic FLM fragment pFLM::gFLM [19], pDP34 was mutagenized by PCR with ULC-12 to replace the sequence ATTGTTCA (bp +632 to bp +640) with a unique AscI restriction site (GGCGCGCC) [71]. The LINE insertion was then amplified from Kil-0 genomic DNA with ULC-16 and ULC-17 and inserted as an AscI fragment into the modified pDP34. All constructs were transformed into Agrobacterium tumefaciens strain GV3101 containing pSOUP and subsequently using floral dip transformation into Col-0, Kil-0, and flm-3 [70, 72]. pDP79 and pDP80 were previously described [19]. T1 transformants were selected by spraying soil-grown plants with 0.1% BASTA. The list of primers is provided in S7 Table.
FLM locus analysis of A. thaliana accessions
To test for the presence of the FLMKil-0 allele in a large collection of accessions, we performed PCR with the primers LP1, LP2, and RP1 on genomic DNA using Phusion (New England Biolabs, Frankfurt, Germany) or TaKaRa LA Taq polymerases (Takara Bio, Saint-Germain-en-Laye, France). Selected PCR products were analyzed by DNA sequencing following gel extraction. Sequencing primers are listed in S7 Table. A complete list of all accessions examined is provided as S6 Table.
Kil-0 genome sequence information was used to identify accessions with a LINE insertion by analyzing genome sequences as determined in the frame of the 1001 Arabidopsis genome sequencing project (www.1001genomes.org). To render read mapping specific for the Kil-0 allele, the insertion at the experimentally retrieved insertion sites flanked by 140 bp sequence was extracted for each of the two insertion breakpoints and defined as target sequences for mapping. Subsequently, all reads of the individual A. thaliana accessions were mapped against the target sequences using SHORE and genomemapper allowing for up to 5% single base pair differences including gaps with regard to the read length [73, 74]. To ensure that each supporting read spans at least 25% of its sequence across the insertion breakpoint, only reads that overlapped an insertion site with the inner 50% of its read sequence were counted as supporting an insertion breakpoint (core-mapping read). Furthermore, only unique mapped reads were included. An insertion was defined as present if there were at least two unique core-mapping reads at the left and right insertion breakpoint.
LINE insert and FLM locus characterization
Several database searches with the Kil-0 LINE insert yielded no highly similar sequences, except for a 559 bp match to the second exon of Col-0 AT1G04625, which is most likely a transposon-related gene based on its ribonuclease H-like description. The LINE sequence within the Kil-0 insert was identified with RepeatMasker (http://www.repeatmasker.org) against PGSP-REdat, v_9.3_Eudicot (http://pgsb.helmholtz-muenchen.de/plant/recat/). The Pfam domains where annotated with hmmsearch of hmmer3 in all 6 reading frames against PfamA v27 [75, 76].
For phylogenetic analyses of the FLM locus, sequences of 88 randomly selected accessions were extracted from the A. thaliana 1001 genome project GEBowser (http://signal.salk.edu/atg1001/3.0/gebrowser.php) and aligned with sequences of the ten FLMLINE accessions and Col-0. Alignments were calculated using ClustalW and Neighbor-Joining trees (Maximum Composite Likelihood method, 1000 bootstrap replicates) were constructed with MEGA5 [77].
3’ RACE PCR
Polyadenylated transcripts were amplified according to [78]. Transcript pools were analyzed on an agarose gel and the upper and lower bands as depicted in S6E Fig were purified from all samples, subcloned into pCR2.1-TOPO (Life Technologies, Carlsbad, CA) and sequenced. Primers sequences are listed in S7 Table.
Determination of FLM pre-mRNA abundance
Nuclei were isolated as previously described [79] with two biological replicates per line. RNA from nuclei pellet was extracted as previously described [80] and DNA was digested using DNaseI (Life Technologies, Carlsbad, CA). cDNA synthesis and qRT-PCR were performed as described above. Primer sequences are provided in S7 Table.
Supporting Information
Zdroje
1. Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–39. Epub 2012/08/18. doi: 10.1038/nrg3291 22898651
2. Song J, Irwin J, Dean C. Remembering the prolonged cold of winter. Curr Biol. 2013;23(17):R807–11. Epub 2013/09/14. doi: 10.1016/j.cub.2013.07.027 24028964
3. Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140(1):136–47. Epub 2010/01/19. doi: 10.1016/j.cell.2009.11.006 20079334
4. Balasubramanian S, Weigel D. Temperature Induced Flowering in Arabidopsis thaliana. Plant Signal Behav. 2006;1(5):227–8. Epub 2006/09/01. 19704664
5. Wigge PA. Ambient temperature signalling in plants. Curr Opin Plant Biol. 2013;16(5):661–6. Epub 2013/09/12. doi: 10.1016/j.pbi.2013.08.004 24021869
6. Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11(5):949–56. Epub 1999/05/20. 10330478
7. Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13(4):935–41. Epub 2001/04/03. 11283346
8. Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290(5490):344–7. Epub 2000/10/13. 11030654
9. Song J, Angel A, Howard M, Dean C. Vernalization—a cold-induced epigenetic switch. J Cell Sci. 2012;125(Pt 16):3723–31. Epub 2012/09/01.
10. Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Curr Opin Plant Biol. 2009;12(2):178–84. Epub 2009/02/07. doi: 10.1016/j.pbi.2008.12.010 19195924
11. Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. The molecular basis of vernalization-induced flowering in cereals. Trends in plant science. 2007;12(8):352–7. Epub 2007/07/17. 17629542
12. Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 2005;1(1):109–18. Epub 2005/08/17. 16103920
13. Strange A, Li P, Lister C, Anderson J, Warthmann N, Shindo C, et al. Major-effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS One. 2011;6(5):e19949. Epub 2011/06/01. doi: 10.1371/journal.pone.0019949 21625501
14. Verhage L, Angenent GC, Immink RG. Research on floral timing by ambient temperature comes into blossom. Trends in plant science. 2014;19(9):583–91. Epub 2014/05/02. doi: 10.1016/j.tplants.2014.03.009 24780095
15. Capovilla G, Schmid M, Pose D. Control of flowering by ambient temperature. J Exp Bot. 2015;66(1):59–69. Epub 2014/10/19. doi: 10.1093/jxb/eru416 25326628
16. Werner JD, Borevitz JO, Warthmann N, Trainer GT, Ecker JR, Chory J, et al. Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc Natl Acad Sci U S A. 2005;102(7):2460–5. Epub 2005/02/08. 15695584
17. Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006;2(7):e106. Epub 2006/07/15. 16839183
18. Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001;26(2):229–36. Epub 2001/06/08. 11389763
19. Pose D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, et al. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature. 2013;503(7476):414–7. Epub 2013/09/27. doi: 10.1038/nature12633 24067612
20. Lee JH, Ryu HS, Chung KS, Pose D, Kim S, Schmid M, et al. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science. 2013;342(6158):628–32. Epub 2013/09/14. doi: 10.1126/science.1241097 24030492
21. Gu X, Le C, Wang Y, Li Z, Jiang D, He Y. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun. 2013;4 : 1947. Epub 2013/06/19. doi: 10.1038/ncomms2947 23770815
22. Wheeler T, von Braun J. Climate change impacts on global food security. Science. 2013;341(6145):508–13. Epub 2013/08/03. doi: 10.1126/science.1239402 23908229
23. Moore FC, Lobell DB. The fingerprint of climate trends on European crop yields. Proc Natl Acad Sci U S A. 2015;112(9):2670–5. Epub 2015/02/19. doi: 10.1073/pnas.1409606112 25691735
24. Mora C, Caldwell IR, Caldwell JM, Fisher MR, Genco BM, Running SW. Suitable Days for Plant Growth Disappear under Projected Climate Change: Potential Human and Biotic Vulnerability. PLoS Biol. 2015;13(6):e1002167. Epub 2015/06/11. doi: 10.1371/journal.pbio.1002167 26061091
25. Thuiller W, Lavorel S, Araujo MB, Sykes MT, Prentice IC. Climate change threats to plant diversity in Europe. Proc Natl Acad Sci U S A. 2005;102(23):8245–50. Epub 2005/05/28. 15919825
26. Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D. FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics. 2005;170(3):1197–207. Epub 2005/05/25. 15911588
27. Weigel D. Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol. 2012;158(1):2–22. Epub 2011/12/08. doi: 10.1104/pp.111.189845 22147517
28. Tsuchiya T, Eulgem T. An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc Natl Acad Sci U S A. 2013;110(37):E3535–43. Epub 2013/08/14. doi: 10.1073/pnas.1312545110 23940361
29. Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ, et al. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci U S A. 2011;108(30):12533–8. Epub 2011/07/13. doi: 10.1073/pnas.1019732108 21746925
30. Duc C, Sherstnev A, Cole C, Barton GJ, Simpson GG. Transcription termination and chimeric RNA formation controlled by Arabidopsis thaliana FPA. PLoS Genet. 2013;9(10):e1003867. Epub 2013/11/10. doi: 10.1371/journal.pgen.1003867 24204292
31. Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100(1):189–92. Epub 2002/12/28. 12502788
32. Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012;40(6):2454–69. Epub 2011/12/01. doi: 10.1093/nar/gkr932 22127866
33. Hori K, Watanabe Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 2005;43(4):530–40. Epub 2005/08/16. 16098107
34. Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2010;107(49):21199–204. Epub 2010/11/17. doi: 10.1073/pnas.1007431107 21078970
35. Ossowski S, Schneeberger K, Lucas-Lledo JI, Warthmann N, Clark RM, Shaw RG, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327(5961):92–4. Epub 2010/01/02. doi: 10.1126/science.1180677 20044577
36. Schultz ST, Lynch M, Willis JH. Spontaneous deleterious mutation in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1999;96(20):11393–8. Epub 1999/09/29. 10500187
37. Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465(7298):627–31. Epub 2010/03/26. doi: 10.1038/nature08800 20336072
38. Hong RL, Hamaguchi L, Busch MA, Weigel D. Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell. 2003;15(6):1296–309. Epub 2003/06/05. 12782724
39. Schauer SE, Schluter PM, Baskar R, Gheyselinck J, Bolanos A, Curtis MD, et al. Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. Plant J. 2009;59(6):987–1000. Epub 2009/05/29. doi: 10.1111/j.1365-313X.2009.03928.x 19473325
40. Yoo SK, Wu X, Lee JS, Ahn JH. AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J. 2011;65(1):62–76. Epub 2010/12/24. doi: 10.1111/j.1365-313X.2010.04402.x 21175890
41. Coustham V, Li P, Strange A, Lister C, Song J, Dean C. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science. 2012;337(6094):584–7. Epub 2012/07/17. doi: 10.1126/science.1221881 22798408
42. Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012;40(Database issue):D1211–5. Epub 2011/11/15. doi: 10.1093/nar/gkr1047 22080561
43. Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–7. Epub 2003/08/02. doi: 10.1126/science.1086391 12893945.
44. Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, et al. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004;37(6):897–905. Epub 2004/03/05. 14996221
45. Lisch D. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol. 2009;60 : 43–66. Epub 2008/11/15. doi: 10.1146/annurev.arplant.59.032607.092744 19007329
46. McCue AD, Slotkin RK. Transposable element small RNAs as regulators of gene expression. Trends Genet. 2012;28(12):616–23. Epub 2012/10/09. doi: 10.1016/j.tig.2012.09.001 23040327
47. Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303(5657):521–3. Epub 2003/11/25. 14631047
48. Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430(6998):471–6. Epub 2004/07/23. 15269773
49. Michaels SD, He Y, Scortecci KC, Amasino RM. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci U S A. 2003;100(17):10102–7. Epub 2003/08/09. 12904584
50. Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci U S A. 2004;101(44):15670–5. Epub 2004/10/27. 15505218
51. He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18(22):2774–84. Epub 2004/11/03. 15520273
52. Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476(7358):105–8. Epub 2011/07/26. doi: 10.1038/nature10241 21785438
53. Castaings L, Bergonzi S, Albani MC, Kemi U, Savolainen O, Coupland G. Evolutionary conservation of cold-induced antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana perennial relatives. Nat Commun. 2014;5 : 4457. Epub 2014/07/18. doi: 10.1038/ncomms5457 25030056
54. Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5(5):e129. Epub 2007/04/19. 17439305
55. Mascarenhas D, Mettler IJ, Pierce DA, Lowe HW. Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol Biol. 1990;15(6):913–20. Epub 1990/12/01. 2103480
56. Rose AB. Intron-mediated regulation of gene expression. Curr Top Microbiol Immunol. 2008;326 : 277–90. Epub 2008/07/18. 18630758
57. Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9(3):355–65. Epub 1997/03/01. 9090880
58. Sanchez-Bermajo E, Zhu W, Tasset C, Eimer H, Sureshkumar S, Singh R, et al. Genetic architecture of natural variation in thermal responses of Arabidopsis thaliana. Plant Physiol. 2015. Epub 2015/07/22.
59. Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol. 2004;55 : 141–72. Epub 2004/09/21.: doi: 10.1146/annurev.arplant.55.031903.141605 15377217.
60. Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, et al. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21(7):1877–96. Epub 2009/07/04. doi: 10.1105/tpc.109.068114 19574434
61. Gazzani S, Gendall AR, Lister C, Dean C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 2003;132(2):1107–14. Epub 2003/06/14. 12805638
62. Pacurar DI, Pacurar ML, Street N, Bussell JD, Pop TI, Gutierrez L, et al. A collection of INDEL markers for map-based cloning in seven Arabidopsis accessions. J Exp Bot. 2012;63(7):2491–501. Epub 2012/01/28. doi: 10.1093/jxb/err422 22282537
63. Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35(Web Server issue):W71–4. Epub 2007/05/09. 17485472
64. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. Epub 2001/05/09. 11328886
65. Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Nat Biotechnol. 2009;27(5):455–7. doi: 10.1038/nbt0509-455 19430453
66. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36 23618408
67. Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. Epub 2014/09/28. doi: 10.1093/bioinformatics/btu638 25260700
68. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. Epub 2014/12/18. 25516281
69. Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22(10):2008–17. Epub 2012/06/23. doi: 10.1101/gr.133744.111 22722343
70. Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42(6):819–32. Epub 2000/07/13. 10890530
71. Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28(16):E78. Epub 2000/08/10. 10931937
72. Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–43. Epub 1999/03/09. 10069079
73. Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, et al. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6(8):550–1. Epub 2009/08/01. doi: 10.1038/nmeth0809-550 19644454
74. Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18(12):2024–33. Epub 2008/09/27. doi: 10.1101/gr.080200.108 18818371
75. Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–37. Epub 2011/05/20. doi: 10.1093/nar/gkr367 21593126
76. Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30. Epub 2013/11/30. doi: 10.1093/nar/gkt1223 24288371
77. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. Epub 2011/05/07. doi: 10.1093/molbev/msr121 21546353
78. Scotto-Lavino E, Du G, Frohman MA. 3' end cDNA amplification using classic RACE. Nat Protoc. 2006;1(6):2742–5. Epub 2007/04/05. 17406530
79. Kaufmann K, Muino JM, Osteras M, Farinelli L, Krajewski P, Angenent GC. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc. 2010;5(3):457–72. Epub 2010/03/06. doi: 10.1038/nprot.2009.244 20203663
80. Box MS, Coustham V, Dean C, Mylne JS. Protocol: A simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods. 2011;7 : 7. Epub 2011/03/15. doi: 10.1186/1746-4811-7-7 21396125
Štítky
Genetika Reprodukční medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 10- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



